The MADS domain factors STK and SEP3 directly regulate VDD expression. STK-SEP3 dimers induce loop formation in the VDD promoter by binding to two nearby CArG boxes, and this loop is essential for VDD expression. In vivo data suggest that the size and/or position of this loop determine the spatial expression profile of VDD. This study provides in vivo evidence for the floral quartet model.
Abstract
MADS domain transcription factors are key regulators of eukaryotic development. In plants, the homeotic MIKC MADS factors that regulate floral organ identity have been studied in great detail. Based on genetic and protein–protein interaction studies, a floral quartet model was proposed that describes how these MADS domain proteins assemble into higher order complexes to regulate their target genes. However, despite the attractiveness of this model and its general acceptance in the literature, solid in vivo proof has never been provided. To gain deeper insight into the mechanisms of transcriptional regulation by MADS domain factors, we studied how SEEDSTICK (STK) and SEPALLATA3 (SEP3) directly regulate the expression of the reproductive meristem gene family transcription factor–encoding gene VERDANDI (VDD). Our data show that STK-SEP3 dimers can induce loop formation in the VDD promoter by binding to two nearby CC(A/T)6GG (CArG) boxes and that this is essential for promoter activity. Our in vivo data show that the size and position of this loop, determined by the choice of CArG element usage, is essential for correct expression. Our studies provide solid in vivo evidence for the floral quartet model.
INTRODUCTION
MADS box genes encode transcriptional regulators involved in diverse and important biological functions. They have been identified in yeast, insects, nematodes, lower vertebrates, mammals, and plants. These transcription factors contain a conserved DNA binding and dimerization domain named the MADS domain (Schwarz-Sommer et al., 1992). In plants, MADS box genes have been highly amplified during evolution; for instance, 107 MADS box genes have been identified in Arabidopsis thaliana and 75 in rice (Oryza sativa; Parenicová et al., 2003; Arora et al., 2007).
The ability of MADS domain proteins to bind DNA as dimers is reflected by the dyad symmetry of their binding sites that are found within promoter and enhancer sequences (Shore and Sharrocks, 1995). Nurrish and Treisman (1995) studied MADS domain protein binding sites and showed that they bind to the consensus sequence CC(A/T)6GG, named the CArG box. Evidence based on in vitro biochemical assays and interaction studies in yeast showed that plant MADS domain proteins form mainly heterodimers, which are thought to assemble into multimeric complexes (Riechmann et al., 1996; Egea-Cortines et al., 1999; Honma and Goto, 2001; de Folter et al., 2005). Many of these studies have been done using MIKC MADS domain factors that regulate floral organ identity in A. thaliana, and their modes of action have been described in the combinatorial genetic ABC model (Coen and Meyerowitz, 1991). These MIKC MADS proteins have a characteristic modular structure. From the N to the C terminus of the protein, four characteristic domains can be identified: the MADS-box (M), intervening (I), keratin-like (K), and C-terminal (C) domains. Importantly, ABC MADS domain factor activity is dependent on another group of MADS domain transcription factors, indicated as class E factors, which are encoded by four largely redundant SEPALLATA genes (SEP1-4) (Pelaz et al., 2000; Ditta et al., 2004). Class E factors establish interactions between A, B, and C class factors, and their combined ectopic expression (A, B, and E or B, C, and E) results in the homeotic conversion of leaves into petals or stamens (Honma and Goto, 2001; Pelaz et al., 2001). These studies resulted into the formulation of a floral quartet model, which suggests that the MADS domain proteins form higher order (quartet) complexes to establish floral organ identity (Theissen and Saedler, 2001).
Similar results were obtained for the factors that regulate ovule development in A. thaliana. The three MADS box genes SEEDSTICK (STK), SHATTERPROOF1 (SHP1), and SHP2 redundantly regulate ovule identity, since ovules are converted into carpel-like structures in the stk shp1 shp2 triple mutant (Pinyopich et al., 2003). Interestingly, the SEP1/sep1 sep2 sep3 triple mutant (in which only one SEP1 allele is active) phenocopied the stk shp1 shp2 triple mutant, showing that the SEP proteins are also important for the development of ovules (Favaro et al., 2003). The role of SEP proteins in the formation of ovules is likely to favor the formation of active complexes, since yeast three-hybrid studies showed that SEP3 was able to bridge interactions among STK, SHP1, and SHP2.
Recently, we identified VERDANDI (VDD), a gene belonging to the REPRODUCTIVE MERISTEM family (Romanel et al., 2009), as a target of the ovule identity factors STK, SHP1, SHP2, and SEP3 (Matias-Hernandez et al., 2010). VDD transcripts are present in the same tissues as these ovule identity genes and silencing of the ovule identity genes, STK, SHP1, and SHP2, led to the complete absence of VDD expression during ovule development. Analysis of the VDD mutant revealed that this gene is important for female gametophyte cell identity determination (Matias-Hernandez et al., 2010).
Studies demonstrated that MADS domain protein complexes often interact with DNA by contacting multiple nearby CArG box sequences, separated by less than 300 bp (Egea-Cortines et al., 1999; Liu et al., 2008). In the regulatory region of VDD, three CArG boxes were identified within a region of 500 bp, and chromatin immunoprecipitation (ChIP) analysis showed that the first and third box were bound by both STK and SEP3 (Matias-Hernandez et al., 2010).
Here, we describe the use of a combination of biophysical, molecular, and in vivo approaches to study the regulation of VDD promoter activity by MADS domain ovule identity factors. In particular, we characterized in vitro and in vivo the interactions of STK and SEP3 with the three CArG boxes and investigated the importance of these interactions for the expression of VDD. Our study provides deeper insight into the mode of action of MADS domain proteins in the regulation of their target genes.
RESULTS
SEP3 and STK Together Mediate DNA Looping in the VDD Promoter Region
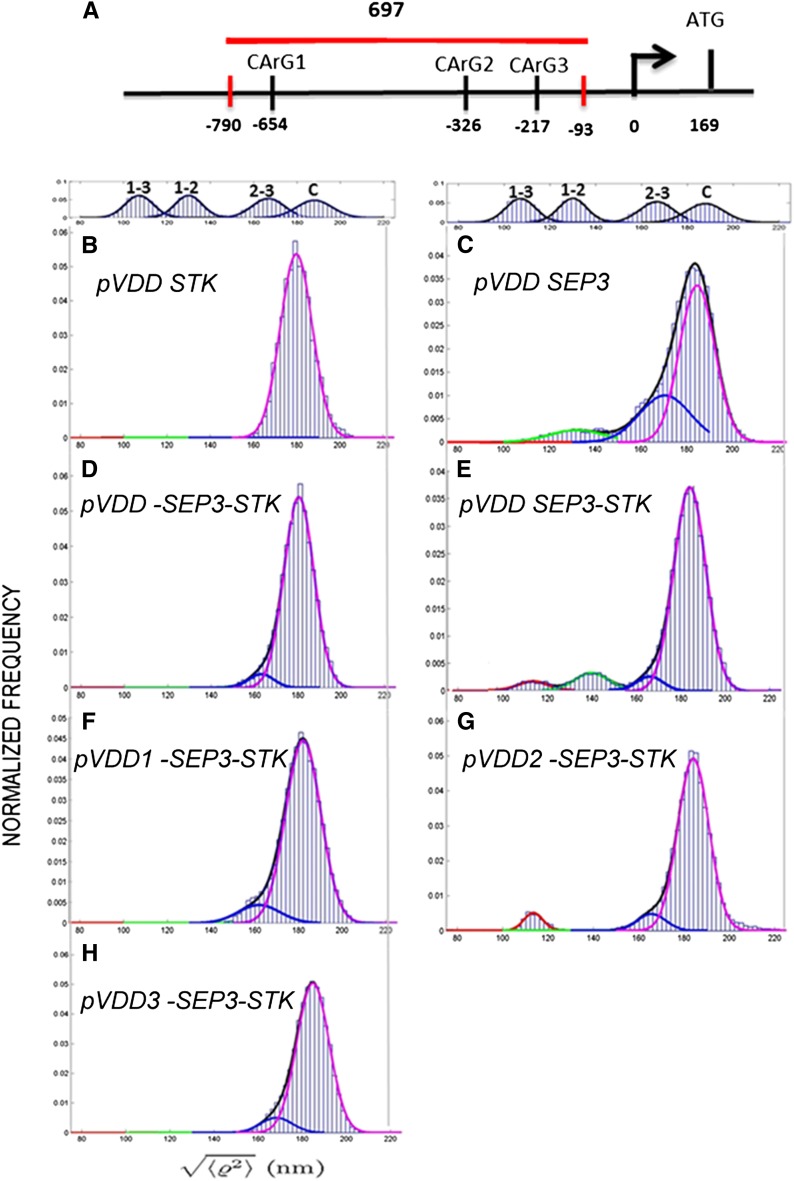
STK and SEP3 form dimers that probably combine into tetrameric complexes (Favaro et al., 2003; Melzer et al., 2008). Furthermore, they regulate the expression of VDD through direct binding to its promoter region (Favaro et al., 2003; Matias-Hernandez et al., 2010). The VDD promoter region contains three CArG boxes within the region 1000 bp upstream of the ATG start codon (Figure 1A; Matias-Hernandez et al., 2010). Cooperative binding of the tetramers (composed of two SEP3-STK heterodimers) to two of the three adjacent CArG boxes would induce the formation of loops within the promoter region, which might have important regulatory functions. To investigate whether SEP3 and STK are indeed able to mediate interactions between elements in the VDD promoter region, tethered particle motion (TPM) analysis (Finzi and Dunlap, 2003; Pouget et al., 2004; Nelson et al., 2006; Dunlap et al., 2011) was performed using a VDD promoter fragment of 697 bp containing all three CArG boxes in the same arrangement found in vivo (Figure 1A). TPM is a powerful, single-molecule technique, which is particularly appropriate to monitor protein-induced DNA conformational changes such as looping, bending, and large-scale compaction (Finzi and Gelles, 1995; Guerra et al., 2007; Zurla et al., 2009; Zaremba et al., 2010).
