Abstract
Poxviruses are large, enveloped viruses that replicate in the cytoplasm and encode proteins for DNA replication and gene expression. Hairpin ends link the two strands of the linear, double-stranded DNA genome. Viral proteins involved in DNA synthesis include a 117-kDa polymerase, a helicase–primase, a uracil DNA glycosylase, a processivity factor, a single-stranded DNA-binding protein, a protein kinase, and a DNA ligase. A viral FEN1 family protein participates in double-strand break repair. The DNA is replicated as long concatemers that are resolved by a viral Holliday junction endonuclease.
Poxviruses replicate in discrete factories in the cytoplasm. Their double-stranded DNA genomes, which encode proteins for DNA replication, are linked at the ends, forming one continuous polynucleotide chain.
Poxviruses are large, enveloped, DNA viruses that infect vertebrate and invertebrate species and replicate entirely in the cytoplasm (Moss 2007). Two poxviruses are human-specific: variola virus and molluscum contagiosum virus. The former causes smallpox, a severe disease with high mortality that was eradicated more than two decades ago; the latter is distributed worldwide and produces discrete benign skin lesions in infants and extensive disease in immunocompromised individuals. Other poxviruses that infect nonhuman species can be transmitted to humans; monkeypox virus, for one, causes lethal disease in Africa and has the potential to spread geographically.
Vaccinia virus (VACV), the vaccine used to prevent smallpox, is the prototype of the family, and cited experiments relate to this virus unless stated otherwise. The VACV genome, together with viral enzymes and factors required for transcription of the early subset of genes, is packaged in the core of infectious virus particles (Moss 2007). The DNA replication proteins, in contrast to those involved in early transcription, are not packaged in virions but are translated from viral early mRNAs. DNA replication occurs following release of the genome from the core, and progeny DNA serves as the template for transcription of intermediate- and late-stage genes (Yang et al. 2011).
TIMING AND LOCATION OF VIRAL DNA REPLICATION
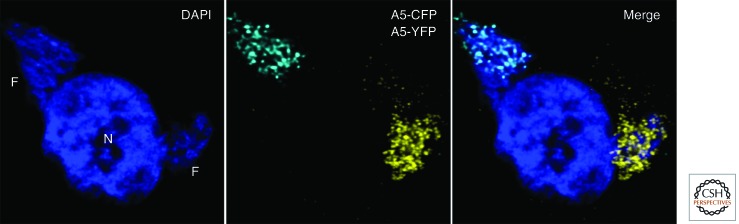
Poxvirus DNA synthesis can usually be detected within 2 h after infection and occurs in the cytoplasm within discrete juxtanuclear sites called factories that can easily be visualized by staining with a fluorescent dye (Fig. 1). A virus factory can form from a single virion, and the number of factories is proportional to the multiplicity of infection (Cairns 1960; Katsafanas and Moss 2007). However, coalescence of individual factories frequently occurs with time (Katsafanas and Moss 2007; Lin and Evans 2010). Factories are initially compact and surrounded by endoplasmic reticulum membranes (Tolonen et al. 2001); a role for membranes in DNA replication has been suggested (Schramm and Krijnse Locker 2005). The factory is also the site of transcription and translation of viral mRNAs in addition to virion assembly (Katsafanas and Moss 2007).
Figure 1.
Poxvirus cytoplasmic factories. HeLa cells were simultaneously infected with two recombinant VACVs. One expresses the A5 core protein fused to cyan fluorescent protein (A5-CFP) and the other expresses the A5 core protein fused to yellow fluorescent protein (A5-YFP). The nuclei (N) and virus factories (F) were stained blue with 4′,6-diamidino-2-phenylindole (DAPI). A confocal microscopy image of a single cell in which individual factories arose from separate viruses is shown. (Reprinted, with permission, from Katsafanas and Moss 2007.)
POXVIRUS GENOME
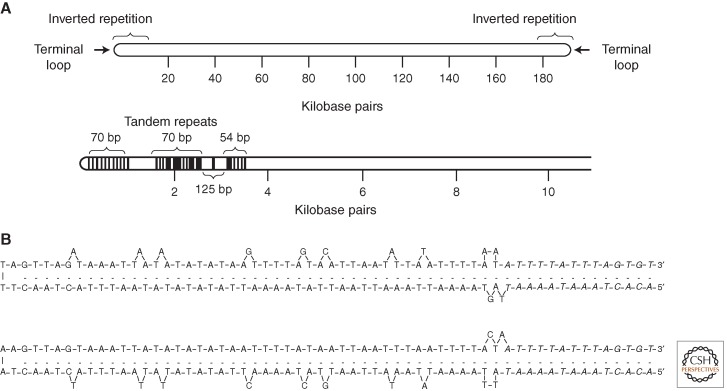
Poxviruses have linear, double-stranded DNA genomes that vary from 130 to 230 kbp. As depicted in Fig. 2A, the two DNA strands are connected at their termini and form a continuous polynucleotide chain (Geshelin and Berns 1974; Baroudy et al. 1982a). The hairpin termini exist in inverted and complementary forms that are incompletely base-paired and AT-rich (Fig. 2B). In the case of VACV, the hairpins are 104 nucleotides in length and contain a 4-nucleotide loop and 10 extrahelical bases on one strand and 2 on the other. The maintenance of the extrahelical bases posits the existence of a double-stranded replication intermediate that is subsequently resolved to form the hairpin. The hairpin is at the end of a long inverted terminal repetition containing sets of short, tandemly repeated sequences (Fig. 2A) (Wittek and Moss 1980; Baroudy and Moss 1982). A region of <100 bp, between the end of the hairpin and start of the short repeats, contains a conserved sequence that is necessary for concatemer resolution (see below).
Figure 2.
Structure of VACV genomic DNA. (A) Depiction of entire genome and an expansion of the 10-kbp inverted terminal repetition showing tandem direct repeats. (B) Nucleotide sequences of inverted and complementary forms of the terminal loops. (A, Reprinted, with permission, from Moss et al. 1983; B, reprinted, with permission, from Baroudy et al. 1982a.)
DNA PRECURSOR METABOLISM
Many poxviruses encode one or more enzymes involved in the synthesis of deoxyribonucleotides, presumably to enhance genome replication in resting cells with suboptimal precursor pools. The list (Table 1) includes thymidine kinase, thymidylate kinase, thymidylate synthetase, ribonucleotide reductase, dUTPase, and deoxycytidine kinase (Moss 2007). The corresponding deletion mutants are viable, although they are generally attenuated in vivo.
