Abstract
In eukaryotic cells, maintenance of genomic stability relies on the coordinated action of a network of cellular processes, including DNA replication, DNA repair, cell-cycle progression, and others. The DNA damage response (DDR) signaling pathway orchestrated by the ATM and ATR kinases is the central regulator of this network in response to DNA damage. Both ATM and ATR are activated by DNA damage and DNA replication stress, but their DNA-damage specificities are distinct and their functions are not redundant. Furthermore, ATM and ATR often work together to signal DNA damage and regulate downstream processes. Here, we will discuss the recent findings and current models of how ATM and ATR sense DNA damage, how they are activated by DNA damage, and how they function in concert to regulate the DDR.
ATM and ATR are the master transducers of DNA damage signals. ATM is primarily activated by double-stranded DNA breaks; ATR responds to a broader spectrum of DNA damage.
During their lifespan, cells are inevitably challenged by extrinsic and intrinsic stresses that threaten the integrity of their genomes. To survive these adverse conditions and pass on intact genetic information to subsequent generations, cells have evolved a highly organized and coordinated effort to ameliorate genotoxic stress called the DNA damage response (DDR). This response underlies the organismal ability to sense and signal problems in its DNA, to arrest cell-cycle progression (cell-cycle checkpoints) and activate appropriate DNA repair mechanisms, or to eliminate cells with unrepairable genomes. The importance of the DDR network for the development and well being of humans is illustrated by the large variety of diseases and cancer-predisposition syndromes that have been linked to mutations of DDR genes (Ciccia and Elledge 2010).
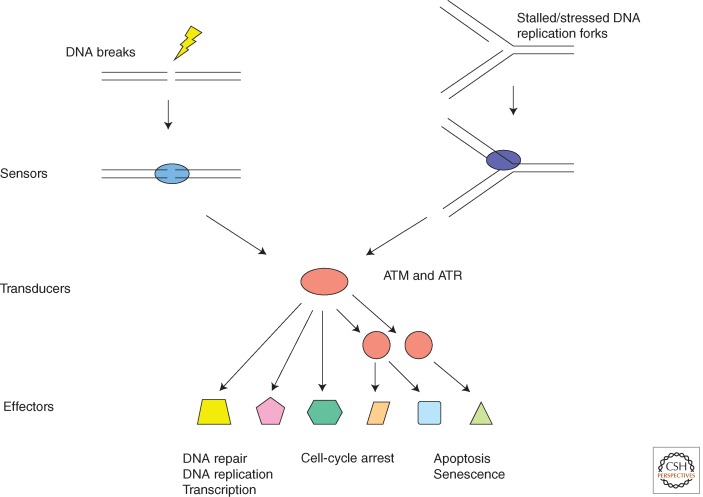
In eukaryotic cells, the cellular response to DNA damage is regulated and coordinated by the DDR signaling pathway. Like classic signal transduction pathways, the DDR uses signal sensors, transducers, and effectors (Fig. 1) (Zhou and Elledge 2000). In contrast to the signal transduction pathways that are activated by ligands of receptor kinases, the DDR signaling pathway is activated by aberrant DNA structures induced by DNA damage or DNA replication stress. The sensors of this pathway are the proteins that directly recognize these aberrant DNA structures and activate the most upstream DDR kinases. The DDR signaling pathway consists of a protein kinase cascade as well as mediator proteins that facilitate the phosphorylation events within the DDR network. The effectors of the DDR signaling pathway are substrates of the DDR kinases that participate in a wide spectrum of cellular processes important for genomic stability, such as DNA replication, DNA repair, and cell-cycle control. Although unique in the way through which it is activated, the DDR is a bona fide signal transduction pathway that is primarily driven by protein phosphorylation.
Figure 1.
The framework of the DDR signaling pathway. Like other signal transduction pathways, the DDR signaling pathway consists of signal sensors, transducers, and effectors. The sensors of this pathway are proteins that recognize DNA structures induced by DNA damage and DNA replication stress. The transducers of this pathway are kinases, including ATM, ATR, and their downstream kinases. The effectors of this pathway are substrates of ATM, ATR, and their downstream kinases. These effectors of ATM and ATR are involved in a broad spectrum of cellular processes that are important for maintenance of genomic stability of organisms.
In mammalian cells, the ATM (ataxia-telangiectasia mutated), ATR (ATM- and Rad3-Related), and DNA-PKcs (DNA-dependent protein kinase) kinases are the most upstream DDR kinases. These large serine/threonine kinases are members of the phosphatidylinositol-3-kinase-like kinase family (PIKKs) (Lempiainen and Halazonetis 2009; Lovejoy and Cortez 2009). In response to DNA damage, hundreds of proteins are phosphorylated at Ser/Thr-Glu motifs and additional sites in an ATM- or ATR-dependent manner, whereas DNA-PKcs appears to regulate a smaller number of targets and play a role primarily in nonhomologous end joining (NHEJ) (Matsuoka et al. 2007; Smolka et al. 2007; Stokes et al. 2007; Bensimon et al. 2010; Beli et al. 2012). ATM and ATR also activate a second wave of phosphorylation through their activation of Chk1, Chk2, and MK2 protein kinases (Matsuoka et al. 1998; Liu et al. 2000; Reinhardt et al. 2007). Although the functional significance of many of these phosphorylation events remains to be understood, the potential ATM/ATR-regulated substrates are involved in a wide variety of cellular processes that may contribute to the DDR. In the picture emerging from these genome-scale studies, ATM and ATR are the master transducers of DNA signals, and they orchestrate a large network of cellular processes to maintain genomic integrity. In vivo and in vitro studies also suggest that the DNA-damage specificities and functions of ATM and ATR are distinct. Whereas ATM is primarily activated by double-stranded DNA breaks (DSBs), ATR responds to a broad spectrum of DNA damage, including DSBs and a variety of DNA lesions that interfere with replication. In this review, we will discuss how ATM and ATR are activated by DNA damage, how they are distinct from each other, and how they function in concert to regulate the DDR.
STRUCTURAL OUTLINES OF ATM AND ATR
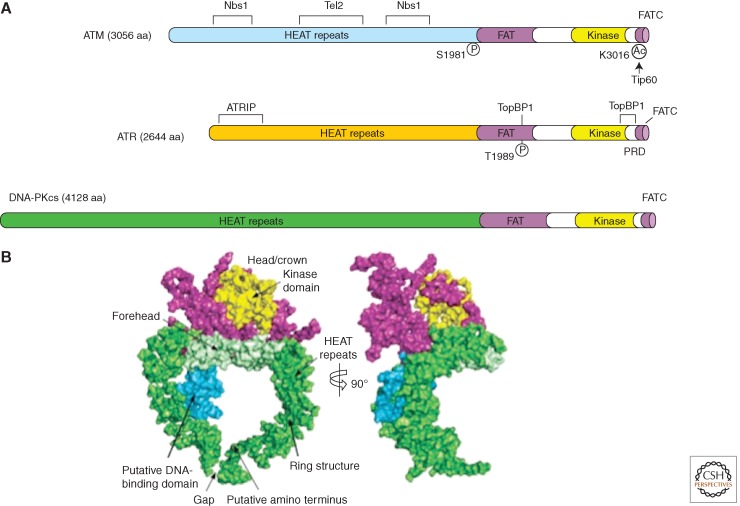
ATM, ATR, and other PIKKs all share a similar domain organization (Fig. 2A). The kinase domains of PIKKs are located near their carboxyl termini, and are flanked by the conserved FAT (FRAP-ATM-TRRAP) and FAT carboxy-terminal (FATC) domains. Large amino terminal and internal regions of ATM and ATR are composed of numerous α-helical HEAT (Huntington-elongation factor 3-protein phosphatase 2A-TOR1) repeats (Perry and Kleckner 2003). ATM was shown to exist as dimers or oligomers in undamaged cells, and is converted into monomers on activation by DNA damage (Bakkenist and Kastan 2003). ATR, on the other hand, forms a stable and obligate complex with ATRIP (ATR interacting protein) (Cortez et al. 2001). ATRIP also dimerizes or oligomerizes through its coiled-coil domain, but this interaction does not appear to be regulated by DNA damage (Ball and Cortez 2005; Itakura et al. 2005).
Figure 2.
Structural outlines of PIKKs. (A) Schematic of the functional domains and selected posttranslational modifications of ATM, ATR, and DNA-PKcs. The bulk of ATM and ATR are composed of a large number (40–50) of α-helical HEAT repeats. Some of these repeats are involved in the interaction with other proteins. The kinase domains of ATM and ATR are located near their carboxyl termini, and are flanked by FAT and FATC domains. ATM and ATR are autophosphorylated in or near their FAT domains after DNA damage, which regulates their activation. The acetylation of the carboxyl terminus of ATM by Tip60 is also involved in ATM activation. A PIKK regulatory domain (PRD) between the kinase and FATC domains of ATR was shown to modulate ATR activation. (B) Overall view of the DNA-PKcs structure. The front view (left) and side view (right) of DNA-PKcs. The various regions of DNA-PKcs are colored and labeled. (Panel B is from Sibanda et al. 2009; reprinted, with permission, from Nature Publishing Group © 2009.)
As a result of their large sizes, high-resolution structures of ATM and ATR have not yet been revealed. However, the crystal structure of DNA-PKcs was obtained at a 6.6-Å resolution (Fig. 2B) (Sibanda et al. 2009). In complex with a carboxyl-terminal fragment of its cofactor Ku80, DNA-PKcs adopts a ring-shaped structure with an extended arm of α-helical HEAT repeats. Sitting atop these repeats is the kinase domain surrounded by the FAT and FATC domains. This overall structure of DNA-PKcs agrees with the previously published structures inferred from electron microscopy studies (Rivera-Calzada et al. 2005; Williams et al. 2008). Interestingly, inside the ring of DNA-PKcs, several HEAT repeats are predicted to form a small globular domain that may interact with DNA to trigger conformational changes. Electron microscopy studies of ATM also revealed an armlike domain suggesting structural resemblance to DNA-PKcs (Llorca et al. 2003).
