This work demonstrates the functional necessity of multisite light-induced phosphorylation of a bHLH transcription factor to signal propagation. It also identifies a novel signal attenuation mechanism whereby the act of forward signal transfer to the receiver is intrinsically linked to a direct, molecular feedback signal that triggers a reduction of overall sensory capacity, via a reduction in abundance of the signal donor.
Abstract
Plants constantly monitor informational light signals using sensory photoreceptors, which include the phytochrome (phy) family (phyA to phyE), and adjust their growth and development accordingly. Following light-induced nuclear translocation, photoactivated phy molecules bind to and induce rapid phosphorylation and degradation of phy-interacting basic Helix Loop Helix (bHLH) transcription factors (PIFs), such as PIF3, thereby regulating the expression of target genes. However, the mechanisms underlying the signal-relay process are still not fully understood. Here, using mass spectrometry, we identify multiple, in vivo, light-induced Ser/Thr phosphorylation sites in PIF3. Using transgenic expression of site-directed mutants of PIF3, we provide evidence that a set of these phosphorylation events acts collectively to trigger rapid degradation of the PIF3 protein in response to initial exposure of dark-grown seedlings to light. In addition, we show that phyB-induced PIF3 phosphorylation is also required for the known negative feedback modulation of phyB levels in prolonged light, potentially through codegradation of phyB and PIF3. This mutually regulatory intermolecular transaction thus provides a mechanism with the dual capacity to promote early, graded, or threshold regulation of the primary, PIF3-controlled transcriptional network in response to initial light exposure, and later, to attenuate global sensitivity to the light signal through reductions in photoreceptor levels upon prolonged exposure.
INTRODUCTION
Being rooted in soil, plants need to adjust their growth and development according to environmental cues. Among these cues, light is one of the most important factors, since it is plants’ only energy source. Plants have evolved different classes of photoreceptors to perceive light information, such as the quality (wavelength) and quantity (intensity) of the incoming signals (Schafer and Nagy, 2006). The phytochrome (phy) family perceives the red (R) and far-red (FR) light information to direct many aspects of plant growth, such as seed germination, seedling deetiolation, shade avoidance, and flowering. The Arabidopsis thaliana genome encodes a small family of five phys (phyA to phyE). PhyA is abundant in young, dark-grown seedlings and plays important roles during early R and continuous FR light–induced seedling deetiolation (Sharrock and Clack, 2002; Tepperman et al., 2006). PhyA is rapidly degraded to very low levels in continuous light. PhyB is more stable in light and plays a major role in mediating hypocotyl inhibition under long-term continuous R light (Somers et al., 1991; Reed et al., 1993). The phys switch reversibly between their biologically inactive Pr and active Pfr conformers, upon sequential absorption of R and FR photons. In dark-germinated seedlings, newly synthesized phys exist in the inactive Pr form and are predominantly in the cytosol. Exposure to R light converts the photoreceptor into its active Pfr form, and this form then translocates into the nucleus, followed by rapid formation of early subnuclear speckles (photobodies) (Sakamoto and Nagatani, 1996; Kircher et al., 1999; Yamaguchi et al., 1999; Kircher et al., 2002; Chen and Chory, 2011). Light-induced nuclear translocation is required for the majority of the biological functions of phyB (Huq et al., 2003; Matsushita et al., 2003). In the nucleus, phys initiate changes in the expression of ∼10% of the genes in the Arabidopsis genome (Tepperman et al., 2006; Jiao et al., 2007; Leivar et al., 2009).
The constitutively nuclear basic Helix Loop Helix (bHLH) transcription factor PIF3 is the founding member of a set of such factors, termed phytochrome interacting factors (PIFs), that interact photoreversibly with the active Pfr form of phy with strong affinity (Ni et al., 1998, 1999). All PIF proteins have a conserved motif in the N-terminal domain, called the active phyB binding motif, that binds phyB with high affinity (Khanna et al., 2004). PIF1 and PIF3 have also been shown to bind phyA, although the active phyA binding site in these two proteins is not conserved, and they have differing affinities for phyA (Huq et al., 2004; Al-Sady et al., 2006; Shen et al., 2008). The constitutively photomorphogenic (cop)-like phenotype of dark-grown pif1 pif3 pif4 pif5 mutant seedlings has provided strong evidence that these PIFs actively promote skotomorphogenesis (repress photomorphogenesis) in the dark, in a partially redundant fashion (Leivar et al., 2008a; Shin et al., 2009). Additional evidence indicates that upon initial light exposure, phy reverses PIF activity by inducing its rapid degradation through the ubiquitin proteasome system (Bauer et al., 2004; Shen et al., 2007, 2008).
Light-induced PIF degradation has been shown, at least for PIF1 and PIF3, to require direct, physical interaction with the photoactivated Pfr form of phy (Al-Sady et al., 2006; Shen et al., 2008). Light also induces rapid colocalization of PIF3 with photoactivated phys in subnuclear speckles or photobodies (Bauer et al., 2004; Chen and Chory, 2011), in a process that likewise requires direct interaction with the photoreceptor (Al-Sady et al., 2006), but whose function is currently unknown. In addition, light induces a rapid mobility shift in PIF3, detectable upon gel electrophoresis, prior to its degradation. This light-induced mobility shift is reversed by phosphatase treatment. Furthermore, mutant PIF3 protein that lacks binding affinity for both phyA and phyB does not show the rapid light-induced mobility shift and degradation (Al-Sady et al., 2006). These results suggest that photoactivated phy induces rapid PIF3 phosphorylation through direct binding and that phosphorylated PIF3 is thereby flagged for ubiquitin-mediated degradation. PIF1, PIF4, and PIF5 are similarly rapidly modified (phosphorylated) prior to light-induced degradation (Shen et al., 2007, 2008; Lorrain et al., 2008). Therefore, it appears that light-induced phosphorylation of PIF proteins represents the primary biochemical mechanism of signal transfer from the photoactivated phy molecule to these transcription factors in vivo. However, the molecular nature of the phosphorylation events and the identity of the specific kinase(s) involved have yet to be determined (Bu et al., 2011a, 2011b). In addition, the mechanism by which light-induced phosphorylation regulates PIF3 degradation is not fully understood.
pif3, pif4, pif5, and pif7 mutants are hypersensitive to R light when grown in prolonged, continuous irradiation, suggesting that these PIFs continue to antagonize photomorphogenesis in fully deetiolated, light-grown seedlings (Huq and Quail, 2002; Kim et al., 2003; Fujimori et al., 2004; Monte et al., 2004; Khanna et al., 2007; Nozue et al., 2007; de Lucas et al., 2008; Feng et al., 2008; Leivar et al., 2008b, 2012; Lorrain et al., 2008, 2009; Shin et al., 2009). The data suggest that an important aspect of this role of the PIFs in prolonged light is imposed through negative feedback regulation of phyB photoreceptor levels (Khanna et al., 2007; Al-Sady et al., 2008; Leivar et al., 2008b, 2012). This modulation of phyB abundance by PIF3 and PIF5 under long-term irradiation has been shown to require direct binding of the bHLH proteins to the photoreceptor (Khanna et al., 2007; Al-Sady et al., 2008). Although recent evidence indicates that the COP1 E3 ligase is involved in ubiquitylation and consequent degradation of phyB, in a PIF-promoted manner (Jang et al., 2010), the role of phy-induced phosphorylation of the PIFs in this process remains to be determined.
To dissect the molecular nature of the signal transfer process from photoactivated phy to the PIF transcription factors, we focused here on the identification and functional characterization of the light-induced in vivo phosphorylation sites in PIF3 during the initial dark-to-light transition of dark-grown seedlings. By introducing missense point mutations at the light-induced phosphosites, we not only reinforced the conclusion that light-induced phosphorylation of PIF3 is required for its subsequent degradation, but also defined the nature of the phosphorylation pattern presumptively recognized by the ubiquitin proteasome system in performing this degradation process. In addition, our data provide insight into the intermolecular transaction underlying the converse PIF3-induced degradation of photoactivated phyB.
RESULTS
Light Induces Multisite Phosphorylation of PIF3 in Vivo
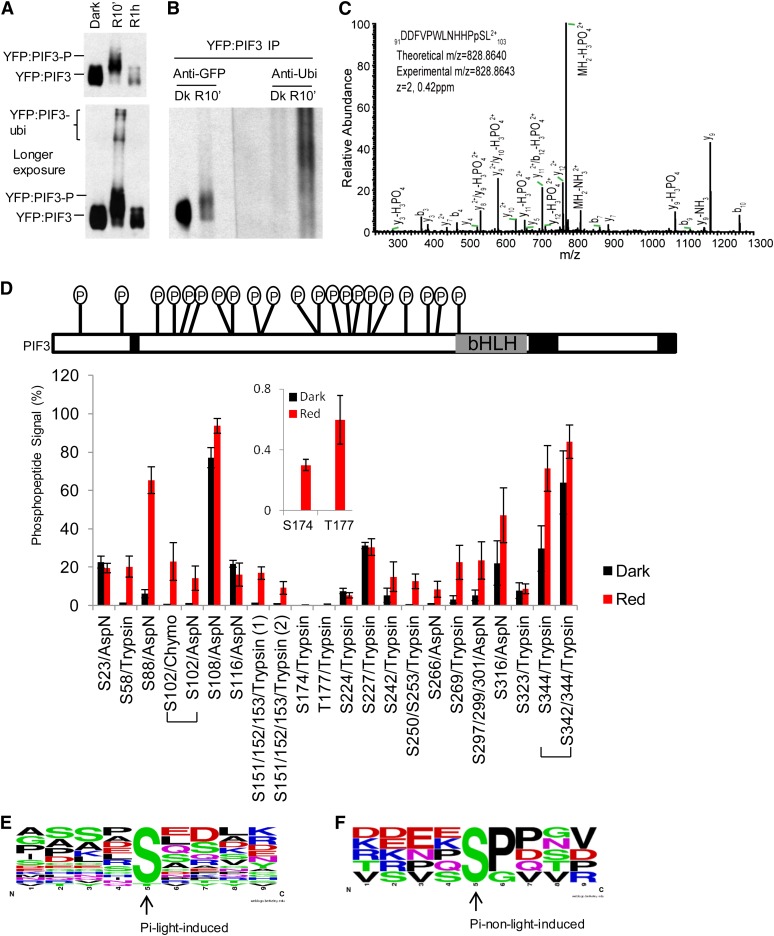
Because of difficulties encountered in producing sufficient quantities of full-length PIF3 (524 amino acids) in planta, suitable for purification and mass spectrometric analysis, we chose to use a more highly expressed derivative, containing amino acids 1 to 507 (i.e., lacking the COOH-terminal 17 residues), as a yellow fluorescent protein (YFP) fusion (YFP:PIF3-N507). Examination of the molecular dynamics of this protein in vivo established that it retains responsiveness to phy-mediated light signals, indistinguishable from those of the endogenous, native PIF3 molecule. Figure 1A (top panel) shows that this YFP:PIF3 fusion protein undergoes a rapid light-induced mobility shift by gel electrophoresis (slower migrating bands) within 10 min of exposure to a saturating R light pulse (R10’), prior to its degradation, which is robust within 1 h of R exposure (R1h). A longer exposure of this immunoblot reveals additional modified YFP:PIF3 bands in the very high molecular weight region of the R10 min sample but lacking in the dark (control) sample (Figure 1A, bottom panel). Immunoblot analysis of affinity-purified YFP:PIF3-N507 from similar samples, using antiubiquitin antibodies, detects a strong signal in this very high molecular weight region, whereas no ubiquitination is detected in the dark sample (Figure 1B). This result suggests that the modified proteins at very high molecular weights in the R10 min sample represent polyubiquitinated YFP:PIF3 protein, whereas the major faster-migrating, but also shifted bands, closer to the mobility of the dark sample bands, are due to a different light-induced posttranslational modification, probably phosphorylation. Collectively, these data indicate that the YFP:PIF3-N507 fusion protein contains the majority, if not all, of the molecular determinants necessary for normal signal transfer from photoactivated phy to PIF3.
Figure 1.
R Light Induces Rapid Enhancement of Multisite Phosphorylation in PIF3 in Vivo.
(A) Top panel: YFP:PIF3-N507 protein from dark (Dk)-grown transgenic Arabidopsis seedlings undergoes rapid light-induced mobility shift (phosphorylation) and degradation similar to native PIF3. Dark-grown YFP:PIF3-N507-expressing transgenic Arabidopsis seedlings were either kept in the dark or given a saturating R light pulse, then moved into darkness for a total time of 10 min or 1 h before protein extraction and immunoblot with GFP antibody. Bottom panel: YFP:PIF3-N507 protein was also detected in the very high molecular weight region (likely polyubiquitinated product) after a longer autoradiograph exposure time.
(B) Immunoblot against ubiquitin shows that light induces rapid YFP:PIF3-N507 phosphorylation (mobility shift; left panel [anti-GFP]) and polyubiquitination (high molecular weight region; right panel [anti-Ubi]) in the transgenic seedlings. YFP:PIF3-N507 protein was affinity purified with polyclonal GFP antibody and then blotted with either monoclonal GFP antibody (anti-GFP) or with ubiquitin antibody (anti-Ubi). Transgenic seedlings were grown and treated with R light (10 min) as in (A). IP, immunoprecipitation.
(C) Collision-induced dissociation mass spectrum showing phosphorylation of Ser-102, a light-induced phosphorylation site in PIF3. YFP:PIF3-N507 protein from transgenic plants was affinity purified as in (B) before being subjected to in-gel digestion with AspN.
(D) The phosphopeptide signal increases rapidly in response to R light exposure of seedlings. Top panel: Distribution of all phosphorylation sites identified in PIF3. The total mass spectrometry peptide coverage for PIF3 is ∼85% (uncovered regions are marked with back boxes). Bottom panel: Comparison of phosphopeptide signal (%) for PIF3 from dark control and R light–exposed (red 10 min) seedlings. The signal of the phosphopeptide containing a given phosphorylation site is expressed as the ion intensity of the phosphopeptide divided by the total ion intensity signal for this peptide (unmodified + phosphorylated) × 100 (see Supplemental Figure 1C online). The values do not represent absolute phosphorylation stoichiometries, but the relative percent values for R versus dark samples do correspond to the change in phosphorylation stoichiometry between these dark and R light–treated samples (red 10 min). Peptides were derived from in-gel digestion with trypsin, chymotrypsin (chymo), or AspN. Data are represented as the mean of biological triplicates ± se. Inset: Two light-induced sites with low phosphopeptide signal values.
