This study uses a screen for suppressors of the repressor of silencing1 mutation, which causes silencing of marker transgenes, to examine global epigenetic regulation, finding that folate polyglutamylation is required for chromatin silencing by maintaining global DNA methylation and histone H3K9 dimethylation.
Abstract
DNA methylation and repressive histone Histone3 Lysine9 (H3K9) dimethylation correlate with chromatin silencing in plants and mammals. To identify factors required for DNA methylation and H3K9 dimethylation, we screened for suppressors of the repressor of silencing1 (ros1) mutation, which causes silencing of the expression of the RD29A (RESPONSE TO DESSICATION 29A) promoter-driven luciferase transgene (RD29A-LUC) and the 35S promoter-driven NPTII (NEOMYCIN PHOSPHOTRANSFERASE II) transgene (35S-NPTII). We identified the folylpolyglutamate synthetase FPGS1 and the known factor DECREASED DNA METHYLATION1 (DDM1). The fpgs1 and ddm1 mutations release the silencing of both RD29A-LUC and 35S-NPTII. Genome-wide analysis indicated that the fpgs1 mutation reduces DNA methylation and releases chromatin silencing at a genome-wide scale. The effect of fpgs1 on chromatin silencing is correlated with reduced levels of DNA methylation and H3K9 dimethylation. Supplementation of fpgs1 mutants with 5-formyltetrahydrofolate, a stable form of folate, rescues the defects in DNA methylation, histone H3K9 dimethylation, and chromatin silencing. The competitive inhibitor of methyltransferases, S-adenosylhomocysteine, is markedly upregulated in fpgs1, by which fpgs1 reduces S-adenosylmethionine accessibility to methyltransferases and accordingly affects DNA and histone methylation. These results suggest that FPGS1-mediated folate polyglutamylation is required for DNA methylation and H3K9 dimethylation through its function in one-carbon metabolism. Our study makes an important contribution to understanding the complex interplay among metabolism, development, and epigenetic regulation.
INTRODUCTION
In plants and mammals, DNA methylation at 5-position of cytosine is an important chromatin feature that is involved in transposon silencing, genome stability, and gene regulation (Slotkin and Martienssen, 2007; Matzke et al., 2009; Law and Jacobsen, 2010; He et al., 2011). DNA methylation is highly correlated with the repressive histone Histone3 Lysine9 dimethylation (H3K9me2) mark at heterochromatic regions. In Arabidopsis thaliana, DNA methylation is present in all three cytosine contexts, which include symmetric CG and CHG sites and asymmetric CHH sites. CG methylation is maintained by the homolog of human DNA METHYLTRANSFERASE1 (MET1) (Ronemus et al., 1996). DECREASED DNA METHYLATION1 (DDM1), a SWItch2/Sucrose NonFermentable2 (SWI2/SNF2) chromatin-remodeling protein, is required for maintaining CG methylation as well as H3K9me2 (Jeddeloh et al., 1999). CHG methylation is catalyzed by a plant-specific CHROMOMETHYLASE3 (CMT3) and depends on H3K9me2 (Cao and Jacobsen, 2002; Du et al., 2012). The direct binding of CMT3 to H3K9me2-containing nucleosomes is required for the interplay between DNA methylation and H3K9me2 (Du et al., 2012). CHH methylation is mainly established by DOMAINS REARRANGED METHYLTRANSFERASE2 (DRM2) and its homologs through the RNA-directed DNA methylation (RdDM) pathway (Henderson et al., 2010). In Arabidopsis, histone H3K9 dimethylation is catalyzed by the Drosophila melanogaster SUPPRESSOR OF VARIEGATION3-9 (SU[VAR]3-9) homologs and related proteins (Baumbusch et al., 2001; Law and Jacobsen, 2010; Veiseth et al., 2011). All these proteins contain a catalytic SET domain. Additionally, an extra SRA (SET- or RING-associated) methyl binding domain is present at the N termini of SU(VAR)3-9 homologs (Baumbusch et al., 2001). The 5-methyl-cytosine binding ability of the SRA domain is required for both DNA methylation and H3K9me2 (Rajakumara et al., 2011). The cooperation between DNA methylation and histone methylation is essential for maintaining stable chromatin silencing in Arabidopsis (Baubec et al., 2010).
S-adenosylmethionine (SAM) serves as the universal methyl-group donor for methyltransferases including DNA and histone methyltransferases (Loenen, 2006; Zhang et al., 2012). SAM originates from folate-dependent one-carbon metabolism. The vitamin B12-dependent methionine (Met) synthase uses 5-methyl-tetrahydrofolate (5-CH3-THF) as the methyl-group donor for the methylation of homocysteine (Hcy) to Met, the precursor of SAM (Friso et al., 2002). After transfer of the methyl group, SAM is converted to S-adenosylhomocysteine (SAH), which is a strong inhibitor of methyltransferases. Removal of SAH is critical to avoid the inhibitory effect of SAH on methylation reactions (Molloy, 2012). S-adenosylhomocysteine hydrolase (SAHH) is the enzyme responsible for the conversion of SAH to Hcy and adenosine. Hcy is further recycled for Met and then SAM biosynthesis (Molloy, 2012). Partial inactivation of SAHH by antisense RNA in tobacco (Nicotiana tabacum) causes morphological changes and DNA hypomethylation (Tanaka et al., 1997). The SAHH/HOMOLOGY-DEPENDENT GENE SILENCING1 (HOG1) knockout mutants in Arabidopsis are embryo lethal, whereas the weak hog1 alleles are leaky and result in genome demethylation (Rocha et al., 2005). In Arabidopsis, the hog1/sahh1 mutants were recovered from several independent forward genetic screens, which suggested that HOG1/SAHH1 plays a critical role in development as well as in DNA and histone methylation and epigenetic silencing (Rocha et al., 2005; Mull et al., 2006; Wu et al., 2009; Baubec et al., 2010; Ouyang et al., 2012). The weak reduction in SAM/SAH ratio is likely correlated with the effect of hog1 on DNA methylation (Rocha et al., 2005).
Tetrahydrofolate and its derivatives act as cofactors in one-carbon metabolism and are required for the transfer of one-carbon units during the synthesis of purines, thymidylate, pantothenate, formyl-methionyl tRNA, and Met (Cossins and Chen, 1997; Lucock, 2000). Met is the direct precursor of SAM, which is required for methylation of DNA, RNA, and proteins. Folates are comprised of pterin, paminobenzoic acid, and Glu moieties. In vivo, folates can be polyglutamylated by sequential conjugation of additional r-linked Glu residues to the first Glu by folylpolyglutamate synthetase (Ravanel et al., 2001). Plants synthesize folates, but mammals must obtain folates from exogenous sources. Therefore, inadequate folate intake in mammals impairs folate-dependent processes and causes several abnormalities, including embryonic defects, neural tube defects, cardiovascular disease, and cancers (Molloy, 2012). The influence of folate status on DNA methylation has been demonstrated in mammals (Giovannucci et al., 1993; Friso et al., 2002). DNA hypomethylation may be one of the important molecular bases for the abnormalities caused by folate deficiency. The methylenetetrahydrofolate reductase catalyzes the conversion of 5,10-methylene THF to 5-methyl THF and plays a central role in folate-mediated one-carbon metabolism (Frosst et al., 1995). An extensively studied polymorphism (C677T) in the enzyme is correlated with DNA hypomethylation and uracil misincorporation when folate intake is inadequate, and C677T may increase the risk of folate deficiency-related abnormalities (Frosst et al., 1995; Shelnutt et al., 2004). The function of folate in DNA and histone methylation in plants was recently demonstrated in Arabidopsis (Zhang et al., 2012). However, further studies are required to fully understand the regulatory mechanism of folate and its role in DNA and histone methylation in vivo.
In our genetic system, both the RD29A (RESPONSE TO DESSICATION29A) promoter-driven luciferase transgene (RD29A-LUC) and the 35S promoter-driven NPTII (NEOMYCIN PHOSPHOTRANSFERASE II) transgene (35S-NPTII) are silenced by mutations in the DNA glycosylase/demethylase REPRESSOR OF SILENCING1 (ROS1) (Gong et al., 2002). We identified a number of components required for transcriptional silencing by screening for suppressors of ros1 based on both luminescence imaging and kanamycin resistance (He et al., 2011; Liu and Gong, 2011). The results showed that RdDM is responsible for the silencing of RD29A-LUC (He et al., 2009), whereas DNA replication and repair proteins are required for maintaining the silencing of 35S-NPTII (Liu and Gong, 2011). The function of DNA replication and repair proteins in transcriptional silencing involves repressive histone H3K9 dimethylation but not DNA methylation (Liu and Gong, 2011). The histone H2B deubiquitination enzyme UBIQUITIN-SPECIFIC PROTEASE 26 (UBP26) and HISTONE DEACETYLASE 6 (HDA6) are required for the silencing of both transgenes (Sridhar et al., 2007; He et al., 2009). In this study, we examined the mechanisms of epigenetic regulation by screening for mutants that release the ros1-based silencing of two transgenes. We identified DDM1, which is known to be important for both DNA methylation and histone H3K9 dimethylation, and also identified FOLYLPOLYGLUTAMATE SYNTHETASE 1 (FPGS1). Our examination of the fpgs1 mutants suggests that folate polyglutamylation plays a key role in the regulation of methyl-group supply for global DNA and histone methylation.
