Natural variation leading to differences in leaf morphology between domesticated tomato and a wild relative is explored in a set of introgression lines. The phenotypic context of leaf morphology with other traits is examined at the whole-plant level, with implications for organ-specific breeding efforts.
Abstract
Introgression lines (ILs), in which genetic material from wild tomato species is introgressed into a domesticated background, have been used extensively in tomato (Solanum lycopersicum) improvement. Here, we genotype an IL population derived from the wild desert tomato Solanum pennellii at ultrahigh density, providing the exact gene content harbored by each line. To take advantage of this information, we determine IL phenotypes for a suite of vegetative traits, ranging from leaf complexity, shape, and size to cellular traits, such as stomatal density and epidermal cell phenotypes. Elliptical Fourier descriptors on leaflet outlines provide a global analysis of highly heritable, intricate aspects of leaf morphology. We also demonstrate constraints between leaflet size and leaf complexity, pavement cell size, and stomatal density and show independent segregation of traits previously assumed to be genetically coregulated. Meta-analysis of previously measured traits in the ILs shows an unexpected relationship between leaf morphology and fruit sugar levels, which RNA-Seq data suggest may be attributable to genetically coregulated changes in fruit morphology or the impact of leaf shape on photosynthesis. Together, our results both improve upon the utility of an important genetic resource and attest to a complex, genetic basis for differences in leaf morphology between natural populations.
INTRODUCTION
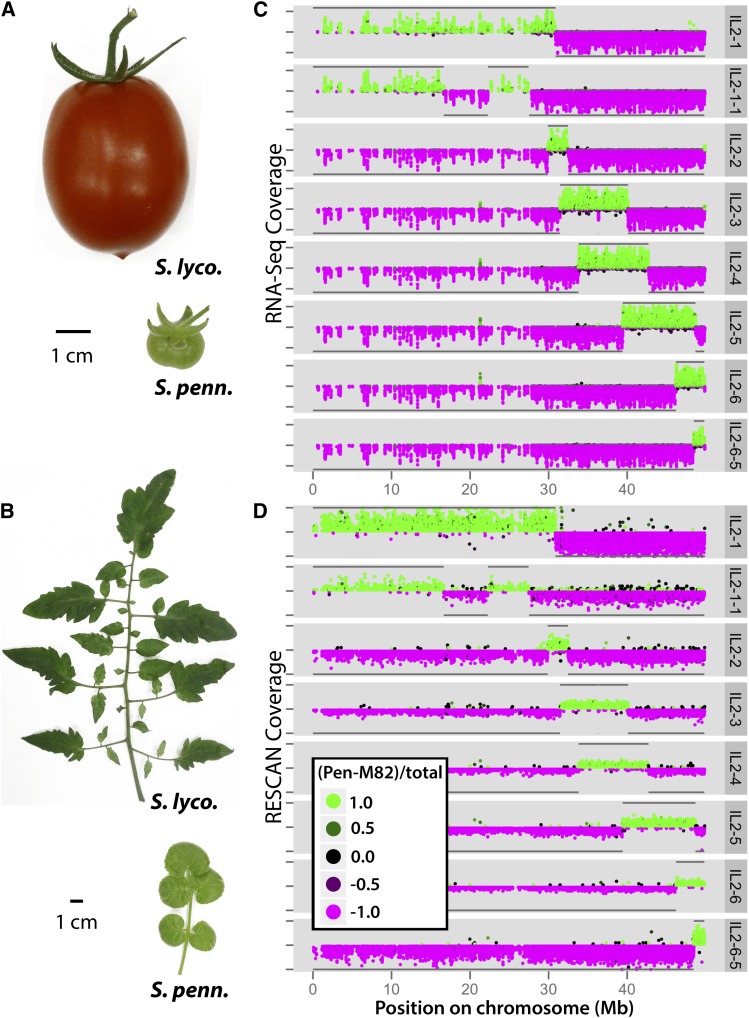
The tomato clade (Solanum sect. Lycopersicon) provides unique opportunities to study natural variation. As one of the world’s most important crops, intense focus has been dedicated to the genetic analysis of fruit size, shape, and sugar content between domesticated tomato (Solanum lycopersicum) and wild relatives (Frary et al., 2000; Fridman et al., 2004; Xiao et al., 2008). One of the most distant wild relatives of domesticated tomato, Solanum pennellii, originated in the deserts of Peru. When comparing tomato to a relative that inhabits such an extreme environment, other phenotypic differences undoubtedly underlie the successes of these two species (Moyle, 2008; Chitwood et al., 2013; Koenig et al., 2013). Changes in drought resistance, disease resistance, and water use efficiency, among many other traits, have enabled these two species to thrive in their respective environments. As much as the fruits of these two species differ (Figure 1A), so do their vegetative organs, such as leaves, which exhibit pronounced differences in size, complexity, and morphology (Figure 1B).
Figure 1.
Phenotypic Differences between IL Parents and RNA-Seq and RESCAN Data for Chromosome 2 ILs.
Phenotypic differences in the fruit (A) and leaves (B) between domesticated tomato (S. lycopersicum) and a wild relative (S. pennellii). Beyond obvious differences in the size, shape, and color of fruits are differences in metabolite content. Leaves between these species vary in size, complexity, and shape and non-cell-autonomously provide the majority of photosynthate to fruits. Shown are the S. pennellii introgression regions for ILs covering chromosome 2 as determined by two methods: RNA-Seq (C) and RESCAN (D). The depth of coverage (distance from midpoint on y axis) and genotype (color and direction on y axis) of each SNP/indel is plotted against chromosomal position (x axis). Polymorphisms that match S. pennellii are colored green and plotted on the top half of each IL panel, while polymorphisms matching cv M82 are plotted in magenta in the bottom halves. The coloring is on a continuum such that the color approaches black as a position’s genotype approaches heterozygosity. The y axis tick marks indicate depths of coverage ranging from 0 to 100 (C) or 0 to 20 (D). Subsequent to genotyping, introgression boundaries consistent between the RNA-Seq and RESCAN analyses were delineated. Using these breakpoints, S. pennellii and cv M82 regions are summarized by horizontal lines at the top and bottom of each IL panel, respectively.
Surprisingly, few studies have explicitly studied quantitative trait loci (QTL) regulating leaf traits, in any species. Such phenotypes are associated with water use efficiency and thermoregulation, traits important to yield (Nicotra et al., 2011; Chitwood et al., 2012a). Studies examining leaf morphology are often limited to analyses of size, dimensions of length and width, and complexity (Jiang et al., 2000; Pérez-Pérez et al., 2002; Holtan and Hake, 2003; Frary et al., 2004). Recently, a genome-wide association study using the maize (Zea mays) nested association mapping population identified liguleless genes as regulators of upright leaf angles. In addition to leaf angle, leaf length and width are regulated by many loci of small effect with little epistasis (Tian et al., 2011). If the complement of genetic changes responsible for differences in leaf shape between species is to be fully understood, similar quantitative genetics approaches will be required in the future. Although length, width, and the dimensions of leaves are important, natural variation in leaf morphology is immense, and methods to quantify the entirety of shape variance are required to determine the full complement of genes regulating differences in populations (Langlade et al., 2005; Chitwood et al., 2012b, 2012c, 2012d). Ultimately, the morphology of leaves is determined at the cellular level, and the genetics underlying natural variation in cellular traits are only now beginning to be examined (Massonnet et al., 2011; Sterken et al., 2012).
Despite the disparate phenotypic differences and ecological habitats occupied by species in the tomato clade, most species are interfertile (Stevens and Rick, 1986), a property that has been exploited to create introgression lines (ILs) between wild tomato species and domesticated cultivars. A unique property of many such lines is that they contain a single, defined, introgressed genomic region from a wild species donor in an otherwise domesticated background (Eshed and Zamir, 1995; Liu and Zamir, 1999; Monforte and Tanksley, 2000; Canady et al., 2005). A set of such ILs that tile genomic segments from desert-adapted S. pennellii into domesticated S. lycopersicum cv M82 (Eshed and Zamir, 1995; Liu and Zamir, 1999) has been extensively phenotyped, amassing a plethora of QTL. The S. pennellii ILs have been used to map QTL for metabolites, enzymatic activity, yield, and fitness traits as well as the genetic basis of heterosis (Rousseaux et al., 2005; Schauer et al., 2006, 2008; Semel et al., 2006; Stevens et al., 2007; Steinhauser et al., 2011). Ultrahigh-density genotyping of the ILs is a first step toward understanding the whole plant relationships that underlie domesticated traits, but our knowledge will always be limited by phenotype (Chitwood and Sinha, 2013; Zamir, 2013).
Here, we precisely define the boundaries of the S. pennellii ILs at both the genomic and transcriptomic levels. Importantly, the combination of ultrahigh-density genotyping with the recently completed tomato genome allows the exact gene content of these ILs to be determined, aiding breeding efforts and the molecular characterization of QTL. Using precisely defined ILs, we undertake a comprehensive phenotyping of leaf traits, from the organ to cellular level. Measuring leaf shape, size, complexity, and serration traits, in addition to pavement cell morphology and stomatal density and patterning, we detect 1035 QTL, 826 toward the direction of S. pennellii and 209 transgressive, beyond the phenotype of the domesticated parent. We observe distinct, highly heritable aspects of leaf shape and show that leaf shape, serration, and complexity can be genetically separated, contrary to previous findings from mutagenesis-based approaches. We additionally observe a relationship between pavement cell size and stomatal density, suggesting that modulation of cell size may be a mechanism to alter the spacing of stomata in natural populations. Finally, we analyze our phenotypes within the context of previously reported metabolic, enzymatic, and whole-plant phenotypes, finding an association between leaf complexity and shape with mono- and disaccharide levels in the fruit pericarp. RNA-Seq analysis of gene expression in the vegetative apices of the ILs demonstrates an association between the expression of developmental and photosynthetic pathways with this constellation of traits. The results suggest that leaf morphology can modulate photosynthetic efficiencies and/or that natural variation regulating the shape of leaves affects fruit morphology, which in turn affects the accumulation of fruit sugar. Our results improve upon an important, stable genetic resource and offer insights into not only the quantitative genetic basis of leaf shape, but also its phenomic context at a whole plant level.
RESULTS
Fine-Scale Genotyping of ILs
For fine-scale genotyping of the 76 ILs, we generated a database of polymorphisms between the domesticated tomato species S. lycopersicum cv M82 and its wild relative, S. pennellii (see Supplemental Data Set 1 online). Single nucleotide polymorphisms (SNPs) and indels between species were identified using RNA-Seq and reduced representation genomic sequencing, hereafter referred to as restriction enzyme sequence comparative analysis (RESCAN; Monson-Miller et al., 2012; Seymour et al., 2012). Taking both approaches together, we identified ∼750,000 polymorphisms between cv M82 and S. pennellii (see Supplemental Table 1 online). Ninety-nine percent of the polymorphisms identified have at most a single gene separating them, saturating coverage at the level of genetic loci.
RNA-Seq and RESCAN data for the 76 ILs and detected SNPs were used to genotype each IL across the entire genome (data available at www-plb.ucdavis.edu/Labs/sinha/TomatoGenome/Resources.htm). A graphical summary of the S. pennellii introgressions for all the ILs shows that they tile over nearly the entire tomato genome (see Supplemental Figure 1 and Supplemental Table 2 online).
For each IL, we plotted the genotype of polymorphisms for all relevant chromosomes (ILs with introgressions on chromosome 2 are shown in Figure 1; see Supplemental Figures 2 and 3 online for all other chromosomes). The RNA-Seq and RESCAN-based genotyping results are consistent with and complement one another. Our RNA-Seq–based genotyping (Figure 1C) has a higher relative depth of coverage, aiding polymorphism identification, although a smaller portion of the genome was sequenced. RESCAN-based genotyping (Figure 1D), which yields a more even distribution of polymorphisms and includes nongenic regions.
