Abstract
Ikaros (Ik) is a critical regulator of hematopoietic gene expression. Here, we established that the Ik interactions with GATA transcription factors and cyclin-dependent kinase 9 (Cdk9), a component of the positive transcription elongation factor b (P-TEFb), are required for transcriptional activation of Ik target genes. A detailed dissection of Ik-GATA and Ik-Cdk9 protein interactions indicated that the C-terminal zinc finger domain of Ik interacts directly with the C-terminal zinc fingers of GATA1, GATA2, and GATA3, whereas the N-terminal zinc finger domain of Ik is required for interaction with the kinase and T-loop domains of Cdk9. The relevance of these interactions was demonstrated in vivo in COS-7 and primary hematopoietic cells, in which Ik facilitated Cdk9 and GATA protein recruitment to gene promoters and transcriptional activation. Moreover, the oncogenic isoform Ik6 did not efficiently interact with Cdk9 or GATA proteins in vivo and perturbed Cdk9/P-TEFb recruitment to Ik target genes, thereby affecting transcription elongation. Finally, characterization of a novel nuclear Ik isoform revealed that Ik exon 6 is dispensable for interactions with Mi2 and GATA proteins but is essential for the Cdk9 interaction. Thus, Ik is central to the Ik-GATA-Cdk9 regulatory network, which is broadly utilized for gene regulation in hematopoietic cells.
INTRODUCTION
The identity of an individual cell is provided by the collection of genes that it expresses, i.e., the transcription program (1). The combined activities of transcription factors along with specific cofactors that they recruit to gene regulatory regions are fundamental for lineage commitment, specification, and/or differentiation of hematopoietic cells (2). A unique family of Kruppel zinc-finger transcription factors includes the key regulators of hematopoiesis, GATA1, GATA2, GATA3, Ikaros (Ik), Aiolos, Helios, Eos, and Pegasus, as well as KLF1, -2, and -3 (3–5). GATA1 is the founding member of the GATA family of DNA binding proteins, which also includes GATA2 and GATA3. These highly related proteins share little homology outside the zinc finger regions (6). GATA1 is critical for the development of erythroid, megakaryocyte, mast cell, and eosinophil lineages (7–11). GATA2 is required for mast cell formation while also contributing to cell homeostasis and survival of hematopoietic stem/progenitor cells (12). GATA3 is important for hematopoietic stem cell maintenance (13), is required at the earliest stages of thymopoiesis, and has been described as a master regulator of T-helper 2 (Th2) cell differentiation (4). GATA proteins contain a C-terminal zinc finger (CF) and an N-terminal zinc finger (NF). The CF is required for DNA binding to the consensus motif A/TGATAA/G (14, 15). Both zinc finger domains are involved in protein-protein interactions with several partners. For instance, GATA1-NF interacts with friend-of-GATA1 (FOG1), TRAP220, and Sp1. GATA1-CF is involved in self-association and participates in protein interactions with PU.1, CBP, KLF1, Sp1, and RBTN2 (3, 16, 17). GATA proteins bind to similar DNA sequences and share common protein partners. They can activate or repress target genes by interacting and recruiting a variety of coregulators to gene regulatory regions (6, 16, 18, 19).
The murine Ikaros (Ikzf1) is composed of seven exons, and several Ik isoforms are generated through alternative splicing. All isoforms share a C-terminal domain containing two zinc fingers involved in Ik homodimerization as well as heterodimerization with various proteins (20–24). However, shorter isoforms differ in the number of N-terminal zinc fingers, which are critical for Ik binding to DNA (25). Ik isoforms with less than two N-terminal zinc fingers either do not bind DNA or do so with low affinity (25, 26), and they exert a dominant negative activity due to their capacity to sequester functional Ik1 or Ik-interacting partners (24, 25). Mice homozygous for the Ik null mutation (Iknull) lack B cells, NK cells, and their earliest described progenitors, show severe defects in T cell differentiation (27), undergo augmented T cell receptor-mediated proliferative responses, and succumb to leukemia and lymphoma with 100% penetrance (28, 29). Nonetheless, Ik function is not limited to lymphocyte commitment, survival, and homeostasis. Ik also provides the appropriate transcription program in early hematopoietic progenitors (30), influences erythroid progenitor cell homeostasis (27), erythroid cell differentiation (31), and dendritic (32) as well as neutrophil (33) cell formation. Ik can bind DNA in a sequence-specific manner either to heterochromatic foci or to proximal and distal gene regulatory regions (20, 34–38). Accordingly, Ik can control chromatin organization (20, 21, 39, 40), transcription initiation (20, 21, 36, 41), and elongation (21).
We previously reported that Ik and GATA1 control gamma globin and Gata2 gene expression during development (20, 21), as well as Notch1 and Hes1 gene expression in erythroid cells (31). In a transgenic mouse model, we showed that Ik enhances transcription initiation and elongation of gamma globin genes in yolk sac primitive erythroid cells by recruiting the cyclin-dependent kinase 9 (Cdk9) to target genes (21). Cdk9 is the catalytic subunit of the serine-threonine kinase multiprotein complex known as positive transcription elongation factor b (P-TEFb), which phosphorylates the polymerase II C-terminal domain (Pol II CTD) at Ser 2 and is presumed to be the main enzyme involved in this activity in mammalian cells (42).
Here, we demonstrate that Ik directly interacts with GATA1, GATA2, and GATA3 as well as Cdk9/P-TEFb through specific protein domains. We establish that in addition to GATA1, the other hematopoietic GATA family members support Ik in regulating the transcription of lineage-specific genes in hematopoietic cells. Altogether, current results reveal that the Ik-GATA protein interaction is a recurrent mechanism of gene expression control in hematopoietic cells and that Ik-dependent transcriptional activation relies on the ability of Ik to interact and recruit Cdk9/P-TEFb to gene promoters for efficient transcription elongation. The latter is further supported by the observation that a dominant-negative isoform of Ik and a novel Ik isoform lacking exon 6 are unable to interact with Cdk9 in vivo.
MATERIALS AND METHODS
Antibodies and primer sets.
GATA1 (N-6), GATA2 (CG2-96), GATA3 (HG3-31), Ik (H-100), HA (Y-11; F7), Cdk9 (C-20), and Mi2 (H-242) antibodies and isotype-matched immunoglobulins were purchased from Santa Cruz Biotechnology and used for Western blotting and immunoprecipitation. Anti-Flag–agarose beads (Sigma) were used for immunoprecipitation. Secondary antibodies used for immunofluorescence studies were purchased from Jackson ImmunoResearch. Primer sequences are listed in Tables 1 and 2.
Table 1.
Oligonucleotides used for qRT-PCR analysis
| Specificity (murine cDNA) | Symbol | Directiona | Sequence (5′–3′) |
|---|---|---|---|
| Actin | Act | F | ATCGTGGGCCGCCCTAGGCACCA |
| R | TCCATGTCGTCCCAGTTGGTAACAA | ||
| Erythropoietin receptor | EpoR | F | TACCTCCCACTCCACCTCAC |
| R | GCTCTGAGTCTGGGACAAGG | ||
| Transferrin receptor 2 | TrR2 | F | CTGCCGCTAGACTTCGGCCG |
| R | CGCCGCACGGATGTAGTCCC | ||
| Granzyme B | GzmB | F | ACAACACTCTTGACGCTGGG |
| R | GATGATCTCCCCTGCCTTTG | ||
| Glycophorin A | GypA | F | GCCGAATGACAAAGAAAAGTTCA |
| R | TCAATAGAACTCAAAGGCACACTGT |
F, forward; R, reverse.
Table 2.
Oligonucleotides used for ChIP analysis
| Specificity (murine genomic DNA) | Symbol | Directiona | Sequence (5′–3′) |
|---|---|---|---|
| Erythropoietin receptor gene promoter | EpoR | F | GTCTCACCAGCCCTGATTGT |
| R | GCAGGAGGACCAGGAGTCTA | ||
| Tamm-Horsfall protein gene promoter | Thp | F | GGTGGATGGTGTGGTCACAAC |
| R | GGTCTTGACACACCAGCTTT | ||
| Pancreatic alpha-amylase promoter | Amy | F | TCAGTTGTAATTCTCCTTGTAGGG |
| R | CCTCCCATCTGAAGTATGTGGGTC | ||
| Transferrin receptor 2 | TrfR2 | F | GCGGTCTAGCATGAACCAGGG |
| R | ACCTAGGCCGCTCATTCCCCAT | ||
| Granzyme B | GzmB | F | AGACCACAAGGGCATAGGGT |
| R | GTCATGCTTGGTCCTGGTACA | ||
| Glycophorin A | GypA | F | AGCCAATGTGTGTCAACGGA |
| R | GCAGCTGTGCTCTCGGTCAT |
F, forward; R, reverse.
In vivo protein interaction study.
Protein coimmunoprecipitation (co-IP) assays were done essentially as previously reported (20, 21), using lysis buffer containing 1 mM dithiothreitol (DTT) and 2 mM β-mercaptoethanol. When indicated, 50 μg/ml of ethidium bromide, 1 μg/ml of DNase I, or 1 μg/ml of RNase I was added to the lysis buffer during protein extraction, antibody incubation, and co-IP washes.
Immunofluorescence studies.
Immunofluorescence (IF) studies were performed as described by Bottardi et al. (21).
In vitro transcription and translation.
In vitro transcription was performed with templates obtained by PCR using T3 RNA polymerase, and in vitro translation was carried out with nuclease-treated rabbit reticulocyte lysates (Promega) with l-[35S]methionine (MP Biomedicals) as detailed by the supplier. Phosphorylation mutants of GATA1 were obtained by site-directed mutagenesis using appropriate primers for the introduction of Ser310Glu, Ser310Asp, and Ser310Ala mutations.
Expression and purification of recombinant proteins (GST fusion proteins).
The entire open reading frame of GATA1, PU.1, as well as Ik1 or the open reading frames corresponding to Ik isoforms 2, 4, and 6 were independently cloned into the pGEX-4T glutathione S-transferase (GST) fusion vector. GATA1-GST was purified from bacteria as described by Doubeikovskaia et al. (43). All the other GST fusion proteins were prepared from Escherichia coli BL21-CodonPlus(DE3) RIL bacteria, which were induced with 400 μM isopropyl-β-d-thiogalactopyranoside. Protein extracts were prepared by lysis of bacteria in ice-cold lysis buffer (50 mM Tris-HCl [pH 7.5], 100 mM NaCl, 1 mM EDTA, 0.5% NP-40, 0.5 mM DTT, 1 mM phenylmethylsulfonyl fluoride [PMSF]) containing proteases inhibitors (Sigma). Lysates were sonicated, centrifuged at 15,000 × g for 15 min at 4°C, and incubated with glutathione (GSH)-Sepharose resin. Proteins were eluted off the resin with GSH, dialyzed into storage buffer (50 mM Tris-HCl [pH 8.0], 100 mM NaCl, and 20% glycerol), and rebound to fresh GST-Sepharose for binding studies. Levels of the various GST fusion proteins were confirmed by Coomassie blue staining. The concentration of each purified protein was estimated by comparison to migration of bovine serum albumin (BSA) on the same gel. Approximately 1 μg of GST-protein or GST alone was immobilized on GST beads and incubated with equal amounts of in vitro-translated 35S-labeled proteins for 6 h at 4°C in binding buffer (50 mM Tris-HCl [pH 7.5], 50 mM NaCl, 0.1% Tween 20, 0.2 mM DTT, 1 mM PMSF). The total amount of GST used per binding reaction mixture was kept constant by adding the appropriate amount of beads without bound proteins. Protein complexes were washed five times in 500 μl of washing buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1 mM EDTA, 0.1% Tween 20, 0.2 mM DTT, 1 mM PMSF). Bound proteins were recovered by boiling for 5 min the GST beads in 25 μl of sample buffer. Proteins were finally resolved on SDS-PAGE and visualized by autoradiography using a Storm 840 PhosphorImager system (GE Healthcare). GST removal from GST-fused Ik1 and GATA1 proteins immobilized on beads was achieved by treatment with thrombin (Amersham). Specifically, 0.6 U of thrombin in phosphate-buffered saline (PBS) was added to approximately 10 μg of purified GST fusion proteins for 4 or 16 h at room temperature.
