Abstract
V(D)J recombination is initiated by the RAG endonuclease, which introduces DNA double-strand breaks (DSBs) at the border between two recombining gene segments, generating two hairpin-sealed coding ends and two blunt signal ends. ATM and DNA-dependent protein kinase catalytic subunit (DNA-PKcs) are serine-threonine kinases that orchestrate the cellular responses to DNA DSBs. During V(D)J recombination, ATM and DNA-PKcs have unique functions in the repair of coding DNA ends. ATM deficiency leads to instability of postcleavage complexes and the loss of coding ends from these complexes. DNA-PKcs deficiency leads to a nearly complete block in coding join formation, as DNA-PKcs is required to activate Artemis, the endonuclease that opens hairpin-sealed coding ends. In contrast to loss of DNA-PKcs protein, here we show that inhibition of DNA-PKcs kinase activity has no effect on coding join formation when ATM is present and its kinase activity is intact. The ability of ATM to compensate for DNA-PKcs kinase activity depends on the integrity of three threonines in DNA-PKcs that are phosphorylation targets of ATM, suggesting that ATM can modulate DNA-PKcs activity through direct phosphorylation of DNA-PKcs. Mutation of these threonine residues to alanine (DNA-PKcs3A) renders DNA-PKcs dependent on its intrinsic kinase activity during coding end joining, at a step downstream of opening hairpin-sealed coding ends. Thus, DNA-PKcs has critical functions in coding end joining beyond promoting Artemis endonuclease activity, and these functions can be regulated redundantly by the kinase activity of either ATM or DNA-PKcs.
INTRODUCTION
Antigen receptor genes are assembled in developing lymphocytes through the process of V(D)J recombination (1). The V(D)J recombination reaction forms the second exon of these genes from component variable (V), joining (J), and, at some loci, diversity (D) gene segments. V(D)J recombination is initiated when the RAG-1 and RAG-2 proteins, which together form the RAG endonuclease, introduce DNA double-strand breaks (DSBs) at the border of two recombining gene segments and their associated RAG recognition sequences, termed recombination signals (RSs) (2). DNA cleavage by RAG results in two broken DNA ends with distinct structures: a blunt signal end and a coding end that is hairpin sealed by a phosphodiester bond connecting the top and bottom strands (2). RAG cleavage occurs only after a synaptic complex forms between two appropriate RSs, resulting in pairs of signal and coding ends that are held in close proximity in a postcleavage complex (2).
RAG DSBs are repaired by nonhomologous end joining (NHEJ), the major DNA DSB repair pathway in all G1-phase cells (3–5). NHEJ normally joins two signal ends, forming a signal join, and two coding ends, forming a coding join, which is essential for completing antigen receptor gene assembly. Nonstandard joins can also form through the rejoining of a signal end to a coding end at one RAG DSB, creating an open-and-shut join, or through the joining of signal and coding ends from distinct RAG DSBs, creating a hybrid join (3, 6, 7). The RAG proteins can catalyze the formation of these joins through a transposition reaction in vitro; in vivo, however, the formation of these nonstandard joins depends on NHEJ (8–12). In normal cells, the formation of these nonstandard joins occurs rarely, presumably due to the architecture of the postcleavage complex determined by the RAG proteins and components of the NHEJ machinery.
The NHEJ factors required for RAG DSB repair include XRCC4, DNA ligase IV, the Artemis endonuclease, Ku70, Ku80 (also referred to as Ku86), and DNA-dependent protein kinase catalytic subunit (DNA-PKcs) (3, 5, 13). In addition, XRCC4-like factor (XLF), also called Cernunnos, functions during V(D)J recombination, but in lymphocytes this function is not critical when other DSB repair proteins are present (3, 14–17). DNA ligase IV, XRCC4, Ku70, and Ku80 are required for coding and signal join formation. DNA-PKcs is a member of the phosphatidylinositol 3-kinase-related protein kinases (PIKK) and is activated by DNA DSBs to phosphorylate serines or threonines followed by glutamine (SQ/TQ), thus regulating the activity of many proteins involved in DNA damage responses (18–20). Together, Ku70, Ku80, and DNA-PKcs form the DNA-PK complex. Ku70 and Ku80 form a ring-like heterodimer that binds to DSBs soon after they are generated and directly recruits DNA-PKcs, which takes up residence at the apex of the broken DNA end (21–24). During V(D)J recombination, DNA-PKcs promotes Artemis endonuclease activity, which is required to open hairpin-sealed coding ends prior to coding join formation (25, 26). As a result, DNA-PKcs-deficient mice exhibit a severe block in lymphocyte development (3). Activation of Artemis by DNA-PKcs in vitro depends both on the association of these two proteins and on DNA-PKcs kinase activity (25, 26).
The ataxia telangiectasia mutated (ATM) kinase is another member of the PIKK family that is activated by RAG DSBs (3). ATM functions to maintain coding ends in postcleavage complexes until they can be joined. Although neither ATM nor DNA-PKcs is absolutely required for signal join formation, one of these kinases must be present and active, suggesting that ATM and DNA-PKcs have common downstream phosphorylation targets required for signal end joining (27–29). Indeed, ATM and DNA-PKcs phosphorylate several common targets in response to DSBs (30, 31). Moreover, while mice deficient in ATM or DNA-PKcs are viable, those with a combined deficiency of ATM and DNA-PKcs exhibit early embryonic lethality, further emphasizing the importance of the shared activities of ATM and DNA-PKcs (32, 33).
ATM and DNA-PKcs functions are regulated by autophosphorylation. ATM autophosphorylation converts inactive ATM dimers into active monomers (34). DNA-PKcs autophosphorylation is not required for its kinase activity but is required to regulate DNA-PKcs functions during DSB repair (18, 35). In addition, DNA-PKcs is phosphorylated by ATM, which may also regulate its function (18, 35). DNA-PKcs has many SQ/TQ motifs, with the best-studied motifs being those that lie in close proximity in the ABCDE (4 TQ and 2 SQ motifs, also referred to as the T2609 cluster) and PQR (5 SQ motifs) clusters (18, 35). The differential phosphorylation of these clusters can have diverse and sometimes opposing effects on DNA-PKcs kinase activity, structure, and association with broken DNA ends (18, 35, 36).
In this study, our analyses of chromosomal V(D)J recombination in lymphoid cells reveal that ATM and DNA-PKcs have overlapping functions during coding join formation. These functions depend on the phosphorylation of DNA-PKcs and are critical for joining coding ends after the hairpins have been opened by Artemis. These studies have important implications for the function of ATM and DNA-PKcs in general NHEJ-mediated DSB repair.
MATERIALS AND METHODS
Mice.
All mice were housed in the specific-pathogen-free facility at Washington University. All animal protocols were approved by the Washington University Institutional Animal Care and Use Committee.
Cell culture.
v-abl-transformed pre-B cells were generated from 3- to 5-week-old mice by culturing bone marrow cells after transduction with the pMSCV v-abl retrovirus as described previously (37). v-abl-transformed pre-B cells were infected with the pMX-DELCJ, pMX-DELSJ, or pMX-INV retroviral recombination substrate, and clones with single integrants were selected as previously described (37). DNA-PKcs3A/3A:Atm−/− and DNA-PKcs3A/3A:LigIV−/− v-abl-transformed pre-B cells were generated by transient expression of the Cre recombinase in DNA-PKcs3A/3A:Atmc/c and DNA-PKcs3A/3A:LigIVloxP/loxP v-abl-transformed pre-B cells, respectively (38, 39). Cells were electroporated with 1 μg of MSCV-IRES-Thy1.1-Cre plasmid DNA per 106 cells at 1,400 V for 20 ms in 4 pulses, using a Neon transfection system (Invitrogen). Thy1.1-expressing cells were isolated using anti-Thy1.1 microbeads and magnetically activated cell sorting (MACS) separation columns according to the manufacturer's instructions (Miltenyi Biotec). After limiting-dilution culture, individual clones were assayed for Cre deletion by Southern blotting as described below.
For RAG induction, v-abl-transformed pre-B cells were treated with 3 μM imatinib for the indicated times at 106 cells/ml. The abl pre-B cells analyzed exhibited similar increases in RAG-1 and RAG-2 gene expression upon treatment with imatinib (data not shown). The ATM kinase inhibitor KU55933 (50% inhibitory concentration [IC50] = 12.9 nM; Tocris) and the DNA-PKcs kinase inhibitors NU7026 (IC50 = 230 nM; Sigma) and NU7441 (IC50 = 14 nM; Tocris) were used at 15 μM, 20 μM, and 5 μM, respectively (27). For analysis of responses to irradiation, v-abl-transformed pre-B cells were treated with imatinib as described above for 1 day prior to exposure to 6 Gy of gamma irradiation using a Gammacell 40 instrument (Kirloskar Technologies).