Noncontiguous Introgressions and Bins
Ultrahigh-density genotyping revealed that seven ILs have multiple introgressions. The majority of the additional introgressions are on the same chromosome as the primary introgression (for example, IL2-1-1 and IL2-3; Figure 1; see Supplemental Figure 4 online); however, we found that IL9-3-1 has an ∼100-kb introgression from S. pennellii at the top of chromosome 12 (see Supplemental Figures 2L and 5 online).
ILs harboring multiple introgressions have important implications for genetic mapping. Unique overlapping regions between introgressions define smaller intervals than the ILs, termed “bins.” The unique combinations of ILs that define a bin can dissect a QTL into considerably smaller intervals than the ILs themselves (Liu and Zamir, 1999; see Supplemental Data Set 2 online). Importantly, because of the additional introgressions, we changed the nomenclature of our bins compared with the original bins. As much of the literature uses the old bin designations and cannot be changed retroactively, we name our bins with a “d-” prefix (as in d-5E), denoting “Davis, CA.” The moniker is critical, as the bin names between the old and new systems do not correspond. Whereas previously the 76 S. pennellii ILs defined 107 bins, precisely defined IL boundaries reveal 112 bins (see Supplemental Figures 4 and 5 online). The majority of bins harbor <500 annotated genes (median = 177 genes, mean = 295.03 genes; see Supplemental Figure 6 and Supplemental Data Set 3 online). Increased bin numbers are caused by ILs with multiple introgressions where previously only one had been detected and slightly different boundaries between borders of ILs that were thought to be shared. In some instances, bins are noncontiguous, especially in the case of ILs harboring multiple introgressions (for example, d-2G follows the IL2-3 split; see Supplemental Figure 4 online) and when a smaller introgression lies completely within a larger introgression (for example, d-10A is divided by IL10-1-1; see Supplemental Figure 5 online). Precise knowledge of IL boundaries allows the gene content of bins to be known with near certainty. A list of annotated genes within the newly defined bins is provided (see Supplemental Data Set 4 online).
Heritability and Detected QTL
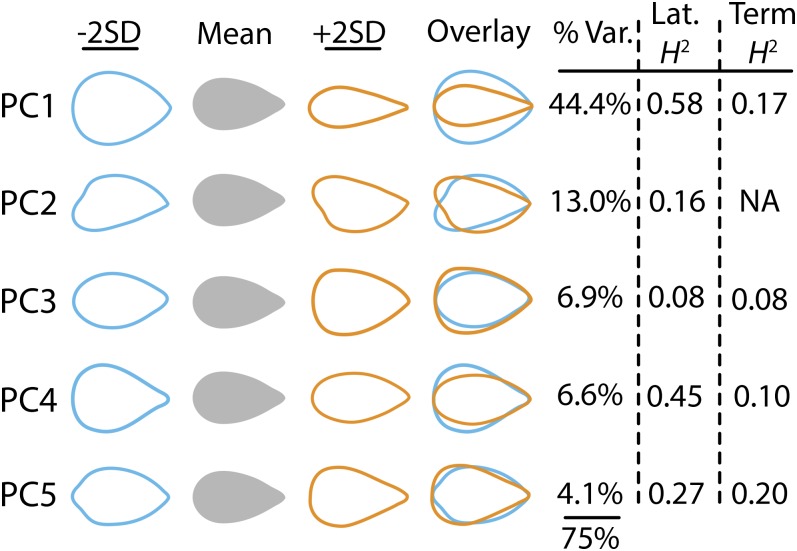
Having precisely defined the introgression boundaries, we next used this resource to determine the genetic basis underlying natural variation in leaf form, measuring a comprehensive suite of leaf traits in the ILs. A number of the traits are derived from outline analyses of terminal (“Term”) and distal lateral (“Lat”) leaflets from field-grown ILs. These include size (TermLfSize and LatLfSize) and measures of serration/lobing (LftCirc and LftSolid). To globally measure the differences in shape between leaflets, we used an elliptical Fourier descriptor (EFD) approach followed by a principal component analysis (Iwata et al., 1998; Iwata and Ukai, 2002). Over 11,000 leaflets were measured, including the terminal and left and right distal lateral leaflets, at a pseudoreplication of 15 leaflets per individual and replication of 10 individuals per IL (raw photos available at www-plb.ucdavis.edu/Labs/sinha/TomatoGenome/Resources.htm). The principal components resulting from the EFD analysis describe intuitive, distinct aspects of shape segregating in the ILs, which we treat as traits (TermPC1-5 and LatPC1-5; Figure 2; Chitwood et al., 2012b, 2012c, 2012d). Together, the five principal components considered in this article explain ∼75% of all leaf shape variance observed.
Figure 2.
Principal Components Resulting from an Elliptical Fourier Descriptor Analysis of Field Leaflet Shapes.
Shown are the first five PCs, explaining 75% of all shape variance from >11,000 field-grown leaflets. Given is the percentage of variance in shape each PC explains and the heritability values of each PC, for lateral (Lat) and terminal (Term) leaflets. PC1 explains a large amount of all shape variance (44.4%) and relates to the length-to-width ratio of leaflets (similar to LftAR). PC2 (13.0%) explains asymmetry relating to the sampling of left and right distal lateral leaflets, which on average are mirror images of each other. The remaining PCs explain shape variance relating to the proximal-distal distribution of blade outgrowth along the leaflet. For all PCs, heritability is greater in the distal lateral leaflets relative to the terminal leaflet. PCs vary in heritability, and the most heritable PCs are 1 and 4. NA, not applicable.
Additionally, we provide measures of leaflet length-to-width ratio (LftAR and LftRound), leaf complexity (CompPri, CompSec, CompInt, CompRachis, and CompAll), pavement cell size (CotPaveArea and CotPaveCnt), pavement cell morphology (CotPaveAR, CotPaveRound, CotPaveSolid, and CotPaveCirc), stomatal density (CotStom, LfAdStom, and LfAbStom), stomatal patterning (CotStomInd), and flowering time (FlowTime) in the ILs (see Supplemental Figure 7 and Supplemental Data Sets 5 and 6 online). An extensive list detailing traits, what they represent, and how they are statistically modeled is provided for reference (see Supplemental Data Set 7 online; trait information has been deposited at Phenom-Networks, www.phenome-networks.com). Correlation analysis and hierarchical clustering suggest that our traits fall into three main categories. The first category includes leaflet size and leaf complexity measures, the second pavement cell-related traits and stomatal density, and the third leaflet length-to-width ratio, serration, and shape (see Supplemental Figure 8 online).
The traits we measure vary widely in their broad-sense heritabilities. Previous reports analyzing measures of leaf length and width in maize (Tian et al., 2011) and shape analysis in Antirrhinum majus (Langlade et al., 2005) suggested a high genetic component to the variance in leaf morphology in these species. Our results are similar; the highest broad-sense heritability values are observed in measures of leaflet serration and length-to-width ratio (see Supplemental Figure 9 online). Specifically, two distinct shape components, PC1 and PC4, from the EFD analysis of leaflet shape are highly heritable, and as discussed later, show unique interactions with metabolite and yield-associated traits. Leaf complexity and size, as well as leaf stomatal density, exhibit intermediate heritability, whereas most of the cellular traits we measured have low heritability. This low heritability may not reflect a small genetic component to these phenotypes, but rather the high variance in these measurements due to the inability to account for micropatterning across the entirety of the leaf surface, and, as we show, robust QTL for such traits can still be determined.
In total, we detect over 1000 QTL at a significance level of <0.05. 829 of these QTL are in the direction toward S. pennellii. Because the directionality of traits was determined by comparisons with S. pennellii grown in the greenhouse (because of the difficulty growing this species under Davis, CA field conditions), whether QTL are transgressive beyond S. pennellii cannot be determined. However, 209 QTL are transgressive beyond cv M82 (see Supplemental Figure 10 online). Of particular note is the numerous QTL that reduce leaf complexity and induce shape changes toward that of S. pennellii. The results suggest that large portions of the genome contribute to natural variation in leaf complexity and shape and that these polygenic contributions can act additively (although a limited role for epistasis in the ILs cannot be discounted).
QTL Regulating Leaf Shape, Complexity, and Serration
No IL comes close to approximating the shape of a S. pennellii leaflet, the longest axis of which lies perpendicular to cv M82 (see Supplemental Figure 11 online). S. pennellii leaflets are also more orbicular and lack the distinct deltoid tip and lobing of a domesticated tomato leaflet. Because the ILs tile the tomato genome and a majority of ILs possess a significant shape QTL (see Supplemental Figure 10 online), this reinforces the idea that leaflet shape is highly polygenic with an important additive component.
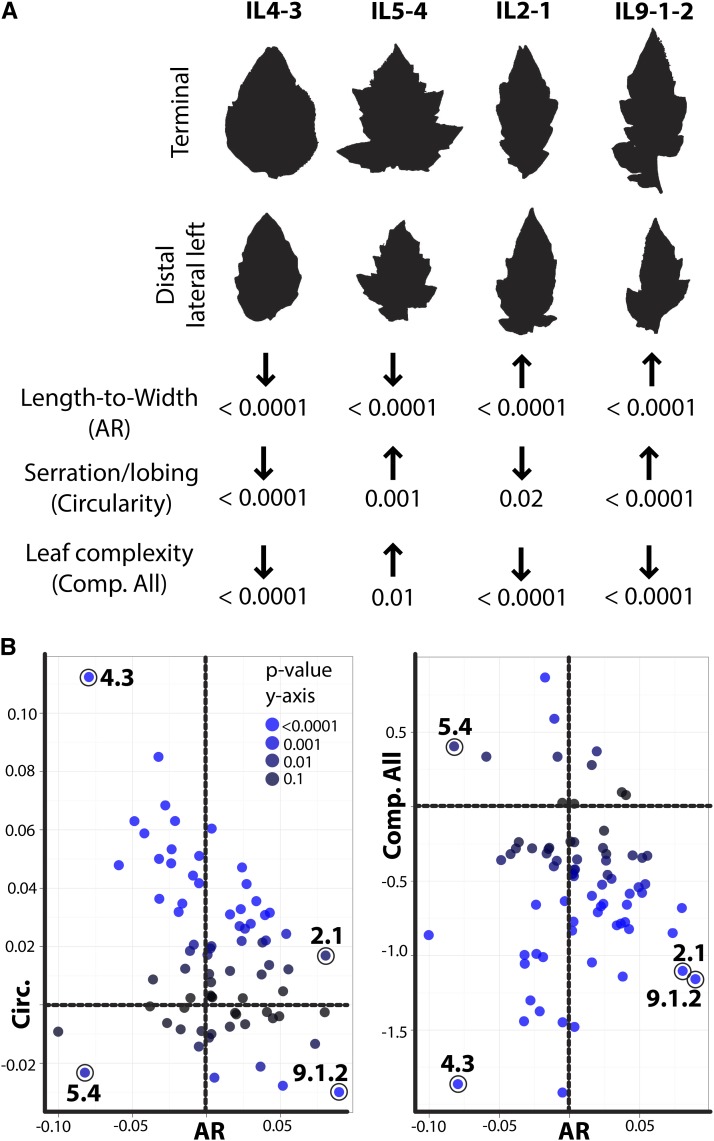
The largest contributing loci to leaflet shape in the IL population include ILs 4-3 and 5-4, which are much wider than cv M82, and ILs 2-1 and 9-1-2, which are transgressively narrower than cv M82 leaflets (see Supplemental Figure 11A online). A close comparison of the averaged leaflet outlines of the two widest and two narrowest ILs against each other reveals that these ILs alter their shape in distinct ways (see Supplemental Figure 11B online). For example, the tip of IL5-4 remains much more distinct than that in IL4-3, while increasing its wideness at the base of the leaflet. Similarly, IL2-1 and 9-1-2 vary distinctly in the degree of constriction at their proximal ends. These four ILs represent four different extremes in PC1-PC4 space. Although a simplification, PC1 explains shape variance relating to overall length-to-width ratio changes, whereas PC4 tends to explain variance relating more to the distinctness in shape between ILs, such as the distribution of laminar outgrowth along the proximal-distal axis and cordate bulges at the base of leaflets (Figure 2).
