Abstract
The study of MYC has led to pivotal discoveries in cancer biology, induced pluripotency, and transcriptional regulation. In this review, continuing advances in our understanding of the function of MYC as a transcription factor and how its transcriptional activity controls normal vertebrate development and contributes to developmental disorders is discussed.
MYC family proteins help to generate functional organ systems during embryonic development. Defects in their regulation disrupt organ formation and homeostasis and can contribute to developmental disorders (e.g., Feingold syndrome).
During embryonic development, a great diversity of cell types are rapidly produced concomitant with their organization into the different functional tissues and organ structures needed to sustain life. A common theme underlying higher-order organ development is the initial establishment of stem cells or multipotent progenitor cells at distinct spatial locations. These stem and progenitor populations then respond to local environmental cues by implementing selective activation and/or silencing of specific transcription programs that drive the generation and ordered proliferative expansion of the different cell types and lineages responsible for organ-specific tissue formation. There is intense interest in how these transcriptional programs are established and maintained, both with respect to the signaling pathways and critical transcription factors involved, because their manipulation may permit the in vivo or ex vivo generation of diverse cell types for therapeutic purposes, and because their misregulation appears to be a root cause of diverse cancers and developmental disorders. Prominent among the transcription factors involved are members of the MYC family of proteins, particularly MYC and MYCN.
MYC and MYCN, together with the other MYC gene family member, MYCL, encode basic-helix-loop-helix-leucine zipper (BHLHZIP) proteins that function primarily as nuclear transcription factors. However, additional activities of MYC have been identified in the control of DNA replication (Dominguez-Sola et al. 2007), and in the cytoplasm where a cleaved form of MYC that lacks the BHLHZIP region can promote differentiation (Conacci-Sorrell et al. 2010). MYC proteins are best known for their frequent involvement in a great variety of cancers and the ability of ectopic MYC to contribute to pluripotency (Cartwright et al. 2005; Takahashi and Yamanaka 2006; Wernig et al. 2007). The role of MYC family proteins in cancer and induced pluripotency is thought to stem from the appropriation of MYC activities that generally, but not universally, tend to maintain cells in a proliferative state and prevent differentiation. In the context of cancer, the activities of MYC are essentially constitutive due to mutations in signaling pathways that regulate MYC, as well as to MYC gene amplification and translocations that prevent the normal down-regulation of MYC expression. But during embryonic development, the activities of MYC are harnessed through Myc’s tightly regulated temporal and spatial expression to perform critical functions in generating functional organ systems and tissues.
TRANSCRIPTIONAL CONTROL BY MYC AND MYC–MAX COMPLEXES
MYC does not bind DNA on its own, but relies on interaction with the protein MAX through shared BHLHZIP motifs to form complexes that bind E-box (CACGTG) and closely related sequences (Blackwood and Eisenman 1991). The original thinking was that MYC–MAX complexes functioned like traditional sequence-specific DNA-binding transcription factors to activate transcription of a restricted set of target genes encoding proteins that then directly promoted cell growth and proliferation (Blackwood et al. 1992; Torres et al. 1992). Although conceptually not far off the mark, the transcriptional activities of MYC have proven to be far more complicated and multifaceted. For example, MYC–MAX heterodimers can associate with the POZ domain/zinc finger transcription factor MIZ, and MYC–MIZ complexes can repress transcription of MIZ target genes (Herkert and Eilers 2010). Additionally, the central tenet that MAX is an obligate partner for MYC’s ability to activate transcription and for its biological activities is under considerable revision. Evidence for MAX-independent functions for MYC in transcriptional regulation is found in Drosophila mutants that lack MAX (Steiger et al. 2008; Gallant and Steiger 2009) and in the rat pheochromocytoma cell line PC12 that lacks MAX (Ribon et al. 1994; Hopewell and Ziff 1995). In both of these settings, MYC can activate transcription, but it is not clear how widespread such MAX-independent transcription might be when MAX is present in cells.
The physiological significance of MAX loss in PC12 cells was reconsidered recently when it was shown that MAX is often deleted in human pheochromocytoma and potentially in other neuronal cancers (Comino-Méndez et al. 2011; Burnichon et al. 2012). These findings, together with previous studies showing that ectopic overexpression of MAX can repress MYC-dependent transcription and suppress oncogenic transformation by MYC (Grandori et al. 2000), are consistent with the idea that MAX has important functions beyond acting to mediate MYC DNA binding and oncogenic activity. Some of the MYC-independent functions of MAX include forming heterodimeric DNA-binding complexes with several other BHLHZIP proteins related to MYC and MAX that function as transcriptional repressors (Ayer et al. 1993; Zervos et al. 1993; Hurlin et al. 1997, 1999, 2003). Additional MAX-independent functions include the potential for direct and indirect association between MYC and transcriptional cofactors already associated with chromatin and between MYC and members of the basal transcriptional machinery and with RNA polymerases as described below.
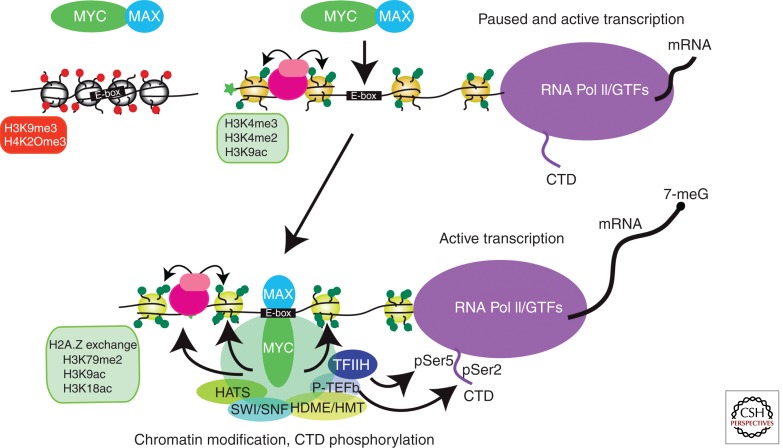
The chromatin landscape of MYC- and MYC–MAX-binding sites suggests that MYC preferentially binds to active promoters or promoters associated with a pre-engaged basal transcription machinery (Fig. 1) (Eberhardy and Farnham 2001; Frank et al. 2001; Guccione et al. 2006; Kidder et al. 2008). MYC associates with a wide assortment of histone- and chromatin-modifying proteins and protein complexes (Adhikary and Eilers 2005; Cowling and Cole 2006; Eilers and Eisenman 2008; He et al. 2013), and once bound to its target promoters, MYC-associated activities generate additional chromatin modifications that create a chromatin environment favorable for DNA and chromatin binding by additional factors that cooperate in stimulating polymerase activity (Frank et al. 2001; Martinato et al. 2008; Lin et al. 2009, 2012; Nie et al. 2012; Soufi et al. 2012). A major effect of MYC binding is attenuation of RNA polymerase II pausing through its recruitment of P-TEFb complexes and P-TEPb-dependent phosphorylation of serine 2 of the RNA polymerase II carboxy-terminal domain (CTD) (Fig. 1) (Eberhardy and Farnham 2001; Rahl et al. 2010; Lin et al. 2012; Nie et al. 2012). MYC also interacts with TFIIH and stimulates TFIIH-dependent phosphorylation of serine 5 of the CTD (Fig. 1) (Cowling and Cole 2007). Serine 5 phosphorylation facilitates translation of mRNAs by functioning as a signal for the recruitment of RNA methyltransferase, the enzyme responsible for 7-methyl guanine capping of nascent mRNA (Cowling and Cole 2010).
Figure 1.
Regulation of RNA polymerase II by MYC. MYC associates with MAX to form DNA-binding complexes that preferentially bind E-box sites that are associated with euchromatic islands marked by specific histone modifications and bound RNA polymerase II. Such euchromatic islands may be formed through the activities of other transcription factors (pink), including “pioneer” transcription factors that can actively modify and open up the local chromatin environment. Chromatin marked by repressive histone modifications (red) and not poised for transcription is generally not accessible to MYC. MYC binding mediates further chromatin modifications that favor active transcription, including representative ones shown, through the recruitment and actions of histone-modifying proteins. These include histone acetyltransferases (HATs), histone demethylases and methyltransferases (HDME/HMT), and SWI/SNF chromatin-remodeling proteins. Although MYC binding amplifies transcription of actively transcribed genes, MYC also attenuates transcriptional pausing by interacting with P-TEFb-promoting pTEFb-dependent phosphorylation at serine 2 of the carboxy-terminal domain of RNA polymerase II. Finally, MYC interacts with the general transcription factor TFIIH to promote CTD phosphorylation at serine 5 and RNA methyltransferase-dependent 5′ methyl capping of nascent transcripts, a function that supports ribosome loading and translation.
In addition to RNA polymerase II, MYC directly or indirectly interacts with and stimulates the activities of RNA polymerases I and III (Gomez-Roman et al. 2003; Arabi et al. 2005; Grandori et al. 2005; Grewal et al. 2005). Finally, the affects of MYC on transcription can be compounded through the induction of various chromatin-modifying and -binding proteins (Knoepfler et al. 2006; Lin et al. 2009). In aggregate, the various proximal promoter-specific functions of MYC that have been defined describe a unique transcription factor, capable of broadly impacting transcription, translation, and the epigenetic state of cells.
