Abstract
The ability to artificially control transcription is essential both to the study of gene function and to the construction of synthetic gene networks with desired properties. Cas9 is an RNA-guided double-stranded DNA nuclease that participates in the CRISPR-Cas immune defense against prokaryotic viruses. We describe the use of a Cas9 nuclease mutant that retains DNA-binding activity and can be engineered as a programmable transcription repressor by preventing the binding of the RNA polymerase (RNAP) to promoter sequences or as a transcription terminator by blocking the running RNAP. In addition, a fusion between the omega subunit of the RNAP and a Cas9 nuclease mutant directed to bind upstream promoter regions can achieve programmable transcription activation. The simple and efficient modulation of gene expression achieved by this technology is a useful asset for the study of gene networks and for the development of synthetic biology and biotechnological applications.
INTRODUCTION
Recombinant DNA technologies have advanced the study of gene function through the development of systems that use naturally occurring gene-regulation mechanisms to control gene expression. Transcription factors bind specific DNA sequences upstream of genes and can either repress or activate their expression, for example by preventing the RNA polymerase (RNAP) from gaining access to the promoter or by facilitating the productive binding of RNAP to the promoter. Many transcription factors have been exploited to modulate gene expression in a controlled fashion (1), but all require the engineering of specific promoter sequences that can be bound by the regulator. On the other hand, the sequence specificity of DNA-binding proteins has been manipulated (2). During the past decade, the relative facility with which Zinc Finger DNA-binding proteins and Transcription Activator–Like Effectors can be reprogrammed to bind specific sequences has been used to direct transcription activation or repression (3–6). However, the engineering of sequence specificity for these proteins remains a time-consuming and cost-intensive process.
Recently, the study of clustered, regularly interspaced, short palindromic repeat (CRISPR) loci, which encode a prokaryotic immune system, has revealed the existence of nucleases whose sequence specificity is determined by a small CRISPR RNA (crRNA) guide (7–9). CRISPR loci constitute an array of short repetitive sequences separated by equally short ‘spacers’ that match the genomes of phages and mobile genetic elements that infect bacteria and archaea (10–13). The repeat-spacer array is transcribed as a long precursor crRNA that is cleaved at repeat sequences by CRISPR-associated (Cas) endoribonucleases, liberating small crRNAs (14–17). The crRNAs are then used as antisense guides for Cas nucleases to find the targets (also known as protospacers) and destroy the genome of the invader (7–9,18,19). CRISPR-Cas systems can be divided into three types depending on the cas gene content and mechanism of immunity (20). In type II systems, the biogenesis of small crRNAs requires the coordinated action of the Cas9 nuclease, the host RNase III and a small trans-activating crRNA (tracrRNA) (21). Cas9 is a crRNA-guided double-stranded DNA endonuclease with two domains, RuvC and HNH, each of which cleaves one strand within the target DNA (7,8). In addition to the match between the guide crRNA and the protospacer sequence, Cas9 also requires the presence of a conserved sequence motif downstream of the protospacer, known as the protospacer-adjacent motif (PAM) (7,8,18,22,23).
One of the best characterized Cas9 nucleases is encoded by Streptococcus pyogenes. After crRNA processing, this enzyme is loaded with a crRNA guide containing 20 nt of the spacer sequence followed by 19–22 nt of repeat sequence (21). Target recognition and cleavage require a match between the target and a ‘seed’ sequence of 12–15 nt at the 3′ end of the guide sequence (upstream mutations do not abrogate DNA cleavage), as well as an NGG PAM (7,8,23). Mutations D10A and H840A in the RuvC and HNH domains, respectively, abolish cleavage but do not impair DNA binding (8). Recently, we and others used the programmable nuclease activity of S. pyogenes Cas9 to direct genome editing both in eukaryotes and prokaryotes (23–28). Here, we exploit the DNA-binding activity of a Cas9 catalytic site mutant, referred to as ‘dead’ Cas9 (dCas9), to engineer a programmable transcription regulator (Figure 1A). We show that directing dCas9 to promoter regions results in transcription repression, presumably by preventing the binding of RNAP. We also show that using crRNA guides that specify dCas9 binding to open reading frames blocks transcription elongation. Finally, we converted dCas9 into a transcription activator by fusing it to the omega subunit of RNAP, a strategy previously shown to enhance transcription (29), and directing it to promoter regions of weakly expressed genes. This technology provides a simple and efficient method for global regulation of gene expression and is predicted to facilitate the study of prokaryotic gene networks and the development of synthetic biology applications.
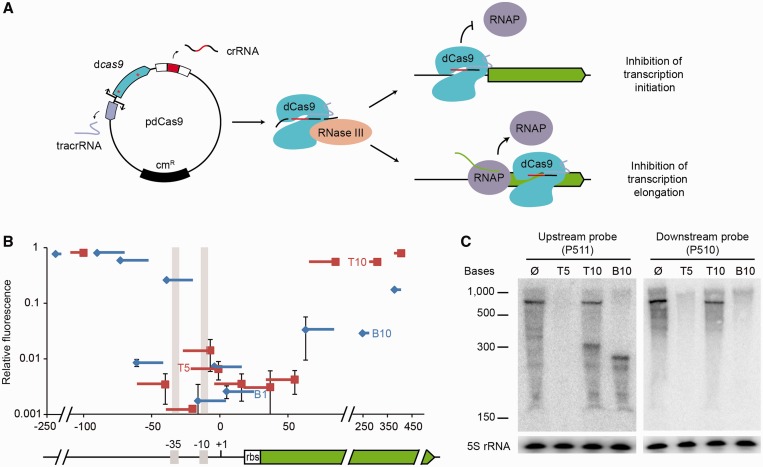
Figure 1.
