Abstract
Eukaryotic chromatin is kept flexible and dynamic to respond to environmental, metabolic, and developmental cues through the action of a family of so-called “nucleosome remodeling” ATPases. Consistent with their helicase ancestry, these enzymes experience conformation changes as they bind and hydrolyze ATP. At the same time they interact with DNA and histones, which alters histone–DNA interactions in target nucleosomes. Their action may lead to complete or partial disassembly of nucleosomes, the exchange of histones for variants, the assembly of nucleosomes, or the movement of histone octamers on DNA. “Remodeling” may render DNA sequences accessible to interacting proteins or, conversely, promote packing into tightly folded structures. Remodeling processes participate in every aspect of genome function. Remodeling activities are commonly integrated with other mechanisms such as histone modifications or RNA metabolism to assemble stable, epigenetic states.
Every dynamic aspect of genome function from replication to transcription requires nucleosome remodeling enzymes (e.g., Swi/Snf). These enzymes also establish stable, epigenetic chromatin states.
OVERVIEW
An unavoidable side effect of the organization of eukaryotic genomes into chromatin is the occlusion of DNA sequence by histones and nonhistone chromatin components. Making use of the genetic information, be it as part of a developmental program or in response to environmental cues, for faithful propagation of chromatin during replication or the repair of damage, necessitates that regulatory factors and complex machineries gain access to DNA sequence. The most fundamental issue of chromatin biology is, therefore, how to ensure access to DNA despite the compact and protective chromatin organization. By its very nature, this organization generates a default state of inaccessibility and, hence, inactivity of the DNA that is subject to it. There are several reasons for this. First, proteins cannot easily associate with DNA sequence that touches the nucleosomal histone surface. Second, nucleosomal DNA is strongly bent during its path around the histone octamer, and many DNA binding proteins find their target sequence distorted and unrecognizable. Finally, nonhistone chromatin components may associate with nucleosomes bearing chemical modifications (i.e., histone posttranslational modifications) and fold the nucleosomal fiber into “higher-order” structures that may be even less accessible.
The inevitable inaccessibility of DNA in chromatin may appear as a problem at first sight, but evolution has turned it into a strong asset by inventing enzymes that are able to “remodel” nucleosomes. This allows access to DNA to be regulated locally and differentially as needed. Nucleosome remodeling involves changing histone–DNA interactions as a means of disrupting, assembling or moving nucleosomes. Nucleosome remodeling enzymes may liberate segments of DNA by complete or partial disassembly of nucleosomes, may alter the composition of nucleosomes with respect to histone variants and, more indirectly, may also affect the folding of the nucleosomal fiber (Workman and Kingston 1998; Kingston and Narlikar 1999; Becker and Horz 2002; Clapier and Cairns 2009; Hargreaves and Crabtree 2011).
Nucleosomes are rather stable entities attributable to the cumulative effect of many weak histone–DNA interactions (Luger and Richmond 1998). It therefore comes as no surprise that nucleosome remodeling reactions require biochemical coupling to ATP hydrolysis. Like all biochemical reactions they are, in principle, reversible and the outcome of remodeling reactions—be it the disruption of histone–DNA interactions or their formation, the sliding of a histone octamer on or off a particular DNA sequence, or alteration of the histone variant composition of a nucleosome—are largely determined by the specification of remodeling enzymes and the involvement of cofactors.
Nucleosome remodeling ATPases are involved in each and every aspect of genome utilization, be it the regulated execution of developmental gene expression programs (Chioda and Becker 2010; Ho and Crabtree 2010) or the fast transcriptional response to environmental signals (Vicent et al. 2010). It also includes their involvement in the scheduled replication of the genome (Falbo and Shen 2006; Neves-Costa and Varga-Weisz 2006; Morettini et al. 2008) or the surveillance of the genome for DNA damage and its repair by a range of strategies, including the recombination of chromosomal segments (Altaf et al. 2007; Bao and Shen 2007; Downs et al. 2007). Defects in genes coding for nucleosome remodeling enzymes can have subtle or rather dramatic consequences, depending on the processes affected and the functional redundancy of the system. Failure of nucleosome remodeling during development may compromise viability or cause morphological defects (Chioda and Becker 2010; Ho and Crabtree 2010). In other cases, failure of remodeling systems may render cells unable to cope with DNA damage and lead to genome instability and cancer (Cairns 2001; Weissman and Knudsen 2009; Hargreaves and Crabtree 2011).
In this article we describe the discovery of nucleosome remodelers through a combination of genetics and biochemistry, their mechanism of action, and the outcomes of remodeling events. We outline the different families of remodeling enzymes, the protein domains that distinguish them, and the diversity of complex machineries they reside in. We describe the roles of remodelers in chromatin assembly, transcriptional regulation, and development. Finally, we discuss the functional crosstalk of nucleosome remodeling with histone variants and posttranslational modifications of both histones and the remodelers themselves.
1. THE DISCOVERY OF NUCLEOSOME REMODELING: A HISTORICAL PERSPECTIVE
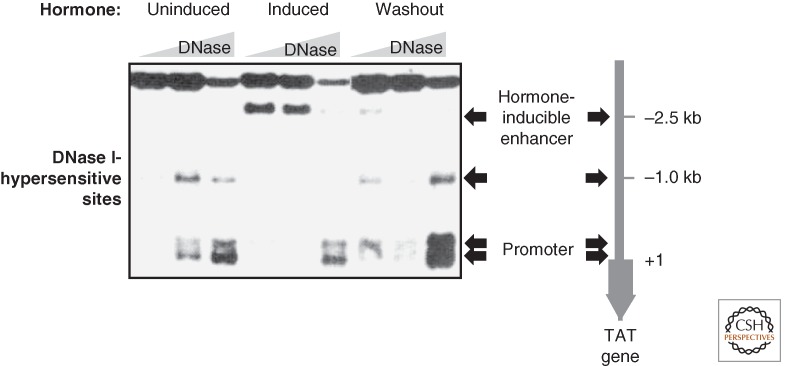
Monitoring the differential accessibility of a complex genome in a eukaryotic nucleus is a first step to understanding its function. Toward this end, probing the accessibility of sequences in intact nuclei with small, nonspecific deoxyribonucleases (DNases), such as DNase I, has been extremely successful (Elgin 1981; Becker and Horz 2002; Elgin and Reuter 2013). In these experiments, intact nuclei are treated mildly with nucleases that digest any DNA they can access within the time of the experiment. In these experiments, chromatin domains harboring active gene loci are an order of magnitude more sensitive toward digestion than inactive domains. Active regulatory elements, such as promoters, enhancers, and replication origins were found to be in even more open chromatin configurations, characterized by hypersensitivity to DNase. DNase I-hypersensitive sites (DHS) have been characterized as regions relatively depleted of canonical nucleosomes and, in turn, are usually occupied by regulatory DNA-binding proteins. The rapid—within minutes—new appearance of a DHS at a regulatory element on induction of transcription by hormones highlighted the dynamic nature of chromatin that enabled fast and very local structural transitions and rendered DNA accessible (Fig. 1). Conversely, the removal of the inducer led to a fast closing of chromatin on dissociation of factors (Reik et al. 1991). These findings motivated the search for the active molecules and processes underlying these transitions, which led to the identification of ATP-dependent nucleosome remodeling factors. These remodeling factors modulate nucleosome organization in a variety of ways (Fig. 2), which will be discussed in this chapter.
Figure 1.
DNase I-hypersensitivity (DH) analysis reveals rapid and reversible local nucleosome remodeling in vivo. The figure shows primary data from a classical DH analysis (Reik et al. 1991). The chromatin organization at the glucocorticoid-responsive enhancer element, 2.5 kb upstream of the promoter of the tyrosine aminotransferase gene, was probed in rat liver cells. Isolated nuclei of cells are digested with increasing amounts of DNase I. Digested genomic DNA is purified, cleaved with a restriction enzyme, resolved by agarose gel electrophoresis, and subjected to Southern blotting. The DH sites are revealed by indirect end-labeling of restriction fragments through hybridization of a small radioactive probe). They are marked with arrows. In the silent, uninduced state there are two DH sites at the promoter and one at −1 kb upstream. When the gene is activated on hormone induction with corticosterone, nucleosomes are remodeled at the enhancer within 15 min. A new DH site appears 2.5 kb upstream of the transcriptional start site, caused by chromatin remodeling (see “induced” columns). This correlates with the binding of glucocorticoid receptors and a complex set of remodeling factors. On removal of the hormone (“washout”), the factors dissociate and canonical nucleosomes reform within 15 min and the −2.5-kb enhancer DH disappears. The enhanced cleavage at the promoter reflects the transcriptional status of the gene.
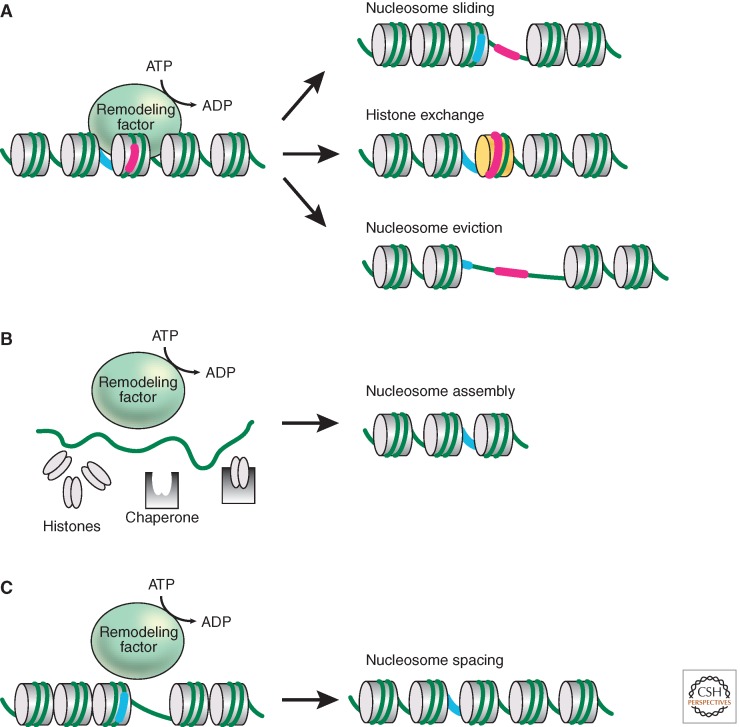
Figure 2.
Consequences of ATP-dependent nucleosome remodeling. (A) Models for nucleosome remodeling are illustrated by showing the change in position or composition of nucleosomes relative to the DNA wrapped around it. The left panel indicates a starting chromatin region with DNA reference points in the linker DNA or on the nucleosome shown in blue and pink, respectively. The right panels show possible outcomes of the remodeling reaction (from top to bottom): translational movement of a nucleosome (sliding) to expose a region that was previously occluded, exchange of a standard histone for a variant histone, and eviction of a nucleosome to expose the associated DNA. (B) Some nucleosome remodeling factors are also able to cooperate with histone chaperones to wrap DNA around histone octamers to generate nucleosomes. (C) Nucleosome remodeling factors may equilibrate the distances between nucleosomes in irregular arrays in a process termed nucleosome “spacing.”
The best known of the remodeling complexes, Swi/Snf, was originally identified genetically in yeast. Its subunits are encoded by several genes required for the expression of the SUC2 invertase gene and the HO endonuclease gene. SUC2 is required for sucrose fermentation, therefore SNF (sucrose nonfermenting) and HO expression is required for mating-type switching, hence SWI (switch; reviewed in Winston and Carlson 1992). The SWI/SNF genes were later shown to be involved in the regulation of a broad set of genes in yeast (Hargreaves and Crabtree 2011). One of the Swi/Snf complex subunits, Swi2/Snf2, bears similarity to helicases and was found to have DNA-stimulated ATPase activity. Further genetic studies suggested that SWI/SNF genes functioned interdependently in positively regulating gene expression, leading to the suggestion that the proteins they encode might function in a multiprotein complex. A connection was made between SWI/SNF function and chromatin when several suppressor mutations of swi/snf phenotypes (called SIN for switch independent) mapped to genes encoding histones or other chromatin components (Vignali et al. 2000; Fry and Peterson 2001). The connection became clearer when the complex was purified from yeast and mammalian cells. Functionally, purified Swi/Snf complex could disrupt nucleosome structure in an ATP-dependent manner and stimulate the binding of transcription factors to nucleosomal DNA in vitro (Fig. 2). Furthermore, these activities generated DNase-hypersensitive sites at transcription factor binding sites within nucleosome arrays (Vignali et al. 2000; Fry and Peterson 2001). Several versions of Swi/Snf-type remodeling complexes have since been found in eukaryotic cells. For example, a second essential and more abundant complex in yeast, RSC (remodels the structure of chromatin), contains many orthologs to Swi/Snf subunits (Clapier and Cairns 2009). In Drosophila melanogaster, the Swi2/Snf2 homolog, Brahma (brm), was identified in genetic screens for suppressors of the transcriptional repressor Polycomb (see Kingston and Tamkun 2014). Brm is also part of multiple versions of Swi/Snf-type complexes in mammalian cells, which play important roles in development and cellular homeostasis (Hargreaves and Crabtree 2011, and see Section 10).
