Abstract
Rifampin is a major drug used in the treatment of tuberculosis infections, and increasing rifampin resistance represents a worldwide clinical problem. Resistance to rifampin is caused by mutations in the rpoB gene, encoding the β-subunit of RNA polymerase. We examined the effect of three different rpoB mutations on the fitness of Mycobacterium tuberculosis. Rifampin-resistant mutants were isolated from a virulent clinical isolate of M. tuberculosis (strain Harlingen) in vitro at a mutation frequency of 2.3 × 10−8. Mutations in the rpoB gene were identified, and the growth rates of three defined mutants were measured by competition with the susceptible parent strain in laboratory medium and by single cultures in a macrophage cell line and in laboratory medium. All of the mutants showed a decreased growth rate in the three assays. The relative fitness of the mutants varied between 0.29 and 0.96 (that of the susceptible strain was set to 1.0) depending on the specific mutant and assay system. Unexpectedly, the relative fitness ranking of the mutants differed between the different assays. In conclusion, rifampin resistance is associated with a cost that is conditional.
Following the introduction of effective antimycobacterial drugs about half a century ago, tuberculosis appeared to have been brought under control. However, tuberculosis has remained a major killer among infectious diseases, and the situation has been worsened by the emergence of Mycobacterium tuberculosis mutants that are resistant to the major drugs (8). As much as antibiotic resistance has hampered effective tuberculosis treatment, it has also caused an interest in understanding the mechanisms of drug action and resistance. Thus, within the last few decades, several chromosomal mutations in M. tuberculosis responsible for resistance to most of the major drugs, including rifampin, isoniazid, pyrazinamide, ethambutol, streptomycin, and fluoroquinolones, have been discovered (for reviews, see references 9, 25, and 27).
It is often believed that a reduced use of antibiotics will lead to a reduction in the frequency of resistant bacteria in the population. The underlying assumption for this idea is that resistance is associated with a reduced bacterial fitness (i.e., reduced transmission between hosts and reduced persistence and growth within and outside hosts) and that in the absence of the drug, resistant bacteria will be outcompeted by susceptible strains and as a result the frequency of resistance will decline. Usually there is a cost associated with resistance, giving credence to this idea (1). However, some resistance mutations appear to confer no cost (<1% reduction in fitness), at least as measured by in vitro assay systems. For example, certain rpsL mutations (streptomycin resistance) in M. tuberculosis (6), Escherichia coli (21, 22), and Salmonella enterica serovar Typhimurium (3, 4), katG mutations (isoniazid resistance) in M. tuberculosis (16), and gyrA and parC mutations (fluoroquinolone resistance) in Streptococcus pneumoniae (10) confer no measurable reduction in growth rate.
These cost measurements are based almost exclusively on one specific type of in vitro assay, and it is often assumed that fitness costs in vitro and in vivo are correlated. Fitness costs conferred by mutations that alter target molecules may also be partly or fully ameliorated by compensatory mutations without loss of resistance. Such compensatory evolution has been observed in vitro (3, 4, 19, 21, 22), in experimental animals (3-5), and in clinical situations (5, 13, 23). Thus, the occurrence of cost-free mutations and compensatory evolution suggests that antibiotic-resistant bacteria will not disappear as a result of restricted use of antibiotics but might instead, as shown by recent clinical studies, persist in the population for a long time even after antibiotic use has been reduced or eliminated (7, 24). Thus, to allow implementation of rational strategies to minimize resistance development, we need knowledge of the resistance mechanisms, the rate at which resistant mutants appear, and relevant determinations of how antibiotic resistance affects the entire bacterial life cycle. In addition, there is an urgent need for the development of new potent drugs and vaccines to prevent the emergence and spread of both susceptible and resistant strains.
Rifampin remains a first-line drug for the treatment of tuberculosis infections. Among clinical isolates, resistance to rifampin is due almost exclusively to amino acid changes in a limited region of the β-subunit of RNA polymerase, encoded by the rpoB gene (26). We isolated rifampin-resistant mutants in vitro and determined the effect of three rpoB mutations on their fitness in vitro and in a macrophage cell line with a virulent clinical isolate of M. tuberculosis.
MATERIALS AND METHODS
Growth and storage of strains.
Strains were grown in Middlebrook 7H9 broth with 10% albumin-dextrose-catalase enrichment and 0.05% Tween 80 (hereafter referred to as 7H9 medium) for 3 weeks. Bacterial cells were stored in glycerol-containing medium at −70°C. Strain Harlingen is a clinical isolate of M. tuberculosis that was isolated as a particularly transmissible and virulent strain in Holland in 1993 (11).