dCas9-mediated repression in E. coli. (A) Plasmid pdCas9 encodes a cas9 mutant containing D10A and H840A substitutions (red asterisks) that abrogate nuclease activity. dCas9 binds to a tracrRNA:precursor crRNA and recruits RNase III to process the precursor and liberate the crRNA. The crRNA directs binding of dCas9 to promoter or open reading frame regions to prevent RNAP binding or elongation, respectively. (B) GFP fluorescence of cells expressing dCas9 guided to different regions of the gfp-mut2 gene, relative to the fluorescence of cells expressing a non-targeting dCas9, as a function of the position of the target sequence within the gene (+1, transcription start). Squares indicate the PAM position, lines the extension of complementarity between the crRNA guide and the reporter gene. Red and blue lines indicate crRNAs sequences identical to top or bottom DNA strand, respectively. Error bars show one standard deviation from the mean of three relative fluorescence values. The gfp-mut2 gene (green), its promoter, including the −35 and −10 elements (gray shade) and the ribosome binding site (rbs) are shown as reference for the localization of the dCas9 binding sites. (C) Nothern blot with probes annealing either upstream or downstream of the T10 and B10 target sites using RNA extracted from cells expressing T5-, T10-, B10-guided dCas9 or a control strain without a target. Detection of 5S RNA serves as control.
MATERIALS AND METHODS
Strains and culture conditions
Escherichia coli cells were grown in Luria-Bertani (LB) broth supplemented, when appropriate, with the following antibiotics: kanamycin (25 µg/ml), chloramphenicol (25 µg/ml) and spectinomycin (50 µg/ml). Liquid cultures of Streptococcus pneumoniae were grown in THYE medium (30 g/l Todd-Hewitt agar, 5 g/l Yeast Extract) and plated on tryptic soy agar supplemented with 5% defibrinated sheep blood. When appropriate, kanamycin (400 µg/ml), spectinomycin (100 µg/ml) or chloramphenicol (5 µg/ml) were added to the media.
Transcription repression experiments were carried out in E. coli strain DH5α and in S. pneumoniae strain DB17. Streptococcus pneumoniae strain DB17 was constructed by introducing the D10A and H840A mutations in the cas9 gene of strain crR6Rk (23) via transformation and recombination of a SOEing PCR product (30) generated with primers L402/B337, B338/B339 and B340/L403. Supplementary Table S1 contains the sequences of all the primers used for plasmid and strain construction in this study.
Transcription activation of the lacZ gene (encoding β-galactosidase) was studied in E. coli KS1ΔZ, a strain carrying the lacZ gene under the control of a weak promoter that can be induced by a cI-ω fusion (29). Green fluorescence protein (GFP) assays were performed in an E. coli MG1655 mutant (JEN202) in which rpoZ, encoding for the ω subunit of RNAP, was replaced by a spectinomycin resistance gene. To construct this strain, plasmid pSWKspec (31) was PCR amplified with primers W573/W574. The PCR product was transformed into E. coli MG1655 harboring plasmid pKOBEG for lambda-red recombination (32). Integrants were selected on LB agar with spectinomycin.
Plasmid construction
Supplementary Table S2 contains a list of the plasmids used in this study. Plasmid pDB98 (23) was used as the source for crRNA guides in S. pneumoniae DB17. This plasmid is a derivative of pLZ12spec (33) that carries a minimal CRISPR array from S. pyogenes SF370 CRISPR02 containing the leader sequence and two repeats separated by a spacer carrying two BsaI restriction sites for the easy cloning of new spacers. Spacers were cloned by digestion with BsaI, and ligation of annealed oligonucleotides designed as follow: 5′-aaac+(target sequence)+g-3′ and 5′-aaaac+(reverse complement of the target sequence)-3′, where the target sequence is 30 nt and is followed by a functional PAM (NGG). A list of all spacers tested in this study is provided in Supplementary Table S3.
Plasmid pCas9 was generated in a previous study (23). Plasmid pdCas9 was constructed by introducing D10A and H840A mutations to cas9 on pCas9 through amplification of this plasmid with primers B337/B340 and B338/B339, followed by Gibson assembly (34) of the two products. Both plasmids contain a minimal CRISPR array with BsaI sites for the cloning of new spacers using complementary oligonucleotides. The in-frame deletion of dcas9 on pdCas9 was achieved by amplification of the plasmid with primers B544/B545 and Gibson assembly of the resulting products.
Fusion of the ω subunit (rpoZ) to dCas9 was achieved by amplification of pdCas9 with primers B441/W551 or B446/W552, and amplification of rpoZ with primers B442/W550 or W553/B448 to create the C- or N-terminal ω fusions, respectively, followed by Gibson assembly. Plasmid pWJ66 carries the C-terminal fusion and pWJ68 the N-terminal fusion. To measure induction in E. coli KS1ΔZ, a chloramphenicol-resistant strain, the plasmid resistance was changed to spectinomycin. Plasmids pWJ66 and pWJ68 were amplified with oligos H001/H002, and the spectinomycin resistance gene was amplified from pSWKspec with oligos H003/H004; Gibson assembly of the PCR products generated plasmids pDB191 and pDB192.
Plasmid pDB127 was constructed by amplification of gfp-mut2 (35) with primers B368/B371, followed by digestion with EcoRI and BamHI, and ligation together with the annealed oligonucleotides B369/B370 in the pZS24-MCS1 (36) vector cut with XhoI and BamHI. The sequence of the PAM-rich promoter carried by pDB127 is provided in the Supplementary Sequences.
Plasmids pWJ89, pWJ96 and pWJ97 were constructed by changing the promoter of gfp-mut2 on pDB127 for biobrick promoters BBa_J23117, BBa_J23116 and BBa_J23110, respectively (http://partsregistry.org). The region upstream of the promoter was changed to include multiple NGG PAM sequences on both strands. Full sequences of these promoters are provided in the Supplementary Sequences.
Fluorescence measurements
Fluorescence was measured in a Tecan microplate reader. In all experiments, background fluorescence, or auto-fluorescence, was measured using a control strain lacking the GFP reporter. Auto-fluorescence was subtracted from the fluorescence readings and relative fluorescence was normalized to cells expressing a non-targeting crRNA (encoded by the BsaI sequences designed for spacer cloning).