Figure 1.
TPM Analysis of STK and SEP3 Interactions with the VDD Promoter.
(A) Schematic representation of the VDD promoter and the distances between CArG boxes in relation to the transcription start site. The fragment used for the TPM experiments is indicated with a red horizontal line (the small vertical red bars indicate the exact position of this fragment). The arrow indicates the transcription start site, and the positions of the CArG boxes and ATG (vertical bars) are relative to the transcription start site (0).
(B) proVDD in the presence of STK.
(C) proVDD in the presence of SEP3.
(D) Negative control of TPM experiment; proVDD with the CArG boxes deleted (proVDD-1-2-3) in the presence of both STK and SEP3.
(E) to (H) proVDD in the presence of both STK and SEP3 (E). proVDD with the first CArG box deleted (F), with the second CArG deleted (G), and with the third CArG deleted (H), in the presence of both proteins.
In principle, binding to CArG boxes and STK-SEP3 protein–protein cooperative interactions could result in three possible loops: between CArG box 1 and 2, between CArG box 2 and 3, or between CArG box 1 and 3 (see Supplemental Figure 1A online).
To facilitate the correct interpretation of the TPM data, we first made a calibration curve using DNA tethers that have lengths that are predicted to be similar to each of the possible looped VDD promoter fragments (see Supplemental Figure 1B online). Therefore, we made tethers 243, 355, and 575 bp long. After analyzing 20 tethers for each DNA fragment, including the 697-bp fragment, we fitted the cumulative frequency distribution of the data with a Gaussian curve (see Supplemental Figure 1C online). The center of the peak of each Gaussian curve indicates the average 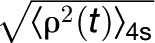 value (TPM signal) for each DNA length. The four values were then plotted as a function of DNA length, and these data were well in agreement with a published calibration curve obtained by Nelson et al. (2006) (see Supplemental Figure 1D online).
value (TPM signal) for each DNA length. The four values were then plotted as a function of DNA length, and these data were well in agreement with a published calibration curve obtained by Nelson et al. (2006) (see Supplemental Figure 1D online).
Subsequently, the effects of STK, SEP3, and STK-SEP3 heterodimers binding to the 697-bp VDD promoter tether were studied by TPM (Figure 1). Furthermore, we also tested STK and SEP3 binding to tethers in which one or all CArG boxes were deleted. When only STK was added to the tether (Figure 1B), no loop formation was observed. However, the unlooped tether was shorter than when no protein was added (compared with the position of the curve with the calibration curve C shown at the top of the panel). This suggests that the addition of STK resulted in a shortening of the tether. This seems to be unrelated to binding of the MADS domain proteins to the CArG boxes, since this shortening of the tether was also observed when we used a tether without CArG boxes (Figure 1D). When only SEP3 was added to the tether, we obtained a more complex pattern (Figure 1C). The experimental histogram displayed a main peak with a shoulder and a tail on the left side. This histogram could be decomposed into three Gaussians: magenta corresponding to the signal from the unlooped tethered; green, which may represent the contribution due to loop formation between CArG box 1 and 2; and blue, which was observed in all the measurements and we believe to be a background effect of SEP3 in these TPM experiments.
Clearer are the results when both STK and SEP3 were added to the tether. The blue curve caused by SEP3 is much less pronounced in these experiments. The control experiments using the tether without CArG boxes (Figure 1D) showed that no loop formation is possible without CArG boxes. Figure 1E shows that two different loops were obtained when STK and SEP3 were added to the wild-type tether. Our interpretation is that the left-most red curve is due to loop formation between CArG box 1 and 3, whereas the green curve might be due to loop formation between CArG box 1 and 2. The experiments with the tethers that contain a single CArG box deletion (Figures 1F to 1H) showed that when CArG box 1 or 3 was deleted, no loop formation is possible. Only when CArG box 2 was deleted the loop between CArG box 1 and 3 was obtained. Interestingly, the putative loop between CArG box 1 and 2 as observed using the wild-type tether (Figure 1E, green curve) was only established when CArG box 1 and 3 were both present, suggesting that in these in vitro TPM experiments the binding to CArG box 1 and 3 might somehow facilitate loop formation between CArG box 1 and 2. In conclusion, these experiments suggest that CArG box 1 and 3 are the boxes in the VDD promoter that are mainly involved in the formation of loops induced by STK and SEP3.
The Role of the CArG Boxes in the Regulation of VDD Expression
The TPM analysis showed that regulators of VDD induced loops into the putative promoter region by binding to two CArG boxes. To investigate the importance of the CArG boxes for the expression of VDD, we performed promoter analysis in which we mutated single or combinations of the three CArG boxes, changing the [A/T]6 into [G/C]6.
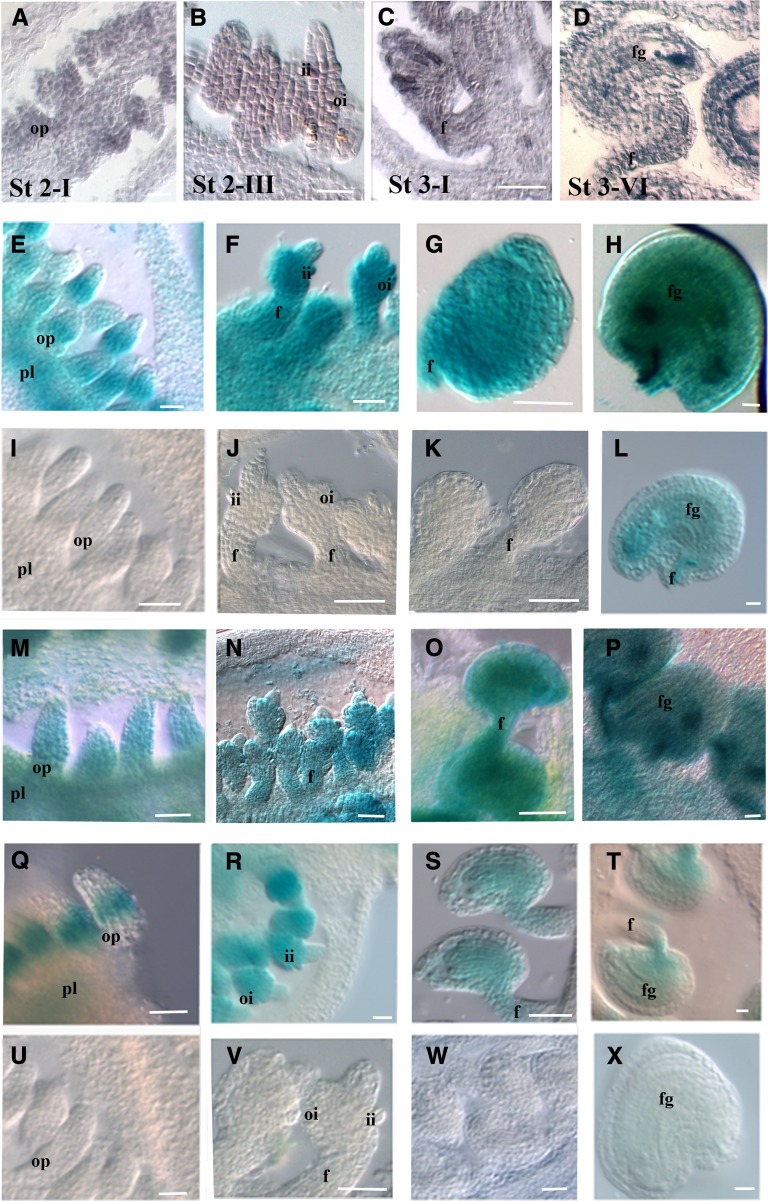
To validate the reporter gene expression profiles, we first performed VDD in situ hybridization expression analysis. This showed that VDD transcripts were first detected at stage 2-I of ovule development (Schneitz et al., 1995; Figure 2A). During subsequent stages of ovule development (until stage 3-VI), VDD expression was observed throughout all tissues of the ovules (Figures 2B to 2D). After fertilization, a strong VDD hybridization signal was observed in embryos at the globular stage, but at heart stage embryos, VDD expression almost disappeared (Figures 3A and 3B).
