Abstract
Retinoids play key roles in development, differentiation, and homeostasis through regulation of specific target genes by the retinoic acid receptor/retinoid X receptor (RAR/RXR) nuclear receptor complex. Corepressors and coactivators contribute to its transcriptional control by creating the appropriate chromatin environment, but the precise composition of these nuclear receptor complexes remains to be elucidated. Using an RNA interference-based genetic screen in mouse F9 cells, we identified the transcriptional corepressor CTBP2 (C-terminal binding protein 2) as a coactivator critically required for retinoic acid (RA)-induced transcription. CTBP2 suppression by RNA interference confers resistance to RA-induced differentiation in diverse murine and human cells. Mechanistically, we find that CTBP2 associates with RAR/RXR at RA target gene promoters and is essential for their transactivation in response to RA. We show that CTBP2 is indispensable to create a chromatin environment conducive for RAR/RXR-mediated transcription by recruiting the histone acetyltransferase p300. Our data reveal an unexpected function of the corepressor CTBP2 as a coactivator for RAR/RXR in RA signaling.
INTRODUCTION
Retinoic acid (RA), the active metabolite of vitamin A, is essential for proper embryonic and adult development. The physiological functions of RA are exerted primarily through its ability to differentially regulate gene expression mediated by the retinoic acid receptors (RARs). RARs are nuclear hormone receptors that function as ligand-dependent transcription factors (1). Their activity requires heterodimerization with the retinoid X receptors (RXRs), which can also associate with several other nuclear hormone receptors. RA signaling through RAR/RXR and the subsequent activation of target genes induce differentiation, cell cycle arrest, and apoptosis in many cell types. Consequently, RA displays distinct anticarcinogenic activities and is currently used and being tested as a therapeutic agent for several human cancers (2, 3).
Gene expression through retinoic acid receptors is regulated through changes in chromatin structure facilitated by chromatin remodeling and modifying complexes (4–7). In the classical view of RA-mediated gene activation, RAR/RXR heterodimers constitutively associate with retinoic acid response elements (RAREs) on promoters of target genes (8). In the absence of ligand, RAR/RXR actively represses transcription through association with corepressors NCoR and SMRT and recruitment of histone deacetylases (HDACs) that prevent opening of the chromatin (9, 10). RA binds to RAR and triggers conformational changes that release the corepressors and in turn promote the assembly of coactivator complexes. Subsequently, transcription of target genes is initiated. Many of the coactivators, including CBP/p300, PCAF, and SRC1 to -3 (NCOA1 to -3), possess histone acetyltransferase activity that promotes transactivation by RAR/RXR (4, 11). In contrast, ligand-dependent corepressors such as LCoR and PRAME recruit HDACs or PcG proteins to ligand-bound RAR/RXR complexes to repress their activities (12–14). Therefore, coactivators/repressors play crucial roles for the context-dependent action of RA. This classical model of gene regulation has been evolving as data from genome-wide studies accumulate. Current models of RA-regulated gene expression emphasize the dynamic nature of corepressor and coactivator complexes and the important role played by histone modifications in maintaining gene repression/activation (15).
The C-terminal binding protein (CTBP) family of proteins are important corepressors involved in several essential cellular processes (16). Usually CTBPs associate with DNA binding transcription factors to repress their targets (17). Vertebrates contain genes that code for two related proteins called CTBP1 and CTBP2. CTBP1, the founding member of the CTBP family, was originally identified and named for its ability to bind the carboxyl terminus of the transforming E1A protein of adenoviruses (18–20). CTBP1 and CTBP2, although very homologous, also exhibit some unique characteristics (21). Most of the roles of CTBP2 described so far are consistent with a major role as a classical corepressor protein.
Functional genetic screens provide a powerful tool to identify novel components of signaling pathways (22, 23). We describe here the use of a large-scale loss-of-function genetic screen to identify genes whose suppression can confer resistance to RA-induced differentiation. Through this work, we identify an unexpected function of CTBP2 as a potent coactivator of RAR/RXR in RA signaling.
MATERIALS AND METHODS
Plasmids.
All retroviral short hairpin RNA (shRNA) vectors were generated by ligating synthetic oligonucleotides (Invitrogen) against the target genes into in the pRetroSuper (pRS) retroviral vector as described previously (24). The following RNA interference (RNAi) target sequences were used for retroviral shRNA vectors for this study: shGFP, GCTGACCCTGAAGTTCATC; shCtbp2_Libr (#1), GTGATCGTGCGAATCGGTA; shCtbp2_A, ACGACAACGTGGACATCAA; shCtbp2_B, TGCAGGACTTGCTATATCA; shCtbp2_C (9), GAACAAGCATCACTAGAGA; shCtbp2_D, GGATCGCTGTGTGCAACAT; shRxrα_Libr, CAAGAGGACAGTACGCAAA; shRxrα#1, AGGACTGCCTGATCGACAA; shRxrα#2, GAAGCGTACTGCAAACACA; shCTBP2#1, GCTATGACAACGTGGACAT; shCTBP2#2, CAATGGTGCCACATACAGA; shRXRα#1, GGACCGGAACGAGAATGAG; and shRXRα#2, CGAACGACCCTGTCACCAA.
All lentiviral shRNA vectors were retrieved from the arrayed TRC (The RNAi Consortium) human genome-wide shRNA collection (TRC-Hs1.0). Additional information about the shRNA vectors can be found at http://www.broadinstitute.org/rnai/public/clone/search using the TRCN number. The following lentiviral shCtbp2 and shCtbp1 vectors were used: shCtbp2#3, TRCN0000109336; shCtbp2#4, TRCN0000109339; shCtbp1#1, TRCN0000085773; and shCtbp1#2, TRCN0000085774.
The human CTBP2 and mouse Ctbp2 expression constructs were generated by the following steps. In step 1, the human CTBP2 and mouse Ctbp2 open reading frames were amplified by PCR using a human MCF-7 cDNA library and a mouse embryo D14 cDNA library and the following primers: CTBP2_5′, ATCCCCCGGGACCATGGCCCTTGTGGATAAGCAC; CTBP2_3′, ATGCTCTAGATTCTCTGCTATTGCTCGTTGG; Ctbp2_5′, ATCCCCCGGGACCATGGCCCTTGTGGATAAGCAC; and Ctbp2_3′, ATGCTCTAGATTCTCTGCTATTGCTCGTTGG.
In step 2, the PCR products were cloned into pcDNA3.1(+) vector by EcoRV and XbaI restriction sites and the sequences were verified.
The expression construct for human p300 was a kind gift from M. Epping.
Reagents and antibodies.
All-trans-retinoic acid (all-trans-RA) and the alkaline phosphatase (AP) staining kit were from Sigma (R2625 and 86R). The following antibodies were used: anti-p300 (C-20, Sc585; Santa Cruz), histone H3 (ab1791; Abcam) and H3 acetyl antibodies (06-599; Upstate Millipore), normal rabbit IgG (Santa Cruz), anti-RXRα (D-20; Santa Cruz), anti-CTBP2 (612044; BD Transduction Laboratories), and anti-CD11b-fluorescein isothiocyanate (FITC) antibody (F2648; Sigma). siRNA for mouse Ctbp2 was purchased from Thermo Scientific (#MU-059787-01-0002; SiGenome mouse Ctbp2).
shRNA bar code screen.
The NKI mouse shRNA library (containing 28,256 shRNA vectors that target 14,128 mouse genes) was constructed into the pRISC retroviral vector, which is derived from pRetroSuper with an additional chloramphenicol resistance marker under regulation of the TET promoter (24). The bar code screen was performed as described previously (25, 26). Additional details can be found at www.screeninc.nki.nl.
Long-term cell proliferation assays.
Cells were seeded into 6-well plates (1 × 104 to 2 ×104 cells/well) and cultured in both the absence and presence of RA as indicated. More details are described in reference 23. All knockdown experiments were done by retroviral infection. The growth curves were performed according to the standard 3T3 protocol. All relevant assays were performed independently at least three times.
Western blotting and coimmunoprecipitation.
The Western blotting and coimmunoprecipitation experiments were performed according to the protocols described previously (27, 28).
Transfections and reporter assays.
Transfections were carried out using calcium phosphate precipitation. RARE-luciferase reporter assays were performed in Dulbecco's modified Eagle's medium (DMEM) supplemented with charcoal-stripped fetal calf serum (FCS) (HyClone, Logan, UT) essentially as described previously (29).
Cell culture and viral transduction.
The mouse F9 and human NTERA2 embryonic teratocarcinoma cell lines were purchased from the American Type Culture Collection (ATCC; Manassas, VA). E14T mouse embryonic stem (ES) cells were kindly obtained from A. Smith (Cambridge, United Kingdom). The HL-60 subclone expressing the murine ecotropic receptor is from the laboratory collection of R. Bernards.
The culture conditions and retroviral infection of E14T mouse ES cells are described in reference 23. HL-60 cells were cultured in RPMI with 8% heat-inactivated fetal bovine serum, penicillin, and streptomycin at 5% CO2. All other cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 8% heat-inactivated fetal calf serum (FCS), penicillin, and streptomycin. F9 cells were cultured on gelatin-treated tissue culture plastic.