β-galactosidase assays
β-galactosidase activity in E. coli and S. pneumoniae was measured as previously described (37).
Northern blot analysis
RNA was extracted from overnight cultures using TRIzol (Invitrogen) following the manufacturer’s protocol. For each sample, 10 µg of RNA were separated on a 5% polyacrylamide gel. The RNA was electro-transferred to a charged membrane and hybridized either to a probe upstream (P511) or downstream (P510) of the T10 and B10 target sites. A probe annealing to the 5 S rRNA (B507) was used as a control.
RESULTS
dCas9-mediated repression
We converted the S. pyogenes Cas9 nuclease into a programmable DNA-binding protein by introducing the D10A and H840A mutations in the RuvC and HNH nuclease domains, respectively (8). This catalytically ‘dead’ version of Cas9, dCas9, was introduced into the pdCas9 plasmid along with the tracrRNA and a minimal CRISPR array designed for the easy cloning of new spacers and expression of crRNA guides (23). To evaluate the effect of dCas9 promoter binding on gene expression in E. coli, we constructed a GFP reporter plasmid (pDB127) carrying the gfp-mut2 gene (35) under the control of a promoter designed to carry several NGG PAM sequences on both strands. Twenty-two different spacers were engineered to express crRNAs guiding dCas9 to different regions of the gfp-mut2 promoter and open reading frame. A greater than 100-fold reduction in fluorescence was observed on targeting of regions overlapping or adjacent to the −35 and −10 promoter elements and to the Shine–Dalgarno sequence (Figure 1B). Targets on both strands showed similar repression levels. These experiments suggest that the binding of dCas9 to any position within the promoter region prevents transcription initiation, presumably through steric inhibition of RNAP binding. To confirm that repression was due to dCas9 binding of the promoter DNA and not an effect of the antisense guide crRNA by itself, we repeated experiments in the absence of dCas9. In all cases tested, the fluorescence levels were identical to a non-targeting dCas9 control (Supplementary Figure S1).
To determine whether dCas9 DNA binding could prevent transcription elongation, we directed it to the open reading frame of gfp-mut2. A reduction in fluorescence was observed when both the coding and non-coding strands were targeted, suggesting that Cas9 binding could block the elongating RNAP (Figure 1B). However, although a ∼20–40% reduction in expression was observed when the non-coding strand (the crRNA has the same sequence as the coding strand) was targeted (red spacers), a range of 6- to 35-fold reduction was observed when dCas9 was directed to the coding strand (blue spacers). To directly determine the effects of dCas9 binding on transcription, we extracted RNA from strains expressing the T5, T10 or B10 crRNA guides or a non-targeting dCas9 and subjected it to northern blot analysis using probes binding before (P511) or after (P510) the B10 and T10 target sites (Figure 1C). Consistent with our fluorescence measurement, no gfp-mut2 transcription was detected when dCas9 was directed to the promoter region (T5 target), and lower levels of transcription were observed after the targeting of the T10 region. Interestingly, a smaller transcript was observed with the P511 probe in cells where dCas9 binds to the T10 or B10 target. These species correspond to the expected size of a transcript that would be interrupted by dCas9 (calculated as ∼250 or 300 nt between the transcription start and the B10 or T10 target sites, respectively). This result is a direct indication that dCas9 causes transcription termination. In accordance with the pronounced decrease in fluorescence caused by B10-bound dCas9, only the truncated gfp transcript, but no full-length transcript, was detected with the P511 probe. Altogether, these results demonstrate that directing dCas9 to different gene regions can prevent both the initiation and elongation of transcription. Interestingly, dCas9 targeting of the coding strand blocks transcription more efficiently than targeting of the non-coding strand, suggesting a more efficient displacement of this protein by the elongating RNAP in this configuration.
To corroborate the generality and broad applicability of the approach, we tested the ability of dCas9 to repress β-galactosidase (bgaA) expression in S. pneumoniae, a Gram-positive organism. To do this, we engineered a strain (DB17) harboring dcas9 and the tracrRNA in the chromosome, whereas crRNA guides were expressed from a plasmid carrying the CRISPR array, pDB98 (23) (Supplementary Figure S2A). Three spacers targeting different positions within the bgaA promoter and two spacers targeting the bgaA open reading frame were tested (Supplementary Figure S2B). We observed up to a 14-fold reduction in β-galactosidase activity depending on the targeted position (Supplementary Figure S2C). Binding of dCas9 to similar regions of the gfp-mut2 and bgaA promoters produced different results. We believe that this could be related to the different regulation of these promoters. Although gfp-mut2 is under the control of a synthetic constitutive promoter, the bgaA promoter is regulated in response to lactose metabolism (37). Alternatively, the binding strength of dCas9 can depend on the guide crRNA sequence. In contrast, dCas9 binding within the open reading frame regions produced similar results in E. coli and S. pneumoniae: targeting the coding strand blocked transcription more efficiently than targeting the non-coding strand. These results suggest that targeting the non-coding strand should be the preferred strategy for the use of dCas9 as a programmable repressor.
Modulation of dCas9-mediated repression levels
Some applications require a precise tuning of gene expression rather than its complete repression. We sought to achieve intermediate repression levels through the introduction of mismatches that will weaken the crRNA/target interactions. We created a series of spacers based on the B1, T5 and B10 constructs that express crRNAs with increasing numbers of mutations in the 5′ end (Figure 2A). Introduction of up to eight mismatches in B1 and T5 did not affect the repression level, and a progressive increase in fluorescence was observed for additional mutations (Figure 2B). A different pattern was observed for the gradual mutagenesis of the B10 crRNA: a gradual decrease in repression was achieved through the introduction of an increasing number of mismatches, with as little as two mismatches providing a lower level of repression than a crRNA fully complementary to the target (Figure 2B). These results demonstrate that the introduction of mismatches in the crRNA guide allows for the modulation of dCas9 repression. However, the number of the mismatches required to achieve this modulation depends on the mode of dCas9 repression, i.e. blocking transcription initiation or elongation.
