In contrast with the prevalent view that supernumerary B chromosomes do not contain genes, this work demonstrates that the B chromosome of rye contributes to the transcriptome and also affects in trans the activity of standard A chromosome–located genes. The authors propose a model for the evolution of B chromosome–located pseudogenes.
Abstract
B chromosomes (Bs) are dispensable components of the genomes of numerous species. In contrast with the prevalent view that Bs do not harbor genes, our recent sequence analysis revealed that Bs of rye (Secale cereale) are rich in gene-derived sequences. We compared these gene-like fragments of the rye B with their ancestral A-located counterparts and confirmed an A chromosomal origin and the pseudogenization of B-located gene-like fragments. About 15% of the pseudogene-like fragments on Bs are transcribed in a tissue-type and genotype-specific manner. In addition, B-located sequences can cause in trans down- or upregulation of A chromosome–encoded genic fragments. Phenotypes and effects associated with the presence of Bs might be explained by the activity of B-located pseudogenes. We propose a model for the evolution of B-located pseudogenes.
INTRODUCTION
Supernumerary B chromosomes (Bs) are optional additions to the basic set of standard A chromosomes (As) and occur in all eukaryotic groups. They differ from the As in inheritance, and the fact that Bs are not required for the normal growth and development of the host organism. Due to the dispensable nature of Bs, they can be present or absent among individuals of the same population in a species (Jones and Rees, 1982; reviewed in Camacho et al., 2000 and Houben et al., 2011).
Although Bs are not essential, some phenotypic effects have been reported. These effects are usually cumulative, depending upon the number and not the presence or absence of Bs. In low numbers, Bs have little if any influence on the phenotype, but in high numbers they often have a negative influence on fitness and fertility of the carrier organism (Jones and Rees, 1982; Carlson, 2009; reviewed in Jones, 1995 and Bougourd and Jones, 1997). Bs are not necessarily inert, although their moderate effects suggest a lack of major functional genes. For instance, under drought stress conditions, seeds carrying Bs had an advantage over seeds not carrying Bs in germination in Allium schoenoprasum (Holmes and Bougourd, 1991); in cichlid fishes, Bs play a role in sex determination (Yoshida et al., 2011); and in the fungus Nectria hematococca, Bs account for antibiotic resistance and pathogenicity (Coleman et al., 2009). Besides the activity of B-located ribosomal genes (Leach et al., 2005), transcription of only few protein encoding genes has so far been associated with Bs (reviewed in Houben et al., 2013).
Recent sequence characterization of the rye (Secale cereale) B (Martis et al., 2012) provides a unique opportunity for the analysis of the evolution and biology of this enigmatic genome component. In contrast with the prevalent view that Bs do not harbor genic sequences, sequencing of sorted As and Bs showed that rye Bs are rich in gene-derived sequences. This allowed us to trace their origin from fragments of As with the largest portions corresponding to rye As 3R and 7R, along with genic sequences derived from all other As (Martis et al., 2012). Around 4000 homologous nonredundant genes were identified after comparison of rye B-derived sequences to Brachypodium distachyon. However, this analysis did not allow conclusions regarding the transcription activity of B-located genic sequences.
Here, we compare selected gene-like fragments of B with their ancestral A-located counterparts. The transcription behavior of B-located gene-like fragments was analyzed in different tissues as well as in different accessions of cultivated rye (S. cereale subsp cereale) and of the subspecies S. cereale subsp afghanicum and subsp segetale. Our data demonstrate that B contributes to the transcriptome and affects in trans the activity of A-located genes. Finally, we propose a model for the evolution of B-located pseudogenes.
RESULTS
Confirmation and Chromosomal Allocation of B-Located Gene-Like Fragments
For in-depth analysis, we selected 15 putative rye B-located gene-like fragments with high similarity to genes of B. distachyon and with a length between 513 and 1654 bp (see Supplemental Table 1 online). First, the in silico predicted sequences were confirmed by PCR with genomic DNA of rye and wheat (Triticum aestivum) with (+B) and without Bs (0B) as well as with DNA of flow-sorted rye Bs (sB). The primers used in this analysis anchored in the outermost flanking regions of gene-like fragments to generate the longest amplicons (hereafter called genomic DNA [gDNA]-primers) (Figure 1A; see Supplemental Table 2 and Supplemental Figure 1 online). Gene-like fragments 1 to 5 were B specific as there was no amplification in rye and wheat without Bs. Fragments 6 to 15 were amplified from rye +B, 0B, and sorted Bs, indicating that these B sequences have counterparts in the As. Fragments 6 and 7 produced extra amplicons in rye +B only, likely due to B-specific size polymorphisms. Only fragment 13 displayed an amplicon of expected size in wheat 0B. Fragment 14 revealed two rye amplicons in 0B and only one in +B, likely due to the preferential amplification of the smaller fragment in +B only.
Figure 1.
Confirmation of B-Located Gene-Like Fragments.
PCR performed using gDNA of rye and wheat with and without Bs and sorted rye B (A) and two wheat-rye B addition lines carrying either short- or long-arm iso-B chromosomes (B). Quality and equal quantity of DNA for different genotypes was shown by amplification using Bilby (a rye-specific sequence) and GAPDH (wheat and rye specific) primer pairs. Lengths of the expected amplicons are indicated.
Next, wheat lines carrying isochromosomes of either the short or the long B arm (Endo et al., 2008) were used to assign the B arm encoding the fragments (Figure 1B). Eight gene-like fragments (1, 2, 3, 5, 7, 8, 9, and 12) were encoded by the long and five (4, 6, 10, 11, and 15) by both B arms. The location of fragments at both chromosome arms suggests multiple copies of the gene-like fragments. Alternatively, these fragments are encoded in the proximity of the centromere and both types of isochromosomes share the same centromeric region. High sequence similarity to wheat did not allow chromosome arm mapping of fragments 13 and 14.
