Abstract
The clinical efficacy of tyrosine kinase inhibitors supports the dependence of distinct subsets of cancers on specific driver mutations for survival, a phenomenon called “oncogene addiction.” We demonstrate that PUMA and BIM are the key apoptotic effectors of tyrosine kinase inhibitors in breast cancers with amplification of the gene encoding human epidermal growth factor receptor 2 (HER2) and lung cancers with epidermal growth factor receptor (EGFR) mutants. The BH3 domain containing proteins BIM and PUMA can directly activate the proapoptotic proteins BAX and BAK to permeabilize mitochondria, leading to caspase activation and apoptosis. We delineated the signal transduction pathways leading to the induction of BIM and PUMA by tyrosine kinase inhibitors. Inhibition of the mitogen-activated or extracellular signal–regulated protein kinase kinase (MEK)–extracellular signal–regulated kinase (ERK) pathway caused increased abundance of BIM, whereas antagonizing the phosphoinositide 3-kinase (PI3K)–AKT pathway triggered nuclear translocation of the FOXO transcription factors, which directly activated the PUMA promoter. In a mouse breast tumor model, the abundance of PUMA and BIM was increased after inactivation of HER2. Moreover, deficiency of Bim or Puma impaired caspase activation and reduced tumor regression caused by inactivation of HER2. Similarly, deficiency of Puma impeded the regression of EGFRL858R-driven mouse lung tumors upon inactivation of the EGFR-activating mutant. Overall, our study identified PUMA and BIM as the sentinels that interconnect kinase signaling networks and the mitochondrion-dependent apoptotic program, which offers therapeutic insights for designing novel cell death mechanism–based anticancer strategies.
INTRODUCTION
A major advancement in cancer therapy over the past decade has been a shift in focus from cytotoxic chemotherapy to targeted cancer therapy (1). Targeted cancer therapy is based on the discovery that distinct subsets of cancers are dependent on specific driver mutations to maintain proliferation and survival such that targeting these driver mutations can provide therapeutic benefit (2). This concept of “oncogene addiction” has been supported by the clinical efficacy of selective tyrosine kinase inhibitors, such as imatinib in treating chronic myeloid leukemia, gefitinib or erlotinib in treating non–small cell lung cancer (NSCLC) harboring activating mutations of EGFR, and lapatinib in treating HER2 (human epidermal growth factor receptor 2)–amplified breast cancer. Induction of cancer cell apoptosis is integral to the success of targeted cancer therapy. However, the underlying mechanism concerning apoptosis induction by targeted cancer therapy is not fully elucidated.
The BCL-2 family proteins control a crucial checkpoint of apoptosis at the mitochondria and can be divided into three subfamilies based on homology shared within the four conserved BCL-2 homology domains (BH1 to BH4) and death regulatory activities: (i) multidomain antiapoptotic BCL-2, BCL-XL, and MCL-1; (ii) multidomain proapoptotic BAX and BAK; and (iii) proapoptotic BH3-only molecules (BH3s) (3). Mitochondria play a key role in mammalian apoptosis, a regulated program of cell suicide (4). Multiple apoptotic stimuli, including many conventional chemotherapy and targeted anticancer agents, culminate in permeabilizing the mitochondrial outer membrane (MOM), resulting in the release of proapoptotic factors such as cytochrome c and SMAC into the cytosol to activate caspases. BAX and BAK are essential effectors that permeabilize MOM, whereas antiapoptotic BCL-2, BCL-XL, and MCL-1 preserve mitochondrial integrity (5–7). BH3s interconnect with the upstream apoptotic signals to promote apoptosis—some BH3s directly activate BAX and BAK, including BID, BIM, and PUMA, and others inactivate BCL-2, BCL-XL, and MCL-1, such as BAD and NOXA (5, 8–12). Although BAX and BAK are essential downstream effectors controlling the mitochondrion-dependent cell death program, they need to be activated by “activator” BH3s (6, 7, 10, 12–16). Genetic deletion of Bid, Bim, and Puma prevents the homo-oligomerization of BAX and BAK and thereby cytochrome c–mediated caspase activation in response to diverse death signals (16). Therefore, activator BH3s are the central initiators of apoptosis that interconnect signal transduction pathways to the mitochondrion-dependent death machinery.
The ErbB or epidermal growth factor receptor (EGFR) family of structurally related receptor tyrosine kinases (RTKs) includes EGFR, ErbB2 (also known as HER2), ErbB3 (also known as HER3), and ErbB4 (also known as HER4) (17). Excessive ErbB signaling induced by HER2 amplification in breast cancer or activating mutations of EGFR in NSCLC initiates several signaling cascades, principally the phosphoinositide 3-kinase (PI3K)–AKT–mammalian target of rapamycin (mTOR) and the mitogen-activated or extracellular signal–regulated protein kinase kinase (MEK)–extracellular signal–regulated kinase (ERK) pathways, leading to cell proliferation and survival. In these RTK-addicted cancers, tyrosine kinase inhibitor treatment disrupts signaling of both PI3K-AKT and MEK-ERK pathways, leading to apoptosis. Although both HER2- and EGFR mutant–addicted cancers share a similar repertoire of signaling cascades, HER2-amplified breast cancers appear to rely more on PI3K signaling for survival. Therefore, these breast cancer cells are exquisitely sensitive to PI3K-mTOR or AKT inhibitors (18, 19). In contrast, inhibition of both PI3K-AKT-mTOR and MEK-ERK signaling is necessary to induce apoptosis in EGFR mutant lung cancers (20).
Among the BCL-2 family proteins, BIM plays a central role in gefitinib- and erlotinib-induced apoptosis in EGFR mutant lung cancers and in lapatinib-induced apoptosis in HER2-amplified breast cancers (20–25). Tyrosine kinase inhibitors transcriptionally activate BIM through uncharacterized mechanism(s), which is probably followed by stabilization of BIM protein through inhibition of the MEK-ERK pathway. Phosphorylation of BIM by the kinases ERK and ribosomal protein S6 kinase (RSK) targets BIM for β-TRCP (β-transducing repeat-containing protein)–mediated ubiquitination and subsequent proteasome-mediated degradation (26, 27). Paradoxically, although both tyrosine kinase and MEK inhibitors increase BIM abundance to comparable amounts, only tyrosine kinase inhibitors induce apoptosis in both HER2- and EGFR-addicted cancers (20). Notably, inhibition of the PI3K-AKT-mTOR pathway does not induce BIM. These findings argue that an apoptotic regulator downstream of the PI3K-AKT-mTOR pathway is required to cooperate with BIM to trigger apoptosis. Inhibition of the PI3K-AKT-mTOR pathway apparently either reduces the abundance or activity of a prosurvival protein or increases the abundance or activity of an apoptotic effector. The abundance of MCL-1, a prosurvival protein, has been reported to be decreased upon inhibition of EGFR and PI3K, but not AKT, in some, but not all, EGFR mutant lung cancers (20, 25). In contrast, PI3K-mTOR and AKT inhibitors neither increase the abundance of BIM nor decrease the abundance of MCL-1 in HER2-amplified breast cancers and yet trigger robust apoptosis (20). Although the proapoptotic activity of BAD can be inhibited by AKT (28, 29), overexpression of BAD alone induces limited apoptosis (10). Therefore, how inhibition of the PI3K-AKT axis induces apoptosis in RTK-addicted cancer cells remains unclear.
Here, we demonstrate that PUMA, a BH3-only BCL-2 family protein, is the apoptotic effector that is activated upon inhibition of the PI3K-AKT pathway in both HER2-amplified breast cancers and EGFR mutant lung cancers. Inhibition of the PI3K-AKT signaling axis triggered nuclear translocation of FOXO transcription factors that targeted the PUMA promoter to transactivate PUMA. Knockdown of PUMA impaired tyrosine kinase inhibitor–induced apoptosis in both HER2- and EGFR mutant–addicted cancer cells. Moreover, knockdown of PUMA protected HER2-amplified breast cancer cells from apoptosis triggered by inhibitors of PI3K-mTOR or AKT. Tyrosine kinase inhibitor–mediated induction of BIM was abrogated by constitutively active MEK but not AKT, whereas transactivation of PUMA was blocked by constitutively active AKT but not MEK. These data position BIM and PUMA downstream of the MEK-ERK pathway and the PI3K-AKT pathway, respectively. Induction of both BIM and PUMA was further demonstrated in a doxycycline-inducible HER2/Neu mouse breast tumor model after the withdrawal of doxycycline to reduce the expression of HER2/Neu (30). Moreover, tumors deficient in either Bim or Puma exhibited defects in caspase activation and thereby displayed impaired tumor regression upon the inactivation of HER2/Neu. Similarly, deficiency of Puma impeded the regression of EGFRL858R-driven mouse lung tumors upon inactivation of the EGFR-activating mutant as assessed by magnetic resonance imaging (MRI) (31). Our studies identify BIM and PUMA as two key apoptotic effectors of tyrosine kinase inhibitors that cooperate to activate BAX- and BAK-dependent mitochondrial apoptosis. Given that activator BH3–only molecules BID, BIM, and PUMA are required for a BAD mimetic ABT-737 to kill cells (16), it is conceivable that induction of PUMA will enhance the apoptotic effect of ABT-737. Indeed, the combination of PI3K inhibitors and ABT-737 provided a synergistic or additive effect in killing HER2- or EGFR-addicted cancer cells even after they acquire resistance to tyrosine kinase inhibitors. Our discovery positions BIM and PUMA as a central node that connects the upstream kinase signaling networks to the downstream mitochondrion-dependent death machinery. We expect that these findings will open new avenues and lay a foundation for further development of cell death mechanism–based anticancer therapy.