Cell culture and transient and stable cell transfections.
The GIE2 cell line (G1E) is a GATA1 null proliferating committed erythroid progenitor cell line. The derived G1E-ER4 cells express a Gata1-estrogen receptor (ER) transgene and upon estradiol or tamoxifen treatment undergo synchronous differentiation recapitulating early stages of erythroid differentiation (44, 45). Cells were maintained in culture as previously described (46). MEL and COS-7 cells were cultured in Dulbecco's modified Eagle's medium, and Jurkat cells were maintained in RPMI containing 1% penicillin-streptomycin and 10% fetal bovine serum and incubated at 37°C and 5% CO2. Expression plasmids carrying genes for murine Ik1, Ik1Δex6, Ik4, Ik6, GATA1, GATA2, GATA3, Cdk9, and Cdk9dn (D167N) were constructed by subcloning the relevant cDNA sequences (with or without Flag-hemagglutinin [HA] tags [FH] at their N-terminal regions) in cytomegalovirus- or simian virus 40-based vectors. COS-7 cells were transiently transfected with FuGene HD reagent (Promega) according to the manufacturer's instructions and harvested after 48 h. Cdk9dn and Ik1Δex6 mutants were obtained by site-directed mutagenesis. To generate G1E cells stably expressing Ik6-FH proteins, 3 million cells were resuspended in 100 μl of Nucleofector solution R and transfected with 10 μg of linearized and endotoxin-free Maxi-prep DNA (Qiagen) by using the G-016 program with the Nucleofector II system (Amaxa Biosystems). Electroporation was carried out with linearized empty pOZN-FH-N (mock) or pOZN-FH-N-Ik6 (pOZN-Ik6) vector (20, 47) containing Ik6 fused to Flag and hemagglutinin tags. Selection of transfected cells was carried out by magnetic affinity sorting with antibody against the interleukin-2 receptor, the surface selection marker, as reported by Nakatani et al. (47), and screened by Western blotting with HA antibody.
Mouse transgenic lines and primary cell isolation.
Homozygous Ikaros null (Iknull) mice (27) and embryonic day 16.5 (e16.5) fetal livers were obtained by crossing heterozygous Iknull mice together. Embryos were genotyped by PCR as previously described (27). Animals were sacrificed by cervical dislocation. Embryos were isolated, and fetal livers were dissected and homogenized in PBS Ca/Mg-free buffer by vigorous pipetting. Bone marrow and spleen samples were isolated from adult Ik wild-type (IkWT) or Iknull mice. To isolate bone marrow cells, femurs and humeri were flushed in PBS Ca/Mg-free buffer with a 30-gauge needle. Cell clumps were mechanically dissociated by passing cells through a 22-gauge needle. Spleens were dissociated in PBS Ca/Mg-free buffer, and the cell suspension was passed through the top of a test tube with a cell strainer cap (Falcon). Animal experiments were conducted in accordance with the Canadian Council on Animal Care (CCAC) guidelines and approved by the Maisonneuve-Rosemont Hospital animal care committee.
Bone marrow lineage negative cell selection and spleen CD4+/CD8+ cell sorting.
Bone marrow-derived lineage negative (lin−) cells were purified using the easySep mouse hematopoietic progenitor enrichment kit (StemCell Technology) according to the manufacturer's instructions. Spleen-derived CD4+/CD8+ lymphoid cells were fractionated by fluorescence-activated cell sorting (FACS). Antibody incubations were carried out on ice for 30 min in PBS supplemented with 5% heat-inactivated FBS. Cells were first incubated with rat anti-CD4 and CD8 antibodies, followed by anti-rat fluorescein isothiocyanate (FITC)-conjugated antibody (BD Biosciences).The cell suspension was passed through a 40-μm nylon mesh filter, and CD4+/CD8+ cells were isolated by FACS on a FACSAria III cell sorter (BD Biosciences). Activation of CD4+/CD8+ cells was obtained by stimulation with 50 ng/ml phorbol myristate acetate and 500 ng/ml ionomycin for 24 h.
ChIP, qPCR, and qRT-PCR assays.
Chromatin immunoprecipitation (ChIP), quantitative real-time PCR (qPCR), and quantitative reverse transcription-PCR (qRT-PCR) procedures were performed as previously described (20, 21).
RESULTS
Ikaros-GATA protein interactions in vivo.
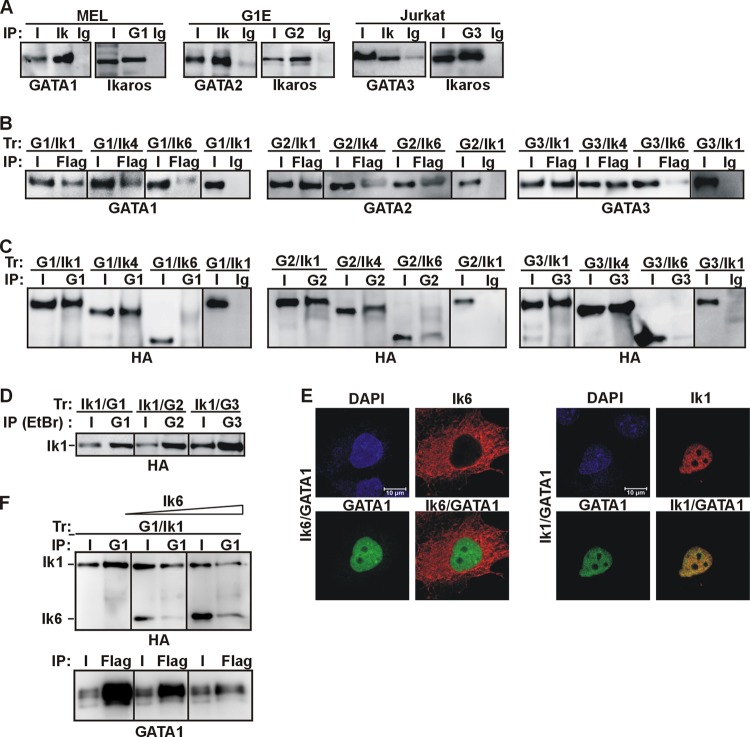
We had previously shown that Ik coimmunoprecipitates with GATA1 in different cellular models (20, 21). Since the zinc finger and protein-protein interaction domains of GATA1, -2, and -3 are highly conserved (8, 18), we assessed whether Ik also interacts with GATA2 and GATA3. Protein co-IP was carried out in GATA1-expressing a mouse erythroleukemia MEL cell line (control), GATA2-expressing and proerythroblast-like G1E cells (46), and the GATA3-expressing and T lymphocyte-like Jurkat cells. Total cell extracts were used for co-IP with Ik, GATA1, GATA2, and GATA3 antibodies or isotype-matched Ig (control Ig). Ik antibody, but not control Ig, immunoprecipitated GATA proteins (Fig. 1A). Likewise, GATA antibodies immunoprecipitated Ik proteins, indicating that, like GATA1, GATA2 and GATA3 are associated with Ik in vivo, in hematopoietic cells.
Fig 1.
Ikaros-GATA protein interactions in vivo. (A) The Ik-GATA interaction in hematopoietic cell lines. Total cell lysates of MEL, G1E, or Jurkat cells were used for protein co-IP with Ik, GATA1 (G1), GATA2 (G2), or GATA3 (G3) antibodies or control Ig (Ig). Input samples (I) represented 2% of total lysates. The co-IP samples were analyzed by Western blotting with GATA or Ik antibodies. (B and C) Total cell lysates of COS-7 cells transfected with expression vectors encoding various combinations of GATA1 (G1), GATA2 (G2), GATA3 (G3), Ik1-FH (Ik1), Ik4-FH (Ik4), or Ik6-FH (Ik6) were immunoprecipitated with Flag or GATA antibodies or control Ig (mouse Ig for Flag co-IP; rat Ig for GATA1 co-IP; mouse Ig for GATA2 and GATA3 co-IP). The co-IP samples were analyzed by Western blotting with GATA or HA antibodies. (D) Total cell lysates of COS-7 cells transfected with expression vectors encoding Ik1-FH and GATA1 (Ik1/G1), Ik1-FH and GATA2 (Ik1/G2), or Ik1-FH and GATA3 (Ik1/G3) were immunoprecipitated with GATA1, GATA2, or GATA3 antibodies in lysis buffers containing 1 mM DTT, 2 mM β-mercaptoethanol, and 50 μg/ml ethidium bromide. Co-IP samples were analyzed by Western blotting with HA antibody. (E) Confocal IF of COS-7 cells expressing Ik1-FH or Ik6-FH together with GATA1. GATA1 (green signal) was detected with rat anti-GATA1 and FITC-conjugated anti-rat antibodies; Ik (red signal) was detected with mouse anti-HA and Texas Red (TR)-conjugated anti-mouse antibodies. Representative COS-7 cells are shown where Ik1-GATA1 colocalized, as yellow signals in merged images. (F) In vivo squelching assay. Total cell lysates of COS-7 cells transfected with Ik1-FH and GATA1 and an increasing concentration of Ik6-FH were immunoprecipitated either with GATA1 or Flag antibodies. Co-IP samples were analyzed by Western blotting with HA or GATA1 antibodies.
To further define the interactions between Ik isoforms and GATA family members, Flag- and hemagglutinin-tagged Ik1, Ik4, or Ik6 (Ik1-FH, Ik4-FH, or Ik6-FH) was independently expressed in combination with untagged GATA proteins in COS-7 cells, and total cell lysates were used in a protein co-IP assay. Ik1 was chosen because it is the most abundantly expressed Ik isoform in normal hematopoietic cells, whereas Ik4 and Ik6 are frequently overexpressed in oncogenic cells (25, 48–52). GATA1, GATA2, and GATA3 were immunoprecipitated with Flag antibody in cells cotransfected with Ik1-FH or Ik4-FH (Fig. 1B). Except for GATA2, which similarly interacted with Ik4-FH and Ik6-FH, GATA protein detection was significantly reduced in cells cotransfected with Ik6-FH, and no interaction could be detected in mock-transfected cells, in cells expressing uniquely Ik or GATA proteins, or in control Ig-immunoprecipitated samples (Fig. 1B and data not shown). Reciprocal co-IP experiments confirmed these results (Fig. 1C). The presence of 50 μg/ml ethidium bromide, 1 mM DTT, and 2 mM β-mercaptoethanol did not significantly change the above-mentioned interactions (Fig. 1D and data not shown), which ruled out possible DNA-mediated interactions as well as disulfide bridge formation or protein aggregates. Altogether, the results obtained with COS-7 cells demonstrate that the interaction between GATA1, GATA2, or GATA3 and Ik1 or Ik4 can occur in nonhematopoietic cells, i.e., without additional hematopoiesis-specific factors.
A weaker GATA-Ik6 interaction could be a consequence of the predominant cytoplasmic distribution of Ik6 (24). Accordingly, although Ik1 and GATA1 colocalized in the nucleus, Ik6 and GATA1 showed divergent localization (Fig. 1E) (21). Thus, GATA1 is not retained in the cytoplasm by Ik6 and does not facilitate Ik6 translocation to the nucleus. However, Ik6 can enter the nucleus when included in protein complexes containing Ik1 or Ik2 (24, 25). Thus, we investigated by in vivo squelching experiments whether Ik6 could interfere with Ik1 for binding to GATA1. In COS-7 cells that expressed Ik1-FH and GATA1, GATA1 antibody immunoprecipitated Ik1 (Fig. 1F). However, GATA1 antibody immunoprecipitated a negligible amount of either Ik1 or Ik6 when Ik1-FH, GATA1, and Ik6-FH was simultaneously expressed. Reciprocal co-IP confirmed these results (Fig. 1F), thus suggesting that Ik6 interferes with Ik1 for interaction with GATA1.
