Abstract
During autophagy, a double membrane envelops cellular material for trafficking to the lysosome. Human beclin-1 and its yeast homologue, Atg6/Vps30, are scaffold proteins bound in a lipid kinase complex with multiple cellular functions, including autophagy. Several different Atg6 complexes exist, with an autophagy-specific form containing Atg14. However, the roles of Atg14 and beclin-1 in the activation of this complex remain unclear. We here addressed the mechanism of beclin-1 complex activation and reveal two critical steps in this pathway. First, we identified a unique domain in beclin-1, conserved in the yeast homologue Atg6, which is involved in membrane association and, unexpectedly, controls autophagosome size and number in yeast. Second, we demonstrated that human Atg14 is critical in controlling an autophagy-dependent phosphorylation of beclin-1. We map these novel phosphorylation sites to serines 90 and 93 and demonstrate that phosphorylation at these sites is necessary for maximal autophagy. These results help clarify the mechanism of beclin-1 and Atg14 during autophagy.
INTRODUCTION
Autophagy is a catabolic membrane trafficking process that turns over cytosolic material following encapsulation by autophagosomes and subsequent degradation in the lysosome (in higher eukaryotes) or the vacuole (in plants and unicellular eukaryotes). Genetic studies in yeast have identified a battery of conserved proteins that are required for starvation-induced autophagy (1). In both higher eukaryotes and yeast, autophagy also occurs constitutively as a cargo-selective quality control process (2). Consistent with its dual role in metabolism and cellular quality control, autophagy has been shown to play a role in a variety of human pathologies, including cancer and neurodegeneration (3).
Many of the proteins identified as essential for autophagy in yeast have homologues in higher eukaryotes that carry out conserved functions. Among these is the tumor suppressor beclin-1, discovered in a two-hybrid screen as a protein that interacts with the antiapoptotic protein Bcl-2 (4, 5). Beclin-1 is a core component of the phosphatidylinositol 3-kinase complex, along with the catalytic subunit Vps34 and the putative protein kinase Vps15 (6). Atg6/Vps30, the yeast homologue of beclin-1, shares 24.4% amino acid homology and functions in selective and nonselective autophagy (7), as well as endosomal trafficking. Similar to beclin-1, additional components bind the core Atg6-lipid kinase complex to direct functions in these different membrane trafficking pathways (8).
Localization of the yeast Atg6/Vps15/Vps34 complex to the preautophagosomal structure is largely dictated by the binding of Atg6 to Atg14 (7). Recently, the human homologue of Atg14, hAtg14/Barkor/Atg14L (here referred to as hAtg14), has been identified by several groups. hAtg14 has been shown to be a member of a beclin-1 complex analogous to that in yeast, although the mechanism of action in autophagy may be distinct (6, 9, 10, 11). hAtg14 interacts with beclin-1 through its coiled-coil domain, and this interaction is required for autophagy and recruits a subset of Vps34 complexes to active sites of autophagosome biogenesis (9). It is also thought that hAtg14 recognizes membrane curvature in the nascent autophagosome (10). Of the additional beclin-1-interacting proteins, UVRAG is notable for its strong binding to beclin-1 and its role in later steps in autophagosome biogenesis and maturation (8).
In this study, we reveal two mechanistic steps in the activation of the beclin-1/Atg6 complex. We identify a 26-amino acid sequence at the C-terminal domain (CTD) of beclin-1 that is critical for hAtg14-independent membrane association. Importantly, this domain is conserved in the yeast Atg6, and yeast lacking the CTD display survival and macroautophagy defects and altered autophagosome size and number. These results define a conserved region in beclin-1/Atg6 that is involved in membrane docking and control of autophagosome morphology. We also find that hAtg14 controls autophagy-dependent phosphorylation of beclin-1, and we map two novel phosphorylation sites at serines 90 and 93. These phosphorylation events are necessary for efficient autophagy, revealing a novel mechanism by which hAtg14 controls beclin-1 function.
MATERIALS AND METHODS
Preparation of TALE nuclease-mediated gene knockout and stable-expression cell lines.
To generate cell lines with knockout (KO) of hAtg14, beclin-1, Atg5, and Atg13, targets were chosen within exons found in all predicted splice variants of each protein. Transcription activator-like effector (TALE) coding regions were created by iterative subcloning steps as described in reference 11, before ligation into a final vector, pcDNA3.1/Zeo-Talen(+63), modified from reference 12, at the NheI site which contains the Fok1 nuclease domain. Cells were cotransfected with TALE nuclease (TALEN) and N1-yellow fluorescent protein (YFP) (Clontech) expression vectors to allow enrichment of high-expression clones by fluorescence-activated cell sorting. Single colonies were isolated and expanded, and genomic DNA preparations collected. Clones were screened by PCR and digested with enzymes. Clones positive for TALEN digestion (i.e., those with PCR products not digested by the enzymes indicated in the figures) were then analyzed by immunoblotting and sequencing to distinguish clones as true knockouts (with nonsense deletions) rather than clones with in-frame deletions. We additionally determined that HCT 116 cells have two copies of beclin-1, Atg5, and Atg13 and three copies of the hAtg14 gene. Beclin-1 knockout lines also exhibited a greater degree of cell aggregation than wild-type (WT) cells (not shown).
To prepare stably transfected cell lines expressing YFP-parkin, hAtg14, or beclin-1 (including YFP-tagged and phosphomutant or phosphomimetic beclin-1 constructs), sequences of interest were subcloned into pBABE-puro (plasmid number 1764; Addgene), and retrovirus prepared in HEK293T cells. Transduction was conducted for 2 days prior to selection with antibiotic and/or cell sorting based on fluorescence.
Analysis of p62 transcript levels.
Low-passage-number cultures of HCT 116 wild-type, hAtg14 KO, and beclin-1 KO cell lines were seeded into 6-well plates at a density of 200,000 cells/well and then incubated for 48 h before total mRNA was extracted from each well with the Qiagen RNeasy plus minikit (Qiagen) according to the manufacturer's instructions. Purified mRNA (500 ng) from each sample was then converted to cDNA by using an Ambion high-capacity RNA-to-cDNA kit (Life Technologies) in 20-μl reaction mixtures following the manufacturer's protocols. Subsequent quantitative reverse transcription (qRT)-PCR toward each target (SQSTM1 [sequestosome 1] or GAPDH [glyceraldehyde-3-phosphate dehydrogenase]) was performed in separate 20-μl reaction mixtures composed of 2 μl of cDNA from these reactions, 10 μl TaqMan gene expression master mix (Life Technologies), 1 μl TaqMan primer-probe set, and 6 μl nuclease-free water. The TaqMan probes used were human GAPDH endogenous control (with FAM dye [6-carboxyfluorescein]; Life Technologies) and human SQSTM1 gene expression assays (assays number Hs01061917_g1 and Hs00177654_m1, 20×; Life Technologies), and 40 cycles of amplification were performed according to the manufacturer's specifications in an Applied Biosystems 7900HT qRT-PCR machine. Three replicates were performed per probe-cDNA combination for qRT-PCR. The relative amounts of mRNA between HCT 116 wild-type, hAtg14 KO, and beclin-1 KO samples were calculated with the comparative cycle threshold (CT) method (13) with normalization to the GAPDH endogenous control CT values.
Antibodies, plasmids, and siRNA.
The following antibodies were used in this work: antibodies to beclin-1 (Proteintech), phospholipase C (PLC) (Upstate), hAtg14 (Cell Signaling), UVRAG (UV radiation resistance-associated gene; Sigma), Tom20 (Santa Cruz), cytochrome c (BD Biosciences), GM-130 (BD Transduction labs), KDEL (Stressgen), green fluorescent protein (GFP) (Invitrogen), Flag M2 (Sigma), Tim23 (BD Biosciences), p62 for immunoblotting (BD Biosciences), p62 for immunofluorescence (Cedarlane), Vps15 (Bethyl Laboratories), Vps34 (Sigma), GAPDH (Sigma), Atg5 (Cell Signaling), Atg13 (Cell Signaling), and EEA1 (BD Transduction Labs). ApeI and carboxypeptidase Y antibodies were described previously (14, 15). The wild-type beclin-1 construct was received from B. Levine, and mutants constructed using standard molecular biology techniques. Vps34 3×FLAG was from the laboratory of Q. Zhong (plasmid number 24398; Addgene) (16). FLAG Vps15 was a gift from the laboratory of J. Hurley, NIDDK. For transient expression of proteins, cells were transfected using FuGENE 6 or HD (Roche) or Lipofectamine LTX (Invitrogen). For small interfering RNA (siRNA) of UVRAG, Silencer select siRNA (Invitrogen) number s14743 was transfected with RNAiMAX (Invitrogen) according to the manufacturer's recommended protocol.
Confocal microscopy and immunocytochemistry.
Cells were fixed in warm 4% paraformaldehyde buffered with phosphate-buffered saline (PBS) for 15 min, permeabilized in 0.1% Triton X-100, and blocked in 3% goat serum for 30 min at room temperature. For immunostaining, cells were incubated with primary antibodies overnight at 4°C, washed, and then incubated with Alexa Fluor-conjugated secondary antibodies (Alexa Fluor 488 or 555; Invitrogen) for 1 h at room temperature. Images were captured with 63× (numerical aperture [NA], 1.4) or 100× (NA, 1.45) objectives using an LSM 510 microscope (Zeiss). For quantification of p62 protein levels in beclin-1 knockout and rescue lines, randomly selected images for each three independent experiments were quantified in ImageJ by taking the integrated intensity and dividing by the cellular area. All images were collected with the same settings determined prior to the experiment to yield nonsaturating conditions.
Cell fractionation and immunoprecipitation of beclin-1.