Table 1.
Poxvirus DNA replication, processing, and packaging proteins
| Protein | Massa (kDa) | Conservationb (VACV ORF)c | Expression | Essential?d |
|---|---|---|---|---|
| Precursor metabolism | ||||
| Thymidine kinase | 20 | S (J2R) | Early | No |
| Thymidylate kinase | 23 | S (A48R) | Early | No |
| Ribonucleotide reductase | 37, 88 | S (F4L, I4L) | Early | No |
| dUTPase | 16 | S (F2L) | Early | No |
| Replication | ||||
| DNA polymerase | 117 | A (E9L) | Early | Yes |
| Helicase–primase | 90 | A (D5R) | Early | Yes |
| Uracil DNA glycosylase | 25 | A (D4R) | Early | Yes |
| Processivity factor | 49 | A (A20R) | Early | Yes |
| Protein kinase | 35 | S (B1R) | Early | Host-dependent |
| Single-stranded DNA-binding protein | 30 | C (I3L) | Early | Yes |
| DNA ligase | 63 | S (A50R) | Early | Host-dependent |
| FEN1-like nuclease | 50 | A (G5R) | Early | Impaired |
| DNA processing | ||||
| Holliday junction resolvase | 21 | A (A22R) | Intermediate | Yes |
| Topoisomerase | 37 | A (H6R) | Late | Impaired |
| DNA packaging | ||||
| ATPase | 34 | A (A32L) | Intermediate | Yes |
| Telomere-binding protein 1 | 36 | C (I1L) | Intermediate | Yes |
aCalculated mass of protein encoded by vaccinia virus (VACV) genome.
bA, all poxviruses; C, all chordopoxviruses; S, some poxviruses.
cOpen reading frame (ORF) names according to Copenhagen strain of VACV (http://www.poxvirus.org).
dIn most cases, essential nature is determined by severe defect of conditional mutant in tissue culture cells, and in some cases by inability to isolate a deletion mutant; severity of defect is host- and temperature-dependent for protein kinase mutant. Topoisomerase and FEN1-like nuclease deletion mutants replicate very poorly. Viral ligase can be complemented by cellular ligase I.
INITIATION OF DNA REPLICATION
Experiments designed to locate putative origins of replication instead showed that any circular DNA molecule transfected into cells infected with VACV or Shope fibroma virus replicates; furthermore, replication is not enhanced by insertion of any viral DNA fragment (DeLange and McFadden 1986; Merchlinsky and Moss 1988). The replicated DNA is in the form of unbranched head-to-tail concatemers, which could form by a rolling circle mechanism. “Origin-independent” plasmid replication occurs within viral DNA factories and depends on each of the known viral proteins needed for genome replication (De Silva and Moss 2005).
Analysis of [3H]thymidine incorporation following a shift from nonpermissive to permissive temperature in cells infected with a VACV DNA-negative mutant suggested that synthesis begins near the genome termini (Pogo et al. 1984). Experiments performed by transfecting linear DNA minichromosomes with hairpin ends indicated the presence of cis-acting replication-enhancing sequences within the terminal 200 bp (Du and Traktman 1996). The latter result suggested a specific replication initiation site within the conserved sequence between the hairpin loop and the direct repeats. However, the latter region contains a well-defined sequence required for concatemer resolution (described below), raising the possibility that cleavage of concatemers enhances replication or recovery of linear minichromosomes. It may be that a random nick can serve as a replication origin but that nicking preferentially occurs near the termini of intact genomes for reasons of specific structure or sequence.
DNA REPLICATION PROTEINS
A characteristic of the replication proteins, as well as the nucleotide metabolism enzymes, is their early synthesis. This feature distinguishes proteins involved in DNA synthesis from those made at intermediate or late times that have roles in DNA processing and packaging. We refer to VACV genes by a letter and a number corresponding to the open reading frame (ORF), followed by R or L indicating the direction of transcription; the direction is omitted when referring to the corresponding protein.
DNA Pol
The VACV E9L gene encodes a 117-kDa DNA polymerase (Pol) (Jones and Moss 1984; Traktman et al. 1984; Earl et al. 1986), orthologs of which are found in all sequenced poxviruses. Poxvirus DNA Pol is homologous with the catalytic subunits of the DNA Pol of other viruses as well as eukaryotic α and δ DNA Pol (Wang et al. 1989). Purified VACV DNA Pol catalyzes primer- and template-dependent DNA synthesis and possesses 3′–5′ proofreading exonuclease activity (Challberg and Englund 1979). The activity of purified DNA Pol is inherently distributive, adding <10 nucleotides per primer–template binding event, although a more processive form exists in the cytoplasm (McDonald et al. 1997). The DNA Pol can also catalyze single-strand DNA annealing, which could generate branched molecules and thereby link DNA synthesis and recombination (Willer et al. 1999, 2000; Hamilton and Evans 2005).
Helicase–Primase
Temperature-sensitive (ts) mutants that map to the D5R gene of VACV show a fast-stop DNA replication phenotype (Roseman and Hruby 1987; Evans and Traktman 1992; Boyle et al. 2007). The D5R ORF encodes a 90-kDa protein that has nucleic-acid-independent ability to catalyze the hydrolysis of ribonucleotide triphosphates (rNTPs) and deoxyribonucleotide triphosphates (dNTPs) (Evans et al. 1995). D5 contains an ATP/GTP-binding motif in the carboxy-terminal segment of the protein, which is conserved in poxviruses and distantly related large DNA viruses (Gorbalenya and Koonin 1989; Iyer et al. 2001). Targeted mutations within the Walker A or B domains, the superfamily III helicase motif C, or the AAA+ motif prevented in vivo complementation of VACV infectivity, and the purified proteins were defective in ATP hydrolysis. Multimerization of D5 appeared to be a prerequisite for enzymatic activity (Boyle et al. 2007).
An additional motif, present in the archaeoeukaryotic primase superfamily, was discovered in the amino-terminal domain of VACV D5 and its orthologs in other poxviruses (Iyer et al. 2005). Individual conserved aspartic acid residues in the predicted primase active site were required for in vivo complementation of infectious virus formation as well as genome and plasmid replication (De Silva et al. 2007). Furthermore, purified recombinant D5 protein was shown to catalyze the synthesis of oligoribonucleotides in vitro without stringent template specificity. Mutagenesis studies showed that the primase and NTPase activities of the recombinant D5 protein could be independently inactivated (De Silva et al. 2007). The absence of stringent template specificity suggests a role for this enzyme in discontinuous lagging-strand DNA synthesis (De Silva et al. 2009).
