Here, we characterize a protein family that forms a complex by oligomerization, which is required for bending of the thylakoid membrane. Without this complex, the typical thylakoid ultrastructure of land plant chloroplasts, composed of grana stacks and stroma lamellae, cannot develop. Strikingly, the loss of this active compartmentalization within thylakoids affects photosynthesis only moderately.
Abstract
Chloroplasts of land plants characteristically contain grana, cylindrical stacks of thylakoid membranes. A granum consists of a core of appressed membranes, two stroma-exposed end membranes, and margins, which connect pairs of grana membranes at their lumenal sides. Multiple forces contribute to grana stacking, but it is not known how the extreme curvature at margins is generated and maintained. We report the identification of the CURVATURE THYLAKOID1 (CURT1) protein family, conserved in plants and cyanobacteria. The four Arabidopsis thaliana CURT1 proteins (CURT1A, B, C, and D) oligomerize and are highly enriched at grana margins. Grana architecture is correlated with the CURT1 protein level, ranging from flat lobe-like thylakoids with considerably fewer grana margins in plants without CURT1 proteins to an increased number of membrane layers (and margins) in grana at the expense of grana diameter in overexpressors of CURT1A. The endogenous CURT1 protein in the cyanobacterium Synechocystis sp PCC6803 can be partially replaced by its Arabidopsis counterpart, indicating that the function of CURT1 proteins is evolutionary conserved. In vitro, Arabidopsis CURT1A proteins oligomerize and induce tubulation of liposomes, implying that CURT1 proteins suffice to induce membrane curvature. We therefore propose that CURT1 proteins modify thylakoid architecture by inducing membrane curvature at grana margins.
INTRODUCTION
Thylakoids, the internal membranes of chloroplasts and cyanobacteria, accommodate the light reactions of photosynthesis. Chloroplasts of land plants contain grana, characteristic cylindrical stacks with a typical diameter of 300 to 600 nm comprising approximately five to 20 layers of thylakoid membrane (Mustárdy and Garab, 2003; Mullineaux, 2005). A single granum consists of a central core of appressed membranes, two stroma-exposed membranes at the top and bottom of the cylindrical structure, and the highly curved margins that merge two grana membranes at their periphery. Grana stacks are interconnected by stroma-exposed membrane pairs of up to a few micrometers in length, the stroma lamellae. All thylakoid membranes within one chloroplast form a continuous network that encloses a single lumenal space (Shimoni et al., 2005). The topography of this network, as well as the precise three-dimensional structure of grana themselves, remains a much debated issue (Allen and Forsberg, 2001; Shimoni et al., 2005; Brumfeld et al., 2008; Mustárdy et al., 2008; Austin and Staehelin, 2011; Daum and Kühlbrandt, 2011). Although grana are ubiquitous in land plants, the fraction of thylakoid membrane found in stroma lamellae appears to be remarkably constant among species (Albertsson and Andreasson, 2004).
Grana and stroma thylakoids differ strikingly in their protein composition (lateral heterogeneity) (Dekker and Boekema, 2005). Photosystem II (PSII) and its light-harvesting complex (LHCII) are concentrated in grana, whereas photosystem I (PSI) and the chloroplast ATP synthase, which protrude extensively from the membrane into the stroma, are excluded from the grana core and reside in the stroma-facing regions. The primary purpose of grana is debated, and suggested functions include prevention of spillover of excitation energy through physical separation of photosystems, fine-tuning of photosynthesis, facilitation of state transitions, and switching between linear and cyclic electron flow and, in particular, enhancing light harvesting under low-light conditions through the formation of large arrays of PSII-LHCII supercomplexes (Trissl and Wilhelm, 1993; Mustárdy and Garab, 2003; Dekker and Boekema, 2005; Mullineaux, 2005; Anderson et al., 2008; Daum and Kühlbrandt, 2011). However, grana formation also imposes constraints on photosynthesis, such as the requirement for long-range diffusion of electron carriers between PSII and PSI (Mullineaux, 2008; Kirchhoff et al., 2011) and the relocation of PSII between appressed and nonappressed regions during the PSII repair cycle (Mulo et al., 2008).
The formation of the intricate network of thylakoid membranes involves forces that mediate the stacking and bifurcation of membranes, as well as mechanisms that promote the curvature of the membranes at the sites of cylindrical grana stacks. Whereas the latter mechanisms are as yet unknown, it is generally accepted that stacking results from the interplay of physicochemical forces of attraction and repulsion between adjacent membranes (Rubin and Barber, 1980; Chow et al., 2005; Anderson et al., 2008). Moreover, lipid composition and lipid–protein interactions are also thought to play a role (Gounaris and Barber, 1983; Webb and Green, 1991; Dekker and Boekema, 2005). Thus, plants adapted to shade and low-light conditions have many more layers of thylakoid membranes per granum than those that prefer bright sunlight (Anderson, 1986). Furthermore, because the diameter of grana increases when thylakoid proteins are less phosphorylated, light-induced PSII phosphorylation has been proposed to increase repulsion between adjacent thylakoid membrane layers (Fristedt et al., 2009).
Here, we show that the CURVATURE THYLAKOID1 (CURT1) protein family, whose members are capable of forming oligomers, can induce membrane curvature in vitro. CURT1 proteins are located in the grana margins and control grana formation in Arabidopsis thaliana, as demonstrated by the analysis of knockout and overexpressor lines. In the absence of CURT1 proteins, chloroplasts contain flat lobe-like thylakoids with considerably fewer grana margins and exhibit a general impairment in photosynthesis, whereas CURT1 overexpression results in taller and slimmer grana stacks with more margins and thylakoid membrane layers. The endogenous CURT1 protein in the cyanobacterium Synechocystis sp PCC6803 can be partially replaced by its Arabidopsis counterpart, indicating that the function of CURT1 proteins is evolutionarily conserved.
RESULTS
Thylakoids Contain CURT1 Oligomers
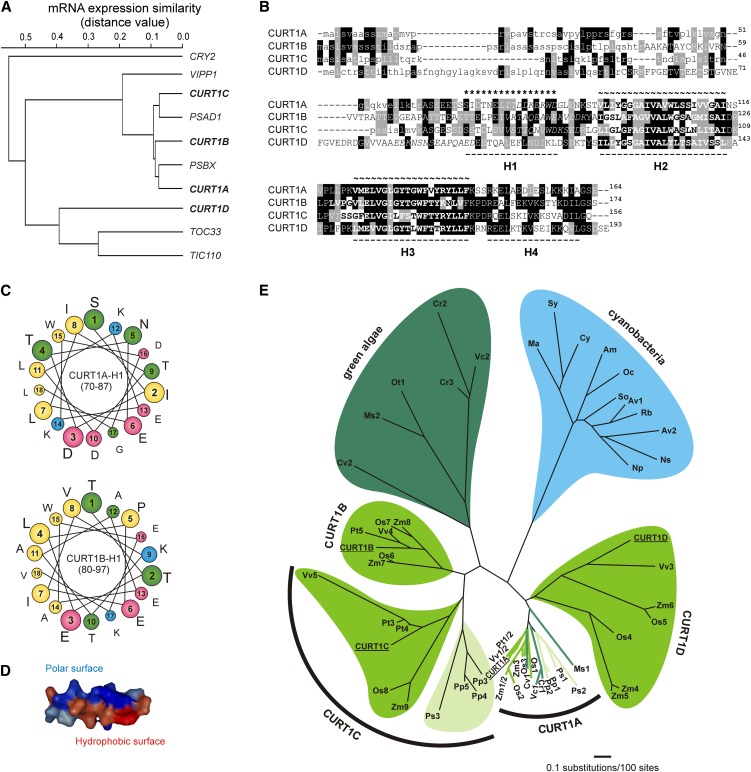
Nuclear photosynthetic genes are often transcriptionally coregulated in Arabidopsis (Biehl et al., 2005), and this transcriptional signature can be used to identify photosynthetic genes (DalCorso et al., 2008). To identify additional components or regulators of photosynthesis, we therefore isolated three Arabidopsis genes of unknown function but featuring this transcriptional signature (Figure 1A), here designated CURT1A, B, and C (because of their function in the curvature of thylakoids; see below). The CURT1 genes belong to a small gene family with four members. Transcripts encoding CURT1D, the fourth member of the family, cluster with transcripts for nonphotosynthetic proteins (Figure 1A). All four CURT1 proteins share a predicted N-terminal chloroplast targeting peptide, two predicted transmembrane domains (TMs), and a tentative N-terminal amphipathic helix and are relatively small (11.0 to 15.7 kD; Figures 1B to 1D). Genes for CURT1 homologs exist in the nuclear genomes of green algae, plants, and, more distantly related, also in cyanobacteria (Figure 1E).
Figure 1.
Characteristics of CURT1 Proteins.
(A) Expression characteristics of CURT1 protein family genes. CURT1A, B, C, and D, together with the chloroplast protein coding genes TIC110, TOC33, PSBX, PSAD1, and VIPP1 and the nonchloroplast protein coding gene CRY2, were hierarchically clustered according to expression profile similarity using the gene coexpression database ATTEDII (http://atted.jp/) (Obayashi et al., 2007). Low distance values indicate high expression similarities.
(B) Sequence alignment of the CURT1 proteins in Arabidopsis. Predicted chloroplast transit peptide sequences are shown in lowercase letters, positions of α-helices (H1-H4) are indicated, and the two TMs (∼) are highlighted in bold. Peptide sequences used for antibody generation are depicted in italics. Identical and closely related amino acids that are conserved in >50% of the sequences are highlighted by black and gray shading, respectively.
(C) CURT1 proteins contain a conserved putative N-terminal amphipathic helix. Helical wheel representations of helix H1 (as defined in [B]) are shown for CURT1A (CURT1A-H1) and CURT1B (CURT1B-H1). In the helical wheel representation, the amino acids are colored according to the physicochemical properties of the side chains (yellow, hydrophobic; blue, polar, positively charged; pink, polar, negatively charged; green, polar, uncharged). Both helical wheel representations show a significant spatial separation of polar and charged from hydrophobic amino acid residues, indicative of an amphipathic helix.
(D) Three-dimensional model of CURT1A-H1. The computed surfaces are gradually colored according to their hydrophobicity from dark red (highly hydrophobic) to deep blue (highly polar).
(E) Phylogenetic analysis of the CURT1 protein family. Branch lengths reflect the estimated number of substitutions per 100 sites. The cyanobacterial and green algal clades are highlighted by blue and dark-green background/branch color, respectively. Bryophytes and gymnosperms are indicated by light-green background/branch color and magnoliophytes (flowering plants) by green color.
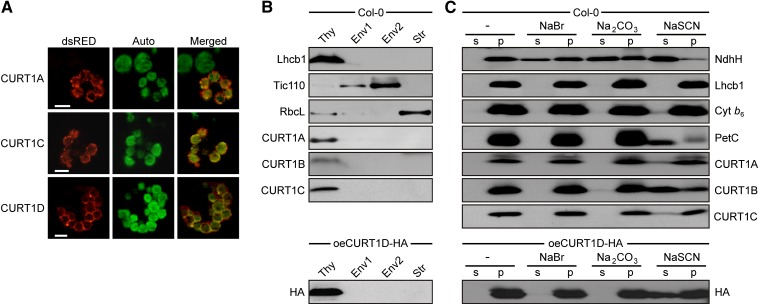
CURT1B was already shown before to be located in thylakoids and designated at this time as “thylakoid membrane phosphoprotein of 14 kD” (TMP14), simply referring to its subcellular location and its molecular mass (Hansson and Vener, 2003). Subsequently, TMP14 was proposed to represent a novel subunit of plant PSI and renamed “PsaP” (Khrouchtchova et al., 2005). Because the protein is not a PSI subunit (see below), we propose to rename “TMP14/PsaP” to “CURT1B” and use this designation in the following. Each of the other three Arabidopsis CURT1 proteins has been detected at least once in chloroplasts in proteomic studies (Friso et al., 2004; Peltier et al., 2004) (see Supplemental Table 1 online). In fact, in vivo subcellular localization of full-length N-terminal fusions of CURT1A, C, and D to the red fluorescent dsRED reporter protein showed that all three proteins are chloroplast located (Figure 2A). When wild-type chloroplasts were fractionated into thylakoids, stroma, and two envelope fractions and subjected to immunoblot analysis, CURT1A, B, and C were exclusively detected in thylakoids (Figure 2B). Overexpression of the low-abundant CURT1D (see Supplemental Figure 1A online) with a C-terminal HA-tag (oeCURT1D-HA) confirmed that CURT1D is also a thylakoid protein (Figure 2B).
