This work reports that the bHLH transcriptional factor BZR1 binds to the promoter of FLD, a putative histone demethylase gene, repressing its transcription to regulate flowering in Arabidopsis. Secondary structure composition changes of BZR1 are mediated by the cyclophilin CYP20-2, which influences protein phosphorylation and subsequent degradation, for flowering regulation.
Abstract
Brassinosteroids (BRs) regulate many physiological processes during plant development, including flowering. However, little is known about the components of BR signaling that mediate flowering. Here, we report that BRASSINAZOLE-RESISTANT1 (BZR1), the conformation of which is altered by a cyclophilin (CYP20-2), binds cis-elements in the FLOWERING LOCUS D (FLD) promoter to regulate flowering. Both bzr1-1D and fld-4 showed delayed flowering. Electrophoretic mobility shift assay and chromatin immunoprecipitation revealed that BZR1 bound to a putative BR response cis-element and suppressed the expression of FLD. Overexpression of FLD partially rescued the late flowering of pBZR1:mBZR1Pro234-Leu-CFP (mx3). Yeast two-hybrid and pull-down assays demonstrated that BZR1 interacts with CYP20-2. Arabidopsis thaliana CYP20-2 had greater peptidyl-prolyl cis-trans isomerase activity than did wheat (Triticum aestivum) CYP20-2. Fourier transform infrared spectroscopy revealed conformation changes in BZR1, dependent on interaction with CYP20-2. Due to differences in activity and substrate preference between CYP20-2 proteins from wheat and Arabidopsis, At-CYP20-2–overexpressing lines showed earlier flowering, whereas Ta CYP20-2 lines flowered later. Immunoblot and chromatin immunoprecipitation assays showed that histone H3 trimethyl Lys4 and H3 acetylation levels were negatively correlated with the transcription of FLD (a putative histone demethylase) in various lines. Therefore, a conformational change of BZR1 mediated by CYP20-2 causes altered flowering through modulation of FLD expression.
INTRODUCTION
Flowering is a major developmental process that initiates the transition from vegetative to reproductive growth in plants. The genetic network mediating flowering is subject to epigenetic regulation and consists of the photoperiod, vernalization, gibberellin (GA), and autonomous pathways. A key component in the autonomous pathway is FLOWERING LOCUS C (FLC), a MADS (for yeast MCM1, plant AGAMOUS and DEFICIENS, and mammal Serum Response Factor) box transcriptional repressor (Rouse et al., 2002). FLC binds and represses floral-promoting genes, such as SUPPRESSOR OF OVEREXPRESSION OF CONSTANS1 (SOC1) (Searle et al., 2006). Targeted 3′ processing of antisense FLC transcripts triggers chromatin silencing to regulate flowering in Arabidopsis thaliana (Liu et al., 2010). Specifically, FLC expression is mediated by Lys and Arg histone modifications (Wang et al., 2007). For example, PROTEIN ARGININE METHYLTRANSFERASE5 (PRMT5)/Shk1 KINASE BINDING PROTEIN1 (SKB1), mediating Arg methylation of histone, is essential for proper pre-mRNA splicing, especially of FLOWERING LOCUS K (FLK). In prmt5 mutants, functional FLK transcript and protein levels are reduced, leading to upregulated FLC and delayed flowering (Deng et al., 2010). SET DOMAIN GROUP25 (SDG25) encodes a histone methyltransferase, and the loss-of-function mutant sdg25-1 confers early flowering associated with suppression of FLC expression (Berr et al., 2009). In addition, FLOWERING LOCUS D (FLD), a plant homolog of the human protein LYSINE-SPECIFIC HISTONE DEMETHYLASE1 (LSD1), represses FLC expression via histone modification (He et al., 2003; Shi et al., 2004). However, little is known about how FLD is regulated.
Brassinosteroids (BRs) regulate multiple processes, including photomorphogenesis and organogenesis, as well as flowering in plants (Bai et al., 2007; Wang et al., 2008; Li et al., 2010b). BR signaling targets the flowering genetic network through two nodes, brassinosteroid insensitive 1 (BRI1) and BRI1-EMS-suppressor 1 (BES1; Domagalska et al., 2007; Yu et al., 2008). BRI1 and BAK1 coperceive BRs at the plasma membrane (Clouse et al., 1996). BSKs mediate signal transduction from the receptor kinase BRI1 to BSU1, which triggers BIN2, a glycogen synthesis kinase 3–like protein (Tang et al., 2008). The bri1 mutants exhibit late flowering, and those null alleles show very strong late flowering phenotypes. The double mutants bri1 fca and bri1 fri have elevated expression of FLC and a more severe flowering phenotype (Domagalska et al., 2007).
The basic helix-loop-helix (bHLH) transcription factors BRASSINAZOLE RESISTANT1 (BZR1) and BRI1 EMS SUPPRESSOR1 (BES1/BZR2) are positive regulators of BR signaling with dual roles in activating BR responses and negative feedback on BR biosynthesis (Wang et al., 2002; Yin et al., 2002). BZR1 and BES1 are functionally regulated by phosphorylation. Degradation of BZR1 via 26S proteasome–mediated proteolysis requires BIN2-mediated phosphorylation (He et al., 2002; Jin et al., 2006). Phosphorylation-dependent nucleocytoplasmic shuttling of BES1 is important for regulating BR signaling output (Ryu et al., 2010b). BSU1 and BIN2 are required for cellular spatial redistribution of phosphorylation-dependent BES1 in BR signaling (Ryu et al., 2010a). The phosphatase PP2A dephosphorylates BZR1 to affect BR signaling (Li and Jin, 2007; Ryu et al., 2007; Tang et al., 2011). In addition, BZR1 signaling is balanced by the transcription factor LIC, a turn-off switch in rice (Oryza sativa) (Wang et al., 2008; Zhang et al., 2012).
Both BZR1 and BES1 bind their target genes to activate BR responses (Yu et al., 2008; Li et al., 2009b; Luo et al., 2010; Sun et al., 2010). BES1 recruits two jumonji N/C (JmjN/C) domain-containing proteins, EARLY FLOWERING6 (ELF6) and RELATIVE OF EARLY FLOWERING6 (REF6), which are histone H3 Lys 27 demethylases (Yu et al., 2008; Lu et al., 2011). ELF6 and REF6 genetically influence flowering (Noh et al., 2004; Lu et al., 2011). However, much less is known about how BZR1 directly regulates flowering at the molecular level.
The cyclophilin (CYP) protein family includes cyclosporin A binding immunophilins as well as FK506 binding proteins (FKBPs) and parvulins (Lu and Armstrong, 2007). Immunophilins with peptidyl-prolyl cis-trans isomerase (PPIase) activity serve as protein folding catalysts by regulating cis and trans peptide-bond conformations of Pro residues in proteins (Matouschek et al., 1995). CYP family members are involved in hormone-mediated plant development. For example, the lack of CYP40 causes defects in the transition from juvenile to adult stages of vegetative development and inflorescence morphological features in Arabidopsis (Berardini et al., 2001). In addition, CYP ROTAMASE CYCLOPHILIN1 (AtCYP18-3) is involved in BR signaling (Trupkin et al., 2012). Previously, we identified CYP20-2 as participating in GA signaling in wheat (Triticum aestivum). Accumulation of Ta CYP20-2 caused repressed GA-mediated DELLA protein degradation, suppressing cell elongation (Li et al., 2010a). DELLAs are mediators of signaling crosstalk between BRs and GAs in the regulation of cell elongation and plant growth (Li et al., 2012). DNA microarray assays of BR signaling revealed expression changes of a series of CYP family genes in Arabidopsis, such as CYP20-2 and FKBP13 (Sun et al., 2010). However, how CYPs function in BR signaling remains unclear.
Here, we report that BZR1 directly regulates FLD to mediate flowering. The conformation of BZR1 is altered by CYP20-2. Differences in activity and substrate preference of CYP20-2 from Arabidopsis and wheat led to altered flowering patterns. Thus, conformational changes of BZR1, mediated by CYP, may directly regulate the expression of FLD to influence flowering.
RESULTS
The FLD Gene Is a Direct Target of BZR1
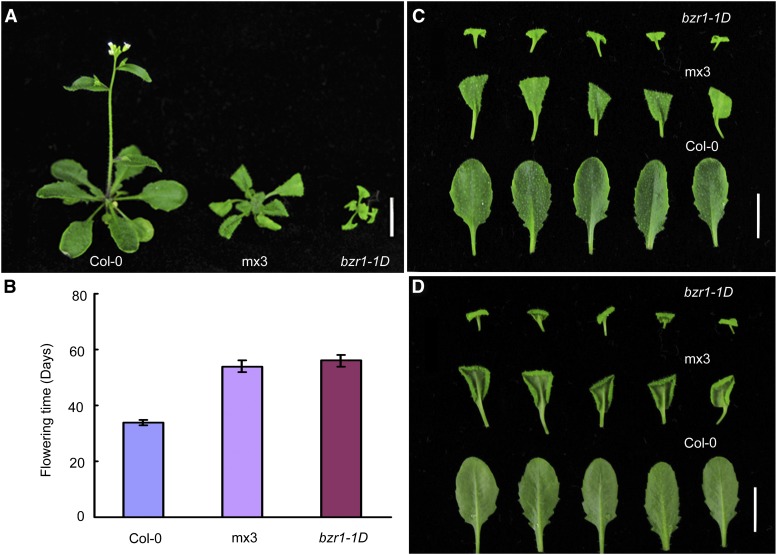
We examined the transgenic Arabidopsis line pBZR1:mBZR1-CFP (for cyan fluorescent protein; mx3), which expresses a version of BZR1 in which Pro-234 is substituted with Leu-234 (Wang et al., 2002). The mx3 line showed a bzr1-1D-like phenotype, with reduced stature, slightly darker green and wider leaves, and shorter petioles and hypocotyls, as well as curly leaves and later flowering, compared with the wild type (Figures 1A to 1D). Both bzr1-1D and mx3 showed delayed flowering compared with the wild type under a long-day photoperiod (Figure 1B).
Figure 1.
Late Flowering Phenotypes of mx3 and bzr1-1D.
(A) and (B) Phenotype (A) and flowering time (B) of mutants (mx3 and bzr1-1D) and the wild type (Col-0) grown under long-day (LD) conditions. Twenty plants were scored. Data are mean ± sd. Bar = 1 cm.
(C) and (D) Phenotypes of rosette leaves in the obverse (C) and reverse (D) view. Bar = 1 cm.
To obtain a global view of potential targets of BZR1 among flowering genes, the sequences 1 kb upstream of transcription start sites in flowering genes (Fornara et al., 2010) were examined for the presence of the key BR response cis-element (BRRE; CGTGNG) (He et al., 2005). In total, 17 of the 174 genes examined displayed sequences with high-confidence identity to BRRE in their promoters. These genes are involved in regulating several flowering pathways (see Supplemental Table 1 online). A putative BRRE, CGTGGG, was identified in the promoters of FLD at -287 to -281 and FY at -11 to -5, but not in any of the other known members of the autonomous pathway. However, the expression of FY was not changed in the transgenic line pBZR1:BZR1-CFP (W2C) nor in mx3 compared with the wild type (see Supplemental Figure 1 online). Therefore, we focused on FLD as a candidate target of the transcription factor BZR1.
