Summary
Animals are often confronted with the decision as to consume a diet that contains competing attractive and aversive compounds. Here, using the fruit fly, Drosophila melanogaster, we describe a mechanism that influences this decision. Addition of bitter compounds to sucrose suppressed feeding behavior, and this inhibition depended on the odorant binding protein, OBP49a. In wild-type flies, bitter compounds suppressed sucrose-induced action potentials, and the inhibition was impaired in Obp49a mutants. However, loss of OBP49a did not affect action potentials in sugar- or bitter-activated gustatory receptor neurons (GRNs) when the GRNs were presented with just one type of tastant. OBP49a was expressed in accessory cells, and acted non-cell autonomously to attenuate nerve firings in sugar-activated GRNs when bitter compounds were combined with sucrose. These findings demonstrate an unexpected role for an OBP in taste, and identify a molecular player involved in the integration of opposing attractive and aversive gustatory inputs.
Introduction
Most foods are comprised of complex mixtures of different tastants, such as sweet and bitter compounds. Consequently, animal food preferences are decided by interactions between multiple constituents, many of which modulate the appeal or aversion of the component tastants. Suppression of the attractiveness of sweet by bitter tasting compounds has a strong survival benefit. Many tastants that are perceived as bitter are toxic, and so inhibition of stimulatory feeding behavior by these chemicals is critical.
When deterrent chemicals are present together with phagostimulatory tastants, they inhibit feeding by acting on two types of gustatory receptor cells. Aversive chemicals in foods not only stimulate deterrent taste cells, but also inhibit taste receptor cells that are activated by awarding compounds. This interaction between bitter and attractive gustatory stimuli has been observed in a wide array of vertebrate and invertebrate animals (Glendinning, 2007). Most studies dealing with the interactions between deterrent and attractive tastants have focused on quinine, a prototypical bitter compound. Electrophysiological recordings in hamsters show that the response to sucrose is inhibited by quinine (Formaker et al., 1997). In the catfish, quinine inhibits the positive gustatory response of several amino acids (Ogawa et al., 1997). Bitter compounds such as quinine are also aversive to flies (Tompkins et al., 1979), and suppress sugar-evoked firings in gustatory receptor neurons (GRNs) (Meunier et al., 2003).
The suppression of the stimulatory effect of attractive tastants by deterrent compounds could take place in the taste receptor cells, or in higher processing central pathways. While both sites might contribute to inhibition of sugar attractiveness by quinine, there is evidence that the afferent taste receptor cells are important for this phenomenon (Formaker et al., 1997; Talavera et al., 2008). Multiple mechanisms have been proposed to account for inhibition of sweet taste by quinine and other bitter compounds within the peripheral region of the gustatory system. The bitter-sweet interaction could be a consequence of lateral inhibition of sugar responsive gustatory receptor cells by bitter-activated neurons, similar to the inhibition of olfactory receptor neurons (ORNs) following activation of neighboring ORNs (Vandenbeuch et al., 2004; Su et al., 2012). Chemical interactions between the sugars and bitter compounds might also inhibit the attractiveness of the sugars. Competition of sugars and bitter chemicals for the same receptor is also plausible. An important advance is the demonstration that the effectiveness of the mammalian TRP channel, TRPM5, which is indirectly activated by sugars, via a G-protein coupled signaling pathway, is inhibited by quinine (Talavera et al., 2008). Thus, TRPM5 may provide one molecular mechanism through which quinine inhibits the attractiveness of sugars.
In Drosophila the molecular mechanism underlying the bitter-sweet interaction is unexplored. In view of an electrophysiological analysis, the site of this interaction is likely to be in the gustatory bristles (sensilla), which house the GRNs and accessory cells, and involve the taste receptors (Meunier et al., 2003). In fly GRNs, the largest class of taste receptors are referred to as gustatory receptors (GRs), which are distantly related to olfactory receptors (ORs) (Clyne et al., 1999; Gao and Chess, 1999; Vosshall et al., 1999; Clyne et al., 2000; Robertson et al., 2003; Scott, 2004; Montell, 2009). The ORs are more extensively characterized than the GRs, and are distinct from mammalian olfactory and taste receptors as the fly ORs are cation channels (Sato et al., 2008; Wicher et al., 2008). Thus, ORs provide the framework for many of the studies focusing on GRs, which may also be cation channels (Sato et al., 2011).
The direct ligand for at least one OR (OR67d) may not be the olfactory cue itself. Rather, there is evidence that the ligand for OR67d is an odorant binding protein (OBP), which is an extracellular protein present in the endolymph (Laughlin et al., 2008). The OBP, referred to as Lush, binds in vitro to OR67d when Lush is bound to a volatile pheromone (Laughlin et al., 2008). The actual receptor complex appears to be comprised of OR67d and a CD36-related protein, SNMP (Laughlin et al., 2008). However, whether Lush serves as the ligand in vivo is unresolved (Gomez-Diaz et al., 2013).
Some OBPs are expressed in gustatory sensilla (McKenna et al., 1994; Pikielny et al., 1994; Ozaki et al., 1995; Galindo and Smith, 2001; Shanbhag et al., 2001; Koganezawa and Shimada, 2002; Sánchez-Gracia et al., 2009; Yasukawa et al., 2010), although the family of 52 OBPs were identified originally in olfactory sensilla and are referred to as “odorant binding proteins”(Vogt and Riddiford, 1981). The roles of most OBPs have not been reported, even in the olfactory system. Mutations affecting two OBPs that are expressed in taste sensilla (OBP57d/e) have been described. However, the contribution of these two OBPs to gustatory behavior appears to be small (Matsuo et al., 2007; Harada et al., 2008). Thus, the functions of OBPs in the gustatory response are largely unknown.
Here, we report an unexpected role for a Drosophila OBP, referred to as OBP49a. Loss of OBP49a had no impact on the production of action potentials in response to any deterrent or attractive compound tested. Rather, OBP49a was expressed in accessory cells and required by sweet activated GRNs for suppression of the attractive sugar responsive by bitter compounds. These findings provide the first molecular handle on the enigmatic phenomenon by which a deterrent compound inhibits the phagostimulatory signal of an attractive tastant in flies.
Results
OBPs are enriched in taste organs
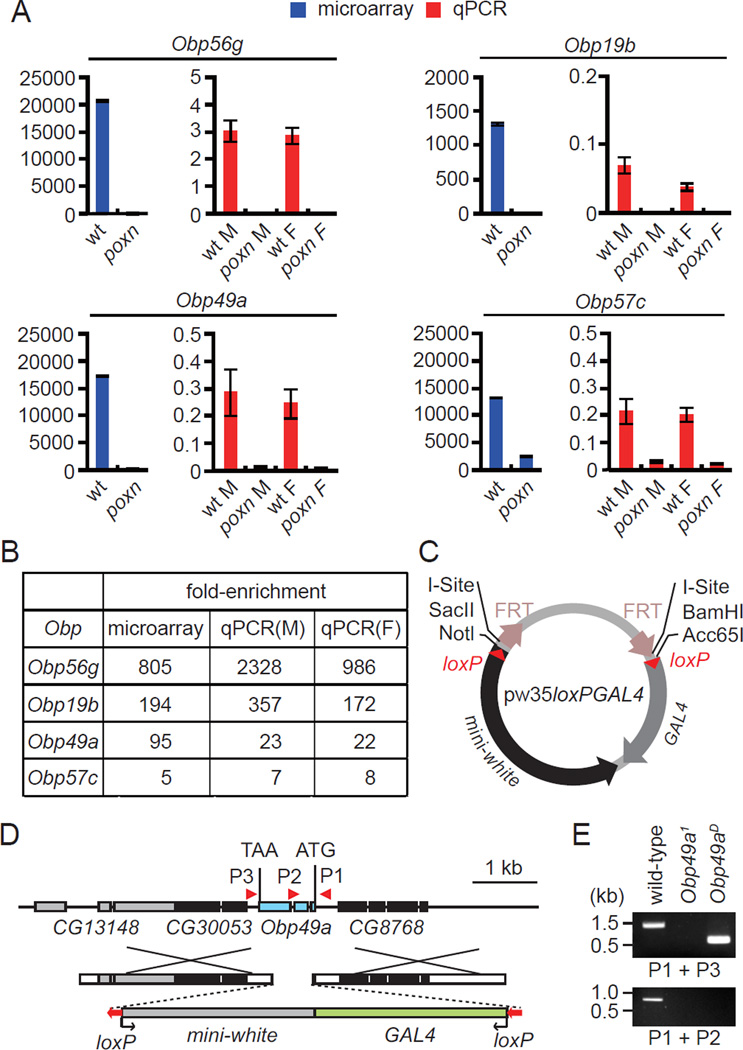
In a previous study, we performed a DNA microarray analysis and identified Drosophila genes that were expressed preferentially in gustatory sensilla on the main taste organ, the labellum (Moon et al., 2009). In this analysis, we found that several genes encoding OBPs were the genes that were the most highly enriched in gustatory sensilla. To evaluate the reliability of the microarray data, we performed quantitative PCR. We prepared total RNA from the labella of control flies (w1118), and from a mutant (poxn) in which the chemosensory bristles were transformed into mechanosensory bristles (Awasaki and Kimura, 1997). The enrichment profiles of each of the four Obp genes examined were similar to the microarray results (Figures 1A–1B). There were no major differences in the expression levels of the Obps in males and females.
Figure 1. Obp genes with enriched expression in the labella, and generation of the Obp49a mutant.
(A) Relative expression of Obp RNAs in the labella of wild-type and poxn mutants. RNA expression was analyzed by microarray analysis (blue bars) and real-time PCR (red bars). The real time PCR analysis was performed using RNA prepared from separate males (M) and females (F). The numbers on the Y-axis are arbitrary units, which indicate relative rather than absolute levels of the various RNAs. Shown are the means ±SEM (n=3, microarray data; n=4—6, real time PCR). (B) Comparison of the enrichment of Obp genes as assessed by microarray and RT-PCR analyses using RNA prepared from males (M) and females (F). (C) The pw35loxPGAL4 vector used to generate the Obp mutants. (D) Schematic of the Obp49a locus and the targeting construct used to generate the Obp49a1 allele. The Obp49aD allele was derived by removing the floxed mini-white and GAL4 genes. The boxes represent exons, and the orientation of the mini-white-GAL4 and loxP are indicated by the arrows. (E) Confirmation of the deletion in Obp49a1 and the excision of the mini-white and the GAL4 in Obp49aD. Genomic DNA was prepared and PCR performed using the primer pairs (P1, P2, and P3) indicated in panel D (see also Figure S1).
