Abstract
The endosomal sorting complexes required for transport (ESCRT) drive multivesicular body (MVB) biogenesis and cytokinetic abscission. Originally identified through genetics and cell biology, more recent work has begun to elucidate the molecular mechanisms of ESCRT-mediated membrane remodeling, with special focus on the ESCRT-III complex. In particular, several light and electron microscopic studies provide high-resolution imaging of ESCRT-III rings and spirals that purportedly drive MVB morphogenesis and abscission. These studies highlight unifying principles to ESCRT-III function, in particular: (1) the ordered assembly of the ESCRT-III monomers into a heteropolymer, (2) ESCRT-III as a dynamic complex, and (3) the role of the AAA ATPase Vps4 as a contributing factor in membrane scission. Mechanistic comparisons of ESCRT-III function in MVB morphogenesis and cytokinesis suggest common mechanisms in membrane remodeling.
The ESCRT pathway mediates certain types of membrane remodeling (e.g., cellular abscission). Recent light and electron microscopy studies focusing on one of the ESCRT complexes, ESCRT-III, are revealing mechanistic details.
THE ESCRT PATHWAY IN MVB BIOGENESIS AND CYTOKINESIS
The endosomal sorting complex required for transport (ESCRT) comprises five distinct protein complexes denoted numerically as ESCRTs -0, -I, -II, -III, and the AAA ATPase Vps4 complex (Hurley, 2010; Babst 2011; Henne et al. 2011). They were initially identified through yeast genetics as vacuolar protein sorting (vps) genes whose loss blocked the delivery of vacuole hydrolases, leading to aberrant endosomal morphology. After their initial identification, further cell biological studies revealed that these gene products constitute separate protein complexes that engage transmembrane cargoes at endosomes and convert these organelles into multivesicular bodies (MVBs) with the cargoes sorted within their internal vesicles.
Since their discovery in 2001 (Katzmann et al. 2001), much has been learned concerning how the ESCRT pathway drives MVB morphogenesis. One particular theme involves the division of labor shared between distinct ESCRT complexes. ESCRTs -0, -I, and -II form stable hetero-oligomers that function together to recognize ubiquitin and the endosome-enriched phospholipid, phosphatidylinositol 3-phosphate (PtdIns(3)P) at the endosomal surface (Fig. 1A) (Raiborg et al. 2001; Katzmann et al. 2003; Teo et al. 2006). In addition, ESCRT-II initiates the assembly of the ESCRT-III complex (Teis et al. 2008). Although also a hetero-oligomer, ESCRT-III is unique for several reasons: (1) it lacks the ability to recognize ubiquitin directly, although recent reports indicate ESCRT-III-interacting protein Bro1/Alix (ALG2-interacting protein X) can directly bind ubiquitin (Dowlatshahi et al. 2012); (2) it displays no lipid specificity for PtdIns(3)P, implying why it can directly function in membrane remodeling at both endosomes as well as the plasma membrane; and (3) ESCRT-III does not form a stable complex, but rather assembles transiently during membrane sculpting and fission events. ESCRT-III is additionally unique because this membrane-remodeling machine induces invaginations that protrude away from the cell cytoplasm. This is topologically inverted from more studied vesicle budding pathways such as clathrin and COP-mediated vesicle formation, and remains a mysterious process whose molecular mechanisms are beginning to be addressed.
Figure 1.
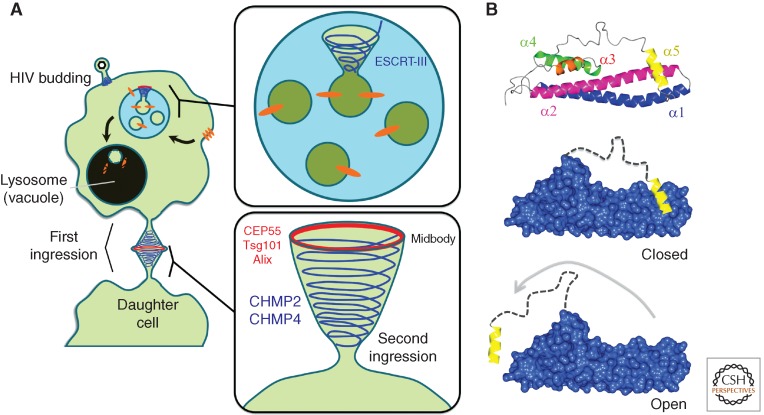
ESCRT-mediated membrane remodeling in cellular physiology and pathophysiology. (A) Schematic of ESCRT-III helices driving MVB cargo sorting, HIV budding, and cell division. (B) Helical model of a Snf7 subunit from Saccharomyces cervisiae (top). ESCRT-III subunits undergo conformational changes from a closed (middle) to open state (bottom).
A remarkable connection between MVB biogenesis and cytokinesis, the process by which the plasma membrane ingresses to divide into separate daughter cells, emerged when it was found that ESCRT proteins are recruited to the cell midbody immediately before final abscission (Garrus et al. 2001; Carlton and Martin-Serrano 2007; Morita et al. 2007). Additionally, ESCRT proteins are directly involved in HIV budding from the plasma membrane, which is topologically consistent with both cell division and MVB morphogenesis (Garrus et al. 2001). For the purposes of this review, we are focusing on the properties of ESCRT-mediated membrane remodeling in MVB morphogenesis and cytokinesis. ESCRT-I subunit Tsg101 (Vps23) and Alix (Bro1) are recruited to the midbody by centrosomal protein of 55 kDa (CEP55), a known cytokinesis effector protein (Fig. 1A). Later studies established that several ESCRT-III proteins also localize to the midbody. Furthermore, depletion of ESCRT-III proteins Snf7/CHMP4B or Vps2/CHMP2, or mutation of the ESCRT-III binding regions on Alix, perturbs late stages of cell division (Table 1) (Carlton and Martin-Serrano 2007).
