Abstract
We have cloned two gibberellin (GA) 3β-hydroxylase genes, OsGA3ox1 and OsGA3ox2, from rice by screening a genomic library with a DNA fragment obtained by PCR using degenerate primers. We have used full-scan GC-MS and Kovats retention indices to show function for the two encoded recombinant fusion proteins. Both proteins show 3β-hydroxylase activity for the steps GA20 to GA1, GA5 to GA3, GA44 to GA38, and GA9 to GA4. In addition, indirect evidence suggests that the OsGA3ox1 protein also has 2,3-desaturase activity, which catalyzes the steps GA9 to 2,3-dehydro-GA9 and GA20 to GA5 (2,3-dehydro GA20), and 2β-hydroxylase activity, which catalyzes the steps GA1 to GA8 and GA4 to GA34. Molecular and linkage analysis maps the OsGA3ox1 gene to the distal end of the short arm of chromosome 5; the OsGA3ox2 gene maps to the distal end of the short arm of chromosome 1 that corresponds to the D18 locus. The association of the OsGA3ox2 gene with the d18 locus is confirmed by sequence and complementation analysis of three d18 alleles. Complementation of the d18-AD allele with the OxGA3ox2 gene results in transgenic plants with a normal phenotype. Although both genes show transient expression, the highest level for OsGA3ox1 is from unopened flower. The highest level for OsGA3ox2 is from elongating leaves.
Gibberellins (GAs) are a large family of tetracyclic, diterpenoid plant hormones that promote various growth and developmental processes in higher plants. A number of GA-responsive dwarf mutants have been isolated from various plant species, including maize (Zea mays), pea (Pisum sativum), tomato (Lycopersicon esculentum), thale cress (Arabidopsis thaliana), and rice (Oryza sativa) (1–7). For example, in maize the d1 mutant is rescued by the exogenous application of GA1, but not by GA20, the immediate precursor of GA1. The mutant accumulates GA20 and metabolizes GA20 to GA1 at a low rate (8, 9). Presumably the D1 gene encodes a 3β-hydroxylase that controls this step. In pea the LE gene has been cloned and shown to encode a GA 3β-hydroxylase (10, 11). The le mutant is rescued by GA1, but not by GA20, and shows low activity in the metabolism of GA20 to GA1 (12, 13).
For rice the d18 mutant has been characterized as a GA-responsive dwarf (5), and three alleles have been identified (2, 5). They are the two strong alleles, d18-Id18h (Hosetsu-waisei dwarf) and d18-AD (Akibare-waisei dwarf), and the weak allele, d18-dy (Waito-C). The strong alleles promote severe dwarf phenotypes; the weak allele promotes a semidwarf phenotype. Physiological and biochemical studies have been carried out with the d18-dy mutant only. The dwarf phenotype of the d18-dy mutant is rescued by the application of GA1, not by GA20 (2). Seedlings of this mutant are deficient in GA1 and accumulate its immediate precursor, GA20 (14). The metabolism of GA20 to GA1 is lower in d18-dy plants than in normal ones (15). It has been proposed that the D18 gene encodes a GA 3β-hydroxylase (15).
Studies have shown that GAs also regulate the development of reproductive organs. For example, a GA-deficient mutant in Arabidopsis, ga1–3, exhibits a male-sterile phenotype (4), and in tomato stamenless-2 and gib-1 arrest anther development at an early stage (16, 17). In tall rice, it has been reported that GA4 accumulates in anthers (18) and that a cell-free extract from anthers metabolizes GA12 to GA34 and GA53 to GA8 (19). The level of GA4 and the activity of GA 3β-hydroxylase from anthers were the same for d18-dy mutants and normals, suggesting that there are at least two GA 3β-hydroxylase genes in rice (14, 19).
Although GA 3β-hydroxylase genes have been recently cloned from several dicot species (10, 11, 20–24), there is no example from monocots. In this study, we report the cloning from rice of two genes, OsGA3ox1 and OsGA3ox2. OsGA3ox1 is a new gene found on chromosome 5. OsGA3ox2 is shown to correspond to the D18 locus, which is known to be located on chromosome 1.
Materials and Methods
Plant Materials.
