Submergence induces the expression of a broad spectrum of genes, including a group of WRKY transcription factors and innate immunity marker genes. This work shows that submergence triggers innate immunity in Arabidopsis to protect plants against a higher probability of pathogen infection either during or after flood.
Abstract
Transcriptional control plays an important role in regulating submergence responses in plants. Although numerous genes are highly induced during hypoxia, their individual roles in hypoxic responses are still poorly understood. Here, we found that expression of genes that encode members of the WRKY transcription factor family was rapidly and strongly induced upon submergence in Arabidopsis thaliana, and this induction correlated with induction of a large portion of innate immunity marker genes. Furthermore, prior submergence treatment conferred higher resistance to the bacterial pathogen Pseudomonas syringae in Arabidopsis. Among the WRKY genes tested, WRKY22 had the highest level of induction during the early stages of submergence. Compared with the wild type, WRKY22 T-DNA insertion mutants wrky22-1 and wrky22-2 had lower disease resistance and lower induction of innate immunity markers, such as FLG22-INDUCED RECEPTOR-LIKE KINASE1 (FRK1) and WRKY53, after submergence. Furthermore, transcriptomic analyses of wrky22-2 and chromatin immunoprecipitation identified several potential targets of WRKY22, which included genes encoding a TIR domain–containing protein, a plant peptide hormone, and many OLIGO PEPTIDE TRANSPORTER genes, all of which may lead to induction of innate immunity. In conclusion, we propose that submergence triggers innate immunity in Arabidopsis via WRKY22, a response that may protect against a higher probability of pathogen infection either during or after flooding.
INTRODUCTION
Due to their immobility, plants have evolved complex sensing mechanisms and response pathways that help them adapt to diverse environmental conditions and ensure their survival (Bailey-Serres and Voesenek, 2008). Transcriptional regulation is one of the mechanisms used by plants to protect against biotic and abiotic stresses. Multiple signaling pathways are triggered transcriptionally in response to flooding (Klok et al., 2002; Fukao and Bailey-Serres, 2008; Hattori et al., 2009). Functional studies of plant flooding and the effects of low oxygen have mainly focused on altering the expression of the genes encoding the enzymes for fermentation and glycolysis (Ellis et al., 1999; Kürsteiner et al., 2003; Bieniawska et al., 2007; Dolferus et al., 2008). Transcriptomic studies have revealed that expression of huge numbers of genes (∼10% genes assayed) is altered upon flooding (Klok et al., 2002; Liu et al., 2005; Kreuzwieser et al., 2009; van Dongen et al., 2009; Hsu et al., 2011), suggesting that global transcriptional regulatory networks play important roles in response to flooding.
Determining the roles of transcription factors (TFs) is key to understanding transcriptional regulatory networks. Several flooding-responsive TFs are known to be involved in hypoxia signaling and/or anoxic tolerance in Arabidopsis thaliana, including a myeloblastosis (MYB) TF, a NAM/ATAF/CUC TF, and several APETALA2/ethylene-responsive factors (AP2/ERFs) (Hoeren et al., 1998; Christianson et al., 2009; Hinz et al., 2010; Licausi et al., 2010a; Yang et al., 2011). In rice (Oryza sativa), several AP2/ERFs play central roles in modulating ethylene signaling in response to submergence (Xu et al., 2006; Fukao and Bailey-Serres, 2008; Hattori et al., 2009). Although the roles of these TFs in flooding tolerance have been characterized (Xu et al., 2006; Fukao and Bailey-Serres, 2008; Hattori et al., 2009), their roles cover only a fraction of the transcriptional regulation seen in transcriptomic data. Therefore, it is important to identify and investigate more TFs and transcriptional regulatory pathways that are associated with submergence at the level of the transcriptome.
In Arabidopsis, the WRKY TF superfamily consists of 74 members (Eulgem, 2005). Members of the WRKY TF family contain at least one conserved DNA binding region, the WRKY domain that comprises the highly conserved WRKYGQK peptide sequence, and a zinc finger motif (Eulgem et al., 2000). WRKY TFs are involved in the regulation of gene expression during biotic stress, abiotic stress, senescence, and several developmental processes (Rushton et al., 2010). Current transcriptomic data indicate that several genes that encode WRKY TF family members are induced to high levels upon flooding (Klok et al., 2002; Liu et al., 2005; Kreuzwieser et al., 2009; Hsu et al., 2011), but the functions of these flood-responsive WRKY TFs during flooding are unclear. Furthermore, WRKY-mediated pathways are also known to have a major role underpinning immune responses in plants (Eulgem and Somssich, 2007; Pandey and Somssich, 2009).
Plant innate immunity is triggered by microbe- or pathogen-associated molecular patterns (MAMPs or PAMPs) and/or effector-triggered immunity. In PAMP-triggered immunity, attack by pathogens is recognized by groups of proteins termed pattern recognition receptors (PRRs) that trigger a defense response (Boller and Felix, 2009). Recognition of the molecular signatures of pathogens activates mitogen-activated protein kinases and calcium-dependent protein kinases to induce innate immunity marker genes, such as FLG22-INDUCED RECEPTOR KINASE1 (At2g19190) (Asai et al., 2002; Boudsocq et al., 2010). These responses can eventually lead to systemic acquired resistance (SAR), which confers immunity against subsequent infections. The SAR defense responses, which are activated at different sites of pathogen attack, are mediated mainly by jasmonic acid, salicylic acid, and ethylene (Thomma et al., 2001). The development of SAR is associated with pathogenesis-related (PR) proteins, proteins encoded by the host plant but specifically induced in pathology-related situations (Van Loon and Van Strien, 1999; van Loon et al., 2006).
In this work, we found that many WRKY and innate immunity marker genes are strongly induced during submergence and that submergence treatment conferred higher resistance to the bacterial pathogen Pseudomonas syringae in Arabidopsis. The correlation between the inductions of these two groups of genes led us to speculate that innate immunity may be induced by submergence. T-DNA insertion mutant studies showed that WRKY22 mediates the bacterial resistance conferred by submergence treatment. Downstream targets of WRKY22 were identified based on differential expression of targets in WRKY22 mutants, and the protein–DNA interactions between WRKY22 protein and promoters of targets. Finally, our results reveal a glimpse of the possible signaling pathways that underlie higher bacterial resistance in plants that have undergone submergence treatment. We speculate that these mechanisms protect plants from pathogen infection that is more likely to occur during or after flooding.
RESULTS
Many WRKY Genes and Marker Genes of Innate Immunity Are Strongly Induced during Submergence
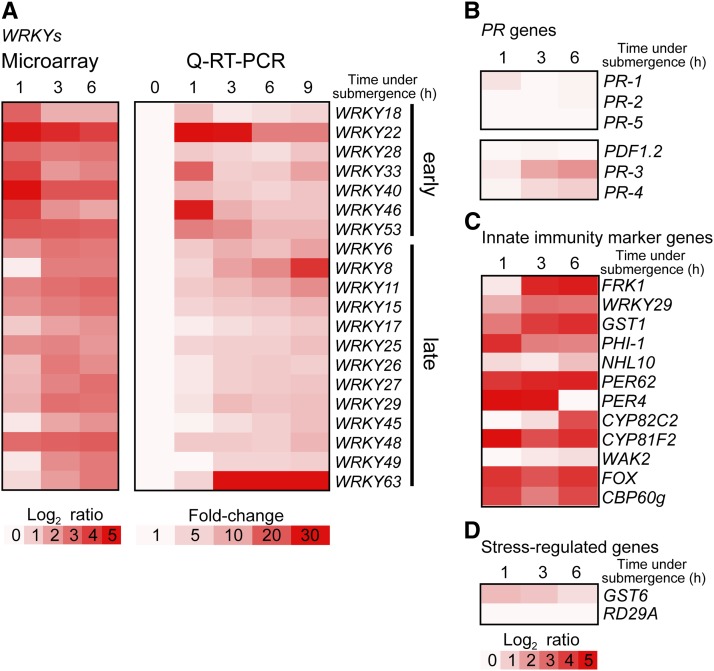
To identify submergence-responsive TFs that had not been previously studied, we dissected transcriptomic profiles in Arabidopsis during submergence via microarray analysis. Using a fourfold induction threshold to filter our microarray data, we found that 20 out of a total of 74 WRKY TF genes in the Arabidopsis genome were strongly induced during submergence (Figure 1A, left). Among these, seven were rapidly induced after 1 h of submergence treatment, implying early roles upstream of signaling pathways. The transcript levels of these WRKY genes were validated by quantitative real-time RT-PCR (qRT-PCR) (Figure 1A, right). Of the PR and stress-related genes tested, only PR-3 and PR-4 were slightly induced within 6 h of submergence, whereas most of the PR genes as well as PLANT DEFENSIN1.2 (PDF1.2; At5g44420) were not dramatically induced (Figure 1B). By contrast, many of the innate immunity marker genes were strongly activated by submergence (Figures 1B to 1D; see Supplemental Table 1 online for gene information). PEP1 RECEPTOR, ELICITOR PEPTIDE PRECURSOR (PROPEP), CELL WALL-ASSOCIATED KINASE (WAK), and RAPID ALKALINIZATION FACTOR1 are considered to be damage-associated molecular pattern (DAMP) marker genes (Boller and Felix, 2009; Tör et al., 2009; De Lorenzo et al., 2011; Ranf et al., 2011; Ma et al., 2012; Ferrari et al., 2013; Logemann et al., 2013). Of the DAMP marker genes tested, PROPEP2 and PROPEP3 were induced upon submergence (see Supplemental Figure 1 online). These results indicate that plant innate immunity may be induced during submergence. We also found that these WRKY genes were coexpressed with innate immunity marker genes following drought, hypoxia, and salt treatments (see Supplemental Figure 2 online; Hruz et al., 2008). These results raise the possibility that the WRKY TFs may mediate immunity in response to various abiotic stresses.
Figure 1.
Induction of Innate Immunity Markers and WRKY Genes in Response to Submergence.
(A) Expression of WRKY genes is induced by submergence. Gene expression was determined by microarray analysis and validated by qRT-PCR in 9-d-old wild-type Arabidopsis (Columbia) seedlings from at least four independent biological replicates.
(B) to (D) Innate immunity marker genes were responsive to submergence (C), but PR genes (B) and stress-regulated genes (D) were not (see Supplemental Table 1 online for gene information). The color scale indicates treatment-to-control ratio of expression in log2 or in fold induction.
