Abstract
Parkinson's disease (PD) is characterized by progressive loss of midbrain dopaminergic neurons resulting in motor dysfunction. While most PD is sporadic in nature, a significant subset can be linked to either dominant or recessive germ line mutations. PARK2, encoding the ubiquitin ligase parkin, is the most frequently mutated gene in hereditary Parkinson's disease. Here, we present evidence for a neuronal ubiquitin ligase cascade involving parkin and the multisubunit ubiquitin ligase SCFFbw7β. Specifically, parkin targets the SCF substrate adapter Fbw7β for proteasomal degradation. Furthermore, we show that the physiological role of parkin-mediated regulation of Fbw7β levels is the stabilization of the mitochondrial prosurvival factor Mcl-1, an SCFFbw7β target in neurons. We show that neurons depleted of parkin become acutely sensitive to oxidative stress due to an inability to maintain adequate levels of Mcl-1. Therefore, loss of parkin function through biallelic mutation of PARK2 may lead to death of dopaminergic neurons through unregulated SCFFbw7β-mediated ubiquitylation-dependent proteolysis of Mcl-1.
INTRODUCTION
Parkinson's disease (PD) is characterized by progressive loss of midbrain dopaminergic neurons resulting in motor dysfunction. While most PD is sporadic in nature, a significant subset can be linked to either dominant or recessive germ line mutations. Of the latter class, mutations in the PARK2 gene account for a large fraction of the cases (1–3). The syndrome caused by biallelic PARK2 mutation is known as autosomal recessive juvenile parkinsonism (ARJP) because it generally presents at a relatively young age compared to sporadic PD. PARK2 encodes a 465-amino-acid protein, known as parkin, with predicted and demonstrated E3 ubiquitin ligase activity (4–6). The ubiquitin ligase-dependent formation of ubiquitin chains on substrate proteins most frequently targets them for degradation by the 26S proteasome, a large multisubunit protease, although other regulatory fates can be regulated by ubiquitylation (7). The finding that parkin is a ubiquitin ligase has led to the hypothesis that that PD, at least the variant caused by PARK2 mutation, results from the abnormal and neurotoxic accumulation of parkin ubiquitin ligase targets because of a failure to target them for proteasomal degradation (8, 9). Although numerous putative parkin substrates have been identified (6, 8, 10–20), there is no consensus concerning which, if any, have a role in PD pathogenesis. Indeed, most of these show no accumulation in the brains of parkin−/− mice, suggesting that biologically relevant substrates of parkin remain to be identified (21–25).
Cullin-ring-ligases (CRLs) are multisubunit E3 ubiquitin ligases characterized by a scaffold protein (Cullin), a ring finger protein, and a substrate binding adapter protein (26, 27). SCF ligases constitute a subfamily of CRLs where the scaffold is composed of Cul-1/Cdc53 and Skp1, the ring finger protein is Rbx1, and the substrate adapter is one of many so-called F-box proteins. F-box proteins contain a motif known as an F box that interacts with Skp1, providing a link to the SCF core (28–30). Fbw7, also known as human Cdc4 (hCdc4) or hSel-10, is one such F-box protein that provides substrate specificity for SCF ubiquitin ligases (31–33). SCFFbw7 has been shown to target a number of important cellular regulatory proteins for ubiquitylation and proteasomal degradation. These include, among others, cyclins E1 (31–33) and E2 (34), c-myc (35–37), c-Jun, SREBP (38), PGC1-α (39), Notch (40, 41), myeloid cell leukemia 1 (Mcl-1) (42, 43), and NF-κB2 (44–46). Since several of these proteins are oncoproteins, it is not surprising that Fbw7 has been found to be mutated in a broad spectrum of human cancers and is therefore considered a tumor suppressor (47, 48).
Fbw7 consists of four known functional domains. The C-terminal half of the protein consists of eight WD-40 repeats forming an eight-bladed β-propeller. This domain recognizes a phosphorylated motif on target proteins known as the CPD (for Cdc4 phosphodegron) (49, 50). In this manner, targeting of substrates by SCFFbw7 is dependent on phosphorylation, and their degradation is linked to protein kinase cascades. Amino terminal to the β-propeller are the F box, which binds the SCF core via Skp1, and a D box, which promotes dimerization of Fbw7 (49, 51). Fbw7 exists as three splice variant isoforms with distinct amino-terminal domains (52). Each isoform is encoded by a unique 5′ exon that specifies the amino terminus and 10 common 3′ exons that determine the rest of the protein. The amino terminus of each isoform specifies its cellular location, with the α isoform being nucleoplasmic (53) but excluded from the nucleolus (35, 54), the β isoform being cytosolic (53) and enriched in the endoplasmic reticulum (55), and the γ isoform being nucleolar (35, 54).
Fbw7 has been reported to bind parkin in neurons and to collaborate with parkin to ubiquitylate and destabilize the Fbw7 target cyclin E1 (18). Excessive cyclin E1 accumulation has been associated with neuronal apoptosis (56), especially under conditions of excitotoxicity, suggesting a neuroprotective role for this parkin-Fbw7 ligase. In the current study, we describe a different relationship between parkin and Fbw7. We find that parkin regulates the activity of SCFFbw7 by targeting Fbw7β, the cytoplasmic isoform, for ubiquitin-dependent proteasomal degradation. The degradation of Fbw7β is important for neuronal survival, particularly under conditions of oxidative stress, because it is necessary for the protection of the antiapoptotic Bcl-1 family member Mcl-1 from ubiquitin-mediated proteolysis.
MATERIALS AND METHODS
Cell culture.
Mixed populations of primary neurons were isolated from day 16 mouse embryos and cultured in high-glucose Dulbecco's modified Eagle's medium (DMEM-Glutamax), supplemented with 10% fetal bovine serum (Atlanta Biologicals) and 100 units/ml penicillin and 100 μg/ml streptomycin (Invitrogen). Cells were plated onto plastic dishes or glass slides (hemacytometer cover glass; Orbeco, Sarasota, FL, USA), coated with poly-l-lysine (Sigma) and mouse laminin (Invitrogen). The day after preparation, cultures were transferred to Neurobasal medium supplemented with B27 (Invitrogen), 1:400 Glutamax (Invitrogen), and 100 units/ml penicillin and 100 μg/ml streptomycin. Primary neurons were grown at 37°C in 3% oxygen and 5% CO2 and cultured for 5 to 7 days before being transduced with adenovirus and/or subjected to oxidative stress. HEK293A and HEK293T cells were maintained in DMEM-Glutamax, supplemented with 10% newborn calf serum and 100 units/ml penicillin and 100 μg/ml streptomycin at 37°C and 5% CO2. SH-SY5Y cells were maintained in DMEM-Glutamax, supplemented with 10% fetal bovine serum and 100 units/ml penicillin and 100 μg/ml streptomycin.
Adenoviral transductions and drug treatments were initiated at day in vitro 6 (DIV6). For oxidative stress experiments, NOC-12 (Calbiochem, La Jolla, CA) was added at 100 μM, and carbonyl cyanide m-chlorophenylhydrazone (CCCP) was added at 10 μM and present in the culture medium for the whole incubation time. tert-Butyl hydroperoxide (tBHP) was added to the culture medium at 50 μM for 1 h, and then the medium was replaced with fresh medium. Adenoviral transductions were carried out two times at 24-h intervals. NOC-12 and other stressors were added after the second transduction, and cells were either fixed for microscopy or harvested for biochemical analysis 16 to 18 h later. For analysis using protein kinase inhibitors, primary neurons were treated for 5 h with either dimethyl sulfoxide (DMSO) or 10 μM SB202190 (p38 mitogen-activated protein kinase [MAPK] inhibitor; Sigma), 50 μM SB216763 (glycogen synthase kinase 3 [GSK3] inhibitor; Sigma), or 100 nM SR3306 (c-Jun N-terminal kinase [JNK] inhibitor; Millipore). For some experiments, 1 h after addition of inhibitors, NOC-12 was added for additional 4 h to create oxidative stress.
Antibodies and reagents.
The following primary antibodies (Abs) were used: antiparkin rabbit Ab (2132; Cell Signaling), antiactin mouse monoclonal Ab (Sigma), anti-Fbw7 rabbit polyclonal (33), anti-glyceraldehyde-3-phosphate dehydrogenase (anti-GAPDH) (9484; Abcam), antiubiquitin (3933; Cell Signaling,) anti-cyclin E mouse monoclonal antibody HE12 (Santa Cruz Biotechnology), anti-cytochrome c oxidase IV (anti-COX IV) rabbit Ab (4844; Cell Signaling,), anti-Mcl-1 rabbit Ab (545; Cell Signaling,), anti-cleaved poly(ADP-ribose) polymerase (PARP) rabbit Ab (9544; Cell Signaling), anti-phospho-S6 (P-S6) ribosomal protein rabbit Ab (4858; Cell Signaling), anti-lysine 48 (K48) linkage-specific polyubiquitin (8081; Cell Signaling), anti-phospho-4E binding protein 1 (P-4EBP1) (2855; Cell Signaling), anti-S6 ribosomal protein (2217; Cell Signaling), anti-eukaryotic translation initiation factor 4E (9742; Cell Signaling), and anti-α-tubulin (DM-1A; Sigma). The following secondary antibodies were used: horseradish peroxidase (HRP)-conjugated anti-mouse and anti-rabbit IgG antibody (Promega). DAPI (4′ 6-diamidino-2-phenylindole) was purchased from Sigma. The mounting medium ProLong Gold antifade reagent (P36930) was purchased from Invitrogen/Molecular probes. NOC-12 was purchased from EMD Chemicals/Calbiochem (487955). MG132 (I-130), PR11, E1, E2 (UbcH7), and ATP-regenerating buffer (B-10) were purchased from Boston Biochem. Cycloheximide was purchased from Sigma (C7698). NOC-12 was used at 200 μM for 2 to 16 h. Cycloheximide was added at 200 μM for 2 to 6 h, and MG132 (Sigma) was applied at 5 μM for 4 h.
Animals.
For neuronal cultures, parkin-nullizygous mice (with a deletion of exon 3) (21) backcrossed 10 times to the C57BL/6 background were obtained from the Jackson Laboratory, Bar Harbor, ME. C57BL/6 mice were used as controls. For experiments with mouse brains, parkin-nullizygous mice (21) were backcrossed to the C57BL/6 background in the laboratory of M. S. Goldberg, and controls were wild-type segregants from subsequent crosses. All animal procedures were performed at the Scripps Research Institute (CA), accredited by the Association for assessment and Accreditation of Laboratory and Animal Care in accordance with protocols approved by the Institutional Animal Care and Use Committee and the principles outlined in the National Institutes of Health Guide for the Care and Use of Laboratory Animals.
Western blotting.
Neurons were harvested by trypsinization (0.125%), collected by centrifugation and washed twice in phosphate-buffered saline (PBS), and cell pellets were stored at −80°C until analyzed. Cell pellets were resuspended in lysis buffer (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 2 mM EGTA, 1% NP-40, protease inhibitor cocktail [Complete Mini, EDTA-free; Roche Diagnostics]). Lysates were incubated on ice for 17 min and sonicated for 10 s, and soluble extracts were obtained by centrifugation using an Eppendorf centrifuge 5415C at the highest speed for 10 min at 4°C. Proteins were separated by SDS-PAGE and, after electrophoresis, transferred using an iBlot (Invitrogen) to nitrocellulose or to polyvinylidene difluoride (PVDF) (Immobilon membrane; Millipore). Membranes were blocked for 1 h with 5% nonfat dry milk in Tris-buffered saline (TBS) containing 0.1% Tween (TBST) and then probed with primary antibody overnight at 4°C.
Mitochondrial fractionation.
Primary neuronal cultures were harvested by scraping in cavitation buffer (5 mM HEPES, 4.3 mM MgCl, 1 mM EGTA, 250 mM sucrose, protease inhibitors) and lysed by 18 strokes with a Dounce homogenizer. Lysates were cleared by centrifugation at 150 × g for 10 min at 4°C. The supernatant was transferred to a fresh tube, and the crude mitochondrial fraction was obtained by centrifugation at 2,500 × g for 10 min at 4°C. The pellet was resuspended in 800 μl of cavitation buffer and subjected to centrifugation at 8,000 × g for 10 min at 4°C; pellets were again resuspended in 800 μl of cavitation buffer and loaded onto a sucrose gradient (5.5 ml each of 1.5 M and 1.0 M sucrose). Samples were centrifuged at 34,000 × g (SW40 rotor) for 45 min at 4°C. The hazy phase was collected and washed three times with dilution buffer (same as the cavitation buffer but without sucrose). Equal amounts of protein were separated by SDS-PAGE and then transferred to a nitrocellulose or Immobilon membrane. Membranes were blocked for 1 h in TBST (10 mM Tris, 500 mM NaCl, 0.1% Tween) containing 5% nonfat milk and incubated overnight with primary antibodies according to the manufacturer's instruction.
Cycloheximide chase.
Cycloheximide was added at a concentration of 50 μM on DIV7. Cells were harvested by trypsinization (0.125%) after 0, 2, 4, and 6 h or other times specified in Fig. 8D and E and 9B and C, collected by centrifugation, and washed twice in PBS; cell pellets were then stored at −80°C until analyzed.
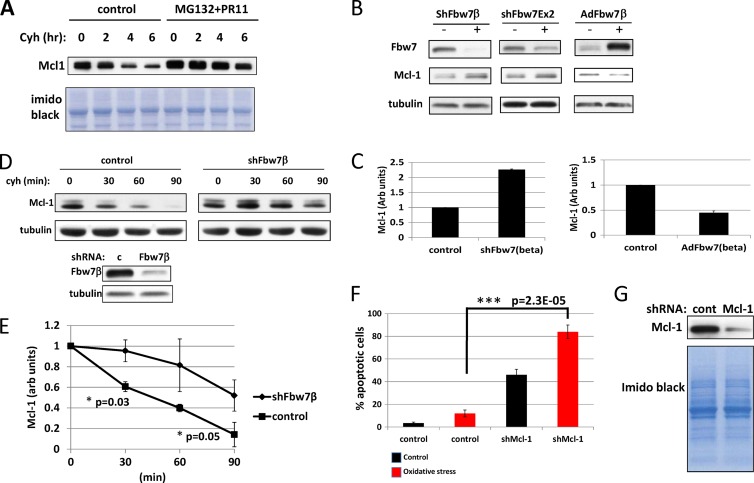
Fig 8.