Figure 2.
CURT1 Proteins Are Intrinsic Thylakoid Proteins.
(A) Subcellular localization of CURT1 proteins. Full-length CURT1A-, CURT1C-, and CURT1D-dsRED fusions were transiently introduced into Arabidopsis protoplasts by polyethylene glycol–mediated DNA uptake and analyzed using fluorescence microscopy (Auto, chloroplasts revealed by chlorophyll autofluorescence; dsRED, fluorescence of the fusion protein; Merged, overlay of both images). Bar = 10 μm.
(B) Suborganellar localization of CURT1 proteins. Chloroplasts from wild-type and oeCURT1D-HA plants were subfractionated into thylakoids (Thy), stroma (Str), and two envelope (Env) fractions. Aliquots (40 µg) of protein from each fraction were subjected to SDS-PAGE, followed by immunoblot analysis using antibodies raised against CURT1A, B, C, or HA. As controls for purity of the different fractions, antibodies recognizing Lhcb1, Tic110, and RbcL, which are located in thylakoids, envelope, and stroma, respectively, were used.
(C) Extraction of thylakoid-associated proteins with chaotropic salt solutions or alkaline pH. Thylakoids from wild-type and oeCURT1D-HA plants were resuspended at 0.5 mg chlorophyll/mL in 10 mM HEPES/KOH, pH 7.5, containing either 2 M NaBr, 0.1 M Na2CO3, 2 M NaSCN, or no additive. After incubation, supernatants containing the extracted proteins (s) and membrane fractions (p) were separated by SDS-PAGE and immunolabeled with antibodies against CURT1A, B, C, and HA. As control for peripheral membrane proteins, antibodies raised against NdhH were used. To control for integral membrane proteins, antibodies specific for Lhcb1, cytochrome b6, and Rieske (PetC) were employed.
To clarify whether the CURT1 proteins constitute integral or peripheral thylakoid proteins, wild-type (Columbia-0 [Col-0]) thylakoids were treated with alkaline and chaotropic (protein disrupting) salts to release membrane-associated proteins (Figure 2C). In this assay, CURT1A and C behaved like the integral proteins LHCII protein 1 (Lhcb1) (with three TMs) and cytochrome b6 (four TMs), whereas CURT1B and CURT1D-HA could be partially extracted from thylakoids with NaSCN, similar to the Rieske protein PetC (with one TM).
To determine the membrane topology of the CURT1 proteins, thylakoids of wild-type plants and of lines expressing HA-tagged versions of CURT1 proteins (see Supplemental Figure 1B online) were subjected to mild digestion with trypsin such that only the stroma-exposed face was accessible to the protease. The fragmentation pattern (see Supplemental Figures 1C and 1D online) and phosphorylation behavior (see Supplemental Table 1 and Supplemental References 1 online) of CURT1 proteins, as well as additional experimental evidence (see legend of Supplemental Figure 1D online), suggest that they are located in the thylakoid membrane with their N- and C termini facing the stroma.
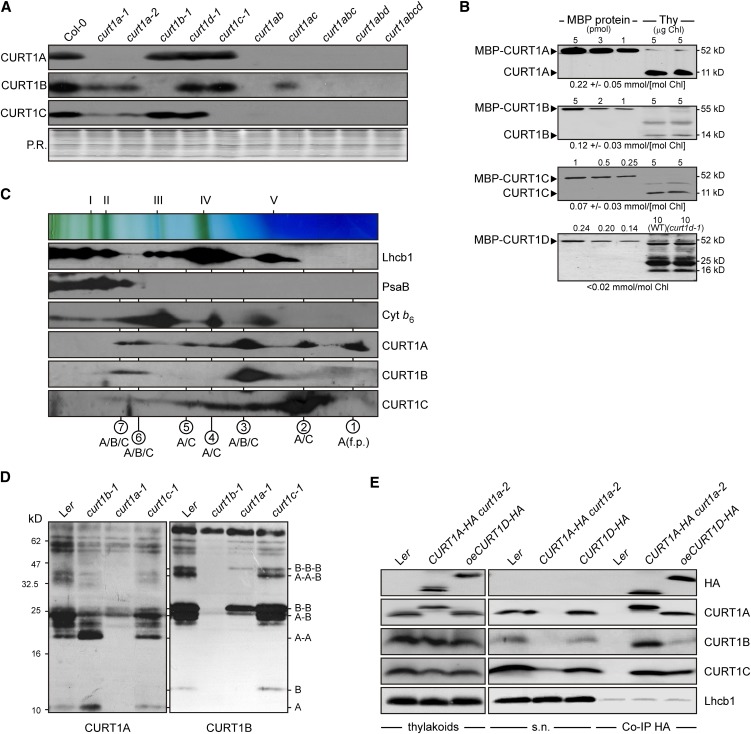
Starting from insertion mutants for the four Arabidopsis CURT1 genes (see Supplemental Figure 2 and Supplemental References 1 online), double and triple mutants and the quadruple mutant were generated. Lack of CURT1D and, in particular, CURT1A decreased the abundance of other CURT1 proteins (Figure 3A; see Supplemental Figures 3A to 3D online). In the four double mutants analyzed, a superadditive effect was observed, whereas in the two triple mutants (curt1abc and curt1abd) and the quadruple mutant (curt1abcd), no CURT1 protein was detectable at all.
Figure 3.
CURT1 Proteins Form Oligomers.
(A) Abundance of CURT1 proteins in curt1 mutant plants. Total protein extracts from Col-0 and curt1 mutant plants corresponding to 3 µg of chlorophyll were fractionated by SDS-PAGE, and blots were probed with antibodies raised against CURT1A, B, and C. The Ponceau Red (P.R.)–stained protein blot served as loading control.
(B) Absolute abundance of CURT1 proteins in wild-type (WT) plants. The four Arabidopsis CURT1 proteins were expressed in Escherichia coli as C-terminal fusions to the MBP, purified by affinity chromatography, and quantified. Adequate quantities of these four MBP fusions, as well as of the MBP fusions of PetC and PsaD as controls (see Supplemental Figure 3E online), were titrated against thylakoid membrane protein preparations and subjected to immunoblot analyses. Representative results from five experiments are shown. The calculated concentration of the respective CURT1 protein in thylakoid preparations is given below each panel.
(C) Two-dimensional BN/SDS-PAGE separation of thylakoid protein complexes. Individual lanes from BN-PA gels like those shown on top were separated in a second dimension by SDS-PAGE. Blots were immunolabeled with antibodies raised against Lhcb1, PsaB, cytochrome b6, and CURT1A, B, and C. The positions of major thylakoid multiprotein complexes are indicated by Roman numerals (top) and the composition of the complexes by circled Arabic numbers (bottom) as in Supplemental Figure 3F online. f.p., free protein.
(D) Chemical cross-linking. Thylakoid proteins from wild-type (Landsberg erecta [Ler]) and mutant (curt1b-1, curt1a-1, and curt1c-1) plants were cross-linked with bis(sulfosuccinimidyl) suberate, separated by SDS-PAGE, and subjected to immunoblot analysis with antibodies raised against CURT1A and B. On the right, the protein composition of cross-linked products is shown. As loading control, blots were stained with Ponceau Red (see Supplemental Figure 3H online).
(E) CoIP. Thylakoid membranes from wild-type, CURT1A-HA curt1a-2, and oeCURT1D-HA plants were solubilized, and HA-tagged proteins were allowed to bind to α-HA affinity matrix. The matrix was recovered and the supernatant was collected. After washing, coimmunoprecipitated proteins were eluted. Thylakoids, supernatant (s.n.), and coimmunoprecipitated proteins (Co-IP HA) were then analyzed by SDS-PAGE followed by immunoblot analysis with antibodies raised against HA, CURT1A, B, and C, and Lhcb1.
To estimate their absolute abundance, known quantities of C-terminal fusions of the four CURT1 proteins to the maltose binding protein (MBP), as well as of MBP fusions of PetC from the cytochrome b6/f complex and the PSI subunit PsaD as controls, were titrated against thylakoid preparations and subjected to immunoblot analysis. Levels of PetC (∼1.35 mmol/mol chlorophyll) and PsaD (∼2.16 mmol/mol chlorophyll) found in thylakoid preparations (see Supplemental Figure 3E online) were in good agreement with published data (Kirchhoff et al., 2002). CURT1A was the most abundant CURT1 protein (∼0.22 mmol/mol chlorophyll), followed by CURT1B (∼0.12 mmol/mol chlorophyll) and CURT1C (∼0.07 mmol/mol chlorophyll), whereas CURT1D was expressed at very low levels (Figure 3B).
The interdependence of CURT1 protein accumulation suggests that the proteins physically interact. To further study these interactions, immunoblot analysis of thylakoid protein complexes separated by Blue-Native PAGE (BN-PAGE) were performed. Seven CURT1A-containing complexes, none of which comigrated with the known high-abundance complexes (Pesaresi et al., 2009b; Armbruster et al., 2010), were identified in wild-type plants (see Supplemental Figure 3F online). The complexes resolved by BN-PAGE were then separated into their subunits by SDS-PAGE in the second dimension and subjected to immunoblot analysis. This analysis showed that three complexes contained all three CURT1 proteins tested, while CURT1A and C are both present in an additional three complexes (Figure 3C). In the single mutants curt1a-1, curt1b-1, and curt1c-1, the remaining two CURT1 proteins still comigrate, forming in curt1b-1 and curt1c-1 some complexes with an even higher molecular weight than in the wild type (see Supplemental Figure 3G online).
To corroborate the interactions between CURT1 proteins, thylakoid proteins were subjected to chemical cross-linking. Comparison of the pattern of signals obtained after SDS-PAGE and immunoblot analysis from cross-linked wild-type thylakoid proteins with the ones obtained from curt1a-1, curt1b-1, and curt1c-1 mutants showed that CURT1A and B are present in several distinct complexes, forming homodimers, heterodimers, homotrimers (CURT1B), and heterotrimers (Figure 3D; see Supplemental Figure 3H online). Moreover, coimmunoprecipitation (CoIP) experiments, performed with thylakoids from plants expressing HA-tagged CURT1A (CURT1A-HA curt1a-2), cmyc-tagged CURT1A, or overexpressing CURT1D-HA (see Supplemental Figure 1 online), confirmed the existence of multiple interactions between different CURT1 proteins, as well as between the same CURT1 proteins (Figure 3E; see Supplemental Figures 3I to 3K online).
