Crystal structure of bacteriophage phi29 pRNA-3WJ motif was solved at 3.05 Å, revealing two divalent metal ions that coordinate 4 nt of the RNA fragments. Subsequent modeling of the phi29 connector/pRNA complex suggests that the pRNA of the phi29 DNA packaging motor exists as a hexameric complex serving as a sheath over the connector. The study helps to elucidate the structural basis for the usual stability of the pRNA-3WJ, as well as gives insights in designing therapeutics RNA nanoparticles using pRNA-3WJ as scaffold.
Keywords: RNA crystal, metal ions, three-way junction, RNA nanotechnology, RNA therapeutics
Abstract
The bacteriophage phi29 DNA packaging motor, one of the strongest biological motors characterized to date, is geared by a packaging RNA (pRNA) ring. When assembled from three RNA fragments, its three-way junction (3WJ) motif is highly thermostable, is resistant to 8 M urea, and remains associated at extremely low concentrations in vitro and in vivo. To elucidate the structural basis for its unusual stability, we solved the crystal structure of this pRNA 3WJ motif at 3.05 Å. The structure revealed two divalent metal ions that coordinate 4 nt of the RNA fragments. Single-molecule fluorescence resonance energy transfer (smFRET) analysis confirmed a structural change of 3WJ upon addition of Mg2+. The reported pRNA 3WJ conformation is different from a previously published construct that lacks the metal coordination sites. The phi29 DNA packaging motor contains a dodecameric connector at the vertex of the procapsid, with a central pore for DNA translocation. This portal connector serves as the foothold for pRNA binding to procapsid. Subsequent modeling of a connector/pRNA complex suggests that the pRNA of the phi29 DNA packaging motor exists as a hexameric complex serving as a sheath over the connector. The model of hexameric pRNA on the connector agrees with AFM images of the phi29 pRNA hexamer acquired in air and matches all distance parameters obtained from cross-linking, complementary modification, and chemical modification interference.
INTRODUCTION
The bacteriophage phi29 DNA packaging motor, one of the strongest biological motors assembled in vitro using recombinant and synthetic materials, is geared by a packaging RNA (pRNA) ring (Guo et al. 1987, 1998; Chen et al. 2000; Shu et al. 2007; Xiao et al. 2008). Since its discovery in 1987 (Guo et al. 1987), the 120-nucleotide (nt) pRNA subunit has been extensively investigated (Reid et al. 1994b,c; Zhang et al. 1994, 1995a,b,c, 1997; Chen and Guo 1997; Chen et al. 2000). The unique features uncovered from the studies of phi29 pRNA include (1) independent folding of the ATPase gp16 interaction domain (Lee and Guo 2006) and the motor binding domain (Reid et al. 1994a,b,c; Zhang et al. 1994, 1995b,c) and (2) the capacity to harbor and escort therapeutic functional modules without causing misfolding or loss of function of the incorporated functionalities. These special properties of phi29 pRNA have led to the development of this RNA as a novel vehicle for applications in nanotechnology and medicine (Hoeprich et al. 2003; Guo et al. 2005a, 2006, 2012a,b; Khaled et al. 2005; Guo 2010; Abdelmawla et al. 2011; Shu et al. 2011a,b,c; Shukla et al. 2011; Zhou et al. 2011). Recently, it was found that the core structure of phi29 pRNA, a 54-nt three-way junction (3WJ) (Fig. 1A), exhibits extreme thermodynamic stability (Shu et al. 2011a). This trifurcate domain of pRNA can be assembled from three short RNA fragments (≤20 mer) with high affinity and low free energy, as demonstrated by its melting curve with a slope of ∼90°. Nevertheless, the fundamental mechanism governing the high thermodynamic stability of the 3WJ has been elusive.
FIGURE 1.
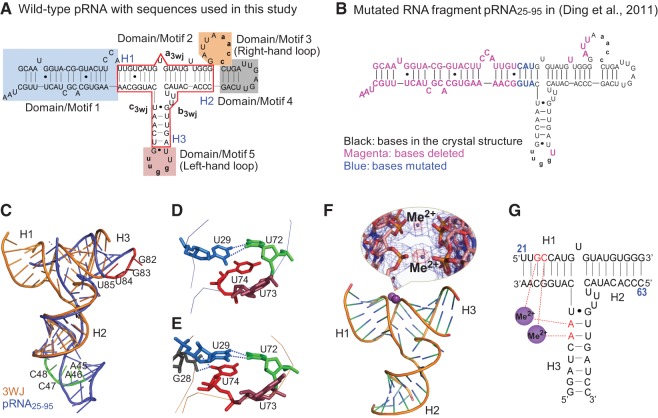
Secondary structure of phi29 pRNA with the 3WJ core and comparison of the two RNA molecules used in crystallization. (A) Full-length pRNA with the 3WJ motif boxed in red. The three individual RNA strands of the 3WJ are shown as a3wj, b3wj, and c3wj. The helical segments are designated as H1, H2, and H3, respectively. The interlocking loop sequences of 5′aacc in Domain/Motif 3 and 3′uugg in Domain/Motif 5 are indicated by lowercase letters. (B) Sequence of the RNA fragment of the published crystal structure pRNA25-95 (Ding et al. 2011), where the nucleotides deleted are in magenta and the nucleotides mutated are in blue. (C) Superposition of the pRNA25-95 (blue) (Ding et al. 2011) and 3WJ domain (gold) crystal structures. The nucleotides in the left-hand (red) and right-hand (green) loops are also highlighted. (D,E) Comparison of the triple U bulge junction (U72U73U74) for the core junction crystal structures of pRNA25-95 (D) (Ding et al. 2011) and the current 3WJ domain (E). Stereoviews are provided in Supplemental Figure S4. (F) Crystal structure of the pRNA 3WJ with two metal binding sites (magenta). A close-up view of the metal binding sites superposed on the 2Fo−Fc electron density map (blue mesh contoured at 1.0 σ) and the anomalous difference map (red mesh contoured at 4.5 σ) is shown in the inset. (G) Schematic representation of the 3WJ with metal coordinating nucleotides in red. Numbers in blue represent the nucleotide locations in the wild-type pRNA sequence.
The phi29 3WJ has been utilized as a useful platform for the construction of stable RNA nanoparticles. Cell receptor-binding ligands, aptamers, siRNAs, or ribozymes fused to individual arms of this 3WJ motif fold into their authentic structures and retain their functions such as specific cancer cell binding, cell entry, gene silencing, catalytic function, and in vivo targeting of cancer cells without accumulating in normal organs and tissues (Abdelmawla et al. 2011; Shu et al. 2011a; Haque et al. 2012), indicating that the 3WJ core with low ΔG guides the folding of the entire RNA particle to produce its global structure. The important application of the 3WJ in nanotechnology and medicine inspired the interest in further studies on pRNA 3WJ crystal structure. Recently a crystal structure of a part (66 nt) of the phi29 pRNA sequence (120 nt) (Fig. 1B; Ding et al. 2011) has been reported. In their studies, one of the key helices of the 3WJ core (5′-U21U22G23U24-3′/5′-A99A98C97G96-3′) was truncated, several bases of the 3WJ motif were mutated, and five other nucleotides at three different locations were also removed (Fig. 1B). As a result, the right-hand interlocking loop motif was changed from 9 nt to 6 nt, and the left interlocking loop motif was changed from 5 nt to 4 nt. More importantly, two of the 4 nt responsible for the coordination of Mg2+ reported here were excluded (Fig. 1B). The truncation, mutation, and alteration of the 3WJ core structure in pRNA molecule may lead to the production of a crystal structure that deviates significantly from the authentic pRNA fold, and therefore, further independent studies on the intact 3WJ crystal structure are necessary.
In this study, we report the crystallization, structure solution, and Mg2+-dependent fluorescence resonance energy transfer (FRET) analysis of the pRNA 3WJ, which likely represents the authentic structure of the metal-bound 3WJ core. Crystals were obtained in the presence of either Mg2+ or Mn2+. The crystal structure revealed two well-coordinated metal ions and a tightly packed core junction structure. To ensure that this result is not a crystal artifact, the effect of the divalent metal ions on the thermodynamic stability and solution structure (via FRET analysis) of pRNA 3WJ was also investigated. Additionally, the crystal structure of metal-coordinated 3WJ was used as a template to build a 3D model of intact, 120-nt monomeric pRNA. Model building indicated that the 120-nt pRNA monomers were capable of assembling into a hexameric pRNA ring, which was constructed and compared with that of AFM images and the previously published hexameric assembly (Hoeprich and Guo 2002), the latter of which was modeled based on secondary structural analysis and distant parameters from numerous biochemical studies. The intact pRNA hexamer model was docked with the 3D structure of the dodecameric ring formed by the phi29 connector protein. The resulting models were further evaluated by comparison with distance parameters obtained from single-molecule FRET (smFRET) and additional measurements by photoaffinity crosslinking (Garver and Guo 1997), chemical modification interference (Trottier et al. 2000; Mat-Arip et al. 2001), complementary modification (Zhang et al. 1997), and atomic force microscopy (Chen et al. 2000; Trottier et al. 2000; Mat-Arip et al. 2001).