Seeds of Japonica-type rice (Oryza sativa L.), tall cultivars Nipponbare and Akibare, and dwarf cultivars Akibare-waisei (d18-AD), Hosetsu-waisei (d18-ID18h), and Waito-C (d18-dy) were surface-sterilized in 1% NaClO for 1 h and then rinsed in sterile distilled water. The seeds were planted in soil and grown in a greenhouse or paddy field. For RNA gel blot analysis, the tissues and organs were collected from tall rice (Akibare). Samples were collected from the following material: young shoots 2 weeks after planting, shoot apices, leaf blades, and elongating leaves (2–3 cm length, no boundary between sheath and blade) 1 month after planting, young panicles (1–2 cm length) approximately 1 week after panicle initiation, stems (nodes plus internodes) at internode elongation stage, and rachises and unopened flowers just before heading stage. Flowers of tall rice (Nipponbare) also were collected at the following anther developmental stages: differentiation stage of pollen mother cells, early stage of pollen differentiation, middle stage of pollen differentiation, late stage of pollen differentiation, and pollen maturing stage. Palea and lemma were removed from the flowers to give anthers and ovaries that were used for RNA gel blot analysis. Flowers at the late stage of pollen differentiation also were used for in situ hybridization. The seeds of rice used for mapping of recombinant inbred lines between the cultivars Asominori (Japonica type) and IR-24 (Indica type) were kindly provided by A. Yoshimura, Kyushu University (Fukuoka, Japan).
Chemicals.
[17,17-2H2]GA19, [17,17-2H2]GA20, [17,17-2H2]GA29, [17,17-2H2]GA44, and [17,17-2H2]GA53 were purchased from L. Mander, Australian National University (Canberra). [15,17,17-2H3]GA9 was synthesized from GA9-norketone and (methyl-d3)triphenylphosphonium-Br by Wittig reaction. All GAs used in this study were analyzed by full-scan GC-MS to show the absence of impurities. Uniconazole, an inhibitor for GA biosynthesis, was obtained from Sumitomo Chemical Company (Tokyo).
GC-MS.
GC-MS was performed with an Auto Mass mass spectrometer (JEOL) connected to a Hewlett–Packard 5890 series II gas chromatograph. The analytical conditions used are described by Kobayashi et al. (25).
Genomic DNA Preparation.
The genomic DNA used for PCR and Southern hybridization was prepared from the seedlings of tall rice (Nipponbare) using the methods described by Murray and Thompson (26). Genomic DNA also was prepared from d18-Id18h, d18-dy, and d18-AD plants by the same method used for the characterization of the d18 alleles.
Isolation of cDNA Clones Encoding Putative GA 3β-Hydroxylases.
The PCR was performed by using genomic DNA from Nipponbare as a template and degenerate primers (sense, 5′-GTNGTNAAYGTNGGNGAYRT-3′, and antisense, 5′-TRRTAYTCRTTCCANGTNAC-3′, indicated by arrows in Fig. 1). The reaction mixture (30 μl) contained 100 ng genomic DNA (Nipponbare), 1× PCR buffer (Applied Biosystems), 1.5 mM MgCl2, 200 μM dNTP, 2 μM each primer, 1.5 μl DMSO, and 1 unit of Taq polymerase (Amplitaq, Applied Biosystems). The reactions were heated to 99°C for 10 min, and then 35 cycles of amplification were performed (96°C for 1 min, 50°C for 2 min, and 72°C for 1 min) followed by a final 5-min incubation at 72°C. The 210-bp DNA fragment obtained was sequenced and used as a probe for screening a rice genomic library (constructed from Nipponbare and IR-36), resulting in the isolation of clones OsGA3ox1 and OsGA3ox2, that contain the entire lengths of putative GA 3β-hydroxylase genes. Full-length cDNA clones for OsGA3ox1 and OsGA3ox2 were obtained by reverse transcription–PCR from the total RNA prepared from rice (Nipponbare) shoot apices and unopened flowers using methods described by Sambrook et al. (27). Reaction mixture (30 μl) for reverse transcription contained 1 μg total RNA, 5 mM MgCl2, 1× PCR buffer, 1 mM dNTP, 0.125 μM oligo(dT) primer, and AMV Reverse Transcriptase XL (RNA-PCR kit, AMV version 2.1, Takara, Kyoto, Japan). The reverse transcription was carried out at 45°C for 1 h, followed by PCR in which 2 μl of the reaction mixture from reverse transcription was used as a template. Other conditions in the PCR were as described above for degenerate primers. The following oligonucleotide primers were used in the PCR: for OsGA3ox1, sense, 5′-CCGGATCCATGACATCGTCGTCGACCTCGCCG-3′ and antisense, 5′-GGAAGCTTCTAACTCTCCTTGTCCTCTTC-3′; for OsGA3ox2, sense, 5′-CCGGATCCATGCCGACGCCGTCGCACTTGAA-3′ and antisense 5′-GGAAGCTTCTATAGCTTATGCGTGGACGT-3′.
Figure 1.
