Abstract
The propagation of bacteriophages and other mobile genetic elements requires exploitation of the phage mechanisms involved in virion assembly and DNA packaging. Here, we identified and characterized four different families of phage-encoded proteins that function as activators required for transcription of the late operons (morphogenetic and lysis genes) in a large group of phages infecting Gram-positive bacteria. These regulators constitute a super-family of proteins, here named late transcriptional regulators (Ltr), which share common structural, biochemical and functional characteristics and are unique to this group of phages. They are all small basic proteins, encoded by genes present at the end of the early gene cluster in their respective phage genomes and expressed under cI repressor control. To control expression of the late operon, the Ltr proteins bind to a DNA repeat region situated upstream of the terS gene, activating its transcription. This involves the C-terminal part of the Ltr proteins, which control specificity for the DNA repeat region. Finally, we show that the Ltr proteins are the only phage-encoded proteins required for the activation of the packaging and lysis modules. In summary, we provide evidence that phage packaging and lysis is a conserved mechanism in Siphoviridae infecting a wide variety of Gram-positive bacteria.
INTRODUCTION
Prophages are temperate bacteriophages that are integrated into the chromosomes of their bacterial hosts. Whole-genome sequencing projects have revealed that prophage sequences are common and widespread among bacterial genomes. Prophages are not only important as genetic elements that impart bacterial genome variability but they may also confer a diverse array of phenotypic traits to their hosts (sometimes referred to as lysogenic conversion factors), including those that govern the course and pathobiology of bacterial infections [reviewed in (1)].
Within the Firmicute division of Gram-positive bacteria, temperate bacteriophages are key vectors for the horizontal transfer of virulence genes. In Staphylococcus aureus, bacteriophages encode and mobilize an impressive array of immune evasion functions as well as clinically relevant toxins, including Panton-Valentine leukocidin, staphylokinase, enterotoxin A and exfoliative toxin (1). In addition to these phage-encoded virulence factors, staphylococcal phages induce replication of S. aureus pathogenicity islands (2,3) and provide the packaging machinery used for these elements to be efficiently transferred [for review see (4)]. Several bacteriophage-encoded virulence determinants also contribute to pathogenesis in group A Streptococcus. Genome sequencing of serotype M1, M3 and M18 strains revealed many virulence factors that were encoded by phages or phage-like elements (5). In Bacillus anthracis, stable infection of strains with environmental bacteriophages conferred survival phenotypes in soil and earthworm intestinal niches (6), again confirming the important role of bacteriophages in bacterial biology.
Although a great deal is known about mechanisms controlling prophage induction and temporal control of bacteriophage gene expression, much of this work has been focussed on a relatively small number of model phages, which primarily infect Escherichia coli (7,8). These phages have been adopted as models with the assumption that their regulatory mechanisms are generally representative in phage biology. However, the diversity of phages and the large number of phage genes of unknown function suggest that there are uncharacterized regulators still to be discovered. A case in point was the recent identification and characterization of RinA, the regulator of the morphogenesis genes in a number of phages infecting Gram-positive bacteria. This regulator had gone unnoticed because it had no homologues among known phage or bacterial transcription factors. We demonstrated that RinA binds to a tightly regulated promoter region, situated upstream of the terS gene, and controls expression of the morphogenetic and lysis modules of the phage by activating their transcription. As expected, rinA deletion eliminated formation of functional phage particles and decreased transfer of phage and phage-dependent mobile genetic elements (9).
RinA homologues were found in phages infecting a wide variety of Gram-positive bacteria (9). Although these proteins had varying degrees of similarity, they shared several common characteristics: all were basic proteins (pI > 8) with a size ∼130 aa. In addition, the rinA gene was generally located just upstream of the gene encoding the small terminase subunit gene (terS). Despite the relatively broad distribution of RinA, however, most of the sequenced phages infecting Gram-positive bacteria did not encode RinA homologues, suggesting the existence of additional regulators or mechanisms involved in activation of the late operons in these phages.
In this work, we explored the hypothesis that most of the phages infecting the Gram-positive Firmicutes control packaging and lysis in a manner similar to that described for RinA. We have now identified five different families of phage-encoded regulators controlling phage-mediated late gene expression. Each family shares the size and charge characteristics previously found for the RinA proteins and a structurally similar C-terminal domain, as well as the location of the gene in the phage genome (Figure 1). These activators constitute a super-family of proteins that we have designated as for late transcriptional regulators (Ltr). Representatives of each family have been shown to control expression of phage-mediated packaging and lysis functions, which are required not only for phage growth but also for phage-mediated horizontal gene transfer.
Figure 1.
Location of the genes controlled by the Ltr proteins. Partial genetic maps of ϕFL2A (Genbank accession number NC_013643) and ϕ55 (accession number AY954963), showing the location of the ltr genes and the genes under their control. Arrows indicate predicted ORFs, numbered as annotated. Grey arrows indicate the regulatory genes deleted in this study, whereas black arrows indicate genes for which expression was analysed by qRT-PCR.
MATERIALS AND METHODS
Bacterial strains and growth conditions
Bacterial strains used in these studies are listed in Supplementary Table S1. Bacteria were grown at 37°C overnight on TSA agar medium, supplemented with antibiotics as appropriate. Broth cultures were grown at 37°C in TSB broth with shaking (240 r.p.m.).
For SOS-dependent prophage induction, bacteria were grown in TSB broth to OD540 = 0.3 (for S. aureus) or OD540 = 0.15 (for Enterococcus faecalis) and induced by adding mitomycin C (MC, 2 μg/ml). Cultures were grown at 32°C with slow shaking (80 r.p.m.). Lysis usually occurred within 3 h. Samples were removed at various time points after phage induction, and standard SDS minilysates were prepared and separated on 0.7% agarose gels, as described previously (10,11). Standard procedures were used for preparation and analysis of phage lysates, lysogens and transduction in S. aureus, as described previously (9).
DNA methods
General DNA manipulations were performed by standard procedures. Labelling of the probes and DNA hybridization were performed according to the protocol supplied with the PCR-DIG DNA-labelling and chemiluminescent detection kit (Roche). To produce the strains carrying mutant prophages, allelic exchange was performed using derivatives of plasmid pMAD or pBT2 carrying the desired mutations, as described previously (11). Plasmid constructs (Supplementary Table S2) were prepared by cloning PCR products obtained from oligonucleotide primers as listed in Supplementary Table S3. All constructs were sequenced by the Institute core sequencing laboratory.
Bioinformatics analysis
Multiple sequences alignment was performed using Praline (12). The nearest neighbour tree of Ltr proteins were generated using MEGA5 (13). The hidden Markov model method SAM-T98 was used for detecting remote protein homologies (14).
Real-time quantitative PCR
Total S. aureus RNA was prepared using the Fast RNA-Blue kit (Bio101) according to the manufacturer’s instructions. Two micrograms of each RNA were subjected, in duplicate, to DNase I (Invitrogen) treatment for 30 min at 37°C. The enzyme was inactivated at 65°C in the presence of EDTA. To verify the absence of genomic DNA in every sample, the RNA duplicates were reverse transcribed in the presence and absence of M-MLV Reverse Transcriptase (Invitrogen). All preparations were purified using QIAquick PCR purification kit (Qiagen). In all, 25 ng of each reaction product was used for a real-time quantitative PCR using the iCycler machine (Biorad) and the LC-DNA Master SYBR Green I mix (Biorad). The different genes were amplified using oligonucleotides listed in Supplementary Table S3. The gyrB transcripts that are constitutively expressed were amplified as an endogenous control. The levels of expression of the different genes were normalized with respect to gyrB expression. Only samples with no amplification of gyrB in the minus reverse transcriptase aliquot were included in the study. To monitor the specificity, the final PCR products were analysed by melting curves and electrophoresis. In each experiment, all the reactions were performed in triplicate. The relative transcriptional levels within distinct experiments were reported as previously indicated (9). The results show the average ± SEM of at least four independent experiments.
