Abstract
The endoplasmic reticulum (ER) is the site of synthesis for nearly one-third of the eukaryotic proteome and is accordingly endowed with specialized machinery to ensure that proteins deployed to the distal secretory pathway are correctly folded and assembled into native oligomeric complexes. Proteins failing to meet this conformational standard are degraded by ER-associated degradation (ERAD), a complex process through which folding-defective proteins are selected and ultimately degraded by the ubiquitin-proteasome system. ERAD proceeds through four tightly coupled steps involving substrate selection, dislocation across the ER membrane, covalent conjugation with polyubiquitin, and proteasomal degradation. The ERAD machinery shows a modular organization with central ER membrane-embedded ubiquitin ligases linking components responsible for recognition in the ER lumen to the ubiquitin-proteasome system in the cytoplasm. The core ERAD machinery is highly conserved among eukaryotes and much of our basic understanding of ERAD organization has been derived from genetic and biochemical studies of yeast. In this article we discuss how the core ERAD machinery is organized in mammalian cells.
Four closely linked steps clear misfolded proteins from the ER: substrate selection, dislocation across the ER membrane, covalent conjugation with polyubiquitin, and proteasomal degradation.
The endoplasmic reticulum (ER) is the entry portal to the secretory pathway and is comprised of a specialized oxidative environment in which nascent polypeptides fold and assemble into native structures with the aid of a unique set of molecular chaperones, folding catalysts, and posttranslational modifications (Helenius and Aebi 2004). An estimated one-third of the mammalian genome encodes proteins destined for the secretory pathway. The ER folding apparatus must therefore be able to accommodate substrates that are highly diverse in terms of structure, oligomeric state, and folding rate. This diversity requires stringent quality control systems to maintain biosynthetic fidelity and to prevent the accumulation or deployment of misfolded proteins that can cause proteotoxicity. The importance of these systems is evidenced by the large number of human diseases that are linked to protein misfolding in the secretory pathway (Guerriero and Brodsky 2012).
ER-associated degradation (ERAD) is the temporally and spatially coordinated surveillance process charged with clearance of aberrant proteins in the ER. Much of what is known about this system has come from studies that have exploited genetic analysis in yeast (reviewed in Vembar and Brodsky 2008; Xie and Ng 2010). The essential features of ERAD are highly conserved among eukaryotes; however, because of the much larger proteome and multicellular lifestyle, the ERAD system in metazoans is considerably more complex than in fungi. In this article, we review the organization and function of the ERAD pathway in mammalian cells.
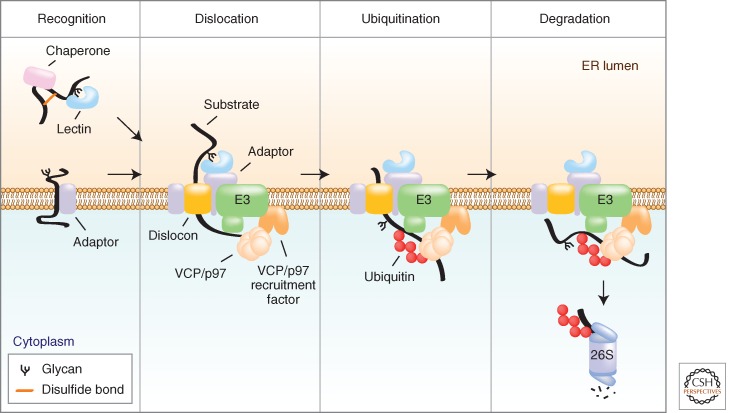
In ERAD, proteins that have been biosynthetically integrated into the ER membrane or translocated into the lumen are ultimately degraded by the ubiquitin-proteasome system (UPS). This imposes a fundamental topological constraint, in that the substrates are not initially present in the same compartment as the proteolytic system that degrades them. Thus the ERAD system necessarily spans the ER bilayer, and degradation must be mechanistically coupled to transfer (dislocation) of substrates to the cytoplasm. ERAD can be envisioned as encompassing four distinct, coupled steps (Fig. 1): (1) substrate recognition; (2) dislocation across the lipid bilayer; (3) addition (and subsequent removal) of polyubiquitin adducts; and (4) degradation by the 26S proteasome.
Figure 1.
Key steps in ERAD. ERAD occurs through a series of temporally ordered steps, which include: Step 1—Recognition: Molecular chaperones and lectins within the ER lumen interact with incompletely folded or unassembled clients. These factors link substrate recognition to the dislocation machinery by binding to membrane-embedded adaptors. Step 2—Dislocation: Substrates are dislocated across the bilayer presumably through proteinaceous pores (dislocons), via a process coupled to the energy derived from ATP hydrolysis by VCP/p97. Step 3—Ubiquitination: On gaining access to the cytosol, substrates are polyubiquitinated by E3 ligases. Step 4—Degradation: Ubiquitinated substrates are degraded by cytosolic 26S proteasomes.
STEP 1: SUBSTRATE RECOGNITION
ERAD Substrates
Substrates of the ERAD system include proteins that have failed to achieve a native structure because of mutation, translational misincorporation, or stochastic inefficiency in acquiring native structure or assembly into protein complexes. ERAD can also be exploited by regulatory pathways to control the abundance of specific ER proteins in response to metabolic signals. Some viruses can even also hijack the ERAD system by encoding effectors that redirect biosynthetic precursors of normal cell-surface proteins toward destruction. Finally, AB toxins like diphtheria, cholera, and ricin appear to use parts of the ERAD system to escape the ER lumen and gain access to their enzymatic substrates in the cytoplasm.
N-Glycans as Sensors of Glycoprotein Folding
The vast majority of proteins synthesized in the ER are cotranslationally modified by covalent addition of high-mannose “core” glycans, with the structure Glc3Man9GlcNAc2 (Glc: glucose, Man: mannose, GlcNAc: N-acetylglucosamine), to consensus Asn residues within canonical N-glycosylation sites (NxS/T) (reviewed in Helenius and Aebi 2004). These glycans play a central role in the quality control system that monitors conformational maturation and directs correctly folded proteins to ER exit sites from which they are deployed to distal compartments of the secretory pathway, or diverts them for destruction by ERAD. Enzymatic deglucosylation, by glucosidases I and II, of core glycans to Glc1Man9GlcNAc2 enables nascent glycoproteins to bind to the lectin-like chaperones calnexin (CNX) or calreticulin (CRT), which form a scaffold that facilitates oxidative folding via recruitment of the protein disulfide isomerase cofactor ERp57. Further deglucosylation removes the final glucose from the N-glycan, thereby preventing further binding of the glycoprotein to CNX/CRT, allowing the protein to progress to ER exit sites. Proteins that have not acquired native structures are substrates for reglucosylation by UDP-glucose/glycoprotein glucosyl transferase (UGGT), an enzyme that preferentially acts on incompletely folded glycoproteins, thereby returning them to CNX/CRT and allowing them to undergo further rounds of oxidative folding (reviewed in Aebi et al. 2010; Hebert et al. 2010). This cycle effectively retains unfolded glycoproteins in contact with the folding machinery in the ER and underlies the mechanism by which oxidative folding can be accomplished for proteins with vastly different folding rates.
Glycoproteins that are unable to fold, as a result of mutation for example, must escape the potentially futile CNX/CRT cycle to be diverted to ERAD. This escape has been proposed to result from the action of mannosidases that progressively remove terminal mannose residues from core glycans, enabling them to interact with a second set of mannose-specific lectins that commit them to ERAD (reviewed in Lederkremer 2009; Aebi et al. 2010). Progressive trimming of terminal mannoses by ERManI (Gonzalez et al. 1999; Tremblay and Herscovics 1999), EDEM1 (Olivari et al. 2006; Hosokawa et al. 2010), EDEM3 (Hirao et al. 2006; Hosokawa et al. 2009), and/or Golgi-resident Man1C1 (Hosokawa et al. 2007) results in substrate oligosaccharides with deglucosylated, demannosylated forms (Man5–Man7) that are incompatible with UGGT-mediated reglucosylation. The resulting oligosaccharide signatures effectively differentiate “terminally misfolded” glycoproteins from their maturation-competent counterparts.
Mannose-Specific Lectins Couple Glycoprotein Structure to ERAD
The soluble ER-resident proteins OS-9 and XTP3-B/Erlectin selectively recognize the trimmed oligosaccharides produced by ERManI/EDEM1-3 through mannose-6-phosphate receptor homology (MRH) domains (Bernasconi et al. 2008; Christianson et al. 2008; Hosokawa et al. 2008). Oligosaccharides trimmed of the C-branch terminal α1,2 (Man8C) expose a terminal α1,6 mannosyl linkage that is preferentially bound by the MRH domain of OS-9 (Quan et al. 2008; Hosokawa et al. 2009). A double tryptophan motif within the MRH domain of OS-9 discriminates untrimmed glycans from those with this signature (Satoh et al. 2010). Beyond the MRH domain, OS-9 and XTP3-B share little sequence homology, suggesting they may not serve as functional paralogs. Knockdown of either lectin has only mild effects in stabilizing ERAD substrates (Christianson et al. 2008; Hosokawa et al. 2009; Bernasconi et al. 2010). However, silencing both genes slowed the degradation of lumenal model substrates, suggesting that they may have some redundant functionality (Bernasconi et al. 2010). Intriguingly, OS-9 interacts with LONP2, a putative Lon protease and XTP3-B interacts with CPVL (carboxypeptidase vitellogenic-like protein) (Christianson et al. 2012). Although the functional significance of these recently discovered interactions is unclear, disruption of LONP2 impairs degradation of ER lumenal substrates, indicating that these putative proteases may have functions in ERAD that warrant further investigation.