Generation of mutations in Obp56g, Obp19b, Obp49a and Obp57c
To survey the roles of OBPs in the gustatory response, we generated mutations affecting four Obp genes. Three of the mutations disrupted Obp56g, Obp19b and Obp49a, which were the genes most enriched in gustatory organs (~100–800-fold; Figure 1A). In addition, we mutated the Obp57c gene, which was enriched in taste sensilla to a lower extent (~5-fold; Figure 1A).
To simultaneously create mutations and gene reporters we used ends-out homologous recombination. We generated a modified targeting vector (pw35loxPGAL4), which included the GAL4 reporter juxtaposed to the mini-white marker gene (Figure 1C). We flanked both genes with loxP sequences to allow for removal of these genes with Cre recombinase. This would provide flexibility in cases in which it would be useful to introduce other reporters in combination with the mutations. We replaced the entire coding regions of Obp56g, Obp19b and Obp49a with the GAL4 and the mini-white coding sequences (Figures 1D and S1A–S1B). To disrupt Obp57c expression, we substituted the start codon with a stop codon so that the neighboring, overlapping gene, Obp57b, would be minimally affected (Figure S1C). We confirmed each Obp knockout by PCR analysis of genomic DNA (Figures 1E and S1A–S1C).
OBP49a deficient flies showed reduced avoidance to sugar/bitter cocktails
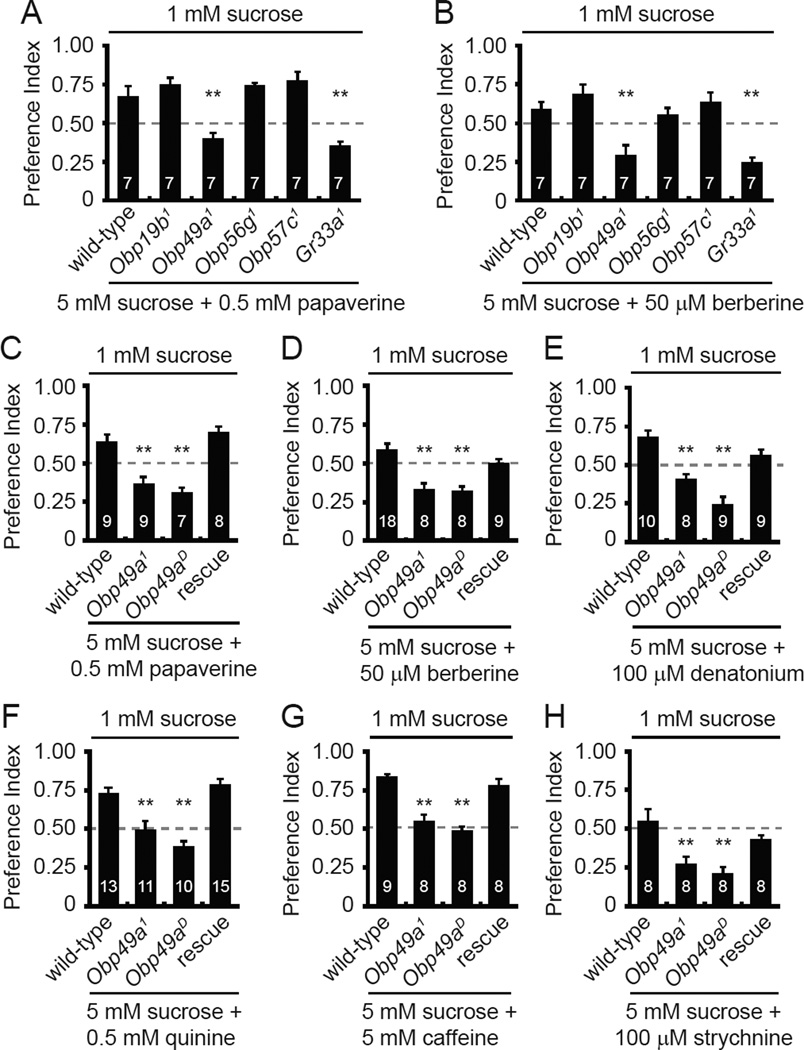
To address whether the Obp mutations affected gustatory behavior, we performed two-way choice assays. The flies were given a choice between 1 mM sucrose and 5 mM sucrose mixed with either red or blue food coloring. After allowing the flies to feed for 90 minutes, we determined the preference indexes. All four mutant flies showed normal preferences for the higher concentration of sucrose (Figure S2A). If bitter compounds are combined with the 5 mM sucrose, wild-type prefer 1 mM sucrose (Moon et al., 2006; Lee et al., 2009). Three of the Obp mutants (Obp56g1, Obp19b1 and Obp57c1) showed normal repulsion to each of the bitter tastants tested (Figures 2A–2B and S2B–S2E). In contrast, mutation of Obp49a impaired the avoidance to a wide array of bitter compounds including papaverine, berberine, denatonium, quinine, caffeine, and strychnine (Figures 2 and S2B–S2E). The only exception was L-canavanine avoidance, which did not depend on any of the Obp mutants tested (Figure S2F). The decreased avoidances to the bitter chemical/5 mM sucrose mixtures were similar to those elicited by mutation of the broadly required gustatory receptor, Gr33a (Figures 2A–2B and S2B–S2D). We obtained Obp49aD flies by excising the GAL4 and white genes. Obp49aD animals displayed the same defects in avoidance to the sucrose/aversive compound cocktails as the Obp49a1 animals (Figures 2C–2H).
Figure 2. Obp49a mutants displayed reduced avoidance to sucrose/bitter mixtures.
Two-way choice behavioral assays were performed by allowing the flies to choose between 1 mM sucrose versus 5 mM sucrose/aversive chemical cocktails. (A and B) Screen for Obps required for gustatory avoidance of papaverine and berberine. Gr33a1 was included as a positive control. (C–H) Testing for requirements for Obp49a for avoidance of a variety of sucrose/bitter cocktails. Both Obp49a1 and Obp49aD showed similar defects in bitter avoidance. To test for rescue of the Obp49aD phenotype, we expressed an Obp49a+ transgene (UAS-Obp49a) in thecogen cells (nompA-GAL4). The numbers of behavioral tests are indicated. Shown are the means ±SEM. The asterisks indicate statistically significant differences (**p<0.01) from wild-type flies (see also Figure S2).
OBP49a is expressed in thecogen cells
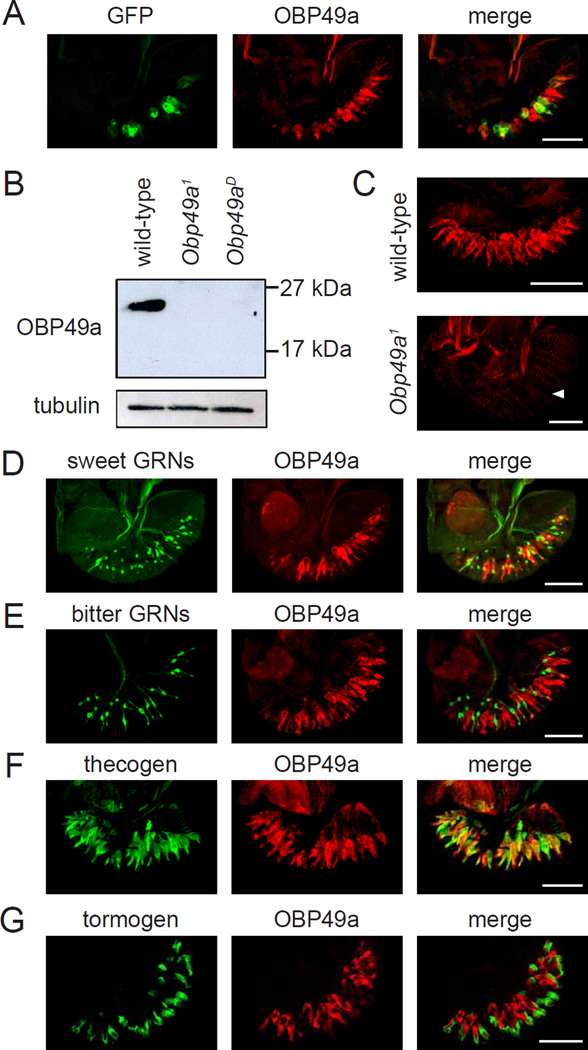
To test for expression of Obp49a in the major taste organ, the labellum, we took advantage of the GAL4 reporter inserted in the Obp49a locus to drive expression of UAS-mCD8::GFP. The GFP was expressed broadly in many cells in the labellum (Figure 3A). To confirm the spatial distribution in the labellum, and to determine the cell type that expressed OBP49a, we raised OBP49a antibodies. The anti-OBP49a recognized a protein of the predicted size (23kDa) in the labellum of wild-type but not in Obp49a1 or Obp49aD (Figure 3B). We performed immunocytochemistry and found that the anti-OBP49a signal was associated with all the chemosensory sensilla in wild-type but not Obp49a1 labella (Figure 3C). We double labeled labella from Obp49a1/UAS-mCD8::GFP flies with anti-GFP and anti-OBP49a. The anti-OBP49a signal was distributed more broadly than the anti-GFP staining (Figure 3A), indicating that the reporter was expressed in a subset of OBP49a positive cells.
Figure 3. Expression of OBP49a in the labellum.