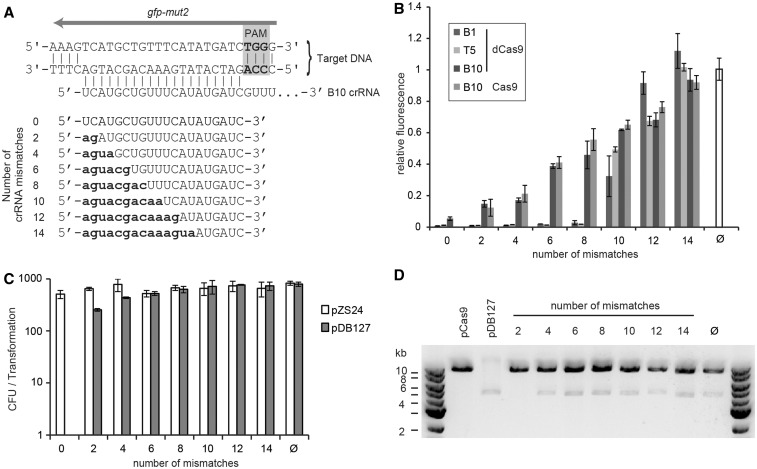
Figure 2.
Effect of mismatches between crRNA guide and target sequence. (A) Protospacer and crRNA-guide sequences for the B10 target site. Mutations in the 5′ region of the crRNA are shown in lower case; Watson–Crick complementary bases were introduced. (B) Effect of an increasing number of mutations in the 5′ end of the B1, T5 and B10 crRNAs on gfp-mut2 repression mediated by dCas9. Repression by wild-type Cas9 guided by mutant versions of the B10 crRNA is also shown. Fluorescence values are normalized to the fluorescence of a strain expressing gfp-mut2 and dCas9, but no crRNA guide (Ø). Error bars show one standard deviation from the mean of three relative fluorescence values. (C) Effect of Cas9 targeting using a B10 crRNA guide with increasing numbers of mutations at the 5′ end on the transformation of the GFP reporter plasmid, pDB127, or an empty vector control, pZS*24. The mean of three independent enumerations of the total number of colony forming units (CFU) per transformation is shown; error bars indicate one standard deviation. (D) Agarose gel electrophoresis of SacI-digested purified plasmids from cells obtained after transformation of the pDB127-target plasmid into cells expressing wild-type Cas9 and different 5′ mutant versions of the B10 crRNA. Individual plasmids are shown to indicate the electrophoretic mobility of each plasmid.
Our results show that 12 nt of complementarity (8 mismatches) between the 3′ end of the crRNA spacer sequence and the target are sufficient to provide strong transcriptional repression by dCas9. In contrast, efficient in vitro cleavage by Cas9 requires 15 nt of homology (5 mismatches) (8). To determine whether this discrepancy is generated by the introduction of the catalytic site mutations in dCas9 or to differences between in vivo and in vitro assays, we tested our mismatch-containing crRNAs for their abilities to direct DNA cleavage by wild-type Cas9. We measured the transformation efficiency of the GFP reporter plasmid pDB127 into cells expressing Cas9 and variants of the B10 crRNA with an increasing number of mutations at the 5′ end (Figure 2C). As expected, no transformants were recovered in the presence of a perfect match between the crRNA and the target, demonstrating Cas9 nuclease activity against pDB127. Two mismatches at the 5′end of the crRNA:target interaction restored transformation efficiency to the same levels obtained with competent cells expressing a non-targeting Cas9. However, transformant colonies were smaller than the control transformation and of variable sizes, suggesting partial CRISPR immunity against pDB127. Consistent with this hypothesis, the targeted plasmid displayed a strong reduction in copy number (Figure 2D). CRISPR immunity against the plasmid was completely eliminated with four mismatches or more. As a minimum of six or seven mismatches are required to abolish Cas9 cleavage in vitro (8), these results suggest that mismatches are less tolerated in vivo than in vitro. Alternatively, the effect of mismatches can be different for different crRNA sequences.
The comparison of Cas9 targeting versus dCas9 repression on the same protospacer (B10) using different crRNAs with increasing numbers of mutations produced another interesting observation. If dCas9 is able to bind and repress gfp expression using a crRNA guide with only 12 nt of complementarity with the target, and Cas9 is not able to cleave the target under the same conditions, this suggests that wild-type Cas9 could bind and repress as dCas9 in the presence of 5′ mismatches between the crRNA and the protospacer. When this was tested, we found not only that wild-type Cas9 can repress gene expression but to the same levels as dCas9 (Figure 2B). These results show that in the presence of mismatches between the 5′ end of the crRNA and the target, wild-type Cas9 is unable to cleave the target but can still bind it with substantial affinity to repress gene expression.
dCas9-mediated activation
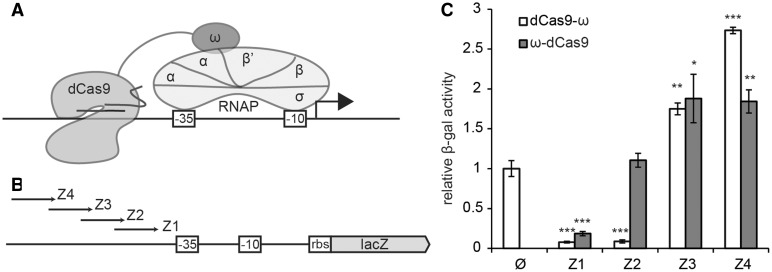
We decided to convert dCas9 into a transcription activator. We relied on previous work that demonstrated that a fusion between the lambda cI repressor and the RNAP omega subunit (ω) can activate transcription by stabilizing the binding of RNAP to a promoter bearing an upstream lambda operator (29). Therefore, we made both C- and N-terminal fusions between the ω subunit and dCas9 (Figure 3A). We also expressed different crRNAs to program the binding of both fusion proteins to four different positions in a constitutive synthetic promoter controlling the lacZ gene (Figure 3B) in an E. coli strain lacking the gene encoding for the ω subunit (rpoZ). We used a fusion of the ω subunit to the C-terminus of dCas9 (dCas9-ω lacking a targeting crRNA guide as a control, and we measured β-galactosidase activity as a reporter of gene expression (Figure 3C). The effect of both fusion proteins on lacZ expression depended on the binding site, still causing gene repression when the binding site was too close to the promoter region. With binding sites more distant from the promoter, we observed a modest increase in β-galactosidase activity; in the best case, we observed 2.8-fold activation for the dCas9-ω C-terminal fusion.
