Blue light–responsive casein kinase 1s (CK1s), CK1.3 and CK1.4, negatively regulate blue light–mediated photomorphogenesis and periodic flowering in Arabidopsis. CK1.3 and CK1.4 directly phosphorylate the blue light receptor Cryptochrome2 (CRY2), which is stimulated by blue light, to promote CRY2 degradation, revealing a blue light signaling regulatory mechanism.
Abstract
Casein kinase1 (CK1) plays crucial roles in regulating growth and development via phosphorylating various substrates throughout the eukaryote kingdom. Blue light is crucial for normal growth of both plants and animals, and blue light receptor cryptochrome2 (CRY2) undergoes blue light–dependent phosphorylation and degradation in planta. To study the function of plant CK1s, systematic genetic analysis showed that deficiency of two paralogous Arabidopsis thaliana CK1s, CK1.3 and CK1.4, caused shortened hypocotyls, especially under blue light, while overexpression of either CK1.3 or CK1.4 resulted in the insensitive response to blue light and delayed flowering under long-day conditions. CK1.3 or CK1.4 act dependently on CRY2, and overexpression of CK1.3 or CK1.4 significantly suppresses the hypersensitive response to blue light by CRY2 overexpression. Biochemical studies showed that CK1.3 and CK1.4 directly phosphorylate CRY2 at Ser-587 and Thr-603 in vitro and negatively regulate CRY2 stability in planta, which are stimulated by blue light, further confirming the crucial roles of CK1.3 and CK1.4 in blue light responses through phosphorylating CRY2. Interestingly, expression of CK1.3 and CK1.4 is stimulated by blue light and feedback regulated by CRY2-mediated signaling. These results provide direct evidence for CRY2 phosphorylation and informative clues on the mechanisms of CRY2-mediated light responses.
INTRODUCTION
Light is important for both animals and plants throughout the life cycle. The quality of light is perceived primarily by photoreceptors. Cryptochromes (CRYs) are conservative blue light sensors in all major evolutionary lineages and regulate multiple developmental processes and the circadian clock (Ahmad and Cashmore, 1993; Ahmad et al., 1998a; Lin et al., 1998; Lin, 2002). Arabidopsis thaliana CRY1 and CRY2 share sequence similarity to photolyases, a family of proteins that catalyze the repair of UV light–damaged DNA (Cashmore et al., 1999), and mediate various light-induced responses in plants, including hypocotyl shortening, cotyledon expansion, photoperiodic flowering, stomatal opening, and anthocyanin production (Liu H. et al., 2011). In addition, CRYs coordinate with other light sensors to modify plant shape under different light conditions, to control flowering time, and to promote stress resistance (Liu H. et al., 2011).
Recent genetic and biochemical studies revealed the molecular mechanism underlying how CRYs sense blue light. The N termini of CRYs can bind flavin adenine dinucleotide and are required for light activation of the photoreceptor activity via dimerization (Wang et al., 2001; Yang et al., 2000, 2001; Sang et al., 2005), and the C termini of CRYs confer the light response by regulating the downstream components (Yang et al., 2001). On the transcriptional level, CRY2 can directly interact with cryptochrome-interacting basic helix-loop-helix (a CRY2-interacting protein) in a blue light–dependent manner to regulate FT expression and photoperiodic flowering (Liu et al., 2008). On the posttranslational level, CRYs suppresses the E3 activity of CONSTITUTIVE PHOTOMORPHOGENIC1 to prevent the degradation of key downstream regulators such as LONG HYPOCOTYL5 (Lian et al., 2011; Liu B. et al., 2011) and CONSTANS (CO; Zuo et al., 2011), through a blue light–dependent interaction with SUPPRESSOR OF PHYTOCHROME A1 to control blue light–mediated photomorphogenesis and photoperiodic flowering. In addition, both CRY1 and CRY2 can be phosphorylated in planta (Ahmad et al., 1998b). CRY1 exhibits autophosphorylation activity in vitro, although there is no sequence similarity with any known kinase domain (Bouly et al., 2003; Shalitin et al., 2003), and CRY2 was hypothesized to be phosphorylated by some other kinases (Shalitin et al., 2002, 2003).
Arabidopsis CRY2 protein is primarily localized in the nucleus (Guo et al., 1998; Kleiner et al., 1999), where it is subjected to blue light–induced conformational changes and phosphorylation, nucleic body formation, and degradation by 26S proteasome (Shalitin et al., 2002; Yu et al., 2007a, 2009). However, the detailed mechanisms, especially which kinase is involved in CRY2 phosphorylation and the contrary roles in both activation and degradation of CRY2 by phosphorylation, remain unclear. In contrast with plant cryptochromes, multiple protein kinases, including a casein kinase 1 (CK1ε), an AMP-activated protein kinase, a glycogen synthase kinase (GSK-3β), and a mitogen-activated protein kinase, have been reported to phosphorylate mammalian cryptochromes (Liu H. et al., 2011). However, the detailed mechanism and effects of these phosphorylations remain unclear.
CK1, a Ser/Thr protein kinase, is multifunctional and has been identified in most eukaryotes (Gross and Anderson, 1998). In mammals, there are six isoforms (α, γ1, γ2, γ3, δ, and ε) of CK1 (Knippschild et al., 2005), which are involved in the regulation of various processes, including vesicle trafficking, growth and morphogenesis, circadian rhythms, DNA repair, and cell cycle progression and cytokinesis (Knippschild et al., 2005). CRY regulates the circadian system by forming a dimer of PERIOD (PER)/CRY to repress Clock/BMAL1 transactivation (Green et al., 2008), and both CK1δ and CK1ε are able to phosphorylate PER and CRY proteins to modulate circadian rhythms in part by a direct regulation on BMAL1-dependent transcription (Vielhaber et al., 2000; Akashi et al., 2002; Eide et al., 2002). In addition, CRY proteins form a complex with PER, and CK1δ/ε protect PER from degradation, leading to the nuclear accumulation of a CRY-PER-CK1δ/ε complex (Vielhaber et al., 2000; Akashi et al., 2002). However, how CRY and CK1 interact to function has not yet been elucidated.
Arabidopsis is a well-established model system for plant biology. There are multiple isoforms of CK1 in Arabidopsis. Previous studies have shown that CKL6 regulates cell-to-cell communication (Lee et al., 2005) and modifies cell shape by phosphorylating tubulin β (Ben-Nissan et al., 2008). In rice (Oryza sativa), CK1 regulates Brassinosteroid signaling (Liu et al., 2003) and Gibberellin signaling (through regulating the stability of SLENDER RICE1 by phosphorylation; Dai and Xue, 2010).
Here, we report that CK1s (CK1.3 and CK1.4) are involved in blue light signaling through phosphorylating Arabidopsis CRY2. Under blue light, CK1.3 and CK1.4 specifically phosphorylate the C terminus of CRY2 to stimulate CRY2 degradation and hence regulate blue light–mediated photomorphogenesis and photoperiodic flowering. These results provide direct evidence for blue light–stimulated CRY2 phosphorylation, which is crucial for the stability of CRY2, and help illustrate the mechanisms of CRY2-mediated blue light responses.
RESULTS
Arabidopsis CK1.3 and CK1.4 Encode Functional CK1s
Analysis of the Arabidopsis genome identified 17 candidate genes encoding CK1s (there are 15 in the rice genome). The much expanded CK1 family in plants may be due to the sessile life among the plant kingdom and suggests the important roles of CK1s during plant growth and development. Biochemical characterization showed that recombinant Arabidopsis CKL3 (At4g28880) and CKL4 (At4g28860) (casein kinase-like proteins; Lee et al., 2005) from Escherichia coli are able to phosphorylate casein (a substrate for CK1), but not BSA, in vitro (Figure 1A), indicating they are active CK1s. Based on the kinase activity against casein and to follow the nomenclature in mammals and rice in the Rice Genome Annotation Project database, we designated them as CK1.3 and CK1.4 to avoid confusion. Phylogenetic analysis indicated that CK1.3 and CK1.4 are the two closest isoforms (see Supplemental Figure 1 online) and share 91% identity in amino acid sequence (see Supplemental Figure 2 online).
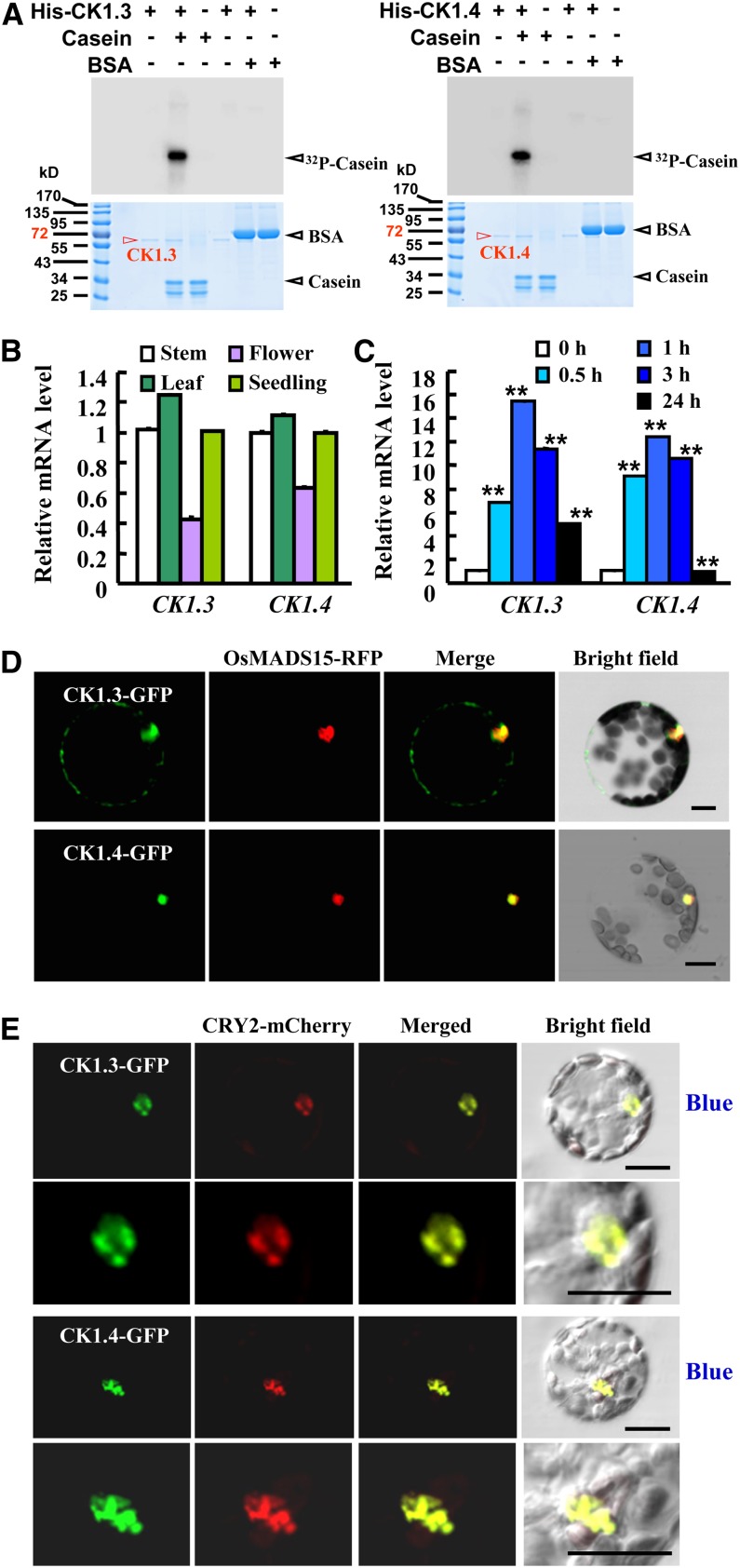
Figure 1.
CK1.3 and CK1.4 Are Expressed in Various Tissues and Encode Active Casein Kinases.
(A) In vitro kinase assays by [γ-32P]ATP autoradiography indicated that recombinant CK1.3 and CK1.4 display CK1 activity and specifically phosphorylate casein in vitro (top panel; BSA was used as control). Coomassie blue (CBB) staining showed that equal quantities of protein (10 μg; bottom panel) were loaded.
