Abstract
Previous studies showed that several miRNAs can regulate pathways involved in UVB-induced premature senescence and response to ultraviolet irradiation. It has also been reported that miR-34c-5p may be involved in senescence-related mechanisms. We propose that miR-34c-5p may play a crucial role in senescence of normal human primary dermal fibroblasts. Here, we explored the roles of miR-34c-5p in UVB-induced premature senescence on dermal fibroblasts. MiR-34c-5p expression was increased in dermal fibroblasts after repeated subcytotoxic UVB treatments. Underexpression of miR-34c-5p in dermal fibroblasts led to a marked delay of many senescent phenotypes induced by repeated UVB treatments. Furthermore, underexpression of miR-34c-5p in dermal fibroblasts can antagonize the alteration of G1-arrested fibroblasts. Moreover, E2F3, which can inactivate p53 pathway and play a role in cell cycle progression, is a down-stream target of miR-34c-5p. Forced down-expression of miR-34c-5p decreased the expression of UVB-SIPS induced P21 and P53 at both mRNA and protein levels. Our data demonstrated that down-regulation of miR-34c-5p can protect human primary dermal fibroblasts from UVB-induced premature senescence via regulations of some senescence-related molecules.
Keywords: miR-34c-5p, UVB, premature senescence, human skin fibroblasts.
1. Introduction
Premature senescence of human dermal fibroblasts can be induced by exposures to a variety of oxidative stress and DNA damaging agents. Debacq-Chainiaux F. et al. developed a robust model of UVB-induced premature senescence of human dermal fibroblasts (UVB-SIPS)1. This model represents an alternative in vitro model in photoaging research for investigating photoaging-related mechanisms2.
MicroRNAs (miRNAs) have emerged recently as a new class of small evolutionarily conserved non-coding RNAs that negatively regulate gene expression. Several miRNAs have been shown to be involved in the regulation of pathways involved in SIPS and response to ultraviolet irradiation3, 4. MiR-34c-5p has been reported to mediate growth arrest and upregulation of the percentage of aging cells in a variety of cell types5. Kyle Lafferty-Whyte et al. also reviewed that miR-34c had the potential to regulate all 4 kinds of senescence induction types (replicative senescence, oxidative stress, oncogene expression, and DNA damage signaling). Their study highlights future potential of miR-34c as novel drug targets for senescence induction6.
Two mature miRNA species are derived from the miR-34c precursor, namely miR-34c-5p and miR-34c-3p. However, they have different seed sequences that regulate different targets. In the present study, we found that miR-34c-5p was overexpressed in UVB-SIPS fibroblasts by means of miRNA microarray, and further confirmed by real-time quantitative PCR (qRT-PCR). In consideration of the functions of miR-34c mentioned above, we speculate that miR-34c-5p might play very important roles in cellular senescence, which needs further study.
2. Experimental Section
Cultivation of primary human dermal fibroblasts and UV light source
Primary human dermal fibroblasts were obtained from four Chinese donors aged 8-12 years by means of a foreskin circumcision, and cells were cultured by the same method previously used 7. For the experiments, human dermal fibroblasts used were between passage 2-4. The source of UVB was BLE-1T158 (Spectronics Corp., Westbury, NY, USA). A Kodacel filter (TA401/407, Kodak, Rochester, USA) was used to block wavelengths of less than 290 nm (ultraviolet C). The UVB dosage was quantified using a Waldmann UV meter (model no. 585100: Waldmann Co., VS-Schwenningen, Germany).
Induction of UVB-SIPS and SA-β-gal activity detection
25mJ/cm2 UVB was performed twice a day for 5 days. Control cells were kept in the same culture conditions without UVB exposure. At 48 hours after the last stress, the SA-β-gal activity was assessed with SA-β-gal Staining kit (Biotime, Haimen, China) by using the method originally described by Oh et al8.
RNA isolation and miRNA microarray
Total RNA isolation and miRNA enrichment were performed with a mirVana miRNA Isolation Kit (Ambion, Austin, TX, USA) according to the manufacturer's instructions. RNA concentration was quantified with a NanoDrop spectrophotometer (Thermo Fisher, Waltham, MA, USA). RNA integrity was evaluated using an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). RNA labeling and hybridization on the Agilent miRNA microarray chips were performed with an miRNA Labeling Reagent and Hybridization Kit (Agilent Technologies, Santa Clara, CA, USA) at 37 °C for 30 min.
Each total RNA sample (100 ng) was treated with calf intestine alkaline phosphatase (TaKaRa, Dalian, China), denatured using 100% dimethyl sulfoxide (Sigma, Taufkirchen, Germany) at 100 °C for 8 min in a thermal cycler, and then transferred to an ice-water bath to prevent RNA from re-annealing. The RNA samples were then labeled with pCp-Cy3 using T4 RNA ligase (Ambion, Austin, TX, USA) via incubation at 16 °C for 2 h. The labeled samples were hybridized to Agilent human miRNA microarrays, which contained probes for 1223 miRNAs cataloged in the Sanger Cambridge database v10.1 (http://microrna.sanger.ac.uk). Hybridizations were performed in SureHyb chambers (Agilent Technologies, Santa Clara, CA, USA) for 24 h at 55 °C. The microarrays were then washed using Agilent-prepared buffers.
The microarray images were scanned with an Agilent microarray scanner, gridded, and analyzed using Agilent Feature Extraction Software version 9.5.1 (Santa, Clara, CA, USA). Normalization was performed using the per-chip median normalization method and the median array 9.
Reverse-transcription real-time PCR analysis
Expression of individual miRNAs was detected by qRT-PCR using miRNA sequence-specific primers (Applied Biosystems, Foster City, CA). Briefly, 10 ng of total RNA was reverse-transcribed using a High-Capacity cDNA Archive kit (Applied Biosystems) followed by amplification in an ABI 7500 Real-Time PCR System (Applied Biosystems, Santa Clara, CA, USA). All RT-PCRs were performed in triplicate. The small nucleolar RNA U6 was used as an endogenous control for the normalization of RNA input. MiRNA expression levels were calculated by relative quantification using ABI 7500 Real-Time PCR SDS 1.2 software (Applied Biosystems, Santa Clara, CA, USA), and the fold change of expression |log2(Control / UVB-SIPS)| ≥ 2 were considered as significantly changes. The PCR reaction without template served as a negative control.