Southern blotting.
Native and denaturing (1.2% agarose and 1 M urea) Southern blot analyses of V(D)J recombination of the retroviral recombination substrates and the endogenous Igk locus were carried out as previously described (27, 37, 40). Cre-mediated deletion of the Atmc allele was assayed by Southern blotting of KpnI-digested DNA and the ATM 3′ probe as previously described (39). Cre-mediated deletion of the DNA ligase IVloxP allele was confirmed by Southern blotting of genomic DNA digested with BamHI and BglII. The DNA ligase IV probe was generated by a PCR using the oligonucleotides 5′-AGGCTTGCTCAGACGCTAGC-3′ and 5′-CACAGCACTCCATTTAATGC-3′. The DNA ligase IVloxP and DNA ligase IV− alleles yield 1.0-kb and 1.5-kb hybridizing bands, respectively. Southern blot analyses of Eb:ZFN cleavage were carried out on HindIII-digested genomic DNA hybridized to an Eb probe generated by PCR amplification using the oligonucleotides 5′-GTTAACCAGGCACAGTAGGAC-3′ and 5′-CCATGGTGCATACTGAAGGC-3′.
PCR analyses.
TdT-assisted PCR of pMX-DELCJ and Jk coding ends was performed as previously described, except that for the Jk1 coding end detection, the Jk double-stranded oligonucleotide 5′-GAAAATCATTCCAACCTCTTGTGGGA-3′ was used as a reverse primer and the Jk1 probe oligonucleotide 5′-GGAGAGTGCCAGAATCTGGTTTCAG-3′ was used as a probe (27, 40). PCR analyses of pMX-DELCJ, pMX-INV, and VkJk coding and hybrid joins were carried out as previously described, except that 42 cycles of amplification were used for pMX-DELCJ hybrid joins (37).
Western blot analysis.
Whole-cell lysates were obtained using RIPA buffer (150 mM NaCl, 10 mM Tris, pH 7.2, 0.1% SDS, 1% Triton X-100, 1% deoxycholate, 5 mM EDTA) supplemented with protease inhibitors (Sigma). Lysates were resolved in 6% to 10% acrylamide gels and transferred onto Immobilon polyvinylidene difluoride (PVDF) membranes (Millipore). The primary antibodies anti-Kap-1 (GeneTex), anti-phospho-Kap-1 (Bethyl), anti-phospho-H2AX (Millipore), anti-DNA-PKcs (NeoMarkers), anti-poly(ADP-ribose) polymerase 1 (Calbiochem), and antiactin (Bethyl) were used. Goat anti-mouse (Invitrogen) and donkey anti-rabbit (Fisher) antibodies were used as secondary antibodies. Detection was carried out using a horseradish peroxidase (HRP) ECL detection kit according to the manufacturer's instructions (Thermo Scientific).
Flow cytometry.
Flow cytometric analysis of green fluorescent protein (GFP) expression was carried out using a BD FACSCalibur flow cytometer (BD Biosciences) and Flow Jo 4.6.2 for Macintosh (Tree Star).
ZFN assay.
A pair of custom zinc finger nucleases (ZFN), Eb1:ZFN1 and Eb2:ZFN2, specific to a region of the mouse T cell receptor beta (Tcrb) locus just downstream of the enhancer (Eb), were obtained (Sigma-Aldrich). Eb1:ZFN1 recognizes the sequence 5′-GTCTGTCTGTCCTGC-3′, and Eb2:ZFN2 recognizes the sequence 5′-TGGCTGGCTTTCGCT-3′. Each zinc finger is fused to a truncated Fok1 nuclease. Eb:ZFN1 and Eb:ZFN2 were cotransfected into v-abl-transformed pre-B cells treated with imatinib as described above for 1 day, using a Neon transfection system at 1,400 V for 15 ms in 4 pulses. Transfected cells were divided into two fractions, with one incubated in imatinib alone and the other in imatinib and NU7026, as described above, for 2 days before cell lysis and analysis by Southern blotting (see above) and Cel-I assay. The Cel-I assays were carried out using a Surveyor mutation detection kit and the manufacturer's protocol (Transgenomic). The genomic DNA was PCR amplified using ZFN primer F (5′-TAACCCCAACTCCAGTCACC-3′) and ZFN primer R (5′-TGGATAGAGCAGGATTGGCT-3′), with the following thermocycler program: 94°C for 2 min and then 26 cycles of 94°C for 15 s, 57°C for 30 s, and 68°C for 1 min, followed by 7 min at 68°C. Heteroduplex formation was performed using 400 ng of PCR products and the following thermocycler program: 95°C for 10 min, 85°C for 1 min, 75°C for 1 min, 65°C for 1 min, 55°C for 1 min, 45°C for 1 min, 35°C for 1 min, 25°C for 1 min, and a 4°C hold. One microliter of Surveyor nuclease was added and incubated for 1 h at 42°C, followed by agarose gel electrophoresis.
RESULTS
Inhibition of DNA-PKcs kinase activity does not prevent coding join formation.
Chromosomal V(D)J recombination can be studied in vivo by using v-abl kinase-transformed murine pre-B cells, referred to here as abl pre-B cells (3, 37). Inhibition of the v-abl kinase by imatinib leads to G1-phase cell cycle arrest, RAG induction, and V(D)J recombination at the endogenous immunoglobulin light chain kappa (Igk) locus and chromosomally integrated retroviral V(D)J recombination substrates such as pMX-DELCJ (37, 41). pMX-DELCJ has an antisense GFP cDNA flanked by a pair of RSs that undergo V(D)J recombination by deletion, forming a chromosomal coding join (Fig. 1A) (37).
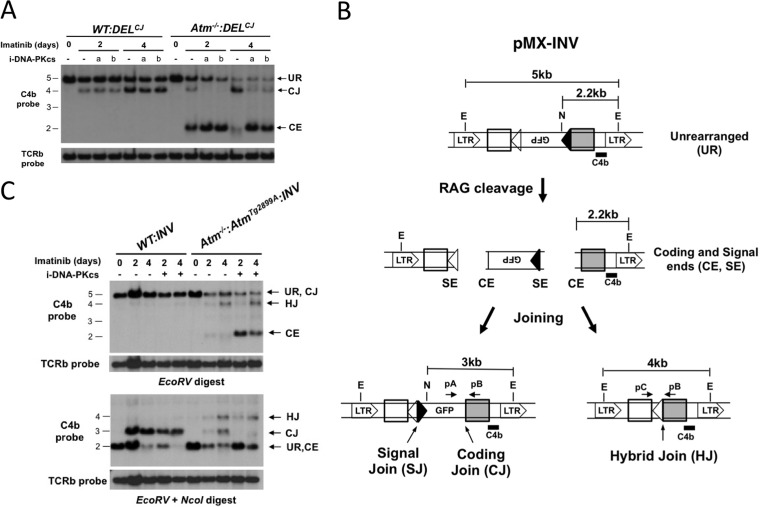
Fig 1.
DNA-PKcs kinase inhibition does not block coding end joining. (A) Schematic of the pMX-DELCJ retroviral recombination substrate showing the two RSs (open and filled triangles) flanking the GFP cDNA in the antisense orientation. The long terminal repeats (LTRs) are shown, as are the relative positions of the EcoRV (E) restriction sites and C4b probe used for Southern blot analysis. (B) Southern blot analysis of pMX-DELCJ in wild-type (WT:DELCJ) and DNA-PKcs-deficient (scid:DELCJ) abl pre-B cells that were untreated (0 days) or treated with imatinib for 2 or 4 days to induce V(D)J recombination. Cells were also treated with vehicle (dimethyl sulfoxide [DMSO]; −) or the DNA-PKcs kinase inhibitor NU7026 (a) or NU7441 (b). Shown are the C4b-hybridizing EcoRV digest bands corresponding to unrear- ranged pMX-DELCJ (UR) and pMX-DELCJ coding joins (CJ) and coding ends (CE). A T cell receptor beta (TCRb) locus probe was used as a DNA loading control. Molecular size markers (in kb) are shown. (C) Western blot analysis of phosphorylated H2AX (γ-H2AX) and KAP-1 (pKAP-1) in WT, Atm−/−, and DNA-PKcs3A/3A:Atm−/− abl pre-B cells treated with imatinib for 1 day, followed by vehicle (DMSO; −) or NU7026 (+) and either no irradiation (IR) (−) or 6 Gy irradiation (+). Actin is shown as a loading control.
