Abstract
The phosphorylation state of pocket proteins during the cell cycle is determined at least in part by an equilibrium between inducible cyclin-dependent kinases (CDKs) and serine/threonine protein phosphatase 2A (PP2A). Two trimeric holoenzymes consisting of the core PP2A catalytic/scaffold dimer and either the B55α or PR70 regulatory subunit have been implicated in the activation of p107/p130 and pRB, respectively. While the phosphorylation state of p107 is very sensitive to forced changes of B55α levels in human cell lines, regulation of p107 in response to physiological modulation of PP2A/B55α has not been elucidated. Here we show that fibroblast growth factor 1 (FGF1), which induces maturation and cell cycle exit in chondrocytes, triggers rapid accumulation of p107-PP2A/B55α complexes coinciding with p107 dephosphorylation. Reciprocal solution-based mass spectrometric analysis identified the PP2A/B55α complex as a major component in p107 complexes, which also contain E2F/DPs, DREAM subunits, and/or cyclin/CDK complexes. Of note, p107 is one of the preferred partners of B55α, which also associates with pRB in RCS cells. FGF1-induced dephosphorylation of p107 results in its rapid accumulation in the nucleus and formation of larger complexes containing p107 and enhances its interaction with E2F4 and other p107 partners. Consistent with a key role of B55α in the rapid activation of p107 in chondrocytes, limited ectopic expression of B55α results in marked dephosphorylation of p107 while B55α knockdown results in hyperphosphorylation. More importantly, knockdown of B55α dramatically delays FGF1-induced dephosphorylation of p107 and slows down cell cycle exit. Moreover, dephosphorylation of p107 in response to FGF1 treatment results in early recruitment of p107 to the MYC promoter, an FGF1/E2F-regulated gene. Our results suggest a model in which FGF1 mediates rapid dephosphorylation and activation of p107 independently of the CDK activities that maintain p130 and pRB hyperphosphorylation for several hours after p107 dephosphorylation in maturing chondrocytes.
INTRODUCTION
The retinoblastoma family of proteins, also called pocket proteins, consists of the product of the retinoblastoma tumor susceptibility gene and the functionally and structurally related proteins p107 and p130. In their active, hypophosphorylated form, these proteins associate with a variety of transcription factors and chromatin-modifying enzymes negatively regulating cell cycle progression and/or inducing differentiation (1–3). In cycling cells, pocket proteins are hypophosphorylated in early to mid-G1 and become hyperphosphorylated in response to mitogenic activation of G1 cyclin/cyclin-dependent kinase (CDK) complexes coinciding with passage through the restriction point. Mitogens stimulate the expression of D-type cyclins and the downregulation of p27, resulting in the sequential activation of D-type cyclin/CDK4/6 and cyclin E/CDK2 complexes that cooperate to phosphorylate pocket proteins at multiple pro-directed Ser/Thr residues (3–5). These phosphorylation events disrupt interactions with other proteins and modulate intramolecular domain interactions and access to docking sites for other protein regulators (1). One major consequence of pocket protein inactivation is the disruption of pocket protein/E2F4 repressor complexes, which results in derepression of E2F-dependent genes, including those encoding G1/S, S, and M cyclins as well as activator E2Fs, as cells commit to a new round of DNA replication (3, 6). Although G1 and G1/S CDKs become inactivated as cells progress through S phase in a cell-type-dependent manner, the activation of the E2F program, the stabilization of S-M cyclins, and phosphorylation/dephosphorylation events targeting CDKs result in the sequential activation of cyclin A/CDK2 and cyclin B/CDK1 complexes that maintain pocket proteins hyperphosphorylated through the remainder of the cell cycle until late mitosis (7–10), when the three pocket proteins are abruptly and coordinately dephosphorylated (11, 12).
While a quite detailed picture of CDK-dependent phosphorylation and its consequences has been built in the past several years (1, 10), the signals and the major players that mediate dephosphorylation and activation of pocket proteins remain poorly understood. Two major classes of Ser/Thr phosphatases, protein phosphatase 1 (PP1) and PP2A, have been implicated in dephosphorylation of pocket proteins (4, 13). PP1 mediates, at least in part, the dephosphorylation of pRB coinciding with abrupt inactivation of CDK activity in late mitosis through mid-G1 (14). During this cell cycle window, PP1α is found to be associated with pRB (15), and recent detailed structural studies have shown that PP1 and CDKs compete for overlapping docking sites present in the C terminus of pRB that are not conserved in p130/p107. In addition, while p130 and p107 exhibit cyclin/CDK binding sites in the spacer region that separates the conserved A and B domains of the pocket region, these motifs do not bind PP1 (16). On the other hand, we have found that sudden inactivation of CDKs in exponentially growing human cell lines results in rapid, concomitant dephosphorylation of the three pocket proteins (17). This dephosphorylation is mediated by PP2A, as it is inhibited by okadaic acid at concentrations that do not inhibit PP1. Furthermore, it is inhibited by the expression of simian virus 40 small t antigen, and both p130 and p107 associate with the catalytic subunit of PP2A. This led to our proposal that the phosphorylation state of pocket proteins is maintained by a dynamic equilibrium between PP2A and CDKs throughout the cell cycle (17).
Although the importance of PP2A activity in modulating the phosphorylation state of pocket proteins during the cell cycle was clear, the identity of the particular PP2A holoenzyme implicated was discovered more recently. PP2A forms trimeric holoenzymes that contain a catalytic (C) and scaffold (A) core dimer that appears to be directed to different substrates by a regulatory B subunit (18). There are two genes encoding two highly similar forms of PP2A/C and PP2A/A and four different families of B subunits, each with multiple members designated B, B′, B′′, and B′′′ (4, 19). The combination of these subunits into trimeric holoenzymes and with additional subunits results in a myriad of different complexes. For instance, proteomic analysis has identified an excess of 200 distinct PP2A complexes in cells (18). We have recently identified B55α as a regulatory B subunit of PP2A that directs p107 and p130 to the PP2A holoenzyme in human cell lines (20). Indeed, B55α remains bound to p107 even when excess small t antigen is expressed in cells, which displaces multiple B subunits from the PP2A core dimer. Loss- and gain-of-function assays demonstrated that the phosphorylation state of p107 is extremely sensitive to limited changes in the expression of B55α. In these assays, p130 appeared less sensitive while pRB appeared unchanged. We reasoned that these differences were likely due to redundancy with other PP2A holoenzymes that preferentially target pRB and p130. Indeed, we found that PR70 preferentially bound pRB and p130 compared to p107 in in vitro pulldown assays. Altogether, this suggested that, as is the case with CDKs, multiple PP2A holoenzymes may be specialized in mediating dephosphorylation of pocket proteins during the cell cycle or in response to specific cellular cues (20). Indeed, PR70 was found to target pRB for dephosphorylation in response to oxidative stress in a calcium-dependent manner (21).
A question that remained unanswered was whether PP2A plays mostly a non-rate-limiting role in the equilibrium with inducible CDKs or whether cellular cues can trigger rapid activation of pocket proteins without altering CDK activity through potent activation of PP2A. A few previous reports had described PP2A-mediated dephosphorylation of p107 or p130 in response to UV (22), retinoic acid (23), and fibroblast growth factor 1 (FGF1) (24, 25), although the particular PP2A holoenzymes had not been defined. Importantly, it has been shown that p107−/− p130−/− mice exhibit defects in chondrocyte cell cycle exit that impair normal endochondral bone formation (26). Given the critical physiological relevance of p107 activation for chondrocyte cell cycle exit and maturation revealed by ablation of p107/p130 in mice, this study is focused on identification of the PP2A holoenzyme induced by FGF1 in chondrocytes. FGF1 induces rapid dephosphorylation of p107 in rat chondrosarcoma (RCS) cells by a mechanism that is associated with complex formation with PP2A, as knockdown of PP2A/A or PP2A/C inhibits p107 dephosphorylation and chondrocyte cell cycle exit (25). Since FGF signaling is critical for endochondral bone formation, as defects in the FGFR3 result in achondroplasia (27), we sought to characterize the trimeric holoenzyme inducible by FGF and the consequences of targeting p107 during chondrocyte maturation and cell cycle exit. Using RCS cells, we identify PP2A/B55α as the critical holoenzyme targeted to p107 following FGF1 stimulation, which results in generation of larger complexes containing p107 as well as changes in p107 complexes. In concert with activation of p107, we found recruitment of p107 to the E2F-regulated promoter of the MYC gene, which has been found to have downregulated expression during this process. In addition, using proteomics analysis, we found that members of the B family (primarily B55α and to a lesser extent B55δ) are the only B subunits detected in association with p107 in these cells and thus mediate recruitment of the PP2A core dimer. This is also consistent with cooperation of B55α and β55δ small interfering RNAs (siRNAs) in inhibiting p107 dephosphorylation. Of note, p107 complexes containing CDKs and/or PP2A exist in cells at comparable relative levels, a finding which suggests a critical competitive equilibrium between these complexes which is apparently disturbed by FGF signaling.
MATERIALS AND METHODS
Cell culture and cell treatments.
RCS cells were grown in Dulbecco's modified Eagle's medium (DMEM) (Cellgro) supplemented with 9% fetal bovine serum (FBS) (Atlanta Biologicals), 100 U/ml penicillin, and 100 μg/ml streptomycin (Gemini) at 37°C in a humidified atmosphere with 9% CO2. Fibroblast growth factor 1 (Peprotech) was reconstituted in 5 mM Na2(HPO4), pH 8.0, and 0.1% bovine serum albumin (BSA) at a concentration of 5 μg/ml. RCS cells were treated at indicated time points at a final FGF concentration of 5 ng/ml culture medium plus 5 μg/ml heparin (Calbiochem). Primary chondrocyte cultures were prepared from metatarsal rudiments isolated from Sprague-Dawley rat fetuses at 20 days postcoitus (dpc) as previously described (28) and maintained in complete DMEM supplemented with 50 μg/ml ascorbic acid.
Flow cytometric cell cycle analysis.
RCS cells were collected at the indicated time points and fixed in methanol. Prior to analysis, the cells were treated with a solution containing 10% propidium iodide (500 μg/ml), 5 mg/ml 10% RNase A and 80% 1× phosphate-buffered saline (PBS) plus 1% FBS, and incubated at 37°C for 30 min. The cells were analyzed with a FACSCalibur (BD) flow cytometer, and the data were analyzed using Cell Quest software (BD).
Protein expression and analysis.