(A) Co-expression of the Obp49a reporter and anti-OBP49a. Obp49a1/UAS-mCD8::GFP labella were stained with GFP and OBP49a antibodies. (B) Western blot using OBP49a antibodies. Extracts were prepared from wild-type, Obp49a1, and Obp49aD labella, and the blot was probed with anti-OBP49a. The blot was also probed with anti-tubulin as a loading control. (C) Staining of labella from wild-type (top) and Obp49a1 flies with anti-OBP49a (bottom). The arrowhead indicates the position of the thecogen cells in the Obp49a1 mutant labellum. (D–G) OBP49a was expressed in thecogen cells. The cellular distribution of OBP49a was examined by comparing the spatial distribution of cell-type-specific markers with anti-OBP49a. (D) Gr5a-GAL4/UAS-mCD8::GFP (sweet GRNs). (E) Gr66a−/−GFP (bitter GRNs). (F) nompA-GAL4/UAS-mCD8::GFP (thecogen). (G) ASE5-GFP (tormogen). Scale bars represent 50 µM
The 31 taste sensilla on each side of the labellum are classified into L-, I-, and S-type depending on their relative position and length (Vosshall and Stocker, 2007; Montell, 2009). L- and S-type sensilla house four GRNs and I-type sensilla contain two GRNs. Each sensillum also has three different types of accessory cells: tricogen (shaft), tormogen (socket), and thecogen (sheath). To address whether OBP49a was expressed in GRNs, we expressed a UAS-mCD8::GFP reporter under control of the Gr5a-GAL4, which labels GRNs that respond to attractive compounds such as sugars (Thorne et al., 2004; Wang et al., 2004). In addition, we used transgenic flies that expressed GFP in GRNs that are activated by aversive compounds such as caffeine and quinine (Gr66a-I-GFP) (Wang et al., 2004). Anti-OBP49a staining did not overlap with either of these markers (Figures 3D–3E) indicating that OBP49a was expressed in other cells in close proximity to cells marked with the Gr5a and Gr66a reporters.
To determine which non-neuronal cell type expressed OBP49a, we used markers that stained either the tormogen (ASE5-GFP) or the thecogen (nompA-GAL4) (Barolo et al., 2000; Chung et al., 2001). Anti-OBP49a was distributed in nompA positive cells, indicating that OBP49a was in thecogen cells, but not in cells expressing the ASE5 reporter (Figures 3F–3G). Moreover, the nompA-GAL4 reporter in combination with UAS-Obp49a restored normal aversion to bitter compound/5 mM sucrose mixtures in an Obp49aD mutant background (Figures 2C–2H).
Requirement for OBP49a for suppression of sucrose-induced action potentials by bitter chemicals
In the olfactory system, the OBP referred to as Lush impacts on the activity of a subset of olfactory receptor neurons (Xu et al., 2005). Thus, in the gustatory system, OBP49a might contribute to the decreased attraction to 5 mM sucrose in the presence of a bitter tastant by affecting the activity of GRNs. If so, OBP49a could act on either of two different types of GRNs. OBP49a could promote the activity of GRNs that respond exclusively to aversive compounds. Alternatively, OBP49a might be needed to suppress GRNs that are activated by sugars.
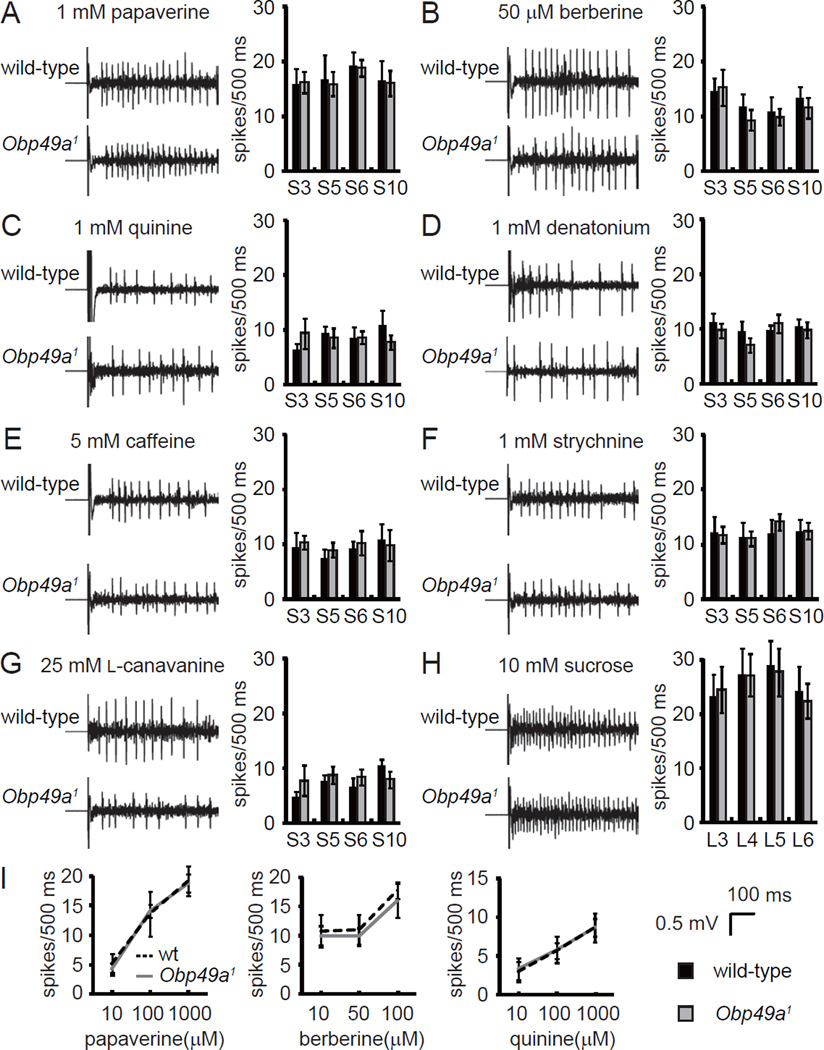
To address whether the activities of either the sugar or bitter-responsive GRNs were altered in Obp49a mutant labella, we performed tip-recordings, which measure nerve firings elicited by tastants. We focused on L-type sensilla to monitor sucrose-induced action potentials, and S-type sensilla to assay the responses to bitter compounds. Application of sucrose to L-type sensilla (L3, L4, L5, and L6), or aversive chemicals to S-type sensilla (S3, S5, S6, and S10), resulted in virtually the same frequencies of action potentials in wild-type and Obp49a1 animals (Figure 4).
Figure 4. Obp49a1 mutant animals showed normal action potential frequencies evoked by bitter tastants.
Tip recordings were performed on the indicated S-type sensilla. The action potential frequencies were based on the number of spikes produced between 50 ms and 550 ms after initiating the stimulation. (A–G) Representative traces of nerve firings from S6 sensilla and quantification of the mean action potentials induced by the indicated concentration of bitter chemicals in S-type sensilla. (A) 1 mM papaverine. (B) 50 µM berberine. (C) 1 mM quinine. (D) 1 mM denatonium. (E) 5 mM caffeine. (F) 1 mM strychnine. (G) 25 mM L-canavanine. (H) Representative traces of nerve firings from L6 sensilla and quantification of the mean action potentials induced by 10 mM sucrose in L-type sensilla. (I) Dose response analysis of the action potential frequencies elicited by papaverine, berberine, and quinine from S6 sensilla. Data are the means ±SEM (n=5—17).
The preceding data indicated that OBP49a was not required for stimulation of GRNs by either sweet or bitter compounds. Therefore, we explored the possibility that OBP49a was required for inhibition of the sweet response by bitter chemicals. L-type sensilla house GRNs that are activated by sugars, water, low salt, and high salt, but they do not respond to bitter chemicals (Hiroi et al., 2004; Weiss et al., 2011), thereby allowing us to assay inhibition of sucrose-elicited spikes by bitter chemicals. As described above, L-type sensilla from either wild-type or Obp49a1 flies displayed robust action potentials in response to 10 mM sucrose. When we exposed wild-type L-type sensilla to 10 mM sucrose combined with bitter chemicals, the responses were inhibited in a dose-dependent manner (Figures 5A–5F). The responses inhibited by bitter chemicals were generated by sugar-activated GRNs rather than water GRNs because we observed the same extent of inhbition by bitter chemicals in flies missing a channel, Pickpocket28 (Ppk28), required for water sensitivity (Figures S3A–S3C) (Cameron et al., 2010; Chen et al., 2010).
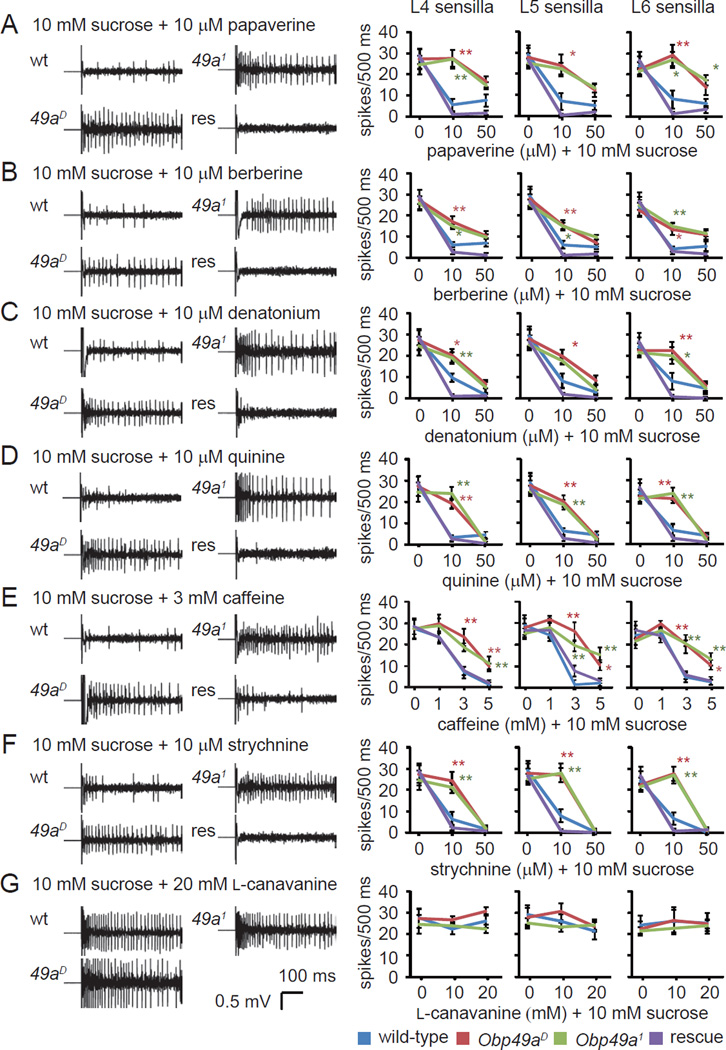
Figure 5. Requirement for Obp49a for inhibition of sucrose-induced action potentials by aversive chemicals.