THE EVOLVING DEFINITION OF A MYC TARGET GENE
During the last decade, the notion that MYC and MYC–MAX complexes regulate a restricted and standard set of target genes that account for its various activities in cell growth, proliferation, apoptosis, and oncogenesis has been largely dispelled. Instead, genome-wide DNA-binding studies reveal that MYC and MAX are highly enriched at thousands of overlapping genomic sites (Fernandez et al. 2003; Li et al. 2003a,b; Orian et al. 2003; Cawley et al. 2004; Lin et al. 2009; Ji et al. 2011). MYC–MAX-binding sites are typically enriched for E-box sequences and most frequently located in the proximal promoters of genes (Fernandez et al. 2003; Zeller et al. 2006; Lin et al. 2012; Nie et al. 2012). Given these findings and the ability of MYC to engage and stimulate RNA polymerases I, II, and III, it is not surprising that MYC induces broad changes in gene expression in the wide variety of cellular settings that have been investigated (Patel et al. 2004; Eilers and Eisenman 2008). However, it remains unresolved whether MYC has a dedicated set of target genes that do not vary from cell type to cell type, or whether it functions as a general transcription factor with no universal targets.
A recent study by Ji et al. (2011) identified a set of approximately 50 MYC target genes that were common to several cancer cell lines and mouse and human embryonic stem cells. Expression of these genes was found to correlate with MYC levels in more than 300 other cell and tissue types (Ji et al. 2011). This core set of target genes highlights a role for MYC in RNA processing, ribosome biogenesis, and macromolecular synthesis (Ji et al. 2011), processes previously identified as controlled by MYC (Eilers and Eisenman 2008; Dang 2012). However, other attempts to identify a core signature of MYC target genes in different cell types have failed (Chandriani et al. 2009; Lee et al. 2012). Indeed, the study by Lee et al. (2012) found genomic MYC binding was highly cell-type specific.
A potential explanation for the difficulty in identifying a core MYC signature comes from two recent studies examining genome-wide binding by MYC and MAX and its relationship to global gene transcription (Lin et al. 2012; Nie et al. 2012). These studies found that MYC, in a largely uniform and dose-dependent manner, increased or “amplified” transcription of genes already undergoing active transcription (Lin et al. 2012; Nie et al. 2012). Moreover, they found that MYC’s amplifier effect was independent of cell type—that is, MYC did not alter the unique diversity of gene transcription found in different, albeit very limited, cell types tested (Lin et al. 2012; Nie et al. 2012). The transcriptional amplification phenomenon was closely associated with MYC-enhanced P-TEFb-dependent CTD serine 2 phosphorylation and pause release (Fig. 1) (Lin et al. 2012; Nie et al. 2012). These findings are in general agreement with the finding that MYC broadly targets both active and transcriptionally poised chromatin (Guccione et al. 2006), and that MYC and RNA polymerase II are often found at the same promoters (Martinato et al. 2008; Lee et al. 2012). Perhaps most importantly, these findings suggest that many, if not all, of the “MYC target genes” identified over the years, including various protein coding genes and nonprotein coding genes (Eilers and Eisenman 2008; Dang 2012), may be largely conditional targets dependent on cellular context and their related epigenetic status.
Thus, although there are a number of issues to reconcile with the amplification model, including how MYC-dependent repression and differential gene activation of MYC targets fits in (Nature Medicine Commentaries 2013), and the seemingly general ability of MYC to promote growth and proliferation (Littlewood et al. 2012), it helps explain the longstanding conundrum of how MYC can elicit such a wide variety of cellular behaviors depending on extracellular inputs and cell types. As discussed further below, this model may also provide considerable insight into how MYC controls embryonic development and disease.
DOSE-DEPENDENT CONTROL OF EMBRYONIC DEVELOPMENT BY MYCN AND MYC
Homozygous null MYCN mouse embryos die at approximately E11.5, with different organ systems showing severe hypoplasia, including the heart, lungs, kidneys, gut, skeleton, central nervous system, and genitourinary system (Charron et al. 1992; Stanton et al. 1992; Sawai et al. 1993). Studies of MYCN heterozygotes and mice engineered to express hypomorphic MYCN alleles (Moens et al. 1992, 1993) show clear dose-dependent effects of MYCN on the development of different organs. For example, mice homozygous for a MYCN hypomorphic allele expressing ∼20%–25% MYCN survived to birth, but showed pronounced lung hypoplasia and associated defects in branching morphogenesis that led to early postnatal lethality (Moens et al. 1992; Nagy et al. 1998). And mice engineered to express ∼15% MYCN showed severe cardiac muscle hypoplasia and lethality between E12.5 and E14.5 (Moens et al. 1993). The essential roles of MYCN in lung and heart development (Okubo et al. 2005; Harmelink and Jiao 2010; Harmelink et al. 2013), as well as in the development of the central nervous system (CNS) (Knoepfler et al. 2002), limbs (Ota et al. 2007), kidneys (Nakhai et al. 2008), and inner ear (Domínguez-Frutos et al. 2011) were confirmed and/or further defined by conditional MYCN deletion at these sites (see below for further discussion).
These findings from mouse studies have significance to human disease as MYCN haploinsufficiency causes Feingold syndrome (van Bokhoven et al. 2005; Cognet et al. 2011). The constellation of organs and tissues affected in Feingold syndrome (Celli et al. 2003) is largely recapitulated by MYCN deficiency in the mouse (Charron et al. 1992; Stanton et al. 1992; Sawai et al. 1993). Defects in intellectual development, deafness, and esophageal and gastrointestinal atresia are additional phenotypes associated with Feingold syndrome (Celli et al. 2003) that were not described in the mouse studies. The atresia malformations can lead to death without surgical intervention (de Jong et al. 2010). In addition to MYCN haploinsufficiency, haploinsufficiency of the microRNA miR-17 ∼ 92 polycistronic cluster causes a human disorder related to Feingold syndrome (de Pontual et al. 2011). Although MYCN can up-regulate miR-17 ∼ 92 transcription and miR-17 ∼ 19 deficiency phenocopies skeletal phenotypes in Feingold syndrome (de Pontual et al. 2011), it remains to be determined whether miR-17 ∼ 92 is broadly controlled by MYCN during development.
Studies of heterozygous and homozygous null MYC embryos together with embryos containing hypomorphic MYC alleles show dose-dependent MYC activities in controlling proliferation, growth, and cellularity of multiple tissues and organs (Davis et al. 1993; Trumpp et al. 2001). MYC null embryos die at approximately E10.5, with lethality associated with a number of abnormalities including defects in heart development and neural tube closure (Davis et al. 1993; Trumpp et al. 2001). However, studies in which MYC was deleted either in the epiblast, such that the extraembryonic placental tissues were spared, or conditionally in all hematopoietic lineages, showed that MYC was dispensable for the development of most organs and tissues up to E11.5 and that lethality at this time was due to a failure in definitive hematopoiesis taking place in the extraembryonic tissues (Dubois et al. 2008; He et al. 2008; reviewed in Laurenti et al. 2009; Delgado and León 2010). The importance of MYC in extraembryonic definitive hematopoiesis corresponds to its strong expression in extraembryonic tissues (Downs et al. 1989).
In contrast to the essential activities of MYC and MYCN, MYCL-deficient mice lacked a discernible phenotype (Hatton et al. 1996). This is most likely explained by redundant expression between family members and evidence that MYC protein functions are largely although not completely redundant (Nesbit et al. 1999; Malynn et al. 2000). The development of conditional MYCL knockout mice (Kopecky et al. 2012; R Eisenman, pers. comm.) will allow the redundancy model to be rigorously examined.
Homozygous deletion of MAX causes lethality early after postimplantation (Shen-Li et al. 2000). The deterioration of MAX-deficient embryos appears to coincide with the exhaustion of maternally supplied stores of MAX (Shen-Li et al. 2000). Mice heterozygous for MAX had no apparent phenotype, raising the possibility that MAX is not limiting for the embryonic functions of MYC and MYCN. Although early embryonic lethality caused by loss of MAX supports the concept that MYC functions are dependent on MAX, given the emerging evidence of separate and independent functions of MYC and MAX as discussed above, it will be important to further evaluate MAX function using conditional, site-, and temporal-specific deletion systems.
