In mammalian cells, the cytoplasmic function of Dicer is well established, but evidence for its roles in the nucleus is fragmentary. This work demonstrates that the C-terminal double-stranded RNA binding domain of human Dicer (hDicer) includes a nuclear localization signal that depends on the importin-β family of import factors. The N-terminal helicase domain also contributes to hDicer localization. The authors conclude that hDicer is a shuttling protein that exhibits cytoplasmic localization at steady state.
Keywords: Dicer, helicase, NLS, RNAi, dsRBD
Abstract
Dicer is a key player in microRNA (miRNA) and RNA interference (RNAi) pathways, processing miRNA precursors and double-stranded RNA into ∼21-nt-long products ultimately triggering sequence-dependent gene silencing. Although processing of substrates in vertebrate cells occurs in the cytoplasm, there is growing evidence suggesting Dicer is also present and functional in the nucleus. To address this possibility, we searched for a nuclear localization signal (NLS) in human Dicer and identified its C-terminal double-stranded RNA binding domain (dsRBD) as harboring NLS activity. We show that the dsRBD-NLS can mediate nuclear import of a reporter protein via interaction with importins β, 7, and 8. In the context of full-length Dicer, the dsRBD-NLS is masked. However, duplication of the dsRBD localizes the full-length protein to the nucleus. Furthermore, deletion of the N-terminal helicase domain results in partial accumulation of Dicer in the nucleus upon leptomycin B treatment, indicating that CRM1 contributes to nuclear export of Dicer. Finally, we demonstrate that human Dicer has the ability to shuttle between the nucleus and the cytoplasm. We conclude that Dicer is a shuttling protein whose steady-state localization is cytoplasmic.
INTRODUCTION
Dicer is a large multidomain endoribonuclease that plays a central role in the microRNA (miRNA) and RNA interference (RNAi) pathways. In metazoa, it is required for the biogenesis of miRNAs and small interfering RNAs (siRNAs) and also is involved in an effector step of RNA silencing by participating in the assembly of the RNA-induced silencing complex (RISC) (Jinek and Doudna 2009; Doyle et al. 2012). Dicer proteins are found in most eukaryotes, with copy number and domain structure varying among different organisms. Four Dicer proteins are present in Arabidopsis thaliana, two in Drosophila melanogaster, and one in mammals, Caenorhabditis elegans, Schizosaccharomyces pombe, and some budding yeast species (Doyle et al. 2012). Metazoan and plant Dicers generally contain ATPase/helicase (hereafter referred to as helicase), DUF283 (domain of unknown function), PAZ (Piwi/Argonaute/Zwille), two RNase III domains, and one or two double-stranded RNA-binding domains (dsRBDs) (see Fig. 1A), but Dicers of lower eukaryotes frequently have a less complex domain organization (MacRae et al. 2006; Weinberg et al. 2011; Doyle et al. 2012). Functions of some individual domains of Dicer are not fully understood. For human Dicer, the PAZ and RNase III domains have been most intensively studied and are involved in RNA binding and/or cleavage together acting as a molecular ruler determining product size (Zhang et al. 2004; MacRae et al. 2006, 2007; Park et al. 2011). The helicase distinguishes between perfect duplex or hairpin RNAs, with a higher affinity for the latter. Furthermore, it plays a role in catalysis keeping Dicer in a semi-repressed state (Ma et al. 2008, 2012; Tsutsumi et al. 2011). This domain also mediates interaction between Dicer and its partner, TRBP, that stimulates full-length Dicer activity, thus suggesting that protein cofactors may modulate helicase inhibition (Haase et al. 2005; Kok et al. 2007; Lau et al. 2009; Chakravarthy et al. 2010). Finally, the roles of the DUF293 and dsRBD domains are less understood (Jinek and Doudna 2009; Doyle et al. 2012; Lau et al. 2012).
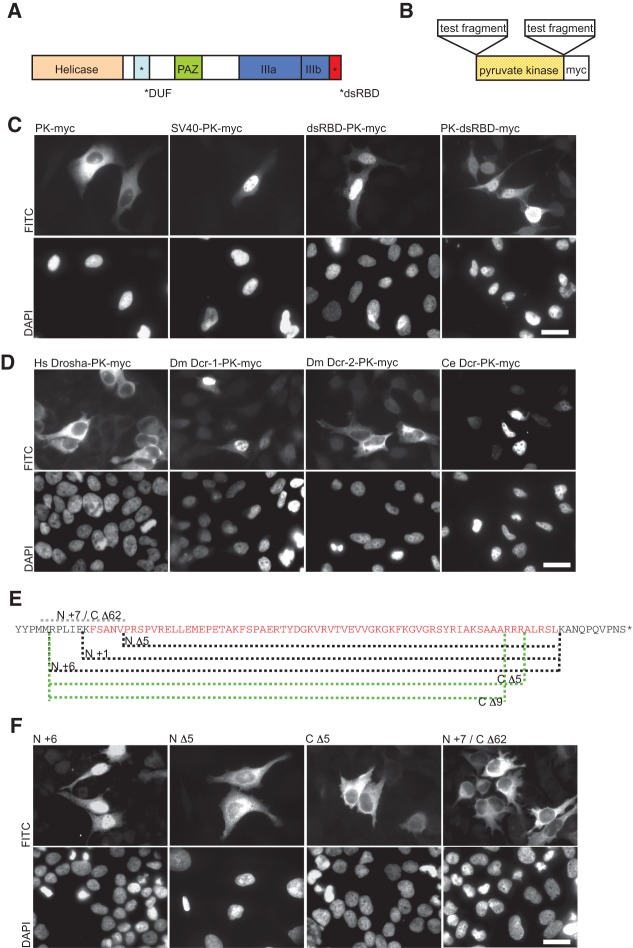
FIGURE 1.
