Abstract
The bronze (bz) locus exhibits the highest rate of recombination of any gene in higher plants. To investigate the possible basis of this high rate of recombination, we have analyzed the physical organization of the region around the bz locus. Two adjacent bacterial artificial chromosome clones, comprising a 240-kb contig centered around the Bz-McC allele, were isolated, and 60 kb of contiguous DNA spanning the two bacterial artificial chromosome clones was sequenced. We find that the bz locus lies in an unusually gene-rich region of the maize genome. Ten genes, at least eight of which are shown to be transcribed, are contained in a 32-kb stretch of DNA that is uninterrupted by retrotransposons. We have isolated nearly full length cDNAs corresponding to the five proximal genes in the cluster. The average intertranscript distance between them is just 1 kb, revealing a surprisingly compact packaging of adjacent genes in this part of the genome. At least 11 small insertions, including several previously described miniature inverted repeat transposable elements, were detected in the introns and 3′ untranslated regions of genes and between genes. The gene-rich region is flanked at the proximal and distal ends by retrotransposon blocks. Thus, the maize genome appears to have scattered regions of high gene density similar to those found in other plants. The unusually high rate of intragenic recombination seen in bz may be related to the very high gene density of the region.
Most of the DNA in complex grass genomes, such as maize and barley, is repetitive and made up of retrotransposons (1, 2). Genes comprise a small percentage of the total DNA and appear to be organized in two types of arrangements relative to the bulk of the DNA: dispersed, in which single genes are scattered among retrotransposons, and clustered, in which genes are grouped together in stretches of DNA uninterrupted by retrotransposons.
Evidence for the scattered type of organization was first obtained by the partial sequence and hybridization analysis of a 280-kb yeast artificial chromosome (YAC) clone of the Adh1F allele in maize (1). That seminal paper established that single genes could be flanked by large blocks of nested retrotransposons, a previously unreported type of genome structure. Evidence for the clustered type of gene organization came first from the gross analysis of the distribution of genes in the Gramineae by density gradient centrifugation (3, 4). This analysis revealed that genes in maize, rice, and barley exist in compositionally homogeneous “gene spaces” of about 100 kb and suggested a genomic organization of gene-rich islands separated by large expanses of repetitive DNA. The subsequent sequence of 225 contiguous kilobases of the Adh1F YAC clone supports both types of organization (5). Of the nine genes identified in that YAC, five exist as single genes separated by retrotransposon blocks ranging in size from 14 to 70 kb. The other four predicted genes occur in a 39.2-kb stretch of low-copy DNA devoid of long terminal repeat (LTR) retrotransposons. Thus, the gene density in this gene cluster is one gene per 9.8 kb, which is five times higher than the calculated average gene density of one gene per 50 kb (5). A shorter length of DNA uninterrupted by retrotransposons has been reported in the 22-kDa zein cluster, where one apparently functional zein gene and four pseudogenes occur within the first 26 kb of a 78-kb cosmid contig (6).
The orthologous Adh1 region of sorghum contains the same nine genes found in maize plus five others in a much tighter space because of the total absence of LTR retrotransposons (5). The 14 genes were found in 78.2 kb of contiguous sequence, representing a gene density of one gene per 5–6 kb. In barley, the sequence of two contigs of over 60 kb each from two different chromosomes provided evidence for similar gene densities in two different parts of the genome (2, 7). Three genes were identified in each of these contigs within regions largely devoid of retrotransposons. The gene density in these regions is one gene per 6–8 kb, which is 20–26 times higher than the genome's average. Slightly higher gene densities have been reported in short (<23 kb) genomic fragments carrying the orthologous Lrk/Tak receptor-like kinase loci in wheat, rice, and barley (8), arguing that both small and large grass genomes contain regions that are highly enriched in genes and have little or no repetitive DNA. These high gene density regions, which have a gene and repetitive DNA composition like Arabidopsis, have yet to be described in maize (9).
The bronze (bz) gene in maize exhibits a very high rate of recombination relative to its size (10, 11). To begin investigating the possible basis of this high rate of recombination, we have analyzed the physical organization of the region around the bz locus. In this study, we show that the bz locus resides in a region of the maize genome that has an even higher gene density than the Arabidopsis average of one gene per 4–5 kb. Ten genes, eight of which are shown to be transcribed, are included in a 32-kb stretch of DNA that is uninterrupted by retrotransposons. Flanking this gene-rich region are blocks of retrotransposons like those described at other loci. Thus, the maize genome appears to have scattered regions of high gene density similar to those described in Arabidopsis and other plants. We suggest that the unusually high rate of intragenic recombination seen in bz may be related to the high gene density of the bz region.
Materials and Methods
DNA Sequencing and Assembly.
Two adjacent bacterial artificial chromosome (BAC) clones of the Bz-McC allele serve as the material for this study (12). They comprise a 240-kb contig spanning an internal NotI site in Bz-McC. Both BAC clones had internal NotI sites that were methylated in the parental genomic DNA. The 125-kb proximal clone contained internal NotI sites at 10, 15, 55, and 58 kb from the NotI site in Bz-McC. Therefore, that BAC clone was subcloned as NotI fragments, and the three NotI subclones on the bz side were sequenced. The 115-kb distal clone contained only one NotI site, 97 kb away from the NotI site in Bz-McC. However, it contained ClaI sites at 8, 23, 29, and 37 kb from the NotI site in Bz-McC. Therefore, that clone was subcloned as a series of ClaI fragments, and the three ClaI subclones on the bz side were sequenced. The partial sequence from a 16-kb clone of the Bz-McC allele (13, 14) provided an overlap of 3 kb with the proximal NotI subclone closest to bz and of 0.2 kb with the distal ClaI subclone closest to bz.