Table 1.
ESCRT proteins in MVB sorting and cytokinesis
| The complexes | Function | Necessary for MVB sorting? | Necessary for cytokinesis? | Disease associations | |
|---|---|---|---|---|---|
| Yeast | Human | ||||
| ESCRT-0 | Binds ubiquitinated cargo and PtdIns(3)P | Cancer | |||
| Vps27 | Hrs | Yes | No | ||
| Hse1 | STAM1/2 | No | No | ||
| ESCRT-I | Binds ESCRT-0,-II, and ubiquitinated cargo; localizes to midbody during cell division | Cancer, AIDS | |||
| Vps23 | Tsg101 | Yes | Yes | ||
| Vps28 | Vps28 | Yes | Yes | ||
| Vps37 | Vps37A-D | Yes | Yes | ||
| Mvb12 | hMvb12A, B | Slight defect | n.d. | ||
| — | UBAP1 | Yes | No | ||
| ESCRT-II | Binds ESCRT-I and nucleates ESCRT-III; binds ubiquitinated cargo and PtdIns(3)P | ||||
| Vps36 | EAP45 | Yes | No | ||
| Vps22 | EAP30 | Yes | No | ||
| Vps25 | EAP20 | Yes | No | ||
| ESCRT-III | Sculpts MVB vesicles and drives cytokinesis | FTD3, AIDS HD Parkinson’s | |||
| Vps20 | CHMP6 | Yes | No? | ||
| Snf7 | CHMP4A-D | Yes | Yes | ||
| Vps24 | CHMP3 | Yes | Yes | ||
| Vps2 | CHMP2A, B | Yes | Yes | ||
| Vps60 | CHMP5 | Yes | Yes | ||
| Bro1 | Alix | Yes | Yes | ||
| Ist1 | IST1 | No | Yes | ||
| Did2 | CHMP1 | Yes | Yes | ||
| AAA ATPases | Drives ESCRT-III disassembly severs MT during abscission | AIDS | |||
| Vps4 | Vps4A and B | Yes | Yes | ||
| — | Spastin | Yes | Yes | HSP | |
| Other MIT domain proteins | |||||
| Vta1 | LIP5 | Slight defect | Yes | ||
| — | MITD1 | No | Yes | ||
| Additional notable proteins | Recruits ESCRTs to midbody phosphorylates CHMP4C; cell division checkpoint | ||||
| — | CEP55 | No | Yes | ||
| — | Aurora B | No | Yes | ||
FTD3, frontotemporal dementia linked to chromosome 3; AIDS, acquired immune deficiency syndrome; HD, Huntington’s disease; HSP, herditary spastic paraplegia; n.d., no data.
Showing that ESCRT proteins play a direct role in cytokinesis was groundbreaking, and highlighted the fact that both MVB sorting and cytokinesis require topologically similar membrane remodeling. However, despite identifying ESCRT-III subunits as multipurpose membrane remodeling effectors, the molecular mechanisms by which ESCRT-III drives this membrane sculpting remained poorly understood. Recent high-resolution light and electron microscopy studies are now providing a working model for how ESCRT-III achieves membrane sculpting. These studies are mechanistically impactful because they provide direct visualization of ESCRT-III assemblies both in vitro and in vivo. The purpose of this review is to highlight the recent advances in the mechanistic understanding of ESCRT-III and discuss their implications for the mechanism of ESCRT-mediated membrane remodeling in different cellular pathways. The review will first discuss ESCRT-III architecture and function in the context of MVB sorting, and then turn to recent advances in understanding its roles in mammalian cell division. Comparison between these two distantly related pathways suggests ancient principles for membrane remodeling that have been adapted over time to function in diverse spatial and temporal scales.
ESCRT-III FUNCTION IN MVB MORPHOGENESIS
Molecular Architecture and Conformational Dynamics of ESCRT-III Subunits
Yeast expresses four “core” ESCRT-III subunits (Vps20, Snf7, Vps24, and Vps2). Humans share these “core” subunits and additionally have four Snf7 paralogues (CHMP4A-D) and two Vps2 paralogues (CHMP2A and B) (CHMP: charged multivesicular body protein). Early studies of ESCRT-III subunits established that they were small, soluble, and electrostatically charged proteins that all share a common architectural fold (Shim et al. 2007). For simplicity, they are often bisected into a basic amino-terminal half and acidic carboxy-terminal half. The amino-terminal half is highly structured and is composed of four α-helices that fold into an elongated bundle approximately 7 nm in length (Muziol et al. 2006; Bajorek et al. 2009). This amino-terminal “core” domain is enriched with basic amino acids that bind acidic lipids within lipid bilayers. The carboxy-terminal half, in contrast, is largely unstructured.
Elegant biochemical work indicated that the acidic carboxy-terminal half of the protein formed intramolecular contacts with the basic amino-terminal half, rendering ESCRT-III subunits in an autoinhibited “closed” conformational state (Fig. 1B) (Zamborlini et al. 2006). X-ray crystallographic structures of VPS24/CHMP3, and IST1 revealed the structural basis for this autoinhibition (Muziol et al. 2006; Bajorek et al. 2009). A conserved carboxy-terminal region folds into an α-helix (denoted here as α5) forming numerous intramolecular contacts along the amino-terminal core domain. More recent work showed that this autoinhibitory mechanism was present in Snf7, indicating that it is a general principle among all “core” ESCRT-III subunits (Henne et al. 2012). Because of this, it is commonly accepted that ESCRT-III subunits exist as “closed” monomers in the cell cytoplasm until their activation and recruitment to membrane surfaces during ESCRT-III assembly.