Figure 3.
Activation of gene expression in E. coli using dCas9 fused to the ω subunit of RNAP. (A) dCas9 is directed to the promoter region and is fused to the ω subunit of RNAP, which recruits the polymerase by interacting with the β′ subunit. A host with a deletion of rpoZ, encoding ω, is used. (B) Either N- or C-terminal fusions of the ω subunit to dCas9 were directed to four regions of the top strand upstream of the −35 element of the lacZ gene. (C) lacZ gene expression levels in the different strains were measured as β-galactosidase activity (Miller units). Activation is reported as the relative Miller units normalized against the units obtained with cells expressing a C-terminal dCas9-ω fusion but no crRNA guide (Ø). The average of three independent experiments is indicated; error bars indicate one standard deviation. Asterisks indicate the P-values associated with each measurement, compared with the no crRNA guide control (Ø). *P ≤ 0.05; **P ≤ 0.005; ***P ≤ 0.001.
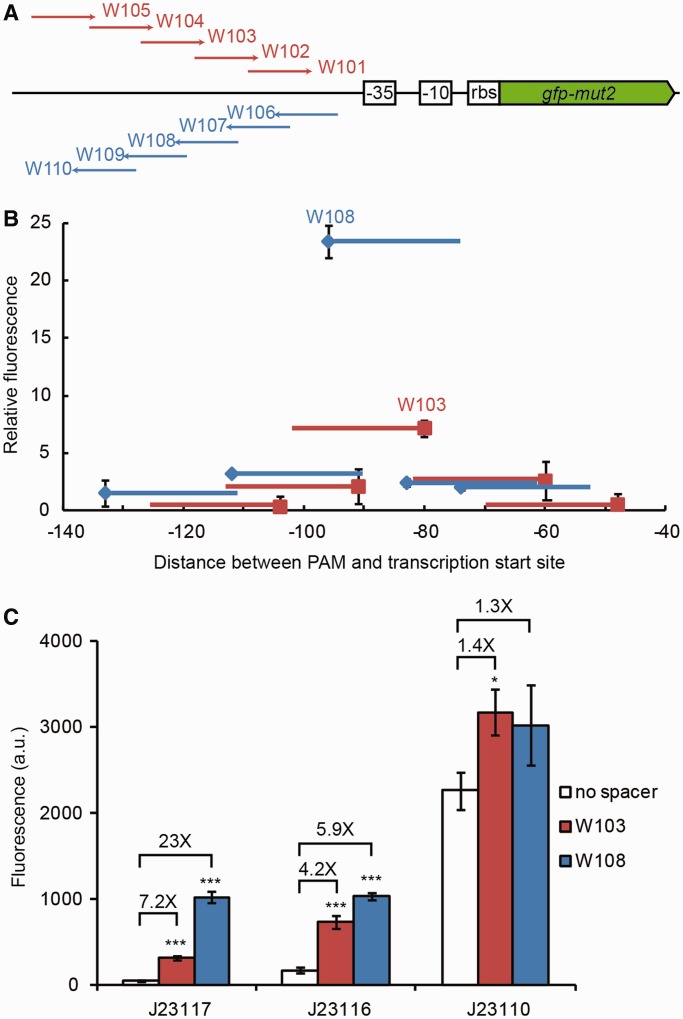
We thus decided to further investigate the activation capabilities of the dCas9-ω fusion when targeted to regions increasingly distant from the −35 promoter element as well as to both DNA strands (Figure 4A). To facilitate measurements, we used a GFP reporter plasmid, pWJ89, with the gfp-mut2 gene under the control of a weak biobrick promoter (BBa_J23117) that is preceded by a sequence rich in NGG PAM sequences on both strands. Among 10 tested binding sites, 2 (specified by W103 and W108 crRNAs, each targeting a different strand) were found to strongly activate gfp-mut2 (Figure 4B). These are located 43 and 59 nt away from the −35 element (80 and 96 nt upstream of the transcription start site) and provide a 7.2- and 23-fold induction, respectively. A similar relative induction was observed when fluorescence is measured at different growth phases (data not shown). These data suggest that a dCas9-ω fusion can induce gene expression when it binds at an optimal distance from the promoter.
Figure 4.
Activation of gene expression using a dCas9-ω fusion. (A) The positions of the different crRNA guides tested (W101–W110), relative to the ribosome binding site (rbs) and the −35 and −10 promoter elements of the gfp-mut2 gene are shown. (B) GFP fluorescence levels, relative to the fluorescence produced by a control strain expressing a non-targeting dCas9-ω, as a function of the position of the target sequence within the gfp-mut2 upstream region (+1, transcription start). Squares indicate the PAM position, lines the extension of complementarity between the crRNA guide and the reporter gene; red lines, top strand targets; blue, bottom strand. Error bars show one standard deviation from the mean of three relative fluorescence values. (C) Activation of three variants gfp-mut2 containing promoters of different strengths (J23117, J23116 and J23110) by W103- or W108-guided dCas9-ω. The relative induction, compared with the fluorescence of cells expressing a non-targeting dCas9-ω, is shown. The average of three independent experiments is indicated; error bars indicate one standard deviation. Asterisks indicate the P-values associated with each measurement, compared with the no spacer control. *P ≤ 0.05; ***P ≤ 0.001.