Design of the tiling microarray
The NA-Staph-b520729F tiling microarray was designed in collaboration with Affymetrix (Santa Clara, CA, USA). Specifically, the microarray (format 49-7875 with 11 µm features) contains a total of 522 406 probes, divided into two parts. The first part corresponds to the tiling sub-array, which contains a total of 384 932 probes (25 mer), which are divided in eight sets. The first set corresponds to 46 310 probes recommended by Affymetrix as controls. The following seven sets are covering both strands of (i) the S. aureus NCTC8325 genome (2 821 347 bp covered by 363 127 probes), the S. aureus pathogenicity islands (ii) SaPI1 (15 229 bp covered by 1892 probes), (iii) SaPI2 (14 753 bp covered by 1897 probes), (iv) SaPIbov1 (15 887 bp covered by 1978 probes), (v) SaPIbov2 (27 031 bp covered by 3356 probes), (vi) the staphylococcal chromosomal cassette SCC mecIV-ACMEI (54 947 bp covered by 6938 probes) and (vii) the phage 80α (43 859 bp covered by 5744 probes). Each 25-mer probe was tiled each 14 nt across the whole genome, resulting in 11-nt overlaps and a 7-nt offset of the tile between strands. This design enables a 7-nt resolution for hybridization of double-stranded targets and a 14-nt resolution for strand-specific targets. The second part of the chip corresponds to the S. aureus gene expression sub-array, which contains 91 164 probes grouped into 3224 probe sets (each probe set includes 14 perfect match and 14 mismatch probes). In all, 3122 probe-sets are targeting unique S. aureus genes, whereas 102 probe-sets are controls recommended by Affymetrix.
cDNA synthesis, fragmentation, labelling and array hybridization and scan
Before cDNA synthesis, RNA integrity from each sample was confirmed on Agilent RNA Nano LabChips (Agilent Technologies). In all, 10 µg RNAs were reverse transcribed using SuperScript II reverse transcriptase (Invitrogen Life Technologies) and processed following the protocol of the Affymetrix GeneChip Expression Analysis Technical Manual (P/N 702232 Rev. 2) in the presence of 6 ng/ml Actinomycin D to avoid spurious second-strand cDNA synthesis during reverse transcription reaction (15). Sense RNA corresponding to Bacillus subtilis poly-A lys, phe, thr, trp, dap genes were spiked into sample RNA as control for labelling and hybridization steps. cDNA was digested by DNase I (PIERCE) in 10× DNAse I buffer (USB-Affymetrix), and the size of digestion products was analysed in the Agilent Bioanalyser 2100 using RNA Nano LabChips to ensure that the fragmentation resulted in a majority of products in the range of 50–200 bp. The fragmented cDNA were then biotinylated using terminal deoxynucleotidyl transferase (Promega) and the GeneChip DNA labelling reagent (Affymetrix) following the manufacturer’s recommendations. Biotinylated cDNA (5 µg per array) were hybridized for 16 h according to the Affymetrix protocol in a total volume of 200 µl per hybridization chamber. Following incubation, the arrays were washed and stained in the Fluidics station 450 (Affymetrix) using the protocol n°FS450_0005. Scanning of the arrays was then performed using the GeneChip scanner 3000 (Affymetrix). A first scan of the chip was carried out with gene expression sub-array parameters followed by a second scan with tiling sub-array parameters. Intensity signals of each probe cells were computed by the GeneChip operating software and stored in cell intensity files (.CEL extension) before preprocessing and analysis.
Microarray data analysis
Data analysis of the tiling sub-array was performed using the Tiling Analysis Software from Affymetrix (http://www.affymetrix.com). Output bar files containing probe signal values were converted in graphic type files (.gr extension file) to be loaded and visualized using the Integrated Genome Browser version 6.2.2 from (http://genoviz.sourceforge.net/) (16).
Mobility shift assays
Electrophoretic mobility shift assays (EMSAs) were performed as described before (17), using purified Ltr proteins and a DIG-labelled DNA fragment listed in Supplementary Table S3.
Rapid amplification of cDNA 5′-ends
Amplification of the terS cDNA 5′-end was performed using the 5′/3′ rapid amplification of cDNA 5′-ends (RACE) Kit (Roche), according to the manufacturer’s protocol.
Enzyme assays
β-Lactamase assays, using nitrocefin as substrate, were performed as described (9), using a Thermomax (Molecular Devices) microtiter plate reader. Cells were obtained in exponential phase. β-Lactamase units are defined as (Vmax)/OD650.
RESULTS
Identification of putative regulators in different phages infecting Gram-positive bacteria
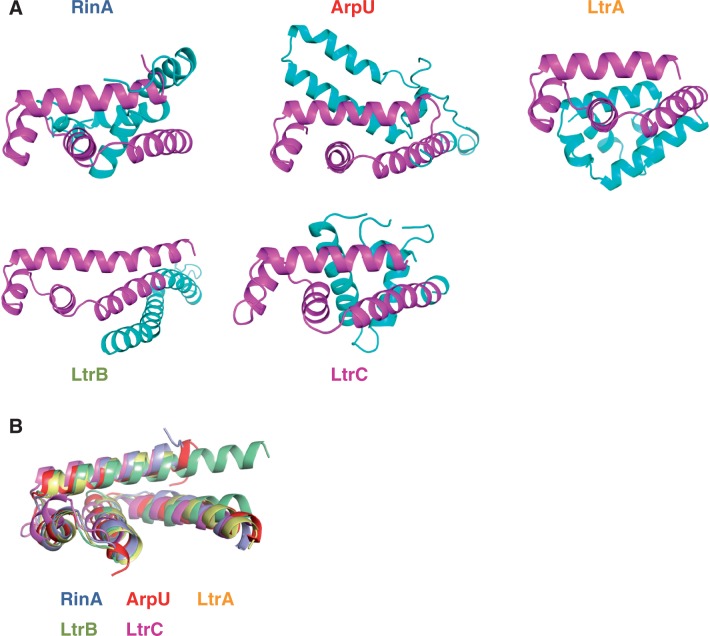
As previously indicated, RinA-like proteins are basic proteins with a length of ∼130 aa, encoded by a gene located upstream of terS. A database search for proteins with similar characteristics encoded at similar locations in other phage and prophage genomes revealed the existence of four additional families of proteins, distinct from RinA at the primary sequence level, encoded by phages infecting Gram-positive bacteria (Figure 2, Table 1). Although the representative members of the different families lack obvious sequence similarity (Supplementary Figure S1), using HMM-profile searches and protein folding-recognition methods, we have demonstrated that they have a conserved domain, with significant structural similarity, encompassing the C-terminal part of the proteins (Figure 3 and Supplementary Table S4). As will be demonstrated later, this domain plays an important role in the function of the proteins by recognizing the DNA repeat sequences present in the promoter region controlled by these regulators. This structural similarity, together with their common biochemical and functional characteristics, supports the idea that these proteins are members of a superfamily of related regulatory proteins. The genes for these proteins were found in phages infecting a variety of different bacterial species, including S. aureus, Staphylococcus carnosus, E. faecalis, Enterococcus faecium, Bacillus thuringiensis, Bacillus cereus, Macrococcus caseolyticus, Streptococcus pnenumoniae, Streptococcus pyogenes, Streptococcus suis, Lactobacillus casei, Brevibacillus brevis, Listeria innocua and Listeria monocytogenes. The discovery of these different families of putative regulators, including RinA, required the need for a generic term for the superfamily. Here, we propose Ltr. In addition, we also propose to designate each one of the previously uncharacterized families as Ltr followed by a capital letter.
Figure 2.
Ltr sequence comparison. Nearest neighbour tree of Ltr proteins generated by MEGA5 (13). Numbers indicate the bootstrap value. Asterisks indicate the regulators characterized in this study.