Tip recordings of L-type sensilla (L4, L5, and L6) showing representative traces and the mean action potentials induced by 10 mM sucrose combined with aversive chemicals. Each stimulant contained 10 mM sucrose, 1 mM KCl as the electrolyte and the indicated concentrations of bitter chemicals. The tip recordings were performed using the indicated sensilla and the following bitter chemicals. (A) Papaverine. (B) Berberine. (C) Denatonium. (D) Quinine. (E) Caffeine. (F) Strychnine. (G) L-canavanine. Shown are the means ±SEM (n=5—13). The genotype of the rescue flies was Obp49aD;nompA-GAL4/UASObp49a. The asterisks denote statistically significant differences (*p<0.05, **p<0.01) from wild-type flies (see also Figure S3).
Of significance here, inhibition of the sucrose-induced action potentials by bitter chemicals was greatly reduced in Obp49a1 and Obp49aD flies (Figures 5A–5F). The impairments in inhibition of sucrose-stimulated nerve firings by aversive chemicals were rescued by expression of wild-type OBP49a in thecogen cells using the GAL4/UAS system (Figures 5A–5F). The contribution of OBP49a to inhibition of the sucrose response by aversive compounds was broad, as the impairments occurred in response to a wide range of aversive tastants. Mutation of Obp49a had no impact on action potentials in L-type sensilla when the sucrose was combined with L-canavanine (Figure 5G). This was expected since L-canavanine did not suppress sucrose-induced nerve firings in wild-type (Figures 5G).
The preceding results suggest that OBP49a is required for inhibiting sucrose-responsive GRs, which may be cation channels (Sato et al., 2011). To test whether bitter compounds and OBP49a might affect the activity of another Drosophila cation channel, we ectopically expressed TRPA1 in sugar responsive GRN under the control of the Gr5a-GAL4. TRPA1 was activated by N-methylmaleimide to the same extent in the presence or absence of either berberine or OBP49a (Figures S3E–S3H).
Since bitter chemicals can inhibit water spikes, we examined whether OBP49a was also involved in the inhibition of water spikes by bitter chemicals (Meunier et al., 2003). Application of 1 mM KCl evoked nerve firings in L-type sensilla from wild-type and Obp49a1 flies but not in the Δppk28 mutant, indicating that the spikes were from water responsive GRNs (Figure S3D). Increasing concentrations of bitter chemicals reduced water spikes in L-type sensiila to the same extent in both wild-type and Obp49a1 flies (Figure S3D). Thus, OBP49a was dispensable for the suppression of the water response by bitter chemicals.
Non-redundant roles for OBP49a and GR33a in sensing bitter chemicals
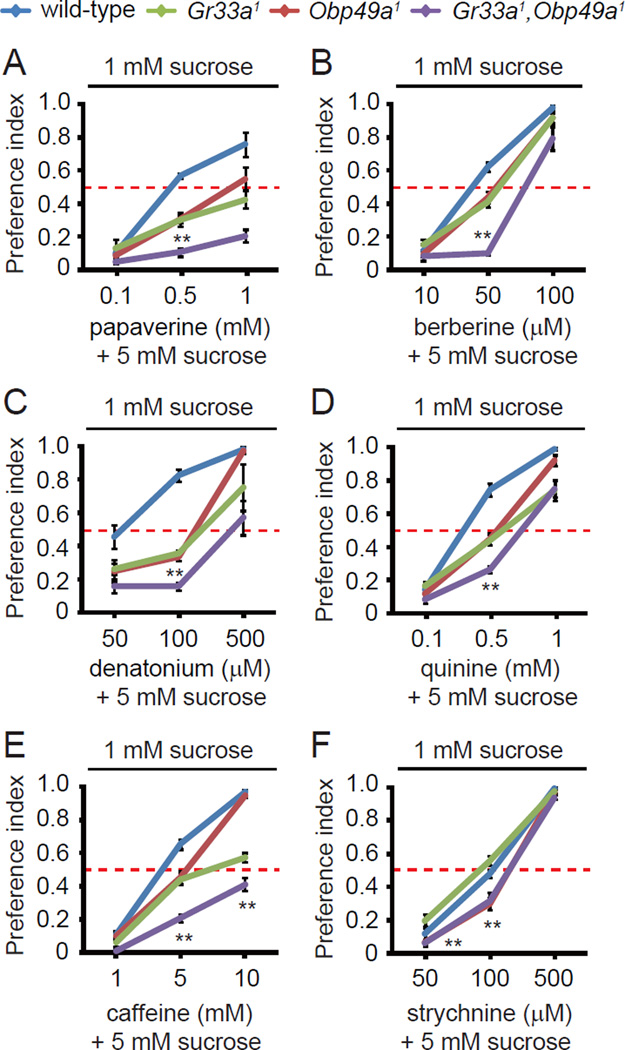
The preceding tip recording analysis indicated that OBP49a participated in suppression of the behavioral attraction to sweet compounds by bitter tastants by attenuating action potentials in sucrose-activated GRNs. The Obp49a1 behavioral phenotype was the same as that displayed by Gr33a1 mutants, even though Gr33a functions in the GRNs in S- and I-type sensilla, which are activated by bitter compounds. Therefore, if Gr33a and Obp49a act on different GRNs, then the Gr33a1,Obp49a1 double mutant should show a more severe phenotype than either the Gr33a1 or Obp49a1 single mutants. Alternatively, if Gr33a and Obp49a acted through a common mechanism in the same GRNs, then the phenotypes of the double and single mutants would be expected to be the same.
We found that the defect in avoidance of the aversive chemical/sucrose cocktail was more severe in the Gr33a1,Obp49a1 double-mutant animals than in Gr33a1 or Obp49a1 flies (Figures 6A–6E). The only exception was with strychnine (Figure 6F), which was consistent with our previous finding that Gr33a1 flies did not display a behavioral defect in strychnine avoidance (Moon et al., 2009). These findings support the conclusion that OBP49a and GR33a are involved in bitter chemical sensing through distinct pathways.
Figure 6. Phenotypes of single and double Obp49a1 and Gr33a1 mutants.
The two-way choice behavioral assays were performed using 1 mM sucrose versus 5 mM sucrose combined with the indicated concentrations of bitter chemicals. (A) Papaverine. (B) Berberine. (C) Denatonium. (D) Quinine. (E) Caffeine. (F) Strychnine. Data are the means ±SEM (n=6—11). The asterisks indicate statistically significant differences between Gr33a1 and Gr33a1,Obp49a1 double mutant flies (**p<0.01).
Rescue of the Obp49a phenotype by tethering OBP49a to GR5a-expressing cells
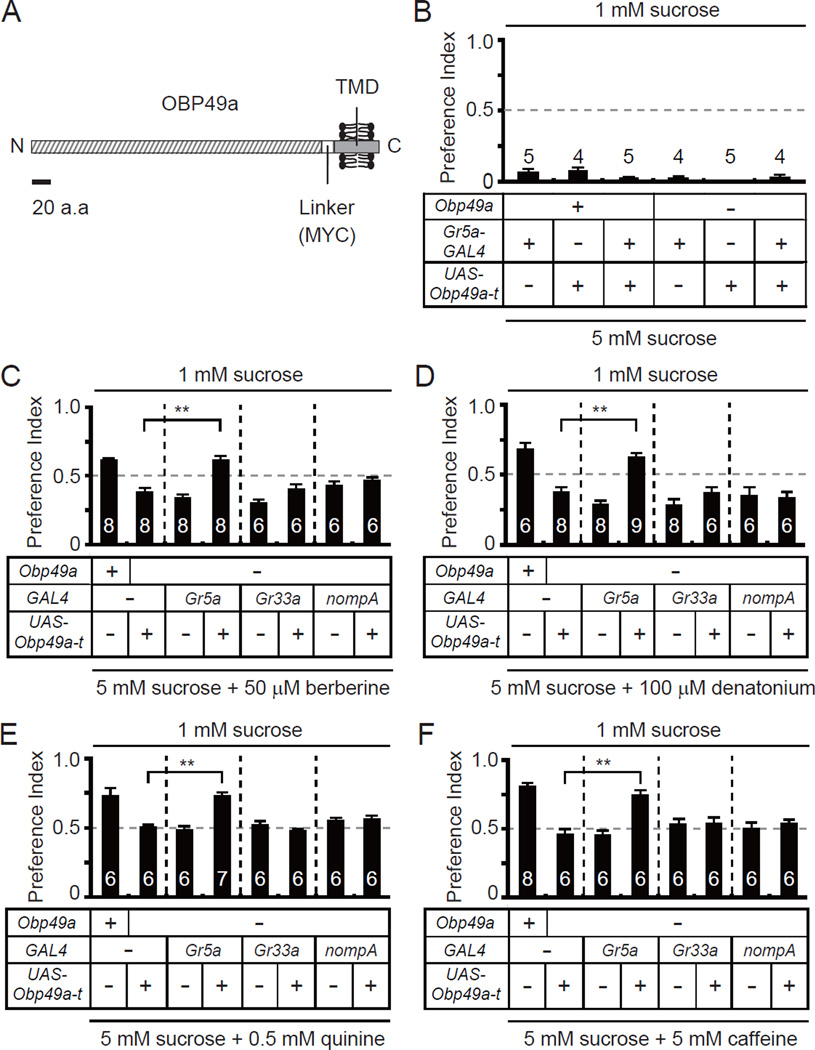
OBPs are secreted into the extracellular endolymph in chemosensory sensilla, and therefore have the potential to function non-cell autonomously. The finding that mutation of Obp49a impaired the suppression of sucrose-induced action potentials by bitter compounds indicated that OBP49a normally acted on sugar-responsive GRNs. To test this proposal, we expressed a membrane-tethered version of OBP49a so that OBP49a would be displayed extracellularly, but remain attached to the expressing cells. To do so, we generated transgenic flies expressing a form of OBP49a that was fused at the C-terminal end to a MYC linker and a transmembrane domain from the platelet derived growth factor receptor (OBP49a-t) (Figure 7A). We used the GAL4/UAS system to express UAS-Obp49a-t in sugar-activated GRNs (Gr5a-GAL4), bitter-activated GRNs (Gr33a-GAL4), or thecogen cells (nompA-GAL4), which synthesize OBP49a.