COORDINATED ACTIONS OF MYCN AND MYC IN LINEAGE DEVELOPMENT AND PROLIFERATIVE EXPANSION
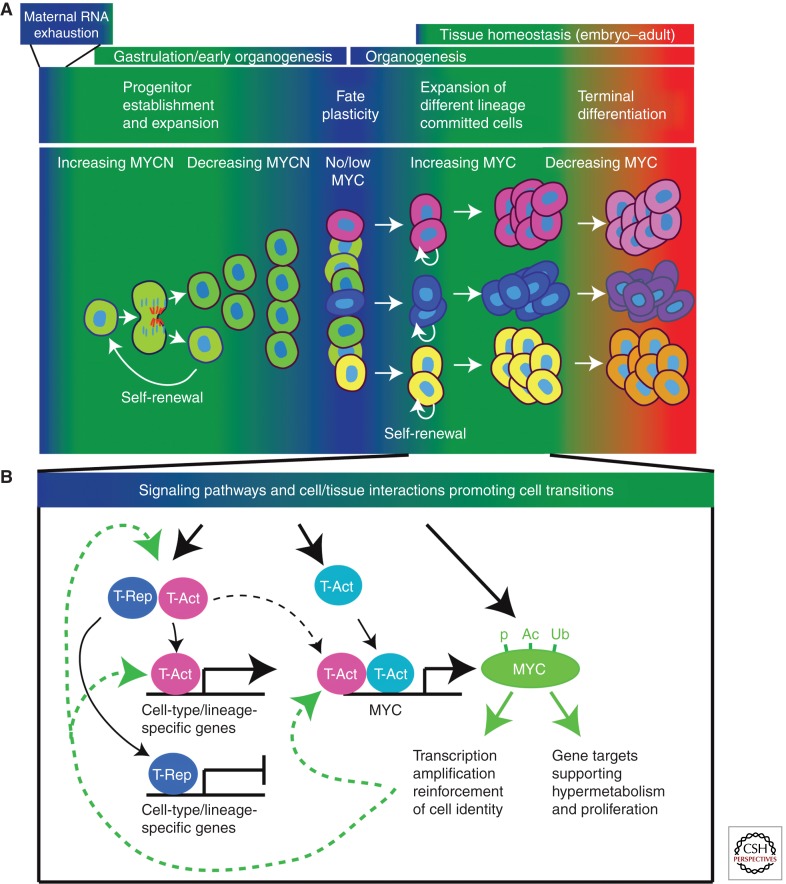
Epiblast deletion of MYC clearly shows that it is not required for proliferation and development of most tissues in the early embryo (up until E11.5) (Dubois et al. 2008). However, this might be expected because MYC RNA appears to be expressed at very low levels in tissues of early mouse, chick, xenopus, and human embryos during the time between exhaustion of maternal supplies of MYC RNA and commencement of organogenesis (Pfeifer-Ohlsson et al. 1985; King et al. 1986; Downs et al. 1989; Jaffredo et al. 1989; Stanton et al. 1992). In contrast to MYC, MYCN is not expressed or present only at low levels in extraembryonic tissues and not required for hematopoiesis (Laurenti et al. 2008), but is widely expressed in the embryo proper at developmental stages immediately before organogenesis (late gastrulation) and during early organogenesis (Mugrauer et al. 1988; Downs et al. 1989; Schmid et al. 1989; Hirvonen et al. 1990; Kato et al. 1991; Stanton et al. 1992; Hurlin et al. 1997). After organogenesis commences, MYCN is down-regulated in most if not all tissues, whereas MYC typically increases (Slamon and Cline 1984; Zimmerman et al. 1986; Mugrauer et al. 1988; Kato et al. 1991; Stanton et al. 1992). These data, together with the widespread failure in organogenesis that occurs in the absence of MYCN (Charron et al. 1992; Stanton et al. 1992; Sawai et al. 1993), are consistent with the notion that MYCN, perhaps with contributions from MYCL in some settings, is instrumental in the initial establishment and expansion of stem and progenitor populations at various strategic positions within developing organ systems (Fig. 2A).
Figure 2.
Speculative model for MYC-dependent regulation of cell plasticity and cell fate transitions during embryonic development. (A) Low levels of MYC expression and activity are proposed to contribute to increased cell plasticity in the early gastrulating embryo and again during early organogenesis. Increasing MYCN during gastrulation and early organogenesis contributes to the establishment and expansion of stem and progenitor populations at discreet locations within emerging organ systems. Following proliferative expansion, declining MYCN levels contribute to increased plasticity of these progenitor populations. Signaling that promotes the formation of new fate-restricted progenitors and stem cells also induces low levels of MYC, which functions to maintain their identity and self-renewal. Strong induction of MYC during commitment to specific lineages or to differentiation states within a given lineage couples reinforcement of cell identity with robust proliferative expansion. Subsequent MYC down-regulation then contributes to terminal differentiation. (B) Signaling that drives the development of cell identity and lineage transitions couples induction of specific transcription activators (T-Act) and repressors (T-Rep) with induction of MYC. The cell- or lineage-specific factors drive transcriptional and epigenetic reprogramming events that confer a unique cell identity (i.e., mesenchymal progenitor) or committed lineage (i.e., B cell). Coordinated MYC induction would act to stimulate and amplify transcription at poised and actively transcripted genes. MYC targets in this context would include the pioneer factors responsible for initiating fate transitions, as well as their transcriptional targets that ultimately define the functional identity of a cell or committed lineage. MYC induction may also couple fate transitions with proliferation by targeting a core set of genes involved in macromolecule biosynthesis and metabolism either undergoing active transcription or that exist in a transcriptionally poised state and therefore subject to regulation by MYC.
As organogenesis proceeds, MYCN levels subside and low levels of MYC appear to support stem and progenitor cell maintenance in a number of different settings, including the skin epidermis (Gandarillas and Watt 1997; Waikel et al. 2001), the intestinal epithelium (Bettess et al. 2005; Muncan et al. 2006), kidney (Couillard and Trudel 2009), pancreas (Nakhai et al. 2008; Bonal et al. 2009), lung (Okubo et al. 2005; Dong et al. 2011), and mammary gland (Stoelzle et al. 2009; Moumen et al. 2012). In these organ systems, MYC induction, which is often controlled by WNT/β-catenin signaling (Waikel et al. 2001; Hu and Rosenblum 2005; Shu et al. 2005; Nakhai et al. 2008), is then associated with the mobilization of stem cells and the proliferative expansion of specific lineages or differentiated cell types, including keratinocytes committed to terminal differentiation (Fig. 2A) (Gandarillas and Watt 1997; Gandarillas et al. 2000; Waikel et al. 2001; Zanet et al. 2005). Additionally, although MYCN does not play an essential role in establishing the hematopoietic system, it is expressed in hematopoietic stem cells and cooperates with MYC in hematopoietic stem cell (HSC) proliferation and self-renewal (Laurenti et al. 2008). Similar to its effects in skin keratinocytes, ectopic MYC in HSCs stimulates their exit from the stem cell niche and promotes the proliferative expansion of differentiated cell types at the expense of self-renewal (Wilson et al. 2004). In contrast to skin epithelial stem cells, the absence of MYC in HSCs leads to their accumulation at the expense of differentiation (Wilson et al. 2004). The latter phenomenon is attributed to defects in maintenance of the HSC niche (Wilson et al. 2004; Laurenti et al. 2008).
In the developing intestine at mid-gestation, MYCN is required for formation of the initial epithelial component of the intestinal tract (Stanton et al. 1992). Subsequent MYCN expression and activities in stem cells of the crypts of Lieberkühn contribute to homeostasis of the organ (Bettess et al. 2005; Muncan et al. 2006). Here, it is interesting to note that MYCN expression is retained in a single cell at the base of the intestinal crypt that lacks MYC and may be the intestinal stem cell (Bettess et al. 2005). Additionally, MYCN protein was found to be abundant in postmitotic villi (Bettess et al. 2005), suggesting potential proliferation-independent activities. Based on these findings, conditional MYCN deletion at these specific locations should be informative in defining how MYCN and MYC coordinate the development and maintain homeostasis of the intestine.
The coordinated actions of MYCN and MYC are particularly apparent in the development of the skeleton and connective tissues of the developing limb. There, undifferentiated mesenchymal progenitors of the emergent limb bud are dependent on MYCN for their proliferative expansion (Sawai et al. 1993; Ota et al. 2007). As the undifferentiated mesenchyme expands, the most centrally located cells, ones furthest from proproliferative WNT and FGF signaling emanating from the surface ectoderm, down-regulate MYCN, condense, and exit the cell cycle (Ten Berge et al. 2008). Chondrogenic progenitors that express little or no MYC and show minimal proliferation emerge from the condensing mesenchyme (Ota et al. 2007; Zhou et al. 2011). Subsequent induction of MYC, which may also be under the control of WNT/β-catenin signaling (Shung et al. 2012), contributes to the proliferative expansion of chondrocytes within the emerging cartilaginous growth plate of the long bones that drives bone growth (Zhou et al. 2011). Osteoblasts and other connective tissue lineages are also produced from the condensing mesenchyme or immediately adjacent cells in the central limb bud, and may also be dependent on the sequential actions of MYCN and MYC (Ota et al. 2007; Zhou et al. 2011). These findings are consistent with the idea that the severe down-regulation or absence of MYC expression and associated cell-cycle exit, and declining RNA polymerase II activity in the prechondrogenic condensation may contribute to or be necessary for epigenetic reprogramming to the chondrogenic lineage and other lineages (Fig. 2) (Zhou et al. 2011).