All protein assays were conducted on ice or at 4°C unless otherwise indicated. Protein extracts were prepared by lysing cells in RCS buffer (50 mM Tris [pH 7.4], 150 mM NaCl, 10 mM KCl, 1 mM EDTA, 1% NP-40, 50 mM NaF, 1 mM Na3VO4, 10 mM sodium pyrophosphate, 10 mM β-glycerophosphate, 1 mM phenylmethylsulfonyl fluoride [PMSF], 10 μg/ml leupeptin, 4 μg/ml pepstatin, 4 μg/ml aprotinin) for 30 min and cleared by centrifugation. Western blot analysis and immunoprecipitations were performed as previously described (17). Proteins were resolved by SDS-PAGE, in which 6% gels were used to determine the phosphorylation status of pocket proteins and 8% to 12% gels were used to assess protein expression. For immunoprecipitation of endogenous proteins, we used ∼1 mg of precleared whole protein lysate per antibody. Lysates were precleared using normal mouse or rabbit IgG and incubated overnight while rocking with specific antibodies and protein A or G beads. Following incubation, the beads were washed 4 to 5 times with RCS buffer. Immunoprecipitates were resolved by SDS-PAGE and analyzed via Western blotting. Subcellular localization studies were performed utilizing the NE-PER nuclear and cytoplasmic extraction kit (Thermo Scientific) according to the manufacturer's directions.
Antibodies.
Anti-p107 (sc-318), anti-cyclin A (sc-596), anti-E2F4 (sc-512), anti-p21 (sc-397), anti-CDK2 (sc-163), and anti-CDK4 (sc-601) rabbit polyclonal antibodies, anti-PP2A/A (sc-6113) and anti-COL2A1 (sc-7764) goat polyclonal antibodies, anti-PP2A-B55α (sc-81606) and anti-cyclin E (sc-248) mouse monoclonal antibodies, and normal rabbit (sc-2027) and normal mouse (sc-2025) IgG were purchased from Santa Cruz Biotechnology. The anti-PP2A-B55α (100C1) rabbit monoclonal and anti-β-actin (4967) rabbit polyclonal antibodies were purchased from Cell Signaling Technology. Anti-p130 (R27020), anti-PP2A/C (1D6), anti-pRB (G3-245), and anti-HSP70 (ADI-SPA-822) monoclonal antibodies were purchased from BD Transduction Laboratories, Upstate Biotech, Millipore, BD Pharmingen, and Enzo Life Sciences, respectively. The anti-PP2A-B55δ (GTX116609) antibody was purchased from GeneTex, and anti-CTCF (ab70303) was purchased from Abcam.
Plasmids and siRNA.
pSG5-puro-Flag-p107 was made by excising the Flag-p107 construct from pCMV-Flag-p107 (20) and inserting it into the pSG5-puro vector. pMSCV-Myc-B55α wild type (WT) and pMSCV-Myc-B55α D197K were described previously (20). pLKO.1 short hairpin RNA (shRNA) Ppp2r2a and scramble plasmids were purchased as a set in bacterial glycerol stock from Sigma (Mission shRNA; SCHLNG-NM_028032). TRCN0000241288 and TRCN0000241290 were used in the shRNA knockdown experiments and renamed shRNA 1 and 2, respectively. Ppp2r2a siRNAs were purchased from Dharmacon, and their sequences are as follows: siRNA 1, 5′-UGACUGGAUCCUACAAUAAUU-3′; siRNA 2, 5′-CAGCAGAUGAUUUGCGAAUUA-3′; siRNA 3, 5′-CAGUAGAGUUUAAUCAUUC-3′. siGENOME nontargeting siRNA pool 2 (D-001206-14) was used as the control. For subcellular localization and cell cycle experiments, Ppp2r2a siRNAs 2 and 3 were used together along with 1 Ppp2r2d siRNA, 5′-GAGACUAUCUGUCGGUGAA-3′. For the rescue experiment, Ppp2r2a siRNAs 2 and 3 were used for open reading frame (ORF) knockdown, while the following 2 siRNAs against the 3′ untranslated region (UTR) were used for UTR knockdown: UTR1, 5′-CCAUGUCUGCUAGCCAUUU-3′; UTR2, 5′-UUGUCUCGGUGGUGGCGUAUU-3′.
Lentiviral infections and transfections.
293T cells were transfected with the pLKO.1 vectors and pCMV-ΔR 8.2 and pCMV-VSVg packaging constructs at a 2:1.3:1 μg DNA ratio, respectively, using the calcium phosphate method. Supernatants were collected 48 h posttransfection and added to the RCS cells, which were selected with medium containing 2 μg/ml puromycin 24 h after infection. For protein expression, all RCS cell transfections were performed using the Fugene HD reagent (Promega) at an 8:1 ratio. siRNA transfections were performed using 37.5 nM siRNA duplex and the reverse transfection protocol with Lipofectamine RNAi Max reagent (Life Technologies) according to the manufacturer's instructions. A total of 9 × 105 cells/10-cm plate or 3 × 105 cells/6-mm plate were transfected twice, 24 h apart, and allowed to grow for 48 to 72 h posttransfection prior to treatment.
Proteomics.
RCS-Flag-p107 and RCS-Myc-B55α WT stable cell lines were made by stably expressing pSG5-puro-Flag-p107 and pMSCV-Myc-B55α WT in RCS cells and selecting with 2 μg/ml puromycin. Colonies expressing levels of tagged protein similar to those of the corresponding endogenous protein were selected for these experiments. About 15 to 20 15-cm plates of exponentially growing cells were collected (4 to 7 mg of protein) and lysed in a buffer containing 50 mM HEPES, pH 7.5, 70 mM potassium acetate [K(OAc)], 5 mM magnesium acetate [Mg(OAc)2], 0.2% maltoside (Thermo Scientific), and a protease inhibitor tablet (Roche). Cell lysates were immunoprecipitated with anti-Flag M2 affinity gel (A2220; Sigma) or anti-Myc affinity gel (A7470; Sigma). The beads were subsequently washed 5 times in lysis buffer and 2 times in MS buffer (0.1 M Tris-HCl; pH 8.5), and the peptides were eluted with saturated urea, treated with Tris(2-carboxyethyl)phosphine (TCEP) (Sigma) and 2-chloracetamide (Sigma), and digested sequentially with endoproteinase Lys-C (Wako) and trypsin (Promega). Liquid chromatography-mass spectrometry experiments were carried out on an EASY-nLC connected to a hybrid LTQ-Orbitrap Classic equipped with a nanoelectrospray ion source (Thermo Scientific). Mass spectrometry spectra were analyzed using Mascot 2.2.0 (Matrix Science) and Scaffold 3 (Proteome Software, Inc.) against human databases essentially as described in reference 29.The resulting false-discovery rate was less than 1%.
FPLC.
RCS cells were treated with FGF at the indicated time points, lysed, and filtered through a 0.22-μm spin column. A total of 600 μg of protein was loaded onto a Superdex 200 10/300 GL column (GE Healthcare) and separated on an AKTA fast protein liquid chromatographer (FPLC) (GE Healthcare) at a flow rate of 0.50 ml/minute and collected in 0.5-ml fractions. The FPLC buffer was filtered and degassed RCS buffer. A total of 40 μl of each protein-containing fraction was resolved using SDS-PAGE and analyzed by Western blotting.
ChIP.
Chromatin immunoprecipitation (ChIP) experiments were carried out essentially as described previously (30, 31). RCS cells (0.5 × 106) were seeded in 15-cm plates, cultured for 36 h, and treated with FGF1-heparin or heparin as indicated above. Treated cells were fixed with 1% formaldehyde-PBS for 15 min at room temperature, and the cross-linking reaction was stopped by adding glycine (0.125 M). Cells were then washed twice with cold PBS and harvested with complete Szak's radioimmunoassay (RIPA) buffer (150 mM NaCl, 1% Nonidet P-40, 0.5% deoxycholate, 0.1% SDS, 50 mM Tris-HCl [pH 8], 5 mM EDTA, protease inhibitor cocktail [Roche], 10 mM PMSF). Cells were then sonicated for 10 cycles of 30 s on and 30 s off (amplitude set at 70) in an ice bath using a Fisher Scientific Model 705 Sonic Dismembrator. Sonicated chromatin was cleared by centrifugation. The protein concentration was quantified by using the bicinchoninic acid (BCA) protein assay kit (Thermo Scientific). Sonicated DNA was about 300 bp. Subsequent steps were performed as described previously (31). After DNA precipitation, the pellet was resuspended in water and analyzed by quantitated PCR (qPCR). SYBR green qPCR mix (Fermentas) was used to determine relative DNA amounts in input chromatin samples and ChIPs (anti-p107, anti-p130, anti-E2F4, and IP mock [IgG]) in 96-well plates using the StepOnePlus PCR instrument (Applied Biosystems). Specific and mock ChIP threshold cycle (CT) values were normalized with input CT values to obtain the percentage of input values separately for each primer set. The percentage of input was calculated, mock ChIP values were subtracted from p107, p130, or E2F4 ChIP values, and results were visualized via Excel graphs. The following antibodies were used for ChIP: anti-p107 (sc-318), anti-E2F4 (sc-1082), and mouse monoclonal IgGs (sc-2025) from Santa Cruz Biotechnology and anti-p130 (R27020) from BD Transduction Laboratories. Reference Sequences (RefSeq) for the rat promoters assayed by ChIP were obtained from the ECR browser (http://ecrbrowser.dcode.org). Primer pairs were designed using Oligo Perfect Designer (www.invitrogen.com) to amplify sequences near E2F elements conserved between rats and humans and/or mice. Primer sequences are as follows: for MYC, GGATCCGGAGTCGCAGTAT and CAAAGCCCTTCTCACTCCAG; for E2F3, GCGTAAACCGTATCCCTTCA and AAAAATAATCGGGGGTCTGG; for CDC6, GTGGGTGTGACTTCGTGTTG and GGAGCTTTGCACTCTTCAGG; and for FGFR1, GTGTGCCTTGGGGTATGATT and CGGAGTGAACCACAAAACCT.