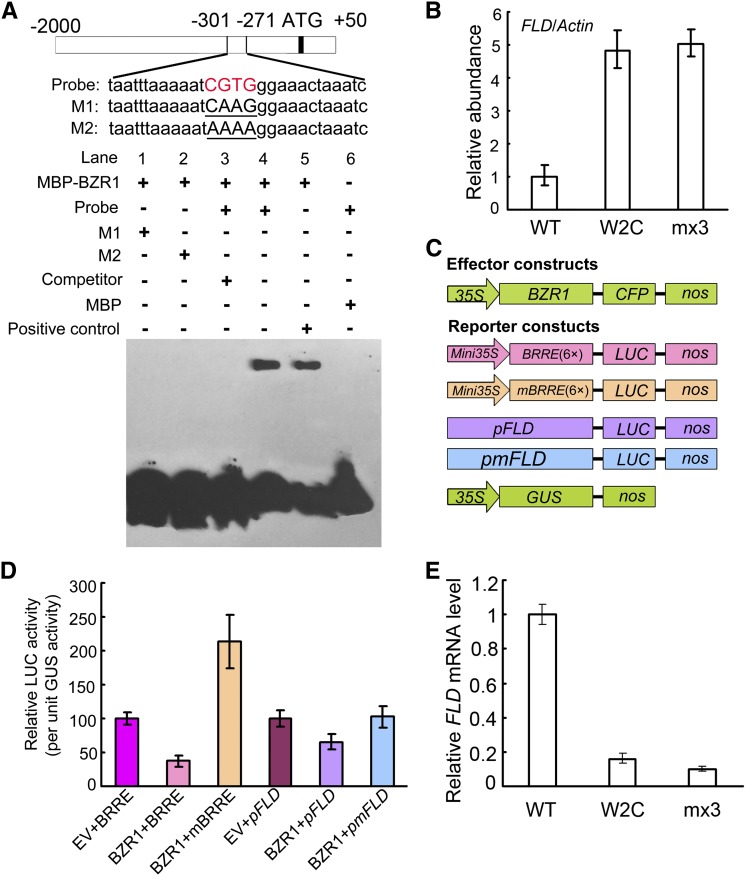
Electrophoretic mobility shift assay (EMSA) revealed that the putative BRRE from the FLD promoter (CGTGGG) was bound by MBP-BZR1, but not by maltose-binding protein (MBP) alone. The unlabeled putative BRRE probe competed away the binding to the labeled BZR1 protein (Figures 2A and 2B). Chromatin immunoprecipitation (ChIP) assays revealed that the promoter sequence of FLD was enriched by immunoprecipitation of CFP-tagged BZR1 (Figure 2C). Transcriptional activity assays in protoplasts revealed that expression of BZR1 reduced the expression of LUC driven by either the native FLD promoter (including the putative BRRE CGTGGG) or repeated BRRE elements. By contrast, LUC expression driven by a mutated FLD promoter with AGAGAG in place of the BRRE was not affected by BZR1 expression. When driven by mBRRE, in which repeated copies of BRRE were each mutated to CATGGG, LUC expression increased with BZR1 expression (Figures 2D and 2E). In both pBZR1:BZR1 (W2C) and pBZR1:mBZR1Pro234-Leu-CFP (mx3), FLD transcript accumulation was substantially decreased compared with the wild type (Figure 2F). Thus, BZR1 binds the putative BRRE in the promoter of FLD to suppress its transcriptional expression.
Figure 2.
BZR1 Binds to the Promoter of FLD.
(A) EMSA assay in vitro showing that the MBP-BZR1 fusion protein, but not MBP alone, binds to the FLD promoter (Probe). The diagram depicts the promoter sequence of the FLD gene. The red letters represent the hypothetical binding element (CGTG). M1, CGTG was mutated as CAAG; M2, CGTG was mutated as AAAA. Competitor indicates 200-fold excess unlabeled competitor probe added to compete with the labeled probe. Positive control, probe designed according to the core binding cis-element of BZR1 with flank sequences deposited in the CONSTITUTIVE PHOTOMORPHOGENIC DWARF (CPD) promoter (CAGAAACCCCCCGTGTGCCC). The amount of MBP-BZR1 was 0.5 μg for lane 5 and 2 μg for the other lanes.
(B) ChIP assay with antibody to GFP. Quantitative real-time PCR analysis of products immunoprecipitated from wild-type (WT), BZR1-CFP (W2C), and mBZR1-CFP (mx3) transgenic plants. Data are mean ± sd of triplicate experiments.
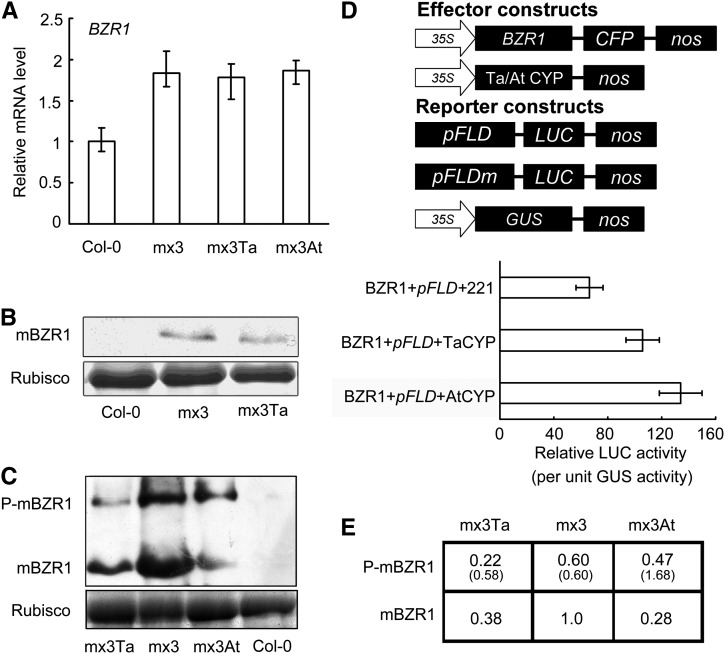
(C) Schematic diagrams of reporter and effector constructs used in the protoplast transcription system.
(D) Expression activity assay of BRRE-, mBRRE-, pFLD-, and pmFLD-LUC reporter genes in Arabidopsis protoplasts transformed with the plasmid 35S:BZR1 or empty vector (EV), with 35S:GUS as an internal control. Data are mean ± sd of triplicate experiments.
(E) FLD transcript levels in the wild-type, pBZR1:BZR1-CFP (W2C), and pBZR1:mBZR1-CFP (mx3) transgenic plants. Data are mean ± sd of triplicate experiments.
Either 35S:FLD or pmFLD:FLD Can Partially Rescue the Late Flowering Phenotype of mx3
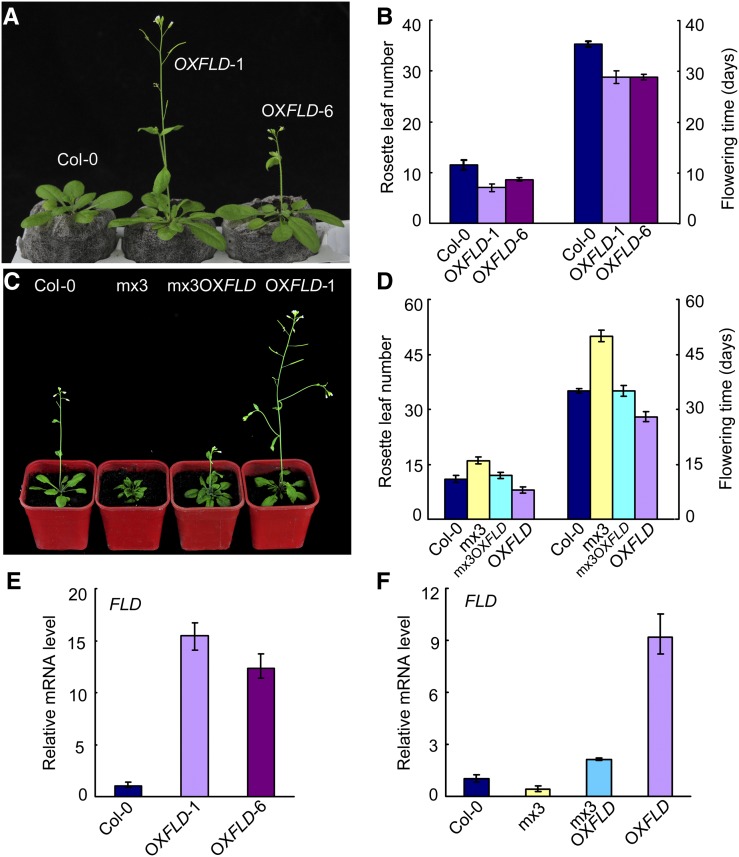
FLD was constitutively expressed under the control of a cauliflower mosaic virus (CaMV) 35S promoter in transgenic Arabidopsis lines (OXFLD). The OXFLD lines showed an early flowering-time phenotype compared with the wild type under a long-day photoperiod (Figures 3A and 3B). When we crossed OXFLD with mx3, all F1 generation plants showed earlier flowering than mx3 but later flowering than OXFLD (Figures 3C and 3D). The transcript level of FLD in mx3 OXFLD was higher than that in mx3 but lower than that in OXFLD (Figures 3E and 3F). Therefore, the late flowering of mx3 was partially rescued by the overexpressed FLD in mx3 OXFLD. These data suggest that BZR1 is functionally dependent on FLD.
Figure 3.
OXFLD Transgenic Arabidopsis Shows an Early Flowering Phenotype and Partially Rescued the Late Flowering Phenotype of mx3.
(A) and (B) Flowering phenotype of Col-0, OXFLD-1, and OXFLD-6 transgenic lines grown in light under long-day conditions for 27 d. Data are mean ± sd. For each line, 25 plants were scored.
(C) and (D) Flowering phenotype of Col-0, mx3, mx3 OXFLD, and OXFLD Arabidopsis grown in light under long-day conditions for 35 d. Data are mean ± sd. For each line, 20 plants were scored.
(E) and (F) Quantitative RT-PCR analysis of FLD transcript in transgenic lines and Col-0. Data are mean ± sd of triplicate experiments.
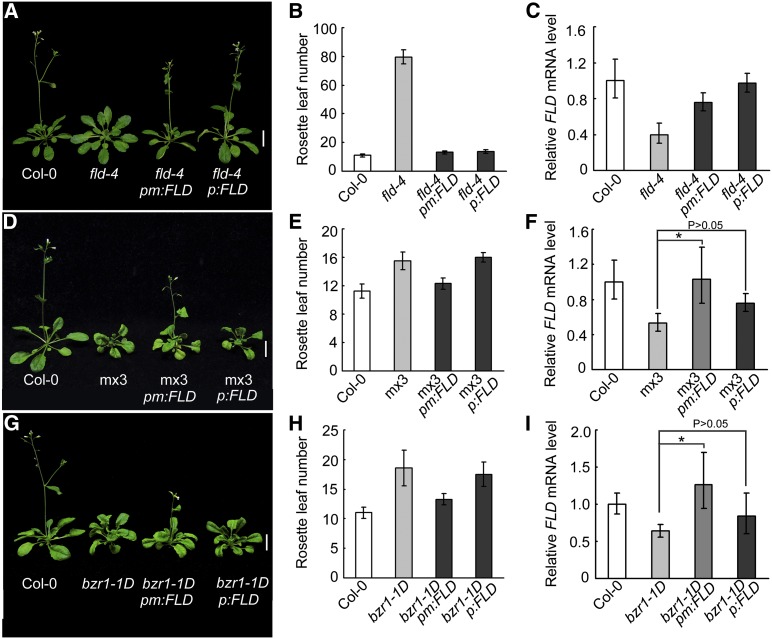
To confirm further the genetic relationship between BZR1 and FLD, FLD was expressed under control of either its native promoter (pFLD:FLD) or the mutated version (pmFLD:FLD) in various genetic backgrounds. Either pFLD:FLD or pmFLD:FLD could rescue the late flowering phenotype of fld-4, which is mutated in a flowering gene homologous to LSD1 (Liu et al., 2007) (Figures 4A to 4C). The double transgenic line mx3 pFLD:FLD showed a similar flowering pattern to that of mx3 (Figures 4D to 4F). By contrast, the mx3 pmFLD:FLD line, with increased FLD transcript, showed earlier flowering compared with mx3. Similar results were obtained in the bzr1-D background. Whereas bzr1-1D pFLD:FLD showed no difference in flowering time compared with bzr1-1D, bzr1-1D pmFLD:FLD flowered earlier (Figures 4G to 4I). Therefore, the late flowering phenotypes of mx3 and bzr1-1D were rescued by pmFLD:FLD. Together, these data indicate that BZR1 contributes to FLD regulation to mediate flowering.
Figure 4.
pmFLD:FLD Partially Rescued the Late Flowering Phenotype of mx3.
