Abstract
APOBEC3G (A3G) is a host-encoded protein that potently restricts the infectivity of a broad range of retroviruses. This can occur by mechanisms dependent on catalytic activity, resulting in the mutagenic deamination of nascent viral cDNA, and/or by other means that are independent of its catalytic activity. It is not yet known to what extent deamination-independent processes contribute to the overall restriction, how they exactly work or how they are regulated. Here, we show that alanine substitution of either tryptophan 94 (W94A) or 127 (W127A) in the non-catalytic N-terminal domain of A3G severely impedes RNA binding and alleviates deamination-independent restriction while still maintaining DNA mutator activity. Substitution of both tryptophans (W94A/W127A) produces a more severe phenotype in which RNA binding and RNA-dependent protein oligomerization are completely abrogated. We further demonstrate that RNA binding is specifically required for crippling late reverse transcript accumulation, preventing proviral DNA integration and, consequently, restricting viral particle release. We did not find that deaminase activity made a significant contribution to the restriction of any of these processes. In summary, this work reveals that there is a direct correlation between A3G’s capacity to bind RNA and its ability to inhibit retroviral infectivity in a deamination-independent manner.
INTRODUCTION
APOBEC3G (A3G) is one of several cell-intrinsic host retroviral restriction factors in humans that potently inhibit the replication of a broad range of viruses, retroviruses and retroelements [reviewed in (1)]. It is currently believed that A3G’s striking ability to deaminate cytidines into uridines in single-stranded retroviral DNA replication intermediates represents the major mechanism responsible for its antiretroviral activity. Extensive mutations, also called hypermutation, can potentially lead to the generation of premature termination codons and dysfunctional proteins resulting in non-infectious viral progeny (2–5). A3G can, however, also restrict the infectivity of retroviruses by means that do not rely on deamination, but these have yet to be clearly understood (6,7).
A3G proteins expressed in retrovirus-infected cells are packaged into the capsids of progeny virions and exert their enzymatic activity during proviral cDNA synthesis in newly infected target cells (1). Packaging of A3G into human immunodeficiency virus type I (HIV-1) virions is RNA dependent and mediated by the interaction of residues in the N-terminal domain (NTD) of A3G and the nucleocapsid region of the retroviral structural protein Gag (8,9). To counteract the deleterious effects of A3G, HIV-1 acquired the ability to prevent its packaging into virions. The viral infectivity factor (Vif) is an HIV-1 accessory protein that binds to A3G before its incorporation into virions and quickly promotes its degradation by the proteasome [reviewed in (10)]. HIV-1 particles that are released from infected cells expressing Vif are devoid of A3G and are thus fully infectious.
A3G can directly bind RNA via its non-catalytic NTD (11–13). Newly translated monomeric A3G rapidly assembles not only in the cytoplasm into RNA-independent dimeric and tetrameric structures but also into larger oligomeric assemblies that require RNA (11,14–17). In actively dividing cells such as activated T cells and cell lines, these oligomeric complexes will further aggregate into large high molecular mass (HMM) ribonucleoprotein complexes, which are estimated to be between 5 and 15 MDa in size (11,18). A3G proteins in these HMM complexes no longer exhibit enzymatic activity and cannot be packaged into HIV-1 virions (19). Thus, only low molecular mass (LMM) oligomeric A3G complexes that have not yet aggregated into HMM complexes are packaged into virions and exert cytidine deaminase activity during proviral DNA synthesis (19). It is still unclear what triggers the formation of HMM complexes in cell lines and activated lymphocytes. Understanding how these large oligomeric structures assemble is of significant importance because binding to RNA is deemed to be required for HIV-1 virion packaging. Paradoxically, RNA also appears to act as a negative regulator of A3G’s catalytic activity by causing its aggregation into ribonucleic complexes (19). A3G binds various RNAs including those coding for itself, GAPDH and HIV-1, as well as several species of non-coding RNAs such as 7SL, hY1, hY3, hY4, hY5 and Alu (18,20–24). It is currently unknown whether binding to any of these RNAs is specifically required for A3G’s antiviral activity.
The catalytic activity of A3G is currently thought to play a dominant role in the inhibition of retroviral infectivity. Notably, in addition to inflicting genetic damage, poor plus-strand transfer and defective proviral integration have also been reported to be caused by DNA editing (25–28). In parallel, several reports show that significant deamination-independent antiretroviral activity is displayed by catalytically inactive A3G enzymes (6,7,28–30). Disruptions in the zinc-binding motif of the C-terminal domain inactivate the catalytic activity of A3G. Deamination-independent mechanisms such as the inhibition of primer annealing, strand transfer, viral transcript accumulation and proviral integration have been described to collectively partake in the overall restriction of infection (28,31). An important component contributing to the deaminase-independent antiretroviral activity appears to be the inhibition of reverse transcript synthesis. This could occur by the direct interaction of A3G with the reverse transcriptase or by creating roadblocks to the processivity of the reverse transcriptase through binding to ssDNA replication intermediates (6,32). In this context, reduced retroviral cDNA synthesis would be one of the causative factors for impaired proviral integration and infection. Despite the identification of numerous antiretroviral mechanisms, it has not yet been established to what extent G-to-A hypermutation and deamination-independent mechanisms contribute to the overall inhibition of infection.
In this study, we investigated the roles of A3G in RNA-binding, HMM complex assembly and cytidine deamination at different stages of the retroviral infection cycle. We found that tryptophans 94 and 127, which are located in the non-catalytic NTD of human A3G, regulate RNA-binding and HMM complex assembly. Interestingly, both W94A and W127A mutants retain the capacity to intensely deaminate proviral DNA but no longer restrict proviral DNA synthesis, integration or viral particle release. These unique features of the mutants have allowed us to measure the direct contribution of deamination and deamination-independent restriction mechanisms on various steps of the infection cycle of three commonly studied retroviruses.
MATERIALS AND METHODS
Velocity sedimentation
The 293T cells transiently expressing FLAG-A2, FLAG-A3G or the various A3G mutants were lysed for 30 min on ice with NP40 lysis buffer [50 mM Tris–HCl (pH 7.4), 150 mM NaCl, 0.1% NP40, 0.1% Na-deoxycholate] supplemented with cOmplete, EDTA-free, protease inhibitor cocktail (Roche). After removing insoluble material by centrifugation at 17 000g for 5 min at 4°C, half the samples were treated with 1 µg/ml of RNase A for 15 min at room temperature. RNase-treated and -untreated samples were then loaded on top of 5–40% sucrose step gradient (5, 10, 20, 30 and 40% in 2.3 ml of fractions) in the following buffer: 10 mM Tris–HCl (pH 7.4), 25 mM KCl and 10 mM MgCl2. Gradients were spun for 6 h at 41 000 rpm (288 000g) at 4°C in a Beckman SW41 Ti rotor. After the spin, 1.4 ml of fractions were sequentially collected from the top of the gradient, and a 75 µl of aliquot of each fraction was mixed with 25 µl of 4× Laemmli loading buffer. Finally samples were resolved by SDS–PAGE and analyzed by immunoblotting.
Retroviral vectors
The pMOV-eGFP expression vector encodes a replicative Moloney murine leukemia virus (MoMLV) with the eGFP reporter gene inserted in the proline-rich region of the ecotropic env gene (33). The single-cycle HIV[p8.9] pseudovirus is generated by a multi-plasmid expression system using a packaging vector for HIV-1 (pCSGW), an expression vector for Gag-Pol-Rev (p8.9) and the pMDG plasmid encoding the envelope glycoprotein of the vesicular stomatitis virus (34,35). The eGFP reporter gene is located in the pCSGW packaging vector and is expressed from an internal spleen focus-forming virus (SFFV) long terminal repeat (LTR) promoter. The pNL4-3-deltaE-eGFP plasmid was obtained from the NIH AIDS Research Reference and Reagent Program (catalog #11100) (36). Two termination codons were introduced in the vif gene to prevent expression as previously described and renamed pHIVΔVif for simplicity (37).
Transfections and virus production
Viruses were produced by transfecting 3 × 105 293T cells seeded in a 6-well plate with 800 ng of pMOV-eGFP (to produce MoMLV) or 500 ng of pHIVΔVif with 200 ng of pMDG (to produce HIVΔVif) using the GeneJuice transfection reagent (Merck). HIV[p8.9] pseudoviruses were produced by co-transfecting 250 ng of p8.9, along with 300 ng of pCSGW and 150 ng of pMDG.
APOBEC virion packaging was done by co-transfecting virus expression plasmids (in the quantities described earlier in the text) along with APOBEC expression vectors: 80 ng for MoMLV and 150 ng for HIV[p8.9] and HIVΔVif. Cells were washed with phosphate buffered saline 16 h following transfection and grown in culture for an additional 48 h. Virus-containing supernatants were then collected, cleared by centrifugation at 800g, and filtered through 0.45 µ cartridge filters. Virus production was quantified and normalized by enzyme-linked immunosorbent assay for p24 or p30 (QuickTiterTM Lentivirus Titer Kit and QuickTiterTM MuLV Core Antigen ELISA Kit, Cell Biolabs Inc.). Viral stocks produced in presence of APOBEC3 proteins were normalized to the p24 or p30 amounts of their respective A2 control.