RESULTS AND DISCUSSION
3WJ overall structure
The 3WJ crystal structure was solved at a resolution of 3.05 Å by piecewise molecular replacement (Table 1; Supplemental Fig. S1B; Robertson and Scott 2008; Robertson et al. 2010). The structure of the 3WJ core is facilitated by “coaxial stacking” of three individual RNA helices. This type of stacking interaction is commonly found in tRNA, ribozyme, and rRNA junction structures (Batey et al. 1999; Laing et al. 2012). The overall structure presented here is similar to the 3WJ region previously reported for the pRNA25-95 crystal structure (Fig. 1C; Ding et al. 2011); however, the H1/H3 interhelical angle is different in the two structures, at least partly due to the presence of metal binding sites that were not included in the previous pRNA25-95 construct (see the next section) (Fig. 1C). Helices joined at the 3WJ largely possessed the characteristics of canonical A-form helix but have distortions from A-form geometry mediated by chelating metal ions. Confirmation that metal binding alters the conformation of the 54-nt 3WJ structure was provided through FRET analysis of 3WJ in the presence and absence of Mg2+ (see the section of smFRET studies). At the core of the 3WJ domain is the junction formed by the intersection of three RNA helices, whose conformation, including that of the UUU bulge (Fig. 1D,E), dictated the relative orientation of three domains connected by RNA helices H1, H2, and H3. The metal-bound 3WJ presented here displayed an overall packing at the core junction with smaller void volumes and additional putative bonding interactions than that of pRNA25-95 (Fig. 1E). In the 3WJ structure, atoms O1P and O2P of nucleotide G28 lie 3.5 Å and 3.6 Å from atom O2 of unpaired base U74, while coaxial packing between U74 and U29 and the U-U (U29-U72) mismatch exhibit near ideal geometries (Fig. 1E).
TABLE 1.
Data collection and refinement statistics
Both crystal structures (3WJ and pRNA25-95) possessed a mismatched base pair between nucleotides U29 and U72 (Fig. 1D,E), and a wobble pair between residues G75 and U91. The conformation of the 3WJ is influenced by remote Mg2+ (or Mn2+) cations, which could induce a conformational shift of the pRNA to comply with the structure requirement for the function of the motor in DNA packaging (Chen et al. 2000).
Divalent metal ion (Mg2+ or Mn2+) coordination of 3WJ
One of the striking features of the 3WJ domain structure was the presence of divalent metal binding sites between helices H1 and H3 (Fig. 1C,F,G). The pRNA 3WJ was coordinated by two divalent metal ions bridging four phosphates (two from helix H1 and two from helix H3) such that the contact distance between phosphates donated by adjacent helices was reduced to 4.5 Å, with a relatively short 2.25 Å average metal-phosphate distance that was consistent with Mn2+/Mg2+ binding (Fig. 1F). Bound metal atoms were initially identified via 10–12 σ peaks in the |Fo|−|Fc| difference density map. Although both Mn2+ and Ca2+ were present in the crystallization conditions, the putative metal sites that bridge adjacent phosphate groups were confirmed as Mn2+ by anomalous difference Fourier analysis at a wavelength where Ca2+ has no measurable signal (Fig. 1F, inset). Therefore, the 3WJ structure deposited in the Protein Data Bank (PDB) contains Mn2+ in the phosphate bridging metal sites. However, low-resolution studies using isomorphous crystals grown with MgCl2 in place of MnCl2 suggest magnesium binding to the same sites in the 3WJ construct (data not shown).
In the final model, one Mn2+/Mg2+ ion was chelated by the phosphates of nucleotides G23 and A90, while the other Mn2+/Mg2+ ion bridges the phosphates of C24 and A89 (Fig. 1G). Although the pRNA 3WJ was similar to the core region of the published pRNA25-95 crystal structure (Fig. 1C), the cation-mediated close contact between helices H1 and H3 facilitates distortion from canonical A-form RNA (Supplemental Fig. S1A). The resulting distortion from A-form RNA, in the context of intact pRNA, resulted in readjustment of left- and right-hand interlocking loops such that a model for an intact pRNA multimer could be assembled while conserving base-pairing between the interlocking loops (see the section of construction of hexamer pRNA model). Two of four metal-chelating nucleotides (G23 and C24) were deleted in the construct of the previously determined crystal structure of pRNA25-95 (Fig. 1B; Ding et al. 2011); thus, the divalent ion coordination was not observed. In the 3WJ structure, a metal-induced conformational change in the angles between helices H1 and H3 reposition the left-hand and right-hand loops such that a continuous hexameric pRNA can be assembled with conserved base-pairing between the loops.
Effect of metal ion on 3WJ structure confirmed by smFRET
Previous results from various studies, including base deletion and mutation (Bailey et al. 1990; Reid et al. 1994c; Zhang et al. 1994, 1995b, 1997), ribonuclease probing (Reid et al. 1994a; Chen and Guo 1997), oligo targeting (Zhang et al. 1995a), competition assays to inhibit phage assembly (Trottier et al. 1996), UV crosslinking to portal protein (Garver and Guo 1997), psoralen crosslinking, and primer extension (Chen and Guo 1997), indicated two functional domains that could fold independently in pRNA. The two interlocking loops, along with the double-helical packaging domain for the binding of the motor ATPase (Lee et al. 2006), are connected through the 3WJ region in pRNA and a “three-arms around a hinge” model was proposed for pRNA function (Fang et al. 2008). Flexibility at the U72U73U74 bulge region was found to be essential for pRNA activity in gearing the DNA packaging motor (Reid et al. 1994c; Zhang et al. 1997), indicating that a conformational change may be required during DNA packaging.
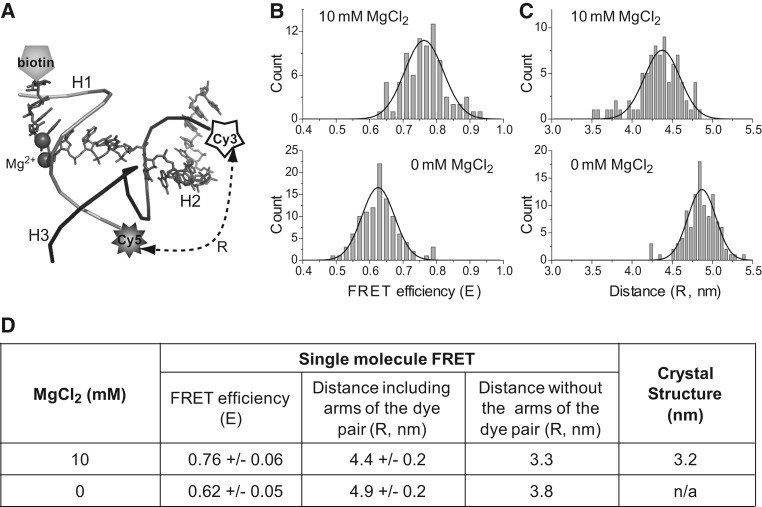
Conformational transitions are crucial to RNA functions (Al-Hashimi and Walter 2008). smFRET has been widely used to study the ion-induced conformational changes of functional RNAs (Zhuang et al. 2000; Kobitski et al. 2007; Qu et al. 2008; Steiner et al. 2008; Xiao et al. 2008; Brenner et al. 2010; McDowell et al. 2010; Haller et al. 2011). In this study, we investigated the effect of Mg2+ on the folding and conformational change of pRNA 3WJ using smFRET. To measure the distance transition, the ends of helices H2 and H3 of the pRNA 3WJ were labeled with a FRET pair (Cy3 and Cy5) and H1 was labeled with biotin for immobilization to the slide (Fig. 2A).
FIGURE 2.
Comparison of distances between the 3WJ H2/H3 distal ends derived from FRET measurements (with and without 10 mM MgCl2) and crystal data. (A) Location of labeling in 3WJ for single-molecule FRET study. (a3wj indicated by gray sticks; b3wj, black backbone; c3wj, gray backbone.) (B,C) Comparison of the FRET efficiency (B) and distance (C) at 0 mM and 10 mM Mg2+, respectively. The Gaussian curve fittings used to locate the peak of the distributions are shown as black lines. (D) Comparison of the distances from FRET and the crystal structure. The distance was corrected by subtracting the arm sizes of the fluorophore pair (Shu et al. 2010).