Mcl-1 in neurons is regulated by Fbw7β. (A) Mcl-1 is degraded by the proteasome in embryonic mouse brain neurons. A cycloheximide chase experiment was carried out on untreated neurons and neurons treated with MG132 and PR11. Cycloheximide was added, and lysates were assayed for Mcl-1 levels at the indicated times. The loading control was the imido black-stained PVDF membrane. (B) Silencing of Fbw7β increases Mcl-1 levels, whereas overexpression of Fbw7β reduces Mcl-1 levels. Primary embryonic mouse brain neurons were transduced with control adenovirus or adenoviruses expressing shRNA targeting specifically Fbw7β (shFbw7β), all Fbw7 isoforms (shFbw7β Ex 2), or overexpressing Fbw7β-Flag (AdFbw7β). Extracts were analyzed by SDS-PAGE and Western blotting for the indicated proteins. (C) Quantitation of two independent experiments where Fbw7β was silenced or overexpressed as described for panel B. Error bars correspond to standard deviations. (D) Cycloheximide chase analyzing stability of Mcl-1 after silencing of Fbw7β. Primary mouse neurons transduced with control (c) adenovirus or adenovirus expressing Fbw7β-specific shRNA were treated with cycloheximide, and extracts prepared at the indicated times were analyzed by SDS-PAGE and Western blotting for Mcl-1 (upper panels). The zero time points were analyzed for Fbw7β (lower panels). (E) Quantitation using ImageJ of the experiment shown in panel D together with an additional biological experiment. Error bars correspond to standard deviations, and P values were determined using Student's t test. Fbw7β was normalized to tubulin. (F) Silencing of neurons for Mcl-1 sensitizes them to apoptosis. Primary mouse neurons transduced with control adenovirus or adenovirus expressing shRNA targeting Mcl-1 were treated with NOC-12 and analyzed for apoptosis as described in the legend of Fig. 7A. Error bars correspond to standard deviations from three independent counts of more than 200 neurons each. P values were determined by Student's t test. (G) Parallel cultures treated with Mcl-1-specific shRNA as described for panel F were analyzed for Mcl-1 levels by SDS-PAGE and Western blotting. Imido black staining of the blot was used as a loading control.
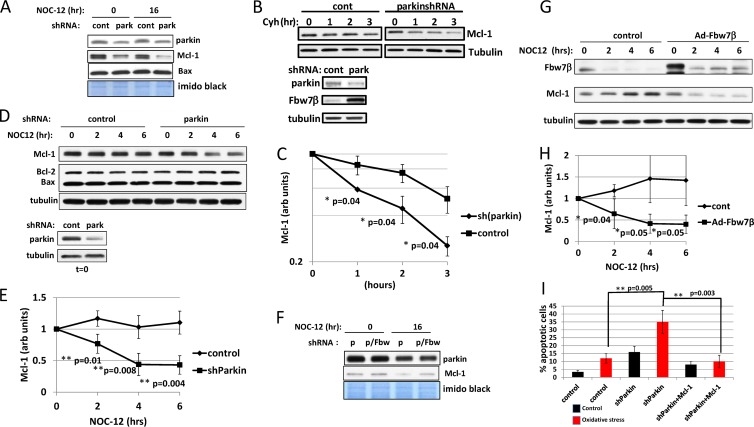
Fig 9.
Parkin regulates Mcl-1 levels through targeting of Fbw7β. (A) Regulation of Mcl-1 levels by parkin. Analysis of Mcl-1 levels after silencing of parkin and in response to oxidative stress was carried out. Protein samples are from the experiment described in the legend of Fig. 7A to C. Bax was also analyzed as a function of parkin silencing. (B) Cycloheximide chase analyzing Mcl-1 levels after silencing of parkin. Primary mouse neurons transduced with control or parkin-targeting shRNA adenovirus were treated with cycloheximide, and extracts prepared at the indicated times were analyzed by SDS-PAGE and Western blotting for Mcl-1 (upper panels). The zero time points were analyzed for parkin and Fbw7β (lower panels). (C) Quantitation using ImageJ of the experiment shown in panel B as well as an additional biological experiment. Mcl-1 was normalized to tubulin. (D) Silencing of parkin destabilizes Mcl-1 in response to oxidative stress. Extracts are from the same experiment shown in Fig. 6C. (E) Quantitation using ImageJ of experiment shown in panel D as well as two additional biological experiments. Mcl-1 was normalized to tubulin. (F) Silencing of Fbw7β rescues Mcl-1 levels after silencing of parkin. Protein samples are from same experiments described in the legend of Fig. 7A, B, and D. (G) Overexpression of Fbw7β destabilizes Mcl-1 in response to oxidative stress. Neurons transduced with control adenovirus or adenovirus expressing Fbw7β-Flag were treated with NOC-12 and analyzed at 2-h intervals for Fbw7β and Mcl-1 by SDS-PAGE and Western blotting. (H) Quantitation using ImageJ of the experiment shown in panel G as well as two additional biological experiments. Mcl-1 was normalized to tubulin. (I) Mcl-1 overexpression rescues the sensitivity to oxidative stress conferred by silencing of parkin. Neurons transduced with control adenovirus, shRNA adenovirus targeting parkin, or shRNA adenovirus targeting parkin simultaneously with an adenovirus expressing Mcl-1 were analyzed for NOC-12-induced apoptosis as described in the legend of Fig. 7. In all cases, error bars correspond to standard deviations, and P values were determined using Student's t test.
In vivo ubiquitylation.
Cells were transduced with adenoviruses expressing FLAG-tagged-Fbw7, hemagglutinin (HA)-ubiquitin, and HA-parkin in the indicated combinations. After 16 h, cells were treated with proteasome inhibitors MG132 and PR11 for 4 h at 10 μM and 70 μM, respectively, and harvested; lysates were immunoprecipitated with anti-Fbw7 antibody or with nonimmune rabbit IgG. Lysates were prepared by resuspending pellets in radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris-HCl, pH 8, 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, supplemented with protease inhibitor cocktail [Complete Mini, EDTA-free; Roche Diagnostics]) and 10 mM N-ethylmaleimide, sonicating for 10 s, and clarifying by 10 min of centrifugation in a microcentrifuge. Antibody incubations were for 4 h at 4°C, and complexes were collected using protein A/G beads. Immunoprecipitates were eluted by boiling in SDS-PAGE sample buffer and analyzed by SDS-PAGE and immunoblotting using anti-HA monoclonal antibody (Upstate). The blot was then stripped and reprobed using anti-Fbw7 antibody.
In vitro ubiquitylation.
Reaction mixtures (30 μl) consisted of buffer (1 mM MgCl2, 50 mM HEPES, pH 7.5, 50 mM NaCl; 1 μM dithiothreitol [DTT]), 3 μl of E1 (Boston Biochem), 2 μl of E2 (UbcH7; Boston Biochem), 3 μl of B-10 ATP-regenerating system (Boston Biochem), 0.3 μl of glutathione S-transferase (GST)–ubiquitin (Boston Biochem), 0.4 μg of Fbw7/Skp1 dimer amino-terminally tagged with maltose binding protein (MBP-Fbw7/Skp1), and 0.1 μg of Flag-parkin. MBP-Fbw7/Skp1 was expressed and purified from Escherichia coli by amylose resin (New England BioLabs) chromatography and elution using maltose, as described by the manufacturer. Baculovirus-expressed Flag-parkin was purified from Sf9 insect cells using anti-Flag (M2) beads (Sigma) and elution with Flag peptide, as described by the manufacturer. After incubation for 1 h at 18°C, reaction mixtures were diluted to 150 μl in cold Tris-buffered saline, pH 7.5 (TBS), and incubated with 30 μl of glutathione-Sepharose beads (GE Healthcare) for 1 h at 4°C. After three washes by centrifugation through 1 ml of TBS, beads were resuspended in 2× SDS-PAGE sample buffer, heated to 100°C for 5 min, and analyzed by SDS-PAGE and Western blotting using anti-Fbw7 antibody. Alternatively, the same reaction mixtures with the modification that 1 μg of MBP-Fbw7/Skp1 and 0.02 μg of Flag-parkin were used were incubated at 30°C for 2 h. Subsequent purification of MBP-Fbw7/Skp1 was accomplished by incubation with amylose resin after dilution into amylose column buffer (10 mM Tris-HCl, pH 7.4, 200 mM NaCl, 1 mM EDTA). Bead washing, sample preparation, and analysis were as described above except that blots were analyzed using antiubiquitin and anti-ubiquitin K48 linkage-specific antibody.
mRNA extraction and quantitative RT-PCR.
Poly(A)+ mRNA was isolated from primary mouse neuronal cultures according to the manufacturer's instructions (RNeasy Minikit; Qiagen), and cDNA was synthesized using SuperScript II RT (Invitrogen). Quantitative PCR was performed using the RT Real-Time SYBR green PCR master mix from SA Bioscience (PA-010-12) in an IQ5 instrument (Bio-Rad). Expression values were normalized against the housekeeping gene actin. The following primers were used: mouse Fbw7β forward, 5′-TGCCTGAGCATGTCCACGTTAGAA-3′; mouse Fbw7β reverse, 5′-TGAACATGGTACAAGGCCAGTGGT-3′; mouse Mcl-1 forward, 5′-GGTTGAGTCCGATTACCGCGTTTCTT-3′; and mouse Mcl-1 reverse, 5′-CAGTTTGTTACGCCGTCGCTGAAA-3′. Gene expression was normalized with respect to the endogenous housekeeping control gene, β-actin, which was not influenced by oxidative stress induced by NOC-12. Relative expression was calculated for each gene using the ΔΔCT (where CT is threshold cycle) method. Statistical analysis of reverse transcription-PCR (RT-PCR) data is based on triplicate samples.
Adenoviruses.
All short hairpin RNA (shRNA)-expressing adenoviruses are based on the mir30 backbone system in the retroviral vector pSHAG-MAGIC2 (57). Targeting sequences for mouse parkin, mouse Fbw7 exon 2, mouse Fbw7β, mouse Mcl-1, and green fluorescent protein (GFP) were amplified by PCR and cloned into pSHAG-MAGIC2 (targeting sequences provided upon request). The entire expression cassette was then removed and recloned into the adenoviral shuttle vector pDC515. Adenovirus was prepared using the AdMax (Microbix Biosystems) system in HEK293A cells and amplified by established procedures. Adenoviruses for expression of human Flag-Fbw7β, human Mcl-1, and human HA-parkin (wild-type and R275W mutant) were made by cloning the respective cDNAs into pDC515 and using established techniques for adenoviral production and amplification. Adenovirus expressing HA-ubiquitin was a gift from Anastasia Kralli (The Scripps Research Institute).
Brain extracts.
Frozen half-brains from adult mice (2 to 8 months of age) were homogenized using a BioPulverizer System I (6911-010 beads) and a FastPrep FP120 (BIO101). Brain tissue (0.2 g) was homogenized with 2.6 g of beads in 750 ml of buffer (50 mM Tris-HCl [pH 7.6], 150 mM NaCl, 1% Triton-X, protease inhibitor cocktail tablet [complete Mini, EDTA-free; Roche Diagnostics]), for 40 s (setting 6) three times. Homogenates were centrifuged at 10,000 × g for 10 min at 4°C, and the supernatant was removed. Lysates were analyzed by SDS-PAGE and Western blotting using anti-Fbw7 and antiparkin antibodies.
Preparation of human brain samples has been described previously (8).
Neuronal apoptosis assay.
Apoptosis was assayed by determining the fraction of cells exhibiting an apoptotic nuclear morphology (apoptotic bodies and pyknotic and fragmented nuclei) (58). Primary mouse embryonic neurons were plated onto glass slides (hemacytometer cover glass; Orbeco, Sarasota, FL, USA) that had been coated with poly-l-lysine and mouse laminin. Cells were transduced with adenovirus or combination of adenoviruses (as indicated in the legends to Fig. 7A, F, and G, Fig. 8F, and Fig. 9I) two times at 24-h intervals prior to treating cells with NOC-12. After 16 additional hours, cells were fixed in 4% formalin (methanol-free 10% formaldehyde solution, ultrapure electron microscopy grade; Polysciences, Inc.) in PBS for 20 min at room temperature, washed three times in PBS, incubated with 0.5 μg/ml DAPI (Invitrogen) in PBS for 10 min, and washed again three times in PBS before being mounted with ProGold mounting medium (Molecular Probes/Invitrogen). To detect cells undergoing apoptosis, the number of fragmented and pyknotic nuclei was determined using a Zeiss Axioscope 2 Plus microscope (Carl Zeiss, Göttingen, Germany). Quantification was based on scoring at least 200 nuclei in each of three fields. Apoptosis was confirmed by PARP cleavage detected by SDS-PAGE and Western blotting.
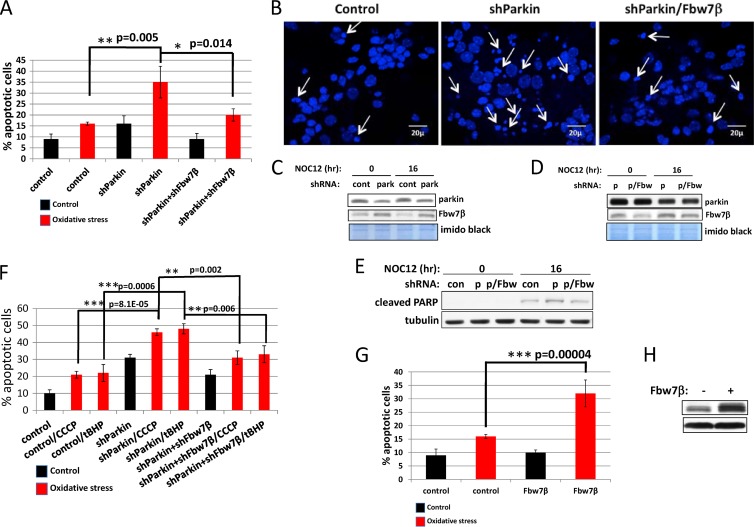
Fig 7.