Figure 2.
VDD Expression and Promoter Analysis during Ovule Development.
(A) to (D) In situ hybridization analysis of VDD during ovule development. Ovule development stage 2-I (A), stage 2-III (B), 3-I (C), and stage 3-VI (D) (these stages should be used as reference for the next lines).
(E) to (H) proVDD:GUS transgenic plants showed a similar expression pattern as observed by the in situ hybridization experiment.
(I) to (L) GUS expression in ovules of proVDDm1:GUS lines.
(M) to (P) GUS expression in ovules of proVDDm2:GUS lines.
(Q) to (T) GUS expression in ovules of proVDDm2:GUS lines.
(U) to (X) Absence of GUS expression as observed in the proVDDm1-2:GUS. Absence of GUS expression was also observed in proVDDm1-3:GUS, proVDDm2-3:GUS, and proVDDm1-2-3:GUS lines.
pl, placenta; op, ovule primordium; f, funiculus; ii, inner integument; oi, outer integument; fg, female gametophyte. Bars = 20 µm.
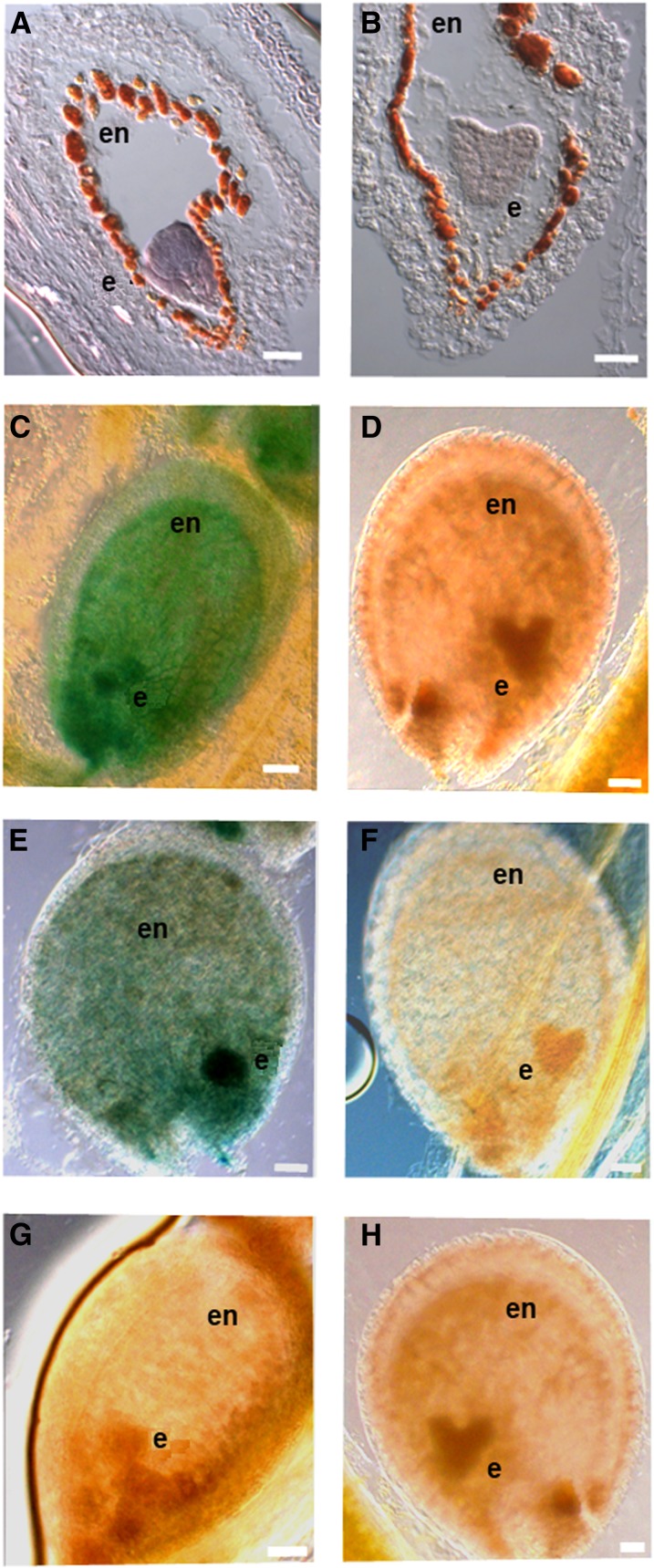
Figure 3.
VDD Expression and Promoter Analysis during Seed Development.
(A) and (B) VDD in situ hybridization analysis in developing seeds with globular (A) and heart stage (B) embryos.
(C) and (D) GUS expression in seeds of proVDD:GUS lines.
(E) and (F) GUS expression in seeds of proVDDm2:GUS lines.
(G) and (H) Absence of GUS expression in proVDDm1:GUS. Absence of GUS expression was also observed in plants containing the following constructs: proVDDm3:GUS, proVDDm1-2:GUS, proVDDm1-3:GUS, proVDDm2-3:GUS, and proVDDm1-2-3:GUS.
e, embryo; en, endothelium. Bars = 20 µm.
To evaluate the importance of the CArG boxes in regulating VDD expression, a putative promoter fragment of 1221 bp upstream of the VDD translation start site was cloned in frame with the uidA reporter gene that encodes β-glucuronidase (GUS). This proVDD:GUS construct was used for A. thaliana transformation. We generated more than 80 transgenic lines containing this construct and 92% of these plants showed similar expression profiles, whereas 8% did not show GUS activity. The GUS expression profile during ovule development perfectly matched the VDD expression that was observed by in situ hybridization experiments (Figures 2E to 2H). In globular stage embryos, GUS expression was observed, whereas at the heart stage, no GUS activity could be detected (Figures 3C and 3D). The in situ profiles confirm the expression in globular stage embryos; however, at the heart stage, they showed some residual VDD expression (Figures 3A and 3B).
Since the proVDD:GUS reporter construct drives GUS expression similar to the endogenous VDD gene, we used this VDD promoter fragment to generate new uidA reporter gene constructs in which single or combinations of the three CArG boxes were mutated. These constructs were all used to transform A. thaliana plants, and at least 80 independent transgenic plants were obtained for each construct. In plants that contained the reporter construct with a single mutated CArG box, expression profiles changed depending on which CArG box was mutated. Mutations in the second CArG (proVDDm2:GUS) box did not affect the expression profile of the reporter gene (Figures 2M to 2P). However, when the first CArG was mutated, the expression of the reporter gene (proVDDm1:GUS) was only detected in developing stage 3-VI ovules (Figures 2I to 2L). When CArG box 3 was mutated (proVDDm3:GUS), GUS expression was visible at stage 2-I, restricted to the chalaza zone (Figure 2Q) of the ovule, and expression levels at later stages were lower than in the wild type (Figures 2R to 2T).