(A) and (B) Flowering phenotype of Col-0, fld-4, fld-4 pm:FLD, and fld-4 p:FLD transgenic lines grown in the light for 35 d under long-day conditions. Data are mean ± sd. For each line, 20 plants were scored. Bar = 1 cm.
(C) Quantitative RT-PCR analysis of FLD transcript in transgenic lines and the wild type.
(D) and (E) Flowering phenotype of Col-0, mx3, mx3 pm:FLD, and mx3 p:FLD Arabidopsis grown in the light under long-day conditions for 35 d. Data are mean ± sd. For each line, 20 plants were scored. Bar = 1 cm.
(F) Quantitative RT-PCR analysis of FLD transcript in transgenic lines and the wild type. Data are mean ± sd of triplicate experiments. *P < 0.05 and P > 0.05 compared with their parent mx3, respectively, as determined by Student’s t test.
(G) and (H) Flowering phenotype of Col-0, bzr1-1D, bzr1-1D pm:FLD, and bzr1-1D p:FLD Arabidopsis grown in the light under long-day conditions for 35 d. Data are mean ± sd. For each line, 20 plants were scored. Bar = 1 cm.
(I) Quantitative RT-PCR analysis of FLD transcript in transgenic lines and Col-0. Data are mean ± sd of triplicate experiments. *P < 0.05 and P > 0.05 compared with their parent bzr1-1D, respectively, as determined by Student’s t test.
pm:FLD, pmFLD:FLD (FLD expression driven by the native promoter of FLD with the BRRE mutated to AGAGAG); p:FLD, pFLD:FLD (FLD expression driven by the native promoter of FLD with a wild-type BRRE). Data are mean ± sd of triplicate experiments.
Interaction of CYP20-2 and BZR1
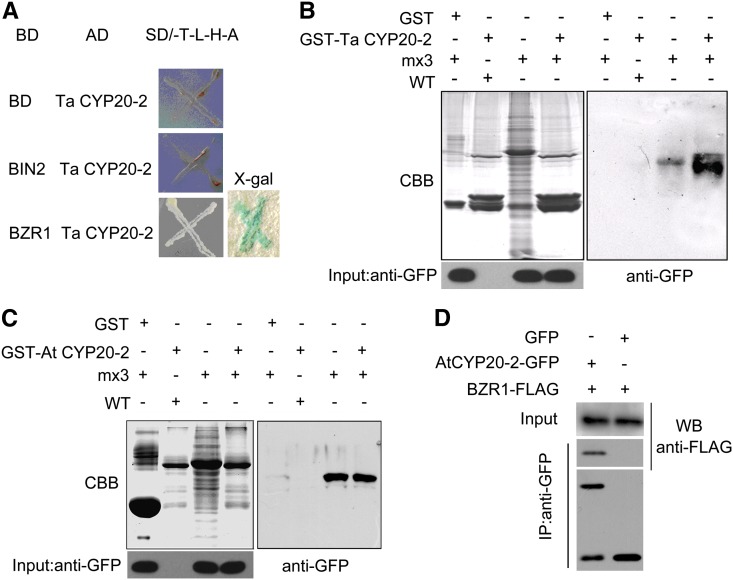
In our previous screen for proteins interacting with BZR1, a cyclophilin was identified as one of the potential targets. In yeast two-hybrid assays performed to confirm the interaction between BZR1 and CYP20-2, yeast cells cotransformed with pGBKT7-BZR1 and pGADT7-TaCYP20-2 were able grow on SD/-His-Trp-Leu-Ade selection medium and showed positive staining for β-galactosidase activity. Yeast cells harboring pGADT7-TaCYP20-2 along with pGBKT7-BIN2 or pGBKT7 failed to grow on the same medium (Figure 5A). To confirm the interaction in vivo, the pBZR1:mBZR1Pro234-Leu-CFP (mx3) transgenic line was used for pull-down assays. CFP-tagged mBZR1 bound to both glutathione S-transferase (GST)-TaCYP20-2 and GST-AtCYP20-2 but not GST alone under the same conditions (Figures 5B and 5C). Furthermore, when CYP20-2-YFP (for yellow fluorescent protein) and BZR1-FLAG fusion proteins were transiently coexpressed in tobacco (Nicotiana tabacum) leaf cells (Figure 5D), BZR1-FLAG could be coimmunoprecipitated with CYP20-2-YFP. Thus, CYP20-2 can interact with BZR1 in vivo.
Figure 5.
Protein Interaction between CYP20-2 and BZR1.
(A) Interaction between Ta CYP20-2 and At BZR1 in yeast two-hybrid assays. Colonies of yeast containing the indicated combination of bait (BD) and prey (AD) vectors were cultured on -Trp-Leu-His-Ade selection medium. β-Galactosidase activity of positive clones was analyzed by use of X-gal.
(B) Pull-down assay to confirm the interaction between Ta CYP20-2 and At BZR1. Glutathione agarose beads containing GST or GST-TaCYP20-2 were incubated with equal amounts of protein extract from mx3. Proteins bound to Sepharose 4B beads were pelleted, subjected to 12% SDS-PAGE, stained with Coomassie blue (CBB; left), and detected by immunoblot analysis with anti-GFP monoclonal antibody (right). WT, the wild type.
(C) Pull-down assay to confirm the interaction between At CYP20-2 and BZR1. Glutathione agarose beads containing GST or GST-AtCYP20-2 were incubated with equal amounts of protein extract from mx3. Proteins bound to Sepharose 4B beads were pelleted, subjected to 12% SDS-PAGE, stained with Coomassie blue (left), and detected by immunoblot analysis with anti-GFP monoclonal antibody (right).
(D) Coimmunoprecipitation assay to confirm the interaction between At CYP20-2 and BZR1. Tobacco leaves cotransformed with 35S:AtCYP20-2-YFP and 35S:BZR1-FLAG were used in immunoprecipitation assays with anti-GFP antibody, and the immunoblot was probed with anti-FLAG antibody.
Overexpression of CYP20-2 Led to a Change in Flowering Time
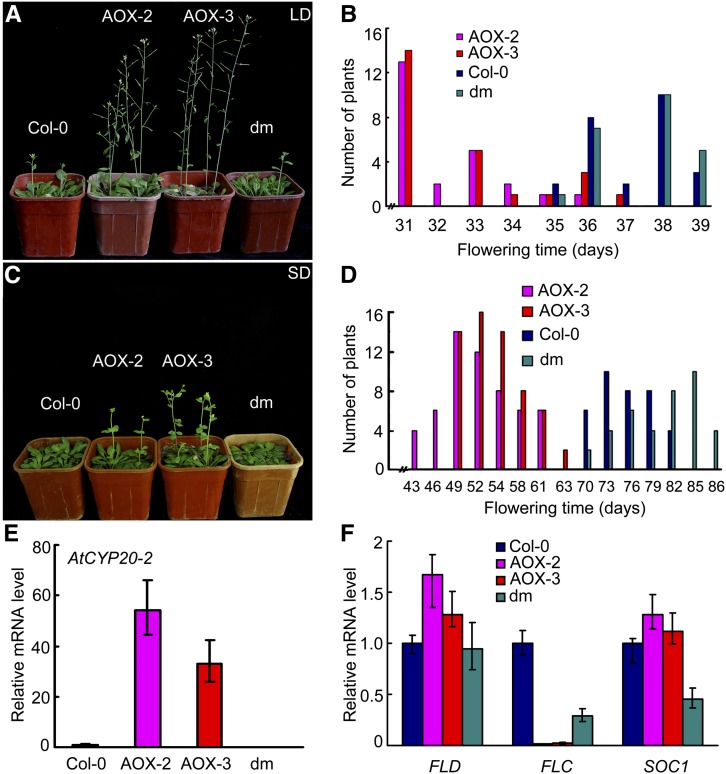
At CYP20-2 overexpression lines (AOX-2 and AOX-3) showed early flowering compared with the wild type either under a long-day or short-day photoperiod (Figures 6A to 6D). By contrast, neither the cyp20-2 and fkbp13 single mutants nor the double mutant cyp20-2 fkbp13 differed from the wild type in flowering time (Figures 6A to 6E), which may be explained by the redundant function of other family members (Edvardsson et al., 2007). In the At CYP20-2–overexpressing lines, FLC expression was extremely downregulated, whereas SOC1 expression was upregulated (Figure 6F). FLD, a negative regulator of FLC, was upregulated in AOX-2 and AOX-3 (Figure 6F). Thus, the expression of FLD to regulate flowering might be mediated by CYP20-2.
Figure 6.
Flowering Phenotypes of 35S:AtCYP20-2 Transgenic Arabidopsis.
(A) to (D) The phenotype and time of flowering of Col-0, 35S:AtCYP20-2 (AOX-2 and AOX-3), and cyp20-2 fkbp13 (dm) Arabidopsis grown under long-day conditions ([A] and [B]) or short-day conditions ([C] and [D]). Twenty-five plants were scored.
(E) and (F) Quantitative RT-PCR analysis of At CYP20-2, FLD, FLC, and SOC1 in Col-0, 35S:AtCYP20-2 overexpression lines, and cyp20-2 fkbp13 (dm). Data are mean ± sd (n = 3).
CYP40, a homolog of CYP20-2 (Li et al., 2010a), promotes the transition from juvenile to adult stages in Arabidopsis (Berardini et al., 2001). To test whether its activity was responsible for the lack of change in flowering time in the cyp20-2 fkbp13 double mutant, CYP40 was knocked down in the cyp20-2 fkbp13 background using RNA interference. The cyp20-2 fkbp13 cyp40RNAi lines showed late flowering compared with their background cyp20-2 fkbp13 (see Supplemental Figure 2 online). These results imply that members of the CYP gene family are required for, and redundantly act in, promotion of flowering.
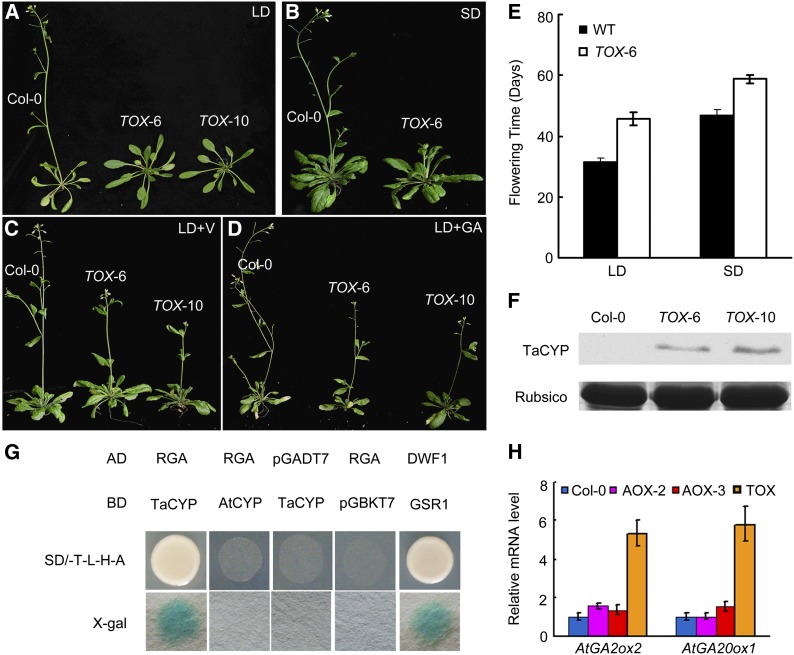
Ectopic overexpression of Ta CYP20-2 in Arabidopsis (TOX-6 and TOX-10) led to delayed flowering under long-day or short-day conditions (Figures 7A, 7B, and 7E). After vernalization treatment (4°C for 40 d), TaCYP20-2–overexpressing lines and the wild type did not differ in flowering time (Figure 7C). It is worth noting that GA3 treatment rescued the delayed flowering of Ta CYP20-2–overexpressing transgenic plants (Figure 7D). These phenotypes were reminiscent of those of autonomous pathway mutants, such as skb1 and fca (Mouradov et al., 2002; Lim et al., 2004; Wang et al., 2007). This result suggests that the GA pathway is involved in the control of flowering time, although BR signaling is involved in this process in the Ta CYP20-2 transgenic lines.