Although glycoproteins represent the vast majority of secretory traffic, and their oligosaccharides provide a common feature for recognition, naturally occurring, folding-defective nonglycosylated proteins are also targeted for ERAD (Sekijima et al. 2005; Okuda-Shimizu and Hendershot 2007; Christianson et al. 2008; Hosokawa et al. 2008). Thus, features in addition to glycan trimming may contribute to diversion of folding-defective proteins to ERAD. Indeed, both OS-9 and XTP3-B have been implicated in ERAD of nonglycosylated proteins (Bernasconi et al. 2008; Christianson et al. 2008). Interestingly, a recent study reported that an amyloidogenic mutant of the naturally nonglycosylated protein transthyretin (TTRD18G) exposes a cryptic glycosylation site that results in posttranslational, STT3B-dependent glycosylation and EDEM3-dependent ERAD (Sato et al. 2012). It is therefore possible that posttranslational glycosylation may provide nonglycosylated proteins with a chance to enter the canonical glycan-dependent ERAD pathway. Alternatively, a bipartite signal provided by both trimmed glycans and misfolded regions has been proposed to contribute to ERAD substrate recognition (Bhamidipati et al. 2005; Spear and Ng 2005; Xie et al. 2009). The presence of the chaperone BiP in complexes with XTP3-B and OS-9, an interaction conserved in yeast (Yos9p-Kar2p), may endow these lectins with the capacity to recognize unfolded segments independent of N-glycan recognition (Christianson et al. 2008; Hosokawa et al. 2008). XTP3-B and OS-9 may also contain an intrinsic polypeptide binding capacity that supplements glycan recognition (Hosokawa et al. 2009). In mammals, the metazoan-specific Hsp90 homolog Grp94/gp96 also binds OS-9 (Christianson et al. 2008). Very few clients requiring this chaperone for protein folding or maturation have been identified (Eletto et al. 2010; Weekes et al. 2012); it is thus possible that Grp94 may play a previously unrecognized role, together with OS-9 and XTP3-B, in ERAD substrate recognition.
STEP 2: DISLOCATION
Adaptors Link Substrate Recognition to Dislocation
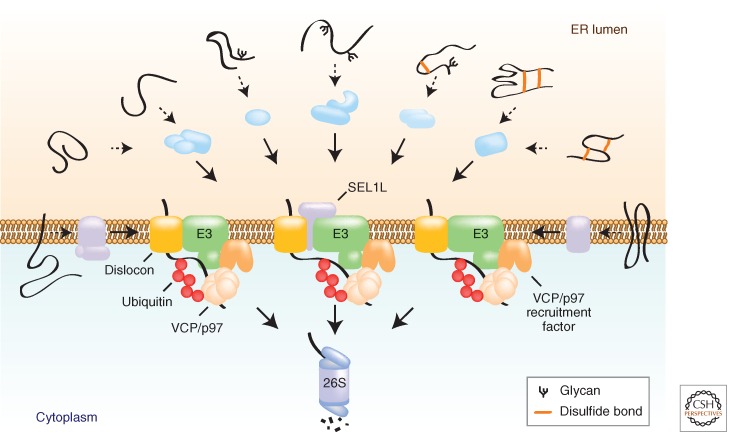
Although ERAD substrates represent a highly diverse class of structurally and topologically dissimilar proteins, they all share the need to be either fully (in the case of soluble proteins within the ER lumen) or partially (in the case of membrane-integrated substrates) translocated across the ER membrane to engage the UPS in the cytoplasm. Moreover, because many ERAD substrates are highly hydrophobic proteins that are likely to aggregate in an aqueous environment, it is essential that the processes of dislocation and degradation be tightly coupled. This coupling is strikingly reflected in the fact that membrane-embedded ubiquitin ligases appear to be “central processing units” around which most of the ERAD machinery is organized, and are likely to comprise part of the proteinaceous channels through which substrates are dislocated (“dislocons”). Recruitment of ERAD substrates to dislocons therefore uses a variety of adaptors that recognize a diverse set of features through which substrates are committed to degradation (Fig. 2).
Figure 2.
Protein adaptors within the ER membrane link substrate recognition to the dislocation apparatus. ERAD substrates (black) differ in topology, the features and location of the folding lesion, and posttranslational modification. To accommodate this diversity, the ERAD system is organized with lumenal substrate recognition factors (blue) and membrane-embedded adaptor proteins (purple) that cooperate to recruit ERAD substrates to a set of E3 ligase-coupled dislocation complexes. The dislocation complexes integrate the coupled processes of substrate ubiquitination, membrane extraction via VCP/p97, and proteolytic destruction by the 26S proteasome.
SEL1L Is an Adaptor that Links Glycan Recognition to the Dislocon
Substrates committed to the ERAD pathway via OS-9/XTP3-B are predominantly soluble glycoproteins that are sequestered within the lumen before dislocation, although some integral membrane glycoproteins with lumen-exposed domains may also be engaged by these lectins. The ER-resident glycoprotein SEL1L scaffolds an array of lumenal substrate recognition factors and links them to Hrd1, a polytopic protein proposed to be a structural component of the dislocon (Carvalho et al. 2010) that possesses a cytoplasmic E3 RING finger ubiquitin ligase domain (discussed below). SEL1L binds to OS-9 and XTP3-B (Bernasconi et al. 2008; Christianson et al. 2008; Hosokawa et al. 2008), to EDEM1 (Cormier et al. 2009) and EDEM3 (Saeed et al. 2011), and to ERFAD-ERp90 (Riemer et al. 2009; Riemer et al. 2011; Christianson et al. 2012). SEL1L also nucleates a complex with integral membrane ERAD components that include Derlin-1, Derlin-2, AUP1, UBXD8, VIMP, and Herp (Mueller et al. 2006; Christianson et al. 2008; Hosokawa et al. 2008; Mueller et al. 2008; Iida et al. 2011; Klemm et al. 2011; Christianson et al. 2012), which in turn recruit the VCP/p97 complexes necessary to drive substrate dislocation. It is thus not surprising that reducing SEL1L expression impairs degradation of both lumenal and integral membrane substrates, independent of their glycosylation state (Mueller et al. 2006; Cattaneo et al. 2008; Christianson et al. 2008; Bernasconi et al. 2010; Christianson et al. 2012). Like its yeast ortholog Hrd3p, SEL1L is necessary to transfer substrates from ER lectins to Hrd1 (Gardner et al. 2000; Carvalho et al. 2006; Denic et al. 2006; Gauss et al. 2006; Christianson et al. 2008). Recently, Bernasconi and colleagues also reported that SEL1L regulates EDEM1 and OS-9 availability through LC3-I-dependent sequestration into ER-derived vesicles (EDEMosomes), in a process proposed to function as an ERAD tuning mechanism (Bernasconi et al. 2012). SEL1L thus serves as an important nexus that coordinates ERAD substrate recruitment, dislocation, and ubiquitination.
Erlins May Be Intramembrane Substrate Adaptors
Erlin1/2 belong to the SPFH (stomatin, prohibitin, flotilin, and HflC/HflK) domain-containing protein family and form a ∼2 MDa heterotetrameric complex in the ER membrane. Erlin1/2 complexes rapidly associate with inositol 1,4,5-triphosphate receptors (IP3R), following the latter’s activation, and link them to the ER-resident E3 ligase RNF170 (Lu et al. 2011). This process results in polyubiquitination and ultimately degradation of IP3Rs in response to calcium-dependent signals that promote receptor down-regulation (Pearce et al. 2007, 2009). Intriguingly, Erlin1/2 were also found in complexes with other E3 ligases, including Hrd1, gp78, and Trc8 (Jo et al. 2011b; Christianson et al. 2012), suggesting that its adaptor role may not be limited to just RNF170 and IP3Rs. TMUB1 (transmembrane and Ub-like domain-containing-1) was identified as an Erlin2 interactor that enhances binding to gp78 (Jo et al. 2011b). Loss of either Erlin2 or TMUB1 impairs sterol-induced ubiquitination and degradation of 3-hydroxy-3-methyl-glutaryl-CoA reductase (HMG-CoAR) (Jo et al. 2011b). Collectively, these findings point toward a general adaptor role for Erlin1/2, bridging E3 ligases with polytopic membrane-integrated substrates through associations with accessory factors.