Extensive developmental and mutagenesis-based approaches in model systems suggest that a suite of leaf morphology features, including serration, complexity, and laminar outgrowth, are under similar genetic regulation, including the activities of auxin and KNOX, CUC, and TCP family members (Barkoulas et al., 2007). How do these leaf features behave within the context of segregating natural variants? At least one IL, IL4-3, possesses significant QTL in the S. pennellii direction for all traits measured, including significantly decreased length-to-width ratio (influenced by laminar outgrowth), decreased serration and lobing (as measured by circularity and solidity), and decreased leaf complexity counts. Nonetheless, serration and shape are genetically separable. For example, IL5-4 is significantly wider than cv M82 but is transgressively more serrated and has increased leaf complexity (Figure 3). Looking at representative leaflets, it becomes apparent that measurements of serration and lobing versus shape impinge upon each other to some degree; for example, the increased lobing in IL5-4 creates proximal lobes that likely contribute toward its increased width. The varying combinations of leaflet shape, serration, and complexity are also present in the transgressively thinner ILs 2-1 and 9-1-2. That these leaf morphology traits segregate independently from each other suggests that either unique genes contribute to natural variation in these features or that the spatiotemporal regulation of known factors is modulated independently from each other.
Figure 3.
Leaflet Shape, Serration, and Leaf Complexity Are Genetically Distinct Components.
(A) Representative leaflets from ILs with significant shape QTL. Given are the direction and significance of length-to-width ratio change relative to cv M82 (AR), serration/lobing (circularity), and leaf complexity. Note that despite considerable accumulated genetic evidence suggesting otherwise, these features do not follow each other, and different ILs exhibit different combinations of these traits.
(B) Graphs demonstrating the breaking between AR, circularity (Circ.), and complexity (Comp.). In each graph, AR is on the x axis for comparison showing IL values for circularity and complexity on the y axis. Colors indicate significance of trait deviations for the y axis.
[See online article for color version of this figure.]
QTL Regulating Cellular Phenotypes
Natural variation in organ shape must arise during development from differences in the patterning, division, and expansion of cells. Additionally, cellular features are important for adaptations to abiotic conditions, such as the patterning and response of stomata to the xerophytic conditions found in the native habitat of S. pennellii (Heichel and Anagnostakis, 1978). To measure natural variation at the cellular level in the ILs, we analyzed the size of pavement cells, their shape characteristics, and stomatal density and patterning on the adaxial side of cotyledons. We additionally measured the density of stomata on the adaxial and abaxial sides of mature, field-grown leaves.
One IL in particular, IL10-3, consistently exhibits extreme phenotypes for a number of cellular features (see Supplemental Figure 12 online). Like S. pennellii, IL10-3 has a significantly lower stomatal density on the adaxial side of cotyledons and true leaves, and it also possesses the largest pavement cell size measured in the ILs. Beyond the developmental interest of adaxial stomatal density, IL10-3 additionally possesses significantly reduced stomatal density on the abaxial side of true leaves, an important adaptive trait considering the desert habitat of S. pennellii (see Supplemental Data Sets 5 and 6 online). Interestingly, the stomatal index of IL10-3 is not significantly different from cv M82 (“CotStomInd”; see Supplemental Data Sets 5 and 6 online), suggesting that the number of stomata per pavement cell is not the major cause of these phenotypes and that the larger pavement cell size pushes the stomata away from each other, reducing their density. The significant correlations between pavement cell size and stomatal density (see Supplemental Figures 8 and 13 online) may represent an evolutionary mechanism to modulate stomatal spacing.
Bin Mapping and Gene Candidates
Bin mapping can be a qualitative endeavor. If one IL possesses a significant phenotypic difference from cv M82 and an overlapping IL does not, then a QTL interval can be narrowed by exclusion. Similarly, a shared region between ILs with similar phenotypes can be used to delimit a QTL by parsimony. Applying exclusion and parsimony can be difficult when dealing with QTLs, though. For example, is an IL not significant enough to use to exclude a region, or conversely is a QTL significant enough to apply parsimony? To help solve this predicament, for each bin, we use a marginal regression approach, regressing bin genotypes of individuals against their trait values (see Supplemental Figures 14A to 14D online). This allows a probability value to be assigned to each bin based on its correlation with a trait. This method is only to be used as an aid in addition to IL-based mapping approaches: Because each bin is defined by, at most, a handful of ILs, the P values assigned to bins in this manner are influenced by the ILs that define them. Graphs simultaneously showing IL and bin mapping results (to compare and integrate these two methods) are provided (see Supplemental Figures 15 to 47 online).
Bin gene content, combined with IL and bin mapping, can lead to candidate genes causal for the QTL of interest. One example is SELF-PRUNING 5G (SP5G), which in our bin map resides on bin d-5E. With respect to flowering time, a QTL can be narrowed down to this bin, as IL5-4 (but not ILs 5-3 or 5-5) takes significantly more days to flower than cv M82 (see Supplemental Figure 14A online). As this bin encompasses only 36 genes and SP5G is a FLOWERING LOCUS T homolog (Carmel-Goren et al., 2003), it is a prime candidate for causing the increased flowering time (see Supplemental Figure 14E online). Moreover, SP5G cosegregates for the pht5.4 QTL regulating plant height and is tightly linked to the OBSCURAVENOSA locus, and it has been suggested that the tight linkage of these two loci contributes to the coincident features of compact plant habit and chloroplast-obscured venation in processing tomato varieties (Jones et al., 2007). Similarly, we observe leaf phenotypes in IL5-4 as well, including wider leaves (see Supplemental Figure 11 online) with more serration and complexity (Figure 3). Given previously demonstrated connections between flowering time pathways and leaf morphology (Willmann and Poethig, 2011), it is not unreasonable to suspect SP5G as a modulator of leaf shape in addition to flowering time.
Not all bins are as small as the aforementioned, but knowing the gene content of an interval can provide a list of potential candidates regulating traits. For example, bin d-9B is inferred to possess a QTL that increases leaflet aspect ratio (AR; see Supplemental Figure 14B and LatPC1 in Supplemental Figure 31 online). An ARF16 homolog, a modulator of leaflet width and hyponasty in Arabidopsis, lies in this interval (Liu et al., 2011). Similarly, d-8F is an interval regulating leaflet AR as well (see Supplemental Figure 14C online). Within this bin lies a homolog of FILAMENTOUS FLOWER/GRAMINIFOLIA, a regulator of laminar outgrowth, as well as an ATHB-2 homolog, which in addition to its role in the shade avoidance response, also modulates leaf width (Siegfried et al., 1999; Steindler et al., 1999; Golz et al., 2004; Eshed et al., 2004). As adaxial-abaxial polarity determinants modulate leaf complexity in tomato (Kim et al., 2003a, 2003b), the inclusion of LEUNIG and KANADI2 homologs (Kerstetter et al., 2001; Cnops et al., 2004; Stahle et al., 2009) on the d-8A interval, which regulates leaf complexity, is interesting (see Supplemental Figure 14D online).
Identifying causal genes for QTL obviously requires fine-mapping, but the delimitation of the exact gene content in bins (see Supplemental Data Set 4 online) provides a powerful starting place to begin studies of natural variation in tomato.
Associations between Leaf Morphology and Fruit Sugar Metabolism
One of the advantages of a true-breeding genetic resource, such as the S. pennellii ILs, is the ability to meta-analyze phenotypic data sets with a common genetic basis (Zamir, 2013). Perhaps the most interesting relationship between phenotypes that has been established by such studies (Schauer et al., 2006, 2008; Steinhauser et al., 2011; Toubiana et al., 2012) is a prominent negative correlation between harvest index (the ratio of fruit yield to overall biomass) and metabolite levels in the pericarp. Generally, the more biomass of a plant dedicated to fruit production, the lower the metabolite concentrations in the fruit. The antagonism between metabolite levels and harvest index obviously bodes badly for breeding efforts to increase both of these critical traits simultaneously. However, the relationship is understandable, especially if viewed from the perspective of limited resources, nutrient allocation, and effects of metabolite dilution at the whole-plant level. Critical to the understanding of these whole plant relationships is detailed knowledge of not only nutrient sinks (i.e., fruits, seeds, and flowers) but the ultimate source (leaves).
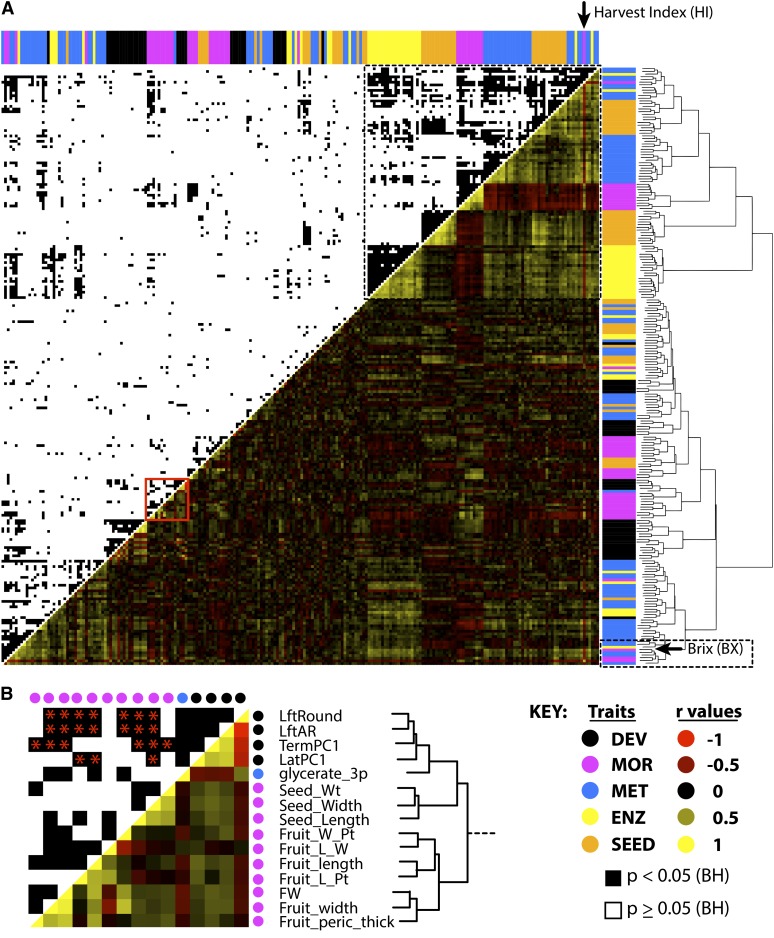
To better understand the role that leaves play in these relationships, we performed a correlation analysis of leaf traits with the existing phenomics database (Phenom-Networks, www.phenome-networks.com; Figure 4; see Supplemental Figures 48 and 49 online). We divide all traits analyzed into five major groups, defined by the studies from which they are reported and the phenotype that they measure. Largely, traits belonging to a group describe related phenotypes, but for consistency they are first defined by the study from which they originate, the authors’ terminology in those studies, and most importantly their class designations in the Phenom-Networks database. “MET” traits are derived from Schauer et al. (2006, 2008) and measure metabolite levels in the fruit pericarp. “MOR” traits (for “morphology,” using the nomenclature of Schauer et al. [2006, 2008] and Phenom-Networks) include both yield-related traits and explicit morphological measurements of fruits and flowers. The term “morphology” is used loosely and in contrast with the “metabolite” traits also measured by Schauer et al. (2006, 2008). For example, “MOR” traits include fruit Brix, earliness (the ratio of red fruit yield to total fruit yield), and plant weight, even though these are not strictly morphological features. “ENZ” traits, derived from Steinhauser et al. (2011), measure enzymatic activities in the fruit pericarp, and “SEED” traits, reported by Toubiana et al. (2012), measure metabolite levels in seeds. Traits described in this article were termed “DEV” traits because of their relevance to leaf development. MET, MOR, ENZ, and SEED traits (represented in blue, magenta, yellow, and orange in figures, respectively) are described in Supplemental Data Set 8 online and their values and correlations with other traits provided in Supplemental Data Sets 9 to 11 online. DEV values are provided in Supplemental Data Set 5 online and described in Supplemental Data Set 7 online.