AMPLIFYING CELL FATE DURING DEVELOPMENT
As illustrated in the above examples, once stem and progenitor cell compartments are established in organ systems and their homeostatic tissue compartments, the basal level of MYC or MYCN and the timing of MYC induction is essential for maintaining the proper balance between stem cell renewal and the production and proliferative expansion of differentiated cell types (Fig. 2). In several developmental settings in which MYCN, MYC, or both MYCN and MYC are deleted, both reduced proliferation and the emergence of differentiated cell types is observed (Knoepfler et al. 2002; Okubo et al. 2005; Nakhai et al. 2008; Couillard and Trudel 2009; Harmelink and Jiao 2010; Kuwahara et al. 2010). Moreover, forced MYC expression has been shown to both promote proliferation and prevent differentiation in a variety of cell types (Eilers and Eisenman 2008). These findings generally support the idea that MYC may function during development to reinforce transcriptional programs that confer functional cell or lineage identities, including stem and progenitor identities. However, it is notable that forced MYC expression is compatible with differentiation within different lineages, including the B-cell (Habib et al. 2007) and T-cell lineages (Link et al. 2012), epidermal keratinocytes (Gandarillas and Watt 1997; Waikel et al. 2001), and more generally in the hematopoietic system (Wilson et al. 2004). Thus, forced MYC expression, or normal regulated induction of endogenous MYC within a specific lineage, does not inherently impede the activity of factors that drive differentiation events, and may even stimulate latent, active, or newly directed differentiation programs.
The finding that MYC has the general effect of amplifying active and stimulating poised transcription in cells (Lin et al. 2012; Nie et al. 2012) provides a new paradigm for viewing how MYC activity might influence cellular plasticity and fate determination. According to the amplification model, the absence of MYC, or MYC below some threshold level, is predicted to increase cell plasticity by restraining or preventing the transcription of factors responsible for installing specific identity or fates (Fig. 2). For the same reason, cells with low MYC might be more responsive to epigenetic reprogramming by factors that drive transitions in cell identity and/or lineage. Many signaling pathways known to strongly impact cell identity and fate during embryonic development, such as the WNT, RAS/MAPK, NOTCH, HEDGEHOG, and NF-κB, include dedicated mechanisms for transcriptional and posttranscriptional MYC induction (Sears 2004; Knoepfler and Kenney 2006; Sharma et al. 2007; Gerondakis and Siebenlist 2010; Clevers and Nusse 2012). The induction of MYC by such pathways may serve to drive or reinforce changes in cell identity by amplifying transcription of both the pioneer factors that initiate fate change and their transcriptional targets (Fig. 2B). In the setting of induced pluripotency, MYC transcription would act, at least in part, by reinforcing the expression and transcriptional targets of pluripotency factors such as Nanog, Sox2, and Oct4 (Lin et al. 2012; Nie et al. 2012; Soufi et al. 2012).
Although largely speculative, support for such a model is suggested by studies examining the relationship between MYC and global gene expression in different cell types where MYC was found to stimulate or maintain cell-type-specific transcriptional programs rather than installing a unique, MYC-specific transcriptional program (Lawlor et al. 2006; Lin et al. 2009, 2012; Ji et al. 2011; Lee et al. 2012; Nie et al. 2012; Pello et al. 2012). Although a number of studies suggest that MYC preferentially stimulates expression of genes involved in ribosome biogenesis, protein synthesis, and metabolism (Patel et al. 2004; Eilers and Eisenman 2008), this may simply reflect classes of genes that typically exist in a transcriptionally poised or active state in most cell types, and are therefore more generally subject to MYC-dependent attenuation of pausing and transcriptional amplification (Fig. 2). The existence of such a class of genes involved in promoting metabolism and macromolecule synthesis might also help explain the competitive advantage that MYC expression confers to cells (de la Cova et al. 2004; Moreno and Basler 2004) and what seems to be a near universal ability of MYC to promote proliferation (Littlewood et al. 2012). However, it is important to note that this model does not yet integrate MYC-dependent repression as a mechanism that contributes to cell identity and lineage development during development (Adhikary et al. 2003; Varlakhanova et al. 2011).
In the great majority of studies in which cells have been subjected to forced MYC expression, the target cells, whether in culture or in vivo, are already in a proliferative state and MYC has the general effect of supporting proliferation and preventing cell-cycle exit (Grandori et al. 2000). But even in postmitotic cells, MYC can induce cell-cycle entry and proliferation. For example, in vivo forced expression of MYC in postmitotic skin keratinocytes, forebrain neurons, adult myocytes, and support cells of adult utricles, led to their reentry into the cell cycle (Pelengaris et al. 1999; Xiao et al. 2001; Lee et al. 2009; Burns et al. 2012). However, while adenoviral-mediated expression of MYCN in cultured postmitotic sympathetic neurons also supported cell-cycle reentry, its expression in postmitotic cortical neurons failed to induce S-phase entry or progression (Wartiovaara et al. 2002). Additionally, forced expression of MYC caused cell-cycle reentry in quiescent epithelial cells of structurally unorganized mammary acini in organotypic culture, but its expression in quiescent cells within mature acini failed to induce cell-cycle reentry (Partanen et al. 2007).
Together with the finding that MYC and MYCN are found in postmitotic cells in vivo (Grady et al. 1987; Mugrauer et al. 1988; Hirvonen et al. 1990; Wakamatsu et al. 1993), these results are consistent with the presence of different classes of postmitotic cells, with some cells more poised to respond to MYC and reenter the cell cycle than others. Moreover, there is evidence of diversity in the epigenetic state of chromatin in postmitotic cells (Srivastava et al. 2010). Superimposing the transcriptional amplification model on these findings leads to the prediction that only postmitotic cells retaining proliferation-associated genes in a transcriptionally poised or low-level active state (but below a threshold needed to support proliferation) and therefore subject to induction by MYC would be capable of reentering the cell cycle in response to MYC. For postmitotic cells that cannot enter the cell cycle in response to forced MYC, their epigenetically repressed state may render them impervious to the actions of MYC (Fig. 1). For the latter cells, the actions of robust pioneer transcription factors capable of initiating epigenetic changes that increase accessibility to DNA binding by MYC (Fig. 1) would be required to make them responsive to the actions of MYC.
MYC IN DEVELOPMENTAL DISORDERS
As discussed above, MYCN haploinsufficiency causes the pleomorphic developmental disorder Feingold syndrome. There exist a number of developmental disorders related to Feingold syndrome referred to as VACTERL association, comprising vertebral defects, anal atresia, cardiac defects, tracheoesophageal fistula, renal malformations, and limb defects (Shaw-Smith 2006). Although MYCN mutations or deletions have not been identified in the wider collection of VACTERL association syndromes, the various causative genes in these syndromes (Shaw-Smith 2006) offer promising leads for upstream regulators of MYCN and perhaps for downstream targets and/or transcriptional programs that MYCN regulates.
There are no developmental disorders currently known that are directly caused by MYC mutations or deletion. However, MYC, as well as MYCN, is regulated by a variety of signaling pathways that not only influence cell identity and fate, but also cause developmental disorders. Thus, as in cancer, where MYC deregulation caused by mutations in pathways such as the RTK/RAS/MAPK and WNT/β-catenin is either proven or implicated to be critical for oncogenesis (Oskarsson et al. 2006; Sansom et al. 2007), it seems highly likely that misregulation of MYC plays an equally important underlying role in developmental disorders caused by defects in these and other signaling systems. For example, cardiac defects associated with the collection of developmental disorders known as rasopathies caused by mutations in the RAS-pathway (Sala et al. 2012) have features that often closely overlap with cardiac defects caused by ectopic MYC expression (Jackson et al. 1990). Additionally, in the developing skeleton, mutations in FGFR3 that cause achondroplasia lead to MYC down-regulation and impaired proliferation through a mechanism involving enhanced β-catenin degradation (Shung et al. 2012). MYC down-regulation appears to be a key mechanism in this disorder because conditional deletion of MYC in chondrocytes caused a similar achondroplasia-like phenotype (Zhou et al. 2011).
Finally, the collection of related developmental disorders known as cohesinopathies that include Cornelia de Lange syndrome and Roberts syndrome are linked to misregulation of MYC (Horsfield et al. 2012). Cohesinopathies are caused by mutations in components of the cohesin complex, including the cohesin subunits NIPPED-B, SMC1 and SMC3, RAD21, and the cohesin acetyltransferase ESCO2 (Horsfield et al. 2012). Cohesinopathies show considerable overlapping pathology with VACTERAL association and Feingold syndromes, suggesting potential involvement of MYCN. For example, these disorders typically feature short stature, dysmorphic facial features including micrognathia, limb defects, hearing loss, gastrointestinal defects including atresia, cardiac defects, genitourinary defects, and mental retardation (van Bokhoven et al. 2005; Shaw-Smith 2006; Horsfield et al. 2012). Interestingly, the cohesin complex, which is best known for its functions in sister chromatid exchange (Horsfield et al. 2012), shares with MYC the property of preferentially binding to genes being actively transcribed (Misulovin et al. 2008). Furthermore, cohesin has been linked to stimulation of transcription elongation through release of paused RNA polymerase II complexes (Fay et al. 2011). These results raise the intriguing possibility that the cohesin complex is mechanistically linked with MYC binding to transcriptionally paused and active genes (Guccione et al. 2006) and to transcription amplification (Lin et al. 2012; Nie et al. 2012).