Isolation of spontaneous rifampin-resistant mutants and calculation of mutation frequency to rifampin resistance.
The parent strain was grown in 7H9 medium to mid-log phase and then used to inoculate 60 independent cultures, which were grown to an optical density at 600 nm of ≈1.00. Approximately 108 cells from each of these 60 independent cultures were spread on Middlebrook 7H10 plates containing 1 μg of rifampin per ml. The plates were incubated at 37°C in a 5% CO2 incubator. The appearance of single colonies was examined periodically over a 4-week period with a dissecting microscope. After 4 weeks, a single colony was picked from each independent plate and inoculated into 7H9 medium. After growth, these bacteria were pelleted by centrifugation, and the pellets were resuspended in glycerol-containing storage medium, aliquoted, and stored at −70°C. The frequency of mutation to rifampin resistance was calculated as the ratio of the median number of resistant mutants from the 60 independent cultures divided by the number of viable cells plated (5).
Characterization of the rifampin-resistant isolates. (i) Determination of MICs.
The MICs for the different isolates were determined with the BACTEC 460 system, following standard procedures. The drug concentrations examined ranged from 0.5 to 32 μg/ml.
(ii) Extraction of DNA.
Mycobacteria were grown on LJ medium. A 10-μl loop was used to transfer cells into a microcentrifuge tube containing 250 μl of 1× TE (Tris-EDTA) buffer. The cells were killed by heating at 80°C for 1 h. The cells were then centrifuged at 13,000 rpm for 2 min. The supernatant was discarded, and the pellet (containing cells with DNA) was resuspended in 500 μl of 150 mM NaCl. This step was followed by another centrifugation. The resuspension and pelleting steps were repeated once more. Finally, the pellet (containing DNA) was resuspended in 25 μl of TE and used for the PCR amplification reactions.
PCR amplification and sequencing of rpoB gene.
A 258-bp region of the rpoB gene which encompasses most of the codons in which mutations lead to rifampin resistance was amplified by PCR (26). The forward primer was 5′-ATCAACATCCGGCCGGTGGT-3′, and the reverse primer was 5′-TACACCGACAGCGAGCCGAT-3′ (12). PCR was run in a 50-μl reaction mix containing 0.5 μM each primer, 200 μM each deoxynucleoside triphosphate, 2 mM MgCl2, 5 μl of 10× PCR buffer, 0.25 μl of AmpliTaq DNA polymerase, and 1 μl of DNA. The PCR conditions were an initial denaturation at 95°C for 5 min, followed by 35 cycles of denaturation at 94°C for 20 s, annealing at 55°C for 20 s, and extension at 72°C for 30 s, and a final extension at 72°C for 7 min. PCR products were purified with GFX PCR and the gel band purification kit (Amersham Pharmacia Biotech). Cycle sequencing was performed according to the Applied Biosystems ABI Prism Big Dye terminator cycle sequencing kit with an ABI Prism 310 or 3100 genetic analyzer.
Competition experiments.
The wild-type and the mutant strains were grown separately in 7H9 medium containing 0.1% Tween 80. The Ser522→Leu (TCG→TTG), His526→Tyr (CAC→TAC), and Ser531→Trp (TCG →TGG) mutants were used in this experiment. For competition cultures, the optical densities of the wild-type and mutant isolates were first adjusted to the same value. Then 30 μl of the mutant and 10 μl of the wild-type bacterial suspensions were mixed in 20 ml of 7H9 medium. Immediately after mixing (day 0), 10-fold serial dilutions were prepared from each mixed culture and plated on both drug-free and drug-containing Middlebrook 7H10 plates with 10% oleic acid-albumin-dextrose-catalase (OADC) enrichment. The plates were incubated at 37°C. This procedure was repeated after 3, 7, and 14 days. The number of CFU was counted after 25 days of incubation. The CFU counts of the wild type on days 0, 3, 7, and 14 were obtained by subtracting the CFU on drug-containing plates from the total CFU on drug-free plates. The data shown in Table 2 represent the averages of two to three independent experiments for each of two independent isolates of the same mutant type. The plating efficiency of the resistant mutants was unaffected by the presence of rifampin.
TABLE 2.