Displacement of the carboxy-terminal autoinhibitory region drives both membrane binding and, for at least Snf7 and Vps24, homo-oligomerization (Shim et al. 2007; Ghazi-Tabatabai et al. 2008; Henne et al. 2012). Numerous observations also suggest that the carboxy-terminal region is critical for stabilizing homo-polymers. The mechanism for this is unclear, but may involve novel intermolecular contacts formed when α5 and the flexible linker region of one subunit reach across to contact a neighboring subunit. When VPS24 crystallized as an antiparallel dimer, each α5 formed intermolecular contacts with its opposing hVps24 monomer. This suggested that these regulatory helices may form stabilizing contacts with neighboring subunits following their release from autoinhibition (Muziol et al. 2006; Bajorek et al. 2009). Similarly, GST-fusion constructs encoding VPS24 α5 were sufficient to interact with VPS24 core domains in in vitro pull down experiments, suggesting they could function as intermolecular contacts (Zamborlini et al. 2006). In another example, deletion or mutation of this helix in yeast Snf7 attenuates polymer formation, although similar manipulation of human Snf7 did not completely halt oligomerization in vivo using mammalian cells (Hanson et al. 2008; Henne et al. 2012).
Thus, it appears that the conformational opening of ESCRT-III subunits tightly coordinates three different activities: (1) subunit release from autoinhibition, (2) promotion of membrane binding, and (3) polymer stabilization. One perplexing question is how this conformational opening exposes the membrane interacting surfaces of ESCRT-III subunits. Previous studies suggest that the primary membrane interacting surfaces of these proteins are composed of positively charged amino acids along the α-helical hairpin (Muziol et al. 2006). How the carboxy-terminal autoinhibitory region occludes these ostensibly large electrostatic contacts requires additional structural studies. One possibility is that the flexible linker that precedes α5 may physically mask the electrostatic interface that contacts membrane. In yeast Snf7, this linker region is highly acidic, and is predicted to form electrostatic contacts with basic residues along the Snf7 core domain (Henne et al. 2012). Another open question is how ESCRT-II and other ESCRT-III subunits trigger the release of ESCRT-III subunit autoinhibition. ESCRT-III subunits recruit one another sequentially to endosomes, indicating that the physical contact between subunits triggers a conformational opening (Teis et al. 2008). How this is structurally coordinated requires additional study.
THE ASSEMBLY OF ESCRT-III POLYMERS: DOME VERSUS HELICAL MODELS
Following its recruitment by ESCRT-I, ESCRT-II directly binds Vps20 and initiates the sequential activation of ESCRT-III subunits at the endosome surface (Teo et al. 2004). Notably, this activation follows a precise sequence of events allowing ESCRT-III to polymerize by a predetermined subunit order, a process termed “ordered assembly.” Initially, Vps20 directly binds Snf7, which in turn homo-oligomerizes. Snf7 in turn recruits Vps24 and Vps2, which contain high-affinity MIM motifs necessary for the recruitment and activation of the AAA ATPase Vps4 (Teis et al. 2008).
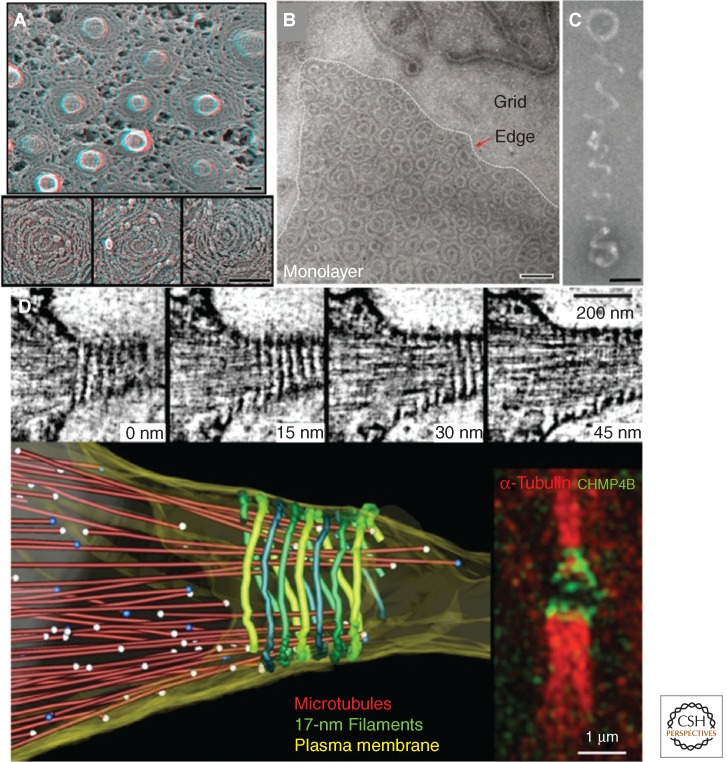
Several studies have begun to visualize this stepwise ESCRT-III assembly with high-resolution techniques. By overexpressing SNF7/CHMP4 truncations that lacked the autoinhibitory carboxy-terminal region, Hanson et al. were the first to visualize ESCRT-III polymers in vivo. Using quick freeze deep-etch EM (DEEM) microscopy, they discovered that SNF7/CHMP4 assembled into circular arrays on the inner leaflet of the plasma membrane of COS-7 cells (Fig. 2A) (Hanson et al. 2008). These arrays were composed of loosely coiled filaments ∼4 nm in diameter that extended across the membrane surface. Strikingly, coexpressing SNF7/CHMP4 with an ATP-hydrolysis defective mutant Vps4 caused the arrays to compact into tightly packed circular lattices. These conditions also generate membrane buds that protruded outward from the plasma membrane in a direction topologically consistent with MVB morphogenesis. Because Vps4 recruitment is thought to follow the assembly of Snf7 polymers, this Vps4-dependent membrane deformation appears consistent with the ordered assembly model for ESCRT-III.
Figure 2.