BAC and plasmid DNAs were isolated as described (12). Inserts were partially digested with 4-bp restriction enzymes to produce a series of randomly cut fragments. The fragments were dephosphorylated, cloned into the vector pBluescript II (Stratagene), and electroporated into Escherichia coli DH10B to generate “shotgun” DNA libraries. DNA minipreps were prepared from randomly picked clones. Sequencing reactions were performed by using the ABI PRISM BigDye Terminator Cycle Sequencing Ready Reaction kit (PE Applied Biosystems) and analyzed on ABI377 sequencing gels. The 10-fold redundant sequence data were assembled with the phred/phrap software (15). Contigs were extended and joined by custom specific primer walking to close the gaps. The reliability of the sequence was confirmed by the location of expected restriction sites.
Sequence Analysis.
The final sequence of the bz region was divided into 5-kb fragments that served as queries to search the GenBank databases with the various blast programs (16). The genscan program (17) was used only to predict the intron–exon structure of genes that had been identified by blast as having homology to other genes in the nucleic acid database but had no matching cDNAs or expressed sequence tags (ESTs). Programs from the lasergene package (DNAStar, Madison, WI) were used for sequence comparisons and alignments.
RNA Isolation, Northern Blotting, and cDNA Library Construction.
Total RNA was extracted from various tissues by using Trizol reagent (Life Technologies, Gaithersburg, MD). Poly(A) RNA was isolated from 1 mg of total RNA by using poly(A) Track (Promega). RNA samples were separated in 1.2% formamide denaturing gels, blotted to nylon membranes, and hybridized to random primer labeled P32 probes. Hybridization and washing conditions were standard (18). A mixed tassel cDNA library was constructed with the UniZAP-XR vector (Stratagene), following the manufacturer's recommendations. Poly(A) RNA was made from W22 Bz-McC tassels collected at four different developmental stages (4–15, 20–27, 29–42, and 42–50 cm). Equivalent amounts of poly(A) RNA from each developmental stage were pooled for first-strand cDNA synthesis. The cDNA library, which consisted of 1.5 × 106 clones, was screened with probes from different genes in the bz region, following standard procedures (18).
Results
The Gene-Rich Region.
We previously isolated two adjacent NotI BAC clones of the bz region, comprising a 240-kb contig centered around the bz locus (12). We have sequenced the adjacent portions of the two BAC clones and have assembled a 10-fold redundant contiguous 60-kb nucleotide sequence (GenBank accession no. AF320086). To identify genes in the bz region, we applied a combination of gene recognition criteria: (i) comparison to DNA sequences in the GenBank databases; (ii) comparison to cDNA sequences from a tassel cDNA library generated from the same stock as the BAC library; and (iii) comparison to DNA sequences corresponding to the sites of insertion of transposed Activator (Ac) elements.
The region contains two previously characterized genes: bz, encoding UDPG-flavonoid 3-O-glucosyl transferase (UF3GT) (13, 19) and stc1, encoding a sesquiterpenoid cyclase (14). In the bz-m2(Ac) mutant, the transposon Activator (Ac) is inserted in the second exon of the gene (13, 20). Several transpositions of Ac from bz-m2(Ac) have been isolated (21), two of which allowed us to originally identify stc1 as a target of transposed Ac elements from the bz locus (14). The demonstration that the stc1 gene had been the target of two independent transpositions of Ac from bz and extensive other evidence from our lab (ref. 22; X.Y., M. Cowperthwaite, S. Maurais, and H.K.D., unpublished observations) argue that Ac can be used reliably as a gene-finding tool in maize.
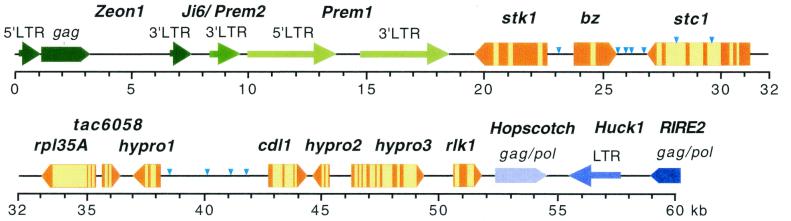
Ten genes were identified in a 32-kb stretch of DNA uninterrupted by retrotransposons, which translates into a gene density of at least one gene per 3.2 kb. The gene-rich region is flanked at the proximal and distal ends by large retrotransposon blocks similar to those first described in the Adh1 region (1). The location of the genes and the retrotransposon blocks in the 60-kb bz region is diagrammed in Fig. 1. The physical map is oriented with its proximal (centromere) end to the left and its distal (telomere) end to the right.
Figure 1.
The organization of the 60-kb gene-rich bz region of maize. The centromere of 9S is on the left. The 10 genes of the bz region are represented as bronze-colored pentagons lying with their apexes pointing in the direction of transcription. Introns are colored yellow. Small insertions in the genic region are shown as turquoise inverted triangles. The retrotransposons in the proximal block are represented in different shades of green and those in the distal cluster, in different shades of blue.