(E) and (F) No strict consensus motif was identified for either light-induced (E) or non-light-induced (F) PIF3 phosphorylation sites using Weblogo. The middle Ser or Thr is the phosphorylation site.
For mass spectrometry analysis of the phosphorylation sites, we affinity purified the YFP:PIF3-N507 protein from either dark-grown or R light–treated seedlings at the 10-min time point (R10 min) and subjected the samples to in-gel proteolytic digestion. For each biological repeat, we excised three gel segments corresponding to the YFP:PIF3 protein bands for the dark and R samples (see Supplemental Figure 1A online). We did three biological repeats with either trypsin or AspN digestion and two biological repeats with chymotrypsin digestion. The use of different digestion enzymes leads to the detection of peptides spanning different subsets of the protein sequence, so by combining results, one achieves increased sequence coverage: Together, we observed ∼85% sequence coverage of PIF3. We identified multiple Ser/Thr phosphorylation sites in the protein, with many of them being identified in both dark- and light-treated samples.
We compared the extent of phosphorylation under dark and light conditions using label-free methods. As an example, Supplemental Figure 1B online shows the ion intensities of unmodified and phosphorylated versions of the peptide DDFVPWLNHHPSL (where phosphorylation was found on the Ser residue) extracted from the mass spectrometry raw data of the three gel fractions. A mass spectrum showing the identification of this phosphorylation site (Ser-102) is shown in Figure 1C. It is not possible to determine absolute stoichiometry of modification from these data, as the addition of a phosphate group affects the intensity of the signal, but it is possible to determine a relative change in phosphorylation between dark and R samples by comparing the phosphopeptide signal between R and dark, after normalizing for differences in total PIF3 protein level, by calculating the total signal for the peptide (sum of unmodified and phosphorylated signal). The top panel in Supplemental Figure 1C online describes how these values, designated the phosphopeptide signal (%), were calculated for each sample. These values were then compared between dark and R light treated samples to determine fold changes in phosphorylation at each phosphosite upon stimulation (Figure 1D). The linearity of the ion intensity was verified with a dilution series of the sample as shown in Supplemental Table 1 online. A summary of the overall distribution of all phosphorylation signals identified in PIF3 is depicted at the top of Figure 1D, and details about the raw data from individual bands that were used to compute these values are in Supplemental Data Set 1 online. To reiterate, the absolute values in this plot do not correspond to phosphorylation stoichiometry, but there will be a correlation between higher values and stoichiometry of phosphorylation. Those sites determined as being most sensitive to R light are those where the ratio of the phosphopeptide signal between R light and dark samples is largest. Some sites showed >20-fold changes, most notably those where there was no detectable signal without light stimulation. As can be seen from the values in Supplemental Data Set 1 online, the phosphopeptide signal in the higher molecular weight fraction from the R sample (R3) is mostly higher than that in the lower molecular weight fraction (R1) (see Supplemental Figure 1B online), consistent with phosphorylation contributing to the slower migration in the gel of this band.
For those sites that were detected in more than one enzyme digest, the results from these different digests agreed on whether a site was light sensitive (see Supplemental Data Set 1 online). For example, the phosphopeptide signals of Ser-102 from two different enzyme digestions are shown in Figure 1D. Using this approach, we defined 16 light-induced phosphorylation sites. Some sites contained more than one Ser or Thr residue, so that the 16 sites correspond to a total of 20 residues. Of these, 19 (Ser-58, Ser-88, Ser-102, Ser-151/Ser-152/Ser-153 [two sites], Ser-174, Thr-177, Ser-242, Ser-250, Ser-253, Ser-266, Ser-269, Ser-297/Ser-299/Ser-301, Ser-316, Ser-342/Ser-344 [where forward slashes indicate a site consisting of the indicated residues, where the actual phosphorylated residue[s] cannot be defined) were identified directly as light-induced phosphopeptides (spectra in Supplemental Figures 1D to 1R online), and one (Ser108) was inferred as follows. Phosphorylation of Ser-108 was detected in Asp-N digestions, but it was not possible to accurately quantify the extent of light induction using our approach, as the phosphorylation appeared to prevent cleavage before Asp-109. Therefore, a missed cleavage was more commonly observed when Ser-108 was phosphorylated, preventing protein level normalization, which requires both modified and unmodified versions of the same peptide. Nevertheless, the phosphopeptide signal was higher in all of the R light than the dark samples, and peptide-level quantification from other peptides indicated that the overall PIF3 levels were lower in the R light samples, suggesting that this site may also be induced by R light. A second method of calculating phosphorylation changes employed ratios between detected signal intensities of phosphorylated and unphosphorylated peptides (bottom panel in Supplemental Figure 1C online) (Steen et al., 2008). The results from this analysis are also presented in Supplemental Data Set 1 and Supplemental Table 2 online and generally agreed with those determined using the percentage phosphopeptide signals.
No strict consensus motif was identified for either the light-induced (Figure 1E) or non-light-induced (Figure1F) PIF3 phosphorylation sites using Weblogo. However, a Pro enriched immediately downstream of the Ser residue in the non-light-induced sites, but lacking in the light-induced sites, might indicate a distinction in target specificity between these two classes of sites.
In Vivo R Light–Induced PIF3 Phosphorylation Is Required for Rapid Degradation
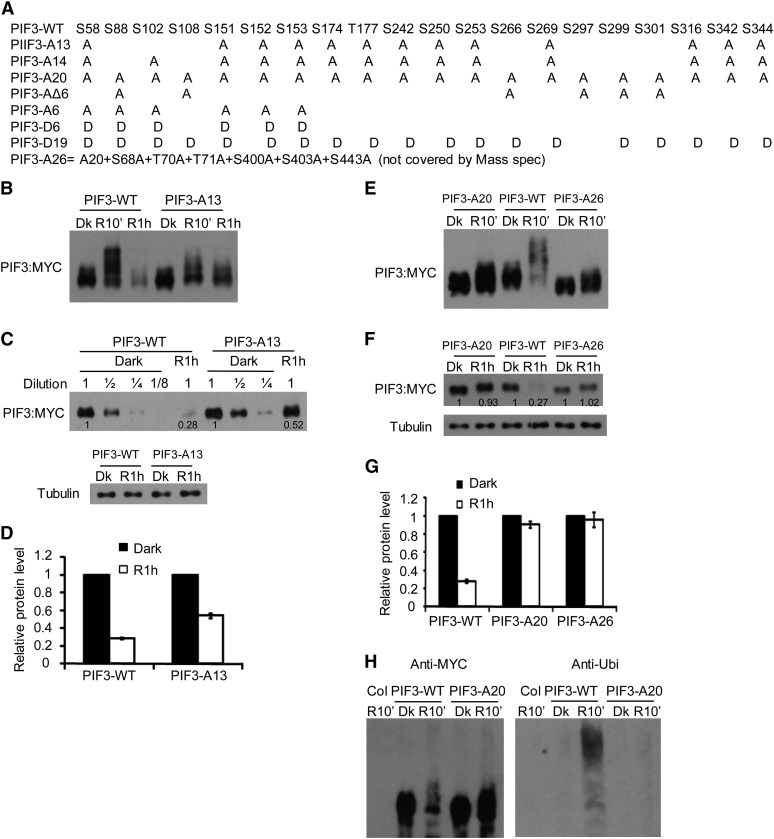
To assess the biological significance of the identified light-induced phosphorylation sites, we introduced point mutations (phospho-dead [S/T to A] or phospho-mimic [S/T to D]) into the full-length wild-type PIF3 polypeptide and generated transgenic lines expressing these proteins, with an epitope tag, driven by the 35S promoter, in the pif3 null mutant background. Figure 2A shows the terminology of all the multisite mutant-PIF3 sequences tested in this study.
Figure 2.
R Light–Induced PIF3 Phosphorylation in Vivo Is Required for Rapid Degradation.
(A) Amino acid substitutions in various mutant PIF3 constructs used for protein expression in vivo. The series of point mutants introduced in the 20 residues at the 15 light-induced phosphorylation sites identified here, shown below the wild-type (WT) sequence. A, Ser/Thr-to-Ala mutation; D, Ser/Thr-to-Asp mutation. Construct designations denote the nature and number of concomitant residue substitutions engineered in each mutant polypeptide. For in vivo functional analysis in Arabidopsis seedlings, all the constructs were transformed into the pif3 mutant and expressed transgenically under the control of the cauliflower mosaic virus 35S promoter.
(B) to (D) Targeted Ser/Thr-to-Ala mutations in a subset of the light-induced phosphorylation sites results in reduced phosphorylation and degradation of PIF3 induced by light in transgenic Arabidopsis.
(B) The PIF3-A13 protein shows a partial reduction in light-induced phosphorylation and degradation. Seedlings of transgenic lines expressing PIF3-WT:MYC or PIF3-A13:MYC fusion proteins were grown for 2.5 d in darkness and then either maintained in the dark (Dk) or given a 1-min saturating R light pulse and returned to darkness for 9 min (R10’) or 59 min (R1h) before extraction into denaturing buffer and immunoblot analysis using antibody against the MYC epitope.
(C) Visual quantitative assessment of the degree of light-induced PIF3-A13 degradation. Immunoblot analysis of the same protein extracts as in (B). The PIF3-WT:MYC and PIF3-A13:MYC proteins at R1h were compared with a dilution series of their corresponding dark samples. Tubulin was used as loading control. Numbers directly on blot image denote the relative PIF3 protein levels normalized to tubulin.
(D) Quantification of the comparative degrees of PIF3-WT and PIF3-A13 degradation at R1h from immunoblot scans. Dark protein levels were set at 1 for each construct. Data are represented as the mean of biological triplicates ± se.
(E) to (H) Targeted Ser/Thr-to-Ala mutations in all identified light-induced sites strongly reduce the phosphorylation, ubiquitination, and degradation of PIF3 induced by light in transgenic plants.
(E) and (F) Immunoblot analysis of R light–induced phosphorylation (R10’) (E) and degradation (R1h) (F) of PIF-A20 and PIF-A26 variants using anti-MYC antibody. Seedlings of transgenic lines expressing PIF3-WT:MYC, PIF3-A20:MYC, or PIF3-A26:MYC fusion proteins were grown and treated with R light as in (B). Immunoblot analysis was similar to (B) and (C). Numbers directly on blot image in (F) denote the relative PIF3 protein levels normalized to tubulin.
(G) Quantification of the comparative degrees of light-induced PIF3-WT, PIF3-A20, and PIF3-A26 degradation at R1h as in (D). Data represent the mean of biological triplicates ± SE.
(H) R light induces a significant accumulation of polyubiquitinated PIF3-WT protein but not PIF3-A20 in transgenic lines. Proteins extracted from Col, PIF3-WT:MYC, and PIF3-A20:MYC transgenic lines were affinity purified with goat anti-MYC antibody and subjected to immunoblot analysis on two duplicate blots, one using monoclonal anti-MYC antibodies and the other antiubiquitin antibodies, for detection of the respective proteins. Seedlings were grown and treated with R light (R10’) as in (B).
We first tested the effect of single phospho-dead point mutations on the light-induced mobility shift and degradation of PIF3. We tested more than half of the light-induced sites individually, including all the strongly induced sites, but found no significant difference between these mutant-PIF3 and wild-type PIF3 proteins in either the light-induced mobility shift at R10 min or protein degradation at R1h. The data for Ser-88 are shown as a representative example in Supplemental Figures 2A and 2B online.
When all 13 light-induced phosphorylation sites identified by trypsin digestion were mutated to Ala, the mutant PIF3 protein (PIF3-A13) showed a clear reduction in light-induced mobility shift (phosphorylation) at R10 min and degradation at R1h in transgenic Arabidopsis (Figures 2B and 2C). As quantified in Figure 2D, the PIF3-WT protein is degraded to ∼25% of the dark level at R1h, whereas PIF3-A13 is only degraded to ∼50% of the dark level.
We then introduced Ala mutations into the residues at all 20 light-induced phosphorylation sites defined in PIF3 (PIF3-A20) and tested this mutant protein in transgenic plants. In parallel, we tested a second mutant, PIF3-A26, that includes additional residues that were not covered by the mass spectrometry analysis (Figures 1D and 2A). As is shown in Figures 2E and 2F, both the R light–induced mobility shift (phosphorylation at R10 min) and degradation (R1h) of PIF3 are strongly reduced in both these mutant PIF3 transgenic lines. Further quantification data suggest that both PIF3-A20 and PIF3-A26 proteins retain >90% of their dark level at R1h (Figure 2G). It is possible that the residual induced mobility shift in PIF3-A20 represents phosphorylation sites that were missed in our mass spectrometry analysis due to a low phosphorylation level in wild-type PIF3 and that these sites were phosphorylated to a higher level in the light in PIF3-A20 due to the strongly increased stability of the mutant protein.
We then assayed light-induced ubiquitination using affinity-purified proteins from these transgenic plants. While PIF3-WT is rapidly ubiquitinated at R10 min, PIF3-A20 ubiquitination is strongly reduced (Figure 2H), indicating that light-induced PIF3 phosphorylation is required for rapid ubiquitin modification in light.
We also assayed the light-induced shift (phosphorylation) and degradation parameters for PIF3-A14 (A13+S102 from chymotrypsin digestion) and PIF3-AΔ6 (six light-induced sites identified by AspN) (Figure 2A). Both PIF3-A14 and PIF3-AΔ6 show reduced light-induced shift and degradation responses, similar to those found in PIF3-A13 (see Supplemental Figures 2C and 2D online).