An alternative biochemical strategy led to the identification of remodelers of the ISWI-type (imitation switch) from Drosophila. Extracts from preblastoderm Drosophila embryos provided a powerful in vitro chromatin assembly system, as they contain abundant histones chaperone and remodeling factors. The reconstitution of chromatin in the test tube, with physiological packaging and repression properties, provided an opportunity to search for factors that were able to increase the accessibility of DNA in an ATP-dependent manner (Becker and Wu 1992; Pazin et al. 1994; Tsukiyama et al. 1994; Varga-Weisz et al. 1995). The activities that were discovered by such strategies were protein complexes containing the ATPase ISWI, including the nucleosome remodeling factor (NURF), the ATP-dependent nucleosome assembly and remodeling factor (ACF), and the chromatin accessibility complex (CHRAC) (Fig. 3) (Tsukiyama et al. 1995; Varga-Weisz et al. 1997; Ito et al. 1999). ISWI had, in fact, been identified earlier because of its sequence similarity to the yeast and Drosophila SWI2/SNF2 proteins (Elfring et al. 1994). The convergence of genetic and biochemical discovery strategies revealed the existence of a novel family of enzymes, the nucleosome remodeling ATPases, dedicated to regulating the access to DNA in chromatin (Flaus et al. 2006; Cairns 2009).
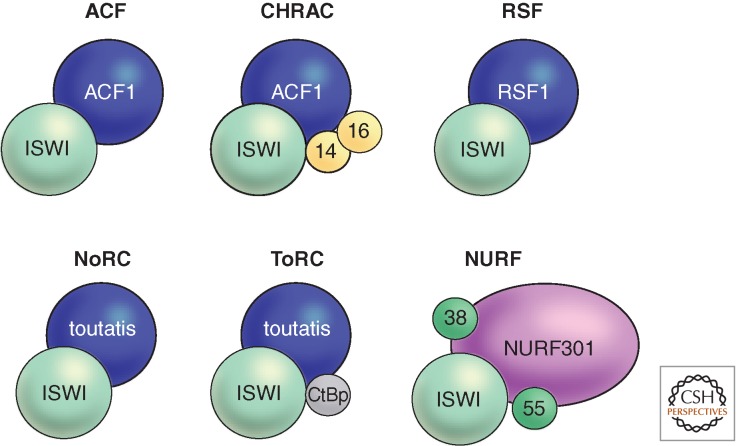
Figure 3.
The ISWI ATPase resides in several remodeling factors. The known ISWI-containing remodeling complexes in Drosophila are schematically shown. The functions of ACF, CHRAC, RSF, and NURF are described in the text. In mammals, the NoRC remodeler is involved in regulating the activity of ribosomal RNA genes (Li et al. 2006). NoRC is defined by the signature factor Tip5. The homologous protein in flies, toutatis, also interacts with ISWI (Vanolst et al. 2005). NoRC interacts with CtBP to form ToRC, which is involved in transcription regulation and nucleosome assembly outside of the nucleolus (Emelyanov et al. 2012). In mammals, additional complexes are known (Bao and Shen 2011; Kasten et al. 2011; Sims and Wade 2011; Yadon and Tsukiyama 2011) and it is likely that further assemblies will be discovered in flies as well.
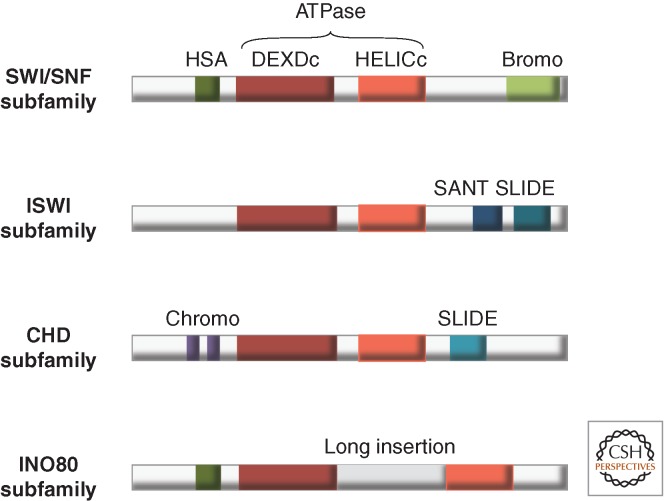
Nucleosome remodeling ATPases related to Swi2/Snf2 (the SNF2 family; Fig. 4) are found in all eukaryotes from yeast to man. They have been refined during evolution, just like the basic chromatin organization itself and can be identified because of their sequence similarity. A comprehensive survey of all known and potential nucleosome remodeling ATPases lists some 1300 SNF2 family members (approximately 30 alone in humans) that can be grouped in no less than 23 subfamilies depending on their domain organization (Fig. 4) (Durr et al. 2006; Flaus et al. 2006; Ryan and Owen-Hughes 2011).
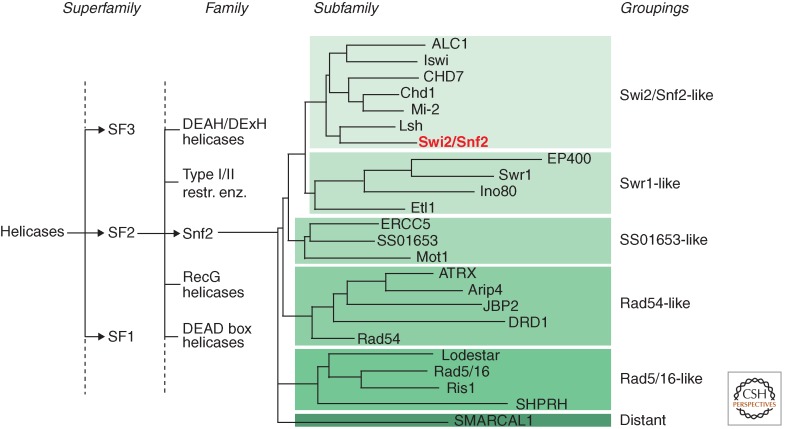
Figure 4.
Sequence relationships for the Snf2 family. Cladogram showing the relationship of the Snf2 family to other helicase-like proteins of superfamily 2 (SF2) (Fairman-Williams et al. 2010). Subfamily relationships within the Snf2 family are indicated based on alignments of the helicase-like region from 1306 members (Flaus et al. 2006). Branch lengths are not to scale. Swi2/Snf2 is the founding member of the Snf2 family of remodelers. (Adapted from Flaus et al. 2006.)
2. THE NUTS AND BOLTS OF NUCLEOSOME REMODELING
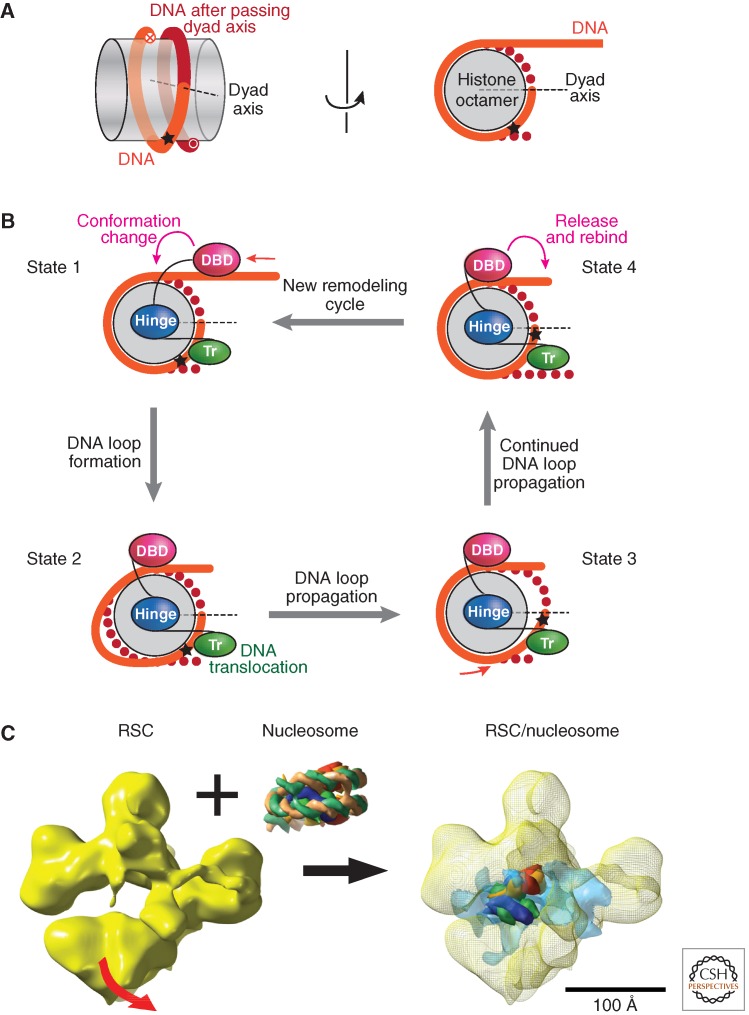
The ATPase domains of all known nucleosome remodelers are characterized by a number of short sequence motifs (I–VI) that reveal their relatedness to the much larger superfamily of nucleic acid helicases and has led to six groupings of Snf2 subfamily proteins (Fig. 4) (Flaus et al. 2006; Flaus and Owen-Hughes 2011). Snf2 family helicases commonly bind a double-stranded nucleic acid and move along one strand in a defined direction, thereby separating the two strands. Detailed mechanistic studies on a small number of selected enzymes revealed that nucleosome remodelers are also DNA translocases—that is, they move along one strand of nucleosomal DNA, yet without separating the two strands (Saha et al. 2006; Gangaraju and Bartholomew 2007). Remodelers engage in other defined contacts with histones and linker DNA, at the same time, which position the ATPase domain at a strategic site within the nucleosomal DNA, ∼2 helical turns off the dyad axis. According to the prevailing model, this anchoring combined with the translocation of the ATPase domain on nucleosomal DNA, leads to the detachment of DNA segments from the histone octamer surface (Saha et al. 2006; Gangaraju and Bartholomew 2007; Racki and Narlikar 2008; Flaus and Owen-Hughes 2011). Cycles of ATP binding, hydrolysis, and product release define a succession of conformation changes of the enzyme, which propel the movement of the enzyme on DNA. This automatically changes the position of DNA relative to the histone surface (see Fig. 5). Once a segment of DNA has been detached, the dislocation of the resulting DNA “bulge” or “loop” on the nucleosomal surface may not require much additional energy input. Another way of illustrating the basic remodeling reaction is to imagine that a remodeling enzyme “pushes” or “pulls” a segment of linker DNA into the nucleosomal domain, which automatically leads to the formation of some kind of a DNA bulge, as the histone octamer surface can only accommodate 147 bp of DNA. This bulge would then be propagated by the transIocase activity over the histone octamer surface. This DNA translocation of the ATPase domain on the nucleosome causes a twisting of the DNA, which induces a local superhelical strain into the DNA (Lia et al. 2006; Cairns 2007). Nucleosome remodeling thus presumably involves a combination of translational and rotational displacement of DNA. A much more detailed mechanical understanding of the remodeling action is currently lacking, but it is likely that the precise mechanism will differ for each of the individual remodeling ATPases, depending on enzyme structure and geometry, the arrangement, affinities, and selectivity of histone and DNA interaction domains, including the translocase domain itself. Unfortunately, it has so far been very difficult to determine the structures of ATPase/translocase domains of remodeling enzymes. The very few notable structures that have been obtained provide an important framework for further mechanistic studies (reviewed in Hauk and Bowman 2011). Electron microscopy studies have generated structures of the Swi/Snf complex and the related RSC complex. The yeast RSC resembles the Swi/Snf complex in subunit composition and overall architecture. Instead of the ATPase Swi2/Snf2 it contains the Sth1 ATPase, which belongs to the same subfamily (Clapier and Cairns 2009). The structure of the RSC complex bound to a nucleosome is particularly insightful (Fig. 5C) (Chaban et al. 2008). This structure reveals that RSC engulfs the nucleosome within a central cavity. Importantly, ATP-independent binding of RSC to the nucleosome appears to alter histone–DNA interactions perhaps to facilitate ATP-dependent remodeling. Nucleosomal DNA within the structure appears relatively unconstrained by the complex and may be able to undergo movement of the kinds described above (Chaban et al. 2008). RSC-nucleosome complexes have been found in vivo: at the yeast UASg locus, RSC positions an apparently partially unwound nucleosome to facilitate transcription factor binding to nearby sites (Floer et al. 2010).