Figure 7. Membrane tethered OBP49a rescued Obp49aD behavioral phenotype when expressed in sugar sensing GRNs.
(A) Structure of the membrane tethered OBP49a (OBP49a-t). TMD, transmembrane domain. A 20 amino acid scale bar is shown. (B) Testing for effects of OBP49a-t expression on sucrose sensing behavior in sweet sensing GRNs (Gr5a GRNs). The flies were given a choice between 1 mM sucrose and 5 mM sucrose. (C–F) Testing for rescue of the impaired avoidance to sucrose/bitter cocktails in Obp49aD flies by expressing UAS-Obp49a-t under the control of the Gr5a-GAL4 (sweet sensing GRNs), Gr33a-GAL4 (bitter sensing GRNs) or the nompA-GAL4 (thecogen cells). The flies were given a choice between 1 mM sucrose and 5 mM sucrose mixed with a bitter tastant. (C) Berberine. (D) Denatonium. (E) Quinine. (F) Caffeine. Shown are the means ±SEMs. The numbers of assays performed are indicated. The asterisks indicate statistically significant differences between Obp49aD+/UAS-Obp49a-t and Obp49aD;Gr5a-GAL4/UAS-Obp49a-t flies (**p<0.01).
We found that Obp49a-t restored normal suppression of the sucrose response in Obp49aD animals, but only if it was expressed in sugar-activated GRNs. This included normal behavioral suppression (Figures 7C–7F), and inhibition of sucrose-induced action potentials by bitter compounds such as berberine, denatonium, quinine and caffeine (Figures 8A–8C and S4A). This was not due to a non-specific effect of OBP49a-t on sugar-activated GRNs because expression of UAS-Obp49a-t under the control of the Gr5a-GAL4 did not alter either the behavioral or electrophysiological responses to sucrose (Figures 7B and S4B). Expression of Obp49a-t either in GRNs that are activated by bitter compounds or in the thecogen cells did not rescue the Obp49aD phenotype (Figure 7).
Figure 8. Cellular requirement for OBP49a-t, and testing for interactions between OBP49a and bitter tastants and GR64a.
(A–C) Tip-recordings were performed using the indicated sensilla and genotypes. Shown are representative traces and the mean numbers of action potentials (n=8—11) induced by 10 mM sucrose plus the indicated bitter tastant. (A) 10 µM berberine. (B) 10 µM denatonium. (C) 10 µM quinine. The asterisks indicate statistically significant differences between Obp49aD+/UAS-Obp49a-t and Obp49aD;Gr5a-GAL4/UAS-Obp49a-t flies (**p<0.01). (D–G) Sensorgrams showing binding between bitter tastants and OBP49a using the BIAcore system. The indicated concentrations of tastants were allowed to interact with the OBP49a, which was bound to CM5 chips. (R.U., relative resonance units). (H–K) Protein complementation assay using the split YFP approach. Labella were dissected from transgenic flies expressing the indicated YFP(1) and YFP(2) fusion proteins in Gr5a GRNs, under control of the GAL4/UAS system. In come cases, 100 µM berberine was applied to sensilla for 1 minute before dissection. The scale bar in (H) represents 10 µm, and applies to all panels in (H–K) (see also Figure S4).
Binding of bitter compounds to OBP49a
The requirement for OBP49a for bitter-induced suppression of the sugar response raised the possibility that it binds to aversive tastants. To test for direct interactions of OBP49a with bitter chemicals, we employed surface plasmon resonance. We ectopically expressed UAS-Obp49a in compounds eyes under the control of the GMR-GAL4, purified OBP49a from head extracts, and coupled the protein to sensor chips. We found that berberine, denatonium and quinine bound to OBP49a in a dosedependent manner (Figures 8D–8F). In contrast, sucrose did not bind to OBP49a (Figure 8G), suggesting that OBP49a specifically interacted with bitter chemicals.
Close association of OBP49a and GR64a
The OBP49a-dependent suppression of the sucrose response by bitter compounds suggested that OBP49a might physically interact with the sucrose receptor. At least two GRs are required for sucrose detection. These include GR64a (Dahanukar et al., 2007; Jiao et al., 2007), as well as GR64f, which is required for sensing nearly all sugars, including sucrose, and may be a co-receptor for sugar-responsive GRs (Jiao et al., 2008).
To test whether OBP49a was in close proximity to GR64a or GR64f (Dahanukar et al., 2007; Jiao et al., 2007) and might therefore associate directly, we employed a YFP-based protein complementation assay. YFP can be split into two complementing fragments, and fluorescence is generated only when the separated parts are brought together. To address whether OBP49a was juxtaposed or interacted with either GR64a or GR64f in vivo, we generated UAS-transgenes encoding the N-terminal YFP fragment, YFP(1), fused to the N-termini of GR64a and GR64f, and the C-terminal YFP fragment YFP(2) linked to the C-termini of OBP49-t. As a control we used a previously described transgene UAS-SNMP1:YFP(2), which encoded YFP(2) linked to a CD36-related receptor, SNMP1 (Benton et al., 2007). SNMP1 functions in pheromone detection in olfactory receptor neurons (Benton et al., 2007). We expressed these constructs in sugar-responsive GRNs under control of the Gr5a-GAL4.
We assayed for YFP-based protein complementation by dissecting labella from the transgenic flies, and performing confocal microscopy. There was no fluorescence visible in labella isolated from flies harboring the transgenes encoding just a single YFP(1) or YFP(2) fusion protein, such as YFP(1):Gr64a or OBP49a-t-YFP(2) (Figure 8H). In contrast, co-expression of YFP(1):GR64a and OBP49a-t-YFP(2) in sugar responsive GRNs produced a strong signal (Figure 8I). To address whether a bitter chemical might enhance the fluorescence, we dipped the labella from immobilized flies in berberine before dissecting the labella. However, exposure to this bitter tastant had no impact on the fluorescence (Figure 8I). We did not detect signals when we expressed OBP49a-t-YFP(2) with YFP(1):GR64f (Figure 8J). The combination of YFP(1):GR64a with SNMP1-YFP(2) also did not produce fluorescence (Figure 8K). These findings support the conclusion that OBP49a either interacts with or is adjacent to GR64a.
Discussion
Many bitter-tasting chemicals are toxic (Glendinning, 2007). Therefore, the ability of animals to suppress their attraction to sugars and other nutritious foods that are laced with bitter tastants is critical for survival. Consequently, this avoidance behavior is conserved throughout the animal kingdom. Nevertheless, the molecules and molecular mechanisms through which positive feeding behavior is inhibited by deterrant compounds are poorly unexplored in most animals, such as the fruit fly.
Potentially, there are multiple neural mechanisms that could explain how aversive tastants suppress the otherwise stimulatory effects of sweeteners and other attractive compounds. Animals ranging from flies to humans have separate taste receptor cells devoted to sensing bitter and sweet tastants, and the suppression of sweet by bitter compounds could take place through integration of separate inputs in the brain. There could also be lateral interactions in the periphery between separate sweet and bitter responsive afferent receptor cells. Alternatively, a bitter compound might directly suppress sweet-activated taste receptor cells.
In this study, we unexpectedly identified a mechanism through which an array of bitter compounds inhibited the stimulatory effects of sucrose in flies. This work emerged from a functional analysis of OBPs in Drosophila taste, and was motivated by the finding that multiple Obp genes were highly enriched in taste sensilla (McKenna et al., 1994; Pikielny et al., 1994; Ozaki et al., 1995; Galindo and Smith, 2001; Shanbhag et al., 2001; Koganezawa and Shimada, 2002; Sánchez-Gracia et al., 2009; Yasukawa et al., 2010), but their roles in the gustatory system were largely unexplored. We found that OBP49a was required for avoiding bitter-tasting compounds in a standard two-way choice assay consisting of 1 mM sucrose alone versus 5 mM sucrose plus bitter tastants. Because wild-type flies find bitter tastants aversive, they prefer the lower concentration of sucrose, when the higher concentration of sucrose is laced with tastants such as berberine, quinine or denatonium. However, the Obp49a mutant animals were impaired in this avoidance behavior. The phenotype was similar to that resulting from elimination of GRs, such as GR33a and GR66a, which are broadly expressed in avoidance GRNs, and are necessary in these GRNs for induction of bitter-induced action potentials (Moon et al., 2006; Lee et al., 2009; Moon et al., 2009). Unlike the Gr33a and Gr66a mutants, the decreased avoidance in the Obp49a1 animals was not due to impairment of aversive tastant-induced action potentials in Gr66a-expressing GRNs.
We conclude that the decreased behavioral avoidance to sucrose/aversive chemicals mixture in Obp49a mutant flies was due to a deficit in the sugar-activated GRNs that express Gr5a, and not due to effects on GRNs activated by bitter compounds. In support of the conclusion, the Gr5a- and Gr66a-expressing GRNs from Obp49a1 sensilla produced normal action potential frequencies in response to sugars and bitter compounds, respectively. However, when sucrose was combined with bitter tastants, the normal inhibition of the sugar-induced nerve firings was strongly impaired. OBP49a is therefore the first molecule shown to promote the inhibition of the sucrose response by aversive chemicals in Drosophila.