For fractionation, cells were resuspended in 20 mM HEPES, pH 7.5, 10 mM KCl, 1.5 mM MgCl2, 1 mM EDTA, 1 mM EGTA, 1 mM dithiothreitol (DTT), 250 mM sucrose, 0.1 mM phenylmethylsulfonyl fluoride (PMSF), passed 7 times through a 25-gauge needle, and clarified at 2,000 × g at 4°C for 20 min. Centrifugation at 10,000 × g yielded cytosol (including light membranes) and pellet (heavy membranes). The supernatant was centrifuged at 100,000 × g for 30 min at 4°C to collect light membranes. Heavy membranes were resuspended in buffer containing 10 mM HEPES, pH 7.5, and 100 mM Na2CO3, incubated at 4°C for 30 min, and centrifuged at 100,000 × g at 4°C for 30 min to achieve carbonate extraction. For isolation of YFP-tagged beclin-1 protein, cell lysates were prepared in 50 mM Tris, pH 7.5, 50 NaCl mM, 1 mM EDTA, 0.5% Triton X-100, clarified, incubated with GFP-Trap beads (Allele Biotechnology) for 2 h at 4°C, washed 5 times in lysis buffer, and eluted by boiling in SDS-PAGE buffer.
Yeast lysates, survival analysis, and electron microscopy.
Standard procedures were used for the cultivation and transformation of yeast cells. For survival analysis, all strains used were of the W303 (MATa ade2-1 can1-100 his3-11 leu2-3 trp1-1 ura3) background. Cells were grown at 26°C in complex XY medium (20 g/liter Bacto peptone, 10 g/liter yeast extract, 10 mM KH2PO4, 0.2 g/liter tryptophan, 0.1 g/liter adenine) supplemented with 2% glucose. For nitrogen starvation of yeast cells, cells were grown overnight in prestarvation medium (1.7 g/liter yeast nitrogen base [YNB] [Y-1251; Sigma], 5 g/liter ammonium sulfate, 2% glucose, 1× DropOut mix, 0.1% auxotrophic requirements), washed two times with water, resuspended at 0.25 optical density (OD) unit per ml at a wavelength of 600 nm (OD600) in starvation medium (1.7 g/liter YNB [Y-1251; Sigma], 2% glucose) and cultivated for 5 days at 30°C. To determine the survival rate, cells were counted using disposable Neubauer counting chambers, diluted to 2,000 cells/ml in water, and plated onto glucose containing XY plates. CFU were scored after 3 days of incubation at 30°C. To delete ATG6 or the C-terminal domain of ATG6, cells of the wild-type strain K699 were transformed with the appropriate PCR fragment for the amplification of the kanamycin marker.
To prepare samples to analyze Cvt autophagy and macroautophagy by immunoblotting, the yeast cells were harvested in 10% cold trichloroacetic acid (TCA), washed three times in cold acetone, and lysed in a BeadBeater for 30 min in a buffer containing 6 M urea, 50 mM Tris, pH 6.8, 1% SDS, 1 mM EDTA. Samples were warmed to 65°C before immunoblotting analysis.
To analyze GFP-carboxypeptidase S (CPS) trafficking, a plasmid expressing the chimeric GFP-CPS protein under the control of the carboxypeptidase Y (CPY) promoter (17) was introduced into the genetic backgrounds indicated below. Correct delivery of the chimera to the vacuolar lumen, which requires a functional vacuolar protein sorting (VPS) trafficking pathway, was assessed by anti-GFP immunoblotting and fluorescence microscopy (17).
To prepare yeast samples for electron microscopy analysis, the strains indicated below were treated as follows: 10 OD600 units were washed twice with warm distilled water and then resuspended with vortexing in fresh 1.5% KMnO4, incubated for 20 min at room temperature, washed repeatedly with distilled water until the supernatant was clear, and then stored as a 1:1 slurry in water until processing.
Mammalian cell starvation and lysate dephosphorylation.
For HCT 116 cell starvation, the following buffer was used: 140 mM NaCl, 1 mM CaCl2, 1 mM MgCl2, 5 mM glucose, 20 mM HEPES, 1% bovine serum albumin (BSA) (wt/vol), pH 7.4. Cells were washed with this buffer 3× before incubation for the time periods indicated below. Inhibition of lysosomal degradation was achieved by concurrent addition of bafilomycin (Sigma) to 50 nM. For dephosphorylation of lysates, samples were lysed in buffer containing 1× NEB3 (New England BioLabs) and 1% Triton X-100 by repeated passage through a 26.5-gauge needle. Half the sample was incubated as a control condition, and calf intestinal phosphatase (New England BioLabs) was added to the other half, with digestion at 37°C for 1 h.
Phos-tag gels and immunoblotting.
For analysis of protein phosphorylation, Phos-tag (Wako) and MnCl2 were added to normal 7% or 8% Tris-glycine minigels at the levels recommended by the manufacturer. Immunoblotting was conducted normally after soaking the gels in 1 mM EDTA for 10 min to remove the Mn2+. All other steps in this analysis were identical to normal SDS-PAGE and immunoblotting protocols. Transfer was conducted onto polyvinyl difluoride membranes, except for the experiment analyzing Atg13 (see Fig. 8E), where we found that Atg13 blotting worked substantially better using nitrocellulose membranes. For quantification of bands in both normal SDS-PAGE and Phos-tag immunoblotting, Image Lab software (Bio-Rad) was used, with manual labeling of the boundaries of all bands of interest.
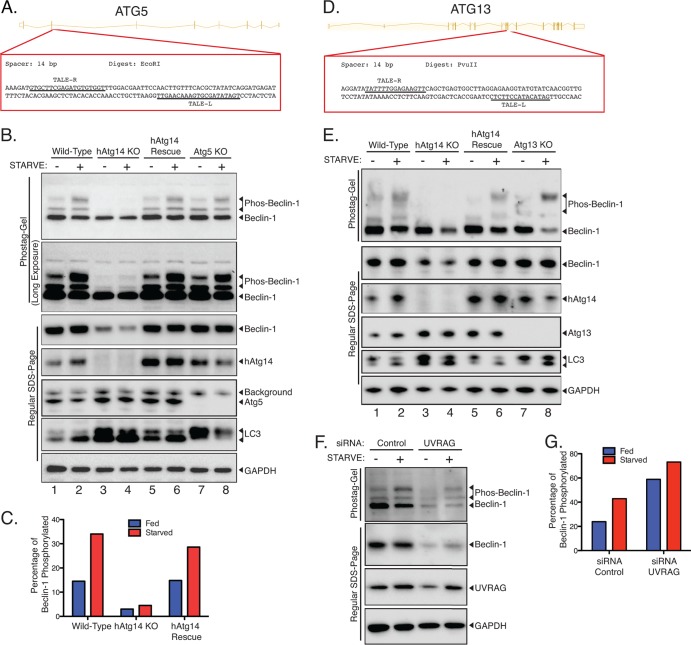
Fig 8.
Beclin-1 phosphorylation is dependent on hAtg14. (A) Strategy for knocking out Atg5 using TALE nucleases. Exon 2 was targeted in the region shown, with the two TALE nuclease binding sequences indicated by underlining. Clones were selected based on EcoRI digest after PCR of the region of interest. (B) Wild-type HCT 116 cells, hAtg14 KO cells, hAtg14 KO cells stably reexpressing hAtg14, or Atg5 KO cells were incubated under nutrient-rich or starvation conditions for 2 h, and cellular lysates analyzed by both Phos-tag and regular SDS-PAGE. A long exposure of a Phos-tag immunoblot of beclin-1 serves to emphasize the very minimal level of beclin-1 phosphorylation in the absence of hAtg14. Immunoblotting for LC3 and GAPDH controlled for autophagy and protein loading, respectively. (C) Quantification of the Phos-tag analysis (nonsaturating exposure) shown in panel B. (D) Strategy for knocking out Atg13 using TALE nucleases. Exon 9 was targeted in the region shown, with the two TALE nuclease binding sequences indicated by underlining. Clones were selected based on PvuII digest after PCR of the region of interest. (E) Analysis of Atg13 KO cells for beclin-1 phosphorylation. Wild-type HCT 116 cells, hAtg14 KO cells, hAtg14 KO cells stably reexpressing hAtg14, or Atg13 KO cells were incubated under nutrient-rich or starvation conditions for 2 h, and cellular lysates were analyzed by both Phos-tag and regular SDS-PAGE. (F) Analysis of beclin-1 phosphorylation after UVRAG siRNA treatment. HCT 116 cells were transfected with siRNA against UVRAG and, after 2 days of expression, incubated under nutrient-rich or starvation conditions for 2 h, and cellular lysates were analyzed by both Phos-tag and regular SDS-PAGE. (G) Quantification of the Phos-tag analysis shown in panel F.
RESULTS
TALE nuclease-mediated knockout of hAtg14 and beclin-1.