A multiple A-chromosomal origin of the B was confirmed by PCR with DNA of the wheat-rye chromosome addition lines 1R-7R (see Supplemental Figure 2A and Supplemental Table 2 online). For the fragments 8 and 10, products of expected size were detected for the rye A pairs 1 and 7, respectively. For fragments 7, 13, and 14, amplicons from several rye chromosome pairs were found. It is likely that the corresponding primers aligned with regions conserved in different members of a gene family that are located in different As of rye. The failure of amplification of expected size could be due to sequence polymorphisms because different rye genotypes were used for sequencing of the B and for generation of the employed wheat-rye addition lines.
To investigate the flanking sequences of a B-located gene-like fragment and its A-chromosomal counterpart, the sequences of fragment 12 and its A-chromosomal counterpart were extended by genome walking. Fragment 12 was selected as an example because of its rye-specific position, which enabled unambiguous extension of B-located sequences by genome walking.
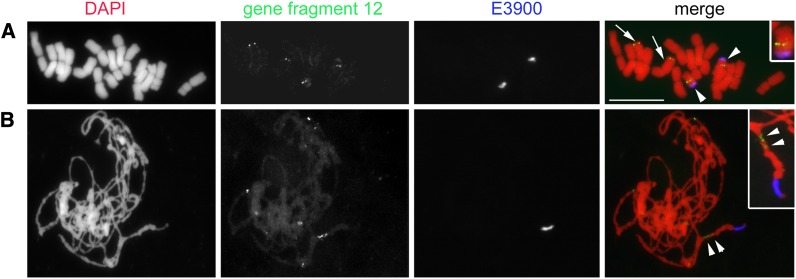
The A and B sequences were extended to 3637 and 1906 bp, respectively (accession numbers KC623498 and KC623499, respectively). In addition to general sequence similarity (see Supplemental Figure 3 online), polymorphisms were observed, including single nucleotide polymorphisms (SNPs) and insertions or deletions in regions of high similarity. The 3′ side of B-located sequences completely differs from A-located sequences. Furthermore, amplification of the region at the 5′ side from sorted B failed, likely due to sequence differences in the restriction site of the enzymes used for genome walking. The BLAST analysis of the extended sequences on A and B did not show any similarity with known repetitive sequences. To confirm the result of genome walking, fluorescence in situ hybridization (FISH) was performed using as probe the A-derived 3.637-bp-long fragment. Distinct hybridization signals were found on Bs and on one A pair in rye mitotic metaphase (Figure 2A) and pachytene cells (Figure 2B). In agreement with the PCR mapping result, fragment 12 is located on the B long arm (Figure 1B). The presence of multiple signals on Bs suggests a secondary amplification of fragment 12.
Figure 2.
Chromosomal Locations of Gene-Like Fragment 12 by FISH.
Mitotic metaphase (A) and meiotic pachytene (B) cells of rye with Bs after single-copy FISH with labeled extended gene-like fragment 12 (in green). FISH with the B-specific E3900 repeat (in blue) allowed the identification of Bs. Note multiple signals (arrow heads) of fragment 12 along the Bs in pachytene chromosome. A-localized FISH signals are indicated by arrows. The inset shows further enlarged B. DAPI, 4′,6-diamidino-2-phenylindole. Bar = 10 µm.
Paralogous Gene-Like Fragments on As and Bs Show Sequence Polymorphisms
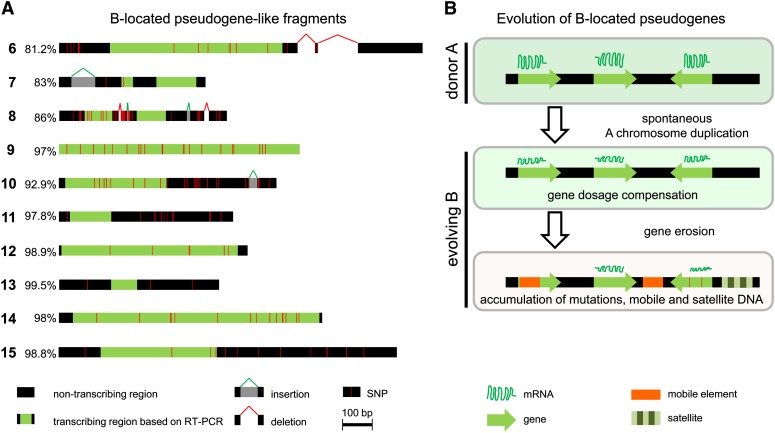
To investigate the similarity between A- and B-located paralogous sequences, sequence comparison was performed for the fragments 6 to 15. Alignment between sequences derived from rye 0B PCR fragments and the next-generation sequencing reads of sorted Bs (accession numbers KC623167 to KC623536) revealed for all fragments polymorphisms that were mainly due to SNPs, insertions, and deletions. The similarity ranged from 81 to 99% (Figure 3A). In addition, the B-derived fragment 6 had a large deletion of around 200 bp, and fragments 7 and 10 had large insertions of 83 and 28 bp in length. Nevertheless, the general exon-intron structure (see Supplemental Figure 1 online) remained conserved between A and B sequences for fragments 7, 8, 13, and 15. Hence, the B-located gene-like fragments could be categorized as nonprocessed pseudogene-like fragments, since an exon-intron structure and high sequence similarity still exists between the duplicated genes and their corresponding parent genes.
Figure 3.
Pseudogenization of B-Located Gene Fragments.
(A) Schemata represent the sequence comparison between rye B-located pseudogene-like fragments (6 to 15) and their A-located parental counterparts. Percentage of similarity, transcribing region, and position of sequence polymorphisms are indicated.
(B) Model for the evolution of B-located pseudogenes. The B of rye descended from As after a spontaneous whole or partial genome duplication. Meiotic recombination of proto-B with donor As became restricted. Proto- B still shows sequence similarity to the parental As. The increased gene dosage may affect gene expression and proto-B-located genes might have been suppressed by, for example, dosage compensation. Finally, B-located gene sequences became pseudogenized by mutations and accumulation of mobile and satellite DNA.