Figure 7.
Flowering Phenotype of 35S:TaCYP20-2 Transgenic Arabidopsis.
(A), (B), and (E) The flowering phenotypes in wild-type (WT; Col-0) and 35S:TaCYP20-2 (TOX-6 and TOX-10) transgenic Arabidopsis grown under long-day (LD) (A) or short-day (SD) conditions (B) for 40 d after germination. Twenty plants were scored. Data are mean ± sd.
(C) and (D) Effect on flowering of vernalization (V) (C) or GA treatment (D).
(C) Col-0 and 35S:TaCYP20-2 transgenic Arabidopsis grown at 4°C for 4 weeks and moved to LD conditions for 30 d.
(D) Plants were treated with GA3 (20 μM) and grown under long-day conditions.
(F) Immunoblot analysis of Ta CYP20-2 protein expression in Col-0 and 35S:TaCYP20-2–overexpressing transgenic Arabidopsis with polyclonal anti-TaCYP20-2 antiserum. Total protein stained with Coomassie blue (ribulose-1,5-bisphosphate carboxylase/oxygenase [Rubisco]) shows equal loading.
(G) Protein interaction between Ta CYP20-2/At CYP20-2 and the DELLA protein RGA was tested by yeast two-hybrid assays. The clones of yeast containing each combination of bait (BD) and prey (AD) vectors were cultured on -Trp-Leu-His-Ade selection medium. β-Galactosidase activity of positive clones was analyzed by use of X-gal.
(H) Real-time RT-PCR analysis of the expression of GA biosynthesis genes At GA2ox2 and At GA20ox1 in 35S:AtCYP20-2 and 35S:TaCYP20-2 transgenic lines. The mean ± sd of three experiments is shown.
To explore the molecular mechanism of late flowering, we examined the transcript levels of critical genes in flowering pathways. Expression of the flowering repressor FLC was upregulated in Ta CYP20-2–overexpressing plants (see Supplemental Figure 3 online). SOC1, which is inhibited in an FLC-dependent manner, was downregulated in the transgenic plants (see Supplemental Figure 3 online). This phenotype corresponded to the Ta CYP20-2 protein level of transgenic lines (Figures 7E and 7F). The flc-3 Ta CYP20-2 line showed a flowering pattern similar to that of the wild type (see Supplemental Figure 3 online). Thus, the function of Ta CYP20-2 in flowering may depend on the FLC-mediated pathway in the transgenic lines. On the other hand, Ta CYP20-2 but not At CYP20-2 interacted with a DELLA (RGA, REPRESSOR OF GA1-3) in yeast two-hybrid assays (Figure 7G). In addition, the expression levels of GA pathway genes GA20ox1 and GA2ox2 were increased in the later-flowering Ta CYP20-2 overexpression lines (Figure 7H). This is consistent with hypothesis that both GA and BR signaling contribute to later flowering in the Ta CYP20-2 overexpression Arabidopsis lines.
CYP20-2 Enhances Phosphorylation of BZR1
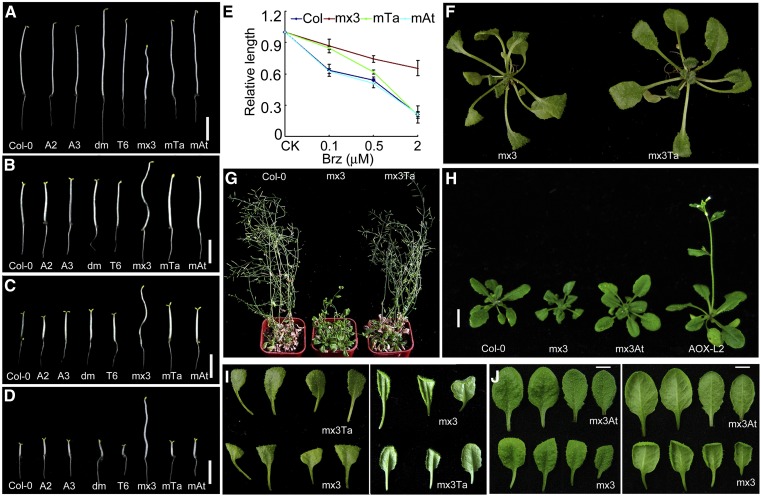
To determine whether there were differences in activity between Ta CYP20-2 and At CYP20-2, we tested BR signaling in various plant lines. We crossed Ta CYP20-2 overexpression lines or At CYP20-2 overexpression lines with mx3 Arabidopsis. Except for mx3, all lines showed hypocotyl lengths similar to that of the wild type in the dark (Figure 8A). Upon treatment with the BR biosynthesis inhibitor brassinazole (Brz) (0.5 μM) in the dark, the mx3 35S:TaCYP20-2 (mx3 Ta) line showed closed cotyledons similar to mx3; however, cotyledons were open in mx3 35S:AtCYP20-2 (mx3 At) under the same conditions, which was similar to the wild type (Figure 8B). When the concentration of Brz was increased to 2 μM, both mx3 Ta and mx3 At showed the same response as the wild type (Figures 8C to 8E). Compared with the wild type, mx3 Ta plants showed coiled leaves. However, the phenotype was weaker and plants flowered earlier than mx3 plants (Figures 8F, 8G, and 8I). In mx3 At, the coiled leaf phenotype was partially rescued (Figures 8H to 8J). Thus, the phenotypes caused by overexpression of mBZR1 were genetically impaired by overexpression of CYP20-2. However, At CYP20-2 made a larger contribution to the BR-signaling phenotype than did Ta CYP20-2 in the transgenic lines.
Figure 8.
35S:CYP20-2 Partially Rescued the BR-Related Phenotype in mx3.
(A) Six-day-old dark-grown seedlings. From left to right: Col-0, A2 (35S:AtCYP20-2 L2), A3 (35S:AtCYP20-2 L3), dm (cyp20-2 fkbp13), T6 (35S:TaCYP20-2 L6), mx3 (pBZR1:mBZR1-CFP), mx3Ta:mx3 35S:TaCYP20-2, and mx3At:mx3 35S:AtCYP20-2. Bar = 1 cm.
(B) to (D) Seedlings were grown in the dark for 6 d with various concentrations of Brz, 0.1 μM (B), 0.5 μM (C), and 2 μM (D). From left to right: Col-0, A2 (35S:AtCYP20-2 L2), A3 (35S:AtCYP20-2 L3), dm (cyp20-2 fkbp13), T6 (35S:TaCYP20-2 L6), mx3 (pBZR1:mBZR1-CFP), mx3Ta:mx3 35S:TaCYP20-2, and mx3At:mx3 35S:AtCYP20-2. Bar = 1 cm.
(E) Relative length of hypocotyls. Twenty plants were measured. Data are mean ± sd.
(F) From left to right: mx3 and mx3 35S:TaCYP20-2 grown in the light for 25 d under long-day conditions.
(G) Col-0, mx3, and mx3 35S:TaCYP20-2 grown in the light for 50 d under long-day conditions.
(H) Col-0, mx3, 35S:AtCYP20-2, and mx3 35S:AtCYP20-2 grown in the light for 25 d under long-day conditions. Bar = 1 cm.
(I) Rosette leaves of mx3 and mx3 35S:TaCYP20-2 plants.
(J) From top to bottom line: Rosette leaves of mx3 35S:At CYP20-2 and mx3 plants. mx3Ta, mx3 35S:Ta CYP20-2; mx3At, mx3 35S:At CYP20-2.
At the transcriptional level, BZR1 expression was greater in mx3 and in the crossed plants than in the wild type (Figure 9A). The relative amount of BZR1 protein was higher in mx3 and mx3 Ta than in the wild type but lower in mx3 Ta and mx3 At than in mx3 plants (Figures 9B and 9C). mx3 was treated with BL, and the amount of unphosphorylated and phosphorylated mBZR1 was monitored by immunoblotting. The result showed that there was more unphosphorylated mBZR1 upon BL treatment, with the intensity of the bands similar to those in Figure 9C (see Supplemental Figure 4 online). The ratio of phosphorylated to unphosphorylated mBZR1 was 0.58, 0.60, and 1.68 in mx3 Ta, mx3, and mx3 At lines, respectively (Figure 9E). Transcriptional activity assays in protoplasts revealed that constitutive expression of either Ta CYP20-2 or At CYP20-2 impaired the function of BZR1 (Figure 9D). Therefore, CYP20-2 may accelerate phosphorylation on mBZR1 for degradation by the ubiquitin-proteasome system, and At CYP20-2 may be more effective in this process than Ta CYP20-2.
Figure 9.
Effects of CYP20-2 Expression on mx3 Arabidopsis.
(A) Real-time PCR of BZR1 transcript in Col-0, mx3, mx3 35S:TaCYP20-2, and mx3 35S:AtCYP20-2. Data are mean ± sd (n = 3).
(B) Immunoblot analysis of mBZR1-CFP in Col-0, mx3, and mx3 35S:TaCYP20-2. Total protein extracts were separated by 12% SDS-PAGE, and mBZR1-CFP was detected with anti-GFP polyclonal antibody. mx3 Ta, mx3 35S:TaCYP20-2. Ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) served as a loading control in (B) and (C).
(C) Immunoblot analysis of phosphorylation of mBZR1-CFP in Col-0, mx3, mx3 35S:TaCYP20-2, and mx3 35S:AtCYP20-2. In order to separate the p-mBZR1 and mBZR1, protein extracts were separated by 10% SDS-PAGE until proteins smaller than 55 kD were running off the gel. The proteins were detected with anti-GFP monoclonal antibody. mx3 Ta, mx3 35S:TaCYP20-2; mx3At, mx3 35S:AtCYP20-2.
(D) Expression activity assay of the pFLD reporter gene in Arabidopsis protoplasts transformed with the plasmid 35:BZR1 in the presence of 35S:AtCYP20-2 or 35S:TaCYP20-2, with 35S:GUS as an internal control. Data are mean ± sd of triplicate experiments.
(E) Both phosphorylated and unphosphorylated forms of BZR1 shown in (C) were quantified. In the table, the numbers represent the relative protein amounts using unphosphorylated forms of BZR1 in mx3 as 1.0. Numbers in parentheses represent the ratio of phosphorylated to unphosphorylated forms of BZR1.
