Abstract
Polycomb group (PcG)-mediated repression is an evolutionarily conserved process critical for cell fate determination and maintenance of gene expression during embryonic development. However, the mechanisms underlying PcG recruitment in mammals remain unclear since few regulatory sites have been identified. We report two novel prospective PcG-dependent regulatory elements within the human HOXB and HOXC clusters and compare their repressive activities to a previously identified element in the HOXD cluster. These regions recruited the PcG proteins BMI1 and SUZ12 to a reporter construct in mesenchymal stem cells and conferred repression that was dependent upon PcG expression. Furthermore, we examined the potential of two DNA-binding proteins, JARID2 and YY1, to regulate PcG activity at these three elements. JARID2 has differential requirements, whereas YY1 appears to be required for repressive activity at all 3 sites. We conclude that distinct elements of the mammalian HOX clusters can recruit components of the PcG complexes and confer repression, similar to what has been seen in Drosophila. These elements, however, have diverse requirements for binding factors, which, combined with previous data on other loci, speaks to the complexity of PcG targeting in mammals.
INTRODUCTION
Recruitment of Polycomb group (PcG) proteins to chromatin is required to maintain the silenced state of genes involved in cell fate decisions and identity. Genome-wide PcG profiling revealed that many of the targets are involved in pluripotency, organogenesis, and senescence (1–4). PcG proteins assemble into two major families of functional complexes, Polycomb repressive complexes 1 and 2 (PRC1 and PRC2). The PRC1 family contains several distinct members that have been proposed to repress transcription by a variety of mechanisms, including compaction of nucleosomes, ubiquitylation of histone H2A, and direct repression of the transcription machinery (5) (see reviews in references 6 and 7). Core proteins in this family of complexes in mammals include RING1A/RING1B, BMI1/MEL18, PHC, and CBX proteins (8, 9). Each mammalian subunit has several homologs, and recent findings have demonstrated that the subunits can assemble to form distinct PRC1 complexes with differing biochemical properties (9, 10). PRC2 is a methyltransferase complex whose primary target is lysine 27 of histone H3 (H3K27). The PRC2 family of complexes appears to have this core activity but interact with a variety of binding partners, including JARID2, one of the proteins studied in this work. In humans, the core PRC2 complex is composed of SUZ12, EED, RpAp48, and EZH2, the subunit that bears methyltransferase activity (11–13).
Recent studies have made it clear that targeting of PcG complexes in mammals does not follow a single set of rules. Paradigms for targeting include a two-step model in which PRC2 binds chromatin and trimethylates H3K27 (H3K27me3) and, subsequently, PRC1 recognizes this histone mark, binds, and mediates gene repression (6, 14). They also include the concept that defined PRE sequences, such as those characterized in Drosophila, would bind a set of sequence-specific binding factors to target the PcG system to mediate repression. As an extension, YY1, the mammalian homolog of PHO, would be important for targeting in mammals, analogous to the key role for PHO in targeting PcG function in Drosophila. Additionally, the role for H3K27me3 as a necessary mark in targeting the PcG system has been shown to be an oversimplification, as it is now known that several PRC1 family complexes do not contain a PC/CBX protein that can bind this mark. Indeed, genetically disrupting PRC2 to eliminate methylation of K27 does not alter certain PRC1 targeting events. Thus, H3K27 methylation is important for targeting but is not necessary for all PcG-regulated events (9, 10).
The DNA elements involved in targeting in mammals also show diversity. In the Drosophila genome, Polycomb response elements (PREs) recruit PRC1 and PRC2 via sequence-specific DNA-binding factors. Mutations in these sequences result in ectopic expression of homeotic genes and improper body patterning that was phenotypically similar to mutations in the PcG genes (6). However, such binding sites are not well defined in mammalian cells and cannot be used reliably for PRE identification. The only clear mammalian homolog to a Drosophila PRE binding protein, the YY1 protein, does not appear to be needed for all PRC2 targeting events, and in fact, its overall binding pattern in the genome is anticorrelated with PcG protein binding, demonstrating that the presence of a YY1 binding site is not sufficient to define a PcG binding event (15). These genome-wide analyses do not provide any information, however, concerning whether or not YY1 might be necessary for the function of individual targeting elements. YY1 can both activate and repress transcription depending on the proteins with which it interacts, hence the name Ying Yang 1 (reviewed in reference 16), and similarly, its ability to function as part of a PRE might depend upon context.
In mammals, individual elements with some characteristics similar to those of PREs that confer PcG-dependent repression have been characterized (17, 18). A putative PRE has also been found in T cells and conferred repression of a reporter gene in Drosophila (19). CpG islands have been shown to recruit PRC2 in the absence of activating sites (15) and might broadly contribute to targeting of PcG proteins. Recent studies have shown that the KDM2B/FBXL10 protein, a component of a subset of PRC1 family complexes, can bind GC-rich sequences and help target both PRC1 components and ubiquitylation of histone H2A.
The two proteins we examine here, YY1 and JARID2, are also both DNA-binding proteins that have been connected to PcG targeting. As described above, the YY1 protein plays complex roles in mammals; it has been implicated via point mutation analysis as being necessary for full function of the D11.12 PRE in the human HOXD cluster (18). Similarly, JARID2, a JMJD family member, was recently identified as a substoichiometric subunit of PRC2 and modulator of PRC2 activity that exhibits DNA-binding activity with loose sequence specificity (20–24). How this protein contributes to PcG recruitment and function, and how mechanisms involving JARID2 coordinate with other mechanisms involved in targeting, is an important question.
We present two novel elements within the human genome that can confer transcriptional repression in a Polycomb-dependent manner. We show that PRC1 and PRC2 are recruited to these elements and are required for transcriptional repression. Moreover, identification of these PcG-dependent regulatory sites has allowed us to compare the roles for the proteins YY1 and JARID2 across three HOX PcG-recruiting elements. We found that these elements all require YY1 binding sites for full function but show differences in potency and differences in requirements for JARID2. Thus, overlapping but distinct mechanisms are used to target PcG repression in the HOX clusters in mammals. The mechanisms contribute to our understanding of the full spectrum of mechanisms that govern PcG targeting in mammals.
MATERIALS AND METHODS
Tissue culture.
Mesenchymal stem cells (MSCs) were derived from H1 and H9 human embryonic stem cells (hESCs) (WA01 and WA09; WiCell) by following a protocol described by Seda Tigli (25). Briefly, the hESCs were grown in Iscove’s modified Dulbecco's medium (IMDM)for 2 weeks and then in mesenchymal stem cell growth medium (MSCGM) (Lonza) for 4 days before fluorescence-activated cell sorting (FACS) for cells that are CD73 positive and CD45 and CD19 negative. To check for stable MSC markers, they were analyzed to express CD105, CD73, and CD90 and lack expression of CD45, CD34, CD14 or CD11b, CD79α or CD19, and HLA-DR surface molecules. Cells were also analyzed for expression of BRACHYURY and VIMENTIN by quantitative real-time-PCR (qRT-PCR). MSCs were maintained in MSCGM (Lonza), and cells with passage numbers below p10 were used.
Lentiviral small interfering RNA (siRNA) knockdown.
293FT cells were transfected with Lipofectamine2000 using short hairpin RNA (shRNA) constructs for BMI1 (TRCN0000020155 and TRCN0000020156), EED (TRCN0000021208 and TRCN0000381067), JARID2 (TRCN0000296776 and TRCN0000358748), and control (SHC002) sequences (Sigma) along with the packaging plasmids Gag-Pol and vesicular stomatitis virus (VSV) (Sigma) to create lentiviral particles. Supernatants containing lentivirus were used to infect cells for 48 h. Infected cells were selected for puromycin resistance for at least 14 days.