Insigs Are Client-Specific Adaptors
HMG-CoAR is targeted for ERAD in a sterol-dependent manner through its interaction with Insig-1. The HMG-CoAR:Insig-1 complex forms in a sterol-dependent manner, which subsequently brings the reductase:Insig-1 complex to gp78 for ubiquitination (Song et al. 2005), dislocation to the cytosol (Hartman et al. 2010) by a VCP/p97-Ufd1-Npl4 complex (Cao et al. 2007), and subsequent degradation by the proteasome. Although a yeast ortholog, Hmg2, undergoes similar sterol-regulated degradation (Hampton and Rine 1994; Hampton et al. 1996), its degradation is mediated by residues within the membrane-embedded domains of Hrd1p that interrogate a state which, in the presence of bound sterols, has been proposed to resemble a loosely folded or misfolded conformation (Bays et al. 2001; Sato et al. 2009) and not via the Insig ortholog Nsg1p (Flury et al. 2005). Although HMG-CoAR also interacts with Hrd1 (Christianson et al. 2012), metazoans appear to have evolved a specialized strategy to tightly regulate sterol biosynthetic enzymes.
F-Box Proteins Capture Dislocated Glycoproteins in the Cytoplasm
The F-box proteins Fbx2/Fbs1 and Fbx6/Fbs2 recognize the chitobiose core of high-mannose N-linked oligosaccharides and act as substrate adaptors for an SCF (Skp1-Cullin-Fbox-Roc1) E3 ligase complex recruited to the ER for ERAD (Yoshida et al. 2002, 2003). Fbx2 and Fbx6 bind only cytoplasmically exposed glycoproteins, as would arise following the dislocation step of ERAD. N-linked glycans are normally removed on cytoplasmic exposure by NGly1/PNGase (Hirsch et al. 2003; Blom et al. 2004), an amidase found in complexes containing components associated with dislocation (Katiyar et al. 2005; Li et al. 2006). Although unlikely to represent a primary ERAD recognition mechanism, Fbx2 and Fbx6 may instead act as backup for cytoplasmically exposed glycoproteins that have evaded NGly1.
Viral-Encoded Adaptors
A number of viral proteins have also evolved to serve as adaptors that redirect correctly folded molecules to ERAD. The human immunodeficiency virus (HIV-1) encodes Vpu, a glycoprotein that binds to and targets newly synthesized CD4 for degradation by recruiting the cytosolic β-TrCP1/2, F-box adaptor proteins of the SCF(β-TrCP) ubiquitin ligase complex (Margottin et al. 1998). The human cytomegalovirus (HCMV) gene products US2 and US11 are classed as immunoevasins because they induce degradation of major histocompatibility complex (MHC) class I heavy chain (HC), enabling virus-infected cells to avoid detection by the immune system (reviewed in Loureiro and Ploegh 2006). MHC class I HC is normally delivered to the plasma membrane, but is also an Hrd1-dependent ERAD substrate if it fails to assemble with β2-microglobulin (Burr et al. 2011). However, the presence of HCMV-encoded US2 causes MHC class I HC to be rerouted and ubiquitinated by a complex containing the ER-resident E3 Trc8, US2, and signal peptide peptidase (Loureiro et al. 2006; Stagg et al. 2009). In contrast, US11 induces MHC class I HC dislocation/degradation in a manner dependent on SEL1L (Mueller et al. 2006), Derlin-1 (Lilley and Ploegh 2004), and other Hrd1 cluster components (e.g., UBE2J1, AUP1, and UBXD8) (Mueller et al. 2008), but depletion of either Hrd1 or gp78, or both, fails to prevent MHC class I HC degradation (Burr et al. 2011). This observation raises questions about the exclusivity of SEL1L as a Hrd1 adaptor and suggests it may be coerced (by US11) into forming alternative E3 complexes.
Considering the strategies viruses have developed, an intriguing possibility lies in the engineering of target-specific ERAD adaptors for therapy. A recent study constructed chimeras of a Hrd1-interacting fragment of SEL1L and target-specific single-chain antibodies/ligands to create “degradins,” which can redirect proteins normally trafficked to the plasma membrane to ERAD (Vecchi et al. 2012). Selective rerouting to ERAD may offer a potential therapeutic strategy to selectively modulate expression of surface and secretory proteins whose elevated abundance correlates with a disease phenotype (e.g., growth factors and their receptors).
The Enigmatic Dislocon
Several candidates have been suggested to function as dislocons based on their requirement for substrate dislocation, polytopic nature, and direct interaction with ERAD substrates, including the Derlin family of proteins (Derlin 1-3) (Lilley and Ploegh 2004; Ye et al. 2004), signal peptide peptidase (Loureiro et al. 2006), Sec61 (Plemper et al. 1997; Scott and Schekman 2008), and the E3 ligase Hrd1 (Carvalho et al. 2010). Ultimately, definitive identification of the proteins that comprise the minimal dislocon will require complete reconstitution of substrate dislocation in synthetic lipid bilayers with isolated and purified ERAD components.
Is Substrate Unfolding Necessary for Dislocation?
Characteristics of the substrates transiting the ERAD system reveal unique features of the dislocation process that distinguish it from other mechanisms of protein translocation. For example, protein import into the ER (via the Sec61 translocon) and mitochondria (via TOM40, TIM22, and TIM23) are initiated by recognition of a signal peptide and involve the transmembrane translocation of a substrate in an unfolded state through a narrow proteinaceous pore (Rapoport 2007). In contrast, ERAD integrates diverse substrate features (e.g., exposed hydrophobicity and glycan status) to distinguish its substrates from folding intermediates in the ER and mediates dislocation of proteins including those with tightly folded (Fiebiger et al. 2002) and ligand-stabilized (Tirosh et al. 2003) domains, with covalently attached glycans (Blom et al. 2004), and even intact virus particles (Lilley et al. 2006; Geiger et al. 2011), the sizes of which preclude passage through the narrow pore of translocons like Sec61.
A Role for Lipid Droplets?
The incompatibility of a traditional protein-based channel with folded ERAD substrates and virus particles led Ploegh to propose an alternative model in which substrate egress is coupled to the formation of lipid droplets (LDs) (Ploegh 2007). LDs are cytoplasmic organelles consisting of a neutral lipid core bounded by phospholipid monolayers that bud from the ER through an as-yet-undefined mechanism (reviewed in Walther and Farese 2012). Established ERAD components, including UBXD8, VCP/p97, AUP1, and UBE2G2, localize to LDs and ERAD substrates HMG-CoA reductase (Hartman et al. 2010) and ApoB (Ohsaki et al. 2006) are present in buoyant LD-enriched fractions under conditions of proteasome impairment. These findings suggest a potential functional relationship between ERAD and LDs. In addition, MHC class I HC, α1-antitrypsin null Hong Kong variant, truncated ribophorin I (Klemm et al. 2011), and HMG-CoAR (Hartman et al. 2010) are stabilized by triacsin c, an inhibitor of acyl-CoA synthetases that prevents triglyceride synthesis and hence LD formation. These findings appear to support the hypothesis that LDs could either be involved in substrate dislocation or could represent an intermediate step for substrates en route to the proteasome (Ploegh 2007). Examination of ERAD function in yeast lacking the enzymes necessary for neutral lipid synthesis and therefore unable to make LDs, however, showed that LD formation is dispensable for degradation of glycosylated soluble lumenal and polytopic membrane ERAD substrates, and also for the dislocation of the plant toxin ricin (Olzmann and Kopito 2011). These data eliminate a role for LD formation in ERAD, but do not exclude the possibility that degradation of specific substrates may be sensitive to lipid rearrangements, be affected by the levels of neutral lipids, or require specialized ER subdomains for efficient degradation.
Reduction of Disulfide Bonds
Despite suggestions that dislocons can accommodate large, folded domains, there is evidence implicating a need to reduce disulfide bonds as a prerequisite for ERAD. ERFAD (ER flavoprotein associated with degradation) and the protein disulfide isomerase (PDI)-family member ERp90/TXD16 (Riemer et al. 2011; Christianson et al. 2012) bind the Hrd1 adaptor SEL1L (Riemer et al. 2009). Although ERFAD shows bona fide oxidoreductase activity (Riemer et al. 2009), ERp90 lacks the typical CXXC active-site motif found in redox-active domains. Association with both OS-9 and SEL1L suggests that an ERFAD-ERp90 complex may serve to reduce disulfide bonds for SEL1L-associated glycoprotein substrates (Riemer et al. 2011). A similar activity was proposed for ERdj5, an EDEM1 interactor also implicated in ERAD of disulfide-containing substrates (Dong et al. 2008; Ushioda et al. 2008). ERdj5 binds BiP through its J domain, whereas its four thioredoxin domains endow it with reductase activity (Ushioda et al. 2008). Like OS-9 and XTP3-B, binding of ERdj5 to BiP suggests it may function at a nexus of protein folding and degradation, perhaps for substrates not recognized through mannose-trimmed glycans.
What Drives Dislocation?
