Abstract
Summary: The Malaria Genome Exploration Tool (MaGnET) is a software tool enabling intuitive ‘exploration-style’ visualization of functional genomics data relating to the malaria parasite, Plasmodium falciparum. MaGnET provides innovative integrated graphic displays for different datasets, including genomic location of genes, mRNA expression data, protein–protein interactions and more. Any selection of genes to explore made by the user is easily carried over between the different viewers for different datasets, and can be changed interactively at any point (without returning to a search).
Availability and Implementation: Free online use (Java Web Start) or download (Java application archive and MySQL database; requires local MySQL installation) at http://malariagenomeexplorer.org
Contact: joanna.sharman@ed.ac.uk or dgerloff@ffame.org
Supplementary information: Supplementary data are available at Bioinformatics online.
Genome sequencing projects targeting malarial parasite species have revealed how little is still known. At least 40% of the ∼5770 genes in Plasmodium falciparum, the most deadly human parasite, remain annotated as ‘unknown function’ according to PlasmoDB (Aurrecoechea et al., 2009). Alongside, large-scale experiments with functional genomic technologies support attempts to shed light on the processes in which all these genes are involved. The use of computational tools helps biologists and bioinformaticians find clues in the emerging data, towards the functions of uncharacterized gene products they pursue experimentally. Tools that enable exploration/browsing-style analysis are particularly appreciated in the biologist community. With increasing volume and types of functional genomics data generously shared by the laboratories that produce them, user interface preferences will also differ, e.g. non-experts (including pupils in science class) will embrace clarity and intuitiveness, while annotators will be accustomed to dense and comprehensive displays.
The readiness of malaria biologists to share data has inspired many valuable community resources alongside the main genomic repository for Plasmodium, PlasmoDB (www.plasmodb.org). Several of these provide derived and related information including the results of local computational annotation and predictions, e.g. at the DFCI Gene Index site (Quackenbush et al., 2001), PlasmoDraft (Brehelin et al., 2008) and MalPort (Joubert et al., 2009). Some resources also emphasize genomic context in coloured linear displays using the familiar ‘genome browser’ formats, e.g. the Malaria Genome Browser at the University of California, Santa Cruz (UCSC) (Chakrabarti et al., 2007) which provides richly annotated versions of such displays. However, navigation of the data hosted at these, or similar, resources is generally limited to a traditional single-gene-at-a-time perspective. Although this is excellent for some uses, investigating groups of genes seems vital, e.g. when investigating how gene products may work together as a system. The currently common formats for multiple-gene-information are large indexed tables and/or textual/link annotation accompanying individual genes. This can be quite effective for screening, e.g. via the PlasmoDB/EuPathDB search interface that allows users to screen secondary information to make more complex selections, but much room for improvement remains, particularly for the laboratory biologist end user (who is not also an annotator). We developed the Malaria Genome Exploration Tool (MaGnET) primarily to enable users to try out a more deeply integrated new user interface, and multiple-gene navigation through highly visual displays (Fig. 1). Some displays were developed specifically to capture relevant aspects of malaria biology (e.g. time-series viewing of mRNA expression, see below), but most of MaGnET's innovative aspects are transferable, and hopefully adoptable by other developers.
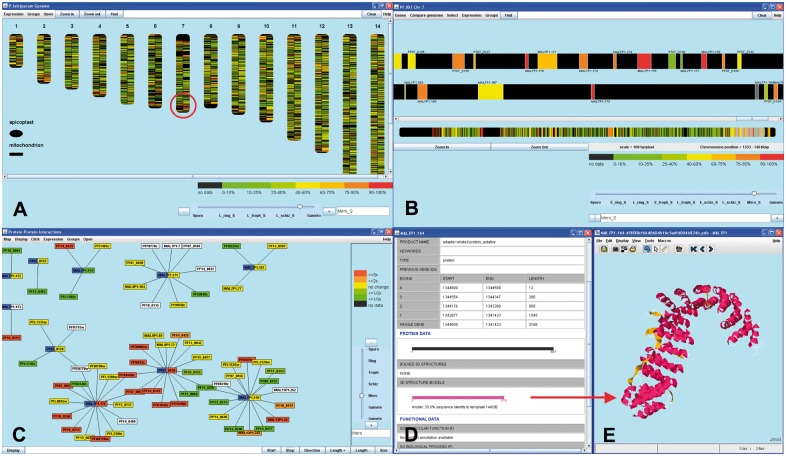
Fig. 1.