RESULTS
Identification of Suppressors of ros1
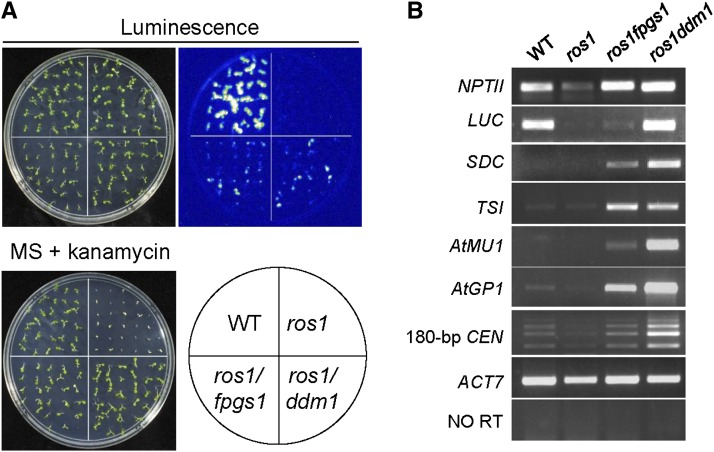
Our genetic screening system uses an Arabidopsis ros1 mutant line that harbors two silenced reporter genes: the RD29A promoter-driven luciferase gene (RD29A-LUC) and the 35S promoter-driven NPTII (35S-NPTII) (Gong et al., 2002). Suppressors of ros1 can be identified by their increased luciferase expression and kanamycin resistance resulting from a release of the ros1 silencing of these two transgenes. Here, by screening for suppressors of ros1, we identified a ddm1 mutant in the ros1 background (see Supplemental Figure 1 online). The silencing of both RD29A-LUC and 35S-NPTII was released by the ddm1 mutation (Figures 1A and 1B), suggesting that DDM1 has a universal role in transcriptional silencing. Moreover, we identified an additional suppressor, which we identified as fpgs1, based on our subsequent characterization (see below) (Figure 1A). The silencing of both Rd29A-LUC and 35S-NPTII was released in fpgs1 as well as in ros1 ddm1 (Figure 1A). The effect of fpgs1 on the silencing of RD29A-LUC was less than that of ddm1. In Arabidopsis, many endogenous genes, transposable elements (TEs), and DNA repeats are silenced by DNA methylation and repressive histone modifications. The DNA repeats flanked protein-coding gene SDC (SUPPRESSOR OF DRM1 DRM2 CMT3), and the TEs At-MU1 and At-GP1 are canonical targets of RdDM (Henderson and Jacobsen, 2008; He et al., 2009), whereas TRANSCRIPTIONALLY SILENT INFORMATION (TSI) is the repetitive DNA sequence targeted by DNA replication and repair proteins but not by RdDM components (Xia et al., 2006). The 180-bp centromeric DNA repeats are not affected by RdDM components or DNA replication and repair proteins (Xia et al., 2006; He et al., 2009b; Yin et al., 2009). Our RT-PCR results indicated that all these endogenous silenced loci are upregulated by ddm1 and to a lesser extent by fpgs1 (Figure 1B). Moreover, the effect of fpgs1 on transcriptional silencing of 35S-NPTII, SDC, TSI, At-MU1, and At-GP1 was confirmed by quantitative RT-PCR (see Supplemental Figure 2 online). The results suggest that both fpgs1 and ddm1 mutations have a universal effect on chromatin silencing.
Figure 1.
The Silencing of Transgenes and Endogenous Genomic Sequences Was Released in Both ros1 fpgs1 and ros1 ddm1 Mutants.
(A) The silencing of RD29A-LUC and 35S-NPTII transgenes was released in ros1 fpgs1and ros1 ddm1. The LUC and NPTII expression levels were evaluated based on luminescence imaging and kanamycin resistance, respectively. The kanamycin resistance of seedlings was determined by growing the seedlings on MS medium supplemented with 150 mg/L kanamycin. The wild type (WT) and ros1 were used as controls.
(B) The transcript levels of indicated loci were examined by RT-PCR. The actin gene ACT7 was amplified as an internal control. “NO RT” indicates the amplification of ACT7 when RNA was used as template without reverse transcription.
[See online article for color version of this figure.]
DNA Methylation and Histone H3K9 Dimethylation Are Reduced in the fpgs1 Mutant
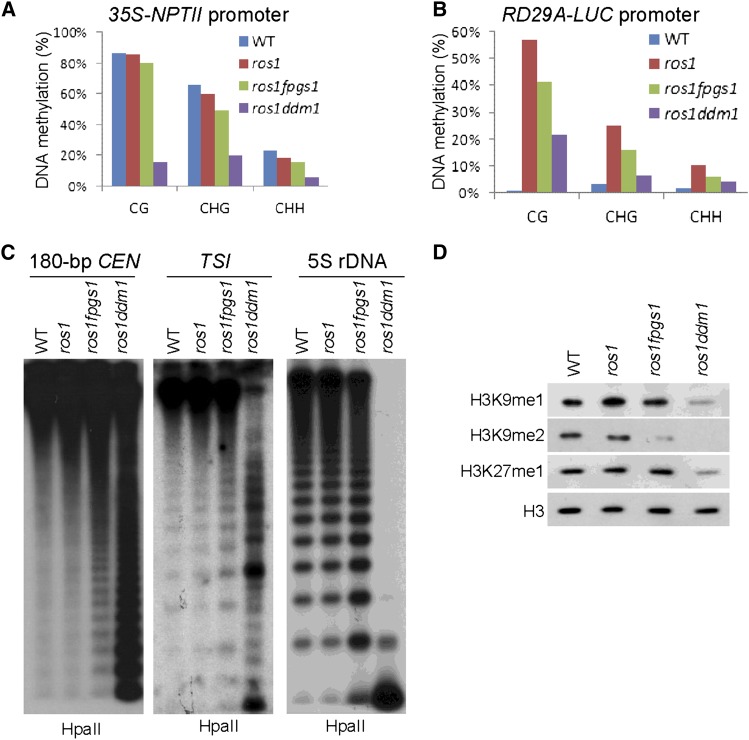
We evaluated the effect of the fpgs1 mutation on DNA methylation by bisulfite sequencing and DNA gel blotting. Bisulfite sequencing suggested that the DNA methylation levels at both the 35S-NPTII transgene promoter and the RD29A-LUC transgene promoter were reduced by fpgs1 but that the effect of fpgs1 was less than that of ddm1 (Figures 2A and 2B; see Supplemental Table 1 online). Similarly, the DNA methylation level at the endogenous RD29A promoter was reduced by ddm1 and to a lesser extent by fpgs1 (see Supplemental Figure 3A and Supplemental Table 1 online). Both ddm1 and fpgs1 affect DNA methylation in all three cytosine contexts, including CG, CHG, and CHH sites, but the effect of fpgs1 is generally weaker than that of ddm1 (Figures 2A and 2B; see Supplemental Table 1 online). The effect of fpgs1 and ddm1 on DNA methylation at endogenous genomic loci was determined by DNA gel blotting. The results indicated that at the three tested genomic loci (180-bp centromeric DNA repeats, TSI, and 5S rDNA), the CG methylation as determined by HpaII cleavage was mildly reduced by fpgs1 and was drastically reduced by ddm1 (Figure 2C). The CHG methylation determined by MspI cleavage was reduced by ddm1 and to a lesser extent by fpgs1 (see Supplemental Figure 3B online). The cytosine methylation at asymmetric CHH sites was not affected by fpgs1 at the 180-bp centromeric DNA repeats, 5S rDNA, and TSI (see Supplemental Figure 3B online). Although the CHH methylation at the 180-bp centromeric DNA repeats and TSI sites was also not affected by ddm1, the CHH methylation at 5S rDNA was clearly upregulated by ddm1 (see Supplemental Figure 3B online). This promoting effect of ddm1 on CHH methylation was consistent with the previous finding that the ddm1 mutation induces de novo DNA methylation through the RdDM pathway (Teixeira et al., 2009).
Figure 2.
The Effect of fpgs1 and ddm1 on DNA and Histone Methylation.
(A) and (B) Detection of DNA methylation at the promoters of 35S-NPTII and RD29A-LUC transgenes by bisulfite sequencing. The DNA methylation levels at CG, CHG, and CHH sites are shown. WT, the wild type.
(C) The DNA methylation of 180-bp centromeric DNA, TSI, and 5S rDNA was evaluated by DNA gel blotting. Genomic DNA was cleaved by the DNA methylation-sensitive endonuclease HpaII followed by DNA gel blotting.
(D) The overall levels of H3K9me1, H3K9me2, and H3K27me1 determined by immunoblotting. The protein extracts were isolated from the wild type, ros1, ros1 fpgs1, and ros1 ddm1. The H3 signal was detected as a loading control.
We conducted immunoblotting assays to determine whether the fpgs1 mutation impairs the different types of repressive histone methylation, including histone H3K9 monomethylation (H3K9me1), H3K9 dimethylation (H3K9me2), and Histone3 Lysine27 monomethylation (H3K27me1) (Johnson et al., 2008; Roudier et al., 2011). As expected, all the three types of repressive histone methylation were reduced by the ddm1 mutation in ros1ddm1 (Figure 2D). Moreover, the H3K9me2 level in ros1 fpgs1 was clearly reduced, while the levels of H3K9me1 and H3K27me1 in ros1 fpgs1 were comparable to that in the wild type and ros1 (Figure 2D).
Map-Based Cloning and Characterization of FPGS1
We identified the crucial mutation in ros1 fpgs1 by map-based cloning. The mutant was crossed to the ros1 mutant (Salk_045303) in the Columbia-0 background, and the F2 plants were selected for map-based cloning based on visualization of luminescence and kanamycin resistance. The mutation was localized to an ∼432-kb interval on chromosome 5 (see Supplemental Figure 4A online). A G-to-A mutation in FPGS1 was identified in ros1 fpgs1 by sequencing candidate genes in the region. The mutation in FPGS1 disrupts the splicing donor site in the eleventh intron of FPGS1 (see Supplemental Figure 4B online). We performed RT-PCR using the primers flanking the eleventh intron to test whether the FPGS1 transcript is affected by fpgs1 (see Supplemental Figures 5A to 5C online). The results indicated that two abnormal FPGS1 transcripts are present in ros1 fpgs1, whereas the normal FPGS1 transcript identified in the wild type and ros1 is absent in ros1 fpgs1 (see Supplemental Figure 5A online). Sequencing of the two abnormal FPGS1 transcripts demonstrated that one transcript retains the full length of the eleventh intron and the other transcript retains a part of the intron (see Supplemental Figures 5B and 5C online). The reading frame shifts in both abnormal FPGS1 transcript variants, suggesting that the functional FPGS1 transcript is disrupted by the fpgs1 mutation in ros1 fpgs1.
The full-length FPGS1 genomic sequence was constructed and transformed into the ros1 fpgs1 mutant for complementation testing. Seedling growth assay showed that the root growth mutant is significantly retarded in ros1 fpgs1 compared with that in the wild type and ros1 (see Supplemental Figure 6A online), which is consistent with the morphological phenotype of fpgs1 as reported previously (Srivastava et al., 2011). We found that the defective root growth of ros1 fpgs1 was complemented by the FPGS1 transgene (see Supplemental Figure 6A online). Moreover, our results demonstrated that the silencing of RD29A-LUC as well as of 35S-NPTII is restored by the PFGS1 transgene in the ros1 fpgs1 background (see Supplemental Figure 6B online). Because the silencing of endogenous genomic target loci At-GP1 and TSI is released by fpgs1 (Figure 1B), we tested whether the FPGS1 transgene restores the silencing of the two loci by RT-PCR. The results demonstrated that both At-GP1 and TSI are silenced by the FPGS1 transgene in ros1 fpgs1 (see Supplemental Figure 6C online). These results suggest that FPGS1 is not only required for root growth but also for transcriptional silencing.