VCP/p97 is a homohexameric enzyme that couples ATP hydrolysis to unfolding and structural reorganization of its client proteins, and plays a critical role in the dislocation of nearly all ERAD substrates. This ATPase is proposed to physically pull the proteins out of the ER, and its essential role in dislocation is supported by the stabilization of nearly all ERAD substrates by RNAi-mediated knockdown, expression of dominant–negative subunits, or chemical inhibition of VCP/p97 (Ye et al. 2001; Rabinovich et al. 2002; Delabarre et al. 2006; Wang et al. 2008b). However, because VCP/p97 is a cytoplasmic protein, it only has the opportunity to bind to a lumenal ERAD substrate after at least part of it has already passed through the membrane. The origin of the energy required to initiate substrate dislocation remains obscure, although it is possible that action of VCP/p97 could be transmitted through the membrane indirectly by inducing conformational changes in membrane-spanning components of the dislocation complex.
A Role for Rhomboid Pseudoproteases in Dislocation
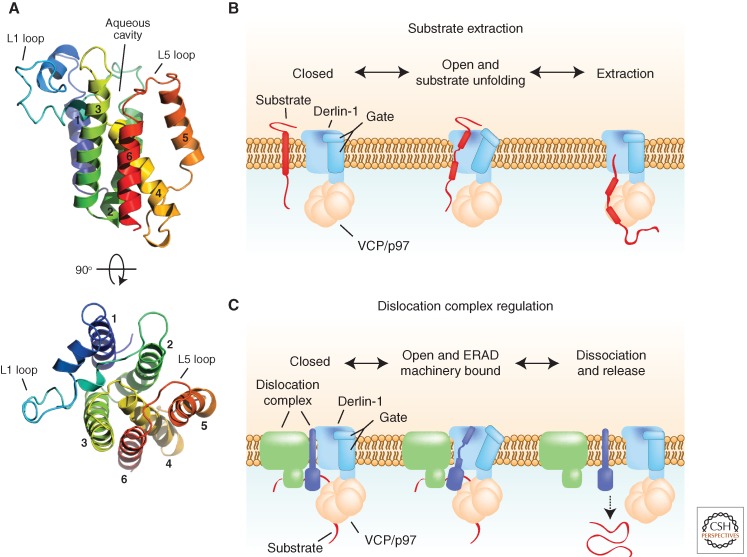
Rhomboids are polytopic intramembrane proteases that use a catalytic Ser/His dyad buried in a hydrophilic cavity within the plane of membrane to cleave substrates within or near their transmembrane domains (Freeman 2008; Urban 2010). The cellular roles of rhomboid pseudoproteases, which are rhomboid family members that lack the catalytic residues necessary for proteolysis, have long been a mystery. Recently, the ER-localized rhomboid pseudoproteases iRhom1 and iRhom2 were found to negatively regulate EGF signaling by targeting a membrane-integrated EGF ligand for degradation via a Hrd1-dependent ERAD pathway (Zettl et al. 2011). A direct role for rhomboid pseudoproteases in ERAD was suggested by the discovery that Derlins and UBAC2, a recently identified UBA-domain-containing ERAD component that interacts with gp78 and UBXD8 (Christianson et al. 2012), are members of the rhomboid pseudoprotease family (Greenblatt et al. 2011). A threading model of Derlin-1, based on the crystal structure of the Escherichia coli rhomboid GlpG, predicts it to form a compact helical bundle of six transmembrane domains with a unique lateral protrusion (L1 loop) at the lumenal membrane interface, and this topology has been experimentally verified (Fig. 3A) (Greenblatt et al. 2011). Although Derlin-1 lacks the catalytic Ser/His residues to make it an active protease, it retains overall rhomboid structure and conserved architectural elements, and mutational analysis of Derlin-1 suggests that the rhomboid domain may play a direct role in substrate extraction from the cytosolic face of the ER membrane (Greenblatt et al. 2011).
Figure 3.
Models of rhomboid pseudoprotease function in ERAD. (A) Structural model of the rhomboid domain of Derlin-1 indicating key conserved features that identify it as a member of the rhomboid family. The L5 loop and TM5 have been proposed to function as a gate that controls the entry of membrane-spanning substrate domains. (B) Rearrangement of TM5 and the L5 loop could function as a gate to allow Derlin-1 to bind and destabilize substrate transmembrane domains, thereby facilitating substrate extraction by VCP/p97. Thinning of the bilayer imposed by the rhomboid structure could enhance this destabilization. (C) Alternatively, Derlin-1 could regulate ERAD machinery by catalyzing the dissociation or cycling of dislocation complexes, which may be coupled to the release of a substrate from the cytosolic face of the ER membrane.
Rhomboid proteases are thought to mediate partial unfolding of their substrates via a lateral gating mechanism mediated by movement of transmembrane domain 5 and the L5 loop to facilitate access of the substrate scissile peptide bond for cleavage by the catalytic site in the central aqueous cavity (Freeman 2008; Urban 2010). The rhomboid structure has been proposed to cause bilayer thinning, which also could contribute to destabilization of transmembrane domains by exposure of normally buried residues to a more aqueous environment (Bondar et al. 2009). By analogy, binding of transmembrane domains within ERAD substrates by Derlin-1 could destabilize the substrate and lower the energy barrier for extraction by VCP/p97 (Fig. 3B), which is directly coupled to Derlin-1 and Derlin-2 through a carboxy-terminal SHP box (Greenblatt et al. 2011) and indirectly coupled to UBAC2 through its interaction with UBXD8 (Christianson et al. 2012). The interaction of the rhomboid domain of Derlin-1 with substrate transmembrane regions could explain its role in the ERAD of the polytopic substrate CFTRΔF508 (Sun et al. 2006; Younger et al. 2006; Wang et al. 2008a).
Alternatively, Derlin-1 could regulate the dynamic state of the ERAD machinery, perhaps facilitating the dissociation of stable dislocation complexes and recycling of components (Fig. 3C). In support of this model, Wahlman et al. observed, using an in vitro reconstituted ERAD assay, that Derlin antibodies blocked ERAD of lumenal substrate only after a short lag period (Wahlman et al. 2007), suggesting Derlins contribute to cycling the dislocation complex for multiple rounds of dislocation. Whether Derlins retain a rhomboid-like gating mechanism, and what the role of the internal substrate-binding cavity is in ERAD, remain to be determined.
STEPS 3–4: UBIQUITINATION AND DEGRADATION
Ubiquitin E3 Ligases (E3s) Implicated in ERAD
Mammalian ERAD has been defined primarily by the activities of two polytopic RING domain ubiquitin ligases, Hrd1/SYVN1 (Nadav et al. 2003; Kikkert et al. 2004) and gp78/AMFR (Fang et al. 2001), whereas other implicated E3s contribute to the destruction of a more limited range of substrates including TEB4/MARCH6 (Hassink et al. 2005), RNF5/Rma-1 (Younger et al. 2006; Morito et al. 2008), TRC8 (Stagg et al. 2009), RFP2/TRIM13 (Lerner et al. 2007; Altier et al. 2011), Kf-1/RNF103 (Maruyama et al. 2008), Nixin (Neutzner et al. 2011), and RNF170 (Lu et al. 2011). Cytoplasmic E3s such as Parkin (Imai et al. 2002), CHIP (Meacham et al. 2001), SCF complexes with the F-box proteins Fbx2, Fbx6 (Yoshida et al. 2002, 2003) and β-TrCP1/2 (Magadan et al. 2010), Smurf1 (Guo et al. 2011), and Nrdp1 (Fry et al. 2011) have also been implicated in ERAD. Additionally, another 15 E3s localized to ER membranes are awaiting confirmation of a role in ERAD (Neutzner et al. 2011). Models for how these ligases might interact to coordinate ERAD substrate dislocation and ubiquitination are discussed in the following section.
Cooperativity among E3 Ubiquitin Ligases
Mounting evidence suggests that multiple ligases are necessary for complete ERAD substrate processing, with possible scenarios that include E3s working (1) in parallel with multiple E3s with equivalent specificity toward a substrate; (2) simultaneously by conjugating ubiquitin to different sites of a single substrate; (3) cooperatively by an initial monoubiquitin conjugation and extension by E4 activity; or (4) via sequential rounds of ubiquitin conjugation and removal. For example, the tandem activities of gp78 and Trc8 cooperate to degrade HMG-CoAR through Insig-1/2 (Jo et al. 2011a). Rma-1/RNF5 may work sequentially with CHIP on different regions of misfolded CFTRΔF508 (Younger et al. 2006) or perhaps as a primer for gp78-mediated chain elongation (Morito et al. 2008). Intriguing results from Ploegh and colleagues suggest that deubiquitination must occur in order for US2-induced dislocation of MHC class I to proceed (Ernst et al. 2009). This observation led to the proposal that two discrete rounds of ubiquitination occur during MHC class I processing: one necessary for dislocation induced by US2 and subsequent deubiquitination by VCP/p97-associated Yod1 (Ernst et al. 2009), and a second interaction of substrate with VCP/p97 to direct substrates to 26S proteasomes (via hHR23B binding). Similar observations were reported for the lumenal substrate RI332 (Sanyal et al. 2012), suggesting that two distinct ubiquitin conjugations steps are required for complete substrate processing.