(B) qRT-PCR analysis revealed the expression of CK1.3 and CK1.4 in stems, leaves, flowers, and seedlings. Expression level of CK1.3 or CK1.4 in stems was set as 1.0. Error bars represent se. The experiments were repeated three times.
(C) qRT-PCR analysis revealed the induced expression of CK1.3 and CK1.4 under blue light. Seedlings were grown under a 3 µmol/m2/s fluence rate of blue light for 0.5, 1, 3, or 24 h. Expression level of CK1.3 or CK1.4 in the dark was set as 1.0. The experiments were repeated three times (**P < 0.01, n = 3).
(D) Transient expression analysis using Arabidopsis protoplasts revealed that both CK1.3-GFP and CK1.4-GFP fusion proteins are mainly localized in the nucleus. A nucleus-localized OsMADS15-RFP fusion protein (Zhang and Xue, 2013) was used as a positive control. Protoplasts were observed after incubation for 9 h. Bars = 20 μm.
(E) CK1.3-GFP and CK1.4-GFP fusion proteins are accumulated in nuclear bodies under blue light treatment. Colocalization analysis by transiently expressing CK1.3-GFP and CK1.4-GFP in Arabidopsis protoplasts expressing CRY2-mCherry indicated the colocalization of CK1.3-GFP and CK1.4-GFP with CRY2-mCherry (enlarged at bottom panels). Protoplasts were exposed to blue light (10 µmol/m2/s) for 5 min before observation. Bars = 20 μm.
Quantitative real-time RT-PCR (qRT-PCR) analysis revealed the expression of CK1.3 and CK1.4 in seedlings, leaves, stems, and flowers (Figure 1B). Further studies by observing β-glucuronidase (GUS) staining in independent transgenic lines harboring the CK1.3 and CK1.4 promoter-GUS gene cassette revealed that CK1.3 and CK1.4 are expressed mainly in the seedling cotyledons and rosette leaves at different developmental stages, as well as flowers and siliques (see Supplemental Figure 3 online). Interestingly, transcription of both CK1.3 and CK1.4 is regulated under blue light treatment (i.e., these genes are rapidly induced by short blue light treatment and suppressed under continuous blue light) (Figure 1C), which implies the special role of CK1.3 and CK1.4 in blue light signaling.
Transient expression studies revealed the mainly nuclear localization of CK1.3-GFP (for green fluorescent protein) or CK1.4-GFP fusion proteins in Arabidopsis protoplasts (Figure 1D; see Supplemental Figure 4A online), which is consistent with a previous report (Akashi et al., 2002). Interestingly, blue light treatment stimulates the nuclear localization of CK1.3 and CK1.4, especially in nuclear bodies (Figure 1E), suggesting that CK1.3 and CK1.4 may modify proteins with similar subcellular localization. This was further confirmed by transient expression analysis through transforming CK1.3-mCherry or CK1.4-mCherry into tobacco (Nicotiana benthamiana) leaves (see Supplemental Figure 4B online).
Knockout Mutants ck1.3-1, ck1.3-2, and ck1.4-1 Are Hypersensitive to Blue Light Irradiation, and Overexpression of CK1.3 or CK1.4 Results in Delayed Flowering under Long Days
To dissect the physiological roles of CK1.3 and CK1.4, three putative mutants, including ck1.3-1 (SALK_016571), ck1.3-2 (SALK_110494), and ck1.4-1 (SALK_043954) (Figure 2A, top panel), were identified from the T-DNA insertion database of the Salk Institute (Alonso et al., 2003). The T-DNA insertions were confirmed by PCR amplification using primers located in the T-DNA and flanked genomic DNA (see Supplemental Figure 5 online), and qRT-PCR analysis of the homozygous lines revealed the deficient transcription of CK1.3 and CK1.4 (Figure 2A, bottom panel), confirming the ck1.3-1, ck1.3-2, and ck1.4-1 knockout mutants.
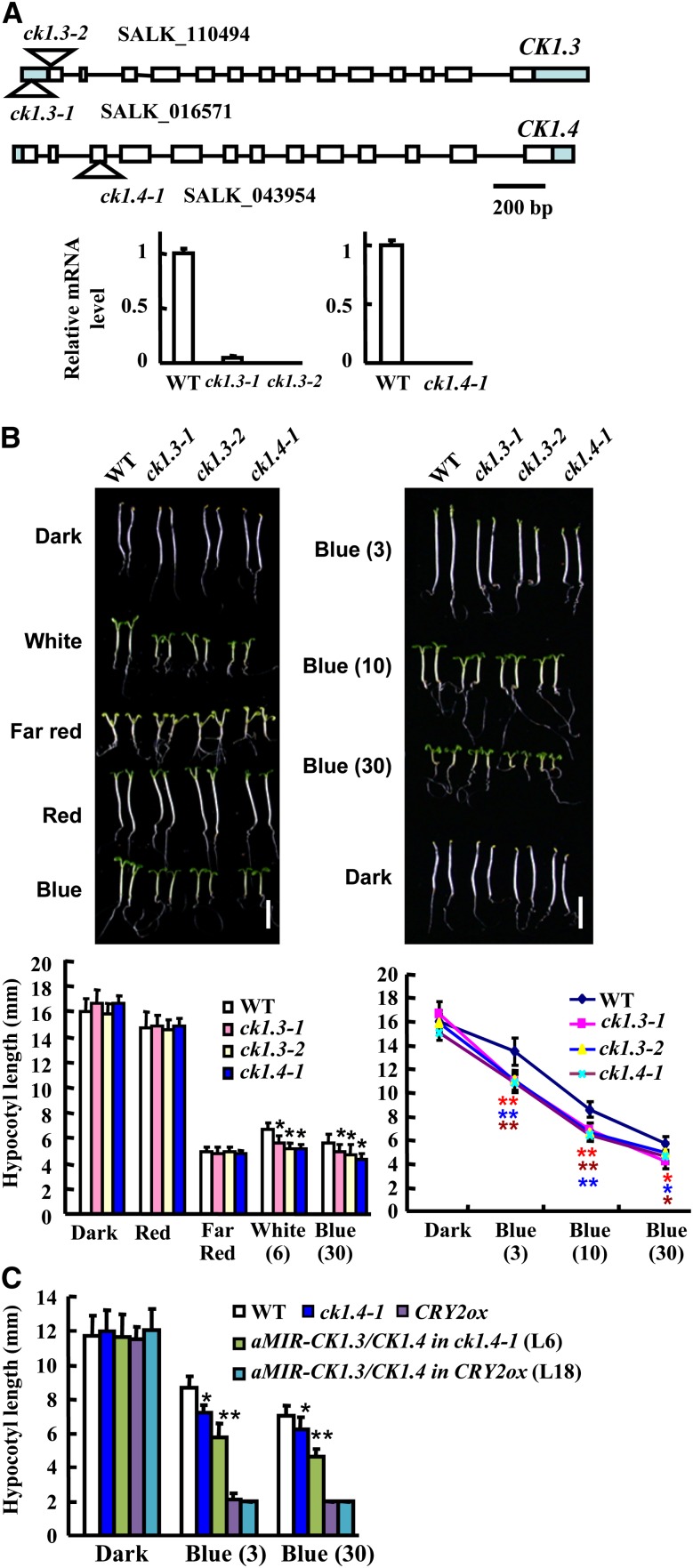
Figure 2.
Identification and Phenotypic Analysis of ck1.3 and ck1.4 Mutants.
(A) A schematic of CK1.3 and CK1.4 genes. Exons (white boxes), untranslated regions (green box), and introns (lines) are shown. T-DNA insertions were located in the 5′-untranslated region, the first exon of CK1.3, and the third exon of CK1.4 (top panel). qRT-PCR analysis showed a deficient expression of CK1.3 or CK1.4 in the corresponding mutants (bottom panel; the expression of the wild type [WT] was set as 1.0). Error bars represent se. The experiments were repeated three times.
(B) ck1.3-1, ck1.3-2, and ck1.4-1 seedlings presented shortened hypocotyls under white light and blue light with various fluence rates (3, 10, and 30 µmol/m2/s; indicated in parentheses, bars = 5 mm). The hypocotyl lengths of 7-d-old seedlings were measured and statistically analyzed (*P < 0.05; **P < 0.01). Error bars represent sd (n > 30).
(C) Transgenic plants harboring the aMIR-CK1.3/CK1.4 construct in the ck1.4-1 background showed significantly shortened hypocotyls under blue light conditions in comparison to the wild type or ck1.4-1 mutant; however, transformation with aMIR-CK1.3/CK1.4 in CRY2ox did not affect the shortened hypocotyls. Blue light intensity (3 or 30 µmol/m2/s) is indicated in parentheses. Hypocotyl lengths of 7-d-old seedlings were measured and statistically analyzed (*P < 0.05; **P < 0.01). Error bars represent sd (n > 30).
As expressions of CK1.3 and CK1.4 are induced by blue light, seedling growth under various light conditions were first observed. All three mutants exhibited shortened hypocotyls under both white and blue light in comparison to those of wild-type seedlings (Figure 2B, top panel). Further analysis showed that ck1.3-1, ck1.3-2, and ck1.4-1 exhibited hypersensitive responses to blue light irradiation and presented significantly shortened hypocotyls under blue light at various fluence rates (3, 10, and 30 µmol/m2/s; Figure 2B, bottom panel). In addition, transgenic plants with suppressed expression of CK1.3 (the expression of CK1.4 is not altered) showed shortened hypocotyls (see Supplemental Figures 6A and 6B online), as did the transgenic plants transformed with aMIR-CK1.3/CK1.4 (an artificial microRNA targeting both CK1.3 and CK1.4) in the ck1.4-1 background (because CK1.3 and CK1.4 are located close to each other on the same chromosome, it is hard to obtain a double mutant; thus, an artificial microRNA strategy was employed). qRT-PCR analysis confirmed the suppressed expression of CK1.3 in transgenic lines (aMIR-CK1.3/CK1.4 in ck1.4-1; see Supplemental Figure 6C online), and suppressed expression of both CK1.3 and CK1.4 resulted in more significantly suppressed hypocotyl elongation under blue light than either the wild type or ck1.4-1 (Figure 2C), indicating that CK1.3 and CK1.4 have redundant functions and exhibit a dose effect in response to blue light. However, when aMIR-CK1.3/CK1.4 was introduced into the CRY2ox background (see Supplemental Figure 6D online), no significant difference was observed, suggesting that CK1.3 and CK1.4 function dependently on CRY2.
Overexpression of CK1.3 or CK1.4 (referred to as CK1.3ox or CK1.4ox; see Supplemental Figure 7A online) in wild-type plants resulted in longer hypocotyls under white and blue light (Figure 3A), further demonstrating the negative effect of CK1.3 and CK1.4 on hypocotyl elongation under blue light. In addition, CK1.3ox or CK1.4ox lines showed delayed flowering under long-day (LD) conditions, with more rosette leaves when bolting (Figure 3B; see Supplemental Figure 7B online), but not under short-day (SD) photoperiod conditions (see Supplemental Figures 7C and 7D online).
