This work describes a novel platform for recombinant protein production that is essentially transient expression from a stably transformed plant. The system provides both activation and amplification of transgene expression in planta and in this study was exemplified using both a therapeutic and industrial protein. The platform is not host limited and can be adapted to large-scale production.
Abstract
In this study, we describe a novel protein production platform that provides both activation and amplification of transgene expression in planta. The In Plant Activation (INPACT) system is based on the replication machinery of tobacco yellow dwarf mastrevirus (TYDV) and is essentially transient gene expression from a stably transformed plant, thus combining the advantages of both means of expression. The INPACT cassette is uniquely arranged such that the gene of interest is split and only reconstituted in the presence of the TYDV-encoded Rep/RepA proteins. Rep/RepA expression is placed under the control of the AlcA:AlcR gene switch, which is responsive to trace levels of ethanol. Transgenic tobacco (Nicotiana tabacum cv Samsun) plants containing an INPACT cassette encoding the β-glucuronidase (GUS) reporter had negligible background expression but accumulated very high GUS levels (up to 10% total soluble protein) throughout the plant, within 3 d of a 1% ethanol application. The GUS reporter was replaced with a gene encoding a lethal ribonuclease, barnase, demonstrating that the INPACT system provides exquisite control of transgene expression and can be adapted to potentially toxic or inhibitory compounds. The INPACT gene expression platform is scalable, not host-limited, and has been used to express both a therapeutic and an industrial protein.
INTRODUCTION
Plants are a cheap source of biomass and as such are an attractive alternative to conventional expression hosts for the manufacture of biologics with clinical, industrial, and research applications (Faye and Gomord, 2010; Fischer et al., 2012). However, the economic viability of plant-based protein production is strongly yield dependent, and considerable research has been dedicated to developing novel approaches to increasing transgene expression. Plant transformation can either be transient, for example, agroinfiltration and microprojectile bombardment, or stable in the case of transgenics. Although both systems may differ greatly with respect to speed, cost, scalability, and host range, both methods commonly use plant virus–derived genetic elements to regulate transcription, enhance translation, amplify transgene copy number, or suppress gene silencing in order to increase recombinant protein accumulation (Mushegian and Shepherd, 1995; Gleba et al., 2005).
Current transient expression systems predominantly use vacuum infiltration to physically deliver recombinant Agrobacterium tumefaciens, harboring the plant expression cassette, systemically into the plant. The expression cassette is often based on the partially deconstructed genome of an RNA virus, commonly Tobacco mosaic virus (TMV) or Potato virus X, and the transferred DNA contains a proviral replicon capable of expressing and accumulating very high recombinant protein levels, of up to 5 g/kg fresh weight biomass or >50% total soluble protein (TSP), within a short period of time (4 to 10 d) (Chapman et al., 1992; Marillonnet et al., 2004). One variation of this, the Cowpea mosaic virus hypertranslatable vector, was completely deconstructed to simply include translation enhancer sequences from Cowpea mosaic virus RNA-2 in the transgene mRNA and to coexpress the Tomato bushy stunt virus P19 suppressor of posttranscriptional gene silencing. This nonreplicating vector alone generated very high recombinant protein yields of up to 1.5 g/kg, corresponding to ∼25 to 30% TSP (Sainsbury and Lomonossoff, 2008). While transient A. tumefaciens–mediated expression using RNA virus-based vectors is rapid and provides high recombinant protein yields, its scale-up can be costly due to the expensive equipment required to deliver the bacteria and the containment facilities required to house the plants (Pogue et al., 2010). Furthermore, these RNA virus vectors appear to be essentially limited to the Solanaceae with greatest emphasis on production in the low biomass host Nicotiana benthamiana (Gleba et al., 2007).
Stably transformed plants provide a sustainable platform for protein production with low production and scale-up costs and, in many cases, established postharvest handling and crop processing practices. From a regulatory perspective, stably transformed plants may be preferred to transient systems as the downstream product can be linked to a fully defined single transgene integration event in a fully characterized transgenic line. Furthermore, stable transgenic lines are a permanent genetic resource that can be harvested with minimal batch-to-batch variation, thus improving GMP compliance (Fischer et al., 2012). To maximize protein accumulation and stability or facilitate purification, transgene expression can be constitutive or limited to various sink or source tissues, such as leaf, tuber, fruit, or seed. While strong constitutive transgene expression is often desired, not all proteins can be ectopically expressed in this manner, in particular those that may be toxic or inhibitory to normal plant growth and development. Therefore, in order to separate plant growth and protein production phases, inducible expression systems are often required for temporal, spatial, and quantitative control of transgene expression.
There are few examples of inducible, transgenic protein production systems capable of low background expression in the absence of the inducer molecule and very high expression upon induction. Perhaps the only system to fulfill these criteria was recently reported by Werner et al. (2011), who adapted the transient TMV-based replicon vector system to transgenic plants by linking its expression to the ethanol-responsive AlcA:AlcR gene switch (Caddick et al., 1998). In the absence of the ethanol inducer molecule, N. benthamiana plants displayed negligible reporter gene expression; however, upon induction, the transgene was amplified and expressed from viral RNA replicons and plants accumulated high absolute levels of recombinant protein comparable to those observed transiently. While this system holds promise, host restrictions associated with such virus vectors and regulatory concerns with respect to the activation and release of viral RNA that is capable of both autonomous replication and cell-to-cell movement will likely limit the application of this technology to a narrow range of plant species and preclude it from field release, respectively.
In this article, we describe a novel, inducible, hyperexpression system for recombinant protein production in plants. The system uses the rolling circle replication machinery of the dicot-infecting mastrevirus, tobacco yellow dwarf virus (TYDV) (Morris et al., 1992), where this single-stranded DNA virus is disaggregated into the In Plant Activation (INPACT) cassette containing the gene of interest, replication recognition sequences, and the replication association cassette, which contains the viral Replication/Replication A (Rep/RepA) genes. The INPACT cassette is uniquely arranged such that the gene of interest is split and only expressed from extrachromosomal, replicating episomes (replisomes) that are released from the host chromosome in the presence of the virus-encoded replication associated proteins, Rep and RepA. Expression of the Rep/RepA activators, in turn, is under the transcriptional control of the AlcA:AlcR gene switch and is responsive to trace levels of the ethanol inducer molecule. This system effectively provides inducible, high-level transient expression from stably transformed plants and is potentially adaptable to a wide host range.
RESULTS
INPACT Vector Design
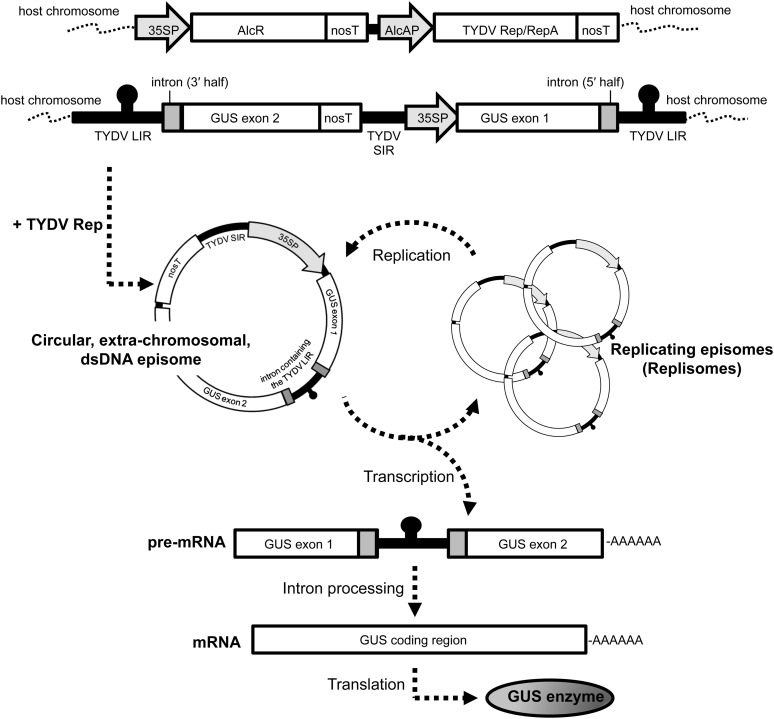
The INPACT expression cassette is uniquely arranged such that the gene of interest is split into two exons, of which the promoter and 5′ end of the split gene is positioned downstream of the 3′ end of the gene and terminator (Figure 1). The split gene cassette is flanked by the TYDV large intergenic regions (LIRs), which contain the viral genomic cis-acting elements necessary for first-strand synthesis, and these LIRs, in turn, are embedded within an intron. The cassette also contains the TYDV small intergenic region (SIR) within which reside cis-acting elements necessary for host-mediated second-strand synthesis. As an integrated sequence, the INPACT cassette cannot express a functional recombinant protein. However, in the presence of the virus-encoded activation proteins, TYDV Rep and RepA, the integrated INPACT cassette serves as a template for duplication by a process known as rolling circle replication, resulting in the production of a circular, extrachromosomal, single-stranded DNA copy of the INPACT cassette. This episome is then converted to a double-stranded DNA molecule via the SIR and host polymerases. This molecular form is both transcriptionally active and can serve as a template for further amplification and as such is termed a replisome (for replicating episome). Finally, the transgene mRNA is processed to remove the intron (and embedded LIR) and is subsequently translated into the protein of interest. Therefore, transgene amplification and expression from the INPACT cassette is strictly dependent on TYDV Rep/RepA abundance. In order to regulate both Rep and RepA proteins, the bicistronic Rep/RepA coding sequence is placed under the transcriptional control of the alcohol-inducible AlcA:AlcR gene switch. This system provides for temporal, spatial, and dose-dependent control of target gene expression.
Figure 1.
Schematic Representation of the INPACT Inducible, Hyperexpression Platform.
35SP, CaMV 35S promoter; AlcR, alcohol receptor gene; nosT, nopaline synthase terminator; AlcAP, AlcA promoter; Rep/RepA, Replicase/Replicase A bicistronic coding region.