RESULTS
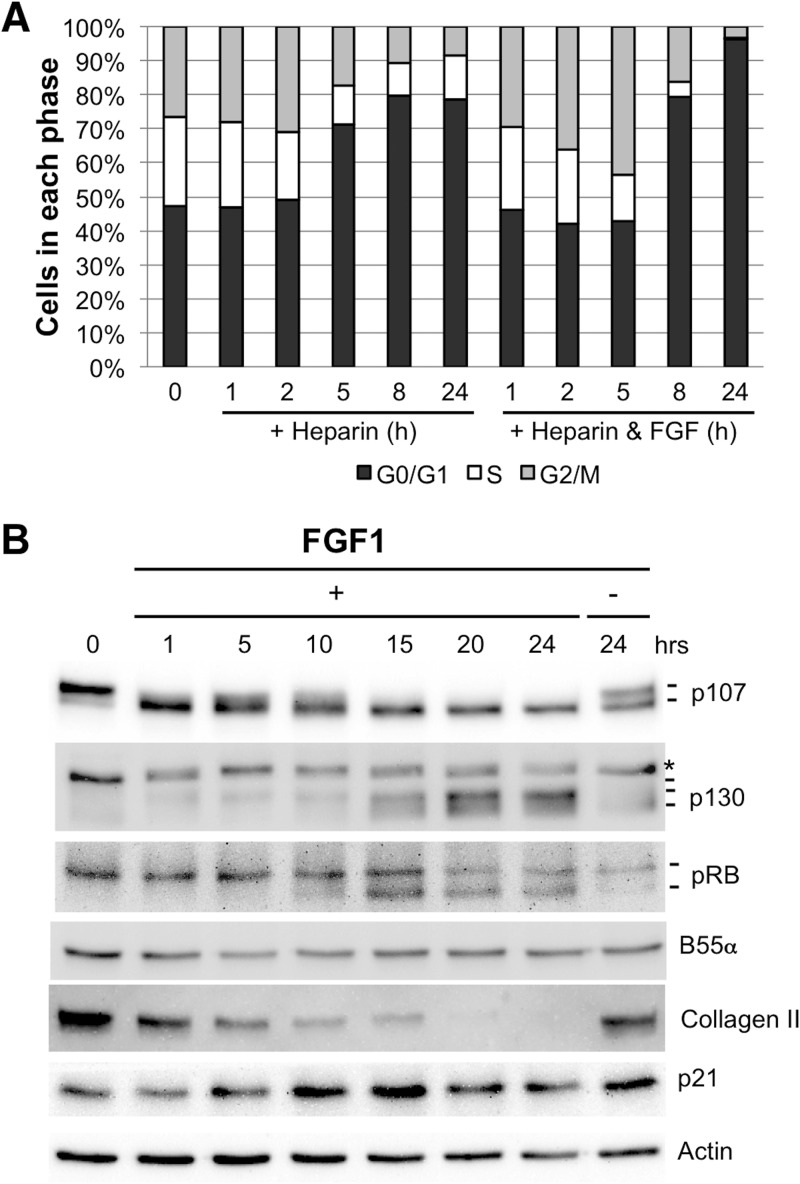
A model of chondrocyte maturation and cell cycle exit in response to treatment with FGFR3 ligands has been extensively characterized (32, 33). RCS cells undergo cell cycle arrest upon treatment with FGF1 (32–35). As shown in Fig. 1A, RCS cells were treated with FGF1 plus heparin or heparin alone at the indicated time points, collected, stained with propidium iodide, and subjected to fluorescence-activated cell sorter (FACS) analysis. A transient accumulation of cells with G2/M DNA content is observed early and subsequently is reduced as cells accumulate with G0/G1 DNA content. The transient arrest at the G2/M transition has previously been shown to be caused by the transient inactivation of cyclin B1/CDK1 complexes following FGF1 stimulation (36). Note that heparin control-treated cells show a much smaller increase in the percentage of cells with G0/G1 DNA content but that the cells continue to proliferate and do not exit the cell cycle. Western blot analysis of lysates of FGF1-treated cells also confirmed the rapid dephosphorylation of p107 1 h after FGF1 treatment, which precedes pRB and p130 dephosphorylation by several hours (Fig. 1B) (24). As we have previously shown that B55α, a PP2A regulatory subunit belonging to the B family, modulates the phosphorylation state of p107 in human cells and that p107 phosphorylation is very sensitive to forced changes in B55α expression, we determined B55α protein levels during the time course (Fig. 1B). B55α protein levels are unaffected by FGF1 treatment, while collagen II, a marker of immature or undifferentiated chondrocytes, gradually decreases. Interestingly, an increase in p21 expression is observed around 10 to 15 h after treatment, coinciding with dephosphorylation of pRB and accumulation of hypophosphorylated p130. The upregulation of p21 in response to FGF1 stimulation in these cells has been reported earlier and coincides with inactivation of cyclin E/CDK2 complexes and pRB hypophosphorylation (34). Since the CDK4/6 inhibitor p16 is also upregulated in response to FGF treatment in these cells (24), dephosphorylation of pRB and p130 is likely a consequence of inhibition of CDK2 and CDK4 while dephosphorylation of p107 is compatible with upregulation of PP2A activity toward p107 as previously reported (25).
Fig 1.
FGF1 induces rapid p107 dephosphorylation prior to pRB dephosphorylation, p130 accumulation, and subsequent G1 arrest and maturation in RCS cells without changes in the expression of B55α. RCS cells were stimulated with FGF1 in the presence of heparin or with heparin alone (control). Cells were collected at the indicated times for cell cycle FACS analysis (A) and Western blot (B). Hyper- and hypophosphorylated forms of p107/p130/pRB and maturation markers are indicated. The asterisk indicates a cross-reacting band.
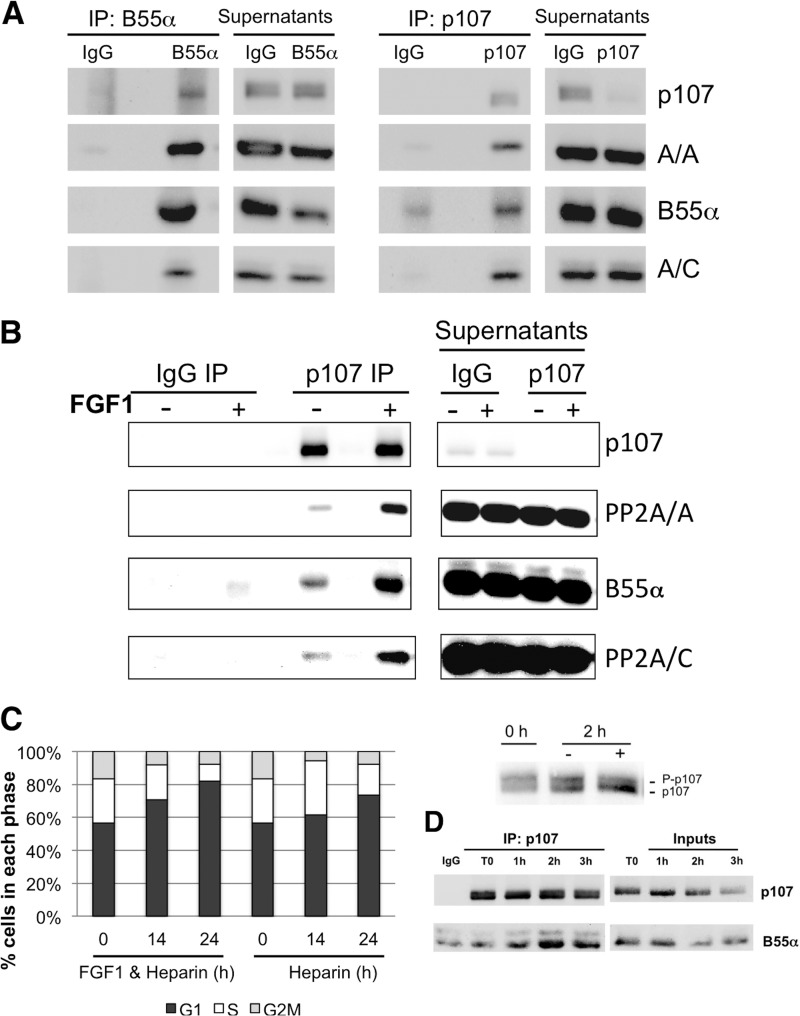
Kolupaeva et al. reported that FGF1 induces transient formation of p107-PP2A/A-PP2A/C complexes in response to FGF1 treatment (25). In this setting, expression of adenovirus E4orf4, which targets primarily subunits of the B55 family (37), blocked FGF-mediated dephosphorylation of p107 (25). While we did not observe modulation of B55α levels by FGF1, it was still conceivable that FGF1 induces complex formation between the PP2A/B55α holoenzyme and p107 via some other mechanism. First, we determined if these complexes are detected in untreated RCS cells, as we have previously detected low levels of these complexes in U2-OS cells. Reciprocal immunoprecipitations with antibodies to p107 and B55α demonstrate the interaction between endogenous p107 and the three subunits of the PP2A/B55α holoenzyme in untreated cells (Fig. 2A). Given that p107 is rapidly dephosphorylated in RCS cells following FGF1 treatment, we next wanted to determine if this signal recruits additional PP2A/B55α holoenzymes to p107. RCS cells were treated with FGF for 30 min (we observe dephosphorylation of p107 as soon as 15 min posttreatment; data not shown), lysed, and immunoprecipitated with p107 antibodies. We detected a marked increase in the association of the three subunits of the PP2A/B55α holoenzyme with p107 without changes in the expression of p107 or the subunits of the B55α/PP2A holoenzyme (Fig. 2B). This upregulation is transient, as it is not maintained at longer time points (data not shown). These data suggest that FGF1 induces formation of a PP2A/B55α-p107 complex via posttranslational modification(s) on any of the subunits involved or the participation of an additional factor regulated by FGF1. We also determined the effects of FGF1 in primary rat chondrocytes obtained as described in Materials and Methods. Treatment of these cells with FGF1 promotes cell cycle arrest at 24 h poststimulation (Fig. 2C). p107 dephosphorylation is clearly detected 2 h poststimulation yet is not as complete as seen in RCS cells, likely due to the unavoidable heterogeneity of the primary cells (compare Fig. 2C, left panel, to Fig. 1B). Immunoprecipitation of endogenous p107 from lysates of FGF1-treated rat chondrocytes with specific antibodies resulted in detection of a B55α/p107 complex, which peaked 2 h poststimulation (Fig. 2D). Thus, the p107/B55α interaction is detectable in a rat cell line and in primary rat chondrocytes, and it is inducible by FGF1.
Fig 2.