Pseudogene-Like Fragments on Bs from Different Rye Accessions Are Largely Conserved
Due to the dispensable nature of B chromosomes, their sequences are less constrained by selection. Therefore, they are prone to rapid sequence evolution via the Muller's Ratchet mechanism as proposed (Green, 1990). To address whether the identified B-located pseudogene-like fragments diverged in Bs of different geographical origins, we examined B-possessing accessions of S. cereale subsp cereale collected from Turkey, Iran, China, Korea, and Japan. In addition, we included two accessions of the weedy rye with and without Bs (S. cereale subsp afghanicum and subsp segetale from Afghanistan and Pakistan, respectively). A monophyletic origin of Bs in the genus Secale was previously suggested (Niwa and Sakamoto, 1995; Marques et al., 2013).
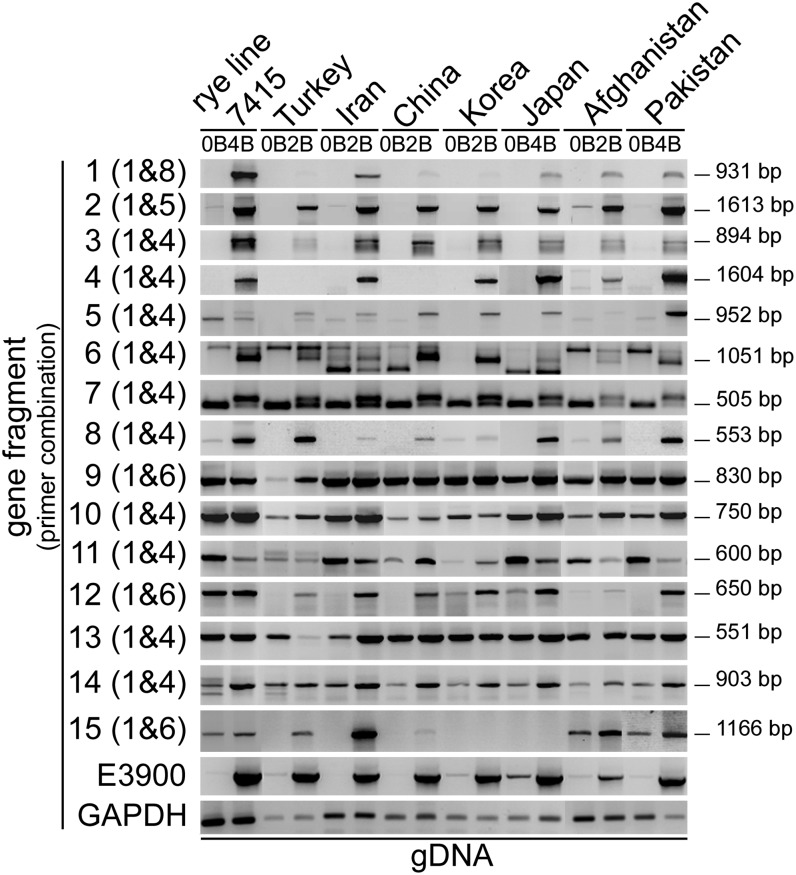
The presence of the pseudogene-like fragments in different rye accessions was tested by PCR with genomic DNA (Figure 4; see Supplemental Table 2 and Supplemental Figure 1 online). All B-specific fragments 1 to 5, identified in reference line 7415, were found only in +B plants of most accessions. In addition, a B-specific pattern was found in some accessions for fragments 6, 7, 8, 12, and 15. Fragments 9, 10, 11, 13, and 14 occurred in +B and 0B accessions, similar to the reference genotype. A B-specific extra band was found in almost all accessions for fragments 6 and 7. For fragments 4 and 15 in few accessions no amplification was found, likely due to sequence differences within the primer region.
Figure 4.
Pseudogene-Like Fragments Are Conserved on the Bs from Different Rye Accessions.
PCR performed with genomic DNA from rye (S. cereale subsp cereale from Turkey, Iran, China, Korea, and Japan, S. cereale subsp afghanicum from Afghanistan, and S. cereale subsp segetale from Pakistan) with and without Bs to determine the presence of the identified pseudogene-like fragments. B-specific repeat E3900 primers were used to demonstrate the presence of Bs. GAPDH-specific primers were used to show equal amounts of genomic DNA. Lengths of the expected amplicons are indicated.
Assuming a weak selection pressure on Bs, gene erosion could differ between Bs of different populations. To address whether the pseudogene-like fragments on Bs from accessions of different geographical regions diverged, we analyzed the B-located sequences of fragments 2, 4, and 8 of different accessions (see Supplemental Figure 4 online). A- and B-located sequences of fragment 8 formed chromosome-type-specific clades. The A-located sequences showed an accession specific pattern, while all B-located fragments displayed a random pattern. Hence, the rye B-located fragments of different accessions did not diverge into clear accession-specific clusters after geographical spreading.
B-Located Pseudogene-Like Fragments Are Transcribed in a Tissue- and Accession-Specific Manner
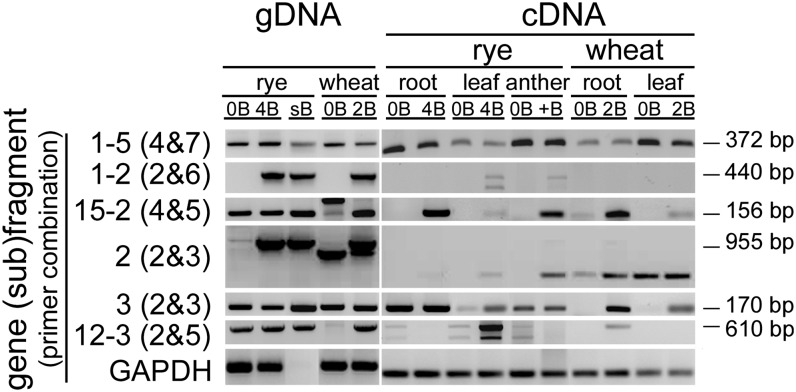
To evaluate the transcription activity of B-located pseudogene-like fragments we designed primers (see Supplemental Table 3 online) for RT-PCR (hereafter cDNA-primers) with cDNA prepared from different tissues of rye and wheat with and without B chromosomes. As none of the genomic DNA (gDNA)-primers were suitable for transcription analysis (see Supplemental Figure 2B online), cDNA-primers were designed based on the predicted intron/exon structures. Therefore, the genomic sequences of all fragments were BLASTed against the rye EST database (Haseneyer et al., 2011) and analyzed using FGENESH software (http://linux1.softberry.com/berry.phtml) (see Supplemental Figure 1 online). Although all fragments revealed regions with similarity to ESTs, a gene structure prediction was possible only for the fragments 1, 2, 7, 8, 9, 13, and 15. The species specificity of the cDNA-primers was tested with 0B and +B genomic DNA of both species. Due to the sequence similarity of wheat and rye transcripts (Haseneyer et al., 2011), rye-specific amplification was only found for the (sub)fragments 6, 9-3, and 12-3 (see Supplemental Figure 5 online). Gene (sub)fragments 1-2, 1-3, 1-6, 2, and 15-3 were B specific as no amplification was detected for 0B rye and 0B wheat.
