Abstract
Improving the knowledge of disease-causing genes is a unique challenge in human health. Although it is known that genes causing similar diseases tend to lie close to one another in a network of protein-protein or functional interactions, the identification of these protein-protein networks is difficult to unravel. Here, we show that Msx1, Snail, Lhx6, Lhx8, Sp3, and Lef1 interact in vitro and in vivo, revealing the existence of a novel context-specific protein network. These proteins are all expressed in the neural crest-derived dental mesenchyme and cause tooth agenesis disorder when mutated in mouse and/or human. We also identified an in vivo direct target for Msx1 function, the cyclin D-dependent kinase (CDK) inhibitor p19ink4d, whose transcription is differentially modulated by the protein network. Considering the important role of p19ink4d as a cell cycle regulator, these results provide evidence for the first time of the unique plasticity of the Msx1-dependent network of proteins in conferring differential transcriptional output and in controlling the cell cycle through the regulation of a cyclin D-dependent kinase inhibitor. Collectively, these data reveal a novel protein network operating in the neural crest-derived dental mesenchyme that is relevant for many other areas of developmental and evolutionary biology.
INTRODUCTION
Organogenesis depends upon a well-ordered series of inductive events involving the coordination of the molecular pathways that regulate the generation and patterning of specific cell types. A key question in organogenesis is to identify the mechanisms by which regulatory networks control cell differentiation and patterning. Transcription factors, for example, control several developmental processes through the selective regulation of target genes, although they display broad DNA-binding specificities in vitro (1). While it is believed that the target gene specificity in vivo is achieved through context-dependent selective protein interactions, the mechanistic details have proved difficult to unravel. Tooth development is an excellent context for investigating this complex problem because of the wealth of information emerging on the molecular mechanisms that govern tooth development from studies of model organisms and human mutations (reviewed in references 2–10, and 11).
Among the different classes of transcription factors, the first member of the Msx homeodomain family of transcription factors, Msx1, is shown to be the first major transcriptional regulator of early tooth development (12). Msx1 mutant mice exhibit, among other phenotypes, a failure (anodontia) and an arrest of tooth development at the bud stage, when Msx1 is required for the expression of several genes, i.e., Bmp4, Lef1, Dlx2, Syndecan-1, Runx2, Fgf3, and p19ink4d, in the dental mesenchyme (13–18). In addition, several loss-of-function mutations in the homologous human MSX1 gene cause nonsyndromic or syndromic tooth agenesis, establishing a similar role for tooth development (19–21).
Despite information about the regulation of Msx1 gene expression during tooth development at the bud stage by epistasis analysis, the downstream targets of Msx1 gene regulation and the molecular mechanism by which Msx1 protein controls transcriptional regulation during early odontogenesis remain unknown. Several in vitro studies have provided some insight into the molecular mechanism by which the Msx gene product exerts its functions in myogenesis and other developmental contexts. It has been shown that Msx1 interacts with different classes of transcription factors and components of the core transcription complex to modify transcription levels and is usually associated with transcriptional repression. In avian cell culture assay, for instance, Pax3 activates the MyoD enhancer gene by binding to its Pax3 DNA binding sites, while Msx1 blocks transcription by binding to its cognate binding sites. When Msx1 and Pax3 are coexpressed, their protein-protein interaction prevents Pax3 from binding to its sites in MyoD, thereby repressing the expression of the enhancer gene and consequently inhibiting muscle cell differentiation (22). Even when target promoters lack cognate Msx1 homeodomain-binding sites, Msx1 is able to repress transcription by associating with core components of the transcriptional machinery, like TBP and Sp1 (23, 24). More recently, it has been shown that Msx1 can interact with a specific isoform of histone, H1b, and thereby bind to the core enhancer element of MyoD, thus inhibiting muscle differentiation through chromatin remodeling (25). On the other hand, functional studies of Msx1 gain-of-function mutations suggest that Msx1 is not invariably a transcriptional repressor. Overexpression of Msx1 in the mammary epithelium, for example, leads to the upregulation of cyclin D1, which prevents the mammary epithelial progenitor cells from terminal differentiation (26). Even though the authors infer that Msx1 indirectly upregulates cyclin D1, their work nevertheless opens the possibility that Msx1 functions as an activator in certain developmental contexts.
The above-described in vitro studies provide a potential regulatory mechanism by which Msx1 exerts its function through interactions with other proteins, but they do not provide information on the tooth-specific Msx1-interacting protein network, nor do they define a function for Msx1 as an activator or repressor in tooth development. It is well documented that transcription factors have exquisite specificities in vivo that reflect their selective regulation of target genes and that their specificity is achieved through selective, context-dependent protein interactions (27). In the context of tooth development, for example, potential candidates for the role of an Msx1-interacting partner are other transcription factors that, like Msx1, play a role during early tooth development (Table 1). Although targeted mouse mutations in either of two members of the distal-less homeobox gene family, Dlx1 and Dlx2, have no effect on tooth development, mice compounded for mutations in both Dlx1 and Dlx2 exhibit a selective absence of upper molars (28–30). Pitx2, a member of the bicoid-like homeobox transcription factor family, is expressed in the epithelium of the tooth bud (31). Interestingly, in patients with Rieger syndrome, which is characterized by skin abnormalities and missing or small teeth, the PITX2 homeodomain gene is mutated (32). Lef1 homozygous mouse mutants exhibit an arrest of both hair and tooth development, with the latter arrested at the bud stage (33). Specificity protein 3 (Sp3) is a ubiquitously expressed protein that belongs to the Sp family of transcription factors. Sp3 is expressed in both dental epithelium and mesenchyme, and Sp3 homozygous null mice exhibit a late tooth phenotype, showing a defective dentine/enamel layer (34). Pax9 homozygous mouse mutants exhibit a cleft of the secondary palate and an arrest of tooth development at the bud stage, the same stage at which tooth development arrests in Msx1, Lef1, and Dlx double mutants (35). The similarity in the tooth phenotype between Msx1 mutants and the other mutants in most cases and the overlapping expression of the genes with Msx1 in the tooth bud mesenchyme raise the possibility that they also interact closely within the molecular regulatory cascade that operates in this organ.
Table 1.
Transcription factors that interact with Msx1 in vitro and in vivo at the bud stage of tooth development
| Gene | Site of expression in tooth bud | Interaction with: | |||
|---|---|---|---|---|---|
| Msx1 |
Msx2 |
||||
| In vitro | In vivo | In vitro | In vivo | ||
| Dlx2 | Dental mesenchyme | + | + | − | − |
| Pax9 | Dental mesenchyme | + | + | − | − |
| Lhx6 | Dental mesenchyme | + | + | − | − |
| Lhx8 | Dental mesenchyme | + | + | − | − |
| Sna | Dental mesenchyme | + | + | − | − |
| Lef-1 | Dental mesenchyme | − | + | − | − |
| Sp3 | Dental mesenchyme | + | + | + | + |
| Prx1 | Dental mesenchyme | − | − | − | − |
| Pitx2 | Dental epithelium | − | − | + | + |
In this study, the murine tooth bud is used as a model system to elucidate the Msx1 homeoprotein's combinatorial interactions with other proteins and how these interactions modulate the transcription of an Msx1 downstream target gene. We show that Msx1 interacts with 5 new transcription factors, providing for the first time a molecular network of transcription factors that operate in early tooth development. We also show that the cyclin D-dependent kinase (CDK) inhibitor p19ink4d promoter is a direct target of Msx1 function in vivo and that Msx1 and its interactors differentially regulate, in a dose-dependent manner, the p19ink4d promoter activity, promoting either functional synergism or antagonism.
MATERIALS AND METHODS
Msx1 baits for yeast two-hybrid screen.
For the yeast two-hybrid screen, the Matchmaker GAL4 two-hybrid system was used (BD Biosciences Clontech). Three different baits were constructed by cloning Msx1 fragments into the GAL4 DNA-BD (binding domain) vector pGBKT7. The baits were (i) full-length Msx1 (Msx1-FL) (bp 1 to 894), (ii) the N-terminal portion of Msx1 (Msx1-NT) (bp 1 to 705), and (iii) the Msx1 homeodomain (Msx1-HD) (bp 466 to 706). The three bait plasmids were transformed into the yeast strain AH109 and tested for self-activation and viability. Of the three baits, Msx1-FL and Msx1-HD did not activate the reporter genes and, hence, were considered potential baits for the screen.
Generation and screening of E13.5 tooth cDNA libraries.
Tooth rudiments were microdissected from wild-type embryonic day 13.5 (E13.5) embryos. Total RNA was extracted using TRIzol (Invitrogen) according to the manufacturer's instructions. Two different cDNA libraries were constructed either by priming with an oligo(dT) primer or by using BD Smart cDNA synthesis technology with a random primer. The two forms of double-stranded cDNAs thus generated were used for recombination with the GALA activation domain vector pGADT7-Rec in vivo to generate a complete GAL4 activation domain (AD) fusion library in yeast. Using the full-length Msx1 bait (Msx1-FL), the two different libraries were screened by selecting for triple dropouts (ADE2, HIS3, and MEL1). From a total of 2 × 105 transformants, 340 colonies were obtained. The size of the clone library was determined by direct yeast colony PCR using Qiagen's Hot Start PCR mix. The four-cutter digestion analysis using restriction enzyme HaeIII was used along with the insert size to classify the library clones into different classes. Clones with identical profiles were grouped in the same class. Two to three individual clones from each class were sequenced using pGADT7 vector-specific primers. The sequence data from each clone were conceptually translated in the reading frame of the library vector, and the sequence identity was determined by searching the mouse genome database at www.ncbi.nlm.nih.gov/blast/Blast.cgi.
Cell culture, transfections, and coimmunoprecipitation assays.
C3H10T1/2 cells (CCL-226; ATCC) were grown in Dulbecco's modified Eagle's medium (DMEM; Invitrogen) supplemented with 10% (vol/vol) fetal bovine serum at 37°C in a 5% CO2 humidified atmosphere. The cells, in 60-mm dishes, were cotransfected with plasmids encoding Dlx2, Lhx6, Lhx8, Snail, Sp3, or Lef1 and pCMV-FLAG-Msx1 or with plasmids encoding Dlx2, Lhx6, Lhx8, Snail, Sp3, or Lef1 and pCMV-Tag2B (Stratagene) using FuGene 6 reagent (Roche) according to the manufacturer's instructions. After 36 h, C3H10T1/2 cells were lysed in radioimmunoprecipitation (RIPA) lysis buffer with protease inhibitors (Roche), and proteins were immunoprecipitated by using EZview red anti-FLAG M2 affinity gel beads (Sigma). For Western blotting of eluted proteins, polyclonal antibodies were used at a 1:500 dilution of primary antibodies against transcription factor Dlx2, Lhx6, Lhx8, Snail, Sp3, or Lef1 (Santa Cruz) or a 1:1,000 dilution of mouse anti-FLAG M2 monoclonal antibody (Sigma). The immunoprecipitated proteins were analyzed by immunoblotting using ECL Western blotting detection reagent (Fisher). All coimmunoprecipitation (co-IP) experiments were independently performed three times, and representative blots are shown.
GST pulldown assay.