After RAG induction with imatinib, wild-type abl pre-B cells containing pMX-DELCJ (WT:DELCJ) underwent robust pMX-DELCJ cleavage and coding join formation (Fig. 1B, 4-kb fragment CJ) (37). As expected, RAG induction in DNA-PKcs-deficient abl pre-B cells (scid:DELCJ) generated from scid mice led to low levels of coding join formation and a significant accumulation of unrepaired coding ends (Fig. 1B, 2-kb fragment CE). In striking contrast, treatment of WT:DELCJ abl pre-B cells with the DNA-PKcs kinase inhibitor NU7026 or NU7441 did not cause an accumulation of unrepaired coding ends or an appreciable block in coding join formation (Fig. 1B). Phosphorylation of KAP-1 and H2AX (forming γ-H2AX) in response to ionizing radiation in Atm−/− abl pre-B cells, which depends on DNA-PKcs, was inhibited by NU7026, demonstrating that NU7026 inhibits DNA-PKcs kinase activity in abl pre-B cells (Fig. 1C) (30, 31). We conclude that a loss of DNA-PKcs protein, but not inhibition of DNA-PKcs kinase activity, leads to a significant block in coding join formation.
ATM can compensate for loss of DNA-PKcs kinase activity during coding end joining.
Although the DNA-PKcs protein is required for coding join formation, we reasoned that ATM kinase activity may compensate for the inhibition of DNA-PKcs kinase activity. Atm−/−:DELCJ abl pre-B cells exhibit a partial block in coding end joining, as evidenced by the accumulation of unrepaired coding ends after RAG induction (Fig. 2A, 2-kb CE band) (37). In contrast to the case with WT:DELCJ abl pre-B cells, treatment of Atm−/−:DELCJ abl pre-B cells with DNA-PKcs kinase inhibitors led to a nearly complete block of pMX-DELCJ coding join formation and a marked accumulation of unrepaired coding ends (Fig. 2A). This was not due to differences in DNA-PKcs levels, as we previously demonstrated that DNA-PKcs is expressed at similar levels in wild-type and Atm−/− abl pre-B cells (27). To determine whether this reflects a requirement for ATM kinase activity, we analyzed rearrangement of the pMX-INV retroviral recombination substrate in Atm−/− abl pre-B cells expressing AtmTgD2899A (Atm−/−:AtmTgD2899A:INV abl pre-B cells) (Fig. 2B and C). pMX-INV is identical to pMX-DELCJ, except that one of the RSs has been inverted so that rearrangement results in inversion of the GFP cDNA and the formation of chromosomal signal and coding joins (Fig. 2B). AtmTgD2899A has a single amino acid mutation that abrogates Atm kinase activity (42). Like Atm deficiency, expression of AtmTgD2899A led to the accumulation of unrepaired pMX-INV coding ends and the formation of pMX-INV hybrid joins (Fig. 2C). However, treatment of Atm−/−:AtmTgD2899A:INV abl pre-B cells with NU7026 led to a dramatic increase in the accumulation of unrepaired coding ends (Fig. 2C). We conclude that the DNA-PKcs protein is required for coding join formation, while DNA-PKcs kinase activity is dispensable when ATM is present and active.
Fig 2.
Redundant DNA-PKcs and ATM kinase activities during coding end joining. (A) Southern blot analysis of pMX-DELCJ rearrangement in WT:DELCJ and Atm−/−:DELCJ abl pre-B cells as described in the legend to Fig. 1B. (B) Schematic of pMX-INV retroviral recombination substrate with components as described for pMX-DELCJ in the legend to Fig. 1A. Unrearranged pMX-INV, coding ends, signal ends, a coding join, a signal join, and a hybrid join are shown. The EcoRV (E) and NcoI (N) sites used for digestion are shown. (C) Southern blot analysis of pMX-INV in WT:INV and Atm−/−:AtmTgD2899A:INV abl pre-B cells treated with imatinib and NU7026 (i-DNA-PKcs), as described in the legend to Fig. 1B. Genomic DNA was digested with either EcoRV or EcoRV and NcoI and probed with the C4b probe (see panel B). Bands generated by unrearranged (UR) pMX-INV, coding joins (CJ), hybrid joins (HJ), and coding ends (CE) are indicated.
DNA-PKcs3A kinase activity is required for coding joins.
Many SQ/TQ motifs in DNA-PKcs are autophosphorylated in response to DNA DSBs. However, there are three TQ motifs in the ABCDE cluster that can be phosphorylated by ATM or DNA-PKcs in response to DNA DSBs (43, 44). Abl pre-B cell lines were generated from mice homozygous for the DNA-PKcs3A allele, in which the threonines of these three TQ motifs have been mutated to alanines (45). DNA-PKcs3A is expressed in DNA-PKcs3A/3A abl pre-B cells at levels comparable to those of DNA-PKcs in wild-type abl pre-B cells (see Fig. S1 in the supplemental material). DNA-PKcs3A kinase activity is intact, as irradiated Atm−/−:DNA-PKcs3A/3A abl pre-B cells exhibited phosphorylation of H2AX and KAP-1 that was inhibited by NU7026 (Fig. 3A) (45). This is also in agreement with previous studies demonstrating that DNA-PKcs proteins with different ABCDE cluster SQ/TQ mutations have robust kinase activity (46).
Fig 3.
Coding join formation is dependent on DNA-PKcs3A kinase activity. (A) Western blot analysis of phosphorylated H2AX and KAP-1 in Atm−/− and Atm−/−:DNA-PKcs3A/3A abl pre-B cells as described in the legend to Fig. 1C. (B) Southern blot analysis of pMX-DELCJ rearrangement in WT:DELCJ and DNA-PKcs3A/3A:DELCJ abl pre-B cells as described in the legend to Fig. 1B. (C) Southern blot analysis of Igk locus rearrangement in WT, Artemis-deficient (Art−/−), and DNA-PKcs3A/3A abl pre-B cells. A schematic of the Igk locus is shown with 3 of the approximately 250 Vk gene segments, the 4 functional Jk gene segments, and the Ck exon (not to scale). The relative positions of the SacI and EcoRI sites and the JkIII probe are also shown, as are schematics for the SacI and EcoRI digest JkIII probe-hybridizing fragments for the germ line (GL) Igk locus and the Jk1, Jk2, Jk3, and Jk4 coding ends (CE). SacI and EcoRI digest JkIII probe-hybridizing fragments from VJk rearrangements vary in size (generating a hybridizing smear in the lane) depending on the Vk and Jk gene segments that are used, as shown in Fig. 2C. (D) Western blot analysis of phosphorylated H2AX and KAP-1 in WT and DNA-PKcs3A/3A abl pre-B cells as described in the legend to Fig. 1C.
After RAG induction, DNA-PKcs3A/3A:DELCJ abl pre-B cells readily formed pMX-DELCJ coding joins similarly to WT:DELCJ abl pre-B cells (Fig. 3B). In striking contrast to WT:DELCJ abl pre-B cells, however, treatment of DNA-PKcs3A/3A:DELCJ abl pre-B cells with NU7026 led to a significant block in coding join formation and accumulation of unrepaired pMX-DELCJ coding ends (Fig. 3B). Analysis of the endogenous Igk locus yielded similar findings (Fig. 3C). Induction of RAG in WT abl pre-B cells led to robust Vk-to-Jk rearrangement at the endogenous Igk locus, as evidenced by Southern blotting revealing hybridizing bands of diverse sizes from rearrangements between approximately 250 Vk gene segments and the 4 functional Jk gene segments in the mouse Igk locus (Fig. 3C; VkJk rearrangements form a hybridizing smear). Induction of RAG in DNA-PKcs3A/3A abl pre-B cells yielded similar results (Fig. 3C). However, upon addition of NU7026, discrete bands reflecting unrepaired Jk coding ends similar in size to those observed in Artemis-deficient (Art−/−) abl pre-B cells were clearly visible for DNA-PKcs3A/3A but not WT abl pre-B cells (Fig. 3C). ATM kinase activity is intact in DNA-PKcs3A/3A abl pre-B cells, as evidenced by H2AX and KAP-1 phosphorylation in response to ionizing radiation in WT and DNA-PKcs3A/3A abl pre-B cells treated with NU7026 (Fig. 3D). We conclude that ATM cannot compensate for the kinase function of DNA-PKcs3A during coding end joining, suggesting that ATM regulates DNA-PKcs activity in RAG DSB repair by phosphorylating one or more of the TQ motifs that are mutated in DNA-PKcs3A.
DNA-PKcs functions downstream of opening hairpin-sealed coding ends.
