Abstract
Template driven chemical ligation of fluorogenic probes represents a powerful method for DNA and RNA detection and imaging. Unfortunately, previous techniques have been hampered by requiring chemistry with sluggish kinetics and background side reactions. We have developed fluorescent DNA probes containing quenched fluorophore-tetrazine and methyl-cyclopropene groups that rapidly react by bioorthogonal cycloaddition in the presence of complementary DNA or RNA templates. Ligation increases fluorescence with negligible background signal in the absence of hybridization template. Reaction kinetics depend heavily on template length and linker structure. Using this technique, we demonstrate rapid discrimination between single template mismatches both in buffer and cell media. Fluorogenic bioorthogonal ligations offer a promising route towards the fast and robust fluorescent detection of specific DNA or RNA sequences.
INTRODUCTION
There is tremendous interest in the use of fluorogenic reactions for detecting and imaging nucleic acids, especially specific DNA and RNA sequences (1–6). Applications include time-resolved imaging of transcription, detection of disease-related single nucleotide polymorphisms, and tracking RNA fragments such as microRNAs (7–10). Despite advances in the use of molecular beacons, aptamers and antisense agents, the rapid detection and imaging of oligonucleotides in live cells and physiologically relevant media remains challenging. Current methods, although powerful, suffer from numerous drawbacks. Molecular beacons require lengthy constructs, are prone to background turn-on reactions owing to non-specific binding, and have difficulty discriminating a single mismatch in the physiological conditions and temperatures required for live-cell reactions (1,3,5,11,12). Additionally, by their design, molecular beacons require a trade-off between selectivity and speed and are likely limited in monitoring temporal dynamics (7,9). Other approaches that use RNA-binding proteins or fluorophore-activating aptamers require large oligonucleotide tags or proteins that may severely influence the distribution of cellular RNA and are likely not amenable to monitoring small RNA fragments (11–14). Although alternatives such as binary, forced intercalation and echo probes provide solutions to some of these problems, there remains a significant need for rapid and robust methods to visualize specific oligonucleotide sequences (15–17).
One particularly useful method for detection of specific nucleic acid sequences has been the template-induced covalent ligation of two reactive DNA fragments (13,14,18). This approach benefits from the use of small probes, and the ability to detect specific native sequences while discriminating single base mismatches. Pioneering work in this field resulted in the development of various reactions, including imidazolide-activated nucleosides (18,19), condensation agent-mediated coupling (19–21), native chemical ligation (20–22), amine/aldehyde coupling (22,23) and reagent-free self-ligations (autoligations) between phosphorothioates and electrophilic groups (23,24). In a seminal study, the Kool group coupled template-driven chemical ligation with loss of a fluorescence quencher enabling imaging and detection of specific sequences (24). Following this work, several groups have developed schemes to adapt chemical ligation to fluorescent-based imaging and detection (25–31). The ligation of short (7–15 nt bases) may be particularly relevant for detecting and imaging microRNAs, which are typically of short length and would be severely impacted by the use of large aptamer or protein-binding RNA tags. However, previous ligation reactions have been hampered by slow kinetics and autohydrolysis, often relying on nucleophilic/electrophilic reactions, which allow cellular or solvent nucleophiles to compete for reactivity (25,32).
We and others have recently explored the use of fluorogenic tetrazine cycloadditions to image small molecules in live cells (33–36). Tetrazines can quench several fluorophores through what is predicted to be a resonance energy transfer mechanism (37). On tetrazine reaction with dienophiles such as norbornenes, trans-cyclooctenes and cyclopropenes, the quenching is removed, and a significant fluorogenic effect is observed. This phenomenon has been used to image the intracellular distribution of small molecules such as taxoids, kinase inhibitors, unnatural amino acids and lipids (33,38–40). Tetrazine bioorthogonal cycloadditions benefit from rapid tunable reaction rates and high stability against hydrolysis in buffer and serum (41–43). Furthermore, tetrazines act as both a fluorescent quencher and a reactive group, minimizing the complexity of fluorogenic ligation probe design (Figure 1). Based on these properties, we developed an oligonucleotide-templated fluorogenic tetrazine ligation approach for the rapid fluorescent detection of specific DNA and RNA sequences.
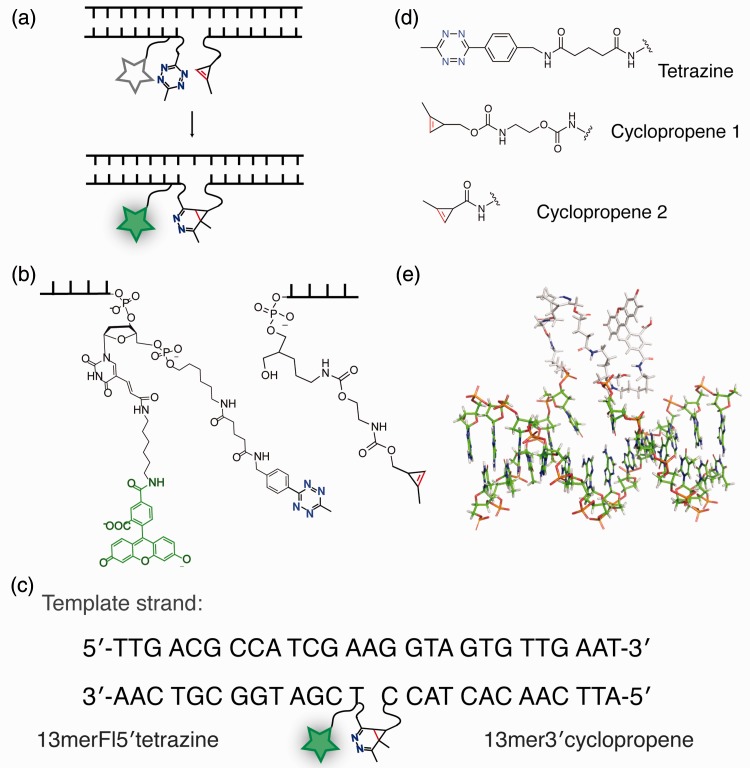
Figure 1.
Antisense oligonucleotide probe design. (a) Modified DNA ligation reaction scheme, catalysed by hybridization against a template strand. (b) Actual reactant structures with linker lengths illustrated. (c) Sequences of the template 27 mer strand and the corresponding 13 mer DNA probes. Probe truncations (to 7 and 5 nt) and template extensions (in the gap region) are indicated in the main text. (d) Structures of tetrazine and cyclopropene reactants used in this study. (e) DNA model of the ligated probe product structure, showing the relative length of the resulting loop, as it connects across a single nucleotide gap in the template of a generated ideal B-type helix.