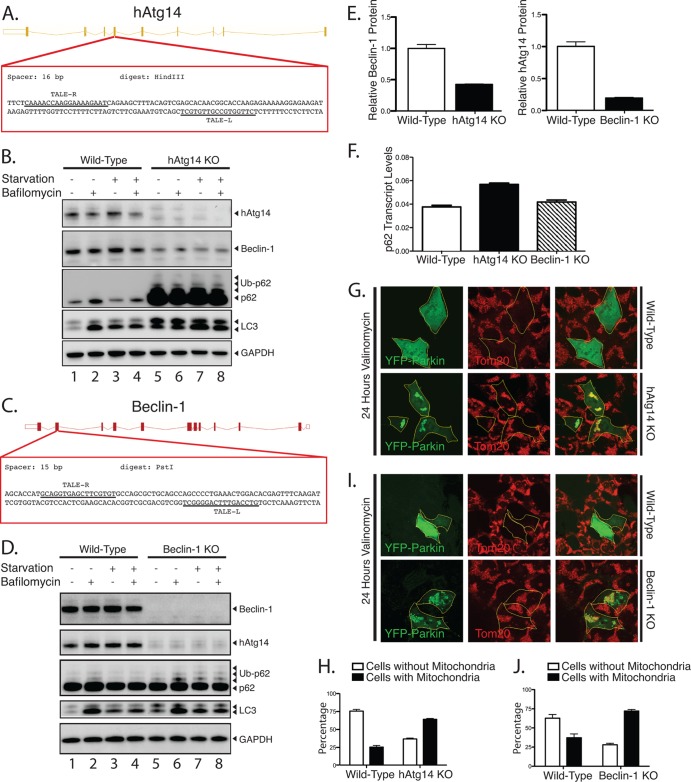
To examine hAtg14 and beclin-1 function, we generated knockout (KO) cell lines for each of these proteins in HCT 116 cells, which are diploid for most genes. We utilized site-directed genetic disruption with engineered TALE nucleases (TALENs). We targeted exon 4 of hAtg14 with a TALEN to efficiently disrupt protein expression (Fig. 1A and B). By immunoblot, we observed defective autophagy in hAtg14 knockout cells, with accumulation of p62 under basal and starvation conditions and accumulation of LC3 relative to the levels in wild-type cells (Fig. 1B). We employed a similar genomic editing strategy to ablate beclin-1 expression in HCT 116 cells (Fig. 1C and D). In this case, we achieved efficient knockout of beclin-1 by targeting exon 2 and isolated two functionally identical beclin-1 knockout clones (not shown; only one clone was used in the remainder of the study). Beclin-1 knockout cells accumulated some ubiquitinated p62, although to a lesser extent than hAtg14 knockouts (Fig. 1D). Consistent with previous results (6), we observed both a large reduction in beclin-1 levels in hAtg14 KO cells and a large reduction in hAtg14 levels in beclin-1 KO cells (Fig. 1B, D, and E). Given the large quantities of p62 we observed in hAtg14 KO cells and that p62 levels have been reported to be transcriptionally regulated in response to stress (18), we measured p62 transcript levels in wild-type, hAtg14 KO, and beclin-1 KO cells (Fig. 1F). Notably, p62 transcript levels increased less than 2-fold in hAtg14 KO cells, which can account for only some of the robust increase observed in p62 protein levels (Fig. 1B).
Fig 1.
Knockout of hAtg14/Barkor and beclin-1 using TALE nuclease technology impairs autophagy. (A) Strategy for knocking out hAtg14 using transcription activator-like effector (TALE) nucleases. Exon 4 was targeted in the region shown, with the two TALE nuclease binding sequences indicated by underlining. Clones were selected based on HindIII digest after PCR of the region of interest. (B) Immunoblot analysis of hAtg14 knockout (KO) cells under normal and treated conditions. Cells were starved and/or treated with bafilomycin A1 (50 nM) for 3 h, and total lysates analyzed by immunoblotting for the proteins indicated. Ub, ubiquinated. (C) Strategy for knocking out beclin-1 using TALE nucleases. Exon 2 was targeted in the region shown, with the two TALE nuclease binding sequences indicated by underlining. Clones were selected based on PstI digest after PCR of the region of interest. (D) Immunoblot analysis of beclin-1 KO cells under normal and treated conditions. Cells were starved and/or treated with bafilomycin A1 (50 nM) for 3 h, and total lysates analyzed by immunoblotting for the proteins indicated. (E) Quantification of beclin-1 levels in hAtg14 KO cells and of hAtg14 levels in beclin-1 KO cells. Levels were normalized to wild-type protein levels found in HCT 116 cells. (F) Quantification of p62/SQSTM1 transcript levels, as assessed by TaqMan assay on triplicate samples from wild-type HCT 116, hAtg14 KO, or beclin-1 KO cells. (G) Analysis of mitophagy in wild-type or hAtg14 KO cells. Cells transfected with YFP-parkin were treated with vehicle or valinomycin at 10 μM for 24 h and analyzed for mitophagy by staining for Tom20. Significant mitochondrial clearance was observed in wild-type HCT 116 but not in hAtg14 KO cells, which mostly displayed trapped mitochondrial clusters. (H) Quantification of results of experiments for which representative images are shown in panel G. (I) Wild-type or beclin-1 KO cells transiently expressing YFP-parkin were treated with valinomycin at 10 μM for 24 h and analyzed for mitophagy by staining for Tom20. Beclin-1 KO cells showed a defect in clearing damaged mitochondria. (J) Quantification of results of experiments for which representative images are shown in panel I. Error bars show standard errors of the mean.
We next examined whether hAtg14 and beclin-1 KO cells are impaired in cargo-specific autophagy. For this, we utilized parkin-mediated mitophagy, a well-characterized form of cargo-specific autophagy in mammalian cells (19). Following transient transfection of YFP-parkin into wild-type and hAtg14 knockout cells, we depolarized mitochondria with valinomycin treatment. At shorter times of incubation with valinomycin (3 h), we observed no defect in YFP-parkin translocation to mitochondria (not shown). A 24-h incubation with valinomycin resulted in the canonical mitochondrial clearance in wild-type cells, whereas mitochondrial clearance was greatly reduced in hAtg14 knockout cells (Fig. 1G and H), indicative of reduced mitophagy. We performed similar analyses for beclin-1. In beclin-1 KO cells transiently expressing YFP-parkin, we observed no defect in parkin translocation at shorter time periods (not shown) but a significant block in mitophagy upon 24-h mitochondrial depolarization (Fig. 1I and J). Thus, TALEN-mediated knockout of hAtg14 and beclin-1 resulted in a defect in specific autophagy, as well as starvation-induced autophagy.
Beclin-1 interaction with cellular membranes.
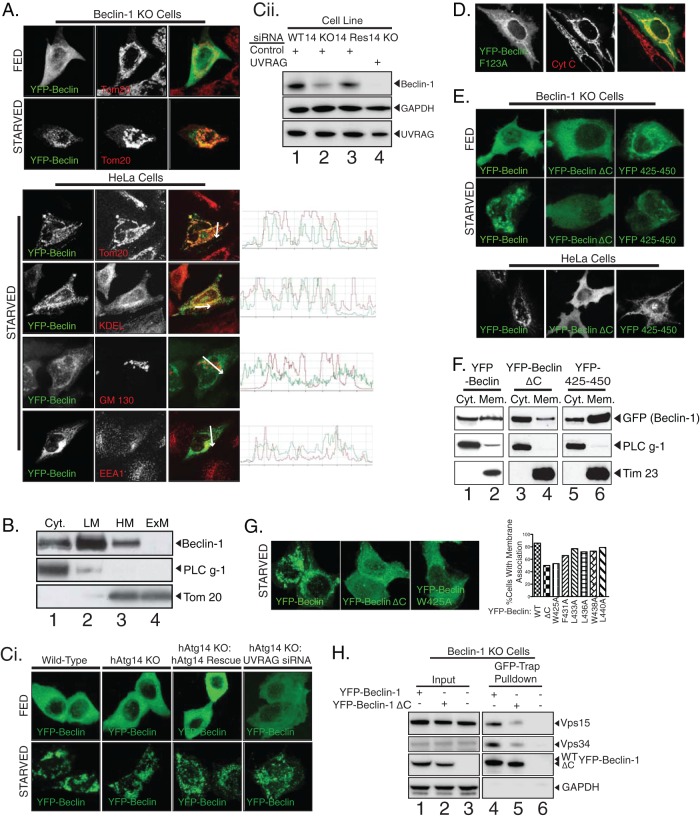
Given that the function of the beclin-1 complex must occur at membranes, we first examined the interaction of beclin-1 with membranes during autophagic processes. To circumvent the absence of a reliable antibody for immunostaining, we investigated beclin-1 subcellular localization by ectopic expression of YFP–beclin-1. YFP–beclin-1 in beclin-1 knockout cells displayed a notably mitochondrial staining under starvation conditions, although in HeLa cells, we also observed significant localization with endoplasmic reticulum (ER) and endosomal markers in some cells (Fig. 2A). We never observed strong colocalization of YFP–beclin-1 with the Golgi marker GM130. Since beclin-1 has no apparent organelle import signal or transmembrane domain, based on Kyte-Doolittle hydropathy analysis (not shown), we performed carbonate extraction experiments to demonstrate that, as expected, beclin-1 is a peripherally bound membrane protein rather than an integral membrane protein (Fig. 2B). We found that this membrane association was not determined by its autophagic binding partner hAtg14, at least in HCT 116 cells, since the pattern of membrane association upon starvation was unchanged in hAtg14 knockout cells (Fig. 2Ci and Cii). Given that UVRAG forms a separate beclin-1 complex and has been reported to be an even tighter binding partner than hAtg14, we knocked down UVRAG in hAtg14 knockout cells to determine whether UVRAG determined beclin-1 membrane association in hAtg14 knockout cells. However, we observed no reduction in the membrane localization of overexpressed YFP–beclin-1 upon starvation in hAtg14 knockout cells with UVRAG knockdown (Fig. 2Ci). Although UVRAG knockdown was incomplete, the combined knockout of hAtg14 and knockdown of UVRAG resulted in very little residual endogenous beclin-1 (Fig. 2Cii), suggesting that these beclin-1 binding partners play a critical role in complex stability. As beclin-1 interacts with Bcl-2, a protein that also localizes to mitochondria and ER (20, 21), we next tested whether this interaction was responsible for membrane localization. A YFP–beclin-1 mutant with phenylalanine mutated to alanine at position 123 (F123A), which is unable to bind Bcl-2 (22), localized to membranes, notably mitochondria (Fig. 2D). Thus, beclin-1 interaction with membranes may include contributions from domains beyond those which associate with Bcl-2 or hAtg14.
Fig 2.