Optimization of infection assays
Transfection and infection assays were carefully optimized for optimal dynamic range and single transduction events using a multiplicity of infection (MOI) of 0.5 (Supplementary Figure S1). During careful optimization of our infection assays, we found that viral infectivity is most adversely affected when virus-producing vectors are co-transfected along with control vectors expressing a protein (e.g. eGFP) rather than an ‘empty vector’ (e.g. pcDNA 3.1), particularly when large quantities are co-transfected (Supplementary Figure S1A and B). For this reason, we chose human A2 as a negative control to establish the maximum infection in all our assays. A2 is a member of the broader APOBEC3 family whose crystal structure enabled the early structural homology models of human A3G (38). A2 harbors a single zinc-binding pseudo-catalytic domain, does not form RNA-dependent oligomers and has no detectable deaminase or antiviral activity (39–42).
Infection assays
Target cells were infected at an MOI of 0.5 with respect to viruses produced in the presence of A2. Infections were carried out by spinoculation at 800g for 1 h in the presence of 8 µg/ml of polybrene (43). Infection levels in human 293T and murine NIH 3T3 cells were monitored by flow cytometry analysis by measuring eGFP fluorescence at 24 h (for MoMLV and HIV[p8.9]) or 48 h (for HIVΔVif) after infection. Infection levels were normalized to A2.
Late reverse transcript and proviral integration quantitation
Methods for quantifying late reverse transcript (LRT) accumulation and proviral integration by quantitative real-time PCR (qPCR) were adapted from the following references (44,45). DNA extractions were performed 12 h post-infection for late LRT analysis and at 24 h post-infection for integration analysis. Primer and probes sequences are given in Supplementary Table S1. For LRT analysis, the primers and probe sets for HIVΔVif or HIV[p8.9] and MoMLV are respectively as follows: qHIV-FWD, qHIV-REV and the qHIV-Probe; qMoMLV-FWD, qMoMLV-REV and the qMLV-Probe. Integrated proviruses were amplified by a first round PCR using primer pairs against eGFP and either Alu (293T) or B1 (NIH 3T3) retroelements. The primers used to amplify integrated sequences were as follows: qeGFP-FWD, qAlu-1 and qAlu-2 for 293T; qeGFP-FWD, qB1-1 and qB1-2 for NIH 3T3. PCR products from the first round were then diluted 1:40 and used for qPCR using nested primers qIN-eGFP-FWD and qIN-eGFP-REV, and the qIN-eGFP-Probe. For each reaction, 0.9 pmol/ml of each primer, 0.25 pmol/ml of the probe and 10 ng of template DNA were used in a 20 ml of reaction volume. Reactions were performed in quadruplicate with TaqMan® Gene Expression master mix, Applied Biosystems (AB). Cycling conditions were 10 min at 95°C, followed by 40 cycles of 15 s at 95°C and 1 min at 60°C carried out on an AB Viii7 System. The copy numbers in each sample were normalized for DNA input using human RNase P or mouse Tfrc copy number assays (TaqMan assays 4 403 326 and 4 458 366, AB). Relative quantitation (RQ) was computed by the ViiA7 data analysis software (AB). Normalizations were performed using corresponding A2 levels for each experiment.
Statistical analysis
Restriction, integration and LRT data were expressed as mean relative values ± SD. All experiments were performed using triplicates values for restriction experiments and quadruplicates for integration and LRT analysis. Experiments were repeated at least three times from completely independent transfection assays. All statistical analyses were performed using Student’s paired t-test using GraphPad Prism software.
RESULTS
Tryptophans 94 and 127 are involved in HMM complex assembly and RNA binding
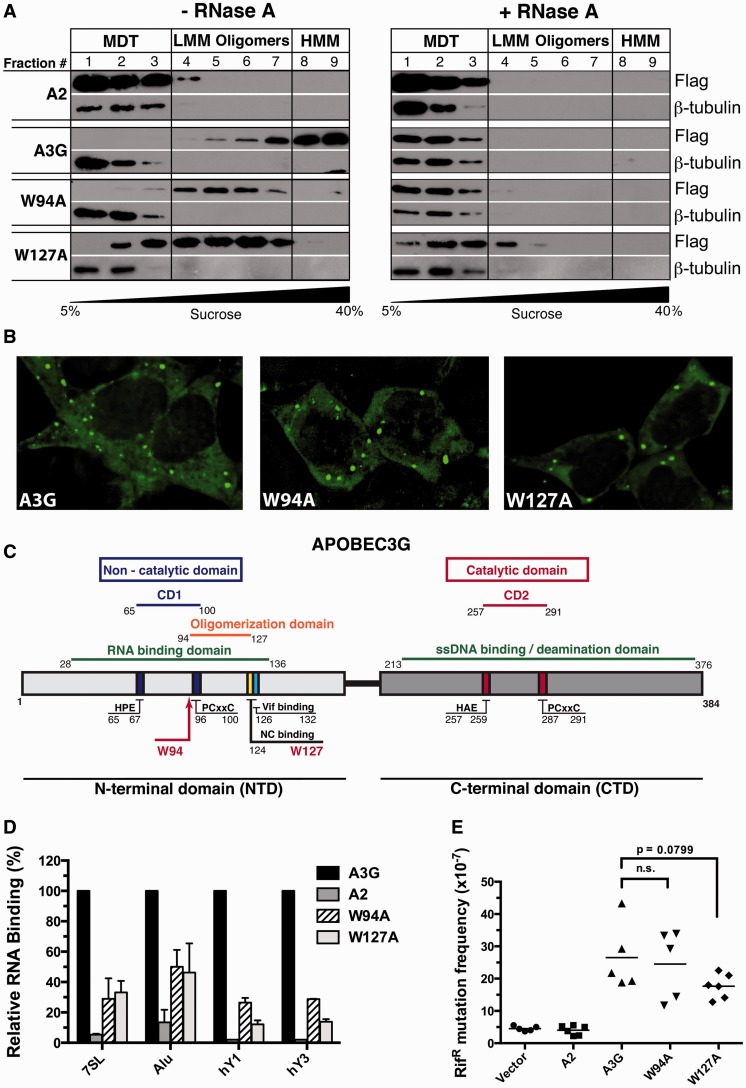
A3G is an RNA-binding enzyme that aggregates into HMM complexes in the cytoplasm of activated CD4 + T lymphocytes and immortalized cell lines (11,18). Here, we have optimized the conditions of velocity sedimentation assays so that HMM complexes consistently accumulate in the bottom two fractions of 5–40% sucrose gradients (fractions 8 and 9), and that RNA-dependent LMM oligomeric forms of A3G consistently accumulate in fractions 4–7. Pretreatment of cell extracts with RNase dissolves HMM and LMM complexes and causes A3G to localize in fractions of the gradient that represent the predicted monomeric, dimeric and tetrameric forms of the protein (labeled as MDT in our figures). The assays were designed so that these RNA-independent forms of A3G consistently accumulate in fractions 1–3. We used endogenous β-tubulin in all our sedimentation assays as a marker for gradient quality control because it exclusively assembles into RNA-independent heterodimers that are consistently detected in fractions 1–3 only.
During the course of a screen to identify the amino acids of A3G that govern its assembly into HMM complexes, we discovered that mutation of tryptophans 94 and 127 to alanine (W94A; W127A) prevented the formation of these complexes (Figure 1A and Supplementary Figure S2). Despite the absence of HMM complexes in fractions 8 and 9, RNA-dependent LMM oligomeric complexes were present throughout the middle fractions of the sucrose gradient (fractions 4–7). Pretreatment of the extracts with RNase resulted in a complete shift toward the top of the gradient populated by the monomeric, dimeric and tetrameric forms of the A3G protein (fractions 1–3). These particular features of the W94A and W127A mutants were not observed with any of the other A3G point mutants that were tested (Supplementary Figure S2).
Figure 1.
Effects of the W94A and W127A mutations on HMM complex assembly, subcellular distribution, RNA-binding and DNA deaminase activity. (A) Lysates of transfected 293T cells, treated (right panels) or untreated (left panels) with RNase, were resolved by velocity sedimentation over a non-denaturing 5–40% sucrose gradient and analyzed by western blot using anti-FLAG and anti-β-tubulin antibodies. (B) Fluorescent imaging of the subcellular localization of 293T cells expressing eGFP-A3G, eGFP-W94A and eGFP-W127A. (C) Schematic representation of the location of important residues and binding domains in the A3G protein sequence. (D) Binding of FLAG-tagged A2, A3G, W94A and W127A to 7SL, Alu, hY1 and hY3 RNAs were determined by qPCR. Relative binding to A3G is depicted. Results represent the mean ± SD of triplicate values from three independent transfection experiments. (E) Evaluation of the intrinsic DNA cytidine deaminase activity using a bacterial mutator assay. Each point represents the mutation frequency (RifR mutants per 107 viable cells) of an independent bacterial culture; median values are indicated.
A3G is a cytoplasmic protein that forms numerous foci. These structures are believed to associate with RNA-processing bodies (P-bodies), which are sites of RNA storage, turnover and decapping (46). We were concerned that altering HMM complex assembly would also affect the cellular localization of the mutant proteins. We therefore transiently expressed eGFP-fusions of the mutant proteins in 293T cells and analyzed their intracellular distribution using fluorescence microscopy. We did not detect any obvious differences in size, intensity or abundance of cellular foci between wild-type A3G and the W94A and W127A mutants (Figure 1B).