CURT1 Depletion Affects Photosynthesis Only Indirectly
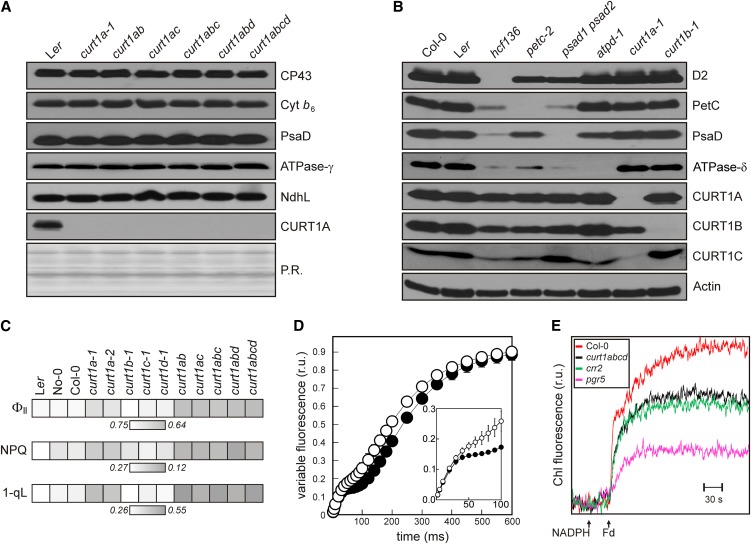
Because of their low abundance, CURT1 proteins are unlikely to be constitutive subunits of the major thylakoid multiprotein complexes. To further test the relationship between CURT1 proteins and other thylakoid proteins, BN- and SDS-PAGE analyses were performed, demonstrating that a lack of even all four CURT1 proteins had no effect on the accumulation of the major thylakoid multiprotein complexes (Figure 4A; see Supplemental Figure 4A online). Vice versa, CURT1 proteins accumulated independently of the major thylakoid protein complexes (Figure 4B). In addition, CURT1B is not a subunit of PSI, contrary to an earlier suggestion (Khrouchtchova et al., 2005) (see Supplemental Figures 4B and 4C online).
Figure 4.
Lack of CURT1 Proteins Does Not Disturb Photosynthetic Complex Accumulation but Affects Photosynthesis Pleiotropically.
(A) The composition of thylakoid complexes from the wild type (Landsberg erecta [Ler]) and curt1 mutants was analyzed as in Figure 3A, except that aliquots corresponding to 5 µg chlorophyll were used. Antibodies specific for the PSII subunit CP43, cytochrome b6 from the cytochrome b6/f complex, the D subunit of PSI (PsaD), the γ-subunit of the chloroplast ATP synthase, NdhL from the NDH complex, and CURT1A were used. Ponceau Red (P.R.) staining served as the loading control.
(B) Immunoblot analysis of leaf proteins (corresponding to 40 µg of total protein) from the wild type (Col-0 and Landsberg erecta), curt1a-1 and curt1b-1 mutants, and mutants devoid of PSII (hcf136), PSI (psad1 psad2), cytochrome b6/f (petc-2), or cpATPase (atpd-1). Antibodies specific for CURT1A, B, and C and the respective thylakoid multiprotein complexes, or actin as control, were used.
(C) Effective quantum yield (ΦII), NPQ, and 1-qL (as listed in Supplemental Table 2 online) for wild-type and curt1 mutant plants are indicated on a gray scale.
(D) Time course of chlorophyll a fluorescence during illumination with actinic light at 100 μmol photons m−2 s−1. Average values ± sd (bars) for three to five different plants are indicated by closed (wild type) or open (curt1abcd) circles. Relative units (r.u.) are shown.
(E) Quantification of cyclic electron transport in situ. Increases in chlorophyll fluorescence were measured in ruptured chloroplasts after the addition of NADPH and Fd as described (Munekage et al., 2002). As controls, the mutants pgr5 (defective in Fd-dependent cyclic electron transport) and crr2 (defective in NDH-dependent cyclic electron transport) were employed.
Mutants lacking one or more CURT1 proteins displayed essentially wild-type growth behavior and leaf coloration, indicating that thylakoid functions are not drastically impaired. Photosynthetic electron flow was not affected in curt1b, curt1c, and curt1d plants (Figure 4C; see Supplemental Table 2 online). However, in curt1a and even more so in double, triple, and quadruple mutants lacking CURT1A, a decrease in the effective quantum yield (ΦII) and nonphotochemical quenching (NPQ) and an increase in excitation pressure (1-qL) (Kramer et al., 2004) were noted. The impairment in photosynthetic electron flow was pronounced at high light intensities (see Supplemental Figure 4D online). On the contrary, the maximum quantum yield of PSII. (Fv/Fm) was wild-type-like for all curt1 mutants (see Supplemental Table 2 online), again indicating that the overall functionality of PSII is not affected. This interpretation was supported by analyses of light-induced fluorescence changes (Figure 4D). Here, the initial fluorescence increase was identical in wild-type and curt1abcd plants, but the plateau following the initial rise was less pronounced in the mutant, indicating that the photochemical efficiency of PSII is unaltered but electron transfer to PSI from the plastoquinone pool (Rappaport et al., 2007) is slightly decreased in curt1abcd plants (Figure 4D). This is consistent with the observation that P700+ rereduction was slightly slower in curt1abcd (see Supplemental Figures 4E and 4F online). Yet, the activity of PSI was not affected in curt1abcd because the oxidation kinetics induced by a saturating light pulse was wild-type-like (see Supplemental Figure 4F online).
To further investigate the perturbation in photosynthesis, cyclic electron transport was analyzed. In curt1abcd, cyclic electron transport was also decreased, as determined by in situ measurements in ruptured chloroplasts and by monitoring the transient induction of NPQ (Figure 4E; see Supplemental Figure 4G online). Because the abundance of the chloroplast NADP(H) dehydrogenase (NDH) subunit L and of the PSI-NDH supercomplex appears to be unchanged in curt1abcd plants (Figure 4A; see Supplemental Figure 4A online), this effect cannot be attributed to overall alterations in the abundance of the NDH complex. Quenching of chlorophyll fluorescence due to state transitions (qT), the reversible association of the mobile pool of LHCII proteins with PSII or PSI, was reduced but not completely abolished in plants lacking CURT1 proteins (the wild type, 0.13 ± 0.01; curt1abcd, 0.04 ± 0.01).
CURT1 Proteins Modify Thylakoid Architecture
The pleiotropic effects on photosynthesis observed in plants without CURT1 proteins suggest that thylakoids might be affected at a more basic level. We therefore examined the thylakoid lipids and found that their composition was not markedly altered in curt1 mutants (see Supplemental Table 3 online). We then investigated the effect of curt1 mutations on the architecture of the thylakoid system by transmission electron microscopy (TEM) (Figure 5A). To this end, transmission electron micrographs of leaf sections from two individual plants of each genotype were analyzed regarding the width and height of their grana stacks. Of each genotype, at least six chloroplasts were surveyed, with a minimum of 12 grana stacks per chloroplast being measured. From these data, average values and standard errors were calculated (see Supplemental Data Set 1 online). In wild-type chloroplasts, the average diameter of grana stacks ranges between 0.41 ± 0.01 and 0.47 ± 0.01 µm (depending on ecotype) and average height is around 0.11 ± 0.01 µm (Figure 5B; see Supplemental Data Set 1 online). The corresponding values for curt1abcd plants, on the other hand, were 1.66 ± 0.08 µm and 0.06 ± 0.00 µm (Figure 5B; see Supplemental Data Set 1 online), as thylakoids were disorganized with extended stretches of unstacked membranes and broader stacks made up of fewer layers than in the wild type (Figure 5A). Moreover, in double, triple, and quadruple mutants, thylakoids were occasionally curved instead of forming horizontal layers and also contained vesicular structures (Figure 5C). The wild-type-like grana phenotype of the three single mutants curt1b-1, curt1c-1, and curt1d-1 and the intermediate phenotype of curt1a plants (average grana stack diameter/height of 1.32 ± 0.05/0.08 ± 0.00 µm) is in accordance with their photosynthetic phenotype (see above). Because thylakoids that lack grana stacks represent the primordial type and are ubiquitous in cyanobacteria, it is not surprising that curt1abcd plants are still capable of performing photosynthesis (see above), although they are virtually devoid of grana. However, the complex perturbations in photosynthesis found in curt1abcd plants are compatible with the fact that important features of photosynthesis in land plants are facilitated by the lateral heterogeneity provided by grana (Trissl and Wilhelm, 1993; Mustárdy and Garab, 2003; Dekker and Boekema, 2005; Mullineaux, 2005; Anderson et al., 2008; Daum and Kühlbrandt, 2011).
Figure 5.
Thylakoid Architecture Depends on CURT1 Protein Levels.
(A) TEM micrographs of ultrathin sections of leaves from the wild type (Landsberg erecta [Ler] and Col-0), curt1 mutants, and lines expressing tagged CURT1A proteins (CURT1A-HA curt1a-2 and CURT1A-cmyc curt1a-2). Note that the CURT1A-HA curt1a-2 and CURT1A-cmyc curt1a-2 lines express ∼50 and 400% of wild-type levels of CURT1A, respectively (see Supplemental Figure 1 online).
(B) Scatterplot indicating the height (y axis) and diameter (x axis) of thylakoid stacks in the genotypes shown in (A) and (D). Average values (n ≥ 30) including the respective se are presented; a complete list of the P values, indicating the significance of the differences in grana stacking between the genotypes, is provided in Supplemental Data Set 1 online. WT, the wild type.
(C) and (D) Sections of chloroplasts as in (A) from curt1ab and curt1abcd (C) and from stn7 stn8 and tap38 mutant plants (D). Circles indicate tubuli and vesicles characteristic for curt1 mutant chloroplasts.
To obtain further evidence for a relationship between grana structure and the abundance of CURT1 proteins, tagged lines expressing varying levels of CURT1A (the major CURT1 protein) were investigated by TEM. CURT1A-HA plants with ∼50% of wild-type CURT1A levels (see Supplemental Figure 1 online) yielded average values for diameter/height of grana stacks of 0.82 ± 0.03/0.09 ± 0.00 µm, intermediate between those measured in curt1a and wild-type plants (Figures 5A and 5B; see Supplemental Data Set 1 online). Plants overexpressing CURT1A-cmyc (oeCURT1a-cmyc curt1a-2), with CURT1A levels about 4 times higher than in wild-type plants (see Supplemental Figure 1 online), had slimmer but higher grana stacks than the wild type (average diameter/height: 0.37 ± 0.01/0.17 ± 0.01 µm) (Figures 5A and 5B; see Supplemental Data Set 1 online).
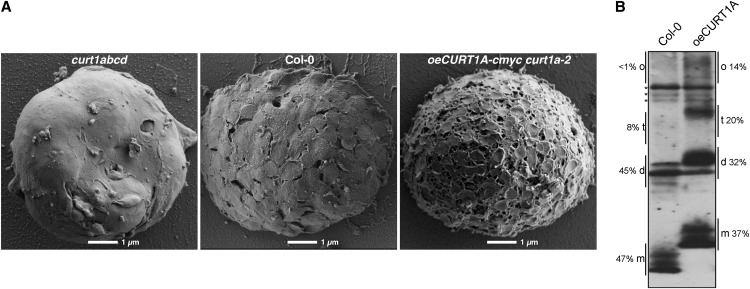
To assess the three-dimensional arrangement of thylakoid membranes, envelope-free chloroplasts from wild-type, the quadruple mutant curt1abcd, and oeCURT1a-cmyc curt1a-2 plants were analyzed by high-resolution scanning electron microscopy. Strikingly, the top view of the thylakoid system confirmed that extended sheets of stroma thylakoids predominate in curt1abcd chloroplasts (Figure 6A). Conversely, in thylakoids from CURT1A-cmyc overexpressors, the grana stacks were more abundant (2.02 ± 1.09 grana/μm2 [the wild type] versus 3.31 ± 0.82 grana/μm2 [oeCURT1A-cmyc]; P = 0.049) but displayed a significantly reduced diameter compared with the wild type (0.57 ± 0.15 μm [the wild type] versus 0.31 ± 0.06 μm [oeCURT1A-cmyc]; P < 0.0001), and stroma lamellae regions adopted more tubular structures (Figure 6A). Moreover, quantification of CURT1A cross-linking products from thylakoids of the wild type and CURT1A-cmyc overexpressors showed that the fraction of interacting CURT1A proteins is increased in the overexpressor (Figure 6B).