Uracil DNA Glycosylase
In eukaryotic and prokaryotic cells, uracil DNA glycosylase (UDG) participates in the removal of uracil from DNA, which can arise through misincorporation of deoxyuridine monophosphate (dUMP) or deamination of cytosine. UDG initiates base-excision repair by hydrolyzing the glycosylic bond linking uracil to a deoxyribose sugar. Shope fibroma virus (Upton et al. 1993) and VACV (Stuart et al. 1993) encode enzymatically active UDGs of 25 kDa, and orthologs of the protein are conserved in all sequenced members of the poxvirus family. Because cellular UDGs function in repair and are typically not essential for viability, it was surprising to find that VACV D4R ts mutants are unable to replicate DNA at the nonpermissive temperature (Stuart et al. 1993; Millns et al. 1994) and that a D4R deletion mutant can only replicate in a complementing cell line expressing the viral protein (Holzer and Falkner 1997). VACV mutants with enzymatically inactive UDG show no defect in DNA replication, however, demonstrating that the essential role of the protein is unrelated to the repair function (De Silva and Moss 2003). Nevertheless, mutants with enzymatically inactive UDG are attenuated in mice, indicating that the repair function is beneficial. D4 interacts with another essential replication protein, A20, suggesting a role as part of a multisubunit replication–repair complex (McCraith et al. 2000; Stanitsa et al. 2006; Boyle et al. 2011).
Processivity Factor
The VACV A20R gene encodes a 49-kDa protein that is conserved in all sequenced poxviruses but has no cellular homolog. A VACV genome-wide yeast two-hybrid analysis showed that the product of the A20R ORF interacts with proteins encoded by the D4R, D5R, and H5R ORFs, two of which were known to be involved in DNA replication (McCraith et al. 2000). Additional coimmunoprecipitation studies showed that nonoverlapping regions of A20 bind D4, D5, and H5, suggesting that these proteins can interact simultaneously to form a multicomponent complex (Ishii and Moss 2002). The early expression of the protein and the phenotype of conditional lethal A20 ts mutants supported a role in DNA replication (Ishii and Moss 2001; Punjabi et al. 2001). Cytoplasmic extracts of cells infected with a ts A20 mutant show a defect in processive DNA polymerase activity, and the A20 protein copurifies with a processive form of DNA Pol (Klemperer et al. 2001; Punjabi et al. 2001). Further studies suggest that A20 collaborates with D4 to enable processivity of DNA Pol (Stanitsa et al. 2006)
Protein Kinase
The VACV B1R ORF encodes a 35-kDa serine/threonine protein kinase that is packaged in virions (Banham and Smith 1992; Lin et al. 1992; Rempel and Traktman 1992). ts mutants that show a DNA replication-negative phenotype were mapped to the B1R gene (Rempel et al. 1990). Comparative genomic analyses identified two sequence-related human putative serine/threonine protein kinases, VRK1 and VRK2 (Nezu et al. 1997; Nichols and Traktman 2004). Human and mouse VRK1 genes can complement the replication defect of a VACV B1 mutant (Boyle and Traktman 2004). A cellular cytoplasmic protein called barrier to autointegration factor (BAF) acts as a potent inhibitor of poxvirus DNA replication unless its DNA-binding activity is blocked by B1-mediated phosphorylation (Nichols et al. 2006; Wiebe and Traktman 2007). The DNA-binding and dimerization capabilities of BAF are essential for its antipoxviral function (Ibrahim et al. 2011). B1 may have an additional role in DNA replication related to its interaction and phosphorylation of the VACV H5 protein (Beaud et al. 1995; McCraith et al. 2000), which interacts with the processivity factor A20 (McCraith et al. 2000; Ishii and Moss 2002).
Single-Stranded DNA-Binding Protein
An early 30-kDa phosphoprotein encoded by the VACV I3L gene preferentially binds single-stranded DNA (Davis and Mathews 1993; Rochester and Traktman 1998). I3 forms octameric complexes on DNA, similar to the Escherichia coli single-stranded DNA-binding protein (Tseng et al. 1999). Participation of I3 in DNA replication is suggested by the presence of the protein in punctate cytoplasmic inclusions containing parental viral genomes (Domi and Beaud 2000; Welsch et al. 2003). The inability to isolate viable I3L deletion mutants indicates that the corresponding protein is essential (Rochester and Traktman 1998), although direct evidence for a role in DNA replication and recombination has not yet been reported.
DNA Ligase
An ATP-dependent DNA ligase encoded by the VACV A50R ORF is conserved in many but not all chordopoxviruses (Kerr and Smith 1989). The ligase repairs nicked duplex DNA substrates consisting of a 5′-phosphate-terminated strand and a 3′-hydroxyl-terminated strand annealed to a bridging template strand (Shuman 1995). Adenylyltransferase and DNA ligation activities are in the amino-terminal one-third of the VACV ligase (Sekiguchi and Shuman 1997). Poxvirus mutants with deleted ligase genes are able to replicate (Colinas et al. 1990; Kerr and Smith 1991) but show host-range defects (Parks et al. 1998) and reduced pathogenicity (Kerr et al. 1991). The ability of VACV to replicate in the absence of the viral DNA ligase was recently shown by small interfering RNA knockdown experiments to depend on cellular DNA ligase I but not other cellular ligases (Paran et al. 2009). Ligase I is recruited to the viral DNA factories in the cytoplasm. Furthermore, replication of ligase-deficient VACV was greatly reduced and delayed in quiescent cells, which have low levels of ligase I. However, synthesis of ligase I was induced within several hours after infection (Paran et al. 2009). By encoding its own ligase, VACV can accelerate DNA synthesis in resting cells and enhance replication.