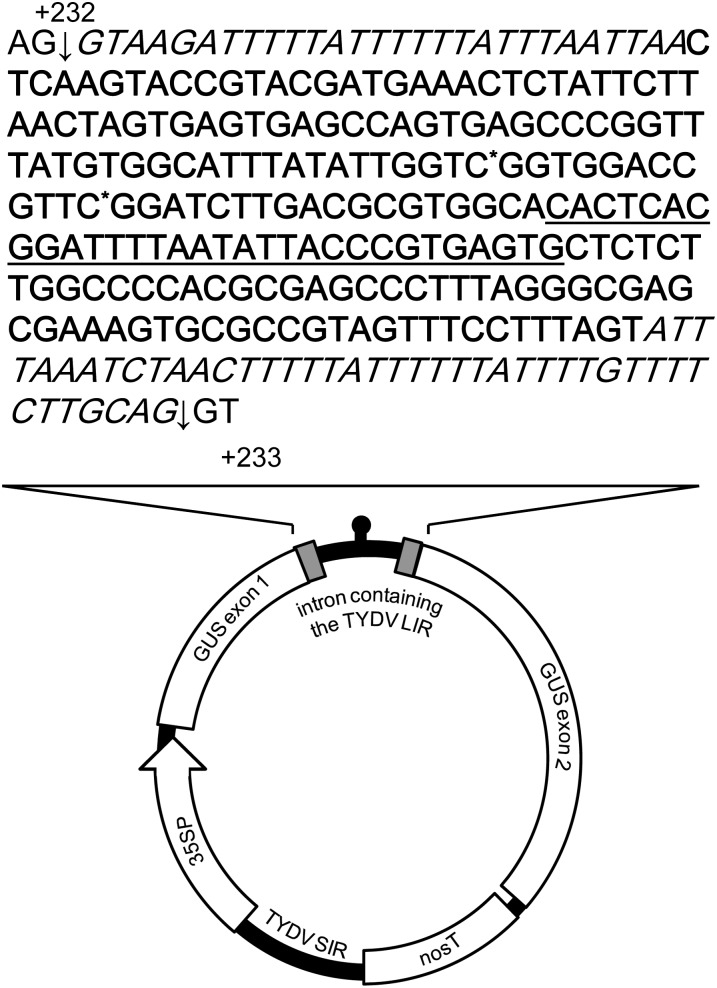
Protein expression from the circular, extrachromosomal, transcriptionally active INPACT cassette is strongly dependent on the correct processing of an intron (containing the TYDV LIR) from the transgene mRNA (Figure 2). As such, the LIR was tailored to remove nonessential sequences by deleting 56 bp from the 3′ end and corrupting two potential intron splice sites. No changes were made to the 5′ end of the LIR, which contains a CA motif, and iterated sequences thought to be essential for cognate Rep recognition/replication nor to the stem-loop sequence, which contains the origin of first strand synthesis. A small synthetic intron, termed a “syntron,” was also designed to specifically house the LIR. The syntron sequence contains consensus plant donor (AG/GTAAG) and acceptor (TGCAG/GT) splice sites (Goodall and Filipowicz, 1989), U-rich tracts repeated within proximity to the 5′ and 3′ splice sites (Baynton et al., 1996), and a branch point consensus sequence appropriately located upstream of the 3′ splice junction. For both the uidA and barnase coding regions, the syntron was introduced within 250 bp 3′ of the ATG start codon and in the case of the uidA gene it was inserted into the first available AG/GT.
Figure 2.
Schematic of the Extrachromosomal INPACT Replisome and Sequence of the Syntron Containing the Modified TYDV LIR.
The 5′ and 3′ intron splice sites are denoted with an arrow, syntron sequence is in italics, and modified TYDV LIR sequence is in bold, with site mutations followed immediately with an asterisk and stem with consensus nonanucleotide loop underlined. The syntron and LIR are located between nucleotides +232 and +233 of the uidA coding region (where the A of the AUG start codon is position +1). Drawing is not to scale.
Production and Analysis of Transgenic Tobacco Parent Lines Containing the TYDV Rep/RepA Coding Sequence under the Control of the AlcA:AlcR Gene Switch
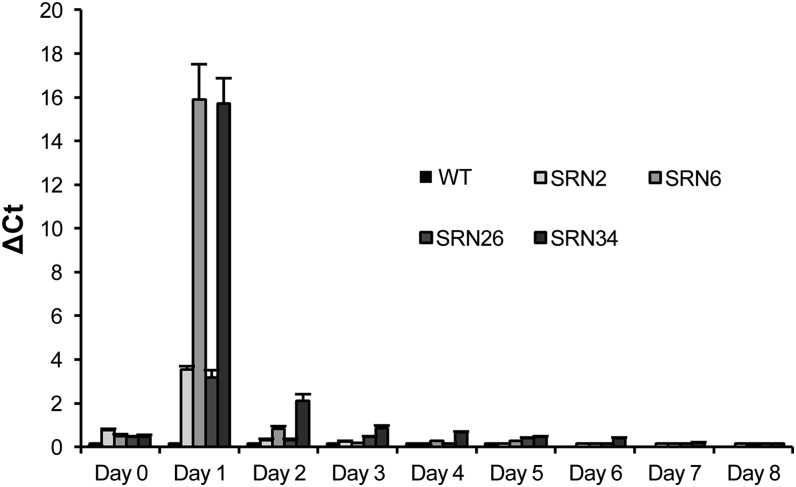
Fifteen independent transgenic tobacco plants were regenerated from leaf disks transformed with the pAlc-Rep/RepA vector, of which four (SRN2, SRN6, SRN26, and SRN34) were selected for analysis. Based on DNA gel blot hybridization profiles, SRN2 and SRN6 were most likely single copy events while SRN26 and SRN34 probably contained two copies each (see Supplemental Figure 1 online). Phenotypic effects of ethanol-induced Rep/RepA expression and the relative abundance of Rep/RepA transcript post-ethanol activation (PEA) in these lines were compared with wild-type controls. Following exposure to ethanol, no changes in gross leaf morphology were observed in wild-type plants. Within 8 d PEA, all transgenic lines showed various phenotypic changes, including degrees of leaf wilting, curling, yellowing, and gloss (Figure 3A). Over time, the severity of these symptoms increased in lines SRN6 and SRN34, ultimately resulting in plant death by day 14. SRN2 and SRN26 plants displayed relatively minor phenotypic changes, such as leaf gloss and mild chlorosis. Plant root systems were compared 8 d PEA. There were no notable differences in root mass or health between wild-type and transgenic SRN2, SRN6, or SRN26 plants, while the roots of SRN34 plants appeared necrotic with dark brown/black pigmentation (Figure 3B). Rep/RepA transcript levels were determined by quantitative real-time PCR (qRT-PCR). Tissue was collected on day 0 and every 24 h for 8 d PEA, from the top, middle, and bottom leaves of transgenic lines SRN 2, 6, 26, and 34 and a wild-type control. RNA was extracted and used as a template for qRT-PCR with primers specific for the Rep/RepA coding sequence. Delta cycle threshold values were calculated and graphed in Figure 4. No Rep/RepA transcript was detected in wild-type tissue at any time point. Low levels of Rep/RepA transcript were detected on day 0 prior to ethanol activation, indicating low leaky expression in the absence of the inducer molecule. Rep/RepA transcript levels were highest in all transgenic lines 1 d PEA, and these levels were significantly higher than all those measured in subsequent days of the time course (P < 0.05). Highest Rep/RepA expression was observed in lines SRN6 and SRN34 and lowest in lines SRN2 and SRN26. On day 3 PEA, Rep/RepA transcript levels in all transgenic lines had reverted to background levels. To investigate the effect of Rep/RepA abundance on INPACT-based expression, lines representing both low and high Rep/RepA expression levels, SRN2 and SRN34, respectively, were selected for supertransformation with an INPACT vector capable of expressing β-glucuronidase (GUS).
Figure 3.
Phenotypic Effects of Rep/RepA Expression in Transgenic Tobacco.
(A) Leaf phenotypes 8 d PEA, from left to right: the wild type, chlorosis (SRN34 parent line), and gloss (SRN2 parent line).
(B) Root system phenotypes 8 d PEA of the wild type (left) and SRN34 parent line (right).
Figure 4.
Quantification of Rep/RepA Levels by qRT-PCR Following Ethanol Activation.
Four independent soil-acclimatized tobacco lines containing the Rep/RepA gene under the control of the ethanol-inducible AlcA:AlcR switch were activated with a 1% ethanol solution by foliar spray and root drench. Tissue was collected at day 0 and every 24 h for 8 d PEA from the top, middle, and bottom leaves. RNA was extracted and used for qRT-PCR. Delta cycle threshold values are shown as the mean transcript levels from the top, middle, and bottom leaf samples from each plant. Samples were analyzed in triplicate. Actin served as a control for normalization. Error bars indicate se. WT, the wild type.
High-Level INPACT-Directed Expression of GUS in Transgenic Tobacco
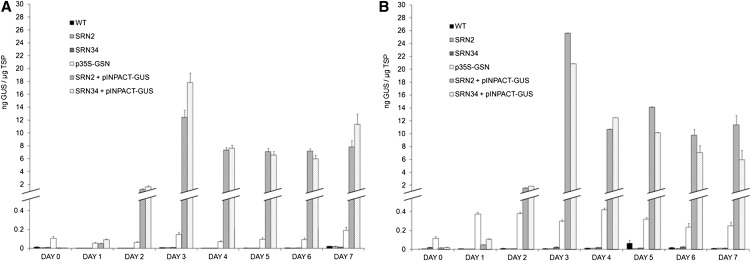
Thirty independent transgenic plants were regenerated from leaf disks of SRN2 and SRN34 parental tobacco lines, transformed with the INPACT-GUS vector. Of these, four from each parental group were selected for further analysis. First, the timing of GUS expression directed by the INPACT system following ethanol activation was investigated. Tissue was collected at day 0 and every 24 h for 7 d PEA, from the top three leaves of plants from two different treatments which involved either a single (Figure 5A) or multiple (Figure 5B) ethanol applications. Plants in the single application group were activated using a foliar spray and root drench with 1% ethanol. Plants in the multiple application group were subjected to additional foliar sprays each day following initial activation. Samples were collected concurrently from transgenic parent lines SRN2 and SRN34, SRN2 and SRN34 supertransformed with an INPACT-GUS cassette, plants transformed with p35S-GSN (containing the uidA reporter gene and syntron under the transcriptional control of the constitutive cauliflower mosaic virus [CaMV] 35S promoter), and wild-type tobacco plants. TSP was extracted from all samples and GUS levels determined by ELISA. Essentially, background levels of GUS were detected in wild-type, SRN2, and SRN34 parent lines on all days on the time course in both treatment groups. On day 0, there was no significant difference between the levels of GUS detected in TSP extracts from INPACT-GUS plants, in either treatment group, relative to their respective parental lines (P < 0.05). This suggests that the background level of Rep/RepA transcript detected in INPACT-GUS plants prior to ethanol activation, due to leaky expression of the AlcA:AlcR switch, was not sufficient to activate the INPACT system. Thus, INPACT provided tightly regulated GUS expression. On day 1 (24 h PEA), an increase in GUS expression was detected in INPACT-GUS plants in both treatment groups. On day 2, the expression of GUS increased dramatically, accumulating to levels of up to 25-fold higher than those detected in p35S-GSN plants. Expression peaked on day 3 in INPACT-GUS plants from both treatment groups. Protein accumulation of up to 25 ng GUS/µg TSP (2.5% TSP) was measured, which was ∼120-fold higher than the levels observed in p35S-GSN plants. Eight weeks later, INPACT-GUS lines were reactivated with a single 1% ethanol foliar spray and root drench. GUS expression was negligible prior to the reactivation (>0.01 ng GUS/µg TSP) but peaked in new growth 3 d PEA, to similar levels obtained previously (26 ± 3.65 ng GUS/µg TSP).