The Secondary Structure Composition of BZR1 Changed upon Interacting with CYP20-2
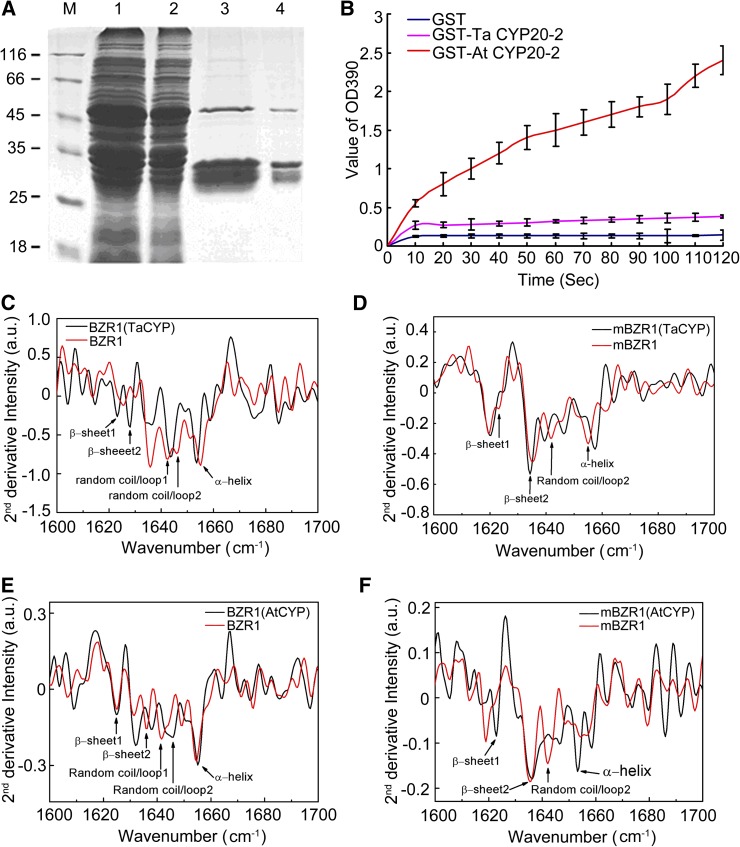
A time course of PPIase enzyme activity of the recombinant proteins GST-TaCYP20-2 (52 kD) and GST-AtCYP20-2 (52 kD) (Figure 10A) clearly showed that GST-AtCYP20-2 had greater PPIase activity than did GST-TaCYP20-2 (Figure 10B). Fourier transform infrared (FTIR) spectroscopy was used to monitor any changes in the secondary structure composition of BZR1 in the presence of GST-TaCYP20-2 or GST-AtCYP20-2. In the secondary derivative spectra, BZR1 showed a number of typical absorption peaks (Figure 10C), which could be assigned to different types of secondary structure (Li et al., 2009a): 1625 cm−1 (β-sheet [β-sheet 1]), 1635 cm−1 (β-sheet [β-sheet 2]), 1642 cm−1 (random coil/loop [random coil/loop 1]), 1646 cm−1 (random coil/loop [random coil/loop 2]), and 1654 cm−1 (α-helices). Upon interaction with Ta CYP20-2, the secondary derivative peaks were 1623 cm−1 (β-sheet 1), 1628 cm−1 (β-sheet 2), 1644 cm−1 (random coil/loop 2), and 1653 cm−1 (α-helices). The assignments of the observed bands in both FTIR and time-resolved infrared spectra are summarized in Table 1. All absorption peaks of secondary structures for BZR1, except random coil 1 at 1642 cm−1, were red shifted after binding with Ta CYP20-2; random coil 1 was blue shifted by 2 cm−1. If we assume that the structure of Ta CYP20-2 is rigid, the secondary structure change would mainly come from BZR1.
Figure 10.
PPIase Activity of Ta CYP20-2 and At CYP20-2 and Their Effects on BZR1 Secondary Structure.
(A) Expression and purification of GST-TaCYP20-2 and GST-AtCYP20-2 recombinant protein. Lane 1, total protein of DH5α cells containing the pGEX4T-1-TaCYP20-2 plasmid. Lane 2, total protein of DH5α cells containing the pGEX4T-1-AtCYP20-2 plasmid. Lane 3, GST-TaCYP20-2 fusion protein purified by glutathione-Sepharose 4B chromatography. Lane 4, GST-AtCYP20-2 fusion protein purified by glutathione-Sepharose 4B chromatography.
(B) PPIase activity of recombinant Ta CYP20-2 (pink), At CYP20-2 (red), and GST (blue) at 10 nM. Data are mean ± sd (n = 3).
(C) Second-derivative infrared spectra of BZR1 with or without GST-TaCYP20-2. Shown are MBP-BZR1 + GST-TaCYP20-2 minus the background absorption of MBP + GST-TaCYP20-2 (black) and MBP-BZR1 + GST minus the background absorption of MBP + GST (red); spectra were obtained in D2O medium. a.u., absorbance units.
(D) Second-derivative infrared spectra of mBZR1 with or without GST-TaCYP20-2. Shown are MBP-mBZR1 + GST-TaCYP20-2 minus the background absorption of MBP + GST-Ta CYP20-2 (black) and MBP-mBZR1 + GST minus the background absorption of MBP + GST (red); spectra were obtained in a D2O medium.
(E) Second-derivative infrared spectra of BZR1 with or without GST-At CYP20-2. Shown are with MBP-BZR1 + GST-AtCYP20-2 minus the background absorption of MBP+GST-AtCYP20-2 (black) and MBP-BZR1 + GST minus the background absorption of MBP + GST (red); spectra were obtained in D2O medium.
(F) Second-derivative infrared spectra of mBZR1 with or without GST-AtCYP20-2. Shown are MBP-mBZR1 + GST-AtCYP20-2 minus the background absorption of MBP + GST-AtCYP20-2 (black) and MBP-mBZR1 + GST minus the background absorption of MBP + GST (red); spectra were obtained in a D2O medium.
Table 1. Assignment of the Observed Amide I’ Band to Its Corresponding Secondary Structures.
| Observed (cm−1) | |||
|---|---|---|---|
| BZR1 | BZR1+CYP20-2 | Assignment | Literature Values |
| 1625 | 1623 | β-Sheet 1 | 1630/1623 ∼1641 |
| 1635 | 1628 | β-Sheet 2 | |
| 1642 | 1640 | Random coil/loop 1 | 1645/1640 ∼1650 |
| 1646 | 1644 | Random coil/loop 2 | |
| 1654 | 1653 | α-Helices | 1652/1650 ∼1656 |
The finding that most of BZR1 has a red shift in its absorption spectrum indicates that the secondary structure of BZR1 is opened to polar D2O, the chemical formula of heavy water, solvent. The presence of a blue-shifted peak (from 1642 to 1644 cm−1) strongly suggests that the Ta CYP20-2 enzyme binds the substrate (BZR1) at this secondary structure (i.e., random coil/loop 1). The blue shift is likely due to hydrophobic interaction between BZR1 and Ta CYP20-2. We suspected that Pro-234 in BZR1 was the core binding site of Ta CYP20-2 and in random coil/loop 1. To test this prediction, we used mBZR1 with Pro-234 substituted with Leu-234. Interestingly, the secondary derivative spectra of mBZR1 alone did not differ from that in the presence of Ta CYP20-2 (Figure 10D). Thus, Ta CYP20-2 may fail to bind mBZR1 as a substrate, which is consistent with our prediction that a mutation at this random coil/loop structure would destroy the binding site.
In contrast with the results with Ta CYP20-2, the presence of At CYP20-2 induced blue shifts in nearly all of the absorption peaks of BZR1, which implies that At CYP20-2 can bind BZR1 and induce opening in the secondary structure of BZR1. However, the structure of BZR1 became even tighter after binding to At CYP20-2 (Figure 10E). In addition, At CYP20-2 induced changes in the secondary derivative spectra of the mutant mBZR1 (Figure 10F). Therefore, there might be a binding site for At CYP20-2 in BZR1 different from that for Ta CYP20-2.
These results suggest that both Ta CYP20-2 and At CYP20-2 can alter the secondary structure conformation of BZR1. However, At CYP20-2 has a stronger and more specific effect than does Ta CYP20-2 in the conformational changes of the targets, which corresponds with its higher PPlase activity in vivo. A model for the function of CYP20-2 in altering the secondary structure of BZR1 is shown in Supplemental Figure 5 online.
Histone Modification on FLC Chromatin Changes Along With FLD Expression
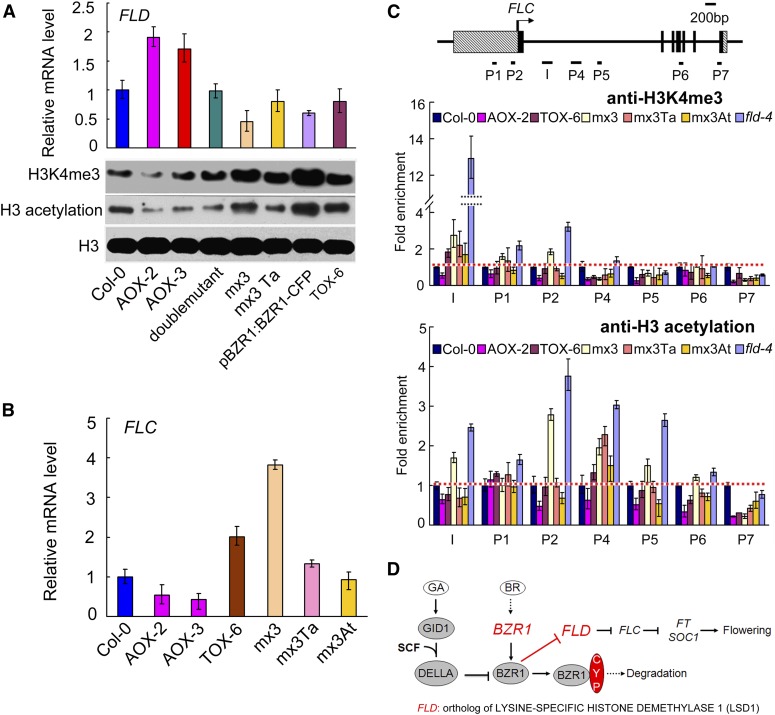
The expression of FLD was higher in At CYP20-2 overexpression lines than in the wild type, but there was less H3K4me3 and H3 acetylation (Figure 11A). By contrast, FLD was downregulated in 35S:TaCYP20-2 lines, and there was more H3K4me3 and H3 acetylation than in the wild type. In pBZR1:BZR1-CFP (W2C) and pBZR1:mBZR1Pro234-Leu-CFP (mx3), FLD transcript was reduced and H3K4me3 and H3 acetylation were dramatically increased. By contrast, H3K4me3 and H3 acetylation was lower in the crossed progeny line mx3 Ta than in mx3 alone (Figure 11A). These data hint that the levels of H3K4me3 and H3 acetylation are negatively correlated with the expression of FLD, a putative demethylase gene, which is targeted by BZR1 via CYP20-2.
Figure 11.
H3K4me3 and H3 Acetylation in FLC Chromatin Is Negatively Correlated with the Transcript Level of FLD.
(A) Quantitative RT-PCR of FLD in Col-0, AOX-2, AOX-3 (35S:AtCYP20-2), double mutant (cyp20-2 fkbp13), mx3 (pBZR1:mBZR1-CFP), mx3Ta (mx3 35S:TaCYP20-2), W2C (pBZR1:BZR1-CFP), and TOX-6 (35S:TaCYP20-2) plants. Data are mean ± sd (n = 3). Immunoblot analysis of histone modification levels. Histone-enriched protein extracts from 2-week-old Col-0, AOX-2, AOX-3, mx3, mx3 35S:TaCYP20-2, W2C, and TOX-6 plants grown under long-day conditions were probed with antibodies to H3K4me3 and H3 acetylation.
(B) Quantitative RT-PCR of FLC in Col-0, AOX-2, AOX-3 (35S:AtCYP20-2), TOX-6 (35S:TaCYP20-2), mx3 (pBZR1:mBZR1-CFP), mx3 Ta (mx3 35S:TaCYP20-2), and mx3At (mx3 35S:AtCYP20-2) plants. The mean ± sd of three experiments is shown.
(C) ChIP assay of the relative levels of trimethylated H3K4 (H3K4me3) and H3 acetylation in FLC chromatin. DNA fragments obtained by ChIP were analyzed by real-time PCR. The primer pairs used in PCR are shown as bars below the FLC gene structure; exons are shown as black boxes, introns as black lines, and untranslated regions as dashed boxes. Data are mean ± sd of triplicate experiments.
(D) Hypothetical model of CYP20-2 and BZR1 function in BR signaling and flowering pathways.
Compared with the wild type, FLC transcription was decreased in the earlier flowering AOX lines, but increased in the later flowering genetic lines, such as mx3 and TOX (Figures 11A and 11B). To examine the relationship between this and the levels of the activating marks H3K4me3 and H3 acetylation, we performed ChIP assays using the fld-4 mutant as a positive control (Figure 11C). Consistent with the FLC expression levels, the 35S:AtCYP20-2 lines showed less H3K4me3 in nearly all regions of FLC chromatin and less H3 acetylation in region I of FLC chromatin compared with the wild type. Conversely, mx3 showed a higher level of chromatin modification than did the wild type, but a similar level to that of 35S:TaCYP20-2 lines. There was more modification in mx3 Ta than in mx3 At lines but less than in mx3 (Figure 11C). Therefore, both H3K4me3 and H3 acetylation in FLC chromatin were changed along with the transcript level of FLD. CYP20-2 modulated the conformation of BZR1, which targeted the promoter of FLD to regulate flowering in Arabidopsis (Figure 11D).