Expression analyses.
Total RNA was isolated from cells using TRIzol (Invitrogen). RNA was treated with DNase I (Roche) before conversion to double-stranded cDNA using the Vilo cDNA kit (Invitrogen). Inventoried primers for qRT-PCR (Applied Biosystems) were used in the Applied Biosystems 7500 system.
MNase mapping, chromatin immunoprecipitation with microarray technology (ChIP-chip), and ChIP-qPCR.
Micrococcal nuclease (MNase) mapping experiments are described by Woo et al. (18). MNase mapping and chromatin immunoprecipitations (ChIPs) were performed for 2 or more biological replicates. Cells were pretreated with detergent for preextraction of proteins for MNase studies or directly cross-linked for ChIP assays. Pretreatment with detergent would allow for the dissociation of loosely associated nonchromatin proteins from the chromatin. The ChIP protocol provided by Agilent was followed. The following antibodies were used: rabbit anti-human BMI1 (Kingston lab), which was validated in references 18 and 26, SUZ12 (ab12073; Abcam) for ChIP (27, 28), H3 for ChIP (ab1791; Abcam), validated in reference 28, H3K27me3 (ab6002, Abcam) for ChIP, validated in reference 29, YY1 (sc-281; Santa Cruz), validated for IP in reference 30, and JARID2 (sc-134548; Santa Cruz), validated for ChIP by JARID2 knockdown experiments (see Fig. 5A and B). After purification, the DNA was amplified with the WGA2 kit (Sigma). NimbleGen custom-tiled microarrays were used for the mapping experiments. qPCR was used to analyze ChIP DNA in triplicate. For the transiently transfected ChIPs, the percent input of the IP (after subtraction of the rabbit IgG control IP) was normalized to the percent input of the histone H3 IP. For the endogenous locus, percent input was determined as described above without normalization.
Fig 5.

JARID2 is enriched at the endogenous regions in MSCs. (A) Western blot showing the expression of JARID2 in bone marrow-derived MSCs (BM-MSCs) and MSCs and the loss of JARID2 in the knockdown MSCs. (B) ChIP-qPCR results showing the relative enrichment levels of JARID2 at the 3 elements in MSCs and in the JARID2 knockdown cells. (C to E) ChIP-qPCR for the relative enrichment of BMI1, SUZ12, H3, and H3K27me3 at the 3 regions in MSCs and JARID2 knockdown MSCs. (F to H) Luciferase activity of the B4.5, C11.12, and D11.12 constructs in MSCs and JARID2 knockdown MSCs. Asterisks indicate statistically significant repression of luciferase activity relative to the activity observed with A2pLuc. Data are represented as the mean (n = 3) ± SEM.
Western blots.
Lysates were prepared using RIPA buffer and protease inhibitor cocktails (Roche) and probed with JARID2 (sc-134548; Santa Cruz) or the beta-actin (Abcam) at 1:1,000 and the secondary horseradish peroxidase (HRP) antibodies (Amersham) at 1:10,000.
Luciferase assay.
The parental pTranslucent (pLuc, HOXA2pLuc) firefly luciferase constructs were used. B4.5, C11.12, and D11.12 were inserted immediately upstream of the HOXA2 promoter of the HOXA2pLuc construct. The Renilla luciferase plasmid (pRL-TK) (Promega) was used as the assay control. Site-directed mutagenesis of the constructs was done with the QuikChange mutagenesis kit (Stratagene).
Cells were transfected with the firefly luciferase and control Renilla plasmids at a ratio of 10:1. At 48 h postnucleofection, both luciferases were measured with the dual-luciferase reporter assay system (Promega). The Monolight 3010 (Pharmigen) luminometer was used for all readings.
To account for variability between experiments, which is common when using transfection protocols, expression from the experimental construct was first normalized to pRL-TK. The relative light units (RLU) were further normalized by setting the value obtained with the pLuc construct to 0% RLU and that obtained with YY1pLuc to 100% RLU.
For cells that carried the integrated plasmids, the cells were transfected with 5 μg plasmids and selected under antibiotic resistance for 2 months before being used for experiments. Cells were transiently transfected with a plasmid containing FLP recombinase and dsRED to obtain cells that no longer carried the repressive element.
Primer sequences.
For the endogenous regions, the following primer/probe sets were used. For B4.5, the forward primer was 5′-GACAACCTGGTGAAAGCAGA-3′ , the reverse primer was 5′-GGCTTACTCCCTCAGATACCC-3′ , and the probe was 5′-CCACCCACCCTGAATCACACCTC-3′ . For C11.12,the forward primer was 5′-AGTGCTTCAACACCCAGGA-3′ , the reverse primer was 5′-TGTTTAACCTGCAAATTCTCTCC-3′ , and the probe was 5′-CCAATTAGCGTTGATGCACATTCCA-3′ . For the luciferase constructs, the following primer/probe sets were used. For B4.5, the forward primer was 5′-GGATGTTACAGCCTCTGCCT-3′ , the reverse primer was 5′-CGCGTAAGGAGCTCGAAG-3′ , and the probe was 5′-CTTTCGGTTTCCCTTCCGCC-3′ . For C11.12, the forward primer was 5′-CCGGGTGCAAGATAAACC-3′ , the reverse primer was 5′-CCAGCTAGCACGCGTAAG-3′ , and the probe was 5′-CCCAAAGCAAGGGCGAATTCTG-3′ . For the control region near B4.5, the following primer/probe set was used: forward, 5′-TGCCACATATCCAATCCCAG-3′ ; reverse, 5′-GTCACTCCTACTTTCACCAGC-3′ ; probe, 5′-GGTGAGCTGAATCAAGGGATGGCA-3′ . For the control region near C11.12, the following primer/probe set was used: forward, 5′-GACAAATAGAAGCCAGGATAGGG-3′ ; reverse, 5′-CCATAGGATTAAGACCACACGG-3′ ; probe, 5′-GGTTGGTGGGCCTAGAGCTTCAA-3′ .
Microarray data accession numbers.
Microarray data are deposited in the NCBI GEO database with the series accession number GSE19046.
RESULTS
Identification of potential PcG regulatory sites.
Polycomb-dependent transcriptional repression of HOX genes is critical for correct positional embryonic development. Pluripotent human embryonic stem cell lines, H1 and H9, multipotent mesenchymal stem cells (MSCs), and differentiated adipocytes and osteoblasts were used to look for alterations in chromatin architecture at the HOX clusters, signifying changes in gene regulation. In earlier studies, each cell type was characterized by histochemical staining and gene expression of lineage-specific genes and the HOX genes (18). In these earlier studies, we digested chromatin prepared from these cell lineages with micrococcal nuclease (MNase) to map sites that were accessible to nuclease activity. Optimized MNase digestion conditions for each cell type were used to obtain mononucleosome-sized fragments of approximately 150 bp, ensuring that most of the accessible DNA had been digested. The DNA was hybridized to high-resolution tiled microarrays that cover the human HOXA, -B, -C, and -D clusters. In parallel, MNase-digested chromatin from the four cell types was used in chromatin immunoprecipitation (ChIP) experiments for the Polycomb group (PcG) proteins BMI1 and SUZ12 and for H3K27me3 (Fig. 1 and 2). PRC1 and PRC2 components, as well as H3K27me3, have been mapped across the genome of mammalian cells (1, 2, 4, 31), and their occupancy correlates well with a transcriptionally silent state. By using MNase-digested chromatin for ChIP experiments and hybridizing the enriched DNA to microarrays with the same design used for MNase mapping, we identified PcG-associated sites within the HOX clusters with high resolution.