Invitrogen's Gateway cloning technology was employed to construct Msx1– and Msx2–glutathione S-transferase (GST) expression plasmids using the vector pDEST-15 (Invitrogen). Using Msx1 or Msx2 cDNA as the template, a PCR product that spans the entire Msx1 or Msx2 open reading frame (ORF) was generated and cloned into the pENTR/D-TOPO vector. Msx1 and Msx2 clones in pENTR/D-TOPO were sequence verified through the PCR-generated regions prior to use in the recombination reaction that transfers the DNA to the GST expression vector, pDEST-15. The GST-Msx1 and Msx2 fusion constructs, as well as the pDEST-15 empty vector that can express just the GST protein, were transformed into the bacterial strain Escherichia coli BL21-S1 and maintained in LB plates without NaCl. The GST fusion proteins were induced at 30°C, using 3 M NaCl. The binding of fusion protein lysates to glutathione-agarose beads was performed according to the manufacturer's instructions (Amersham-Pharmacia). The effective binding of GST-Msx1 and GST-Msx2 fusion proteins to glutathione-agarose beads was detected by Western blot analysis using an anti-Msx1/Msx2 antiserum (Hybridoma Bank). Equimolar amounts of GST-Msx1, GST-Msx2, and GST proteins, as well as glutathione-agarose beads, were incubated with 2 to 10 μl of 35S-labeled proteins in a binding buffer containing 20 mM Tris-HCl, pH 8.0, 0.2 mM EDTA, pH 8.0, 0.1 M NaCl, 1 mM dithiothreitol (DTT), 1 mM phenylmethylsulfonyl fluoride (PMSF), and 0.2% NP-40. The binding reactions were carried out at 4°C for 12 to 16 h. After binding, the beads were washed 3 times with 1 ml of the buffer with a higher concentration of NP-40 (0.4%). The beads were resuspended in 25 μl of 1× SDS loading buffer and boiled for 5 min. The supernatant was resolved by SDS-PAGE. On completion of electrophoresis, the gel was dried under vacuum and subjected to autoradiography.
In vitro transcription/translation reactions.
All in vitro transcription/translation reactions were carried out using Promega's TNT coupled reticulocyte lysate system. Full-length cDNAs of all the genes tested (Dlx2, Lhx6, Lhx8, Lef1, mHox, Pax9, Pitx2, Sna, and Sp3, cloned into pBluescript-SK, pGEM3zf, or pcDNA3) were used as templates in the in vitro transcription/translation reactions to generate [35S]methionine-labeled proteins (Amersham Pharmacia). The efficiency of the translation reaction was determined by SDS-PAGE followed by autoradiography.
Construction of mammalian cell expression vectors.
The full-length Msx1 ORF was cloned into FLAG-tagged mammalian expression vector 2B (Stratagene) or V5 epitope-tagged mammalian expression vector pcDNA3.1/nV5-DEST (Invitrogen). The Dlx2 and Lhx8 ORFs were cloned into the mammalian expression vector pDEST26 (Invitrogen) that contains a polyhistidine (6×His) epitope tag. Full-length Pax9 was cloned in the FLAG- or Myc-tagged mammalian expression vector pCMV-Ta (Stratagene). The Lhx6, Sna, and Lef1 ORFs were cloned into the Myc-tagged mammalian expression vector pCMV-Tb (Stratagene). Sp3 was cloned into pcDNA3.
Immunofluorescence.
Primary antibodies against 8 transcription factors (Msx1, Pax9, Dlx2, Snail, Sp3, Lef1, Lhx6, and Lhx8) and secondary antibodies (goat anti-rabbit IgG-fluoresceinisothiocyanate [FITC] and rhodamine-conjugated IgGs [IgG-R] goat anti-rabbit IgG-R and bovine anti-goat IgG-R) were purchased from Santa Cruz. They were used at a 1:200 or 1:400 dilution, respectively. Cells were sequentially stained with triple labeling to examine their colocalization. First, cells were fixed in 4% paraformaldehyde (PFA) in phosphate-buffered saline (PBS) for 8 min at room temperature, followed by washing in PBS twice. Next, fixed cells were permeabilized by incubation with 0.2% Triton X-100 in PBS for 5 min. Then, cells were blocked in 10% normal goat serum in PBS for 1 h or overnight prior to 1 h of incubation with the respective primary antibody (1:200 dilution in PBS containing 1.5% normal serum). After three changes of PBS, cells were incubated with FITC or tetramethyl rhodamine isocyanate (TRITC)-conjugated secondary antibodies (1:400 dilution) and washed three times with PBS in the dark to avoid photobleaching, followed by the second round of the sequential staining procedure in the same manner. Nuclei were counterstained with DRAQ5 at a 1:1,000 dilution (Cell Signaling). Culture slides with triple labeling were mounted on coverslips with mounting medium for fluorescence (VectaShield) and sealed with nail polish. Stained cells were observed and images were captured with a Leica TCS SP2 laser confocal microscope. All imaging experiments (sequential scanning mode) were performed three times at room temperature, and representative images are shown.
Coimmunoprecipitation assay for Msx2.
Two cell lines, C3H10T1/2 and C2C12, were grown in 60-mm dishes and cotransfected with plasmid constructs of Pax9, Dlx2, Lhx6, Lhx8, Snail, Sp3, or Lef1 and pCMV-FLAG-Msx2 using TurboFect in vitro transfection reagent (Fermentas). After 36 h, cells were lysed in 200 μl lysis buffer (20 mM Na-phosphate buffer, pH 7.4, 100 mM NaCl, 1 mM MgCl2, 1% NP-40, 0.1% SDS, and Complete protease inhibitor [Roche]) for 30 min on ice. Cell lysates were centrifuged at 16,000 × g for 15 min, supernatants were transferred to clean microcentrifuge tubes, and coimmunoprecipitations were performed overnight at 4°C by using anti-FLAG M2 agarose beads (Sigma). Proteins were washed twice with the lysis buffer, eluted using 3× FLAG peptide, and analyzed by Western blotting as described previously (36). For each coimmunoprecipitation assay, at least 0.5 mg of total protein extract was immunoprecipitated with anti-FLAG agarose (Sigma-Aldrich) and immunoblotted with individual antibodies against transcription factors Pax9, Dlx2, Lhx6, Lhx8, Snail, Sp3, and Lef1. All co-IP experiments were independently performed three times, and representative blots are shown.
ChIP assay.
Chromatin immunoprecipitation (ChIP) assays were performed with anti-FLAG antibody (clone M2; Sigma) and anti-Msx1 antibody (M0944; Sigma) according to the ChIP assay protocol (Upstate/Millipore). Briefly, C3H10T1/2 cells transfected with pCMV-FLAG-Msx1 or untransfected cells were cross-linked with 1% formaldehyde for 5 to 10 min at room temperature, neutralized with glycine (125 mM), and washed twice with chilled 1× PBS containing Complete protease inhibitor. Cell lysates were sonicated to shear the DNA to 200 to 1,000 bp and centrifuged at 12,000 × g for 10 min. A small aliquot (10 ml) was saved as input DNA for later PCR analysis. Chromatins were immunoprecipitated overnight at 4°C by mild agitation with 5 mg anti-FLAG antibody or 5 mg anti-Msx1 antibody. For a negative control, the supernatant solutions were incubated with 5 mg of normal mouse or rabbit IgG. After digestion with proteinase K for 1 h at 55°C, immunoprecipitated DNAs were purified by spin column, eluted in 50 ml of elution buffer, and stored at −20°C until PCR analysis.
PCR amplification and real time-quantitative PCR analysis.
To analyze the target regions containing the Msx1 binding site in the proximal promoter of the p19INK4d gene, the DNA samples were amplified by PCR that generated ∼200-bp PCR products spanning two potential binding sites using two pairs of primers. The primers for binding site 1 (forward, 5′-TAA AAA GGA GTC AGG CAG TAG TGG-3′, and reverse, 5′-TTT TTC TAC GTT TTT CAA GAT AGG G-3′) and site 2 (forward, 5′-CAT TGA TCC ATC TTT CAG GTT CTA-3′, and reverse, 5′-GGC CTG AAA CTT ATT TGA TCT GTT-3′) were synthesized by Integrated DNA Technologies. The PCR cycle was 94°C for 3 min and 40 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 45 s, followed by 2 min of incubation at 72°C. DNA samples were quantified with iQ software, version 3.0, using an iCycler quantitative PCR (qPCR) system (Bio-Rad) and PerfeCTa SYBR Green I supermix for iQ (Quanta BioScience). For real-time qPCR analysis, the cycling parameters were 95°C for 3 min and 45 cycles of 95°C for 15 s, 55°C for 45 s (to collect and analyze data), followed by a dissociation stage for the melt curve. Real-time qPCR data analysis was performed by the percentage-of-input method (Invitrogen).
RESULTS
Msx1 interacts with Lhx6 and Sna in vivo.
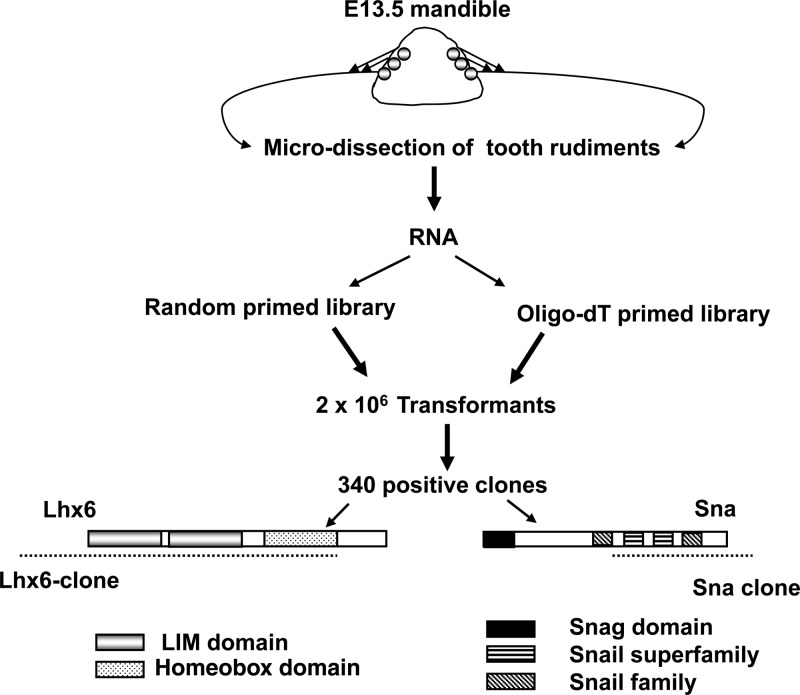
To identify protein interactors of the Msx1 homeoprotein in vivo, we performed a yeast two-hybrid screen using the full-length Msx1 homeoprotein as the bait. We used a tooth cDNA library obtained from microdissected E13.5 mouse tooth buds to restrict our screen to the tissue and developmental stage of our interest. After analyzing 340 clones, we obtained two clones that corresponded to the ORFs of the Lhx6 and Sna genes (Fig. 1). From our randomly primed tooth cDNA library screen, we obtained an Lhx6 clone that included the two LIM domains and the homeodomain (Fig. 1). Lhx6 belongs to a distinct subclass of homeobox genes called the LIM homeobox gene family. The LIM homeodomain proteins (Lhx) contain, in addition to a classical homeodomain, two cysteine-rich motifs called LIM domains that mediate protein-protein interactions (37–39). By using an oligo(dT)-primed cDNA library to perform the yeast two-hybrid screen, we obtained a clone of Sna that included the C-terminal zinc finger region (Fig. 1). Sna is a zinc finger-containing transcription factor that belongs to the snail superfamily of genes. The zinc finger motif is known to bind DNA, as well as to interact with other proteins (40).