We also analyzed reporter constructs in which two or all three CArG boxes were mutated (proVDDm1-2:GUS, proVDDm1-3:GUS, proVDDm2-3:GUS, proVDDm1-2-3:GUS). In all these transgenic plants, no GUS expression was observed, showing that the presence of two CArG boxes is essential for VDD promoter activity. An example of the obtained results is shown for proVDDm1-2:GUS in Figures 2U to 2X.
These experiments were all done with mutated CArG boxes. However, in the TPM analyses described above, we used promoter fragments in which CArG boxes were deleted. To verify the consistency of these data, we also prepared reporter gene constructs in which CArG boxes were completely deleted as described in the TPM experiments. This showed that exactly the same results were obtained as when mutated CArG boxes were used (see Supplemental Figure 2 online).
Interestingly, whereas during ovule development there seems to be flexibility in the use of the CArG boxes, in seeds this was different. Only inactivation of CArG box 2 did not result in a complete loss of VDD promoter activity during seed development (Figures 3E and 3F), whereas inactivation of CArG box 1, CArG box 3, and all other combinations did eliminate GUS expression (Figures 3G and 3H), showing that the presence of both CArG box 1 and 3 are critical for correct VDD expression in developing seeds.
In Vivo Binding of STK and SEP3 to the Three CArG Boxes in the VDD Promoter
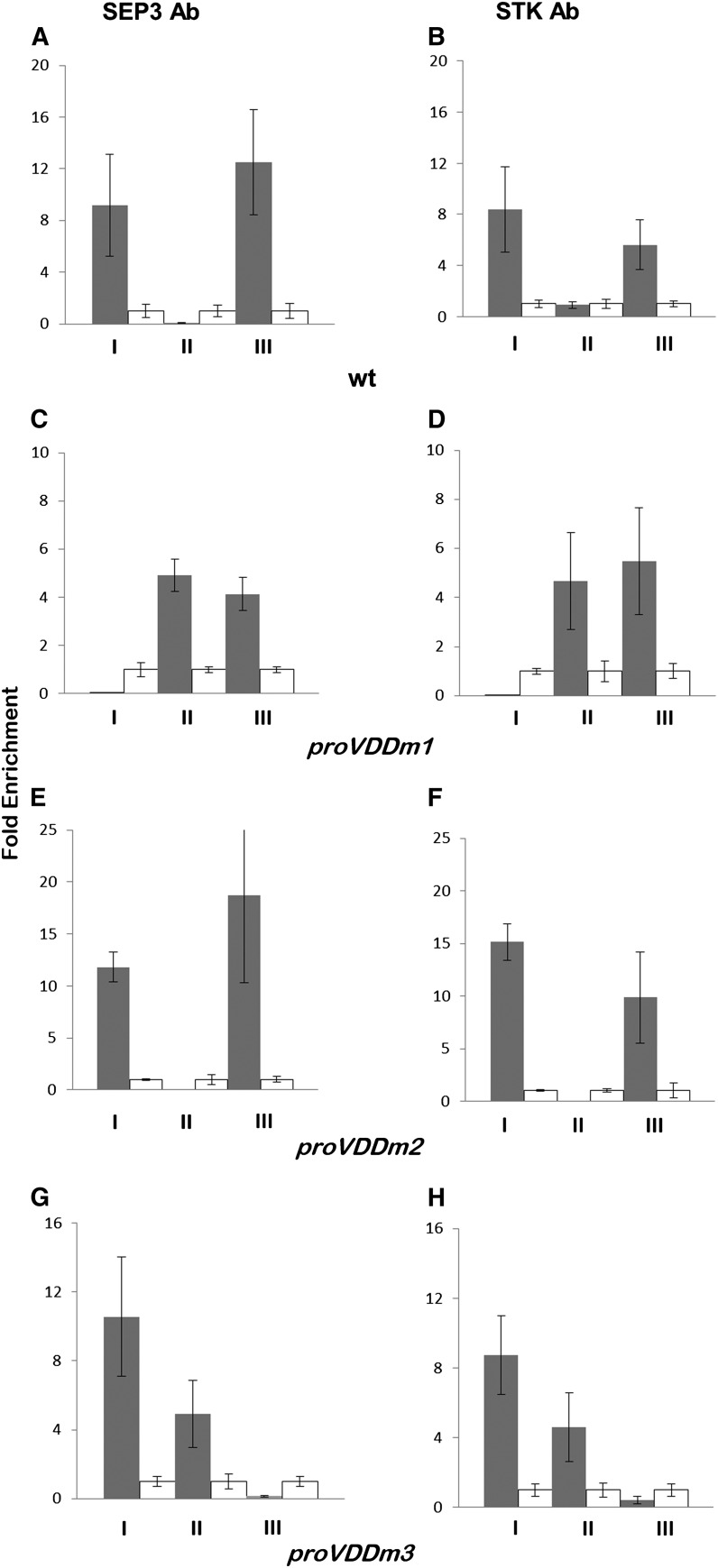
Previously published ChIP data showed that CArG box 1 and CArG box 3 in the VDD promoter region are directly bound by SEP3 and STK, whereas no binding to CArG box 2 was observed (Matias-Hernandez et al., 2010). Repeating this experiment resulted in exactly the same observation (Figures 4A and 4B). Subsequently, we performed ChIP assays combined with real-time PCR analysis using chromatin extracted from unfertilized flowers from reporter lines that contain VDD promoter constructs with one of the three CArG boxes mutated. Specific primers for the mutated CArG boxes were used to discriminate binding to the endogenous promoter from binding to the exogenous DNA constructs. These experiments showed that when CArG box 1 was mutated (proVDDm1:GUS), CArG box 2 was used by STK and SEP3 (Figures 4C and 4D). A similar result was obtained when we performed ChIP analysis using chromatin extracted from inflorescences of the proVDDm3:GUS reporter line; in this case, CArG box 1 and 2 were bound by STK and SEP3 (Figures 4G and 4H). As expected, when CArG box 2 was mutated, the MADS domain factors bound normally to CArG box 1 and 3 (Figures 4E and 4F).
Figure 4.
In Vivo Binding of SEP3 and STK to CArG Boxes in the VDD Promoter Region.
ChIP experiments using SEP3 (A, C, E, and G) and STK (B, D, F, and H) antibodies to investigate binding to the CArG boxes (I, CArG box 1; II, CArG box 2; III, CArG box 3) in the VDD promoter. Negative controls (white bars) for SEP3 ChIP assays were done using wild-type leaf tissues, and for STK ChIP negative controls, flowers of the stk mutant were used.
(A) and (B) SEP3 and STK binding to the endogenous VDD promoter.
(C) to (H) For wild-type CArG boxes, we cannot distinguish between the endogenous and heterologous VDD promoter regions. ChIP assays to test SEP3 and STK binding to the wild-type and mutated CArG boxes using proVDDm1:GUS-containing lines ([C] and [D]), proVDDm2:GUS lines ([E] and [F]), and proVDDm:GUS lines ([G] and [H]). Fold enrichments were calculated over the negative controls. Error bars represent the propagated error value using three replicates.
Interestingly, in plants containing the reporter line in which CArG box 1 and 3 were mutated (proVDDm1-3:GUS), no enrichment was observed on any of the CArG boxes, suggesting that binding to CArG box 1 or 3 facilitates binding of the SEP3-STK dimer to CArG box 2. We also performed ChIP experiments using plants containing the reporter lines proVDDm1-2:GUS and proVDDm2-3:GUS. These experiments showed that the mutated CArG boxes are never bound by STK and SEP3 (see Supplemental Figures 3A to 3F online).