We wanted to determine whether DNA-PKcs3A kinase activity is required to promote opening of hairpin-sealed coding ends, which is a critical function of DNA-PKcs during coding join formation. To this end, the DNA end structure was analyzed by TdT-assisted PCR (Fig. 4A; see Fig. S2 in the supplemental material) (40). TdT adds a poly(A) tract to open but not hairpin-sealed DNA ends, and this poly(A) tract can be used for PCR amplification (Fig. 4A; see Fig. S2). LigIV−/− abl pre-B cells have a block in coding join formation, but the unrepaired coding ends have open hairpins due to the activity of Artemis. These open pMX-DELCJ and Jk coding ends were readily amplified from LigIV−/−:DELCJ abl pre-B cells (Fig. 4B and C). In contrast, very low levels of products were observed in Artemis-deficient abl pre-B cells (Art−/−:DELCJ), in which coding ends are predominantly hairpin sealed (Fig. 4B and C). Analysis of pMX-DELCJ and Jk coding ends in DNA-PKcs3A/3A:DELCJ abl pre-B cells treated with NU7026 revealed robust TdT-assisted PCR products indicative of open hairpins (Fig. 4B and C).
Fig 4.
Coding ends have open hairpins after inhibition of DNA-PKcs3A kinase activity. (A) Schematic of TdT-assisted PCR. Shown are open and hairpin-sealed coding ends. TdT adds poly(A) to open but not hairpin-sealed coding ends. The T17 forward primer is shown with the reverse primer specific for either the pMX-DELCJ or Jk1 coding end. These primers amplify products from open coding ends where poly(A) has been added. (B and C) TdT-assisted PCRs of pMX-DELCJ (B) and Jk1 (C) coding ends in Art−/−, LigIV−/−, and DNA-PKcs3A/3A abl pre-B cells treated with NU7026 (+ i-DNA-PKcs). Genomic DNA from cells treated with imatinib for 4 days was incubated with dATP, with (+) or without (−) TdT, followed by PCR amplification as shown in panel A. PCR amplification was performed on 5-fold dilutions of genomic DNA treated with TdT and the single highest level of genomic DNA not treated with TdT. Interleukin-2 (IL-2) gene PCR on 5-fold genomic DNA dilutions is shown as a DNA loading control. (D) Denaturing Southern blot analysis of pMX-DELCJ coding ends was carried out on genomic DNA that was digested with EcoRI and probed with C4b (Fig. 1A). Art−/−:DELCJ, LigIV−/−:DELCJ, and DNA-PKcs3A/3A:DELCJ abl pre-B cells not treated (−) or treated with imatinib for 2 days (+), in the presence (+) or absence (−) of the DNA-PKcs inhibitor NU7026 (i-DNA-PKcs), were assayed. The hybridizing bands representing unrearranged pMX-DELCJ and a pMX-DELCJ coding join are shown, as are bands from hairpin-sealed and open coding ends.
Coding end structure was also analyzed by denaturing Southern blot analyses (Fig. 4D; see Fig. S3 in the supplemental material) (40). After restriction enzyme digestion and denaturation, open coding ends form single-stranded DNA fragments that migrate at a lower molecular weight than the native duplex (see Fig. S3). However, hairpin-sealed coding ends cannot be denatured into single-stranded components due to the phosphodiester bond connecting the top and bottom strands (see Fig. S3). Thus, denatured hairpin-sealed coding ends migrate at the same molecular weight as that of the native duplex (see Fig. S2 in the supplemental material). As expected, denaturing Southern blot analysis revealed that most of the unrepaired pMX-DELCJ coding ends in Art−/−:DELCJ abl pre-B cells were hairpin sealed, whereas those in LigIV−/−:DELCJ abl pre-B cells were primarily open (Fig. 4D; see Fig. S3). Analyses of coding ends in DNA-PKcs3A/3A:DELCJ abl pre-B cells treated with NU7026 revealed that most had open hairpins (Fig. 4D). These findings demonstrate that inhibition of the kinase activity of DNA-PKcs3A does not prevent the opening of hairpin-sealed coding ends. Thus, we conclude that DNA-PKcs has functions in joining open coding ends that are compromised by the TQ motif mutations in DNA-PKcs3A.
DNA-PKcs3A exhibits defects in general NHEJ-mediated DSB repair.
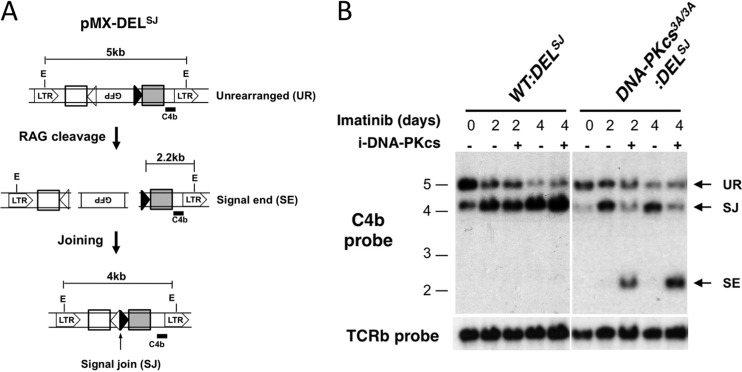
As DNA-PKcs3A kinase activity is required to join open coding ends, we reasoned that this mutation may affect a general function of DNA-PKcs in the repair of DNA ends that are not hairpin sealed. We first determined whether DNA-PKcs3A/3A abl pre-B cells exhibit defects in the joining of blunt signal ends. pMX-DELSJ is identical to pMX-DELCJ, except that the RSs have been inverted, such that rearrangement leads to a chromosomal signal join (Fig. 5A). Robust signal join formation was observed after RAG induction in DNA-PKcs3A/3A:DELSJ abl pre-B cells, whereas inhibition of DNA-PKcs kinase activity in these cells led to a severe block in signal join formation (Fig. 5B). Inhibition of DNA-PKcs kinase activity had no effect on signal joining in WT:DELSJ abl pre-B cells. Thus, DNA-PKcs3A kinase activity is required for the joining of blunt signal ends.
Fig 5.
DNA-PKcs3A kinase activity is required for signal end joining. (A) Schematic of pMX-DELSJ with components as described for pMX-DELCJ in the legend to Fig. 1A. (B) Southern blot analysis of pMX-DELSJ rearrangement in WT:DELSJ and DNA-PKcs3A/3A:DELSJ abl pre-B cells as described in the legend to Fig. 1B. The bands reflecting unrearranged pMX-DELSJ (UR) and a pMX-DELSJ signal join (SJ) and signal end (SE) are indicated.
In order to assay DSBs other than those generated by RAG cleavage, we assayed chromosomal DSBs generated by Fok-1 zinc finger endonuclease fusion proteins targeted to sequences in the T cell receptor beta (Tcrb) locus, which we refer to as Eb:ZFN proteins (see Fig. S4 in the supplemental material). Eb:ZFN targets a sequence immediately 3′ of the Tcrb enhancer (Eb) (Fig. 6A). Transient expression of Eb:ZFN results in Fok-1 cleavage at Eb, creating a DSB with broken DNA ends having 4-bp 5′ single-stranded overhangs. These DNA ends were readily observed by Southern blotting after transient expression of Eb:ZFN in LigIV−/− abl pre-B cells (Fig. 6B). In WT and DNA-PKcs3A/3A abl pre-B cells, DNA DSBs are generated by Eb:ZFN expression, as evidenced by the Cel-I assay, which measures imprecise NHEJ-mediated DSB repair after DNA cleavage at the target site (Fig. 6C and D). However, in WT and DNA-PKcs3A/3A abl pre-B cells, these DSBs are all rapidly repaired, as there were no detectable DNA ends (Fig. 6B). Treatment of WT abl pre-B cells with NU7026 had no effect on the repair of Eb:ZFN-induced chromosomal DNA DSBs (Fig. 6B). However, treatment of the DNA-PKcs3A/3A abl pre-B cells with NU7026 led to the accumulation of unrepaired DSBs, similar to what was observed in LigIV−/− abl pre-B cells (Fig. 6B). We conclude that DNA-PKcs3A kinase activity is required for DNA-PKcs function in rejoining broken DNA ends during general NHEJ-mediated DSB repair.
Fig 6.