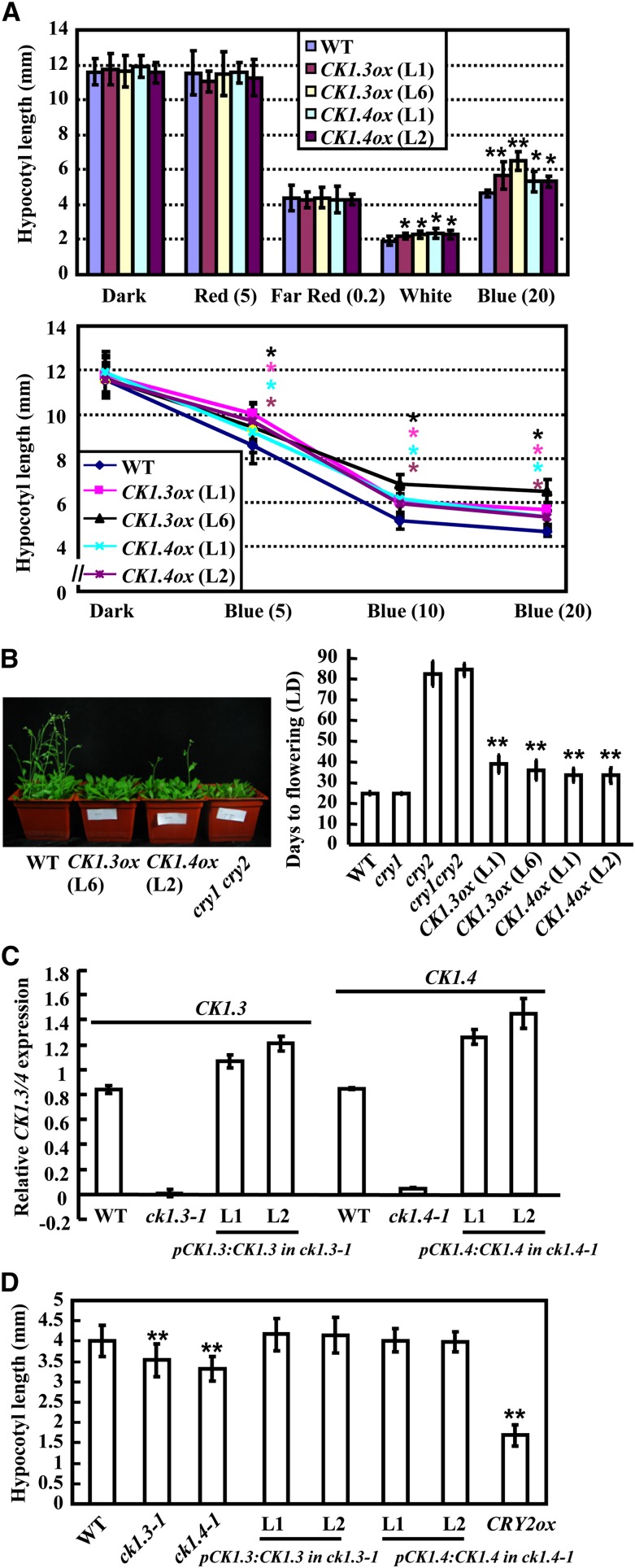
Figure 3.
Enhanced Overexpression of CK1.3 or CK1.4 Results in Shortened Hypocotyls and Delayed Flowering.
(A) Overexpression of CK1.3 or CK1.4 in wild-type (WT) plants resulted in longer hypocotyls under white light (50 µmol/m2/s) and blue light with various fluence rates (5, 10, and 20 µmol/m2/s; bottom panel). Hypocotyl lengths of 7-d-old seedlings were measured and statistically analyzed (*P < 0.05; bottom panel). Error bars represent sd (n > 30). Light intensity with different various fluence rates (µmol/m2/s) is indicated in parentheses.
(B) Observation of 35-d-old adult plants revealed the delayed flowering of CK1.3ox and CK1.4ox plants compared with the wild type (left panel). The flowering times were recorded when bolting with inflorescences (∼1 cm high) was observed and were statistically analyzed (**P < 0.01). Error bars represent sd (n > 15).
(C) Complementary expression of CK1.3 and CK1. 4 driven by their own native promoters rescued the defective expression of CK1.3 and CK1.4 in the ck1.3-1 and ck1.4-1 mutants. The relative transcription levels of CK1.3 and CK1.4 were detected by qRT-PCR using ACTIN7 as an internal positive reference. The expression level of CK1.3 or CK1.4 in wild-type seedling was set as 1.0. The data are presented as the average ± se. The experiments were repeated three times.
(D) Complementary expression of CK1.3 and CK1.4 rescued the normal hypocotyl elongation of the ck1.3-1 and ck1.4-1 mutants. The data are presented as average ± se (**P < 0.01). The experiments were repeated three times.
Previous studies showed that the expression of the florigen gene FT is regulated by CRY2 in an LD-specific manner (Lin et al., 1998; Zuo et al., 2011) and CO mediates the blue light signal (Song et al., 2012). Consistent with this observation, qRT-PCR analysis revealed that overexpression of either CK1.3 or CK1.4 resulted in significantly reduced FT transcription before dark (Zeitgeber time 13 h [ZT13], 13 h after light-on in a 16-h-light/8-h-dark cycle) and an additional peak at ZT4 (see Supplemental Figure 7E online). In addition, overexpression of either CK1.3 or CK1.4 caused reduced CO expression at ZT13 (see Supplemental Figure 7F online). The altered expression of CO and FT, especially the reduced FT expression at dawn, might account for the delayed photoperiodic flowering of the CK1.3ox and CK1.4ox lines.
As expected, complementary expression of CK1.3 or CK1.4 driven by their own native promoter rescued the defective expression of CK1.3 or CK1.4 in ck1.3-1 or ck1.4-1, respectively (Figure 3C), and recovered the hypocotyl elongation (Figure 3D), confirming the negative effects of CK1.3 and CK1.4 on blue light–mediated photomorphogenesis.
CRY2 Is Genetically Epistatic to CK1.3 and CK1.4
CK1.3/4 expression is induced by blue light, and altered CK1.3/4 expression resulted in blue light–related growth changes. Considering that CRY2 serves as an important blue light receptor (Lin et al., 1998), the genetic relationship between CK1.3/4 and CRY2 was then studied. Double mutants ck1.3-1 cry2 and ck1.4-1 cry2 were generated by a genetic cross, and comparison of hypocotyl growth showed that under white and blue light, ck1.3-1 cry2 and ck1.4-1 cry2 double mutants exhibited longer hypocotyls, which was similar to cry2 (Figure 4A). A deficiency of CK1.3 or CK1.4 under CRY2 overexpression (ck1.3-1 CRY2ox or ck1.4-1 CRY2ox, aMIR-CK1.3/CK1.4 in CRY2ox) still presented the short hypocotyls, which was similar to CRY2ox (Figures 2C and 4A), suggesting that CRY2 is necessary for CK1.3/4 function and CK1.3/4 may act dependently on CRY2.
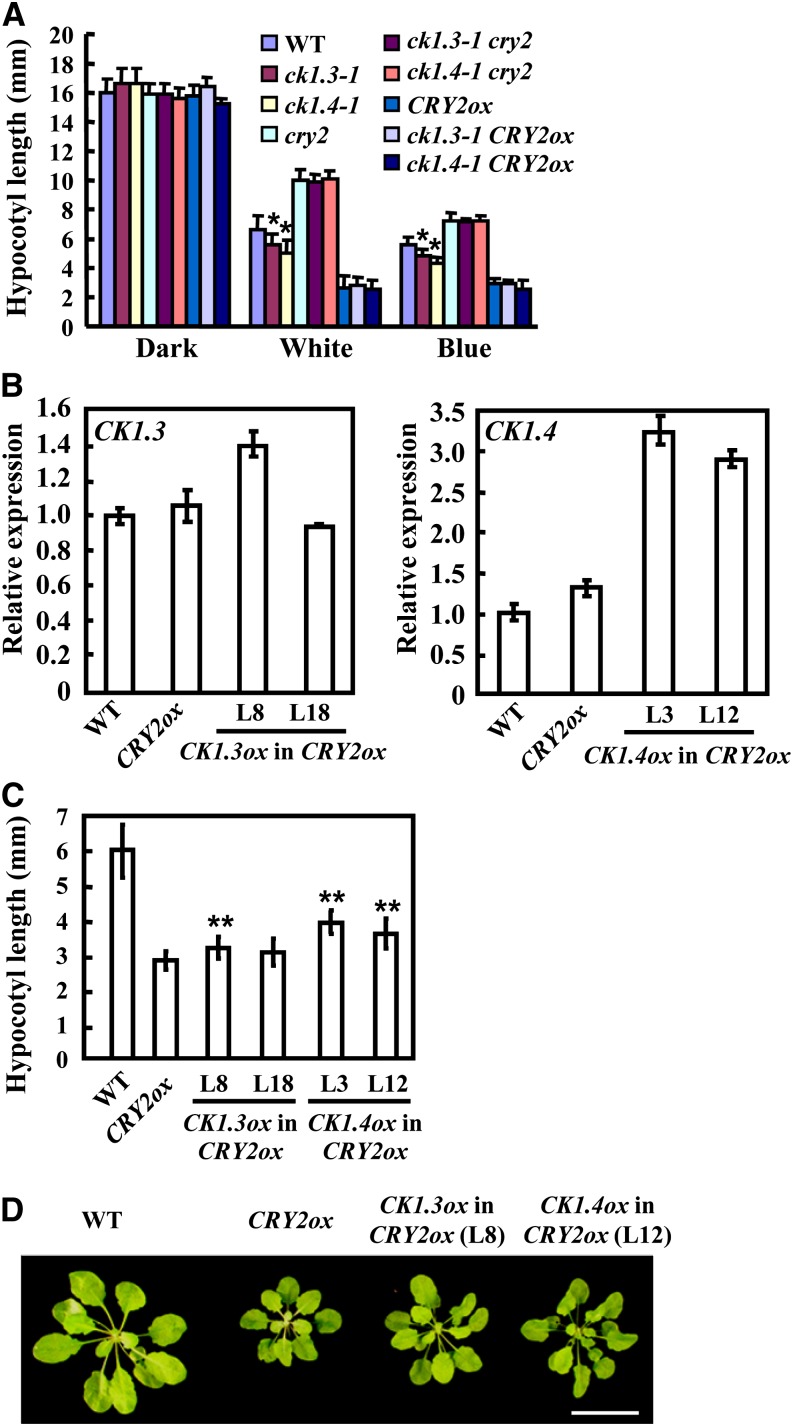
Figure 4.
CK1.3 and CK1.4 Act Dependently on CRY2 in Blue Light Signaling.
(A) Analysis of the hypocotyl lengths of wild-type (WT), ck1.3-1, ck1.4-1, ck1.3-1 cry2, ck1.4-1 cry2, ck1.3-1 CRY2ox, and ck1.4-1 CRY2ox seedlings grown under dark, white light, or blue light for 7 d showed that deficiency of CK1.3 or CK1.4 did not affect the elongated hypocotyls of cry2 or the shortened hypocotyls resulting from CRY2 overexpression (CRY2ox). Hypocotyl lengths were measured and statistically analyzed (*P < 0.05). Error bars represent sd (n > 30).
(B) Overexpression of CK1.3 (left) or CK1.4 (right) in CRY2ox seedlings. The relative transcription level of CK1.3 and CK1.4 in CRY2ox seedlings was detected by qRT-PCR using ACTIN7 as an internal positive control. Expression level of CK1.3 or CK1.4 in wild-type seedlings was set as 1.0. The data are presented as the average ± se. The experiments were repeated three times.
(C) Overexpression of CK1.3 or CK1.4 in CRY2ox partially suppresses the shortened hypocotyls under blue light. Hypocotyl lengths of 7-d-old seedlings were measured and statistically analyzed (**P < 0.01). Error bars represent sd (n > 30).
(D) Phenotypic observations of 4-week-old wild-type, CRY2ox, and CRY2ox plants overexpressing CK1.3 or CK1.4 showed that the shortened petiole of CRY2ox under short-day conditions (9 h light/15 h dark) was suppressed by overexpression of CK1.3 or CK1.4. Bar = 2 cm.
CRY2 plays important roles in regulating photoperiodic flowering, which was also examined under altered CK1.3/4. The cry2 mutant showed delayed flowering especially under LD conditions, which is similar to the phenotype observed in ck1.3-1 cry2 or ck1.4-1 cry2 double mutants. However, in contrast with the hypersensitive response to blue light of seedlings, CRY2ox lines took a similar number of days to flower under either LD or SD conditions, while neither the single mutants (ck1.3-1, ck1.3-2, or ck1.4-1) nor the aMIR-CK1.3/CK1.4 (ck1.4-1) transgenic lines exhibited any differences in flowering time compared with the wild type (see Supplemental Figures 8A to 8D online). These results suggest that wild-type plants may harbor enough CRY2 to sense the change of blue light and adapt to the environment for the floral transition to occur, and more CRY2 does not stimulate this process but may suppress hypocotyl and petiole elongation through different downstream events (Figures 2C and 4D).
In addition, overexpression of CK1.3 or CK1.4 in CRY2ox seedlings (Figure 4B) suppressed the shortened hypocotyl phenotype under blue light (Figure 4C) and the shortened petiole phenotype of CRY2ox rosette leaves under SD conditions (Figure 4D), further confirming that CRY2 is genetically epistatic to CK1.3/4 and that CK1.3/4 functions as negative regulators of blue light responses.