FPGS1 encodes a conserved folylpolyglutamate synthetase isoform that catalyzes folate polyglutamylation on the initial Glu of folate in chloroplasts (Ravanel et al., 2001; Mehrshahi et al., 2010; Srivastava et al., 2011). To confirm the function of FPGS1 in DNA methylation, we determined the DNA methylation levels of At-GP1 and TSI in two individual fpgs1 T-DNA mutants, Salk_015472 and Salk_007791 (see Supplemental Figure 7A online). The results indicated that DNA methylation of At-GP1 and TSI is markedly decreased in the two individual fpgs1 mutants relative to the wild type (see Supplemental Figure 7B online), confirming that FPGS1 is required for DNA methylation. The At-GP1 and TSI transcript levels were determined by RT-PCR (see Supplemental Figure 7C online). The results showed that transcriptional silencing of At-GP1 and TSI is significantly released in the two fpgs1 T-DNA mutants. These results confirm that FPGS1 is required for DNA methylation and transcriptional silencing. Arabidopsis has three folylpolyglutamate synthetases, FPGS1, FPGS2, and FPGS3 (Mehrshahi et al., 2010). The fpgs2 and fpgs3 T-DNA mutants, Salk_008883 and SAIL_580_H10, were used to determine the possible function of FPGS2 and FPGS3 in DNA methylation by chop-PCR (see Supplemental Figure 7A online). In chop-PCR, genomic DNA from each genotype was digested by the methylated DNA-specific restriction enzyme McrBC, followed by quantitative PCR; therefore, an increased signal in chop-PCR indicates lower levels of methylation. The results suggested that the DNA methylation levels of At-GP1 and TSI were not affected by fpgs2 and fpgs3 (see Supplemental Figure 7B online). Moreover, we tested the effect of fpgs2 and fpgs3 on transcriptional silencing of At-GP1 and TSI by RT-PCR and found that neither fpgs2 nor fpgs3 affects transcriptional silencing (see Supplemental Figure 7C online), which is consistent with the finding that fpgs2 and fpgs3 have no effect on DNA methylation at the loci (see Supplemental Figure 7B online). These results imply that either FPGS2 or FPGS3 is dispensable for DNA methylation and transcriptional silencing, but functional redundancy between FPGS2 and FPGS3 cannot be excluded.
Genome-Wide Effect of fpgs1 on DNA Methylation
We conducted whole-genome bisulfite sequencing to analyze the global effect of fpgs1 on DNA methylation. The effect of ddm1 on DNA methylation was evaluated as a control. From the whole-genome bisulfite sequencing, we obtained 11,323,625, 14,196,929, and 9,592,891 unique mapped ∼40-nucleotide reads for ros1, ros1 ddm1, and ros1 fpgs1, respectively (see Supplemental Table 2 online). Calculations of the average CG, CHG, and CHH methylation across the whole genome indicated that the DNA methylation at all the three cytosine contexts is reduced by ddm1 and to a lesser extent by fpgs1 (Table 1). To evaluate the quality of the whole-genome bisulfite sequencing results, we checked the DNA methylation levels of the 35S-NPTII and RD29A-LUC promoters determined by the whole-genome bisulfite sequencing (see Supplemental Figures 8A and 8B online). The effect fpgs1 and ddm1 on DNA methylation of the 35S-NPTII and RD29A-LUC promoters as determined by the whole-genome bisulfite sequencing was highly similar to the effect that was determined by sequence-specific bisulfite sequencing (Figures 2A and 2B; see Supplemental Figures 8A and 8B online), suggesting the whole-genome bisulfite sequencing generated high-quality data.
Table 1. Average Cytosine Methylation Levels at CG, CHG, and CHH and Total Cytosine Sites.
| Genotypes | CG | CHG | CHH | Total |
|---|---|---|---|---|
| ros1 | 18.40% | 5.30% | 1.70% | 4.85% |
| ros1 fpgs1 | 15.60% | 3.10% | 1.30% | 3.73% |
| ros1 ddm1 | 8.00% | 1.80% | 1.00% | 2.23% |
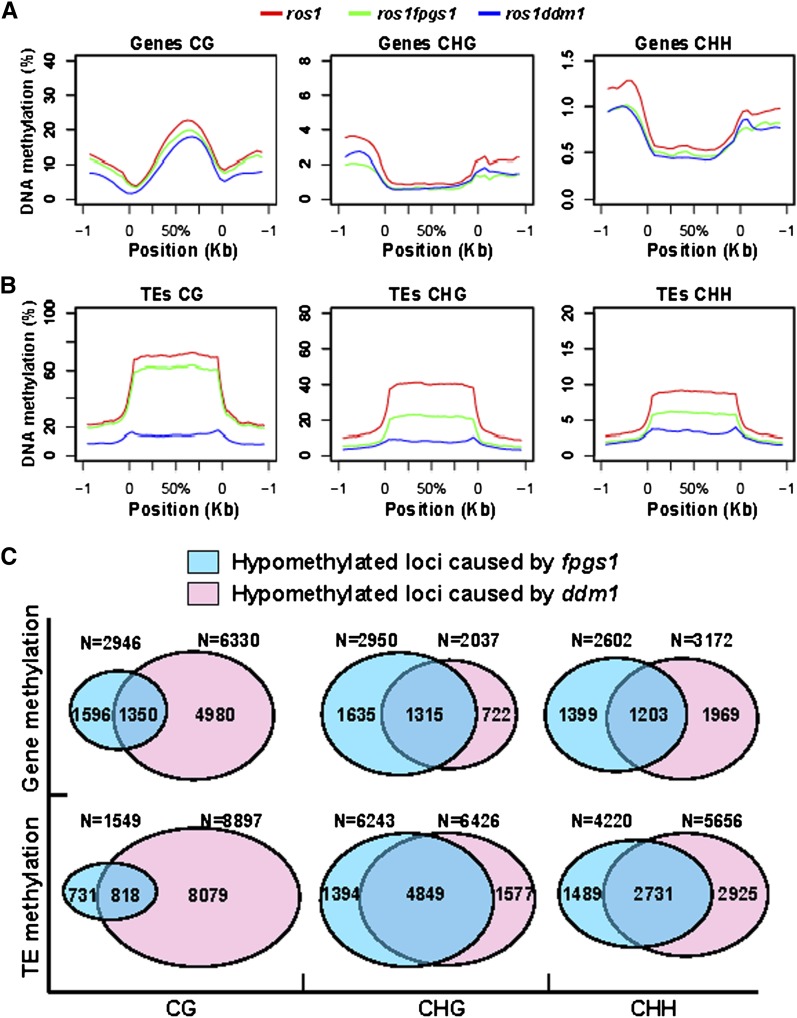
The average CG, CHG, and CHH methylation was determined at genes and TEs, respectively. Consistent with previous reports (Wierzbicki et al., 2012), our result suggested that gene body methylation was predominantly present at CG sites (Figure 3A). Gene body CG methylation was clearly reduced by ddm1 as well as by fpgs1. At the regions surrounding gene bodies, CG methylation was clearly reduced by ddm1 but only marginally reduced by fpgs1, whereas CHG and CHH methylation was reduced by ddm1 as well as by fpgs1 (Figure 3A). In contrast with genes, TE bodies showed higher DNA methylation than their surrounding regions at CG, CHG, and CHH sites (Figure 3B). Not only ddm1 but also fpgs1 drastically reduced TE body methylation at all the three cytosine contexts, although the effect of fpgs1 is less than ddm1 (Figure 3B). At TE surrounding regions, ddm1 causes a decrease of DNA methylation at CG, CHG, and CHH sites, but fpgs1 can only affect DNA methylation at CHG and CHH sites (Figure 3B). The effects of fpgs1 on DNA methylation across Arabidopsis chromosomes indicated that fpgs1 reduces CHG and CHH methylation substantially at centromeric and pericentromeric regions and weakly at euchromatic regions. However, the effect of fpgs1 on CG methylation is always weak throughout whole Arabidopsis chromosomes, which is different from the substantial effects of ddm1 on all cytosine contexts at centromeric and pericentromeric regions (see Supplemental Figures 9A to 9J online).
Figure 3.
The Effect of fpgs1 and ddm1 on DNA Methylation at Genes and TEs.
(A) and (B) Diagrams show the levels of CG, CHG, and CHH methylation at genes (A), TEs (B), and their 1-kb upstream and downstream regions in ros1 (read line), ros1 fpgs1 (green line), and ros1 ddm1 (blue line).
(C) Shown are the numbers of hypomethylated genes and TEs caused by fpgs1 and ddm1 as well as the numbers of their overlapping loci. The numbers of hypomethylated loci at CG, CHG, and CHH sites are separately indicated.
The number of differentially methylated genes and TEs was determined; loci were selected that showed a significant (P < 0.05) change of more than 1.5-fold compared with the control ros1 plants (see Supplemental Data Sets 1 to 4 online). The results indicated that 2946 genes and 1549 TEs are hypomethylated at CG sites in fpgs1 mutants, compared with 6330 genes and 8897 TEs that are hypomethylated in ddm1 mutants (Figure 3C). In all, 45.8% of hypomethylated genes (1350/2946) and 52.8% of TEs (818/1549) caused by fpgs1 at CG sites are also hypomethylated by ddm1 (Figure 3C), whereas 21.3% of ddm1-mediated hypomethylated genes (1350/6330) and 9.2% of TEs (818/8897) at CG sites are simultaneously affected by fpgs1. The numbers of hypomethylated genes and TEs at CHG sites caused by fpgs1 are 2950 and 6243, respectively. Among them, 1315 genes and 4849 TEs are also hypomethylated by ddm1 at CHG sites (Figure 3C). The numbers of hypomethylated genes and TEs at CHH sites caused by fpgs1 are comparable to those caused by ddm1 (Figure 3C). There are 2602 genes and 4220 TEs that are hypomethylated in ros1 fpgs1 at CHH sites, compared with 3172 genes and 5656 TEs that are hypomethylated in ros1 ddm1 (Figure 3C). Among them, 1203 genes and 2731 TEs are overlapped between ros1 fpgs1 and ros1 ddm1 (Figure 3C), suggesting that the CHH sites in a large number of genes and TEs are common targets of FPGS1 and DDM1. Together, FPGS1 generally contributes to DNA methylation in all three cytosine contexts, including CG, CHG, and CHH sites.