CONCATEMER RESOLUTION
Head-to-head and tail-to-tail telomere junctions, detected by restriction enzyme analysis of viral DNA from infected cells, indicated the presence of concatemers (Moyer and Graves 1981; Baroudy et al. 1982b). The junction contains a precise duplex copy of the hairpin loop at the ends of the mature viral genome (Merchlinsky et al. 1988). When transfected into cells infected with VACV or Shope fibroma virus, circular plasmids containing concatemer junctions are resolved into linear minichromosomes with hairpin termini (Delange et al. 1986; Merchlinsky and Moss 1986). The sequence 5′-T6-N7–9-T/C-A3-T/A-3′ in an inverted repeat orientation on either side of a concatemer junction was found to be required for resolution (DeLange and McFadden 1987; Merchlinsky 1990). In supercoiled plasmids, the junction can form a cruciform structure resembling a four-way Holliday junction (HJ) recombination intermediate (Dickie et al. 1987; Merchlinsky et al. 1988). Bioinformatic analyses led to the discovery of motifs and structural elements that are critical for activity of E. coli RuvC HJ resolvase in ORFs that are conserved in all poxvirus genomes (Aravind et al. 2000; Garcia et al. 2000). The RuvC homolog encoded by the VACV A22R gene was found to cleave four- and three-stranded junctions and also have branch nuclease activity (Garcia et al. 2000, 2006; Culyba et al. 2006, 2007). Mutation of either of two conserved acidic amino acids abrogated the catalytic activity of the viral protein without affecting HJ DNA binding (Garcia et al. 2000). Like RuvC, the A22 protein is a dimer in solution and when bound to HJ structures (Garcia et al. 2006). A VACV-inducible A22 null mutant was defective in processing concatemers into unit-length genomes with hairpin ends under nonpermissive conditions, indicating that the enzyme is required for resolution, although additional proteins may also be involved in determining site specificity (Garcia and Moss 2001).
The VACV-encoded type 1 topoisomerase (Shaffer and Traktman 1987; Shuman and Moss 1987) cleaves and ligates a variety of DNA structures in vitro, including an HJ (Sekiguchi et al. 1996; Palaniyar et al. 1999). Studies with a VACV topoisomerase deletion mutant suggest a role for the enzyme in enhancing early gene expression within the confines of the virus core (Da Fonseca and Moss 2003).
DNA REPLICATION MODELS
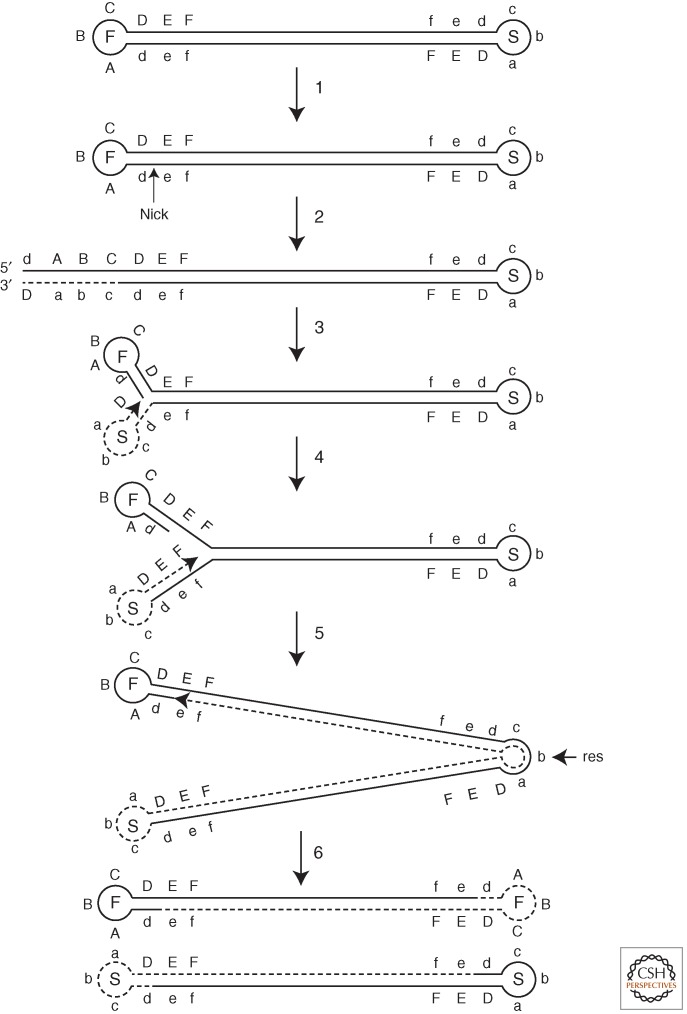
Self-priming, primer-dependent, and recombination models of poxvirus DNA replication have been considered. DNA hairpins at the ends of the poxvirus genome suggest a self-priming model of DNA replication (Fig. 3), resembling the rolling hairpin strand-displacement mechanism for parvoviruses (Muzyczka and Berns 2001). In the poxvirus variation of this model, a nick on one strand proximal to the hairpin by an unidentified nuclease generates a 3′ OH end to which deoxynucleotides can be added (Moyer and Graves, 1981; Baroudy et al. 1982a,b). Because of self-complementarity, the strands fold back and the replication complex continues deoxynucleotide addition to the distal hairpin and around it. The result is the formation of a concatemer, and reiteration of the process could lead to higher-order concatemers. Finally, resolution of the concatemers by the HJ resolvase results in unit genomes.
Figure 3.
Self-priming model of poxvirus DNA replication. F and S within the hairpin loops refer to the fast and slow electrophoretic mobilities of DNA fragments containing inverted and complementary hairpin sequences. Dashed lines indicate newly synthesized DNA with arrowheads at the 3′ OH ends. Complementary sequences are depicted by upper- and lowercase letters. The resolution site (res) within the concatemer junction is indicated. (Modified from Moss et al. 1983.)
Reports of VACV DNA covalently linked to RNA and the chasing of short DNA into larger molecules raises the possibilities of RNA priming and semidiscontinuous DNA replication (Olgiati et al. 1976; Esteban and Holowczak 1977). This model has acquired new interest with the recent findings of a poxvirus primase (De Silva et al. 2007) and the requirement for either a viral or cellular DNA ligase (Paran et al. 2009).
DNA RECOMBINATION
There is evidence that the DNA polymerase participates directly in recombination and that the 3′ exonuclease and DNA-joining activities are involved (Hamilton and Evans 2005; Hamilton et al. 2007; Gammon and Evans 2009). In addition, the protein encoded by the G5R gene, which is conserved in all poxviruses and expressed early in infection, belongs to the FEN1 family of exo-/endonucleases (Da Silva et al. 2006; Iyer et al. 2006). Although the amount of viral DNA produced in the absence of G5 is similar to that made by wild-type virus, the mean size is approximately one-fourth of the genome length and the defective virions package little or no DNA (Senkevich et al. 2009). Experiments with transfected plasmids show that G5 is required for double-strand break repair by homologous recombination, suggesting a similar role during VACV genome replication (Senkevich et al. 2009). Double-strand breaks commonly occur at replication forks, consistent with the semidiscontinuous model of poxvirus DNA replication.