Table 1.
Ltr homologs in phages from Gram-positive bacteria
| Species | Accession number |
|---|---|
| Family I: ArpU homologs | |
| E. hirae | CAA90711 |
| E. faecalis phage ϕFL2A | YP_003347324a |
| E. faecium TX1330 | EEI59987 |
| B. cereus ATCC 10876 | ZP_04319074 |
| L. casei BL23 | CAQ66402 |
| L. monocytogenes EGD-e | NP_465827 |
| L. innocua Clip11262 | NP_471075 |
| Listeria phage A118 | NP_463531 |
| Streptococcus pneumoniae phage EJ-1 | NP_945275 |
| S. suis 89/1591 | ZP_03625181a |
| S. pyogenes MGAS315 | NP_664754 |
| Family II: Sigma 70_r4 (LtrA) | |
| Staphylococcus phage 37 | YP_240149a |
| E. faecalis phage ϕFL4A | ACZ64178 |
| E. faecalis V583 | NP_814123 |
| E. faecium C68 | ZP_05832578 |
| B. cereus ATCC 10876 | ZP_04318786 |
| Bacillus turingensis ATCC 35646 | ZP_00741150 |
| L. casei BL23 | YP_001986923 |
| E. faecium C68 | ZP_05832578 |
| Family III: DUF1492 (LtrB) | |
| S. pneumoniae | ZP_01822960 |
| S. suis 89/1591 | ZP_03625554a |
| S. pyogenes NZ131 | YP_002285386 |
| S. pyogenes M1 GAS | NP_269550 |
| Streptococcus dysgalactiae | YP_002996849 |
| Streptococcus infantarius | ZP_02920657 |
| Streptococcus gallolyticus | ZP_07463829 |
| Streptococcus urinalis | ZP_08727397 |
| Family IV: Homologs to ORF34 ϕ55 (LtrC) | |
| S. aureus phage 55 | YP_240527a |
| Staphylococcus phage PVL | NP_058499 |
| S. carnosus TM300 | YP_002633590a |
| Staphylococcus warneri L37603 | ZP_04679056 |
| Staphylococcus haemolyticus JCSC1435 | YP_253693 |
| Macrococcus caseolyticus JCSC5402 | YP_002560360 |
| Brevibacillus brevis NBRC 100599 | YP_002774582 |
aProteins characterised in this study.
Figure 3.
Modelling of Ltr proteins. (A) The predicted structure of representative members of the different Ltr families is shown using a schematic (cartoon) representation coloured according to structure assignments by SAM-T98 (14). The two domains are clearly visible, with the conserved C-terminal domain coloured in magenta, and the N-terminal domain in cyan. (B) Superposition of the C-terminal domain of the different Ltr proteins. The structure of this domain is virtually identical in the different Ltr proteins. RinA (S. aureus phage 80α; YP_001285353); ArpU (E. faecalis phage ϕFL2A; YP_003347324); LtrA (S. aureus phage 37; YP_240149); LtrB (S. suis 89/1591; ZP_03625554); LtrC (S. aureus phage 55; YP_240527).
Family I corresponded to homologues of autolysin regulatory protein (ArpU) (Table 1), and we have retained that designation. ArpU was described as a regulator of cellular muramidase-2 of Enterococcus hirae (18), and no additional work with this protein has been reported. Although initially described as a chromosomally encoded gene, analyses of the sequences in the database clearly demonstrate that ArpU homologues are phage-encoded (Supplementary Figure S2). Family II (LtrA) is a group of proteins with limited similarity to a portion of several sigma factors corresponding to conserved region 4 (Sigma 70_r4). This region of sigma is involved in recognition of the −35 element of promoters (19), and we hypothesise that this region is involved in DNA binding and possibly specific recognition of the terS promoter. Family III (LtrB) corresponds to proteins carrying a conserved domain designated DUF1492. As specified in the description of this domain, this family consists of several hypothetical, highly conserved Streptococcal and related phage proteins of ∼100 residues in length, the function of which is unknown. Family IV (LtrC) contained proteins homologous to the ORF34 gene product from S. aureus phage 55. This family has not been previously identified or characterized.
Deletion of the genes encoding the Ltr proteins eliminated production of phage particles
To examine the functionality of the Ltr family of proteins, initial analysis of candidates from two families was carried out: ArpU (Family I, from enterococcal phage ϕFL2A, YP_003347324) and LtrC (Family IV, from S. aureus phage ϕ55, YP_240527). We chose these proteins because both are encoded by transducing phages, capable of the transfer of mobile genetic elements, and they belong to the families of predicted regulators that were the most divergent from the previously characterized RinA (Figure 2). The gene for each of these putative regulatory proteins was inactivated in the corresponding ϕFL2A and ϕ55 prophages (Figure 1). The different phage mutants were induced with mitomycin C, screening lysates prepared 60 and 90 min after induction were examined by Southern blot with a phage-specific probe, and the induced cultures were assayed for phage titers.
Deletion of the genes encoding the Ltr regulators did not impair phage replication (data not shown) but eliminated the production of functional phage particles (Table 2). Furthermore, there was no lysis of the mutants after induction (data not shown), suggesting that these mutants were defective in expression of the lysis module. This defect was partially restored by complementation; when plasmids pJP1115 (pBT2-arpUϕFL2A) and pJP1054 (pCN51-LtrCϕ55), carrying the ltr genes from ϕFL2A and ϕ55, respectively, under the control of the Pcad promoter, were introduced into strains carrying the corresponding mutant prophage, phage titers were partially restored on mitomycin C induction (Table 2), confirming the regulatory role of these proteins in expression of the packaging and lysis functions. The complementing plasmids carry no flanking homologous DNA, and the observed phage titers for the deletion mutants depended on the presence of the complementing plasmid in the indicator strain, confirming that this was complementation and not marker rescue. We attribute the partial complementation observed in these strains to differences in timing and levels of Ltr expression from the plasmid as compared with a normal phage infection. Constitutive expression of Ltr would lead to premature expression of the phage late genes, and therefore, assembly and packaging before sufficient replication for a normal burst had occurred.
Table 2.
Effect of mutations on phage titre
| Donor strain | Phage | Plasmid | Phage titrea |
|---|---|---|---|
| E. faecalis | |||
| LIV1040 | ϕFL2A wt | 1.3 × 107 | |
| JP8039 | ϕFL2A ΔarpU | <100 | |
| JP8841 | ϕFL2A ΔarpU | pBT2-arp UϕFL2A | 7 × 104 |
| S. aureus | |||
| JP2348 | ϕ55 wt | 1.1 × 108 | |
| JP8071 | ϕ55 ΔltrC | <10 | |
| JP8172 | ϕ55 ΔltrC | pCN51-ltrCϕ55 | 9.6 × 104 |
| JP8636 | ϕ55 ΔltrC | pCN51-rinAϕ29 | <10 |
| JP8635 | ϕ55 ΔltrC | pCN51-arp UϕFL2A | <10 |
aPfu/ml of induced culture, using JH2-2 pCN51-arpUϕFL2A (for ϕFL2A) or RN4220 pCN51-ltrCϕ55 (for ϕ55) as recipient strains.
As previously reported for RinA, the phenotype of the ltr mutants suggested that the Ltr proteins might control phage packaging by regulating expression of the morphogenetic genes. The mRNA levels of several ϕFL2A and ϕ55 late genes (see map in Figure 1) were measured by quantitative real-time PCR at various times (0, 60 and 90 min) after MC induction, comparing expression levels in the wild-type phages and the ltr mutants. As a control, we also measured expression of the ssb and dnaC genes, which belong to the early transcript. The ltr mutation resulted in a significant decrease in the expression of genes involved in DNA packaging, capsid assembly and cell lysis, but did not affect the expression of the early genes (Figure 4). These results indicate that the Ltr proteins are transcriptional activators of the phage-encoded packaging and lysis modules.