Whereas their ability to bind DNA remains to be tested, the HEAT repeats of ATM and ATR are known to mediate protein–protein interactions important for the function and regulation of theses kinases. ATM interacts with Nbs1, a regulator of ATM activation, via specific internal HEAT repeats (Falck et al. 2005; You et al. 2005). ATR interacts with ATRIP through its amino-terminal HEAT repeats (Ball et al. 2005). It was recently found that the Tel2–Tti1–Tti2 (TTT) complex interacts with the HEAT repeats of ATM and other PIKKs (Takai et al. 2007, 2010; Hurov et al. 2010). Depletion of different components of the TTT complex not only reduced the levels of ATM, ATR, and other PIKKs, but also compromised the interaction between ATR and ATRIP (Takai et al. 2010). The TTT complex associates with Hsp90 and may act as a PIKK-specific cochaperone to promote proper folding of these large HEAT repeat-containing proteins.
The FAT and FATC domains of ATM and ATR are important regulatory domains. In human cells, the FAT and kinase domains of ATM interact with each other in the absence of DNA damage, which inhibits ATM activity (Bakkenist and Kastan 2003). This interaction between the FAT and kinase domains of ATM is disrupted by the autophosphorylation of Ser1981 in response to DNA damage, alleviating the inhibition of ATM kinase. In mouse cells, however, the autophosphorylation of ATM at Ser1987 (the equivalent of Ser1981 in human ATM) is dispensable for ATM activation (Pellegrini et al. 2006), suggesting that the inhibitory function of the FAT domain may not be evolutionarily conserved. The FAT domain of ATR is phosphorylated at Thr1989 in response to DNA damage and it promotes the stimulation of ATR by TopBP1 (Liu et al. 2011a). The FATC domain of ATM interacts with the acetyltransferase Tip60 and is required for efficient ATM activation through an unknown mechanism (Sun et al. 2005). The FATC domain of ATR is indispensable for its basal kinase activity. Additionally, the PIKK-regulatory domain, a short region located between the FATC and kinase domains of ATR, was shown to interact with TopBP1 and facilitate ATR activation (Mordes and Cortez 2008).
SENSING OF DOUBLE-STRANDED DNA ENDS BY ATM
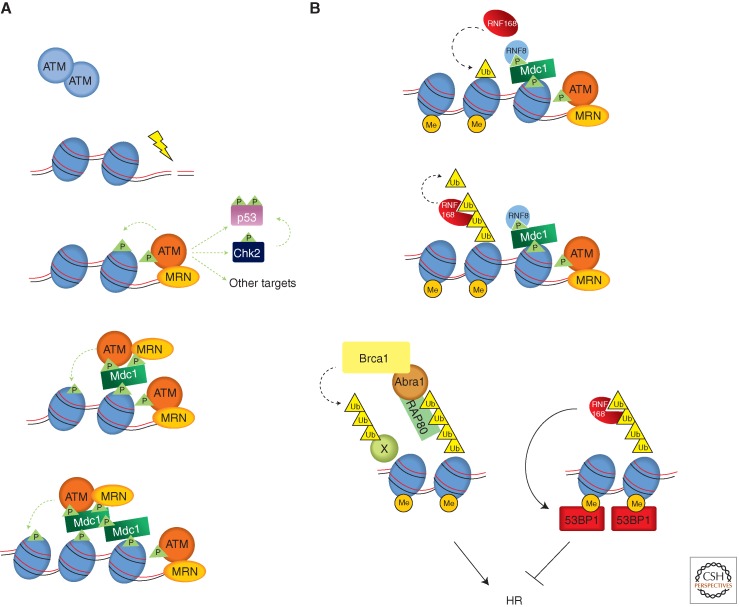
DSBs are one of the most deleterious forms of DNA damage. ATM-deficient cells derived from ataxia-telangiectasia (AT) patients are hypersensitive to radiation, and are defective in DSB repair as well as the G1/S, intra-S, and G2/M checkpoints. In response to DSBs, ATM is rapidly localized to sites of DNA damage and its kinase activity is elevated (Fig. 3A). The activation of ATM by DSBs leads to the phosphorylation of a number of substrates, such as Brca1, Chk2, and p53, mediating the effects of ATM on DNA repair, cell-cycle arrest, apoptosis, and other downstream processes (Shiloh 2003; Lavin 2008).
Figure 3.
Activation of ATM by DSBs. (A) Recognition of DNA ends and chromatin by ATM. The MRN complex functions as a sensor of DNA ends and activates ATM. The ATM activated by DNA ends (red) phosphorylates substrates such as Chk2 and p53, and the H2AX in flanking nucleosomes. Phosphorylated H2AX (γH2AX) is recognized by Mdc1, which triggers a feed-forward loop that spreads activated ATM and γH2AX over large chromatin domains. (B) A ubiquitination cascade at DSBs that promotes the recruitment of Brca1 and 53BP1. The E3 ubiquitin ligase RNF8 is recruited to DSBs via phosphorylated Mdc1. RNF8 promotes another E3 ligase RNF168 to ubiquitinate substrates including histones H2A and H2AX. With the help of the HECT domain ligase HERC2 (not depicted here), RNF168 extends the K63-linked ubiquitin chains on substrates. These ubiquitin chains are recognized by RAP80, which forms a complex with Brca1 through Abraxas. Brca1, which is also a ubiquitin ligase, is critical for HR. RNF168 also promotes the recruitment of 53BP1, which binds the dimethylated lysine 20 of histone H4 (H4K20me2) exposed around DSBs. In contrast to Brca1, 53BP1 antagonizes HR. How the recruitment of Brca1 and 53BP1 is coordinated remains unknown. The dashed green and black lines represent phosphorylation and ubiquitylation events, respectively.
One key regulator of ATM activation is the Mre11-Rad50-Nbs1 (MRN) complex. In cells, MRN is one of the first factors recruited to DSBs. In vitro, purified MRN complex can directly bind to double-stranded DNA (dsDNA) ends. Furthermore, MRN is required for the rapid localization of ATM to DSBs and its activation in cells, and purified MRN complex is sufficient to directly stimulate the dimeric/oligomeric ATM kinase in the presence of dsDNA (Uziel et al. 2003; Lee and Paull 2005). All of this evidence suggests that MRN is a sensor of DSBs that activates ATM (Fig. 3A).
Although MRN is clearly involved in ATM activation at DSBs, how it activates ATM in a DSB-dependent manner is still not fully understood. In vitro studies using purified proteins suggest that the activation of ATM by MRN requires free dsDNA ends (Lee and Paull 2005). Furthermore, the function of MRN in ATM activation is stimulated by dsDNA in a length-dependent manner (Lee and Paull 2005). Similar observations were also made in Xenopus and human cell extracts (You et al. 2007; Shiotani and Zou 2009). In Xenopus egg extracts, MRN and ATM not only bind to dsDNA ends but also dsDNA regions flanking the ends (You et al. 2007). In human cell extracts, ATM is not only activated by blunt dsDNA ends, but also by dsDNA ends with short single-stranded DNA (ssDNA) overhangs (Shiotani and Zou 2009). In the presence of short ssDNA overhangs, the junctions of ssDNA and dsDNA are critical for ATM activation. Together, these results suggest that, at DSBs, MRN recognizes dsDNA ends or dsDNA/ssDNA junctions and may move into flanking dsDNA regions to activate ATM. The DNA end-unwinding and exo-/endonuclease activities of MRN were implicated in ATM activation by in vitro studies (Lee and Paull 2005; Jazayeri et al. 2008), although mouse cells expressing a nuclease-deficient mutant of Mre11 were still able to activate ATM (Buis et al. 2008). The DNA end-tethering activity of MRN may also facilitate ATM activation by increasing the local concentration of DNA ends (Dupre et al. 2006). In vitro, MRN directly stimulates the binding of ATM to its substrates even in the absence of DNA (Lee and Paull 2004). The carboxyl terminus of Nbs1 is known to interact with ATM (Falck et al. 2005; You et al. 2005). However, in mouse cells, the carboxyl terminus of Nbs1 is only important for phosphorylation of some but not all ATM substrates (Stracker et al. 2007). Recent crystallographic data of a dimeric catalytic domain of Mre11 and a short Mre11-interacting peptide of NBS1 led to the proposal of a model whereby changes in MRN complex conformation on binding to DNA and ATP might be relayed to ATM through changes in the carboxyl terminus of Nbs1 (Schiller et al. 2012).
CHROMATIN-MEDIATED DNA-BREAK RECOGNITION BY ATM
Following the initial activation of ATM by DNA ends, ATM triggers a cascade of DDR events on the chromatin flanking DSBs. The key event driving this process is the phosphorylation of the histone variant H2AX by ATM. The DSB-induced phosphorylation of H2AX occurs within minutes after DNA damage, and it rapidly spreads over large chromatin domains (>500 kb) flanking the DNA breaks (Meier et al. 2007; Savic et al. 2009). Whereas H2AX phosphorylation is not critical for phosphorylation of ATM substrates such as Chk2 and p53 (Fernandez-Capetillo et al. 2002; Kang et al. 2005), it is required for accumulation of numerous DNA repair proteins and chromatin-remodeling complexes around DSBs. Cumulative evidence suggests that ATM is part of the chromatin-mediated program at DSBs that promotes DSB repair and coordinates multiple cellular processes in the chromatin domains flanking DSBs (Lukas et al. 2011).
To perform its chromatin-mediated functions, ATM has to specifically recognize the chromatin flanking DSBs. The protein Mdc1 is a key mediator of this process. Through its BRCT domains, Mdc1 directly binds to phosphorylated H2AX (known as γH2AX) and is required for full-sized γH2AX foci formation (Stewart et al. 2003; Lee et al. 2005; Stucki et al. 2005). Mdc1 also interacts with ATM through its FHA domain (Lou et al. 2006). In addition, Mdc1 interacts with Nbs1, which provides an indirect means to associate with ATM (Chapman and Jackson 2008; Melander et al. 2008; Spycher et al. 2008; Wu et al. 2008). The initial activation of ATM by DNA ends may result in phosphorylation of the H2AX immediately adjacent to DSBs. The ability of Mdc1 to bind both γH2AX and ATM enables it to target ATM to the nucleosomes containing γH2AX, providing a mechanism for ATM to recognize the chromatin flanking DSBs and to propagate H2AX phosphorylation along chromatin (Fig. 3A).