Submergence Treatment Conferred Higher Resistance to P. syringae
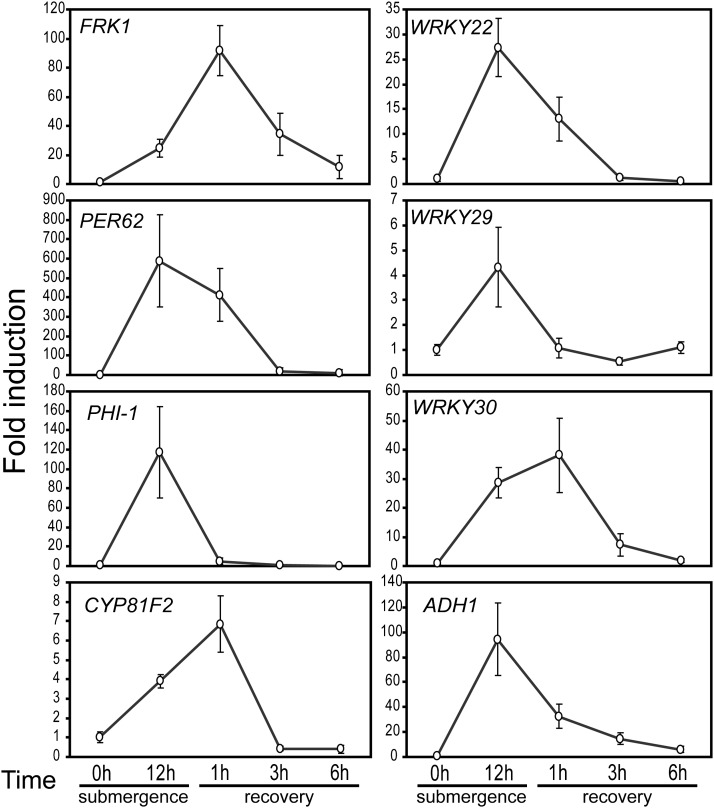
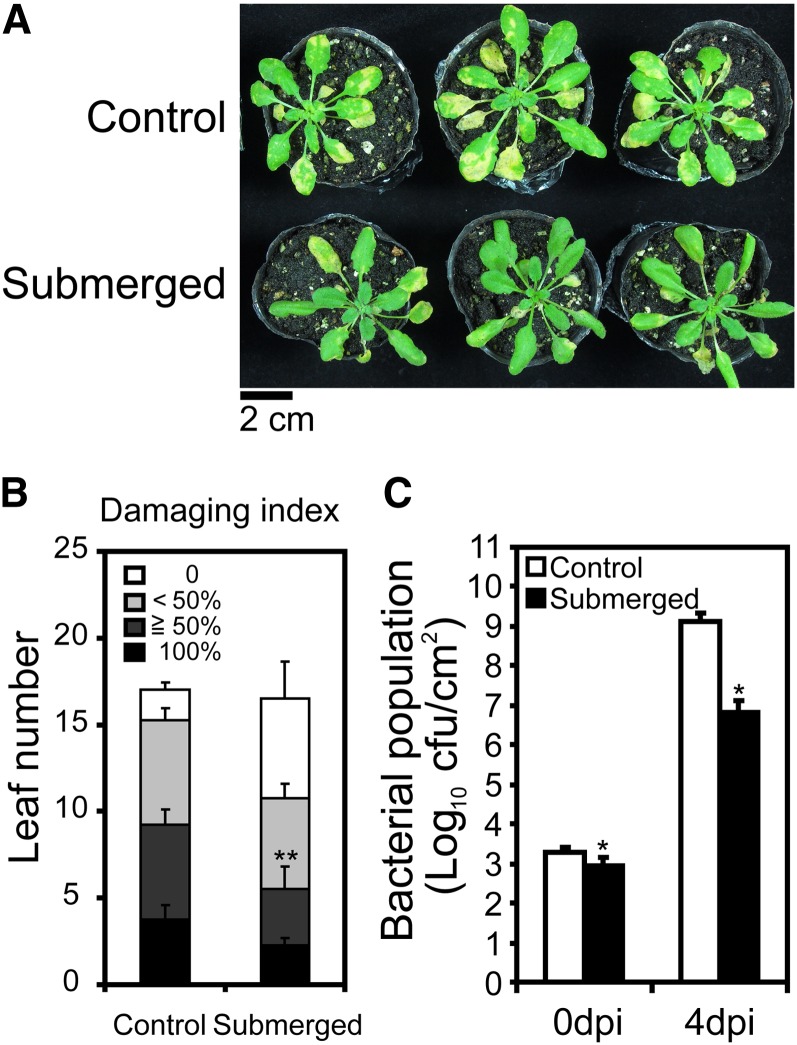
As submergence upregulated many innate immunity markers and WRKY genes, we speculated that plant immunity may be increased by submergence. To determine whether plant immunity was induced, we first submerged Arabidopsis plants and then assayed their immunity through inoculation with the virulent bacterial pathogen P. syringae pv tomato DC3000 (Pst DC3000). To minimize interference from damage caused by submergence, the length of time plants were submerged was carefully chosen to ensure that submergence was long enough to induce innate immunity markers and WRKY genes, but not so long as to damage plants. A recent report defined the median lethal time of complete submergence in the dark to be around 8 d for Arabidopsis ecotype Columbia-0 (Vashisht et al., 2011). We found that submergence treatment for 12 h followed by 1 h of recovery under light in air induced high levels of transcripts for innate immunity marker genes FRK1, PEROXIDASE62, CYP81F2, WRKY22, and WRKY30 (Figure 2). We therefore used these parameters for submergence prior to inoculation for subsequent experiments. Next, presubmerged Arabidopsis were dip inoculated with Pst DC3000 and disease progression was evaluated. The presubmerged plants developed fewer disease symptoms and had lower bacterial populations than did nonsubmerged control plants (Figure 3). Since some innate immunity markers and WRKY genes were induced rapidly (Figure 1), such rapid induction could contribute to certain level of submergence-triggered plant immunity. Thus, we also tested whether shorter period of submergence triggered plant immunity. The plants treated with 2 h of presubmergence developed fewer disease symptoms than did nonsubmerged control plants (see Supplemental Figure 3 online), suggesting that those rapidly induced innate immunity markers and WRKY genes contribute to part of the submergence-triggered immunity. Collectively, these molecular and phenotypic data indicate that prior submergence conferred higher resistance to pathogens in plants than that found in nonsubmerged plants.
Figure 2.
Expression of Innate Immunity Marker Genes after Submergence.
Five-week-old Columbia plants were submerged for 12 h and allowed to recover for up to 9 h, with samples collected at the indicated time points. Transcript levels were detected by qRT-PCR using specific primers. TUBULIN mRNA was used as an internal control. The data represent means ± sd from four to seven independent biological replicates.
Figure 3.
Plant Immunity Is Triggered by Submergence.
(A) Disease symptoms assessed 4 d postinoculation (dpi) with P. syringae in submerged and control Columbia plants. Bar = 2 cm.
(B) The levels of resistance were defined using a damage index based on the necrotic and chlorotic area of leaves (black, 100% leaf area; dark gray, equal to or >50% leaf area; light gray, <50% leaf area; white, no damage observed). The data represent means ± sd from four independent repeats. Statistical differences between submerged and control were determined using Student’s t test for the sum of 100% and ≥50% indexes. **P < 0.01.
(C) Bacterial population at 0 and 4 d postinoculation in submerged and control Columbia plants. The data represent means ± sd from five independent repeats. Statistical differences between submerged and control plants were determined by Student’s t test. *P < 0.05. cfu, colony-forming units.
WRKY22 Is Rapidly Induced by Submergence
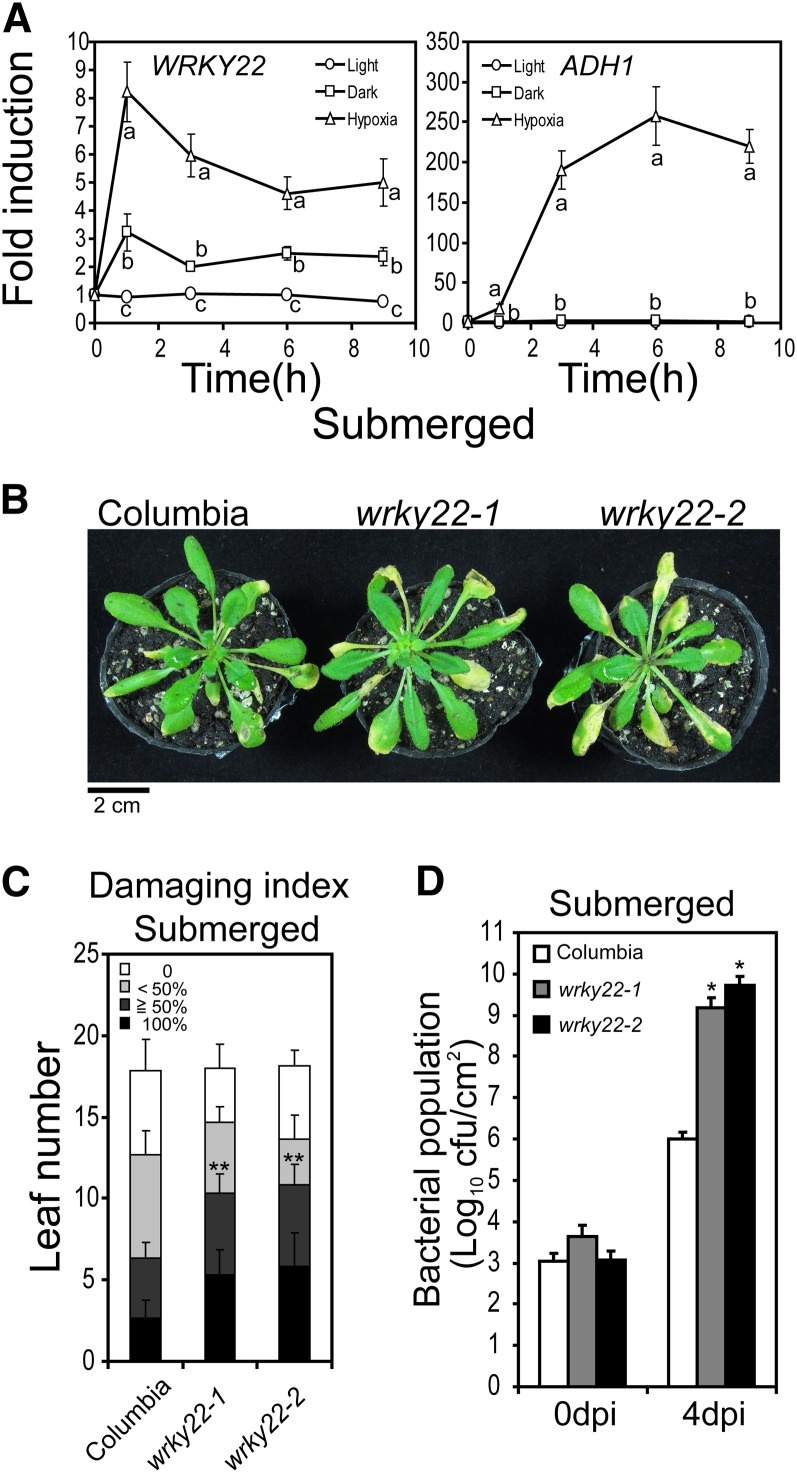
As we had found that submergence could induce expression of innate immunity markers and WRKY genes and confer higher resistance to a virulent bacterial pathogen, we next investigated whether the induction of innate immunity markers and activation of disease resistance during submergence was mediated by the WRKY TFs. We thus tested whether mutations in WRKY could affect submergence-triggered resistance and induction of innate immunity markers upon submergence. We selected WRKY22 for functional characterization, as it was rapidly induced after 1 h of submergence treatment and had the highest raw intensity fold induction of all the submergence-responsive WRKY genes in our microarray data (Figure 1A, left). As determined by qRT-PCR, WRKY22 was upregulated 30-fold after 1 h of submergence treatment, and this upregulation gradually decreased to >10-fold after 6 h (Figure 1A, right). Although WRKY22 is known to be induced by dark treatment (Zhou et al., 2011), our qRT-PCR data indicated that WRKY22 was induced threefold after 1 h of dark treatment and eightfold after 1 h of hypoxic (0.5% of oxygen) treatment (Figure 4A), whereas it was induced >30-fold after 1 h of submergence treatment (Figure 1A, right; see Supplemental Figure 4C online). This strong and rapid induction of WRKY22 suggests that WRKY22 may play a role upstream of submergence signaling. Two independent mutant lines, designated wrky22-1 (SALK_047120) and wrky22-2 (SALK_098205), carrying T-DNA insertions in the first and third exons of the WRKY22 gene, respectively, were obtained (see Supplemental Figures 4A and 4B online). Levels of T-DNA disrupted WRKY22 transcript in these knockout lines were reduced to ∼50% of intact WRKY22 transcript in Columbia plants (see Supplemental Figure 4C online), suggesting that in addition to disrupting the coding sequences, the T-DNA insertions also affected the mRNA levels.