The dsRBD of human Dicer functions as a nuclear localization signal (NLS). (A) Schematic representation of human Dicer with individual protein domains indicated by different colors. (B) The pyruvate kinase (PK-myc) fusion constructs used to assess NLS activity. (C) In transfected HeLa cells, PK-myc localizes to the cytoplasm but can be driven to the nucleus by an SV40 NLS (SV40-PK-myc). dsRBD-PK-myc and PK-dsRBD-myc show strong nuclear accumulation. Staining with DAPI is shown in the lower panel. (D) dsRBDs from various proteins were tested for their ability to function as a NLS in the PK reporter assay as indicated. The dsRBDs from human Drosha (Hs Drosha-PK-myc) and Drosophila Dcr-2 (Dm Dcr-2-PK-myc) were found not to function as a NLS, whereas those of Drosophila Dcr-1 (Dm Dcr-1-PK-myc) and C. elegans Dcr (Ce Dcr-PK-myc) functioned as a NLS. Bar, 20 µm. (E) Sequence of Dicer’s dsRBD (amino acids 1845 to 1912, in red) and its flanking amino acids (in black). The indicated deletions made at either the N or C terminus of the dsRBD are shown by dashed lines. The protein fragments of each dsRBD reporter tested are as follows: N Δ5, 1850 to 1912; N +1, 1844 to 1912; N +6, 1839 to 1912; C Δ5, 1839 to 1907; C Δ9, 1839 to 1903. N +7/C Δ62, including only residues 1837 to 1849, served as an additional control to test if residues spanning gaps between other deletion constructs can function as a NLS. (F) In transfected HeLa cells, Dicer dsRBD N +6 and N +1 (data not shown for N +1) accumulated in the nucleus. N- or C-terminal deletions (N Δ5, C Δ5, and C Δ9 [data not shown for C Δ9]) localize to the cytoplasm. The additional control N +7/C Δ62 also localizes to the cytoplasm. Staining with DAPI is shown in the lower panel. Representative images are shown from a minimum of three independent experiments. Bar, 20 µm.
The molecular mechanism of how Dicer processes its substrates has been studied intensively. In a model based on mutagenic studies of human Dicer (Zhang et al. 2004) and crystallographic structure of the Giardia intestinalis Dicer (MacRae et al. 2006), the enzyme functions as an intra-molecular pseudodimer of RNase IIIa and IIIb domains containing two independent catalytic sites, each capable of cutting one strand of RNA duplex to generate products with 2-nt 3′ overhangs. The end of the dsRNA is recognized by the PAZ domain, and the substrate is placed in the positively charged valley on the surface of the RNase III domains. This model has also been extended to S. pombe (Colmenares et al. 2007). It was further demonstrated that human Dicer not only anchors the 3′ end of the RNA but also the 5′ end, with the position of cleavage being determined by the ∼22-nt distance from the 5′ end (5′ counting rule) (Park et al. 2011). Recently, several studies described the electron microscopy (EM) reconstructions of human Dicer that all reported an L-shaped molecule with morphologically discrete regions (Lau et al. 2009, 2012; Wang et al. 2009). Site-specific tagging positioned the helicase domain at the very base occupying the horizontal arm of the L. Both the RNase III and PAZ domains occupied the vertical arm of the L, with the PAZ at the top and the RNase III at the bottom (Lau et al. 2012). Such an arrangement supports biochemical observations that the distance between the PAZ and RNase III domains acts as a molecular ruler. Importantly, it also accommodates the observed RNA binding by the helicase domain in human Dicer, as its position below the catalytic core allows it to bind both dsRNA and loop segments of its substrates (Ma et al. 2008, 2012; Soifer et al. 2008).
In mammalian cells, the miRNA pathway starts in the nucleus with the transcription of primary miRNA precursors (pri-miRNAs), most of which are initially cleaved by the Drosha/DGCR8 complex to generate precursor-miRNAs (pre-miRNAs). These are then exported to the cytoplasm via Exportin 5 (XPO5). Once in the cytoplasm, processing by Dicer gives rise to the double-stranded siRNA-like form of miRNA (Kim et al. 2009). One strand of the duplex, corresponding to mature miRNA, is then incorporated in the RISC-like miRNP complex, which mediates translational repression of mRNA and/or its deadenylation and degradation (Fabian et al. 2010; Huntzinger and Izaurralde 2011). However, there has been growing—albeit mostly indirect—evidence that Dicer in mammalian cells may also have additional roles in the nucleus. Loss-of-function studies have revealed nuclear phenotypes, including heterochromatic defects such as reduced epigenetic silencing of centromeric repeats and increased telomere recombination and elongation in Dicer-deficient mouse embryonic stem (ES) cells (Kanellopoulou et al. 2005; Benetti et al. 2008). Studies of Dicer-deficient ES cells have also implicated the enzyme as having a role in X chromosome inactivation, although this remains controversial (Nesterova et al. 2008; Ogawa et al. 2008; Kanellopoulou et al. 2009). In the chicken-human hybrid DT40 cells, loss of Dicer leads to premature sister chromatid separation due to abnormalities in heterochromatin formation (Fukagawa et al. 2004). Dicer has also been implicated in regulating intergenic transcription at the human β-globin locus (Haussecker and Proudfoot 2005). Additionally, in mouse oocytes where Dicer was conditionally deleted, phenotypes including multiple disorganized spindles and severe chromosome congression defects were observed (Murchison et al. 2007). More recently, fragments excised in a Dicer-dependent manner from small nucleolar RNAs (snoRNAs) that have properties of miRNAs were described, raising the possibility that Dicer may be active in the nucleus (Ender et al. 2008; Taft et al. 2009). Although well documented, these studies do not address the key question of whether the observed phenotypes result from a direct loss of Dicer from the nucleus or are a consequence of a failure in miRNA biogenesis in the cytoplasm.