Alignment of deduced amino acid sequences of GA 3β-hydroxylases and other GA dioxygenases from the monocot, rice, and several dicots. Alterations in the sequences from d18-Id18h and d18-dy are indicated. Light/dark shading indicates similar/identical residues. The three residues that are invariant throughout the plant 2-oxoglutarate-dependent dioxygenases (whose side chains act as iron ligands) are marked with ○. ▾ indicates the position of the common intron in the rice 3β-hydroxylase genes; ▿ indicates the second intron in the OsGA3ox1 gene. Dashes indicate gaps introduced to optimize alignment. Arrows indicate the regions used in the design of degenerate primers. GenBank accession numbers are as follows: OsGA3ox1, AB054084; OsGA3ox2, AB056519; NtGA3ox, AB032198; AtGA3ox1, L37126; PsGA3ox, AF001219; AtGA20ox1, X83379; AtGA2ox1, AJ132435. Nucleotide sequences of OsGA3ox2 for d18-Id18h (accession number AB056517) and d18-dy (accession number AB056518) mutants are in GenBank. Recently, the nucleotide sequence of a P1 artificial chromosome clone that harbors OsGA3ox2 gene is registered in GenBank (accession number AP002523).
Sequence Analysis.
Nucleotide sequences were determined by the dideoxynucleotide chain-termination method using an automated sequencing system (model ABI373A, Applied Biosystems). Sequences were analyzed with genetyx computer software (Software Development, Tokyo).
Heterologous Expression and Enzyme Assays.
The entire coding region of the OsGA3ox1 and OsGA3ox2 cDNA were inserted into a suitable restriction site of pMAL-c2 and transformed into Escherichia coli strain JM109. A fresh overnight culture (1 ml) of E. coli was added to 100 ml of 2× YT medium [NaCl (1 g/liter), yeast extract (1 g/liter), and tryptone (1.6 g/liter)] containing ampicillin (0.1 mg/liter). The culture was incubated at 30°C for 4 h. Isopropyl β-d-thiogalactoside (1 mg/liter) was added, and the culture was further incubated at 17°C for 18 h. The cells were harvested, homogenized by sonication, and centrifuged to give a crude extract. The presence of the OsGA3ox1 recombinant protein in the crude extract was confirmed by SDS/PAGE, and this crude extract was used in the enzyme assay. Because much less amount of the OsGA3ox2 recombinant protein was observed in the crude extract, it was concentrated by an affinity column packed with 100 μl amylose resin according to the manufacturer's instruction (New England Biolabs). The eluate from the column was used in the enzyme assay; 2-ketoglutarate (final concentration, 5 mM), ascorbate (5 mM), and FeSO4 (0.5 mM) were added to the eluate (200 μl) to give a reaction mixture, and it was incubated with substrate GA at 30°C for 1 h. The reaction mixture was adjusted to pH 2 with HCl and extracted with ethyl acetate. The extract was analyzed by full-scan GC-MS and Kovats retention indices to identify product GAs (25).
DNA and RNA Gel Blot Analyses.
Gene-specific probes were prepared by α-[32P]dCTP labeling of a KpnI–PvuII fragment (369 bp) of OsGA3ox1 cDNA and a BssHII–PvuII fragment (429 bp) of OsGA3ox2 cDNA according to the manufacturer's instruction (Ready-to-Go DNA Labeling Beads, Amersham Pharmacia). Each specific probe was used in genomic Southern blot hybridization to confirm the absence of cross-hybridization. Rice genomic DNA (from Nipponbare; 1 μg per lane) was digested with 10 units of EcoRV (for OsGA3ox1 probe) or ApaI (for OsGA3ox2 probe) at 37°C overnight, separated by 0.7% agarose gel electrophoresis, and transferred to Hybond N+ nylon membrane (Amersham Pharmacia) (27). Hybridization was performed at 65°C in 0.25 M Na2HPO4, 1 mM EDTA, and 7% SDS. Filters were washed twice in 2× SSC, 0.1% SDS at 65°C for 15 min, and once in 0.2× SSC, 0.1% SDS at 65°C for 15 min.
In RNA gel blot analyses, total RNA was prepared as described by Sambrook et al. (27). RNA (10 μg per sample) was electrophoresed and transferred to Hybond N+ nylon membrane. Hybridization was performed in 5× SSC, 10% (wt/vol) dextran sulfate, 0.5% (wt/vol) SDS, and 0.1 mg/ml denatured salmon sperm DNA at 65°C. Filters were washed with the procedure mentioned above.
To investigate the effects of uniconazole and GA3 on the expression of the rice GA 3β-hydroxylase genes, seeds of Nipponbare were sterilized, as described above, placed in Petri dishes, and imbibed in 10 ml of water. An ethanol solution (100 μl) of either uniconazole (final concentration, 10 μM) or GA3 (final concentration, 10 μM) was added to the culture. For a control experiment, only ethanol (100 μl) was added to the culture. Twenty-four hours later, the total RNA was isolated from the embryos.
Characterization of the d18 Alleles.