Figure 4.
WRKY22 Mediates Submergence-Triggered Immunity.
(A) Transcript levels of WRKY22 and ADH1 were detected by qRT-PCR. Nine-day-old Columbia seedlings were in light, dark, or hypoxia (0.5% oxygen gas balanced with nitrogen in dark) for up to 9 h and were collected at specific time points (0, 1, 3, 6, and 9 h). TUBULIN mRNA was used as an internal control. The data represent means ± sd from seven independent biological replicates and were subjected to an one-way analysis of variance and Tukey’s honestly significant difference tests (P < 0.05). Data from the same time point with different lowercase letters were significantly different from each other.
(B) Disease symptoms assessed 4 d postinoculation in submerged Columbia, wrky22-1, and wrky22-2 plants. Bar = 2 cm.
(C) Levels of resistance in submerged Columbia, wrky22-1, and wrky22-2 plants. The levels of resistance were defined using a damage index based on the necrotic and chlorotic area of leaves, as described in Figure 3. The data represent means ± sd from eight independent repeats. Pairwise statistical differences between Columbia and WRKY22 mutants were determined by Student’s t test for the sum of 100% and ≥50% indexes. **P < 0.01.
(D) Bacterial populations at 0 and 4 d postinoculation in submerged Columbia, wrky22-1, and wrky22-2 plants. The data represent means ± sd from five to six independent repeats. Statistical differences between Columbia and WRKY22 mutants were identified using Student’s t test. *P < 0.05.
Submergence-Triggered Resistance Is Affected in WRKY22 Mutants
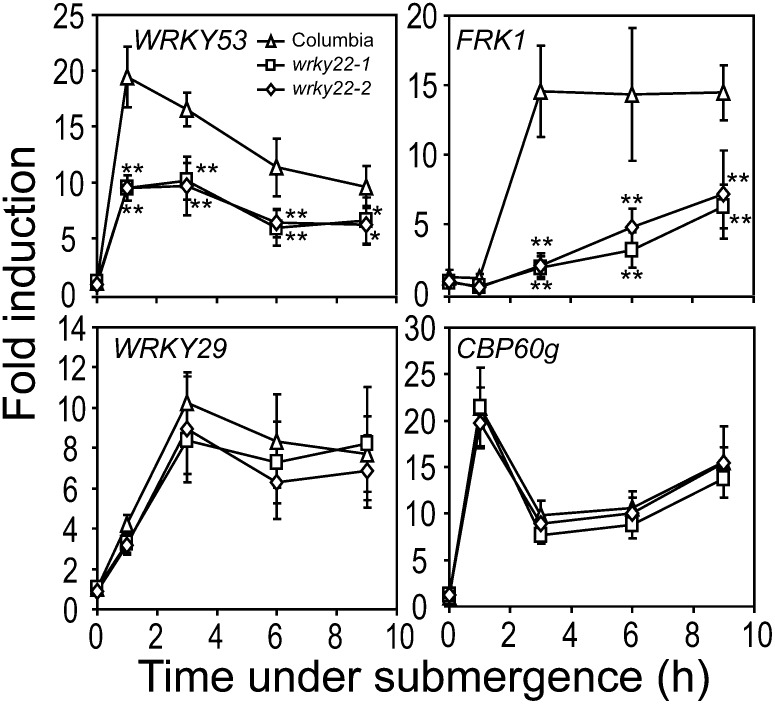
To determine whether WRKY22 is important for submergence-triggered resistance, wrky22-1 and wrky22-2 were subjected to submergence followed by inoculation with Pst DC3000. After submergence, wrky22-1 and wrky22-2 developed more disease symptoms and higher bacterial populations than did wild-type plants (Figures 4B to 4D), suggesting that WRKY22 mediates submergence-triggered resistance. In nonsubmerged plants, disease symptoms in wrky22-1 and wrky22-2 could not be visually distinguished by the naked eye (see Supplemental Figure 5A online), but the degrees of the symptoms in wrky22-1 and wrky22-2, as measured by the damage index that used in Figure 3B, were statistically higher than in the wild type (see Supplemental Figure 5B online). This result suggested that WRKY22 also mediates plant basal innate immunity. To further characterize the molecular function of WRKY22 in submergence-triggered resistance, we examined the effects of WRKY22 mutations on the submergence-responsive innate immunity markers. Expression of two defense-related WRKY genes, WRKY29 and WRKY53, and two innate immunity markers, FRK1 and CAM-BINDING PROTEIN60-LIKE G, were examined in wrky22-1 and wrky22-2 with qRT-PCR. Induction of WRKY53 and FRK1 transcripts were significantly lower in wrky22-1 and wrky22-2 (Figure 5). This result indicates that WRKY22 mediates a portion of the signaling pathway activating innate immunity in response to submergence.
Figure 5.
Innate Immunity Markers FRK1 and WRKY53 Are Transcriptionally Regulated by WRKY22.
Transcript levels were detected by qRT-PCR using specific primers. TUBULIN mRNA was used as an internal control. The data represent means ± sd from four to eight independent biological replicates. *P < 0.05 and **P < 0.01 in Student’s t test.
Identification of WRKY22 Targets in Response to Submergence
To investigate the WRKY22-mediated signaling pathway in response to submergence, we used two approaches to identify WRKY22 target genes: (1) a time course study using Agilent Arabidopsis arrays to identify differentially expressed genes in wrky22-2 lines, and (2) chromatin immunoprecipitation followed by microarray hybridization (ChIP-chip) to screen for WRKY22 direct targets.
To identify differential expression of submergence-responsive genes, we first generated a list of submergence-responsive genes that showed more than twofold induction in expression with raw intensity >100 at any one time point from 1 to 6 h under submergence in the wild-type Columbia strain. The expression of the selected submergence-responsive genes in the wild type was then compared with the expression in the wrky22-2 line. Genes with raw intensities or normalized ratios >1.5 (Columbia/wrky22-2 ratio) were identified as genes downregulated in the wrky22-2 line (see Supplemental Data Set 1 online), among which were potential targets of WRKY22. The top 20 genes according to the ratios from each time point were taken. These genes were then categorized based on functional classification. Differentially regulated genes not included in the top 20 lists but within the same family or classification were also taken (see Supplemental Table 2 online). Expression of many submergence-responsive genes was affected in wrky22-2 line. These genes were functionally categorized as immunity related, TFs, carbohydrate and amino acid metabolism related, redox related, transporters, heat shock proteins, protein kinases, U-box proteins, and hormone biosynthesis related, suggesting that these functional categories were mediated by WRKY22 in response to submergence.
To identify direct targets of WRKY22, we created a transgenic Arabidopsis line that expresses a c-myc epitope-tagged WRKY22 and used ChIP-chip to screen for candidates and validate the in vivo protein–DNA interactions with ChIP followed by quantitative PCR (ChIP-qPCR). The WRKY22 and c-myc epitope tag fusion construct was generated and transformed into wrky22-2 plants. The resulting transgenic lines should have better ChIP efficiency than the wild-type background, due to the reduced competition for WRKY22 binding sites from endogenous WRKY22. RT-PCR and immunoblot analyses were performed to validate that the induction patterns of transcript and protein levels of c-myc–tagged WRKY22 during hypoxia were similar to those of endogenous WRKY22 (see Supplemental Figure 6 online). ChIP-enriched DNA fragments were identified using criteria of a window of +300 to −1200 of a gene for a promoter, a width of four probes or more, and a false discovery rate < 0.1. The ChIP-chip experiments were repeated six times (i.e., six biological replicates). Candidates were defined by the presence of the promoter in three out of six biological replicates. Candidates were then classified based on their hypoxic responsiveness with a positive response defined as gene expression levels exhibiting more than twofold or <0.5-fold induction in any time point under submergence treatments in expression array data. Three candidate lists of upregulated, downregulated, and unchanged targets in response to submergence were generated (see Supplemental Table 3 online). Among 569 candidates, 29 were upregulated, 22 were downregulated, and 518 were not significantly regulated. To determine the submergence induction of gene expression that is directly mediated by WRKY22, we focused on the 29 upregulated candidates (see Supplemental Table 3 online). Twenty-two out of 29 promoters in the list harbor a canonical W-box, which is defined by the sequence TTGACY. A degenerate W-box (TTGACN) sequence, which is used as a lenient W-box definition (Ciolkowski et al., 2008), can be found in six of the seven promoters without a canonical W-box in the upregulated candidate list. ChIP-qPCR was then used to validate these in vivo protein–DNA interactions. Fourteen candidates that were randomly selected from the upregulated candidate list were tested using ChIP-qPCR. Twelve candidates (∼86%) passed the validation. Enrichment of many promoter regions was observed, including the promoters of At5g44910, 1-AMINO-CYCLOPROPANE-1-CARBOXYLATE SYNTHASE7 (ACS7), ENDOPLASMIC RETICULUM OXIDOREDUCTINS2 (ERO2), PLANT U-BOX 24 (PUB24), and TREHALASE1 (TRE1) (see Supplemental Table 3 online). This result suggests that WRKY22 directly targets the promoter regions of these submergence-responsive genes.
Innate Immunity Conferred by Submergence Is Mediated by WRKY22
Plant innate immunity is usually triggered through PRRs in response to pathogen-produced MAMPs. For instance, exogenous applications of MAMPs triggered coinduction patterns of WRKYs and innate immunity marker genes (see Supplemental Figure 2B online). Surprisingly, MAMP-free submergence conditions were also able to induce coexpression of WRKYs and innate immunity marker genes (Figures 1A and 1C). Thus, we speculated that submergence may activate Arabidopsis endogenous elicitors that trigger defense mechanisms similar to those elicited by MAMPs. The WRKY22 target FRK1 is known to be triggered through receptor-like kinase (RLK) signaling of FLAGELLIN-SENSITIVE2 (FLS2) in response to flagellin or flg22 (a peptide fragment of flagellin) from microbes (Gómez-Gómez and Boller, 2000). A recent report showed that FLS2 not only recognizes flg22 peptide originally from microbes, but also interacts with a plant endogenous peptide hormone, CLAVATA3 peptide (Lee et al., 2011), suggesting that PRRs can recognize both MAMPs and other molecular patterns to trigger similar signaling pathways.