In addition to H2AX phosphorylation, the kinase activity of ATM contributes to its chromatin binding in several other ways. Autophosphorylation of ATM has been implicated in Mdc1 binding (Lou et al. 2006). The activation of ATM promotes its association with Mdc1 and its accumulation around DSBs (Berkovich et al. 2007; So et al. 2009). It is interesting to note that ionizing radiation (IR) not only induces DSBs but also reactive oxygen species (ROS), which may directly trigger ATM autophosphorylation and promote its localization to DSBs (Guo et al. 2010). Recent studies also revealed that Mdc1 is a substrate of ATM (Matsuoka et al. 2007; Luo et al. 2011; Jungmichel et al. 2012; Liu et al. 2012a). The phosphorylation of Mdc1 by ATM promotes oligomerization of Mdc1 and may facilitate the spreading of Mdc1 and ATM on chromatin. Interestingly, studies using mouse cells suggest that whereas Mdc1 is important for the spreading of ATM on chromatin near DSBs, ATM is able to phosphorylate H2AX on chromatin distal to DSBs in the absence of Mdc1 (Savic et al. 2009). How ATM reaches and acts on chromatin distal from DSBs remains to be elucidated.
The ATM recruited to chromatin may be regulated by additional mechanisms. The Tip60 acetyltransferase interacts constitutively with ATM and may be recruited to DSBs with ATM (Sun et al. 2005). In response to DSBs, Tip60 acetylates ATM near the FATC domain, which promotes ATM monomerization and activation (Sun et al. 2007). The activation of Tip60 at damage sites requires its chromodomain through which it interacts with histone H3 trimethylated on lysine 9 (H3K9me3), a histone mark associated with heterochromatin (Sun et al. 2009). The initial activation of ATM and its effector kinase Chk2 by DNA ends leads to phosphorylation of KAP-1 and dissociation of heterochromatin protein HP1-β from H3K9me3 around DSBs, which may expose the histone mark to Tip60 and further stimulate the function of ATM in heterochromatin (Ayoub et al. 2008; Goodarzi et al. 2008; Bolderson et al. 2012).
Recently, a flurry of studies revealed that the phosphorylation of H2AX by ATM around DSBs sets off elaborate ubiquitylation and SUMOylation cascades to promote recruitment of Brca1 and 53BP1 (Fig. 3B) (Huen et al. 2007; Kim et al. 2007a,b; Kolas et al. 2007; Mailand et al. 2007; Sobhian et al. 2007; Wang and Elledge 2007; Wang et al. 2007; Doil et al. 2009; Galanty et al. 2009; Morris et al. 2009; Stewart 2009; Bekker-Jensen et al. 2010; Meerang et al. 2011; Mattiroli et al. 2012). Brca1 is an important regulator of homologous recombination (HR) of DSBs. The HR defect in the absence of Brca1 is overcome by the removal of 53BP1 (Bunting et al. 2010), suggesting that the balance between the two proteins is important for the regulation of HR. Depletion of RNF168, a E3 ubiquitin ligase required for 53BP1 recruitment, also suppressed the HR defect in Brca1-deficient cells (Munoz et al. 2012). In addition to their functions in regulating DSB repair, Brca1 and 53BP1 contribute to the efficient phosphorylation of some ATM substrates and have been implicated in checkpoint activation (DiTullio et al. 2002; Wang et al. 2002; Fabbro et al. 2004; Kitagawa et al. 2004). Interestingly, both Brca1 and 53BP1 were also recently shown to enhance ATM kinase activity in vitro when MRN function was limiting (Lee et al. 2010).
AN ATM-TO-ATR SWITCH AT DNA BREAKS
DSBs not only activate ATM, but also turn on the ATR kinase. Both ATM and ATR are critical for DSB-induced checkpoint responses and DSB repair, suggesting that they have nonredundant functions in these processes (Brown and Baltimore 2003; Wang et al. 2004). ATM and ATR are known to have distinct DNA-damage specificities. Unlike ATM, ATR is activated by a broad spectrum of DNA damage in addition to DSBs. The localization of ATR-ATRIP to sites of DNA damage is dependent on the direct interaction between ATRIP and long stretches of RPA-coated ssDNA, suggesting that ssDNA may be the key structure that elicits the ATR response at DSBs (Zou and Elledge 2003).
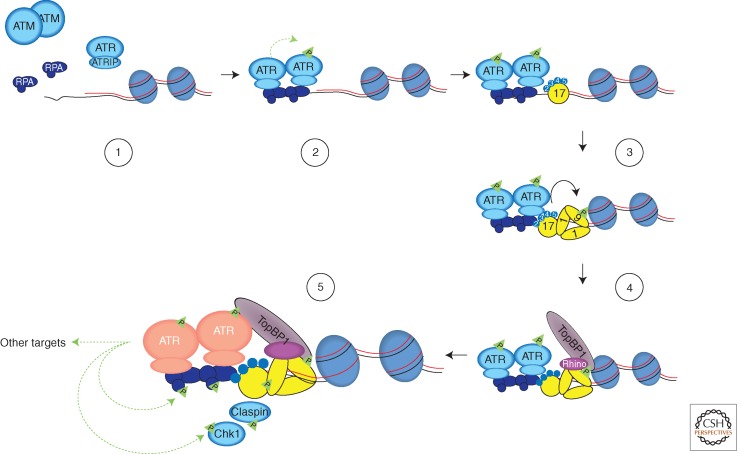
How is ssDNA generated at DSBs? When DSBs arise in mammalian cells before DNA replication, the first line of defense is usually NHEJ. Although it tends to be error-prone, NHEJ is a quick and robust way to rejoin DNA ends throughout the cell cycle. ATM has been shown to contribute to NHEJ (Zha et al. 2011). When DSBs are generated during S phase at DNA replication forks or after replication in the G2 phase of the cell cycle, a second DSB repair pathway, HR, can come into action. In contrast to NHEJ, HR is an error-free repair pathway. HR not only requires homologous DNA sequences (usually from sister chromatids) as repair templates, but also ssDNA ends to search for homologous sequences. Based on studies using yeast, Xenopus, and human cells, several conserved nucleases have been implicated in the generation of ssDNA at DSBs through a process called resection (Symington and Gautier 2011). These nucleases include the MRN complex and its associated protein CtIP (Limbo et al. 2007; Sartori et al. 2007; You et al. 2009), Exo1 (Tomimatsu et al. 2012), and Dna2 (Wawrousek et al. 2010; Peng et al. 2012). ATM is also required for efficient resection, possibly through CtIP and Exo1 regulation (You et al. 2009; Bolderson et al. 2010). Consistent with the idea that DSB resection is important for ATR activation, depletion, or inactivation of ATM, Mre11, CtIP, Exo1, and Dna2 all lead to diminished ATR responses to DSBs (Fig. 4) (Adams et al. 2006; Jazayeri et al. 2006; Myers and Cortez 2006; Sartori et al. 2007; Peng et al. 2012; Tomimatsu et al. 2012).
Figure 4.
A fail-safe, multistep mechanism for ATR activation. Increased amounts of ssDNA are generated by resection of DNA ends or by uncoordinated DNA unwinding and synthesis at replication forks. Extensively resected DNA ends are no longer recognized by ATM efficiently. Once coated by RPA, ssDNA recruits the ATR-ATRIP complex (1), and promotes ATR trans-autophosphorylation (2). RPA-ssDNA also promotes the recruitment of the Rad17-Rfc2-5 clamp loader to junctions between ssDNA and dsDNA, and the loading of Rad9-Rad1-Hus1 (9-1-1) checkpoint clamps onto dsDNA (3). TopBP1 interacts with phosphorylated Rad9 and with Rhino, which associates with 9-1-1 (4). The TopBP1 recruited to dsDNA by 9-1-1 and Rhino engages the ATR-ATRIP complex on RPA-ssDNA through the ATR autophosphorylation site T1989. This process enables TopBP1 to stimulate ATR-ATRIP to its full capacity (pink) on ssDNA (5). TopBP1 may also function as a scaffold to facilitate ATR substrate recognition. This multistep process for ATR activation ensures that ATR is only activated when both ssDNA and ssDNA/dsDNA junctions are present at sites of DNA damage and are recognized by DNA damage sensors, providing a fail-safe but versatile mechanism to signal DNA damage. The dashed green lines represent phosphorylation events, and the solid black line represents the loading of 9-1-1 by the Rad17-RFC2-5 complex.
DSB resection not only promotes ATR activation, but also coordinates the functions of ATM and ATR at DSBs. Using an in vitro assay that monitors the activation of both ATM and ATR by dsDNA, resection of DNA ends was found to induce ATR activation, but progressively attenuates the ability of dsDNA to activate ATM (Shiotani and Zou 2009). Thus, as ssDNA is gradually lengthened during DSB resection, DSB ends switch from an ATM-activating mode to an ATR-activating mode. Consistent with this resection-driven ATM-to-ATR switch observed in vitro, two specific substrates of ATM and ATR, Chk2 and Chk1, are consecutively phosphorylated in cells after IR. Furthermore, enhancing DSB resection by overexpressing CtIP and Exo1 accelerates the ATM-to-ATR switch, whereas reduction of Exo1 levels delays the switch (Shiotani and Zou 2009; Tomimatsu et al. 2012). Interestingly, depletion of CtIP delays the phosphorylation of RPA but not Chk1 by ATR, suggesting that the impact of removing different resection factors on ATR activation may be different (Kousholt et al. 2012). The activation of ATR by resected DSBs enables ATR to activate its effector kinase Chk1, which is critical for intra-S and G2/M checkpoint responses. It was also suggested that when DSBs arise at stalled DNA replication forks, Chk1 phosphorylates the recombinase Rad51 and promotes repair of the break through HR (Sorensen et al. 2005).