Transcription activity of all pseudogene-like fragments was found in a species- and tissue-dependent manner. The highest activity was found in anthers with Bs (23 of 26 primer pairs tested) (see Supplemental Figure 5 online). The results are summarized in Figure 5 based on presence or absence of gene (sub)fragments on gDNA and their expression pattern. While the (sub)fragment 1-5 was expressed in all tested tissue types and species, all other 25 (sub)fragment showed tissue- and/or species-specific activity. Regardless of the B specificity at the genomic level, 77% of all (sub)fragments (20 out of 26) revealed transcription activity only in B-containing samples in a species- and tissue-type-specific manner. Importantly, four of these [15% of all (sub)fragments; i.e., 1-2, 1-3, 2, and 15-3] are encoded by Bs as no amplification was found in genomic 0B DNA. The B-derived transcription was sequence confirmed for (sub)fragments 1-2 and 2 (see Supplemental Figures 6A and 6B online). Six (sub)fragments (i.e., 1-4, 9-1, 9-3, 12-1, 13, and 15-2) were only found in +B rye in a tissue-specific manner even though amplification was found also in 0B genomic DNA. Similar was found for four (sub)fragments in +B wheat [i.e., the (sub)fragments 1-1, 3, 10, 12-3, and 15-1].
Figure 5.
B-Located Pseudogene-Like Fragments Are Transcribed in a Tissue- and Species-Specific Manner.
RT-PCR performed with cDNA derived from roots, leaves, and anthers of rye and of root and leaves of wheat with and without B chromosomes. Representative (sub)fragments are shown. PCR with genomic DNA was used as control. The presence of equal amounts of cDNA was tested by primers specific to GAPDH. Lengths of the expected amplicons from gDNA are indicated.
Sequencing of 12-1 and 15-2 transcripts revealed their A- and B-derived origin, respectively (see Supplemental Figures 6C and 6D online). Hence, upregulation of 12-1 is triggered in trans by the presence of Bs. The B-located pseudogene-like (sub)fragment 12-3 of rye downregulated in root and anthers in trans the expression of its corresponding A-located sequence. The A origin of the identified transcripts was sequence confirmed. In addition, a low level of unspliced B-derived transcripts was only found in anthers (see Supplemental Figure 6E online). Sequencing was performed for the gene fragments 1-2, 2, and 12-3 and splicing of A- and B-derived transcripts was confirmed. Subfragments 1-2, 2, and 12-3 revealed splicing of A- and B-derived transcripts because smaller products compared with gDNA PCR were found after RT-PCR and subsequent sequencing (see Supplemental Figures 6A, 6B, and 6E online). In silico translation of transcripts for (sub)fragments 1-2, 12-1, and 12-3 revealed, regardless of their chromosome origin and splicing patterns, several premature stop codons. No stop codon was predicted for the (sub)fragments 2 and 15-2.
Finally, we investigated whether the transcriptional activity of B pseudogene–like fragments in leaf tissue is conserved across different rye accessions. Again, first the presence of the corresponding sequences was confirmed by PCR with cDNA-primers and genomic DNA (see Supplemental Table 3 and Supplemental Figure 7 online). Except for (sub)fragment 15-3, which showed no amplification, all other analyzed pseudogene-like fragments had amplification patterns similar to reference rye in all accessions (see Supplemental Figure 7 online). Nevertheless, the sporadic absence of a gene (sub)fragment in some accessions was observed. The expression pattern of the confirmed (sub)fragments in different accessions could be classified in four classes (Figure 6; see Supplemental Figure 8 online). In the first class, (sub)fragments (2, 8, and 15-2) were preferentially expressed in +B plants, although for (sub)fragments 8 and 15-2 (see Supplemental Figure 7 online), sequences were present on As, too. In the second class, (sub)fragments were not transcribed in 0B and +B rye (1-1, 1-2, 1-3, 1-6, 9-3, 13, and 15-3). The (sub)fragments of the third class were transcriptionally active in 0B and +B plants of all accessions [(sub)fragments 1-5, 3, 4, 7, 11, and 15-1]. Class II and III were not informative because B originated transcripts were either absent or could not be discriminated from A-originated transcripts. Class IV showed a complex pattern. In some accessions, no transcription was found either in 0B and +B background (1-4, 5, 9-1, 9-2, 12-3, and 14), in 0B (1-4, 6, 10, 12-2, 12-3, and 14) or in +B plants (5, 12-2, and 12-3). The absence of transcription in +B material indicates a trans-modulation of A-located gene expression by B transcripts. Hence, our results indicate that some B-located pseudogene-like fragments are transcriptionally active and are able to modulate the expression level of A-located genes in trans. These features are conserved among Bs of different rye accessions.
Figure 6.
Transcription Activity of B Pseudogene-Like Fragments across Different Rye Accessions.