Figure 5.
Mechanism of nucleosome remodeling. (A) Nucleosome view emphasizing the left hand wrapping of DNA. (Left) Side view of the nucleosome core with the histone octamer represented as a gray transparent cylinder and the DNA in orange (before the dyad axis) and red (after the dyad axis). (Right) Top view of the nucleosome (rotated 90º) in which the DNA after the dyad axis is represented by red dots. The star represents a reference point on the DNA sequence. (B) Model for DNA movement across the histone octamer during a remodeling event by the ISWI-type enzymes. Successive steps in a remodeling event are represented by States I–IV. In State I the DNA binding domain (DBD) is bound to the linker DNA and the translocase (Tr) domain is bound near the nucleosome dyad. A hypothetical “hinge” mediates the changes in conformation. In State II a conformational change between the DBD and the Tr “pulls in” DNA, which becomes visible as a bulge on the histone octamer surface. The Tr activity propagates this bulge across the surface of the histone octamer beyond the dyad axis (State III). The DNA loop continues to diffuse across the octamer surface and is released into the distal linker DNA (State IV). Loop diffusion thus effectively repositions the histone octamer relative to the DNA sequence (i.e., the star has moved closer to the dyad axis). A further conformational change triggered by aspects of the ATPase cycle lead to a resetting of the remodeler relative to the histone octamer (compare in States IV and I). The remodeler now engages with a different segment of linker DNA to start another cycle of remodeling. (A,B, Adapted from Clapier and Cairns 2009.) (C) Cryo-electron microscopy (EM) analysis of the RSC structure and nucleosome interaction. The yeast RSC resembles the Swi/Snf complex in subunit composition and overall architecture. Instead of the ATPase Swi2/Snf2, it contains the Sth1ATPase, which belongs to the same subfamily (Clapier and Cairns 2009). A 25-Å cryo-EM map of RSC (left) shows a central cavity that closely matches the shape and dimensions of a nucleosome core particle. Movement (indicated by the red arrow) of the bottom RSC domain appears to control access to the central cavity. Incubation of RSC with nucleosome core particles (NCPs) results in formation of a RSC-NCP complex (right panel) in which NCP density is apparent in the central RSC cavity. Interestingly, interaction with RSC in the absence of any ATP hydrolysis appears to result in extensive changes in NCP organization. Histone density can be identified, but nucleosomal DNA appears disordered (semitransparent blue density). This loosening of DNA may facilitate DNA translocation during remodeling. (Image and interpretation provided by Francisco Asturias, Scripps Research Institute, La Jolla.)
The current classification of the many SNF2-type ATPases into subfamilies rests on sequence features within the ATPase domain (Flaus and Owen-Hughes 2011), but the more closely related ATPases also share particular domains and sequence signatures outside of the ATPase domains. A notable feature of the members of the Ino80 or SWR1 subfamilies is a major insertion of some 300 amino acids in front of helicase motif IV (Fig. 6). The best-studied remodeling ATPases—Swi2/Snf2, ISWI, Chd1, Mi-2, Ino80, and Swr1—are prototypic enzymes that represent subfamilies of enzymes (Fig. 4) (Bao and Shen 2011; Flaus and Owen-Hughes 2011; Kasten et al. 2011; Sims and Wade 2011; Yadon and Tsukiyama 2011). The hallmark of SWI2-type ATPases (synonymous with SWI2/SNF2) is, among other features, a bromodomain. This domain “reads” acetylated lysine marks of histone and nonhistone proteins (for more structural information, see Cheng 2014; Patel 2014). ISWI-type enzymes feature carboxy-terminal SANT-SLIDE domains and CHD-type enzymes bear amino-terminal chromodomains, which recognize and bind methylated lysine residues of histone H3 (Fig. 6). It is likely that this classification lessens our appreciation of more fundamental similarities between remodelers. For example, the resolution of the structure of the carboxyl terminus of CHD1 revealed that it contains a domain that resembles the “SLIDE” domain, a DNA binding domain that so far was thought to be characteristic of enzymes belonging to the ISWI subfamily. This common structural motif had not been found by simple sequence comparison (Ryan et al. 2011). Also, some of the associated domains may be involved in auto-regulation of the ATPase, and may thus render the ATPase conditional on substrate availability. Mechanistically this may occur through allosteric conformational changes on substrate binding, or through direct competition between a regulatory domain and the substrate for access to the active site (Hauk et al. 2010; Flaus and Owen-Hughes 2011; Hauk and Bowman 2011).
Figure 6.
Four ATPase subfamilies: Signatures of SNF2, ISWI, CHD, and Ino80. The grouping of the remodeling ATPase of the Snf2 family is defined by signature motifs within the ATPase domain, whereas additional domains define the subfamilies. Members of the INO80 (and Swr1) subfamily of ATPases have a longer insertion between the two ATPase subdomains than other remodelers (Fig. 5). These subfamilies also contain a HSA (helicase-SANT) domain. The SWI/SNF family of ATPases contains a HSA domain, but is further defined by a carboxy-terminal bromodomain (capable of binding acetylated lysine residues). The ISWI and CHD family of ATPases each have SANT-SLIDE modules (blue) whereas only the CHD family has tandem chromodomains. (Adapted from Clapier and Cairns 2009.)
In addition to the signature domains mentioned above, illustrated in Figure 6, other structures can be described that are found in ATPases of different subfamilies. For example, the HSA (helicase-SANT-associated) domain, which binds actin-related proteins (ARPs) is present in the ATPases of the SWI/SNF complex, the related RSC complex, the SWR1 complex, and the Ino80 complex (Fig. 6) (Dion et al. 2010).
Conceivably, the specific outcome of a basic remodeling reaction depends not only on the geometry and enzymatic parameters of the ATPases themselves, but also on associated proteins. The unpeeling of DNA segments from the histone surface may lead to a delocalization of nucleosomes (their “sliding”) if the detached DNA segment is propagated around the octamer and expelled on the other side. However, it is conceivable that a very similar remodeling reaction may lead to the removal or replacement of histones, or to the complete eviction of a nucleosome (Fig. 2). An interesting variation of the theme has been suggested, whereby a nucleosome is used as a “wedge” to destabilize an adjacent particle. If nucleosomes are moved to collide with one another, DNA may be peeled off one or both of them, yielding unconventional remodeling intermediates (Chaban et al. 2008; Engeholm et al. 2009; Dechassa et al. 2010). This possibility is consistent with the fact that RSC engulfs the nucleosome to which it is bound (Fig. 5C), as it may be difficult for a remodeler to displace the histone octamer under those circumstances (Chaban et al. 2008). Whether histone octamers are moved intact or whether partial disassembly occurs may be largely determined by the cooperation of remodeling ATPases with associated histone chaperones that may either scavenge the histones as they are liberated from the toroidal DNA supercoil or perhaps deliver variant histones for exchange.
3. DIVERSITY OF NUCLEOSOME REMODELING COMPLEXES
Although isolated remodeling ATPases can alter histone–DNA interactions in vitro, they usually associate with other proteins to form defined multisubunit complexes. These complexes are commonly referred to as “remodeling factors.” Some remodeling factors are made of only a few subunits, such as those organized around ISWI-type ATPases, which typically only consist of two to four subunits (Fig. 3). The large Ino80- and SWI2- type complexes, which contain more than a dozen subunits, mark the other end of the scale (for examples, see Figs. 7 and 8). The associated subunits frequently contribute additional domains (e.g., plant homeodomain [PHD] fingers, bromodomains) that bind modified epitopes on histones and nonhistone proteins. In most cases, however, the precise role of the ATPase-associated subunits is unknown, but is likely to target the remodeling ATPase to specific sites of action, to regulate their activity, and the precise outcome of the remodeling reaction as well as to integrate the remodeling reaction into the physiological context of a nuclear process, such as transcription, replication, or DNA repair.
Figure 7.
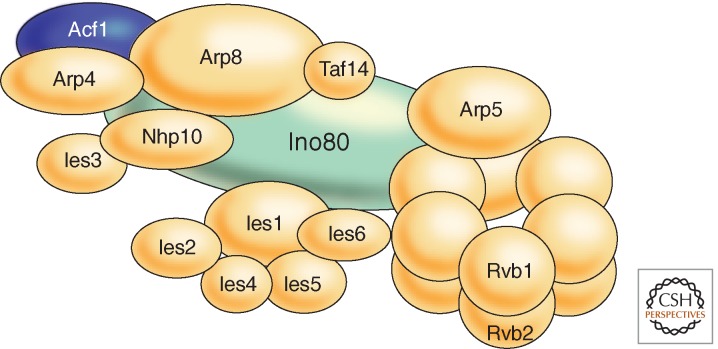
Example of a complex remodeling machine: INO80. The Saccharomyces cerevisiae INO80 complex provides an example of the subunit composition of a complex nucleosome-remodeling machine. INO80 subunits include the core ATPase, Ino80 (INOsitol requiring), Rvb1 (RuVB-like), Rvb2, Act1 (actin), Arp4 (actin-related protein), Arp5, Arp8, Nhp10 (nonhistone protein), Taf14 (TATA-binding protein-associated factor), Ies1 (Ino eighty subunit), Ies2, Ies3, Ies4, Ies5, and Ies6. (Adapted from Bao and Shen 2011.)
Figure 8.
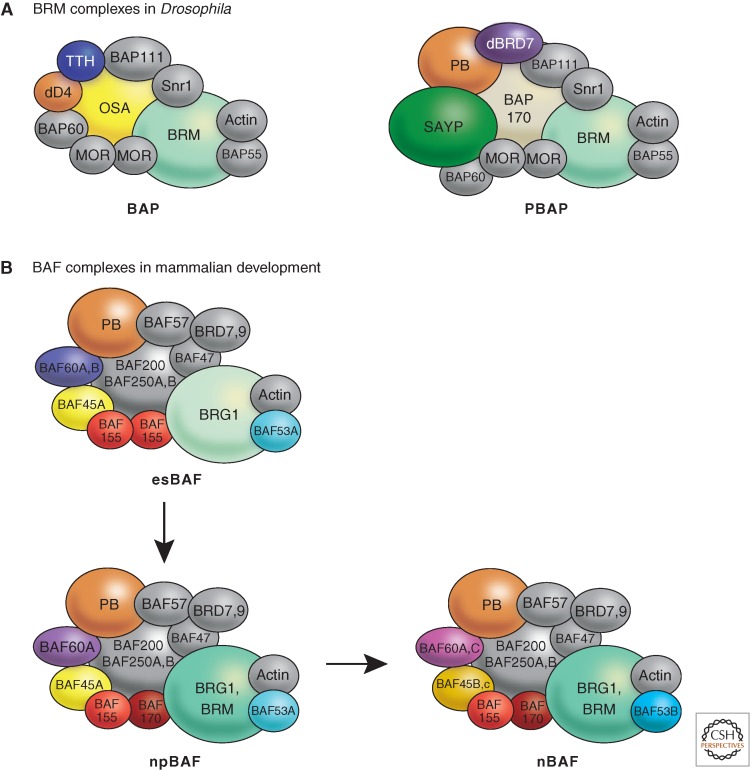
Diversity of Swi/Snf complexes in metazoa. (A) BAP and PBAP complexes in Drosophila. There are two distinct Swi/Snf-type complexes in Drosophila: the BRM-associated proteins complex (BAP) and the polybromo-containing BAP complex (PBAP). Although these complexes share multiple subunits including the ATPase BRM, they each have distinct subunits. The OSA, dD4, and TTH subunits are only found in the BAP complex and not in the PBAP complex. In contrast, Polybromo, BAF170, dBRD7, and SAYP are subunits of the PBAP complex but not the BAP complex. (Adapted from Ho and Crabtree 2010, with updates from Moshkin et al. 2012.) (B) Cell- and tissue-specific versions of BAF complexes in mammals. These complexes contain either the BRG1 or BRM ATPases. They may also contain polybromo (PB) and BAF200 (PBAF complexes) or BAF250A/B (BAF complexes). Shown here are composite representations of those variants. This figure serves to illustrate tissue-specific assemblies of BAF complexes, which have distinct functions in specific cell types. The subunits present in BAF complexes for each tissue are indicated (e.g., BAF60A or C). PB is colored the same as in A. The other subunits shaded with color are those whose presence varies in the different tissue, and define the tissue-specific complexes. (Adapted from Ho and Crabtree 2010.)