Phoenix cells were used as producers of retroviral supernatants as described at http://www.stanford.edu/group/nolan/retroviral_systems/phx.html.
HEK 293T cells were used as producers of lentiviral supernatants as described at http://www.broadinstitute.org/rnai/public/resources/protocols.
The calcium phosphate method was used for the transfection of Phoenix and 293T cells. Infected cells were selected for successful retroviral integration using 2 μg/ml of puromycin.
Flow cytometric analysis.
Flow cytometric analysis was carried out according to the standard protocol. Briefly, HL-60 cells were incubated with anti-CD11b-FITC antibody according to the manufacturer's protocol for 20 min on ice, washed three times with RPMI medium, and analyzed by the FACSCalibur system (Becton, Dickinson), and the data were analyzed using CellQuest software (Becton, Dickinson).
Quantitative RT-PCR (qRT-PCR).
Quantitative reverse transcription-PCR (qRT-PCR) assays were carried out to measure mRNA levels of genes using the 7500 fast real-time PCR system (Applied Biosystems) as described previously (30). Relative mRNA levels of each gene shown were normalized to the expression of the housekeeping gene GAPDH (coding for glyceraldehyde-3-phosphate dehyrdrogenase). The sequences of the primers for assays using SYBR green master mix (Roche) are as follows: Gapdh_Forward, AGGTCGGTGTGAACGGATTTG; Gapdh_Reverse, TGTAGACCATGTAGTTGAGGTCA; Ctbp1_Forward, GTGCCCTGATGTACCATACCA; Ctbp1_Reverse, TGATGTCGATATTGTCAAACCCG; Ctbp2_Forward, GGCAGCGATTGGACAGGATTT; Ctbp2_Reverse, AGGATGGGCATCTCCACAGT; Rxrα_Forward, ATGGACACCAAACATTTCCTGC; Rxrα_Reverse, CCAGTGGAGAGCCGATTCC; Cyp26a1_Forward, AAGCTCTGGGACCTGTACTGT; Cyp26a1_Reverse, CTCCGCTGAAGCACCATCT; Crabp2_Forward, CCTCCACCACTGTGCGAAC; Crabp2_Reverse, ACTCTCCCATTTCACCAAACTCT; Hoxb5_Forward, CCTTCTCGGGGCGTTATCC; Hoxb5_Reverse, CCTGAAGCGGGGTTCCTTG; Cyp26a1_RARE_Forward, TTCACTGAGATGTCACGGTCC; Cyp26a1_RARE_Reverse, TTCCCAATCCTTTAGCCTGA; Crabp2_RARE_Forward, AACGTTGTTTCTGGCTTTGG; Crabp2_RARE_Reverse, CTCCGAGGCTGATTTTTACG; Hoxb5_RARE_Forward, TCCCCCTTCTCACTTTCTCC; Hoxb5_RARE_Reverse, GGGAATCACGTGCTTTTGTT; Cyp26a1_6kbds_Forward, CAGATGTGTCACTGGGGATG; Cyp26a1_6kbds_Reverse, TTGTCACGAGGAGACAGCAC; Hoxb5_6kbds_Forward, CAGATGTGTCACTGGGGATG; Hoxb5_6kbds_Reverse, TTGTCACGAGGAGACAGCAC; GAPDH_Forward, AAGGTGAAGGTCGGAGTCAA; GAPDH_Reverse, AATGAAGGGGTCATTGATGG; CTBP2_Forward, TTAAAAAGACGGACAGCCCA; CTBP2_Reverse, AGGGGAACTTGCAGGAGTCT; RXRα_Forward, GGACTGCCTGATTGACAAGC; RXRα_Reverse, TTCAGCCCCATGTTTGCCTC; CYP26A1_Forward, TCTGATCACTTACCTGGGGC; CYP26A1_Reverse, TTCCAAAATTTCCATGTCCAA; RARβ_Forward, TGAGTCCTGGGCAAATCCTG; and RARβ_Reverse, CGGTTTGGGTCAATCCACTGA.
ChIP.
Chromatin immunoprecipitation (ChIP) was performed as described in reference 31 F9 cells at 80% confluence were cross-linked with 1% formaldehyde in medium for 10 min at room temperature. After quenching of formaldehyde with 125 mM glycine, whole-cell extracts were prepared for use in chromatin immunoprecipitation assays as described. The chromatin was sonicated to get fragment sizes of around 400 to 1,000 bp as analyzed on agarose gels. Four hundred micrograms of fragmented chromatin was used in immunoprecipitations with IgG, CTBP2 (612044; BD Transduction Laboratories), p300 (C-20, Sc585), H3 (ab1791), or H3 acetyl antibodies (06-599), and DNA of the immunoprecipitates and control input DNA (4 μg fragmented chromatin) were analyzed by regular and real-time PCR. The sequences of the primer sets used for ChIP-qPCR analysis are listed in the supplemental material.
In vitro binding assays.
Plasmids encoding glutathione S-transferase (GST) fusions of RXRα, RARα, and peroxisome proliferator-activated receptor γ (PPARγ) were expressed and purified from bacteria using standard protocols. 35S-labeled Ctbp2 and Rxr were generated using the Promega TNT coupled transcription-translation system as described by the manufacturer. Two microliters of the reaction mixture was incubated with the glutathione-Sepharose beads loaded with GST alone or GST-RXRα, GST-RARα, or GST-PPARγ (23) for 2 h in a cold room in buffer containing 25 mM HEPES-KOH (pH 7.6), 0.1 mM EDTA, 12.5 mM MgCl2, 10% glycerol, 0.1% NP-40, and 150 mM KCl. After 5 washes with the same buffer, the beads were boiled in sample buffer and separated by 10% SDS-PAGE. Bound proteins were detected using autoradiography.
RNA-Seq gene expression analysis.
Total mRNA of each sample was converted into a library of template molecules suitable for subsequent cluster generation using the reagents provided in the Illumina TruSeq RNA sample preparation kit, following the manufacturer's protocol. Sequence reads were generated using Illumina HiSeq 2000 with TruSeq v3 reagent kits and software.
The reads (between 13 and 25 million 51-bp single-end reads per sample) were mapped to the mouse reference genome (build 37) using TopHat (v.1.4.0) (32) which allows the spanning of exon-exon splice junctions. The open-source tool HTSeq-count (EMBL, v.0.5.3p3) was then used to generate a list of the total number of uniquely mapped reads (between 7 and 15 million reads per sample) for each gene that is present in the provided Gene Transfer Format (GTF) file (Ensembl version 64). Of those reads, over 95% were assigned to protein coding genes.
A random sampling model in the R package DEGseq (33) was then used to determine which genes were differentially expressed between the samples, taking a P value of 0.05 as a cutoff. The input of this method is the absolute number of reads for a gene, which is the output of HTSeq-count. Genes with no expression for all samples in the comparison were discarded from the data set. The expression levels of the remaining genes were added to 1 in order to avoid negative values after log2 transformation during the normalization step within this method. Normalization to 10 million reads was performed.
Genes were considered differentially regulated if they showed at least a 2-fold change between the two tested conditions. Specifically, genes were defined as upregulated when the log2-transformed ratio of the absolute number of reads between the two compared conditions was ≥1. Genes with a log2-transformed ratio of ≤−1 were considered downregulated; the remaining genes were defined as unchanged. We employed the hypergeometric test to determine the significance in the overlap analyses.
Microarray data accession number.
The gene expression data for F9 cells expressing shCtbp2, shRxrα, or the shGFP control are available at the NCBI Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) under accession no. GSE47708.
RESULTS
A large-scale RNA interference bar code screen identifies Ctbp2 as a critical regulator of RA signaling.
The mouse F9 embryonic teratocarcinoma cell line is highly sensitive to RA and is used extensively as a model to investigate RA signaling. We have used this model system previously to identify novel components of the RA signaling pathway through large-scale RNA interference-based genetic screens (23). In this study, we used a collection of 28,256 short hairpin (shRNA) vectors targeting 14,128 mouse genes to identify genes whose suppression causes resistance to RA. In that study, the shRNA inserts were recovered by PCR from individual RA-resistant colonies and subjected to DNA sequence analysis to reveal their identities. Using this time-consuming approach, we identified Zfp423 as a new component of RA signaling (23).