Figure 6.
CURT1A Levels Determine Thylakoid Architecture.
(A) Topographic view of envelope-free chloroplasts from curt1abcd, wild-type (Col-0), and CURT1A-cmyc curt1a-2 leaves (that express tagged CURT1A proteins at ∼400% of wild-type CURT1A levels) by scanning electron microscopy.
(B) Wild-type and CURT1A-cmyc curt1a-2 (oeCURT1A) thylakoids were cross-linked and analyzed as in Figure 3D, and CURT1A signals were quantified. Intensities for bands representing monomers (m), dimers (d), tetramers (t), and oligomers (o) are given in percentages next to the corresponding genotype. Asterisks indicate unspecific bands.
Taken together, the electron microscopy data clearly show a striking association between the dose of CURT1 proteins and the architecture of thylakoid membranes. In the absence of CURT1 proteins, grana consist of fewer but broader layers of membrane, whereas overexpression of CURT1A is characterized by higher stacks of smaller grana discs, tubular stroma lamellae, and increased oligomerization of CURT1 proteins. The negative correlation between the diameter and height of grana must be attributed to the fact that a fixed proportion of thylakoid membrane is incorporated into the grana stacks (Albertsson and Andreasson, 2004). Therefore, an increase in the diameter of grana must be compensated for by a decrease in the number of stacks and vice versa.
CURT1 Proteins Are Enriched at Grana Margins
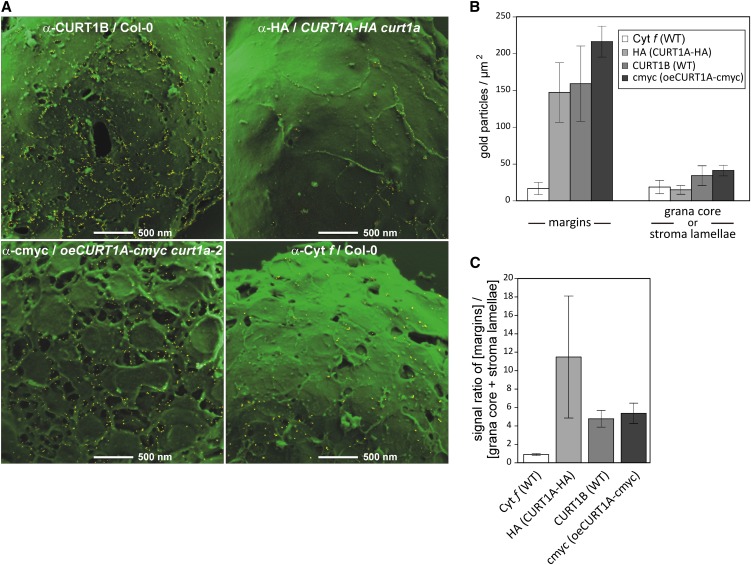
The localization of the CURT1A and B proteins was determined by immunogold labeling of envelope-free chloroplasts followed by scanning electron microscopy. To this end, envelope-free chloroplasts of wild-type, CURT1A-HA, and oeCURT1A-cmyc plants were first immunodecorated with specific antibodies and then treated with the appropriate gold-labeled secondary antibodies (Figure 7A). Signals for CURT1B (in the wild type), HA (in CURT1A-HA), and cmyc (in oeCURT1A-cmyc) were quantified (Figures 7B and 7C) and found to be highly enriched at the lateral boundaries of the thylakoid membranes that face the stroma, where the membrane curves to form grana margins. In fact, signals for CURT1A-HA were almost exclusively associated with margins, whereas CURT1B- and cmyc-specific antibodies also labeled a few sites outside of the margins. In particular, signals were also detected in the tubular structures branching from the cylindrical grana stack in oeCURT1A-cmyc plants. When envelope-free chloroplasts from the wild type were immunolabeled with antibodies recognizing cytochrome f as control, signals were dispersed over the entire thylakoid system, as expected (Figure 7). The predominant, if not exclusive, localization of the CURT1 proteins in thylakoids regions of high curvature (in particular grana margins), together with the fact that the dose of CURT1 proteins influences the extent of grana stacking, leads us to conclude that CURT1 proteins are directly involved in causing the localized membrane curvature found in grana margins.
Figure 7.
CURT1 Proteins Are Enriched at Grana Margins.
(A) Scanning electron microscopy images of envelope-free chloroplasts from wild-type (Col-0), CURT1A-HA curt1a-2, and oeCURT1A-cmyc curt1a-2 plants, following immunogold labeling with antibodies raised against CURT1B, cytochrome f, HA, and cmyc. Bars = 500 nm.
(B) Quantification of the distribution of immunogold-labeled CURT1 proteins. At least eight independent scanning electron microscopy pictures of each experiment, as exemplarily shown in (A), were analyzed for the distribution of signals from gold particles. “Margins” were defined as the area covering around 5 nm in both directions of the visual membrane bending zones. The residual surface was combined to represent the grana core and stroma lamellae regions. Note that the minor amounts of CURT1 proteins in Col-0 and oeCURT1A-cmyc plants assigned to the grana core or stroma lamellae regions were also mainly detected close to the curved membrane borders but just outside of the region arbitrarily defined as “margins.” Error bars represent the sd between the independent scanning electron microscopy images. WT, the wild type.
(C) The distribution ratios of the CURT1 proteins and, as control, cytochrome f were determined based on the specific accumulation of gold particles in the margins or the remaining membrane area (grana core + stroma lamellae) of the same scanning electron microscopy images analyzed in (B). Error bars represent sd as in (B).
Relationship between CURT1 Proteins and Thylakoid Phosphorylation
The extent of thylakoid protein phosphorylation had previously been associated with changes in grana architecture (Fristedt et al., 2009). Therefore, TEM analysis was performed on chloroplasts displaying either hypophosphorylation (stn7 stn8 double mutant; Bellafiore et al., 2005; Bonardi et al., 2005) or hyperphosphorylation (tap38 mutant; Pribil et al., 2010; Shapiguzov et al., 2010) of thylakoid proteins. Grana from stn7 stn8 plants had fewer but broader membrane discs as reported (Fristedt et al., 2009) and were comparable to grana from CURT1A-HA plants expressing CURT1A at ∼50% of wild-type levels (Figures 5B and 5D; see Supplemental Data Set 1 online). By contrast, grana stacks in tap38 plants had more membrane layers than seen in the wild type (Figures 5B and 5D; see Supplemental Data Set 1 online).
Because a decrease in thylakoid phosphorylation appears to have a similar effect on grana architecture to that of decreasing CURT1 levels, we tested whether reduced thylakoid phosphorylation might cause the changes in grana architecture observed in the curt1a and curt1abcd mutants. However, the levels of PSII core and LHCII phosphorylation were actually higher in the two curt1 mutants (see Supplemental Figure 5 online), implying (1) that modulation of thylakoid membrane curvature by CURT1 proteins does not involve PSII core phosphorylation and (2) that loss of CURT1-dependent membrane curvature overrides the influence of PSII core phosphorylation.
Intrinsic Membrane-Bending Property of CURT1
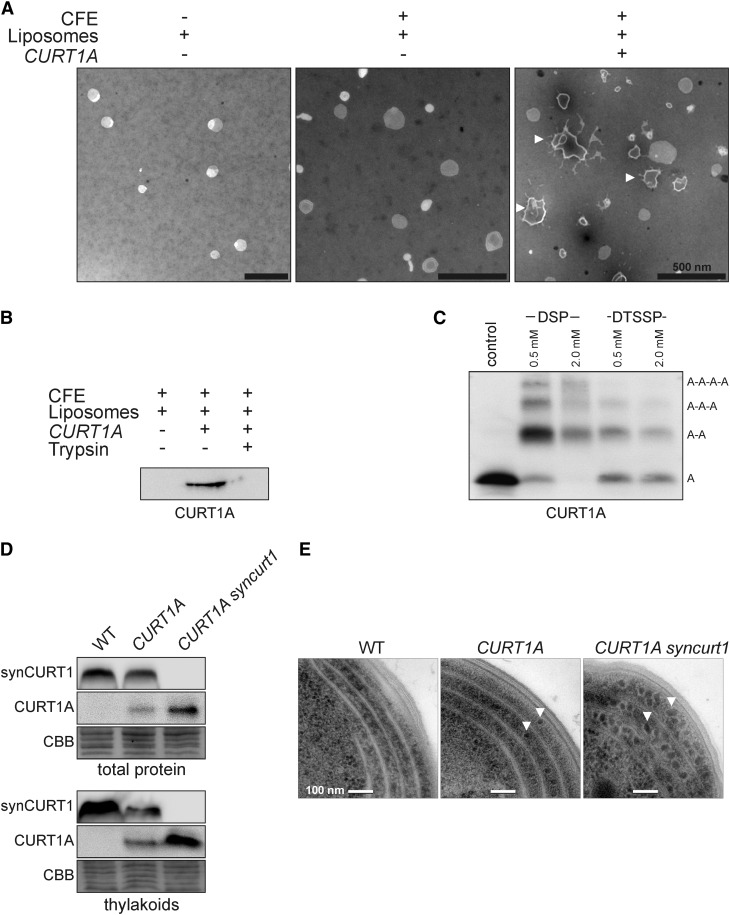
To test whether CURT1 proteins can directly induce membrane curvature, CURT1A was translated in a cell-free system in the presence of liposomes with a thylakoid-like lipid composition. After purification of liposomes, TEM analysis revealed tubulation of CURT1A-containing liposomes (Figure 8A), demonstrating that CURT1A can directly induce membrane curvature in vitro. CURT1A was found to be integrated into liposomes, as shown by protein gel blot analysis of purified liposomes (Figure 8B). Moreover, trypsination experiments, employing an antibody that recognizes the N-terminal domain of CURT1A (and which contains a trypsination site; see Supplemental Figure 1C online), indicated that the N terminus of CURT1A protrudes from the liposomal surface and is accessible to trypsination (Figure 8B). Thus, the topology of CURT1A in liposomes is similar to that of natural CURT1A, with a stromal orientation of its N terminus. To test whether liposomal CURT1A also forms oligomers, CURT1A proteoliposomes were cross-linked. Both membrane-permeable and impermeable cross-linkers were capable of producing cross-linking products, indicating that liposomal CURT1 proteins form oligomers, providing a link between the membrane-bending activity of CURT1 proteins and their capability to spontaneously oligomerize (Figure 8C).
Figure 8.
CURT1A Oligomers Tubulate Liposomes and Their Functions Are Evolutionary Conserved.
(A) Liposomes with thylakoid-like lipid composition were incubated with E. coli cell-free extracts (CFEs) either containing a template (CURT1A) for de novo CURT1A synthesis or not. After purification, liposomes were analyzed by TEM. Tubular structures are indicated by white arrowheads.
(B) Liposomal integration of CURT1A. Liposomes from reaction mixtures with indicated composition were purified and subjected to SDS-PAGE and protein gel blot analysis. Because the CURT1A antibody recognizes a stroma-exposed region of CURT1A (see Supplemental Figure 1C online), trypsination of proteoliposomes confirmed the surface-exposed and thylakoid-like topology of CURT1A.
(C) CURT1A proteoliposomes were cross-linked with membrane-permeable [dithiobis(succinimidylpropionate) (DSP)] and -impermeable [3,3′-dithiobis(sulfosuccinimidylpropionate) (DTSSP)] cross-linkers. Cross-linking products (A-A to A-A-A-A) were detected by SDS-PAGE and protein gel blot analysis.
(D) Heterologous expression of Arabidopsis CURT1A in Synechocystis alters thylakoid membrane structure. Different Synechocystis mutant strains that express CURT1A in the presence (CURT1A) or absence (CURT1A syncurt1) of endogenous Synechocystis CURT1 (synCURT1) were generated. Total proteins and thylakoid proteins of wild-type (WT) and mutant Synechocystis strains were analyzed by SDS-PAGE and immunoblot analysis, employing antibodies specific for CURT1A and synCURT1. Coomassie blue staining (CBB) served as the loading control.