Mutations in the Light-Induced PIF3 Phosphorylation Sites Do Not Affect Its Binding Affinity for phyB
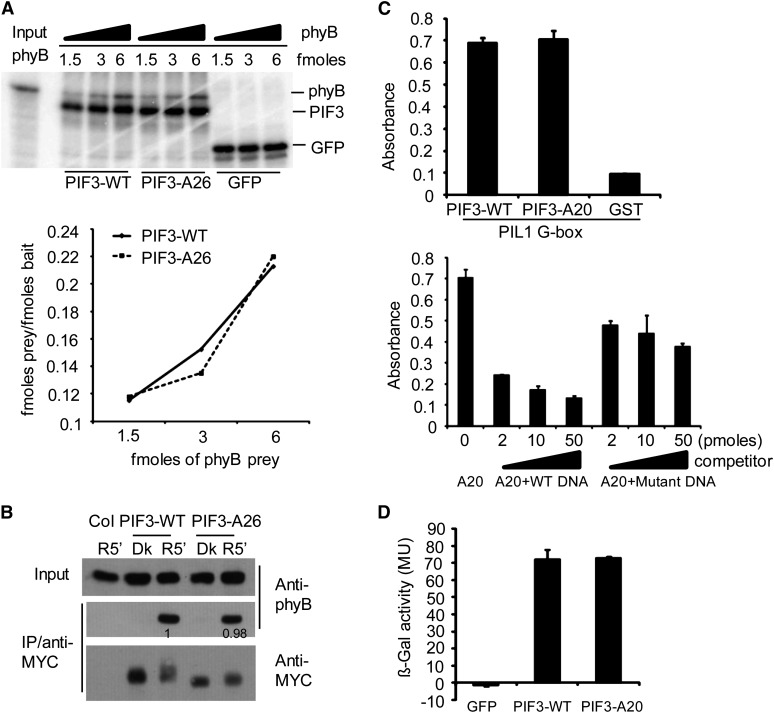
One possible reason for the observed reduction in light-induced phosphorylation of the mutagenized PIF3 might be a reduced capacity to bind to photoactivated phyB. We showed previously that such interaction with either phyA and/or phyB is necessary for light-induced PIF3 phosphorylation and degradation using PIF3 containing targeted mutations in the binding sites for these phys that abrogated binding to the photoreceptors (Al-Sady et al., 2006). We therefore tested here whether our multisite mutant PIF3 proteins can bind normally to phyB. In vitro binding analysis using TNT-produced, isotope-labeled protein indicates that the PIF3-WT and PIF3-A26 sequences have similar binding affinities for active phyB Pfr (Figure 3A). Similarly, normal in vitro binding affinity for phyB Pfr was also obtained for PIF3-A14 (see Supplemental Figure 3 online).
Figure 3.
Multisite Mutations at the Light-Induced PIF3 Phosphorylation Sites Do Not Affect Its Binding Affinity for the phyB Protein, Recognition of a Target Gene Promoter in Vitro, or Transcriptional Activation Activity in Yeast.
(A) PIF3-WT and PIF3-A26 have similar binding affinity for Pfr phyB in vitro. Top panel: Autoradiographs showing interactions of PIF3 with different fold dilutions of phyB prey. All proteins were produced in a TNT in vitro expression system labeled with [35S]Met. PIF3-WT, PIF3-A26, and GFP were first immunoprecipitated with an antibody against the MYC epitope present in each of these constructs and then used as baits to coimmunoprecipitate Pfr phyB. Bottom panel: Quantitative analysis of the data obtained in the top panel. The amount of each bait and prey used was calculated from a standard curve using a known amount of [35S]Met. The femtomoles of prey/femtomoles of bait are plotted against increasing amount of the phyB prey used.
(B) PIF3-WT and PIF3-A26 have similar conformer-specific, apparent binding affinity for Pfr phyB upon coextraction from R light–irradiated seedlings. Proteins were extracted from dark-grown transgenic seedlings that had been irradiated (R5′) or not (Dk) with R light for 5 min. Transgenically expressed MYC epitope–tagged PIF3 was immunoprecipitated (IP) with goat polyclonal MYC antibody and subjected to immunoblot analysis using monoclonal MYC (PIF3 bait) or phyB (prey) antibody. Proteins from nontransgenic Col seedlings were used as the negative control. Numbers directly on the blot image denote the relative phyB protein levels normalized to the PIF3:MYC bait.
(C) PIF3-WT and PIF3-A20 show similar apparent binding affinity for a G-box DNA sequence motif from the PIL1 gene promoter in vitro using a DPI-ELISA assay. Top panel: In vitro binding assay with a PIL1 G-box DNA probe using recombinant GST:PIF3:WT or GST:PIF3:A20 proteins, and GST protein as a negative control. Bottom panel: Competition binding assay of GST:PIF3-A20 with different concentrations of wild-type or mutant G-box probes. Data represent the means of independent duplicates ± se. WT DNA, wild-type competitor probes; mutant DNA, competitor probes mutated in the G-box motif at positions known to eliminate sequence-specific binding by the PIFs.
(D) PIF3-WT and PIF3-A20 have similar transcriptional activation activity in yeast. PIF3-WT and PIF3-A20 were fused to the LexA DNA binding domain and tested for autonomous transcriptional activation activity in yeast using a standard liquid ONPG assay. A GFP fusion protein was used as negative control. MU, miller units. Data represent the means of independent duplicates ± se.
In addition, both PIF3-WT and PIF3-A26 were found to bind conformer-specifically to the active Pfr form of phyB with similar apparent affinities in coimmunoprecipitation experiments, using extracts from the abovementioned lines transgenically expressing these proteins (Figure 3B). This result is consistent with normal phyB-PIF3 interactions having been retained in vivo. Therefore, the strongly reduced phosphorylation and degradation in our multisite mutant PIF3 proteins are likely due to the lack of phosphate accepting sites rather than an indirect effect of reduced binding affinity for the photoactivated phy molecules.
Mutations in the Light-Induced Phosphorylation Sites Affect neither the Sequence-Specific DNA Binding nor Transcriptional Activation Activities of PIF3
We also tested the binding of our multisite mutant PIF3 to a target gene promoter in vitro. Since PIL1 is a well-documented direct target gene of PIF3, we tested the in vitro binding of both wild-type and mutant PIF3 sequences to two of the G-box motifs in the PIL1 promoter that we recently identified by chromatin immunoprecipitation–sequencing analysis (Zhang et al., 2013). Data from a DNA protein interaction (DPI)–ELISA assay (Brand et al., 2010), using recombinant protein, suggest that both PIF3-WT and PIF3-A20 bind to these G-boxes with similarly strong affinity (Figure 3C).
We found previously that PIF3 has significant intrinsic transcriptional activation activity in yeast. We therefore compared the activities of PIF3-WT and PIF3-A20, fused to the LexA DNA binding domain in yeast using a standard liquid o-nitrophenyl β-d-galactopyranoside (ONPG) assay. The data show that the two PIF sequences exhibit similar transcriptional activity in yeast, indicating that the multisite point mutations we introduced into the PIF3 protein do not appear to affect its general structure (Figure 3D).
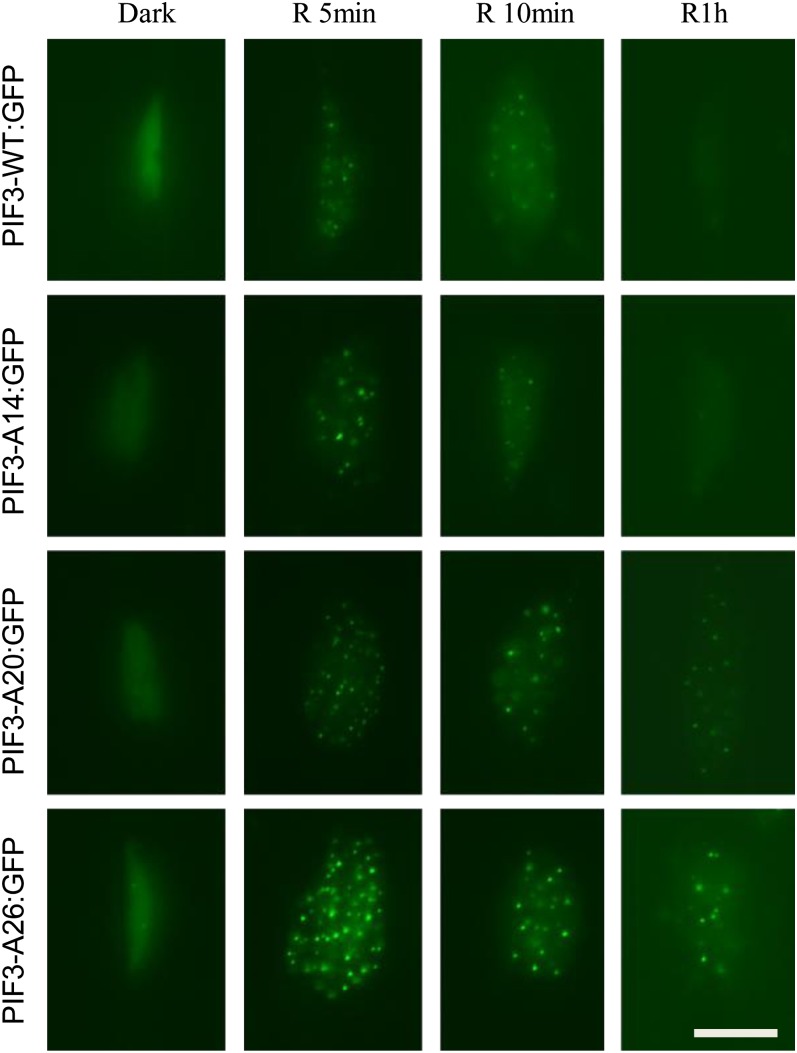
Phosphorylation Is Not Required for Light-Induced Localization of PIF3 in Subnuclear Speckles
As described above, following R light induction, PIF3 is rapidly colocalized with phyB in subnuclear structures called speckles or photobodies. It has been shown that mutant PIF3 that is unable to bind either phyA or phyB is defective in light-induced photobody localization, as well as phosphorylation and degradation (Al-Sady et al., 2006). One possible component of light-induced PIF3 degradation could involve phosphorylation-mediated subnuclear localization. Therefore, we asked if light-induced phophorylation of PIF3 is required for photobody localization, using green fluorescent protein (GFP)-tagged, PIF3-expressing transgenic lines. PIF3-A14, -A20, and -A26 were all able to translocate into photobodies rapidly in light in a manner similar to PIF3-WT. Fluorescent speckles were detectable within 1 min of light exposure for all four fusion proteins and remained visible for the next 5 to 10 min as shown in Figure 4. Visibly observable wild-type PIF3 declined rapidly over the next 1 h, as expected. However, whereas PIF3-A14 behaved similarly to PIF3-WT, visible levels and photobody localization were robustly retained in the PIF3-A26 and PIF3-A20 proteins over the entire 1-h period (Figure 4). Immunoblot analysis shows that the mutant PIF3:GFP fusion proteins, PIF3-A20 and PIF3-A26, also exhibit strongly reduced light-induced phosphorylation and degradation (see Supplemental Figures 4A and 4B online), similar to that of the comparable PIF3:MYC fusion proteins (Figure 2; see Supplemental Figure 2 online). These results suggest both that light-induced phosphorylation is not required for PIF3 photobody localization and that PIF3 photobody localization is not sufficient for degradation.
Figure 4.
Light-Induced PIF3 Phosphorylation Is Not Required for Its Subnuclear Localization in Photobodies (Speckles).
Epifluorescent imaging of GFP fluorescence in hypocotyl cell nuclei of transgenic seedlings expressing the various PIF3:GFP fusions of the PIF3 variants indicated. Seedlings were grown for 3 d in darkness and then either maintained in the dark or given a 1-min saturating R light pulse and returned to darkness for 4, 9, or 59 min before imaging. Bar = 10 µm.
Phosphomimic Mutations in the Light-Induced Phosphorylation Sites in PIF3 Promote Its Degradation in Vivo in the Absence of Light
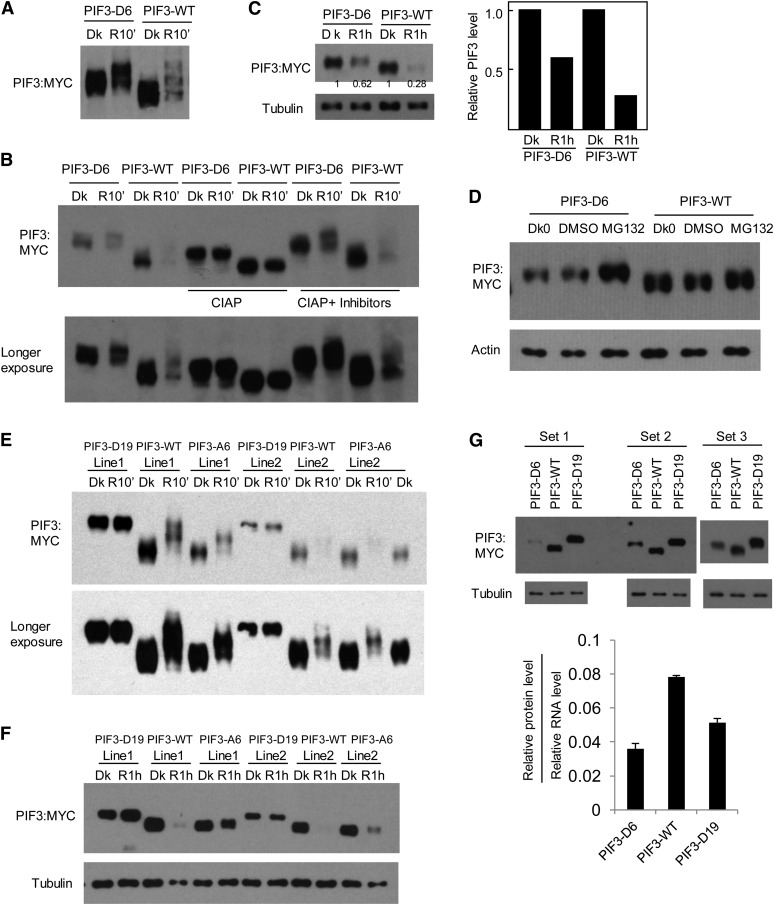
To investigate whether phosphorylation is sufficient to induce PIF3 degradation, we introduced phosphomimic mutations into PIF3. First we introduced Ser-to-Asp mutations for six strongly light-induced residues (five sites) in PIF3, Ser-58, Ser-88, Ser-102, Ser-151, and Ser-152/Ser-153 (PIF3-D6). Most of these phosphorylation sites show a phosphopeptide signal of 20% or more in the R light–treated samples and a large fold increase in the ratio of this signal in the R samples relative to dark samples (Figure 1D; see Supplemental Data Set 1 online). When expressed in plants, PIF3-D6 already displays a mobility shift in the dark compared with PIF3-WT, and light induces an additional mobility shift at R10 min, while retaining an apparently higher detectable protein level than that of PIF3-WT (Figure 5A; see Supplemental Figure 5A online). In vitro phosphatase treatment increases the mobility of both the PIF3-WT and PIF3-D6 proteins in the dark control samples, while retaining the mobility difference between the two (Figure 5B). This observation is consistent with the evidence that PIF3-WT is already phosphorylated in the dark (Figure 1) and indicates that the dephosphorylated PIF3-D6 phosphomimic protein has an intrinsically different mobility from that of the wild type, as a result of the amino acid substitutions. The additional mobility shift induced by light in both PIF3-D6 and PIF3-WT proteins is also reversed by in vitro phosphatase treatment (Figure 5B). The effects of the phosphatase treatment on both dark- and light-treated samples are eliminated by phosphatase inhibitors (Figure 5B), confirming that the observed effects on mobility are due to phosphorylation of the respective proteins. The PIF3-D6 protein level at R1h is reduced to ∼60% of the dark level, whereas PIF3-WT is reduced to ∼30% of the dark level (Figure 5C; see Supplemental Figure 5B online). These results show that light can still enhance the degradation rate of PIF3-D6 through phosphorylation, but to a lesser extent than for PIF3-WT, indicating that residues in addition to the six mutated in the PIF3-D6 phosphomimic remain subject to light-induced phosphorylation in this variant.