DNA-PKcs3A kinase activity is required to repair chromosomal DSBs generated by a zinc finger nuclease. (A) Schematic of the Tcrb locus showing the constant region (Cb) and the enhancer (Eb). The location of the Eb:ZFN cleavage site is indicated, as are the HindIII sites and Eb probe used for Southern blot analysis of Eb:ZFN cleavage. (B) Southern blot analysis of WT, LigIV−/−, and DNA-PKcs3A/3A abl pre-B cells after treatment with imatinib, Eb:ZFN introduction, and treatment with either DMSO (−) or NU7026 (+; i-DNA-PKcs). Genomic DNA was digested with HindIII and probed with the Eb probe. The hybridizing band generated by an Eb:ZFN DSB is indicated, as is the hybridizing band generated by the Tcrb locus, where a DSB was either not generated or was generated and repaired (*). (C) Schematic of the Cel-I assay. NHEJ-mediated repair of Eb-ZFN DSBs can lead to nucleotide loss (boxes). PCR amplification across the Eb-ZFN site followed by denaturation and reannealing will lead to mismatches that can be cleaved by Cel-I. The level of Cel-I cleavage is an underestimate of Eb:ZFN cleavage, as it detects only Eb:ZFN DSBs that have nucleotide deletions upon repair. (D) Cel-I assay of DNA samples from panel B. Full-length and Cel-I-digested products are shown. Note that there were no Cel-I products in the LigIV−/− cells or the DNA-PKcs3A/3A abl pre-B cells treated with NU7026 (+; i-DNA-PKcs), as the Eb:ZFN DSBs generated in these cells were not repaired (see panel B).
DNA-PKcs3A catalyzes aberrant DNA end joining during V(D)J recombination.
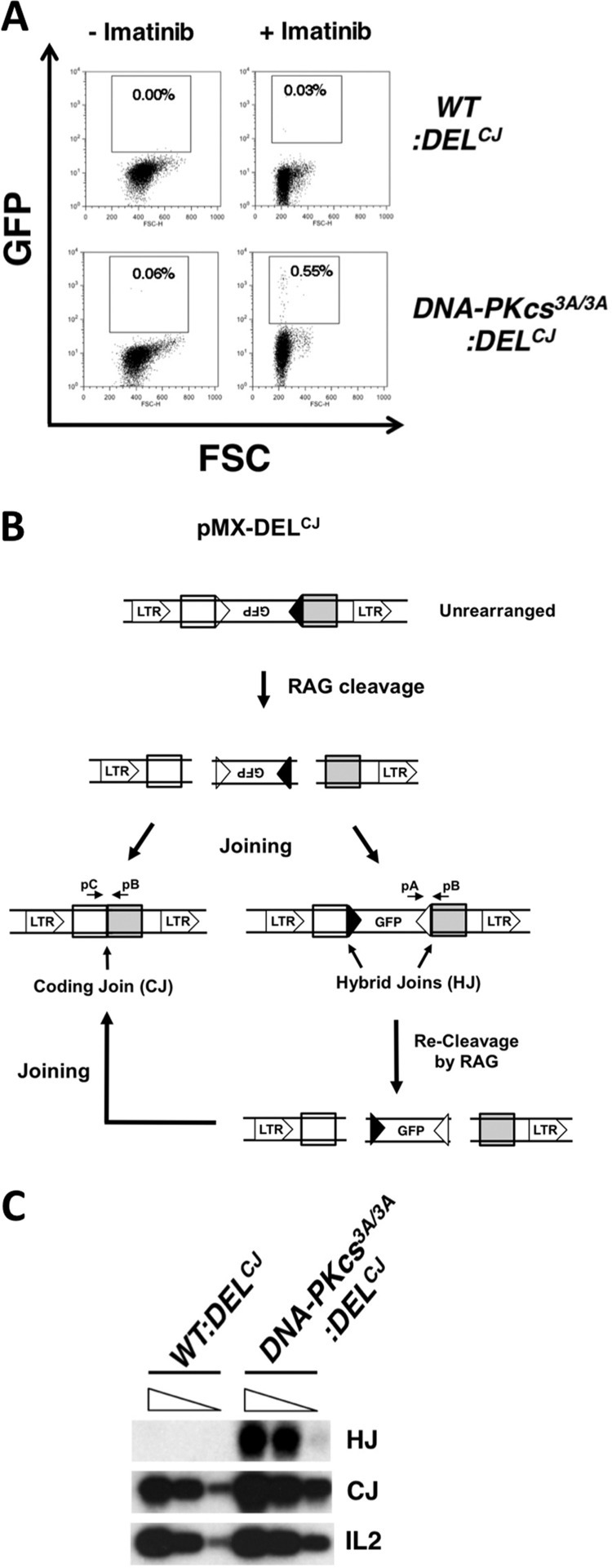
DNA-PKcs3A/3A:DELCJ abl pre-B cells exhibited a low but highly reproducible increase in the fraction of GFP-expressing cells after RAG induction compared to WT:DELCJ abl pre-B cells (Fig. 7A). The pMX-DELCJ GFP cDNA is inverted and expressed only if both coding ends are aberrantly joined to the signal end at the opposite RAG DSB, forming two hybrid joins (Fig. 7B). Indeed, PCR analysis revealed that pMX-DELCJ hybrid joins formed in DNA-PKcs3A/3A:DELCJ but not WT:DELCJ abl pre-B cells (Fig. 7C). pMX-DELCJ hybrid joins can be recleaved by RAG, leading to pMX-DELCJ coding join formation and an underestimate of hybrid joining in DNA-PKcs3A/3A abl pre-B cells (Fig. 7B). To determine the frequency of hybrid join formation, we generated DNA-PKcs3A/3A:INV abl pre-B cells. The aberrant joining of a pMX-INV signal end at one RAG DSB to the coding end at the other forms a single chromosomal hybrid join that cannot be recleaved by RAG (Fig. 2B). Thus, any pMX-INV hybrid joins formed will be “trapped,” allowing for the accurate quantification of hybrid join formation. Southern blot analysis of DNA-PKcs3A/3A:INV abl pre-B cells after RAG induction revealed robust pMX-INV hybrid join formation, comprising about 30% of total joins (coding and hybrid) (Fig. 8A to C). In contrast, pMX-INV hybrid joins were not observed by either PCR or Southern blotting after RAG induction in WT:INV abl pre-B cells (Fig. 8A to C). Hybrid join formation was also observed during Vk-to-Jk rearrangement at the endogenous Igk locus in DNA-PKcs3A/3A but not WT abl pre-B cells (Fig. 8D) and splenic B cells (Fig. 8E). Hybrid joins were not found in DNA-PKcs3A/3A:LigIV−/−:INV abl pre-B cells, demonstrating that they are formed by NHEJ, not RAG-mediated transposition (Fig. 8F). Thus, although RAG DSBs are rapidly repaired in DNA-PKcs3A/3A abl pre-B cells, coding ends are frequently aberrantly joined to signal ends, forming nonfunctional hybrid joins.
Fig 7.
Hybrid join formation during deletional rearrangements in DNA-PKcs3A/3A abl pre-B cells. (A) Flow cytometric analysis of WT:DELCJ and DNA-PKcs3A/3A:DELCJ abl pre-B cells either untreated or treated with imatinib for 4 days. GFP expression (y axis) and forward scatter (FSC; x axis) are shown. (B) Schematic of pMX-DELCJ as described in the legend to Fig. 1A. Shown is the resolution of pMX-DELCJ RAG DSBs as a coding join or two hybrid joins that lead to expression of GFP. Also shown is how pMX-DELCJ with two hybrid joins can be recleaved by RAG and repaired to form a coding join. (C) PCR analysis of pMX-DELCJ hybrid join (HJ) and coding join (CJ) formation, using oligonucleotide primers pA plus pB and pC plus pB, shown in panel B. PCR was carried out on 5-fold dilutions of genomic DNA from cells treated with imatinib for 4 days. IL-2 gene PCR is shown as a control.
Fig 8.
Hybrid join formation during inversional rearrangements in DNA-PKcs3A/3A abl pre-B cells. (A) Southern blot analysis of pMX-INV in imatinib-treated WT:INV and DNA-PKcs3A/3A:INV abl pre-B cells, carried out as described in the legend to Fig. 1B on genomic DNA digested with EcoRV and NcoI and probed with the C4b probe. Bands generated by unrearranged (UR) pMX-INV or a coding end (CE) overlap in size and are indicated. Also indicated are bands from a pMX-INV coding join (CJ) and hybrid join (HJ). (B) Quantification of hybrid join formation as a percentage of total (coding plus hybrid) joining in three independently derived WT:INV and DNA-PKcs3A/3A:INV abl pre-B cell lines treated for 4 days with imatinib. (C) PCR analysis of pMX-INV hybrid join formation in genomic DNA from cells treated with imatinib for 4 days, using oligonucleotide primers pC and pB. (D and E) PCR analysis of Igk locus Vk6-23-to-Jk1 hybrid join (HJ) and coding join (CJ) formation in genomic DNAs (5-fold dilutions) from WT and DNA-PKcs3A/3A abl pre-B cells treated with imatinib for 4 days (D) or genomic DNAs isolated from WT and DNA-PKcs3A/3A splenic B cells (E). All PCR analyses (C to E) were carried out on 5-fold dilutions of genomic DNA, and IL-2 gene PCR is shown as a DNA quantity control. (F) Southern blot analysis of pMX-INV rearrangement in DNA-PKcs3A/3A:INV and DNA-PKcs3A/3A:LigIV−/−:INV abl pre-B cells as described for panel B.