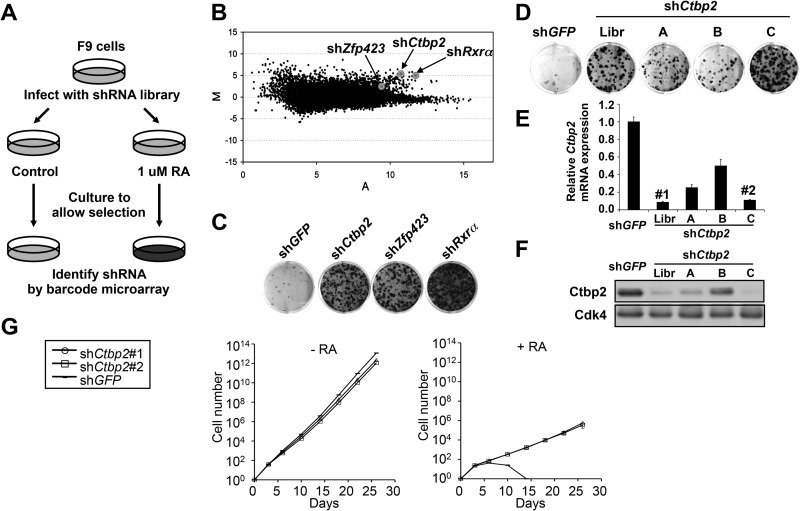
In the present study, we repeated the genetic screen for genes whose suppression confers resistance to RA in mouse F9 cells, now using the far more efficient bar coding approach to identify genes that confer RA resistance (26). Using retroviral infection, we introduced the entire 28,256-member shRNA library polyclonally into F9 cells and plated the cells at low density with or without 1 μM all-trans-RA (Fig. 1A). After 4 weeks of incubation with RA, genomic DNA was isolated from both cultures. The stably integrated shRNA vectors were then recovered by PCR amplification. The relative abundance of individual shRNA vectors was quantified by hybridization of the PCR products to microarrays harboring all 28,256 bar code sequences. The bar code screen was performed in triplicate, and the combined results are shown in Fig. 1B. Each dot in the M/A plot represents one individual shRNA vector in the library. M and A values reflect relative enrichment and hybridization signal intensity. Reproducible outliers that confer drug resistance are located in the right upper corner of the M/A plot. Therefore, we prioritized our candidates by applying M/A cutoff values (M > 4.0 and A > 10.0). Among the 24 candidates that met these criteria, we identified a vector targeting Rxrα (Fig. 1B), which validates the screen, as RXR is an obligatory heterodimerization partner of RAR. The vector targeting Zfp423 did not meet these stringent criteria, but it was nevertheless positively selected (Fig. 1B). Each of the selected genes was then retested for the ability to confer resistance to RA. Of the shRNA vectors tested, the strongest effects were seen with the positive control, shRxrα, and, to a lesser extent, shZfp423 (Fig. 1C). In addition, the shRNA targeting Ctbp2 was also able to confer resistance to RA. Empty vector (pRS) and vector targeting the green fluorescent protein gene (shGFP) served as controls throughout the study.
Fig 1.
A genome-wide RNAi screen identifies Ctbp2 as a component of the RA signaling pathway. (A) Schematic outline of the RA resistance screen performed in mouse F9 cells. Mouse shRNA library polyclonal virus was produced to infect F9 cells. The infected cells were treated with 1 μM all-trans-retinoic acid (RA) for 4 weeks, and shRNA inserts were recovered from the resistant colonies as described previously (25). (B) Analysis of the relative abundance of the recovered shRNA cassettes from the RA bar code experiment. Averaged data from three independent experiments were normalized and log2 transformed. Among the 24 top shRNA candidates (M > 4.0, A > 10.0), shCtbp2 and shRxrα were identified. shZfp423 (M = 9.3, A = 2.1) was also positively selected. (C) Individual shRNAs from the library targeting Rxrα, Ctbp2, and Zfp423 conferred RA resistance. F9 cells expressing shCtbp2, shRxrα, shZfp423, or the control, shGFP, were treated with 1 μM RA for 3 weeks, after which the cells were photographed, fixed, and stained. (D) Validation of independent shRNAs targeting Ctbp2. The functional phenotypes of nonoverlapping shRNAs targeting each gene are indicated by the colony formation assay in 1 μM RA. Lib, library. (E and F) The knockdown ability of each of the shRNAs was measured by examining the mRNA levels of the intended target gene by qRT-PCR (E) and the protein levels by Western blotting (F). Error bars represent standard deviations (SD) of triplicate independent experiments. (G) Proliferation curves according to the 3T3 protocol of F9 cells expressing shRNAs targeting Ctbp2 or GFP in the absence and presence of RA (1 μM). Error bars represent SD of triplicate independent experiments.
We focused on Ctbp2 as this gene is mostly known as a transcriptional repressor and we could not readily explain how loss of a repressor could confer resistance to RA (16). To validate Ctbp2, additional nonoverlapping shRNAs were generated against the gene and tested for their ability to confer RA resistance. Figure 1D to F show that four shCtbp2 vectors have a clear correlation between the degree of Ctbp2 knockdown of the vectors and their ability to confer RA resistance, validating that Ctbp2 is a genuine hit from the screen. Two of the most potent retroviral shRNAs, shCtbp2_Libr and shCtbp2_C, are referred to as shCtbp2#1 and -#2, respectively. For the remaining candidate genes tested, we did not observe a correlation between knockdown of the intended gene and the ability to confer resistance to RA when additional shRNA vectors were tested (data not shown), and these genes were not studied further.
Next, we examined whether the RA resistance mediated by suppression of Ctbp2 is due to the inhibition of RA signaling or due to a general growth advantage unrelated to RA. We performed a long-term proliferation assay of F9 cells expressing shCtbp2 vectors (#1 and #2) or the control shGFP in the absence or presence of exogenous RA, using the 3T3 protocol (Fig. 1G). In the absence of RA, no significant growth difference was detected in all cell lines. When exposed to 1 μM RA, the cells expressing shRNAs targeting Ctbp2 continued to proliferate, while the control cells were drastically inhibited. These data argue against a generalized growth advantage for the Ctbp2 knockdown F9 cells and suggest a specific resistance to RA-induced growth arrest.
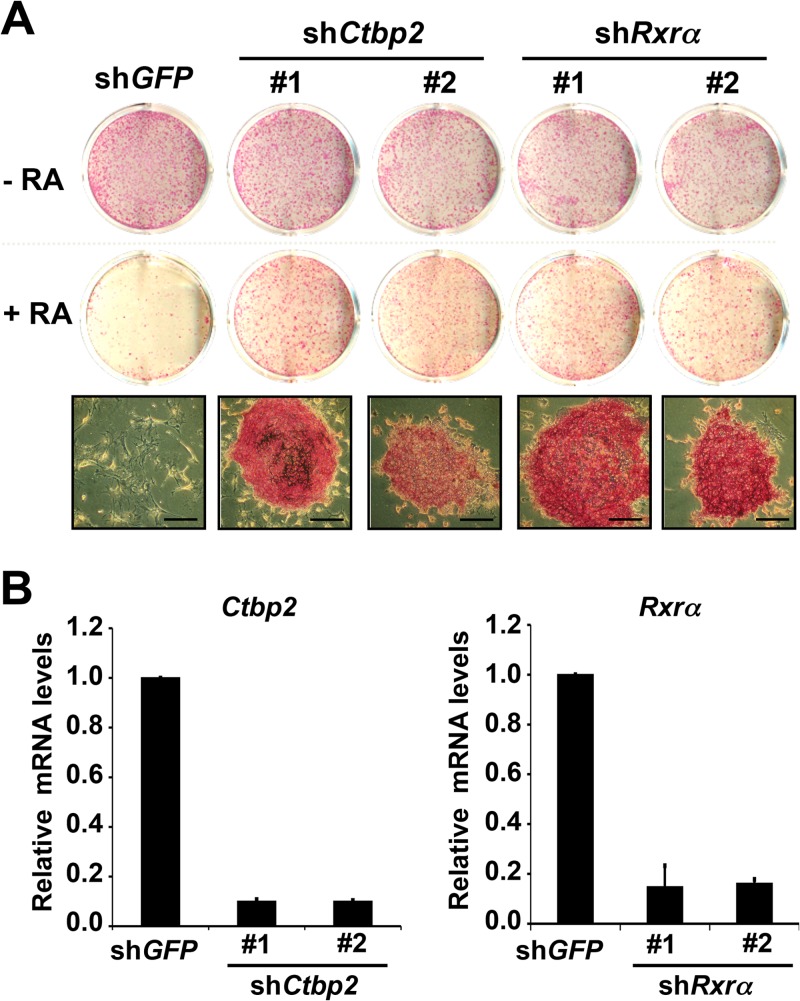
In addition to F9 cells, we also tested whether Ctbp2 is required for the differentiation of the mouse embryonic stem (ES) cells. Mouse ES cells are very responsive to retinoic acid and undergo differentiation when treated with RA. We introduced shRNAs targeting Ctbp2, Rxrα, or GFP into E14T mouse ES cells by retroviral infection. The knockdown efficiencies of these shRNAs in E14T cells were comparable to those seen in F9 cells (Fig. 2B). The infected cells were plated at low density and cultured in the absence or presence of 1 μM retinoic acid for a week. The surviving cells were stained for alkaline phosphatase (AP), which is a marker of undifferentiated ES cells. As shown in Fig. 2A, there were no significant changes in growth and maintenance of the undifferentiated cells in the absence of RA. However, in the presence of RA, proliferation of control cells was severely compromised and the remaining differentiated cells failed to stain positive for AP. In contrast, cells stably expressing shRNA against Ctbp2 or Rxrα continued to proliferate and stained positively for AP.
Fig 2.
Ctbp2 is also required for RA-induced differentiation in mouse ES cells. (A) Downregulation of Ctbp2 by RNAi in E14T mouse ES cells confers resistance to RA-induced differentiation. E14T cells expressing shRNAs against Ctbp2, Rxrα, or GFP (control) were grown in the absence or presence of RA (1 μM) for 1 week, after which cells were fixed, stained for alkaline phosphatase (AP), and photographed. Bars, 50 mm. (B) mRNA levels of Ctbp2and Rxrα in E14T mouse ES cells expressing shRNAs targeting Ctbp2, Rxrα, or GFP. Error bars denote SD. (Control images reprinted from reference 23 with permission of the publisher.)