CK1.3 and CK1.4 Interact with and Phosphorylate CRY2 to Stimulate CRY2 Degradation
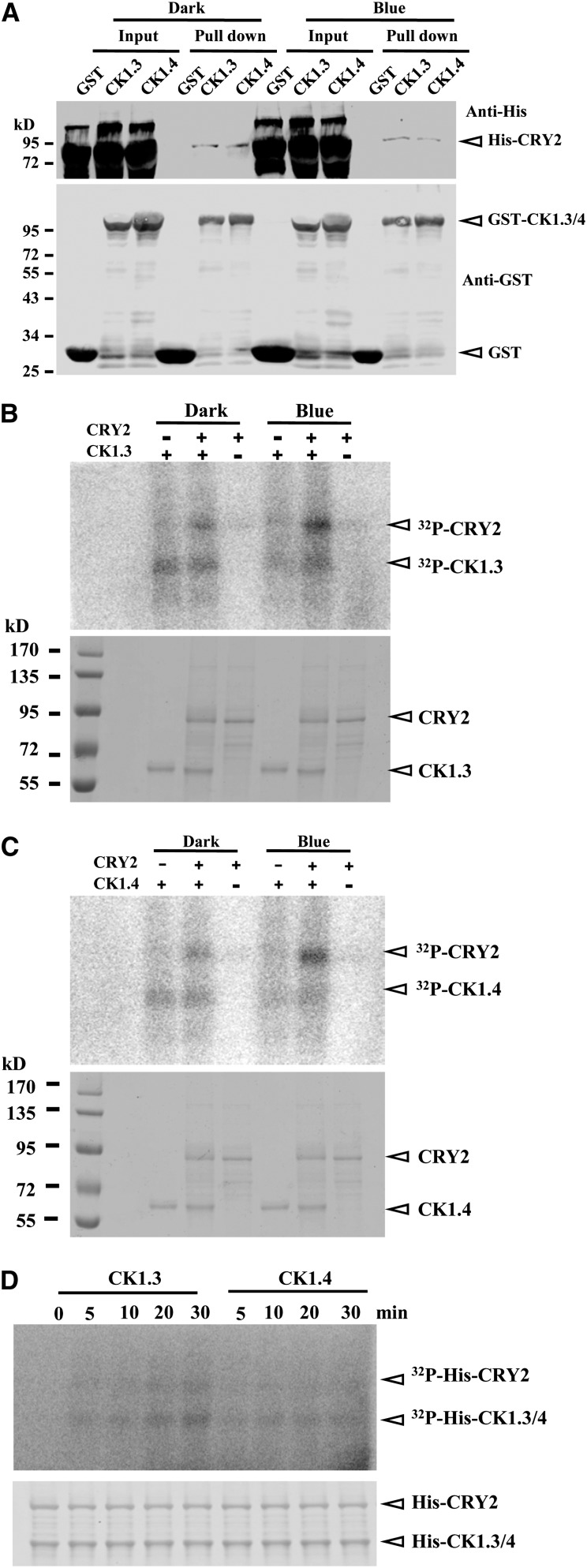
CRY2 undergoes blue light–dependent phosphorylation in the nucleus (Shalitin et al., 2002). Whether CK1.3/4 could directly phosphorylate CRY2 was then investigated using purified recombinant His-CRY2 (from Baculovirus-infected insect cells), glutathione S-transferase (GST), GST-CK1.3, and GST-CK1.4 (from E. coli). The pull-down assay showed that both GST-CK1.3 and GST-CK1.4 were coprecipitated with His-CRY2 in the dark or under constant blue light (GST as negative control; Figure 5A). However, we failed to detect the in vivo CRY2-CK1.3 or CRY2-CK1.4 interactions by coimmunoprecipitation after numerous attempts, which may be due to the transient and weak interaction between CRY2 and CK1.3 or CK1.4. Indeed, similar phenomena (weak interaction between kinases and substrates) were reported in other studies on kinases, confirming the transient “kiss-and-run” nature of the enzyme–substrate interactions in vivo (Gudesblat et al., 2012; Pusch et al., 2012).
Figure 5.
CK1.3 and CK1.4 Interact with and Phosphorylate CRY2, Which Is Stimulated by Blue Light.
(A) In vitro GST pull-down assay revealed the interaction of CK1.3 and CK1.4 proteins with CRY2 under dark or blue light exposure (10 µmol/m2/s). The quantitative assay of the input proteins is shown. GST was used as a negative control (bottom panel). GST-CK1.3 and GST-CK1.4 were expressed and purified from E. coli and His-CRY2 from insect cells. CK1.3, CK1.4, and CRY2 were detected by a His tag antibody (top panel) or a GST tag antibody (bottom panel), respectively. Positions of CK1.3, CK1.4, and CRY2 are highlighted by arrowheads.
(B) and (C) In vitro kinase assay indicated that phosphorylation of CRY2 by CK1.3 (B) and CK1.4 (C) is stimulated by blue light exposure. All the reactions were initiated by adding kinases under red light and then transferred to dark or blue light (10 µmol/m2/s) for 3 h. Kinase activity was detected by [γ-32P]ATP autoradiography (top panels), and equal input of His-CK1.3, His-CK1.4, and His-CRY2 (10 μg) was shown by Coomassie blue staining (bottom panels).
(D) In vitro kinase assay by [γ-32P]ATP autoradiography revealed that CK1.3 and CK1.4 rapidly phosphorylated CRY2 under blue light (top panel). Purified recombinant His-CRY2 protein from insect cells was used for the assay, and all reactions were initiated by adding CK1.3 or CK1.4 under red light before blue light exposure. Blue light (10 µmol/m2/s) treatment was applied for 0, 5, 10, 15, or 30 min. The input of His-CRY2, His-CK1.3, and His-CK1.4 were detected by Coomassie blue staining (bottom panel).
CRY2 is a highly phosphorylated protein after exposure to blue light (Kleiner et al., 1999; Yu et al., 2007b), and analysis showed that both CK1.3 and CK1.4 could phosphorylate CRY2 in vitro, relatively weakly in the dark but much enhanced after exposure to blue light, revealing a blue light–stimulated phosphorylation by CK1.3 and CK1.4 (Figures 5B and 5C). Additionally, the blue light–stimulated phosphorylation of CRY2 could be observed within 5 min (Figure 5D), processing as quickly as occurs in planta. Considering the blue light–independent interaction of CK1.3/4 with CRY2, revealed by the pull-down assay, it is hypothesized that blue light might activate CRY2 to unfold the sites subjected to phosphorylation by CK1.
Previous studies showed that CRY2 proteins become localized in nuclear bodies to undergo the ubiquitin-26S proteasome degradation when exposed to blue light. Indeed, subcellular localization analysis using Arabidopsis protoplasts confirmed that CRY2-mCherry fusion protein is localized in nuclear bodies under blue light and is colocalized with CK1.3-GFP or CK1.4-GFP (Figure 1E). Transient expression analysis through cotransforming CRY2-GFP with CK1.3-mCherry or CK1.4-mCherry into tobacco leaves further confirmed this result (see Supplemental Figure 4B online). These results further support the hypothesis that interaction with CK1.3/CK1.4 may be involved in the regulation of CRY2 degradation under blue light.
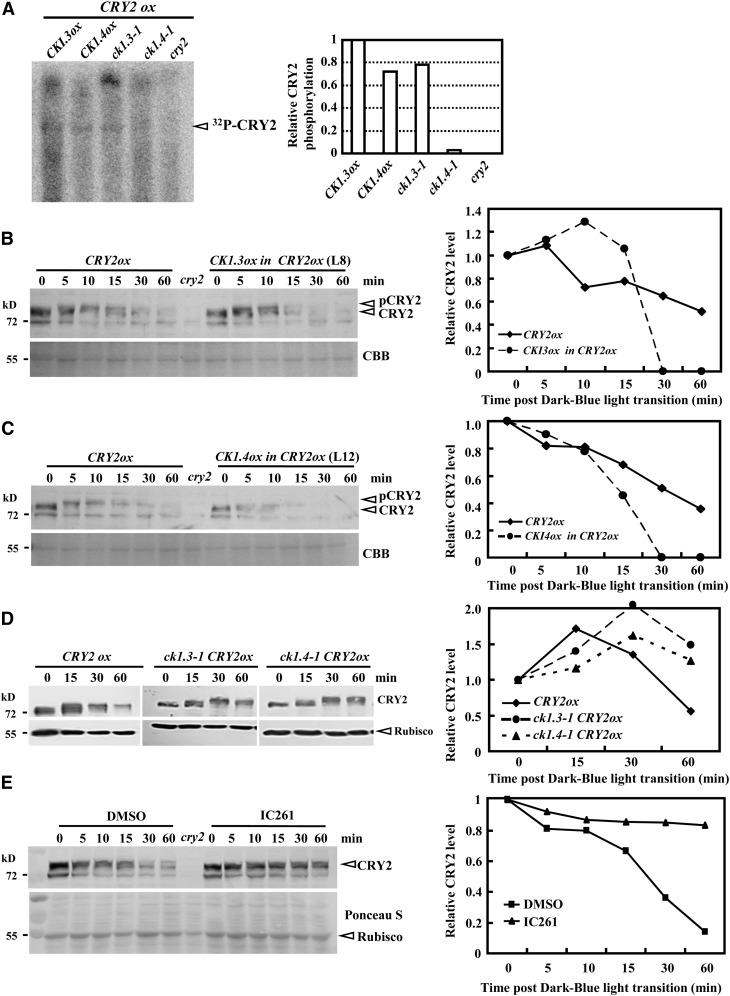
CRY2 undergoes blue light–dependent degradation mainly through phosphorylation. Indeed, in vivo assay of CRY2 protein by immunoprecipitation revealed the reduced/suppressed CRY2 phosphorylation under CK1.3 or CK1.4 deficiency (Figure 6A), and observations using CRY2ox plants showed that ∼10 min is required for CRY2 to be fully phosphorylated and ∼60 min for CRY2 to be completely degraded (Figure 6B). By contrast, overexpression of CK1.3 (Figure 6A) or CK1.4 (Figure 6C) resulted in the significantly accelerated degradation of CRY2 under the same conditions. Interestingly, overexpression of CK1.3 or CK1.4 did not enhance the phosphorylation or reduce the quantity of CRY2 under dark conditions, indicating that CK1.3 and CK1.4 stimulate CRY2 degradation in a blue light–dependent manner. Consistently, CRY2 degradation was significantly reduced in ck1.3-1 or ck1.4-1 when exposed to blue light (10 µmol/m2/s; Figure 6D).
Figure 6.
CK1.3 and CK1.4 Stimulate the Blue Light–Dependent Degradation of CRY2.
(A) In vivo assay revealed rescued/suppressed CRY2 phosphorylation under CK1.3 or CK1.4 deficiency. Seven-day-old seedlings under red light (referred to published methods; Shalitin et al., 2002) were incubated with 200 μCi [32P]H3PO4 overnight and then exposed to blue light (6 μ mol/m2/s) for 15 min. Seedlings (1 g) were collected to extract crude protein for immunoprecipitation and autoradiography (left panel). The relative CRY2 phosphorylation level was calculated by comparing the signal strength of the CRY2 band (right panel; calculated by the Tanon Gis 1D program). The quantity of CRY2 in CK1.3ox was set as 1.0.
(B) and (C) Protein gel blot analysis revealed a significant increase of CRY2 degradation when exposed to blue light (10 µmol/m2/s) under enhanced expression of CK1.3 (B) or CK1.4 (C). Equal amounts of proteins (∼10 μg) were used for blotting (confirmed by Coomassie blue [CBB] staining). The relative protein level was calculated by comparing the signal strength of the CRY2 band to the ribulose-1,5-bis-phosphate carboxylase/oxygenase band in the Coomassie blue PAGE gel (right panels; calculated by the Tanon Gis 1D program). The quantity at time “0” was set as 1.0, similarly in (D) and (E).
(D) Protein gel blot analysis (left panel) revealed that CRY2 degradation was significantly reduced in ck1.3-1 or ck1.4-1 when exposed to blue light (10 µmol/m2/s). Equal amounts of proteins (∼10 μg) were used for blotting (confirmed by the similar intensities of a nonspecific band).