Design and subcloning of short hairpin siRNA template into lentiviral vector
A third generation of self-inactivating lentivirus vector containing a CMV-driven GFP reporter and a H1 promoter upstream of cloning restriction sites (ClaI and mluI) to allow the introduction of oligonucleotides encoding short hairpin RNAs (shRNAs) was provided by Genechem (Shanghai, China). The hairpin consists of a T, a 21 nt sense sequence, a short spacer (TTCAAGAGA), an antisense sequence, 6 Ts (a stop signal for RNA polymerase III) and an mluI site. Oligos were annealed and inserted between the mluI and ClaI sites of the plasmid. The shDNA sequence for constructing lentiviruses against human pre-miR-34c-5p (Lenti-siR-miR-34c-5p) was 5′- AGGCAGUGUAGUUAGCUGAUUGC-3′ and 5′- AAUCACUAACCACACGGCCAGG-3'. Correct insertions of shRNA cassettes were confirmed by restriction mapping and direct DNA sequencing.
Lentivirus production
Recombinant lentiviruses were produced by co-transfecting 293T cells with the lentivirus expression plasmid and packaging plasmids using calciumphosphate method. Infectious lentiviruses were harvested at 48 and 72 h post transfection, centrifuged to eliminate cell debris, and then filtered through 0.22-μm cellulose acetate filters. Infectious titer was determined by fluorescence-activated cell sorting analysis of green fluorescent protein (GFP) positive in 293T cells. Virus titers were in the range of 108 transducing units/mL medium.
Lentiviral vector transductions
On the day of transduction, NHSFs were replated at 2×104 cells/well in 12-well plates along with recombinant lentivirus Lenti-siR-miR-34c-5p at different multiplicities of infection (MOIs) in serum-free growth medium containing 5 mg/mL polybrene at 37 °C and 5% CO2. After 4 h, serum containing growth medium was added to the cells, and there was complete replacement of growth medium after 48 h. GFP expression was examined by fluorescent microscopy after 8 days. NHSFs were plated in six-well plates at a density of 1×105 cells per well and transfected with Lenti-siR-miR-34c-5p at an MOI of 100 as described above. Empty virus vector control plasmids was employed as vector control and equal volume saline as negative control.
Assay for E2F3 over-expression
Eukaryote pIRES-E2F3 expression vector was generated by inserting the open reading frame of E2F3a cDNA into the Cla I/EcoR I site of the pIRES1 neo vector (Clontech Laboratories, Inc., Mountain View, CA, USA). Fibroblasts were seeded into 6-well plates at a concentration of 0.5×105/well and transfected with 0.5 μg/well of pIRES1 neo as a control vector or pIRES-E2F3 expression vector by using Lipofectamine RNAiMAX. The effects manifested by E2F3 over-expression were assayed at 7 days after the transfection with plasmids.
Cell cycle detection
Fibroblasts were fixed with 70% alcohol, washed twice with PBS, digested with RNase, and stained with propidium iodide (PI). A flow cytometer (FAC-Scan, BD, NJ, USA) was used to gather data and images, to analyze the cell cycle, and to calculate the percentage of cells in the G1 phase.
Bioinformatic analysis of miR-34c-5p target genes
Putative miR-34c-5p targets were predicted using several different algorithms, including TargetScan (http://www.targetscan.org/), Pictar (http://pictar.bio.nyu.edu/) and miRanda (http://microrna.sanger.ac.uk/). An interaction between miR-34c-5p and the 3' UTR of its target gene was predicted by RNAhybrid (http://bibiserv.techfak.uni-bielefeld.de/rnahybrid/).
Dual-luciferase assay
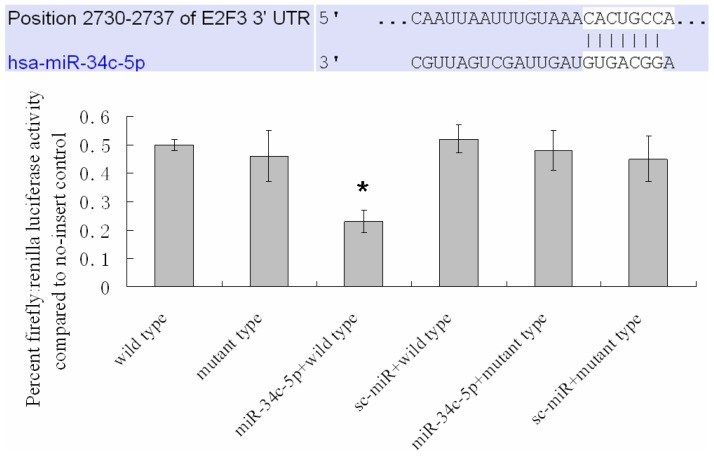
The 3' UTR of human E2F3, with one predicted miR-34c-5p binding site mutations (2730-2737, Figure 4), was inserted into the multicloning site of the pMir-Luc-target vector plasmid (Shanghai Bioladder Co., Ltd, Shanghai, China). The miR-34c-5p expression clone (miR-34c-5p) was constructed downstream a CMV promoter, and a luciferase assay was performed (Luc-Pair miR Luciferase Assay Kit; GeneCopoeia). The cells were plated in six wells and incubated until 70% confluent and transfected with one of the following 6 combinations: (1) E2F3 3' UTR (2730-2737); (2) E2F3 3' UTR with miR-34c-5p 2730-2737 seed-matching mutation (E2F3 3' UTR (2730-2737) mt); (3) E2F3 3' UTR (2730-2737) + sc-miR (miRNA scramble control); (4) E2F3 3' UTR (2730-2737) mt + sc-miR; (5) E2F3 3' UTR (2730-2737) + miR-34c-5p; (6) E2F3 3' UTR (2730-2737) mt + miR-34c-5p. The cells were transferred to a 96-well plate 18 hours after transfection and cultured for another 24 h. Both firefly luciferase and Renilla luciferase activities were determined in the NHSFs. Firefly luciferase activity was then normalized with Renilla luciferase activity in the same well.
Figure 4.
MiR-34c-5p targets the E2F3 3' UTR. Mutation of a predicted miR-34c-5p binding site in the E2F3 3' UTR (2730-2737) abrogated the repressive effect of miR-34c-5p on the luciferase activity of a E2F3 3' UTR-luciferase reporter. Results are shown as means±SD (n = 3). *P < 0.05 versus wild type.