Growth rates of susceptible and resistant M. tuberculosis strains during growth in vitro in single cultures and competitionsa
| Strain | Mutation | MIC (μg/ml) | Single culture
|
Competition culture
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Doubling time (h) | No. of divisions | Relative fitness | Doubling time (h)
|
No. of divisions
|
Relative fitness | |||||
| Mutant | Wild type | Mutant | Wild type | |||||||
| Harlingen | None | 0.25 | 22.4 | 15.0 | 1.00 | |||||
| Ser531→Trp (TCG→TGG) | >32 | 31.6 | 10.6 | 0.71 ± 0.09 | 36.3 | 24.3 | 9.4 | 13.9 | 0.67 ± 0.06 | |
| His526→Tyr (CAC→TAC) | >32 | 26.2 | 12.8 | 0.86 ± 0.03 | 30.6 | 27.2 | 11.1 | 12.5 | 0.89 ± 0.05 | |
| Ser522→Leu (TCG→TTG) | >16 | 23.6 | 14.2 | 0.95 ± 0.02 | 44.4 | 23.8 | 7.7 | 13.7 | 0.54 ± 0.03 | |
| H37Ra | None (wild type) | 0.25 | 24.5 | 13.7 | 0.91 ± 0.05 | |||||
| H37Rv | None (wild type) | 0.25 | 24.8 | 13.5 | 0.90 ± 0.05 | |||||
Data shown represent the average of two to three independent replicate experiments for each of two independent isolates of the same mutant type. Standard errors for relative fitness are indicated. In single and competition cultures, the doubling time was estimated from a plot of ln(CFU) = f(time), where the slope is ln2/doubling time (hours). The relative fitness is the ratio of doubling time (wild type)/doubling time (mutant).
Macrophage infection experiments. (i) Growth of bacteria and cell lines.
The susceptible parent strain and resistant mutants were grown in 7H9 medium. The bacterial cultures were centrifuged and washed once with fresh 7H9 medium. The pellets were resuspended in 3 ml of 7H9 medium and then vortexed with sterile glass beads for 30 min, sonicated twice (30 s each), and then filtered through a 5-μm-pore-size filter unit (Millipore) to disrupt clumps in the bacterial suspensions. Finally, the optical densities of the suspensions were adjusted to give a 1:1 infection ratio.
A frozen ampoule of U937 cells was thawed at 37°C and washed in warm complete RPMI medium containing 10% fetal bovine serum (Gibco, Invitrogen Corporation), and cell viability was determined. Cells were cultured in complete RPMI medium for 3 days, fresh medium was added, and incubation was continued for 4 days. The viability and the number of viable cells per milliliter were determined, phorbol myristate acetate was added, and approximately 105 cells were dispensed into each well of eight-well slide flasks (Lab-Tek II chamber slide system). Cells were incubated to mature for 3 days.
(ii) Infection of macrophages.
For a 1:1 multiplicity of infection, 200 μl of each bacterial suspension was mixed with RPMI. Prior to infection, the medium from each well was removed and the wells were washed with RPMI. Then 0.5 ml of the mixture of bacteria and medium was pipetted into each well. For day 0, two wells each for microscopy and CFU were infected. For days 3, 5, and 7, three wells each for microscopy and CFU were infected. After a 4-h infection period, infecting medium from each well was removed and each well was vigorously washed twice with 0.5 ml of RPMI. Day 0 slide flasks were processed immediately. Day 3, 5, and 7 slide flasks were filled with 0.5 ml of fresh complete medium and returned to the incubator.
CFU determinations and microscopy.
Previous experiments have shown that the number of bacteria removed with the medium on day 3 is less than 2% of the total, while the number of bacteria removed with the medium on day 7 can be 20% or more (because of macrophage lysis, where intracellular bacteria are released into the growth medium). Thus, when we quantitated the CFU within lysates from the wells on days 0 and 3, the medium was discarded, whereas on days 5 and 7, the medium was combined with the lysate from the well to quantitate the total CFU. Lysis was performed with a 10-min incubation in sterile distilled water containing 0.036% sodium dodecyl sulfate. After the lysis period, 50 μl of 20% bovine serum albumin was added to the lysate (15, 32). Then 10-fold dilutions were prepared and plated on Middlebrook 7H10 medium with 10% OADC. The plates were incubated in a 37°C incubator with 5% CO2. The CFU were counted after 3 weeks. For microscopy, about 2,500 macrophages were counted for each time point and strain. Twenty different fields from each well were counted, and the number of bacteria present in each macrophage was counted.