Figure 4.

Ltr proteins control expression of the morphogenetic and lysis clusters of ϕ55 and ϕFL2A. Real time quantification of expression of different genes in ϕFL2A wild-type and the arpU mutant (A) or in ϕ55 wild-type and the ltrC mutant (B), at different times (0, 60 or 90 min) after MC induction of the phage lytic cycle. Expression was normalized to gyrB.
Identification of an Ltr-inducible promoter upstream of terS
As previously reported for RinA, and based on genome organization (Figure 1), we predicted the existence of at least one promoter directly controlled by the Ltr proteins, located just upstream of terS, the first gene in the morphogenetic gene cluster. To determine the transcription start sites and locate these putative promoters in the ϕFL2A and ϕ55 phages, 5′ RACE analysis was performed with total RNA isolated after induction of the corresponding prophages. One transcriptional start site was identified for each phage, which mapped to a G located 28 (ϕ55) or 118 bp (ϕFL2A) upstream of the ATG initiation codon for terS.
Ltr proteins are the only phage-encoded proteins required for terS expression
To confirm the activity of the promoters upstream of the terS genes, and to test whether expression of the Ltr regulators was enough to induce expression of the terS gene in the absence of other phage-encoded proteins, we made use of plasmid pCN42, which contains a Pcad promoter and a β-lactamase reporter (see Figure 5A). We cloned into this plasmid the arpU (ϕFL2A) or ltrC (ϕ55) coding sequences (expression of which depends on the Pcad promoter present in the plasmid), the terS promoter region and the 5′ coding region of the terS gene corresponding to each phage, which was fused to the β-lactamase reporter (see Figure 5A). As controls, we generated plasmids with the same structure except that they carried mutant copies of the respective ltr genes (Figure 5A). These plasmids were introduced into the staphylococcal strain RN4220, and the expression of the β-lactamase reporter was measured. As shown in Figure 5B, both the staphylococcal ϕ55 and enterococcal ϕFL2A promoter regions were expressed in S. aureus, and expression of the reporter required the presence of the corresponding Ltr protein. This indicates that these regulators are the only phage-encoded proteins required for the expression of terS. In addition, the absence of read-through expression from Pcad in the absence of the Ltr proteins demonstrated the existence of a transcriptional terminator in the intergenic region between the ltr gene and the terS promoter.
Figure 5.
Ltr proteins control terS expression. (A) Schematic representation of the different blaZ transcriptional fusions. (B) Derivatives of strain RN4220 containing each of the indicated plasmids were assayed at mid-exponential phase for β-lactamase activity under standard conditions. Samples were normalized for total cell mass. ϕ55 LtrC (YP_240527); ϕFL2A ArpU (YP_003347324); ϕ S. suis ArpU (ZP_03625181); ϕ S. carnosus LtrC (YP_002633590); ϕ37 LtrA (YP_240149); ϕ S. suis LtrB (ZP_03625554).
Specificity and minimal DNA requirements for Ltr binding
The numerous genome sequences of phages encoding Ltr proteins allowed us to investigate both the specificity as well as the minimal DNA requirement that support Ltr binding. We have observed that phages expressing different Ltr proteins sometimes carry identical packaging modules. This is the case with staphylococcal phages ϕ55 and ϕ29 (Supplementary Figure S3). The ϕ29 regulator is a RinA homologue (YP_240603), whereas the ϕ55 ORF34 protein exemplifies regulators in family IV (LtrC). We tested for possible cross-reactivity between these two regulators by determining whether RinA from phage ϕ29 could activate terS expression from ϕ55. We cloned ϕ29 rinA under Pcad control in pCN51 and introduced this plasmid into the ϕ55 ΔltrC mutant. We also complemented this mutant with the aforementioned plasmid pJP1050, a pCN51 derivative expressing ArpU. As shown in Table 2, the different regulators did not cross-react but activated expression only of their specific terS genes. The intergenic region between the gene for the Ltr protein and the terS gene from phages ϕ55 and ϕ29, which includes the terminator for their respective ltr genes and the terS promoter, is nearly identical except for the 5′ region just after the ltr (ltrC or rinA) coding sequence (Supplementary Figure S3). These regions contain in the ϕ55 and ϕ29 genomes several direct repeats immediately distal to the coding sequences for their respective activator genes (Supplementary Figure S3), which we speculate are the recognition sequences for Ltr protein binding.
Initially, to determine whether the repeat regions are absolutely required for Ltr binding, we analysed this region from three different regulators (RinAϕ29, LtrCϕ55 and ArpUϕFL2A; Figure 6A). The fragment spanning the rinA-terS junction from phage ϕ29, diagrammed in Supplementary Figure S3, was fused to the β-lactamase reporter gene present in plasmid pCN41, generating plasmid pJP1214. As a control, we generated plasmids pJP1216, pJP1217 and pJP1218, in which the nucleotides encompassing the first, the second or both repeats were changed to a stretch of As (Figure 6A). These plasmids were introduced into strains JP7904 (ϕ29 lysogen), generating strains JP9585 (wt), JP9769 (first repeat mutated), JP9770 (second repeat mutated) and JP9771 (both repeats mutated), respectively. In absence of phage induction (required to provide the Ltr protein), no transcription of the reporter gene was observed in any strain (data not shown). However, prophage induction significantly increased the expression of the β-lactamase reporter uniquely in the strain carrying plasmid pJP1214 (wt repeats), but not in the others (Figure 6B). Identical results were obtained when we analysed, using pCN41 derivative plasmids, the role of the four repeats present in phage ϕ55. Thus, although induction of the phage ϕ55 increased β-lactamase expression in strain carrying the plasmid pJP1221 (wt), we did not observe any β-lactamase expression from plasmid pJP1227, in which the DNA sequence encompassing the four repeats were changed to T (Figure 6A and B).
Figure 6.
The repeat containing region is required for Ltr binding. (A) Identification of the repeat regions from phages ϕ29, ϕ55 and ϕFL2A, proposed to be involved in Ltr binding. (B) Derivatives of strains JP7904 (lysogenic for ϕ29) and JP2348 (lysogenic for ϕ55) containing each of the indicated plasmids were SOS induced and assayed for β-lactamase activity. Plasmids containing the repeats sequence from phage ϕFL2A were introduced in RN4220 and assayed at mid-exponential phase for β-lactamase activity under standard conditions. Samples were normalized for total cell mass. (C) Lineup of RinA protein sequences from phages 80α and ϕ29, coloured according to relative sequence conservation at each position. Adapted from lineup generated by PRALINE. The scoring scheme works from 0 for the least conserved alignment position, up to 10 (asterisk) for the most conserved alignment position. (D) Derivatives of strains RN10359 (lysogenic for 80α) and JP7904 (lysogenic for ϕ29), containing each of the indicated wt or mutated plasmids, were SOS induced and assayed for β-lactamase activity under standard conditions.
Finally, we observed that the phage ϕFL2A, encoding an ArpU family member, also contained three direct repeats in the same position and with the same length (7 nt) that were found for the previously characterized regulators RinA and LtrC (Figure 6 and Supplementary Figure S3). To generalize our results, we analysed the putative binding region for the ArpU protein. To do that, we made use of the aforementioned plasmid pJP1007 (see Figure 5), in which the arpU coding sequence under the control of the Pcad promoter, the terS promoter region, and the 5′ coding region of the terS gene was fused to the β-lactamase reporter. As a control, we generated plasmid pJP1228, in which the nucleotides corresponding to the three repeats were replaced with adenines. As expected, the three repeats were absolutely required for the ArpU-dependent activation of terS transcription (Figure 6B). Taken together, all these results strongly suggest that the direct repeats present immediately distal to the coding sequences of their respective activator genes are the recognition sequences for Ltr protein binding.