DNA DAMAGE SENSING BEYOND DNA BREAKS
In contrast to ATM, ATR is essential for the survival of proliferating cells and its deletion leads to early embryonic lethality in mouse and cell lethality in human cells (Brown and Baltimore 2000, 2003; de Klein et al. 2000; Cortez et al. 2001). This essentiality is likely caused by the fact that ATR monitors replication fork progression and is required to protect cells from intrinsic replication stress that occurs during unperturbed S phase (Casper et al. 2002; McNees et al. 2010). Studies in yeast and metazoans have shown that ATR maintains genome stability by phosphorylating a large number of substrates including proteins that function at replication forks and origins. These phosphorylation events may protect the genome during DNA synthesis by regulating nucleotide levels, fork progression, origin firing, and cell-cycle transitions, and activating specific DNA repair pathways to promote complete and accurate replication of the genome (Cimprich and Cortez 2008; Flynn and Zou 2011).
The key structure that recruits the ATR-ATRIP kinase complex to sites of DNA damage and stressed replication forks is RPA-coated ssDNA (Zou and Elledge 2003). RPA-ssDNA is an important structure for many DNA repair pathways. In addition to HR, RPA-ssDNA is also involved in nucleotide excision repair, mismatch repair, long-patch base excision repair, postreplication repair, interstrand cross-link repair, and replication fork restart. Furthermore, increasing amounts of RPA-ssDNA is generated at or behind replication forks when the coordination between helicase and DNA polymerases is compromised by DNA damage and other impediments (Byun et al. 2005; Lopes et al. 2006). The ability of ATR-ATRIP to recognize RPA-ssDNA gives it its unusual versatility in sensing DNA damage and replication stress (Zou and Elledge 2003; Ball et al. 2005; Namiki and Zou 2006; Shigechi et al. 2012).
ATR ACTIVATION THROUGH A FAIL-SAFE, MULTISTEP PROCESS
Although RPA-ssDNA plays a key role in recruiting ATR-ATRIP to sites of DNA damage in which it can recognize and phosphorylate some substrates, this structure alone is not sufficient to fully activate the ATR signaling pathway. In vivo and in vitro studies suggest that at least three regulatory mechanisms contribute to the full activation of the ATR pathway. First, ATR and some of its substrates are independently localized to damaged DNA, increasing the local concentrations of these proteins at sites of DNA damage. Second, the kinase activity of ATR is stimulated by specific regulators on damaged DNA. Third, through a series of phosphorylation-mediated protein–protein interactions (e.g., the ATR-TopBP1, TopBP1-Rad9, and Rad17-Claspin interactions), ATR forms a dynamic signaling complex with its substrates. All these mechanisms are regulated by damaged DNA and are intertwined with each other. Only when these mechanisms operate in concert is ATR able to signal DNA damage or DNA replication stress at its full capacity.
In addition to RPA-ssDNA, the junction of ssDNA and dsDNA is also a key structure for ATR activation (Fig. 4). The junctions of ssDNA/ dsDNA, which always accompany ssDNA, are recognized by the Rad17-RFC2-5 clamp loader. With assistance from the RPA complex, the ring-shaped Rad9-Rad1-Hus1 (9-1-1) clamp is loaded onto dsDNA by the Rad17-RFC2-5 complex (Ellison and Stillman 2003; Zou et al. 2003). In budding yeast, the yeast ortholog of the 9-1-1 clamp (Ddc1-Rad17-Mec3) on DNA can directly activate the yeast ATR (Mec1) (Majka et al. 2006). It is still unclear whether the 9-1-1 clamp directly activates ATR in vertebrates. However, it is clear that both TopBP1 in vertebrates and its ortholog Dpb11 in yeast stimulate the kinase activity of ATR-ATRIP even in the absence of DNA or other proteins (Kumagai et al. 2006; Mordes et al. 2008a,b; Navadgi-Patil and Burgers 2008). Vertebrate TopBP1 and yeast Dpb11, as well as the yeast Ddc1 subunit of 9-1-1, stimulate ATR or Mec1 through a bipartite ATR/Mec1-activation domain (Kumagai et al. 2006; Navadgi-Patil and Burgers 2009; Navadgi-Patil et al. 2011), suggesting that the basic mechanism for ATR stimulation may be evolutionarily conserved.
Although TopBP1 and Dpb11 can directly stimulate ATR and Mec1 in vitro, their functions in vivo are tightly regulated by DNA damage, suggesting that at their endogenous levels the ATR/Mec1-activation domains of TopBP1 and Dpb11 alone cannot engage ATR/Mec1 efficiently. Both TopBP1 and Dpb11 contain multiple BRCT domains, some of which can function as phosphopeptide binding modules. Furthermore, both TopBP1 and Dpb11 are known to interact with 9-1-1 in both a phosphorylation-dependent (Furuya et al. 2004; Delacroix et al. 2007; Lee et al. 2007; Puddu et al. 2008) and phosphorylation-independent manner (Lee and Dunphy 2010). A recently identified human protein called RHINO facilitates the binding of TopBP1 to 9-1-1 independently of 9-1-1 phosphorylation (Cotta-Ramusino et al. 2011). In addition to its recruitment, TopBP1 may be regulated by Rad17 and 9-1-1 on damaged DNA (Lee and Dunphy 2010). Such a mechanism may ensure that TopBP1 only activates ATR when it encounters ATR at sites of DNA damage (Fig. 4).
Recent studies have shed light on how TopBP1 engages and stimulates the ATR-ATRIP complex on RPA-ssDNA. The binding of ATR-ATRIP to RPA-ssDNA not only provides a mechanism to recruit the kinase to sites of DNA damage, but also a means to promote ATR autophosphorylation in trans. Two studies showed that ATR is phosphorylated at Thr1989 in the FAT domain after DNA damage (Liu et al. 2011a; Nam et al. 2011), and one of the studies demonstrated that this phosphorylation is critical for ATR activation (Liu et al. 2011a). The phosphorylation of ATR at Thr1989 is dependent on RPA, ATRIP, and ATR kinase activity, but independent of Rad17 and TopBP1, suggesting that it is directly driven by the binding of ATR-ATRIP to RPA-ssDNA (Liu et al. 2011a). The phosphorylation of Thr1989 creates a docking site for the BRCT domains 7 and 8 of TopBP1. Because the Thr1989 of human ATR is not conserved in lower vertebrates, whether the regulation of TopBP1 by ATR autophosphorylation is evolutionarily conserved is still unclear. In Xenopus extracts, the phosphorylation of TopBP1 by ATM promotes the binding of TopBP1 to ATR (Yoo et al. 2009). Apart from these phosphorylation-mediated interactions, TopBP1 also interacts with ATR-ATRIP through its ATR-activating domain (Kumagai et al. 2006; Mordes et al. 2008a). These interactions between TopBP1 and the ATR-ATRIP complex may lead to conformational changes of the kinase that increase the activity of its kinase domain and/or its binding to substrates (Mordes and Cortez 2008).
As stated above, the recruitment of TopBP1 and the engagement of TopBP1 with ATR-ATRIP require multiple DNA damage sensors on both ssDNA and dsDNA. The recognition of RPA-ssDNA by ATR-ATRIP, as well as the loading of 9-1-1 at junctions of ssDNA and dsDNA, are both required for TopBP1 to stimulate ATR at sites of DNA damage. Even after TopBP1 is recruited to sites of DNA damage, the ability of TopBP1 to stimulate ATR may be regulated by its interaction with 9-1-1 and its phosphorylation by ATR or ATM. These mechanisms may act together to ensure that ATR is only activated in which both ssDNA and ssDNA/dsDNA junctions are present, but not by ssDNA alone, making ATR activation a unique “fail-safe” event that responds only in the presence of a specific spectrum of DNA damage. This mechanism ensures that the key events controlled by ATR activation (i.e., activation of specific repair pathways, induction of apoptosis, or senescence, do not occur spontaneously).
The full activation of ATR signaling requires not only ATR itself and DNA damage sensors, but also proteins that function as signal transducers and effectors. Many of the proteins that participate in ATR signaling, such as RPA, Rad17, TopBP1, Claspin, and Chk1, are substrates of ATR. Importantly, many of these proteins have been shown to interact with each other in an ATR- and phosphorylation-mediated manner, suggesting a critical role for ATR in the assembly of a signaling complex. Interestingly, through its multiple BRCT domains, TopBP1 may be able to simultaneously interact with ATR-ATRIP and some of its substrates. This unique ability of TopBP1 may allow it to function as a kinase-substrate scaffold that facilitates the recognition of ATR substrates (Liu et al. 2011a). The protein–protein interactions within this signaling complex may regulate either the magnitude or the timing of ATR responses. For example, an interaction between the carboxyl terminus of Rad9 and Claspin is specifically required for the early phase of Chk1 activation during the ATR response (Liu et al. 2012b).
In addition to the core players in the ATR pathways, a long list of proteins has been shown to influence ATR signaling to various extents (Nam and Cortez 2011). For example, evidence for a possible cross talk between RNA-processing proteins and the DDR has surfaced from a number of genome-scale studies (Matsuoka et al. 2007; Paulsen et al. 2009; Hurov et al. 2010; Slabicki et al. 2010; Adamson et al. 2012). Intriguingly, the Cdc5L component of the Prp19 mRNA splicing complex was found to interact with ATR and regulate the phosphorylation of Chk1 and Rad17 (Zhang et al. 2009). Another interesting link has been made between the proapoptotic protein Bid and ATR activation. Bid interacts with both ATRIP and RPA and facilitates the accumulation of ATR-ATRIP on chromatin during replicative stress (Liu et al. 2011b). Understanding the mechanisms through which ATR signaling is regulated by these and other modulators may provide a more complete vision of the interplay between the ATR pathway and other cellular processes.