Ikaros-GATA protein interactions in vitro.
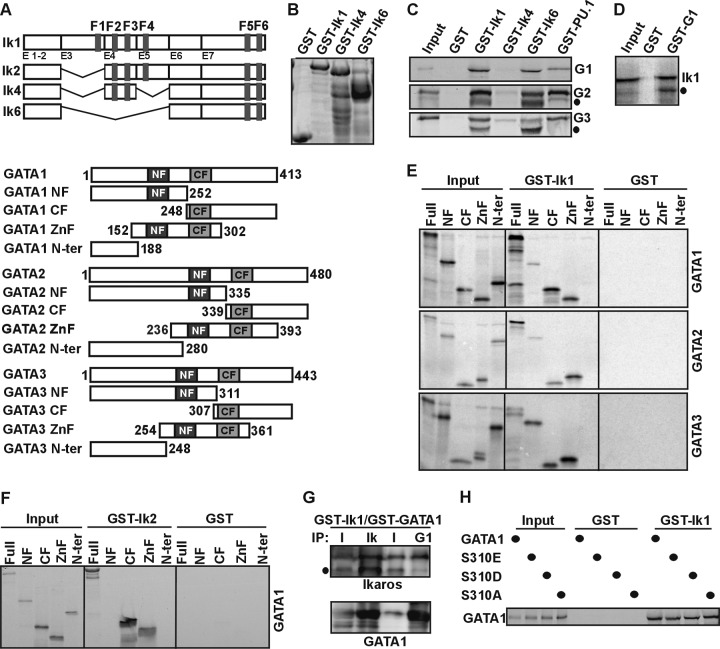
The potential direct interaction between Ik and GATA proteins was assessed by in vitro pulldown experiments using bacterially synthesized Ik isoforms immobilized on glutathione-Sepharose beads (GST-Ik1, GST-Ik4, or GST-Ik6) and in vitro-synthesized 35S-labeled GATA1, GATA2, or GATA3 (Fig. 2A and B). Full-length GATA proteins were retained by GST-Ik1, GST-Ik6, and GST-PU.1 but not by GST-Ik4 or GST alone, indicating that Ik1 and Ik6 directly interact with GATA proteins (Fig. 2C). An Ik1-GATA1 direct interaction was confirmed by GST pulldown with bacterially synthesized GATA1 and in vitro-synthesized 35S-labeled Ik1 proteins (Fig. 2D). Ik C-terminal zinc fingers mediate self-interaction as well as interactions with other proteins (20–24). The fact that GST-Ik6 contains only the C-terminal zinc fingers and yet interacts with GATA1, GATA2, and GATA3 suggests that this domain is required for the interaction with GATA proteins.
Fig 2.
Ikaros-GATA protein interactions in vitro. (A) Schematic view of Ik1, Ik2, Ik4, Ik6, GATA1, GATA2, and GATA3 protein structures and deletion mutants. (B) Coomassie staining of bacterially purified Ik1-, Ik4-, and Ik6-GST proteins. (C) GST pulldown assay results: [35S]methionine-labeled GATA1 (G1), GATA2 (G2), and GATA3 (G3) proteins were tested for interactions with GST, GST-Ik1, GST-Ik4, GST-Ik6, and GST-PU.1 proteins. Filled circles, nonspecific bands. (D) GST pulldown assay results: [35S]methionine-labeled Ik1 proteins were tested for interactions with GST or GST-GATA1 (GST-G1) proteins. Filled circle, nonspecific band. (E) GST pulldown assay results: [35S]methionine-labeled truncated GATA1, GATA2, and GATA3 proteins, as indicated above each panel, were tested for interactions with GST or GST-Ik1 proteins. Full, full length; NF, N-terminal zinc finger; CF, C-terminal zinc finger; ZnF, C- and N-terminal zinc fingers; N-ter, constructs lacking both zinc fingers. (F) GST pulldown assay results: [35S]methionine-labeled truncated GATA1 fragments were tested for interactions with GST or GST-Ik2 proteins. Full, full length; NF, N-terminal zinc finger; CF, C-terminal zinc finger; ZnF, C- and N-terminal zinc fingers; N-ter, constructs lacking both zinc fingers. (G) In vitro immunoprecipitation assay results: GST-Ik1 and GST-GATA1 proteins were purified from bacteria, and the GST moiety was eliminated by thrombin digestion. Co-IP experiments were carried out with Ikaros (Ik) or GATA1 (G1) antibodies, and co-IP samples were analyzed by Western blotting with Ik or GATA1 antibodies. Filled circle, nonspecific band. (H) GST pulldown assay results: [35S]methionine-labeled wild type or phosphomimetic mutants of GATA1 protein were tested for interaction with GST or GST-Ik1 proteins.
GATA1, GATA2, and GATA3 are modular proteins. To identify which region is necessary for interaction with Ik, several GATA deletions were tested for in vitro interactions with GST-Ik1. Full-length GATA, the GATA CF, and GATA C- and N-terminal zinc finger (ZnF) fragments were similarly retained by GST-Ik1 (Fig. 2E). GATA3 NF also retained a significant amount of GST-Ik1, whereas the GATA1 NF or GATA2 NF association with GST-Ik1 was weaker (Fig. 2E), and no association could be detected between GATA1 NF and GST-Ik2 (Fig. 2F). As expected, GATA constructs lacking both zinc fingers (GATA N-ter) or GST alone did not interact with either GST-Ik1 or GST-Ik2 (Fig. 2E and F). Thus, GATA CF is sufficient for in vitro interaction with Ik1 and Ik2. The fact that Ik1-GATA1 complexes could be formed when we used both proteins purified from bacteria (Fig. 2G) further indicates that they can interact directly in vitro.
GATA1 can be phosphorylated at Ser310 upon erythropoietin receptor activation (53). Since Ser310 is retained in the GATA1 CF construct, which efficiently interacts with GST-Ik1, we investigated whether GATAl Ser310 phosphorylation could modify the strength of interaction with Ik. Two phosphomimetic (GATA1S310E and GATA1S310D) and a phosphorylation-deficient (GATA1S310A) GATA1 protein were labeled with [35S]methionine and tested for in vitro interactions with GST-Ik1. The single mutation did not significantly affect the interaction with Ik (Fig. 2H), indicating that Ser310 phosphorylation levels do not influence the Ik-GATA1 interaction in vitro.
Collectively, these results indicate that the direct interaction with Ik is a general feature of GATA transcription factors and occurs between the C-terminal zinc finger domain of Ik and the two zinc fingers of GATA proteins, in particular, the CF that is more efficiently retained by GST-Ik.
Ikaros-Cdk9 protein interaction in vivo.
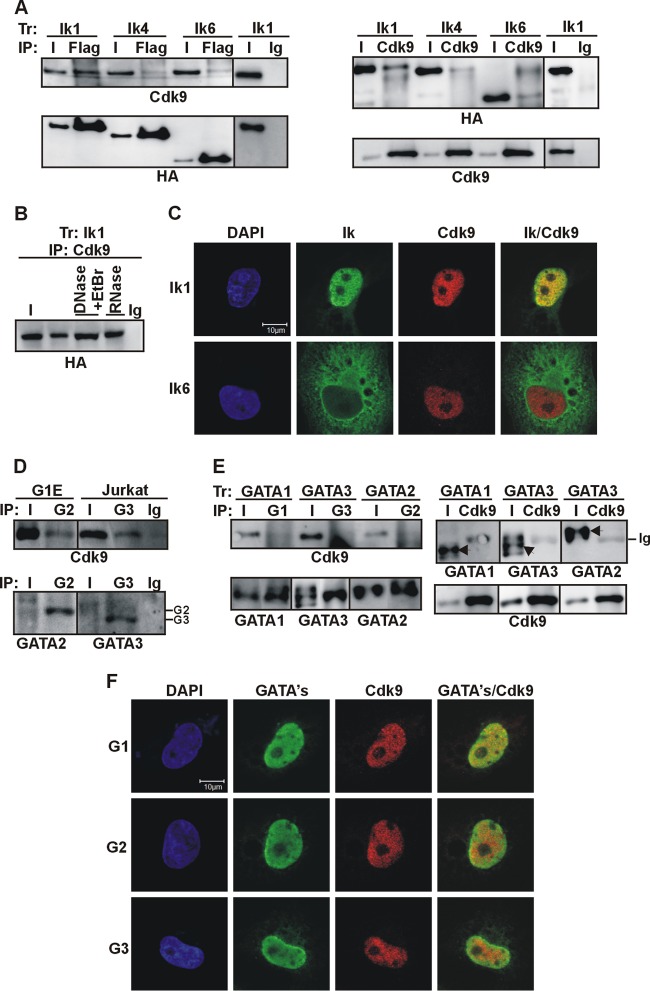
We recently showed that Ik1 can interact with Cdk9, the catalytic subunit of the active P-TEFb complex in Jurkat and mouse yolk sac erythroid cells (21). Here, we questioned whether Cdk9 equally interacts with different Ik isoforms. Total cell lysates of COS-7 cells transfected with Ik1-FH, Ik4-FH, or Ik6-FH were immunoprecipitated with Flag or Cdk9 antibodies. Cdk9 was detected upon Flag co-IP in lysates of Ik1-FH-transfected cells, and in a reciprocal co-IP, Ik1 was immunoprecipitated by Cdk9 antibody (Fig. 3A). The interaction between Cdk9 and Ik4-FH or Ik6-FH, though detectable, was weaker, and no specific bands could be detected in mock-transfected cells or in co-IPs performed with control Ig (Fig. 3A and data not shown). The weaker interaction between Cdk9 and Ik4 or Ik6 was not significantly modified by the addition of GATA1 (data not shown). DNase I and ethidium bromide or RNase did not affect the Ik1-Cdk9 protein interaction (Fig. 3B). Thus, Cdk9 interacts in vivo with Ik1, whereas it hardly interacts with the shorter dominant-negative isoforms, Ik4 and Ik6.
Fig 3.
Ikaros-Cdk9 protein interactions in vivo. (A) Total cell lysates of COS-7 cells, transfected (Tr) with vectors expressing Ik1-FH (Ik1), Ik4-FH (Ik4), or Ik6-FH (Ik6) were immunoprecipitated with Flag or Cdk9 antibodies or control Ig. Co-IP samples were analyzed by Western blotting with the indicated antibodies. (B) Total cell lysates of COS-7 cells transfected with Ik1-FH were immunoprecipitated with Cdk9 antibody in co-IP buffers containing DNase and ethidium bromide or RNase, as detailed in Materials and Methods. Co-IP samples were analyzed by Western blotting with HA antibody. (C) Confocal IF of COS-7 cells expressing Ik1-FH or Ik6-FH. Ik (green signals) was detected with mouse anti-HA and Alexa Fluor 488-conjugated anti-mouse antibodies; Cdk9 (red signals) was detected with rabbit anti-Cdk9 and Cy3-conjugated anti-rabbit antibodies. Representative COS-7 cells are shown where Ik1-Cdk9 colocalization was identified as yellow signals in the merged image. (D) Total cell lysates of G1E or Jurkat cells were immunoprecipitated with GATA2 (G2) or GATA3 (G3) antibodies or control Ig. Input samples (I) represented 2% of total lysates. Co-IP samples were analyzed by Western blotting; GATA2- or GATA3-specific bands are indicated on the right side of the panel. (E) Total cell lysates of COS-7 cells transfected with GATA1-, GATA2-, or GATA3-expressing vectors were immunoprecipitated with GATA1 (G1), GATA2 (G2), GATA3 (G3), or Cdk9 antibodies. Samples were analyzed by Western blotting with the indicated antibodies. Arrows indicate specific bands. (F) Confocal IF of COS-7 transfected cells. GATA1 (green signals) was detected with rat anti-GATA1 and FITC-conjugated anti-rat antibodies; GATA2 and GATA3 (green signals) were detected with mouse anti-GATA and Alexa Fluor 488-conjugated anti-mouse antibodies; Cdk9 (red signals) was detected with rabbit Cdk9 and Cy3-conjugated anti-rabbit antibodies. Representative COS-7 cells are shown; GATA-Cdk9 colocalization was identified as yellow signals in merged images.