(E) The same genotypes as in (D) were analyzed regarding their thylakoid structure by TEM. Exemplary detached phycobilisomes are indicated by white arrowheads.
CURT1 Function Is Evolutionary Conserved
Cyanobacteria have CURT1 proteins (Figure 1E) but do not form grana. To clarify the impact of CURT1 proteins on cyanobacterial thylakoid structure, strains of the cyanobacterium Synechocystis were generated displaying altered levels of the endogenous (synCURT1) or Arabidopsis (CURT1A) CURT1 protein. In fact, loss of synCURT1 had serious consequences for cell viability because we failed to obtain fully segregating knockout strains for synCURT1. However, we could generate strains that express CURT1A instead of synCURT1 (strain: CURT1A syncurt1), indicating that Arabidopsis CURT1 can replace Synechocystis CURT1 (Figure 8D). Interestingly, strains that express CURT1A in addition to endogenous synCURT1 (strain: CURT1A), express less synCURT1 than the wild type and less CURT1A than CURT1A syncurt1 (Figure 8D), suggesting that mechanisms might operate in Synechocystis that prevent overaccumulation of CURT1 proteins. The growth rate of the two mutant strains, in particular CURT1A syncurt1, was reduced compared with wild-type cells, indicating that Arabidopsis CURT1A only partially replaces Synechocystis CURT1. When thylakoid architecture was studied by TEM, multiple changes were observed in the strains with altered CURT1 levels (Figure 8E). The thylakoids of the mutant strains had a crumpled appearance with a decreased average lumen width (the wild type, 23.8 ± 2.9 nm; CURT1A, 19.5 ± 1.5 nm; CURT1A syncurt1, 17.3 ± 2.9 nm; n > 30 cells). Moreover, phycobilisomes were associated with wild-type thylakoids but detached from the thylakoid membranes of the mutant strains.
Taken together, these data suggest that even in cyanobacteria, the progenitors of chloroplasts, CURT1 proteins are involved in establishing thylakoid architecture. Strikingly, Arabidopsis CURT1A can partially replace Synechocystis CURT1.
DISCUSSION
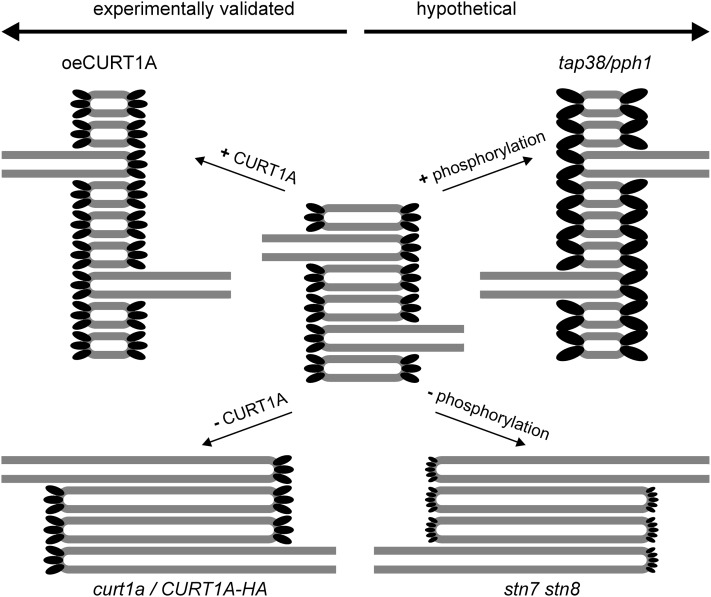
Multiple lines of evidence point to the conclusion that CURT1 proteins determine thylakoid architecture by inducing membrane curvature (Figure 9). (1) In Arabidopsis, levels of CURT1 proteins and of the number of membrane layers in grana stacks (and in turn of the area of grana margins, which are the sites of highest membrane curvature) are directly correlated. (2) CURT1 proteins are predominantly located in grana margins. (3) CURT1 proteins are capable of inducing membrane curvature in vitro. Expression of Arabidopsis CURT1A in Synechocystis cells alters thylakoid structure, although whether reduced levels of endogenous synCURT1 or the action of the Arabidopsis protein is responsible remains to be clarified. In either case, the increased membrane curvature revealed by the crumpled appearance of thylakoids in these cells and the decrease in lumen width, might lead to detachment of phycobilisomes due to steric hindrance of the protruding extrinsic antenna complexes. Moreover, because this steric hindrance due to phycobilisomes does also prevent the direct interaction of thylakoid membranes, grana formation by CURT1 overexpression might be actually feasible in Synechocystis strains without phycobilisomes.
Figure 9.
Hypothetic Model for the Effects of CURT1 Proteins and Their Phosphorylated Forms on Grana Stacking.
CURT1 proteins are symbolized by black ellipses. Hypothetical phosphorylation-induced changes in CURT1 structure or oligomerization state are indicated by changes in ellipse size. Note that changes in grana stacking in the stn7 stn8 and tap38 mutants might alternatively be caused by changes in repulsion between membrane layers due to altered PSII phosphorylation (Fristedt et al., 2009), although this effect is overridden in curt1 mutants with increased thylakoid phosphorylation.
A number of eukaryotic proteins are known to share the in vitro vesicle/tubule-forming activity of CURT1A. Many of these possess either an ENTH (epsin N-terminal homology) domain (Kay et al., 1999; De Camilli et al., 2002; Ford et al., 2002) or a BAR (Bin-amphiphysin-Rvs) domain (Farsad and De Camilli, 2003; Zimmerberg and Kozlov, 2006), both of which include an N-terminal amphipathic helix. Epsin’s ability to curve membranes is dependent upon insertion of this helix into one leaflet of the membrane bilayer (Ford et al., 2002) and formation of highly ordered aggregates via self-association of the ENTH domain (Yoon et al., 2010). The membrane-deforming activity of N-terminal amphipathic helices in the prokaryotic MinE protein (involved in bacterial cell division) and the peroxisomal protein required for peroxisome biogenesis 11 has been directly demonstrated (Opaliński et al., 2011; Shih et al., 2011). With ∼100 amino acids, CURT1 proteins are considerably smaller than canonical ENTH (∼150 amino acids) or BAR (∼200 amino acids) domains. However, CURT1 proteins appear to contain an amphipathic helix, which might be involved in its membrane-deforming activity (Figures 1B to 1D). Moreover, their capacity for oligomerization in vivo in thylakoids and in vitro in liposomes (Figures 3D and 8C) suggests that aggregation of CURT1 might play a pivotal role in inducing membrane curvature, analogous to the mechanisms suggested for MinE (Shih et al., 2011) or epsin (Yoon et al., 2010).
The role of protein phosphorylation in modulating thylakoid architecture remains enigmatic. Our results clearly show that CURT1-dependent membrane curvature overrides the influence of PSII core phosphorylation on grana stacking (Figures 5A and 5B; see Supplemental Figure 5 online). Moreover, CURT1 proteins also seem to act in determining thylakoid structure in cyanobacteria, where no PSII core phosphorylation occurs (Figures 8D and 8E). Our hypothesis is that dephosphorylation of CURT1 proteins (in stn7 stn8 double mutants) might alter the structure of the predicted N-terminal amphipathic helix and/or its oligomerization behavior. This, in turn, could modify the membrane-bending activity of CURT1 proteins in the grana margins, leading to more extensive but fewer membrane layers in grana (Figure 9). This model is supported by the fact that phosphorylated forms of CURT1A, B, and C have been found in phosphorylomes (see Supplemental Table 1 and Supplemental References 1 online).
Taken together, our data identify the CURT1 proteins as a family of proteins that form oligomers, can bend membranes in vitro, and determine the architecture of grana in plants. Their function in defining thylakoid structure can be traced back to the endosymbiotic precursor of chloroplasts and, in land plants, the regulation of their activity might involve their posttranslational modification by phosphorylation.
METHODS
Plant Material and Propagation
Insertion lines for CURT1 genes originate from six different mutant collections (see Supplemental Figure 2 and Supplemental References 1 online). Arabidopsis thaliana plants were either grown on soil in a growth chamber or, when nonphotoautotrophic, on Murashige and Skoog medium supplemented with Suc (Pesaresi et al., 2009a).
Intracellular Localization of dsRED Fusions in Arabidopsis Protoplasts
The red fluorescent protein from the reef coral Discosoma sp. (dsRED) was used as a reporter to determine the intracellular location of CURT1 proteins as described (Jach et al., 2001; DalCorso et al., 2008).
Nucleic Acid Analysis
DNA and RNA analyses were performed as described (Armbruster et al., 2010). Primers for PCRs are listed in Supplemental Table 4 online. For RT-PCR, total RNA was extracted from leaf tissue with the RNeasy plant mini kit (Qiagen) according to the manufacturer's instructions. cDNA was prepared from 1 µg of total RNA applying the iScript cDNA synthesis kit (Bio-Rad) following the manufacturer's instructions. For RT-PCR, cDNA was diluted 10-fold, and 3 μL of the dilution was used in a 20-μL reaction. Thermal cycling consisted of an initial step at 95°C for 3 min, followed by 30 cycles of 10 s at 95°C, 30 s at 55°C, and 10 s at 72°C. All reactions were performed in three biological replicates.
Generation of Tagged CURT1 Proteins
Plants expressing CURT1A and D with a C-terminal HA or cmyc tag were generated by introducing the respective coding region into the vectors pPCV812ΔNotI-Pily (for hemagglutinin [HA] fusions) or pPCV812ΔNotI-Lola (for cmyc fusions) under control of the 35S promoter and then transforming curt1a-2 (CURT1A-HA or CURT1A-cmyc) and wild-type (CURT1D-HA) flowers with Agrobacterium tumefaciens (strain GV3101) containing the constructs (Clough and Bent, 1998). Individual transgenic plants were identified as described (Armbruster et al., 2010).
Generation of MBP Fusions and Protein Titration
Sequences coding for the mature proteins PetC, PsaD1, and CURT1A, B, C, and D were amplified from Arabidopsis cDNA using sequence-specific primer combinations containing either an EcoRI or XbaI restriction site at the 5′ terminus (see Supplemental Table 4 online). Following restriction digests, PCR products were directionally inserted into the corresponding polylinker region of pMal-c2 (New England Biolabs) downstream of the MBP coding sequence. Fusion proteins were expressed in the Escherichia coli strain BL21 and purified under native conditions with amylose-resin (New England Biolabs). Purified proteins were quantified (protein assay kit; Bio-Rad), and concentrations of recombinant proteins were calculated. Total amounts of the respective proteins in thylakoid membrane preparations were estimated as described (Hertle et al., 2013).
Protein Isolation, PAGE, and Immunoblot Analyses
Leaves harvested from 4-week-old plants were used for the preparation of thylakoids (Bassi et al., 1985) and total proteins (Martínez-García et al., 1999). PSI isolation was performed as described (DalCorso et al., 2008).
For BN-PAGE, thylakoid samples equivalent to 30 µg (n-dodecyl β-d-maltoside [β-DM]; Sigma-Aldrich) or 100 µg (digitonin; Sigma-Aldrich) chlorophyll were solubilized in 750 mM 6-aminocaproic acid, 5 mM EDTA, pH 7, and 50 mM NaCl in the presence of 1.0% (w/v) β-DM or 1.6% (w/v) digitonin for 10 min (β-DM) or 60 min (digitonin) at 4°C. Following centrifugation (15 min [β-DM] or 60 min [digitonin]; 21,000g), the solubilized material was fractionated using nondenaturing BN-PAGE (Schägger et al., 1988). Designation of supercomplexes was as follows: PSI-LHCII and PSI-LHCI-LHCII supercomplex; PSImono and PSI-LHCI monomers; PSIIdi and PSII dimers; cytochrome b6/fdi and dimeric cytochrome b6/f complexes; and LHCIImono/tri and monomeric/trimeric LHCII. Two-dimensional PAGE analysis and immunoblot analysis with appropriate antibodies, signal detection, and quantification were performed as described (Armbruster et al., 2010).