(E) Compared with the degradation of CRY2 under blue light (10 µmol/m2/s; 5, 10, 15, 30, and 60 min after dark-blue light transition), IC261 treatment (50 µM) significantly suppressed CRY2 degradation. Equal amounts of proteins (∼10 μg) were used for blotting (confirmed by Coomassie blue staining). DMSO was used as a control.
In addition, similar to the reduced CRY2 degradation in ck1.3-1 or ck1.4-1, treatment with the widely used CK1-specific inhibitor IC261 almost completely blocked CRY2 proteolysis and extended the half-life of CRY2 (Figure 6E), further confirming the negative role of CK1 in CRY2 stability.
Phosphorylation of Ser-587 and Thr-603 Is Crucial for CRY2 Function
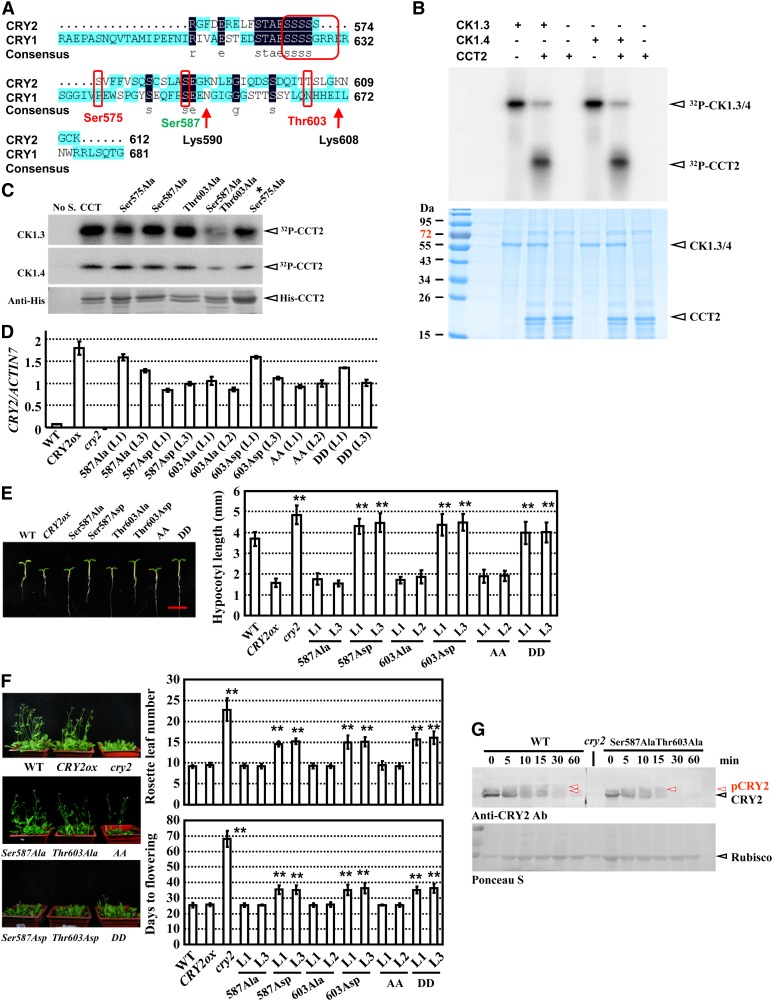
Several candidate CK1 phosphorylation sites were predicted at the C terminus of CRY2 (CCT2) and the C terminus of CRY1 (CCT1) (Figure 7A; for details, see Supplemental Table 1 online) by computational analysis (http://scansite.mit.edu/motifscan_seq.phtml); indeed, an in vitro kinase assay indicated that both CK1.3 and CK1.4 can phosphorylate CCT2 (Figure 7B).
Figure 7.
Phosphorylation of Ser-587 and Thr-603 by CK1.3 and CK1.4 Suppresses CRY2-Mediated Photomorphogenesis.
(A) Presence of multiple candidate CK1 phosphorylation sites in the C terminus of CRY2 and CCT2, including Ser570-575, Ser-587, and Thr-603 (highlighted by red circles). Prediction analysis was performed via integrating two programs (http://www.cbs.dtu.dk/services/NetPhosK and http://scansite.mit.edu/motifscan_seq.phtml). Lys-590 and Lys-607 are two candidate ubiquitination sites in CCT2 (not in CCT1; i.e., the C terminus of CRY1). The same amino acids are indicated with a dark-blue background, and highlighted (consensus), and similar amino acids are indicated with a light-blue background.
(B) Kinase activity assay by [γ-32P]ATP autoradiography indicated that CK1.3 and CK1.4 phosphorylated CCT2 in vitro (top panel). Purified recombinant His-CCT2 fusion protein was used for the assay. Input of His-CCT2 was detected by Coomassie blue staining (bottom panel). Arrows indicate the positions of CK1.3 and CK1.4 or CCT2, respectively.
(C) Kinase assay by [γ-32P]ATP autoradiography indicated that Ser-587 and Thr-603 are the phosphorylation sites of CK1.3 and CK1.4 in vitro (top panels). Purified recombinant 6XHis-CCT2s (Ser575Ala, Ser587Ala, Thr603Ala, Ser587Ala Thr603Ala, and Ser570-575Ala as *Ser575Ala) proteins were used for the assay. Input of 6XHis-CCT2 was detected with an antibody against His-tag (bottom panel). No S, no substrate.
(D) qRT-PCR analysis confirmed the expression of CRY2 (different versions, including dephosphorylated status Ser587Ala, Thr603Ala, and AA [Ser587Ala and Thr603Ala] and constitutively phosphorylated status Ser587Asp, Thr603Asp, and DD [Ser587Asp and Thr603Asp]) in transgenic lines. ACTIN7 was used as a positive internal reference. The data are presented as the average ± se. The experiments were repeated three times. WT, the wild type.
(E) Growth of 7-d-old seedlings of wild-type, CRY2ovx, and seedlings overexpressing different point-mutated CRY2s under blue light (left panel, representative images are shown, bar = 5 mm). Hypocotyl lengths were measured and statistically analyzed (**P < 0.01; right panel). Error bars represent sd (n > 15).
(F) Growth of 40-d-old plants of wild-type, CRY2ox, cry2 mutant, and transgenic seedlings as described in (E) under LD conditions (left panel; representative images are shown). Days of flowering were measured and statistically analyzed (**P < 0.01; right panel). Error bars represent sd (n > 15).
(G) Protein gel blot analysis revealed that mutations at Ser587Ala and Thr603Ala resulted in a significant decrease of CRY2 phosphorylation when exposed to blue light. Seven-day-old etiolated seedlings of CRY2ox or CRY2 Ser587AlaThr603Alaox lines were exposed to blue light (10 μmol/m2/s) and used for protein extraction. Equal amounts of proteins (∼10 μg) were used for blotting (confirmed by Ponceau S staining; bottom panel). Rubisco, ribulose-1,5-bisphosphate carboxylase/oxygenase.
Detailed analysis showed that many Ser and Thr are located in the CCT2 domain, possibly contributing to CRY2 phosphorylation. Amino acids Ser-575, Ser-587, Thr-603, and Ser570-575 (abbreviated as *Ser-575) were then selected and point mutated to Ala to examine the exact phosphorylation sites of CK1.3 or CK1.4. An in vitro kinase assay showed that single mutation of Ser-575, Ser-587, and Thr-603 did not significantly change the phosphorylation status of CCT2, while mutation of both Ser-587 and Thr-603 evidently reduced this phosphorylation (Figure 7C), indicating that these two sites may be the sites that are phosphorylated by CK1.3 or CK1.4.
The physiological function of CK1 by modifying CRY2 in planta was further studied by producing different phosphorylated states for CRY2. The dephosphorylated state of CRY2, CRY2Ser587Ala, CRY2Thr603Ala, CRY2Ser587AlaThr603Ala (abbreviated as CRY2AA), and constitutively phosphorylated mimic CRY2, CRY2Ser587Asp, CRY2Thr603Asp, and CRY2Ser587AspThr603Asp (abbreviated as CRY2DD) were transformed into wild-type plants, respectively (Figure 7D). Analysis of the transgenic plants showed that compared with CRY2ox seedlings, CRY2Ser587Ala, CRY2Thr603Ala, and CRY2AA exhibited similar hypocotyls elongation under blue light and flowering time under LD conditions; by contrast, CRY2Ser587Asp, CRY2Thr603Asp, and CRY2DD showed elongated hypocotyls and much delayed flowering (Figures 7E and 7F). These results indicate that CRY2 phosphorylated by CK1.3 or CK1.4 at Ser-587 and Thr-603 is unstable in planta and demonstrate the crucial roles of CK1.3 and CK1.4 in regulating CRY2 stability through phosphorylation.
Protein gel blot analysis showed that mutation of Ser587Ala and Thr603Ala in CRY2 resulted in a significant decrease of CRY2 phosphorylation, when exposed to blue light (Figure 7G), suggesting that Ser-587 and Thr-603 are two important sites responsible for blue light–induced CRY2 phosphorylation by CK1. 3 and CK1.4. In addition, of the phosphorylation mimicry transgenic plants, CRY2DDox, the CRY2 protein level is very low in the dark and unstable when exposed to blue light, compared with CRY2ox or CRY2AAox (see Supplemental Figure 9 online). Combined with the delayed and enhanced CRY2 degradation in ck1.3-1/ck1.4-1 mutants or CK1.3ox/CK1.4ox lines, it is concluded that CK1.3/CK1.4 promotes CRY2 degradation through phosphorylation in planta. However, considering the enhanced degradation of CRY2 harboring mutations of Ser587Ala and Thr603Ala, which results in the suppressed phosphorylation of CRY2, other modifications (including phosphorylation at other sites) may exist and induce CRY2 degradation; further studies will help dissect the detailed mechanisms. Considering the role of CCT2 in binding downstream factors, it is also possible that mutations of Ser-587 and Thr-603 might lead to conformational changes and block its interaction with other proteins/factors. How CK1.3 and CK1.4 functions in this regulation requires further investigation.
DISCUSSION
Arabidopsis CK1s Regulate Blue Light Signaling by Stimulating CRY2 Degradation
How CRYs, which are conservative light receptors for eukaryotes, are posttranslationally regulated through phosphorylation is still unclear, and the related kinases remain to be identified. In mouse livers, nutrient-responsive adenosine monophosphate-activated protein kinase can phosphorylate and destabilize mouse CRY1, revealing the interaction between nutrition and circadian rhythm (Lamia et al., 2009). Arabidopsis CRY1 exhibits autophosphorylation activity in vitro (Bouly et al., 2003; Shalitin et al., 2003), which is different from CRY2 (Shalitin et al., 2002, 2003). In addition, Arabidopsis CRY1 and CRY2 can be phosphorylated by phytochrome A–associated kinase activity in vitro and exhibit a red light responsive manner (Ahmad et al., 1998b). Although in vivo studies demonstrated that both CRY1 and CRY2 undergo blue light–dependent phosphorylation (Shalitin et al., 2002, 2003), how CRY2 is phosphorylated to mediate the blue light signaling remains unclear. Our studies demonstrate that CK1 is involved in blue light signaling through stimulating the degradation of CRY2.
Recent studies revealed that CRY2 protein accumulates in the dark and is subjected to rapid and blue light–stimulated phosphorylation and degradation by 26S proteasome in the nucleus (Kleiner et al., 1999; Shalitin et al., 2002; Yu et al., 2007a; Yu et al., 2009). Localization of CK1.3 and CK1.4 in the nucleus, especially in the nuclear bodies after blue light illumination, may contribute to the blue light–induced CRY2 degradation by phosphorylation.
Considering that CK1.3 and CK1.4 stimulate CRY2 degradation in a blue light–dependent manner, while it is difficult to detect the transient CK1.3/4-CYR2 interaction in vivo, it is suggested that CK1.3 and CK1.4 are necessary for the blue light–dependent degradation of CRY2, and other factors may be involved in the CK1.3/4-stimulated degradation of CRY2 (for example, additional interacting E3 ligases). In addition, the induced expression of CK1.3 and CK1.4 by blue light may contribute to the rapid phosphorylation of CRY2 under blue light.