To date, most studies have shown that Dicer localizes to the cytoplasm in mammalian cells. However, these studies do not exclude the possibility that Dicer is present, either at low or transient levels, also in the nucleus (Billy et al. 2001; Provost et al. 2002). Supporting this possibility is the observation that Dicer is associated with ribosomal DNA (rDNA) chromatin in mammalian cells (Sinkkonen et al. 2010). Furthermore, use of advanced fluorescence cross correlation microscopy has also suggested that Dicer may be present in the mammalian nucleus (Ohrt et al. 2012). Recently, mammalian Dicer has been implicated in the transcriptional silencing of genes expressing convergent transcripts and in processing of dsRNA in the nucleus (Gullerova and Proudfoot 2012).
In contrast to mammals, Dicer proteins from several lower organisms have been shown to have well-defined roles in the nucleus. One of the best examples applies to S. pombe, where Dicer (Dcr1) is involved in heterochromatin formation at centromeres by processing, in the nucleus, heterochromatic siRNAs that are subsequently loaded into the RNA-induced transcriptional silencing complex (RITS) (Lejeune et al. 2010). Moreover, recent work in S. pombe demonstrated that the extended C-terminal dsRBD of Dcr1 mediates nuclear accumulation as well as nuclear retention of the protein, thus providing a mechanistic insight into the localization of the fission yeast Dicer (Emmerth et al. 2010; Barraud et al. 2011). In Drosophila, Dcr-2 was found associated with transcriptionally active euchromatic regions and shown to interact with core transcription machinery including Pol II. Loss of Dcr-2 was shown to perturb the positioning of Pol II on promoters, which affected transcription (Cernilogar et al. 2011).
In this study, we set out to investigate Dicer localization in mammalian cells. We found that human Dicer dsRBD functions as a nonclassical nuclear localization signal (NLS) that can mediate the nuclear localization of a reporter construct. Mutational analysis identified a basic region on the surface of the dsRBD as essential for its nuclear accumulation. We further demonstrate that Dicer’s dsRBD binds importins β, 7, and 8. Unlike in S. pombe Dicer, the dsRBD-NLS of the human enzyme appears to be inhibited from driving nuclear localization of the full-length protein. However, upon deletion of the helicase domain, Dicer is able to accumulate in the nucleus after CRM1-dependent nuclear export is blocked with the drug leptomycin B (LMB). These observations, together with the results of heterokaryon assays, suggest that human Dicer is a shuttling protein whose steady state localization is cytoplasmic.
RESULTS
dsRBD of human Dicer functions as an atypical NLS
We investigated whether vertebrate Dicer proteins contain conserved sequences with a potential to act as NLSs. Using in silico tools predicting classical mono- and bipartite NLSs, we identified three putative signals starting at residues 740, 788, and 1349 (see Materials and Methods for sequences tested). Minimal and extended versions of each predicted NLS as well as the characterized Simian virus 40 (SV40) NLS were cloned upstream of a myc-tagged chicken pyruvate kinase (PK) open reading frame (ORF) (Fig. 1B). PK is a bona fide cytoplasmic protein that has previously been used as a cargo to test sequences for NLS activity; as PK is over 60 kDa in size, it cannot passively diffuse across the nuclear envelope. We found that none of the three predicted NLSs of human Dicer was able to drive nuclear localization of the PK-myc reporter in HeLa cells (data not shown). The PK fusion with the SV40 NLS, a positive control, localized to the nucleus, while a control PK-myc protein showed cytoplasmic localization (Fig. 1C).
Not all experimentally documented NLSs correspond to classical mono- or bipartite stretches of amino acids, underscoring the divergent nature of nuclear import signals. One example of an atypical NLS is the third dsRBD of the human RNA-editing enzyme adenosine deaminase that acts on RNA 1 (ADAR1), as well as the extended dsRBD of the S. pombe Dicer (Eckmann et al. 2001; Strehblow et al. 2002; Emmerth et al. 2010). As human Dicer also contains a canonical dsRBD at the C terminus, we tested its potential to act as a NLS using the PK-myc reporter. As shown in Figure 1C, the fusion protein, containing the dsRBD fused to either the N or C terminus of PK, efficiently accumulated in the nucleus, indicating that the dsRBD of human Dicer can function as a NLS in a position-independent way.