RESULTS
BIM and PUMA are induced upon HER2 inhibition in HER2-amplified breast cancers to execute apoptosis
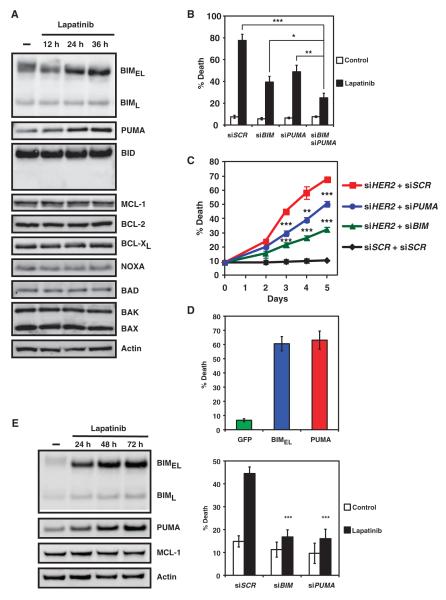
Given that the BCL-2 family proteins control critical checkpoints of intrinsic apoptosis (3), we sought to explore whether and how BCL-2 family proteins are involved in tyrosine kinase inhibitor–induced apoptosis. We are especially interested in identifying the apoptotic effector that is activated upon inhibition of the PI3K-AKT axis. We first examined the effects of inhibition of HER2 on the abundance of BCL-2 family proteins. Lapatinib treatment of HER2-amplified BT474 breast cancer cells resulted in apparent increases in the abundance of BH3-only molecules BIM and PUMA (Fig. 1A). By contrast, the abundance of antiapoptotic members BCL-2, BCL-XL, and MCL-1 or proapoptotic BH3-only molecules BID, NOXA, and BAD did not obviously change (Fig. 1A). In agreement with a previous report (20), MCL-1 degradation was not seen in HER2-addicted cancer cells. To determine the functional importance of BIM and PUMA induction upon HER2 inhibition, we performed small interfering RNA (siRNA)–mediated knockdown of BIM, PUMA, or both to determine whether BIM and PUMA are required for tyrosine kinase inhibitor–induced apoptosis. Indeed, knockdown of BIM or PUMA protected BT474 cells from lapatinib-induced apoptosis (Fig. 1B and fig. S1). Cells with combined knockdown of BIM and PUMA displayed increased resistance to lapatinib-induced apoptosis, compared to those with single knockdown of either BH3-only molecule. To further determine the role of BIM and PUMA in RTK inactivation–induced breast cancer cell death, we examined the impact of BIM or PUMA deficiency on apoptosis induced by knockdown of HER2 in BT474 cells (Fig. 1C and fig. S2). Similar to lapatinib, the full apoptotic response initiated by HER2 depletion required both BIM and PUMA (Fig. 1C).
Fig. 1.
BIM and PUMA are required for lapatinib-induced apoptosis of HER2-amplified breast cancer cells. (A) BT474 cells, mock-treated or treated with lapatinib (0.5 μM), were immunoblotted with the indicated antibodies (n = 3 independent experiments). (B) Quantification of cell death by fluorescence-activated cell sorting (FACS) analysis following annexin V staining of BT474 cells transfected with scramble siRNA (siSCR) or siRNA against BIM and/or PUMA and left untreated or treated with lapatinib. Data are mean percentages of annexin V–positive cells ± SD from three independent experiments. *P < 0.05; **P < 0.01; ***P < 0.001. (C) Quantification of cell death by FACS analysis following propidium iodide staining of BT474 cells transfected with scramble siRNA (siSCR) or siRNA against BIM or PUMA in combination with siRNA against HER2. Data are mean percentages of propidium iodide–positive cells ± SD from three independent experiments. **P < 0.01; ***P < 0.001. (D) BT474 cells were infected with retrovirus expressing green fluorescent protein (GFP), BIMEL, or PUMA, and cell death was quantified by FACS analysis following propidium iodide staining. Data are mean percentages of propidium iodide–positive cells ± SD from three independent experiments. (E) HCC1419 cells, untreated or treated with lapatinib (1 μM), were immunoblotted with the indicated antibodies (left panel, n = 3 independent experiments). Cell death was quantified by FACS analysis after propidium iodide staining after lapatinib treatment (right panel). Data are mean percentages of propidium iodide–positive cells ± SD from three independent experiments. ***P < 0.001.
Exogenous introduction of BIM or PUMA by retrovirus-mediated gene transduction resulted in breast cancer cell death (Fig. 1D). Furthermore, BIM and PUMAwere both induced and required to execute apoptosis upon HER2 inhibition in another HER2-amplified breast cancer cell line, HCC1419 (Fig. 1E). Together, our data support a central role of activator BH3–only molecules BIM and PUMA in HER2 inactivation–induced apoptosis in HER2-addicted breast cancer cells.
BIM and PUMA are key downstream apoptotic effectors that mediate MEK and PI3K-AKT inhibition–induced cell death, respectively
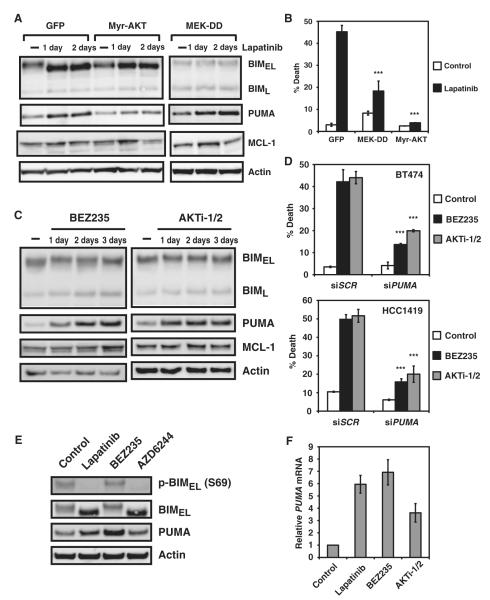
The key signaling pathways downstream of HER2 include the RAS-RAF-MEK-ERK and PI3K-AKT pathways. To molecularly dissect the signaling cascades leading to the increased abundance of BIM and PUMA upon HER2 inhibition, we undertook a gain-of-function approach. Specifically, constitutively active AKT, namely, myristoylated AKT (Myr-AKT), or a constitutively active mutant of MEK (MEK-DD) was overexpressed in BT474 cells to determine their impact on tyrosine kinase inhibitor–induced activation of BIM and PUMA. Lapatinib treatment of BT474 cells reduced the phosphorylation of ERK and AKT, which was abrogated by overexpression of MEK-DD and Myr-AKT, respectively (fig. S3). In MEK-DD–expressing cells, lapatinib treatment resulted in blunted BIM yet apparently normal PUMA induction (Fig. 2A). In contrast, in cells expressing Myr-AKT, lapatinib failed to efficiently induce PUMA, whereas the induction of BIM seemed to be intact (Fig. 2A and fig. S4). These data position BIM and PUMA downstream of the MEK and PI3K-AKT pathways, respectively (Fig. 2A). Consistent with the ineffective induction of either BIM or PUMA in respective MEK-DD and Myr-AKT cells, these cells were resistant to lapatinib-induced apoptosis (Fig. 2B). Hence, the abundance of BIM and PUMA are tightly suppressed by distinct survival signaling cascades in HER2-addicted breast cancers.
Fig. 2.
Inhibition of the MEK-ERK pathway induces BIM, whereas blockade of the PI3K-AKT pathway induces PUMA. (A) BT474 cells stably expressing GFP, Myr-AKT, or a constitutively active mutant of MEK (MEK-DD) were untreated or treated with lapatinib and immunoblotted with the indicated antibodies (n = 2 independent experiments). (B) Quantification of cell death by FACS analysis after propidium iodide staining of BT474 cells stably expressing GFP, Myr-AKT, or a constitutively active mutant of MEK (MEK-DD) that were untreated or treated with lapatinib. Data are mean percentages of propidium iodide–positive cells ± SD from three independent experiments. ***P < 0.001. (C) BT474 cells, untreated or treated with BEZ235 or AKTi-1/2, were immunoblotted with the indicated antibodies (n = 2 independent experiments). (D) Quantification of cell death by FACS analysis after annexin V staining of BT474 cells or propidium iodide staining of HCC1419 cells transfected with scramble siRNA (siSCR) or siRNA against PUMA and left untreated or treated with BEZ235 or AKTi-1/2. Data are mean percentages of annexin V– or propidium iodide–positive cells ± SD from three independent experiments. ***P < 0.001. (E) BT474 cells were untreated or treated with lapatinib, BEZ235, or AZD6244 and immunoblotted with the indicated antibodies (n = 2 independent experiments). (F) PUMA mRNA abundance was assessed in BT474 cells untreated or treated with lapatinib, BEZ235, or AKTi-1/2. Data are normalized against glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and presented as means ± SD of three independent experiments.
Whereas lapatinib treatment induced both BIM and PUMA in HER2-amplified breast cancer cells, our gain-of-function experiments using Myr-AKT and MEK-DD support separable signaling pathways leading to the respective induction of BIM or PUMA. To complement our genetic approaches, we used pathway-specific pharmacological inhibitors (fig. S5). The regulation of PUMA abundance by the PI3K-AKT pathway was further investigated using individual PI3K-AKT pathway inhibitors, including BEZ235 (a dual PI3K-mTOR inhibitor), GDC0941 (a class I PI3K inhibitor), and AKTi-1/2 (an allosteric AKT1/2 inhibitor). All three inhibitors apparently induced PUMA but not BIM, indicating that the abundance of PUMA, but not BIM, is regulated by AKT signaling (Fig. 2C and fig. S6). Moreover, knockdown of PUMA protected BT474 and HCC1419 from both BEZ235- and AKTi-1/2–induced cell death (Fig. 2D). Together, our findings support a thesis that PUMA functions as an important effector mediating the PI3K-AKT inhibitor–triggered apoptosis in HER2-addicted breast cancer cells, linking PUMA regulation and PI3K-AKT signaling.
The observation that lapatinib treatment in MEK-DD cells failed to induce BIM is consistent with previous reports that position BIM downstream of the MEK-ERK pathway (20, 26, 27). Phosphorylation of BIMEL by ERK marks BIMEL for proteasome-mediated degradation (26, 27, 32). However, a report indicated that the in vivo relevance of ERK-regulated degradation of BIMEL in BIM-induced apoptosis appears to be context-dependent (32). Furthermore, ERK inhibition also induces the transcription of BIM (20–25). Accordingly, both lapatinib and AZD6244 treatments resulted in a loss of Ser69 phosphorylation of BIMEL and increased abundance of BIM protein and message (Fig. 2E and fig. S7). However, BIM induction alone under such a scenario is insufficient to trigger apoptosis in HER2-addicted breast cancer cells (20). On the other hand, both BEZ235 and AKTi-1/2 treatments induced PUMA and triggered apoptosis (Fig. 2, C and D), supporting a more prominent role of PUMA in HER2 inactivation–induced breast cancer cell death. Moreover, the increased abundance of PUMA upon treatment with lapatinib, BEZ235, or AKTi-1/2 occurred primarily through transcription induction (Fig. 2, E and F).