Detection of E2F3, P21 WAF-1, and P53 expression
The primer sequence and size of PCR products of target genes are listed in Table 1. U6 primers were used as internal RNA loading and amplification controls. Primary antibodies against E2F3 (Santa Cruz Biotechnology, Santa Cruz, CA), p21WAF-1 (Cell Signaling Technology, California, USA), P53 (Cell Signaling Technology, California, USA), and β-actin (Biotime, Haimen, China) were used in western blot analysis.
Table 1.
The primer sequence of target genes.
| Gene name | primer sequences | |
|---|---|---|
| E2F3 | Forward primer: Reverse primer: |
5`- CGGTCATCAGTACCTCTCAGA-3` 5`- AGACGTATCATACCGCGTTTTT -3` |
| p53 | Forward primer: Reverse primer: |
5`- CCGCAGTCAGATCCTAGCG -3` 5`- AATCATCCATTGCTTGGGACG -3` |
| p21 | Forward primer: Reverse primer: |
5`- CCTCATCCACCCCATCCCT -3` 5`- GTCCATAGCCTCTACTGCCA -3` |
| GAPDH | Forward primer: Reverse primer: |
5`-TGTTGCCATCAATGACCCCTT-3` 5`-CTCCACGACGTACTCAGCG -3` |
| MiR-34c-5p | Forward primer: | 5`-AGGCAGTGTAGTTAGCTGATTGC-3` |
| U6 | Forward primer: Reverse primer: |
5`-ATTGGAACGATACAGAGAAGATT-3` 5`-GGAACGCTTCACGAATTTG-3` |
Statistical analysis
All analyses were performed with SPSS 13.0 (SPSS Corporation, Chicago, CA, USA). Statistical significance of multiple treatments was determined by analysis of variance (ANOVA) test. P <0.05 was considered to be significant.
3. Results and Discussion
In order to screen the differentially expressed miRNA in UVB-SIPS, we compared the miRNA expression pattern in UVB-SIPS with untreated control in dermal fibroblast. In total, 8 miRNAs were up-regulated and 5 miRNAs were down-regulated after a total of 250 mJ/cm2 UVB irradiation at 48 hours after last irradiation (Table 2).
Table 2.
Differently expressed miRNAs in UVB-induced premature senescence of human dermal fibroblasts
| miRNAs | Normalization | log2 (Control / UVB-SIPS) | |
|---|---|---|---|
| Control | UVB-SIPS | ||
| hsa-miR-1224-3p | 419.357776 | 1692.797989 | 2.013156 |
| hsa-miR-197-3p | 2149.208603 | 10773.654995 | 2.32563 |
| hsa-miR-1976 | 1103.044789 | 4707.197797 | 2.093377 |
| hsa-miR-23a-5p | 1033.895368 | 4379.935474 | 2.082819 |
| hsa-miR-34c-5p | 639.074483 | 2801.90345 | 2.132351 |
| hsa-miR-4701-5p | 658.034808 | 3367.663795 | 2.355512 |
| hsa-miR-574-5p | 327.902091 | 1370.018711 | 2.062859 |
| hsa-miR-766-3p | 1964.066606 | 8691.728655 | 2.145799 |
| hsa-miR-1185-1-3p | 1599.359178 | 311.123359 | -2.361935 |
| hsa-miR-1185-2-3p | 1502.326927 | 262.706468 | -2.515675 |
| hsa-miR-4638-5p | 1734.31208 | 375.679215 | -2.20679 |
| hsa-miR-4695-5p | 5210.743432 | 979.097142 | -2.411965 |
| hsa-miR-933 | 9073.073163 | 1658.726843 | -2.451515 |
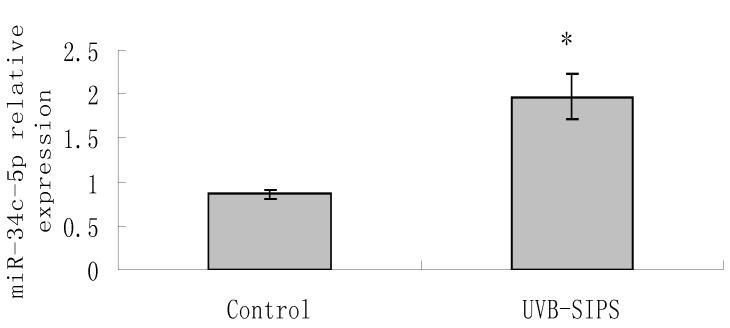
MiR-34c-5p expression was further confirmed to be increased significantly (P < 0.05) after UVB treatment by qRT-PCR detection (Figure 1). Then, we transfected Lenti-siR-miR-34c-5p or Empty Virus Vector into fibroblasts and named them as fibroblasts-siR-miR-34c-5p and fibroblasts-vector, respectively. We used qRT-PCR to confirm the expression level of miR-34c-5p (Figure 2).
Figure 1.
qRT-PCR confirmation of miR-34c-5p expression in UVB-induced premature senescence of human dermal fibroblasts. Results are shown as means±SD (n = 3). *P<0.05 compared with control group. Bars, mean ± standard error.
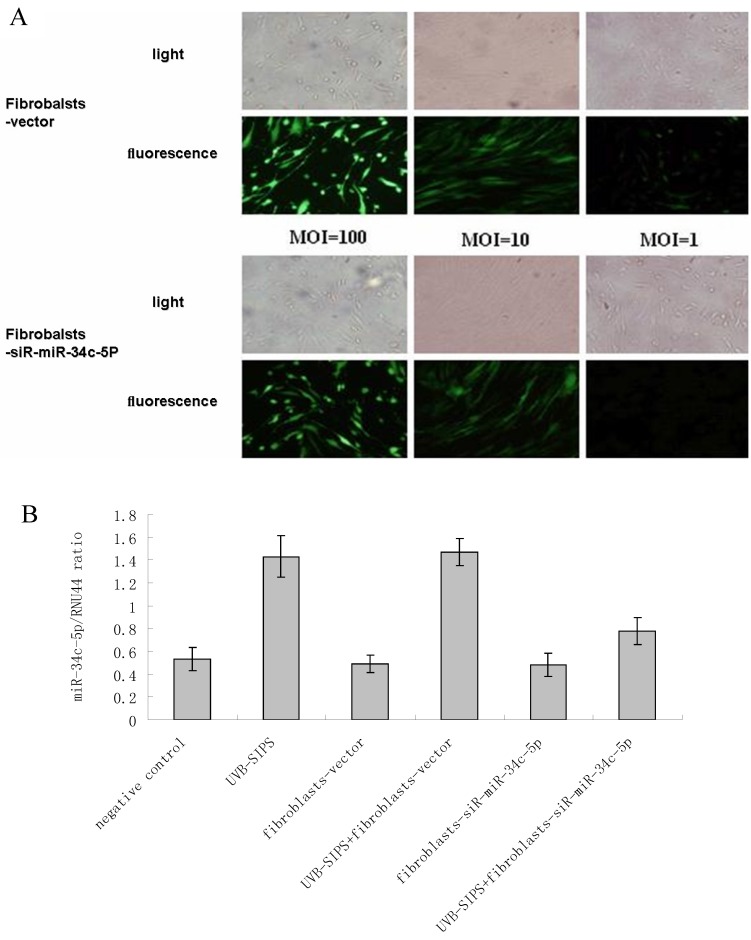
Figure 2.