Figure 4.
Relationship between Leaf Morphology and Previously Measured IL Traits.
(A) Hierarchical clustering of leaf traits with previously studied traits. DEV (black), leaf development traits from this study; MOR (magenta), whole-plant, yield, and reproductive morphological traits as described by Schauer et al. (2006, 2008); MET (blue), metabolic traits described in the same studies; ENZ (yellow), enzymatic activities, as measured by Steinhauser et al. (2011); SEED (orange), seed metabolites, as described by Toubiana et al. (2012). Hierarchical clustering is based on absolute correlation values, with red denoting negative Pearson correlation coefficients and yellow positive. The top half of the plot shows significant correlations (<0.05) between traits after global multiple test adjustment, indicated in black. Trait identities are indicated as a marginal rug plot along the sides of the graph. The large group of highly correlated traits (in the top right-hand corner, indicated by the dotted line) is consistent with previous reports of negative correlation between MOR traits (including harvest index, [HI], indicated by an arrow) with fruit metabolite levels. DEV traits cluster away from this previously described relationship and closely associate with Brix, plant weight, and mono- and disaccharide levels, indicated by the dotted lined box toward the bottom of the graph. A more detailed view of the hierarchical clustering is found in Supplemental Figure 50 online.
(B) Detailed analysis of the clustering reveals unexpected whole-plant relationships between traits. For example, LftAR, LftRound, and PC1 (all highly heritable traits describing leaflet length-to-width ratio) most closely cluster with traits relating to the dimensions of seeds and fruit, suggesting that the morphology of disparate organ types is regulated by common genetic elements. Relevant significant correlations, as multiple test–adjusted for the traits shown, are shown with red asterisks. The traits represented in (B) are indicated in (A) by a red box.
Hierarchical clustering of the mean z-scores for traits reveals a strong negative relationship of harvest index and yield-associated (MOR) traits with metabolite levels (MET; dotted box, upper right-hand corner of Figure 4A), demonstrating the robustness of this previously described relationship (Schauer et al., 2006, 2008; Toubiana et al., 2012). The metabolites exhibiting the strongest negative correlation with harvest index and MOR traits are related to nitrogen and amino acid metabolism in both the fruit and seed (for a close-up of the trait identities in Figure 4A, see Supplemental Figure 50 online; amino acids indicated by asterisk).
Leaf traits (DEV; black) cluster exclusively outside of the aforementioned complex of phenotypes (i.e., the strong negative relationship between harvest index and metabolites; Figure 4A). As DEV traits were not considered in previous analyses, the traits from other classes that cluster with DEV traits are informative as to the importance of leaf traits as correlates of metabolism and yield. For example, a small group of highly heritable DEV traits explaining the length-to-width ratio of leaflets (LftAR, LftRound, LatPC1, and TermPC1) cocluster and are significantly correlated with MOR traits related to length-to-width ratio in fruit and seeds (Figure 4B, red box in Figure 4A). Length-to-width ratio is the major source of shape variance (>40%) in field-grown leaflets (PC1; Figure 2). Such correlation suggests that leaf shape is not independent from the genetic basis of morphology in disparate organs, with implications for the independent modulation of organ shapes during evolution. As we discuss below, changes in either leaf or fruit morphology can explain correlations we observed with fruit sugar, demonstrating the difficulty of organ specific breeding efforts.
Brix and pericarp levels of Suc, Glc, Fru, Gal, mannose, and trehalose (small dotted box in lower right-hand corner in Figure 4A; see Supplemental Figure 50 online) cluster away from the previously described constellation of harvest index and metabolite antagonisms. Also included in this cluster are plant weight and earliness. This suggests a more prominent relationship between carbon metabolism and leaves than the previously found connection between nitrogen metabolism and harvest index. A more detailed analysis confirms the special relationship between leaf complexity and shape with sugar metabolism in the fruit (see Supplemental Figure 48 online), and jackknifing suggests that the correlations are robust and not artifacts resulting from undue influence of a few ILs (see Supplemental Figure 51 and Supplemental Data Set 12 online). Additionally, if only correlation between leaf development traits with traits from other classes are considered, then not only do Brix and sugar levels in the fruit exhibit the highest connectivity with leaf complexity and shape, but they are among the most significant correlations (see Supplemental Figure 49 online).
Association between Photosynthetic Gene Expression and Leaf Morphology
Leaf morphology may correlate with sugar accumulation in the fruit due to a variety of factors, indirectly and directly involving leaves. (1) The coregulation of fruit and leaf morphology through similar gene regulatory networks (especially considering that fruits are modified leaves) may lead to shape changes in the fruit that affect the accumulation of sugars. (2) Natural variation in leaf morphology may affect photosynthetic efficiencies through physiological parameters (Nicotra et al., 2011; Chitwood et al., 2012a). The latter hypothesis is particularly intriguing considering that >80% of sugars in the fruit are produced directly by photosynthesis in leaves and subsequently translocated through the phloem (Heatherington et al., 1998; Lytovchenko et al., 2011). Nonetheless, fruit photosynthesis has been demonstrated to significantly affect the accumulation of fruit sugars (Powell et al., 2012), and the role of fruit morphology in this process remains to be more fully explored.
To explore these hypotheses, we correlated the gene expression levels in the 76 ILs, as measured in the vegetative apex using RNA-Seq, against other IL trait values (Figure 5A; see Supplemental Data Sets 13 to 15 online). After hierarchically clustering traits and the expression profiles of those genes significantly correlated with at least one trait (after multiple test correction; see Supplemental Data Set 16 online), a distinct group (indicated by an asterisk in Figure 5A) contained genes with numerous correlations to leaf development (DEV) traits. In addition to leaf complexity, LftCirc/LftSolid (measures of serration), and PC1/LftAR (length-to-width ratio), fruit Glc is represented among the traits significantly correlated with this group of genes (see Supplemental Figure 52 online).
Figure 5.
An Association between Photosynthetic Gene Expression and Leaf Morphology.
(A) Hierarchical clustering of traits and gene expression profiles in the vegetative apex measured across the 76 ILs. Gene expression profiles across ILs were regressed against traits and only those genes with at least one significant correlation with a trait were considered. Colors indicate significant correlation after multiple test adjustment with a gene expression profile and the class to which the correlated trait belongs (DEV, black/white; MOR, magenta; MET, blue; ENZ, yellow; and SEED, orange). One cluster of genes (indicated by an asterisk) significantly correlate with numerous DEV traits related to leaf development.
(B) Gene Ontology enrichment analysis of the gene group with an asterisk reveals numerous significant categories related to photosynthesis.
As might be predicted, potential regulators of leaf morphology are present in this group of genes, including ARF3/ETTIN, ARF4, AGO1, SAW1, BELL1, PIN5, and GRF7 homologs (see Supplemental Data Set 17 online), which regulate laminar outgrowth, patterning, indeterminacy, and cell expansion. Support for an intimate association between the genetic coregulation of leaf and fruit morphology with fruit sugar is apparent in genes such as AUXIN-RESPONSE FACTOR4 (ARF4), which modulate not only leaf shape through auxin and adaxial-abaxial pathways, but also the morphology of the fruit (Jones et al., 2002; Yifhar et al., 2012; Sagar et al., 2013).
A Gene Ontology enrichment analysis for genes within this group reveals numerous significantly enriched categories related to photosynthesis (Figures 5A and 5B; see Supplemental Data Set 18 online). It is unlikely that leaf complexity and shape modulate the levels of photosynthetic genes via changes in overall blade area given the negative correlation between leaf complexity and leaflet area (see Supplemental Figure 13 online). Rather, the correlation of photosynthetic gene expression with leaf development traits may reflect the influence of leaf morphology on photosynthetic efficiency physiologically. It is also possible that developmental gene regulatory networks impinge upon photosynthesis pathways more directly, independent of leaf shape. ARF4 again provides a striking example: Not only does this gene regulate both leaf and fruit morphology (Yifhar et al., 2012), but it also regulates the accumulation of chloroplasts and the greening of fruits (Jones et al., 2002), which were recently shown to affect sugar levels (Sagar et al., 2013). Considering the relationship between leaf and fruit shape, a fuller understanding of the causative factors underlying sugar accumulation will require a similar gene expression analysis in fruits to that performed here in vegetative apices.
DISCUSSION
Although breeding traits from wild relatives into domesticated lines is important in and of itself, the knowledge of the identities of genes regulating these traits can be incredibly powerful and explanatory (Frary et al., 2000; Fridman et al., 2004; Xiao et al., 2008; Kimura et al., 2008; Li and Chetelat, 2010). In this study, the high-density genotyping we perform at the genetic and transcriptomic levels elaborates upon previous genetic maps of the S. pennellii ILs and provides outstanding resolution of the recombination breakpoints that define their introgressions. In many instances, the increased resolution resulting from our genotyping yielded insights into the bin structure and the complement of genes harbored by the ILs. Precise knowledge of the genetic content of each IL is a prerequisite for positional cloning of QTL, reverse genetics, and genetic genomics. Together with the recently completed tomato genome (Tomato Genome Consortium, 2012), the means to begin understanding the genetic basis of natural variation in the tomato complex are coming into place (Moyle, 2008; Ranjan et al., 2012).
Despite the exhaustive study of harvest and fruit-related phenotypes, little has been done to study leaves in the S. pennellii ILs. Indeed, approaches utilizing natural variation to study leaf development are rare in any species (Jiang et al., 2000; Holtan and Hake, 2003; Kimura et al., 2008; Tian et al., 2011). Even rarer are studies of variation in leaf morphology by a quantitative means capable of describing total shape variance (Iwata et al., 1998; Iwata and Ukai, 2002; Langlade et al., 2005; Chitwood et al., 2012b, 2012c, 2012d). Our analyses reveal that leaf morphology is highly heritable and that the S. pennellii ILs are a valuable resource to study natural variation in leaves (Figure 2; see Supplemental Figure 9 online). Moreover, the drastic, but superficially simple shape differences in leaves between S. pennellii and S. lycopersicum cv M82 are regulated through a complex, polygenic genetic basis (see Supplemental Figure 10 online). For example, in addition to the simple regulation of length-to-width ratio, different QTL can impart distinct shapes by which leaflets modulate their width (see Supplemental Figure 11 online). Additionally, we demonstrate that leaflet serration and leaf complexity segregate independently from shape characteristics and even each other (Figure 3). This observation goes against the prevailing wisdom that leaf morphology is regulated by common genetic elements (Barkoulas et al., 2007) and suggests that only a small fraction of the genes regulating the tremendous variation of leaf morphology in natural populations has been discovered.
What mechanisms could explain the disparate association between leaf morphology and sugar levels in the fruit? A purely developmental connection between leaves and fruit is likely part of the explanation. Among other correlations we detect is a relationship between traits describing the length-to-width ratio of leaflets with traits explaining similar dimensions of fruit and seeds (Figure 4B). Considering the extensive analysis of fruit size and morphology in tomato (Frary et al., 2000; Cong et al., 2008; Xiao et al., 2008), it will be interesting to analyze genetic perturbations modulating fruit phenotypes for their effect on vegetative development and vice versa (Wu et al., 2011). Another possibility is that leaf development is altered by genetic changes in overall metabolism (Hackel et al., 2006) or that manipulation of carbon metabolism itself induces morphological changes in leaves (Tsai et al., 1997; Geigenberger et al., 2004; Lawson et al., 2006; Raines and Paul, 2006).
