Abstract
Plant genomes are the source of large numbers of small RNAs, generated via a variety of genetically separable pathways. Several of these pathways converge in the production of phased, secondary, small interfering RNAs (phasiRNAs), originally designated as trans-acting small interfering RNAs or tasiRNAs. PhasiRNA biogenesis requires the involvement of microRNAs as well as the cellular machinery for the production of siRNAs. PhasiRNAs in Arabidopsis thaliana have been well described for their ability to function in trans to suppress target transcript levels. Plant genomic data from an expanding set of species have demonstrated that Arabidopsis is relatively sparing in its use of phasiRNAs, while other genomes contain hundreds or even thousands of phasiRNA-generating loci. In the dicots, targets of those phasiRNAs include several large or conserved families of genes, such as those encoding disease resistance proteins or transcription factors. Suppression of nucleotide-binding, leucine-rich repeat (NB-LRR) disease resistance genes by small RNAs is particularly unusual because of a high level of redundancy. In this review, we discuss plant phasiRNAs and the possible mechanistic significance of phasiRNA-based regulation of the NB-LRRs.
INTRODUCTION
Plant small RNAs are in the size range of ∼21 to 24 nucleotides; these short, processed transcripts play crucial roles in a variety of biological regulation processes, such as development, plant defense, and epigenetic modifications. Small RNAs in plants can be categorized into several major classes, including microRNAs (miRNAs); heterochromatic small interfering RNAs (hc-siRNAs); phased, secondary, small interfering RNAs (phasiRNAs); and natural antisense transcript small interfering RNAs (NAT-siRNAs). These categories are defined according to their origin and biogenesis (Axtell, 2013), with functions at both transcriptional and posttranscriptional levels. Common features of all small RNAs are that members of the DICER-LIKE (DCL) family are employed to cut longer RNAs into specific smaller lengths, and the resulting small RNAs are thereafter incorporated into ARGONAUTE (AGO) family proteins to target complementary nucleotide sequences, functioning in a suppressive manner. In addition, recent data demonstrate that plant small RNAs are mobile, so that they can have effects over a long distance, including causing posttranscriptional silencing or epigenetic changes (Chitwood and Timmermans, 2010; Dunoyer et al., 2010; Molnar et al., 2010).
miRNAs are typically processed from a hairpin-like secondary structure of a noncoding mRNA (ncRNA), with a precursor mRNA generated by RNA polymerase II (Pol II). The RNase III enzyme DCL1 is responsible for the biogenesis of the mature miRNA via processing of the mRNA precursor. DCL1 is one of four Dicer proteins encoded in a typical dicot genome or one of five encoded a typical monocot genome (see below). miRNAs function in a homology-dependent manner against target mRNAs to typically either (1) direct cleavage at highly specific sites or (2) suppress translation; these modes of action depend largely on the miRNA complementarity with target sequences (Valencia-Sanchez et al., 2006; Voinnet, 2009). Small interfering RNAs (siRNAs) are defined by the dependency of their biogenesis on an RNA-dependent RNA polymerase (RDR). The activity of at least three of the six RDRs (RDR1/2/6) encoded in the Arabidopsis thaliana genome is believed to generate a double-stranded RNA (dsRNA) intermediary that is recognized and cleaved by a Dicer enzyme to generate different classes of siRNAs; so far, little is known about the function of the triplicated paralogs RDR3/4/5 (Willmann et al., 2011). hc-siRNAs are ∼24 nucleotides in length, generated from DCL3 activity from intergenic or repetitive regions of genome via the activity of the plant-specific RNA polymerases Pol IV and possibly Pol V (Matzke et al., 2009; Law and Jacobsen, 2010; Lee et al., 2012). The function of hc-siRNAs is largely to maintain genome integrity, by maintenance of suppressive levels and types of DNA methylation on transposable elements. PhasiRNAs are derived from an mRNA converted to dsRNA by RDR6 and processed by DCL4, exemplified by the category of Arabidopsis trans-acting siRNAs (tasiRNAs) (Vazquez et al., 2004). In an exceptional case, phasiRNAs may also be 24-nucleotide products of DCL5 (previously known as DCL3b) in grass reproductive tissues (Song et al., 2012). The trans-acting name (tasiRNAs) of some phasiRNAs comes from their ability to function like miRNAs in a homology-dependent manner, directing AGO1-dependent slicing of mRNAs from genes other than that of their source mRNA (see below). NAT-siRNAs are a narrowly described, unusual, and perhaps questionable category of small RNAs purportedly derived from two distinct, homologous, and interacting mRNAs (Borsani et al., 2005). While hc-siRNAs play a crucial role in chromatin modifications, miRNAs, phasiRNAs, and NAT-siRNAs function mainly at the posttranscriptional level by either cleavage or translational suppression of target transcripts, although a few instances have been described in which they can direct DNA methylation (Wu et al., 2010, 2012).
In the last few years, as a result of extensive genome sequencing in plants coupled with small RNA analysis, many new small RNAs have been described. Typically, with each new genome, a new cohort of miRNAs is described along with their mRNA targets. In parallel to these miRNA studies, one of the most interesting findings of recent years in these new genomes has been the identification of a set of loci generating phased, secondary siRNAs, larger in number in most non-Brassica plant genomes than described for Arabidopsis. These secondary siRNAs are in many cases derived from a variety of protein-coding transcripts and in other cases from newly described long, noncoding mRNAs. In the first half of this review, we focus on the biogenesis, characterization, and roles of these phased siRNAs in plants, and we discuss the potential regulatory roles of phasiRNA-producing loci (PHAS loci). In the second half of this review, we focus on the largest gene family in plants known to be regulated by miRNAs that trigger secondary siRNAs, nucleotide-binding, leucine-rich repeat (NB-LRR)–encoding genes. The function of NB-LRRs is almost exclusively in defense, and the proteins and the gene families that encode them are well described, comprising a useful case study of small RNA-based suppression of an extensive gene family. The mechanistic and functional basis for this suppression, as well as that observed for the MYB, PENTATRICOPEPTIDE REPEAT (PPR), and other genes or gene families, is still largely unknown, but we will review several speculative ideas for the role of this highly redundant suppressive activity.
OVERVIEW OF tasiRNA BIOGENESIS, FUNCTION, AND DIVERSIFICATION IN PLANTS
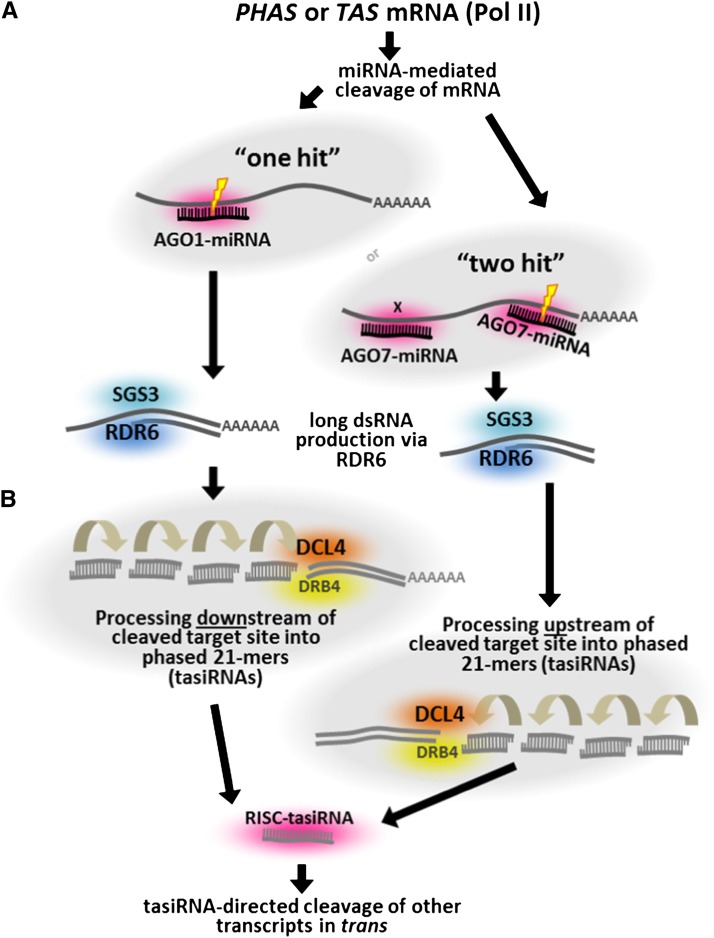
TasiRNA are a class of secondary siRNAs generated from noncoding TAS transcripts by miRNA triggers in a phased pattern (Peragine et al., 2004; Vazquez et al., 2004; Allen et al., 2005; Yoshikawa et al., 2005). The term “phased” indicates simply that the small RNAs are generated precisely in a head-to-tail arrangement, starting from a specific nucleotide; this arrangement results from miRNA-triggered initiation followed by DCL4-catalyzed cleavage (Figure 1). The primary proteins that participate in tasiRNA biogenesis include RDR6, SUPPRESSOR OF GENE SILENCING3 (SGS3), DCL4, AGO1, AGO7, and DOUBLE-STRANDED RNA BINDING FACTOR4 (Peragine et al., 2004; Vazquez et al., 2004; Xie et al., 2005; Adenot et al., 2006; Montgomery et al., 2008a; Fukudome et al., 2011). While the roles of RDR6 and DCL4 are relatively clear, the role of SGS3 has not been well described until recently. An in vitro analysis demonstrated that SGS3 can be recruited to RNA-induced silencing complex (RISC) bound with AGO1, via the 3′ nucleotides of the 22-nucleotide miR173 paired with the TAS2 target RNA; the function of SGS3 may be to stabilize the 3′ target fragment resulting from miRNA-directed cleavage (Yoshikawa et al., 2013). There may be other proteins involved in this process that are yet to be described or that have minor roles, while yet other proteins may participate less directly via partially redundant roles; for example, DCL2 and DCL3 have partial redundancy with DCL4 in tasiRNA biogenesis (Gasciolli et al., 2005; Henderson et al., 2006). Most importantly, there are two mechanisms by which 21-nucleotide tasiRNAs are produced, known as the “one-hit” or “two-hit” pathways (Figure 1A). In the one-hit mechanism, a single miRNA directs cleavage of the mRNA target triggering the production of phasiRNAs in the fragment 3′ to (or downstream of) the target site (Figure 1B) (Allen et al., 2005). We now know that this one-hit miRNA trigger is typically 22 nucleotides in length (Figure 2A) (Chen et al., 2010; Cuperus et al., 2010). In the two-hit model, a pair of 21-nucleotide miRNA target sites is employed, of which cleavage occurs at only the 3′ target site, triggering the production of phasiRNAs in the fragment 5′ to (or upstream of) the target site (Figures 1B and 2B) (Axtell et al., 2006).
Figure 1.
Pathways for the Biogenesis of PhasiRNAs Modeled on Arabidopsis.
(A) As the first step in secondary siRNA biogenesis, mRNA targets are cleaved by a miRNA. In the one-hit model, exemplified in Arabidopsis by TAS1, TAS2, and TAS4, a 22-nucleotide miRNA targets a single site. In the two-hit model, there are two target sites for a 21-nucleotide miRNA, exemplified in Arabidopsis by TAS3 transcripts cleaved by an AGO7-loaded miR390. Activity of the trigger miRNA recruits RDR6 and SGS3, resulting in production of a second strand of the target mRNA.
(B) The dsRNA is successively processed by DCL4 and other components to generate 21-nucleotide tasiRNAs; the direction of processing depends on the miRNA trigger mechanism. The secondary siRNAs are loaded onto an AGO protein and go on to function against other mRNAs.
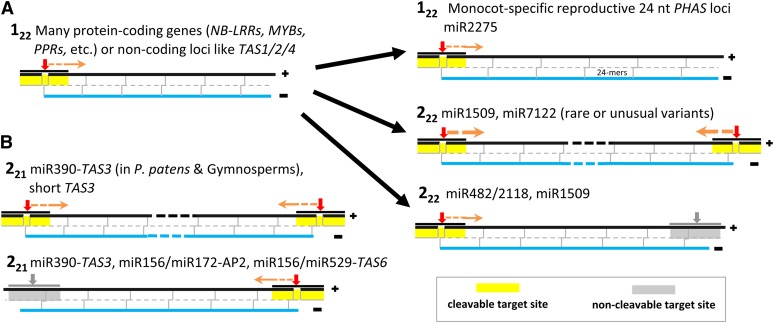
Figure 2.
Triggers and Processing Mechanisms of PhasiRNAs
The primary mechanisms of processing for plant phasiRNAs are described along with prototypical loci and the miRNAs that trigger siRNA biogenesis at these loci. Red arrows indicate cleavage sites, and orange arrows indicate the direction of precursor processing into phasiRNAs, which are indicated by gray lines in the double-stranded black/blue precursors.