Determination of lentiviral transduction efficiency and lentivirus-mediated miR-34c-5p down-expression in fibroblasts. (A) Transduction efficiency was estimated 8 days after infecting at indicated MOIs. GFP expression was observed under light microscopy (up), fluorescence microscopy (down). The highest GFP expression of NHSFs appeared with MOI of 100. (B) MiR-34c-5p was underexpressed by lentivirus transfection in dermal fibroblasts and confirmed by qRT-PCR. Results are shown as means±SD (n = 3).
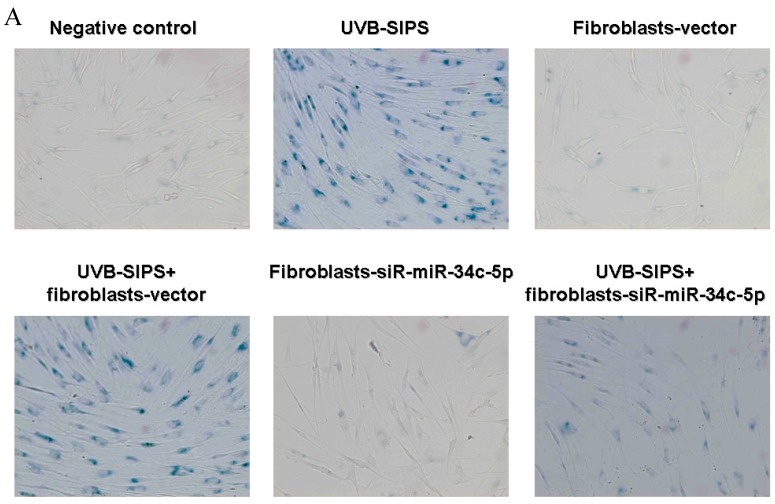
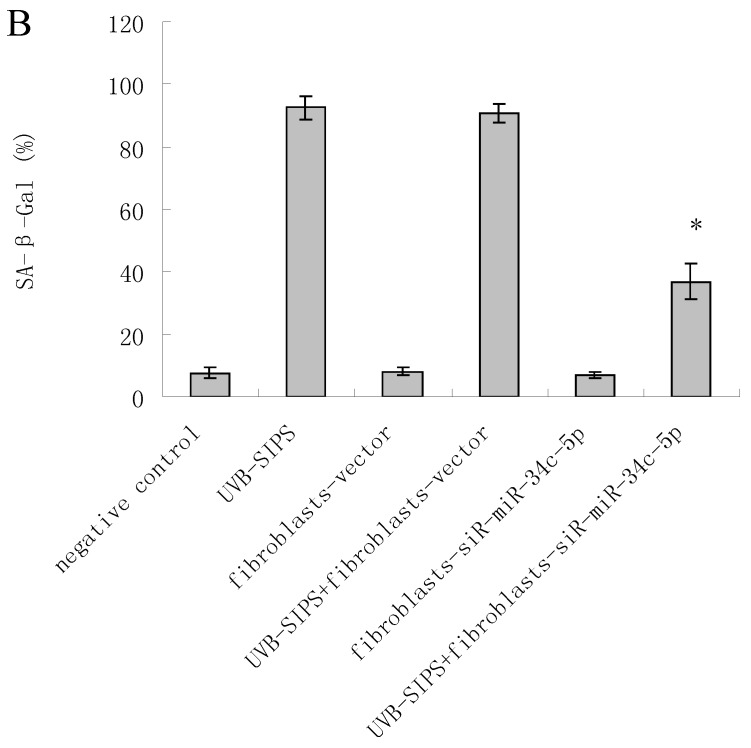
To explore the role of miR-34c-5p in cellular senescence induced by UVB, fibroblasts-vector and fibroblasts-siR-miR-34c-5p were treated with repeated UVB treatments. As expected, after treatment, fibroblasts-vector irradiated cells became enlarged and irregular in shape, and the percentage of SA-β-gal positive cells increased (Figure 3b; P < 0.05. fibroblasts-vector: 8.03±1.32%, fibroblasts-vector+UVB-SIPS: 90.83±2.92%). However, underexpression of miR-34c-5p alleviated the morphological changes induced by repeated UVB treatments (Figure 3a). There were less SA-β-gal positive cells in fibroblasts-siR-miR-34c-5p than that in fibroblasts-vector after UVB treatment (Figure 3b; P < 0.05. fibroblasts-siR-miR-34c-5p+UVB-SIPS: 36.88±5.53%). Our data provided evidence that the down-regulation of miR-34c-5p in human dermal fibroblasts can delay UVB-induced premature senescence. In good agreement with our report, Kumamoto et al. showed that after the administration of the MDM2 inhibitor, Nutlin-3, to human diploid fibroblasts, induction of miR-34c as well as senescence was observed10.
Figure 3.