The FRET efficiency between donor Cy3 and acceptor Cy5 at H2 and H3 were compared in the absence and presence of Mg2+ (Fig. 2B,C). A reduction in FRET efficiency of 0.14 (±0.07), corresponding to an estimated decrease of ∼5 Å in the distance, occurred when a buffer containing 10 mM Mg2+ was introduced to the sample, indicating a structural change of the 3WJ between the two helices H2 and H3 upon the interaction with Mg2+. This agreed with previous finding that Mg2+ induces a conformational change in pRNA (Chen and Guo 1997). Furthermore, the distances measured from smFRET were closer to the ones observed in the crystal structure of the 3WJ core presented here than to those in pRNA25-95 (Fig. 2D).
Effect of metal ion on 3WJ structure confirmed by DNA packaging and viral assembly assay
Divalent metal ions play a critical role in stabilizing the tertiary structure of RNA and in facilitating RNA functions (Carter and Holbrook 2002; Draper 2004; Scott 2007; Johnson-Buck et al. 2011). The positively charged metal ions shield the negatively charged phosphate backbone of RNA, rendering the RNA tertiary structure more compact. This interaction between metal ions and the RNA could lead either to the formation of a positively charged cloud around the RNA molecule or to site-specific binding (Draper 2004; Johnson-Buck et al. 2011). The latter effect is often observed in various catalytic RNA structures (Cate et al. 1996, 1997; Cate and Doudna 1996; Murray et al. 1998; Kazantsev et al. 2005, 2009; Vicens and Cech 2006).
Metal ions are required to stimulate an appropriate pRNA conformation for dimer formation, procapsid binding, and DNA packaging (Chen and Guo 1997; Chen et al. 2000). It was found that the optimal Mg2+ concentration for phi29 DNA packaging was >3 mM (Supplemental Fig. S2). Although the pRNA 3WJ can be assembled from three individual RNA fragments in the absence of metal ions (Shu et al. 2011a), the divalent metal ion sites observed in the 3WJ crystal structure implied that these phosphate-bridged metal coordination may be involved in pRNA function. The specific coordination sites for the Mg2+ ions on RNA can be confirmed through the rescue of the activity of phosphothioate modified RNA by thiophilic metal ions such as Mn2+ or Cd2+ (Dahm and Uhlenbeck 1991; Peracchi et al. 1997; Basu and Strobel 1999; Kovacheva et al. 2004). However, evaluation by “manganese rescue” of the pRNA activity in DNA packaging and phi29 virion assembly is not feasible since Mg2+ is involved in multiple steps of DNA packaging and virion assembly other than facilitating the proper pRNA conformation, as Mn2+ can replace Mg2+ for the motor to package DNA, albeit at lower efficacy (Chen et al. 2000). In general, the increased stability observed for Mg2+-mediated stabilization of RNA over other cations (such as Na+ or Ca2+) is facilitated by its small size and high charge density. In addition to favorable electrostatic interactions, the relatively short interatomic distances between the phosphate groups and Mn2+/Mg2+ in the 3WJ structure (2.17–2.35 Å) can stabilize RNA oxyanion/Mg2+ interactions via charge transfer, polarization, and exchange, thus providing a special case of bidentate RNA-Mg2+ clamps (Petrov et al. 2011). These findings are consistent with a model in which the pRNA 3WJ (and by extension, intact pRNA hexamers) are stabilized by Mg2+ binding at specific sites between the phosphates of helices H1 and H3, optimally orienting the left- and right-hand loops for multimer assembly.
Effect of metal ion on 3WJ structure confirmed by gel shift assay
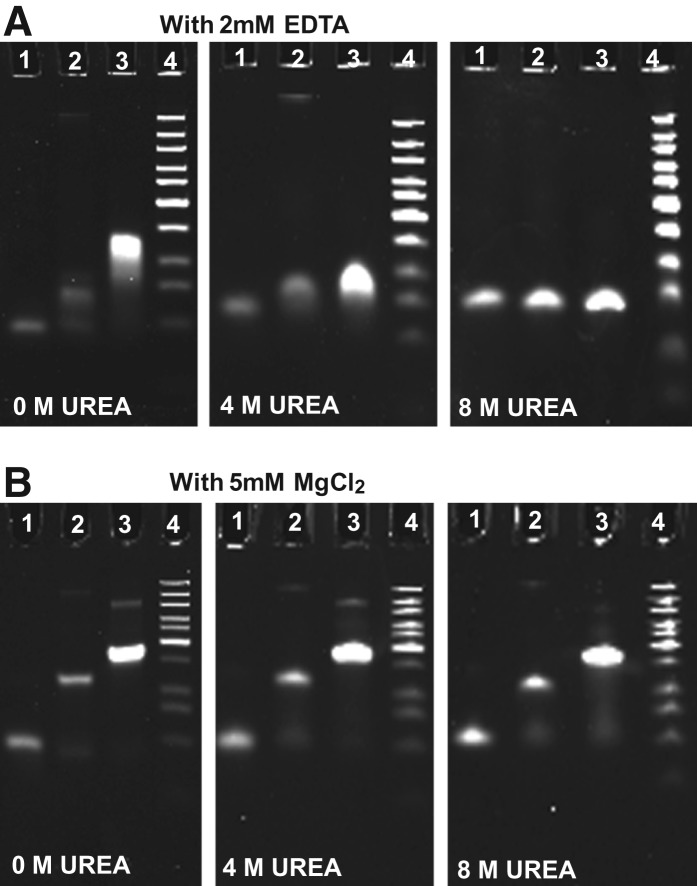
Urea is a common reagent used to denature DNA and RNA (Carlson et al. 1975; Pagratis 1996). Previous report has compared the assembly of 11 3WJ cores from different biological RNAs and has shown that in the presence of 5 mM MgCl2, the pRNA-3WJ assembly stayed associated in the presence of 8 M urea, while majority of others dissociated under the same condition (Shu et al. 2011a). The stability of the 3WJ complex was further assessed using 15% PAGE gels containing 0 M, 4 M, and 8 M urea both in the presence and in the absence of Mg2+ (Fig. 3). Without urea, the 3WJ is stable in both the presence and the absence of 5 mM MgCl2. However, in the presence of 4 M urea, the complex remained intact in the presence of 5 mM MgCl2 while it dissociated in the absence of MgCl2. Moreover, in the presence of 5 mM Mg2+, the pRNA 3WJ remained intact up to 8 M urea. The result demonstrated that Mg2+ is critical for the resistance to urea denaturation of this pRNA-3WJ core, while it failed to promote the urea resistance in several other 3WJ cores in the previous report (Shu et al. 2011a).
FIGURE 3.
Effect of magnesium ions on the resistance of pRNA 3WJ to urea denaturation, assayed by 15% PAGE gels with (A) 2 mM EDTA and (B) 5 mM MgCl2. (Lane 1) Single RNA oligo b3wj; (lane 2) sample annealed from two oligos a3wj and b3wj; (lane 3) 3WJ assembled from three oligos a3wj, b3wj, and c3wj; and (lane 4) DNA ladder.
Construction of a hexamer pRNA model using the 3WJ crystal structure and comparison with AFM images
Extensive investigations on the function and folding of pRNA have been carried out (Reid et al. 1994b,c; Zhang et al. 1994, 1995a,b,c, 1997; Chen and Guo 1997; Chen et al. 2000). Based on previous reports, the pRNA can be divided into the following domains (Fig. 1A), including domain 1, a 5′/3′-end double-helix for the binding of the motor ATPase gp16 (Zhang et al. 1994; Lee et al. 2006); domain 2, a stably folded trifurcate domain (3WJ) (Shu et al. 2011a); domain 3, the right-hand interlocking loop for inter-RNA interaction in RNA ring formation via hand-in-hand interactions (Guo et al. 1998; Zhang et al. 1998; Chen et al. 2000); domain 4, the head loop (Reid et al. 1994a,b,c; Guo et al. 1998; Chen et al. 2000; Hoeprich and Guo 2002; Kitamura et al. 2008); and domain 5, the left-hand interlocking loop for inter-RNA interaction in RNA ring formation via hand-in-hand interactions (Guo et al. 1998; Zhang et al. 1998; Chen et al. 2000). The availability of the structure of individual domains makes the assembly of the entire pRNA structure and the ring feasible.