Genome-Wide Effect of fpgs1 on Chromatin Silencing
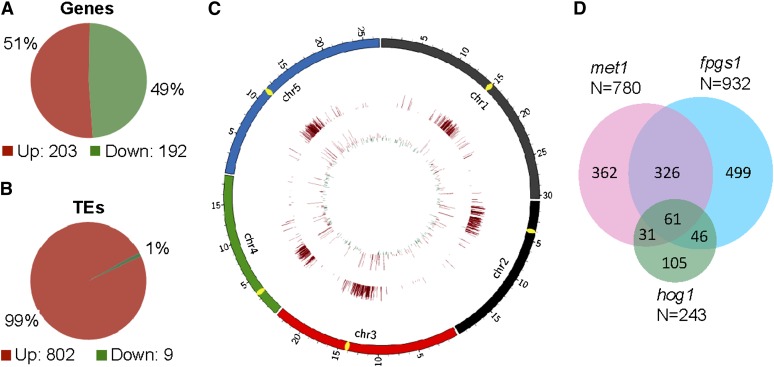
We performed RNA deep sequencing to detect the genome-wide effect of fpgs1 on chromatin silencing. We obtained 60,999,740 ∼40-nucleotide RNA reads for ros1 and 58,592,351 for ros1 fpgs1. More than 90% of these RNA reads can be uniquely matched on the Arabidopsis genome, suggesting the high quality of the RNA libraries (see Supplemental Table 3 online). By comparing the RNA data between ros1 and ros1 fpgs1, we found that 395 protein-coding genes and 811 TEs were significantly affected by fpgs1. The numbers of upregulated versus downregulated genes in ros1 fpgs1 relative to ros1 were similar (203 upregulated versus 192 downregulated) (Figure 4A; see Supplemental Data Set 5 online). However, the number of upregulated TEs (802) was much higher than the number of downregulated TEs (9) (Figure 4B; see Supplemental Data Set 6 online). From the diagram of the genome-wide effect of fpgs1 on RNA transcript levels, we found that protein-coding genes are equivalently affected by fpgs1 throughout chromosomes (Figure 4C). However, the TEs induced by fpgs1 are enriched in the centromeric and pericentromeric regions (Figure 4C), which is consistent with the distribution of TEs on chromosomes. The results suggest that TEs rather than genes are preferentially silenced by FPGS1-dependent mechanisms.
Figure 4.
Characterization of FPGS1-Dependent Loci by RNA Deep Sequencing.
(A) and (B) Diagrams show the numbers of differentially expressed protein-coding genes (A) and TEs (B) caused by fpgs1.
(C) The distribution of differentially expressed protein-coding genes and TEs caused by fpgs1. The transcript levels of genes and TEs are shown in the inside circle and the outside circle, respectively. The outward bars and inward bars represent fpgs1-upregulated loci and -downregulated loci, respectively. The lengths of bars represent the fold changes of differences.
(D) The fpgs1-upregulated Arabidopsis Genome Initiative annotated genes and transposons were compared with the previously reported met1- and hog1-upregulated loci. The numbers of the upregulated loci as well as the number of overlapping loci are shown.
Our RNA deep sequencing identified a total of 932 annotated genes and transposons that are upregulated by fpgs1 (see Supplemental Data Set 7 online). Two previous reports demonstrated that MET1 and HOG1 are required for the silencing of a number of annotated genes and transposons, including 780 loci upregulated by met1 and 243 loci upregulated by hog1 (To et al., 2011; Ouyang et al., 2012). In the loci upregulated by met1 and hog1, 387 (49.6% of 780) and 107 (44.0% of 243) loci, respectively, overlap with the loci upregulated by fpgs1 (Figure 4D). The rates of overlapping loci are significantly higher than expected by chance (P = 0 for the overlap between met1 and fpgs1; P < 3.65e-91 for the overlap between hog1 and fpgs1). The results demonstrated that FPGS1-dependent silencing shares a number of common targets with MET1 and HOG1 at the whole-genome level, suggesting that like MET1 and HOG1, FPGS1 is probably required for maintaining DNA methylation and chromatin silencing at the whole-genome level. The identified upregulated loci were confirmed by quantitative RT-PCR (see Supplemental Figure 10 online), suggesting that the RNA deep sequencing results are reliable.
The Effect of fpgs1 on TE Silencing Is Correlated with DNA Methylation
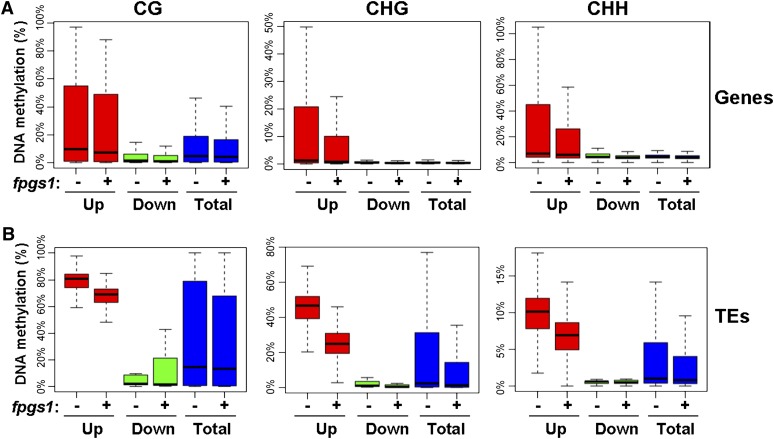
We analyzed the genome-wide effect of fpgs1 on DNA methylation and RNA transcripts, respectively. To determine whether the effect of fpgs1 on DNA methylation and RNA transcripts is correlated, we analyzed the DNA methylation status of those differentially expressed genes and TEs caused by fpgs1. From the RNA deep sequencing data, the number of upregulated genes (203) caused by fpgs1 is equivalent to the number of downregulated genes (192) (Figure 4A). We calculated the overall DNA methylation levels of upregulated genes, downregulated genes, and all genes (Figure 5A). The results indicated that the DNA methylation level of upregulated genes is slightly higher than that of downregulated genes as well as of all genes whether FPGS1 is mutated or not, and the fpgs1 mutation has no much effect on DNA methylation at all genes (Figure 5A). Our RNA deep sequencing data suggested that the number of upregulated TEs (802) caused by fpgs1 is much more than the number of downregulated TEs (9) (Figure 4B). Our whole-genome DNA methylation analysis indicated that the overall DNA methylation level of upregulated TEs is much higher than that of downregulated TEs and all TEs (Figure 5B), suggesting that the hypermethylated TEs are preferentially affected by fpgs1. Moreover, the DNA methylation level of the upregulated TEs is markedly reduced by fpgs1 at all the three cytosine contexts CG, CHG, and CHH (Figure 5B). These results demonstrated that the role of FPGS1 in TE silencing is affected with its function in DNA methylation.
Figure 5.
Box Plot of DNA Methylation Levels in fpgs1-Mediated Differentially Expressed Genes and TEs.
DNA methylation of differentially expressed genes (A) and TEs (B) in ros1 and ros1fpgs1. “−”, ros1; “+”, ros1 fpgs1. “Up” and “Down” represent genes or TEs that are upregulated and downregulated by fpgs1, respectively. “Total” represents total genes or TEs in Arabidopsis. DNA methylation at CG, CHG, and CHH sites was separately indicated.
[See online article for color version of this figure.]
5-formyltetrahydrofolate Complements the Defect of Chromatin Silencing Caused by fpgs1
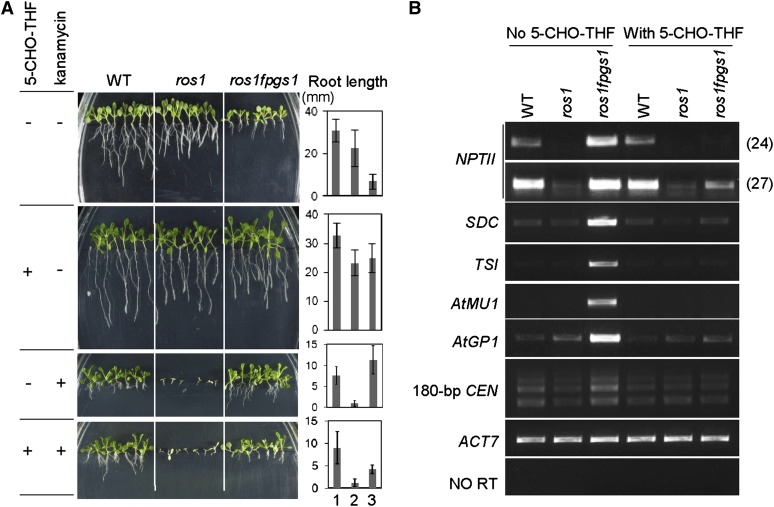
Previous studies showed that mutation of FPGS1 significantly reduced total folate abundance (especially in chloroplasts) and led to short primary roots (Mehrshahi et al., 2010; Srivastava et al., 2011). Exogenous application of 5-formyltetrahydrofolate (5-CHO-THF), a stable form of folate, could rescue the root defect as well as the other developmental defects of the fpgs1 mutant (Srivastava et al., 2011). We investigated whether application of 5-CHO-THF complements the defects in chromatin silencing caused by fpgs1. A root growth assay indicated that the roots of ros1 fpgs1 plants are much shorter than the wild type and ros1 on Murashige and Skoog (MS) medium without extra supplementation (Figure 6A). The root defect of ros1 fpgs1 was mostly complemented by supplementation with 250 μM 5-CHO-THF to the MS medium (Figure 6A). We next examined the effect of 5-CHO-THF on kanamycin resistance of ros1 fpgs1 mutant plants. Because the silencing of the 35S-NPTII transgene is released by the fpgs1 mutation in ros1 fpgs1 (Figures 1B and 6B; see Supplemental Figures 2 and 6 online), the ros1 fpgs1 mutant plants are resistant to 150 mg/L kanamycin on MS medium (Figures 1A and 6A). However, the kanamycin resistance of ros1 fpgs1 was markedly reduced when 250 μM 5-CHO-THF was supplemented in MS medium (Figure 6A). Quantitative RT-PCR demonstrated that addition of 5-CHO-THF specifically suppressed 35S-NPTII transgene expression in ros1 fpgs1 but had no effect on the wild type or ros1 (see Supplemental Figure 2 online).
Figure 6.
Exogenous Application of 5-CHO-THF Rescues the Defect in Chromatin Silencing.
(A) Addition of 250 μM 5-CHO-THF complements the short root phenotype of ros1 fpgs1 on MS medium and compromises the kanamycin resistance of ros1fpgs1 on MS medium supplemented with 150 mg/L kanamycin. The MS medium that contains or does not contain the indicated reagents is indicated by “+” and “−,” respectively. Root length for each sample is shown in the right panel. 1, 2, and 3 represent the wild type (WT), ros1, and ros1 fpgs1, respectively.
(B) Exogenous application of 5-CHO-THF rescues the defect in chromatin silencing in ros1 fpgs1. The transcript levels of the indicated loci were evaluated by RT-PCR. ACT7 was amplified as an internal control.
[See online article for color version of this figure.]
We further checked whether supplementation with 5-CHO-THF could also restore the silencing of endogenous genomic loci that is released in ros1 fpgs1. The silencing of SDC, TSI, At-MU1, At-GP1, and the 180-bp centromeric DNA repeats was suppressed in ros1 fpgs1 on MS control medium, but the silencing of all these loci was restored when 5-CHO-THF was added to the medium (Figure 6B; see Supplemental Figure 2 online). These results suggest that the chromatin silencing defect in ros1 fpgs1 can be complemented by exogenous application of 5-CHO-THF. Our RNA deep sequencing analysis identified a number of TEs that are upregulated by fpgs1 (Figure 4B). By quantitative RT-PCR, we found that the high transcript levels of these TEs caused by fpgs1 were markedly decreased when 5-CHO-THF was added to MS medium (see Supplemental Figure 10 online). The folate derivative may restore the chromatin silencing of ros1 fpgs1 at the whole-genome level.