The induction of PUMA by HER2 inactivation is transduced through FOXO transcription factors
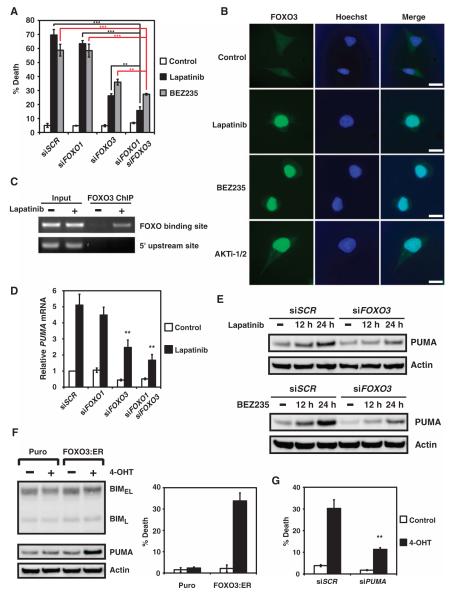
Although PUMAwas first cloned as a transcription target of p53 and functions as a key apoptotic mediator upon p53 activation, subsequent studies have demonstrated that PUMA can also be induced through p53-independent mechanisms to execute apoptosis in response to glucose withdrawal, cytokine withdrawal, and endoplasmic reticulum stress (12, 33, 34). Knockdown of p53 neither prevented lapatinib-mediated induction of PUMA nor protected BT474 cells from lapatinib-induced apoptosis (fig. S8), suggestive of a p53-independent mechanism. Because FOXO3a can transactivate PUMA upon inhibition of the PI3K-AKT pathway in response to cytokine or growth factor deprivation (33), we investigated the potential participation of FOXO1 and FOXO3 in tyrosine kinase inhibitor–mediated PUMA induction. Knockdown of FOXO3 protected against lapatinib-induced apoptosis, and combined knockdown of FOXO3 and FOXO1 offered the most protection (Fig. 3A and fig. S2). AKT phosphorylates FOXO proteins, which suppresses the nuclear localization of FOXO proteins, thereby preventing the transactivation of their downstream targets (35). Inhibition of the PI3K-AKT signaling pathway by lapatinib, BEZ235, or AKTi-1/2 all induced nuclear translocation of FOXO3 (Fig. 3B). Furthermore, chromatin immunoprecipitation (ChIP) assays demonstrated that lapatinib treatment resulted in a direct binding of FOXO3 to its cognate DNA binding sequences within the PUMA promoter (Fig. 3C). Moreover, lapatinib-induced PUMA transcription and thus protein accumulation were largely blunted when FOXO3 was deficient (Fig. 3, D and E). Consistent with these findings, BEZ235-mediated PUMA induction was also compromised in cells with knockdown of FOXO3 (Fig. 3E). To link FOXO3 activation to PUMA induction, we used a 4-hydroxytamoxifen (4-OHT)–inducible, constitutively active mutant of FOXO3 (FOXO3:ER) that targets to the nucleus upon treatment with 4-OHT (36). In response to 4-OHT, FOXO3:ER-expressing BT474 cells appeared to display PUMA, but not BIM induction, and underwent apoptosis (Fig. 3F). Consequently, knockdown of PUMA impaired the ability of FOXO3:ER to induce apoptosis (Fig. 3G). Although FOXO transcription factors can regulate BIM transcription in neurons and hematopoietic cells (37–39), they do not regulate BIM abundance in HER2-amplified breast cancer cells, implicating context-dependent mechanism(s) such as epigenetic control and tissue-specific coactivators or corepressors. Together, we have delineated a PI3K-AKT-FOXO cascade through which PUMA is activated upon inhibition of HER2 or PI3K-AKT.
Fig. 3.
Inhibition of PI3K-AKT triggers nuclear translocation of FOXO and transactivation of PUMA. (A) Quantification of cell death by FACS analysis after annexin V staining of BT474 cells transfected with scramble siRNA (siSCR) or siRNAs against FOXO1, FOXO3, or both and left untreated or treated with lapatinib or BEZ235. Data are mean percentages of annexin V–positive cells ± SD from three independent experiments. **P < 0.01; ***P < 0.001. (B) Fluorescence microscopy of BT474 cells untreated or treated with lapatinib, BEZ235, or AKTi-1/2. Green, FOXO3; blue, Hoechst staining of DNA (n = 2 independent experiments). Scale bars, 25 μm. (C) BT474 cells, untreated or treated with lapatinib, were subjected to ChIP with the anti-FOXO3 antibody, followed by polymerase chain reaction (PCR) amplification of the PUMA promoter. A 5′ sequence located 4 kb upstream of the transcription start site of PUMA served as a negative control (n = 2 independent experiments). (D) PUMA mRNA abundance was assessed in BT474 cells that were transfected with scramble siRNA (siSCR) or siRNA against FOXO1 and/or FOXO3 and left untreated or treated with lapatinib. Data are normalized against GAPDH and are means ± SD of three independent experiments. **P < 0.01. (E) BT474 cells, transfected with scramble siRNA (siSCR) or siRNA against FOXO3, were untreated or treated with lapatinib or BEZ235 and immunoblotted with the indicated antibodies (n = 2 independent experiments). (F) Immunoblot analysis of BT474 stable cell lines that were generated with a control retrovirus (puro) or a retrovirus expressing a 4-OHT–inducible, constitutively active mutant of FOXO3 (FOXO3:ER) (left panel). Cell death was quantified by FACS analysis after annexin V staining (right panel). Data are mean percentages of annexin V–positive cells ± SD from three independent experiments. (G) Cell death was quantified by FACS analysis after annexin V staining in BT474 cells stably expressing FOXO3:ER that were transfected with scramble siRNA (siSCR) or siRNA against PUMA and left untreated or treated with 4-OHT. Data are mean percentages of annexin V–positive cells ± SD from three independent experiments. **P < 0.01.
BIM and PUMA are induced upon EGFR inhibition in EGFR mutant lung cancer cells
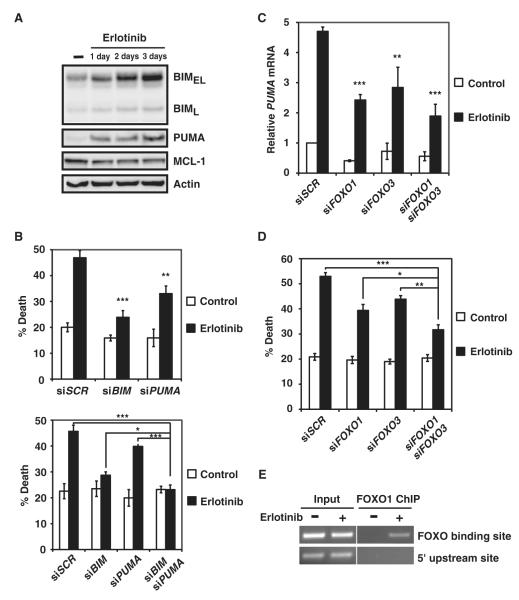
Our study thus far uncovered an important role of PUMA induction in mediating HER2 inactivation–orchestrated apoptosis, which prompted us to investigate whether PUMA also plays a role in EGFR inactivation–induced apoptosis of EGFR-addicted lung cancer cells. Previous reports using EGFR-addicted lung cancer cells have demonstrated the accumulation of BIM in response to gefitinib and erlotinib, two clinically efficacious EGFR inhibitors (21–25). In line with these studies, we observed BIM induction upon EGFR inhibition in HCC827 and PC9 cells (Fig. 4A and fig. S9A). PUMA was also induced in these cells (Fig. 4A and fig. S9A). Notably, the abundance of MCL-1 appeared to be reduced in erlotinib-treated HCC827, but not PC9 cells (20, 25) (Fig. 4A and fig. S9A). The functional importance of PUMA induction by erlotinib in triggering apoptosis was demonstrated by knockdown of PUMA in HCC827 and PC9 cells (Fig. 4B and fig. S9B). Although BIM knockdown conferred stronger resistance to erlotinib-initiated apoptosis than PUMA knockdown, combined deficiency in BIM and PUMA resulted in the most resistance (Fig. 4B and fig. S9B). We then determined the signaling details leading to the PUMA induction in erlotinib-treated lung cancer cells and examined the possibility of a similar AKT-FOXO-PUMA axis operating in EGFR-addicted cells. Indeed, knockdown of FOXO1 or FOXO3 disrupted the erlotinib-mediated PUMA induction and significantly reduced erlotinib-induced apoptosis. Deficiency in both FOXO1 and FOXO3 resulted in the lowest amount of PUMA induction and accordingly offered the best protection (Fig. 4, C and D). It appeared that FOXO1 has a more prominent role in EGFR-addicted lung cancer cells, whereas FOXO3 plays a more important role in HER2-addicted breast cancer cells (Figs. 3 and 4). This selectivity is likely due to the different abundance of FOXO family members among individual cancer cell types. Last, the direct targeting of PUMA promoter by FOXO1 upon erlotinib treatment was demonstrated by ChIP assays (Fig. 4E).
Fig. 4.
BIM and PUMA are required for erlotinib-induced apoptosis of EGFR-addicted lung cancer cells. (A) HCC827 cells, untreated or treated with erlotinib, were immunoblotted with the indicated antibodies (n = 3 independent experiments). (B) Quantification of cell death by FACS analysis after annexin V staining of HCC827 cells transfected with scramble siRNA (siSCR) or siRNA against BIM and/or PUMA and left untreated or treated with erlotinib. Data are mean percentages of annexin V–positive cells ± SD from three independent experiments. *P < 0.05; **P < 0.01; ***P < 0.001. (C) PUMA mRNA abundance was assessed in HCC827 cells transfected with scramble siRNA (siSCR) or siRNA against FOXO1 and/or FOXO3 and left untreated or treated with erlotinib. Data are normalized against GAPDH and are means ± SD of three independent experiments. **P < 0.01; ***P < 0.001. (D) Quantification of cell death by FACS analysis after annexin V staining of HCC827 cells transfected with scramble siRNA (siSCR) or siRNA against FOXO1 and/or FOXO3 and left untreated or treated with erlotinib. Data are mean percentages of annexin V–positive cells ± SD from three independent experiments. *P < 0.05; **P < 0.01; ***P < 0.001. (E) HCC827 cells, untreated or treated with erlotinib, were subjected to ChIP with the anti-FOXO1 antibody, followed by PCR amplification of the PUMA promoter. A 5′ sequence located 4 kb upstream of the transcription start site of PUMA served as a negative control (n = 2 independent experiments).