Fig 1.
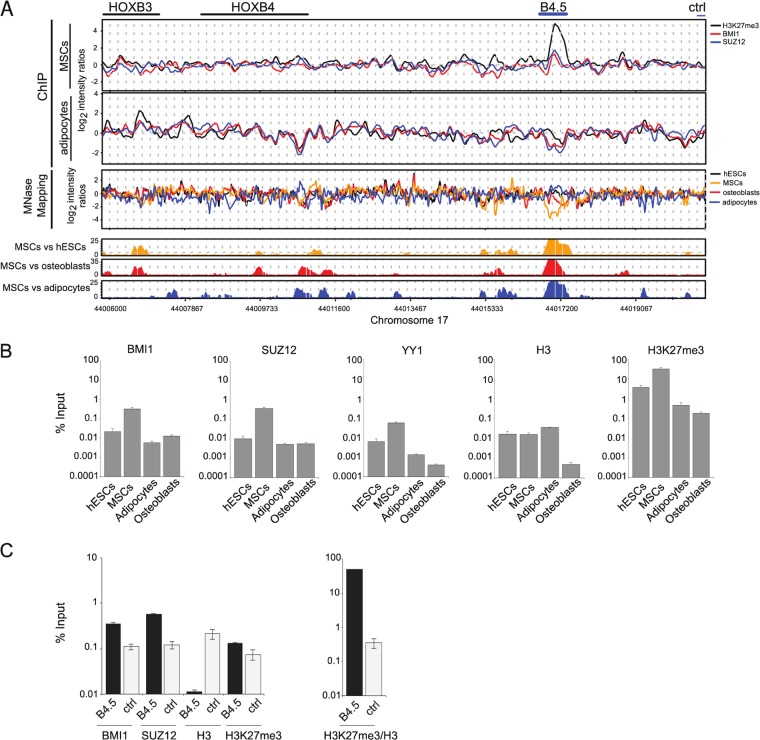
Identification of B4.5, an intergenic region associated with PcG proteins and MNase hypersensitivity. (A) ChIP-microarray results for BMI1, SUZ12, and H3K27me3 from MSCs and adipocytes along the HOXB cluster between HOXB4 and HOXB5 (top) and MNase mapping results for hESCs, MSCs, adipocytes, and osteoblasts across the same region (bottom). The graphs below the MNase mapping data illustrate the statistical significance of the differences between the MSC mapping data and the other cell types. (B) ChIP-quantitative PCR for BMI1, SUZ12, H3, and H3K27me3 from endogenous B4.5 from the hESCs, MSCs, adipocytes, and osteoblasts. Data are represented as the mean (n = 3) ± the standard error of the mean (SEM). (C) ChIP-qPCR from MSCs at the endogenous sites of B4.5 and a control region (chromosome 17, 44021287 to 44021388), also located between HOXB4 and HOXB5. Data are represented as the mean (n = 3) ± SEM.
Fig 2.
Identification of C11.12, an intergenic region associated with PcG proteins and MNase hypersensitivity. (A) ChIP-microarray results for BMI1, SUZ12, and H3K27me3 from MSCs and adipocytes along the HOXC cluster between HOXC11 and HOXC12 (top) and MNase mapping results for hESC, MSCs, adipocytes, and osteoblasts across the region (bottom). The graphs below the MNase mapping data show the statistical significance of the differences between the MSC mapping data and the other cell types. (B) ChIP-quantitative PCR for the enrichment of BMI1, SUZ12, H3, and H3K27me3 from endogenous C11.12 from the hESCs, MSCs, adipocytes, and osteoblasts. Data are represented as the mean (n = 3) ± SEM. (C) ChIP-qPCR from MSCs at the endogenous sites of C11.12 and a control region (chromosome 12, 52639367 to 52639504), also located between HOXC11 and HOXC12. Data are represented as the mean (n = 3) ± SEM.
These data allowed us to identify a region, termed D11.12, that showed low nucleosome density and enrichment for PcG marks and whose function we have previously reported (18). We examined the same large data set and identified two new regions in the HOX clusters that had characteristics similar to those of D11.12 (Fig. 1 and 2). We report here the characterization of these two loci to determine whether they share PRE characteristics with D11.12 and to determine how they and D11.12 compare in their requirements for JARID2 and YY1.
The regions that were identified reside between HOXB4 and HOXB5 and between HOXC11 and HOXC12; they are referred to below as B4.5 and C11.12, respectively. The B4.5 region was identified as a possible PRE based upon MNase cleavage patterns and increased binding by BMI1 and SUZ12, as measured by ChIP experiments (Fig. 1A). The top two panels compare ChIP enrichments for H3K27me3, SUZ12, and BMI1 from the MSCs and adipocytes. These data were confirmed by ChIP-quantitative PCR, which showed that the largest enrichment for these marks occurred in MSCs (Fig. 1B).
The bottom panel of Fig. 1A shows the MNase mapping patterns from the four cell types, and statistically significant differences between the MSCs and the other cell types are plotted below them. The region encompassing B4.5 showed marked MNase mapping differences among all cell types, with the greatest sensitivity to MNase, and, hence, inferred low nucleosome occupancy seen in MSCs. We did not, however, observe depletion of histone H3 at B4.5, as measured by ChIP-qPCR, when MSCs were compared to embryonic stem (ES) cells (Fig. 1B), a finding which contrasts with the expectation based upon the MNase cleavage analysis (see below). When a control region residing between HOXB4 and HOXB5 was analyzed by qPCR in MSCs and compared to B4.5, there was lower enrichment of PcG proteins than at B4.5 (Fig. 1C). The relative enrichment of BMI1, SUZ12, and H3K27me3 at B4.5 compared to the control region was 4.5-fold, 15-fold, and 10-fold, respectively. In contrast, there was an approximately 10-fold-greater enrichment of H3 at the control region relative to B4.5, indicating that B4.5 is, in fact, nucleosome depleted relative to a nearby region in these cells.
C11.12 was uncovered as a potential PcG-mediated regulatory region using identical methods (Fig. 2). As was observed for B4.5, there was enrichment of BMI1, SUZ12, and H3K27me3 at C11.12 by ChIP-microarray (Fig. 2A, top) and by ChIP-qPCR (Fig. 2B). A control region within HOXC11 and HOXC12 did not enrich for BMI1 and SUZ12 (Fig. 2C). The relative enrichment of BMI1, SUZ12, and H3K27me3 at C11.12 compared to the control region was 3-fold, 5-fold, and 2-fold, respectively. Differences in MNase mapping patterns were observed among the four cell types, and the region with the most statistically significant changes was at C11.12 (Fig. 2A). We observed some depletion of H3 at C11.12, as measured by ChIP-qPCR, although the extent of depletion as measured by this technique did not mirror the MNase cleavage patterns observed across the different cell types by microarray analysis. ChIP-qPCR for the control region between HOXC11 and HOXC12 showed a 19-fold-greater amount of H3 than at C11.12 (Fig. 2C). Based on data from the ENCODE project, the region corresponding to B4.5 displays DNase I hypersensitivity in fibroblast cell types, and the region corresponding to C11.12 is hypersensitive to DNase I in several different cell types, consistent with our analysis, indicating that these two regions might confer regulation. We conclude that B4.5 and C11.12 are regions of interest for potential PcG activity based upon increased PcG binding, increased methylation of K27 of histone H3, and significant changes in MNase mapping patterns during differentiation.
