Abstract
LINE1s occupy 17% of the human genome and are its only active autonomous mobile DNA. L1s are also responsible for genomic insertion of processed pseudogenes and >1 million non-autonomous retrotransposons (Alus and SVAs). These elements have significant effects on gene organization and expression. Despite the importance of retrotransposons for genome evolution, much about their biology remains unknown, including cellular factors involved in the complex processes of retrotransposition and forming and transporting L1 ribonucleoprotein particles. By co-immunoprecipitation of tagged L1 constructs and mass spectrometry, we identified proteins associated with the L1 ORF1 protein and its ribonucleoprotein. These include RNA transport proteins, gene expression regulators, post-translational modifiers, helicases and splicing factors. Many cellular proteins co-localize with L1 ORF1 protein in cytoplasmic granules. We also assayed the effects of these proteins on cell culture retrotransposition and found strong inhibiting proteins, including some that control HIV and other retroviruses. These data suggest candidate cofactors that interact with the L1 to modulate its activity and increase our understanding of the means by which the cell coexists with these genomic ‘parasites’.
INTRODUCTION
A recent study proposes that over two-thirds of the human genome are repetitive DNA, and that most of this derives from transposable elements (1). Long INterspersed Element-1s (LINE1s) are the major class of retrotransposons, and they move in a ‘copy and paste’ manner involving reverse transcription of an RNA intermediate and insertion of its cDNA copy at a new chromosomal location. RNA of the element and its associated proteins are transported into the nucleus where it is believed reverse transcription and integration occurs in a single step on the DNA itself by a process known as target-primed reverse transcription. In this model, element-encoded endonuclease nicks the bottom strand of target DNA to expose a 3′-hydoxyl that primes reverse transcription of the L1 RNA template. Second-strand cDNA synthesis follows and the integrant is resolved in a manner poorly understood (2). Short target site duplications flank the element at the insertion site.
LINE1 (L1) retrotransposons, the only remaining active autonomous mobile DNA in humans, underwent a massive mammalian expansion, and ∼500 000 copies occupy at least 17% of our genome. L1-encoded proteins have also been responsible for the genomic insertion in trans of thousands of processed pseudogenes, and >1 million Alu and 3000 SINE-VNTR-Alu (SVA) non-autonomous retrotransposons (3–5). Most L1s are dead molecular fossils, truncated, rearranged or mutated. However, at least 100 remain potentially active in any human individual (6,7). Up to 5% of newborn children are estimated to have a new retrotransposon insertion, and to date, there are 97 known human disease-causing germ line insertions of L1s, Alus and SVAs (8,9). Recent investigations, driven in part by advances in high-throughput sequencing, suggest that rates of retrotransposition may be much higher during early embryogenesis and in certain somatic cell types, such as neuronal progenitor cells, stem cells and some cancers (10–16).
Retrotransposons are active participants in reorganizing their resident genomes. Retrotransposition occasionally generates target site deletions, or adds non-retrotransposon DNA to the genome by processes termed 5′- and 3′-transduction. Recombination between non-homologous retrotransposons causes deletions, duplications or rearrangements of gene sequence. Ongoing retrotransposition salts genomes with novel splice sites, polyadenylation signals and promoters, and so builds new transcription modules. Transposable elements have also greatly contributed to chromosome architecture and cell evolution [see reviews in (9,17,18)].
The L1 expresses a 6-kb bicistronic RNA that encodes the 40 kDa Open Reading Frame-1 RNA-binding protein (ORF1p) of essential but uncertain function, and a 150 kDa ORF2 protein with endonuclease and reverse transcriptase (RT) activities (Figure 1A). Retrotransposition is a complex process involving transcription of the L1, transport of its RNA to the cytoplasm, translation of the bicistronic RNA, formation of a ribonucleoprotein (RNP) particle, its re-import to the nucleus and target-primed reverse transcription at the integration site.
Figure 1.
pc-L1-1FH immunoprecipitates basal L1 RNP complexes from 293T cell lysates after α-FLAG agarose purification. (A) Structure of FLAG-HA-tagged pc-L1-1FH cloned in vector pcDNA6 myc/his B. RT: ORF2 reverse transcriptase domain; EN: endonuclease domain; PCMV: CMV promoter; BGH An: bovine growth hormone polyadenylation signal. (B) FLAG-tagged ORF1p expressed from the construct pc-L1-1FH binds α-FLAG agarose independent of RNase digestion (lanes 5 and 8), but untagged ORF1p (construct pc-L1-RP) will not bind (lane 6). (C and D) Detection of L1 proteins in the RNP IP. Lanes 1–4: input lysates; lanes 6–9: immunoprecipitates; lanes 1, 2, 6 and 7: cytoplasmic fractions; lanes 3, 4, 8 and 9: nuclear fractions. (C) FLAG-HA-tagged ORF1p, detected by α-FLAG antibody. Putative ORF1p dimer and trimer bands are visible in IP samples (lanes 7 and 9). The reason for their absence in lysate samples is unclear (lanes 2 and 4). IP purification factors were determined for cytoplasmic (lanes 2 versus 7, 26-fold) and nuclear (lanes 4 versus 9, 42-fold) fractions and are presented in Supplementary Table S3. Lane labels are at the bottom of panel D. (D) ORF2p detected by α-ORF2-N (154–167) antibody (lanes 7 and 9). ORF2p in nuclear lysate samples is below the level of detection (lanes 3 and 4). (E) ORF2p reverse transcriptase activity detected in both nuclear and cytoplasmic IP reactions containing pc-L1-1FH (lanes 3 and 5), but not in reactions with the empty vector (lanes 2 and 4). RT- control: the RT incubation step was omitted and 2 µl of pc-L1-1FH immunoprecipitate was added directly to the PCR reaction. No PCR product was detected (lanes 1 and 6). The assay is described in Kulpa and Moran (51). (F) L1 RNA detected by RT–PCR (lanes 3 and 5). RT-: RT enzyme was omitted from the cDNA synthesis step using pc-L1-1FH immunoprecipitates (lanes 1 and 6). (G) FLAG-HA-tagged ORF2p is detected in nuclear and cytoplasmic extracts after IP of pc-L1-2FH. (H) The purity of nuclear and cytoplasmic whole-cell lysate fractions is shown by western blotting. α-HDAC1 is a strictly nuclear protein (54) and α-MEK1/2 is cytoplasmic (55). (I) Immunoprecipitated samples resolved on silver-stained polyacrylamide gels. To support protein identification data from complex IP samples, selected prominent band regions were excised for additional MS sequencing. Both cytoplasmic (left) and nuclear (right) IP fractions are shown.
The complexities of retrotransposition, and the various known mechanisms of transposable element control, predict a complex interaction between L1s and cellular proteins. These proteins and their roles in retrotransposition are poorly understood. A few transcription factors that interact with L1s have been identified (19–23). Non-homologous end-joining repair proteins, such as XRCC1, Ku70 and DNA-PK, have been implicated in resolution of the L1 integrate at the time of insertion (24–26). Yeast two hybrid, affinity purification and shRNA screening studies have identified a few proteins that interact with the L1 and affect retrotransposition, including HNRNPL, NCL, MOV10 and PABPC1 (23,27–29). In addition, the cell has evolved a number of proteins that stand against unrestricted retrotransposition, including the APOBEC3 family of cytosine deaminases, adenosine deaminase ADAR1, chromatin-remodeling factors and members of the piRNA pathway for post-transcription gene silencing that functions in the male germ line [reviewed in (30–32)].
The L1 RNP complex is the fundamental unit of retrotransposition. Ultra-centrifugation, immunofluorescence (IF), immunoprecipitation (IP) and cell culture assays indicate that the basal L1 RNP comprises ORF1p and ORF2p preferentially bound to their own encoding L1 RNA (33–35). Cellular factors modulating retrotransposition are likely associated with the basal RNP. We, therefore, predicted that valuable information might be gleaned about the many steps of retrotransposition by identifying cellular macromolecules that interact with the L1 RNP and affect its efficiency of retrotransposition. We epitope-tagged ORF1p in an L1 construct and, following its IP from human cells, identified associated proteins by mass spectrometry (MS). Preliminary examination of these results identified MOV10 helicase as interacting with the L1 complex to inhibit its retrotransposition (29). We now present a detailed investigation of the protein interactome of ORF1p and its L1 RNP complex. Association of many of these proteins with the L1/ORF1p RNP was confirmed by direct co-IP and subcellular co-localization in cytoplasmic granules. When overexpressed, many of these proteins significantly reduce retrotransposition in cell culture. Our results will guide future investigations of the L1 life cycle, from transcription to reinsertion in the genome, and of strategies the cell has evolved to coexist with these genomic mutagens.
MATERIALS AND METHODS
Cloning of plasmid constructs
To generate pc-L1-1FH, we introduced by Kunkel mutagenesis (36) tandem FseI-PacI-SgrAI restriction enzyme sites to replace the stop codon of ORF1 of L1RP (37) cloned in the modified pBS KS- vector jCC5-RPS. A double FLAG-Hemagluttinin (HA)-tag was extracted by polymerase chain reaction (PCR) from plasmid pOZ-FH-N [gift of Y. Nakatani, Harvard (38)] and cloned between the FseI and SgrAI sites. The tagged L1 was then inserted into the vector pcDNA6 myc/his B (Invitrogen) between the NotI and SacII sites.
pCEP-EGFP, pc-L1-RP, ORF1-GFP-L1-RP, FLAG-tagged FL-CDSA (DBPA) and RFP-tagged HNRNPA1 have been described (27,34). The 6myc-p72 (full-length DDX17 isoform 1), mouse pUB-GFP-ZBP1 (GFP-mIGF2BP1) and pcDNA 3.1/V5-His CycT1 (CCNT) were gifts from R. Janknecht [University of Oklahoma; (39)], S. Sim [Yale University; (40)] and A. Gatignol (Lady Davis Institute for Medical Research, Montréal), respectively. Ultimate ORF cDNA clones (Invitrogen), available to us from a cDNA library maintained by the ChemCore of Johns Hopkins University School of Medicine, were V5-tagged on their N-termini by shuttling them from pENTR221 vector into pcDNA3.1/nV5-DEST using Gateway Technology (Invitrogen). Ultimate ORF Clone ID numbers are shown in Supplementary Table S2. Selected cDNAs (SRP14, PURA, IVNS1ABP, NAT10 and DDX5) were amplified by PCR from a CytoTrap XR human testes library (Stratagene), cloned into the Gateway pDONR vector and shuttled into pcDNA3.1/nV5-DEST.
Immunoprecipitation
For MS sequence determination, HEK293T cells were transfected in T75 flasks with 15 µg of pc-L1-1FH or pcDNA6 myc/his B empty vector and expanded for 44 h. For each IP reaction, ∼3 × 107 cells were harvested with phosphate-buffered saline (PBS)/2 mM ethylenediaminetetraacetic acid and pelleted, and 2.0 ml of cytoplasmic extracts was prepared with NE-PER Nuclear and Cytoplasmic Extraction Kit (Pierce) according to the manufacturer’s protocol, with the exception that the recommended detergent concentration was reduced by half (0.25% NP40). Nuclear pellets were washed three times with Dulbecco’s phosphate-buffered saline (DPBS; Invitrogen), lysed in 900 μl of nuclear extraction reagent and final nuclear extracts were diluted 1.5 times with DPBS. Buffers were supplemented with protease inhibitor cocktail, phosphatase inhibitor cocktails II and III, vanadyl ribonucleoside complexes, phenylmethylsulfonyl fluoride (Sigma) and RNasin (Promega). Extracts were incubated with 60 μl of anti-FLAG M2 affinity gel (Sigma) with rotation for 3 h, washed 5× (5 min each) with buffer A [160 mM NaCl/50 mM Tris–HCl (pH 7.5)/1 mM ethylenediaminetetraacetic acid/0.2% NP40], and eluted with 150 μg/ml FLAG peptide (Sigma). Eluates (90 μl per sample) were recovered with Costar Spin-X Centrifuge Tube Filters (Corning) and mixed with 3× sodium dodecyl sulfate (SDS)/β-mercaptoethanol loading buffer. A purification factor table for immunoprecipitation of FLAG-tagged ORF1p expressed from pc-L1-1FH is shown in Supplementary Table S3. Total protein concentrations were determined by bicinchoninic acid protein assay. A standard curve was calculated from dilutions of bovine serum albumin. ORF1p purification was assessed by ImageJ software (NIH) analysis of band intensities on western blots.
For each co-immunoprecipitation of Figure 3A, extracts from 3.5 × 106 293T cells transfected with L1 and test protein constructs in T75 flasks were prepared in 370 μl of buffer A supplemented as aforementioned. Lysates containing test proteins of predominantly nuclear localization were sonicated. Reactions transfected with pc-L1-1FH were immunoprecipitated with 30 μl of anti-FLAG M2 affinity gel and eluted as aforementioned. RNase inhibitors were omitted from samples treated with 15 μg/ml DNase-free RNase (Roche). To detect their interaction with endogenous ORF1p, V5-tagged proteins were transfected in 7 × 106 2102Ep embryonal carcinoma cells (Figure 3B), immunoprecipitated on Protein G Agarose/Salmon Sperm DNA (Upstate) and eluted in SDS loading buffer with heating at 95°C, followed by addition of β-mercaptoethanol.