RESULTS
A previous study of rifampin-resistant M. tuberculosis showed that some mutations in rpoB impose a cost in vitro, as determined by competition experiments (2). Here, we extended this study and examined other types of rpoB mutations. In addition, and importantly, we determined whether two different in vitro assays and an assay system nearer the in vivo situation, i.e., a macrophage cell line, gave similar results in terms of the fitness costs of resistance. It is usually assumed that the results from in vitro assays can be extrapolated to the in vivo situation, although very few studies comparing different assay systems with regard to cost determinations have been done.
Several independent spontaneous rifampin-resistant colonies were isolated on agar plates containing rifampin. The mutation frequency, as calculated from the median number of mutants, was 2.3 × 10−8 (5, 28). In total, 27 rifampin-resistant mutants were examined for rifampin MIC and rpoB sequence. The MICs of rifampin for the isolates were >32 μg/ml except for a few isolates which had MICs of 16 μg/ml. DNA was prepared from the 27 mutants, and the rpoB gene was sequenced. Unexpectedly, for a third of these isolates (10 of 27) no mutations were found in the rpoB gene. The mechanism of resistance in these isolates is unknown. The identified rpoB mutations were Ser522→Leu (TCG→TTG) (four isolates), His526→Tyr (CAC→TAC) (11 isolates), and Ser531→Trp (TCG →TGG) (two isolates) (Table 1). All mutations were single nucleotide substitutions and included both transition and transversion mutations.
TABLE 1.
Characteristics of rifampin-resistant mutants
| No. of independent isolates | MIC (μg/ml) | Mutation in rpoB |
|---|---|---|
| 2 | >32 | Ser531→Trp (TCG→TGG) |
| 11 | >32 | His526→Tyr (CAC→TAC) |
| 4 | >16 | Ser522→Leu (TCG→TTG) |
| 10 | >32 | None |
For each specific mutant type, two independent isolates were examined. First, fitness was measured by competition experiments in vitro between the wild type and the resistant mutants. The results of the pairwise competition experiments showed that the wild type outcompeted all of the mutant isolates (Table 2). The fitness of each mutant was calculated relative to that of the parent, for which a fitness value of 1.0 was assigned. The mutants had fitness values ranging from 0.54 to 0.89. The fitness data showed that each of the two independent isolates containing the same resistance mutation behaved similarly in the competitions. Moreover, the extent of fitness loss was dependent on the specific mutation in the rpoB gene. Thus, the mutants showed decreasing fitness, His526→Tyr > Ser531→Trp > Ser522→Leu, in the in vitro competitions.
In competition experiments, it is usually assumed that the competing strains do not affect each other and that they compete only by their intrinsic growth rate and efficiency in utilizing available nutrients. To exclude a possible interaction between the bacteria in the pairwise competitions, fitness was also measured in single cultures. Similarly to the competition experiments, all of the mutants grew more slowly than the wild type in single cultures (Table 2), and the extent of the decrease in fitness according to the single-culture assay was similar to that in the competitions. However, the ranking of the mutants was different, implying that the competing strains might affect each other's growth. Thus, in the single cultures, mutant fitness was ranked Ser522→Leu > His526→Tyr > Ser531→Trp.