MaGnET screenshots. (A) Genome Viewer displaying mRNA expression at the merozoite stage of the P.falciparum 3D7 life cycle (Le Roch et al., 2003). Genes in the circled region were noted to be highly expressed at this stage. (B) Chromosome Viewer display of the circled region on Chromosome 7. (C) Protein–Protein Interaction Viewer showing primary interactions of the selected gene products (highlighted by partly blue labels); the directionality of change in protein abundance from the previous life cycle stage (Le Roch et al., 2004) is indicated by label colour. (D) Gene fact sheet for one gene from (B) with genomic and structural information. (E) Double clicking on the model bar (magenta) opens the 3-D structure in Jmol (http://www.jmol.org)
‘Exploring’ the information for multiple genes is most productive when the interfaces to different types of data (including those that cannot be displayed in linear format, e.g. protein–protein interactions) are integrated not only in navigation but also visually, e.g. by overlaying information from one type of data (e.g. gene expression) onto visual displays of another data type. It is difficult to effectively integrate, and display, some higher-level data types (e.g. protein–protein interaction, mRNA expression experiments and pathway information) in the familiar browser templates. Accordingly some malaria resources that offer more specialized information for P.falciparum, e.g. PlasmoID (Rao et al., 2009) (primarily protein–protein interactions), plasmoMAP (Date and Stoeckert, 2006) (pathways) and MPMP (Ginsburg, 2006) (pathways), provide specifically developed displays for individual data types. This imposes limitations on how much integration between different data types is offered to the users visually. Some nice examples of displays integrating several types of functional genomics data exist in resources that are not organism-specific. These include primarily viewers that can map expression data (or other features) onto molecular pathway displays, e.g. the Omics viewer in the Pathway Tools package (Karp et al., 2002), GenMAPP (Mlecnik et al., 2005) and the PathwayExplorer web service for mapping of user-owned expression data (Mlecnik et al., 2005). From a user perspective, it would be desirable to emphasize integrated visualization much more strongly and systematically than is realized in current publicly available software and across more different types of experimental functional genomics data.
Previous work in our group yielded a more integrated visualization tool for the budding yeast Saccharomyces cerevisiae, YETI (Yeast Exploration Tool Integrator) (Orton et al., 2004). Here we present a similar development for model strain of the malaria-causing parasite P.falciparum 3D7: the Malaria Genome Exploration Tool (MaGnET). MaGnET offers a simple and highly visual ‘workbench’-style environment, as a supplement to existing public genome databases. Additionally, some biologists may appreciate analyzing preliminary or confidential data prior to depositing it at a public repository.
MaGnET consists of a light-weight Java programme for effective visualization linked to a MySQL database. It can be used in two different ways: (i) online as a Java Web Start version that connects to an online database server. Two database mirrors are currently available at UCSC and the University of Edinburgh (further mirrors are planned); (ii) as a stand-alone version where both the Java programme and the database are run (and administered) by the user (Supplementary Fig. S1). MaGnET requires Java Runtime Environment 1.6 or higher on Windows, Mac OSX and UNIX systems; performance is best with Java Runtime Environment 1.7 and ≥4 GB of RAM.
MaGnET features specifically designed visualization of different types of functional genomic data. Its four main viewers are:
The Genome Viewer visualizes the location of genomic features. Groups of selected genes can be viewed in their genomic context on a genome schematic showing the 14 nuclear chromosomes of P. falciparum, its mitochondrial and its plastid chromosome. Advanced functionality allows to overlay gene expression from stored data; users can ‘step through’ a time series via a scroll bar and follow changes during the parasite’s life cycle (Fig. 1A). Zooming in on a chromosome allows to closely examine the genes within a region of interest (Fig. 1B).
The Protein–Protein Interaction Viewer allows to explore networks of proteins predicted to interact by yeast two-hybrid experiments. Maps containing all primary interactions for selected proteins of interest are easily created and expanded to include secondary interactions and so forth. Similarly to above, protein and mRNA expression data from a selected experiment can be overlaid visually onto the interaction network (Fig. 1C).
The Expression Data Viewer visualizes time-series expression profiles for individual genes/proteins and small groups. Time-series graphs are constructed using the JFreeChart library (http://www.jfree.org/jfreechart). This section also features a Query Builder utility allowing complex querying.
The Data Analysis Viewer enables keyword-based searches of the database, e.g. by Gene Ontology term. A list of matching genes is returned in an interactive table. The whole table, or parts of it, can be carried forward for exploration.
All viewers are screen-size windows, to maximize their graphics and (customizable) use of colours to overlay information from other data types. Displays update automatically when the gene selections are modified. A navigation pane with clickable thumbnails (‘Window Control Panel’) facilitates switching between viewers. See Supplementary Materials for an overview diagram and data references.