Figure 3.
Ectopically expressed and endogenous proteins associate with L1 complexes in multiple cell lines. (A) V5-, 6xMyc- or GFP-tagged proteins exogenously expressed in 293T cells specifically co-immunoprecipitate with pc-L1-1FH, but not empty vector (pcDNA6 myc/his B) [IP: α-FLAG affinity gel, western blotting (WB): α-V5, α-Myc or α-GFP]. IP reactions were in the presence or absence of 15 μg/ml RNase (lanes 3–5). Lysate input samples are also shown (lanes 1 and 2). Several protein panels are reproduced from Goodier et al. (29). GFP-mIGF2BP1 is derived from mouse.
The bottom-most panel is representative of tagged ORF1p in the input and IP fractions and confirms that RNase treatment does not affect ORF1p immunoprecipitation on α-FLAG agarose. Molecular weights shown include the epitope tag. The protein standard is See Blue Plus 2 (Invitrogen). (B) Co-IP of endogenous ORF1p from 2102Ep cells by selected V5-tagged proteins (IP: α-V5/IgG affinity gel). Upper rows: detection of ORF1p (WB: α-ORF1 AH40). Asterisk indicates proteins that strongly co-IP endogenous ORF1p. ‘o’ marks proteins that clearly associate with ORF1p on gel overexposure. Lower rows confirm successful IP of the test proteins (WB: α-V5). Exposure times are not necessarily the same for each lane. Input lysate fractions are shown in Supplementary Figure S1. (C) Co-IP of selected endogenous proteins by pc-L1-1FH from 293T cells (IP: α-FLAG affinity gel, WB: various antibodies). The antibody name is followed by the expected protein molecular weight. NCL has an expected weight of 77 kDa, but observed molecular weight of ∼100 kDa. As previously reported (27), an antibody against DDX39B [UAP56; (50)] detects a dominant band of 55 kDa in cytoplasmic lysates, and a smaller isoform of 49 kDa (the expected size for DDX39B) that co-IPs with tagged ORF1p. In total, 12 antibodies were tested; α-PCBP2 (Abnova), α-FBL (Santa Cruz) and α-TOP1 (Spring) failed to detect their endogenous targets in pc-L1-1FH immunoprecipitates. The bottom-most panel shows efficient immunoprecipitation of tagged ORF1p detected by α-FLAG antibody.
MS sequencing and database analyses
MS sequencing was performed by the Johns Hopkins Mass Spectrometry and Proteomics Facility. Proteins in solution were reduced and alkylated with iodacetamide and proteolyzed with sequencing grade modified porcine trypsin (Promega). Proteins in gel bands were proteolyzed with trypsin as previously described (41). Protein identification by liquid chromatography tandem mass spectrometry (LCMS/MS) was performed using LTQ ion trap MS (Thermo Fisher Scientific) interfaced with a 2D nanoLC system (Eksigent). Peptides were fractionated by reverse-phase high-performance liquid chromatography on a 75 μm × 100 mm C18 column with a 10 μm emitter using a 0–60% acetonitrile/0.1% formic acid gradient >90 min at 300 nl/min. Peptide sequences were identified using Proteome Discoverer and Mascot software (Matrix Science) to search the NCBInr 167 database, with oxidation on methionine and carbamidomethylation on cysteine as variable modifications. Mascot search result *.dat files were processed in Scaffold (Proteome Software, Inc.) to validate protein and peptide identifications. A protein required ≥90% probability for peptide annotation and the identification of at least two unique peptides to be included in this study. Any protein also detected in the empty vector reaction was discarded as a false positive. Core ribosomal proteins and suspected contaminant proteins (including keratins, annexin A1, plakoglobin, desmoplakin, peroxiredoxin2 and methylosome subunit plCln) detected in pre-run or post-run bovine serum albumin controls were also omitted from analyses.
Additional analyses used DAVID v6.7 [Database for Annotation, Visualization and Integrated Discovery; (42)] and STRING v9 [Search Tool for the Retrieval of Interacting Genes/Proteins; (43)]. The Nucleolar Proteome Database is available at http://www.lamondlab.com/NoPDB (44).
Yeast two-hybrid analyses
The Cytotrap system (Stratagene) screen of a human testis cDNA library has been described (27). A human HeLa cDNA library (HL4000AA) was also screened in yeast strain Y187 using the GAL4 Matchmaker Two Hybrid System II according to the manufacturer’s protocols (Clontech). As baits, full-length L1 ORF1 and the N-terminal 717 nucleotides of ORF2 (EN domain) were cloned in the vector pAS2-1.
Cell culture and retrotransposition assays
Human 2102Ep embryonal carcinoma cells (a gift from P. K. Andrews, University of Sheffield), HeLa-HA cells (a gift from J. Moran, University of Michigan) and HEK293T cells (ATCC) were grown in Dulbecco’s modified Eagle’s medium with 10% FBS (Hyclone), GlutaMax and Pen-Strep (Invitrogen). All transfections used FuGENE HD reagent (Promega).
The EGFP (enhanced green fluorescent protein) cell culture L1 retrotransposition assay was conducted as previously described (45). Briefly, 2.5 × 105 293T cells/well were seeded in 6-well plates. The following day, 1.0 μg of 99-PUR-RPS-EGFP, a plasmid containing L1RP and EGFP retrotransposition reporter cassette, was co-transfected together with 0.5 μg of empty vector (pcDNA3, Invitrogen) or test plasmid (four replicate wells each). Five days post-transfection, cells having a retrotransposition event, and hence expressing EGFP, were assayed by flow cytometry. Gating exclusions were based on background fluorescence of plasmid 99-PUR-JM111-EGFP, which has a double-point mutation in ORF1 that abolishes retrotransposition (46). Within each experiment, results were normalized to fluorescence of 99-PUR-RPS-EGFP co-transfected with pcDNA3 empty vector.
To control for any differences in transfection efficiencies of test proteins or off-target effects on EGFP reporter expression, we followed the strategy of Wei et al. (47). Each test plasmid (0.5 μg) was co-transfected in quadruplicate wells of 12-well plates with pCEP-EGFP (0.5 μg), a construct that constitutively expresses EGFP from a cytomegalovirus (CMV) promoter. Four days post-transfection, EGFP fluorescence was determined by flow cytometry. These results were then used to correct retrotransposition rates (by dividing each retrotransposition reading by the average of four replicate pCEP-EGFP readings for the corresponding test protein; Figure 6B).
Figure 6.
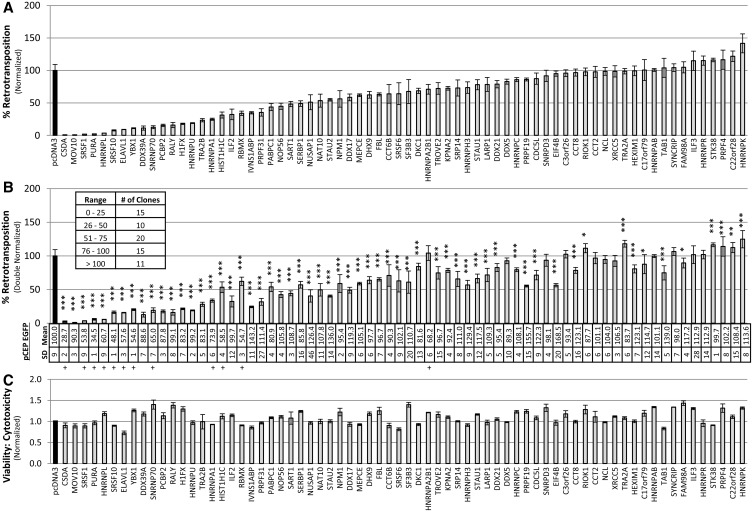
Some L1-associated proteins strongly inhibit L1 retrotransposition in 293T cells. (A) The 99-PUR-RPS-EGFP was co-transfected in 293T cells with empty vector (pcDNA3) or test constructs expressing tagged proteins. Five days later, percentages of EGFP-positive cells were determined by flow cytometry. Each plasmid pair was transfected in four replicate wells, and results are normalized to pcDNA3 vector control (black bar). Proteins are ordered by their effect on retrotransposition. (B) To control for any off-target effects, test constructs were co-transfected with pCEP-EGFP, a plasmid that constitutively expresses EGFP. Four days later, cells were assayed for gain or loss of fluorescent cells (panel below). Fluorescence is normalized to pcDNA3 control (table, top row). Readings of ≤80% are marked below with ‘+’. Standard deviation for four replicates is shown (bottom row). These results were then used to adjust the retrotransposition levels of Figure 1A by dividing by the average of the pCEP-EGFP expression readings. P-values were calculated by two-tailed t-test and are indicated above each histogram bar (*P < 0.05, **P < 0.01, ***P < 0.001). The inset table summarizes the number of proteins that fall into each retrotransposition percent range. (C) Results of MultiTox-Fluor Multiplex Cytotoxicity assay (Promega) for potential cell toxicity caused by overexpression of test proteins. Test constructs were transfected in 96-well plates and assayed at 3 days. The histogram shows ratios of live to dead cell readings normalized to empty vector control.
To determine potential cell toxicity because of transfection of test proteins, 15,000 293T cells were seeded in 75 μl of Dulbecco’s modified Eagle’s complete medium in 96-well plates. The next day, transfection reactions prepared with 100 ng of test plasmid, 0.3 μl of Fugene HD and 25 μl of Opti-MEM Reduced Serum Medium (Invitrogen) were added to each well. After 3 days, a MultiTox-Fluor Multiplex Cytotoxicity Assay kit (Promega) was used to assay cell toxicity. This assay simultaneously measures cell viability and cytotoxicity in a single-reagent reaction, permitting ratios of live to dead cell readings to be calculated.
Immunofluorescence and Western blotting
Immunofluorescence techniques have been described previously (27). Purified AH40.1 polyclonal antibody against ORF1 protein was a gift from T. Fanning (Armed Forces Institute of Pathology, MD, USA) and M. Singer (Carnegie Institution of Washington) (48). Polyclonal affinity-purified antibody α-ORF2-N (154–167) was described (27). Other antibodies included ms α-FLAG-tag (Sigma), ms α-V5-tag (Invitrogen), ms α-Myc-tag (Invitrogen), ms α-fibrillarin (38F3; Santa Cruz), rb α-HEXIMI [a gift from J. Yik, Berkeley; (49)], gt α-LARP1 (SDIX), ms α-nucleolin/C23 (4E2; Santa Cruz), ms α-nucleophosmin/B23 (Zymed), ms α-PABPC1 (clone 10E10; G. Dreyfuss, University of Pennsylvania), ms α-Ro60 (S. Wolin, Yale University) and rb α-UAP56 [M. Green, University of Massachusetts Medical Center; (50)]. Donkey Cy3-, DyLight 488- or DyLight 549-conjugated and horseradish peroxidase-conjugated secondary antibodies were from Jackson ImmunoResearch Laboratories. Slides were examined with a Nikon A1 confocal laser microscope system, and images were analyzed with the NIS-Elements software platform.
All protein samples were resolved on NuPAGE 4–12% Bis–Tris polyacrylamide gels (Invitrogen) using 3 × SDS/β-mercaptoethanol loading buffer. Western blot antibody incubation was in DPBS/0.05% Tween 20/5% dry milk overnight at 4°C. Membranes were washed 3× for 10 min with DPBS/0.05% Tween 20, exposed to secondary antibody at room temperature for 2 h, and washed again. Detection used SuperSignal West Pico Chemiluminescent Substrate (Pierce) and Hyperfilm ECL (GE Healthcare).
Reverse transcriptase assay and RT–PCR
Reverse transcriptase analysis followed the LEAP protocol of Kulpa and Moran (51). Two microliters of IP sample was added to each cDNA extension reaction. cDNA PCR amplification used Expand Long Template PCR System (Roche) and 2 μl of the extension reaction. Products were separated on 2% agarose gels and confirmed by sequencing. Primers used in the assay are noted in Supplementary Table S1.
For RT PCR analyses, RNA from total cell lysates was extracted using the RNA Easy Mini kit (Qiagen). Immunoprecipitates were used without extraction. Samples were treated with TURBO DNase (Invitrogen), and cDNA was generated using an iScript Select cDNA Synthesis Kit for L1 RNA (Figure 1) or iScript cDNA Synthesis Kit (Bio-Rad) for small RNAs and mRNAs (Figure 4). Standard PCR reactions used the Expand Long Template PCR System (Roche) and primers listed in Supplementary Table S1.