Genomic DNA was prepared from the d18 dwarf mutants (d18-Id18h, d18-dy, and d18-AD) and used for PCR as a template to amplify the 1.6-kb fragment that include the entire coding region of OsGA3ox2. The fragments obtained from the d18-Id18h (Hosetsu-waisei) and d18-dy (Waito-C) alleles were sequenced.
Complementation Analysis.
A binary vector, pBI-Hm, was prepared from pBI-H1 (kindly provided by K. Nakamura, Nagoya University, Nagoya, Japan) by incorporation of the hygromycin-resistant gene into pBI-H1. A genomic fragment (8 kbp) harboring the OsGA3ox2 locus was cloned into the HindIII site of the pBI-Hm. The binary vector was introduced into Agrobacterium tumefaciens EHA101 (28) by electroporation. Transformation of rice with A. tumefaciens was performed as described by Hiei et al. (29).
In Situ Hybridization.
Flowers were fixed overnight at 4°C in 4% (wt/vol) paraformaldehyde plus 0.25% (vol/vol) glutaraldehyde in 0.1 M sodium phosphate buffer (pH 7.2). The fixed material was dehydrated by a graded concentration of ethanol, then t-butanol, and embedded in Paraplast Plus (Sherwood Medical, Norfolk, NE). Microtome sections (9 μm thick) were mounted on silanized glass slides (Matsunami Glass, Osaka, Japan). Paraffin was removed by using a graded concentration of ethanol, and samples were dried for 1 h.
In situ hybridization was performed by using digoxygenin-labeled sense or antisense RNA prepared from cDNA of OsGA3ox1 and OsGA3ox2. Hybridization and detection of the probes were performed by using methods described by Kouchi and Hata (30).
Results and Discussions
Isolation of GA 3β-Hydroxylase Genes.
Using rice genomic DNA as a template, we performed PCR with degenerate primers to give a DNA fragment that had relatively high homology with previously reported GA 3β-hydroxylase genes (10, 11, 20–24). A rice genome library was screened with this fragment as a probe to isolate full-length clones. Sequence analysis of the clones revealed the occurrence of two genes for putative GA 3β-hydroxylase. They were designated as OsGA3ox1 and OsGA3ox2 according to the nomenclature used by Coles et al. (31).
Based on the sequences of the clones, reverse transcription–PCR was performed to obtain the cDNA clones. Full-length cDNA fragments of OsGA3ox1 and OsGA3ox2 contained an ORF encoding polypeptides of 379 and 373 aa, respectively (Fig. 1). The deduced amino acid sequences of both clones suggested that they belonged to a family of 2-oxoglutarate-dependent dioxygenases. Such dioxygenases contain sequences that are conserved within the class of enzyme, including two histidine and one asparatic acid residues at cofactor binding sites. Their sequences had the greatest similarity to GA 3β-hydroxylase sequences of enzymes that catalyze the later steps of GA biosynthesis (Fig. 1). However, the degree of similarity was less than those observed among dicot GA 3β-hydroxylases. Like other GA 3β-hydroxylases, OsGA3ox2 had one intron in its coding region, whereas OsGA3ox1 had two introns.
Functional Analyses.
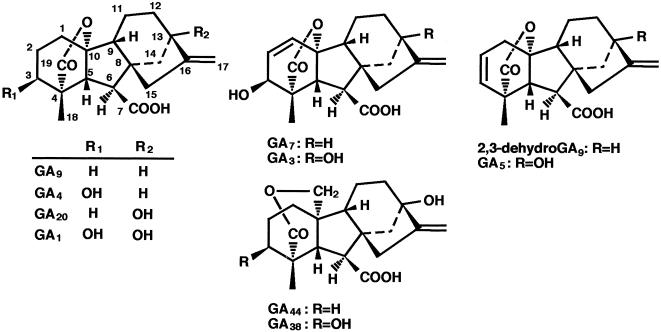
To demonstrate that OsGA3ox1 and OsGA3ox2 encode active GA 3β-hydroxylases, we subcloned the coding regions in expression vector pMAL-c2 to produce in-frame fusion proteins and expressed them in E. coli. The recombinant proteins were used for functional assays, and the results are summarized in Table 1. Both recombinant proteins catalyzed the conversions of GA5 to GA3, GA9 to GA4, GA20 to GA1 and GA44 to GA38. The metabolic step of GA5 to GA3 indicates a 3β-hydroxylation with rearrangement of double bond. The metabolism of GA5 to GA3 has been observed in rice (32). In maize, the dwarf-1 gene (putative GA 3β-hydroxylase) was suggested to catalyze this step (33). Thus, it is not surprising that the rice GA 3β-hydroxylase can catalyzes the metabolism of GA5 to GA3. In addition, it was suggested that OsGA3ox1 protein catalyzes 2,3-desaturation and 2β-hydroxylation. For example, GA9 was metabolized not only to GA4, but also to GA7 and GA34 (Fig. 2). As determined by total ion current, GA4 occupied 65% of the total amount of metabolites, whereas 29% was occupied for GA7 and 6% for GA34. The identification of GA7 suggests that the OsGA3ox1 protein has 2,3-desaturation activity (34); this 2,3-desaturation activity is consistent with the fact that GA7 has been identified in rice anthers (35). The identification of GA34 revealed that OsGA3ox1 protein also catalyzes 2β-hydroxylation. No metabolite was identified from GA53, GA19, and GA29. These results show that OsGA3ox2 encodes an active GA 3β-hydroxylase and that OsGA3ox1 encodes a novel type of GA 2β,3β-hydroxylase, whose substrate specificity is different from pumpkin GA 2β,3β-hydroxylase (21). It is noteworthy that partially purified GA 3β-hydroxylase from bean catalyzed 2β-hydroxylation of GA20 to produce GA29 (36). However, the 2β-hydroxylated GAs, namely GA34 from GA9 and GA8 from GA1, were minor products of the reactions, and the role of 2β-hydroxylation activity of OsGA3ox1 protein in growth regulation is unclear.
