Abstract
Autophagy is a highly conserved self-digestion pathway involved in various physiological and pathophysiological processes. Recent studies have implicated a pivotal role of autophagy in adipocyte differentiation, but the molecular mechanism for its role and how it is regulated during this process are not clear. Here, we show that CCAAT /enhancer-binding protein β (C/EBPβ), an important adipogenic factor, is required for the activation of autophagy during 3T3-L1 adipocyte differentiation. An autophagy-related gene, Atg4b, is identified as a de novo target gene of C/EBPβ and is shown to play an important role in 3T3-L1 adipocyte differentiation. Furthermore, autophagy is required for the degradation of Klf2 and Klf3, two negative regulators of adipocyte differentiation, which is mediated by the adaptor protein p62/SQSTM1. Importantly, the regulation of autophagy by C/EBPβ and the role of autophagy in Klf2/3 degradation and in adipogenesis are further confirmed in mouse models. Our data describe a novel function of C/EBPβ in regulating autophagy and reveal the mechanism of autophagy during adipocyte differentiation. These new insights into the molecular mechanism of adipose tissue development provide a functional pathway with therapeutic potential against obesity and its related metabolic disorders.
INTRODUCTION
Adipose tissue is not only a storage depot of fat but also an endocrine organ influencing whole-body energy homeostasis (1–3). The overexpansion of adipose tissue mass plays a central role in obesity-related complications, such as type 2 diabetes, hypertension, hyperlipidemia, and arteriosclerosis (4–6). Therefore, a comprehensive investigation into the molecular mechanisms underlying adipogenesis is of both fundamental and clinical relevance for the development of novel therapeutics for obesity and associated metabolic syndromes.
The 3T3-L1 cell line is an invaluable cellular model in studying adipogenesis (Fig. 1A). Upon addition of adipogenic inducers, these cells undergo one to two rounds of mitotic clonal expansion (MCE) followed by terminal adipocyte differentiation (7). During adipogenesis, CCAAT /enhancer-binding protein β (C/EBPβ) is induced very early and plays a crucial role in initiating the differentiation program by activating the expression of peroxisome proliferator-activated receptor γ (PPARγ) and C/EBPα, two key adipogenic transcription factors (8). These two factors serve as pleiotropic transactivators of many adipocyte-specific genes to promote and maintain the terminally differentiated phenotype. Besides its role in transactivation of PPARγ and C/EBPα, C/EBPβ is also involved in regulating mitotic clonal expansion, a cell proliferation process required for terminal adipocyte differentiation (9–11).
Fig 1.
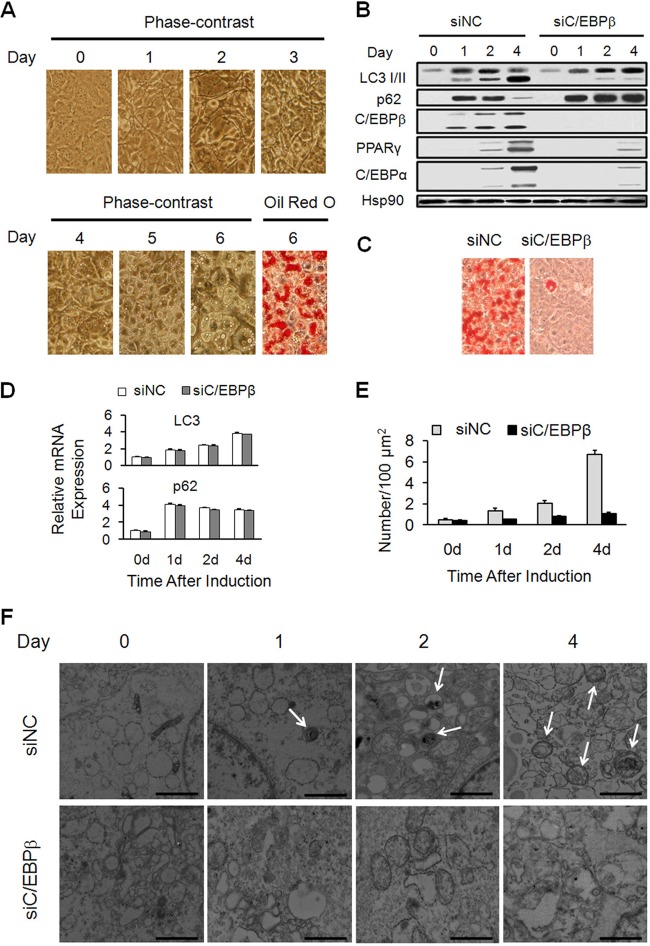
C/EBPβ is required for the activation of autophagy and adipogenesis during 3T3-L1 adipocyte differentiation. (A) The growth-arrested 3T3-L1 cells were induced to differentiation. On the indicated day, cells were subjected to phase-contrast microscopy, and Oil red O staining was performed on day 6. Lipid droplets begin to appear on day 3, the amount and size of which increase day by day. On day 6, mature adipocytes are formed. (B to F) 3T3-L1 cells were transfected with control siRNA (siNC) or C/EBPβ siRNA (siC/EBPβ) followed by adipogenic induction. (B) The protein levels of autophagic markers (LC3 I/II and p62), C/EBPβ, and adipogenic markers (PPARγ and C/EBPα) during differentiation were analyzed by Western blotting. Hsp90 serves as a loading control. (C) Oil red O staining of 3T3-L1 cells on day 6 of differentiation was performed. (D) The mRNA levels of LC3 and p62 during differentiation were analyzed by RT-qPCR. (E) Transmission electron microscopic analyses were conducted. The quantification of autophagosome abundance is shown. (F) Transmission electron micrographs of the cells on the indicated days are shown (bars, 1 μm). Note the presence of autophagosomes (arrows).
Adipocyte differentiation is controlled by the interplay of a series of positive and negative effectors. Pref-1, Wnt1, Wnt10b, TRB3, GATA2/3, and Klf2/3 are among the well-characterized negative regulators of adipogenesis (12–17). The timely decline of these negative regulators is required for the successful progression of adipocyte differentiation. However, the mechanisms governing the downregulation of these negative effectors, either at mRNA levels or at protein levels, remain largely unknown.
Autophagy is a catabolic process to form the autophagosome in which the cell packages organelles and proteins and delivers the cargo to the lysosomes for degradation and recycling (18, 19). It is a cellular pathway crucial for the maintenance of cellular homeostasis and normal mammalian physiology. Most of the genes involved in autophagy, named autophagy-related genes (Atg), have been identified. Targeted ablation of critical autophagy genes in mice unravels important roles of autophagy in neuronal development, tumor suppression, β-cell protection, erythrocyte differentiation, and so on (19). Recent studies have shown that autophagy regulates adipose mass and differentiation. Depletion of Atg5 or Atg7, two essential autophagy genes, significantly inhibits adipocyte differentiation in 3T3-L1 cells and mouse embryo fibroblasts (MEFs). Furthermore, white adipose tissue (WAT) from the adipocyte-specific Atg7 knockout mice exhibits a brown adipose tissue (BAT)-like phenotype. Thus, autophagy regulates lipid accumulation in adipose tissue by controlling adipocyte differentiation as well as determining the balance between white and brown fat (20–22). While the transdifferentiation of WAT to BAT by the autophagy defect is attributed to impaired mitochondrial turnover during the maturation of adipocytes (23, 24), little is known about the mechanism underlying the role of autophagy in adipocyte differentiation. In addition, although the molecular components that participate in various steps of autophagy have been extensively studied, factors regulating the program of autophagy gene expression remain poorly understood in adipogenesis as well as in other physiological/pathophysiological processes.
Because both C/EBPβ and autophagy are crucial for adipogenesis, we ask whether there is a connection between them during this process. In this study, it is shown that C/EBPβ specifically promotes autophagy through transactivation of the expression of Atg4b, a cysteine proteinase, which cleaves LC3 precursor at its C terminus to form LC3 I. Moreover, p62-mediated autophagic degradation of the two negative regulators Klf2 and Klf3 is required for adipocyte differentiation.
MATERIALS AND METHODS
Cell culture and induction of differentiation.