Starting from the proximal end, the following genes are found in the region: stk1 (serine threonine kinase 1), bz (bronze), stc1 (sesquiterpene cyclase 1), rpl35A (large ribosomal subunit protein 35A), tac6058 (transposed Ac6058 insertion site), hypro1 (hypothetical protein 1), cdl1 (cell division-like protein 1), hypro2 (hypothetical protein 2), hypro3 (hypothetical protein 3), and rlk (receptor-like kinase). Two of these 10 genes, bz and stc1, have already been defined genetically and molecularly. Two others, rpl35A and cdl1, share cross-kingdom homology with genes of known function and, therefore, are almost certain to encode equivalent ribosomal and cell division proteins in maize. A fifth one, stk1, is highly similar to many plant genes that encode predicted or confirmed serine–threonine kinase (STK) domains and most likely encodes an enzyme with protein kinase activity in maize as well. The rlk gene is homologous to several genes encoding receptor-like kinases, mostly from Arabidopsis. The remaining four have no predicted function. The three hypro genes have homology to hypothetical genes from Arabidopsis that were identified in genomic sequences by gene prediction programs. The tenth gene, tac6058, has no homology to any sequences in the databases and was initially identified simply as the site of insertion of the transposed Ac element Ac6058 (21). Eight of the 10 genes have been shown to be expressed on the basis of Northern blot analysis and/or cDNA sequencing. Only cdl1 and hypro1 are lacking confirmation of expression. The hypro and tac designations are meant to be provisional and are used in this paper simply for identification purposes.
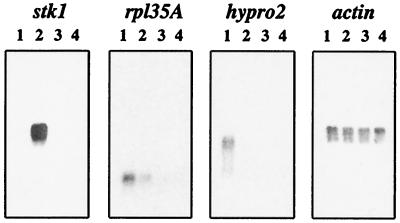
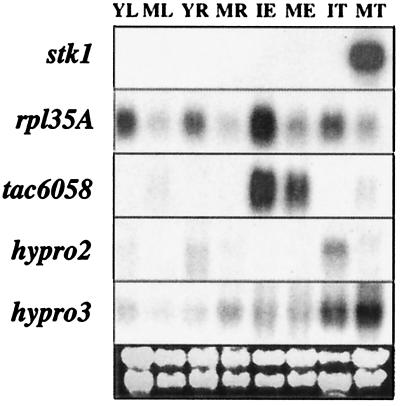
We have analyzed expression of all of the genes in the bz region by Northern blots. To ensure that the signal detected was produced by a transcript from the bz region, we used the deletion mutation sh-bz-X2 as a negative control. sh-bz-X2 is an x-ray-induced deletion of a large chromosomal segment that includes the sh and bz loci situated 2 cM apart in 9S (23). Although the deletion is viable in homozygous condition, it has clearly deleterious properties: sh-bz-X2 homozygotes differ from wild type in overall plant vigor, and sh-bz-X2 pollen shows reduced transmission relative to wild type in heterozygotes. We have confirmed by Southern blot that all 10 genes in the bz region are also deleted in this mutation (refs. 14 and 24; data not shown). Thus, any transcript from this region should either be absent or, in the case of a family of transcripts of similar size, significantly reduced in the deletion. For eight of the nine detectable transcripts in Northern blots, we obtained evidence that the signal was specific to the gene in the bz region. Representative data for tassel Northern blots probed with stk1, rpl35A, and hypro2 are shown in Fig. 2. The exceptional gene was cdl1, which is a member of a gene family and did not appear to be differentially expressed in wild type and sh-bz-X2 in any tissue (data not shown). To obtain a general picture of the expression pattern of the bz region genes during development, we monitored their transcripts in four different tissues (roots, leaves, tassel, and endosperms) at an early and a late developmental stage. Those Northern blot data are shown in Fig. 3.
Figure 2.
Expression of three genes in the bz region at different stages of tassel development in wild type and a deletion mutant. W22 inbred lines carrying either Bz-McC or sh-bz-X2 were used in this comparison. One microgram of poly(A) RNA was loaded in each lane, and the membrane was hybridized sequentially with probes from genes within the bz region (stk1, rpl35A, and hypro2) and outside the bz region (actin). Lanes 1 and 2, Bz-McC immature and mature tassel, respectively; lanes 3 and 4, sh-bz-X2 immature and mature tassel, respectively.
Figure 3.
Expression patterns of five genes in the bz region at an early and a late developmental stage of four different tissues. Thirty micrograms of total RNA were loaded in each lane, and the membrane was hybridized sequentially with probes from stk1, rpl35A, tac6058, hypro2, and hypro3. YL, young leaf (5-day-old seedling); ML, mature leaf (2-month-old plant); YR, young root (5-day-old seedling); MR, mature root (2-month-old plant); IE, immature endosperm (2 weeks after pollination); ME, mature endosperm (5 weeks after pollination); IT, immature tassel (just before emergence); MT, mature tassel (at anthesis).
To define intron–exon junctions for the genes in the bz region, we compared the respective genomic DNA sequences with sequences from either cDNA clones or the EST database. We previously defined the intron–exon junctions of bz and stc1 with cDNA clones isolated from husks and juvenile leaves, respectively (13, 14). As shown in Figs. 2 and 3, at least six of the other genes are expressed at various levels in developing or mature tassels. Therefore, a cDNA library was constructed from tassel mRNA, normalized for different developmental stages, and candidate clones for several genes were isolated and sequenced. Introns of genes without matching cDNAs were predicted from the genomic sequence with the program genscan (17). Features of the introns of the 10 genes are summarized in Table 1.
Table 1.
Properties of the genes in the bz region
| Name | Size and composition
|
Intron features
|
||
|---|---|---|---|---|
| Size, kb | % GC | Number | Average size, bp | |
| stk1 | 3.11* | 63 | 3 | 96 |
| 2.36† | 67 | |||
| bz | 1.87* | 70 | 1 | 100 |
| 1.42† | 75 | |||
| stc1 | 4.48* | 43 | 6 | 407 |
| 1.78† | 51 | |||
| rpl35A | 2.39* | 46 | 3 | 591 |
| 0.34† | 60 | |||
| tac6058 | 0.76* | 46 | 2 | 85 |
| 0.41† | 39 | |||
| hypro1 | 0.87† | 64 | 3 | 176 |
| cdl1 | 0.59† | 54 | 3 | 341 |
| hypro2 | 0.52† | 60 | 2 | 83 |
| hypro3 | 1.03† | 55 | 8 | 255 |
| rlk | 0.56† | 61 | 2 | 106 |
Transcribed region.