Conservation of the VDD CArG Boxes in Species Related to A. thaliana
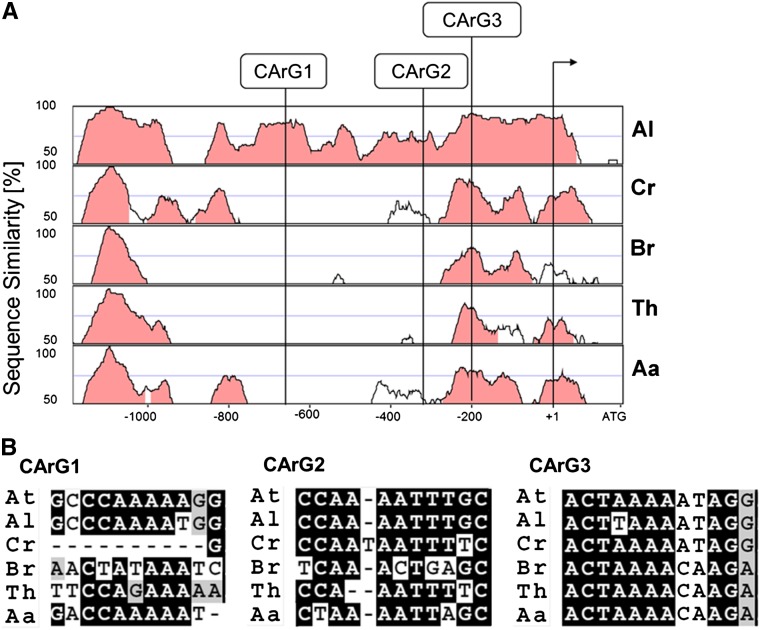
There are three CArG boxes in the A. thaliana VDD promoter region, but only CArG box 1 and 3 seem to be important for proper VDD expression in ovules and seeds. The question therefore arose if there might be conservation of all three boxes or just two of them. We investigated by phylogenetic shadowing, using orthologous promoters of Arabidopsis lyrata, Arabis alpina, Brassica rapa, Capsella rubella, and Thellungiella halophila, if there is conservation of the position of all three CArG boxes in these species (Figure 5). This analysis showed that in A. lyrata, all three CArG boxes are located in more or less the same position, suggesting that in the genus Arabidopsis the regulatory mechanism to regulate VDD expression is probably conserved.
Figure 5.
Phylogenetically Conserved Regions in the VDD Promoter and Multiple Sequence Alignment Analysis.
(A) Pairwise alignments of the A. thaliana VDD promoter to putatively orthologous sequences of A. lyrata, C. rubella, B. rapa, T. halophila, and A. alpina, respectively, shown as VISTA plots. Light red indicates regions where a sliding window of at least 75 bp has >70% identity. Vertical lines indicate the position of the three CArG boxes within the A. thaliana VDD promoter relative to the transcriptional start site (arrow).
(B) Multiple sequence alignment of the three putative CArG box or CArG box-like sequences found within the same position of the VDD promoter of A. thaliana and orthologous genes in the five Brassicaceae, not strictly considering the CC(A/T)6-8GG (allowing one mismatch) rule.
CArG box 3 is located in a highly conserved region in six species analyzed, whereas CArG box 1 and 2 are not (Figure 5A). The conservation in CArG box sequences also confirms this, since CArG box 3 is the only one that is most conserved in sequence between these species (Figure 5B). However, if we consider a consensus sequence of CC(A/T)6-8GG (Nurrish and Treisman,1995; Wang et al., 2004) with maximum one deviation from this consensus, then CArG box 3 is only conserved in A. lyrata and C. rubella. Searching the promoter sequences of the VDD orthologs of all these species showed that in A. alpina and T. halophila, CArG boxes that fulfill the consensus sequence could be identified in the same region where the three CArG boxes are in A. thaliana and A. lyrata. However, the position and spacing of these are different.
DISCUSSION
MADS domain proteins that regulate flower development have been shown to interact with each other, preferentially forming heterodimers (de Folter et al., 2005). This model, based on biochemical and genetic studies, predicts that these floral homeotic MADS domain protein dimers bind to two CArG boxes as a quartet and establish DNA loops in the promoters of target genes (Egea-Cortines et al., 1999; Melzer and Theissen, 2009; Smaczniak et al., 2012). The class E or SEP proteins are important in this model for the establishment of these higher order MADS domain protein complexes (Honma and Goto, 2001; Pelaz et al., 2001).
Our TPM in vitro experiments using a fragment of the VDD promoter region containing three adjacent CArG boxes strongly support the idea that loop formation can be established by a STK-SEP3 MADS domain complex. Especially CArG box 1 and 3 seem to be most important for the establishment of loop formation. This is in agreement with ChIP experiments that showed that in vivo STK and SEP3 only use CArG box 1 and 3 in the VDD promoter (Matias-Hernandez et al., 2010). Interestingly, when we tested by TPM assays the VDD promoter with a single CArG box deletion, loop formation was only observed when the CArG box 2 was deleted. Using tethers in which CArG box 1 or 3 were deleted, no loop formation was observed. This indicates that loop formation between CArG box 1 and 2 or between 2 and 3 is not possible in these in vitro assays. An exception to this rule might be when all three CArG boxes are available and the binding to CArG box 1 and 3 somehow also favors the establishment of a loop between CArG box 1 and 2. These TPM data are not completely in agreement with our in vivo data that suggest that the SEP3-STK dimer used CArG box 2 in the absence of CArG box 1 or 3.
Loop formation in the DNA facilitated by specific cis-elements has shown to be important for establishing the interaction of distantly related enhancers for correct regulation of transcription (Bulger and Groudine, 2011), as for instance described for the well studied Abdominal-B gene of Drosophila melanogaster (Cléard et al., 2006; Ho et al., 2011). However, most of the loop formations that have been intensively studied are related to long-range DNA looping. In the case of the VDD promoter region, CArG box 1 and 3 are only 444 bp apart. The function of these short-range loops has been poorly studied and understood. The general idea is that DNA looping is a conformational state in which cis-elements are brought in close vicinity to each other and create locally a high concentration of transcription factors close to the transcription start site of genes to initiate transcription (Dekker et al., 2002). This suggests that long- and short-range looping events might have the same function. A study of short-range loop formation in the murine iNOS promoter region also points in this direction (Guo et al., 2008). Our studies in flowers showed that when one of the three CArG boxes was mutated in the VDD promoter, transcriptional activation of VDD still occurred. However, the promoter was inactive when two of the three CArG boxes were eliminated. This suggests that loop formation is essential for the transcriptional activation of VDD. Furthermore, the loop size or its location also seemed to be critical. This became clear from the ChIP and in vivo expression studies using the reporter lines. The ChIP experiments showed that normally CArG box 1 and 3 are used. However, when one of these two CArG boxes was mutated, CArG box 2 was occupied by SEP3 and STK. This shows that all three boxes have affinity for the SEP3-STK dimer, but there seems to be an affinity difference between them, with CArG box 2 having the lowest affinity. When CArG box 2 was used in combination with CArG box 1 or 3, a change in the DNA loop position and/or size is expected to occur (Figure 6). This change in the predicted loop structure was shown to have an effect on the expression of VDD, as evidenced by the reporter gene studies. Loop formation between CArG box 1 and 2 or between CArG box 2 and 3 did activate expression of the reporter gene, but the timing and domain of its expression were altered. These results suggest that there might be a mechanistic difference between long-range and short-range loop formation and that the size and position of the loop are important. For instance, a loop between CArG box 2 and 3 has the same position relative to the transcription start site, but the loop is smaller (Figure 6). It is difficult to imagine that loop size is critical for long-range loop formation, where loops can be thousands of base pairs. Therefore, short-range loops might be important to give locally structure to the chromatin to recruit or stabilize specific transcriptional complexes. However, we cannot completely exclude another scenario in which the stability of the MADS domain protein complex on CArG box 2 is less stable due to weak binding to this site and that, therefore, transcription of VDD is deregulated. The idea that loop formation in the VDD promoter is important for its activity was further strengthened by studying reporter gene constructs in which two out of three CArG boxes were mutated. In these transgenic GUS reporter lines, all these promoter fragments were inactive. ChIP experiments showed that when CArG box 1 and 3 were both mutated, CArG box 2 did not bind STK or SEP3, suggesting that binding of SEP3 and STK to CArG box 2 was only possible when CArG box 1 or 3 was available, supporting the cooperative assembly of the MADS quartet on the VDD promoter.