Given that there are few examples of dsRBDs acting as NLSs and the fact that this domain features prominently in proteins involved in RNA silencing pathways, we investigated whether NLS activity is a more general feature of the dsRBD. Using the PK-myc reporter, we tested dsRBDs from D. melanogaster and C. elegans Dicers, and human Drosha. The dsRBDs from D. melanogaster Dcr-1 and C. elegans Dcr functioned as NLSs, whereas the dsRBDs of D. melanogaster Dcr-2 and human Drosha did not (Fig. 1D). Hence, not all dsRBDs act as NLSs, indicating that this is not a general property of the domain. Multiple sequence alignments were performed to look for any conserved residues that may be shared between those dsRBDs which function as NLSs (see Supplemental Fig. S1A). Overall similarity was poor, with the highest degree of conservation being at the C-terminal end of the domain. No obvious residues were identified that could explain the NLS activity of certain dsRBDs. We also aligned the dsRBDs from various vertebrate Dicers and, strikingly, found 100% conservation at the amino acid level, indicating that all have the potential to function as a NLS (see Supplemental Fig. S1B).
dsRBD integrity, but not RNA-binding potential, is essential for NLS activity
The dsRBD is a structurally conserved motif of ∼70 aa in length found in a wide range of RNA-binding proteins, including Dicer, and other proteins involved in RNA silencing pathways. We investigated whether the integrity of human Dicer’s dsRBD is required for it to function as a NLS. We found that deletions of amino acids flanking the domain had no effect on its ability to target PK-myc to the nucleus (data not shown). However, deletions extending into the predicted minimal dsRBD, either N- or C-terminal, abolished NLS activity (Fig. 1E,F). Hence, the structural integrity of the dsRBD appears to be critical for NLS function.
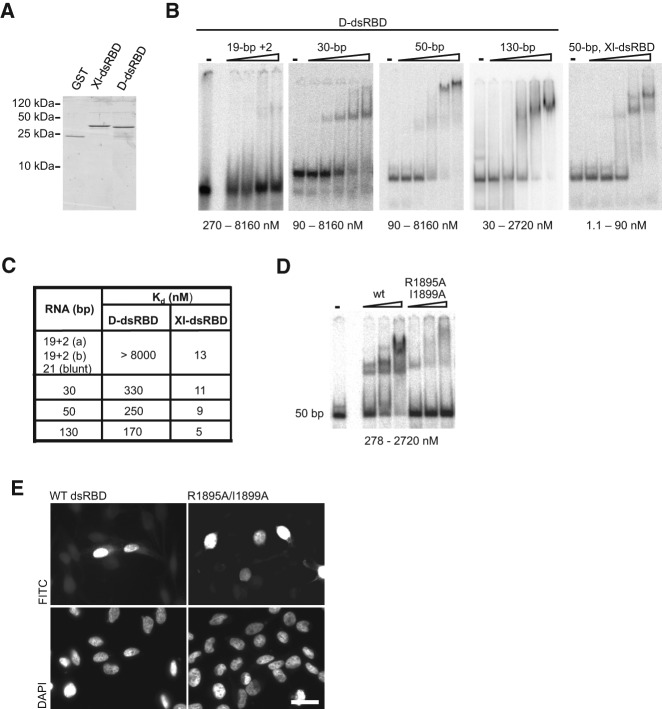
We investigated whether the RNA binding potential of the dsRBD contributes to its NLS activity. First, we assessed the RNA binding properties of the minimal dsRBD that functions as a NLS. Using gel mobility shift assays, the ability of the purified recombinant dsRBD fused to GST (Fig. 2A) to bind dsRNAs of 130, 70, 50, 30, 21, and 19 bp in length was tested (Fig. 2B). The two used 19-bp dsRNAs contained 2-nt 3′ overhangs, thus they mimicked siRNAs. Activity of Dicer’s dsRBD was compared with that of the dsRBD2 of Xenopus laevis RNA-binding protein A (XlrbpA), previously shown to bind dsRNA very robustly (Fig. 2A; St. Johnston et al. 1992; Krovat and Jantsch 1996). Both analyzed dsRBDs formed complexes with 130-, 70-, 50- and 30-bp dsRNAs, although XlrbpA dsRBD2 bound dsRNAs with an affinity ∼30 times higher than Dicer’s dsRBD (Fig. 2B,C). XlrbpA dsRBD2 was also able to bind 21- and 19-bp dsRNAs with affinity comparable to that of longer substrates. In contrast, Dicer dsRBD showed much lower affinity for the siRNA-like molecules, irrespective of them containing 3′ overhangs or blunt termini (Fig. 2B,C). GST alone did not bind any of the tested dsRNAs (data not shown). Recently, another study reported the RNA-binding characteristics of the human Dicer dsRBD as an isolated domain, comparing it to the first dsRBD of DGCR8 (Wostenberg et al. 2012). The reported Kd values for Dicer-dsRBD ranged from ∼2 to 10 μM, with the affinity of binding decreasing with decrease of the substrate length, which is similar to our findings. However, the difference between a 33- or 44-bp dsRNA and the siRNA-like 22-bp substrate was less dramatic than that revealed by our experiments. Additionally, the Dicer-dsRBD was also shown to be incapable of differentiating between pre-miRNAs and dsRNA substrates (Wostenberg et al. 2012).
FIGURE 2.
RNA-binding potential of Dicer dsRBD is not required for NLS activity. (A) Dicer’s dsRBD (D-dsRBD) and the second dsRBD from the Xenopus laevis RNA-binding protein A (XlrbpA) (Xl-dsRBD) were expressed as GST-fusion proteins and purified from Escherichia coli. (B) To analyze the D-dsRBD RNA-binding properties, native gel mobility shift assays were performed three or more times with increasing amounts of recombinant D-dsRBD and substrates of differing length as indicated. D-dsRBD bound 30-, 50-, and 130-bp-long dsRNAs, but only little binding of the shorter 21- or 19-bp substrates was observed at protein concentrations tested. Two independent sequences for the 19-bp substrates containing 2-nt 3′-overhangs, 19 + 2 (a) and 19 + 2 (b), were tested. (C) Apparent binding affinities for the different substrates were measured for D-dsRBD and the Xl-dsRBD using the representative experiments shown in panel B. D-dsRBD bound longer dsRNAs with an affinity ∼30 times lower compared to the Xl-dsRBD. (D) Mutational analysis of D-dsRBD was performed (also see Supplemental Fig. S2) to identify residues important for RNA-binding. Introduction of the double point mutation (R1895A/I1899A) severely abrogated RNA-binding. (E) The NLS activity of the double point mutant (R1895A/I1899A) was tested in the PK-myc reporter assay and compared to the wild-type (WT) domain. Both fusion proteins efficiently accumulated in the nucleus. Staining with DAPI is shown in the lower panel. Representative images are shown from a minimum of three independent experiments. Bar, 20 µm.