Reduction in parkin levels or overexpression of Fbw7β sensitizes neurons to oxidative stress, whereas reduction of Fbw7β levels compensates for loss of parkin. (A) Measurement of apoptosis based on scoring of pyknotic nuclei. Primary mouse neurons on glass coverslips were transduced with control (GFP), parkin-specific, or parkin- and Fbw7β-specific shRNA-expressing adenoviruses and then treated with NOC-12 for 16 h. Primary mouse neurons were then fixed, stained with DAPI, and scored for apoptotic phenotypes (condensed, fragmented nuclei). Error bars represent the standard deviations from three independent counts of more than 200 neurons each. (B) Fluorescence micrographs of representative fields corresponding to the experiment quantified in panel A. Arrows indicate representative apoptotic nuclei. (C and D) Analysis of parkin and Fbw7β levels in parallel cultures treated as described for panel A. Imido black staining of the blot served as a loading control. p, parkin; p/Fbw, parkin and Fbw7β. (E) Analysis of cleaved PARP, an apoptotic marker, in parallel cultures treated as described in panel A. (F) The response to CCCP and tBHP is similar to the response to NOC-12. Primary mouse neurons transduced with control adenovirus or adenoviruses targeting parkin or parkin and Fbw7β. Neurons were untreated or treated with CCCP or tBHP and analyzed for apoptotic nuclei as described for panel A. (G) Primary mouse neurons on glass coverslips were transduced with control or Fbw7β-Flag-expressing adenoviruses and then treated with NOC-12 for 16 h and processed as described for panel A. Error bars correspond to standard deviations from three independent counts of more than 200 neurons each. (H) Analysis of Fbw7β in a parallel culture treated as described for panel G. Lanes correspond to nonstressed control samples. All error bars correspond to standard deviations, and P values were determined using Student's t test.
Lentiviruses.
The human parkin and Fbw7β cDNAs were cloned into the pLVX-GFP and pLVX-DsRed vectors (Lenti-X Expression System; Clontech), respectively. Lentiviruses were produced in HEK293T cells, as described by the manufacturer.
Confocal microscopy.
Neurons transduced with lentiviruses expressing Fbw7β-DsRed and parkin-GFP were fixed and analyzed by laser scanning confocal microscopy using a Zeiss LSM 710 instrument and the accompanying software.
Reproducibility and statistical analysis.
All experiments were repeated at least two times, giving comparable results to those reported. Error bars represent standard deviations. Student's t test was used for statistical analysis.
RESULTS
Parkin associates with Fbw7β in neurons.
It has been reported previously that the parkin ubiquitin ligase associates with the F-box protein Fbw7 in neurons (18). We confirmed this in parkin−/− embryonic mouse brain neurons using exogenously expressed parkin (Fig. 1A). Fbw7 exists in three splice-variant isoforms, only one of which, Fbw7β, is cytosolic (53, 55). Based on mobility, the Fbw7 isoform associated with parkin is Fbw7β, as expected. However, when overexpressed in HEK293A cells, all Fbw7 isoforms can associate with parkin (Fig. 1B). When primary embryonic mouse brain neurons were transduced with adenoviruses expressing GFP-parkin and Fbw7β-DsRed, laser-scanning confocal microscopy detected significant intracellular colocalization of Fbw7β and parkin (Fig. 1C). Therefore, parkin can physically associate with Fbw7β and coexists with Fbw7β at the same cytoplasmic sites.
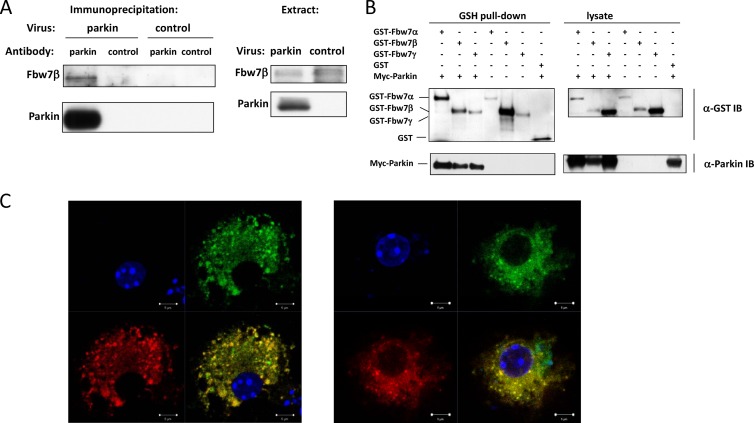
Fig 1.
Parkin associates with Fbw7β in primary neurons. (A) Coprecipitation of exogenous parkin with endogenous Fbw7β in primary neurons. Parkin−/− primary embryonic mouse brain neurons were transduced with either a control adenovirus or an adenovirus expressing N-terminally myc-tagged parkin. Lysates were then immunoprecipitated with a mouse monoclonal antibody specific for parkin or equivalent amounts of mouse IgG. After SDS-PAGE and Western blotting, membranes were probed with Fbw7- and parkin-specific antibodies, respectively. (B) Parkin can form complexes with all Fbw7 isoforms. Retrovirus-transduced HEK293A cell lines stably expressing each of the three Fbw7 isoforms as GST fusions were transfected with a plasmid expressing myc-parkin. Fbw7 complexes were precipitated using glutathione (GSH)-Sepharose beads and analyzed by SDS-PAGE and Western blotting using anti-GST or anti-myc tag antibody. (C) Colocalization of parkin and Fbw7β in primary neurons. GFP-parkin and Fbw7β-DsRed were expressed in primary embryonic mouse brain neurons by lentiviral transduction. Proteins were then detected by laser scanning confocal microscopy. Each of the two panels shows the following: upper left, DNA (DAPI); upper right, GFP-parkin; lower left, Fbw7β-DsRed; lower right, merged image. Scale bar, 5 μm. α, anti; IB, immunoblotting.
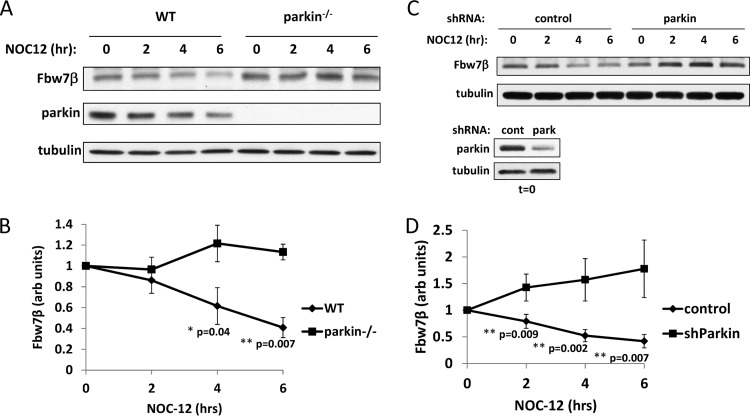
Loss of parkin in neurons leads to increased levels of Fbw7β.
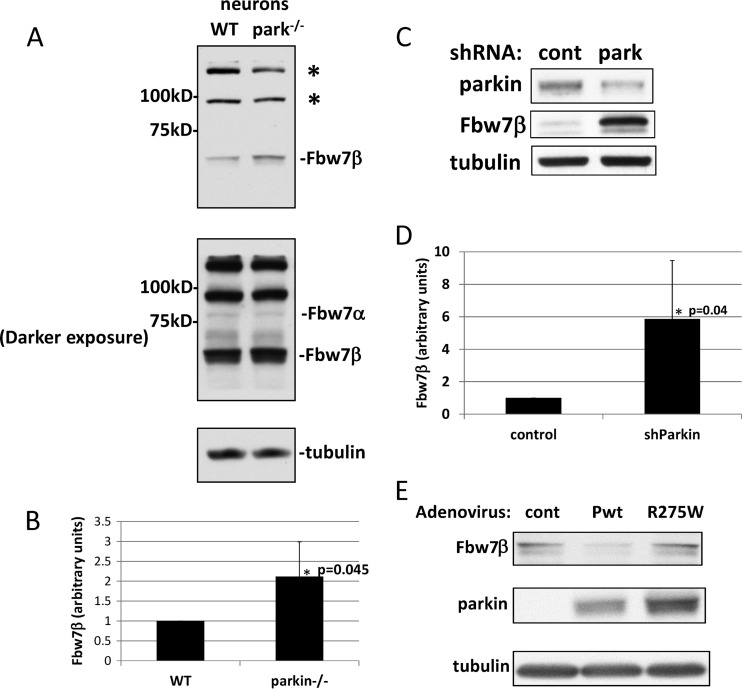
It has been reported that Fbw7 forms a novel, neuron-specific ubiquitin ligase with parkin that targets cyclin E1 under conditions of excitotoxicity (18). However, we could find no effect on cyclin E levels when parkin was eliminated from nontreated embryonic mouse brain neurons by germ line mutation (see Fig. S1A in the supplemental material). Under these conditions, cyclin E1 was expressed ectopically using a recombinant adenovirus because the endogenous protein is not detectable in postmitotic neurons. Similar results were obtained for endogenous cyclin E1 in SH-SY5Y neuroblastoma cells after siRNA-mediated silencing of parkin (see Fig. S1B). However, we found that Fbw7β levels, but not those of the nuclear isoform Fbw7α, were significantly increased in parkin-null embryonic brain neurons (Fig. 2A and B). Similar results were obtained when parkin levels in wild-type embryonic brain neurons were reduced by RNA interference (RNAi)-mediated silencing (Fig. 2C and D). Restoring wild-type parkin to parkin−/− neurons by adenoviral transduction reduced Fbw7β to wild-type levels, whereas introduction of a mutant allele of parkin associated with hereditary parkinsonism (parkin R275W; tryptophan substituted for arginine at position 275) (59) had no effect on Fbw7β levels (Fig. 2E).
Fig 2.
Loss or silencing of parkin leads to increased levels of Fbw7β in mouse neurons. (A) Extracts prepared from primary embryonic mouse brain neurons of the indicated genotype were analyzed by SDS-PAGE and Western blotting for Fbw7. Upper panel, light exposure. Lower panel, dark exposure. Asterisks indicate non-Fbw7 proteins recognized by the antibody. (B) Quantitation using ImageJ of three independent biological experiments equivalent to the experiment shown in panel A. In each case, Fbw7β was normalized to tubulin in the same blot. (C) Parkin was silenced in primary embryonic mouse brain neurons by transduction with an adenovirus expressing a parkin-specific shRNA (park) or a control shRNA (cont) specific for GFP. Extracts were analyzed by SDS-PAGE and Western blotting for Fbw7β and parkin. (D) Quantitation using ImageJ of three independent biological experiments equivalent to the experiment shown in panel C. In each case, Fbw7β was normalized to tubulin in the same blot. (E) Parkin-nullizygous primary embryonic mouse brain neurons were transduced with a control adenovirus (cont) or an adenovirus expressing wild-type parkin (Pwt) or a mutant parkin protein associated with hereditary Parkinson's disease (R275W). Extracts were analyzed by SDS-PAGE and Western blotting for Fbw7β and parkin. All error bars correspond to standard deviations. P values were determined using Student's t test. WT, wild type.
Fbw7β levels are elevated in the cortexes of human ARJP patients with mutated parkin.
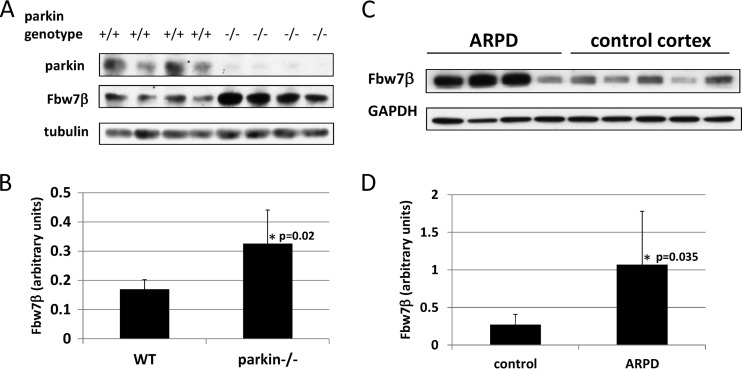
Fbw7β levels were elevated in whole brain extracts of parkin−/− mice (Fig. 3A and B). Fbw7β levels were then determined in cortex samples from Parkinson's patients carrying homozygous recessive germ line mutations in the PARK2 gene encoding parkin and compared to those of nondiseased controls (Fig. 3C and D) (8). Two of the ARJP patients carried a deletion of exon 4, which causes truncation of parkin at amino acid 143, whereas the other two carried a deletion of exon 3, causing termination at amino acid 96 (2, 60, 61). Three of the four ARJP samples exhibited highly elevated levels of Fbw7β, relative to the controls, similar to what was observed in extracts from parkin-nullizygous mouse brains. Therefore, loss or reduction of parkin in both mouse and human brains correlates with elevated levels of Fbw7β. Since parkin is a ubiquitin ligase, these results suggest that parkin may target Fbw7β for ubiquitin-mediated proteasomal degradation.
Fig 3.
Analysis of Fbw7β levels in mouse and human brains. (A) Extracts from whole brains of age-matched wild-type (+/+) and parkin-nullizygous (−/−) mice were analyzed by SDS-PAGE and Western blotting for Fbw7β and parkin. (B) Quantitation using ImageJ of experiment shown in panel A. Fbw7β was normalized to tubulin. (C) Brain cortex extracts from four autosomal-recessive Parkinson's disease patients with a biallelic PARK2 mutation and five cortical extracts from normal brains were analyzed by SDS-PAGE and Western blotting for Fbw7β. The third and fifth control samples correspond to different extracts prepared from the same cortex. The loading control was glyceraldehyde phosphate dehydrogenase (GAPDH). (D) Quantitation using ImageJ of experiment shown in panel C. Fbw7β was normalized to GAPDH. All error bars correspond to standard deviations. P values were determined using Student's t test.
The half-life of Fbw7β is increased in parkin-null neurons.
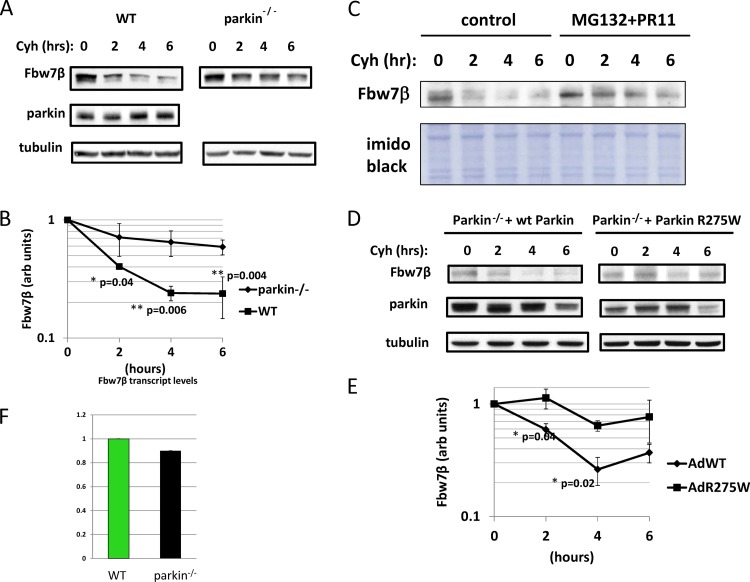
In order to determine whether increased levels of Fbw7β in parkin-null neurons are due to decreased rates of proteolysis, cycloheximide chase experiments were carried out in parallel on wild-type and parkin−/− embryonic brain neurons (Fig. 4A and B). Fbw7β is an unstable protein, decaying with the half-life of approximately 2 h in wild-type neurons. Degradation of Fbw7β is mediated by the proteasome since its half-life is significantly increased in response to proteasomal inhibition (Fig. 4C). The half-life of Fbw7β was significantly extended in parkin−/− neurons, consistent with a role for parkin in Fbw7β proteolysis (Fig. 4 A and B). However, when wild-type but not mutant (R275W) parkin was added back to parkin−/− neurons by adenoviral transduction, the Fbw7β half-life characteristic of wild-type neurons was restored (Fig. 4D and E). No effect on Fbw7β mRNA levels was observed (Fig. 4F), ruling out a transcriptional mechanism to explain elevated Fbw7β levels in parkin−/− neurons. Therefore, the increase in Fbw7β levels in response to reduction or loss of parkin is due to a decrease in the rate of Fbw7β proteolysis. Note that endogenous parkin levels do no change during the course of the cycloheximide chase (Fig. 4A), indicating that parkin, when expressed at normal levels, is a stable protein in embryonic brain neurons. Indeed, this is likely the reason that shRNA-mediated silencing of parkin in postmitotic neurons has only limited efficacy (Fig. 2C).