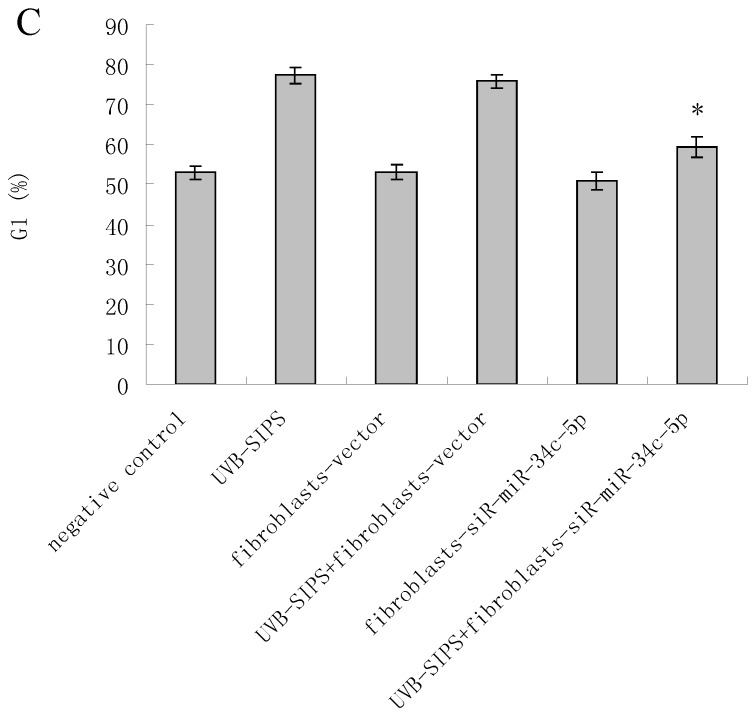
MiRNA underexpression delayed premature senescence induced by repeated subcytotoxic UVB treatments in normal human dermal fibroblasts. (a) The SA-β-gal staining in fibroblasts-vector and fibroblasts-siR-miR-34c-5p after repeated UVB treatments. (b) The SA-β-gal positive cells were increased significantly in non-transfected and fibroblasts-vector after repeated UVB treatments, and there were less SA-β-gal positive cells in fibroblasts-siR-miR-34c-5p than that in fibroblasts-vector after repeated UVB treatments. Results are shown as means±SD (n = 3). (c) The G1 phase cells were increased significantly in non-transfected and fibroblasts-vector after repeated UVB treatments, and there were less G1 phase cells in fibroblasts-siR-miR-34c-5p than that in fibroblasts-vector after repeated UVB treatments. Results are shown as means±SD (n = 3). *P < 0.05, compared with UVB-SIPS and UVB-SIPS+fibroblasts-vector.
To assess whether the over-expression of miR-34c-5p affects the G1 arrest in fibroblasts after UVB-SIPS, the percentage of cells in G1 phase were measured. The level of G1 phase was increased in fibroblasts-vector+UVB-SIPS. However, down expression of miR-34c-5p antagonized the above change (Figure 3c).
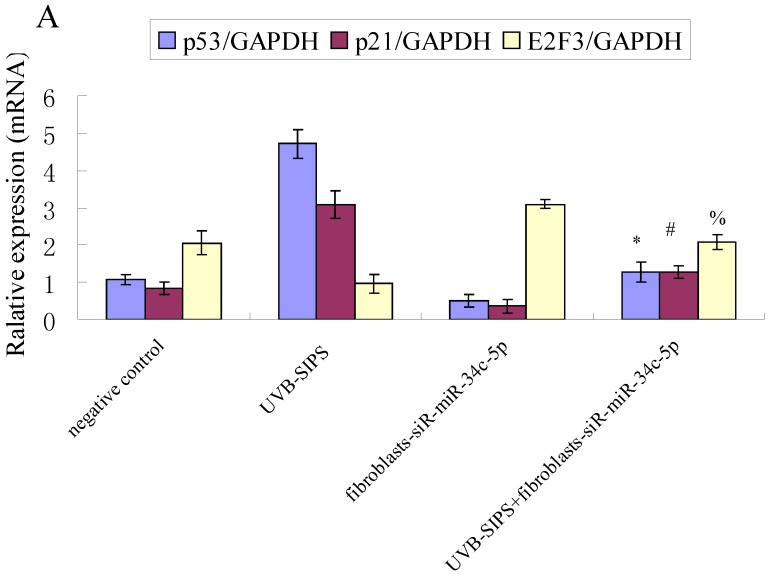
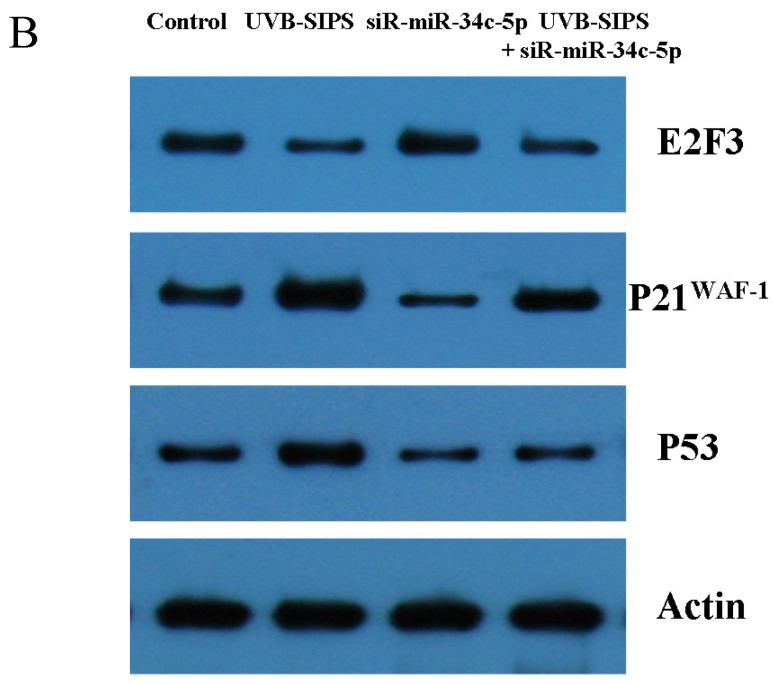
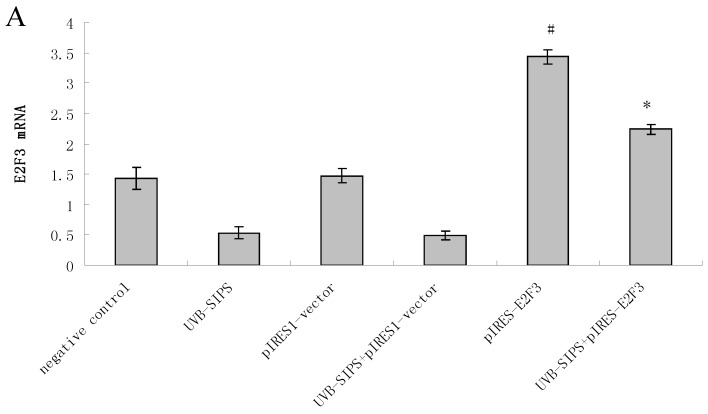
Compared with classical transcriptional factors, miRNAs act mainly via regulation of their target genes at the mRNA level. For example, regulation of microtubule-associated protein tau by miR-34c-5p determines the chemosensitivity of gastric cancer to paclitaxel 11. miR-34c-5p directly targets sGCbeta under hypoxia 12. Gluconeogenesis is severely compromised in hepatocellular carcinoma (HCC) by IL6-Stat3-mediated activation of miR-23a, which directly targets PGC-1α and G6PC, leading to decreased glucose production 13. With the help of current bioinformatics software, we proposed that E2F3, which play an essential role in cell cycle progression, proliferation, and development, is the target of miR-34c-5p. Moreover, dual-luciferase assay analysis indicated that the predicted 2730-2737 binding site was functional in vitro (Figure 4). To further confirm the effect of miR-34c-5p on E2F3 expression, we detected the mRNA and protein levels of E2F3 under miR-34c-5p down-expression condition after UVB-SIPS. We found that the expression of E2F3 decreased significantly in UVB-SIPS and then they were rescued by miR-34c-5p down-expression in fibroblasts-siR-miR-34c-5p after repeated UVB treatments (Figure 5; P < 0.05). These results indicated that E2F3 is a real down-stream target of miR-34c-5p.
