Abstract
Developmental studies on wing colour patterns have been performed in nymphalid butterflies, but efficient genetic manipulations, including mutagenesis, have not been well established. Here, we have performed mutagenesis experiments in a lycaenid butterfly, the pale grass blue Zizeeria maha, to produce colour-pattern mutants. We fed the P-generation larvae an artificial diet containing the mutagen ethyl methane sulfonate (EMS), and the F1- and F2-generation adults showed various aberrant colour patterns: dorsoventral transformation, anterioposterior background colouration gap, weak contrast, disarrangement of spots, reduction of the size of spots, loss of spots, fusion of spots, and ectopic spots. Among them, the disarrangement, reduction, and loss of spots were likely produced by the coordinated changes of many spots of a single wing around the discal spot in a system-dependent manner, demonstrating the existence of the central symmetry system. The present study revealed multiple genetic regulations for system-dependent and wing-wide colour-pattern determination in lycaenid butterflies.
The diverse colour patterns of butterfly wings are considered an excellent system to study the development and evolution of biological patterns1. As a foundation for understanding the wing colour-pattern diversity of butterflies, Schwanwitsch2 and Süffert3 proposed a general scheme called the nymphalid groundplan, which was extended later by Nijhout1,4. This groundplan, which specifies the positions of colour-pattern elements on a two-dimensional plane in nymphalid butterfly wings, was recently revised and further extended5,6. The revised groundplan6 states that the overall wing colour pattern comprises five elemental systems: the central, basal, and border symmetry systems, the wing-root band system, and the marginal band system4. A single system is basically composed of a core element and a pair of paracore elements that are remotely positioned from the core element. That is, a core element and a pair of paracore elements may be regulated together in a system-dependent manner. The developmental mechanisms that produce this basic system configuration have been proposed to be a reaction-diffusion mechanism with additional components6,7,8,9,10.
Among the colour-pattern elements defined by the groundplan is the eyespot, a conspicuous element that belongs to the border symmetry system in nymphalid butterflies. The eyespots of the nymphalid butterfly Junonia coenia, a species that has large dorsal eyespots, have been intensively studied surgically and physiologically1,11,12,13,14,15. Our group has been using related nymphalid butterflies such as J. orithya16,17, J. almana9,18, Vanessa indica19,20, and V. cardui21,22, all of which belong to the subfamily Nymphalinae, for physiological and evolutionary studies.
A distinct nymhalid butterfly, Bicyclus anynana, which belongs to the subfamily Satyrinae, has been considered a model butterfly system for molecular biological studies23,24. Compared with the previously mentioned species, the eyespot structures and overall wing colour patterns of B. anynana are relatively simple, making this species useful as a model organism. Partly due to this characteristic, B. anynana has been subjected to genome-wide molecular analyses25,26,27. Likewise, some genetic manipulation methods have been devised for this and other butterflies. For example, transgenic butterflies have been produced28,29,30, gene delivery using vaccinia virus31,32, sindbis virus33, and electroporation34 have been reported, and RNAi methods have been evaluated35. Despite some success, however, a fundamental lack of efficient gene manipulation techniques for butterflies has hampered the molecular functional analysis of candidate genes for colour-pattern development such as distal-less, engrailed, and spalt15,36,37,38. An X-ray-induced mutagenesis study using B. anynana revealed compartment-dependent modular control of eyespot development39. The mutant phenotype, i.e., the loss of two specific eyespots, suggested that each eyespot is genetically controlled independently. Curiously, no other laboratory-induced mutants have been reported, although a few different spontaneous mutants have been analysed intensively40,41.
Philosophically speaking, despite the apparent advantage of the B. anynana system, focusing on a single simple system when attempting to study biological diversity might be somewhat misleading. Thus, studies of other butterfly systems may be encouraged. Consistent with this notion, a comparison of the expression patterns of several genes in different species of butterflies revealed developmental diversity at the molecular level38,42. Recent genomic studies focusing on other nymphalid butterflies, such as the monarch butterfly, Danaus plexippus43, and Heliconius butterflies44,45,46,47,48, could eventually reveal the molecular diversity of colour-pattern development in butterflies. In addition, it is interesting to study not only a single eyespot or another element but also a symmetry system that covers a large wing area as a whole because the symmetry systems are basic features of the nymphalid groundplan1,2,3,4,5,6.
Lycaenid colour patterns have been analysed based on the nymphalid groundplan1, but lycaenid butterflies have largely been beyond the scope of the field of butterfly biology except for some physiological and evolutionary studies19,49,50, although their colour patterns are almost as diverse as those of nymphalid butterflies. We hypothesised that lycaenid colour patterns are constructed based on the core-paracore principle5,6, similarly to nymphalid colour patterns, despite possible differences in elemental positioning between the lycaenid and nymphalid butterflies. That is, we may be able to find lycaenid mutants in which many wing spots may “move” or “disappear” together in a system-dependent manner. In the present study, we performed mutagenesis experiments, focusing on the pale grass blue Zizeeria maha (Fig. 1a, b), a common lycaenid butterfly distributed widely in Japan. Its rearing method, including how to make and use the artificial diet, has already been established51. The ventral wings contain well-defined arrays of black spots against light background colouration, which makes it easy to analyse any colour-pattern aberrations.
Figure 1. Terminology for colour-pattern elements and non-mutant phenotypes of Z. maha.

(a) The ventral side of the fore- and hindwings, defined as “normal type”. (b) The terminology of the spot arrays in the ventral hindwing. From the peripheral to the basal arrays, the arrays are referred to as the first, second, third, and fourth (basal) spot arrays. (c) A panda-type adult, indicating genetic deterioration due to sibling crosses. The second spot array almost entirely disappeared, and the first spot array was also reduced in size. In contrast, the third spot array was distinct. Background was paler than usual. Overall, scale development appeared to be impaired. This individual was an F3 adult from Experiment 1 in the control experiments (no EMS). (d) A crescent-type adult. A spot in the third array showed a crescent shape (an orange arrow). Overall paler background was also conspicuous. This individual is an F3 adult from Experiment 1 in the control experiments (no EMS). (e, f) An adult with wing-size asymmetry. Right wings are smaller than left wings. Wing-size difference is indicated by two red arrows. This individual is an F1b adult from the mutagenesis experiments, but this phenotype was also observed in an F3 individual from Experiment 1 in the control experiments (no EMS).
Here, we used ethyl methane sulfonate (EMS), a mutagen that efficiently induces point mutations52,53,54. It has been known that genetic mosaics were known to be obtained in the F1 generation in the EMS-induced mutagenesis experiments52,53,54. Mosaic mutants are produced because a single-base damage in one strand of the post-mitotic sperm DNA is repaired after the first zygotic division52,53,54. We took advantage of the mosaic analysis, because colour patterns of one wing side (i.e., right or left wings) or a part of a wing may be modified in the mosaic individuals, which can be compared directly to the non-modified wings of the same individual. This comparison helped us to identify mutants and to understand how colour patterns are developmentally determined. We successfully obtained several aberrant colour-pattern mutants in the F1 and F2 generations. These mutant phenotypes revealed system-dependent regulation of spot development, an important aspect of how wing colour patterns are determined in lycaenid butterflies.
Results
No spontaneous colour-pattern modification without EMS
To assess frequency of spontaneous colour-pattern changes without EMS treatment, we performed three sets of rearing experiments, in which many individuals were reared and their colour patterns were examined (Supplementary Table 1). In the first, second, and third sets of experiments, we obtained 550, 402, and 107 adults, respectively. Among these 1,059 adults, we found no individual with color-pattern modification (MR [modification rate] = 0%). On the other hand, we found a small number of the so-called panda type (Fig. 1c, Supplementary Table 1), which is an indication of genetic deterioration due to sibling crosses51, and hence were excluded from our “modification” category. We also found a small number of the so-called crescent type (Fig. 1d, Supplementary Table 1), which is often accompanied by the panda-type traits but is also relatively frequently seen in normal populations51, and hence were excluded from our “modification” category. In addition, we found an individual that exhibited wing-size asymmetry between right and left wings (Fig. 1e, f), although its interpretation has not been established. These results demonstrated that our rearing system itself did not produce any colour-pattern modification without EMS treatment.