MATERIALS AND METHODS
Starting materials
All chemicals were received from commercial sources and used without further purification. End-modified DNA oligonucleotides were purchased from Integrated DNA Technologies, Inc. (IDT) and used after HPLC purification, lyophilization and resuspension in ddi H2O. DNA and RNA templates of 27–37 nt length were purchased from IDT and used on resuspension in ddi H2O without further purification. Synthesis of the cyclopropene and tetrazine tags is described in the Supplementary Information.
DNA template: 5′-TTG ACG CCA TCG AAG G[T]A GTG TTG AAT-3′ (linker region underlined, single mismatch position in brackets); 13 mer Fl5′tet: 5′-tetrazine –/5AmMC6/ /iFluorT/ CG ATG GCG TCA A-3′ (modified nucleotides indicated following the manufacturer’s nomenclature); 13 mer 3′cyclopropene: 5′- ATT CAA CAC TAC C /3AmMO/ – cyclopropene-3′; 7 mer 3′cyclopropene: 5′-CAC TAC C /3AmMO/ – cyclopropene-3′; 5 mer 3′cyclopropene: 5′-CTA CC /3AmMO/ – cyclopropene-3′.
Modified oligonucleotide synthesis and characterization
Probe linker length was chosen on first confirming that ligating partner connectivity is possible across several probe gap lengths, based on a model structure. PolyA-polyT DNA double helix (using B-DNA torsion angles at the C2′-endo sugar-backbone) was generated by 3D-DART web server (44) and was used as the initial backbone for further modelling using Maestro 9.3 (Schrödinger LLC, New York, NY, 2012). Final reacted probe structures were initially built in manually, followed by several rounds of energy minimization of the selected added structures using the OPLS_2005 all-atom force field with default parameters (45) for up to 1000 iterations per round until the RMSD between iterations of <0.1 Å was reached. Representative model with a single nucleotide gap between the hybridized probes is shown in Figure 1e.
Amine-modified oligonucleotide sequences at selected terminus were modified by reacting with excess cyclopropene or tetrazine carboxylic acid NHS ester in 50 mM sodium bicarbonate buffer containing 100–1000 mM NaCl. Main product formed without significant side reactions. Cyclopropene or tetrazine carboxylic acid NHS esters were dissolved in dimethylformamide at 100 mM concentration, and added at 5× excess to the aqueous reaction solution containing 0.1–1.5 mM oligonucleotide. On partial reaction or NHS ester hydrolysis, another 5× excess was added 30–60 min later, and the process was repeated until no more reactant oligonucleotide was detected by HPLC.
Oligonucleotide detection by HPLC was done using Agilent 1260 Infinity LC/MS system with Phenomenox Oligo-MS 150–4.6 mm column. About 20 pmol samples were injected and eluted with a gradient of 6–18% acetonitrile in 5 mM ammonium acetate at pH 7.25.
Final prepared oligonucleotide molecular weights were confirmed by MALDI-TOF MS using a Biflex IV system (Bruker Daltronics). Oligonucleotides at ∼50 µM were mixed with equal volumes of 3-hydroxypicolinic acid (3-HPA) matrix. Each construct molecular weights were located at the main m/z peaks. Representative spectra are shown in Supplementary Figure S1. Ligation reaction product weights were determined by time-of-flight mass spectrometer (TOF MS) on separation by LC using the same gradient elution described earlier in the text using a 6230 TOF MS (Agilent Technologies). Detected ligation product molecular weights are show in Supplementary Figure S2. An example HPLC reaction (Supplementary Figure S3) shows the typical ligation reaction on completion, with overlaid reactant elution peaks using Agilent Technologies 1260 Infinity LC/MS (equipped with a 6120 quadrupole MS ionization-spray detector) with the gradient elution described earlier in the text.
Melting temperature of probes and ligation products in hybridization buffer [50 mM MOPS (pH 7.5), 250 mM NaCl] were measured using a Beckman-Coulter DU 640 spectrophotometer equipped with a high performance temperature controller and micro auto six-cell holder. Samples at 1 µM concentration of each oligonucleotide were heated to 90°C and slowly cooled to RT over 1–1.5 h before the measurements. Melting temperature values were obtained by cooling samples from 90 to 20°C at the rate of 1°C/min and measuring at 1°C increments with each read averaged >1 s. Data were fitted to the first order derivative by using the manufacturer’s software, and resulting values are compiled in Supplementary Table S1.
Ligations of modified oligonucleotides
Tetrazine and cyclopropene-modified oligonucleotide probes were ligated in the presence of a DNA or RNA template at a 1:1:1 ratio. Reactions were done in varying solution and temperature conditions, as indicated. Main hybridization buffer was 50 mM MOPS at pH 7.5, 250 mM NaCl. Ligation reactions with a mismatched DNA template were performed in 5 mM MgCl2 (final concentration on accounting for the presence of EDTA) in standard 1× Tris-borate buffer [purchased as 10× UltraPure TBE formulation from Invitrogen: 1.0 M Tris (pH 8.4), 0.9 M Boric acid, 10 mM EDTA].
Tetrazine stability was tested by taking Nanodrop UV-Vis absorbance measurements over time. Characteristic tetrazine absorbance peak intensity at 520 nm was measured by first subtracting the background level intensity at each point, estimated as the trendline between the intensity levels immediately preceding and following the tetrazine peak. Tetrazine and cyclopropene modification analogs used for this reaction are indicated next to the corresponding data trends in Supplementary Figure S4. Reactions were done in the standard hybridization buffer at room temperature of ∼22°C at 1 mM tetrazine and 10 mM cyclopropene concentrations. Data for the tetrazine-cyclopropene reaction were fit to a one-phase exponential decay of the tetrazine absorption peak using GraphPad Prism 6.0a for Mac (GraphPad Software, La Jolla, CA, USA, www.graphpad.com) by the following relationship: y = (y0 – yplateau) * exp(–kobs * x) + yplateau. Data were fit without value constraints. The resultant kobs values were converted to the second-order rate constants (k2) as: k2 = kobs/(cyclopropene)0. Reaction half-times were calculated as t1/2 = 1/[k2*(cyclopropene)0].