3T3-L1 preadipocytes were propagated and maintained in Dulbecco's modified Eagle's medium (DMEM) containing 10% calf serum. To induce differentiation, 2-day postconfluent 3T3-L1 preadipocytes (day 0) were fed with DMEM containing 10% fetal bovine serum (FBS), 1 μg/ml insulin, 1 μM dexamethasone, and 0.5 mM 3-isobutyl-1-methylxanthine (MIX) until day 2. Cells were then fed with DMEM supplemented with 10% FBS and 1 μg/ml insulin for 2 days, after which they were fed every other day with DMEM containing 10% FBS. For autophagy inhibition, cells were treated with 0.5 mM 3-methyladenine (3-MA) (Sigma-Aldrich) from the indicated day.
Oil red O and Nile red staining for lipid.
In vitro differentiated cells were fixed for 20 min in buffered formalin and stained with Oil red O for 60 min. Isolated cells from WAT were resuspended in phosphate-buffered saline (PBS), and Nile red (stock, 0.5 mg/ml in acetone) was added into cells with a l:100 dilution. After 5 min of incubation, the cells were subjected to flow cytometry.
ChIP.
Proteins bound to DNA were cross-linked with 1% formaldehyde at 4°C for 20 min. After sonication, the protein-DNA complexes were immunoprecipitated using control IgG or anti-C/EBPβ antibody (Santa Cruz). After reversal of the cross-links at 65°C for 6 h, DNA was purified on DNA purification columns (Qiagen). The primers (5′ to 3′) for chromatin immunoprecipitation-quantitative PCR (ChIP-qPCR) or ChIP-PCR are as follows: Atg4b forward (F), TACCAGGGAGATTTCAGT , and reverse (R), TTGAGATGTCATTGTGGC ; Atg2a F, CTGGGTATCAAAGGCTCA , and R, TCTCACAGTCATTGTAGGGA ; Atg7 F, TTGAGCGGCGGTAAGTAAG , and R, CAGAATGAGCAACCAGAGGC ; Atg9a F, AGGCTTCTGAGGGAGGGT , and R, CAGTTCTGCGGTAAATACG ; Atg10 F, TGTAGGAGTCTTAGGGGTTA , and R, CATTTTGCCTGTTTCTTT ; PPARγ2 F, TTCAGATGTGTGATTAGGAG , and R, AGACTTGGTACATTACAAGG ; and insulin, F, CTTCAGCCCAGTTGACCAAT ; and R, AGGGAGGAGGAAAGCAGAAC .
RNA interference.
The small interfering RNAs (siRNAs) were designed and synthesized by GenePharma. The sequences (5′ to 3′) for successful siRNAs were as follows: siC/EBPβ, GCCCTGAGTAATCACTTAAAG ; siAtg4b, GCTGCACTTCCTACTGATT ; siAtg4b′, CCACTACTTTATTGGCTAT ; siKlf2, GCGGCAAGACCTACACCAA ; siKlf3, GCAATAAGGTCTACACTAA ; siP62, GGTTGACATTGATGTGGAA ; and siNC, TTCTCCGAACGTGTCACGT . 3T3-L1 cells were transfected at ∼50% confluence with siRNAs using Lipofectamine RNAiMAX (Invitrogen). In the case of siP62-post, cells were transfected with p62 siRNA at 4 h after adipogenic induction.
RNA isolation and RT-qPCR.
Total RNAs were extracted with TRIzol (Invitrogen) and transcribed to cDNA using the Superscript III kit (Invitrogen). The cDNAs were analyzed using the Power SYBR green PCR kit on the ABI Prism 7300 qPCR machine (Applied Biosystems). All qPCR data were normalized to 18S rRNA. Data were further normalized to day 0 of control cells. The primers (5′ to 3′) for RT-qPCR are as follows: Atg4b F, TATGATACTCTCCGGTTTGCTGA , and R, GTTCCCCCAATAGCTGGAAAG ; Atg2a F, GTGTGGTACTACGGGAGGTCT , and R, CCTGGTGTTGCCGTCCAAT ; Atg7 F, TTCTGCAATGATGTGGTGGC , and R, TGTGCACTGCTGGTCCAGAG ; Atg9a F, CAGTTTGACACTGAATACCAGCG , and R, AATGTGGTGCCAAGGTGATTT ; Atg10 F, GTAGTTACCAAGTGCCGGTTC , and R, AGCTAACGGTCTCCCATCTAAA ; Atg4a F, GAAGGAAGTTTTCCCCGATTGG , and R, GGGTTGTTCTTTTTGTCTCTCCC ; Atg4c F, GATGAAAGCAAGATGTTGCCTG , and R, TCTTCCCTGTAGGTCAGCCAT ; Atg4d F, GGAACAACGTCAAGTATGGTTGG , and R, CTCCCTCGAAATGGTAGCATC ; Klf2 F, ACCTAAAGGCGCATCTGCGTAC , and R, TTCGGTAGTGGCGGGTAAGC ; Klf3 F, GGATACCCAAAGGAAGCG , and R, TCATCAGACCGAGCGAAC ; LC3 F, ACCAAGCCTTCTTCCTCC , and R, TGTCCCGAATGTCTCCTG ; p62 F, CCTCTAGGCATTGAGGTTG , and R, GCTGCTTGGCTGAGTGTTA ; and 18S rRNA F, CGCCGCTAGAGGTGAAATTCT , and R, CATTCTTGGCAAATGCTTTCG .
Western blotting analyses.
Cells were lysed in an SDS buffer, and samples were resolved on an SDS-polyacrylamide gel. The expression of different proteins was analyzed by Western blotting with antibodies to the following: p62, Pref-1, Wnt1, Wnt10b, Klf2, and heat shock protein 90 (Hsp90) from Abcam; C/EBPβ, C/EBPα, PPARγ, GATA-2, and ubiquitin from Santa Cruz; LC3, extracellular signal-regulated kinase (ERK), and pERK from Cell Signaling; TRB3 from Millipore; 422/aP2 (prepared in M. D. Lane's lab); Klf3 (prepared in our lab); β-catenin from BD, Biosciences; Flag from Sigma-Aldrich; and Myc from Invitrogen.
Luciferase reporter assays.
The distal promoter regions of mouse Atg4b and its artificial mutants were amplified via PCR and cloned into PGL3-Promoter (Promega). Cells were transfected with Lipofectamine 2000 (Invitrogen). Luciferase activity was measured using the dual-luciferase reporter assay (Promega), with normalization of firefly luciferase to Renilla luciferase activity.
His pulldown assay for the interaction between Klf2/3 and p62.
Cells were transfected with His-tagged Klf2/3 and treated with 0.5 mM 3-MA for 8 h prior to harvest. The cells then were lysed in the cell lysis buffer (50 mM Tris-HCl [pH 8.0], 150 mM NaCl, 1% Triton X-100, 20 mM imidazole) plus protease cocktail and phosphatase inhibitors from Roche. The lysates were clarified by centrifugation followed by precipitation with Ni-nitrilotriacetic acid (Ni-NTA) beads (Qiagen). After three washes, the precipitates were subjected to Western blotting.
Ubiquitination assays.
Cells were transfected with His-tagged Klf2/3 and treated with 0.5 mM 3-MA for 8 h prior to harvest. Cells were resuspended in buffer A (6 M guanidine hydrochloride, 50 mM Tris-HCl [pH 8.0], 250 mM NaCl, 20 mM imidazole) and sonicated. The lysates were clarified by centrifugation followed by precipitation with Ni-NTA beads (Qiagen). Beads were washed twice with buffer B (3 M guanidine hydrochloride, 50 mM Tris-HCl [pH 8.0], 250 mM NaCl, 20 mM imidazole) and twice with buffer C (50 mM Tris-HCl [pH 8.0], 250 mM NaCl, 20 mM imidazole, 0.2% Triton X-100) and subjected to Western blotting.
Confocal microscopic analyses.
Cells were fixed in 4% formaldehyde for 30 min at room temperature prior to cell permeabilization with 0.1% Triton X-100 (4°C, 10 min). Cells were blocked with PBS containing 2% bovine serum albumin (BSA) for 1 h at room temperature and processed for immunostaining with anti-LC3 antibody (Novus) and DAPI (4′,6-diamidino-2-phenylindole) (Sigma-Aldrich). Fluorescence images were taken and analyzed with a Leica confocal microscope (Leica TCS SP5, Leica, Germany).