(A) The one-hit pathway is typified by a single target site for a 22-nucleotide miRNA that results in downstream processing of the target transcript into ∼21-nucleotide phasiRNAs. This is denoted as a 122 locus. There are at least three notable variations on the one-hit model, including (1) the reproductive lncRNAs of monocots that are processed by DCL5 into 24-nucleotide phasiRNAs, triggered by miR2275 and thus also 122 loci, but with different biogenesis components; (2) and (3) are both 222 loci, but the 3′ site can be either cleaved or not cleaved.
(B) The two-hit pathway is typified by two target sites of a 21-nucleotide miRNA that results in processing upstream of the 3′ site. This is denoted as a 221 locus, and the best characterized examples are TAS3 and related loci, although a few other examples have been described. The 5′ site may be cleaved, which may result in processing from both directions, or the 5′ site may be noncleaved, as originally described for the Arabidopsis TAS3 locus.
TasiRNAs like miRNAs and other siRNAs, are usually incorporated into the RISC, leading to silencing of corresponding targets. TasiRNA functions have been well described from extensive work in Arabidopsis, which has a set of TAS genes that represent a core set of loci varying in their levels of conservation compared with other plants. Four families of TAS genes comprising eight loci have been identified in the Arabidopsis Columbia-0 genome (Table 1), among which miR173 targets both TAS1a/b/c family and the TAS2 locus, miR390 targets the TAS3a/b/c family, while miR828 triggers the production of TAS4-derived tasiRNAs (Allen et al., 2005; Yoshikawa et al., 2005; Rajagopalan et al., 2006). TAS3 is unique for several reasons: (1) it’s the only well-described two-hit locus in Arabidopsis, and (2) the 21-nucleotide miR390 trigger is exclusively loaded to a specialized Argonaute, AGO7 (Axtell et al., 2006; Montgomery et al., 2008a). A subset of TAS3a-derived tasiRNAs (tasi-ARFs) are involved in auxin responses, such as determining phase change or regulating root development, by altering transcript levels of auxin response factor (ARF) members, including ARF2, ARF3/ETT, and ARF4 (Allen et al., 2005; Williams et al., 2005; Fahlgren et al., 2006; Hunter et al., 2006; Marin et al., 2010). These tasi-ARFs form a concentration gradient from the adaxial side to the abaxial side of the leaf, suggesting they can move intercellularly as a regulator of ARF3-involved development (Chitwood et al., 2009). The functions of the other Arabidopsis TAS genes are not well described. TAS1 tasiRNAs target both PPR-encoding transcripts as well as approximately five genes of unknown functions; TAS2-derived tasiRNAs target PPR-encoding transcripts as well (Allen et al., 2005; Yoshikawa et al., 2005). TAS4 tasiRNAs increase in the shoot under phosphate-deficient conditions and perhaps participate in anthocyanin biosynthesis by targeting a group of MYB transcription factors (Rajagopalan et al., 2006; Hsieh et al., 2009). Another function of tasiRNAs is to mediate DNA methylation in cis at the TAS loci (Wu et al., 2012), which is unusual, given that these are 21-nucleotide small RNAs. However, since this methylation does not obviously suppress the expression level of TAS genes, the functional importance of this observation is not yet clear.
Table 1. Well-Described Noncoding PHAS Loci in Plants.
| Category | TAS1 and TAS2 Relatives | TAS3 | TAS4 | TAS6 | Reproductive lncRNAs |
|---|---|---|---|---|---|
| Plant families in which orthologs have been described (to date) | Eudicots | Land plants | Arabidopsis | P. patens | Gramineae |
| miRNA triggers | miR7122 superfamily (includes miR173) | miR390 | miR828 | miR529, miR156 | miR2118 (21-nucleotide PHAS loci); miR2275 (24-nucleotide PHAS loci)a |
| Numbers of phased loci | 4b | 3b | 1b | 3 | >1000 in rice |
| Key references | Allen et al. (2005), Yoshikawa et al. (2005), R. Xia et al. (2013) | Allen et al. (2005), Axtell et al. (2006) | Rajagopalan et al. (2006) | Arif et al. (2012), Cho et al. (2012) | Johnson et al. (2009), Song et al. (2012) |
miR2118 and miR2275 were shown to trigger 21- and 24-nucleotide phasiRNAs, respectively (Johnson et al., 2009; Song et al., 2012).
Number of loci in Arabidopsis.
TasiRNAs have been characterized in mosses, indicating that the utilization of tasiRNAs for gene regulatory functions is an ancient pathway in plants. In Physcomitrella patens, miR390, TAS3a, and the resulting tasi-ARFs have all been described (Axtell et al., 2006; Talmor-Neiman et al., 2006), as well as additional TAS loci, some of which are not conserved with Arabidopsis (Arif et al., 2012). The P. patens TAS6 is a two-hit locus like TAS3 but is targeted by different conserved miRNAs, and it has important roles in development, including bud formation (Cho et al., 2012). TAS3 is believed to be the most well-conserved TAS locus, as it has been identified across a broad range of species, from P. patens to monocots, such as rice (Oryza sativa) and maize (Zea mays) (Williams et al., 2005; Heisel et al., 2008), and including gymnosperms such as pine (Pinus taeda) (Axtell et al., 2006). In Arabidopsis, only the 3′ miRNA target site in the TAS3 transcript is cleaved, as in other flowering plants, while both miR390 complementary sites in moss and pine showed cleavage (Figure 2B) (Axtell et al., 2006). A shorter variant of TAS3 than that found in Arabidopsis is also conserved in many eudicots and is found alongside the canonical TAS3 locus; this variant has cleavable target sites at both 5′ and 3′ positions and includes only a single tasiARF (Figure 2A) (Krasnikova et al., 2009; Xia et al., 2012). A recent study of phasiRNA trigger evolution (e.g., miRNAs) across a broad range of plant species demonstrated that after the appearance of the miR390 family ∼450 million years ago, duplication, divergence, and neofunctionalization gave rise to at least seven families of miRNAs in two major superfamilies, the miR7122 and miR4376 superfamilies, while still maintaining miR390 as an important miRNA (R. Xia et al., 2013). These three closely related miRNA groups share a common origin and regulate distinct gene families (R. Xia et al., 2013). Thus, land plants have widely exploited the regulatory functions of phasiRNA to regulate large gene families.
Studies characterizing tasi- or phasiRNAs in many plant genomes utilized bioinformatics methods for genome-wide scans. Due to the precise 21-nucleotide phasing of tasiRNAs (Allen et al., 2005), genome-wide analysis with computational algorithms can identify candidate phased loci (Chen et al., 2007; Howell et al., 2007). These scans empirically define a specific P value or phasing score as a threshold or cutoff; in Arabidopsis, this approach identified the known tasiRNAs (see above) as well as several protein-coding genes, such as PPR transcripts, with 21-nucleotide phased siRNAs (Chen et al., 2007; Howell et al., 2007). It was even shown that one tasiRNA, tasiR2140, plays a role in triggering tertiary tasiRNAs, as part of an expanded cascade of tasiRNA regulation (Chen et al., 2007). Interestingly, PPR transcripts were shown to generate 21-nucleotide secondary phased siRNAs, some of which were triggered by tasiRNAs and some triggered by other miRNAs (Chen et al., 2007; Howell et al., 2007), but with the important observation that phased, secondary siRNAs are generated not only from noncoding TAS loci but also from protein-coding transcripts. In more recent work, we attempted to clarify the “tasi” versus “phasi” nomenclature (Zhai et al., 2011); trans-acting function often is not confirmed experimentally coincident with the identification of phased siRNAs; thus, “phasiRNAs” are loci merely identified as phased, whereas “tasiRNAs” have been demonstrated to act in trans (Zhai et al., 2011). In addition, the “TAS” name has been given only to noncoding transcripts with no function other than to give rise to secondary siRNAs. Recent work has described the TAS6 locus (Arif et al., 2012) and many TAS-like (TASL) loci (R. Xia et al., 2013), as well as an as yet unnamed TAS-like ncRNA locus (Zhai et al., 2011), indicating that additional ncRNA-derived TAS loci will continue to be described and named, some of which may be lineage specific. TAS5 has also been described (Li et al., 2012a), but we believe it is inappropriately named, as it appears to be an incorrectly annotated protein coding (NB-LRR) transcript. With the proliferation of sequenced plant genomes in recent years, an integral part of genome annotation is to identify the full complement of phasiRNA-generating loci.
As a consequence of the relatively well understood biogenesis pathway for tasiRNA and mechanism of their function, several labs have exploited this effective RNA silencing method for the study of gene function. Montgomery et al. used a synthetic TAS3a and TAS1c in Arabidopsis to produce artificial tasiRNAs targeting the PDS gene, resulting in photobleaching at the site of activity (Montgomery et al., 2008a, 2008b). In separate work, silencing of the CHLORINA42 gene produced photobleaching and was achieved by use of a modified TAS1a transcript (Felippes and Weigel, 2009). Finally, a TAS1c silencing system containing anywhere from a single FAD2-specific siRNA to a 210-bp fragment of FAD2 could successfully phenocopy the FAD2 loss-of-function mutant (de la Luz Gutiérrez-Nava et al., 2008). Presumably, in any of these systems or using other phased siRNA-producing transcripts, multiple artificial tasiRNAs could be developed to silencing several genes at once. Thus, artificial tasiRNAs are a powerful tool for gene functional analysis.
miRNA TRIGGERS OF PHASED, SECONDARY siRNAs
As mentioned above, an intriguing early observation was that either one or two miRNA target sites can trigger tasiRNA biogenesis. The two-hit model provided the first mechanistic insights into the process for tasiRNA biogenesis, describing TAS3 as the prototypical two-hit locus (Axtell et al., 2006). Early experimental examination of the two TAS3 target sites demonstrated that miR390 has an unusual association with AGO7 that is important for tasiRNA biogenesis and that the 5′ proximal miR390 target site must not be cleaved (Montgomery et al., 2008a), although outside of Arabidopsis, TAS3 variants may be cleaved at the 5′ position (Axtell et al., 2006; Krasnikova et al., 2009; Xia et al., 2012). However, more recent data from other plant genomes describe two-hit loci for which the miRNA triggers are believed to be AGO1 associated and for which the 5′ proximal site may be cleaved. For example, in Medicago, in addition to TAS3, another 221 TAS locus was described (identified as such because it is a two-hit locus with two 21-nucleotide miRNA target sites); like TAS3, the 5′ proximal site is not cleaved and the 3′ site is cleaved, but the triggers are miR172 and miR156, two well-conserved miRNAs that are AGO1 loaded in Arabidopsis (Figure 2B) (Zhai et al., 2011). In both Medicago and apple (Malus domestica), 222 loci have been described, with cleavage by two 22-nucleotide miRNAs at both 5′ and 3′ proximal target sites resulting in bidirectional processing into phasiRNAs of the fragment between the target sites (Figure 2A) (Zhai et al., 2011; R. Xia et al., 2013). In yet another 222 variant, a cleavable 5′ site and noncleavable 3′ site trigger phasiRNAs (Figure 2A) (Shivaprasad et al., 2012; R. Xia et al., 2013). More recent work in P. patens emphasized the diversity of two-hit loci, confirming via a newly described TAS6 locus that 221 PHAS loci can be triggered by a pair of presumably AGO1-loaded 21-nucleotide miRNAs that can be different from one another (Figure 2B) (Cho et al., 2012). Thus, our current understanding is that the noncleaving 5′ proximal miRNA target site is apparently a unique feature of AGO7-loaded miR390 for some TAS3 loci, with other two-hit loci utilizing cleaved 5′ proximal sites via AGO1-loaded miRNAs.
Analysis of one-hit triggers of PHAS loci and experiments using these miRNAs have also produced intriguing findings. In 2010, a pair of articles described that a shared feature of one-hit loci is that the triggers are 22-nucleotide and not 21-nucleotide miRNAs (Chen et al., 2010; Cuperus et al., 2010). This led to the hypothesis that 22-nucleotide miRNAs have special properties: the ability to trigger the production of phased siRNAs, confirmed via experiments employing a variety of constructs to generate miRNAs of specific lengths (Chen et al., 2010; Cuperus et al., 2010). In these experiments, canonical 21-nucleotide miRNAs, known not to trigger phased siRNAs, triggered the production of secondary siRNAs when produced as 22-nucleotide variants. Consistent with these results, tasiR2140, an unusual 22-nucleotide tasiRNA, triggers phasiRNA biogenesis from its target transcripts (Chen et al., 2010). More recent work has demonstrated that alterations in the 3′ nucleotide of the trigger miRNA can disrupt phasiRNA biogenesis, indicative of a role specifically for the small RNA length or target interactions (Zhang et al., 2012a).