Coding region.
stk1.
The stk1 gene is highly similar to several plant genes encoding putative serine–threonine kinases (STKs). The highest similarity (E = 0.0) is to two Arabidopsis genes (AC006403 and T01061). The stk1 gene also shares homology with several maize genes encoding putative receptor kinases, such as CRI4 (25) and KIK (26). In the absence of a proposed gene nomenclature for the different members of the STK-encoding gene family in maize, we have chosen to identify the gene next to bz in 9S as stk1. As seen in Fig. 3, stk1 shows a highly specific pattern of expression. Among the tissues examined, it is expressed only in mature tassels. The intron–exon junction of stk1 was determined from a comparison of the genomic sequence with the sequence of an almost full length cDNA isolated from our tassel cDNA library. stk1 consists of four exons encoding a protein of 787 amino acids. stk1 is transcribed centripetally (i.e., toward the centromere), and its start of transcription is only 280 bp proximal to the insertion Ins1 located in the bz upstream region (13).
bz.
The bz gene has been characterized previously (13, 19). It consists of two exons and encodes a 471-aa protein (UF3GT). bz is transcribed centrifugally (i.e., away from the centromere) from a start point located 484 bp downstream of the insertion Ins1.
stc1.
The stc1 gene has also been characterized previously (14). It consists of seven exons and encodes a 592-aa protein. Two of the three longer introns contain small insertions (discussed below). stc1 is transcribed centripetally, its polyA addition site being separated from that of bz by less than 1.3 kb.
rpl35A.
A gene encoding a protein with high homology to the 60S ribosomal protein L35A from a wide range of organisms, consequently designated rpl35A, is located about 1.7 kb distal to and in the same transcriptional orientation as stc1. This protein is highly conserved in species ranging from yeast to humans. The highest similarity (E = 2e−33 − 2e−32) is to four Arabidopsis genes (AL161667, AC008046, AC020579, and AC067971). As can be seen in Fig. 3, rpl35A1 is expressed predominantly in young tissues (young leaf, 2-week-old endosperms, and immature tassel). Expression declines in seedling shoots and roots between 4 and 8 days, in developing endosperms and embryos between 2 and 5 weeks, and in immature ears between 3 and 9 cm (data not shown). A matching EST from a W23 root cDNA library (AW037029) was used to define intron–exon junctions in rpl35A1. The gene consists of four exons, separated by two average-size introns (101 and 92 bp) in the coding region and by a surprisingly long intron (1,581 bp) just after the stop codon in the 3′ untranslated region (UTR). rpl35A appears to be a member of a small gene family (≥3), as determined from the different 3′ UTR sequences of several cDNA clones isolated from a leaf library. Like other large ribosomal subunit 35A proteins in the database, the maize protein is short, consisting of only 112 amino acids.
tac6058.
The gene that we have provisionally designated as tac6058 does not have homology to any sequences in the database. It was identified initially as the site of insertion of Ac6058 (21). A probe of the sequence adjacent to Ac6058 (tac6058) detects a small transcript in endosperms, tassel, and leaf (Fig. 3). The transcript is particularly abundant in 2-week old endosperms. An apparently full length cDNA was isolated from our tassel cDNA library and used to define the intron–exon structure of the gene. tac6058 consists of three exons, separated by two average-size introns (85 and 84 bp). Assuming that the transcript is translated, the putative protein encoded by tac6058 would be 135 aa long, the 85-bp intron would lie in the 5′ UTR, and the 84-bp intron would lie in the coding region. However, the putative start codon is in a poor sequence context for an initiator ATG (27), and the putative stop codon occurs only 4 bp upstream of the polyA addition site. Therefore, the tac6058 transcript may correspond to a noncoding RNA. The transcription initiation sites of tac6058 and rpl35A are just 200 bp apart, suggesting that their divergent promoters could either overlap or be a part of the transcribed region of the other gene.
hypro1.
A putative gene encoding a protein with homology to a hypothetical protein from Arabidopsis (BAB08785; E = 2e−26) is located 0.5 kb distal and in opposite transcriptional orientation to tac6058. For purposes of discussion in this paper, we have called it hypro1. This is only one of two genes in the bz region for which we lack evidence confirming that it is expressed. We have used the gene prediction program genscan to identify the location of two introns. Conceptual translation of the hypro1 genomic sequence in maize yields a protein of 288 aa, compared with 318 aa for the BAB08785 protein in Arabidopsis.
cdl1.
The longest DNA stretch in the bz region without identifiable genes lies distal to hypro1. It is 4.5 kb in length and contains several insertion sequences (discussed below). The next obvious gene after hypro1 encodes a protein with homology to cell division proteins from humans and fission yeast and to a putative cell division like protein from Arabidopsis (AL137189; E = 4e−25). We have termed it, therefore, cdl1 (for cell division-like 1). A cdl1 probe fails to detect a difference between wild type and the sh-bz-X2 deletion in Northern blots of different tissues, so we cannot be sure that this gene is expressed. However, a pathogen-induced cDNA from Sorghum bicolor is highly similar to cdl1. In addition, we have isolated several cDNAs from our tassel cDNA library that are related, but not identical, to cdl1. Together with the genscan program, these cDNAs have helped to define the intron–exon junctions of cdl1. The predicted gene contains four exons and encodes a protein of 197 aa.
hypro2.