We propose that OBP49a, which is synthesized and secreted by thecogen cells into the endolymph, acts directly on sugar-activated (Gr5a-expressing) cells to inhibit sugarinduced action potentials, in response to bitter compounds. Further supporting this proposal, we rescued the Obp49aD mutant phenotype with a membrane tagged version of OBP49a (OBP49a-t) that was restricted to the external surface of Gr5a-expressing cells, but not if OBP49a-t was confined to thecogen cells or Gr66a-expressing cells. Even though OBP49a was normally produced in thecogen cells, we were not able to rescue the Obp49a mutant phenotype if we expressed OBP49a-t in these accessory cells. Conversely, expression of an untagged version of OBP49a in thecogen cells fully restored normal inhibition of sucrose-induced action potentials by bitter compounds. These findings demonstrate that OBP49a was required non-cell autonomously by sugar-responsive cells.
Our findings indicate that at least one important cellular mechanism through which bitter and sweet taste is integrated, occurs in the taste receptor neurons. However, the current study does not exclude that there may also be integration of bitter and sugar activated signals in the brain. While Obp49a mutants showed a significant deficit in suppression of the sugar response by bitter compounds, the inhibition was not eliminated at the highest concentrations of the noxious tastants. Thus, there is an additonal mechanism that contributes to inhibition of the sucrose response by bitter chemicals.
Mammals appear to use a similar cellular strategy for suppressing the appeal of sugars, when they are mixed with the bitter compound, quinine. In mammalian taste buds, sugars activate G-protein coupled receptors, and the signaling pathway culminates with activation of the TRPM5 channel (Pérez et al., 2002; Zhang et al., 2003). Quinine has a profound effect on inhibiting TRPM5, thereby reducing the attraction to sugars (Talavera et al., 2008). However, this TRPM5 mechanism may be relatively specific to quinine, since another bitter compound, denatonium, is ~100-fold less effective at inhibiting TRPM5 (Talavera et al., 2008). By contrast, the OBP49a-dependent mechanism described here participates in the inhibition of the sucrose response to a wide array of aversive tastants.
There are at least two possible molecular mechanisms through which localization of OBP49a at the cell surface of sugar-responsive GRNs inhibits these neurons. OBP49a binds directly to bitter compounds and either interacts with or lies in close proximity to the sucrose receptor, GR64a. According to one possibility, OBP49a might deliver bitter chemicals to the cell surface of sugar-activated GRs, thereby greatly increasing the local concentration of bitter chemicals. The bitter chemicals might then bind to sugar-activated GRs, causing them to change from a high affinity state to low affinity state for sugars. Alternatively, the bitter chemicals might not bind direclty to sugar-activated GRs, even at very high concentrations. Rather, once bound to bitter tastants, OBP49a might undergo a conformational change that in turn inhibits the GR64a complex. Since GRs may be cation channels (Sato et al., 2011), OBP49a might provide insects a mechanism by which bitter compounds suppress sugar-activated cation conductances.
Experimental Procedures
Fly stocks
All fly stocks were maintained on conventional cornmeal-agar-molasses medium, 12-h light/12-h dark cycles at 25°C and 60 % humidity. 70FLP,70I-Scel/CyO, Sco/CyO,P[w+,Cre], UAS-mCD8::GFP flies were obtained from the Bloomington Stock Center. Gr5a-GAL4 and Gr66a−/−GFP were from K. Scott. ASE5-GFP, nompA-GAL4, and UAS-SNMP1-YFP(2) were provided by J.W. Posakony, Y.D. Chung, and L. Vosshall, respectively.
Construction of the pw35loxPGAL4 vector
To generate pw35loxPGAL4, we modified the pw35GAL4 vector (Moon et al, 2009) We inserted loxP oligonucleotides into the NotI and Acc65I sites. Each oligonucleotide also included portions of the NotI and Acc65I sites so that these two restriction sites were preserved. The orientation of the loxP sequences were in same orientation so that we could remove the floxed mini-white and the GAL4 coding sequences after genetically introducing the Cre recombinase.
Generation of the Obp mutants
To generate the Obp19b1, Obp49a1, and Obp56g1 alleles, we PCR amplified 3 kb genomic DNAs encompassing both the 5’ and 3’ ends of the Obp coding sequences from isogenic w1118 flies. The genomic fragments were selected to introduce deletions of 930, 759 and 465 bp, respectively. To produce the OBP57c1 allele, we PCR amplified from isogenic w1118 flies a 3 kb of genomic DNA extending from the 5’ end of the start codon, and a 3 kb genomic DNA extending from the 3’side of the start codon. This latter DNA included a stop codon at codon position one. Each homologous arm was subcloned into the pw35loxPGAL4 vector. The transgenic flies were generated by first obtaining random insertions of the transgenes (BestGene Inc., Chino Hills, CA), and then by mobilizing the transgenes and screening for targeted insertions as described previously (Gong and Golic, 2003). Each Obp mutation was confirmed by genomic PCR. The primer information used to confirm the deletions are: 5’-ACATCGTCGAGATGGTGCTGAACA-3’ and 5’-TCCGCATCTGGAATACACTTCGCT-3’ for the Obp19b1 deletion. 5’-TGACTTGCACACGCTGTGAATACG-3’ (P1), 5’-TACCCACAATCCGTTCAGT-3’ (P2), and 5’-ATACAGCCAGCATTTCCTTTGCGG-3’ (P3) for the Obp49a1 deletion. 5’-TGTCCAAGGAACTGGTGACGGATT-3’ and 5’-TGGCTGCCTGGAAGCAGGATAATA-3’ for the Obp56g1 deletion. 5’-TTCAGAACCGCCTTTCTGCGAGTA-3’ and 5’-TGGCCAGGCAATCTGACACGTAAT-3’ for the Obp57c1 deletion. To generate the Obp49aD allele, the Obp49a1 flies were crossed to flies containing the P[w+,Cre] transgene. The mosaic-eyed progeny were collected and crossed to balancer flies, and the white-eyed flies progeny of this latter cross were subjective to genomic PCR analysis using primers P1 and P3.
Creation of flies expressing a membrane tethered OBP49a-t
To generate the UAS-Obp49a-t transgenic flies, we first amplified the Obp49a coding sequence lacking the translation stop codon from w1118 labellar cDNA using the Hifidelity PCR kit (Roche), and cloned the cDNA into the pUAST vector. Sequences encoding the 10 amino acid MYC linker (EQKLISEEDL) and the transmembrane domain from the platelet derived growth factor receptor were amplified from the pDisplay vector (Invitrogen), and cloned in-frame 3’ to the coding region for Obp49a. We also subcloned the cDNA encoding Obp49a with a normal stop codon and without the sequences encoding MYC and the membrane-tethered tag (UAS-Obp49a) into the pUAST vector. The transgenic flies were generated by BestGene Inc. (Chino Hills, CA).
Real time PCR analysis
We extracted total RNA from the labella of adult male and female wild-type and poxn flies using the Trizol reagent (Invitrogen, Carlsbad, CA), and generated cDNAs from 0.5 µg RNA using the SuperScript III First Strand Synthesis System (Invitrogen). Quantitative PCR was performed using an ABI7500 real-time PCR machine (Applied Biosystems, Foster City, CA) and the ABI SYBR green system. Transcript levels were normalized to rp49 as an internal control and the ΔΔCT (CT = threshold cycle) method was used to calculate the relative amount of mRNAs. We repeated the experiments at least four times.
Anti-OBP49a antibodies and Western blots
Rabbit polyclonal OBP49a antibodies were raised to a synthetic peptide (CKPPRGPPPSAEDM; amino acid 199–212). 20 labella were dissected from wild-type, Obp49a1, and Obp49aD flies and homogenized in 1X SDS sample buffer with a pellet pestles (Kimble-Kontes). The extracts were subjected to electrophoresis by SDS-PAGE and transferred to PVDF membranes (Millipore). The membranes were probed with primary antibodies against OBP49a (1:1,000) and Tubulin (1:3,000, 12G10 from Hybridoma Bank), and then with peroxidase-conjugated anti-mouse or rabbit IgG secondary antibodies (1:5,000, Sigma).
Immunohistochemistry
Whole mount fly labellar immunostaining was performed as described previously (Moon et al., 2009) using anti-OBP49a (1:400) and mouse anti-GFP (1:400, Molecular Probes) primary antibodies, and anti-mouse-Alexa488 (1:400, Molecular Probes) and anti-rabbit-Alexa568 (1:400, Molecular Probes) secondary antibodies. The stained samples were mounted with Vectashield (Vector Laboratories, Burlingame, CA) and visualized with a Zeiss LSM700 confocal microscope (Jena, Germany).
Chemicals
Sucrose, denatonium, quinine, papaverine, caffeine, strychnine, L-canavanine, sulforhodamine B and KCl were purchased from Sigma-Aldrich (Saint Louis, MO). Berberine sulfate trihydrate, and Brilliant Blue FCF were obtained from Wako Pure Chemical Industries, Ltd (Osaka, Japan).
Two-way choice behavioral tests
The binary food choice assays were performed as described previously (Meunier et al., 2003; Moon et al., 2006). Briefly 3–6 days old flies were starved for 18 hours and then placed in 72 well microtiter dishes. Each alternating well was filled with 1% agarose combined with one of two types of test mixtures. To perform the sucrose test, the wells contained either 5 mM or 1 mM sucrose. The aversion to bitter chemicals was assayed by comparing the preferences for 1 mM sucrose versus 5 mM sucrose plus the indicated concentrations of aversive compounds. To monitor food intake, one test mixture contained blue dye (Brilliant Blue FCF, 0.125 mg/ml) while the other contained red dye (sulforhodamine B, 0.2 mg/ml). After allowing the flies to feed for 90 minutes at room temperature in the dark, the animals were frozen at −20°C. The numbers of flies that were blue (NB), red (NR) or purple (NMIX) were determined under a dissection microscope and the preference index (P.I.) values were calculated according to the following equation: (NR+0.5NMIX)/(NR+NB+NMIX). P.I.s=1.0 and 0 indicated complete preferences for one or the other food alternative. A P.I.=0.5 indicated no preference.