Figure 4.
snRNAs, scRNAs (left) and selected mRNAs (right) are detected in L1 RNP immunoprecipitates by RT–PCR. Results are for whole-cell lysates (lanes 1–4) and immunoprecipitates (lanes 5–8) from empty vector (pcDNA6 myc/his B; lanes 1, 3, 5 and 7) and pc-L1-1FH (lanes 2, 4, 6 and 8) transfected cells. RT enzyme was omitted (lanes 1, 2, 5 and 6) or included (lanes 3, 4, 7 and 8).
RESULTS
Tagged L1 constructs successfully immunoprecipitate the basal L1 RNP
Endogenous full-length Short INterspersed Element (SINE) and LINE transcripts are difficult to detect in most cell types, although expression levels increase under some conditions of stress (52). L1-encoded proteins, especially ORF2p, are also suppressed in most cells (53). Low expressivity and the confounding presence of numerous retrotransposons within transcribed mRNAs make biochemical study of endogenous L1 elements difficult.
Faced with the difficulties of investigating RNA and proteins encoded by endogenous L1s, we previously developed expression constructs of L1RP [an L1 highly active in cell culture, (37)] containing ORF1 fused with T7, TAP or FLAG epitope tags (27,53). For the present study, L1RP with tandem HA-FLAG tag fused to the C-terminus of ORF1 (construct pc-L1-1FH) or ORF2 (pc-L1-2FH) was cloned into a vector that replicates in HEK293T cells (Figure 1A). Tagging ORF1 in this manner diminishes (to 30%) but maintains cell culture retrotransposition of L1RP in an EGFP-based reporter assay (29,45).
pc-L1-1FH allows the immunoprecipitation on α-FLAG agarose of basal L1 RNP complexes from cell lysates; untagged ORF1p expressed from an otherwise identical construct (pc-L1-RP) does not bind agarose (Figure 1B, lanes 5, 6 and 8). After transfection in 293T cells and purification, we detected in cytoplasmic and nuclear immunoprecipitates both ORF1 and ORF2 proteins (Figures 1C and 1D), robust ORF2p RT activity (Figure 1E) as determined by an in vitro PCR-based assay (51), and L1 RNA (Figure 1F). Western blot analyses of immunoprecipitated tagged ORF1p consistently yielded an expected band of 45 kDa plus minor bands of ∼90 and 140 kDa (Figure 1C, lanes 7 and 9), sizes consistent with persistent multimers resistant to SDS loading buffer containing β-mercaptoethanol. ORF1p trimerization has been characterized (56,57), and Hohjoh and Singer (58) indicated that ORF1p multimers might form by forces other than disulfide bonds. Multimers of ORF1p explain the detection of ORF1p peptides in excised gel fragments >45 kDa (Figure 1I).
Western blot analyses of RNP complexes formed by pc-L1-2FH detected tagged ORF2 protein in large-scale preparations of cytoplasmic and nuclear immunoprecipitates (Figure 1G). However, it was not possible using pc-L1-2FH to purify sufficient complexes to reliably analyze their protein composition by MS; therefore, we used pc-L1-1FH to co-IP cellular proteins associated with LINE1.
MS analysis of the L1 RNP reveals a tight network of RNA-binding proteins
We transfected pc-L1-1FH and empty vector in parallel in 293T cells and performed α-FLAG-tag IP from nuclear and cytoplasmic fractions. Western blotting confirmed significant fraction purity (Figure 1H). Both complex samples and eight selected band regions excised from preparative gels were analyzed by liquid chromatography tandem MS. Silver-stained electrophoretic gels showed relatively few bands in empty vector lanes compared with those in pc-L1-1FH lanes (Figure 1I). Excluding 36 ribosomal proteins (listed in Supplementary Table S2), 96 proteins with two or more predicted peptides and unique to pc-L1-1FH isolates were identified. Forty proteins were identified in the nuclear fraction only, 34 in the cytoplasmic fraction only and 22 in both fractions (Table 1 and Supplementary Table S2). As expected, exogenous ORF1 protein expressed from pc-L1-1FH was predominant in MS analyses of both cytoplasmic and nuclear IP reactions (Figure 1C and Supplementary Table S2). Although ORF2p is clearly visible on Western blots (Figure 1D), its peptide sequences were not found among the MS reads.
Table 1.
Summary of analyses of proteins that co-IP with the L1
| Gene symbol | Alternate symbol | Protein name | Co-IPs with pc-L1-1FH | With ORF1p in granules | Nuclear (Nu) / cytoplasmic (C) extracts |
|---|---|---|---|---|---|
| C3orf26 | MGC4308 | Chromosome 3 open reading frame 26 | Y(a) | N | Nu |
| C17orf79 | COPR5 | Chromosome 17 open reading frame 79, cooperator of PRMT5 | Beads | nd | Nu |
| C22orf28 | HSPC117 | Chromosome 22 open reading frame 28, tRNA-splicing ligase RtcB homolog | N | Y | C |
| CAT | Catalase | nc | nc | C | |
| CCNT1 | CYCT1 | Cyclin T1 | ? | N | |
| CCT2 | TCP-1-β | Chaperonin containing TCP1, subunit 2 (β) | N | N | C |
| CCT4 | TCP1 Δ | Chaperonin containing TCP1, subunit 4 (Δ)/stimulator of TAR RNA binding, | nc | nc | C |
| CCT6B | TCP-1-ζ-2 | Chaperonin containing TCP1, subunit 6B (ζ 2) | N | nd | C |
| CCT8 | TCP-1-θ | Chaperonin containing TCP1, subunit 8 (θ) | N | nd | C |
| CDC5L | CDC5 cell division cycle 5-like (S. pombe) | ? | N | Nu | |
| CSDA | DBPA | Cold shock domain protein A | Y(27) | Y | Nu/C |
| DARS | Aspartyl-tRNA synthetase | nc | nc | C | |
| DDX17 | p82 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 17, isoform p82 variant | Y(a) | N | Nu |
| DDX21 | RH-II/GuA | DEAD (Asp-Glu-Ala-Asp) box polypeptide 21 | Y(a) | N | Nu |
| DDX23 | PRPF28 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 23 | nc | nc | C |
| DDX39A | BAT1, URH49 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 39 | Y(a,b,c) | N | Nu |
| DDX5 | p68 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 5 | Y(a) | N | Nu |
| DHX9 | DDX9, RHA | DEAH (Asp-Glu-Ala-His) box polypeptide 9 | ? | o | Nu |
| DKC1 | DKC1 dyskeratosis congenita 1, dyskerin | N | nd | C | |
| EIF4B | Eukaryotic translation initiation factor 4B | beads | Y | Nu/C | |
| ELAVL1 | HUR | Embryonic lethal abnormal vision n, Drosophila like 1/Hu antigen R | Y(a,b) | Y | Nu |
| FAM120A | C9orf10 | Family with sequence similarity 120A/oxidative stress-associated Src activator | nc | nc | Nu |
| FAM98A | Family with sequence similarity 98, member A | Y(a) | Y | C | |
| FBL | Fibrillarin | ? | N | Nu | |
| H1FX | Histone H1x | Y(a,b) | N | Nu | |
| HEXIM1 | HIS1 | Hexamethylene bis-acetamide inducible 1 | Y(a,c) | N | C |
| HIST1H1B | H1B | Histone cluster 1, H1b | nc | nc | Nu/C |
| HIST1H1C | H1.2, H1C | Histone 1 H1C | Y(a) | N | Nu/C |
| HIST1H1E | H1.4, H1E | Histone cluster 1, H1e | nc | nc | Nu |
| HNRNPA1 | Heterogeneous nuclear ribonucleoprotein A1 | Y(a,b) | Y | Nu | |
| HNRNPA2B1 | Heterogeneous nuclear ribonucleoprotein A2/B1, transcript variant B1 | Y(a) | N | Nu | |
| HNRNPAB | ABBP1 | Heterogeneous nuclear ribonucleoprotein A/B/APOBEC1-binding protein 1 | Y(a) | Y | Nu |
| HNRNPC | HNRNP C1/C2 | Heterogeneous nuclear ribonucleoprotein C (C1/C2) | Y(a,b) | N | Nu |
| HNRNPH3 | Heterogeneous nuclear ribonucleoprotein H3 | ? | N | Nu | |
| HNRNPK | Heterogeneous nuclear ribonucleoprotein K | ? | N | Nu/C | |
| HNRNPL | Heterogeneous nuclear ribonucleoprotein L, | Y(a) | N | Nu | |
| HNRNPR | Heterogeneous nuclear ribonucleoprotein R | ? | N | Nu | |
| HNRNPU | SAF-A | Heterogeneous nuclear ribonucleoprotein U (scaffold attachment factor A) | Y(a,b) | N | Nu |
| IGF2BP1 | IMP1, ZBP1 | Insulin-like growth factor 2 mRNA binding protein 1 | Y(a) | Y | Nu/C |
| IGF2BP2 | IMP2 | Insulin-like growth factor-binding protein 2, 36 kDa | nc | nc | C |
| ILF2 | NF45 | Interleukin enhancer-binding factor 2 | Y(a) | o | Nu |
| ILF3 | NFAR, NF90 | Interleukin enhancer-binding factor 3 | Y(a,b) | N | Nu |
| IVNS1ABP | Influenza virus NS1A-binding protein | Beads | Y | C | |
| KPNA2 | Karyopherin α 2 (RAG cohort 1, importin α 1) | Y(a) | nd | C | |
| KRI1 | KRI1 homolog (S.cerevisiae) | nc | nc | Nu | |
| LARP1 | La ribonucleoprotein domain family, member 1 | Y(c) | Y | Nu/C | |
| LUC7L3 | CROP | Cisplatin resistance-associated-overexpressed protein | nc | nc | C |
| MARS | Methionyl-tRNA synthetase | nc | nc | C | |
| MATR3 | Matrin 3 | nc | nc | Nu | |
| MEPCE | BCDIN3 | 7SK snRNA methylphosphate capping enzyme | Y(a) | N | C |
| MOV10 | KIAA1631 | Moloney leukemia virus 10, homolog (mouse) | Y(a,b) | Y | Nu/C |
| NAT10 | N-acetyltransferase 10 (GCN5-related) | ? | N | Nu | |
| NCL | Nucleolin | Y(a,c) | N | Nu/C | |
| NOP56 | NOL5A | Nucleolar protein 56 | Y(a) | N | Nu |
| NPM1 | B23 | Nucleophosmin 1 | Y(c) | N | Nu |
| NUSAP1 | Nucleolar and spindle-associated protein 1 | ?,Y(b) | N | C | |
| PABPC1 | Poly(A)-binding protein, cytoplasmic 1 | Y(a,c) | Y | Nu/C | |
| PABPC4 | Poly(A)-binding protein, cytoplasmic 4 | nc | nc | Nu/C | |
| PCBP2 | HNRNPE2 | Poly(rC)-binding protein 2 | Y(a) | Y | Nu/C |
| PDIA3 | Protein disulfide isomerase family A, member 3 | nc | nc | C | |
| PPAN | Peter pan homolog (Drosophila) | nc | nc | C | |
| PPM1B | Protein phosphatase, Mg2+/Mn2+ dependent, 1B | nc | nc | C | |
| PRPF4 | PRP4 pre-mRNA processing factor 4 homolog (yeast)/ U4/U6 small nuclear ribonucleoprotein Prp4 | N | nd | C | |
| PRPF19 | PRP19/PSO4 pre-mRNA processing factor 19 homolog (S.cerevisiae) | N | N | C | |
| PRPF31 | PRP31 pre-mRNA processing factor 31 homolog (S.cerevisiae) /U4/U6 small nuclear ribonucleoprotein Prp31 | beads | Y | C | |
| PTBP1 | HNRNPI | Polypyrimidine tract binding protein 1 | nc | nc | C |
| PURA | Purine-rich element-binding protein A | Y(a,b) | N | Nu | |
| RALY | HNRNPCL2 | RNA-binding protein, autoantigenic (hnRNP-associated with lethal yellow homolog (mouse)) | Y(a,b) | N | Nu |
| RBMX | HNRNPG | RNA-binding motif protein, X-linked | Y(a) | N | Nu |
| RIOK1 | RIO kinase 1 (yeast) | Y(a) | Y | C | |
| RNMT | RG7MT1 | RNA (guanine-7-) methyltransferase | nc | nc | Nu |
| SART1 | Squamous cell carcinoma antigen recognized by Tcells/U4/U6.U5 tri-snRNP-associated protein 1 | Beads | N | Nu | |
| SERBP1 | PAIRBP1, CGI-55 | SERPINE1 mRNA-binding protein 1 | Y(27) | Y | Nu |
| SF3B1 | PRPF10 | Splicing factor 3B, subunit 1 | nc | nc | Nu/C |
| SF3B3 | SAP130 | Splicing factor 3B, subunit 3 | ?,Y(b) | N | Nu/C |
| SNRNP70 | U1 Small nuclear ribonucleoprotein 70 kDa | Y(a) | N | C | |
| SNRPD3 | Small nuclear ribonucleoprotein polypeptide D3 polypeptide 18 kDa | Beads | N | C | |
| SNX8 | Sorting nexin 8 | nc | nc | C | |
| SPIN1 | Spindlin 1 | nc | nc | Nu/C | |
| SR140 | U2SURP | U2 snRNP-associated SURP domain containing | Beads | N | Nu |
| SRP14 | Signal recognition particle 14 kDa | Y(a) | N | Nu | |
| SRSF1 | ASF, SF2, SF2p33 | Serine/arginine-rich splicing factor 1 isoform 1 | Y(a,b) | Y | Nu/C |
| SRSF6 | SFRS6, SRP55 | Splicing factor, arginine/serine-rich 6 | Y(a,b) | N | C |
| SRSF10 | FUSIP1, SRp38 | Serine/arginine-rich splicing factor 10 | Y(a) | Y | Nu |
| SSB | La, LARP3 | Sjogren syndrome antigen B (autoantigen La) | Y(c) | N | Nu/C |
| STAU1 | Staufen, RNA-binding protein, homolog 1 (Drosophila) | Y(a) | Y | C | |
| STAU2 | Staufen, RNA-binding protein, homolog 2 (Drosophila) | Y(a,b) | Y | C | |
| STK38 | Serine/threonine protein kinase 38 | Beads | Y | Nu/C | |
| SYNCRIP | hnRNPQ | Synaptotagmin binding, cytoplasmic RNA interacting protein | N | Y | Nu/C |
| TAB1 | MAP3K7IP1 | TGF-β activated kinase 1/MAP3K7-binding protein 1 | ? | N | C |
| TOP1 | DNA topoisomerase I | nc | nc | Nu | |
| TRA2A | Transformer-2 α homolog (Drosophila) | Y(a) | N | Nu | |
| TRA2B | SFRS10 | Transformer 2 β homolog (Drosophila) | Y(a) | N | Nu |
| TROVE2 | Ro60, SSA | 60 kDa Ro protein, Sjogren syndrome antigen A2 | Y(a,c) | Y | Nu/C |
| XRCC5 | KU80 | X-ray repair complementing defective repair in Chinese hamster cells 5 (double-strand-break rejoining) | Beads | N | Nu/C |
| YBX1 | YB1, NSEP1 | Y box-binding protein 1 | Y(a,b,c) | o | Nu/C |
The list of 96 proteins was used to query the Gene Ontogeny (GO) database (Figure 2A and Supplementary Table S4). A majority of these proteins bind nucleic acids. The L1 ORF1p interactome contains many heterogeneous nuclear ribonucleoproteins (hnRNPs), splicing factors, chaperones, transport and localization proteins, and transcription/translation factors. Querying the STRING database of experimentally confirmed protein–protein interactions returns a tight network of interactions dominated by RNP proteins (Figure 2B).