The results described earlier in the text do not determine whether the upstream DNA repeats are sufficient to support Ltr binding. To address this question, we initially compared activation of gene expression using plasmids pJP1207, pJP1214 and pJP1221. These pCN41 derivative plasmids contain the terS promoter region (including the repeats) and the 5′ coding region of the terS gene from phages 80α, ϕ29 and ϕ55, respectively. Phages 80α and ϕ29 encode a RinA protein, and as shown in Supplementary Figure S3, the putative binding regions for the RinA proteins (which we have defined as a block of 59 nt just distal to rinA, including the direct repeats) are basically identical in these two phages. In contrast, the DNA just downstream of this block of nucleotides, including the intergenic region and the terS gene, is completely different. Phage ϕ55 encodes LtrC, and as shown in Supplementary Figure S3, has a repeat-containing region different from that present in phages 80α and ϕ29. However, as indicated previously, the cloned intergenic region from phage ϕ55 is basically identical to that present in ϕ29 and completely different from that of phage 80α (Supplementary Figure S3). Plasmids pJP1207 (80α), pJP1214 (ϕ29) and pJP1221 (ϕ55) were introduced into lysogenic strains RN10359 (80α), JP7904 (ϕ29) and JP2348 (ϕ55), the resident prophages were SOS induced and the β-lactamase reporter was measured. As shown in Figure 7A, the β-lactamase reporter was uniquely expressed in those strains carrying cognate pair phage–plasmid with the DNA repeat region corresponding to the resident prophage, confirming the specificity of the Ltr proteins for their corresponding repeats. Although the RinA-expressing phages 80α and ϕ29 cross-reacted, expression of the reporter gene was higher in those strains containing the specific phage and its cognate cloned DNA sequence. An explanation for this is given later in the text.
Figure 7.
DNA requirements for Ltr binding. (A) Derivatives of strains RN10359 (lysogenic for 80α), JP7904 (lysogenic for ϕ29) and JP2348 (lysogenic for ϕ55), containing each of the indicated plasmids, were SOS induced and assayed for β-lactamase activity under standard conditions. (B) Scheme of the chimeric structures generated to analyse the role of the different regions contained in the terS promoter regions form three different phages (80α, ϕ29 and ϕ55). (C) Derivatives of strains RN10359 (lysogenic for 80α) and JP2348 (lysogenic for ϕ55) containing each of the indicated plasmids were SOS induced and assayed for β-lactamase activity under standard conditions.
The previous results suggested that the repeat containing region is necessary and sufficient to confer Ltr binding specificity. This was further confirmed by generating chimeric structures in plasmid pCN41 in which we interchanged the repeat-containing regions of phages 80α and ϕ55. As mentioned previously, although both are S. aureus infecting phages, the sequence of this region is completely different among these phages (Supplementary Figure S3). We generated plasmid pJP1232, which contains the repeat region from phage 80α fused to the sequence present in ϕ55, and plasmid pJP1234, which contains the repeat region from ϕ55 fused to the sequence present in phage 80α (see scheme in Figure 7B). A similar approach was used to generate chimeras between phages 80α and ϕFL2A and ϕ55 and ϕFL2A. We generated plasmids pJP1233 and pJP1235, carrying the repeat region from phages 80α and ϕ55 fused to the intergenic and terS DNA regions (without the repeat containing region) from the enterococcal infecting phage ϕFL2A, which encodes an ArpU family activator. The rationale was to fuse the repeat regions from the staphylococcal phages to DNA from other genera, which is completely different (both in sequence and in size) and is not related to that present in the staphylococcal phages (Supplementary Figure S3). These plasmids were introduced into the lysogenic strains RN10359 (80α) and JP2348 (ϕ55), and the expression of the β-lactamase reporter was measured after induction of the resident prophage present in each strain. As shown in Figure 7C, phage 80α induced the expression of the reporter genes uniquely from all those plasmids containing its cognate repeats. Identical results were obtained with ϕ55 (Figure 7C). In summary, all these results support the hypothesis that the repeat containing region is necessary and sufficient for specific Ltr binding.
Phages ϕ29 and 80α encode RinA proteins that are identical except in their C-terminal regions, which contain a few substitutions (Figure 6C). As mentioned previously, when examining the repeats where these proteins bind we observed that they were identical except for a base (Supplementary Figure S3). In view of these observations, we speculated that the C-terminal region of these proteins were different to increase their specificity for their cognate repeat sequences. To test this, we made use of the aforementioned plasmids pJP1207 and pJP1214, in which the terS promoter region and the 5′ coding region of the terS gene from phages 80α and ϕ29, respectively, were fused to the β-lactamase reporter. In addition, we generated plasmid pJP1219, which is identical to pJP1214 except that it carries repeat sequences from 80α, and plasmid pJP1212, which is identical to pJP1207 except that it contains the ϕ29 repeats. Each of these four plasmids was introduced into strain RN10359 (80α) and JP7904 (ϕ29), the resident prophages were induced and the β-lactamase reporter was measured. As shown in Figure 6D, expression of the reporter was increased in those strains carrying the cognate pairs of phage–activator and repeats. In conclusion, our results suggest that the C-terminal region of the RinA proteins is involved specifically in recognizing the DNA binding repeats. As shown in Figure 3, the putative structure of this region was conserved in all the representative members of the Ltr superfamily analysed.
Ltr proteins bind to their cognate terS promoters
The results from the reporter plasmids suggested that the Ltr proteins bind specifically and uniquely to their cognate repeat-containing regions present at the end of the ltr coding sequences. To investigate this possibility further, representative His-tagged ArpU family (ϕFL2A) and LtrC family (ϕ55) proteins were expressed in E. coli and purified to homogeneity. Purified recombinant proteins were incubated with an end-labelled ∼100-bp PCR fragment containing the entire presumptive Ltr binding region (see shaded regions in Supplementary Figure S3), and complexes were examined by electrophoretic mobility shift assays. As shown in Figure 8, the recombinant proteins retarded mobility of their cognate terS promoter fragments. This binding was eliminated in the presence of an excess (500-fold) of unlabeled terS fragment, but not when a 500-fold excess of non-specific unlabeled DNA fragment was used as a competitor (data not shown). Moreover, although replacement of a single repeat region did not eliminate binding, the ArpU protein was unable to bind mutant derivative probes containing substitutions of two or more repeat regions (Figure 8C), further confirming that the Ltr proteins activate terS transcription by binding specifically to the repeat-containing region upstream of the late promoter.
Figure 8.
Binding of the Ltr proteins to their cognate terS promoters. Electrophoretic mobility of DIG-labelled repeat containing region from ϕFL2A (ArpU) and ϕ55 (LtrC) was measured in the presence or absence of increasing amounts of purified ArpU (A) or LtrC (B) proteins, as indicated on the top line. (C) Mobility of the wild-type (wt) or mutant (mut) DIG-labelled DNA probes in the presence of purified ArpU protein. Direct repeats are underlined. Asterisk indicated DIG-labelled probe; arrows indicate the shifted complexes.
The phage-encoded cI repressors control expression of the Ltr proteins
The ltr genes lie just downstream of the end of the phage replication module (Figure 1). The precise boundaries of the replication module have not been determined, as this region includes a number of genes of unknown function. However, it has been proposed that this module begins with ssb, the gene encoding a predicted single-stranded DNA-binding protein (20). It would be expected that expression of these genes, as well as ltr, should temporally precede expression of the morphogenetic operon and be under control of the phage cI repressor. In an attempt to characterize the phage transcription units, independent northern blot analyses were performed on RNA isolated after SOS induction of phages 80α, ϕ55 and ϕFL2A. In all cases, although specific hybridization was obtained at appropriate time points, only small RNAs in the 2–4 kb size range could be detected, using probes from several different genes across this region (data not shown). This was also observed in the characterization of the morphogenetic clusters of phages 80α and ϕ11 (9). This suggests that either there are multiple transcription start sites throughout this region or that the RNA is extensively processed.