Colour-pattern modifications in the P generation
In the P generation adults, no pattern changes were identified in those animals that were fed the artificial diet containing 0 mM, 2.5 mM, and 25 mM EMS (MR = 0) (Fig. 2a and Supplementary Fig. 1; All aberrations found in the P, F1, F2, and F3 generations in the mutagenesis experiments were listed in Supplementary Table 2). The group fed a diet containing 50 mM EMS produced 28 adults and only one of them showed a modified wing colour pattern (MR = 3.6%) (Fig. 2a); this pattern was called vague-spot (VS) type (Fig. 2b). Two groups that were fed a diet containing 100 mM EMS diet produced a total of 40 eclosed adults, including 8 individuals with modifications (MR = 20.0%) (Fig. 2a). Among them, 7 adults (87.5%) and 1 adult (12.5%) had asymmetric (either right or left wing was modified or both were modified differently) and symmetric (both right and left wings were modified in a similar way) modifications, respectively. Loss-of-spot (LS), ectopic-spot (ES), and weak-contrast (WC) phenotypes were observed (Fig. 2c–e). Among these individuals with asymmetric defects, right wing modifications were found in 29% of the modified adults, left wing modifications were found in 29% of the modified adults, and bilateral modifications were found in 42% of the modified adults, suggesting that modified wing-side was random. These aberrant individuals were likely to have somatic cell mutations and not germ-line mutations, because they were the P-generation individuals that directly consumed EMS. Alternatively, they may be phenocopies that were produced by a functional inhibition of proteins by chemical stress.
Figure 2. Wing modifications obtained in the P generation.
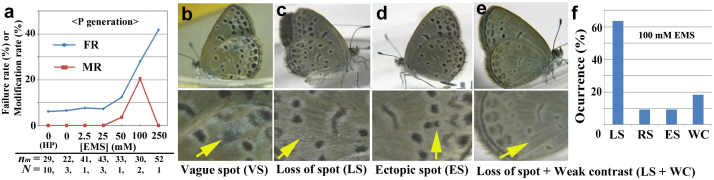
Note that these modified individuals were rare and were not used to generate offspring. Unique two-letter codes are assigned to all of the modification types. (a) The failure rate (FR) and the modification rate (MR). The number of trials (N) and the mean number of individuals (larvae) per trial (nm) are indicated (thus, these numbers are different from those of normal eclosion shown in Supplementary Table 2). HP indicates the host plant (i.e., natural diet, or ND). The MR increases dose-dependently up to 100 mM, but the FR also increases. (b) Vague-spot (VS) type. This individual was obtained by feeding 50 mM EMS. (c) Loss-of-spot (LS) type. In this individual, a particular spot was lost. This and other individuals shown here (c–e) were obtained by feeding 100 mM EMS. (d) Ectopic-spot (ES) type. (e) Loss-of-spot (LS) and weak-contrast (WC) types. (f) The occurrence of modification types obtained by feeding 100 mM EMS. RS indicates reduction-of-spot type (see phenotype shown in Figure 4). Total number of modified individuals was 8, but because double count was allowed when an individual shows two modifications traits, total number of counts was 11, as shown in Supplementary Tables 2 and 3.
Regarding the individuals reared with a diet containing 100 mM EMS, most of the modifications were categorised as LS (loss-of-spot) type, but ES (ectopic-spot), RS (reduction-of-spot), and WC (weak-contrast) types were also identified (Fig. 2f, Supplementary Table 3). Notably, all modifications were specific to a single spot, except the WC type. At the highest concentration (i.e., 250 mM), we obtained no modified individuals (Fig. 2a). This finding is largely due to high toxicity, resulting in a small number of surviving individuals, as indicated by the dose-dependent increase in the failure rate of eclosion (Fig. 2a).
Colour-pattern modifications in the F1 and F2 generations
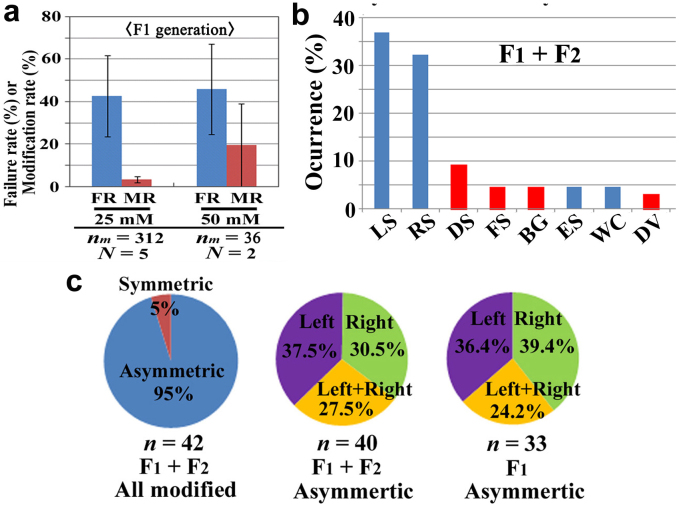
We obtained the F1, F2, and F3 generations from the P generation animals that were fed 25 mM or 50 mM EMS and that had normal (non-modified) wing colour patterns (Supplementary Figs. 1 and 2; Supplementary Tables 4–8). These F1-generation individuals did not consume EMS, but a small number of them exhibited colour-pattern modifications, suggesting that they were germ-line mutants for F1 screens. High failure rates of the F1 generation (Fig. 3a) were also consistent with this view. We detected no significant difference in the failure rates or modification rates between groups whose parents were fed 25 mM and 50 mM EMS (Fig. 3a). The EMS concentration required to obtain a reasonable number of modified individuals for the F1 screens was thus considered to be 25 mM. The failure and modification rates varied among groups, and the total modification rate of the F1 generation was 3.9% (n = 895), and mean modification rate per mating trial was 7.9% (Supplementary Fig. 1, Supplementary Tables 2, 4). Similarly, failure and modification rates of the F2 generation varied among groups (Supplementary Fig. 1), and the total modification rate of the F2 generation was 4.7% (n = 149), and mean modification rate per mating trial was 3.9% (Supplementary Table 2). Because there was no difference in the colour-pattern changes between these generations, we report their modification types together below.
Figure 3. Failure rate, modification rate, occurrence of modification types, and asymmetry of modifications in the F1 and F2 generations.
(a) The failure rate (FR) and the modification rate (MR) in the F1 generation. The number of trials (N) and the mean number of individuals (larvae) per trial (nm) are indicated (thus, these numbers are different from those of normal eclosion shown in Supplementary Table 2). The relatively large MR at 50 mM may be due to the small number of individuals obtained. (b) The occurrence of the modification types in the F1 and F2 generations. Those not obtained in the P generation are shown in red bars. Total number of modified individuals was 42 (Supplementary Table 2), but because a given individual was classified into one or two categories for modification type(s), total number of counts was 65 (Supplementary Table 5). (c) Symmetric and asymmetric modifications in the F1 and F2 generations. Most mutant individuals were asymmetric (left graph) as expected in the EMS mutagenesis. Among the asymmetric individuals obtained (middle and right graphs), right- and left-wing modifications occurred more or less equally.
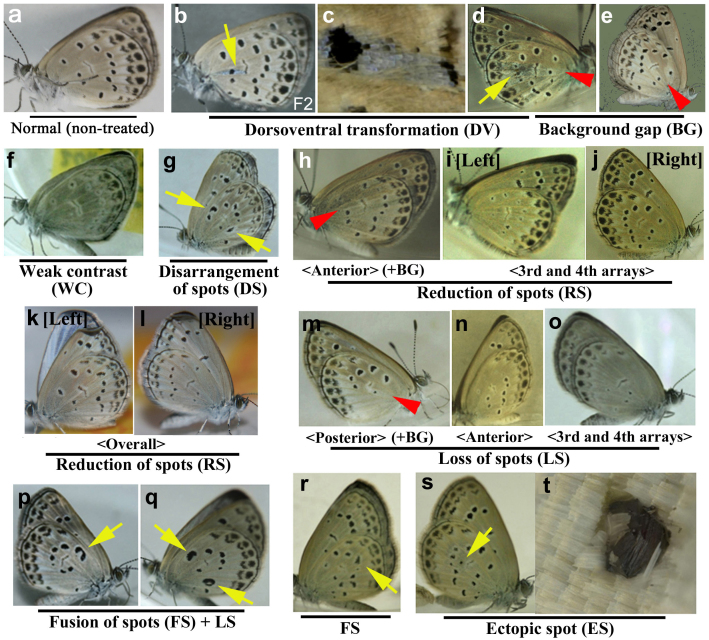
Together with a normal type (Fig. 4a), representative colour-pattern modifications that were obtained in the F1 and F2 generations are shown in Fig. 4b–t and Supplementary Fig. 2. Two types of background modifications were obtained. First, dorsoventral transformations (DV) were identified, in which the blue dorsal scale were found on the ventral side (Fig. 4b–d). Second, we identified defects in which the anterior area of a wing showed darker background colouration than the posterior area with a sharp boundary, which was termed the background-gap (BG) type (Fig. 4d, e).
