Abstract
In spite of the important regulatory functions of antisense transcripts in gene expression, it remains unknown how antisense transcription is initiated. Recent studies implicated RNA polymerase II in initiation of antisense transcription. However, how RNA polymerase II is targeted to initiate antisense transcription has not been elucidated. Here, we have analyzed the association of RNA polymerase II with the antisense initiation site at the 3′ end of the GAL10 coding sequence in dextrose-containing growth medium that induces antisense transcription. We find that RNA polymerase II is targeted to the antisense initiation site at GAL10 by Reb1p activator as well as general transcription factors (e.g., TFIID, TFIIB, and Mediator) for antisense transcription initiation. Intriguingly, while GAL10 antisense transcription is dependent on TFIID, its sense transcription does not require TFIID. Further, the Gal4p activator that promotes GAL10 sense transcription is dispensable for antisense transcription. Moreover, the proteasome that facilitates GAL10 sense transcription does not control its antisense transcription. Taken together, our results reveal that GAL10 sense and antisense transcriptions are regulated differently and shed much light on the mechanisms of antisense transcription initiation.
INTRODUCTION
Eukaryotic transcription is an extensively regulated, complex process (1–7). It is controlled by the targeted association of specific and general transcription factors (GTFs), where typically an activator binds to a gene promoter and initiates a specific protein regulatory network leading to the formation of a preinitiation complex (PIC) for transcription initiation. Transcription is also highly influenced by the epigenetic landscape of the gene being transcribed, which includes cytosine DNA methylation, histone modifications, and RNA-mediated gene regulation (3, 8–10). In fact, regulatory RNAs have a complex interplay with DNA methylation and histone modifications for controlling gene expression (8, 11–23). Regulatory RNAs consist of noncoding RNAs (ncRNAs), which include microRNAs (miRNAs), small nuclear RNAs (snRNAs), small interfering RNAs (siRNAs), Piwi-interacting RNAs (piRNAs), and natural antisense transcripts (NATs) (24).
The ncRNAs have been implicated extensively in X-chromosome inactivation, gene imprinting, dosage compensation, and heterochromatin formation in the heterochromatic regions of the genome (8, 17–23, 25, 26). However, we are now beginning to understand the functions of ncRNAs in the euchromatic genome (8, 17, 24). In Saccharomyces cerevisiae, for example, IME4 and SER3 genes are regulated by a cis mechanism of transcriptional interference (27, 28, 29). The IME4 gene is required for entry into meiosis, and its expression is blocked in haploids by IME4 antisense transcripts. Analogous to IME4, the transcription of a 3-phosphoglycerate dehydrogenase-encoding gene, SER3, is repressed by ncRNA via blocking activator binding to the SER3 promoter (28, 29). On the other hand, a repressive mechanism distinct from transcriptional interference has been demonstrated for the PHO84 gene, where stabilization of antisense transcripts represses gene expression (30). In this model, aging cells lose a component of the exosome, Rrp6p, which leads to stable PHO84 antisense transcript (30). The PHO84 antisense transcript facilitates the recruitment of the deacetylase, Hda1p, which represses the PHO84 sense transcription (30). Recently, a similar mode of regulation has been shown for the GAL1-GAL10 gene cluster, where the antisense transcript favors a repressive deacetylated chromatin state, leading to transcriptional inhibition of sense transcription (31, 32). Thus, antisense transcripts or ncRNAs play important roles in gene regulation.
Transcriptome analysis of various eukaryotes has revealed the existence of a much larger pool of ncRNAs than previously thought, further corroborating their significance in the regulatory processes (33). Seven to 30% of all genes in plants and animals are associated with antisense transcripts, and up to 72% of transcripts in human and mouse have antisense partners (8, 17, 34, 35). Antisense transcripts have crucial regulatory roles in gene expression and have been associated with controlling cell fate, developmental processes, stress adaptation, response to viral infections, and oncogenesis (8, 11–23, 27, 36–47). Despite the abundance and physiological and pathological significance of antisense transcripts, how the synthesis of antisense RNA itself is regulated has not been clearly elucidated.
So far, there has not been any systematic study to decipher the mechanism of antisense transcription initiation. Recent studies (31, 32) in yeast have demonstrated that antisense transcription is originated from the 3′ end of the GAL10 coding sequence in dextrose-containing growth medium (which is repressive to sense transcription). Further, it has been demonstrated that the initiation of antisense transcription from GAL10 is dependent on the activator Reb1p or Reb1p-binding site which is present at the 3′ end of the GAL10 coding sequence (31, 32). However, it remains unknown how Reb1p initiates antisense transcription. Further, it has not been analyzed whether TATA-binding protein (TBP) and other GTFs are involved in antisense transcription initiation. Moreover, previous studies (31, 32) have implicated RNA polymerase II (Pol II) in antisense transcription based on capping and polyadenylation of antisense RNA, but how RNA polymerase II is targeted to the antisense initiation site for antisense transcription has not yet been elucidated. Furthermore, the direct role of RNA polymerase II in antisense transcription has not been demonstrated. Here, using chromatin immunoprecipitation (ChIP) (48–53) and transcriptional analyses in various mutants, we have studied RNA polymerase II associated with antisense transcription initiation at the 3′ end of the GAL10 coding sequence in dextrose-containing growth medium. We find that RNA polymerase II associated with sense transcription from the 5′ end of the GAL10 gene falls off the coding sequence when the carbon source in growth medium is switched from galactose (that induces sense transcription) to dextrose, and, subsequently, RNA polymerase II associates with the 3′ end of the GAL10 coding sequence for antisense transcription. Such association of RNA polymerase II is dependent on the activator Reb1p-binding site at the 3′ end of the GAL10 coding sequence, hence implicating the role of an activator in targeting RNA polymerase II for antisense transcription. Further, we find that the association of RNA polymerase II with the 3′ end of the GAL10 coding sequence in dextrose-containing growth medium requires TBP and TBP-associated factors (TAFs), thus supporting the role of TFIID (a complex of TBP and TAFs) in targeting antisense RNA polymerase II. Likewise, Mediator as well as TFIIB is also required for the association of RNA polymerase II with the 3′ end of the GAL10 coding sequence in dextrose-containing growth medium. Collectively, our results demonstrate for the first time the roles of TBP, TAFs, TFIIB, and Mediator as well as the activator-binding site in facilitation of the recruitment of RNA polymerase II to the antisense initiation site at the 3′ end of the GAL10 coding sequence (and hence antisense transcription). Intriguingly, we find that GAL10 antisense transcription is dependent on TFIID, while its sense transcription does not require TFIID. Further, GAL10 antisense transcription does not depend on the proteasome complex that is required for GAL sense transcription. Moreover, Gal4p activator, which is essential for sense transcription of GAL genes, is dispensable for GAL10 antisense transcription. These results support the idea that GAL10 sense and antisense transcriptions are independent of each other and are regulated differently. Collectively, these results provide significant insights on the mechanisms of antisense transcription initiation in vivo.
MATERIALS AND METHODS
Plasmids.
The plasmid pFA6a-13Myc-KanMX6 (54) was used for genomic tagging of the largest subunit (Rpb1p) of RNA polymerase II. The plasmid PRS406 was used for PCR-based disruption of PDR5 and GAL1-GAL10 promoter. PRS416 was used as a reporter plasmid to test the transcriptional activity of the RPS5 promoter or GAL10 3′ end. The GAL10 3′ end and RPS5 promoter were inserted into the pRS416 plasmid using BamHI and XbaI sites.
Strains.
A yeast strain (Saccharomyces cerevisiae) harboring temperature-sensitive (TS) mutations in TAF11p, TAF13p, and TFIIB and isogenic wild-type equivalents were obtained from the Green laboratory (Michael R. Green, University of Massachusetts Medical School) (55, 56). A yeast strain harboring TS mutations in TBP and its isogenic wild-type equivalent were obtained from the Struhl laboratory (Kevin Struhl, Harvard Medical School) (57). Wild-type and TS mutant strains of Srb4p were obtained from the Young laboratory (Richard A. Young, Whitehead Institute for Biomedical Research) (58). Rpb1p wild-type and TS mutant strains were also obtained from the Young laboratory. The yeast strain carrying mutations in the Reb1p-binding site at the 3′ end of the GAL10 coding sequence and its isogenic wild-type equivalent were obtained from the Tollervey laboratory (David Tollervey, University of Edinburg, United Kingdom) (31). The yeast strain bearing a TS mutation in Rpt4p (rpt4-ts or sug2-13) (Sc677) and its isogenic wild-type equivalent (Sc599) were obtained from the Kodadek and Johnston laboratories (59). Multiple Myc epitope tags were added to the original chromosomal locus of RBP1 in strain W303a to generate the ZDY4 strain, using the pFA6a-13Myc-KanMX6 plasmid. The endogenous GAL4 gene of NSY1 (49) was disrupted using a PCR-based gene disruption method to generate NSY9 (49). The PDR5 gene was deleted from the wild-type strain by the PCR-based gene disruption method to generate SLY16a (52). The GAL1-GAL10 promoter was deleted from the ZDY4 strain to generate GDY90.
Growth media.
Yeast cells were grown in yeast extract-peptone plus 2% dextrose (YPD) to an optical density at 600 nm (OD600) of 1.0 at 30°C prior to formaldehyde-based in vivo cross-linking for analysis of antisense RNA polymerase II at the GAL10 locus. For kinetic analysis of RNA polymerase II association, yeast cells were grown in yeast extract-peptone plus 2% galactose (YPG) to an OD600 of 0.8 at 30°C and then transferred to YPD medium for different time periods prior to cross-linking. For experiments in the wild-type and TS mutant strains, yeast cells were grown in YPD medium at 23°C to an OD600 of 0.85 and then transferred to 37°C for 1 h before cross-linking or harvesting for RNA analysis. For MG132-based experiments, a yeast strain that has a null mutation of PDR5 was initially grown in synthetic complete medium (yeast nitrogen base and complete amino acid mixture plus 2% dextrose) at 30°C to an OD600 of 0.7 and then treated with MG132 (75 μM) for 2 h.
ChIP assay.
For analysis of the recruitment of antisense RNA polymerase II, the ChIP protocol was modified as described previously (48–53). Such modification has been done in order to detect low-abundant RNA polymerase II associated with antisense transcription along with proper controls such as nonspecific antibody and nonspecific DNA. Basically, this modified assay is almost the same as our previous ChIP protocol (55, 56, 60–62) except for the amount of whole-cell extract used in the immunoprecipitation step and the volume in which immunoprecipitated DNA was dissolved. In this modified ChIP protocol, we have used four times more whole-cell extract in the immunoprecipitation step, and immunoprecipitated DNA was dissolved in half the volume used in our previous ChIP protocol. By doing this, we have enriched immunoprecipitated DNA, and thus the association of low-abundant antisense RNA polymerase II can be detected, if present. Briefly, a total of 800 μl of lysate was prepared from 100 ml of yeast culture. Following sonication, 400 μl of lysate was used for each immunoprecipitation using 10 μl of anti-Myc antibody (2 μg; Santa Cruz Biotechnology, Inc.) and 100 μl of protein A/G plus agarose beads. Ten microliters of antihemagglutinin (anti-HA) (2 μg; Santa Cruz Biotechnology, Inc.) was also used as a nonspecific antibody control. Similarly, we carried out the ChIP assay using 2 μl of 8WG16 (Covance) antibody (which targets to the carboxy-terminal domain of the largest subunit of RNA polymerase II) in 200 μl of lysate. Immunoprecipitated DNA sample was dissolved in 10 μl of Tris-EDTA (TE) buffer, pH 8.0, of which 1 μl was used for the PCR analysis. In parallel, the PCR analysis for input DNA was performed using 1 μl of DNA that was prepared by dissolving purified DNA from 5 μl of lysate in 100 μl TE buffer, pH 8.0. The PCR analysis was carried out within a linear range using 23 PCR cycles, as done previously (62, 63). The primer pairs used for PCR analysis were as follows: region A, 5′-CTATGTTCAGTTAGTTTGGCTAGC-3′ and 5′-TTGATGCTCTGCATAATAATGCCC-3′; region B, 5′-TTAATGCGAATCATAGTAGTATCG-3′ and 5′-TTACCAATAGATCACCTGGAAATTC-3′; region C, 5′-CGCTTAACTGCTCATTGCTATATTG-3′ and 5′-TTGTTCGGAGCAGTGCGGCGC-3′; Pol I, 5′-GAGTCCTTGTGGCTCTTGGC-3′ and 5′-AATACTGATGCCCCCGACC-3′.
For the location of the A, B, and C regions, see the schematic in Fig. 1A. The primer pair represented by Pol I is located at the 18S ribosomal DNA (rDNA) region (64). This primer pair has been used previously as a nonspecific DNA control in the ChIP analysis of RNA polymerase II genes. Autoradiograms were scanned and quantitated by the National Institutes of Health ImageJ (version 1.62) program. Immunoprecipitated DNAs were quantitated as the ratio of immunoprecipitate to input in the autoradiogram. The ChIP experiments were carried out three or more times. These experiments are biologically independent. The average ChIP signal of the biologically independent experiments is reported with standard deviation (SD) (Microsoft Office Excel 2003).
Fig 1.