Next, we generated a panel of mutants expected to affect dsRNA binding by Dicer’s dsRBD (see Supplemental Fig. S2A). Design of the mutants was originally based on structural data for the XlrbpA dsRBD2 complexed with dsRNA (Ryter and Schultz 1998) and an extensive mutational analysis of this dsRBD (Krovat and Jantsch 1996). However, as the crystal structure of the mouse Dicer dsRBD (sequences of mouse and human Dicer dsRBDs are identical) became available (Du et al. 2008), the relevant mutations shown in Supplemental Figure S2B are placed on a backbone of the mouse Dicer dsRBD docked to dsRNA. Five single point mutant proteins were purified as GST fusions and tested for their ability to bind 50- and 130-bp dsRNAs. Binding of S1896A was unaffected, while binding activity of mutants R1895A, Y1897A, R1898A, and I1899A was reduced by approximately two- to ninefold (see Supplemental Fig. S2C; data not shown). We also tested activity of proteins containing multiple mutations and found that the double mutant R1895A/I1899A severely abrogated RNA-binding (Fig. 2D; data not shown).
We compared NLS activity of dsRBD containing the double mutation R1895A/I1899A with that of the wild-type (wt) domain. Both efficiently drove nuclear localization of the PK-myc reporter (Fig. 2E), indicating that RNA-binding potential of the dsRBD plays no major role in the NLS function of the domain. We conclude that the human Dicer dsRBD has two separable functions: dsRNA-binding and NLS activity.
The dsRBD of human Dicer binds the nuclear transport receptors Impβ, Imp7, and Imp8
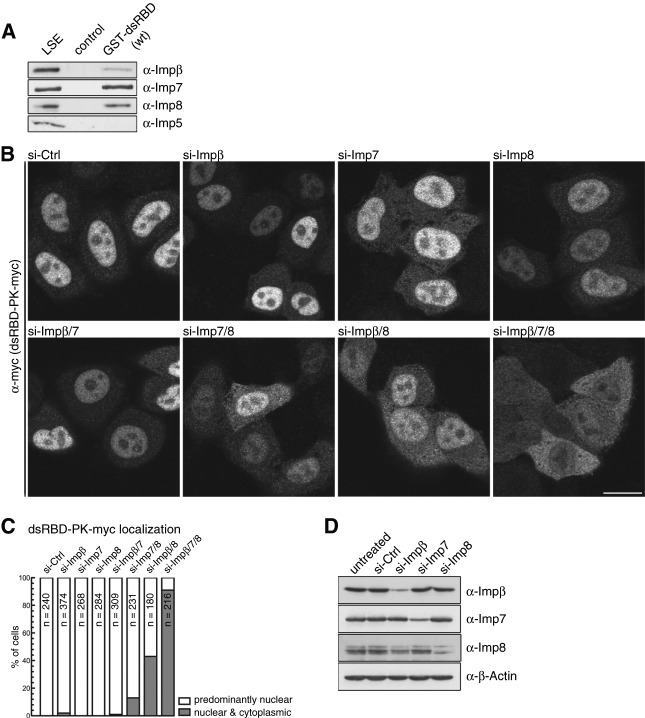
Our demonstration that Dicer's dsRBD can function as a NLS led us to test if any of the known nuclear import receptors (importins) interact with the domain. Importins recognize their cargo NLS and mediate translocation of the bound protein to the nucleus through the nuclear pore complex (NPC). Given that the superfamily of importin β-related proteins mediates the majority of nuclear transport processes in metazoa and yeast, we tested members of this family for their ability to interact with the dsRBD. The overexpressed recombinant Dicer dsRBD fused to GST was immobilized on glutathione (GSH)-sepharose and incubated with HeLa cell extract. Bound proteins were eluted and analyzed by Western blotting using antibodies specific for human Impβ, Imp7, Imp8, and Imp5. As shown in Figure 3A, Impβ, Imp7, and Imp8 associated with the Dicer dsRBD, whereas Imp5 did not. The observation that Imp5 does not interact with Dicer's dsRBD indicated that not all importin β-related importins bind Dicer's dsRBD. This was also the case for transportin 1 and transportin 2, which were found not to interact with the Dicer dsRBD in vitro (data not shown).
FIGURE 3.
The dsRBD of human Dicer binds the nuclear transport receptors Impβ, Imp7, and Imp8. (A) Recombinant, purified GST-dsRBD (wt) was immobilized on glutathione (GSH)-sepharose beads and incubated with low-salt HeLa cell extract (LSE). Bound proteins were eluted and separated by SDS-PAGE, followed by immunoblotting using antibodies directed to human Impβ, Imp7, Imp8, and Imp5. Control, GSH-sepharose beads alone. (B) Simultaneous depletion of Impβ, Imp7, and Imp8 results in a cytoplasmic accumulation of dsRBD-PK-myc. Tetracycline-inducible dsRBD-PK-myc reporter cells were transfected with indicated siRNAs. Sixty hours after transfection, PK-dsRBD-myc expression was induced for 12 h. Cells were then fixed, followed by immunofluorescence (IF) using an α-myc antibody. Bar, 20 μm. (C) Quantification of the experiment shown in B. Cells displaying predominantly nuclear or nuclear and cytoplasmic localization of dsRBD-PK-myc were counted. (N) Number of counted cells. (D) HeLa cells were transfected with siRNAs targeting different importins. Seventy-two hours post-transfection, cells were harvested, and the protein levels were analyzed by Western blotting. β-actin served as a loading control.