5-CHO-THF Complements the Defects of DNA Methylation and Histone H3K9 Dimethylation Caused by fpgs1
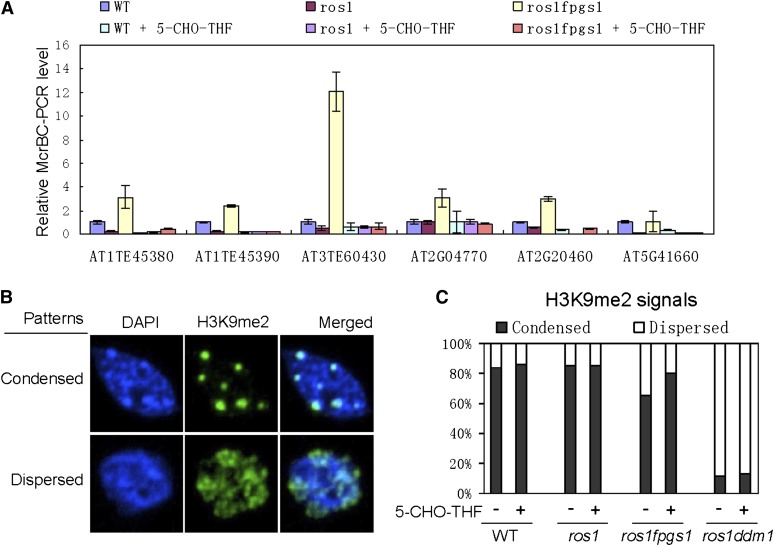
Because DNA methylation is reduced by fpgs1 (Figures 2 and 3; see Supplemental Figures 3 and 9 online), we tested whether the silencing defect of ros1 fpgs1 at the loci identified by RNA deep sequencing is affected with the reduction of DNA methylation. By using the unmethylated DNA-sensitive endonuclease McrBC followed by chop-PCR, we found that DNA methylation was drastically reduced by fpgs1 at the newly identified loci upregulated by fpgs1 (Figure 7A; see Supplemental Figure 11 online). We tested whether exogenous application of 5-CHO-THF complements the defect of DNA methylation in ros1 fpgs1 in vivo. Our chop-PCR results showed that the DNA methylation defect was markedly rescued by application of 5-CHO-THF at the loci identified by RNA deep sequencing (Figure 7A; see Supplemental Figure 11 online). Moreover, DNA gel blotting assays indicated that the reduction of DNA methylation at the 180-bp centromeric DNA and TSI was partially rescued by 5-CHO-THF (see Supplemental Figure 12 online). Therefore, exogenous application of 5-CHO-THF complements the defects of DNA methylation caused by fpgs1. Previous studies indicated that the total folate level is significantly reduced by fpgs1 (Mehrshahi et al., 2010; Srivastava et al., 2011). A proper level of folate is likely required for maintaining DNA methylation in vivo. The role of FPGS1 in the maintenance of DNA methylation is related to its function in folate metabolism. Moreover, we found that even in wild-type plants, application of 5-CHO-THF increases DNA methylation at AT1TE45380, AT1TE45390. AT2G20460, and AT5G41660 (Figure 7A), suggesting that the folate-mediated one-carbon metabolism may act as a limiting factor for maintaining DNA methylation during the development of wild-type plants.
Figure 7.
Effect of 5-CHO-THF on DNA Methylation and Histone H3K9 Dimethylation.
(A) Genomic DNA was cleaved by the unmethylated DNA-sensitive endonuclease McrBC followed by quantitative PCR. DNA methylation was evaluated at the identified fpgs1-upregulated genes and TEs in the wild type (WT), ros1, and ros1 fpgs1. 5-CHO-THF (500 μM) was added to MS medium to determine the effect of 5-CHO-THF on DNA methylation. Relative PCR levels and standard deviations are shown in the chart.
(B) Shown are the nuclei that have condensed H3K9me2 signals and dispersed H3K9me2 signals. The nuclei were counterstained by 4′,6-diamidino-2-phenylindole (DAPI).
(C) The percentages of the nuclei with condensed and dispersed H3K9me2 signals are separately indicated for the wild type, ros1, and ros1 fpgs1. The effect of 5-CHO-THF on the H3K9me2 signals is shown.
We demonstrated the effect of fpgs1 on histone H3K9 dimethylation (Figure 2D). By immunofluorescence assays, we investigated whether 5-CHO-THF can complement the defect of histone H3K9me2 caused by fpgs1. Without 5-CHO-THF treatment, 65.4% of nuclei show condensed H3K9me2 signals in ros1 fpgs1, which is significantly less than the 84.1% in the wild type and the 85.7% in ros1 (Figures 7B and 7C), suggesting that fpgs1 reduces H3K9 dimethylation in vivo. However, the effect of fpgs1 on H3K9 dimethylation is much less than that of ddm1 because the percentage of nuclei that show condensed H3K9 dimethylation signals was reduced to 11.5% in ros1 ddm1 (Figures 7B and 7C). When 5-CHO-THF was applied, the percentage of nuclei showing condensed H3K9me2 signals was increased to 80.0% in ros1 fpgs1, whereas the percentages of such nuclei in the wild type, ros1, and ros1 ddm1 were not significantly affected (Figures 7B and 7C). The results indicated that application of 5-CHO-THF specifically rescued the defect of H3K9me2 caused by fpgs1. Examining these data together, we conclude that the functioning of FPGS1 in folate metabolism is required for both DNA methylation and H3K9 dimethylation in vivo.
The fpgs1 Mutation Causes Accumulation of Hcy and SAH
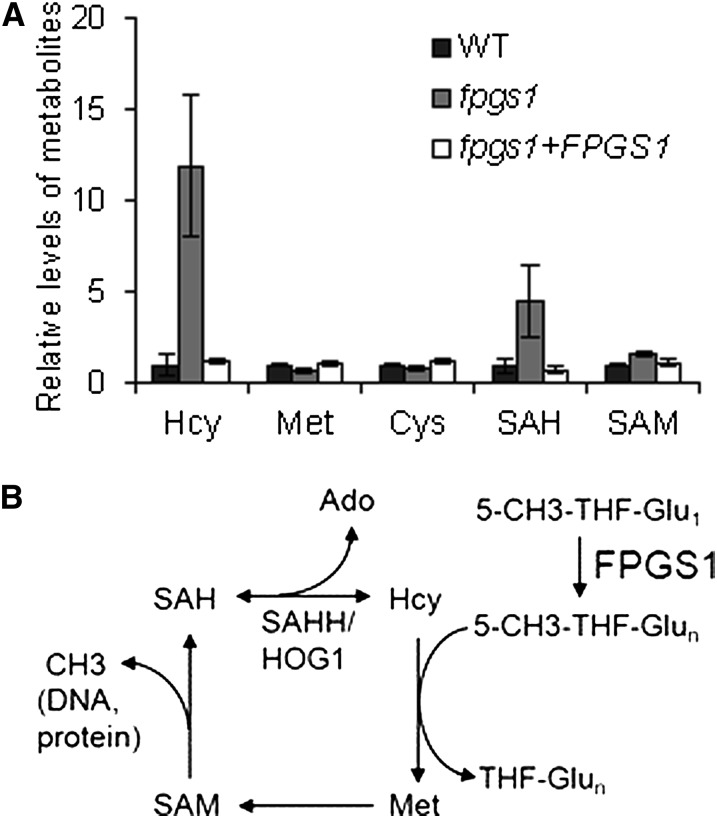
Folate-mediated one-carbon metabolism produces SAM, the universal methyl-donor required for the methylation of DNA, RNA, and proteins. In one-carbon metabolism, 5-CH3-THF is the methyl donor for Met. Met synthase catalyzes the transfer of one methyl-group from 5-CH3-THF to Hcy to form Met, which is then converted to SAM by Met adenosyltransferase. SAM is converted to SAH after its methyl-group is transferred to substrates. SAH can compete with SAM to bind to the active site of methyltransferases (Molloy et al., 2012). It is possible that fpgs1 affects DNA methylation by disrupting one-carbon metabolism. We measured SAM and its related metabolites SAH, Hcy, Met, and Cys in the fpgs1T-DNA mutant, SALK_015472, as well as in the wild type. The results indicated that SAH and Hcy are markedly upregulated in fpgs1 compared with that in the wild type, whereas Met, Cys, and SAM are not significantly affected in fpgs1 (Figure 8A). The high levels of SAH and Hcy were restored to the wild-type levels when the FPGS1 transgene was expressed in fpgs1 (Figure 8A), indicating that fpgs1 is responsible for upregulation of SAH and Hcy.
Figure 8.
The fpgs1 Mutation Causes Accumulation of Hcy and SAH and Reduces SAM Accessibility to DNA Methyltransferases.
(A) The metabolites related to one-carbon metabolism were tested in the wild type (WT), the fpgs1 T-DNA mutant SALK_015472, and the FPGS1 transgenic plants in the fpgs1 mutant background. Standard deviation of three independent biological repeats is shown.
(B) Model for the function of FPGS1 on methyl-group supply in one-carbon metabolism. FPGS1 converts 5-CH3-THF-Glu1 to 5-CH3-THF-Glun, which can transfer its active methyl group to Hcy for Met synthesis. Met are converted to SAM, which can act as a cofactor to provide methyl-group for many methyltransferases. SAH is the product of methylation reaction and is a competitor inhibitor of methyltransferases. SAHH can be cleaved by SAH hydrolase (SAHH1/HOG1) to generate adenosine (Ado) and Hcy.
Folate polyglutamination directly affects one-carbon metabolism because various folate-dependent enzymes in the one-carbon metabolism preferentially utilize polyglutamated folate rather than monoglutamated folate (Shane, 1989; Ravanel et al., 2001). Moreover, folate polyglutamation prevents the transfer of folate between different cellular compartments, which may be critical for the regulation of specific folate-mediated metabolism in the compartments (Matherly and Goldman, 2003). Given that DNA methylation is markedly reduced by fpgs1, it is possible that 5-CH3-THF can be utilized as a methyl-group donor only when it is polyglutamated (Figure 8B). The induction of Hcy caused by fpgs1 is likely caused by a feedback effect of the reduced level of polyglutamated 5-CH3-THF. The high level of Hcy is easily converted to SAH and results in SAH accumulation in fpgs1 (Figures 8A and 8B). SAH can compete with SAM and act as a competitive inhibitor of DNA methyltransferases. Thus, the high level of SAH caused by fpgs1 is directly related to the reduction of DNA methylation.