Table 1.
Identification of the metabolites from GAs incubated with recombinant rice GA 3β-hydroxylases by full-scan GC-MS and Kovats retention indices (KRI)
| Recombinant protein | Substrate GA | Product GA* | KRI | Characteristic ions at m/z (% relative intensity of base peak) |
|---|---|---|---|---|
| OsGA3ox1 | 2H3-GA9 | GA4 | 2610 | 479 (17), 464 (31), 389 (36), 227 (100) |
| GA7 | 2627 | 477 (4), 462 (11), 389 (6), 225 (100) | ||
| GA34 | 2750 | 567 (100), 552 (14), 477 (4), 232 (18) | ||
| 2H2-GA20 | GA1 | 2748 | 566 (100), 551 (31), 449 (33), 209 (25) | |
| GA3 | 2766 | 564 (100), 549 (42), 447 (26), 210 (48) | ||
| GA8 | 2880 | 654 (13), 639 (5), 537 (100), 313 (11) | ||
| 2H0-GA5 | GA3 | 2768 | 562 (100), 547 (47), 445 (38), 208 (79) | |
| 2H2-GA44 | GA38 | 2939 | 522 (73), 507 (6), 432 (5), 209 (100) | |
| OsGA3ox2 | 2H3-GA9 | GA4 | 2608 | 479 (12), 464 (21), 389 (25), 227 (100) |
| 2H2-GA20 | GA1 | 2745 | 566 (100), 551 (30), 449 (60), 209 (45) | |
| 2H0-GA5 | GA3 | 2766 | 562 (87), 547 (38), 445 (48), 208 (100) | |
| 2H2-GA44 | GA38 | 2940 | 522 (84), 507 (8), 432 (4), 209 (100) |
For the identification of GA38, the sample was derivatized to the methyl ester trimethylsilyl ether. Other samples were trimethylsilylated.
Figure 2.
Numbering system and structures of the four GAs used in the metabolic studies. See Table 1 for specific feeds and metabolites.
Genetic Analysis.
The D18 locus maps to the top of the short arm of chromosome 1, flanking the FS-2 locus (37, 38). To test whether OsGA3ox1 and/or OsGA3ox2 maps to the D18 locus, we performed restriction fragment length polymorphism analysis. DNA gel blot analysis with gene-specific probes for OsGA3ox1 and OsGA3ox2 revealed EcoRV (OsGA3ox1) and ApaI (OsGA3ox2) polymorphisms between Asominori and IR 24. Linkage analysis was performed by using recombinant inbred lines prepared by crossing of Asominori and IR 24 (39). OsGA3ox1 mapped to the distal end of the short arm of chromosome 5; OsGA3ox2 mapped to the distal end of the short arm of chromosome 1. The mapping position of OsGA3ox1 was between R3166 (18.5 centimorgans, cM) and C119 (27.5 cM) on chromosome 5, whereas that of OsGA3ox2 was between R1613 (16.1 cM) and R1944 (24.1 cM) on chromosome 1. Thus OsGA3ox2 corresponds to the D18 locus.