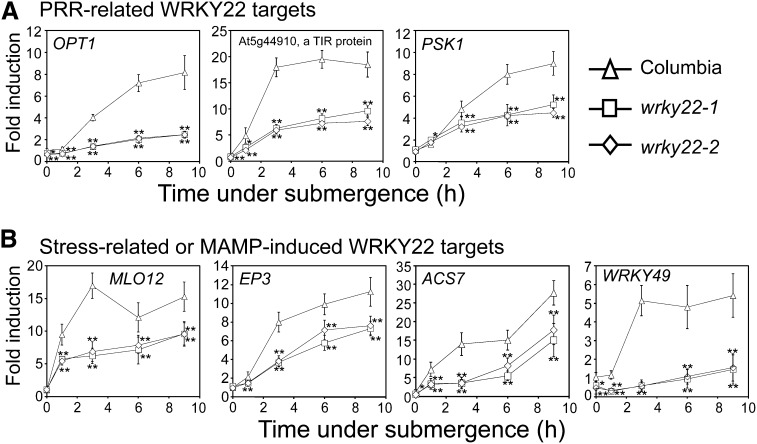
By integrating our results of WRKY22 targets derived from expression array and ChIP with validation by qRT-PCR (Figure 6; see Supplemental Figure 7 online), we found that WRKY22 mediates PRR-related and stress-related signals in response to submergence. Notably, a toll/interleukin-1 receptor domain–containing protein, encoded by a WRKY22 direct target At5g44910 (Figure 6A), is also a RLK, which is a potential peptide receptor. An indirect target PSK1 (At1g13590; Figure 6A), which encodes a plant peptide hormone, could generate or amplify signals. OLIGO PEPTIDE TRANSPORTER1 (OPT1; At5g55930) and several OPT genes, which were found to be WRKY22 targets (Figure 6A; see Supplemental Table 2 online), could transport peptide- or amino acid–related signals. Another type of RLK, wall-associated kinases, can recognize oligogalacturonides that are released from plant cell wall fragments as DAMPs (Brutus et al., 2010; De Lorenzo et al., 2011). Several members of the WAK-like (WAKL) family (i.e., WAKL2, WAKL6, and WAKL10) were identified as putative targets of WRKY22 upon submergence (see Supplemental Table 2 online). Moreover, several biotic stress–related or MAMP-induced genes were also identified as targets of WRKY22 in response to submergence (Figure 6B). In addition to WRKY22-mediated induction of FRK1 under submergence (Figure 5; see Supplemental Table 2 online), these characteristics of WRKY22 targets strengthen the idea that submergence may lead to PRR-mediated signaling through the mediation of WRKYs to trigger innate immunity.
Figure 6.
Several PRR-Related and MAMP-Induced Genes Are WRKY22 Targets under Submergence.
Transcript levels of PRR-related (A) and MAMP-induced (B) genes were detected by qRT-PCR using specific primers. TUBULIN mRNA was used as an internal control. The data represent means ± sd from four to eight independent biological replicates. *P < 0.05 and **P < 0.01 in Student’s t test.
WRKY22-Regulated Networks upon Submergence Are Associated with Pathogen Resistance
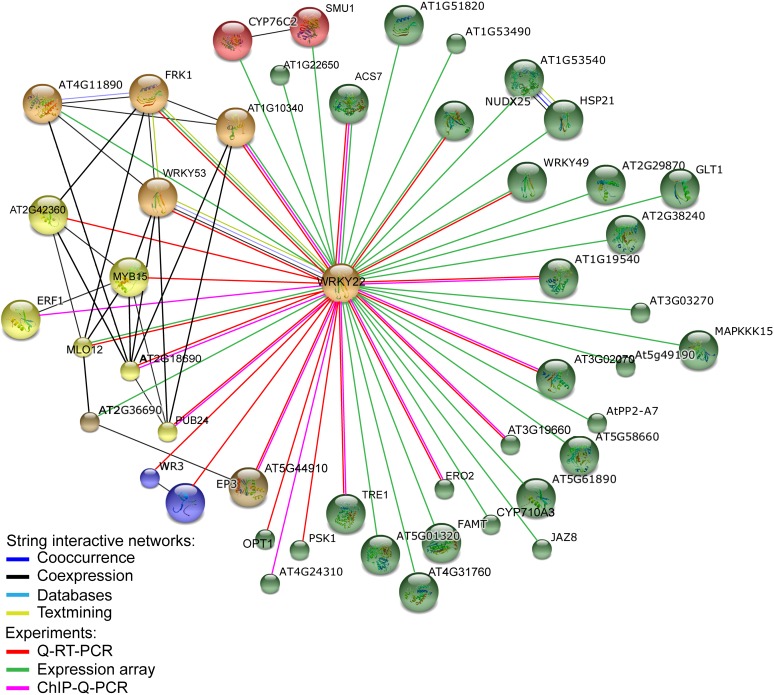
To uncover WRKY22-regulated transcriptional networks upon submergence, we integrated our experimentally derived lists of WRKY22 targets from qRT-PCR results (Figures 5 and 6; see Supplemental Figure 7 online), microarray analysis (asterisk-labeled genes in Supplemental Table 2 online), and ChIP-qPCR results (see Supplemental Table 3 online) and then used the STRING Web-based database (Szklarczyk et al., 2011) to build functional interaction networks based on available experimental evidence (Figure 7). In addition to the previously mentioned PRR-related genes affected by WRKY22, many pathogen-associated genes are highly connected as subnetworks. One subnetwork includes two WRKY22-regulated innate immunity markers, FRK1 and WRKY53, and an RLK gene (At4g11890) that is upregulated during downy mildew disease infection (Hok et al., 2011). Another subnetwork contains MILDEW RESISTANCE LOCUS O 12 (MLO12), which is a member of a conserved protein family in plants required for powdery mildew fungi pathogenesis (Panstruga, 2005; Consonni et al., 2006). Another WRKY22 target, EP3 (At3g54420), encoding a class IV chitinase, is rapidly induced by MAMPs (e.g., flg22) as well as bacterial and viral pathogens (de A. Gerhardt et al., 1997; Whitham et al., 2003; Navarro et al., 2004). In qRT-PCR analysis, both MLO12 and EP3 were expressed at a lower level in wrky22 mutants (Figure 6B). In a WRKY22-regulated subnetwork, MYB15 (At3g23250) and PUB24 (At3g11840), encoding a TF and a U-box E3 ubiquitin ligase, respectively, have been shown to respond to the plant defense elicitor chitin (Libault et al., 2007). Collectively, the existence of WRKY22-regulated networks associated with pathogen responses suggests that WRKY22 plays a role in regulating submergence-triggered pathogen resistance.
Figure 7.
Integrated Networks of WRKY22 Downstream Targets upon Submergence.
Networks were constructed with a Web-based analysis tool STRING (Szklarczyk et al., 2011). Gene lists of WRKY22 downstream targets are integrated from qRT-PCR results (Figures 5 and 6; see Supplemental Figure 7 online), microarray analysis (asterisk-labeled genes in Supplemental Table 2 online), and ChIP-qPCR results (see Supplemental Table 3 online). These three types of experimental evidence are represented by three different line colors. The STRING database assembles information about co-occurrence, coexpression, databases, and text mining, which are also marked with different line colors. Nodes of genes were clustered to six groups by KMEANS in STRING and marked by a different color code.
WRKY22-regulated networks are also associated with abiotic stress. Notably, ethylene biosynthesis is known to be regulated through transcriptional control of a key enzyme, 1-amino-cyclopropane-1-carboxylate synthase, during flooding (Van Der Straeten et al., 2001; Peng et al., 2005; Rieu et al., 2005). We found that ACS7 was transcriptionally regulated by WRKY22 via directly binding to the ACS7 promoter under submergence (Figures 6B and 7; see Supplemental Table 3 online). A recent article shows activation of the ACS7 promoter by abiotic stresses and further suggests ACS7 is involved in the crosstalk between ethylene and abscisic acid for the abiotic stress adaptation of plants (Dong et al., 2011). Submergence induction of another WRKY22 direct target, TRE1, was negatively regulated by WRKY22 (Figure 7; see Supplemental Figure 7B and Supplemental Table 3 online). Expression of TRE1 is involved in stomatal function and leads to increased drought tolerance (Van Houtte et al., 2013). Responsiveness of WOUND-RESPONSIVE3/NITRATE TRANSPORTER3.1 (WR3/NRT3.1), a wound-induced marker gene (León et al., 1998), to submergence was reduced in wrky22 mutants (Figure 7; see Supplemental Figure 7B and Supplemental Table 2 online). Interestingly, other than the involvement of EP3 in defense responses, the WRKY22 target EP3 has also been shown to be induced by an abiotic stress, phosphate starvation (Hammond et al., 2003). Taken together, these WRKY22-regulated networks associated with abiotic stresses suggest that WRKY22 could act as a universal node in the mechanisms commonly responsive to a spectrum of abiotic stresses, including wounding, drought, and submergence.
DISCUSSION
Here, we report previously unknown regulatory pathways that mediate responses in submergence and pathogen resistance in Arabidopsis. We demonstrated that submergence can activate innate immunity markers and WRKY TFs to confer higher disease resistance to plants. Using mutant analysis, we were able to demonstrate that a submergence-inducible WRKY22 acts as a regulator to mediate the activation of innate immunity. This example lends genetic support to the hypothesis that other members of submergence-inducible WRKYs in the family could also play roles in submergence-triggered disease resistance. Via identifying downstream targets of WRKY22, we further demonstrated potential genes and their molecular functions involved in the underlying signaling pathways.
Interconnection between Biotic and Abiotic Stresses
Stress research is typically focused on a single environmental challenge. However, in natural environments, plants are simultaneously or sequentially exposed to multiple stresses. To survive stresses, plants have evolved sophisticated defense mechanisms against both biotic and abiotic stresses. Submergence is a complex stress that consists of multiple environmental changes, including light intensity, temperature, pH, and dissolved oxygen concentration. In fields, water logging may increase the incidence of root rot diseases caused by plant pathogens (Davison and Tay, 1987; Walker, 1991; Yanar et al., 1997). The increased incidence of disease could be due to higher probability of pathogen infection after the flood and/or faster development of the pathogen under higher humidity (Huber and Gillespie, 1992). Although a current report shows two rice chromosomal regions associated with biotic stress and submergence tolerance based on in silico data (Kottapalli et al., 2006), defense mechanisms against biotic and submergence stresses are largely unknown.
Defense responses in abiotic and biotic stresses are regulated by groups of cross-communicating signal transduction pathways. Hormones, such as abscisic acid, jasmonic acid, salicylic acid, and gibberellic acid, mediate signaling pathways required for both pathogen resistance and tolerance of abiotic stresses, including wounding, drought, salt, and cold (Ding et al., 2002; Xiong and Yang, 2003; Chini et al., 2004; Tanaka et al., 2006). A rice mitogen-activated protein kinase gene, MAPK5, negatively regulates pathogen defenses and positively mediates abiotic stresses (Xiong and Yang, 2003). An Arabidopsis disease resistance protein, ACTIVATED DISEASE RESISTANCE1, which processes N-terminal kinase subdomains, enhances drought tolerance (Chini et al., 2004). In addition, many TFs mediate responses for both disease defense and abiotic stresses (Guo et al., 2004; Cao et al., 2006; Sohn et al., 2006; Zhang et al., 2007; Liu et al., 2008; Seo et al., 2010; Zhang et al., 2009). A recent review discussed the central roles of some WRKY TFs in mediating both abiotic and biotic stresses from the point of view of systems biology (Friedel et al., 2012). Together, these data suggest that common signaling pathways interconnect biotic stress and some abiotic stresses. Our study describes such a signaling pathway that specifically connects biotic stress and submergence.