Fig 1.
A schematic representation of the yeast two-hybrid screen using E13.5 tooth cDNA. Microdissected tooth rudiments from E13.5 embryos were used to make two cDNA sources, one oligo(dT) primed and the other random primed. The cDNAs were transformed into the yeast library vector by homologous recombination. From screening 2 × 106 transformants and analyzing 340 clones, two clones that corresponded to the ORFs of Lhx6 and Sna were obtained. The Lhx6 clone contained the LIM domains and the homeodomain shown, in comparison with the full-length ORF. The Sna clone, obtained from the oligo(dT)-primed library, included the 3′ untranslated region and the C-terminal zinc fingers.
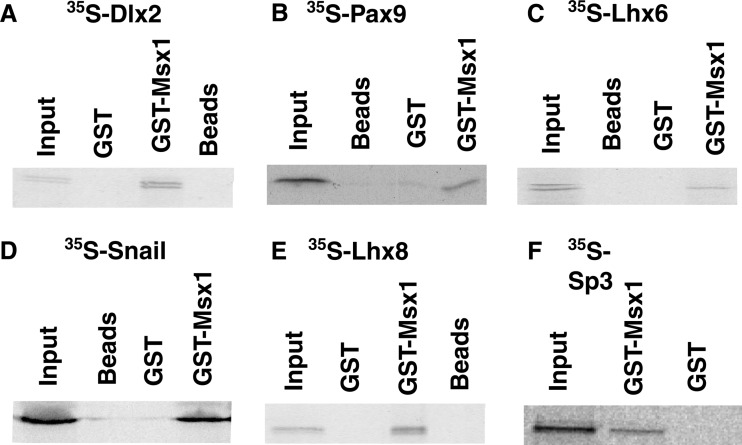
To further characterize the interaction of Msx1 with Lhx6 and Sna, we generated 35S-labeled Lhx6 and Sna proteins in vitro and bacterially expressed a GST-Msx1 fusion protein. Msx1 interacted physically with both Lhx6 and Sna (Fig. 2C and D). The interaction was specific to GST-Msx1, since neither GST nor beads showed a nonspecific interaction when incubated with 35S-labeled Lhx6 or 35S-labeled Sna (Fig. 2).
Fig 2.
Msx1 interacts directly with tooth-specific transcription factors in vitro. GST pulldown assays with in vitro-translated, 35S-labeled Dlx2 (A), Pax9 (B), Lhx6 (C), Snail (D), Lhx8 (E), and Sp3 (F) were performed using the Msx1-GST fusion protein. GST-bound beads and unbound beads were used as controls. Immobilized proteins were eluted in the SDS buffer, resolved by SDS-PAGE, and visualized by autoradiography.
Transcription factors that play an important role in tooth development interact with Msx1.
Several transcription factors are known to spatially and temporally overlap with Msx1 during the bud stage of tooth development and are essential for odontogenesis (Table 1). To determine whether this set of transcription factors may constitute potential Msx1 interactors, along with Lhx6 and Snail identified from our yeast two-hybrid screen, we performed, at first, a simple GST pulldown assay between the Msx1 protein and these transcription factors. We identified a physical interaction between Msx1 and two different new transcription factors: Lhx8, another member of the Lhx gene family, and Sp3 (Fig. 2E and F). Known Msx1 interactors, such as Dlx2 and Pax9, were used as positive controls (Fig. 2A and B) (41, 42). In contrast, we did not detect any interaction between GST-Msx1 and Lef1, Pitx2, or Prx1 (data not shown). The latter results are of particular significance for the following reasons. Functional studies have shown that Pitx2 mutant mice have a tooth phenotype very similar to that of Msx1 mutant mice; however, the expression of Pitx2 is restricted to the dental epithelium. Since Msx1 is expressed exclusively in the dental mesenchyme, the lack of a physical interaction between GST-Msx1 and 35S-labeled Pitx2 emphasizes the specificity of Msx1-mediated interactions only with a subset of transcription factors that are expressed in the dental mesenchyme. Moreover, we could not identify a physical interaction between Msx1 and either Lef1 or Prx1, both of which were considered ideal candidates for interaction with Msx1, due to their overlapping expression with Msx1 in the dental mesenchyme and the similarity in their odontogenic phenotypes. Although these observations strengthen the specificity of the physical interactions detected in our GST pulldown assay, they cannot exclude the possibility of an indirect interaction.
Msx1 protein interactors coimmunoprecipitate with Msx1 in living cells.
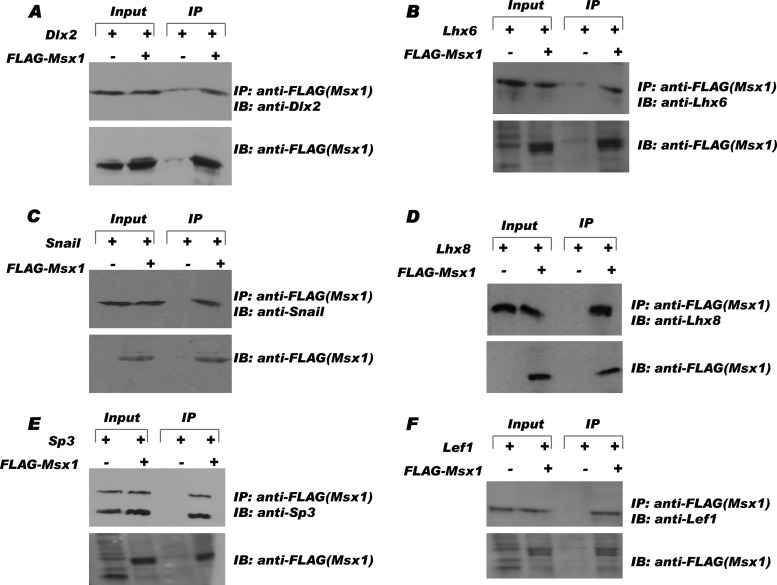
To investigate whether the interactions we identified from our yeast two-hybrid screen and GST pulldown assay could be reproduced in living cells, we performed coimmunoprecipitation assays in C3H10T1/2, a pluripotent embryonic mesenchymal cell line, and C2C12, a premesenchymal cell line (Fig. 3 and data not shown). We expressed Msx1 exogenously as a FLAG-tagged fusion protein (Fig. 3), Dlx2 and Lhx8 as His-tagged fusion proteins, and Lhx6, Snail, and Lef1 as Myc-tagged fusion proteins for immunoprecipitation analysis using anti-FLAG antibody. Western blot analyses detected all of the transcription factors in the anti-FLAG immunoprecipitates from cells cotransfected with FLAG-Msx1 (Fig. 3A to F, fourth lanes) but not in those from cells cotransfected with empty control vector (Fig. 3A to F, third lanes). As controls, equal protein levels (20%) of each transcription factor were present in both input samples (Fig. 3A to F, first and second lanes). In contrast, we were unable to coimmunoprecipitate exogenous Pitx2 and/or Prx1 with V5-Msx1 in the C2C12 cell line or FLAG-Msx1 in the C3H10T1/2 cell line under different conditions (data not shown), further suggesting that Msx1 is unlikely to interact with these transcription factors physically or in vivo. In sharp contrast to the results for Prx1 and Pitx2, Lef1 is able to interact with Msx1 in living cells.
Fig 3.
Msx1 interacts with tooth-specific transcription factors in vivo. pCMV-FLAG-Msx1 or control vector (pCMV-FLAG-Tag2B) was coexpressed with transcription factors Dlx2 (A), Lhx6(B), Snail (C), Lhx8 (D), Sp3 (E), and Lef1 (F) in C3H10T1/2 cells. After 36 h, cell lysate (20%) of each was taken as the input sample to verify the level of expression (first and second lanes). All the transcription factors tested were detected only in the presence of FLAG-Msx1 in the immunoprecipitation samples (fourth lanes) and not in the control samples (third lanes). IP, immunoprecipitation; IB, immunoblotting.
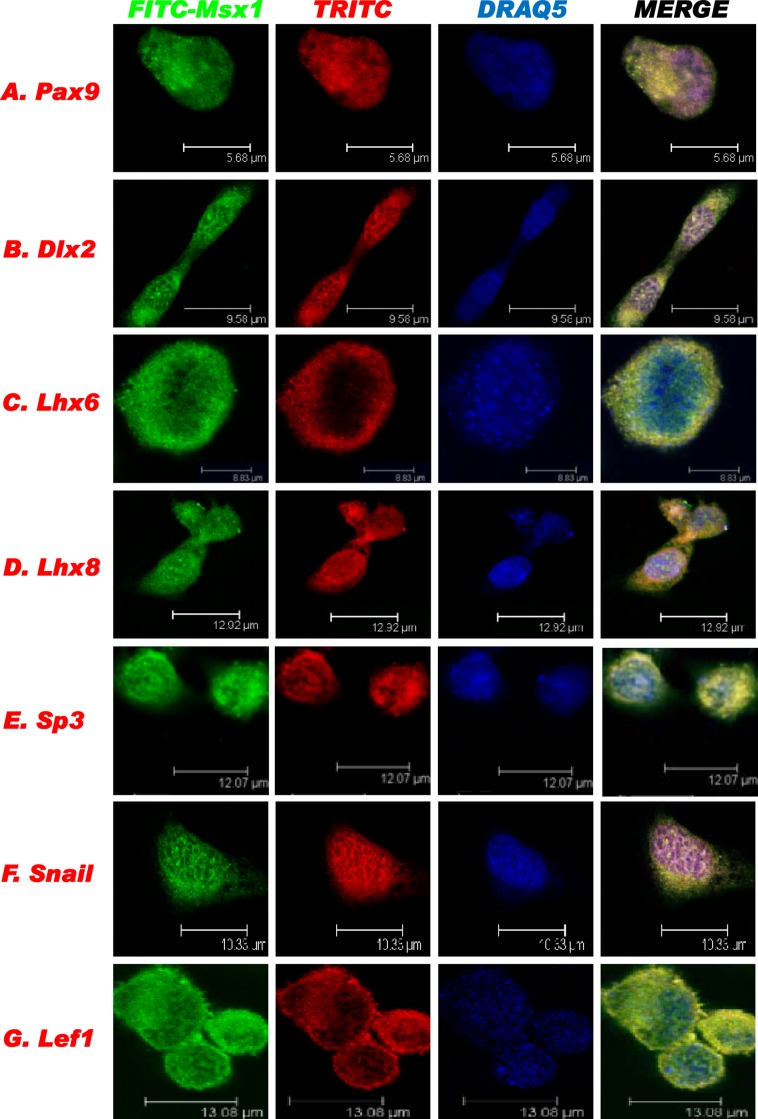
We also performed protein colocalization analysis using confocal microscopy (Fig. 4). C3H10T1/2 cells were used for the immunofluorescence staining to confirm the endogenous localization of Msx1 and its interacting proteins. Endogenous Msx1 was visualized by green fluorescence (FITC immunostaining), and individual interacting transcription factors were visualized by red fluorescence (TRITC immunostaining). The overlay images (Fig. 4, yellow) indicated that all transcription factors tested were endogenously coexpressed in the nuclei of C3H10T1/2 cells.