Ligation reactions were done in standard 96 fluorescence well plates (with data compiled in Table 1) using SpectraMax GeminiXS (Molecular Devices). Temperature was set and controlled by the instrument. Where indicated by standard deviations, experiments were performed multiple times. To avoid any secondary structure artifacts, DNA or RNA template with added tetrazine probe was first heated to 90°C and crash-cooled on ice for 2 min right before the experiment. Each well was filled with a total of 100 µl of reaction solution, and reactions were timed from the addition of the cyclopropene probe as the last step before starting the measurements. Wells were scanned every 20–60 s (depending on the speed of the reactions and the number of columns of simultaneously measured data) by using 485 nm excitation and 538 nm emission monochromators with the emission cutoff filter at 495 nm. Control reactions without template were done in parallel each time. Buffer-only containing well intensities were subtracted from the measured raw data before analysis. Data were analysed using GraphPad Prism 6.0a by applying an exponential fit of a one-phase association to each curve where fluorescein signal during reaction was measured, to determine the fitted kobs values from: y = y0 + (yplateau – y0) * [1 – exp(–kobs*x)]. Reported half-times (t1/2) for 1 µM reactions were calculated as: t1/2 = ln(2)/kobs.
Table 1.
Reaction rates of DNA and RNA templates in varying temperature and media ionic strength conditions
| Template | Cyclopropene | Rxn conditions | t½ (s) |
|---|---|---|---|
| 27 mer | 13 mer 3′cycp1 | 250 mM NaCl, MOPSa | 36 ± 2c |
| 27 mer | 7 mer 3′cycp1 | 250 mM NaCl, MOPSa | 130 ± 40c |
| 27 mer | 5 mer 3′cycp1 | 250 mM NaCl, MOPSa | 9700 ± 500c |
| 27 mer | 13 mer 3′cycp2 | 250 mM NaCl, MOPSa | 1900 ± 100 c |
| 27 mer | 7 mer 3′cycp1 | C-DMEM, seruma | 240 ± 80 |
| 27 mer n = 1 mism. | 7 mer 3′cycp1 | C-DMEM, seruma | 1000 ± 90 |
| 27 mer | 13 mer 3′cycp1 | 5 mM MgCl2, TBb | 29 ± 2 |
| 27 mer n = 1 mism. | 13 mer 3′cycp1 | 5 mM MgCl2, TBb | 59 ± 6 |
| 27 mer | 7 mer 3′cycp1 | 5 mM MgCl2, TBb | 161 ± 2 |
| 27 mer n = 1 mism. | 7 mer 3′cycp1 | 5 mM MgCl2, TBb | 4000 ± 200 |
| 27 mer | 7 mer 3′cycp1 | C-DMEM, serumb | 152 ± 7 |
| 27 mer RNA | 13 mer 3′cycp1 | 250 mM NaCl, MOPSb | 40 ± 3 |
Regular buffer conditions were maintained at pH 7.5 with 50 mM MOPS. TB denotes Tris–Borate buffer.
aExperiments were performed at 25°C.
bExperiments were performed at 37°C.
cStandard deviations were calculated for experiments done in triplicate, otherwise, error was estimated from the fitted values.
RESULTS AND DISCUSSION
For ligation partners, we required a tetrazine–dienophile pair that would not appreciably react at micromolar concentrations for a period of days, but when brought into close proximity by an oligonucleotide template, would react rapidly and produce a fluorescence signal. Based on preliminary experiments, we chose to explore the reaction between methyl-terminated tetrazines and methyl-cyclopropene dienophiles (Figure 1b). For the fluorogenic tetrazine probe, we synthesized an asymmetric methyl-tetrazine from an alkyl nitrile and hydrazine using a recently disclosed metal catalyzed reaction (46). Previous work has demonstrated that methyl modification lowers the reactivity of tetrazines while simultaneously greatly increasing their stability in physiological media (47). We appended the tetrazine to the 5′ position of a 13 mer oligonucleotide using an amide coupling reaction. Adjacent to the tetrazine, we included an internal fluorescein off a terminal thymine base (Figure 1c and e). Despite the flexible linker distance between the resultant tetrazine and fluorescein, significant quenching was observed. For the dienophile probe, we appended methyl-cyclopropene derivatives at the 3′ end of oligonucleotides owing to their moderate reactivity, high stability and small size (Figure 1d). We have previously shown that methyl-cyclopropene derivatives are stable reaction partners with fluorogenic tetrazines (40). We compared a reactive methyl-cyclopropene carbamate (cycp1) and a less reactive methyl-cyclopropene carboxyamide (cycp2) to study the influence of intermolecular reactivity on oligonucleotide-templated intramolecular reaction rate. Reaction of the tetrazine probe with the cyclopropene probe in the presence of a DNA template led to a 9.3-fold increase in the peak emission intensity (Figure 2). The quenched tetrazine probes are extremely stable in buffer and cell media at room temperature and 37°C (Supplementary Figures S5 and S6), in line with previous reports on related methyl-terminated tetrazines (47).
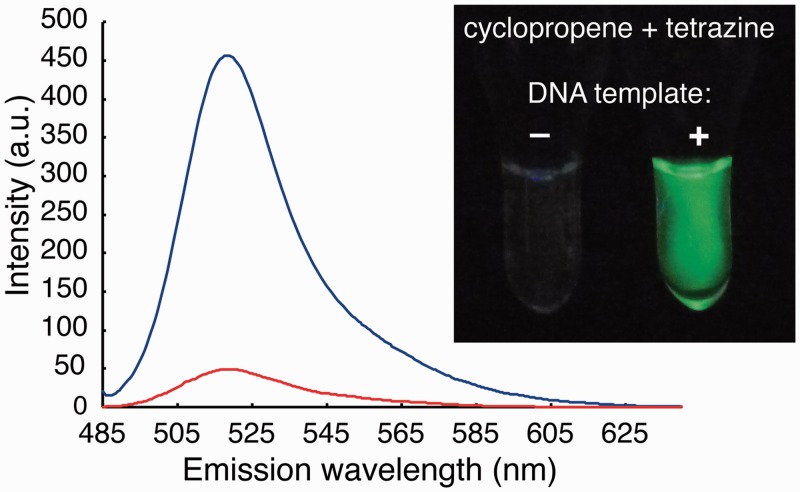
Figure 2.