Repression of reporter constructs.
Since these regions were associated with PcG components, we used a luciferase reporter assay to determine whether these sequences were able to confer gene repression that was dependent upon PRC1 and PRC2 components. The parental plasmid used, pLuc, has a low level of luciferase activity and serves as the background control. The test construct for repression studies used approximately 1 kb of the human HOXA2 promoter to drive Luc expression (A2pLuc). We chose the HOXA2 promoter because HOXA2 is regulated by PcG activity, HOXA2 is expressed in MSCs, and its promoter was able to drive luciferase expression in these cells. To test the effect of B4.5 on expression from the A2pLuc construct, an approximately 1.4-kb region encompassing B4.5 was cloned upstream of the HOXA2 promoter (B4.5). Cells were transfected with the plasmids, and luciferase activity was measured 48 h posttransfection. Background levels from pLuc were subtracted from all readings, and activity was subsequently normalized relative to the luciferase activity observed with A2pLuc. B4.5 repressed luciferase activity approximately 3-fold relative to the activity of A2pLuc in MSCs with statistical significance (Fig. 3A). Previous work on the D11.12 element indicated that the protein YY1, the mammalian homolog of the Drosophila PcG gene PHO, contributed to the repressive effect conferred by this element. The B4.5 region contains four predicted YY1 binding sites, as diagrammed in Fig. 3B, along with the point mutations made to abrogate YY1 binding (mutB4.5). We confirmed the association of YY1 with the wild-type B4.5 construct; there was no significant detectable binding of YY1 to mutB4.5 using ChIP-qPCR (Fig. 3C). When the mutB4.5 construct was tested in the luciferase assay, the results indicated that mutation of the YY1 sites compromised the ability of B4.5 to repress luciferase activity (Fig. 3A).
Fig 3.
B4.5, C11.12, and D11.12 repress luciferase activity in MSCs. (A, D, and G) Luciferase assays to test the repressive potential of B4.5, C11.12, and D11.12. Constructs are represented on the left of each graph. The parental construct, pLuc, places the firefly luciferase gene under the control of the minimal thymidine kinase promoter. The HOXA2pLuc construct, A2pLuc, places the firefly luciferase gene under the control of 1 kb of the HOXA2 promoter. These plasmids were used in all luciferase assay experiments. Renilla luciferase was cotransfected with the firefly luciferase constructs. Luciferase readings were measured 48 h posttransfection. Results from the luciferase assay are displayed as percent relative light units (%RLU). (A) Luciferase activity of the B4.5 region. (D) Luciferase activity of the C11.12 region. (G) Luciferase activity of the D11.12 region. Asterisks indicate statistically significant repression of luciferase activity relative to the activity observed with A2pLuc calculated using a t test. (B, E, and H) Cloned regions for B4.5, C11.12, and D11.12 shown as schematics for the locations of YY1 sites and their mutations for mutB4.5, C11.12, and D11.12. The upper sequence is the wild-type sequence, and the bottom sequence is the mutant sequence, with the mutations in lowercase letters. (C, F, and I) ChIP for YY1 from the wild-type plasmids and the YY1 plasmids (n = 2). (G to I) Luciferase activity of the B4.5, C11.12, and D11.12 constructs when BMI1 and EED are knocked down in MSCs. Scrambled shRNA was used as a negative control.
We characterized the activity of the C11.12 region using similar reporter assays and compared these effects to the previously characterized D11.12 region. A 0.6-kb region that spans the MNase hypersensitive region of C11.12 was cloned upstream of the HOX2A promoter in the A2pLuc plasmid and transfected into MSCs. We observed an almost-complete repression of luciferase activity with C11.12 (Fig. 3D). Mutation of the single YY1 site (mutC11.12), as depicted in Fig. 3E, from the cloned region resulted in the loss of YY1 binding to the C11.12 sequence (Fig. 3F) as well as the loss of repression (Fig. 3D). Previously, we characterized the repressive activity of D11.12 on a reporter construct that harbored multiple YY1 sites upstream of the TK promoter (18). Here, we tested the ability of D11.12 to confer repression on the HOXA2 promoter contained in the A2pLuc-based construct and saw similar results (Fig. 3G). In addition, when the four YY1 sites of D11.12 were mutated (mutD11.12), as shown in Fig. 3H, we observed an increase in luciferase activity to a level near that seen with the A2pLuc construct (Fig. 3G). As we observed with both mutB4.5 and mutC11.12, mutD11.12 displayed a loss of YY1 binding as measured using ChIP, consistent with the proposal that the loss of repression is due to the loss of YY1 binding (Fig. 3I).
When each of the three elements was tested using reporter assays, each repressed luciferase activity relative to the parental constructs. The association of BMI1, SUZ12, and H3K27me3 with the putative PREs suggested that the elements repressed luciferase activity in a PcG-dependent manner. We tested this hypothesis by performing knockdown experiments on key components of PRC1 and PRC2; specifically, we used shRNA constructs to knock down individual components of these PcG complexes as well as a scrambled negative-control shRNA in MSCs and measured the impact on expression from the luciferase constructs (Fig. 3J to L). Previously, we showed that the shRNA constructs used here are effective at reducing the levels of BMI1 and EED in MSCs (18). When the B4.5 construct was transfected into the BMI1 knockdown cells, we saw an increase to a level of luciferase activity that was similar to the activity of the parental A2pLuc construct (Fig. 3J). When EED expression was knocked down in MSCs, luciferase activity was also derepressed. In addition, derepression increased with the mutB4.5 construct in the BMI1 and EED knockdown cells relative to the levels observed with mutB4.5 in the MSCs. We conclude that these PcG proteins are required to observe repression by the B4.5 element and that a substantial portion of the PcG-dependent repressive function of B4.5 appears to be independent of the YY1 binding sites.
The C11.12 construct exhibited a loss of repressive activity in the BMI1 and EED knockdown cells and became expressed to levels similar to that observed with A2pLuc in these knockdown cells (Fig. 3K). Knocking down either EED or BMI1 expression caused a decrease in activity of the mutC11.12 construct that does not bind YY1, although this effect was not statistically significant. Knockdown with the control shRNA did not result in a derepression of luciferase activity. We conclude that repression by the C11.12 element depends upon both PcG proteins and the YY1 binding site.
In addition, we observed derepression of luciferase activity from D11.12_A2pLuc following knockdown to levels that were considerably greater than the expression level of A2pLuc (Fig. 3L). As proposed previously, we suggest that D11.12 contains activating elements that are unmasked when PcG proteins are removed. YY1 may also serve to recruit transcriptional activating factors in the absence of PcG proteins. Luciferase expression from the mutD11.12 construct in the PcG knockdown cells was also derepressed, but to a lesser extent than seen with the D11.12 construct. No derepression was observed when the cells were transduced with the control shRNA construct. Taking these data together, we conclude that the repressive activities observed by B4.5, C11.12, and D11.12 are PcG dependent in MSCs.
Repression following stable integration of reporter constructs.