Fig 4.
Endogenous colocalization of Msx1 and interacting transcription factors in living cells. Msx1 was labeled with FITC-conjugated antibody and the other transcription factors, at their endogenous levels in C3H10T1/2 cells, were labeled with TRITC for confocal imaging analysis. DRAQ5 was used to stain DNA.
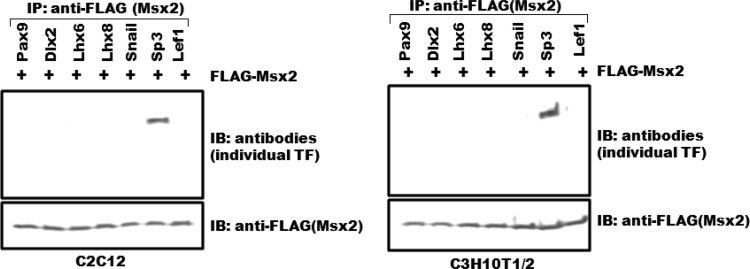
In sum, by employing several approaches, we identified 5 new transcription factors that mediate protein-protein interactions with Msx1, consistent with their functional relationship during early tooth development. These interactions of Msx1 with Lhx6, Lhx8, Snail, Sp3, and Lef1 are highly specific, since no physical interaction was observed except for Sp3 with Msx2, the second member of the Msx family of transcription factors, which shares a high degree of sequence homology with Msx1 (Table 1 and Fig. 5). In addition, transcription factors that would be considered candidates for interaction with Msx1 due to their overlapping expression with Msx1 in the dental mesenchyme (such as Prx1) or their odontogenic phenotype (such as Pitx2) do not interact with Msx1 either physically or in living cells, further highlighting the specificity of Msx1's interactions (Table 1 and data not shown). Moreover, these results also suggest that Msx1 works in concert with other transcription factors to control gene expression during early tooth morphogenesis.
Fig 5.
Msx2 interacts with Sp3 only. FLAG-Msx2 was cotransfected with transcription factors (TF) Pax9, Dlx2, Lhx6, Lhx8, Snail, Sp3, and Lef1 into C2C12 and C3H10T1/2 cells. Only Sp3 and not the other Msx1 interactors tested was coimmunoprecipitated with Msx2 both in C2C12 and C3H10T1/2 cells.
Msx1 binds in vivo to the p19 Ink4d promoter.
How do these interactions mediate the ability of Msx1 to regulate a key downstream target gene at the bud stage of tooth development? In the context of myogenesis, for example, Msx1 functions as a repressor of a myogenic regulatory gene, the MyoD gene, and it does so in concert with other proteins, like Pax3, a known activator of MyoD gene expression (22, 43). In vivo studies indicate that the repressive activity of Msx1 is mediated by direct binding to a key regulatory element in the MyoD promoter known as CER (25). In the context of tooth development, the downstream direct targets of Msx1 gene regulation remain unknown. The next question, therefore, is to identify a key target gene whose activity is directly regulated by Msx1 and to determine how the Msx1 target promoter is further regulated by the combinatorial action of Msx1 and Msx1-interacting protein complex(es) during tooth development.
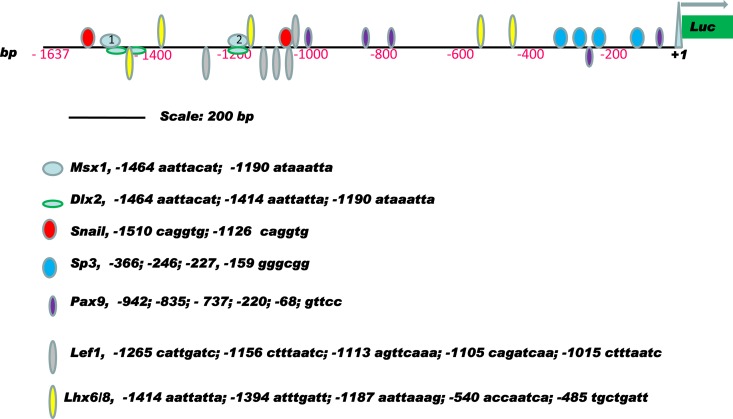
Previous studies revealed that in the Msx1 knockout mouse, arrest of tooth development occurs at the bud stage, when Msx1 is required for the expression of several downstream genes, including p19ink4d. It was shown that loss of Msx1 function results in elevated p19ink4d expression and significant reduction of cell proliferation in the dental mesenchyme (15, 18). These results suggest that, under physiological conditions, the Msx1 homeoprotein promotes cell proliferation in the dental mesenchyme by repressing the expression of the CDK inhibitor (p19ink4d), thus facilitating cell cycle progression. Computational sequence analysis of the nucleotides in the proximal 1.6 kb of the murine p19ink4d promoter region revealed the presence of two fully conserved Msx1 binding sites 1,464 bp and 1,190 bp upstream from the transcription initiation site in mouse that are conserved in the human p19INK4d promoter as well (UniProbe database, http://thebrain.bwh.harvard.edu/uniprobe/). Additionally, this promoter sequence contains putative binding sites for the Msx1-interacting transcription factors Snail, Lhx6/8, Pax9, Dlx2, Lef1, and Sp3, suggesting that this sequence may contain an important cis-regulatory element regulating the expression of p19ink4d in the dental mesenchyme (Fig. 6).
Fig 6.
Schematic representation of the transcription factor binding sites in the murine p19ink4d proximal promoter region. All the binding loci are based on computer analysis and UniProbe database profiling (http://thebrain.bwh.harvard.edu/uniprobe/). Msx1 possesses two binding clusters, labeled 1 (nt 1464) and 2 (nt 1190) in pale blue ovals.
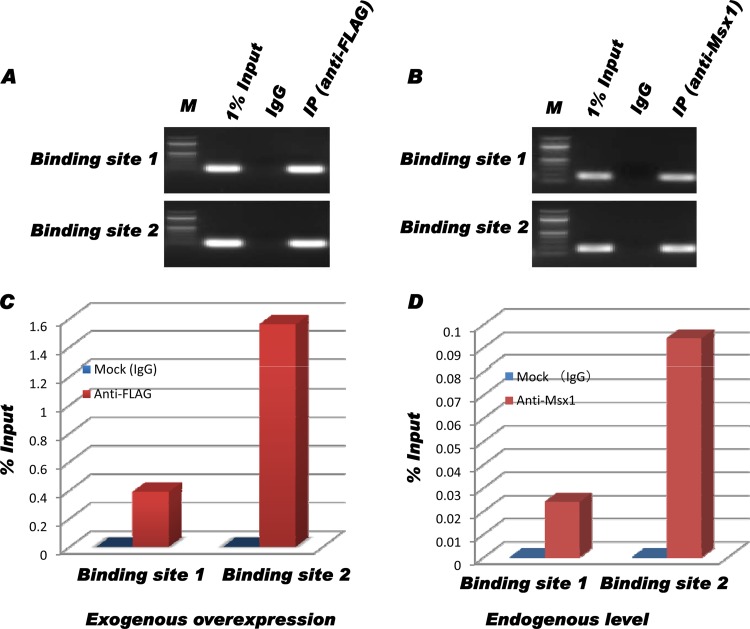
To determine whether Msx1 binds to any of these sites and, therefore, directly regulates p19ink4d, in vivo chromatin immunoprecipitation was performed with either exogenously expressed Msx1-FLAG or endogenous Msx1. Immunoprecipitated chromatin fragments (IP samples) and nonimmunoprecipitated samples (1% input) were subjected to PCR and real-time qPCR analysis using specific primers spanning the two binding sites. PCR amplifications showed that Msx1 binds directly to the conserved motif (TAAT) in the endogenous promoter of the mouse p19ink4d gene (Fig. 7). In each set, the promoter regions were specifically amplified in samples from C3H10T1/2 cells transfected with pCMV-Msx1-FLAG (Fig. 7A and C) and from nontransfected C3H10T1/2 cells (Fig. 7B and D). Furthermore, real-time quantitative PCR indicated a binding preference of Msx1 to site 2 (nucleotide [nt] 1190) that was 3-fold greater than its affinity for site 1 (nt 1464), with differential association frequency (Fig. 7C and D). This result demonstrates that Msx1 binds to the proximal p19ink4d promoter in vivo.
Fig 7.
The p19ink4d proximal promoter region is a direct target of Msx1. (A) After chromatin immunoprecipitation, samples from C3H10T1/2 cells transfected with pCMV-Msx1-FLAG were PCR amplified; the binding region was directly amplified prior to immunoprecipitation (1% Input) and specifically amplified in the immunoprecipitated sample (anti-FLAG). No amplification was detected in the nonspecific IgG (normal mouse serum IgG)-immunoprecipitated sample (IgG) due to low background under the exogenous overexpression of Msx1 in cells. (B) After chromatin immunoprecipitation, samples from untransfected C3H10T1/2 cells were PCR amplified; the binding region was directly amplified prior to immunoprecipitation (1% Input) and specifically amplified in the immunoprecipitated sample (anti-Msx1). (C and D) Quantitative analysis of Msx1 binding preference from transfected cells (exogenous expression) (C) and untransfected cells (endogenous expression) (D). In both situations, there was a 3-fold-higher preference of Msx1 for binding site 2 (nt 1190) over binding site 1 (nt 1464) in vivo. M, DNA marker.
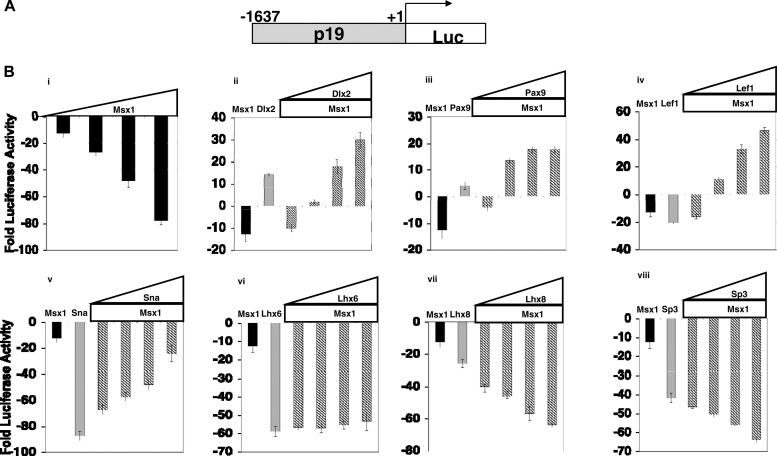
To determine whether the p19ink4d promoter is repressed or activated by Msx1, C3H10T1/2 cells were cotransfected with a p19-luciferase reporter plasmid and increasing amounts of the Msx1 expression plasmid (see Fig. 9B, panel i). We found that transfection of Msx1 alone resulted in repression of the p19-luciferase reporter in a concentration-dependent manner. Consistent with this, loss of Msx1 function in mice results in elevated p19ink4d expression and significant reduction of cell proliferation in the dental mesenchyme (15, 18). These results suggest that, under physiological conditions, the Msx1 homeoprotein promotes cell proliferation in the dental mesenchyme by repressing the expression of the CDK inhibitor p19ink4d, thus facilitating cell cycle progression.