Stomatal Immunity Could Contribute to Part of Submergence-Triggered Immunity
Stomatal closure is a part of innate immunity responses that mediated by the FLS2 receptor against bacterial invasion (Melotto et al., 2006; Zeng and He, 2010). Recent studies demonstrate the involvement of L-type lectin receptor kinases (LecRK) in stomatal immunity. LecRK-VI.2 mediates PAMP-triggered immunity response and acts as a positive mediator of stomatal immunity (Singh et al., 2012). Mechanisms reversing such bacteria-mediated stomatal closure involve negative regulation of stomatal immunity by LecRK-V.5 (Desclos-Theveniau et al., 2012). Our data showed that the bacterial population was lower in submerged plants than in nonsubmerged control plants (Figure 3C). Interestingly, this bacterial titer in submerged plants was also statistically lower at 0 days after inoculation, suggesting that stomatal immunity could be involved in submergence-triggered immunity. Notably, transcripts of both positive and negative mediators of stomatal immunity, LecRK-VI.2 and LecRK-V.5, were highly accumulated under submergence (see Supplemental Figure 8 online), suggesting that stomatal immunity is transcriptionally regulated under submergence. Additionally, a chloroplastic enzyme, ASPARTATE OXIDASE (AO), is required for stomatal immunity (Macho et al., 2012). Our data showed transcripts of AO and several other LecRK genes were regulated under submergence (see Supplemental Figure 8 online). Collectively, stomatal immunity could represent a part of the mechanism for submergence-triggered immunity against bacterial invasion. However, further evidence associated with stomatal immunity, such as oxidative burst, stomatal aperture, and regulatory mechanisms of LecRKs, is required from future studies. Understanding of such submergence-triggered stomatal immunity is important to elucidate defense mechanisms of submergence-triggered immunity.
WRKY TFs May Modulate Regulatory Networks in Response to Submergence
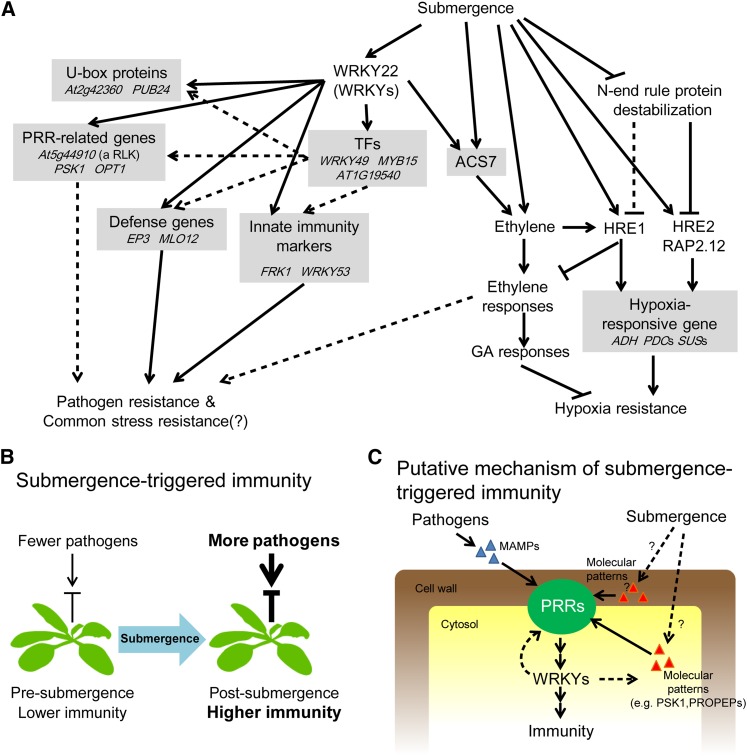
A number of WRKY TFs were highly induced by submergence (Figure 1), suggesting the existence of WRKY-mediated transcriptional regulation in submergence signaling. Some of these submergence-responsive WRKYs were induced during early stages of submergence treatments, while others were induced later. Due to the different temporal expression patterns among induced WRKYs, it is likely that WRKYs could modulate early and late submergence signals and might be positioned upstream and downstream within submergence signaling pathways. Some WRKYs are regulated by mitogen-activated protein kinase cascades in plant defense signaling through protein phosphorylation and/or transcriptional regulation (Asai et al., 2002; Miao et al., 2007; Qiu et al., 2008). Several WRKYs are known to regulate their own expression or expression of other WRKY genes, indicating the occurrence of WRKY auto/cross-regulation (Miao et al., 2007; Qiu et al., 2008; Skibbe et al., 2008). Given the auto- and cross-regulation observed for some WRKY TFs, one may further speculate that WRKY proteins regulate themselves or with other TFs to form regulatory networks in submergence signaling. Through identification of WRKY22 targets, we indeed found that many WRKYs and other families of TFs are targets of WRKY22 to form regulatory networks (Figures 5 to 8; see Supplemental Tables 2 and 3, Supplemental Figure 7, and Supplemental Data Set 1 online).
Figure 8.
Models of WRKY22-Mediated Submergence-Triggered Responses.
(A) Summary of WRKY22-mediated and -independent pathways under submergence.
(B) Model of submergence-triggered immunity.
(C) Putative molecular mechanism of submergence-triggered immunity. Solid lines indicate validated events. Dotted lines and question marks indicate predicted events.
Submergence Triggers Hypoxia-Specific Pathways and Pathways Commonly Induced by Stresses
Oxygen deficiency induces a wide spectrum of TFs in Arabidopsis (Licausi et al., 2010b; Hsu et al., 2011). Among these TFs, some are specifically induced by oxygen deficiency and some are commonly induced by stresses. For example, we previously studied HRE1 (At1g72360), an AP2/ERF that is specifically induced by hypoxic treatments (Yang et al., 2011). In this study, we here show that a group of WRKY TFs were strongly induced by submergence (Figure 1) and other stresses, including drought and salt (see Supplemental Figure 2 online). We not only found that these WRKY genes were coexpressed with innate immunity marker genes (Figure 1; see Supplemental Figure 2 online), but also determined that a representative WRKY TF, WRKY22, regulates innate immunity genes, defense genes, and PRR-relative genes (summarized in Figure 8A). Submergence-triggered responses of several signaling components, such as TFs, U-box proteins, and ACS7, were also mediated by WRKY22. Regarding pathways specifically responsive to hypoxia, HRE1 and HRE2 are specifically and strongly induced by hypoxia and mediate induction of hypoxic core genes, including ADH, PDCs, and SUSs (Licausi et al., 2010a; Yang et al., 2011). The induction of HRE1 can be triggered by ethylene and then plays a negative regulatory role in modulating ethylene responses (Yang et al., 2011). In addition to regulation on the transcript level, HRE2 protein stability is enhanced under hypoxia through an N-end rule protein destabilization mechanism (Gibbs et al., 2011). Through identification of WRKY22-regulated networks, we here provide an example of regulatory pathways upon submergence for TFs that are commonly induced by stresses.
Activation of Innate Immunity Could Have Coevolved with Submergence
A recent report demonstrated that plant immune responses are under the control of a circadian regulator, CIRCADIAN CLOCK-ASSOCIATED1, and suggested that this temporal control of plant defense allows plants to anticipate infection at the time of day when a pathogen normally disperses the spores and time immune responses accordingly (Wang et al., 2011). This example supports the notion that plants have evolved mechanisms that are intimately intertwined with environmental conditions. Our study showed that submergence equips Arabidopsis with higher innate immunity through WRKY22. In natural conditions, submergence commonly leads to a higher probability of pathogen infection and faster development of pathogens (Davison and Tay, 1987; Huber and Gillespie, 1992; Kottapalli et al., 2006). Therefore, we suggest that plants have evolved disease defense mechanisms in response to submergence in anticipation of a higher risk of pathogen attack (Figure 8B).
Summary
The establishment of a link between submergence stress and plant defense highlights a hitherto unrevealed aspect of the arsenal plants evolved against environmental variation. Although the underlying signaling networks have yet to be fully elucidated, this study gives a glimpse into the possible signaling pathways involved in submergence-triggered disease resistance (Figures 8B and 8C). Understanding the perception and upstream signaling that primes immunity in response to submergence will be the key to revealing the signaling networks. The next challenge is to elucidate the specific cues, particularly the roles of related PRRs and molecular patterns that trigger plant immunity under submergence.
METHODS
Plant Materials
Experiments were performed on Arabidopsis thaliana ecotype Columbia-0. Two T-DNA insertion mutant lines of WRKY22, wrky22-1 (SALK_047120) and wrky22-2 (SALK_098205), were obtained from the ABRC, Ohio State University. For ChIP, transgenic lines expressing c-myc–tagged WRKY22 in wrky22-2 background plants were generated using a T-DNA insertion containing 1000 bp of the upstream promoter sequence and WRKY22 full-length genomic DNA in frame with 18 copies of c-myc sequence at 3′ end in a pMDC110-derived binary vector. The fragment of WRKY22 promoter sequence and full-length genomic DNA were amplified from Columbia-0 genomic DNA with a set of primers fused with attB sites, AtWRKY22p-1attB1(5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTTTTCCAAGTGTGTTCATACT-3′) and AtWRKY22-2 w/o-stop-attB2 (5′-GGGGACCACTTTGTACAAGAAAGCTGGGTTATTCCTCCGGTGGTAGTGG-3′), for Gateway cloning.
Growth Conditions
All seeds were surface sterilized with 0.5% sodium hypochlorite for 20 min and washed at least five times with sterilized water. Seeds were sown on plates with 0.55% Phytagel (Sigma-Aldrich) in half-strength Murashige and Skoog (MS) medium (Duchefa Biochemie) containing 0.5% Suc at pH 5.7 and kept at 4°C in the dark for 3 d to achieve uniform germination. The plates were then transferred to a growth chamber and placed vertically. Plants were grown at 22°C under a 16-h-light (81 μmol s−1 m−2)/8-h-dark cycle until the indicated ages. To obtain 5-week-old plants for inoculation assays, the plates with 5-d-old seedlings were transferred to a growth chamber and grown under a 9-h-light (81 μmol s−1 m−2, 09:00 to 18:00 h; at 22°C)/15-h-dark (at 18°C) cycle for 2 d. These 7-d-old seedlings were then transplanted onto a 6:2:1 mixture of peat moss, vermiculite, and perlite in pots until they were 5 weeks old. To obtain 14-d-old seedlings for ChIP assay, 5-d-old seedlings were transplanted onto fresh plates, and the plates were placed vertically to prevent roots from growing into medium. The transplanted seedlings were grown in the growth chamber until they were 14 d old.
Submergence and Low Oxygen Treatments
For presubmergence treatment of 5-week-old plants for inoculation, pots with plants were placed into distilled water that was bubbled with 3% oxygen balanced with nitrogen at a depth of at least 3 cm from the water surface. To prevent the pot and medium floating during submergence, four glass beads (1.6 cm diameter) were loaded into the bottom of pots before the seedlings were transplanted. For submergence treatments of 9-d-old seedlings, plates with plants on the surface of the medium were placed into half-strength MS liquid medium that was bubbled with 3% oxygen balanced with nitrogen for the indicated times. The liquid MS medium was pretreated with 3% oxygen for 1 h before use. For gas treatments, plates with plants on the surface of the medium were placed into air-tight jars. For hypoxic treatments, the gas in the jars was replenished with at least 10× volume of premixed 0.5% oxygen balanced with nitrogen for the time indicated. For anoxic treatments, the gas in the jars was replenished with at least 10× volume of pure nitrogen and supplemented with a pack of oxygen absorbent (GasPak EZ Anaerobe Container System; Becton, Dickinson and Company) for the indicated times. All treatments were performed in the dark.