A gene encoding a protein with homology to another small Arabidopsis protein, the 190-aa hypothetical protein T01953 (E = 3e−29), is located less than 200 bp distal and in opposite transcriptional orientation to cdl1. We have called this gene hypro2. The highest level of expression of hypro2 is in immature tassels (Fig. 3). A homologous maize leaf primordium EST (AI991425) is too short to help identify intron–exon junctions. We have isolated two different cDNAs from our tassel cDNA library that are homologous, but not identical, to hypro2 and thus define a gene family. Because intron position is conserved among homologous genes (28), those cDNA sequences served to identify two possible introns in the hypro2 gene. Conceptual translation of the spliced genomic sequence yields a protein of 174 aa, which differs from the Arabidopsis hypothetical protein mostly in exon 1.
hypro3.
Another gene encoding a protein sharing homology to several hypothetical proteins from Arabidopsis (BAB11289; E = 6e−53), Drosophila, and Caenorhabditis elegans is located about 900 bp distal and in opposite transcriptional orientation to hypro2. We have called this gene hypro3. It is expressed preferentially in the tassel, at both early and late stages of development (Fig. 3). A short maize endosperm EST (AI665084) is homologous to hypro3. We have isolated two different cDNAs from our tassel cDNA library that appear to belong to the hypro3 gene family. Their sequences enabled us to identify three introns in the 5′ half of the hypro3 gene. Because the remaining hypro3 and cDNA sequences are highly divergent, we used the genscan program to identify the rest of the coding sequence in hypro3. The program recognized a 3.1-kb gene consisting of nine exons, including the three exons present in our tassel cDNAs (exons 2–4, which encode the part of the protein homologous to other eukaryotic hypothetical proteins) and the exon present in the maize endosperm EST (exon 6). Although we know that hypro3 is expressed, we cannot confirm the makeup of the predicted gene, because we were unable to isolate its corresponding cDNA.
rlk1.
The tenth gene in the bz region lies 1.2 kb distal to and in the same transcriptional orientation as hypro3. It encodes a protein that is homologous to several plant receptor-like kinases, hence its rlk1 designation. Its highest similarity is to a putative receptor-like kinase from rice (BAA94519; E = 3e−22). rlk1 is expressed preferentially in immature ears and 1-week-old seeds (data not shown). The program genscan predicts a 1.1-kb gene containing two introns and encoding a protein of 186 aa.
The Small Insertions in the Gene-Rich Region.
Miniature inverted repeat transposable element (MITEs) (29) and other small sequences without typical MITE features were uncovered by two types of sequence comparisons: either between alleles of different genes in the region (bz and stc1) or between our BAC sequence and other gene sequences in the database. Table 2 summarizes the properties of the 11 insertions identified in the bz region.
Table 2.
Properties of the insertions in the bz genic region
| Insertion name | Location | Size, bp | FDR sequence | TIR* size | Ref. |
|---|---|---|---|---|---|
| Ins1 | stk1-bz IG | 271 | CTAA | 19 (6) | 13 |
| Ins3 | bz 3′UTR | 62 | ACAT | 6 (1) | 13, this study |
| Tour-Zm7 | bz-stc1 IG | 136 | ND | 14 (3) | 13, 30 |
| Ins4 | bz-stc1 IG | 163 | ND | ND | 13, this study |
| Ins5 | bz-stc1 IG | 283 | TTA | 14 (3) | This study |
| Tour-Zm1 | stc1 intron | 119 | GT/CA | 14 (3) | 30, this study |
| Ins6 | stc1 intron | 168 | TAA | ND | This study |
| Stow-Zm3 | hypro1-cdl | 80 | TA | 35 (1) | 32, this study |
| Tour-Zm20 | hypro1-cdl | 178 | TTA | 87 (8) | 31, this study |
| Tour-Zm3 | hypro1-cdl | 126 | T/GAA | 14 (1) | 30, this study |
| Ins7 | hypro1-cdl | 408 | TTA | 15 (3) | This study |
Tour, Tourist; Stow, Stowaway. IG, intergenic. FDR, flanking direct repeat; ND, not detected.
TIR, in base pairs; number of mismatches in parentheses.
Several of the insertions in the bz region have already been described as insertion polymorphisms among three sequenced bz alleles (13). They include the 271-bp Ins1 insertion between stk1 and bz, a 62-bp insertion in the bz 3′UTR, and two insertions in the bz-stc1 intergenic region: a 136-bp insertion that was subsequently found to be a member of the Tourist family of small elements (30) and a 163-bp insertion that lacks the terminal inverted repeat (TIR) and flanking direct repeat typical of MITE elements (31). Because the 62- and 163-bp insertion lack homology to previously described MITEs, we have opted to name them Ins3 and Ins4, respectively, following our earlier designation for other insertions in the bz region (13).
By sequencing two different stc1 alleles, Stc1-McC and Stc1-W22, and the bz-stc1 intergenic region flanking them (ref. 14; H.F. and H.K.D., unpublished results), we have now identified several other insertion polymorphisms in the region. A 283-bp insertion with typical MITE features but no homology to the sequence databases occurs only between the Bz-McC and Stc1-McC alleles. We have named this insertion Ins5. It has a 14-bp TIR with three single base-pair mismatches and is flanked by a 3-bp direct repeat. Two other insertions occur within two of the three long introns of the Stc1-McC allele. They are a 119-bp Tourist-Zm1 element in intron 5 and a previously unreported 168-bp insertion element in intron 3 that we have named Ins6. Like a MITE, Ins6 is flanked by a 3-bp direct repeat, but it lacks an obvious TIR.