A single complex containing multiple different E3s has not yet been reported for ERAD; however, yeast Hrd1p is able to form Usa1p-dependent oligomers (Horn et al. 2009; Carvalho et al. 2010), raising the question of whether rounds of ubiquitin conjugation that generate polyubiquitin chains are mediated by a single RING domain or result from coordinated activity of multiple E3s. Along with substrates, there is evidence that ERAD E3s ubiquitinate each other, possibly constituting a negative feedback loop (Morito et al. 2008; Shmueli et al. 2009; Ballar et al. 2010; Jo et al. 2011a). Regulated ubiquitination of E3s may represent a form of ERAD tuning, in which monoubiquitin is able to modulate activity of an existing E3 pool and polyubiquitin is able to modulate the size of the available E3 pool through degradation.
Multiple Adaptors Recruit VCP/p97 to Components of the ERAD Machinery
VCP/p97 is coupled to a myriad of cellular processes through its association with specific recruitment factors and is tightly coupled to proteasome-mediated substrate degradation (Meyer et al. 2012). In mammalian cells at least seven different membrane-embedded mammalian ERAD components have VCP/p97-binding motifs. These include UBX domains (UBXD2 [Liang et al. 2006] and UBXD8 [Suzuki et al. 2012]), VIM motifs (gp78 [Ballar et al. 2006] and VIMP [Ye et al. 2004]), SHP boxes (Derlin-1 and Derlin-2 [Greenblatt et al. 2011]), and undefined cytosolic regions of Hrd1 and VIMP (Ye et al. 2005). Why might multiple VCP/p97-binding sites be required for a single dislocation complex? One possibility is that multiple binding events could serve to regulate or orient VCP/p97 at the dislocon for direct substrate dislocation or, alternatively, VCP/p97 recruitment could play a role in the regulation of dislocon dynamics and organization.
In addition to its role in substrate extraction, VCP/p97 functions as a scaffold that links dislocated substrates to cytoplasmic cofactors involved in substrate modification and processing. The cytoplasmic deglycosylating enzyme NGly1 is recruited to dislocation complexes by direct binding to VCP/p97 through its PUB domain and functions to facilitate proteasomal clearance of substrates by removing N-linked glycans from dislocating substrates (Kim et al. 2006; Li et al. 2006). VCP/p97 also associates with a host of Ub-binding proteins (Ufd1 and Npl4), deubiquitinating enzymes (Yod1, VCIP135, Usp19, and Ataxin-3), and an E4 ubiquitin extension enzyme (Ube4a) that have been implicated in ERAD. Recent data indicate that impairment of VCP/p97-associated deubiquitination by treatment with Eeyarestatin or expression of dominant–negative Yod1 abrogates dislocation, whereas expression of a VCP/p97-binding deubiquitinating enzyme restores it, although it blocks proteasomal clearance (Ernst et al. 2009). These data suggest that multiple rounds of ubiquitination, and a round of deubiquitination, are required for efficient ERAD. One model posits that the initial ubiquitin chain promotes VCP/p97 binding, but must be removed to allow the substrate to pass through the narrow VCP/p97 pore. However, the crystal structure of VCP/p97 solved for all four ATP binding/hydrolysis states indicates a narrow ∼7 Å pore in the D1 domain occluded by a Zn2+ ion that coordinates ATP cycles around the hexamer (Delabarre et al. 2006). Although these structures argue against passage of the substrate through the entire pore, it should be noted that they were solved in the absence of VCP/p97 cofactors, which may allosterically influence the conformation (and function) of VCP/p97.
CONCLUDING REMARKS
Although considerable progress has been made toward identifying the core components of the mammalian ERAD machinery, many questions remain unanswered. Key outstanding challenges include the molecular mechanism of dislocation, the source of energy for this process, and the manner in which these steps are coupled to ensure that hydrophobic substrates of this process remain soluble as they move from a lipidic environment to the cytosol. Furthermore, large gaps remain in our understanding of how different types of substrates are recognized and delivered from their diverse environments to meet a common fate at the proteasome. Solving these complex problems will require more extensive systems-level analysis of protein networks, and ultimately biochemical reconstitution of key steps in cell-free systems.
ACKNOWLEDGMENTS
We thank the members of the Kopito and Christianson labs for helpful discussions. R.R.K. and J.A.O. are supported by grants from the National Institutes of Health. J.C.C. is supported by funding from the Ludwig Institute for Cancer Research.
Footnotes
Editors: Susan Ferro-Novick, Tom A. Rapoport, and Randy Schekman
Additional Perspectives on The Endoplasmic Reticulum available at www.cshperspectives.org
REFERENCES
- Aebi M, Bernasconi R, Clerc S, Molinari M 2010. N-glycan structures: Recognition and processing in the ER. Trends Biochem Sci 35: 74–82 [DOI] [PubMed] [Google Scholar]
- Altier C, Garcia-Caballero A, Simms B, You H, Chen L, Walcher J, Tedford HW, Hermosilla T, Zamponi GW 2011. The Cavβ subunit prevents RFP2-mediated ubiquitination and proteasomal degradation of L-type channels. Nat Neurosci 14: 173–180 [DOI] [PubMed] [Google Scholar]
- Ballar P, Shen Y, Yang H, Fang S 2006. The role of a novel p97/valosin-containing protein-interacting motif of gp78 in endoplasmic reticulum-associated degradation. J Biol Chem 281: 35359–35368 [DOI] [PubMed] [Google Scholar]
- Ballar P, Ors AU, Yang H, Fang S 2010. Differential regulation of CFTRΔF508 degradation by ubiquitin ligases gp78 and Hrd1. Int J Biochem Cell Biol 42: 167–173 [DOI] [PubMed] [Google Scholar]
- Bays NW, Gardner RG, Seelig LP, Joazeiro CA, Hampton RY 2001. Hrd1p/Der3p is a membrane-anchored ubiquitin ligase required for ER-associated degradation. Nat Cell Biol 3: 24–29 [DOI] [PubMed] [Google Scholar]
- Bernasconi R, Pertel T, Luban J, Molinari M 2008. A dual task for the Xbp1-responsive OS-9 variants in the mammalian endoplasmic reticulum: Inhibiting secretion of misfolded protein conformers and enhancing their disposal. J Biol Chem 283: 16446–16454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernasconi R, Galli C, Calanca V, Nakajima T, Molinari M 2010. Stringent requirement for HRD1, SEL1L, and OS-9/XTP3-B for disposal of ERAD-LS substrates. J Cell Biol 188: 223–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernasconi R, Galli C, Noack J, Bianchi S, de Haan CA, Reggiori F, Molinari M 2012. Role of the SEL1L:LC3-I complex as an ERAD tuning receptor in the mammalian ER. Mol Cell 46: 1–11 [DOI] [PubMed] [Google Scholar]
- Bhamidipati A, Denic V, Quan EM, Weissman JS 2005. Exploration of the topological requirements of ERAD identifies Yos9p as a lectin sensor of misfolded glycoproteins in the ER lumen. Mol Cell 19: 741–751 [DOI] [PubMed] [Google Scholar]
- Blom D, Hirsch C, Stern P, Tortorella D, Ploegh HL 2004. A glycosylated type I membrane protein becomes cytosolic when peptide: N-glycanase is compromised. EMBO J 23: 650–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bondar AN, del Val C, White SH 2009. Rhomboid protease dynamics and lipid interactions. Structure 17: 395–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burr ML, Cano F, Svobodova S, Boyle LH, Boname JM, Lehner PJ 2011. HRD1 and UBE2J1 target misfolded MHC class I heavy chains for endoplasmic reticulum-associated degradation. Proc Natl Acad Sci 108: 2034–2039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J, Wang J, Qi W, Miao HH, Ge L, DeBose-Boyd RA, Tang JJ, Li BL, Song BL 2007. Ufd1 is a cofactor of gp78 and plays a key role in cholesterol metabolism by regulating the stability of HMG-CoA reductase. Cell Metab 6: 115–128 [DOI] [PubMed] [Google Scholar]
- Carvalho P, Goder V, Rapoport TA 2006. Distinct ubiquitin-ligase complexes define convergent pathways for the degradation of ER proteins. Cell 126: 361–373 [DOI] [PubMed] [Google Scholar]
- Carvalho P, Stanley AM, Rapoport TA 2010. Retrotranslocation of a misfolded luminal ER protein by the ubiquitin-ligase Hrd1p. Cell 143: 579–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cattaneo M, Otsu M, Fagioli C, Martino S, Lotti LV, Sitia R, Biunno I 2008. SEL1L and HRD1 are involved in the degradation of unassembled secretory Ig-mu chains. J Cell Physiol 215: 794–802 [DOI] [PubMed] [Google Scholar]