Pseudomonas syringae Inoculation
Strain DC 3000 of P. syringae pv tomato (Pst DC3000) was cultivated at 28°C, 200 rpm in King’s medium B (King et al., 1954) containing rifampicin (50 mg/L). For bacterial inoculation, Pst DC3000 was collected by centrifugation and resuspended in 10 mM MgCl2 at A600 = 0.2, corresponding to a titer of 108 colony-forming units/mL. Bacteria were then diluted to 107 colony-forming units/mL in 10 mM MgCl2 and 0.02% Silwet L-77 for dip inoculation. After inoculation, plants were kept in 100% relative humidity.
Quantification of Bacterial Population
For quantification of bacterial populations, three leaf discs (0.38-cm diameter) from one individual plant were pooled as a single data point. Leaf discs were collected from leaves washed twice with sterile water and homogenized in King’s B containing rifampicin, followed by plating appropriate dilutions on solid medium.
Microarray Analysis
Total RNA isolation, DNase treatment, and RNA integrity assays were conducted as previously described (Hsu et al., 2011). Preparation of fluorescence-labeled cDNA and microarray experiments were performed at the DNA Microarray Core Facility, Institute of Plant and Microbial Biology, Academia Sinica, Taiwan, as described at http://ipmb.sinica.edu.tw/microarray/protocol.htm. Arrays in this study were performed with the Agilent Arabidopsis (V4) Gene Expression Microarray, 4× 44k, based on the manufacturer’s two-color microarray protocols. Four independent biological replicates were performed, of which two included a dye-swap. Array signals were detected and analyzed using the Agilent DNA Microarray Scanner G2565CA and Agilent Feature Extraction 10.7.1.1 software, respectively. The acquired results were then imported into GeneSpring 11.5 (Agilent Technologies) using lowess normalization. The data discussed in this publication have been deposited in the National Center for Biotechnology Information (NCBI)’s Gene Expression Omnibus (GEO) (Edgar et al., 2002) and are accessible through GEO Series accession number GSE40139 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE40139).
qRT-PCR
Reverse Transcription and qRT-PCR were conducted as previously described (Hsu et al., 2011). Briefly, qRT-PCR was performed using 1 μL cDNA (from 2 μg total RNA to 100 μL cDNA), 0.2 μM each primer, and SYBR Green PCR Master Mix (Applied Biosystems) on an ABI 7500 real-time PCR machine (Applied Biosystems) using the default settings. Sequences of primers used can be found in Supplemental Table 4 online. TUB3 (AT5G62700) was used as an internal control for normalization. Relative expression levels were compared by calculating the expression of the gene at a certain time point to a common reference sample from the same tissues obtained at time zero from Columbia.
ChIP
To immunoprecipitate the WRKY22 protein, transgenic plants expressing c-myc epitope-tagged WRKY22 were used. To minimize the competition of protein–DNA interaction with endogenous WRKY22, this tagged WRKY22 was expressed in wrky22-2 plants. The antibody for the ChIP experiments was a monoclonal antibody against c-myc (9E10), which was obtained from the Developmental Studies Hybridoma Bank, University of Iowa. ChIP protocols were as previously described by Kaufmann et al. (2010). Briefly, 1.5 g of 14-d-old seedlings was chemically cross-linked by the addition of 1% formaldehyde for 15 min at room temperature. The fixed seedlings were rinsed five times with MC buffer (10 mM sodium phosphate, pH 7, 50 mM NaCl, and 0.1 M Suc) and frozen in liquid nitrogen. The frozen samples were homogenized, filtered, centrifuged, and washed to isolate nuclei. The nuclear pellets were resuspended, lysed, and sonicated to shear cross-linked DNA. The sonication was performed with a Bioruptor UCD-200 (Diagenode) until the average size of sheared DNA was ∼500 bp. The sonicated chromatin was precleared with protein-A agarose beads for 1 h and immunoprecipitated with 2 μg anti-c-myc antibody for 2 h followed by addition of protein-A agarose beads for 3 h. The beads were washed five times with IP buffer (50 mM HEPES, pH 7.5, 150 mM NaCl, 5 mM MgCl2, 10 μM ZnSO4, 1% Triton X-100, and 0.05% SDS). The bound IP complexes were eluted with elution buffer (0.1 M Gly, 0.5 M NaCl, and 0.05% Tween 20, pH 2.8) and neutralized with 1 M Tris, pH 9.0. The eluates were digested with proteinase K overnight at 37°C to remove proteins followed by incubation at 65°C for at least 6 h to reverse cross-linking. The reverse cross-linked DNA was then purified by extraction with phenol:chloroform:isoamyl alcohol and ethanol precipitation in the presence of glycogen. The DNA pellets were washed with 70% ethanol, air dried, and resuspended in TE 8.0.
ChIP-Chip and ChIP-qPCR
One hundred picograms of DNA were amplified using the GenomePlex Whole Genome Amplification Kit (Sigma-Aldrich) according to the manufacturer’s instructions. The amplified DNA was then purified using the purification kit (Qiagen). The purified DNA was quantified using a Nanodrop spectrophotometer and labeled with Cy3 (for Columbia) and Cy5 (for WRKY22-18×c-myc in wrky22-2) fluorescent dyes. The labeled DNA was hybridized to NimbleGen A. thaliana ChIP-chip 385K Minimal Promoter Arrays (NimbleGen). The hybridization and washing of arrays were performed following the Nimblegen standard operating protocols (www.nimblegen.com). Array images were acquired with Axon GenePix 4000B and GenePix 6.0 software (Axon Instruments). The images were then converted to scaled log2 ratio of the probes for peak finding analysis according to the genomic position in NimbleScan software (NimbleGen). Peaks of enriched DNA fragments were identified and mapped to promoters in the Arabidopsis genome using the default setting in the NimbleScan software (NimbleGen) according to the manufacturer’s description. The data discussed in this publication have been deposited in NCBI’s GEO (Edgar et al., 2002) and are accessible through GEO Series accession number GSE40138 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE40138). For ChIP-qPCR, Cy dye labeling was omitted. The DNA was diluted with TE 8.0 to 2 ng/μL. ChIP-qPCR was performed using 1 μL of the diluted DNA using the methods described above for qRT-PCR. The primer sequences used can be found in Supplemental Table 4 online.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: At1g10340, At1g19540, At1g22650, At1g51820, At1g53490, At1g53540, At2g18690, At2g29870, At2g36690, At2g38240, At2g42360, At3g03270, At3g19660, At3g20270, At4g11890, At4g24310, At4g31760, At5g01320, At5g44910, At5g49190, At5g58660, At5g61890, ACS7 (At4g26200), ADH1 (At1g77120), AO (At5g14760), At-PP2-A7 (At5g45090), CBP60g (At5g26920), CYP710A3 (At2g28850), CYP76C2 (At2g45570), CYP81F2 (At5g57220), EP3 (At3g54420), ERF1 (At3g23240), ERO2 (At2g38960), FAMT (At3g44860), FRK1 (At2g19190), GLT1 (At5g53460), HSP21 (At4g27670), JAZ8 (At1g30135), LecRK-V.5 (At3g59700), LecRK-VI.2 (At5g01540), MAPKKK15 (At5g55090), MLO12 (At2g39200), MYB15 (At3g23250), NUDX25 (At1g30110), OPT1 (At5g55930), PDF1.2 (At5g44420), PER62 (At5g39580), PHI-1(At1g35140), PR-3 (At3g12500), PR-4 (At3g04720), PROPEP2 (At5g64890), PROPEP3 (At5g64905), PSK1 (At1g13590), PUB24 (At3g11840), RALF1 (At1g02900), SMU1 (At1g73720), TRE1 (At4g24040), WAKL10 (At1g79680), WAKL2 (At1g16130), WAKL6 (At1g16110), WR3/NRT3.1 (At5g50200), WRKY22 (At4g01250), WRKY29 (At4g23550) WRKY30 (At5g24110), WRKY49 (At5g43290), and WRKY53 (At4g23810). Microarray data from this article can be found in NCBI’s GEO (Edgar et al., 2002) under GEO series accession numbers GSE40139 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE40139) and GSE40138 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE40138).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Induction of DAMP Markers, PROPEP2 and PROPEP3, in Response to Submergence.
Supplemental Figure 2. Expression of Innate Immunity Marker Genes, PR Genes, Submergence Early-Induced WRKYs, and Submergence Late-Induced WRKYs upon Abiotic and Biotic Stresses.
Supplemental Figure 3. Plant Immunity Is Triggered by Short Submergence.
Supplemental Figure 4. Isolation of WRKY22 T-DNA Insertion Mutants.
Supplemental Figure 5. WRKY22 Mediates Basal Immunity and Submergence-Triggered Immunity to Pseudomonas syringae.
Supplemental Figure 6. Transgenic c-myc–Tagged WRKY22 Are Induced by Submergence in Transcript and Protein Levels.
Supplemental Figure 7. Effects of WRKY22 Knockout on Transcript Levels of WRKY22 Target Genes.
Supplemental Figure 8. Regulation of LecRKs and Stomatal Immunity-Associated Genes under Submergence.
Supplemental Table 1. List of Innate Immunity and Stress-Regulated Marker Genes.
Supplemental Table 2. Differential Expression of Selected Submergence-Responsive Genes in WRKY22 Knockout Plants.
Supplemental Table 3. Potential Direct Targets of WRKY22 by ChIP-Chip and ChIP-Q-PCR Experiments.
Supplemental Table 4. Primer Lists.
Supplemental Data Set 1. Submergence-Responsive Genes That Are Differentially Expressed in WRKY22 Knockout Plants.
Acknowledgments
This work was supported by the National Science Council (Grant NSC 98-2321-B-001-027-MY3) and Academia Sinica, Taiwan. We thank Laurent Zimmerli of the Institute of Plant Biology, National Taiwan University, for providing the Pst DC3000 strain. We also thank Chih-Cheng Chien of the Agricultural Biotechnology Research Center, Academia Sinica, for comments and discussion.
AUTHOR CONTRIBUTIONS
F.-C.H.designed the research, performed research, analyzed data, and wrote the article. M.-Y.C. performed qRT-PCR. S.-J.C. performed the expression microarray and ChIP-chip microarray hybridization. Y.-R.L. contributed to the construction of transgenic plants. H.-P.P. performed ChIP. M.-C.S. designed the research and wrote the article.