Other insertions in the 32-kb gene-rich region were identified by comparison to maize sequences in the GenBank databases. All of them are located in the 4.5 kb of DNA separating hypro1 and cdl1, the longest intergenic interval in the region. An 80-bp Stowaway-Zm3 element (32) is inserted just 140 bp distal to the putative start codon of hypro1. It resembles a foldback element in having long 35-bp TIRs (with one mismatch), and it is flanked by a direct repeat of the dinucleotide TA. Less than 1.7 kb distal to this MITE is a 178-bp Tourist-Zm20 element (31), which also has foldback features. Its TIRs cover almost half of the element's length and are flanked by the trinucleotide TTA. One kilobase further distal is a 126-bp Tourist-Zm3 element (30). It has 14-bp TIRs and is flanked by an imperfect trinucleotide (TAA/GAA). Peculiarly, Tourist-Zm3 and Tourist-Zm20 elements are also found close to each other in the Adh1-Cm allele (33).
The last insertion in the region is located 320 bp distal to the Tourist-Zm3 element. We have named this 408-bp insertion Ins7. Ins7 is larger than most maize MITEs and lacks homology to previously described MITEs. It has 15-bp TIRs (with three mismatches) and is flanked by a 3-bp direct repeat. Interestingly, Ins7 is also found inserted in the Ds element of the sh2-m1 allele (34), where it is flanked by the same TTA trinucleotide. No other Ds element in the database shares this sequence.
The Flanking Retrotransposon Clusters.
The 32-kb gene-rich sequence in the bz region appears to be flanked on either side by large retrotransposon blocks similar to those found in the Adh1 (1) and 22-kDa zein regions (6). To obtain a general picture of the organization of these blocks, we sequenced 18 kb of the proximal cluster and 8 kb of the distal cluster.
The proximal retrotransposon cluster begins 1.3 kb from the 3′ end of the stk1 gene with an 8.2-kb internally deleted Prem1 element (35). The ends of Prem1 are delimited by direct repeats of the host pentanucleotide CACAT that were generated on insertion. The element consists of two almost identical 3.5-kb LTRs separated by a 1.2-kb sequence that contains a primer binding site and a polypurine tract (PPT) immediately internal to the two LTRs. These LTRs, which are more than twice the size of the longest Prem1 LTR currently in the database (U03684), are probably complete. The 1.2-kb 3′ LTR of a Ji-6/Prem2 type of retrotransposon (36), with its adjacent PPT, is located within 400 bp of this Prem1 element. Next to this LTR, and interrupting the continuity of the retrotransposon, is a complete 7.3-kb Zeon1 element (37) flanked by the pentanucleotide repeat CAAGT. Thus, at least part of the retrotransposon cluster proximal to the bz gene-rich region exhibits the same type of nesting organization first described at Adh1 (1, 5).
The distal retrotransposon cluster begins 0.5 kb beyond the putative stop codon of rlk. Extending distally, this cluster contains a 2.0-kb truncated gag/pol domain from the retrotransposon Hopscotch (38), a 2.7-kb sequence, including a 1.4-kb Huck1 LTR, that is also found in the Adh1-F region (36), and a 1.0-kb sequence that is homologous to the gag/pol domain of the rice retrotransposon RIRE2 (39). Thus, the limited sequence data from the distal retroelement cluster suggest a fragmented, rather than nested, type of organization. Further sequencing of this cluster should clarify this point.
Discussion
The High Genic Content of the bz Region.
We have isolated two adjacent NotI BAC clones comprising a 240-kb contig of the bz region in maize and have sequenced 60 kb of DNA spanning the bz gene. We find that the highly recombinogenic bz locus lies in an unusually gene-rich region. Ten genes are distributed over 32 kb of sequence uninterrupted by retrotransposon insertions, which corresponds to a gene density 16 times higher than the maize average (5). This high gene density, higher than the Arabidopsis average and highest of any grass genome region sequenced to date (40), may account for the high recombination found in bz. Most genes are hypomethylated in maize (41). Methylated cytosines are known to bind specifically to nuclear proteins that influence the state of chromatin condensation (42, 43). Therefore, a gene cluster containing largely unmethylated DNA would adopt a less condensed chromatin configuration and be more accessible to the recombination machinery during meiosis. Flanking the gene-rich region are two large retrotransposon blocks of the type described at the Adh1 locus (1). Preliminary sequence data of the two adjacent BACs from the unmethylated external NotI sites indicate that clusters of genes also occur in those regions. Thus, the overall organization of this segment of 9S may be one of alternating clusters of gene-rich regions and retrotransposon blocks.
Of the 10 genes in the bronze region, bz is the only one to have been first defined mutationally (44). Two other genes were originally identified as sites into which the transposable element Ac had transposed from the nearby bz mutant bz-m2(Ac). One of them, stc1, has been studied in detail (14). The second one, tac6058, lacks homology to any sequence in the sequence databases. It is well expressed in developing endosperms, but the Ac insertion homozygote has no obvious mutant phenotype.
The seven other genes in the region have been identified from a comparison of the DNA sequence to the existing databases. For stk1 and rpl35A, we have defined exon–intron structures on the basis of a full-length cDNA sequence isolated from a tassel cDNA library of the same genotype and an allelic EST in the sequence database, respectively. stk1 encodes a protein with homology to plant serine–threonine kinases and rpl35A, a protein with homology to the ribosomal protein 35A from several organisms. The remaining five genes are homologous to genes predicted from the Arabidopsis and rice genome sequencing projects. cdl1 and rlk1 encode proteins with homology to a cell division-like protein and a receptor-like kinase, respectively. The three genes encoding proteins with homology to hypothetical proteins from Arabidopsis have been provisionally designated hypro1, hypro2, and hypro3. We confirmed that three of these five, hypro2, hypro3, and rlk, are expressed in maize. The first two are members of small gene families, for which we succeeded in isolating cDNA clones from a tassel cDNA library. We used these clones to partially define exon–intron structures of the genes, on the basis of extensive conservation of intron position among homologous genes in plants (28).