Transmission electron microscopic analyses.
Cells were fixed with 2% paraformaldehyde and 2% glutaraldehyde in 0.1 mol/liter phosphate buffer (pH 7.4), followed by 1% OsO4 treatment. After dehydration, thin sections were stained with uranyl acetate and lead citrate for observation under a JEM 1011CX electron microscope (JEOL). Images were acquired digitally from a randomly selected pool of 20 fields under each condition. The quantification of autophagosomes was performed as previously described (25).
Adenoviral expression vectors and infection.
The adenoviral expression vector pBlock-it (Invitrogen) encoding short hairpin RNA (shRNA) of Atg4b was constructed according to the manufacturer's protocols. Adenovirus was amplified and purified using Sartorius adenovirus purification kits (Sartorius, Germany). Adenovirus solution was weekly injected subcutaneously adjacent to inguinal fat pad in mice from 4 weeks old for 4 weeks. The sequences (5′ to 3′) for shRNAs are as follows: shAtg4b, GGGACTGACAGACATCAATGA ; and shLacZ, AATTTAACCGCCAGTCAGGCT . All studies involving animal experimentation were approved by the Animal Care and Use Committee of the Fudan University Shanghai Medical College and followed the National Institute of Health guidelines on the care and use of animals.
H&E staining and cell size quantification.
Standard hematoxylin and eosin (H&E) staining was performed on 5-μm paraffin sections of white adipose tissue. Cell diameter was measured with Image J from the H&E-staining section of 3 individual samples in each group.
RD-fed and HFD-fed mice.
Mice were fed with regular diet (RD) or high-fat diet (HFD) (51% kcal in fat) at the age of 4 weeks. Mice were sacrificed 8 weeks later, and inguinal WAT was subjected to Western blotting.
Statistical analyses.
Results are expressed as means ± standard errors (SE). Comparisons between groups were made by unpaired two-tailed Student's t test, where P < 0.05 was considered statistically significant. All experiments were repeated at least three times, and representative data are shown.
RESULTS
C/EBPβ plays important roles in autophagy and adipogenesis during 3T3-L1 adipocyte differentiation.
C/EBPβ has been shown to play a pivotal role during the early stage of adipogenesis, and the inhibition of autophagy causes the dysfunction of adipocyte differentiation. To test whether the autophagy is regulated by C/EBPβ in adipocyte differentiation, 3T3-L1 preadipocytes were induced to differentiation upon transfection with small interfering RNA (siRNA) against either control sequence or C/EBPβ. The knockdown efficiency of C/EBPβ siRNA and its effect on adipogenesis were confirmed as indicated by Western blotting, adipocyte marker gene expression (PPARγ and C/EBPα), and Oil red O staining (Fig. 1B and C). Autophagy was analyzed by molecular characterization of autophagy markers as well as by transmission electron microscopy.
The level of phosphatidylethanolamine (PE)-conjugated form of microtubule-associated protein 1 light chain 3 (LC3), LC3 II, is a useful marker for autophagy. In the control siRNA-treated cells, the expression of LC3 was upregulated at both the protein level and the mRNA level (Fig. 1B and D). Meanwhile, the abundance of LC3 II dramatically increased as differentiation progressed, especially from day 2 to day 4, which reflects the increase of autophagosomes (Fig. 1B). The degradation of p62, which manifests the autophagic activity (26), was also examined. The protein level of p62 increased and peaked on day 1 after induction (Fig. 1B), which is consistent with the expression change in its mRNA level (Fig. 1D). However, in contrast to the slight decline of p62 mRNA afterwards (Fig. 1D), there was a sharp decline of p62 protein from day 2 to day 4 (Fig. 1B), indicating an increase of autophagy flux in control siRNA-treated cells. Consistently, in the control siRNA-treated cells, the number of autophagosomes, which is characterized by double-membrane structures, increased after adipogenic induction, with a substantial increase from day 2 to day 4 (Fig. 1E and F). In C/EBPβ siRNA-treated cells, however, the activation of autophagy was significantly inhibited, as indicated by reduced LC3 II production and impaired degradation of p62 protein (Fig. 1B) and the decrease of autophagosomes (Fig. 1E and F). In contrast, the mRNA expression profiles of LC3 and p62 were not affected by C/EBPβ siRNA (Fig. 1D). These data illustrate an important role of C/EBPβ in activation of autophagy during 3T3-L1 adipocyte differentiation.
Transcriptional activation of Atg4b by C/EBPβ.
A promoter-wide chromatin immunoprecipitation (ChIP) coupled with microarrays (ChIP-chip) at an early stage of 3T3-L1 adipocyte differentiation was recently conducted by us (9), which identified putative C/EBPβ binding sites on the promoters of five autophagy-related genes—Atg2a, Atg4b, Atg7, Atg9a, and Atg10 (Table 1). After adipogenic induction, the mRNA levels of all these five genes were upregulated, as analyzed by reverse transcription (RT)-qPCR, as shown in Fig. 2A. However, among these genes, only the expression of Atg4b was significantly inhibited by C/EBPβ siRNA, suggesting that Atg4b might be subjected to transcriptional regulated by C/EBPβ. Furthermore, ChIP-qPCR confirmed the significant binding of C/EBPβ to the promoter of Atg4b in differentiating cells after induction (day 2) but not in quiescent cells before induction (day 0), as shown in Fig. 2B. Relatively weak binding of C/EBPβ was detected on the promoters of the other four Atg genes (Fig. 2C), which might explain why the expression of these four genes was not significantly affected by C/EBPβ siRNA.
Table 1.
Summary of the five autophagy-related genes that were identified to be potentially targeted by C/EBPβ in our previously reported ChIP-chip dataa
| Chr no. | Start site | End | FDR (%) | Peak position (bp) | Length (bp) | RefSeq | Gene | Strand | TSS |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 95579030 | 95579454 | 1.02 | 212 | 424 | NM_174874 | Atg4b | + | 95585438 |
| 1 | 75075153 | 75075828 | 2.39 | 315 | 675 | NM_001003917 | Atg9a | − | 75075070 |
| 6 | 114608472 | 114609249 | 0.91 | 382 | 777 | NM_028835 | Atg7 | + | 114608743 |
| 13 | 91698421 | 91699057 | 0.97 | 386 | 636 | NM_025770 | Atg10 | − | 91698226 |
| 19 | 6236295 | 6236788 | 3.25 | 281 | 493 | NM_194348 | Atg2a | + | 6241667 |
The previously reported ChIP-chip data are given in reference 9. For each region of C/EBPβ enrichment, the mm8 chromosomal coordinates are given, including the chromosome (Chr) number, the start site, and the end of the region (End). “Length” refers to the size of the continuous region across which C/EBPβ signal was significantly enriched, while “Peak position” refers to the location of highest signal within the region. The false discovery rate (FDR) is also shown. Genes near the C/EBPβ binding sites are shown in the “RefSeq” and “Gene” columns. TSS, transcription start site.
Fig 2.

C/EBPβ induces Atg4b expression. (A) 3T3-L1 cells were transfected with the indicated siRNAs followed by adipogenic induction. The mRNA levels of the autophagy genes were determined by RT-qPCR on the indicated days. For each gene, data were normalized to the day 0 point of siNC-treated cells. *, P < 0.05 versus siNC-treated cells on day 2. (B) 3T3-L1 cells were induced to differentiation. At the times indicated, ChIP-qPCR was performed. Data were normalized to the IgG controls at each time point. (C) C/EBPβ enrichment at the promoters of the five autophagy-related genes was performed by ChIP-qPCR on day 2 of adipocyte differentiation. Data were normalized to the IgG control of each gene. One region of the insulin gene serves as a negative control. (D) Schematic representation of Atg4b distal promoter constructs used for luciferase assays. Asterisks and lowercase letters indicate mutations in the (−6408 Mut) Luc construct. (E) 3T3-L1 cells were transfected with control vector or C/EBPβ-expressing vector, together with the indicated reporter constructs. Luciferase activities were measured 36 h later and plotted. (F) Cells were treated as in panel A, and cell lysates were collected on the indicated days and subjected to Western blotting. Hsp90 serves as a loading control. (G and H) 3T3-L1 cells were infected with empty vector or with retrovirus expressing Flag-tagged C/EBPβ and induced to differentiation. (G) The mRNA level of Atg4b was determined by RT-qPCR on the indicated days. (H) Cell lysates were collected on day 2 for Western blotting.