CONCLUDING REMARKS
Although much is known about the activities of MYC and MYCN during embryonic development, much less is known about how these activities are coordinated by developmental cues. This is complicated by the great variety of signaling pathways that are implicated in controlling MYC abundance in cells and the multitude of ways that MYC activities can be regulated. In addition to regulation at the transcriptional, translational, and posttranslational levels, MYC activity has the potential to be significantly impacted by members of the extended network of MAX and MAX-like factor (MLX)-interacting transcription factors (Hurlin and Huang 2006; Dang 2012). At one level, establishing the temporal relationship between MYC induction or suppression in different developmental settings with emergent epigenetic signatures and gene regulation may help clarify how MYC impacts cell fate. However, a major challenge going forward is to integrate these relationships with a more complete understanding of how MYC expression and activity is determined. The promise of such work is the recognition of effective strategies for the therapeutic manipulation of MYC and its key activities in diverse disease and injury settings.
ACKNOWLEDGMENTS
P.J.H. is supported by grants from Shriners Hospitals for Children.
Footnotes
Editors: Chi V. Dang and Robert N. Eisenmann
Additional Perspectives on MYC and the Pathway to Cancer available at www.perspectivesinmedicine.org
REFERENCES
- Adhikary S, Eilers M 2005. Transcriptional regulation and transformation by Myc proteins. Nat Rev Mol Cell Biol 6: 635–645 [DOI] [PubMed] [Google Scholar]
- Adhikary S, Peukert K, Karsunky H, Beuger V, Lutz W, Elsässer H-P, Möröy T, Eilers M 2003. Miz1 is required for early embryonic development during gastrulation. Mol Cell Biol 23: 7648–7657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabi A, Wu S, Ridderstråle K, Bierhoff H, Shiue C, Fatyol K, Fahlén S, Hydbring P, Söderberg O, Grummt I, et al. 2005. c-Myc associates with ribosomal DNA and activates RNA polymerase I transcription. Nat Cell Biol 7: 303–310 [DOI] [PubMed] [Google Scholar]
- Ayer DE, Kretzner L, Eisenman RN 1993. Mad: A heterodimeric partner for Max that antagonizes Myc transcriptional activity. Cell 72: 211–222 [DOI] [PubMed] [Google Scholar]
- Bettess MD, Dubois N, Murphy MJ, Dubey C, Roger C, Robine S, Trumpp A 2005. c-Myc is required for the formation of intestinal crypts but dispensable for homeostasis of the adult intestinal epithelium. Mol Cell Biol 25: 7868–7878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackwood EM, Eisenman RN 1991. Max: A helix-loop-helix zipper protein that forms a sequence-specific DNA-binding complex with Myc. Science 251: 1211–1217 [DOI] [PubMed] [Google Scholar]
- Blackwood EM, Kretzner L, Eisenman RN 1992. Myc and Max function as a nucleoprotein complex. Curr Opin Genet Dev 2: 227–235 [DOI] [PubMed] [Google Scholar]
- Bonal C, Thorel F, Ait-Lounis A, Reith W, Trumpp A, Herrera PL 2009. Pancreatic inactivation of c-Myc decreases acinar mass and transdifferentiates acinar cells into adipocytes in mice. Gastroenterology 136: 309–319.e9 [DOI] [PubMed] [Google Scholar]
- Burnichon N, Cascón A, Schiavi F, Morales NP, Comino-Méndez I, Abermil N, Inglada-Pérez L, De Cubas AA, Amar L, Barontini M, et al. 2012. MAX mutations cause hereditary and sporadic pheochromocytoma and paraganglioma. Clin Cancer Res 18: 2828–2837 [DOI] [PubMed] [Google Scholar]
- Burns JC, Yoo JJ, Atala A, Jackson JD 2012. MYC gene delivery to adult mouse utricles stimulates proliferation of postmitotic supporting cells in vitro. PLoS ONE 7: e48704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartwright P, McLean C, Sheppard A, Rivett D, Jones K, Dalton S 2005. LIF/STAT3 controls ES cell self-renewal and pluripotency by a Myc-dependent mechanism. Development 132: 885–896 [DOI] [PubMed] [Google Scholar]
- Cawley S, Bekiranov S, Ng HH, Kapranov P, Sekinger EA, Kampa D, Piccolboni A, Sementchenko V, Cheng J, Williams AJ, et al. 2004. Unbiased mapping of transcription factor binding sites along human chromosomes 21 and 22 points to widespread regulation of noncoding RNAs. Cell 116: 499–509 [DOI] [PubMed] [Google Scholar]
- Celli J, Van Bokhoven H, Brunner HG 2003. Feingold syndrome: Clinical review and genetic mapping. Am J Med Genet A 122A: 294–300 [DOI] [PubMed] [Google Scholar]
- Chandriani S, Frengen E, Cowling VH, Pendergrass SA, Perou CM, Whitfield ML, Cole MD 2009. A core MYC gene expression signature is prominent in basal-like breast cancer but only partially overlaps the core serum response. PLoS ONE 4: e6693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charron J, Malynn BA, Fisher P, Stewart V, Jeannotte L, Goff SP, Robertson EJ, Alt FW 1992. Embryonic lethality in mice homozygous for a targeted disruption of the N-myc gene. Genes Dev 6: 2248–2257 [DOI] [PubMed] [Google Scholar]
- Clevers H, Nusse R 2012. Wnt/β-catenin signaling and disease. Cell 149: 1192–1205 [DOI] [PubMed] [Google Scholar]
- Cognet M, Nougayrede A, Malan V, Callier P, Cretolle C, Faivre L, Genevieve D, Goldenberg A, Heron D, Mercier S, et al. 2011. Dissection of the MYCN locus in Feingold syndrome and isolated oesophageal atresia. Eur J Hum Genet 19: 602–606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comino-Méndez I, Gracia-Aznárez FJ, Schiavi F, Landa I, Leandro-García LJ, Letón R, Honrado E, Ramos-Medina R, Caronia D, Pita G, et al. 2011. Exome sequencing identifies MAX mutations as a cause of hereditary pheochromocytoma. Nat Genet 43: 663–667 [DOI] [PubMed] [Google Scholar]
- Conacci-Sorrell M, Eisenman RN 2011. Post-translational control of Myc function during differentiation. Cell Cycle 10: 604–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conacci-Sorrell M, Ngouenet C, Eisenman RN 2010. Myc-nick: A cytoplasmic cleavage product of Myc that promotes α-tubulin acetylation and cell differentiation. Cell 142: 480–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couillard M, Trudel M 2009. C-myc as a modulator of renal stem/progenitor cell population. Dev Dyn 238: 405–414 [DOI] [PubMed] [Google Scholar]
- Cowling VH, Cole MD 2006. Mechanism of transcriptional activation by the Myc oncoproteins. Semin Cancer Biol 16: 242–252 [DOI] [PubMed] [Google Scholar]
- Cowling VH, Cole MD 2007. The Myc transactivation domain promotes global phosphorylation of the RNA polymerase II carboxy-terminal domain independently of direct DNA binding. Mol Cell Biol 27: 2059–2073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowling VH, Cole MD 2010. Myc regulation of mRNA cap methylation. Genes Cancer 1: 576–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang CV 2012. MYC on the path to cancer. Cell 149: 22–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis AC, Wims M, Spotts GD, Hann SR, Bradley A 1993. A null c-myc mutation causes lethality before 10.5 days of gestation in homozygotes and reduced fertility in heterozygous female mice. Genes Dev 7: 671–682 [DOI] [PubMed] [Google Scholar]
- de Jong EM, Felix JF, De Klein A, Tibboel D 2010. Etiology of esophageal atresia and tracheoesophageal fistula: “Mind the gap.” Curr Gastroenterol Rep 12: 215–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Cova C, Abril M, Bellosta P, Gallant P, Johnston LA 2004. Drosophila myc regulates organ size by inducing cell competition. Cell 117: 107–116 [DOI] [PubMed] [Google Scholar]
- Delgado MD, León J 2010. Myc roles in hematopoiesis and leukemia. Genes Cancer 1: 605–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Pontual L, Yao E, Callier P, Faivre L, Drouin V, Cariou S, Van Haeringen A, Geneviève D, Goldenberg A, Oufadem M, et al. 2011. Germline deletion of the miR-17 ∼ 92 cluster causes skeletal and growth defects in humans. Nat Genet 43: 1026–1030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domínguez-Frutos E, López-Hernández I, Vendrell V, Neves J, Gallozzi M, Gutsche K, Quintana L, Sharpe J, Knoepfler PS, Eisenman RN, et al. 2011. N-myc controls proliferation, morphogenesis, and patterning of the inner ear. J Neurosci 31: 7178–7189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez-Sola D, Ying CY, Grandori C, Ruggiero L, Chen B, Li M, Galloway DA, Gu W, Gautier J, Dalla-Favera R 2007. Non-transcriptional control of DNA replication by c-Myc. Nature 448: 445–451 [DOI] [PubMed] [Google Scholar]