A recent publication indicates that the secondary structure of the miRNA duplex, rather than the 22-nucleotide length, is the primary determinant of activity in triggering secondary siRNAs (Manavella et al., 2012). Manavella et al. observed that not only 22-nucleotide miRNAs but also 21-nucleotide miRNAs with 22-nucleotide miRNA* sequences can trigger secondary siRNA biogenesis. This led them to identify the characteristic shared by these miRNAs as an asymmetric duplex in the precursor, with the asymmetry resulting from a bulge or unpaired nucleotide. While AGO7-associated miRNAs (like miR390) are known to trigger secondary siRNAs (AGO2 may be similar), some miRNA triggers that they examined typically are loaded into AGO1, which is not associated predominantly with secondary siRNA production; this suggests that the same RISC components (AGO proteins) can either produce or not produce secondary siRNAs (Manavella et al., 2012). To test the role of an asymmetric duplex, they created a synthetic version of miR173 with two asymmetric bulges, which was shown to give rise to 21-nucleotide versions of both the miRNA and miRNA* and yet still triggered secondary siRNAs as effectively as a 22/21-nucleotide asymmetric precursor (Manavella et al., 2012). They thus inferred that RISC is reprogrammed upon interaction with an asymmetric duplex (i.e., a bulge caused by an unpaired base), and this reprogrammed RISC recruits proteins for secondary siRNA biogenesis. Manavella et al. also showed that the siRNAs were produced through via RDR6/SGS3/DCL4, the cofactors likely recruited by the RISC.
While these data are quite convincing, other data suggest that duplex asymmetry cannot entirely explain the ability of some plant miRNAs to trigger secondary siRNAs. In recent work, we examined miRNAs in an Arabidopsis hen1 mutant background; HEN1 is an enzyme that adds a 2′-O-methyl group to the 3′ terminal nucleotide of miRNAs and siRNAs. We observed that in hen1, two targets of miR170 and miR171a, miRNAs produced from symmetric precursors, give rise to phasiRNAs (Zhai et al., 2013). In the wild type, no phasiRNAs are produced from the targets, and these miRNAs are 21 nucleotides, but in the hen1 mutant, 3′ uridylation of the miRNAs after biogenesis gives rise to 22-nucleotide variants. We inferred that in this case, it is the 22-nucleotide length that confers the triggering activity for secondary siRNAs. Consistent with our observations, there are several reports of secondary siRNAs that, for inexplicable reasons, consistently are generated as 22-nucleotide siRNAs and themselves then trigger secondary siRNAs at targets in trans; these include the Arabidopsis tasiR2140 (Chen et al., 2007) as well as tasiRNAs from several TASL loci in multiple species (R. Xia et al., 2013). Also inconsistent with the requirement of an asymmetric precursor, miR828 is produced from a symmetrical stem-loop precursor yet triggers phasiRNA production from TAS4 and many MYB genes (Rajagopalan et al., 2006; Xia et al., 2012). Finally, mismatches between the 3′-terminus of miRNA triggers and their TAS targets reduce the stability of the interaction between the cleavage fragment and RISC complex, inhibiting tasiRNA production (Zhang et al., 2012a). This is likely because a mismatched 3′ end would fail to recruit SGS3 and thus fail to stabilize the 3′ mRNA fragment (Yoshikawa et al., 2013). These results suggest the importance of miRNA-target interactions in generating tasiRNAs.
PHASED SECONDARY siRNAs FROM LONG NONCODING RNAs IN GRASSES
Recent work has demonstrated an abundance of loci producing phasiRNAs in monocots, with examples in rice, maize, and Brachypodium (Johnson et al., 2009; International Brachypodium Initiative, 2010; Song et al., 2012). In addition to the handful of tasiRNA loci conserved with Arabidopsis, these grasses have two classes of phasiRNAs specifically expressed in reproductive tissues, including an unusual class of 24-nucleotide PHAS loci as well as a large number of 21-nucleotide PHAS loci (Table 1). Both classes of reproductive phasiRNAs are derived predominantly from long noncoding RNAs (lncRNAs) that are not repetitive (Johnson et al., 2009). One study of reproductive phasiRNAs in rice showed that the processing of 21-nucleotide phased siRNAs is largely dependent on Os-DCL4, the ortholog of Arabidopsis DCL4 (Song et al., 2012). Interestingly, at least the cereals and perhaps most other monocots have an extra Dicer relative to dicots, known as either DCL3b, for the similarity in its PAZ domain to DCL3, or DCL5 based on the diversity of its dsRNA binding domain (Margis et al., 2006). We prefer the DCL5 name because of the apparently unique role of this protein, functioning to produce the reproductive 24-nucleotide phased siRNAs (Song et al., 2012). These two classes of reproductive phasiRNAs are triggered by distinct miRNA triggers; miR2118 and miR2275 were identified as the triggers of phased 21- and 24-nucleotide siRNAs, respectively (Johnson et al., 2009; Song et al., 2012). Although the functions of 21- and 24-nucleotide reproductive phasiRNAs have not been reported, their specificity in reproductive organs is quite intriguing. In fact, PHAS loci are functionally important, as shown by identification via forward genetics of a lncRNA, subsequently processed to 21-nucleotide phasiRNAs, required for rice male fertility (Ding et al., 2012a, 2012b; Zhou et al., 2012). Based on the functions of 21- and 24-nucleotide siRNAs in Arabidopsis, the grass reproductive phasiRNAs likely function in posttranscriptional regulation and chromatin modifications, respectively. In addition, based on the specificity of their expression in reproductive tissues, we might speculate that their roles include reproductive development, meiosis, or gamete formation. Thus, studies of the functions of phased siRNAs in grasses and their lncRNA precursors hold great promise for exciting discoveries.
PHASED SECONDARY siRNAs AS A REGULATORY MECHANISM FOR PROTEIN CODING GENES
miRNA-triggered secondary siRNAs are also generated from protein-coding loci in many plant genomes, first described in Arabidopsis (Howell et al., 2007). A significant number of PPR, NB-LRR, and MYB families were shown to generate phasiRNAs in Arabidopsis, Medicago, apple, and peach (Prunus persica) (Table 2) (Howell et al., 2007; Zhai et al., 2011; Xia et al., 2012; Zhu et al., 2012). The PPR family is one of the largest gene families in Arabidopsis, containing ∼450 members in total, some of which have been shown to be involved with organelle RNA processes (Lurin et al., 2004; O’Toole et al., 2008). In Arabidopsis, a small number of miRNAs and tasiRNAs have been shown collectively to target ∼40 PPRs, among which 28 are closely related (Howell et al., 2007). A comparative analysis across plant species demonstrated conservation of the ability of this subgroup of PPRs to spawn secondary siRNAs, targeting a broader group of PPRs in many but not all plants (R. Xia et al., 2013). The triggers of these PPR-derived secondary siRNAs are a superfamily of miRNAs, as well as unusual, 22-nucleotide secondary siRNAs that function in trans (Chen et al., 2007; R. Xia et al., 2013). These small RNAs target variable sites within the PPR domains (Figure 3A). Since this regulatory network includes both miRNAs and tasiRNAs, it represents a highly redundant, interconnected set of PPR-targeting small RNAs. It was proposed that this regulation could be beneficial to the evolutionary expansion of PPR genes (Howell et al., 2007). The superfamily of miRNAs that trigger secondary siRNAs from PPRs is unusual because (1) it is derived from the prototypical phasiRNA trigger, miR390, and (2) it gave rise to a different superfamily of miRNAs that target Ca2+ ATPases, some of which (but perhaps not all) also generate phasiRNAs (Wang et al., 2011; R. Xia et al., 2013).
Table 2. Well-Described PHAS Loci from Protein-Coding Gene Families in Plants.
| Category | MYBs | NB-LRRs | PPRs | Ca2+-ATPase | AFB |
|---|---|---|---|---|---|
| Plant families in which phasiRNA-producing members have been described (to date) | Rosaceae | Brassicaceae, Coniferae, Fabaceae, Rosaceae, Solanaceae, Vitaceae | Brassicaceae, Fabaceae, Rosaceae, Solanaceae, Vitaceae | Seed plants | Flowering plants |
| Encoded protein type | MYB | NB-LRR | PPR | Ca2+-ATPase | TIR/AFB |
| miRNA triggers | miR159, miR828, miR858 | miR482, miR2118, miR1507, miR2109/miR5213 | miR161, miR7122a | miR4376a | miR393 |
| Numbers of phased loci | >10 in apple | >100 in Medicago | Variable, but few | Variable, but few | Variable, but few |
| Key references | Xia et al. (2012), Zhu et al. (2012). | Zhai et al. (2011), Li et al. (2012b), Shivaprasad et al. (2012) | Howell et al. (2007), Xia et al. (2012), R. Xia et al. (2013) | Wang et al. (2011), R. Xia et al. (2013) | Si-Ammour et al. (2011), Zhai et al. (2011) |
This miRNA belongs to a superfamily of related miRNAs; see description by R. Xia et al. (2013).
These miRNAs are more abundant in fruit and flower tissues of the Rosaceae, but they are not exclusively expressed in these tissues (Xia et al., 2012).
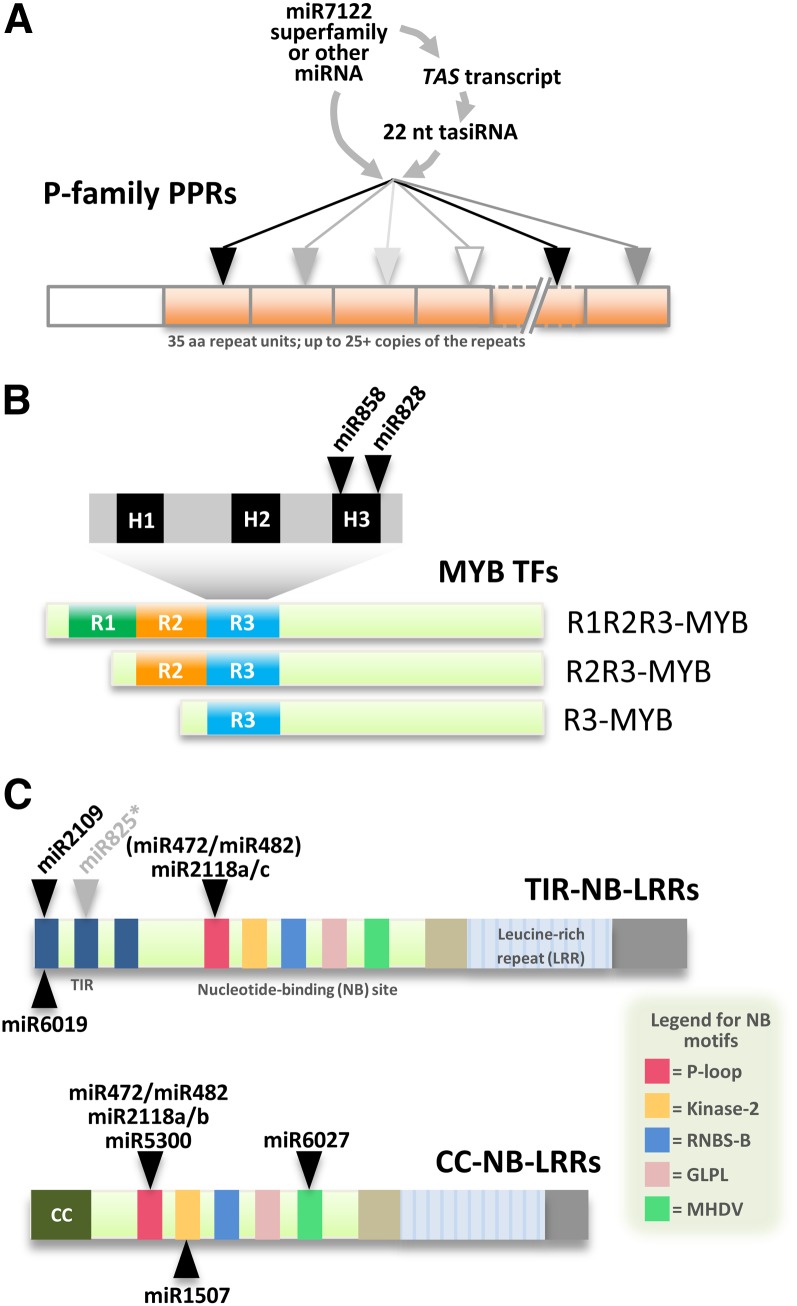
Figure 3.
miRNAs Target Nucleotides Encoding Conserved Protein Motifs of Several Gene Families.
In each case, miRNAs, designated by a black arrowhead, target sites that encode protein motifs or domains as indicated in each panel.
(A) PPR genes encoding the P subclass of PPRs are targeted by both miRNAs and tasiRNAs. Each gray box represents one degenerate repeat of ∼35 amino acids. PPR proteins have a widely varying number of these repeat units (indicated by the broken repeat unit). Gray or outlined arrowheads indicate that miRNA or tasiRNA target sites may exist at varying levels or may not exist at all in some repeats, due to the degeneracy of the repeat sequences.