Glossary
- TF
transcription factor
- MAMP
microbe-associated molecular pattern
- PRR
pattern recognition receptor
- SAR
systemic acquired resistance
- qRT-PCR
quantitative real-time RT-PCR
- DAMP
damage-associated molecular pattern
- ChIP-chip
chromatin immunoprecipitation followed by microarray hybridization
- RLK
receptor-like kinase
- MS
Murashige and Skoog
- NCBI
National Center for Biotechnology Information
- GEO
Gene Expression Omnibus
References
- Asai T., Tena G., Plotnikova J., Willmann M.R., Chiu W.-L., Gomez-Gomez L., Boller T., Ausubel F.M., Sheen J. (2002). MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 415: 977–983 [DOI] [PubMed] [Google Scholar]
- Bailey-Serres J., Voesenek L.A.C.J. (2008). Flooding stress: Acclimations and genetic diversity. Annu. Rev. Plant Biol. 59: 313–339 [DOI] [PubMed] [Google Scholar]
- Bieniawska Z., Paul Barratt D.H., Garlick A.P., Thole V., Kruger N.J., Martin C., Zrenner R., Smith A.M. (2007). Analysis of the sucrose synthase gene family in Arabidopsis. Plant J. 49: 810–828 [DOI] [PubMed] [Google Scholar]
- Boller T., Felix G. (2009). A renaissance of elicitors: Perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu. Rev. Plant Biol. 60: 379–406 [DOI] [PubMed] [Google Scholar]
- Boudsocq M., Willmann M.R., McCormack M., Lee H., Shan L., He P., Bush J., Cheng S.-H., Sheen J. (2010). Differential innate immune signalling via Ca(2+) sensor protein kinases. Nature 464: 418–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brutus A., Sicilia F., Macone A., Cervone F., De Lorenzo G. (2010). A domain swap approach reveals a role of the plant wall-associated kinase 1 (WAK1) as a receptor of oligogalacturonides. Proc. Natl. Acad. Sci. USA 107: 9452–9457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y., Song F., Goodman R.M., Zheng Z. (2006). Molecular characterization of four rice genes encoding ethylene-responsive transcriptional factors and their expressions in response to biotic and abiotic stress. J. Plant Physiol. 163: 1167–1178 [DOI] [PubMed] [Google Scholar]
- Chini A., Grant J.J., Seki M., Shinozaki K., Loake G.J. (2004). Drought tolerance established by enhanced expression of the CC-NBS-LRR gene, ADR1, requires salicylic acid, EDS1 and ABI1. Plant J. 38: 810–822 [DOI] [PubMed] [Google Scholar]
- Christianson J.A., Wilson I.W., Llewellyn D.J., Dennis E.S. (2009). The low-oxygen-induced NAC domain transcription factor ANAC102 affects viability of Arabidopsis seeds following low-oxygen treatment. Plant Physiol. 149: 1724–1738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciolkowski I., Wanke D., Birkenbihl R.P., Somssich I.E. (2008). Studies on DNA-binding selectivity of WRKY transcription factors lend structural clues into WRKY-domain function. Plant Mol. Biol. 68: 81–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consonni C., Humphry M.E., Hartmann H.A., Livaja M., Durner J., Westphal L., Vogel J., Lipka V., Kemmerling B., Schulze-Lefert P., Somerville S.C., Panstruga R. (2006). Conserved requirement for a plant host cell protein in powdery mildew pathogenesis. Nat. Genet. 38: 716–720 [DOI] [PubMed] [Google Scholar]
- Davison E.M., Tay F.C.S. (1987). The effect of waterlogging on infection of Eucalyptus marginata seedlings by Phytophthora cinnamomi. New Phytol. 105: 585–594 [Google Scholar]
- de A Gerhardt L.B., Sachetto-Martins G., Contarini M.G., Sandroni M., de P Ferreira R., de Lima V.M., Cordeiro M.C., de Oliveira D.E., Margis-Pinheiro M. (1997). Arabidopsis thaliana class IV chitinase is early induced during the interaction with Xanthomonas campestris. FEBS Lett. 419: 69–75 [DOI] [PubMed] [Google Scholar]
- De Lorenzo G., Brutus A., Savatin D.V., Sicilia F., Cervone F. (2011). Engineering plant resistance by constructing chimeric receptors that recognize damage-associated molecular patterns (DAMPs). FEBS Lett. 585: 1521–1528 [DOI] [PubMed] [Google Scholar]
- Desclos-Theveniau M., Arnaud D., Huang T.-Y., Lin G.J.-C., Chen W.-Y., Lin Y.-C., Zimmerli L. (2012). The Arabidopsis lectin receptor kinase LecRK-V.5 represses stomatal immunity induced by Pseudomonas syringae pv. tomato DC3000Arabidopsis. PLoS Pathog. 8: e1002513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding C.-K., Wang C.Y., Gross K.C., Smith D.L. (2002). Jasmonate and salicylate induce the expression of pathogenesis-related-protein genes and increase resistance to chilling injury in tomato fruit. Planta 214: 895–901 [DOI] [PubMed] [Google Scholar]
- Dolferus R., Wolansky M., Carroll R., Miyashita Y., Ismond K., Good A. (2008). Functional analysis of lactate dehydrogenase during hypoxic stress in Arabidopsis. Funct. Plant Biol. 35: 131–140 [DOI] [PubMed] [Google Scholar]
- Dong H., Zhen Z., Peng J., Chang L., Gong Q., Wang N.N. (2011). Loss of ACS7 confers abiotic stress tolerance by modulating ABA sensitivity and accumulation in Arabidopsis. J. Exp. Bot. 62: 4875–4887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R., Domrachev M., Lash A.E. (2002). Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30: 207–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis M.H., Dennis E.S., Peacock W.J. (1999). Arabidopsis roots and shoots have different mechanisms for hypoxic stress tolerance. Plant Physiol. 119: 57–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulgem T. (2005). Regulation of the Arabidopsis defense transcriptome. Trends Plant Sci. 10: 71–78 [DOI] [PubMed] [Google Scholar]
- Eulgem T., Rushton P.J., Robatzek S., Somssich I.E. (2000). The WRKY superfamily of plant transcription factors. Trends Plant Sci. 5: 199–206 [DOI] [PubMed] [Google Scholar]
- Eulgem T., Somssich I.E. (2007). Networks of WRKY transcription factors in defense signaling. Curr. Opin. Plant Biol. 10: 366–371 [DOI] [PubMed] [Google Scholar]
- Ferrari, S., Savatin, D.V., Sicilia, F., Gramegna, G., Cervone, F., and De Lorenzo, G.(March 13, 2013). Oligogalacturonides Plant damage-associated molecular patterns and regulators of growth and development. Front. Plant Sci. (online) / 10.3389/fpls.2013.00049. [DOI] [PMC free article] [PubMed]
- Friedel, S., Usadel, B., von Wirén, N., and Sreenivasulu, N.(December 31, 2012). Reverse engineering A key component of systems biology to unravel global abiotic stress cross-talk. Front. Plant Sci. (online) / 10.3389/fpls.2012.00294. [DOI] [PMC free article] [PubMed]
- Fukao T., Bailey-Serres J. (2008). Submergence tolerance conferred by Sub1A is mediated by SLR1 and SLRL1 restriction of gibberellin responses in rice. Proc. Natl. Acad. Sci. USA 105: 16814–16819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs D.J., Lee S.C., Isa N.M., Gramuglia S., Fukao T., Bassel G.W., Correia C.S., Corbineau F., Theodoulou F.L., Bailey-Serres J., Holdsworth M.J. (2011). Homeostatic response to hypoxia is regulated by the N-end rule pathway in plants. Nature 479: 415–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Gómez L., Boller T. (2000). FLS2: An LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol. Cell 5: 1003–1011 [DOI] [PubMed] [Google Scholar]
- Guo Z.-J., Chen X.-J., Wu X.-L., Ling J.-Q., Xu P. (2004). Overexpression of the AP2/EREBP transcription factor OPBP1 enhances disease resistance and salt tolerance in tobacco. Plant Mol. Biol. 55: 607–618 [DOI] [PubMed] [Google Scholar]
- Hammond J.P., Bennett M.J., Bowen H.C., Broadley M.R., Eastwood D.C., May S.T., Rahn C., Swarup R., Woolaway K.E., White P.J. (2003). Changes in gene expression in Arabidopsis shoots during phosphate starvation and the potential for developing smart plants. Plant Physiol. 132: 578–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattori Y., et al. (2009). The ethylene response factors SNORKEL1 and SNORKEL2 allow rice to adapt to deep water. Nature 460: 1026–1030 [DOI] [PubMed] [Google Scholar]
- Hinz M., Wilson I.W., Yang J., Buerstenbinder K., Llewellyn D., Dennis E.S., Sauter M., Dolferus R. (2010). Arabidopsis RAP2.2: An ethylene response transcription factor that is important for hypoxia survival. Plant Physiol. 153: 757–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeren F.U., Dolferus R., Wu Y., Peacock W.J., Dennis E.S. (1998). Evidence for a role for AtMYB2 in the induction of the Arabidopsis alcohol dehydrogenase gene (ADH1) by low oxygen. Genetics 149: 479–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hok S., Danchin E.G.J., Allasia V., Panabières F., Attard A., Keller H. (2011). An Arabidopsis (malectin-like) leucine-rich repeat receptor-like kinase contributes to downy mildew disease. Plant Cell Environ. 34: 1944–1957 [DOI] [PubMed] [Google Scholar]
- Hruz T., Laule O., Szabo G., Wessendorp F., Bleuler S., Oertle L., Widmayer P., Gruissem W., Zimmermann P. (2008). Genevestigator v3: A reference expression database for the meta-analysis of transcriptomes. Adv. Bioinforma. 2008: 420747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu F.-C., Chou M.-Y., Peng H.-P., Chou S.-J., Shih M.-C. (2011). Insights into hypoxic systemic responses based on analyses of transcriptional regulation in Arabidopsis. PLoS ONE 6: e28888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber L., Gillespie T.J. (1992). Modeling leaf wetness in relation to plant-disease epidemiology. Annu. Rev. Phytopathol. 30: 553–577 [Google Scholar]
- Kaufmann K., Muiño J.M., Østerås M., Farinelli L., Krajewski P., Angenent G.C. (2010). Chromatin immunoprecipitation (ChIP) of plant transcription factors followed by sequencing (ChIP-SEQ) or hybridization to whole genome arrays (ChIP-CHIP). Nat. Protoc. 5: 457–472 [DOI] [PubMed] [Google Scholar]
- King E.O., Ward M.K., Raney D.E. (1954). Two simple media for the demonstration of pyocyanin and fluorescin. J. Lab. Clin. Med. 44: 301–307 [PubMed] [Google Scholar]
- Klok E.J., Wilson I.W., Wilson D., Chapman S.C., Ewing R.M., Somerville S.C., Peacock W.J., Dolferus R., Dennis E.S. (2002). Expression profile analysis of the low-oxygen response in Arabidopsis root cultures. Plant Cell 14: 2481–2494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kottapalli K.R., Sarla N., Kikuchi S. (2006). In silico insight into two rice chromosomal regions associated with submergence tolerance and resistance to bacterial leaf blight and gall midge. Biotechnol. Adv. 24: 561–589 [DOI] [PubMed] [Google Scholar]
- Kreuzwieser J., Hauberg J., Howell K.A., Carroll A., Rennenberg H., Millar A.H., Whelan J. (2009). Differential response of gray poplar leaves and roots underpins stress adaptation during hypoxia. Plant Physiol. 149: 461–473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kürsteiner O., Dupuis I., Kuhlemeier C. (2003). The pyruvate decarboxylase1 gene of Arabidopsis is required during anoxia but not other environmental stresses. Plant Physiol. 132: 968–978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H., Chah O.-K., Sheen J. (2011). Stem-cell-triggered immunity through CLV3p-FLS2 signalling. Nature 473: 376–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- León J., Rojo E., Titarenko E., Sánchez-Serrano J.J. (1998). Jasmonic acid-dependent and -independent wound signal transduction pathways are differentially regulated by Ca2+/calmodulin in Arabidopsis thaliana. Mol. Gen. Genet. 258: 412–419 [DOI] [PubMed] [Google Scholar]