To confirm that Impβ, Imp7, and Imp8 play a role in mediating the import of Dicer dsRBD in vivo, we generated a HeLa cell line inducibly expressing a dsRBD-PK-myc reporter. RNAi was used to deplete each of these importins in this cell line, and localization of the reporter was assessed by counting cells and classifying them as having either predominantly nuclear or both nuclear and cytoplasmic localization of the reporter. As shown in Figure 3B (top panel), depletion of Impβ, Imp7, or Imp8 individually had no obvious effect on the nuclear accumulation of the dsRBD-PK-myc reporter. The specificity of each siRNA targeting individual importins was confirmed by Western blotting (Fig. 3D). As many importins work redundantly with each other (Jakel and Gorlich 1998), we tested the effect of depleting combinations of Impβ, Imp7, and Imp8. Simultaneous depletion of Impβ and Imp7 had no effect on nuclear accumulation of the reporter, but depletion of Imp7 and Imp8 resulted in a low level of its cytoplasmic localization (∼10% of cells showing both nuclear and cytoplasmic localization of dsRBD-PK-myc) (Fig. 3B, lower panel; Fig. 3C). Upon depletion of Impβ together with Imp8, 40% of cells showed nuclear and cytoplasmic localization, and depletion of all three importins had the strongest effect, with 90% of cells showing nuclear and cytoplasmic localization of the reporter (Fig. 3B,C). Therefore, it appears that Impβ, Imp7, and Imp8 all play a role in mediating the import of Dicer dsRBD to the nucleus.
Dicer dsRBD contains a basic region critical for NLS activity
We sought biochemical evidence supporting the interaction of the selected importins with Dicer dsRBD. First, we analyzed the interaction of wt GST-dsRBD with recombinant Impβ or Imp7 (Imp8 was not tested, as it was unavailable) expressed in E. coli. As shown in Figure 4A, GST-dsRBD efficiently pulled down both Impβ and Imp7 from the bacterial lysates supplemented with the purified proteins. We also tested if the interaction of GST-dsRBD with either Impβ or Imp7 is sensitive to the presence of RanGTP. In general, importins have a high affinity for the GTP form of the GTPase Ran, resulting in dissociation of importins from their substrates in the nucleus. We added a mutant form of Ran locked in the GTP bound state (RanQ69L) to the pull-down assay. The presence of RanQ69L(GTP) prevented the interaction of both Impβ and Imp7 with GST-dsRBD, indicating that the interaction is disrupted by RanGTP (Fig. 4A). Notably, Impβ and Imp7 can either work independently (Jakel and Gorlich 1998) or heterodimerize to support nuclear import of certain cargo in a cooperative manner (Jakel et al. 1999). Binding of either Impβ or Imp7 alone to histone H1 is inefficient compared to the interaction of the heterodimer (Jakel et al. 1999). In the case of the dsRBD, binding of Impβ and Imp7 was efficient in the absence of the other partner, indicating that heterodimerization is not a prerequisite for interaction with the dsRBD (Fig. 4A).
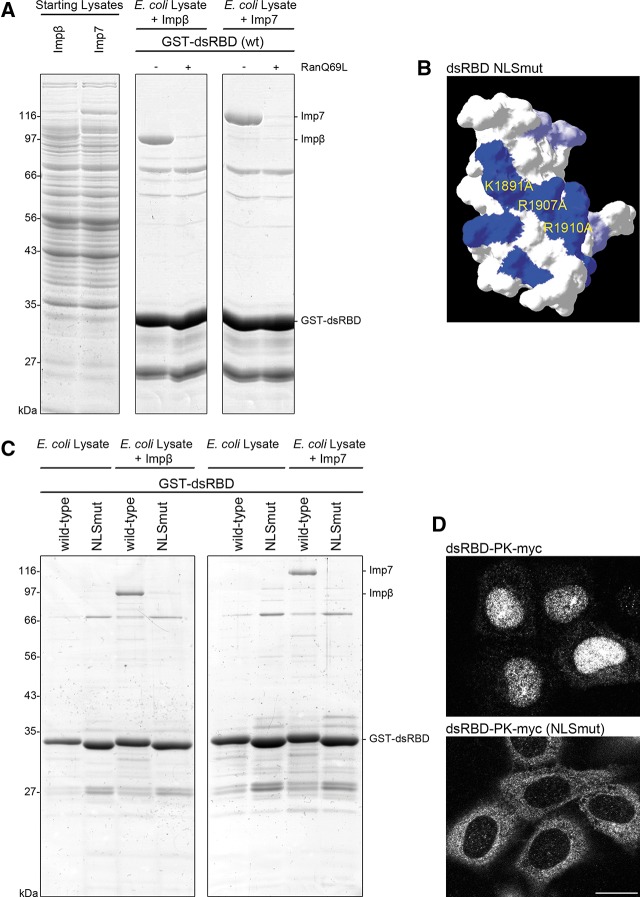
FIGURE 4.