(B) miRNAs target nucleotides encoding H3 motifs in the conserved R3 domains of MYB transcription factors in plants.
(C) Numerous miRNAs target nucleotides encoding conserved motifs of NB-LRRs in many plant species. The NB domain has five conserved motifs indicated by colored boxes; other conserved domains and motifs characterize these proteins, as indicated. Considering many plant species, multiple encoded motifs of NB-LRRs are targeted, including the TIR1, TIR2, P-loop, kinase-2, and MHDV motifs. miR472 and miR482 are nearly identical (see Figure 4A) and indicated parenthetically for TNLs, as CNLs are the preferential targets (with TNLs as less frequent targets). miR825* is indicated in gray, as it is observed to target an encoded TIR2 only in Arabidopsis.
Genes encoding MYB transcription factors are also rich sources of miRNA-triggered secondary siRNAs. MYBs are a family of DNA binding proteins that play important roles in a variety of transcriptionally regulated processes, such as cellular morphogenesis, meristem formation, cell cycle, and anthocyanin biosynthesis (Jin and Martin, 1999; Petroni and Tonelli, 2011). MYB transcription factors are encoded by one of the largest of gene families in many plant genomes (Feller et al., 2011). Several MYBs have been identified to be responsible (among other activities) for anthocyanin biosynthesis both in fruit development of apple and other Rosaceae species, as well as in maize kernels (Takos et al., 2006; Lin-Wang et al., 2010; Feller et al., 2011). In the case of apple, phasiRNAs are produced from a number of MYB-coding genes; for example, miR828 and miR858 target the conserved motifs of up to 81 MYB transcripts (Figure 3B) (Xia et al., 2012). A comparative phylogenetic analysis revealed that those MYB genes containing target sites of both miR858 and miR828 are conserved across a broad range of plants with the miRNAs found only in eudicots thus far (Xia et al., 2012); perhaps phasiRNA regulation of MYBs is an adaptation specific to the eudicots. The apple MYB-derived phasiRNAs are predicted to target a variety of genes with distinct functions, potentially expanding this miRNA-mediated regulatory network (Xia et al., 2012). Similar to apple, peach also produces a large number of MYB-derived phasiRNAs (Zhu et al., 2012). As with PPRs, MYBs are encoded by a large and complex gene family in plant genomes, but the functional or evolutionary role of phasiRNA transcriptional suppression of the family is unclear.
NB-LRR–encoding genes comprise one of the largest families found to be targeted by small RNAs. Compared with the PPR- and MYB-encoding gene families in other plant genomes, a much larger number of NB-LRRs were found to be PHAS loci in the Medicago genome (Zhai et al., 2011). Many phasiRNAs target NB-LRR transcripts either in cis or in trans at other NB-LRR loci, representing a self-reinforcing regulatory network (Zhai et al., 2011). As with the PPR family in Arabidopsis and other plants (Howell et al., 2007; R. Xia et al., 2013), NB-LRRs can be targeted redundantly by both miRNAs and secondary siRNAs (Zhai et al., 2011). NB-LRR regulation by secondary siRNAs has also been reported to exist widely in the Solanaceae (Zhai et al., 2011; Li et al., 2012b; Shivaprasad et al., 2012). Most recently, an examination of numerous NB-LRRs in a wide variety of plant species demonstrated significant levels of secondary siRNAs in Norway spruce (Picea abies; a gymnosperm), Amborella (a basal angiosperm), cotton (Gossypium hirsutum), poplar (Populus spp), grapevine (Vitis vinifera), apple, and peach, indicating broad conservation and an ancient origin for the role of phasiRNAs in regulation of NB-LRRs (Källman et al., 2013). Possible reasons for phasiRNA regulation of NB-LRR transcripts are discussed in more detail below.
Transcripts of protein-coding genes other than PPRs, MYBs, or NB-LRRs also generate phasiRNAs, but thus far, outside of these three large gene families, these protein-coding PHAS loci are solitary or very small families. For example, in soybean (Glycine max), the small RNA biogenesis machinery is itself subject to phasiRNA regulation, evidenced by secondary siRNAs mapping to both DCL2 and SGS3 transcripts, and a number of other single- or low-copy genes are sources of phasiRNAs (Zhai et al., 2011). Likewise in peach, phasiRNAs are produced from many single- or low-copy genes, including (among others) those encoding TIR/AFB, ARF, and a Ca2+-ATPase (R. Xia et al., 2013). The PHAS characteristic of many low-copy protein-coding genes is conserved across species; for example, the tomato ortholog of the peach Ca2+-ATPase is also a PHAS locus (Wang et al., 2011). Computational analysis of grape small RNAs identified nearly 50 phased loci in total, among which at least 20 are protein-coding genes and some of which are members of the NB-LRR family (Zhang et al., 2012b). These results indicate phasiRNA-associated regulatory networks are utilized by many low-copy genes and gene families involved in diverse biological processes and pathways, although the evidence of this is thus far mainly or exclusively from eudicots. It’s possible that phasiRNAs perform regulatory functions such as tuning or heavily suppressing transcript levels that are equally important for single-copy genes, although in such cases, these secondary siRNAs are presumably functioning in cis since there may be no trans targets. Alternatively, phasiRNA could function in some sort of positional manner related to their mobility, such as defining a concentration gradient across cell layers, like the tasi-ARFs (Chitwood et al., 2009).
OVERVIEW OF PLANT NB-LRRs
Plants possess a two-layer process of immune defenses to respond to pathogenic stress (Jones and Dangl, 2006). In the first layer, transmembrane pattern recognition receptors are responsible for recognizing pathogen-associated molecular patterns (PAMPs), activating PAMP-triggered immunity (Medzhitov and Janeway, 1997; Monaghan and Zipfel, 2012). In the second layer, the intracellular disease resistance (R) proteins play a crucial role in pathogenic effector (avirulence, or AVR protein) detection leading to effector-triggered immunity in plants, either by interacting directly with effectors or in a manner described by the guard hypothesis, in which an endogenous AVR target is monitored for signs of pathogen attack (Van der Biezen and Jones, 1998; Dangl and Jones, 2001; Axtell and Staskawicz, 2003; Mackey et al., 2003; Jones and Dangl, 2006). Many plant R proteins are characterized by having a NB domain and an LRR domain (Dangl and Jones, 2001; DeYoung and Innes, 2006; Jones and Dangl, 2006). NB-LRRs can be categorized into two major groups according to their N-terminal domains: TIR-NB-LRRs (TNLs) possess a Toll/Interleukin-1 receptor homology domain, and CC-NB-LRRs (CNLs) possess a domain which includes a coiled-coil motif (Meyers et al., 1999, 2003; Pan et al., 2000; McHale et al., 2006). Within these domains (TIR, CC, NB, and LRR) there are numerous highly conserved motifs, and the nucleotides encoding some of these motifs are important targets of miRNAs, as described below. Genes encoding the CNL-type (CNLs) are found in both dicots and monocots, whereas TNL-encoding genes (TNLs) are found exclusively in dicots (Kim et al., 2012). Both CNLs and TNLs are found in mosses, early land plants, as well as two other types of NB-LRRs that apparently were not retained in angiosperms (Xue et al., 2012). It is believed that TNLs were lost in the monocots (Pan et al., 2000) and perhaps some dicot lineages such as Mimulus guttatus (Kim et al., 2012). Most plant genomes encode hundreds of R proteins, ranging from the reported low of ∼55 NB-containing proteins in papaya (Carica papaya) (Ming et al., 2008) and ∼62 in cucumber (Cucumis sativus; Yang et al., 2013), to ∼150 in Arabidopsis (Meyers et al., 2003) and more than 500 in rice and 700 in Medicago truncatula, a model species for legumes (Zhou et al., 2004; Young et al., 2011). Within the ∼80% complete genomic sequence of Medicago, ∼540 NB-LRR–coding genes were identified of which ∼14% are predicted pseudogenes (Young et al., 2011). In sequenced Solanaceae genomes such as potato (Solanum tuberosum), ∼400 NB-LRRs were identified, many (∼41%) of which may be pseudogenes (Xu et al., 2011; Lozano et al., 2012). These pseudogenes might be left over from an ongoing evolutionary arms race between the host and pathogens.
NB-LRRs ARE TARGETED BY NETWORKS OF HIGHLY REDUNDANT SMALL RNAs
As mentioned above, analysis of siRNAs matched to NB-LRRs in Medicago and several Solanaceous species identified many phased, secondary siRNAs (Zhai et al., 2011; Li et al., 2012b; Shivaprasad et al., 2012). In Medicago, transcripts encoding NB-LRRs are targeted by miRNAs at several conserved motifs, triggering phasiRNA production from these genes, which we refer to as “phasi-NB-LRRs” or pNLs (Zhai et al., 2011). While more than 114 phasiRNA-producing NB-LRRs were identified, >60% of Medicago genomic NB-LRRs had significant levels of 21-nucleotide small RNAs, suggesting that most members of this gene family are targeted by 22-nucleotide miRNAs (Zhai et al., 2011). Because phasiRNAs can also function both in cis and in trans, targeting other related transcripts, a limited number of miRNA triggers can dramatically amplify their suppressive functions through the production of secondary phasiRNAs, and these two kinds of small RNAs seem to have a joint effect in regulating the great majority of NB-LRRs in Medicago. Thus, miRNAs act as master regulators of the NB-LRR gene family via the production of phasiRNAs (Zhai et al., 2011). Compared with Medicago, the numbers of PHAS loci are relatively smaller in other legume species, such as soybean (Zhai et al., 2011). However, due to the synergistic effect by miRNAs and secondary phasiRNAs, a significant proportion of NB-LRRs could be targeted and downregulated in legumes other than Medicago. In the Solanaceous species (tomato [Solanum lycopersicum], potato, and tobacco [Nicotiana tabacum]), numerous pNLs have been described, although a lower proportion of genomic NB-LRRs in Solanaceous genomes are pNLs than reported in Medicago (Zhai et al., 2011; Li et al., 2012b; Shivaprasad et al., 2012).
In the relationship between miRNAs and their NB-LRR targets, there is an unusual level of redundancy (Figure 3C). In Medicago, three families of 22-nucleotide miRNA (miR1507, miR2109, and miR2118a/b/c) target the sequences encoding highly conserved protein motifs, such as TIR-1, P-loop, and Kinase-2, triggering phasiRNA production (Zhai et al., 2011). These three unrelated families of miRNAs show no specialization for clades or subgroups within the NB-LRRs, implying that any one of these miRNAs is capable of targeting diverse members of the NB-LRR family. Together with additional NB-LRR–targeting miRNAs from the Solanaceae, there are at least six miRNA families that target NB-LRRs (Figure 3C). Typically, plant miRNAs and their target families of genes show a one-to-one relationship, with a single miRNA (or family) that targets a single set of genes. For example, there are five copies of the miR172 family in Arabidopsis, which all target members of the APETALA2 gene family (Aukerman and Sakai, 2003). Thus, the case of NB-LRRs, targeted independently by as many as six different miRNA families, appears to be highly unusual. There are two additional levels of redundancy in NB-LRR–miRNA interactions worth considering: (1) The phasiRNAs generated via miRNA cleavage may function in trans to silence related targets. This trans-acting activity was confirmed in Medicago (Zhai et al., 2011), and given the tremendous abundance of phasiRNAs produced from NB-LRRs in many species, this is likely a significant mechanism for silencing within the family. (2) An additional level of redundancy is represented by the diversity of miRNAs that target nucleotides encoding conserved protein motifs. The most extreme case of this is the superfamily of miRNAs that target the encoded P-loop, a group which includes miR472, miR482, miR2089, miR2118, and miR5300. While some of this variation in naming is simply a historical artifact (i.e., miR472 and miR482 are nearly identical), there is substantial sequence variation in members of this superfamily such that the members wouldn’t fit the definition of a single family (Figure 4A) (Meyers et al., 2008). This superfamily could be known by its inclusion of both the miR482 type, more predominant in the Solanaceae (Shivaprasad et al., 2012), or the miR2118 type, more predominant in the Fabaceae (Zhai et al., 2011). The TIR-1 motif is similarly targeted by two unrelated miRNAs, miR2109 and miR6019, which target nonoverlapping nucleotides that encode the motif (Figure 4B). It’s possible that future miRNA annotation in more diverse species will identify even more divergent members of these families or superfamilies that may contribute further to the high level of redundancy of in the suppression of NB-LRR transcripts.