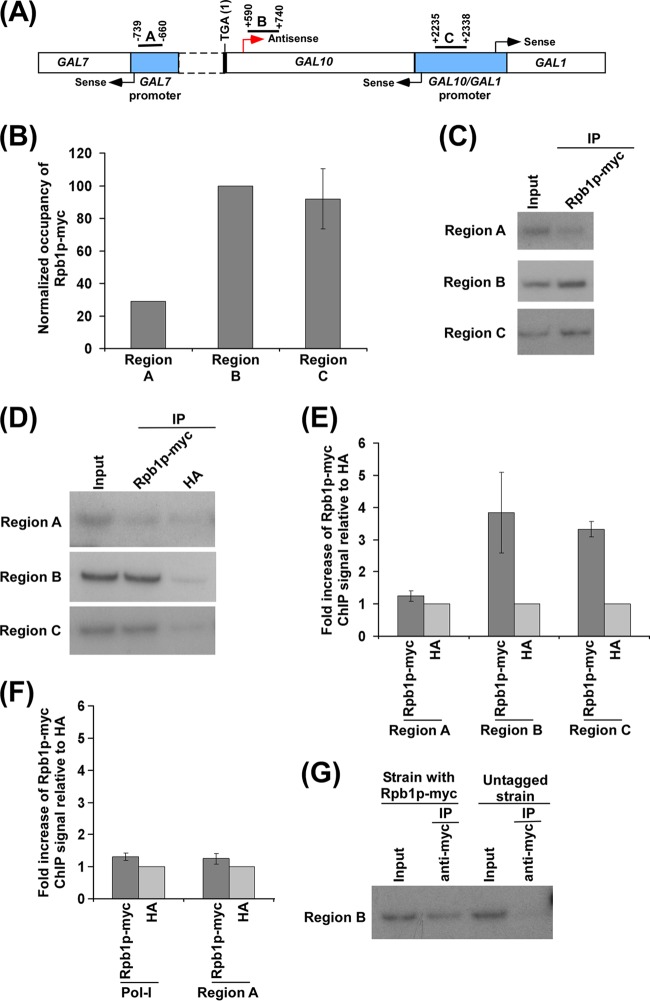
Analysis of RNA polymerase II association with the antisense start site at the 3′ end of the GAL10 coding sequence. (A) A schematic diagram showing the locations of the primer pairs (regions A, B, and C) at the GAL1, GAL10, and GAL7 loci for the ChIP analysis. Regions A, B, and C are located at the GAL7 core promoter, the 3′ end of the GAL10 coding sequence, and the upstream activating sequence (UAS) at the GAL10-GAL1 bidirectional promoter, respectively. The numbers are presented with respect to the position of the translational stop codon of GAL10. (B) RNA polymerase II is associated with the 3′ end of the GAL10 coding sequence (region B) in dextrose-containing growth medium. A yeast strain expressing the Myc epitope-tagged largest subunit (Rpb1p) of RNA polymerase II was grown in YPD medium to an OD600 of 1.0 at 30°C prior to formaldehyde-based in vivo cross-linking. The ChIP assay was performed as described in Materials and Methods. Immunoprecipitation was carried out using an anti-Myc antibody (9E10; Santa Cruz Biotechnology, Inc.) against Myc-tagged Rpb1p. Immunoprecipitated DNA was analyzed by PCR using the primer pairs encompassing regions A, B, and C as described for panel A. The ratio of the immunoprecipitate over the input in the autoradiogram (termed a ChIP signal) was measured. The maximum ChIP signal was set to 100, and other ChIP signals were normalized with respect to 100 (represented as normalized or relative occupancy). Normalized occupancy was plotted in the form of a histogram. (C) Autoradiograms for the ChIP data presented in panel B. IP, immunoprecipitation. (D) Analysis of RNA polymerase II at the 3′ end of the GAL10 coding sequence (region B), GAL7 core promoter (region A), and GAL1-GAL10 UAS (region C) along with anti-HA as a nonspecific antibody in dextrose-containing growth medium. A yeast strain expressing Myc-tagged Rpb1p was grown in YPG medium to an OD600 of 0.8 at 30°C and then transferred to YPD medium for 3 h prior to cross-linking. (E) The ratio of the ChIP signal of Myc-tagged Rpb1p to anti-HA (i.e., fold increase of Rpb1p-Myc ChIP signal relative to HA) in panel D is plotted in the form of a histogram. A ratio of 1 indicates no association of RNA polymerase II. (F) Analysis of association of RNA polymerase II with the RNA polymerase I gene (Pol I) relative to anti-HA in dextrose-containing growth medium. The ratio of the ChIP signal of Myc-tagged Rpb1p to anti-HA was plotted in the form of a histogram. (G) The ChIP analysis of RNA polymerase II association at the 3′ end of the GAL10 coding sequence (region B) in the yeast strain expressing Rpb1p with or without a Myc epitope tag. Yeast strains were grown as described for panel D. Immunoprecipitation was carried out using an anti-Myc antibody.
Total RNA preparation.
Total RNA was prepared from yeast cell culture as described by Peterson et al. (65). Briefly, 10 ml of yeast culture was harvested and then suspended in 100 μl of RNA preparation buffer (500 mM NaCl, 200 mM Tris-HCl, 100 mM Na2-EDTA, and 1% SDS) along with 100 μl of phenol-chloroform-isoamyl alcohol and a 100-μl volume-equivalent of glass beads (acid washed) (Sigma). Subsequently, the yeast cell suspension was vortexed at maximum speed (10 in a VWR Mini Vortex Mixer) (catalog number 58816-121; VWR) five times (30 s each). The cell suspension was put on ice for 30 s between pulses. After vortexing, 150 μl of RNA preparation buffer and 150 μl of phenol-chloroform-isoamyl alcohol were added to the yeast cell suspension, followed by vortexing for 15 s at maximum speed on a VWR Mini-Vortex mixer. The aqueous phase was collected following a 5-min centrifugation at maximum speed in a microcentrifuge machine. The total RNA was isolated from the aqueous phase by ethanol precipitation.
RT-PCR analysis of GAL10 antisense transcripts.
Reverse transcription-PCR (RT-PCR) analysis of GAL10 antisense transcript was performed according to standard protocols (66). Briefly, total RNA was prepared from yeast culture as described above. Equal amounts (15 to 30 μg) of total RNA was used in the reverse transcription assay for both wild-type and mutant strains. RNA was treated with RNase-free DNase (M610A; Promega) and then reverse transcribed into cDNA using primer P1 (see Fig. 2A) or P2 (see Fig. 4A) targeted to the GAL10 antisense transcript (P1, 5′-CTACGAGATTCCCAAATATGATTCC-3′; P2, 5′-GCTAAGATAATGGGGCTCTTTACAT-3′) as described in the protocol supplied by Promega (A3800; Promega). Reverse transcription was carried out at 42°C using avian myeloblastosis virus (AMV) reverse transcriptase (RTase). PCR was performed using synthesized first-strand cDNA (or the extended P1 or P2 primer shown in Fig. 2A and 4A) as the template and the primer pairs targeted to the GAL10 core promoter and coding sequence (represented as regions L and M, respectively, in Fig. 2A and 4A) and GAL7 coding sequence (marked as region N in Fig. 2A and 4A). Primer pairs targeted to the GAL1, ADH1, and ACT1 coding sequences were also used to amplify the above cDNAs generated by the P1 or P2 primer. RT-PCR products were separated by 2.2% agarose gel electrophoresis and visualized by ethidium bromide staining. The RT-PCR analysis was carried out in biological triplicates. The average value of these biologically independent experiments is reported with standard deviation (SD) (Microsoft Office Excel 2003). The primer pairs used in the PCR analysis of cDNAs were as follows: GAL7 (N), 5′-AAAGTGCAATCTGTGAGAGGCAATT-3′ and 5′-TTTTCTCTTGCTTCTCTGGAGAGAT-3′; GAL10 (M), 5′-CTACGAGATTCCCAAATATGATTCC-3′ and 5′-TAACGCAAGATAGCAAACTTCCAAC-3′; GAL10 (L), 5′-GCTAAGATAATGGGGCTCTTTACAT-3′ and 5′-TTTCACTTTGTAACTGAGCTGTCAT-3′; GAL1, 5′-CAGTGGATTGTCTTCTTCGGCCGC-3′ and 5′-GGCAGCCTGATCCATACCGCCATT-3′; ACT1, 5′-TCCACCACTGCTGAAAGAGAAATTG-3′ and 5′-AATAGTGATGACTTGACCATCTGGA-3′; ADH1, 5′-CGGTAACAGAGCTGACACCAGAGA-3′ and 5′-ACGTATCTACCAACGATTTGACCC-3′; RPS5, 5′-AGGCTCAATGTCCAATCATTGAAAG-3′ and 5′-CAACAACTTGGATTGGGTTTTGGTC-3′.
Fig 2.
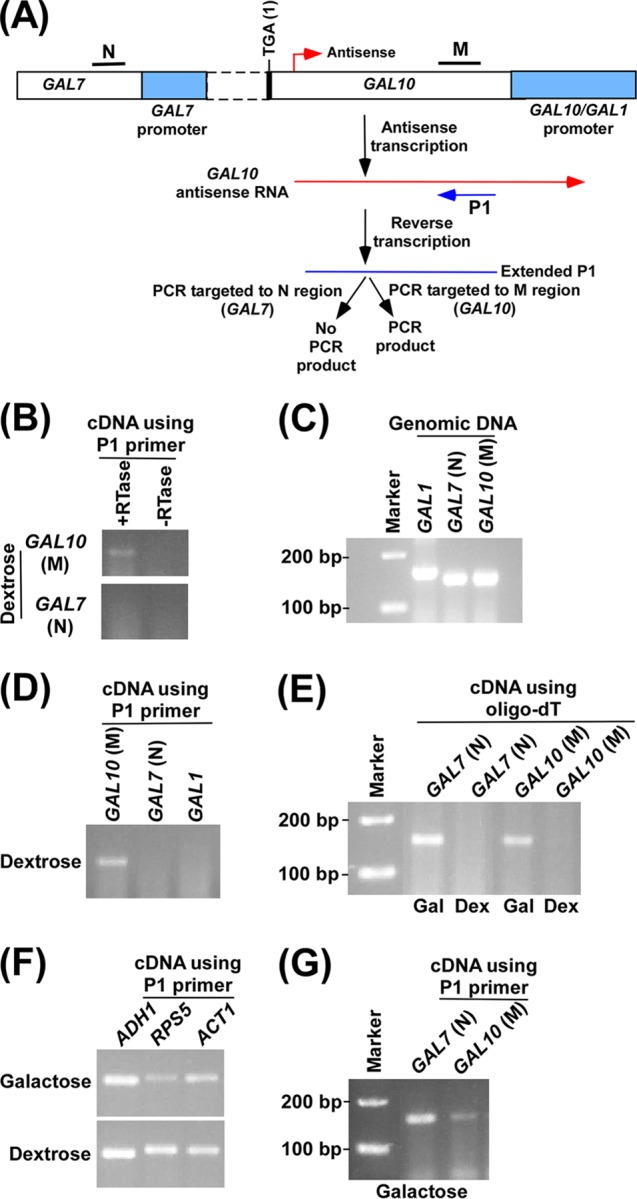
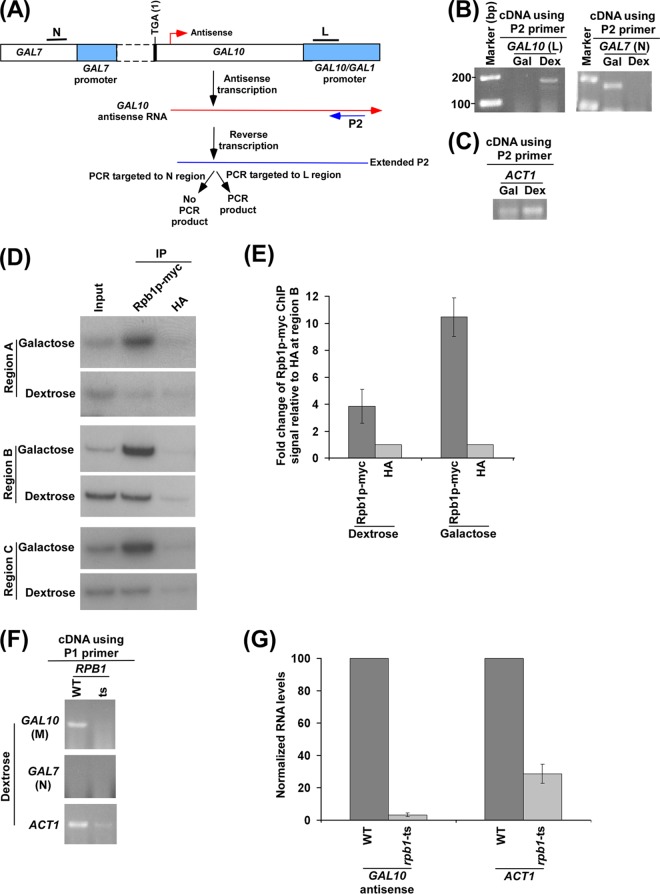
Analysis of GAL10 antisense transcript. (A) Schematic diagram showing the experimental strategy for the analysis of GAL10 antisense transcript. The P1 primer targeted toward the 5′ end of the GAL10 antisense transcript was extended by AMV reverse transcriptase-based reverse transcription at 42°C, and subsequently the extended primer was amplified by primer pairs targeted to the coding regions, M and N, of GAL10 and GAL7, respectively. (B) GAL10 antisense transcription in dextrose-containing growth medium. A yeast strain expressing Myc-tagged Rpb1p was grown in YPG medium to an OD600 of 0.8 at 30°C and then switched to YPD medium for 3 h. Total RNA was isolated and analyzed following the experimental strategy as described in panel A. The primer pair targeted to the GAL10 coding sequence (region M) generated PCR product from cDNA synthesized by the P1 primer. Reverse transcriptase was not used in cDNA synthesis in the −RTase lane. (C) Amplification of genomic DNA using PCR primer pairs targeted to the GAL7, GAL10, and GAL1 coding sequences. The same primer pairs were used as in panels A and B. (D) RT-PCR analysis in dextrose-containing growth medium as described in panel B. (E) RT-PCR analysis in dextrose-containing growth medium as described in panel B using oligo(dT) primer. A yeast strain expressing Myc-tagged Rpb1p was grown in YPG medium to an OD600 of 0.8 at 30°C and then transferred to YPG or YPD medium for 3 h. Gal, galactose; Dex, dextrose. (F) RT-PCR analysis as described in panel B in dextrose- and galactose-containing growth medium, using the P1 primer in cDNA synthesis. A yeast strain expressing Myc-tagged Rpb1p was grown as described in panel E. (G) RT-PCR analysis as described in panel B in galactose-containing growth medium.
Fig 4.
RNA polymerase II is essential for GAL10 antisense transcription. (A) Schematic diagram showing the experimental strategy for the analysis of GAL10 antisense transcript using the P2 primer. The P2 primer targeted toward the 5′ end of the GAL10 antisense transcript, but encompassing a region corresponding to the GAL10-GAL1 bidirectional promoter, was extended by reverse transcription at 42°C, and subsequently the extended primer was amplified by primer pairs targeted to the core promoter region of GAL10 (region L) and the coding sequence region of GAL7 (region N). (B) GAL10 antisense transcription in dextrose-containing growth medium. The primer pair targeted to the GAL10 core promoter region (region L) but not the GAL7 coding sequence (region N) generated PCR product from cDNA synthesized by the P2 primer in dextrose-containing growth medium. A yeast strain expressing Myc-tagged Rpb1p was grown as described in the legend of Fig. 2E. (C) Amplification of cDNAs shown in panel B using a PCR primer pair targeted to the ACT1 coding sequences. (D) Analysis of RNA polymerase II at the 3′ end of the GAL10 coding sequence (region B) (Fig. 1A), GAL7 core promoter (region A) (Fig. 1A), and GAL10-GAL1 UAS (region C) (Fig. 1A) along with anti-HA as a nonspecific antibody control in dextrose- and galactose-containing growth media. A yeast strain expressing Myc-tagged Rpb1p was grown as described in the legend of Fig. 2E prior to cross-linking. Immunoprecipitations were performed as described in the legend of Fig. 1B. (E) The ratio of the ChIP signal of Myc-tagged Rpb1p to that of anti-HA (i.e., fold increase of Rpb1p-myc ChIP signal relative to HA) at region B in panel D is plotted in the form of a histogram. A ratio of 1 indicates no association of RNA polymerase II. (F) RNA polymerase II is essential for GAL10 antisense transcription in dextrose-containing growth medium. Wild-type (WT) and TS mutant strains of Rpb1p were grown in YPD medium at 23°C up to an OD600 of 0.85 and then switched to 37°C for 1 h prior to harvesting. Total RNA was prepared from the wild-type and TS mutant strains of Rpb1p and then analyzed for GAL10 antisense RNA using the P1 primer in cDNA synthesis. (G) The transcription data shown in panel F were plotted in the form of a histogram. The RNA level in the wild-type strain was set to 100, and the level of RNA in the mutant strain was normalized with respect to 100.