High-resolution imaging of ESCRT-III assemblies. (A) Electron micrograph of circular array protein scaffolds on membrane tubules in COS-7 cells expressing Snf7-1/CHMP4A and VPS4B(E235Q)-GFP. (Image from Hanson et al. 2008; adapted, with permission, from The Rockefeller University Press © 2008.) (B) Electron micrograph of reconstituted yeast Snf7 spirals assembled on lipid monolayers. (C) A helical polymer of reconstituted yeast Snf7, Vps24, and Vps2, imaged by electron microscopy. (Panels B and C are from Henne et al. 2012; adapted, with permission, from Cell Press © 2012.) (D) Electron tomography of high-pressure frozen cells, which show 17-nm filaments spiraling along the constriction zone. (This image is from Guizetti et al. 2011; adapted, with permission, from the author.) Scale bars, 100 nm, unless stated otherwise.
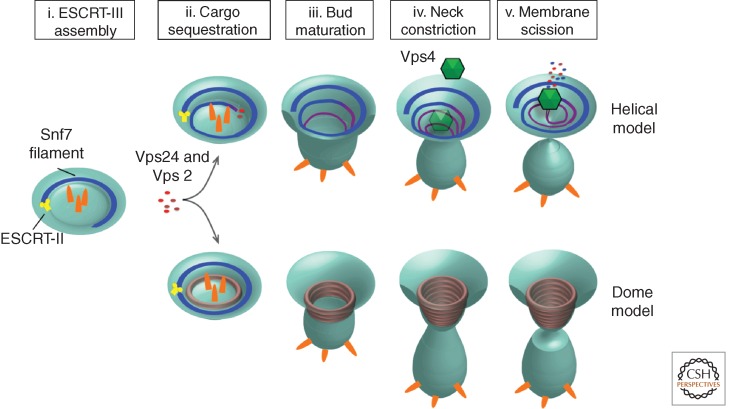
Although visually striking, the imaging of Snf7 circular arrays did not provide an intuitive role for Vps24 and Vps2 in ESCRT-III-mediated membrane remodeling. In vitro studies using purified VPS24/CHMP3 and VPS2/CHMP2 soon provided additional details. Using a similar strategy, studies by Lata et al. (2008) used negative stain electron microscopy to show that truncated VPS24 and VPS2 coassemble into helical cylinders in vitro. The outsides of these cylinders adhered to liposomes, suggesting a membrane-binding interface that was topologically consistent with MVB budding (Lata et al. 2008). Notably, these cylinders terminated with domelike caps. These observations have given rise to the “dome model” of ESCRT-III catalyzed membrane scission (Fig. 3) (Fabrikant et al. 2009). In this model, a Vps24/Vps2 dome lines the interior of a membrane bud. The protein dome naturally tapers the lipid bilayers into close proximity at the dome base, promoting spontaneous fission. This tapering alone may be sufficient for the fission, or it may be catalyzed by the recruitment of Vps4 and subsequent disassembly of the Vps24/Vps2 dome (Lata et al. 2008).
Figure 3.
Helical and dome models for ESCRT-III-mediated membrane remodeling and vesicle scission.
These studies provided important insights into the molecular architecture of individual ESCRT-III polymers. However, it was unclear how Snf7, Vps24, and Vps2 worked together to drive membrane remodeling. More recent work now suggests that these proteins coassemble into helical polymers that drive vesicle budding by a springlike mechanism (Henne et al. 2012). These helices are structurally distinct from the flat spirals and circular arrays that Snf7 forms as a homopolymer. Thus, Vps24 and Vps2 may modulate Snf7 polymer architecture by either incorporating into a growing Snf7 polymer or binding to a preassembled polymer and modulating its architecture, inducing a transition from a two-dimensional spiral into a three-dimensional helix (Figs. 2B, 2C, and 3). This transition may generate the mechanical force necessary for vesicle budding (Henne et al. 2012). This model also explains why vesicles could bud into giant unilamellar vesicles (GUVs) after addition of Snf7, but was more efficient when Snf7, Vps24, and Vps2 were sequentially added to GUVs (Wollert et al. 2009; Henne et al. 2012).
Vps4 INVOLVEMENT IN ESCRT-MEDIATED MEMBRANE SCULPTING
The dome and helical models provide structural explanations for ESCRT-III-induced membrane invagination, but do not necessarily explain membrane fission. In the dome model, the protein shell causes the membrane to taper down to a highly curved termination. This has been proposed to promote spontaneous fission (Fabrikant et al. 2009), although electrostatically charged protein scaffolds have been proposed to stabilize high curvature and inhibit membrane fission (Fig. 3) (Boucrot et al. 2012). An alternative model would be that Vps4-mediated disassembly of the ESCRT-III dome would release membrane tension, promoting buckling and subsequent fission.
Vps4 directly modulates the architecture of ESCRT-III helices in vivo (Hanson et al. 2008). Expression of an ATP hydrolysis-defective Vps4E235Q caused hSnf7 spirals to condense into tightly bundled circular arrays with membrane tubules sprouting from their centers. This suggested that Vps4 contributed directly to ESCRT-III-mediated membrane remodeling. In contrast, in vitro experiments on GUVs indicated that Vps4 was not required for ESCRT-III-mediated vesicle biogenesis, but rather functioned to disassemble ESCRT-III following vesicle fission (Wollert et al. 2009). However, at least two recent observations suggest a direct role for Vps4 in vesicle formation. First, live-cell total internal reflection fluorescence microscopy (TIRFM) of HIV budding from the plasma membrane revealed that fluorescently tagged Vps4A was recruited to HIV buds before their release, indicating a prefission role for Vps4 (Baumgartel et al. 2011). Second, more recent real-time imaging of GUVs suggests that ESCRT-III can drive membrane invaginations, but vesicle release was often inefficient (Henne et al. 2012). Several studies have noted that GUVs spontaneously deform and generate internal vesicles independent of ESCRT-III-mediated remodeling (Mayers and Audhya 2011). The addition of ESCRT proteins may stabilize or even promote these spontaneous deformations, completing vesicle scission in a reconstituted, but not physiologically relevant, manner. Thus, further experiments are needed to definitively show whether Vps4 is directly involved in completing ESCRT-mediated membrane remodeling and fission.