Fig 4.
Parkin regulates Fbw7β proteolysis in primary neurons. (A) Wild-type and parkin−/− primary embryonic mouse brain neurons were treated with cycloheximide (Cyh) at time zero, and extracts were prepared initially and at 2-h intervals up to 6 h (cycloheximide chase). Extracts were analyzed by SDS-PAGE and Western blotting for Fbw7β and parkin. (B) Quantitation (in arbitrary [arb] units) using ImageJ of the experiment shown in panel A as well as of two additional independent biological experiments. Fbw7β is normalized to tubulin. (C) Fbw7β is targeted by the proteasome. Untreated cultures of mouse embryonic mouse brain neurons or cultures treated with the proteasome inhibitors MG132 and PR11 were analyzed by a cycloheximide chase. Cycloheximide was added, and lysates were prepared at 2-h intervals up to 6 h and analyzed by SDS-PAGE and Western blotting for Fbw7β. The loading control was imido black staining of the polyvinylidene difluoride membrane. (D) Parkin−/− primary embryonic mouse brain neurons were transduced with adenoviruses expressing either wild-type (wt) parkin or mutant parkin (R275W) and then subjected to a cycloheximide chase and analyzed as described for panel A. Data are from two independent biological experiments. (E) Quantitation using ImageJ of the experiment shown in panel D. Fbw7β is normalized to tubulin. AdWT, adenovirus expressing wild-type parkin; AdR275W, adenovirus expressing mutant parkin. (F) The parkin genotype does not affect Fbw7β transcript levels. Total RNA was prepared from wild-type and parkin−/− neurons and analyzed by quantitative real-time PCR. Levels of Fbw7β mRNA were normalized to levels of β-actin mRNA. Error bars correspond to standard deviations from quadruplicate PCRs. For cycloheximide chases, error bars correspond to standard deviations; P values were determined using Student's t test.
Parkin polyubiquitylates Fbw7 in vivo and in vitro.
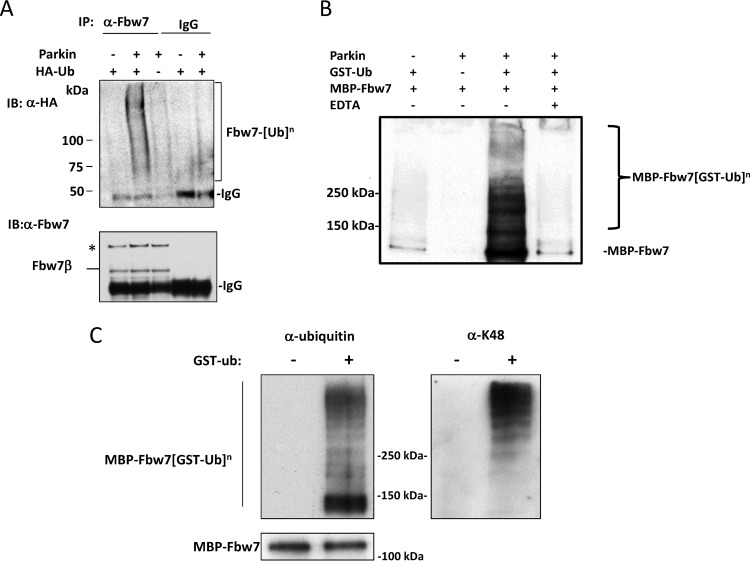
The facts that the rate of proteolysis of Fbw7β is increased when parkin is present and that the two proteins form a complex suggest that parkin may polyubiquitylate Fbw7β, promoting its degradation by the proteasome. To test this idea, we first increased the level of parkin in embryonic brain neurons by adenoviral transduction to determine if this would increase the level of Fbw7β ubiquitylation in vivo. Cells were also transduced with an adenovirus expressing HA-tagged ubiquitin. After inhibiting proteasome activity, lysates were prepared and immunoprecipitated using anti-Fbw7 antibody. Blots of immunoprecipitates were probed first with anti-HA and then with anti-Fbw7 antibodies. In Fig. 5A high-molecular-mass ubiquitin conjugates are visible above background only in the lane corresponding to parkin overexpression. In order to determine whether parkin can directly ubiquitylate Fbw7, we reconstituted a parkin ubiquitin ligase assay in vitro using purified recombinant proteins prepared in E. coli and insect cells. Fbw7 tends to form soluble high-molecular-weight aggregates under typical ubiquitin ligase assay conditions; these aggregates do not dissociate in SDS-PAGE sample buffer or urea but resemble ubiquitin conjugates on blots (see Fig. S2 in the supplemental material). We therefore used GST-ubiquitin in the reaction mixture so that genuine ubiquitin conjugates could be purified prior to analysis by SDS-PAGE and blotting. Maltose binding protein (MBP)-Fbw7 ubiquitin conjugates were detected only after incubation with parkin and ubiquitin (Fig. 5B). In order to determine if parkin decorates Fbw7 with K48-linked ubiquitin chains, associated with proteasomal degradation (62), MBP-Fbw7 was purified after ubiquitylation, as described above, using amylose-Sepharose beads that capture MBP-tagged proteins. Parallel blots were developed with antiubiquitin and anti-polyubiquitin K48 linkage-specific antibodies, respectively (Fig. 5C). High-molecular weight MBP-Fbw7 ubiquitin conjugates were enriched for chains containing K48 linkages, consistent with a role for parkin in targeting Fbw7β for proteasomal degradation.
Fig 5.
Parkin ubiquitylates Fbw7. (A) Parkin stimulates ubiquitylation of Fbw7 in neurons. Primary embryonic mouse brain neurons were transduced with adenoviruses expressing either parkin, HA-ubiquitin, or both. Prior to preparation of extracts, neurons were treated with proteasome inhibitors MG132 and PR11. Extracts were immunoprecipitated with either anti-Fbw7 antibody or nonimmune rabbit IgG. Immune complexes were then analyzed by SDS-PAGE and Western blotting using anti-HA antibody (upper panel). The blot was then stripped and developed using anti-Fbw7 antibody (lower panel). The asterisk indicates nonspecific species recognized by anti-Fbw7 antibody. (B) In vitro reconstitution of ubiquitylation of Fbw7 by parkin. Reaction mixtures containing the indicated purified proteins were incubated with a cocktail containing the ubiquitin-activating enzyme (E1), the ubiquitin-conjugating enzyme (E2) UbcH7, and an ATP-regenerating system. GST-ubiquitin conjugates were then purified using glutathione-Sepharose beads and analyzed by SDS-PAGE and Western blotting using anti-Fbw7 antibody. Note that unmodified MBP-Fbw7 is copurified most likely because Fbw7 forms dimers via a dimerization domain amino terminal to its F box, allowing nonubiquitylated Fbw7 to copurifiy with ubiquitylated species. (C) Parkin forms lysine 48 (K48)-linked chains on Fbw7. MBP-Fbw7 from reactions analogous to those described in panel D was purified using amylose-Sepharose beads and analyzed by SDS-PAGE and Western blotting. Parallel samples were analyzed using antiubiquitin antibody or anti-K48-linked ubiquitin antibody. α, anti; Ub, ubiquitin; IP, immunoprecipitation; IB, immunoblotting. The superscript “n” indicates a variable number of ubiquitin molecules conjugated to Fbw7.
Fbw7β undergoes parkin-dependent proteolysis in response to oxidative stress.
One of the proposed roles of parkin is to protect neurons from oxidative stress (63, 64). We hypothesized that degradation of Fbw7β might be part of the neuroprotective function of parkin. We therefore determined how parkin affects Fbw7β levels in neurons responding to acute oxidative stress. Parkin−/− and wild-type neurons were treated with the oxidant NOC-12 [1-hydroxy-2-oxo-3-(N-ethyl-2-aminoethyl)-3-ethyl-1-triazene], a compound that generates nitric oxide free radicals, and then Fbw7β levels were determined at 2-h intervals for 6 h (Fig. 6A and B). Parkin-dependent reduction of Fbw7β levels was observed in response to the oxidative stress. Similar results were observed in neurons where parkin levels were reduced by RNAi-mediated silencing (Fig. 6C and D).
Fig 6.
Parkin promotes reduction in Fbw7β levels under conditions of oxidative stress. (A) Wild-type and parkin−/− neurons were treated with the oxidant NOC-12 and harvested at 2-h intervals for 6 h. Extracts were analyzed by SDS-PAGE and Western blotting using Fbw7 and parkin antibodies. (B) Quantitation using ImageJ of the experiment shown in panel A and two additional biological experiments. Fbw7β was normalized to tubulin. (C) Neurons were transduced with either control (GFP-specific; cont) or parkin-specific shRNA (park) adenovirus, treated with NOC-12, and processed as described in the legend of Fig. 2C. Parkin silencing was confirmed in the time zero (t = 0) samples. (D) Quantitation using ImageJ of the experiment shown in panel C and two additional biological experiments. Fbw7β was normalized to tubulin. Error bars correspond to standard deviations, and P values were determined using Student's t test. shParkin, shRNA targeting parkin.
Parkin-dependent neuroprotection in response to oxidative stress requires reduction of Fbw7β levels.
In order to determine the role of Fbw7β reduction in the neuronal response to oxidative stress, we measured levels of apoptosis 16 h after the addition of NOC-12. Wild-type and parkin−/− neurons were scored for apoptosis based on the percentage of fragmented and pyknotic nuclei. Surprisingly, parkin−/− neurons were no more sensitive to oxidative stress than wild-type neurons (see Fig. S3A and B in the supplemental material). However, wild-type neurons subjected to specific RNAi-mediated silencing of parkin exhibited increased sensitivity to oxidative stress as scored by counting apoptotic nuclei (Fig. 7A to C) and by measuring levels of cleaved poly(ADP-ribose) polymerase (PARP) (Fig. 7E). Off-target effects of the parkin shRNA could be ruled out because expression of the parkin shRNA in parkin−/− neurons had no effect on sensitivity to oxidative stress or Fbw7β levels (see Fig. S3C and D). We conclude that germ line ablation of parkin leads to physiological compensations that allow neurons to resist oxidative stress in the chronic absence of parkin (see below). This lack of a strong oxidative stress phenotype in parkin−/− neurons is consistent with the lack of a neurodegenerative phenotype in the parkin−/− mouse (21–24). Therefore, most subsequent experiments were carried out under conditions where parkin levels were reduced acutely by RNAi-mediated silencing. In order to evaluate the role of Fbw7β reduction in neuroprotection from oxidative stress, parkin and Fbw7β levels were reduced simultaneously in neurons that were then treated with NOC-12 (Fig. 7A, B, D, and E). Reduction of Fbw7β almost completely reversed the sensitization to oxidative stress mediated by parkin reduction, as measured by scoring apoptotic nuclei and by measuring levels of cleaved PARP. Similar results were obtained using another oxidant, tert-butyl hydroperoxide (tBHP), and a mitochondrial depolarizing agent that causes intrinsic oxidative stress, carbonyl cyanide m-chlorophenylhydrazone (CCCP) (Fig. 7F). Conversely, overexpression of Fbw7β sensitized neurons to oxidative stress, similarly to neurons with reduced parkin levels (Fig. 7G and H). These results are consistent with Fbw7β reduction being a critical downstream effector of parkin-mediated neuroprotection. Furthermore, it suggests that a target (or targets) of the SCFFbw7β ubiquitin ligase is important for neuronal survival under conditions of oxidative stress.
The target of parkin-mediated neuroprotection from oxidative stress is the prosurvival Bcl-2 family member myeloid cell leukemia-1 (Mcl-1).