Figure 4. Wing modifications obtained in the F1 and F2 generations.
An individual in (b) and (c) is from the F2 generation. All others are from the F1 generation. The yellow arrows indicate a modified portion of the hindwings. The red arrowheads indicate gaps of background colouration. (a) Normal type. (b) Dorsoventral transformation (DV). (c) High magnification of (b). (d) DV with a background gap (BG) between the anterior and posterior areas. (e) BG. (f) Weak contrast (WC) between the spots and the background. (g) Disarrangement of spots (DS). (h) Reduction of spots (RS) with BG. The spots located in the anterior part are reduced. (i, j) RS (an identical individual). The third and fourth spot arrays are reduced. (k, l) RS (an identical individual). All spots throughout the hindwing are reduced. (m) Loss of spots (LS) with BG. The spots located in the posterior part are lost. (n) LS. The spots located in the anterior part are lost. (o) LS. The third and fourth spots are almost completely lost. (p, q) Fusion of spots (FS) and LS. (r) FS. (s) Ectopic spot (ES). (t) High magnification of the ectopic spot in (s).
Almost all of the other types that we obtained were modifications of the third and fourth spot arrays. In other words, the first and second spot arrays were modified much less frequently than the third and fourth spot arrays. The following six modification types were found. First, we obtained the weak-contrast (WC) type, in which the contrast between the spots and the background was rather weak (Fig. 4f). Second, we identified defects where the spot arrays were disarranged in a single wing in the disarrangement-of-spot (DS) type (Fig. 4g). Thus, the spots were not aligned regularly in the DS type. Third, we frequently observed a reduction of spot (RS) size (Fig. 4h–l). The pattern of reduction was unique to an individual. For example, in an RS type individual, only the anterior spots were reduced with a background gap (BG) (Fig. 4h), but in another individual, all of the spots of the third and fourth arrays were reduced (Fig. 4i, j). Yet, in another individual, all of the spots including the first and second arrays were reduced (Fig. 4k, l). A fourth type of defect, similar to the reduction type, was the loss-of-spot (LS) type. Various LS subtypes were observed (Fig. 4m–o). For example, in one LS type individual, the posterior spots of the third and fourth arrays were completely lost with the BG phenotype (Fig. 4m), but in another individual, the anterior spots were completely lost (Fig. 4n), and a third individual showed almost complete loss of the third and fourth arrays (Fig. 4o). A fifth type of defect, fusion of spots (FS) was observed (Fig. 4p–r). This defect was often accompanied by the LS phenotype (Fig. 4p, q). The sixth type of defect, the ectopic spot (ES) occurred (Fig. 4s) when an ectopic spot was constructed by black scales of disorganised directions (Fig. 4t).
The LS and RS types were observed most frequently (Fig. 3b, Supplementary Table 5). The DS, FS, BG, and DV types of modifications were observed in the F1 and F2 generations but not in the P generation. Notably, almost all of the modifications, including the LS and RS types and excluding the ES type, were not specific to a single spot. That is, a group of spots were subjected to similar modifications at the same time on a given wing surface. These modifications are likely system-dependent modifications. Collectively, most of the modifications were asymmetric (Fig. 3c, left). Only two individuals showed symmetric modifications: the LS type and the RS type. Among the asymmetric modifications, no preference between right and left was observed (Fig. 3c, middle and right), suggesting that the modification sidedness was random.
Morphometric evaluations of colour-pattern modifications
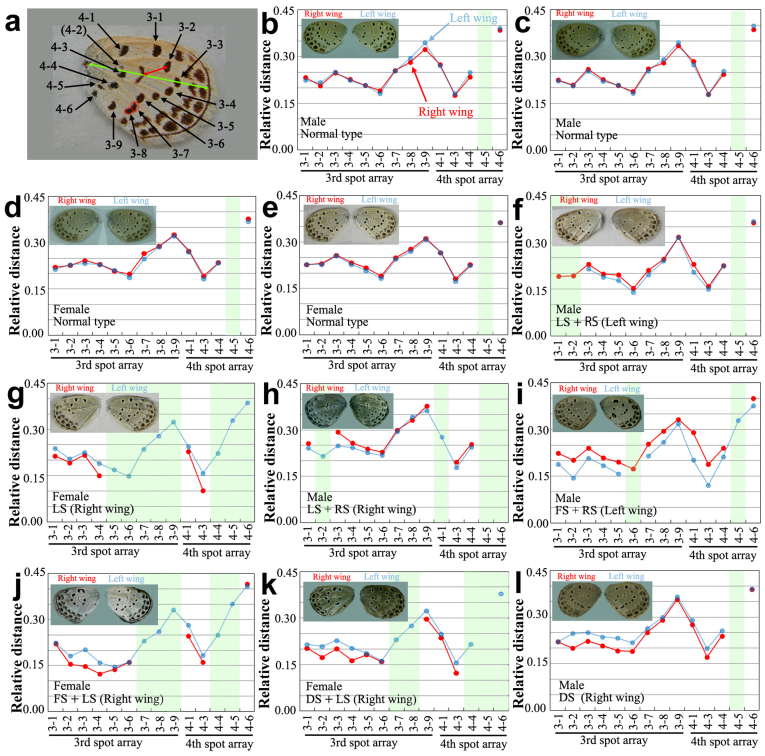
To quantitatively demonstrate systematic spot modifications in the mutants of the LS, RS, FS, and DS types, we here measured relative distances of spots that belong to the third and fourth spot arrays from the center of the system, the discal spot (Fig. 5). In a normal individual, relative distances of spots in the right and left wings were almost identical, demonstrating the symmetry of the right and left wings in each individual (Fig. 5a–e). In contrast, in the F1 mosaic mutants that belong to the LS, RS, FS and DS types, many spots appeared to be asymmetrically positioned together (Fig. 5f–l), demonstrating a systematic dislocation of consecutive spots. Furthermore, in the LS-type individuals, a few consecutive spots typically disappeared together (Fig. 5f–k).
Figure 5. Morphometric analysis of the spot distances.
Spots that belong to the third and fourth arrays on the right (shown in red in graphs) and left (shown in blue in graphs) wings were measured. In these graphs, the portions of “lost spots” were shaded in light green. The spot 4-2 was not shown in the graphs, because it is not found even in the normal type. (a) Nomenclature of spots that belong to the third and fourth spot arrays and the measurements of their relative distances. Spots were numbered from the anterior side to the posterior side. Spots that belong to the first and second arrays were similarly named (see Supplementary Fig. 3). Distances were measured between the centre of the discal spot and the centre of each spot (in this example, the spot 3-2), as indicated by a red line. The wingspan was measured from the wing base to the end of the M1 vein, as indicated by a green line. The spots 3-7 and 3-8 (circled) often fuse together, but they were considered independent. (b-e) Normal types. The right and left values coincided in most spots. (f) An LS+RS type. The spots 3-1, 3-2, and 4-5 were lost, and spots 3-3, 3-4, 3-5, 3-6, and 4-1 of the right wings were slightly dislocated away from the discal spot, compared to those of the left wing. Similar dislocations are more apparent in (g–l). (g) An LS type. The spots 3-5 to 3-9 and 4-4 to 4-6 were lost, and many spots were disarranged. (h) An LS+RS type. (i) An FS+RS type. (j) An FS+LS type. (k) A DS+LS type. (l) A DS type.