ESCRT COMPLEXES IN CYTOKINESIS
The observations that ESCRT components are required for completion of cytokinesis in plants (Spitzer et al. 2006) and in cultured mammalian cells (Carlton and Martin-Serrano 2007) have opened new avenues for understanding the membrane-sculpting properties of ESCRTs, because cytokinesis is topologically similar to but spatially distinct from MVB budding. The mammalian cytokinesis process overlaps with telophase in the cell division cycle and begins with the formation of an actomyosin-driven cleavage furrow. As the cleavage furrow ingresses, an intercellular bridge is formed between the two daughter cells, and eventually the daughter cells become completely separated when this bridge resolves at one or two secondary ingressions—constriction zones—that form on either side of the electron-dense midbody during the process known as cytokinetic abscission. The intercellular bridge typically has a diameter of more than 1 μm just before abscission and is packed with interdigitating spindle microtubules, so one of the enigmas in the field of cell division has been how secondary ingression, microtubule severing, and membrane scission are coordinated and executed. Proposed models for membrane scission have ranged from constriction through intracellular vesicle fusion and assembly of cortical membrane severing machineries. The involvement of ESCRTs in cytokinetic abscission offers mechanistic insight that has led to new views of how microtubule severing and membrane scission are executed.
A key factor in mammalian cytokinesis is the centrosomal protein CEP55, which is recruited to the midbody during mitotic exit as the activity of an inhibitory kinase, Plk1, ceases (Bastos and Barr 2010). CEP55 in turn recruits ESCRT-I to the midbody via recognition of motif containing the amino acid sequence glycine-proline-proline (GPP). Interestingly, the ESCRT-associated protein Alix is also recruited to the midbody by a GPP-mediated interaction with CEP55 (Carlton and Martin-Serrano 2007; Morita et al. 2007). Recruitment of ESCRT-I and Alix to the midbody functions as a platform for recruitment and activation of ESCRT-III without the need for ESCRT-II. The specific function of ESCRT-I in cytokinesis remains unresolved to date. Depletion of TSG101 profoundly inhibits cytokinesis, but it is not clear whether other subunits of ESCRT-I are also involved in cytokinesis.
The details of ESCRT-III activation during cytokinesis are not known, but it has been shown that Alix interacts directly with the ESCRT-III subunit SNF7/CHMP4, and that this interaction is required for cytokinesis (Morita et al. 2007). An interaction between the Bro1 domain of Alix and the autoinhibitory carboxyl-terminus of SNF7/CHMP4 might release autoinhibition and thereby activate ESCRT-III oligomerization. On the other hand, VPS20/CHMP6 also seems to play a role in cytokinesis, although it is less essential than the other 11 human ESCRT-III subunits (Table 1) (Morita et al. 2011). This raises the possibility that ESCRT-III activation during cytokinesis could be mechanistically related to ESCRT-III activation during MVB biogenesis.
Electron microscopy of cytokinetic bridges has shown the presence of 17-nm helical filaments that spiral from the midbody toward the constriction zones (Fig. 2D) (Guizetti et al. 2011). Because these filaments are CHMP4B-dependent and because helices of CHMP4B can be detected by structured illumination microscopy, it is tempting to speculate that the 17-nm filaments consist of ESCRT-III subunits, although this remains to be formally shown. Intriguingly, filaments composed of yeast Snf7 measure ∼4 nm in diameter, but bundle together in pairs, forming protofilaments ∼9–12 nm in diameter (Henne et al. 2012). In further support, spiraling CHMP4B filaments ∼5–6 nm diameter have also been observed when overexpressed in COS-7 cells (Hanson et al. 2008).
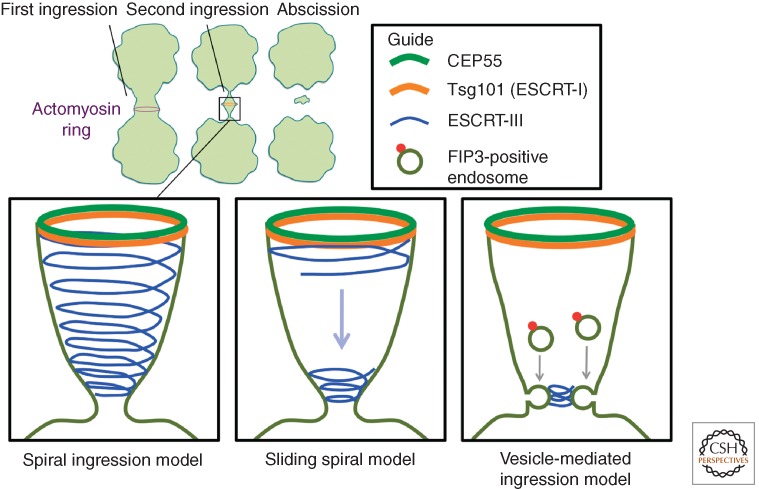
Three alternative models have been proposed for ESCRT-mediated formation of the constriction zone during cytokinesis (Fig. 4). According to the “spiral-ingression” model, ESCRT-III-containing spirals wind from the midbody to define the abscission zone at the end of the narrowing spiral. Here, Spastin is recruited by CHMP1B to sever microtubules, and membrane severing is mediated directly by the ESCRT filaments (Yang et al. 2008; Guizetti et al. 2011). A variant of this model is the “sliding spiral” model in which ESCRT-III filaments start to form at the midbody, and a spiral filament slides along the intracellular bridge to form an abscission zone about 1 µm from the midbody (Elia et al. 2011). A third alternative is the “vesicle-mediated ingression” model, in which FIP3-containing vesicles derived from recycling endosomes define the abscission zone by causing narrowing of the intercellular bridge through fusion with the plasma membrane (Schiel et al. 2012). According to this latter model, ESCRT-III filaments are recruited after the abscission zone is formed. Common to the three models is that ESCRTs mediate the final abscission between the two daughter cells. As is the case with scission of MVB vesicles, several alternative explanations have been put forward, including membrane constriction caused by inward-directed curvature of ESCRT-III filaments, constriction caused by remodeling of ESCRT-III filaments, or ESCRT-III-mediated promotion of spontaneous lipid-driven membrane bulging (Fededa and Gerlich 2012). These models for cytokinesis are in agreement with current models for ESCRT-mediated MVB morphogenesis. In particular, the “spiral ingression” model is consistent with the visualization of yeast ESCRT-III spirals and helices in vitro.