It is also possible that leaf morphology affects sugar metabolism in the fruit. Not only are leaves the ultimate source of most photoassimilates, but overwhelming evidence suggests that sugars are apoplastically unloaded from the phloem into the fruit (Fridman et al., 2004; Baxter et al., 2005; Hackel et al., 2006; Zanor et al., 2009), suggesting a directionality to the correlations we observe. The traits that correlate with fruit sugar levels are not related to ratios of biomass to yield (Do et al., 2010) or the size of leaves, but rather, sensu stricto, leaf shape and complexity. That the expression profiles of photosynthetic genes correlate with leaf morphology traits (Figures 5A and 5B) only bolsters the idea that leaf shape can modulate fruit sugar levels via a photosynthetic mechanism. Similar analyses to those presented here of photosynthetic gene expression in IL fruits are required to better resolve the roles of vegetative and reproductive organs in sugar accumulation.
Regardless of the mechanism, our results highlight an often overlooked fact: Leaves, as the major source of photoassimilate in the fruit (Heatherington et al., 1998; Lytovchenko et al., 2011; Powell et al., 2012) and as organs with a shared developmental foundation with carpels, are an important consideration for any breeding effort. Substantial theoretical and empirical evidence has accumulated that leaf shape and size play major roles in water use efficiency and thermoregulation (Parkhurst and Loucks, 1972; Givnish and Vermeij, 1976; Poorter et al., 2010; Nicotra et al., 2011; Chitwood et al., 2012a). Additionally, the relationship in the fossil record between temperature and precipitation with leaf serration and size further supports a functional significance of leaf shape (Bailey and Sinnott, 1915; Wolfe, 1971; Greenwood, 1992; Wilf et al., 1998). The environment imposed by agriculture upon domesticated species, including tomato, is radically different from that encountered by their wild ancestors. If the morphology of leaves matters with respect to photosynthetic efficiency, then leaf size and shape may have been bred as much as the crop, even when the crop is not the leaf, as in tomato. Just as leaf angle has been responsible for most yield increases in maize over the past century (Duvick, 2005; Tian et al., 2011), we propose that leaves may have affected the tomato fruit through developmental and photosynthetic mechanisms.
METHODS
Plant Materials, Growth Conditions, and Experimental Design
Second-generation Solanum pennellii ILs (Eshed and Zamir, 1995; Liu and Zamir, 1999) and Solanum lycopersicum cv M82 seeds were obtained from the Tomato Genetics Resource Center (University of California, Davis) and Dani Zamir (Hebrew University, Rehovot, Israel). For information about the seed stocks used for different lines and experiments, please see Supplemental Data Set 19 online.
In mid April, seed were washed in 50% bleach for ∼2 min, rinsed, and placed onto water-soaked paper towels in Phytatrays (Sigma-Aldrich) in preparation for field planting. Seeds were placed in darkness for 3 d before moving to a 16:8 light cycle in growth chambers for 4 d. Seedlings were then transplanted into 5 × 10 subdivided trays (11 × 22 inches) in Sunshine Mix soil (Sun Gro) in a greenhouse. Importantly, seedlings were transplanted in trays in the same randomized block design used in the field: Not only did this assist field transplanting, but it allowed cellular trait measurements to be taken in the lath house. Twenty-one days after plating, seedlings were then transferred to a lath house (early May). In both the greenhouse and lath house, seedlings were vigorously top watered and allowed to completely dry between waterings to harden for the field.
Thirty-five days after plating, seedlings were hand transplanted to the field (late May). Transplanted seedlings were initially sprinkler watered followed by ditch irrigated. Ditch irrigation was used as needed throughout the season. ILs and cv M82 were arranged in a block design with 10 replicates. Each block consisted of two rows. Arrangement within each block was randomized.
Anthesis began in early June, and field measures of leaf morphology were taken in early July from mature leaves. Harvest of fruit began late August and continued until mid September.
All traits (except flowering time), whether cellular traits measured in the lath house or field measurements of mature leaves, were studied in a 2010 field in Davis, CA. Additionally, leaf complexity traits were measured in a 2011 field in Davis, CA and the results incorporated together with 2010 data in statistical models. Flowering time was measured exclusively from the 2011 field season.
For the RNA-Seq–based genotyping and expression analysis experiments, seeds of the ILs and two parents were washed in 50% bleach for ∼2 min. Afterwards, seeds were placed in darkness for 3 d before moving to a 16:8 light cycle in growth chambers for 5 d. Seedlings were then transplanted into 2 × 5 pots per tray in Sunshine Mix soil (Sun Gro). For each replicate, six seedlings of each parent or IL were planted per pot. The 76 ILs (and two replicates each of cv M82 and S. pennellii) were divided into four cohorts of 20 randomly assigned genotypes. These cohorts sampled different shelves and regions of shelves across four temporal replicates in a Latin square design to account for positional effects on growth. Within a cohort’s assigned space for each of four temporal replicates, pots were randomly distributed. The seedlings were harvested 5 d after transplanting (13 d of growth in total). Cotyledons and mature leaves >1 cm in total length were excluded, and remaining tissues (including the shoot apical meristem) above the midpoint of the hypocotyl were pooled, for all individuals in a pot, into 2-mL microcentrifuge tubes and immediately frozen in liquid nitrogen.
RNA-Seq Library Preparation
mRNA isolation and RNA-Seq library preparation were performed from 80 samples at a time using a high-throughput RNA-Seq protocol (Kumar et al., 2012). The prepared libraries were sequenced in pools of 12 for replicates 1 and 2 (one lane each) and in pools of 80 for replicates 3 and 4 (seven lanes) at the UC Davis Genome Centre Expression Analysis Core using the HiSequation 2000 platform (Illumina).
Preprocessing RNA-Seq and RESCAN Sequence Data
Preprocessing of reads involved removal of low quality reads (phred score < 20), trimming of low-quality bases from the 3′ ends of the reads, and removal of adapter contamination using custom Perl scripts. The quality-filtered reads were sorted into individual libraries based on barcodes and then barcodes were trimmed using the Fastx toolkit.
RNA-Seq Mapping
RNA-Seq reads were initially mapped to the Heinz reference genome using BWA (parameters: -e 15 -i 10 -k 1 -l 25 -n 0.05; Li and Durbin, 2009). Nonuniquely mapped reads and reads with a mapping quality <20 were discarded. Unmapped reads were subsequently remapped using TopHat (parameters: -m 1 -g 1–segment-length 22 I = 5000–library-type fr-unstranded–solexa1.3-quals–butterfly-search; Trapnell et al., 2009). Mapping and intermediate processing (including sorting, filtering, and duplicate removal using samtools [Li et al., 2009] and Picard [http://picard.sourceforge.net/]) were automated using a Perl script available at http://github.com/mfcovington/RNaseq_mapping.
To remove reads originating from repeat-rich genomic regions, RESCAN sequencing reads were initially mapped to the Sol Genomic Network's tomato repeat database using BWA (BWA parameters: -e 15 -i 10 -k 1 -l 25 -n 0.05) (we created the fasta file for this from the gff3 file available at ftp://ftp.sgn.cornell.edu/genomes/Solanum_lycopersicum/annotation/ITAG2.3_release/ITAG2.3_repeats.gff3). Reads not mapped to the repeat database were extracted using bam2fastq program (http://www.hudsonalpha.org/gsl/software/bam2fastq.php). Subsequently, these repeat-filtered reads were mapped to the Heinz reference genome using the same BWA parameters. Samtools (with the'–bq 1' option) was used to retain the reads that mapped uniquely to the reference genome.
Polymorphism Identification for RNA-Seq
Polymorphisms between cv M82 and Heinz or S. pennellii and Heinz were identified using a set of Perl scripts (available at http://github.com/mfcovington/snp_identification). These scripts identify potential SNPs/indels based on pileup data extracted from the sequence alignments. RNA-Seq reads that encroach upon introns can lead to the identification of false SNPs/indels if the portion of the read that protrudes into the intron is not long enough to be recognized by TopHat as containing an intron junction. To eliminate these false polymorphisms (and maintain actual polymorphisms that exist within the exon near an exon-intron junction as well as those at the beginning or end of a transcript), we developed a filter based on the ratios of coverage at the putative polymorphism position and flanking positions (offset by eight nucleotides). We used two types of coverage: one that only counts actual reads and another that includes gaps in reads that represent introns (CIGAR score of'N'). Both gap and no-gap depth of coverage was calculated with a samtools-based Perl module (available at http://github.com/mfcovington/coverage_calc).
A preliminary list of cv M82 versus S. pennellii polymorphisms was generated by combining the filtered SNP/indel lists for the individual parents versus Heinz. Shared polymorphisms were discarded as well as polymorphisms at chromosomal positions for which the opposite parent had a depth of coverage less than four reads. These were performed using a set of Perl and R scripts (available at http://github.com/mfcovington/snp_identification).
Polymorphism Identification for RESCAN
VarScan (v2.2.7), a software for variant detection from next generation sequencing data, was used to call SNPs for generating parental references (Koboldt et al., 2012). To this end, the pileup2snp command was used (parameters: –min-coverage 4–min-reads 2–min-avg-qual 20–min-var-frequation 0.9–p-value 0.05). Subsequently, the compare function of VarScan (with the “unique” option) was used to discard common SNPs between cv M82 and S. pennellii, compared with the Heinz genome. The S. pennellii SNP reference was further refined by removing SNPs represented in more than five of the 76 ILs, since the reads contributing to these SNPs are likely derived from unannotated repetitive regions. These steps yielded preliminary RESCAN polymorphism list.
Polymorphism Noise Reduction
The preliminary RNA-Seq and RESCAN polymorphism lists were filtered to remove spurious SNPs/indels. This was done by genotyping the parental lines that were used for initial polymorphism identification (cv M82 and S. pennellii) and removing any SNPs/indels that return an unexpected genotype (using a Perl script available at http://github.com/mfcovington/snp_identification). This noise reduction step results in final RNA-Seq and RESCAN versions of the database of polymorphisms between cv M82 and S. pennellii (see Supplemental Data Set 1 online).
IL Genotyping and Plotting
IL sequence data was mapped to the Heinz reference genome. Mpileup information from each alignment was interrogated for every chromosomal position in the relevant version of the SNP database (i.e., RNA-Seq versus RESCAN) to determine the number of reads matching cv M82 versus S. pennellii (using a set of Perl scripts available at http://github.com/mfcovington/genotyping). The genotype of every SNP/indel for all ILs can be found at www-plb.ucdavis.edu/Labs/sinha/TomatoGenome/Resources.htm.
These data were plotted with a ggplot2-based (Wickham, 2009) approach that conveys genotype and depth of coverage information for each polymorphism across the genome (using an R script available at http://github.com/mfcovington/geno_plot). The technique we developed to plot genotypes is also able to show heterozygous regions. Polymorphism positions that are S. pennellii or cv M82 for every sequencing read are shown as bright green or magenta, respectively. As the ratio of S. pennellii to cv M82 evens out, the color of the data point approaches black. For example, our stock of IL1-2 is segregating and the introgressed region is clearly heterozygous for the pool we sequenced (see Supplemental Figures 2A and 3A online).
Trait Measurement
Cellular traits were measured from the adaxial side of cotyledons from plants in the lath house or from the adaxial and abaxial side of true leaves from the field. In all cases, dental impression (Provil Novo Light Standard Fast; Pearson Dental Supplies) was applied using an application gun and allowed to dry before archiving. Fingernail polish (Sally Hanson Double Duty) was applied to impressions, allowed to dry completely, removed from the impression, and floated on microscope slides with water. Water was removed and the nail polish remained affixed to the slide. Micrographs of samples were taken using a standard compound microscope. For each individual impression, two micrographs were taken to ensure representative measures. For each micrograph, four cotyledon pavement cells were traced using Bamboo Tablets (Wacom) in ImageJ (Abramoff et al., 2004) and the area and shape descriptors recorded. For stomata and epidermal cell counts, Bamboo Tablets were used to quickly place dots in ImageJ over the feature of interest, followed by custom macros that would count and record the number of features. Pseudoreplication was averaged.
Leaf complexity measurements were taken in the field. Pairs of measurers would measure two leaves per plant, including primary, secondary, and intercalary leaflet numbers. Leaf complexity was measured in both 2010 and 2011 field seasons. Pseudoreplication was averaged.