Cdk9 (54) and Ik6 (24) can localize to the cytoplasm where, in principle, they could interact together. To elucidate this issue, Cdk9 and Ik localization was investigated by IF staining of COS-7 cells transfected with Ik isoforms. Merged images of Ik1-FH and Cdk9 produced yellow nuclear signals per focal plane, revealing that these proteins colocalized in the nucleus; however, Ik6-FH did not colocalize with Cdk9 either in the nucleus or in the cytoplasm (Fig. 3C) (21). In conclusion, only Ik1 can efficiently interact with Cdk9 both in vitro and in vivo.
The interplay between certain GATA family members and the P-TEFb components Cdk9 and/or CycT1 has been reported (21, 55–58). We previously showed that (i) GATA1 and Cdk9 coimmunoprecipitate in MEL but not in COS-7 cells, (ii) GATA1 can enhance the strength of interaction between Ik and Cdk9, and (iii) Ik is essential for the GATA1-Cdk9 interaction (21). Thus, we asked whether GATA2 and GATA3 could interact with Cdk9 in G1E, Jurkat cells, or in COS-7 cells where Ik is not expressed. Figure 3D shows that GATA2 interacted with Cdk9 in G1E cells and GATA3 interacted with Cdk9 in Jurkat cells. However, in COS-7 cells transfected with GATA1 (as a control), GATA2, or GATA3, co-IP experiments could not demonstrate any significant interaction between Ik and GATA proteins (Fig. 3E). This issue was further supported by IF analysis of COS-7 cells expressing GATA1, GATA2, or GATA3 (Fig. 3F). GATA proteins diffusely occupied the nucleus, whereas Cdk9 exhibited a more punctuate nuclear distribution. Merged images of Cdk9 and GATA staining scarcely produced yellow signals per focal plane. Thus, in Ik-deficient nonhematopoietic cells, Cdk9 does not efficiently interact with GATA1, GATA2, or GATA3.
Ikaros-Cdk9 protein interaction in vitro.
The direct protein interaction between Ik and Cdk9 was assessed by in vitro GST pulldown with GST-Ik1, GST-Ik4, or GST-Ik6 and 35S-labeled full-length Cdk9 or Cdk9 deletion fragments (Fig. 4A and B). Cdk9 was efficiently retained by GST-Ik1 and GST-Ik4. However, deletion of the central part of the protein in the oncogenic Ik6 isoform severely perturbed the interaction with Cdk9 (Fig. 4C). In a parallel set of experiments, GST-Ik1 was incubated with 35S-labeled Cdk9 deletion fragments. These deletions eliminated Cdk9 protein domains required for interaction with other Cdk9-interacting partners (59). Cdk9Δ82N, Cdk9Δ129N, Cdk9Δ289C, and Cdk9Δ82N289C all interacted to a similar degree with GST-Ik1, whereas Cdk9Δ83C could not pull down GST-Ik1 (Fig. 4D). These results suggest that the second and third zinc fingers of Ik (included in Ik1 and Ik4 but absent in Ik6) are required for Ik interaction with the kinase and T-loop domains of Cdk9 in vitro.
Fig 4.
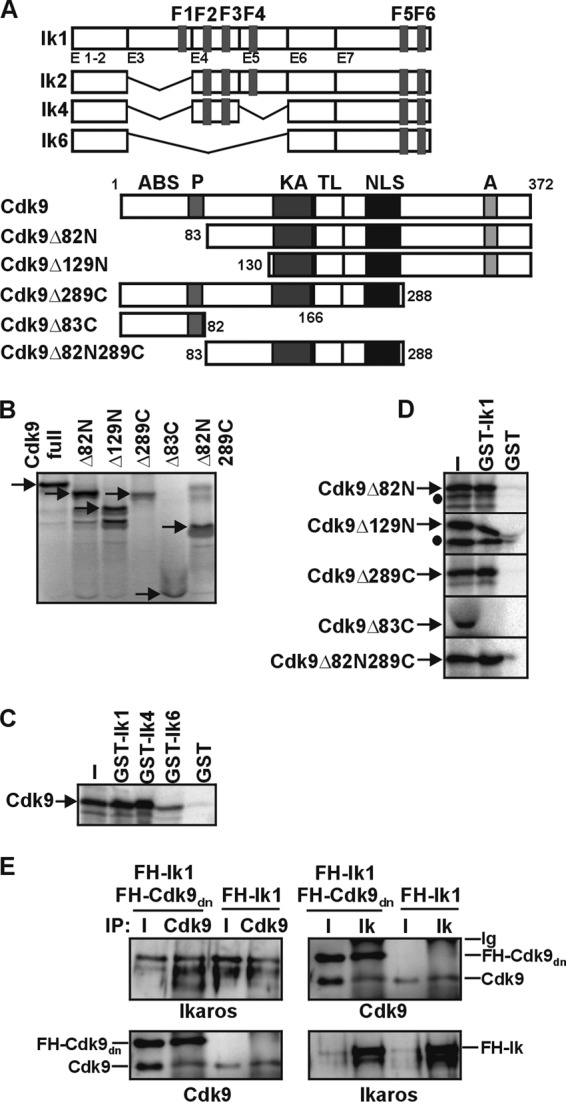
Ikaros-Cdk9 protein interaction in vitro. (A) Schematic of Ik and Cdk9 protein structures and deletion mutants. Functional domains are designated according to Cdk9 amino acid residues. (B) [35S]methionine-labeled full-length and truncated Cdk9 proteins were resolved on SDS-PAGE gels; arrows indicate predicted molecular weights. (C and D) GST pulldown assay result: [35S]methionine-labeled full-length Cdk9 (C) or truncated Cdk9 (D) proteins were prepared by coupled transcription-translation reactions and tested for interactions with equal amounts of bacterial GST, GST-Ik1, GST-Ik4, or GST-Ik6 (C) or with GST or GST-Ik1 (D) by GST pulldown assay. Bound proteins were analyzed by SDS-PAGE and autoradiography. Input samples represent 20% of binding reaction mixtures. Filled circles, nonspecific bands. (E) Total cell lysates of COS-7 cells transfected with vectors expressing the FH-tagged kinase-dead Cdk9 mutant (FH-Cdk9dn) together with Ik1-FH (Ik1) or Ik1-FH alone were immunoprecipitated with Cdk9 or Ikaros (Ik) antibodies. Co-IP samples were analyzed by Western blotting with Ik or Cdk9 antibodies.
The dominant-negative form of Cdk9 (Cdk9dn) is a kinase-dead mutant that carries an Asp-to-Asn substitution at position 167 (D167N) of the T-loop (60), e.g., within the Ik interaction domain (Fig. 4D). Cdk9dn competes with endogenous Cdk9 for interaction with cyclins and forms inactive protein complexes (60). Cdk9dn was tested for its ability to interact with Ik. Co-IP experiments showed that Ik immunoprecipitated endogenous as well as FH-tagged Cdk9dn in COS-7 cells (Fig. 4E). Thus, the Cdk9 kinase activity and, more precisely, the Asp residue at position 167 are not essential for interaction with Ik.
Ikaros/Cdk9 and GATA proteins are transcriptional activators of several hematopoiesis-specific genes.
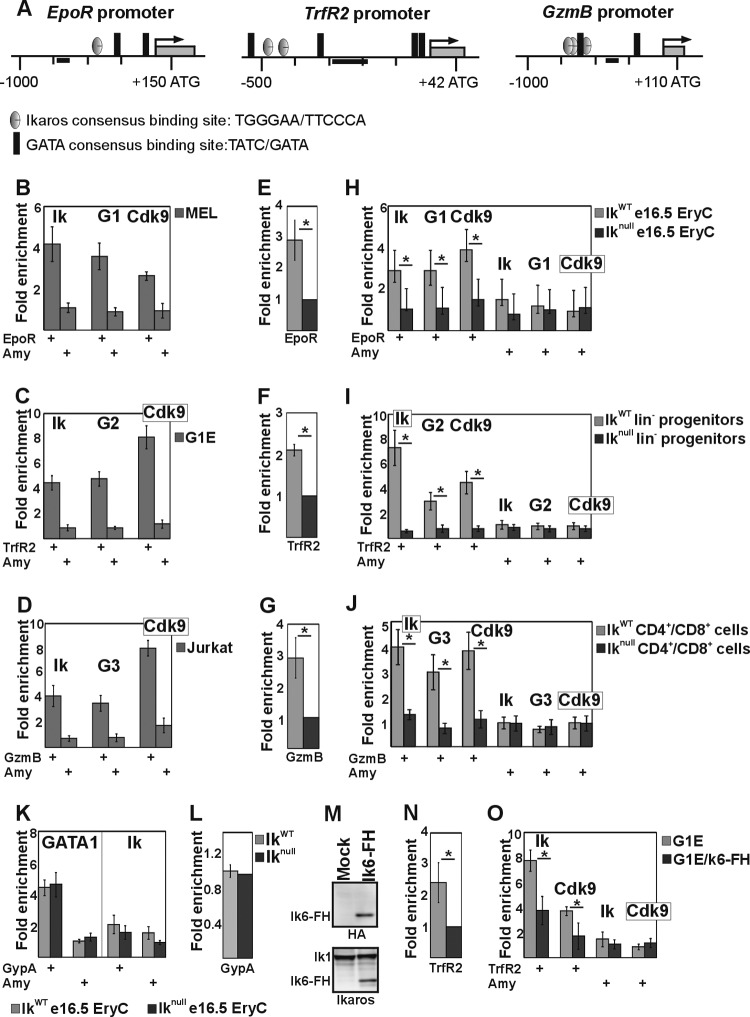
The finding that Ik interacts in vitro and in vivo with GATA and Cdk9 proteins prompted us to investigate whether Ik and/or Ik-GATA combined action is required for lineage-specific gene expression in primary cells. In erythroid cells, both Ik (61) and GATA1 (62, 63) have been linked to Transferrin (TrfR) and Erythropoietin (EpoR) receptor gene expression. Notably, TrfR2, a TrfR homolog that encodes the transferrin receptor 2 (TRFR2), is expressed, together with Ik and GATA2, in hematopoietic progenitor/stem cells (64–66) and silenced upon erythroid differentiation of G1E progenitor cells (67). In T cells, Ik (68) and probably GATA3 (69) activate the expression of the Granzyme B (GzmB) gene by binding to upstream regulator elements (70). To validate a functional role of Ik-GATA cooperativity and to gain further insight into the biological significance of the Ik-Cdk9 interaction, we studied the expression of EpoR, TrfR2, and GzmB genes in erythroid (MEL), progenitor (G1E), and T-lymphoid (Jurkat) cells, respectively. The promoters of these genes contain Ik and GATA consensus sequences (Fig. 5A). Expression of these genes was confirmed in the appropriate cell lines (data not shown). Next, Ik and GATA protein recruitment to gene regulatory regions was assessed by ChIP. Ik and GATA1 were detected at the EpoR promoter in MEL cells (Fig. 5B); Ik and GATA2 were found at the TrfR2 promoter in G1E progenitor cells (Fig. 5C); Ik and GATA3 were detected at the GzmB promoter in Jurkat cells (Fig. 5D). Interestingly, Cdk9 was also recruited to the EpoR, TrfR2, and GzmB promoter regions (Fig. 5B to D). To further define the role of Ik for GATA and Cdk9 recruitment to chromatin regulatory regions and gene transcription, qRT-PCR and ChIP experiments were performed in primary cells in the presence or absence of Ik proteins. Specifically, erythroid cells (e16.5 fetal liver erythroid cells [e16.5 EryC]), hematopoietic progenitors (bone marrow lineage-negative cells [lin−]), or activated T lymphocytes (spleen-derived CD4+ and CD8+ cells [CD4+/CD8+]) were isolated from IkWT or Iknull mice. The EpoR, TrfR2, and GzmB genes were, respectively, expressed in e16.5 EryC (Fig. 5E), lin− (Fig. 5F), and CD4+/CD8+ (Fig. 5G) IkWT cells. Their expression levels decreased in Iknull cells (Fig. 5E to G). Ik, Cdk9, and GATA proteins were efficiently recruited to gene regulatory regions in IkWT primary cells, whereas chromatin occupancy severely decreased in cells isolated from Iknull mice (Fig. 5H to J). These results suggest that Ik facilitates Cdk9 as well as GATA1, GATA2, and GATA3 protein recruitment to hematopoiesis-specific genes, thus providing appropriate gene expression.