DISCUSSION
BZR1 Directly Represses FLD to Regulate Flowering in Arabidopsis
Our results show that core BRREs (CGTGT/CG) are bound by BZR1, which represses the transcription of FLD in bzr1-1D and mx3 Arabidopsis. bzr1-1D and mx3 transgenic Arabidopsis showed extremely late flowering. BZR1 may directly repress FLD expression to modulate FLC and regulate flowering. However, it is worth noting that the late flowering phenotype seen in bzr1-1D may have nothing to do with BR signal transduction from BRI1 and BAK1. The mutants of BR positive regulators, such as bri1 and bak1, display late flowering phenotypes only when the expression levels of those regulators are decreased. Neither the expression level of FLD nor that of BZR1 was altered in bri1-5 (see Supplemental Figure 6 online) (Sun et al., 2010), even though it shows a late flowering phenotype. Therefore, the late flowering phenotype of bzr1-1D is not likely caused by BR signaling from BRI1 and BAK1. Although BES1 and BZR1 are important components in BR signaling, these two transcription factors are also regulated by other signaling molecules, such as GAs (Wang et al., 2002; Yin et al., 2002, 2005; He et al., 2005; Bai et al., 2012). Therefore, the molecular mechanism of flowering regulation by BZR1 is distinct from that of BRI1.
Since the expression of FLD was decreased in mx3 and pBZR1:BZR1, late flowering was expected in both mx3 and pBZR1:BZR1. However, pBZR1:BZR1 did not cause obvious phenotypes on flowering time in the wild-type background (Wang et al., 2002) (see Supplemental Figure 7 online), but pBZR1:mBZR1-CFP (mx3) showed late flowering (Figure 1). Similarly, pBZR1:BZR1 showed the same response to Brz in a hypocotyl elongation assay compared with the wild type, but the expression of CPD was decreased in pBZR1:BZR1 (Gampala et al., 2007). All of these results may be due to the sustained BZR1 accumulation conferred by the bzr1-1D mutation in mx3 that is absent in pBZR1:BZR1.
Arabidopsis overexpressing At CYP20-2 showed early flowering with decreased FLC transcript and increased FLD (Figure 6). Loss of function of FLD increases the levels of di- and trimethylated H3K4 and core histone tail acetylation in FLC chromatin (He et al., 2003; Jiang et al., 2007; Liu et al., 2007). Di- and trimethylated H3K4 are associated with activation of FLC (He et al., 2004). The increased FLD expression in At CYP20-2–overexpressing lines was consistent with the reduced level of trimethylated H3K4 and H3 acetylation in FLC chromatin (Figure 11). FLD is transcriptionally suppressed by BZR1. Consistent with this, trimethylated H3K4 and H3 acetylation were greatly increased in pBZR1:mBZR1Pro234-Leu-CFP and pBZR1:BZR1-CFP transgenic plants but returned to a normal level in mx3Ta Arabidopsis (Figure 11). On the other hand, FLD interacts with FCA and FPA for RNA 3′ processing to regulate flowering (Sonmez et al., 2011). Therefore, FLC may be one of the targets of FLD to regulate flowering. CYP20-2 therefore represents a component in the autonomous pathway of flowering.
CYP20-2 Alters the Conformation of BZR1 to Regulate Flowering
BZR1 may be involved in flowering via posttranscriptional regulation. The BZR1 transcript level did not differ between mx3 35S:CYP20-2 and mx3 alone (Figure 9), whereas the mBZR1 protein level in the crossed lines was markedly lower than in mx3. Furthermore, the expression of CYP20-2 caused accumulation of the phosphorylated form of mBZR1, which could be degraded by the proteasome machinery in mx3 35S:CYP20-2. The CYP-like domain is specifically associated with subunit S1 of the 19S regulatory particle in the 26S proteasome to selectively promote the accumulation of properly folded targets of the ubiquitin-proteasome system for degradation (Yi et al., 2007). Our data suggest that CYP20-2 interacts with BZR1 (Figure 5) and alters its secondary structure to affect its phosphorylation status (Figures 9 and 10). Therefore, CYP20-2 may catalyze cis-trans isomerization of the peptide bond N-terminal to Pro residues in BZR1 to promote phosphorylation of BZR1, which can be degraded by a ubiquitin-proteasome system.
Since the PPIase activity of Ta CYP20-2 was weaker than that of At CYP20-2 (Figure 10B), Ta CYP20-2 overexpression lines with increased FLC transcript showed late flowering (Figure 7; see Supplemental Figure 3 online). FLD transcript was slightly decreased in Ta CYP20-2–overexpressing lines, but trimethylated H3K4 and H3 acetylation in FLC chromatin was increased (Figure 11). The level of trimethylated H3K4 and H3 acetylation was lower in mx3 35S:TaCYP20-2 than in mx3 (Figure 11). However, mx3 35S:TaCYP20-2 was more insensitive to Brz than was mx3 35S:AtCYP20-2, which is more like mx3 (Figure 8). The alteration of signaling was weaker with Ta CYP20-2 than with At CYP20-2. Increased BZR1 protein levels result in reduced expression of CPD during feedback inhibition of BR biosynthesis (Wang et al., 2002). The expression of BR biosynthesis genes, such as BRASSINOSTEROID-6-OXIDASE, CPD, and DWARF4, was lower in transgenic Arabidopsis mx3 than the wild type but increased in mx3 35S:CYP20-2 lines. However, their expressions were more enhanced with At CYP20-2 compared with Ta CYP20-2 (see Supplemental Figure 8 online), which suggests feedback regulation of the biosynthesis in the transgenic lines. Therefore, our findings of Ta CYP20-2 as a PPIase with weak activity further suggest that CYP20-2 modulates the conformation of BZR1, which binds to FLD to regulate flowering in Arabidopsis (Figure 11D).
It is worth noting that CYP20-2 proteins from different species differentially act in GA and BR signaling. In wheat, Ta CYP20-2 is a negative regulator in the GA signaling pathway of DELLA protein degradation (Li et al., 2010a). Ta CYP20-2 interacted with a DELLA protein (RGA) in Arabidopsis, but At CYP20-2 did not (Figure 7G). On the other hand, expression of GA biosynthesis genes was altered in the 35S:TaCYP20-2 lines, which may represent a GA signaling response (Xu et al., 1995). The expressions of At GA20ox1 and At GA2ox2 were increased in 35S:TaCYP20-2 but not changed in 35S:AtCYP20-2 (Figure 7H). Our conformational analysis suggests that At CYP20-2 has a much stronger and more specific effect on the conformation of BZR1 than does Ta CYP20-2. It is possible that the later flowering of the 35S:TaCYP20-2 lines can be explained by GA signaling that occurs via Ta CYP20-2 interaction with DELLA independent of BZR1. By contrast, the early flowering of 35S:AtCYP20-2 could be mediated by CYP20-2 action on BZR1.
In summary, our studies shed light on BZR1-mediated flowering and help elucidate the protein-level mechanism of CYP-mediated conformational changes in BZR1 to regulate its function. CYP20-2 acts as a molecular chaperone to negatively regulate posttranscriptional modification of BZR1 in Arabidopsis. FLD is a direct target of BZR1 and mediates the autonomous pathway for flowering.
METHODS
Generation of Transgenic Arabidopsis Overexpression Lines
The sequences of genes such as FLD and Ta CYP20-2 were cloned into the pSN1301 vector (modified from vector pCambia1301) (Clontech) containing the CaMV 35S promoter and a β-glucuronidase (GUS) gene as a marker. In pFLD:FLD (with wild-type BRRE in its promoter) and pmFLD:FLD (with the BRRE in its promoter mutated to AGAGAG), the genome sequence of FLD with its native promoter was cloned into the vector pCAMBIA1300 (Clontech). At CYP20-2 was cloned into the pBIN vector (gift from Paul Dupree), which carried a YFP gene as a marker. The genes were driven by the CaMV 35S promoter.
Arabidopsis thaliana (Columbia-0 [Col-0]) plants were transformed by the floral dip method (Clough and Bent, 1998). Seeds from Agrobacterium tumefaciens–transfected Arabidopsis were screened on one-half-strength Murashige and Skoog medium (MS) containing 20 mg/L hygromycin (for OXFLD, mx3 pFLD:FLD, mx3 pmFLD:FLD, and 35S:TaCYP20-2) or 50 mg/L kanamycin (for 35S:AtCYP20-2). Arabidopsis was grown at 22°C under long-day (16:8 h, light:dark) or short-day (8:16 h, light:dark) conditions.
Quantitative Real-Time PCR
Total RNA was extracted from Arabidopsis seedlings with TRNzol A+ reagent (Tiangen). cDNA synthesis was performed with Moloney Murine Leukemia Virus Reverse Transcriptase (Promega). For real-time PCR, 2 μg RNA was used for reverse transcription. The cDNA samples were diluted to 2 to 8 ng/mL. Triplicate quantitative assays were performed with 5 μL of each cDNA dilution with the SYBR Green Real-time PCR Master mix (Toyobo) and an MX3000P real-time PCR system (Stratagene). The relative quantification method (Δ-Δ CT) was used to evaluate quantitative variation between the replicates.
Bioinformatics Analysis
Flowering-related genes have been well documented (Fornara et al., 2010). For bioinformatics analysis, Mochiview (Homann and Johnson, 2010) was used to identify putative BZR1-targeted genes, with a BRRE cis-element in the promoter (within 1 kb of the transcription start site), and then flowering-related genes were selected from among those putative target genes.
EMSA
EMSA was performed as described (Ma et al., 2009). Briefly, oligonucleotide probes were synthesized, annealed, and labeled by use of the Biotin 3′ End DNA labeling kit (Pierce). Escherichia coli BL21 cells transformed with pMAL-ER1-BZR1 and pMAL-C2 plasmids were induced by isopropyl β-D-1-thiogalactopyranoside as described previously. Cells were lysed in lysate buffer (50 mM Tris-HCl, pH 8.0, 1 mM EDTA, and 100 mM NaCl) using an ultrasonic cell crusher. After centrifugation, the supernatant was purified by Amylose Resin affinity chromatography (NEB). The binding reactions were performed according to the manufacturer’s protocol. The chemiluminescence of biotin-labeled DNA was detected using the Light Shift Chemiluminescent EMSA kit (Pierce).
ChIP
ChIP experiments were performed as described (Bowler et al., 2004) with 2-week-old plants. Immunoprecipitation was performed with rabbit polyclonal antibodies against GFP (1:1000) (G1544; Sigma-Aldrich), H3 (1:1000) (17-10046; Millipore), acetylated H3 (1:1000) (17-10241; Millipore), and trimethyl H3K4 (1:300) (17-614; Millipore). Primers and PCR detection of FLC regions were as described (Pazhouhandeh et al., 2011). ChIP products were analyzed by quantitative real-time PCR. Data from ChIP experiments are expressed as mean ± sd from three biological replicates.
Protoplast Transient Expression Assay
Isolation of Arabidopsis protoplasts was as described (Yoo et al., 2007) with some modifications. The enzyme solution contained 0.3% (w/v) macerozyme R10 (Yakult Pharmaceutical). The isolated protoplasts were transformed with effector, reporter, and reference plasmids at a ratio of 5:3:2 by the polyethylene glycol method. The pFLD-LUC reporter gene construct contains the native sequence of the FLD promoter fused to luciferase (LUC). pmFLD-LUC was generated by replacing CGTGGG in the promoter sequence with AGAGAG. The BRRE-LUC reporter gene construct contains a repeat of the BRRE sequence of the FLD promoter fused to LUC driven by a mini 35S promoter. mBRRE-LUC was generated by replacing CGTGGG with CATGGG. 35S:GUS contained a GUS gene driven by the CaMV 35S promoter, which was used as an internal control. LUC and GUS enzymatic assays were performed as described (Lin et al., 2007).