Apoptotic cell death is regulated in part by the balance between pro- and antiapoptotic members of Bcl-2 (or BH domain) family of proteins. Ultimately, these proteins regulate mitochondrial outer membrane permeability, which is a trigger for apoptosis. Interestingly, one prosurvival member of the Bcl-2 family, Mcl-1, has been shown to be a target of SCFFbw7 in tumor cells (42, 43). Therefore, we investigated the relationship between parkin, Fbw7β, and Mcl-1 in promoting neuronal survival in response to oxidative stress. Mcl-1 is an unstable protein in neurons, and its half-life is significantly increased by proteasomal inhibition, consistent with ubiquitin-mediated proteasomal degradation (Fig. 8A). We then determined whether Mcl-1 levels are regulated by Fbw7β in primary neurons. When Fbw7β levels were reduced using shRNAs targeting either the β isoform specifically or all isoforms (targeting exon 2), levels of Mcl-1 were increased (Fig. 8B and C). Conversely, when Fbw7β levels were increased by adenoviral transduction, Mcl-1 levels were reduced (Fig. 8B and C). To determine whether the increase in Mcl-1 levels observed when Fbw7β levels were reduced was due to a change in Mcl-1 half-life, a cycloheximide chase was carried out under conditions of Fbw7β silencing (Fig. 8D and E). Reduction in Fbw7β levels significantly stabilized Mcl-1, consistent with a role for SCFFbw7β in ubiquitin-mediated proteosomal degradation of Mcl-1 in neurons. To determine whether Mcl-1 is important for neuronal survival, RNAi-mediated silencing was used to reduce Mcl-1 levels, and neurons were assayed for survival in response to oxidative stress (Fig. 8F and G). Silencing of Mcl-1 increased the basal level of neuronal apoptosis and severely sensitized neurons to oxidative stress. Therefore, changes in Mcl-1 levels are likely to have an impact on neuronal survival. When parkin levels were decreased in neurons by RNAi-mediated silencing, Mcl-1 levels were reduced in parallel with an increase in Fbw7β levels (Fig. 7C and 9A). Furthermore, the half-life of Mcl-1 was reduced in response to RNAi-mediated silencing of parkin (Fig. 9B and C). When neurons silenced for parkin were subjected to oxidative stress, Mcl-1 was destabilized (Fig. 9A, D, and E), consistent with increased neuronal apoptosis, whereas levels of apoptotic regulators Bcl-2 and Bax remained unchanged (Fig. 9A and D). However, silencing of Fbw7β in parallel with parkin rescued Mcl-1 levels (Fig. 9F) and neuronal apoptosis (Fig. 7A and E), confirming that parkin-mediated reduction in Fbw7β levels is important for maintaining Mcl-1 during oxidative stress. Expressing parkin-specific shRNA in parkin−/− neurons had no effect on Mcl-1 levels or Fbw7β levels (see Fig. S3D in the supplemental material), consistent with a lack of impact on neuronal survival (see Fig. S3C) and ruling out off-target effects of the parkin shRNA on Mcl-1. Overexpression of Fbw7β destabilized Mcl-1 in response to oxidative stress (Fig. 9G and H) and increased neuronal apoptosis (Fig. 7G), similarly to silencing of parkin (Fig. 7A). Finally, Mcl-1 overexpression rescued the sensitivity to oxidative stress conferred by parkin silencing (Fig. 9I). These data taken together are consistent with parkin promoting neuronal survival during acute oxidative stress by targeting Fbw7β for proteasomal degradation, thereby stabilizing the prosurvival factor Mcl-1 and preventing apoptosis.
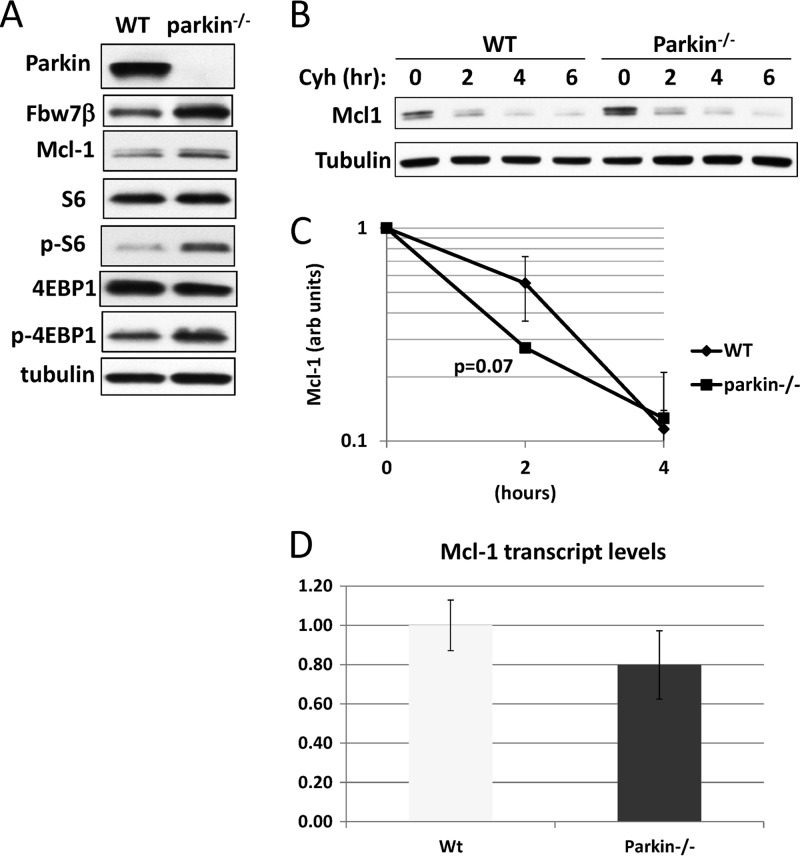
Mcl-1 levels are increased in parkin−/− neurons by constitutive activation of the mTORC1 pathway.
Since we did not observe increased sensitivity to oxidative stress in parkin−/− relative to wild-type neurons, we compared levels of Mcl-1 (Fig. 10A). Surprisingly, Mcl-1 levels were higher in parkin−/− neurons than in wild-type neurons, thereby explaining their lack of sensitivity to oxidative stress. However, importantly, Mcl-1 turned over more rapidly in parkin−/− neurons than in wild-type neurons, consistent with parkin still having a role in maintaining Mcl-1 stability (Fig. 10B and C). Expression of Mcl-1 could be increased at the transcriptional or translational level. Quantitative real-time PCR analysis revealed no difference in Mcl-1 mRNA levels (Fig. 10D). However, Mcl-1 has been reported to be regulated at the level of translation by the mTORC1 pathway (65). We therefore assayed for markers of upregulation of mTORC1. Phosphorylation of both ribosomal protein S6 (S6) and initiation factor 4E binding protein 1 (4EBP1) was significantly increased in parkin−/− neurons, consistent with activation of mTORC1 and increased Mcl-1 translation (Fig. 10A). These results suggest that germ line ablation of parkin may lead to compensatory activation of mTORC1 and a concomitant increase in Mcl-1 translation in order to maintain neuronal survival.
Fig 10.
Analysis of Mcl-1 in parkin−/− neurons. (A) Germ line ablation of parkin increases Mcl-1 levels and activates the mTORC1 pathway. Extracts from wild-type and parkin−/− primary mouse neurons were analyzed by SDS-PAGE and Western blotting for the indicated proteins. Antibodies specific for the phosphorylated forms of ribosomal protein S6 (p-S6) and 4EBP1 (p-4EBP1) were used to detect activation of the mTORC1 pathway. (B) Although expressed at a higher level, Mcl-1 is more unstable in parkin−/− neurons. Mcl-1 was analyzed by cycloheximide chase in wild-type and parkin−/− neurons. (C) Quantitation using ImageJ of the experiment shown in panel B as well as an additional biological experiment. Mcl-1 was normalized to tubulin. (D) Mcl-1 mRNA levels are unaffected by parkin genotype. Mcl-1 mRNA levels were compared in quadruplicate samples from wild-type and parkin−/− neurons by quantitative real-time PCR. Mcl-1 mRNA levels were normalized to β-actin mRNA. All error bars shown represent standard deviations.
Regulation of Mcl-1 in response to oxidative stress.
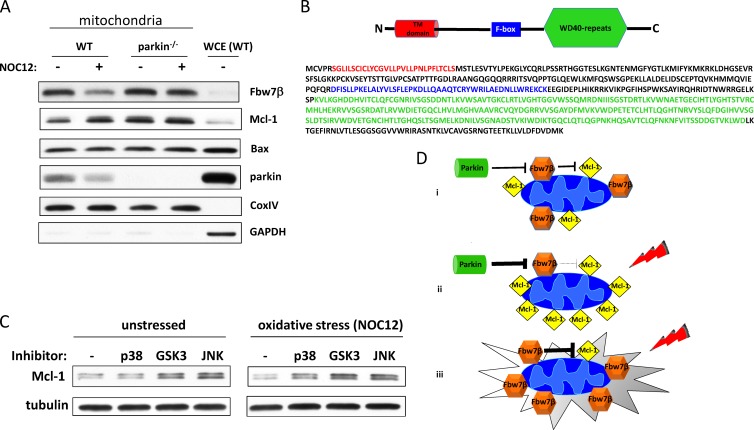
Mcl-1 carries out antiapoptotic functions at the mitochondrial outer membrane, where it inactivates proapoptotic proteins such as Bax and Bak (66). In order to determine how parkin and Fbw7β regulate the functional pool of Mcl-1, we purified mitochondria from unstressed wild-type and parkin−/− neurons and from neurons treated with NOC-12 for 80 min (longer treatments fragmented mitochondria such that they could not be purified by differential sedimentation protocols). Both Fbw7β and Mcl-1 were highly enriched in the mitochondrial fraction compared to levels in equivalent amounts of whole-cell extract (Fig. 11A). Fbw7β has an amino-terminal predicted transmembrane domain (Fig. 11B), possibly accounting for its tight association with mitochondria. The colocalization of Fbw7β and Mcl-1, presumably on the mitochondrial outer membrane, is consistent with a role for Fbw7β in the regulation of Mcl-1 turnover. It has been previously reported that Fbw7β is associated with the endoplasmic reticulum (ER) (55). However, localization experiments were carried out in nonneuronal cells ectopically expressing Fbw7β. We cannot exclude the possibility that some neuronal Fbw7β is associated with the ER since our mitochondrial fraction is contaminated with ER membranes (see Fig. S4 in the supplemental material). However, the enrichment of Fbw7β in the mitochondrial fraction is consistent with mitochondrial localization of a significant fraction of Fbw7β. Only a relatively small fraction of parkin was stably associated with mitochondria under these conditions. However, this does not preclude a weak association that did not survive the stringency of the purification. After 80 min of oxidative stress, parkin-dependent changes in the mitochondrial pools of Fbw7β and Mcl-1 are dramatic. Rapid loss of Fbw7β correlates with an equally rapid increase in Mcl-1. These early responses are not observed for the mitochondrial pools of Fbw7β and Mcl-1 in parkin−/− neurons although Mcl-1 levels are intrinsically higher (Fig. 11A), presumably due to enhanced translation. Therefore, parkin is required to increase mitochondrial Mcl-1 in response to oxidative stress, presumably promoting survival. Although Bax levels may be slightly elevated on parkin−/− mitochondria relative to the wild type, these levels do not change rapidly in response to oxidative stress at 80 min.
Fig 11.
Regulation of Mcl-1 in response to oxidative stress. (A) Parkin regulates the mitochondrial pool of Fbw7β and Mcl-1. Mitochondria were purified from wild-type and parkin−/− primary embryonic mouse brain neurons before and after treatment with NOC-12 for 80 min. Mitochondrial extracts (20 μg) were analyzed by SDS-PAGE and Western blotting for the indicated proteins. Twenty micrograms of whole-cell extract (WCE) from wild-type neurons was run in the last lane for comparison. (B) Topology of Fbw7β. A schematic representation (not to scale) and primary structure of Fbw7β are shown. The putative transmembrane (TM) domain is in red, the F box is in blue, and WD-40 repeats that form an eight-bladed β-propeller (substrate binding domain) are in green. (C) p38 MAPK, GSK3, and JNK all contribute to Mcl-1 degradation. Primary embryonic mouse brain neurons were treated for 5 h with either vehicle (DMSO), a p38 MAPK inhibitor (SB202190), a GSK3 inhibitor (SB216763), or a JNK inhibitor (SR3306) (left panels). Alternatively, neurons were treated with the same inhibitors for 1 h, after which NOC-12 was added for an additional 4 h (total incubation with inhibitors, 5 h). Extracts were analyzed by SDS-PAGE and Western blotting for Mcl-1 levels. (D) A model for the parkin–Fbw7β–Mcl-1 axis. (i) Under unstressed conditions, basal levels of parkin-mediated degradation of Fbw7β create a homeostasis between Fbw7β and Mcl-1 levels on the mitochondrial outer membrane, respectively. (ii) Under conditions of oxidative stress, parkin-mediated degradation of Fbw7β increases, allowing mitochondrial Mcl-1 to accumulate and protect neurons from apoptosis. This may be a necessary adjustment to increased Mcl-1 degron phosphorylation, possibly due to activation of p38 MAPK. (iii) In the absence of parkin, Fbw7β levels are not regulated in response to oxidative stress, Mcl-1 is degraded, and neuronal apoptosis ensues.
GSK3, JNK, and p38 MAPK contribute to Mcl-1 phosphodegron activation in neurons.
SCFFbw7 ubiquitin ligases recognize a consensus phosphodegron in targeted substrates. Three protein kinases have been reported to phosphorylate the Mcl-1 phosphodegron in different nonneuronal cell types: glycogen synthase kinase 3 (GSK3), c-Jun N-terminal kinase (JNK), and p38 mitogen-activated protein kinase (p38 MAPK) (42, 43). In order to determine the role of these protein kinases in regulating Mcl-1 stability in neurons, we treated neurons with kinase-specific inhibitors and compared Mcl-1 levels to those of untreated controls. Mcl-1 levels were increased by treatment with an inhibitor of either GSK3 or JNK (Fig. 11C), suggesting that both of these kinases contribute to Mcl-1 phosphodegron activation. An inhibitor of the stress-activated kinase p38 MAPK had no effect. However, when neurons were subjected to oxidative stress by treatment with NOC-12, inhibitors of all three protein kinases stabilized Mcl-1 (Fig. 11C), suggesting that stress destabilizes Mcl-1 by promoting increased phosphorylation of its phosphodegron through the activation of p38 MAPK. This may explain the role of parkin-mediated reduction of Fbw7β levels in maintaining survival in response to stress.
DISCUSSION
Relevance to Parkinson's disease and potential clinical implications.
Homozygous mutation of the PARK2 locus, which encodes parkin, leads to an early-onset neurodegenerative syndrome known as ARJP, or autosomal recessive juvenile parkinsonism (1–3). The manifestations of this movement disorder are, in many respects, indistinguishable from those associated with the more common sporadic form of Parkinson's disease (67, 68). Both syndromes are characterized by a spectrum of motor symptoms that are caused by progressive death of dopaminergic neurons in a midbrain region known as the substantia nigra pars compacta. However, although both ARJP and sporadic PD are caused by loss of the same spectrum of neurons, it is not at all clear that they share the same etiology. There are two schools of thought on the cause of neuronal death in PD. Since sporadic PD is characterized by the accumulation of cytoplasmic protein aggregates known as Lewy bodies, it has been proposed that neuronal death is associated with possible cytotoxicity of these aggregates (8, 9). The strongest argument in favor of this hypothesis is the fact that one of the major constituents of Lewy bodies is the protein α-synuclein, and mutations in α-synuclein that promote spontaneous aggregation cause PD in humans and neurodegenerative disease in mice (67, 69, 70). However, ARJP caused by PARK2 mutation is only rarely associated with Lewy body formation (3, 59, 71), arguing that loss of parkin is likely to cause neuronal death by another mechanism. A second hypothesis is that PD is caused by an inherited or acquired inability to resist neuronal oxidative and/or mitochondrial stress. In support of this view, parkin-null mice and Parkinson's disease patients exhibit defects in both antioxidant production and mitochondrial functions (21, 72–76), important for protection from oxidative stress. In this regard, an attractive explanation for selective loss of dopaminergic neurons is that dopamine metabolism is associated with excessive production of reactive oxygen and nitrogen species, which might render such neurons particularly vulnerable if defense mechanisms are compromised (77–79). Therefore, it is significant that we have linked parkin to a key pathway required for neuronal survival in response to oxidative stress. Our experiments were carried out with a mixed population of neurons derived from whole brains, suggesting that the link between parkin and neuronal survival mediated by stabilizing Mcl-1 is general and not restricted to dopaminergic neurons, at least under conditions of acute stress. At this point, we cannot say whether this pathway is important in neuroprotection from low levels of chronic oxidative stress, such as might occur in dopaminergic neurons undergoing dopamine metabolism. However, it is reasonable to speculate that the chronic stress conferred by metabolism of dopamine in dopaminergic neurons may render them more dependent on parkin and cause the selective attrition of these neurons over time in individuals where parkin is mutationally inactivated. Whether loss of parkin and the pathway it controls play a role in other forms of PD is uncertain. However, it has been found that parkin in at least some brains from PD patients has been inactivated by chemical modification, dopamine quinone modification, and nitrosylation, possibly linking sporadic PD to loss of parkin activity (78, 80, 81). Another model linking parkin to stress sensitivity is its demonstrated role in mitochondrial quality control, specifically, in clearance via mitophagy of damaged mitochondria (82–87). Neurons that accumulate nonfunctional mitochondria are expected to be sensitive to various forms of stress (88).