Leaf shape traits were derived from photographs. More than 11,000 leaflets were measured for this study. For each individual, five leaves were collected into plastic Ziploc bags and transported back to lab. For each leaf, the terminal and two distal lateral leaflets were dissected and arranged under nonreflective glass (a total of 15 leaflets per individual were measured). Olympus SP-500 UZ cameras were mounted on copy stands (Adorama 36-inch Deluxe Copy Stand) and controlled remotely by computer using Cam2Com software (Sabsik). Using custom ImageJ (Abramoff et al., 2004) macros, individual leaflets were extracted and named appropriately to denote individual and leaflet type (terminal, distal lateral left, and distal lateral right). Leaflet outlines were then batch processed in ImageJ to measure circularity, solidity, AR, and roundness.
Global analysis of leaflet shape was conducted using EFDs followed by principal component analysis using the program SHAPE (Iwata and Ukai, 2002). Object contours were extracted as chain-code. Chain-code was subsequently used to calculate normalized EFDs. Normalization was based upon manual orientation with respect to the proximal-distal axis of the leaflet. Principal component analysis was performed on the EFDs resulting from the first 20 harmonics of Fourier coefficients. Coefficients of EFDs were calculated at −2 and +2 standard deviations for each principal component and the respective contour shapes reconstructed from an inverse Fourier transformation. Principal components resulting from terminal and lateral leaflets were considered separately, and the remaining pseudoreplication was averaged.
Raw data of leaflet photos and micrographs of epidermal impressions can be found at the following database: www-plb.ucdavis.edu/Labs/sinha/TomatoGenome/Resources.htm. Trait information has also been deposited at Phenom-Networks (www.phenome-networks.com).
Statistical Modeling and QTL Analysis
Traits were modeled using mixed-effect linear models with the lme4 package (http://CRAN.R-project.org/package=lme4) in R (R Development Core Team, 2011). Before modeling, the distribution of the trait was checked to determine if it was normal, and if not, it was appropriately transformed. A thorough description of transformations applied to traits, whether model terms were treated as fixed or random, and the significance of terms is provided in Supplemental Data Set 7 online. Models were selected through a process of backward selection, in which two models differing by only the presence of a single term were compared to determine the significance of the term in explaining variance in the data. The process was repeated for all terms (replacing the previously tested term and testing another), and at the end of the process the most nonsignificant term (using a P value threshold of 0.05) was removed from the model. This process was iterated until only significant terms remained in the model. We then performed a forward selection check of the resulting minimal model, adding terms previously removed back to the model and comparing to the minimal model to ensure that the nonsignificance of removed terms persists. That the distribution of residuals in the model was normal was verified. Model fitted values were used for subsequent analysis. P values from models for significant differences between ILs and cv M82 were extracted using the pvals.fnc function from the language R package (http://CRAN.R-project.org/package=languageR).
For bin analysis, each individual was factored as to whether it did or did not possess a particular bin. This process was repeated for each bin. Marginal regression was performed by fitting a linear model between each trait as a function of the presence for each bin. Resulting significance values for each bin with respect to a given trait were then multiple test adjusted using the Holm method to control the family-wise error rate at 0.05 level.
Meta-Analysis of Traits, Hierarchical Clustering, and Network Analysis
Traits from other studies used in meta-analysis were downloaded from www.phenome-networks.com. The studies from which traits are derived and whether or not they are included in this study are detailed in Supplemental Data Set 8 online. Only those traits for which data were collected for >60 ILs were considered, so as to not unduly bias results. In reality, this means that 67 ILs were measured for the trait with the fewest recorded values used in this study. For each trait in a data set, data were z-score normalized, and z-scores were averaged across replicates (see Supplemental Data Set 9 online). A correlation matrix (Pearson) was then created between all traits, both those measured in this study and those from others (see Supplemental Data Set 10 online). Significance values for correlations were determined and the false discovery rate controlled using the Benjamini and Hochberg method (see Supplemental Data Set 11 online; Benjamini and Hochberg, 1995). Subsequently, subsets of the correlation matrix would be analyzed; for example, only those correlations between a DEV trait with a trait of another class that are significant after multiple test adjustment.
Hierarchical clustering on data was performed using the hclust function from the stats package in R (R Development Core Team, 2011), clustering by the absolute value of the Pearson correlation coefficient using Ward’s minimum variance method. Hive plots, as previously conceived (Krzywinski et al., 2011), were implemented using a Web interface developed by the Wodak Lab (wodaklab.org). Jackknifing was performed using custom scripts.
Unless otherwise noted, visualization of statistical results was performed with the ggplot2 package in R (Wickham, 2009).
Gene Expression Analysis
Mapping and normalization were done on the iPLANT Atmosphere cloud server (Goff et al., 2011). S. lycopersicum reads were mapped to 34,727 tomato cDNA sequences predicted from the gene models from the ITAG2.4 genome build (downloadable from http://solgenomics.net/itag/release/2.3/list_files; Tomato Genome Consortium, 2012). A pseudo reference list was constructed for S. pennellii using the homologous regions between S. pennellii scaffolds v.1.9 and S. lycopersicum cDNA references above. Using the defined boundaries of ILs, custom R scripts were used to prepare IL-specific references that had the S pennellii sequences in the introgressed region and S. lycopersicum sequences outside the introgressed region. The reads were mapped using BWA (Li and Durbin, 2009) using default parameters except for the following that were changed: bwa aln: -k 1 -l 25 -e 15 -i 10 and bwa samse: -n 0. Nonuniquely mapped reads were discarded. Raw counts for each gene were then tabulated using a Perl script, and the counts table was then filtered in R using the Bioconductor package EdgeR version 2.6.10 (Robinson and Oshlack, 2010) such that only genes that have more than two reads per million in at least three of the samples were kept. Normalization factors were then calculated using the trimmed mean of M-values method (Robinson and Oshlack, 2010), and this was multiplied with the library size of each sample to get the effective library size. The reads per million was then calculated for each gene of a sample as (gene counts × 1,000,000)/effective library size. The average of all the normalized replicates of each IL or parent was then calculated, and this average, in normalized reads per million, was used for the gene expression analysis.
Averaged, normalized reads for each IL, representing 20,332 genes, were regressed against 222 trait profiles as a linear model. After multiple test adjustment using the Benjamini and Hochberg method, 12,501 correlations were significant out of the 4,513,704 correlations tested. The 12,501 significant correlations between gene expression profiles and traits represented 3951 genes. These genes were hierarchically clustered based on their expression profile across ILs using the hclust function from the stats package in R, clustering by the absolute value of the Pearson correlation coefficient using Ward’s minimum variance method. Independently clustered genes and traits were then plotted against each other as a matrix shown in Figure 5A. Clusters of genes were then analyzed for enrichment of Gene Ontology terms at a 0.05 false discovery rate cutoff (goseq Bioconductor package; Young et al., 2010).
Accession Numbers
Sequence data from this article are presented in the supplemental data. All supplemental materials from this article are deposited in the DRYAD repository: http://dx.doi.org/10.5061/dryad.rm5v5.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. S. pennellii Introgression Summary for All 76 ILs.
Supplemental Figure 2. RNaseq-Based Genotyping of All Chromosomes.
Supplemental Figure 3. RESCAN-Based Genotyping of All Chromosomes.
Supplemental Figure 4. Map of S. pennellii Introgression Lines, Chromosomes 1-6.
Supplemental Figure 5. Map of S. pennellii Introgression Lines, Chromosomes 7-12.
Supplemental Figure 6. Distribution of Genes per Bin.
Supplemental Figure 7. Z-Score Values of ILs Relative to cv M82.
Supplemental Figure 8. Correlation between Leaf Developmental Traits.
Supplemental Figure 9. Broad-Sense Heritability for Leaf Developmental Traits.
Supplemental Figure 10. Detected Leaf Development QTLs.
Supplemental Figure 11. Leaflet Shape QTL.
Supplemental Figure 12. IL10-3 Exhibits QTL Affecting Pavement Cell Size and Stomatal Density.
Supplemental Figure 13. Developmentally Insightful Correlations between Leaf Development Traits.
Supplemental Figure 14. Bin Mapping and Gene Candidates.
Supplemental Figure 15. Bin Mapping Result Legend.
Supplemental Figure 16. Bin Mapping Results for CompAll.
Supplemental Figure 17. Bin Mapping Results for CompInt.
Supplemental Figure 18. Bin Mapping Results for CompPri.
Supplemental Figure 19. Bin Mapping Results for CompRachis.
Supplemental Figure 20. Bin Mapping Results for CompSec.
Supplemental Figure 21. Bin Mapping Results for CotPaveAR.
Supplemental Figure 22. Bin Mapping Results for CotPaveArea.
Supplemental Figure 23. Bin Mapping Results for CotPaveCirc.
Supplemental Figure 24. Bin Mapping Results for CotPaveCnt.
Supplemental Figure 25. Bin Mapping Results for CotPaveRound.
Supplemental Figure 26. Bin Mapping Results for CotPaveSolid.
Supplemental Figure 27. Bin Mapping Results for CotStom.
Supplemental Figure 28. Bin Mapping Results for CotStomInd.
Supplemental Figure 29. Bin Mapping Results for FlowTime.
Supplemental Figure 30. Bin Mapping Results for LatLfSize.
Supplemental Figure 31. Bin Mapping Results for LatPC1.
Supplemental Figure 32. Bin Mapping Results for LatPC2.
Supplemental Figure 33. Bin Mapping Results for LatPC3.
Supplemental Figure 34. Bin Mapping Results for LatPC4.
Supplemental Figure 35. Bin Mapping Results for LatPC5.
Supplemental Figure 36. Bin Mapping Results for LfAbStom.
Supplemental Figure 37. Bin Mapping Results for LfAdStom.
Supplemental Figure 38. Bin Mapping Results for LfStomRatio.
Supplemental Figure 39. Bin Mapping Results for LftAR.
Supplemental Figure 40. Bin Mapping Results for LftCirc.
Supplemental Figure 41. Bin Mapping Results for LftRound.
Supplemental Figure 42. Bin Mapping Results for LftSolid.
Supplemental Figure 43. Bin Mapping Results for TermLfSize.
Supplemental Figure 44. Bin Mapping Results for TermPC1.
Supplemental Figure 45. Bin Mapping Results for TermPC3.
Supplemental Figure 46. Bin Mapping Results for TermPC4.
Supplemental Figure 47. Bin Mapping Results for TermPC5.
Supplemental Figure 48. Significant Correlations between Leaf Complexity and Shape with Fruit Sugar Levels.
Supplemental Figure 49. Network Analysis Reveals a Relationship between Leaf Complexity and Shape with Sugars, Brix, and Biomass.
Supplemental Figure 50. Hierarchical Clustering of Traits Analyzed in This Study.
Supplemental Figure 51. Jackknifing Results Indicate Stable Correlations between Leaf Complexity and Shape with Sugar Metabolism and Yield Traits.
Supplemental Figure 52. Traits Significantly Correlated with a Distinct Cluster of Genes.
Supplemental Table 1. Summary of cv M82 Versus S. pennellii SNP/Indel Distribution for RNaseq and RESCAN Analyses.
Supplemental Table 2. Chromosomal Positions of the S. pennellii Introgression Boundaries for All 76 ILs.
Supplemental Data Set 1. Polymorphism Database.
Supplemental Data Set 2. Unique IL Combinations Define Bins.
Supplemental Data Set 3. Genes per Bin.
Supplemental Data Set 4. Annotation of Genes Present in Each Bin.
Supplemental Data Set 5. Modeled Trait Values and Significance Values.
Supplemental Data Set 6. Graphs of Fitted Values for Each Trait, Their Distributions, and Significance Values.
Supplemental Data Set 7. Descriptions of Traits and Modeling.
Supplemental Data Set 8. Traits Used from Other Studies.
Supplemental Data Set 9. Matrix of Averaged z-Scores.
Supplemental Data Set 10. Pairwise Pearson Correlation Coefficient Values between Traits.
Supplemental Data Set 11. Significance Values for Pairwise Correlations between Traits.