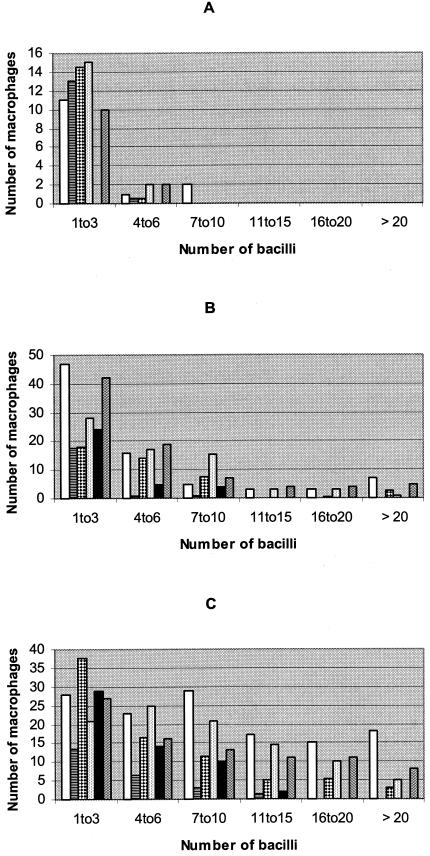
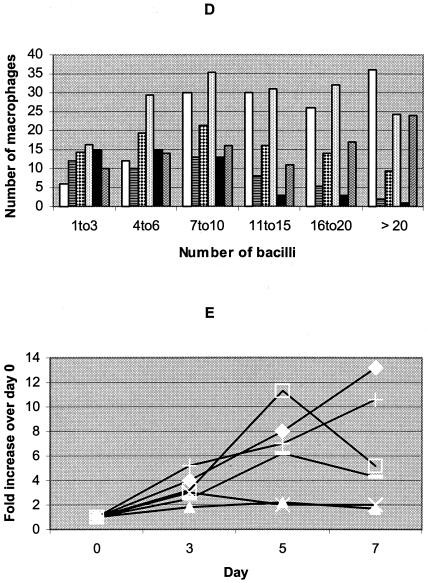
To further examine the effect of these mutations on fitness, we measured growth rates in a macrophage cell line. Initially, we determined the number of bacteria by plating and microscopy. These preliminary data (not shown) indicated that microscopy was easier and faster to perform than viable-count determinations by plating. Microscopy was thus chosen for bacterial enumeration in these experiments. Figure 1A to D shows the number of macrophages containing variable numbers of bacteria on days 0 (4 h after infection), 3, 5, and 7. On day 0, most of the infected macrophages were infected with one to three bacteria (Fig. 1A). A much smaller number of macrophages were infected with four to six bacteria. This indicates that the infecting bacterial suspension contained predominantly single bacteria. The number of macrophages infected and the number of bacteria per macrophage increased as the culture period increased from day 0 to day 7 (Fig. 1B to D). This is an indication that the macrophages were initially infected with single bacteria, but as the infection progressed, the bacteria replicated within the macrophages, resulting in macrophage lysis and infection of previously uninfected neighboring macrophages.
FIG. 1.
Number of macrophages (y axis) containing the indicated number of bacteria (x axis) on days 0 (A), 3 (B), 5 (C), and 7 (D) and increases in the number of bacteria over that at day 0 (E) of the macrophage cultures for the various rifampin-resistant mutants and the susceptible control strains. (A to D) The strains used, from left to right within each group of bars, were the wild type (Harlingen), the Ser531→Trp (TCG→TGG), His526→Tyr (CAC→TAC), and Ser522→Leu (TCG→TTG) mutants, H37Ra, and H37Rv. (E) ⧫, Harlingen; ▴, Ser531→Trp (TCG→TGG) mutant; ▪, His526→Tyr (CAC→TAC) mutant; —, Ser522→Leu (TCG→TTG) mutant; ×, H37Ra; +, H37Rv.
Figure 1E shows the number of bacteria obtained from the macrophage cultures of the different mutants and the control strains. Since the number of bacteria in the original suspensions were not equivalent, data are presented as increases over the number on day 0 for each isolate. The His526→Tyr mutant grew from day 0 to day 5, and then its growth declined by day 7. A similar trend was observed for the Ser522→Leu mutant. However, the Ser531→Trp mutant showed very limited growth which eventually decreased by day 7. Thus, the relative growth rate ranking of these mutants in macrophages was His526→Tyr > Ser522→Leu > Ser531→Trp. The avirulent strain H37Ra showed a slight increase by day 3 and then declined, with no change in the number of bacteria between days 5 and 7. The number of bacteria for both H37Rv and Harlingen continued to increase until day 7.
DISCUSSION
In this study, we compared the fitness of spontaneous rifampin-resistant isolates of M. tuberculosis with that of an isogenic susceptible parent strain. The parental strain (Harlingen) was isolated as a highly virulent and transmissible strain (11). The rifampin-resistant mutants studied contained mutations at codons 522, 526, and 531 in the rpoB gene, and all of these mutations conferred a fitness cost, seen as a decreased growth rate in vitro and in macrophages. However, when comparing the different mutants in the three different assay systems, there was no strong quantitative correlation in fitness reduction for the different assay systems. In fact, the three assay systems all gave different relative rankings of the mutants. This could possibly be a reflection of physiological trade-offs, where good adaptation under one condition results in maladaptation under another. Which assay gives the value most relevant for the in vivo situation is debatable, but we would argue that single cultures in macrophages are likely to give the truest value, since these conditions are closest to the growth conditions in the human host.