Ctbp2 but not Ctbp1 is required for RA-induced differentiation.
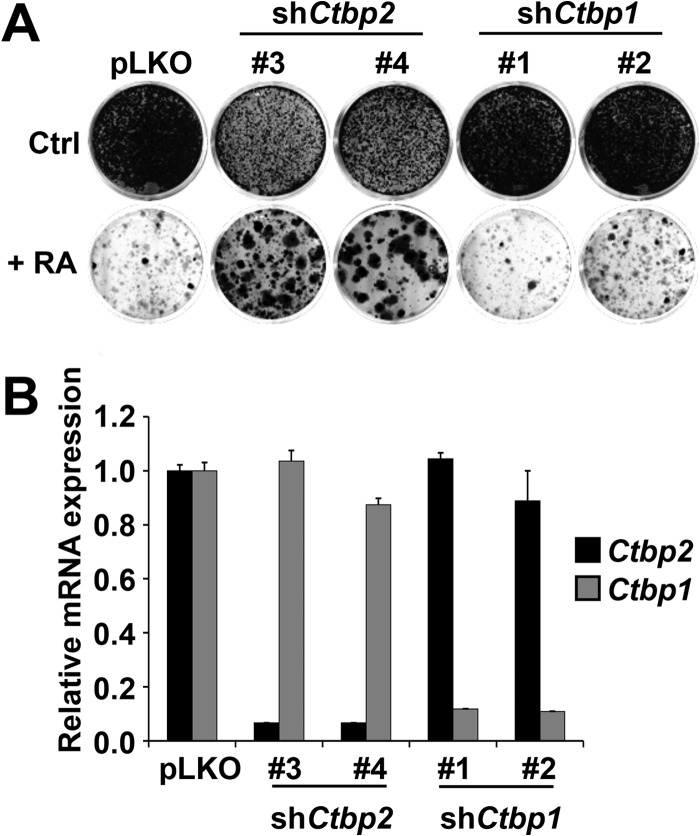
The vertebrate genome codes for two related CTBPs referred to as CTBP1 and CTBP2 (16). These two proteins appear to have redundant as well as unique functions, as judged by mouse knockout studies (21). To address whether Ctbp1 is also involved in RA signaling, we targeted Ctbp1 in F9 cells using lentiviral shRNA vectors (Fig. 3). Empty vector pLKO and lentiviral shRNAs targeting Ctbp2 (#3 and #4) were used as controls. As shown in Fig. 3A, all of the shCtbp1 vectors failed to confer RA resistance and Ctbp1 knockdown also had no effect on RA target gene induction when expressed, even though the knockdown efficiency of Ctbp1 was around 90% for all vectors (Fig. 3B). In contrast, knockdown of Ctbp2 conferred RA resistance. These results indicate that Ctbp2, but not Ctbp1, is required for RA-induced differentiation.
Fig 3.
Ctbp1 is not required for RA-induced differentiation. (A) Two different nonoverlapping shRNAs against Ctbp1 did not confer resistance in the presence of 1 μM RA in a colony formation assay. Two independent shRNAs against Ctbp2 conferred resistance. The cells were plated at low density and grown for 3 weeks, after which the cells were photographed, fixed, and stained. Ctrl, control. (B) The knockdown ability of each of the shRNAs against Ctbp1 and Ctbp2 was measured by examining the mRNA levels of the intended target gene by qRT-PCR. Error bars denote SD.
CTBP2 is a transcriptional cofactor of RA receptors.
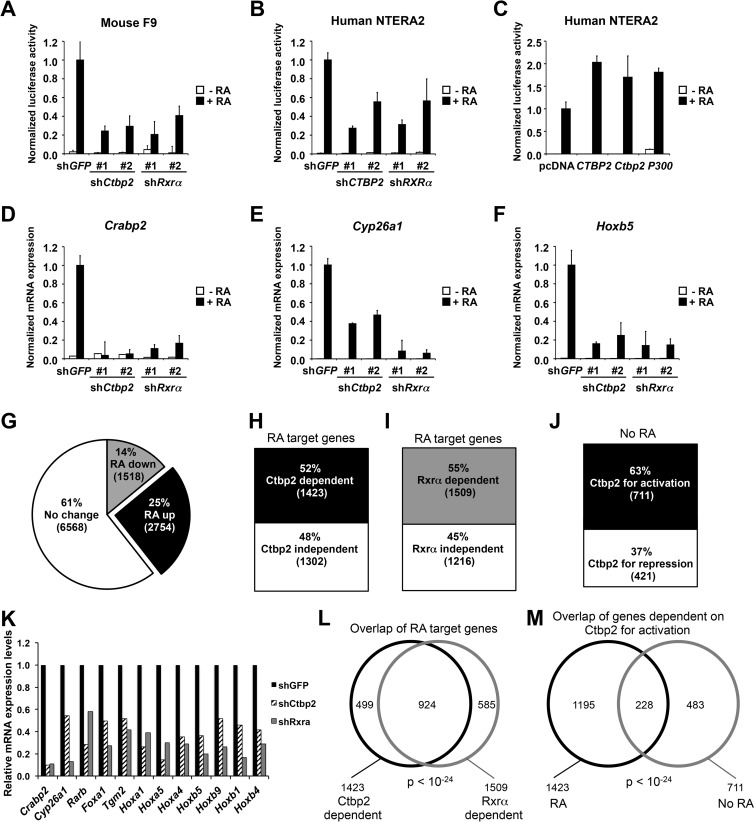
Since suppression of Ctbp2 impaired RA-induced differentiation, we reasoned that Ctbp2 could play a positive role in Rar/Rxr-mediated transactivation. To address this hypothesis, shRNA vectors targeting Ctbp2, Rxrα, and GFP were cotransfected with a reporter gene containing consensus retinoic acid response elements (RAREs) linked to luciferase (RARE-Luc) into F9 cells. Both shCtbp2 vectors impaired the reporter gene activation by RA to a similar extent, as seen with the shRNAs targeting Rxrα, indicating that Ctbp2 is important for transcriptional activation of Rar/Rxr in response to RA (Fig. 4A). As an independent cellular system, we performed the RARE-Luc reporter assays in NTERA2 human embryonic teratocarcinoma cells. As shown in Fig. 4B, knockdown of CTBP2 also impaired the RA-induced expression of the same reporter construct in these cells. Conversely, overexpression of CTBP2 hyperactivated the RARE-Luc reporter in response to RA. A similar result was observed with expression of the histone acetyltransferase p300, a known coactivator of RARα/RXRα (Fig. 4C). These results suggest that CTBP2 functions as a coactivator for RARα/RXRα transactivation.
Fig 4.
CTBP2 is a transcriptional cofactor for RXRα/RARα. (A to C) Activation of a RARE-luciferase (RARE-Luc) reporter gene by RXRα/RARα requires CTBP2. The normalized luciferase activities shown represent ratios between luciferase values and Renilla internal control values and are the averages ± SD from three independent transfections. (A) Downregulation of Ctbp2 by RNAi inhibited the transcriptional induction of the RARE-Luc reporter gene in mouse F9 cells in response to 24 h of 1 μM RA stimulation. (B) Suppression of CTBP2 by RNAi inhibited the activation of the RARE-Luc reporter gene in human NTERA2 cells in response to 1 μM RA treatment. (C) Overexpression of human CTBP2, mouse Ctbp2, and human p300 histone acetyltransferase was able to hyperactivate the luciferase expression induced by 1 μM RA in the NTERA2 cell line. (D to F) Ctbp2 is required for transcriptional regulation of endogenous RA target genes in response to RA. Shown are the results from mRNA expression analysis of RA target genes Crabp2 (D), Cyp26a1 (E), and Hoxb5 (F) in F9 cells expressing shRNAs targeting Ctbp2, Rxrα, or GFP cultured in the absence or presence of 1 μM RA for 48 h. Error bars represent SD of triplicate independent experiments. (G) Pie chart showing the results from the RNA sequencing analysis in F9 shGFP control cells cultured in the absence or presence of RA for 48 h. Twenty-five percent (2,754) of the genes are upregulated in an RA-dependent manner, 14% (1,518) of the genes are downregulated in an RA-dependent manner, and 61% (6,568) of the genes remain unchanged. (H and I) Effect of Ctbp2 and Rxr knockdown on RA-upregulated genes by whole-genome transcriptome analysis in F9 cells. A bar chart shows that 52% of the upregulated genes are dependent on Ctbp2 (H) and 55% of the genes are dependent on Rxr (I) for transcription. (J) Ctbp2-dependent genes for activation and repression in the absence of RA (J). (K) Representation of the major bona fide RA target genes and their dependence on Ctbp2 and Rxrα for activation. (See Materials and Methods for details.) (L and M) Venn diagram representation of the overlap of genes between Ctbp2 and Rxr dependency (L). Shown is the overlap (228 genes) of Ctbp2-dependent genes for activation (711 genes) with the RA-dependent Ctbp2-dependent genes for activation (1,423 genes) (M).