Figure 5.
Quantification of GUS Expression in Transgenic Tobacco.
Time course of GUS expression following single (A) and multiple (B) ethanol applications. Plants were activated using a 1% ethanol foliar spray and root drench. Plants within the multiple ethanol treatment group (B) were subjected to additional 1% ethanol foliar sprays each day following initial activation. Tissue was collected concurrently at day 0 and every 24 h for 7 d PEA, from the top three leaves of each plant. GUS levels were quantified by ELISA. Four independent transformation events representing (1) SRN2 + pINPACT-GUS, (2) SRN34 + pINPACT-GUS, and (3) p35S-GSN were used. Samples were collected from three biological replicates of each transgenic event and the wild type (WT) and analyzed in triplicate. Error bars indicate se.
The effect of Rep/RepA expression (parental background) and ethanol dose on INPACT-directed GUS expression was examined. GUS expression 3 d PEA in INPACT-GUS plants with an SRN2 background (low Rep/RepA expression) and INPACT-GUS plants with an SRN34 background (high Rep/RepA expression) was compared between treatment groups. In the single ethanol treatment group, INPACT-directed GUS expression was significantly higher in plants with a high Rep/RepA-expressing parental line, relative to those with the low Rep/RepA background (P < 0.05). Interestingly, in the multiple ethanol treatment group, no significant difference between the two parental lines was observed (P > 0.05). The level of GUS expression 3 d PEA in INPACT-GUS plants with the same parental background, but subjected to different ethanol treatments, was also compared. The level of GUS expression was significantly higher in INPACT-GUS plants with low Rep/RepA background (SRN2 parent) in the multiple ethanol treatment group relative to plants with the same parent but in the single ethanol treatment group (P < 0.05). No significant difference was detected between GUS levels in INPACT-GUS plants with the high Rep/RepA background (SRN34 parent) from either ethanol treatment group (P > 0.05). These combined results suggest that high Rep/RepA expression PEA provides better activation of the INPACT-GUS system and subsequently higher GUS accumulation. Hence, in the single treatment group, greater GUS expression was detected in high Rep/RepA-expressing INPACT-GUS plants, relative to low Rep/RepA INPACT-GUS plants. The use of multiple ethanol treatments to enhance Rep/RepA expression appears to overcome the constraints of lower Rep/RepA expression, as demonstrated by the increase in GUS expression from INPACT-GUS plants with low Rep/RepA expression in the multiple, relative to single, ethanol treatment group. Notably, Rep/RepA expression appears to reach a threshold above which Rep/RepA levels do not further enhance activation of the INPACT system or recombinant protein levels. This threshold seems to have been achieved in the high Rep/RepA, SRN34 background as multiple ethanol treatments did not increase GUS production from an INPACT-GUS cassette in this parent line.
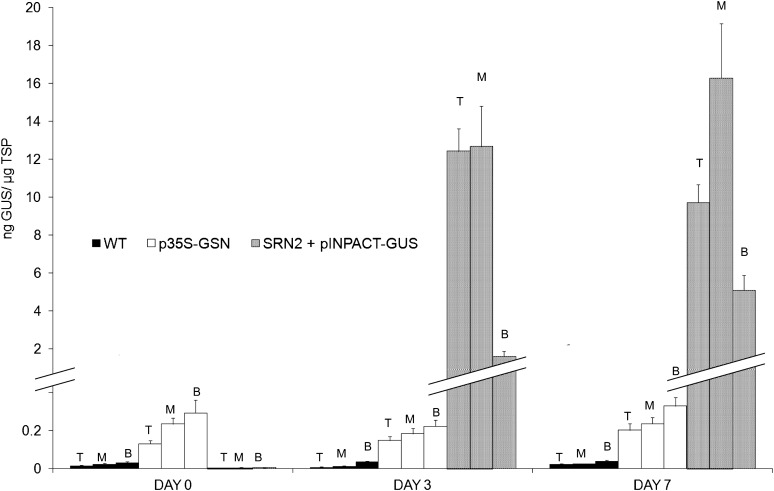
Next, we analyzed GUS distribution following ethanol activation in INPACT transgenic plants relative to plants transformed with p35S-GSN. Tissue was collected at days 0, 3, and 7 PEA, from the top, middle, and bottom three leaves of plants activated using a 1% ethanol foliar spray and root drench. Samples were collected concurrently from wild-type, p35S-GUS plants, and the SRN2 parent line supertransformed with an INPACT-GUS cassette. TSP was extracted from all leaf samples and GUS levels measured by ELISA (Figure 6). Essentially, background-only levels of GUS were detected in all sampling positions (top, middle, and bottom) in wild-type controls. No significant differences in GUS accumulation were detected in tissues from the different sampling positions of p35S-GSN plants at any time during the time course (P > 0.05). No significant differences were observed in GUS accumulation in the different sampling positions from INPACT-GUS plants on day 0 (P > 0.05), which were equivalent to the wild type (i.e., negligible GUS expression). However, 3 d PEA, expression in the INPACT plants had dramatically increased, and there was a significant difference observed between the bottom leaves of INPACT-GUS plants relative to the top and middle. Seven days PEA, a significant difference was observed between each of the three sampling positions (P < 0.05), with the middle leaves having the highest GUS levels. These results suggest that in plants constitutively expressing GUS (p35S-GSN lines), protein production is uniform in leaves of different ages. By contrast, INPACT-directed GUS expression was lower in older leaves relative to middle and young leaves. Notably, GUS levels driven by INPACT were significantly higher (10- to 120-fold), in leaves of all ages, compared with those directed by the constitutive CaMV 35S promoter.
Figure 6.
INPACT-Directed GUS Accumulation throughout Transgenic Tobacco Plants Following Ethanol Activation.
Soil-acclimatized INPACT plants were activated using a 1% ethanol foliar spray and root drench. Tissue was collected on days 0, 3, and 7 PEA, from the top (T), middle (M), and bottom (B) three leaves of all plants. GUS levels were quantified by ELISA. Four independent transformation events representing (1) SRN2 + pINPACT-GUS and (2) p35S-GSN were used. Samples were collected from three biological replicates of each transgenic event and the wild type (WT) and analyzed in triplicate. Error bars indicate se.
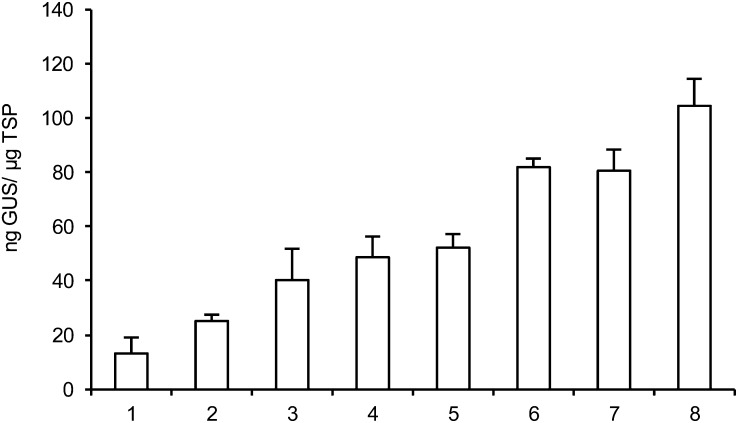
To determine the maximum levels of GUS expression afforded by the INPACT expression platform, eight independent INPACT-GUS transgenic plants were activated using the optimized parameters above (i.e., 1% ethanol foliar spray and root drench with daily foliar applications). Amounts of GUS were measured in the top leaves from three biological replicates of each line, 3 d PEA (Figure 7). GUS levels ranged between 13.05 and 104.31 ng GUS/µg TSP, the highest expressing line representing ∼10% TSP.
Figure 7.
Maximum GUS Levels Afforded by the INPACT Expression Platform in Transgenic Tobacco Plants.
Eight independent INPACT transgenic lines (SRN2 + pINPACT-GUS; 1 to 8) were activated using a 1% ethanol foliar spray and root drench with daily foliar applications. Tissue was collected 3 d PEA from the top three leaves of all plants and GUS levels quantified by ELISA. Samples were collected from three biological replicates of each transgenic event and analyzed in triplicate. Error bars indicate se.
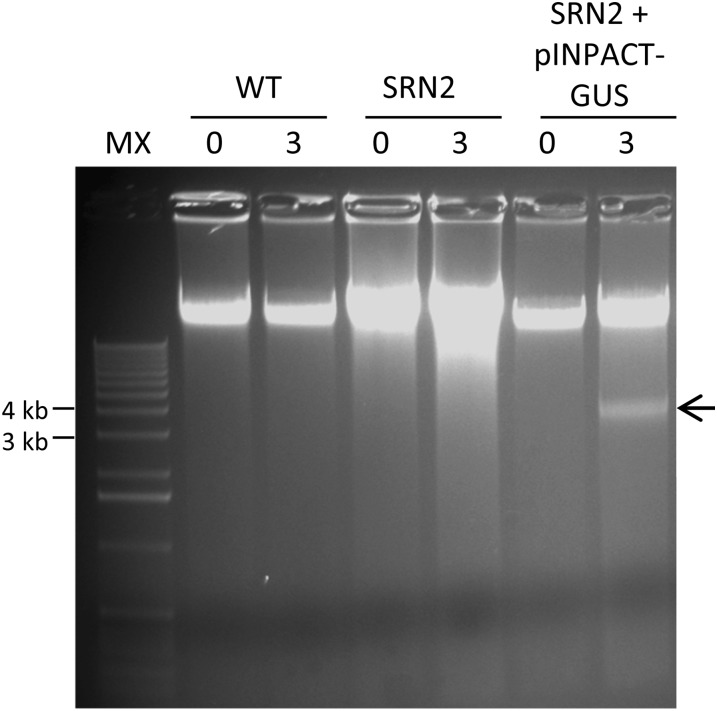
INPACT-GUS Replisomes Accumulate to Large Numbers in Transgenic Tobacco Plants Following Ethanol Activation
Wild-type tobacco plants and transgenic lines SRN2 and SRN2 + INPACT-GUS were grown to the 10 leaf stage and activated with a single 1% ethanol root drench and foliar application. Five days PEA, total genomic DNA was extracted from the top leaf of each plant and electrophoresed through agarose (Figure 8). A single band of between 3 and 4 kb was unique to genomic DNA isolated from the SRN2 + INPACT-GUS line 3 d PEA. The identity of this molecule as the extrachromosomal, double-stranded, closed circular form of the INPACT-GUS replisome was confirmed by cloning and sequencing. These replisomes were estimated to constitute ∼1% of the total nuclear DNA based on staining intensity in comparison to the total genomic DNA. Furthermore, these replisomes were confirmed to be predominantly unmethylated by comparative digestion with methylation-sensitive restriction enzymes DpnI and DpnII (see Supplemental Figure 2 online).