The chordopoxviruses, except for avipoxviruses, encode homologs of serine recombinases (Afonso et al. 2006). However, only the crocodile poxvirus contains all amino acids that comprise the catalytic active site, and deletion of the VACV F16 homolog had no effect on replication in cultured cells (Senkevich et al. 2011).
DNA PACKAGING
Electron microscopic images suggest that DNA nucleoids enter the spherical immature virion before their closure (Morgan 1976; Griffiths et al. 2001). The protein encoded by the A32L gene has sequence similarity to the products of gene I of filamentous single-stranded DNA bacteriophages and to the IVa2 gene of adenovirus, both of which are ATPases involved in DNA packaging (Koonin et al. 1993). Repression of A32 synthesis prevents genome packaging (Cassetti et al. 1998). DNA packaging also fails to occur under nonpermissive conditions in cells infected with a ts mutant mapped to the I6 telomere-binding protein of VACV (DeMasi et al. 2001; Grubisha and Traktman 2003). Although concatemer resolution is required for DNA packaging (Garcia and Moss 2001), the two processes are not coupled (Merchlinsky and Moss 1989) and neither the A32 nor I1 mutant has a defect in concatemer resolution. It seems likely that binding of I6 occurs after resolution and precedes interaction of the genome with A32 or other packaging proteins.
DNA REPLICATION TARGETS FOR ANTIVIRAL THERAPY
Poxvirus-encoded DNA replication proteins are prime targets for antivirals. Cidofovir and oral derivatives are acyclic nucleoside analogs that are incorporated into the growing DNA strand and inhibit the 5′-to-3′ chain extension and 3′-to-5′ exonuclease activities of poxvirus DNA polymerase (Andrei and Snoeck 2010; Rice et al. 2011).
FUTURE DIRECTIONS
The following are but a few of the many outstanding questions. (1) Does DNA replication initiate by a self-priming mechanism at a nick, via a primer, or both? If nicking occurs, is there sequence or structural specificity to the site of the nick and what enzyme catalyzes nick formation? (2) Is the entire genome replicated by strand displacement, or does discontinuous synthesis occur, as suggested by the findings of a role for primase and DNA ligase? (3) Does DNA replication become recombination-dependent at late stages? (4) Is the HJ endonuclease sufficient for concatemer resolution? (5) How is DNA packaged?
ACKNOWLEDGMENTS
I thank George Katsafanas for help with the figures. The review was written with the support of the Division of Intramural Research, National Institute of Allergy and Infectious Diseases, National Institutes of Health.
Footnotes
Editors: Stephen D. Bell, Marcel Méchali, and Melvin L. DePamphilis
Additional Perspectives on DNA Replication available at www.cshperspectives.org
REFERENCES
- Afonso CL, Tulman ER, Delhon G, Lu Z, Viljoen GJ, Wallace DB, Kutish GF, Rock DL 2006. Genome of crocodilepox virus. J Virol 80: 4978–4991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrei G, Snoeck R 2010. Cidofovir activity against poxvirus infections. Viruses 2: 2803–2830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravind L, Makarova KS, Koonin EV 2000. Holliday junction resolvases and related nucleases: Identification of new families, phyletic distribution and evolutionary trajectories. Nucleic Acids Res 28: 3417–3432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banham A, Smith GL 1992. Vaccinia virus gene B1R encodes a 34-kDa serine/threonine protein kinase that localizes in cytoplasmic factories and is packaged into virions. Virology 191: 803–812 [DOI] [PubMed] [Google Scholar]
- Baroudy BM, Moss B 1982. Sequence homologies of diverse length tandem repetitions near ends of vaccinia virus genome suggest unequal crossing over. Nucleic Acids Res 10: 5673–5679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baroudy BM, Venkatesan S, Moss B 1982a. Incompletely base-paired flip-flop terminal loops link the two DNA strands of the vaccinia virus genome into one uninterrupted polynucleotide chain. Cell 28: 315–324 [DOI] [PubMed] [Google Scholar]
- Baroudy BM, Venkatesan S, Moss B 1982b. Structure and replication of vaccinia virus telomeres. Cold Spring Harb Symp Quant Biol 47: 723–729 [DOI] [PubMed] [Google Scholar]
- Beaud G, Beaud R, Leader DP 1995. Vaccinia virus gene H5R encodes a protein that is phosphorylated by the multisubstrate vaccinia virus B1R protein kinase. J Virol 69: 1819–1826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle KA, Traktman P 2004. Members of a novel family of mammalian protein kinases complement the DNA-negative phenotype of a vaccinia virus ts mutant defective in the B1 kinase. J Virol 78: 1992–2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle KA, Arps L, Traktman P 2007. Biochemical and genetic analysis of the vaccinia virus D5 protein: Multimerization-dependent ATPase activity is required to support viral DNA replication. J Virol 81: 844–859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle KA, Stanitsa ES, Greseth MD, Lindgren JK, Traktman P 2011. Evaluation of the role of the vaccinia virus uracil DNA glycosylase and A20 proteins as intrinsic components of the DNA polymerase holoenzyme. J Biol Chem 286: 24702–24713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns J 1960. The initiation of vaccinia infection. Virology 11: 603–623 [DOI] [PubMed] [Google Scholar]
- Cassetti MC, Merchlinsky M, Wolffe EJ, Weisberg AS, Moss B 1998. DNA packaging mutant: Repression of the vaccinia virus A32 gene results in noninfectious, DNA-deficient, spherical, enveloped particles. J Virol 72: 5769–5780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Challberg MD, Englund PT 1979. Purification and properties of the deoxyribonucleic acid polymerase induced by vaccinia virus. J Biol Chem 254: 7812–7819 [PubMed] [Google Scholar]
- Colinas RJ, Goebel SJ, Davis SW, Johnson GP, Norton EK, Paoletti E 1990. A DNA ligase gene in the Copenhagen strain of vaccinia virus is nonessential for viral replication and recombination. Virology 179: 267–275 [DOI] [PubMed] [Google Scholar]
- Culyba MJ, Harrison JE, Hwang Y, Bushman FD 2006. DNA cleavage by the A22R resolvase of vaccinia virus. Virology 352: 466–476 [DOI] [PubMed] [Google Scholar]
- Culyba MJ, Minkah N, Hwang Y, Benhamou OM, Bushman FD 2007. DNA branch nuclease activity of vaccinia A22 resolvase. J Biol Chem 282: 34644–34652 [DOI] [PubMed] [Google Scholar]