Figure 2.
Proteins identified with the L1 form a tight network of interactions dominated by RNA-binding proteins. (A) Pie chart of results of DAVID (Database for Annotation, Visualization and Integrated Discovery) analysis showing selected functional categories for the 96 candidate proteins (42). Protein counts for each category are shown within the slices. Protein names are listed in Supplementary Table S4. (B) STRING (Search tool for the retrieval of interacting genes/proteins)-derived network of known protein–protein interactions among the 96 candidate proteins. The confidence view is shown in Jensen et al. (43).
cDNAs for 69 of the 96 putative L1-interacting proteins were subcloned from a cDNA library into expression vectors or were obtained as gifts. Most cDNAs-encoded proteins with N-terminal V5-epitope-tags. After their co-transfection in 293T cells, 41 of the 69 proteins tested were found to co-IP specifically with the FLAG-tagged ORF1p L1 RNP (Figure 3A). Nine additional proteins bound non-specifically to the affinity agarose. Nine other proteins were expressed at significantly higher levels in the presence of co-transfected pc-L1-1FH compared with the empty vector; therefore, their specific co-IP with the L1 could not reliably be confirmed (marked as ‘?’ in Table 1). Predictably, most interactions were RNA-dependent and disappeared with RNase treatment (Figure 3A).
We next assayed the ability of a subset of the V5-tagged proteins to co-IP endogenous ORF1p from 2102Ep cells, a human embryonal carcinoma line that expresses L1 ORF1p at particularly high levels (48). After IP on α-V5-bound agarose, overexposure of Western blots showed that many proteins tested (16/21), but not empty vector, obviously co-IPed endogenous ORF1p (Figure 3B). Finally, IP of pc-L1-1FH on α-FLAG agarose was also able to pull-down selected endogenous proteins from 293T cells (Figure 3C). Collectively, the co-IP data strongly support the MS sequencing results and validate the use of tagged-L1 constructs for discovering cellular proteins that potentially associate with the L1.
Further confirming their association with the L1, a number of the proteins identified in the present study were also detected during yeast two-hybrid cDNA library screens. As previously described (27), a CytoTrap human testes cDNA library (Stratagene) was queried using ORF1p as bait. This system targets test proteins to the cell membrane for detection. Six of nine proteins previously recovered by the CytoTrap screen were also found by the present study to associate with the L1 (CSDA, DDX21, DDX39A, HNRNPA1, SERBP1 and YBX1). Although recovered from yeast, TLS-FUS, signal recognition particle (SRP)72 and SRSF7 were not detected by L1 IP; however, SRP14 and several other serine/arginine-rich (SR) splicing factors (SRSF1, SRSF6 and SFRS10) were found [(27); J. Goodier, unpublished data]. We also performed a GAL4 Matchmaker II (Clontech) two-hybrid analysis of a HeLa cell cDNA library using ORF1p or the endonuclease domain (EN) of ORF2p as bait. The GAL4 system queries interactions by targeting proteins to the nucleus. The strongest and most frequently interacting ‘hit’ was HEXIM1, which was recovered with ORF2p EN. HEXIM1 negatively regulates transcription by sequestering positive transcription elongation factor b (P-TEFb), together with 7SK small nuclear (sn) RNA, CDK9, CCNT1 (cyclin T1), LARP7 and MEPCE in an inactive RNP (49,59). Notably, HEXIM1, MEPCE, CCNT1 and 7SK RNA were all also recovered with pc-L1-1FH during the present study (Table 1 and Figure 4). ORF1p bait mainly bound endogenous ORF1-related clones from the Matchmaker II library (five unique clones from a total of eight). No other hits overlapped with the present study.
Non-L1 RNAs associate with the ORF1p complex
Some proteins detected by our MS sequencing preferentially associate with small RNAs that are overrepresented in the human genome as pseudogene families. For example, SR140, SART1 and SR splicing factors bind small spliceosomal RNAs, U1 to U6: these small nuclear (sn) RNAs all have processed pseudogenes, >60% of which are related to U6 (60,61). Autoantigens TROVE2 (Ro60) and SSB (La) form a core complex with hY small cytoplasmic (sc) RNAs, which also comprise pseudogene families (60,61). hnRNPK and nucleolin are subunits of TROVE2 particles containing hY1 and hY3 RNAs (62,63). TROVE2, SSB and SRP14 also bind RNA of Alu retrotransposons, which comprise 10% of the human genome, and HEXIM1, MEPCE and CCNT1 complex with 7SK snRNA (49,59). The human 7SK pseudogene family (64) includes 160 members with obvious short target site duplications, a hallmark of L1-mediated insertion (J. Goodier, unpublished data).
We performed conventional RT–PCR to assay for the presence of selected RNA species in tagged-ORF1p RNP immunoprecipitates. L1 RNA and all small RNAs tested were associated with pc-L1-1FH, but were absent or barely detectable in empty vector control samples (Figure 4). Alu RNA was also detected in RNP samples; however, it is difficult to conclude these represent bona fide Pol III-derived Alu transcripts, as many mRNAs also contain embedded Alus. Some but not all mRNAs tested were detected in the pc-L1-1FH sample, including α-actin and Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) mRNA. Interestingly, humans have 62 and mice 331 GAPDH-associated pseudogenes, one of the largest numbers among all proteins, and a phenomenon linked with high-mRNA expression and recent bursts of mammalian retrotransposition (4,65).
It is reasonable to suggest that the association of RNA-binding proteins with the L1 RNP make their bound RNAs preferred targets for pseudogene formation. Of course, expression levels of these RNAs may differ significantly between tumor cells, normal somatic cells and the germ line where retrotransposition acts to establish pseudogenes in the inherited human genome. RNA-binding proteins are also known to associate with abundant RNA species in cell extracts after cell lysis; therefore, observed interactions may not recapitulate the in vivo state of ORF1p RNP complexes (66). Furthermore, some small RNAs may be overrepresented as retrogenes simply because of high-RNA copy number. Application to multiple cell lines of recently developed techniques such as PAR-CLIP (67), which combines photo-activatable cross-linking and high-throughput RNA-sequencing, will yield a more comprehensive and quantitative enumeration of the RNA composition of human L1 RNPs.
ORF1p co-localizes in cytoplasmic granules with many RNP-associated proteins
Stress granules (SGs) are cytoplasmic aggregates that contain stalled 48S pre-initiation complexes and are induced by a range of stresses (68). SGs frequently associate with P-bodies (PBs), another class of cytoplasmic granule rich in factors of RNA decay, including those of the RNA-induced silencing complex (RISC). We previously demonstrated that exogenously expressed ORF1p co-localizes with L1 RNA and ORF2p in SGs [(34), confirmed by (35)]. Endogenous ORF1p also forms cytoplasmic granules in the absence of external stress applied to the cell (53).
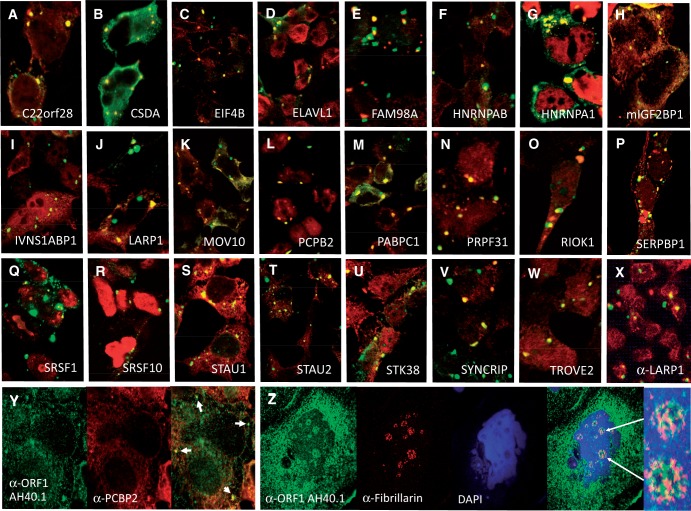
As described earlier in the text, a majority of proteins we tested directly co-IP with the ORF1p-tagged L1 (Figure 3). We next co-transfected in 293T cells tagged cDNA constructs and ORF1-EGFP-L1-RP, a plasmid containing CMV promoter, ORF1 C-terminally tagged with EGFP, followed by intact downstream L1 sequence (34). One-third of the tagged proteins tested (23/69) at least partially co-localized with ORF1p-EGFP in cytoplasmic granules in the absence of any applied stress (Figure 5A–W). Panels X and Y show co-localization in granules of ORF1p with endogenous LARP1 and PCBP2. Similarly, we previously reported that endogenous ORF1p co-localizes with endogenous HNRNPA1 and PABPC1 in granules of chemically stressed 2102Ep cells (27). Several additional proteins we report here as associated with the ORF1p RNP have also been detected in cytoplasmic granules by other studies but not this one, perhaps because of differences in cell type (Supplementary Table S5). Indeed, many protein components of SGs and PBs have been identified [reviewed in (68,69)]. To the best of our knowledge, seven proteins of Figure 5 have not been previously reported in cytoplasmic granules, including C22orf28, FAM98A, HNRNPAB, IVNS1APB1, PRPF31, RIOK1 and STK38.
Figure 5.
Many L1-associated proteins co-localize with EGFP-ORF1p in cytoplasmic granules of unstressed 293T cells. (A–W) Construct ORF1-EGFP-L1-RP was co-transfected with V5-tagged proteins in all cases except (B) FL-CSDA, (G) RFP-HNRNPA1 and (H) mouse GFP-mIGF2BP1 (the latter being co-transfected with pc-L1-1FH, which was detected by α-FLAG antibody). Only overlaid confocal micrographs are shown. (X) Endogenous LARP1 protein co-localizes with ORF1-EGFP-L1-RP in 293T cells. (Y) Endogenous ORF1p and PCBP2 co-localize in some cytoplasmic granules of 2102Ep cells (shown by arrows). (Z) Endogenous ORF1p and fibrillarin (FBL) co-localize in nucleoli of 2102Ep cells. ORF1p is typically found in nucleoli of only a minor percentage of cells (53). Enlargement of two nucleoli are shown. In Y and Z endogenous, ORF1p is detected by α-ORF1 AH40.1 antibody.