Fluorogenic template driven probe ligation. Fluorescein fluorescence signal is quenched in the presence of a tetrazine modification on the oligonucleotide probe. Fluorescence scans were done before (red) and after (blue) the reaction of 27 mer DNA template with 13 mer Fl5′tet + 13 mer 3′cycp1 in 250 mM NaCl, 50 mM MOPS buffer (pH 7.5). Emission was scanned on excitation at 480/5 nm. Insert shows UV irradiated 1 micromolar solutions of fluorogenic tetrazine and cyclopropene probes in the absence (−) and presence (+) of oligonucleotide template after a 5 min incubation.
The DNA template was optimized with respect to the template nucleotide gap length between the ligating tetrazine and cyclopropene probe binding sites. Based on the template, ligation required reactive probes to bridge nucleotide gaps of varying length. A gap of a single nucleotide resulted in the fastest ligation kinetics (Figures 1c, 3a, full data in Supplementary Figure S7 and Supplementary Table S2) and was used for all further experiments. We incorporated flexible linkers to minimize possible steric constraints during ligation (Figure 1b and e). Interestingly, the observed distance dependency of the fluorogenic reaction kinetics provides information on the proximity of bound probes, functioning as a molecular ruler (Figure 3a, full data in Supplementary Figure S4 and Supplementary Table S2).
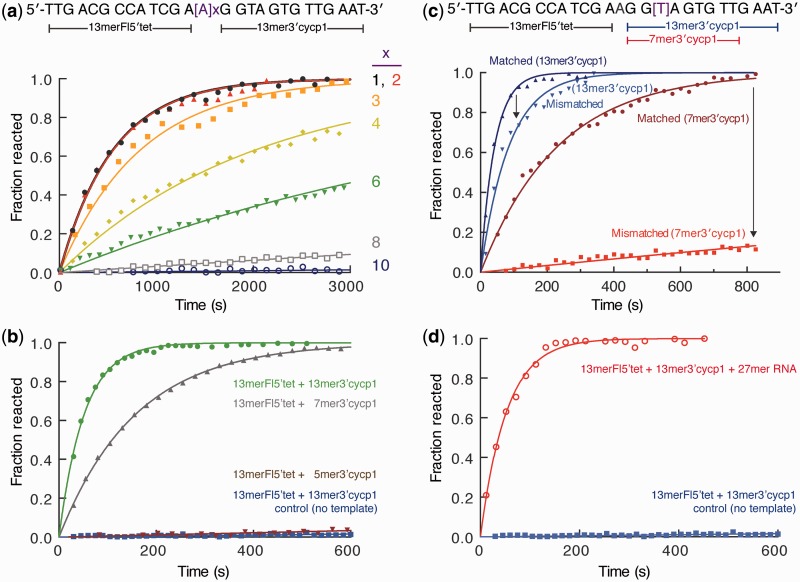
Figure 3.
Effect of the probe and template structure on tetrazine ligation reaction. (a) Probe binding site separation affects the apparent reaction rate. DNA templates (shown above the data graph) with increasing polyA insert between the probe binding sequences were compared by reacting 13 mer Fl5′tet + 13 mer 3′cycp1 in the hybridization buffer at 25°C [250 mM NaCl, 50 mM MOPS (pH 7.5)]. The number, x, of inserted nucleotides are indicated following each fitted curve. Apparent reaction rates and half lives are listed in Supplementary Table S2, and complete 1–10 linker data are shown in Supplementary Figure S7. (b) DNA probe length optimization leads to the most effective probes. Fluorescence data of the 13 mer Fl5′tet + 13 mer 3′cycp1 reaction with 27 mer DNA template [1 µM each in 150 mM MOPS (pH 7.5), 25°C] is shown as measured data points and fitted curves in green and red, respectively. DNA-templated reaction of 13 mer Fl5′tet with shorter 7 mer 3′cycp1 is shown in grey, with 5 mer 3′cycp1 in brown. Control reaction of 13 mer Fl5′tet and 13 mer 3′cycp1 with no template is plotted in blue. (c) Effect of a single nucleotide mismatch on the reaction kinetics depends on the length of the hybridization probe. The mismatch was introducted on the side of the template where the 3′cyclopropene-modified probe binds; therefore, reactant cyclopropene oligomers were varied. Hybridization reactions against the fully matched 27 mer template (labelled ‘Matched’) and against the singly mismatched 27 mer template (labelled ‘Mismatched’) were done with 13 mer Fl5′tet reacting with 13 mer 3′cycp1 or with 7 mer 3′cycp1 (labelled accordingly). Solution conditions were Tris–borate buffer (pH 8.4), 5 mM MgCl2 at 37°C. (d) Short oligonucleotide probes are equally effective in ligating with an RNA template. The corresponding 27 mer RNA template was used in ligating 13 mer Fl5′tet and 13 mer 3′cycp1 as in (c). Reaction kinetics data are listed in Table 1.
Probe ligation kinetics were highly dependent on the presence and amount of a matching sequence template (Supplementary Figure S9). Hybridization buffer containing 1 µM 13 mer fluorescein-tetrazine (13 mer Fl5′tet) and 13 mer-cyclopropene1 (13 mer3′cycp1) showed no appreciable change in fluorescence over time (control data in Figure 3b, Supplementary Figures S5 and S9). This is expected, as the reaction rate between the corresponding tetrazine and cyclopropene precursors was measured to be 0.37 ± 0.05 M−1s−1 with a predicted half-life of 32 days at 1 µM (Supplementary Figure S4). At 1 µM concentration of each oligonucleotide probe, the reaction half-life is predicted to be ∼5 days without template. However, addition of 1 µM of a 27 mer template strand resulted in a rapid increase in fluorescence owing to template-driven ligation and unquenching of the fluorescein probe (Figure 3). The reaction half-life for the fastest reaction with 13 mer 3′cycp1 along the DNA template was 36 ± 2 s, with the measured fluorescence signal plateauing after ∼200 s, indicating that the reaction was driven to completion. HPLC and high-resolution mass spectrometry confirmed that the reaction led to the ligation products (Supplementary Figures S1–S3). Based on the disappearance of 13 mer Fl5′tet and the appearance of product by LC/MS, we estimate the reaction yield to be ∼92% (Supplementary Figure S3). Reactant 13 mer Fl5′tet and 13 mer 3′cycp1 probe melting temperatures were significantly lower (57.4 and 50.4°C, respectively) than that of the ligation product (65.8°C). However, the tetrazine/cyclopropene product melting temperature is 9.7°C lower than the melting temperature of a 27 mer matching DNA sequence (75.5°C), indicating that the internal reaction product loop has a slight destabilization effect (Supplementary Table S1).