Figure 8.
INPACT Tobacco Lines Accumulate Abundant INPACT-GUS Replisomes Following Ethanol Activation.
A wild-type (WT) negative control plant and transgenic events representing (1) SRN2 and (2) SRN2 + INPACT-GUS were acclimatized in soil and activated with a 1% ethanol foliar spray and root drench. Leaf samples were taken on 0 and 3 d PEA. Total DNA was extracted using the CTAB method and electrophoresed through a 1% agarose gel. The gel was stained with SYBR Safe DNA gel stain. Arrow denotes the double-stranded DNA circular replisome. MX, molecular weight marker X (Roche).
The INPACT Gene Expression System Is Not Host Limited
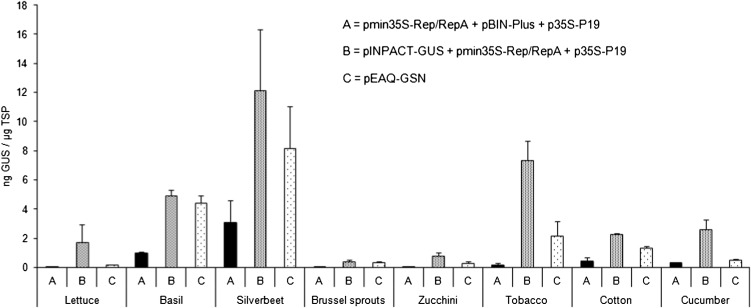
Transient expression using agroinfiltration was used to determine whether the INPACT platform was functional in alternative plant hosts. Eight crop species from seven different plant families were selected: lettuce (Cos) (Asteraceae; Lactuca sativa), basil (Lamiaceae; Ocimum basilicum), silverbeet (Fordhook Giant) (Chenopodiaceae; Beta vulgaris), Brussels sprouts (Brassicaceae; Brassica oleracea), zucchini (cv Blackjack) (Cucurbitaceae; Cucurbita pepo), tobacco (Solanaceae; Nicotiana tabacum), cotton (Malvaceae; Gossypium hirsutum), and cucumber (Cucurbitaceae; Cucumis sativus). An EAQ-HT vector (Sainsbury et al., 2009) encoding the GUS reporter was included as a comparative control, as this vector system is considered a benchmark for high-level transient expression. The vector contains the Tomato bushy stunt virus P19 silencing suppressor gene under the transcriptional control of the CaMV 35S in its T-DNA, which most likely contributes to this high level expression. As a result of this function, the EAQ-HT vector cannot be used for stable transformation as constitutive expression of the P19 protein is detrimental to normal plant growth and development. For this reason, the CaMV 35S promoter alone was used for GUS expression comparisons in stably transformed plants (Figures 5 and 6). Plants were coinfiltrated with agrobacteria harboring plasmids as follows: (1) pmin35S-Rep/RepA + p35S-P19 + pBIN-Plus (empty vector), (2) pINPACT-GUS + p35S-p19 + pmin35S-Rep/RepA, and (3) pEAQ-GSN. Leaf tissue was collected 4 d postinfiltration and GUS levels determined by ELISA (Figure 9). Low levels of GUS were detected in the negative control treatment (1), representing background from infiltration of vectors that do not express GUS. GUS expression directed by the INPACT vector was detected in all species 4 d postinfiltration, and these levels were consistently higher than GUS levels directed by the EAQ-HT hypertranslatable expression vector. While INPACT-directed GUS expression varied greatly between plant species and these levels were generally lower than those observed in stably transformed tobacco plants, these findings suggest that, unlike other virus-based expression platforms, the INPACT system is not solely limited to members of the Solanaceae but has the potential to be adapted to a broad range of plant species.
Figure 9.
INPACT Directs High-Level Transient GUS Expression in D24iverse Crop Species.
Various plant species were grown in soil to 1 month of age and infiltrated with recombinant agrobacteria containing vector combinations pmin35S-Rep/RepA + p35S-p19 + pBIN-Plus (A), pmin35S-Rep/RepA + p35S-p19 + pINPACT-GUS (B), or pEAQ-GSN (C). Leaves were collected 4 d postinfiltration and GUS levels quantified by ELISA. Samples were analyzed in triplicate. Error bars indicate se.
INPACT-Directed Expression of a Lethal Ribonuclease in Transgenic Tobacco
Ten and eight independent transgenic tobacco plants were regenerated from leaf disks of the SRN2 parental line transformed with pINPACT-cytoB (encoding cytosolic barnase) and pINPACT-apoB (encoding apoplast-targeted barnase), respectively. These lines, and two wild-type tobacco plants, were grown to the 10 to 12 leaf stage and activated with a single 1% ethanol root drench and foliar application. Plants were visually monitored over 8 d PEA for phenotypic changes. Within 3 d PEA, significant morphological changes, including leaf discoloration, curling, and wilting, were evident in seven out of ten tobacco lines containing pINPACT-cytoB (Figure 10). Eight days PEA, leaves of these plants showed obvious hallmarks of necrosis, including severe browning and wilting. By contrast, transgenic lines containing the pINPACT-apoB cassette showed only mild discoloration (yellowing) 8 d PEA. Wild-type tobacco plants appeared phenotypically normal over the entire time course.
Figure 10.
Effects of INPACT-Based Expression of Cytosolic Versus Apoplast-Targeted Barnase in Transgenic Tobacco Plants Following Ethanol Activation.
Transgenic lines containing SRN2 + INPACT-cytoB (A) and SRN2 + INPACT-apoB (B) were soil acclimatized and activated with a 1% ethanol foliar spray and root drench. Phenotypic effects of intracellular barnase expression were monitored at day 0 and days 3, 5, and 8 PEA. Significant leaf discoloration was observed in the upper leaves of plants expressing cytosolic barnase 3 d PEA, and this necrosis was evident throughout the plant 8 d PEA. Plants expressing apoplast-targeted barnase showed no evidence of barnase-associated necrosis; however, 8 d PEA leaves of these lines did appear mildly chlorotic.
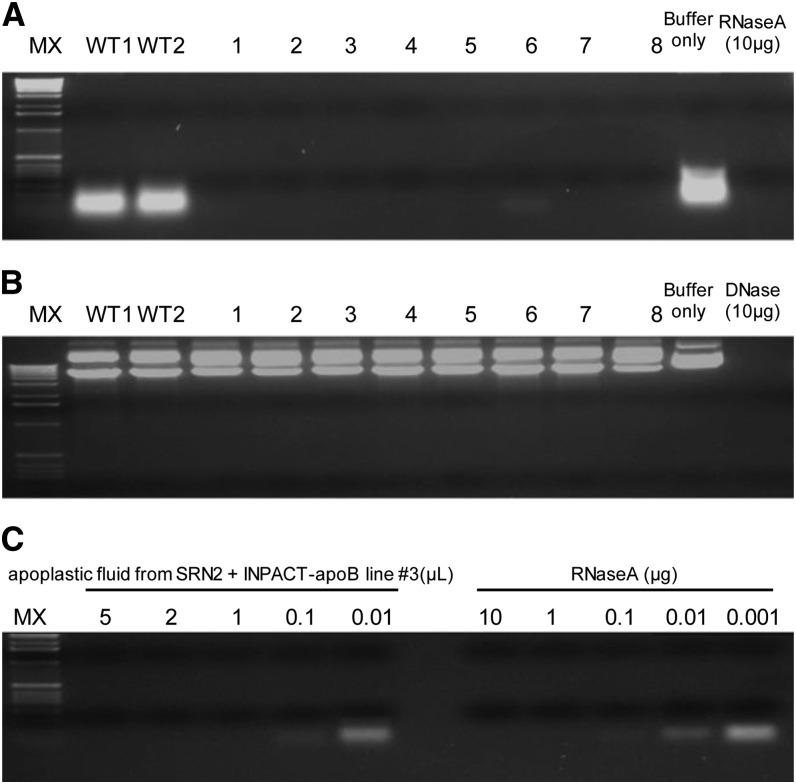
Apoplastic fluid was extracted from the top two leaves of two wild-type plants and the eight transgenic lines representing SRN2 + pINPACT-apoB, 4 d PEA. Specificity and activity of the barnase-containing apoplastic fluid was assayed by incubation with either plant-derived RNA or Escherichia coli–derived plasmid DNA (Figure 11). Interstitial fluid isolated from all eight transgenic lines specifically degraded RNA but not DNA, indicating that these fluids contained an RNA-specific nuclease. The RNase potency of the fluid from one transgenic line (#3) was determined by comparing its activity to that of RNase A. One microliter of this fluid was determined to have an equal activity to 1 µg of RNase A. Interstitial fluid from the leaves of two wild-type plants did not degrade RNA or DNA.
Figure 11.
Activity and Potency of Barnase in Apoplastic Fluid Derived from Ethanol-Activated INPACT Tobacco Plants.
Apoplastic fluid was extracted from eight independent transgenic events (1 to 8) representing SRN2 + INPACT-apoB and two wild-type plants (WT1 and WT2) 4 d PEA. Substrate specificity of the fluid was assessed by incubation with plant-derived RNA (A) and plasmid double-stranded DNA (B). Reactions were incubated at 37°C for 30 min, electrophoresed through a 1% agarose gel, and stained with SYBR Safe DNA gel stain. (C) To determine its RNase potency, apoplastic fluid from transgenic line #3 was serial diluted and incubated with RNA and its activity compared with known quantities of RNase A. MX, molecular weight marker X (Roche).