RESULTS
Analysis of RNA polymerase II associated with antisense transcription initiation at the 3′ end of the GAL10 gene.
Although RNA polymerase II has been implicated in antisense transcription based on capping and polyadenylation of antisense transcript (31, 32), a thorough analysis of antisense RNA polymerase II and its regulation have not yet been elucidated. Here, we have taken the advantage of the GAL gene cluster in yeast (Saccharomyces cerevisiae) to analyze the association of RNA polymerase II and its regulation at the antisense initiation site. The GAL gene cluster consists of three genes, GAL1, GAL7, and GAL10, and is a highly regulated galactose-inducible genetic unit (Fig. 1A). While GAL1 and GAL10 are divergent genes sharing a common bidirectional promoter, GAL10 and GAL7 are tandem genes (Fig. 1A). This genetic organization has significant implications in transcriptional regulation of the GAL cluster through transcriptional interference (67). Previous studies (31, 32) have demonstrated the existence of 2.6-, 4-, and 6-kb-long noncoding antisense transcripts initiated from the 3′ end of the GAL10 coding sequence under conditions repressive to GAL gene sense transcription (i.e., dextrose-containing growth medium). Such antisense transcription from the 3′ end of the GAL10 coding sequence leads to GAL10 ncRNAs, which attenuate GAL1-GAL10 sense transcription in dextrose-containing growth medium. With this system, we asked if one can actually track the association of RNA polymerase II with GAL10 for antisense transcription in dextrose-containing growth medium. To this end, we grew a yeast strain expressing the largest subunit of RNA polymerase II (Rpb1p) bearing a Myc epitope tag in dextrose-containing growth medium and then performed formaldehyde-based in vivo cross-linking followed by a ChIP assay (Materials and Methods) to analyze the association of RNA polymerase II with the 3′ end of the GAL10 coding region (Fig. 1A, region B). As a control, the association of RNA polymerase II with the GAL7 core promoter (Fig. 1A, region A) was also analyzed. Since the antisense transcription of GAL10 has been shown to start from the 3′ end of the GAL10 coding sequence toward GAL1 but not GAL7 (Fig. 1A) (31, 32), the association of antisense RNA polymerase II would not be observed with the GAL7 core promoter. Our ChIP assay revealed a robust association of RNA polymerase II with the 3′ end of the GAL10 coding sequence in dextrose-containing growth medium (Fig. 1B and C, region B). However, such an association of RNA polymerase II was not observed at the GAL7 core promoter (Fig. 1B and C, region A). This is as expected since GAL7 is inactive in dextrose-containing growth medium. Further, sense RNA polymerase II does not associate with the GAL10 coding region in dextrose-containing growth medium because of Mig1p-mediated repression as well as the masking of the Gal4p activation domain by the repressor, Gal80p (60, 61, 68–71). Thus, the significantly high level of RNA polymerase II at the 3′ end of the GAL10 coding sequence in dextrose-containing growth medium supports the association of RNA polymerase II with the antisense transcription initiation site. Further, since antisense transcription is initiated from the 3′ end of the GAL10 coding sequence and continues through the GAL10-GAL1 bidirectional promoter, the association of antisense RNA polymerase II would also be observed at the GAL10-GAL1 bidirectional promoter in dextrose-containing growth medium. Indeed, we find a significantly high level of RNA polymerase II at the GAL10-GAL1 bidirectional promoter in dextrose-containing growth medium (Fig. 1B and C, region C), similar to the level of RNA polymerase II at the 3′ end of the GAL10 coding sequence (Fig. 1B and C, region B).
Next, we analyzed the association of RNA polymerase II at the 3′ end of the GAL10 coding sequence (Fig. 1A, region B) along with nonspecific anti-HA antibody in dextrose-containing growth medium in order to determine whether the level of RNA polymerase II signal at the 3′ end of the GAL10 coding sequence is just a background. We find that the level of RNA polymerase II at the GAL10 antisense initiation site is significantly higher than the background signal generated by nonspecific anti-HA antibody in dextrose-containing growth medium (Fig. 1D and E). Interestingly, the level of RNA polymerase II at the GAL7 core promoter (Fig. 1A, region A) in dextrose-containing growth medium is almost same as the background signal (Fig. 1D and E). Further, we demonstrate that there is no enrichment of RNA polymerase II signal at the RNA polymerase I (Pol I) gene with respect to anti-HA antibody signal in dextrose-containing growth medium (Fig. 1F). Furthermore, as a control, we demonstrate the absence of the ChIP signal (using an anti-myc antibody in the immunoprecipitation step) at the 3′ end of the GAL10 coding sequence in dextrose-containing growth medium in the yeast strain that does not have Myc-tagged Rpb1p (Fig. 1G). Thus, the RNA polymerase II signal observed at the antisense initiation site of GAL10 in dextrose-containing growth medium is not background but, rather, RNA polymerase II associated with antisense transcription.
Taken together, our results support the occupancy of RNA polymerase II associated with antisense transcription at GAL10 under the growth condition that is repressive to sense transcription. Consistently, we observed antisense transcription from the 3′ end of the GAL10 coding sequence in dextrose-containing growth medium (Fig. 2A and B). For analysis of GAL10 antisense transcript, we used a specific primer (P1) targeted toward the 5′ end of the GAL10 antisense transcript in synthesizing cDNA, which was subsequently amplified by a primer pair targeted to the GAL10 coding sequence (region M), as schematically shown in Fig. 2A. Using such an assay, we detected GAL10 antisense transcript in dextrose-containing growth medium (Fig. 2B). As a control, a primer pair targeted to the GAL7 coding sequence (region N) was also used in the above PCR analysis of cDNA generated by the P1 primer. Such analysis did not generate PCR signal (Fig. 2B) as the GAL7 primer pair was located at the left side (or upstream) of the GAL10 antisense initiation site (Fig. 2A). The absence of the PCR signal at GAL7 (Fig. 2B) is not due to the fact that the GAL7 primer pair did not work in the PCR assay as the same primer pair was successfully used in our previous studies (49, 53). Further, we show here that the primer pair targeted to the GAL7 coding sequence (region N) amplified yeast genomic DNA (Fig. 2C). The absence of the PCR signal using the primer pair targeted to the GAL7 coding sequence (region N) also supported the idea that there was no residual DNA contamination, similar to results in the absence of reverse transcriptase (−RTase), which served as the control. Thus, GAL7 served as a control for the absence of residual DNA contamination. Further, we supported the absence of residual DNA contamination by performing the above RT-PCR analysis in the absence of RTase. We found the absence of PCR signal when RTase was not used in the above RT-PCR analysis (Fig. 2B). Since the specific P1 primer was used in cDNA synthesis, the primer pair located at the right side (or downstream) of the P1 primer in the PCR analysis of GAL10 antisense cDNA would not generate PCR signal. Indeed, the PCR signal was not observed when the primer pair targeted to the GAL1 coding sequence was used in the PCR analysis (Fig. 2D). This primer pair (targeted to the GAL1 coding sequence) has been shown to work in our previous studies (49, 53) and has also been found here to amplify yeast genomic DNA (Fig. 2C). Thus, our RT-PCR analysis using a specific P1 primer targeted to the 5′ end of the GAL10 antisense RNA in the cDNA synthesis supported the presence of GAL10 antisense RNA in dextrose-containing growth medium (Fig. 2B), consistent with the association of RNA polymerase II (Fig. 1). However, the use of an oligo(dT) primer (Invitrogen) did not produce GAL10 antisense RNA in the above RT-PCR analysis in dextrose-containing growth medium (Fig. 2E, last lane). However, sense transcription of GAL10 and GAL7 was observed using the oligo(dT) primer in cDNA synthesis in galactose-containing growth medium as these GAL genes undergo sense transcription in galactose but not dextrose-containing growth medium (60, 61, 68–71). A long GAL10 antisense transcript may not be completely reverse transcribed by oligo(dT) or may not be efficiently polyadenylated, leading to the absence of PCR signal in the RT-PCR analysis using an oligo(dT) primer. Thus, a specific P1 primer targeted to the 5′ end of the GAL10 antisense RNA was used for cDNA synthesis in dextrose-containing growth medium to efficiently detect GAL10 antisense transcripts.
Intriguingly, the use of the specific P1 primer in cDNA synthesis also detected sense transcripts of ADH1, ACT1, and RPS5 in dextrose-containing growth medium (Fig. 2F). This is possibly due to hybridization of the P1 primer with the poly(A) tails of mRNAs via matched (A·T) and mismatched (A·G, A·C, and A·A) base pairs at 42°C during cDNA synthesis by AMV reverse transcriptase. In support of this possibility, previous studies have demonstrated and characterized A·G, A·C, and A·A mismatched base pairs (72–78). Further, the P1 primer may also randomly hybridize to RNAs (including mRNAs, rRNAs, and tRNAs) at 42°C during cDNA synthesis via matched (A·T and G·C) and previously demonstrated mismatched (G·T, G·A, A·C, T·C, G·G, A·A, T·T, and C·C) base pairs (72–84). Moreover, random primers target RNAs and, hence, have been used to analyze a broad spectrum of RNAs. Thus, the use of the P1 primer in cDNA synthesis in RT-PCR analysis is likely to detect RNA transcripts from RNA polymerase I, II, and III genes, similar to the random primers. Indeed, we find that the use of the P1 primer in cDNA synthesis detected GAL7 and GAL10 mRNAs in galactose-containing growth medium (Fig. 2G). Likewise, ADH1, RPS5, and ACT1 transcripts were also detected in galactose- and dextrose-containing growth media using the P1 primer in cDNA synthesis as these genes are known to be expressed in these growth media. Thus, similar to the oligo(dT) primer, the P1 primer can be used in cDNA synthesis to study the sense transcription of ACT1, ADH1, RPS5, and other genes (that are expressed in dextrose- and/or galactose-containing growth medium). Since sense transcription does not occur at the GAL gene cluster in dextrose-containing growth medium (60, 61, 68–71), the aforementioned RT-PCR analysis using a specific P1 primer would be useful in analyzing antisense transcripts from the GAL10 locus in dextrose-containing growth medium. Further, sense transcripts of ACT1 or ADH1 in dextrose-containing growth medium can be used as controls in the RT-PCR analysis of GAL10 antisense transcription. Indeed, these genes have been used as controls in the GAL10 antisense transcription analysis described below (Fig. 3D; see also the P1 primer results shown in Fig. 4 to 8).
Fig 3.
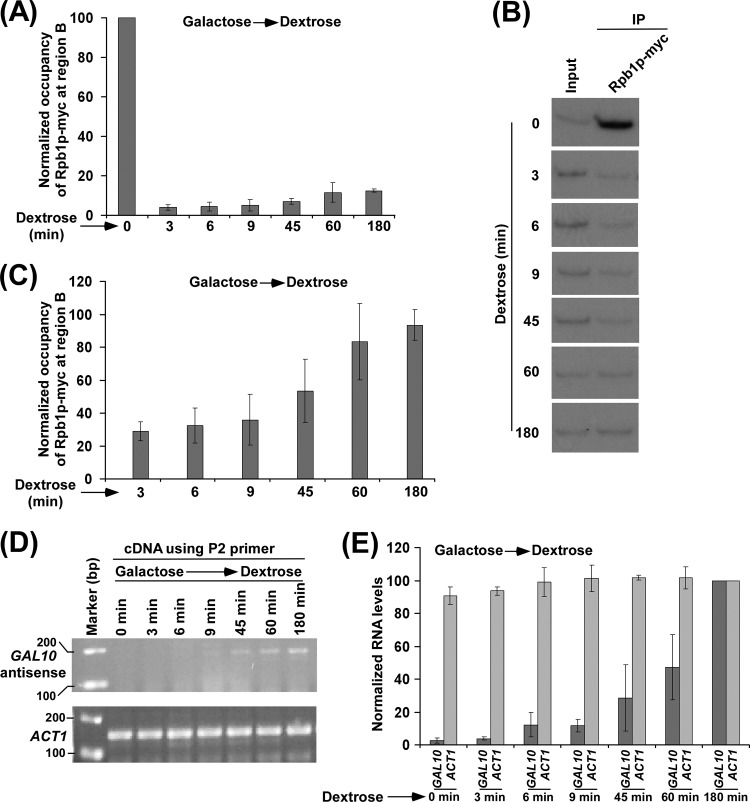
Kinetic analysis of RNA polymerase II association with the 3′ end of the GAL10 coding sequence. (A) Kinetic analysis of RNA polymerase II association with the 3′ end of the GAL10 coding sequence following the switch of the growth medium from YPG to YPD. A yeast strain expressing Myc-tagged Rpb1p was grown in YPG medium to an OD600 of 0.8 at 30°C and then transferred to YPD medium for different time periods prior to cross-linking. Immunoprecipitation was performed as described in the legend of Fig. 1B. The ChIP signal at 0 min was set to 100, and the ChIP signals at other time points were normalized with respect to 100. The normalized (or relative) occupancy of Rpb1p was plotted in the form of a histogram. (B) Autoradiograms for the ChIP data in panel A. (C) Association of RNA polymerase II with the 3′ end of the GAL10 coding sequence in YPD medium (from 3 to 180 min) described in panel A was presented separately. The maximum ChIP signal in YPD medium was set to 100, and other ChIP signals were normalized with respect to 100. In one experiment, the 60-min time point shows slightly higher ChIP signal than the 180-min time point and, thus, was set to 100. In other experiments, the 180-min time point shows slightly higher ChIP signal than the 60-min time point and was set to 100. The normalized (or relative) occupancy of Rpb1p was plotted in the form of a histogram. (D) RT-PCR analysis. Generation of GAL10 antisense RNA following the switch of the growth medium from YPG to YPD. A yeast strain was grown as described in the legend of panel A. GAL10 antisense RNA was analyzed using the P2 primer in cDNA synthesis and the primer pair targeted to the GAL10 core promoter in PCR amplification of cDNA (as schematically described in Fig. 4A). The levels of ACT1 transcripts were monitored as controls. (E) The results shown in panel D were plotted in the form of a histogram. Maximum signal of GAL10 antisense transcript was set to 100, and the levels of GAL10 antisense transcripts at other time points were normalized with respect to 100. Likewise, the maximum ACT1 transcript level was set to 100, and the levels of ACT1 transcripts at other time points were normalized with respect to 100.