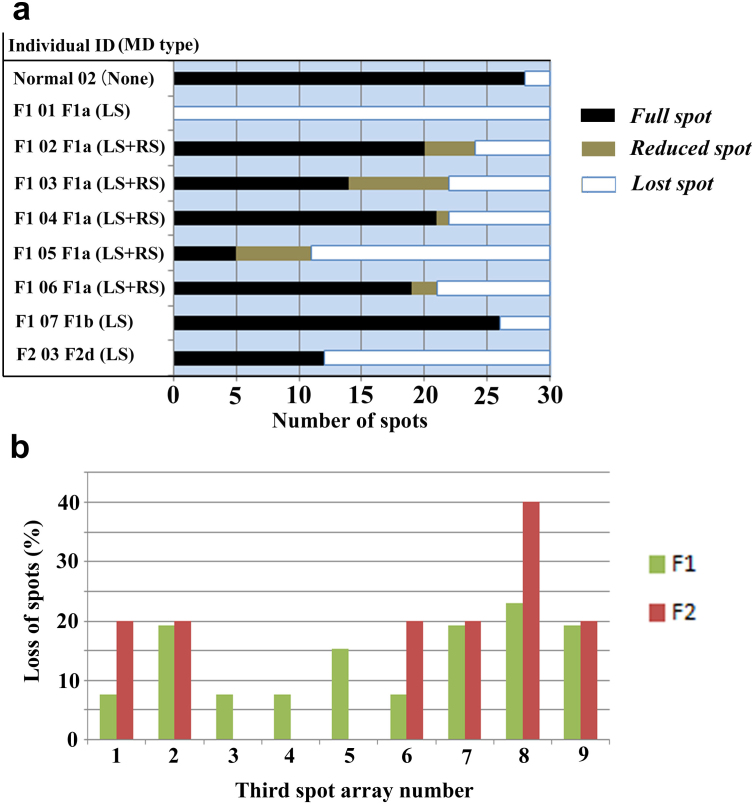
To further aid our understanding of the LS and RS phenotypes, we constructed tables that indicate the existence of the full spot, the reduced spot, or no spot in a wing (Supplementary Tables 9–20; Supplementary Fig. 3). Nine representative individuals were then shown in a graph that indicates the number of the full, reduced, and lost spots among 30 spots both in the right and left wings (Fig. 6a). Furthermore, using all the modified individuals listed in Supplementary Tables 9–18, the percentages of the full, reduced and lost spots in each location were obtained (Fig. 6b; Supplementary Tables 19, 20). Whereas the normal-type individuals always contained the full spots in the third and fourth arrays except the spot 4–2, 4–5, and 4–6 (n = 50), the LS-type individuals lacked a few or several spots together (Fig. 6). In the RS-type individuals, many spots were reduced in size (Supplementary Table 16). These results demonstrated that the spots that belong to the third and fourth arrays were dislocated or disappeared together in a system-dependent manner.
Figure 6. Full, reduced, and lost spots in the F1 and F2 mutants.
(a) The three-state spot assignment in representative individuals. There are 30 spots in the third and fourth spot arrays in the right and left hindwings in total (15 spots each). Number of full (black), reduced (gray), and lost (white) spots are counted and graphically shown. The ID number corresponds to the list of Supplementary Tables 9–18. Note the diverse patterns in these individuals. (b) Percentages of loss of each spot in the right hindwings among the F1 and F2 individuals. See Fig. 5a for spot number identification, and see Supplementary Table 20 for more detail. Note that the percentages are not even among the third array spots, and that the relatively high percentages of loss are seen together in the high-number spots (i.e., spots 7, 8, and 9).
Relationship between the forewing and hindwing
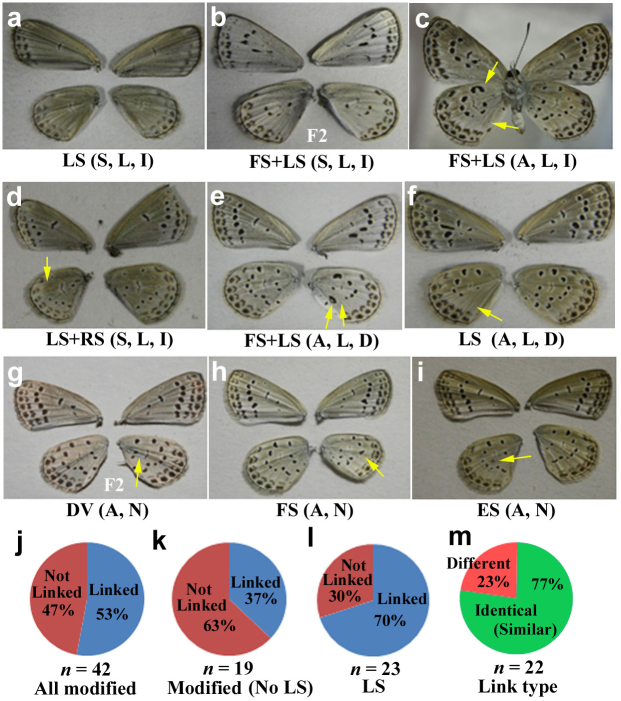
Thus far, we have focused on the ventral hindwing patterns. Here, we examined whether modifications of the ventral hindwing are linked with those of the ventral forewing to understand the mosaic nature of the EMS treatment. The representative wings are shown in Fig. 7a–i. Although there was a group of individuals that showed only hindwing modifications, the modification linkage between the forewing and hindwing were relatively apparent. That is, among the modified individuals that were examined including the LS type (n = 42), 53% of them showed some modifications on the forewings (Fig. 7j). These modifications were one-sided when they were observed in both the forewing and the hindwing. For example, a right hindwing modification was always found together with a right (not left) forewing modification. Among the modified individuals excluding the LS type, the majority were not linked (Fig. 7k). However, when focusing on the most frequent type (n = 23), the LS type, the percentage of linked individuals was 70% (Fig. 7l). In the LS type individuals, there were two linkage types between the fore- and hindwings: identical (or similar) (77%) and different (23%) (Fig. 7m).
Figure 7. Modification linkages between the fore- and hindwings.
(a–i) Each individual was evaluated in terms of the modification types. First, the defect was classified as symmetric (S) or asymmetric (A), based on whether the modification was on a single wing (A) or on both wings (S). Next, each defect was identified as linked (L) or non-linked (N), depending on if a mutant phenotype was found on the forewing and hindwing (L) or just the hindwing (N). Finally, linked defects were classified as identical (I) or different (D), depending on whether modifications of the forewing and hindwing were identical (or similar) (I) or different (D). All individuals shown are from the F1 generation, except those in panels b and g, which are from the F2 generation. Hindwing modifications are indicated by yellow arrows. (j) The percentages of linked or non-linked individuals among all of the modified individuals. (k) The percentages of linked or non-linked individuals among all of the modified individuals, except those with LS defects. (l) The percentages of linked or non-linked individuals among LS mutants. (m) The percentages of each linkage type among all individuals with linked defects.
Successive breeding
Because the aberrant phenotypes we obtained in the F1 generation have never been observed in our breeding system before, they are most likely genetic mutants that resulted from the EMS treatment of the parents. To test the possibility of maintaining these mutations in successive lines, we obtained F2 offspring from these aberrant F1 individuals (Supplementary Fig. 1, Supplementary Tables 2, 7).
In multiple-pair crosses, we crossed an F1b male (FS+RS) with F1b females (normal), resulting in F2d with an MR = 13.0%. That is, we obtained 3 modified individuals: an ES type (asymmetric), an RS+LS type (asymmetric), and an LS type (asymmetric). Among them, the RS+LS type was similar to its F1 male parent. The heritable modification rate was thus 33.3%. Similarly, we crossed an F1b male (DV+BG) and an F1c male (LS) with two F1b females (normal) and two F1c females (normal), resulting in F2e with an MR = 0%. In single-pair crosses, an F1a male or female (LS) was crossed with a wild-type female or male, producing F2a, F2b, and F2c offspring. Only F2a contained modified individuals with an MR = 6.3%. That is, we obtained 4 modified individuals: a DV-type individual (asymmetric, shown in Fig. 4b, c), 2 LS-type individuals (asymmetric with abnormal venation), and an RS+DS-type individual (asymmetric). One of the LS-type individuals was similar to the LS-type F1 parents in terms of particular spot loss (Supplementary Fig. 1). In this sense, the heritable modification rate was thus 25.0%. However, because both one of the parents and its 2 progenies (out of 4) were categorised as the LS type irrespective of locations of lost spots, the heritable modification rate of LS type could be calculated as 50.0%.