MSCs were transfected for stable integration of B4.5_A2pLuc and C11.12_A2pLuc to assess whether stably integrated constructs would better reflect the nature of chromatin biology (Fig. 4A). To prevent the possible effects of neighboring elements after random integration into the genome, beta-globin insulators were placed up- and downstream of the B4.5 or C11.12 construct. Furthermore, FRT sites flanking the B4.5 or C11.12 element were inserted with the purpose of being able to excise the elements after stable integration of the constructs. A second construct containing the lacZ gene and a drug selection marker was randomly integrated into the genome along with B4.5_A2pLuc or C11.12_A2pLuc. After 2 months of drug selection, a plasmid containing FLP recombinase in tandem with dsRED was transiently transfected into the stable cell lines to remove B4.5 and C11.12 by targeting the FRT sites. Transfected cells that had gained plasmid DNA were selected by FACS for dsRED expression. This enabled us to study the effects of stably integrated constructs with or without the putative repressive elements, B4.5(+) or B4.5(−) and C11.12(+) or C11.12(−), respectively, minimizing the possibility of effects from variables such as copy number and integration site. Luciferase measurements taken from the B4.5(+) and B4.5(−) or C11.12(+) and C11.12(−) cells were normalized to beta-galactosidase activity from the lacZ gene (Fig. 4B). The removal of the repressive elements from the integrated constructs resulted in increased luciferase activity, demonstrating the repressive effect of the elements present in the B4.5(+) and C11.12(+) cells. ChIP-qPCR experiments displayed enrichments for BMI1, SUZ12, and H3K27me3 with the promoter and with the luciferase gene for both the integrated B4.5 and C11.12 constructs (Fig. 4C and D). The primer set used to amplify the promoter is designed to amplify part of the FRT sequence and the HOXA2 promoter so that the PCR product would specifically amplify the construct and neither the endogenous HOXA2 promoter nor the endogenous B4.5 sequence. The PCR target is immediately adjacent to the B4.5 sequence, so the enrichment of YY1 by ChIP is from the B4.5 portion of the integrated construct. ChIP-qPCR was also performed in the cells with the integrated constructs at control regions between HOXB4 and HOXB5 and between HOXC11 and HOXC12. The B4.5(−) and C11.12(−) cells were analyzed for the association of BMI1, SUZ12, and H3K27me3 by ChIP-qPCR. The amounts of PcG proteins and the histone methylation mark on the promoter region or on the luciferase gene were reduced compared to the amounts seen on the B4.5(+) and C11.12(+) cells (Fig. 4C and D). Taken together, these results demonstrate that the defined elements confer repression and recruit PcG proteins to an integrated construct in cells.
Fig 4.
B4.5 and C11.12 confer Polycomb-mediated repression from integrated genomic sites. (A) Schematic of plasmids used for genomic integration. B4.5(+) and C11.12(+) denote constructs containing the repressive elements. B4.5(−) and C11.12(−) are the integrated constructs that no longer bear the repressive elements. (B) Luciferase activity of the integrated B4.5 and C11.12 elements before and after FLP recombinase activity. (C and D) ChIP-qPCR for BMI1, SUZ12, YY1, H3, and H3K27me3 from the integrated constructs. The PCR primers target the promoter, luciferase gene, and endogenous control regions relevant to either B4.5 or C11.12. The H3K27me3/H3 graph calculates the enrichment of H3K27me3 relative to the abundance of H3 present at the sites. Data are represented as the mean (n = 3) ± SEM.
Dependence on the JARID2 protein in repression.
Recent studies that implicate JARID2 in PcG function led us to test whether this protein is needed for function of these elements. JARID2, a JMJD protein with DNA-binding activity, associates with PRC2 complexes at substoichiometric levels in MSCs and other cell types (20–24). Depletion of JARID2 results in the loss of differentiation potential in ESCs, in particular, the mesoderm, and in changes in expression of PcG target genes (20, 22, 23). We found that JARID2 is expressed in MSCs. To test whether it plays a role in repression conferred by the elements characterized here, we identified shRNAs that are capable of efficiently knocking down JARID2 (Fig. 5A). We determined that JARID2 binds to the endogenous B4.5, C11.12, and D11.12 loci in MSCs using ChIP and that binding is reduced approximately 100-fold in JARID2 knockdown cells (Fig. 5B). We conclude that JARID2 associates with the regulatory elements that are characterized here when those elements are in their normal genomic location.
The association of JARID2 with these sites suggested that it might be involved in recruiting PRC2 components, such as SUZ12. To test this hypothesis, the occupancy of BMI1, SUZ12, histone H3, and H3K27me3 was measured using ChIP in cells that had stable knockdown of JARID2 expression. The association of BMI1 with the endogenous B4.5 site was largely unchanged in the MSCs that did not express JARID2, while association of SUZ12 and H3K27me3 at B4.5 was decreased more than 10-fold in these cells (Fig. 5C). In addition, we observed an increase in the level of histone H3 associated with B4.5. At the endogenous C11.12 site, no significant differences were seen with BMI1 and SUZ12 when JARID2 was knocked down relative to the wild-type MSCs (Fig. 5D). However, we observed a 10-fold decrease of H3K27me3 at the C11.12 element in the JARID2 knockdown cells. The functional significance of this decrease is unclear, since the association of BMI1, a PRC1 subunit whose targeting is expected to be in part caused by methylation of H3K27, was largely unchanged. Concomitantly, an increase in H3 association with C11.12 was also observed. Interestingly, at the endogenous D11.12 site, we observed a considerable decrease in the association with SUZ12 and BMI1 in the JARID2 knockdown cells (Fig. 5E). There was also a significant decrease in the association of H3K27me3 at D11.12 when JARID2 was depleted. Taken together, the data indicate that although JARID2 is associated with each of these three endogenous elements, the loss of JARID2 has various effects upon the association of SUZ12, BMI1, H3, and H3K27me3 with each element.
These binding data suggested that depletion of JARID2 might have differing impacts on the repressive ability of each of these elements. The binding analysis led us to predict that JARID2 depletion would have greater impact on function for B4.5 and D11.12 and lesser impact on C11.12. To test this, we measured activity of the luciferase constructs in the MSCs and JARID2 knockdown MSCs (Fig. 5F to H). When the activity of the B4.5 and mutB4.5 constructs was measured using a transient-transfection protocol in the JARID2 knockdown cells, we observed derepression of luciferase activity similar to the extent we observed with the PcG knockdown cells (Fig. 5F). Thus, despite the limited depletion of BMI1 at this site in the JARID2 knockdown cells (Fig. 5C), B4.5 no longer has the ability to repress on a plasmid when JARID2 is absent. When the luciferase assay was performed with the D11.12 and mutD11.12 constructs in the JARID2 knockdown cells, derepression was also observed (Fig. 5E). While the derepression with D11.12 in the JARID2 knockdown cells was sizeable, we also observed a trend toward further depression with mutD11.12 in the JARID2 knockdown cells.
Taken together, our results suggest that B4.5 and D11.12 repressive activity is compromised by knockdown of JARID2 in MSCs, presumably due to a decreased SUZ12 association as seen with the endogenous loci. In contrast to the derepression of B4.5 and D11.12 observed with knockdown of JARID2, there was very little impact on C11.12 in these knockdown cells. While JARID2 was associated with C11.12 in MSC cells, depletion of JARID2 did not impact BMI1 or SUZ12 occupancy, consistent with the lack of impact on the ability of C11.12 to regulate expression on a plasmid (Fig. 5D). We conclude that JARID2 localizes to potential regulatory elements within the HOX clusters and that it affects the ability of a subset of these elements to function as PcG-mediated repressive elements in a manner that correlates with its impact on PcG protein binding to the endogenous locations of these elements.