Fig 8.
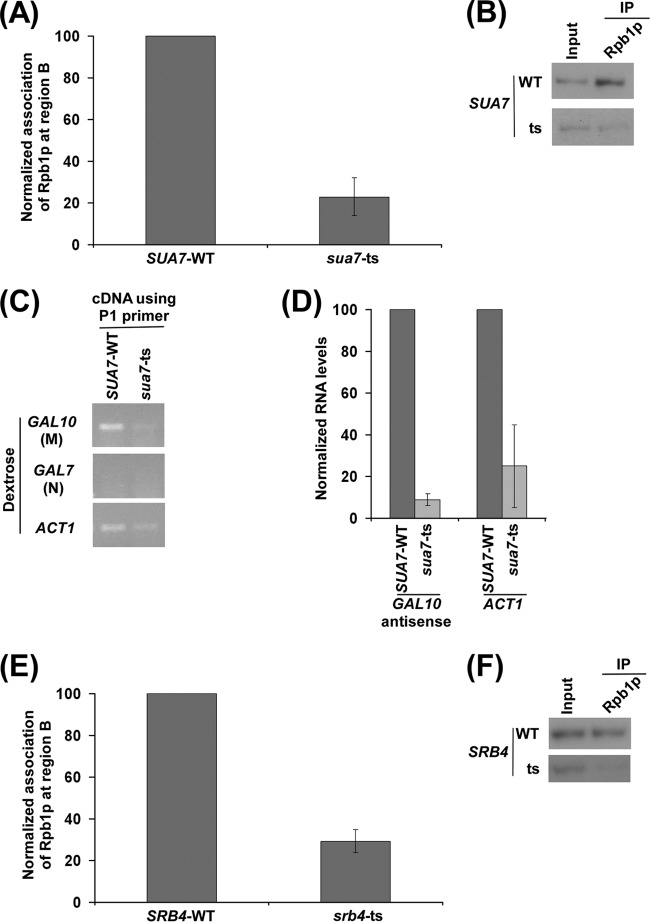
TFIIB and Mediator are required for recruitment of antisense RNA polymerase II to the 3′ end of the GAL10 coding sequence. (A) Analysis of recruitment of RNA polymerase II to the 3′ end of the GAL10 coding sequence in the wild-type and mutant strains of TFIIB (SUA7-WT and sua7-ts) in dextrose-containing growth medium. Both the wild-type and mutant strains were grown, cross-linked, and immunoprecipitated as described in the legend of Fig. 6A. (B) The autoradiograms for the ChIP data shown in panel A. (C) RT-PCR analysis of GAL10 antisense RNA in the SUA7 wild-type and TS mutant strains in dextrose-containing growth medium as described in the legend of Fig. 2B. Yeast cells were grown as described in the legend of Fig. 6A. (D) The transcription data shown in panel C were plotted in the form of a histogram. The PCR signal of GAL10 antisense transcript in wild-type strain was set to 100, and the level of GAL10 antisense transcript in the mutant strain was normalized with respect to 100. Likewise, the ACT1 transcript level was set to 100 in the wild-type strain, and the level of ACT1 transcript in the mutant strain was normalized with respect to 100. (E) Analysis of recruitment of RNA polymerase II to the 3′ end of the GAL10 coding sequence in the wild-type and mutant strains of Mediator (SRB4-WT and srb4-ts) in dextrose-containing growth medium. Both the wild-type and mutant strains were grown, cross-linked, and immunoprecipitated as described in the legend of Fig. 6A. (F) The autoradiograms for the ChIP data shown in panel E.
Collectively, we find that RNA polymerase II is associated with GAL10 for antisense transcription under the growth condition that is repressive to sense transcription. In order to better distinguish RNA polymerase II associated with antisense transcription from that of sense transcription at GAL10, we also performed a kinetic analysis of RNA polymerase II association with the 3′ end of the GAL10 coding sequence following sense transcription repression. To this end, we first grew the yeast strain expressing Myc epitope-tagged Rpb1p in galactose-containing growth medium (which induces GAL10 sense transcription) and then transferred the yeast cells to dextrose-containing growth medium (which represses GAL10 sense transcription and favors antisense transcription) for different time periods to kinetically analyze the association of RNA polymerase II with the 3′ end of the GAL10 coding sequence. We find a strong association of RNA polymerase II with the 3′ end of the GAL10 coding sequence at the 0-min time point (Fig. 3A and B). This is expected as RNA polymerase II was involved in sense transcription of the GAL10 gene in galactose-containing growth medium. Thus, a significantly high level of RNA polymerase II association with the 3′ end of the GAL10 coding sequence was observed at 0 min (Fig. 3A and B). However, when yeast cells were transferred to dextrose-containing growth medium, the association of RNA polymerase II involved in sense transcription dropped dramatically within 3 min (Fig. 3A and B), consistent with previous studies (51, 85). Interestingly, the association of RNA polymerase II with the 3′ end of the GAL10 coding sequence increased significantly at later time points in dextrose-containing growth medium (Fig. 3A, B, and C). Thus, our results demonstrate that RNA polymerase II involved in GAL10 sense transcription in galactose-containing growth medium dissociates from GAL10 in dextrose-containing growth medium within 3 min. Subsequently, RNA polymerase II associates with the 3′ end of the GAL10 coding sequence to initiate the antisense transcription. Consistently, we find increased synthesis of GAL10 antisense RNA upon switching the carbon source in the growth medium from galactose to dextrose (Fig. 3D and E). For kinetic analysis of GAL10 antisense transcription, we used the specific primer P2 in cDNA synthesis (Fig. 4A) since the use of the P1 primer in cDNA synthesis detected GAL10 sense transcript in galactose-containing growth medium following amplification of cDNA by a primer pair targeted to the GAL10 coding sequence (region M) (Fig. 2G). The P2 primer is targeted to the promoter region of GAL10-GAL1, as schematically shown in Fig. 4A, and the cDNA generated by the P2 primer was amplified by PCR using a primer pair targeted to the GAL10 core promoter (Fig. 4, region L). Such analysis would not detect GAL10 sense transcript in galactose-containing growth medium as sense transcript does not complement the nucleotide sequence of the promoter but, rather, that of the coding sequence. Indeed, we detected GAL10 antisense transcript in dextrose but not galactose-containing growth medium using the P2 primer in cDNA synthesis (Fig. 4B, left). As a control, we showed the absence of PCR signal for the GAL7 coding sequence (region N) in dextrose-containing growth medium using the same cDNA generated by the P2 primer (Fig. 4B, right). However, we observed GAL7 sense transcript in galactose-containing growth medium (Fig. 4B, right). Again, this is possibly due to hybridization of the P2 primer with poly(A) tails of the sense transcripts or RNAs via matched and mismatched base pairs at 42°C during cDNA synthesis by AMV reverse transcriptase, similar to the use of the P1 primer described above. Thus, like the use of the P1 primer, the use of the P2 primer in cDNA synthesis would detect sense transcripts. Indeed, the use of the P2 primer in cDNA synthesis detected ACT1 sense transcripts in both galactose- and dextrose-containing growth media (Fig. 4C). These results indicate that, using the P2 primer in cDNA synthesis, one can study the generation of GAL10 antisense RNA upon switching the carbon source in the growth medium from galactose to dextrose. In such an assay, PCR signal would be observed from the GAL10 locus not in galactose-containing growth medium but in dextrose-containing growth medium, hence supporting the synthesis of GAL10 antisense RNA. While the use of the P2 primer in cDNA synthesis does not detect GAL10 sense transcripts in galactose-containing growth medium, GAL10 sense transcripts can be detected using the P1 primer in cDNA synthesis in galactose-containing growth medium. Thus, the P2 primer was used in the cDNA synthesis in the kinetic analysis of GAL10 antisense transcription. Nonetheless, both the P1 and P2 primers can be used for GAL10 antisense transcription analysis in dextrose-containing growth medium. In fact, the P1 primer has been used in GAL10 antisense transcription analysis in dextrose-containing growth medium as described below (Fig. 4F; see also the P1 primer results shown in Fig. 5 to 7).
Fig 5.
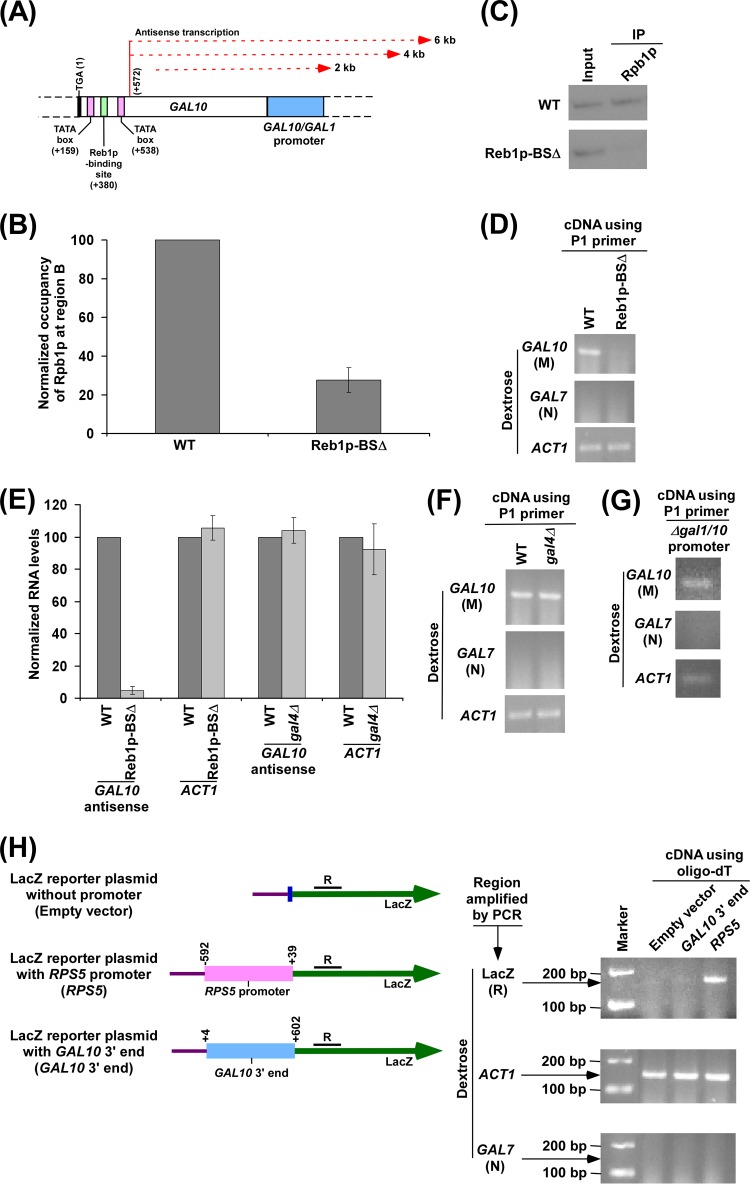
Reb1p-binding site is essential for recruitment of RNA polymerase II at the 3′ end of the GAL10 coding sequence in dextrose-containing growth medium for GAL10 antisense transcription. (A) Schematic diagram showing the Reb1p-binding site, TATA box, and antisense transcription initiation site at the 3′ end of the GAL10 coding sequence. The numbers are presented with respect to the position of the translational stop codon of GAL10. (B) Analysis of antisense RNA polymerase II recruitment to the 3′ end of the GAL10 coding sequence in the wild-type (WT) strain and mutant carrying mutations in the Reb1p-binding site (Reb1p-BSΔ). Both the wild-type and mutant strains were grown as described in the legend of Fig. 1B. Immunoprecipitation was performed using 8WG16 antibody (Covance, Inc.) against the carboxy-terminal domain of the largest subunit (Rpb1p) of RNA polymerase II. Immunoprecipitated DNA was analyzed by PCR using a primer pair targeted to the 3′ end of the GAL10 coding sequence (region B) (Fig. 1A). The ChIP signal of the wild-type strain was set to 100, and the ChIP signal of the mutant strain was normalized with respect to 100 (represented as normalized or relative occupancy). (C) The autoradiograms for the ChIP data presented in panel B. (D) Analysis of GAL10 antisense transcription in the wild-type and Reb1p-BSΔ strains in dextrose-containing growth medium using the P1 primer in cDNA synthesis. Both wild-type and mutant strains were grown as described in panel B. (E) The transcription data in panels D and F were plotted in the form of a histogram. The PCR signal of GAL10 antisense transcript in the wild-type strain was set to 100, and the level of GAL10 antisense transcript in the mutant strain was normalized with respect to 100. Likewise, the ACT1 transcript level was set to 100 in the wild-type strain, and the level of ACT1 transcript in the mutant strain was normalized with respect to 100. (F) Analysis of GAL10 antisense transcription in the wild-type and Δgal4 strains in dextrose-containing growth medium. Both wild-type and mutant strains were grown as described in panel B. (G) Analysis of GAL10 antisense transcription in dextrose-containing growth medium in the yeast strain that does not have GAL1-GAL10 promoter. A yeast strain without a GAL1-GAL10 promoter was grown as described in panel B. (H) The GAL10 3′ end alone cannot drive transcription from a reporter plasmid. Schematic diagrams (left) show the LacZ reporter plasmid (pRS416) with RPS5 promoter or GAL10 3′ end. The numbers above RPS5 promoter (sense) are presented with respect to the first nucleotide of the transcription start site of RPS5. The numbers above GAL10 3′ end are presented with respect to the last nucleotide of the translational stop codon of GAL10. The region R at the LacZ coding sequence was amplified using cDNA generated by oligo(dT). Analysis of LacZ transcript under the RPS5 promoter or GAL10 3′ end was performed (right) The above constructs were transformed into wild-type yeast cells and then grown in dextrose-containing medium to an OD600 of 1.0 at 30°C prior to harvesting for RNA analysis. The coding regions of ACT1 and GAL7 were amplified as loading and no-DNA-contamination controls, respectively, using cDNA generated by oligo(dT).
Fig 7.