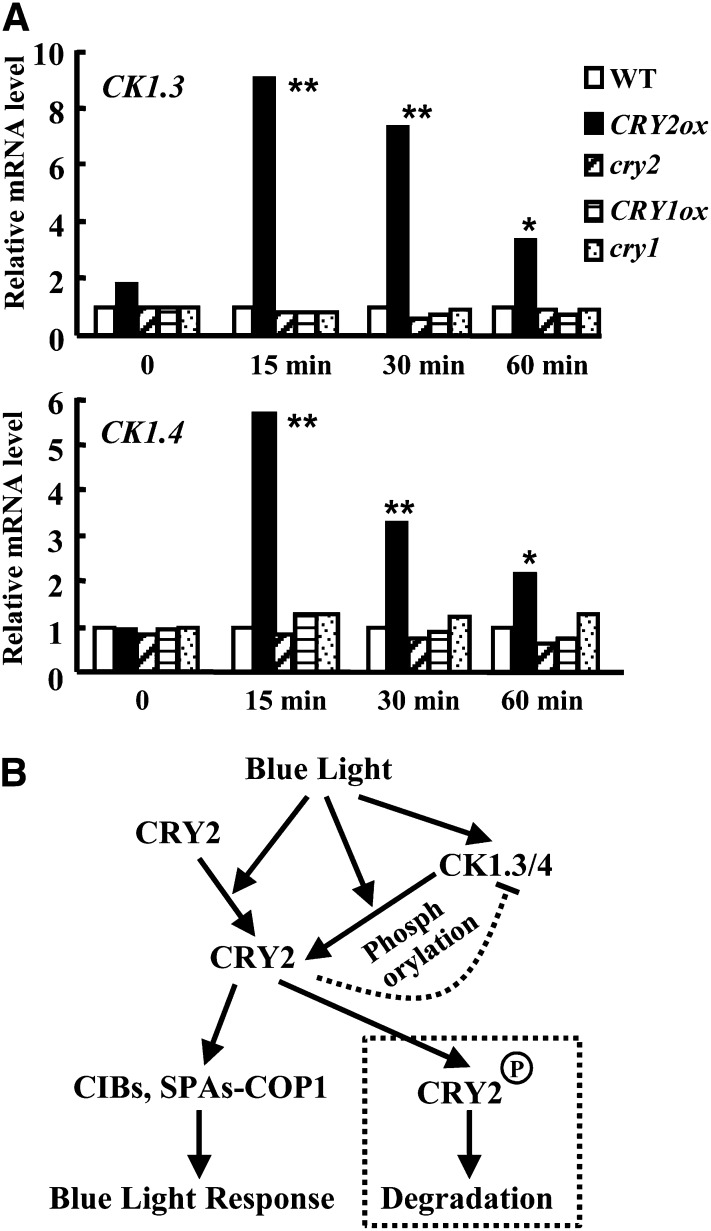
Two major CK1 phosphorylation sites, Ser-587 and Thr-603, at the C terminus of CRY2, are crucial for the phosphorylation and function of CRY2. In addition, it is interesting to note that the expression of CK1.3 and CK1.4 is induced by blue light rapidly, while it is suppressed by continuous blue light. CRY2, rather than CRY1, is the major receptor mediating blue light–regulated expression of CK1.3 and CK1.4 (Figure 8A). Combined with these results, a model is proposed to illustrate the mechanism of how CK1.3/4 regulates blue light signaling (Figure 8B). In the dark, unphosphorylated CRY2 resides at the nucleus in a resting state, which might be maintained by PP7 phosphatase (Møller et al., 2003); when exposed to blue light, CRY2 exhibits an open conformation and gains activity to extend the active motif (NC80) (Yu et al., 2007b) to bind the downstream signaling components, resulting in activated blue light signaling. At the same time, the increasing CK1.3 and CK1.4 activity confers phosphorylation of CRY2 at Ser-587 and Thr-603 and stimulates the ubiquitin-26S proteasome degradation of CRY2. In addition, continuous blue light exposure suppresses the expression of CK1.3 and CK1.4, which results in restoration of CRY2, revealing the molecular mechanism of how CRY2 quantity is regulated by the light-dark cycle. CRY2 is only accumulated under an LD photoperiod, and the feedback regulation of CK1.3/4 may help to restrict Arabidopsis, an LD plant. Whether CK1 and PP7 phosphatase are two competing factor that function as a switch to regulate the phosphorylation status of CRY2 requires further investigation.
Figure 8.
Expression of CK1.3 and CK1.4 Is Mediated by CRY2 and a Hypothetical Model of How CK1s Are Involved in CRY2-Mediated Blue Light Signaling.
(A) qRT-PCR analysis revealed that the expression of CK1.3 (top panel) or CK1.4 (bottom panel) is rapidly induced under blue light (3 µmol/m2/s fluence rate) for 15, 30, and 60 min in a CRY2 overexpression background. In addition, expression of CK1.3 and CK1.4 were reduced in cry2 mutant, and there was no significant change in either cry1 mutant or CRY1ox (CK1.4 expression was reduced in cry1). ACTIN7 was used as a positive internal reference. Expression of CK1.3 or CK1.4 in the wild type (WT) under blue light condition (3 µmol/m2/s fluence rate) for 0, 15, 30, and 60 min was set as 1.0. Statistical analysis indicated the significant differences (*P < 0.05; **P < 0.01). The experiments were repeated three times.
(B) Hypothetical model of the role of CK1.3 and CK1.4 in CRY2-mediated blue light signaling. As an important blue light receptor, the resting CRY2 under dark is activated by blue light and undergoes conformational changes to expand the active motif to bind downstream partners, including CIBs and SUPPRESSOR OF PHYTOCHROME As-CONSTITUTIVE PHOTOMORPHOGENIC1, to confer the blue light responses. At the same time, expression of CK1.3 and CK1.4 is rapidly induced in a CRY2-dependent manner. The conformational change of CRY2 may expose the candidate phosphorylation sites, Ser-587 and Thr-603, recognized by CK1.3 and CK1.4, which results in blue light–stimulated CRY2 degradation through phosphorylation. The exquisite regulation of expression of CK1s and CRY2 degradation reveals the complex and finely controlled blue light signaling.
In addition to the Ser-587 and Thr-603 at the CCT2 domain, which are crucial for CRY2 phosphorylation (mutations of these result in reduced phosphorylation), other phosphorylation sites of CRY2 may exist and regulate CRY2 degradation. Under blue light exposure, the CCT2 domain, precisely the NC80 motif (Yang et al., 2001; Yu et al., 2007b), is released to bind downstream factors to confer the blue light responses. It is possible that Ser-587 and Thr-603 might play roles in this process through regulating the conformational change of CCT2 by phosphorylation. Additionally, we noticed that there are two candidate ubiquitinated Lys residues (Lys-590 and Lys-608, close to CK1 phosphorylation sites) in CCT2 but not in CCT1 (Figure 7A). Whether the phosphorylation of Ser-587 and Thr-603 leads to stimulated ubiquitination at Lys-590 and Lys-608 of CRY2 requires further investigation.
CK1 is a multifunctional protein kinase involved in various signaling pathways throughout the plant and animal kingdoms, and CRYs exist in almost all evolutionary clades. In addition to CCT2, our preliminary results showed that both CK1.3 and CK1.4 could also phosphorylate the C terminus of CRY1, suggesting that CRY1 and CRY2 might share similar posttranslational regulation, although they do show distinct characteristics: CRY2 can be trans-phosphorylated and degraded, while CRY1 is stable and shows autophosphorylation activity. This study expands our knowledge of CRY regulation and will be helpful for studies of the underlying mechanisms in mammals.
Circadian Regulation of CK1.3 and CK1.4 Coordinated with CRY2 Accumulation
The rigorous regulation of the dose and activity of CRY2 by CK1.3 and CK1.4 is crucial for sensing blue light, regulating photomorphogenesis, and the floral transition. In addition to CRY2-mediated induction, the expression of CK1.3 and CK1.4 shows a pattern reminiscent of a circadian rhythm (see Supplemental Figure 10A online), suggesting that CK1.3 and CK1.4 may execute their role through a specific pattern. Indeed, both CK1.3 and CK1.4 show a high expression peak before dark under LD conditions (see Supplemental Figure 10B online), similar to the expression pattern of FT, indicating that the expression of CK1.3 and CK1.4 is strictly regulated by a circadian rhythm.
Previous studies showed that blue light–dependent degradation leads to an oscillation in the quantity of CRY2, which only occurs under LD conditions, contributing to the strict control of photoperiodic flowering (Mockler et al., 1999). Expression of CK1.3 and CK1.4 is consistent with the observations that CRY2 protein accumulates under continuous blue light and that short-term blue light under SD conditions cannot maintain sufficient CRY2 levels. The feedback regulation of CK1.3 and CK1.4 by CRY2 will promote studies of how downstream components, especially the CRY2 partners, regulate the expression of CK1.3 and CK1.4, which may facilitate the understanding of underlying mechanisms of CRY2 modulation and blue light signaling.
Both CK1.3 and CK1.4 possess autophosphorylation activity (Figures 5B to 5D and 7B), which is also detected in many types of protein kinases. Further studies of how the activity of CK1s is regulated will help elucidate the upstream regulatory mechanism of CK1.
METHODS
Plant Materials, Growth Conditions, and Measurement of Hypocotyl Lengths
Arabidopsis thaliana and tobacco (Nicotiana benthamiana) plants were grown in soil under 16 h light/8 h dark in a phytotron at 22°C under white light. Arabidopsis ecotype Columbia-0 was used in the experiments as the wild-type control. Seeds of the wild type, various mutants, and transgenic lines were surface sterilized and sown on plates containing Murashige and Skoog (MS) medium (Duchefe Biochemie). After stratification at 4°C for 4 d, the plates were incubated under LD conditions (16 h light/8 h dark, 22°C).
The seedlings’ hypocotyl lengths were measured 7 d after germination under different light conditions, including white light (15 µmol/m2/s), far-red light (5 µmol/m2/s), red light (5 µmol/m2/s), blue light (10 µmol/m2/s), or blue light of different fluence rates (3, 10, or, 30 µmol/m2/s), with the help of the Image J program (National Institutes of Health). A heteroscedastic t test was performed to analyze the significant differences. All experiments were performed in triplicate, and the data represent mean values ± sd. For statistical analysis of photoperiodic flowering, the LD condition was set as a 16-h-light/8-h-dark cycle and SD was set as 9 h light/15 h dark.
Identification of ck1.3-1, ck1.3-2, and ck1.4-1 Mutants
The CK1.3 and CK1.4 genomic sequence was used to query the flanking sequence database of the Arabidopsis mutation population (SALK; http://signal.salk.edu/cgi-bin/tdnaexpress). T-DNA insertion mutants ck1.3-1 (SALK_016571C), ck1.3-2 (SALK_110494), and ck1.4-1 (SALK_043954C) were obtained from the SALK collection.
A PCR-based approach was used to identify the mutants. Primers, CK1.3-1, CK1.3-2, CK1.4-1, CK1.4-2, and LBb1 (see Supplemental Table 2 online) were used to confirm the insertion of T-DNA and to identify the homozygous lines. The homozygous lines were used for further analysis.
Protein Expression and in Vitro Kinase Assay
The coding regions of CK1.3 and CK1.4 were amplified with primers CK1.3-3, CK1.3-4, CK1.4-3, and CK1.4-4 and subcloned into vectors pET28a (Novagen) and pGEX-4T-1 (GE Healthcare). Proteins were recombinantly expressed in Escherichia coli (strain BL21) by supplementing with 1 mM isopropyl-β-D-thiogalactopyranoside (28°C, 3 h).
The coding region of CRY2 was subcloned into vector pFastBac HTB (Invitrogen). The resultant vector was transformed into E. coli (strain DH10Bac) for recombination of Bacmid, which was used for infection of insect cells (Sf9) according to the Bac-to-Bac Baculovirus Expression System (Invitrogen). The His tag protein was purified using Ni-NTA His binding resin (Novagen) and GST tag protein by glutathione sepharose (Novagen).