We also previously reported that both ORF1p and ORF2p enter nuclei and concentrate in nucleoli of a minor percentage of cells. A functional nucleolar localization signal was mapped to the EN domain of ORF2p (34,53). These data predict that some nucleolar proteins will associate with those L1 RNPs residing in nucleoli. It is also possible some protein(s) bind L1 RNPs to cause their active transport to nucleoli. We examined the Nucleolar Proteome Database (44) and DAVID analyses and found that 47 of the 96 non-ribosomal proteins that co-IP with the L1 RNP have been reported in nucleoli. These include such canonical nucleolar proteins as nucleolin, fibrillarin and nucleophosmin (Supplementary Table S6). Figure 5Z illustrates co-localization of endogenous fibrillarin and ORF1p in nucleoli of 2102Ep cells. High-resolution microscopy suggests that ORF1p and fibrillarin partially partition into different nucleolar compartments (inset). Fibrillarin concentrates in the dense fibrillar compartment (70).
L1 RNP-associated proteins inhibit cell culture retrotransposition
Cellular proteins with roles in retrotransposition, either in its promotion or inhibition, likely associate with the L1 RNP during some stage of its lifecycle. Therefore, we assayed 70 tagged cDNA clones for effect of their overexpression on L1 retrotransposition in a widely used cell culture assay developed in our laboratory (45,46). An EGFP reporter cassette, interrupted by an intron in opposite transcriptional orientation and inserted into the 3′-UTR of L1RP (construct 99-PUR-RPS-EGFP), is expressed only when the L1 transcript is spliced, reverse transcribed, its cDNA inserted in the genome and the EGFP reporter gene expressed from its own promoter. We co-transfected in 293T cells 99-PUR-RPS-EGFP, together with empty vector or tagged cDNA constructs, and 5 days post-transfection assayed for fluorescent (i.e. retrotransposition-positive) cells by flow cytometry (Figure 6A).
The duration of the cell culture retrotransposition assay is necessarily long: in 293T cells first retrotransposition events appear after 24 hours, and slowly accumulate over the course of the experiment. Therefore, it is important to determine whether co-expression of proteins have off-target effects that might bias results. Potential effects include cytotoxicity or altered EGFP reporter gene expression caused by ectopic expression of the test protein, or differences in plasmid transfection efficiencies. To reveal any biases, we followed the approach of Wei et al. (47) and, in conjunction with retrotransposition assays, co-transfected pCEP-EGFP, a vector that constitutively expresses EGFP, together with empty vector or tagged test constructs, and after 4 days performed flow cytometry. Twelve constructs reduced or increased expression from pCEP-EGFP by ≥20%. We, therefore, adjusted the retrotransposition assay results to account for loss/gain of EGFP expression caused by the test-proteins (Figure 6B).
We also used the MultiTox-Fluor Multiplex Cytotoxicity Assay kit (Promega) to directly assay cell toxicity caused by expression of the test proteins. This dual-detection assay generates a ratio of live to dead cell readings, so normalizing for cell number. Most test proteins showed little effect on the viability of 293T cells, and cell toxicity cannot generally account for observed decreases in retrotransposition (Figure 6C).
Proteins tested inhibited retrotransposition or left it little changed (Figure 6B). Fifteen reduced retrotransposition to <25% of wild-type levels. Together these proteins are involved in a range of cellular functions, including RNA splicing modulation, gene expression and RNA transport.
DISCUSSION
Little is known of the cellular proteins involved in LINE1 retrotransposition. Our work is the first to comprehensively identify macromolecules associated with the L1 RNP and to assess effects of their overexpression on cell culture retrotransposition. We report the isolation and analysis of human LINE1 RNP complexes and reveal candidate interacting non-ribosomal proteins. Associations with the L1 immunoprecipitates of many of the detected proteins were confirmed by co-IP and subcellular co-localization.
A significant percentage of the proteins identified may be common to many RNP complexes. For example, 18 of the proteins shown in Table 1 are shared with proteins associated with Ago1 and Ago2 RISC RNP complexes (71). We predict that some proteins we have identified are integral components of the L1 RNP with roles in retrotransposition function and control. Other proteins may be non-specific binders, or tethered to non-L1 RNAs captured within the RNP. As with any affinity purification, association of certain proteins may be indirect because of their presence in other co-purifying multi-protein complexes that contain only one or two proteins that directly interact with the L1 RNP. Some interactions may be cell-line specific or may not represent what occurs during in vivo development. Moreover, an unknown amount of tagged ORF1p, free in solution and not part of retrotransposition-competent RNPs, will have co-purified with immunoprecipitates. Only ongoing functional analyses of individual members of the L1 interactome will ascertain their relevance to L1 biology. However, the discovery of numerous interacting proteins suggests that the processes of L1 translation, RNP formation, RNA import and export, and genomic insertion recruit a wide spectrum of cellular cofactors.
It is of considerable interest that many of the proteins that co-IP with the L1 also co-localize with ORF1p in cytoplasmic RNA granules. The observation that ORF1p foci form constitutively in the absence of exogenous stress prompted us to suggest that the cell recognizes overexpression of the L1 itself as a stress and targets L1 RNPs to granules where they can be sequestered or shunted to P-bodies for degradation, thereby mitigating mutagenic effects of retrotransposition (27). The data of Figure 5 describe candidate proteins that may mediate sequestration of L1 RNPs [one possible example being MOV10, which closely co-localizes with exogenous and endogenous ORF1p in cytoplasmic granules and strongly restricts retrotransposition; (29)]. However, a direct role for cytoplasmic granules in retrotransposition remains unclear, and studies to date have dealt only with retroviral-like elements. P-body components are in fact important for retrotransposition of Ty1 and Ty3 yeast Long terminal repeat (LTR) retrotransposons (72,73). Contrarily, P-body disruption increases retrotransposition of rodent intracisternal A particle (IAP) elements (74).
The Nucleolar Proteome Database and DAVID reveal that one-half of pc-L1-1FH-associated non-ribosomal proteins can be found in nucleoli (Supplementary Table S6). As with stress granules, a role for nucleoli in L1 retrotransposition is unknown. Both ORF1 and ORF2 proteins have been detected in nucleoli. Many small RNAs that form retropseudogene families spend at least part of their life cycle in nucleoli (53,75); perhaps it is in nucleoli that these small RNAs are captured in L1 RNPs and carried to sites of genome insertion. Interestingly, arthropod non-LTR R1 and R2 retrotransposons have evolved to insert into nucleolar 28S rRNA genes (76). Nucleolar proteins that associate with the L1 RNP may provide clues to investigate the role of nucleoli in retrotransposition.
Functional categories within our list of L1 RNP-associated proteins erect signposts to further research. For example, enzymes involved in post-translational modification were detected. Little is known of modifications to retrotransposon proteins and their effects on retrotransposition. Phosphorylation of Ty5 integrase regulates its integration into heterochromatin (77). Hohjoh and Singer (58) reported phosphorylation of L1 ORF1p, but they did not identify the modified residue. We detected with the ORF1p RNP one Ser/Thr protein phosphatase, PPM1B and two Ser/Thr kinases: RIOK1, a protein of unknown function, and STK38, a negative regulator of MAP3K1/2 signaling (78).
Several proteins are involved in the chromosome organization. NAT10 acetylates histones and microtubules and may affect telomerase subunit assembly and localization (79). NUSAP1 is essential for bundling and stabilizing spindle microtubules and may promote their targeting to chromosomes (80). Multiple isoforms of H1 linker histone also co-IP with ORF1p RNPs and inhibit cell culture retrotransposition when overexpressed (Figures 3 and 6B). Recently it was shown that inhibition of Drosophila dH1 increases expression of all classes of mobile DNA, including R1 and R2 retrotransposons (81).
In association with the pc-L1-1FH, we detected RNA and RNP transport proteins, including DDX39A, HNRNPA2B1, HNRNPA1, KPNA2 (importin α1), staufens (STAU1 and STAU2), SRP14, YBX1 and zipcode-binding proteins (IGF2BP1 and IGF2BP2) (Supplementary Table S4). Mouse IGF2BP1, for example, has been shown to mediate nuclear export of the TROVE2 (Ro60)/Y3 snRNA complex (40); both Ro60 and hY3 RNA were detected with the L1 RNP (Figures 3 and 4). Staufens are double-stranded RNA-binding proteins required for microtubule-dependent transport of neuronal RNA through dendrites, and may be involved in RNA export from the nucleus (82). Importin α, in association with importin β, binds proteins at their nuclear localization signals and facilitates RNP transport to the nuclear pore and across the nuclear envelope (83). And SRP14, as part of the signal recognition particle, targets selected proteins to the endoplasmic reticulum. Binding of SRP 9/14 proteins to RNA of Alu elements [themselves evolved from SRP (7SL) RNA] precedes and is likely necessary for efficient L1-mediated Alu retrotransposition (84,85). By studying transport proteins, we may begin to understand how the cell controls movement of L1 RNPs out of the nucleus, into granules, or back into the nucleus to sites of genomic insertion.
We also identified factors involved in alternative mRNA splicing (Supplementary Table S4). TRA2B, for example, regulates tissue-specific and alternative splicing and is essential for mouse development (86). Recently, it was found that HNRNPC, a global suppressor of splicing, binds splice sites within Alu RNAs and competes with splicing factor U2AF65 to prevent aberrant exonization (87). Several SR proteins, and especially SRSF1, mediate not only constitutive and alternative splicing, but other aspects of mRNA elongation, export, translation and decay (88). It is not known whether these factors associate directly with L1 RNA or rather with other mRNAs co-purifying with the L1 RNP complex. However, although the L1 lacks introns, its mRNA contains numerous, although mostly weakly predicted splice sites, can undergo extensive and complex splicing in cells and produces products that differ significantly in abundance and pattern between different tumor cell lines. It has been proposed that internal splicing of full-length L1 transcripts adds another level of control against retrotransposon insertion (89,90).
Phylogenetic analyses suggest that eukaryote non-LTR retrotransposons predate all LTR retrotransposons, which in turn gave rise to retroviruses through the acquisition of an envelope (env) gene (91–94). Predictably, some cellular mechanisms of retroviral and retrotransposon inhibition are shared. MOV10 helicase, for example, originally identified as an inhibitor of HIV, strongly restricts L1 retrotransposition in cell culture (29,95). APOBEC3 family members, also first known to inhibit some HIV strains by hypermutation of minus-strand DNA, were subsequently found to be potent inhibitors of retrotransposition in cells. Intriguingly, suppression of LI and Alu retrotransposons appears not to be accompanied by APOBEC-induced mutation, but it may involve sequestering retrotransposon RNAs in high-molecular weight complexes, possibly stress granules [reviewed in (31,96)]. These multiple modes of activity predict the existence of auxiliary proteins that modulate APOBEC3 function. Our study did not detect APOBECs, probably because they are expressed at low levels in 293T cells (97). However, 35 proteins are shared between the APOBEC3G and our immunoprecipitated L1 complexes (98). APOBEC1, which deaminates apolipoprotein B mRNA, also reduces mobility of mouse L1s by a deamination-independent mechanism (99). Interestingly, two APOBEC1-associated proteins were detected in our L1 RNP reactions: (i) Apobec1-binding protein-2 (ABBP-2/HNRPAB), a Class II DnaJ homolog required for efficient apoB mRNA editing (100) and (ii) SYNCRIP (HNRPQ), which inhibits apoB mRNA editing (101). Perhaps one of these proteins recruits the L1 to APOBEC complexes.
Figure 6B shows 19 proteins whose expression reduces cell culture retrotransposition by >60%. In some cases, protein overexpression may disrupt other cellular processes, and only indirectly limit L1 activity. Some retrotransposition effects may be cell line specific. With these caveats in mind, this study presents candidate factors worthy of further investigation for key roles in the biology of retrotransposition. We have shown that RISC pathway–associated helicase MOV10 strongly restricts all active human non-LTR retrotransposons in cells (29). Dai et al. (23) reported that PolyA-binding protein C1 (PABPC1) is part of the L1 RNP complex, and important for its formation and retrotransposition. Recently Peddigari et al. (28) used affinity capture to show that four proteins, hnRNPL, hnRNPR, SYNCRIP and NCL, specifically bind a cis-acting ORF2 IRES sequence in mouse L1 RNA. Notably, their knockdown of endogenous hnRNPL increased retrotransposition up to 10-fold in the cell culture assay, whereas knockdown of NCL decreased retrotransposition; SYNCRIP and hnRNPR had no effect. All four of these proteins were also identified in our study, and overexpression of human hnRNPL reduced retrotransposition almost 95% in 293T cells (the three other proteins had no effect; Figure 6B). Such studies justify our approach of cataloguing proteins interacting with the L1 RNP as a way to discover cellular factors involved in modulating retrotransposition.