Rifampin resistance has been shown to be associated with a fitness reduction for several bacterial species (Table 3). Billington et al. compared the relative fitness of laboratory-isolated spontaneous rifampin-resistant mutants of M. tuberculosis H37Rv with that of a susceptible parent (2). The fitness of most (eight of nine) of the mutants examined was found to be lower than that of the susceptible parent. A detailed study by Reynolds showed that most rpoB mutants of E. coli had decreased fitness measured as growth rate in batch cultures, even though a few mutants showed unaltered or slightly increased fitness under the in vitro conditions used (19). Finally, Wichelhaus et al. studied rifampin resistance in Staphylococcus aureus and noted that fitness was reduced in all 18 rpoB mutants examined (relative fitness ranged from 0.60 to 0.96) as determined by competition assays in vitro (29). Thus, from the data shown here and from previous studies, it is clear that rpoB mutations are generally associated with a cost and that these costs are in a similar range for different mutations, species, and assay systems (Table 3).
TABLE 3.
Rifampin resistance mutations and fitness of various bacterial species assayed in competition experiments in culturesa
| Bacterial species | rpoB mutation | Relative fitness | Reference |
|---|---|---|---|
| E. coli | His526→Tyr | 0.91 | 19 |
| His526→Leu | 0.94 | ||
| S. aureus | His481→Tyr (corresponds to E. coli codon 526) | 0.93 | 29 |
| His481→Asn (corresponds to E. coli codon 526) | 1.00 | ||
| Ser486→Leu (corresponds to E. coli codon 531) | 0.86 | ||
| M. tuberculosis | His526→Tyr | 0.79 | 2 |
| Ser531→Leu | 0.84* | ||
| M. tuberculosis | His526→Tyr | 0.89 | This work |
| Ser531→Trp | 0.67 |
Only the subset of mutations that have been identified in all three species are shown. *, calculated as the average of four independent mutants (2).
Strains carrying the specific point mutation Ser531→Trp are rarely isolated from patients (0 to 14% of rifampin-resistant isolates) (12, 14, 17, 18, 30, 31), although they are associated with high-level resistance. Here, this mutant also had the lowest fitness in two assay systems (macrophages and single cultures). Another point mutation (not analyzed here) at the same site involves the change Ser531→Leu. Among clinical isolates from several different geographic locations, this mutation is found at a much higher frequency (86 to 100% of all rifampin resistance mutations) (12, 14, 17, 18, 30, 31). The reason for this difference in clinical recovery frequency is probably associated with the relative fitness cost of these different mutations. As seen by Billington et al. (2), the Ser531→Leu mutation confers a relatively low fitness cost (the average relative fitness in competitions is 0.84), whereas the Ser531→Trp mutation analyzed here confers a high cost both in vitro (relative fitness, 0.71 and 0.67; Table 2) and in macrophages (relative fitness, 0.28; Table 4). Thus, one would predict if the various mutants form with similar rates, the frequencies at which they are found in the population correlate to their overall fitness. Such correlations have previously been observed for mutations causing resistance to streptomycin (6, 20), isoniazid (16), and rifampin (10). However, as shown here, fitness estimates are dependent on assay conditions, and quantitative correlations between fitness measured in vitro and frequencies determined clinically should be interpreted with care.
TABLE 4.
Growth rates of resistant mutants and susceptible M. tuberculosis in U937 macrophages
| Strain | Mutation | MIC (μg/ml) | Doubling time (h) | No. of divisions | Relative fitness |
|---|---|---|---|---|---|
| Harlingen | None (wild type) | 0.25 | 46.0 | 3.6 | 1.00 |
| Ser531→Trp (TCG→TGG) | >32 | 166.0 | 1.0 | 0.28 ± 0.06 | |
| His526→Tyr (CAC→TAC) | >32 | 72.9 | 2.4 | 0.63 ± 0.02 | |
| Ser522→Leu (TCG→TTG) | >16 | 91.3 | 1.9 | 0.50 ± 0.16 | |
| H37Ra | None (wild type) | 0.25 | 111.2 | 1.5 | 0.41 ± 0.12 |
| H37Rv | None (wild type) | 0.25 | 50.6 | 3.3 | 0.91 ± 0.06 |
Data shown represent the average of two to three independent replicate experiments for each of two independent isolates of the same mutant type. Standard errors for relative fitness are indicated. The doubling time was estimated from a plot of ln(visible cells) = f(time), where the slope is ln2/doubling time (hours). The relative fitness is the ratio of doubling time (wild type)/doubling time (mutant).