To investigate further the regulation of ltr gene expression, we carried out transcriptional profiling using phage 80α as a model. We isolated RNA before or 30 and 60 min after SOS induction of strain RN10359, which is lysogenic for phage 80α, and this RNA was analysed using tiling microarrays. The tiling microarrays were designed to analyse the genomic landscape of transcription in a strand-specific manner at 14 nt resolution (see ‘Materials and Methods’ section). We expected that the tiling array format would permit us to map the position of phage 80α transcripts in a complementary way to the conventional molecular analysis. Figure 9A shows the tiling-array hybridization signals of phage 80α, using RNA obtained at different times after induction of the phage cycle. The first observation from this experiment is that phage 80α showed basal transcription of most of its genes in absence of induction. This basal transcription can be explained by the spontaneous low level of prophage induction that occurs in lysogenic cells. Second, there is a clear temporal difference between transcription of rinA, which is seen at 30 min post-induction, and the late (morphogenetic) operon, which does not increase until the 60 min time point. This is consistent with an essential role for rinA in activating late transcription. Finally, 30 min after induction of the phage lytic cycle, we observed what appears to be fairly uniform transcription on the top strand of a region encompassing the cro to the rinA genes. This region includes two predicted open reading frames (ORF11 and ORF19) that lie in the opposite orientation and would be expressed from the other strand, which is also confirmed by the tiling array. Such a distribution of mapped reads in ‘overlapping operons’ has been described previously in genome-wide profiling of S. aureus transcription (21). In that study, these overlapping reads appear to correlate with regions in which short antisense RNAs also map; therefore, it is not clear whether the apparent continuous transcription across these divergent open reading frames reflects a polycistronic mRNA that spans this region or the synthesis of an additional short transcript from the same strand that is an antisense RNA to the divergent ORF. Thus, there are two alternative interpretations of the results we obtained with 80α. One possibility is that that all the genes from cro to rinA belong to a single early operon, which is controlled by the phage-encoded cI repressor. An alternative interpretation is that there are multiple promoters, all co-temporally regulated, and that there are overlapping short RNAs leading to the appearance of uniform transcription across this region.
Figure 9.
The phage-encoded cI repressor controls rinA expression. (A) Transcriptional tiling map showing expression of the early (including rinA) and late genes from phage 80α. Strain RN10359, lysogenic for 80α, was SOS induced, and samples were analysed at the indicated times. (B) Schematic representation of the different blaZ transcriptional fusions obtained to analyse the control of the rinA gene. Plasmids were introduced into strain RN4220 and assayed at mid-exponential phase for β-lactamase activity under standard conditions. In addition, expression of the β-lactamase reporter was also assayed after infection with phage 80α. ND; not determined.
To further explore rinA regulation and the role of the phage repressor, we generated plasmid pJP1195, which contains the DNA region of phage 80α encompassing the cI repressor to the rinA gene, which was fused to the β-lactamase reporter gene. As controls, we constructed plasmids pJP1196, which lacks the cI repressor and pJP1197, which is deleted for both the cI repressor and the cI-cro promoter region (see scheme in Figure 9B). Each of these plasmids was introduced into the non-lysogenic strain RN4220, and the expression of the β-lactamase reporter gene was measured. As expected, rinA transcription was observed in the strain carrying the cI-defective plasmid pJP1196, but not in strains carrying plasmids pJP1195 (encoding the cI repressor) and pJP1197 (mutant in the promoter region controlled by the cI repressor), confirming that the cI repressor controls rinA expression. To determine whether this was a direct or indirect effect, we infected the strain containing plasmid pJP1197 with phage 80α and measured β-lactamase expression. If a specific promoter other than that directly controlled by cI controlled rinA, 80α infection should be able to restore transcription of the rinA gene from plasmid pJP1197 (lacking cI or the cI-cro promoter region). This was not the case (Figure 9B), consistent with a model in which rinA expression is under the direct control of the cI-cro promoter region and therefore part of a single polycistronic early transcript. Similar results were obtained analysing expression of ltrC from ϕ55 (data not shown), suggesting that the proposed mechanism controlling ltr transcription is widespread. It should be pointed out, however, that we cannot conclusively rule out an alternative mechanism in which cI regulates expression of a cis-acting factor that is required to stimulate rinA transcription. Regardless of the mechanism, however, these results demonstrate that rinA is temporally coregulated with other early genes in a cI-dependent fashion, consistent with its role in regulating late transcription.
Our previous studies of 80α transcription indicated that the entire 80α late gene cluster was expressed from a single RinA-activated promoter upstream of terS (9). This is consistent with what was observed in the tiling analysis of the RNA samples obtained 60 min after induction of phage 80α (Figure 9A). To test whether this could be generalized to activation of the morphogenetic gene clusters of different phages by their respective Ltr proteins, we replaced the terS upstream region (ltrC and PterS) in strain JP2348, which is lysogenic for ϕ55, with the region encompassing rinA and the PterS from phage 80α. We also replaced in strain RN10359, lysogenic for 80α, the region encompassing rinA and PterS with the region that contained the ltrC (ORF34) and PterS from ϕ55. We also introduced SaPI1 into these strains; this SaPI is induced by both phages (3) and requires the correct expression of the entire phage-encoded late operon to be transferred (22,23). We predicted that if all late genes are co-transcribed from the promoter situated 5′ of the terS gene, these strains will produce functional phage and SaPI1 particles and lyse. This was the case: the cells lysed after SOS induction and SaPI1 and phage particles were produced basically to the same extent as seen after induction of strains carrying wt phages (data not shown). We noted, however, that the size of the plaques was reduced in the chimeric 80α phage expressing LtrC. A possible explanation for this is discussed later in the text.
Ltr homologs control late gene expression in other phages from Gram-positive bacteria
In addition to the ArpU (ϕFL2A) and LtrC (ϕ55) regulators described earlier in the text, we tested whether other Ltr proteins, from the same family or from other families, worked similarly. Using plasmid pCN42, as previously described (Figure 5), we examined the functionality of ArpU and LtrC family homologues present in the prophages carried in S. suis 89/1591 (ArpU; ZP_03625181) or S. carnosus TM300 (LtrC; YP_002633590). Additionally, we analysed a member from each of the families not previously studied: the regulator present in phage 37 (YP_240149) as an example of the LtrA (sigma70_r4) family, and the regulator present in a S. suis 89/1591 prophage (ZP_03625554) as an example of the LtrB (DUF1492) family (see scheme in Figure 5A). As a control, we generated plasmids carrying mutations in each of the different ltr genes. We introduced these plasmids into S. aureus strain RN4220, and measured β-lactamase expression. As shown in Figure 5B, all of the different Ltr proteins controlled terS expression, confirming that activation of transcription by members of this superfamily represents a widespread strategy used by phages infecting G+ bacteria to control phage morphogenesis and lysis.
DISCUSSION
Transcriptional regulation in bacteriophages infecting the low GC Gram-positive bacteria remains poorly characterized, despite the clear importance of phages in the evolution of these medically and economically important organisms. The immunity repressors of temperate phages of these bacteria have been experimentally analysed in just a few cases, including B. subtilis phage ϕ105 (24), S. aureus phage ϕ11 (25) and Lactococcus lactis phage TP901-1 (26). Analysis of the lysogeny modules of a collection of temperate Siphoviridae from low GC content Gram-positive bacteria revealed a common genetic organization that differs clearly from established genera of temperate Siphoviridae, as well as a novel genomic location for this module (27).