MaGnET makes two steps forward in navigation that we believe are vital for the targeted non-expert users to take better advantage of what is offered by the data, and the stand-alone version enables users with technical knowledge (or support) to include their own data. From a developer's perspective, we also hope that our interface developments are realized also in other resources that feature varied data types. A first advantage of MaGnET over most other tools is the degree of integration over all its viewers. Second, when exploring different datasets, users can carry two selected groups of genes across and modify them interactively [add/remove any gene(s)] in any viewer. More standard but practical features include an informative ‘fact sheet’ that can be brought up at any point (Fig. 1D) with genomic, functional, structural, ortholog and sequence information, and link-outs to other relevant databases (Supplementary Table S1). Through inclusion of the open third-party programme Jmol (www.jmol.org), MaGnET also visualizes 3-dimensional (3D) protein structures or models at one click (Fig. 1E). The MaGnET database collates primarily experimental data from various public Plasmodium resources (e.g. PlasmoDB) and from published functional genomic studies (Supplementary Table S2). Individual data points from large-scale experiments should not be presumed accurate without further corroboration (e.g. the validity of protein interactions may depend on correct refolding). We will aim to include the best quality data currently available in our updates and welcome user input. Like other resources cited above, we include some predicted annotation, e.g. Gene Ontology and InterPro functional assignments from PlasmoDB after careful filtering and labelling. With our non-expert users in mind, we aim to only deliver data of at least reasonable quality, i.e. only conservative predictions (Supplementary Material).
Supplementary Material
ACKNOWLEDGMENTS
We thank Simon Tomlinson (University of Edinburgh) and Richard Orton (University of Glasgow) for helpful input concerning MaGnET’s design and development, and Thomas Juettemann (EBI) for valuable technical support. The University of Edinburgh and UCSC were sites of development, and UCSC remains a data mirror for MaGnET. We are indebted to the providers of the data sources we use to populate the MaGnET database for making their results publicly available, and the anonymous reviewers for their suggestions for software improvements.
Funding: Medical Research Council (PhD studentship to J.L.S); National Institute of General Medical Sciences (R21GM100303 to D.L.G.).
Conflict of Interest: none declared.
REFERENCES
- Aurrecoechea C, et al. PlasmoDB: a functional genomic database for malaria parasites. Nucleic Acids Res. 2009;37:D539–D543. doi: 10.1093/nar/gkn814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brehelin L, et al. PlasmoDraft: a database of Plasmodium falciparum gene function predictions based on postgenomic data. BMC Bioinformatics. 2008;9:440. doi: 10.1186/1471-2105-9-440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarti K, et al. Structural RNAs of known and unknown function identified in malaria parasites by comparative genomics and RNA analysis. RNA. 2007;13:1923–1939. doi: 10.1261/rna.751807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Date SV, Stoeckert CJ., Jr Computational modeling of the Plasmodium falciparum interactome reveals protein function on a genome-wide scale. Genome Res. 2006;16:542–549. doi: 10.1101/gr.4573206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginsburg H. Progress in in silico functional genomics: the malaria Metabolic Pathways database. Trends Parasitol. 2006;22:238–240. doi: 10.1016/j.pt.2006.04.008. [DOI] [PubMed] [Google Scholar]
- Joubert F, et al. Discovery: an interactive resource for the rational selection and comparison of putative drug target proteins in malaria. Malar. J. 2009;8:178. doi: 10.1186/1475-2875-8-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karp PD, et al. The Pathway Tools software. Bioinformatics. 2002;18(Suppl. 1):S225–S232. doi: 10.1093/bioinformatics/18.suppl_1.s225. [DOI] [PubMed] [Google Scholar]
- Le Roch KG, et al. Discovery of gene function by expression profiling of the malaria parasite life cycle. Science. 2003;301:1503–1508. doi: 10.1126/science.1087025. [DOI] [PubMed] [Google Scholar]
- Le Roch KG, et al. Global analysis of transcript and protein levels across the Plasmodium falciparum life cycle. Genome Res. 2004;14:2308–2318. doi: 10.1101/gr.2523904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mlecnik B, et al. PathwayExplorer: web service for visualizing high-throughput expression data on biological pathways. Nucleic Acids Res. 2005;33:W633–W637. doi: 10.1093/nar/gki391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orton RJ, et al. YETI: Yeast Exploration Tool Integrator. Bioinformatics. 2004;20:284–285. doi: 10.1093/bioinformatics/btg408. [DOI] [PubMed] [Google Scholar]
- Quackenbush J, et al. The TIGR Gene Indices: analysis of gene transcript sequences in highly sampled eukaryotic species. Nucleic Acids Res. 2001;29:159–164. doi: 10.1093/nar/29.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao A, et al. PlasmoID: A P. falciparum protein information discovery tool. In Silico Biol. 2009;9:195–202. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.