Fig 9.
Functional consequences of the interaction of Msx1 with tooth-specific transcription factors for the transcriptional regulation of the p19 promoter. C3H10T1/2 cells were cotransfected with 1 μg of p19-Luc reporter plasmid along with increasing amounts of expression plasmid Msx1-FLAG (0.25 μg, 0.50 μg, 0.75 μg, and 1.0 μg). Cells were harvested 24 h after the transfections for reporter gene assays. All the transfection experiments were performed three times, and the bars indicate the means ± standard deviations. Msx1 used alone repressed the p19 promoter. Pax9, Dlx2, Lef1, and Snail in conjugation with Msx1 acted as activators of transcription at the p19 promoter, whereas Lhx8 and Sp3 used with Msx1 resulted in repression of p19. The repression by Lhx6 at the p19 promoter did not change in the presence of Msx1.
Thus, we provide clear evidence that Msx1 represses p19ink4d transcriptional activity in vivo and does so by direct binding to Msx1 recognition sites on the p19ink4d promoter (Fig. 6 and 7; see also Fig. 9B, panel i). The p19ink4d promoter constitutes an Msx1 target promoter during tooth development.
The interaction between Msx1 and its protein partners promotes either functional antagonism or synergism.
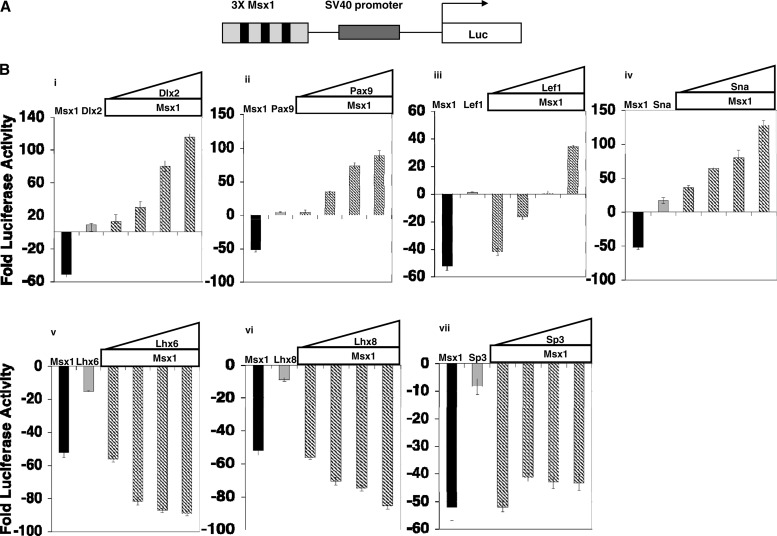
To determine the functional consequences of the Msx1 heterodimeric interactions, we tested the transcriptional activity of Msx1 in the presence of its interacting partners using transient cotransfection assays (Fig. 8 and 9). At first, we used C3H10T1/2 cells and a reporter plasmid encoding a synthetic element containing 3 Msx1-specific binding sites upstream from the simian virus 40 (SV40) promoter and a luciferase reporter gene (Fig. 8A). We found that at equimolar concentrations, Msx1 was a potent repressor of this reporter, while transfection of Dlx2, Pax9, Lef1, and Snail resulted in modest or minimal activation of the promoter in a dose-dependent manner compared with the results for the control (empty expression plasmid) (Fig. 8B, panels i to iv). The Msx1-mediated repression, however, was blocked in a concentration-dependent manner by cotransfection with Dlx2, Pax9, Lef1, and Snail, leading to activation of the Msx1-dependent promoter (Fig. 8B, panels i to iv). Interestingly, the ability of these transcription factors to block repression by Msx1 is more potent than their ability to activate the promoter when transfected alone.
Fig 8.
Functional consequences of the interaction of Msx1 with tooth-specific transcription factors for the transcriptional regulation of an artificial Msx1-dependent promoter. (A) Schematic representation of reporter plasmid containing 3 Msx1 binding sites (3× Msx1). (B) C3H10T1/2 cells were cotransfected with 1 μg of reporter plasmid containing 3× Msx1-Luc, along with increasing amounts of expression plasmid Msx1-FLAG (0.25 μg, 0.50 μg, 0.75 μg, and 1.0 μg). Cells were harvested 24 h after transfection for reporter gene assays. Transcription efficiencies were determined using Renilla luciferase plasmid. For this and subsequent experiments, the levels of luciferase activity were normalized to Renilla luciferase activity and expressed as fold luciferase activity relative to the level of luciferase activity from cells transfected with the reporter construct and empty expression plasmid. Effects of Dlx2 (i), Pax9 (ii), Lef1 (iii), Sna (iv), Lhx6 (v), Lhx8 (vi), and Sp3 (vii) on Msx1-mediated transcription are shown. One microgram of the 3× Msx1-Luc reporter plasmid was cotransfected with 0.25 μg of Msx1-FLAG or 0.25 μg of the indicated transcription factor, or increasing amounts of transcription factors were added (0.25 μg, 0.50 μg, 0.75 μg, and 1.0 μg) along with 0.25 μg of Msx1-FLAG. All the transfection experiments were performed three times, and results are shown as means ± standard deviations. Pax9, Dlx2, Sna, and Lef1 acted as activators of transcription at the 3× Msx1 promoter in conjugation with Msx1, whereas Lhx8 and Lhx6 with Msx1 resulted in repression. The repression by Sp3 at the 3× Msx1 promoter did not change in the presence of Msx1.
These results further confirm previous studies showing that Msx1 acts as a repressor and that Dlx2 and Pax9 act as activators that, upon interaction with Msx1, antagonize and thus alleviate Msx1 repression in a concentration-dependent manner (41, 42, 44). More importantly, they show that the newly identified Msx1 interactors Lef1 and Snail alone did not show any effect on the transcription of the Msx1-dependent promoter. However, upon interaction with Msx1, both Lef1 and Snail antagonize transcriptional repression by Msx1 in a concentration-dependent manner.
When we performed similar experiments to determine the transcriptional activity of Msx1 in the presence of Lhx6, Lhx8, and Sp3, we found that, at equimolar concentrations, transfection of Lhx6, Lhx8, and Sp3 resulted in potent repression of the Msx1-dependent promoter (Fig. 8B, panels v to vii). We also found that the Msx1-mediated repression was further increased in a concentration-dependent manner by cotransfection with Lhx6 and Lhx8 but remained unaffected by cotransfection with Sp3 (Fig. 8B, panels v to vii). These results show that Lhx6 and Lhx8 act synergistically with Msx1 in controlling transcriptional repression by Msx1. They also show that, although Sp3 is able to interact with Msx1 in vitro and in vivo, it is not able to modulate the properties of Msx1-mediated transcription on the Msx1-dependent promoter in the absence of any Sp3 binding site.
Msx1 heterodimeric interactions differentially modulate p19ink4d transcriptional activity, promoting either functional antagonism or synergism.
Using the artificial promoter containing Msx1 binding sites, we showed that the heterodimeric interactions of Msx1 with its protein partners promote either functional antagonism or synergism, depending on its protein partner.
To determine whether similar functional consequences occur in the presence of the relevant p19ink4d promoter in vivo, we tested the transcriptional activity of the p19ink4d promoter in the presence of Msx1 and its interacting partners. We tested them alone (Fig. 9B, panel i) or in combination using transient cotransfection assays (Fig. 9, panels ii to viii).
We found that the Msx1-mediated repression was blocked in a concentration-dependent manner by cotransfection with Dlx2, Pax9, Lef1, and Snail, leading to activation of the p19ink4d promoter similar to what we observed with the artificial Msx1-dependent promoter (Fig. 9, panels ii to v). These results further confirm our previous observations that, upon interaction with Msx1, the Msx1-interacting proteins Dlx2, Pax9, Lef1, and Snail antagonize transcriptional repression by Msx1 (Fig. 8). Of interest, Snail alone, in contrast to what we observed with the artificial Msx1-dependent promoter, results in robust transcriptional repression of the p19ink4d promoter. A potential explanation for this difference comes from previous studies showing that Snail acts as a repressor on promoters containing Snail binding sites (45). The p19ink4d promoter contains several Snail binding sites, and in the absence of Msx1, Snail is free to bind to these sites, resulting in transcriptional repression of the p19ink4d promoter (Fig. 6). Upon interaction with Msx1, however, the repression is relieved, resulting in transcriptional activation similar to what we observed with the artificial Msx1-dependent promoter. It is also of note that the interaction of Lef1 with other homeoproteins, like Dlx2 and Pitx2 (46, 47), leads to the synergistic activation of downstream promoters, which in the case of its interaction with Msx1 leads to the activation of both the artificial and the p19ink4d promoter but through functional antagonism of Msx1 repression (46, 47; this study).
We also found that the Msx1-mediated repression was further increased in a concentration-dependent manner by cotransfection with Lhx8 and Sp3 but remained unaffected by cotransfection with Lhx6 (Fig. 9B, panels vii, viii, and vi, respectively). These results show that Lhx8 and Sp3 act synergistically with Msx1 in controlling the transcriptional repression of p19ink4d promoter by Msx1, whereas Lhx6 is not able to modulate the properties of Msx1-mediated transcription on the p19ink4d promoter.
Interestingly, in contrast to what we observed with the artificial promoter, Sp3 is able to modulate the properties of Msx1-mediated transcription when Sp3 binding sites are present (Fig. 6). The p19ink4d promoter contains several Sp3 binding sites, in addition to its Msx1 binding sites. Therefore, it is plausible that, upon interaction, binding of Sp3 to its recognition site may contribute to increased repression of the p19ink4d promoter, implying a synergistic mode of repression.
DISCUSSION
An Msx1-interacting network of transcription factors operates during early tooth development.
Organogenesis is the result of precise spatial and temporal regulation of genes engaged by several regulatory circuits. Of the numerous factors involved, homeodomain-containing transcription factors play a pivotal role in this process. In this study, we used the tooth as a developmental context to identify protein interactors of the Msx1 homeoprotein. We have identified five novel transcription factors, Lhx6, Lhx8, Lef1, Sp3, and Snail, that mediate protein-protein interactions with Msx1. Msx1 and these proteins are encoded by genes known to cause tooth agenesis disorders, so this reveals for the first time the existence of an Msx1-interacting network of transcription factors that may operate during early tooth development.
Msx1 homeoprotein interacts with LIM homeodomain proteins and the Sp family of transcription factors to synergistically repress transcription.