CROSS TALK BETWEEN ATM AND ATR
Although ATM and ATR have distinct DNA-damage specificities, they are known to cross talk with each other to a certain extent. This cross talk occurs at multiple levels. First, ATM and ATR can influence the localization of each other to sites of DNA damage. As described above, at DSBs, ATM promotes the activation of ATR by enhancing DNA end resection (Jazayeri et al. 2006; Myers and Cortez 2006; Shiotani and Zou 2009). On the other hand, ATR was shown to phosphorylate H2AX in response to DNA replication stress, which may recruit ATM to the chromatin adjacent to stressed replication forks (Ward and Chen 2001). Second, ATM and ATR may directly phosphorylate each other. It was shown that ATM is phosphorylated by ATR at Ser1981 in response to DNA replication stress (Stiff et al. 2006). Third, ATM and ATR may affect the function and localization of each other’s regulators. As described above, in Xenopus extracts, ATM phosphorylates TopBP1 and promotes its interaction with ATR (Yoo et al. 2007, 2009). ATM may also promote the recruitment of TopBP1 to sites of DNA damage through γH2AX and Mdc1 (Wang et al. 2011). Additionally, a recent study suggested that the phosphorylation of Rad9 by ATM is important for the maintenance of genomic stability (Shin et al. 2012). Fourth, ATM and ATR may provide functional redundancy during the DDR. Even in the absence of ATM, slow resection of DSBs still activates ATR (Tomimatsu et al. 2009), albeit with delayed kinetics. Similarly, when ATR signaling is compromised, ATM is activated by collapsed replication forks (Chanoux et al. 2009). Finally, ATM and ATR may cross talk at the substrate level. During the DDR, many proteins are phosphorylated in an ATM-/ATR-dependent manner (Matsuoka et al. 2007). There is evidence that ATM and ATR function redundantly in the phosphorylation of some of these proteins. ATM and ATR may phosphorylate the same sites or different sites in these substrates. The two kinases may phosphorylate the substrates in response to different types of DNA damage, or in different phases of the DDR. Despite the extensive cross talk at all these levels, genetic evidence suggests that ATM and ATR clearly have nonredundant functions, and that each of the kinases can operate in the absence of the other (Brown and Baltimore 2003; Chanoux et al. 2009; Balmus et al. 2012).
CONCLUDING REMARKS
Here, we have presented our current view of ATM and ATR activation. It is important to note that multiple additional layers of complexity have not yet been incorporated into the current models, and they have to be taken into account as we work toward a more complete understanding of the function and regulation of ATM and ATR. In the last few years, tremendous progress has been made in our understanding of the initiating events of the DDR. Although many of the key proteins involved in sensing and signaling DNA damage have been identified, new ones are still being discovered. Additionally, different types of posttranslational modifications (PTMs) have been implicated in coordinating the ordered assembly and disassembly of protein complexes during the DDR. Identification of targets of PTMs and understanding their functional significance in the DDR will be a critical step forward. A better understanding of the regulation of the DDR may provide new opportunities to exploit the genomic instability of cancer cells in targeted therapy.
The recent discovery that ATM and ATR are activated by oncogenic stresses suggests that cancer cells may rely on the DDR pathways to survive genomic instability (Halazonetis et al. 2008). Importantly, as exemplified by the use of PARP inhibitors in the treatment of BRCA-deficient breast and ovarian cancers, inhibition of specific DDRs may confer “synthetic lethality” to the cancer cells with specific types of genomic instability (Bouwman and Jonkers 2012). Development of highly specific inhibitors of ATM, Chk1, and Chk2 has made promising progress (Smith et al. 2010). More recently, ATR-specific inhibitors have also become available and were shown to enhance the sensitivity of cancer cells to cisplatin and irradiation (Charrier et al. 2011; Reaper et al. 2011; Pires et al. 2012). In addition to its potential clinical applications, the specific ATR inhibitors may facilitate the identification of ATR substrates and allow a better understanding of the distinct functions of ATM and ATR during the DDR. Thus, now is an exciting time for research on the ATM and ATR signaling pathways, and we are on the verge of making major breakthroughs in both basic and translational research.
ACKNOWLEDGMENTS
We are especially grateful to Dr. Stephen Elledge for his insightful comments on the manuscript. A.M. is supported by a postdoctoral fellowship from the Fonds de Recherche du Québec–Santé (FRQS). L.Z. is a Jim and Ann Orr Massachusetts General Hospital Research Scholar and is supported by NIH grant GM076388. We apologize to colleagues whose work we could not discuss because of space constraints.
Footnotes
Editors: Errol C. Friedberg, Stephen J. Elledge, Alan R. Lehmann, Tomas Lindahl, and Marco Muzi-Falconi
Additional Perspectives on DNA Repair, Mutagenesis, and Other Responses to DNA Damage available at www.cshperspectives.org
REFERENCES
- Adams KE, Medhurst AL, Dart DA, Lakin ND 2006. Recruitment of ATR to sites of ionising radiation-induced DNA damage requires ATM and components of the MRN protein complex. Oncogene 25: 3894–3904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adamson B, Smogorzewska A, Sigoillot FD, King RW, Elledge SJ 2012. A genome-wide homologous recombination screen identifies the RNA-binding protein RBMX as a component of the DNA-damage response. Nat Cell Biol 14: 318–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayoub N, Jeyasekharan AD, Bernal JA, Venkitaraman AR 2008. HP1-β mobilization promotes chromatin changes that initiate the DNA damage response. Nature 453: 682–686 [DOI] [PubMed] [Google Scholar]
- Bakkenist CJ, Kastan MB 2003. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature 421: 499–506 [DOI] [PubMed] [Google Scholar]
- Ball HL, Cortez D 2005. ATRIP oligomerization is required for ATR-dependent checkpoint signaling. J Biol Chem 280: 31390–31396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ball HL, Myers JS, Cortez D 2005. ATRIP binding to replication protein A-single-stranded DNA promotes ATR-ATRIP localization but is dispensable for Chk1 phosphorylation. Mol Biol Cell 16: 2372–2381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balmus G, Zhu M, Mukherjee S, Lyndaker AM, Hume KR, Lee J, Riccio ML, Reeves AP, Sutter NB, Noden DM, et al. 2012. Disease severity in a mouse model of ataxia telangiectasia is modulated by the DNA damage checkpoint gene Hus1. Hum Mol Genet 21: 3408–3420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekker-Jensen S, Rendtlew Danielsen J, Fugger K, Gromova I, Nerstedt A, Lukas C, Bartek J, Lukas J, Mailand N 2010. HERC2 coordinates ubiquitin-dependent assembly of DNA repair factors on damaged chromosomes. Nat Cell Biol 12: 80–86 [DOI] [PubMed] [Google Scholar]
- Beli P, Lukashchuk N, Wagner SA, Weinert BT, Olsen JV, Baskcomb L, Mann M, Jackson SP, Choudhary C 2012. Proteomic investigations reveal a role for RNA processing factor THRAP3 in the DNA damage response. Mol Cell 46: 212–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensimon A, Schmidt A, Ziv Y, Elkon R, Wang SY, Chen DJ, Aebersold R, Shiloh Y 2010. ATM-dependent and -independent dynamics of the nuclear phosphoproteome after DNA damage. Sci Signal 3: rs3. [DOI] [PubMed] [Google Scholar]
- Berkovich E, Monnat RJ Jr, Kastan MB 2007. Roles of ATM and NBS1 in chromatin structure modulation and DNA double-strand break repair. Nat Cell Biol 9: 683–690 [DOI] [PubMed] [Google Scholar]
- Bolderson E, Tomimatsu N, Richard DJ, Boucher D, Kumar R, Pandita TK, Burma S, Khanna KK 2010. Phosphorylation of Exo1 modulates homologous recombination repair of DNA double-strand breaks. Nucleic Acids Res 38: 1821–1831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolderson E, Savage KI, Mahen R, Pisupati V, Graham ME, Richard DJ, Robinson PJ, Venkitaraman AR, Khanna KK 2012. Kruppel-associated Box (KRAB)-associated co-repressor (KAP-1) Ser-473 phosphorylation regulates heterochromatin protein 1β (HP1-β) mobilization and DNA repair in heterochromatin. J Biol Chem 287: 28122–28131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouwman P, Jonkers J 2012. The effects of deregulated DNA damage signalling on cancer chemotherapy response and resistance. Nat Rev Cancer 12: 587–598 [DOI] [PubMed] [Google Scholar]
- Brown EJ, Baltimore D 2000. ATR disruption leads to chromosomal fragmentation and early embryonic lethality. Genes Dev 14: 397–402 [PMC free article] [PubMed] [Google Scholar]
- Brown EJ, Baltimore D 2003. Essential and dispensable roles of ATR in cell cycle arrest and genome maintenance. Genes Dev 17: 615–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buis J, Wu Y, Deng Y, Leddon J, Westfield G, Eckersdorff M, Sekiguchi JM, Chang S, Ferguson DO 2008. Mre11 nuclease activity has essential roles in DNA repair and genomic stability distinct from ATM activation. Cell 135: 85–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunting SF, Callen E, Wong N, Chen HT, Polato F, Gunn A, Bothmer A, Feldhahn N, Fernandez-Capetillo O, Cao L, et al. 2010. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell 141: 243–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byun TS, Pacek M, Yee MC, Walter JC, Cimprich KA 2005. Functional uncoupling of MCM helicase and DNA polymerase activities activates the ATR-dependent checkpoint. Genes Dev 19: 1040–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casper AM, Nghiem P, Arlt MF, Glover TW 2002. ATR regulates fragile site stability. Cell 111: 779–789 [DOI] [PubMed] [Google Scholar]