Fitting the increase in fluorescence over time to an exponential growth curve, an observed first order rate constant of 0.019 ± 0.001 s−1 was determined for the DNA-templated reaction of the two 13 mer probes at 1 µM. Assuming that the reaction is limited by ligation (given the high concentration of DNA), we can estimate the effective molarity of the intramolecular reaction to be 52 mM (calculated as the ratio of the intramolecular rate constant over the intermolecular second-order rate of 0.366 ± 0.005 M−1s−1) (48). This is in line with previous estimates of DNA template driven covalent reactions which, to the authors’ knowledge, are not typically fully elucidated (49). Reaction kinetics did not differ between DNA or RNA templates, adding to the versatility of this reaction (Table 1 and Figure 3b and d). In comparison with the 13 mer 3′cycp1, carboxamide construct 13 mer 3′cycp2 reacts slower with the 13 mer Fl5′tet in the presence of template. This is expected based on previous studies and the lower second-order rate constant of cycp2 and tet in the absence of DNA (0.003 M−1s−1) compared with cycp1. In the presence of DNA template, the first-order reaction rate was determined to be (3.7 ± 0.3) × 10−4 s−1, with a half-life of 1882 ± 132 s (31 min) and an estimated effective molarity of 123 mM (calculated as the ratio of intramolecular over the intermolecular rate constant). These reactions may be compared with the action of DNA/RNA ligases that connect immediately adjoining templated oligonucleotides (50,51). Apparent reaction rates at 0.05 µM human ligase are in the range of 0.2–6.6 min−1 for phosphodiester backbone bond formation, as determined by a single fluorophore DNA probe system (52). These rates are, remarkably, on the order of the observed kinetics of the tetrazine–cyclopropene ligation (kobs 1.1 min−1 for two 13 mer probe ligation at 1 µM). Owing to linker flexibility, the tetrazine ligation setup offers template position gap variability, a situation where the ligase-mediated reaction could not proceed.
We also determined the effect of shortening the oligonucleotide length of cycp1 probes. The 7 mer 3′cycp1 bound less avidly to the template (ligation product melting temperature was 59.9°C), but, at 1 µM, still reacted rapidly with 13 mer Fl5′tet with a half-life of 129 ± 38 s (Table 1). The rate constant increased by increasing the concentration of the reaction components, indicating that the reaction did not behave like a true first-order intramolecular reaction, and that the binding of the 7mer with the template likely influenced the reaction rate. Further reduction of the cyclopropene probe length to a 5 mer resulted in weak template binding (Table 1 and Figure 3b).
Importantly, these probes worked robustly in cell media containing serum (Table 1). Preincubation of the probes for several hours in serum containing media did not diminish the intensity of the signal on template hybridization, demonstrating the utility of stable bioorthogonal handles (Supplementary Figure S6). This points to the possibility of diagnostic and imaging application in live cells or tissue samples. As a proof of concept, we have performed imaging studies using the synthetic probes and live mammalian cell hosts (Figure 4). Templates were able to trigger turn-on in live cells and significantly increase fluorescence compared with cells where matching template sequence was absent. Probe stability is required for tracking cellular processes over time and for limiting background turn-on when detecting less abundant targets. The general practicality of such probes in complex environments could enable exciting opportunities in therapeutic and diagnostic settings. In addition, other environments not amenable to enzymatic approaches, like gels, solvents or non-physiological conditions, may be accessible to the minimal probes as described here.
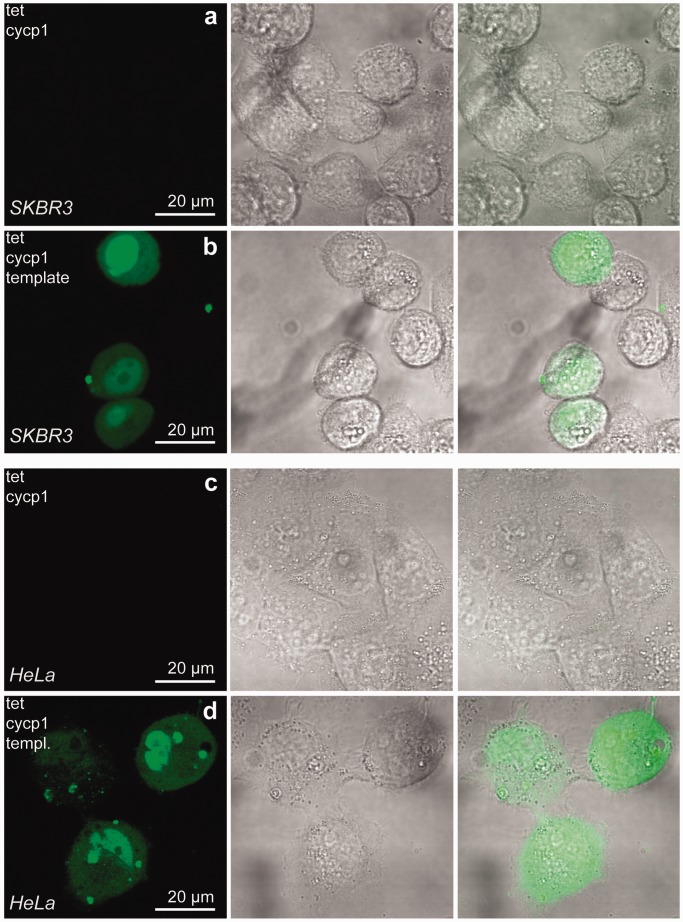
Figure 4.
Oligonucleotide probe reaction in live mammalian cells. SKBR3 (rows a–b) and HeLa (rows c–d) human breast and cervical cancer cells, respectively, were incubated with 0.5 µM DNA probes and template pre-equilibrated with transfection agents [reaction product signals shown in row (b) SKBR3, row (d) HeLa cells]. Images were obtained using an Olympus FV1000 confocal microscope. No reaction was observed in cells not exposed to a matched 27 nt template [row (a) SKBR3, row (c) HeLa cells]. First column shows the 488 nm excitation Argon laser signal, followed by a differential interference contrast image of cells and an overlay. Scale bars are indicated.