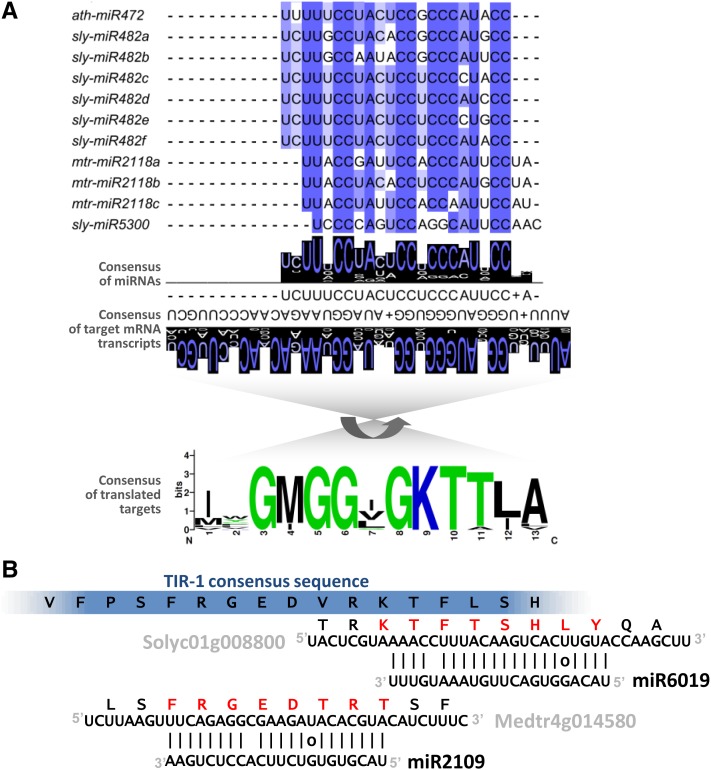
Figure 4.
miRNAs Target Conserved Sequences in Members of the NB-LRR Gene Families.
(A) The miR482/miR2118 superfamily of miRNAs is a relatively diverse group (above) that typically targets nucleotides encoding the P-loop motif of NB-LRR proteins (below). Consensus sequences of either the miRNAs or their targets demonstrate a high degree of conservation (illustrated by WebLogo). In this figure, for illustrative purposes, we’ve randomly selected a diverse set of miRNA superfamily members (listed by their names in miRBase) and ∼16 targets from the same source species. In this case, the targets were predominantly CNLs, but miR2118 also targets many TNLs. For ease of alignment, the miRNAs are shown 5′ to 3′ in the consensus, the mRNA targets are shown 3′ to 5′, and the translated protein motif is inverted relative to the target mRNAs (indicated by the curved arrow).
(B) Redundancy in miRNA targeting at the encoded TIR-1 motif of NB-LRRs. miR6019 and miR2109 aligned to their TNL-encoding targets show they target the same encoded motif (blue bar at top), but at adjacent, nonoverlapping sites. The consensus at the top is from Meyers et al. (1999). Red letters indicate the amino acids encoded by the target region. Example targets from tomato (“Soly…”) and Medicago (“Medtr…”) are indicated aligned to the miRNA sequences.
The functional relevance of endogenous NB-LRR silencing is unknown, yet the data undeniably demonstrate that it is widespread within the gene family, robust, and found in many diverse angiosperms and as far back evolutionarily as the gymnosperms. Although the Poaceae apparently lack this NB-LRR–suppressive regulatory machinery (i.e., the miRNAs and therefore the phasiRNAs), and it’s greatly reduced in the Brassicaceae, the phenomenon of pNLs is so prevalent and redundant that it seems they must have an important function. Furthermore, understanding the mechanistic importance of phasiRNAs in NB-LRR regulation may provide insights into the analogous miRNA/phasiRNA suppression of the PPRs and MYB transcription factors, or other protein-coding genes described above and listed in Table 2. In the following sections, we outline four hypotheses for the biological significance of small RNA–based NB-LRR suppression.
HYPOTHESIS #1: miRNA-BASED SUPPRESSION OF NB-LRRs IS IMPORTANT IN BENEFICIAL MICROBIAL INTERACTIONS
Beneficial symbiotic interactions between microbes and plant hosts is common, including the most ancient form represented by various levels of plant–fungal (mycorrhizal) interactions, up to perhaps the most intricate form in which legume species have co-opted bacteria (rhizobia) for nitrogen fixation in specialized host tissues (nodules). During any form of symbiosis, the host plant must recognize and accept the beneficial microbes, a process using sophisticated signaling networks and molecular dialogues (Deakin and Broughton, 2009). In legumes, signaling molecules secreted by rhizobia, including nod factors, are required for recognition and nodule initiation (Geurts et al., 2005; Oldroyd and Downie, 2008; Madsen et al., 2010). A connection between nodulation and miRNAs was observed as several miRNAs are modulated dynamically after rhizobial inoculation (Li et al., 2010); this work also demonstrated that the overexpression of certain miRNAs could promote soybean nodulation, including miR482, which targets NB-LRRs. Also in soybean, a TNL was demonstrated to confer symbiotic specificity, and this gene was later identified as a PHAS locus (Yang et al., 2010; Zhai et al., 2011). In addition to rhizobial symbiosis, studies have revealed that miRNAs also participate in mycorrhizal symbiosis with Medicago (Devers et al., 2011; Lauressergues et al., 2012). Thus, there are numerous evolved pathways in plants and particularly legumes to facilitate beneficial microbial interactions, and there is indirect evidence for the participation of miRNAs (including the NB-LRR–targeting miR482).
The most obvious hypothesis for the role of phasiRNAs from NB-LRR transcripts is that legumes could globally suppress NB-LRR levels to limit defense responses in the presence of rhizobia to facilitate nodulation. Are the data consistent with this hypothesis? We documented that legumes, relative to other families of plants, have very high abundances of the miRNA master regulators of NB-LRR transcript levels (Zhai et al., 2011). Our analysis of different Medicago tissues has shown no evidence of substantial regulation of the miRNAs or phasiRNAs, both of which are found at robust levels at many NB-LRR loci, even in aerial tissues far from the site of rhizobial interactions (Zhai et al., 2011). It is possible that miRNAs and their NB-LRR targets are regulated in a highly localized, cell-type-specific fashion not detectable by sequencing-based methods. And a point incongruous with this hypothesis is that legumes are only one of many plant families that have pNLs (including many plants that neither nodulate nor are known to have specialized symbiotic interactions), so the role of small RNA–based regulation of NB-LRRs may not be specific to legumes. An intriguing observation is that the miRNA families that target NB-LRRs are almost entirely missing from Arabidopsis (see below) (Zhai et al., 2011), and it is well known that Arabidopsis does not undergo mycorrhizal interactions, leading us to wonder if there is a relationship between silencing of NB-LRRs and symbiotic interactions. A large number of nonlegume species participate in beneficial symbiosis with mycorrhizal fungi (Finlay, 2008) or endophytes. Our analysis of miR2118 demonstrates that it is present in gingko, a plant that dates to ∼265 million years ago (a time at which plant–mycorrhizal interactions were becoming more refined), and other NB-LRR-targeting miRNA families are present in early angiosperms (Zhai et al., 2011). Consistent with this, recent data show extraordinarily high levels of NB-LRR–derived phased siRNAs in Norway spruce, another gymnosperm (Källman et al., 2013). These observations lead us to speculate that perhaps these NB-LRR–targeting miRNAs evolved early in plants to promote beneficial microbial interactions, ultimately facilitating the evolution of the complex structures (nodules) that develop in legumes to house nitrogen-fixing rhizobia.
Beyond the lack of data showing regulation of NB-LRRs, there are additional problems with this hypothesis, such as the observation that although rice and maize are known to undergo arbuscular mycorrhizal interactions, the NB-LRR families in these grasses show no signs of miRNA targeting or secondary siRNAs. There are >500 CNL-type NB-LRRs encoded in the rice genome, yet apparently no phasiRNAs arise from these transcripts. In the grasses, as described above, miR2118 targets noncoding RNAs not NB-LRRs. In Arabidopsis (Columbia-0), perhaps four pNLs have been identified, including three CNLs and one TNL (Howell et al., 2007; Chen et al., 2010); these four pNLs could comprise the evolutionary vestiges of the more extensive system observed in other plant families. Thus, Arabidopsis may be a good system in which to test the role of miRNA suppression of NB-LRRs. We are left wondering how plant lineages can lose major, suppressive regulatory small RNA networks like the pNLs, or perhaps those comprising PPRs or MYBs, and whether this suggests that the function of those networks is entirely dispensable. To return to the central question of this hypothesis: Are the data consistent with a hypothetical role in globally modulating plant defenses via suppression of NB-LRRs to promote beneficial microbial interactions? We conclude that there is not yet significant support for this hypothesis, yet future experiments to test this will be quite important.
HYPOTHESIS #2: SMALL RNA SUPPRESSION OF NB-LRRs IS IMPORTANT FOR PLANT DEFENSES
An alternative to hypothesis #1 is that the regulation of NB-LRRs is important for plant immunity to pathogens. This idea emerged from publications describing pNLs in tomato, tobacco, and potato (Zhai et al., 2011; Li et al., 2012b; Shivaprasad et al., 2012). Separate work by the Baulcombe and Baker labs demonstrated that the miRNA triggers of pNLs are regulated during plant defense responses. B. Baker and colleagues, working in Nicotiana benthamiana, reported that nta-miR6019 and nta-miR6020 are able to cleave transcripts of the N gene, producing 21-nucleotide phasiRNAs (Li et al., 2012b). The N gene encodes a member of the TNL class of NB-LRRs, imparting resistance against tobacco mosaic virus (Whitham et al., 1994). Attenuation of N-mediated resistance was observed when nta-miR6019 and nta-miR6020 were overexpressed in tobacco, consistent with miRNA-mediated suppression of this immunity pathway (Li et al., 2012b). Using bioinformatics pipelines, more miRNAs and PHAS loci of NB-LRRs were identified by the Baker lab in tobacco, implying that the N gene is far from unique. The Baulcombe lab conducted similar analyses in tomato, identifying miRNA-directed phasiRNA production at NB-LRR loci (Shivaprasad et al., 2012). The miR482 family in tomato, which has a close relationship with the miR2118 family described above for Medicago, targets the P-loop motif of CNL-encoding transcripts, triggering phased secondary siRNAs. Intriguingly, miR482 levels were slightly reduced (approximately by half) after inoculation with the bacterial pathogen (Pseudomonas syringae DC3000), whereas with some viruses, miR482 levels were reduced by up to 80% (Shivaprasad et al., 2012). These changes resulted in increased target NB-LRR transcript levels, although there was no known role for those encoded NB-LRRs in responses to the tested pathogens (Shivaprasad et al., 2012). These data suggest that plants could boost R gene expression as a consequence of sensing pathogen effectors by modulating miRNA levels.
What is still missing from these studies to directly implicate miRNAs in the regulation of NB-LRR levels during defense responses? The Baker lab showed miRNA overexpression can decrease N transcript levels, but they didn’t show regulation of the miRNAs during endogenous defense responses (Li et al., 2012b). The Baulcombe lab showed reductions of miR482 levels and increases of NB-LRR levels for a small number of genes of unknown specificity but didn’t show a direct connection between those RNA changes and immunity (Shivaprasad et al., 2012). Neither group showed global, coordinate changes in NB-LRR (or secondary siRNA) levels as a result of altered miRNA levels resulting from interactions with pathogens. As yet, there is no clear example of pathogen response that directly suppresses a miRNA and results in an increased NB-LRR of known specificity which in turn improves immunity. It is clear that the Solanaceous species have numerous NB-LRR–targeting miRNAs (Zhai et al., 2011; Li et al., 2012b; Shivaprasad et al., 2012), but is this indicative of greater redundancy of control for individual NB-LRRs or more specialization of targeting by individual miRNAs? Based on the trans-acting activity of the legume NB-LRR secondary siRNAs, we would expect that the Solanaceous phasiRNAs and miRNAs could comprise a widespread, interconnected suppressive network targeting NB-LRRs, but this has yet to be shown for the Solanaceous genomes. If such a network exists, is it regulated dynamically in response to microbes? And are there subnetworks interconnected based on pathogen specificities? And if the role in immunity is proven for these miRNAs in the Solanaceous species, is the role the same for these miRNAs in the legumes? One could even imagine the diversity of miRNAs targeting NB-LRRs could reflect a diversity of functions, perhaps with some NB-LRRs differentially regulated during microbial interactions and others constitutively expressed.