For model structure construction, the crystal structure of the pRNA 3WJ and the published structural data (Hoeprich and Guo 2002; Harris and Schroeder 2010; Ding et al. 2011) were used to build the intact 120-nt pRNA monomer as well as a hexameric pRNA ring (see Materials and Methods) (Figs. 1A, 4A). Briefly, the entire 3WJ structure, including the two metal ions, was used as a central building block for grafting the structures of the other four domains from three PDB files (PDB codes 1L4O, 2KVN, 3R4F) (Hoeprich and Guo 2002; Harris et al. 2010; Ding et al. 2011) to generate an intact 120-nt pRNA (Fig. 1A). The 3WJ served as domain 2, and all other domains were connected to the 3WJ. Domain 1 was grafted from the previous hexameric pRNA model (PDB 1L4O) (Hoeprich and Guo 2002). Domains 3, 4, and 5 were grafted from the crystal structure of pRNA25-95 (PDB 3R4F) (Ding et al. 2011), with missing residues inserted to complete the entire 120-nt pRNA monomer.
FIGURE 4.
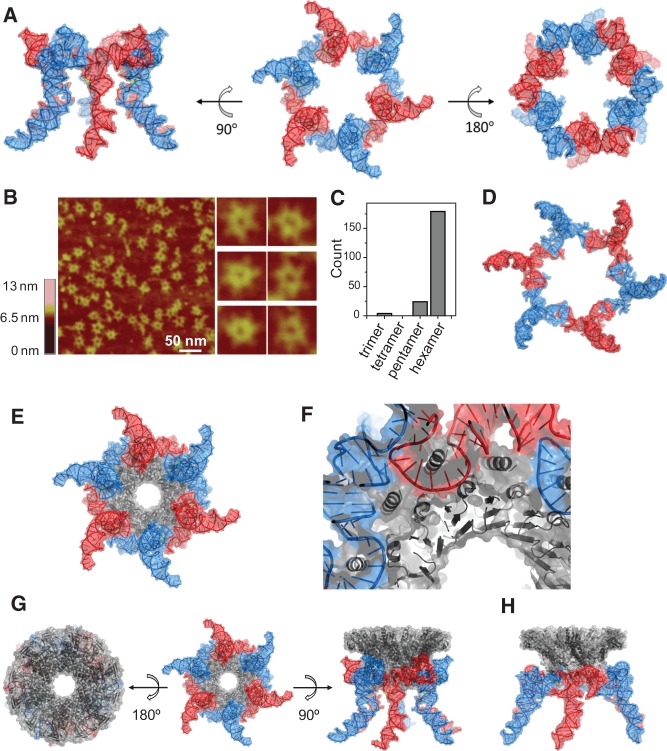
Structure of a pRNA hexamer ring and docking of the hexameric pRNA model with the dodecameric connector ring of phi29. (A) A 3D model of hexameric pRNA based on the crystal structure of 3WJ with views from three mutually perpendicular angles. (B) AFM images of hexameric re-engineered pRNA rings show strong correlation in size and shape with the 3D computer models for the intact, full-length pRNA hexamers. (C) Histogram of RNA particles in AFM images with discernible stoichiometry. (D) Top view of the published 3D computer model of hexameric pRNA constructed based on biochemical data (PDB 1L4O) (Hoeprich and Guo 2002). (E) Model of a pRNA hexamer complexed with a dodecameric connector ring based on the 3WJ and connector (PDB 1H5W) (Guasch et al. 2002) crystal structures. (For an animation movie of the phi29 pRNA-connector model, see Supporting Video S1.) (F) A close-up view of proper anchoring of the connector N-terminal helices for optimal interactions with pRNA. (G) Three mutually perpendicular views of the hexameric pRNA-connector assembly. (H) A side-view of the 3D model of the hexameric pRNA-connector assembly based on biochemical data (PDB 1L4P) (Hoeprich and Guo 2002).
The pRNA hexamer was assembled using pRNA with 3-nt extensions at the interlocking loops (Shu et al. 2013b) (see Materials and Methods), which enhances the stability of RNA hexamer, while the wild-type pRNA hexamer is relatively unstable. This re-engineered pRNA binds to procapsid specifically with the same efficiency as the wild-type pRNA. The assembled pRNA hexamer was confirmed by AFM imaging (Fig. 4B), which showed a strong resemblance to the model structure (Fig. 4A). The pRNA ring showed an outer contour diameter of ∼30 nm formed through the loop–loop interactions, with a center ring of ∼10 nm in diameter to sheath on the connector that has an outer contour diameter of 9.4 nm for its narrower end and 13.8 nm for its wider end (Guo et al. 2005b). A comparison of the AFM images with the pRNA hexamer model showed a strong correlation in shape and size, including sixfold rotational axes and protrusions every 60° (these correspond to helix H1, essential for contacting the ATPase). The inner diameter of the hexamer (∼9.7 nm) was in good correspondence with the diameter of the pRNA hexamer determined by AFM, supporting the model of the pRNA hexameric ring. This model structure was very similar to the previously published computer model of the pRNA hexamer based on biochemical distance constraints (Fig. 4D).
Docking of the hexamer pRNA model to the motor connector
Connector protein, the foothold for pRNA binding on the phi29 motor, contains a wide end and a narrow end. The wide end is embedded inside the capsid, and the narrow end is exposed for interaction with the pRNA (Jimenez et al. 1986; Simpson et al. 2000; Guo et al. 2005b). The interaction between pRNA and the connector was demonstrated by crosslinking of the two (Garver and Guo 1997). It was found that the N-terminal 14 amino acids of gp10 are essential for pRNA binding since deletion of these 14 amino acids rendered the connector incompetent for RNA binding (Xiao et al. 2005) and mutation in R3K4R5 led to a similar result (Atz et al. 2007). Particularly, these three positively charged amino acids (R3K4R5) are believed to be responsible for the binding to pRNA with the negatively charged phosphate backbone.
Modeling the connector/pRNA complex was initiated by alignment of the sixfold and 12-fold rotational axes of the molecules. The inner diameter of the pRNA hexamer was matched with the variable outer diameter of the connector to establish the depth of sheathing of the pRNA onto the connector. A limited rotational search (by definition, the largest error can be no more than 15°) identified one angle that allowed persistence of the N-terminal helices and their putative recognition of the pRNA (Fig. 4E–H).
Ten residues missing from the N terminus of the connector crystal structure (PDB 1H5W) (Guasch et al. 2002) were incorporated as a 10 residue extension of the N-terminal α helix (Fig. 4F). Of the newly modeled N-terminal 10 amino acids of the connector, all amino acids previously determined to interact with the pRNA lie near phosphates or bases of the pRNA hexamer. The close proximity of nucleotides 28–32, 69–72, 46–50, 62, and 81–84 of the pRNA to connector residue S170 agreed with results from Fe-BABE probing of the motor structure (Supplemental Fig. S3; Atz et al. 2007). In addition, the procapsid binding domain of the pRNA (bases 26–83) was in close contact with the connector protein, in agreement with the results that this region is protected from nuclease degradation and chemical modification upon motor assembly (Reid et al. 1994a; Zhang et al. 2001). (For an animation movie of the phi29 pRNA/connector model, see Supporting Video S1.)
Verification of distances between nucleotides in the hexameric RNA ring with distance parameters obtained by biochemical methods
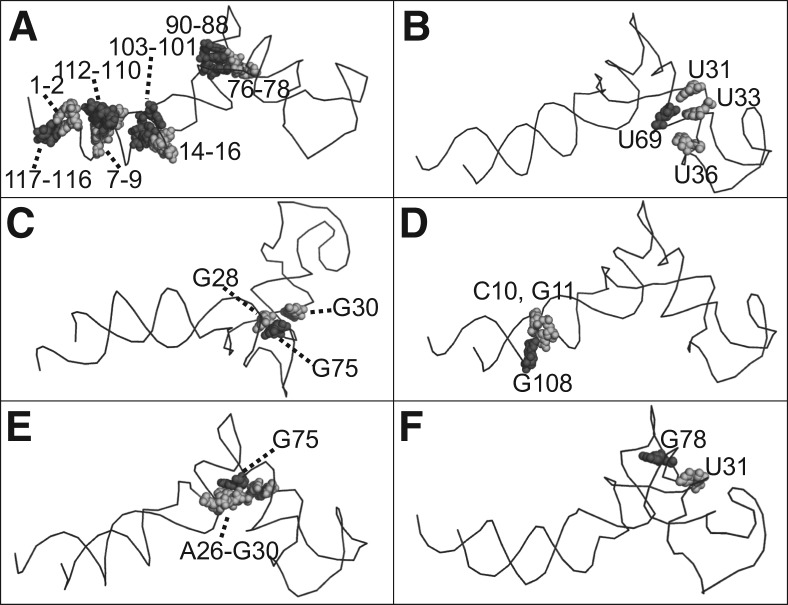
Previously, various biochemical methods were used to study the tertiary structure of pRNA. The presence of helical regions was predicted by phylogenetics and later confirmed by complementary modification experiments (Reid et al. 1994c; Zhang et al. 1995b, 1997). Disruptions of the helical regions 1–2/117–116, 7–9/112–110, 14–16/103–101, and 76–78/90–88 were found to inactivate pRNA activity in DNA packaging, while activity can be restored by compensation mutations. These base-pairing interactions are reflected in the constructed hexameric pRNA model (bases 1–2 are paired with bases 117–116; bases 7–9 are paired with bases 112–110; bases 14–16 are paired with bases 103–101; and bases 76–78 are paired with bases 90–88) (Fig. 5A).