- Dong J, Sutor S, Jiang G, Cao Y, Asmann YW, Wigle DA 2011. c-Myc regulates self-renewal in bronchoalveolar stem cells. PLoS ONE 6: e23707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downs KM, Martin GR, Bishop JM 1989. Contrasting patterns of myc and N-myc expression during gastrulation of the mouse embryo. Genes Dev 3: 860–869 [DOI] [PubMed] [Google Scholar]
- Dubois NC, Adolphe C, Ehninger A, Wang RA, Robertson EJ, Trumpp A 2008. Placental rescue reveals a sole requirement for c-Myc in embryonic erythroblast survival and hematopoietic stem cell function. Development 135: 2455–2465 [DOI] [PubMed] [Google Scholar]
- Eberhardy SR, Farnham PJ 2001. c-Myc mediates activation of the cad promoter via a post-RNA polymerase II recruitment mechanism. J Biol Chem 276: 48562–48571 [DOI] [PubMed] [Google Scholar]
- Eilers M, Eisenman RN 2008. Myc’s broad reach. Genes Dev 22: 2755–2766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay A, Misulovin Z, Li J, Schaaf CA, Gause M, Gilmour DS, Dorsett D 2011. Cohesin selectively binds and regulates genes with paused RNA polymerase. Curr Biol 21: 1624–1634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez PC, Frank SR, Wang L, Schroeder M, Liu S, Greene J, Cocito A, Amati B 2003. Genomic targets of the human c-Myc protein. Genes Dev 17: 1115–1129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank SR, Schroeder M, Fernandez P, Taubert S, Amati B 2001. Binding of c-Myc to chromatin mediates mitogen-induced acetylation of histone H4 and gene activation. Genes Dev 15: 2069–2082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallant P, Steiger D 2009. Myc’s secret life without Max. Cell Cycle 8: 3848–3853 [DOI] [PubMed] [Google Scholar]
- Gandarillas A, Watt FM 1997. c-Myc promotes differentiation of human epidermal stem cells. Genes Dev 11: 2869–2882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandarillas A, Davies D, Blanchard JM 2000. Normal and c-Myc-promoted human keratinocyte differentiation both occur via a novel cell cycle involving cellular growth and endoreplication. Oncogene 19: 3278–3289 [DOI] [PubMed] [Google Scholar]
- Gerondakis S, Siebenlist U 2010. Roles of the NF-κB pathway in lymphocyte development and function. Cold Spring Harb Perspect Biol 2: a000182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Roman N, Grandori C, Eisenman RN, White RJ 2003. Direct activation of RNA polymerase III transcription by c-Myc. Nature 421: 290–294 [DOI] [PubMed] [Google Scholar]
- Grady EF, Schwab M, Rosenau W 1987. Expression of N-myc and c-src during the development of fetal human brain. Cancer Res 47: 2931–2936 [PubMed] [Google Scholar]
- Grandori C, Cowley SM, James LP, Eisenman RN 2000. The Myc/Max/Mad network and the transcriptional control of cell behavior. Annu Rev Cell Dev Biol 16: 653–699 [DOI] [PubMed] [Google Scholar]
- Grandori C, Gomez-Roman N, Felton-Edkins ZA, Ngouenet C, Galloway DA, Eisenman RN, White RJ 2005. c-Myc binds to human ribosomal DNA and stimulates transcription of rRNA genes by RNA polymerase I. Nat Cell Biol 7: 311–318 [DOI] [PubMed] [Google Scholar]
- Grewal SS, Li L, Orian A, Eisenman RN, Edgar BA 2005. Myc-dependent regulation of ribosomal RNA synthesis during Drosophila development. Nat Cell Biol 7: 295–302 [DOI] [PubMed] [Google Scholar]
- Guccione E, Martinato F, Finocchiaro G, Luzi L, Tizzoni L, Dall’ Olio V, Zardo G, Nervi C, Bernard L, Amati B 2006. Myc-binding-site recognition in the human genome is determined by chromatin context. Nat Cell Biol 8: 764–770 [DOI] [PubMed] [Google Scholar]
- Habib T, Park H, Tsang M, De Alborán IM, Nicks A, Wilson L, Knoepfler PS, Andrews S, Rawlings DJ, Eisenman RN, et al. 2007. Myc stimulates B lymphocyte differentiation and amplifies calcium signaling. J Cell Biol 179: 717–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmelink CM, Jiao K 2010. Critical functions of myocardial Mycn in the developing mouse heart. Dev Biol 356: 153 [Google Scholar]
- Harmelink C, Peng Y, DeBenedittis P, Chen H, Shou W, Jiao K 2013. Myocardial Mycn is essential for mouse ventricular wall morphogenesis. Dev Biol 373: 53–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatton KS, Mahon K, Chin L, Chiu FC, Lee HW, Peng D, Morgenbesser SD, Horner J, DePinho RA 1996. Expression and activity of L-Myc in normal mouse development. Mol Cell Biol 16: 1794–1804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He C, Hu H, Braren R, Fong S-Y, Trumpp A, Carlson TR, Wang RA 2008. c-myc in the hematopoietic lineage is critical for its angiogenic function in the mouse embryo. Development 135: 2467–2477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He S, Liu Z, Oh D-Y, Thiele CJ 2013. MYCN and the epigenome. Front Oncol 3: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herkert B, Eilers M 2010. Transcriptional repression: The dark side of myc. Genes Cancer 1: 580–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirvonen H, Mäkelä TP, Sandberg M, Kalimo H, Vuorio E, Alitalo K 1990. Expression of the myc proto-oncogenes in developing human fetal brain. Oncogene 5: 1787–1797 [PubMed] [Google Scholar]
- Hopewell R, Ziff EB 1995. The nerve growth factor-responsive PC12 cell line does not express the Myc dimerization partner Max. Mol Cell Biol 15: 3470–3478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horsfield JA, Print CG, Mönnich M 2012. Diverse developmental disorders from the one ring: Distinct molecular pathways underlie the cohesinopathies. Front Genet 3: 171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu MC, Rosenblum ND 2005. Smad1, β-catenin and Tcf4 associate in a molecular complex with the Myc promoter in dysplastic renal tissue and cooperate to control Myc transcription. Development 132: 215–225 [DOI] [PubMed] [Google Scholar]
- Hurlin PJ, Huang J 2006. The MAX-interacting transcription factor network. Semin Cancer Biol 16: 265–274 [DOI] [PubMed] [Google Scholar]
- Hurlin PJ, Quéva C, Eisenman RN 1997. Mnt, a novel Max-interacting protein is coexpressed with Myc in proliferating cells and mediates repression at Myc binding sites. Genes Dev 11: 44–58 [DOI] [PubMed] [Google Scholar]
- Hurlin PJ, Steingrìmsson E, Copeland NG, Jenkins NA, Eisenman RN 1999. Mga, a dual-specificity transcription factor that interacts with Max and contains a T-domain DNA-binding motif. EMBO J 18: 7019–7028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurlin PJ, Zhou Z-Q, Toyo-oka K, Ota S, Walker WL, Hirotsune S, Wynshaw-Boris A 2003. Deletion of Mnt leads to disrupted cell cycle control and tumorigenesis. EMBO J 22: 4584–4596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson T, Allard MF, Sreenan CM, Doss LK, Bishop SP, Swain JL 1990. The c-myc proto-oncogene regulates cardiac development in transgenic mice. Mol Cell Biol 10: 3709–3716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffredo T, Vandenbunder B, Dieterlen-Lièvre F 1989. In situ study of c-myc protein expression during avian development. Development 105: 679–695 [DOI] [PubMed] [Google Scholar]
- Ji H, Wu G, Zhan X, Nolan A, Koh C, De Marzo A, Doan HM, Fan J, Cheadle C, Fallahi M, et al. 2011. Cell-type independent MYC target genes reveal a primordial signature involved in biomass accumulation. PLoS ONE 6: e26057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato K, Kanamori A, Wakamatsu Y, Sawai S, Kondoh H 1991. Tissue distribution of N-myc expression in the early organogenesis period of the mouse embryo. Dev Growth Differ 33: 29–39 [DOI] [PubMed] [Google Scholar]
- Kidder BL, Yang J, Palmer S 2008. Stat3 and c-Myc genome-wide promoter occupancy in embryonic stem cells. PLoS ONE 3: e3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King MW, Roberts JM, Eisenman RN 1986. Expression of the c-myc proto-oncogene during development of Xenopus laevis. Mol Cell Biol 6: 4499–4508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoepfler PS, Kenney AM 2006. Neural precursor cycling at sonic speed: N-Myc pedals, GSK-3 brakes. Cell Cycle 5: 47–52 [DOI] [PubMed] [Google Scholar]