To confirm the relation of OsGA3ox2 to the D18 locus, we directly analyzed the entire coding sequences of OsGA3ox2 isolated from d18 alleles and compared the sequences with the wild type (Nipponbare) (Fig. 1). In the strong allele, d18-Id18h, deletion of a guanine base at position 750 (counted from the adenine in the start codon) shifts the reading frame. In the weak allele, d18-dy, an in-frame 9-base deletion at positions 181–189 deleted three aa resides, Val61 to Arg63. In a strong allele, d18-AD, no PCR product was generated. Genomic Southern analysis of d18-AD indicated the occurrence of 7-kbp deletion in the genomic DNA fragment that include D18 locus. Based on this evidence, it was concluded that at least a part of the coding sequence of OsGA3ox2 was lost in the d18-AD mutant. Further analysis has not been done on this allele. We also carried out the complementation of d18-AD by the introduction of the wild-type OsGA3ox2 gene (8.2 kbp) containing 5′ and 3′ flanking sequences (6 kbp and 1 kbp), all exons (1.1 kbp), and an intron (0.1 kbp). A normal phenotype appeared in eight of 10 transgenic plants. These results confirmed that OsGA3ox2 corresponds to the D18 locus. The in vitro assay with GA20 as a substrate showed that d18-ld18h protein had no activity; a slight activity was observed from the d18-dy protein (data not shown).
Gene Expression.
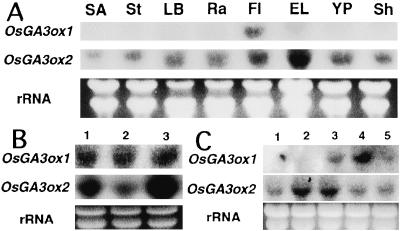
The expression of OsGA3ox1 and OsGA3ox2 was examined in several organs using RNA gel blot analysis with gene-specific probes (Fig. 3A). The expression of OsGA3ox1 was highest in unopened flowers. The expression of OsGA3ox2 was highest in elongating leaves and absent in shoot apices.
Figure 3.
(A) RNA gel blot analyses of OsGA3ox1 and OsGA3ox2 in various rice organs (cultivar Akibare). Total RNA was isolated from shoot apices (SA), stems (St), leaf blades (LB), rachis (Ra), unopened flowers (Fl), elongating leaves (EL), young panicles (YP), and shoots (Sh). (B) Effects of GA3 and uniconazol on the expression of OsGA3ox1 and OsGA3ox2 in germinating seeds. Lane 1, control; lane 2, 10 μM GA3; lane 3, 10 μM uniconazole. (C) Gene expression of GA 3β-hydroxylases in anthers and ovaries at different stages of development. Lane 1, stage differentiation of pollen mother cells; lane 2, early-stage pollen differentiation; lane 3, middle-stage pollen differentiation; lane 4, late-stage pollen differentiation; lane 5, mature-stage pollen. Blots were hybridized with probes for GA 3β-hydroxylases (OsGA3ox1, OsGA3ox2). Approximately 10 μg of total RNA from each sample was loaded onto the gel and stained with ethidium bromide (rRNA).
It has been reported that the expression of GA4 (AtGA3ox1) in Arabidopsis and LE (PsGA3ox1) in pea is under feedback regulation (11, 20). We used germinating seeds to examine the expression of OsGA3ox1 and OsGA3ox2 in the presence of GA3 and the GA biosynthetic inhibitor, uniconazol (Fig. 3B). Both OsGA3ox1 and OsGA3ox2 were expressed in the germinating seed (Fig. 3B, lane 1). Neither GA3 nor uniconazole affected the expression level of OsGA3ox1. The expression of OsGA3ox2 was down-regulated by GA3 (Fig. 3B, lane 2) and up-regulated by uniconazole (Fig. 3B, lane 3).
Kobayashi et al. (19) have reported that mature anthers of rice metabolize GAs. We have examined the expression of OsGA3ox1 and OsGA3ox2 in the anthers and ovaries at five developmental stages (Fig. 3C). The level of the OsGA3ox1 transcript was high at late-stage pollen differentiation only, whereas the level of the OsGA3ox2 transcript was relatively high at early-stage pollen differentiation and middle-stage pollen differentiation.
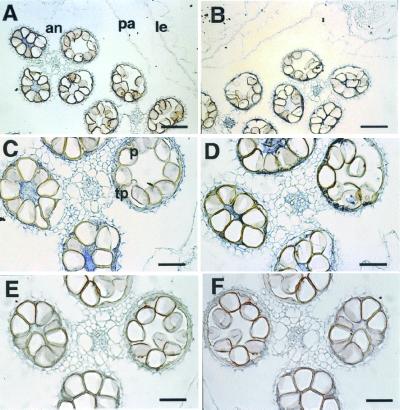
To investigate the expression site of OsGA3ox1 and OsGA3ox2 in the anther, in situ hybridization was performed with digoxygenin-labeled RNA probes. Blue staining by an antisense probe for OsGA3ox1 and OsGA3ox2 were observed in the tapetum of anthers (Fig. 4 A–D). No staining was observed with the sense-strand probes (Fig. 4 E and F), which indicates that the blue staining by antisense probes was not an artifact.
Figure 4.