One of the benefits of in situ probe ligation is the ability to discriminate sequences containing a single nucleotide mismatch in the template oligonucleotide. This is due to the relatively greater impact of a base mismatch on the binding of shorter versus longer probes (53,54). We tested whether tetrazine ligation is sensitive enough to discriminate between a single mismatch on the template using previously tested conditions (55), optimized to 5 mM MgCl2 in Tris-borate buffer at 37°C. One mismatch (T to G) was introduced in the template portion that hybridizes with a cyclopropene probe. We observed a reaction rate reduction by an order of magnitude in the case of a 7 mer 3′cycp1 (Table 1), but, similar to previous work, only a moderate difference in the 13 mer 3′cycp1 (Figure 3c). Additionally, there is 4.5-fold decrease in the reaction rate at 25°C in cell media/serum conditions using 7 mer 3′cycp1 (Table 1), indicating that a single-mismatch discrimination may be possible in physiological media. With respect to signal intensity, this corresponded to a ∼10-fold drop in signal intensity after 13 min of reaction with mismatched template, at which time the matched template elicited full reaction in buffer (Figure 3c). Probes could also be used to detect a matched sequence in the presence of a competing mismatch introduced earlier in the text (Supplementary Figure S8). Generally, this ligation reaction with a fully matched template or one containing one nucleotide mismatch was similarly robust in hybridization buffers and physiologically relevant cell media when 7 mer 3′cycp1 probe was used (Table 1). Such selectivity, although likely substitution-specific, highlights one potential advantage of in situ ligation of oligonucleotides, the ability to discriminate between single mismatches, which may be useful for applications such as single nucleotide polymorphism discrimination.
Despite the rich history of oligonucleotide-templated ligation reactions, we believe that bioorthogonal tetrazine chemistry offers unique advantages. The application of bioorthogonal chemistry allows the use of rapidly reacting chemoselective coupling partners without an obligatory increase of background reactions in aqueous buffers or physiological media. Other popular ligation strategies that rely on nucleophilic displacement or redox reactions are limited in their ability to increase ligation rates owing to a concomitant increase in background side reactions, as biological media contains high concentrations of nucleophiles such as amines and thiols, as well as redox active agents. Indeed, the minimal autohydrolytic turn-on of the 13 mer Fl5′tet over a 35 h period (Supplementary Figures S4 and S5) points to this benefit. Also, previous approaches typically rely on separate fluorescent quenchers and reactive probes. The dual use of tetrazine as both a fluorescence quencher and reactant uniquely simplifies experimental design.
Rapid reaction kinetics are important not only in the presence of off-pathway side reactions or cellular degradation but also because the reaction rate determines the limit of temporal resolution of target oligonucleotide detection. For instance, there is significant interest in monitoring the transcriptional dynamics of RNA synthesis and degradation (56). Optimal ligation kinetics will match the timescales of such processes, which can be on the order of minutes in live cells (56). As these ligations are tuneable, optimization of linker length and coupling partners may further improve kinetics.
CONCLUSIONS
We have developed quenched oligonucleotide tetrazine probes as well as cyclopropene reaction partners that undergo rapid DNA/RNA template-dependent ligation with a resultant increase in fluorescence. By confining tetrazine and cyclopropene probes in close proximity without steric hindrance, the oligonucleotide template enforces an effective molarity in the 50–120 mM range, depending on the linker constructs used. This large effective concentration allows reactions to proceed rapidly only in the presence of sequence-specific targets, with minimal background reaction in the absence of templates. Bioorthogonal reaction-dependent fluorescence quenching benefits detection as tetrazines are not prone to autohydrolysis. Reactivity of the system can be tuned by varying the tetrazine, dienophile, oligonucleotide probe length, linker length and template gap width spanning the reaction space. Future work will further optimize probe properties such as fluorogenic turn-on ratio and reaction rate. We predict that short oligonucleotide-based templated tetrazine cycloadditions, owing to their stability, rapid tunable reaction rates and bioorthogonal reactivity, could have a myriad of applications not only in the detection and imaging of DNA and RNA in live cells or biological samples but also in therapeutic applications such as the delivery and in situ assembly of antisense probes and therapeutic ribozymes.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1 and 2, Supplementary Figures 1–9, Supplementary Methods, and Supplementary NMR Spectra.
FUNDING
NIH [K01EB010078] (in part) and the University of California, San Diego. Equipment used for collecting data was funded by the NSF under [CHE-0741968], by the NIH under [SRR0256362] supported by Yongxuan Su, and by NIH [P30 NS047101] supported by Jennifer Santini. Funding for open access charge: University of California, San Diego.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors greatly acknowledge Prof. Simpson Joseph, Prof. Russell Doolittle, Prof. Navtej Toor and Dr Ralph Mazitschek for helpful discussions. Melting temperature measurements were done in Prof. Y. Tor laboratory at UCSD.
REFERENCES
- 1.Stojanovic MN, de Prada P, Landry DW. Catalytic molecular beacons. ChemBioChem. 2001;2:411–415. doi: 10.1002/1439-7633(20010601)2:6<411::AID-CBIC411>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 2.Liu J, Cao Z, Lu Y. Functional nucleic acid sensors. Chem. Rev. 2009;109:1948–1998. doi: 10.1021/cr030183i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dave N, Liu J. Fast molecular beacon hybridization in organic solvents with improved target specificity. J. Phys. Chem. B. 2010;114:15694–15699. doi: 10.1021/jp106754k. [DOI] [PubMed] [Google Scholar]