FIGURE 5.
Confirmation of the close proximity between two nucleotide groups (black and gray) in the pRNA model based on the 3WJ crystal structure, in agreement with the published data revealed by (A) complementary modification; (B) psoralen crosslinking; (C) Phenphi crosslinking; and (D–F) azidophenacyl crosslinking.
Photoaffinity crosslinking has been applied to probe bases that are in close proximity in the pRNA structure (Chen and Guo 1997). Crosslinking using psoralen, a small photoactive probe that can covalently link uridines upon UV irradiation (Cimino et al. 1985; Tyc and Steitz 1992), showed that U69 is in close proximity to U31, U33, and U36 in the presence of Mg2+ (Chen and Guo 1997). Additional photoaffinity crosslinking using phenphi to crosslink guanosine bases in close proximity showed that base G75 is close to G28 and G30 (Mohammad et al. 1999). Similar data using azidophenacyl (APA) crosslinking also indicates that base G75 is in close proximity to bases 26–30, while G78 is close to U31, and G108 is proximal to bases 10–11 (Mat-Arip et al. 2001). These are all in good agreement with the pRNA model reported here, as shown in Figure 5, B through F.
Chemical modification revealed that nucleotides 18–20, 42–48, 55–57, and 82–86 are solvent accessible in monomer, dimer as well as procapsid-bound pRNA forms (Trottier et al. 2000; Zhang et al. 2001), indicating that these regions are present as single-stranded loops (Zhang et al. 2001). When three bases C18C19A20 are deleted (Reid et al. 1994b; Zhang et al. 2001) or paired with 3′GGU5′ inserted between A99 and A100 (Zhang et al. 1997), the resulting pRNA loses its packaging activity while retaining the ability to form dimers and bind procapsid, suggesting CCA is not involved in procapsid binding. Comparison of these regions in the newly modeled hexameric pRNA reported here agrees with these data.
CONCLUSIONS
The 3WJ structure serves as a core of the phi29 hexameric motor pRNA. Crystal structure analysis, as well as biophysical and biochemical studies, indicates that the binding of divalent metal ions promotes a structural change of the 3WJ of pRNA. Analysis of the 3WJ core structure provides a better understanding in the overall pRNA structure and multimerization, which are crucial for pRNA function. In recent years, RNA nanotechnology has emerged to play an important role in cancer therapy and disease treatment, serving as a vehicle to deliver therapeutics (Guo 2010; Guo et al. 2012a). Studies on 3D structures of RNA lead to identifications of useful RNA motifs (Leontis et al. 2006), which can be used to carry therapeutics for delivery (Shu et al. 2011a, 2013a,b; Haque et al. 2012). Several self-assembling, stable, and polyvalent RNA nanoparticles have been produced based on computational designs (Afonin et al. 2010; Dibrov et al. 2011; Grabow et al. 2011), in which information of the geometry of the individual RNA motifs as building blocks is essential. The pRNA 3WJ core has been demonstrated to be a useful platform to generate therapeutic RNA nanoparticles for targeted delivery (Shu et al. 2011a, 2013a,b; Haque et al. 2012). Our analysis of this 3WJ’s crystal structure of core will give insights in the designs of more effective therapeutics RNA nanoparticles.
MATERIALS AND METHODS
RNA synthesis and crystal screening
Each of the pRNA 3WJ strands (Fig. 1A), a3wj, b3wj, and c3wj (Shu et al. 2011a, 2013a,b; Haque et al. 2012), was purchased from IDT. Individual RNA strands were designed to produce blunt ends when they are annealed to facilitate crystal packing through base stacking interactions (Reyes et al. 2009). The nucleotide U24 was replaced with C24 in strand a3wj, and this change did not affect the pRNA’s biological activity in DNA packaging. The three strands were self-assembled to form the 3WJ in DEPC water simply by mixing them at equimolar concentrations. Initial crystallization trials were carried out at 22°C in 96-well plates with the hanging drop vapor diffusion method using a “Mosquito” nano-crystallization robot. Drops consisting of 0.1 μL RNA solution were mixed with an equal volume of reservoir solution and equilibrated against 100 μL reservoir solution. Lead conditions were optimized to 100 mM acetate (pH 4.6), 24%–26% 2-methyl-2,4-pentanediol (MPD), 20 mM CaCl2, and 10 mM MnCl2.
3WJ crystallization and structure determination
The initial crystallization trials were carried out at 295 K in 24-well plates using the hanging-drop vapor-diffusion method with commercially available screening kits for nucleic acids or nucleic acid/protein complexes, including Natrix, Nucleix (Qiagen), and Nuc-Pro (Jena Biosciences). Drops consisting of 0.5 µL RNA solution in DEPC-treated water (7 mg/mL) were mixed with an equal volume of reservoir solution and equilibrated against 500 µL reservoir solution. Initial hits were identified, and conditions yielding small crystals were further optimized by variation of crystallization parameters and additives. The final conditions that produced the best diffraction quality crystals (Supplemental Fig. S1C) contained 24%–26% (v/v) MPD, 20 mM CaCl2, 10 mM MnCl2, and 100 mM sodium acetate (pH 4.6). Crystals appeared within a week and continued to grow until they reached average dimensions of 0.03 × 0.03 × 0.3 mm (Supplemental Fig. S1C). The crystals were mounted on cryoloops and directly plunged into liquid nitrogen without additional cryoprotectant prior to data collection.
Native data sets were collected from single crystals cryocooled to 100 K on a MAR225 CCD detector at the SER-CAT 22BM beamline at APS. The data sets were processed with HKL2000 and scaled and merged with Scalepack (Otwinowski and Minor 1997). The best crystals, space group I4, diffract to 3.05 Å (Supplemental Fig. S1D) and have unit-cell parameters a = b = 125.01 Å and c = 26.99 Å. Final data collection statistics are summarized in Table 1.
The structure was determined by piecewise molecular replacement using three copies of a seven base-paired poly-adenine standard A-form double helix (stem) as a model. Solvent content analysis indicated one 3WJ molecule in the asymmetric unit (VM= 3.07 Å3/Da, 68.2% solvent content assuming RNA density of 1.7 g/cm3). The molecular replacement search for three stems (which potentially constitute a monomeric 3WJ structure) was carried out with Phaser (McCoy et al. 2007). The best solution contained three stems and had a Z score of 7.7 for the translation function, 12% higher than the next highest potential solution. Subsequent rigid body refinement using CNS (Brunger et al. 1998) resulted in R and Rfree values of 46.1% and 49.6%, respectively. The phases were improved through rounds of manual rebuilding with Coot (Emsley and Cowtan 2004) and composite omit map calculation implemented in CNS, which enabled building of >80% of the structure. Subsequent rounds of model building and refinement using Phenix (Adams et al. 2002) identified the proper asymmetric unit and sequence register. The metal and water sites were identified and validated by normal and anomalous difference Fourier maps as well as bonding geometry.
The structure was refined with Phenix (Adams et al. 2010) to final Rwork and Rfree values of 18.2% and 19.7%, respectively. Four TLS groups matching the three helices, i.e., (1) residues A1-8/C9-16, (2) residues C1-8/B13-20, (3) residues B1-5/A14-18, and (4) the central region of the 3WJ with residues B6-12/A9-13 were defined for the refinement of atomic displacement parameters. The final model displays a good stereochemistry according to MolProbity (Chen et al. 2010) and consists of 54 nucleotides, three Mn2+ ions, and two solvent molecules. Refinement statistics are given in Table 1, and the final 2Fo-Fc electron density map is shown in Supplemental Figure S1B. The coordinates and the structure factors have been deposited in the PDB (4KZ2).
