Abstract
Histone methylation is a key element of the eukaryotic epigenome. Since the discovery of the first histone demethylase (HDM) in 2004, more than 20 demethylases have been identified and characterized. They belong to either the LSD family or the JmjC family, demonstrating the reversibility of all methylation states at almost all major histone lysine methylation sites. These findings ended decades of debate about the reversibility of histone methylation, representing a major breakthrough that shifts our understanding of epigenetic inheritance and regulation of genome function. Here, we summarize the discovery of HDMs and more recent advances, challenges, and future prospects of HDM research.
The discovery of LSD1 in 2004 ended decades of debate about the reversibility of histone methylation. Since then, more than 20 demethylases, belonging to either the LSD family or the JmjC family, have been identified.
Histone methylation at lysine and arginine residues are key covalent histone modifications in epigenetic regulation. Together with DNA methylation, they constitute hallmarks of epigenetic inheritance. Although other histone modifications such as acetylation and phosphorylation have been known to be reversible for some time, the reversibility of histone methylation was in question. It was not until 2004, with the discovery of the first histone demethylase (HDM), that this issue was resolved.
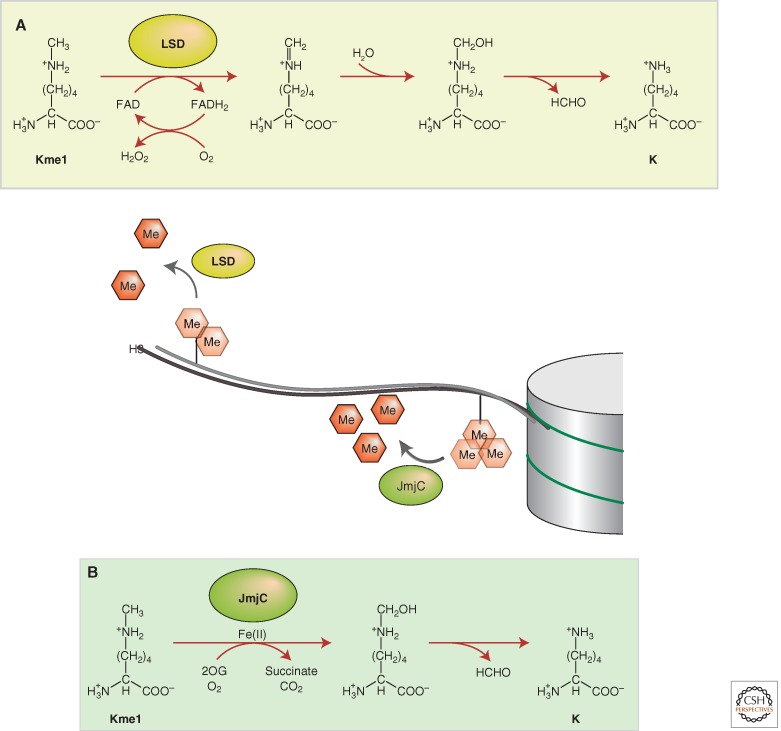
As is often the case, the first HDM, LSD1 (Gene ID: KDM1A), was discovered in an unexpected way. While studying how metabolic enzymes and their homologs and cofactors are involved in epigenetic gene regulation, Drs. Yang Shi and Yujiang Geno Shi became curious about how a metabolic enzyme homolog named nPAO (nuclear polyamine oxidase; also called KIAA0601/BHC110), which they had previously identified from the transcriptional CtBP complex, might function in epigenetic gene regulation (Shi et al. 2003). Based on nPAO’s homology with known polyamine oxidases and given that polyamines are also minor components of chromatin, it was hypothesized that nPAO might regulate chromatin structure through a polyamine oxidation mechanism. Despite months of attempts, experiments aimed at detecting the putative polyamine oxidase activity of nPAO were unsuccessful. Although it is chemically possible that oxidation of methylated lysine could lead to demethylation of lysine through an amine oxidation reaction, such a reaction mechanism has never been discovered or reported. When the substrate was switched from polyamine to histone H3 dimethylated at lysine 4 (H3K4me2), Y. G. Shi was finally able to successfully detect the nPAO-mediated histone demethylation. Dr. Yang Shi's laboratory thus discovered the first lysine demethylase (KDM1) (Shi et al. 2004). nPAO was then renamed LSD1 for lysine-specific histone demethylase 1, and the LSD family of histone demethylases has since been defined. This discovery ended decades of debate about the reversibility of histone methylation, representing a major breakthrough and a paradigm