In situ localization of OsGA3ox1 (A, C, and E) and OsGA3ox2 (B, D, and F) mRNA during the stage of late pollen differentiation (same stage as in Fig. 3C, lane 4). (A and B) Cross section of a flower hybridized with the antisense probe. The signal (blue staining) can be seen in the anthers. (C and D) Higher-magnification view of a cross section of anthers stained with the antisense probes. Note the staining in the tapetum. (E and F) Anthers at the same stage, stained with the sense probe. Morphological features of tissues: an, anther; pa, palea; le, lemma; p, pollen; tp, tapetum. (Bar indicates 50 μm in A and B; 100 μm in C–F.)
Although OsGA3ox1 and OsGA3ox2 share high homology in their deduced amino acid sequences, their expression patterns and in vitro activities are different from each other. We show that OsGA3ox2 corresponds to the D18 gene, a gene that is known to control the elongation of the vegetative shoot. The feedback regulation of the expression of OsGA3ox2 supports this position. The relatively high level of expression of OsGA3ox2 in immature flower tissues suggests that OsGA3ox2 also may be involved in the regulation of the growth of reproductive tissues. Several reports suggest that GA biosynthesis is required for anther development (4, 16, 17, 40). However, the d18 mutants are not male-sterile, which suggests that OsGA3ox2 may have a limited role in the regulation of anther development.
OsGA3ox1 was expressed mainly in the tapetum at late-stage pollen differentiation. Although OsGA3ox2 also was expressed in the tapetum at this stage, the expression level was relatively low. These observations suggest that the OsGA3ox1 gene is responsible for GA4 biosynthesis in the anthers, which would explain the previous observation that GA4 accumulates in the anthers of d18-dy plant (14). Hasegawa et al. (41) studied the localization of GA4 in anthers by immunohistochemistry and found that GA4 apparently accumulated in the tapetum and pollen grains. The expression of OsGA3ox1 and OsGA3ox2 in the tapetum partially explains their results, although the metabolic origin of the GA4 observed in pollen grains is unknown.
Acknowledgments
We thank Dr. Soon-Kwan Hong for his advice and help on in situ hybridization experiments. This work was supported by the program Isolation and Functional Analysis of Useful Rice Genes and Development of Techniques for their Utilization from the Ministry of Agriculture, Forestry and Fishery, and by the Special Coordination Funds for Promoting Science and Technology from Research Fellowship of the Japan Society for the Promotion of Young Scientists (to H.I.).
Abbreviation
- GA
gibberellin
Footnotes
References
- 1.Phinney B O. Proc Natl Acad Sci USA. 1956;42:185–189. doi: 10.1073/pnas.42.4.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Murakami Y. In: Plant Growth Substances 1970. Carr D J, editor. Berlin: Springer; 1972. pp. 166–174. [Google Scholar]
- 3.Koornneef M. Arabidopsis Inf Serv. 1978;15:17–20. [Google Scholar]
- 4.Koornneef M, van der Veen J H. Theor Appl Genet. 1980;58:257–263. doi: 10.1007/BF00265176. [DOI] [PubMed] [Google Scholar]
- 5.Kinoshita T, Shinbashi N. Jpn J Breed. 1982;32:219–231. [Google Scholar]
- 6.Koornneef M, Bosma T D G, Hanhart C J, van der Veen J H, Zeevaart J A D. Theor Appl Genet. 1990;80:852–857. doi: 10.1007/BF00224204. [DOI] [PubMed] [Google Scholar]
- 7.Reid J B, Ross J J. Int J Plant Sci. 1993;154:22–34. [Google Scholar]
- 8.Spray C, Phinney B O, Gaskin P, Gilmour S J, MacMillan J. Planta. 1984;160:464–468. doi: 10.1007/BF00429764. [DOI] [PubMed] [Google Scholar]
- 9.Fujioka S, Yamane H, Spray C R, Gaskin P, MacMillan J, Phinney B O, Takahashi N. Plant Physiol. 1988;88:1367–1372. doi: 10.1104/pp.88.4.1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martin D N, Proebsting W M, Hedden P. Proc Natl Acad Sci USA. 1997;94:8907–8911. doi: 10.1073/pnas.94.16.8907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lester D R, Ross J J, Davies P J, Reid J B. Plant Cell. 1997;9:1435–1443. doi: 10.1105/tpc.9.8.1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ingram T J, Reid J B, Potts W C, Murfet I C. Physiol Plant. 1983;59:607–616. [Google Scholar]