Some ATPase-associated subunits appear as evolutionary conserved “recurring themes,” as they are found in several remodeling factors. For example, the large SWI/SNF, RSC, Ino80, and SWR1 complexes contain actin-related proteins (ARPs) and even nuclear actin itself (Figs. 7 and 8). Different ARPs contribute to complex assembly and remodeling activity in various ways. In some cases, ARPs and nuclear actin may bind particularly modified histones or variants and thus mediate association of remodeling complexes to chromatin. In other cases, they may provide interfaces to the transcription machinery or to nuclear pores (Dion et al. 2010). Another example of a recurring theme is the hexameric rings of the RVB1 and RVB2 ATPases from the AAA family, which are important functional components of SWR1 and Ino80 subfamily of complexes (Fig. 7). These enzymes resemble the bacterial RuvB helicase that is involved in the resolution of holiday junctions during homologous recombination (Jha and Dutta 2009). The assembly of SWR1 and Ino80 complexes is compromised in the absence of the RVB hexamers, which suggest a scaffolding function. Nonetheless, the fact that these rings themselves have ATPase activity suggests more interesting, dynamic activities.
An important concept in the study of chromatin remodelers is that one particular ATPase may associate with alternative sets of proteins to form distinct types of remodeling complexes. For example, the ATPase ISWI in Drosophila can serve as the “engine” for at least five different remodeling factors with nuclear functions ranging from transcription control to DNA repair and chromosome organization (Fig. 3) (Yadon and Tsukiyama 2011).
The complexity of remodeling factors has increased dramatically during metazoan evolution and multiple “variations on a theme” have been described for the mammalian complexes, including subunit isoforms, variants, and posttranslational modifications. This is best illustrated for the 11-subunit yeast SWI/SNF complex. SWI/SNF is mainly involved in regulating transcription initiation and it is targeted to promoters through direct interactions with sequence-specific DNA binding factors (see Section 4). The Drosophila homolog of the SWI2/SNF2 ATPase is called Brm (Brahma) for historical reasons (see Kingston and Tamkun 2014). Brm associates with seven to eight brahma-associated proteins (BAPs) to form complexes that loosely resemble the yeast SWI/SNF complex. Interestingly, two related complexes, BAP and PBAP, share six subunits but differ in the signature subunits: Osa, dD4, and TTH for BAP; PB, BAP170, SAYP, and dBRD7 for PBAP (Fig. 8A) (Moshkin et al. 2007; Moshkin et al. 2012). The ‘core complex” in the absence of these signature subunits is not functional in vivo. At the same time, they specify the preferential recruitment to distinct sets of target genes. BRM is a global regulator of transcription in Drosophila as the majority of transcriptionally active genes was bound by BRM in polytene chromosomes and required the ATPase for activity (Hargreaves and Crabtree 2011). BAP subunits function as trithorax gene products and oppose the function of polycomb proteins in many developmental pathways (see Kingston and Tamkun 2014).
The mammalian orthologs of Brahma are the highly related ATPases BRM and BRG1. The proteins that associate with them, termed “BRG/BRM-associated factors” (BAFs), form a family of related remodeling machineries (Hargreaves and Crabtree 2011). The mammalian BAF complexes resemble the yeast SWI/SNF complexes vaguely: Eight subunits of both complexes are clearly evolutionary related, but yeast and mammalian complexes also contain several species-specific subunits (Ho and Crabtree 2010). A hallmark of these complexes is their diversity, as several subunits have cell-type-specific isoforms, which associate with BAF complexes in mutually exclusive and combinatorial ways (Fig. 8B). This explains why BAF complexes are involved in regulating very different gene expression programs in different tissues, from the maintenance of the pluripotent state of embryonic stem (ES) cells to the highly differentiated profile of postmitotic neurons (see Section 10).
4. NUCLEOSOME REMODELING FACTORS AS REGULATORS OF TRANSCRIPTION
Promoters can be tentatively sorted into different classes according to their chromatin architecture and arrangement of binding sites. The transcription start sites of many constitutively expressed “housekeeping” promoters tend to be depleted of nucleosomes and so depend less on nucleosome remodeling. Tightly regulated genes, in contrast, rely on remodeling factors to clear their promoters of repressive nucleosomes for expression (Cairns 2009; Rach et al. 2011). Promoter architecture is nonetheless more complex and diverse than this, and nucleosome remodeling factors are equally involved in setting up promoter organization as well as in its subsequent remodeling.
Nucleosome remodelers play important roles in regulating the initiation and elongation of transcription. They are often recruited to target genes through interactions with sequence-specific transcription factors to serve as coactivators or corepressors (Clapier and Cairns 2009). In yeast, for example, many transcription factors are able to recruit the Swi/Snf complex to target genes by direct interaction (Fig. 9) (Vignali et al. 2000; Fry and Peterson 2001). The yISW2 complex is also targeted to meiotic genes by Ume6, to participate in repression (Clapier and Cairns 2009). In an interesting variation of the theme, the Hir1 and Hir2 corepressors recruit Swi/Snf to histone gene loci, but it is required there for subsequent transcription activation (Dimova et al. 1999).
Figure 9.
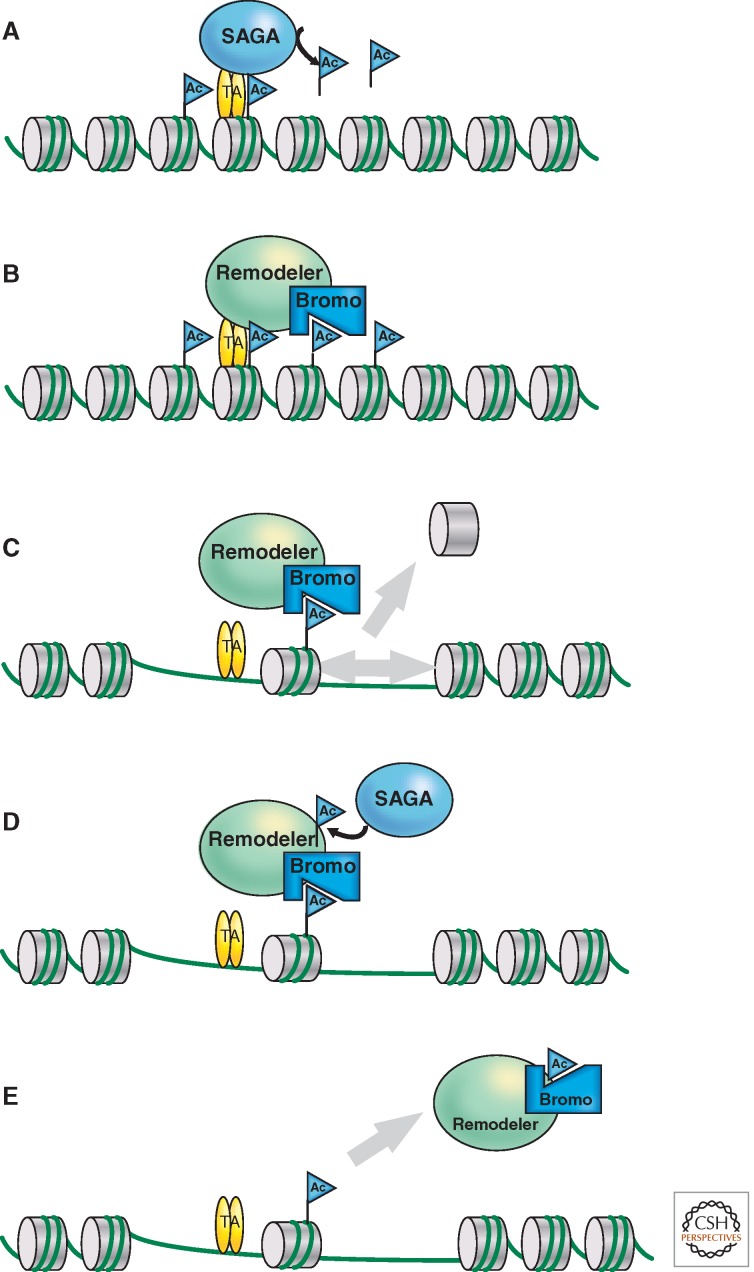
Model for the action of Swi/Snf-type nucleosome remodelers at promoters. and their regulation by acetylation. (A) SAGA or other histone acetyltransferase (HAT) complexes can be recruited to gene promoters by interacting with sequence-specific DNA-binding transcription activators (TA). Once recruited these HATs acetylate (blue Ac flag) nucleosomes in proximity of the activator recognition site. (B) The Swi/Snf or RSC nucleosome remodeling complexes (remodeler) can be recruited to promoters by interactions with transcription activators. Bromodomains (bromo) within subunits of these complexes then interact with the acetylated (Ac) nucleosomes at the promoter. (C) ATP-dependent remodeling and/or displacement (gray arrows) are preferentially directed at the acetylated nucleosomes bound by the bromodomains in the remodeling complex. (D) SAGA or other Gcn5-containing complexes acetylate specific lysines within subunits of the remodeling complex. These acetylated lysines compete for interaction with the bromodomains, which (E) leads to dissociation of the remodeler from the acetylated nucleosomes (gray arrow). (Adapted from Suganuma and Workman 2011.)
In mammals, Swi/Snf interacts with a number of transcription factors, including steroid receptors, tumor suppressors, and oncogenes like RB, BRCA-1, c-Myc, and MLL (Hargreaves and Crabtree 2011). In Drosophila, the NURF complex interacts with several transcription factors including GAGA factor, heat shock factor, ecdysone receptor, and the dKen repressor through the NURF301 subunit (Alkhatib and Landry 2011).
Once recruited to target gene promoters, chromatin remodelers alter the local chromatin organization by nucleosome movement or displacement, which can facilitate either gene activation (Li et al. 2007) or repression. In combination with histone chaperones, complexes like Swi/Snf and RSC are able to displace nucleosomes in trans either by forcing the histone octamer onto another piece of DNA or by moving the histones onto chaperones (Workman 2006). Nucleosome displacement by Swi/Snf is facilitated by histone acetylation (see Fig. 9, and Section 7) and by interactions of the complexes with transcription factors (Gutierrez et al. 2007).
Nucleosome remodeling complexes are involved in both the activation and repression of gene expression. Mutation of their subunits in yeast often leads to as many genes showing an increase in expression as those whose expression decreases (Hargreaves and Crabtree 2011). Although some of these effects will be indirect, it is not difficult to imagine a chromatin remodeler activating or repressing genes through the same mechanism (for example, moving nucleosomes on or off a promoter). Nonetheless, most chromatin remodelers have been characterized based on their activity in either activating or repressing gene expression.
In yeast, both Swi/Snf and the related RSC complex are required for expression of nonoverlapping subsets of genes (Hargreaves and Crabtree 2011). Whereas Swi/Snf regulates many inducible genes, RSC is more involved in controlling essential, constitutively expressed genes, such as genes encoding ribosomal protein subunits (Hargreaves and Crabtree 2011). Importantly, only a small minority of yeast genes absolutely requires Swi/Snf or RSC for expression. This does not negate that Swi/Snf or RSC may still function at many other genes. RSC was, in fact, found to bind to >700 target genes in the yeast genome, including histone genes, genes regulated by stress, and several genes transcribed by RNA polymerase III (Hargreaves and Crabtree 2011). Moreover, 20% of genes in yeast grown in glucose require Snf2 to keep their promoters nucleosome-depleted (Tolkunov et al. 2011). These data suggest that Swi/Snf and RSC function more broadly than initially indicated by gene expression analysis alone.
Other nucleosome remodeling factors also function in gene activation (Clapier and Cairns 2009). The yeast INO80 complex activates genes regulated by the inositol/choline response element and is required for activation of the PHO84 gene. In the absence of INO80, the expression levels of 150 genes changed (approximately half up and half down; Morrison and Shen 2009). The mammalian INO80 complex is recruited to target genes by the YY1 transcription factor (Cai et al. 2007).
Several nucleosome remodeling complexes appear to function primarily in repression of transcription, as noted above, be it directly or indirectly. Chromatin immunoprecipitation studies showed that Swi/Snf physically occupies some genes where it functions as a repressor (Martens and Winston 2003; Hargreaves and Crabtree 2011). Similarly, most Brg1 binding in ES cells occurs at enhancers and intragenic regulatory elements where it functions as a repressor (Hargreaves and Crabtree 2011). An instructive example of an indirect repressive effect is provided by action of the Swi/Snf on the SER3 gene in yeast. Swi/Snf activates the upstream SRG1 promoter, which leads to transcription of a noncoding RNA. The transcription extends through the downstream SER3 gene promoter and interferes with its transcription initiation (Martens et al. 2005).