The situation may be even more complex, due to the involvement of pathogen-encoded suppressors of RNA silencing. Previous reports have demonstrated a wide variety of viral suppressors of RNA silencing, bacterial suppressors of RNA silencing, and, most recently, oomycete suppressors of RNA silencing, all of which can suppress the immunity of plants by inhibiting RNA silencing pathway of their hosts (Li and Ding, 2006; Navarro et al., 2008; Qiao et al., 2013). In the latter example, Qiao et al. (2013) demonstrated two oomycete suppressors of silencing function specifically to suppress RNA interference (shown via release of transgene silencing), miRNA levels, and secondary siRNA levels. In the context of miRNAs and phasiRNAs that suppress NB-LRRs, it may seem counterintuitive that a pathogen would reduce small RNA levels, thereby increasing NB-LRR levels; Qiao et al. point out that substantial upregulation of defense components could be beneficial to the pathogen during its necrotrophic phase, presumably by triggering cell death (Qiao et al., 2013). In other cases, the depression of NB-LRR levels triggered by pathogen-derived RNA silencing suppressors may represent host exploitation of the pathogen activity to upregulate R genes and increase host immunity. But as described above, regulation of miRNAs and NB-LRR-derived phasiRNAs in host responses are as yet relatively poorly characterized.
HYPOTHESIS #3: SMALL RNAs ACT AS DAMPERS OR BUFFERS OF NB-LRR TRANSCRIPT LEVELS
The constitutively high levels of NB-LRR–targeting miRNAs in the legumes and Solanaceous species (Zhai et al., 2011; Shivaprasad et al., 2012) suggest a role in steady state regulation of the target transcripts. One function for these small RNAs may be simply to buffer the NB-LRR transcript levels, dampening any dramatic changes in their levels, particularly large increases that could cause deleterious accumulations of NB-LRR proteins. At the same time, the very low steady state NB-LRR transcript levels resulting from constitutive suppression would restrict steady state protein levels to a bare minimum, although presumably still a level at which NB-LRR–mediated responses are fully functional. There should also be feedback regulation: In the presence of an excess of 22-nucleotide miRNAs, any increased NB-LRR transcript levels would result in increased secondary siRNAs, which would in turn reduce NB-LRR levels. This could prevent NB-LRR proteins accumulating to a deleterious level. This dampening effect could be important to protect from variation in promoters that would otherwise lead to variation in transcript levels beyond some desirable limit.
This hypothesis is consistent with the observation that plants actively regulate both R transcript levels and R protein levels. One of the best described examples of this is the regulation of SUPPRESSOR OF NPR1-1 CONSTITUTIVE1 (SNC1), a TNL-type R gene that has been well studied due to the identification of an autoactive mutant allele (Zhang et al., 2003). Suppressor screens using this allele have identified a variety of regulatory genes that function to modulate both transcript and protein levels of SNC1 (Zhang and Li, 2005). Recent work has identified MOS9, a transcriptional regulator of SNC1 that also modulates transcript levels of the SNC1 paralog, RPP4, a functional R gene (S. Xia et al., 2013). The functional RPP5 gene is found at the same locus, and multiple members of this gene family show altered expression levels in response to modified SNC1 levels (Yi and Richards, 2007). Small RNAs play a role in transcript regulation at the SNC1/RPP4/RPP5 cluster (Yi and Richards, 2007), as does epigenetic control (S. Xia et al., 2013), but there is no clear connection to miRNAs or secondary siRNAs that we describe above. Thus, work on SNC1 demonstrates the importance of fine-tuning R gene transcript levels to avoid levels which are either too high or too low (S. Xia et al., 2013). This is consistent with our hypothesis that miRNAs and phasiRNAs may function to modulate NB-LRR transcript levels, although it still begs the questions of how the grasses perform these functions in the absence of pNLs and how Arabidopsis gets by with just a handful of pNLs.
HYPOTHESIS #4: SMALL RNA REGULATION OF NB-LRRs HAS LONG-TERM EVOLUTIONARY BENEFITS
Another function proposed for the phasiRNAs produced from NB-LRRs is that they may have long-term benefits for R gene evolution. This was proposed by both the Baulcombe and Baker labs in the context of their work on Solanaceous pNLs (Li et al., 2012b; Shivaprasad et al., 2012). Plants are under pressure from pathogens to maintain diverse recognition capabilities as defenses against pathogens deploying an array of effectors. Yet, even a single poorly functioning R gene can have high fitness costs, most well described for RPM1 (Tian et al., 2003). The Baulcombe and Baker labs speculate that these fitness costs could be mitigated by phasiRNA-mediated transcriptional suppression of NB-LRRs, facilitating diversification within the family that could lead to R gene amplification, diversification, and neofunctionalization, thereby allowing new resistance specificities to emerge (Li et al., 2012b; Shivaprasad et al., 2012). This hypothesis, also suggested for the PPR gene family (Howell et al., 2007), would indicate that plants balance the benefits and costs of the multiplication and diversification of R genes via posttranscriptional controls. This is similar to the buffering hypothesis above, in that there is a bulk control of NB-LRRs to minimize their presence, but with the benefits manifest in an evolutionary time frame. One unclear aspect of this hypothesis is if or how beneficial NB-LRRs might be released from this transcriptional suppression; the redundancy of the miRNAs that target conserved sequences and the trans-acting activity of the phasiRNAs apparently creates a suppressive network that would extend to nearly all members of the gene family, whether they’re beneficial or deleterious. Another unclear aspect of this hypothesis is that the number of NB-LRRs in a genome seems unrelated to the existence of the pNL controls. For example, the rice genome has 500+ NB-LRRs (Zhou et al., 2004), yet like the other grasses, it is missing the activity of miRNAs that trigger pNL formation. In summary, we believe that evolutionary benefits of pNLs have yet to be clearly defined, and while they might exist, could result from a simple buffering of NB-LRR levels to reduce the fitness cost of these genes (hypothesis #3).
SUMMARY
The deeper we examine plant small RNAs, the more extensive we find are their regulatory roles; this is particularly apparent in species beyond Arabidopsis, many of which demonstrate complex regulatory circuits that are missing or only barely extant in Arabidopsis. Phased, secondary siRNAs are among the most interesting of these circuits, as they’re involved in regulation of numerous protein-coding genes and gene families or of a variety of long noncoding RNAs. Some of the largest protein-encoding gene families in plants are regulated by phasiRNAs, including the NB-LRR, MYB, and PPR families. Phased siRNAs produced from NB-LRRs are particularly interesting to us because of (1) the extensive level of redundancy, (2) the presence/absence variation that observed for this regulatory network among plant families, and (3) elucidation of their function may provide insights into the importance of phasiRNAs in regulation of MYB, PPR, and other protein-coding gene families. We described at least four hypotheses for the functional relevance of phasiRNA regulation of NB-LRRs, but it is entirely possible that their role is something that we haven’t yet considered. Alternatively, it’s possible that more than one of these hypotheses is correct or even that pNLs may have different functions in different lineages, like a role in symbiotic interactions in legumes, but defenses in Solanaceous species.
One question that has been asked about pNLs is whether they may play a role to suppress autoimmune responses resulting from outcrossing. Hybrid necrosis is a classically defined phenomenon in which incompatible allelic interactions in a cross results in hybrid sterility or lethality. In Arabidopsis, studies of such genetic incompatibilities across different strains identified autoimmune responses for which the underlying causative locus was an allele of an NB-LRR (Bomblies et al., 2007). These autoimmune responses may function as a barrier to gene flow. Could pNLs function in this interaction between genomes? The example explored by Bomblies et al. (2007) and in their review of classic examples of hybrid necrosis (Bomblies and Weigel, 2007) suggests that the phenomenon results from autoactivated R proteins, with scant evidence of a role for transcript levels in hybrid necrosis. miRNAs and phasiRNAs are dominant suppressors and thus would suppress the deleterious phenotype typically associated with NB-LRR overexpression. In fact, Bomblies et al. (2007) used artificial miRNAs against an autoactive NB-LRR to suppress the incompatible phenotype; we would expect a similar beneficial role from an endogenous pNL network in prevention of NB-LRR overexpression. Thus, it seems unlikely that pNL regulation plays a role in promoting hybrid necrosis, although theoretically it could suppress hybrid necrosis.
Given the importance of highly diversified NB-LRRs for plant defenses and the extensive, redundant network of small RNAs found in some species for their regulation, the small RNAs may exist for the purpose of enhancing aspects of interactions between plants and microbes. Yet after consideration of a variety of possibilities for the function of these small RNAs, we are left with a number of questions: (1) Why posttranscriptionally suppress NB-LRRs at all? Why not use more conventional transcriptional controls, such as transcription factors and promoters? (2) Have legumes enhanced the pNL network because the miRNAs do something better or different in legumes? (3) Why do Arabidopsis and the grasses have few or no pNLs? And, in an evolutionary context, how do lineages transition from extensive utilization of pNLs to rare or absent pNLs? Future studies will need to address these questions by examination, at a cellular level, of the regulation of small RNAs in the presence of both beneficial and pathogenic microbes.
The most appealing hypothesis to us for the function of miRNAs and secondary siRNAs targeting NB-LRRs is the idea of a buffer or a damper that maintains transcript levels at a basal level, only high enough to be minimally functional. Maybe the promoters and transcription factors that drive expression of these genes confer only a very crude or on/off level of specificity for cell or tissue types and thus are poor regulators of transcript abundance. Like their miRNA triggers, secondary siRNAs serving as a damper could tune transcript levels, even reducing them below a minimum level possible via transcription factors. Such an activity could also be important for phasiRNAs found at other gene families, such as the PPRs or MYB transcription factors. The fact that these gene families share large sizes and redundancy in the miRNA/phasiRNA suppression suggests that this regulatory mechanism also may coordinately balance transcript levels for many related genes within the family to ensure similar activity of all members of the gene family. Yet, many protein-coding PHAS loci are single or low copy, and their PHAS characteristic is conserved across lineages, indicating that phasiRNA-based gene regulation is selectively advantageous and may have an important cis function. Clearly, there are many important experiments yet to be performed on the miRNA triggers of these phased siRNAs, and the gene families that they target, to test and assess the functional roles of phasiRNA-based gene regulation.
In a 2006 article describing the role of tasiRNAs in plant development, Poethig and colleagues postulated that “Given that ta-siRNAs are ancient and effective mechanisms for simultaneously regulating a large number of related and unrelated genes, one might imagine that this mechanism would have been strongly selected in the evolution of regulatory pathways” (Poethig et al., 2006). While at that time, there was scant support for this conclusion, more recent evidence suggests that Arabidopsis may be unusual in its narrow use of tasiRNAs for gene regulation, while other plants have exploited this novel regulatory pathway to a much greater degree. And given that only a relatively small number of plant genomes have been characterized, we are likely to discover other pathways and gene families in which secondary siRNAs play regulatory roles.
Acknowledgments
Q.F. was supported by a graduate fellowship from the China Scholarship Council. Research on plant phasiRNAs in the Meyers lab is supported in part by a grant from the United Soybean Board. We thank members of the Meyers lab for many helpful discussions, particularly Jixian Zhai, Pingchuan Li, and Siwaret Arikit.
AUTHOR CONTRIBUTIONS
All authors contributed to the writing of this article.