(A) Endogenous PP2A/B55α complexes coimmunoprecipitate with p107 in chondrocytes, and the association is markedly increased upon FGF1 treatment in RCS cells. Whole-cell lysates were immunoprecipitated with the indicated antibodies and resolved by Western blot analysis with antibodies to the indicated proteins (A, B, and C). (B) FGF1 induces rapid transient formation of p107/B55α PP2A complexes coinciding with p107 dephosphorylation in RCS cells. RCS cells were stimulated with FGF1 in the presence of heparin for 30 min. (C) FGF1 induces p107 dephosphorylation and growth arrest in primary rat chondrocytes. Primary rat chondrocytes were treated with FGF1 (20 ng/ml) and heparin or heparin alone, and cells were processed as described for Fig. 1A (left) or for Western blot analysis (right). (D) B55α coimmunoprecipitates with p107 in primary rat chondrocytes.
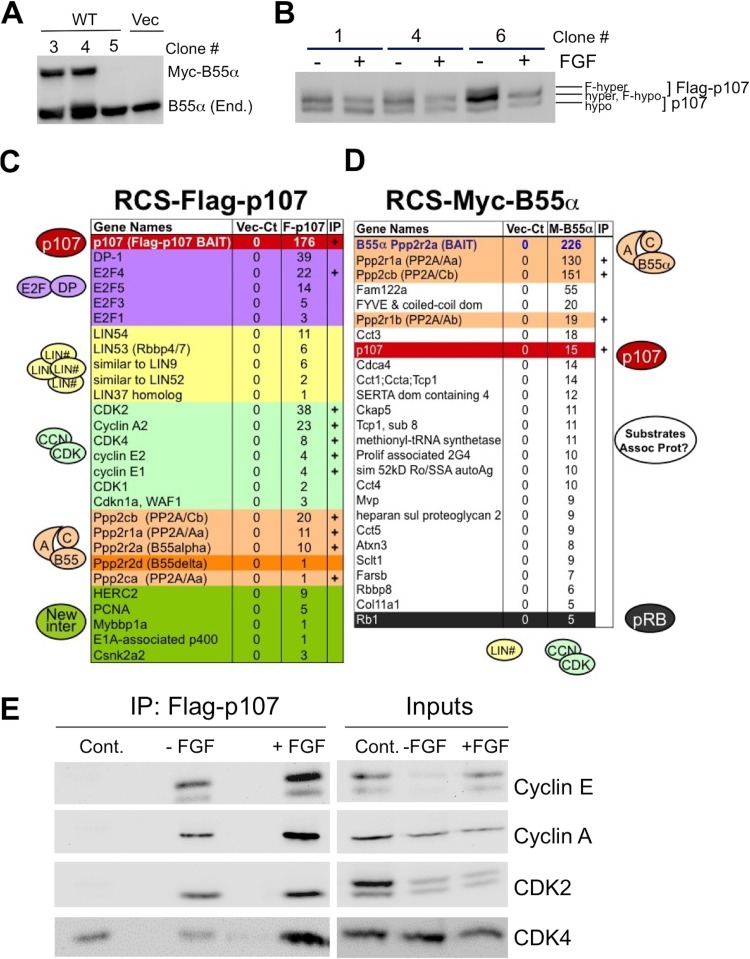
To determine if B55α was the primary B regulatory subunit targeting the PP2A/A-C dimer to p107, we generated RCS cell lines ectopically expressing Myc-tagged B55α and Flag-tagged p107. Myc-B55α was expressed at about 75% of the level of the endogenous protein (Fig. 3A, clone 3), while Flag-p107 was expressed at around ∼120% of the level of endogenous p107, and its phosphorylation state is modulated by FGF1 (Fig. 3B, clone 6). Both cell lines grew efficiently and maintained the transgene stably. We next performed immunoprecipitations from lysates of about 2.25 × 107 cells using antibodies against each tag. Immunoprecipitates were extensively washed, and tryptic peptides were subjected to liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis. As shown in Fig. 3C, the Flag-p107 proteomics analysis yielded peptides that allowed identification of multiple known partners of p107, including E2F/DP1 transcription factors, cyclins and CDKs, multiple subunits of the DREAM complex (LIN proteins) (38), and a number of peptides corresponding to proteins not previously found as partners of p107 (Mybbp1a, p400, and Herc2). Importantly, we detected peptides corresponding to the catalytic B subunit, scaffold, and two members of the B family of regulatory PP2A subunits (B55α and B55δ). Based on the number of spectral hits, B55α is the preferred B subunit partner of p107, with B55δ being significantly less abundant (one peptide unique to B55δ and two peptides common to B55α and B55δ). Of note, no other PP2A/B subunits or subunits of any other phosphatase were detected in the proteomic analysis, suggesting a major role of B55α in modulation of p107 in chondrocytes. In striking agreement with these results, Fig. 3D shows that one of the major partners of the PP2A/B55α holoenzyme in these cells is p107, which was ranked as the 4th most abundant partner of B55α other than PP2A subunits by number of spectral hits. Interestingly, we also detected several peptides corresponding to pRB, suggesting that pRB is also a substrate of this holoenzyme (see Discussion). To confirm these results, we performed immunoprecipitations using Flag-p107 cells and were able to verify many of the binding partners with more spectral hits, including cyclins A and E, CDK2, and CDK4 (Fig. 3E) and PP2A subunits (data not shown). Due to the high sensitivity of the LC-MS/MS and the unavailability of adequate antibodies for detection of endogenous interactions, some of the minor partners have yet to be confirmed via immunoprecipitation followed by Western blot analysis.
Fig 3.
Generation of stable RCS-Flag-p107 and RCS-Myc-B55α cell lines and analysis of Myc-B55α and Flag-p107 complexes in RCS cells via solution-based proteomics analysis. Stable RCS-Myc-B55α and RCS-Flag-p107 clones were generated as described in Materials and Methods (A and B). (A) RCS-Myc-B55α clone 3 was selected for proteomics analysis. Vec, vector. (B) Clone 6 was selected for proteomics analysis. Note that the hypophosphorylated form of Flag-p107 (F-hypo) comigrates with endogenous hyperphosphorylated p107 (hyper) (differentially phosphorylated forms are indicated). (C and D) RCS-Flag-p107 (F-p107), RCS-Myc-B55α (M-B55α), and control RCS-vector (Vec-Ct) cells were lysed, and p107 and B55α complexes were immunoprecipitated with corresponding antitag antibodies and processed for proteomics analysis as described in Materials and Methods. Numbers indicate the numbers of spectral hits and are ranked. (C) Proteins are grouped based on function as E2F/DP, LIN members of DREAM complex, cyclin/CDKs, and B55-PP2A holoenzymes. The last group (green) contains potential unvalidated novel interactions. (D) B55α holoenzyme and potential substrates are indicated on the right. A few spectral hits were also detected for two LIN proteins, a cyclin, and a CDK and are indicated on the right but not shown in this truncated table. If confirmed, these proteins may be associated with p107 or forming independent complexes with B55α. The plus sign indicates confirmation by IP/Western blot (WB) analysis. (E) Confirmation of select binding partners from proteomics analysis. RCS Flag-p107 cells were treated with FGF1 for 2 h, collected, immunoprecipitated using the M2 Flag antibody, and subjected to WB analysis. Cont., the parental RCS line.
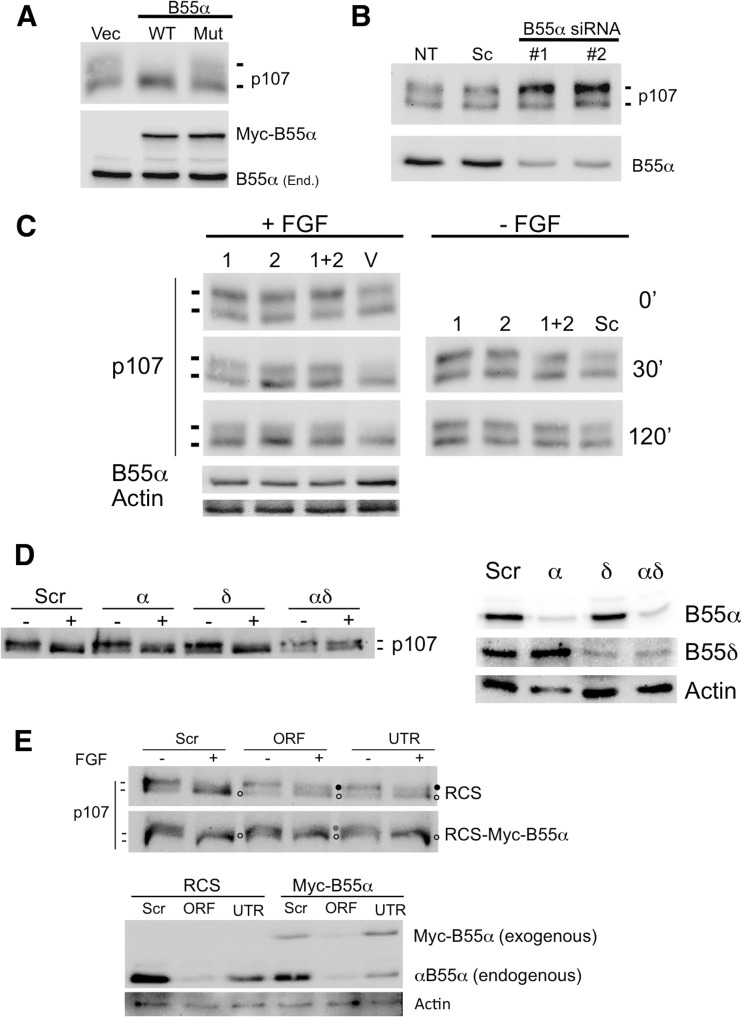
We previously demonstrated that limited ectopic expression of B55α in U2-OS cells results in accumulation of hypophosphorylated p107, and this is not observed with a D197K point mutant that fails to bind p107 (20). To determine whether limited expression of B55α in RCS cells induces p107 dephosphorylation, RCS cells were transfected with Myc-B55α-WT, Myc-B55α-D197K, or an empty vector. Figure 4A shows that limited expression of B55α-WT results in marked accumulation of hypophosphorylated p107, which does not occur in the cells expressing the empty vector or the Myc-B55α-D197K mutant. Conversely, cells transfected with two different B55α siRNAs exhibit increased levels of hyperphosphorylated p107 relative to the untransfected and scramble siRNA controls (Fig. 4B), demonstrating that relative small changes in the expression of B55α protein markedly affect p107 phosphorylation in RCS cells. To determine whether B55α is important for FGF1-mediated dephosphorylation of p107, we used two different shRNAs alone or in combination to knock down B55α and subsequently treated the virally transduced cells with FGF1 for 30 min or 2 h. We have noticed that shRNA knockdown of B55α in these cells is not very efficient. Nevertheless, knockdown of B55α by ∼50% markedly delays FGF1-induced p107 dephosphorylation (Fig. 4C). At 30 min, p107 dephosphorylation is mostly complete in the FGF1-treated shRNA vector control cells but not in the cells with B55α knockdown. This delay is still clearly detected by 2 h of FGF1 treatment. Subsequently, we used multiple siRNAs and double reverse transfection that resulted in more-effective knockdown of B55α and delayed, but not completely blocked, FGF1-induced dephosphorylation (Fig. 4D and E). It is possible that complete and/or more-sustained knockdown of B55α is required to fully block FGF1-induced dephosphorylation of p107. However, since B55δ is likely a minor partner of p107 (Fig. 3C), it is also conceivable that B55δ compensates in a setting in which B55α expression is reduced. Figure 4D shows that although B55δ siRNAs had little effect on p107 dephosphorylation, B55α/B55δ double knockdowns were more effective at delaying p107 dephosphorylation than the B55α knockdown. Moreover, we also determined if the delays in FGF1-dependent p107 dephosphorylation resulting from siRNAs targeting either the ORF or the UTR of B55α can be attenuated by limited expression of a B55α transgene lacking its natural UTR. As expected (Fig. 4E, lower panel), expression of exogenous B55α resulted in more-effective dephosphorylation of p107 and effectively prevented a dephosphorylation delay by the siRNAs targeting the UTR, a result which is clearly seen in the upper panel. However, the siRNA targeting the ORF can still prevent complete dephosphorylation of p107. Considering (i) that multiple different shRNAs and siRNAs targeting B55α but not the vector/scramble controls promote p107 hyperphosphorylation and delay FGF1-dependent p107 dephosphorylation and (ii) the observation that FGF1 induces rapid formation of a B55α/PP2A holoenzyme complex with p107 coinciding with p107 dephosphorylation, we conclude that B55α is the major modulator of the p107 phosphorylation state in RCS cells and the key upstream activator of p107 in FGF signaling in chondrocytes.