- 4.Armitage BA. Imaging of RNA in live cells. Curr. Opin. Chem. Biol. 2011;15:806–812. doi: 10.1016/j.cbpa.2011.10.006. [DOI] [PubMed] [Google Scholar]
- 5.Chen AK, Davydenko O, Behlke MA, Tsourkas A. Ratiometric bimolecular beacons for the sensitive detection of RNA in single living cells. Nucleic Acids Res. 2010;38:e148. doi: 10.1093/nar/gkq436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Silverman AP, Kool ET. Detecting RNA and DNA with templated chemical reactions. Chem. Rev. 2006;106:3775–3789. doi: 10.1021/cr050057+. [DOI] [PubMed] [Google Scholar]
- 7.Wang K, Tang Z, Yang CJ, Kim Y, Fang X, Li W, Wu Y, Medley CD, Cao Z, Li J, et al. Molecular engineering of DNA: molecular beacons. Angew Chem. Int. Ed. Engl. 2009;48:856–870. doi: 10.1002/anie.200800370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marti AA, Jockusch S, Stevens N, Ju J, Turro NJ. Fluorescent hybridization probes for sensitive and selective DNA and RNA detection. Acc. Chem. Res. 2007;40:402–409. doi: 10.1021/ar600013q. [DOI] [PubMed] [Google Scholar]
- 9.Tsourkas A, Behlke MA, Rose SD, Bao G. Hybridization kinetics and thermodynamics of molecular beacons. Nucleic Acids Res. 2003;31:1319–1330. doi: 10.1093/nar/gkg212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guo J, Ju J, Turro NJ. Fluorescent hybridization probes for nucleic acid detection. Anal. Bioanal. Chem. 2012;402:3115–3125. doi: 10.1007/s00216-011-5526-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tyagi S. Imaging intracellular RNA distribution and dynamics in living cells. Nat. Methods. 2009;6:331–338. doi: 10.1038/nmeth.1321. [DOI] [PubMed] [Google Scholar]
- 12.Mhlanga MM, Vargas DY, Fung CW, Kramer FR, Tyagi S. tRNA-linked molecular beacons for imaging mRNAs in the cytoplasm of living cells. Nucleic Acids Res. 2005;33:1902–1912. doi: 10.1093/nar/gki302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gryaznov SM, Schultz R, Chaturvedi SK, Letsinger RL. Enhancement of selectivity in recognition of nucleic acids via chemical autoligation. Nucleic Acids Res. 1994;22:2366–2369. doi: 10.1093/nar/22.12.2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kumar R, El-Sagheer A, Tumpane J, Lincoln P, Wilhelmsson LM, Brown T. Template-directed oligonucleotide strand ligation, covalent intramolecular DNA circularization and catenation using click chemistry. J. Am. Chem. Soc. 2007;129:6859–6864. doi: 10.1021/ja070273v. [DOI] [PubMed] [Google Scholar]
- 15.Kolpashchikov DM. A binary DNA probe for highly specific nucleic Acid recognition. J. Am. Chem. Soc. 2006;128:10625–10628. doi: 10.1021/ja0628093. [DOI] [PubMed] [Google Scholar]
- 16.Socher E, Jarikote DV, Knoll A, Roglin L, Burmeister J, Seitz O. FIT probes: peptide nucleic acid probes with a fluorescent base surrogate enable real-time DNA quantification and single nucleotide polymorphism discovery. Anal. Biochem. 2008;375:318–330. doi: 10.1016/j.ab.2008.01.009. [DOI] [PubMed] [Google Scholar]
- 17.Okamoto A. ECHO probes: a concept of fluorescence control for practical nucleic acid sensing. Chem. Soc. Rev. 2011;40:5815–5828. doi: 10.1039/c1cs15025a. [DOI] [PubMed] [Google Scholar]
- 18.Rohatgi R, Bartel DP, Szostak JW. Nonenzymatic, template-directed ligation of oligoribonucleotides is highly regioselective for the formation of 3′−5′ phosphodiester bonds. J. Am. Chem. Soc. 1996;118:3340–3344. doi: 10.1021/ja9537134. [DOI] [PubMed] [Google Scholar]
- 19.Sarkar T, Conwell CC, Harvey LC, Santai CT, Hud NV. Condensation of oligonucleotides assembled into nicked and gapped duplexes: potential structures for oligonucleotide delivery. Nucleic Acids Res. 2005;33:143–151. doi: 10.1093/nar/gki156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bruick RK, Dawson PE, Kent SB, Usman N, Joyce GF. Template-directed ligation of peptides to oligonucleotides. Chem. Biol. 1996;3:49–56. doi: 10.1016/s1074-5521(96)90084-8. [DOI] [PubMed] [Google Scholar]
- 21.Peng X, Li H, Seidman M. A template-mediated click-click reaction: PNA-DNA, PNA-PNA (or peptide) ligation, and single nucleotide discrimination. Eur. J. Org. Chem. 2010;2010:4194–4197. doi: 10.1002/ejoc.201000615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peelen D, Smith LM. Immobilization of amine-modified oligonucleotides on aldehyde-terminated alkanethiol monolayers on gold. Langmuir. 2005;21:266–271. doi: 10.1021/la048166r. [DOI] [PubMed] [Google Scholar]
- 23.Xu Y, Karalkar NB, Kool ET. Nonenzymatic autoligation in direct three-color detection of RNA and DNA point mutations. Nat. Biotechnol. 2001;19:148–152. doi: 10.1038/84414. [DOI] [PubMed] [Google Scholar]
- 24.Sando S, Kool ET. Quencher as leaving group: efficient detection of DNA-joining reactions. J. Am. Chem. Soc. 2002;124:2096–2097. doi: 10.1021/ja017328s. [DOI] [PubMed] [Google Scholar]
- 25.Silverman AP, Kool ET. Quenched probes for highly specific detection of cellular RNAs. Trends Biotechnol. 2005;23:225–230. doi: 10.1016/j.tibtech.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 26.Shibata A, Abe H, Ito Y. Oligonucleotide-templated reactions for sensing nucleic acids. Molecules. 2012;17:2446–2463. doi: 10.3390/molecules17032446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.El-Sagheer AH, Brown T. Click chemistry with DNA. Chem. Soc. Rev. 2010;39:1388–1405. doi: 10.1039/b901971p. [DOI] [PubMed] [Google Scholar]
- 28.Gorska K, Keklikoglou I, Tschulena U, Winssinger N. Rapid fluorescence imaging of miRNAs in human cells using templated Staudinger reaction. Chem. Sci. 2011;2:1969. [Google Scholar]
- 29.Furukawa K, Abe H, Tamura Y, Yoshimoto R, Yoshida M, Tsuneda S, Ito Y. Fluorescence detection of intron lariat RNA with reduction-triggered fluorescent probes. Angew. Chem. Int. Ed. Engl. 2011;50:12020–12023. doi: 10.1002/anie.201104425. [DOI] [PubMed] [Google Scholar]
- 30.Chen XH, Roloff A, Seitz O. Consecutive signal amplification for DNA detection based on de novo fluorophore synthesis and host-guest chemistry. Angew. Chem. Int. Ed. Engl. 2012;51:4479–4483. doi: 10.1002/anie.201108845. [DOI] [PubMed] [Google Scholar]