A DNA fragment containing the distal promoter of Atg4b (bp −6408 to ∼−5984 upstream of the transcription start site), as identified in the ChIP-chip analysis, was subcloned into the luciferase reporter construct pGL3-Promoter (Fig. 2D) and transfected into 3T3-L1 preadipocytes with or without the C/EBPβ expression vector. Luciferase activity was increased about 6-fold by C/EBPβ (Fig. 2E). Consistently, deletion or mutation of the putative C/EBP binding site (bp −6143 to −6130), as shown in Fig. 2D, almost totally abolished the transactivation by C/EBPβ (Fig. 2E).
Consistent with the mRNA profile, the Atg4b protein level was also increased after adipogenic induction, which was drastically inhibited by C/EBPβ siRNA (Fig. 2F). To further validate the role of C/EBPβ in Atg4b induction, C/EBPβ was overexpressed in 3T3-L1 cells. Ectopic expression of C/EBPβ enhanced the expression of Atg4b at both mRNA levels and protein levels (Fig. 2G and H). These results demonstrate that Atg4b is a bona fide target of C/EBPβ during adipogenesis.
Transactivation of Atg4b is required for autophagy and adipogenesis during 3T3-L1 adipocyte differentiation.
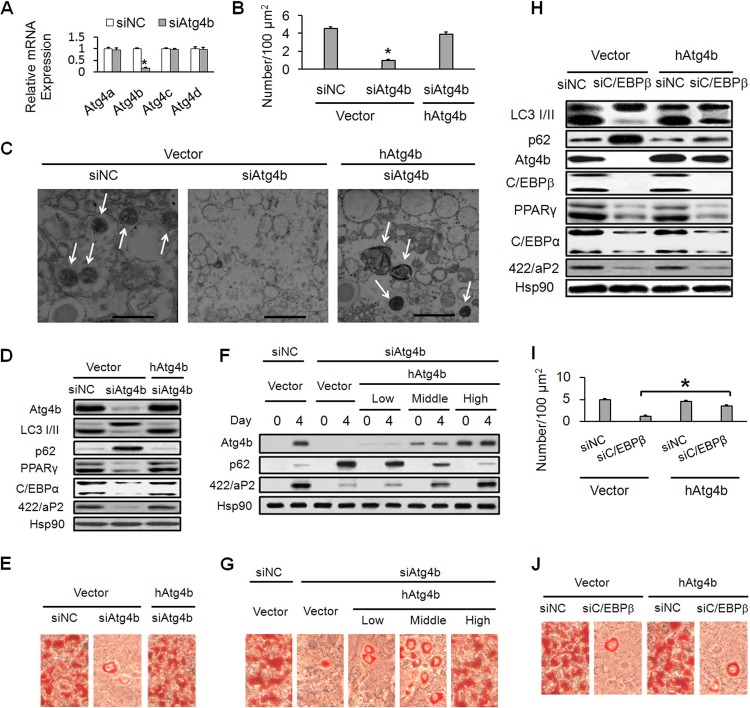
Atg4b exposes glycine from the LC3 precursor at its C terminus to form LC3 I (27). This initial proteolytic processing is required for the subsequent lipidation of LC3 to form LC3 II, which is essential for autophagosome formation (28). Experiments were then conducted to test the role of Atg4b in adipocyte differentiation. The efficiency and specificity of Atg4b siRNA were confirmed by RT-qPCR (Fig. 3A). Similarly to C/EBPβ siRNA, knockdown of Atg4b by siRNA significantly inhibited autophagy, as indicated by the decreased autophagosomes (Fig. 3B and C), impaired LC3 II conversion, and accumulation of p62 protein (Fig. 3D). Furthermore, Atg4b knockdown blocked the terminal differentiation of 3T3-L1 cells, as indicated by adipocyte marker gene expression (genes coding for PPARγ, C/EBPα, and 422/aP2) as well as Oil red O staining (Fig. 3D and E).
Fig 3.
Transactivation of Atg4b is required for autophagy and adipogenesis during 3T3-L1 adipocyte differentiation. (A) 3T3-L1 cells were transfected with control siRNA (siNC) or Atg4b siRNA (siAtg4b). The mRNA levels of the Atg4 genes on the indicated days were determined by RT-qPCR. *, P < 0.05 versus siNC-treated cells. (B to E) 3T3-L1 cells were infected with empty vector or retroviruses expressing human Atg4b (hAtg4b). The cells then were transfected with siNC or siAtg4b. Postconfluence, cells were induced to differentiation. (B) Transmission electron microscopic analyses were conducted on day 4. The quantification of autophagosome abundance is shown. *, P < 0.05 versus siNC-treated cells. (C) Transmission electron micrographs of the cells are shown on the indicated days (bars, 1 μm). Note the presence of autophagosomes (arrows). (D) Cell lysates were collected on day 4 and subjected to Western blotting. (E) Oil red O staining of 3T3-L1 cells on day 6 of differentiation was performed. (F and G) 3T3-L1 cells were infected with empty vector or retroviruses expressing different levels of hAtg4b (low, middle, and high). The cells then were transfected with siNC or siAtg4b. Postconfluence, cells were induced to differentiation. (F) Cell lysates were collected on the indicated days and subjected to Western blotting. (G) Oil red O staining of 3T3-L1 cells on day 6 of differentiation was performed. (H to J) 3T3-L1 cells were infected with empty vector or retroviruses expressing hAtg4b. The cells then were transfected with siNC or siC/EBPβ. Postconfluence, cells were induced to differentiation. (H) Cell lysates were collected on day 4 and subjected to Western blotting. (I) Transmission electron microscopic analyses were conducted on day 4. The quantification of autophagosome abundance is shown. *, P < 0.05. (J) Oil red O staining of 3T3-L1 cells on day 6 of differentiation was performed.
Rescue experiments were performed to further confirm the above observation. Since 3T3-L1 preadipocytes are of mouse origin, human Atg4b was ectopically expressed in Atg4b siRNA-treated 3T3-L1 cells by retrovirus transduction. Western blotting demonstrated that the human Atg4b gene is resistant to mouse Atg4b siRNA knockdown and was expressed at a similar level to the postinduction (day 4) endogenous Atg4b gene in the parental control cells (Fig. 3D). Forced expression of human Atg4b rescued autophagy (Fig. 3B, C, and D) and adipogenesis (Fig. 3D and E), which reinforces the idea that Atg4b is required for autophagy and adipogenesis. In addition, the rescue effect was dependent on the ectopic expression level of Atg4b (Fig. 3F and G). When the ectopic expression level of human Atg4b was low, which was only slightly higher than the preinduction (day 0) basal Atg4b level in the parental control cells, the rescue effect was not obvious. With the gradual increase of the ectopic expression level of human Atg4b (middle level), the rescue effect was dramatically improved, and when the ectopic expression level reached the postinduction endogenous Atg4b level in the parental control cells (high level), it almost completely rescued autophagy and adipogenesis. These data, together with those shown in Fig. 2, indicate that the expression level of Atg4b is highly regulated during adipogenesis and the transactivation of Atg4b by C/EBPβ is required for the efficient activation of autophagy and adipogenesis.
Furthermore, forced expression of Atg4b led to significant rescue of autophagy, which was inhibited by C/EBPβ siRNA (Fig. 3H and I). However, it could not restore C/EBPβ siRNA-mediated inhibition of adipogenesis (Fig. 3H and J). This is understandable, because C/EBPβ is directly involved in the transactivation of adipogenic master genes (coding for PPARγ and C/EBPα), a process that could not be rescued simply by ectopic expression of Atg4b upon depletion of C/EBPβ. Taken together, these results indicate that Atg4b is a very important downstream target gene of C/EBPβ in promoting autophagy during adipocyte differentiation.
Autophagy plays an important role between days 2 and 3 during 3T3-L1 adipocyte differentiation.