We performed a literature search to ascertain whether any of the proteins shown in Table 1 also interact with HIV retrovirus. Twenty-one of the 150 top-ranked cellular proteins identified by a proteomics screen as associated with HIV-1 Rev also co-IPed with pc-L1-1FH, including RNA helicases DDX5, DHX9, DDX17 and DDX21. Silencing of DDX17 in HeLa CD4+ cells greatly reduced HIV p24 release and cellular Gag and Env, whereas conversely, silencing DDX5 increased levels of p24 and Gag (102). A functional genomic screen for host proteins that affect HIV-1 infection duplicates three proteins from our list, CCNT, PURA and SSB (103). A second genome-scale siRNA screen for cellular factors associated with HIV-1 replication identified only CCT2 (104), whereas a third (105) identified none of our proteins. In addition to MOV10, several proteins that strongly inhibit retrotransposition (Figure 6) also interact with infectious viruses. PURA binds the HIV-1 trans-activation response (TAR) element to activate transcription and interacts with Rev to enhance nuclear export of unspliced viral RNA (106,107). YBX1 also binds TAR and Tat protein to increase viral promoter activity (108). ELAVL1 binds HIV-1 RT and modulates reverse transcription in infected cells (109). A dominant negative truncation mutant of HNRNPU binds the 3′ LTR of HIV-1 mRNA and blocks its accumulation in the cytoplasm (110). Inhibition of HNRNPA1 in T lymphocytes causes an increase of proviral transcription and increased viral production (111). PCBP2 downregulates vesicular stomatitis virus gene expression and is involved in the interferon-α response against hepatitis C virus (112). Finally, IVNS1APB, a protein of unknown function, interacts with the influenza A virus NS1 protein (113). We suggest that by studying factors controlling endogenous retrotransposition, new insights may also emerge into the control of viral infections.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–6, Supplementary Figure 1 and Supplementary References [114–131].
FUNDING
National Institutes of Health (NIH) [R01 HL083017-01A2 and R01 GM099875-01A1 to H.H.K.]. Funding for open access charge: NIGMS and NIH.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
Special thanks to Dr Robert Cole and the Mass Spectrometry and Proteomics Facility, and the HIT Center ChemCore of Johns Hopkins University. The authors also thank the S. Leach laboratory for use of their confocal microscope.
REFERENCES
- 1.De Koning APJ, Gu W, Castoe TA, Batzer MA, Pollock DD. Repetitive elements may comprise over two-thirds of the human genome. PLoS Genet. 2011;7:e1002384. doi: 10.1371/journal.pgen.1002384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Luan DD, Korman MH, Jakubczak JL, Eickbush TH. Reverse transcription of R2Bm RNA is primed by a nick at the chromosomal target site: a mechanism for non-LTR retrotransposition. Cell. 1993;72:595–605. doi: 10.1016/0092-8674(93)90078-5. [DOI] [PubMed] [Google Scholar]
- 3.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 4.Zhang Z, Harrison PM, Liu Y, Gerstein M. Millions of years of evolution preserved: a comprehensive catalog of the processed pseudogenes in the human genome. Genome Res. 2003;13:2541–2558. doi: 10.1101/gr.1429003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang H, Xing J, Grover D, Hedges DJ, Han K, Walker JA, Batzer MA. SVA elements: a hominid-specific retroposon family. J. Mol. Biol. 2005;354:994–1007. doi: 10.1016/j.jmb.2005.09.085. [DOI] [PubMed] [Google Scholar]
- 6.Brouha B, Schustak J, Badge RM, Lutz-Prigge S, Farley AH, Moran JV, Kazazian HH., Jr Hot L1s account for the bulk of retrotransposition in the human population. Proc. Natl Acad. Sci. USA. 2003;100:5280–5285. doi: 10.1073/pnas.0831042100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beck CR, Collier P, Macfarlane C, Malig M, Kidd JM, Eichler EE, Badge RM, Moran JV. LINE-1 retrotransposition activity in human genomes. Cell. 2010;141:1159–1170. doi: 10.1016/j.cell.2010.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cordaux R, Hedges DJ, Herke SW, Batzer MA. Estimating the retrotransposition rate of human Alu elements. Gene. 2006;373:134–137. doi: 10.1016/j.gene.2006.01.019. [DOI] [PubMed] [Google Scholar]
- 9.Hancks DC, Kazazian HH., Jr Active human retrotransposons: variation and disease. Curr. Opin. Genet. Dev. 2012;22:191–203. doi: 10.1016/j.gde.2012.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Muotri AR, Chu VT, Marchetto MCN, Deng W, Moran JV, Gage FH. Somatic mosaicism in neuronal precursor cells mediated by L1 retrotransposition. Nature. 2005;435:903–910. doi: 10.1038/nature03663. [DOI] [PubMed] [Google Scholar]
- 11.Garcia-Perez JL, Marchetto MCN, Muotri AR, Coufal NG, Gage FH, O’Shea KS, Moran JV. LINE-1 retrotransposition in human embryonic stem cells. Hum. Mol. Genet. 2007;16:1569–1577. doi: 10.1093/hmg/ddm105. [DOI] [PubMed] [Google Scholar]
- 12.Solyom S, Ewing AD, Rahrmann E, Doucet T, Nelson H, Burns B, Harris R, Sigmon D, Casella A, Erlanger B, et al. Extensive somatic L1 retrotransposition in colorectal tumors. Genome Res. 22:2328–2238. doi: 10.1101/gr.145235.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baillie JK, Barnett MW, Upton KR, Gerhardt DJ, Richmond TA, De Sapio F, Brennan PM, Rizzu P, Smith S, Fell M, et al. Somatic retrotransposition alters the genetic landscape of the human brain. Nature. 2011;479:534–537. doi: 10.1038/nature10531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee E, Iskow R, Yang L, Gokcumen O, Haseley P, Luquette LJ, 3rd, Lohr JG, Harris CC, Ding L, Wilson RK, et al. Landscape of somatic retrotransposition in human cancers. Science. 2012;337:967–971. doi: 10.1126/science.1222077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Evrony GD, Cai X, Lee E, Hills LB, Elhosary PC, Lehmann HS, Parker JJ, Atabay KD, Gilmore EC, Poduri A, et al. Single-neuron sequencing analysis of l1 retrotransposition and somatic mutation in the human brain. Cell. 2012;151:483–496. doi: 10.1016/j.cell.2012.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iskow RC, McCabe MT, Mills RE, Torene S, Pittard WS, Neuwald AF, Van Meir EG, Vertino PM, Devine SE. Natural mutagenesis of human genomes by endogenous retrotransposons. Cell. 2010;141:1253–1261. doi: 10.1016/j.cell.2010.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Beck CR, Garcia-Perez JL, Badge RM, Moran JV. LINE-1 elements in structural variation and disease. Annu. Rev. Genomics Hum. Genet. 2011;12:187–215. doi: 10.1146/annurev-genom-082509-141802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Burns KH, Boeke JD. Human transposon tectonics. Cell. 2012;149:740–752. doi: 10.1016/j.cell.2012.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Becker KG, Swergold GD, Ozato K, Thayer RE. Binding of the ubiquitous nuclear transcription factor YY1 to a cis regulatory sequence in the human LINE-1 transposable element. Hum. Mol. Genet. 1993;2:1697–1702. doi: 10.1093/hmg/2.10.1697. [DOI] [PubMed] [Google Scholar]
- 20.Tchénio T, Casella JF, Heidmann T. Members of the SRY family regulate the human LINE retrotransposons. Nucleic Acids Res. 2000;28:411–415. doi: 10.1093/nar/28.2.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang N, Zhang L, Zhang Y, Kazazian HH., Jr An important role for RUNX3 in human L1 transcription and retrotransposition. Nucleic Acids Res. 2003;31:4929–4940. doi: 10.1093/nar/gkg663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harris CR, Dewan A, Zupnick A, Normart R, Gabriel A, Prives C, Levine AJ, Hoh J. p53 responsive elements in human retrotransposons. Oncogene. 2009;28:3857–3865. doi: 10.1038/onc.2009.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dai L, Taylor MS, O’Donnell KA, Boeke JD. Poly(A) binding protein C1 is essential for efficient L1 retrotransposition and affects L1 RNP formation. Mol. Cell. Biol. 2012;32:4323–4336. doi: 10.1128/MCB.06785-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morrish TA, Gilbert N, Myers JS, Vincent BJ, Stamato TD, Taccioli GE, Batzer MA, Moran JV. DNA repair mediated by endonuclease-independent LINE-1 retrotransposition. Nat. Genet. 2002;31:159–165. doi: 10.1038/ng898. [DOI] [PubMed] [Google Scholar]
- 25.Gasior SL, Roy-Engel AM, Deininger PL. ERCC1/XPF limits L1 retrotransposition. DNA Repair (Amst.) 2008;7:983–989. doi: 10.1016/j.dnarep.2008.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Suzuki J, Yamaguchi K, Kajikawa M, Ichiyanagi K, Adachi N, Koyama H, Takeda S, Okada N. Genetic evidence that the non-homologous end-joining repair pathway is involved in LINE retrotransposition. PLoS Genet. 2009;5:e1000461. doi: 10.1371/journal.pgen.1000461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goodier JL, Zhang L, Vetter MR, Kazazian HH., Jr LINE-1 ORF1 protein localizes in stress granules with other RNA-binding proteins, including components of RNA interference RNA-induced silencing complex. Mol. Cell. Biol. 2007;27:6469–6483. doi: 10.1128/MCB.00332-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peddigari S, Li PWL, Rabe JL, Martin SL. hnRNPL and nucleolin bind LINE-1 RNA and function as host factors to modulate retrotransposition. Nucleic Acids Res. 2013;41:575–585. doi: 10.1093/nar/gks1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goodier JL, Cheung LE, Kazazian HH. MOV10 RNA helicase is a potent inhibitor of retrotransposition in cells. PLoS Genet. 2012;8:e1002941. doi: 10.1371/journal.pgen.1002941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goodier JL, Kazazian HH., Jr Retrotransposons revisited: the restraint and rehabilitation of parasites. Cell. 2008;135:23–35. doi: 10.1016/j.cell.2008.09.022. [DOI] [PubMed] [Google Scholar]
- 31.Koito A, Ikeda T. Intrinsic restriction activity by AID/APOBEC family of enzymes against the mobility of retroelements. Mob. Genet. Elements. 2011;1:197–202. doi: 10.4161/mge.1.3.17430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bao J, Yan W. Male germline control of transposable elements. Biol. Reprod. 2012;86:162, 1–14. doi: 10.1095/biolreprod.111.095463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wei W, Gilbert N, Ooi SL, Lawler JF, Ostertag EM, Kazazian HH, Boeke JD, Moran JV. Human L1 retrotransposition: cis preference versus trans complementation. Mol. Cell. Biol. 2001;21:1429–1439. doi: 10.1128/MCB.21.4.1429-1439.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Goodier JL, Mandal PK, Zhang L, Kazazian HH., Jr Discrete subcellular partitioning of human retrotransposon RNAs despite a common mechanism of genome insertion. Hum. Mol. Genet. 2010;19:1712–1725. doi: 10.1093/hmg/ddq048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Doucet AJ, Hulme AE, Sahinovic E, Kulpa DA, Moldovan JB, Kopera HC, Athanikar JN, Hasnaoui M, Bucheton A, Moran JV, et al. Characterization of LINE-1 ribonucleoprotein particles. PLoS Genet. 2010;6:e1001150. doi: 10.1371/journal.pgen.1001150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kunkel TA. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc. Natl Acad. Sci. USA. 1985;82:488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kimberland ML, Divoky V, Prchal J, Schwahn U, Berger W, Kazazian HH., Jr Full-length human L1 insertions retain the capacity for high frequency retrotransposition in cultured cells. Hum. Mol. Genet. 1999;8:1557–1560. doi: 10.1093/hmg/8.8.1557. [DOI] [PubMed] [Google Scholar]
- 38.Nakatani Y, Ogryzko V. RNA Polymerases and Associated Factors, Part C. Vol. 370. Academic Press; 2003. Immunoaffinity purification of mammalian protein complexes; pp. 430–444. [DOI] [PubMed] [Google Scholar]
- 39.Mooney SM, Grande JP, Salisbury JL, Janknecht R. Sumoylation of p68 and p72 RNA helicases affects protein stability and transactivation potential. Biochemistry. 2010;49:1–10. doi: 10.1021/bi901263m. [DOI] [PubMed] [Google Scholar]