Electrophysiology
Tip recordings (Hodgson et al., 1955; Wieczorek and Wolff, 1989) were performed as described previously (Moon et al., 2006). 1-day-old flies, which were kept on fresh fly food after eclosion, were immobilized by inserting a glass capillary, which was filled with Ringer’s solution, into the abdomen through to the head. This electrode also served as a reference electrode. The indicated labellar sensilla were stimulated with a recording electrode (10—20 µm tip diameter) containing the test tastants in 1mM KCl as the electrolyte. The recording electrode was connected to a preamplifier (TastePROBE, Syntech, Hilversum, The Netherlands). The signals were collected and amplified (10×) using a signal connection interface box (Syntech) in conjunction with a 100—3000 Hz band-pass filter. The inputs were also linked to a loudspeaker to facilitate audio monitoring. Recordings of action potentials were acquired at a 12-kHz sampling rate and analyzed with Autospike 3.1 software (Syntech). The spikes were sorted based on their amplitude for further quantitative analyses.
OBP49a expression and purification
OBP49a was expressed in fly eyes under the control of the long GMR-GAL4 (Wernet et al., 2003). The fly heads expressing OBP49a in eyes were separated from bodies by agitation of frozen flies. 10 ml of collected fly heads were homogenized in 25 ml of 10 mM Tris-HCl, pH 7.4, 10 % glycerol, using a motor-driven homogenizer and further homogenized with a Dounce homogenizer. The extracts were centrifuged for 10 min at 3,000 ×g, and the supernatants were ultracentrifuged at 4°C for 60 min at 100,000 ×g. OBP49a was purified by serial use of the HiTrap SP XL 5 ml and HiTrap Q XL 5 ml columns (GE Healthcare), followed by affinity purification with OBP49a antibodies. The purity of OBP49a was assessed by fractionation of the protein by SDS-PAGE and silver staining (Figures S4C and S4D).
Surface Plasmon Resonance (SPR) binding experiments
SPR was conducted using a BIAcore 3000 (GE Healthcare) at 25°C. Coupling of OBP49a to CM5 chips (GE Healthcare) was performed by injecting 0.1 µg/ml of protein with 10 mM sodium acetate, pH 4.0 at a 5 µl/min flow rate, and confirmed by an increase of 10,000 resonance units on the sensor chip. The chemicals were diluted to the indicated concentrations in the continuous flow buffer HBS-P (10 mM HEPES pH 7.4, 150 mM NaCl, 0.005 % Surfactant P20). Each analytic run was performed at 30 µl/min flow rate. The chip matrix was regenerated using 20 mM NaOH after each binding analysis.
Protein complementation assay (PCA)
To obtain the UAS-YFP(1):Gr64a, UAS-YFP(1):Gr64f transgenic flies for the PCA, we first generated pUAST-YFP(1) by PCR amplifying a 462 base pair YFP(1) fragment from pAKAR3EV (Komatsu et al., 2011) that extended from the Kozak sequence. This fragment was subcloned between the EcoRI and KpnI sites of pUAST. We then inserted the coding sequences of Gr64a and Gr64f into pUAST-YFP(1), so that YFP(1) was linked to the N-termini of the GRs. To produce the pUAST-OBP49a-t-YFP(2) construct, we used pUAST-Obp49a-t to PCR amplify the OBP49a-t coding sequence that lacked the stop codon, and then inserted the fragment into pUAST. We then used pAKAR3EV as the template to PCR amplify two pieces of DNA fragments encoding a 116 amino acid long flexible EV linker and YFP(2), which encoded residues 155 to 237 of YFP. We inserted these DNA fragments adjacent to the 3’ end of the OBP49a-t coding region. We expressed these transgenes, as well as UAS-Snmp1-YFP(2) (Benton et al., 2007), under the control of the Gr5a-GAL4. To apply berberine to the sensilla, we immobilized the flies with a glass capillary and dipped the labella into a solution containing 100 µM berberine for one minute before dissecting the labella. We also immersed labella in 100 µM berberine/100 mM sucrose solutions, and obtained indistinguishable results from that generated with untreated labella, or labella dipped in berberine only (data not shown). The labella were fixed with 4% paraformaldehyde in PBS-T (0.2% Triton X-100 in PBS) for 20 min. Fixed labella were washed with PBS-T three times and cut in half with a razor blade and then, mounted in VECTASHELD (Vector Laboratories). Fluorescence was viewed in whole mounts of labella using a Zeiss LSM700 confocal microscope.
Data analyses
All error bars represent SEMs. Unpaired Student’s t-tests were used to compare two sets of data. ANOVA with the Tukey post hoc tests were used to compare multiple sets of data. Asterisks indicate statistical significance (*p<0.05, **p<0.01).
Supplementary Material
Highlights.
Control of sweet/bitter taste interaction by an odorant binding protein.
Inhibition of sweet-activated gustatory neurons by an odorant binding protein.
Bitter compounds inhibit the sweet response by binding to OBP49a.
Acknowledgments
We thank the Bloomington Stock Center, and Drs. K. Scott, J.W. Posakony and Y.D. Chung for fly stocks, and Dr. C.Y. Park and Mr. J. Song at Surface Plasmon Resonance Facility, UNIST for technical help. The tubulin monoclonal antibody developed by Drs. Frankel and Nelsen was obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the NICHD and maintained by The University of Iowa, Department of Biology, Iowa City, IA 52242. This work was supported by a grant to C.M. from the NIDCD (DC007864) and by a grant to S.J.M. from the Converging Research Center Program funded by the Ministry of Education, Science, and Technology (2012K001350) and from the Basic Science Research Program through the National Research Foundation of Korea funded by the Ministry of Education, Science, and Technology (2009-0075341 and 2012R1A1A1012081).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Awasaki T, Kimura K. pox-neuro is required for development of chemosensory bristles in Drosophila . J. Neurobiol. 1997;32:707–721. doi: 10.1002/(sici)1097-4695(19970620)32:7<707::aid-neu6>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- Barolo S, Walker RG, Polyanovsky AD, Freschi G, Keil T, Posakony JW. A Notch-independent activity of Suppressor of Hairless is required for normal mechanoreceptor physiology. Cell. 2000;103:957–969. doi: 10.1016/s0092-8674(00)00198-7. [DOI] [PubMed] [Google Scholar]
- Benton R, Vannice KS, Vosshall LB. An essential role for a CD36-related receptor in pheromone detection in Drosophila. Nature. 2007;450:289–293. doi: 10.1038/nature06328. [DOI] [PubMed] [Google Scholar]
- Cameron P, Hiroi M, Ngai J, Scott K. The molecular basis for water taste in Drosophila. Nature. 2010;465:91–95. doi: 10.1038/nature09011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Wang Q, Wang Z. The amiloride-sensitive epithelial Na+ channel PPK28 is essential for Drosophila gustatory water reception. J. Neurosci. 2010;30:6247–6252. doi: 10.1523/JNEUROSCI.0627-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung YD, Zhu J, Han Y, Kernan MJ. nompA encodes a PNS-specific, ZP domain protein required to connect mechanosensory dendrites to sensory structures. Neuron. 2001;29:415–428. doi: 10.1016/s0896-6273(01)00215-x. [DOI] [PubMed] [Google Scholar]
- Clyne PJ, Warr CG, Carlson JR. Candidate taste receptors in Drosophila . Science. 2000;287:1830–1834. doi: 10.1126/science.287.5459.1830. [DOI] [PubMed] [Google Scholar]
- Clyne PJ, Warr CG, Freeman MR, Lessing D, Kim J, Carlson JR. A novel family of divergent seven-transmembrane proteins: candidate odorant receptors in Drosophila . Neuron. 1999;22:327–338. doi: 10.1016/s0896-6273(00)81093-4. [DOI] [PubMed] [Google Scholar]
- Dahanukar A, Lei YT, Kwon JY, Carlson JR. Two Gr genes underlie sugar reception in Drosophila . Neuron. 2007;56:503–516. doi: 10.1016/j.neuron.2007.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Formaker BK, MacKinnon BI, Hettinger TP, Frank ME. Opponent effects of quinine and sucrose on single fiber taste responses of the chorda tympani nerve. Brain Res. 1997;772:239–242. doi: 10.1016/s0006-8993(97)00845-7. [DOI] [PubMed] [Google Scholar]
- Galindo K, Smith DP. A large family of divergent Drosophila odorant-binding proteins expressed in gustatory and olfactory sensilla. Genetics. 2001;159:1059–1072. doi: 10.1093/genetics/159.3.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Q, Chess A. Identification of candidate Drosophila olfactory receptors from genomic DNA sequence. Genomics. 1999;60:31–39. doi: 10.1006/geno.1999.5894. [DOI] [PubMed] [Google Scholar]
- Glendinning JI. How do predators cope with chemically defended foods? Biol Bull. 2007;213:252–266. doi: 10.2307/25066643. [DOI] [PubMed] [Google Scholar]
- Gomez-Diaz C, Reina JH, Cambillau C, Benton R. Ligands for pheromone-sensing neurons are not conformationally activated odorant binding proteins. PLoS Biol. 2013;11:e1001546. doi: 10.1371/journal.pbio.1001546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong WJ, Golic KG. Ends-out, or replacement, gene targeting in Drosophila . Proc. Natl. Acad. Sci. USA. 2003;100:2556–2561. doi: 10.1073/pnas.0535280100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada E, Haba D, Aigaki T, Matsuo T. Behavioral analyses of mutants for two odorant-binding protein genes, Obp57d and Obp57e, in Drosophila melanogaster . Genes Genet. Syst. 2008;83:257–264. doi: 10.1266/ggs.83.257. [DOI] [PubMed] [Google Scholar]