Yeast Two-Hybrid Test
Yeast two-hybrid assays for Ta CYP20-2 interaction with At BZR1 and RGA were performed using the Matchmaker system (Clontech). The open reading frames of Ta CYP20-2 and RGA were inserted in-frame into both pGADT7 and pGBKT7 vectors. Yeast strain AH109 (Clontech) was transformed with pGADT7-TaCYP20-2 and pGBKT7-BZR1, pGADT7-RGA and pGBKT7-TaCYP20-2 plasmids by the lithium acetate–mediated method. Transformants were plated on synthetic dropout selection medium lacking adenine, His, Leu, and Trp. Colonies with a positive signal were examined by activating the lacZ reporter gene.
In Vitro Pull-Down Assay
The fusion proteins from E. coli DH5α cells harboring pGEX4T-1-CYP20-2 and pGEX4T-1 plasmids were purified with glutathione Sepharose 4B beads. The beads bound with GST-CYP20-2 and GST were washed with PBS (0.14 M NaCl, 2.7 mM KCl, 10.1 mM Na2HPO4, and 1.8 mM KH2PO4) for GST pull-down assays. Each 300-μL reaction contained total protein (150 μg) from mx3 in 2 mM DTT, 5% glycerol, 1% Triton X-100, and 1 mM phenylmethylsulfonyl fluoride. After being added to glutathione Sepharose 4B beads carrying a GST fusion protein, reaction mixtures were incubated for at least 1 h at 4°C under gentle rotation. After being washed five times with PBS, beads were resuspended with 2× loading buffer (100 mM Tris-HCl, pH 6.8, 200 mM DTT, 4% SDS, 20% glycerol, and 0.2% bromophenol blue). Proteins were eluted at 95°C for 10 min and separated by 12% SDS-PAGE for Coomassie Brilliant Blue staining and immunodetection with anti-GFP antiserum at 1:5000 dilution.
Coimmunoprecipitation Assay
Coimmunoprecipitation assays were performed as described (Zhang et al., 2009) with some modifications. Tobacco (Nicotiana tabacum) leaves coexpressing At CYP20-2-YFP and BZR1-FLAG were ground in immunoprecipitation buffer and centrifuged at 20,000g for 20 min. Supernatant (1.5 mL) was incubated with anti-GFP coupled to Protein G Sepharose beads overnight under gentle rotation. Beads were washed four times with wash buffer, and the proteins were eluted at 95°C for 10 min in 2× loading buffer (100 mM Tris-HCl, pH 6.8, 200 mM DTT, 4% SDS, 20% glycerol, and 0.2% bromophenol blue).
Fusion Protein Preparation and PPIase Activity Assay
The cDNA of CYP20-2 was cloned into the pGEX4T-1 vector to generate pGEX4T-1-CYP20-2 containing a GST-CYP20-2 fusion construct driven by a lac I promoter. The recombined plasmid was transformed into E. coli DH5α cells and grown at 37°C. The GST-CYP20-2 fusion protein was induced by 1 mM isopropyl β-D-1-thiogalactopyranoside for 5 h and purified by glutathione affinity chromatography as described in the Bulk and RediPack GST purification kit (Pharmacia).
PPIase activity assays were performed as described (Fischer et al., 1989). The typical assay mixture consisted of 35 mM HEPES buffer, pH 8.0, 100 μM N-succinyl-Ala-Ala-Pro-Phe-p-nitroanilide dissolved in 60% (v/v) aqueous dimethyl sulfoxide, 10 μM chymotrypsin, and GST-CYP20-2 or GST at 10 nM. Components of the assay mixture other than chymotrypsin were combined and preincubated at room temperature. Chymotrypsin was added and quickly mixed to initiate the reaction, and absorbance was read at 390 nm every 0.5 s for 2 min with a nucleic acid and protein analyzer (Beckman Coulter DU640).
FTIR
FTIR spectroscopy was performed as described (Li et al., 2009a). The fusion proteins MBP-BZR1, MBP-mBZR1, GST-CYP20-2, MBP, and GST were purified and dissolved in PBS as described previously. PBS was replaced with D2O buffer containing 50 mM K2DPO4/KD2PO4 (pD 7.0) five times using a Microcon YM-3 centrifugal filter unit (Millipore) and Amicon Ultra-0.5 mL, 10-kD Centrifugal Filter Unit (Millipore). The final concentration of MBP-BZR1 and MBP was 12 mg/mL, that of MBP-mBZR1 was 1.25 mg/mL, and that of GST and GST-CYP20-2 was 0.8 mg/mL. FTIR spectra were collected with a spectrometer (ABB-BOMEM; Bureau) equipped with a broad-band Mercury-Cadmium-Telluride detector cooled by liquid nitrogen. A two-compartment CaF2 sample cell with a 56-mm-thick Teflon spacer was used for the protein sample and reference D2O or proteins. We collected FTIR for MBP, MBP-BZR1, and MBP-mBZR1 with GST-CYP20-2 or GST in D2O with a volume ratio of 2:1. On average, 50 scans were taken for each spectrum. The assignments of the observed bands in both FTIR and time-resolved IR spectra were performed as described (Ye et al., 2007; Li et al., 2009a).
Isolation of Nucleoprotein and Immunoblot Analysis
Histone-enriched nucleoprotein extraction from 2-week-old Arabidopsis seedlings was performed as described (Bowler et al., 2004). The nuclear and cytosolic fractions were separated as described (Gampala et al., 2007). Proteins were analyzed by 15% SDS-PAGE and transferred to polyvinylidene difluoride membranes. Membranes containing the proteins were incubated with antibodies against GFP (1:10,000; G1544; Sigma-Aldrich), H3K4 dimethylation (1:2000), H3K4 trimethylation (1:3000; 17-614; Millipore), H3 acetylation (1:10,000; 17-10241; Millipore), and H3 (1:10,000; 17-10046; Millipore).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: At CYP20-2, At5G13120; FKBP13, At5g45680; FLC, AT5G10140; FLD, AT3G10390; SOC1, AT2G45660; BZR1, AT1G75080; CPD, AT5G05690; DWF4, At3g50660; BR6OX, AT5G38970, Ta CYP20-2, AAP44537; At GA20ox1, At4G25420; and At GA2ox2, At1G30040. Sequences of PCR primers used in this study are shown in Supplemental Table 2 online.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Real-Time PCR Analysis of Autonomous Pathway Gene Expression in Col, 35S:AtCYP20-2 L2 (AOXL2), 35S:TaCYP20-2 (TOXL6), pBZR1:mBZR1-CFP (mx3), and pBZR1:BZR1-CFP Transgenic Arabidopsis.
Supplemental Figure 2. cyp20-2 fkbp13 cyp40RNAi Lines Showed Late Flowering.
Supplemental Figure 3. flc-3 Can Rescue the Late Flowering Phenotype of Overexpression Ta CYP20-2 Lines.
Supplemental Figure 4. Immunoblot Analysis of Phosphorylation and Unphosphorylation of mBZR1-CFP with or without BL Treatment in mx3.
Supplemental Figure 5. Schematic Diagram to Elaborate the Effect of CYP20-2 on the Secondary Structure of BZR1.
Supplemental Figure 6. Real-Time PCR Analysis of the Expression of FLD and FLC in bri1-5 and bzr1-1D.
Supplemental Figure 7. pBZR1:BZR1 Transgenic Arabidopsis Did Not Show Obvious Flowering Phenotypes Compared with Col-0.
Supplemental Figure 8. Real-Time PCR Analysis of BR Biosynthetic Genes in mx3, mx3 35S:TaCYP20-2, and mx3 35S:AtCYP20-2 Arabidopsis.
Supplemental Table 1. Putative BZR1-Targeted Flowering-Related Genes.
Supplemental Table 2. Sequences of PCR Oligos Used in This Study.
Acknowledgments
We thank Zhiyong Wang for comments on the project and for gifts of the plant materials pBZR1:mBZR1Pro234-Leu-CFP (mx3), pBZR1:BZR1-CFP (W2C) transgenic Arabidopsis, and bzr1-1D mutant, Fuquan Liu for the gift of fld-4, and Paul Dupree for the gift of the pBIN vector. This work was supported by the National Natural Science Foundation of China (31070677) and the Foundation for Innovative Research Groups of the National Natural Science Foundation of China (31121065).
AUTHOR CONTRIBUTIONS
K.C., Y.X., Y.Z., B.L., and C.Y. designed the research. Y.Z., B.L., D.Z., and S.G. performed the research. K.C., Y.Z., and B.L. analyzed the data. Z.M. performed bioinformatics analysis. Y.W., H.L., and S.L. analyzed the protein secondary structure on FTIR spectroscopy. K.C., Y.Z., Y.X., C.Y., and Y.W. wrote the article.
Glossary
- GA
gibberellin
- BR
brassinosteroid
- PPIase
peptidyl-prolyl cis-trans isomerase
- BRRE
BR response cis-element
- EMSA
electrophoretic mobility shift assay
- ChIP
chromatin immunoprecipitation
- CaMV
cauliflower mosaic virus
- Brz
brassinazole
- FTIR
Fourier transform infrared
- D2O
the chemical formula of heavy water
References
- Bai M.Y., Shang J.X., Oh E., Fan M., Bai Y., Zentella R., Sun T.P., Wang Z.Y. (2012). Brassinosteroid, gibberellin and phytochrome impinge on a common transcription module in Arabidopsis. Nat. Cell Biol. 14: 810–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai M.Y., Zhang L.Y., Gampala S.S., Zhu S.W., Song W.Y., Chong K., Wang Z.Y. (2007). Functions of OsBZR1 and 14-3-3 proteins in brassinosteroid signaling in rice. Proc. Natl. Acad. Sci. USA 104: 13839–13844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berardini T.Z., Bollman K., Sun H., Poethig R.S. (2001). Regulation of vegetative phase change in Arabidopsis thaliana by cyclophilin 40. Science 291: 2405–2407 [DOI] [PubMed] [Google Scholar]
- Berr A., Xu L., Gao J., Cognat V., Steinmetz A., Dong A., Shen W.H. (2009). SET DOMAIN GROUP25 encodes a histone methyltransferase and is involved in FLOWERING LOCUS C activation and repression of flowering. Plant Physiol. 151: 1476–1485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowler C., Benvenuto G., Laflamme P., Molino D., Probst A.V., Tariq M., Paszkowski J. (2004). Chromatin techniques for plant cells. Plant J. 39: 776–789 [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Clouse S.D., Langford M., McMorris T.C. (1996). A brassinosteroid-insensitive mutant in Arabidopsis thaliana exhibits multiple defects in growth and development. Plant Physiol. 111: 671–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X., Gu L., Liu C., Lu T., Lu F., Lu Z., Cui P., Pei Y., Wang B., Hu S., Cao X. (2010). Arginine methylation mediated by the Arabidopsis homolog of PRMT5 is essential for proper pre-mRNA splicing. Proc. Natl. Acad. Sci. USA 107: 19114–19119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domagalska M.A., Schomburg F.M., Amasino R.M., Vierstra R.D., Nagy F., Davis S.J. (2007). Attenuation of brassinosteroid signaling enhances FLC expression and delays flowering. Development 134: 2841–2850 [DOI] [PubMed] [Google Scholar]
- Edvardsson A., Shapiguzov A., Petersson U.A., Schröder W.P., Vener A.V. (2007). Immunophilin AtFKBP13 sustains all peptidyl-prolyl isomerase activity in the thylakoid lumen from Arabidopsis thaliana deficient in AtCYP20-2. Biochemistry 46: 9432–9442 [DOI] [PubMed] [Google Scholar]
- Fischer G., Wittmann-Liebold B., Lang K., Kiefhaber T., Schmid F.X. (1989). Cyclophilin and peptidyl-prolyl cis-trans isomerase are probably identical proteins. Nature 337: 476–478 [DOI] [PubMed] [Google Scholar]
- Fornara, F., de Montaigu, A., and Coupland, G. (2010). SnapShot: Control of flowering in Arabidopsis Cell 141: 550. [DOI] [PubMed]
- Gampala S.S., et al. (2007). An essential role for 14-3-3 proteins in brassinosteroid signal transduction in Arabidopsis. Dev. Cell 13: 177–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J.X., Gendron J.M., Sun Y., Gampala S.S., Gendron N., Sun C.Q., Wang Z.Y. (2005). BZR1 is a transcriptional repressor with dual roles in brassinosteroid homeostasis and growth responses. Science 307: 1634–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J.X., Gendron J.M., Yang Y., Li J., Wang Z.Y. (2002). The GSK3-like kinase BIN2 phosphorylates and destabilizes BZR1, a positive regulator of the brassinosteroid signaling pathway in Arabidopsis. Proc. Natl. Acad. Sci. USA 99: 10185–10190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y., Doyle M.R., Amasino R.M. (2004). PAF1-complex-mediated histone methylation of FLOWERING LOCUS C chromatin is required for the vernalization-responsive, winter-annual habit in Arabidopsis. Genes Dev. 18: 2774–2784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y., Michaels S.D., Amasino R.M. (2003). Regulation of flowering time by histone acetylation in Arabidopsis. Science 302: 1751–1754 [DOI] [PubMed] [Google Scholar]