Acknowledgments
This work was supported by grants from the Swedish Research Council (D.I.A.), Swedish Institute for Infectious Disease Control (D.I.A. and S.H.), the Swedish Heart-Lung Foundation (D.I.A. and S.H.), and Sida-ISP (SAREC).
We thank Sophie Maisnier-Patin and Cecilia Dahlberg for comments and critical reading and Anna Syk, Ingela Hedenstrom, and Juan Carlos Toro for material and technical assistance.
REFERENCES
- 1.Andersson, D. I. 2003. Persistence of antibiotic resistant bacteria. Curr. Opin. Microbiol. 6:452-456. [DOI] [PubMed] [Google Scholar]
- 2.Billington, O. J., T. D. McHugh, and S. H. Gillespie. 1999. Physiological cost of rifampin resistance induced in vitro in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 43:1866-1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Björkman, J., D. Hughes, and D. I. Andersson. 1998. Virulence of antibiotic-resistant Salmonella typhimurium. Proc. Natl. Acad. Sci. USA 95:3949-3953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Björkman, J., I. Nagaev, O. G. Berg, D. Hughes, and D. I. Andersson. 2000. Effects of environment on compensatory mutations to ameliorate costs of antibiotic resistance. Science 287:1479-1482. [DOI] [PubMed] [Google Scholar]
- 5.Björkholm, B., M. Sjölund, P. G. Falk, O. Berg, L. Engstrand, and D. I. Andersson. 2001. Mutation frequency and biological cost of antibiotic resistance in Helicobacter pylori. Proc. Natl. Acad. Sci. USA 98:14607-14612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Böttger, E. C., B. Springer, M. Pletschette, and P. Sander. 1998. Fitness of antibiotic-resistant microorganisms and compensatory mutations. Nat. Med. 12:1343-1344. [DOI] [PubMed] [Google Scholar]
- 7.Enne, V. I., D. M. Livermore, P. Stephens, and L. M. Hall. 2001. Persistence of sulphonamide resistance in Escherichia coli in the UK despite national prescribing restriction. Lancet 357:1325-1328. [DOI] [PubMed] [Google Scholar]
- 8.Espinal, M. A., A. Laszlo, L. Simonsen, F. Boulahbal, S. J. Kim, A. Reniero, S. Hoffner, H. L. Rieder, N. Binkin, C. Dye, R. Williams, M. C. Raviglione, et al. 2001. Global trends in resistance to antituberculosis drugs. N. Engl. J. Med. 344:1294-1303. [DOI] [PubMed] [Google Scholar]
- 9.Gillespie, S. H. 2002. Evolution of drug resistance in Mycobacterium tuberculosis: clinical and molecular perspective. Antimicrob. Agents Chemother. 46:267-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gillespie, S. H., L. L. Voelker, and A. Dickens. 2002. Evolutionary barriers to quinolone resistance in Streptococcus pneumoniae. Microb. Drug Resist. 8:79-84. [DOI] [PubMed] [Google Scholar]
- 11.Kiers, A., A. P. Drost, D. van Soolingen, and J. Veen. 1997. Use of DNA fingerprinting in international source case finding during a large outbreak of tuberculosis in the Netherlands. Int. J. Tuberc. Lung Dis. 1:239-245. [PubMed] [Google Scholar]
- 12.Morlock, G. P., B. B. Plikaytis, and J. T. Crawford. 2000. Characterization of spontaneous, rifampin-resistant mutants of Mycobacterium tuberculosis strain H37Rv. Antimicrob. Agents Chemother. 44:3298-3301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nagaev, I., J. Björkman, D. I. Andersson, and D. Hughes. 2001. Biological cost and compensatory evolution in fusidic acid-resistant Staphylococcus aureus. Mol. Microbiol. 40:433-439. [DOI] [PubMed] [Google Scholar]
- 14.Ohno, H., H. Koga, S. Kohno, T. Tashiro, and K. Hara. 1996. Relationship between rifampin MICs for and rpoB mutations of Mycobacterium tuberculosis strains isolated in Japan. Antimicrob. Agents Chemother. 40:1053-1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Paul, S., P. Laochumroonvorapong, and G. Kaplan. 1996. Comparable growth of virulent and avirulent Mycobacterium tuberculosis in human macrophages in vitro. J. Infect. Dis. 174:105-112. [DOI] [PubMed] [Google Scholar]