Fig 4.
The phosphorylation state of p107 is sensitive to changes in the expression of B55α, and loss of B55α function delays p107 dephosphorylation in response to FGF signaling RCS cells. (A) Overexpression of B55α results in a relative accumulation of hypophosphorylated p107, which is not observed with a D197K point mutant (Mut). Vec, vector; WT, wild type. (B) siRNA-mediated knockdown of B55α results in a relative increase of hyperphosphorylated p107 not seen in cells transfected with scrambled (Sc) siRNA. NT, not transfected. (C) RCS cells were transduced with lentiviruses encoding B55α or control shRNA vectors, selected with puromycin for 3 days, and then stimulated with heparin alone (−FGF) or FGF and heparin (+FGF) for the times indicated. Proteins were detected via WB analysis. V, vector. (D) siRNA-mediated knockdowns of B55α and B55δ cooperate to prevent FGF1-induced p107 dephosphorylation (right). Levels of B55α and B55δ are shown on the right. Scr, scrambled. (E) Expression of B55α resistant to siB55α directed to the UTR promotes p107 dephosphorylation even in the presence of B55α UTR siRNA. p107 hypophosphorylated (white bullets) and hyperphosphorylated (black bullets) forms are indicated. ORF, open reading frame; UTR, untranslated region.
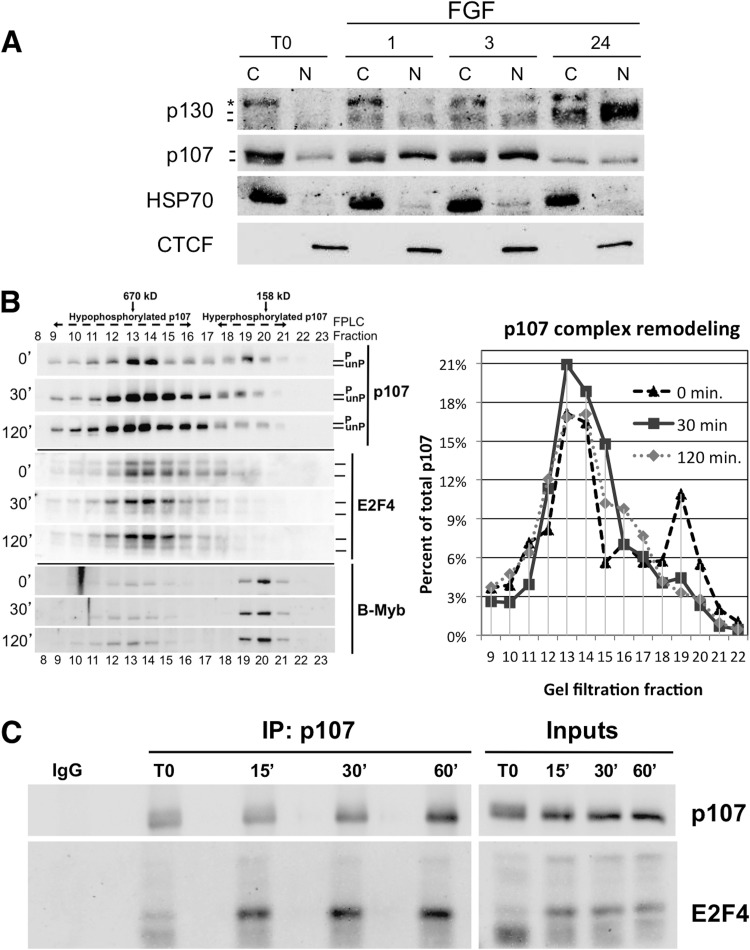
Because FGF1 induces rapid dephosphorylation of p107 and thus appears to generate active p107, we next determined if FGF1 changed the localization of p107. As shown in Fig. 5A, stimulation of RCS cells with FGF1 leads to a rapid relative increase in the localization of p107 to the nucleus by 1 h which is subsequently downregulated as p130 levels accumulate in both the nuclear and cytoplasmic fractions and cells exit the cell cycle. While cytoplasmic fractions consist of hyperphosphorylated and hypophosphorylated p107, the nuclear fraction apparently contains only hypophosphorylated p107. Of note, while knockdown of B55α and B55α/B55δ clearly delayed FGF1-induced dephosphorylation in the cytoplasmic fraction, it had only modest effects in the fraction of p107 relocated to the nucleus (data not shown). This indicates that either relocalization of p107 to the nucleus is not solely dependent on p107 dephosphorylation or, alternatively, the level of knockdown is insufficient to drastically affect p107 accumulation in the nucleus.
Fig 5.
FGF1 induces changes in p107 localization and protein complexes. (A) RCS cells were treated with FGF1 and collected at the indicated time points. Cytoplasmic and nuclear lysates were obtained and resolved by SDS-PAGE for Western blot analysis. The asterisk indicates a cross-reacting band. Hypo- and hyperphosphorylated p107 forms are indicated. (B) Hypophosphorylated p107 elutes with larger complexes, and relative accumulation of these complexes is stimulated by FGF1. Lysates of RCS cells stimulated with FGF1 at the indicated time points were analyzed via gel filtration FPLC followed by a Western blot with p107 and E2F4 antibodies. (C) FGF1 induces rapid formation of p107/E2F4 complexes coinciding with p107 dephosphorylation in RCS cells. RCS cells were stimulated with FGF1 in the presence of heparin for the indicated time points. Whole-cell lysates were immunoprecipitated with p107 antibodies and resolved by Western blot analysis with antibodies to p107 and E2F4.
Next we determined if FGF1 affected the mass of p107 complexes. To this end, we fractionated lysates of cells treated with FGF1 for 30 min and 2 h by FPLC/gel filtration. As seen in Fig. 5B, FGF1 stimulation resulted in decreased detection of hyperphosphorylated p107 in fractions 19 and 20 (the smallest p107 complexes) and a relative increase in the detection of p107 in larger complexes with a peak in fractions of about 670 kDa after 30 min and 2 h. The levels of p107 in each fraction relative to the level of p107 in all fractions combined are shown graphically (Fig. 5B, right), clearly showing the downregulation of the peak containing hyperphosphorylated p107 in small complexes in FGF1-treated cells. We also determined effects of FGF1 on E2F4 and found a major change in the mobility of E2F4 isoforms as well as a relative accumulation of E2F4 in the larger fractions. The changes in E2F4 isoform mobility may be due to posttranslational modifications. No major changes were detected in the fractionation of Myb (Fig. 5A) or the subunits of the PP2A/B55α holoenzyme (data not shown). To verify our FPLC data, we performed p107 IPs on RCS cells treated with FGF1 for 15, 30, and 60 min. p107 dephosphorylation is observed as early as 15 min after the addition of FGF1, and this coincides with an increase in E2F4 association (Fig. 5B). These data show that FGF1-mediated dephosphorylation of p107 results in remodeling of p107 complexes.