Tryptophans 94 and 127 are located in the NTD of the protein in a region predicted to be involved in RNA binding, protein oligomerization, Vif interaction and cellular localization (Figure 1C) (47). W127 was first identified as a residue essential for the packaging of A3G into HIV virions (9). It is also required for binding to Alu, 7SL and various hY RNAs, and these RNA-binding features of A3G correlate with its ability to inhibit Alu retrotransposition (20,24). Direct in vitro binding assays performed using purified protein also confirmed the reduced affinity of the W127A mutant for RNA (48). Other studies revealed that this residue was critical for cytoplasmic localization and N-terminal oligomerization (17,47). W94 was also reported to influence A3G packaging into HIV virions, but to a lesser extent than W127 (24). There are however discordant reports as to whether W94 can bind 7SL RNA (13,22).
W94A and W127A have reduced RNA-binding capacity, but DNA editing is mostly unaffected
Here, we independently investigated the binding of the A3G mutants to a selection of RNAs: Alu, 7SL, hY1, hY3 and β-actin (Figure 1D). We measured the relative capacity of the mutants to bind RNA compared with wild-type A3G by carrying out qPCR analysis on RNA isolated from immunoprecipitates of the A3G variants transiently expressed in 293T cells. We found that in agreement with earlier studies (20,24), the W94A and W127A mutants associated 50–90% less efficiently with Alu, 7SL, hY1 and hY3 RNAs compared with A3G (Figure 1D). A2 non-specifically bound RNA to similar levels as the bead-only control and was thus used as a negative binding control in all our subsequent assays (Supplementary Figure S3A). β-actin mRNA did not significantly bind to any of the APOBEC proteins, which is in line with previous studies (18,21), and was excluded from the graphs to improve clarity.
Before further characterizing these mutants, we wanted to ascertain whether they retained enzymatic activity on DNA by using a bacterial mutator assay commonly used to measure the catalytic activity of cytidine deaminases (49). The results of this assay revealed that the enzymatic activities of the mutants were similar to the wild-type A3G protein, whereby both these proteins were capable of mutating Escherichia coli genomic DNA and giving rise to a relatively large number of rifampicin-resistant colonies (RifR) (Figure 1E). In summary, our results show that the W94A and W127A mutants both have severely diminished RNA-binding properties compared with wild-type A3G, but this had no significant impact on the catalytic activity of the proteins.
RNA-binding mutants are packaged with different efficiencies into HIV-1 ΔVif and MoMLV virions
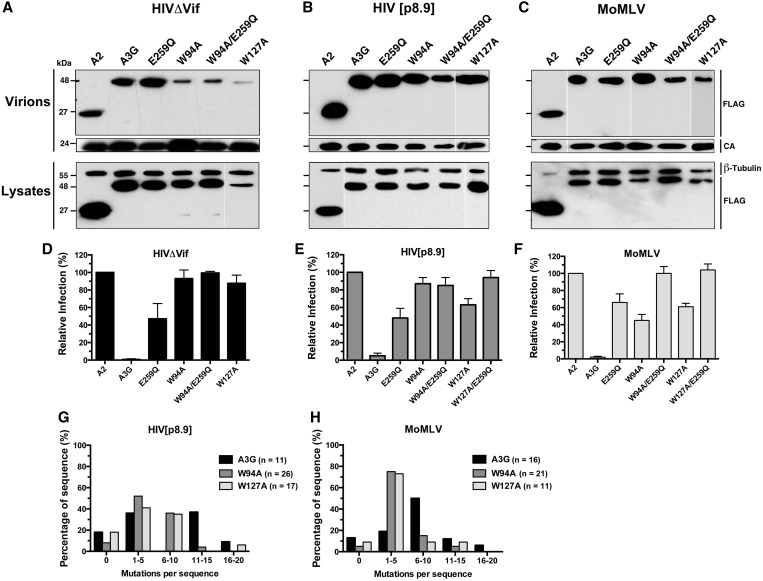
Here, we compared the virion packaging efficiency of the wild-type A3G protein to that of W94A, W127A, an inactive catalytic mutant E259Q, and corresponding W94A or W127A compound mutants: W94A/E259Q and W127A/E259Q. Three retroviruses were tested: HIVΔVif (NL4-3-derived with the env gene substituted for eGFP), HIV[p.8.9] (a self-inactivating HIV-derived pseudovirus expressing eGFP from an internal SFFV promoter) and replicative ecotropic MoMLV expressing an Env-eGFP fusion protein (see ‘Materials and Methods’ section for details). As previously described by others, we found that W94A and W127A were poorly packaged into HIVΔVif particles (Figure 2A). Surprisingly, all A3G variants were packaged efficiently into HIV[p8.9] and MoMLV virions (Figure 2B and C). These results indicate that the factors that govern virion encapsidation are different for HIV-1ΔVif and MoMLV. Our reasoning as to why the mutants proteins are packaged efficiently into HIV[p8.9] virions is presented in the discussion.
Figure 2.
Virus-dependent packaging of W94A and W127A, and retroviral restriction analysis. (A–C) Viral particles were purified from cell supernatants, lysed and assayed for the presence of FLAG-tagged APOBEC proteins by western blot analysis (top panels). Lysates of virus-producing cells are shown in the bottom panels. (D–F) Restriction analysis of virus infectivity as measured by eGFP expression in target cells infected with HIVΔVif (D), HIV[p8.9] (E) or MoMLV (F). Results represent the mean ± SD of triplicate values from at least three independent transfection experiments. (G and H) Analysis of mutations in proviral DNA. Histograms depict the proportion of total sequences containing the indicated number of mutations. The total number of clones sequenced is indicated in the chart legend. Sequences were compiled from one experiment for A3G and W127A and from two independent experiments for W94A.
RNA binding is required for retroviral restriction
Infection assays show that both W94A and W127A mutants displayed little or no antiretroviral activity on HIVΔVif as would be expected because of the packaging defect, whereas the catalytic mutant, E259Q, reduced the relative number of eGFP-positive target cells by ∼40–50% for all viruses tested (Figure 2D, E and F). Although the W94A and W127A mutants were ineffective in restricting the infection of HIV[p8.9] (Figure 2E), they reduced the infectivity of MoMLV by 55 and 40%, respectively (Figure 2F). Double mutants for both RNA-binding and catalytic activity, W94A/E259Q and W127A/E259Q, were completely ineffective in restricting the infection of all the viruses tested.
We next asked whether W94A and W127A could mutate HIV[p8.9] and MoMLV, despite having defective RNA-binding properties. As predicted by the bacterial mutator assay, both W94A and W127A mutants introduced high levels of hypermutation in both retroviruses tested, with the vast majority (82–92%) of all sequences analyzed being mutated (Table 1). Also, we found no evidence of DNA editing by mutant proteins containing the E259Q substitution. Analysis of the DNA context specificity for the deamination revealed a strong preference for the targeting of 5′-CCC-′3 trinucleotides for all A3G variants, indicating that reduced RNA binding did not impact DNA-targeting specificity (Table 2).
Table 1.
Editing of HIV[p8.9] and MoMLV by A3G and variants
| Virus | APOBEC3 protein | Base pairs sequenced* | Sequences mutated (%) | Total number of G-to-A mutations | G-to-A mutation frequency (mutations/kb) |
|---|---|---|---|---|---|
| HIV[p8.9] | |||||
| A3G | 7887 | 82 | 82 | 12.7 | |
| E259Q | 4302 | 0 | 0 | ||
| W94A | 18 642 | 92 | 132 | 7.1 | |
| W94A/E259Q | 4302 | 0 | 0 | ||
| W127A | 12 189 | 82 | 88 | 7.2 | |
| MoMLV | |||||
| A3G | 11 472 | 88 | 111 | 9.7 | |
| E259Q | 4302 | 0 | 0 | ||
| W94A | 15 057 | 95 | 85 | 5.6 | |
| W94A/E259Q | 4302 | 0 | 0 | ||
| W127A | 7887 | 91 | 38 | 4.8 |
*A 717-bp segment of integrated viral DNA was amplified by PCR and cloned. Independent clones were sequenced, and mutations computed on the plus-strand DNA were analyzed.
Table 2.
Local sequence preference for deamination on the minus-strand retroviral DNA*
| APOBEC3 | HIV[p8.9] |
MoMLV |
||
|---|---|---|---|---|
| −2 | −1 | −2 | −1 | |
| A3G | ||||
| A | 20 | 2 | 20 | 0 |
| C | 48 | 77 | 39 | 68 |
| G | 9 | 1 | 7 | 3 |
| T | 23 | 20 | 37 | 29 |
| W94A | ||||
| A | 12 | 1 | 8 | 0 |
| C | 60 | 90 | 68 | 89 |
| G | 4 | 1 | 4 | 1 |
| T | 24 | 8 | 20 | 10 |
| W127A | ||||
| A | 33 | 3 | 14 | 2 |
| C | 44 | 82 | 53 | 83 |
| G | 5 | 3 | 3 | 0 |
| T | 18 | 13 | 29 | 16 |
*Values were computed with respect to the deaminated cytidine (position zero) on the viral minus strand. Values represent the percentage of occurrence of each base at positions −1 and −2 respective to the deaminated cytidine. Sequences compiled in Table 1 were used for the analysis.
Because wild-type and mutant A3G proteins appear to be packaged with the same efficiency in MoMLV and HIV[p8.9] virions, differences in mutation rates could be explained by reduced deamination efficiency. To evaluate this, we calculated the mutation frequency in each individual sequence examined (Figure 2G and H). Our analysis shows that W94A and W127A introduced on average between 1 and 10 mutations per sequence for HIV[p8.9] and 1–5 for MoMLV. Wild-type A3G introduced slightly more mutations per sequence on both viruses, which explains the results of Table 1.