DISCUSSION
Prior studies have established that ATM and DNA-PKcs have unique functions during coding end joining (3, 28). ATM promotes the stability of coding ends in postcleavage complexes, and DNA-PKcs activates Artemis to open hairpin-sealed coding ends. Here we show that ATM and DNA-PKcs have additional functions during coding join formation that are regulated by the kinase activity of either of these proteins, suggesting that ATM and DNA-PKcs phosphorylate common targets required for coding end joining. Inhibition of DNA-PKcs kinase activity has no demonstrable effect on coding end joining in cells where ATM is present and active. In contrast, inhibition of DNA-PKcs3A kinase activity leads to a severe block in coding end joining when ATM is present and active. Thus, the TQ motifs that are mutated in DNA-PKcs3A must be intact for ATM kinase activity to compensate for the loss of DNA-PKcs kinase activity during coding end joining.
What are the common targets of ATM and DNA-PKcs that are important for coding join formation? It is possible that ATM is prevented from phosphorylating proteins other than DNA-PKcs in DNA-PKcs3A/3A abl pre-B cells. However, the three TQ motifs mutated in DNA-PKcs3A are ATM targets, suggesting that ATM can regulate coding end joining through the direct phosphorylation of DNA-PKcs (43, 44). The ATM-mediated phosphorylation of these TQ motifs may have regulatory effects similar to those of the autophosphorylation of other DNA-PKcs SQ/TQ motifs. In this regard, there are three other SQ/TQ motifs in the ABCDE cluster of DNA-PKcs. Thus, ATM-mediated phosphorylation of the three TQ motifs mutated in DNA-PKcs3A and autophosphorylation of other SQ/TQ motifs in the ABCDE cluster may provide similar thresholds of ABCDE cluster phosphorylation needed to regulate specific DNA-PKcs functions in DSB repair (18, 35). It is also possible that autophosphorylation of other SQ/TQ motifs in DNA-PKcs has effects on DNA-PKcs function similar to those of phosphorylation of the TQ motifs mutated in DNA-PKcs3A.
The loss of DNA-PKcs in vivo leads to a block in coding end joining even when ATM is present. Indeed, DNA-PKcs, but not ATM, activates Artemis endonuclease activity in vitro (25, 26). However, whether ATM can activate Artemis in vitro when DNA-PKcs is present but its kinase function is inactivated was not determined. In this regard, we found that inhibition of DNA-PKcs kinase activity in abl pre-B cells has no effect on coding join formation when ATM is present and active. Thus, opening of hairpin-sealed coding ends by Artemis in vivo relies on the presence of the DNA-PKcs protein and either ATM or DNA-PKcs kinase activity, likely due to the ability of ATM to phosphorylate DNA-PKcs and/or Artemis. Notably, this would have to be at sites other than the three TQ motifs mutated in DNA-PKcs3A, as unrepaired coding ends in DNA-PKcs3A/3A abl pre-B cells treated with a DNA-PKcs kinase inhibitor are open.
In addition to the DNA-PKcs mutation in SCID mice, naturally occurring equine and canine DNA-PKcs mutations have been described, as have DNA-PKcs mutations in cell lines selected for DSB repair defects (47–56). These different DNA-PKcs mutations have various effects on V(D)J recombination, with most affecting coding join formation and some affecting both signal and coding join formation. Some of the DNA-PKcs mutations lead to diminished kinase activity, but this is usually coupled with reduced DNA-PKcs protein expression; thus, the defects in V(D)J recombination in cells expressing these mutants could be due to alterations in DNA-PKcs kinase activity, protein level, or both. DNA-PKcs point mutants that have disrupted kinase activity but are expressed at normal levels have been generated (57, 58). Cells expressing these mutants are defective in the ability to repair RAG DSBs generated on extrachromosomal plasmid V(D)J recombination substrates (57, 58). This is in contrast to our findings, which suggest that if ATM is present and active, it will compensate for mutations that inactivate DNA-PKcs kinase activity. It is possible that DNA-PKcs inhibitors permit a low level of kinase activity that, along with ATM activity, can promote normal coding end joining. It has been suggested that V(D)J recombination on extrachromosomal plasmid substrates requires minimal DNA-PKcs kinase activity (57). However, it is also possible that the mutations that inactivate DNA-PKcs kinase activity have additional effects on DNA-PKcs functions required for RAG DSB repair (57, 58). In this regard, some of these mutants exhibit defects in coding and signal join formation, whereas others are defective only in coding join formation (57, 58). Finally, it is possible that RAG DSBs generated on extrachromosomal plasmid substrates cannot activate ATM and thus rely on DNA-PKcs kinase activity for joining. Indeed, no differences were observed in comparing V(D)J recombination of extrachromosomal plasmid substrates in wild-type and ATM-deficient fibroblasts, whereas ATM deficiency clearly leads to defects in the repair of chromosomal RAG DSBs in lymphoid cells (3, 37, 59).
The coding ends that accumulate in DNA-PKcs3A/3A abl pre-B cells treated with DNA-PKcs kinase inhibitors have open hairpins, indicating that DNA-PKcs has functions during coding end joining in addition to promoting Artemis activity. Indeed, the kinase activity of DNA-PKcs3A is also important for joining blunt signal ends and chromosomal DSBs with 4-bp overhangs generated by a zinc finger nuclease fusion protein. Thus, the defect in RAG DSB repair in DNA-PKcs3A/3A abl pre-B cells reflects a function of DNA-PKcs in general DSB repair. After recruitment by Ku70/80, DNA-PKcs binds at the apex of the broken DNA ends and must dissociate from these DNA ends before they can be joined (18, 35). Mutations in DNA-PKcs that promote its retention at broken DNA ends, such as mutations of several of the SQ/TQ motifs of the ABCDE cluster, inhibit DSB repair (60). Thus, it is possible that ATM-mediated phosphorylation of the TQ motifs mutated in DNA-PKcs3A or the autophosphorylation of other ABCDE cluster SQ/TQ motifs functions similarly to promote the dissociation of DNA-PKcs from broken DNA ends.
In addition to their role in promoting the overlapping activities of ATM and DNA-PKcs during coding end joining, our findings suggest that phosphorylation of the TQ motifs mutated in DNA-PKcs3A regulates unique DNA-PKcs activities during DSB repair. Although RAG DSBs are efficiently repaired in DNA-PKcs3A/3A abl pre-B cells, they are frequently misrepaired, with coding ends ligated to signal ends, forming hybrid joins. These hybrid joins are formed by NHEJ, as evidenced by their dependence on DNA ligase IV. Hybrid join formation is also observed in cells that express truncated RAG proteins or are deficient in ATM or components of the Mre11-Rad50-Nbs1 (MRN) complex, which is required to recruit ATM to DSBs (12, 37, 61, 62). Increased hybrid join formation in these settings is observed only during inversional rearrangements, possibly due to instability of the postcleavage complex. In contrast, hybrid join formation in DNA-PKcs3A/3A abl pre-B cells is also observed during deletional rearrangements. In this case, two chromosomal hybrid joins must be generated in order for these cells to express GFP, making it unlikely that the mechanistic basis of hybrid join formation is instability of the postcleavage complex.
DNA-PKcs could function to appropriately pair DNA ends in the RAG postcleavage complex, with DNA-PKcs3A being defective in this function. DNA-PKcs could promote the activity of RAG or other NHEJ proteins to appropriately pair and join RAG DSBs in the postcleavage complex. However, DNA-PKcs could also have direct functions in the pairing of DNA ends generated by RAG cleavage. In this regard, DNA-PKcs can form dimers upon binding to DNA in vitro (63, 64). Notably, DNA-PKcs dimer structure is influenced by the structure of the bound DNA end (64). Thus, if DNA-PKcs forms dimers when bound to DNA in vivo, these dimers may serve to appropriately pair hairpin-sealed coding ends and blunt signal ends for joining in the RAG postcleavage complex.
Supplementary Material
ACKNOWLEDGMENTS
We thank Tom Ellenberger and Gaya Amarasinghe for helpful discussions and reviews of the manuscript.
This work was supported by National Institutes of Health grants CA136470 (B.P.S.), AI074953 (B.P.S.), AI47829 (B.P.S.), CA92584 (S.L.-M. and M.H.), and GM105404 (M.H.). M.H. was also supported by the Lawrence Berkeley National Lab IDAT program, and S.L.-M. was supported by CIHR grant 691369.