If the association of JARID2 with the HOX clusters is important for regulating gene expression from the endogenous HOX loci, depletion of JARID2 in MSCs would be expected to alter the pattern of expression of endogenous HOX genes. Based upon the data presented above, we anticipate that different repressive elements would have distinct requirements for JARID2 function and that HOX genes across the four clusters would not all respond in a similar manner to JARID2 knockdown. We determined the expression profile of the HOX genes in the JARID2 knockdown MSCs (Fig. 6A). Expression from HOXA1, -A2, -A10, -A11, -B13, -D1, -D12, and -D13 was increased more than 2-fold in the JARID2 knockdown MSCs compared to expression in wild-type MSCs. We conclude that JARID2 is important for maintaining repression of HOX genes in MSCs and that this impact is greatest on the distal HOX genes in the HOXA and HOXD clusters and minimal in the HOXC cluster.
Fig 6.

HOX gene expression profile of JARID2 knockdown MSCs. (A) qRT-PCR for HOX gene expression in MSCs and JARID2 knockdown MSCs. (B) qRT-PCR for HOX gene expression in MSCs and BMI1 and EED knockdown MSCs. (C) Organization of the HOXA, -B, -C, and -D clusters in humans. Genes that were upregulated more than 2-fold in expression in the JARID2 knockdown cells are boxed.
The elements examined all required PRC1 and PRC2 components for function yet had differential requirements for JARID2 function, predicting that the impact on endogenous HOX expression of knocking down PcG components would be more general than that on knocking down JARID2. We examined this hypothesis in MSCs and found that EED or BMI1 knockdown increased expression from all of the genes impacted by JARID2 knockdown and also increased expression from other genes, such as HOXC10 and HOXC13 (Fig. 6B). We conclude that PcG components are more broadly required than JARID2 for repression of HOX genes in MSCs, consistent with JARID2 being involved in a subset of PcG-regulated repressive mechanisms in the human HOX clusters.
DISCUSSION
Polycomb-dependent transcriptional repression of HOX genes is critical for correct positional embryonic development, and its requirement is highly conserved throughout evolution. The mechanisms that target PcG function in mammals, however, appear to be context dependent and intricate. With the very limited number of characterized mammalian DNA elements that can confer transcriptional repression via the recruitment of Polycomb, it is difficult to begin to understand how transcriptional expression profiles of key regulatory and developmental genes are set and maintained. Here, we present the identification of additional elements located in the human HOX clusters that confer PcG-dependent repression and describe their modes of recruitment. They exhibit MNase sensitivity and harbor YY1 binding sites. While these elements exhibit such similarities, they differ in their requirement for JARID2. Our findings suggest that the combination of different recruitment factors may recruit subtly different PcG complexes to mediate a nuanced repression repertoire to affect a context-dependent outcome. Mechanisms of PcG-dependent repression in mammalian systems will very likely be varied, as reflected by the many subcomplexes of the PcG proteins. These studies increase our understanding of how PcG complexes are targeted to an important region of the human genome, the HOX clusters, and demonstrate a varied role for JARID2 in their function.
We previously employed human embryonic stem cells (hESCs) and their mesenchymal derivatives (i.e., mesenchymal stem cells [MSCs], adipocytes, and osteoblasts) to look for alterations in chromatin architecture at the HOX clusters that signify changes in gene regulation (18). We reasoned that in order for DNA binding to occur, regulatory proteins must have access to their target sequence. Hence, DNA accessible to enzymatic cleavage—and, by inference, potentially accessible to DNA-binding factors—was identified. In Drosophila, PREs are characteristically DNase I- and MNase-sensitive regions, so we looked for regions that were MNase hypersensitive in human ES cells. We had previously identified a sequence located between HOXD11 and HOXD12 (D11.12) that bound PcG components and conferred repression in a cell-type-specific manner (18). Here, we present two additional regions in the human HOX clusters, B4.5 and C11.12, with similar characteristics: (i) significant depletion of nucleosome occupancy as measured by MNase mapping and (ii) enrichment for PcG proteins and for the H3K27me3 mark produced by PRC2.
One distinguishing feature of the putative PRE sites is the hypersensitivity to MNase cleavage. In the Drosophila homeotic cluster, bona fide PREs are hypersensitive to enzymatic cleavage and are associated with the histone variant H3.3, where high histone turnover occurs (32, 33). While we observed differences in MNase cleavage patterns between the cell types, these differences were less striking by ChIP-qPCR for histone H3. This might be explained by the different qualities measured by these protocols: the MNase cleavage protocol measures the amount of mononucleosome-sized DNA following MNase cleavage, while the ChIP protocol measures the amount of DNA associated with histone H3. The possibility of high rates of histone H3 turnover at these sites, as with PREs found in Drosophila, may increase the accessibility of this region to MNase cleavage, creating a snapshot of apparent nucleosome depletion. The dynamic histone activity would also result in an apparent decrease of histone H3 and the associated DNA at B4.5 relative to other non-PRE regions. We are still able to observe greater enrichment of H3K27me3 at B4.5, most likely because the vast majority of associated histone H3s are trimethyated at lysine 27. Another option is that there might be a local chromatin structure refractory to MNase cutting that produces products of a different size than the mononucleosome-sized bands that were isolated for hybridization to the microarray. In either case, the results indicate that the local chromatin architecture associated with the putative PREs is unique.
Upon identifying B4.5 and C11.12, we found that these elements were capable of repressing gene expression. Due to the limited number of putative mammalian PREs that have been characterized, very little is known about how these elements function. The chromatins associated with PRC1 and PRC2 complexes at promoters are the targets of transcriptional repression. However, the promoters themselves are not PREs since they do not confer PcG-mediated repression, per se. PREs at intergenic regions may serve as regulatory elements to control transcriptional repression from tens of kilobases, as observed for PREs in flies. The identification of mammalian PREs is significant because it extends our understanding of how cell-specific gene silencing of key developmental regulators, such as the HOX genes, works.
The factors that recruit PcG complexes to specific sites to mediate repression remain largely unknown. The three putative PREs that we have characterized in the HOX clusters contain potential DNA binding sites for both activating and repressive proteins. Interestingly, all contain YY1 binding sites that are important for the PcG recruitment and necessary to mediate repression. The mouse PRE, PRE-kr, was also found to have a YY1 DNA-binding motif within it (17). Other work has shown recruitment of PcG proteins to DNA by YY1 and found this to require the REPO domain (34). YY1 can also bind Xist RNA and serve as a docking protein for PRC2 at the inactive X chromosome (35), indicating that YY1 plays several roles in targeting PcG function.
Although YY1 appears to be important for the repressive capabilities of the three putative PREs described here, it is unlikely that YY1 is a defining component of all mammalian PREs. In Drosophila, in silico prediction of PREs, based upon known PRE binding motifs, determined that the simple presence of sequences, such as PHO binding sites, was not sufficient to identify PREs (36, 37). Genome-wide PcG association studies demonstrated that YY1 binding sites do not correlate well with PcG protein binding sites in mouse ES cells (15). YY1 has been shown to have other functions outside PcG function (38). YY1 has many binding partners and can mediate both transcriptional activation and repression depending upon its associated factors, so it is expected that YY1 location alone will not predict PcG location. Based upon studies of PREs in Drosophila and work by our group and others (reviewed by Simon and Kingston [6]), several mechanisms using different DNA-binding factors will contribute to PcG recruitment in mammals, some likely independent of YY1 function.
GC-rich sequences have been shown to recruit PRC2 protein, EZH2, and its methylation activity to chromatin in the absence of activating factors. This might contribute to PcG recruitment, although, as commented upon by Mendenhall et al. (15), there is a distinction between the recruitment of PcG proteins and their repressive function, which was not determined. While D11.12 contains a CpG island within it, B4.5 and C11.12 do not. However, they are still able to recruit PRC1 and PRC2 when cloned into a reporter construct, and moreover, they repressed luciferase activity. Thus, while CpG islands can recruit PRC2, their presence is not necessary for two of the elements examined here.