We produced the F3 generation by single-pair crosses using wild-type individuals as a paternal or maternal parent (Supplementary Fig. 1, Supplementary Tables 2, 8). However, the FR for the F3 generation was more than 90%, and modified individuals were not obtained.
Non-colour-pattern aberrations
We noticed morphological defects other than colour-pattern changes in the F1 generation. We obtained a small number of individuals that exhibited aberrations in foreleg and compound-eye morphology (Supplementary Fig. 4, Supplementary Table 2). We also obtained relatively frequent cases of individuals with wing-size asymmetry between right and left wings (Fig. 1e, f, Supplementary Table 2).
Discussion
We performed mutagenesis experiments using a lycaenid butterfly, the pale grass blue Z. maha. This study is methodologically unique in three ways. First, this study employed a lycaenid butterfly to understand the developmental regulation of colour patterns. The use of a lycaenid butterfly for developmental biology and genetics has been challenging, partly because of the lack of a rearing method. In this sense, this study opens up a new field of lycaenid developmental biology and genetics. Second, this is a rare mutagenesis study in butterflies. We obtained various mutants that show insightful phenotypes. To the best of our knowledge, most of the phenotypes that were obtained in this study have never been reported before, even in the field collection of spontaneous mutants (see literature survey in Otaki et al.50). Third, we took advantage of the mosaic nature of the EMS-induced F1 screens, although the fact that many mosaic individuals are obtained in the F1 generation is considered one of the “drawbacks” of using EMS for mutagenesis in Drosophila52,53,54. Many modifications were specific to one wing or a single symmetry system (i.e., third and fourth spot arrays), which helped us to differentiate between normal and abnormal colour patterns without fail. Importantly, this fact also suggests the existence of system-dependent regulations.
In addition to the fact that the various mosaic phenotypes were obtained, the following points indicate that we successfully obtained genetic mutants. First, no colour-pattern modification was observed in non-treated adults in our rearing system. The aberrant phenotypes are difficult, if not impossible, to obtain at this high frequency by spontaneous mutation or physiological plasticity, based on our control experiments and our previous studies50,51. Second, no similar phenotypes have been found in breeding lines without any mutagen in our laboratory. Third, only the P-generation (not F1-, F2-, and F3-generation) adults consumed EMS, and fourth, we used the P generation adults that did not show any colour-pattern aberrations to generate the F1 generation. Because the F1, F2, and F3 generations all ate natural host plant leaves without any mutagen, the aberrant phenotypes observed in these generations are highly likely caused by genetic damage. Fifth, aberrations found in forelegs, compound eyes, and wing size are likely indications of genetic mutants.
Sixth, most of the mutant phenotypes obtained in this study were asymmetric and were likely genetically mosaic, as expected from the nature of EMS. For example, the cells that carry mutated genes were found in the modified right wing, but the cells in the left wing did not have the mutation. The random nature of right and left modifications and the linkage between the hindwing and forewing support this notion. Seventh, in many cases, the modifications of the hindwing and forewing (not the right and left wings) were identical or similar, suggesting that the cells in both wings carry the same mutation, and not caused by toxic effects of EMS.
The possibility of epigenetic changes55 cannot be completely excluded, but epigenetic influence is often observed in stressed parents from whom offspring are produced. Unfortunately, we failed to obtain mutants in the F3 generation, and this could be interpreted as a non-genetic nature of the aberrant phenotypes. However, this failure is probably not to be attributed to our technical mistake (see below). Importantly, this failure does not disvalue the various phenotypes we obtained in the F1 and F2 generations, because, whatever the case may be, these phenotypes support our hypothesis, i.e., the systematic nature of the spot development. Although the higher EMS concentration (i.e., 100 mM) caused aberrations in the P generation that might be similar to mutant phenotypes of the F1 and F2 generations, this finding may simply suggests somatic mutations in the P generation. Alternatively, the physiological aberrations at the higher EMS concentration may be phenocopies of the corresponding mutants, which are produced by a functional inhibition of proteins. The optimal concentration of EMS for feeding the P generation larvae was 25 mM EMS, which is consistent with the standard mutagenesis protocol used in Drosophila genetics52,53,54.
In the F2 generation, at least two individuals had modifications similar to those of the F1 parents. One of them had extensive symmetric modifications, and thus this individual was likely a non-mosaic mutant. Unfortunately, this non-mosaic candidate for a mutant line could not produce offspring; we were unable to establish mutant lines due to a high failure rate. The reason for this failure is not known, but it may be because this species is vulnerable to sibling crosses51. We think that genetically distinct males and females must be crossed to produce healthy offspring. Although we used wild-type individuals that were supposed to be genetically different from the mutants we obtained, we cannot exclude the possibility that they were collected from a genetically similar population. This problem of line establishment should be overcome in the future if one wants to use this species of butterfly in genetics. We plan to use an isolated population of this species that might be resistant against sibling crosses.
We conclude that the mutagenesis was successful, and colour-pattern changes resulted from the genetic mutations. We obtained a 3.9% F1 mutation rate in this study. Although we focused only on wing colour-pattern mutants, this mutation rate was reasonably high. This rate is similar to that of a previous mutagenesis study with B. anynana39. However, only eyespot-dependent changes have been reported in B. anynana using an X-ray mutagenesis method39. In contrast, we observed multiple types of defects. This discrepancy may simply be due to the differences in species and methodology.
Despite a failure to establish a mutant line, we successfully obtained many cases of aberrant colour patterns that directly helped us to understand how the normal colour patterns develop. We obtained dorsoventral transformation mutants in which the dorsal colouration was expressed in the ventral surface. This result suggests that a dorsoventral regulatory gene was mutated. Indeed, the apterous gene product can specify dorsoventral axes in Drosophila wings56,57,58. Similarly, we identified mutants that had an anterioposterior background gap (the BG type). An anterioposterior regulatory gene that is functionally associated with background colouration was likely mutated in these animals.
The ectopic spot addition may be caused by a misplacement of organising centres for spot development. This misplacement may be considered a type of spot-dependent change. In contrast, other phenotypes found in the F1 and F2 generations likely resulted from system-dependent changes or wing-wide changes. We obtained several subtypes of reduction-of-spot (RS) and loss-of-spots (LS) mutants in the F1 and F2 generations, all of which indicate certain levels of system-dependent regulation, as several spots were reduced together in size or lost together in individual wings. This finding is in sharp contrast to the LS type defects found in the P generation, which expressed the lack of a single spot. Furthermore, we observed the disarrangement-of-spot (DS) type where the third and fourth arrays of spots were disarranged. Similar disarrangement was also observed in the fusion-of-spot (FS) type, where many, sometimes all, of the spots of the third and fourth arrays appeared to be dislocated toward the discal spot. We did not score such mutants as DS, nor did we set a distinct category for these displacement mutants, because the spot dislocation was found in the majority of the mutant types (the LS, RS, FS, and DS types) and because the spot dislocation was revealed only after the morphometric analysis (because when all or many of the spots moved toward the discal spot in a coordinated manner, it is difficult to distinguish this phenotype from the normal phenotype). It appears that, in many mutants that belong to the LS, RS, FS, and DS types, the spots are closer to the discal spot than they are in normal wings. This spot re-arrangement increases the proximity of spots, resulting in the fusion of spots and the loss of spots in particular wing compartments.