RNA binding is required for the inhibition of proviral DNA accumulation and integration
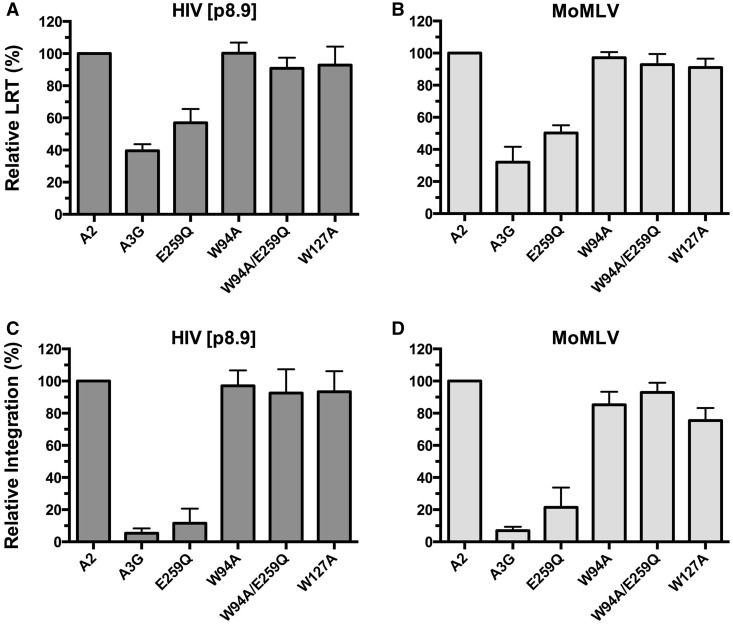
Retroviruses produced in the presence of A3G display reduced levels of LRT and proviral integration (50). Here, we sought to examine how the W94A and W127A mutations impact LRT accumulation and proviral integration. Results show that neither W94A nor W127A significantly hinder LRT accumulation, whereas wild-type A3G and E259Q reduced these levels by ∼40–60% for both viruses (Figure 3A and B). A3G and E259Q had much more dramatic effects on integration with measured reductions of 94 and 89% for HIV[p8.9] and 92 and 81% for MoMLV, respectively (Figure 3C and D). These results clearly reveal the marginal role of deamination in preventing these two early steps of the infection. On the other hand, W94A had no significant effect on reducing the proviral integration of either MoMLV or HIV[p8.9]. Equally, W127A did not reduce the integration of HIV[p8.9], but appeared to have a slight effect on MoMLV. Inactivation of the deaminase activity of the W94A RNA-binding mutant had no detectable impact on LRT accumulation or integration, which again supports that deamination is not a major contributor in preventing these specific processes.
Figure 3.
Effects of W94A and W127A on proviral DNA synthesis and integration. (A and B) Analysis of LRT accumulation. (C and D) Analysis of proviral DNA integration. DNA from infected 293T cells was collected at 12 h post-infection for LRT analysis and at 24 h for integration analysis. Infections were performed on 293T cells with HIV[p8.9] (A and C), or on NIH 3T3 cells with MoMLV (B and D). The results reflect the mean RQ ± SD of three independent experiments each performed in quadruplicate. Data were normalized to A2 values.
Hypermutation does not affect MoMLV particle release
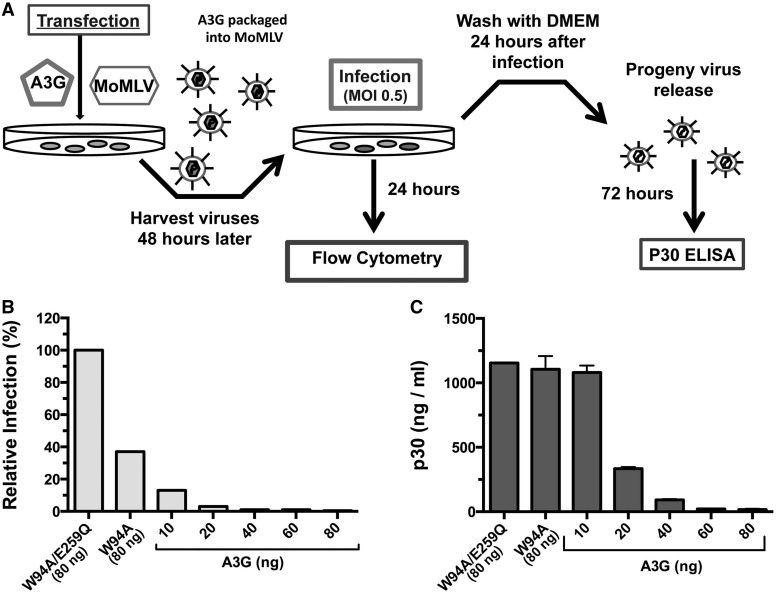
We were curious to determine whether viral particle release was affected by the DNA mutator activity of the RNA-binding mutant W94A. Because W94A is not adequately packaged into HIVΔVif particles, we performed this experiment on MoMLV. Deamination-induced damage that can affect particle release includes: mutational damage to the retroviral promoter, loss of protein function or localization, or the generation of stop codons that halt protein synthesis. A3G variants and MoMLV expression plasmids were co-transfected at increasing A3G-to-virus ratios into 293T cells, and NIH 3T3 target cells were infected with MoMLV particles at an MOI of 0.5. Virus-containing supernatants were then collected 72 h later, and p30 levels were measured by enzyme-linked immunosorbent assay (Figure 4A).
Figure 4.
Effect of deamination on MoMLV particle release. (A) Diagram of the experimental method. (B) Relative infection of NIH 3T3 cells by MoMLV measured after 24 h. Viral particles were produced in presence of a 1:10 A3G (80 ng)-to-MoMLV (800 ng) proviral plasmid ratio for W94A and W94A/E259Q, and increasing amounts of wild-type A3G as indicated. (C) p30 levels in cell supernatants were measured by enzyme-linked immunosorbent assay 72 h after infection. Data represent the mean ± SD of three independent protein measurements.
Despite W94A reducing the apparent infection by ∼60% (Figure 4B), the amount of p30 particles released was nearly identical compared with the enzymatically inactive W94A/E259Q control (Figure 4C). On the other hand, A3G had a dramatic effect on MoMLV infection at all co-transfection ratios tested. However, reduction of particle release was only observed at a 1:40 ratio (20 ng) and above (Figure 4C). Overall, these experiments indicate that mutations inflicted by W94A had no detectable impact on MoMLV particle release.
Vpr14–88 polypeptide fusions rescue RNA-binding and deamination-independent restriction
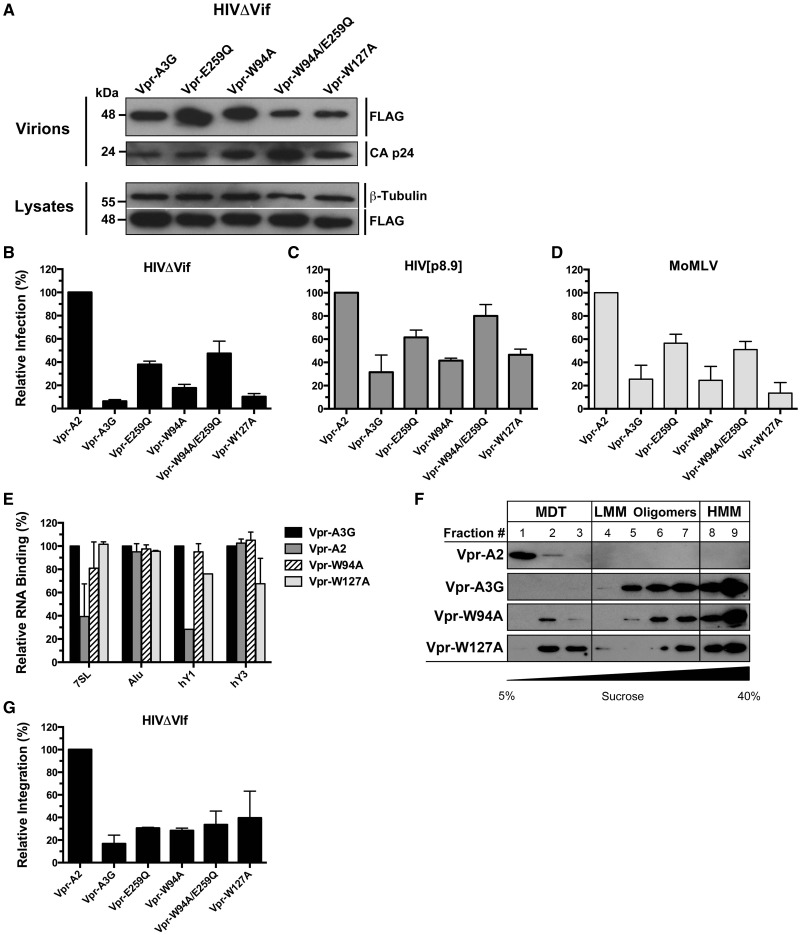
Here, we returned our attention to HIVΔVif restriction by the A3G RNA-binding mutants. It has been shown that fusing a Vpr polypeptide to proteins of interest can enable their packaging into HIV virions (13,51). Improving virion packaging of the A3G mutant proteins would allow us to determine whether RNA binding is also required for HIVΔVif restriction. To investigate this issue, we generated fusion proteins with the Vpr14–88 polypeptide (referred to as Vpr for simplicity) with all our A3G variants and performed virion packaging and restriction assays. As expected, we observed vastly improved packaging of both Vpr-W94A and Vpr-W127A into HIVΔVif, and a recovery of the antiretroviral activities of both mutants (Figure 5A and B). Surprisingly, both Vpr-W94A and Vpr-W127A now restricted HIV[p8.9] and MoMLV to levels comparable with Vpr-A3G (Figure 5C and D). This was unexpected because we had not detected a packaging defect with the mutants on HIV[p8.9] and MoMLV (Figure 2B and C).