Hexamer model building
The 3WJ crystal structure was used as a template to build a 120-nt pRNA monomer. Three regions of duplex RNA, designated 1, 2, and 5 (the nomenclature follows a previous publication) (Hoeprich and Guo 2002) are connected at the 3WJ, while regions 3 and 4 comprise the right-hand loop and head loop, respectively. Region 1 was grafted from the hexamer pRNA model and includes the ATPase interaction surface. Region 2 displays the left-hand loop, and region 5 comprises the RNA helix from the 3WJ and includes region 3, designated the right-hand loop.
The head loop and right-hand loop (regions 3 and 4) were positioned by superposing our 3.05 Å 3WJ crystal structure with the crystal structure of pRNA25-95 (PDB 3RF4) (Ding et al. 2011) on region 5 using the sugar-phosphate backbone for the 7 bp prior to the right-hand loop. Residues from these regions were grafted onto the 3WJ crystal structure. The left-hand loop was aligned by superposing 3WJ and PDB 3RF4 for 6 bp of region 2 leading up to the left-hand loop, followed by grafting the loop from 3RF4 onto the 3WJ core. The RNA helix containing region 1, including the ATPase interaction domain, came from PDB 1L4O, previously published 3D model for the pRNA hexamer (Hoeprich and Guo 2002). Residues 1–20 and 100–117 were grafted from 1L4O onto the 3WJ structure after superposition of the sugar-phospate backbone for 4 bp leading up to the CCA bulge. Finally, two single-residue insertions missing from any current model in the database were built into the pRNA monomer, including a “flipped out” conformation for U35 and an in-line conformation for residue U81 of the left-hand loop. To complete the monomer model, three unpaired residues (118–120) were added to the terminus of region 1. For all molecular “grafts,” bonds and angles were energy minimized using the “regularize” feature in Coot. All duplex regions from our 3WJ structure were included in the initial model.
To facilitate building a symmetric hexameric pRNA, the monomer model was put into a P6 pseudo-cell, and the orientation was optimized to obtain the best alignment of the 4 bp of the left- and right-hand loops within a hexamer. The resulting hexameric assembly was energy minimized using AMBER force fields (with no symmetry restraints) implemented with Chimera (UCSF). When the six individual monomers output from energy minimization were analyzed as symmetric hexameric assemblies, one model displayed superior packing of left- and right-hand loops relative to the other five. This model was subjected to another round of model building and energy regularization with Coot before expansion into the final sixfold symmetric pRNA hexamer model (Fig. 4).
AFM imaging of re-engineered pRNA ring
Loop-extended pRNAs were constructed by extending the 4-nt complementary loops to 7-nt loops and synthesized through in vitro transcription (Shu et al. 2013b). Six different re-engineered pRNA molecules containing intermolecular complementary loops were mixed at equal concentrations to form stable pRNA hexamers in solution, and the hexameric pRNA was purified from a native PAGE gel. The sample was fixed to the specially modified mica surface (APS) (Lyubchenko and Shlyakhtenko 2009) for AFM imaging. The AFM images were acquired in air using Super Sharp DLC probes (K-Tek Nanotechnology) and a MultiMode AFM NanoScope IV system (Bruker Nano/Veeco) operating in Tapping mode.
smFRET of pRNA 3WJ
Individual biotin-labeled or fluorophore (Cy3 or Cy5)-labeled RNA strands were purchased from IDT, and the labeled 3WJ was produced by mixing the three strands at equimolar concentrations in DEPC water. The 3WJ was then diluted in buffers containing 50 mM Tris (pH 8) and 100 mM NaCl, with 0 mM or 10 mM MgCl2 for comparison in the absence or presence of Mg2+. The sample was immobilized at the biotin-BSA–coated surface of the sample chamber via biotin/streptavidin interaction according to methods previously described (Shu et al. 2010). Concentration of the samples was carefully picked to ensure the observation of individual fluorescent spots while keeping sufficient number of these spots in each field of view. Prism-type total internal reflection fluorescence (TIRF) imaging was performed according to methods previously described (Shu et al. 2010). A laser beam of wavelength 532 nm was used to excite the FRET pair, and dual-color fluorescence images were taken. The images were analyzed to obtain FRET efficiencies and to calculate distances between the FRET pairs.
Effect of Mg2+ ion on the resistance of 3WJ to urea denaturation
The 3WJ samples were assembled from the individual RNA strands in a buffer containing 25 mM Tris (pH 8), 50 mM NaCl, and 5 mM MgCl2 and were assayed by 15% PAGE gels. The gels were made in 1× TB buffer (89 mM Tris-borate at pH 8) containing 5 mM MgCl2 (TBM) or 2 mM EDTA (TBE) to compare the stability of 3WJ with or without Mg2+. Different concentrations of urea (0 M, 4 M, and 8 M) were mixed into the gel to test the stability of 3WJ. The gels were run in their corresponding TBM or TBE buffers.
DATA DEPOSITION
The coordinates and the structure factors of the pRNA 3WJ crystal structure have been deposited in the PDB (accession code 4KZ2).
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
ACKNOWLEDGMENTS
We thank Eric Westhof for critical review on the manuscript and the crystal structure as well as assistance in refinement of the final pRNA structure model. The research was supported by NIH grants EB003730 and CA151648 to P.G. and ADA grant 7-08-CD-03 to Y.C. AFM images were obtained at Nanoimaging Core Facility supported by NIH SIG program and UNMC Program of ENRI to Y.L. P.G. is a cofounder of Kylin Therapeutics, Inc., and Biomotor and Nucleic Acid Nanotechnology Development Corp. Ltd.
Footnotes
Freely available online through the RNA Open Access option.
REFERENCES
- Abdelmawla S, Guo S, Zhang L, Pulukuri S, Patankar P, Conley P, Trebley J, Guo P, Li QX 2011. Pharmacological characterization of chemically synthesized monomeric pRNA nanoparticles for systemic delivery. Mol Ther 19: 1312–1322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC 2002. PHENIX: Building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr 58: 1948–1954 [DOI] [PubMed] [Google Scholar]
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. 2010. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66: 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afonin KA, Bindewald E, Yaghoubian AJ, Voss N, Jacovetty E, Shapiro BA, Jaeger L 2010. In vitro assembly of cubic RNA-based scaffolds designed in silico. Nat Nanotechnol 5: 676–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Hashimi HM, Walter NG 2008. RNA dynamics: It is about time. Curr Opin Struct Biol 18: 321–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atz R, Ma S, Gao J, Anderson DL, Grimes S 2007. Alanine scanning and Fe-BABE probing of the bacteriophage φ29 prohead RNA–connector interaction. J Mol Biol 369: 239–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey S, Wichitwechkarn J, Johnson D, Reilly B, Anderson D, Bodley JW 1990. Phylogenetic analysis and secondary structure of the Bacillus subtilis bacteriophage RNA required for DNA packaging. J Biol Chem 265: 22365–22370 [PubMed] [Google Scholar]
- Basu S, Strobel SA 1999. Thiophilic metal ion rescue of phosphorothioate interference within the Tetrahymena ribozyme P4-P6 domain. RNA 5: 1399–1407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batey RT, Rambo RP, Doudna JA 1999. Tertiary motifs in RNA structure and folding. Angew Chem Int Ed Engl 38: 2326–2343 [DOI] [PubMed] [Google Scholar]
- Brenner MD, Scanlan MS, Nahas MK, Ha T, Silverman SK 2010. Multivector fluorescence analysis of the xpt guanine riboswitch aptamer domain and the conformational role of guanine. Biochemistry 49: 1596–1605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. 1998. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr 54: 905–921 [DOI] [PubMed] [Google Scholar]
- Carlson RD, Olins AL, Olins DE 1975. Urea denaturation of chromatin periodic structure. Biochemistry 14: 3122–3125 [DOI] [PubMed] [Google Scholar]
- Carter RJ, Holbrook SR 2002. RNA structure: Roles of Me2+. In Encyclopedia of Life Sciences, pp. 1–7 John Wiley & Sons, Ltd., Hoboken, NJ [Google Scholar]
- Cate JH, Doudna JA 1996. Metal-binding sites in the major groove of a large ribozyme domain. Structure 4: 1221–1229 [DOI] [PubMed] [Google Scholar]