Next, we analyzed the effect of Ctbp2 knockdown on mRNA expression levels of a panel of bona fide RA target genes in F9 cells, including Cyp26a1, Crabp2, and Hoxb5. As expected, these target genes were strongly induced in the control cells following exposure to RA (Fig. 4D to F). However, induction of these genes was significantly inhibited in the cells expressing shCtbp2 or shRxrα. To extend this study further on a genome-wide scale, we performed transcriptome gene expression analysis in F9 cells expressing shCtbp2, shRxrα, or the shGFP control cultured in absence or presence of RA for 48 h, using next-generation sequencing (RNA-Seq).
A total of 2,754 genes were found to be upregulated (>2-fold) and 1,518 genes were downregulated (>2-fold) in response to RA treatment in the control cells (Fig. 4G; see Table S2 in the supplemental material). To examine the effect of Ctbp2 on transactivation by the Rar/Rxr complex, we focused on the 2,754 upregulated RA target genes in shGFP control cells, of which 2,725 could also be mapped to the sequence data from cells expressing shCtbp2 or shRxrα (see Table S3 in the supplemental material). Consistent with the notion that Ctbp2 functions as a coactivator for Rar/Rxr transactivation, totals of 1,423 (52%) (see Table S4 in the supplemental material) and 1,509 (55%) (see Table S5 in the supplemental material) genes out of 2,725 genes were dependent on Ctbp2 and Rxrα, respectively, for activation (Fig. 4H and I). Importantly, ∼65% of the genes overlapped between Ctbp2 and Rxr knockdown (P < 10−24), indicating that the majority of genes that depend on Rxr for transactivation in response to RA also require Ctbp2 as a cofactor (Fig. 4L; see Table S6 in the supplemental material). As expected, many of the bona fide RA target genes require both Ctbp2 and Rxrα for activation in response to RA treatment (Fig. 4K). In addition to the shRNA-based knockdown used throughout this study, we also used siRNAs to transiently knock down Ctbp2 and obtained similar results (data not shown). These results further support the role of Ctbp2 as a transcriptional cofactor of RA receptors. We also looked at the effect of Ctbp2 knockdown on the overall gene expression changes in the absence of RA. Interestingly, out of 6,028 significantly expressed genes, a total of 711 genes were downregulated (genes dependent on Ctbp2 for activation) and 421 genes were upregulated (genes dependent on Ctbp2 for repression), suggesting that Ctbp2 predominantly serves as an activator of transcription (Fig. 4J; see Tables S7, S8, and S9 in the supplemental material). Furthermore, we looked at the overlap among the genes that require Ctbp2 for activation (a total of 711) with the RA pathway. Strikingly, 228 (32%) genes were indeed targets of RA signaling (Fig. 4M).
CTBP2 is also involved in RA-induced differentiation in human promyelocytic leukemia cells.
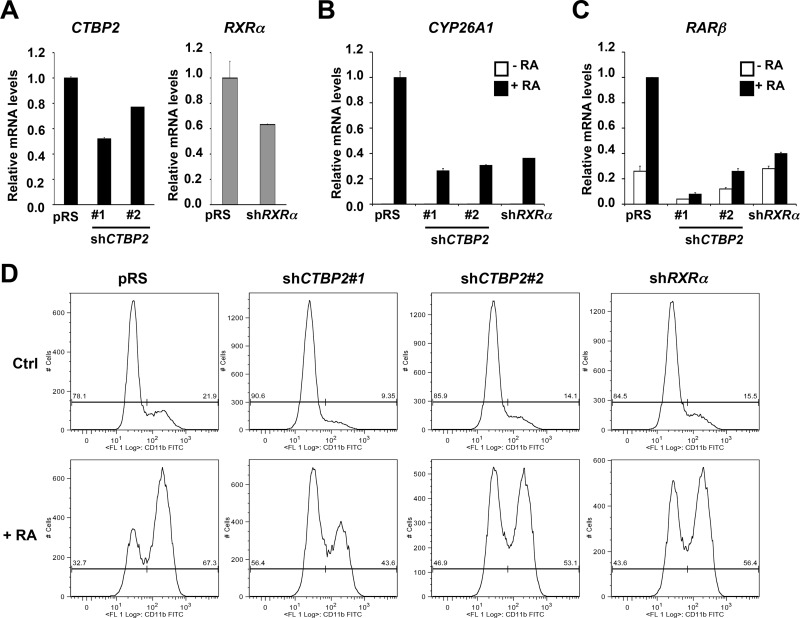
To further substantiate the role of CTBP2 in RA signaling, we asked if CTBP2 is also required for RA-induced differentiation in human HL-60 promyelocytic leukemia cells, another well-established model for studies of differentiation in response to RA. As expected, HL-60 control cells induced expression of RA target genes, such as CYP26A1 and RARβ, in response to RA treatment (Fig. 5B and C). In contrast, induction of these RA target genes in HL-60 cells expressing shRNA vectors targeting CTBP2 or RXRα was impaired (Fig. 5A to C), in spite of the fact that gene knockdown was modest due to the poor infection efficiency of these cells with retroviral vectors (Fig. 5A). This suggests that CTBP2 is also required for transcriptional regulation of RA target genes in human promyelocytic leukemia cells.
Fig 5.
CTBP2 is also involved in RA-induced differentiation of human promyelocytic leukemia cells. (A) mRNA expression of CTBP2 and RXRα in HL-60 cells expressing pRS control or vectors targeting CTBP2 or RXRα was examined by qRT-PCR. Error bars denote SD. (B and C) CTBP2 is required for transcriptional induction of RA target genes in HL-60 cells in response to RA. Shown are the results from mRNA expression analysis of CYP26A1 (B) and RARβ (C) in HL-60 cells expressing the pRS control or vectors targeting CTBP2 or RXRα cultured in the absence or presence of 1 μM RA for 48 h. Error bars denote SD. (D) CTBP2 is involved in RA-induced differentiation in human promyelocytic leukemia cells. Shown are results from HL-60 cells expressing the pRS control or vectors targeting CTBP2 or RXRα cultured in the absence or presence of 1 μM RA for 48 h. Flow cytometric analysis of CD11b expression was used to assess differentiation.
As a result of RA-induced differentiation, HL-60 cells acquire some of the characteristics of the granulocytic lineages, for example, in the appearance of CD11b myeloid surface marker. Flow cytometric analysis (fluorescence-activated cell sorter [FACS]) of CD11b expression in HL-60 cells shows the increase of CD11b-positive cells in response to RA treatment (Fig. 5D, bottom). This RA-induced increase of CD11b expression in HL-60 cells expressing shRNA vectors targeting CTBP2 or RXRα was significantly lower than that in the parental cells (Fig. 5D). We also observed the basal CD11b expression in normal medium (without exogenous RA) was also lower in cells expressing shCTBP2 or shRXRα than that in control cells (Fig. 5D, top). This is likely due to a small amount of RA in the serum component of the culture medium. Together, these results indicate that CTBP2 is also required for RA-induced differentiation in human leukemia cells.
Ctbp2 physically interacts with RA receptor complex and the RARE region of the RA target gene promoters.
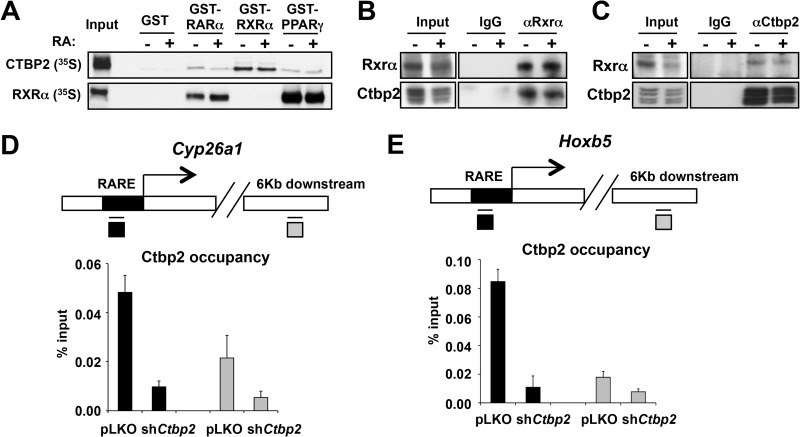
Having established that CTBP2 functions as a coactivator of RA receptors, we investigated a possible physical interaction between CTBP2 and RA receptors using in vitro GST pulldown and coimmunoprecipitation assays. 35S-labeled CTBP2 or RXR was incubated with GST fusions of RARα, RXRα, and PPARγ. As shown in Fig. 6A, CTBP2 preferentially interacted with GST-RXR and with lower affinity to GST-RAR and GST-PPARγ. Addition of the all-trans-RA to the in vitro reaction had no effect on the binding. These results suggest that CTBP2 may also be required for activation by other nuclear receptor combinations, such as the RXRα-PPARγ and RXRα-RXRα dimers. Indeed, loss of CTBP2 also impaired activation of peroxisome proliferator hormone response element (PPRE)-luciferase reporter as well as the DR1-luciferase reporter genes (data not shown).
Fig 6.