Figure 5.
Vpr14–88 polypeptide (Vpr) fusions restore virion packaging, RNA-binding, HMM complex assembly and antiretroviral properties of W94A and W127A. (A) Packaging of all Vpr-A3G fusion proteins into HIVΔVif virions. (B–D) Antiretroviral activities of Vpr-A3G fusion proteins on HIVΔVif (B), HIV[p8.9] (C) and MoMLV (D). (E) Evaluation of the RNA-binding properties of Vpr fusion proteins. Data represent the mean ± SD of triplicate values from three independent experiments. (F) Lysates of 293T cells transfected with Vpr expression vectors analyzed by velocity sedimentation over non-denaturing sucrose gradients. (G) Effect of Vpr fusion proteins on HIVΔVif proviral integration. The results reflect the mean RQ ± SD of three independent experiments each performed in quadruplicate. Data were normalized to Vpr-A2 values.
Vpr is an HIV-1 accessory protein known to directly bind RNA (52). Proteins fused to Vpr would hence be expected to display overall increased RNA-binding properties. To determine whether RNA binding is restored with the mutants, we measured the binding of Vpr fusion proteins to Alu, 7SL, hY1, hY3 and β-actin RNAs using a similar approach as in Figure 1D. We found that binding to RNA was vastly improved in all cases except for β-actin that remained at background levels and again was not plotted on the graph. Strikingly, Vpr-A2 also displayed RNA-binding properties similar to Vpr-A3G for Alu and hY3 and greatly improved binding to 7SL and hY1 (Figure 5E). Comparative binding of the RNAs with Vpr-A3G, Vpr-A2 and agarose beads is shown in Supplementary Figure S3B. To verify whether increased RNA binding also impacted the intracellular oligomeric forms of the mutant APOBEC3 proteins, velocity sedimentation assays were carried out on the Vpr fusion proteins. Both Vpr-W94A and Vpr-W127A had their ability to assemble into HMM complexes restored (Figure 5F). Finally, we tested whether the Vpr fusion proteins restricted proviral integration of HIVΔVif (Figure 5G). Integration was compromised to a similar extent by all Vpr-A3G variants, but not by Vpr-A2 that was now also capable of binding RNA.
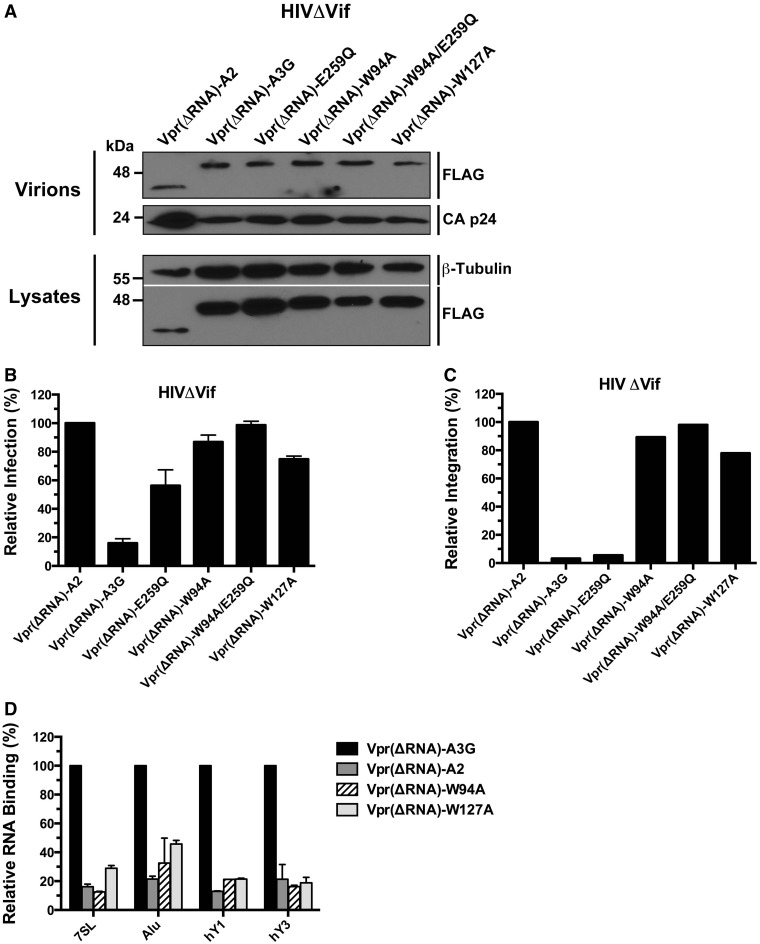
To ensure that the observed phenotype of the Vpr fusion proteins was conferred specifically by the RNA-binding properties of Vpr, we deleted amino acids 87 and 88 of the Vpr14–88 polypeptide that have been previously been shown to mediate RNA binding (52) and repeated experiments depicted in Figure 5. We found that W94A and W127A fusions with the Vpr14–86 polypeptide defective in RNA binding, Vpr(ΔRNA), were efficiently packaged into HIVΔVif virions (Figure 6A), did not restrict the infection (Figure 6B), were unable to inhibit proviral integration (Figure 6C) and displayed RNA-binding defects (Figure 6D). In summary, these data show that RNA binding is an essential property for A3G to be able to restrict Vif-deficient HIV-1 infection.
Figure 6.
Fusion to the RNA-binding defective Vpr14–86 polyprotein [Vpr(ΔRNA)] does not restore the restriction potential of the W94A and W127A mutants. (A) Analysis of the packaging of all Vpr(ΔRNA)-A3G fusion proteins into HIVΔVif virions. (B) Antiretroviral activities of Vpr(ΔRNA)-A3G fusion proteins on HIVΔVif. (C) Evaluation of the RNA-binding properties of VprΔRNA) fusion proteins. Data represent the mean ± SD of triplicate values from three independent experiments. (D) Effect of Vpr(ΔRNA) fusion proteins on HIVΔVif proviral integration. The results reflect the mean RQ of one experiment performed in quadruplicate. Data were normalized to Vpr-A2 values.
Residues W94 and W127 cooperate to bind RNA
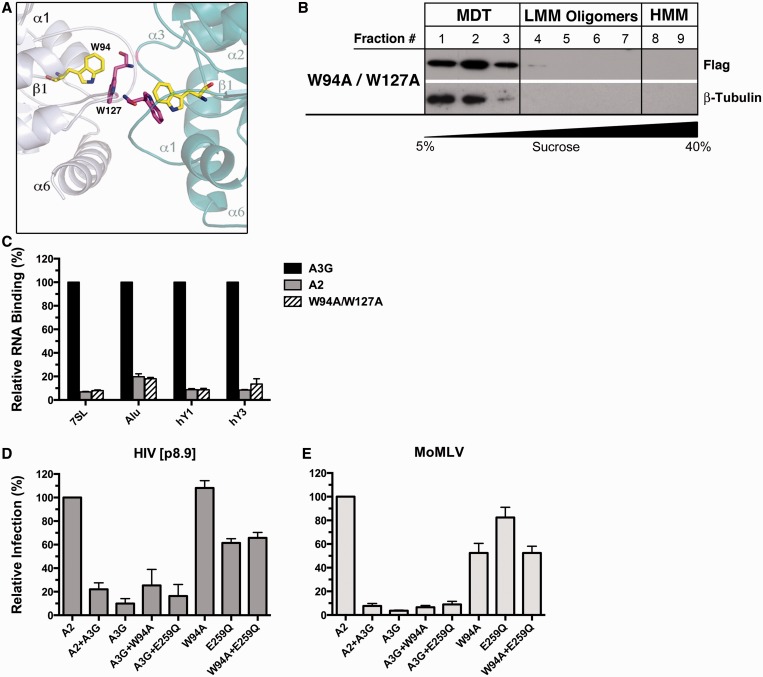
To gain further insight into how W94 and W127 enable A3G to interact with RNA, we conducted homology modeling of the A3G head-to-head NTD dimer (Supplementary Figure S4). In our model, the two A3G NTD monomers make extensive contacts, including the loops connecting the α1-β1 and β4-α4 with the corresponding β4-α4 and α1-β1 loops of the reciprocal protomer. Interestingly, close inspection of the NTD dimer shows that W94 of the first monomer is in close proximity to W127 in the other monomer (Figure 7A and Supplementary Figure S4A), a result also observed by Lavens et al. (53). The structure shows that on dimerization, there is a significant increase in the size of the positively charged patch that extends to the C-terminal end of α6 of the reciprocal dimer’s subunit (Supplementary Figure S4B). Overall, our modeling study suggests that A3G dimerization generates a large surface for RNA binding, and that W94A and W127A substitutions would strongly disfavor the binding of RNA. An A3G mutant carrying a double W94A/W127A substitution should therefore potentiate the RNA-binding defect. To validate this prediction, we generated the double mutant and analyzed its RNA-binding properties. We found that the RNA-dependent oligomerization of W94A/W127A was completely abolished (Figure 7B). Additionally, the double mutant did not significantly bind to any of the RNAs tested (Figure 7C).