Figure 5.
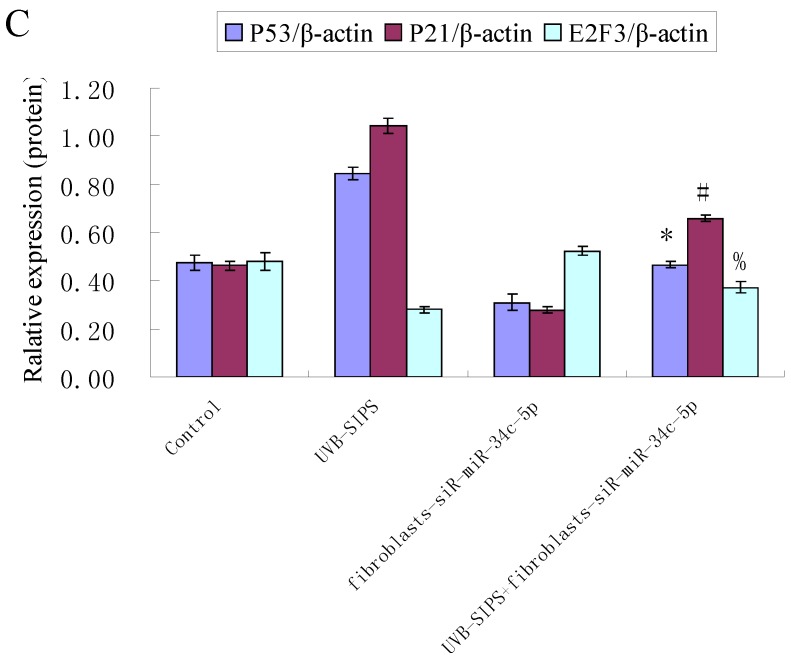
Effect of miR-34c-5p downexpression on E2F3, p21WAF-1 and P53 expression in UVB-SIPS fibroblasts. (a) qRT-PCR analysis of E2F3, p21WAF-1 and P53 in control, UVB-SIPS, fibroblasts-siR-miR-34c-5p and UVB-SIPS+ fibroblasts-siR-miR-34c-5p. Results are shown as means±SD (n = 3). (b) Western blotting analysis of E2F3, p21WAF-1 and P53 level in control, UVB-SIPS, fibroblasts-siR-miR-34c-5p and UVB-SIPS+ fibroblasts-siR-miR-34c-5p. (c) Blots from triplicate experiments were scanned, and densitometric values corrected for actin were plotted as mean±SD. *P < 0.05, #P < 0.05, %P < 0.05 compared with UVB-SIPS.
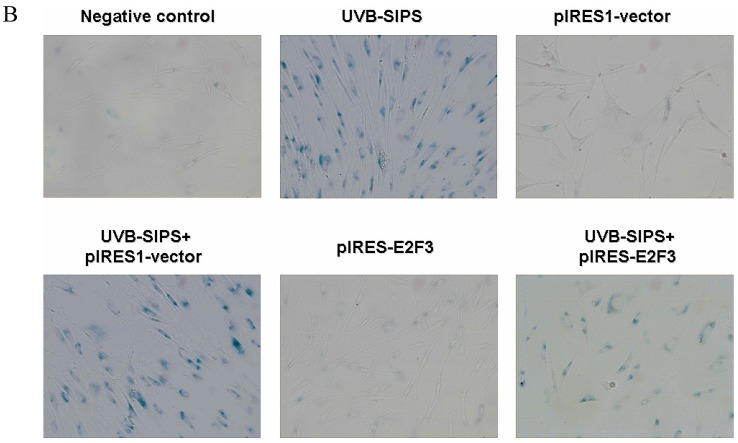
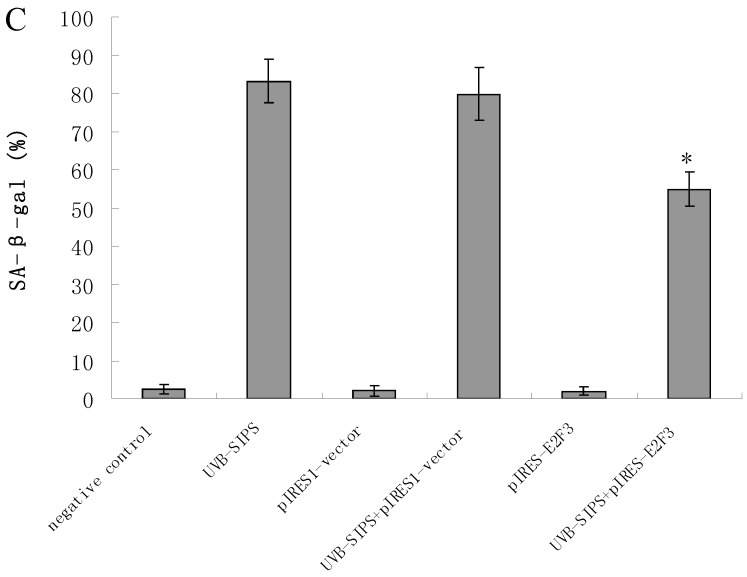
It is reported that activation of the p53 pathway could be caused as a consequence of the repression of the E2F3 as in the case of fibroblast with knockout of E2F3 14-16. It is well-established that the p53 pathway is involved in aging and photoaging 17. Active p53 is known to trigger p21WAF-1 overexpression 18. P21WAF-1 is a cyclin dependent kinase inhibitor that blocks the cell cycle in G1 phase 19, which corresponds to the cell cycle analysis results. Supporting these assumptions, we confirmed that UVB-SIPS increased the expression of p21WAF-1 and P53 expression in cultured fibroblasts. To further investigate the mechanism of miR-34c-5p in UVB-induced senescence, we explored the effects of miR-34c-5p on p21WAF-1 and P53 expression in UVB-SIPS fibroblasts. We found that the expression of p21WAF-1 and P53 were decreased by miR-34c-5p under-expression in fibroblasts-siR-miR-34c-5p after repeated UVB treatments (Figure 5; P < 0.05). Moreover, over-expressed E2F3, which is confirmed to be the direct target of miR-34c-5p, can rescue fibroblasts from UVB induced premature senescence. As indicated by our results, there were less SA-β-gal positive cells in pIRES-E2F3 than that in pIRES1-vector after UVB treatment (Figure 6c; P < 0.05, UVB-SIPS+pIRES-E2F3: 54.82±4.55%). In light of our results, it seems reasonable to conclude that decreased levels of miR-34c-5p may contribute to up-regulated E2F3 which sequentially lead to inactivation of p53-p21 pathway thus interfering with UVB-SIPS induced senescence. It has been shown that active p53 directly induces miR-34c-5p expression 20, 21. In this consideration, miR-34c-5p in turn indirectly increases p53 activity, thus establishing a positive feedback loop leading to cell-cycle arrest. However, additional experiments will be required to confirm this interpretation.
Figure 6.