CTBP2 physically associates with RARα/RXRα and the RARE region of RA target gene promoters. (A) In vitro interaction of CTBP2 and RXRα. A GST pulldown assay was used to determine the binding between GST fusions of RARα or RXRα or PPARγ with 35S-labeled CTBP2. 35S-labeled CTBP2 or RXRα was incubated with equal amounts of GST fusion proteins with and without all-trans-RA at 4°C for 2 h in buffer containing 25 mM HEPES-KOH (pH 7.6), 0.1 mM EDTA, 12.5 mM MgCl2, 10% glycerol, 0.1% NP-40, and 150 mM KCl. After 5 washes, bound proteins were subjected to SDS-PAGE followed by autoradiography. Note that CTBP2 interacts significantly more with GST-RXRα. (B) In vivo interaction of CTBP2 and RXRα complex in mouse F9 cells. RXRα was immunoprecipitated (46) from F9 total cell lysates using RXR-specific antibodies, and coimmunoprecipitated proteins were immunoblotted for RXRα and CTBP2. Note that RXRα coimmunoprecipitated CTBP2 and normal rabbit IgG did not. (C) In vivo interaction of CTBP2 and RXRα was further confirmed by reverse immunoprecipitation. CTBP2 was immunoprecipitated using CTBP2 antibodies, and coimmunoprecipitated proteins were immunoblotted for CTBP2 and RXRα. (D and E) CTBP2 is associated with the RAREs of two different RA target gene promoters. Chromatin immunoprecipitation (ChIP) was followed by qPCR analysis of CTBP2 binding to RAREs of Cyp26a1 and Hoxb5. Note the loss of CTBP2 binding in Ctbp2 knockdown cells. The data presented here are shown after subtraction of IgG background and are the average of four PCRs from two independent chips. Error bars denote SD.
Consistent with the in vitro findings, immunoprecipitation of endogenous Rxrα from F9 cell extracts coimmunoprecipitated endogenous Ctbp2 (Fig. 6B). Conversely, when we immunoprecipitated Ctbp2 from the same extracts, we coimmunoprecipitated Rxrα (Fig. 6C). A similar binding to Rar was not observed with the antibodies available with us, most likely due to the lower affinity of Ctbp2 for Rarα than for Rxrα.
We also examined if Ctbp2 is present on RAREs within the promoters of bona fide RA target genes in chromatin. Using antibody directed against Ctbp2 or control IgG, we performed chromatin immunoprecipitation assays on F9 cells expressing pLKO control or shCtbp2. Figure 6D and E clearly show significant enrichment of Ctbp2 on RAREs of the RA target genes Cyp26a1 and Hoxb5 in control cells. Less enrichment of Ctbp2 was observed on a nonspecific region 6 kb downstream of the RARE. Most importantly, marked reduction of Ctbp2 occupancy on RAREs was observed in F9 cells expressing shCtbp2, indicating the genuine Ctbp2 binding at RA target gene promoters. Taken together, these biochemical data demonstrate that Ctbp2 is directly involved in receptor-mediated activation of RA target genes.
Loss of Ctbp2 compromises p300 recruitment to RAREs of RA target gene promoters.
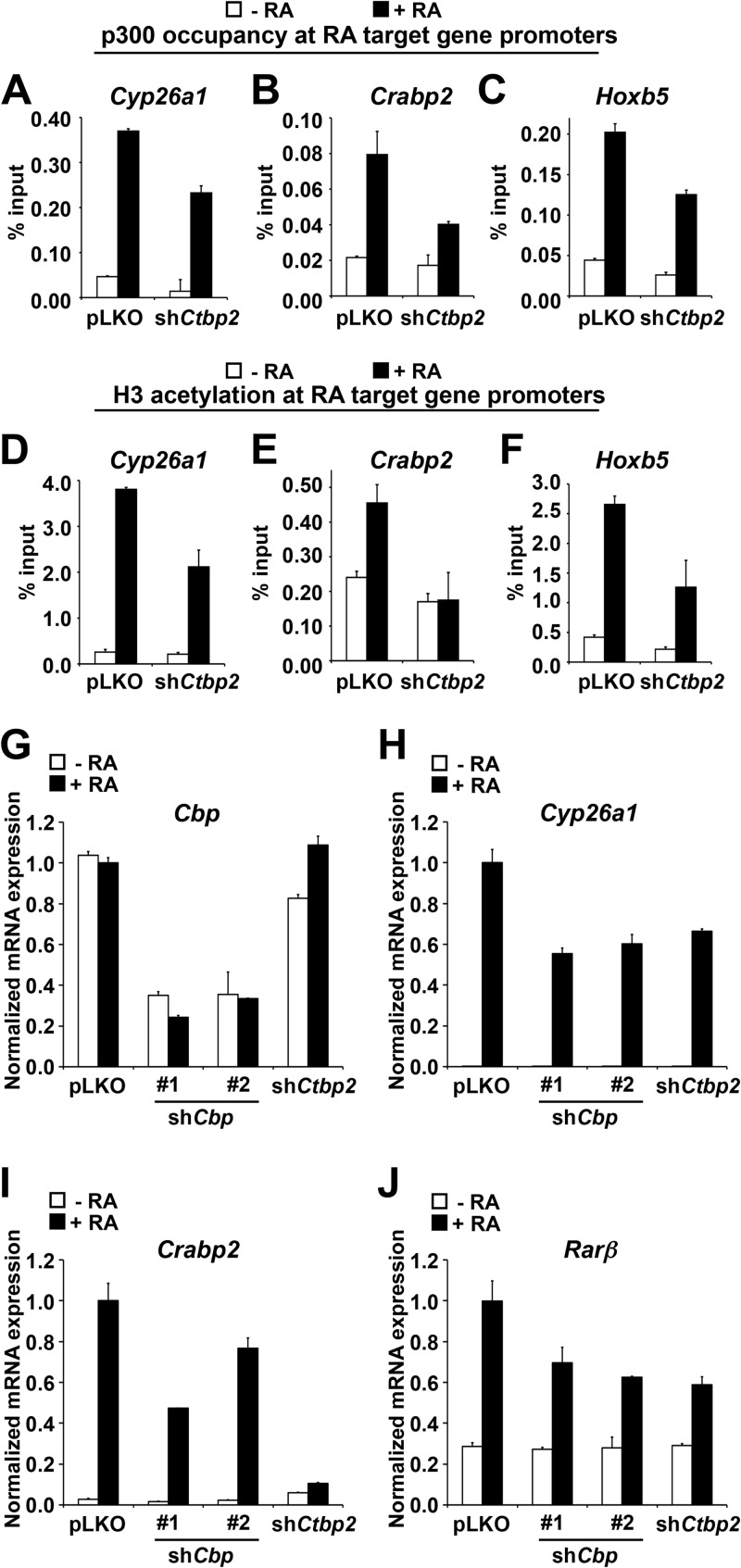
One possible scenario is that CTBP2 transactivates RA receptors in making the chromatin environment conducive for transcription. Histone acetylation mediated by p300/CBP plays a crucial role in activation of RA target genes. Coincidentally, both CTBP1 and CTBP2 have been previously shown to interact with p300 (34–36). We explored the possibility that CTBP2 recruits p300 to RA target gene promoters using the chromatin immunoprecipitation approach on F9 cells expressing pLKO control or shCtbp2. As expected, upon RA treatment, we detected an increase in p300 occupancy on the promoters of RA target genes in control cells, including Cyp26a1, Crabp2, and Hoxb5 (Fig. 7A to C). Upon knockdown of Ctbp2, p300 recruitment to these target gene promoters was hampered. Moreover, loss of p300 on RA target gene promoters led to a concomitant reduction in histone H3 acetylation on the same promoters (Fig. 7D to F). Consistently, we found that knockdown of Cbp also impaired the RA target gene activation, similar to knockdown of Ctbp2 (Fig. 7G to J). From these experiments, we conclude that Ctbp2 is required for the recruitment of p300/Cbp onto RA target gene promoters, thus creating a conductive chromatin environment necessary for RA-induced gene activation.
Fig 7.
CTBP2 recruits p300 histone acetyltransferase to RA target genes. (A to C) Chromatin immunoprecipitation (ChIP) followed by qPCR analysis of p300 binding to RAREs of Crabp2, Hoxb5, and Cyp26a1 in control knockdown (shCtr) and Ctbp2 knockdown (shCtbp2) cells. (D to F) ChIP followed by qPCR analysis of H3 acetylation on RAREs of Crabp2, Hoxb5, and Cyp26a1 in control knockdown (shCtr) and Ctbp2 knockdown (shCtbp2) cells. Note that loss of Ctbp2 leads to concomitant loss of p300 (A to C) and H3 (K9 and K14) acetylation (D to F). The data presented here are shown after subtraction of IgG background and are the average of four PCRs from two independent chips, and error bars denote SD. (G to J) mRNA expression analysis of Cbp (G) and RA target genes Cyp26a1 (H) and Crabp2 (I) and RARβ (J) in F9 cells expressing shRNAs targeting Cbp, Ctbp2, or empty vector cultured in the absence or presence of 1 μM RA for 48 h. Error bars represent SD of triplicate independent experiments.