- 40.Sim S, Yao J, Weinberg DE, Niessen S, Yates JR, 3rd, Wolin SL. The zipcode-binding protein ZBP1 influences the subcellular location of the Ro 60-kDa autoantigen and the noncoding Y3 RNA. RNA. 2012;18:100–110. doi: 10.1261/rna.029207.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- 42.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 43.Jensen LJ, Kuhn M, Stark M, Chaffron S, Creevey C, Muller J, Doerks T, Julien P, Roth A, Simonovic M, et al. STRING 8–a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res. 2009;37:D412–D416. doi: 10.1093/nar/gkn760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Leung AKL, Trinkle-Mulcahy L, Lam YW, Andersen JS, Mann M, Lamond AI. NOPdb: nucleolar proteome database. Nucleic Acids Res. 2006;34:D218–D220. doi: 10.1093/nar/gkj004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ostertag EM, Prak ET, DeBerardinis RJ, Moran JV, Kazazian HH., Jr Determination of L1 retrotransposition kinetics in cultured cells. Nucleic Acids Res. 2000;28:1418–1423. doi: 10.1093/nar/28.6.1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Moran JV, Holmes SE, Naas TP, DeBerardinis RJ, Boeke JD, Kazazian HH., Jr High frequency retrotransposition in cultured mammalian cells. Cell. 1996;87:917–927. doi: 10.1016/s0092-8674(00)81998-4. [DOI] [PubMed] [Google Scholar]
- 47.Wei W, Morrish TA, Alisch RS, Moran JV. A transient assay reveals that cultured human cells can accommodate multiple LINE-1 retrotransposition events. Anal. Biochem. 2000;284:435–438. doi: 10.1006/abio.2000.4675. [DOI] [PubMed] [Google Scholar]
- 48.Leibold DM, Swergold GD, Singer MF, Thayer RE, Dombroski BA, Fanning TG. Translation of LINE-1 DNA elements in vitro and in human cells. Proc. Natl Acad. Sci. USA. 1990;87:6990–6994. doi: 10.1073/pnas.87.18.6990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yik JHN, Chen R, Nishimura R, Jennings JL, Link AJ, Zhou Q. Inhibition of P-TEFb (CDK9/Cyclin T) kinase and RNA polymerase II transcription by the coordinated actions of HEXIM1 and 7SK snRNA. Mol. Cell. 2003;12:971–982. doi: 10.1016/s1097-2765(03)00388-5. [DOI] [PubMed] [Google Scholar]
- 50.Fleckner J, Zhang M, Valcárcel J, Green MR. U2AF65 recruits a novel human DEAD box protein required for the U2 snRNP-branchpoint interaction. Genes Dev. 1997;11:1864–1872. doi: 10.1101/gad.11.14.1864. [DOI] [PubMed] [Google Scholar]
- 51.Kulpa DA, Moran JV. Cis-preferential LINE-1 reverse transcriptase activity in ribonucleoprotein particles. Nat. Struct. Mol. Biol. 2006;13:655–660. doi: 10.1038/nsmb1107. [DOI] [PubMed] [Google Scholar]
- 52.Li TH, Schmid CW. Differential stress induction of individual Alu loci: implications for transcription and retrotransposition. Gene. 2001;276:135–141. doi: 10.1016/s0378-1119(01)00637-0. [DOI] [PubMed] [Google Scholar]
- 53.Goodier JL, Ostertag EM, Engleka KA, Seleme MC, Kazazian HH., Jr A potential role for the nucleolus in L1 retrotransposition. Hum. Mol. Genet. 2004;13:1041–1048. doi: 10.1093/hmg/ddh118. [DOI] [PubMed] [Google Scholar]
- 54.De Ruijter AJM, van Gennip AH, Caron HN, Kemp S, van Kuilenburg ABP. Histone deacetylases (HDACs): characterization of the classical HDAC family. Biochem. J. 2003;370:737–749. doi: 10.1042/BJ20021321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fukuda M, Gotoh I, Gotoh Y, Nishida E. Cytoplasmic localization of mitogen-activated protein kinase kinase directed by its NH2-terminal, leucine-rich short amino acid sequence, which acts as a nuclear export signal. J. Biol. Chem. 1996;271:20024–20028. doi: 10.1074/jbc.271.33.20024. [DOI] [PubMed] [Google Scholar]
- 56.Martin SL, Branciforte D, Keller D, Bain DL. Trimeric structure for an essential protein in L1 retrotransposition. Proc. Natl Acad. Sci. USA. 2003;100:13815–13820. doi: 10.1073/pnas.2336221100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Khazina E, Truffault V, Büttner R, Schmidt S, Coles M, Weichenrieder O. Trimeric structure and flexibility of the L1ORF1 protein in human L1 retrotransposition. Nat. Struct. Mol. Biol. 2011;18:1006–1014. doi: 10.1038/nsmb.2097. [DOI] [PubMed] [Google Scholar]
- 58.Hohjoh H, Singer MF. Cytoplasmic ribonucleoprotein complexes containing human LINE-1 protein and RNA. EMBO J. 1996;15:630–639. [PMC free article] [PubMed] [Google Scholar]
- 59.Peterlin BM, Brogie JE, Price DH. 7SK snRNA: a noncoding RNA that plays a major role in regulating eukaryotic transcription. Wiley Interdiscip. Rev. RNA. 2012;3:92–103. doi: 10.1002/wrna.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Perreault J, Noël JF, Brière F, Cousineau B, Lucier JF, Perreault JP, Boire G. Retropseudogenes derived from the human Ro/SS-A autoantigen-associated hY RNAs. Nucleic Acids Res. 2005;33:2032–2041. doi: 10.1093/nar/gki504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wolin SL, Steitz JA. The Ro small cytoplasmic ribonucleoproteins: identification of the antigenic protein and its binding site on the Ro RNAs. Proc. Natl Acad. Sci. USA. 1984;81:1996–2000. doi: 10.1073/pnas.81.7.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fabini G, Raijmakers R, Hayer S, Fouraux MA, Pruijn GJ, Steiner G. The heterogeneous nuclear ribonucleoproteins I and K interact with a subset of the ro ribonucleoprotein-associated Y RNAs in vitro and in vivo. J. Biol. Chem. 2001;276:20711–20718. doi: 10.1074/jbc.M101360200. [DOI] [PubMed] [Google Scholar]
- 63.Fouraux MA, Bouvet P, Verkaart S, van Venrooij WJ, Pruijn GJM. Nucleolin associates with a subset of the human Ro ribonucleoprotein complexes. J. Mol. Biol. 2002;320:475–488. doi: 10.1016/s0022-2836(02)00518-1. [DOI] [PubMed] [Google Scholar]
- 64.Murphy S, Altruda F, Ullu E, Tripodi M, Silengo L, Melli M. DNA sequences complementary to human 7 SK RNA show structural similarities to the short mobile elements of the mammalian genome. J. Mol. Biol. 1984;177:575–590. doi: 10.1016/0022-2836(84)90038-x. [DOI] [PubMed] [Google Scholar]
- 65.Liu YJ, Zheng D, Balasubramanian S, Carriero N, Khurana E, Robilotto R, Gerstein MB. Comprehensive analysis of the pseudogenes of glycolytic enzymes in vertebrates: the anomalously high number of GAPDH pseudogenes highlights a recent burst of retrotrans-positional activity. BMC Genomics. 2009;10:480. doi: 10.1186/1471-2164-10-480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mili S, Steitz JA. Evidence for reassociation of RNA-binding proteins after cell lysis: Implications for the interpretation of immunoprecipitation analyses. RNA. 2004;10:1692–1694. doi: 10.1261/rna.7151404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, Ascano M, Jr, Jungkamp AC, Munschauer M, et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010;141:129–141. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Anderson P, Kedersha N. Stress granules: the Tao of RNA triage. Trends Biochem. Sci. 2008;33:141–150. doi: 10.1016/j.tibs.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 69.Zheng D, Chen CYA, Shyu AB. Unraveling regulation and new components of human P-bodies through a protein interaction framework and experimental validation. RNA. 2011;17:1619–1634. doi: 10.1261/rna.2789611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ochs RL, Lischwe MA, Spohn WH, Busch H. Fibrillarin: a new protein of the nucleolus identified by autoimmune sera. Biol. Cell. 1985;54:123–133. doi: 10.1111/j.1768-322x.1985.tb00387.x. [DOI] [PubMed] [Google Scholar]
- 71.Höck J, Weinmann L, Ender C, Rüdel S, Kremmer E, Raabe M, Urlaub H, Meister G. Proteomic and functional analysis of Argonaute-containing mRNA-protein complexes in human cells. EMBO Rep. 2007;8:1052–1060. doi: 10.1038/sj.embor.7401088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Checkley MA, Nagashima K, Lockett SJ, Nyswaner KM, Garfinkel DJ. P-body components are required for Ty1 retrotransposition during assembly of retrotransposition-competent virus-like particles. Mol. Cell. Biol. 2010;30:382–398. doi: 10.1128/MCB.00251-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dutko JA, Kenny AE, Gamache ER, Curcio MJ. 5′ to 3′ mRNA decay factors colocalize with Ty1 gag and human APOBEC3G and promote Ty1 retrotransposition. J. Virol. 2010;84:5052–5066. doi: 10.1128/JVI.02477-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lu C, Contreras X, Peterlin BM. P bodies inhibit retrotransposition of endogenous intracisternal a particles. J. Virol. 2011;85:6244–6251. doi: 10.1128/JVI.02517-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Buzdin A, Gogvadze E, Lebrun MH. Chimeric retrogenes suggest a role for the nucleolus in LINE amplification. FEBS Lett. 2007;581:2877–2882. doi: 10.1016/j.febslet.2007.05.034. [DOI] [PubMed] [Google Scholar]
- 76.Xiong Y, Burke WD, Jakubczak JL, Eickbush TH. Ribosomal DNA insertion elements R1Bm and R2Bm can transpose in a sequence specific manner to locations outside the 28S genes. Nucleic Acids Res. 1988;16:10561–10573. doi: 10.1093/nar/16.22.10561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Dai J, Xie W, Brady TL, Gao J, Voytas DF. Phosphorylation regulates integration of the yeast Ty5 retrotransposon into heterochromatin. Mol. Cell. 2007;27:289–299. doi: 10.1016/j.molcel.2007.06.010. [DOI] [PubMed] [Google Scholar]
- 78.Enomoto A, Kido N, Ito M, Morita A, Matsumoto Y, Takamatsu N, Hosoi Y, Miyagawa K. Negative regulation of MEKK1/2 signaling by serine-threonine kinase 38 (STK38) Oncogene. 2008;27:1930–1938. doi: 10.1038/sj.onc.1210828. [DOI] [PubMed] [Google Scholar]
- 79.Fu D, Collins K. Purification of human telomerase complexes identifies factors involved in telomerase biogenesis and telomere length regulation. Mol. Cell. 2007;28:773–785. doi: 10.1016/j.molcel.2007.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ribbeck K, Raemaekers T, Carmeliet G, Mattaj IW. A role for NuSAP in linking microtubules to mitotic chromosomes. Curr. Biol. 2007;17:230–236. doi: 10.1016/j.cub.2006.11.050. [DOI] [PubMed] [Google Scholar]
- 81.Vujatovic O, Zaragoza K, Vaquero A, Reina O, Bernués J, Azorín F. Drosophila melanogaster linker histone dH1 is required for transposon silencing and to preserve genome integrity. Nucleic Acids Res. 2012;40:5402–5414. doi: 10.1093/nar/gks224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Miki T, Takano K, Yoneda Y. The role of mammalian Staufen on mRNA traffic: a view from its nucleocytoplasmic shuttling function. Cell Struct. Funct. 2005;30:51–56. doi: 10.1247/csf.30.51. [DOI] [PubMed] [Google Scholar]
- 83.Goldfarb DS, Corbett AH, Mason DA, Harreman MT, Adam SA. Importin alpha: a multipurpose nuclear-transport receptor. Trends Cell Biol. 2004;14:505–514. doi: 10.1016/j.tcb.2004.07.016. [DOI] [PubMed] [Google Scholar]
- 84.Hsu K, Chang DY, Maraia RJ. Human signal recognition particle (SRP) Alu-associated protein also binds Alu interspersed repeat sequence RNAs. Characterization of human SRP9. J. Biol. Chem. 1995;270:10179–10186. doi: 10.1074/jbc.270.17.10179. [DOI] [PubMed] [Google Scholar]
- 85.Bennett EA, Keller H, Mills RE, Schmidt S, Moran JV, Weichenrieder O, Devine SE. Active Alu retrotransposons in the human genome. Genome Res. 2008;18:1875–1883. doi: 10.1101/gr.081737.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Elliott DJ, Best A, Dalgliesh C, Ehrmann I, Grellscheid S. How does Tra2β protein regulate tissue-specific RNA splicing? Biochem. Soc. Trans. 2012;40:784–788. doi: 10.1042/BST20120036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zarnack K, König J, Tajnik M, Martincorena I, Eustermann S, Stévant I, Reyes A, Anders S, Luscombe NM, Ule J. Direct competition between hnRNP C and U2AF65 protects the transcriptome from the exonization of alu elements. Cell. 2013;152:453–466. doi: 10.1016/j.cell.2012.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Twyffels L, Gueydan C, Kruys V. Shuttling SR proteins: more than splicing factors. FEBS J. 2011;278:3246–3255. doi: 10.1111/j.1742-4658.2011.08274.x. [DOI] [PubMed] [Google Scholar]