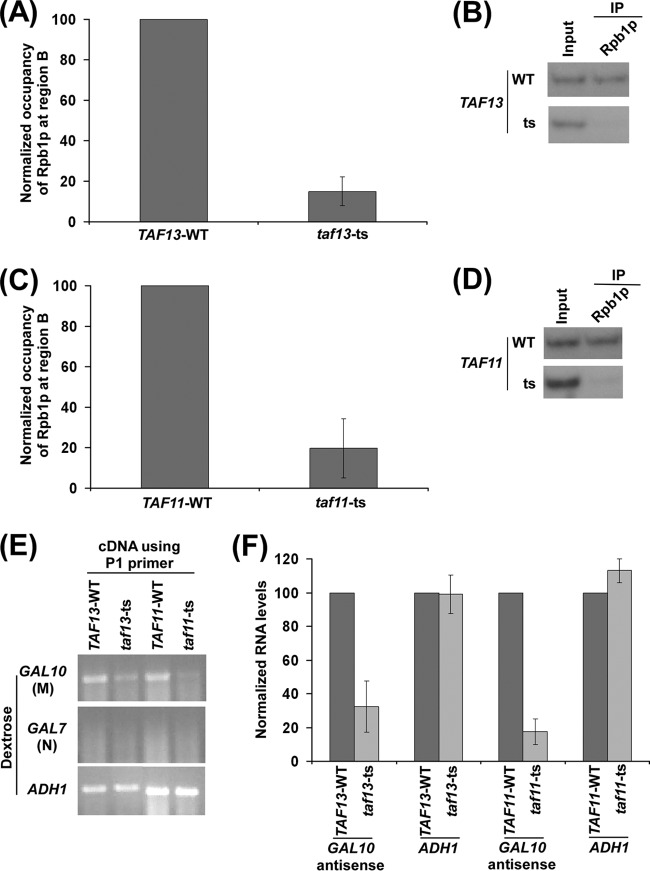
Analysis of recruitment of antisense RNA polymerase II to the 3′ end of the GAL10 coding sequence in the wild-type and mutant strains of TAF11p and TAF13p in dextrose-containing growth medium for antisense transcription. (A) TAF13p is required for recruitment of RNA polymerase II to the 3′ end of the GAL10 coding sequence in dextrose-containing growth medium. Both the wild-type and mutant strains were grown, cross-linked, and immunoprecipitated as described in the legend of Fig. 6A. (B) Autoradiograms for the ChIP data shown in panel A. (C) TAF11p is required for recruitment of RNA polymerase II to the 3′ end of the GAL10 coding sequence in dextrose-containing growth medium. Both the wild-type and mutant strains were grown, cross-linked, and immunoprecipitated as described in the legend of Fig. 6A. (D) Autoradiograms for the ChIP data shown in panel C. (E) RT-PCR analysis of GAL10 antisense transcription in the taf11-ts and taf13-ts mutant strains and their wild-type equivalents in dextrose-containing growth medium as described in the legend of Fig. 2B. Yeast cells were grown as described in the legend of Fig. 6A. (F) The data in panel E were plotted in the form of a histogram. The PCR signal of GAL10 antisense transcript in the wild-type strain was set to 100, and the level of GAL10 antisense transcript in the mutant strain was normalized with respect to 100. Likewise, the ADH1 transcript level was set to 100 in the wild-type strain, and the level of ADH1 transcript in the mutant strain was normalized with respect to 100.
As presented above, our data demonstrate the association of RNA polymerase II with the antisense transcription initiation site upon switching the carbon source in the growth medium from galactose to dextrose, and, consistently, antisense transcripts are synthesized. Although previous studies (31, 32) suggested a role of RNA polymerase II in antisense transcription based on capping and polyadenylation, there was no demonstration of the association of RNA polymerase II with the antisense transcription initiation site. Our results demonstrate the association of RNA polymerase II with the antisense transcription initiation site at GAL10 in dextrose-containing growth medium. Intriguingly, we find that the association of RNA polymerase II with the 3′ end of the GAL10 coding sequence is significantly less in dextrose-containing growth medium than in galactose-containing growth medium (Fig. 4D and E). This is consistent with the fact that antisense transcription is less abundant or frequent than sense transcription. Since RNA polymerase II is associated with the GAL10 antisense transcription initiation site in dextrose-containing growth medium, it would be required for GAL10 antisense transcription. To test this, we analyzed the level of GAL10 antisense transcription in the wild-type and TS mutant strains of the largest subunit of RNA polymerase II following a 1-h inactivation at the nonpermissive temperature in dextrose-containing growth medium. We found that GAL10 antisense transcription was impaired in the rpb1-ts mutant strain in dextrose-containing growth medium (Fig. 4F and G), thus supporting the direct role of RNA polymerase II in antisense transcription, similar to the function of RNA polymerase II in sense transcription (for example, ACT1 transcription) (Fig. 4F and G).
Reb1p-binding site at the 3′ end of the GAL10 coding sequence facilitates the targeting of RNA polymerase II for antisense transcription.
Sense transcription of the protein-coding genes by RNA polymerase II is a highly targeted process and requires GTFs for the formation of the PIC to initiate transcription. Typically, an activator binds to the upstream activating sequence (UAS) of a gene and facilitates the formation of the PIC, and, hence, recruitment of RNA polymerase II holoenzyme to initiate sense transcription. However, there has not been any systematic study to analyze whether activators and GTFs perform similar roles to promote the targeting of antisense RNA polymerase II. Therefore, we have analyzed below whether the targeting of antisense RNA polymerase II to the 3′ end of the GAL10 coding sequence requires these factors.
Genome-wide studies in humans analyzing the binding of transcriptional activators SP1, c-Myc, and p53 have revealed that they bind in close proximity to the 3′ end of the protein-coding genes on chromosomes 21 and 22 (86), thus suggesting that they might be regulating antisense transcription. Similarly, in yeast, the Reb1p activator has been shown to bind within several protein-coding genes (87), suggesting its role in controlling antisense transcription. Indeed, previous studies have shown that Reb1p, a Myb-related protein which plays important roles in RNA polymerase I- and II-mediated transcription and polymerase I transcription termination, associates with the 3′ end of the GAL10 coding sequence and promotes GAL10 ncRNA transcription (31, 32). We thus hypothesized that Reb1p might be acting as an activator to promote the targeting of RNA polymerase II to the 3′ end of the GAL10 coding sequence for antisense transcription. To test this hypothesis, we analyzed the role of Reb1p in recruitment of RNA polymerase II to the 3′ end of the GAL10 coding sequence in dextrose-containing growth medium. For this experiment, we have used the yeast strain (from the Houseley laboratory) (31) that carries mutations in the 4 Reb1p-binding sites at the 3′ end of the GAL10 coding sequence (Fig. 5A), and such mutations impaired the binding of Reb1p to the 3′ end of the GAL10 coding sequence (31) (and, hence, antisense transcription) (31). Association of RNA polymerase II with the 3′ end of the GAL10 coding sequence was analyzed in this mutant strain along with its isogenic wild-type equivalent in dextrose-containing growth medium. We found that the association of RNA polymerase II with the 3′ end of the GAL10 coding sequence was significantly impaired in the yeast strain carrying mutations in the Reb1p-binding sites compared to the wild-type equivalent (Fig. 5B and C). Thus, our results support the role of the Reb1p-binding site (or presumably Reb1p) in facilitating the targeting of RNA polymerase II to the site of antisense transcription initiation. Therefore, GAL10 antisense transcription would be impaired in the yeast strain carrying mutations in the Reb1p-binding site. Indeed, we found that GAL10 antisense transcription in dextrose-containing growth medium was impaired in the yeast strain carrying mutations in the Reb1p-binding site (Fig. 5D and E), consistent with previous studies (31). As a control, we demonstrate that ACT1 transcription was not altered in the yeast strain carrying mutations in the Reb1p-binding site in comparison to the wild-type equivalent (Fig. 5D and E). As mentioned above, the absence of PCR signal using the primer pair targeted to the GAL7 coding sequence (Fig. 2A, region N) in dextrose-containing growth medium (Fig. 5D) demonstrates that there was no DNA contamination in the RT-PCR analysis. Collectively, our data support the role of the activator Reb1p-binding site in targeting RNA polymerase II for GAL10 antisense transcription. Intriguingly, the Gal4p activator that is involved in GAL10 sense transcription does not regulate GAL10 antisense transcription in dextrose-containing growth medium (Fig. 5E and F). GAL10 sense transcription does not occur in dextrose-containing growth medium due to Mig1p-mediated repression and masking of the Gal4p activation domain by Gal80p (60, 61, 68–71). Thus, GAL10 antisense transcription is independent of its sense transcription. This is further substantiated by the fact that GAL10 antisense transcription occurs in dextrose-containing growth medium in the absence of the GAL1-GAL10 promoter (Fig. 5G). Intriguingly, the 3′ end of the GAL10 coding sequence alone cannot drive transcription from a reporter plasmid containing a LacZ coding sequence (Fig. 5H). Likewise, the promoterless vector did not generate LacZ transcript (Fig. 5H). The RPS5 sense promoter was used as a positive control and drove LacZ transcription from the reporter plasmid (Fig. 5H). These results indicate the requirement of additional DNA sequence elements or factors in addition to the GAL10 3′ end to drive antisense transcription from its own chromosomal locus.
TBP promotes the recruitment of RNA polymerase II to the 3′ end of the GAL10 coding sequence for antisense transcription.
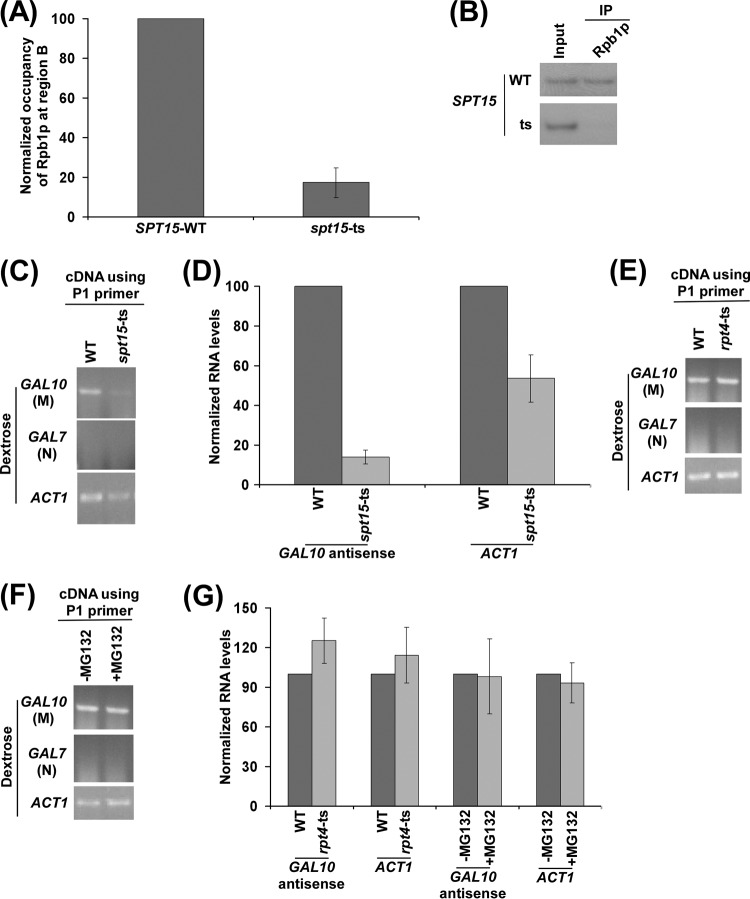
Next, we analyzed the role of TBP in targeting the recruitment of RNA polymerase II involved in antisense transcription initiation at the 3′ end of the GAL10 coding sequence. TBP is an essential GTF and is required for recruitment of RNA polymerase II (2). Likewise, RNA polymerase II involved in antisense transcription may also require TBP for its recruitment to the 3′ end of the GAL10 coding sequence under repressive growth conditions. In support of this idea, two TATA boxes (that bind with TBP) are found at the 3′ end of the GAL10 coding sequence (32). One TATA box is present upstream of the Reb1p-binding site, and another one is located downstream of the Reb1p-binding site (Fig. 5A) (32). Further, recent global genome-wide location analyses revealed the association of TBP to the 3′ ends of genes, hence suggesting the role of TBP in antisense transcription (88–91). To mechanistically determine the role of TBP in antisense transcription, we analyzed the recruitment of RNA polymerase II to the 3′ end of the GAL10 coding sequence in dextrose-containing growth medium in the wild-Type and TS mutant strains of TBP. We found that the recruitment of RNA polymerase II to the 3′ end of the GAL10 coding sequence was significantly decreased in the TS mutant strain of TBP (spt15-ts) compared to the wild-type equivalent (Fig. 6A and B). Thus, RNA polymerase II requires TBP for its targeting to the antisense start site to initiate antisense transcription. Although the presence of a TATA box at the 3′ end of the GAL10 coding sequence and association of TBP at the 3′ ends of the genes (88–91) suggested the role of TBP in antisense transcription initiation, there has not been any study that demonstrates this fact. Our results demonstrate that TBP is required for recruitment of RNA polymerase II associated with antisense transcription. Thus, GAL10 antisense transcription would be altered in the spt15-ts mutant strain. Indeed, we find that GAL10 antisense transcription in dextrose-containing growth medium was impaired in the spt15-ts mutant strain (Fig. 6C and D), consistent with the role of TBP in regulation of sense transcription (for example, ACT1 transcription) (Fig. 6C and D). As mentioned above, the absence of the PCR signal in cDNA amplification using the primer pair targeted to the GAL7 coding sequence in dextrose-containing growth medium (Fig. 6C) demonstrated that there was no DNA contamination in the RT-PCR analysis. Collectively, our data support the idea that TBP promotes the targeting of RNA polymerase II for antisense transcription. Intriguingly, the recruitment of TBP and RNA polymerase II to the promoter during GAL sense transcription is facilitated by the proteasome (1, 2, 49, 92). Such regulation has recently been attributed to occur via degradation of Gal80p (92), which specifically controls GAL1-GAL10 sense transcription (60, 61, 68–71). However, our RT-PCR analysis revealed that, unlike its sense transcription, GAL10 antisense transcription in dextrose-containing growth medium was not regulated by the proteasome (Fig. 6E, F, and G). The levels of ACT1 mRNAs were monitored as controls since previous studies (93) demonstrated that ACT1 transcription was not regulated by the proteasome. Indeed, we found that ACT1 transcription was not altered by the proteasome (Fig. 6E, F, and G). Further, the absence of the PCR signal in cDNA amplification using the primer pair targeted to the GAL7 coding sequence (Fig. 6E and F) demonstrated the absence of DNA contamination in the RT-PCR analysis. Collectively, these results support the idea that GAL10 antisense transcription is not regulated by the proteasome. This is as expected since GAL10 antisense transcription in dextrose-containing growth medium is not regulated by Gal4p (and, hence, Gal80p) (Fig. 5E and F). Thus, the function of the proteasome may be specific in the regulation of GAL1-GAL10 sense transcription in the presence of galactose. Taken together, our data reveal that GAL10 sense and antisense transcriptions are regulated differently.
Fig 6.