Supplemental Data Set 12. Jackknifing Results for Significant Correlations between DEV and Traits of Other Classes.
Supplemental Data Set 13. IL Expression Values for Genes Residing on Chromosomes 1-3 and Unassembled Genes.
Supplemental Data Set 14. IL Expression Values for Genes Residing on Chromosomes 4-8.
Supplemental Data Set 15. IL Expression Values for Genes Residing on Chromosomes 9-12.
Supplemental Data Set 16. Gene Expression Profiles Significantly Correlated with Traits.
Supplemental Data Set 17. A Group of Genes with Similar Expression Profiles.
Supplemental Data Set 18. GO Categories Enriched for a Group of Genes with Distinct Expression Profiles.
Supplemental Data Set 19. Seed Sources Used in This Study.
Acknowledgments
We thank numerous undergraduate researchers (University of California, Davis) for their contributions, including Tandis Arani, Andy Buck, Jessica Cruz, Michael Davis, Nora Downs, Tommy Hatcher, Max Mumbach, Dan Naylor, Nataly Raymundo, Katy Rush, Paradee Thammapichai, Thinh Thiem, Alaha Wahab, Jennifer Weil, Choua Yang, and Sharon Zimmerman. We thank Jim Jackson (University of California, Davis) for field care, and Dani Zamir (Hebrew University, Rehovot, Israel) and the Tomato Genetics Resource Center (University of California, Davis) for gifts of germplasm. D.H.C. is a fellow of the Life Sciences Research Foundation funded through the Gordon and Betty Moore Foundation. This work is supported through a National Science Foundation grant (IOS-0820854) awarded to N.R.S., J.N.M., and J.P.
AUTHOR CONTRIBUTIONS
D.H.C., R.K., L.R.H., A.R., M.F.C., Y.I., J.P., J.N.M., and N.R.S. designed the research. D.H.C., R.K., L.R.H., A.R., M.F.C., Y.I., and D.F. performed the research. A.R., M.F.C., and J.M.J.-G. contributed new analytic and computational tools. D.H.C., R.K., L.R.H., A.R., M.F.C., D.F., and J.M.J.-G. analyzed the data. D.H.C., R.K., A.R., M.F.C., J.P., J.N.M., and N.R.S. wrote the article.
Glossary
- QTL
quantitative trait loci
- IL
introgression line
- SNP
Single nucleotide polymorphism
- RESCAN
restriction enzyme sequence comparative analysis
- EFD
elliptical Fourier descriptor
- AR
aspect ratio
References
- Abramoff M.D., Magelhaes P.J., Ram S.J. (2004). Image processing with ImageJ. Biophotonics International 11: 36–42 [Google Scholar]
- Bailey I.W., Sinnott E.W. (1915). A botanical index of Cretaceous and Tertiary climates. Science 41: 831–834 [DOI] [PubMed] [Google Scholar]
- Barkoulas M., Galinha C., Grigg S.P., Tsiantis M. (2007). From genes to shape: Regulatory interactions in leaf development. Curr. Opin. Plant Biol. 10: 660–666 [DOI] [PubMed] [Google Scholar]
- Baxter C.J., Carrari F., Bauke A., Overy S., Hill S.A., Quick P.W., Fernie A.R., Sweetlove L.J. (2005). Fruit carbohydrate metabolism in an introgression line of tomato with increased fruit soluble solids. Plant Cell Physiol. 46: 425–437 [DOI] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y. (1995). Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57: 289–300 [Google Scholar]
- Canady M.A., Meglic V., Chetelat R.T. (2005). A library of Solanum lycopersicoides introgression lines in cultivated tomato. Genome 48: 685–697 [DOI] [PubMed] [Google Scholar]
- Carmel-Goren L., Liu Y.S., Lifschitz E., Zamir D. (2003). The SELF-PRUNING gene family in tomato. Plant Mol. Biol. 52: 1215–1222 [DOI] [PubMed] [Google Scholar]
- Chitwood D.H., Headland L.R., Filiault D.L., Kumar R., Jiménez-Gómez J.M., Schrager A.V., Park D.S., Peng J., Sinha N.R., Maloof J.N. (2012a). Native environment modulates leaf size and response to simulated foliar shade across wild tomato species. PLoS ONE 7: e29570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitwood D.H., Headland L.R., Kumar R., Peng J., Maloof J.N., Sinha N.R. (2012b). The developmental trajectory of leaflet morphology in wild tomato species. Plant Physiol. 158: 1230–1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitwood D.H., Headland L.R., Ranjan A., Martinez C.C., Braybrook S.A., Koenig D.P., Kuhlemeier C., Smith R.S., Sinha N.R. (2012c). Leaf asymmetry as a developmental constraint imposed by auxin-dependent phyllotactic patterning. Plant Cell 24: 2318–2327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitwood D.H., Maloof J.N., Sinha N.R. (2013). Dynamic transcriptomic profiles between tomato and a wild relative reflect distinct developmental architectures. Plant Physiol. 162: 537–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitwood D.H., Naylor D.T., Thammapichai P., Weeger A.C.S., Headland L.R., Sinha N.R. (2012d). Conflict between intrinsic leaf asymmetry and phyllotaxis in the resupinate leaves of Alstroemeria psittacina. Front. Plant Sci. 3: 182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitwood D.H., Sinha N.R. (2013). A census of cells in time: Quantitative genetics meets developmental biology. Curr. Opin. Plant Biol. 16: 92–99 [DOI] [PubMed] [Google Scholar]
- Cnops G., Jover-Gil S., Peters J.L., Neyt P., De Block S., Robles P., Ponce M.R., Gerats T., Micol J.L., Van Lijsebettens M. (2004). The rotunda2 mutants identify a role for the LEUNIG gene in vegetative leaf morphogenesis. J. Exp. Bot. 55: 1529–1539 [DOI] [PubMed] [Google Scholar]
- Cong B., Barrero L.S., Tanksley S.D. (2008). Regulatory change in YABBY-like transcription factor led to evolution of extreme fruit size during tomato domestication. Nat. Genet. 40: 800–804 [DOI] [PubMed] [Google Scholar]
- Do P.T., Prudent M., Sulpice R., Causse M., Fernie A.R. (2010). The influence of fruit load on the tomato pericarp metabolome in a Solanum chmielewskii introgression line population. Plant Physiol. 154: 1128–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duvick D.N. (2005). Genetic progress in yield of United States maize (Zea mays L.). Maydica 50: 193–202 [Google Scholar]
- Eshed Y., Izhaki A., Baum S.F., Floyd S.K., Bowman J.L. (2004). Asymmetric leaf development and blade expansion in are mediated by KANADI and YABBY activities. Development 131: 2997–3006 [DOI] [PubMed] [Google Scholar]
- Eshed Y., Zamir D. (1995). An introgression line population of Lycopersicon pennellii in the cultivated tomato enables the identification and fine mapping of yield-associated QTL. Genetics 141: 1147–1162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frary A., Fritz L.A., Tanksley S.D. (2004). A comparative study of the genetic bases of natural variation in tomato leaf, sepal, and petal morphology. Theor. Appl. Genet. 109: 523–533 [DOI] [PubMed] [Google Scholar]
- Frary A., Nesbitt T.C., Grandillo S., Knaap E., Cong B., Liu J., Meller J., Elber R., Alpert K.B., Tanksley S.D. (2000). fw2.2: A quantitative trait locus key to the evolution of tomato fruit size. Science 289: 85–88 [DOI] [PubMed] [Google Scholar]
- Fridman E., Carrari F., Liu Y.S., Fernie A.R., Zamir D. (2004). Zooming in on a quantitative trait for tomato yield using interspecific introgressions. Science 305: 1786–1789 [DOI] [PubMed] [Google Scholar]
- Geigenberger P., Regierer B., Lytovchenko A., Leisse A., Schauer N., Springer F., Kossmann J., Fernie A.R. (2004). Heterologous expression of a ketohexokinase in potato plants leads to inhibited rates of photosynthesis, severe growth retardation and abnormal leaf development. Planta 218: 569–578 [DOI] [PubMed] [Google Scholar]
- Givnish T.J., Vermeij G.J. (1976). Sizes and shapes of liane leaves. Am. Nat. 110: 743–778 [Google Scholar]
- Goff S.A., et al. (2011). The iPlant Collaborative: Cyberinfrastructure for plant biology. Front. Plant Sci. 2: 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golz J.F., Roccaro M., Kuzoff R., Hudson A. (2004). GRAMINIFOLIA promotes growth and polarity of Antirrhinum leaves. Development 131: 3661–3670 [DOI] [PubMed] [Google Scholar]
- Greenwood D.R. (1992). Taphonomic constraints on foliar physiognomic interpretations of Late Cretaceous and Tertiary palaeoclimates. Rev. Palaeobot. Palynol. 71: 149–190 [Google Scholar]
- Hackel A., Schauer N., Carrari F., Fernie A.R., Grimm B., Kühn C. (2006). Sucrose transporter LeSUT1 and LeSUT2 inhibition affects tomato fruit development in different ways. Plant J. 45: 180–192 [DOI] [PubMed] [Google Scholar]
- Heatherington S.E., Smillie R.M., Davies W.J. (1998). Photosynthetic activites of vegetative and fruiting tissues of tomato. J. Exp. Bot. 49: 1173–1181 [Google Scholar]
- Heichel G.H., Anagnostakis S.L. (1978). Stomatal response to light of Solanum pennellii, Lycopersicon esculentum, and a graft-induced chimera. Plant Physiol. 62: 387–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtan H.E.E., Hake S. (2003). Quantitative trait locus analysis of leaf dissection in tomato using Lycopersicon pennellii segmental introgression lines. Genetics 165: 1541–1550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata H., Niikura S., Matsuura S., Takano Y., Ukai Y. (1998). Evaluation of variation of root shape of Japanese radish (Raphanus sativus L.) based on image analysis using elliptic Fourier descriptors. J. Hered. 93: 384–38512547931 [Google Scholar]
- Iwata H., Ukai Y. (2002). SHAPE: A computer program package for quantitative evaluation of biological shapes based on elliptic Fourier descriptors. J. Hered. 93: 384–385 [DOI] [PubMed] [Google Scholar]
- Jiang C., Wright R.J., Woo S.S., DelMonte T.A., Paterson A.H. (2000). QTL analysis of leaf morphology in tetraploid Gossypoium (cotton). Theor. Appl. Genet. 100: 409–418 [Google Scholar]
- Jones B., Frasse P., Olmos E., Zegzouti H., Li Z.G., Latché A., Pech J.C., Bouzayen M. (2002). Down-regulation of DR12, an auxin-response-factor homolog, in the tomato results in a pleiotropic phenotype including dark green and blotchy ripening fruit. Plant J. 32: 603–613 [DOI] [PubMed] [Google Scholar]
- Jones C.M., Rick C.M., Adams D., Jernstedt J., Chetelat R.T. (2007). Genealogy and fine mapping of obscuravenosa, a gene affecting the distribution of chloroplasts in leaf veins, and evidence of selection during breeding of tomatoes (Lycopersicon esculentum; Solanaceae). Am. J. Bot. 94: 935–947 [DOI] [PubMed] [Google Scholar]
- Kerstetter R.A., Bollman K., Taylor R.A., Bomblies K., Poethig R.S. (2001). KANADI regulates organ polarity in Arabidopsis. Nature 411: 706–709 [DOI] [PubMed] [Google Scholar]
- Kim M., McCormick S., Timmermans M., Sinha N. (2003a). The expression domain of PHANTASTICA determines leaflet placement in compound leaves. Nature 424: 438–443 [DOI] [PubMed] [Google Scholar]
- Kim M., Pham T., Hamidi A., McCormick S., Kuzoff R.K., Sinha N. (2003b). Reduced leaf complexity in tomato wiry mutants suggests a role for PHAN and KNOX genes in generating compound leaves. Development 130: 4405–4415 [DOI] [PubMed] [Google Scholar]
- Kimura S., Koenig D., Kang J., Yoong F.Y., Sinha N. (2008). Natural variation in leaf morphology results from mutation of a novel KNOX gene. Curr. Biol. 18: 672–677 [DOI] [PubMed] [Google Scholar]
- Koboldt D.C., Zhang Q., Larson D.E., Shen D., McLellan M.D., Lin L., Miller C.A., Mardis E.R., Ding L., Wilson R.K. (2012). VarScan 2: Somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 22: 568–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koenig D., et al. (2013). Comparative transcriptomics reveals patterns of selection in domesticated and wild tomato. Proc. Natl. Acad. Sci. USA 110: E2655-E2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzywinski, M., Birol, I., Jones, S., and Marra, M.(2011). Hive plots–Rational approach to visualizing networks. Brief. Bioinform. 10.1093/bib/bbr069. [DOI] [PubMed]