- Christianson JC, Shaler TA, Tyler RE, Kopito RR 2008. OS-9 and GRP94 deliver mutant α1-antitrypsin to the Hrd1-SEL1L ubiquitin ligase complex for ERAD. Nat Cell Biol 10: 272–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christianson JC, Olzmann JA, Shaler TA, Sowa ME, Bennett EJ, Richter CM, Tyler RE, Greenblatt EJ, Harper JW, Kopito RR 2012. Defining human ERAD networks through an integrative mapping strategy. Nat Cell Biol 14: 93–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cormier JH, Tamura T, Sunryd JC, Hebert DN 2009. EDEM1 recognition and delivery of misfolded proteins to the SEL1L-containing ERAD complex. Mol Cell 34: 627–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delabarre B, Christianson J, Kopito R, Brunger A 2006. Central pore residues mediate the p97/VCP activity required for ERAD. Mol Cell 22: 451–462 [DOI] [PubMed] [Google Scholar]
- Denic V, Quan EM, Weissman JS 2006. A luminal surveillance complex that selects misfolded glycoproteins for ER-associated degradation. Cell 126: 349–359 [DOI] [PubMed] [Google Scholar]
- Dong M, Bridges JP, Apsley K, Xu Y, Weaver TE 2008. ERdj4 and ERdj5 are required for endoplasmic reticulum-associated protein degradation of misfolded surfactant protein C. Mol Biol Cell 19: 2620–2630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eletto D, Dersh D, Argon Y 2010. GRP94 in ER quality control and stress responses. Semin Cell Dev Biol 21: 479–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst R, Mueller B, Ploegh HL, Schlieker C 2009. The otubain YOD1 is a deubiquitinating enzyme that associates with p97 to facilitate protein dislocation from the ER. Mol Cell 36: 28–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang S, Ferrone M, Yang C, Jensen JP, Tiwari S, Weissman AM 2001. The tumor autocrine motility factor receptor, gp78, is a ubiquitin protein ligase implicated in degradation from the endoplasmic reticulum. Proc Natl Acad Sci 98: 14422–14427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiebiger E, Story C, Ploegh HL, Tortorella D 2002. Visualization of the ER-to-cytosol dislocation reaction of a type I membrane protein. EMBO J 21: 1041–1053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flury I, Garza R, Shearer A, Rosen J, Cronin S, Hampton RY 2005. INSIG: A broadly conserved transmembrane chaperone for sterol-sensing domain proteins. EMBO J 24: 3917–3926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman M 2008. Rhomboid proteases and their biological functions. Annu Rev Genet 42: 191–210 [DOI] [PubMed] [Google Scholar]
- Fry WH, Simion C, Sweeney C, Carraway KL 3rd 2011. Quantity control of the ErbB3 receptor tyrosine kinase at the endoplasmic reticulum. Mol Cell Biol 31: 3009–3018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner RG, Swarbrick GM, Bays NW, Cronin SR, Wilhovsky S, Seelig L, Kim C, Hampton RY 2000. Endoplasmic reticulum degradation requires lumen to cytosol signaling. Transmembrane control of Hrd1p by Hrd3p. J Cell Biol 151: 69–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauss R, Jarosch E, Sommer T, Hirsch C 2006. A complex of Yos9p and the HRD ligase integrates endoplasmic reticulum quality control into the degradation machinery. Nat Cell Biol 8: 849–854 [DOI] [PubMed] [Google Scholar]
- Geiger R, Andritschke D, Friebe S, Herzog F, Luisoni S, Heger T, Helenius A 2011. BAP31 and BiP are essential for dislocation of SV40 from the endoplasmic reticulum to the cytosol. Nat Cell Biol 13: 1305–1314 [DOI] [PubMed] [Google Scholar]
- Gonzalez DS, Karaveg K, Vandersall-Nairn AS, Lal A, Moremen KW 1999. Identification, expression, and characterization of a cDNA encoding human endoplasmic reticulum mannosidase I, the enzyme that catalyzes the first mannose trimming step in mammalian Asn-linked oligosaccharide biosynthesis. J Biol Chem 274: 21375–21386 [DOI] [PubMed] [Google Scholar]
- Greenblatt EJ, Olzmann JA, Kopito RR 2011. Derlin-1 is a rhomboid pseudoprotease required for the dislocation of mutant α-1 antitrypsin from the endoplasmic reticulum. Nat Struct Mol Biol 18: 1147–1152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerriero CJ, Brodsky JL 2012. The delicate balance between secreted protein folding and endoplasmic reticulum-associated degradation in human physiology. Physiol Rev 92: 537–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X, Shen S, Song S, He S, Cui Y, Xing G, Wang J, Yin Y, Fan L, He F, et al. 2011. The E3 ligase Smurf1 regulates Wolfram syndrome protein stability at the endoplasmic reticulum. J Biol Chem 286: 18037–18047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampton RY, Rine J 1994. Regulated degradation of HMG-CoA reductase, an integral membrane protein of the endoplasmic reticulum, in yeast. J Cell Biol 125: 299–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampton RY, Gardner RG, Rine J 1996. Role of 26S proteasome and HRD genes in the degradation of 3-hydroxy-3-methylglutaryl-CoA reductase, an integral endoplasmic reticulum membrane protein. Mol Biol Cell 7: 2029–2044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman IZ, Liu P, Zehmer JK, Luby-Phelps K, Jo Y, Anderson RG, DeBose-Boyd RA 2010. Sterol-induced dislocation of 3-hydroxy-3-methylglutaryl coenzyme A reductase from endoplasmic reticulum membranes into the cytosol through a subcellular compartment resembling lipid droplets. J Biol Chem 285: 19288–19298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassink G, Kikkert M, van Voorden S, Lee SJ, Spaapen R, van Laar T, Coleman CS, Bartee E, Fruh K, Chau V, et al. 2005. TEB4 is a C4HC3 RING finger-containing ubiquitin ligase of the endoplasmic reticulum. Biochem J 388: 647–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert DN, Bernasconi R, Molinari M 2010. ERAD substrates: Which way out? Semin Cell Dev Biol 21: 526–532 [DOI] [PubMed] [Google Scholar]
- Helenius A, Aebi M 2004. Roles of N-linked glycans in the endoplasmic reticulum. Annu Rev Biochem 73: 1019–1049 [DOI] [PubMed] [Google Scholar]
- Hirao K, Natsuka Y, Tamura T, Wada I, Morito D, Natsuka S, Romero P, Sleno B, Tremblay LO, Herscovics A, et al. 2006. EDEM3, a soluble EDEM homolog, enhances glycoprotein endoplasmic reticulum-associated degradation and mannose trimming. J Biol Chem 281: 9650–9658 [DOI] [PubMed] [Google Scholar]
- Hirsch C, Blom D, Ploegh HL 2003. A role for N-glycanase in the cytosolic turnover of glycoproteins. EMBO J 22: 1036–1046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn SC, Hanna J, Hirsch C, Volkwein C, Schutz A, Heinemann U, Sommer T, Jarosch E 2009. Usa1 functions as a scaffold of the HRD-ubiquitin ligase. Mol Cell 36: 782–793 [DOI] [PubMed] [Google Scholar]
- Hosokawa N, You Z, Tremblay LO, Nagata K, Herscovics A 2007. Stimulation of ERAD of misfolded null Hong Kong α1-antitrypsin by Golgi α1,2-mannosidases. Biochem Biophys Res Commun 362: 626–632 [DOI] [PubMed] [Google Scholar]
- Hosokawa N, Wada I, Nagasawa K, Moriyama T, Okawa K, Nagata K 2008. Human XTP3-B forms an endoplasmic reticulum quality control scaffold with the HRD1-SEL1L ubiquitin ligase complex and BiP. J Biol Chem 283: 20914–20924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosokawa N, Kamiya Y, Kamiya D, Kato K, Nagata K 2009. Human OS-9, a lectin required for glycoprotein endoplasmic reticulum-associated degradation, recognizes mannose-trimmed N-glycans. J Biol Chem 284: 17061–17068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosokawa N, Tremblay LO, Sleno B, Kamiya Y, Wada I, Nagata K, Kato K, Herscovics A 2010. EDEM1 accelerates the trimming of α1,2-linked mannose on the C branch of N-glycans. Glycobiology 20: 567–575 [DOI] [PubMed] [Google Scholar]
- Iida Y, Fujimori T, Okawa K, Nagata K, Wada I, Hosokawa N 2011. SEL1L protein critically determines the stability of the HRD1-SEL1L endoplasmic reticulum-associated degradation (ERAD) complex to optimize the degradation kinetics of ERAD substrates. J Biol Chem 286: 16929–16939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai Y, Soda M, Hatakeyama S, Akagi T, Hashikawa T, Nakayama KI, Takahashi R 2002. CHIP is associated with Parkin, a gene responsible for familial Parkinson’s disease, and enhances its ubiquitin ligase activity. Mol Cell 10: 55–67 [DOI] [PubMed] [Google Scholar]