- Chanoux RA, Yin B, Urtishak KA, Asare A, Bassing CH, Brown EJ 2009. ATR and H2AX cooperate in maintaining genome stability under replication stress. J Biol Chem 284: 5994–6003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman JR, Jackson SP 2008. Phospho-dependent interactions between NBS1 and MDC1 mediate chromatin retention of the MRN complex at sites of DNA damage. EMBO Rep 9: 795–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charrier JD, Durrant SJ, Golec JM, Kay DP, Knegtel RM, MacCormick S, Mortimore M, O’Donnell ME, Pinder JL, Reaper PM, et al. 2011. Discovery of potent and selective inhibitors of ataxia telangiectasia mutated and Rad3 related (ATR) protein kinase as potential anticancer agents. J Med Chem 54: 2320–2330 [DOI] [PubMed] [Google Scholar]
- Ciccia A, Elledge SJ 2010. The DNA damage response: Making it safe to play with knives. Mol Cell 40: 179–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimprich KA, Cortez D 2008. ATR: An essential regulator of genome integrity. Nat Rev Mol Cell Biol 9: 616–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortez D, Guntuku S, Qin J, Elledge SJ 2001. ATR and ATRIP: Partners in checkpoint signaling. Science 294: 1713–1716 [DOI] [PubMed] [Google Scholar]
- Cotta-Ramusino C, McDonald ER III, Hurov K, Sowa ME, Harper JW, Elledge SJ 2011. A DNA damage response screen identifies RHINO, a 9-1-1 and TopBP1 interacting protein required for ATR signaling. Science 332: 1313–1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Klein A, Muijtjens M, van Os R, Verhoeven Y, Smit B, Carr AM, Lehmann AR, Hoeijmakers JH 2000. Targeted disruption of the cell-cycle checkpoint gene ATR leads to early embryonic lethality in mice. Curr Biol 10: 479–482 [DOI] [PubMed] [Google Scholar]
- Delacroix S, Wagner JM, Kobayashi M, Yamamoto K, Karnitz LM 2007. The Rad9-Hus1-Rad1 (9-1-1) clamp activates checkpoint signaling via TopBP1. Genes Dev 21: 1472–1477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiTullio RA Jr, Mochan TA, Venere M, Bartkova J, Sehested M, Bartek J, Halazonetis TD 2002. 53BP1 functions in an ATM-dependent checkpoint pathway that is constitutively activated in human cancer. Nat Cell Biol 4: 998–1002 [DOI] [PubMed] [Google Scholar]
- Doil C, Mailand N, Bekker-Jensen S, Menard P, Larsen DH, Pepperkok R, Ellenberg J, Panier S, Durocher D, Bartek J, et al. 2009. RNF168 binds and amplifies ubiquitin conjugates on damaged chromosomes to allow accumulation of repair proteins. Cell 136: 435–446 [DOI] [PubMed] [Google Scholar]
- Dupre A, Boyer-Chatenet L, Gautier J 2006. Two-step activation of ATM by DNA and the Mre11-Rad50-Nbs1 complex. Nat Struct Mol Biol 13: 451–457 [DOI] [PubMed] [Google Scholar]
- Ellison V, Stillman B 2003. Biochemical characterization of DNA damage checkpoint complexes: Clamp loader and clamp complexes with specificity for 5′ recessed DNA. PLoS Biol 1: E33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabbro M, Savage K, Hobson K, Deans AJ, Powell SN, McArthur GA, Khanna KK 2004. BRCA1-BARD1 complexes are required for p53Ser–15 phosphorylation and a G1/S arrest following ionizing radiation-induced DNA damage. J Biol Chem 279: 31251–31258 [DOI] [PubMed] [Google Scholar]
- Falck J, Coates J, Jackson SP 2005. Conserved modes of recruitment of ATM, ATR and DNA-PKcs to sites of DNA damage. Nature 434: 605–611 [DOI] [PubMed] [Google Scholar]
- Fernandez-Capetillo O, Chen HT, Celeste A, Ward I, Romanienko PJ, Morales JC, Naka K, Xia Z, Camerini-Otero RD, Motoyama N, et al. 2002. DNA damage-induced G2-M checkpoint activation by histone H2AX and 53BP1. Nat Cell Biol 4: 993–997 [DOI] [PubMed] [Google Scholar]
- Flynn RL, Zou L 2011. ATR: A master conductor of cellular responses to DNA replication stress. Trends Biochem Sci 36: 133–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuya K, Poitelea M, Guo L, Caspari T, Carr AM 2004. Chk1 activation requires Rad9 S/TQ-site phosphorylation to promote association with C-terminal BRCT domains of Rad4TOPBP1. Genes Dev 18: 1154–1164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galanty Y, Belotserkovskaya R, Coates J, Polo S, Miller KM, Jackson SP 2009. Mammalian SUMO E3-ligases PIAS1 and PIAS4 promote responses to DNA double-strand breaks. Nature 462: 935–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodarzi AA, Noon AT, Deckbar D, Ziv Y, Shiloh Y, Lobrich M, Jeggo PA 2008. ATM signaling facilitates repair of DNA double-strand breaks associated with heterochromatin. Mol Cell 31: 167–177 [DOI] [PubMed] [Google Scholar]
- Guo Z, Kozlov S, Lavin MF, Person MD, Paull TT 2010. ATM activation by oxidative stress. Science 330: 517–521 [DOI] [PubMed] [Google Scholar]
- Halazonetis TD, Gorgoulis VG, Bartek J 2008. An oncogene-induced DNA damage model for cancer development. Science 319: 1352–1355 [DOI] [PubMed] [Google Scholar]
- Huen MS, Grant R, Manke I, Minn K, Yu X, Yaffe MB, Chen J 2007. RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly. Cell 131: 901–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurov KE, Cotta-Ramusino C, Elledge SJ 2010. A genetic screen identifies the Triple T complex required for DNA damage signaling and ATM and ATR stability. Genes Dev 24: 1939–1950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itakura E, Sawada I, Matsuura A 2005. Dimerization of the ATRIP protein through the coiled-coil motif and its implication to the maintenance of stalled replication forks. Mol Biol Cell 16: 5551–5562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jazayeri A, Falck J, Lukas C, Bartek J, Smith GC, Lukas J, Jackson SP 2006. ATM- and cell cycle-dependent regulation of ATR in response to DNA double-strand breaks. Nat Cell Biol 8: 37–45 [DOI] [PubMed] [Google Scholar]
- Jazayeri A, Balestrini A, Garner E, Haber JE, Costanzo V 2008. Mre11-Rad50-Nbs1-dependent processing of DNA breaks generates oligonucleotides that stimulate ATM activity. EMBO J 27: 1953–1962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jungmichel S, Clapperton JA, Lloyd J, Hari FJ, Spycher C, Pavic L, Li J, Haire LF, Bonalli M, Larsen DH, et al. 2012. The molecular basis of ATM-dependent dimerization of the Mdc1 DNA damage checkpoint mediator. Nucleic Acids Res 40: 3913–3928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J, Ferguson D, Song H, Bassing C, Eckersdorff M, Alt FW, Xu Y 2005. Functional interaction of H2AX, NBS1, and p53 in ATM-dependent DNA damage responses and tumor suppression. Mol Cell Biol 25: 661–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Chen J, Yu X 2007a. Ubiquitin-binding protein RAP80 mediates BRCA1-dependent DNA damage response. Science 316: 1202–1205 [DOI] [PubMed] [Google Scholar]
- Kim H, Huang J, Chen J 2007b. CCDC98 is a BRCA1-BRCT domain-binding protein involved in the DNA damage response. Nat Struct Mol Biol 14: 710–715 [DOI] [PubMed] [Google Scholar]
- Kitagawa R, Bakkenist CJ, McKinnon PJ, Kastan MB 2004. Phosphorylation of SMC1 is a critical downstream event in the ATM-NBS1-BRCA1 pathway. Genes Dev 18: 1423–1438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolas NK, Chapman JR, Nakada S, Ylanko J, Chahwan R, Sweeney FD, Panier S, Mendez M, Wildenhain J, Thomson TM, et al. 2007. Orchestration of the DNA-damage response by the RNF8 ubiquitin ligase. Science 318: 1637–1640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kousholt AN, Fugger K, Hoffmann S, Larsen BD, Menzel T, Sartori AA, Sorensen CS 2012. CtIP-dependent DNA resection is required for DNA damage checkpoint maintenance but not initiation. J Cell Biol 197: 869–876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumagai A, Lee J, Yoo HY, Dunphy WG 2006. TopBP1 activates the ATR-ATRIP complex. Cell 124: 943–955 [DOI] [PubMed] [Google Scholar]
- Lavin MF 2008. Ataxia-telangiectasia: From a rare disorder to a paradigm for cell signalling and cancer. Nat Rev Mol Cell Biol 9: 759–769 [DOI] [PubMed] [Google Scholar]
- Lee J, Dunphy WG 2010. Rad17 plays a central role in establishment of the interaction between TopBP1 and the Rad9-Hus1-Rad1 complex at stalled replication forks. Mol Biol Cell 21: 926–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Paull TT 2004. Direct activation of the ATM protein kinase by the Mre11/Rad50/Nbs1 complex. Science 304: 93–96 [DOI] [PubMed] [Google Scholar]
- Lee JH, Paull TT 2005. ATM activation by DNA double-strand breaks through the Mre11-Rad50-Nbs1 complex. Science 308: 551–554 [DOI] [PubMed] [Google Scholar]
- Lee MS, Edwards RA, Thede GL, Glover JN 2005. Structure of the BRCT repeat domain of MDC1 and its specificity for the free COOH-terminal end of the γ-H2AX histone tail. J Biol Chem 280: 32053–32056 [DOI] [PubMed] [Google Scholar]
- Lee J, Kumagai A, Dunphy WG 2007. The Rad9-Hus1-Rad1 checkpoint clamp regulates interaction of TopBP1 with ATR. J Biol Chem 282: 28036–28044 [DOI] [PubMed] [Google Scholar]
- Lee JH, Goodarzi AA, Jeggo PA, Paull TT 2010. 53BP1 promotes ATM activity through direct interactions with the MRN complex. EMBO J 29: 574–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lempiainen H, Halazonetis TD 2009. Emerging common themes in regulation of PIKKs and PI3Ks. EMBO J 28: 3067–3073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limbo O, Chahwan C, Yamada Y, de Bruin RA, Wittenberg C, Russell P 2007. Ctp1 is a cell-cycle-regulated protein that functions with Mre11 complex to control double-strand break repair by homologous recombination. Mol Cell 28: 134–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Guntuku S, Cui XS, Matsuoka S, Cortez D, Tamai K, Luo G, Carattini-Rivera S, DeMayo F, Bradley A, et al. 2000. Chk1 is an essential kinase that is regulated by Atr and required for the G2/M DNA damage checkpoint. Genes Dev 14: 1448–1459 [PMC free article] [PubMed] [Google Scholar]