The activity assay was performed according to a previous report (Klimczak and Cashmore, 1993) with minor modifications. The assay was initiated by adding 6 μL of kinase solution (1 µg protein) containing recombinant His-CK1.3 and His-CK1.4 in a total volume of 40 μL containing 50 mM Tris-HCl, pH 8.0, 10 mM MgCl2, 1 mM DTT, 10 mM ATP, 10 μCi [γ-32P]ATP (NEC902A; Perkin-Elmer), and 10 µg of substrate (recombinant His-CCT2, His-CCT2S575A, His-CCT2S587A, His-CCT2T603A, His-CCT2S587AT603A, His-CCT2S*575A, His-CRY2, BSA [A0332, Sangon], and partially dephosphorylated casein [C4032; Sigma-Aldrich]). Reactions were incubated at room temperature (25°C) for 3 h unless otherwise noted and terminated by adding 2× SDS loading buffer. After boiling for 5 min, the reaction products were fractionated by SDS-PAGE and transferred to a polyvinylidene difluoride (PVDF) membrane (Perkin-Elmer) by semidry blotting. After washing with TBST buffer (0.1 M Tris-HCl, 0.15 M NaCl, and 0.05% Tween 20, pH 7.5) three times, the blot was examined by autoradiography (Fujifilm FLA 9000 plus DAGE).
Primers used for constructs harboring point-mutated CCT2 protein are listed in Supplemental Table 2.
qRT-PCR Analysis
qRT-PCR analyses were performed to examine the transcription of CK1.3 and CK1.4 in various tissues or under different light treatments or of ck1.3-1, ck1.4-1, cry1, cry2, CRY1ox, CRY2ox, and transgenic CRY2ox plants overexpressing CK1.3 or CK1.4. Total RNAs were extracted from Arabidopsis seedlings of the wild type, various mutants, and transgenic plants growing under different conditions using TRIzol reagent (Invitrogen) according to the manufacturer’s manual. Reverse transcription was performed using the reverse transcription kits (Toyobo), and the resultant single-strand cDNAs were used as templates for qRT-PCR using a SYBR Green qPCR kit (real-time PCR Master Mix; Toyobo) with a Rotor-Gene real-time thermocycler R3000 (Corbett Research). Gradient dilutions for each PCR product were use as templates to generate a linear standard curve.
Primers CK1.3-1 and CK1.3-2, and CK1.4-1 and CK1.4-2 were used to examine the expression of CK1.3 or CK1.4. The ACTIN7 gene was amplified and used as an internal positive control. Primers FT-F and FT-R, and CO-F and CO-R were used to examine the expression of FT or CO, respectively. A heteroscedastic t test was performed for statistical analysis.
Constructs and Plant Transformation
The following vectors were constructed: pCambia1300-CK1.3pro-GUS, pCambia1300-CK1.4pro-GUS, pCambia1301-CK1.3-antisense (p-A-CK1.3), pBA-35S-CK1.3, pBA-35S-CK1.4, pCambia2301-CK1.3, pCambia2301-CK1.4, pCambia2301-CK1.3-YFP, pCambia2301-CK1.4-YFP, pHB-CRY2S587A, pHB-CRY2S587D, pHB-CRY2T603A, pHB-CRY2T603D, pHB-CRY2AA, pHB-CRY2DD, and pCambia2300s-aMIR-CK1.3/4 (aMIR-CK1.3/4 was generated based on pRS300). The point mutated CRY2 was constructed using the QuickChange II site-directed mutagenesis kit (200524; Agilent Stratagene) and confirmed by sequencing. Primers are listed in Supplemental Table 2 online. All constructs were introduced into the Agrobacterium tumefaciens strain GV3101 and transformed into Arabidopsis by the floral dip method (Clough and Bent, 1998). Transgenic seeds were screened on MS plates containing kanamycin, hygromycin, or glufosinate-ammonium. At least 10 independent transgenic lines were characterized for each construct.
Construction of the Double Mutants
The following double mutants were generated: cry2 ck1.3-1, cry2 ck1.4-1, CRY2ox ck1.3-1, and CRY2ox ck1.4-1. The parental mutants used for crosses are listed in Supplemental Table 3 online and the PCR primers used for genotyping are listed in Supplemental Table 2 online.
Immunoblot Analysis
For the CRY2 degradation assay, seedlings were grown on MS medium in the dark for 7 d and then all the seedlings were transferred to a new dish filled with liquid MS medium containing 100 μM CHX (C7698; Sigma-Aldrich) to block the synthesis of new proteins. For CK1 inhibitor treatment, 50 μM (final concentration, DMSO as control) IC261 (I0658; Sigma-Aldrich) was added into MS. The dishes were exposed to blue light (10 µmol/m2/s) for 0, 5, 10, 15, 30, or 60 min, and protein extraction was performed by grinding the seedlings in liquid nitrogen. The ground tissues were resuspended in extraction buffer (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.5% Tween 20, 1 mM EDTA, and 1 mM DTT) containing a protease inhibitor cocktail (one tablet for 50 mL, Complete 04693116001; Roche). After addition of an equal volume of 2× sample buffer (1× sample buffer: 67.5 mM Tris-HCl, pH 6.8, 2% [w/v] SDS, 10% [w/v] glycerol, 0.01% [w/v] bromophenol blue, and 0.1 M DTT), the samples were boiled for 5 min, separated by 10% SDS-PAGE, and transferred to a PVDF membrane by semidry blotting. The blots were incubated with a rabbit anti-CRY2 antiserum (1:5000) and then with a goat anti-rabbit IgG AP-conjugated secondary antibody (1:3000; Santa Cruz Biotechnology). AP activity was detected by the BCIP/NBT detection reagents (826858A; Invitrogen) on the PVDF membrane. A prestained protein ladder (Thermo Scientific; 26617) was used to mark the molecular weight of every band.
Subcellular Localization Studies of CK1.3 and CK1.4 and Colocalization with CRY2
To express the CK1.3-GFP and CK1.4-GFP fusion proteins, the coding regions of CK1.3 and CK1.4 were amplified with primers CK1.3-3 and CK1.3-4, and CK1.4-3 and CK1.4-4, respectively, and subcloned into vector pA7 (C terminus fusion). For CRY2-mCherry fusion protein, the coding region of CRY2 was subcloned into a modified pCambia1300 containing mCherry reporter (Shaner et al., 2004). The resultant plasmids were transiently expressed in Arabidopsis protoplasts using the polyethylene glycol/CaCl2 method (Yoo et al., 2007). OsMADS15–red fluorescent protein (Zhang and Xue, 2013) was used as a positive control.
For colocalization analysis, equal amounts of CK1.3-GFP or CK1.4-GFP and CRY2-mCherry constructs were mixed for transformation. Protoplasts were incubated in the dark for 9 h (23°C) before observation. For blue light treatment, incubated protoplasts were first exposed to blue light (10 µmol/m2/s) for 5 min. Fluorescence was observed by confocal laser scanning microscopy (Olympus FV1000).
For transient expression of fusion proteins in tobacco leaf epidermal cells, CK1.3-mCherry and CK1.4-mCherry were subcloned into a modified pCambia1300 vector containing mCherry. The transformation was performed according to a previous method (Van den Ackerveken et al., 1996).
In Vitro Binding Assays
GST-CK1.3 and GST-CK1.4 proteins (5 µg) were incubated separately with 15 μL of glutathione Sepharose 4B beads (Novagen) in 300 μL binding buffer (20 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1 mM EDTA, 0.2% Triton X-100, 10% glycerol, 0.5 mM phenylmethylsulfonyl fluoride, and 1 mM DTT) containing 1× complete protease inhibitor cocktail at 4°C for 1 h. The beads were then washed three times with binding buffer (1 mL). His-CRY2 proteins (5 µg) were then added, and the samples were incubated on a rotating shaker at 25°C for 3 h under dark or blue light. The beads were pelleted by centrifugation and washed five times with ice-cold washing buffer (50 mM Tris-HCl, pH 8.0, 140 mM NaCl, 1 mM EDTA, and 0.1% Triton X-100). The washed pellet was resuspended in 2× SDS sample buffer, and proteins were separated by SDS-PAGE and transferred to PVDF membrane for protein gel blot analysis. The prey (His-CRY2) was detected by a mouse His-tag antibody (1:3000; Santa Cruz Biotechnology), and equal bait (GST-CK1.3 and GST-CK1.4) input was confirmed by a mouse GST-tag antibody (1:3000; Santa Cruz Biotechnology).
GUS Staining Assays
Histochemical GUS assays were performed according to standard protocols (Sessions et al., 1999) with minor modifications. Tissues were placed directly into GUS reaction buffer (0.5 mg/mL X-glucuronic acid, 10 mM EDTA, 0.1% Triton X-100, and 2 mM potassium ferri/ferrocyanide in 50 mM phosphate buffer, pH 7.0) and incubated overnight at 37°C after vacuumization for 20 min. Stained tissues were cleared in 70% ethanol and observed using differential interference contrast microscopy (Nikon SMZ1500).
Phylogenetic Analysis
Sequences of homologous CK1 from Arabidopsis, rice (Oryza sativa), and Homo sapiens were obtained from the National Center for Biotechnology Information website (http://www.ncbi.nlm.nih.gov/) and the Rice Genome Annotation Project (http://rice.plantbiology.msu.edu/). The conserved domains of proteins were analyzed using the website http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi. As all CK1 homologs have Ser/Thr protein kinase domains, these conserved domains were used for phylogenetic analyses. Sequence alignments were generated with CLUSTAL_X 1.81, and the alignments were adjusted prior to construction of the phylogenetic tree. Neighbor-joining analyses were performed using MEGA3.1 (Kumar et al., 2004) with the pairwise deletion option, with a Poisson correction set for distance model and 1000 bootstrap replicates selected.
Sequence alignments data are provided in Supplemental Data Set 1 online.
Accession Numbers
Sequence data for the Arabidopsis genes described in this study can be found in the Arabidopsis Genome Initiative database under the following accession numbers: CK1.3, At4g28880; CK1.4, At4g28860; CRY2, At1g04400; FT, At1g65480; CO, At5g15840; and ACTIN7, At5g09810. The accession numbers of peptide sequences for phylogenetic analysis were as follows: from H. sapiens, CK1α, NP_001020276.1; CK1ε, NP_001885.1; CK1δ, ABM64211.1; CK1γ, NP_071331.2; from rice, EL1, NP_001051531; CKI, NP_001053309; Os01g51200, BAB92346; Os01g13060, NP_001042496; Os01g38950, NP_001043372; Os02g40860, NP_001047465; Os02g56560, EAZ25030; Os02g17910, NP_001046556; Os05g51560, NP_001056503; Os10g33650, NP_001064847; Os05g11140, Os01g10150, Os01g54100, Os01g56580, and Os07g07560 from the Rice Genome Annotation Project; from Arabidopsis, CKL1, NP_193170; CKL2, AAY24533; CK1.3, NP_194617; CK1.4, NP_194615; CKL5, NP_179537; CKL6, NP_567812; CKL7, NP_199223; CKL8, NP_199146; CKL9a, AAY24537; CKL9b, AAY24538; CKL10, NP_188976; CKL11, AAY24540; CKL12, NP_680447; CKL13, NP_171939; At3g03940, NP_187044.1; At3g13670, NP_187977.1; At2g25760, NP_973532.1; At5g18190, and NP_197320.1.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Phylogenetic Analysis of CK1.3 and CK1.4.
Supplemental Figure 2. Amino Acid Sequence Alignment of CK1.3 and CK1.4.
Supplemental Figure 3. Expression Pattern Analysis of CK1.3 and CK1.4.
Supplemental Figure 4. CK1.3-mCherry and CK1.4-mCherry Are Colocalized with CRY2-YFP at Nuclear Bodies after Exposure to Blue Light.
Supplemental Figure 5. Genotyping Analysis of the T-DNA Insertion Mutants of CK1.3 and CK1.4.
Supplemental Figure 6. Phenotypic Analysis of Transgenic Lines with Suppressed Expression of CK1.3.
Supplemental Figure 7. Overexpression of CK1.3 or CK1.4 Results in Delayed Flowering under LD Conditions.
Supplemental Figure 8. The Flowering Time and Rosette Leaf Number of Different Mutants and Transgenic Plants under LD or SD Conditions.
Supplemental Figure 9. Phosphorylation Mimic CRY2Ser587AspThr603Asp Version Shows Significantly Increased Degradation When Exposed to Blue Light.
Supplemental Figure 10. Expression of CK1.3 and CK1.4 under Long-Day Conditions.
Supplemental Table 1. Candidate CK1 Phosphorylation Sites of CRY2, Predicted by the Scansite Program.
Supplemental Table 2. Primers Used for Mutant Genotyping and Plasmid Construction.