INPACT-Directed Expression of an Industrial Enzyme, Bovine Trypsinogen, in Transgenic Tobacco
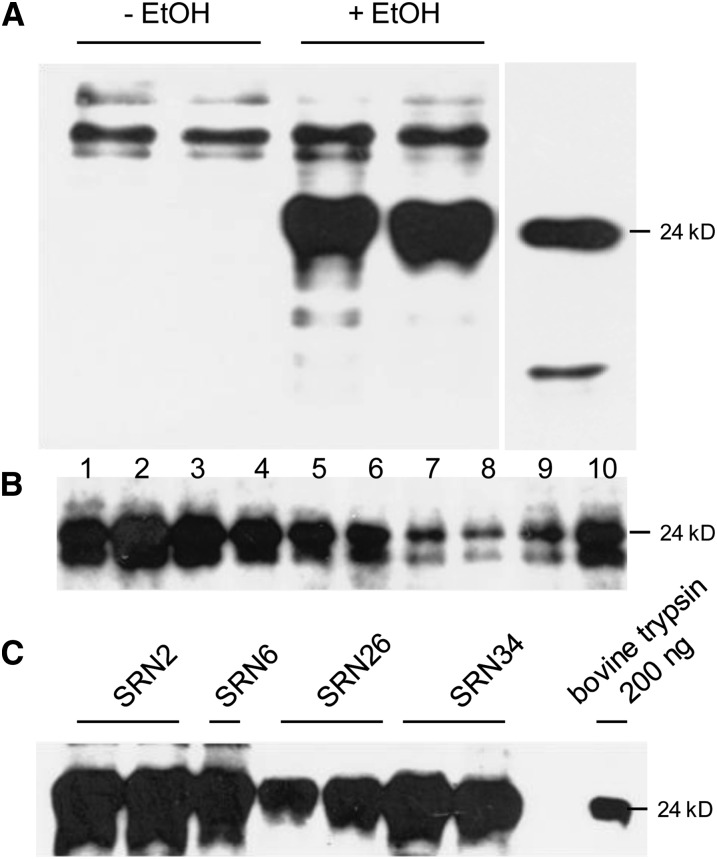
Leaf disks from SRN2, -6, -26, and -34 tobacco plants were transformed with recombinant agrobacteria harboring the pINPACT-ApoTrp vector and between eight and 12 independent transgenic events obtained per parental line. Plants were grown to the 10 to 12 leaf stage and activated with a single 1% ethanol root drench and foliar application. Apoplastic fluid was collected on day 0 and 3 d PEA, and recombinant trypsinogen was detected by immunoblotting. Antibodies raised to bovine trypsin cross-reacted with very high molecular mass proteins in samples from both time points; however, a major protein of ∼24 kD (similar to that of the bovine trypsin control) was only detected following ethanol application (Figure 12A). Highest trypsinogen expression was observed in the top four to five leaves, although recombinant protein expression was distributed throughout the plant and easily detected in the top 10 leaves, 3 d PEA (Figure 12B). Transgenic lines capable of expressing high levels of bovine trypsinogen were identified from each of the four AlcA:AlcR Rep/RepA parent lines (Figure 12C), and highest yields of trypsinogen were estimated at ∼196 mg/kg (dry weight).
Figure 12.
INPACT-Directed Expression of Bovine Trypsinogen in Transgenic Tobacco.
Transgenic plants containing the INPACT-apoTryp cassette were acclimatized in soil and activated with a 1% ethanol foliar spray and root drench.
(A) Apoplastic fluid was collected on day 0 (-EtOH) and 3 d PEA (+EtOH) in duplicate.
(B) TSP was extracted from the first 10leaves (1 = top), 3 d PEA.
(C) TSP was extracted from the top leaf of the best expressing lines representing each of the four SRN parent lines, 3 d PEA. Samples were electrophoresed through a polyacrylamide gel and recombinant trypsinogen detected by immunoblot analysis using a rabbit anti-bovine trypsin polyclonal antibody (US Biological). A positive control lane was included containing 200 ng of purified bovine trypsin (Sigma-Aldrich).
INPACT-Directed Expression of a Therapeutic Protein, Human Vitronectin, in Transgenic N. benthamiana
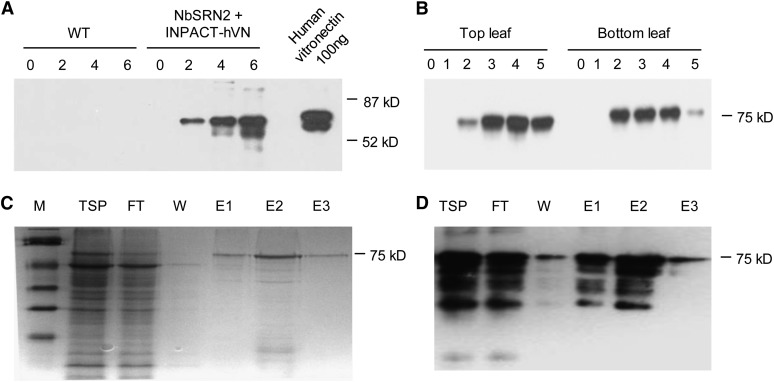
Four independent transgenic N. benthamiana plants were regenerated from leaf disks transformed with the pAlc-Rep/RepA vector, of which one line, NbSRN2, was selected for further work. NbSRN2 was supertransformed with agrobacteria harboring the pINPACT-hVN vector and six independent transgenic events regenerated. Detached leaves of transgenic plants were activated in a 1% ethanol liquid suspension and TSP extracted 4 d PEA. Monoclonal antibodies specific for human Vitronectin reacted with a major protein of ∼75 kD only in leaf samples activated with ethanol (Figure 13A), and the highest yields were estimated at ∼100 mg/kg (fresh weight). In soil acclimatized plants, recombinant Vitronectin expression was abundant in the top and bottom leaves, and protein accumulation peaked 3 d following a foliar application of 1% ethanol (Figure 13B). Adhesive properties of plant-made Vitronectin were demonstrated by binding the recombinant protein to heparin sepharose under denaturing conditions and eluting the protein using high salt concentrations (Figures 13C and 13D). Using this approach, the recombinant plant Vitronectin was extracted to up to 80% purity and was easily visible as a protein band of ∼75 kD, following PAGE, Coomassie blue staining, and immunoblotting (Figures 13C and 13D).
Figure 13.
INPACT-Directed Expression of Human Vitronectin in Transgenic N. benthamiana.
(A) Time course of Vitronectin expression in detached leaves of wild-type (WT) and transgenic plants containing the INPACT-hVN cassette (NbSRN2 + INPACT-hVN) activated in a Murashige and Skoog suspension containing 0.5% ethanol. Samples were taken at days 0, 2, 4, and 6 PEA. TSP was separated by PAGE and analyzed by immunoblotting with an anti-human Vitronectin monoclonal antibody (The Antibody Shop). A positive control lane was included containing 100 ng of purified human Vitronectin (Promega).
(B) Time course of Vitronectin expression in soil-acclimated transgenic plants containing the INPACT-hVN cassette. Samples were taken from the top and bottom leaves over a 5-d period (0 to 5) following activation with a 1% ethanol solution and analyzed by immunoblotting.
(C) and (D) Coomassie blue–stained PAGE (C) and immunoblot analysis (D) tracking partial purification of plant-made Vitronectin using heparin sepharose and HIS-tag affinity columns. M, prestained SDS-PAGE standards, low range (Bio-Rad); FT, flow-through; W, wash; E, elution (E1-5 represent fractions 1 to 5).
DISCUSSION
The expression of exogenous proteins in plants, particularly therapeutic proteins and vaccines, has and continues to be an area of great interest and development. The current strong trend is the application of disaggregated virus vector systems for transient expression in, almost exclusively, N. benthamiana, via agroinfiltration (Giritch et al., 2006; Regnard et al., 2010; Sainsbury et al., 2010). These systems provide for (1) quick, high-level expression and (2) rapid development from design and assembly of an expression cassette to expressed protein, the rapid response concept being of particular relevance to the deployment of vaccines for new infectious diseases. These expression systems are particularly attractive for low to moderate volume, high-value proteins and, thus, the expensive infrastructure and containment required for production via agroinfiltration can be easily justified. N. benthamiana is also well suited to this production system, as it supports very high transient expression levels and is easily grown in contained facilities (Goodin et al., 2008). However, it has relatively low biomass and therefore is not suitable for high volume protein production.
By contrast, we developed a plant expression system that is adaptable to both small-scale confined production of high-value proteins in Solanaceous hosts as well as field production of lower value proteins (or expression of traits) in a wide range of plant species. The technology, INPACT, is based on a two-cassette system both of which are stably integrated into the host nuclear genome where one expression cassette encodes the Rep/RepA activators under the transcriptional control of the AlcA:AlcR promoter system (Caddick et al., 1998) and the second activatable cassette contains the split gene of interest. Expression of the gene of interest is activated by an ethanol spray, and, importantly, there is negligible expression in the absence of the ethanol inducer molecule due to the unique split gene arrangement. We developed the technology in tobacco (N. tabacum cv Samsun) rather than N. benthamiana because of the potential to use high biomass tobacco for field production. In our best stably transformed tobacco line, INPACT-directed GUS levels reached up to 10% TSP following ethanol activation. By contrast, Conley et al. (2011) stably expressed four different recombinant proteins in a range of tobacco species and cultivars, using a conventional cassette and the dual enhancer CaMV 35S promoter. In the tobacco cultivar Samsun, expression levels of the four proteins varied greatly, the highest being the APA protein (a novel synthetic human antibody against Pseudomonas aeruginosa), averaging just under 1% TSP. Importantly, GUS in INPACT lines was abundant throughout the entire plant, albeit at lower levels in the oldest leaves compared with the younger leaves. GUS levels returned to background after ethanol application, but the plants could be reactivated with ethanol 2 months later. Reactivation produced GUS levels in regrowth equal to those obtained previously. This indicates that the system is amenable to multiple applications and harvests, further reducing production costs.
An essential characteristic of a broad plant expression system is its ability to be exploited in a wide range of different plant species. This is of particular importance for applications in field-grown crops and of lesser importance for the production of high value proteins in contained systems. We demonstrated that the INPACT vector expresses and replicates in a range of dicot species from seven different families. It is highly likely that the INPACT system could be adapted to any plant species as its mechanism of replication, rolling circle replication, is probably not host specific; supporting this, it has previously been reported that some geminiviruses are capable of replicating in nonhost plant cells (Stenger et al., 1992; Teng et al., 2010). Furthermore, the INPACT cassette does not encode the cognate virus movement protein, a gene product known to influence geminivirus host range properties (Ingham and Lazarowitz, 1993). Also, TYDV naturally infects species from four different dicot families, including tobacco (Solanaceae), Phaseolus vulgaris (Fabaceae), Amaranthus retroflexus (Amaranthaceae), and Raphanus raphanistrum (Brassicaceae) (Trebicki et al., 2010). The application of these viruses in a range of plant species has been exploited with at least one other geminivirus vector system. In this case, a BeYDV-based vector system was successfully adapted for the transient overexpression of two pharmaceutical biologics in lettuce, a nonhost to BeYDV (Lai et al., 2012). By contrast, RNA virus-based vector systems, such as those using TMV or Potato virus X incorporate both the relevant viral replicase and movement proteins, and this results in severe host range limitations, essentially confining these systems to the Solanaceae. Similarly, high expression afforded by the nonreplicating, hypertranslation system described by Sainsbury and Lomonossoff (2008), has been limited to members of the Nicotiana genus (Sainsbury and Lomonossoff, 2008; Sun et al., 2011).