Dicer’s dsRBD contains a basic region critical for NLS activity. (A) Recombinant GST-dsRBD (wt) was immobilized on GSH-sepharose beads and mixed with the indicated purified factors (Impβ, Imp7) added to E. coli lysate. Bound proteins were eluted and separated by SDS-PAGE followed by Coomassie staining. (B) Representation of the basic patch in Dicer’s dsRBD (blue, with positions of mutated residues indicated). Mutation of its amino acid residues to alanines destroyed the dsRBD NLS activity. Mutation of two additional basic residues, K1887 and K1889, to alanines had no appreciable effect on nuclear localization of the dsRBD-PK reporter (data not shown). (C) Binding of Impβ and Imp7 to Dicer’s dsRBD is dependent on the integrity of the basic patch. The experiment was performed as described in A, comparing Impβ and Imp7 recruitment to the wt and NLSmut dsRBD. (D) HeLa cells were transiently transfected with dsRBD-PK-myc or dsRBD-PK-myc NLSmut (K1891A/R1907A/R1910A). Twenty-four hours post-transfection, cells were fixed and immunofluorescence (IF) was performed with an α-myc antibody. Representative images are shown from a minimum of three independent experiments. Bar, 20 μm.
As discussed above, sequence alignments of metazoan Dicer dsRBDs having NLS activity failed to highlight any obvious consensus that would explain its NLS function. Classical mono- and bipartite NLSs contain stretches of structurally exposed basic residues that are recognized by the import machinery (Fried and Kutay 2003). To identify residues or structural elements that could explain the NLS function of human Dicer dsRBD, its three-dimensional structure (Du et al. 2008) was compared with the structures of XlrbpA dsRBD2 (Ryter and Schultz 1998) and Trbp dsRBD2 (Yamashita et al. 2011), neither of which functions as an NLS (M Jantsch, pers. comm.; M Doyle et al., unpubl.). Inspection of the non-RNA-binding surfaces of these dsRBDs revealed that Dicer dsRBD contains a patch of basic amino acids which are not present in the other dsRBD domains (Fig. 4B; see Supplemental Fig. S3). We mutated three residues in the basic patch, K1891, R1907, and R1910, to alanines (NLSmut) and compared the ability of the wt GST-dsRBD and the NLSmut GST-dsRBD to interact with either Impβ or Imp7. Purified GST-dsRBD fusions were incubated with E. coli lysates, either control or expressing the indicated importins. As shown in Figure 4C, the wt GST-dsRBD fusion could effectively interact with both Impβ and Imp7, whereas the NLSmut GST-dsRBD fusion had a reduced ability to interact with Impβ and did not interact with Imp7.
To further verify the importance of K1891, R1907, and R1910 residues for NLS function, we mutated them to alanines in the dsRBD-PK reporter protein and tested the effect on its localization. As shown in Figure 4D, the wt reporter protein localized to the nucleus (top panel), whereas the NLSmut reporter showed cytoplasmic localization. Two other basic residues found close to but not within the patch (K1887 and K1889) had no appreciable effect on nuclear localization of the dsRBD-PK reporter protein when mutated to alanines (data not shown).
dsRBD is occluded from functioning as a NLS when present in full-length Dicer
Although Dicer’s dsRBD can function as a NLS when placed either N- or C-terminally to pyruvate kinase and can interact with Impβ, Imp7, and Imp8, a similar role in the context of a full-length Dicer protein remains undefined. Previously, we showed that both the endogenous and ectopically expressed Dicer appear to localize to the cytoplasm of different mammalian cells (Billy et al. 2001). We considered three different scenarios to explain this paradox: (1) one of Dicer’s domains interacts with cytoplasmic components, effectively retaining the protein in the cytoplasm; (2) in the context of a full-length protein, the dsRBD is masked either by another Dicer domain and/or RNA bound to the enzyme, preventing the Impβ, Imp7, and Imp8 interaction; or (3) Dicer shuttles between the cytoplasm and the nucleus, and its cytoplasmic localization simply reflects the steady-state status of the protein.
To start to address these different possibilities, we first fused an SV40 NLS to the C terminus of the full-length Dicer protein. In another construct, Dicer’s dsRBD was duplicated at the end of the protein (Fig. 5A). Localization of wt Dicer and the two fusion proteins was then compared. While wt Dicer localized to the cytoplasm, both fusion proteins, Dicer-SV40 and Dicer-dsRBD, accumulated in the nucleus (Fig. 5B). The nuclear localization of Dicer-SV40 and Dicer-dsRBD argues against a retention signal or trans-acting factors preventing Dicer import to the nucleus and, instead, suggests that the dsRBD NLS is masked or inhibited either by another Dicer domain or associated RNA. To address the latter possibility, we first removed Dicer domains N-terminal to the RNase IIIa domain. Deletion up to the start of this domain did not result in nuclear localization of the remaining C-terminal fragment of Dicer (Fig. 6A, upper left panel). No further deletions were performed since removal of RNase IIIa would disrupt the integrity of the RNase IIIa/b pseudodimer catalytic domain as well as create a protein small enough to passively diffuse into the nucleus.
FIGURE 5.