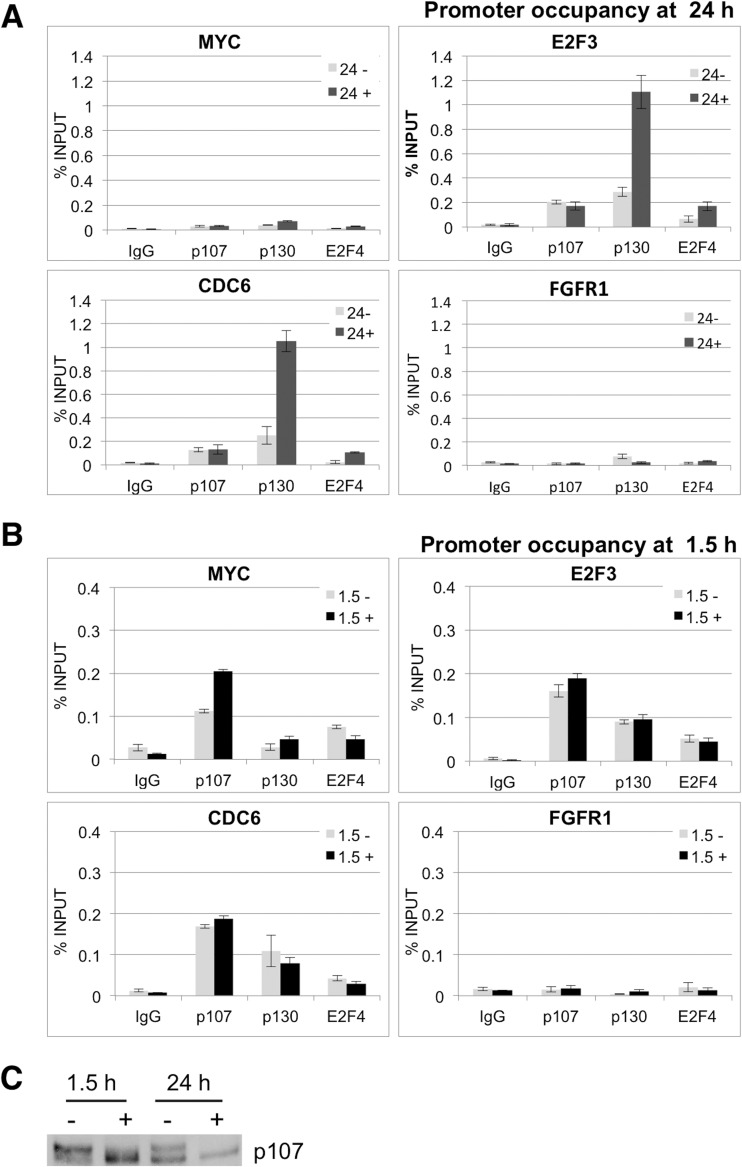
To ascertain the consequences of activation of p107, we determined if recruitment of p107 to promoters of E2F-regulated genes, whose expression is often regulated by FGF1 (24), is modulated. First, we determined recruitment of p107, p130, and E2F4 to the promoters of various genes containing canonical E2F elements, including a subset of genes whose products are downregulated by FGF, 24 h after FGF stimulation via chromatin immunoprecipitation (ChIP) assays. RCS cells were treated in the presence or absence of FGF1 for 24 h, and ChIP assays were performed as described previously (31). Consistent with accumulation of active hypophosphorylated p130 forms known to interact with E2F4 (Fig. 1) (39), we observed increased recruitment of both p130 and E2F4 to the E2F3, CDC6, and CDK2 promoters (Fig. 6A and data not shown). Of note, while p107 was clearly detected at these promoters (Fig. 6A and data not shown), there was no modulation of p107 occupancy by 24 h of FGF1 stimulation. Interestingly, while MYC expression is downregulated by FGF (24) and contains an E2F element in its promoter, we observed little recruitment, if any, of p107/p130 at this time point. As a control, we also determined occupancy at a region of the FGFR1 promoter that, although it contains a consensus E2F sequence, is not known to be functional. None of these factors were detected at this promoter region. Since p107 is activated several hours prior to activation of p130 (Fig. 1B), we next determined recruitment of p107 to the same promoters 1.5 h after FGF stimulation via ChIP analysis. Figure 6B shows that an increase in p107 promoter occupancy is observed at the MYC promoter after 1.5 h of FGF treatment. p107 was also clearly detected at the E2F3 and CDC6 promoters, but p107 occupancy increased only slightly at these promoters. These data are consistent with an FGF1-stimulated repressive role for p107 in the expression of MYC and perhaps other genes that are downregulated early during chondrocyte maturation and the initiation of cell cycle exit. Also of note, while E2F4 is detected at the MYC, E2F3, and CDC6 promoters, E2F4 occupancy is not induced early by FGF1, suggesting that p107 may regulate the MYC promoter in cooperation with factors other than E2F4. De-cross-linked lysates were analyzed by Western blot to monitor p107 dephosphorylation as a control (Fig. 6C).
Fig 6.
FGF1-regulated p107 occupancy at promoters containing canonical E2F elements. (A and B) RCS cells were stimulated with FGF1 and heparin or heparin alone and fixed with formaldehyde 24 (A) or 1.5 (B) h later. ChIPs were performed as described in the experimental design section. (C) p107 dephosphorylation was determined by a Western blot (as described above).
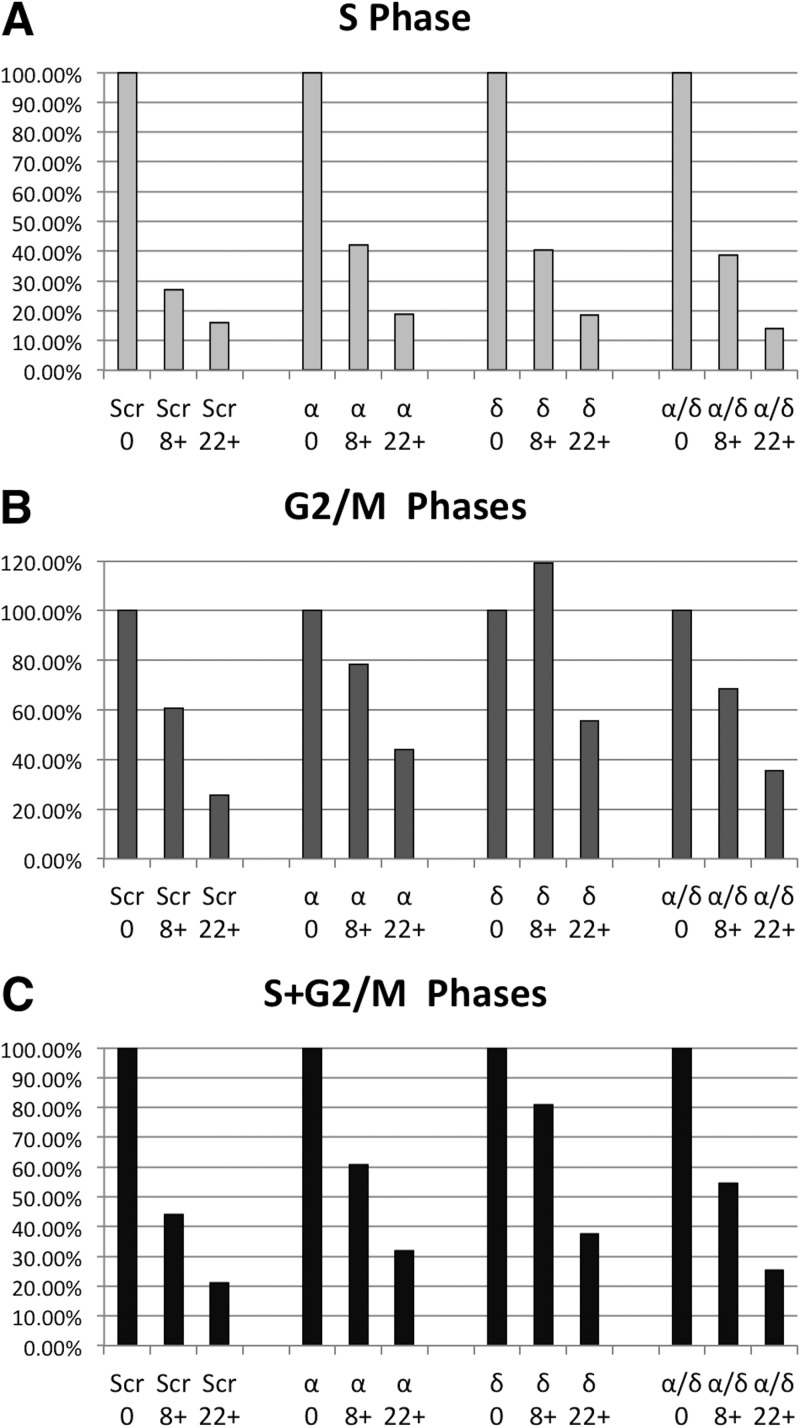
Lastly, we wanted to determine if B55α and/or B55δ is required for FGF1-induced cell cycle exit in RCS cells. siRNAs to both B55α and B55δ delay but do not block cell cycle exit. The effects are seen early, as the reduction in the number of cells in S phase induced by FGF1 is significantly slower in cells with reduced B55α and/or B55δ (8 h) (Fig. 7A). However, these effects are not additive, and most cells appear to exit the cell cycle. In addition, we observe more clear effects in the accumulation of cells with G2/M DNA contents (Fig. 7B), a finding which is consistent with the recently proposed role of these phosphatases in counteracting the action of CDK1 in modulating the phosphorylation state of mitotic substrates (4, 8, 40). Even when considering cells accumulating from S phase to mitosis (Fig. 7C), it is clear that cells efficiently exit the cell cycle by 22 h. It is likely that even in B55α/B55δ knockdown cells, there is enough remaining PP2A activity to activate p107 when CDK activities are downregulated at later times by CKIs coinciding with the activation of p130 and pRB.
Fig 7.
Effects of B55α and/or B55δ loss of function in FGF1-induced cell cycle exit. RCS cells were transfected as described in Fig. 4D and 4 days later were stimulated for the indicated times, collected, and processed for DNA content/flow cytometric analysis. The percentage of cells remaining in S (A), G2/M (B), and S+G2/M (C) phases versus untreated cells is represented at the indicated times.
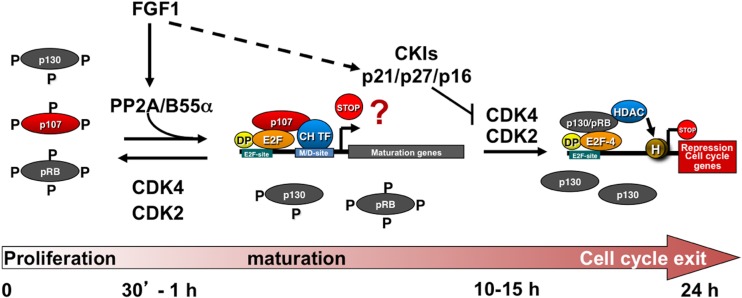
Altogether, our data show that B55α is implicated in the rapid and selective activation of p107 during chondrocyte maturation and cell cycle exit. Active stimulation of B55α/p107 complexes mediates p107 dephosphorylation prior to inactivation of CDK activity that reportedly occurs several hours later. The preferential association of p107 with B55α compared to p130 and pRB (20) (Fig. 3D) likely determines the differential timing of activation of pocket proteins during chondrocyte maturation and cell cycle exit (Fig. 8).
Fig 8.
Differential effects of FGF1 on pocket proteins are mediated by the B55α-PP2A holoenzyme. In this model, FGF1 activates B55α PP2A-mediated dephosphorylation of p107, leading to rapid formation of p107 complexes that target genes regulated early in this process. p130 and pRB remain at least partially hyperphosphorylated and do not play an initial role in controlling gene expression. FGF-dependent upregulation of p21, which results in inactivation of CDK2, and p16, which presumably inactivates CDK4, is likely responsible for the delayed dephosphorylation of these two pocket proteins. When p130 is activated, it may start substituting for p107 in repression of E2F-dependent genes and/or may target new sets of genes. p107 typically forms repressor complexes with E2Fs, but other possibilities, including positive transcriptional regulation with fate-specific transcription factors, are conceivable.