- Kumar R., Ichihashi Y., Kimura S., Chitwood D.H., Headland L.R., Peng J., Maloof J.N., Sinha N.R. (2012). A high-throughput method for Illumina RNA-Seq library preparation. Front. Plant Sci. 3: 202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langlade N.B., Feng X., Dransfield T., Copsey L., Hanna A.I., Thébaud C., Bangham A., Hudson A., Coen E. (2005). Evolution through genetically controlled allometry space. Proc. Natl. Acad. Sci. USA 102: 10221–10226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson T., Bryant B., Lefebvre S., Lloyd J.C., Raines C.A. (2006). Decreased SBPase activity alters growth and development in transgenic tobacco plants. Plant Cell Environ. 29: 48–58 [DOI] [PubMed] [Google Scholar]
- Li H., Durbin R. (2009). Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., 1000 Genome Project Data Processing Subgroup (2009). The sequence alignment/map format and SAM tools. Bioinformatics 25: 2078–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Chetelat R.T. (2010). A pollen factor linking inter- and intraspecific pollen rejection in tomato. Science 330: 1827–1830 [DOI] [PubMed] [Google Scholar]
- Liu Y.-S., Zamir D. (1999). Second generation L. pennellii introgression lines and the concept of bin mapping. Report of the Tomato Genetics Cooperative 49: 26–30 [Google Scholar]
- Liu Z., Jia L., Wang H., He Y. (2011). HYL1 regulates the balance between adaxial and abaxial identity for leaf flattening via miRNA-mediated pathways. J. Exp. Bot. 62: 4367–4381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lytovchenko A., et al. (2011). Tomato fruit photosynthesis is seemingly unimportant in primary metabolism and ripening but plays a considerable role in seed development. Plant Physiol. 157: 1650–1663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massonnet C., Tisné S., Radziejwoski A., Vile D., De Veylder L., Dauzat M., Granier C. (2011). New insights into the control of endoreduplication: endoreduplication could be driven by organ growth in Arabidopsis leaves. Plant Physiol. 157: 2044–2055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monforte A.J., Tanksley S.D. (2000). Development of a set of near isogenic and backcross recombinant inbred lines containing most of the Lycopersicon hirsutum genome in a L. esculentum genetic background: a tool for gene mapping and gene discovery. Genome 43: 803–813 [PubMed] [Google Scholar]
- Monson-Miller J., Sanchez-Mendez D.C., Fass J., Henry I.M., Tai T.H., Comai L. (2012). Reference genome-independent assessment of mutation density using restriction enzyme-phased sequencing. BMC Genomics 13: 72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyle L.C. (2008). Ecological and evolutionary genomics in the wild tomatoes (Solanum sect. Lycopersicon). Evolution 62: 2995–3013 [DOI] [PubMed] [Google Scholar]
- Nicotra A.B., Leigh A., Boyce C.K., Jones C.S., Niklas K.J., Royer D.L., Tsukaya H. (2011). The evolution and functional significance of leaf shape in the angiosperms. Funct. Plant Biol. 38: 535–552 [DOI] [PubMed] [Google Scholar]
- Parkhurst D.F., Loucks O.L. (1972). Optimal leaf size in relation to environment. J. Ecol. 60: 505–537 [Google Scholar]
- Pérez-Pérez J.M., Serrano-Cartagena J., Micol J.L. (2002). Genetic analysis of natural variations in the architecture of Arabidopsis thaliana vegetative leaves. Genetics 162: 893–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poorter H., Niinemets U., Walter A., Fiorani F., Schurr U. (2010). A method to construct dose-response curves for a wide range of environmental factors and plant traits by means of a meta-analysis of phenotypic data. J. Exp. Bot. 61: 2043–2055 [DOI] [PubMed] [Google Scholar]
- Powell A.L.T., et al. (2012). Uniform ripening encodes a Golden 2-like transcription factor regulating tomato fruit chloroplast development. Science 336: 1711–1715 [DOI] [PubMed] [Google Scholar]
- R Development Core Team (2011). R: A Language and Environment for Statistical Computing. (Vienna, Austria: R Foundation for Statistical Computing). [Google Scholar]
- Raines C.A., Paul M.J. (2006). Products of leaf primary carbon metabolism modulate the developmental programme determining plant morphology. J. Exp. Bot. 57: 1857–1862 [DOI] [PubMed] [Google Scholar]
- Ranjan A., Ichihashi Y., Sinha N.R. (2012). The tomato genome: Implications for plant breeding, genomics and evolution. Genome Biol. 13: 167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M.D., Oshlack A. (2010). A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 11: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousseaux M.C., Jones C.M., Adams D., Chetelat R., Bennett A., Powell A. (2005). QTL analysis of fruit antioxidants in tomato using Lycopersicon pennellii introgression lines. Theor. Appl. Genet. 111: 1396–1408 [DOI] [PubMed] [Google Scholar]
- Sagar M., et al. (2013). SlARF4, an auxin response factor involved in the control of sugar metabolism during tomato fruit development. Plant Physiol. 161: 1362–1374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauer N., Semel Y., Balbo I., Steinfath M., Repsilber D., Selbig J., Pleban T., Zamir D., Fernie A.R. (2008). Mode of inheritance of primary metabolic traits in tomato. Plant Cell 20: 509–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauer N., et al. (2006). Comprehensive metabolic profiling and phenotyping of interspecific introgression lines for tomato improvement. Nat. Biotechnol. 24: 447–454 [DOI] [PubMed] [Google Scholar]
- Semel Y., Nissenbaum J., Menda N., Zinder M., Krieger U., Issman N., Pleban T., Lippman Z., Gur A., Zamir D. (2006). Overdominant quantitative trait loci for yield and fitness in tomato. Proc. Natl. Acad. Sci. USA 103: 12981–12986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seymour D.K., Filiault D.L., Henry I.M., Monson-Miller J., Ravi M., Pang A., Comai L., Chan S.W., Maloof J.N. (2012). Rapid creation of Arabidopsis doubled haploid lines for quantitative trait locus mapping. Proc. Natl. Acad. Sci. USA 109: 4227–4232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegfried K.R., Eshed Y., Baum S.F., Otsuga D., Drews G.N., Bowman J.L. (1999). Members of the YABBY gene family specify abaxial cell fate in Arabidopsis. Development 126: 4117–4128 [DOI] [PubMed] [Google Scholar]
- Stahle M.I., Kuehlich J., Staron L., von Arnim A.G., Golz J.F. (2009). YABBYs and the transcriptional corepressors LEUNIG and LEUNIG_HOMOLOG maintain leaf polarity and meristem activity in Arabidopsis. Plant Cell 21: 3105–3118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steindler C., Matteucci A., Sessa G., Weimar T., Ohgishi M., Aoyama T., Morelli G., Ruberti I. (1999). Shade avoidance responses are mediated by the ATHB-2 HD-zip protein, a negative regulator of gene expression. Development 126: 4235–4245 [DOI] [PubMed] [Google Scholar]
- Steinhauser M.C., Steinhauser D., Gibon Y., Bolger M., Arrivault S., Usadel B., Zamir D., Fernie A.R., Stitt M. (2011). Identification of enzyme activity quantitative trait loci in a Solanum lycopersicum x Solanum pennellii introgression line population. Plant Physiol. 157: 998–1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterken R., Kiekens R., Boruc J., Zhang F., Vercauteren A., Vercauteren I., De Smet L., Dhondt S., Inzé D., De Veylder L., Russinova E., Vuylsteke M. (2012). Combined linkage and association mapping reveals CYCD5;1 as a quantitative trait gene for endoreduplication in Arabidopsis. Proc. Natl. Acad. Sci. USA 109: 4678–4683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens, M.A., and Rick, C.M. (1986). Genetics and breeding. In The Tomato Crop, J.G. Atherton and J. Rudich, eds (London, New York: Chapman and Hall), pp. 36–109. [Google Scholar]
- Stevens R., Buret M., Duffé P., Garchery C., Baldet P., Rothan C., Causse M. (2007). Candidate genes and quantitative trait loci affecting fruit ascorbic acid content in three tomato populations. Plant Physiol. 143: 1943–1953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian F., Bradbury P.J., Brown P.J., Hung H., Sun Q., Flint-Garcia S., Rocheford T.R., McMullen M.D., Holland J.B., Buckler E.S. (2011). Genome-wide association study of leaf architecture in the maize nested association mapping population. Nat. Genet. 43: 159–162 [DOI] [PubMed] [Google Scholar]
- Tomato Genome Consortium (2012). The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485: 635–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toubiana D., Semel Y., Tohge T., Beleggia R., Cattivelli L., Rosental L., Nikoloski Z., Zamir D., Fernie A.R., Fait A. (2012). Metabolic profiling of a mapping population exposes new insights in the regulation of seed metabolism and seed, fruit, and plant relations. PLoS Genet. 8: e1002612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Pachter L., Salzberg S.L. (2009). TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics 25: 1105–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai C.H., Miller A., Spalding M., Rodermel S. (1997). Source strength regulates an early phase transition of tobacco shoot morphogenesis. Plant Physiol. 115: 907–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham, H. (2009). ggplot2: Elegant Graphics for Data Analysis. (New York: Springer). [Google Scholar]
- Wilf P., Wing S.L., Greenwood D.R., Greenwood C.L. (1998). Using fossil leaves as paleoprecipitation indicators: An Eocene example. Geology 26: 203–206 [Google Scholar]
- Willmann M.R., Poethig R.S. (2011). The effect of the floral repressor FLC on the timing and progression of vegetative phase change in Arabidopsis. Development 138: 677–685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe J.A. (1971). Tertiary climatic fluctuations and methods of analysis of tertiary floras. Palaeogeogr. Palaeoclimatol. Palaeoecol. 9: 27–57 [Google Scholar]
- Wu S., Xiao H., Cabrera A., Meulia T., van der Knaap E. (2011). SUN regulates vegetative and reproductive organ shape by changing cell division patterns. Plant Physiol. 157: 1175–1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao H., Jiang N., Schaffner E., Stockinger E.J., van der Knaap E. (2008). A retrotransposon-mediated gene duplication underlies morphological variation of tomato fruit. Science 319: 1527–1530 [DOI] [PubMed] [Google Scholar]
- Yifhar T., Pekker I., Peled D., Friedlander G., Pistunov A., Sabban M., Wachsman G., Alvarez J.P., Amsellem Z., Eshed Y. (2012). Failure of the tomato trans-acting short interfering RNA program to regulate AUXIN RESPONSE FACTOR3 and ARF4 underlies the wiry leaf syndrome. Plant Cell 24: 3575–3589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young M.D., Wakefield M.J., Smyth G.K., Oshlack A. (2010). Gene ontology analysis for RNA-seq: Accounting for selection bias. Genome Biol. 11: R14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamir D. (2013). Where have all the crop phenotypes gone? PLoS Biol. 11: e1001595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanor M.I., Rambla J.L., Chaïb J., Steppa A., Medina A., Granell A., Fernie A.R., Causse M. (2009). Metabolic characterization of loci affecting sensory attributes in tomato allows an assessment of the influence of the levels of primary metabolites and volatile organic contents. J. Exp. Bot. 60: 2139–2154 [DOI] [PMC free article] [PubMed] [Google Scholar]