- Da Fonseca F, Moss B 2003. Poxvirus DNA topoisomerase knockout mutant exhibits decreased infectivity associated with reduced early transcription. Proc Nat Acad Sci 100: 11291–11296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Da Silva M, Shen L, Tcherepanov V, Watson C, Upton C 2006. Predicted function of the vaccinia virus G5R protein. Bioinformatics 22: 2846–2850 [DOI] [PubMed] [Google Scholar]
- Davis RE, Mathews CK 1993. Acidic C terminus of vaccinia virus DNA-binding protein interacts with ribonucleotide reductase. Proc Natl Acad Sci 90: 745–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLange AM, McFadden G 1986. Sequence-nonspecific replication of transfected plasmid DNA in poxvirus-infected cells. Proc Natl Acad Sci 83: 614–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLange AM, McFadden G 1987. Efficient resolution of replicated poxvirus telomeres to native hairpin structures requires two inverted symmetrical copies of a core target DNA sequence. J Virol 61: 1957–1963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delange AM, Reddy M, Scraba D, Upton C, McFadden G 1986. Replication and resolution of cloned poxvirus telomeres in vivo generates linear minichromosomes with intact viral hairpin termini. J Virol 59: 249–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeMasi J, Du S, Lennon D, Traktman P 2001. Vaccinia virus telomeres: Interaction with the viral I1, I6, and K4 proteins. J Virol 75: 10090–10105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Silva FS, Moss B 2003. Vaccinia virus uracil DNA glycosylase has an essential role in DNA synthesis that is independent of its glycosylase activity: Catalytic site mutations reduce virulence but not virus replication in cultured cells. J Virol 77: 159–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Silva FS, Moss B 2005. Origin-independent plasmid replication occurs in vaccinia virus cytoplasmic factories and requires all five known poxvirus replication factors. Virol J 2: 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Silva FS, Lewis W, Berglund P, Koonin EV, Moss B 2007. Poxvirus DNA primase. Proc Natl Acad Sci 104: 18724–18729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Silva FS, Paran N, Moss B 2009. Products and substrate/template usage of vaccinia virus DNA primase. Virology 383: 136–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickie P, Morgan AR, McFadden G 1987. Cruciform extrusion in plasmids bearing the replicative intermediate configuration of a poxvirus telomere. J Mol Biol 196: 541–558 [DOI] [PubMed] [Google Scholar]
- Domi A, Beaud G 2000. The punctate sites of accumulation of vaccinia virus early proteins are precursors of sites of viral DNA synthesis. J Gen Virol 81: 1231–1235 [DOI] [PubMed] [Google Scholar]
- Du S, Traktman P 1996. Vaccinia virus DNA replication: Two hundred base pairs of telomeric sequence confer optimal replication efficiency on minichromosome templates. Proc Natl Acad Sci 93: 9693–9698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earl PL, Jones EV, Moss B 1986. Homology between DNA polymerases of poxviruses, herpesviruses, and adenoviruses: Nucleotide sequence of the vaccinia virus DNA polymerase gene. Proc Natl Acad Sci 83: 3659–3663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteban M, Holowczak JA 1977. Replication of vaccinia DNA in mouse L cells. I. In vivo DNA synthesis. Virology 78: 57–75 [DOI] [PubMed] [Google Scholar]
- Evans E, Traktman P 1992. Characterization of vaccinia virus DNA replication mutants with lesions in the D5 gene. Chromosoma 102: S72–S82 [DOI] [PubMed] [Google Scholar]
- Evans E, Klemperer N, Ghosh R, Traktman P 1995. The vaccinia virus D5 protein, which is required for DNA replication, is a nucleic acid-independent nucleotide triphosphatase. J Virol 69: 5353–5361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gammon DB, Evans DH 2009. The 3′-to-5′ exonuclease activity of vaccinia virus DNA polymerase is essential and plays a role in promoting virus genetic recombination. J Virol 83: 4236–4250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia AD, Moss B 2001. Repression of vaccinia virus Holliday junction resolvase inhibits processing of viral DNA into unit-length genomes. J Virol 75: 6460–6471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia AD, Aravind L, Koonin EV, Moss B 2000. Bacterial-type DNA Holliday junction resolvases in eukaryotic viruses. Proc Natl Acad Sci 97: 8926–8931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia AD, Otero J, Lebowitz J, Schuck P, Moss B 2006. Quartenary structure and cleavage specificity of a poxvirus Holliday junction resolvase. J Biol Chem 281: 11618–11626 [DOI] [PubMed] [Google Scholar]
- Geshelin P, Berns KI 1974. Characterization and localization of the naturally occurring cross-links in vaccinia virus DNA. J Mol Biol 88: 785–796 [DOI] [PubMed] [Google Scholar]
- Gorbalenya AE, Koonin EV 1989. Viral proteins containing the purine NTP-binding sequence pattern. Nucleic Acids Res 17: 8413–8440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths G, Roos N, Schleich S, Krijnse Locker J 2001. Structure and assembly of intracellular mature vaccinia virus: Thin-section analyses. J Virol 75: 11056–11070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grubisha O, Traktman P 2003. Genetic analysis of the vaccinia virus I6 telomere-binding protein uncovers a key role in genome encapsidation. J Virol 77: 10929–10942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton MD, Evans DH 2005. Enzymatic processing of replication and recombination intermediates by the vaccinia virus DNA polymerase. Nucleic Acids Res 33: 2259–2268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton MD, Nuara AA, Gammon DB, Buller RM, Evans DH 2007. Duplex strand joining reactions catalyzed by vaccinia virus DNA polymerase. Nucleic Acids Res 35: 143–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holzer G, Falkner FG 1997. Construction of a vaccinia virus deficient in the essential DNA repair enzyme uracil DNA glycosylase by a complementing cell line. J Virol 71: 4997–5002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahim N, Wicklund A, Wiebe MS 2011. Molecular characterization of the host defense activity of the barrier to autointegration factor against vaccinia virus. J Virol 85: 11588–11600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii K, Moss B 2001. Role of vaccinia virus A20R protein in DNA replication: Construction and characterization of temperature-sensitive mutants. J Virol 75: 1656–1663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii K, Moss B 2002. Mapping interaction sites of the A20R protein component of the vaccinia virus DNA replication complex. Virology 303: 232–239 [DOI] [PubMed] [Google Scholar]