Association of beclin-1 with membranes involves the C-terminal domain. (A) Beclin-1 KO cells expressing YFP–beclin-1 and starved for 2 h were counterstained for mitochondria (Tom20) (top). HeLa cells expressing YFP–beclin-1 and counterstained for cytochrome c to mark mitochondria (Tom20), endoplasmic reticulum (KDEL), Golgi body (GM130), or early endosomes (EEA1) under starvation conditions (bottom). (B) HeLa cells were fractionated into cytosol (Cyt.), light membranes (LM), heavy membranes (HM), and then carbonate-extracted heavy membranes (ExM) and analyzed for endogenous beclin-1. PLC γ-1 and Tom20 serve as markers of the cytosolic and heavy membrane fractions, respectively. (Ci) YFP–beclin-1 was expressed ectopically in wild-type cells, hAtg14 KO cells, hAtg14 KO cells stably reexpressing hAtg14, or hAtg14 KO cells transfected with UVRAG siRNA and analyzed under fed and starved conditions. (Cii) Prior to transfection, immunoblotting for endogenous beclin-1 and UVRAG was performed on the samples used in the experiments for which representative images are shown in panel Ci, with immunoblotting for GAPDH serving as a loading control. (D) Membrane binding of YFP–beclin-1 still occurs in Bcl-2 binding-deficient beclin-1 F123A. HeLa cells expressing beclin-1 F123A were fixed and counterstained for the mitochondrial marker cytochrome c (Cyt C). (E) Confocal imaging of ectopically expressed YFP–beclin-1, YFP–beclin-1 lacking the C-terminal domain, or the YFP-tagged beclin-1 C-terminal domain alone (YFP 425–450) in beclin-1 KO cells under nutrient-rich or starvation conditions. Similar results were seen in HeLa cells. (F) Fractionation of cellular lysates from HeLa cells expressing the constructs used in the experiments whose results are shown in panel E was analyzed by immunoblotting. PLC γ-1 and Tim23 serve as markers of cytosolic and membrane fractions, respectively. (G) Alanine scanning mutagenesis of C-terminal large hydrophobic residues. YFP–beclin-1 W425A and YFP–beclin-1 ΔC had similar reductions in the degree of membrane association upon starvation. (H) Immunoprecipitation with GFP-Trap of beclin-1 and beclin-1 ΔC from cells co-overexpressing YFP–beclin-1 (or ΔC), Vps34-Flag, and Flag-Vps15.
The C-terminal domain in beclin-1 is involved in membrane association.
To identify novel domains involved in the membrane association of beclin-1, we examined the subcellular localization of serial N-terminal and C-terminal deletion mutants of YFP–beclin-1 by confocal microscopy. Deletion of the C-terminal 26 amino acids from full-length YFP–beclin-1 (beclin-1 ΔC) greatly reduced membrane localization, as shown by immunofluorescence in beclin-1 KO cells, HeLa cells (Fig. 2E), and wild-type HCT 116 cells (not shown). Conversely, the C-terminal 26 amino acids (425 to 450) of beclin-1 were sufficient to localize YFP to membranes in a subset of cells (Fig. 2E). Subcellular fractionation of cells revealed that YFP fused to the beclin-1 C-terminal 26 amino acids localizes to membranes to a greater extent than YFP–beclin-1 ΔC and comparably to YFP–beclin-1 (Fig. 2F). Given the preponderance of large hydrophobic amino acids in the C-terminal domain, we next sought to determine individual residues in the C-terminal 26 amino acids that contributed strongly to membrane association. We performed alanine scanning mutagenesis of conserved residues and identified tryptophan 425 as a major contributor to the observed starvation-induced membrane localization of YFP–beclin-1 (Fig. 2G). To examine the effect of C-terminal deletion of beclin-1 on protein complex formation, we performed immunoprecipitation experiments of overexpressed YFP–beclin-1 or YFP–beclin-1 ΔC in beclin-1 KO lines. We observed that Vps34 and Vps15 still bound to YFP–beclin-1 ΔC, although to a lesser extent than wild-type YFP–beclin-1 (Fig. 2H), suggesting that membrane association may help to stabilize the beclin-1 complex.
The C-terminal domain is required for yeast Atg6 membrane association.
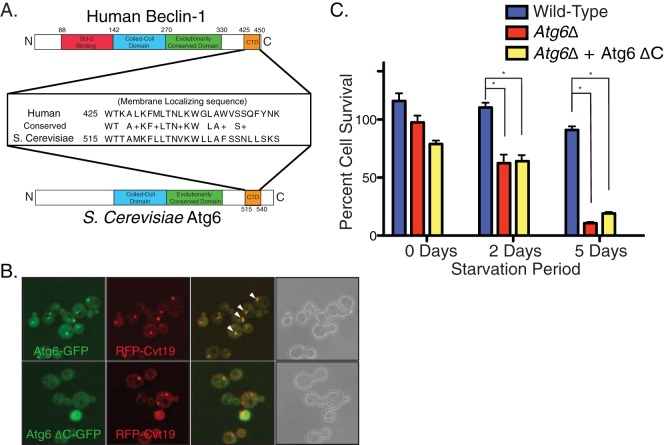
Since the C-terminal region of beclin-1 is one of the most evolutionarily conserved regions in the protein (Fig. 3A), we examined the role of this domain in the yeast homologue, Atg6/Vps30. Full-length Atg6-GFP localized to discrete puncta, some of which correspond to the pre-autophagosomal structure (PAS), as indicated by partial colocalization with the known PAS marker red fluorescent protein (RFP)-Cvt19 (Fig. 3B) (23) and as has been observed previously (7). Deletion of the C-terminal region of Atg6 (Atg6 ΔC-GFP) abrogated the punctate distribution and colocalization with RFP-Cvt19, and the fluorescence was observed only in the cytosol (Fig. 3B). Therefore, although the subcellular localization of the yeast homologue is distinct from that of beclin-1, the role of the C-terminal domain (CTD) in membrane association appears conserved.
Fig 3.
The C-terminal domain (CTD) of beclin-1 is conserved in the yeast Atg6 and required for membrane association and cell survival in response to starvation. (A) Sequence alignment of the conserved CTD (orange) of human beclin-1 and yeast Atg6, in schematic with other known domains. (B) Confocal imaging of yeast cells expressing Atg6-GFP or Atg6 ΔC-GFP from the endogenous promoter and coexpressing plasmid-borne RFP-Cvt19 as a PAS marker. Colocalizing regions of GFP-Atg6 puncta with RFP-Cvt19 are indicated by arrowheads. No significant difference was observed upon nutrient deprivation (not shown). (C) Atg6Δ cells or cells expressing wild-type Atg6 or Atg6 ΔC were plated out and starved for the times indicated, and the cell survival rate quantified. Error bars show standard standard errors of the mean. Unpaired t test. * denotes P < 0.05.
The C-terminal domain is required for survival of yeast under nitrogen starvation.
Because both beclin-1 ΔC and Atg6 ΔC are defective in membrane association, we explored the function of CTD in autophagy. Considering the mild phenotype of beclin-1 KO cells, we examined the role of the CTD for the yeast Atg6. More than 70% of yeast with wild-type Atg6 survived 5 days of incubation under starvation conditions, whereas less than 10% of Atg6Δ yeast survived (Fig. 3C). Less than 20% of Atg6 ΔC cells survived following 5 days in starvation medium, comparable to the survival of Atg6Δ cells (Fig. 3C). Thus, the CTD is required for the autophagic function of Atg6 during starvation.
Loss of the Atg6 C-terminal domain results in a Cvt pathway lesion.
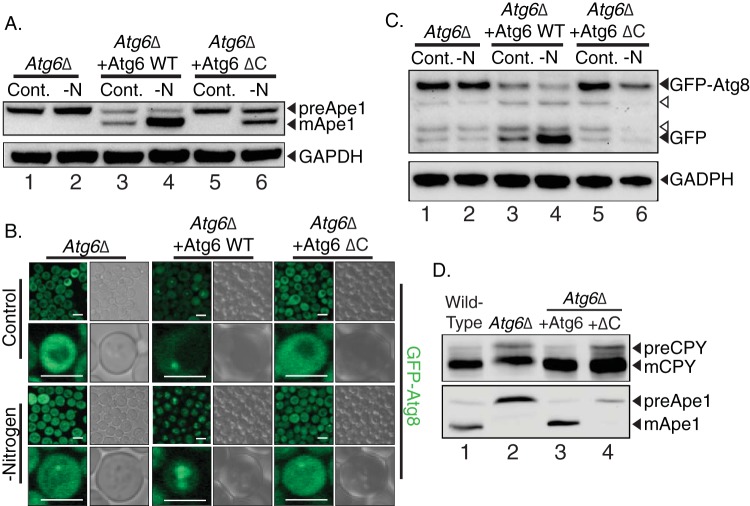
To elucidate the mechanism of the CTD in autophagy, we analyzed the effect of truncating this region on Cvt pathway autophagy. Atg6Δ cells were unable to process aminopeptidase I (Ape1) into its mature form, whereas the expression of wild-type Atg6 restored this process (Fig. 4A). Conversely, the expression of Atg6 ΔC did not rescue Ape1 maturation when cells were incubated under nitrogen-replete conditions, under which the Cvt pathway is the dominant form of cellular autophagy in yeast (22). Under nitrogen starvation conditions, however, where macroautophagy predominates, Atg6 ΔC partially rescued Ape1 processing (Fig. 4A, lane 6). This phenotype is typical of Cvt pathway-specific lesions, in which Ape1 is matured only upon the induction of macroautophagy by starvation. However, the fact that Atg6 ΔC does not allow starvation-induced Atg8 trafficking to the vacuole (Fig. 4B) argues that, while Atg6 ΔC can recapitulate some aspects of the Cvt-to-macroautophagy transition, it does not support a fully normal macroautophagic response (24).