To test whether the dCas9-ω fusion can induce expression of gfp-mut2 under the control of stronger promoters, we replaced the weak promoter in pWJ89 (BBa_J23117) with promoters of intermediate (BBa_J23116) and high (BBa_J23110) strength (Figure 4C). We compared dCas9-ω activation of gfp expression from the three constructs using the W103 and W108 crRNA guides. All promoters could be induced by the targeting of dCas9-ω to both binding sites; however, the relative induction diminishes as the promoter gets stronger. Altogether, these results show that dCas9 can be used to activate gene expression, with the possibility of achieving different levels of activation depending on the strength of the targeted promoter (the best induction being obtained with weak promoters).
DISCUSSION
The manipulation of gene expression in prokaryotes is usually achieved through the use of promoters whose activity can be modulated using small molecules (38). This requires genetic engineering to place the gene of interest under the control of inducible promoters. Given the lack of RNAi (39) in prokaryotes, methods that allow for simple and global regulation of gene expression are limited (40). Here, we describe the use of an RNA-guided DNA-binding protein, dCas9, to either repress or activate genes in E. coli and S. pneumoniae. In this method, dCas9 can be directed to any region of the bacterial chromosome that is specified by the base-pair complementarity between the RNA guide and the cognate genomic sequence, i.e. without the need to modify the promoter sequence of the gene whose expression is manipulated.
Repression is achieved by directing dCas9 to either promoter or open-reading frame regions. Although binding of dCas9 to promoters prevents transcription initiation, binding to the open-reading frame prevents elongation, especially when the coding strand is targeted. While this article was in review, Qi and colleagues (41) also showed that dCas9 can act as a transcription repressor by preventing initiation or elongation, both in E. coli and human (HEK293) cells. As opposed to our system, which uses the natural CRISPR array and tracrRNA, Qi et al. used a chimeric crRNA as a guide. Chimeric crRNAs combine critical crRNA and tracrRNA moieties into a single small RNA that can be loaded into dCas9 without the need for processing by RNAse III (8). In this study, dCas9 was under the control of an inducible promoter, demonstrating that repression can be reversible if the inducer is withdrawn from the culture. By using several chimeric guide RNAs, Qi et al. demonstrated that this methodology can be applied for repression of multiple loci at the same time. Multiplexing will also be possible with our system, as the CRISPR array can be engineered to contain multiple spacers encoding different crRNA guides.
Target specificity is an important aspect of all the recent Cas9-based technologies. Qi et al. analyzed this by truncating the 5′ end of a chimeric guide RNA that prevents transcription elongation. They observed that repression is noticeable with at least 12 nt of homology between the chimeric RNA and the protospacer (41). In addition, the authors performed RNAseq in the presence and absence of dCas9 targeting and determined that the only transcript with a significant change in abundance was the one specified by the RNA guide. We also determined the effects of the accumulation of mismatches at the 5′ end of crRNAs that prevent both transcription initiation and elongation. In accordance with the results of Qi et al., we found that a 12 nt match between crRNA and protospacer produces a weak but significant repression if the coding strand of an open-reading frame is targeted. In theory, as in addition to the crRNA:protospacer matches a perfect PAM is required for dCas9 repression, such off-target sequence occurs randomly about once every 414 bp, 268 Mbp, and thus is unlikely to be found in bacterial genomes [the largest size to date is 13 Mbp (42)], but more likely to be present in larger eukaryotic genomes. Nonetheless, off target effects can happen, especially during studies that require the design of many crRNA guides. For example, during the course of this study, we were unable to clone one of the designed spacers on the pdCas9 vector. We later found that this spacer showed a 12 nt perfect match next to a good PAM in the essential murC gene (43) (Supplementary Figure S3). Such off-target effect could easily be avoided by a systematic blast of the engineered spacers. By analyzing the effect of mismatches between the crRNA and its target on dCas9 repression, we found that wild-type Cas9 can repress gene expression when directed by a crRNA with at least 4 nt of mismatch at the 5′end that prevents it from efficiently cleaving its target. This finding suggests that endogenous CRISPR systems could actively repress gene expression when an imperfect match exists between the crRNA and its targets. The inhibition of biofilm formation by the type I CRISPR system of Pseudomonas aeruginosa, a phenomenon that requires mismatches between the crRNA and its target, could be an example of this side effect of CRISPR immunity on transcription (44).
Finally, we demonstrated that dCas9 can be directed to promoter regions to activate gene expression. This requires the addition of an activation domain, in this study the ω subunit of RNAP, which was previously shown to provide effective recruitment of RNAP (29). Activation levels depended on the distance between the dCas9-binding sequence and the −35 promoter element. A maximum of a 23-fold induction was achieved with a bottom strand target with its PAM positioned 59 nt upstream of the −35 element. This level of activation is low when compared with the activation achieved by a cI-ω fusion, which provides 70-fold induction (29). We believe that activation can be further optimized by changing the protein linker between dCas9 and the activation domain and/or testing different activation domains.
Results presented here and in Qi et al. (41) reveal a new powerful technology to regulate gene expression in prokaryotes with an exciting future. The use of larger CRISPR arrays generating multiple crRNA guides can provide the possibility of affecting many genes at the same time and will allow genetic network organization to be probed. dCas9 gene regulation can also be applied to the engineering of synthetic gene networks in living cells for biotechnological applications. There is also the possibility of tethering dCas9 fused to fluorescent proteins to specific loci to investigate the influence of gene function and expression on the subcellular positioning of chromosomal loci in bacteria (45). We anticipate that dCas9-based technologies will contribute to the success of these applications.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–3, Supplementary Figures 1–3 and Supplementary Sequences.