- Iyer LM, Aravind L, Koonin EV 2001. Common origin of four diverse families of large eukaryotic DNA viruses. J Virol 75: 11720–11734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer LM, Koonin EV, Leipe DD, Aravind L 2005. Origin and evolution of the archaeo-eukaryotic primase superfamily and related palm-domain proteins: Structural insights and new members. Nucleic Acids Res 33: 3875–3896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer LM, Balaji S, Koonin EV, Aravind L 2006. Evolutionary genomics of nucleo-cytoplasmic large DNA viruses. Virus Res 117: 156–184 [DOI] [PubMed] [Google Scholar]
- Jones EV, Moss B 1984. Mapping of the vaccinia virus DNA polymerase gene by marker rescue and cell-free translation of selected mRNA. J Virol 49: 72–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsafanas GC, Moss B 2007. Colocalization of transcription and translation within cytoplasmic poxvirus factories coordinates viral expression and subjugates host functions. Cell Host Microbe 2: 221–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr SM, Smith GL 1989. Vaccinia virus encodes a polypeptide with DNA ligase activity. Nucleic Acids Res 17: 9039–9050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr SM, Smith GL 1991. Vaccinia virus DNA ligase is nonessential for virus replication: Recovery of plasmids from virus-infected cells. Virology 180: 625–632 [DOI] [PubMed] [Google Scholar]
- Kerr SM, Johnston LH, Odell M, Duncan SA, Law KM, Smith GL 1991. Vaccinia DNA ligase complements Saccharomyces cerevisiae Cdc9, localizes in cytoplasmic factories and affects virulence and virus sensitivity to DNA damaging agents. EMBO J 10: 4343–4350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klemperer N, McDonald W, Boyle K, Unger B, Traktman P 2001. The A20R protein is a stoichiometric component of the processive form of vaccinia virus DNA polymerase. J Virol 75: 12298–12307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV, Senkevich TG, Chernos VI 1993. Gene A32 protein product of vaccinia virus may be an ATPase involved in viral DNA packaging as indicated by sequence comparisons with other putative viral ATPases. Virus Genes 7: 89–94 [DOI] [PubMed] [Google Scholar]
- Lin YCJ, Evans DH 2010. Vaccinia virus particles mix inefficiently, and in a way that would restrict viral recombination, in coinfected cells. J Virol 84: 2432–2443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S, Chen W, Broyles SS 1992. The vaccinia virus B1R gene product is a serine/threonine protein kinase. J Virol 66: 2717–2723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCraith S, Holtzman T, Moss B, Fields S 2000. Genome-wide analysis of vaccinia virus protein–protein interactions. Proc Natl Acad Sci 97: 4879–4884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald WF, Klemperer N, Traktman P 1997. Characterization of a processive form of the vaccinia virus DNA polymerase. Virology 234: 168–175 [DOI] [PubMed] [Google Scholar]
- Merchlinsky M 1990. Mutational analysis of the resolution sequence of vaccinia virus DNA: Essential sequence consists of two separate AT-rich regions highly conserved among poxviruses. J Virol 64: 5029–5035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchlinsky M, Moss B 1986. Resolution of linear minichromosomes with hairpin ends from circular plasmids containing vaccinia virus concatemer junctions. Cell 45: 879–884 [DOI] [PubMed] [Google Scholar]
- Merchlinsky M, Moss B 1988. Sequence-independent replication and sequence-specific resolution of plasmids containing the vaccinia virus concatemer junction: Requirements for early and late trans-acting factors. In Cancer cells 6: Eukaryotic DNA replication (ed. Kelly T, Stillman B), pp. 87–93 Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Merchlinsky M, Moss B 1989. Resolution of vaccinia virus DNA concatemer junctions requires late gene expression. J Virol 63: 1595–1603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchlinsky M, Garon C, Moss B 1988. Molecular cloning and sequence of the concatemer junction from vaccinia virus replicative DNA: Viral nuclease cleavage sites in cruciform structures. J Mol Biol 199: 399–413 [DOI] [PubMed] [Google Scholar]
- Millns AK, Carpenter MS, DeLange AM 1994. The vaccinia virus-encoded uracil DNA glycosylase has an essential role in viral DNA replication. Virology 198: 504–513 [DOI] [PubMed] [Google Scholar]
- Morgan C 1976. The insertion of DNA into vaccinia virus. Science 193: 591–592 [DOI] [PubMed] [Google Scholar]
- Moss B 2007. Poxviridae: The viruses and their replication. In Fields virology (ed. Knipe DM, Howley PM), pp. 2905–2946 Lippincott Williams & Wilkins, Philadelphia [Google Scholar]
- Moss B, Winters E, Jones EV 1983. Replication of vaccinia virus. In Mechanics of DNA replication and recombination (ed. Cozzarelli N), pp. 449–461 A. Liss, New York [Google Scholar]
- Moyer RW, Graves RL 1981. The mechanism of cytoplasmic orthopoxvirus DNA replication. Cell 27: 391–401 [DOI] [PubMed] [Google Scholar]
- Muzyczka N, Berns KI 2001. Parvoviridae: The viruses and their replication. In Fields virology (ed. Knipe DM, Howley PM), pp. 2327–2359 Lippincott, Williams & Wilkins, Philadelphia [Google Scholar]
- Nezu J-I, Oku A, Jones MH, Shimane M 1997. Identification of two novel human putative serine/threonine kinases, VRK1 and VRK2, with structural similarity to vaccinia virus B1R kinase. Genomics 45: 327–331 [DOI] [PubMed] [Google Scholar]
- Nichols RJ, Traktman P 2004. Characterization of three paralogous members of the mammalian vaccinia related kinase family. J Biol Chem 279: 7934–7946 [DOI] [PubMed] [Google Scholar]
- Nichols RJ, Wiebe MS, Traktman P 2006. The vaccinia-related kinases phosphorylate the N′ terminus of BAF, regulating its interaction with DNA and its retention in the nucleus. Mol Biol Cell 17: 2451–2464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olgiati DD, Pogo BG, Dales S 1976. Evidence for RNA linked to nascent DNA in HeLa cells. J Cell Biol 68: 557–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palaniyar N, Gerasimopoulos E, Evans DH 1999. Shope fibroma virus DNA topoisomerase catalyses Holliday junction resolution and hairpin formation in vitro. J Mol Biol 287: 9–20 [DOI] [PubMed] [Google Scholar]