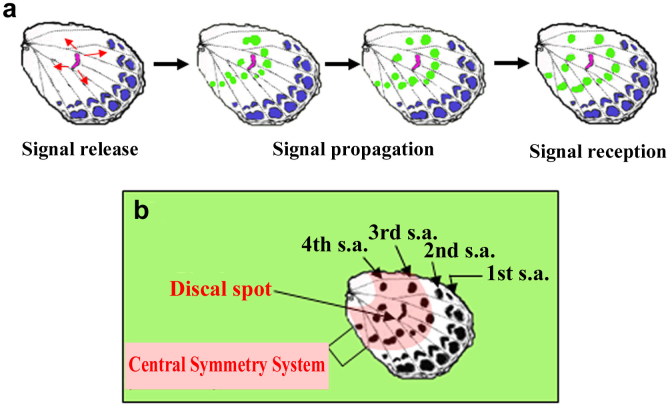
Thus, the third and fourth spot arrays that surround the discal spot can constitute a single symmetry system in lycaenid butterflies, regardless of the physical separation between the spot arrays and the discal spot (Fig. 8). The discal spot corresponds to the core element (sensu Otaki6) of the system in nymphalid butterflies, and the spots of the third and fourth arrays may correspond to the paracore elements (sensu Otaki6). This report represents the first demonstration of a symmetry system (which we suggest to be called the lycaenid central symmetry system) that covers a relatively large area in lycaenid butterflies by colour-pattern changes. This means that spot-inducing signals are released from the prospective discal spot and drive formation of the third and fourth spot arrays (Fig. 8).
Figure 8. A model for the system-dependent spot determination of the third and fourth arrays on the ventral hindwing of Zizeeria maha.
(a) Temporal sequence of events of dynamic signals driving spot determination. Signals are released from the prospective discal spot (red). Propagating signals from the discal spot are drawn in red arrows and green dots. (b) The lycaenid central symmetry system of the ventral hindwing defined by this species in reference to mutant phenotypes. The central symmetry system, shaded in pink, is composed of the discal spot (core element) and its surrounding third and fourth spot arrays (paracore elements). Spot array is abbreviated as s.a. in this figure.
In contrast to the third and fourth spot arrays, the discal spot and the first and second spot arrays were relatively stable. The reason for this result is not known, but we speculate that it is related to the spatial and temporal regulations of spot development. Development of the third and fourth spot arrays may require sophisticated regulations to release and propagate signals in both spatially and temporarily coordinated manners. In contrast, formation of the first and second spot arrays may not need such sophisticated regulatory mechanisms. In other words, because the first and second spot arrays are positioned closely in a small area, small regulatory fluctuations may not cause apparent changes.
Although the nature of the long-range signalling involved in this process is unknown, it seems that the positions of the spots in the third and fourth arrays are determined by the signals released from the primary organising centre, which later becomes the discal spot (Fig. 8). Then, the settled signals from the discal spot may induce secondary organising centres that later become independent spots. The weak primary signals from the discal spot may arrange the spots of the third and fourth arrays closer to the discal spot, whereas the strong primary signals may produce spots that are farther from the discal spot. This mechanistic explanation for the mutant phenotypes is consistent with the induction model proposed for nymphalid eyespots and parafocal elements8,9,10. Analyses for the developmental process of the normal and abnormal spots in the pupal wings may be required in the future, using methods that have been developed for nymphalid butterflies59,60.
One clue to understand the mechanistic basis of the mutant phenotypes obtained in the F1 and F2 generations comes from the fact that the F2 modifications were not necessarily similar to their parental F1 modifications. It seems that the master regulatory gene was mutated and several different phenotypes were produced with low expressivity, depending on the genetic background. For example, the master gene may regulate the intensity of the primary signals from the discal spot, but the final colour patterns are also influenced by other genes.
Curiously, we did not obtain mutants with spot-enlargement phenotypes. The spot-enlargement phenotypes (temperature-shock type or TS type) are physiologically plastic phenotypes induced by temperature treatments, and the phenotypes were likely genetically fixed in the northern range margin population50. Our results imply that the random mutation process did not frequently generate mutations that cause the spot-enlargement phenotypes. This supports the previously described plasticity-oriented evolution of colour patterns in the northern range margin population50. However, more extensive F1 screens may produce the spot-enlargement mutants in lycaenid butterflies.
Finally, we note that the mutant phenotypes identified in this study are similar to those found in butterflies that were caught in the Fukushima area and their offspring61. Additionally, they are dissimilar to the field-caught TS phenotypes that were obtained in the northern range margin and to the laboratory-induced TS phenotypes by cold-shock treatment50. The diversity of phenotypes in the present study is also consistent with the Fukushima study61. These facts mutually ensured that at least some of aberrant individuals that were obtained in the present study and in the previous study61 were genetic mutants.
In conclusion, in addition to spot-specific regulations, there appear to be several regulatory mechanisms for colour-pattern determination. Fate specifications could involve dorsoventral sides and anterioposterior areas. Importantly, the positions of the groups of spots that belong to a single symmetry system are regulated coordinately. The molecular nature of these regulatory mechanisms is unknown but is consistent with the formal model10. Such wing-wide regulatory mechanisms may be an important topic for future investigation6,7,8,9,10,18,59,62,63, which may be assisted not only by further mutagenesis studies but also by newly developed methods for efficient gene transfer to butterflies60,64.
Methods
Butterflies
We used the pale grass blue Zizeeria maha okinawana (Matsumura, 1929) (Fig. 1a) caught in Okinawa-jima Island, Japan, except a control experiment (Experiment 2) in which case Z. maha argia (Ménétriès, 1857) caught in Kobe, Hyogo Prefecture, Japan, was used. This species is monophagous and eats the wood sorrel, Oxalis corniculata. We followed the methods of a previous study47 for rearing this species with an artificial diet under summer-morph conditions (25 ± 1°C, 16L-8D). We used the conventional terminology to identify spots and spot arrays of this butterfly (Fig. 1b).
Rearing system
To demonstrate that our rearing system itself does not produce any colour-pattern modification, three sets of experiments were performed (Supplementary Table 1). In the first set of experiments (Experiment 1), 10 males and 10 females were field-collected in Okinawa-jima Island, housed together in a large tank (45 cm H × 120 cm W × 45 cm D), and allowed to lay eggs on potted natural host plant leaves. The hatched larvae were transferred to petri-dishes or other plastic containers and reared with the natural host plant leaves, according to our previous study51. They were defined as the P generation. Among the P generation adults, 10 males and 10 females were selected randomly and housed together to collect eggs. These processes were repeated up to the F3 generation, producing progeny by sibling crosses. In the second set of experiments (Experiment 2), non-sibling crosses and rearing were performed to obtain the P and F1 generations, using host plant leaves. To obtain the P generation, 3 males and 5 females were housed together in each of three small tanks (30 cm H × 30 cm W × 30 cm D) named A, B, and C (In total, 9 males and 15 females were used). These founding parents were caught in Kobe. To obtain the F1 generation, 3 male from A, 4 females from B, and 2 females from C were housed together in a small tank. Similarly, in the third set of experiments (Experiment 3), non-sibling crosses and rearing were performed, partially using artificial diet to obtain the P and F1 generations. To obtain P(ND) (i.e., the P generation reared with natural diet) or P(AD) (i.e., the P generation reared with artificial diet), 1 male (wild) and 3 females (wild) were housed together in a small tank (In total, 2 males and 6 females were used). These founding parents were caught in Okinawa-jima Island. To obtain F1(ND) or F1(AD), 4 males and 2 females from the P generation were housed together in a small tank (In total, 8 males and 4 females were used). A part of the third set of experiments (Experiment 3) has already been reported51.
Throughout this paper, the panda type and the crescent type were not considered “modified” in our screens, because the panda type is an indication of genetic deterioration due to sibling crosses51 and because the crescent type can be found relatively frequently in normal populations (and also in sibling crosses), being consistent with our results obtained in Experiment 1 (see Supplementary Table 1). In this paper, we define “wild type” as adult individuals caught in the field or as adult individuals reared from eggs of field-caught females with the natural host plant. If no colour-pattern aberration was detected, such an individual was scored as “normal”.
Mutagenesis
The females were caught in a field, and eggs were collected on potted host plant leaves. The hatched larvae were transferred to petri-dishes or other plastic containers with natural host plant leaves. When the larvae reached the third or fourth instar stage, they were fed an artificial diet containing EMS (Sigma, Tokyo, Japan). EMS is a highly efficient mutagen that causes single-base changes in DNA and often results in mosaic phenotypes in the F1 generation52,53,54. The EMS solution was prepared in ddH2O at concentrations of 2.5 mM, 25 mM, 50 mM, 100 mM, and 250 mM and was added to the artificial diet immediately before feeding (25.8 μL of solution per 1.0 g of diet).