FUNDING
Harvey L. Karp Discovery Award and the Bettencourt Schuller Foundation (to D.B.); Helmsley Postdoctoral Fellowship for Basic and Translational Research on Disorders of the Digestive System at The Rockefeller University (to P.S.); NIH grant [R01 GM044025 to A.H.]; NIH Director’s Pioneer Award [DP1MH100706], Transformative R01, the Keck, McKnight, Gates, Damon Runyon, Searle Scholars, Klingenstein, and Simons Foundations, Bob Metcalfe, Mike Boylan and Jane Pauley (to F.Z.); Searle Scholars Program, the Rita Allen Scholars Program, an Irma T. Hirschl Award and a NIH Director’s New Innovator Award [1DP2AI104556-01 to L.A.M.]. Funding for open access charge: NIH Director’s New Innovator Award [1DP2AI104556-01].
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank members of the Nussenzweig’s laboratory at The Rockefeller University for technical assistance with GFP fluorescence measurements. They are grateful to Seth Darst for his advice.
REFERENCES
- 1.Auslander S, Fussenegger M. From gene switches to mammalian designer cells: present and future prospects. Trends Biotechnol. 2012;31:155–168. doi: 10.1016/j.tibtech.2012.11.006. [DOI] [PubMed] [Google Scholar]
- 2.Khalil AS, Lu TK, Bashor CJ, Ramirez CL, Pyenson NC, Joung JK, Collins JJ. A synthetic biology framework for programming eukaryotic transcription functions. Cell. 2012;150:647–658. doi: 10.1016/j.cell.2012.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beerli RR, Segal DJ, Dreier B, Barbas CF., 3rd Toward controlling gene expression at will: specific regulation of the erbB-2/HER-2 promoter by using polydactyl zinc finger proteins constructed from modular building blocks. Proc. Natl Acad. Sci. USA. 1998;95:14628–14633. doi: 10.1073/pnas.95.25.14628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cong L, Zhou R, Kuo YC, Cunniff M, Zhang F. Comprehensive interrogation of natural TALE DNA-binding modules and transcriptional repressor domains. Nat. Commun. 2012;3:968. doi: 10.1038/ncomms1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miller JC, Tan S, Qiao G, Barlow KA, Wang J, Xia DF, Meng X, Paschon DE, Leung E, Hinkley SJ, et al. A TALE nuclease architecture for efficient genome editing. Nat. Biotechnol. 2011;29:143–148. doi: 10.1038/nbt.1755. [DOI] [PubMed] [Google Scholar]
- 6.Zhang F, Cong L, Lodato S, Kosuri S, Church GM, Arlotta P. Efficient construction of sequence-specific TAL effectors for modulating mammalian transcription. Nat. Biotechnol. 2011;29:149–153. doi: 10.1038/nbt.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gasiunas G, Barrangou R, Horvath P, Siksnys V. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc. Natl Acad. Sci. USA. 2012;109:E2579–E2586. doi: 10.1073/pnas.1208507109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Westra ER, van Erp PB, Kunne T, Wong SP, Staals RH, Seegers CL, Bollen S, Jore MM, Semenova E, Severinov K, et al. CRISPR immunity relies on the consecutive binding and degradation of negatively supercoiled invader DNA by Cascade and Cas3. Mol. Cell. 2012;46:595–605. doi: 10.1016/j.molcel.2012.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bikard D, Marraffini LA. Innate and adaptive immunity in bacteria: mechanisms of programmed genetic variation to fight bacteriophages. Curr. Opin. Immunol. 2012;24:15–20. doi: 10.1016/j.coi.2011.10.005. [DOI] [PubMed] [Google Scholar]
- 11.Fineran PC, Charpentier E. Memory of viral infections by CRISPR-Cas adaptive immune systems: acquisition of new information. Virology. 2012;434:202–209. doi: 10.1016/j.virol.2012.10.003. [DOI] [PubMed] [Google Scholar]
- 12.Westra ER, Swarts DC, Staals RH, Jore MM, Brouns SJ, van der Oost J. The CRISPRs, they are a-changin': how prokaryotes generate adaptive immunity. Annu. Rev. Genet. 2012;46:311–339. doi: 10.1146/annurev-genet-110711-155447. [DOI] [PubMed] [Google Scholar]
- 13.Wiedenheft B, Sternberg SH, Doudna JA. RNA-guided genetic silencing systems in bacteria and archaea. Nature. 2012;482:331–338. doi: 10.1038/nature10886. [DOI] [PubMed] [Google Scholar]
- 14.Brouns SJ, Jore MM, Lundgren M, Westra ER, Slijkhuis RJ, Snijders AP, Dickman MJ, Makarova KS, Koonin EV, van der Oost J. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 2008;321:960–964. doi: 10.1126/science.1159689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carte J, Wang R, Li H, Terns RM, Terns MP. Cas6 is an endoribonuclease that generates guide RNAs for invader defense in prokaryotes. Genes Dev. 2008;22:3489–3496. doi: 10.1101/gad.1742908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hatoum-Aslan A, Maniv I, Marraffini LA. Mature clustered, regularly interspaced, short palindromic repeats RNA (crRNA) length is measured by a ruler mechanism anchored at the precursor processing site. Proc. Natl Acad. Sci. USA. 2011;108:21218–21222. doi: 10.1073/pnas.1112832108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haurwitz RE, Jinek M, Wiedenheft B, Zhou K, Doudna JA. Sequence- and structure-specific RNA processing by a CRISPR endonuclease. Science. 2010;329:1355–1358. doi: 10.1126/science.1192272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garneau JE, Dupuis ME, Villion M, Romero DA, Barrangou R, Boyaval P, Fremaux C, Horvath P, Magadan AH, Moineau S. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature. 2010;468:67–71. doi: 10.1038/nature09523. [DOI] [PubMed] [Google Scholar]