Although autophagy is required for adipocyte differentiation, its underlying mechanism is still not clear. As shown in Fig. 1E, a substantial increase of autophagic activity was detected from day 2 to day 4 of 3T3-L1 adipocyte differentiation, which was in concert with the marked increase of the key adipogenic factors PPARγ and C/EBPα from day 2 to day 4 (Fig. 1B). This time course correlation suggests an important role of autophagy during this period. To confirm this concept, a commonly used autophagy inhibitor, 3-methyladenine (3-MA), which blocks the formation of autophagosomes (29), was used to further investigate the function of autophagy during adipocyte differentiation.
The scheme for 3-MA treatment is illustrated in Fig. 4A. The autophagy inhibitor was added on the indicated time point, without interrupting the adipogenic differentiation process. All of the analyses were performed on day 6. As expected, the treatment of the cells with 3-MA significantly inhibited autophagy, as indicated by impaired conversion of LC3 and accumulation of p62 (Fig. 4B). Little cytotoxic effect of 3-MA on the cells was detected (Fig. 4C). Moreover, inhibition of autophagy from days 0, 1, and 2 blocked terminal adipocyte differentiation to a similar level, while inhibition of autophagy from day 3 or 4 had little effect on adipocyte differentiation (Fig. 4B and D). To preclude the potential off-target effect of 3-MA, wortmannin, another autophagy inhibitor, was applied and similar results were obtained (data not shown). These results demonstrate a critical role of autophagy between days 2 and 3 of 3T3-L1 adipocyte differentiation.
Fig 4.

Autophagy plays an important role between days 2 and 3 of 3T3-L1 adipocyte differentiation. (A) Scheme showing the treatment of 3T3-L1 cells with autophagy inhibitor 3-MA. 3T3-L1 cells were induced to differentiation with or without 3-MA treatment from the indicated day (arrowheads). Control (Ctrl) cells were treated with vehicle from day 0. 3-MA-treated cells were treated with 3-MA from days 0 (a), 1 (b), 2 (c), 3 (d), and 4 (e). (B) Cells were treated as in panel A. On day 6, Western blotting was conducted using the indicated antibodies. (C) 3T3-L1 cells were induced to differentiation in the presence or absence of 3-MA. The percentages of apoptotic (Apop) and necrotic (Nec) cells were determined after acridine orange-ethidium bromide costaining by fluorescence microscopy on day 2. Acridine orange-stained cells were evaluated for morphological characteristics of apoptosis, and cells staining positive for ethidium bromide were considered necrotic. (D) Cells were treated as in panel A. On day 6, Oil red O staining was performed.
Degradation of Klf2 and Klf3 by autophagy facilitates adipocyte differentiation.
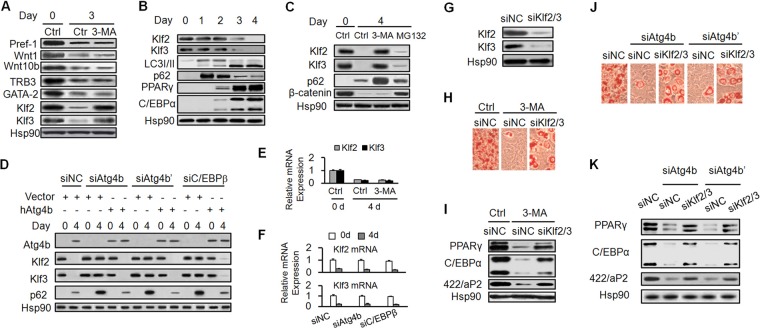
As mentioned above, the timely downregulation of negative regulators is essential for adipocyte differentiation. Because autophagy is one of the main pathways utilized by cells to degrade intracellular proteins, it is hypothesized that autophagy promoted adipocyte differentiation via controlling the degradation of negative regulators. To address this possibility, the effect of autophagy inhibition on the degradation of a series of well-characterized negative regulators was examined. The 3-MA treatment impeded the turnover of Klf2 and Klf3 but had little effect on the degradation of the other factors (Fig. 5A). Of interest, Klf2 and Klf3 have been shown to inhibit the transactivation of PPARγ and C/EBPα, respectively, through directly binding to their promoters (12, 15). The time course correlation between the marked PPARγ and C/EBPα upregulation and substantial autophagy activation prompted us to further investigate the role of autophagy in the degradation of Klf2 and Klf3.
Fig 5.
Klf2 and Klf3 are degraded by autophagy. (A) 3T3-L1 cells were induced to differentiation with or without 3-MA. Cell lysates were collected on the indicated day for Western blotting using the indicated antibodies. (B) Cell lysates were collected on the indicated days and subjected to Western blotting. (C) 3T3-L1 cells were induced to differentiation in the presence of vehicle (control [Ctrl]), 3-MA, or MG132. Western blotting was then conducted. β-Catenin serves as a positive control of proteasome-mediated degradation. (D) 3T3-L1 cells were infected with empty vector or retroviruses expressing human Atg4b (hAtg4b). The cells then were transfected with siNC, siAtg4b, siAtg4b′ (having one additional Atg4b siRNA), or siC/EBPβ. Cell lysates were collected on the indicated days and subjected to Western blotting. (E) 3T3-L1 cells were induced to differentiation with or without 3-MA. The mRNA levels of Klf2 and Klf3 were determined by RT-qPCR. (F) 3T3-L1 cells were transfected with the indicated siRNAs and induced to differentiation. The mRNA levels of Klf2 and Klf3 were determined by RT-qPCR. (G) Klf2 and Klf3 were efficiently knocked down by siRNA. (H to K) Knockdown of Klf2 and Klf3 significantly restores adipogenesis upon autophagy being inhibited. Preconfluent 3T3-L1 cells were transfected with the indicated siRNAs. Cells were then induced to differentiation in the presence or absence of 3-MA. On day 6, cells were subjected to Oil red O staining (H and J) or Western blotting (I and K).
As shown in Fig. 5B, the Klf2 and Klf3 proteins were at expressed at high levels from day 0 to day 2, but the levels fell abruptly on day 3 and were barely detectable on day 4. Accordingly, the expression of PPARγ and C/EBPα occurred on day 2, with a marked increase of expression on day 3. In concert with these expression changes, autophagy activity was substantially increased from day 2 to day 3, as indicated by dramatic conversion of LC3 and turnover of p62 protein. Inhibition of autophagy by 3-MA prevented the degradation of Klf2 and Klf3 (Fig. 5C). In contrast, MG132, an inhibitor of proteasomes, had a minor role in the degradation of Klf2 and Klf3 (Fig. 5C). Similarly, inhibition of autophagy by the knockdown of C/EBPβ (siC/EBPβ) or Atg4b (siAtg4b and siAtg4b′, two different siRNAs against Atg4b) impairs the decline of Klf2 and Klf3, which could be restored by the ectopic expression of human Atg4b (Fig. 5D). Moreover, little effect on the mRNA expression profiles of Klf2 and Klf3 was detected upon the inhibition of autophagy (Fig. 5E and F).
To test whether the degradation of Klf2 and Klf3 is important for the action of autophagy in adipocyte differentiation, a rescue experiment using Klf2 and Klf3 siRNAs was performed. Knockdown of Klf2 and Klf3 resulted in significant reversal of the inhibition of adipogenesis in the presence of the autophagy inhibitor 3-MA or siAtg4b/siAtg4b′ (Fig. 5G to K). Collectively, these results indicate that the turnover of Klf2 and Klf3 by autophagy plays an important role in adipocyte differentiation.
p62 mediates the degradation of Klf2 and Klf3 during adipocyte differentiation.
The p62 protein, also called sequestosome 1 (SQSTM1), is an autophagy-specific substrate (30). A recent study indicated that p62 bound to and recruited the polyubiquitinated oncoprotein PML-retinoic acid receptor α (RARα) to autophagosomes for degradation, resulting in myeloid cell differentiation (25). Therefore, the potential role of p62 in regulating the turnover of Klf2 and Klf3 and in adipocyte differentiation was explored. A significant interaction between p62 and Klf2/3 was detected on day 3 of differentiation, rather than on day 0, 1, or 2 (Fig. 6A and B). Since p62 is a polyubiquitin-binding protein (30), the ubiquitination of Klf2 and Klf3 was investigated. In contrast to the low level of Klf2/3 polyubiquitination on days 0, 1, and 2, significant polyubiquitination of Klf2/3 was detected on day 3 (Fig. 6A and B), which is consistent with the significant interaction between p62 and Klf2/3 on day 3.
Fig 6.