The parkin-null mouse as a model for Parkinson's disease.
Parkin-nullizygous mice exhibit only very mild phenotypes and do not experience neurodegeneration, unlike humans with biallelic mutation at the PARK2 locus (21–25). This finding has presented a conundrum in the PD research field and has led to the suggestion that the mouse is not an appropriate organism for investigating the function of parkin or the etiology of PD. One possibility is that the approximately 24-month life span of the mouse provides insufficient time for chronic low-level oxidative stress to damage and eliminate dopaminergic neurons. In humans with a mutation at the PARK2 locus, neurodegenerative phenotypes are not observed until young adulthood. Another possible explanation lies in the finding presented above that germ line mutation of parkin in the mouse leads to a compensatory increase in survival signaling. Specifically, significant upregulation of the mTORC1 pathway most likely promotes increased translation of the prosurvival Bcl2 family member Mcl-1 (65). This compensates for the increased turnover of Mcl-1 caused by loss of parkin-dependent regulation of Fbw7β levels. As a result, parkin−/− neurons are no more sensitive to acute oxidative stress than are wild-type neurons. It is possible that human neurons, or a subset of human neurons, do not have this germ line reprogramming capacity and are therefore more susceptible to loss of parkin function than mouse neurons. It will be informative to determine whether mutationally or chemically attenuating the mTORC1 pathway can sensitize the mouse to germ line mutation of parkin, leading to a PD-like phenotype. Developmental compensation for germ line mutation of parkin in the mouse has been suggested previously in that conditional mutation of parkin in adult mice produces a neurodegeneration phenotype not observed in germ line-mutated mice (17).
Other substrates and functions of the parkin ubiquitin ligase.
Numerous proteins have been reported to be targeted by the parkin ubiquitin ligase for ubiquitin-mediated proteasomal degradation. Indeed, a recent proteomic analysis has shown that parkin ubiquitylated numerous mitochondrial and cytoplasmic proteins, especially under conditions of mitochondrial depolarization (89). Previously reported targets have included CDCrel-1 (6), α-synuclein (8), Pael R (13), synphilin-1 (10), cyclin E1 (18), and p38/JTV-1 (11, 15). Of these, the first five were shown not to be altered in the brains of parkin−/− mice. p38/JTV-1 showed a modest increase in the brains of parkin−/− mice and ARJP human brains (15). Overexpression of p38/JTV-1 in cultured cells led to cell death, consistent with a potential role for this putative parkin target in neuronal pathology associated with parkin loss (15).
More recently, three additional substrates of parkin have been identified, Bax, PARIS, and NEMO. It was shown that parkin can ubiquitylate the proapoptotic factor Bax in vivo and in vitro and that although this does not cause a change in steady-state cellular levels of Bax, parkin overexpression prevents accumulation of Bax in the mitochondrial fraction in response to stress in a cell line with dopaminergic neuron-like properties (14). We did not observe an increase in Bax recruitment to parkin−/− mitochondria in response to oxidative stress although we utilized a shorter treatment time and primary neurons rather than a cell line. However, we did observe a slight increase in steady-state mitochondrial Bax levels, consistent with the model, although the increase could be due to elevated mitochondrial Mcl-1, a Bax binding partner. The transcriptional repressor PARIS (for parkin-interacting substrate), shown to be ubiquitylated and degraded in a parkin-dependent manner, accumulates in the brains of patients with homozygous parkin mutation and in the striatum and ventral midbrain of conditional parkin-nullizygous mice (17). The target of PARIS-mediated repression is the transcriptional coactivator PGC-1α, which promotes expression of genes required for mitochondrial function and survival from stress (90, 91). Consistent with a role in PD etiology, parkin appears to regulate PARIS only in midbrain dopaminergic neurons (17). Parkin has also been shown to carry out positive regulatory ubiquitylation of the NF-κB essential modulator (NEMO), essential for canonical NF-κB signaling (92). The resulting transcriptional upregulation of OPA1, the mitochondrial guanosine triphosphatase, is important for protection from stress-induced cell death.
Parkin has been shown, both in cultured cells and by in vitro reconstitution, to monoubiquitylate the PDZ protein PICK1, a synaptic scaffolding protein that interacts with neurotransmitter receptors, transporters, and ion channels (93). Although ubiquitylation of PICK1 by parkin affected neither its steady-state level nor its turnover kinetics, it suppressed PICK1-dependent activation of ion currents mediated by the ASIC2a proton-gated channel, and, as a result, parkin−/− hippocampal neurons showed excessive potentiation of such currents in response to protein kinase C (PKC) activation. This mechanism may be relevant to PD in that parkin may protect neurons from ion channel overactivity and, ultimately, excitotoxicity.
Roles have also been attributed to parkin in mitochondrial fission-fusion homeostasis and in the clearance of damaged depolarized mitochondria via the mitophagy pathway. In the former process, the relevant targets appear to be the mitofusins, Mfn1 and Mfn2, which promote mitochondrial fusion. Although there are no data showing that parkin directly ubiquitylates Mfn, the data support parkin-dependent ubiquitylation of Mfn in cells (94, 95). Whether ubiquitylation modifies Mfn function or increases its rate of turnover by the proteasome remains to be clarified as the data are not conclusive. However, acute loss of parkin by transient gene silencing leads to mitochondrial fragmentation, consistent with parkin-dependent inactivation or turnover of Mfn (96). On the other hand, cells and neurons experiencing chronic parkin loss by germ line mutation, e.g., parkin−/− neurons and ARJP patient-derived fibroblasts, show no overt phenotype in mitochondrial morphology (94). Therefore, it appears, as with the parkin-dependent regulation of Mcl-1 reported here, that cells chronically experiencing parkin loss can normalize mitochondrial homeostasis via compensatory mechanisms.
With respect to the role of parkin in mitophagy, it has been shown that parkin overexpression can promote the clearance of mitochondria (mitophagy) damaged by depolarizing agents such CCCP (82–87). This is an attractive idea since aggregated aberrant mitochondria have been detected in the neurons of PD patients. The model proposed is that upon depolarization, the kinase PINK1 activates parkin and recruits it to the mitochondrial surface, where proteins are then stably ubiquitylated by the E3 ligase activity of parkin. This ubiquitylation then is a signal for recruitment of the autophagic machinery. Recent proteomic analysis of parkin ubiquitylation targets in response to mitochondrial depolarization strongly supports this model (89). However, the role of this function of parkin in neurodegeneration is not yet clear. Indeed, it has been recently demonstrated using an in vivo mouse model that parkin is not recruited to damaged mitochondria and that the absence of parkin does not affect either the clearance of damaged mitochondria or the neuropathology associated with mitochondrial dysfunction (97). Furthermore, a detailed analysis of mitochondrial structure and integrity in the midbrain and striatum of 2-year-old mice that were deficient in the products of all three recessive PD genes, namely, parkin, DJ-1, and PINK1, showed that there was no difference in size, number, or appearance of mitochondria in the triple knockout mouse (98).
We emphasize that none of the functions proposed for parkin is mutually exclusive of its role in stabilizing Mcl-1.
A role for Mcl-1 in neuronal survival.
Germ line ablation of Mcl-1 in the mouse causes preimplantation lethality (99). However, in the heterozygous mouse, the threshold for apoptosis in hippocampal neurons after drug-induced seizure was significantly lowered, suggesting a role for Mcl-1 in neuronal survival (100). More recently, it was shown that ablation of Mcl-1 in vivo in neuronal progenitor cells during development causes widespread cell-autonomous apoptosis (101). We show in the present study that acute reduction in Mcl-1 levels by RNAi-mediated silencing significantly increases basal levels of apoptosis in cultured neurons and renders them extremely sensitive to oxidative stress, indicating that Mcl-1 is critical for neuronal survival. In another study, Mcl-1 was shown to regulate survival of cerebellar granule neurons (102). Proteasome-dependent degradation of Mcl-1 in these neurons is required for apoptosis induced by removal of serum and KCl from the growth medium. However, the ubiquitin ligase shown to be responsible for Mcl-1 ubiquitylation in that study was Trim17 rather than SCFFbw7. The results of our experiments, carried out with a mixed population of neurons derived from whole brains, suggest that SCFFbw7 is responsible for Mcl-1 regulation in the most abundant neuron types but does not preclude other ubiquitin ligases, such as Trim17, from having that role in a subset of neuron types.
We found that in neurons, at least three protein kinases are responsible for promoting Mcl-1 ubiquitylation: GSK3, JNK, and p38 MAPK. Based on prior studies on Mcl-1, we assume that all three of these kinases target residues corresponding the phosphodegron recognized by SCFFbw7 (42, 43). However, we cannot exclude the possibility that some of the effects of the inhibitors of these kinases are indirect, for example, the activation of one kinase impaired by another. However, it is interesting that p38 MAPK becomes important for Mcl-1 degradation only under conditions of stress, suggesting that stress may prime neurons for apoptosis through p38-dependent phosphorylation of Mcl-1, superimposed on constitutive phosphorylation by GSK3 and JNK. In such situations, parkin appears to play a prosurvival role by reducing Fbw7β levels, thereby protecting phosphorylated Mcl-1 from ubiquitylation and degradation, a mechanism that would be unavailable to neurons lacking parkin (Fig. 11D). How increased parkin-mediated degradation of Fbw7β occurs upon stress remains to be determined. It is possible that recruitment of parkin to the mitochondrial surface under conditions of oxidative stress increases degradation of Fbw7β associated with the mitochondrial outer membrane. Under the experimental conditions employed in this study, we did not observe an accumulation of parkin in purified mitochondria after oxidative stress. However, a weak association might not survive the multiple sedimentations associated with the purification protocol utilized.
Therapeutic implications.
Recently, one of the potentially neuroprotective avenues of disease-modifying intervention for Parkinson's disease has been small-molecule inhibition of JNK (103). Furthermore, in a mouse model for PD involving administration of a dopaminergic neuron-specific poison, MPTP (1,2,3,6-methylphenyltetrahydropyridine), a JNK inhibitor was shown to be protective against dopaminergic neuron loss in the midbrain. Our results suggest that the therapeutic efficacy of JNK inhibitors may be mediated by restricting Mcl-1 degron phosphorylation and concomitant promotion of neuronal survival by stabilization of Mcl-1. In a similar vein, it is also possible that small-molecule inhibition of Fbw7β substrate binding would have therapeutic benefit for patients suffering from Parkinson's disease and other neurodegenerative disorders.
Supplementary Material
ACKNOWLEDGMENTS
S.I.R. acknowledges support from NIH grants NS059904 and CA078343. M.G.S. acknowledges support from the Government of Canada (CIHR; CRC) and the Bhargava Family Research Chair in Neurodegeneration. M.S.G. acknowledges generous gifts from the David M. Crowley Foundation and Parkinson's Benefactors.
We are grateful to H. Shimura and N. Hattori for tissue specimens. We thank Zhuohua Zhang, Sanford-Burnham Research Institute, for parkin-expressing baculovirus and Raman Kumar for construction of MBP-Fbw7.
Footnotes
Published ahead of print 15 July 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.00535-13.