Figure 7.
Retroviral restriction by A3G requires RNA but not a protein co-factor. (A) Cropped field from the homology model of the head-to-head NTD dimer of A3G (see Supplementary Figure S4 for the complete model and details). The W94 and W127 interaction domain at the interface of the monomers is shown, W94 (yellow); W127 (pink). (B) Lysates of 293T cells transfected with the W94A/W127A mutant resolved by non-denaturing velocity sedimentation. (C) Evaluation of the RNA-binding properties of the W94A/W127A mutant. (D and E) Viruses for the complementation assays were produced by co-transfecting a total of 80 ng of APOBEC expression plasmids (40 ng each) for MoMLV restrictions assays, or 150 ng for HIV[p8.9] (75 ng each). Data represent the mean ± SD of triplicate values from three independent experiments.
Co-expression of W94A with E259Q does not restore the restriction defect
The inability of the W94A and W127A mutants to prevent viral cDNA accumulation and integration could potentially be explained by the absence of a cofactor that normally binds to these tryptophan residues on wild-type A3G. Here, we sought to establish whether we could restore restriction to its full potency by producing viruses in the presence of equal quantities of W94A and E259Q. Only the W94A mutant was used in these assays because it has the ability to self-associate as opposed to W127A that does not (Supplementary Figure S5A and B). E259Q is efficiently packaged into HIV[p8.9] and MoMLV virions and can assemble into HMM complexes (Supplementary Figure S2). Our complementation assays on HIV[p8.9] and MoMLV indicate that E259Q and W94A do not complement each other’s function, which would have resulted in an increase of the overall restriction (Figure 7D and E). These results weigh against the possibility that a virion-packaged trans-acting cofactor is required for enabling A3G to restrict retroviral infection.
DISCUSSION
We initially set out to identify the residues in A3G that are responsible for HMM complex assembly to gain further insight into the protein’s regulation. Careful optimization of velocity sedimentation assays facilitated consistent and well-defined separation between HMM complexes, oligomeric LMM complexes and the RNA-independent forms of the protein (Figure 1A). We used this method to screen A3G point mutants and identified W94 and W127 as critical residues for HMM assembly. These two amino acids have been the object of previous studies that have focused on RNA binding, protein oligomerization and packaging of A3G into HIV-1 virions. In agreement with previous work, we found that both mutants associated much less efficiently with various RNAs (Figure 1D) (17,22,24,54,55).
Although W94A or W127A substitutions are known to have detrimental effects on HIV-1 virion packaging, we were surprised to see that the packaging of these mutants into MoMLV and HIV[p8.9] was largely unaffected (Figure 2B and C). We do not fully understand why HIV[p8.9] did not also suffer from the same packaging defects as HIVΔVif because both viruses express identical Gag NC sequences. HIV[p8.9] does however contain numerous non-HIV elements in its genome that we believe were responsible for rescuing the packaging defect. Further studies are required to identify the determinants that govern the packaging of A3G into MoMLV and HIV[p8.9].
Perhaps the most controversial part of this study was the finding that A3G’s deaminase activity had little or no detectable impact on LRT accumulation and proviral integration for all three viruses tested. Even MoMLV progeny virus release was unaffected by the inactivation of the catalytic activity of the W94A mutant. These results do not however mean that A3G-mutated viruses are infectious; they are most likely highly compromised in their replicative fitness. But yet again, it is well documented that the infected cells of HIV-1 patients contain multiple copies of integrated virus that can potentially recombine and complement each other’s function (56–61). In a similar manner, sublethally mutated proteins from one provirus could complement the function of lethally mutated proteins from another (61–63). This therefore raises questions about long-term protection that is actually conferred by hypermutation during the course of a natural retroviral infection.
Although the deaminase activity of the W94A and W127A mutants did not impair the early stages of HIVΔVif or HIV[p8.9] infection, it reduced the infection levels of MoMLV by ∼50–60% (Figures 2E and F and 6B). Infection levels measured in our single-round assays reflect the number of target cells expressing a reporter protein (eGFP) driven by the promoter of the integrated provirus. Reporter gene expression is only possible if the provirus has successfully integrated into the target genome. Apparent antiretroviral activity in these systems is therefore a reflection that processes such as eGFP mRNA expression, splicing, translation and protein fluorescence have been affected by the mutations. In the case of HIVΔVif and HIV[p8.9], eGFP is expressed from a monocistronic mRNA driven by either the LTR promoter (HIVΔVif) or an internal SFFV promoter (HIV[p8.9]). In MoMLV, however, eGFP is expressed as a fusion protein with Env. Upstream of the eGFP coding sequence are 296 amino acids of the N-terminus of Env. This sequence contains 72 putative deamination target sites that can potentially yield 13 termination codons (Supplementary Table S2). The eGFP coding sequence within all three viruses is identical and contains only a single site that can generate a termination codon. We therefore believe that the reduced apparent infection of MoMLV by W94A and W127A could be caused in part by the generation of premature termination codons in the N-terminal Env segment thereby preventing the expression of the eGFP reporter protein. Another possibility that may contribute to explain our observations is that a portion of deaminated proviral cDNA is degraded before integration via a uracil DNA glycosylase base excision pathway (64).
It has been debated whether the E259Q substitution, which eliminates the proton donor in the catalytic site required for the deamination process, could affect intrinsic properties of the A3G protein other than catalytic activity alone, such as DNA binding for instance. To address this controversial issue, we compared the effects of the E259Q mutant with that of the C-terminal domain DNA-binding mutant R313A (65). We found no differences between the two mutants in their capacities to assemble into HMM complexes, restrict HIVΔVif infection or inhibit proviral integration (Supplementary Figure S6). Equally, we did not find any hypermutated proviral sequences when HIVΔVif was produced with either mutant (data not shown).
Another important question that emerged from this study was whether the W94 and W127 residues of A3G recruit a virion-packaged cofactor required for deamination-independent viral restriction. To answer this, we co-expressed the W94A and E259Q mutants and performed retroviral restriction assays. Our hypothesis was that if a co-factor was involved, association of W94A and E259Q mutants would improve overall restriction levels. Our results showed however that restriction was not restored, therefore weighing against the existence of such a co-factor (Figure 7D and E). Nonetheless, although RNA binding is essential for deamination-independent restriction, it is not alone sufficient to provide maximum restriction potential. Specific RNA species that bind to A3G may be required as supported by the absence of detectable restriction of infection with the RNA-binding Vpr-A2 fusion protein (Figure 5). Clues to the identity of these RNAs could be obtained from differential analyses of the RNA content of HMM and LMM complexes. Additionally, the RNA-binding affinity of A3G and the manner by which both its protein domains interact with RNA may also be of capital importance to prevent retroviral cDNA synthesis and integration.
In summary, the current work illustrates the essential and direct role of RNA in the deamination-independent restriction of retroviruses by A3G. Proviral DNA synthesis and integration are potently inhibited by processes that do not require the cytidine deaminase activity of the protein. Deamination-independent restriction mechanisms therefore appear to be important contributors in preventing irreversible and potentially harmful proviral integration into the host’s genomic DNA. Although abundant A3G-induced G-to-A mutations had only a minor impact on restricting the early stages of the infection, they most likely play a major role in limiting the infectivity, fitness and spread of progeny retroviruses in physiological conditions.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1 and 2, Supplementary Figures 1–6, Supplementary Materials and Methods and Supplementary References [66,67].
FUNDING
Canadian Institutes of Health Research [#89774]; Early Researcher Award from the Ontario Ministry of Research and Innovation (to M.-A.L.). Funding for open access charge: Canadian Institutes of Health Research [#89774 to M.-A.L.].
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Tara Read, Lionel Filion, Raed Hanania and Kristin Kemmerich for technical support. They are especially grateful to Reuben Harris, Linda Chelico and Ilona Skerjanc for helpful discussions and comments on the manuscript, and the NIH AIDS Research and Reference Reagent Program for reagents. K.B. holds an Ontario Graduate Scholarship. J.-F.C. holds a Canada Research Chair in Structural Biology and Epigenetics. M.-A.L. holds a Canada Research Chair in Molecular Virology and Intrinsic Immunity.