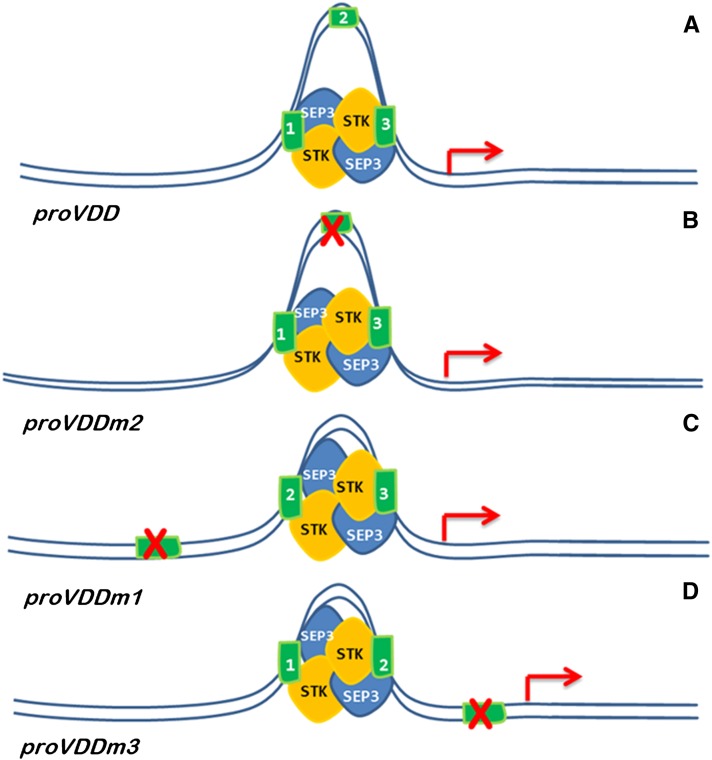
Figure 6.
Schematic Representation of the Predicted Mode of Action of the STK-SEP3 MADS Domain Complex on the VDD Promoter.
Illustration of loop formation in the VDD promoter (A), VDD promoter with the second CArG mutated (B), first CArG mutated (C), and third CArG mutated (D). Cartoon clarifies the putative changes in loop size and position with respect to the transcription start site (red arrow).
Our data strongly suggest that loop formation between two CArG boxes is important for VDD promoter activity. Nevertheless, based on our in vivo results, an alternative hypothesis to explain our observations might also be considered. It could be that for VDD promoter activity SEP-STK dimers have to bind to at least two of the three CArG boxes without the necessity that they also loop the DNA. However, our TPM studies and evidence coming from other studies strongly support the looping hypothesis (Egea-Cortines et al., 1999; Melzer and Theissen, 2009; Melzer et al., 2009; Smaczniak et al., 2012).
Another important consideration is that STK and SEP3 might bind as homodimers to the CArG boxes. However, STK homodimers were not observed in yeast two-hybrid assays (Favaro et al., 2003; de Folter et al., 2005), although this technique cannot completely exclude such a possibility. Our ChIP analyses showed that both STK and SEP3 bound to both CArG box 1 and 3, and the TPM data showed that SEP3 or STK by themselves were unable to induce loop formation between CArG box 1 and 3.
These data suggest that VDD promoters, in which STK (or SEP3) homodimers are bound to two CArG boxes, will probably not be active. However, a loop induced by a homodimer of STK and a homodimer of SEP3 binding to two CArG boxes cannot be completely excluded. Nevertheless, we favor a model in which STK-SEP3 heterodimers regulate VDD expression because SEP3 and STK have high affinity for each other in the yeast assay and the formation of this heterodimer seems to be highly conserved in plants (Favaro et al., 2002).
The phylogenetic shadowing experiments showed that in A. lyrata all three CArG boxes were in almost the same position as in A. thaliana, suggesting that the regulatory mechanism that we describe here is at least conserved in the genus Arabidopsis. It remains to be determined why three CArG boxes are conserved when only two of them seem to be used. A possible explanation might be that other MADS domain proteins bind to box 2; however, under the controlled greenhouse conditions that we used, these interactions seem not to be important for correct VDD expression, since mutations in CArG box 2 did not alter the expression profile of VDD. This might be different when plants are grown under more unfavorable climatic conditions. In the more distantly related species, it is difficult to conclude whether VDD orthologs are regulated in a similar way. When we only strictly consider the CArG consensus, then we can find in some promoters alternative binding sites, although they are not exactly in the same positions as observed in the Arabidopsis VDD promoter. When more deviated CArG box-like sequences would also bind SEP3 and STK, then this regulatory mechanism might be conserved in more distantly related species. ChIP and reporter gene studies will be necessary to test this interesting hypothesis in the future.
During seed development, both CArG box 1 and 3 seemed to be essential for VDD expression, since CArG box 2 was not able to compensate for the loss of one of these two CArG boxes. This suggests that the composition of the MADS domain complexes that bind to these CArG boxes during seed development are different and that these do not have enough affinity for CArG box 2. This is supported by the observation that VDD is highly expressed in the embryo, a tissue in which STK mRNAs were never detected by in situ hybridization.
In conclusion, a combination of in vitro and in vivo data strongly support the hypothesis that MADS domain protein dimers composed of SEP3 and STK (or SHP1/SHP2, which are considered to be redundant with STK in the regulation of VDD) can bind the DNA at nearby CArG boxes and that by forming higher order (quartet) complexes, they loop the DNA. This loop formation is important for target gene expression and both the size and position of these small loops influence gene expression. This is the first in vivo example that shows the importance of MADS domain quartets for target gene regulation and the importance of loop formation for gene expression in plants.
METHODS
Plant Material and Growth Conditions
Arabidopsis thaliana wild-type (ecotype Columbia) and stk-2 mutant plants (Pinyopich et al., 2003) were grown at 22°C under short-day (8 h light/16 h dark) or long-day (16 h light/8 h dark) conditions.
Plasmid Construction and A. thaliana Transformation
For VDD promoter analysis, the region 1221 bp upstream of the VDD translation start site was amplified by PCR and fused to the GUS reporter gene. Seven other constructs were cloned containing different combinations of site-specifically mutagenized CArG boxes (see Supplemental Table 1 online). All the constructs were made using Gateway technology (Invitrogen). In the first amplification step, we used the Gateway vector pDONOR207 and then recombined the constructs into pBGWFS7 (Karimi et al., 2002). Wild-type plants were transformed with all constructs using the floral dip method (Clough and Bent, 1998). Seeds of the transformed plants were harvested upon maturation, and the seeds germinated on soil and the transgenic plants were selected by spraying with 0.1% BASTA herbicide.
Cytological Assays
For in situ hybridization analysis, A. thaliana flowers were fixed and embedded in paraffin as described previously (Huijser et al., 1992). Sections of plant tissue were probed with digoxigenin-labeled VDD antisense RNA corresponding to nucleotides 240 to 557 (Matias-Hernandez et al., 2010). Hybridization and immunological detection were performed as described previously (Coen et al., 1990).
All GUS assays were performed overnight as described previously (Liljegren et al., 2000). Samples were incubated in clearing solution, dissected, and observed using a Zeiss Axiophot D1 microscope equipped with differential interference contrast optics. Images were captured on an AxiocamMRc5 camera (Zeiss) using the Axiovision program (version 4.1). For each construct, we analyzed more than 80 independent transformants.
ChIP Assays
For ChIP experiments, chromatin was extracted from wild-type, stk mutant, and transgenic (proVDDm1:GUS, proVDDm2:GUS, and proVDDm3:GUS) flowers (maximum flower developmental stage 12 and before fertilization occurs; Smyth et al., 1990). Wild-type plants were grown under short-day conditions for 2 weeks, and chromatin was extracted. The stk single mutant and wild-type leaves were used as negative controls for STK and SEP3 ChIPs, respectively. STK and SEP3 binding to the DNA was only considered when the analyzed fragments were significantly enriched compared with the controls, in at least three independent experiments. For further details, see Supplemental Methods 1 online.
Quantitative Real-Time RT-PCR
Enrichment folds were detected using a SYBR Green assay (Bio-Rad). The real-time PCR assay was performed in triplicate using a Bio-Rad C1000 thermal cycler optical system. For ChIP experiments, relative enrichment was calculated as described in Supplemental Methods online. Primers used for ChIP experiments are listed in Supplemental Table 2 online.
SEP3 and STK Purification
The SEP3 coding sequence was amplified using the primers 5′-CCATATGGGAAGAGGGAGAGTAGAATTG-3′ and 5′-CGCTCGAGAATAGAGTTGGTGTCATAAGG-3′. The STK coding sequence was amplified using the primers 5′-CCCATATGGGAAGAGGAAAGATAGAAATAAAG-3′ and 5′-CCCTCGAGTCCGAGATGAAGAATTTTCTTG-3′.
The two fragments were digested with NdeI and XhoI and cloned into pET-23(+) (Novagen). The recombinant proteins were produced in Escherichia coli BL21(DE3) (Novagen). The cultures were grown at 37°C at A600nm 0.6 for STK and A600nm 0.8 for SEP3. For protein induction, isopropyl β-d-thio-galactopyranoside (supplied by Roche) was added to a final concentration of 0.1 mM, and after induction, the cultures were incubated at 18°C for 15 h. The His-tagged recombinant proteins were purified using affinity nickel-nitrilotriacetic acid agarose columns (Qiagen), and for cell lysis, the pellet of 1 liter culture was sonicated.