Figure 5.
Phosphomimic Mutations of a Subset of Strongly Light-Induced Phosphorylation Sites in PIF3 Promote Its Degradation in Vivo in the Absence of Light.
(A) PIF3-D6 exhibits a mobility shift in the dark (Dk) in transgenic Arabidopsis compared with the PIF3-WT control. Seedlings of transgenic lines expressing PIF3-D6:MYC or PIF3-WT:MYC fusion proteins were grown for 2.5 d in darkness, then either kept in the dark or given a saturating R light pulse and returned to darkness for 10 min before extraction into denaturing buffer and immunoblot analysis using MYC antibody.
(B) The same protein extracts as in (A) were treated with calf intestine alkaline phosphatase (CIAP) or phosphatase plus phosphatase inhibitors (CIAP + inhibitors) before immunoblot analysis.
(C) PIF3-D6 exhibits less light-induced degradation than the PIF3-WT control. Seedling growth, light treatment, and immunoblot analysis were as in (A), except that proteins were extracted at 1 h after light treatment and compared with those from dark samples. Tubulin was used as a control. Numbers directly on the blot image denote the relative PIF3 protein levels normalized to tubulin. Right panel shows quantification of relative PIF3 protein levels (normalized to the dark level set as 1).
(D) PIF3-D6 proteins are degraded through the 26S proteasome–mediated pathway in the absence of light. The 2.5-d-old dark-grown (Dk0) PIF3-D6:MYC– or PIF3-WT:MYC–expressing seedlings were treated with proteasome inhibitor MG132 or DMSO for a further 4 h in the dark before extraction and immunoblot analysis using antibodies against the MYC-epitope tag or against actin as a control.
(E) and (F) Light-induced mobility shift and degradation analysis of the PIF3-D19 and PIF3-A6 proteins from transgenic plants.
(E) Phosphomimic mutations of all light-induced sites identified in PIF3 (PIF3-D19) generate a mobility shift of the protein in the dark, similar to that induced by light at 10 min in the PIF3-WT protein, and no additional light-induced phosphorylation is observed. By contrast, the PIF3-A6 mutant protein exhibits reduced light-induced phosphorylation compared with PIF3-WT.
(F) The PIF3-D19 protein shows no apparent light-induced degradation, whereas the PIF3-A6 variant exhibits reduced light-induced degradation. Light treatment and immunoblot analysis were as in (A) and (C).
(G) PIF3-D6 and PIF3-D19 protein levels are lower than that of PIF3-WT after normalization to their corresponding RNA levels. Top panels: Triplicates of biological protein samples from 3-d-old dark-grown seedlings of the PIF3-D6, PIF3-WT, and PIF3-D19 were assayed by immunoblot using MYC antibody. Tubulin was used as a loading control. Bottom panel: Relative PIF3 protein levels after normalization to the corresponding RNA levels. Data represent the mean of biological triplicates ± se.
To examine whether the apparently reduced rate of light-induced degradation of PIF3-D6 relative to the initial dark control level is due to preexisting accelerated degradation of this variant in the dark through the 26S proteasome system, we treated dark-grown PIF3-D6 and PIF3-WT seedlings (Dk0) with proteasome inhibitor MG132 in the dark before protein extraction and immunoblot analysis. While we did not observe much change in protein abundance for PIF3-WT with or without MG132 treatment when compared with the actin control, significant protein accumulation was observed for PIF3-D6 when treated with MG132 (Figure 5D). Examination of the relative levels of the PIF3-D6 and PIF3-WT proteins (quantified from the immunoblots), normalized to the respective mRNA levels (quantified by RT-qPCR) in the dark, shows that the apparent lower levels of the PIF3-D6 than the PIF3-WT protein result from differences in posttranslational stability, rather than any differences in the corresponding transgenically expressed transcripts (Figure 5G). These results suggest that PIF3-D6 is indeed degraded constitutively through the ubiquitin/26S proteasome pathway in the dark and that light may act additively to increase the rate of this degradation through phosphorylation of additional residues. The reduced relative extent of light-induced degradation of PIF3-D6 in this line compared with PIF3-WT (Figure 5C) is thus likely due to the already lower initial PIF3-D6 than PIF3-WT protein level in the dark that results from partial constitutive degradation.
To explore this possibility more thoroughly, we introduced phosphomimic mutations at the majority of the light-induced phosphorylation sites identified in PIF3 (PIF3-D19; Figures 1D and 2A). When expressed in plants, PIF3-D19 is strongly shifted in the dark already, similar to the maximum shift for PIF3-WT in the light (Figure 5E). Light did not induce an additional mobility shift at R10 min or degradation at R1h for PIF3-D19 (Figures 5E and 5F; see Supplemental Figure 5C online). These results suggest either that PIF3-D19 is already degraded at the maximal light-induced rate in the dark through the ubiquitin/26S proteasome pathway or that this level of residue substitution interferes with either the additional ubiquitylation and/or degradation processes in the light. Comparison of the relative levels of the PIF3-D19, PIF3-D6, and PIF3-WT proteins, normalized to the respective mRNA levels in the dark, suggests that the latter conclusion may be correct, as the reduction in PIF3-D6 levels is not further enhanced in the PIF3-D19 variant in dark-grown seedlings (Figure 5G). Further consistent with this conclusion, we found that MG132 did not significantly increase PIF3-D19 protein levels in the dark (data not shown), indicating that the phosphomimic-induced constitutive degradation of PIF3-D6 is apparently negated by the additional residue substitutions in PIF3-D19.
Separately, we also made Ser-to-Ala mutations for the six residues displaying the most strongly light-induced phosphorylation in PIF3 (PIF3-A6; Figure 2A) and expressed the protein in plants. Immunoblot results show that PIF3-A6 is significantly reduced in its light-induced mobility shift and degradation (Figures 5E and 5F; see Supplemental Figure 5C online).
Light-Induced PIF3 Phosphorylation and Degradation Are Necessary for Feedback Regulation of phyB Levels
The pattern of seedling morphogenic phenotypes displayed by monogenic pif mutants and PIF protein overexpressors is complex (Leivar and Quail, 2011). Monogenic null mutants in each of the four quartet PIFs exhibit little or no visible deviation from normal wild-type skotomorphogenic growth and development when seedlings are grown in the dark (Huq and Quail, 2002; Kim et al., 2003; Fujimori et al., 2004; Huq et al., 2004; Monte et al., 2004; Oh et al., 2004; Leivar et al., 2008a; Lorrain et al., 2009; Shin et al., 2009; Stephenson et al., 2009). By contrast, such mutants display hypersensitivity to R light under prolonged irradiation conditions (Huq and Quail, 2002; Kim et al., 2003; Fujimori et al., 2004; Monte et al., 2004; Khanna et al., 2007; Nozue et al., 2007; de Lucas et al., 2008; Leivar et al., 2008b, 2012; Lorrain et al., 2008, 2009; Shin et al., 2009). This latter response is due, at least in part, to hyperaccumulation of phyB and the consequent enhanced global photosensitivity resulting from higher levels of this photoreceptor molecule, which is known to have the dominant role in the seedling deetiolation response to R light (Somers et al., 1991; Reed et al., 1993; Leivar et al., 2012). This phyB hyperaccumulation is due to a reduction, in these pif mutants, of the feedback degradation of the photoactivated phyB molecule that results normally from interaction with the PIF proteins (Khanna et al., 2007; Al-Sady et al., 2008; Leivar et al., 2008b, 2012). PIF overexpressor lines display converse phenotypic patterns. In dark-grown seedlings, increasingly higher levels of transgenically driven PIF5 overexpression were found to result in increasingly severe inhibition of hypocotyl elongation and promotion of apical hook curvature, compared with the wild type, as a result of induction of high levels of ethylene synthesis (Khanna et al., 2007). In light-grown seedlings, by contrast, PIF overexpressors display longer hypocotyls, reduced hook opening, and reduced cotyledon separation compared with the wild type. This phenomenon is due, at least in part, to decreased phyB abundance (and, thus, decreased global photosensitivity), as a result of enhanced PIF interaction–mediated degradation of the photoactivated phyB molecule (Khanna et al., 2007; Al-Sady et al., 2008; Leivar et al., 2008b, 2012).
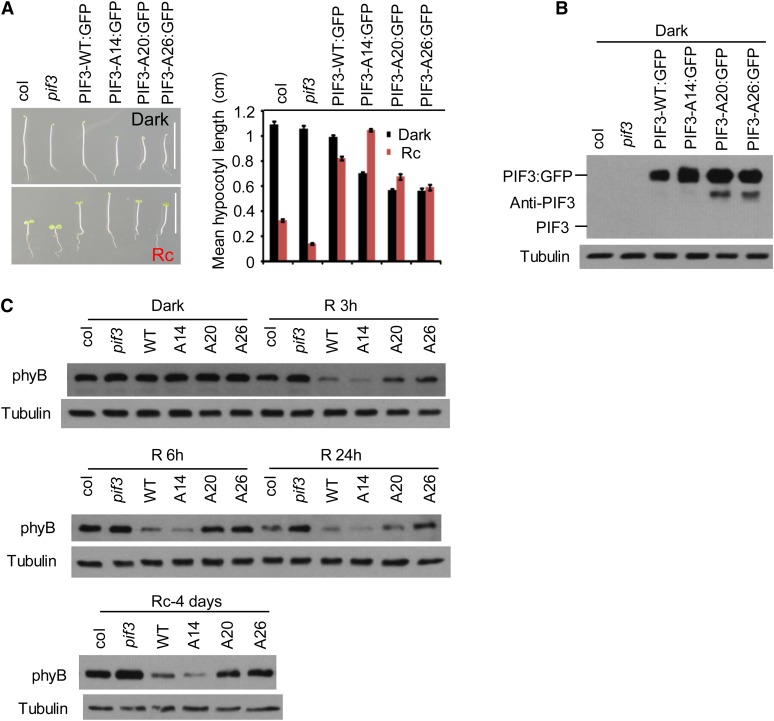
To determine whether the enhanced light stability of the PIF3-A14, PIF3-A20, and PIF3-A26 proteins compared with PIF3-WT (Figures 2 and 4; see Supplemental Figure 4 online) results in an overexpression phenotype, we first examined the hypocotyl lengths of the PIF3:GFP fusion transgenic seedlings (used for Figure 4) after growth in continuous R light (see below for discussion of the dark-grown seedling phenotypes; see also Supplemental Figure 6 online). The data show that all four transgenic lines display long hypocotyl phenotypes compared with the wild-type Arabidopsis (Columbia [Col]) seedlings (Figure 6A), presumably reflecting the relatively high levels of PIF protein expressed in each of these lines (Figure 6B). However, whereas the longer hypocotyls of the PIF3-A14–expressing line than the PIF3-WT–expressing line appears to correlate with the greater light stability of PIF3-A14, the shorter hypocotyls of the PIF3-A20– and PIF3-A26–expressing lines do not correlate with the equally greater light stability of PIF3-A20 and PIF3-A26. These hypocotyl differences are accompanied by a corresponding reciprocal visible difference in cotyledon expansion, diagnostic of a normal, overall coordinated photomorphogenic response (Quail, 2002; Khanna et al., 2006), and indicating that the effects are central to the signaling pathway. Because, as described above, these mutant proteins display normal site-specific DNA binding to target gene promoters and retain transcriptional activation activity, the possibility that this apparent discrepancy in the PIF3-A20– and PIF3-A26–expressing lines is due to a reduced intrinsic ability to promote the transcription of target genes involved in driving skotomorphogenic-like growth does not appear likely.
Figure 6.
Light-Induced PIF3 Phosphorylation and Degradation Are Necessary for Negative Feedback Modulation of phyB Levels.
(A) The PIF3-A20 and PIF3-A26 transgenic lines are not as tall as the PIF3-WT and PIF3-A14 lines in prolonged continuous R light. Left panel: Four-day-old seedlings transgenically expressing PIF3:GFP fusions of the various PIF3 variants indicated in the pif3 mutant are shown in comparison with the Col wild type and the pif3 mutant. Seedlings were grown either in the dark (top image) or continuous R light (Rc) (bottom image) for 4 d. Bars = 1 cm. Right panel: Mean hypocotyl lengths of the genotypes shown on the left. Approximately 30 seedlings of each genotype were used for each analysis. Error bars represent se.
(B) PIF3 protein levels in the various lines shown in (A). Proteins were extracted from dark-grown seedlings and subjected to immunoblot analysis using affinity-purified anti-PIF3 antibody. A longer exposure would be required to observe endogenous PIF3 (see Supplemental Figure 6C online). Tubulin was used as control.