In the absence of the ethanol inducer molecule, there is no expression of the gene of interest from the integrated INPACT cassette making the system applicable to the production of proteins that are toxic or inhibitory to plant growth and development or where timing of expression is crucial. This was demonstrated with the expression of barnase, an extremely active and toxic ribonuclease (Hartley, 1989). In the absence of activation, transgenic lines harboring an INPACT cassette capable of expressing cytosolic barnase grew normally; however, ethanol activation resulted in rapid, barnase-associated necrosis throughout these plants within 3 d. This ability to stably integrate then activate a gene encoding a toxic or inhibitory protein should have wide applications. For instance, we are currently investigating the application of INPACT for the production of various cellulolytic enzymes in plants to facilitate the conversion of cellulose to fermentable sugars. This would ultimately be exploited in very high biomass crops, such as sugarcane (Saccharum officinarum).
As a demonstration of the practical exploitation of the INPACT platform for the production of useful biologics, we chose to express two very different animal-derived proteins, the low-value industrial protease, bovine trypsinogen, and the high-value, therapeutic, human Vitronectin. Trypsin is a pancreatic Ser protease, and due to its targeted polypeptide degrading ability (Kemmler et al., 1971), has applications in cell culture and is an important component of many industrial processes in the food and pharmaceutical industries. Using INPACT to direct the expression of an apoplast-targeted zymogen form of the enzyme, we were able to achieve levels of up to 196 mg/kg (dry weight) in tobacco leaves, 3 d PEA. This yield is more than comparable to levels achieved in maize (Zea mays) seed (58 mg/kg dry weight; Woodard et al., 2003). Human Vitronectin is a large, multifunctional glycoprotein found in blood, some tissues, and the extracellular matrix. It promotes cell adhesion and migration via an RGD integrin binding site and associated motifs (Chillakuri et al., 2010), and as such has applications in cell culture and in wound healing therapies (Upton et al., 2008). Using the INPACT platform, highest yields of plant-made Vitronectin were ∼100 mg/kg (fresh weight). In addition, we partially purified the recombinant protein using a single-step process based on the molecule’s natural affinity for heparin, a fundamental function of Vitronectin in regulating blood coagulation (Preissner, 1991).
Chemical induction, particularly using the AlcA:AlcR system, has been used with plant virus vectors previously. Zhang and Mason (2006) controlled the expression of BeYDV Rep/RepA and, in turn, virus vector replication using the ethanol inducible gene switch for the accumulation of Norwalk virus capsid protein in both tobacco NT-1 cells and potato (Solanum tuberosum) plants. They demonstrated up to a 10-fold increase in recombinant protein levels postethanol induction; however, in this instance, Norwalk virus capsid protein was already constitutively expressed prior to ethanol induction. More recently, Werner et al. (2011) reported the adaptation of the TMV-based proviral expression system to transgenic N. benthamiana. In this case, viral RNA release and cell-to-cell movement were separately placed under the control of the AlcA:AlcR gene switch. After ethanol induction, green fluorescent protein was expressed to levels of up to 4.3 g/kg fresh weight, rivaling those levels obtained with transient agroinfiltration. However, the limitation of the TMV system continues to be that it is restricted to a small number of Solanaceous hosts.
Extrachromosomal, circular, replicating DNA forms of the INPACT cassette or replisomes were only observed in transgenic plants following ethanol activation. Closed circular, double-stranded rolling circle replication intermediates accumulated to very high levels 3 d PEA and were readily visible in plant genomic DNA extracts by electrophoresis. The amount of replisomes positively correlated with the level of GUS expression, and in some transgenic lines, we estimated the INPACT replisomes constituted ∼1% of the total nuclear DNA. INPACT replisomes were not methylated, which may preclude these DNA forms from the negative effects of transcriptional gene silencing, as DNA methylation is a common effector of the transcriptional gene silencing process (Razin, 1998; Wolffe and Matzke, 1999). At this time, we have no evidence to suggest that INPACT-based expression in planta is subject to posttranscriptional gene silencing; however, the rapid and high levels of protein expression afforded by INPACT and its ability to be reactivated suggest the system may not be greatly affected by this phenomenon.
The INPACT system is essentially an inducible transient expression system from a stably transformed plant. From a regulatory perspective, its advantages are twofold: First, the INPACT vector lacks both virus-derived movement and coat proteins and the cassette encoding the genes involved in replication, Rep/RepA, does not contain the viral replication recognition sequences. Second, the platform has the demonstrated advantage of high-level expression, but from a single defined integration event that can be fully genetically characterized. The technology can be exploited for the production of large quantities of exogenous proteins in a wide range of different plant species and can be used to express “difficult” proteins. It is suitable for both small-scale glasshouse confined production or, importantly, for field cultivation, where production is limited only by the amount of transgenic biomass necessary to produce the desired quantity of recombinant protein, which could be in the kilogram to ton range. With high-intensity agricultural practices in combination with high biomass hosts, such as tobacco or sugarcane, the technology has the potential for large-scale production of industrial proteins in plants.
METHODS
Construction of Vectors
The 84-bp syntron (see Supplemental Figure 3 online) was fused between nucleotides 232 and 233 in the uidA reporter gene using overlapping primers and PCR. The resulting uidA-syntron sequence was inserted between a 530-bp CaMV 35S promoter and a nos terminator using NcoI and BamHI restriction sites. The resulting expression cassette was mobilized into pCAMBIA1300 as an EcoRI restriction fragment to generate the binary plasmid p35S-GSN.
Tobacco (Nicotiana tabacum) leaf material infected with TYDV was obtained from Myrtleford, Victoria. Full-length TYDV genomic DNA was isolated from this material using PCR and the cloned sequence showed ∼95% similarity to that of GenBank accession number M81103.1 (Morris et al., 1992). TYDV genomic DNA was used as a template to PCR amplify the TYDV LIR (nucleotides 8 to 212) and SIR (nucleotides 1324 to 1483) and the complementary strand Rep/RepA bicistronic gene sequence (nucleotides 2850 to 1481). The TYDV Rep/RepA coding region was ligated between the AlcA promoter and nos terminator in pACN using PstI restriction sites. The resulting cassette was excised as a HindIII restriction fragment and ligated into similarly digested pSRNAGS. The resulting binary plasmid contained a nos-NPTII-nos selection cassette, a CaMV35S-ALCR-nos expression cassette, and the AlcA-Rep/RepA-nos cassette located between the left and right T-DNA borders in pBIN19. This plasmid was called pAlc-Rep/RepA.
To corrupt two potential intron splice sites (AGGT and AGGA) in the TYDV LIR, adenines at nucleotides 88 and 100 (relative to GenBank accession number M81103.1) were changed to cytosines using PCR. An INPACT cassette containing the split uidA reporter gene was assembled by sequentially ligating the first TYDV LIR (EcoRI-SwaI fragment), the 3′ syntron half and uidA exon 2 (SwaI-SacI fragment), nos terminator (SacI-XhoI fragment), TYDV SIR (XhoI-AscI fragment), CaMV 35S promoter (AscI-NcoI fragment), uidA exon 1 and 5′ syntron half (NcoI-PacI fragment), and the second TYDV LIR (PacI-HindIII fragment) into pCAMBIA1300. The resulting INPACT binary plasmid was called pINPACT-GUS. The Bacillus amyloliquifaciens 90–amino acid barstar protein was back-translated using human codon bias and the coding sequence chemically synthesized (see Supplemental Figure 4 online). This resulting codon modified 273-bp barstar gene was ligated between a CaMV 35S promoter and CaMV 35S terminator using NcoI-SacI restriction sites. This plasmid was called p35S-barstar. The B. amyloliquifaciens 111–amino acid barnase protein was back-translated using human codon bias and the coding sequence chemically synthesized (see Supplemental Figure 5 online). However, in this case, the 336-bp codon modified barnase gene was engineered to contain a syntron between nucleotides 179 and 180. The uidA exon 1 and 5′ syntron half in pINPACT-GUS was replaced with barnase exon 1 and the 5′ syntron half using NcoI and PacI restriction sites. The 3′ syntron half and uidA exon 2 were then replaced with the 3′ syntron half and barnase exon 2 using SwaI and SacI restriction sites. The nos terminator was replaced with a 1-kb Chrysanthemum spp RbcS terminator using SacI and XhoI restriction sites. The CaMV 35S promoter was replaced with the 1-kb Chrysanthemum spp RbcS promoter ± the sea anemone equistatin secretion signal (Outchkourov et al., 2003). The barstar expression cassette in p35S-barstar was excised as a HindIII fragment and ligated into the unique HindIII site located downstream of both INPACT cassettes generating two vectors (1) pINPACT-cytoB for cytosolic barnase expression and (2) pINPACT-apoB for apoplast-targeted barnase expression.
The bovine trypsinogen protein without native secretion signal (amino acids 19 to 246) was back-translated using maize (Zea mays) codon bias and the coding sequence chemically synthesized. Again, this sequence was engineered to contain the syntron between nucleotides 66 and 67 (see Supplemental Figure 6 online). The barnase exon 1 and 5′ syntron half in pINPACT-apoB was replaced with trypsinogen exon 1 and the 5′ syntron half using NcoI and PacI restriction sites. The 3′ syntron half and barnase exon 2 were then replaced with the 3′ syntron half and trypsinogen exon 2 using SwaI and SacI restriction sites. The resulting binary plasmid was called pINPACT-apoTrp. A cDNA clone of human Vitronectin was obtained from Invitrogen (clone ID 4040317). Vitronectin exon 1 (nucleotides 1 to 276, where 1 is the A of the ATG start codon) was PCR amplified to contain the 5′ syntron half and PacI restriction site. This sequence replaced the uidA exon 1 and 5′ syntron half in pINPACT-GUS using NcoI and PacI restriction sites. Vitronectin exon 2 (nucleotides 277 to 1437), including sequence encoding a C-terminal KDEL retention signal and 6XHIS tag, was PCR amplified to contain the 3′ syntron half and a SwaI restriction site. This sequence replaced the uidA exon 2 and 3′ syntron half using SwaI and SacI restriction sites. The resulting binary plasmid was called pINPACT-hVN.
For transient agroinfiltration assays, three additional plasmids were prepared. A vector for low-level expression of Rep/RepA was constructed by first ligating the TYDV Rep/RepA gene between the CaMV 35S promoter and CaMV 35S terminator in pDH51 using BamHI-SalI restriction sites. The 35S-Rep/RepA-35S cassette (containing a truncated CaMV 35S promoter) was excised by EcoRV-EcoRI digestion and ligated into SmaI-EcoRI–digested pBIN-Plus. The resulting plasmid was called pmin35S-Rep/RepA. A silencing suppressor cassette was constructed by excising the Tomato bushy stunt virus P19 gene from EAQ-HT (Sainsbury et al., 2009) using BamHI-SacI and ligating it between the CaMV 35S promoter and nos terminator in pBIN-Plus. The resulting plasmid was called p35S-P19. For GUS expression comparisons, the EAQ-HT hypertranslation vector described by Sainsbury et al. (2009) was selected. The GUS coding region and syntron were excised from p35S-GSN as a blunt-ended NcoI-SalI fragment and ligated into blunt-ended AgeI-XhoI–digested EAQ-HT. The resulting plasmid was called pEAQ-GSN.