DISCUSSION
We identify here CTBP2 as a critical component of the RAR/RXR nuclear receptor complex that is required for the transcriptional response to RA. CTBP2 is mostly known as a transcriptional repressor. Our study provides several lines of evidence demonstrating a role for CTBP2 as a coactivator of gene expression. Knockdown of CTBP2 confers resistance to RA-induced differentiation in F9 teratocarcinoma cells and mouse embryonic stem cells, as well as in human promyelocytic leukemia cells. Loss of CTBP2 also compromises activation of endogenous RA target genes and exogenous expression of CTBP2 hyperactivated the RARE-Luc reporter in response to RA, indicating that CTBP2 is a cofactor for RAR/RXR transactivation. Genome-wide transcriptome analysis revealed a high degree of dependence on CTBP2 for RA-mediated gene activation. Mechanistically, CTBP2 physically interacts with the RA receptor and is also present on the RAREs on the promoters of target genes. Moreover, we show that CTBP2 is important for recruitment of p300 histone acetyltransferase to the promoters of RA target genes required for gene activation.
CTBP family proteins are well-known corepressors involved in several essential cellular processes (16). CTBP is part of the corepressor complex, CoREST and is involved in repression of tumor suppressor genes such as CDH1 (encoding E-cadherin), PTEN, and CDKN2A (37–41). In the present study, we found that suppression of Ctbp2, but not of Ctbp1, confers RA resistance, suggesting the unique attribute of Ctbp2 as a coactivator in RA signaling (Fig. 3). Additionally, exogenous expression of CTBP2 hyperactivated RARE-Luc reporter activity (Fig. 4C). Consistently, Ctbp1 knockdown had no effect on RA target genes (data not shown). Even though CTBP1 and CTBP2 are both part of the Co-REST complex, our data suggest that CTBP2 in addition, has a unique role as a coactivator of the RA receptor complex. This is in line with the previous suggestions by others that Ctbp can activate Wingless signaling (21, 42). In Drosophila Kc cells and fly tissues, Ctbp is both required for activation and repression of a subset of Wingless target genes (43). Interestingly, the authors describe in an independent study that the activation/repression is mainly mediated by the dimerization status of Ctbp2 (42). Ctbp2 dimers mediate repression, whereas Ctbp2 monomers mediate activation. Using the RARE-Luc reporter system in the NTERA2 cells, we found that CTBP2 mutants, including the delta-N mutant (deletion of the unique N-terminus region of CTBP2), GG mutant [G189A and G192A; abolishes NAD(H) binding], RR mutant (R147L and R169L; abolishes CTBP2 dimerization), and A58E mutant (mutation in the PLDLS binding cleft of CTBP2) (47) all appeared to activate the RARE-Luc (data not shown). Further study is needed to pinpoint the functional domain of CTBP2 required for transactivation of RA receptors. In addition, we found that CTBP2 is also required for transcriptional activation of RXR-PPARγ heterodimers as well as RXR-RXR homodimers, arguing for a wider role of CTBP2 in the activation of the nuclear hormone receptors (data not shown). Together, these studies point to a model in which CTBP2 can be a potent activator of signaling pathways. Consistent with this, we see that in the absence of RA stimulation, Ctbp2 is required for expression of 711 genes, of which 228 (32%) are also responding to RA signaling (Fig. 4J and M). This indicates that Ctbp2 also has major RA-independent roles in transcription activation.
It has also been described by others that transcriptional cofactors can have a dual function in both gene activation and repression (15). For example, dual-specificity histone demethylase LSD1 mediates repression of neuronal genes by removal of H3K4 methylation (an activation mark) in nonneuronal cells, but it acts as a coactivator in androgen receptor signaling by reversing H3K9 methylation (a repression mark) (44). Similarly, coactivator GRIP1 can be a coactivator or a corepressor of glucocorticoid receptor, using distinct domains for activation and repression (45).
The transition from gene repression to gene activation is an important transition that requires a set of different activities on the chromatin. Identification of the key factors that mediate this change is of paramount importance. Our study demonstrates that CTBP2 serves as a critical mediator in recruiting p300 for histone acetylation of RA target gene promoters upon gene activation. As a key coactivator of transcription, p300 also acetylates transcription regulators and corecruits other cofactors during transcriptional activation. Interestingly, CTBP2 has a unique N-terminal 20-amino-acid region containing three lysine residues (Lys-6, -8, and -10) that can be acetylated by p300, and acetylation of Lys-10 appears to be required for nuclear retention of CTBP2 (36). Our present results indicating that CTBP2 is required for p300 function during RA-induced transcriptional activation (Fig. 7) may indicate that acetylation of CTBP2 by p300 is part of a feed-forward loop that contributes to maximal activation of transcription in response to RA. Moreover, these findings highlight the dynamic functional interaction between CTBP2 and p300.
In conclusion, we have taken an unbiased functional genetic approach and discovered an unexpected role for CTBP2, a well-known corepressor protein, as a key activator of the RA signaling pathway. Our findings challenge the conventional view that the RAR/RXR complex is bound either to repressors (in the absence of ligand) or to activators (in the presence of ligand). As such, our findings shed a new light on the dynamics of corepressor-coactivator interactions in the RA signaling pathway.
Supplementary Material
ACKNOWLEDGMENTS
We thank the NKI Genomics Core Facility for support. We also thank Andreas Schlicker for constructive suggestions on analysis of the data.
This work was supported by a grant from the Dutch Cancer Society (KWF) and The Netherlands Genomics Initiative (NGI).
Footnotes
Published ahead of print 17 June 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.01213-12.
REFERENCES
- 1. Rochette-Egly C, Germain P. 2009. Dynamic and combinatorial control of gene expression by nuclear retinoic acid receptors (RARs). Nucl. Recept. Signal. 7:e005. 10.1621/nrs.07005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Altucci L, Gronemeyer H. 2001. The promise of retinoids to fight against cancer. Nat. Rev. Cancer 1:181–193 [DOI] [PubMed] [Google Scholar]
- 3. Freemantle SJ, Spinella MJ, Dmitrovsky E. 2003. Retinoids in cancer therapy and chemoprevention: promise meets resistance. Oncogene 22:7305–7315 [DOI] [PubMed] [Google Scholar]
- 4. Chen H, Lin RJ, Schiltz RL, Chakravarti D, Nash A, Nagy L, Privalsky ML, Nakatani Y, Evans RM. 1997. Nuclear receptor coactivator ACTR is a novel histone acetyltransferase and forms a multimeric activation complex with P/CAF and CBP/p300. Cell 90:569–580 [DOI] [PubMed] [Google Scholar]
- 5. McKenna NJ, O'Malley BW. 2002. Nuclear receptor coactivators—an update. Endocrinology 143:2461–2465 [DOI] [PubMed] [Google Scholar]
- 6. Spencer TE, Jenster G, Burcin MM, Allis CD, Zhou J, Mizzen CA, McKenna NJ, Onate SA, Tsai SY, Tsai MJ, O'Malley BW. 1997. Steroid receptor coactivator-1 is a histone acetyltransferase. Nature 389:194–198 [DOI] [PubMed] [Google Scholar]
- 7. Wei LN. 2003. Retinoid receptors and their coregulators. Annu. Rev. Pharmacol. Toxicol. 43:47–72 [DOI] [PubMed] [Google Scholar]
- 8. Chambon P. 1996. A decade of molecular biology of retinoic acid receptors. FASEB J. 10:940–954 [PubMed] [Google Scholar]
- 9. Chen JD, Evans RM. 1995. A transcriptional co-repressor that interacts with nuclear hormone receptors. Nature 377:454–457 [DOI] [PubMed] [Google Scholar]
- 10. Horlein AJ, Naar AM, Heinzel T, Torchia J, Gloss B, Kurokawa R, Ryan A, Kamei Y, Soderstrom M, Glass CK, Rosenfeld MG. 1995. Ligand-independent repression by the thyroid hormone receptor mediated by a nuclear receptor co-repressor. Nature 377:397–404 [DOI] [PubMed] [Google Scholar]
- 11. Onate SA, Tsai SY, Tsai MJ, O'Malley BW. 1995. Sequence and characterization of a coactivator for the steroid hormone receptor superfamily. Science 270:1354–1357 [DOI] [PubMed] [Google Scholar]
- 12. Cavailles V, Dauvois S, L'Horset F, Lopez G, Hoare S, Kushner PJ, Parker MG. 1995. Nuclear factor RIP140 modulates transcriptional activation by the estrogen receptor. EMBO J. 14:3741–3751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Epping MT, Wang L, Edel MJ, Carlee L, Hernandez M, Bernards R. 2005. The human tumor antigen PRAME is a dominant repressor of retinoic acid receptor signaling. Cell 122:835–847 [DOI] [PubMed] [Google Scholar]