Crosses
The mating methods used were described previously51. EMS-fed individuals were defined as the P generation. The details of the crosses for F1 screens we performed are described in Supplementary Fig. 1 and Supplementary Table 4. Two types of crossing methods were performed; multiple-pair crosses, in which several adults were confined in a single tank, and single-pair crosses, in which only a male and a female were confined in a single tank. Two types of multiple-pair crosses were performed. First, both male and female P individuals fed the EMS-containing artificial diet were crossed with each other. Second, males fed the EMS-containing artificial diet were crossed with females reared with the natural host plant. The offspring obtained from these crosses were defined as the F1 generation, which were reared only with the natural host plant. The P individuals that contained no detectable colour-pattern modifications were used to obtain the F1 offspring.
We selected some F1 adults that had wing colour-pattern modifications to obtain the F2 generation with single-pair and multiple-pair crosses. The F2 larvae were reared only with the natural host plant. Similarly, we performed single-pair crosses between the F2 and wild-type adults to obtain the F3 offspring. The F3 larvae were reared only with the natural host plant.
Qualitative and quantitative evaluations
To qualitatively evaluate wing colour-pattern changes, we focused on the ventral surface of the hindwings, which are readily observable in live adults. The modifications were often one-sided, meaning that only the right or left wing was modified. Additionally, two-sided modifications were often asymmetric, meaning that the right and left wings were differently modified. The changes in both wings were often classified into the same category. Also note that a genetically mosaic individual usually shows an asymmetric phenotype; however, a genetically non-mosaic (homogeneous) individual may also show an asymmetric phenotype due to insufficient expression of a given trait (i.e., low expressivity).
We quantitatively evaluated the results of the crosses as follows. First, we defined the failure rate (FR) of eclosion: FR (%) = [{(number of pupae) – (number of normally eclosed pupae)}/(number of pupae)] × 100. This means that the FR indicates a percentage of the dead pupae, the pupae that failed eclosion, and the eclosed individuals that could not fly at all due to severe wing deformation. Second, we defined the modification rate (MR) for wing colour pattern: MR (%) = {(number of modified adults)/(normally eclosed adults)} × 100.
To quantitatively evaluate wing colour-pattern changes, we measured relative distances of spots that belong to the third and fourth spot arrays from the centre of the system, the discal spot, using a Keyence digital microscope VHX-1000 (Osaka, Japan). Numbers were assigned to each spots of the hindwing (Fig. 5a; Supplementary Fig. 3), and relative distance was calculated as a ratio of the discal-spot distance (from the centre of the discal spot to the centre of each spot) to the hindwing length (from the wing base to the wing edge at the end of the M1 vein) (Fig. 5a). Both the right and left wings of an individual were subjected to measurements, and results were expressed in a graph to indicate positional differences between the right and left wings.
The tables for describing the presence or absence of spots were constructed for the normal, LS, and RS types (Supplementary Tables 9–20). We selected normal male (n = 25) and female (n = 25) individuals from diverse sources of our specimens not to bias this process. Note that the identification of the LS type depends not only on the third- and fourth-array spots but also on the first- and second-array spots. All spots were visually checked for their three states, i.e., presence (the full normal spot), reduction (the reduced spot in size), or absence (no spot).
Author Contributions
J.M.O. designed and coordinated the study and performed a part of rearing experiments, M.I. performed most experiments, A.H. performed a part of rearing experiments and contributed to establishment of the rearing system, and J.M.O. wrote the manuscript.
Supplementary Material
Supplementary Information
Acknowledgments
We are grateful to T. Gushi, S. Mahdi, C. Nohara, W. Taira, S. Kinjo and other members of the BCPH Unit of Molecular Physiology for technical help and for discussion. This work was partly supported by the Research Foundation for Opto-Science and Technology, Hamamatsu, Japan and by the Incentive Project from University of the Ryukyus.
References
- Nijhout H. F. The Development and Evolution of Butterfly Wing Patterns. (Smithsonian Institution Press, Washington, 1991). [Google Scholar]
- Schwanwitsch B. N. On the groundplan of wing-pattern in nymphalids and certain other families of rhopalocerous Lepidoptera. Proc. Zool. Soc. Lond. B 34, 509–528 (1924). [Google Scholar]
- Süffert F. Zur vergleichende Analyse der Schmetterlingszeichnung. Biologisches Zentralblatt 47, 385–413 (1927). [Google Scholar]
- Nijhout H. F. Elements of butterfly wing patterns. J. Exp. Zool. 291, 213–225 (2001). [DOI] [PubMed] [Google Scholar]
- Otaki J. M. Colour-pattern analysis of parafocal elements in butterfly wings. Entomol. Sci. 2009, 12, 74–83. [Google Scholar]
- Otaki J. M. Colour pattern analysis of nymphalid butterfly wings: Revision of the nymphalid groundplan. Zool. Sci. 29, 568–576 (2012). [DOI] [PubMed] [Google Scholar]
- Otaki J. M. Colour-pattern analysis of eyespots in butterfly wings: a critical examination of morphogen gradient models. Zool. Sci. 28, 403–413 (2011). [DOI] [PubMed] [Google Scholar]
- Otaki J. M. Generation of butterfly wing eyespot patterns: a model for morphological determination of eyespot and parafocal element. Zool. Sci. 28, 817–827 (2011). [DOI] [PubMed] [Google Scholar]
- Otaki J. M. Artificially induced changes of butterfly wing colour patterns: dynamic signal interactions in eyespot development. Sci. Rep. 1, 111 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otaki J. M. Structural analysis of eyespots: dynamics of morphogenic signals that govern elemental positions in butterfly wings. BMC Syst. Biol. 6, 17 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nijhout H. F. Pattern formation on lepidopteran wings: determination of an eyespot. Dev. Biol. 80, 267–274 (1980). [DOI] [PubMed] [Google Scholar]
- Nijhout H. F. Ontogeny of the colour pattern on the wings of Precis coenia (Lepidoptera: Nymphalidae). Dev. Biol. 80, 275–288 (1980). [DOI] [PubMed] [Google Scholar]
- Nijhout H. F. Colour pattern modification by coldshock in Lepidoptera. J. Embryol. Exp. Morphol. 81, 287–305 (1984). [PubMed] [Google Scholar]
- Nijhout H. F. Cautery-induced colour patterns in Precis coenia (Lepidoptera: Nymphalidae). J. Embryol. Exp. Morphol. 86, 191–203 (1985). [PubMed] [Google Scholar]
- Brakefield P. M. et al. Development, plasticity and evolution of butterfly eyespot patterns. Nature 384, 236–242 (1996). [DOI] [PubMed] [Google Scholar]
- Otaki J. M., Ogasawara T. & Yamamoto H. Tungstate-induced colour-pattern modifications of butterfly wings are independent of stress response and ecdysteroid effect. Zool. Sci. 22, 635–644 (2005). [DOI] [PubMed] [Google Scholar]
- Mahdi S. H., Gima S., Tomita Y., Yamasaki H. & Otaki J. M. Physiological characterization of the cold-shock-induced humoral factor for wing colour-pattern changes in butterflies. J. Insect Physiol. 56, 1022–1031 (2010). [DOI] [PubMed] [Google Scholar]
- Otaki J. M. Reversed type of colour-pattern modifications of butterfly wings: a physiological mechanism of wing-wide colour-pattern determination. J. Insect Physiol. 53, 526–537 (2007). [DOI] [PubMed] [Google Scholar]
- Otaki J. M. & Yamamoto H. Species-specific colour-pattern modifications of butterfly wings. Dev. Growth Differ. 46, 1–14 (2004). [DOI] [PubMed] [Google Scholar]
- Otaki J. M. Phenotypic plasticity of wing colour patterns revealed by temperature and chemical applications in a nymphalid butterfly Vanessa indica. J. Therm. Biol. 33, 128–139 (2008). [Google Scholar]
- Otaki J. M. Colour-pattern modifications of butterfly wings induced by transfusion and oxyanions. J. Insect Physiol. 44, 1181–1190 (1998). [DOI] [PubMed] [Google Scholar]