Chemical Cross-Linking
Thylakoid preparations (300 µg/mL chlorophyll) were washed five times with 20 mM HEPES/KOH, pH 7.5, to remove EDTA. After addition of bis(sulfosuccinimidyl) suberate (final concentration 4 mM; Thermo Scientific) and incubation for 1 h at 0°C, the reaction was quenched by adding 150 mM Tris/HCl, pH 7.5.
CURT1A proteoliposomes were cross-linked for 1 h on ice with either 0.5 and 2.0 mM dithiobis(succinimidylpropionate) or 0.5 mM and 2.0 mM 3,3′-dithiobis(sulfosuccinimidylpropionate) (Pierce, Thermo Scientific). Subsequently, the reaction was quenched at a final concentration of 30 mM Tris, pH 7.6. Samples were resuspended in nonreducing Laemmli buffer and heated for 10 min at 100°C prior to Tris-Tricine PAGE.
CoIP
For CoIP experiments with HA- or cmyc-tagged proteins, thylakoid membranes were solubilized with 1.6% digitonin in CoIP buffer (50 mM HEPES/KOH, pH 8.0, 330 mM sorbitol, 150 mM NaCl, 0.5% [w/v] BSA, and 1 mM PMSF), and the supernatant was recovered after centrifugation (60 min, 16,000g, 4°C). The supernatant was coincubated with HA or cmyc affinity beads overnight on a wheel at 4°C. The beads were then collected by centrifugation for 2 min at 3000g and washed six times with Co-IP buffer containing 0.5% digitonin. Proteins were eluted from the matrix by incubation with reducing SDS loading dye buffer. For CoIP experiments with CURT1B, thylakoids were incubated for 2 h with a CURT1B-specific antibody before solubilization, and Protein-A beads were used instead of HA or cmyc beads.
Generation and Characterization of Synechocystis Mutant Strains
Synechocystis sp PCC 6803 Glc-tolerant wild-type and mutant strains were grown at 30°C in BG11 medium containing 5 mM Glc, under continuous illumination at 30 µmol m−2 s−1. For growing on plates, 1.5% (w/v) agar and 0.3% (w/v) sodium thiosulfate were added. In both cases, BG-11 was supplemented with antibiotics appropriate for the particular strain.
To generate the CURT1A strain, the coding region of CURT1A without cTP was introduced into the Synechocystis slr0168 open reading frame (Kunert et al., 2000) together with an nptI-sacB double-selection cassette (Cai and Wolk, 1990). To generate the CURT1A syncurt1 strain, the endogenous synCURT1 gene (slr0483) was replaced by the coding region of CURT1A without cTP, together with the double-selection cassette. In both strains, CURT1A was put under the control of the Synechocystis psbA2 promoter (Eriksson et al., 2000). Vectors used for transformation were assembled with the Golden Gate Shuffling method (Engler et al., 2009), using the pL2-1 destination vector (Weber et al., 2011). The Synechocystis slr0168 upstream and downstream regions, the psbA2 promoter, and synCURT1 were PCR amplified from Synechocystis genomic DNA. The CURT1A coding sequence was amplified from Arabidopsis cDNA, while the nptI-sacB cartridge was amplified from plasmid pRL250. All the PCRs were performed using Phusion high-fidelity DNA polymerase (New England Biolabs) and sequence-specific primers containing an additional BsaI restriction site at their 5′ termini. During the Golden Gate restriction-ligation reaction, cleavage of the fragments by BsaI endonuclease generated the complementary 5′ protruding ends necessary for the assembly of the final constructs.
Transformation of Synechocystis sp PCC 6803 was performed as described (Vermaas et al., 1987), and transformants were selected based on kanamycin resistance. Complete segregation of the introduced gene was confirmed by PCR.
For protein gel blot analyses, cells were harvested by centrifugation (5000g, 10 min, 4°C). Cells were then resuspended in 50 mM HEPES-NaOH, pH 7.8, 10 mM MgCl2, 5 mM CaCl2, and 25% glycerol and ruptured by five disrupting strokes of 20 s using 0.25/0.3-mm glass beads. After centrifugation (5000g, 5 min, 4°C), the supernatant constitutes the total protein fraction. After subsequent centrifugation (45,000g, 1.5 h, 4°C), the pellet, representing the membrane fraction, was recovered and suspended in 1 volume of the above mentioned buffer. Samples were solubilized at 70°C for 10 min in 2% (w/v) SDS in presence of 100 mM DTT prior to separation via 15% PA SDS Tris-Tricine gel. For protein gel blot analysis, antibodies raised against CURT1A or the peptide CysThrAspValGlyProIleThrThrProAsnProGlnLysSer of synCURT1 (BioGenes) were used.
Preparation of Proteoliposomes
For proteoliposome generation, unilamellar vesicles were prepared as described (Haferkamp and Kirchhoff, 2008). Briefly, a lipid mixture of 25% monogalactosyl diacylglycerol, 42% digalactosyl diacylglycerol, 16% sulfoquinovosyl diacylglycerol, and 17% phosphatidylglycerol dissolved in chloroform:methanol (1:1) (Lipid Products) was dried under a stream of nitrogen and stored under vacuum overnight. The lipid film was slowly dissolved in diethylpyrocarbonate-treated bidest. H2O to a concentration of 20 mg/ml and subjected to five freeze-thaw cycles in liquid nitrogen. To obtain the final vesicle size, the lipid suspension was extruded 21 times at 250 kPa and 60°C through a 0.1-µm Nuclepore membrane (Whatman).
CURT1A protein was expressed using a S12 cell free expression system as described (Kim et al., 2006) with minor modifications. The reaction mix contained 20% (v/v) cell extract, no polyethylene glycol 8000, and 200 μL of the liposome preparation. After 3 h of incubation in a 37°C shaker, the mix was separated by a discontinuous Suc gradient (100,000g, 18 h) and the liposome containing fraction extracted. For topology studies, the liposome fraction was mixed with trypsin (Sigma Aldrich) and CaCl2 to a final concentration of 0.25 mg/mL and 25mM, respectively, and incubated for 20 min on ice. Samples were then analyzed by SDS-PAGE and protein gel blot analysis.
Electron Microscopy
For TEM, plant material (pieces of tissue from 5th to 7th true leaves of 28-d-old plants at the beginning of the light period) and Synechocystis cells were fixed with 2.5% glutaraldehyde in 75 mM sodium cacodylate and 2 mM MgCl2, pH 7.0, for 1 h at 25°C, rinsed several times in fixative buffer, and postfixed (2 h) with 1% osmium tetroxide in fixative buffer at 25°C. After two washing steps in distilled water, the pieces were stained en bloc with 1% uranyl acetate in 20% acetone (30 min), followed by dehydration and embedding in Spurr’s low-viscosity resin. Ultrathin sections of 50 to 70 nm were cut with a diamond knife and mounted on uncoated copper grids. The sections were poststained with aqueous lead citrate (100 mM, pH 13.0). Micrographs were taken with an EM-912 electron microscope (Zeiss) equipped with an integrated OMEGA energy filter operated in the zero-loss mode.
For scanning electron microscopy, isolated chloroplasts were made envelope-free by one freeze-thaw cycle in liquid nitrogen. Incubation with specific antibodies was performed in a 1:1 dilution with 50 mM HEPES/KOH, pH 8.0, 330 mM sorbitol, 150 mM NaCl, and 0.5% (w/v) BSA (overnight, 4°C). Unbound antibody was removed by washing with the same buffer (centrifugation at 0.2g for 2.5 min), and gold-coated secondary antibody was added and incubated (60 min, 4°C), followed by removal of unbound secondary antibody by repeated washing. Drops of chloroplast suspensions were placed on pieces of conductive silicon wafer, covered with a cover slip, and rapidly frozen with liquid nitrogen. The cover slip was removed, and the slide was immediately fixed with 2.5% glutaraldehyde in iso-osmotic cacodylate buffer, pH 7.0, postfixed with osmium tetroxide, dehydrated in a graded series of acetone solutions, and critical-point dried from liquid CO2, mounted on stubs, and examined uncoated at 1 kV with the Zeiss AURIGA high-resolution field-emission scanning electron microscope. Topographic images were taken with the chamber SE detector. For immunogold labeling detection, back-scattered electron images were taken with the energy-selective in-lens (EsB) Detector, with the EsB grid set at 900 V. The immunogold signal was colored in yellow and superimposed on the green-colored topographic image of the chloroplasts.
For electron microscopy of liposomes, liposomes were negatively stained by placing a drop of the sample on a carbon-coated copper grid, freshly hydrophilized by glow discharge. After incubation for 2 min, the drop was quickly removed with a Pasteur pipette, and the grid was stained with 2% phosphotungstic acid and 0.05% Glc.
Bioinformatic Analyses
Amino acid sequences were aligned using ClustalW (v2.0.12; http://mobyle.pasteur.fr/) (Thompson et al., 1994) and BoxShade (v3.21; http://www.ch.embnet.org). Chloroplast transit peptide sequences were predicted with ChloroP (http://www.cbs.dtu.dk/services/ChloroP/) (Emanuelsson et al., 1999) and TMs with TMHMM (http://www.cbs.dtu.dk/services/TMHMM-2.0/) (Krogh et al., 2001). α-Helices were predicted by I-TASSER (Zhang, 2008; Roy et al., 2010) and Jpred (Cole et al., 2008). Amphipathic properties of α-helices were illustrated by helical wheel representations using a script for helical wheel plotting (http://rzlab.ucr.edu/scripts/wheel/wheel.cgi). Representation of the CURT1A-H1 sequence as a three-dimensional model of an α-helix was computed by I-TASSER and processed by 3D-Mol Viewer (Invitrogen).
Phylogenetic Analyses
For the phylogenetic analysis of CURT1 sequences, the C-terminal 99 amino acids of each protein (for a complete list, see the Accession Numbers section) were aligned using ClustalW (v2.0.12; http://mobyle.pasteur.fr/; default settings, gap extension: 1). For sequence alignment, see Supplemental Figure 6 online. The program Phylip (v3.67; http://mobyle.pasteur.fr/) was used, using as distance model the Jones-Taylor-Thornton matrix and bootstrapping (1000 repetitions) to compute a consensus tree (Boolean). TreeView (v1.6.6; http://taxonomy.zoology.gla.ac.uk/rod/treeview.html) was used for the unrooted phylogenetic tree visualization. The neighbor-joining method was employed for tree building.
Determination of the Topology of CURT1 Proteins
Intact Arabidopsis chloroplasts were isolated and purified from leaves of 4- to 5-week-old plants (Aronsson and Jarvis, 2002), broken open in 10 mM Tris/HCl, pH 8.0, and 1 mM EDTA, pH 8.0, at a chlorophyll concentration of 2 mg/mL, and loaded onto a three-step Suc gradient (Li et al., 1991). After centrifugation (30,000g; 4°C), the upper phase (containing the stroma), both interphases (with the envelope membranes), and the pellet (thylakoids) were collected, and all three membrane fractions were washed with TE buffer.
Salt washes of thylakoids were performed as described (Karnauchov et al., 1997). Soluble and membrane proteins were separated by centrifugation for 10 min (10,000g, 4°C), and immunoblot analysis was performed on both fractions.
For tryptic proteolysis experiments, thylakoid membranes were resuspended in 50 mM HEPES/KOH, pH 8.0, and 300 mM sorbitol at a chlorophyll concentration of 0.5 mg/mL. Trypsin (Sigma-Aldrich) was added to a concentration of 10 µg/mL. Samples were taken 30 min later, and proteolysis was stopped by adding soybean trypsin inhibitor (50 μg/mL; Sigma-Aldrich).