Supplemental Table 3. Double Mutants and the Parents Used for Genetic Crosses.
Supplemental Data Set 1. Sequence Alignments Used for Phylogenetic Analysis.
Acknowledgments
The study was supported by the National Science Foundation of China (91117009 and 31130060). We thank Hong-Quan Yang (Shanghai JiaoTong University, China) for kindly providing the Arabidopsis seeds cry1, cry2, and CRY2ox, the pHB vector, and the CRY2 antibody. We also thank Detlef Weigel (Max-Planck-Institute for Developmental Biology, Germany) for kindly providing the vector pRS300 and Peng Zhang (Shanghai Institute of Plant Physiology and Ecology, Chinese Academy of Sciences) for help with expressing CRY2 in insect cells.
AUTHOR CONTRIBUTIONS
S.-T.T. performed the experiments and helped to write the article. C.D. performed the experiments. H.-T.L. analyzed the results. H.-W.X. designed the experiments and wrote the article.
Glossary
- qRT-PCR
quantitative real-time RT-PCR
- GUS
β-glucuronidase
- LD
long-day
- SD
short-day
- GST
glutathione S-transferase
- MS
Murashige and Skoog
- PVDF
polyvinylidene difluoride
References
- Ahmad M., Cashmore A.R. (1993). HY4 gene of A. thaliana encodes a protein with characteristics of a blue-light photoreceptor. Nature 366: 162–166 [DOI] [PubMed] [Google Scholar]
- Ahmad M., Jarillo J.A., Smirnova O., Cashmore A.R. (1998a). Cryptochrome blue-light photoreceptors of Arabidopsis implicated in phototropism. Nature 392: 720–723 [DOI] [PubMed] [Google Scholar]
- Ahmad M., Jarillo J.A., Smirnova O., Cashmore A.R. (1998b). The CRY1 blue light photoreceptor of Arabidopsis interacts with phytochrome A in vitro. Mol. Cell 1: 939–948 [DOI] [PubMed] [Google Scholar]
- Akashi M., Tsuchiya Y., Yoshino T., Nishida E. (2002). Control of intracellular dynamics of mammalian period proteins by casein kinase I ε (CKIepsilon) and CKIdelta in cultured cells. Mol. Cell. Biol. 22: 1693–1703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Ben-Nissan G., Cui W., Kim D.J., Yang Y., Yoo B.C., Lee J.Y. (2008). Arabidopsis casein kinase 1-like 6 contains a microtubule-binding domain and affects the organization of cortical microtubules. Plant Physiol. 148: 1897–1907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouly J.P., Giovani B., Djamei A., Mueller M., Zeugner A., Dudkin E.A., Batschauer A., Ahmad M. (2003). Novel ATP-binding and autophosphorylation activity associated with Arabidopsis and human cryptochrome-1. Eur. J. Biochem. 270: 2921–2928 [DOI] [PubMed] [Google Scholar]
- Cashmore A.R., Jarillo J.A., Wu Y.J., Liu D. (1999). Cryptochromes: Blue light receptors for plants and animals. Science 284: 760–765. [DOI] [PubMed] [Google Scholar]
- Clough, S.J., and Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Dai C., Xue H.W. (2010). Rice early flowering1, a CKI, phosphorylates DELLA protein SLR1 to negatively regulate gibberellin signalling. EMBO J. 29: 1916–1927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eide E.J., Vielhaber E.L., Hinz W.A., Virshup D.M. (2002). The circadian regulatory proteins BMAL1 and cryptochromes are substrates of casein kinase Iepsilon. J. Biol. Chem. 277: 17248–17254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green C.B., Takahashi J.S., Bass J. (2008). The meter of metabolism. Cell 134: 728–742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross S.D., Anderson R.A. (1998). Casein kinase I: Spatial organization and positioning of a multifunctional protein kinase family. Cell. Signal. 10: 699–711 [DOI] [PubMed] [Google Scholar]
- Gudesblat G.E., Schneider-Pizoń J., Betti C., Mayerhofer J., Vanhoutte I., van Dongen W., Boeren S., Zhiponova M., de Vries S., Jonak C., Russinova E. (2012). SPEECHLESS integrates brassinosteroid and stomata signalling pathways. Nat. Cell Biol. 14: 548–554 [DOI] [PubMed] [Google Scholar]
- Guo H., Yang H., Mockler T.C., Lin C. (1998). Regulation of flowering time by Arabidopsis photoreceptors. Science 279: 1360–1363 [DOI] [PubMed] [Google Scholar]
- Kleiner O., Kircher S., Harter K., Batschauer A. (1999). Nuclear localization of the Arabidopsis blue light receptor cryptochrome 2. Plant J. 19: 289–296 [DOI] [PubMed] [Google Scholar]
- Klimczak L.J., Cashmore A.R. (1993). Purification and characterization of casein kinase I from broccoli. Biochem. J. 293: 283–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knippschild U., Gocht A., Wolff S., Huber N., Löhler J., Stöter M. (2005). The casein kinase 1 family: Participation in multiple cellular processes in eukaryotes. Cell. Signal. 17: 675–689 [DOI] [PubMed] [Google Scholar]
- Kumar S., Tamura K., Nei M. (2004). MEGA3: Integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Brief. Bioinform. 5: 150–163 [DOI] [PubMed] [Google Scholar]
- Lamia K.A., Sachdeva U.M., DiTacchio L., Williams E.C., Alvarez J.G., Egan D.F., Vasquez D.S., Juguilon H., Panda S., Shaw R.J., Thompson C.B., Evans R.M. (2009). AMPK regulates the circadian clock by cryptochrome phosphorylation and degradation. Science 326: 437–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.Y., Taoka K.I., Yoo B.C., Ben-Nissan G., Kim D.J., Lucas W.J. (2005). Plasmodesmal-associated protein kinase in tobacco and Arabidopsis recognizes a subset of non-cell-autonomous proteins. Plant Cell 17: 2817–2831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian H.L., He S.B., Zhang Y.C., Zhu D.M., Zhang J.Y., Jia K.P., Sun S.X., Li L., Yang H.Q. (2011). Blue-light-dependent interaction of cryptochrome 1 with SPA1 defines a dynamic signaling mechanism. Genes Dev. 25: 1023–1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C. (2002). Blue light receptors and signal transduction. Plant Cell 14 (suppl.): S207–S225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C., Yang H., Guo H., Mockler T.C., Chen J., Cashmore A.R. (1998). Enhancement of blue-light sensitivity of Arabidopsis seedlings by a blue light receptor cryptochrome 2. Proc. Natl. Acad. Sci. USA 95: 2686–2690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B., Zuo Z., Liu H., Liu X., Lin C. (2011). Arabidopsis cryptochrome 1 interacts with SPA1 to suppress COP1 activity in response to blue light. Genes Dev. 25: 1029–1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Liu B., Zhao C., Pepper M., Lin C. (2011). The action mechanisms of plant cryptochromes. Trends Plant Sci. 16: 684–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Yu X., Li K., Klejnot J., Yang H., Lisiero D., Lin C. (2008). Photoexcited CRY2 interacts with CIB1 to regulate transcription and floral initiation in Arabidopsis. Science 322: 1535–1539 [DOI] [PubMed] [Google Scholar]
- Liu W., Xu Z.H., Luo D., Xue H.W. (2003). Roles of OsCKI1, a rice casein kinase I, in root development and plant hormone sensitivity. Plant J. 36: 189–202 [DOI] [PubMed] [Google Scholar]
- Mockler T.C., Guo H., Yang H., Duong H., Lin C. (1999). Antagonistic actions of Arabidopsis cryptochromes and phytochrome B in the regulation of floral induction. Development 126: 2073–2082 [DOI] [PubMed] [Google Scholar]
- Møller S.G., Kim Y.S., Kunkel T., Chua N.H. (2003). PP7 is a positive regulator of blue light signaling in Arabidopsis. Plant Cell 15: 1111–1119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pusch S., Harashima H., Schnittger A. (2012). Identification of kinase substrates by bimolecular complementation assays. Plant J. 70: 348–356 [DOI] [PubMed] [Google Scholar]
- Sang Y., Li Q.H., Rubio V., Zhang Y.C., Mao J., Deng X.W., Yang H.Q. (2005). N-terminal domain-mediated homodimerization is required for photoreceptor activity of Arabidopsis CRYPTOCHROME1. Plant Cell 17: 1569–1584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessions A., Weigel D., Yanofsky M.F. (1999). The Arabidopsis thaliana MERISTEM LAYER 1 promoter specifies epidermal expression in meristems and young primordia. Plant J. 20: 259–263 [DOI] [PubMed] [Google Scholar]
- Shalitin D., Yang H., Mockler T.C., Maymon M., Guo H., Whitelam G.C., Lin C. (2002). Regulation of Arabidopsis cryptochrome 2 by blue-light-dependent phosphorylation. Nature 417: 763–767 [DOI] [PubMed] [Google Scholar]
- Shalitin D., Yu X., Maymon M., Mockler T., Lin C. (2003). Blue light-dependent in vivo and in vitro phosphorylation of Arabidopsis cryptochrome 1. Plant Cell 15: 2421–2429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaner N.C., Campbell R.E., Steinbach P.A., Giepmans B.N.G., Palmer A.E., Tsien R.Y. (2004). Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat. Biotechnol. 22: 1567–1572 [DOI] [PubMed] [Google Scholar]
- Song Y.H., Smith R.W., To B.J., Millar A.J., Imaizumi T. (2012). FKF1 conveys timing information for CONSTANS stabilization in photoperiodic flowering. Science 336: 1045–1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van den Ackerveken G., Marois E., Bonas U. (1996). Recognition of the bacterial avirulence protein AvrBs3 occurs inside the host plant cell. Cell 87: 1307–1316 [DOI] [PubMed] [Google Scholar]
- Vielhaber E., Eide E., Rivers A., Gao Z.H., Virshup D.M. (2000). Nuclear entry of the circadian regulator mPER1 is controlled by mammalian casein kinase I epsilon. Mol. Cell. Biol. 20: 4888–4899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Ma L.G., Li J.M., Zhao H.Y., Deng X.W. (2001). Direct interaction of Arabidopsis cryptochromes with COP1 in light control development. Science 294: 154–158 [DOI] [PubMed] [Google Scholar]
- Yang H.Q., Tang R.H., Cashmore A.R. (2001). The signaling mechanism of Arabidopsis CRY1 involves direct interaction with COP1. Plant Cell 13: 2573–2587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H.Q., Wu Y.J., Tang R.H., Liu D., Liu Y., Cashmore A.R. (2000). The C termini of Arabidopsis cryptochromes mediate a constitutive light response. Cell 103: 815–827 [DOI] [PubMed] [Google Scholar]
- Yoo S.D., Cho Y.H., Sheen J. (2007). Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2: 1565–1572 [DOI] [PubMed] [Google Scholar]
- Yu X., Klejnot J., Zhao X., Shalitin D., Maymon M., Yang H., Lee J., Liu X., Lopez J., Lin C. (2007a). Arabidopsis cryptochrome 2 completes its posttranslational life cycle in the nucleus. Plant Cell 19: 3146–3156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X., Sayegh R., Maymon M., Warpeha K., Klejnot J., Yang H., Huang J., Lee J., Kaufman L., Lin C. (2009). Formation of nuclear bodies of Arabidopsis CRY2 in response to blue light is associated with its blue light-dependent degradation. Plant Cell 21: 118–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X., Shalitin D., Liu X., Maymon M., Klejnot J., Yang H., Lopez J., Zhao X., Bendehakkalu K.T., Lin C. (2007b). Derepression of the NC80 motif is critical for the photoactivation of Arabidopsis CRY2. Proc. Natl. Acad. Sci. USA 104: 7289–7294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J.J., Xue H.W. (2013). OsLEC1/OsHAP3E participates in the determination of meristem identity in both vegetative and reproductive developments of rice. J. Integr. Plant Biol. 55: 232–249 [DOI] [PubMed] [Google Scholar]
- Zuo Z., Liu H., Liu B., Liu X., Lin C. (2011). Blue light-dependent interaction of CRY2 with SPA1 regulates COP1 activity and floral initiation in Arabidopsis. Curr. Biol. 21: 841–847 [DOI] [PMC free article] [PubMed] [Google Scholar]