JARID2, whose depletion shows differential impact on the PREs reported here and on HOX regulation, was previously shown to interact with PRC2 and to thus be involved in regulation of PcG-mediated repression (20–24). The results described here indicate that JARID2 is engaged in a subset of PcG-regulated events despite broader localization profiles, as observed by others (21, 22). Our conclusions are based upon functional assessment of three elements residing in three different HOX clusters. Knockdown of JARID2 resulted in the derepression of B4.5 and D11.12 activity in the luciferase assay. The partial loss of SUZ12 and decreased H3K27me3 levels in the absence of JARID2 are in agreement with the observation that JARID2 is a component of the PRC2 complex. While we do observe decreased association of H3K27me3 at these sites, we do not claim that there are global decreases of H3K27me3. This is consistent with the observation made that JARID2 knockdown in embryonic stem cells shows no global changes in H3K27me3 but, rather, a reduction at target genes (21). Notably, the change in BMI1 association with B4.5 and D11.12 is not as substantial as the decrease in SUZ12 association. It is possible that PRC1 is recruited to these elements in a PRC2-independent manner, such as via interaction with YY1 binding sites or via RYBP-containing PRC1 complexes. RYBP-PRC1 complexes are able to associate with chromatin independently (39) and contain unique compositions that are associated with different PcG targets (9). It has also been demonstrated that different PRC1 complexes can be observed at the same locus, such as at the INK4A/ARF locus, although the functional significance of these different complexes coexisting at promoters has not been fully characterized (40).
The expression levels of the HOXB and -C genes in the different cell types of our differentiation model were described earlier (18). In the case of the genes neighboring HOXB4.5, HOXB4 and HOXB5 are very lowly or not expressed in the hESCs and MSCs. For C11.12, HOXC11 and HOXC12 are also not expressed in either cell type. The knockdown of the PcG proteins resulted in upregulation of the nearest neighbors of B4.5, suggesting that B4.5 may confer this repression on these genes. Interestingly, knockdown of the PcG genes did not result in the upregulation of expression of HOXC11 and HOXC12 but instead upregulated HOXC10 and HOXC13. While we have identified and characterized B4.5 and C11.12 to have the ability to confer repression, the underlying mechanism of PRE-mediated targeting remains to be studied.
Taken together, these studies indicate that the recruitment of PcG complexes to regulatory regions and their resulting repressive activities in mammalian cells will be complex and context dependent. Given the large number of PRC1 family complexes recently characterized, it is likely that there will be diverse mechanisms of PcG-mediated repression. In vivo and in vitro experiments have demonstrated that distinct aspects of PcG function, such as compaction (41) and ubiquitylation of H2A (42), are coupled to transcriptional silencing. It is likely that different loci in different cell types at different stages of the developing embryo will have specific requirements. A thorough catalog of the classes of targeting and function of PcG complexes is needed to fully understand how gene expression patterns of master regulators are maintained in humans, a central aspect of human biology that governs both development and disease processes.
ACKNOWLEDGMENTS
This work was supported by grants GM43901 and HG003141 from the National Institutes of Health for R.E.K. and C.J.W. C.J.W. was supported by NRSA GM072265. P.J.K. and P.V.K. were supported by NIH grant GM082798.
We thank Sara Miller and Behfar Alderahi for discussions and critical reading of the manuscript.
Footnotes
Published ahead of print 17 June 2013
REFERENCES
- 1. Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, Bell GW, Otte AP, Vidal M, Gifford DK, Young RA, Jaenisch R. 2006. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature 441: 349– 353 [DOI] [PubMed] [Google Scholar]
- 2. Lee TI, Jenner RG, Boyer LA, Guenther MG, Levine SS, Kumar RM, Chevalier B, Johnstone SE, Cole MF, Isono K, Koseki H, Fuchikami T, Abe K, Murray HL, Zucker JP, Yuan B, Bell GW, Herbolsheimer E, Hannett NM, Sun K, Odom DT, Otte AP, Volkert TL, Bartel DP, Melton DA, Gifford DK, Jaenisch R, Young RA. 2006. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell 125: 301– 313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bracken AP, Dietrich N, Pasini D, Hansen KH, Helin K. 2006. Genome-wide mapping of Polycomb target genes unravels their roles in cell fate transitions. Genes Dev. 20: 1123– 1136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Squazzo SL, O'Geen H, Komashko VM, Krig SR, Jin VX, Jang SW, Margueron R, Reinberg D, Green R, Farnham PJ. 2006. Suz12 binds to silenced regions of the genome in a cell-type-specific manner. Genome Res. 16: 890– 900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lehmann L, Ferrari R, Vashisht AA, Wohlschlegel JA, Kurdistani SK, Carey M. 2012. Polycomb repressive complex 1 (PRC1) disassembles RNA polymerase II preinitiation complexes. J. Biol. Chem. 287: 35784– 35794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Simon JA, Kingston RE. 2009. Mechanisms of polycomb gene silencing: knowns and unknowns. Nat. Rev. Mol. Cell Biol. 10: 697– 708 [DOI] [PubMed] [Google Scholar]
- 7. Luis NM, Morey L, Di Croce L, Benitah SA. 2012. Polycomb in stem cells: PRC1 branches out. Cell Stem Cell 11: 16– 21 [DOI] [PubMed] [Google Scholar]
- 8. Francis NJ, Saurin AJ, Shao Z, Kingston RE. 2001. Reconstitution of a functional core polycomb repressive complex. Mol. Cell 8: 545– 556 [DOI] [PubMed] [Google Scholar]
- 9. Gao Z, Zhang J, Bonasio R, Strino F, Sawai A, Parisi F, Kluger Y, Reinberg D. 2012. PCGF homologs, CBX proteins, and RYBP define functionally distinct PRC1 family complexes. Mol. Cell 45: 344– 356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Morey L, Pascual G, Cozzuto L, Roma G, Wutz A, Benitah SA, Di Croce L. 2012. Nonoverlapping functions of the Polycomb group Cbx family of proteins in embryonic stem cells. Cell Stem Cell 10: 47– 62 [DOI] [PubMed] [Google Scholar]
- 11. Cao R, Zhang Y. 2004. SUZ12 is required for both the histone methyltransferase activity and the silencing function of the EED-EZH2 complex. Mol. Cell 15: 57– 67 [DOI] [PubMed] [Google Scholar]
- 12. Pasini D, Bracken AP, Jensen MR, Lazzerini Denchi E, Helin K. 2004. Suz12 is essential for mouse development and for EZH2 histone methyltransferase activity. EMBO J. 23: 4061– 4071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kirmizis A, Bartley SM, Kuzmichev A, Margueron R, Reinberg D, Green R, Farnham PJ. 2004. Silencing of human polycomb target genes is associated with methylation of histone H3 Lys 27. Genes Dev. 18: 1592– 1605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Stock JK, Giadrossi S, Casanova M, Brookes E, Vidal M, Koseki H, Brockdorff N, Fisher AG, Pombo A. 2007. Ring1-mediated ubiquitination of H2A restrains poised RNA polymerase II at bivalent genes in mouse ES cells. Nat. Cell Biol. 9: 1428– 1435 [DOI] [PubMed] [Google Scholar]