Even less work has been done on other aspects of transcriptional control, particularly in the temperate phages that play such key roles in transduction and lysogenic conversion. A promoter conserved between several lactococcal phages was shown to be regulated by a transcriptional activator from lactococcal bacteriophage ϕ31 (28,29). Although this was described as a middle promoter, it is located near the phage cos site and lies just upstream of a gene cluster that includes a portal gene homologue, indicating that it is actually a late promoter, and analysis of the activator shows that it is a member of the RinA family. An activator of late gene transcription (Alt) has also been identified in the L. lactis phage TP901-1 (30). Like Ltr proteins, Alt binds to a promoter region located upstream of the terS gene (31), but Alt homologues appear to be much more narrowly distributed than the Ltr proteins described here; a BLAST search reveals them only in a group of related lactococcal phages. Transcriptional mapping of S. thermophilus phage Sϕ21, based on northern blot analysis, was consistent with a role for phage-encoded regulators for middle and late transcription (32), but these were not identified.
In a previous study, we identified and characterized a family of late transcription regulators, exemplified by the RinA proteins of staphylococcal phages 80α and ϕ11, which are required for expression of phage packaging and lysis functions (9). Although RinA homologues are widely distributed in phages of low GC Gram-positive bacteria, even many staphylococcal phages did not have a homologue of RinA. This prompted an in silico analysis of genes in the equivalent location in the genomes of these other phages, and further characterization of their function as reported here. We have now identified four additional families of transcriptional regulators that play an equivalent role in a large number of phages. All these families, including RinA, constitute a new super-family of Ltr. The ltr genes encode small proteins with predicted molecular weight of ∼15 kDa that show no similarity to other previously characterized transcriptional regulatory proteins. All of these proteins are encoded by genes located at the end of the early gene cluster, immediately upstream of the late promoters and upstream of the late structural gene clusters. In all cases tested, these proteins appear to be activators that stimulate transcription from a promoter upstream of their respective terS gene, they show specificity for their cognate control regions, and they appear to be the only factor necessary for expression of the entire late gene cluster from a single promoter. It is interesting that S. thermophilus phage Sϕ21 encodes a DUF1492 homologue (Class IV) at the equivalent position in its genome. This suggests that the multiple transcription starts reported for this phage on the basis of northern blot and primer extension analysis (32) were actually 5′-ends that arose from processing of the late transcript, as was found to be the case for ϕ11 and 80α (9).
An understanding of the evolution of global transcription regulators is essential for comprehending the complex networks of cellular metabolism that have developed among related organisms. Bacteriophages live under intense selection pressure, where the primary competitive advantage is predicted to be a high replication, packaging and transfer rate (33). This situation suggests that the evolutionary selection of these organisms has optimized expression of their components to produce a maximal growth rate. Tightly orchestrated temporal control is necessary to prevent premature packaging of the bacteriophage DNA before it has been completely replicated. This temporal control is produced in most Gram-positive infecting phages by the Ltr proteins. As it is predicted that activation of the phage late promoter will occur once the Ltr protein accumulates, linking expression of the ltr genes to the early transcript assures that this increased expression of the ltr genes will be uniquely produced after efficient phage replication. Interestingly, and as discussed earlier in the text, we noted that the chimeric 80α phage expressing the family IV regulator from ϕ55 produced smaller plaques than wt 80α phage. However, this chimeric phage produced a small number of normal plaques. Sequence analysis of three of these plaques revealed changes in the ribosome-binding site for the terS gene. In view of this result, we hypothesize that the different activators are optimized for different replication rates or different levels of expression of the phage-encoded proteins, and that differences in these features among the phages infecting Gram-positive bacteria could be the major force responsible for the diversification and specialization observed in the Ltr family of regulators.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–4, Supplementary Figures 1–3 and Supplementary References [34–39].
FUNDING
Funding for open access charge: Ministerio de Ciencia e Innovación (MICINN) [Consolider-Ingenio CSD2009-00006, BIO2011-30503-C02-01 and Eranet-pathogenomics PIM2010EPA-00606 to J.R.P]; Cardenal Herrera-CEU University [Copernicus-Santander program to J.R.P.]; Insituto Nacional de Investigaciones Agrarias (INIA) [DR08-0093 to M.A.T-M.]; National Institute of Health [R56AI081837 to G.E.C, R01AI022159-23A2 to R.P.N.].
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors are thankful to Alberto Marina (IBV-CSIC, Valencia) for providing technical help in the modelling the Ltr proteins. J.R.P. conceived and designed the study; N.Q.-P. and M.A.T.-M. characterized the mutants and performed gene expression experiments; S.C. and V.M. performed mobility shift assay experiments; A.T.-A., M.A.T.-M. and I.L. performed the tiling array experiment; J.R.P., G.E.C., R.P.N., I.L., N.Q.-P. and M.A.T.-M. analysed the data; J.R.P. and G.E.C. wrote the manuscript; J.R.P., M.A.T.-M., G.E.C., I.L. and R.P.N. obtained funding.
REFERENCES
- 1.Brüssow H, Canchaya C, Hardt W-D. Phages and the evolution of bacterial pathogens: from genomic rearrangements to lysogenic conversion. Microbiol. Mol. Biol. Rev. 2004;68:560–602. doi: 10.1128/MMBR.68.3.560-602.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tormo-Más MÁ, Donderis J, García-Caballer M, Alt A, Mir-Sanchis I, Marina A, Penadés JR. Phage dUTPases control transfer of virulence genes by a proto-oncogenic G protein-like mechanism. Mol. Cell. 2013;49:947–958. doi: 10.1016/j.molcel.2012.12.013. [DOI] [PubMed] [Google Scholar]
- 3.Tormo-Más MÁ, Mir I, Shrestha A, Tallent SM, Campoy S, Lasa Í, Barbé J, Novick RP, Christie GE, Penadés JR. Moonlighting bacteriophage proteins derepress staphylococcal pathogenicity islands. Nature. 2010;465:779–782. doi: 10.1038/nature09065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Novick RP, Christie GE, Penadés JR. The phage-related chromosomal islands of Gram-positive bacteria. Nat. Rev. Microbiol. 2010;8:541–551. doi: 10.1038/nrmicro2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Banks DJ, Porcella SF, Barbian KD, Beres SB, Philips LE, Voyich JM, DeLeo FR, Martin JM, Somerville GA, Musser JM. Progress toward characterization of the group A Streptococcus metagenome: complete genome sequence of a macrolide-resistant serotype M6 strain. J. Infect. Dis. 2004;190:727–738. doi: 10.1086/422697. [DOI] [PubMed] [Google Scholar]
- 6.Schuch R, Fischetti VA. The secret life of the anthrax agent Bacillus anthracis: bacteriophage-mediated ecological adaptations. PLoS One. 2009;4:e6532. doi: 10.1371/journal.pone.0006532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Court DL, Oppenheim AB, Adhya SL. A new look at bacteriophage genetic networks. J. Bacteriol. 2006;189:298–304. doi: 10.1128/JB.01215-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Campbell A. Comparative molecular biology of lambdoid phages. Annu. Rev. Microbiol. 1994;48:193–222. doi: 10.1146/annurev.mi.48.100194.001205. [DOI] [PubMed] [Google Scholar]