- Libault M., Wan J., Czechowski T., Udvardi M., Stacey G. (2007). Identification of 118 Arabidopsis transcription factor and 30 ubiquitin-ligase genes responding to chitin, a plant-defense elicitor. Mol. Plant Microbe Interact. 20: 900–911 [DOI] [PubMed] [Google Scholar]
- Licausi F., van Dongen J.T., Giuntoli B., Novi G., Santaniello A., Geigenberger P., Perata P. (2010a). HRE1 and HRE2, two hypoxia-inducible ethylene response factors, affect anaerobic responses in Arabidopsis thaliana. Plant J. 62: 302–315 [DOI] [PubMed] [Google Scholar]
- Licausi F., Weits D.A., Pant B.D., Scheible W.R., Geigenberger P., Van Dongen J.T. (2010b). Hypoxia responsive gene expression is mediated by various subsets of transcription factors and miRNAs that are determined by the actual oxygen availability. New Phytol. 190: 442–456. [DOI] [PubMed] [Google Scholar]
- Liu F., Vantoai T., Moy L.P., Bock G., Linford L.D., Quackenbush J. (2005). Global transcription profiling reveals comprehensive insights into hypoxic response in Arabidopsis. Plant Physiol. 137: 1115–1129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Zhang H., Yang Y., Li G., Yang Y., Wang X.e., Basnayake B.M., Li D., Song F. (2008). Functional analysis reveals pleiotropic effects of rice RING-H2 finger protein gene OsBIRF1 on regulation of growth and defense responses against abiotic and biotic stresses. Plant Mol. Biol. 68: 17–30 [DOI] [PubMed] [Google Scholar]
- Logemann, E., Birkenbihl, R.P., Rawat, V., Schneeberger, K., Schmelzer, E., and Somssich, I.E. (2013). Functional dissection of the PROPEP2 and PROPEP3 promoters reveals the importance of WRKY factors in mediating microbe-associated molecular pattern-induced expression. New Phytol. 198: 1165–1177. [DOI] [PubMed]
- Ma Y., Walker R.K., Zhao Y., Berkowitz G.A. (2012). Linking ligand perception by PEPR pattern recognition receptors to cytosolic Ca2+ elevation and downstream immune signaling in plants. Proc. Natl. Acad. Sci. USA 109: 19852–19857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macho A.P., Boutrot F., Rathjen J.P., Zipfel C. (2012). Aspartate oxidase plays an important role in Arabidopsis stomatal immunity. Plant Physiol. 159: 1845–1856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melotto M., Underwood W., Koczan J., Nomura K., He S.Y. (2006). Plant stomata function in innate immunity against bacterial invasion. Cell 126: 969–980 [DOI] [PubMed] [Google Scholar]
- Miao Y., Laun T.M., Smykowski A., Zentgraf U. (2007). Arabidopsis MEKK1 can take a short cut: It can directly interact with senescence-related WRKY53 transcription factor on the protein level and can bind to its promoter. Plant Mol. Biol. 65: 63–76 [DOI] [PubMed] [Google Scholar]
- Navarro L., Zipfel C., Rowland O., Keller I., Robatzek S., Boller T., Jones J.D.G. (2004). The transcriptional innate immune response to flg22. Interplay and overlap with Avr gene-dependent defense responses and bacterial pathogenesis. Plant Physiol. 135: 1113–1128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey S.P., Somssich I.E. (2009). The role of WRKY transcription factors in plant immunity. Plant Physiol. 150: 1648–1655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panstruga R. (2005). Serpentine plant MLO proteins as entry portals for powdery mildew fungi. Biochem. Soc. Trans. 33: 389–392 [DOI] [PubMed] [Google Scholar]
- Peng H.P., Lin T.Y., Wang N.N., Shih M.C. (2005). Differential expression of genes encoding 1-aminocyclopropane-1-carboxylate synthase in Arabidopsis during hypoxia. Plant Mol. Biol. 58: 15–25 [DOI] [PubMed] [Google Scholar]
- Qiu J.-L., et al. (2008). Arabidopsis MAP kinase 4 regulates gene expression through transcription factor release in the nucleus. EMBO J. 27: 2214–2221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranf S., Eschen-Lippold L., Pecher P., Lee J., Scheel D. (2011). Interplay between calcium signalling and early signalling elements during defence responses to microbe- or damage-associated molecular patterns. Plant J. 68: 100–113 [DOI] [PubMed] [Google Scholar]
- Rieu I., Cristescu S.M., Harren F.J.M., Huibers W., Voesenek L.A.C.J., Mariani C., Vriezen W.H. (2005). RP-ACS1, a flooding-induced 1-aminocyclopropane-1-carboxylate synthase gene of Rumex palustris, is involved in rhythmic ethylene production. J. Exp. Bot. 56: 841–849 [DOI] [PubMed] [Google Scholar]
- Rushton P.J., Somssich I.E., Ringler P., Shen Q.J. (2010). WRKY transcription factors. Trends Plant Sci. 15: 247–258 [DOI] [PubMed] [Google Scholar]
- Seo P.J., Kim M.J., Park J.-Y., Kim S.-Y., Jeon J., Lee Y.-H., Kim J., Park C.-M. (2010). Cold activation of a plasma membrane-tethered NAC transcription factor induces a pathogen resistance response in Arabidopsis. Plant J. 61: 661–671 [DOI] [PubMed] [Google Scholar]
- Singh P., Kuo Y.-C., Mishra S., Tsai C.-H., Chien C.-C., Chen C.-W., Desclos-Theveniau M., Chu P.-W., Schulze B., Chinchilla D., Boller T., Zimmerli L. (2012). The lectin receptor kinase-VI.2 is required for priming and positively regulates Arabidopsis pattern-triggered immunity. Plant Cell 24: 1256–1270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skibbe M., Qu N., Galis I., Baldwin I.T. (2008). Induced plant defenses in the natural environment: Nicotiana attenuata WRKY3 and WRKY6 coordinate responses to herbivory. Plant Cell 20: 1984–2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohn K.H., Lee S.C., Jung H.W., Hong J.K., Hwang B.K. (2006). Expression and functional roles of the pepper pathogen-induced transcription factor RAV1 in bacterial disease resistance, and drought and salt stress tolerance. Plant Mol. Biol. 61: 897–915 [DOI] [PubMed] [Google Scholar]
- Szklarczyk D., Franceschini A., Kuhn M., Simonovic M., Roth A., Minguez P., Doerks T., Stark M., Muller J., Bork P., Jensen L.J., von Mering C. (2011). The STRING database in 2011: Functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 39 (Database issue): D561–D568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka N., Matsuoka M., Kitano H., Asano T., Kaku H., Komatsu S. (2006). gid1, a gibberellin-insensitive dwarf mutant, shows altered regulation of probenazole-inducible protein (PBZ1) in response to cold stress and pathogen attack. Plant Cell Environ. 29: 619–631 [DOI] [PubMed] [Google Scholar]
- Thomma B.P.H.J., Penninckx I.A.M.A., Broekaert W.F., Cammue B.P. (2001). The complexity of disease signaling in Arabidopsis. Curr. Opin. Immunol. 13: 63–68 [DOI] [PubMed] [Google Scholar]
- Tör M., Lotze M.T., Holton N. (2009). Receptor-mediated signalling in plants: Molecular patterns and programmes. J. Exp. Bot. 60: 3645–3654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Der Straeten D., Zhou Z., Prinsen E., Van Onckelen H.A., Van Montagu M.C. (2001). A comparative molecular-physiological study of submergence response in lowland and deepwater rice. Plant Physiol. 125: 955–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dongen J.T., Fröhlich A., Ramírez-Aguilar S.J., Schauer N., Fernie A.R., Erban A., Kopka J., Clark J., Langer A., Geigenberger P. (2009). Transcript and metabolite profiling of the adaptive response to mild decreases in oxygen concentration in the roots of Arabidopsis plants. Ann. Bot. (Lond.) 103: 269–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Houtte H., Vandesteene L., López-Galvis L., Lemmens L., Kissel E., Carpentier S., Feil R., Avonce N., Beeckman T., Lunn J.E., Van Dijck P. (2013). Overexpression of the trehalase gene AtTRE1 leads to increased drought stress tolerance in Arabidopsis and is involved in abscisic acid-induced stomatal closure. Plant Physiol. 161: 1158–1171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Loon L.C., Rep M., Pieterse C.M. (2006). Significance of inducible defense-related proteins in infected plants. Annu. Rev. Phytopathol. 44: 135–162 [DOI] [PubMed] [Google Scholar]
- Van Loon L.C., Van Strien E.A. (1999). The families of pathogenesis-related proteins, their activities, and comparative analysis of PR-1 type proteins. Physiol. Mol. Plant Pathol. 55: 85–97 [Google Scholar]
- Vashisht D., Hesselink A., Pierik R., Ammerlaan J.M.H., Bailey-Serres J., Visser E.J.W., Pedersen O., van Zanten M., Vreugdenhil D., Jamar D.C.L., Voesenek L.A., Sasidharan R. (2011). Natural variation of submergence tolerance among Arabidopsis thaliana accessions. New Phytol. 190: 299–310 [DOI] [PubMed] [Google Scholar]
- Walker G. (1991). Chemical, physical and biological control of carrot seedling diseases. Plant Soil 136: 31–39 [Google Scholar]
- Wang W., Barnaby J.Y., Tada Y., Li H., Tör M., Caldelari D., Lee D.U., Fu X.-D., Dong X. (2011). Timing of plant immune responses by a central circadian regulator. Nature 470: 110–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitham S.A., Quan S., Chang H.-S., Cooper B., Estes B., Zhu T., Wang X., Hou Y.-M. (2003). Diverse RNA viruses elicit the expression of common sets of genes in susceptible Arabidopsis thaliana plants. Plant J. 33: 271–283 [DOI] [PubMed] [Google Scholar]
- Xiong L., Yang Y. (2003). Disease resistance and abiotic stress tolerance in rice are inversely modulated by an abscisic acid-inducible mitogen-activated protein kinase. Plant Cell 15: 745–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu K., Xu X., Fukao T., Canlas P., Maghirang-Rodriguez R., Heuer S., Ismail A.M., Bailey-Serres J., Ronald P.C., Mackill D.J. (2006). Sub1A is an ethylene-response-factor-like gene that confers submergence tolerance to rice. Nature 442: 705–708 [DOI] [PubMed] [Google Scholar]
- Yanar Y., Lipps P.E., Deep I.W. (1997). Effect of soil saturation duration and soil water content on root rot of maize caused by Pythium arrhenomanes. Plant Disease 81: 475–480 [Google Scholar]
- Yang C.-Y., Hsu F.-C., Li J.-P., Wang N.-N., Shih M.-C. (2011). The AP2/ERF transcription factor AtERF73/HRE1 modulates ethylene responses during hypoxia in Arabidopsis. Plant Physiol. 156: 202–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng W., He S.Y. (2010). A prominent role of the flagellin receptor FLAGELLIN-SENSING2 in mediating stomatal response to Pseudomonas syringae pv tomato DC3000 in Arabidopsis. Plant Physiol. 153: 1188–1198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G., Chen M., Li L., Xu Z., Chen X., Guo J., Ma Y. (2009). Overexpression of the soybean GmERF3 gene, an AP2/ERF type transcription factor for increased tolerances to salt, drought, and diseases in transgenic tobacco. J. Exp. Bot. 60: 3781–3796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Li W., Chen J., Yang Y., Zhang Z., Zhang H., Wang X.-C., Huang R. (2007). Transcriptional activator TSRF1 reversely regulates pathogen resistance and osmotic stress tolerance in tobacco. Plant Mol. Biol. 63: 63–71 [DOI] [PubMed] [Google Scholar]
- Zhou X., Jiang Y., Yu D. (2011). WRKY22 transcription factor mediates dark-induced leaf senescence in Arabidopsis. Mol. Cells 31: 303–313 [DOI] [PMC free article] [PubMed] [Google Scholar]