SEP3 was soluble and purified in native condition following Bellorini et al. (1997). E. coli BL21(DE3) cells expressing the recombinant STK protein were resuspended in 20 mM Tris, 500 mM NaCl, 2 M urea, 50 μg/mL lysozyme, and 0.5% Tween 20, pH 8.0, and one protease inhibitor cocktail tablet completely EDTA-free (Roche) was added. Five milliliters of lysis buffer per gram of wet cells was used. Samples were disrupted by sonication (eight 15-s pulses, with a 15-s pause between pulses) and centrifuged in a Sorvall superspeed RC2-B at 13,500 rpm for 30 min at room temperature. After centrifugation, the supernatant was removed. The pellet containing the inclusion bodies was solubilized in 4 mL of 20 mM Tris, 500 mM NaCl, 10 mM imidazole, and 6 M urea, pH 8, per gram of wet cells. The sample was subsequently loaded onto a nickel-nitrilotriacetic acid column, and after washing and elution, the bound recombinant STK protein was subsequently solubilized using a linear 6 to 0 M urea gradient. SEP3 purification was evaluated by SDS page and STK purification was evaluated by an immune blot using anti\x{2013}STK antibodies (see Supplemental Figure 4).
DNA Constructs for TPM Analysis
The VDD promoter (proVDD) fragment, containing the three CArG boxes, was obtained by PCR using a 5′ digoxigenin- and a 3′ biotin-labeled primer (Oligos Etc.). The proVDDdel1-2-3 fragment, in which the three CArG boxes were deleted, was produced by PCR with specific primers carrying CArG box deletions. The final fragment was recombined into the pGEM-T-Easy plasmid (Promega) and amplified with labeled primers as described above. Fragments used to obtain a calibration curve, corresponding to 243, 355, and 575 bp, were amplified from proVDD (primers are listed in Supplemental Table 3 online).
Tethered Particle Motion Assay
TPM analysis was performed as described previously (Finzi and Dunlap, 2003). About 50 DNA-tethered beads were tracked for each of the following experimental conditions: proVDD incubated with SEP3, STK, or both. We also tested the mutated promoter without any of the three CArG boxes, proVDDdel1-2-3-, in the presence of both proteins (concentration of 700 nM each).
Phylogenetic Shadowing
Sequences from Arabidopsis lyrata, Brassica rapa, and Thellungiella halophila were obtained from Phytozome. The Capsella rubella sequence was assembled from raw sequence reads (http://trace.ncbi.nlm.nih.gov/Traces/sra/sra.cgi). The Arabis alpina sequence was obtained from an internal genome-sequencing project at the MPIPZ Köln. Pairwise alignments and VISTA plots (Mayor et al., 2000) were made as described previously (Herrero et al., 2012), but with a calculation window of 75 bp and a consensus identity of 70%. Multiple sequence alignments were performed with ClustalW (Larkin et al., 2007). The CArG box consensus used was CC(A/T)6-8GG, allowing one mismatch. However, the base preceding the (A/T)s should be a C and the base after the (A/T)s should be a G (Nurrish and Treisman,1995; Wang et al., 2004).
Accession Number
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under accession number AT5G18000 (VDD).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Summary of TPM Analysis.
Supplemental Figure 2. proVDD Deletion Studies.
Supplemental Figure 3. In Vivo Binding of SEP3 and STK to CArG Boxes with Two CArG Boxes Mutated in the VDD Promoter Region.
Supplemental Figure 4. STK and SEP3 Protein Purification from E. coli.
Supplemental Table 1. Primers Used for Mutated Plasmid Construction.
Supplemental Table 2. Primers Used for ChIP Experiment.
Supplemental Table 3. Tethered Particle Motion Fragments.
Supplemental Methods 1. Chromatin Immunoprecipitation Assays.
Acknowledgments
We thank George Coupland for the scientific discussion. M.A.M. was funded by Marie Curie European Union Initial Training Network (ITN) SYSFLO project, and L.F. was supported by National Institutes of Health Grant RGM084070A.
AUTHOR CONTRIBUTIONS
M.M.K. and L.C. designed research. R.F.G. and M.A.M. performed research. S.M., L.F., and C.M. contributed new reagents/analytic tools. M.A.M., R.F.G., M.C.B., M.M.K., and L.C. analyzed data. M.A.M., M.M.K., and L.C. wrote the article.
Glossary
- ChIP
chromatin immunoprecipitation
- TPM
tethered particle motion
- GUS
β-glucuronidase
References
- Arora R., Agarwal P., Ray S., Singh A.K., Singh V.P., Tyagi A.K., Kapoor S. (2007). MADS-box gene family in rice: Genome-wide identification, organization and expression profiling during reproductive development and stress. BMC Genomics 8: 242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellorini M., Lee D.K., Dantonel J.C., Zemzoumi K., Roeder R.G., Tora L., Mantovani R. (1997). CCAAT binding NF-Y-TBP interactions: NF-YB and NF-YC require short domains adjacent to their histone fold motifs for association with TBP basic residues. Nucleic Acids Res. 25: 2174–2181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulger M., Groudine M. (2011). Functional and mechanistic diversity of distal transcription enhancers. Cell 144: 327–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cléard F., Moshkin Y., Karch F., Maeda R.K. (2006). Probing long-distance regulatory interactions in the Drosophila melanogaster bithorax complex using Dam identification. Nat. Genet. 38: 931–935 [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Coen E.S., Meyerowitz E.M. (1991). The war of the whorls: Genetic interactions controlling flower development. Nature 353: 31–37 [DOI] [PubMed] [Google Scholar]
- Coen E.S., Romero J.M., Doyle S., Elliott R., Murphy G., Carpenter R. (1990). floricaula: A homeotic gene required for flower development in Antirrhinum majus. Cell 63: 1311–1322 [DOI] [PubMed] [Google Scholar]
- de Folter S., Immink R.G.H., Kieffer M., Parenicová L., Henz S.R., Weigel D., Busscher M., Kooiker M., Colombo L., Kater M.M., Davies B., Angenent G.C. (2005). Comprehensive interaction map of the Arabidopsis MADS box transcription factors. Plant Cell 17: 1424–1433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker R.J., van Soest S., Fontijn R.D., Salamanca S., de Groot P.G., VanBavel E., Pannekoek H., Horrevoets A.J.G. (2002). Prolonged fluid shear stress induces a distinct set of endothelial cell genes, most specifically lung Krüppel-like factor (KLF2). Blood 100: 1689–1698 [DOI] [PubMed] [Google Scholar]
- Ditta G., Pinyopich A., Robles P., Pelaz S., Yanofsky M.F. (2004). The SEP4 gene of Arabidopsis thaliana functions in floral organ and meristem identity. Curr. Biol. 14: 1935–1940 [DOI] [PubMed] [Google Scholar]
- Dunlap, D., Zurla, C., Manzo, C., and Finzi, L. (2011). Probing DNA topology using tethered particle motion. In Single Molecule Analysis: Methods and Protocols, E.J.G. Peterman and G.J.L. Wuite, eds, J. Walker series ed. Vol. 783 (New York: Springer) pp. 295–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egea-Cortines M., Saedler H., Sommer H. (1999). Ternary complex formation between the MADS-box proteins SQUAMOSA, DEFICIENS and GLOBOSA is involved in the control of floral architecture in Antirrhinum majus. EMBO J. 18: 5370–5379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favaro R., Pinyopich A., Battaglia R., Kooiker M., Borghi L., Ditta G., Yanofsky M.F., Kater M.M., Colombo L. (2003). MADS-box protein complexes control carpel and ovule development in Arabidopsis. Plant Cell 15: 2603–2611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favaro R., Immink R.G., Ferioli V., Bernasconi B., Byzova M., Angenent G.C., Kater M., Colombo L. (2002). Ovule-specific MADS-box proteins have conserved protein-protein interactions in monocot and dicot plants. Molecular Genet Genomics 268(2): 152–159 [DOI] [PubMed] [Google Scholar]