- Homann O.R., Johnson A.D. (2010). MochiView: Versatile software for genome browsing and DNA motif analysis. BMC Biol. 8: 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang D., Yang W., He Y., Amasino R.M. (2007). Arabidopsis relatives of the human lysine-specific Demethylase1 repress the expression of FWA and FLOWERING LOCUS C and thus promote the floral transition. Plant Cell 19: 2975–2987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H., Li S., Villegas A., Jr (2006). Down-regulation of the 26S proteasome subunit RPN9 inhibits viral systemic transport and alters plant vascular development. Plant Physiol. 142: 651–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Xu W., Xu Y., Zhang Y., Wang T., Bai Y., Han C., Zhang A., Xu Z., Chong K. (2010a). Integrative study on proteomics, molecular physiology, and genetics reveals an accumulation of cyclophilin-like protein, TaCYP20-2, leading to an increase of Rht protein and dwarf in a novel GA-insensitive mutant (gaid) in wheat. J. Proteome Res. 9: 4242–4253 [DOI] [PubMed] [Google Scholar]
- Li H., Ke H., Ren G., Qiu X., Weng Y.X., Wang C.C. (2009a). Thermal-induced dissociation and unfolding of homodimeric DsbC revealed by temperature-jump time-resolved infrared spectra. Biophys. J. 97: 2811–2819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Jin H. (2007). Regulation of brassinosteroid signaling. Trends Plant Sci. 12: 37–41 [DOI] [PubMed] [Google Scholar]
- Li J., Li Y., Chen S., An L. (2010b). Involvement of brassinosteroid signals in the floral-induction network of Arabidopsis. J. Exp. Bot. 61: 4221–4230 [DOI] [PubMed] [Google Scholar]
- Li L., Yu X., Thompson A., Guo M., Yoshida S., Asami T., Chory J., Yin Y. (2009b). Arabidopsis MYB30 is a direct target of BES1 and cooperates with BES1 to regulate brassinosteroid-induced gene expression. Plant J. 58: 275–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q.F., Wang C., Jiang L., Li S., Sun S.S., He J.X. (2012). An interaction between BZR1 and DELLAs mediates direct signaling crosstalk between brassinosteroids and gibberellins in Arabidopsis. Sci. Signal. 5: ra72. [DOI] [PubMed] [Google Scholar]
- Lim M.H., Kim J., Kim Y.S., Chung K.S., Seo Y.H., Lee I., Kim J., Hong C.B., Kim H.J., Park C.M. (2004). A new Arabidopsis gene, FLK, encodes an RNA binding protein with K homology motifs and regulates flowering time via FLOWERING LOCUS C. Plant Cell 16: 731–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin R., Ding L., Casola C., Ripoll D.R., Feschotte C., Wang H. (2007). Transposase-derived transcription factors regulate light signaling in Arabidopsis. Science 318: 1302–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F., Marquardt S., Lister C., Swiezewski S., Dean C. (2010). Targeted 3′ processing of antisense transcripts triggers Arabidopsis FLC chromatin silencing. Science 327: 94–97 [DOI] [PubMed] [Google Scholar]
- Liu F., Quesada V., Crevillén P., Bäurle I., Swiezewski S., Dean C. (2007). The Arabidopsis RNA-binding protein FCA requires a lysine-specific demethylase 1 homolog to downregulate FLC. Mol. Cell 28: 398–407 [DOI] [PubMed] [Google Scholar]
- Lu C., Armstrong J.S. (2007). Role of calcium and cyclophilin D in the regulation of mitochondrial permeabilization induced by glutathione depletion. Biochem. Biophys. Res. Commun. 363: 572–577 [DOI] [PubMed] [Google Scholar]
- Lu F., Cui X., Zhang S., Jenuwein T., Cao X. (2011). Arabidopsis REF6 is a histone H3 lysine 27 demethylase. Nat. Genet. 43: 715–719 [DOI] [PubMed] [Google Scholar]
- Luo X.M., et al. (2010). Integration of light- and brassinosteroid-signaling pathways by a GATA transcription factor in Arabidopsis. Dev. Cell 19: 872–883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Q., Dai X., Xu Y., Guo J., Liu Y., Chen N., Xiao J., Zhang D., Xu Z., Zhang X., Chong K. (2009). Enhanced tolerance to chilling stress in OsMYB3R-2 transgenic rice is mediated by alteration in cell cycle and ectopic expression of stress genes. Plant Physiol. 150: 244–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matouschek A., Rospert S., Schmid K., Glick B.S., Schatz G. (1995). Cyclophilin catalyzes protein folding in yeast mitochondria. Proc. Natl. Acad. Sci. USA 92: 6319–6323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouradov A., Cremer F., Coupland G. (2002). Control of flowering time: Interacting pathways as a basis for diversity. Plant Cell 14 (suppl.): S111–S130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noh B., Lee S.H., Kim H.J., Yi G., Shin E.A., Lee M., Jung K.J., Doyle M.R., Amasino R.M., Noh Y.S. (2004). Divergent roles of a pair of homologous jumonji/zinc-finger-class transcription factor proteins in the regulation of Arabidopsis flowering time. Plant Cell 16: 2601–2613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazhouhandeh M., Molinier J., Berr A., Genschik P. (2011). MSI4/FVE interacts with CUL4-DDB1 and a PRC2-like complex to control epigenetic regulation of flowering time in Arabidopsis. Proc. Natl. Acad. Sci. USA 108: 3430–3435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouse D.T., Sheldon C.C., Bagnall D.J., Peacock W.J., Dennis E.S. (2002). FLC, a repressor of flowering, is regulated by genes in different inductive pathways. Plant J. 29: 183–191 [DOI] [PubMed] [Google Scholar]
- Ryu H., Kim K., Cho H., Hwang I. (2010a). Predominant actions of cytosolic BSU1 and nuclear BIN2 regulate subcellular localization of BES1 in brassinosteroid signaling. Mol. Cells 29: 291–296 [DOI] [PubMed] [Google Scholar]
- Ryu H., Cho H., Kim K., Hwang I. (2010b). Phosphorylation dependent nucleocytoplasmic shuttling of BES1 is a key regulatory event in brassinosteroid signaling. Mol. Cells 29: 283–290 [DOI] [PubMed] [Google Scholar]
- Ryu H., Kim K., Cho H., Park J., Choe S., Hwang I. (2007). Nucleocytoplasmic shuttling of BZR1 mediated by phosphorylation is essential in Arabidopsis brassinosteroid signaling. Plant Cell 19: 2749–2762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Searle I., He Y., Turck F., Vincent C., Fornara F., Kröber S., Amasino R.A., Coupland G. (2006). The transcription factor FLC confers a flowering response to vernalization by repressing meristem competence and systemic signaling in Arabidopsis. Genes Dev. 20: 898–912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y., Lan F., Matson C., Mulligan P., Whetstine J.R., Cole P.A., Casero R.A., Shi Y. (2004). Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 119: 941–953 [DOI] [PubMed] [Google Scholar]
- Sonmez C., Bäurle I., Magusin A., Dreos R., Laubinger S., Weigel D., Dean C. (2011). RNA 3′ processing functions of Arabidopsis FCA and FPA limit intergenic transcription. Proc. Natl. Acad. Sci. USA 108: 8508–8513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., et al. (2010). Integration of brassinosteroid signal transduction with the transcription network for plant growth regulation in Arabidopsis. Dev. Cell 19: 765–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W., Kim T.W., Oses-Prieto J.A., Sun Y., Deng Z., Zhu S., Wang R., Burlingame A.L., Wang Z.Y. (2008). BSKs mediate signal transduction from the receptor kinase BRI1 in Arabidopsis. Science 321: 557–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W., et al. (2011). PP2A activates brassinosteroid-responsive gene expression and plant growth by dephosphorylating BZR1. Nat. Cell Biol. 13: 124–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trupkin S.A., Mora-García S., Casal J.J. (2012). The cyclophilin ROC1 links phytochrome and cryptochrome to brassinosteroid sensitivity. Plant J. 71: 712–723 [DOI] [PubMed] [Google Scholar]
- Wang L., Xu Y., Zhang C., Ma Q., Joo S.H., Kim S.K., Xu Z., Chong K. (2008). OsLIC, a novel CCCH-type zinc finger protein with transcription activation, mediates rice architecture via brassinosteroids signaling. PLoS ONE 3: e3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Zhang Y., Ma Q., Zhang Z., Xue Y., Bao S., Chong K. (2007). SKB1-mediated symmetric dimethylation of histone H4R3 controls flowering time in Arabidopsis. EMBO J. 26: 1934–1941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z.Y., Nakano T., Gendron J., He J., Chen M., Vafeados D., Yang Y., Fujioka S., Yoshida S., Asami T., Chory J. (2002). Nuclear-localized BZR1 mediates brassinosteroid-induced growth and feedback suppression of brassinosteroid biosynthesis. Dev. Cell 2: 505–513 [DOI] [PubMed] [Google Scholar]
- Xu Y.L., Li L., Wu K., Peeters A.J., Gage D.A., Zeevaart J.A. (1995). The GA5 locus of Arabidopsis thaliana encodes a multifunctional gibberellin 20-oxidase: Molecular cloning and functional expression. Proc. Natl. Acad. Sci. USA 92: 6640–6644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye M., Zhang Q.L., Li H., Weng Y.X., Wang W.C., Qiu X.G. (2007). Infrared spectroscopic discrimination between the loop and alpha-helices and determination of the loop diffusion kinetics by temperature-jump time-resolved infrared spectroscopy for cytochrome c. Biophys. J. 93: 2756–2766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi H., Friedman J.L., Ferreira P.A. (2007). The cyclophilin-like domain of Ran-binding protein-2 modulates selectively the activity of the ubiquitin-proteasome system and protein biogenesis. J. Biol. Chem. 282: 34770–34778 [DOI] [PubMed] [Google Scholar]
- Yin Y., Vafeados D., Tao Y., Yoshida S., Asami T., Chory J. (2005). A new class of transcription factors mediates brassinosteroid-regulated gene expression in Arabidopsis. Cell 120: 249–259 [DOI] [PubMed] [Google Scholar]
- Yin Y., Wang Z.Y., Mora-Garcia S., Li J., Yoshida S., Asami T., Chory J. (2002). BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell 109: 181–191 [DOI] [PubMed] [Google Scholar]
- Yoo S.D., Cho Y.H., Sheen J. (2007). Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat. Protoc. 2: 1565–1572 [DOI] [PubMed] [Google Scholar]
- Yu X., Li L., Li L., Guo M., Chory J., Yin Y. (2008). Modulation of brassinosteroid-regulated gene expression by Jumonji domain-containing proteins ELF6 and REF6 in Arabidopsis. Proc. Natl. Acad. Sci. USA 105: 7618–7623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C., Xu Y., Guo S., Zhu J., Huan Q., Liu H., Wang L., Luo G., Wang X., Chong K. (2012). Dynamics of brassinosteroid response modulated by negative regulator LIC in rice. PLoS Genet. 8: e1002686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L.Y., et al. (2009). Antagonistic HLH/bHLH transcription factors mediate brassinosteroid regulation of cell elongation and plant development in rice and Arabidopsis. Plant Cell 21: 3767–3780 [DOI] [PMC free article] [PubMed] [Google Scholar]