- Otaki J. M. Stress-induced colour-pattern modifications and evolution of the Painted Lady butterflies Vanessa cardui and Vanessa kershawi. Zool. Sci. 24, 811–819 (2007). [DOI] [PubMed] [Google Scholar]
- Beldade P. & Brakefield P. M. The genetics and evo-devo of butterfly wing patterns. Nat. Rev. Genet. 3, 442–452 (2002). [DOI] [PubMed] [Google Scholar]
- Brakefield P. M., Beldade P. & Zwaan B. J. The African butterfly Bicyclus anynana: a model for evolutionary genetics and evolutionary developmental biology. Cold Spring Harbor Protocols 2009, pdb.emo122 (2009). [DOI] [PubMed] [Google Scholar]
- Beldade P., Rudd S., Gruber J. D. & Long A. D. A wing expressed tag resource for Bicyclus anynana butterflies, an evo-devo model. BMC Genomics 7, 130 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beldade P., Saenko S. V., Pul N. & Long A. D. A gene-based linkage map for Bicyclus anynana butterflies allows for a comprehensive analysis of synteny with the lepidopteran reference genome. PLoS Genet. 5, e1000366 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conceição I. C., Long A. D., Gruber J. D. & Beldade P. Genomic sequence around butterfly wing development genes: annotation and comparative analysis. PLoS One 6, e23778 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcus J. M., Ramos D. M. & Monteiro A. Germline transformation of the butterfly Bicyclus anynana. Proc. R. Soc. Lond. B (Suppl.) 271, S263–S265 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos D. M., Kamal F., Wimmer E. A., Cartwright A. N. & Monteiro A. Temporal and spatial control of transgene expression using laser induction of the hsp70 promoter. BMC Dev. Biol. 6, 55 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos D. M. & Monteiro A. Transgenic approaches to study wing colour pattern development in Lepidoptera. Mol. Biosyst. 3, 530–535 (2007). [DOI] [PubMed] [Google Scholar]
- Li Y., Yuan S. & Moyer R. W. The non-permissive infection of insect (gypsy moth) LD-652 cells by vaccinia virus. Virology 248, 74–82 (1998). [DOI] [PubMed] [Google Scholar]
- Lewis D. L. & Brunetti C. R. Ectopic transgene expression in butterfly imaginal wing discs using vaccinia virus. BioTechniques 40, 48–54 (2006). [DOI] [PubMed] [Google Scholar]
- Lewis D. L. et al. 1999. Ectopic gene expression and homeotic transformations in arthropods using recombinant sindbis virus. Curr. Biol. 9, 1279–1287 (1999). [DOI] [PubMed] [Google Scholar]
- Golden K., Sagi V., Markwarth N., Chen B. & Monteiro A. In vivo electroporation of DNA into the wing epidermis of the butterfly, Bicyclus anynana. J. Insect Sci. 7, 1–8 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terenius O. et al. RNA interference in Lepidoptera: an overview of successful and unsuccessful studies and implications for experimental design. J. Insect Physiol. 57, 231–245 (2011). [DOI] [PubMed] [Google Scholar]
- Keys D. N. et al. Recruitment of a hedgehog regulatory circuit in butterfly eyespot evolution. Science 283, 532–534 (1999). [DOI] [PubMed] [Google Scholar]
- Brunetti C. R. et al. The generation and diversification of butterfly eyespot color patterns. Curr. Biol. 11, 1578–1585 (2011). [DOI] [PubMed] [Google Scholar]
- Monteiro A., Glaser G., Stockslager S., Glansdorp N. & Ramos D. Comparative insights into questions of lepidopteran wing pattern homology. BMC Dev. Biol. 6, 52 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteiro A., Prijs J., Bax M., Hakkaart T. & Brakefield P. M. Mutants highlight the modular control of butterfly eyespot patterns. Evol. Dev. 5, 180–187 (2003). [DOI] [PubMed] [Google Scholar]
- Monteiro A. et al. The combined effect of two mutations that alter serially homologous colour pattern elements on the fore and hindwings of a butterfly. BMC Genet. 8, 22 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saenko S. V., Brakefield P. M. & Beldade P. Single locus affects embryonic segment polarity and multiple aspects of an adult evolutionary novelty. BMC Biol. 8, 111 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirai L. T. et al. Evolutionary history of the recruitment of conserved developmental genes in association to the formation and diversification of a novel trait. BMC Evol. Biol. 12, 21 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan S., Merlin C., Boore J. L. & Reppert S. M. The monarch butterfly genome yields insight into long-distance migration. Cell 147, 1171–1185 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papa R. et al. Highly conserved gene order and numerous novel repetitive elements in genomic regions linked to wing pattern variation in Heliconius butterflies. BMC Genomics 9, 345 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joron M. et al. Chromosomal rearrangements maintain a polymorphic supergene controlling butterfly mimicry. Nature 477, 203–206 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Heliconius Genome Consortium. Butterfly genome reveals promiscuous exchange of mimicry adaptations among species. Nature 487, 94–98 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed R. D. et al. optix drives the repeated convergent evolution of butterfly wing pattern mimicry. Science 333, 1137–1141 (2011). [DOI] [PubMed] [Google Scholar]
- Martin A. et al. Diversification of complex butterfly wing patterns by repeated regulatory evolution of a Wnt ligand. Proc. Natl. Acad. Sci. USA 109, 12632–12637 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otaki J. M. & Yamamoto H. Colour-pattern modifications and speciation in lycaenid butterflies. Trans. Lepidopterol. Soc. Jpn. 54, 197–205 (2003). [Google Scholar]
- Otaki J. M., Hiyama A., Iwata M. & Kudo T. Phenotypic plasticity in the range-margin population of the lycaenid butterfly Zizeeria maha. BMC Evol. Biol. 10, 252 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiyama A., Iwata M. & Otaki J. M. Rearing the pale grass blue Zizeeria maha (Lepidoptera, Lycaenidae): Toward the establishment of a lycaenid model system for butterfly physiology and genetics. Entomol. Sci. 13, 293–302 (2010). [Google Scholar]
- Lewis E. B. & Bacher F. Method of feeding ethyl methane sulfonate (EMS) to Drosophila melanogaster males. Drosophila Inform. Serv. 43, 193 (1968). [Google Scholar]
- St Johnston D. The art and design of genetic screens: Drosophila melanogaster. Nat. Rev. Genet. 3, 176–188 (2002). [DOI] [PubMed] [Google Scholar]
- Grigliatti T. A. Mutagenesis. In Roberts D. B. (ed), Drosophila: A Practical Approach. Second edition. IRL Press, Oxford, 55–84 (1998). [Google Scholar]
- Hiyama A., Taira W. & Otaki J. M. Colour-pattern evolution in response to environmental stress in butterflies. Front. Genet. 3, 15 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zecca M. & Struhl G. Subdivision of the Drosophila wing imaginal disc by EGFR-mediated signaling. Development 129, 1357–1368 (2002). [DOI] [PubMed] [Google Scholar]
- Blair S. S. Compartments and appendage development in Drosophila. BioEssays 17, 299–309 (1995). [DOI] [PubMed] [Google Scholar]
- Milán M. & Cohen S. M. Temporal regulation of Apterous activity during development of the Drosophila wing. Development 127, 3069–3078 (2000). [DOI] [PubMed] [Google Scholar]
- Kusaba K. & Otaki J. M. Positional dependence of scale size and shape in butterfly wings: Wing-wide phenotypic coordination of colour-pattern elements and background. J. Insect Physiol. 55, 174–182 (2009). [DOI] [PubMed] [Google Scholar]
- Dhungel B., Ohno Y., Matayoshi R. & Otaki J. M. Baculovirus-mediated gene transfer in butterfly wings in vivo: An efficient expression system with anti-gp64 antibody. BMC Biotechnol. 13, 27 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiyama A. et al. The biological impacts of the Fukushima nuclear accident on the pale grass blue butterfly. Sci. Rep. 2, 570 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otaki J. M. Physiologically induced colour-pattern changes in butterfly wings: Mechanistic and evolutionary implications. J. Insect Physiol. 54, 1099–1112 (2008). [DOI] [PubMed] [Google Scholar]
- Ohno Y. & Otaki J. M. Eyespot colour pattern determination by serial induction in fish: Mechanistic convergence with butterfly eyespots. Sci. Rep. 2, 290 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ando T. & Fujiwara H. Electroporation-mediated somatic transgenesis for rapid functional analysis in insects. Development 140, 454–458 (2013). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information