Figure 4.
Three models for ESCRT-III-mediated abscission.
As with MVB biogenesis, the ATPase activity of VPS4 is essential during cytokinesis (Carlton and Martin-Serrano 2007; Morita et al. 2007). VPS4 is recruited at a late stage during the abscission process through interaction with the ESCRT-III like IST1 protein (Agromayor et al. 2009; Bajorek et al. 2009), which is negatively regulated by another MIT-domain-containing protein, MITD1 (Hadders et al. 2012; Lee et al. 2012). It is interesting to note that IST1, while being essential for mammalian cytokinesis, is dispensable for viral budding (Agromayor et al. 2009), illustrating a point of divergence between these two ESCRT-III-dependent membrane scission processes. The recruitment of VPS4 occurs after that of CHMP4B and coincides with membrane cutting (Elia et al. 2011). This is consistent with the possibility that VPS4 could provide the mechanochemical force for membrane severing as such, but also with the scenario that it could serve in ESCRT-III disassembly and recycling.
A relatively frequent event during mammalian cell division is the entrapment of lagging chromosomes in the intercellular bridge. To prevent chromosomal damage during cytokinesis, the cells are equipped with an abscission checkpoint termed NoCut, which senses trapping of lagging chromosomes and ensures that the time of abscission is delayed sufficiently to allow removal of residual chromatin from the cytokinetic bridge. The chromosomal passenger complex (CPC), which consists of Aurora B kinase, INCENP, Survivin, and Borealin, is the central player in the NoCut checkpoint. It has been unclear how CPC activation can be translated into cytokinesis arrest, but recent studies of the ESCRT-III isoform CHMP4C have revealed an intriguing link between CPC and the abscission machinery (Carlton et al. 2012). Whereas depletion of CHMP4B and other ESCRT-III components inhibits or delays cytokinetic abscission, depletion of CHMP4C reduces abscission time by about 30 min in HeLa cells. CHMP4C associates with the Borealin component of CPC and contains a specific insertion, which includes a site for Aurora B-mediated phosphorylation. Aurora B-mediated phosphorylation of this site, which is conserved in mammals but not in lower eukaryotes, promotes recruitment of CHMP4C to the midbody and halts abscission. The exact mechanism of abscission inhibition by CHMP4C remains to be identified, but it is tempting to speculate that phosphorylated CHMP4C engages in nonproductive ESCRT-III filaments.
CONCLUDING REMARKS: REMAINING QUESTIONS AND FUTURE STUDIES
Recent studies have highlighted the remarkable ability of ESCRT-III to remodel membranes in diverse cellular pathways. Using high-resolution methodologies, the mechanisms that govern this membrane remodeling are beginning to be understood at extreme spatial and temporal detail. However, several outstanding questions remain to be addressed, and will require new and clever techniques.
The most pressing question is how the final membrane fission event is coordinated. Whether ESCRT-III itself is sufficient to drive this fission, or whether it requires additional factors like the energy released through ATP hydrolysis from Vps4 is a closely linked question that requires further investigation. We also need a better understanding of ESCRT-III dynamics during membrane remodeling. The high spatial resolution provided by transmission electron microscopy fails to provide real-time imaging of ESCRT dynamics. Similarly, live-cell imaging of ESCRT-mediated cytokinesis and viral budding provides excellent temporal resolution, but is limited by the spatial resolution of visible light. New techniques are required to better understand ESCRT-III dynamics, which will hopefully reveal how these dynamics generate membrane-bending forces. Such techniques may also provide insights into how ESCRT-III subunits coassemble into helices, including whether Vps24 and Vps2 integrate with Snf7 into a heteropolymer, or bind to the surface of a Snf7 homopolymer.
Further high-resolution experiments may also reveal other mechanistic distinctions unique to the ESCRT pathway. ESCRT-III drives membrane remodeling that pushes away from the cytoplasm, yet it is unclear how it avoids consuming itself in this process. Likewise, the vesicles that bud into the endosomal lumen by ESCRTs contain no outer vesicle coat, yet show consistent diameters, suggesting ESCRT complexes regulate vesicle size. How this is achieved remains a mystery, but could be attributed to the uniform size of ESCRT-III rings, which may set the size of vesicles by surrounding the membrane surface that is transformed into the vesicle (Henne et al. 2012). Other models suggest that raftlike membrane domains that are induced by ESCRT-binding are sufficient to stabilize vesicle budding and potentially regulate vesicle diameter (Rozycki et al. 2012). Another possibility is that the transmembrane cargoes themselves contribute to vesicle morphogenesis through crowding. Here, the physical sterics of cargo packing may promote the membrane bending initially generated by ESCRTs. In line with this, experiments using high concentrations of GFP molecules tethered to GUVs show that clustered proteins are sufficient to drive membrane bending, although the significance of this needs to be shown in living cells (Stachowiak et al. 2012). Indeed, how ubiquitinated cargoes are initially captured, sequestered, and loaded into vesicles by ESCRTs requires additional study. Recent work shows that EGF receptors are transferred from ESCRT-0 to ESCRT-III by the coordinated action of HD-PTP/PTPN23 and UBPY (Ali et al. 2013). This implies a “hand-off” in which cargoes are simultaneously deubiquitinated and loaded into forming vesicles, and should be investigated further.