REFERENCES
- 1.Gasser T. 2007. Update on the genetics of Parkinson's disease. Mov. Disord. 22(Suppl 17):S343–S350 [DOI] [PubMed] [Google Scholar]
- 2.Kitada T, Asakawa S, Hattori N, Matsumine H, Yamamura Y, Minoshima S, Yokochi M, Mizuno Y, Shimizu N. 1998. Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature 392:605–608 [DOI] [PubMed] [Google Scholar]
- 3.Pramstaller PP, Schlossmacher MG, Jacques TS, Scaravilli F, Eskelson C, Pepivani I, Hedrich K, Adel S, Gonzales-McNeal M, Hilker R, Kramer PL, Klein C. 2005. Lewy body Parkinson's disease in a large pedigree with 77 Parkin mutation carriers. Ann. Neurol. 58:411–422 [DOI] [PubMed] [Google Scholar]
- 4.Imai Y, Soda M, Takahashi R. 2000. Parkin suppresses unfolded protein stress-induced cell death through its E3 ubiquitin-protein ligase activity. J. Biol. Chem. 275:35661–35664 [DOI] [PubMed] [Google Scholar]
- 5.Shimura H, Hattori N, Kubo S, Mizuno Y, Asakawa S, Minoshima S, Shimizu N, Iwai K, Chiba T, Tanaka K, Suzuki T. 2000. Familial Parkinson disease gene product, parkin, is a ubiquitin-protein ligase. Nat. Genet. 25:302–305 [DOI] [PubMed] [Google Scholar]
- 6.Zhang Y, Gao J, Chung KK, Huang H, Dawson VL, Dawson TM. 2000. Parkin functions as an E2-dependent ubiquitin-protein ligase and promotes the degradation of the synaptic vesicle-associated protein, CDCrel-1. Proc. Natl. Acad. Sci. U. S. A. 97:13354–13359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Komander D, Rape M. 2012. The ubiquitin code. Annu. Rev. Biochem. 81:203–229 [DOI] [PubMed] [Google Scholar]
- 8.Shimura H, Schlossmacher MG, Hattori N, Frosch MP, Trockenbacher A, Schneider R, Mizuno Y, Kosik KS, Selkoe DJ. 2001. Ubiquitination of a new form of alpha-synuclein by parkin from human brain: implications for Parkinson's disease. Science 293:263–269 [DOI] [PubMed] [Google Scholar]
- 9.Sriram SR, Li X, Ko HS, Chung KK, Wong E, Lim KL, Dawson VL, Dawson TM. 2005. Familial-associated mutations differentially disrupt the solubility, localization, binding and ubiquitination properties of parkin. Hum. Mol. Genet. 14:2571–2586 [DOI] [PubMed] [Google Scholar]
- 10.Chung KK, Zhang Y, Lim KL, Tanaka Y, Huang H, Gao J, Ross CA, Dawson VL, Dawson TM. 2001. Parkin ubiquitinates the alpha-synuclein-interacting protein, synphilin-1: implications for Lewy-body formation in Parkinson disease. Nat. Med. 7:1144–1150 [DOI] [PubMed] [Google Scholar]
- 11.Corti O, Hampe C, Koutnikova H, Darios F, Jacquier S, Prigent A, Robinson JC, Pradier L, Ruberg M, Mirande M, Hirsch E, Rooney T, Fournier A, Brice A. 2003. The p38 subunit of the aminoacyl-tRNA synthetase complex is a Parkin substrate: linking protein biosynthesis and neurodegeneration. Hum. Mol. Genet. 12:1427–1437 [DOI] [PubMed] [Google Scholar]
- 12.Hattori N, Mizuno Y. 2004. Pathogenetic mechanisms of parkin in Parkinson's disease. Lancet 364:722–724 [DOI] [PubMed] [Google Scholar]
- 13.Imai Y, Soda M, Inoue H, Hattori N, Mizuno Y, Takahashi R. 2001. An unfolded putative transmembrane polypeptide, which can lead to endoplasmic reticulum stress, is a substrate of Parkin. Cell 105:891–902 [DOI] [PubMed] [Google Scholar]
- 14.Johnson BN, Berger AK, Cortese GP, Lavoie MJ. 2012. The ubiquitin E3 ligase parkin regulates the proapoptotic function of Bax. Proc. Natl. Acad. Sci. U. S. A. 109:6283–6288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ko HS, von Coelln R, Sriram SR, Kim SW, Chung KK, Pletnikova O, Troncoso J, Johnson B, Saffary R, Goh EL, Song H, Park BJ, Kim MJ, Kim S, Dawson VL, Dawson TM. 2005. Accumulation of the authentic parkin substrate aminoacyl-tRNA synthetase cofactor, p38/JTV-1, leads to catecholaminergic cell death. J. Neurosci. 25:7968–7978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Poole AC, Thomas RE, Yu S, Vincow ES, Pallanck L. 2010. The mitochondrial fusion-promoting factor mitofusin is a substrate of the PINK1/parkin pathway. PLoS One 5:e10054. 10.1371/journal.pone.0010054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shin JH, Ko HS, Kang H, Lee Y, Lee YI, Pletinkova O, Troconso JC, Dawson VL, Dawson TM. 2011. PARIS (ZNF746) repression of PGC-1α contributes to neurodegeneration in Parkinson's disease. Cell 144:689–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Staropoli JF, McDermott C, Martinat C, Schulman B, Demireva E, Abeliovich A. 2003. Parkin is a component of an SCF-like ubiquitin ligase complex and protects postmitotic neurons from kainate excitotoxicity. Neuron 37:735–749 [DOI] [PubMed] [Google Scholar]
- 19.Wang X, Winter D, Ashrafi G, Schlehe J, Wong YL, Selkoe D, Rice S, Steen J, LaVoie MJ, Schwarz TL. 2011. PINK1 and Parkin target Miro for phosphorylation and degradation to arrest mitochondrial motility. Cell 147:893–906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang Y, Nishimura I, Imai Y, Takahashi R, Lu B. 2003. Parkin suppresses dopaminergic neuron-selective neurotoxicity induced by Pael-R in Drosophila. Neuron 37:911–924 [DOI] [PubMed] [Google Scholar]
- 21.Goldberg MS, Fleming SM, Palacino JJ, Cepeda C, Lam HA, Bhatnagar A, Meloni EG, Wu N, Ackerson LC, Klapstein GJ, Gajendiran M, Roth BL, Chesselet MF, Maidment NT, Levine MS, Shen J. 2003. Parkin-deficient mice exhibit nigrostriatal deficits but not loss of dopaminergic neurons. J. Biol. Chem. 278:43628–43635 [DOI] [PubMed] [Google Scholar]
- 22.Itier JM, Ibanez P, Mena MA, Abbas N, Cohen-Salmon C, Bohme GA, Laville M, Pratt J, Corti O, Pradier L, Ret G, Joubert C, Periquet M, Araujo F, Negroni J, Casarejos MJ, Canals S, Solano R, Serrano A, Gallego E, Sanchez M, Denefle P, Benavides J, Tremp G, Rooney TA, Brice A, Garcia de Yebenes J. 2003. Parkin gene inactivation alters behaviour and dopamine neurotransmission in the mouse. Hum. Mol. Genet. 12:2277–2291 [DOI] [PubMed] [Google Scholar]
- 23.Perez FA, Palmiter RD. 2005. Parkin-deficient mice are not a robust model of parkinsonism. Proc. Natl. Acad. Sci. U. S. A. 102:2174–2179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Periquet M, Corti O, Jacquier S, Brice A. 2005. Proteomic analysis of parkin knockout mice: alterations in energy metabolism, protein handling and synaptic function. J. Neurochem. 95:1259–1276 [DOI] [PubMed] [Google Scholar]
- 25.Von Coelln R, Thomas B, Savitt JM, Lim KL, Sasaki M, Hess EJ, Dawson VL, Dawson TM. 2004. Loss of locus coeruleus neurons and reduced startle in parkin null mice. Proc. Natl. Acad. Sci. U. S. A. 101:10744–10749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Deshaies RJ, Joazeiro CA. 2009. RING domain E3 ubiquitin ligases. Annu. Rev. Biochem. 78:399–434 [DOI] [PubMed] [Google Scholar]
- 27.Sarikas A, Hartmann T, Pan ZQ. 2011. The cullin protein family. Genome Biol. 12:220. 10.1186/gb-2011-12-4-220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Deshaies RJ. 1999. SCF and Cullin/ring H2-based ubiquitin ligases. Annu. Rev. Cell Dev. Biol. 15:435–467 [DOI] [PubMed] [Google Scholar]
- 29.Feldman RM, Correll CC, Kaplan KB, Deshaies RJ. 1997. A complex of Cdc4p, Skp1p, and Cdc53p/cullin catalyzes ubiquitination of the phosphorylated CDK inhibitor Sic1p. Cell 91:221–230 [DOI] [PubMed] [Google Scholar]
- 30.Skowyra D, Craig KL, Tyers M, Elledge SJ, Harper JW. 1997. F-box proteins are receptors that recruit phosphorylated substrates to the SCF ubiquitin-ligase complex. Cell 91:209–219 [DOI] [PubMed] [Google Scholar]
- 31.Koepp DM, Schaefer LK, Ye X, Keyomarsi K, Chu C, Harper JW, Elledge SJ. 2001. Phosphorylation-dependent ubiquitination of cyclin E by the SCFFbw7 ubiquitin ligase. Science 294:173–177 [DOI] [PubMed] [Google Scholar]
- 32.Moberg KH, Bell DW, Wahrer DC, Haber DA, Hariharan IK. 2001. Archipelago regulates cyclin E levels in Drosophila and is mutated in human cancer cell lines. Nature 413:311–316 [DOI] [PubMed] [Google Scholar]
- 33.Strohmaier H, Spruck CH, Kaiser P, Won KA, Sangfelt O, Reed SI. 2001. Human F-box protein hCdc4 targets cyclin E for proteolysis and is mutated in a breast cancer cell line. Nature 413:316–322 [DOI] [PubMed] [Google Scholar]
- 34.Klotz K, Cepeda D, Tan Y, Sun D, Sangfelt O, Spruck C. 2009. SCF(Fbxw7/hCdc4) targets cyclin E2 for ubiquitin-dependent proteolysis. Exp. Cell Res. 315:1832–1839 [DOI] [PubMed] [Google Scholar]
- 35.Welcker M, Orian A, Grim JE, Eisenman RN, Clurman BE. 2004. A nucleolar isoform of the Fbw7 ubiquitin ligase regulates c-Myc and cell size. Curr. Biol. 14:1852–1857 [DOI] [PubMed] [Google Scholar]
- 36.Welcker M, Orian A, Jin J, Grim JE, Harper JW, Eisenman RN, Clurman BE. 2004. The Fbw7 tumor suppressor regulates glycogen synthase kinase 3 phosphorylation-dependent c-Myc protein degradation. Proc. Natl. Acad. Sci. U. S. A. 101:9085–9090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yada M, Hatakeyama S, Kamura T, Nishiyama M, Tsunematsu R, Imaki H, Ishida N, Okumura F, Nakayama K, Nakayama KI. 2004. Phosphorylation-dependent degradation of c-Myc is mediated by the F-box protein Fbw7. EMBO J. 23:2116–2125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wei W, Jin J, Schlisio S, Harper JW, Kaelin WG., Jr 2005. The v-Jun point mutation allows c-Jun to escape GSK3-dependent recognition and destruction by the Fbw7 ubiquitin ligase. Cancer Cell 8:25–33 [DOI] [PubMed] [Google Scholar]
- 39.Olson BL, Hock MB, Ekholm-Reed S, Wohlschlegel JA, Dev KK, Kralli A, Reed SI. 2008. SCFCdc4 acts antagonistically to the PGC-1α transcriptional coactivator by targeting it for ubiquitin-mediated proteolysis. Genes Dev. 22:252–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tetzlaff MT, Yu W, Li M, Zhang P, Finegold M, Mahon K, Harper JW, Schwartz RJ, Elledge SJ. 2004. Defective cardiovascular development and elevated cyclin E and Notch proteins in mice lacking the Fbw7 F-box protein. Proc. Natl. Acad. Sci. U. S. A. 101:3338–3345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tsunematsu R, Nakayama K, Oike Y, Nishiyama M, Ishida N, Hatakeyama S, Bessho Y, Kageyama R, Suda T, Nakayama KI. 2004. Mouse Fbw7/Sel-10/Cdc4 is required for notch degradation during vascular development. J. Biol. Chem. 279:9417–9423 [DOI] [PubMed] [Google Scholar]
- 42.Inuzuka H, Shaik S, Onoyama I, Gao D, Tseng A, Maser RS, Zhai B, Wan L, Gutierrez A, Lau AW, Xiao Y, Christie AL, Aster J, Settleman J, Gygi SP, Kung AL, Look T, Nakayama KI, DePinho RA, Wei W. 2011. SCF(FBW7) regulates cellular apoptosis by targeting MCL1 for ubiquitylation and destruction. Nature 471:104–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wertz IE, Kusam S, Lam C, Okamoto T, Sandoval W, Anderson DJ, Helgason E, Ernst JA, Eby M, Liu J, Belmont LD, Kaminker JS, O'Rourke KM, Pujara K, Kohli PB, Johnson AR, Chiu ML, Lill JR, Jackson PK, Fairbrother WJ, Seshagiri S, Ludlam MJ, Leong KG, Dueber EC, Maecker H, Huang DC, Dixit VM. 2011. Sensitivity to antitubulin chemotherapeutics is regulated by MCL1 and FBW7. Nature 471:110–114 [DOI] [PubMed] [Google Scholar]
- 44.Arabi A, Ullah K, Branca RM, Johansson J, Bandarra D, Haneklaus M, Fu J, Aries I, Nilsson P, Den Boer ML, Pokrovskaja K, Grander D, Xiao G, Rocha S, Lehtio J, Sangfelt O. 2012. Proteomic screen reveals Fbw7 as a modulator of the NF-κB pathway. Nat. Commun. 3:976. 10.1038/ncomms1975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Busino L, Millman SE, Scotto L, Kyratsous CA, Basrur V, O'Connor O, Hoffmann A, Elenitoba-Johnson KS, Pagano M. 2012. Fbxw7α- and GSK3-mediated degradation of p100 is a pro-survival mechanism in multiple myeloma. Nat. Cell Biol. 14:375–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fukushima H, Matsumoto A, Inuzuka H, Zhai B, Lau AW, Wan L, Gao D, Shaik S, Yuan M, Gygi SP, Jimi E, Asara JM, Nakayama K, Nakayama KI, Wei W. 2012. SCFFbw7 modulates the NFkB signaling pathway by targeting NFkB2 for ubiquitination and destruction. Cell Rep. 1:434–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tan Y, Sangfelt O, Spruck C. 2008. The Fbxw7/hCdc4 tumor suppressor in human cancer. Cancer Lett. 271:1–12 [DOI] [PubMed] [Google Scholar]
- 48.Welcker M, Clurman BE. 2008. FBW7 ubiquitin ligase: a tumour suppressor at the crossroads of cell division, growth and differentiation. Nat. Rev. Cancer 8:83–93 [DOI] [PubMed] [Google Scholar]
- 49.Hao B, Oehlmann S, Sowa ME, Harper JW, Pavletich NP. 2007. Structure of a Fbw7-Skp1-cyclin E complex: multisite-phosphorylated substrate recognition by SCF ubiquitin ligases. Mol. Cell 26:131–143 [DOI] [PubMed] [Google Scholar]
- 50.Orlicky S, Tang X, Willems A, Tyers M, Sicheri F. 2003. Structural basis for phosphodependent substrate selection and orientation by the SCFCdc4 ubiquitin ligase. Cell 112:243–256 [DOI] [PubMed] [Google Scholar]
- 51.Welcker M, Clurman BE. 2007. Fbw7/hCDC4 dimerization regulates its substrate interactions. Cell Div. 2:7. 10.1186/1747-1028-2-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Spruck CH, Strohmaier H, Sangfelt O, Muller HM, Hubalek M, Muller-Holzner E, Marth C, Widschwendter M, Reed SI. 2002. hCDC4 gene mutations in endometrial cancer. Cancer Res. 62:4535–4539 [PubMed] [Google Scholar]