- Knoepfler PS, Cheng PF, Eisenman RN 2002. N-myc is essential during neurogenesis for the rapid expansion of progenitor cell populations and the inhibition of neuronal differentiation. Genes Dev 16: 2699–2712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoepfler PS, Zhang X, Cheng PF, Gafken PR, McMahon SB, Eisenman RN 2006. Myc influences global chromatin structure. EMBO J 25: 2723–2734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopecky BJ, Decook R, Fritzsch B 2012. N-Myc and L-Myc are essential for hair cell formation but not maintenance. Brain Res 1484: 1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwahara A, Hirabayashi Y, Knoepfler PS, Taketo MM, Sakai J, Kodama T, Gotoh Y 2010. Wnt signaling and its downstream target N-myc regulate basal progenitors in the developing neocortex. Development 137: 1035–1044 [DOI] [PubMed] [Google Scholar]
- Laurenti E, Varnum-Finney B, Wilson A, Ferrero I, Blanco-Bose WE, Ehninger A, Knoepfler PS, Cheng P-F, MacDonald HR, Eisenman RN, et al. 2008. Hematopoietic stem cell function and survival depend on c-Myc and N-Myc activity. Cell Stem Cell 3: 611–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurenti E, Wilson A, Trumpp A 2009. Myc’s other life: Stem cells and beyond. Curr Opin Cell Biol 21: 844–854 [DOI] [PubMed] [Google Scholar]
- Lawlor ER, Soucek L, Brown-Swigart L, Shchors K, Bialucha CU, Evan GI 2006. Reversible kinetic analysis of Myc targets in vivo provides novel insights into Myc-mediated tumorigenesis. Cancer Res 66: 4591–4601 [DOI] [PubMed] [Google Scholar]
- Lee H, Casadesus G, Nunomura A, Zhu X, Castellani RJ, Richardson SL, Perry G, Felsher DW, Petersen RB, Smith MA 2009. The neuronal expression of MYC causes a neurodegenerative phenotype in a novel transgenic mouse. Am J Pathol 174: 891–897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee B-K, Bhinge AA, Battenhouse A, McDaniell RM, Liu Z, Song L, Ni Y, Birney E, Lieb JD, Furey TS, et al. 2012. Cell-type specific and combinatorial usage of diverse transcription factors revealed by genome-wide binding studies in multiple human cells. Genome Res 22: 9–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Ohgi KA, Zhang J, Krones A, Bush KT, Glass CK, Nigam SK, Aggarwal AK, Maas R, Rose DW, et al. 2003a. Eya protein phosphatase activity regulates Six1–Dach–Eya transcriptional effects in mammalian organogenesis. Nature 426: 247–254 [DOI] [PubMed] [Google Scholar]
- Li Z, Van Calcar S, Qu C, Cavenee WK, Zhang MQ, Ren B 2003b. A global transcriptional regulatory role for c-Myc in Burkitt’s lymphoma cells. Proc Natl Acad Sci 100: 8164–8169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C-H, Lin C, Tanaka H, Fero ML, Eisenman RN 2009. Gene regulation and epigenetic remodeling in murine embryonic stem cells by c-Myc. PLoS ONE 4: e7839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CY, Lovén J, Rahl PB, Paranal RM, Burge CB, Bradner JE, Lee TI, Young RA 2012. Transcriptional amplification in tumor cells with elevated c-Myc. Cell 151: 56–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Link JM, Ota S, Zhou Z-Q, Daniel CJ, Sears RC, Hurlin PJ 2012. A critical role for Mnt in Myc-driven T-cell proliferation and oncogenesis. Proc Natl Acad Sci 109: 19685–19690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Littlewood TD, Kreuzaler P, Evan GI 2012. All things to all people. Cell 151: 11–13 [DOI] [PubMed] [Google Scholar]
- Malynn BA, De Alboran IM, O’Hagan RC, Bronson R, Davidson L, DePinho RA, Alt FW 2000. N-myc can functionally replace c-myc in murine development, cellular growth, and differentiation. Genes Dev 14: 1390–1399 [PMC free article] [PubMed] [Google Scholar]
- Martinato F, Cesaroni M, Amati B, Guccione E 2008. Analysis of Myc-induced histone modifications on target chromatin. PLoS ONE 3: e3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misulovin Z, Schwartz YB, Li X-Y, Kahn TG, Gause M, MacArthur S, Fay JC, Eisen MB, Pirrotta V, Biggin MD, et al. 2008. Association of cohesin and Nipped-B with transcriptionally active regions of the Drosophila melanogaster genome. Chromosoma 117: 89–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moens CB, Auerbach AB, Conlon RA, Joyner AL, Rossant J 1992. A targeted mutation reveals a role for N-myc in branching morphogenesis in the embryonic mouse lung. Genes Dev 6: 691–704 [DOI] [PubMed] [Google Scholar]
- Moens CB, Stanton BR, Parada LF, Rossant J 1993. Defects in heart and lung development in compound heterozygotes for two different targeted mutations at the N-myc locus. Development 119: 485–499 [DOI] [PubMed] [Google Scholar]
- Moreno E, Basler K 2004. dMyc transforms cells into super-competitors. Cell 117: 117–129 [DOI] [PubMed] [Google Scholar]
- Moumen M, Chiche A, Deugnier M-A, Petit V, Gandarillas A, Glukhova MA, Faraldo MM 2012. The proto-oncogene Myc is essential for mammary stem cell function. Stem Cells 30: 1246–1254 [DOI] [PubMed] [Google Scholar]
- Mousavi K, Sartorelli V 2010. Myc-nick: The force behind c-Myc. Sci Signal 3: e49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mugrauer G, Alt FW, Ekblom P 1988. N-myc proto-oncogene expression during organogenesis in the developing mouse as revealed by in situ hybridization. J Cell Biol 107: 1325–1335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muncan V, Sansom OJ, Tertoolen L, Phesse TJ, Begthel H, Sancho E, Cole AM, Gregorieff A, De Alboran IM, Clevers H, et al. 2006. Rapid loss of intestinal crypts upon conditional deletion of the Wnt/Tcf-4 target gene c-Myc. Mol Cell Biol 26: 8418–8426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy DJ, Junttila MR, Pouyet L, Karnezis A, Shchors K, Bui DA, Brown-Swigart L, Johnson L, Evan GI 2008. Distinct thresholds govern Myc’s biological output in vivo. Cancer Cell 14: 447–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy A, Moens C, Ivanyi E, Pawling J, Gertsenstein M, Hadjantonakis A-K, Pirity M, Rossant J 1998. Dissecting the role of N-myc in development using a single targeting vector to generate a series of alleles. Curr Biol 8: 661–666 [DOI] [PubMed] [Google Scholar]
- Nakhai H, Siveke JT, Mendoza-Torres L, Schmid RM 2008. Conditional inactivation of Myc impairs development of the exocrine pancreas. Development 135: 3191–3196 [DOI] [PubMed] [Google Scholar]
- Nature Medicine Commentaries 2013. Unlocking the mysterious mechanisms of MYC. Nat Med 19: 26–27 [DOI] [PubMed] [Google Scholar]
- Nesbit CE, Tersak JM, Prochownik EV 1999. MYC oncogenes and human neoplastic disease. Oncogene 18: 3004–3016 [DOI] [PubMed] [Google Scholar]
- Nie Z, Hu G, Wei G, Cui K, Yamane A, Resch W, Wang R, Green DR, Tessarollo L, Casellas R, et al. 2012. c-Myc is a universal amplifier of expressed genes in lymphocytes and embryonic stem cells. Cell 151: 68–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okubo T, Knoepfler PS, Eisenman RN, Hogan BLM 2005. Nmyc plays an essential role during lung development as a dosage-sensitive regulator of progenitor cell proliferation and differentiation. Development 132: 1363–1374 [DOI] [PubMed] [Google Scholar]
- Orian A, Van Steensel B, Delrow J, Bussemaker HJ, Li L, Sawado T, Williams E, Loo LWM, Cowley SM, Yost C, et al. 2003. Genomic binding by the Drosophila Myc, Max, Mad/Mnt transcription factor network. Genes Dev 17: 1101–1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oskarsson T, Essers MAG, Dubois N, Offner S, Dubey C, Roger C, Metzger D, Chambon P, Hummler E, Beard P, et al. 2006. Skin epidermis lacking the c-myc gene is resistant to Ras-driven tumorigenesis but can reacquire sensitivity upon additional loss of the p21Cip1 gene. Genes Dev 20: 2024–2029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ota S, Zhou Z-Q, Keene DR, Knoepfler P, Hurlin PJ 2007. Activities of N-Myc in the developing limb link control of skeletal size with digit separation. Development 134: 1583–1592 [DOI] [PubMed] [Google Scholar]
- Partanen JI, Nieminen AI, Mäkelä TP, Klefstrom J 2007. Suppression of oncogenic properties of c-Myc by LKB1-controlled epithelial organization. Proc Natl Acad Sci 104: 14694–14699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel JH, Loboda AP, Showe MK, Showe LC, McMahon SB 2004. Analysis of genomic targets reveals complex functions of MYC. Nat Rev Cancer 4: 562–568 [DOI] [PubMed] [Google Scholar]