The yeast Ume6 repressor recruits the ISW2 complex to meiotic promoters in which it organizes repressive chromatin. Isw2 collaborates with the Rpd3 deacetylase complexes to repress numerous Ume6-dependent and -independent genes (Fazzio et al. 2001). This was corroborated by a widespread analysis in which loss of ISWI function in Drosophila 3rd instar larvae indicated that 75% of the 500 genes with altered expression profiles showed increased expression (out of 15,000 genes tested; Hargreaves and Crabtree 2011). The Mi2 ATPase of higher eukaryotes functions as part of the repressive nucleosome remodeler/deacetylase complexes (NuRD). NuRD represents a heterogeneous group of factors of variable, cell-specific subunit composition (Bowen et al. 2004), and is part of the MeCP1 complex that connects chromatin remodeling, histone deacetylation, and DNA methylation. More recently, Mi2 was also found as part of a distinct complex, dMec, which functions as a SUMO-dependent corepressor (Kunert and Brehm 2009).
In addition to regulating transcription initiation by activation or repression, chromatin remodelers also affect the process of transcription elongation. Elongating RNA polymerase II has to confront nucleosomes along the entire gene body and a number of elongation factors, histone chaperones, histone modifications, and chromatin remodelers have been implicated in facilitating its progress (Li et al. 2007; Selth et al. 2010). In general, histones are acetylated and methylated during transcription elongation and all or part of the histones in the octamer are chaperoned around the elongating polymerase to be reassembled and deacetylated behind the polymerase (Li et al. 2007; Clapier and Cairns 2009). Remodelers then might play a role in disrupting nucleosomes in front of the polymerase and in reassembling and spacing the nucleosomes in its wake. Examples for such action come from all subfamilies of remodelers and several species. Human Swi/Snf was shown early on to help RNA polymerase pass a nucleosome-induced transcriptional pause in vitro (Vignali et al. 2000). Yeast Swi/Snf has been implicated in histone eviction during polymerase elongation in vivo (Schwabish and Struhl 2007). The yeast RSC complex can associate with RNA polymerase II (Soutourina et al. 2006) in vitro and stimulate transcription through a nucleosome in a manner enhanced by histone acetylation (Li et al. 2007). Yeast Chd1 colocalizes with RNA polymerase II and interacts with a number of factors involved in elongation, such as the PAF complex, the Spt4/5 elongation factors, and the FACT histone chaperone complex (Simic et al. 2003). The Drosophila CHD family remodeler Kismet associates with actively transcribed regions on polytene chromosomes and apparently functions in an early step in transcription initiation or elongation (Srinivasan et al. 2005; Murawska et al. 2008). Mutants in Kismet lead to reduced levels of elongating RNA polymerase II and reduced levels of the histone chaperone Spt6 on polytene chromosomes (Srinivasan et al. 2005).
To summarize, chromatin remodelers are involved in the entire transcription process from promoting or blocking transcription initiation to active transcription elongation. They are recruited to target genes by DNA-binding transcription factors, RNA polymerases, and elongation factors. They serve in disrupting nucleosomes to facilitate initiation and elongation of transcription, and reassembling and spacing nucleosomes in the wake of passing RNA polymerase. They are the engines that both deny RNA polymerase access to a gene (repression) and help RNA polymerase contend with nucleosomes (activation).
5. NUCLEOSOME REMODELING IN CHROMATIN ASSEMBLY AND ORGANIZATION
The previous paragraphs emphasized the roles of nucleosome remodeling factors in nucleosome disassembly as a means to access the genetic information. Less intuitive, but equally important is the involvement of nucleosome remodeling in the assembly and organization of chromatin with repressive properties as well as for the diversification of structures that are collectively called “the epigenome.” Nucleosome remodeling is effective at all levels of chromatin organization: the de novo assembly of nucleosomes (Fig. 2B), their positioning relative to the underlying DNA, and the generation of fibers with defined distances between the nucleosomes (“nucleosome spacing”; Fig. 2C), which profoundly affects the folding of chromatin. Finally, nucleosome remodeling also affects the association of linker histones and perhaps even the interaction of nonhistone proteins with the nucleosome fiber.
The dual capacity of remodeling factors to detach segments of DNA from the histone surface and, conversely, to wrap DNA around histones during nucleosome assembly is best illustrated by the process of histone variant exchange. For example, nucleosome remodeling factors of the SWR1 family remove a “canonical” H2A/H2B dimer from a nucleosome and then replace it by a dimer containing the H2A.Z variant (for more detail, see Section 6 and Henikoff and Smith 2014).
Although nucleosomes form spontaneously by mixing DNA and histone in high salt solutions and gradual removal of the salt, nucleosome assembly under physiological conditions requires the cooperation of histone chaperones and remodeling activities (Fig. 2B). Although histone chaperones keep the highly charged histones soluble and assure their ordered deposition onto DNA (see also Almouzni and Cedar 2014), the example of ACF reveals that remodelers can catalyze the supercoiling of DNA around histone octamers (Torigoe et al. 2011). Indeed, nucleosome remodeling factors of the ISWI family appear to be particularly involved in de novo nucleosome assembly. In vitro, ACF and RSF are able to promote nucleosome formation, but whereas ACF requires the assistance of histone chaperones, RSF profits from “built-in” chaperone activity (Loyola et al. 2003; Lusser et al. 2005). In Drosophila, genetic analysis of the respective signature subunits of these remodeling factors (i.e., ACF1 and RSF1 [Fig. 3]), confirms their roles in chromatin assembly in vivo. Those animals that survive an ACF1 deficiency are characterized by “sloppy,” irregular chromatin, and defects in heterochromatin specification as well as in chromatin-mediated gene silencing (Chioda and Becker 2010). In contrast, an RSF1 deficiency does not affect the basal chromatin organization, but leads to a more specific defect in the incorporation of the histone variant H2A.V and selective impairment of chromatin structures that depend on this variant, like heterochromatin (Hanai et al. 2008).
The bulk of nucleosome assembly occurs during S phase and accordingly a number of remodeling factors are targeted to sites of active DNA replication (Morettini et al. 2008; Rowbotham et al. 2011). Their physiological roles are difficult to assess because of their dual potential in opening chromatin to facilitate DNA synthesis, and in subsequent nucleosome assembly to reconstitute the integrity of the nucleosome fiber. Nucleosome assembly also occurs in interphase during DNA damage repair, and many different remodelers are recruited to sites of DNA damage repair (Altaf et al. 2007; Bao and Shen 2007; Downs et al. 2007; Lan et al. 2010).
The generation of repressive chromatin may, in the simplest case, involve the positioning of nucleosomes onto regulatory DNA sequences. The energy input of ATP hydrolysis allows the remodelers to move nucleosomes onto DNA sequences that are intrinsically unfavorable. For example, the yeast ISWI-type remodeler isw2 can slide nucleosomes onto a promoter to repress its activity (Whitehouse and Tsukiyama 2006). Genome-wide chromatin remodeling by ISW2 near promoters and in coding regions increases nucleosome density and enforces the accuracy of transcription (Yadon et al. 2010). More specifically, the isw2 and isw1 complexes contribute to the positioning of nucleosomes in intergenic regions or in the middle of genes, respectively, which suppresses cryptic, antisense transcription that would otherwise pose a risk of interfering with sense transcription (Whitehouse et al. 2007; Tirosh et al. 2010). The positioning and phasing of nucleosomes around transcription start sites in yeast, which frequently carry information in the form of histone variants and specific modifications, are brought about by the RSC complex in S. cerevisiae (Wippo et al. 2011; Zhang et al. 2011). In fission yeast the CHD-type remodeler, Mit1, has been shown to be involved in aligning nucleosomal arrays with respect to promoter boundaries, a process termed nucleosome “phasing” (Lantermann et al. 2010).
The nucleosome assembly activity of remodeling factors improves the integrity of the chromatin fiber by filling gaps in the succession of nucleosomes. The sliding of nucleosomes enables remodelers to improve the regularity of nucleosomal fibers by equalizing the inter-nucleosome distances (i.e., their “spacing”) (Fig. 2C) (Becker and Horz 2002). The more regular a nucleosome array, the better it will fold into a variety of “next level” chromatin fibers characterized by diameters of ∼30 nm. The folding of nucleosomal arrays into fibers is further promoted by the association of linker histones. The presence of linker histones affects the activity of remodeling factors differentially; some enzymes are unable to remodel nucleosomes that are “locked in” by H1 association, but others, notably the ISWI-containing ACF, are able to move nucleosomes even with H1 bound. ACF can also facilitate the incorporation of H1 into chromatin in vitro (Lusser et al. 2005; Torigoe et al. 2011). These biochemical observations may be physiologically significant as ISWI-containing remodelers are suggested to modulate the steady-state association of H1 with chromosomes (Chioda and Becker 2010). In summary, nucleosome remodeling factors are likely to regulate the folding of chromatin by modulating the integrity, regularity, and spacing parameters of nucleosomal arrays, thereby laying the foundations for higher-order chromatin structures (Korber and Becker 2010). This hypothesis is supported by observations in yeast and flies. In Schizosaccharomyces pombe the formation of silenced heterochromatin correlates with the regularity of nucleosome positioning, which is promoted by ATPase Mit1 in the context of the SHREC complex (Sugiyama et al. 2007). Drosophila lacking the nucleosome assembly and spacing factor, ACF, show defects in heterochromatin formation and polycomb-dependent silencing (Chioda and Becker 2010). These examples show that nucleosome remodeling factors not only serve to catalyze fast, local, and reversible changes of nucleosome organization, but that their action may also promote the assembly of stable and epigenetically silenced chromatin domains.
6. RECOGNITION OF HISTONE MODIFICATIONS BY CHROMATIN REMODELERS
Most remodeling complexes contain domains that mediate their specific association with modified histones, in addition to interaction with sequence-specific DNA-bound transcription factors. For example, CHD family members often contain two tandem chromodomains (Fig. 6) (Brehm et al. 2004). The tandem chromodomains of human CHD1 allows it to specifically recognize a methylated lysine 4 residue (H3K4me) of a histone H3 tail (Hargreaves and Crabtree 2011). Because of their inherent weakness, interactions with modified histones are not likely to be primary targeting determinants, but may modulate the activity of the enzyme once it has been recruited by other means. As H3K4 methylation is an active mark at the 5′ end of genes, chromodomain-histone interactions are believed to modulate the activity of CHD1 at promoters. In agreement with this idea, the chromodomains of yeast and Drosophila CHD1 appear to be important for their ATPase and nucleosome remodeling activities, but did not significantly affect their chromosomal localization (Hauk et al. 2010; Morettini et al. 2011). Thus, the CHD1 chromodomain interaction with methylated H3K4 may play a regulatory role of the remodeling activity beyond its recruitment to active genes.
The plant homeodomain (PHD domain) is a versatile zinc finger domain that reads histone modifications along the amino-terminal tail of histone H3 (Sanchez and Zhou 2011). A PHD domain in the BPTF subunit of the NURF complex recognizes H3K4me3 and couples this modification to nucleosome remodeling (Hargreaves and Crabtree 2011). The CHD4 subunit of the NuRD complex contains two PHD domains, the second of which recognizes H3K9me3, and the first one interacts preferentially with the amino terminus of H3 if unmodified at K4 (Mansfield et al. 2011). Remarkably, DPF3b, a factor associated with human BAF, recognizes H3K14 acetylation through tandem PHD fingers (Zeng et al. 2010). Previously, recognition of acetylated histones was primarily attributed to bromodomains.
Bromodomains have the potential to facilitate the recruitment, retention, and/or activity of chromatin remodelers on acetylated nucleosomes. The presence of acetyl-lysine-binding bromodomains in several remodeling ATPases and associated subunits suggests that some remodelers are sensitive to the acetylation state of target nucleosomes (Horn and Peterson 2001; Workman 2006). All Snf2-like ATPases contain at least one bromodomain (Fig. 6) and additional ones may be found on other subunits of the Swi/Snf type complexes (Clapier and Cairns 2009).