Fig 4.
The CTD is required for specific (Cvt) autophagy, macroautophagy, and vacuolar protein sorting. (A) Analysis of Cvt pathway autophagy by aminopeptidase-I processing. Atg6Δ cells or those cells rescued with Atg6 or Atg6 ΔC were depleted of nitrogen (-N) for 3 h, and the cellular lysates analyzed by immunoblotting for Ape1. GAPDH serves as a loading control. (B) Analysis of autophagy in yeast using an Atg8-GFP strain. Atg6Δ cells, Atg6Δ cells expressing Atg6, or Atg6 ΔC cells were subjected to control or nitrogen-depleted conditions and analyzed by confocal microscopy. Scale bars are 5 μm. (C) Immunoblot for GFP-Atg8 processing in the same strains used in the experiments for which representative results are shown in panel B. GAPDH serves as a loading control. Open arrowheads denote background bands. (D) Wild-type cells, Atg6Δ cells, or Atg6Δ cells expressing Atg6-RFP (+Atg6) and Atg6 ΔC-RFP (+ΔC) from the genomic Atg6 locus were grown to mid-log phase and analyzed for their capacity to carry out vacuolar protein sorting, as assessed by carboxypeptidase Y (CPY) cleavage. Immunoblotting for Ape1 cleavage serves to control for defects in specific autophagy due to Atg6 deletion. preApe1, precursor Ape1; mApe1, mature Ape1; preCPY, precursor CPY; mCPY, mature CPY.
The Atg6 C-terminal domain is required for macroautophagy and vacuolar protein sorting.
To determine the effect of CTD truncation on macroautophagy, we analyzed GFP-Atg8 trafficking. Atg6Δ cells failed to deliver GFP-Atg8 to the vacuole upon nitrogen starvation (Fig. 4B, left). The expression of plasmid-borne Atg6 variants in this deletion background showed a clear difference in the GFP-Atg8 patterns in cells expressing wild-type Atg6 (Fig. 4B, middle) and cells expressing Atg6 ΔC (Fig. 4B, right). Under nitrogen-replete conditions, cells expressing Atg6 show a cytosolic distribution of GFP-Atg8 with some puncta. In contrast, cells expressing Atg6 ΔC lack punctate GFP-Atg8 and show only cytosolic staining. Upon the induction of macroautophagy by nitrogen starvation, cells expressing Atg6 deliver GFP-Atg8 to the vacuole. However, in yeast cells expressing Atg6 ΔC, no vacuolar GFP-Atg8 is observed, indicating a defect in macroautophagy. These observations were corroborated by immunoblotting for GFP. Upon starvation, Atg6Δ cells expressing wild-type Atg6 cleave GFP-Atg8 to yield free GFP (Fig. 4C, lane 4), reflecting the transport of GFP-Atg8 into the vacuolar lumen. However, in extracts from Atg6Δ cells expressing Atg6 ΔC, the release of GFP was not observed, similar to the results in Atg6Δ cells (Fig. 4C). This confirmed a defect in macroautophagy in Atg6 ΔC cells. As Atg6 (also known as Vps30) is known to function in vacuolar protein sorting, we tested whether the Atg6 CTD deletion impacted this pathway as well. We analyzed the trafficking of carboxypeptidase Y (CPY), a known marker of vacuolar protein sorting functions in yeast (25). Whereas wild-type and Atg6Δ cells both demonstrated a complete conversion of CPY to the mature, vacuolar form, Atg6Δ cells and Atg6Δ cells expressing Atg6 ΔC showed a defect in CPY conversion, retaining some of the higher-molecular-weight precursor form (Fig. 4D). The blockage in CPY conversion was relatively minor compared with the complete ablation of Cvt pathway sorting of aminopeptidase. These results indicate that the CTD of Atg6 is required for both the Cvt pathway and macroautophagy and has a lesser effect on vacuolar protein sorting.
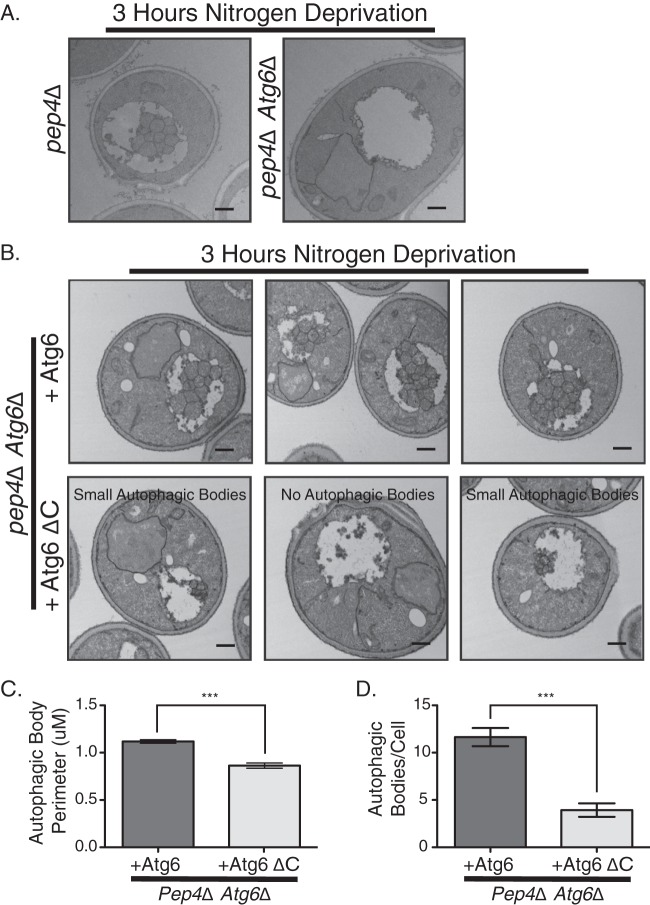
The C-terminal domain of Atg6 is required for normal autophagosome morphology.
Based on the relatively large Cvt pathway lesion and macroautophagy defects we observed with Atg6 ΔC cells, we speculated that autophagosome morphology might be altered, as observed for Atg8 defects (26, 27). We visualized autophagic bodies by electron microscopy in a Pep4Δ background, where autophagic bodies accumulate in the vacuolar lumen rather than being degraded. As expected, under starvation conditions, Pep4Δ cells accumulate normal autophagic bodies (26), whereas Pep4Δ Atg6Δ cells were unable to accumulate autophagic bodies (Fig. 5A). Normal autophagic bodies were restored upon the introduction of plasmid-borne wild-type Atg6 (Fig. 5B, top), whereas the expression of Atg6 ΔC only partially restored the formation of autophagic bodies, and these displayed a novel morphological phenotype. There were far fewer autophagic bodies per cell in the Atg6 ΔC cells (3.9/cell compared with 11.6/cell for the wild type) (Fig. 5D), and those present were 25% smaller than in Atg6 cells, as measured by tracing of the perimeter of the autophagic body (Fig. 5C). This morphological phenotype is consistent with a Cvt pathway lesion and the macroautophagy defects observed in Atg6 ΔC cells. Autophagic bodies were never observed in nitrogen-replete cells (not shown), and no unusual vacuolar structures were observed in Atg6 ΔC cells, again indicating that the CTD specifically influences the autophagy function of Atg6. Taken together with our observations of a Cvt pathway lesion and a defect in macroautophagy, these results demonstrate that deletion of the CTD in yeast Atg6 induces a specific defect in autophagic morphology.
Fig 5.
The Atg6 C-terminal domain controls autophagic body size and number. (A) Pep4Δ or Pep4Δ Atg6Δ cells were starved and analyzed by electron microscopy for autophagic body formation. (B) Electron microscopy analysis of Pep4Δ Atg6Δ cells expressing either wild-type Atg6 or Atg6 ΔC. Pep4Δ Atg6Δ cells expressing Atg6 had autophagic bodies morphologically similar to those in Pep4Δ cells, whereas cells expressing Atg6 ΔC had either small autophagic bodies or no autophagic bodies, as shown in the three sample images. (C) Quantification of autophagic body perimeters in cells used in the experiments for which representative images are shown in panel B. (D) Quantification of autophagic body numbers in cells used in the experiments for which representative images are shown in panel B. Scale bar is 500 nm in all images. Error bars show standard errors of the mean. Unpaired t test. *** denotes P < 0.0001.
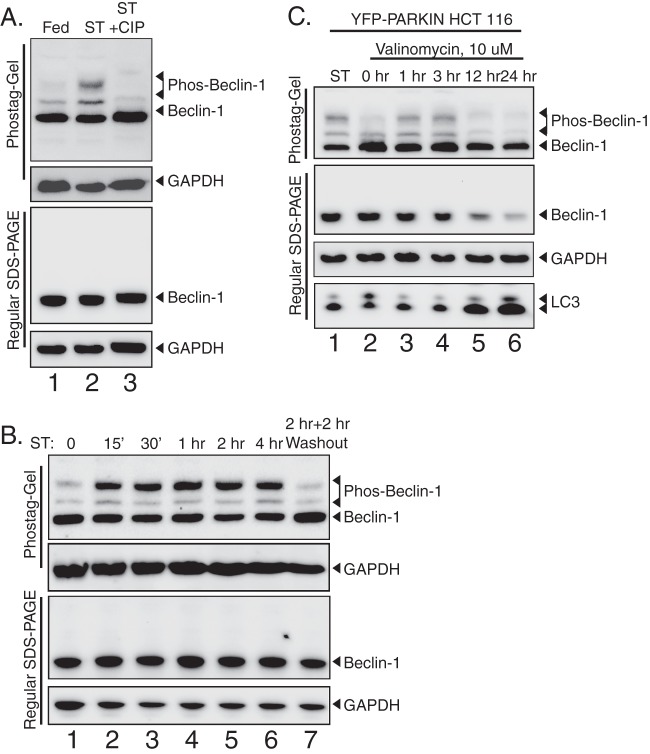
Beclin-1 is phosphorylated in response to autophagic stimuli.