- Pelengaris S, Littlewood T, Khan M, Elia G, Evan G 1999. Reversible activation of c-Myc in skin: Induction of a complex neoplastic phenotype by a single oncogenic lesion. Mol Cell 3: 565–577 [DOI] [PubMed] [Google Scholar]
- Pello OM, Pizzol MD, Mirolo M, Soucek L, Zammataro L, Amabile A, Doni A, Nebuloni M, Swigart LB, Evan GI, et al. 2012. Role of c-MYC in alternative activation of human macrophages and tumor-associated macrophage biology. Blood 119: 411–421 [DOI] [PubMed] [Google Scholar]
- Pfeifer-Ohlsson S, Rydnert J, Goustin AS, Larsson E, Betsholtz C, Ohlsson R 1985. Cell-type-specific pattern of myc protooncogene expression in developing human embryos. Proc Natl Acad Sci 82: 5050–5054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahl PB, Lin CY, Seila AC, Flynn RA, McCuine S, Burge CB, Sharp PA, Young RA 2010. c-Myc regulates transcriptional pause release. Cell 141: 432–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribon V, Leff T, Saltiel AR 1994. c-Myc does not require max for transcriptional activity in PC-12 cells. Mol Cell Neurosci 5: 277–282 [DOI] [PubMed] [Google Scholar]
- Sala V, Gallo S, Leo C, Gatti S, Gelb BD, Crepaldi T 2012. Signaling to cardiac hypertrophy: Insights from human and mouse RASopathies. Mol Med 18: 938–947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sansom OJ, Meniel VS, Muncan V, Phesse TJ, Wilkins JA, Reed KR, Vass JK, Athineos D, Clevers H, Clarke AR 2007. Myc deletion rescues Apc deficiency in the small intestine. Nature 446: 676–679 [DOI] [PubMed] [Google Scholar]
- Sawai S, Shimono A, Wakamatsu Y, Palmes C, Hanaoka K, Kondoh H 1993. Defects of embryonic organogenesis resulting from targeted disruption of the N-myc gene in the mouse. Development 117: 1445–1455 [DOI] [PubMed] [Google Scholar]
- Schmid P, Schulz WA, Hameister H 1989. Dynamic expression pattern of the myc protooncogene in midgestation mouse embryos. Science 243: 226–229 [DOI] [PubMed] [Google Scholar]
- Sears RC 2004. The life cycle of C-myc: From synthesis to degradation. Cell Cycle 3: 1133–1137 [PubMed] [Google Scholar]
- Sharma VM, Draheim KM, Kelliher MA 2007. The Notch1/c-Myc pathway in T cell leukemia. Cell Cycle 6: 927–930 [DOI] [PubMed] [Google Scholar]
- Shaw-Smith C 2006. Oesophageal atresia, tracheo-oesophageal fistula, and the VACTERL association: Review of genetics and epidemiology. J Med Genet 43: 545–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen-Li H, O’Hagan RC, Hou H, Horner JW, Lee H-W, DePinho RA 2000. Essential role for Max in early embryonic growth and development. Genes Dev 14: 17–22 [PMC free article] [PubMed] [Google Scholar]
- Shu W, Guttentag S, Wang Z, Andl T, Ballard P, Lu MM, Piccolo S, Birchmeier W, Whitsett JA, Millar SE, et al. 2005. Wnt/β-catenin signaling acts upstream of N-myc, BMP4, and FGF signaling to regulate proximal-distal patterning in the lung. Dev Biol 283: 226–239 [DOI] [PubMed] [Google Scholar]
- Shung C-Y, Ota S, Zhou Z-Q, Keene DR, Hurlin PJ 2012. Disruption of a Sox9-β-catenin circuit by mutant Fgfr3 in thanatophoric dysplasia type II. Hum Mol Genet 21: 4628–4644 [DOI] [PubMed] [Google Scholar]
- Slamon DJ, Cline MJ 1984. Expression of cellular oncogenes during embryonic and fetal development of the mouse. Proc Natl Acad Sci 81: 7141–7145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soufi A, Donahue G, Zaret KS 2012. Facilitators and impediments of the pluripotency reprogramming factors’ initial engagement with the genome. Cell 151: 994–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava S, Mishra RK, Dhawan J 2010. Regulation of cellular chromatin state. Organogenesis 6: 37–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanton BR, Perkins AS, Tessarollo L, Sassoon DA, Parada LF 1992. Loss of N-myc function results in embryonic lethality and failure of the epithelial component of the embryo to develop. Genes Dev 6: 2235–2247 [DOI] [PubMed] [Google Scholar]
- Steiger D, Furrer M, Schwinkendorf D, Gallant P 2008. Max-independent functions of Myc in Drosophila melanogaster. Nat Genet 40: 1084–1091 [DOI] [PubMed] [Google Scholar]
- Stoelzle T, Schwarb P, Trumpp A, Hynes NE 2009. c-Myc affects mRNA translation, cell proliferation and progenitor cell function in the mammary gland. BMC Biol 7: 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S 2006. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126: 663–676 [DOI] [PubMed] [Google Scholar]
- ten Berge D, Brugmann SA, Helms JA, Nusse R 2008. Wnt and FGF signals interact to coordinate growth with cell fate specification during limb development. Development 135: 3247–3257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres R, Schreiber-Agus N, Morgenbesser SD, DePinho RA 1992. Myc and Max: A putative transcriptional complex in search of a cellular target. Curr Opin Cell Biol 4: 468–474 [DOI] [PubMed] [Google Scholar]
- Trumpp A, Refaeli Y, Oskarsson T, Gasser S, Murphy M, Martin GR, Bishop JM 2001. c-Myc regulates mammalian body size by controlling cell number but not cell size. Nature 414: 768–773 [DOI] [PubMed] [Google Scholar]
- van Bokhoven H, Celli J, Van Reeuwijk J, Rinne T, Glaudemans B, Van Beusekom E, Rieu P, Newbury-Ecob RA, Chiang C, Brunner HG 2005. MYCN haploinsufficiency is associated with reduced brain size and intestinal atresias in Feingold syndrome. Nat Genet 37: 465–467 [DOI] [PubMed] [Google Scholar]
- Varlakhanova N, Cotterman R, Bradnam K, Korf I, Knoepfler PS 2011. Myc and Miz-1 have coordinate genomic functions including targeting Hox genes in human embryonic stem cells. Epigenetics Chromatin 4: 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waikel RL, Kawachi Y, Waikel PA, Wang XJ, Roop DR 2001. Deregulated expression of c-Myc depletes epidermal stem cells. Nat Genet 28: 165–168 [DOI] [PubMed] [Google Scholar]
- Wakamatsu Y, Watanabe Y, Shimono A, Kondoh H 1993. Transition of localization of the N-Myc protein from nucleus to cytoplasm in differentiating neurons. Neuron 10: 1–9 [DOI] [PubMed] [Google Scholar]
- Wartiovaara K, Barnabe-Heider F, Miller FD, Kaplan DR 2002. N-myc promotes survival and induces S-phase entry of postmitotic sympathetic neurons. J Neurosci 22: 815–824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wernig M, Meissner A, Foreman R, Brambrink T, Ku M, Hochedlinger K, Bernstein BE, Jaenisch R 2007. In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature 448: 318–324 [DOI] [PubMed] [Google Scholar]
- Wilson A, Murphy MJ, Oskarsson T, Kaloulis K, Bettess MD, Oser GM, Pasche A-C, Knabenhans C, Macdonald HR, Trumpp A 2004. c-Myc controls the balance between hematopoietic stem cell self-renewal and differentiation. Genes Dev 18: 2747–2763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao G, Mao S, Baumgarten G, Serrano J, Jordan MC, Roos KP, Fishbein MC, MacLellan WR 2001. Inducible activation of c-Myc in adult myocardium in vivo provokes cardiac myocyte hypertrophy and reactivation of DNA synthesis. Circ Res 89: 1122–1129 [DOI] [PubMed] [Google Scholar]
- Zanet J, Pibre S, Jacquet C, Ramirez A, de Alborán IM, Gandarillas A 2005. Endogenous Myc controls mammalian epidermal cell size, hyperproliferation, endoreplication and stem cell amplification. J Cell Sci 118: 1693–1704 [DOI] [PubMed] [Google Scholar]
- Zeller KI, Zhao X, Lee CWH, Chiu KP, Yao F, Yustein JT, Ooi HS, Orlov YL, Shahab A, Yong HC, et al. 2006. Global mapping of c-Myc binding sites and target gene networks in human B cells. Proc Natl Acad Sci 103: 17834–17839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zervos AS, Gyuris J, Brent R 1993. Mxi1, a protein that specifically interacts with Max to bind Myc-Max recognition sites. Cell 72: 223–232 [DOI] [PubMed] [Google Scholar]
- Zhou Z-Q, Shung C-Y, Ota S, Akiyama H, Keene DR, Hurlin PJ 2011. Sequential and coordinated actions of c-Myc and N-Myc control appendicular skeletal development. PLoS ONE 6: e18795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman KA, Yancopoulos GD, Collum RG, Smith RK, Kohl NE, Denis KA, Nau MM, Witte ON, Toran-Allerand D, Gee CE 1986. Differential expression of myc family genes during murine development. Nature 319: 780–783 [DOI] [PubMed] [Google Scholar]