The genes in the bz region do not share any obvious common features. Half are transcribed toward the centromere and half, away from the centromere (Fig. 1). Except for the two protein kinases, the identifiable proteins do not appear to be functionally related. The eight genes that have been transcriptionally analyzed have different spatial and temporal expression patterns (Fig. 3). Some DNA properties of the 10 genes are compared in Table 1. The size of the coding region ranges 8-fold, from 0.34 to 2.65 kb, and averages 1.0 kb. The length of the transcribed region in the five genes with matching cDNAs ranges from 0.76 to 4.48 kb. Fortuitously, these five genes are next to each other on the proximal half of the cluster, making it possible to measure the length of the nontranscribed space separating adjacent genes. This space may include 0, 1, or 2 promoters, depending on the transcriptional orientation of neighboring genes, and clearly represents an overestimate of the real intergenic distance. The average intertranscript space for the five most proximal genes in the bz cluster is just 1.0 kb, revealing a surprisingly compact packaging of adjacent genes in this part of the genome.
Carels and Bernardi (28) have recently uncovered two classes of genes in maize: a GC-rich class (55–75% GC) with no or few short introns and a GC-poor class (40–55%) with numerous long introns. Four of the five genes for which we have nearly full length cDNAs appear to fall into these two classes (Table 1). Thus, bz and stk1 fall in the first class, whereas stc1 and rpl35A fall in the second. The only anomaly is tac6058, which may not code for a protein. All 19 intron–exon junctions derived directly from a comparison between the genomic and cDNA sequences of seven genes have the invariant GT and AG dinucleotides at the intronic 5′ and 3′ ends. The exon sequence flanking the introns appears to be more conserved at the 5′ end than at the 3′ end: the two terminal exon nucleotides tend to be A and G at the 5′ end (A12G15), but they are so variable at the 3′ end that there is no clear majority dinucleotide (GenBank accession no. AF320086).
The Insertions of the bz Region.
As in other maize genomic regions (5, 6), the two predominant types of insertions in the bz region are MITEs and retrotransposons. These insertions occupy totally nonoverlapping domains. Whereas retrotransposons occur in clusters flanking the gene-rich region, MITEs are scattered within the genic region.
Five different MITEs were found located within introns or between genes in the region: Tourist-Zm1, Tourist-Zm3, Tourist-Zm7, Tourist-Zm20 (31), and Stowaway-Zm3 (32). The remaining insertions have no sequence similarity to MITEs in the databases, and some of them lack typical MITE features, but they occupy the same genomic domain as MITEs (Table 2). Ins4 lacks TIR and flanking direct repeat (FDR) sequences altogether. Ins3 and Ins6 have much shorter or no TIRs but are flanked by either 3- or 4-bp direct repeats, respectively. Possibly, these are MITEs in which the TIR sequences have diverged to the point of being unrecognizable. Ins1, Ins5, and Ins7 are larger than most maize MITEs but have TIRs and FDRs. Again, in the absence of a functional test for their mobilization, these could be considered MITEs at the larger end of the size spectrum.
The retrotransposon insertions occur in blocks at either end of the genic region. Partial sequence of the proximal cluster revealed a nested type of organization, similar to that first described in the Adh1 region (1, 5). A complete Zeon1 element (37) is inserted in a retrotransposon of the Ji6-Prem2 family (36). Adjacent to the Ji6-Prem2 3′ LTR and within 1.3 kb of the stk1 gene are the complete LTRs of an internally deleted, but uninterrupted, Prem1 element. In contrast, partial sequence of the distal retrotransposon cluster suggests a fragmented, rather than nested, type of organization. Fragments of three apparently unrelated retrotransposons, Hopscotch, Huck1, and RIRE2, occur adjacent to each other, just 0.5 kb distal to the rlk gene.
Acknowledgments
We thank Matt Cowperthwaite for comments on the manuscript and Victor Llaca for help with the sequence analysis software. This research was supported by National Science Foundation Grants MCB 99–04646 and DBI 98–13364.
Abbreviations
- Ac
Activator
- BAC
bacterial artificial chromosome
- bz
bronze
- EST
expressed sequence tag
- LTR
long terminal repeat
- MITE
miniature inverted repeat transposable element
- TIR
terminal inverted repeat
Footnotes
References
- 1.SanMiguel P, Tikhonov A, Jin Y K, Motchoulskaia N, Zakharov D, Melake-Berhan A, Springer P S, Edwards K J, Lee M, Avramova Z, Bennetzen J L. Science. 1996;274:765–768. doi: 10.1126/science.274.5288.765. [DOI] [PubMed] [Google Scholar]
- 2.Shirasu K, Schulman A H, Lahaye T, Schulze-Lefert P. Genome Res. 2000;10:908–915. doi: 10.1101/gr.10.7.908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carels N, Barakat A, Bernardi G. Proc Natl Acad Sci USA. 1995;92:11057–11060. doi: 10.1073/pnas.92.24.11057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barakat A, Carels N, Bernardi G. Proc Natl Acad Sci USA. 1997;94:6857–6861. doi: 10.1073/pnas.94.13.6857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tikhonov A P, SanMiguel P J, Nakajima Y, Gorenstein N M, Bennetzen J L, Avramova Z. Proc Natl Acad Sci USA. 1999;96:7409–7414. doi: 10.1073/pnas.96.13.7409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Llaca V, Messing J. Plant J. 1998;15:211–220. doi: 10.1046/j.1365-313x.1998.00200.x. [DOI] [PubMed] [Google Scholar]