P62 mediates the degradation of Klf2 and Klf3 through autophagy. (A and B) 3T3-L1 preadipocytes were transfected with Klf2/3-Myc/His vectors and induced to differentiation. On the indicated days, cells were collected and the Klf2/3-Myc/His proteins were precipitated with Ni-nitrilotriacetic acid (NTA) beads, followed by Western blotting. Cells were pretreated with 3-MA for 8 h before collection. Ub, ubiquitin. The whole-cell lysates (WCL) were also subjected to Western blotting. (C to F) Preconfluent 3T3-L1 cells were transfected with control siRNA or p62 siRNA (siP62-pre). Postconfluence, the cells were induced to differentiation. (C) The knockdown efficiency of p62 siRNA was confirmed by Western blotting. (D) The activation of ERK (pERK1/2) was detected at the indicated time point. Oil red O staining (E) and Western blotting (F) were performed on day 6. (G to L) Transfection of p62 siRNA at 4 h after adipogenic induction (siP62-post) blocked Klf2/3 decline and adipogenesis. The p62 siRNA was transfected 4 h after adipogenic induction (siP62-post), and cell lysates were collected at the indicated time points for Western blotting (G and H). On day 6, cells were subjected to Oil red O staining (I) and Western blotting (J). (K) Cells were transfected with GFP-tagged Klf2/3 and then treated with the indicated siRNAs. At 60 h after induction, cells were subjected to confocal analyses. In each analysis, the cells with colocalization between GFP-tagged proteins and LC3 in 50 GFP-positive cells were counted. The results are shown as percentage of cells with colocalization. (L) The confocal images for panel K are shown. (M and N) 3T3-L1 cells were infected with empty vector or retroviruses expressing wild-type human p62 (hP62-Wt) or the mutant p62 (hP62-Mt), which lacks the LIR motif (amino acids 321 to 342 of hP62). The cells then were treated with the indicated siRNAs. On day 6, the cells were subjected to Oil red O staining (M) and Western blotting (N).
It has been reported that knockdown of p62 by short hairpin RNA (shRNA) in 3T3-L1 preadipocytes enhances adipogenesis through increasing the early activation of ERK (0 to ∼4 h after induction) without affecting the second increase of ERK activation (day 3 after induction) (31, 32). In our study, p62 siRNA was transfected into preconfluent 3T3-L1 cells (siP62-pre) and cells were induced to differentiation postconfluence. In keeping with the previous report, treatment of siP62-pre increased the early activation of ERK (Fig. 6C and D) and enhanced adipogenesis (Fig. 6E and F). A modification of the siRNA treatment was conducted to avoid the effect of p62 knockdown on ERK activity. The p62 siRNA was transfected into 3T3-L1 cells at 4 h after induction (siP62-post), which dramatically inhibited the p62 protein level by day 2 of adipogenic differentiation (Fig. 6G), without affecting ERK activation (Fig. 6H). In this case, adipocyte differentiation was blocked, accompanied by accumulation of Klf2 and Klf3 (Fig. 6I and J).
Confocal microscopic analysis was also conducted. Because the antibodies against Klf2/3 were not suitable for immunofluorescence staining, GFP-tagged Klf2/3 was transfected into 3T3-L1 cells. Colocalization between GFP-tagged Klf2/3 and LC3 puncta (autophagosome marker) was detected during adipocyte differentiation, which was significantly impaired by the treatment siP62-post (Fig. 6K and L). Consistently, the colocalization between green fluorescent protein (GFP)-tagged Klf2/3 and LAMP2 (lysosomal marker) was detected, which was also blocked by siP62-post (data not shown).
Previous studies revealed that p62 recognizes polyubiquitinated protein aggregates and binds to LC3 to target aggregates to autophagosomes for degradation (30). Ectopic expression of wild-type human p62 (hP62-Wt), which is resistant to p62 siRNA of mouse origin, rescued the inhibition of adipogenesis and impairment of Klf2/3 decline mediated by siP62-post. In contrast, ectopic expression of mutant human p62 (hP62-Mt), which lacks the LC3-interacting region (LIR), failed to rescue it, supporting the critical role of the p62-LC3 interaction in mediating the removal of Klf2/3 (Fig. 6M and N). Collectively, these studies implicate an important role of p62 in promoting the degradation of Klf2 and Klf3 by autophagy during adipocyte differentiation.
C/EBPβ-regulated autophagy plays an important role in adipogenesis in vivo.
To verify the above-described findings in a more physiological context, experiments were further performed in mice. C/EBPβ binding to the distal promoter of Atg4b was confirmed with ChIP analysis on mouse white adipose tissue (WAT) (Fig. 7A).
Fig 7.

C/EBPβ-regulated autophagy plays an important role in adipogenesis in vivo. (A) Inguinal white adipose tissue (WAT) from mice was subjected to ChIP using C/EBPβ antibody. Enrichment of C/EBPβ on the promoters of the indicated genes was analyzed by PCR. One promoter region of PPARγ and that of insulin serve as a positive control and a negative control, respectively. (B to G) Adenovirus expressing Atg4b shRNA (shAtg4b) was weekly injected subcutaneously adjacent to one side of inguinal fat pad from 4 weeks of age for an additional 4 weeks. LacZ shRNA (shLacZ) was simultaneously injected at the contralateral site as a control. (B) Inguinal WAT samples from adenovirus-treated mice were subjected to Western blotting with the indicated antibodies. (C and D) A reduction in size and weight was seen in inguinal WAT when Atg4b was knocked down. The inguinal WAT was dissected and analyzed. *, P < 0.05. (E) The inguinal WAT was subjected to H&E staining. (F) The size distribution of adipocytes was analyzed and plotted based on H&E staining. (G) Isolated cells from WAT were stained with Nile red (used for staining adipocytes) followed by flow cytometry analysis. *, P < 0.05. (H) Mice fed the high-fat diet (HFD) have enhanced autophagy and adipogenesis. Inguinal WAT samples from mice fed the regular diet (RD) or HFD were subjected to Western blotting using the indicated antibodies.
To examine the role of Atg4b in autophagy and adipogenesis in vivo, recombinant adenovirus expressing Atg4b shRNA was injected subcutaneously adjacent to one inguinal fat pad site. Recombinant adenovirus expressing LacZ shRNA was injected in the contralateral site as a control. Injection of Atg4b shRNA into the white fat pads resulted in impaired LC3 processing and accumulation of p62, indicating autophagy inhibition (Fig. 7B). In line with the results obtained from the cellular model, knockdown of Atg4b by shRNA caused an increase in Klf2/3 and a decrease in PPARγ and C/EBPα (Fig. 7B). Moreover, the amount of WAT was significantly reduced by Atg4b knockdown (Fig. 7C and D). Histological analyses showed the diminished size of adipocytes in fat pads injected with Atg4b shRNA, indicating decreased lipid content in adipocytes (Fig. 7E and F). In addition, the percentage of adipocytes in WAT was reduced in Atg4b shRNA-treated fat pads, suggesting impaired adipocyte differentiation (Fig. 7G).
The high-fat-diet (HFD)-fed C57BL/6J mouse is an obese mouse model characterized by enhanced adipogenesis. Compared with the WAT of regular-diet (RD)-fed mice, the elevated level of C/EBPβ, Atg4b, autophagy, PPARγ, and C/EBPα and decreased level of Klf2/3 were detected in WAT from HFD-fed mice (Fig. 7H). These data correlate well with the results described above and further confirm the important function of autophagy during adipocyte differentiation, which is regulated by C/EBPβ.
DISCUSSION
C/EBPβ is a basic leucine zipper transcription factor involved in a variety of physiological processes, including metabolism regulation, inflammation, cell differentiation, and proliferation (10, 11, 33, 34). In this study, we identified a novel role of C/EBPβ in transcriptional control of autophagy during adipocyte differentiation (Fig. 1 and 2). Thus, our data implicate C/EBPβ as an important regulator that orchestrates the activation of autophagy.
Autophagy originally had been considered to be a bulk degradation pathway with little or no selectivity. However, growing lines of evidence indicate the selectivity of autophagy in sorting vacuolar enzymes and in the removal of protein aggregates, specific proteins, unwanted organelles, and microbes (35). p62 is a substrate degraded by autophagy and has been recently been implicated as a mediator of selective autophagy, which targets the oncoprotein PML-RARα for autophagic degradation, thereby promoting myeloid cell differentiation (25). Our work shows that Klf2 and Klf3 are also the cargos that are to be selectively degraded by autophagy in a p62-dependent manner.