- Jo Y, Lee PC, Sguigna PV, DeBose-Boyd RA 2011a. Sterol-induced degradation of HMG CoA reductase depends on interplay of two Insigs and two ubiquitin ligases, gp78 and Trc8. Proc Natl Acad Sci 108: 20503–20508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo Y, Sguigna PV, DeBose-Boyd RA 2011b. Membrane-associated ubiquitin ligase complex containing gp78 mediates sterol-accelerated degradation of 3-hydroxy-3-methylglutaryl-coenzyme A reductase. J Biol Chem 286: 15022–15031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katiyar S, Joshi S, Lennarz WJ 2005. The retrotranslocation protein Derlin-1 binds peptide:N-glycanase to the endoplasmic reticulum. Mol Biol Cell 16: 4584–4594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikkert M, Doolman R, Dai M, Avner R, Hassink G, van Voorden S, Thanedar S, Roitelman J, Chau V, Wiertz E 2004. Human HRD1 is an E3 ubiquitin ligase involved in degradation of proteins from the endoplasmic reticulum. J Biol Chem 279: 3525–3534 [DOI] [PubMed] [Google Scholar]
- Kim I, Ahn J, Liu C, Tanabe K, Apodaca J, Suzuki T, Rao H 2006. The Png1-Rad23 complex regulates glycoprotein turnover. J Cell Biol 172: 211–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klemm EJ, Spooner E, Ploegh HL 2011. Dual role of ancient ubiquitous protein 1 (AUP1) in lipid droplet accumulation and endoplasmic reticulum (ER) protein quality control. J Biol Chem 286: 37602–37614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lederkremer GZ 2009. Glycoprotein folding, quality control and ER-associated degradation. Curr Opin Struct Biol 19: 515–523 [DOI] [PubMed] [Google Scholar]
- Lerner M, Corcoran M, Cepeda D, Nielsen ML, Zubarev R, Pontén F, Uhlén M, Hober S, Grandér D, Sangfelt O 2007. The RBCC gene RFP2 (Leu5) encodes a novel transmembrane E3 ubiquitin ligase involved in ERAD. Mol Biol Cell 18: 1670–1682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Zhao G, Zhou X, Schindelin H, Lennarz WJ 2006. The AAA ATPase p97 links peptide N-glycanase to the endoplasmic reticulum-associated E3 ligase autocrine motility factor receptor. Proc Natl Acad Sci 103: 8348–8353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang J, Yin C, Doong H, Fang S, Peterhoff C, Nixon RA, Monteiro MJ 2006. Characterization of erasin (UBXD2): A new ER protein that promotes ER-associated protein degradation. J Cell Sci 119: 4011–4024 [DOI] [PubMed] [Google Scholar]
- Lilley BN, Ploegh HL 2004. A membrane protein required for dislocation of misfolded proteins from the ER. Nature 429: 834–840 [DOI] [PubMed] [Google Scholar]
- Lilley BN, Gilbert JM, Ploegh HL, Benjamin TL 2006. Murine polyomavirus requires the endoplasmic reticulum protein Derlin-2 to initiate infection. J Virol 80: 8739–8744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loureiro J, Ploegh HL 2006. Antigen presentation and the ubiquitin-proteasome system in host-pathogen interactions. Adv Immunol 92: 225–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loureiro J, Lilley BN, Spooner E, Noriega V, Tortorella D, Ploegh HL 2006. Signal peptide peptidase is required for dislocation from the endoplasmic reticulum. Nature 441: 894–897 [DOI] [PubMed] [Google Scholar]
- Lu JP, Wang Y, Sliter DA, Pearce MM, Wojcikiewicz RJ 2011. RNF170 protein, an endoplasmic reticulum membrane ubiquitin ligase, mediates inositol 1,4,5-trisphosphate receptor ubiquitination and degradation. J Biol Chem 286: 24426–24433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magadan JG, Perez-Victoria FJ, Sougrat R, Ye Y, Strebel K, Bonifacino JS 2010. Multilayered mechanism of CD4 downregulation by HIV-1 Vpu involving distinct ER retention and ERAD targeting steps. PLoS Pathog 6: e1000869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margottin F, Bour SP, Durand H, Selig L, Benichou S, Richard V, Thomas D, Strebel K, Benarous R 1998. A novel human WD protein, h-β TrCp, that interacts with HIV-1 Vpu connects CD4 to the ER degradation pathway through an F-box motif. Mol Cell 1: 565–574 [DOI] [PubMed] [Google Scholar]
- Maruyama Y, Yamada M, Takahashi K 2008. Ubiquitin ligase Kf-1 is involved in the endoplasmic reticulum-associated degradation pathway. Biochem Biophys Res Commun 374: 737–741 [DOI] [PubMed] [Google Scholar]
- Meacham GC, Patterson C, Zhang W, Younger JM, Cyr DM 2001. The Hsc70 co-chaperone CHIP targets immature CFTR for proteasomal degradation. Nat Cell Biol 3: 100–105 [DOI] [PubMed] [Google Scholar]
- Meyer H, Bug M, Bremer S 2012. Emerging functions of the VCP/p97 AAA-ATPase in the ubiquitin system. Nat Cell Biol 14: 117–123 [DOI] [PubMed] [Google Scholar]
- Morito D, Hirao K, Oda Y, Hosokawa N, Tokunaga F, Cyr DM, Tanaka K, Iwai K, Nagata K 2008. Gp78 cooperates with RMA1 in endoplasmic reticulum-associated degradation of CFTRΔF508. Mol Biol Cell 19: 1328–1336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller B, Lilley BN, Ploegh HL 2006. SEL1L, the homologue of yeast Hrd3p, is involved in protein dislocation from the mammalian ER. J Cell Biol 175: 261–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller B, Klemm EJ, Spooner E, Claessen JH, Ploegh HL 2008. SEL1L nucleates a protein complex required for dislocation of misfolded glycoproteins. Proc Natl Acad Sci 105: 12325–12330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadav E, Shmueli A, Barr H, Gonen H, Ciechanover A, Reiss Y 2003. A novel mammalian endoplasmic reticulum ubiquitin ligase homologous to the yeast Hrd1. Biochem Biophys Res Commun 303: 91–97 [DOI] [PubMed] [Google Scholar]
- Neutzner A, Neutzner M, Benischke AS, Ryu SW, Frank S, Youle RJ, Karbowski M 2011. A systematic search for endoplasmic reticulum (ER) membrane-associated RING finger proteins identifies Nixin/ZNRF4 as a regulator of calnexin stability and ER homeostasis. J Biol Chem 286: 8633–8643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohsaki Y, Cheng J, Fujita A, Tokumoto T, Fujimoto T 2006. Cytoplasmic lipid droplets are sites of convergence of proteasomal and autophagic degradation of apolipoprotein B. Mol Biol Cell 17: 2674–2683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuda-Shimizu Y, Hendershot LM 2007. Characterization of an ERAD pathway for nonglycosylated BiP substrates, which require Herp. Mol Cell 28: 544–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivari S, Cali T, Salo KE, Paganetti P, Ruddock LW, Molinari M 2006. EDEM1 regulates ER-associated degradation by accelerating de-mannosylation of folding-defective polypeptides and by inhibiting their covalent aggregation. Biochem Biophys Res Commun 349: 1278–1284 [DOI] [PubMed] [Google Scholar]
- Olzmann JA, Kopito RR 2011. Lipid droplet formation is dispensable for endoplasmic reticulum-associated degradation. J Biol Chem 286: 27872–27874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce MM, Wang Y, Kelley GG, Wojcikiewicz RJ 2007. SPFH2 mediates the endoplasmic reticulum-associated degradation of inositol 1,4,5-trisphosphate receptors and other substrates in mammalian cells. J Biol Chem 282: 20104–20115 [DOI] [PubMed] [Google Scholar]
- Pearce M, Wormer D, Wilkens S, Wojcikiewicz R 2009. An ER membrane complex composed of SPFH1 and SPFH2 mediates the ER-associated degradation of IP3 receptors. J Biol Chem 284: 10433–10445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plemper RK, Bohmler S, Bordallo J, Sommer T, Wolf DH 1997. Mutant analysis links the translocon and BiP to retrograde protein transport for ER degradation. Nature 388: 891–895 [DOI] [PubMed] [Google Scholar]
- Ploegh HL 2007. A lipid-based model for the creation of an escape hatch from the endoplasmic reticulum. Nature 448: 435–438 [DOI] [PubMed] [Google Scholar]
- Quan EM, Kamiya Y, Kamiya D, Denic V, Weibezahn J, Kato K, Weissman JS 2008. Defining the glycan destruction signal for endoplasmic reticulum-associated degradation. Mol Cell 32: 870–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinovich E, Kerem A, Frohlich KU, Diamant N, Bar-Nun S 2002. AAA-ATPase p97/Cdc48p, a cytosolic chaperone required for endoplasmic reticulum-associated protein degradation. Mol Cell Biol 22: 626–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapoport TA 2007. Protein translocation across the eukaryotic endoplasmic reticulum and bacterial plasma membranes. Nature 450: 663–669 [DOI] [PubMed] [Google Scholar]