- 7.Panstruga R, Buschges R, Piffanelli P, Schulze-Lefert P. Nucleic Acids Res. 1998;26:1056–1062. doi: 10.1093/nar/26.4.1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Feuillet C, Keller B. Proc Natl Acad Sci USA. 1999;96:8265–8270. doi: 10.1073/pnas.96.14.8265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bennetzen J L. Plant Cell. 2000;12:1021–1029. doi: 10.1105/tpc.12.7.1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dooner H K. Genetics. 1986;113:1021–1036. doi: 10.1093/genetics/113.4.1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dooner H K, Martinez-Ferez I M. Plant Cell. 1997;9:1633–1646. doi: 10.1105/tpc.9.9.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fu H, Dooner H K. Genome Res. 2000;10:866–873. doi: 10.1101/gr.10.6.866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ralston E J, English J, Dooner H K. Genetics. 1988;119:185–197. doi: 10.1093/genetics/119.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shen B, Zheng Z, Dooner H K. Proc Natl Acad Sci USA. 2000;97:14807–14812. doi: 10.1073/pnas.240284097. . (First Published December 5, 2000, 10.1073/pnas.240284097) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Green P. DOE Human Genome Program Contractor–Grantee Workshop V. Office of Science, Office of Biological and Environmental Research, Washington, DC: U.S. Department of Energy; 1996. p. 157. [Google Scholar]
- 16.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Burge C, Karlin S. J Mol Biol. 1997;268:78–94. doi: 10.1006/jmbi.1997.0951. [DOI] [PubMed] [Google Scholar]
- 18.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 19.Furtek D B, Schiefelbein J W, Johnston F, Nelson O E. Plant Mol Biol. 1988;11:473–481. doi: 10.1007/BF00039028. [DOI] [PubMed] [Google Scholar]
- 20.McClintock B. Carnegie Inst Wash Ybk. 1955;54:245–255. [Google Scholar]
- 21.Dooner H K, Belachew A. Genetics. 1989;122:447–457. doi: 10.1093/genetics/122.2.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yan X, Zhan C, Cowperthwaite M, Maurais S, Dooner H K. Plant Biology '99. Baltimore, MD: Am. Soc. Plant Physiol.; 1999. p. 143. [Google Scholar]
- 23.Mottinger J P. Genetics. 1970;64:259–271. doi: 10.1093/genetics/64.2.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dooner H K, Weck E, Adams S, Ralston E, Favreau M, English J. Mol Gen Genet. 1985;200:240–246. [Google Scholar]
- 25.Becraft P W, Stinard P S, McCarty D R. Science. 1996;273:1406–1409. doi: 10.1126/science.273.5280.1406. [DOI] [PubMed] [Google Scholar]
- 26.Braun D M, Stone J M, Walker J C. Plant J. 1997;12:83–95. doi: 10.1046/j.1365-313x.1997.12010083.x. [DOI] [PubMed] [Google Scholar]
- 27.Kozak M. Nucleic Acids Res. 1981;9:5233–5252. doi: 10.1093/nar/9.20.5233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carels N, Bernardi G. Genetics. 2000;154:1819–1825. doi: 10.1093/genetics/154.4.1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bureau T E, Ronald P C, Wessler S R. Proc Natl Acad Sci USA. 1996;93:8524–8529. doi: 10.1073/pnas.93.16.8524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bureau T E, Wessler S R. Plant Cell. 1992;4:1283–1294. doi: 10.1105/tpc.4.10.1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bureau T E, Wessler S R. Proc Natl Acad Sci USA. 1994;91:1411–1415. doi: 10.1073/pnas.91.4.1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bureau T E, Wessler S R. Plant Cell. 1994;6:907–916. doi: 10.1105/tpc.6.6.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Osterman J C, Dennis E S. Plant Mol Biol. 1989;13:203–212. doi: 10.1007/BF00016138. [DOI] [PubMed] [Google Scholar]
- 34.Giroux M J, Clancy M, Baier J, Ingham L, McCarty D, Hannah L C. Proc Natl Acad Sci USA. 1994;91:12150–12154. doi: 10.1073/pnas.91.25.12150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Turcich M P, Mascarenhas J P. Sex Plant Reprod. 1994;7:2–11. [Google Scholar]
- 36.SanMiguel P, Gaut B S, Tikhonov A, Nakajima Y, Bennetzen J L. Nat Genet. 1998;20:43–45. doi: 10.1038/1695. [DOI] [PubMed] [Google Scholar]
- 37.Hu W, Das O P, Messing J. Mol Gen Genet. 1995;248:471–480. doi: 10.1007/BF02191647. [DOI] [PubMed] [Google Scholar]
- 38.White S E, Habera L F, Wessler S R. Proc Natl Acad Sci USA. 1994;91:11792–11796. doi: 10.1073/pnas.91.25.11792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ohtsubo H, Kumekawa N, Ohtsubo E. Genes Genet Syst. 1999;74:83–91. doi: 10.1266/ggs.74.83. [DOI] [PubMed] [Google Scholar]
- 40.Keller B, Feuillet C. Trends Plant Sci. 2000;5:246–251. doi: 10.1016/s1360-1385(00)01629-0. [DOI] [PubMed] [Google Scholar]
- 41.Antequera F, Bird A P. EMBO J. 1988;7:2295–2299. doi: 10.1002/j.1460-2075.1988.tb03072.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jones P L, Veenstra G J, Wade P A, Vermaak D, Kass S U, Landsberger N, Strouboulis J, Wolffe A P. Nat Genet. 1998;19:187–191. doi: 10.1038/561. [DOI] [PubMed] [Google Scholar]
- 43.Wade P A, Gegonne A, Jones P L, Ballestar E, Aubry F, Wolffe A P. Nat Genet. 1999;23:62–66. doi: 10.1038/12664. [DOI] [PubMed] [Google Scholar]
- 44.Rhoades M M. Am Nat. 1952;86:105–108. [Google Scholar]