(C) PIF3-A20 and PIF3-A26 transgenic lines retain more phyB protein in the R light than PIF3-WT and PIF3-A14 lines. phyB protein level in the same PIF3 transgenic lines as shown in (A) were analyzed by immunoblot using a monoclonal anti-phyB antibody or antitubulin antibody as a control. Top two panels: Three-day-old dark-grown seedlings were either kept in the dark or irradiated with continuous R light for 3, 6, or 24 h before protein extraction. Bottom panel: Seeds were grown in continuous R light for 4 d before protein extraction.
Instead, examination of the phyB levels in these transgenic lines suggests that differences in the degree of feedback modulation of phyB stability among these different PIF3 variants may explain this phenotypic pattern. The data show that the phyB levels in the PIF3-WT:GFP– and PIF3-A14:GFP–expressing seedlings in prolonged R light are lower than the Col control, consistent with the known PIF overexpression phenotype (Figure 6C). Unexpectedly, by contrast, the presumptively higher levels of the robustly light-stable PIF3-A20:GFP and PIF3-A26:GFP proteins, rather than leading to still lower levels of phyB, instead result in higher levels of the photoreceptor (Figure 6C). The resultant higher global photosensitivity of these lines then in turn correlates with their shorter hypocotyls and larger cotyledons under prolonged irradiation. Although PIF3-A14 appears to degrade intrinsically somewhat more slowly than PIF3-WT (see Supplemental Figure 4B online), it is more abundant than the PIF3-WT protein in the transgenic lines used for analyzing phyB degradation here (Figure 6B; see Supplemental Figure 4B online). We suggest that this lower intrinsic degradation rate of PIF3-A14 is offset by its greater abundance (Figure 6B), leading to more comparable absolute phyB degradation rates in the PIF3-A14 and PIF3-WT lines used (Figure 6C). Mechanistically, the above data collectively suggest that the negative feedback modulation of phyB levels by the PIFs is not simply linked to the absolute levels of the interacting PIFs but rather also to the intrinsic rate of light-induced degradation of these PIFs.
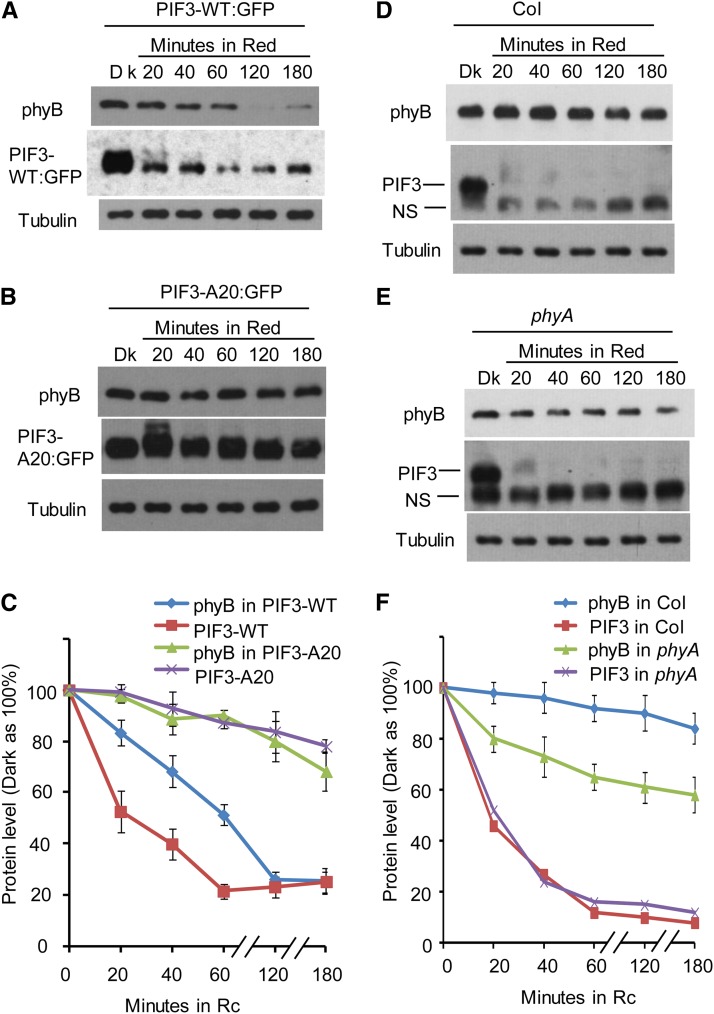
Comparative kinetic analysis of the early light-induced degradation rates of phyB, PIF3-WT, and PIF3-A20 in the respective transgenic lines supports this conclusion. We found that in the PIF3-WT overexpression line, phyB degradation is detectable within 20 min of light exposure, with a half-life of ∼1 h, correlating with rapid PIF3-WT protein degradation (Figures 7A and 7C). This rate of phyB degradation is strikingly more rapid than that of phyB in the nontransgenic Col wild type (Figures 7D and 7F), consistent with the above noted acceleration of phyB degradation induced by elevated PIF-WT protein levels (Khanna et al., 2007; Al-Sady et al., 2008; Leivar et al., 2008b, 2012). However, in the PIF3-A20 line, both light-induced phyB and PIF3 degradation are much slower than that in the PIF3-WT line (Figure 7B). Quantification of the protein gel blot data from three biological repeats suggests that in the PIF3-WT–expressing line, the degradation of PIF3-WT protein reaches a maximum within 1 h of light exposure, and the degradation of phyB is close to maximum within 2 h of light exposure (Figures 7C; see Supplemental Figures 7A and 7B online). On the other hand, in the PIF3-A20–expressing line, the degradation kinetics of both PIF3-A20 and phyB are much slower: after 3 h of R light, PIF3-A20 is reduced to ∼80% of the dark level, and phyB is reduced to ∼70% of the dark level (Figures 7C; see Supplemental Figures 7C and 7D online).
Figure 7.
Light-Induced phyB Degradation Kinetics Correlate Robustly with the Differential Rates of Degradation Displayed by the Wild-Type and Phosphorylation Refractory A20 PIF3 Mutant Proteins.
(A) The PIF3-WT overexpression transgenic line shows rapid light-induced degradation of both phyB and PIF3-WT. Three-day-old, dark-grown PIF3-WT:GFP–expressing seedlings were either kept in the dark (Dk) or exposed to continuous R light for the periods indicated. Extracted proteins were subjected to immunoblot analysis using either anti-phyB, anti-GFP, or antitubulin antibodies.
(B) The PIF3-A20 overexpression line shows comparatively slower light-induced degradation of both phyB and PIF3-A20. Seedlings were grown, exposed to light, and analyzed as in (A).
(C) Quantification of phyB and PIF3-GFP degradation kinetics in the PIF3-WT:GFP and PIF3-A20:GFP lines, from replicated immunoblot scans. Dark protein levels were set at 100% for each protein. Data are represented as the mean of biological triplicates ± se. Rc, continuous R light.
(D) Light-induced phyB degradation in wild-type Col is relatively slow. Col seedlings were grown and treated with R light as in (A) before protein extraction and immunoblot analysis using antibodies against phyB, PIF3, or tubulin.
(E) R light–induced phyB degradation in the phyA mutant is faster than that in Col. phyA seedling growth, R light treatment, and immunoblot analysis were the same as for the Col seedlings in (D).
(F) Quantification of phyB and PIF3 degradation kinetics in Col and the pif3 mutant. Dark protein levels were set at 100% for each protein. Data are represented as the mean of biological triplicates ± se.
[See online article for color version of this figure.]
In wild-type Col seedlings, the initial rate of light-induced phyB degradation is much slower than that of the PIF proteins (Figures 7D and 7F). One possible reason for this might be competitive binding of the PIFs by the phyA molecule. Although phyA has up to a 50-fold lower apparent affinity for PIF3 than does phyB (Huq et al. 2004), it is 10-fold more abundant than phyB in dark-grown seedlings (Somers et al., 1991; Sharrock and Clack, 2002), thus potentially limiting the level of PIFs available for phyB binding and degradation upon initial exposure of these seedlings to light. On the other hand, this competition would be expected to be transient, declining with increasing time in the light, because the phyA molecule is itself subject to relatively rapid degradation to a level ∼20-fold lower than that initially present in the dark (Somers et al., 1991; Sharrock and Clack, 2002). If this suggested mechanism is operative, we might expect to see a better correlation of light-induced phyB and PIF degradation after light-induced phyA degradation to a low level in the Col wild type (see Supplemental Figure 7I online), as well as more immediately upon light exposure of a phyA mutant. To test this idea, we performed a time-course analysis of R light–induced phyB degradation in a phyA mutant in comparison to wild-type Col. Quantification of the immunoblot data from three biological repeats shows that in the Col wild type, phyB degradation slowly increases over the first 3 h of light exposure, as expected, such that the phyB protein drops to ∼80% of the dark level by the end of this treatment (Figures 7D and 7F; see Supplemental Figures 7E and 7F online). Compellingly, on the other hand, the phyA mutant exhibits a significantly increased initial rate of light-induced phyB degradation, such that the phyB level in this mutant drops to ∼80% of the dark level within the first 20 min of light exposure and continues to drop to ∼60% of the dark level by the end of 3 h of R light exposure (Figures 7E and 7F; see Supplemental Figures 7G and 7H online). The degradation kinetics of native PIF3 in both wild-type Col and the phyA mutant are very similar (Figures 7D to 7F), indicating that phyB alone is sufficient to saturate the maximum rate of PIF3 degradation normally induced collectively by phyA and phyB combined (Al-Sady et al., 2006). Consistent with previous data, the phyA protein is degraded with a half-life of ∼1 h in the Col wild type under our conditions (see Supplemental Figure 7I online). Collectively, these results suggest that light-induced phyB degradation is intimately linked to endogenous native PIF degradation in the wild-type cell, in a manner similar to that observable in transgenic overexpressing lines.
The dark-grown phenotypes of the PIF3-WT, -A14, -A20, and -A26 GFP fusion overexpressing lines analyzed in Figures 6 and 7 are consistent with those described above for prolonged R light irradiations. These lines display marginal (PIF3-WT) to more severe (PIF3-A14, -A20, and -A26) short hypocotyl phenotypes, relative to the Col wild-type seedlings, more or less in parallel with the level of overexpression of the relevant PIF3 variant (Figures 6A and 6B; see Supplemental Figures 4B, 6A, and 6C online). This phenotype is correlated with a 20-fold elevation of ACS8 expression (the rate-limiting step in ethylene biosynthesis) in the PIF3-A20 line (see Supplemental Figure 6B online) and is rescued by exogenously provided silver (which blocks ethylene action) in all the transgenic lines (see Supplemental Figure 6A online). These results are consistent with the known effects of PIF5 overexpression in dark-grown seedlings referred to above (Khanna et al., 2007). In addition, seedlings of the PIF3-A26:MYC line are normal in the dark but shorter than those of the PIF3-A26:GFP line in R light (cf. Figure 6A with Supplemental Figure 6D online), correlating with the approximately twofold lower protein level in the PIF3-A26:MYC line than in the PIF3-A26:GFP line (see Supplemental Figure 6E online). These data further support the conclusion that the short hypocotyl phenotypes of the PIF3-A20 and PIF3-A26 lines in R light (Figure 6A) are likely due to a higher stability of phyB in these lines compared with the PIF3-WT line. However, it remains possible that other molecular functions of PIF3, such as the capacity to interact with DELLA proteins (de Lucas et al., 2008; Feng et al., 2008), are also affected in the PIF3-A20 and PIF3-A26 lines, thereby influencing hypocotyl elongation.
Comparison of the seedling phenotypes of PIF3-D6:MYC phosphomimic and PIF3-WT:MYC lines that express similar initial protein levels in the dark (Figure 5C) shows that the PIF3-D6:MYC seedlings are normal in the dark, and similar to, or slightly taller than, those of the PIF3-WT line in the light (see Supplemental Figure 6F online).
The slower light-induced degradation rate of PIF3-D6 than of PIF3-WT (Figure 5C) might have been predicted to lead to less PIF-induced phyB degradation, higher phyB levels, and, thus, hypersensitivity to light compared with the PIF3-WT line. One possible explanation for the absence of such hypersensitivity might be that the underlying higher, light-independent, degradation rate of PIF3-D6 might compensate for the lower light-dependent rate, reinstating an elevated net PIF3-D6 degradation rate that results in a correspondingly higher degradation rate, and consequently lower levels, of phyB than predicted because photoactivated-phyB can then bind to PIF3-D6.
DISCUSSION
In this study, we defined the molecular determinants in the PIF3 protein that participate in the biochemical mechanism of signal transfer from light-activated phyB molecules to this bHLH factor, with consequent regulation of target gene transcription. In parallel, we have shown that these signaling determinants are concomitantly and integrally necessary for the negative feedback regulation that is exerted by PIF3 on phyB abundance.