TBP is required for recruitment of RNA polymerase II to the 3′ end of the GAL10 coding sequence in dextrose-containing growth medium for GAL10 antisense transcription. (A) Analysis of recruitment of antisense RNA polymerase II to the 3′ end of the GAL10 coding sequence in the wild-type and mutant strains of TBP (SPT15-WT and spt15-ts) in dextrose-containing growth medium. Both the wild-type and mutant strains were grown in YPD medium at 23°C to an OD600 of 0.85 and then transferred to 37°C for 1 h before cross-linking. Immunoprecipitation was performed as described in the legend of Fig. 5B. (B) Autoradiograms for the ChIP data shown in panel A. (C) Analysis of GAL10 antisense transcription in the wild-type and spt15-ts mutant strains in dextrose-containing growth medium using the P1 primer in cDNA synthesis. Both the wild-type and mutant strains were grown as described in panel A. (D) The transcription data in panel C were plotted in the form of a histogram. The PCR signal of GAL10 antisense transcript in the wild-type strain was set to 100, and the level of GAL10 antisense transcript in the mutant strain was normalized with respect to 100. Likewise, the ACT1 transcript level was set to 100 in the wild-type strain, and the level of ACT1 transcript in the mutant strain was normalized with respect to 100. (E) Analysis of GAL10 antisense transcription in the wild-type and rpt4-ts mutant strains in dextrose-containing growth medium using the P1 primer in cDNA synthesis. Yeast strains were grown as described in panel A. (F) Analysis of GAL10 antisense transcription in the presence and absence of MG132 in dextrose-containing growth medium using the P1 primer in cDNA synthesis. Yeast cells carrying null mutation of PDR5 were grown in YPD medium at 30°C to an OD600 of 0.7 and then treated with MG132 (75 μM) for 2 h prior to harvesting. (G) The GAL10 antisense transcription data shown in panels E and F were plotted in the form of a histogram. The PCR signal of GAL10 antisense transcript in the wild-type strain was set to 100, and the level of GAL10 antisense transcript in the mutant strain was normalized with respect to 100. Likewise, the ACT1 transcript level was set to 100 in wild-type strain, and the level of ACT1 transcript in the mutant strain was normalized with respect to 100. Similarly, the PCR signals were normalized in the presence of MG132 with respect to the absence of MG132.
TAFs facilitate the targeting of RNA polymerase II to the 3′ end of the GAL10 coding sequence for antisense transcription.
TBP and a set of TAFs form the TFIID complex that is an important transcription factor. In yeast, about 90% of genes are TAF dependent (2). TAFs are highly conserved transcription factors and are required for the association of TBP (and hence RNA polymerase II) with the core promoter of a large number of protein-coding genes (2). We thus asked whether TAFs are also required for recruitment of RNA polymerase II to the antisense transcription initiation site at GAL10, similar to their roles in targeting RNA polymerase II involved in sense transcription. To this end, we analyzed the association of RNA polymerase II with the 3′ end of the GAL10 coding sequence in the TAF11p and TAF13p TS mutant and wild-type strains in dextrose-containing growth medium. These TAFs are specific to the TFIID complex and are not shared with the SAGA complex (2). TAF11p and TAF13p have been shown to form a heterodimer and interact with TBP for sense transcription (2, 55). Interestingly, our ChIP analysis revealed that the association of RNA polymerase II with the 3′ end of the GAL10 coding sequence in dextrose-containing growth medium was severely impaired in both the TAF11 and TAF13 TS mutant strains compared to the wild-type equivalent (Fig. 7A to D). Thus, the targeting of RNA polymerase II to the antisense transcription initiation site is dependent on TFIID-specific TAFs. Consistently, we found that GAL10 antisense transcription in dextrose-containing growth medium was significantly impaired in the TS mutant strains of TAF11p and TAF13p (Fig. 7E and F). The sense transcription of the TAF-independent ADH1 gene (94) was monitored in the above taf11-ts and taf13-ts mutant strains as controls. We found that ADH1 mRNA levels were not altered in these TS mutant strains in comparison to the wild-type equivalents (Fig. 7E and F). The absence of PCR signal in cDNA amplification using the primer pair targeted to the GAL7 coding sequence (Fig. 7E) served as a “no-DNA-contamination” control in this set of RT-PCR analyses in dextrose-containing growth medium. Taken together, our results demonstrate that RNA polymerase II requires TAFs and TBP for its association with the antisense transcription initiation site at GAL10 for antisense transcription (Fig. 6A to D and 7A to F). These observations support for the first time the role of TFIID in promoting the recruitment of RNA polymerase II (and hence antisense transcription) to the antisense initiation site.
TFIIB and Mediator facilitate the targeting of RNA polymerase II to the 3′ end of the GAL10 coding sequence for antisense transcription.
Transcription initiation at the sense strand requires several other GTFs such as TFIIA, TFIIB, TFIIE, TFIIF, and TFIIH. An ordered and proper assembly of these factors at the core promoter ensures the appropriate transcription initiation complex or PIC ready for producing the transcript. These GTFs are essential for accurate sense transcription (6, 95). Likewise, these GTFs may also regulate the association of RNA polymerase II with the antisense transcription initiation site to control antisense transcription. To test this, we analyzed the role of TFIIB in recruitment of antisense RNA polymerase II to the antisense start site at GAL10. During sense transcription, TFIIB clamps TBP to the promoter DNA. TFIIB makes intimate contact with RNA polymerase II and helps in the transcription start site selection (96–98). Thus, TFIIB may be enabling the proper binding of RNA polymerase II to the 3′ end of the GAL10 coding sequence for antisense transcription in dextrose-containing growth medium. Indeed, we found that the recruitment of RNA polymerase II to the antisense initiation site at GAL10 was significantly reduced in the TFIIB TS mutant strain (sua7-ts) compared to its wild-type equivalent in dextrose-containing growth medium (Fig. 8A and B). Consistently, GAL10 antisense transcription in dextrose-containing growth medium was impaired in the sua7-ts mutant strain (Fig. 8C and D), similar to the role of TFIIB in sense transcription (for example, ACT1 transcription) (Fig. 8C and D). The absence of the PCR signal in cDNA amplification using the primer pair targeted to the GAL7 coding sequence (Fig. 8C) served as a no-DNA-contamination control in this set of RT-PCR analyses. Collectively, our data reveal that TFIIB plays an important role in targeting RNA polymerase II to the antisense initiation site at GAL10 for antisense transcription.
Next, we analyzed the role of the Mediator complex in targeting the recruitment of RNA polymerase II to the antisense initiation site at GAL10. The Mediator complex acts as a bridge between the activator and the basal transcriptional machinery to promote sense transcription (99, 100). Mediator is a multisubunit complex and consists of three modules: the Srb4 module (Srb2p, Srb4p, Srb5p, Srb6p, Rox3p, Med8p, Med11p, and Med6p), the Gal11/Sin4 module (Gal11p, Rgr1p, Sin4p, Pgd1p, and Med2p), and the Med9/Med10 module (Med1p, Med4p, Med7p, Srb7p, Med9p, and Med10p). The Srb4 module of the Mediator complex interacts with the C-terminal domain of RNA polymerase II. Srb4p is essential to maintain the global structural and functional integrity of the Mediator complex (61, 101, 102). Mediator is required for recruitment of RNA polymerase II involved in sense transcription. Likewise, the Mediator complex may also be required to promote the recruitment of RNA polymerase II to the antisense initiation site. To test this, we analyzed the recruitment of RNA polymerase II to the antisense transcription initiation site at GAL10 in dextrose-containing growth medium in the wild-type and TS mutant strains of Srb4p at the nonpermissive temperature. Interestingly, we found that the association of RNA polymerase II with the antisense initiation site at GAL10 was significantly decreased in the srb4-ts mutant strain compared to the wild-type equivalent in dextrose-containing growth medium (Fig. 8E and F). Thus, our results implicate the role of the Mediator complex in targeting RNA polymerase II (and hence antisense transcription) to the antisense initiation site at GAL10.
DISCUSSION
The ncRNAs have been increasingly implicated in oncogenesis and pathogenesis of a number of diseases (37, 38, 40–46, 103–105). Genome-wide studies have revealed extensive existence of ncRNAs in all eukaryotes, and the roles of ncRNAs in various cellular processes are significant (17, 18). Antisense RNAs form a major fraction of ncRNAs and play crucial roles in gene expression (8, 17, 18, 36, 106). Previous studies have demonstrated that antisense RNAs from GAL10 are capped at the 5′ end and polyadenylated at the 3′ end (31). Further, antisense transcription from GAL10 is associated with histone H3 K36 (lysine 36) methylation that is carried out by RNA polymerase II via its interaction with Set2p methyltransferase (31). These observations support the idea that RNA polymerase II holoenzyme is involved in antisense transcription (31, 32). Although RNA polymerase II has been implicated in antisense transcription, there has not been any study that demonstrated its association and regulation at the 3′ end of the gene to initiate antisense transcription. Further, the role of RNA polymerase II in antisense transcription has not been directly demonstrated. Here, we have demonstrated the association of RNA polymerase II with the antisense transcription initiation site at GAL10 in dextrose-containing growth medium (Fig. 1, 2, and 3). Such association is essential for antisense transcription (Fig. 4F and G) and serves as an efficient readout for GAL10 ncRNA transcription. Importantly, we find that the association of RNA polymerase II is dependent on the Reb1p activator-binding site at the 3′ end of the GAL10 coding sequence in dextrose-containing growth medium (Fig. 5B and C), hence supporting the role of the activator in targeting RNA polymerase II to the antisense transcription initiation site. Further, TFIID, TFIIB, and Mediator are also required for recruitment of RNA polymerase II to the site of antisense transcription initiation at GAL10 (Fig. 6, 7, and 8). Collectively, our data support the idea that antisense transcription requires an elaborate regulatory network of activator and GTFs to target RNA polymerase II to the 3′ end of the gene to initiate antisense transcription, thus shedding much light on the mechanisms of antisense transcription initiation.
Unlike analysis of sense transcription, delineating the mechanism of antisense transcription initiation is a challenging problem, primarily due to low and infrequent antisense transcription. To overcome this problem, we have used our modified ChIP protocol (48–53) and analyzed the association of RNA polymerase II with the 3′ end of the GAL10 coding sequence in dextrose-containing growth medium, which favors antisense transcription. Using the ChIP assay, we have successfully analyzed for the first time the association of RNA polymerase II with the 3′ end of the GAL10 coding sequence in dextrose-containing growth medium (Fig. 1B to G). Further, our kinetic analysis revealed that RNA polymerase II is associated with the GAL10 coding sequence in galactose-containing growth medium (Fig. 3A and B) and dissociates within 3 min in dextrose-containing growth medium (Fig. 3A and B). However, at later time points in dextrose-containing growth medium, RNA polymerase II associates with the 3′ end of the GAL10 coding sequence to initiate antisense transcription (Fig. 3A to E). Thus, RNA polymerase II that is involved in GAL10 sense transcription does not get engaged in antisense transcription as sense RNA polymerase II falls off, and subsequently RNA polymerase II associates with the 3′ end of the GAL10 coding sequence in dextrose-containing growth medium to initiate antisense transcription (Fig. 3). Further, the association of RNA polymerase II with the 3′ end of the GAL10 coding sequence in dextrose-containing growth medium is not influenced by the activator Gal4p involved in GAL10 sense transcription (data not shown). Consistently, our RT-PCR analysis revealed that Gal4p is dispensable for GAL10 antisense transcription in dextrose-containing growth medium (Fig. 5E and F). Moreover, as mentioned above, the growth in the dextrose-containing medium switches off GAL10 sense transcription. Furthermore, we observed the occurrence of GAL10 antisense transcription in the absence of GAL1-GAL10 promoter (Fig. 5G). These results support the idea that GAL10 antisense transcription is independent of its sense transcription. Intriguingly, GAL10 sense transcription is not dependent on TFIID (60, 61, 94), while TFIID is required for GAL10 antisense transcription (Fig. 7). In addition, GAL10 antisense transcription is not regulated by the proteasome (Fig. 6E to G). However, GAL sense transcription is dependent on the proteasome (1, 2, 49). Thus, GAL10 antisense transcription is regulated differently from sense transcription.
Our kinetic analysis of RNA polymerase II association with the 3′ end of the GAL10 coding sequence in dextrose-containing growth medium reveals a time delay in association of RNA polymerase II involved in antisense transcription following the dissociation of RNA polymerase II involved in sense transcription (Fig. 3). This time delay indicates that there might be some factors which initially associate with the 3′ end of the GAL10 coding sequence and finally target RNA polymerase II to initiate antisense transcription in dextrose-containing growth medium. Indeed, we find that in the absence of activator Reb1p binding at the 3′ end of the GAL10 coding sequence, the association of RNA polymerase II with the antisense initiation site is significantly impaired (Fig. 5B and C). These observations support the role of Reb1p in promoting the recruitment of RNA polymerase II to the antisense initiation site, analogous to the role of the activator in recruitment of RNA polymerase II involved in sense transcription. Consistently, GAL10 antisense transcription is significantly impaired in the reb1p-ts mutant strain or Reb1p-binding site mutant (31, 32) (Fig. 5D and E).
The activator may be promoting the recruitment of RNA polymerase II associated with antisense transcription by facilitating the assembly of GTFs such as TBP, TAFs, TFIIB, and Mediator, which subsequently target RNA polymerase II, analogous to the role of the activator in targeting RNA polymerase II during sense transcription (2). TAFs have been implicated as targets of activators (2, 94, 107–109) in sense transcription, and such interaction has been demonstrated to be functionally important to assemble GTFs and RNA polymerase II to initiate sense transcription. Thus, Reb1p may be targeting TAFs, which are essential for TBP binding, to promote the recruitment of RNA polymerase II at the 3′ end of the GAL10 coding sequence in dextrose-containing growth medium, similar to the roles of TAFs in sense transcription. Indeed, we observe a strong dependence of RNA polymerase II recruitment to the antisense initiation site at GAL10 on TAFs and TBP (Fig. 6A and B and 7A to D). Further, TAFs and TBP have been recently shown to be associated with the 3′ ends of genes (88–91), suggesting their possible roles in antisense transcription. In fact, our results demonstrate the role of TAFs and TBP in recruitment of RNA polymerase II (and hence antisense transcription) to the antisense transcription initiation site at GAL10 (Fig. 6A to D and 7). Like TBP and TAFs, Mediator and TFIIB also facilitate the targeting of RNA polymerase II to the antisense initiation site at GAL10 (Fig. 8). Thus, our study demonstrates for the first time the roles of activator and GTFs in promoting the targeting of RNA polymerase II to the antisense initiation site at the 3′ end of the GAL10 coding sequence for initiation of antisense transcription, hence suggesting the formation of the PIC at the 3′ end of the GAL10 coding sequence. In support of this, previous studies (88–91) have also implicated the formation of the PIC at the 3′ ends of genes. Although previous studies (88–91) suggested the formation of the PIC at the 3′ ends of genes, the function of such a complex was not analyzed. Our data demonstrate that PIC components such as TBP, TAFs, TFIIB, and Mediator are essential for recruitment of RNA polymerase II (and hence antisense transcription) to the site of antisense transcription initiation, thus supporting the functionality of the PIC in antisense transcription.