- Cate JH, Gooding AR, Podell E, Zhou K, Golden BL, Kundrot CE, Cech TR, Doudna JA 1996. Crystal structure of a group I ribozyme domain: Principles of RNA packaging. Science 273: 1678–1685 [DOI] [PubMed] [Google Scholar]
- Cate JH, Hanna RL, Doudna JA 1997. A magnesium ion core at the heart of a ribozyme domain. Nat Struct Biol 4: 553–558 [DOI] [PubMed] [Google Scholar]
- Chen C, Guo P 1997. Magnesium-induced conformational change of packaging RNA for procapsid recognition and binding during phage φ29 DNA encapsidation. J Virol 71: 495–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Sheng S, Shao Z, Guo P 2000. A dimer as a building block in assembling RNA: A hexamer that gears bacterial virus phi29 DNA-translocating machinery. J Biol Chem 275: 17510–17516 [DOI] [PubMed] [Google Scholar]
- Chen VB, Arendall WB III, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC 2010. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr 66: 12–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimino GD, Gamper HB, Isaacs ST, Hearst JE 1985. Psoralens as photoactive probes of nucleic acid structure and function: Organic chemistry, photochemistry, and biochemistry. Ann Rev Biochem 54: 1151–1193 [DOI] [PubMed] [Google Scholar]
- Dahm SC, Uhlenbeck OC 1991. Role of divalent metal ions in the hammerhead RNA cleavage reaction. Biochemistry 30: 9464–9469 [DOI] [PubMed] [Google Scholar]
- Dibrov SM, McLean J, Parsons J, Hermann T 2011. Self-assembling RNA square. Proc Natl Acad Sci 108: 6405–6408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding F, Lu C, Zhao W, Rajashankar KR, Anderson DL, Jardine PJ, Grimes S, Ke A 2011. Structure and assembly of the essential RNA ring component of a viral DNA packaging motor. Proc Natl Acad Sci 108: 7357–7362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draper DE 2004. A guide to ions and RNA structure. RNA 10: 335–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K 2004. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60: 2126–2132 [DOI] [PubMed] [Google Scholar]
- Fang Y, Shu D, Xiao F, Guo P, Qin PZ 2008. Modular assembly of chimeric phi29 packaging RNAs that support DNA packaging. Biochem Biophys Res Commun 372: 589–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garver K, Guo P 1997. Boundary of pRNA functional domains and minimum pRNA sequence requirement for specific connector binding and DNA packaging of phage φ29. RNA 3: 1068–1079 [PMC free article] [PubMed] [Google Scholar]
- Grabow WW, Zakrevsky P, Afonin KA, Chworos A, Shapiro BA, Jaeger L 2011. Self-assembling RNA nanorings based on RNAI/II inverse kissing complexes. Nano Lett 11: 878–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guasch A, Pous J, Ibarra B, Gomis-Rüth FX, Valpuesta JM, Sousa N, Carrascosa JL, Coll M 2002. Detailed architecture of a DNA translocating machine: The high-resolution structure of the bacteriophage φ29 connector particle. J Mol Biol 315: 663–676 [DOI] [PubMed] [Google Scholar]
- Guo P 2010. The emerging field of RNA nanotechnology. Nat Nanotechnol 5: 833–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo P, Erickson S, Anderson D 1987. A small viral RNA is required for in vitro packaging of bacteriophage φ29 DNA. Science 236: 690–694 [DOI] [PubMed] [Google Scholar]
- Guo P, Zhang C, Chen C, Trottier M, Garver K 1998. Inter-RNA interaction of phage φ29 pRNA to form a hexameric complex for viral DNA transportation. Mol Cell 2: 149–155 [DOI] [PubMed] [Google Scholar]
- Guo S, Tschammer N, Mohammed S, Guo P 2005a. Specific delivery of therapeutic RNAs to cancer cells via the dimerization mechanism of phi29 motor pRNA. Hum Gene Ther 16: 1097–1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Blocker F, Guo P 2005b. Construction of connector arrays with tetragonal to decagonal transition induced by pRNA of phi29 DNA-packaging motor. J Nanosci Nanotechnol 5: 856–863 [DOI] [PubMed] [Google Scholar]
- Guo S, Huang F, Guo P 2006. Construction of folate-conjugated pRNA of bacteriophage phi29 DNA packaging motor for delivery of chimeric siRNA to nasopharyngeal carcinoma cells. Gene Ther 13: 814–820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo P, Haque F, Hallahan B, Reif R, Li H 2012a. Uniqueness, advantages, challenges, solutions, and perspectives in therapeutics applying RNA nanotechnology. Nucleic Acid Ther 22: 226–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo P, Shu Y, Binzel D, Cinier M 2012b. Synthesis, conjugation, and labeling of multifunctional pRNA nanoparticles for specific delivery of siRNA, drugs and other therapeutics to target cells. Methods Mol Biol 928: 197–219 [DOI] [PubMed] [Google Scholar]
- Haller A, Rieder U, Aigner M, Blanchard SC, Micura R 2011. Conformational capture of the SAM-II riboswitch. Nat Chem Biol 7: 393–400 [DOI] [PubMed] [Google Scholar]
- Haque F, Shu D, Shu Y, Shlyakhtenko L, Rychahou P, Evers M, Guo P 2012. Ultrastable synergistic tetravalent RNA nanoparticles for targeting to cancers. Nano Today 7: 245–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris S, Schroeder SJ 2010. Nuclear magnetic resonance structure of the prohead RNA E-loop hairpin. Biochemistry 49: 5989–5997 [DOI] [PubMed] [Google Scholar]
- Hoeprich S, Guo P 2002. Computer modeling of three-dimensional structure of DNA-packaging RNA (pRNA) monomer, dimer, and hexamer of phi29 DNA Packaging motor. J Biol Chem 277: 20794–20803 [DOI] [PubMed] [Google Scholar]
- Hoeprich S, Zhou Q, Guo S, Qi G, Wang Y, Guo P 2003. Bacterial virus phi29 pRNA as a hammerhead ribozyme escort to destroy hepatitis B virus. Gene Ther 10: 1258–1267 [DOI] [PubMed] [Google Scholar]
- Jimenez J, Santisteban A, Carazo JM, Carrascosa JL 1986. Computer graphic display method for visualizing three-dimensional biological structures. Science 232: 1113–1115 [DOI] [PubMed] [Google Scholar]
- Johnson-Buck AE, McDowell SE, Walter NG 2011. Metal ions: Supporting actors in the playbook of small ribozymes. Met Ions Life Sci 9: 175–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazantsev AV, Krivenko AA, Harrington DJ, Holbrook SR, Adams PD, Pace NR 2005. Crystal structure of a bacterial ribonuclease P RNA. Proc Natl Acad Sci 102: 13392–13397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazantsev AV, Krivenko AA, Pace NR 2009. Mapping metal-binding sites in the catalytic domain of bacterial RNase P RNA. RNA 15: 266–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaled A, Guo S, Li F, Guo P 2005. Controllable self-assembly of nanoparticles for specific delivery of multiple therapeutic molecules to cancer cells using RNA nanotechnology. Nano Lett 5: 1797–1808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura A, Jardine PJ, Anderson DL, Grimes S, Matsuo H 2008. Analysis of intermolecular base pair formation of prohead RNA of the phage φ29 DNA packaging motor using NMR spectroscopy. Nucleic Acids Res 36: 839–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobitski AY, Nierth A, Helm M, Jaschke A, Nienhaus GU 2007. Mg2+-dependent folding of a Diels-Alderase ribozyme probed by single-molecule FRET analysis. Nucleic Acids Res 35: 2047–2059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacheva YS, Tzokov SB, Murray IA, Grasby JA 2004. The role of phosphate groups in the VS ribozyme-substrate interaction. Nucleic Acids Res 32: 6240–6250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laing C, Wen D, Wang JT, Schlick T 2012. Predicting coaxial helical stacking in RNA junctions. Nucleic Acids Res 40: 487–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TJ, Guo P 2006. Interaction of gp16 with pRNA and DNA for genome packaging by the motor of bacterial virus phi29. J Mol Biol 356: 589–599 [DOI] [PubMed] [Google Scholar]
- Leontis NB, Lescoute A, Westhof E 2006. The building blocks and motifs of RNA architecture. Curr Opin Struct Biol 16: 279–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyubchenko YL, Shlyakhtenko LS 2009. AFM for analysis of structure and dynamics of DNA and protein–DNA complexes. Methods 47: 206–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mat-Arip Y, Garver K, Chen C, Sheng S, Shao Z, Guo P 2001. Three-dimensional interaction of Phi29 pRNA dimer probed by chemical modification interference, cryo-AFM, and cross-linking. J Biol Chem 276: 32575–32584 [DOI] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ 2007. Phaser crystallographic software. J Appl Crystallogr 40: 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDowell SE, Jun JM, Walter NG 2010. Long-range tertiary interactions in single hammerhead ribozymes bias motional sampling toward catalytically active conformations. RNA 16: 2414–2426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammad T, Chen C, Guo P, Morrison H 1999. Photoinduced cross-linking of RNA by cis-Rh(phen)2Cl2+ and cis-Rh(phen)(phi)Cl2+: A new family of light activatable nucleic acid cross-linking agents. Bioorg Med Chem Lett 9: 1703–1708 [DOI] [PubMed] [Google Scholar]