Encoded by the SQSTM1 gene, p62 is a scaffold and adaptor protein that modulates protein-protein interactions. Apart from its role in regulating autophagy, p62 is also a signaling organizer that regulates essential cellular functions, such as cell signaling, receptor internalization, and gene transcription (36). It is reported that loss of p62 leads to an enhanced basal ERK activity that is essential for adipocyte differentiation (32). As is known, ERK activation should be tightly and temporally controlled to ensure adipogenesis. There are two bursts of ERK activation during adipocyte differentiation. The early increase of ERK activity occurred within several hours after adipogenic induction, which is proposed to enhance the activity of factors that regulate both PPARγ and C/EBPα expression (37). The second increase of ERK activation appears on days 2 and 3 after induction, which is thought to have a negative role in adipogenesis (38). Depletion of p62 by shRNA affects the early activation of ERK but not its late stimulation, thereby promoting adipogenesis (32). The reason for this selective effect of p62 is not clear. Transfection of p62 siRNA was performed at 4 h after induction to avoid the effect of p62 knockdown on ERK activation (Fig. 6H). In this way, adipogenesis was inhibited and the degradation of Klf2/3 by autophagy was impaired. Thus, p62 plays at least two roles during adipocyte differentiation: restricting the early ERK activity to keep an appropriate level of adipogenesis and facilitating the removal of some negative regulators such as Klf2/3 to ensure adipocyte differentiation.
C/EBPβ activates the expression of PPARγ and C/EBPα by directly binding to their promoters. Although C/EBPβ is induced very early in adipocyte differentiation, the expression of PPARγ and C/EBPα occurs much later. This lag appears necessary, because PPARγ and C/EBPα are both antimitotic, and their premature expression would otherwise prevent the mitotic clonal expansion (MCE) required for adipocyte differentiation (8). In our work, Klf2 and Klf3, two negative regulators of PPARγ and C/EBPα expression, were shown to be efficiently degraded by autophagy from day 2 to day 4 of 3T3-L1 adipocyte differentiation, thereby providing a mechanistic explanation for the delayed expression of PPARγ and C/EBPα. It should be noted that no obvious degradation of Klf2 and Klf3 was observed from day 0 to day 2, despite the existence of basal autophagic activity during this period (Fig. 5B). Our data showed that significant polyubiquitination of Klf2/3 was detected on day 3, which is in contrast to the low level of Klf2/3 polyubiquitination on days 0, 1, and 2 (Fig. 6A and B). The polyubiquitination has been reported to be required for the binding of autophagy cargos to p62 (30, 39), thereby explaining the delayed removal of Klf2/3.
Although autophagy plays a pivotal role in adipogenesis, this does not mean that all of the autophagy-related genes are required for adipogenesis. For example, our results indicated that knockdown of either Atg2a or Atg9a had no obvious effect on 3T3-L1 adipocyte differentiation (data not shown), possibly due to functional redundancy by other genes. Before our work, only Atg5 and Atg7, among the many autophagy-related genes, had been proven to be essential for adipogenesis (20, 21). In mammalian cells, four Atg4 homologues have been reported (40), among which Atg4b has the strongest cysteine proteinase activity for the cleavage of LC3 precursor (19). Although a critical role of Atg4b in inner ear development has been reported (41), the involvement of Atg4b in the development of adipose tissue has not been investigated. In this study, specific depletion of Atg4b without affecting the expression of the other Atg4 family members demonstrates its important role in autophagy and adipocyte differentiation (Fig. 3 and Fig. 7). Therefore, Atg4b, like Atg5 and Atg7, is an important autophagy-related gene involved in the regulation of adipogenesis.
In summary, the present study describes a direct link between C/EBPβ and autophagy during adipocyte differentiation. Based upon the work presented here and previous reports, we propose the model that is illustrated in Fig. 8. C/EBPβ transactivates the expression of Atg4b to promote autophagy, which mediates the degradation of Klf2/3 in a p62- and ubiquitination-dependent manner, thereby relieving the repression of PPARγ and C/EBPα transcription. Meanwhile, C/EBPβ is involved in the transactivation of PPARγ and C/EBPα. These two effects coordinately promote the expression of PPARγ and C/EBPα and facilitate adipogenesis. The findings presented in this study provide new insights into the role of C/EBPβ and shed light on the mechanism of autophagy during adipocyte differentiation. Modulation of this regulatory pathway may be of therapeutic value for intervening in the overexpansion of adipose mass, thereby preventing obesity and its related metabolic complications.
Fig 8.

Proposed model of the role of C/EBPβ and autophagy in adipocyte differentiation. Adipogenesis is a process strictly controlled by the interplay of positive and negative regulators. On one hand, C/EBPβ transactivates the expression of Atg4b, which, like other autophagy genes, such as Atg5 and Atg7, is required for the activation of autophagy. The adipogenic inhibitors Klf2/3 then are degraded by autophagy in a p62- and ubiquitination-dependent manner, thereby relieving the repression of PPARγ and C/EBPα transcription. One the other hand, C/EBPβ directly binds to the promoters of PPARγ and C/EBPα and activates their transcription. These two effects coordinately promote the expression of PPARγ and C/EBPα and facilitate adipogenesis.
ACKNOWLEDGMENTS
This research is partially supported by National Key Basic Research Project grants 2011CB910201 and 2013CB530601, the State Key Program of National Natural Science Foundation 31030048C120114, Shanghai Key Science and Technology Research Project 10JC1401000 (for Q.-Q.T.), National Natural Science Foundation grant 30870510 (for X.L.), National Natural Science Foundation grant 31000603 (for L.G.), and the Fudan University Ming-Dao Project for Graduate Students EZF101336 (for J.-X.H.). The Department of Biochemistry and Molecular Biology is supported by Shanghai Leading Academic Discipline Project B110 and 985 Project 985 III-YFX0302.
We have no potential conflicts of interest relevant to this article.