Several studies indicate that the LIM domains of the Lhx proteins function as protein-protein interaction components that modulate the binding of the LIM homeodomain to DNA and thereby regulate the overall transcriptional activity of the Lhx homeoprotein (48–51). In the developing embryo, Lhx6 and Lhx8 are important for the development of the brachial arch, the basal forebrain, and molar dentition (52, 53). During molar tooth development, both Lhx6 and Lhx8 are expressed in the dental mesenchyme at the bud stage, and like Msx1 mutant mice, Lhx6 Lhx8 double mutant mice lack molar teeth due to the failure of specification of the molar mesenchyme (Table 1) (52). In vitro studies have shown that Msx1 can physically interact with Lhx2 in the context of limb development, similar to their Drosophila melanogaster orthologues, muscle segment homeobox (msh) and apterous (ap) (54).
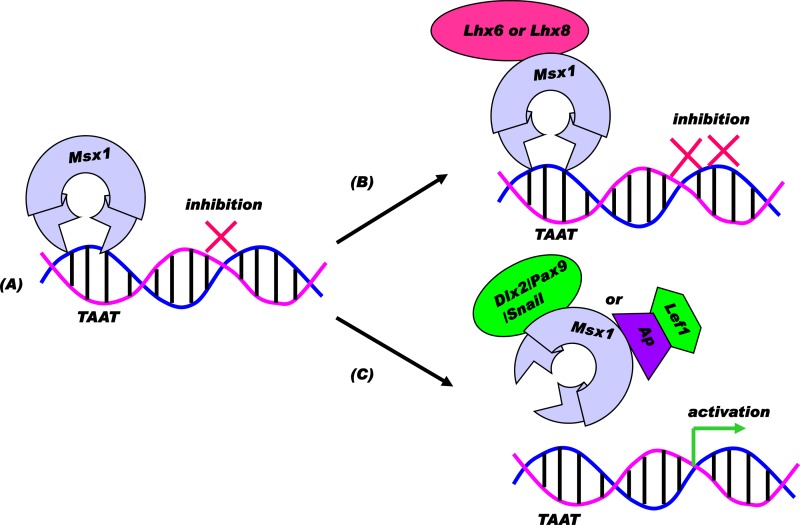
We show that Msx1 interacts with both Lhx6 and Lhx8 at the molecular level and that this interaction results in the enhanced repression of a direct Msx1 target promoter, suggesting a synergistic repression model (Fig. 10B). This increase in repression in the presence of the Msx1-Lhx6 or Msx1-Lhx8 complex could be due to several possible causes. The Msx1-Lhx complex may prevent the recruitment of a transcriptional activator to the promoter or could associate with a corepressor, leading to a transcriptional repression. The latter possibility holds true in the case of a LIM protein that interacts with the zinc finger protein Rlim, leading to the recruitment of a Sin3A/histone deacetylase corepressor complex (49). Similarly, we can hypothesize that Lhx6 or Lhx8 interaction with Msx1 results in enhancement of the repression, probably by recruiting a corepressor.
Fig 10.
Model for transcriptional consequences of Msx1-dependent combinatorial interactions. (A) Direct repression model: repressor Msx1 may bind a target promoter region containing the core motif TAAT via the homeodomain to exert repression. (B) Synergistic repression model: corepressor Sp3, Lhx8, or Lhx6 can interact with Msx1 to enhance repression via functional synergism. (C) Functional antagonism model: activator Dlx2, Pax9, or Snail or Lef1 via an adaptor protein can interact with Msx1 to relieve repression, leading to downstream gene activation via functional antagonism.
The synergistic repression model is also observed when Msx1 interacts with specificity protein 3 (Sp3), a ubiquitously expressed protein that belongs to the Sp family of transcription factors. Although structurally all Sp family members exhibit common features, like a glutamine-rich domain at the N-terminal region of the protein that is known to function as an activation domain, numerous studies indicate that Sp3 functions as a transcriptional repressor and only occasionally as a weak transcriptional activator (55–59). In the context of tooth development, we show that Sp3 alone acts as a transcriptional repressor and that the Msx1-Sp3 interaction further leads to increased repression of the p19ink4d promoter, implying a synergistic mode of repression.
During tooth development, Sp3 is expressed in both dental mesenchyme and epithelium, including the tooth bud mesenchyme. However, Sp3 homozygous null mice exhibit a late tooth phenotype, in contrast to the early tooth phenotype observed in Msx1 null mice, leaving the question of the functional importance of this interaction open (34). What we show, however, is that Sp3 is the only protein to interact with Msx2, another member of the Msx transcription factor family that shares high homology with Msx1. Interestingly, Msx2 homozygous null mice exhibit a late tooth phenotype, similar to the one observed in Sp3 null mice (60). The lack of an early tooth phenotype in Sp3 null mice may be due to functional redundancy with Sp1, another member of the Sp family. Comparative functional studies of Sp1 and Sp3 indicate that these proteins are uniformly expressed during development. Nevertheless, the ratio of Sp1 and Sp3 varies between different cell types during the course of differentiation or exposure to exogenous stimulus. Actually, this relative difference in the amounts of Sp1/Sp3 could account for functional redundancy and also for the differential transcription of several genes, especially given the fact that both Sp1 and Sp3 can recognize and compete for identical DNA-binding sites (61–63). In this context, we can speculate that the amount of Msx1-Sp3 heteromeric complex, together with the relative amount of Sp1 protein in the cell, could potentially account for the selective late tooth phenotype in the Sp3 null mice. Taking into consideration that, like Sp1 and Sp3, Msx1 and Msx2 also show functional redundancy, future studies involving conditional mutants of these genes will further illustrate the functional relationship and significance of these genes during the early and late stages of odontogenesis.
Msx1 homeoprotein interacts with Lef1 homeodomain protein and a Snail superfamily zinc finger transcription factor to activate transcription through functional antagonism of Msx1 repression.
The murine Snail (Sna) contains a zinc finger transcription factor that belongs to the Snail superfamily of C2H2-type zinc finger proteins, and members of this family play a central role in mesoderm formation, in cell movement processes like epithelial-mesenchymal transitions, and in the regulation of cell cycle events (64–67). As a transcription factor, Snail is known to act as a repressor. In Drosophila, for example, Sna mediates protein interactions with the well-known corepressor protein CtBP (carboxy-terminal binding protein) and thereby represses downstream target genes (68). In mammals, Snail represses the E-cadherin promoter by interacting with the corepressor Sin3A/histone deacetylase (HDAC1)/HDAC2 complex (45). On the other hand, Snail induces several genes, such as Slug, Zic5, FoxD3, and Twist; however, the molecular mechanism governing the activator function of Snail is still unknown (69).
Until this study, there has been no report of Snail mediating protein interactions with any homeoprotein. In the context of tooth development, we show that Msx1 interacts with Snail at the molecular level and that this interaction results in the transcriptional activation of a direct Msx1 target promoter through a functional antagonism of Msx1 repression (Fig. 10C). This activation of transcription in the presence of the Msx1-Snail complex could be due to several possible causes. One explanation could be that the Msx1 and Snail interaction could prevent Msx1 binding to the promoter, resulting in relieving the repressor function of Msx1. Another possibility to consider is that the Msx1-Snail complex formation may lead to the recruitment of a transcriptional coactivator, leading to transcriptional activation.
Snail is expressed in the dental mesenchyme at the bud stage; however, as Snail mutant mice die early in gestation due to severe gastrulation defects, it is difficult to assess its particular functional role during tooth development (http://bite-it.helsinki.fi/) (64). There are studies, however, indicating that Snail plays a role in cell cycle regulation, where a functional overlap between Msx1 and Snail may exist. Snail was found to impair cell cycle progression by repressing cyclin D2 transcription by binding directly to the E-box sites within the cyclin D2 promoter. Additionally, Sna provides resistance to cell death induced by developmental cues by activating the mitogen-activated protein kinase (MAPK) and phosphatidylinositol 3-kinase (PI3K) pathways (70). Under physiological conditions, the Msx1 homeoprotein promotes cell proliferation in the dental mesenchyme by repressing the expression of the CDK inhibitor (p19ink4d), thus facilitating cell cycle progression (18). Thus, it is conceivable that, upon interaction with Snail, the repression of the p19ink4d promoter is relieved, leading to activation of the promoter and thereby to the potential inhibition of cell proliferation and cell cycle progression (this study). Based on these results, we may hypothesize that the combinatorial interactions between Msx1 and Snail might help maintain a stringent control over cell proliferation and differentiation during early odontogenesis.
Transcriptional activation of a direct Msx1 target promoter through functional antagonism of Msx1 repression is also observed when Msx1 interacts with Lef1, another homeodomain protein. In the developing embryo, Lef1 is important for the development of hair and tooth (33, 71). During molar tooth development, Lef1 is expressed in the tooth bud mesenchyme, and Lef1 mutant mice exhibit an arrest of tooth development at the bud stage, like Msx1 mutant mice (Table 1) (33). This activation of transcription in the presence of the Msx1-Lef1 complex could be due either to prevention of Msx1 binding to the promoter, resulting in relieving the repressor function of Msx1, or to the recruitment of a transcriptional coactivator, leading to transcriptional activation. Previous studies have shown, for example, that when Lef1 physically interacts with another homeobox transcription factor, Dlx2, the Lef1-Dlx2 interaction synergistically activates the Msx2 promoter, whereas Lef1 alone has no effect on the Msx2 promoter (46). In a similar manner, when Lef1 interacts with Pitx2, the protein-protein interaction between them results in the synergistic activation of the Lef1 promoter (47). In this study, we show that Msx1 interacts indirectly with Lef1 at the molecular level and that, although this interaction results in transcriptional activation of the Msx1 direct target promoter p19ink4D, it does so not through synergism but through functional antagonism of Msx1 repression.
p19ink4d, a cell cycle regulator, is a direct target for the Msx1-dependent protein network.
To determine the physiological relevance of the Msx1 interactions, we have also identified an Msx1 in vivo target, the p19ink4d promoter, that serves as a powerful tool to test how its transcription is regulated by the combinatorial interaction of Msx1 and its partners. We show that, depending on its partner, Msx1 repression of p19ink4d is either blocked, leading to activation, or further enhanced, leading to increased repression in a concentration-dependent manner. Considering the important role of p19ink4d as a cell cycle regulator, these results provide evidence for the first time of the unique plasticity of the Msx1-dependent subnetwork of proteins in conferring differential transcriptional output and in controlling the cell cycle through the regulation of a cyclin D-dependent kinase inhibitor.
Specifically, a key step of the cell cycle is the strict control of the G1/S transition. In the early and late G1 phase of the cell cycle, several mitogenic signals lead to the sequential accumulation and activation of cyclins and cyclin D/E kinases. The assembly of the cyclin/cyclin D-dependent kinase (CDK) complexes is negatively regulated by small polypeptides, the CDK inhibitors (CKIs). Thus, cell cycle progression requires an appropriate balance of positive and negative regulatory factors, such as cyclins, CDKs, and CDK inhibitors (CKIs). These cell cycle regulators must complete each phase of the cycle and, at the same time, must ensure that phase transitions are nonreversible. However, the mechanisms involved in the regulation of this process remain largely unknown.