The identification of Ser and Thr residues that are the targets of light-induced phosphorylation of PIF3 and the demonstration by targeted mutagenesis that these residues are functionally necessary for the induced degradation of the protein robustly establishes that photoactivated phyB-induced transphosphorylation of this transcription factor is the direct biochemical mechanism of signal transfer. In addition, however, the multisite nature of the phosphorylation events provides insight into the mechanism by which this covalent modification is subsequently sensed by the ubiquitin-proteasome system in initiating degradation. The phyB-PIF3 signaling process fits an established paradigm, whereby phosphorylation of the target protein provides a recognition signal (termed a phosphodegron) for a cognate E3-ubiquitin ligase, which then ubiquitylates that protein, directing it to the 26S proteasome for proteolytic degradation (Hunter, 2007). The necessity for multisite phosphorylation of substrate proteins for E3-ligase recognition has been demonstrated for numerous signaling systems (Deshaies and Ferrell, 2001; Deshaies and Joazeiro, 2009; Bao et al., 2010; Varedi K et al., 2010). Most notable perhaps is Sic1, involved in cell cycle regulation in yeast, where phosphorylation of a minimum of six residues appears to set a threshold for SCF E3-ligase recognition, thereby providing a sensitive switch mechanism for the induction of the G1/S transition (Deshaies and Ferrell, 2001; Nash et al., 2001). The molecular basis for this multisite recognition in this and other systems has not been resolved, but possibilities include the cooperative action of multiple low-affinity sites that bind to multiple basic cavities in the E3-ligase substrate binding subunit (Nash et al., 2001; Orlicky et al., 2003; Hao et al., 2007; Deshaies and Joazeiro, 2009; Bao et al., 2010; Varedi K et al., 2010; Zhao et al., 2010; Kõivomägi et al., 2011; Tang et al., 2012). Although the number of phosphorylation sites apparently functionally involved in light-induced PIF3 degradation, identified here by targeted mutagenesis, appears large compared with these other reported systems (Varedi K et al., 2010), our data suggest that a smaller subset of these residues (e.g., those identified in PIF3-A6: Ser-58, Ser-88, Ser-102, Ser-151, Ser-152, and Ser-153) may contribute more strongly to the collective activity of the sites than other residues (Figures 5E and 5F). This observation might be consistent with the proposition that phosphor-dependent recognition by the cognate E3-ligase may involve a hierarchy of low-affinity binding sites that collaboratively constitute the phosphodegron directing PIF3 ubiquitylation and degradation (Deshaies and Joazeiro, 2009; Varedi K et al., 2010). This configuration might impart switch-like responsiveness of PIF3 activity to incoming light signals, as proposed by Varedi K et al. (2010) for such proteins subject to regulation by multisite phosphorylation or function more as a rheostat, incrementally tuning PIF3 interaction affinity for, and corresponding ubiquitylation by, its cognate E3 ligase (Rubin, 2013).
Neither the protein kinase(s) responsible for light-induced phosphorylation of PIF3 nor the E3 ubiquitin ligase(s) responsible for its ubiquitylation has been identified. The phosphorylation site motifs identified here do not appear to provide any obvious kinase candidates, consistent with the known difficulty of assigning newly documented phosphorylation sites to individual kinases (Ubersax and Ferrell, 2007; Santamaria et al., 2011; Sörensson et al., 2012; Rubin, 2013; Zulawski et al., 2013). Moreover, the absence of any apparent consensus phosphorylation site motif among the light-induced sites here (Figure 1E) leaves open the question of whether one broadly promiscuous kinase or multiple motif-specific kinases catalyze these events, an apparently common issue (Nash et al., 2001; Holt et al., 2009; Kõivomägi et al., 2011; Rubin, 2013). This question is relevant to the mechanism by which photoactivated phyB can stimulate the phosphorylation of multiple PIF family members because the phosphorylation site motifs identified in PIF3 here do not appear to be conserved in these other PIFs. On the other hand, the presence of a Pro on the C-terminal side of the phosphorylated residue in a significant proportion of the non-light-induced phosphosites (Figure 1F), coupled with the absence of this residue from the light-induced sites (Figure 1E), does suggest a potential dichotomy between the kinases that phosphorylate these two classes of sites. There are some classes of kinases, such as the CDKs and ERK2, that display selectivity toward phosphosite motifs containing a Pro in this position, whereas many others discriminate against this configuration (Ubersax and Ferrell, 2007).
A recent study has shown that casein kinase 2 (CK2) can phosphorylate seven specific Ser/Thr residues in PIF1 in vitro, without light stimulation or phy involvement, and that three of these sites are necessary for efficient degradation in vivo (Bu et al., 2011a, 2011b). However, site-directed PIF1 protein mutants lacking these in vitro CK2 phosphoacceptor sites still display robustly light-induced phosphorylation, albeit with less efficient degradation, in vivo. These data have been interpreted to indicate that multiple kinases may be involved in PIF1 degradation in vivo (Bu et al., 2011a, 2011b). In principle, this proposal is consistent with our identification here of both light-dependent and light-independent phophosites in PIF3 (Figure 1D). However, in contrast with the PIF1 data, the light-independent phosphosites we detected in PIF3 do not match the known CK2 consensus motif (Meggio and Pinna, 2003; de la Fuente van Bentem et al., 2008). One possible reason for this difference is that different kinases are responsible for the light-independent phosphorylation of PIF1 and PIF3. Alternatively, because there is no direct genetic evidence that CK2 phosphorylates PIF1 in vivo (Bu et al., 2011a, 2011b; Mulekar et al., 2012), it is possible that an as yet unidentified kinase is responsible for light-independent phosphorylation of both these PIFs.
For some well-characterized systems, phosphorylated phosphodegron motifs have been shown to be recognized by their cognate E3 ubiquitin ligase in a sequence-specific manner (Hunter, 2007). However, although the possibility of using synthetic versions of phosphodegrons, such as those identified here for PIF3, to identify E3 ligases for specific protein substrates, like PIF3, has been proposed (Jin et al., 2005), successful use of this approach does not appear to have been widely reported.
In an earlier study, we found that, in addition to the rapid, light-induced phosphorylation of PIF3, the rapid, concomitant translocation of PIF3 and photoactivated phyB into nuclear speckles (photobodies; Chen and Chory, 2011) is dependent on functional binding sites for the photoreceptor within the PIF3 protein (Al-Sady et al., 2006). These data raised the possibility that phy-induced phosphorylation is a prerequisite for this translocation. Our finding here that the nonphosphorylated higher-order PIF3 mutants display normal, rapid, light-induced migration into these speckles (Figure 4), but greatly reduced degradation (Figure 2; see Supplemental Figures 2 and 4 online), indicates both that this migration does not require phosphorylation of PIF3 and that speckle localization per se is not sufficient for degradation. This conclusion raises the possibility that phyB physically transports bound PIF3 to the photobodies where the bHLH factor becomes phosphorylated, ubiquitylated, and degraded. However, despite some recent advances (Chen and Chory, 2011; Galvão et al., 2012), the functional role of the photobodies in these coupled processes remains to be defined.
Previously, we showed that the interaction of photoactivated phyA or phyB with PIF3 is necessary, not only for the light-induced phosphorylation and degradation of PIF3 in vivo, but also for the negative feedback degradation of phyB that is observed under prolonged irradiation conditions (Al-Sady et al., 2008). Our present data show that interaction per se (without concomitant phosphorylation) is not sufficient for this negative feedback regulation. This conclusion is based on the observation that the higher-order, nonphosphomodified, multisite PIF3 mutants (PIF3-A20 and PIF3-A26) have a strongly attenuated capacity to induce reductions in phyB levels compared with the PIF3-WT sequence (Figures 6 and 7), despite retaining apparently normal binding affinity for photoactivated phyB (Figure 3). These data indicate that the light-induced degradation of phyB is dependent on the concomitant phosphorylation and degradation of PIF3. Together with the strong temporal relationships between the light-induced phyB and PIF3 degradation curves in Figures 7A to 7C, these data indicate that these processes are concurrent and suggest the possibility of codegradation of the two molecules. This appears to represent a novel signaling configuration, whereby the act of signaling directly results in feedback attenuation of receptor activity.
At first glance, one caveat with this suggestion is the markedly different degradation kinetics of PIF3 and phyB upon initial exposure of dark-grown wild-type seedlings to light. Whereas PIF3 is rapidly degraded (half-time of 10 to 15 min) in response to the light signal, phyB remains relatively unchanged in level for up to several hours (Khanna et al., 2007; Monte et al., 2007; Al-Sady et al., 2008; Leivar et al., 2008b) (Figures 7D and 7F). However, a possible explanation for this much slower rate of phyB degradation might be that phyB is considerably more abundant than PIF3 in the dark-grown wild-type seedling, such that the absolute rate of phyB depletion would appear to be minimal compared with that of the much less abundant PIF3. Consistent with this possibility, our data indicate that the rate of phyB degradation depends on the level of degradable PIF3, suggesting that PIF3 is the rate-limiting factor in this process. Similarly consistent with this possibility is the demonstration that the rate of light-induced phyB degradation is increased in the phyA null mutant (Figures 7D to 7F). This finding suggests that the initially even more abundant phyA competes for binding to the limited levels of PIF3 available, thereby reducing concomitant phyB-PIF3 degradation. A second complication with the potential codegradation mechanism is that whereas there is evidence that COP1 functions as an E3 ligase that ubiquitylates phyB in a manner promoted by the PIF proteins (Jang et al., 2010), cop1 mutants display reduced levels of PIF3, rather than the higher levels that might be expected if COP1 is the E3 ligase responsible for PIF3 degradation (Bauer et al., 2004). It is also notable that our data indicate that simple physical binding of PIF3 to phyB, without concomitant phosphorylation, is insufficient to induce presumptive COP1 E3-ligase-mediated phyB degradation in vivo (Figures 3, 6, and 7), as would be suggested by the data of Jang et al. (2010), showing that recombinant PIF3 promotes COP1-mediated ubiquitylation of recombinant phyB in in vitro assays. The molecular basis for these apparent discrepancies remains to be determined, but one possibility is that different ligases ubiquitylate phyB and PIF3 in parallel.
Taken together, our data establish that multisite phosphorylation of PIF3, induced by the physical, intranuclear binding of photoactivated phyB to the transcription factor, is not only necessary for optimal light-induced ubiquitylation and degradation of PIF3, but is also intimately involved in the concomitant, converse, negative regulation of phyB abundance exerted by PIF3 on the photoreceptor. Because the simple light-induced interaction of phyB with PIF3 does not alone induce degradation of either molecule in the absence of phosphorylation, some form of concurrent codegradation of the interacting signaling partners appears to be a strong possibility. Overall, the apparent mechanistic coupling of these two phenomena suggests complexity in the short signaling pathway, from the activated photoreceptor to the primary transcriptional network, involving a rapid, graded, or switch-like transduction process immediately upon initial light exposure, followed later, under prolonged irradiation, by a slower, negative feedback attenuation process that provides modulated desensitization of the system to the incoming light signal.
METHODS
Plant Materials and Growth Conditions
The Columbia-0 wild type and pif3-3 and phyA211 mutants in Columbia-0 were previously described (Reed et al., 1994; Monte et al., 2004). All transgenic plants were in the pif3-3 background. The 35S:YFP:PIF3-507 transgenic line for mass spectrometry analysis was generated as follows: PIF3 cDNA was amplified with primers NLS-F and PIF3-507AA-R (see Supplemental Table 3 online) using NLS:PIF3 as template. The product was cloned into a pENTR/D-TOPO vector (Invitrogen) and transferred into a Gateway binary vector (pGWB42) containing a 35S promoter and YFP fusion at the N terminus. For phosphorylation site mutations, point mutations were introduced into full-length PIF3 cDNA entry vector (Al-Sady et al., 2006) by site-directed mutagenesis (Stratagene) using primers listed in Supplemental Table 3 online. Wild-type and mutant PIF3 in the entry vector were then transferred into the Gateway binary vector B17 for MYC fusion, or B7FWG for GFP fusion, at the C terminus. Both B17 and B7FWG contain a 35S promoter.
Seeds were stratified for 5 d at 4°C in darkness and induced to germinate with 3 h white light before exposure to a terminal 5-min FR pulse (60 µmol/m2/s) on growth medium as reported (Leivar et al., 2008a). For immunoprecipitation, immunoblot, and quantitative PCR analysis, seeds were then grown in darkness for 2.5 to 3 d at 21°C before R light treatment as indicated. For MG132 treatment, 3-d-old dark-grown seedlings were treated with 50 µM MG132 or 1% DMSO in liquid growth medium for 2 to 4 h. For seedling phenotypes, seeds were grown either in darkness or continuous R light (8 µmol/m2/s) for 4 d at 21°C. Some of the plates contained 20 µM Silver Thiosulfate added to the medium (as indicated). Measurements were typically done with at least 30 seedlings with biological repeats.
Transgenic Line Selection and Analysis
At least 15 independent transgenic lines were examined for seedling phenotypes and protein expression levels in the dark, and at least two independent lines per construct were analyzed for light-induced protein mobility shift and degradation. For the light-induced mutant PIF3:MYC protein mobility shift and degradation analysis, we chose lines that express a similar protein level to that of PIF3-WT:MYC in the dark for comparison. For subcellular localization of the mutant PIF3:GFP and phyB degradation analysis, we chose GFP fusion lines with a high expression level.
Identification of Phosphorylation Sites and Quantification
Proteins were extracted into MOPS buffer (100 mM MOPS, pH 7.6, 150 mM NaCl, 0.1% Nonidet P-40, 1% Triton X-100, 20 mM iodoacetamide, 1 mM phenylmethylsulfonyl fluoride (PMSF), 2 µg/L aprotinin, 5 µg/L leupeptin, 1 µg/L pepstatin, 2× Complete protease inhibitor cocktail, and PhosStop cocktail from Roche), centrifuged, and filtered through two layers of Miracloth, then incubated with rabbit anti-GFP antibody for 1 h at 4°C. YFP:PIF3-507 fusion proteins were then captured with protein A agarose beads, washed five times with immunoprecipitation buffer, and eluted with 2% SDS. Eluted proteins were then concentrated using a spin column (Millipore) and separated by SDS-PAGE. After Coomassie Brilliant Blue staining, YFP:PIF3 protein bands were excised and subjected to in-gel digestion with either trypsin, AspN, or chymotrypsin.
Peptide mixtures were desalted using C18 ZipTips (Millipore) and then analyzed by liquid chromatography–tandem mass spectrometry on a Nanoacquity ultraperformance liquid chromatography system connected to an LTQ-Orbitrap XL. Peptides were loaded onto a trapping column (NanoAcquity UPLC 180 μm × 20 mm; Waters) and then washed with 0.1% formic acid. The analytical column was a BEH130 C18 100 μm × 100 mm (Waters). The flow rate was 400 nL/min, and a 90-min gradient was used; peptides were eluted by a gradient from 2 to 30% solvent B (acetonitrile/0.1%formic acid) over 65 min, followed by a short wash at 50% solvent B. After a precursor scan was measured in the Orbitrap by scanning from mass-to-charge ratio 350 to 1400, the six most intense multiply charged precursors were selected for collision-induced dissociation in the linear ion trap.