RT-PCR with cDNA of leaves from different rye accessions (S. cereale subsp cereale from Turkey, Iran, China, Korea, and Japan, S. cereale subsp afghanicum from Afghanistan, and S. cereale subsp segetale from Pakistan) with and without Bs. The transcription patterns were classified into four classes (I to IV). Class (I) (sub)fragments are expressed in +B plants only. Class (II) are not transcribed in 0B and +B plants. Class (III) are transcribed in all 0B and +B plants. Class (IV) shows complex expression patterns. Representative (sub)fragments for each group are shown. Lengths of the expected amplicons from gDNA are indicated.
DISCUSSION
B-Located Pseudogene-Like Fragments Descended from Different As
The presence and transcriptional activity of 15 rye B chromosome-located pseudogene-like fragments were tested in different accessions of S. cereale subsp cereale, S. cereale subsp afghanicum and subsp segetale, as well as in wheat with and without Bs from different geographical origins. The identification of A-located sequences with high similarity to the B-located pseudogene-like fragments 6 to 15 confirms the A chromosome origin of the rye B. No A-derived counterparts for the B-specific fragments 1 to 5 were found, likely due to mutations in the primer recognition sites. Amplification of the fragments 8 and 10 from wheat-rye addition lines harboring 1R and 7R supports the hypothesis that rye B-located pseudogene-like fragments originated from different A chromosomes.
Comparison of the fragments 6 to 15 shared by As and Bs revealed polymorphisms typical for pseudogenization, such as SNPs and large insertions or deletions, indicating a reduced selection pressure for B-located genic sequences. A higher mutation rate is also known for transcribed genes ectopically duplicated in the rice genome (Wang et al., 2009). Beside these mutations, amplification of B-located sequences occurred as shown for fragment 12. After FISH, Bs displayed multiple hybridization signals, while only one signal was observed on an A chromosome pair.
Comparison of the A-located parental genes with their B-located pseudogenes showed that the rate of mutation is not the same among analyzed fragments (Figure 3). A high degree of polymorphism may indicate a B location of genic fragments already at the onset of B evolution. Alternatively, these sequences belonged to genes whose activity interfered with that of their A counterparts. Therefore, rapid inactivation occurred after gene duplication by B chromosome formation (or later insertion into the already existing B) in accordance with the gene balance hypothesis (Birchler et al., 2005).
More recent integration of A-derived genes after proto-B formation might explain fragments characterized by fewer polymorphisms. Movement of genes is shown to be frequent in grass genomes, resulting from transposition of mobile elements and/or DNA double-strand break repair (Wicker et al., 2010, 2011). However, different A-chromosomal regions also evolve at different rates (Matassi et al., 1999). Furthermore, different mutation rates are suggested to correlate negatively with the level of transcription (Sharp et al., 1989).
Depending on the analyzed species, the half-life of an active duplicated gene to become mutated or lost was estimated to be two to seven million years (Lynch and Conery, 2000). Assuming that Bs are prone to accumulate mutations, one could expect distinct patterns of pseudogenization for the Bs of different populations. However, the comparison of pseudogene-like fragments revealed no accession-specific mutation patterns in contrast with their A-located parental genes. The absence of accession-specific B types can be explained by the very recent spreading of rye across geographic regions by human agricultural activity and the reduced selection pressure. Rye is a young cultivated plant; the earliest remains of wild rye are 11,000 to 12,000 years old (Hillman et al., 2001; Willcox et al., 2009). Evidence for S. cereale grown as crop in its own right was found in c. 6600 BC old records from Turkey (Hillmann, 1978; Fairbairn et al., 2002). From East Turkey the crop spread into other countries, including the countries used for our comparison of Bs. Thus, the separation of the different genotypes used for our analyses evolved rather recently in evolutionary terms.
B-Located Pseudogene-Like Fragments Contribute to the Transcriptome
In contrast with the prevalent notation that B chromosomes are silent, we demonstrate that 15% of the pseudogene-like fragments on Bs are transcribed in a tissue-type and genotype-specific manner. Therefore, our previous estimation of a 20-fold lower transcriptional activity of Bs compared with As (Carchilan et al., 2009) was an underestimate due to a different approach. The first study was based on amplified fragment length polymorphism, and only differences in PCR fragment size and restriction site sequence allowed the detection of B-specific transcripts. Because of the high sequence similarity of A- and B-derived transcripts, this method likely resulted in an underestimation of B-derived transcripts. In the case of high A and B sequence similarity, our approach also could not distinguish between A- and B-derived transcripts. Furthermore, the total activity of identical A and B sequences does not necessarily increase if dosage compensation occurs as observed for extra copies of an A (reviewed in Birchler, 2010; Prestel et al., 2010).
Trans-modulation of A-derived transcripts by Bs [i.e.(sub)fragment 12-1 and 12-3] was found to occur in a tissue-type-specific manner. A similar effect of an almost gene deficient chromosome has been shown for Drosophila melanogaster (Lemos et al., 2008). The Y chromosome of D. melanogaster regulates the activity of hundreds of genes located on other chromosomes. B-caused downregulation of A-located genes is likely mediated by mechanisms that have been postulated for pseudogenes of other species. A growing body of evidence strongly suggests a role for pseudogenes in regulating parental gene expression by serving as a source of endogenous siRNA and antisense transcripts as competitive inhibitors of translation of wild-type transcripts and of sequestering microRNAs (reviewed in Poliseno, 2012). Another possibility for how Bs could exert control on As is their effect on the spatial interphase organization of the genome itself. It is suggested that spatial positioning of genes and chromosomes can influence gene expression (Misteli, 2007). Due to the lack of complete sequence coverage, the potential translation of B-derived transcripts could not be determined.
Thus, B-derived transcripts provide additional complexity of gene expression by themselves and in combination with their related A-located wild-type genes. Phenotypes and effects associated with the presence of Bs (Jimenez et al., 1994; Bougourd and Jones, 1997) might become explainable by the activity of B-located pseudogenes. Transcriptionally active pseudogene(s) embedded in the B nondisjunction control region could, in addition to the previously identified noncoding transcripts of repeats (Carchilan et al., 2007), be candidates contributing to non-Mendelian accumulation of the rye B.