REFERENCES
- 1.Albin JS, Harris RS. Interactions of host APOBEC3 restriction factors with HIV-1 in vivo: implications for therapeutics. Expert Rev. Mol. Med. 2010;12:e4. doi: 10.1017/S1462399409001343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jern P, Russell RA, Pathak VK, Coffin JM. Likely role of APOBEC3G-mediated G-to-A mutations in HIV-1 evolution and drug resistance. PLoS Pathog. 2009;5:e1000367. doi: 10.1371/journal.ppat.1000367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Suspene R, Rusniok C, Vartanian JP, Wain-Hobson S. Twin gradients in APOBEC3 edited HIV-1 DNA reflect the dynamics of lentiviral replication. Nucleic Acids Res. 2006;34:4677–4684. doi: 10.1093/nar/gkl555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yu Q, Konig R, Pillai S, Chiles K, Kearney M, Palmer S, Richman D, Coffin JM, Landau NR. Single-strand specificity of APOBEC3G accounts for minus-strand deamination of the HIV genome. Nat. Struct. Mol. Biol. 2004;11:435–442. doi: 10.1038/nsmb758. [DOI] [PubMed] [Google Scholar]
- 5.Armitage AE, Deforche K, Chang CH, Wee E, Kramer B, Welch JJ, Gerstoft J, Fugger L, McMichael A, Rambaut A, et al. APOBEC3G-induced hypermutation of human immunodeficiency virus type-1 is typically a discrete “all or nothing” phenomenon. PLoS Genet. 2012;8:e1002550. doi: 10.1371/journal.pgen.1002550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bishop KN, Verma M, Kim EY, Wolinsky SM, Malim MH. APOBEC3G inhibits elongation of HIV-1 reverse transcripts. PLoS Pathog. 2008;4:e1000231. doi: 10.1371/journal.ppat.1000231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Newman EN, Holmes RK, Craig HM, Klein KC, Lingappa JR, Malim MH, Sheehy AM. Antiviral function of APOBEC3G can be dissociated from cytidine deaminase activity. Curr. Biol. 2005;15:166–170. doi: 10.1016/j.cub.2004.12.068. [DOI] [PubMed] [Google Scholar]
- 8.Burnett A, Spearman P. APOBEC3G multimers are recruited to the plasma membrane for packaging into human immunodeficiency virus type 1 virus-like particles in an RNA-dependent process requiring the NC basic linker. J. Virol. 2007;81:5000–5013. doi: 10.1128/JVI.02237-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huthoff H, Malim MH. Identification of amino acid residues in APOBEC3G required for regulation by human immunodeficiency virus type 1 Vif and Virion encapsidation. J. Virol. 2007;81:3807–3815. doi: 10.1128/JVI.02795-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wissing S, Galloway NL, Greene WC. HIV-1 Vif versus the APOBEC3 cytidine deaminases: an intracellular duel between pathogen and host restriction factors. Mol. Aspects Med. 2010;31:383–397. doi: 10.1016/j.mam.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chelico L, Pham P, Calabrese P, Goodman MF. APOBEC3G DNA deaminase acts processively 3′ –> 5′ on single-stranded DNA. Nat. Struct. Mol. Biol. 2006;13:392–399. doi: 10.1038/nsmb1086. [DOI] [PubMed] [Google Scholar]
- 12.Iwatani Y, Takeuchi H, Strebel K, Levin JG. Biochemical activities of highly purified, catalytically active human APOBEC3G: correlation with antiviral effect. J. Virol. 2006;80:5992–6002. doi: 10.1128/JVI.02680-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bulliard Y, Turelli P, Rohrig UF, Zoete V, Mangeat B, Michielin O, Trono D. Functional analysis and structural modeling of human APOBEC3G reveal the role of evolutionarily conserved elements in the inhibition of human immunodeficiency virus type 1 infection and Alu transposition. J. Virol. 2009;83:12611–12621. doi: 10.1128/JVI.01491-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wedekind JE, Gillilan R, Janda A, Krucinska J, Salter JD, Bennett RP, Raina J, Smith HC. Nanostructures of APOBEC3G support a hierarchical assembly model of high molecular mass ribonucleoprotein particles from dimeric subunits. J. Biol. Chem. 2006;281:38122–38126. doi: 10.1074/jbc.C600253200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bennett RP, Salter JD, Liu X, Wedekind JE, Smith HC. APOBEC3G subunits self-associate via the C-terminal deaminase domain. J. Biol. Chem. 2008;283:33329–33336. doi: 10.1074/jbc.M803726200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Salter JD, Krucinska J, Raina J, Smith HC, Wedekind JE. A hydrodynamic analysis of APOBEC3G reveals a monomer-dimer-tetramer self-association that has implications for anti-HIV function. Biochemistry. 2009;48:10685–10687. doi: 10.1021/bi901642c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huthoff H, Autore F, Gallois-Montbrun S, Fraternali F, Malim MH. RNA-dependent oligomerization of APOBEC3G is required for restriction of HIV-1. PLoS Pathog. 2009;5:e1000330. doi: 10.1371/journal.ppat.1000330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chiu YL, Witkowska HE, Hall SC, Santiago M, Soros VB, Esnault C, Heidmann T, Greene WC. High-molecular-mass APOBEC3G complexes restrict Alu retrotransposition. Proc. Natl Acad. Sci. USA. 2006;103:15588–15593. doi: 10.1073/pnas.0604524103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Soros VB, Yonemoto W, Greene WC. Newly synthesized APOBEC3G is incorporated into HIV virions, inhibited by HIV RNA, and subsequently activated by RNase H. PLoS Pathog. 2007;3:e15. doi: 10.1371/journal.ppat.0030015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Khan MA, Goila-Gaur R, Opi S, Miyagi E, Takeuchi H, Kao S, Strebel K. Analysis of the contribution of cellular and viral RNA to the packaging of APOBEC3G into HIV-1 virions. Retrovirology. 2007;4:48. doi: 10.1186/1742-4690-4-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kozak SL, Marin M, Rose KM, Bystrom C, Kabat D. The anti-HIV-1 editing enzyme APOBEC3G binds HIV-1 RNA and messenger RNAs that shuttle between polysomes and stress granules. J. Biol. Chem. 2006;281:29105–29119. doi: 10.1074/jbc.M601901200. [DOI] [PubMed] [Google Scholar]
- 22.Zhang W, Du J, Yu K, Wang T, Yong X, Yu XF. Association of potent human antiviral cytidine deaminases with 7SL RNA and viral RNP in HIV-1 virions. J. Virol. 2010;84:12903–12913. doi: 10.1128/JVI.01632-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang T, Tian C, Zhang W, Luo K, Sarkis PT, Yu L, Liu B, Yu Y, Yu XF. 7SL RNA mediates virion packaging of the antiviral cytidine deaminase APOBEC3G. J. Virol. 2007;81:13112–13124. doi: 10.1128/JVI.00892-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bach D, Peddi S, Mangeat B, Lakkaraju A, Strub K, Trono D. Characterization of APOBEC3G binding to 7SL RNA. Retrovirology. 2008;5:54. doi: 10.1186/1742-4690-5-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mbisa JL, Barr R, Thomas JA, Vandegraaff N, Dorweiler IJ, Svarovskaia ES, Brown WL, Mansky LM, Gorelick RJ, Harris RS, et al. Human immunodeficiency virus type 1 cDNAs produced in the presence of APOBEC3G exhibit defects in plus-strand DNA transfer and integration. J. Virol. 2007;81:7099–7110. doi: 10.1128/JVI.00272-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mbisa JL, Bu W, Pathak VK. APOBEC3F and APOBEC3G inhibit HIV-1 DNA integration by different mechanisms. J. Virol. 2010;84:5250–5259. doi: 10.1128/JVI.02358-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Browne EP, Allers C, Landau NR. Restriction of HIV-1 by APOBEC3G is cytidine deaminase-dependent. Virology. 2009;387:313–321. doi: 10.1016/j.virol.2009.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Luo K, Wang T, Liu B, Tian C, Xiao Z, Kappes J, Yu XF. Cytidine deaminases APOBEC3G and APOBEC3F interact with human immunodeficiency virus type 1 integrase and inhibit proviral DNA formation. J. Virol. 2007;81:7238–7248. doi: 10.1128/JVI.02584-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bishop KN, Holmes RK, Malim MH. Antiviral potency of APOBEC proteins does not correlate with cytidine deamination. J. Virol. 2006;80:8450–8458. doi: 10.1128/JVI.00839-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Iwatani Y, Chan DS, Wang F, Maynard KS, Sugiura W, Gronenborn AM, Rouzina I, Williams MC, Musier-Forsyth K, Levin JG. Deaminase-independent inhibition of HIV-1 reverse transcription by APOBEC3G. Nucleic Acids Res. 2007;35:7096–7108. doi: 10.1093/nar/gkm750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li XY, Guo F, Zhang L, Kleiman L, Cen S. APOBEC3G inhibits DNA strand transfer during HIV-1 reverse transcription. J. Biol. Chem. 2007;282:32065–32074. doi: 10.1074/jbc.M703423200. [DOI] [PubMed] [Google Scholar]
- 32.Wang X, Ao Z, Chen L, Kobinger G, Peng J, Yao X. The cellular antiviral protein APOBEC3G interacts with HIV-1 reverse transcriptase and inhibits its function during viral replication. J. Virol. 2012;86:3777–3786. doi: 10.1128/JVI.06594-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sliva K, Erlwein O, Bittner A, Schnierle BS. Murine leukemia virus (MLV) replication monitored with fluorescent proteins. Virol. J. 2004;1:14. doi: 10.1186/1743-422X-1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Langlois MA, Beale RC, Conticello SG, Neuberger MS. Mutational comparison of the single-domained APOBEC3C and double-domained APOBEC3F/G anti-retroviral cytidine deaminases provides insight into their DNA target site specificities. Nucleic Acids Res. 2005;33:1913–1923. doi: 10.1093/nar/gki343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Naldini L, Blomer U, Gallay P, Ory D, Mulligan R, Gage FH, Verma IM, Trono D. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science. 1996;272:263–267. doi: 10.1126/science.272.5259.263. [DOI] [PubMed] [Google Scholar]