References
- Adenot X., Elmayan T., Lauressergues D., Boutet S., Bouché N., Gasciolli V., Vaucheret H. (2006). DRB4-dependent TAS3 trans-acting siRNAs control leaf morphology through AGO7. Curr. Biol. 16: 927–932 [DOI] [PubMed] [Google Scholar]
- Allen E., Xie Z., Gustafson A.M., Carrington J.C. (2005). MicroRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 121: 207–221 [DOI] [PubMed] [Google Scholar]
- Arif M.A., Fattash I., Ma Z., Cho S.H., Beike A.K., Reski R., Axtell M.J., Frank W. (2012). DICER-LIKE3 activity in Physcomitrella patens DICER-LIKE4 mutants causes severe developmental dysfunction and sterility. Mol. Plant 5: 1281–1294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aukerman M.J., Sakai H. (2003). Regulation of flowering time and floral organ identity by a microRNA and its APETALA2-like target genes. Plant Cell 15: 2730–2741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axtell M.J. (2013). Classification and comparison of small RNAs from plants. Annu. Rev. Plant Biol. 64: 137–159 [DOI] [PubMed] [Google Scholar]
- Axtell M.J., Jan C., Rajagopalan R., Bartel D.P. (2006). A two-hit trigger for siRNA biogenesis in plants. Cell 127: 565–577 [DOI] [PubMed] [Google Scholar]
- Axtell M.J., Staskawicz B.J. (2003). Initiation of RPS2-specified disease resistance in Arabidopsis is coupled to the AvrRpt2-directed elimination of RIN4. Cell 112: 369–377 [DOI] [PubMed] [Google Scholar]
- Bomblies K., Lempe J., Epple P., Warthmann N., Lanz C., Dangl J.L., Weigel D. (2007). Autoimmune response as a mechanism for a Dobzhansky-Muller-type incompatibility syndrome in plants. PLoS Biol. 5: e236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bomblies K., Weigel D. (2007). Hybrid necrosis: Autoimmunity as a potential gene-flow barrier in plant species. Nat. Rev. Genet. 8: 382–393 [DOI] [PubMed] [Google Scholar]
- Borsani O., Zhu J., Verslues P.E., Sunkar R., Zhu J.K. (2005). Endogenous siRNAs derived from a pair of natural cis-antisense transcripts regulate salt tolerance in Arabidopsis. Cell 123: 1279–1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H.M., Chen L.T., Patel K., Li Y.H., Baulcombe D.C., Wu S.H. (2010). 22-Nucleotide RNAs trigger secondary siRNA biogenesis in plants. Proc. Natl. Acad. Sci. USA 107: 15269–15274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H.M., Li Y.H., Wu S.H. (2007). Bioinformatic prediction and experimental validation of a microRNA-directed tandem trans-acting siRNA cascade in Arabidopsis. Proc. Natl. Acad. Sci. USA 104: 3318–3323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitwood D.H., Nogueira F.T., Howell M.D., Montgomery T.A., Carrington J.C., Timmermans M.C. (2009). Pattern formation via small RNA mobility. Genes Dev. 23: 549–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitwood D.H., Timmermans M.C. (2010). Small RNAs are on the move. Nature 467: 415–419 [DOI] [PubMed] [Google Scholar]
- Cho S.H., Coruh C., Axtell M.J. (2012). miR156 and miR390 regulate tasiRNA accumulation and developmental timing in Physcomitrella patens. Plant Cell 24: 4837–4849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuperus J.T., Carbonell A., Fahlgren N., Garcia-Ruiz H., Burke R.T., Takeda A., Sullivan C.M., Gilbert S.D., Montgomery T.A., Carrington J.C. (2010). Unique functionality of 22-nt miRNAs in triggering RDR6-dependent siRNA biogenesis from target transcripts in Arabidopsis. Nat. Struct. Mol. Biol. 17: 997–1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dangl J.L., Jones J.D.G. (2001). Plant pathogens and integrated defence responses to infection. Nature 411: 826–833 [DOI] [PubMed] [Google Scholar]
- Deakin W.J., Broughton W.J. (2009). Symbiotic use of pathogenic strategies: Rhizobial protein secretion systems. Nat. Rev. Microbiol. 7: 312–320 [DOI] [PubMed] [Google Scholar]
- de la Luz Gutiérrez-Nava M., Aukerman M.J., Sakai H., Tingey S.V., Williams R.W. (2008). Artificial trans-acting siRNAs confer consistent and effective gene silencing. Plant Physiol. 147: 543–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devers E.A., Branscheid A., May P., Krajinski F. (2011). Stars and symbiosis: MicroRNA- and microRNA*-mediated transcript cleavage involved in arbuscular mycorrhizal symbiosis. Plant Physiol. 156: 1990–2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeYoung B.J., Innes R.W. (2006). Plant NBS-LRR proteins in pathogen sensing and host defense. Nat. Immunol. 7: 1243–1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding J., Lu Q., Ouyang Y., Mao H., Zhang P., Yao J., Xu C., Li X., Xiao J., Zhang Q. (2012b). A long noncoding RNA regulates photoperiod-sensitive male sterility, an essential component of hybrid rice. Proc. Natl. Acad. Sci. USA 109: 2654–2659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding J., Shen J., Mao H., Xie W., Li X., Zhang Q. (2012a). RNA-directed DNA methylation is involved in regulating photoperiod-sensitive male sterility in rice. Mol. Plant 5: 1210–1216 [DOI] [PubMed] [Google Scholar]
- Dunoyer P., Schott G., Himber C., Meyer D., Takeda A., Carrington J.C., Voinnet O. (2010). Small RNA duplexes function as mobile silencing signals between plant cells. Science 328: 912–916 [DOI] [PubMed] [Google Scholar]
- Fahlgren N., Montgomery T.A., Howell M.D., Allen E., Dvorak S.K., Alexander A.L., Carrington J.C. (2006). Regulation of AUXIN RESPONSE FACTOR3 by TAS3 ta-siRNA affects developmental timing and patterning in Arabidopsis. Curr. Biol. 16: 939–944 [DOI] [PubMed] [Google Scholar]
- Felippes F.F., Weigel D. (2009). Triggering the formation of tasiRNAs in Arabidopsis thaliana: The role of microRNA miR173. EMBO Rep. 10: 264–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feller A., Machemer K., Braun E.L., Grotewold E. (2011). Evolutionary and comparative analysis of MYB and bHLH plant transcription factors. Plant J. 66: 94–116 [DOI] [PubMed] [Google Scholar]
- Finlay R.D. (2008). Ecological aspects of mycorrhizal symbiosis: With special emphasis on the functional diversity of interactions involving the extraradical mycelium. J. Exp. Bot. 59: 1115–1126 [DOI] [PubMed] [Google Scholar]
- Fukudome A., Kanaya A., Egami M., Nakazawa Y., Hiraguri A., Moriyama H., Fukuhara T. (2011). Specific requirement of DRB4, a dsRNA-binding protein, for the in vitro dsRNA-cleaving activity of Arabidopsis Dicer-like 4. RNA 17: 750–760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasciolli V., Mallory A.C., Bartel D.P., Vaucheret H. (2005). Partially redundant functions of Arabidopsis DICER-like enzymes and a role for DCL4 in producing trans-acting siRNAs. Curr. Biol. 15: 1494–1500 [DOI] [PubMed] [Google Scholar]
- Geurts R., Fedorova E., Bisseling T. (2005). Nod factor signaling genes and their function in the early stages of Rhizobium infection. Curr. Opin. Plant Biol. 8: 346–352 [DOI] [PubMed] [Google Scholar]
- Heisel S.E., Zhang Y., Allen E., Guo L., Reynolds T.L., Yang X., Kovalic D., Roberts J.K. (2008). Characterization of unique small RNA populations from rice grain. PLoS ONE 3: e2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson I.R., Zhang X., Lu C., Johnson L., Meyers B.C., Green P.J., Jacobsen S.E. (2006). Dissecting Arabidopsis thaliana DICER function in small RNA processing, gene silencing and DNA methylation patterning. Nat. Genet. 38: 721–725 [DOI] [PubMed] [Google Scholar]
- Howell M.D., Fahlgren N., Chapman E.J., Cumbie J.S., Sullivan C.M., Givan S.A., Kasschau K.D., Carrington J.C. (2007). Genome-wide analysis of the RNA-DEPENDENT RNA POLYMERASE6/DICER-LIKE4 pathway in Arabidopsis reveals dependency on miRNA- and tasiRNA-directed targeting. Plant Cell 19: 926–942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh L.C., Lin S.I., Shih A.C., Chen J.W., Lin W.Y., Tseng C.Y., Li W.H., Chiou T.J. (2009). Uncovering small RNA-mediated responses to phosphate deficiency in Arabidopsis by deep sequencing. Plant Physiol. 151: 2120–2132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter C., Willmann M.R., Wu G., Yoshikawa M., de la Luz Gutiérrez-Nava M., Poethig S.R. (2006). Trans-acting siRNA-mediated repression of ETTIN and ARF4 regulates heteroblasty in Arabidopsis. Development 133: 2973–2981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Brachypodium Initiative (2010). Genome sequencing and analysis of the model grass Brachypodium distachyon. Nature 463: 763–768 [DOI] [PubMed] [Google Scholar]
- Jin H., Martin C. (1999). Multifunctionality and diversity within the plant MYB-gene family. Plant Mol. Biol. 41: 577–585 [DOI] [PubMed] [Google Scholar]
- Johnson C., Kasprzewska A., Tennessen K., Fernandes J., Nan G.L., Walbot V., Sundaresan V., Vance V., Bowman L.H. (2009). Clusters and superclusters of phased small RNAs in the developing inflorescence of rice. Genome Res. 19: 1429–1440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones J.D., Dangl J.L. (2006). The plant immune system. Nature 444: 323–329 [DOI] [PubMed] [Google Scholar]
- Källman T., Chen J., Gyllenstrand N., Lagercrantz U. (2013). A significant fraction of 21-nucleotide small RNA originates from phased degradation of resistance genes in several perennial species. Plant Physiol. 162: 741–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Lim C.J., Lee B.W., Choi J.P., Oh S.K., Ahmad R., Kwon S.Y., Ahn J., Hur C.G. (2012). A genome-wide comparison of NB-LRR type of resistance gene analogs (RGA) in the plant kingdom. Mol. Cells 33: 385–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasnikova M.S., Milyutina I.A., Bobrova V.K., Ozerova L.V., Troitsky A.V., Solovyev A.G., Morozov S.Y. (2009). Novel miR390-dependent transacting siRNA precursors in plants revealed by a PCR-based experimental approach and database analysis. J. Biomed. Biotechnol. 2009: 952304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauressergues D., Delaux P.M., Formey D., Lelandais-Brière C., Fort S., Cottaz S., Bécard G., Niebel A., Roux C., Combier J.P. (2012). The microRNA miR171h modulates arbuscular mycorrhizal colonization of Medicago truncatula by targeting NSP2. Plant J. 72: 512–522 [DOI] [PubMed] [Google Scholar]
- Law J.A., Jacobsen S.E. (2010). Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat. Rev. Genet. 11: 204–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T.F., Gurazada S.G., Zhai J., Li S., Simon S.A., Matzke M.A., Chen X., Meyers B.C. (2012). RNA polymerase V-dependent small RNAs in Arabidopsis originate from small, intergenic loci including most SINE repeats. Epigenetics 7: 781–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., Ding S.W. (2006). Virus counterdefense: Diverse strategies for evading the RNA-silencing immunity. Annu. Rev. Microbiol. 60: 503–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., Orban R., Baker B. (2012a). SoMART: A web server for plant miRNA, tasiRNA and target gene analysis. Plant J. 70: 891–901 [DOI] [PubMed] [Google Scholar]
- Li F., Pignatta D., Bendix C., Brunkard J.O., Cohn M.M., Tung J., Sun H., Kumar P., Baker B. (2012b). MicroRNA regulation of plant innate immune receptors. Proc. Natl. Acad. Sci. USA 109: 1790–1795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Deng Y., Wu T., Subramanian S., Yu O. (2010). Misexpression of miR482, miR1512, and miR1515 increases soybean nodulation. Plant Physiol. 153: 1759–1770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin-Wang K., Bolitho K., Grafton K., Kortstee A., Karunairetnam S., McGhie T.K., Espley R.V., Hellens R.P., Allan A.C. (2010). An R2R3 MYB transcription factor associated with regulation of the anthocyanin biosynthetic pathway in Rosaceae. BMC Plant Biol. 10: 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozano R., Ponce O., Ramirez M., Mostajo N., Orjeda G. (2012). Genome-wide identification and mapping of NBS-encoding resistance genes in Solanum tuberosum group phureja. PLoS ONE 7: e34775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lurin C., et al. (2004). Genome-wide analysis of Arabidopsis pentatricopeptide repeat proteins reveals their essential role in organelle biogenesis. Plant Cell 16: 2089–2103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackey D., Belkhadir Y., Alonso J.M., Ecker J.R., Dangl J.L. (2003). Arabidopsis RIN4 is a target of the type III virulence effector AvrRpt2 and modulates RPS2-mediated resistance. Cell 112: 379–389 [DOI] [PubMed] [Google Scholar]
- Madsen L.H., Tirichine L., Jurkiewicz A., Sullivan J.T., Heckmann A.B., Bek A.S., Ronson C.W., James E.K., Stougaard J. (2010). The molecular network governing nodule organogenesis and infection in the model legume Lotus japonicus. Nat. Commun. 1: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manavella P.A., Koenig D., Weigel D. (2012). Plant secondary siRNA production determined by microRNA-duplex structure. Proc. Natl. Acad. Sci. USA 109: 2461–2466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margis R., Fusaro A.F., Smith N.A., Curtin S.J., Watson J.M., Finnegan E.J., Waterhouse P.M. (2006). The evolution and diversification of Dicers in plants. FEBS Lett. 580: 2442–2450 [DOI] [PubMed] [Google Scholar]