BIM and PUMA are required for HER2 inactivation–mediated apoptosis in vivo
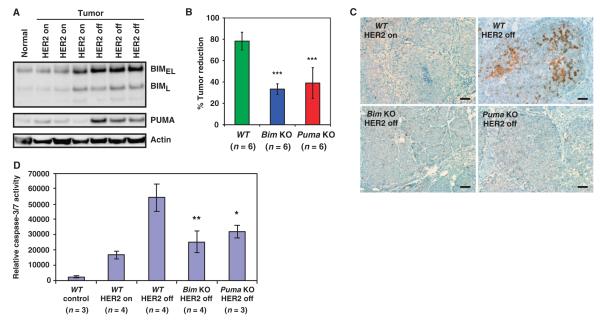
To elucidate the importance of BIM and PUMA induction in HER2 inactivation–induced apoptosis in vivo, we used the MMTV-rtTA; TetO-NeuNT (MTB+TAN+) murine breast cancer model, in which the spatiotemporal expression of HER2/NeuNT (TetO-NeuNT) is controlled by a tetracycline-inducible transcription factor that is specifically expressed in mammary gland (MMTV-rtTA) (30). In the MTB+TAN+ model, HER2/Neu-addicted breast tumors are induced in female mice after the administration of doxycycline in water, and tumor regression is initiated upon the withdrawal of doxycycline (30). Both BIM and PUMA were induced in the HER2/Neu-addicted tumors undergoing regression (Fig. 5A and fig. S10), which concurred with an increased activation of effector caspase-3 and caspase-7 (Fig. 5, B to D). To individually interrogate the role of BIM and PUMA in HER2 inactivation–induced apoptosis and subsequent tumor regression, we performed the same experiments in MTB+TAN+Bim−/− and MTB+TAN+Puma−/− female mice. Bim or Puma deficiency significantly impeded the induction of apoptosis caused by HER2 deinduction as determined by activation of caspases and hence slowed tumor regression (Fig. 5, B and C, and fig. S11). The degree of caspase activation was further quantified in tumors derived from wild-type, Bim knockout, or Puma knockout mice (Fig. 5D). Doxycycline withdrawal to turn off HER2 increased activation of caspase-3 and caspase-7 (caspase-3/7) (Fig. 5D). Consistent with slower tumor reduction, caspase-3/7 activities were significantly reduced in Bim knockout and Puma knockout breast tumors upon doxycycline withdrawal (Fig. 5D). Collectively, these in vivo findings along with our in vitro data demonstrate important roles for both BIM and PUMA in HER2 inactivation–induced apoptosis and tumor regression.
Fig. 5.
BIM and PUMA mediate HER2 inactivation–induced apoptosis in vivo. (A) MTB+TAN+ female mice were administered doxycycline water to induce HER2. When mice developed tumors measuring 1 cm in the longest dimension, doxycycline was withdrawn for 24 hours to turn off HER2. Tumor lysates were immunoblotted with the indicated antibodies (n = 3 independent experiment). A mammary gland from a mouse that was never given doxycycline served as a control. (B) MTB+TAN+ (WT), MTB+TAN+Bim−/− (Bim KO), and MTB+TAN+Puma−/− (Puma KO) female mice were administered doxycycline to induce tumor formation. When mice developed tumors measuring 1 cm in the longest dimension, doxycycline was withdrawn for 3 days and tumor volume was measured using calipers. Data are mean percentages of tumor reduction ± SD. ***P < 0.01. WT, wild type; KO, knockout. (C) Immunohistochemistry for cleaved caspase-3 in tumors obtained from MTB+TAN+ (WT), MTB+TAN+Bim−/− (Bim KO), or MTB+TAN+Puma−/− (Puma KO) female mice 24 hours after doxycycline withdrawal (n = 2 independent experiments). Scale bars, 50 μm. (D) Normal mammary glands or 1-cm tumors obtained from female mice with the indicated genotypes in the presence of doxycycline (HER2 on) or 24 hours after doxycycline withdrawal (HER2 off) were assessed for caspase-3/7 activity using a luciferase-based assay. Data are means ± SD. *P < 0.05; **P < 0.01.
PUMA is required for EGFR inactivation–mediated apoptosis in vivo
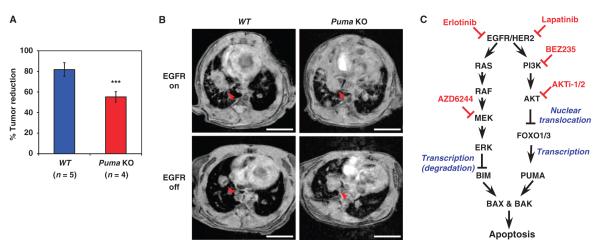
Because the role of BIM in EGFR inactivation–initiated apoptosis has been intensively studied (20–25, 40), we focused on elucidating the importance of PUMA induction in lung tumor regression. To this end, we used the TetO-EGFRL858R; CCSP-rtTA mouse NSCLC model, in which a constitutively active EGFR mutant (EGFRL858R) can be spatiotemporally induced and removed in the lung tissue upon the addition and removal of doxycycline, respectively (fig. S12) (31). The tumor progression and regression were assessed by MRI. The extent of tumor reduction was significantly lower in TetO-EGFRL858R; CCSP-rtTA; Puma−/− (Puma knockout) mice than in TetO-EGFRL858R; CCSP-rtTA (WT) mice (Fig. 6, A and B, and fig. S13), demonstrating the importance of PUMA induction in mediating EGFR inactivation–induced lung cancer cell death.
Fig. 6.
PUMA mediates EGFR inactivation–induced apoptosis in vivo. (A) TetO-EGFRL858R; CCSP-rtTA (WT) or TetO-EGFRL858R; CCSP-rtTA; Puma−/− (Puma KO) mice were fed doxycycline-impregnated food to induce lung tumors. Tumor growth was monitored by MRI. When mice developed tumors measuring 0.5 cm in the longest dimension detected by MRI, doxycycline was withdrawn for 72 hours to turn off EGFRL858R. Tumor reduction was assessed by MRI. Tumor volumes were calculated using ImageJ software. Data are mean percentages of tumor reduction ± SD. ***P < 0.05. (B) Representative magnetic resonance images of lung tumors from TetO-EGFRL858R; CCSP-rtTA (WT) or TetO-EGFRL858R; CCSP-rtTA; Puma−/− (Puma KO) mice before (EGFR on) and after doxycycline withdrawal for 3 days (EGFR off) (n = 4 independent experiments). Arrowheads indicate lung tumors. Scale bars, 5 mm. (C) Signal transduction pathways leading to the activation of BIM and PUMA upon inhibition of EGFR or HER2.
In total, despite the molecular differences between HER2 inactivation–and EGFR inactivation–induced apoptosis, we demonstrated that both BIM and PUMA can be induced through a shared, dual pathway inhibition through in vitro cell culture systems and in vivo mouse tumor models (Fig. 6C). Our studies have delineated the individual signaling pathways responsible for the activation of BIM and PUMA upon tyrosine kinase inhibitor treatment.
PI3K inhibitors and ABT-737 synergize to kill tyrosine kinase inhibitor–resistant cancer cells
Despite the clinical efficacy of gefitinib and erlotinib in treating EGFR mutant lung cancers, there is a wide spectrum of clinical response and benefit among patients (41). Pretreatment abundance of BIM mRNA is a factor that contributes to the heterogeneous clinical response (40). It is conceivable that if the abundance of BIM message is low at baseline, the amount of BIM induced by tyrosine kinase inhibitors will not be sufficient to break the antiapoptotic/proapoptotic BCL-2 rheostat. Our discovery of PUMA as a central apoptotic effector upon inhibition of PI3K-AKT in RTK-addicted cancer cells prompted us to investigate whether targeting the PI3K-PUMA axis can provide an alternative strategy to kill EGFR mutant lung cancers with low abundance of BIM mRNA. PUMA induced by PI3K inhibitors alone cannot alter the balance between antiapoptotic and proapoptotic BCL-2 members because induced PUMA would be sequestered by antiapoptotic BCL-2 members, including BCL-2, BCL-XL, and MCL-1, such that BAX and BAK cannot be activated. We and others have previously demonstrated that BAD and its mimetics ABT-737 or ABT-263 (navitoclax) (also known as BCL-2 and /BCL-XL inhibitors) activate BAX and BAK indirectly by displacing BIM, PUMA, or truncated BID from antiapoptotic BCL-2 and BCL-XL (10, 11, 16, 42, 43). It is conceivable that increased abundance of tBID, BIM, and PUMA could increase the sensitivity of cells to ABT-737. Therefore, the combination of PI3K inhibitors and ABT-737 might effectively induce apoptosis because induced PUMAwould be displaced from BCL-2 and BCL-XL by ABT-737 to activate BAX and BAK. Furthermore, induced PUMA can bind and thereby inactivate MCL-1, which confers resistance to ABT-737.