- Liu S, Shiotani B, Lahiri M, Marechal A, Tse A, Leung CC, Glover JN, Yang XH, Zou L 2011a. ATR autophosphorylation as a molecular switch for checkpoint activation. Mol Cell 43: 192–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Vaithiyalingam S, Shi Q, Chazin WJ, Zinkel SS 2011b. BID binds to replication protein A and stimulates ATR function following replicative stress. Mol Cell Biol 31: 4298–4309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Luo S, Zhao H, Liao J, Li J, Yang C, Xu B, Stern DF, Xu X, Ye K 2012a. Structural mechanism of the phosphorylation-dependent dimerization of the MDC1 forkhead-associated domain. Nucleic Acids Res 40: 3898–3912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Song N, Zou L 2012b. The conserved C terminus of Claspin interacts with Rad9 and promotes rapid activation of Chk1. Cell Cycle 11: 2711–2716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llorca O, Rivera-Calzada A, Grantham J, Willison KR 2003. Electron microscopy and 3D reconstructions reveal that human ATM kinase uses an arm-like domain to clamp around double-stranded DNA. Oncogene 22: 3867–3874 [DOI] [PubMed] [Google Scholar]
- Lopes M, Foiani M, Sogo JM 2006. Multiple mechanisms control chromosome integrity after replication fork uncoupling and restart at irreparable UV lesions. Mol Cell 21: 15–27 [DOI] [PubMed] [Google Scholar]
- Lou Z, Minter-Dykhouse K, Franco S, Gostissa M, Rivera MA, Celeste A, Manis JP, van Deursen J, Nussenzweig A, Paull TT, et al. 2006. MDC1 maintains genomic stability by participating in the amplification of ATM-dependent DNA damage signals. Mol Cell 21: 187–200 [DOI] [PubMed] [Google Scholar]
- Lovejoy CA, Cortez D 2009. Common mechanisms of PIKK regulation. DNA Repair (Amst) 8: 1004–1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukas J, Lukas C, Bartek J 2011. More than just a focus: The chromatin response to DNA damage and its role in genome integrity maintenance. Nat Cell Biol 13: 1161–1169 [DOI] [PubMed] [Google Scholar]
- Luo K, Yuan J, Lou Z 2011. Oligomerization of MDC1 protein is important for proper DNA damage response. J Biol Chem 286: 28192–28199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mailand N, Bekker-Jensen S, Faustrup H, Melander F, Bartek J, Lukas C, Lukas J 2007. RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell 131: 887–900 [DOI] [PubMed] [Google Scholar]
- Majka J, Niedziela-Majka A, Burgers PM 2006. The checkpoint clamp activates Mec1 kinase during initiation of the DNA damage checkpoint. Mol Cell 24: 891–901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka S, Huang M, Elledge SJ 1998. Linkage of ATM to cell cycle regulation by the Chk2 protein kinase. Science 282: 1893–1897 [DOI] [PubMed] [Google Scholar]
- Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER III, Hurov KE, Luo J, Bakalarski CE, Zhao Z, Solimini N, Lerenthal Y, et al. 2007. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science 316: 1160–1166 [DOI] [PubMed] [Google Scholar]
- Mattiroli F, Vissers JH, van Dijk WJ, Ikpa P, Citterio E, Vermeulen W, Marteijn JA, Sixma TK 2012. RNF168 ubiquitinates K13-15 on H2A/H2AX to drive DNA damage signaling. Cell 150: 1182–1195 [DOI] [PubMed] [Google Scholar]
- McNees CJ, Tejera AM, Martinez P, Murga M, Mulero F, Fernandez-Capetillo O, Blasco MA 2010. ATR suppresses telomere fragility and recombination but is dispensable for elongation of short telomeres by telomerase. J Cell Biol 188: 639–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meerang M, Ritz D, Paliwal S, Garajova Z, Bosshard M, Mailand N, Janscak P, Hubscher U, Meyer H, Ramadan K 2011. The ubiquitin-selective segregase VCP/p97 orchestrates the response to DNA double-strand breaks. Nat Cell Biol 13: 1376–1382 [DOI] [PubMed] [Google Scholar]
- Meier A, Fiegler H, Munoz P, Ellis P, Rigler D, Langford C, Blasco MA, Carter N, Jackson SP 2007. Spreading of mammalian DNA-damage response factors studied by ChIP-chip at damaged telomeres. EMBO J 26: 2707–2718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melander F, Bekker-Jensen S, Falck J, Bartek J, Mailand N, Lukas J 2008. Phosphorylation of SDT repeats in the MDC1 N terminus triggers retention of NBS1 at the DNA damage-modified chromatin. J Cell Biol 181: 213–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mordes DA, Cortez D 2008. Activation of ATR and related PIKKs. Cell Cycle 7: 2809–2812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mordes DA, Glick GG, Zhao R, Cortez D 2008a. TopBP1 activates ATR through ATRIP and a PIKK regulatory domain. Genes Dev 22: 1478–1489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mordes DA, Nam EA, Cortez D 2008b. Dpb11 activates the Mec1-Ddc2 complex. Proc Natl Acad Sci 105: 18730–18734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris JR, Boutell C, Keppler M, Densham R, Weekes D, Alamshah A, Butler L, Galanty Y, Pangon L, Kiuchi T, et al. 2009. The SUMO modification pathway is involved in the BRCA1 response to genotoxic stress. Nature 462: 886–890 [DOI] [PubMed] [Google Scholar]
- Munoz MC, Laulier C, Gunn A, Cheng A, Robbiani DF, Nussenzweig A, Stark JM 2012. Ring finger nuclear factor RNF168 is important for defects in homologous recombination caused by loss of the breast cancer susceptibility factor BRCA1. J Biol Chem 287: 40618–40628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers JS, Cortez D 2006. Rapid activation of ATR by ionizing radiation requires ATM and Mre11. J Biol Chem 281: 9346–9350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam EA, Cortez D 2011. ATR signalling: More than meeting at the fork. Biochem J 436: 527–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam EA, Zhao R, Glick GG, Bansbach CE, Friedman DB, Cortez D 2011. Thr-1989 phosphorylation is a marker of active ataxia telangiectasia-mutated and Rad3-related (ATR) kinase. J Biol Chem 286: 28707–28714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namiki Y, Zou L 2006. ATRIP associates with replication protein A-coated ssDNA through multiple interactions. Proc Natl Acad Sci 103: 580–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navadgi-Patil VM, Burgers PM 2008. Yeast DNA replication protein Dpb11 activates the Mec1/ATR checkpoint kinase. J Biol Chem 283: 35853–35859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navadgi-Patil VM, Burgers PM 2009. The unstructured C-terminal tail of the 9-1-1 clamp subunit Ddc1 activates Mec1/ATR via two distinct mechanisms. Mol Cell 36: 743–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navadgi-Patil VM, Kumar S, Burgers PM 2011. The unstructured C-terminal tail of yeast Dpb11 (human TopBP1) protein is dispensable for DNA replication and the S phase checkpoint but required for the G2/M checkpoint. J Biol Chem 286: 40999–41007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulsen RD, Soni DV, Wollman R, Hahn AT, Yee MC, Guan A, Hesley JA, Miller SC, Cromwell EF, Solow-Cordero DE, et al. 2009. A genome-wide siRNA screen reveals diverse cellular processes and pathways that mediate genome stability. Mol Cell 35: 228–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrini M, Celeste A, Difilippantonio S, Guo R, Wang W, Feigenbaum L, Nussenzweig A 2006. Autophosphorylation at serine 1987 is dispensable for murine Atm activation in vivo. Nature 443: 222–225 [DOI] [PubMed] [Google Scholar]
- Peng G, Dai H, Zhang W, Hsieh HJ, Pan MR, Park YY, Tsai RY, Bedrosian I, Lee JS, Ira G, et al. 2012. Human nuclease/helicase DNA2 alleviates replication stress by promoting DNA end resection. Cancer Res 72: 2802–2813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry J, Kleckner N 2003. The ATRs, ATMs, and TORs are giant HEAT repeat proteins. Cell 112: 151–155 [DOI] [PubMed] [Google Scholar]
- Pires IM, Olcina MM, Anbalagan S, Pollard JR, Reaper PM, Charlton PA, McKenna WG, Hammond EM 2012. Targeting radiation-resistant hypoxic tumour cells through ATR inhibition. Br J Cancer 107: 291–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puddu F, Granata M, Di Nola L, Balestrini A, Piergiovanni G, Lazzaro F, Giannattasio M, Plevani P, Muzi-Falconi M 2008. Phosphorylation of the budding yeast 9-1-1 complex is required for Dpb11 function in the full activation of the UV-induced DNA damage checkpoint. Mol Cell Biol 28: 4782–4793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reaper PM, Griffiths MR, Long JM, Charrier JD, Maccormick S, Charlton PA, Golec JM, Pollard JR 2011. Selective killing of ATM- or p53-deficient cancer cells through inhibition of ATR. Nat Chem Biol 7: 428–430 [DOI] [PubMed] [Google Scholar]
- Reinhardt HC, Aslanian AS, Lees JA, Yaffe MB 2007. p53-deficient cells rely on ATM- and ATR-mediated checkpoint signaling through the p38MAPK/MK2 pathway for survival after DNA damage. Cancer Cell 11: 175–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera-Calzada A, Maman JD, Spagnolo L, Pearl LH, Llorca O 2005. Three-dimensional structure and regulation of the DNA-dependent protein kinase catalytic subunit (DNA-PKcs). Structure 13: 243–255 [DOI] [PubMed] [Google Scholar]
- Sartori AA, Lukas C, Coates J, Mistrik M, Fu S, Bartek J, Baer R, Lukas J, Jackson SP 2007. Human CtIP promotes DNA end resection. Nature 450: 509–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savic V, Yin B, Maas NL, Bredemeyer AL, Carpenter AC, Helmink BA, Yang-Lott KS, Sleckman BP, Bassing CH 2009. Formation of dynamic γ-H2AX domains along broken DNA strands is distinctly regulated by ATM and MDC1 and dependent upon H2AX densities in chromatin. Mol Cell 34: 298–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiller CB, Lammens K, Guerini I, Coordes B, Feldmann H, Schlauderer F, Mockel C, Schele A, Strasser K, Jackson SP, et al. 2012. Structure of Mre11-Nbs1 complex yields insights into ataxia-telangiectasia-like disease mutations and DNA damage signaling. Nat Struct Mol Biol 19: 693–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigechi T, Tomida J, Sato K, Kobayashi M, Eykelenboom JK, Pessina F, Zhang Y, Uchida E, Ishiai M, Lowndes NF, et al. 2012. ATR-ATRIP kinase complex triggers activation of the Fanconi anemia DNA repair pathway. Cancer Res 72: 1149–1156 [DOI] [PubMed] [Google Scholar]