Footnotes
Published ahead of print 8 July 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.00308-13.
REFERENCES
- 1.Tonegawa S. 1983. Somatic generation of antibody diversity. Nature 302:575–581 [DOI] [PubMed] [Google Scholar]
- 2.Schatz DG, Swanson PC. 2011. V(D)J recombination: mechanisms of initiation. Annu. Rev. Genet. 45:167–202 [DOI] [PubMed] [Google Scholar]
- 3.Helmink BA, Sleckman BP. 2012. The response to and repair of RAG-mediated DNA double-strand breaks. Annu. Rev. Immunol. 30:175–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lieber MR. 2010. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu. Rev. Biochem. 79:181–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rooney S, Chaudhuri J, Alt FW. 2004. The role of the non-homologous end-joining pathway in lymphocyte development. Immunol. Rev. 200:115–131 [DOI] [PubMed] [Google Scholar]
- 6.Lewis SM, Hesse JE, Mizuuchi K, Gellert M. 1988. Novel strand exchanges in V(D)J recombination. Cell 55:1099–1107 [DOI] [PubMed] [Google Scholar]
- 7.Morzycka-Wroblewska E, Lee FE, Desiderio SV. 1988. Unusual immunoglobulin gene rearrangement leads to replacement of recombinational signal sequences. Science 242:261–263 [DOI] [PubMed] [Google Scholar]
- 8.Agrawal A, Eastman QM, Schatz DG. 1998. Transposition mediated by RAG1 and RAG2 and its implications for the evolution of the immune system. Nature 394:744–751 [DOI] [PubMed] [Google Scholar]
- 9.Hiom K, Gellert M. 1998. Assembly of a 12/23 paired signal complex: a critical control point in V(D)J recombination. Mol. Cell 1:1011–1019 [DOI] [PubMed] [Google Scholar]
- 10.Melek M, Gellert M, van Gent DC. 1998. Rejoining of DNA by the RAG1 and RAG2 proteins. Science 280:301–303 [DOI] [PubMed] [Google Scholar]
- 11.Roth DB, Craig NL. 1998. VDJ recombination: a transposase goes to work. Cell 94:411–414 [DOI] [PubMed] [Google Scholar]
- 12.Sekiguchi JA, Whitlow S, Alt FW. 2001. Increased accumulation of hybrid V(D)J joins in cells expressing truncated versus full-length RAGs. Mol. Cell 8:1383–1390 [DOI] [PubMed] [Google Scholar]
- 13.Lieber MR, Ma Y, Pannicke U, Schwarz K. 2004. The mechanism of vertebrate nonhomologous DNA end joining and its role in V(D)J recombination. DNA Repair (Amst.) 3:817–826 [DOI] [PubMed] [Google Scholar]
- 14.Li G, Alt FW, Cheng HL, Brush JW, Goff PH, Murphy MM, Franco S, Zhang Y, Zha S. 2008. Lymphocyte-specific compensation for XLF/cernunnos end-joining functions in V(D)J recombination. Mol. Cell 31:631–640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu X, Jiang W, Dubois RL, Yamamoto K, Wolner Z, Zha S. 2012. Overlapping functions between XLF repair protein and 53BP1 DNA damage response factor in end joining and lymphocyte development. Proc. Natl. Acad. Sci. U. S. A. 109:3903–3908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oksenych V, Alt FW, Kumar V, Schwer B, Wesemann DR, Hansen E, Patel H, Su A, Guo C. 2012. Functional redundancy between repair factor XLF and damage response mediator 53BP1 in V(D)J recombination and DNA repair. Proc. Natl. Acad. Sci. U. S. A. 109:2455–2460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zha S, Guo C, Boboila C, Oksenych V, Cheng HL, Zhang Y, Wesemann DR, Yuen G, Patel H, Goff PH, Dubois RL, Alt FW. 2011. ATM damage response and XLF repair factor are functionally redundant in joining DNA breaks. Nature 469:250–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meek K, Dang V, Lees-Miller SP. 2008. DNA-PK: the means to justify the ends? Adv. Immunol. 99:33–58 [DOI] [PubMed] [Google Scholar]
- 19.Shiloh Y. 2003. ATM and related protein kinases: safeguarding genome integrity. Nat. Rev. Cancer 3:155–168 [DOI] [PubMed] [Google Scholar]
- 20.Smith GC, Jackson SP. 1999. The DNA-dependent protein kinase. Genes Dev. 13:916–934 [DOI] [PubMed] [Google Scholar]
- 21.Falck J, Coates J, Jackson SP. 2005. Conserved modes of recruitment of ATM, ATR and DNA-PKcs to sites of DNA damage. Nature 434:605–611 [DOI] [PubMed] [Google Scholar]
- 22.Walker JR, Corpina RA, Goldberg J. 2001. Structure of the Ku heterodimer bound to DNA and its implications for double-strand break repair. Nature 412:607–614 [DOI] [PubMed] [Google Scholar]
- 23.Yoo S, Dynan WS. 1999. Geometry of a complex formed by double strand break repair proteins at a single DNA end: recruitment of DNA-PKcs induces inward translocation of Ku protein. Nucleic Acids Res. 27:4679–4686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yoo S, Kimzey A, Dynan WS. 1999. Photocross-linking of an oriented DNA repair complex. Ku bound at a single DNA end. J. Biol. Chem. 274:20034–20039 [DOI] [PubMed] [Google Scholar]
- 25.Goodarzi AA, Yu Y, Riballo E, Douglas P, Walker SA, Ye R, Harer C, Marchetti C, Morrice N, Jeggo PA, Lees-Miller SP. 2006. DNA-PK autophosphorylation facilitates Artemis endonuclease activity. EMBO J. 25:3880–3889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ma Y, Pannicke U, Schwarz K, Lieber MR. 2002. Hairpin opening and overhang processing by an Artemis/DNA-dependent protein kinase complex in nonhomologous end joining and V(D)J recombination. Cell 108:781–794 [DOI] [PubMed] [Google Scholar]
- 27.Gapud EJ, Dorsett Y, Yin B, Callen E, Bredemeyer A, Mahowald GK, Omi KQ, Walker LM, Bednarski JJ, McKinnon PJ, Bassing CH, Nussenzweig A, Sleckman BP. 2011. Ataxia telangiectasia mutated (Atm) and DNA-PKcs kinases have overlapping activities during chromosomal signal joint formation. Proc. Natl. Acad. Sci. U. S. A. 108:2022–2027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gapud EJ, Sleckman BP. 2011. Unique and redundant functions of ATM and DNA-PKcs during V(D)J recombination. Cell Cycle 10:1928–1935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zha S, Jiang W, Fujiwara Y, Patel H, Goff PH, Brush JW, Dubois RL, Alt FW. 2011. Ataxia telangiectasia-mutated protein and DNA-dependent protein kinase have complementary V(D)J recombination functions. Proc. Natl. Acad. Sci. U. S. A. 108:2028–2033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Callen E, Jankovic M, Wong N, Zha S, Chen HT, Difilippantonio S, Di Virgilio M, Heidkamp G, Alt FW, Nussenzweig A, Nussenzweig M. 2009. Essential role for DNA-PKcs in DNA double-strand break repair and apoptosis in ATM-deficient lymphocytes. Mol. Cell 34:285–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tomimatsu N, Mukherjee B, Burma S. 2009. Distinct roles of ATR and DNA-PKcs in triggering DNA damage responses in ATM-deficient cells. EMBO Rep. 10:629–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gurley KE, Kemp CJ. 2001. Synthetic lethality between mutation in Atm and DNA-PK(cs) during murine embryogenesis. Curr. Biol. 11:191–194 [DOI] [PubMed] [Google Scholar]
- 33.Sekiguchi J, Ferguson DO, Chen HT, Yang EM, Earle J, Frank K, Whitlow S, Gu Y, Xu Y, Nussenzweig A, Alt FW. 2001. Genetic interactions between ATM and the nonhomologous end-joining factors in genomic stability and development. Proc. Natl. Acad. Sci. U. S. A. 98:3243–3248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bakkenist CJ, Kastan MB. 2003. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature 421:499–506 [DOI] [PubMed] [Google Scholar]
- 35.Weterings E, Chen DJ. 2007. DNA-dependent protein kinase in nonhomologous end joining: a lock with multiple keys? J. Cell Biol. 179:183–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dobbs TA, Tainer JA, Lees-Miller SP. 2010. A structural model for regulation of NHEJ by DNA-PKcs autophosphorylation. DNA Repair (Amst.) 9:1307–1314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bredemeyer AL, Sharma GG, Huang CY, Helmink BA, Walker LM, Khor KC, Nuskey B, Sullivan KE, Pandita TK, Bassing CH, Sleckman BP. 2006. ATM stabilizes DNA double-strand-break complexes during V(D)J recombination. Nature 442:466–470 [DOI] [PubMed] [Google Scholar]