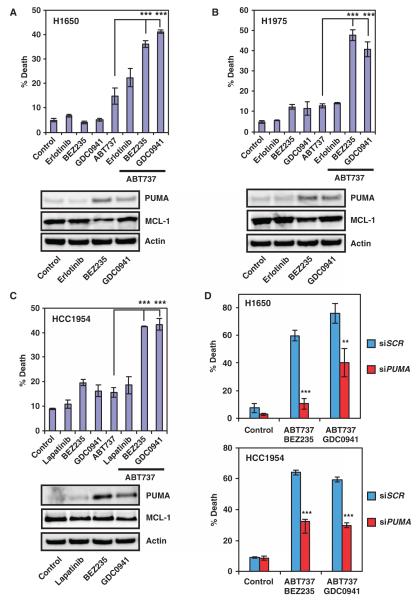
Indeed, both BEZ235 and GDC0941 induced PUMA and synergized with ABT-737 to kill tyrosine kinase inhibitor–resistant H1650 lung cancer cells that have low abundance of BIM mRNA, which is likely because of heterozygous loss of a BIM allele (40) (Fig. 7A). Erlotinib failed to induce PUMA in H1650 cells (Fig. 7A), which might be due to its PTEN (phosphatase and tensin homolog) deficiency. A common mechanism that underlies acquired resistance is the emergence of second-site mutation T790M in the kinase domain of EGFR. Consequently, we tested the combination of PI3K inhibitors and ABT-737 in another tyrosine kinase inhibitor–resistant lung cancer cell line H1975, which harbors both EGFRL858R and EGFRT790M mutations. BEZ235 and GDC0941, but not erlotinib, induced PUMA in these cells (Fig. 7B). In support of our treatment strategy, both BEZ235 and GDC0941 synergized with ABT-737 to kill these cells (Fig. 7B).
Fig. 7.
PI3K inhibitors and ABT-737 synergize to kill tyrosine kinase inhibitor–resistant cancer cells. (A) Quantification of cell death by FACS analysis after annexin V staining of H1650 cells untreated or treated with the indicated chemicals. Protein lysates were immunoblotted with the indicated antibodies (n = 2 independent experiments). (B) Quantification of cell death by FACS analysis after annexin V staining of H1975 cells untreated or treated with the indicated chemicals. Protein lysates were immunoblotted with the indicated antibodies (n = 2 independent experiments). (C) Quantification of cell death by FACS analysis after annexin V staining of HCC1954 cells untreated or treated with the indicated chemicals. Protein lysates were immunoblotted with the indicated antibodies (n = 2 independent experiments). (D) Quantification of cell death by FACS analysis after annexin V staining of H1650 or HCC1954 cells transfected with scramble siRNA (siSCR) or siRNA against PUMA and left untreated or treated with the indicated chemicals. Data are mean percentages of annexin V–positive cells ± SD from three independent experiments. **P < 0.01; ***P < 0.001.
Given the synergistic killing of tyrosine kinase inhibitor–resistant lung cancer cells by PI3K inhibitors and ABT-737, we examined the efficacy of such combination in treating HCC1954 breast cancer cells, which harbor a PIK3CA H1047R mutation along with HER2 amplification. Activating mutations of PI3KCA often make HER2-amplified breast cancer cells less responsive to tyrosine kinase inhibitors. Similarly, both BEZ235 and GDC0941 induced PUMA, and the combination of BEZ235 or GDC0941 with ABT-737 provided an additive effect in killing HCC1954 breast cancer cells (Fig. 7C and fig. S14).
We next investigated whether reduction of MCL-1 abundance contributes to the synergistic or additive effect of ABT-737 and BEZ235 or GDC0941 because inhibition of PI3K can reduce MCL-1 abundance in some cell lines (20). MCL-1 abundance was reduced to a slightly greater extent in H1650 and H1975 lung cancer cell lines by BEZ235 and in the HCC1954 breast cancer cell line by GDC0941 (Fig. 7, A to C). However, we could not find a clear association between the reduction of MCL-1 abundance and the response to the combination therapy. We performed siRNA-mediated knockdown of PUMA, which protected against apoptosis induced by either combination strategy, thus indicating the functional importance of PUMA in this combination therapy (Fig. 7D and fig. S15). Overall, the combination of PI3K inhibitors and ABT-737 offers a novel strategy for the treatment of tyrosine kinase inhibitor–resistant lung or breast cancers.
DISCUSSION
The successful development and application of specific kinase inhibitors underscore the oncogene addiction hypothesis (2). Driven by distinct oncogenes, individual cancer types exhibit preferential dependency on specific signaling cascades. Whereas HER2-amplified breast cancers can be successfully targeted by PI3K-AKT inhibition alone, EGFR mutant NSCLC and K-RAS mutant lung and colon cancers require combination therapy to simultaneously inhibit multiple survival pathways (20, 44, 45). Combining PI3K and MEK inhibitors serves as a common strategy showing efficacy against numerous cancer types. The failure of single-agent therapy has unveiled intrinsic feedback mechanisms, in which inhibition of a single node in a pathway inadvertently strengthens the same or a parallel pathway. For example, inhibition of TORC1 or AKT can result in PI3K activation through increased abundance of RTKs (46, 47). Moreover, activation of mitogen-activated protein kinase (MAPK) signaling occurs upon PI3K/mTORC1 inhibition (48). Consequently, PI3K and MEK combination therapy has been theorized to bypass such negative feedback loops to inhibit both MAPK and PI3K-AKT survival pathways. Yet, how this corresponds to an increase in apoptosis has not been fully elucidated.
Here, we identify BIM and PUMA as key sentinels interconnecting kinase signaling networks and the mitochondrion-dependent apoptotic program (Fig. 6C). The induction of activator BH3–only molecules BID, BIM, and PUMA provides a direct mechanism engaging cell death, as outlined by the BCL-2 hierarchical model (10). Because BIM and PUMA can directly trigger homo-oligomerization of BAX and BAK, leading to MOM permeabilization and cytochrome c release, they constitute a critical node of the BCL-2 network (10, 12, 16). Although their abundance is normally under tight control, the induction of BIM and PUMA through pharmacological means may serve as an effective strategy to eliminate cancer cells. By dissecting the molecular network that connects oncogenic kinases and the BCL-2 family, our work provides a plausible mechanistic explanation for how a combined MAPK and PI3K-AKT inhibitory strategy kills cells. Furthermore, our studies also present an opportunity to further exploit cancer-dependent signaling cascades for anticancer treatment.
The previously characterized role of BIM in EGFR and HER2 inhibition–induced cell death (20–25) is consistent with our in vitro and in vivo studies. However, BIM induction alone is not sufficient to explain oncogene inactivation–induced apoptosis. The MEK inhibitor AZD6244 alone potently increases BIM abundance yet fails to kill HER2- or EGFR-addicted cancer cells (20) (Fig. 2E). Furthermore, PI3K-AKT inhibition can trigger apoptosis in HER2-addicted breast cancer cells without induction of BIM (Fig. 2, C to E, and fig. S2). Although degradation of MCL-1 has been proposed to cooperate with BIM to mediate tyrosine kinase inhibitor–induced apoptosis, it does not hold true for all the EGFR- and HER2-addicted cancers. Similar to MCL-1, the degradation of survivin, a member of the inhibitor of apoptosis protein (IAP) family, has been suggested as a PI3K-dependent mechanism that elicits oncogene inactivation–induced apoptosis (49, 50). However, siRNA-mediated knockdown of survivin was insufficient to induce apoptosis in PI3K inhibitor–sensitive cells, and survivin overexpression did not prevent lapatinib- or BEZ235-induced cell death (50).
Here, we highlight the role of PUMA in both HER2 inactivation– and EGFR inactivation–mediated apoptosis. Our in vitro and in vivo data position PUMA downstream of PI3K-AKT and support PUMA as a critical “missing link” bridging tyrosine kinase inhibitors and BAX- and BAK-dependent apoptosis. Moreover, knockout of Puma impeded both caspase activation and tumor regression in genetically engineered mouse models of HER2 inactivation– and EGFR inactivation–induced apoptosis. We further delineated FOXO transcription factors as the mediators of PUMA induction upon tyrosine kinase inhibitor treatment. Primarily known for their roles in modulating BIM transcription in neurons and hematopoietic cells (37–39), FOXO transcription factors do not regulate BIM abundance in HER2-amplified and EGFR mutant cells, implicating the regulation as cell- or tissue context–dependent processes. Our data indicate that PUMA is largely responsible for the apoptosis observed in HER2-addicted breast cancer cells upon PI3K-AKT inhibition, and MEK-ERK inhibition activates BIM through both transcriptional and posttranslational mechanisms. Together, an efficacious combination of PI3K and MEK inhibitors would result in the activation of both BIM and PUMA via simultaneously suppressing parallel pathways (Fig. 6C).
The BH3 mimetic ABT-737 works in conjunction with targeted agents and chemotherapeutics to kill cancer cells (51). The inclusion of PUMA to the arsenal of death effectors responsible for oncogene inactivation–mediated apoptosis could offer novel strategies for targeting malignant cells. We presented one such strategy: PI3K inhibitors efficiently induce PUMA that works in concert with ABT-737 to trigger apoptosis in tyrosine kinase inhibitor–resistant cancer cell lines (Fig. 7). Our studies not only help further delineate and explain how the BCL-2 family proteins BIM and PUMA execute apoptosis in response to tyrosine kinase inhibition in oncogene-addicted cancer cells, but also provide clues concerning drug resistance mechanisms. With a clearer cell death blueprint that specifically operates in cancer cells such as oncogene addiction, a better and yet less toxic cell death mechanism–based rational design can thus be envisioned and possibly executed to benefit cancer patients.
MATERIALS AND METHODS
Cell lines and reagents
HCC827, NCI-H1650, and NCI-H1975 were obtained from the American Type Culture Collection. BT474 and SKBR3 cells were obtained from R. Bose at Washington University in St. Louis. HCC1419 cells were obtained from M. Ellis at Washington University in St. Louis. HCC1954 cells were obtained from S. Chandarlapaty at Memorial Sloan-Kettering Cancer Center. PC9 cells were obtained from D. Scheinberg at Memorial Sloan-Kettering Cancer Center. BT474, SKBR3, and HCC1954 were maintained in DMEM-F12 (Dulbecco’s modified Eagle’s medium–Ham’s F12) (Invitrogen) supplemented with 10% fetal bovine serum (Invitrogen), glutamine (Invitrogen), and penicillin-streptomycin (Invitrogen). NCI-H1650, NCI-H1975, HCC1419, and PC9 were maintained in RPMI 1640 supplemented with 10% fetal bovine serum, glutamine, nonessential amino acids (Invitrogen), sodium pyruvate (Invitrogen), and penicillin-streptomycin. HCC827 were maintained in IMDM (Iscove’s modified Dulbecco’s medium) (Invitrogen) supplemented with 10% fetal bovine serum, glutamine, nonessential amino acids, sodium pyruvate, and penicillin-streptomycin. Lapatinib, erlotinib, BEZ235, GDC0941, AKTi-1/2, and AZD6244 were obtained from Selleck Chemicals. ABT-737 was obtained from Abbott Laboratories. Inhibitors were used at the following concentrations: lapatinib (0.5 μM for BT474 and HCC1954 and 1 μM for HCC1419), erlotinib (1 μM), BEZ235 (1 μM), GDC0941 (1 μM), AKTi-1/2 (1 μM), AZD6244 (1 μM), and ABT-737 (1 μM).