7. REGULATION OF REMODELERS BY POSTTRANSLATIONAL MODIFICATION
There is increasing evidence that chromatin remodelers themselves are modified and regulated by enzymes that are better known for modifying histones. Gcn5, the acetyltransferase subunit of the SAGA complex, has long been linked to the function of chromatin remodelers like Swi/Snf. Thus, SAGA affects remodeling activity indirectly through acetylating histones and directly through remodeler acetylation (Fig. 9) (Clapier and Cairns 2009). The Rsc4 subunit of the SWI/SNF-type remodeler RSC contains tandem bromodomains one of which binds H3K14ac. Remarkably, the adjacent bromodomain is able to bind acetylated lysine 25 of Rsc4 itself (VanDemark et al. 2007). Binding of this bromodomain to K25ac of Rsc4 inhibits interaction of the other bromodomain with H3K14ac, thereby hampering the interaction of RSC with acetylated nucleosomes. Gcn5 is responsible for acetylation of both H3K14 and Rsc4K25, and hence, has the ability to promote RSC association with nucleosomes through H3 acetylation or inhibit RSC association with nucleosomes through Rsc4 acetylation (VanDemark et al. 2007). These opposing reactions may serve to fine-tune the local activity of remodeling enzyme. An analogous, but somewhat different mechanism has been described for the Swi/Snf complex. The bromodomain-containing Snf2 ATPase is acetylated by Gcn5 (Kim et al. 2010). The acetylated sites on the ATPase itself are in competition with similar epitopes on the histone substrates, a fact that suggests complex regulatory interactions (Fig. 9) (Kim et al. 2010). The mammalian Swi/Snf ATPase subunit Brm is also regulated through acetylation by the acetyltransferase PCAF (the metazoan Gcn5) (Bourachot et al. 2003). Moreover, a proteomic screen identified three sites of acetylation on the other mammalian Snf2-like ATPase, Brg1 (SMARCA4) (Choudhary et al. 2009). Regulation of chromatin remodelers by acetylation is not limited to the Swi/Snf type complexes; the Drosophila ISWI ATPase can be acetylated by the Gcn5 and p300 acetyltransferases on lysine 753, a region of the protein that bears similarity to the H3 tail (Ferreira et al. 2007). These examples show that acetylation may serve to regulate the properties of both components of nucleosome remodeling reaction, the nucleosome substrate, and the remodeling enzyme.
PARP-1 (poly[adenosine diphosphate-ribose] polymerase-1) has emerged as a major regulator of genome function. PARP-1 also modifies histones and proteins involved in chromatin modification, transcription, and DNA repair (Krishnakumar and Kraus 2010), most notably the ISWI ATPase. PARylation of ISWI inhibits its ATPase activity and nucleosome binding. This is corroborated by PARP-1 counteracting the function of ISWI in flies (Krishnakumar and Kraus 2010).
Phosphorylation is a modification that is widely used to coordinate processes during cell cycle progression and during the DNA damage response. In this context it is not surprising that Swi/Snf-type remodeling complexes are also regulated by phosphorylation in diverse ways, which is illustrated by the following, selected examples (Vignali et al. 2000). The Sfh1 subunit of the yeast RSC complex is phosphorylated in G1 phase of the cell cycle, and temperature-sensitive alleles of Sfh1 arrest at the G2/M transition. In humans, the hbrm and Brg1 proteins (Snf2 homologs) are phosphorylated during mitosis, which leads to their dissociation from mitotic chromatin. The Drosophila Mi-2 subunit of the NuRD complex is constitutively phosphorylated by the CK2 kinase increasing its affinity for nucleosomes and its nucleosome remodeling activity (Bouazoune and Brehm 2005). The Ies4 subunit of the INO80 complex is phosphorylated in response to DNA damage by the Mec1/Tel1 kinases (ataxia telangiectasia mutated [ATM]/ATM and Rad3-related [ATR] homologs) in yeast. Phosphorylation of Ies4 does not appear to be required for DNA repair, but is required for the DNA damage checkpoint responses (Morrison and Shen 2009). As multiple chromatin remodelers have been implicated in double-strand break repair (Polo and Jackson 2011), it would not be surprising if additional complexes are found to be regulated by phosphorylation during DNA repair.
In summary, known modifications involved in the regulation of chromatin remodeling complexes include acetylation, PARylation, and phosphorylation. Modification of remodeler subunits can activate or inhibit the activity of the complexes and their interactions with nucleosomes. Such modifications appear to “fine tune” the regulation of remodeling complexes.
8. INTERACTION OF CHROMATIN REMODELERS WITH DNA METHYLATION
DNA methylation has long been implicated in gene silencing and as it has a clear potential to be inherited through cell divisions, it is a prime component of epigenetic regulation (Clouaire and Stancheva 2008; Li and Zhang 2014). In eukaryotes, DNA methylation occurs exclusively at CG dinucleotides and its effects on gene silencing are manifested through meCG binding proteins (MBDs). MBD proteins associate with a number of transcriptional corepressor complexes including histone methylases, histone deacetylases, and chromatin remodelers (Clouaire and Stancheva 2008). The chromatin remodeling complex with the most studied connection to DNA methylation is the Mi-2/NuRD complex. NuRD contains MBD2 or MBD3 as core subunits and preferentially remodels and deacetylates nucleosomes containing methylated DNA. MBD2 and MBD3 are in distinct versions of the NuRD complex and as MBD3 does not actually bind methylated DNA presumably only the MBD2 form of NuRD contributes to DNA methylation-mediated repression (Clouaire and Stancheva 2008).
9. CHROMATIN REMODELERS AND HISTONE VARIANTS
Nucleosome remodelers are commonly involved in incorporating histone variants into chromatin. In turn, the nucleosome remodeling activity can be affected by the presence of histone variants in nucleosomal substrates. The most prominent example is the function of the SWR1 complex in the incorporation of variant histone Htz1 (yeast H2A.Z) into chromatin. H2A.Z is a highly conserved variant that is implicated in gene activation, protection from heterochromatin spreading and chromosome segregation (Altaf et al. 2009; Morrison and Shen 2009). Htz1 is inserted into chromatin by the SWR1 complex in yeast and by the SRCAP or p400/Tip60 complexes in humans (Billon and Cote 2011). Swr1, SRCAP, and p400 are the ATPase subunits of the respective complexes and catalyze the ATP-dependent replacement of H2A with H2A.Z. Htz1 is inserted at promoters of silent genes, but assists in their subsequent activation (Workman 2006). The SWR complex contains a bromodomain protein, Bdf1, which may bring SWR to acetylated nucleosomes at promoters for the insertion of Htz1. In mammals, incorporation of H2A.Z at some promoters requires SRCAP whereas others require p400. p400 is part of the TIP60 complex in flies that can directly interact with transcription factors at promoters.
The histone variant H2A.X has been suggested to poise chromatin for remodeling at sites of DNA breaks (Talbert and Henikoff 2010). H2A.X is phosphorylated on a specific serine in the carboxy-terminal tail of the protein by the ATM kinase in response to DNA breaks. Phosphorylated H2A.X (γ-H2AX) is important for recruiting or retaining a number of chromatin modifying and repair proteins to the site of the double-strand break (Talbert and Henikoff 2010). γ-H2AX is bound by Arp4, a subunit of the NuA4 HAT complex as well as the SWR1 and INO80 chromatin remodeling complexes, and is thought to play a role in the recruitment of all three of these complexes to DNA breaks, where they contribute to the repair process (Altaf et al. 2009).
Other H2A variants also affect the function of nucleosome remodeling complexes. MacroH2A, the largest of the histone variants, contains a 30-kDa carboxy-terminal globular tail called “macrodomain” (Gamble and Kraus 2010). Macrodomains bind metabolites of NAD generated by PARP or Sirtuin-type deacetylases. MacroH2A is most characterized in its involvement in facultative heterochromatin and transcriptional repression at the inactive X chromosome. More recently, its presence in nucleosomes was shown to inhibit recruitment of or remodeling by the Swi/Snf complex (Gamble and Kraus 2010). In contrast to macroH2A, histone H2A.Bbd (Barr body deficient) appears to associate with active acetylated chromatin and is excluded from the inactive X chromosome (Gonzalez-Romero et al. 2008). This short mammalian-specific histone variant forms nucleosomes that are less stable than those containing canonical H2A (Gonzalez-Romero et al. 2008). Surprisingly, the presence of H2A.Bbd in nucleosomes reduced their ability to be remodeled/mobilized by the Swi/Snf and ACF complexes in vitro. At the same time, H2A.Bbd-containing nucleosome arrays were more active templates in p300-activated transcription (Gonzalez-Romero et al. 2008). Thus, the contribution of H2A.Bbd to gene expression most likely is attributable to inherently destabilizing nucleosomes rather than making nucleosomes more sensitive to nucleosome remodeling.
10. NUCLEOSOME REMODELING DURING DEVELOPMENT
Because of their versatile activities nucleosome remodeling factors affect every aspect of genome function, even those that are characterized by epigenetic stability. Nucleosome remodeling may affect the assembly of such stable states directly by laying the foundations of higher-order chromatin organization or by strategic placement of histone variants as described in Section 9. Alternatively, they may help to establish and maintain stable, cell lineage-specific gene expression programs by serving as essential coregulators of the transcription machinery. A case in point is the versatile BAF complex, involved in orchestrating a number of rather diverse gene expression programs ranging from embryonic stem cells to postmitotic neurons (Yoo and Crabtree 2009; Ho and Crabtree 2010; Hargreaves and Crabtree 2011).
The explanation for the extraordinary versatility of the BAF complexes lies in the diversity of their subunit composition. The BAF remodelers of different cells all share a “core complex” of identical subunits that may be considered the engine that drives the fundamental, BAF-specific remodeling reaction. In addition to these core subunits, BAF complexes purified from different cells contain additional, cell-specific subunits (Fig. 8B). For example, the BAF complex purified from ES cells (esBAF) never contains the ATPase isoform BRM or the associated BAF170 subunit, but features BAF53a and BAF45a subunits. In contrast, the BAF complex isolated from postmitotic neurons (nBAF), may contain BRM in place of BRG1, BAF170 instead of BAF155, and is characterized by the presence of the variants BAF53b and BAF45b/c. The closely related BAF45 variants differ in their amino-terminal Krüppel-like domain. Remarkably, the proliferating and self-renewing neural progenitor cells contain BAF complexes of intermediate subunit composition (npBAF) (Fig. 8B) (Yoo and Crabtree 2009; Ho and Crabtree 2010; Hargreaves and Crabtree 2011). The most likely scenario is that the specificity subunits mediate interactions with cell-type-specific transcription factors that recruit the remodeling coactivators to alternative sets of genes. The switch between the different BAF complexes is brought about by repression of BAF53a through microRNAs that are specifically activated in postmitotic neurons and suppressed by the action of the transcription repressor REST in precursor cells. Continued expression of the progenitor signature subunits BAF45a and BAF53a prevents neuronal differentiation (Yoo and Crabtree 2009; Ho and Crabtree 2010; Hargreaves and Crabtree 2011). In agreement with their roles as regulators of differentiation programs, many BAF proteins function as tumor suppressors and their expression is lost or reduced in several forms of cancer (Hargreaves and Crabtree 2011). In conclusion the BAF complexes provide a striking example of a novel mechanism regulating patterns of gene expression during development. Developmental patterns of gene expression are not only dictated by the presence or absence of tissue specific transcription factors and epigenetic marks on chromatin but also by the subunit composition of a nucleosome remodeling complex.
11. CONCLUSIONS
Nucleosome remodeling enzymes, despite the dynamic nature of the chromatin transitions they catalyze, are involved in the assembly and propagation of stable and lasting—epigenetic—chromatin states. Canonical roles for nucleosome remodeling factors involve local nucleosome remodeling at regulatory elements to affect specific gene expression programs, as well as assuring the integrity of the chromatin fiber by nucleosome assembly and spacing, and finally their role in the exchange of histone variants. Remodeling factors may have other, less explored functions as well. We know of isolated examples in which remodelers use their ATP-dependent DNA translocase activities to modulate the chromatin association of nonhistone substrates (Kia et al. 2008; Wollmann et al. 2011). We also contemplate their impact on chromatin organization independent of their enzymatic remodeling reactions, given the large number of remodeling machines associated with chromatin (Varga-Weisz and Becker 2006). A lot remains to be discovered.
ACKNOWLEDGMENTS
We thank Joanne Chatfield for assistance in preparing this article. Work in the laboratory of J.L.W. is funded by National Institute of General Medical Sciences (NIGMS) grants GM047867 and GM99945 and the Stowers Institute for Medical Research. Work on nucleosome remodeling factors in the laboratory of P.B. was funded by Deutsche Forschungsgemeinschaft SFB594, SPP1356, and the European Union through the Network of Excellence “The Epigenome” (FP6-503433).
Footnotes
Editors: C. David Allis, Marie-Laure Caparros, Thomas Jenuwein, and Danny Reinberg
Additional Perspectives on Epigenetics available at www.cshperspectives.org
REFERENCES
*Reference is also in this collection.