Finally, further studies are needed to reveal the molecular architecture of ESCRT-III complexes at atomic resolution. Numerous electron micrographs exist for ESCRT-III assemblies, but these cannot reveal details like protein–protein interfaces. Currently, crystal structures exist for portions of VPS24/CHMP3, SNF7/CHMP4, and IST1 (Muziol et al. 2006; Bajorek et al. 2009; Martinelli et al. 2012). However, there are no structures of either full-length ESCRT-III subunits, or ESCRT-III hetero-oligomers. Solving such structures will provide insights into the molecular interfaces along subunit–subunit contacts. Such structures may even be able to be fitted into existing protein densities derived from electron micrographs, revealing the molecular architecture of ESCRT-III in higher resolution. No doubt future studies will continue to reveal new and exciting properties of this novel membrane sculpting machinery.
ACKNOWLEDGMENTS
The authors thank members of the Emr and Stenmark labs for helpful discussions. W.M.H. is sponsored by a Sam and Nancy Fleming Research Fellowship. S.D.E. is supported by a Cornell University Research Grant. H.S. is supported by the Research Council of Norway and the European Research Council.
Footnotes
Editors: Sandra L. Schmid, Alexander Sorkin, and Marino Zerial
Additional Perspectives on Endocytosis available at www.cshperspectives.org
REFERENCES
- Agromayor M, Carlton JG, Phelan JP, Matthews DR, Carlin LM, Ameer-Beg S, Bowers K, Martin-Serrano J 2009. Essential role of hIST1 in cytokinesis. Mol Biol Cell 20: 1374–1387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali N, Zhang L, Taylor S, Mironov A, Urbe S, Woodman P 2013. Recruitment of UBPY and ESCRT exchange drive HD-PTP-dependent sorting of EGFR to the MVB. Curr Biol 23: 453–461 [DOI] [PubMed] [Google Scholar]
- Babst M 2011. MVB vesicle formation: ESCRT-dependent, ESCRT-independent and everything in between. Curr Opin Cell Biol 23: 452–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajorek M, Schubert HL, McCullough J, Langelier C, Eckert DM, Stubblefield WM, Uter NT, Myszka DG, Hill CP, Sundquist WI 2009. Structural basis for ESCRT-III protein autoinhibition. Nat Struct Mol Biol 16: 754–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastos RN, Barr FA 2010. Plk1 negatively regulates Cep55 recruitment to the midbody to ensure orderly abscission. J Cell Biol 19: 751–760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumgartel V, Ivanchenko S, Dupont A, Sergeev M, Wiseman PW, Krausslich HG, Brauchle C, Muller B, Lamb DC 2011. Live-cell visualization of dynamics of HIV budding site interactions with an ESCRT component. Nat Cell Biol 13: 469–474 [DOI] [PubMed] [Google Scholar]
- Boucrot E, Pick A, Camdere G, Liska N, Evergren E, McMahon HT, Kozlov MM 2012. Membrane fission is promoted by insertion of amphipathic helices and is restricted by crescent BAR domains. Cell 149: 124–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlton JG, Martin-Serrano J 2007. Parallels between cytokinesis and retroviral budding: A role for the ESCRT machinery. Science 316: 1908–1912 [DOI] [PubMed] [Google Scholar]
- Carlton JG, Caballe A, Agromayor M, Kloc M, Martin-Serrano J 2012. ESCRT-III governs the Aurora B-mediated abscission checkpoint through CHMP4C. Science 336: 220–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowlatshahi DP, Sandrin V, Vivona S, Shaler TA, Kaiser SE, Melandri F, Sundquist WI, Kopito RR 2012. ALIX is a Lys63-specific polyubiquitin binding protein that functions in retrovirus budding. Dev Cell 23: 1247–1254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elia N, Sougrat R, Spurlin TA, Hurley JH, Lippincott-Schwartz J 2011. Dynamics of endosomal sorting complex required for transport (ESCRT) machinery during cytokinesis and its role in abscission. Proc Natl Acad Sci 108: 4846–4851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabrikant G, Lata S, Riches JD, Briggs JA, Weissenhorn W, Kozlov MM 2009. Computational model of membrane fission catalyzed by ESCRT-III. PLoS Comput Biol 5: e1000575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fededa JP, Gerlich DW 2012. Molecular control of animal cell cytokinesis. Nat Cell Biol 14: 440–447 [DOI] [PubMed] [Google Scholar]
- Garrus JE, von Schwedler UK, Pornillos OW, Morham SG, Zavitz KH, Wang HE, Wettstein DA, Stray KM, Cote M, Rich RL, et al. 2001. Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell 107: 55–65 [DOI] [PubMed] [Google Scholar]
- Ghazi-Tabatabai S, Saksena S, Short JM, Pobbati AV, Veprintsev DB, Crowther RA, Emr SD, Egelman EH, Williams RL 2008. Structure and disassembly of filaments formed by the ESCRT-III subunit Vps24. Structure 16: 1345–1356 [DOI] [PubMed] [Google Scholar]
- Guizetti J, Schermelleh L, Mantler J, Maar S, Poser I, Leonhardt H, Muller-Reichert T, Gerlich DW 2011. Cortical constriction during abscission involves helices of ESCRT-III-dependent filaments. Science 331: 1616–1620 [DOI] [PubMed] [Google Scholar]
- Hadders MA, Agromayor M, Obita T, Perisic O, Caballe A, Kloc M, Lamers MH, Williams RL, Martin-Serrano J 2012. ESCRT-III binding protein MITD1 is involved in cytokinesis and has an unanticipated PLD fold that binds membranes. Proc Natl Acad Sci 109: 17424–17429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson PI, Roth R, Lin Y, Heuser JE 2008. Plasma membrane deformation by circular arrays of ESCRT-III protein filaments. J Cell Biol 180: 389–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henne WM, Buchkovich NJ, Emr SD 2011. The ESCRT pathway. Dev Cell 21: 77–91 [DOI] [PubMed] [Google Scholar]