Plasmids and siRNA
FOXO3(TA)ERT2 construct was obtained from R. DePinho (36). The following siRNA oligos were purchased from Ambion Silencer Select oligos (Applied Biosystems): BIM, 5′-CAACCACUAUCUCAGUGCAtt-3′; PUMA #1, 5′-GCCUGUAAGAUACUGUAUAtt-3′; PUMA #2, 5′-GGAGGGUCCUGUACAAUCUtt-3′; HER2, 5′-GUUGGAUGAUUGACUCUGAtt-3′; FOXO1, 5′-CCAUGGACAACAACAGUAAtt-3′; FOXO3 #1, 5′-GGCUCCUCCUUGUACUCAAtt-3′. The scramble siRNA was purchased from Applied Biosystems (part number 4390844). A second siFOXO3 oligo was purchased from Dharmacon: 5′-UGCAUUAACUUGCGGUAUUtt-3′. siRNA oligos were reversetransfected with Lipofectamine RNAiMAX (Invitrogen) to a final concentration of 10 nM.
Viability assays
Cell death was quantified by annexin V (BioVision) or propidium iodide (Sigma) staining, followed by flow cytometric analyses. Viability assays were performed at the following times: BT474 or HCC1419 treated with lapatinib at 72 hours; BT474 or HCC1419 treated with BEZ235 at 48 or 72 hours, respectively; BT474 treated with GDC0941 at 36 hours; BT474 or HCC1419 treated with AKTi-1/2 at 96 hours; BT474 expressing GFP, BIMEL, or PUMA at 48 hours; BT474 expressing FOXO3:ER treated with 4-OHT at 72 hours; HCC827 or PC9 treated with erlotinib at 72 or 96 hours, respectively; H1650 and H1975 treated with inhibitors at 48 hours; and HCC1954 treated with inhibitors at 24 hours. Flow cytometry was performed with a FACSCalibur or an LSRFortessa (BD Biosciences). Data were analyzed with CellQuest Pro (BD Biosciences), FACSDiva (BD Biosciences), or FlowJo software. P values for statistical analyses were obtained with Student’s t test.
Reverse transcription and quantitative PCR
Total RNA was extracted from cells with TRIzol (Invitrogen) according to the manufacturer’s instruction. Total RNA was harvested at the following times: BT474, HCC1419, and PC9 cells treated with inhibitors at 24 hours; BT474 transfected with siRNA treated with lapatinib at 18 hours; and HCC827 treated with erlotinib at 12 hours. Reverse transcription was performed with oligo-dT plus random decamer primers (Ambion) with SuperScript II (Invitrogen). Quantitative PCR was performed with SYBR Green Master Mix (Applied Biosystems) in duplicate with the indicated gene-specific primers. Quantitative PCR was performed on an ABI Prism 7300 sequence detection system (Applied Biosystems). Data were analyzed as described previously by normalization against GAPDH (52). GAPDH was detected with a rodent-specific GAPDH TaqMan probe (Applied Biosystems). The primers for quantitative reverse transcription PCR are listed below: BIM, 5′-CCAGCACCCATGAGTTGTGACAA-3′ and 5′-GCGTTAAACTCGTCTCCAATACGCC-3′; PUMA, 5′-ACGACCTCAACGCACAGTACG-3′ and 5′-GTAAGGGCAGGAGTCCCATGATG-3′.
Antibodies and immunoblot analysis
Antibodies used for immunoblots are listed as follows: anti-BAK (NT, Millipore), anti-BAX (N-20, Santa Cruz Biotechnology), anti-BIM (Calbiochem), anti–phospho-BIM (Ser69) (Cell Signaling Technology), anti-PUMA (Sigma), anti-BAD (C-20, Santa Cruz Biotechnology), anti–BCL-2 (6C8), anti–BCL-XL (Cell Signaling Technology), anti–MCL-1 (S-19, Santa Cruz Biotechnology), anti-FOXO1 (Cell Signaling Technology), anti-FOXO3 (Millipore), anti–phospho-ERK (Cell Signaling Technology), anti–phospho-AKT (Ser473) (Cell Signaling Technology), anti–phospho-FOXO (Cell Signaling Technology), anti–phospho-S6K (Cell Signaling Technology), anti-S6K (Cell Signaling Technology), anti-HER2 (Calbiochem), anti-EGFR (Santa Cruz Biotechnology), and anti-actin (Sigma). For immunoblot analyses, cells were treated with the indicated inhibitors for 24 hours unless otherwise stated. Cells were lysed in radioimmunoprecipitation assay buffer, and protein concentration was determined by BCA kit (Pierce). Proteins (25 to 40 μg) were resolved by NuPAGE (Invitrogen) and transferred onto polyvinylidene difluoride membranes (Immobilon-P, Millipore). Antibody detection was accomplished with enhanced chemiluminescence (Western Lightning, PerkinElmer) and the LAS-3000 Imaging System (Fujifilm).
Indirect immunofluorescence microscopy
BT474 cells were treated with indicated inhibitors for 12 hours, then fixed in 4% paraformaldehyde, and permeabilized with 0.1% Triton X-100. Cells were sequentially incubated with anti-FOXO3 antibody (Cell Signaling Technology), Alexa Fluor 488–conjugated goat anti-rabbit secondary antibody (Invitrogen), and Hoechst 33342 (Invitrogen). Images were acquired with a SPOT camera (Diagnostics Instruments) mounted on an Olympus IX51 microscope (Olympus).
Chromatin immunoprecipitation
BT474 or HCC827 cells, treated with lapatinib or erlotinib for 12 hours, were subjected to ChIP as previously described with either anti-FOXO1 (H-128, Santa Cruz Biotechnology) or anti-FOXO3 (Millipore) antibodies (53). Precipitated DNA was amplified by PCR with PUMA-specific primers. The PCR products were analyzed by agarose gel electrophoresis (2%) and visualized with ethidium bromide under ultraviolet light. Primers used to amplify the PUMA promoter containing the FOXO binding site were as follows: forward, 5′-GTGAGAGGCGGCTCGGCGTT-3′; reverse, 5′-CCTGGGGTCGACCCTCTTCC-3′. Primers used to amplify the 5′ sequence located at 4 kb upstream of the transcription start site of PUMA were as follows: forward, 5′-ACCCTGCAATGGGCAGTCTGGA-3′; reverse, 5′-TGAGAGAGGGGCAGTCTCTAGCCC-3′.
Mouse models
Bim−/− and Puma−/− mice were described previously (54, 55). TetO-NeuNT (TAN) and MMTV-rtTA (MTB) mice were described previously (30). TetO-EGFRL858R and CCSP-rtTA mice were described previously (31). To generate MTB+TAN+ Bim−/− mice, we crossed TAN+/+Bim+/− mice with MTB+Bim+/− mice. To generate MTB+TAN+Puma−/− mice, we crossed TAN+/+Puma+/− mice with MTB+Puma+/− mice. MTB+TAN+ mice were also generated from each of these crosses. To generate TetO-EGFRL858R; CCSP-rtTA or TetO-EGFRL858R; CCSP-rtTA; Puma−/− mice, we crossed EGFRL858R; Puma+/− mice with CCSP-rtTA; Puma+/− mice. Animal experiments were performed according to the institutional guidelines. Animals were administered with doxycycline (2 mg/ml; Sigma) plus sucrose (50 mg/ml) in their drinking water to induce HER2. When mice developed tumors measuring 1 cm in the longest dimension, doxycycline was withdrawn for 3 days and tumor volume was measured with calipers. To induce EGFRL858R, we administered doxycycline by feeding mice with doxycycline-impregnated food pellets [625 parts per million (ppm); Harlan-Teklad]. Tumor growth was monitored by MRI. When mice developed tumors measuring 0.5 cm in the longest dimension as detected by MRI, doxycycline was withdrawn for 72 hours to turn off EGFRL858R. Tumor reduction was assessed by MRI. Tumor volumes were calculated with ImageJ software. P values for statistical analyses were obtained with Mann-Whitney test with Bonferroni correction.
Immunohistochemistry and caspase activity assays
Formalin-fixed, paraffin-embedded tumor tissues were sectioned at 7 μm in thickness and subjected to immunohistochemistry against cleaved caspase-3 (Cell Signaling Technology) with the DISCOVERY ULTRA instrument (Ventana) according to the manufacturer’s instructions. Caspase activities were quantified with Caspase-Glo 3/7 Assay System (Promega) according to the manufacturer’s protocol.
Supplementary Material
Acknowledgments
We thank H.-F. Chen for technical assistance and I. Ostrovnaya for assistance in statistical analysis.
Funding: This work was supported by grants to E.H.-Y.C. from the National Cancer Institute/NIH (R01CA125562) and the American Cancer Society (118518-RSG-10-030-01-CCG).
Footnotes
SUPPLEMENTARY MATERIALS www.sciencesignaling.org/cgi/content/full/6/268/ra20/DC1
Fig. S1. Immunoblot analysis of siRNA-mediated knockdown of BIM and PUMA.
Fig. S2. Immunoblot analysis of siRNA-mediated knockdown.
Fig. S3. Regulation of ERK and AKT phosphorylation by constitutively active AKT and MEK.
Fig. S4. Inhibition of lapatinib-mediated induction of PUMA mRNA by Myr-AKT.
Fig. S5. Inhibition of ERK and AKT phosphorylation by various kinase inhibitors.
Fig. S6. PUMA is required for GDC0941-induced apoptosis of BT474 cells.
Fig. S7. BIM mRNA is induced upon treatment with tyrosine kinase inhibitors.
Fig. S8. Knockdown of p53 fails to prevent lapatinib-mediated PUMA induction and apoptosis.