Enforced expression of E2F3 delayed premature senescence induced by repeated subcytotoxic UVB treatments in normal human dermal fibroblasts. (a) E2F3 was over-expressed by pIRES-E2F3 vector transfection in dermal fibroblasts after repeated UVB treatments and confirmed by qRT-PCR. (b) The SA-β-gal staining in pIRES-E2F3 or pIRES1-vector transfected fibroblasts after repeated UVB treatments. (c) The SA-β-gal positive cells were increased significantly in non-transfected and pIRES1-vector after repeated UVB treatments, and there were less SA-β-gal positive cells in pIRES-E2F3 transfected fibroblasts than that of in pIRES1-vector group after repeated UVB treatments. Results are shown as means±SD (n = 3). #P < 0.05, compared with pIRES1-vector and negative control. *P < 0.05, compared with UVB-SIPS and UVB-SIPS+pIRES1-vector.
The major damage of photoaged skin is related to the connective tissue of the dermal compartment with quantitative and qualitative alterations of the dermal extracellular matrix 22. It has been reported that following chronic ultraviolet stress treatment, the expression of interstitial collagenase/matrix metalloproteinase-1 is induced. This induction is prolonged in the period of growth arrest phase in dermal fibroblasts, while tissue inhibitor of metalloproteinase-1, the major inhibitor of matrix-metalloproteinase-1, is only slightly induced 23. This imbalance between matrix-degrading metalloproteinases and their inhibitors may lead to connective tissue damage, a hallmark of premature aging. These findings, together with our observation that miR-34c-5p plays an important role in cellular senescence, suggesting that miR-34c-5p may also be involved in photoaging of the skin.
4. Conclusions
We found that several miRNAs, including miR-34c-5p, are dysregulated in UVB-SIPS, and down-regulation of miR-34c-5p can delay senescence of human dermal fibroblasts induced by UVB treatment. Further study elucidate that E2F3 is the downstream target of miR-34c-5p. E2F3 regulated P53-P21 pathway play a role in regulating the procedure of UVB-SIPS. We believe that the relationships of miR-34c-5p and some senescence-related proteins, and the exact mechanisms of miR-34c-5p in senescence might be a novel and potent way to study cellular senescence and photoaging.
Acknowledgments
This work was supported by grant from the China National Natural Science Foundation (81000700 and 81171518), science project from traditional Chinese medicine Bureau of Jiangsu Province (LZ11084), and Jiangsu National Natural Science Foundation (BK2012877 and BK2012168).