Tandem mass spectrometry peak lists were extracted using an in-house script PAVA, and data were searched using Protein Prospector against the UniProtKB database Arabidopsis thaliana entries (downloaded March 21, 2012 with a total of 54,896 entries) to which randomized sequence versions were concatenated to allow estimation of false discovery rate (Elias and Gygi, 2007). A precursor mass tolerance of 20 ppm and a fragment mass error tolerance of 0.6 D were allowed. Carbamidomethylcysteine was searched as a constant modification. Variable modifications include protein N-terminal acetylation, peptide N-terminal Gln conversion to pyroglutamate, and Met oxidation. Subsequent searches were performed to find those peptides modified by phosphorylation. The search parameters were as above, but this time allowing for phosphorylation on Ser, Thr, and Tyr. Assignments of all modified peptides were checked manually and are illustrated in Supplemental Figures 1D to 1R online.
Relative quantification of the phosphorylation at a given site was calculated from the maximum peak heights of the extracted ion chromatograms, corresponding to the phosphorylated and unmodified versions of the peptides, using two different procedures (see Supplemental Figure 1C online). In the first method, a value termed the phosphopeptide signal (%) was defined as the relative level of phosphorylated peptide detected at a given site, expressed as a percentage of the combined total signal of phosphorylated and nonphosphorylated peptide containing that site (see Supplemental Figure 1C online, top panel). This value is compared directly for dark control and R light–treated seedlings at each site in Figure 1D. In the second method, a value termed the relative phospho ratio was defined as the ratio of two values, each of which are themselves ratios (Steen et al., 2008). In this procedure, the ratio of the abundance of phosphorylated to nonphosphorylated residues, detected at a given site, is calculated separately for dark control and R-treated seedlings, and then the ratio of these two derived values is expressed as the ratio of R to dark values (see Supplemental Figure 1C, bottom panel, Supplemental Data Set 1, and Supplemental Table 2 online). However, it should be noted that neither procedure provides absolute stoichiometry of the phosphorylation because the addition of a phosphate group changes the ionization efficiency of the phosphopeptide compared with the corresponding unmodified peptide and therefore affects the intensity of the signal (Steen et al., 2008).
Immunoblots and Quantification
Proteins were extracted directly from pulverized tissue into boiling denaturing buffer (100 mM MOPS, pH 7.6, 100 mM NaCl, 10% glycerol, 40 mM 2-mercaptoethanol, 5% SDS, 1× protease inhibitor cocktail from Roche, and 2 mM PMSF), cleared by centrifugation, separated by SDS-PAGE, and transferred to polyvinylidene difluoride membrane. Mouse monoclonal antibodies against MYC (Convance 9E10 and Cell Signaling 9B11), GFP (Clontech, JL-8), α-tubulin (Sigma-Aldrich), actin (Sigma-Aldrich A0480), phyB (B1) (Somers et al., 1991), phyA (073D), and affinity-purified rabbit polyclonal antibody against PIF3 (Al-Sady et al., 2006) were used for immunodetection. Anti-rabbit-horseradish peroxidase (HRP) and anti-mouse-HRP were used as secondary antibodies (Promega), and an ECL plus chemiluminescence kit (Amersham) was used for detection. Immunoblot bands were quantified using Image J software (NIH) and normalized to tubulin. The linearity of the signal was assessed by running a parallel protein extract dilution curve. The relative protein level from each genotype in R light is compared with the corresponding protein level in the dark.
In Vitro Pull-Down Assays
GFP:MYC, PIF3-WT:MYC, PIF3-A14:MYC, or PIF3-A26:MYC in the pENTR/D-TOPO vector were transferred into a Gateway vector pDEST24 (Invitrogen) containing a T7 promoter and glutathione S-transferase (GST) fusion at the C terminus, and the proteins were synthesized using a TNT kit (Promega). In vitro pull-down assays were performed as described (Khanna et al., 2004) with modification: Goat polyclonal anti-MYC antibody (Abcam 9132) and protein G beads (Millipore) were used to pull down the PIF3:MYC bait and the GFP:MYC control protein. The phyB prey preparation was precleared with protein G beads before light treatment.
Coimmunoprecipitation from Seedling Extracts
PIF3:MYC fusion proteins were extracted the same way as for mass spectrometry analysis and then incubated with goat polyclonal anti-MYC antibody for 1 h at 4°C. Protein G beads (Millipore) were used to capture the complex. Immunoprecipitation products were separated by SDS-PAGE, blotted, and probed with mouse monoclonal anti-MYC, anti-phyB (B1), or anti-ubiquitin antibody (Cell Signaling P4D1). Anti-mouse-HRP antibodies were used as secondary antibodies (Promega), and an ECL plus chemiluminescence kit (Amersham) was used for detection.
In Vitro Protein-DNA Binding Assays
Recombinant GST-PIF3 and GST proteins were purified according to the manufacturer’s instructions (GE). DNA probes were generated by annealing a 5′-biotinylated oligonucleotide to a complementary unmodified oligonucleotide (IDT; see Supplemental Table 3 online). The DPI-ELISA assays were performed as described (Brand et al., 2010) using HRP-labeled anti-GST antibody (GenScript A00866). Detailed methods are described in Supplemental Methods 1 online.
Yeast Transcription Activity Assays
GFP, PIF3-WT, or PIF3-A20 in a pENTR D-TOPO vector (Invitrogen) was transferred into a Gateway vector pEG202 containing the lexA DNA binding domain at the N terminus. All constructs were transformed into yeast strain EGY48, and transcription activity was measured using a standard liquid ONPG assay as described (Clontech Yeast Protocols Handbook).
Epifluorescence Microscopy
Epifluorescence microscopy analysis was performed as described (Al-Sady et al., 2006). Images were processed for optimal presentation using Photoshop software (Adobe).
Alkaline Phosphatase Treatment
Proteins were extracted into hot extraction buffer (100 mM MOPS, pH 7.6, 100 mM NaCl, 10% glycerol, 40 mM 2-mercaptoethanol, 2% SDS, 1× protease inhibitor cocktail, and 2 mM PMSF), boiled for 3 min, and then cleared by centrifugation. Extracted proteins in the supernatant were then diluted 20-fold into calf intestinal alkaline phosphatase buffer (50 mM Tris, pH 8.8, 1% Triton X-100, 50 mM MgCl2, 100 mM NaCl, and 1× cocktail) with or without phosphatase inhibitors (Phosstop from Roche). Calf-intestinal alkaline phosphatase (Roche) was added at a concentration of 400 units/mL, and the samples were incubated at 37°C for 2 h.
Quantitative RT-PCR
Quantitative RT-PCR was performed as described (Leivar et al., 2009). Each PCR reaction was repeated at least twice, and the mean value of technical replicates was recorded for each biological replicate. Data from biological triplicates were collected, and the mean value with standard error is represented in the bar graph. PP2A (AT1G13320) was used as a normalization control. Primers are listed in Supplemental Table 3 online.
Quantification of Relative Protein to RNA Level
The relative protein to RNA levels for PIF3-D6 and PIF3-D19 in dark-grown samples were calculated as follows: For each biological repeat, the relative PIF3/tubulin protein level was quantified from immunoblot analysis by setting the PIF3-WT/Tubulin level as 1. The corresponding relative PIF3 RNA level was quantified by quantitative RT-PCR using PP2A as a normalization control. Primer efficiency was tested by a dilution curve. The ratio of the relative PIF3 protein to RNA level was then calculated for each biological repeat.
Accession Numbers
The Arabidopsis Genome Initiative locus identifier for PIF3 is AT1G09530, for PHYB is AT2G18790, for PHYA is AT1G09570, and for ACS8 is AT4G37770.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Identification and Quantification of in Vivo Phosphorylation Sites in PIF3.
Supplemental Figure 2. Light-Induced PIF3 Mobility Shift and Degradation Analysis of Additional PIF3 Mutant Variants as Defined in Figure 2A.
Supplemental Figure 3. PIF3-WT and PIF3-A14 Have Similar Apparent Binding Affinity for Pfr phyB in Vitro.
Supplemental Figure 4. GFP Fusions of the Various PIF3 Mutant Proteins Behave Similarly to Their Respective PIF3:MYC Fusion-Protein Counterparts as Regards Light-Induced Mobility Shift (Phosphorylation) and Degradation Responses.
Supplemental Figure 5. Light-Induced PIF3 Mobility Shift and Degradation Analysis of Mutant PIF3 Protein Variants from Additional Transgenic Lines.
Supplemental Figure 6. Seedling Phenotypes of Transgenic Plants Expressing PIF3 Constructs with Mutated Phosphorylation Sites.
Supplemental Figure 7. Additional Biological Repeats of the Analysis in Figure 7 Comparing the Light-Induced Degradation Kinetics of the phyB and PIF3-A20 Mutant Proteins.
Supplemental Table 1. Dilution Experiments Showed a Linear Change in Ion Intensity as Quantified by Using Extracted Ion Chromatograms.
Supplemental Table 2. List of Phosphopeptides Identified from PIF3 in Dark- or Light-Treated Samples.
Supplemental Table 3. Primer Sequences.
Supplemental Methods 1. DPI-ELISA assay.
Supplemental Data Set 1. Quantitation of Phosphorylation Detected by Mass Spectrometry of PIF3 from Dark- and Light-Treated Seedlings.
Acknowledgments
We thank Bassem Al-Sady for critical reading of the article; Scott C. Peck, Nevan Krogan, Gerard Cagney, Bassem Al-Sady, and Kathy Li for initial help with mass spectrometry analysis; Anne Pfeiffer for the protocol and primers used for the DPI-ELISA assay; and all lab members for technical support and critical discussion. This work was supported by National Institutes of Health Grant 2R01 GM-047475, Department of Energy Grant DEFG03-87ER13742, and USDA/Agriculture Research Service Current Research Information System Grant 5335-21000-027-00D to P.H.Q.; by Department of Energy Grant DEFG02-08ER15973 to Z.-Y.W.; and by National Institutes of Health Grant 8P41GM103481 to A.L.B.
AUTHOR CONTRIBUTIONS
W.N. and P.H.Q. designed research. W.N., S.-L.X., R.J.C., T.N.D.P., S.G., D.A.M., A.L.B., and Z.-Y.W. performed research and contributed new analytical tools. W.N. and S.-L.X. analyzed data. W.N. and P.H.Q. wrote the article.