Some studies have shown that p19ink4d expression is regulated by transcription factors, such as Egr1 that represses p19ink4d in prostate cancer cells, leading to increased cell proliferation (72). Other studies, however, have shown that p19ink4d is induced by FOXO during G1 arrest, Stat3 in macrophage proliferation inhibition, Sp1 in multiple cell lines treated with HDAC inhibitors, and AML-1 in megakaryocytes, leading to cell cycle arrest (73–76). Interestingly, p19ink4d is the only member of the INK4 family of CKIs whose mRNA and protein levels fluctuate periodically during the cell cycle, and recently, it has been shown that E2F1 mediates this periodicity (77, 78). It has been shown that E2F1 has both a proliferative function through the induction of cyclin E and an antiproliferative function through the induction of p19ink4d. Thus, the induction of p19ink4d by E2F1 is an event that takes place during cell cycle progression, and this step could be part of a regulatory network contributing to the fine-tuning of the cell cycle.
Our data provide evidence that the Msx1 transcription factor alone represses the CKI p19ink4d through direct binding to its promoter. When in combination with its interactors, however, and depending on its interactor, Msx1 might induce cell proliferation by further repressing p19ink4d or arrest cell proliferation by activating p19ink4d, thus proving an additional mechanism for controlling the G1/S phase of cell cycle progression. Collectively, these data allows us to speculate that Msx1 through its interactions acts as a switch to control cell cycle progression and may, like E2F1, be part of a regulatory network that contributes to the fine-tuning of the cell cycle progression. In vivo, loss of Msx1 function in mice results in elevated p19ink4d expression and significant reduction of cell proliferation in the dental mesenchyme (15, 18). This suggests that, under physiological conditions, the Msx1 homeoprotein promotes cell proliferation in the dental mesenchyme by controlling the expression of the CDK inhibitor p19ink4d through a timely, balanced control of its repression or activation, thus facilitating cell cycle progression.
In sum, understanding the genetic background of diseases is critical to medical research, with implications in diagnosis, treatment, and drug development. Most of the approaches use only local network information and, thus, are restricted to inferring only a few gene associations. The network of transcription factors identified from our analysis plays a critical role during early tooth development and could facilitate an integrative systems biology approach to elucidating the cellular networks that contribute to tooth agenesis, for example. It would be interesting in the future to provide a global, network-based approach for prioritizing tooth development genes and inferring protein complex associations.
ACKNOWLEDGMENTS
We thank Annette Neübuser (Research Institute of Molecular Pathology, Austria) for Pax9, John Rubenstein (University of California, San Francisco, CA) for Dlx2, and Vassilis Pachnis (National Institute for Medical Research, London, United Kingdom) for Lhx6 and -8 cDNA clones. We are thankful to Martine Roussel (St. Jude Children's Research Hospital, Memphis, TN) for sharing the p19ink4d promoter construct. We thank Jinyun Chen, Erin Howe, Hui Xuan, and Yan Xia for their invaluable assistance.
The study was supported by funds from NIH (grants K22 DE 14230, RO3 DE 018415, and RO1 DE 19226), Harvard Medical School (Milton Fund Award), and Shiseido, Inc., to M.B. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
We have no financial, personal, or professional interests that could be construed to have influenced this paper.
Footnotes
Published ahead of print 10 June 2013
REFERENCES
- 1.Thesleff I, Vaahtokari A, Partanen AM. 1995. Regulation of organogenesis. Common molecular mechanisms regulating the development of teeth and other organs. Int. J. Dev. Biol. 39:35–50 [PubMed] [Google Scholar]
- 2.Bei M, Peters H, Maas RL. 2002. The role of PAX and MSX genes in craniofacial development, p 101–112 In Lin KY, Ogle RC, Jane JA. (ed), Craniofacial surgery: science & surgical technique. W. B. Saunders Company, Philadelphia, PA [Google Scholar]
- 3.Bei M. 2009. Molecular genetics of ameloblast cell lineage. J. Exp. Zool. B Mol. Dev. Evol. 312B:437–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bei M. 2009. Molecular genetics of tooth development. Curr. Opin. Genet. Dev. 19:504–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hayashi S, Scott MP. 1990. What determines the specificity of action of Drosophila homeodomain proteins? Cell 63:883–894 [DOI] [PubMed] [Google Scholar]
- 6.Maas R, Bei M. 1997. The genetic control of early tooth development. Crit. Rev. Oral Biol. Med. 8:4–39 [DOI] [PubMed] [Google Scholar]
- 7.Peters H, Balling R. 1999. Teeth: where and how to make them. Trends Genet. 15:59–65 [DOI] [PubMed] [Google Scholar]
- 8.Thesleff I, Mikkola M. 2002. The role of growth factors in tooth development. Int. Rev. Cytol. 217:93–135 [DOI] [PubMed] [Google Scholar]
- 9.Thesleff I, Sharpe P. 1997. Signaling networks regulating dental development. Mech. Dev. 67:111–123 [DOI] [PubMed] [Google Scholar]
- 10.Tucker A, Sharpe P. 2004. The cutting-edge of mammalian development: how the embryo makes teeth. Nat. Rev. Genet. 5:499–508 [DOI] [PubMed] [Google Scholar]
- 11.Veltmaat JM, Mailleux AA, Thiery JP, Bellusci S. 2003. Mouse embryonic mammogenesis as a model for the molecular regulation of pattern formation. Differentiation 71:1–17 [DOI] [PubMed] [Google Scholar]
- 12.Satokata I, Maas R. 1994. Msx-1 deficient mice exhibit cleft palate and abnormalities of craniofacial and tooth development. Nat. Genet. 6:348–356 [DOI] [PubMed] [Google Scholar]
- 13.Åberg T, Wang XP, Kim JH, Yamashiro T, Bei M, Rice R, Ryoo HM, Thesleff I. 2004. Runx2 mediates FGF signaling from epithelium to mesenchyme during tooth morphogenesis. Dev. Biol. 270:76–93 [DOI] [PubMed] [Google Scholar]
- 14.Bei M, Maas R. 1998. FGFs and BMP4 induce Msx1-dependent and Msx1-independent signaling pathways in early tooth development. Development 125:4325–4333 [DOI] [PubMed] [Google Scholar]
- 15.Bei M, Kratochwil K, Maas R. 2000. BMP4 rescues a non-cell-autonomous function of Msx1 in tooth development. Development 127:4711–4718 [DOI] [PubMed] [Google Scholar]
- 16.Bei M, Chen Y, Woo I, Satokata I, Mass RL. 1996. Control of murine tooth development by Msx1 gene, p 431–440 In Davidovitch Z. (ed), The biological mechanisms of tooth eruption, resorption and replacement by implants. Harvard Society for the Advancement of Orthodontics, Boston, MA [Google Scholar]
- 17.Chen Y, Bei M, Woo I, Satokata I, Maas R. 1996. Msx1 controls inductive signaling in mammalian tooth morphogenesis. Development 122:3035–3044 [DOI] [PubMed] [Google Scholar]
- 18.Han J, Ito Y, Yeo JY, Sucov HM, Maas R, Chai Y. 2003. Cranial neural crest-derived mesenchymal proliferation is regulated by Msx1-mediated p19 (INK4d) expression during odontogenesis. Dev. Biol. 261:183–196 [DOI] [PubMed] [Google Scholar]
- 19.Jumlongras D, Bei M, Stimson JM, Wang WF, DePalma SR, Seidman CE, Felbor U, Maas R, Seidman JG, Olsen BR. 2001. A nonsense mutation in MSX1 causes Witkop syndrome. Am. J. Hum. Genet. 69:67–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van den Boogaard MH, Dorland M, Beemer FA, Ploos van Amstel HK. 2000. MSX1 mutation is associated with orofacial clefting and tooth agenesis in humans. Nat. Genet. 24:342–343 [DOI] [PubMed] [Google Scholar]
- 21.Vastardis H, Karimboux N, Guthua SW, Seidman JG, Seidman CE. 1996. A human MSX1 homeodomain missense mutation causes selective tooth agenesis. Nat. Genet. 13:417–421 [DOI] [PubMed] [Google Scholar]
- 22.Bendall AJ, Ding J, Hu G, Shen MM, Abate-Shen C. 1999. Msx1 antagonizes the myogenic activity of Pax3 in migrating limb muscle precursors. Development 126:4965–4976 [DOI] [PubMed] [Google Scholar]
- 23.Shetty S, Takahashi T, Matsui H, Ayengar R, Raghow R. 1999. Transcriptional autorepression of Msx1 gene is mediated by interactions of Msx1 protein with a multi-protein transcriptional complex containing TATA binding protein, Sp1 and cAMP-response-element-binding protein-binding protein (CBP/p300). Biochem. J. 339:751–758 [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang H, Catron KM, Abate-Shen C. 1996. A role for the Msx-1 homeodomain in transcriptional regulation: residues in the N-terminal arm mediate TATA binding protein interaction and transcriptional repression. Proc. Natl. Acad. Sci. U. S. A. 93:1764–1769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee H, Habas R, Abate-Shen C. 2004. Msx1 cooperates with histone H1b for inhibition of transcription and myogenesis. Science 304:1675–1678 [DOI] [PubMed] [Google Scholar]
- 26.Hu G, Lee H, Price SM, Shen MM, Abate-Shen C. 2001. Msx homeobox genes inhibit differentiation through upregulation of cyclin D1. Development 128:2373–2384 [DOI] [PubMed] [Google Scholar]
- 27.Mann RS, Affolter M. 1998. Hox proteins meet more partners. Curr. Opin. Genet. Dev. 8:423–429 [DOI] [PubMed] [Google Scholar]
- 28.Qiu M, Bulfone A, Martinez S, Meneses S, Shimamura K, Pedersen RA, Rubenstein JLR. 1995. Null mutation of Dlx-2 results in abnormal morphogenesis of proximal first and second branch arch derivatives and abnormal differentiation in the forebrain. Genes Dev. 9:2523–2538 [DOI] [PubMed] [Google Scholar]
- 29.Qiu M, Bulfone A, Ghattas I, Meneses JJ, Sharpe PT, Presley R, Pedersen RA, Rubenstein JLR. 1997. Role of the Dlx-1 and -2 alter morphogenesis of proximal skeletal and soft tissue structures derived from the first and second arches. Dev. Biol. 185:165–184 [DOI] [PubMed] [Google Scholar]
- 30.Thomas BL, Tucker AS, Qui M, Ferguson CA, Hardcastle Z, Rubenstein JLR, Sharpe PT. 1997. Role of Dlx1 and Dlx2 genes in patterning of murine dentition. Dentition 124:4811–4818 [DOI] [PubMed] [Google Scholar]
- 31.Mucchielli ML, Mitsiadis TA, Raffo S, Brunet JF, Proust JP, Goridis C. 1997. Mouse Otlx2/RIEG expression in the odontogenic epithelium precedes tooth initiation and requires mesenchyme-derived signals for its maintenance. Dev. Biol. 189:275–284 [DOI] [PubMed] [Google Scholar]