Fig. S9. BIM and PUMA are activated upon erlotinib-induced apoptosis of PC9 cells.
Fig. S10. Immunoblot analysis of HER2 in HER2/Neu-driven mammary tumors.
Fig. S11. Immunoblot analysis of BIM and PUMA in HER2/Neu-driven mammary tumors.
Fig. S12. Immunoblot analysis of EGFR in the tetracycline-inducible EGFRL858R transgenic mouse model.
Fig. S13. Immunoblot analysis of PUMA in EGFRL858R-driven mouse lung tumors.
Fig. S14. Immunoblot analysis of phosphorylation of AKT and S6K.
Fig. S15. Immunoblot analysis of siRNA-mediated knockdown of PUMA.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: The PUMA knockout mice require a material transfer agreement from St. Jude Children’s Research Hospital.
Author contributions: G.R.B. designed and performed the experiments and analyzed the data; E.H.-Y.C. designed the research, analyzed the data, and supervised the project; Y.T.G., Y.D., S.T., H.L., and Y.H. performed some experiments; J.J.-D.H. analyzed the data; and P.M.C., L.A.C., and G.P.Z. generated essential reagents.
REFERENCES AND NOTES
- 1.Sellers WR. A blueprint for advancing genetics-based cancer therapy. Cell. 2011;147:26–31. doi: 10.1016/j.cell.2011.09.016. [DOI] [PubMed] [Google Scholar]
- 2.Weinstein IB. Cancer. Addiction to oncogenes—The Achilles heal of cancer. Science. 2002;297:63–64. doi: 10.1126/science.1073096. [DOI] [PubMed] [Google Scholar]
- 3.Youle RJ, Strasser A. The BCL-2 protein family: Opposing activities that mediate cell death. Nat. Rev. Mol. Cell Biol. 2008;9:47–59. doi: 10.1038/nrm2308. [DOI] [PubMed] [Google Scholar]
- 4.Wang X. The expanding role of mitochondria in apoptosis. Genes Dev. 2001;15:2922–2933. [PubMed] [Google Scholar]
- 5.Letai A, Bassik MC, Walensky LD, Sorcinelli MD, Weiler S, Korsmeyer SJ. Distinct BH3 domains either sensitize or activate mitochondrial apoptosis, serving as prototype cancer therapeutics. Cancer Cell. 2002;2:183–192. doi: 10.1016/s1535-6108(02)00127-7. [DOI] [PubMed] [Google Scholar]
- 6.Wei MC, Zong WX, Cheng EH, Lindsten T, Panoutsakopoulou V, Ross AJ, Roth KA, MacGregor GR, Thompson CB, Korsmeyer SJ. Proapoptotic BAX and BAK: A requisite gateway to mitochondrial dysfunction and death. Science. 2001;292:727–730. doi: 10.1126/science.1059108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheng EH, Wei MC, Weiler S, Flavell RA, Mak TW, Lindsten T, Korsmeyer SJ. BCL-2, BCL-XL sequester BH3 domain-only molecules preventing BAX- and BAK-mediated mitochondrial apoptosis. Mol. Cell. 2001;8:705–711. doi: 10.1016/s1097-2765(01)00320-3. [DOI] [PubMed] [Google Scholar]
- 8.Kuwana T, Bouchier-Hayes L, Chipuk JE, Bonzon C, Sullivan BA, Green DR, Newmeyer DD. BH3 domains of BH3-only proteins differentially regulate Bax-mediated mitochondrial membrane permeabilization both directly and indirectly. Mol. Cell. 2005;17:525–535. doi: 10.1016/j.molcel.2005.02.003. [DOI] [PubMed] [Google Scholar]
- 9.Certo M, Del Gaizo Moore V, Nishino M, Wei G, Korsmeyer S, Armstrong SA, Letai A. Mitochondria primed by death signals determine cellular addiction to anti-apoptotic BCL-2 family members. Cancer Cell. 2006;9:351–365. doi: 10.1016/j.ccr.2006.03.027. [DOI] [PubMed] [Google Scholar]
- 10.Kim H, Rafiuddin-Shah M, Tu HC, Jeffers JR, Zambetti GP, Hsieh JJ, Cheng EH. Hierarchical regulation of mitochondrion-dependent apoptosis by BCL-2 subfamilies. Nat. Cell Biol. 2006;8:1348–1358. doi: 10.1038/ncb1499. [DOI] [PubMed] [Google Scholar]
- 11.Deng J, Carlson N, Takeyama K, Cin P. Dal, Shipp M, Letai A. BH3 profiling identifies three distinct classes of apoptotic blocks to predict response to ABT-737 and conventional chemotherapeutic agents. Cancer Cell. 2007;12:171–185. doi: 10.1016/j.ccr.2007.07.001. [DOI] [PubMed] [Google Scholar]
- 12.Kim H, Tu HC, Ren D, Takeuchi O, Jeffers JR, Zambetti GP, Hsieh JJ, Cheng EH. Stepwise activation of BAX and BAK by tBID, BIM, and PUMA initiates mitochondrial apoptosis. Mol. Cell. 2009;36:487–499. doi: 10.1016/j.molcel.2009.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Desagher S, Osen-Sand A, Nichols A, Eskes R, Montessuit S, Lauper S, Maundrell K, Antonsson B, Martinou JC. Bid-induced conformational change of Bax is responsible for mitochondrial cytochrome c release during apoptosis. J. Cell Biol. 1999;144:891–901. doi: 10.1083/jcb.144.5.891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lovell JF, Billen LP, Bindner S, Shamas-Din A, Fradin C, Leber B, Andrews DW. Membrane binding by tBid initiates an ordered series of events culminating in membrane permeabilization by Bax. Cell. 2008;135:1074–1084. doi: 10.1016/j.cell.2008.11.010. [DOI] [PubMed] [Google Scholar]
- 15.Gavathiotis E, Suzuki M, Davis ML, Pitter K, Bird GH, Katz SG, Tu HC, Kim H, Cheng EH, Tjandra N, Walensky LD. BAX activation is initiated at a novel interaction site. Nature. 2008;455:1076–1081. doi: 10.1038/nature07396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ren D, Tu HC, Kim H, Wang GX, Bean GR, Takeuchi O, Jeffers JR, Zambetti GP, Hsieh JJ, Cheng EH. BID, BIM, and PUMA are essential for activation of the BAX- and BAK-dependent cell death program. Science. 2010;330:1390–1393. doi: 10.1126/science.1190217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kolch W, Pitt A. Functional proteomics to dissect tyrosine kinase signalling pathways in cancer. Nat. Rev. Cancer. 2010;10:618–629. doi: 10.1038/nrc2900. [DOI] [PubMed] [Google Scholar]
- 18.She QB, Chandarlapaty S, Ye Q, Lobo J, Haskell KM, Leander KR, DeFeo-Jones D, Huber HE, Rosen N. Breast tumor cells with PI3K mutation or HER2 amplification are selectively addicted to Akt signaling. PLoS One. 2008;3:e3065. doi: 10.1371/journal.pone.0003065. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 19.Brachmann SM, Hofmann I, Schnell C, Fritsch C, Wee S, Lane H, Wang S, Garcia-Echeverria C, Maira SM. Specific apoptosis induction by the dual PI3K/mTor inhibitor NVP-BEZ235 in HER2 amplified and PIK3CA mutant breast cancer cells. Proc. Natl. Acad. Sci. U.S.A. 2009;106:22299–22304. doi: 10.1073/pnas.0905152106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Faber AC, Li D, Song Y, Liang MC, Yeap BY, Bronson RT, Lifshits E, Chen Z, Maira SM, Garcia-Echeverría C, Wong KK, Engelman JA. Differential induction of apoptosis in HER2 and EGFR addicted cancers following PI3K inhibition. Proc. Natl. Acad. Sci. U.S.A. 2009;106:19503–19508. doi: 10.1073/pnas.0905056106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cheng EH, Sawyers CL. In cancer drug resistance, germline matters too. Nat. Med. 2012;18:494–496. doi: 10.1038/nm.2725. [DOI] [PubMed] [Google Scholar]
- 22.Gong Y, Somwar R, Politi K, Balak M, Chmielecki J, Jiang X, Pao W. Induction of BIM is essential for apoptosis triggered by EGFR kinase inhibitors in mutant EGFR-dependent lung adenocarcinomas. PLoS Med. 2007;4:e294. doi: 10.1371/journal.pmed.0040294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cragg MS, Kuroda J, Puthalakath H, Huang DC, Strasser A. Gefitinib-induced killing of NSCLC cell lines expressing mutant EGFR requires BIM and can be enhanced by BH3 mimetics. PLoS Med. 2007;4:1681–1689. doi: 10.1371/journal.pmed.0040316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Costa DB, Halmos B, Kumar A, Schumer ST, Huberman MS, Boggon TJ, Tenen DG, Kobayashi S. BIM mediates EGFR tyrosine kinase inhibitor-induced apoptosis in lung cancers with oncogenic EGFR mutations. PLoS Med. 2007;4:1669–1679. doi: 10.1371/journal.pmed.0040315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Deng J, Shimamura T, Perera S, Carlson NE, Cai D, Shapiro GI, Wong KK, Letai A. Proapoptotic BH3-only BCL-2 family protein BIM connects death signaling from epidermal growth factor receptor inhibition to the mitochondrion. Cancer Res. 2007;67:11867–11875. doi: 10.1158/0008-5472.CAN-07-1961. [DOI] [PubMed] [Google Scholar]
- 26.Ley R, Ewings KE, Hadfield K, Cook SJ. Regulatory phosphorylation of Bim: Sorting out the ERK from the JNK. Cell Death Differ. 2005;12:1008–1014. doi: 10.1038/sj.cdd.4401688. [DOI] [PubMed] [Google Scholar]
- 27.Dehan E, Bassermann F, Guardavaccaro D, Vasiliver-Shamis G, Cohen M, Lowes KN, Dustin M, Huang DC, Taunton J, Pagano M. βTrCP- and Rsk1/2-mediated degradation of BimEL inhibits apoptosis. Mol. Cell. 2009;33:109–116. doi: 10.1016/j.molcel.2008.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.del Peso L, González-García M, Page C, Herrera R, Nuñez G. Interleukin-3-induced phosphorylation of BAD through the protein kinase Akt. Science. 1997;278:687–689. doi: 10.1126/science.278.5338.687. [DOI] [PubMed] [Google Scholar]