- Paran N, De Silva FS, Senkevich TG, Moss B 2009. Cellular DNA ligase I is recruited to cytoplasmic vaccinia virus factories and masks the role of the vaccinia ligase in viral DNA replication. Cell Host Microbe 6: 563–569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks RJ, Winchcombe-Forhan C, DeLange AM, Xing X, Evans DH 1998. DNA ligase gene disruptions can depress viral growth and replication in poxvirus-infected cells. Virus Res 56: 135–147 [DOI] [PubMed] [Google Scholar]
- Pogo BG, Berkowitz EM, Dales S 1984. Investigation of vaccinia virus DNA replication employing a conditional lethal mutant defective in DNA. Virology 132: 436–444 [DOI] [PubMed] [Google Scholar]
- Punjabi A, Boyle K, DeMasi J, Grubisha O, Unger B, Khanna M, Traktman P 2001. Clustered charge-to-alanine mutagenesis of the vaccinia virus A20 gene: Temperature-sensitive mutants have a DNA-minus phenotype and are defective in the production of processive DNA polymerase activity. J Virol 75: 12308–12318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rempel RE, Traktman P 1992. Vaccinia virus B1 kinase: Phenotypic analysis of temperature-sensitive mutants and enzymatic characterization of recombinant proteins. J Virol 66: 4413–4426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rempel RE, Anderson MK, Evans E, Traktman P 1990. Temperature-sensitive vaccinia virus mutants identify a gene with an essential role in viral replication. J Virol 64: 574–583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice AD, Adams MM, Lampert B, Foster S, Lanier R, Robertson A, Painter G, Moyer RW 2011. Efficacy of CMX001 as a prophylactic and presymptomatic antiviral agent in New Zealand white rabbits infected with rabbitpox virus, a model for orthopoxvirus infections of humans. Viruses 3: 63–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochester SC, Traktman P 1998. Characterization of the single-stranded DNA binding protein encoded by the vaccinia virus I3 gene. J Virol 72: 2917–2926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roseman NA, Hruby DE 1987. Nucleotide sequence and transcript organization of a region of the vaccinia virus genome which encodes a constitutively expressed gene required for DNA replication. J Virol 61: 1398–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schramm B, Krijnse Locker J 2005. Cytoplasmic organization of poxvirus DNA replication. Traffic 6: 839–846 [DOI] [PubMed] [Google Scholar]
- Sekiguchi J, Shuman S 1997. Domain structure of vaccinia DNA ligase. Nucleic Acids Res 25: 727–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekiguchi J, Seeman NC, Shuman S 1996. Resolution of Holliday junctions by eukaryotic DNA topoisomerase I. Proc Natl Acad Sci 93: 785–789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senkevich TG, Koonin EV, Moss B 2009. Predicted poxvirus FEN1-like nuclease required for homologous recombination, double-strand break repair and full-size genome formation. Proc Natl Acad Sci 106: 17921–17926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senkevich TG, Koonin EV, Moss B 2011. Vaccinia virus F16 protein, a predicted catalytically inactive member of the prokaryotic serine recombinase superfamily, is targeted to nucleoli. Virology 417: 334–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaffer R, Traktman P 1987. Vaccinia virus encapsidates a novel topoisomerase with the properties of a eucaryotic type I enzyme. J Biol Chem 262: 9309–9315 [PubMed] [Google Scholar]
- Shuman S 1995. Vaccinia virus DNA ligase: Specificity, fidelity, and inhibition. Biochemistry 34: 16138–16147 [DOI] [PubMed] [Google Scholar]
- Shuman S, Moss B 1987. Identification of a vaccinia virus gene encoding a type I DNA topoisomerase. Proc Natl Acad Sci 84: 7478–7482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanitsa ES, Arps L, Traktman P 2006. Vaccinia virus uracil DNA glycosylase interacts with the A20 protein to form a heterodimeric processivity factor for the viral DNA polymerase. J Biol Chem 281: 3439–3451 [DOI] [PubMed] [Google Scholar]
- Stuart DT, Upton C, Higman MA, Niles EG, McFadden G 1993. A poxvirus-encoded uracil DNA glycosylase is essential for virus viability. J Virol 67: 2503–2512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolonen N, Doglio L, Schleich S, Krijnse Locker J 2001. Vaccinia virus DNA replication occurs in endoplasmic reticulum-enclosed cytoplasmic mini-nuclei. Mol Biol Cell 12: 2031–2046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traktman P, Sridhar RC, Roberts BE 1984. Transcriptional mapping of the DNA polymerase gene of vaccinia virus. J Virol 49: 125–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng M, Palaniyar N, Zhang WD, Evans DH 1999. DNA binding and aggregation properties of the vaccinia virus I3L gene product. J Biol Chem 274: 21637–21644 [DOI] [PubMed] [Google Scholar]
- Upton C, Stuart DT, McFadden G 1993. Identification of a poxvirus gene encoding a uracil DNA glycosylase. Proc Natl Acad Sci 90: 4518–4522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang TS, Wong SW, Korn D 1989. Human DNA polymerase α: Predicted functional domains and relationships with viral DNA polymerases. FASEB J 3: 14–21 [DOI] [PubMed] [Google Scholar]
- Welsch S, Doglio L, Schleich S, Krijnse Locker J 2003. The vaccinia virus I3L gene product is localized to a complex endoplasmic reticulum-associated structure that contains the viral parental DNA. J Virol 77: 6014–6028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiebe MS, Traktman P 2007. Poxviral B1 kinase overcomes barrier to autointegration factor, a host defense against virus replication. Cell Host Microbe 1: 187–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willer DO, Mann MJ, Zhang WD, Evans DH 1999. Vaccinia virus DNA polymerase promotes DNA pairing and strand-transfer reactions. Virology 257: 511–523 [DOI] [PubMed] [Google Scholar]
- Willer DO, Yao XD, Mann MJ, Evans DH 2000. In vitro concatemer formation catalyzed by vaccinia virus DNA polymerase. Virology 278: 562–569 [DOI] [PubMed] [Google Scholar]
- Wittek R, Moss B 1980. Tandem repeats within the inverted terminal repetition of vaccinia virus DNA. Cell 21: 277–284 [DOI] [PubMed] [Google Scholar]
- Yang Z, Reynolds SE, Martens CA, Bruno DP, Porcella SF, Moss B 2011. Expression profiling of the intermediate and late stages of poxvirus replication. J Virol 85: 9899–9908 [DOI] [PMC free article] [PubMed] [Google Scholar]