shift in our understanding of the dynamics of histone methylation. The chemical reaction that LSD1/KDM1A catalyzes is an amine oxidation by oxidative cleavage of the α-carbon bond of the methylated lysine to form an imine intermediate, which is hydrolyzed to form formaldehyde, releasing one molecule of H2O2 and the demethylated lysine (see Fig. 1). Notably, because the formation of an imine intermediate requires protonated nitrogen, the LSD family of demethylases can only demethylate mono- and dimethylated (me1, me2), but not the trimethylated (me3) lysine residues. This raised the possibility of other classes of HDMs that were yet to be discovered. Soon afterward, the Shi laboratories began to investigate the Jumonji C (JmjC) domain–containing proteins as potential histone demethylase candidates (Falnes et al. 2002; Trewick et al. 2002). Separately, Dr. Yi Zhang’s laboratory took an unbiased activity-based biochemical purification approach to search for new histone demethylases, whereas Dr. Kristian Helin’s and Dr. Thomas Jenuwein’s laboratories were also searching for additional histone demethylases, zeroing in on the JmjC domain–containing proteins.
Figure 1.
Histone demethylation mediated by (A) an LSD family demethylase through an FAD-dependent amine oxidase reaction, and (B) a JmjC domain family demethylase through a 2OG-Fe(II)-dependent dioxygenase reaction. LSD demethylases can demethylate the mono- and dimethylated states of histone lysine residues. The JmjC domain family demethylases can demethylate mono-, di-, and trimethylated histone lysine residues. For simplicity, only monomethyl-lysine is illustrated.
By the end of 2005, Tsukada succeeded in detecting HDM activity by developing a 2-oxoglutarate (2OG)-Fe(II)-dependent dioxygenase assay system based on the reaction used by the DNA repair demethylase AlkB and led Dr. Yi Zhang’s laboratory to successfully identify the first JmjC domain–containing HDM (JHDM1/KDM2), which demethylates histone H3K36 (Tsukada et al. 2006). It turns out that the mammalian genome has 30 different proteins that contain a JmjC domain. The chemical reaction catalyzed by the JmjC domain–containing HDMs is the oxygenation of a methyl group by a radical attack of a highly reactive oxoferryl species to form an unstable carbinolamine intermediate; subsequent release of formaldehyde from the carbinolamine produces demethylated lysine (see Fig. 1). Unlike the LSD1-mediated chemical reaction, this reaction is compatible with trimethylated (me3) lysine residues. Thus, in addition to the LSD family of demethylases (containing LSD1 and LSD2), the discovery of the JmjC domain–containing demethylase family (containing more than 20 HDMs) has further shown the reversibility of all histone methylation states (mono-, di-, and trimethylation) at almost all major histone methylation modified sites. Other laboratories, including those of Yang Shi, Kristian Helin, and Thomas Jenuwein, have made pioneering contributions in defining this class of HDMs (Cloos et al. 2006; Fodor et al. 2006; Whetstine et al. 2006).