Fig 5.
Ikaros/Cdk9 and GATA proteins are transcriptional activators of several hematopoiesis-specific genes. (A) Schematic representation of the Erythropoietin receptor (EpoR), Transferrin receptor 2 (TrfR2), and Granzyme B (GzmB) murine promoters. Black bars, amplicons. (B to D, H to J, K, and O) ChIP assay results with MEL (B), G1E (C), Jurkat (D), and G1E cell lines carrying Flag- and HA-tagged Ik6 (O) or e16.5 fetal liver erythroid cells (e16.5 EryC [H and K]), bone marrow-derived lineage-negative progenitor cells (lin− [I]), or spleen-derived CD4+/CD8+ cells (J) purified from IkWT or Iknull mice. ChIP experiments were carried out with the antibodies indicated on the top of each panel. Ik, Ikaros; G1, G2, or G3, GATA1, -2, or -3. Immunoprecipitated and input chromatin samples were used as the templates for qPCR; quantification was carried out according to the 2−ΔΔCT method, using the mouse kidney-specific Tamm-Horsfall protein (Thp) promoter as an internal control. The fold enrichment values (y axis) for EpoR, TrfR2, GzmB, Glycophorin A (GypA), and Amylase (Amy) promoters relative to the control and the input samples are plotted as means and standard deviations. A value of 1 indicates no enrichment. *, P ≤ 0.05 by Student's t test (n ≥ 4). (E to G, L, and N) Quantitative RT-PCR results performed on equal numbers of cells. Mature transcripts were retrotranscribed with oligo(dT) primers; transcript quantification was performed according to the Pfaffl method (100) using specific qPCR primers and mouse Actin as an endogenous control. y axis, relative enrichment levels. The ratios are plotted as means ± standard deviations. *, P ≤ 0.05 by Student's t test (n ≥ 4). (M) Semiquantitative Western blot assay of total cell lysates of mock-transfected or Ik6-FH-transfected G1E cells. (Upper panel) Detection of Ik6-FH with HA antibody; (bottom panel) detection of endogenous Ik1and Ik6-FH with Ik antibody.
GATA factor occupancy was then investigated at promoters of genes regulated by GATA but not Ik proteins. In erythroid cells, Glycophorin A (GypA) gene expression is enhanced by GATA1 (71) but not by Ik (61). A DNA binding site search did not reveal any Ik consensus sequence across the GypA promoter (data not shown). As expected, ChIP showed GATA1 but not Ik occupancy at GypA promoter in IkWT e16.5 fetal liver erythroid cells (Fig. 5K). Interestingly, in Iknull cells, GATA1 occupancy at the GypA promoter did not vary (Fig. 5K), and GypA expression did not decrease (Fig. 5L). Thus, GATA proteins can act as transcriptional activators independently of Ik at genes that are not direct Ik targets.
We also attempted overexpression of the dominant-negative isoform of Ik6 in G1E cells. Flag- and HA-tagged Ik6 (Ik6-FH, detected by Western blotting with HA antibody) was expressed at lower levels than endogenous Ik (Fig. 5M) and did not induce major phenotypic abnormalities (data not shown). In G1E/Ik6 cells, the level of TrfR2 expression decreased (Fig. 5N), and Ik as well as Cdk9 were recruited less efficiently to the TrfR2 promoter (Fig. 5O). These results strongly suggest that Ik6 interferes with longer Ik isoforms for binding to Cdk9, thereby impeding Cdk9 recruitment to Ik target genes.
The Ikaros Δexon 6 isoform does not interact with Cdk9.
While cloning Ik isoforms from wild-type mouse thymus cDNA, a novel Ik isoform whereby exon 6 is missing (Ik1Δex6) (Fig. 6A) was isolated. To investigate whether this Ik mutant is capable of interacting with Cdk9, COS-7 cells were transfected with FH-tagged Ik1Δex6, and total cell lysates were immunoprecipitated with Flag or Cdk9 antibodies. Surprisingly, Cdk9 was not significantly precipitated by Flag antibody and FH-Ik1Δex6 was not precipitated by Cdk9 antibody (Fig. 6B). However, Ik1Δex6 could efficiently interact with the chromatin-remodeling factor Mi2 (Fig. 6B), an important Ik-interacting partner, and with GATA proteins (Fig. 6C). Notably, the Ik1 and Ik1Δex6 cellular distributions closely resembled each other because both proteins could be detected in the nucleoplasm of transfected COS-7 cells by IF (Fig. 6D). These results indicate that the novel Ik1Δex6 isoform selectively abrogates the Ik interaction with Cdk9/P-TEFb. Thus, among the Ik isoforms tested (Ik1, Ik4, Ik6, and Ik1Δex6), only full-length Ik1 can engage in a stable interaction with Cdk9.
Fig 6.
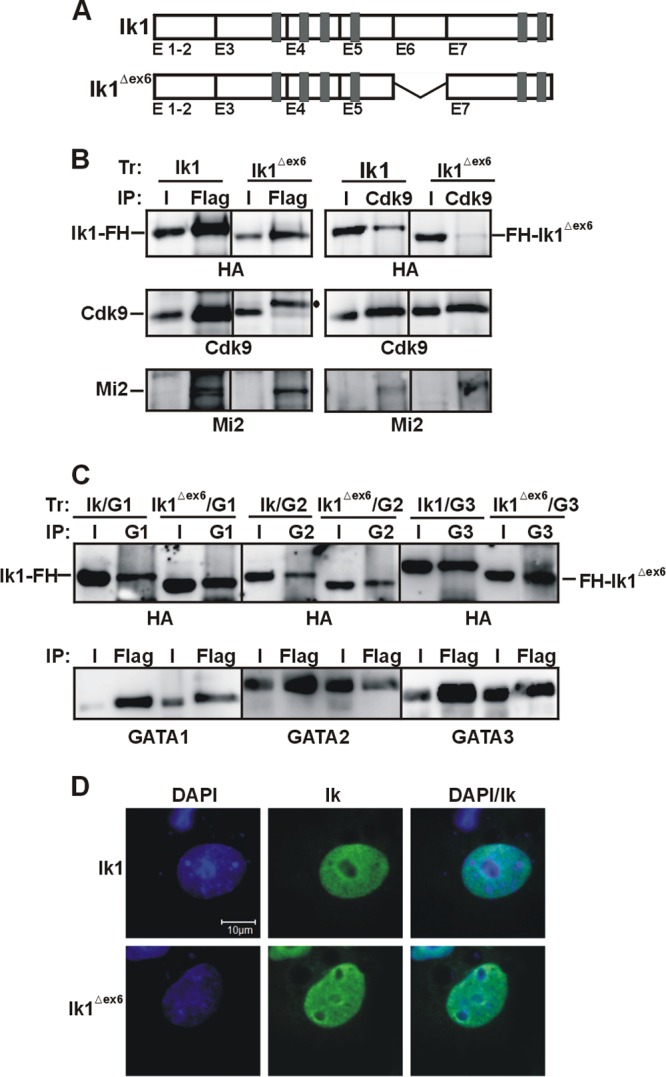
The Ikaros Δexon 6 isoform does not interact with Cdk9. (A) Schematic view of Ik1 and the Ik1Δex6 isoform. (B and C) Total cell lysates of COS-7 cells transfected (Tr) with expression vectors encoding Ik1-FH (Ik1), FH-Ik1Δex6 (Ik1Δex6), GATA1 (G1), GATA2 (G2), or GATA3 (G3) were immunoprecipitated with Flag, Cdk9, GATA1, GATA2, or GATA3 antibodies. Co-IP samples were analyzed by Western blotting with HA, Cdk9, Mi2, GATA1, GATA2, or GATA3 antibodies. •, nonspecific band. (D) Confocal IF of COS-7 cells expressing Ik1-FH (Ik1) or FH-Ik1Δex6 (Ik1Δex6); Ik was detected with mouse anti-HA and FITC-conjugated anti-mouse antibodies. Results with representative COS-7 cells are shown.
DISCUSSION
In this study, we have demonstrated that Ik directly interacts with the hematopoietic GATA family members and the P-TEFb catalytic subunit Cdk9. Our results revealed that Ik1, but not the shorter Ik isoforms, facilitates GATA1, GATA2, and GATA3 as well as Cdk9 recruitment to chromatin and is required for appropriate expression of several hematopoietic genes. Thus, the Ik-GATA-Cdk9 protein network characterized in the manuscript is likely to be required for modulating gene expression in all hematopoietic cells.
Ikaros-GATA protein interactions.
The biological function of Ik depends on the cellular context and/or developmental stage (72). Cooperative binding of transcription factors and cofactors to gene regulatory regions often requires direct interaction between transcription factors and, conversely, multiprotein complexes formed on DNA are stabilized by protein-protein interactions (73). Even though GATA proteins can act without Ik at the GypA promoter, which only contains GATA consensus sequences (Fig. 5), we and others have emphasized the combined requirement of Ik and GATA proteins for transcriptional control of multiple genes in different hematopoietic cell types (20, 21, 31, 74–78). Here, we have shown that, in addition to interact with GATA1 in erythroid cells, Ik interacts with GATA2 in hematopoietic progenitor cells and with GATA3 in lymphoid cells. Our results suggest that the CF of GATA proteins interacts with the C-terminal zinc-finger domain of Ik with no requirement for additional factors, as opposed to, for instance, GATA1 interaction with the MeCP2 complex, which requires FOG1 (17). In accordance with our results, it is known that the GATA CF is needed for self-association and for interaction with PU.1, CBP, Sp1, and KLF1 (3), and the C-terminal zinc finger domain of Ik is required for interaction with self, family members, and other transcriptional regulators (5).
By using epitope-tagged variants of Ik isoforms, we found that dominant-negative Ik isoforms (Ik4 and Ik6) interact with GATA proteins less efficiently that Ik1, although Ik6 and GATA proteins interact with Ik in vitro. The absence of detectable interaction in vivo could be explained by Ik6 and GATA cellular localization, because Ik6 is predominantly cytoplasmic, whereas GATA factors exhibit typical nuclear distribution. However, squelching experiments demonstrated that when Ik6 enters the nucleus upon heterodimerization with Ik1, it hinders the Ik1-GATA1 interaction without engaging itself in a stable interaction with GATA1. Thus, in vivo, Ik6 possesses higher binding affinity for Ik1 than for GATA partners. Since dominant-negative Ik isoforms are related to the prognosis of several solid tumors (79) and are frequently overrepresented in malignant hematopoietic cells (80, 81), disruption of the combined action of Ik and GATA factors is likely to be highly relevant for neoplastic transformation.