- 29.Datta SR, Dudek H, Tao X, Masters S, Fu H, Gotoh Y, Greenberg ME. Akt phosphorylation of BAD couples survival signals to the cell-intrinsic death machinery. Cell. 1997;91:231–241. doi: 10.1016/s0092-8674(00)80405-5. [DOI] [PubMed] [Google Scholar]
- 30.Moody SE, Sarkisian CJ, Hahn KT, Gunther EJ, Pickup S, Dugan KD, Innocent N, Cardiff RD, Schnall MD, Chodosh LA. Conditional activation of Neu in the mammary epithelium of transgenic mice results in reversible pulmonary metastasis. Cancer Cell. 2002;2:451–461. doi: 10.1016/s1535-6108(02)00212-x. [DOI] [PubMed] [Google Scholar]
- 31.Politi K, Zakowski MF, Fan PD, Schonfeld EA, Pao W, Varmus HE. Lung adenocarcinomas induced in mice by mutant EGF receptors found in human lung cancers respond to a tyrosine kinase inhibitor or to down-regulation of the receptors. Genes Dev. 2006;20:1496–1510. doi: 10.1101/gad.1417406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Clybouw C, Merino D, Nebl T, Masson F, Robati M, O’Reilly L, Hubner A, Davis RJ, Strasser A, Bouillet P. Alternative splicing of Bim and Erk-mediated BimEL phosphorylation are dispensable for hematopoietic homeostasis in vivo. Cell Death Differ. 2012;19:1060–1068. doi: 10.1038/cdd.2011.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.You H, Pellegrini M, Tsuchihara K, Yamamoto K, Hacker G, Erlacher M, Villunger A, Mak TW. FOXO3a-dependent regulation of Puma in response to cytokine/growth factor withdrawal. J. Exp. Med. 2006;203:1657–1663. doi: 10.1084/jem.20060353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhao Y, Coloff JL, Ferguson EC, Jacobs SR, Cui K, Rathmell JC. Glucose metabolism attenuates p53 and Puma-dependent cell death upon growth factor deprivation. J. Biol. Chem. 2008;283:36344–36353. doi: 10.1074/jbc.M803580200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Greer EL, Brunet A. FOXO transcription factors at the interface between longevity and tumor suppression. Oncogene. 2005;24:7410–7425. doi: 10.1038/sj.onc.1209086. [DOI] [PubMed] [Google Scholar]
- 36.Gan B, Lim C, Chu G, Hua S, Ding Z, Collins M, Hu J, Jiang S, Fletcher-Sananikone E, Zhuang L, Chang M, Zheng H, Wang YA, Kwiatkowski DJ, Kaelin WG, Jr., Signoretti S, DePinho RA. FoxOs enforce a progression checkpoint to constrain mTORC1-activated renal tumorigenesis. Cancer Cell. 2010;18:472–484. doi: 10.1016/j.ccr.2010.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dijkers PF, Medema RH, Lammers JW, Koenderman L, Coffer PJ. Expression of the pro-apoptotic Bcl-2 family member Bim is regulated by the forkhead transcription factor FKHR-L1. Curr. Biol. 2000;10:1201–1204. doi: 10.1016/s0960-9822(00)00728-4. [DOI] [PubMed] [Google Scholar]
- 38.Gilley J, Coffer PJ, Ham J. FOXO transcription factors directly activate bim gene expression and promote apoptosis in sympathetic neurons. J. Cell Biol. 2003;162:613–622. doi: 10.1083/jcb.200303026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stahl M, Dijkers PF, Kops GJ, Lens SM, Coffer PJ, Burgering BM, Medema RH. The forkhead transcription factor FoxO regulates transcription of p27Kip1 and Bim in response to IL-2. J. Immunol. 2002;168:5024–5031. doi: 10.4049/jimmunol.168.10.5024. [DOI] [PubMed] [Google Scholar]
- 40.Faber AC, Corcoran RB, Ebi H, Sequist LV, Waltman BA, Chung E, Incio J, Digumarthy SR, Pollack SF, Song Y, Muzikansky A, Lifshits E, Roberge S, Coffman EJ, Benes CH, Gómez HL, Baselga J, Arteaga CL, Rivera MN, Dias-Santagata D, Jain RK, Engelman JA. BIM expression in treatment-naïve cancers predicts responsiveness to kinase inhibitors. Cancer Discov. 2011;1:352–365. doi: 10.1158/2159-8290.CD-11-0106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pao W, Miller VA, Politi KA, Riely GJ, Somwar R, Zakowski MF, Kris MG, Varmus H. Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS Med. 2005;2:e73. doi: 10.1371/journal.pmed.0020073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Del Gaizo Moore V, Brown JR, Certo M, Love TM, Novina CD, Letai A. Chronic lymphocytic leukemia requires BCL2 to sequester prodeath BIM, explaining sensitivity to BCL2 antagonist ABT-737. J. Clin. Invest. 2007;117:112–121. doi: 10.1172/JCI28281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Garrison SP, Phillips DC, Jeffers JR, Chipuk JE, Parsons MJ, Rehg JE, Opferman JT, Green DR, Zambetti GP. Genetically defining the mechanism of Puma- and Bim-induced apoptosis. Cell Death Differ. 2012;19:642–649. doi: 10.1038/cdd.2011.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Engelman JA, Chen L, Tan X, Crosby K, Guimaraes AR, Upadhyay R, Maira M, McNamara K, Perera SA, Song Y, Chirieac LR, Kaur R, Lightbown A, Simendinger J, Li T, Padera RF, García-Echeverría C, Weissleder R, Mahmood U, Cantley LC, Wong KK. Effective use of PI3K and MEK inhibitors to treat mutant Kras G12D and PIK3CA H1047R murine lung cancers. Nat. Med. 2008;14:1351–1356. doi: 10.1038/nm.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sos ML, Fischer S, Ullrich R, Peifer M, Heuckmann JM, Koker M, Heynck S, Stückrath I, Weiss J, Fischer F, Michel K, Goel A, Regales L, Politi KA, Perera S, Getlik M, Heukamp LC, Ansén S, Zander T, Beroukhim R, Kashkar H, Shokat KM, Sellers WR, Rauh D, Orr C, Hoeflich KP, Friedman L, Wong KK, Pao W, Thomas RK. Identifying genotype-dependent efficacy of single and combined PI3K- and MAPK-pathway inhibition in cancer. Proc. Natl. Acad. Sci. U.S.A. 2009;106:18351–18356. doi: 10.1073/pnas.0907325106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.O’Reilly KE, Rojo F, She QB, Solit D, Mills GB, Smith D, Lane H, Hofmann F, Hicklin DJ, Ludwig DL, Baselga J, Rosen N. mTOR inhibition induces upstream receptor tyrosine kinase signaling and activates Akt. Cancer Res. 2006;66:1500–1508. doi: 10.1158/0008-5472.CAN-05-2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chandarlapaty S, Sawai A, Scaltriti M, Rodrik-Outmezguine V, Grbovic-Huezo O, Serra V, Majumder PK, Baselga J, Rosen N. AKT inhibition relieves feedback suppression of receptor tyrosine kinase expression and activity. Cancer Cell. 2011;19:58–71. doi: 10.1016/j.ccr.2010.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Carracedo A, Ma L, Teruya-Feldstein J, Rojo F, Salmena L, Alimonti A, Egia A, Sasaki AT, Thomas G, Kozma SC, Papa A, Nardella C, Cantley LC, Baselga J, Pandolfi PP. Inhibition of mTORC1 leads to MAPK pathway activation through a PI3K-dependent feedback loop in human cancer. J. Clin. Invest. 2008;118:3065–3074. doi: 10.1172/JCI34739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Okamoto K, Okamoto I, Okamoto W, Tanaka K, Takezawa K, Kuwata K, Yamaguchi H, Nishio K, Nakagawa K. Role of survivin in EGFR inhibitor–induced apoptosis in non–small cell lung cancers positive for EGFR mutations. Cancer Res. 2010;70:10402–10410. doi: 10.1158/0008-5472.CAN-10-2438. [DOI] [PubMed] [Google Scholar]
- 50.Tanizaki J, Okamoto I, Fumita S, Okamoto W, Nishio K, Nakagawa K. Roles of BIM induction and survivin downregulation in lapatinib-induced apoptosis in breast cancer cells with HER2 amplification. Oncogene. 2011;30:4097–4106. doi: 10.1038/onc.2011.111. [DOI] [PubMed] [Google Scholar]
- 51.Chonghaile TN, Letai A. Mimicking the BH3 domain to kill cancer cells. Oncogene. 2008;27(suppl. 1):S149–S157. doi: 10.1038/onc.2009.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tu HC, Ren D, Wang GX, Chen DY, Westergard TD, Kim H, Sasagawa S, Hsieh JJ, Cheng EH. The p53-cathepsin axis cooperates with ROS to activate programmed necrotic death upon DNA damage. Proc. Natl. Acad. Sci. U.S.A. 2009;106:1093–1098. doi: 10.1073/pnas.0808173106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu H, Takeda S, Kumar R, Westergard TD, Brown EJ, Pandita TK, Cheng EH, Hsieh JJ. Phosphorylation of MLL by ATR is required for execution of mammalian S-phase checkpoint. Nature. 2010;467:343–346. doi: 10.1038/nature09350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jeffers JR, Parganas E, Lee Y, Yang C, Wang J, Brennan J, MacLean KH, Han J, Chittenden T, Ihle JN, McKinnon PJ, Cleveland JL, Zambetti GP. Puma is an essential mediator of p53-dependent and -independent apoptotic pathways. Cancer Cell. 2003;4:321–328. doi: 10.1016/s1535-6108(03)00244-7. [DOI] [PubMed] [Google Scholar]
- 55.Takeuchi O, Fisher J, Suh H, Harada H, Malynn BA, Korsmeyer SJ. Essential role of BAX,BAK in B cell homeostasis and prevention of autoimmune disease. Proc. Natl. Acad. Sci. U.S.A. 2005;102:11272–11277. doi: 10.1073/pnas.0504783102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.