- Riemer J, Appenzeller-Herzog C, Johansson L, Bodenmiller B, Hartmann-Petersen R, Ellgaard L 2009. A luminal flavoprotein in endoplasmic reticulum-associated degradation. Proc Natl Acad Sci 106: 14831–14836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riemer J, Hansen HG, Appenzeller-Herzog C, Johansson L, Ellgaard L 2011. Identification of the PDI-family member ERp90 as an interaction partner of ERFAD. PLoS ONE 6: e17037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeed M, Suzuki R, Watanabe N, Masaki T, Tomonaga M, Muhammad A, Kato T, Matsuura Y, Watanabe H, Wakita T, et al. 2011. Role of the endoplasmic reticulum-associated degradation (ERAD) pathway in degradation of hepatitis C virus envelope proteins and production of virus particles. J Biol Chem 286: 37264–37273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanyal S, Claessen JH, Ploegh HL 2012. A viral deubiquitylating enzyme restores dislocation of substrates from the ER in semi-intact cells. J Biol Chem 287: 23594–23603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato BK, Schulz D, Do PH, Hampton RY 2009. Misfolded membrane proteins are specifically recognized by the transmembrane domain of the Hrd1p ubiquitin ligase. Mol Cell 34: 212–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T, Sako Y, Sho M, Momohara M, Suico MA, Shuto T, Nishitoh H, Okiyoneda T, Kokame K, Kaneko M, et al. 2012. STT3B-Dependent posttranslational N-glycosylation as a surveillance system for secretory protein. Mol Cell 47: 99–110 [DOI] [PubMed] [Google Scholar]
- Satoh T, Chen Y, Hu D, Hanashima S, Yamamoto K, Yamaguchi Y 2010. Structural basis for oligosaccharide recognition of misfolded glycoproteins by OS-9 in ER-associated degradation. Mol Cell 40: 905–916 [DOI] [PubMed] [Google Scholar]
- Scott DC, Schekman R 2008. Role of Sec61p in the ER-associated degradation of short-lived transmembrane proteins. J Cell Biol 181: 1095–1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekijima Y, Wiseman RL, Matteson J, Hammarstrom P, Miller SR, Sawkar AR, Balch WE, Kelly JW 2005. The biological and chemical basis for tissue-selective amyloid disease. Cell 121: 73–85 [DOI] [PubMed] [Google Scholar]
- Shmueli A, Tsai YC, Yang M, Braun MA, Weissman AM 2009. Targeting of gp78 for ubiquitin-mediated proteasomal degradation by Hrd1: Cross-talk between E3s in the endoplasmic reticulum. Biochem Biophys Res Commun 390: 758–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song B-L, Sever N, DeBose-Boyd RA 2005. Gp78, a membrane-anchored ubiquitin ligase, associates with Insig-1 and couples sterol-regulated ubiquitination to degradation of HMG CoA reductase. Mol Cell 19: 829–840 [DOI] [PubMed] [Google Scholar]
- Spear ED, Ng DTW 2005. Single, context-specific glycans can target misfolded glycoproteins for ER-associated degradation. J Cell Biol 169: 73–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stagg HR, Thomas M, van den Boomen D, Wiertz EJ, Drabkin HA, Gemmill RM, Lehner PJ 2009. The TRC8 E3 ligase ubiquitinates MHC class I molecules before dislocation from the ER. J Cell Biol 186: 685–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun F, Zhang R, Gong X, Geng X, Drain PF, Frizzell RA 2006. Derlin-1 promotes the efficient degradation of the cystic fibrosis transmembrane conductance regulator (CFTR) and CFTR folding mutants. J Biol Chem 281: 36856–36863 [DOI] [PubMed] [Google Scholar]
- Suzuki M, Otsuka T, Ohsaki Y, Cheng J, Taniguchi T, Hashimoto H, Taniguchi H, Fujimoto T 2012. Derlin-1 and UBXD8 are engaged in dislocation and degradation of lipidated ApoB-100 at lipid droplets. Mol Biol Cell 23: 800–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirosh B, Furman MH, Tortorella D, Ploegh HL 2003. Protein unfolding is not a prerequisite for endoplasmic reticulum-to-cytosol dislocation. J Biol Chem 278: 6664–6672 [DOI] [PubMed] [Google Scholar]
- Tremblay LO, Herscovics A 1999. Cloning and expression of a specific human α1,2-mannosidase that trims Man9GlcNAc2 to Man8GlcNAc2 isomer B during N-glycan biosynthesis. Glycobiology 9: 1073–1078 [DOI] [PubMed] [Google Scholar]
- Urban S 2010. Taking the plunge: Integrating structural, enzymatic and computational insights into a unified model for membrane-immersed rhomboid proteolysis. Biochem J 425: 501–512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ushioda R, Hoseki J, Araki K, Jansen G, Thomas DY, Nagata K 2008. ERdj5 is required as a disulfide reductase for degradation of misfolded proteins in the ER. Science 321: 569–572 [DOI] [PubMed] [Google Scholar]
- Vecchi L, Petris G, Bestagno M, Burrone OR 2012. Selective targeting of proteins within the secretory pathway for endoplasmic reticulum-associated degradation. J Biol Chem 287: 20007–20015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vembar SS, Brodsky JL 2008. One step at a time: Endoplasmic reticulum-associated degradation. Nat Rev Mol Cell Biol 9: 944–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahlman J, DeMartino GN, Skach WR, Bulleid NJ, Brodsky JL, Johnson AE 2007. Real-time fluorescence detection of ERAD substrate retrotranslocation in a mammalian in vitro system. Cell 129: 943–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther TC, Farese RV Jr 2012. Lipid droplets and cellular lipid metabolism. Annu Rev Biochem 81: 687–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Heath-Engel H, Zhang D, Nguyen N, Thomas DY, Hanrahan JW, Shore GC 2008a. BAP31 interacts with Sec61 translocons and promotes retrotranslocation of CFTRΔF508 via the derlin-1 complex. Cell 133: 1080–1092 [DOI] [PubMed] [Google Scholar]
- Wang Q, Li L, Ye Y 2008b. Inhibition of p97-dependent protein degradation by Eeyarestatin I. J Biol Chem 283: 7445–7454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weekes MP, Antrobus R, Talbot S, Hor S, Simecek N, Smith DL, Bloor S, Randow F, Lehner PJ 2012. Proteomic plasma membrane profiling reveals an essential role for gp96 in the cell surface expression of LDLR family members, including the LDL receptor and LRP6. J Proteome Res 11: 1475–1484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie W, Ng DT 2010. ERAD substrate recognition in budding yeast. Semin Cell Dev Biol 21: 533–539 [DOI] [PubMed] [Google Scholar]
- Xie W, Kanehara K, Sayeed A, Ng DT 2009. Intrinsic conformational determinants signal protein misfolding to the Hrd1/Htm1 endoplasmic reticulum-associated degradation system. Mol Biol Cell 20: 3317–3329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y, Meyer HH, Rapoport TA 2001. The AAA ATPase Cdc48/p97 and its partners transport proteins from the ER into the cytosol. Nature 414: 652–656 [DOI] [PubMed] [Google Scholar]
- Ye Y, Shibata Y, Yun C, Ron D, Rapoport TA 2004. A membrane protein complex mediates retro-translocation from the ER lumen into the cytosol. Nature 429: 841–847 [DOI] [PubMed] [Google Scholar]
- Ye Y, Shibata Y, Kikkert M, van Voorden S, Wiertz E, Rapoport TA 2005. Inaugural article: Recruitment of the p97 ATPase and ubiquitin ligases to the site of retrotranslocation at the endoplasmic reticulum membrane. Proc Natl Acad Sci 102: 14132–14138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida Y, Chiba T, Tokunaga F, Kawasaki H, Iwai K, Suzuki T, Ito Y, Matsuoka K, Yoshida M, Tanaka K, et al. 2002. E3 ubiquitin ligase that recognizes sugar chains. Nature 418: 438–442 [DOI] [PubMed] [Google Scholar]
- Yoshida Y, Tokunaga F, Chiba T, Iwai K, Tanaka K, Tai T 2003. Fbs2 is a new member of the E3 ubiquitin ligase family that recognizes sugar chains. J Biol Chem 278: 43877–43884 [DOI] [PubMed] [Google Scholar]
- Younger JM, Chen L, Ren HY, Rosser MF, Turnbull EL, Fan CY, Patterson C, Cyr DM 2006. Sequential quality-control checkpoints triage misfolded cystic fibrosis transmembrane conductance regulator. Cell 126: 571–582 [DOI] [PubMed] [Google Scholar]
- Zettl M, Adrain C, Strisovsky K, Lastun V, Freeman M 2011. Rhomboid family pseudoproteases use the ER quality control machinery to regulate intercellular signaling. Cell 145: 79–91 [DOI] [PMC free article] [PubMed] [Google Scholar]