As mentioned above, TBP is required for recruitment of RNA polymerase II at the 3′ end of the GAL10 coding sequence in dextrose-containing growth medium. Therefore, it is likely that the 3′ end of the GAL10 coding sequence would contain a TATA box for TBP binding. Indeed, there are two TATA boxes flanking the activator Reb1p-binding site (Fig. 5A). Since antisense transcription is initiated downstream of the Reb1p-binding site of GAL10 (31, 32), TBP would probably bind to the TATA box downstream of the Reb1p-binding site to form the PIC and recruit RNA polymerase II for initiation of antisense transcription. The binding of TBP and subsequently formation of the PIC assembly would require a nucleosome-free region. Indeed, previous studies (89, 90, 110, 111) have shown that a majority of antisense transcripts are synthesized from the nucleosome-free region at the 3′ ends of protein-coding genes.
Recent studies (31, 32) demonstrated the role of RNA polymerase II for histone H3 K4 methylation at the 3′ end of the GAL10 coding sequence in dextrose-containing growth medium. Like RNA polymerase II, Reb1p or the Reb1p-binding site has also been implicated in histone H3 K4 methylation at the 3′ end of the GAL10 coding sequence (31, 32). However, it was not known how Reb1p facilitates histone H3 K4 methylation at the 3′ end of the GAL10 coding sequence in dextrose-containing growth medium. Our results support a link between Reb1p and histone H3 K4 methylation via RNA polymerase II (i.e., Reb1p promotes recruitment of RNA polymerase II that facilitates histone H3 K4 methylation at the 3′ end of the GAL10 coding sequence in dextrose-containing growth medium).
Intriguingly, a recent study (91) demonstrated a long-range interaction of the 3′ end region of GAL10 with the GAL1 3′ end via a Reb1-binding site during antisense transcription in dextrose-containing growth medium. However, such a long-range interaction, or gene looping, was not observed during sense transcription in galactose-containing growth medium (91). These results implicate the role of gene looping or long-range chromosomal interaction in initiating GAL10 antisense transcription. Thus, it is likely that the insertion of the 3′ end region of GAL10 into a reporter-containing plasmid would not initiate transcription. Indeed, we find that the 3′-end region of GAL10 alone was unable to drive transcription from a reporter plasmid (Fig. 5H).
In summary, we demonstrate here the occupancy of RNA polymerase II at the antisense initiation site at the 3′ end of the GAL10 coding sequence in dextrose-containing growth medium for antisense transcription. Such association of RNA polymerase II is dependent on the activator-binding site as well as GTFs. Intriguingly, GAL10 sense and antisense transcriptions are not similarly regulated. The collective results of this study significantly advance our current understanding of the regulation of antisense transcription initiation, and such knowledge will be useful in understanding disease pathogenesis as aberrant antisense transcription is associated with a large number of human diseases (37, 38, 40–46, 103–105).
ACKNOWLEDGMENTS
We thank Michael R. Green, Kevin Struhl, Richard A. Young, David Tollervey, Stephen A. Johnston, and Thomas Kodadek for yeast strains.
The work in the Bhaumik laboratory was supported by a National Institutes of Health grant (1R15GM088798-01), a grant-in-aid (10GRNT4300059) from the American Heart Association (Greater Midwest Affiliate), a Mallinckrodt Foundation grant, and Excellence in Academic Medicine awards of Southern Illinois University School of Medicine.
Footnotes
Published ahead of print 8 July 2013
REFERENCES
- 1.Bhaumik SR, Mailk S. 2008. Diverse regulatory mechanisms of eukaryotic transcriptional activation by the proteasome complex. Crit. Rev. Biochem. Mol. Biol. 43:419–433 [DOI] [PubMed] [Google Scholar]
- 2.Bhaumik SR. 2011. Distinct regulatory mechanisms of eukaryotic transcriptional activation by SAGA and TFIID. Biochim. Biophys. Acta 1809:97–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bhaumik SR, Smith E, Shilatifard A. 2007. Covalent modifications of histones during development and disease pathogenesis. Nat. Struct. Mol. Biol. 14:1008–1016 [DOI] [PubMed] [Google Scholar]
- 4.Venters BJ, Pugh BF. 2009. How eukaryotic genes are transcribed. Crit. Rev. Biochem. Mol. Biol. 44:117–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roeder RG. 1998. Role of general and gene-specific cofactors in the regulation of eukaryotic transcription. Cold Spring Harbor Symp. Quant. Biol. 63:201–218 [DOI] [PubMed] [Google Scholar]
- 6.Maston GA, Evans SK, Green MR. 2006. Transcriptional regulatory elements in the human genome. Annu. Rev. Genomics Hum. Genet. 7:29–59 [DOI] [PubMed] [Google Scholar]
- 7.Hochheimer A, Tjian R. 2003. Diversified transcription initiation complexes expand promoter selectivity and tissue-specific gene expression. Genes Dev. 17:1309–1320 [DOI] [PubMed] [Google Scholar]
- 8.Su WY, Xiong H, Fang JY. 2010. Natural antisense transcripts regulate gene expression in an epigenetic manner. Biochem. Biophys. Res. Commun. 396:177–181 [DOI] [PubMed] [Google Scholar]
- 9.Shukla A, Chaurasia P, Bhaumik SR. 2009. Histone methylation and ubiquitination with their cross-talk and roles in gene expression and stability. Cell. Mol. Life Sci. 66:1419–1433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Malik S, Bhaumik SR. 2010. Mixed lineage leukemia: histone H3 lysine 4 methyltransferases from yeast to human. FEBS J. 277:1805–1821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Imamura T, Yamamoto S, Ohgane J, Hattori N, Tanaka S, Shiota K. 2004. Non-coding RNA directed DNA demethylation of Sphk1 CpG island. Biochem. Biophys. Res. Commun. 322:593–600 [DOI] [PubMed] [Google Scholar]
- 12.Werner A, Berdal A. 2005. Natural antisense transcripts: sound or silence? Physiol. Genomics 23:125–131 [DOI] [PubMed] [Google Scholar]
- 13.Tufarelli C, Stanley JA, Garrick D, Sharpe JA, Ayyub H, Wood WG, Higgs DR. 2003. Transcription of antisense RNA leading to gene silencing and methylation as a novel cause of human genetic disease. Nat. Genet. 34:157–165 [DOI] [PubMed] [Google Scholar]
- 14.Ohhata T, Hoki Y, Sasaki H, Sado T. 2008. Crucial role of antisense transcription across the Xist promoter in Tsix-mediated Xist chromatin modification. Development 135:227–235 [DOI] [PubMed] [Google Scholar]
- 15.Bernstein E, Allis CD. 2005. RNA meets chromatin. Genes Dev. 19:1635–1655 [DOI] [PubMed] [Google Scholar]
- 16.Bernstein E, Duncan EM, Masui O, Gil J, Heard E, Allis CD. 2006. Mouse polycomb proteins bind differentially to methylated histone H3 and RNA and are enriched in facultative heterochromatin. Mol. Cell. Biol. 26:2560–2569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Werner A, Sayer JA. 2009. Naturally occurring antisense RNA: function and mechanisms of action. Curr. Opin. Nephrol. Hypertens. 18:343–349 [DOI] [PubMed] [Google Scholar]
- 18.Lapidot M, Pilpel Y. 2006. Genome-wide natural antisense transcription: coupling its regulation to its different regulatory mechanisms. EMBO Rep. 7:1216–1222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lavorgna G, Dahary D, Lehner B, Sorek R, Sanderson CM, Casari G. 2004. In search of antisense. Trends Biochem. Sci. 29:88–94 [DOI] [PubMed] [Google Scholar]
- 20.Werner A. 2005. Natural antisense transcripts. RNA Biol. 2:53–62 [DOI] [PubMed] [Google Scholar]
- 21.Vanhee-Brossollet C, Vaquero C. 1998. Do natural antisense transcripts make sense in eukaryotes? Gene 211:1–9 [DOI] [PubMed] [Google Scholar]
- 22.Sun M, Hurst LD, Carmichael GG, Chen J. 2005. Evidence for a preferential targeting of 3′-UTRs by cis-encoded natural antisense transcripts. Nucleic Acids Res. 33:5533–5543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang XJ, Gaasterland T, Chua NH. 2005. Genome-wide prediction and identification of cis-natural antisense transcripts in Arabidopsis thaliana. Genome Biol. 6:R30. 10.1186/gb-2005-6-4-r30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tisseur M, Kwapisz M, Morillon A. 2011. Pervasive transcription: lessons from yeast. Biochimie 93:1889–18896 [DOI] [PubMed] [Google Scholar]
- 25.Lee JT. 2009. Lessons from X-chromosome inactivation: long ncRNA as guides and tethers to the epigenome. Genes Dev. 23:1831–1842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tian D, Sun S, Lee JT. 2010. The long noncoding RNA, Jpx, is a molecular switch for X chromosome inactivation. Cell 143:390–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hongay CF, Grisafi PL, Galitski T, Fink GR. 2006. Antisense transcription controls cell fate in Saccharomyces cerevisiae. Cell 127:735–745 [DOI] [PubMed] [Google Scholar]
- 28.Martens JA, Laprade L, Winston F. 2004. Intergenic transcription is required to repress the Saccharomyces cerevisiae SER3 gene. Nature 429:571–574 [DOI] [PubMed] [Google Scholar]
- 29.Martens JA, Wu PY, Winston F. 2005. Regulation of an intergenic transcript controls adjacent gene transcription in Saccharomyces cerevisiae. Genes Dev. 19:2695–26704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Camblong J, Iglesias N, Fickentscher C, Dieppois G, Stutz F. 2007. Antisense RNA stabilization induces transcriptional gene silencing via histone deacetylation in S. cerevisiae. Cell 131:706–717 [DOI] [PubMed] [Google Scholar]
- 31.Houseley J, Rubbi L, Grunstein M, Tollervey D, Vogelauer M. 2008. A ncRNA modulates histone modification and mRNA induction in the yeast GAL gene cluster. Mol. Cell 32:685–695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pinskaya M, Gourvennec S, Morillon A. 2009. H3 lysine 4 di- and tri-methylation deposited by cryptic transcription attenuates promoter activation. EMBO J. 28:1697–1707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yazgan O, Krebs JE. 2007. Noncoding but nonexpendable: transcriptional regulation by large noncoding RNA in eukaryotes. Biochem. Cell Biol. 85:484–496 [DOI] [PubMed] [Google Scholar]
- 34.Katayama S, Tomaru Y, Kasukawa T, Waki K, Nakanishi M, Nakamura M, Nishida H, Yap CC, Suzuki M, Kawai J. 2005. Antisense transcription in the mammalian transcriptome. Science 309:1564–1566 [DOI] [PubMed] [Google Scholar]
- 35.He Y, Vogelstein B, Velculescu VE, Papadopoulos N, Kinzler KW. 2008. The antisense transcriptomes of human cells. Science 322:1855–1857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wery M, Kwapisz M, Morillon A. 2011. Noncoding RNAs in gene regulation. Wiley Interdiscip. Rev. Syst. Biol. Med. 3:728–738 [DOI] [PubMed] [Google Scholar]
- 37.Faghihi MA, Modarresi F, Khalil AM, Wood DE, Sahagan BG, Morgan TE, Finch CE, St Laurent G, III, Kenny PJ, Wahlestedt C. 2008. Expression of a noncoding RNA is elevated in Alzheimer's disease and drives rapid feed-forward regulation of beta-secretase. Nat. Med. 14:723–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Seitz A, Gourevitch D, Zhang XM, Clark L, Chen P, Kragol M, Levenkova N, Rux J, Samulewicz S, Heber-Katz E. 2005. Sense and antisense transcripts of the apolipoprotein E gene in normal and ApoE knockout mice, their expression after spinal cord injury and corresponding human transcripts. Hum. Mol. Genet. 14:2661–2670 [DOI] [PubMed] [Google Scholar]
- 39.Kumar M, Carmichael GG. 1998. Antisense RNA: function and fate of duplex RNA in cells of higher eukaryotes. Microbiol. Mol. Biol. Rev. 62:1415–1434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reis EM, Nakaya HI, Louro R, Canavez FC, Flatschart AV, Almeida GT, Egidio CM, Paquola AC, Machado AA, Festa F, Yamamoto D, Alvarenga R, da Silva CC, Brito GC, Simon SD, Moreira-Filho CA, Leite KR, Camara-Lopes LH, Campos FS, Gimba E, Vignal GM, El-Dorry H, Sogayar MC, Barcinski MA, da Silva AM, Verjovski-Almeida S. 2004. Antisense intronic non-coding RNA levels correlate to the degree of tumor differentiation in prostate cancer. Oncogene 23:6684–6692 [DOI] [PubMed] [Google Scholar]
- 41.Grigoriadis A, Oliver GR, Tanney A, Kendrick H, Smalley MJ, Jat P, Neville AM. 2009. Identification of differentially expressed sense and antisense transcript pairs in breast epithelial tissues. BMC Genomics 10:324. 10.1186/1471-2164-10-324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Aartsma-Rus A, van Ommen GJ. 2010. Progress in therapeutic antisense applications for neuromuscular disorders. Eur. J. Hum. Genet. 18:146–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Crooke RM. 2005. Antisense oligonucleotides as therapeutics for hyperlipidaemias. Expert Opin. Biol. Ther. 5:907–917 [DOI] [PubMed] [Google Scholar]
- 44.Luther HP. 2005. Role of endogenous antisense RNA in cardiac gene regulation. J. Mol. Med. (Berl.) 83:26–32 [DOI] [PubMed] [Google Scholar]
- 45.Ulanova M, Schreiber AD, Befus AD. 2006. The future of antisense oligonucleotides in the treatment of respiratory diseases. BioDrugs 20:1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Popescu FD. 2005. Antisense- and RNA interference-based therapeutic strategies in allergy. J. Cell. Mol. Med. 9:840–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mattick JS, Makunin IV. 2006. Non-coding RNA. Hum. Mol. Genet. 15:R17–29 [DOI] [PubMed] [Google Scholar]