- 38.Shull ER, Lee Y, Nakane H, Stracker TH, Zhao J, Russell HR, Petrini JH, McKinnon PJ. 2009. Differential DNA damage signaling accounts for distinct neural apoptotic responses in ATLD and NBS. Genes Dev. 23:171–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zha S, Sekiguchi J, Brush JW, Bassing CH, Alt FW. 2008. Complementary functions of ATM and H2AX in development and suppression of genomic instability. Proc. Natl. Acad. Sci. U. S. A. 105:9302–9306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Helmink BA, Tubbs AT, Dorsett Y, Bednarski JJ, Walker LM, Feng Z, Sharma GG, McKinnon PJ, Zhang J, Bassing CH, Sleckman BP. 2011. H2AX prevents CtIP-mediated DNA end resection and aberrant repair in G1-phase lymphocytes. Nature 469:245–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Muljo SA, Schlissel MS. 2003. A small molecule Abl kinase inhibitor induces differentiation of Abelson virus-transformed pre-B cell lines. Nat. Immunol. 4:31–37 [DOI] [PubMed] [Google Scholar]
- 42.Daniel JA, Pellegrini M, Lee BS, Guo Z, Filsuf D, Belkina NV, You Z, Paull TT, Sleckman BP, Feigenbaum L, Nussenzweig A. 2012. Loss of ATM kinase activity leads to embryonic lethality in mice. J. Cell Biol. 198:295–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen BP, Uematsu N, Kobayashi J, Lerenthal Y, Krempler A, Yajima H, Lobrich M, Shiloh Y, Chen DJ. 2007. Ataxia telangiectasia mutated (ATM) is essential for DNA-PKcs phosphorylations at the Thr-2609 cluster upon DNA double strand break. J. Biol. Chem. 282:6582–6587 [DOI] [PubMed] [Google Scholar]
- 44.Meek K, Douglas P, Cui X, Ding Q, Lees-Miller SP. 2007. trans-Autophosphorylation at DNA-dependent protein kinase's two major autophosphorylation site clusters facilitates end processing but not end joining. Mol. Cell. Biol. 27:3881–3890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang S, Yajima H, Huynh H, Zheng J, Callen E, Chen HT, Wong N, Bunting S, Lin YF, Li M, Lee KJ, Story M, Gapud E, Sleckman BP, Nussenzweig A, Zhang CC, Chen DJ, Chen BP. 2011. Congenital bone marrow failure in DNA-PKcs mutant mice associated with deficiencies in DNA repair. J. Cell Biol. 193:295–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ding Q, Reddy YV, Wang W, Woods T, Douglas P, Ramsden DA, Lees-Miller SP, Meek K. 2003. Autophosphorylation of the catalytic subunit of the DNA-dependent protein kinase is required for efficient end processing during DNA double-strand break repair. Mol. Cell. Biol. 23:5836–5848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Araki R, Fujimori A, Hamatani K, Mita K, Saito T, Mori M, Fukumura R, Morimyo M, Muto M, Itoh M, Tatsumi K, Abe M. 1997. Nonsense mutation at Tyr-4046 in the DNA-dependent protein kinase catalytic subunit of severe combined immune deficiency mice. Proc. Natl. Acad. Sci. U. S. A. 94:2438–2443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Blunt T, Gell D, Fox M, Taccioli GE, Lehmann AR, Jackson SP, Jeggo PA. 1996. Identification of a nonsense mutation in the carboxyl-terminal region of DNA-dependent protein kinase catalytic subunit in the scid mouse. Proc. Natl. Acad. Sci. U. S. A. 93:10285–10290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Danska JS, Holland DP, Mariathasan S, Williams KM, Guidos CJ. 1996. Biochemical and genetic defects in the DNA-dependent protein kinase in murine scid lymphocytes. Mol. Cell. Biol. 16:5507–5517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ding Q, Bramble L, Yuzbasiyan-Gurkan V, Bell T, Meek K. 2002. DNA-PKcs mutations in dogs and horses: allele frequency and association with neoplasia. Gene 283:263–269 [DOI] [PubMed] [Google Scholar]
- 51.Fukumura R, Araki R, Fujimori A, Mori M, Saito T, Watanabe F, Sarashi M, Itsukaichi H, Eguchi-Kasai K, Sato K, Tatsumi K, Abe M. 1998. Murine cell line SX9 bearing a mutation in the DNA-PKcs gene exhibits aberrant V(D)J recombination not only in the coding joint but also in the signal joint. J. Biol. Chem. 273:13058–13064 [DOI] [PubMed] [Google Scholar]
- 52.Fukumura R, Araki R, Fujimori A, Tsutsumi Y, Kurimasa A, Li GC, Chen DJ, Tatsumi K, Abe M. 2000. Signal joint formation is also impaired in DNA-dependent protein kinase catalytic subunit knockout cells. J. Immunol. 165:3883–3889 [DOI] [PubMed] [Google Scholar]
- 53.Peterson SR, Kurimasa A, Oshimura M, Dynan WS, Bradbury EM, Chen DJ. 1995. Loss of the catalytic subunit of the DNA-dependent protein kinase in DNA double-strand-break-repair mutant mammalian cells. Proc. Natl. Acad. Sci. U. S. A. 92:3171–3174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Peterson SR, Stackhouse M, Waltman MJ, Chen F, Sato K, Chen DJ. 1997. Characterization of two DNA double-stranded break repair-deficient cell lines that express inactive DNA-dependent protein kinase catalytic subunits. J. Biol. Chem. 272:10227–10231 [DOI] [PubMed] [Google Scholar]
- 55.Shin EK, Perryman LE, Meek K. 1997. A kinase-negative mutation of DNA-PK(CS) in equine SCID results in defective coding and signal joint formation. J. Immunol. 158:3565–3569 [PubMed] [Google Scholar]
- 56.Wiler R, Leber R, Moore BB, VanDyk LF, Perryman LE, Meek K. 1995. Equine severe combined immunodeficiency: a defect in V(D)J recombination and DNA-dependent protein kinase activity. Proc. Natl. Acad. Sci. U. S. A. 92:11485–11489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kienker LJ, Shin EK, Meek K. 2000. Both V(D)J recombination and radioresistance require DNA-PK kinase activity, though minimal levels suffice for V(D)J recombination. Nucleic Acids Res. 28:2752–2761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kurimasa A, Kumano S, Boubnov NV, Story MD, Tung CS, Peterson SR, Chen DJ. 1999. Requirement for the kinase activity of human DNA-dependent protein kinase catalytic subunit in DNA strand break rejoining. Mol. Cell. Biol. 19:3877–3884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hsieh CL, Arlett CF, Lieber MR. 1993. V(D)J recombination in ataxia telangiectasia, Bloom's syndrome, and a DNA ligase I-associated immunodeficiency disorder. J. Biol. Chem. 268:20105–20109 [PubMed] [Google Scholar]
- 60.Uematsu N, Weterings E, Yano K, Morotomi-Yano K, Jakob B, Taucher-Scholz G, Mari PO, van Gent DC, Chen BP, Chen DJ. 2007. Autophosphorylation of DNA-PKCS regulates its dynamics at DNA double-strand breaks. J. Cell Biol. 177:219–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Deriano L, Chaumeil J, Coussens M, Multani A, Chou Y, Alekseyenko AV, Chang S, Skok JA, Roth DB. 2011. The RAG2 C terminus suppresses genomic instability and lymphomagenesis. Nature 471:119–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Helmink BA, Bredemeyer AL, Lee BS, Huang CY, Sharma GG, Walker LM, Bednarski JJ, Lee WL, Pandita TK, Bassing CH, Sleckman BP. 2009. MRN complex function in the repair of chromosomal Rag-mediated DNA double-strand breaks. J. Exp. Med. 206:669–679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.DeFazio LG, Stansel RM, Griffith JD, Chu G. 2002. Synapsis of DNA ends by DNA-dependent protein kinase. EMBO J. 21:3192–3200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hammel M, Yu Y, Mahaney BL, Cai B, Ye R, Phipps BM, Rambo RP, Hura GL, Pelikan M, So S, Abolfath RM, Chen DJ, Lees-Miller SP, Tainer JA. 2010. Ku and DNA-dependent protein kinase dynamic conformations and assembly regulate DNA binding and the initial non-homologous end joining complex. J. Biol. Chem. 285:1414–1423 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.