- Murray JB, Terwey DP, Maloney L, Karpeisky A, Usman N, Beigelman L, Scott WG 1998. The structural basis of hammerhead ribozyme self-cleavage. Cell 92: 665–673 [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W 1997. Processing of X-ray diffraction data collected in oscillation mode. Macromolecular Crystallography, Pt A 276: 307–326 [DOI] [PubMed] [Google Scholar]
- Pagratis NC 1996. Rapid preparation of single stranded DNA from PCR products by streptavidin induced electrophoretic mobility shift. Nucleic Acids Res 24: 3645–3646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peracchi A, Beigelman L, Scott EC, Uhlenbeck OC, Herschlag D 1997. Involvement of a specific metal ion in the transition of the hammerhead ribozyme to its catalytic conformation. J Biol Chem 272: 26822–26826 [DOI] [PubMed] [Google Scholar]
- Petrov AS, Bowman JC, Harvey SC, Williams LD 2011. Bidentate RNA-magnesium clamps: On the origin of the special role of magnesium in RNA folding. RNA 17: 291–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu X, Smith GJ, Lee KT, Sosnick TR, Pan T, Scherer NF 2008. Single-molecule nonequilibrium periodic Mg2+-concentration jump experiments reveal details of the early folding pathways of a large RNA. Proc Natl Acad Sci 105: 6602–6607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid RJD, Bodley JW, Anderson D 1994a. Characterization of the prohead-pRNA interaction of bacteriophage φ29. J Biol Chem 269: 5157–5162 [PubMed] [Google Scholar]
- Reid RJD, Bodley JW, Anderson D 1994b. Identification of bacteriophage φ29 prohead RNA domains necessary for in vitro DNA-gp3 packaging. J Biol Chem 269: 9084–9089 [PubMed] [Google Scholar]
- Reid RJD, Zhang F, Benson S, Anderson D 1994c. Probing the structure of bacteriophage φ29 prohead RNA with specific mutations. J Biol Chem 269: 18656–18661 [PubMed] [Google Scholar]
- Reyes FE, Garst AD, Batey RT 2009. Strategies in RNA crystallography. Methods Enzymol 469: 119–139 [DOI] [PubMed] [Google Scholar]
- Robertson MP, Scott WG 2008. A general method for phasing novel complex RNA crystal structures without heavy-atom derivatives. Acta Crystallogr D Biol Crystallogr D64: 738–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson MP, Chi YI, Scott WG 2010. Solving novel RNA structures using only secondary structural fragments. Methods 52: 168–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott WG 2007. Ribozymes. Curr Opin Struct Biol 17: 280–286 [DOI] [PubMed] [Google Scholar]
- Shu D, Zhang H, Jin J, Guo P 2007. Counting of six pRNAs of phi29 DNA-packaging motor with customized single molecule dual-view system. EMBO J 26: 527–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu D, Zhang H, Petrenko R, Meller J, Guo P 2010. Dual-channel single-molecule fluorescence resonance energy transfer to establish distance parameters for RNA nanoparticles. ACS Nano 4: 6843–6853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu D, Shu Y, Haque F, Abdelmawla S, Guo P 2011a. Thermodynamically stable RNA three-way junctions for constructing multifuntional nanoparticles for delivery of therapeutics. Nat Nanotechnol 6: 658–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu Y, Cinier M, Fox SR, Ben-Johnathan N, Guo P 2011b. Assembly of therapeutic pRNA-siRNA nanoparticles using bipartite approach. Mol Ther 19: 1304–1311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu Y, Cinier M, Shu D, Guo P 2011c. Assembly of multifunctional phi29 pRNA nanoparticles for specific delivery of siRNA and other therapeutics to targeted cells. Methods 54: 204–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu Y, Shu D, Haque F, Guo P 2013a. Fabrication of pRNA nanoparticles to deliver therapeutic RNAs and bioactive compounds into tumor cells. Nat Protoc (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu Y, Haque F, Shu D, Li W, Zhu Z, Kotb M, Lyubchenko Y, Guo P 2013b. Fabrication of 14 different RNA nanoparticles for specific tumor targeting without accumulation in normal organs. RNA 19: 767–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla GC, Haque F, Tor Y, Wilhelmsson LM, Toulme JJ, Isambert H, Guo P, Rossi JJ, Tenenbaum SA, Shapiro BA 2011. A boost for the emerging field of RNA nanotechnology. ACS Nano 5: 3405–3418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson AA, Tao Y, Leiman PG, Badasso MO, He Y, Jardine PJ, Olson NH, Morais MC, Grimes S, Anderson DL, et al. 2000. Structure of the bacteriophage φ29 DNA packaging motor. Nature 408: 745–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner M, Karunatilaka KS, Sigel RK, Rueda D 2008. Single-molecule studies of group II intron ribozymes. Proc Natl Acad Sci 105: 13853–13858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trottier M, Zhang CL, Guo P 1996. Complete inhibition of virion assembly in vivo with mutant pRNA essential for phage φ29 DNA packaging. J Virol 70: 55–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trottier M, Mat-Arip Y, Zhang C, Chen C, Sheng S, Shao Z, Guo P 2000. Probing the structure of monomers and dimers of the bacterial virus phi29 hexamer RNA complex by chemical modification. RNA 6: 1257–1266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyc K, Steitz JA 1992. A new interaction between the mouse 5′ external transcribed spacer of pre-rRNA and U3 snRNA detected by psoralen crosslinking. Nucleic Acids Res 20: 5375–5382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicens Q, Cech TR 2006. Atomic level architecture of group I introns revealed. Trends Biochem Sci 31: 41–51 [DOI] [PubMed] [Google Scholar]
- Xiao F, Moll D, Guo S, Guo P 2005. Binding of pRNA to the N-terminal 14 amino acids of connector protein of bacterial phage phi29. Nucleic Acids Res 33: 2640–2649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao F, Zhang H, Guo P 2008. Novel mechanism of hexamer ring assembly in protein/RNA interactions revealed by single molecule imaging. Nucleic Acids Res 36: 6620–6632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang CL, Lee C-S, Guo P 1994. The proximate 5′ and 3′ ends of the 120-base viral RNA (pRNA) are crucial for the packaging of bacteriophage φ29 DNA. Virology 201: 77–85 [DOI] [PubMed] [Google Scholar]
- Zhang CL, Garver K, Guo P 1995a. Inhibition of phage φ29 assembly by antisense oligonucleotides targeting viral pRNA essential for DNA packaging. Virology 211: 568–576 [DOI] [PubMed] [Google Scholar]
- Zhang CL, Tellinghuisen T, Guo P 1995b. Confirmation of the helical structure of the 5′/3′ termini of the essential DNA packaging pRNA of phage φ29. RNA 1: 1041–1050 [PMC free article] [PubMed] [Google Scholar]
- Zhang CL, Trottier M, Guo PX 1995c. Circularly permuted viral pRNA active and specific in the packaging of bacteriophage φ29 DNA. Virology 207: 442–451 [DOI] [PubMed] [Google Scholar]
- Zhang CL, Tellinghuisen T, Guo P 1997. Use of circular permutation to assess six bulges and four loops of DNA-Packaging pRNA of bacteriophage φ29. RNA 3: 315–322 [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Lemieux S, Wu X, St.-Arnaud S, McMurray CT, Major F, Anderson D 1998. Function of hexameric RNA in packaging of bacteriophage φ29 DNA in vitro. Mol Cell 2: 141–147 [DOI] [PubMed] [Google Scholar]
- Zhang C, Trottier M, Chen C, Guo P 2001. Chemical modification patterns of active and inactive as well as procapsid-bound and unbound DNA-packaging RNA of bacterial virus Phi29. Virology 281: 281–293 [DOI] [PubMed] [Google Scholar]
- Zhou J, Shu Y, Guo P, Smith D, Rossi J 2011. Dual functional RNA nanoparticles containing phi29 motor pRNA and anti-gp120 aptamer for cell-type specific delivery and HIV-1 inhibition. Methods 54: 284–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang X, Bartley LE, Babcock HP, Russell R, Ha T, Herschlag D, Chu S 2000. A single-molecule study of RNA catalysis and folding. Science 288: 2048–2051 [DOI] [PubMed] [Google Scholar]