A Model for B-Located Pseudogene Evolution
We conclude that in analogy to sex chromosome evolution, rye B evolution is characterized by erosion of its gene content and by accumulation of repetitive DNA (Klemme et al., 2013). However, in contrast with sex chromosomes, the rye B is nonessential for the host and therefore requires an accumulation mechanism to counteract purifying selection. The sequence of events leading to degeneration of its gene content is not yet known, but it seems that each B has its own evolutionary pathway. We propose a multistep model for the fate of B-located pseudogenes (Figure 3B). Initially, a proto-B was formed by segmental or during whole-genome duplication, followed by unbalanced segregation of this chromosome, which was likely small. Meiotic recombination with donor As became restricted, probably due to multiple rearrangements involving different As. However, this B-chromosome-to-be would still show sequence similarity to the parental As. Because an increased gene dosage may affect gene expression, the expression of paralogs on the B might have been reprogrammed early during the evolution of the Bs. Thus, proto-B genes might have been suppressed by silencing mechanisms and have degenerated by accumulating mutations. Additional gene fragments might have been inserted from A-chromosomal regions and organellar genomes into Bs via transposition events and/or error-prone double-strand break repair. Exceptions are those coding and/or noncoding sequences responsible for the drive as an advantage for the maintenance of Bs.
In summary, we provide evidence that B-located pseudogene-like fragments contribute to the transcriptional landscape of the host genome. Some of these nonprocessed pseudogenes have the potential to regulate the expression of A-located genes. Considering the coexistence of almost sequence-identical A- and B-derived transcripts, it is likely that transcript regulation via dosage compensation occurs in rye, resulting in a constant transcript number regardless of the gene copy number.
METHODS
Plant Materials
Rye (Secale cereale subsp cereale) plants with and without Bs of the fertile inbred line 7415 (Jimenez et al., 1994), of cultivated rye (S. cereale subsp cereale) collected from Turkey, Iran, Korea, Japan (Niwa and Sakamoto, 1995), and China (No. 37; in Niwa and Sakamoto, 1996), and of weedy rye collected from Pakistan (subsp segetale, No. 34; in Niwa and Sakamoto, 1996) and Afghanistan (subsp afghanicum; Niwa and Sakamoto, 1995) were grown at the same temperature, humidity, and light conditions (16 h light, 22°C day/16°C night). In addition, hexaploid wheat (Triticum aestivum) cv Lindström with and without Bs (Lindström, 1965) and hexaploid wheat cv Chinese Spring with B short-arm (line BS-2) or long-arm (line BS-3) isochromosome (Endo et al., 2008) as well as wheat-rye addition lines (1R-7R; wheat cv ‘Chinese spring’ with added chromosome pairs from rye cultivar ‘Imperial’) were cultivated under the same growth conditions.
gDNA and RNA Extraction, PCR, and RT-PCR
gDNA was extracted from leaf tissue using a DNeasy plant mini kit (Qiagen). DNA of flow-sorted rye Bs (sB) was prepared as described by Martis et al. (2012). Total RNA was isolated from roots, leaves, and anthers (microscopically staged between meiosis and development of mature pollen) using the Trizol method (Chomczynski and Sacchi, 1987). Absence of DNA contamination was confirmed by PCR using primers (see Supplemental Table 2 online) specific for Bilby (Francki, 2001) or CRW2 repeats (Liu et al., 2008). The cDNA was synthesized with DNase-treated total RNA, using a RevertAid H Minus first-strand cDNA synthesis kit (Fermentas). PCR reactions were performed in 25 μL reaction volume containing 100 ng of gDNA or cDNA, 1 mM primers (see Supplemental Tables 2 and 3 online), PCR buffer, deoxynucleotide triphosphates, and 1 unit of Taq polymerase (Qiagen). Thirty-five amplification cycles (45 s at 95°C, 1 min at corresponding annealing temperature [see Supplemental Tables 2 and 3 online] and 1 min at 72°C) were run. Primers specific for the constitutively expressed GAPDH gene (see Supplemental Table 2 online) were used as control for equal amount of gDNA and cDNA.
Sequence Analysis
PCR and RT-PCR products were cloned with a StrataClone PCR cloning kit (Agilent) according to manufacturer’s instructions and sequenced. The quality of sequencing was checked using Sequencher 4.7, and sequences were processed by Editseq and aligned using MegAlign (Lasergene 8). Intron/exon structure prediction was performed using the FGENESH 2.6 program (Salamov and Solovyev, 2000). The in silico translation of sequences was performed using the in silico translation tool (http://web.expasy.org/translate/).
Phylogenetic Analysis
After aligning the sequences with ClustalW, phylogenetic trees were calculated using the boot-strapped neighbor-joining algorithm in MEGA 5.05 (http://www.megasoftware.net/; see Supplemental Data Sets 1-3 online) with 1000 trials.
Genome Walking
To extend the flanking sequences of a selected gene-like fragment, genome walking was performed using the GenomeWalker Universal Kit (Clontech) according to manufacturer’s instructions. Briefly, a library was made from gDNA of inbred line 7415 (Jimenez et al., 1994) with and without B and hexaploid wheat cv Lindström with added standard rye Bs (Lindström 1965) using four different restriction enzymes. Nested primer pairs were designed for the candidate gene-like fragment (see Supplemental Table 4 online), and PCR products were cloned and sequenced.
FISH
FISH probes were labeled with ChromaTide Texas Red-12-dUTP, Alexa Fluor 488-5-dUTP (Invitrogen), or Cy5-dUTP (GE Healthcare Life Sciences) by nick translation. FISH was performed as described (Ma et al., 2010). Imaging was performed using an Olympus BX61 microscope and an ORCA-ER CCD camera (Hamamatsu). All images were collected in gray scale and pseudocolored with Adobe Photoshop 6.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers KC623167 to KC623536.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Gene Structure Model for Selected Gene-Like Fragments.
Supplemental Figure 2. Identification of Parental A Chromosomes for B-Located Pseudogene-Like Fragments.
Supplemental Figure 3. Comparison between the Extended Sequences of Gene-Like Fragment 12 Derived from Rye A and B Chromosomes.