Footnotes
Published ahead of print 10 June 2013
REFERENCES
- 1. Rajala MW, Scherer PE. 2003. The adipocyte—at the crossroads of energy homeostasis, inflammation, and atherosclerosis. Endocrinology 144: 3765– 3773 [DOI] [PubMed] [Google Scholar]
- 2. Rosen ED, Spiegelman BM. 2006. Adipocytes as regulators of energy balance and glucose homeostasis. Nature 444: 847– 853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Schaffler A, Muller-Ladner U, Scholmerich J, Buchler C. 2006. Role of adipose tissue as an inflammatory organ in human diseases. Endocr. Rev. 27: 449– 467 [DOI] [PubMed] [Google Scholar]
- 4. Calle EE, Kaaks R. 2004. Overweight, obesity and cancer: epidemiological evidence and proposed mechanisms. Nat. Rev. Cancer 4: 579– 591 [DOI] [PubMed] [Google Scholar]
- 5. Haslam DW, James WP. 2005. Obesity. Lancet 366: 1197– 1209 [DOI] [PubMed] [Google Scholar]
- 6. Kopelman PG. 2000. Obesity as a medical problem. Nature 404: 635– 643 [DOI] [PubMed] [Google Scholar]
- 7. Green H, Kehinde O. 1975. An established preadipose cell line and its differentiation in culture. II. Factors affecting the adipose conversion. Cell 5: 19– 27 [DOI] [PubMed] [Google Scholar]
- 8. Tang QQ, Lane MD. 1999. Activation and centromeric localization of CCAAT/enhancer-binding proteins during the mitotic clonal expansion of adipocyte differentiation. Genes Dev. 13: 2231– 2241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Guo L, Li X, Huang JX, Huang HY, Zhang YY, Qian SW, Zhu H, Zhang YD, Liu Y, Wang KK, Tang QQ. 2012. Histone demethylase Kdm4b functions as a co-factor of C/EBPbeta to promote mitotic clonal expansion during differentiation of 3T3-L1 preadipocytes. Cell Death Differ. 19: 1917– 1927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tang QQ, Otto TC, Lane MD. 2003. CCAAT/enhancer-binding protein beta is required for mitotic clonal expansion during adipogenesis. Proc. Natl. Acad. Sci. U. S. A. 100: 850– 855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhang YY, Li X, Qian SW, Guo L, Huang HY, He Q, Liu Y, Ma CG, Tang QQ. 2011. Transcriptional activation of histone H4 by C/EBPβ during the mitotic clonal expansion of 3T3-L1 adipocyte differentiation. Mol. Biol. Cell 22: 2165– 2174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Banerjee SS, Feinberg MW, Watanabe M, Gray S, Haspel RL, Denkinger DJ, Kawahara R, Hauner H, Jain MK. 2003. The Kruppel-like factor KLF2 inhibits peroxisome proliferator-activated receptor-gamma expression and adipogenesis. J. Biol. Chem. 278: 2581– 2584 [DOI] [PubMed] [Google Scholar]
- 13. Bezy O, Vernochet C, Gesta S, Farmer SR, Kahn CR. 2007. TRB3 blocks adipocyte differentiation through the inhibition of C/EBPβ transcriptional activity. Mol. Cell. Biol. 27: 6818– 6831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ross SE, Hemati N, Longo KA, Bennett CN, Lucas PC, Erickson RL, MacDougald OA. 2000. Inhibition of adipogenesis by Wnt signaling. Science 289: 950– 953 [DOI] [PubMed] [Google Scholar]
- 15. Sue N, Jack BH, Eaton SA, Pearson RC, Funnell AP, Turner J, Czolij R, Denyer G, Bao S, Molero-Navajas JC, Perkins A, Fujiwara Y, Orkin SH, Bell-Anderson K, Crossley M. 2008. Targeted disruption of the basic Kruppel-like factor gene (Klf3) reveals a role in adipogenesis. Mol. Cell. Biol. 28: 3967– 3978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sul HS. 2009. Pref-1: role in adipogenesis and mesenchymal cell fate. Mol. Endocrinol. 23: 1717– 1725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tong Q, Dalgin G, Xu H, Ting CN, Leiden JM, Hotamisligil GS. 2000. Function of GATA transcription factors in preadipocyte-adipocyte transition. Science 290: 134– 138 [DOI] [PubMed] [Google Scholar]
- 18. Cecconi F, Levine B. 2008. The role of autophagy in mammalian development: cell makeover rather than cell death. Dev. Cell 15: 344– 357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mizushima N. 2007. Autophagy: process and function. Genes Dev. 21: 2861– 2873 [DOI] [PubMed] [Google Scholar]
- 20. Baerga R, Zhang Y, Chen PH, Goldman S, Jin S. 2009. Targeted deletion of autophagy-related 5 (atg5) impairs adipogenesis in a cellular model and in mice. Autophagy 5: 1118– 1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Singh R, Xiang Y, Wang Y, Baikati K, Cuervo AM, Luu YK, Tang Y, Pessin JE, Schwartz GJ, Czaja MJ. 2009. Autophagy regulates adipose mass and differentiation in mice. J. Clin. Invest. 119: 3329– 3339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhang Y, Goldman S, Baerga R, Zhao Y, Komatsu M, Jin S. 2009. Adipose-specific deletion of autophagy-related gene 7 (atg7) in mice reveals a role in adipogenesis. Proc. Natl. Acad. Sci. U. S. A. 106: 19860– 19865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Goldman S, Zhang Y, Jin S. 2010. Autophagy and adipogenesis: implications in obesity and type II diabetes. Autophagy 6: 179– 181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zhang Y, Zeng X, Jin S. 2012. Autophagy in adipose tissue biology. Pharmacol. Res. 66: 505– 512 [DOI] [PubMed] [Google Scholar]
- 25. Wang Z, Cao L, Kang R, Yang M, Liu L, Zhao Y, Yu Y, Xie M, Yin X, Livesey KM, Tang D. 2011. Autophagy regulates myeloid cell differentiation by p62/SQSTM1-mediated degradation of PML-RARalpha oncoprotein. Autophagy 7: 401– 411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mizushima N, Yoshimori T, Levine B. 2010. Methods in mammalian autophagy research. Cell 140: 313– 326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Fujita N, Hayashi-Nishino M, Fukumoto H, Omori H, Yamamoto A, Noda T, Yoshimori T. 2008. An Atg4B mutant hampers the lipidation of LC3 paralogues and causes defects in autophagosome closure. Mol. Biol. Cell 19: 4651– 4659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, Kominami E, Ohsumi Y, Yoshimori T. 2000. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 19: 5720– 5728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhou L, Zhang J, Fang Q, Liu M, Liu X, Jia W, Dong LQ, Liu F. 2009. Autophagy-mediated insulin receptor down-regulation contributes to endoplasmic reticulum stress-induced insulin resistance. Mol. Pharmacol. 76: 596– 603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, Overvatn A, Bjorkoy G, Johansen T. 2007. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J. Biol. Chem. 282: 24131– 24145 [DOI] [PubMed] [Google Scholar]
- 31. Lee SJ, Pfluger PT, Kim JY, Nogueiras R, Duran A, Pages G, Pouyssegur J, Tschop MH, Diaz-Meco MT, Moscat J. 2010. A functional role for the p62-ERK1 axis in the control of energy homeostasis and adipogenesis. EMBO Rep. 11: 226– 232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Rodriguez A, Duran A, Selloum M, Champy MF, Diez-Guerra FJ, Flores JM, Serrano M, Auwerx J, Diaz-Meco MT, Moscat J. 2006. Mature-onset obesity and insulin resistance in mice deficient in the signaling adapter p62. Cell Metab. 3: 211– 222 [DOI] [PubMed] [Google Scholar]
- 33. Elefanty AG, Stanley EG. 2012. Efficient generation of adipocytes in a dish. Nat. Cell Biol. 14: 126– 127 [DOI] [PubMed] [Google Scholar]
- 34. Rahman SM, Janssen RC, Choudhury M, Baquero KC, Aikens RM, de la Houssaye BA, Friedman JE. 2012. CCAAT/enhancer-binding protein beta (C/EBPbeta) expression regulates dietary-induced inflammation in macrophages and adipose tissue in mice. J. Biol. Chem. 287: 34349– 34360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kraft C, Peter M, Hofmann K. 2010. Selective autophagy: ubiquitin-mediated recognition and beyond. Nat. Cell Biol. 12: 836– 841 [DOI] [PubMed] [Google Scholar]
- 36. Moscat J, Diaz-Meco MT. 2011. Feedback on fat: p62-mTORC1-autophagy connections. Cell 147: 724– 727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Prusty D, Park BH, Davis KE, Farmer SR. 2002. Activation of MEK/ERK signaling promotes adipogenesis by enhancing peroxisome proliferator-activated receptor gamma (PPARgamma) and C/EBPalpha gene expression during the differentiation of 3T3-L1 preadipocytes. J. Biol. Chem. 277: 46226– 46232 [DOI] [PubMed] [Google Scholar]
- 38. Font de Mora J, Porras A, Ahn N, Santos E. 1997. Mitogen-activated protein kinase activation is not necessary for, but antagonizes, 3T3-L1 adipocytic differentiation. Mol. Cell. Biol. 17: 6068– 6075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gao C, Cao W, Bao L, Zuo W, Xie G, Cai T, Fu W, Zhang J, Wu W, Zhang X, Chen YG. 2010. Autophagy negatively regulates Wnt signalling by promoting Dishevelled degradation. Nat. Cell Biol. 12: 781– 790 [DOI] [PubMed] [Google Scholar]
- 40. Marino G, Salvador-Montoliu N, Fueyo A, Knecht E, Mizushima N, Lopez-Otin C. 2007. Tissue-specific autophagy alterations and increased tumorigenesis in mice deficient in Atg4C/autophagin-3. J. Biol. Chem. 282: 18573– 18583 [DOI] [PubMed] [Google Scholar]
- 41. Marino G, Fernandez AF, Cabrera S, Lundberg YW, Cabanillas R, Rodriguez F, Salvador-Montoliu N, Vega JA, Germana A, Fueyo A, Freije JM, Lopez-Otin C. 2010. Autophagy is essential for mouse sense of balance. J. Clin. Invest. 120: 2331– 2344 [DOI] [PMC free article] [PubMed] [Google Scholar]