- Shiloh Y 2003. ATM and related protein kinases: Safeguarding genome integrity. Nat Rev Cancer 3: 155–168 [DOI] [PubMed] [Google Scholar]
- Shin MH, Yuan M, Zhang H, Margolick JB, Kai M 2012. ATM-dependent phosphorylation of the checkpoint clamp regulates repair pathways and maintains genomic stability. Cell Cycle 11: 1796–1803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiotani B, Zou L 2009. Single-stranded DNA orchestrates an ATM-to-ATR switch at DNA breaks. Mol Cell 33: 547–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibanda BL, Chirgadze DY, Blundell TL 2009. Crystal structure of DNA-PKcs reveals a large open-ring cradle comprised of HEAT repeats. Nature 463: 118–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slabicki M, Theis M, Krastev DB, Samsonov S, Mundwiller E, Junqueira M, Paszkowski-Rogacz M, Teyra J, Heninger AK, Poser I, et al. 2010. A genome-scale DNA repair RNAi screen identifies SPG48 as a novel gene associated with hereditary spastic paraplegia. PLoS Biol 8: e1000408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J, Tho LM, Xu N, Gillespie DA 2010. The ATM-Chk2 and ATR-Chk1 pathways in DNA damage signaling and cancer. Adv Cancer Res 108: 73–112 [DOI] [PubMed] [Google Scholar]
- Smolka MB, Albuquerque CP, Chen SH, Zhou H 2007. Proteome-wide identification of in vivo targets of DNA damage checkpoint kinases. Proc Natl Acad Sci 104: 10364–10369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- So S, Davis AJ, Chen DJ 2009. Autophosphorylation at serine 1981 stabilizes ATM at DNA damage sites. J Cell Biol 187: 977–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobhian B, Shao G, Lilli DR, Culhane AC, Moreau LA, Xia B, Livingston DM, Greenberg RA 2007. RAP80 targets BRCA1 to specific ubiquitin structures at DNA damage sites. Science 316: 1198–1202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorensen CS, Hansen LT, Dziegielewski J, Syljuasen RG, Lundin C, Bartek J, Helleday T 2005. The cell-cycle checkpoint kinase Chk1 is required for mammalian homologous recombination repair. Nat Cell Biol 7: 195–201 [DOI] [PubMed] [Google Scholar]
- Spycher C, Miller ES, Townsend K, Pavic L, Morrice NA, Janscak P, Stewart GS, Stucki M 2008. Constitutive phosphorylation of MDC1 physically links the MRE11-RAD50-NBS1 complex to damaged chromatin. J Cell Biol 181: 227–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart GS 2009. Solving the RIDDLE of 53BP1 recruitment to sites of damage. Cell Cycle 8: 1532–1538 [DOI] [PubMed] [Google Scholar]
- Stewart GS, Wang B, Bignell CR, Taylor AM, Elledge SJ 2003. MDC1 is a mediator of the mammalian DNA damage checkpoint. Nature 421: 961–966 [DOI] [PubMed] [Google Scholar]
- Stiff T, Walker SA, Cerosaletti K, Goodarzi AA, Petermann E, Concannon P, O’Driscoll M, Jeggo PA 2006. ATR-dependent phosphorylation and activation of ATM in response to UV treatment or replication fork stalling. EMBO J 25: 5775–5782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokes MP, Rush J, Macneill J, Ren JM, Sprott K, Nardone J, Yang V, Beausoleil SA, Gygi SP, Livingstone M, et al. 2007. Profiling of UV-induced ATM/ATR signaling pathways. Proc Natl Acad Sci 104: 19855–19860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stracker TH, Morales M, Couto SS, Hussein H, Petrini JH 2007. The carboxy terminus of NBS1 is required for induction of apoptosis by the MRE11 complex. Nature 447: 218–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stucki M, Clapperton JA, Mohammad D, Yaffe MB, Smerdon SJ, Jackson SP 2005. MDC1 directly binds phosphorylated histone H2AX to regulate cellular responses to DNA double-strand breaks. Cell 123: 1213–1226 [DOI] [PubMed] [Google Scholar]
- Sun Y, Jiang X, Chen S, Fernandes N, Price BD 2005. A role for the Tip60 histone acetyltransferase in the acetylation and activation of ATM. Proc Natl Acad Sci 102: 13182–13187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Xu Y, Roy K, Price BD 2007. DNA damage-induced acetylation of lysine 3016 of ATM activates ATM kinase activity. Mol Cell Biol 27: 8502–8509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Jiang X, Xu Y, Ayrapetov MK, Moreau LA, Whetstine JR, Price BD 2009. Histone H3 methylation links DNA damage detection to activation of the tumour suppressor Tip60. Nat Cell Biol 11: 1376–1382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symington LS, Gautier J 2011. Double-strand break end resection and repair pathway choice. Annu Rev Genet 45: 247–271 [DOI] [PubMed] [Google Scholar]
- Takai H, Wang RC, Takai KK, Yang H, de Lange T 2007. Tel2 regulates the stability of PI3K-related protein kinases. Cell 131: 1248–1259 [DOI] [PubMed] [Google Scholar]
- Takai H, Xie Y, de Lange T, Pavletich NP 2010. Tel2 structure and function in the Hsp90-dependent maturation of mTOR and ATR complexes. Genes Dev 24: 2019–2030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomimatsu N, Mukherjee B, Burma S 2009. Distinct roles of ATR and DNA-PKcs in triggering DNA damage responses in ATM-deficient cells. EMBO Rep 10: 629–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomimatsu N, Mukherjee B, Deland K, Kurimasa A, Bolderson E, Khanna KK, Burma S 2012. Exo1 plays a major role in DNA end resection in humans and influences double-strand break repair and damage signaling decisions. DNA Repair (Amst) 11: 441–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uziel T, Lerenthal Y, Moyal L, Andegeko Y, Mittelman L, Shiloh Y 2003. Requirement of the MRN complex for ATM activation by DNA damage. EMBO J 22: 5612–5621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Elledge SJ 2007. Ubc13/Rnf8 ubiquitin ligases control foci formation of the Rap80/Abraxas/Brca1/Brcc36 complex in response to DNA damage. Proc Natl Acad Sci 104: 20759–20763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Matsuoka S, Carpenter PB, Elledge SJ 2002. 53BP1, a mediator of the DNA damage checkpoint. Science 298: 1435–1438 [DOI] [PubMed] [Google Scholar]
- Wang H, Powell SN, Iliakis G, Wang Y 2004. ATR affecting cell radiosensitivity is dependent on homologous recombination repair but independent of nonhomologous end joining. Cancer Res 64: 7139–7143 [DOI] [PubMed] [Google Scholar]
- Wang B, Matsuoka S, Ballif BA, Zhang D, Smogorzewska A, Gygi SP, Elledge SJ 2007. Abraxas and RAP80 form a BRCA1 protein complex required for the DNA damage response. Science 316: 1194–1198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Gong Z, Chen J 2011. MDC1 collaborates with TopBP1 in DNA replication checkpoint control. J Cell Biol 193: 267–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward IM, Chen J 2001. Histone H2AX is phosphorylated in an ATR-dependent manner in response to replicational stress. J Biol Chem 276: 47759–47762 [DOI] [PubMed] [Google Scholar]
- Wawrousek KE, Fortini BK, Polaczek P, Chen L, Liu Q, Dunphy WG, Campbell JL 2010. Xenopus DNA2 is a helicase/nuclease that is found in complexes with replication proteins And-1/Ctf4 and Mcm10 and DSB response proteins Nbs1 and ATM. Cell Cycle 9: 1156–1166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams DR, Lee KJ, Shi, J, Chen DJ, Stewart PL 2008. Cryo-EM structure of the DNA-dependent protein kinase catalytic subunit at subnanometer resolution reveals α helices and insight into DNA binding. Structure 16: 468–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Luo K, Lou Z, Chen J 2008. MDC1 regulates intra-S-phase checkpoint by targeting NBS1 to DNA double-strand breaks. Proc Natl Acad Sci 105: 11200–11205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo HY, Kumagai A, Shevchenko A, Dunphy WG 2007. Ataxia-telangiectasia mutated (ATM)-dependent activation of ATR occurs through phosphorylation of TopBP1 by ATM. J Biol Chem 282: 17501–17506 [DOI] [PubMed] [Google Scholar]
- Yoo HY, Kumagai A, Shevchenko A, Dunphy WG 2009. The Mre11-Rad50-Nbs1 complex mediates activation of TopBP1 by ATM. Mol Biol Cell 20: 2351–2360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- You Z, Chahwan C, Bailis J, Hunter T, Russell P 2005. ATM activation and its recruitment to damaged DNA require binding to the C terminus of Nbs1. Mol Cell Biol 25: 5363–5379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- You Z, Bailis JM, Johnson SA, Dilworth SM, Hunter T 2007. Rapid activation of ATM on DNA flanking double-strand breaks. Nat Cell Biol 9: 1311–1318 [DOI] [PubMed] [Google Scholar]
- You Z, Shi LZ, Zhu Q, Wu P, Zhang YW, Basilio A, Tonnu N, Verma IM, Berns MW, Hunter T 2009. CtIP links DNA double-strand break sensing to resection. Mol Cell 36: 954–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zha S, Guo C, Boboila C, Oksenych V, Cheng HL, Zhang Y, Wesemann DR, Yuen G, Patel H, Goff PH, et al. 2011. ATM damage response and XLF repair factor are functionally redundant in joining DNA breaks. Nature 469: 250–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N, Kaur R, Akhter S, Legerski RJ 2009. Cdc5L interacts with ATR and is required for the S-phase cell-cycle checkpoint. EMBO Rep 10: 1029–1035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou BB, Elledge SJ 2000. The DNA damage response: Putting checkpoints in perspective. Nature 408: 433–439 [DOI] [PubMed] [Google Scholar]
- Zou L, Elledge SJ 2003. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science 300: 1542–1548 [DOI] [PubMed] [Google Scholar]
- Zou L, Liu D, Elledge SJ 2003. Replication protein A-mediated recruitment and activation of Rad17 complexes. Proc Natl Acad Sci 100: 13827–13832 [DOI] [PMC free article] [PubMed] [Google Scholar]