- 31.Meguellati K, Koripelly G, Ladame S. DNA-templated synthesis of trimethine cyanine dyes: a versatile fluorogenic reaction for sensing G-quadruplex formation. Angew Chem. Int. Ed. Engl. 2010;49:2738–2742. doi: 10.1002/anie.201000291. [DOI] [PubMed] [Google Scholar]
- 32.Sando S, Abe H, Kool ET. Quenched auto-ligating DNAs: multicolor identification of nucleic acids at single nucleotide resolution. J. Am. Chem. Soc. 2004;126:1081–1087. doi: 10.1021/ja038665z. [DOI] [PubMed] [Google Scholar]
- 33.Devaraj NK, Hilderbrand S, Upadhyay R, Mazitschek R, Weissleder R. Bioorthogonal turn-on probes for imaging small molecules inside living cells. Angew. Chem. Int. Ed. Engl. 2010;49:2869–2872. doi: 10.1002/anie.200906120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Devaraj NK, Weissleder R. Biomedical applications of tetrazine cycloadditions. Acc. Chem. Res. 2011;44:816–827. doi: 10.1021/ar200037t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu DS, Tangpeerachaikul A, Selvaraj R, Taylor MT, Fox JM, Ting AY. Diels-Alder cycloaddition for fluorophore targeting to specific proteins inside living cells. J. Am. Chem. Soc. 2012;134:792–795. doi: 10.1021/ja209325n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Plass T, Milles S, Koehler C, Szymanski J, Mueller R, Wiessler M, Schultz C, Lemke EA. Amino acids for Diels-Alder reactions in living cells. Angew. Chem. Int. Ed. Engl. 2012;51:4166–4170. doi: 10.1002/anie.201108231. [DOI] [PubMed] [Google Scholar]
- 37.Dumas-Verdes C, Miomandre F, Lépicier E, Galangau O, Vu TT, Clavier G, Méallet-Renault R, Audebert P. BODIPY-Tetrazine multichromophoric derivatives. Eur. J. Org. Chem. 2010;2010:2525–2535. [Google Scholar]
- 38.Budin G, Yang KS, Reiner T, Weissleder R. Bioorthogonal probes for polo-like kinase 1 imaging and quantification. Angew. Chem. Int. Ed. Engl. 2011;50:9378–9381. doi: 10.1002/anie.201103273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lang K, Davis L, Torres-Kolbus J, Chou C, Deiters A, Chin JW. Genetically encoded norbornene directs site-specific cellular protein labelling via a rapid bioorthogonal reaction. Nat. Chem. 2012;4:298–304. doi: 10.1038/nchem.1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang J, Seckute J, Cole CM, Devaraj NK. Live-cell imaging of cyclopropene tags with fluorogenic tetrazine cycloadditions. Angew. Chem. Int. Ed. Engl. 2012;51:7476–7479. doi: 10.1002/anie.201202122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sletten EM, Bertozzi CR. From mechanism to mouse: a tale of two bioorthogonal reactions. Acc. Chem. Res. 2011;44:666–676. doi: 10.1021/ar200148z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Blackman ML, Royzen M, Fox JM. Tetrazine ligation: fast bioconjugation based on inverse-electron-demand diels-alder reactivity. J. Am. Chem. Soc. 2008;130:13518–13519. doi: 10.1021/ja8053805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Devaraj NK, Weissleder R, Hilderbrand SA. Tetrazine-based cycloadditions: application to pretargeted live cell imaging. Bioconjug. Chem. 2008;19:2297–2299. doi: 10.1021/bc8004446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van Dijk M, Bonvin AM. 3D-DART: a DNA structure modelling server. Nucleic Acids Res. 2009;37:W235–W239. doi: 10.1093/nar/gkp287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Banks JL, Beard HS, Cao Y, Cho AE, Damm W, Farid R, Felts AK, Halgren TA, Mainz DT, Maple JR, et al. Integrated modeling program, applied chemical theory (IMPACT) J. Comput. Chem. 2005;26:1752–1780. doi: 10.1002/jcc.20292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang J, Karver MR, Li W, Sahu S, Devaraj NK. Metal-catalyzed one-pot synthesis of tetrazines directly from aliphatic nitriles and hydrazine. Angew. Chem. Int. Ed. Engl. 2012;51:5222–5225. doi: 10.1002/anie.201201117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Karver MR, Weissleder R, Hilderbrand SA. Synthesis and evaluation of a series of 1,2,4,5-tetrazines for bioorthogonal conjugation. Bioconjug. Chem. 2011;22: 2263–2270. doi: 10.1021/bc200295y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cacciapaglia R, Di Stefano S, Mandolini L. Effective molarities in supramolecular catalysis of two-substrate reactions. Acc. Chem. Res. 2004;37:113–122. doi: 10.1021/ar020076v. [DOI] [PubMed] [Google Scholar]
- 49.Li X, Liu DR. DNA-templated organic synthesis: nature's strategy for controlling chemical reactivity applied to synthetic molecules. Angew. Chem. Int. Ed. Engl. 2004;43:4848–4870. doi: 10.1002/anie.200400656. [DOI] [PubMed] [Google Scholar]
- 50.Landegren U, Kaiser R, Sanders J, Hood L. A ligase-mediated gene detection technique. Science. 1988;241:1077–1080. doi: 10.1126/science.3413476. [DOI] [PubMed] [Google Scholar]
- 51.Ellenberger T, Tomkinson AE. Eukaryotic DNA ligases: structural and functional insights. Annu. Rev. Biochem. 2008;77:313–338. doi: 10.1146/annurev.biochem.77.061306.123941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen X, Ballin JD, Della-Maria J, Tsai MS, White EJ, Tomkinson AE, Wilson GM. Distinct kinetics of human DNA ligases I, IIIalpha, IIIbeta, and IV reveal direct DNA sensing ability and differential physiological functions in DNA repair. DNA Repair. 2009;8:961–968. doi: 10.1016/j.dnarep.2009.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Herschlag D. Implications of ribozyme kinetics for targeting the cleavage of specific RNA molecules in vivo: more isn't always better. Proc. Natl Acad. Sci. USA. 1991;88:6921–6925. doi: 10.1073/pnas.88.16.6921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Elghanian R, Storhoff JJ, Mucic RC, Letsinger RL, Mirkin CA. Selective colorimetric detection of polynucleotides based on the distance-dependent optical properties of gold nanoparticles. Science. 1997;277:1078–1081. doi: 10.1126/science.277.5329.1078. [DOI] [PubMed] [Google Scholar]
- 55.Xu Y, Kool ET. High sequence fidelity in a non-enzymatic DNA autoligation reaction. Nucleic Acids Res. 1999;27:875–881. doi: 10.1093/nar/27.3.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Darzacq X, Shav-Tal Y, de Turris V, Brody Y, Shenoy SM, Phair RD, Singer RH. In vivo dynamics of RNA polymerase II transcription. Nat. Struct. Mol. Biol. 2007;14:796–806. doi: 10.1038/nsmb1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.