- Henne WM, Buchkovich NJ, Zhao Y, Emr SD 2012. The endosomal sorting complex ESCRT-II mediates the assembly and architecture of ESCRT-III helices. Cell 151: 356–371 [DOI] [PubMed] [Google Scholar]
- Hurley JH 2010. The ESCRT complexes. Crit Rev Biochem Mol Biol 45: 463–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzmann DJ, Babst M, Emr SD 2001. Ubiquitin-dependent sorting into the multivesicular body pathway requires the function of a conserved endosomal protein sorting complex, ESCRT-I. Cell 106: 145–155 [DOI] [PubMed] [Google Scholar]
- Katzmann DJ, Stefan CJ, Babst M, Emr SD 2003. Vps27 recruits ESCRT machinery to endosomes during MVB sorting. J Cell Biol 162: 413–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lata S, Schoehn G, Jain A, Pires R, Piehler J, Gottlinger HG, Weissenhorn W 2008. Helical structures of ESCRT-III are disassembled by VPS4. Science 321: 1354–1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Chang J, Renvoise B, Tipirneni A, Yang S, Blackstone C 2012. MITD1 is recruited to midbodies by ESCRT-III and participates in cytokinesis. Mol Biol Cell 23: 4347–4361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinelli N, Hartlieb B, Usami Y, Sabin C, Dordor A, Miguet N, Avilov SV, Ribeiro EA Jr, Gottlinger H, Weissenhorn W 2012. CC2D1A is a regulator of ESCRT-III CHMP4B. J Mol Biol 419: 75–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayers JR, Audhya A 2011. Vesicle formation within endosomes: An ESCRT marks the spot. Commun Integr Biol 5: 50–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita E, Sandrin V, Chung HY, Morham SG, Gygi SP, Rodesch CK, Sundquist WI 2007. Human ESCRT and ALIX proteins interact with proteins of the midbody and function in cytokinesis. EMBO J 26: 4215–4227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita E, Sandrin V, McCullough J, Katsuyama A, Baci Hamilton I, Sundquist WI 2011. ESCRT-III protein requirements for HIV-1 budding. Cell Host Microbe 9: 235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muziol T, Pineda-Molina E, Ravelli RB, Zamborlini A, Usami Y, Gottlinger H, Weissenhorn W 2006. Structural basis for budding by the ESCRT-III factor CHMP3. Dev Cell 10: 821–830 [DOI] [PubMed] [Google Scholar]
- Raiborg C, Bremnes B, Mehlum A, Gillooly DJ, D’Arrigo A, Stang E, Stenmark H 2001. FYVE and coiled-coil domains determine the specific localisation of Hrs to early endosomes. J Cell Sci 114: 2255–2263 [DOI] [PubMed] [Google Scholar]
- Rozycki B, Boura E, Hurley JH, Hummer G 2012. Membrane-elasticity model of Coatless vesicle budding induced by ESCRT complexes. PLoS Comput Biol 8: e1002736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiel JA, Simon GC, Zaharris C, Weisz J, Castle D, Wu CC, Prekeris R 2012. FIP3-endosome-dependent formation of the secondary ingression mediates ESCRT-III recruitment during cytokinesis. Nat Cell Biol 14: 1068–1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim S, Kimpler LA, Hanson PI 2007. Structure/function analysis of four core ESCRT-III proteins reveals common regulatory role for extreme C-terminal domain. Traffic 8: 1068–1079 [DOI] [PubMed] [Google Scholar]
- Spitzer C, Schellmann S, Sabovljevic A, Shahriari M, Keshavaiah C, Bechtold N, Herzog M, Muller S, Hanisch FG, Hulskamp M 2006. The Arabidopsis elch mutant reveals functions of an ESCRT component in cytokinesis. Development 133: 4679–4689 [DOI] [PubMed] [Google Scholar]
- Stachowiak JC, Schmid EM, Ryan CJ, Ann HS, Sasaki DY, Sherman MB, Geissler PL, Fletcher DA, Hayden CC 2012. Membrane bending by protein-protein crowding. Nat Cell Biol 14: 944–949 [DOI] [PubMed] [Google Scholar]
- Teis D, Saksena S, Emr SD 2008. Ordered assembly of the ESCRT-III complex on endosomes is required to sequester cargo during MVB formation. Dev Cell 15: 578–589 [DOI] [PubMed] [Google Scholar]
- Teo H, Perisic O, Gonzalez B, Williams RL 2004. ESCRT-II, an endosome-associated complex required for protein sorting: Crystal structure and interactions with ESCRT-III and membranes. Dev Cell 7: 559–569 [DOI] [PubMed] [Google Scholar]
- Teo H, Gill DJ, Sun J, Perisic O, Veprintsev DB, Vallis Y, Emr SD, Williams RL 2006. ESCRT-I core and ESCRT-II GLUE domain structures reveal role for GLUE in linking to ESCRT-I and membranes. Cell 125: 99–111 [DOI] [PubMed] [Google Scholar]
- Wollert T, Wunder C, Lippincott-Schwartz J, Hurley JH 2009. Membrane scission by the ESCRT-III complex. Nature 458: 172–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D, Rismanchi N, Renvoise B, Lippincott-Schwartz J, Blackstone C, Hurley JH 2008. Structural basis for midbody targeting of spastin by the ESCRT-III protein CHMP1B. Nat Struct Mol Biol 15: 1278–1286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamborlini A, Usami Y, Radoshitzky SR, Popova E, Palu G, Gottlinger H 2006. Release of autoinhibition converts ESCRT-III components into potent inhibitors of HIV-1 budding. Proc Natl Acad Sci 103: 19140–19145 [DOI] [PMC free article] [PubMed] [Google Scholar]