Whereas Atg6 yeast is constitutively localized to the PAS via the C-terminal domain, human beclin-1 binding to membranes via the CTD increases under starvation conditions, suggesting that there may exist specific signals that regulate this process (Fig. 2). Searching for posttranslational modifications of beclin-1 and hAtg14 that might regulate subcellular location and autophagic function, we observed autophagy-dependent phosphorylation of beclin-1 (Fig. 6). To assess phosphorylation, we utilized Phos-tag gels, on which phosphorylated bands migrate as supershifted species (25). As shown (Fig. 6A), upon starvation, beclin-1 migrates as three distinct species on a Phos-tag gel but as only one species on a normal SDS-PAGE gel, as analyzed by immunoblotting. We confirmed that these higher bands represented phosphorylated species of beclin-1 by treatment with phosphatase, which reduced beclin-1 to a single band (Fig. 6A, lane 3). Given that in previous studies using Phos-tag analysis, each individual band usually corresponded to a single phosphorylation event (28), we considered it most likely that these two bands represented two sites of phosphorylation which occur additively with respect to the supershifting on the gel. As shown (Fig. 6B), this phosphorylation is strongly reversed in response to nutrient replenishment, further linking this phosphorylation event to autophagy induction and nutrient stress. In order to confirm that the observed phosphorylation event occurs in response to multiple forms of autophagic stimuli, we examined beclin-1 phosphorylation during mitophagy. For this, we utilized cells stably expressing YFP-parkin and treated with the mitochondrion-uncoupling agent valinomycin (19, 29). As shown (Fig. 6C), beclin-1 phosphorylation is an early event in mitophagy, confirming the importance of this modification in multiple autophagic pathways. At longer time points, we observed degradation of beclin-1 during mitophagy compared with that of the loading control GAPDH, which is not surprising given the high level of autophagic flux, as indicated by LC3 conversion.
Fig 6.
Beclin-1 is phosphorylated during autophagy and mitophagy. (A) Phos-tag immunoblot analysis of beclin-1 during starvation. Wild-type HCT 116 cells were maintained in nutrient-rich medium (fed) or starved for 2 h (ST). Starved lysates were treated with calf intestinal phosphatase (ST+CIP) to remove phosphate groups and analyzed by immunoblotting after Phos-tag or regular SDS-PAGE. (B) Reversal of autophagic stimuli removes beclin-1 phosphorylation. Lysates were collected and analyzed after incubation in starvation medium for the indicated times or after 2 h under starvation conditions followed by a 2-h chase in normal growth medium (lane 6). (C) Beclin-1 phosphorylation occurs during mitochondrial autophagy. HCT 116 cells stably expressing YFP-parkin, an effector of mitochondrial autophagy, were treated with the mitochondrial uncoupler valinomycin (10 μM) for the indicated times, and the lysates analyzed by Phos-tag or normal SDS-PAGE, followed by immunoblotting for the indicated proteins.
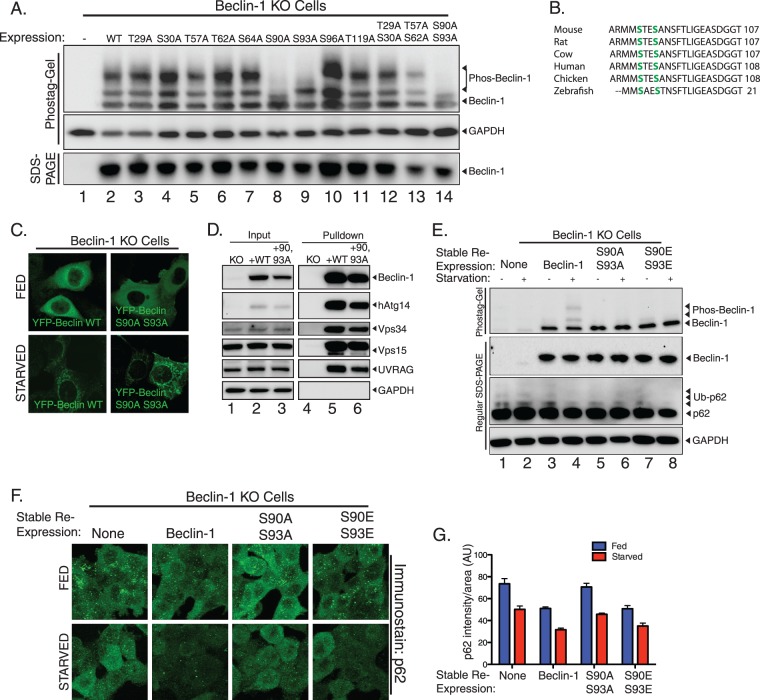
Serines 90 and 93 are autophagy-dependent beclin-1 phosphorylation sites.
In order to determine the function of beclin-1 phosphorylation during autophagy, we sought to identify the sites of phosphorylation. Previous literature reported that threonine 119 is phosphorylated during autophagy by death-associated protein kinase (DAPK) (30), and a more-recent report demonstrated two autophagy inhibitory events at serines 234 and 295 (31). Since the latter report suggested that these events correlated inversely with autophagy, we examined mass spectrometry studies which determined a number of different serines and threonines which are potentially modified by phosphorylation (32, 33). We undertook alanine scanning mutagenesis of these serine and threonine residues to identify phosphorylation sites. In order to ascertain which site is phosphorylated, we transiently expressed mutants of beclin-1 in beclin-1 knockout cells (Fig. 7A), in which phosphorylation occurs at high levels even in the absence of autophagic stimuli, perhaps as compensation for prolonged lack of the beclin-1 activity. As shown (Fig. 7), we observed that, while the majority of point mutants retained the phosphorylation pattern of wild-type beclin-1, mutation of two neighboring serines, residues 90 and 93, ablated the phosphorylation (Fig. 7A, lanes 8, 9, and 14). These sites are conserved throughout vertebrates (Fig. 7B), although only one of these sites is conserved in yeast. Notably, while the S93A mutation removed only the higher-shifted phosphorylation band, the S90A mutation and the S90A/S93A double mutation completely ablated phosphorylation (Fig. 7A, lanes 8, 9, and 14). Based on the observation that mutation of S90 blocks all beclin-1 phosphorylation, it is possible that S90 phosphorylation occurs prior to phosphorylation of S93 and that either monophosphorylated beclin-1 at S90 or diphosphorylated beclin-1 at S90 and S93 represents the activated, autophagic form of beclin-1.
Fig 7.
Phosphorylation of serines 90 and 93 in beclin-1 contributes to autophagic function. (A) Beclin-1 KO cells or beclin-1 KO cells expressing beclin-1 wild type or indicated mutant were analyzed by immunoblotting after Phos-tag or regular SDS-PAGE. This approach identified serines 90 and 93 as responsible for the supershifting observed when performing Phos-tag immunoblotting of beclin-1 (lanes 8, 9, and 14). (B) Beclin-1 phosphorylation sites are conserved in higher eukaryotes. Sequence alignment of beclin-1 phosphorylation sites and surrounding amino acids of the indicated species. Phosphorylated serines are highlighted in green. (C) Beclin-1 KO cells were transfected with YFP–beclin-1 wild type or YFP–beclin-1 S90A/S93A, incubated under nutrient-rich or starvation conditions, and analyzed by confocal microscopy. (D) Complex formation in beclin-1 KO cells stably reexpressing wild-type or S90A/S93A YFP–beclin-1. Cell lysates were prepared, and complexes purified by incubation with GFP-Trap beads and analyzed by immunoblotting for the indicated proteins. Equal retention of binding partners was observed regardless of mutation. (E) Beclin-1 KO cells stably reexpressing wild-type, S90A/S93A, or S90E/S93E beclin-1 were compared with KO cells for the ability to turn over p62 in response to starvation. (F) p62 turnover in the cell lines used in the experiments for which results are shown in panel E was assessed by immunofluorescence detection of endogenous p62 under both nutrient-rich and starvation conditions; n = 3 separate experiments. (G) Quantification of results from experiments for which representative images are shown in panel F. Error bars show standard errors of the mean. AU, arbitrary units.
Beclin-1 phosphorylation does not alter complex formation or membrane association.
We examined whether beclin-1 phosphorylation during autophagy is required for binding to membranes during autophagy. Under starvation conditions, YFP–beclin-1 S90A/S93A still revealed a membrane-bound pattern, similar to that of wild-type YFP–beclin-1 (Fig. 7C), suggesting that ablation of beclin-1 phosphorylation does not impair membrane association. Thus, membrane binding appears to be upstream from beclin-1 phosphorylation. We also tested the ability of beclin-1 S90A/S93A to form its normal complex with Vps15 and Vps34. For these experiments, we utilized stable rescue lines of beclin-1 knockout cells expressing a low level of YFP–beclin-1 or YFP–beclin-1 S90A/S93A. As shown (Fig. 7D), pulldown of beclin-1 resulted in similar retention of the binding partners Vps15 and Vps34 independent of mutation of the phosphorylation sites. Both hAtg14 and UVRAG, which are known to form mutually exclusive beclin-1 complexes (6), were bound to YFP–beclin-1 S90A/S93A, indicating that beclin-1 phosphorylation is not required for the formation of these beclin-1 complexes.
Phosphorylation of beclin-1 is involved in autophagy.