Although it took more than 40 years to identify LSD1, the first HDM, in less than 4 years since the initial discovery we have seen the rapid progress in our knowledge of histone demethylation, such that it is now a major focus of epigenetic research. More than 20 HDMs actively catalyze the demethylation of almost all major histone lysine methylation sites and a number of arginine methylation sites. Yet, the following fundamental questions pertaining to enzymatic action, regulation, and biological function remain unaddressed: First, there is the possibility that a third class of HDMs is yet to be discovered. This is largely based on the premise that there is no known demethylase for H3K79 methylation. This modification is unique in that it is the only residue to be methylated by a non-SET domain histone methyltransferase, Dot1/Dot1L, and thus it is tempting to speculate that H3K79 demethylation may use a new and different class of HDMs. Also, new arginine demethylases may constitute a new class of HDMs. Second, the functional characterization of demethylases has been mostly limited to aspects of gene transcription and, in particular, promoter transcription initiation. This is a narrow focus that cannot fully explain the wide range of involvement of HDMs in biological and pathological processes. It will be worthwhile to explore how HDMs are involved in transcriptional elongation, cotranscriptional messenger RNA processing, DNA replication, and/or repair processes. Third, how HDMs are themselves regulated has yet to be explored. For example, it will be worthwhile to know how they are specifically targeted to their functional sites. How do demethylases exert their functions in the context of larger complexes containing associated cofactors? How are their expression and activity regulated? And because the action of these enzymes requires metabolic cofactors, it will be important to understand how cellular metabolism is linked to the intracellular function and regulation of HDMs.
The impact of the discovery of histone demethylases has extended beyond the regulation of histone methylation. It has also lent mechanistic insight into potential DNA demethylation, as exemplified by the recent discovery of the ten-eleven translocation family of DNA 5mC dioxygenases. We expect to see more discoveries along this line that will greatly impact the field of epigenetics. Most importantly, HDMs are involved in many normal and pathological processes including gene transcription, stem cell self-renewal, development, and tumorigenesis. Now that we know that histone methylation is reversible, there is great hope that demethylases will be promising therapeutic targets in the future of “epigenetic medicines.” Answers to the fundamental questions raised above are an absolute requisite not only for understanding the biological functions of histone demethylation, but for the progression of further clinical translational research on complex human diseases.
Footnotes
Editors: C. David Allis, Marie-Laure Caparros, Thomas Jenuwein, and Danny Reinberg
Additional Perspectives on Epigenetics available at www.cshperspectives.org
REFERENCES
- Cloos PA, Christensen J, Agger K, Maiolica A, Rappsilber J, Antal T, Hansen KH, Helin K 2006. The putative oncogene GASC1 demethylates tri- and dimethylated lysine 9 on histone H3. Nature 442: 307–311 [DOI] [PubMed] [Google Scholar]
- Falnes PO, Johansen RF, Seeberg E 2002. AlkB-mediated oxidative demethylation reverses DNA damage in Escherichia coli. Nature 419: 178–182 [DOI] [PubMed] [Google Scholar]
- Fodor BD, Kubicek S, Yonezawa M, O'Sullivan RJ, Sengupta R, Perez-Burgos L, Opravil S, Mechtler K, Schotta G, Jenuwein T 2006. Jmjd2b antagonizes H3K9 trimethylation at pericentric heterochromatin in mammalian cells. Genes Dev 20: 1557–1562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi YJ, Sawada J-I, Sui GC, Affar EB, Whetstine J, Lan F, Ogawa H, Luke MP-S, Nakatani Y, Shi Y 2003. Coordinated histone modifications mediated by a CtBP co-repressor complex. Nature 422: 735–738 [DOI] [PubMed] [Google Scholar]
- Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, Casero RA, Shi Y 2004. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 119: 941–953 [DOI] [PubMed] [Google Scholar]
- Trewick SC, Henshaw TF, Hausinger RP, Lindahl T, Sedgwick B 2002. Oxidative demethylation by Escherichia coli AlkB directly reverts DNA base damage. Nature 419: 174–178 [DOI] [PubMed] [Google Scholar]
- Tsukada Y, Fang J, Erdjument-Bromage H, Warren ME, Borchers CH, Tempst P, Zhang Y 2006. Histone demethylation by a family of JmjC domain-containing proteins. Nature 439: 811–816 [DOI] [PubMed] [Google Scholar]
- Whetstine JR, Nottke A, Lan F, Huarte M, Smolikov S, Chen Z, Spooner E, Li E, Zhang G, Colaiacovo M, Shi Y 2006. Reversal of histone lysine trimethylation by the JMJD2 family of histone demethylases. Cell 125: 467–481 [DOI] [PubMed] [Google Scholar]