Glossary
- bHLH
basic Helix Loop Helix
- R
red
- FR
far-red
- DPI
DNA protein interaction
- ONPG
o-nitrophenyl β-d-galactopyranoside
- Col
Columbia
- PMSF
phenylmethylsulfonyl fluoride
- HRP
horseradish peroxidase
- GST
glutathione S-transferase
References
- Al-Sady B., Kikis E.A., Monte E., Quail P.H. (2008). Mechanistic duality of transcription factor function in phytochrome signaling. Proc. Natl. Acad. Sci. USA 105: 2232–2237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Sady B., Ni W., Kircher S., Schäfer E., Quail P.H. (2006). Photoactivated phytochrome induces rapid PIF3 phosphorylation prior to proteasome-mediated degradation. Mol. Cell 23: 439–446 [DOI] [PubMed] [Google Scholar]
- Bao M.Z., Shock T.R., Madhani H.D. (2010). Multisite phosphorylation of the Saccharomyces cerevisiae filamentous growth regulator Tec1 is required for its recognition by the E3 ubiquitin ligase adaptor Cdc4 and its subsequent destruction in vivo. Eukaryot. Cell 9: 31–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer D., Viczián A., Kircher S., Nobis T., Nitschke R., Kunkel T., Panigrahi K.C., Adám E., Fejes E., Schäfer E., Nagy F. (2004). Constitutive photomorphogenesis 1 and multiple photoreceptors control degradation of phytochrome interacting factor 3, a transcription factor required for light signaling in Arabidopsis. Plant Cell 16: 1433–1445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand L.H., Kirchler T., Hummel S., Chaban C., Wanke D. (2010). DPI-ELISA: A fast and versatile method to specify the binding of plant transcription factors to DNA in vitro. Plant Methods 6: 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bu Q., Zhu L., Dennis M.D., Yu L., Lu S.X., Person M.D., Tobin E.M., Browning K.S., Huq E. (2011b). Phosphorylation by CK2 enhances the rapid light-induced degradation of phytochrome interacting factor 1 in Arabidopsis. J. Biol. Chem. 286: 12066–12074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bu Q., Zhu L., Huq E. (2011a). Multiple kinases promote light-induced degradation of PIF1. Plant Signal. Behav. 6: 1119–1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M., Chory J. (2011). Phytochrome signaling mechanisms and the control of plant development. Trends Cell Biol. 21: 664–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Fuente van Bentem S., et al. (2008). Site-specific phosphorylation profiling of Arabidopsis proteins by mass spectrometry and peptide chip analysis. J. Proteome Res. 7: 2458–2470 [DOI] [PubMed] [Google Scholar]
- de Lucas M., Davière J.M., Rodríguez-Falcón M., Pontin M., Iglesias-Pedraz J.M., Lorrain S., Fankhauser C., Blázquez M.A., Titarenko E., Prat S. (2008). A molecular framework for light and gibberellin control of cell elongation. Nature 451: 480–484 [DOI] [PubMed] [Google Scholar]
- Deshaies R.J., Ferrell J.E., Jr (2001). Multisite phosphorylation and the countdown to S phase. Cell 107: 819–822 [DOI] [PubMed] [Google Scholar]
- Deshaies R.J., Joazeiro C.A. (2009). RING domain E3 ubiquitin ligases. Annu. Rev. Biochem. 78: 399–434 [DOI] [PubMed] [Google Scholar]
- Elias J.E., Gygi S.P. (2007). Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods 4: 207–214 [DOI] [PubMed] [Google Scholar]
- Feng S., et al. (2008). Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature 451: 475–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimori T., Yamashino T., Kato T., Mizuno T. (2004). Circadian-controlled basic/helix-loop-helix factor, PIL6, implicated in light-signal transduction in Arabidopsis thaliana. Plant Cell Physiol. 45: 1078–1086 [DOI] [PubMed] [Google Scholar]
- Galvão R.M., Li M., Kothadia S.M., Haskel J.D., Decker P.V., Van Buskirk E.K., Chen M. (2012). Photoactivated phytochromes interact with HEMERA and promote its accumulation to establish photomorphogenesis in Arabidopsis. Genes Dev. 26: 1851–1863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao B., Oehlmann S., Sowa M.E., Harper J.W., Pavletich N.P. (2007). Structure of a Fbw7-Skp1-cyclin E complex: multisite-phosphorylated substrate recognition by SCF ubiquitin ligases. Mol. Cell 26: 131–143 [DOI] [PubMed] [Google Scholar]
- Holt L.J., Tuch B.B., Villén J., Johnson A.D., Gygi S.P., Morgan D.O. (2009). Global analysis of Cdk1 substrate phosphorylation sites provides insights into evolution. Science 325: 1682–1686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter T. (2007). The age of crosstalk: Phosphorylation, ubiquitination, and beyond. Mol. Cell 28: 730–738 [DOI] [PubMed] [Google Scholar]
- Huq E., Al-Sady B., Hudson M., Kim C., Apel K., Quail P.H. (2004). Phytochrome-interacting factor 1 is a critical bHLH regulator of chlorophyll biosynthesis. Science 305: 1937–1941 [DOI] [PubMed] [Google Scholar]
- Huq E., Al-Sady B., Quail P.H. (2003). Nuclear translocation of the photoreceptor phytochrome B is necessary for its biological function in seedling photomorphogenesis. Plant J. 35: 660–664 [DOI] [PubMed] [Google Scholar]
- Huq E., Quail P.H. (2002). PIF4, a phytochrome-interacting bHLH factor, functions as a negative regulator of phytochrome B signaling in Arabidopsis. EMBO J. 21: 2441–2450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang I.C., Henriques R., Seo H.S., Nagatani A., Chua N.H. (2010). Arabidopsis PHYTOCHROME INTERACTING FACTOR proteins promote phytochrome B polyubiquitination by COP1 E3 ligase in the nucleus. Plant Cell 22: 2370–2383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y., Lau O.S., Deng X.W. (2007). Light-regulated transcriptional networks in higher plants. Nat. Rev. Genet. 8: 217–230 [DOI] [PubMed] [Google Scholar]
- Jin J., Ang X.L., Shirogane T., Wade Harper J. (2005). Identification of substrates for F-box proteins. Methods Enzymol. 399: 287–309 [DOI] [PubMed] [Google Scholar]
- Khanna R., Huq E., Kikis E.A., Al-Sady B., Lanzatella C., Quail P.H. (2004). A novel molecular recognition motif necessary for targeting photoactivated phytochrome signaling to specific basic helix-loop-helix transcription factors. Plant Cell 16: 3033–3044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna R., Shen Y., Marion C.M., Tsuchisaka A., Theologis A., Schäfer E., Quail P.H. (2007). The basic helix-loop-helix transcription factor PIF5 acts on ethylene biosynthesis and phytochrome signaling by distinct mechanisms. Plant Cell 19: 3915–3929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna R., Shen Y., Toledo-Ortiz G., Kikis E.A., Johannesson H., Hwang Y.S., Quail P.H. (2006). Functional profiling reveals that only a small number of phytochrome-regulated early-response genes in Arabidopsis are necessary for optimal deetiolation. Plant Cell 18: 2157–2171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Yi H., Choi G., Shin B., Song P.S., Choi G. (2003). Functional characterization of phytochrome interacting factor 3 in phytochrome-mediated light signal transduction. Plant Cell 15: 2399–2407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kircher S., Gil P., Kozma-Bognár L., Fejes E., Speth V., Husselstein-Muller T., Bauer D., Adám E., Schäfer E., Nagy F. (2002). Nucleocytoplasmic partitioning of the plant photoreceptors phytochrome A, B, C, D, and E is regulated differentially by light and exhibits a diurnal rhythm. Plant Cell 14: 1541–1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kircher S., Kozma-Bognar L., Kim L., Adam E., Harter K., Schafer E., Nagy F. (1999). Light quality-dependent nuclear import of the plant photoreceptors phytochrome A and B. Plant Cell 11: 1445–1456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kõivomägi M., Valk E., Venta R., Iofik A., Lepiku M., Balog E.R., Rubin S.M., Morgan D.O., Loog M. (2011). Cascades of multisite phosphorylation control Sic1 destruction at the onset of S phase. Nature 480: 128–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P., Monte E., Al-Sady B., Carle C., Storer A., Alonso J.M., Ecker J.R., Quail P.H. (2008b). The Arabidopsis phytochrome-interacting factor PIF7, together with PIF3 and PIF4, regulates responses to prolonged red light by modulating phyB levels. Plant Cell 20: 337–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P., Monte E., Cohn M.M., Quail P.H. (2012). Phytochrome signaling in green Arabidopsis seedlings: Impact assessment of a mutually negative phyB-PIF feedback loop. Mol. Plant 5: 734–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P., Monte E., Oka Y., Liu T., Carle C., Castillon A., Huq E., Quail P.H. (2008a). Multiple phytochrome-interacting bHLH transcription factors repress premature seedling photomorphogenesis in darkness. Curr. Biol. 18: 1815–1823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P., Quail P.H. (2011). PIFs: Pivotal components in a cellular signaling hub. Trends Plant Sci. 16: 19–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P., Tepperman J.M., Monte E., Calderon R.H., Liu T.L., Quail P.H. (2009). Definition of early transcriptional circuitry involved in light-induced reversal of PIF-imposed repression of photomorphogenesis in young Arabidopsis seedlings. Plant Cell 21: 3535–3553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorrain S., Allen T., Duek P.D., Whitelam G.C., Fankhauser C. (2008). Phytochrome-mediated inhibition of shade avoidance involves degradation of growth-promoting bHLH transcription factors. Plant J. 53: 312–323 [DOI] [PubMed] [Google Scholar]
- Lorrain S., Trevisan M., Pradervand S., Fankhauser C. (2009). Phytochrome interacting factors 4 and 5 redundantly limit seedling de-etiolation in continuous far-red light. Plant J. 60: 449–461 [DOI] [PubMed] [Google Scholar]
- Matsushita T., Mochizuki N., Nagatani A. (2003). Dimers of the N-terminal domain of phytochrome B are functional in the nucleus. Nature 424: 571–574 [DOI] [PubMed] [Google Scholar]
- Meggio F., Pinna L.A. (2003). One-thousand-and-one substrates of protein kinase CK2? FASEB J. 17: 349–368 [DOI] [PubMed] [Google Scholar]
- Monte E., Al-Sady B., Leivar P., Quail P.H. (2007). Out of the dark: How the PIFs are unmasking a dual temporal mechanism of phytochrome signalling. J. Exp. Bot. 58: 3125–3133 [DOI] [PubMed] [Google Scholar]
- Monte E., Tepperman J.M., Al-Sady B., Kaczorowski K.A., Alonso J.M., Ecker J.R., Li X., Zhang Y., Quail P.H. (2004). The phytochrome-interacting transcription factor, PIF3, acts early, selectively, and positively in light-induced chloroplast development. Proc. Natl. Acad. Sci. USA 101: 16091–16098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulekar J.J., Bu Q., Chen F., Huq E. (2012). Casein kinase II α subunits affect multiple developmental and stress-responsive pathways in Arabidopsis. Plant J. 69: 343–354 [DOI] [PubMed] [Google Scholar]
- Nash P., Tang X., Orlicky S., Chen Q., Gertler F.B., Mendenhall M.D., Sicheri F., Pawson T., Tyers M. (2001). Multisite phosphorylation of a CDK inhibitor sets a threshold for the onset of DNA replication. Nature 414: 514–521 [DOI] [PubMed] [Google Scholar]
- Ni M., Tepperman J.M., Quail P.H. (1998). PIF3, a phytochrome-interacting factor necessary for normal photoinduced signal transduction, is a novel basic helix-loop-helix protein. Cell 95: 657–667 [DOI] [PubMed] [Google Scholar]
- Ni M., Tepperman J.M., Quail P.H. (1999). Binding of phytochrome B to its nuclear signalling partner PIF3 is reversibly induced by light. Nature 400: 781–784 [DOI] [PubMed] [Google Scholar]
- Nozue K., Covington M.F., Duek P.D., Lorrain S., Fankhauser C., Harmer S.L., Maloof J.N. (2007). Rhythmic growth explained by coincidence between internal and external cues. Nature 448: 358–361 [DOI] [PubMed] [Google Scholar]
- Oh E., Kim J., Park E., Kim J.I., Kang C., Choi G. (2004). PIL5, a phytochrome-interacting basic helix-loop-helix protein, is a key negative regulator of seed germination in Arabidopsis thaliana. Plant Cell 16: 3045–3058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlicky S., Tang X., Willems A., Tyers M., Sicheri F. (2003). Structural basis for phosphodependent substrate selection and orientation by the SCFCdc4 ubiquitin ligase. Cell 112: 243–256 [DOI] [PubMed] [Google Scholar]
- Quail P.H. (2002). Phytochrome photosensory signalling networks. Nat. Rev. Mol. Cell Biol. 3: 85–93 [DOI] [PubMed] [Google Scholar]
- Reed J.W., Nagatani A., Elich T.D., Fagan M., Chory J. (1994). Phytochrome A and phytochrome B have overlapping but distinct functions in Arabidopsis development. Plant Physiol. 104: 1139–1149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed J.W., Nagpal P., Poole D.S., Furuya M., Chory J. (1993). Mutations in the gene for the red/far-red light receptor phytochrome B alter cell elongation and physiological responses throughout Arabidopsis development. Plant Cell 5: 147–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin S.M. (2013). Deciphering the retinoblastoma protein phosphorylation code. Trends Biochem. Sci. 38: 12–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto K., Nagatani A. (1996). Nuclear localization activity of phytochrome B. Plant J. 10: 859–868 [DOI] [PubMed] [Google Scholar]
- Santamaria, A., Wang, B., Elowe, S., Malik, R., Zhang, F., Bauer, M., Schmidt, A., Sillje, H.H., Korner, R., and Nigg, E.A. (2011). The Plk1-dependent phosphoproteome of the early mitotic spindle. Mol. Cell Proteomics 10: M110 004457. [DOI] [PMC free article] [PubMed]
- Schafer, E., and Nagy, F. (2006). Photomorphogenesis in Plants and Bacteria. (Springer, Dordrecht, The Netherlands). [Google Scholar]
- Sharrock R.A., Clack T. (2002). Patterns of expression and normalized levels of the five Arabidopsis phytochromes. Plant Physiol. 130: 442–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H., Zhu L., Castillon A., Majee M., Downie B., Huq E. (2008). Light-induced phosphorylation and degradation of the negative regulator PHYTOCHROME-INTERACTING FACTOR1 from Arabidopsis depend upon its direct physical interactions with photoactivated phytochromes. Plant Cell 20: 1586–1602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y., Khanna R., Carle C.M., Quail P.H. (2007). Phytochrome induces rapid PIF5 phosphorylation and degradation in response to red-light activation. Plant Physiol. 145: 1043–1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J., Kim K., Kang H., Zulfugarov I.S., Bae G., Lee C.H., Lee D., Choi G. (2009). Phytochromes promote seedling light responses by inhibiting four negatively-acting phytochrome-interacting factors. Proc. Natl. Acad. Sci. USA 106: 7660–7665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somers D.E., Sharrock R.A., Tepperman J.M., Quail P.H. (1991). The hy3 long hypocotyl mutant of Arabidopsis is deficient in phytochrome B. Plant Cell 3: 1263–1274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sörensson C., Lenman M., Veide-Vilg J., Schopper S., Ljungdahl T., Grøtli M., Tamás M.J., Peck S.C., Andreasson E. (2012). Determination of primary sequence specificity of Arabidopsis MAPKs MPK3 and MPK6 leads to identification of new substrates. Biochem. J. 446: 271–278 [DOI] [PubMed] [Google Scholar]
- Steen J.A., Steen H., Georgi A., Parker K., Springer M., Kirchner M., Hamprecht F., Kirschner M.W. (2008). Different phosphorylation states of the anaphase promoting complex in response to antimitotic drugs: A quantitative proteomic analysis. Proc. Natl. Acad. Sci. USA 105: 6069–6074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephenson P.G., Fankhauser C., Terry M.J. (2009). PIF3 is a repressor of chloroplast development. Proc. Natl. Acad. Sci. USA 106: 7654–7659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X., Orlicky S., Mittag T., Csizmok V., Pawson T., Forman-Kay J.D., Sicheri F., Tyers M. (2012). Composite low affinity interactions dictate recognition of the cyclin-dependent kinase inhibitor Sic1 by the SCFCdc4 ubiquitin ligase. Proc. Natl. Acad. Sci. USA 109: 3287–3292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tepperman J.M., Hwang Y.S., Quail P.H. (2006). phyA dominates in transduction of red-light signals to rapidly responding genes at the initiation of Arabidopsis seedling de-etiolation. Plant J. 48: 728–742 [DOI] [PubMed] [Google Scholar]
- Ubersax J.A., Ferrell J.E., Jr (2007). Mechanisms of specificity in protein phosphorylation. Nat. Rev. Mol. Cell Biol. 8: 530–541 [DOI] [PubMed] [Google Scholar]
- Varedi K S.M., Ventura A.C., Merajver S.D., Lin X.N. (2010). Multisite phosphorylation provides an effective and flexible mechanism for switch-like protein degradation. PLoS ONE 5: e14029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi R., Nakamura M., Mochizuki N., Kay S.A., Nagatani A. (1999). Light-dependent translocation of a phytochrome B-GFP fusion protein to the nucleus in transgenic Arabidopsis. J. Cell Biol. 145: 437–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Mayba O., Pfeiffer A., Shi H., Tepperman J.M., Speed T.P., Quail P.H. (2013). A quartet of PIF bHLH factors provides a transcriptionally centered signaling hub that regulates seedling morphogenesis through differential expression-patterning of shared target genes in Arabidopsis. PLoS Genet. 9: e1003244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B., Li L., Tumaneng K., Wang C.Y., Guan K.L. (2010). A coordinated phosphorylation by Lats and CK1 regulates YAP stability through SCF(beta-TRCP). Genes Dev. 24: 72–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zulawski M., Braginets R., Schulze W.X. (2013). PhosPhAt goes kinases—Searchable protein kinase target information in the plant phosphorylation site database PhosPhAt. Nucleic Acids Res. 41 (Database issue): D1176–D1184 [DOI] [PMC free article] [PubMed] [Google Scholar]