- 16.Pym, A. S., B. Saint-Joanis, and S. T. Cole. 2002. Effect of katG mutations on the virulence of Mycobacterium tuberculosis and the implication for transmission in humans. Infect. Immun. 70:4955-4960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qian, L., C. Abe, T.-P. Lin, M.-C. Yu, S.-N. Cho, S. Wang, and J. T. Douglas. 2002. rpoB genotypes of Mycobacterium tuberculosis Beijing family isolates from east Asian countries. J. Clin. Microbiol. 40:1091-1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ramaswamy, S., and J. M. Musser. 1998. Molecular genetic basis of anti-microbial agent resistance in Mycobacterium tuberculosis. Tuberc. Lung Dis. 79:3-29. [DOI] [PubMed] [Google Scholar]
- 19.Reynolds, M. G. 2000. Compensatory evolution in rifampin-resistant Escherichia coli. Genetics 156:1471-1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sander, P., B. Springer, T. Prammananan, A. Sturmfels, M. Kappler, M. Pletschette, and E. C. Böttger. 2002. Fitness cost of chromosomal drug resistance-conferring mutations. Antimicrob. Agents Chemother. 46:1204-1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schrag, S. J., and V. Perrot. 1996. Reducing antibiotic resistance. Nature 381:120-121. [DOI] [PubMed] [Google Scholar]
- 22.Schrag, S. J., V. Perrot, and B. R. Levin. 1997. Adaptation to the fitness costs of antibiotic resistance in Escherichia coli. Proc. R. Soc. London B 264:1287-1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sherman, D. R., K. Mdluli, M. J. Hickey, T. M. Arain, C. E. Barry III, and K. C. Stover. 1996. Compensatory ahpC gene expression in isoniazid-resistant Mycobacterium tuberculosis. Science 272:1641-1643. [DOI] [PubMed] [Google Scholar]
- 24.Sjölund, M., K. Wreiber, D. I. Andersson, M. J. Blaser, and L. Engstrand. 2003. Long-term persistence of resistant Enterococcus species after antibiotics to eradicate Helicobacter pylori. Ann. Intern. Med. 139:483-487. [DOI] [PubMed] [Google Scholar]
- 25.Somoskovi, A., L. M. Parsons, and M. Salfinger. 2001. The molecular basis of resistance to isoniazid, rifampin, and pyrazinamide in Mycobacterium tuberculosis. Respir. Res. 2:164-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Telenti, A., P. Imboden, F. Marchesi, D. Lowrie, S. T. Cole, M. J. Colston, L. Matter, K. Schoolfer, and T. Bodmer. 1993. Detection of rifampicin-resistant mutations in Mycobacterium tuberculosis. Lancet 341:647-650. [DOI] [PubMed] [Google Scholar]
- 27.Victor, T. C., P. D. van Helden, and R. Warren. 2002. Prediction of drug resistance in M. tuberculosis: molecular mechanisms, tools, and applications. IUBMB Life 53:231-237. [DOI] [PubMed] [Google Scholar]
- 28.Werngren, J., and S. E. Hoffner. 2003. Drug-susceptible Mycobacterium tuberculosis Beijing genotype does not develop mutation-conferred resistance to rifampin at an elevated rate. J. Clin. Microbiol. 41:1520-1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wichelhaus, T. A., B. Böddinghaus, S. Besier, V. Schäfer, V. Brade, and A. Ludwig. 2002. Biological cost of rifampin resistance from the perspective of Staphylococcus aureus. Antimicrob. Agents Chemother. 46:3381-3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang, B., H. Koga, H. Ohno, K. Ogawa, M. Fukuda, Y. Hirakata, S. Maesaki, K. Tomono, T. Tashiro, and S. Kohno. 1998. Relationship between antimycobacterial activities of rifampicin, rifabutin and KRM-1648 and rpoB mutations of Mycobacterium tuberculosis. J. Antimicrob. Chemother. 42:621-628. [DOI] [PubMed] [Google Scholar]
- 31.Yuen, L. K., D. Leslie, and P. J. Coloe. 1999. Bacteriological and molecular analysis of rifampin-resistant Mycobacterium tuberculosis strains isolated in Australia. J. Clin. Microbiol. 37:3844-3850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang, M., J. Gong, Y. Lin, and P. F. Barnes. 1998. Growth of virulent and avirulent Mycobacterium tuberculosis strains in human macrophages. Infect. Immun. 66:794-799. [DOI] [PMC free article] [PubMed] [Google Scholar]