Measurement of Photosynthetic Parameters
Maximum (Fv/Fm) and effective (ΦII) quantum yields of PSII, excitation pressure (1-qL), NPQ, and state transitions were measured as described (Varotto et al., 2000; Kramer et al., 2004; DalCorso et al., 2008; Pribil et al., 2010).
Fast light-induced absorption and fluorescence changes were measured with a Joliot type spectrometer (JTS10; Biologic). Cut plant leaves, dark-adapted for at least 1 h, were used, and all experiments were performed with at least three leaves from independent plants. The absorption changes reflecting the redox changes of P700 were measured at 810 nm. The actinic light was provided by an orange light-emitting diode, and the light intensity was 7000 and 100 μmol photons m−2 s−1 for the absorption and fluorescence experiments, respectively.
In Situ Assay of Ferredoxin-Dependent Plastoquinone Reduction Activity
Ferredoxin-dependent plastoquinone reduction activity was measured in ruptured chloroplasts as described (Munekage et al., 2002).
Leaf Lipid Quantification
Total lipids were extracted from whole leaves with chloroform/methanol. After removal of solvent with nitrogen gas in a sample concentrator, total leaf lipids were transmethylated with methanolic HCl in the presence of pentadecanoic acid (15:0) as an internal standard. After extraction with hexane, fatty acid methyl esters were quantified by gas chromatography as described (Browse et al., 1986).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative (http://www.Arabidopsis.org/), GenBank (http://www.ncbi.nlm.nih.gov), or EMBL (http://www.ebi.ac.uk/embl/) databases under the following accession numbers: Arabidopsis: CURT1A, At4g01150; CURT1B, At2g46820; CURT1C, At1g52220; CURT1D, At4g38100; Tic110, At1g06950; Toc33, At1g02280; PsbX, At2g06520; PsaD1, At4g02770; Vipp1, At1g65260; and CRY2, At1g04400. Vitis vinifera: Vv1, 225454426; Vv2, 225462673; Vv3, 225469778; Vv4, 225453634; and Vv5, 225442615. Populus trichocarpa: Pt1, 224130462; Pt2, 224067952; Pt3, 224070909; Pt4, 224054368; and Pt5, 224064380. Zea mays: Zm1, 226504766; Zm2, 226500194; Zm3, 226496830; Zm4, 226499766; Zm5, 226529623; Zm6, 226528453; Zm7, 212275151; Zm8, 226509242; and Zm9, GRMZM2G302639. Oryza sativa subsp japonica: Os1, 115476522; Os2, 115467478; Os3, 115448483; Os4, 115467106; Os5, 115479645; Os6, 115483154; Os7, 115472001; and Os8, 297597647. Picea sitchensis: Ps1, 116782153; Ps2, 116781331; and Ps3, 116783046. Physcomitrella patens subsp patens: Pp1, 168018075; Pp2, 168048886; Pp3, 168030972; Pp4, 168019666; and Pp5, 168004237. Chlamydomonas reinhardtii: Cr1, 159488417; Cr2, 159490118; and Cr3, 159466615. Volvox carteri: Vc1, 302843342; and Vc2, 302846248. Ostreococcus tauri: Ot, 308798659. Micromonas spusilla: Ms1, 303278702; and Ms2, 303273852. Chlorella variabilis: Cv1, 307104959; and Cv2, 307106927. Synechocystis sp PCC 6803: Sy, 16331518. Nostoc punctiforme PCC 73102: Np, 186683920. Cyanothece sp CCY0110: Cy, 126659752. Oscillatoria sp PCC 6506: Oc, 300337383. Arthrospira maxima CS-328: Am, 209493929. Nodularia spumigena CCY9414: Ns, 119510999. Raphidiopsis brookii D9: Rb, 281196613. Microcystis aeruginosa PCC 7806: Ma, 159030779. Anabaena variabilis: Av1, 75907007; Av2, 75906820. Synechococcus sp PCC7002: So, 170079043.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Analysis of Accumulation and Topology of CURT1 Proteins.
Supplemental Figure 2. Mutations of the CURT1 Genes.
Supplemental Figure 3. Analysis of CURT1 Protein Abundance and Oligomerization.
Supplemental Figure 4. CURT1 Proteins Are Not Constitutive Components of the Major Thylakoid Photosynthesis Complexes and Their Lack Affects Photosynthesis Pleiotropically.
Supplemental Figure 5. Quantification of Thylakoid Phosphorylation in curt1 Mutants.
Supplemental Figure 6. Alignment of Orthologous CURT1 Protein Sequences for Phylogenetic Analyses.
Supplemental Table 1. Proteomics-Based Data on Subcellular Localization and Phosphorylation of CURT1 Proteins.
Supplemental Table 2. Chlorophyll Fluorescence Parameters Measured in Wild-Type and curt1 Mutant Leaves.
Supplemental Table 3. Fatty Acid Composition of Arabidopsis Leaf Lipids.
Supplemental Table 4. Oligonucleotides Used for PCR and RT-PCR.
Supplemental Data Set 1. Diameter and Height of Grana in Different Mutants and Wild-Type Plants.
Supplemental References 1. Supplemental References for Supplemental Figures and Tables.
Acknowledgments
We thank the Deutsche Forschungsgemeinschaft (Grant B8 of SFB-TR1) for support, Paul Hardy for critical reading of the article, Elina Makarenko, Steffen Heinz, Alma Tursic, and Christine Hümmer for technical assistance, and Henrik Vibe Scheller, Ute Vothknecht, Dominique Rumeau, and Jörg Meurer for providing antibodies.
AUTHOR CONTRIBUTIONS
U.A., M.P., M.L., S.V., F.R., P.J., P.D., G.W., and D.L. designed the research. U.A., M.P., M.L., S.V., W.X., M.S., A.P.H., U.R., P.E.J., F.R., P.J., and G.W. performed the research. U.A., M.P., F.R., P.D., G.W., and D.L. prepared the article. D.L. supervised the study.
Glossary
- PSI
photosystem I
- PSII
Photosystem II
- LHCII
light-harvesting complex II
- TM
transmembrane domain
- Col-0
Columbia-0
- BN-PAGE
Blue-Native PAGE
- NPQ
nonphotochemical quenching
- TEM
transmission electron microscopy
- β-DM
n-dodecyl β-d-maltoside
- CoIP
coimmunoprecipitation
References
- Albertsson P.A., Andreasson E. (2004). The constant proportion of grana and stroma lamellae in plant chloroplasts. Physiol. Plant. 121: 334–342 [DOI] [PubMed] [Google Scholar]
- Allen J.F., Forsberg J. (2001). Molecular recognition in thylakoid structure and function. Trends Plant Sci. 6: 317–326 [DOI] [PubMed] [Google Scholar]
- Anderson J.M. (1986). Photoregulation of the composition, function, and structure of thylakoid membranes. Annu. Rev. Plant Physiol. Plant Mol. Biol. 37: 93–136 [Google Scholar]
- Anderson J.M., Chow W.S., De Las Rivas J. (2008). Dynamic flexibility in the structure and function of photosystem II in higher plant thylakoid membranes: The grana enigma. Photosynth. Res. 98: 575–587 [DOI] [PubMed] [Google Scholar]
- Armbruster U., Zühlke J., Rengstl B., Kreller R., Makarenko E., Rühle T., Schünemann D., Jahns P., Weisshaar B., Nickelsen J., Leister D. (2010). The Arabidopsis thylakoid protein PAM68 is required for efficient D1 biogenesis and photosystem II assembly. Plant Cell 22: 3439–3460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aronsson H., Jarvis P. (2002). A simple method for isolating import-competent Arabidopsis chloroplasts. FEBS Lett. 529: 215–220 [DOI] [PubMed] [Google Scholar]
- Austin J.R., II, Staehelin L.A. (2011). Three-dimensional architecture of grana and stroma thylakoids of higher plants as determined by electron tomography. Plant Physiol. 155: 1601–1611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassi R., dal Belin Peruffo A., Barbato R., Ghisi R. (1985). Differences in chlorophyll-protein complexes and composition of polypeptides between thylakoids from bundle sheaths and mesophyll cells in maize. Eur. J. Biochem. 146: 589–595 [DOI] [PubMed] [Google Scholar]
- Bellafiore S., Barneche F., Peltier G., Rochaix J.D. (2005). State transitions and light adaptation require chloroplast thylakoid protein kinase STN7. Nature 433: 892–895 [DOI] [PubMed] [Google Scholar]
- Biehl A., Richly E., Noutsos C., Salamini F., Leister D. (2005). Analysis of 101 nuclear transcriptomes reveals 23 distinct regulons and their relationship to metabolism, chromosomal gene distribution and co-ordination of nuclear and plastid gene expression. Gene 344: 33–41 [DOI] [PubMed] [Google Scholar]
- Bonardi V., Pesaresi P., Becker T., Schleiff E., Wagner R., Pfannschmidt T., Jahns P., Leister D. (2005). Photosystem II core phosphorylation and photosynthetic acclimation require two different protein kinases. Nature 437: 1179–1182 [DOI] [PubMed] [Google Scholar]
- Browse J., McCourt P.J., Somerville C.R. (1986). Fatty acid composition of leaf lipids determined after combined digestion and fatty acid methyl ester formation from fresh tissue. Anal. Biochem. 152: 141–145 [DOI] [PubMed] [Google Scholar]
- Brumfeld V., Charuvi D., Nevo R., Chuartzman S., Tsabari O., Ohad I., Shimoni E., Reich Z. (2008). A note on three-dimensional models of higher-plant thylakoid networks. Plant Cell 20: 2546–2549, author reply 2549–2551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Y.P., Wolk C.P. (1990). Use of a conditionally lethal gene in Anabaena sp. strain PCC 7120 to select for double recombinants and to entrap insertion sequences. J. Bacteriol. 172: 3138–3145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow W.S., Kim E.H., Horton P., Anderson J.M. (2005). Granal stacking of thylakoid membranes in higher plant chloroplasts: the physicochemical forces at work and the functional consequences that ensue. Photochem. Photobiol. Sci. 4: 1081–1090 [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Cole C., Barber J.D., Barton G.J. (2008). The Jpred 3 secondary structure prediction server. Nucleic Acids Res. 36 (Web Server issue): W197–W201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DalCorso G., Pesaresi P., Masiero S., Aseeva E., Schünemann D., Finazzi G., Joliot P., Barbato R., Leister D. (2008). A complex containing PGRL1 and PGR5 is involved in the switch between linear and cyclic electron flow in Arabidopsis. Cell 132: 273–285 [DOI] [PubMed] [Google Scholar]
- Daum B., Kühlbrandt W. (2011). Electron tomography of plant thylakoid membranes. J. Exp. Bot. 62: 2393–2402 [DOI] [PubMed] [Google Scholar]
- De Camilli P., Chen H., Hyman J., Panepucci E., Bateman A., Brunger A.T. (2002). The ENTH domain. FEBS Lett. 513: 11–18 [DOI] [PubMed] [Google Scholar]
- Dekker J.P., Boekema E.J. (2005). Supramolecular organization of thylakoid membrane proteins in green plants. Biochim. Biophys. Acta 1706: 12–39 [DOI] [PubMed] [Google Scholar]
- Emanuelsson O., Nielsen H., von Heijne G. (1999). ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci. 8: 978–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engler C., Gruetzner R., Kandzia R., Marillonnet S. (2009). Golden gate shuffling: A one-pot DNA shuffling method based on type IIs restriction enzymes. PLoS ONE 4: e5553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson J., Salih G.F., Ghebramedhin H., Jansson C. (2000). Deletion mutagenesis of the 5′ psbA2 region in Synechocystis 6803: Identification of a putative cis element involved in photoregulation. Mol. Cell Biol. Res. Commun. 3: 292–298 [DOI] [PubMed] [Google Scholar]
- Farsad K., De Camilli P. (2003). Mechanisms of membrane deformation. Curr. Opin. Cell Biol. 15: 372–381 [DOI] [PubMed] [Google Scholar]