- 89.Belancio VP, Hedges DJ, Deininger P. Mammalian non-LTR retrotransposons: for better or worse, in sickness and in health. Genome Res. 2008;18:343–358. doi: 10.1101/gr.5558208. [DOI] [PubMed] [Google Scholar]
- 90.Belancio VP, Hedges DJ, Deininger P. LINE-1 RNA splicing and influences on mammalian gene expression. Nucleic Acids Res. 2006;34:1512–1521. doi: 10.1093/nar/gkl027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Xiong Y, Eickbush TH. Origin and evolution of retroelements based upon their reverse transcriptase sequences. EMBO J. 1990;9:3353–3362. doi: 10.1002/j.1460-2075.1990.tb07536.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Doolittle RF, Feng DF. Tracing the origin of retroviruses. Curr. Top. Microbiol. Immunol. 1992;176:195–211. doi: 10.1007/978-3-642-77011-1_13. [DOI] [PubMed] [Google Scholar]
- 93.Malik HS, Eickbush TH. Phylogenetic analysis of ribonuclease H domains suggests a late, chimeric origin of LTR retrotransposable elements and retroviruses. Genome Res. 2001;11:1187–1197. doi: 10.1101/gr.185101. [DOI] [PubMed] [Google Scholar]
- 94.Malik HS, Henikoff S, Eickbush TH. Poised for contagion: evolutionary origins of the infectious abilities of invertebrate retroviruses. Genome Res. 2000;10:1307–1318. doi: 10.1101/gr.145000. [DOI] [PubMed] [Google Scholar]
- 95.Arjan-Odedra S, Swanson CM, Sherer NM, Wolinsky SM, Malim MH. Endogenous MOV10 inhibits the retrotransposition of endogenous retroelements but not the replication of exogenous retroviruses. Retrovirology. 2012;9:53. doi: 10.1186/1742-4690-9-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Holmes RK, Malim MH, Bishop KN. APOBEC-mediated viral restriction: not simply editing? Trends Biochem. Sci. 2007;32:118–128. doi: 10.1016/j.tibs.2007.01.004. [DOI] [PubMed] [Google Scholar]
- 97.Vetter ML, Johnson ME, Antons AK, Unutmaz D, D’Aquila RT. Differences in APOBEC3G expression in CD4+ T helper lymphocyte subtypes modulate HIV-1 infectivity. PLoS Pathog. 2009;5:e1000292. doi: 10.1371/journal.ppat.1000292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Chiu Y-L, Witkowska HE, Hall SC, Santiago M, Soros VB, Esnault C, Heidmann T, Greene WC. High-molecular-mass APOBEC3G complexes restrict Alu retrotransposition. Proc. Natl Acad. Sci. USA. 2006;103:15588–15593. doi: 10.1073/pnas.0604524103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ikeda T, Abd El Galil KH, Tokunaga K, Maeda K, Sata T, Sakaguchi N, Heidmann T, Koito A. Intrinsic restriction activity by apolipoprotein B mRNA editing enzyme APOBEC1 against the mobility of autonomous retrotransposons. Nucleic Acids Res. 2011;39:5538–5554. doi: 10.1093/nar/gkr124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Lau PP, Villanueva H, Kobayashi K, Nakamuta M, Chang BH, Chan L. A DnaJ protein, apobec-1-binding protein-2, modulates apolipoprotein B mRNA editing. J. Biol. Chem. 2001;276:46445–46452. doi: 10.1074/jbc.M109215200. [DOI] [PubMed] [Google Scholar]
- 101.Blanc V, Navaratnam N, Henderson JO, Anant S, Kennedy S, Jarmuz A, Scott J, Davidson NO. Identification of GRY-RBP as an apolipoprotein B RNA-binding protein that interacts with both apobec-1 and apobec-1 complementation factor to modulate C to U editing. J. Biol. Chem. 2001;276:10272–10283. doi: 10.1074/jbc.M006435200. [DOI] [PubMed] [Google Scholar]
- 102.Naji S, Ambrus G, Cimermančič P, Reyes JR, Johnson JR, Filbrandt R, Huber MD, Vesely P, Krogan NJ, Yates JR, 3rd, et al. Host cell interactome of HIV-1 Rev includes RNA helicases involved in multiple facets of virus production. Mol. Cell Proteomics. 2012;11 doi: 10.1074/mcp.M111.015313. M111.015313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Brass AL, Dykxhoorn DM, Benita Y, Yan N, Engelman A, Xavier RJ, Lieberman J, Elledge SJ. Identification of host proteins required for HIV infection through a functional genomic screen. Science. 2008;319:921–926. doi: 10.1126/science.1152725. [DOI] [PubMed] [Google Scholar]
- 104.Zhou H, Xu M, Huang Q, Gates AT, Zhang XD, Castle JC, Stec E, Ferrer M, Strulovici B, Hazuda DJ, et al. Genome-scale RNAi screen for host factors required for HIV replication. Cell Host Microbe. 2008;4:495–504. doi: 10.1016/j.chom.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 105.Liu L, Oliveira NMM, Cheney KM, Pade C, Dreja H, Bergin A-MH, Borgdorff V, Beach DH, Bishop CL, Dittmar MT, et al. A whole genome screen for HIV restriction factors. Retrovirology. 2011;8:94. doi: 10.1186/1742-4690-8-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Chepenik LG, Tretiakova AP, Krachmarov CP, Johnson EM, Khalili K. The single-stranded DNA binding protein, Pur-alpha, binds HIV-1 TAR RNA and activates HIV-1 transcription. Gene. 1998;210:37–44. doi: 10.1016/s0378-1119(98)00033-x. [DOI] [PubMed] [Google Scholar]
- 107.Kaminski R, Darbinian N, Sawaya BE, Slonina D, Amini S, Johnson EM, Rappaport J, Khalili K, Darbinyan A. Puralpha as a cellular co-factor of Rev/RRE-mediated expression of HIV-1 intron-containing mRNA. J. Cell. Biochem. 2008;103:1231–1245. doi: 10.1002/jcb.21503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ansari SA, Safak M, Gallia GL, Sawaya BE, Amini S, Khalili K. Interaction of YB-1 with human immunodeficiency virus type 1 Tat and TAR RNA modulates viral promoter activity. J. Gen. Virol. 1999;80(Pt 10):2629–2638. doi: 10.1099/0022-1317-80-10-2629. [DOI] [PubMed] [Google Scholar]
- 109.Lemay J, Maidou-Peindara P, Bader T, Ennifar E, Rain J-C, Benarous R, Liu LX. HuR interacts with human immunodeficiency virus type 1 reverse transcriptase, and modulates reverse transcription in infected cells. Retrovirology. 2008;5:47. doi: 10.1186/1742-4690-5-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Valente ST, Goff SP. Inhibition of HIV-1 gene expression by a fragment of hnRNP U. Mol. Cell. 2006;23:597–605. doi: 10.1016/j.molcel.2006.07.021. [DOI] [PubMed] [Google Scholar]
- 111.Kress E, Baydoun HH, Bex F, Gazzolo L, Duc Dodon M. Critical role of hnRNP A1 in HTLV-1 replication in human transformed T lymphocytes. Retrovirology. 2005;2:8. doi: 10.1186/1742-4690-2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Dinh PX, Beura LK, Panda D, Das A, Pattnaik AK. Antagonistic effects of cellular poly(C) binding proteins on vesicular stomatitis virus gene expression. J. Virol. 2011;85:9459–9471. doi: 10.1128/JVI.05179-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wolff T, O’Neill RE, Palese P. NS1-Binding protein (NS1-BP): a novel human protein that interacts with the influenza A virus nonstructural NS1 protein is relocalized in the nuclei of infected cells. J. Virol. 1998;72:7170–7180. doi: 10.1128/jvi.72.9.7170-7180.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Low WK, Dang Y, Schneider-Poetsch T, Shi Z, Choi NS, Merrick WC, Romo D, Liu JO. Inhibition of eukaryotic translation initiation by the marine natural product pateamine A. Mol. Cell. 2005;20:709–722. doi: 10.1016/j.molcel.2005.10.008. [DOI] [PubMed] [Google Scholar]
- 115.Gallouzi IE, Brennan CM, Stenberg MG, Swanson MS, Eversole A, Maizels N, Steitz JA. HuR binding to cytoplasmic mRNA is perturbed by heat shock. Proc. Natl Acad. Sci. USA. 2000;97:3073–3078. doi: 10.1073/pnas.97.7.3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Guil S, Long JC, Cáceres JF. hnRNP A1 relocalization to the stress granules reflects a role in the stress response. Mol. Cell. Biol. 2006;26:5744–5758. doi: 10.1128/MCB.00224-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Stöhr N, Lederer M, Reinke C, Meyer S, Hatzfeld M, Singer RH, Hüttelmaier S. ZBP1 regulates mRNA stability during cellular stress. J. Cell Biol. 2006;175:527–534. doi: 10.1083/jcb.200608071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Nykamp K, Lee MH, Kimble J. C. elegans La-related protein, LARP-1, localizes to germline P bodies and attenuates Ras-MAPK signaling during oogenesis. RNA. 2008;14:1378–1389. doi: 10.1261/rna.1066008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Meister G, Landthaler M, Peters L, Chen PY, Urlaub H, Lührmann R, Tuschl T. Identification of novel argonaute-associated proteins. Curr. Biol. 2005;15:2149–2155. doi: 10.1016/j.cub.2005.10.048. [DOI] [PubMed] [Google Scholar]
- 120.Kedersha NL, Gupta M, Li W, Miller I, Anderson P. RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2 alpha to the assembly of mammalian stress granules. J. Cell Biol. 1999;147:1431–1442. doi: 10.1083/jcb.147.7.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Fujimura K, Kano F, Murata M. Identification of PCBP2, a facilitator of IRES-mediated translation, as a novel constituent of stress granules and processing bodies. RNA. 2008;14:425–431. doi: 10.1261/rna.780708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Delestienne N, Wauquier C, Soin R, Dierick JF, Gueydan C, Kruys V. The splicing factor ASF/SF2 is associated with TIA-1-related/TIA-1-containing ribonucleoproteic complexes and contributes to post-transcriptional repression of gene expression. FEBS J. 2010;277:2496–2514. doi: 10.1111/j.1742-4658.2010.07664.x. [DOI] [PubMed] [Google Scholar]
- 123.Thomas MG, Martinez Tosar LJ, Loschi M, Pasquini JM, Correale J, Kindler S, Boccaccio GL. Staufen recruitment into stress granules does not affect early mRNA transport in oligodendrocytes. Mol. Biol. Cell. 2005;16:405–420. doi: 10.1091/mbc.E04-06-0516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Quaresma AJC, Bressan GC, Gava LM, Lanza DCF, Ramos CHI, Kobarg J. Human hnRNP Q re-localizes to cytoplasmic granules upon PMA, thapsigargin, arsenite and heat-shock treatments. Exp. Cell Res. 2009;315:968–980. doi: 10.1016/j.yexcr.2009.01.012. [DOI] [PubMed] [Google Scholar]
- 125.Jønson L, Vikesaa J, Krogh A, Nielsen LK, Hansen TV, Borup R, Johnsen AH, Christiansen J, Nielsen FC. Molecular composition of IMP1 ribonucleoprotein granules. Mol. Cell Proteomics. 2007;6:798–811. doi: 10.1074/mcp.M600346-MCP200. [DOI] [PubMed] [Google Scholar]
- 126.Abdelhaleem MM, Hameed S, Klassen D, Greenberg AH. Leukophysin: an RNA helicase A-related molecule identified in cytotoxic T cell granules and vesicles. J. Immunol. 1996;156:2026–2035. [PubMed] [Google Scholar]
- 127.Bortz E, Westera L, Maamary J, Steel J, Albrecht RA, Manicassamy B, Chase G, Martínez-Sobrido L, Schwemmle M, García-Sastre A. Host- and strain-specific regulation of influenza virus polymerase activity by interacting cellular proteins. mBio. 2011;2:e00151–11. doi: 10.1128/mBio.00151-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Fukuda T, Naiki T, Saito M, Irie K. hnRNP K interacts with RNA binding motif protein 42 and functions in the maintenance of cellular ATP level during stress conditions. Genes Cells. 2009;14:113–128. doi: 10.1111/j.1365-2443.2008.01256.x. [DOI] [PubMed] [Google Scholar]
- 129.Fujimura K, Suzuki T, Yasuda Y, Murata M, Katahira J, Yoneda Y. Identification of importin alpha1 as a novel constituent of RNA stress granules. Biochim. Biophys. Acta. 2010;1803:865–871. doi: 10.1016/j.bbamcr.2010.03.020. [DOI] [PubMed] [Google Scholar]
- 130.Yang WH, Bloch DB. Probing the mRNA processing body using protein macroarrays and ‘autoantigenomics’. RNA. 2007;13:704–712. doi: 10.1261/rna.411907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Gallois-Montbrun S, Kramer B, Swanson CM, Byers H, Lynham S, Ward M, Malim MH. Antiviral protein APOBEC3G localizes to ribonucleoprotein complexes found in P bodies and stress granules. J. Virol. 2007;81:2165–2178. doi: 10.1128/JVI.02287-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.