References
- 1.Debacq-Chainiaux F, Borlon C, De Hertogh B, Remacle J, Morvan PY, Vallee R, Toussaint O. Identification of potential anti-photoageing algal compounds using an in-vitro model of photoageing. J Pharm Pharmacol. 2006;58(12):1577–83. doi: 10.1211/jpp.58.12.0003. [DOI] [PubMed] [Google Scholar]
- 2.Chainiaux F, Magalhaes JP, Eliaers F, Remacle J, Toussaint O. UVB-induced premature senescence of human diploid skin fibroblasts. Int J Biochem Cell Biol. 2002;34(11):1331–9. doi: 10.1016/s1357-2725(02)00022-5. [DOI] [PubMed] [Google Scholar]
- 3.Zhou BR, Xu Y, Permatasari F, Liu WL, Li W, Guo XF, Huang QH, Guo Z, Luo D. Characterization of the miRNA profile in UVB-irradiated normal human keratinocytes. Exp Dermatol. 2012;21(4):317–9. doi: 10.1111/j.1600-0625.2012.01465.x. [DOI] [PubMed] [Google Scholar]
- 4.Li G, Luna C, Qiu J, Epstein DL, Gonzalez P. Alterations in microRNA expression in stress-induced cellular senescence. Mech Ageing Dev. 2009;130(11-12):731–41. doi: 10.1016/j.mad.2009.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.He L, He X, Lim LP, de Stanchina E, Xuan Z, Liang Y, Xue W, Zender L, Magnus J, Ridzon D, Jackson AL, Linsley PS, Chen C, Lowe SW, Cleary MA, Hannon GJ. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447(7148):1130–4. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lafferty-Whyte K, Cairney CJ, Jamieson NB, Oien KA, Keith WN. Pathway analysis of senescence-associated miRNA targets reveals common processes to different senescence induction mechanisms. Biochim Biophys Acta. 2009;1792(4):341–52. doi: 10.1016/j.bbadis.2009.02.003. [DOI] [PubMed] [Google Scholar]
- 7.Zhou BR, Xu Y, Wu D, Permatasari F, Gao YY, Luo D. Ginsenoside Rg1 protects human fibroblasts against psoralen- and UVA-induced premature senescence through a telomeric mechanism. Arch Dermatol Res. 2012;304(3):223–8. doi: 10.1007/s00403-012-1221-9. [DOI] [PubMed] [Google Scholar]
- 8.Oh S, Lee E, Lee J, Lim Y, Kim J, Woo S. Comparison of the effects of 40% oxygen and two atmospheric absolute air pressure conditions on stress-induced premature senescence of normal human diploid fibroblasts. Cell Stress Chaperones. 2008;13(4):447–58. doi: 10.1007/s12192-008-0041-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu CG, Calin GA, Meloon B, Gamliel N, Sevignani C, Ferracin M, Dumitru CD, Shimizu M, Zupo S, Dono M, Alder H, Bullrich F, Negrini M, Croce CM. An oligonucleotide microchip for genome-wide microRNA profiling in human and mouse tissues. Proc Natl Acad Sci USA. 2004;101(26):9740–4. doi: 10.1073/pnas.0403293101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kumamoto K, Spillare EA, Fujita K, Horikawa I, Yamashita T, Appella E, Nagashima M, Takenoshita S, Yokota J, Harris CC. Nutlin-3a activates p53 to both down-regulate inhibitor of growth 2 and up-regulate mir-34a, mir-34b, and mir-34c expression, and induce senescence. Cancer Res. 2008;68(9):3193–203. doi: 10.1158/0008-5472.CAN-07-2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu H, Huang M, Lu M, Zhu W, Shu Y, Cao P, Liu P. Regulation of microtubule-associated protein tau (MAPT) by miR-34c-5p determines the chemosensitivity of gastric cancer to paclitaxel. Cancer Chemother Pharmacol. 2013 May;71(5):1159–71. doi: 10.1007/s00280-013-2108-y. [DOI] [PubMed] [Google Scholar]
- 12.Xu X, Wang S, Liu J, Dou D, Liu L, Chen Z, Ye L, Liu H, He Q, Raj JU, Gao Y. Hypoxia induces downregulation of soluble guanylyl cyclase beta1 by miR-34c-5p. J Cell Sci. 2012;125(Pt 24):6117–26. doi: 10.1242/jcs.113381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang B, Hsu SH, Frankel W, Ghoshal K, Jacob ST. Stat3-mediated activation of microRNA-23a suppresses gluconeogenesis in hepatocellular carcinoma by down-regulating glucose-6-phosphatase and peroxisome proliferator-activated receptor gamma, coactivator 1 alpha. Hepatology. 2012;56(1):186–97. doi: 10.1002/hep.25632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sharma N, Timmers C, Trikha P, Saavedra HI, Obery A, Leone G. Control of the p53-p21CIP1 Axis by E2f1, E2f2, and E2f3 is essential for G1/S progression and cellular transformation. J Biol Chem. 2006;281(47):36124–31. doi: 10.1074/jbc.M604152200. [DOI] [PubMed] [Google Scholar]
- 15.Wu L, Timmers C, Maiti B, Saavedra HI, Sang L, Chong GT, Nuckolls F, Giangrande P, Wright FA, Field SJ, Greenberg ME, Orkin S, Nevins JR, Robinson ML, Leone G. The E2F1-3 transcription factors are essential for cellular proliferation. Nature. 2001;414(6862):457–62. doi: 10.1038/35106593. [DOI] [PubMed] [Google Scholar]
- 16.Timmers C, Sharma N, Opavsky R, Maiti B, Wu L, Wu J, Orringer D, Trikha P, Saavedra HI, Leone G. E2f1, E2f2, and E2f3 control E2F target expression and cellular proliferation via a p53-dependent negative feedback loop. Mol Cell Biol. 2007;27(1):65–78. doi: 10.1128/MCB.02147-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kosmadaki MG, Gilchrest BA. The role of telomeres in skin aging/photoaging. Micron. 2004;35(3):155–9. doi: 10.1016/j.micron.2003.11.002. [DOI] [PubMed] [Google Scholar]
- 18.Li GZ, Eller MS, Firoozabadi R, Gilchrest BA. Evidence that exposure of the telomere 3' overhang sequence induces senescence. Proc Natl Acad Sci USA. 2003;100(2):527–31. doi: 10.1073/pnas.0235444100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sherr CJ, Roberts JM. CDK inhibitors: positive and negative regulators of G1-phase progression. Genes Dev. 1999;13(12):1501–12. doi: 10.1101/gad.13.12.1501. [DOI] [PubMed] [Google Scholar]
- 20.Corney DC, Hwang CI, Matoso A, Vogt M, Flesken-Nikitin A, Godwin AK, Kamat AA, Sood AK, Ellenson LH, Hermeking H, Nikitin AY. Frequent downregulation of miR-34 family in human ovarian cancers. Clin Cancer Res. 2010;16(4):1119–28. doi: 10.1158/1078-0432.CCR-09-2642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hermeking H. The miR-34 family in cancer and apoptosis. Cell Death Differ. 2010;17(2):193–9. doi: 10.1038/cdd.2009.56. [DOI] [PubMed] [Google Scholar]
- 22.Yaar M, Gilchrest BA. Skin aging: postulated mechanisms and consequent changes in structure and function. Clin Geriatr Med. 2001;17(4):617–30. doi: 10.1016/s0749-0690(05)70089-6. [DOI] [PubMed] [Google Scholar]
- 23.Wlaschek M, Ma W, Jansen-Durr P, Scharffetter-Kochanek K. Photoaging as a consequence of natural and therapeutic ultraviolet irradiation--studies on PUVA-induced senescence-like growth arrest of human dermal fibroblasts. Exp Gerontol. 2003;38(11-12):1265–70. doi: 10.1016/j.exger.2003.09.006. [DOI] [PubMed] [Google Scholar]