- 14. Fernandes I, Bastien Y, Wai T, Nygard K, Lin R, Cormier O, Lee HS, Eng F, Bertos NR, Pelletier N, Mader S, Han VK, Yang XJ, White JH. 2003. Ligand-dependent nuclear receptor corepressor LCoR functions by histone deacetylase-dependent and -independent mechanisms. Mol. Cell 11:139–150 [DOI] [PubMed] [Google Scholar]
- 15. Perissi V, Jepsen K, Glass CK, Rosenfeld MG. 2010. Deconstructing repression: evolving models of co-repressor action. Nat. Rev. Genet. 11:109–123 [DOI] [PubMed] [Google Scholar]
- 16. Chinnadurai G. 2009. The transcriptional corepressor CtBP: a foe of multiple tumor suppressors. Cancer Res. 69:731–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chinnadurai G. 2007. Transcriptional regulation by C-terminal binding proteins. Int. J. Biochem. Cell Biol. 39:1593–1607 [DOI] [PubMed] [Google Scholar]
- 18. Boyd JM, Subramanian T, Schaeper U, La Regina M, Bayley S, Chinnadurai G. 1993. A region in the C-terminus of adenovirus 2/5 E1a protein is required for association with a cellular phosphoprotein and important for the negative modulation of T24-ras mediated transformation, tumorigenesis and metastasis. EMBO J. 12:469–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Schaeper U, Boyd JM, Verma S, Uhlmann E, Subramanian T, Chinnadurai G. 1995. Molecular cloning and characterization of a cellular phosphoprotein that interacts with a conserved C-terminal domain of adenovirus E1A involved in negative modulation of oncogenic transformation. Proc. Natl. Acad. Sci. U. S. A. 92:10467–10471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Subramanian T, La Regina M, Chinnadurai G. 1989. Enhanced ras oncogene mediated cell transformation and tumorigenesis by adenovirus 2 mutants lacking the C-terminal region of E1a protein. Oncogene 4:415–420 [PubMed] [Google Scholar]
- 21. Hildebrand JD, Soriano P. 2002. Overlapping and unique roles for C-terminal binding protein 1 (CtBP1) and CtBP2 during mouse development. Mol. Cell. Biol. 22:5296–5307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Holzel M, Huang S, Koster J, Ora I, Lakeman A, Caron H, Nijkamp W, Xie J, Callens T, Asgharzadeh S, Seeger RC, Messiaen L, Versteeg R, Bernards R. 2010. NF1 is a tumor suppressor in neuroblastoma that determines retinoic acid response and disease outcome. Cell 142:218–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Huang S, Laoukili J, Epping MT, Koster J, Holzel M, Westerman BA, Nijkamp W, Hata A, Asgharzadeh S, Seeger RC, Versteeg R, Beijersbergen RL, Bernards R. 2009. ZNF423 is critically required for retinoic acid-induced differentiation and is a marker of neuroblastoma outcome. Cancer Cell 15:328–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Brummelkamp TR, Bernards R, Agami R. 2002. A system for stable expression of short interfering RNAs in mammalian cells. Science 296:550–553 [DOI] [PubMed] [Google Scholar]
- 25. Berns K, Hijmans EM, Mullenders J, Brummelkamp TR, Velds A, Heimerikx M, Kerkhoven RM, Madiredjo M, Nijkamp W, Weigelt B, Agami R, Ge W, Cavet G, Linsley PS, Beijersbergen RL, Bernards R. 2004. A large-scale RNAi screen in human cells identifies new components of the p53 pathway. Nature 428:431–437 [DOI] [PubMed] [Google Scholar]
- 26. Brummelkamp TR, Fabius AW, Mullenders J, Madiredjo M, Velds A, Kerkhoven RM, Bernards R, Beijersbergen RL. 2006. An shRNA barcode screen provides insight into cancer cell vulnerability to MDM2 inhibitors. Nat. Chem. Biol. 2:202–206 [DOI] [PubMed] [Google Scholar]
- 27. Bajpe PK, van der Knaap JA, Demmers JA, Bezstarosti K, Bassett A, van Beusekom HM, Travers AA, Verrijzer CP. 2008. Deubiquitylating enzyme UBP64 controls cell fate through stabilization of the transcriptional repressor Tramtrack. Mol. Cell. Biol. 28:1606–1615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Reddy BA, Bajpe PK, Bassett A, Moshkin YM, Kozhevnikova E, Bezstarosti K, Demmers JA, Travers AA, Verrijzer CP. 2010. Drosophila transcription factor Tramtrack69 binds MEP1 to recruit the chromatin remodeler NuRD. Mol. Cell. Biol. 30:5234–5244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Epping MT, Wang L, Plumb JA, Lieb M, Gronemeyer H, Brown R, Bernards R. 2007. A functional genetic screen identifies retinoic acid signaling as a target of histone deacetylase inhibitors. Proc. Natl. Acad. Sci. U. S. A. 104:17777–17782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kortlever RM, Higgins PJ, Bernards R. 2006. Plasminogen activator inhibitor-1 is a critical downstream target of p53 in the induction of replicative senescence. Nat. Cell Biol. 8:877–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Martens JH, Verlaan M, Kalkhoven E, Dorsman JC, Zantema A. 2002. Scaffold/matrix attachment region elements interact with a p300-scaffold attachment factor A complex and are bound by acetylated nucleosomes. Mol. Cell. Biol. 22:2598–2606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Trapnell C, Pachter L, Salzberg SL. 2009. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25:1105–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wang L, Feng Z, Wang X, Zhang X. 2010. DEGseq: an R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 26:136–138 [DOI] [PubMed] [Google Scholar]
- 34. Kim JH, Cho EJ, Kim ST, Youn HD. 2005. CtBP represses p300-mediated transcriptional activation by direct association with its bromodomain. Nat. Struct. Mol. Biol. 12:423–428 [DOI] [PubMed] [Google Scholar]
- 35. Zhao LJ, Subramanian T, Chinnadurai G. 2006. Changes in C-terminal binding protein 2 (CtBP2) corepressor complex induced by E1A and modulation of E1A transcriptional activity by CtBP2. J. Biol. Chem. 281:36613–36623 [DOI] [PubMed] [Google Scholar]
- 36. Zhao LJ, Subramanian T, Zhou Y, Chinnadurai G. 2006. Acetylation by p300 regulates nuclear localization and function of the transcriptional corepressor CtBP2. J. Biol. Chem. 281:4183–4189 [DOI] [PubMed] [Google Scholar]
- 37. Grooteclaes M, Deveraux Q, Hildebrand J, Zhang Q, Goodman RH, Frisch SM. 2003. C-terminal-binding protein corepresses epithelial and proapoptotic gene expression programs. Proc. Natl. Acad. Sci. U. S. A. 100:4568–4573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Mroz EA, Baird AH, Michaud WA, Rocco JW. 2008. COOH-terminal binding protein regulates expression of the p16INK4A tumor suppressor and senescence in primary human cells. Cancer Res. 68:6049–6053 [DOI] [PubMed] [Google Scholar]
- 39. Paliwal S, Kovi RC, Nath B, Chen YW, Lewis BC, Grossman SR. 2007. The alternative reading frame tumor suppressor antagonizes hypoxia-induced cancer cell migration via interaction with the COOH-terminal binding protein corepressor. Cancer Res. 67:9322–9329 [DOI] [PubMed] [Google Scholar]
- 40. Pena C, Garcia JM, Garcia V, Silva J, Dominguez G, Rodriguez R, Maximiano C, Garcia de Herreros A, Munoz A, Bonilla F. 2006. The expression levels of the transcriptional regulators p300 and CtBP modulate the correlations between SNAIL, ZEB1, E-cadherin and vitamin D receptor in human colon carcinomas. Int. J. Cancer 119:2098–2104 [DOI] [PubMed] [Google Scholar]
- 41. Thiery JP. 2002. Epithelial-mesenchymal transitions in tumour progression. Nat. Rev. Cancer 2:442–454 [DOI] [PubMed] [Google Scholar]
- 42. Bhambhani C, Chang JL, Akey DL, Cadigan KM. 2011. The oligomeric state of CtBP determines its role as a transcriptional co-activator and co-repressor of Wingless targets. EMBO J. 30:2031–2043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Fang M, Li J, Blauwkamp T, Bhambhani C, Campbell N, Cadigan KM. 2006. C-terminal-binding protein directly activates and represses Wnt transcriptional targets in Drosophila. EMBO J. 25:2735–2745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, Casero RA. 2004. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 119:941–953 [DOI] [PubMed] [Google Scholar]
- 45. Rogatsky I, Luecke HF, Leitman DC, Yamamoto KR. 2002. Alternate surfaces of transcriptional coregulator GRIP1 function in different glucocorticoid receptor activation and repression contexts. Proc. Natl. Acad. Sci. U. S. A. 99:16701–16706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Echeverri CJ, Beachy PA, Baum B, Boutros M, Buchholz F, Chanda SK, Downward J, Ellenberg J, Fraser AG, Hacohen N, Hahn WC, Jackson AL, Kiger A, Linsley PS, Lum L, Ma Y, Mathey-Prevot B, Root DE, Sabatini DM, Taipale J, Perrimon N, Bernards R. 2006. Minimizing the risk of reporting false positives in large-scale RNAi screens. Nat. Methods 3:777–779 [DOI] [PubMed] [Google Scholar]
- 47. Zhao LJ, Kuppuswamy M, Vijayalingam S, Chinnadurai G. 2009. Interaction of ZEB and histone deacetylase with the PLDLS-binding cleft region of monomeric C-terminal binding protein 2. BMC Mol Biol 10:89. 10.1186/1471-2199-10-89 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.