Stable Transformation of Nicotiana spp
All plasmids for stable transformation were mobilized into Agrobacterium tumefaciens (strain LBA4404) by electroporation. Agrobacterium-mediated transformation of N. tabacum (cv Samsun) and Nicotiana benthamiana leaf disks and their regeneration were performed as described by Horsch et al. (1985). Transgenic plants containing the pAlc-Rep/RepA cassette were regenerated first. Rep/RepA expression was analyzed in tobacco plants transformed with pAlc-Rep/RepA, by qRT-PCR, following ethanol activation. Lines with high and low Rep/RepA expression (SRN34 and SRN2, respectively) were selected as parental lines and supertransformed with pINPACT-GUS. For barnase studies, only SRN2 was supertransformed with pINPACT-cytoB and pINPACT-apoB. For bovine trypsinogen studies, all four parental lines were supertransformed with pINPACT-apoTrp. For human Vitronectin studies, a single N. benthamiana line (NbSRN2) containing the pAlc-Rep/RepA cassette was selected and supertransformed with pINPACT-hVN. Tissue culture plants were soil acclimated and transferred to growth cabinets with a 16-h photoperiod and constant temperature of 27°C. Plants were grown until the 10 to 12 leaf stage prior to assaying.
Agroinfiltration
Plasmids pmin35S-Rep/RepA, pINPACT-GUS, p35S-GSN, pEAQ-GSN, and p35S-p19 and empty vector control pBIN-Plus were mobilized into Agrobacterium (strain Agl1) by electroporation. Cultures of Agrobacterium were grown overnight in Luria-Bertani medium with the appropriate antibiotic selection agent. Cells were collected by low-speed centrifugation (4000g) and resuspended in MMA solution containing 10 mM MES, pH 5.5, 10 mM MgCl2, and 100 μM acetosyringone. Cultures were incubated at room temperature with gentle agitation for 3 h after which they were diluted to an OD600 of 1.0. Three leaves of selected plants were infiltrated on the abaxial side until saturated, using a syringe without a needle. When mixtures of Agrobacterium were infiltrated, bacterial cultures were prepared separately in induction media and combined immediately before infiltration at a ratio of 1:1. Leaf samples from each treatment were harvested 4 d postinfiltration and GUS protein quantified by ELISA.
Ethanol Activation
Activation of soil acclimatized plants was achieved using a foliar spray and root drench with 1% ethanol. Plants were not watered for 3 d prior to ethanol activation. For multiple ethanol treatments, additional 1% ethanol foliar sprays were applied on each day following initial activation. Reactivation was achieved by treating plants with a foliar spray and root drench of 1% ethanol, 8 weeks after the initial ethanol treatment. In one case, detached leaves of N. benthamiana tissue culture plants were activated by immersing the leaves in Murashige and Skoog liquid media containing 0.5% ethanol and incubating at 27°C with a 16-h photoperiod and gentle agitation.
Isolation of Nucleic Acids from Leaf Samples
Leaf samples were immediately snap frozen in liquid nitrogen following harvesting. Tri reagent (Sigma-Aldrich) was used to extract total RNA from tissue samples according to the manufacturer’s instructions and the method of Azevedo et al. (2003). Total DNA was isolated using the CTAB method described by Stewart and Via (1993). To visualize INPACT replisomes, ∼5 to 10 µg of total DNA was electrophoresed through a 1% agarose gel and stained with SYBR Safe DNA gel stain.
qRT-PCR
RNA was extracted using the Qiagen RNeasy reagents and protocol, including on-column DNaseI digestion. First-strand DNA was synthesized from total RNA treated with M-MLV reverse transcriptase (Promega). Real-time quantitative PCR was performed using GoTaq qPCR Master Mix (Promega) containing SYBR green. Reactions were run on a Rotor-Gene Q (Qiagen) and analyzed with the associated software version 1.7.94. Actin served as a control for normalization. Primer sequences were as follows (5′–3′): Actin-F (CTATTCTCCGCTTTGGACTTGGCA), Actin-R (AGGACCTCAGGACAACGGAAACG), Rep/RepA-F (GAACTATTATCCAGACTGCGACTTC), and Rep/RepA-R (TGAGGAAATTCATCCTTTATCATGT). Thermocycling conditions were 50°C for 2 min and 95°C for 2 min followed by 40 cycles of 95°C for 15 s, 55°C for 30 s, 72°C for 5 s, and 82°C for 5 s. Melt curve analysis verified primer specificity. qRT-PCR was performed in triplicate. Delta cycle threshold values were used to calculate transcript levels as described by Levy et al. (2004).
GUS ELISA
For quantification of GUS, 10-mg samples (three disks) of fresh leaf tissue were homogenized using a Tissuelyser (Qiagen) for 2 min at 20 Hz. Samples were suspended in 600 μL of GUS extraction buffer (100 mM Na2HPO4, 1 mM Na2EDTA, pH 8, 1 mM DTT, 0.01% SDS, and 0.05% Tween 20) and further homogenized for 2 min at 10 Hz. Extracts were cleared of debris by centrifugation (4000g for 30 min at 4°C). TSP content was measured using the Bradford assay. To quantify GUS in TSP extracts, sandwich ELISAs were performed. High binding 96-well plates (Nunc Maxisorp) were coated overnight at 4°C with 100 μL/well of 0.5 μg/mL rabbit anti-GUS IgG (Sigma-Aldrich G5545) in BBS (25 mM borate and 75 mM NaCl, pH 8.5), and 1 h at room temperature in blocking buffer (1% BSA in BBS). TSP extracts (100 µL/well) were added and the plates incubated for 1 h at room temperature. Plates were then incubated with 100 μL/well of 0.5 μg/mL horseradish peroxidase–labeled rabbit anti-GUS IgG (Invitrogen A5790 conjugated to horseradish peroxidase) in blocking buffer for 1 h at room temperature and developed with TMB (Sigma-Aldrich). Plates were read at 650 nm, using 620 nm as a reference, with an absorbency plate reader. Purified GUS (GUS Type VII-A; Sigma-Aldrich G7646) was used to develop a standard curve of reactivity.
Statistical Analysis
Data were expressed as means ± se. Student's t tests were used to compare two groups. For qRT-PCR, a one-way analysis of variance was performed with Tukey post test for each day of the time course. P < 0.05 was considered significant.
Isolation of Apoplastic Fluid Containing Barnase and Assessment of Ribonuclease Specificity and Dose
Apoplastic fluid was isolated from tobacco leaves by vacuum infiltration and low-speed centrifugation. Briefly, leaves were submerged in 50 mM phosphate buffer, pH 7.0, and subjected to a vacuum pressure of −80 kPa in a closed chamber. Leaves were blotted dry, placed in 15-mL Falcon tubes, and centrifuged for 10 to 30 min at low speed (100g). Approximately 20 to 50 μL of apoplastic fluid was obtained for each leaf sample. Apoplastic fluid (5 µL) was incubated with 1 µg of closed circular, double-stranded plasmid DNA derived from Escherichia coli, or 5 µg of total RNA derived from N. tabacum in a 20-μL reaction volume. Reactions were incubated at 37°C for 30 min, electrophoresed through a 1% agarose gel, and stained with SYBR Safe DNA gel stain. RNase A from bovine pancreas (Roche) and RQ1 RNase-free, DNase (Promega) were used as enzyme controls.
Recombinant Protein Isolation and Purification
Plant-made recombinant trypsinogen was detected in apoplastic fluid or TSP extracted from whole tobacco leaves. Apoplastic fluid was isolated as described above except using 150 mM citric acid, pH 2.0. TSP was extracted by grinding freeze-dried leaf material in 150 mM citric acid, pH 2.0, and clarification by low-speed centrifugation. Protein concentrations were measured using the BCA assay (Thermo Scientific). Plant-made Vitronectin was partially purified by grinding N. benthamiana leaf material to a fine powder under liquid nitrogen and incubation in 4 volumes of VN extraction buffer (50 mM phosphate buffer, 8 M urea, and 10 mM β-mercaptoethanol, pH 7) for 1 h at room temperature with gentle agitation. The mixture was filtered through cheese cloth and clarified by centrifugation at 11, 000 rpm for 20 min at 4°C. TSP was loaded onto a column containing 1 mL packed volume of Heparin Sepharose Fast Flow (GE Healthcare) and washed with 5 volumes of VN extraction buffer. Bound Vitronectin was eluted from the column with VN extraction buffer containing 300 mM NaCl.
PAGE and Immunoblotting
Protein samples were electrophoresed through a 12.5% polyacrylamide gel and either visualized by Coomassie Brilliant Blue staining or transferred to positively charged nylon membranes (Roche) by electroblotting and blocked with 5% skim milk powder in TBST (20 mM Tris, pH 7.5, 150 mM NaCl, and 0.1% [v/v]Tween20). For trypsinogen immunodetection, membranes were incubated with a rabbit primary antibody raised against bovine trypsin (US Biological; 1:3000 dilution), followed by horseradish peroxidase–linked goat anti-rabbit secondary antibody (Thermo Scientific; 1:3000 dilution). For Vitronectin immunodetection, membranes were incubated with a mouse primary monoclonal antibody raised against human Vitronectin (Antibody Shop; 1:3000 dilution), followed by horseradish peroxidase–linked goat anti-mouse secondary antibody (Thermo Scientific; 1:3000 dilution). All antibodies were visualized using the Supersignal West Femto Maximum Sensitivity Substrate (Thermo Scientific) system, according to the manufacturer’s guidelines. As controls, 200 ng of bovine trypsin (Sigma-Aldrich) and 100 ng of human Vitronectin (Promega) were included and all gels contained prestained SDS-PAGE standards, low range (Bio-Rad) for molecular mass estimation.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: B. amyloliquifaciens barstar protein (P11540), B. amyloliquifaciens barnase protein (CAA31365.1), Bovine trypsinogen protein (P00760.3), Human Vitronectin gene (BC005046), and N. tabacum actin gene (X69885.1)
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Estimation of Copy Number in Alcohol-Inducible Rep/RepA Parent Tobacco Lines.
Supplemental Figure 2. Restriction Endonuclease Analysis of INPACT Replisomes.
Supplemental Figure 3. Nucleotide Sequence of the Synthetic Intron (Syntron).