DISCUSSION
Folate and its derivatives are crucial for the homeostasis of the one-carbon pool. Plants, fungi, and bacteria can synthesize folate, but mammals must take up exogenous folate. In humans, folate deficiency is related to DNA hypomethylation and uracil misincorporation and causes diverse abnormalities (Giovannucci et al., 1993; Molloy, 2012). Therefore, studying the crucial enzymes in folate metabolism is important for understanding the role folate metabolism in humans as well as in plants and other organisms.
In Arabidopsis, folylpolyglutamate synthetases have three members, FPGS1, FPGS2, and FPGS3, which are present in the chloroplast, mitochondria, and cytosol, respectively (Ravanel et al., 2001). However, the functions of the three FPGS proteins are redundant and can be partially rescued by each other (Mehrshahi et al., 2010). The fpgs1 mutant was previously identified based on its morphological phenotypes (Srivastava et al., 2011). The primary root of the fpgs1 mutant is significantly shorter than the wild type (Srivastava et al., 2011). The fpgs2 and fpgs3 single mutants have no obvious morphological phenotypes, but the fpgs2 fpgs3 double mutant exhibits seedling lethality (Mehrshahi et al., 2010). Moreover, the fpgs1 fpgs2 double mutant is embryo-lethal, and the fpgs1 fpgs3 double mutant causes more severe morphological defects than the fpgs1 single mutant, with dwarfed leaves, late flowering, and reduced fertility (Mehrshahi et al., 2010). These results underscore the essential role of folate polyglutamylation in Arabidopsis. Although the importance of folate polyglutamylation in Arabidopsis development has been demonstrated, the mechanism by which folate polyglutamylation affects gene expression and development remains to be elucidated.
We identified the folylpolyglutamate synthetase FPGS1 by screening for suppressors of ros1 and demonstrated that FPGS1 is required for DNA and histone methylation and chromatin silencing. HOG1/SAHH1 encodes an SAHH that catalyzes the conversion of SAH to Hcy, which is further utilized for Met and then SAM synthesis. Moreover, because SAH is a strong competitive inhibitor of methyltransferases, removal of SAH by SAHH is also critical for the activity of methyltransferases (Bacolla et al., 1999; Rocha et al., 2005). In Arabidopsis, the hog1/sahh1 mutations suppress DNA methylation and transcriptional silencing (Rocha et al., 2005; Mull et al., 2006; Baubec et al., 2010; Ouyang et al., 2012). Moreover, the hog1 mutant also shows developmental abnormalities, including delayed germination, slow growth, short primary roots, and reduced fertility (Rocha et al., 2005; Wu et al., 2009). These developmental phenotypes are highly similar to the phenotypes found in the previously described fpgs1 fpgs3 double mutant (Mehrshahi et al., 2010). It is possible that the developmental phenotypes of fpgs1 fpgs3 and hog1 might be related to the global defects in DNA methylation and chromatin silencing. In comparing fpgs1-upregulated loci with the hog1-upregulated loci reported previously (Ouyang et al., 2012), we found that 107 loci are upregulated by both fpgs1 and hog1, which is significantly greater than expected by chance. Upregulation of these loci might account for the developmental defects caused by both fpgs1 and hog1.
Our RNA deep sequencing suggested that the fpgs1 mutation releases the silencing of a number of TEs (Figure 4B). We observed that TEs are predominantly upregulated by fpgs1, whereas the number of upregulated versus downregulated genes caused by fpgs1 is similar (Figures 4A and 4B). Upregulation of TEs is highly correlated with reduced DNA methylation in ros1fpgs1 relative to ros1, whereas upregulation of genes is not significantly correlated with DNA methylation changes caused by fpgs1 (Figures 5A and 5B). The results suggest that the effect of fpgs1 on transcriptional silencing of TEs is directly through its effect on DNA methylation. By contrast, the effect of fpgs1 on differentially expressed genes is mostly independent of DNA methylation changes.
Our genome-wide bisulfite sequencing indicated that the fpgs1 mutation reduces DNA methylation especially at centromeric and pericentromeric regions on each Arabidopsis chromosome (see Supplemental Figure 9 online). Because TEs are predominantly present at centromeric and pericentromeric regions, the derepression effect of fpgs1 on TEs relates to the reduction of DNA methylation at the whole-genome level. Moreover, we found that the fpgs1 mutation upregulates and downregulates similar numbers of protein-coding genes, although fpgs1 predominantly upregulates TEs. However, the upregulation of protein-coding genes is significantly stronger in centromeric and pericentromeric regions than in the two arms of chromosomes (Figure 4C), which is consistent with the stronger effect of fpgs1 on the DNA methylation at centromeric and pericentromeric regions (see Supplemental Figure 9 online). Our genome-wide bisulfite sequencing indicated that the average DNA methylation level at the promoter regions of protein-coding genes is markedly reduced by fpgs1 especially at CHG and CHH sites (Figure 3A), supporting that the derepression effect of fpgs1 on the genes in centromeric and pericentromeric regions is related to the reduction of DNA methylation at their promoter regions.
The effect of fpgs1 on DNA methylation and chromatin silencing is less than that of ddm1 (Figures 1B, 2A to 2C, 3A, 3B, and 4; see Supplemental Figure 9 online). In Arabidopsis, the chloroplast, the mitochondrion, and the cytosol each has one kind of folylpolyglutamate synthetase, FPGS1, FPGS2, and FPGS3, respectively (Ravanel et al., 2001). The weak effect of fpgs1 is probably due to a functional redundancy between FPGS1 and its homologs FPGS2 and FPGS3 (Ravanel et al., 2001). A previous study indicated that the fpgs1 fpgs2, fpgs1 fpgs3, and fpgs2 fpgs3 double mutants have much more severe developmental defects than any of the single mutants, suggesting that FPGS1, FPGS2, and FPGS3 can partially replace each other (Mehrshahi et al., 2010). Although the effect of fpgs2 and fpgs3 on DNA methylation and transcriptional silencing was not found in each single mutant (see Supplemental Figures 7B and 7C online), we cannot exclude that FPGS2 and FPGS3 may function redundantly in DNA and histone methylation and transcriptional silencing.
During Arabidopsis development, DNA methylation at symmetric cytosine sites CG and CHG can be maintained by the DNA methyltransferases MET1 and CMT3, respectively, whereas unsymmetric CHH methylation can be established on unmethylated DNA by the DNA methyltransferase DRM2 and its homologs (Ronemus et al., 1996; Cao and Jacobsen, 2002; Henderson et al., 2010). Our study indicated that the DNA methylation defect at symmetric CG and CHG sites caused by fpgs1 can be rescued by application of 5-CHO-THF, suggesting that DNA methylation at symmetric cytosine sites can be immediately established even if the cytosine in parent cells is unmethylated. The results revealed that DNA methylation can be effectively reestablished during development, which is in contrast with previous findings that disruption of DNA methylation caused by ddm1 can only be gradually complemented after several generations (Teixeira et al., 2009). We found that the overall DNA methylation level is higher in the fpgs1 mutant than in ddm1, although both fpgs1 and ddm1 significantly reduced DNA methylation at the genome-wide level (Table 1, Figures 2A to 2C). It is possible that the relatively higher basal DNA methylation level in the fpgs1 mutant enables exogenous 5-CHO-THF to rapidly restore DNA methylation to the wild-type level.
Our results suggest that the defect of folate polyglutamylation caused by fpgs1 affects folate-mediated one-carbon metabolism (Figure 8A). One-carbon metabolism is required for the biosynthesis of SAM, a crucial methyl-group donor for methylation of DNA, histone, and other substrates (Loenen, 2006). SAH is a competitive inhibitor of methyltransferases and reduces the accessibility of SAM to methyltransferases. A proper SAM/SAH ratio is required for the activity of methyltransferases (Molloy, 2012). Our results suggest that SAH is drastically induced by fpgs1, which results in a low level of SAM/SAH ratio (Figure 8A). Different cytosine contexts and histone methyl marks are catalyzed by diverse DNA and histone methyltransferases. We hypothesize that the sensitivity of various DNA and histone methyltransferases to SAM/SAH reduction is different. In the fpgs1 mutant, the reduction of SAM/SAH can only effectively affect a subset of DNA and histone methyltransferases, leading to reduced DNA and histone methylation at the part of cytosine contexts and histone marks. Alternatively, it is also possible that histone H3K9 dimethylation is indirectly affected by the reduction of DNA methylation caused by fpgs1 because of the coupling of DNA methylation and H3K9 dimethylation ((Rajakumara et al., 2011; Du et al., 2012). Future studies are needed to understand this preferential effect of folate polyglutamylation on DNA methylation and H3K9 dimethylation.
METHODS
Plant Materials, Mutant Identification, and Cloning
Arabidopsis thaliana C24 and its derivative ros1 mutant that harbors the RD29A-LUC and 35S-NPTII transgenes were used in this study. We mutagenized the ros1 mutant seeds with ethyl methanesulfonate to screen for suppressors of ros1 as previously described (Liu et al., 2011). The selfed T2 seedlings were screened based on luminescence imaging or kanamycin resistance. The positive mutants were confirmed based on the luminescence strength and kanamycin resistance of their offspring. The confirmed mutant was crossed to the ros1 mutant in the Columbia-0 background (Salk_045303), and the selfed F2 population was used for map-based cloning to localize the chromosome region of the mutation. The candidate genes of the region were sequenced to find the mutation. The FPGS1 genomic sequence was cloned into the modified pCAMBIA1305 vector with its 3′-terminal in frame with the 3xFlag epitope sequence. The primers used for FPGS1 cloning are indicated in Supplemental Data Set 8 online. The construct was transformed into ros1 pfgs1 for complementation testing.
Analysis of RNA Transcript Levels
We performed quantitative RT-PCR to measure the transcript levels of protein-coding genes and TEs. Reverse primers used in this study include oligo(dT), random DNA oligos, and sequence-specific DNA oligos. ReverTra Ace qPCR RT Master Mix with gDNA remover (FSQ301; Toyobo) was used for reverse transcription. For quantitative RT-PCR, SYBR Premix Ex Taq II (Tli RNaseH Plus) (RR420A; Takara) was used on an Applied Biosystems 7500 Fast real-time PCR system. The RNA transcript levels were determined by three independent biological replicates. RNA transcripts were also subjected to RT-PCR and examined on ethidium bromide–stained agarose gels; the gel-based RT-PCR results were confirmed by quantitative RT-PCR in this study. The primer sequences are listed in Supplemental Data Set 8 online. ACT7 was used as an internal control. To confirm the absence of DNA contamination in the RNA samples, we amplified ACT7 by directly using RNA as templates. The amplification was shown as “No RT.” The absence of amplification indicated the absence of DNA contamination.