- Alkhatib SG, Landry JW 2011. The nucleosome remodeling factor. FEBS Lett 585: 3197–3207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Almouzni G, Cedar H 2014. Maintenance of epigenetic information. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a019372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altaf M, Auger A, Covic M, Cote J 2009. Connection between histone H2A variants and chromatin remodeling complexes. Biochem Cell Biol 87: 35–50 [DOI] [PubMed] [Google Scholar]
- Altaf M, Saksouk N, Cote J 2007. Histone modifications in response to DNA damage. Mutat Res 618: 81–90 [DOI] [PubMed] [Google Scholar]
- Bao Y, Shen X 2007. Chromatin remodeling in DNA double-strand break repair. Curr Opin Genet Dev 17: 126–131 [DOI] [PubMed] [Google Scholar]
- Bao Y, Shen X 2011. SnapShot: Chromatin remodeling: INO80 and SWR1. Cell 144: 158–158. e2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker PB, Horz W 2002. ATP-dependent nucleosome remodeling. Annu Rev Biochem 71: 247–273 [DOI] [PubMed] [Google Scholar]
- Becker PB, Wu C 1992. Cell-free system for assembly of transcriptionally repressed chromatin from Drosophila embryos. Mol Cell Biol 12: 2241–2249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billon P, Cote J 2011. Precise deposition of histone H2A.Z in chromatin for genome expression and maintenance. Biochim Biophys Acta 1819: 290–302 [DOI] [PubMed] [Google Scholar]
- Bouazoune K, Brehm A 2005. dMi-2 chromatin binding and remodeling activities are regulated by dCK2 phosphorylation. J Biol Chem 280: 41912–41920 [DOI] [PubMed] [Google Scholar]
- Bourachot B, Yaniv M, Muchardt C 2003. Growth inhibition by the mammalian SWI-SNF subunit Brm is regulated by acetylation. EMBO J 22: 6505–6515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowen NJ, Fujita N, Kajita M, Wade PA 2004. Mi-2/NuRD: Multiple complexes for many purposes. Biochim Biophys Acta 1677: 52–57 [DOI] [PubMed] [Google Scholar]
- Brehm A, Tufteland KR, Aasland R, Becker PB 2004. The many colours of chromodomains. Bioessays 26: 133–140 [DOI] [PubMed] [Google Scholar]
- Cai Y, Jin J, Yao T, Gottschalk AJ, Swanson SK, Wu S, Shi Y, Washburn MP, Florens L, Conaway RC, et al. 2007. YY1 functions with INO80 to activate transcription. Nat Struct Mol Biol 14: 872–874 [DOI] [PubMed] [Google Scholar]
- Cairns BR 2001. Emerging roles for chromatin remodeling in cancer biology. Trends Cell Biol 11: S15–21 [DOI] [PubMed] [Google Scholar]
- Cairns BR 2007. Chromatin remodeling: Insights and intrigue from single-molecule studies. Nat Struct Mol Biol 14: 989–996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns BR 2009. The logic of chromatin architecture and remodelling at promoters. Nature 461: 193–198 [DOI] [PubMed] [Google Scholar]
- Chaban Y, Ezeokonkwo C, Chung WH, Zhang F, Kornberg RD, Maier-Davis B, Lorch Y, Asturias FJ 2008. Structure of a RSC-nucleosome complex and insights into chromatin remodeling. Nat Struct Mol Biol 15: 1272–1277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Cheng X 2014. Structural and functional coordination of DNA and histone methylation. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a018747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chioda M, Becker PB 2010. Soft skills turned into hard facts: Nucleosome remodelling at developmental switches. Heredity 105: 71–79 [DOI] [PubMed] [Google Scholar]
- Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, Mann M 2009. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science 325: 834–840 [DOI] [PubMed] [Google Scholar]
- Clapier CR, Cairns BR 2009. The biology of chromatin remodeling complexes. Annu Rev Biochem 78: 273–304 [DOI] [PubMed] [Google Scholar]
- Clouaire T, Stancheva I 2008. Methyl-CpG binding proteins: Specialized transcriptional repressors or structural components of chromatin? Cell Mol Life Sci 65: 1509–1522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dechassa ML, Sabri A, Pondugula S, Kassabov SR, Chatterjee N, Kladde MP, Bartholomew B 2010. SWI/SNF has intrinsic nucleosome disassembly activity that is dependent on adjacent nucleosomes. Mol Cell 38: 590–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimova D, Nackerdien Z, Furgeson S, Eguchi S, Osley MA 1999. A role for transcriptional repressors in targeting the yeast Swi/Snf complex. Mol Cell 4: 75–83 [DOI] [PubMed] [Google Scholar]
- Dion V, Shimada K, Gasser SM 2010. Actin-related proteins in the nucleus: Life beyond chromatin remodelers. Curr Opin Cell Biol 22: 383–391 [DOI] [PubMed] [Google Scholar]
- Downs JA, Nussenzweig MC, Nussenzweig A 2007. Chromatin dynamics and the preservation of genetic information. Nature 447: 951–958 [DOI] [PubMed] [Google Scholar]
- Durr H, Flaus A, Owen-Hughes T, Hopfner KP 2006. Snf2 family ATPases and DExx box helicases: Differences and unifying concepts from high-resolution crystal structures. Nucleic Acids Res 34: 4160–4167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elfring LK, Deuring R, McCallum CM, Peterson CL, Tamkun JW 1994. Identification and characterization of Drosophila relatives of the yeast transcriptional activator SNF2/SWI2. Mol Cell Biol 14: 2225–2234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elgin SC 1981. DNAase I-hypersensitive sites of chromatin. Cell 27: 413–415 [DOI] [PubMed] [Google Scholar]
- *.Elgin SCR, Reuter G 2013. Position-effect variegation, heterochromatin formation, and gene silencing in Drosophila. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a017780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emelyanov AV, Vershilova E, Ignatyeva MA, Pokrovsky DK, Lu X, Konev AY, Fyodorov DV 2012. Identification and characterization of ToRC, a novel ISWI-containing ATP-dependent chromatin assembly complex. Genes Dev 26: 603–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engeholm M, de Jager M, Flaus A, Brenk R, van Noort J, Owen-Hughes T 2009. Nucleosomes can invade DNA territories occupied by their neighbors. Nat Struct Mol Biol 16: 151–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairman-Williams ME, Guenther UP, Jankowsky E 2010. SF1 and SF2 helicases: Family matters. Curr Opin Struct Biol 20: 313–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falbo KB, Shen X 2006. Chromatin remodeling in DNA replication. J Cell Biochem 97: 684–689 [DOI] [PubMed] [Google Scholar]
- Fazzio TG, Kooperberg C, Goldmark JP, Neal C, Basom R, Delrow J, Tsukiyama T 2001. Widespread collaboration of Isw2 and Sin3-Rpd3 chromatin remodeling complexes in transcriptional repression. Mol Cell Biol 21: 6450–6460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira R, Eberharter A, Bonaldi T, Chioda M, Imhof A, Becker PB 2007. Site-specific acetylation of ISWI by GCN5. BMC Mol Biol 8: 73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flaus A, Martin DM, Barton GJ, Owen-Hughes T 2006. Identification of multiple distinct Snf2 subfamilies with conserved structural motifs. Nucleic Acids Res 34: 2887–2905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flaus A, Owen-Hughes T 2011. Mechanisms for ATP-dependent chromatin remodelling: The means to the end. FEBS J 278: 3579–3595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floer M, Wang X, Prabhu V, Berrozpe G, Narayan S, Spagna D, Alvarez D, Kendall J, Krasnitz A, Stepansky A, et al. 2010. A RSC/nucleosome complex determines chromatin architecture and facilitates activator binding. Cell 141: 407–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry CJ, Peterson CL 2001. Chromatin remodeling enzymes: Who’s on first? Curr Biol 11: R185–R197 [DOI] [PubMed] [Google Scholar]
- Gamble MJ, Kraus WL 2010. Multiple facets of the unique histone variant macroH2A: From genomics to cell biology. Cell Cycle 9: 2568–2574 [DOI] [PubMed] [Google Scholar]
- Gangaraju VK, Bartholomew B 2007. Mechanisms of ATP dependent chromatin remodeling. Mutat Res 618: 3–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Romero R, Mendez J, Ausio J, Eirin-Lopez JM 2008. Quickly evolving histones, nucleosome stability and chromatin folding: All about histone H2A.Bbd. Gene 413: 1–7 [DOI] [PubMed] [Google Scholar]
- Gutierrez JL, Chandy M, Carrozza MJ, Workman JL 2007. Activation domains drive nucleosome eviction by SWI/SNF. EMBO J 26: 730–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanai K, Furuhashi H, Yamamoto T, Akasaka K, Hirose S 2008. RSF governs silent chromatin formation via histone H2Av replacement. PLoS Genet 4: e1000011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hargreaves DC, Crabtree GR 2011. ATP-dependent chromatin remodeling: Genetics, genomics and mechanisms. Cell Res 21: 396–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauk G, Bowman GD 2011. Structural insights into regulation and action of SWI2/SNF2 ATPases. Curr Opin Struct Biol 21: 719–727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauk G, McKnight JN, Nodelman IM, Bowman GD 2010. The chromodomains of the Chd1 chromatin remodeler regulate DNA access to the ATPase motor. Mol Cell 39: 711–723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Henikoff S, Smith M 2014. Histone variants and epigenetics. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a019364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho L, Crabtree GR 2010. Chromatin remodelling during development. Nature 463: 474–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn PJ, Peterson CL 2001. The bromodomain: A regulator of ATP-dependent chromatin remodeling? Front Biosci 6: D1019–D1023 [DOI] [PubMed] [Google Scholar]
- Ito T, Levenstein ME, Fyodorov DV, Kutach AK, Kobayashi R, Kadonaga JT 1999. ACF consists of two subunits, Acf1 and ISWI, that function cooperatively in the ATP-dependent catalysis of chromatin assembly. Genes Dev 13: 1529–1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jha S, Dutta A 2009. RVB1/RVB2: Running rings around molecular biology. Mol Cell 34: 521–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasten MM, Clapier CR, Cairns BR 2011. SnapShot: Chromatin remodeling: SWI/SNF. Cell 144: 310.e1. [DOI] [PubMed] [Google Scholar]
- Kia SK, Gorski MM, Giannakopoulos S, Verrijzer CP 2008. SWI/SNF mediates polycomb eviction and epigenetic reprogramming of the INK4b-ARF-INK4a locus. Mol Cell Biol 28: 3457–3464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Saraf A, Florens L, Washburn M, Workman JL 2010. Gcn5 regulates the dissociation of SWI/SNF from chromatin by acetylation of Swi2/Snf2. Genes Dev 24: 2766–2771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingston RE, Narlikar GJ 1999. ATP-dependent remodeling and acetylation as regulators of chromatin fluidity. Genes Dev 13: 2339–2352 [DOI] [PubMed] [Google Scholar]
- *.Kingston R, Tamkun J 2014. Transcriptional regulation by Trithorax group proteins. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a019349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korber P, Becker PB 2010. Nucleosome dynamics and epigenetic stability. Essays Biochem 48: 63–74 [DOI] [PubMed] [Google Scholar]
- Krishnakumar R, Kraus WL 2010. The PARP side of the nucleus: Molecular actions, physiological outcomes, and clinical targets. Mol Cell 39: 8–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunert N, Brehm A 2009. Novel Mi-2 related ATP-dependent chromatin remodelers. Epigenetics 4: 209–211 [DOI] [PubMed] [Google Scholar]
- Lan L, Ui A, Nakajima S, Hatakeyama K, Hoshi M, Watanabe R, Janicki SM, Ogiwara H, Kohno T, Kanno S, et al. 2010. The ACF1 complex is required for DNA double-strand break repair in human cells. Mol Cell 40: 976–987 [DOI] [PubMed] [Google Scholar]
- Lantermann AB, Straub T, Stralfors A, Yuan GC, Ekwall K, Korber P 2010. Schizosaccharomyces pombe genome-wide nucleosome mapping reveals positioning mechanisms distinct from those of Saccharomyces cerevisiae. Nat Struct Mol Biol 17: 251–257 [DOI] [PubMed] [Google Scholar]
- *.Li E, Zhang Y 2014. DNA methylation in mammals. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a019133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Carey M, Workman JL 2007. The role of chromatin during transcription. Cell 128: 707–719 [DOI] [PubMed] [Google Scholar]