- 36.Zhang H, Zhou Y, Alcock C, Kiefer T, Monie D, Siliciano J, Li Q, Pham P, Cofrancesco J, Persaud D, et al. Novel single-cell-level phenotypic assay for residual drug susceptibility and reduced replication capacity of drug-resistant human immunodeficiency virus type 1. J. Virol. 2004;78:1718–1729. doi: 10.1128/JVI.78.4.1718-1729.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Simon JH, Malim MH. The human immunodeficiency virus type 1 Vif protein modulates the postpenetration stability of viral nucleoprotein complexes. J. Virol. 1996;70:5297–5305. doi: 10.1128/jvi.70.8.5297-5305.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Prochnow C, Bransteitter R, Klein MG, Goodman MF, Chen XS. The APOBEC-2 crystal structure and functional implications for the deaminase AID. Nature. 2007;445:447–451. doi: 10.1038/nature05492. [DOI] [PubMed] [Google Scholar]
- 39.Krzysiak TC, Jung J, Thompson J, Baker D, Gronenborn AM. APOBEC2 is a monomer in solution: implications for APOBEC3G models. Biochemistry. 2012;51:2008–2017. doi: 10.1021/bi300021s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sato Y, Probst HC, Tatsumi R, Ikeuchi Y, Neuberger MS, Rada C. Deficiency in APOBEC2 leads to a shift in muscle fiber type, diminished body mass, and myopathy. J. Biol. Chem. 2010;285:7111–7118. doi: 10.1074/jbc.M109.052977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Conticello SG, Langlois MA, Yang Z, Neuberger MS. DNA deamination in immunity: AID in the context of its APOBEC relatives. Adv. Immunol. 2007;94:37–73. doi: 10.1016/S0065-2776(06)94002-4. [DOI] [PubMed] [Google Scholar]
- 42.Langlois MA, Kemmerich K, Rada C, Neuberger MS. The AKV murine leukemia virus is restricted and hypermutated by mouse APOBEC3. J. Virol. 2009;83:11550–11559. doi: 10.1128/JVI.01430-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.O’Doherty U, Swiggard WJ, Malim MH. Human immunodeficiency virus type 1 spinoculation enhances infection through virus binding. J. Virol. 2000;74:10074–10080. doi: 10.1128/jvi.74.21.10074-10080.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Butler SL, Hansen MS, Bushman FD. A quantitative assay for HIV DNA integration in vivo. Nat. Med. 2001;7:631–634. doi: 10.1038/87979. [DOI] [PubMed] [Google Scholar]
- 45.Brussel A, Delelis O, Sonigo P. Alu-LTR real-time nested PCR assay for quantifying integrated HIV-1 DNA. Methods Mol. Biol. 2005;304:139–154. doi: 10.1385/1-59259-907-9:139. [DOI] [PubMed] [Google Scholar]
- 46.Wichroski MJ, Robb GB, Rana TM. Human retroviral host restriction factors APOBEC3G and APOBEC3F localize to mRNA processing bodies. PLoS Pathog. 2006;2:e41. doi: 10.1371/journal.ppat.0020041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stenglein MD, Matsuo H, Harris RS. Two regions within the amino-terminal half of APOBEC3G cooperate to determine cytoplasmic localization. J. Virol. 2008;82:9591–9599. doi: 10.1128/JVI.02471-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Feng Y, Chelico L. Intensity of deoxycytidine deamination of HIV-1 proviral DNA by the retroviral restriction factor APOBEC3G is mediated by the noncatalytic domain. J. Biol. Chem. 2011;286:11415–11426. doi: 10.1074/jbc.M110.199604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Harris RS, Petersen-Mahrt SK, Neuberger MS. RNA editing enzyme APOBEC1 and some of its homologs can act as DNA mutators. Mol. Cell. 2002;10:1247–1253. doi: 10.1016/s1097-2765(02)00742-6. [DOI] [PubMed] [Google Scholar]
- 50.Mariani R, Chen D, Schrofelbauer B, Navarro F, Konig R, Bollman B, Munk C, Nymark-McMahon H, Landau NR. Species-specific exclusion of APOBEC3G from HIV-1 virions by Vif. Cell. 2003;114:21–31. doi: 10.1016/s0092-8674(03)00515-4. [DOI] [PubMed] [Google Scholar]
- 51.Ao Z, Yu Z, Wang L, Zheng Y, Yao X. Vpr14-88-Apobec3G fusion protein is efficiently incorporated into Vif-positive HIV-1 particles and inhibits viral infection. PloS One. 2008;3:e1995. doi: 10.1371/journal.pone.0001995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang S, Pointer D, Singer G, Feng Y, Park K, Zhao LJ. Direct binding to nucleic acids by Vpr of human immunodeficiency virus type 1. Gene. 1998;212:157–166. doi: 10.1016/s0378-1119(98)00178-4. [DOI] [PubMed] [Google Scholar]
- 53.Lavens D, Peelman F, Van der Heyden J, Uyttendaele I, Catteeuw D, Verhee A, Van Schoubroeck B, Kurth J, Hallenberger S, Clayton R, et al. Definition of the interacting interfaces of Apobec3G and HIV-1 Vif using MAPPIT mutagenesis analysis. Nucleic Acids Res. 2010;38:1902–1912. doi: 10.1093/nar/gkp1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bulliard Y, Narvaiza I, Bertero A, Peddi S, Rohrig UF, Ortiz M, Zoete V, Castro-Diaz N, Turelli P, Telenti A, et al. Structure-function analyses point to a polynucleotide-accommodating groove essential for APOBEC3A restriction activities. J. Virol. 2011;85:1765–1776. doi: 10.1128/JVI.01651-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chelico L, Prochnow C, Erie DA, Chen XS, Goodman MF. Structural model for deoxycytidine deamination mechanisms of the HIV-1 inactivation enzyme APOBEC3G. J. Biol. Chem. 2010;285:16195–16205. doi: 10.1074/jbc.M110.107987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Levy DN, Aldrovandi GM, Kutsch O, Shaw GM. Dynamics of HIV-1 recombination in its natural target cells. Proc. Natl Acad. Sci. USA. 2004;101:4204–4209. doi: 10.1073/pnas.0306764101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gelderblom HC, Vatakis DN, Burke SA, Lawrie SD, Bristol GC, Levy DN. Viral complementation allows HIV-1 replication without integration. Retrovirology. 2008;5:60. doi: 10.1186/1742-4690-5-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Iwabu Y, Mizuta H, Kawase M, Kameoka M, Goto T, Ikuta K. Superinfection of defective human immunodeficiency virus type 1 with different subtypes of wild-type virus efficiently produces infectious variants with the initial viral phenotypes by complementation followed by recombination. Microbes Infect. 2008;10:504–513. doi: 10.1016/j.micinf.2008.01.012. [DOI] [PubMed] [Google Scholar]
- 59.Charpentier C, Nora T, Tenaillon O, Clavel F, Hance AJ. Extensive recombination among human immunodeficiency virus type 1 quasispecies makes an important contribution to viral diversity in individual patients. J. Virol. 2006;80:2472–2482. doi: 10.1128/JVI.80.5.2472-2482.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wodarz D, Levy DN. Multiple HIV-1 infection of cells and the evolutionary dynamics of cytotoxic T lymphocyte escape mutants. Evolution. 2009;63:2326–2339. doi: 10.1111/j.1558-5646.2009.00727.x. [DOI] [PubMed] [Google Scholar]
- 61.Russell RA, Moore MD, Hu WS, Pathak VK. APOBEC3G induces a hypermutation gradient: purifying selection at multiple steps during HIV-1 replication results in levels of G-to-A mutations that are high in DNA, intermediate in cellular viral RNA, and low in virion RNA. Retrovirology. 2009;6:16. doi: 10.1186/1742-4690-6-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Refsland EW, Hultquist JF, Harris RS. Endogenous origins of HIV-1 G-to-A hypermutation and restriction in the nonpermissive T cell line CEM2n. PLoS Pathog. 2012;8:e1002800. doi: 10.1371/journal.ppat.1002800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sadler HA, Stenglein MD, Harris RS, Mansky LM. APOBEC3G contributes to HIV-1 variation through sublethal mutagenesis. J. Virol. 2010;84:7396–7404. doi: 10.1128/JVI.00056-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Weil AF, Ghosh D, Zhou Y, Seiple L, McMahon MA, Spivak AM, Siliciano RF, Stivers JT. Uracil DNA glycosylase initiates degradation of HIV-1 cDNA containing misincorporated dUTP and prevents viral integration. Proc. Natl Acad. Sci. USA. 2013;110:E448–E457. doi: 10.1073/pnas.1219702110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shindo K, Li M, Gross PJ, Brown WL, Harjes E, Lu Y, Matsuo H, Harris RS. A comparison of two single-stranded dna binding models by mutational analysis of APOBEC3G. Biology. 2012;1:260–276. doi: 10.3390/biology1020260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chen KM, Harjes E, Gross PJ, Fahmy A, Lu Y, Shindo K, Harris RS, Matsuo H. Structure of the DNA deaminase domain of the HIV-1 restriction factor APOBEC3G. Nature. 2008;452:116–119. doi: 10.1038/nature06638. [DOI] [PubMed] [Google Scholar]
- 67.Dapp MJ, Holtz CM, Mansky LM. Concomitant lethal mutagenesis of human immunodeficiency virus type 1. J. Mol. Biol. 2012;419:158–170. doi: 10.1016/j.jmb.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.