- Hiroi M, Meunier N, Marion-Poll F, Tanimura T. Two antagonistic gustatory receptor neurons responding to sweet-salty and bitter taste in Drosophila . J. Neurobiol. 2004;61:333–342. doi: 10.1002/neu.20063. [DOI] [PubMed] [Google Scholar]
- Hodgson ES, Lettvin JY, Roeder KD. Physiology of a primary chemoreceptor unit. Science. 1955;122:417–418. doi: 10.1126/science.122.3166.417-a. [DOI] [PubMed] [Google Scholar]
- Jiao Y, Moon SJ, Montell C. A Drosophila gustatory receptor required for the responses to sucrose, glucose, and maltose identified by mRNA tagging. Proc. Natl. Acad. Sci. USA. 2007;104:14110–14115. doi: 10.1073/pnas.0702421104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y, Moon SJ, Wang X, Ren Q, Montell C. Gr64f is required in combination with other gustatory receptors for sugar detection in Drosophila . Curr. Biol. 2008;18:1797–1801. doi: 10.1016/j.cub.2008.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koganezawa M, Shimada I. Novel odorant-binding proteins expressed in the taste tissue of the fly. Chem. Senses. 2002;27:319–332. doi: 10.1093/chemse/27.4.319. [DOI] [PubMed] [Google Scholar]
- Komatsu N, Aoki K, Yamada M, Yukinaga H, Fujita Y, Kamioka Y, Matsuda M. Development of an optimized backbone of FRET biosensors for kinases and GTPases. Mol. Biol. Cell. 2011;22:4647–4656. doi: 10.1091/mbc.E11-01-0072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laughlin JD, Ha TS, Jones DN, Smith DP. Activation of pheromone-sensitive neurons is mediated by conformational activation of pheromone-binding protein. Cell. 2008;133:1255–1265. doi: 10.1016/j.cell.2008.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Moon SJ, Montell C. Multiple gustatory receptors required for the caffeine response in Drosophila . Proc. Natl. Acad. Sci. USA. 2009;106:4495–4500. doi: 10.1073/pnas.0811744106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo T, Sugaya S, Yasukawa J, Aigaki T, Fuyama Y. Odorant-binding proteins OBP57d and OBP57e affect taste perception and host-plant preference in Drosophila sechellia . PLoS Biol. 2007;5:e118. doi: 10.1371/journal.pbio.0050118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna MP, Hekmat-Scafe DS, Gaines P, Carlson JR. Putative Drosophila pheromone-binding proteins expressed in a subregion of the olfactory system. J. Biol. Chem. 1994;269:16340–16347. [PubMed] [Google Scholar]
- Meunier N, Marion-Poll F, Rospars JP, Tanimura T. Peripheral coding of bitter taste in Drosophila . J. Neurobiol. 2003;56:139–152. doi: 10.1002/neu.10235. [DOI] [PubMed] [Google Scholar]
- Montell C. A taste of the Drosophila gustatory receptors. Curr. Opin. Neurobiol. 2009;19:345–353. doi: 10.1016/j.conb.2009.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon SJ, Köttgen M, Jiao Y, Xu H, Montell C. A taste receptor required for the caffeine response in vivo. Curr. Biol. 2006;16:1812–1817. doi: 10.1016/j.cub.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Moon SJ, Lee Y, Jiao Y, Montell C. A Drosophila gustatory receptor essential for aversive taste and inhibiting male-to-male courtship. Curr. Biol. 2009;19:1623–1627. doi: 10.1016/j.cub.2009.07.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa K, Marui T, Caprio J. Quinine suppression of single facial taste fiber responses in the channel catfish. Brain Res. 1997;769:263–272. doi: 10.1016/s0006-8993(97)00729-4. [DOI] [PubMed] [Google Scholar]
- Ozaki M, Morisaki K, Idei W, Ozaki K, Tokunaga F. A putative lipophilic stimulant carrier protein commonly found in the taste and olfactory systems. A unique member of the pheromone-binding protein superfamily. Eur. J. Biochem. 1995;230:298–308. doi: 10.1111/j.1432-1033.1995.0298i.x. [DOI] [PubMed] [Google Scholar]
- Pérez CA, Huang L, Rong M, Kozak JA, Preuss AK, Zhang H, Max M, Margolskee RF. A transient receptor potential channel expressed in taste receptor cells. Nat. Neurosci. 2002;5:1169–1176. doi: 10.1038/nn952. [DOI] [PubMed] [Google Scholar]
- Pikielny CW, Hasan G, Rouyer F, Rosbash M. Members of a family of Drosophila putative odorant-binding proteins are expressed in different subsets of olfactory hairs. Neuron. 1994;12:35–49. doi: 10.1016/0896-6273(94)90150-3. [DOI] [PubMed] [Google Scholar]
- Robertson HM, Warr CG, Carlson JR. Molecular evolution of the insect chemoreceptor gene superfamily in Drosophila melanogaster . Proc. Natl. Acad. Sci. USA. 2003;2(100 Suppl):14537–14542. doi: 10.1073/pnas.2335847100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Gracia A, Vieira FG, Rozas J. Molecular evolution of the major chemosensory gene families in insects. Heredity. 2009;103:208–216. doi: 10.1038/hdy.2009.55. [DOI] [PubMed] [Google Scholar]
- Sato K, Pellegrino M, Nakagawa T, Nakagawa T, Vosshall LB, Touhara K. Insect olfactory receptors are heteromeric ligand-gated ion channels. Nature. 2008;452:1002–1006. doi: 10.1038/nature06850. [DOI] [PubMed] [Google Scholar]
- Sato K, Tanaka K, Touhara K. Sugar-regulated cation channel formed by an insect gustatory receptor. Proc. Natl. Acad. Sci. USA. 2011;108:11680–11685. doi: 10.1073/pnas.1019622108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott K. The sweet and the bitter of mammalian taste. Curr. Opin. Neurobiol. 2004;14:423–437. doi: 10.1016/j.conb.2004.06.003. [DOI] [PubMed] [Google Scholar]
- Shanbhag SR, Park SK, Pikielny CW, Steinbrecht RA. Gustatory organs of Drosophila melanogaster: fine structure and expression of the putative odorant-binding protein PBPRP2. Cell Tissue Res. 2001;304:423–437. doi: 10.1007/s004410100388. [DOI] [PubMed] [Google Scholar]
- Su CY, Menuz K, Reisert J, Carlson JR. Non-synaptic inhibition between grouped neurons in an olfactory circuit. Nature. 2012;492:66–71. doi: 10.1038/nature11712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talavera K, Yasumatsu K, Yoshida R, Margolskee RF, Voets T, Ninomiya Y, Nilius B. The taste transduction channel TRPM5 is a locus for bittersweet taste interactions. FASEB J. 2008;22:1343–1355. doi: 10.1096/fj.07-9591com. [DOI] [PubMed] [Google Scholar]
- Thorne N, Chromey C, Bray S, Amrein H. Taste perception and coding in Drosophila . Curr. Biol. 2004;14:1065–1079. doi: 10.1016/j.cub.2004.05.019. [DOI] [PubMed] [Google Scholar]
- Tompkins L, Cardosa MJ, White FV, Sanders TG. Isolation and analysis of chemosensory behavior mutants in Drosophila melanogaster . Proc. Natl .Acad. Sci. USA. 1979;76:884–887. doi: 10.1073/pnas.76.2.884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenbeuch A, Pillias AM, Faurion A. Modulation of taste peripheral signal through interpapillar inhibition in hamsters. Neurosci. Lett. 2004;358:137–141. doi: 10.1016/j.neulet.2004.01.013. [DOI] [PubMed] [Google Scholar]
- Vogt RG, Riddiford LM. Pheromone binding and inactivation by moth antennae. Nature. 1981;293:161–163. doi: 10.1038/293161a0. [DOI] [PubMed] [Google Scholar]
- Vosshall LB, Amrein H, Morozov PS, Rzhetsky A, Axel R. A spatial map of olfactory receptor expression in the Drosophila antenna. Cell. 1999;96:725–736. doi: 10.1016/s0092-8674(00)80582-6. [DOI] [PubMed] [Google Scholar]
- Vosshall LB, Stocker RF. Molecular architecture of smell and taste in Drosophila. Annu. Rev. Neurosci. 2007;30:505–533. doi: 10.1146/annurev.neuro.30.051606.094306. [DOI] [PubMed] [Google Scholar]
- Wang Z, Singhvi A, Kong P, Scott K. Taste representations in the Drosophila brain. Cell. 2004;117:981–991. doi: 10.1016/j.cell.2004.06.011. [DOI] [PubMed] [Google Scholar]
- Weiss LA, Dahanukar A, Kwon JY, Banerjee D, Carlson JR. The molecular and cellular basis of bitter taste in Drosophila . Neuron. 2011;69:258–272. doi: 10.1016/j.neuron.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wernet MF, Labhart T, Baumann F, Mazzoni EO, Pichaud F, Desplan C. Homothorax switches function of Drosophila photoreceptors from color to polarized light sensors. Cell. 2003;115:267–279. doi: 10.1016/s0092-8674(03)00848-1. [DOI] [PubMed] [Google Scholar]
- Wicher D, Schafer R, Bauernfeind R, Stensmyr MC, Heller R, Heinemann SH, Hansson BS. Drosophila odorant receptors are both ligand-gated and cyclic-nucleotide-activated cation channels. Nature. 2008;452:1007–1011. doi: 10.1038/nature06861. [DOI] [PubMed] [Google Scholar]
- Wieczorek H, Wolff G. The labellar sugar receptor of Drosophila. J. Comp. Physiol. A. 1989;164:825–834. [Google Scholar]
- Xu P, Atkinson R, Jones DN, Smith DP. Drosophila OBP LUSH is required for activity of pheromone-sensitive neurons. Neuron. 2005;45:193–200. doi: 10.1016/j.neuron.2004.12.031. [DOI] [PubMed] [Google Scholar]
- Yasukawa J, Tomioka S, Aigaki T, Matsuo T. Evolution of expression patterns of two odorant-binding protein genes, Obp57d and Obp57e, in Drosophila . Gene. 2010 doi: 10.1016/j.gene.2010.07.006. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Hoon MA, Chandrashekar J, Mueller KL, Cook B, Wu D, Zuker CS, Ryba NJ. Coding of sweet, bitter, and umami tastes: different receptor cells sharing similar signaling pathways. Cell. 2003;112:293–301. doi: 10.1016/s0092-8674(03)00071-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.