DISCUSSION
The data reported here support a model in which FGF1 promotes assembly of PP2A/B55α holoenzymes with p107 by a mechanism that does not involve upregulation of B55α protein levels (Fig. 8). The relative increase in PP2A/B55α holoenzymes targeting p107 shifts the equilibrium with CDKs toward rapid and selective dephosphorylation and activation of p107 without affecting pRB and p130, which remain hyperphosphorylated. Dephosphorylation of p107 results in its transient accumulation in the nucleus, remodeling of complexes, including an increase in p107/E2F4 and p107/cyclin/CDK complexes, and p107 targeting to the MYC promoter, which is downregulated early in this process, as well as potentially other cell cycle and/or maturation genes. Because p130 and pRB remain hyperphosphorylated early on and, thus, inactive, they do not seem to play an initiating role in controlling gene expression during the very early steps of chondrocyte maturation that culminate in cell cycle exit. Hours after activation of p107, the CDK inhibitors p16 and p21 are upregulated (24, 34), and in the case of p21, this coincides with cyclin E/CDK2 inactivation and pRB dephosphorylation (34), which occurs in parallel with p130 dephosphorylation. This two-tier, sequential activation of pocket proteins and the demonstrated requirement of p107/p130 for chondrocyte cell cycle exit and endochondral bone formation (26, 41) are likely in place to coordinate repression of cell cycle genes and perhaps induction of maturation genes (Fig. 8). Importantly, the proposal that p107 and p130/pRB are activated by FGF through separate pathways, B55α or CKI activation, respectively, is consistent with the finding that simultaneous disruption of p107 and p27 or p57 is analogous to disruption of p107 and p130 in chondrocyte cell cycle exit and endochondral bone formation (26, 42, 43). In this regard, coablation of p107 and p27 in mice strongly suggests that p27 and p130 act in analogous pathways (43).
An equilibrium between cell cycle-induced CDKs and maturation/differentiation-induced PP2A may modulate cell cycle exit decisions.
We have previously shown that an equilibrium exists between CDKs and PP2A to modulate the phosphorylation state of the three pocket proteins during the cell cycle (17). In this equilibrium, sudden pharmacologic inhibition of CDK activity results in immediate dephosphorylation of pocket proteins by PP2A. Thus, cell cycle exit cues that work through accumulation of CDK inhibitors are limited in time by the rate of accumulation of the CKIs that stoichiometrically bind CDKs. In this scenario, a reduction of CDK activity will require sufficient accumulation of the upregulated CKI(s) to warrant a shift in the PP2A/CDK equilibrium toward pocket protein dephosphorylation. Because accumulation of CKIs requires protein synthesis and/or stabilization, this process is not very fast. However, increased recruitment of PP2A holoenzymes to pocket proteins in response to rapid signaling provides a mechanism for very fast activation of particular pocket proteins. In this study, we demonstrate that rapid FGF1 signaling promotes formation of a complex of p107 and PP2A/B55α holoenzymes that mediate p107 dephosphorylation despite the availability of CDK activity that maintains pRB and p130 hyperphosphorylation. Relative shifts in the PP2A/CDK activity ratio due to selective PP2A recruitment to pocket proteins may represent a mechanistic paradigm for rapid activation of pocket proteins. This idea is supported by the finding that oxidative stress induces PR70, a member of the B″ family of PP2A regulatory subunits, resulting in rapid dephosphorylation of pRB, and perhaps also p130/p107, in the absence of CDK inhibition (21).
This equilibrium operating in pocket proteins in which both CDKs and PP2A are regulated is reminiscent of a similar equilibrium between CDK1 and PP2A/B55 holoenzymes that regulates mitotic entry and exit (44). In vertebrates, activation of CDK1 at the G2/M transition appears inadequate to trigger phosphorylation of CDK1 substrates in the absence of downregulation of specific PP2A holoenzymes directed to substrates by B55α and/or B55δ (45–47). As cells enter mitosis, PP2A activity drops as a result of activation of a kinase designated MASTL/GWL, which phosphorylates and activates direct inhibitors (ARPP19 and ENSA) of PP2A/B55 holoenzymes (48, 49). As cells complete mitotic events and anaphase is triggered, CDK1 becomes inactivated and PP2A activity increases and is required for mitotic exit (50, 51). Whether B55α is regulated via inactivation of ARPP19/ENSA or other inhibitors in response to FGF signaling is unclear. In this regard, we detected both ARPP19 and ENSA in B55α immunoprecipitates via proteomics analysis. However, commercial antibodies to ARPP19 and ENSA failed to detect association of these inhibitors with B55α in RCS cells treated or untreated with FGF1 despite these proteins being detected in lysates (data not shown). Further studies are under way to determine the mechanism that promotes the formation of the p107-PP2A/B55α complex.
Rapid activation of p107 preceding pRB dephosphorylation.
p107/p130 double knockout mice die shortly after birth as result of defects on endochondral bone ossification that result in shortened limbs and limit the size of the rib cage interfering with normal breathing (26). The bone defect is due to increased chondrocyte proliferation and cell density in epiphyseal centers, while chondrocytes in the zone of flattened cells exhibit delays in cell cycle exit and hypertrophy. These studies also showed a subtle defect in the thickness of certain forelimb bones in p107−/− mice that was not observed in p130−/− mice (26). Consistent with this finding, p107−/− chondrocytes in micromass cultures fail to exit the cell cycle in response to FGF signaling while pRB−/− chondrocytes exit the cell cycle as wild-type cells in this setting. Thus, early activation of p107 appears to play a unique role in the initiation of cell cycle exit in maturing chondrocytes. Using RCS cells, we observe that p107 is found mostly hyperphosphorylated in exponentially growing cells, and hyperphosphorylated p130 is hardly detectable under these conditions. Several hours after FGF treatment, active hypophosphorylated p130 (forms 1 and 2) (39) becomes readily visible coinciding with pRB dephosphorylation. Since we and others have shown that hyperphosphorylated p130 (form 3) is unstable because it is targeted for proteosomal degradation by the ubiquitin ligase SCFSKP2 (52, 53), inactivation of CDKs concomitant with dephosphorylation of pRB may set the conditions for accumulation of active hypophosphorylated p130, which at this point may cooperate with p107 to control late events in chondrocyte maturation and cell cycle exit. This is also consistent with detecting active recruitment of p107 at the MYC promoter, which has been previously shown to be downregulated following FGF stimulation (41). Of note, although HES1 has also been found to be potently downregulated by FGF1 (data not shown), we found relatively low p107 occupancy following stimulation by FGF1, a finding which reinforces the idea of p107 selectivity and evidences the need for follow-up unbiased ChIP sequencing analysis to identify the key p107 regulated genes in this process.
ACKNOWLEDGMENTS
We thank Victoria Kolupaeva and Claudio Basilico for helpful discussions and reagents. We thank Geoffrey T. Smith for technical assistance.
This work was supported, in part, by National Institutes of Health grant MH083585 to X.G. This work was also supported by a grant from the Pennsylvania Department of Health to X.G. S.H. and M.J.S. were supported by the Beckman Institute and the Gordon and Betty Moore Foundation through grant GBMF775.
Footnotes
Published ahead of print 17 June 2013
REFERENCES
- 1. Dick FA. 2007. Structure-function analysis of the retinoblastoma tumor suppressor protein—is the whole a sum of its parts? Cell Div. 2:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Graña X, Garriga J, Mayol X. 1998. Role of the retinoblastoma protein family, pRB, p107 and p130 in the negative control of cell growth. Oncogene 17:3365–3383 [DOI] [PubMed] [Google Scholar]
- 3. Sotillo E, Graña X. 2010. Escape from cellular quiescence, p 3–22 In Enders GH. (ed), Cell cycle deregulation in cancer. Springer Publishing, New York, NY [Google Scholar]
- 4. Kurimchak A, Graña X. 2012. PP2A holoenzymes negatively and positively regulate cell cycle progression by dephosphorylating pocket proteins and multiple CDK substrates. Gene 499:1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sherr CJ, Roberts JM. 1999. CDK inhibitors: positive and negative regulators of G1-phase progression. Genes Dev. 13:1501–1512 [DOI] [PubMed] [Google Scholar]
- 6. Rowland BD, Bernards R. 2006. Re-evaluating cell-cycle regulation by E2Fs. Cell 127:871–874 [DOI] [PubMed] [Google Scholar]
- 7. Fisher RP. 2012. The CDK network: linking cycles of cell division and gene expression. Genes Cancer 3:731–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kurimchak A, Graña X. 2012. PP2A counterbalances phosphorylation of pRB and mitotic proteins by multiple CDKs: potential implications for PP2A disruption in cancer. Genes Cancer 3:739–748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Enders GH. 2012. Mammalian interphase cdks: dispensable master regulators of the cell cycle. Genes Cancer 3:614–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. MacDonald J, Dick FA. 2012. Posttranslational modifications of the retinoblastoma tumor suppressor protein as determinants of function. Genes Cancer 3:619–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Calbó J, Parreño M, Sotillo E, Yong T, Mazo A, Garriga J, Graña X. 2002. G1 cyclin/CDK coordinated phosphorylation of endogenous pocket proteins differentially regulates their interactions with E2F4 and E2F1 and gene expression. J. Biol. Chem. 277:50263–50274 [DOI] [PubMed] [Google Scholar]
- 12. Graña X. 2008. Downregulation of the phosphatase nuclear targeting subunit (PNUTS) triggers pRB dephosphorylation and apoptosis in pRB positive tumor cell lines. Cancer Biol. Ther. 7:842–844 [DOI] [PubMed] [Google Scholar]