- 9.Ferrer MD, Quiles-Puchalt N, Harwich MD, Tormo-Mas MA, Campoy S, Barbe J, Lasa I, Novick RP, Christie GE, Penadés JR. RinA controls phage-mediated packaging and transfer of virulence genes in Gram-positive bacteria. Nucleic Acids Res. 2011;39:5866–5878. doi: 10.1093/nar/gkr158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Úbeda C, Maiques E, Knecht E, Lasa Í, Novick RP, Penadés JR. Antibiotic-induced SOS response promotes horizontal dissemination of pathogenicity island-encoded virulence factors in staphylococci. Mol. Microbiol. 2005;56:836–844. doi: 10.1111/j.1365-2958.2005.04584.x. [DOI] [PubMed] [Google Scholar]
- 11.Úbeda C, Maiques E, Barry P, Matthews A, Tormo MÁ, Lasa Í, Novick RP, Penadés JR. SaPI mutations affecting replication and transfer and enabling autonomous replication in the absence of helper phage. Mol. Microbiol. 2008;67:493–503. doi: 10.1111/j.1365-2958.2007.06027.x. [DOI] [PubMed] [Google Scholar]
- 12.Simossis VA, Heringa J. PRALINE: a multiple sequence alignment toolbox that integrates homology-extended and secondary structure information. Nucleic Acids Res. 2005;33:W289–W294. doi: 10.1093/nar/gki390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Karplus K, Barrett C, Hughey R. Hidden Markov models for detecting remote protein homologies. Bioinformatics. 1998;14:846–856. doi: 10.1093/bioinformatics/14.10.846. [DOI] [PubMed] [Google Scholar]
- 15.Perocchi F, Xu Z, Clauder-Münster S, Steinmetz LM. Antisense artifacts in transcriptome microarray experiments are resolved by actinomycin D. Nucleic Acids Res. 2007;35:e128. doi: 10.1093/nar/gkm683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nicol JW, Helt GA, Blanchard SG, Raja A, Loraine AE. The Integrated Genome Browser: free software for distribution and exploration of genome-scale datasets. Bioinformatics. 2009;25:2730–2731. doi: 10.1093/bioinformatics/btp472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Úbeda C, Maiques E, Tormo M, Campoy S, Lasa Í, Barbé J, Novick RP, Penadés JR. SaPI operon I is required for SaPI packaging and is controlled by LexA. Mol. Microbiol. 2007;65:41–50. doi: 10.1111/j.1365-2958.2007.05758.x. [DOI] [PubMed] [Google Scholar]
- 18.Lleò MM, Fontana R, Solioz M. Identification of a gene (arpU) controlling muramidase-2 export in Enterococcus hirae. J. Bacteriol. 1995;177:5912–5917. doi: 10.1128/jb.177.20.5912-5917.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dombroski AJ, Walter WA, Record MT, Siegele DA, Gross CA. Polypeptides containing highly conserved regions of transcription initiation factor sigma 70 exhibit specificity of binding to promoter DNA. Cell. 1992;70:501–512. doi: 10.1016/0092-8674(92)90174-b. [DOI] [PubMed] [Google Scholar]
- 20.Christie GE, Matthews AM, King DG, Lane KD, Olivarez NP, Tallent SM, Gill SR, Novick RP. The complete genomes of Staphylococcus aureus bacteriophages 80 and 80α—Implications for the specificity of SaPI mobilization. Virology. 2010;407:381–390. doi: 10.1016/j.virol.2010.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lasa Í, Toledo-Arana A, Dobin A, Villanueva M, de Los Mozos IR, Vergara-Irigaray M, Segura V, Fagegaltier D, Penadés JR, Valle J, et al. Genome-wide antisense transcription drives mRNA processing in bacteria. Proc. Natl Acad. Sci. USA. 2011;108:20172–20177. doi: 10.1073/pnas.1113521108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tormo MA, Ferrer MD, Maiques E, Ubeda C, Selva L, Lasa I, Calvete JJ, Novick RP, Penadés JR. Staphylococcus aureus pathogenicity island DNA is packaged in particles composed of phage proteins. J. Bacteriol. 2008;190:2434–2440. doi: 10.1128/JB.01349-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tallent SM, Langston TB, Moran RG, Christie GE. Transducing particles of Staphylococcus aureus pathogenicity island SaPI1 are comprised of helper phage-encoded proteins. J. Bacteriol. 2007;189:7520–7524. doi: 10.1128/JB.00738-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dhaese P, Seurinck J, De Smet B, Van Montagu M. Nucleotide sequence and mutational analysis of an immunity repressor gene from Bacillus subtilis temperate phage phi 105. Nucleic Acids Res. 1985;13:5441–5455. doi: 10.1093/nar/13.15.5441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Das M, Ganguly T, Chattoraj P, Chanda PK, Bandhu A, Lee CY, Sau S. Purification and characterization of repressor of temperate S. aureus phage phi11. J. Biochem. Mol. Biol. 2007;40:740–748. doi: 10.5483/bmbrep.2007.40.5.740. [DOI] [PubMed] [Google Scholar]
- 26.Pedersen M, Ligowska M, Hammer K. Characterization of the CI repressor protein encoded by the temperate lactococcal phage TP901-1. J. Bacteriol. 2010;192:2102–2110. doi: 10.1128/JB.01387-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lucchini S, Desiere F, Brüssow H. Similarly organized lysogeny modules in temperate Siphoviridae from low GC content gram-positive bacteria. Virology. 1999;263:427–435. doi: 10.1006/viro.1999.9959. [DOI] [PubMed] [Google Scholar]
- 28.Walker SA, Klaenhammer TR. Molecular characterization of a phage-inducible middle promoter and its transcriptional activator from the lactococcal bacteriophage phi31. J. Bacteriol. 1998;180:921–931. doi: 10.1128/jb.180.4.921-931.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Walker SA, Dombroski CS, Klaenhammer TR. Common elements regulating gene expression in temperate and lytic bacteriophages of Lactococcus species. Appl. Environ. Microbiol. 1998;64:1147–1152. doi: 10.1128/aem.64.3.1147-1152.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brøndsted L, Pedersen M, Hammer K. An activator of transcription regulates phage TP901-1 late gene expression. Appl. Environ. Microbiol. 2001;67:5626–5633. doi: 10.1128/AEM.67.12.5626-5633.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pedersen M, Kilstrup M, Hammer K. Identification of DNA-binding sites for the activator involved in late transcription of the temperate lactococcal phage TP901-1. Virology. 2006;345:446–456. doi: 10.1016/j.virol.2005.10.007. [DOI] [PubMed] [Google Scholar]
- 32.Ventura M, Bruttin A, Canchaya C, Brüssow H. Transcription analysis of Streptococcus thermophilus phages in the lysogenic state. Virology. 2002;302:21–32. doi: 10.1006/viro.2002.1571. [DOI] [PubMed] [Google Scholar]
- 33.Rohwer F, Thurber RV. Viruses manipulate the marine environment. Nature. 2009;459:207–212. doi: 10.1038/nature08060. [DOI] [PubMed] [Google Scholar]
- 34.Kreiswirth BN, Löfdahl S, Betley MJ, O'Reilly M, Schlievert PM, Bergdoll MS, Novick RP. The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature. 1983;305:709–712. doi: 10.1038/305709a0. [DOI] [PubMed] [Google Scholar]
- 35.Yasmin A, Kenny JG, Shankar J, Darby AC, Hall N, Edwards C, Horsburgh MJ. Comparative genomics and transduction potential of Enterococcus faecalis temperate bacteriophages. J. Bacteriol. 2010;192:1122–1130. doi: 10.1128/JB.01293-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Úbeda C, Barry P, Penadés JR, Novick RP. A pathogenicity island replicon in Staphylococcus aureus replicates as an unstable plasmid. Proc. Natl Acad. Sci. USA. 2007;104:14182–14188. doi: 10.1073/pnas.0705994104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Arnaud M, Chastanet A, Débarbouillé M. New vector for efficient allelic replacement in naturally nontransformable, low-GC-content, gram-positive bacteria. Appl. Environ. Microbiol. 2004;70:6887–6891. doi: 10.1128/AEM.70.11.6887-6891.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Charpentier E, Anton AI, Barry P, Alfonso B, Fang Y, Novick RP. Novel cassette-based shuttle vector system for gram-positive bacteria. Appl. Environ. Microbiol. 2004;70:6076–6085. doi: 10.1128/AEM.70.10.6076-6085.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Söding J. Protein homology detection by HMM-HMM comparison. Bioinformatics. 2005;21:951–960. doi: 10.1093/bioinformatics/bti125. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.