It is unclear how the interaction between Ik and specific GATA factors is controlled. Interference or assistance by Ik family members, protein posttranslational modifications, and fluctuations of protein levels could possibly play roles.
Ik family members, i.e., Aiolos, Helios, Eos, and Pegasus, can form homodimers or interact with one another. Like Ik, these proteins are reported to be alternatively spliced; thus, different isoforms can be produced in normal or malignant cells (5). Deregulation of Ik and Aiolos, particularly their splice variants, can result in cellular hyperproliferation and development of lymphomas and leukemia, whereas constitutive expression of mutant forms of Helios leads to aggressive T cell lymphoma (5, 81). It is not known whether any of the Ik family members interacts with Cdk9 or GATA proteins, but their binding to the wild type and Ik variants might indirectly modulate Ik-Cdk9 and Ik-GATA interactions.
Ik and GATA proteins can be acetylated, sumoylated, and phosphorylated, and these posttranslational modifications might affect the strengths of their interactions. For instance, GATA1 is phosphorylated at Ser310 upon erythropoietin receptor activation (53). However, single phosphorylation-deficient and phosphomimetic mutations do not affect the strength of interaction with Ik in vitro, thus suggesting that Ser310 phosphorylation does not influence the Ik-GATA1 interaction. Accordingly, Ser310 phosphorylation is dispensable for the function of GATA1 in hematopoiesis (82). Whether protein acetylation, sumoylation, or ubiquitination can modify the strength of Ik-GATA interactions remains unknown.
Spatiotemporal regulation of GATA proteins has been shown to be more important than their identity, and GATA2 and GATA3 can rescue a GATA1 null phenotype when expressed under the control of Gata1 regulatory sequences (83). Thus, it is possible that the ability of Ik to interact with GATA1, GATA2, or GATA3 depends on the protein concentration. During terminal differentiation of erythroid and megakaryocyte cells, GATA2 levels decline and GATA1 levels increase (84, 85). At the same time, there is an exchange of GATA factors recruited on gene regulatory regions, with consequent modifications of expression levels. The GATA switch can be reproduced in the G1E-ER4 cells upon tamoxifen treatment. In these cells, TrfR2 expression decreases and GATA1 is then recruited to the TrfR2 promoter together with Ik (data not shown). Thus, we speculate that genes regulated by Ik and GATA2 in hematopoietic progenitors will be controlled afterwards by Ik and GATA1, in erythroid and megakaryocytic cells, or Ik and GATA3 in lymphoid cells. However, the results of Malinge et al. suggested that this regulatory loop could be more complex, since during megakaryopoiesis but not erythropoiesis, GATA1/2 can directly affect Ik gene expression by binding to Ik regulatory regions, with GATA2 activating and GATA1 repressing Ik expression (76).
The Ikaros interaction with the P-TEFb complex.
In lymphoid cells, Ik is primarily involved in gene repression. Indeed, most of the Ik proteins are present in the NuRD complex, and a small amount is associated with the corepressors Sin3A, Sin3B, Sin3BSF, CtIP, and Rb (23, 86, 87). Ik is also a transcriptional activator (24, 25, 88). It can interact with cofactors promoting gene transcription, such as BRG1 (20, 89, 90), and numerous hematopoiesis-specific genes are downregulated in Iknull cells (91). Interestingly, an Ik transcriptional effect at target genes can be dynamic during development and/or cellular specification (20, 21, 37, 76). The interplay of Ik with Cdk9 and, moreover, GATA factors that are differentially expressed during hematopoietic lineage specification might account for the Ik capacity to both repress and activate target genes according to the cell type and stage of differentiation and/or development.
The current results indicate that Ik enhances transcription elongation by interacting directly with Cdk9/P-TEFb. Under normal growth conditions, more than half of Cdk9/P-TEFb located in the nucleus of HeLa cells is sequestered in a catalytically inactive complex termed the 7SK snRNP, which also contains the 7SK snRNA and the nuclear proteins LARP7, MePCE, and HEXIM1/2, which block Cdk9 kinase activity (42, 54, 92). Cdk9/P-TEFb cycles between inactive and active complexes during gene transcription (93). A variety of conditions that globally affect cell growth and differentiation can trigger deregulation of the P-TEFb transcription cycle (54). Results presented here suggest that Ik4 and/or Ik6 overexpression could perturb P-TEFb activity at Ik target genes, since these dominant-negative isoforms sequestered Ik1 and prevented its interaction with other factors (Fig. 1F). We also demonstrated that Ik1Δex6 does not interact with Cdk9 but translocates to the nucleus and retains the ability to interact with GATA and Mi2 proteins. This suggests that Ik exon 6 is specifically required for interaction with Cdk9/P-TEFb and, hence, transcription elongation. Thus, these Ik isoforms that are overexpressed in leukemic cells might counteract the transcription elongation activity of wild-type Ik proteins, thereby altering transcription of hematopoiesis-specific genes.
Ikaros and leukemia.
Ik mutations have been observed in different cases of human leukemia as well as lymphoma. Mice haploinsufficient for Ik also develop these diseases at high frequency (28). Based on observations made with different leukemic clones, it is proposed that the ratio between the different Ik isoforms could be critical for the development of leukemia. Short Ik isoforms are frequently overexpressed in childhood acute lymphoblastic leukemia (ALL) (52). In particular, childhood ALL clones frequently express Ik4 (52) which, as we showed here, does not efficiently interact with any of the GATA proteins. Ik deletions are also highly represented in BCR-ABL ALL and in chronic myeloid leukemia (CML) with lymphoid blast crisis cases but not in chronic-phase CML, in acute myeloid leukemia, or in CML with myeloid blast crisis (48, 50, 51). Klein et al. (49) suggested that the overexpression of aberrant Ik isoforms occurred at a posttranscriptional level, but Mullighan et al. (50) and Iacobucci et al. (48) demonstrated that intragenic Ik deletions can account for overexpression of aberrant isoforms. Deregulation and mutations of hematopoietic GATA factors have also been associated with hematopoietic malignancies (94). For instance, the GATA1-short variant, which lacks the N-terminal domain, has been linked to transient abnormal myelopoiesis (95) as well as acute megakaryoblastic leukemia (96, 97) in children with Down syndrome. Finally, physiological GATA2 activity suppresses the development of myelodysplastic syndrome (MDS) and leukemia (98), and disruption of GATA3 has been found among the somatic mutations associated with early T cell precursor ALL (99).
In conclusion, we have provided evidence that Ik directly interacts with Cdk9/P-TEFb and the hematopoietic GATA factors. We show that Ik regulates gene expression in combination with GATA1, -2, or -3 in hematopoietic cells. Since these proteins are important for hematopoietic cell fate, disruption of the Ik-GATA-Cdk9 regulatory network is likely to perturb hematopoiesis and, hence, promote hematopoietic malignancies. In support of this assumption, we have demonstrated that Ik6 and the newly identified isoform Ik1Δex6 are unable to interact with Cdk9/P-TEFb. It can be argued that overexpression of these isoforms interferes with the Ik capacity to promote transcription elongation of hematopoietic target genes and, thereby, facilitates neoplasia.
ACKNOWLEDGMENTS
We thank M. J. Weiss for G1E cells and K. Georgopoulos for the Ikaros null mice.
This work was supported by a grant from the Canadian Institute of Health Research (CIHR) held by E.M. Work in E.B.A.'s lab was supported by a grant from the Natural Sciences and Engineering Research Council. E.M. is senior scholar of the Fond de la Recherche en Santé du Québec (FRSQ); N.M. was supported by a FQRNT doctoral training award; E.B.A. is a scholar of CIHR and FRSQ.
Footnotes
Published ahead of print 3 June 2013
REFERENCES
- 1.Pertea M. 2012. The human transcriptome: an unfinished story. Genes 3:344–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dillon N. 2012. Factor mediated gene priming in pluripotent stem cells sets the stage for lineage specification. Bioessays 34:194–204 [DOI] [PubMed] [Google Scholar]
- 3.Bresnick EH, Lee HY, Fujiwara T, Johnson KD, Keles S. 2010. GATA switches as developmental drivers. J. Biol. Chem. 285:31087–31093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hosoya T, Maillard I, Engel JD. 2010. From the cradle to the grave: activities of GATA-3 throughout T-cell development and differentiation. Immunol. Rev. 238:110–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.John LB, Ward AC. 2011. The Ikaros gene family: transcriptional regulators of hematopoiesis and immunity. Mol. Immunol. 48:1272–1278 [DOI] [PubMed] [Google Scholar]
- 6.Patient RK, McGhee JD. 2002. The GATA family (vertebrates and invertebrates). Curr. Opin. Genet. Dev. 12:416–422 [DOI] [PubMed] [Google Scholar]
- 7.Pevny L, Simon MC, Robertson E, Klein WH, Tsai SF, D'Agati V, Orkin SH, Costantini F. 1991. Erythroid differentiation in chimaeric mice blocked by a targeted mutation in the gene for transcription factor GATA-1. Nature 349:257–260 [DOI] [PubMed] [Google Scholar]
- 8.Weiss MJ, Orkin SH. 1995. Transcription factor GATA-1 permits survival and maturation of erythroid precursors by preventing apoptosis. Proc. Natl. Acad. Sci. U. S. A. 92:9623–9627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Whyatt D, Lindeboom F, Karis A, Ferreira R, Milot E, Hendriks R, de Bruijn M, Langeveld A, Gribnau J, Grosveld F, Philipsen S. 2000. An intrinsic but cell-nonautonomous defect in GATA-1-overexpressing mouse erythroid cells. Nature 406:519–524 [DOI] [PubMed] [Google Scholar]
- 10.Whyatt DJ, Karis A, Harkes IC, Verkerk A, Gillemans N, Elefanty AG, Vairo G, Ploemacher R, Grosveld F, Philipsen S. 1997. The level of the tissue-specific factor GATA-1 affects the cell-cycle machinery. Genes Funct. 1:11–24 [DOI] [PubMed] [Google Scholar]
- 11.Yamamoto M, Takahashi S, Onodera K, Muraosa Y, Engel JD. 1997. Upstream and downstream of erythroid transcription factor GATA-1. Genes Cells 2:107–115 [DOI] [PubMed] [Google Scholar]