- 13. Kolupaeva V, Janssens V. 2013. PP1 and PP2A phosphatases—cooperating partners in modulating retinoblastoma protein activation. FEBS J. 280:627–643 [DOI] [PubMed] [Google Scholar]
- 14. Ludlow JW, Glendening CL, Livingston DM, DeCarprio JA. 1993. Specific enzymatic dephosphorylation of the retinoblastoma protein. Mol. Cell. Biol. 13:367–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Durfee T, Becherer K, Chen PL, Yeh SH, Yang Y, Kilburn AE, Lee WH, Elledge SJ. 1993. The retinoblastoma protein associates with the protein phosphatase type 1 catalytic subunit. Genes Dev. 7:555–569 [DOI] [PubMed] [Google Scholar]
- 16. Hirschi A, Cecchini M, Steinhardt RC, Schamber MR, Dick FA, Rubin SM. 2010. An overlapping kinase and phosphatase docking site regulates activity of the retinoblastoma protein. Nat. Struct. Mol. Biol. 17:1051–1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Garriga J, Jayaraman AL, Limon A, Jayadeva G, Sotillo E, Truongcao M, Patsialou A, Wadzinski BE, Graña X. 2004. A dynamic equilibrium between CDKs and PP2A modulates phosphorylation of pRB, p107 and p130. Cell Cycle 3:1320–1330 [DOI] [PubMed] [Google Scholar]
- 18. Virshup DM, Shenolikar S. 2009. From promiscuity to precision: protein phosphatases get a makeover. Mol. Cell 33:537–545 [DOI] [PubMed] [Google Scholar]
- 19. Eichhorn PJ, Creyghton MP, Bernards R. 2009. Protein phosphatase 2A regulatory subunits and cancer. Biochim. Biophys. Acta 1795:1–15 [DOI] [PubMed] [Google Scholar]
- 20. Jayadeva G, Kurimchak A, Garriga J, Sotillo E, Davis AJ, Haines DS, Mumby M, Graña X. 2010. B55alpha PP2A holoenzymes modulate the phosphorylation status of the retinoblastoma-related protein p107 and its activation. J. Biol. Chem. 285:29863–29873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Magenta A, Fasanaro P, Romani S, Di Stefano V, Capogrossi MC, Martelli F. 2008. Protein phosphatase 2A subunit PR70 interacts with pRb and mediates its dephosphorylation. Mol. Cell. Biol. 28:873–882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Voorhoeve PM, Watson RJ, Farlie PG, Bernards R, Lam EW. 1999. Rapid dephosphorylation of p107 following UV irradiation. Oncogene 18:679–688 [DOI] [PubMed] [Google Scholar]
- 23. Vuocolo SC, Purev E, Zhang D, Bartek J, Hansen K, Soprano DR, Soprano KJ. 2003. Protein phosphatase 2A associates with Rb2/p130 and mediates retinoic acid-induced growth suppression of ovarian carcinoma cells. J. Biol. Chem. 278:41881–41889 [DOI] [PubMed] [Google Scholar]
- 24. Dailey L, Laplantine E, Priore R, Basilico C. 2003. A network of transcriptional and signaling events is activated by FGF to induce chondrocyte growth arrest and differentiation. J. Cell Biol. 161:1053–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kolupaeva V, Laplantine E, Basilico C. 2008. PP2A-mediated dephosphorylation of p107 plays a critical role in chondrocyte cell cycle arrest by FGF. PLoS One 3:e3447. 10.1371/journal.pone.0003447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cobrinik D, Lee MH, Hannon G, Mulligan G, Bronson RT, Dyson N, Harlow E, Beach D, Weinberg RA, Jacks T. 1996. Shared role of the pRb-related p130 and p107 proteins in limb development. Genes Dev. 10:1633–1644 [DOI] [PubMed] [Google Scholar]
- 27. Ornitz DM, Marie PJ. 2002. FGF signaling pathways in endochondral and intramembranous bone development and human genetic disease. Genes Dev. 16:1446–1465 [DOI] [PubMed] [Google Scholar]
- 28. Wu S, Morrison A, Sun H, De Luca F. 2011. Nuclear factor-κB (NF-κB) p65 interacts with Stat5b in growth plate chondrocytes and mediates the effects of growth hormone on chondrogenesis and on the expression of insulin-like growth factor-1 and bone morphogenetic protein-2. J. Biol. Chem. 286:24726–24734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Haines DS, Lee JE, Beauparlant SL, Kyle DB, den Besten W, Sweredoski MJ, Graham RLJ, Hess S, Deshaies RJ. 2012. Protein interaction profiling of the p97 adaptor UBXD1 uncovers a role for the complex in ERGIC-53 trafficking. Mol. Cell. Proteomics 11:M111.016444. 10.1074/mcp.M111.016444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Donner AJ, Ebmeier CC, Taatjes DJ, Espinosa JM. 2010. CDK8 is a positive regulator of transcriptional elongation within the serum response network. Nat. Struct. Mol. Biol. 17:194–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Keskin H, Garriga J, Georlette D, Graña X. 2012. Complex effects of flavopiridol on the expression of primary response genes. Cell Div. 7:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Krejci P, Bryja V, Pachernik J, Hampl A, Pogue R, Mekikian P, Wilcox WR. 2004. FGF2 inhibits proliferation and alters the cartilage-like phenotype of RCS cells. Exp. Cell Res. 297:152–164 [DOI] [PubMed] [Google Scholar]
- 33. Sahni M, Ambrosetti DC, Mansukhani A, Gertner R, Levy D, Basilico C. 1999. FGF signaling inhibits chondrocyte proliferation and regulates bone development through the STAT-1 pathway. Genes Dev. 13:1361–1366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Aikawa T, Segre GV, Lee K. 2001. Fibroblast growth factor inhibits chondrocytic growth through induction of p21 and subsequent inactivation of cyclin E-Cdk2. J. Biol. Chem. 276:29347–29352 [DOI] [PubMed] [Google Scholar]
- 35. Rozenblatt-Rosen O, Mosonego-Ornan E, Sadot E, Madar-Shapiro L, Sheinin Y, Ginsberg D, Yayon A. 2002. Induction of chondrocyte growth arrest by FGF: transcriptional and cytoskeletal alterations. J. Cell Sci. 115:553–562 [DOI] [PubMed] [Google Scholar]
- 36. Tran T, Kolupaeva V, Basilico C. 2010. FGF inhibits the activity of the cyclin B1/CDK1 kinase to induce a transient G(2) arrest in RCS chondrocytes. Cell Cycle 9:4379–4386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Li S, Brignole C, Marcellus R, Thirlwell S, Binda O, McQuoid MJ, Ashby D, Chan H, Zhang Z, Miron MJ, Pallas DC, Branton PE. 2009. The adenovirus E4orf4 protein induces G2/M arrest and cell death by blocking protein phosphatase 2A activity regulated by the B55 subunit. J. Virol. 83:8340–8352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Litovchick L, Sadasivam S, Florens L, Zhu X, Swanson SK, Velmurugan S, Chen R, Washburn MP, Liu XS, DeCaprio JA. 2007. Evolutionarily conserved multisubunit RBL2/p130 and E2F4 protein complex represses human cell cycle-dependent genes in quiescence. Mol. Cell 26:539–551 [DOI] [PubMed] [Google Scholar]
- 39. Mayol X, Garriga J, Graña X. 1996. G1 cyclin/Cdk-independent phosphorylation and accumulation of p130 during the transition from G1 to G0 lead to its association with E2F-4. Oncogene 13:237–246 [PubMed] [Google Scholar]
- 40. Lorca T, Castro A. 2012. Deciphering the new role of the Greatwall/PP2A pathway in cell cycle control. Genes Cancer 3:712–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Laplantine E, Rossi F, Sahni M, Basilico C, Cobrinik D. 2002. FGF signaling targets the pRb-related p107 and p130 proteins to induce chondrocyte growth arrest. J. Cell Biol. 158:741–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yan Y, Frisen J, Lee MH, Massague J, Barbacid M. 1997. Ablation of the CDK inhibitor p57Kip2 results in increased apoptosis and delayed differentiation during mouse development. Genes Dev. 11:973–983 [DOI] [PubMed] [Google Scholar]
- 43. Yeh N, Miller JP, Gaur T, Capellini TD, Nikolich-Zugich J, de la Hoz C, Selleri L, Bromage TG, van Wijnen AJ, Stein GS, Lian JB, Vidal A, Koff A. 2007. Cooperation between p27 and p107 during endochondral ossification suggests a genetic pathway controlled by p27 and p130. Mol. Cell. Biol. 27:5161–5171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lorca T, Castro A. 2013. The Greatwall kinase: a new pathway in the control of the cell cycle. Oncogene 32:537–543 [DOI] [PubMed] [Google Scholar]
- 45. Castilho PV, Williams BC, Mochida S, Zhao Y, Goldberg ML. 2009. The M phase kinase Greatwall (Gwl) promotes inactivation of PP2A/B55delta, a phosphatase directed against CDK phosphosites. Mol. Biol. Cell 20:4777–4789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Mochida S, Ikeo S, Gannon J, Hunt T. 2009. Regulated activity of PP2A-B55 delta is crucial for controlling entry into and exit from mitosis in Xenopus egg extracts. EMBO J. 28:2777–2785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Vigneron S, Brioudes E, Burgess A, Labbe JC, Lorca T, Castro A. 2009. Greatwall maintains mitosis through regulation of PP2A. EMBO J. 28:2786–2793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Gharbi-Ayachi A, Labbe JC, Burgess A, Vigneron S, Strub JM, Brioudes E, Van-Dorsselaer A, Castro A, Lorca T. 2010. The substrate of Greatwall kinase, Arpp19, controls mitosis by inhibiting protein phosphatase 2A. Science 330:1673–1677 [DOI] [PubMed] [Google Scholar]
- 49. Mochida S, Maslen SL, Skehel M, Hunt T. 2010. Greatwall phosphorylates an inhibitor of protein phosphatase 2A that is essential for mitosis. Science 330:1670–1673 [DOI] [PubMed] [Google Scholar]
- 50. Manchado E, Guillamot M, de Carcer G, Eguren M, Trickey M, Garcia-Higuera I, Moreno S, Yamano H, Canamero M, Malumbres M. 2010. Targeting mitotic exit leads to tumor regression in vivo: modulation by Cdk1, Mastl, and the PP2A/B55α,δ phosphatase. Cancer Cell 18:641–654 [DOI] [PubMed] [Google Scholar]
- 51. Schmitz MH, Held M, Janssens V, Hutchins JR, Hudecz O, Ivanova E, Goris J, Trinkle-Mulcahy L, Lamond AI, Poser I, Hyman AA, Mechtler K, Peters JM, Gerlich DW. 2010. Live-cell imaging RNAi screen identifies PP2A-B55alpha and importin-beta1 as key mitotic exit regulators in human cells. Nat. Cell Biol. 12:886–893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Bhattacharya S, Garriga J, Calbo J, Yong T, Haines DS, Graña X. 2003. SKP2 associates with p130 and accelerates p130 ubiquitylation and degradation in human cells. Oncogene 22:2443–2451 [DOI] [PubMed] [Google Scholar]
- 53. Tedesco D, Lukas J, Reed SI. 2002. The pRb-related protein p130 is regulated by phosphorylation-dependent proteolysis via the protein-ubiquitin ligase SCF(Skp2). Genes Dev. 16:2946–2957 [DOI] [PMC free article] [PubMed] [Google Scholar]