- Li J, Langst G, Grummt I 2006. NoRC-dependent nucleosome positioning silences rRNA genes. EMBO J 25: 5735–5741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lia G, Praly E, Ferreira H, Stockdale C, Tse-Dinh YC, Dunlap D, Croquette V, Bensimon D, Owen-Hughes T 2006. Direct observation of DNA distortion by the RSC complex. Mol Cell 21: 417–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loyola A, Huang JY, LeRoy G, Hu S, Wang YH, Donnelly RJ, Lane WS, Lee SC, Reinberg D 2003. Functional analysis of the subunits of the chromatin assembly factor RSF. Mol Cell Biol 23: 6759–6768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luger K, Richmond TJ 1998. DNA binding within the nucleosome core. Curr Opin Struct Biol 8: 33–40 [DOI] [PubMed] [Google Scholar]
- Lusser A, Urwin DL, Kadonaga JT 2005. Distinct activities of CHD1 and ACF in ATP-dependent chromatin assembly. Nat Struct Mol Biol 12: 160–166 [DOI] [PubMed] [Google Scholar]
- Mansfield RE, Musselman CA, Kwan AH, Oliver SS, Garske AL, Davrazou F, Denu JM, Kutateladze TG, Mackay JP 2011. Plant homeodomain (PHD) fingers of CHD4 are histone H3-binding modules with preference for unmodified H3K4 and methylated H3K9. J Biol Chem 286: 11779–11791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens JA, Winston F 2003. Recent advances in understanding chromatin remodeling by Swi/Snf complexes. Curr Opin Genet Dev 13: 136–142 [DOI] [PubMed] [Google Scholar]
- Martens JA, Wu PY, Winston F 2005. Regulation of an intergenic transcript controls adjacent gene transcription in Saccharomyces cerevisiae. Genes Dev 19: 2695–2704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morettini S, Podhraski V, Lusser A 2008. ATP-dependent chromatin remodeling enzymes and their various roles in cell cycle control. Front Biosci 13: 5522–5532 [DOI] [PubMed] [Google Scholar]
- Morettini S, Tribus M, Zeilner A, Sebald J, Campo-Fernandez B, Scheran G, Worle H, Podhraski V, Fyodorov DV, Lusser A 2011. The chromodomains of CHD1 are critical for enzymatic activity but less important for chromatin localization. Nucleic Acids Res 39: 3103–3115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison AJ, Shen X 2009. Chromatin remodelling beyond transcription: The INO80 and SWR1 complexes. Nat Rev Mol Cell Biol 10: 373–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moshkin YM, Chalkley GE, Kan TW, Reddy BA, Ozgur Z, van Ijcken WF, Dekkers DH, Demmers JA, Travers AA, Verrijzer CP 2012. Remodelers organize cellular chromatin by counteracting intrinsic histone-DNA sequence preferences in a class-specific manner. Mol Cell Biol 32: 675–688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moshkin YM, Mohrmann L, van Ijcken WF, Verrijzer CP 2007. Functional differentiation of SWI/SNF remodelers in transcription and cell cycle control. Mol Cell Biol 27: 651–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murawska M, Kunert N, van Vugt J, Langst G, Kremmer E, Logie C, Brehm A 2008. dCHD3, a novel ATP-dependent chromatin remodeler associated with sites of active transcription. Mol Cell Biol 28: 2745–2757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neves-Costa A, Varga-Weisz P 2006. The roles of chromatin remodelling factors in replication. Results Probl Cell Differ 41: 91–107 [DOI] [PubMed] [Google Scholar]
- *.Patel D 2014. A structural perspective on readout of epigenetic H and DM marks. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a018754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazin MJ, Kamakaka RT, Kadonaga JT 1994. ATP-dependent nucleosome reconfiguration and transcriptional activation from preassembled chromatin templates. Science 266: 2007–2011 [DOI] [PubMed] [Google Scholar]
- Polo SE, Jackson SP 2011. Dynamics of DNA damage response proteins at DNA breaks: A focus on protein modifications. Genes Dev 25: 409–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rach EA, Winter DR, Benjamin AM, Corcoran DL, Ni T, Zhu J, Ohler U 2011. Transcription initiation patterns indicate divergent strategies for gene regulation at the chromatin level. PLoS Genet 7: e1001274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Racki LR, Narlikar GJ 2008. ATP-dependent chromatin remodeling enzymes: Two heads are not better, just different. Curr Opin Genet Dev 18: 137–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reik A, Schutz G, Stewart AF 1991. Glucocorticoids are required for establishment and maintenance of an alteration in chromatin structure: Induction leads to a reversible disruption of nucleosomes over an enhancer. EMBO J 10: 2569–2576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowbotham SP, Barki L, Neves-Costa A, Santos F, Dean W, Hawkes N, Choudhary P, Will WR, Webster J, Oxley D, et al. 2011. Maintenance of silent chromatin through replication requires SWI/SNF-like chromatin remodeler SMARCAD1. Mol Cell 42: 285–296 [DOI] [PubMed] [Google Scholar]
- Ryan DP, Owen-Hughes T 2011. Snf2-family proteins: Chromatin remodellers for any occasion. Curr Opin Chem Biol 15: 649–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan DP, Sundaramoorthy R, Martin D, Singh V, Owen-Hughes T 2011. The DNA-binding domain of the Chd1 chromatin-remodelling enzyme contains SANT and SLIDE domains. EMBO J 30: 2596–2609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha A, Wittmeyer J, Cairns BR 2006. Chromatin remodelling: The industrial revolution of DNA around histones. Nat Rev Mol Cell Biol 7: 437–447 [DOI] [PubMed] [Google Scholar]
- Sanchez R, Zhou MM 2011. The PHD finger: A versatile epigenome reader. Trends Biochem Sci 36: 364–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwabish MA, Struhl K 2007. The Swi/Snf complex is important for histone eviction during transcriptional activation and RNA polymerase II elongation in vivo. Mol Cell Biol 27: 6987–6995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selth LA, Sigurdsson S, Svejstrup JQ 2010. Transcript elongation by RNA polymerase II. Annu Rev Biochem 79: 271–293 [DOI] [PubMed] [Google Scholar]
- Simic R, Lindstrom DL, Tran HG, Roinick KL, Costa PJ, Johnson AD, Hartzog GA, Arndt KM 2003. Chromatin remodeling protein Chd1 interacts with transcription elongation factors and localizes to transcribed genes. EMBO J 22: 1846–1856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims JK, Wade PA 2011. SnapShot: Chromatin remodeling: CHD. Cell 144: 626–626. e1 [DOI] [PubMed] [Google Scholar]
- Soutourina J, Bordas-Le Floch V, Gendrel G, Flores A, Ducrot C, Dumay-Odelot H, Soularue P, Navarro F, Cairns BR, Lefebvre O, et al. 2006. Rsc4 connects the chromatin remodeler RSC to RNA polymerases. Mol Cell Biol 26: 4920–4933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan S, Armstrong JA, Deuring R, Dahlsveen IK, McNeill H, Tamkun JW 2005. The Drosophila trithorax group protein Kismet facilitates an early step in transcriptional elongation by RNA Polymerase II. Development 132: 1623–1635 [DOI] [PubMed] [Google Scholar]
- Suganuma T, Workman JL 2011. Signals and combinatorial functions of histone modifications. Annu Rev Biochem 80: 473–499 [DOI] [PubMed] [Google Scholar]
- Sugiyama T, Cam HP, Sugiyama R, Noma K, Zofall M, Kobayashi R, Grewal SI 2007. SHREC, an effector complex for heterochromatic transcriptional silencing. Cell 128: 491–504 [DOI] [PubMed] [Google Scholar]
- Talbert PB, Henikoff S 2010. Histone variants—Ancient wrap artists of the epigenome. Nat Rev Mol Cell Biol 11: 264–275 [DOI] [PubMed] [Google Scholar]
- Tirosh I, Sigal N, Barkai N 2010. Widespread remodeling of mid-coding sequence nucleosomes by Isw1. Genome Biol 11: R49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolkunov D, Zawadzki KA, Singer C, Elfving N, Morozov AV, Broach JR 2011. Chromatin remodelers clear nucleosomes from intrinsically unfavorable sites to establish nucleosome-depleted regions at promoters. Mol Biol Cell 22: 2106–2118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torigoe SE, Urwin DL, Ishii H, Smith DE, Kadonaga JT 2011. Identification of a rapidly formed nonnucleosomal histone-DNA intermediate that is converted into chromatin by ACF. Mol Cell 43: 638–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukiyama T, Becker PB, Wu C 1994. ATP-dependent nucleosome disruption at a heat-shock promoter mediated by binding of GAGA transcription factor. Nature 367: 525–532 [DOI] [PubMed] [Google Scholar]
- Tsukiyama T, Daniel C, Tamkun J, Wu C 1995. ISWI, a member of the SWI2/SNF2 ATPase family, encodes the 140 kDa subunit of the nucleosome remodeling factor. Cell 83: 1021–1026 [DOI] [PubMed] [Google Scholar]
- VanDemark AP, Kasten MM, Ferris E, Heroux A, Hill CP, Cairns BR 2007. Autoregulation of the rsc4 tandem bromodomain by gcn5 acetylation. Mol Cell 27: 817–828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanolst L, Fromental-Ramain C, Ramain P 2005. Toutatis, a TIP5-related protein, positively regulates Pannier function during Drosophila neural development. Development 132: 4327–4338 [DOI] [PubMed] [Google Scholar]
- Varga-Weisz PD, Becker PB 2006. Regulation of higher-order chromatin structures by nucleosome-remodelling factors. Curr Opin Genet Dev 16: 151–156 [DOI] [PubMed] [Google Scholar]
- Varga-Weisz PD, Blank TA, Becker PB 1995. Energy-dependent chromatin accessibility and nucleosome mobility in a cell-free system. EMBO J 14: 2209–2216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varga-Weisz PD, Wilm M, Bonte E, Dumas K, Mann M, Becker PB 1997. Chromatin-remodelling factor CHRAC contains the ATPases ISWI and topoisomerase II. Nature 388: 598–602 [DOI] [PubMed] [Google Scholar]
- Vicent GP, Nacht AS, Zaurin R, Ballare C, Clausell J, Beato M 2010. Minireview: Role of kinases and chromatin remodeling in progesterone signaling to chromatin. Mol Endocrinol 24: 2088–2098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vignali M, Hassan AH, Neely KE, Workman JL 2000. ATP-dependent chromatin-remodeling complexes. Mol Cell Biol 20: 1899–1910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissman B, Knudsen KE 2009. Hijacking the chromatin remodeling machinery: Impact of SWI/SNF perturbations in cancer. Cancer Res 69: 8223–8230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehouse I, Rando OJ, Delrow J, Tsukiyama T 2007. Chromatin remodelling at promoters suppresses antisense transcription. Nature 450: 1031–1035 [DOI] [PubMed] [Google Scholar]
- Whitehouse I, Tsukiyama T 2006. Antagonistic forces that position nucleosomes in vivo. Nat Struct Mol Biol 13: 633–640 [DOI] [PubMed] [Google Scholar]
- Winston F, Carlson M 1992. Yeast SNF/SWI transcriptional activators and the SPT/SIN chromatin connection. Trends Genet 8: 387–391 [DOI] [PubMed] [Google Scholar]
- Wippo CJ, Israel L, Watanabe S, Hochheimer A, Peterson CL, Korber P 2011. The RSC chromatin remodelling enzyme has a unique role in directing the accurate positioning of nucleosomes. EMBO J 30: 1277–1288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollmann P, Cui S, Viswanathan R, Berninghausen O, Wells MN, Moldt M, Witte G, Butryn A, Wendler P, Beckmann R, et al. 2011. Structure and mechanism of the Swi2/Snf2 remodeller Mot1 in complex with its substrate TBP. Nature 475: 403–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Workman JL 2006. Nucleosome displacement in transcription. Genes Dev 20: 2009–2017 [DOI] [PubMed] [Google Scholar]
- Workman JL, Kingston RE 1998. Alteration of nucleosome structure as a mechanism of transcriptional regulation. Annu Rev Biochem 67: 545–579 [DOI] [PubMed] [Google Scholar]
- Yadon AN, Tsukiyama T 2011. SnapShot: Chromatin remodeling: ISWI. Cell 144: 453–453.e1 [DOI] [PubMed] [Google Scholar]
- Yadon AN, Van de Mark D, Basom R, Delrow J, Whitehouse I, Tsukiyama T 2010. Chromatin remodeling around nucleosome-free regions leads to repression of noncoding RNA transcription. Mol Cell Biol 30: 5110–5122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo AS, Crabtree GR 2009. ATP-dependent chromatin remodeling in neural development. Curr Opin Neurobiol 19: 120–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng L, Zhang Q, Li S, Plotnikov AN, Walsh MJ, Zhou MM 2010. Mechanism and regulation of acetylated histone binding by the tandem PHD finger of DPF3b. Nature 466: 258–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Wippo CJ, Wal M, Ward E, Korber P, Pugh BF 2011. A packing mechanism for nucleosome organization reconstituted across a eukaryotic genome. Science 332: 977–980 [DOI] [PMC free article] [PubMed] [Google Scholar]