DNA Methylation Analysis
DNA methylation was analyzed by bisulfite sequencing, DNA gel blotting, and chop-PCR. For bisulfite sequencing, 2 μg of genomic DNA was treated and purified according to the protocol for the EpiTect bisulfite kit (Qiagen). The purified DNA was amplified, and the amplification product was cloned into the pMD18-T vector (Takara) for sequencing. The DNA methylation levels at CG, CHG, and CHH sites were separately calculated. For DNA gel blotting, genomic DNA was cleaved with the DNA methylation-sensitive endonucleases HpaII, MspI, and HaeIII at 37°C for 2 d and run on 1% agarose gels. The probes of 180-bp centromeric DNA repeats, TSI, and 5S rDNA were labeled by [α-32P]dCTP with Klenow enzyme. The endonuclease McrBC cleaves DNA containing methylated cytosine but has no action on unmethylated DNA. For chop-PCR, genomic DNA was cleaved with McrBC, and the products were subjected to PCR and quantitative PCR.
Whole-Genome Bisulfite Sequencing
Raw sequencing data was mapped to TAIR10 reference genome modified by the single nucleotide polymorphisms between C24 and Columbia-0. Only the sequences mapped to unique positions on the Arabidopsis genome were retained for DNA methylation analysis. DNA methylation was calculated when cytosine sites have at least fivefold coverage. The methylation level for each cytosine was evaluated by the percentage of reads reporting a C in the total number of reads at the site. Gene and TE annotations were downloaded from The Arabidopsis Information Resource. One-kilobase upstream and downstream surrounding regions were included to calculate the DNA methylation levels of genes and TEs. The methylation levels of genes and TEs were estimated by pooling the read counts that show at least fivefold coverage. CG, CHG, and CHH methylation was calculated.
Annotated genes or transposons including 1-kb upstream and downstream flanking sequences were aligned to the modified TAIR10 reference genome. The average methylation level for each 100-bp interval was plotted. The lengths of genes or transposons were adjusted to a common sum. DNA methylation levels in 200-kb windows were plotted across each chromosome to indicate the genome-wide DNA methylation status in ros1, ros1 fpgs1, and ros1 ddm1. The fold change of DNA methylation between ros1 fpgs1 and ros1 in the 200-kb window was calculated and shown across each chromosome. To evaluate the correlation between differentially expressed genes or TEs and DNA methylation, we determined the DNA methylation levels of different classes of differentially expressed genes and TEs using the method described previously (Zhang et al., 2013).
Immunofluorescence Assay
Histone H3K9 dimethylation was immunolocalized as previously described (Onodera et al., 2005). In brief, after nuclei were blocked with 3% BSA in PBS, the primary antibody H3K9me2 (ab1220; Abcam) was diluted at 1:50 and incubated overnight at 4°C. Secondary anti-mouse fluorescein isothiocyanate (Invitrogen) or anti-rabbit fluorescein isothiocyanate (Invitrogen) was used at 1:200 dilutions and incubated for 2 h at 37°C. Chromatin was counterstained with 4′,6-diamidino-2-phenylindole in mounting medium. Images were acquired with a spinning-disk confocal microscope and processed with Adobe Photoshop (Adobe).
RNA Deep Sequencing and Data Analysis
Total RNA was extracted from 2-week-old Arabidopsis seedlings and sent to Beijing Genomics Institute for RNA library preparation and deep sequencing. Arabidopsis genome sequences and annotated gene models were downloaded from TAIR10 (http://www.Arabidopsis.org/). Tophat v2.0.6 was used to align the raw RNA reads to the Arabidopsis genome (Trapnell et al., 2009), allowing up to two mismatches. The edgeR package (Robinson et al., 2010) was used in the differential expression analysis for genes and TEs. Significance of expression differences was calculated using Fisher’s exact test. The distribution of differentially expressed genes and TEs throughout the Arabidopsis genome was plotted by circos (Krzywinski et al., 2009).
Measurement of Metabolites in Arabidopsis Seedlings
Two-week-old Arabidopsis seedlings from MS medium plates were used for measurement of SAM, SAH, Hcy, Cys, and Met. The methods for sample preparation and metabolite measurement were previously described (Nikiforova et al., 2005). The experiments were biologically repeated three times.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: AT5G66750 (DDM1), AT5G05980 (FPGS1), AT3G10160 (FPGS2), AT3G55630 (FPGS3), AT4G13940 (HOG1), AT2G36490 (ROS1), AT5G49160 (MET1), AT5G52310 (RD29A), AT2G17690 (SDC), At4g03650 (At-GP1), At4g08680 (AtMU1), BD298459.1 (TSI), and AT5G09810 (ACT7).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Identification and Characterization of DDM1.
Supplemental Figure 2. Quantitative RT-PCR of NPTII, SDC, TSI, AtMU1, and AtGP1.
Supplemental Figure 3. The Effect of fpgs1 and ddm1 on DNA Methylation.
Supplemental Figure 4. Identification and Characterization of FPGS1.
Supplemental Figure 5. Identification of Abnormal FPGS1 Transcript Variants in ros1 fpgs1 Compared with That in the Wild Type and ros1.
Supplemental Figure 6. Complementation Assay for fpgs1.
Supplemental Figure 7. DNA Methylation and Transcriptional Silencing in the T-DNA fpgs1, fpgs2, and fpgs3 Mutants.
Supplemental Figure 8. The Effect of fpgs1 and ddm1 on DNA Methylation at the Promoters of the 35S-NPTII and RD29A-LUC Transgenes.
Supplemental Figure 9. The Effect of fpgs1 and ddm1 on DNA Methylation across Arabidopsis Chromosomes.
Supplemental Figure 10. Quantitative RT-PCR of the Genes and TEs Identified by RNA Deep Sequencing.
Supplemental Figure 11. The DNA Methylation Levels of fpgs1 Upregulated Genes and TEs Were Determined by Chop-PCR.
Supplemental Figure 12. The Effect of 5-CHO-THF on DNA Methylation at 180-bp Centromeric DNA and TSI Sites in the Wild type, ros1, and ros1 fpgs1.
Supplemental Table 1. Primary Data for Locus-Specific Bisulfite Sequencing.
Supplemental Table 2. Numbers of Obtained Reads from Whole-Genome Bisulfite Sequencing.
Supplemental Table 3. Numbers of Obtained RNA Reads from RNA Deep Sequencing.
Supplemental Data Set 1. Differentially Methylated Genes Caused by fpgs1.
Supplemental Data Set 2. Differentially Methylated Transposable Elements Caused by fpgs1.
Supplemental Data Set 3. Differentially Methylated Genes Caused by ddm1.
Supplemental Data Set 4. Differentially Methylated Transposable Elements Caused by ddm1.
Supplemental Data Set 5. List of Differentially Expressed Genes Affected by fpgs1.
Supplemental Data Set 6. List of Differentially Expressed Transposable Elements Affected by fpgs1.
Supplemental Data Set 7. List of AGI Annotated Genes and Transposable Elements Upregulated by fpgs1.
Supplemental Data Set 8. List of DNA Oligos Used in This Study.
Acknowledgments
We thank Zhirong Shen at the Metabolomics Center of National Institute of Biological Sciences, Beijing, for technical assistance in metabolite analysis. This work was supported by the National Basic Research Program of China (973 Program; 2012CB910900) and the 973 Program (2011CB812600) from the Chinese Ministry of Science and Technology.
AUTHOR CONTRIBUTIONS
H.-R.Z. performed research, analyzed the data, and wrote the article. F.-F.Z. performed research and analyzed the data. Z.-Y.M. and L.J. performed research. H.-W.H., T.C., J.-K.Z., and C.Z. analyzed the data. X.-J.H. designed the research, analyzed the data, and wrote the article.
Glossary
- SAM
S-adenosylmethionine
- 5-CH3-THF
5-methyl-tetrahydrofolate
- SAH
S-adenosylhomocysteine
- SAHH
S-adenosylhomocysteine hydrolase
- RdDM
RNA-directed DNA methylation
- TE
transposable element
- 5-CHO-THF
5-formyltetrahydrofolate
- MS
Murashige and Skoog
- Hcy
homocysteine
References
- Bacolla A., Pradhan S., Roberts R.J., Wells R.D. (1999). Recombinant human DNA (cytosine-5) methyltransferase. II. Steady-state kinetics reveal allosteric activation by methylated DNA. J. Biol. Chem. 274: 33011–33019 [DOI] [PubMed] [Google Scholar]
- Baubec T., Dinh H.Q., Pecinka A., Rakic B., Rozhon W., Wohlrab B., von Haeseler A., Mittelsten Scheid O. (2010). Cooperation of multiple chromatin modifications can generate unanticipated stability of epigenetic states in Arabidopsis. Plant Cell 22: 34–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumbusch L.O., Thorstensen T., Krauss V., Fischer A., Naumann K., Assalkhou R., Schulz I., Reuter G., Aalen R.B. (2001). The Arabidopsis thaliana genome contains at least 29 active genes encoding SET domain proteins that can be assigned to four evolutionarily conserved classes. Nucleic Acids Res. 29: 4319–4333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X., Jacobsen S.E. (2002). Locus-specific control of asymmetric and CpNpG methylation by the DRM and CMT3 methyltransferase genes. Proc. Natl. Acad. Sci. USA 99 (Suppl 4): 16491–16498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cossins E.A., Chen L. (1997). Folates and one-carbon metabolism in plants and fungi. Phytochemistry 45: 437–452 [DOI] [PubMed] [Google Scholar]
- Du J., et al. (2012). Dual binding of chromomethylase domains to H3K9me2-containing nucleosomes directs DNA methylation in plants. Cell 151: 167–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friso S., Choi S.W., Girelli D., Mason J.B., Dolnikowski G.G., Bagley P.J., Olivieri O., Jacques P.F., Rosenberg I.H., Corrocher R., Selhub J. (2002). A common mutation in the 5,10-methylenetetrahydrofolate reductase gene affects genomic DNA methylation through an interaction with folate status. Proc. Natl. Acad. Sci. USA 99: 5606–5611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frosst P., et al. (1995). A candidate genetic risk factor for vascular disease: A common mutation in methylenetetrahydrofolate reductase. Nat. Genet. 10: 111–113 [DOI] [PubMed] [Google Scholar]
- Giovannucci E., Stampfer M.J., Colditz G.A., Rimm E.B., Trichopoulos D., Rosner B.A., Speizer F.E., Willett W.C. (1993). Folate, methionine, and alcohol intake and risk of colorectal adenoma. J. Natl. Cancer Inst. 85: 875–884 [DOI] [PubMed] [Google Scholar]
- Gong Z., Morales-Ruiz T., Ariza R.R., Roldan-Arjona T., David L., Zhu J.K. (2002). ROS1, a repressor of transcriptional gene silencing in Arabidopsis, encodes a DNA glycosylase/lyase. Cell 111: 803–814 [DOI] [PubMed] [Google Scholar]