Supplemental Figure 4. Phylogenetic Analysis of Gene-Like Fragments.
Supplemental Figure 5. B-Located Pseudogene-Like Fragments Are Transcribed in a Tissue- and Species-Specific Manner.
Supplemental Figure 6. Detail Analysis of Genomic and Transcribed Sequences of the Representative Pseudogene-Like (Sub)fragments 1-2, 2, 12-1, 15-2, and 12-3.
Supplemental Figure 7. Validation of Putative Transcribing Regions on B Chromosomes of Rye Accessions of Different Geographical Origin.
Supplemental Figure 8. Transcription Activity of B Pseudogene-Like Fragments across Different Rye Accessions.
Supplemental Table 1. Selected Rye B Gene-Like Fragments.
Supplemental Table 2. List of gDNA-Primer Sequences.
Supplemental Table 3. List of cDNA-Primer Sequences (Excluding the Intron-Exon Junctions).
Supplemental Table 4. List of Primer Sequences for Genome Walking.
Supplemental Data Set 1. Text File of Alignment Output for Gene-Like Fragment 2 from MEGA 5.05 Software Used to Construct Phylogenetic Tree in Supplemental Figure 4 Online.
Supplemental Data Set 2. Text File of Alignment Output for Gene-Like Fragment 4 from MEGA 5.05 Software Used to Construct Phylogenetic Tree in Supplemental Figure 4 Online.
Supplemental Data Set 3. Text File of Alignment Output for Gene-Like Fragment 8 from MEGA 5.05 Software Used to Construct Phylogenetic Tree in Supplemental Figure 4 Online.
Acknowledgments
We thank Katsumasa Niwa (Tokyo University of Agriculture, Japan), Takashi Ryu Endo (Kyoto University, Japan), and Maria J. Puertas (Universidad Complutense, Spain) for providing valuable plant material. We also thank Katrin Kumke (Leibniz Institute of Plant Genetics and Crop Plant Research, Germany) and Oda Weiß (Leibniz Institute of Plant Genetics and Crop Plant Research, Germany) for excellent technical assistance as well as Ingo Schubert (Leibniz Institute of Plant Genetics and Crop Plant Research, Germany) and Frank R. Blattner (Leibniz Institute of Plant Genetics and Crop Plant Research) for fruitful discussion. This work was supported by the Deutsche Forschungsgemeinschaft Germany (HO 1779/10-1/14-1). We appreciate constructive comments made by anonymous reviewers.
AUTHOR CONTRIBUTIONS
A.M.B.-M. and A.H designed the research. A.M.B.-M., K.M., and R.K.-A. performed experiments. A.M.B.-M. and A.H. wrote the article.
Glossary
- B
B chromosome
- A
A chromosome
- gDNA
genomic DNA
- SNP
single nucleotide polymorphism
- FISH
fluorescence in situ hybridization
References
- Birchler J.A. (2010). Reflections on studies of gene expression in aneuploids. Biochem. J. 426: 119–123 [DOI] [PubMed] [Google Scholar]
- Birchler J.A., Riddle N.C., Auger D.L., Veitia R.A. (2005). Dosage balance in gene regulation: Biological implications. Trends Genet. 21: 219–226 [DOI] [PubMed] [Google Scholar]
- Bougourd S.M., Jones R.N. (1997). B chromosomes: A physiological enigma. New Phytol. 137: 43–54 [Google Scholar]
- Camacho J.P.M., Sharbel T.F., Beukeboom L.W. (2000). B-chromosome evolution. Philos. Trans. R. Soc. Lond. B Biol. Sci. 355: 163–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carchilan M., Delgado M., Ribeiro T., Costa-Nunes P., Caperta A., Morais-Cecílio L., Jones R.N., Viegas W., Houben A. (2007). Transcriptionally active heterochromatin in rye B chromosomes. Plant Cell 19: 1738–1749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carchilan M., Kumke K., Mikolajewski S., Houben A. (2009). Rye B chromosomes are weakly transcribed and might alter the transcriptional activity of A chromosome sequences. Chromosoma 118: 607–616 [DOI] [PubMed] [Google Scholar]
- Carlson, W. (2009). The B chromosome of maize. In Handbook of Maize J.L. Bennetzen and S. Hake, eds (New York: Springer), pp. 459–480. [Google Scholar]
- Chomczynski P., Sacchi N. (1987). Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162: 156–159 [DOI] [PubMed] [Google Scholar]
- Coleman J.J., et al. (2009). The genome of Nectria haematococca: Contribution of supernumerary chromosomes to gene expansion. PLoS Genet. 5: e1000618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo T.R., Nasuda S., Jones N., Dou Q., Akahori A., Wakimoto M., Tanaka H., Niwa K., Tsujimoto H. (2008). Dissection of rye B chromosomes, and nondisjunction properties of the dissected segments in a common wheat background. Genes Genet. Syst. 83: 23–30 [DOI] [PubMed] [Google Scholar]
- Fairbairn A., Asouti, E., Near, J., and Martinoli, D.(2002). Macro-botanical evidence for plant use at Neolithic Çatalhöyük, south-central Anatolia, Turkey. Veg. Hist. Archaeobot. 11: 41–54 [Google Scholar]
- Francki M.G. (2001). Identification of Bilby, a diverged centromeric Ty1-copia retrotransposon family from cereal rye (Secale cereale L.). Genome 44: 266–274 [DOI] [PubMed] [Google Scholar]
- Green D.M. (1990). Muller's ratchet and the evolution of supernumerary chromosomes. Genome 33: 818–824 [Google Scholar]
- Haseneyer G., Schmutzer T., Seidel M., Zhou R., Mascher M., Schön C.C., Taudien S., Scholz U., Stein N., Mayer K.F., Bauer E. (2011). From RNA-seq to large-scale genotyping - Genomics resources for rye (Secale cereale L.). BMC Plant Biol. 11: 131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillmann G.C. (1978). On the origins of domesticated rye – Secale cereale: The finds from Aceramic Can Hasan III in Turkey. Anatolian Stud. 28: 157–174 [Google Scholar]