- Finzi L., Dunlap D. (2003). Single-molecule studies of DNA architectural changes induced by regulatory proteins. Methods Enzymol. 370: 369–378 [DOI] [PubMed] [Google Scholar]
- Finzi L., Gelles J. (1995). Measurement of lactose repressor-mediated loop formation and breakdown in single DNA molecules. Science 267: 378–380 [DOI] [PubMed] [Google Scholar]
- Guerra R.F., Imperadori L., Mantovani R., Dunlap D.D., Finzi L. (2007). DNA compaction by the nuclear factor-Y. Biophys. J. 93: 176–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H., Mi Z., Kuo P.C. (2008). Characterization of short range DNA looping in endotoxin-mediated transcription of the murine inducible nitric-oxide synthase (iNOS) gene. J. Biol. Chem. 283: 25209–25217 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Herrero E., et al. (2012). EARLY FLOWERING4 recruitment of EARLY FLOWERING3 in the nucleus sustains the Arabidopsis circadian clock. Plant Cell 24: 428–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huijser P., Klein J., Lönnig W.E., Meijer H., Saedler H., Sommer H. (1992). Bracteomania, an inflorescence anomaly, is caused by the loss of function of the MADS-box gene squamosa in Antirrhinum majus. EMBO J. 11: 1239–1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho, M.C.W., Schiller, B.J., Akbari, O.S., Bae, E., and Drewell, R.A. (2011). Disruption of the abdominal-B promoter tethering element results in a loss of long-range enhancer-directed Hox gene expression in Drosophila. PLoS One 6: e16283. [DOI] [PMC free article] [PubMed]
- Honma T., Goto K. (2001). Complexes of MADS-box proteins are sufficient to convert leaves into floral organs. Nature 409: 525–529 [DOI] [PubMed] [Google Scholar]
- Karimi M., Inzé D., Depicker A. (2002). GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Larkin M.A., et al. (2007). Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947–2948 [DOI] [PubMed] [Google Scholar]
- Liljegren S.J., Ditta G.S., Eshed Y., Savidge B., Bowman J.L., Yanofsky M.F. (2000). SHATTERPROOF MADS-box genes control seed dispersal in Arabidopsis. Nature 404: 766–770 [DOI] [PubMed] [Google Scholar]
- Liu C., Chen H., Er H.L., Soo H.M., Kumar P.P., Han J.-H., Liou Y.C., Yu H. (2008). Direct interaction of AGL24 and SOC1 integrates flowering signals in Arabidopsis. Development 135: 1481–1491 [DOI] [PubMed] [Google Scholar]
- Matias-Hernandez L., Battaglia R., Galbiati F., Rubes M., Eichenberger C., Grossniklaus U., Kater M.M., Colombo L. (2010). VERDANDI is a direct target of the MADS domain ovule identity complex and affects embryo sac differentiation in Arabidopsis. Plant Cell 22: 1702–1715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayor C., Brudno M., Schwartz J.R., Poliakov A., Rubin E.M., Frazer K.A., Pachter L.S., Dubchak I. (2000). VISTA: Visualizing global DNA sequence alignments of arbitrary length. Bioinformatics 16: 1046–1047 [DOI] [PubMed] [Google Scholar]
- Melzer R., Theissen G. (2009). Reconstitution of ‘floral quartets’ in vitro involving class B and class E floral homeotic proteins. Nucleic Acids Res. 37: 2723–2736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melzer R., Verelst W., Theissen G. (2009). The class E floral homeotic protein SEPALLATA3 is sufficient to loop DNA in ‘floral quartet’-like complexes in vitro. Nucleic Acids Res. 37: 144–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melzer S., Lens F., Gennen J., Vanneste S., Rohde A., Beeckman T. (2008). Flowering-time genes modulate meristem determinacy and growth form in Arabidopsis thaliana. Nat. Genet. 40: 1489–1492 [DOI] [PubMed] [Google Scholar]
- Nelson P.C., Zurla C., Brogioli D., Beausang J.F., Finzi L., Dunlap D. (2006). Tethered particle motion as a diagnostic of DNA tether length. J. Phys. Chem. B 110: 17260–17267 [DOI] [PubMed] [Google Scholar]
- Nurrish S.J., Treisman R. (1995). DNA binding specificity determinants in MADS-box transcription factors. Mol. Cell. Biol. 15: 4076–4085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parenicová L., de Folter S., Kieffer M., Horner D.S., Favalli C., Busscher J., Cook H.E., Ingram R.M., Kater M.M., Davies B., Angenent G.C., Colombo L. (2003). Molecular and phylogenetic analyses of the complete MADS-box transcription factor family in Arabidopsis: New openings to the MADS world. Plant Cell 15: 1538–1551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelaz S., Ditta G.S., Baumann E., Wisman E., Yanofsky M.F. (2000). B and C floral organ identity functions require SEPALLATA MADS-box genes. Nature 405: 200–203 [DOI] [PubMed] [Google Scholar]
- Pelaz S., Gustafson-Brown C., Kohalmi S.E., Crosby W.L., Yanofsky M.F. (2001). APETALA1 and SEPALLATA3 interact to promote flower development. Plant J. 26: 385–394 [DOI] [PubMed] [Google Scholar]
- Pinyopich A., Ditta G.S., Savidge B., Liljegren S.J., Baumann E., Wisman E., Yanofsky M.F. (2003). Assessing the redundancy of MADS-box genes during carpel and ovule development. Nature 424: 85–88 [DOI] [PubMed] [Google Scholar]
- Pouget, N., Dennis, C., Turlan, C., Grigoriev, M., Chandler, M., and Salome, L. (2004). Single-particle tracking for DNA tether length monitoring. Nucleic Acids Res. 32: e73. [DOI] [PMC free article] [PubMed]
- Riechmann J.L., Wang M.Q., Meyerowitz E.M. (1996). DNA-binding properties of Arabidopsis MADS domain homeotic proteins APETALA1, APETALA3, PISTILLATA and AGAMOUS. Nucleic Acids Res. 24: 3134–3141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanel E.A.C., Schrago C.G., Couñago R.M., Russo C.A.M., Alves-Ferreira M. (2009). Evolution of the B3 DNA binding superfamily: New insights into REM family gene diversification. PLoS ONE 4: e5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneitz K., Hulskamp M., Pruitt R.E. (1995). Wild-type ovule development in Arabidopsis thailana - A light-microscope study of cleared whole-mount tissue. Plant J. 7: 731–749 [Google Scholar]
- Schwarz-Sommer Z., Hue I., Huijser P., Flor P.J., Hansen R., Tetens F., Lönnig W.E., Saedler H., Sommer H. (1992). Characterization of the Antirrhinum floral homeotic MADS-box gene deficiens: Evidence for DNA binding and autoregulation of its persistent expression throughout flower development. EMBO J. 11: 251–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shore P., Sharrocks A.D. (1995). The MADS-box family of transcription factors. Eur. J. Biochem. 229: 1–13 [DOI] [PubMed] [Google Scholar]
- Smaczniak C., Immink R.G.H., Angenent G.C., Kaufmann K. (2012). Developmental and evolutionary diversity of plant MADS-domain factors: Insights from recent studies. Development 139: 3081–3098 [DOI] [PubMed] [Google Scholar]
- Smyth D.R., Bowman J.L., Meyerowitz E.M. (1990). Early flower development in Arabidopsis. Plant Cell 2: 755–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theissen G., Saedler H. (2001). Plant biology. Floral quartets. Nature 409: 469–471 [DOI] [PubMed] [Google Scholar]
- Wang H., Caruso L.V., Downie A.B., Perry S.E. (2004). The embryo MADS domain protein AGAMOUS-Like 15 directly regulates expression of a gene encoding an enzyme involved in gibberellin metabolism. Plant Cell 16: 1206–1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaremba M., Owsicka A., Tamulaitis G., Sasnauskas G., Shlyakhtenko L.S., Lushnikov A.Y., Lyubchenko Y.L., Laurens N., van den Broek B., Wuite G.J.L., Siksnys V. (2010). DNA synapsis through transient tetramerization triggers cleavage by Ecl18kI restriction enzyme. Nucleic Acids Res. 38: 7142–7154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zurla C., Manzo C., Dunlap D., Lewis D.E.A., Adhya S., Finzi L. (2009). Direct demonstration and quantification of long-range DNA looping by the lambda bacteriophage repressor. Nucleic Acids Res. 37: 2789–2795 [DOI] [PMC free article] [PubMed] [Google Scholar]