- He X.J., Chen T., Zhu J.K. (2011). Regulation and function of DNA methylation in plants and animals. Cell Res. 21: 442–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X.J., Hsu Y.F., Pontes O., Zhu J., Lu J., Bressan R.A., Pikaard C., Wang C.S., Zhu J.K. (2009). NRPD4, a protein related to the RPB4 subunit of RNA polymerase II, is a component of RNA polymerases IV and V and is required for RNA-directed DNA methylation. Genes Dev. 23: 318–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X.J., Hsu Y.F., Zhu S., Wierzbicki A.T., Pontes O., Pikaard C.S., Liu H.L., Wang C.S., Jin H., Zhu J.K. (2009b). An effector of RNA-directed DNA methylation in Arabidopsis is an ARGONAUTE 4- and RNA-binding protein. Cell 137: 498–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson I.R., Deleris A., Wong W., Zhong X., Chin H.G., Horwitz G.A., Kelly K.A., Pradhan S., Jacobsen S.E. (2010). The de novo cytosine methyltransferase DRM2 requires intact UBA domains and a catalytically mutated paralog DRM3 during RNA-directed DNA methylation in Arabidopsis thaliana. PLoS Genet. 6: e1001182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson I.R., Jacobsen S.E. (2008). Tandem repeats upstream of the Arabidopsis endogene SDC recruit non-CG DNA methylation and initiate siRNA spreading. Genes Dev. 22: 1597–1606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeddeloh J.A., Stokes T.L., Richards E.J. (1999). Maintenance of genomic methylation requires a SWI2/SNF2-like protein. Nat. Genet. 22: 94–97 [DOI] [PubMed] [Google Scholar]
- Johnson L.M., Law J.A., Khattar A., Henderson I.R., Jacobsen S.E. (2008). SRA-domain proteins required for DRM2-mediated de novo DNA methylation. PLoS Genet. 4: e1000280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzywinski M., Schein J., Birol I., Connors J., Gascoyne R., Horsman D., Jones S.J., Marra M.A. (2009). Circos: An information aesthetic for comparative genomics. Genome Res. 19: 1639–1645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law J.A., Jacobsen S.E. (2010). Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat. Rev. Genet. 11: 204–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Bai G., Zhang C., Chen W., Zhou J., Zhang S., Chen Q., Deng X., He X.J., Zhu J.K. (2011). An atypical component of RNA-directed DNA methylation machinery has both DNA methylation-dependent and -independent roles in locus-specific transcriptional gene silencing. Cell Res. 21: 1691–1700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q., Gong Z. (2011). The coupling of epigenome replication with DNA replication. Curr. Opin. Plant Biol. 14: 187–194 [DOI] [PubMed] [Google Scholar]
- Loenen W.A. (2006). S-adenosylmethionine: Jack of all trades and master of everything? Biochem. Soc. Trans. 34: 330–333 [DOI] [PubMed] [Google Scholar]
- Lucock M. (2000). Folic acid: Nutritional biochemistry, molecular biology, and role in disease processes. Mol. Genet. Metab. 71: 121–138 [DOI] [PubMed] [Google Scholar]
- Matherly L.H., Goldman D.I. (2003). Membrane transport of folates. Vitam. Horm. 66: 403–456 [DOI] [PubMed] [Google Scholar]
- Matzke M., Kanno T., Daxinger L., Huettel B., Matzke A.J. (2009). RNA-mediated chromatin-based silencing in plants. Curr. Opin. Cell Biol. 21: 367–376 [DOI] [PubMed] [Google Scholar]
- Mehrshahi P., et al. (2010). Functional analysis of folate polyglutamylation and its essential role in plant metabolism and development. Plant J. 64: 267–279 [DOI] [PubMed] [Google Scholar]
- Molloy A.M. (2012). Genetic aspects of folate metabolism. Subcell. Biochem. 56: 105–130 [DOI] [PubMed] [Google Scholar]
- Mull L., Ebbs M.L., Bender J. (2006). A histone methylation-dependent DNA methylation pathway is uniquely impaired by deficiency in Arabidopsis S-adenosylhomocysteine hydrolase. Genetics 174: 1161–1171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikiforova V.J., Kopka J., Tolstikov V., Fiehn O., Hopkins L., Hawkesford M.J., Hesse H., Hoefgen R. (2005). Systems rebalancing of metabolism in response to sulfur deprivation, as revealed by metabolome analysis of Arabidopsis plants. Plant Physiol. 138: 304–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onodera Y., Haag J.R., Ream T., Costa Nunes P., Pontes O., Pikaard C.S. (2005). Plant nuclear RNA polymerase IV mediates siRNA and DNA methylation-dependent heterochromatin formation. Cell 120: 613–622 [DOI] [PubMed] [Google Scholar]
- Ouyang B., Fei Z., Joung J.G., Kolenovsky A., Koh C., Nowak J., Caplan A., Keller W.A., Cui Y., Cutler A.J., Tsang E.W. (2012). Transcriptome profiling and methyl homeostasis of an Arabidopsis mutant deficient in S-adenosylhomocysteine hydrolase1 (SAHH1). Plant Mol. Biol. 79: 315–331 [DOI] [PubMed] [Google Scholar]
- Rajakumara E., Law J.A., Simanshu D.K., Voigt P., Johnson L.M., Reinberg D., Patel D.J., Jacobsen S.E. (2011). A dual flip-out mechanism for 5mC recognition by the Arabidopsis SUVH5 SRA domain and its impact on DNA methylation and H3K9 dimethylation in vivo. Genes Dev. 25: 137–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravanel S., Cherest H., Jabrin S., Grunwald D., Surdin-Kerjan Y., Douce R., Rébeillé F. (2001). Tetrahydrofolate biosynthesis in plants: Molecular and functional characterization of dihydrofolate synthetase and three isoforms of folylpolyglutamate synthetase in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 98: 15360–15365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M.D., McCarthy D.J., Smyth G.K. (2010). edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha P.S., Sheikh M., Melchiorre R., Fagard M., Boutet S., Loach R., Moffatt B., Wagner C., Vaucheret H., Furner I. (2005). The Arabidopsis HOMOLOGY-DEPENDENT GENE SILENCING1 gene codes for an S-adenosyl-L-homocysteine hydrolase required for DNA methylation-dependent gene silencing. Plant Cell 17: 404–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronemus M.J., Galbiati M., Ticknor C., Chen J., Dellaporta S.L. (1996). Demethylation-induced developmental pleiotropy in Arabidopsis. Science 273: 654–657 [DOI] [PubMed] [Google Scholar]
- Roudier F., et al. (2011). Integrative epigenomic mapping defines four main chromatin states in Arabidopsis. EMBO J. 30: 1928–1938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shane B. (1989). Folylpolyglutamate synthesis and role in the regulation of one-carbon metabolism. Vitam. Horm. 45: 263–335 [DOI] [PubMed] [Google Scholar]
- Shelnutt K.P., Kauwell G.P., Gregory J.F., III, Maneval D.R., Quinlivan E.P., Theriaque D.W., Henderson G.N., Bailey L.B. (2004). Methylenetetrahydrofolate reductase 677C—>T polymorphism affects DNA methylation in response to controlled folate intake in young women. J. Nutr. Biochem. 15: 554–560 [DOI] [PubMed] [Google Scholar]
- Slotkin R.K., Martienssen R. (2007). Transposable elements and the epigenetic regulation of the genome. Nat. Rev. Genet. 8: 272–285 [DOI] [PubMed] [Google Scholar]
- Sridhar V.V., Kapoor A., Zhang K., Zhu J., Zhou T., Hasegawa P.M., Bressan R.A., Zhu J.K. (2007). Control of DNA methylation and heterochromatic silencing by histone H2B deubiquitination. Nature 447: 735–738 [DOI] [PubMed] [Google Scholar]
- Srivastava A.C., Ramos-Parra P.A., Bedair M., Robledo-Hernández A.L., Tang Y., Sumner L.W., Díaz de la Garza R.I., Blancaflor E.B. (2011). The folylpolyglutamate synthetase plastidial isoform is required for postembryonic root development in Arabidopsis. Plant Physiol. 155: 1237–1251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka H., Masuta C., Uehara K., Kataoka J., Koiwai A., Noma M. (1997). Morphological changes and hypomethylation of DNA in transgenic tobacco expressing antisense RNA of the S-adenosyl-L-homocysteine hydrolase gene. Plant Mol. Biol. 35: 981–986 [DOI] [PubMed] [Google Scholar]
- Teixeira F.K., et al. (2009). A role for RNAi in the selective correction of DNA methylation defects. Science 323: 1600–1604 [DOI] [PubMed] [Google Scholar]
- To T.K., et al. (2011). Arabidopsis HDA6 regulates locus-directed heterochromatin silencing in cooperation with MET1. PLoS Genet. 7: e1002055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Pachter L., Salzberg S.L. (2009). TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics 25: 1105–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veiseth S.V., Rahman M.A., Yap K.L., Fischer A., Egge-Jacobsen W., Reuter G., Zhou M.M., Aalen R.B., Thorstensen T. (2011). The SUVR4 histone lysine methyltransferase binds ubiquitin and converts H3K9me1 to H3K9me3 on transposon chromatin in Arabidopsis. PLoS Genet. 7: e1001325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wierzbicki A.T., Cocklin R., Mayampurath A., Lister R., Rowley M.J., Gregory B.D., Ecker J.R., Tang H., Pikaard C.S. (2012). Spatial and functional relationships among Pol V-associated loci, Pol IV-dependent siRNAs, and cytosine methylation in the Arabidopsis epigenome. Genes Dev. 26: 1825–1836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X., Li F., Kolenovsky A., Caplan A., Cui Y.H., Cutler A., Tsang E.W.T. (2009). A mutant Deficient in S-adenosylhomocysteine hydrolase in Arabidopsis shows defects in root hair development. Botany 87: 571–584 [Google Scholar]
- Xia R., Wang J., Liu C., Wang Y., Wang Y., Zhai J., Liu J., Hong X., Cao X., Zhu J.K., Gong Z. (2006). ROR1/RPA2A, a putative replication protein A2, functions in epigenetic gene silencing and in regulation of meristem development in Arabidopsis. Plant Cell 18: 85–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin H., Zhang X., Liu J., Wang Y., He J., Yang T., Hong X., Yang Q., Gong Z. (2009). Epigenetic regulation, somatic homologous recombination, and abscisic acid signaling are influenced by DNA polymerase epsilon mutation in Arabidopsis. Plant Cell 21: 386–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Deng X., Miki D., Cutler S., La H., Hou Y.J., Oh J., Zhu J.K. (2012). Sulfamethazine suppresses epigenetic silencing in Arabidopsis by impairing folate synthesis. Plant Cell 24: 1230–1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., et al. (2013). DTF1 is a core component of RNA-directed DNA methylation and may assist in the recruitment of Pol IV. Proc. Natl. Acad. Sci. USA 110: 8290–8295 [DOI] [PMC free article] [PubMed] [Google Scholar]