- 48.Shukla A, Stanojevic N, Duan Z, Sen P, Bhaumik SR. 2006. Ubp8p, a histone deubiquitinase whose association with SAGA is mediated by Sgf11p, differentially regulates lysine 4 methylation of histone H3 in vivo. Mol. Cell. Biol. 26:3339–3352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Malik S, Shukla A, Sen P, Bhaumik SR. 2009. The 19S proteasome subcomplex establishes a specific protein interaction network at the promoter for stimulated transcriptional initiation in vivo. J. Biol. Chem. 284:35714–35724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Malik S, Chaurasia P, Lahudkar S, Durairaj G, Shukla A, Bhaumik SR. 2010. Rad26p, a transcription-coupled repair factor, is recruited to the site of DNA lesion in an elongating RNA polymerase II-dependent manner in vivo. Nucleic Acids Res. 38:1461–1477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lahudkar S, Shukla A, Bajwa P, Durairaj G, Stanojevic N, Bhaumik SR. 2011. The mRNA cap-binding complex stimulates the formation of pre-initiation complex at the promoter via its interaction with Mot1p in vivo. Nucleic Acids Res. 39:2188–2209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Uprety B, Lahudkar S, Malik S, Bhaumik SR. 2012. The 19S proteasome subcomplex promotes the targeting of NuA4 HAT to the promoters of ribosomal protein genes to facilitate the recruitment of TFIID for transcriptional initiation in vivo. Nucleic Acids Res. 40:1969–1983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Malik S, Chaurasia P, Lahudkar S, Uprety B, Bhaumik SR. 2012. Rad26p regulates the occupancy of histone H2A-H2B dimer at the active genes in vivo. Nucleic Acids Res. 40:3348–3363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Longtine MS, McKenzie A, III, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pingle JR. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 14:953–961 [DOI] [PubMed] [Google Scholar]
- 55.Shen WC, Bhaumik SR, Causton HC, Simon I, Zhu X, Jennings EG, Wang TH, Young RA, Green MR. 2003. Systematic analysis of essential yeast TAFs in genome-wide transcription and preinitiation complex assembly. EMBO J. 22:3395–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li XY, Virbasius A, Zhu X, Green MR. 1999. Enhancement of TBP binding by activators and general transcription factors. Nature 399:605–609 [DOI] [PubMed] [Google Scholar]
- 57.Cormack BP, Struhl K. 1992. The TATA-binding protein is required for transcription by all three nuclear RNA polymerases in yeast cells. Cell 69:685–696 [DOI] [PubMed] [Google Scholar]
- 58.Thompson CM, Young RA. 1995. General requirement for RNA polymerase II holoenzymes in vivo. Proc. Natl. Acad. Sci. 92:4587–4590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Russell SJ, Johnston SA. 2001. Evidence that proteolysis of Gal4 cannot explain the transcriptional effects of proteasome ATPase mutations. J. Biol. Chem. 276:9825–9831 [DOI] [PubMed] [Google Scholar]
- 60.Bhaumik SR, Green MR. 2001. SAGA is an essential in vivo target of the yeast acidic activator Gal4p. Genes Dev. 15:1935–1945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bhaumik SR, Raha T, Aiello DP, Green MR. 2004. In vivo target of a transcriptional activator revealed by fluorescence resonance energy transfer. Genes Dev. 18:333–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bhaumik SR, Green MR. 2003. Interaction of Gal4p with components of transcription machinery in vivo. Methods Enzymol. 370:445–454 [DOI] [PubMed] [Google Scholar]
- 63.Bhaumik SR, Green MR. 2002. Differential requirement of SAGA components for recruitment of TATA-box-binding protein to promoters in vivo. Mol. Cell. Biol. 22:7365–7371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Merz K, Hondele M, Goetze H, Gmelch K, Stoeckl U, Griesenbeck J. 2008. Actively transcribed rRNA genes in S. cerevisiae are organized in a specialized chromatin associated with the high-mobility group protein Hmo1 and are largely devoid of histone molecules. Genes Dev. 22:1190–1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Peterson CL, Kruger W, Herskowitz I. 1991. A functional interaction between the C-terminal domain of RNA polymerase II and the negative regulator SIN1. Cell 64:1135–1143 [DOI] [PubMed] [Google Scholar]
- 66.Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Struhl K. 2001. Current protocols in molecular biology. Wiley, New York, NY [Google Scholar]
- 67.Gullerova M, Proudfoot NJ. 2010. Transcriptional interference and gene orientation in yeast: noncoding RNA connections. Cold Spring Harbor Symp. Quant. Biol. 75:299–311 [DOI] [PubMed] [Google Scholar]
- 68.Johnston M. 1987. A model fungal gene regulatory mechanism: the GAL genes of Saccharomyces cerevisiae. Microbiol. Rev. 51:458–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Johnston M, Carlson M. 1992. Regulation of carbon and phosphate utilization, p 193–281 In Jones EW, Pringle JR, Broach JR. (ed), The molecular and cellular biology of yeast Saccharomyces: gene expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 70.Ozcan S, Johnston M. 1996. Two different repressors collaborate to restrict expression of the yeast glucose transporter genes HXT2 and HXT4 to low levels of glucose. Mol. Cell. Biol. 16:5536–5545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Campbell RN, Leverentz MK, Ryan LA, Reece RJ. 2008. Metabolic control of transcription: paradigms and lessons from Saccharomyces cerevisiae. Biochem. J. 414:177–187 [DOI] [PubMed] [Google Scholar]
- 72.Bhaumik SR, Chary KV, Govil G, Liu K, Miles HT. 1997. Homopurine and homopyrimidine strands complementary in parallel orientation form an antiparallel duplex at neutral pH with A-C, G-T, and T-C mismatched base pairs. Biopolymers 41:773–784 [DOI] [PubMed] [Google Scholar]
- 73.Sau AK, Chary KV, Govil G, Chen CQ, Howard FB, Miles HT. 1995. Evidence for A+(anti)-G(syn) mismatched base-pairing in d-GGTAAGCGTACC. FEBS Lett. 377:301–305 [DOI] [PubMed] [Google Scholar]
- 74.Tikhomirova A, Beletskaya IV, Chalikian TV. 2006. Stability of DNA duplexes containing GG, CC, AA, and TT mismatches. Biochemistry 45:10563–10571 [DOI] [PubMed] [Google Scholar]
- 75.Cheng JW, Chou SH, Reid BR. 1992. Base pairing geometry in GA mismatches depends entirely on the neighboring sequence. J. Mol. Biol. 228:1037–1041 [DOI] [PubMed] [Google Scholar]
- 76.de los Santos C, Kouchakdjian M, Yarema K, Basu A, Essigmann J, Patel DJ. 1991. NMR studies of the exocyclic 1,N6-ethenodeoxyadenosine adduct (epsilon dA) opposite deoxyguanosine in a DNA duplex. Epsilon dA(syn) · dG(anti) pairing at the lesion site. Biochemistry 30:1828–1835 [DOI] [PubMed] [Google Scholar]
- 77.Cisse II, Kim H, Ha T. 2012. A rule of seven in Watson-Crick base-pairing of mismatched sequences. Nat. Struct. Mol. Biol. 19:623–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bhaumik SR, Chary KV. 2002. Molecular dynamics and mechanics calculations on a DNA duplex with A+-C, G-T and T-C mispairs. J. Biomol. Struct. Dyn. 20:199–206 [DOI] [PubMed] [Google Scholar]
- 79.Parvathy VR, Bhaumik SR, Chary KV, Govil G, Liu K, Howard FB, Miles HT. 2002. NMR structure of a parallel-stranded DNA duplex at atomic resolution. Nucleic Acids Res. 30:1500–1511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yang XL, Wang AH. 1997. Structural analysis of Z-Z DNA junctions with A:A and T:T mismatched base pairs by NMR. Biochemistry 36:4258–4267 [DOI] [PubMed] [Google Scholar]
- 81.Gantchev TG, Cecchini S, Hunting DJ. 2005. Dynamic conformational states of DNA containing T · T or BrdU · T mispaired bases: wobble H-bond pairing versus cross-strand inter-atomic contacts. J. Mol. Model. 11:141–159 [DOI] [PubMed] [Google Scholar]
- 82.Moe JG, Russu IM. 1992. Kinetics and energetics of base-pair opening in 5′-d(CGCGAATTCGCG)-3′ and a substituted dodecamer containing G · T mismatches. Biochemistry 31:8421–8428 [DOI] [PubMed] [Google Scholar]
- 83.Guest CR, Hochstrasser RA, Sowers LC, Millar DP. 1991. Dynamics of mismatched base pairs in DNA. Biochemistry 30:3271–3279 [DOI] [PubMed] [Google Scholar]
- 84.Fazakerley GV, Quignard E, Woisard A, Guschlbauer W, van der Marel GA, van Boom JH, Jones M, Radman M. 1986. Structures of mismatched base pairs in DNA and their recognition by the Escherichia coli mismatch repair system. EMBO J. 5:3697–3703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Schwabish MA, Struhl K. 2007. The Swi/Snf complex is important for histone eviction during transcriptional activation and RNA polymerase II elongation in vivo. Mol. Cell. Biol. 27:6987–6995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Cawley S, Bekiranov S, Ng HH, Kapranov P, Sekinger EA, Kampa D, Piccolboni A, Sementchenko V, Cheng J, Williams AJ, Wheeler R, Wong B, Drenkow J, Yamanaka M, Patel S, Brubaker S, Tammana H, Helt G, Struhl K, Gingeras TR. 2004. Unbiased mapping of transcription factor binding sites along human chromosomes 21 and 22 points to widespread regulation of noncoding RNAs. Cell 116:499–509 [DOI] [PubMed] [Google Scholar]
- 87.Rhee HS, Pugh BF. 2011. Comprehensive genome-wide protein-DNA interactions detected at single-nucleotide resolution. Cell 147:1408–1419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Rhee HS, Pugh BF. 2012. Genome-wide structure and organization of eukaryotic pre-initiation complexes. Nature 483:295–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mavrich TN, Ioshikhes IP, Venters BJ, Jiang C, Tomsho LP, Qi J, Schuster SC, Albert I, Pugh BF. 2008. A barrier nucleosome model for statistical positioning of nucleosomes throughout the yeast genome. Genome Res. 18:1073–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Venters BJ, Pugh BF. 2009. A canonical promoter organization of the transcription machinery and its regulators in the Saccharomyces genome. Genome Res. 19:360–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Murray SC, Serra Barros A, Brown DA, Dudek P, Ayling J, Mellor J. 2012. A pre-initiation complex at the 3′-end of genes drives antisense transcription independent of divergent sense transcription. Nucleic Acids Res. 40:2432–2444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ang K, Ee G, Ang E, Koh E, Siew WL, Chan YM, Nur S, Tan YS, Lehming N. 2012. Mediator acts upstream of the transcriptional activator Gal4. PLoS Biol. 10:e1001290. 10.1371/journal.pbio.1001290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lipford JR, Smith GT, Chi Y, Deshaies RJ. 2005. A putative stimulatory role for activator turnover in gene expression. Nature 438:113–116 [DOI] [PubMed] [Google Scholar]
- 94.Li XY, Bhaumik SR, Green MR. 2000. Distinct classes of yeast promoters revealed by differential TAF recruitment. Science 288:1242–1244 [DOI] [PubMed] [Google Scholar]
- 95.Orphanides G, Lagrange T, Reinberg D. 1996. The general transcription factors of RNA polymerase II. Genes Dev. 10:2657–2683 [DOI] [PubMed] [Google Scholar]
- 96.Boeger H, Bushnell DA, Davis R, Griesenbeck J, Lorch Y, Strattan JS, Westover KD, Kornberg RD. 2005. Structural basis of eukaryotic gene transcription. FEBS Lett. 579:899–903 [DOI] [PubMed] [Google Scholar]
- 97.Pardee TS, Bangur CS, Ponticelli AS. 1998. The N-terminal region of yeast TFIIB contains two adjacent functional domains involved in stable RNA polymerase II binding and transcription start site selection. J. Biol. Chem. 273:17859–17864 [DOI] [PubMed] [Google Scholar]
- 98.Li Y, Flanagan PM, Tschochner H, Kornberg RD. 1994. RNA polymerase II initiation factor interactions and transcription start site selection. Science 263:805–807 [DOI] [PubMed] [Google Scholar]
- 99.Kornberg RD. 2005. Mediator and the mechanism of transcriptional activation. Trends Biochem. Sci. 30:235–239 [DOI] [PubMed] [Google Scholar]
- 100.Myers LC, Kornberg RD. 2000. Mediator of transcriptional regulation. Annu. Rev. Biochem. 69:729–749 [DOI] [PubMed] [Google Scholar]
- 101.Kang JS, Kim SH, Hwang MS, Han SJ, Lee YC, Kim YJ. 2001. The structural and functional organization of the yeast mediator complex. J. Biol. Chem. 276:42003–42010 [DOI] [PubMed] [Google Scholar]
- 102.Koh SS, Ansari AZ, Ptashne M, Young RA. 1998. An activator target in the RNA polymerase II holoenzyme. Mol. Cell. 1:895–904 [DOI] [PubMed] [Google Scholar]
- 103.Huarte M, Rinn JL. 2010. Large non-coding RNAs: missing links in cancer? Hum. Mol. Genet. 19:R152–R161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, Wang Y, Brzoska P, Kong B, Li R, West RB, van de Vijver MJ, Sukumar S, Chang HY. 2010. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 464:1071–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Aguilo F, Zhou MM, Walsh MJ. 2011. Long noncoding RNA, polycomb, and the ghosts haunting INK4b-ARF-INK4a expression. Cancer Res. 71:5365–5369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Guttman M, Rinn JL. 2012. Modular regulatory principles of large non-coding RNAs. Nature 482:339–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kuras L, Kosa P, Mencia M, Struhl K. 2000. TAF-Containing and TAF-independent forms of transcriptionally active TBP in vivo. Science 288:1244–1248 [DOI] [PubMed] [Google Scholar]
- 108.Garbett KA, Tripathi MK, Cencki B, Layer JH, Weil PA. 2007. Yeast TFIID serves as a coactivator for Rap1p by direct protein-protein interaction. Mol. Cell. Biol. 27:297–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Layer JH, Miller SG, Weil PA. 2010. Direct transactivator-transcription factor IID (TFIID) contacts drive yeast ribosomal protein gene transcription. J. Biol. Chem. 285:15489–15499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Xu Z, Wei W, Gagneur J, Perocchi F, Clauder-Münster S, Camblong J, Guffanti E, Stutz F, Huber W, Steinmetz LM. 2009. Bidirectional promoters generate pervasive transcription in yeast. Nature 457:1033–1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kaplan N, Moore IK, Fondufe-Mittendorf Y, Gossett AJ, Tillo D, Field Y, LeProust EM, Hughes TR, Lieb JD, Widom J, Segal E. 2009. The DNA-encoded nucleosome organization of a eukaryotic genome. Nature 458:362–366 [DOI] [PMC free article] [PubMed] [Google Scholar]