- 53.Ye X, Nalepa G, Welcker M, Kessler BM, Spooner E, Qin J, Elledge SJ, Clurman BE, Harper JW. 2004. Recognition of phosphodegron motifs in human cyclin E by the SCF(Fbw7) ubiquitin ligase. J. Biol. Chem. 279:50110–50119 [DOI] [PubMed] [Google Scholar]
- 54.Bhaskaran N, van Drogen F, Ng HF, Kumar R, Ekholm-Reed S, Peter M, Sangfelt O, Reed SI. 2013. Fbw7α and Fbw7γ collaborate to shuttle cyclin E1 into the nucleolus for multiubiquitylation. Mol. Cell. Biol. 33:85–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Matsumoto A, Tateishi Y, Onoyama I, Okita Y, Nakayama K, Nakayama KI. 2011. Fbxw7β resides in the endoplasmic reticulum membrane and protects cells from oxidative stress. Cancer Sci. 102:749–755 [DOI] [PubMed] [Google Scholar]
- 56.Padmanabhan J, Park DS, Greene LA, Shelanski ML. 1999. Role of cell cycle regulatory proteins in cerebellar granule neuron apoptosis. J. Neurosci. 19:8747–8756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Paddison PJ, Silva JM, Conklin DS, Schlabach M, Li M, Aruleba S, Balija V, O'Shaughnessy A, Gnoj L, Scobie K, Chang K, Westbrook T, Cleary M, Sachidanandam R, McCombie WR, Elledge SJ, Hannon GJ. 2004. A resource for large-scale RNA-interference-based screens in mammals. Nature 428:427–431 [DOI] [PubMed] [Google Scholar]
- 58.Steckley D, Karajgikar M, Dale LB, Fuerth B, Swan P, Drummond-Main C, Poulter MO, Ferguson SS, Strasser A, Cregan SP. 2007. Puma is a dominant regulator of oxidative stress induced Bax activation and neuronal apoptosis. J. Neurosci. 27:12989–12999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Farrer M, Chan P, Chen R, Tan L, Lincoln S, Hernandez D, Forno L, Gwinn-Hardy K, Petrucelli L, Hussey J, Singleton A, Tanner C, Hardy J, Langston JW. 2001. Lewy bodies and parkinsonism in families with parkin mutations. Ann. Neurol. 50:293–300 [DOI] [PubMed] [Google Scholar]
- 60.Hattori N, Kitada T, Matsumine H, Asakawa S, Yamamura Y, Yoshino H, Kobayashi T, Yokochi M, Wang M, Yoritaka A, Kondo T, Kuzuhara S, Nakamura S, Shimizu N, Mizuno Y. 1998. Molecular genetic analysis of a novel Parkin gene in Japanese families with autosomal recessive juvenile parkinsonism: evidence for variable homozygous deletions in the parkin gene in affected individuals. Ann. Neurol. 44:935–941 [DOI] [PubMed] [Google Scholar]
- 61.Shimura H, Hattori N, Kubo S, Yoshikawa M, Kitada T, Matsumine H, Asakawa S, Minoshima S, Yamamura Y, Shimizu N, Mizuno Y. 1999. Immunohistochemical and subcellular localization of Parkin protein: absence of protein in autosomal recessive juvenile parkinsonism patients. Ann. Neurol. 45:668–672 [DOI] [PubMed] [Google Scholar]
- 62.Hershko A, Ciechanover A. 1992. The ubiquitin system for protein degradation. Annu. Rev. Biochem. 61:761–807 [DOI] [PubMed] [Google Scholar]
- 63.Jiang H, Ren Y, Zhao J, Feng J. 2004. Parkin protects human dopaminergic neuroblastoma cells against dopamine-induced apoptosis. Hum. Mol. Genet. 13:1745–1754 [DOI] [PubMed] [Google Scholar]
- 64.Thomas B, Beal MF. 2007. Parkinson's disease. Hum. Mol. Genet. 16(Spec no 2):R183–R194 [DOI] [PubMed] [Google Scholar]
- 65.Mills JR, Hippo Y, Robert F, Chen SM, Malina A, Lin CJ, Trojahn U, Wendel HG, Charest A, Bronson RT, Kogan SC, Nadon R, Housman DE, Lowe SW, Pelletier J. 2008. mTORC1 promotes survival through translational control of Mcl-1. Proc. Natl. Acad. Sci. U. S. A. 105:10853–10858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Youle RJ, Strasser A. 2008. The BCL-2 protein family: opposing activities that mediate cell death. Nat. Rev. Mol. Cell Biol. 9:47–59 [DOI] [PubMed] [Google Scholar]
- 67.Baba Y, Markopoulou K, Putzke JD, Whaley NR, Farrer MJ, Wszolek ZK, Uitti RJ. 2006. Phenotypic commonalities in familial and sporadic Parkinson disease. Arch. Neurol. 63:579–583 [DOI] [PubMed] [Google Scholar]
- 68.Kitada T, Tomlinson JJ, Ao HS, Grimes DA, Schlossmacher MG. 2012. Considerations regarding the etiology and future treatment of autosomal recessive versus idiopathic Parkinson disease. Curr. Treat Options Neurol. 14:230–240 [DOI] [PubMed] [Google Scholar]
- 69.Luk KC, Kehm V, Carroll J, Zhang B, O'Brien P, Trojanowski JQ, Lee VM. 2012. Pathological alpha-synuclein transmission initiates Parkinson-like neurodegeneration in nontransgenic mice. Science 338:949–953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Spillantini MG, Schmidt ML, Lee VM, Trojanowski JQ, Jakes R, Goedert M. 1997. Alpha-synuclein in Lewy bodies. Nature 388:839–840 [DOI] [PubMed] [Google Scholar]
- 71.Hardy J. 2003. The relationship between Lewy body disease, Parkinson's disease, and Alzheimer's disease. Ann. N. Y. Acad. Sci. 991:167–170 [DOI] [PubMed] [Google Scholar]
- 72.Bindoff LA, Birch-Machin M, Cartlidge NE, Parker WD, Jr, Turnbull DM. 1989. Mitochondrial function in Parkinson's disease. Lancet ii:49. [DOI] [PubMed] [Google Scholar]
- 73.Bindoff LA, Desnuelle C, Birch-Machin MA, Pellissier JF, Serratrice G, Dravet C, Bureau M, Howell N, Turnbull DM. 1991. Multiple defects of the mitochondrial respiratory chain in a mitochondrial encephalopathy (MERRF): a clinical, biochemical and molecular study. J. Neurol. Sci. 102:17–24 [DOI] [PubMed] [Google Scholar]
- 74.Keeney PM, Xie J, Capaldi RA, Bennett JP., Jr 2006. Parkinson's disease brain mitochondrial complex I has oxidatively damaged subunits and is functionally impaired and misassembled. J. Neurosci. 26:5256–5264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Palacino JJ, Sagi D, Goldberg MS, Krauss S, Motz C, Wacker M, Klose J, Shen J. 2004. Mitochondrial dysfunction and oxidative damage in parkin-deficient mice. J. Biol. Chem. 279:18614–18622 [DOI] [PubMed] [Google Scholar]
- 76.Parker WD, Jr, Parks JK, Swerdlow RH. 2008. Complex I deficiency in Parkinson's disease frontal cortex. Brain Res. 1189:215–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hattoria N, Wanga M, Taka H, Fujimura T, Yoritaka A, Kubo S, Mochizuki H. 2009. Toxic effects of dopamine metabolism in Parkinson's disease. Parkinsonism Relat. Disord. 15(Suppl 1):S35–S38 [DOI] [PubMed] [Google Scholar]
- 78.LaVoie MJ, Ostaszewski BL, Weihofen A, Schlossmacher MG, Selkoe DJ. 2005. Dopamine covalently modifies and functionally inactivates parkin. Nat. Med. 11:1214–1221 [DOI] [PubMed] [Google Scholar]
- 79.Lotharius J, Brundin P. 2002. Impaired dopamine storage resulting from alpha-synuclein mutations may contribute to the pathogenesis of Parkinson's disease. Hum. Mol. Genet. 11:2395–2407 [DOI] [PubMed] [Google Scholar]
- 80.Chung KK, Thomas B, Li X, Pletnikova O, Troncoso JC, Marsh L, Dawson VL, Dawson TM. 2004. S-nitrosylation of parkin regulates ubiquitination and compromises parkin's protective function. Science 304:1328–1331 [DOI] [PubMed] [Google Scholar]
- 81.Yao D, Gu Z, Nakamura T, Shi ZQ, Ma Y, Gaston B, Palmer LA, Rockenstein EM, Zhang Z, Masliah E, Uehara T, Lipton SA. 2004. Nitrosative stress linked to sporadic Parkinson's disease: S-nitrosylation of parkin regulates its E3 ubiquitin ligase activity. Proc. Natl. Acad. Sci. U. S. A. 101:10810–10814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Batlevi Y, La Spada AR. 2011. Mitochondrial autophagy in neural function, neurodegenerative disease, neuron cell death, and aging. Neurobiol. Dis. 43:46–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.de Vries RL, Przedborski S. 2013. Mitophagy and Parkinson's disease: be eaten to stay healthy. Mol. Cell Neurosci. 55:37–43 [DOI] [PubMed] [Google Scholar]
- 84.Deas E, Wood NW, Plun-Favreau H. 2011. Mitophagy and Parkinson's disease: the PINK1-parkin link. Biochim. Biophys. Acta 1813:623–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Imai Y, Lu B. 2011. Mitochondrial dynamics and mitophagy in Parkinson's disease: disordered cellular power plant becomes a big deal in a major movement disorder. Curr. Opin. Neurobiol. 21:935–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Narendra D, Walker JE, Youle R. 2012. Mitochondrial quality control mediated by PINK1 and Parkin: links to parkinsonism. Cold Spring Harb. Perspect. Biol. 4:a011338. 10.1101/cshperspect.a011338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Youle RJ, Narendra DP. 2011. Mechanisms of mitophagy. Nat. Rev. Mol. Cell Biol. 12:9–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.MacDonald ME, Ambrose CM, Duyao MP, Myers RH, Lin C, Srinidhi L, Barnes G, Taylor SA, James M, Groot N, MacFarlane H, Jenkins B, Anderson MA, Wexler NS, Gusella JF.1993. A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington's disease chromosomes. Cell 72:971–983 [DOI] [PubMed] [Google Scholar]
- 89.Sarraf SA, Raman M, Guarani-Pereira V, Sowa ME, Huttlin EL, Gygi SP, Harper JW. 2013. Landscape of the PARKIN-dependent ubiquitylome in response to mitochondrial depolarization. Nature 496:372–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lin J, Handschin C, Spiegelman BM. 2005. Metabolic control through the PGC-1 family of transcription coactivators. Cell Metab. 1:361–370 [DOI] [PubMed] [Google Scholar]
- 91.St-Pierre J, Drori S, Uldry M, Silvaggi JM, Rhee J, Jager S, Handschin C, Zheng K, Lin J, Yang W, Simon DK, Bachoo R, Spiegelman BM. 2006. Suppression of reactive oxygen species and neurodegeneration by the PGC-1 transcriptional coactivators. Cell 127:397–408 [DOI] [PubMed] [Google Scholar]
- 92.Muller-Rischart AK, Pilsl A, Beaudette P, Patra M, Hadian K, Funke M, Peis R, Deinlein A, Schweimer C, Kuhn PH, Lichtenthaler SF, Motori E, Hrelia S, Wurst W, Trumbach D, Langer T, Krappmann D, Dittmar G, Tatzelt J, Winklhofer KF. 2013. The E3 ligase parkin maintains mitochondrial integrity by increasing linear ubiquitination of NEMO. Mol. Cell 49:908–921 [DOI] [PubMed] [Google Scholar]
- 93.Joch M, Ase AR, Chen CX, MacDonald PA, Kontogiannea M, Corera AT, Brice A, Seguela P, Fon EA. 2007. Parkin-mediated monoubiquitination of the PDZ protein PICK1 regulates the activity of acid-sensing ion channels. Mol. Biol. Cell 18:3105–3118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Rakovic A, Grunewald A, Kottwitz J, Bruggemann N, Pramstaller PP, Lohmann K, Klein C. 2011. Mutations in PINK1 and parkin impair ubiquitination of mitofusins in human fibroblasts. PLoS One 6:e16746. 10.1371/journal.pone.0016746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Tanaka A, Cleland MM, Xu S, Narendra DP, Suen DF, Karbowski M, Youle RJ. 2010. Proteasome and p97 mediate mitophagy and degradation of mitofusins induced by Parkin. J. Cell Biol. 191:1367–1380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lutz AK, Exner N, Fett ME, Schlehe JS, Kloos K, Lammermann K, Brunner B, Kurz-Drexler A, Vogel F, Reichert AS, Bouman L, Vogt-Weisenhorn D, Wurst W, Tatzelt J, Haass C, Winklhofer KF. 2009. Loss of parkin or PINK1 function increases Drp1-dependent mitochondrial fragmentation. J. Biol. Chem. 284:22938–22951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Sterky FH, Lee S, Wibom R, Olson L, Larsson NG. 2011. Impaired mitochondrial transport and Parkin-independent degeneration of respiratory chain-deficient dopamine neurons in vivo. Proc. Natl. Acad. Sci. U. S. A. 108:12937–12942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kitada T, Tong Y, Gautier CA, Shen J. 2009. Absence of nigral degeneration in aged parkin/DJ-1/PINK1 triple knockout mice. J. Neurochem. 111:696–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Rinkenberger JL, Horning S, Klocke B, Roth K, Korsmeyer SJ. 2000. Mcl-1 deficiency results in peri-implantation embryonic lethality. Genes Dev. 14:23–27 [PMC free article] [PubMed] [Google Scholar]
- 100.Mori M, Burgess DL, Gefrides LA, Foreman PJ, Opferman JT, Korsmeyer SJ, Cavalheiro EA, Naffah-Mazzacoratti MG, Noebels JL. 2004. Expression of apoptosis inhibitor protein Mcl1 linked to neuroprotection in CNS neurons. Cell Death Differ. 11:1223–1233 [DOI] [PubMed] [Google Scholar]
- 101.Arbour N, Vanderluit JL, Le Grand JN, Jahani-Asl A, Ruzhynsky VA, Cheung EC, Kelly MA, MacKenzie AE, Park DS, Opferman JT, Slack RS. 2008. Mcl-1 is a key regulator of apoptosis during CNS development and after DNA damage. J. Neurosci. 28:6068–6078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Magiera MM, Mora S, Mojsa B, Robbins I, Lassot I, Desagher S. 2013. Trim17-mediated ubiquitination and degradation of Mcl-1 initiate apoptosis in neurons. Cell Death Differ. 20:281–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Chambers JW, Pachori A, Howard S, Ganno M, Hansen D, Jr, Kamenecka T, Song X, Duckett D, Chen W, Ling YY, Cherry L, Cameron MD, Lin L, Ruiz CH, Lograsso P. 2011. Small molecule c-jun-N-terminal kinase (JNK) inhibitors protect dopaminergic neurons in a model of Parkinson's disease. ACS Chem. Neurosci. 2:198–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.