- 19.Hale CR, Zhao P, Olson S, Duff MO, Graveley BR, Wells L, Terns RM, Terns MP. RNA-guided RNA cleavage by a CRISPR RNA-Cas protein complex. Cell. 2009;139:945–956. doi: 10.1016/j.cell.2009.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Makarova KS, Haft DH, Barrangou R, Brouns SJ, Charpentier E, Horvath P, Moineau S, Mojica FJ, Wolf YI, Yakunin AF, et al. Evolution and classification of the CRISPR-Cas systems. Nat. Rev. Microbiol. 2011;9:467–477. doi: 10.1038/nrmicro2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Deltcheva E, Chylinski K, Sharma CM, Gonzales K, Chao Y, Pirzada ZA, Eckert MR, Vogel J, Charpentier E. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. 2011;471:602–607. doi: 10.1038/nature09886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Deveau H, Barrangou R, Garneau JE, Labonte J, Fremaux C, Boyaval P, Romero DA, Horvath P, Moineau S. Phage response to CRISPR-encoded resistance in Streptococcus thermophilus. J. Bacteriol. 2008;190:1390–1400. doi: 10.1128/JB.01412-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jiang W, Bikard D, Cox D, Zhang F, Marraffini LA. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat. Biotechnol. 2013;31:233–239. doi: 10.1038/nbt.2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cho SW, Kim S, Kim JM, Kim JS. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat. Biotechnol. 2013;31:230–232. doi: 10.1038/nbt.2507. [DOI] [PubMed] [Google Scholar]
- 25.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hwang WY, Fu Y, Reyon D, Maeder ML, Tsai SQ, Sander JD, Peterson RT, Yeh JR, Joung JK. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat. Biotechnol. 2013;31:227–229. doi: 10.1038/nbt.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jinek M, East A, Cheng A, Lin S, Ma E, Doudna J. RNA-programmed genome editing in human cells. Elife. 2013;2:e00471. doi: 10.7554/eLife.00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mali P, Yang L, Esvelt KM, Aach J, Guell M, Dicarlo JE, Norville JE, Church GM. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dove SL, Hochschild A. Conversion of the omega subunit of Escherichia coli RNA polymerase into a transcriptional activator or an activation target. Genes Dev. 1998;12:745–754. doi: 10.1101/gad.12.5.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Horton RM. In vitro recombination and mutagenesis of DNA : SOEing together tailor-made genes. Methods Mol. Biol. 1993;15:251–261. doi: 10.1385/0-89603-244-2:251. [DOI] [PubMed] [Google Scholar]
- 31.Demarre G, Guerout AM, Matsumoto-Mashimo C, Rowe-Magnus DA, Marliere P, Mazel D. A new family of mobilizable suicide plasmids based on broad host range R388 plasmid (IncW) and RP4 plasmid (IncPalpha) conjugative machineries and their cognate Escherichia coli host strains. Res. Microbiol. 2005;156:245–255. doi: 10.1016/j.resmic.2004.09.007. [DOI] [PubMed] [Google Scholar]
- 32.Chaveroche MK, Ghigo JM, d'Enfert C. A rapid method for efficient gene replacement in the filamentous fungus Aspergillus nidulans. Nucleic Acids Res. 2000;28:E97. doi: 10.1093/nar/28.22.e97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Husmann LK, Scott JR, Lindahl G, Stenberg L. Expression of the Arp protein, a member of the M protein family, is not sufficient to inhibit phagocytosis of Streptococcus pyogenes. Infect. Immun. 1995;63:345–348. doi: 10.1128/iai.63.1.345-348.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA, 3rd, Smith HO. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods. 2009;6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 35.Cormack BP, Valdivia RH, Falkow S. FACS-optimized mutants of the green fluorescent protein (GFP) Gene. 1996;173:33–38. doi: 10.1016/0378-1119(95)00685-0. [DOI] [PubMed] [Google Scholar]
- 36.Lutz R, Bujard H. Independent and tight regulation of transcriptional units in Escherichia coli via the LacR/O, the TetR/O and AraC/I1-I2 regulatory elements. Nucleic Acids Res. 1997;25:1203–1210. doi: 10.1093/nar/25.6.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zahner D, Hakenbeck R. The Streptococcus pneumoniae beta-galactosidase is a surface protein. J. Bacteriol. 2000;182:5919–5921. doi: 10.1128/jb.182.20.5919-5921.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Keasling JD. Gene-expression tools for the metabolic engineering of bacteria. Trends Biotechnol. 1999;17:452–460. doi: 10.1016/s0167-7799(99)01376-1. [DOI] [PubMed] [Google Scholar]
- 39.Hannon GJ, Rossi JJ. Unlocking the potential of the human genome with RNA interference. Nature. 2004;431:371–378. doi: 10.1038/nature02870. [DOI] [PubMed] [Google Scholar]
- 40.Na D, Yoo SM, Chung H, Park H, Park JH, Lee SY. Metabolic engineering of Escherichia coli using synthetic small regulatory RNAs. Nat. Biotechnol. 2013;31:170–174. doi: 10.1038/nbt.2461. [DOI] [PubMed] [Google Scholar]
- 41.Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, Lim WA. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013;152:1173–1183. doi: 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chang YJ, Land M, Hauser L, Chertkov O, Del Rio TG, Nolan M, Copeland A, Tice H, Cheng JF, Lucas S, et al. Non-contiguous finished genome sequence and contextual data of the filamentous soil bacterium Ktedonobacter racemifer type strain (SOSP1-21) Stand. Genomic. Sci. 2011;5:97–111. doi: 10.4056/sigs.2114901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gerdes SY, Scholle MD, Campbell JW, Balazsi G, Ravasz E, Daugherty MD, Somera AL, Kyrpides NC, Anderson I, Gelfand MS, et al. Experimental determination and system level analysis of essential genes in Escherichia coli MG1655. J. Bacteriol. 2003;185:5673–5684. doi: 10.1128/JB.185.19.5673-5684.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zegans ME, Wagner JC, Cady KC, Murphy DM, Hammond JH, O'Toole GA. Interaction between bacteriophage DMS3 and host CRISPR region inhibits group behaviors of Pseudomonas aeruginosa. J. Bacteriol. 2009;191:210–219. doi: 10.1128/JB.00797-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Libby EA, Roggiani M, Goulian M. Membrane protein expression triggers chromosomal locus repositioning in bacteria. Proc. Natl Acad. Sci. USA. 2012;109:7445–7450. doi: 10.1073/pnas.1109479109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.