- 15. Mendenhall EM, Koche RP, Truong T, Zhou VW, Issac B, Chi AS, Ku M, Bernstein BE. 2010. GC-rich sequence elements recruit PRC2 in mammalian ES cells. PLoS Genet. 6: e1001244. 10.1371/journal.pgen.1001244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Atchison M, Basu A, Zaprazna K, Papasani M. 2011. Mechanisms of Yin Yang 1 in oncogenesis: the importance of indirect effects. Crit. Rev. Oncog. 16: 143– 161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sing A, Pannell D, Karaiskakis A, Sturgeon K, Djabali M, Ellis J, Lipshitz HD, Cordes SP. 2009. A vertebrate Polycomb response element governs segmentation of the posterior hindbrain. Cell 138: 885– 897 [DOI] [PubMed] [Google Scholar]
- 18. Woo CJ, Kharchenko PV, Daheron L, Park PJ, Kingston RE. 2010. A region of the human HOXD cluster that confers polycomb-group responsiveness. Cell 140: 99– 110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cuddapah S, Roh TY, Cui K, Jose CC, Fuller MT, Zhao K, Chen X. 2012. A novel human polycomb binding site acts as a functional polycomb response element in Drosophila. PLoS One 7: e36365. 10.1371/journal.pone.0036365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Li G, Margueron R, Ku M, Chambon P, Bernstein BE, Reinberg D. 2010. Jarid2 and PRC2, partners in regulating gene expression. Genes Dev. 24:368– 380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pasini D, Cloos PA, Walfridsson J, Olsson L, Bukowski JP, Johansen JV, Bak M, Tommerup N, Rappsilber J, Helin K. 2010. JARID2 regulates binding of the Polycomb repressive complex 2 to target genes in ES cells. Nature 464: 306– 310 [DOI] [PubMed] [Google Scholar]
- 22. Peng JC, Valouev A, Swigut T, Zhang J, Zhao Y, Sidow A, Wysocka J. 2009. Jarid2/Jumonji coordinates control of PRC2 enzymatic activity and target gene occupancy in pluripotent cells. Cell 139: 1290– 1302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Shen X, Kim W, Fujiwara Y, Simon MD, Liu Y, Mysliwiec MR, Yuan GC, Lee Y, Orkin SH. 2009. Jumonji modulates polycomb activity and self-renewal versus differentiation of stem cells. Cell 139: 1303– 1314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Landeira D, Sauer S, Poot R, Dvorkina M, Mazzarella L, Jorgensen HF, Pereira CF, Leleu M, Piccolo FM, Spivakov M, Brookes E, Pombo A, Fisher C, Skarnes WC, Snoek T, Bezstarosti K, Demmers J, Klose RJ, Casanova M, Tavares L, Brockdorff N, Merkenschlager M, Fisher AG. 2010. Jarid2 is a PRC2 component in embryonic stem cells required for multi-lineage differentiation and recruitment of PRC1 and RNA polymerase II to developmental regulators. Nat. Cell Biol. 12: 618– 624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Seda Tigli R, Ghosh S, Laha MM, Shevde NK, Daheron L, Gimble J, Gumusderelioglu M, Kaplan DL. 2009. Comparative chondrogenesis of human cell sources in 3D scaffolds. J. Tissue Eng. Regen. Med. 3: 348– 360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jagani Z, Wiederschain D, Loo A, He D, Mosher R, Fordjour P, Monahan J, Morrissey M, Yao YM, Lengauer C, Warmuth M, Sellers WR, Dorsch M. 2010. The Polycomb group protein Bmi-1 is essential for the growth of multiple myeloma cells. Cancer Res. 70: 5528– 5538 [DOI] [PubMed] [Google Scholar]
- 27. Kia SK, Gorski MM, Giannakopoulos S, Verrijzer CP. 2008. SWI/SNF mediates polycomb eviction and epigenetic reprogramming of the INK4b-ARF-INK4a locus. Mol. Cell. Biol. 28: 3457– 3464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gheldof N, Tabuchi TM, Dekker J. 2006. The active FMR1 promoter is associated with a large domain of altered chromatin conformation with embedded local histone modifications. Proc. Natl. Acad. Sci. U. S. A. 103: 12463– 12468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kanhere A, Viiri K, Araujo CC, Rasaiyaah J, Bouwman RD, Whyte WA, Pereira CF, Brookes E, Walker K, Bell GW, Pombo A, Fisher AG, Young RA, Jenner RG. 2010. Short RNAs are transcribed from repressed polycomb target genes and interact with polycomb repressive complex-2. Mol. Cell 38: 675– 688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ai W, Liu Y, Wang TC. 2006. Yin yang 1 (YY1) represses histidine decarboxylase gene expression with SREBP-1a in part through an upstream Sp1 site. Am. J. Physiol. Gastrointest. Liver Physiol. 290: G1096– G1104 [DOI] [PubMed] [Google Scholar]
- 31. Kallin EM, Cao R, Jothi R, Xia K, Cui K, Zhao K, Zhang Y. 2009. Genome-wide uH2A localization analysis highlights Bmi1-dependent deposition of the mark at repressed genes. PLoS Genet. 5: e1000506. 10.1371/journal.pgen.1000506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mito Y, Henikoff JG, Henikoff S. 2007. Histone replacement marks the boundaries of cis-regulatory domains. Science 315:1408– 1411 [DOI] [PubMed] [Google Scholar]
- 33. Deal RB, Henikoff JG, Henikoff S. 2010. Genome-wide kinetics of nucleosome turnover determined by metabolic labeling of histones. Science 328: 1161– 1164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wilkinson F, Pratt H, Atchison ML. 2010. PcG recruitment by the YY1 REPO domain can be mediated by Yaf2. J. Cell. Biochem. 109: 478– 486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Jeon Y, Lee JT. 2011. YY1 tethers Xist RNA to the inactive X nucleation center. Cell 146: 119– 133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ringrose L, Rehmsmeier M, Dura JM, Paro R. 2003. Genome-wide prediction of Polycomb/Trithorax response elements in Drosophila melanogaster. Dev. Cell 5: 759– 771 [DOI] [PubMed] [Google Scholar]
- 37. Okulski H, Druck B, Bhalerao S, Ringrose L. 2011. Quantitative analysis of polycomb response elements (PREs) at identical genomic locations distinguishes contributions of PRE sequence and genomic environment. Epigenetics Chromatin 4: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Vella P, Barozzi I, Cuomo A, Bonaldi T, Pasini D. 2012. Yin Yang 1 extends the Myc-related transcription factors network in embryonic stem cells. Nucleic Acids Res. 40:3403– 3418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tavares L, Dimitrova E, Oxley D, Webster J, Poot R, Demmers J, Bezstarosti K, Taylor S, Ura H, Koide H, Wutz A, Vidal M, Elderkin S, Brockdorff N. 2012. RYBP-PRC1 complexes mediate H2A ubiquitylation at Polycomb target sites independently of PRC2 and H3K27me3. Cell 148: 664– 678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Maertens GN, El Messaoudi-Aubert S, Racek T, Stock JK, Nicholls J, Rodriguez-Niedenfuhr M, Gil J, Peters G. 2009. Several distinct polycomb complexes regulate and co-localize on the INK4a tumor suppressor locus. PLoS One 4: e6380. 10.1371/journal.pone.0006380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Francis NJ, Kingston RE, Woodcock CL. 2004. Chromatin compaction by a polycomb group protein complex. Science 306:1574– 1577 [DOI] [PubMed] [Google Scholar]
- 42. Eskeland R, Leeb M, Grimes GR, Kress C, Boyle S, Sproul D, Gilbert N, Fan Y, Skoultchi AI, Wutz A, Bickmore WA. 2010. Ring1B compacts chromatin structure and represses gene expression independent of histone ubiquitination. Mol. Cell 38: 452– 464 [DOI] [PMC free article] [PubMed] [Google Scholar]