To assess the effects of beclin-1 phosphorylation at serines 90 and 93 on autophagy, we generated additional stable rescues of beclin-1 knockout cells using untagged wild-type beclin-1, S90A/S93A beclin-1, or the phosphomimetic S90E/S93E beclin-1. In response to starvation, cells stably expressing wild-type beclin-1 displayed an increase in beclin-1 phosphorylation, whereas cells stably expressing the phosphomutant S90A/S93A beclin-1 or the phosphomimetic S90E/S93E beclin-1 did not (Fig. 7E, top). Importantly, we observed a reduction in the clearance of the ubiquitinated form of the autophagy adaptor protein p62 in response to starvation in cells expressing S90A/S93A beclin-1, whereas cells expressing wild-type beclin-1 were able to clear p62 at an accelerated rate relative to its clearance in knockout cells in response to starvation. S90E/S93E beclin-1-expressing cells displayed clearance similar to that observed in wild-type beclin-1-expressing cells. To further investigate the functional importance of beclin-1 phosphorylation at serines 90 and 93, we conducted immunofluorescence analysis for p62 under normal and starvation conditions. In agreement with our immunoblotting results, the p62 levels, as measured by total integrated fluorescence per unit area, were higher under both nutrient-replete and starvation conditions in beclin-1 KO cells, a condition that was rescued by reexpression of wild-type but not S90A/S93A beclin-1. As in our immunoblotting analysis, S90E/S93E beclin-1 displayed p62 levels similar to those for wild-type beclin-1. These results indicate that phosphorylation of beclin-1 at serines 90 and 93 is critical for maximally efficient autophagy.
Beclin-1 phosphorylation depends on hAtg14.
We next asked whether autophagy-dependent beclin-1 phosphorylation is controlled by other autophagic proteins, including proteins of the autophagy-specific beclin-1 complex. We analyzed beclin-1 phosphorylation levels during starvation in wild-type and hAtg14 KO cells. To control for the possibility that beclin-1 phosphorylation is nonspecifically dependent on autophagy, we prepared Atg5 KO cells using TALEN-mediated gene disruption (strategy is shown in Fig. 8A). Atg5 KO cells had normal beclin-1 phosphorylation levels, although autophagy was impaired as analyzed by LC3 immunoblotting (Fig. 8B, lanes 7 and 8). Importantly, we found that no phosphorylation of beclin-1 occurred under basal or starvation conditions in hAtg14 KO cells (Fig. 8B, lanes 3 and 4), indicating that phosphorylation is completely dependent on hAtg14. Stable reexpression of hAtg14 in these cells returned beclin-1 phosphorylation to normal levels and restored LC3 conversion (Fig. 8B, lanes 5 and 6). As cells with reduced hAtg14 have reduced beclin-1 levels (Fig. 1E) (6), we quantified the levels of phosphorylated beclin-1 relative to total beclin-1 in our Phos-tag analysis (Fig. 8C) to ensure that the phenomenon we observed was a reduction in beclin-1 phosphorylation rather than simply an overall reduction of beclin-1 levels. Relative to total beclin-1 protein levels, very minimal beclin-1 phosphorylation was observed in hAtg14 KO cells compared with the levels in wild-type HCT116 cells or hAtg14 KO cells stably reexpressing hAtg14 (Fig. 8C). To further corroborate the specificity of hAtg14 in controlling autophagy-dependent beclin-1 phosphorylation, we also prepared Atg13 KO cells using TALEN-mediated gene cleavage (Fig. 8D). As with Atg5 KO cells, no reduction in beclin-1 phosphorylation was observed (Fig. 8E, lanes 7 and 8), although an accumulation of LC3 was observed. hAtg14 KO cells demonstrated a failure to phosphorylate beclin-1 (Fig. 8E, lanes 3 and 4). We tested whether UVRAG, which interacts with beclin-1 in a second Vps34 complex, was also involved in beclin-1 phosphorylation. While siRNA of UVRAG greatly depleted total beclin-1 levels, consistent with a role in stabilization of beclin-1 by UVRAG, it did not ablate the capacity of beclin-1 to be phosphorylated in a starvation-dependent manner, despite considerably less total beclin-1 (Fig. 8F and G). Taken together, these data suggest that, while both UVRAG and hAtg14 bind beclin-1, the starvation-dependent phosphorylation we observed specifically requires hAtg14 function and not autophagy per se.
DISCUSSION
This study describes two features of human beclin-1 and its yeast homologue Atg6 that contribute to its autophagic function. First, we identify a C-terminal region in beclin-1 that is conserved in its yeast homologue, Atg6, and is involved in membrane association and controls autophagosome morphology. Second, we identify an autophagy-dependent phosphorylation of beclin-1 at serines 90 and 93 that is controlled by the binding partner hAtg14 and is required for full autophagic activity.
We show that in mammalian cells, the C-terminal domain of beclin-1 is necessary and sufficient for membrane association. Similarly, the homologous region of Atg6 is required for normal subcellular localization of the protein in yeast cells. Two other groups recently reported partial crystal structures of beclin-1 (34) and the yeast Atg6 (35) which suggested roles for C-terminal sequences of the respective proteins in membrane association and autophagy, results we herein confirm and expand upon. For the yeast Atg6, we found that deletion of the C-terminal domain results in a Cvt pathway defect in aminopeptidase I trafficking, which is partially rescued by starvation. Furthermore, autophagic delivery of GFP-Atg8 is fully blocked by the Atg6ΔC mutation, implying a significant defect in macroautophagy. Consistent with these results, electron microscopy analyses of autophagic bodies show that Atg6ΔC cells generate aberrantly small autophagic bodies. Similar defects in autophagosome formation have been observed in Atg8 mutant cells (26, 27). Taken together, it is tempting to speculate that the autophagosomes formed in yeast expressing Atg6ΔC do not properly recruit Atg8, as reflected by the block in GFP-Atg8 trafficking under starvation conditions. This also suggests a novel relationship between Atg6 and Atg8 function and implicates the C-terminal domain in mediating these effects on autophagosome expansion. One possible explanation is that the C-terminal domain plays a role in stabilizing Atg6 at sites of membrane curvature, rather than acting in the initial targeting of the protein to membranes. This interpretation is also reminiscent of a recent report of curvature-dependent membrane binding by the C terminus of hAtg14 (10). Although yeast Atg6 and mammalian beclin-1 diverge greatly in primary sequence, it is interesting to note that the two most-conserved domains are the domain involved in association with Vps34 and the C-terminal domain (Fig. 3A) (36). These two core functions of beclin-1 represent a possible scaffold from which Atg6/beclin-1 evolved to adapt to the variety of phosphatidylinositol 3-phosphate-dependent membrane trafficking functions mediated in eukaryotic cells.
We also describe an autophagy-dependent phosphorylation of beclin-1. We observed a marked increase in the phosphorylation of beclin-1 upon the induction of autophagy by at least two independent triggers: induction of starvation-induced autophagy and selective mitophagy, both correlated with enhanced phosphorylation of beclin-1. Using high-throughput mass spectrometry studies as a guide, we mapped this phosphorylation to serines 90 and 93, two sites of beclin-1 in a region of the protein with unclear function. Although we found that both sites were phosphorylated, it appears that S90 phosphorylation is probably the initial event, as S93A beclin-1 still presented an additional phosphorylated species, whereas S90A beclin-1 did not. The sequence surrounding S90 also suggests the possibility of methylation at R87 as another modification of beclin-1 which could affect S90 phosphorylation and protein function (37).
We examined the functional significance of beclin-1 phosphorylation at serines 90 and 93 by reexpression of S90A/S93A phospho-null or S90E/S93E phosphomimetic constructs in beclin-1 knockout cells. Rescue of beclin-1 knockouts with S90A/S93A beclin-1 did not restore maximal autophagy, whereas rescue with S90E/S93E displayed autophagy similar to that of the wild type, suggesting that phosphorylation at serines 90 and 93 is required for autophagy but not sufficient. We observed no contribution of these sites to either the autophagy-dependent relocalization of beclin-1 to membranes or the normal formation of either the hAtg14-containing or the UVRAG-containing Vps34 complexes.
Importantly, we demonstrated that beclin-1 phosphorylation is specifically dependent on hAtg14 by examining beclin-1 phosphorylation in hAtg14, Atg5, and Atg13 knockout cells, as well as under UVRAG knockdown conditions. Among our TALEN-derived KO lines, only hAtg14 was required for beclin-1 phosphorylation, and while knockdown of UVRAG greatly reduced beclin-1 protein expression levels, it did not ablate starvation-induced beclin-1 phosphorylation. This suggests that beclin-1 phosphorylation is a key early event in autophagy specifically controlled by hAtg14. Although the kinase for this event remains unclear given that multiple beclin-1 kinases have been proposed (30, 31), while this paper was under review, Kim et al. (38) identified the same phosphorylation sites in beclin-1, also reported dependence on hAtg14, and provided evidence that AMP-activated protein kinase is the responsible kinase. Their study used different techniques and was conducted largely in HEK and MEF cell lines, whereas the present study was conducted largely in HCT 116 cells. Taken together with this study and two previous large-scale proteomics studies that identified S90 and S93 as possible sites of beclin-1 phosphorylation (32, 33), the evidence suggests that these sites are major targets of phosphorylation-mediated regulation of beclin-1 activity.
This phosphorylation event also likely reflects only one of multiple posttranslational modifications occurring on beclin-1 during any given cellular context. It is likely that cross-talk of different posttranslational modifications, including both ubiquitination and phosphorylation, occurs on beclin-1 to regulate autophagy, as it represents a critical hub in this process. In this regard, it is interesting to note a recent study describing two phosphorylation events within the Vps34-binding domain of beclin-1 which repress autophagy (31). It will be critical in future studies to understand how all these modifications integrate to control beclin-1 function.
ACKNOWLEDGMENTS
We thank S. Cheng and V. Crocker for assistance with electron microscopy. We are also grateful to W. A. Dunn for advice on yeast electron microscopy. We thank B. Levine for the beclin-1 construct and the beclin-1 F123A point mutant. We also thank other members of the Youle and Abeliovich laboratories for helpful discussions.