- 12.Cantor AB, Iwasaki H, Arinobu Y, Moran TB, Shigematsu H, Sullivan MR, Akashi K, Orkin SH. 2008. Antagonism of FOG-1 and GATA factors in fate choice for the mast cell lineage. J. Exp. Med. 205:611–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ku CJ, Hosoya T, Maillard I, Engel JD. 2012. GATA-3 regulates hematopoietic stem cell maintenance and cell-cycle entry. Blood 119:2242–2251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Crossley M, Merika M, Orkin SH. 1995. Self-association of the erythroid transcription factor GATA-1 mediated by its zinc finger domains. Mol. Cell. Biol. 15:2448–2456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martin DI, Orkin SH. 1990. Transcriptional activation and DNA binding by the erythroid factor GF-1/NF-E1/Eryf 1. Genes Dev. 4:1886–1898 [DOI] [PubMed] [Google Scholar]
- 16.Tsiftsoglou AS, Vizirianakis IS, Strouboulis J. 2009. Erythropoiesis: model systems, molecular regulators, and developmental programs. IUBMB Life 61:800–830 [DOI] [PubMed] [Google Scholar]
- 17.Rodriguez P, Bonte E, Krijgsveld J, Kolodziej KE, Guyot B, Heck AJ, Vyas P, de Boer E, Grosveld F, Strouboulis J. 2005. GATA-1 forms distinct activating and repressive complexes in erythroid cells. EMBO J. 24:2354–2366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bresnick EH, Martowicz ML, Pal S, Johnson KD. 2005. Developmental control via GATA factor interplay at chromatin domains. J. Cell Physiol. 205:1–9 [DOI] [PubMed] [Google Scholar]
- 19.Weiss MJ, Orkin SH. 1995. GATA transcription factors: key regulators of hematopoiesis. Exp. Hematol. 23:99–107 [PubMed] [Google Scholar]
- 20.Bottardi S, Ross J, Bourgoin V, Fotouhi-Ardakani N, Affar EB, Trudel BM, Milot E. 2009. Ikaros and GATA-1 combinatorial effect is required for silencing of human gamma-globin genes. Mol. Cell. Biol. 29:1526–1537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bottardi S, Zmiri FA, Bourgoin V, Ross J, Mavoungou L, Milot E. 2011. Ikaros interacts with P-TEFb and cooperates with GATA-1 to enhance transcription elongation. Nucleic Acids Res. 39:3505–3519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koipally J, Georgopoulos K. 2000. Ikaros interactions with CtBP reveal a repression mechanism that is independent of histone deacetylase activity. J. Biol. Chem. 275:19594–19602 [DOI] [PubMed] [Google Scholar]
- 23.Koipally J, Renold A, Kim J, Georgopoulos K. 1999. Repression by Ikaros and Aiolos is mediated through histone deacetylase complexes. EMBO J. 18:3090–3100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sun L, Liu A, Georgopoulos K. 1996. Zinc finger-mediated protein interactions modulate Ikaros activity, a molecular control of lymphocyte development. EMBO J. 15:5358–5369 [PMC free article] [PubMed] [Google Scholar]
- 25.Molnar A, Georgopoulos K. 1994. The Ikaros gene encodes a family of functionally diverse zinc finger DNA-binding proteins. Mol. Cell. Biol. 14:8292–8303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hahm K, Ernst P, Lo K, Kim GS, Turck C, Smale ST. 1994. The lymphoid transcription factor LyF-1 is encoded by specific, alternatively spliced mRNAs derived from the Ikaros gene. Mol. Cell. Biol. 14:7111–7123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang JH, Nichogiannopoulou A, Wu L, Sun L, Sharpe AH, Bigby M, Georgopoulos K. 1996. Selective defects in the development of the fetal and adult lymphoid system in mice with an Ikaros null mutation. Immunity 5:537–549 [DOI] [PubMed] [Google Scholar]
- 28.Avitahl N, Winandy S, Friedrich C, Jones B, Ge Y, Georgopoulos K. 1999. Ikaros sets thresholds for T cell activation and regulates chromosome propagation. Immunity 10:333–343 [DOI] [PubMed] [Google Scholar]
- 29.Winandy S, Wu P, Georgopoulos K. 1995. A dominant mutation in the Ikaros gene leads to rapid development of leukemia and lymphoma. Cell 83:289–299 [DOI] [PubMed] [Google Scholar]
- 30.Ng SY, Yoshida T, Georgopoulos K. 2007. Ikaros and chromatin regulation in early hematopoiesis. Curr. Opin. Immunol. 19:116–122 [DOI] [PubMed] [Google Scholar]
- 31.Ross J, Mavoungou L, Bresnick EH, Milot E. 2012. GATA-1 utilizes Ikaros and polycomb repressive complex 2 to suppress Hes1 and to promote erythropoiesis. Mol. Cell. Biol. 32:3624–3638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu L, Nichogiannopoulou A, Shortman K, Georgopoulos K. 1997. Cell-autonomous defects in dendritic cell populations of Ikaros mutant mice point to a developmental relationship with the lymphoid lineage. Immunity 7:483–492 [DOI] [PubMed] [Google Scholar]
- 33.Dumortier A, Kirstetter P, Kastner P, Chan S. 2003. Ikaros regulates neutrophil differentiation. Blood 101:2219–2226 [DOI] [PubMed] [Google Scholar]
- 34.Brown KE, Guest SS, Smale ST, Hahm K, Merkenschlager M, Fisher AG. 1997. Association of transcriptionally silent genes with Ikaros complexes at centromeric heterochromatin. Cell 91:845–854 [DOI] [PubMed] [Google Scholar]
- 35.Georgopoulos K. 2002. Haematopoietic cell-fate decisions, chromatin regulation and ikaros. Nat. Rev. Immunol. 2:162–174 [DOI] [PubMed] [Google Scholar]
- 36.Harker N, Naito T, Cortes M, Hostert A, Hirschberg S, Tolaini M, Roderick K, Georgopoulos K, Kioussis D. 2002. The CD8α gene locus is regulated by the Ikaros family of proteins. Mol. Cell 10:1403–1415 [DOI] [PubMed] [Google Scholar]
- 37.Sabbattini P, Lundgren M, Georgiou A, Chow C, Warnes G, Dillon N. 2001. Binding of Ikaros to the λ5 promoter silences transcription through a mechanism that does not require heterochromatin formation. EMBO J. 20:2812–2822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Trinh LA, Ferrini R, Cobb BS, Weinmann AS, Hahm K, Ernst P, Garraway IP, Merkenschlager M, Smale ST. 2001. Down-regulation of TDT transcription in CD4(+)CD8(+) thymocytes by Ikaros proteins in direct competition with an Ets activator. Genes Dev. 15:1817–1832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Keys JR, Tallack MR, Zhan Y, Papathanasiou P, Goodnow CC, Gaensler KM, Crossley M, Dekker J, Perkins AC. 2008. A mechanism for Ikaros regulation of human globin gene switching. Br. J. Haematol. 141:398–406 [DOI] [PubMed] [Google Scholar]
- 40.Kim J, Sif S, Jones B, Jackson A, Koipally J, Heller E, Winandy S, Viel A, Sawyer A, Ikeda T, Kingston R, Georgopoulos K. 1999. Ikaros DNA-binding proteins direct formation of chromatin remodeling complexes in lymphocytes. Immunity 10:345–355 [DOI] [PubMed] [Google Scholar]
- 41.Yoshida T, Ng SY, Georgopoulos K. 2010. Awakening lineage potential by Ikaros-mediated transcriptional priming. Curr. Opin. Immunol. 22:154–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou Q, Li T, Price DH. 2012. RNA polymerase II elongation control. Annu. Rev. Biochem. 81:119–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Doubeikovskaia Z, Aries A, Jeannesson P, Morle F, Doubeikovski A. 2001. Purification of human recombinant GATA-1 from bacteria: implication for protein-protein interaction studies. Protein Express. Purif. 23:426–431 [DOI] [PubMed] [Google Scholar]
- 44.Cheng Y, Wu W, Kumar SA, Yu D, Deng W, Tripic T, King DC, Chen KB, Zhang Y, Drautz D, Giardine B, Schuster SC, Miller W, Chiaromonte F, Zhang Y, Blobel GA, Weiss MJ, Hardison RC. 2009. Erythroid GATA1 function revealed by genome-wide analysis of transcription factor occupancy, histone modifications, and mRNA expression. Genome Res. 19:2172–2184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Welch JJ, Watts JA, Vakoc CR, Yao Y, Wang H, Hardison RC, Blobel GA, Chodosh LA, Weiss MJ. 2004. Global regulation of erythroid gene expression by transcription factor GATA-1. Blood 104:3136–3147 [DOI] [PubMed] [Google Scholar]
- 46.Weiss MJ, Yu C, Orkin SH. 1997. Erythroid-cell-specific properties of transcription factor GATA-1 revealed by phenotypic rescue of a gene-targeted cell line. Mol. Cell. Biol. 17:1642–1651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nakatani Y, Ogryzko V. 2003. Immunoaffinity purification of mammalian protein complexes. Methods Enzymol. 370:430–444 [DOI] [PubMed] [Google Scholar]
- 48.Iacobucci I, Storlazzi CT, Cilloni D, Lonetti A, Ottaviani E, Soverini S, Astolfi A, Chiaretti S, Vitale A, Messa F, Impera L, Baldazzi C, D'Addabbo P, Papayannidis C, Lonoce A, Colarossi S, Vignetti M, Piccaluga PP, Paolini S, Russo D, Pane F, Saglio G, Baccarani M, Foa R, Martinelli G. 2009. Identification and molecular characterization of recurrent genomic deletions on 7p12 in the IKZF1 gene in a large cohort of BCR-ABL1-positive acute lymphoblastic leukemia patients: on behalf of Gruppo Italiano Malattie Ematologiche dell'Adulto Acute Leukemia Working Party (GIMEMA AL WP). Blood 114:2159–2167 [DOI] [PubMed] [Google Scholar]
- 49.Klein F, Feldhahn N, Herzog S, Sprangers M, Mooster JL, Jumaa H, Muschen M. 2006. BCR-ABL1 induces aberrant splicing of IKAROS and lineage infidelity in pre-B lymphoblastic leukemia cells. Oncogene 25:1118–1124 [DOI] [PubMed] [Google Scholar]
- 50.Mullighan CG, Miller CB, Radtke I, Phillips LA, Dalton J, Ma J, White D, Hughes TP, Le Beau MM, Pui CH, Relling MV, Shurtleff SA, Downing JR. 2008. BCR-ABL1 lymphoblastic leukaemia is characterized by the deletion of Ikaros. Nature 453:110–114 [DOI] [PubMed] [Google Scholar]
- 51.Nakayama H, Ishimaru F, Avitahl N, Sezaki N, Fujii N, Nakase K, Ninomiya Y, Harashima A, Minowada J, Tsuchiyama J, Imajoh K, Tsubota T, Fukuda S, Sezaki T, Kojima K, Hara M, Takimoto H, Yorimitsu S, Takahashi I, Miyata A, Taniguchi S, Tokunaga Y, Gondo H, Niho Y, Harada M, et al. 1999. Decreases in Ikaros activity correlate with blast crisis in patients with chronic myelogenous leukemia. Cancer Res. 59:3931–3934 [PubMed] [Google Scholar]
- 52.Sun L, Goodman PA, Wood CM, Crotty ML, Sensel M, Sather H, Navara C, Nachman J, Steinherz PG, Gaynon PS, Seibel N, Vassilev A, Juran BD, Reaman GH, Uckun FM. 1999. Expression of aberrantly spliced oncogenic Ikaros isoforms in childhood acute lymphoblastic leukemia. J. Clin. Oncol. 17:3753–3766 [DOI] [PubMed] [Google Scholar]
- 53.Crossley M, Orkin SH. 1994. Phosphorylation of the erythroid transcription factor GATA-1. J. Biol. Chem. 269:16589–16596 [PubMed] [Google Scholar]
- 54.Romano G, Giordano A. 2008. Role of the cyclin-dependent kinase 9-related pathway in mammalian gene expression and human diseases. Cell Cycle 7:3664–3668 [DOI] [PubMed] [Google Scholar]
- 55.Elagib KE, Mihaylov IS, Delehanty LL, Bullock GC, Ouma KD, Caronia JF, Gonias SL, Goldfarb AN. 2008. Cross-talk of GATA-1 and P-TEFb in megakaryocyte differentiation. Blood 112:4884–4894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kaichi S, Takaya T, Morimoto T, Sunagawa Y, Kawamura T, Ono K, Shimatsu A, Baba S, Heike T, Nakahata T, Hasegawa K. 2011. Cyclin-dependent kinase 9 forms a complex with GATA4 and is involved in the differentiation of mouse ES cells into cardiomyocytes. J. Cell Physiol. 226:248–254 [DOI] [PubMed] [Google Scholar]