- Marin E., Jouannet V., Herz A., Lokerse A.S., Weijers D., Vaucheret H., Nussaume L., Crespi M.D., Maizel A. (2010). miR390, Arabidopsis TAS3 tasiRNAs, and their AUXIN RESPONSE FACTOR targets define an autoregulatory network quantitatively regulating lateral root growth. Plant Cell 22: 1104–1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzke M., Kanno T., Daxinger L., Huettel B., Matzke A.J. (2009). RNA-mediated chromatin-based silencing in plants. Curr. Opin. Cell Biol. 21: 367–376 [DOI] [PubMed] [Google Scholar]
- McHale L., Tan X., Koehl P., Michelmore R.W. (2006). Plant NBS-LRR proteins: Adaptable guards. Genome Biol. 7: 212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medzhitov R., Janeway C.A., Jr (1997). Innate immunity: The virtues of a nonclonal system of recognition. Cell 91: 295–298 [DOI] [PubMed] [Google Scholar]
- Meyers B.C., et al. (2008). Criteria for annotation of plant microRNAs. Plant Cell 20: 3186–3190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers B.C., Dickerman A.W., Michelmore R.W., Sivaramakrishnan S., Sobral B.W., Young N.D. (1999). Plant disease resistance genes encode members of an ancient and diverse protein family within the nucleotide-binding superfamily. Plant J. 20: 317–332 [DOI] [PubMed] [Google Scholar]
- Meyers B.C., Kozik A., Griego A., Kuang H., Michelmore R.W. (2003). Genome-wide analysis of NBS-LRR-encoding genes in Arabidopsis. Plant Cell 15: 809–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ming R., et al. (2008). The draft genome of the transgenic tropical fruit tree papaya (Carica papaya Linnaeus). Nature 452: 991–996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molnar A., Melnyk C.W., Bassett A., Hardcastle T.J., Dunn R., Baulcombe D.C. (2010). Small silencing RNAs in plants are mobile and direct epigenetic modification in recipient cells. Science 328: 872–875 [DOI] [PubMed] [Google Scholar]
- Monaghan J., Zipfel C. (2012). Plant pattern recognition receptor complexes at the plasma membrane. Curr. Opin. Plant Biol. 15: 349–357 [DOI] [PubMed] [Google Scholar]
- Montgomery T.A., Howell M.D., Cuperus J.T., Li D., Hansen J.E., Alexander A.L., Chapman E.J., Fahlgren N., Allen E., Carrington J.C. (2008a). Specificity of ARGONAUTE7-miR390 interaction and dual functionality in TAS3 trans-acting siRNA formation. Cell 133: 128–141 [DOI] [PubMed] [Google Scholar]
- Montgomery T.A., Yoo S.J., Fahlgren N., Gilbert S.D., Howell M.D., Sullivan C.M., Alexander A., Nguyen G., Allen E., Ahn J.H., Carrington J.C. (2008b). AGO1-miR173 complex initiates phased siRNA formation in plants. Proc. Natl. Acad. Sci. USA 105: 20055–20062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro L., Jay F., Nomura K., He S.Y., Voinnet O. (2008). Suppression of the microRNA pathway by bacterial effector proteins. Science 321: 964–967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldroyd G.E., Downie J.A. (2008). Coordinating nodule morphogenesis with rhizobial infection in legumes. Annu. Rev. Plant Biol. 59: 519–546 [DOI] [PubMed] [Google Scholar]
- O’Toole N., Hattori M., Andres C., Iida K., Lurin C., Schmitz-Linneweber C., Sugita M., Small I. (2008). On the expansion of the pentatricopeptide repeat gene family in plants. Mol. Biol. Evol. 25: 1120–1128 [DOI] [PubMed] [Google Scholar]
- Pan Q., Wendel J., Fluhr R. (2000). Divergent evolution of plant NBS-LRR resistance gene homologues in dicot and cereal genomes. J. Mol. Evol. 50: 203–213 [DOI] [PubMed] [Google Scholar]
- Peragine A., Yoshikawa M., Wu G., Albrecht H.L., Poethig R.S. (2004). SGS3 and SGS2/SDE1/RDR6 are required for juvenile development and the production of trans-acting siRNAs in Arabidopsis. Genes Dev. 18: 2368–2379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petroni K., Tonelli C. (2011). Recent advances on the regulation of anthocyanin synthesis in reproductive organs. Plant Sci. 181: 219–229 [DOI] [PubMed] [Google Scholar]
- Poethig R.S., Peragine A., Yoshikawa M., Hunter C., Willmann M., Wu G. (2006). The function of RNAi in plant development. Cold Spring Harb. Symp. Quant. Biol. 71: 165–170 [DOI] [PubMed] [Google Scholar]
- Qiao Y., et al. (2013). Oomycete pathogens encode RNA silencing suppressors. Nat. Genet. 45: 330–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopalan R., Vaucheret H., Trejo J., Bartel D.P. (2006). A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes Dev. 20: 3407–3425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivaprasad P.V., Chen H.M., Patel K., Bond D.M., Santos B.A., Baulcombe D.C. (2012). A microRNA superfamily regulates nucleotide binding site-leucine-rich repeats and other mRNAs. Plant Cell 24: 859–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Si-Ammour A., Windels D., Arn-Bouldoires E., Kutter C., Ailhas J., Meins F., Vazquez F. (2011). miR393 and secondary siRNAs regulate expression of the TIR1/AFB2 auxin receptor clade and auxin-related development of Arabidopsis leaves. Plant Physiol. 157: 683–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X., et al. (2012). Roles of DCL4 and DCL3b in rice phased small RNA biogenesis. Plant J. 69: 462–474 [DOI] [PubMed] [Google Scholar]
- Takos A.M., Jaffé F.W., Jacob S.R., Bogs J., Robinson S.P., Walker A.R. (2006). Light-induced expression of a MYB gene regulates anthocyanin biosynthesis in red apples. Plant Physiol. 142: 1216–1232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talmor-Neiman M., Stav R., Klipcan L., Buxdorf K., Baulcombe D.C., Arazi T. (2006). Identification of trans-acting siRNAs in moss and an RNA-dependent RNA polymerase required for their biogenesis. Plant J. 48: 511–521 [DOI] [PubMed] [Google Scholar]
- Tian D., Traw M.B., Chen J.Q., Kreitman M., Bergelson J. (2003). Fitness costs of R-gene-mediated resistance in Arabidopsis thaliana. Nature 423: 74–77 [DOI] [PubMed] [Google Scholar]
- Valencia-Sanchez M.A., Liu J., Hannon G.J., Parker R. (2006). Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev. 20: 515–524 [DOI] [PubMed] [Google Scholar]
- Van der Biezen E.A., Jones J.D. (1998). Plant disease-resistance proteins and the gene-for-gene concept. Trends Biochem. Sci. 23: 454–456 [DOI] [PubMed] [Google Scholar]
- Vazquez F., Vaucheret H., Rajagopalan R., Lepers C., Gasciolli V., Mallory A.C., Hilbert J.L., Bartel D.P., Crété P. (2004). Endogenous trans-acting siRNAs regulate the accumulation of Arabidopsis mRNAs. Mol. Cell 16: 69–79 [DOI] [PubMed] [Google Scholar]
- Voinnet O. (2009). Origin, biogenesis, and activity of plant microRNAs. Cell 136: 669–687 [DOI] [PubMed] [Google Scholar]
- Wang Y., Itaya A., Zhong X., Wu Y., Zhang J., van der Knaap E., Olmstead R., Qi Y., Ding B. (2011). Function and evolution of a MicroRNA that regulates a Ca2+-ATPase and triggers the formation of phased small interfering RNAs in tomato reproductive growth. Plant Cell 23: 3185–3203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitham S., Dinesh-Kumar S.P., Choi D., Hehl R., Corr C., Baker B. (1994). The product of the tobacco mosaic virus resistance gene N: Similarity to toll and the interleukin-1 receptor. Cell 78: 1101–1115 [DOI] [PubMed] [Google Scholar]
- Williams L., Carles C.C., Osmont K.S., Fletcher J.C. (2005). A database analysis method identifies an endogenous trans-acting short-interfering RNA that targets the Arabidopsis ARF2, ARF3, and ARF4 genes. Proc. Natl. Acad. Sci. USA 102: 9703–9708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willmann M.R., Endres M.W., Cook R.T., Gregory B.D. (2011). The functions of RNA-dependent RNA Polymerases in Arabidopsis. The Arabidopsis Book 9: e0146, doi/10.199/tab.0146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L., Mao L., Qi Y. (2012). Roles of dicer-like and argonaute proteins in TAS-derived small interfering RNA-triggered DNA methylation. Plant Physiol. 160: 990–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L., Zhou H., Zhang Q., Zhang J., Ni F., Liu C., Qi Y. (2010). DNA methylation mediated by a microRNA pathway. Mol. Cell 38: 465–475 [DOI] [PubMed] [Google Scholar]
- Xia, R., Meyers, B.C., Liu, Z., Beers, E.P., and Ye, S. (2013). MicroRNA superfamilies descended from miR390 and their roles in secondary small interfering RNA biogenesis in eudicots. Plant Cell 25: 1555–1572. [DOI] [PMC free article] [PubMed]
- Xia R., Zhu H., An Y.Q., Beers E.P., Liu Z. (2012). Apple miRNAs and tasiRNAs with novel regulatory networks. Genome Biol. 13: R47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia S., Cheng Y.T., Huang S., Win J., Soards A., Jinn T.L., Jones J., Kamoun S., Chen S., Zhang Y., Li X. (2013). Regulation of transcription of NB-LRR-encoding genes SNC1 and RPP4 via H3K4 tri-methylation. Plant Physiol. 162: 1694–1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z., Allen E., Wilken A., Carrington J.C. (2005). DICER-LIKE 4 functions in trans-acting small interfering RNA biogenesis and vegetative phase change in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 102: 12984–12989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., et al. Potato Genome Sequencing Consortium (2011). Genome sequence and analysis of the tuber crop potato. Nature 475: 189–195 [DOI] [PubMed] [Google Scholar]
- Xue J.Y., Wang Y., Wu P., Wang Q., Yang L.T., Pan X.H., Wang B., Chen J.Q. (2012). A primary survey on bryophyte species reveals two novel classes of nucleotide-binding site (NBS) genes. PLoS ONE 7: e36700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., Li D., Li Y., Gu X., Huang S., Garcia-Mas J., Weng Y. (2013). A 1,681-locus consensus genetic map of cultivated cucumber including 67 NB-LRR resistance gene homolog and ten gene loci. BMC Plant Biol. 13: 53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S., Tang F., Gao M., Krishnan H.B., Zhu H. (2010). R gene-controlled host specificity in the legume-rhizobia symbiosis. Proc. Natl. Acad. Sci. USA 107: 18735–18740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi H., Richards E.J. (2007). A cluster of disease resistance genes in Arabidopsis is coordinately regulated by transcriptional activation and RNA silencing. Plant Cell 19: 2929–2939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa M., Iki T., Tsutsui Y., Miyashita K., Poethig R.S., Habu Y., Ishikawa M. (2013). 3′ Fragment of miR173-programmed RISC-cleaved RNA is protected from degradation in a complex with RISC and SGS3. Proc. Natl. Acad. Sci. USA 110: 4117–4122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa M., Peragine A., Park M.Y., Poethig R.S. (2005). A pathway for the biogenesis of trans-acting siRNAs in Arabidopsis. Genes Dev. 19: 2164–2175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young N.D., et al. (2011). The Medicago genome provides insight into the evolution of rhizobial symbioses. Nature 480: 520–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai J., et al. (2011). MicroRNAs as master regulators of the plant NB-LRR defense gene family via the production of phased, trans-acting siRNAs. Genes Dev. 25: 2540–2553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai J., et al. (2013). Plant microRNAs display differential 3′-trancation and tailing, modifications which are ARGONAUTE1-dependent and conserved across species. Plant Cell, doi/10.1105/tpc.113.114603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C., Li G., Wang J., Fang J. (2012b). Identification of trans-acting siRNAs and their regulatory cascades in grapevine. Bioinformatics 28: 2561–2568 [DOI] [PubMed] [Google Scholar]
- Zhang C., Ng D.W., Lu J., Chen Z.J. (2012a). Roles of target site location and sequence complementarity in trans-acting siRNA formation in Arabidopsis. Plant J. 69: 217–226 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Goritschnig S., Dong X., Li X. (2003). A gain-of-function mutation in a plant disease resistance gene leads to constitutive activation of downstream signal transduction pathways in suppressor of npr1-1, constitutive 1. Plant Cell 15: 2636–2646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Li X. (2005). A putative nucleoporin 96 Is required for both basal defense and constitutive resistance responses mediated by suppressor of npr1-1,constitutive 1. Plant Cell 17: 1306–1316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H., et al. (2012). Photoperiod- and thermo-sensitive genic male sterility in rice are caused by a point mutation in a novel noncoding RNA that produces a small RNA. Cell Res. 22: 649–660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T., Wang Y., Chen J.Q., Araki H., Jing Z., Jiang K., Shen J., Tian D. (2004). Genome-wide identification of NBS genes in japonica rice reveals significant expansion of divergent non-TIR NBS-LRR genes. Mol. Genet. Genomics 271: 402–415 [DOI] [PubMed] [Google Scholar]
- Zhu H., Xia R., Zhao B., An Y.Q., Dardick C.D., Callahan A.M., Liu Z. (2012). Unique expression, processing regulation, and regulatory network of peach (Prunus persica) miRNAs. BMC Plant Biol. 12: 149. [DOI] [PMC free article] [PubMed] [Google Scholar]