This work was supported by the NINDS, National Institutes of Health, and by Israel Science Fund grant 1016/07 (to H.A.).
A.I.F., D.P.S, H.A. and A.N. performed analysis of the CTD in yeast. A.I.F., B.J.D. and S.-W.R. performed analysis of the CTD in mammalian cells. S.A.H. performed TaqMan analysis of p62. A.I.F. made TALEN-generated knockout cell lines and performed beclin-1 phosphorylation analysis. R.J.Y and A.I.F. wrote the manuscript.
Footnotes
Published ahead of print 22 July 2013
REFERENCES
- 1.Nakatogawa H, Suzuki K, Kamada Y, Ohsumi Y. 2009. Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nat. Rev. Mol. Cell Biol. 10:458–467 [DOI] [PubMed] [Google Scholar]
- 2.Komatsu M, Ichimura Y. 2010. Selective autophagy regulates various cellular functions. Genes Cells 15:923–933 [DOI] [PubMed] [Google Scholar]
- 3.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. 2008. Autophagy fights disease through cellular self-digestion. Nature 451:1069–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liang XH, Jackson S, Seaman M, Brown K, Kempkes B, Hibshoosh H, Levine B. 1999. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature 402:672–676 [DOI] [PubMed] [Google Scholar]
- 5.Yue Z, Jin S, Yang C, Levine AJ, Heintz N. 2003. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc. Natl. Acad. Sci. U. S. A. 100:15077–15082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Itakura E, Kishi C, Inoue K, Mizushima N. 2008. Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with mammalian Atg14 and UVRAG. Mol. Biol. Cell 19:5360–5372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Obara K, Sekito T, Ohsumi Y. 2006. Assortment of phosphatidylinositol 3-kinase complexes–Atg14p directs association of complex I to the pre-autophagosomal structure in Saccharomyces cerevisiae. Mol. Biol. Cell 17:1527–1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Funderburk SF, Wang QJ, Yue Z. 2010. The Beclin 1-VPS34 complex—at the crossroads of autophagy and beyond. Trends Cell Biol. 20:355–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Matsunaga K, Morita E, Saitoh T, Akira S, Ktistakis NT, Izumi T, Noda T, Yoshimori T. 2010. Autophagy requires endoplasmic reticulum targeting of the PI3-kinase complex via Atg14L. J. Cell Biol. 190:511–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fan W, Nassiri A, Zhong Q. 2011. Autophagosome targeting and membrane curvature sensing by Barkor/Atg14(L). Proc. Natl. Acad. Sci. U. S. A. 108:7769–7774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang P, Xiao A, Zhou M, Zhu Z, Lin S, Zhang B. 2011. Heritable gene targeting in zebrafish using customized TALENs. Nat. Biotechnol. 29:699–700 [DOI] [PubMed] [Google Scholar]
- 12.Miller JC, Tan S, Qiao G, Barlow KA, Wang J, Xia DF, Meng X, Paschon DE, Leung E, Hinkley SJ, Dulay GP, Hua KL, Ankoudinova I, Cost GJ, Urnov FD, Zhang HS, Holmes MC, Zhang L, Gregory PD, Rebar EJ. 2011. A TALE nuclease architecture for efficient genome editing. Nat. Biotechnol. 29:143–148 [DOI] [PubMed] [Google Scholar]
- 13.Schmittgen TD, Livak KJ. 2008. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 3:1101–1108 [DOI] [PubMed] [Google Scholar]
- 14.Klionsky DJ, Cueva R, Yaver DS. 1992. Aminopeptidase I of Saccharomyces cerevisiae is localized to the vacuole independent of the secretory pathway. J. Cell Biol. 119:287–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klionsky DJ, Banta LM, Emr SD. 1988. Intracellular sorting and processing of a yeast vacuolar hydrolase: proteinase A propeptide contains vacuolar targeting information. Mol. Cell. Biol. 8:2105–2116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sun Q, Fan W, Chen K, Ding X, Chen S, Zhong Q. 2008. Identification of Barkor as a mammalian autophagy-specific factor for Beclin 1 and class III phosphatidylinositol 3-kinase. Proc. Natl. Acad. Sci. U. S. A. 105:19211–19216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Odorizzi G, Babst M, Emr SD. 1998. Fab1p PtdIns(3)P 5-kinase function essential for protein sorting in the multivesicular body. Cell 95:847–858 [DOI] [PubMed] [Google Scholar]
- 18.Jain A, Lamark T, Sjottem E, Larsen KB, Awuh JA, Overvatn A, McMahon M, Hayes JD, Johansen T. 2010. p62/SQSTM1 is a target gene for transcription factor NRF2 and creates a positive feedback loop by inducing antioxidant response element-driven gene transcription. J. Biol. Chem. 285:22576–22591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Youle RJ, Narendra DP. 2011. Mechanisms of mitophagy. Nat. Rev. Mol. Cell Biol. 12:9–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martinou JC, Youle RJ. 2011. Mitochondria in apoptosis: Bcl-2 family members and mitochondrial dynamics. Dev. Cell 21:92–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heath-Engel HM, Chang NC, Shore GC. 2008. The endoplasmic reticulum in apoptosis and autophagy: role of the BCL-2 protein family. Oncogene 27:6419–6433 [DOI] [PubMed] [Google Scholar]
- 22.Pattingre S, Tassa A, Qu X, Garuti R, Liang XH, Mizushima N, Packer M, Schneider MD, Levine B. 2005. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell 122:927–939 [DOI] [PubMed] [Google Scholar]
- 23.Shintani T, Huang WP, Stromhaug PE, Klionsky DJ. 2002. Mechanism of cargo selection in the cytoplasm to vacuole targeting pathway. Dev. Cell 3:825–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Abeliovich H, Klionsky DJ. 2001. Autophagy in yeast: mechanistic insights and physiological function. Microbiol. Mol. Biol. Rev. 65:463–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Suriapranata I, Epple UD, Bernreuther D, Bredschneider M, Sovarasteanu K, Thumm M. 2000. The breakdown of autophagic vesicles inside the vacuole depends on Aut4p. J. Cell Sci. 113(Pt 22):4025–4033 [DOI] [PubMed] [Google Scholar]
- 26.Abeliovich H, Dunn WA, Jr, Kim J, Klionsky DJ. 2000. Dissection of autophagosome biogenesis into distinct nucleation and expansion steps. J. Cell Biol. 151:1025–1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xie Z, Nair U, Klionsky DJ. 2008. Atg8 controls phagophore expansion during autophagosome formation. Mol. Biol. Cell 19:3290–3298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kinoshita E, Kinoshita-Kikuta E, Takiyama K, Koike T. 2006. Phosphate-binding tag, a new tool to visualize phosphorylated proteins. Mol. Cell. Proteomics 5:749–757 [DOI] [PubMed] [Google Scholar]
- 29.Narendra D, Tanaka A, Suen DF, Youle RJ. 2009. Parkin-induced mitophagy in the pathogenesis of Parkinson disease. Autophagy 5:706–708 [DOI] [PubMed] [Google Scholar]
- 30.Zalckvar E, Berissi H, Mizrachy L, Idelchuk Y, Koren I, Eisenstein M, Sabanay H, Pinkas-Kramarski R, Kimchi A. 2009. DAP-kinase-mediated phosphorylation on the BH3 domain of beclin 1 promotes dissociation of beclin 1 from Bcl-XL and induction of autophagy. EMBO Rep. 10:285–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang RC, Wei Y, An Z, Zou Z, Xiao G, Bhagat G, White M, Reichelt J, Levine B. 2012. Akt-mediated regulation of autophagy and tumorigenesis through Beclin 1 phosphorylation. Science 338:956–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Daub H, Olsen JV, Bairlein M, Gnad F, Oppermann FS, Korner R, Greff Z, Keri G, Stemmann O, Mann M. 2008. Kinase-selective enrichment enables quantitative phosphoproteomics of the kinome across the cell cycle. Mol. Cell 31:438–448 [DOI] [PubMed] [Google Scholar]
- 33.Oppermann FS, Gnad F, Olsen JV, Hornberger R, Greff Z, Keri G, Mann M, Daub H. 2009. Large-scale proteomics analysis of the human kinome. Mol. Cell. Proteomics 8:1751–1764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang W, Choi W, Hu W, Mi N, Guo Q, Ma M, Liu M, Tian Y, Lu P, Wang FL, Deng H, Liu L, Gao N, Yu L, Shi Y. 2012. Crystal structure and biochemical analyses reveal Beclin 1 as a novel membrane binding protein. Cell Res. 22:473–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Noda NN, Kobayashi T, Adachi W, Fujioka Y, Ohsumi Y, Inagaki F. 2012. Structure of the novel C-terminal domain of vacuolar protein sorting 30/autophagy-related protein 6 and its specific role in autophagy. J. Biol. Chem. 287:16256–16266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Furuya N, Yu J, Byfield M, Pattingre S, Levine B. 2005. The evolutionarily conserved domain of Beclin 1 is required for Vps34 binding, autophagy and tumor suppressor function. Autophagy 1:46–52 [DOI] [PubMed] [Google Scholar]
- 37.Rust HL, Thompson PR. 2011. Kinase consensus sequences: a breeding ground for crosstalk. ACS Chem. Biol. 6:881–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim J, Kim YC, Fang C, Russell RC, Kim JH, Fan W, Liu R, Zhong Q, Guan KL. 2013. Differential regulation of distinct Vps34 complexes by AMPK in nutrient stress and autophagy. Cell 152:290–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Abrahamsen H, Stenmark H, Platta HW. 2012. Ubiquitination and phosphorylation of Beclin 1 and its binding partners: tuning class III phosphatidylinositol 3-kinase activity and tumor suppression. FEBS Lett. 586:1584–1591 [DOI] [PubMed] [Google Scholar]