- 32.Semina EV, Reiter R, Leysens NJ, Alward WL, Small KW, Datson NA, Siegel-Bartelt J, Bierke-Nelson D, Bitoun P, Zabel BU, Carey JC, Murray JC. 1996. Cloning and characterization of a novel bicoid-related homeobox transcription factor gene, RIEG, involved in Rieger syndrome. Nat. Genet. 14:392–399 [DOI] [PubMed] [Google Scholar]
- 33.van Genderen C, Okamura RM, Fariñas I, Quo RG, Parslow TG, Bruhn L, Grosschedl R. 1994. Development of several organs that require inductive epithelial-mesenchymal interactions is impaired in LEF-1-deficient mice. Genes Dev. 8:2691–2703 [DOI] [PubMed] [Google Scholar]
- 34.Bouwman P, Gollner H, Elsasser HP, Eckhoff G, Karis A, Grosveld F, Philipsen S, Suske G. 2000. Transcription factor Sp3 is essential for post-natal survival and late tooth development. EMBO J. 19:655–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peters H, Neubüser A, Kratochwil K, Balling R. 1998. Pax9 deficient mice lack pharyngeal pouch derivatives and teeth and exhibit craniofacial and limb abnormalities. Genes Dev. 12:2735–2747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gupta V, Bei M. 2006. Modification of Msx1 by SUMO-1. Biochem. Biophys. Res. Commun. 345:74–77 [DOI] [PubMed] [Google Scholar]
- 37.Feuerstein R, Wang X, Song D, Cooke NE, Liebhaber SA. 1994. The LIM/double zinc-finger motif functions as a protein dimerization domain. Proc. Natl. Acad. Sci. U. S. A. 91:10655–10659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schmeichel KL, Beckerle MC. 1994. The LIM domain is a modular protein-binding interface. Cell 79:211–219 [DOI] [PubMed] [Google Scholar]
- 39.Taira M, Evrard JL, Steinmetz A, Dawid IB. 1995. Classification of LIM proteins. Trends Genet. 11:431–432 [DOI] [PubMed] [Google Scholar]
- 40.Nieto MA. 2002. The Snail superfamily of zinc finger transcription factors. Nat. Rev. Mol. Cell. Biol. 3:155–166 [DOI] [PubMed] [Google Scholar]
- 41.Ogawa T, Kapadia H, Wang B, D'Souza RN. 2005. Studies on Pax9-Msx1 protein interactions. Arch. Oral Biol. 50:141–145 [DOI] [PubMed] [Google Scholar]
- 42.Zhang H, Hug G, Wang H, Sciavolino P, Iler N, Shen MM, Abate-Shen C. 1997. Heterodimerization of Msx and Dlx homeoproteins results in functional antagonism. Mol. Cell. Biol. 17:2920–2932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Woloshin P, Song K, Degnin C, Killary AM, Goldhamer DJ, Sassoon D, Thayer MJ. 1995. MSX1 inhibits myoD expression in fibroblast × 10T1/2 cell hybrids. Cell 82:611–620 [DOI] [PubMed] [Google Scholar]
- 44.Givens ML, Rave-Harel N, Goonewardena VD, Kurotani R, Berdy SE, Swan CH, Rubenstein JL, Robert B, Mellon PL. 2005. Developmental regulation of gonadotropin-releasing hormone gene expression by the MSX and DLX homeodomain protein families. J. Biol. Chem. 280:19156–19165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Peinado H, Ballestar E, Esteller M, Cano A. 2004. Snail mediates E-cadherin repression by the recruitment of the Sin3A/histone deacetylase 1 (HDAC1)/HDAC2 complex. Mol. Cell. Biol. 24:306–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Diamond E, Amen M, Hu Q, Espinoza HM, Amendt BA. 2006. Functional interactions between Dlx2 and lymphoid enhancer factor regulate Msx2. Nucleic Acids Res. 34:5951–5965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vadlamudi U, Espinoza HM, Ganga M, Martin DM, Liu X, Engelhardt JF, Amendt BA. 2005. PITX2, β-catenin and LEF-1 interact to synergistically regulate the LEF-1 promoter. J. Cell Sci. 118:1129–1137 [DOI] [PubMed] [Google Scholar]
- 48.Arber S, Caroni P. 1996. Specificity of single LIM motifs in targeting and LIM/LIM interactions in situ. Genes Dev. 10:289–300 [DOI] [PubMed] [Google Scholar]
- 49.Bach I, Carriere C, Ostendorff HP, Andersen B, Rosenfeld MG. 1997. A family of LIM domain-associated cofactors confer transcriptional synergism between LIM and Otx homeodomain proteins. Genes Dev. 11:1370–1380 [DOI] [PubMed] [Google Scholar]
- 50.Breen JJ, Agulnick AD, Westphal H, Dawid IB. 1998. Interactions between LIM domains and the LIM domain-binding protein Ldb1. J. Biol. Chem. 273:4712–4717 [DOI] [PubMed] [Google Scholar]
- 51.Dawid IB, Breen JJ, Toyama R. 1998. LIM domains: multiple roles as adapters and functional modifiers in protein interactions. Trends Genet. 14:156–162 [DOI] [PubMed] [Google Scholar]
- 52.Denaxa M, Sharpe PT, Pachnis V. 2009. The LIM homeodomain transcription factors Lhx6 and Lhx7 are key regulators of mammalian dentition. Dev. Biol. 333:324–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Grigoriou M, Tucker AS, Sharpe PT, Pachnis V. 1998. Expression of Lhx-6 and Lhx-7, a novel subfamily of Lim homeodomain genes, suggests a role in mammalian head development. Development 125:2063–2074 [DOI] [PubMed] [Google Scholar]
- 54.Lu CH, Rincón-Limas DE, Botas J. 2000. Conserved overlapping and reciprocal expression of msh/Msx1 and apterous/Lhx2 in Drosophila and mice. Mech. Dev. 99:177–181 [DOI] [PubMed] [Google Scholar]
- 55.Dennig J, Beato M, Suske G. 1996. An inhibitor domain in Sp3 regulates its glutamine-rich activation domains. EMBO J. 15:5659–5667 [PMC free article] [PubMed] [Google Scholar]
- 56.Ihn H, Trojanowska M. 1997. Sp3 is a transcriptional activator of the human 2(I) collagen gene. Nucleic Acids Res. 25:3712–3717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kadonaga JT, Carner KR, Masiarz FR, Tjian R. 1987. Isolation of cDNA encoding transcription factor Sp1 and functional analysis of the DNA binding domain. Cell 51:1079–1090 [DOI] [PubMed] [Google Scholar]
- 58.Kennett SB, Udvadia AJ, Horowitz JM. 1997. Sp3 encodes multiple proteins that differ in their capacity to stimulate or repress transcription. Nucleic Acids Res. 25:3110–3117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Suske G. 1999. The Sp-family of transcription factors. Gene 238:291–300 [DOI] [PubMed] [Google Scholar]
- 60.Bei M, Stowell S, Maas R. 2004. Msx2 controls ameloblast terminal differentiation. Dev. Dyn. 231:758–765 [DOI] [PubMed] [Google Scholar]
- 61.Hagen G, Muller S, Beato M, Suske G. 1994. Sp1-mediated transcriptional activation is repressed by Sp3. EMBO J. 13:3843–3851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kumar AP, Butler AP. 1997. Transcription factor Sp3 antagonizes activation of the ornithine decarboxylase promoter by Sp1. Nucleic Acids Res. 25:2012–2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhao L, Chang LS. 1997. The human POLD1 gene. Identification of an upstream activator sequence, activation by Sp1 and Sp3 and cell cycle regulation. J. Biol. Chem. 272:4869–4882 [PubMed] [Google Scholar]
- 64.Carver EA, Jiang R, Lan Y, Oram KF, Gridley T. 2001. The mouse snail gene encodes a key regulator of the epithelial-mesenchymal transition. Mol. Cell. Biol. 21:8184–8188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Manzanares M, Locascio A, Nieto MA. 2001. The increasing complexity of the Snail superfamily in metazoan evolution. Trends Genet. 17:178–181 [DOI] [PubMed] [Google Scholar]
- 66.Nieto MA, Bennet MF, Sargent MG, Wilkinson DG. 1992. Cloning and developmental expression of Sna, a murine homologue of the Drosophila snail gene. Development 116:227–237 [DOI] [PubMed] [Google Scholar]
- 67.Smith DE, Del Amo FF, Gridley T. 1992. Isolation of Sna, a mouse gene homologous to the Drosophila genes snail and escargot: its expression pattern suggests multiple roles during postimplantation development. Development 116:1033–1039 [DOI] [PubMed] [Google Scholar]
- 68.Nibu Y, Zhang H, Bajor E, Barolo S, Small S, Levine M. 1998. dCtBP mediates transcriptional repression by Knirps, Kruppel and Snail in the Drosophila embryo. EMBO J. 17:7009–7020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Aybar MJ, Nieto MA, Mayor R. 2003. Snail precedes slug in the genetic cascade required for the specification and migration of the Xenopus neural crest. Development 130:483–494 [DOI] [PubMed] [Google Scholar]
- 70.Vega S, Morales AV, Ocaña OH, Valdés F, Fabregat I, Nieto MA. 2004. Snail blocks the cell cycle and confers resistance to cell death. Genes Dev. 18:1131–1143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kratochwil K, Dull M, Farinas I, Galceran J, Grosschedl R. 1996. Lef1 expression is activated by BMP-4 and regulates inductive tissue interactions in tooth and hair development. Genes Dev. 10:1382–1394 [DOI] [PubMed] [Google Scholar]
- 72.Virolle T, Krones-Herzig A, Baron V, De Gregorio G, Adamson ED, Mercola D. 2003. Egr1 promotes growth and survival of prostate cancer cells. Identification of novel Egr1 target genes. J. Biol. Chem. 278:11802–11810 [DOI] [PubMed] [Google Scholar]
- 73.Gilles L, Guieze R, Bluteau D, Cordette-Lagarde V, Lacout C, Favier R, Larbret F, Debili N, Vainchenker W, Raslova H. 2008. P19INK4D links endomitotic arrest and megakaryocyte maturation and is regulated by AML-1. Blood 111:4081–4091 [DOI] [PubMed] [Google Scholar]
- 74.Katayama K, Nakamura A, Sugimoto Y, Tsuruo T, Fujita N. 2008. FOXO transcription factor-dependent p15(INK4b) and p19(INK4d) expression. Oncogene 27:1677–1686 [DOI] [PubMed] [Google Scholar]
- 75.O'Farrell AM, Parry DA, Zindy F, Roussel MF, Lees E, Moore KW, Mui AL. 2000. Stat3-dependent induction of p19INK4D by IL-10 contributes to inhibition of macrophage proliferation. J. Immunol. 164:4607–4615 [DOI] [PubMed] [Google Scholar]
- 76.Yokota T, Matsuzaki Y, Miyazawa K, Zindy F, Roussel MF, Sakai T. 2004. Histone deacetylase inhibitors activate INK4d gene through Sp1 site in its promoter. Oncogene 23:5340–5349 [DOI] [PubMed] [Google Scholar]
- 77.Carcagno AL, Marazita MC, Ogara MF, Ceruti JM, Sonzogni SV, Scassa ME, Giono LE, Canepa ET. 2011. E2F1-mediated upregulation of p19INK4d determines its periodic expression during cell cycle and regulates cellular proliferation. PLoS One 6:e21938. 10.1371/journal.pone.0021938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ceruti JM, Scassa ME, Flo JM, Varone CL, Canepa ET. 2005. Induction of p19INK4d in response to ultraviolet light improves DNA repair and confers resistance to apoptosis in neuroblastoma cells. Oncogene 24:4065–4080 [DOI] [PubMed] [Google Scholar]