- 57.Meier N, Krpic S, Rodriguez P, Strouboulis J, Monti M, Krijgsveld J, Gering M, Patient R, Hostert A, Grosveld F. 2006. Novel binding partners of Ldb1 are required for haematopoietic development. Development 133:4913–4923 [DOI] [PubMed] [Google Scholar]
- 58.Song SH, Kim A, Ragoczy T, Bender MA, Groudine M, Dean A. 2010. Multiple functions of Ldb1 required for beta-globin activation during erythroid differentiation. Blood 116:2356–2364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Claudio PP, Cui J, Ghafouri M, Mariano C, White MK, Safak M, Sheffield JB, Giordano A, Khalili K, Amini S, Sawaya BE. 2006. Cdk9 phosphorylates p53 on serine 392 independently of CKII. J. Cell Physiol. 208:602–612 [DOI] [PubMed] [Google Scholar]
- 60.Garriga J, Mayol X, Grana X. 1996. The CDC2-related kinase PITALRE is the catalytic subunit of active multimeric protein complexes. Biochem. J. 319:293–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lopez RA, Schoetz S, DeAngelis K, O'Neill D, Bank A. 2002. Multiple hematopoietic defects and delayed globin switching in Ikaros null mice. Proc. Natl. Acad. Sci. U. S. A. 99:602–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shimizu R, Trainor CD, Nishikawa K, Kobayashi M, Ohneda K, Yamamoto M. 2007. GATA-1 self-association controls erythroid development in vivo. J. Biol. Chem. 282:15862–15871 [DOI] [PubMed] [Google Scholar]
- 63.Zon LI, Youssoufian H, Mather C, Lodish HF, Orkin SH. 1991. Activation of the erythropoietin receptor promoter by transcription factor GATA-1. Proc. Natl. Acad. Sci. U. S. A. 88:10638–10641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ivanova NB, Dimos JT, Schaniel C, Hackney JA, Moore KA, Lemischka IR. 2002. A stem cell molecular signature. Science 298:601–604 [DOI] [PubMed] [Google Scholar]
- 65.Mansson R, Hultquist A, Luc S, Yang L, Anderson K, Kharazi S, Al-Hashmi S, Liuba K, Thoren L, Adolfsson J, Buza-Vidas N, Qian H, Soneji S, Enver T, Sigvardsson M, Jacobsen SE. 2007. Molecular evidence for hierarchical transcriptional lineage priming in fetal and adult stem cells and multipotent progenitors. Immunity 26:407–419 [DOI] [PubMed] [Google Scholar]
- 66.Ramalho-Santos M, Yoon S, Matsuzaki Y, Mulligan RC, Melton DA. 2002. “Stemness”: transcriptional profiling of embryonic and adult stem cells. Science 298:597–600 [DOI] [PubMed] [Google Scholar]
- 67.Subramaniam VN, Summerville L, Wallace DF. 2002. Molecular and cellular characterization of transferrin receptor 2. Cell. Biochem. Biophys. 36:235–239 [DOI] [PubMed] [Google Scholar]
- 68.Wargnier A, Lafaurie C, Legros-Maida S, Bourge JF, Sigaux F, Sasportes M, Paul P. 1998. Down-regulation of human granzyme B expression by glucocorticoids. Dexamethasone inhibits binding to the Ikaros and AP-1 regulatory elements of the granzyme B promoter. J. Biol. Chem. 273:35326–35331 [DOI] [PubMed] [Google Scholar]
- 69.Wilson BJ. 2008. Does GATA3 act in tissue-specific pathways? A meta-analysis-based approach. J. Carcinog. 7:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ronni T, Payne KJ, Ho S, Bradley MN, Dorsam G, Dovat S. 2007. Human Ikaros function in activated T cells is regulated by coordinated expression of its largest isoforms. J. Biol. Chem. 282:2538–2547 [DOI] [PubMed] [Google Scholar]
- 71.Lahlil R, Lecuyer E, Herblot S, Hoang T. 2004. SCL assembles a multifactorial complex that determines glycophorin A expression. Mol. Cell. Biol. 24:1439–1452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yoshida T, Ng SY, Zuniga-Pflucker JC, Georgopoulos K. 2006. Early hematopoietic lineage restrictions directed by Ikaros. Nat. Immunol. 7:382–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ptashne M. 2005. Regulation of transcription: from lambda to eukaryotes. Trends Biochem. Sci. 30:275–279 [DOI] [PubMed] [Google Scholar]
- 74.Gregory GD, Raju SS, Winandy S, Brown MA. 2006. Mast cell IL-4 expression is regulated by Ikaros and influences encephalitogenic Th1 responses in EAE. J. Clin. Invest. 116:1327–1336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hatton RD, Harrington LE, Luther RJ, Wakefield T, Janowski KM, Oliver JR, Lallone RL, Murphy KM, Weaver CT. 2006. A distal conserved sequence element controls Ifng gene expression by T cells and NK cells. Immunity 25:717–729 [DOI] [PubMed] [Google Scholar]
- 76.Malinge S, Thiollier C, Chlon TM, Dore LC, Diebold L, Bluteau O, Mabialah V, Vainchenker W, Dessen P, Winandy S, Mercher T, Crispino JD. 2013. Ikaros inhibits megakaryopoiesis through functional interaction with GATA-1 and NOTCH signaling. Blood 121:2440–2451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Quirion MR, Gregory GD, Umetsu SE, Winandy S, Brown MA. 2009. Cutting edge: Ikaros is a regulator of Th2 cell differentiation. J. Immunol. 182:741–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Umetsu SE, Winandy S. 2009. Ikaros is a regulator of Il10 expression in CD4+ T cells. J. Immunol. 183:5518–5525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yang L, Luo Y, Wei J. 2010. Integrative genomic analyses on Ikaros and its expression related to solid cancer prognosis. Oncol. Rep. 24:571–577 [DOI] [PubMed] [Google Scholar]
- 80.Jager R, Gisslinger H, Passamonti F, Rumi E, Berg T, Gisslinger B, Pietra D, Harutyunyan A, Klampfl T, Olcaydu D, Cazzola M, Kralovics R. 2010. Deletions of the transcription factor Ikaros in myeloproliferative neoplasms. Leukemia 24:1290–1298 [DOI] [PubMed] [Google Scholar]
- 81.Rebollo A, Schmitt C. 2003. Ikaros, Aiolos and Helios: transcription regulators and lymphoid malignancies. Immunol. Cell Biol. 81:171–175 [DOI] [PubMed] [Google Scholar]
- 82.Rooke HM, Orkin SH. 2006. Phosphorylation of Gata1 at serine residues 72, 142, and 310 is not essential for hematopoiesis in vivo. Blood 107:3527–3530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ferreira R, Wai A, Shimizu R, Gillemans N, Rottier R, von Lindern M, Ohneda K, Grosveld F, Yamamoto M, Philipsen S. 2007. Dynamic regulation of Gata factor levels is more important than their identity. Blood 109:5481–5490 [DOI] [PubMed] [Google Scholar]
- 84.Grass JA, Boyer ME, Pal S, Wu J, Weiss MJ, Bresnick EH. 2003. GATA-1-dependent transcriptional repression of GATA-2 via disruption of positive autoregulation and domain-wide chromatin remodeling. Proc. Natl. Acad. Sci. U. S. A. 100:8811–8816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Jing H, Vakoc CR, Ying L, Mandat S, Wang H, Zheng X, Blobel GA. 2008. Exchange of GATA factors mediates transitions in looped chromatin organization at a developmentally regulated gene locus. Mol. Cell 29:232–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hung HL, Lau J, Kim AY, Weiss MJ, Blobel GA. 1999. CREB-binding protein acetylates hematopoietic transcription factor GATA-1 at functionally important sites. Mol. Cell. Biol. 19:3496–3505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Koipally J, Georgopoulos K. 2002. Ikaros-CtIP interactions do not require C-terminal binding protein and participate in a deacetylase-independent mode of repression. J. Biol. Chem. 277:23143–23149 [DOI] [PubMed] [Google Scholar]
- 88.Georgopoulos K, Moore DD, Derfler B. 1992. Ikaros, an early lymphoid-specific transcription factor and a putative mediator for T cell commitment. Science 258:808–812 [DOI] [PubMed] [Google Scholar]
- 89.O'Neill D, Yang J, Erdjument-Bromage H, Bornschlegel K, Tempst P, Bank A. 1999. Tissue-specific and developmental stage-specific DNA binding by a mammalian SWI/SNF complex associated with human fetal-to-adult globin gene switching. Proc. Natl. Acad. Sci. U. S. A. 96:349–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.O'Neill DW, Schoetz SS, Lopez RA, Castle M, Rabinowitz L, Shor E, Krawchuk D, Goll MG, Renz M, Seelig HP, Han S, Seong RH, Park SD, Agalioti T, Munshi N, Thanos D, Erdjument-Bromage H, Tempst P, Bank A. 2000. An ikaros-containing chromatin-remodeling complex in adult-type erythroid cells. Mol. Cell. Biol. 20:7572–7582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Nichogiannopoulou A, Trevisan M, Neben S, Friedrich C, Georgopoulos K. 1999. Defects in hemopoietic stem cell activity in Ikaros mutant mice. J. Exp. Med. 190:1201–1214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Saunders A, Core LJ, Lis JT. 2006. Breaking barriers to transcription elongation. Nat. Rev. Mol. Cell Biol. 7:557–567 [DOI] [PubMed] [Google Scholar]
- 93.Zhou Q, Yik JH. 2006. The yin and yang of P-TEFb regulation: implications for human immunodeficiency virus gene expression and global control of cell growth and differentiation. Microbiol. Mol. Biol. Rev. 70:646–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zheng R, Blobel GA. 2010. GATA Transcription factors and cancer. Genes Cancer 1:1178–1188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Greene ME, Mundschau G, Wechsler J, McDevitt M, Gamis A, Karp J, Gurbuxani S, Arceci R, Crispino JD. 2003. Mutations in GATA1 in both transient myeloproliferative disorder and acute megakaryoblastic leukemia of Down syndrome. Blood Cells Mol. Dis. 31:351–356 [DOI] [PubMed] [Google Scholar]
- 96.Wechsler J, Greene M, McDevitt MA, Anastasi J, Karp JE, Le Beau MM, Crispino JD. 2002. Acquired mutations in GATA1 in the megakaryoblastic leukemia of Down syndrome. Nat. Genet. 32:148–152 [DOI] [PubMed] [Google Scholar]
- 97.Zipursky A. 2003. Transient leukaemia: a benign form of leukaemia in newborn infants with trisomy 21. Br. J. Haematol. 120:930–938 [DOI] [PubMed] [Google Scholar]
- 98.Bresnick EH, Katsumura KR, Lee HY, Johnson KD, Perkins AS. 2012. Master regulatory GATA transcription factors: mechanistic principles and emerging links to hematologic malignancies. Nucleic Acids Res. 40:5819–5831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zhang J, Ding L, Holmfeldt L, Wu G, Heatley SL, Payne-Turner D, Easton J, Chen X, Wang J, Rusch M, Lu C, Chen SC, Wei L, Collins-Underwood JR, Ma J, Roberts KG, Pounds SB, Ulyanov A, Becksfort J, Gupta P, Huether R, Kriwacki RW, Parker M, McGoldrick DJ, Zhao D, Alford D, Espy S, Bobba KC, Song G, Pei D, Cheng C, Roberts S, Barbato MI, Campana D, Coustan-Smith E, Shurtleff SA, Raimondi SC, Kleppe M, Cools J, Shimano KA, Hermiston ML, Doulatov S, Eppert K, Laurenti E, Notta F, Dick JE, Basso G, Hunger SP, Loh ML, Devidas M, Wood B, Winter S, Dunsmore KP, Fulton RS, Fulton LL, Hong X, Harris CC, Dooling DJ, Ochoa K, Johnson KJ, Obenauer JC, Evans WE, Pui CH, Naeve CW, Ley TJ, Mardis ER, Wilson RK, Downing JR, Mullighan CG. 2012. The genetic basis of early T-cell precursor acute lymphoblastic leukaemia. Nature 481:157–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Pfaffl MW. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45. [DOI] [PMC free article] [PubMed] [Google Scholar]