- Ford M.G., Mills I.G., Peter B.J., Vallis Y., Praefcke G.J., Evans P.R., McMahon H.T. (2002). Curvature of clathrin-coated pits driven by epsin. Nature 419: 361–366 [DOI] [PubMed] [Google Scholar]
- Friso G., Giacomelli L., Ytterberg A.J., Peltier J.B., Rudella A., Sun Q., Wijk K.J. (2004). In-depth analysis of the thylakoid membrane proteome of Arabidopsis thaliana chloroplasts: New proteins, new functions, and a plastid proteome database. Plant Cell 16: 478–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fristedt R., Willig A., Granath P., Crèvecoeur M., Rochaix J.D., Vener A.V. (2009). Phosphorylation of photosystem II controls functional macroscopic folding of photosynthetic membranes in Arabidopsis. Plant Cell 21: 3950–3964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gounaris K., Barber J. (1983). Monogalactosyldiacylglycerol: The most abundant polar lipid in nature. Trends Biochem. Sci. 8: 378–381 [Google Scholar]
- Haferkamp S., Kirchhoff H. (2008). Significance of molecular crowding in grana membranes of higher plants for light harvesting by photosystem II. Photosynth. Res. 95: 129–134 [DOI] [PubMed] [Google Scholar]
- Hansson M., Vener A.V. (2003). Identification of three previously unknown in vivo protein phosphorylation sites in thylakoid membranes of Arabidopsis thaliana. Mol. Cell. Proteomics 2: 550–559 [DOI] [PubMed] [Google Scholar]
- Hertle A.P., Blunder T., Wunder T., Pesaresi P., Pribil M., Armbruster U., Leister D. (2013). PGRL1 is the elusive ferredoxin-plastoquinone reductase in photosynthetic cyclic electron flow. Mol. Cell 49: 511–523 [DOI] [PubMed] [Google Scholar]
- Jach G., Binot E., Frings S., Luxa K., Schell J. (2001). Use of red fluorescent protein from Discosoma sp. (dsRED) as a reporter for plant gene expression. Plant J. 28: 483–491 [DOI] [PubMed] [Google Scholar]
- Karnauchov I., Herrmann R.G., Klösgen R.B. (1997). Transmembrane topology of the Rieske Fe/S protein of the cytochrome b6/f complex from spinach chloroplasts. FEBS Lett. 408: 206–210 [DOI] [PubMed] [Google Scholar]
- Kay B.K., Yamabhai M., Wendland B., Emr S.D. (1999). Identification of a novel domain shared by putative components of the endocytic and cytoskeletal machinery. Protein Sci. 8: 435–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khrouchtchova A., Hansson M., Paakkarinen V., Vainonen J.P., Zhang S., Jensen P.E., Scheller H.V., Vener A.V., Aro E.M., Haldrup A. (2005). A previously found thylakoid membrane protein of 14kDa (TMP14) is a novel subunit of plant photosystem I and is designated PSI-P. FEBS Lett. 579: 4808–4812 [DOI] [PubMed] [Google Scholar]
- Kim T.W., Keum J.W., Oh I.S., Choi C.Y., Park C.G., Kim D.M. (2006). Simple procedures for the construction of a robust and cost-effective cell-free protein synthesis system. J. Biotechnol. 126: 554–561 [DOI] [PubMed] [Google Scholar]
- Kirchhoff H., Hall C., Wood M., Herbstová M., Tsabari O., Nevo R., Charuvi D., Shimoni E., Reich Z. (2011). Dynamic control of protein diffusion within the granal thylakoid lumen. Proc. Natl. Acad. Sci. USA 108: 20248–20253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchhoff H., Mukherjee U., Galla H.J. (2002). Molecular architecture of the thylakoid membrane: Lipid diffusion space for plastoquinone. Biochemistry 41: 4872–4882 [DOI] [PubMed] [Google Scholar]
- Kramer D.M., Johnson G., Kiirats O., Edwards G.E. (2004). New fluorescence parameters for the determination of q(a) redox state and excitation energy fluxes. Photosynth. Res. 79: 209–218 [DOI] [PubMed] [Google Scholar]
- Krogh A., Larsson B., von Heijne G., Sonnhammer E.L. (2001). Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J. Mol. Biol. 305: 567–580 [DOI] [PubMed] [Google Scholar]
- Kunert A., Hagemann M., Erdmann N. (2000). Construction of promoter probe vectors for Synechocystis sp. PCC 6803 using the light-emitting reporter systems Gfp and LuxAB. J. Microbiol. Methods 41: 185–194 [DOI] [PubMed] [Google Scholar]
- Li H.M., Moore T., Keegstra K. (1991). Targeting of proteins to the outer envelope membrane uses a different pathway than transport into chloroplasts. Plant Cell 3: 709–717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-García J.F., Monte E., Quail P.H. (1999). A simple, rapid and quantitative method for preparing Arabidopsis protein extracts for immunoblot analysis. Plant J. 20: 251–257 [DOI] [PubMed] [Google Scholar]
- Mullineaux C.W. (2005). Function and evolution of grana. Trends Plant Sci. 10: 521–525 [DOI] [PubMed] [Google Scholar]
- Mullineaux C.W. (2008). Factors controlling the mobility of photosynthetic proteins. Photochem. Photobiol. 84: 1310–1316 [DOI] [PubMed] [Google Scholar]
- Mulo P., Sirpiö S., Suorsa M., Aro E.M. (2008). Auxiliary proteins involved in the assembly and sustenance of photosystem II. Photosynth. Res. 98: 489–501 [DOI] [PubMed] [Google Scholar]
- Munekage Y., Hojo M., Meurer J., Endo T., Tasaka M., Shikanai T. (2002). PGR5 is involved in cyclic electron flow around photosystem I and is essential for photoprotection in Arabidopsis. Cell 110: 361–371 [DOI] [PubMed] [Google Scholar]
- Mustárdy L., Buttle K., Steinbach G., Garab G. (2008). The three-dimensional network of the thylakoid membranes in plants: Quasihelical model of the granum-stroma assembly. Plant Cell 20: 2552–2557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustárdy L., Garab G. (2003). Granum revisited. A three-dimensional model—where things fall into place. Trends Plant Sci. 8: 117–122 [DOI] [PubMed] [Google Scholar]
- Obayashi T., Kinoshita K., Nakai K., Shibaoka M., Hayashi S., Saeki M., Shibata D., Saito K., Ohta H. (2007). ATTED-II: A database of co-expressed genes and cis elements for identifying co-regulated gene groups in Arabidopsis. Nucleic Acids Res. 35 (Database issue): D863–D869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opaliński L., Kiel J.A., Williams C., Veenhuis M., van der Klei I.J. (2011). Membrane curvature during peroxisome fission requires Pex11. EMBO J. 30: 5–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peltier J.B., Ytterberg A.J., Sun Q., van Wijk K.J. (2004). New functions of the thylakoid membrane proteome of Arabidopsis thaliana revealed by a simple, fast, and versatile fractionation strategy. J. Biol. Chem. 279: 49367–49383 [DOI] [PubMed] [Google Scholar]
- Pesaresi P., Hertle A., Pribil M., Kleine T., Wagner R., Strissel H., Ihnatowicz A., Bonardi V., Scharfenberg M., Schneider A., Pfannschmidt T., Leister D. (2009b). Arabidopsis STN7 kinase provides a link between short- and long-term photosynthetic acclimation. Plant Cell 21: 2402–2423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesaresi P., Scharfenberg M., Weigel M., Granlund I., Schröder W.P., Finazzi G., Rappaport F., Masiero S., Furini A., Jahns P., Leister D. (2009a). Mutants, overexpressors, and interactors of Arabidopsis plastocyanin isoforms: Revised roles of plastocyanin in photosynthetic electron flow and thylakoid redox state. Mol. Plant 2: 236–248 [DOI] [PubMed] [Google Scholar]
- Pribil M., Pesaresi P., Hertle A., Barbato R., Leister D. (2010). Role of plastid protein phosphatase TAP38 in LHCII dephosphorylation and thylakoid electron flow. PLoS Biol. 8: e1000288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappaport F., Béal D., Joliot A., Joliot P. (2007). On the advantages of using green light to study fluorescence yield changes in leaves. Biochim. Biophys. Acta 1767: 56–65 [DOI] [PubMed] [Google Scholar]
- Roy A., Kucukural A., Zhang Y. (2010). I-TASSER: A unified platform for automated protein structure and function prediction. Nat. Protoc. 5: 725–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin B.T., Barber J. (1980). The role of membrane surface charge in the control of photosynthetic processes and the involvement of electrostatic screening. Biochim. Biophys. Acta 592: 87–102 [DOI] [PubMed] [Google Scholar]
- Schägger H., Aquila H., Von Jagow G. (1988). Coomassie blue-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for direct visualization of polypeptides during electrophoresis. Anal. Biochem. 173: 201–205 [DOI] [PubMed] [Google Scholar]
- Shapiguzov A., Ingelsson B., Samol I., Andres C., Kessler F., Rochaix J.D., Vener A.V., Goldschmidt-Clermont M. (2010). The PPH1 phosphatase is specifically involved in LHCII dephosphorylation and state transitions in Arabidopsis. Proc. Natl. Acad. Sci. USA 107: 4782–4787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih Y.L., Huang K.F., Lai H.M., Liao J.H., Lee C.S., Chang C.M., Mak H.M., Hsieh C.W., Lin C.C. (2011). The N-terminal amphipathic helix of the topological specificity factor MinE is associated with shaping membrane curvature. PLoS ONE 6: e21425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimoni E., Rav-Hon O., Ohad I., Brumfeld V., Reich Z. (2005). Three-dimensional organization of higher-plant chloroplast thylakoid membranes revealed by electron tomography. Plant Cell 17: 2580–2586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J.D., Higgins D.G., Gibson T.J. (1994). CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22: 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trissl H.W., Wilhelm C. (1993). Why do thylakoid membranes from higher plants form grana stacks? Trends Biochem. Sci. 18: 415–419 [DOI] [PubMed] [Google Scholar]
- Varotto C., Pesaresi P., Meurer J., Oelmüller R., Steiner-Lange S., Salamini F., Leister D. (2000). Disruption of the Arabidopsis photosystem I gene psaE1 affects photosynthesis and impairs growth. Plant J. 22: 115–124 [DOI] [PubMed] [Google Scholar]
- Vermaas W.F.J., Williams J.G.K., Arntzen C.J. (1987). Sequencing and modification of psbB, the gene encoding the CP-47 protein of Photosystem II, in the cyanobacterium Synechocystis 6803. Plant Mol. Biol. 8: 317–326 [DOI] [PubMed] [Google Scholar]
- Webb M.S., Green B.R. (1991). Biochemical and biophysical properties of thylakoid acyl lipids. Biochim. Biophys. Acta 1060: 133–158 [Google Scholar]
- Weber E., Engler C., Gruetzner R., Werner S., Marillonnet S. (2011). A modular cloning system for standardized assembly of multigene constructs. PLoS ONE 6: e16765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon Y., Tong J., Lee P.J., Albanese A., Bhardwaj N., Källberg M., Digman M.A., Lu H., Gratton E., Shin Y.K., Cho W. (2010). Molecular basis of the potent membrane-remodeling activity of the epsin 1 N-terminal homology domain. J. Biol. Chem. 285: 531–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. (2008). I-TASSER server for protein 3D structure prediction. BMC Bioinformatics 9: 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerberg J., Kozlov M.M. (2006). How proteins produce cellular membrane curvature. Nat. Rev. Mol. Cell Biol. 7: 9–19 [DOI] [PubMed] [Google Scholar]