- Hillman G., Hedges R., Moore A., Colledge S., Pettitt P. (2001). New evidence of Lateglacial cereal cultivation at Abu Hureyra on the Euphrates. Holocene 11: 383–393 [Google Scholar]
- Holmes D.S., Bougourd S.M. (1991). B-chromosome selection in Allium schoenoprasum. 2. Experimental populations. Heredity 67: 117–122 [Google Scholar]
- Houben, A., Banaei-Moghaddam, A., and Klemme, S. (2013). Biology and evolution of B chromosomes. In Plant Genome Diversity, Vol. 2., J. Greilhuber, J. Dolezel, and J.F. Wendel, eds (Vienna: Springer Press) pp. 149–166. [Google Scholar]
- Houben A., Nasuda S., Endo T.R. (2011). Plant B chromosomes. Methods Mol. Biol. 701: 97–111 [DOI] [PubMed] [Google Scholar]
- Jimenez M.M., Romera F., Puertas M.J., Jones R.N. (1994). B-chromosomes in inbred lines of rye (Secale cereale L). 1. Vigor and fertility. Genetica 92: 149–154 [Google Scholar]
- Jones R.N. (1995). Tansley Review No 85, B chromosomes in plants. New Phytol. 131: 411–434 [DOI] [PubMed] [Google Scholar]
- Jones, R.N., and Rees, H. (1982). B Chromosomes, 1st ed. (London, New York: Academic Press). [Google Scholar]
- Klemme S., Banaei-Moghaddam A.M., Macas J., Wicker T., Novák P., Houben A. (2013). High-copy sequences reveal distinct evolution of the rye B chromosome. New Phytol. 199: 550–558 [DOI] [PubMed] [Google Scholar]
- Leach C.R., Houben A., Field B., Pistrick K., Demidov D., Timmis J.N. (2005). Molecular evidence for transcription of genes on a B chromosome in Crepis capillaris. Genetics 171: 269–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemos B., Araripe L.O., Hartl D.L. (2008). Polymorphic Y chromosomes harbor cryptic variation with manifold functional consequences. Science 319: 91–93 [DOI] [PubMed] [Google Scholar]
- Lindström J. (1965). Transfer to wheat of accessory chromosomes from rye. Hereditas 54: 149–155 [Google Scholar]
- Liu Z., Yue W., Li D., Wang R.R., Kong X., Lu K., Wang G., Dong Y., Jin W., Zhang X. (2008). Structure and dynamics of retrotransposons at wheat centromeres and pericentromeres. Chromosoma 117: 445–456 [DOI] [PubMed] [Google Scholar]
- Lynch, M., and Conery, J.S. (2000). The evolutionary fate and consequences of duplicate genes. Science 290: 1151–1155. [DOI] [PubMed] [Google Scholar]
- Ma L., Vu G.T.H., Schubert V., Watanabe K., Stein N., Houben A., Schubert I. (2010). Synteny between Brachypodium distachyon and Hordeum vulgare as revealed by FISH. Chromosome Res. 18: 841–850 [DOI] [PubMed] [Google Scholar]
- Marques A., Banaei-Moghaddam A.M., Klemme S., Blattner F.R., Niwa K., Guerra M., Houben A. (June 5, 2013). B chromosomes of rye are highly conserved and accompanied the development of early agriculture. Ann. Bot. 112: 527–534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martis M.M., et al. (2012). Selfish supernumerary chromosome reveals its origin as a mosaic of host genome and organellar sequences. Proc. Natl. Acad. Sci. USA 109: 13343–13346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matassi G., Sharp P.M., Gautier C. (1999). Chromosomal location effects on gene sequence evolution in mammals. Curr. Biol. 9: 786–791 [DOI] [PubMed] [Google Scholar]
- Misteli T. (2007). Beyond the sequence: Cellular organization of genome function. Cell 128: 787–800 [DOI] [PubMed] [Google Scholar]
- Niwa K., Sakamoto S. (1995). Origin of B chromosomes in cultivated rye. Genome 38: 307–312 [DOI] [PubMed] [Google Scholar]
- Niwa K., Sakamoto S. (1996). Detection of B chromosomes in rye collected from Pakistan and China. Hereditas 124: 211–215 [Google Scholar]
- Poliseno L. (2012). Pseudogenes: Newly discovered players in human cancer. Sci. Signal. 5: re5. [DOI] [PubMed] [Google Scholar]
- Prestel M., Feller C., Becker P.B. (2010). Dosage compensation and the global re-balancing of aneuploid genomes. Genome Biol. 11: 216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salamov A.A., Solovyev V.V. (2000). Ab initio gene finding in Drosophila genomic DNA. Genome Res. 10: 516–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp P.M., Shields D.C., Wolfe K.H., Li W.H. (1989). Chromosomal location and evolutionary rate variation in enterobacterial genes. Science 246: 808–810. [DOI] [PubMed] [Google Scholar]
- Wang X.F., Yu Z.H., Yang X.Z., Deng X.W., Li L. (2009). Transcriptionally active gene fragments derived from potentially fast-evolving donor genes in the rice genome. Bioinformatics 25: 1215–1218 [DOI] [PubMed] [Google Scholar]
- Wicker T., Buchmann J.P., Keller B. (2010). Patching gaps in plant genomes results in gene movement and erosion of colinearity. Genome Res. 20: 1229–1237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicker T., et al. (2011). Frequent gene movement and pseudogene evolution is common to the large and complex genomes of wheat, barley, and their relatives. Plant Cell 23: 1706–1718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willcox G., Buxo R., Herveux L. (2009). Late Pleistocene and early Holocene climate and the beginnings of cultivation in northern Syria. Holocene 19: 151–158 [Google Scholar]
- Yoshida K., Terai Y., Mizoiri S., Aibara M., Nishihara H., Watanabe M., Kuroiwa A., Hirai H., Hirai Y., Matsuda Y., Okada N. (2011). B chromosomes have a functional effect on female sex determination in Lake Victoria cichlid fishes. PLoS Genet. 7: e1002203. [DOI] [PMC free article] [PubMed] [Google Scholar]