Supplemental Figure 4. Nucleotide Sequence of the Codon Modified Barstar Gene.
Supplemental Figure 5. Nucleotide Sequence of the Codon Modified Barnase Gene.
Supplemental Figure 6. Nucleotide Sequence of the Codon Modified Bovine Trypsinogen Gene.
Acknowledgments
We thank the Australian Research Council for funding this research.
AUTHOR CONTRIBUTIONS
B.D., C.L.M., M.K., and T.A.J. contributed to designing and performing this research, assisting in data analysis, and writing the article. R.M.H. and J.L.D. contributed to experimental design, data analysis, and writing the article.
Glossary
- TMV
Tobacco mosaic virus
- TSP
total soluble protein
- TYDV
tobacco yellow dwarf virus
- INPACT
In Plant Activation
- LIR
large intergenic region
- SIR
small intergenic region
- PEA
postethanol activation
- qRT-PCR
quantitative real-time PCR
- GUS
β-glucuronidase
- CaMV
cauliflower mosaic virus
References
- Azevedo H., Lino-Neto T., Tavares R.M. (2003). An improved method for high-quality RNA isolation from needles of adult maritime pine trees. Plant Mol. Biol. Rep. 21: 333–338 [Google Scholar]
- Baynton C.E., Potthoff S.J., McCullough A.J., Schuler M.A. (1996). U-rich tracts enhance 3′ splice site recognition in plant nuclei. Plant J. 10: 703–711 [DOI] [PubMed] [Google Scholar]
- Caddick M.X., Greenland A.J., Jepson I., Krause K.P., Qu N., Riddell K.V., Salter M.G., Schuch W., Sonnewald U., Tomsett A.B. (1998). An ethanol inducible gene switch for plants used to manipulate carbon metabolism. Nat. Biotechnol. 16: 177–180 [DOI] [PubMed] [Google Scholar]
- Chapman S., Kavanagh T., Baulcombe D. (1992). Potato virus X as a vector for gene expression in plants. Plant J. 2: 549–557 [DOI] [PubMed] [Google Scholar]
- Chillakuri C.R., Jones C., Mardon H.J. (2010). Heparin binding domain in vitronectin is required for oligomerization and thus enhances integrin mediated cell adhesion and spreading. FEBS Lett. 584: 3287–3291 [DOI] [PubMed] [Google Scholar]
- Conley A.J., Zhu H., Le L.C., Jevnikar A.M., Lee B.H., Brandle J.E., Menassa R. (2011). Recombinant protein production in a variety of Nicotiana hosts: A comparative analysis. Plant Biotechnol. J. 9: 434–444 [DOI] [PubMed] [Google Scholar]
- Faye L., Gomord V. (2010). Success stories in molecular farming-a brief overview. Plant Biotechnol. J. 8: 525–528 [DOI] [PubMed] [Google Scholar]
- Fischer R., Schillberg S., Hellwig S., Twyman R.M., Drossard J. (2012). GMP issues for recombinant plant-derived pharmaceutical proteins. Biotechnol. Adv. 30: 434–439 [DOI] [PubMed] [Google Scholar]
- Giritch A., Marillonnet S., Engler C., van Eldik G., Botterman J., Klimyuk V., Gleba Y. (2006). Rapid high-yield expression of full-size IgG antibodies in plants coinfected with noncompeting viral vectors. Proc. Natl. Acad. Sci. USA 103: 14701–14706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleba Y., Klimyuk V., Marillonnet S. (2005). Magnifection—A new platform for expressing recombinant vaccines in plants. Vaccine 23: 2042–2048 [DOI] [PubMed] [Google Scholar]
- Gleba Y., Klimyuk V., Marillonnet S. (2007). Viral vectors for the expression of proteins in plants. Curr. Opin. Biotechnol. 18: 134–141 [DOI] [PubMed] [Google Scholar]
- Goodall G.J., Filipowicz W. (1989). The AU-rich sequences present in the introns of plant nuclear pre-mRNAs are required for splicing. Cell 58: 473–483 [DOI] [PubMed] [Google Scholar]
- Goodin M.M., Zaitlin D., Naidu R.A., Lommel S.A. (2008). Nicotiana benthamiana: Its history and future as a model for plant-pathogen interactions. Mol. Plant Microbe Interact. 21: 1015–1026 [DOI] [PubMed] [Google Scholar]
- Hartley R.W. (1989). Barnase and barstar: Two small proteins to fold and fit together. Trends Biochem. Sci. 14: 450–454 [DOI] [PubMed] [Google Scholar]
- Horsch R.B., Fry J.E., Hoffmann N.L., Eichholtz D., Rogers S.G., Fraley R.T. (1985). A simple and general method for transferring genes into plants. Science 227: 1229–1231 [DOI] [PubMed] [Google Scholar]
- Ingham D.J., Lazarowitz S.G. (1993). A single missense mutation in the BR1 movement protein alters the host range of the squash leaf curl geminivirus. Virology 196: 694–702 [DOI] [PubMed] [Google Scholar]
- Kemmler W., Peterson J.D., Steiner D.F. (1971). Studies on the conversion of proinsulin to insulin. I. Conversion in vitro with trypsin and carboxypeptidase B. J. Biol. Chem. 246: 6786–6791 [PubMed] [Google Scholar]
- Lai H.F., He J.Y., Engle M., Diamond M.S., Chen Q. (2012). Robust production of virus-like particles and monoclonal antibodies with geminiviral replicon vectors in lettuce. Plant Biotechnol. J. 10: 95–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy M., Edelbaum O., Sela I. (2004). Tobacco mosaic virus regulates the expression of its own resistance gene N. Plant Physiol. 135: 2392–2397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marillonnet S., Giritch A., Gils M., Kandzia R., Klimyuk V., Gleba Y. (2004). In planta engineering of viral RNA replicons: Efficient assembly by recombination of DNA modules delivered by Agrobacterium. Proc. Natl. Acad. Sci. USA 101: 6852–6857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris B.A.M., Richardson K.A., Haley A., Zhan X.C., Thomas J.E. (1992). The nucleotide sequence of the infectious cloned DNA component of tobacco yellow dwarf virus reveals features of geminiviruses infecting monocotyledonous plants. Virology 187: 633–642 [DOI] [PubMed] [Google Scholar]
- Mushegian A.R., Shepherd R.J. (1995). Genetic elements of plant viruses as tools for genetic engineering. Microbiol. Rev. 59: 548–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Outchkourov N.S., Peters J., de Jong J., Rademakers W., Jongsma M.A. (2003). The promoter-terminator of chrysanthemum rbcS1 directs very high expression levels in plants. Planta 216: 1003–1012 [DOI] [PubMed] [Google Scholar]
- Pogue G.P., et al. (2010). Production of pharmaceutical-grade recombinant aprotinin and a monoclonal antibody product using plant-based transient expression systems. Plant Biotechnol. J. 8: 638–654 [DOI] [PubMed] [Google Scholar]
- Preissner K.T. (1991). Structure and biological role of vitronectin. Annu. Rev. Cell Biol. 7: 275–310 [DOI] [PubMed] [Google Scholar]
- Razin A. (1998). CpG methylation, chromatin structure and gene silencing-a three-way connection. EMBO J. 17: 4905–4908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regnard G.L., Halley-Stott R.P., Tanzer F.L., Hitzeroth I.I., Rybicki E.P. (2010). High level protein expression in plants through the use of a novel autonomously replicating geminivirus shuttle vector. Plant Biotechnol. J. 8: 38–46 [DOI] [PubMed] [Google Scholar]
- Sainsbury F., Lomonossoff G.P. (2008). Extremely high-level and rapid transient protein production in plants without the use of viral replication. Plant Physiol. 148: 1212–1218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sainsbury F., Sack M., Stadlmann J., Quendler H., Fischer R., Lomonossoff G.P. (2010). Rapid transient production in plants by replicating and non-replicating vectors yields high quality functional anti-HIV antibody. PLoS ONE 5: e13976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sainsbury F., Thuenemann E.C., Lomonossoff G.P. (2009). pEAQ: versatile expression vectors for easy and quick transient expression of heterologous proteins in plants. Plant Biotechnol. J. 7: 682–693 [DOI] [PubMed] [Google Scholar]
- Stenger D.C., Davis K.R., Bisaro D.M. (1992). Limited replication of tomato golden mosaic virus DNA in explants of nonhost species. Mol. Plant Microbe Interact. 5: 525–527 [Google Scholar]
- Stewart C.N., Jr., andVia L.E. (1993). A rapid CTAB DNA isolation technique useful for RAPD fingerprinting and other PCR applications. Biotechniques 14: 748–750 [PubMed] [Google Scholar]
- Sun Q.Y., Ding L.W., Lomonossoff G.P., Sun Y.B., Luo M., Li C.Q., Jiang L.W., Xu Z.F. (2011). Improved expression and purification of recombinant human serum albumin from transgenic tobacco suspension culture. J. Biotechnol. 155: 164–172 [DOI] [PubMed] [Google Scholar]
- Teng K.L., Chen H., Lai J.B., Zhang Z.H., Fang Y.Y., Xia R., Zhou X.P., Guo H.S., Xie Q. (2010). Involvement of C4 protein of beet severe curly top virus (family Geminiviridae) in virus movement. PLoS ONE 5: e11280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trebicki P., Harding R.M., Rodoni B., Baxter G., Powell K.S. (2010). Vectors and alternative hosts of tobacco yellow dwarf virus in southeastern Australia. Ann. Appl. Biol. 157: 13–24 [Google Scholar]
- Upton Z., Cuttle L., Noble A., Kempf M., Topping G., Malda J., Xie Y., Mill J., Harkin D.G., Kravchuk O., Leavesley D.I., Kimble R.M. (2008). Vitronectin: Growth factor complexes hold potential as a wound therapy approach. J. Invest. Dermatol. 128: 1535–1544 [DOI] [PubMed] [Google Scholar]
- Werner S., Breus O., Symonenko Y., Marillonnet S., Gleba Y. (2011). High-level recombinant protein expression in transgenic plants by using a double-inducible viral vector. Proc. Natl. Acad. Sci. USA 108: 14061–14066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolffe A.P., Matzke M.A. (1999). Epigenetics: Regulation through repression. Science 286: 481–486 [DOI] [PubMed] [Google Scholar]
- Woodard S.L, et al. (2003). Maize (Zea mays)-derived bovine trypsin: Characterization of the first large-scale, commercial protein product from transgenic plants. Biotechnol. Appl. Biochem. 38: 123–130 [DOI] [PubMed] [Google Scholar]
- Zhang X.R., Mason H. (2006). Bean Yellow Dwarf Virus replicons for high-level transgene expression in transgenic plants and cell cultures. Biotechnol. Bioeng. 93: 271–279 [DOI] [PubMed] [Google Scholar]