- 13.Ingram T J, Reid J B, Murfet I C, Gaskin P, Willis C L, MacMillan J. Planta. 1984;160:455–463. doi: 10.1007/BF00429763. [DOI] [PubMed] [Google Scholar]
- 14.Kobayashi M, Sakurai A, Saka H, Takahashi N. Plant Cell Physiol. 1989;30:963–969. [Google Scholar]
- 15.Kobayashi M, Gaskin P, Spray C R, Phinney B O, MacMillan J. Plant Physiol. 1994;106:1367–1372. doi: 10.1104/pp.106.4.1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sawhney V K, Greyson R I. Can J Bot. 1973;51:2473–2479. [Google Scholar]
- 17.Jacobsen S E, Olszewski N E. Plant Physiol. 1991;97:409–414. doi: 10.1104/pp.97.1.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kobayashi M, Yamaguchi I, Murofushi N, Ota Y, Takahashi N. Agric Biol Chem. 1988;52:1189–1194. [Google Scholar]
- 19.Kobayashi M, Kamiya Y, Sakurai A, Saka H, Takahashi N. Plant Cell Physiol. 1990;31:289–293. [Google Scholar]
- 20.Chiang H-H, Hwang I, Goodman H M. Plant Cell. 1995;7:195–201. doi: 10.1105/tpc.7.2.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lange T, Robatzek S, Frisse A. Plant Cell. 1997;9:1459–1467. doi: 10.1105/tpc.9.8.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Toyomasu T, Kawaide H, Mitsuhashi W, Inoue Y, Kamiya Y. Plant Physiol. 1998;118:1517–1523. doi: 10.1104/pp.118.4.1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yamaguchi S, Smith M W, Brown R G S, Kamiya Y, Sun T-P. Plant Cell. 1998;10:2115–2126. doi: 10.1105/tpc.10.12.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Itoh H, Ueguchi M T, Kawaide H, Chen X, Kamiya Y, Matsuoka M. Plant J. 1999;20:15–24. doi: 10.1046/j.1365-313x.1999.00568.x. [DOI] [PubMed] [Google Scholar]
- 25.Kobayashi M, Yoshizawa K, Sakurai A, Nakamura T. Biosci Biotechnol Biochem. 1996;60:159–160. [Google Scholar]
- 26.Murray M G, Thompson W F. Nucleic Acids Res. 1980;8:4321–4325. doi: 10.1093/nar/8.19.4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 28.Hood E E, Helmer G L, Fraley R T, Chilton M D. J Bacteriol. 1986;168:1291–1301. doi: 10.1128/jb.168.3.1291-1301.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hiei Y, Ohta S, Komari T, Kumashiro T. Plant J. 1994;6:271–282. doi: 10.1046/j.1365-313x.1994.6020271.x. [DOI] [PubMed] [Google Scholar]
- 30.Kouchi H, Hata S. Mol Gen Genet. 1993;238:106–119. doi: 10.1007/BF00279537. [DOI] [PubMed] [Google Scholar]
- 31.Coles J P, Phillips A L, Croker S J, García-Lepe R, Lewis M J, Hedden P. Plant J. 1999;17:547–556. doi: 10.1046/j.1365-313x.1999.00410.x. [DOI] [PubMed] [Google Scholar]
- 32.Kobayashi M, Kwak S-S, Kamiya Y, Yamane H, Takahashi N, Sakurai A. Agric Biol Chem. 1991;55:249–251. [Google Scholar]
- 33.Spray C R, Kobayashi M, Suzuki Y, Phinney B O, Gaskin P, MacMillan J. Proc Natl Acad Sci USA. 1996;93:10515–10518. doi: 10.1073/pnas.93.19.10515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Albone K S, Gaskin P, MacMillan J, Phinney B O, Willis C L. Plant Physiol. 1990;94:132–142. doi: 10.1104/pp.94.1.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hasegawa M, Nakajima M, Takeda K, Yamaguchi I, Murofushi N. Biosci Biotechnol Biochem. 1995;59:1716–1720. [Google Scholar]
- 36.Smith V A, Gaskin P, MacMillan J. Plant Physiol. 1990;94:1390–1401. doi: 10.1104/pp.94.3.1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kinoshita T. Rice Gen News. 1995;12:9–154. [Google Scholar]
- 38.Harushima Y, Yano M, Shomura A, Sato M, Shimano T, Kuboki Y, Yamamoto T, Lin S-Y, Antonio B A, Parco A, et al. Genetics. 1998;148:479–494. doi: 10.1093/genetics/148.1.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tsunematsu H, Yoshimura A, Harushima Y, Nagamura Y, Kurata N, Yano M, Sasaki T, Iwata N. Breeding Sci. 1996;46:279–284. [Google Scholar]
- 40.Silverstone A L, Chang C-W, Krol E, Sun T-P. Plant J. 1997;12:9–19. doi: 10.1046/j.1365-313x.1997.12010009.x. [DOI] [PubMed] [Google Scholar]
- 41.Hasegawa M, Hashimoto N, Zhang J, Nakajima M, Takeda K, Yamaguchi I, Murofushi N. Biosci Biotechnol Biochem. 1995;59:1925–1929. [Google Scholar]