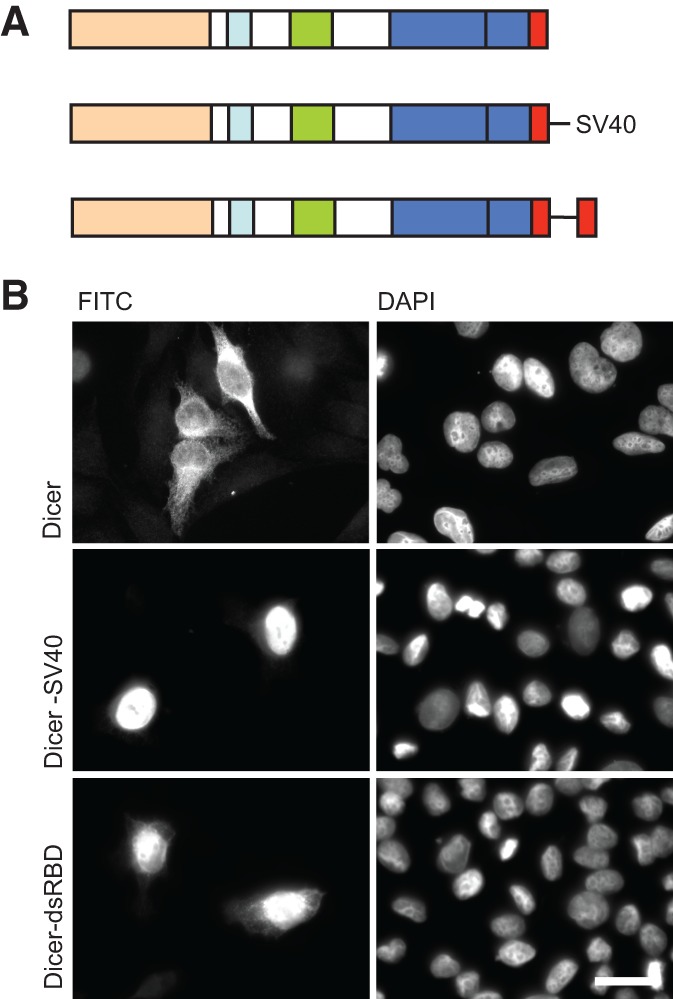
Dicer dsRBD is occluded in the full-length protein. (A) Schematic representation of human Dicer and constructs which have been tested for localization. (B) Transfection of HeLa cells reveals that nuclear localization of full-length Dicer (Dicer) can be achieved either by the addition of an SV40 NLS at the C terminus (Dicer-SV40) or by duplicating the dsRBD (Dicer-dsRBD). Representative images are shown from a minimum of three independent experiments. Bar, 20 µm.
FIGURE 6.
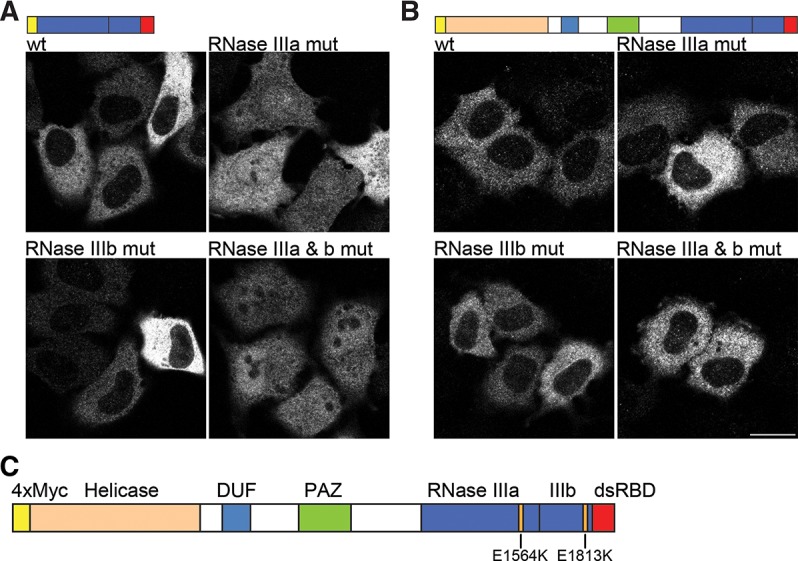
RNA-binding influences Dicer localization. (A) Transfection of HeLa cells with the C-terminal fragment of Dicer (RNase III domains plus dsRBD). The wild-type (wt) construct fails to localize to the nucleus. When E1564K is introduced into RNase IIIa catalytic core (RNase IIIa mut), nuclear accumulation can be observed. A similar mutation (E1813K) in the catalytic core of RNase IIIb (RNase IIIb mut) also leads to partial localization in the nucleus, although the effect is much weaker compared to RNase IIIa mut. The double mutation E1564K/E1813K showed strong nuclear accumulation. (B) Transfection of HeLa cells with full-length Dicer constructs. Wild-type (wt) Dicer localizes to the cytoplasm. Introduction of mutations E1564K or E1813K into the RNase III domains either individually (RNase IIIa mut and RNase IIIb mut) or together (RNase IIIa & b mut) showed cytoplasmic staining similar to wt protein. Representative images are shown from a minimum of three independent experiments. Bar, 20 µm. (C) Schematic representation of human Dicer with individual protein domains indicated by different colors and the position of each RNase III mutation shown.
Taken together, the data indicate that the failure of full-length Dicer to accumulate in the nucleus is not due to the protein being retained in the cytoplasm and that RNase III domains have an intrinsic negative effect on the NLS function of the dsRBD.
RNA binding potential influences localization of the RNase III/dsRBD Dicer fragment
As described above, the presence of the catalytic RNase III domain adjacent to the dsRBD is sufficient to prevent its NLS function. The catalytic domain of Dicer contains an extended surface, greatly contributing to the substrate interaction with the enzyme, and we reasoned that RNA-binding could influence NLS activity, possibly by making the dsRBD inaccessible to Impβ, Imp7, and Imp8. Since the dsRBD showed NLS activity as a separated entity, we reasoned that binding of RNA to the RNase III domains of Dicer might possibly interfere with nuclear import in the context of Dicer. Thus, we generated single point mutations in the RNase IIIa and RNase IIIb domains, E1564K and E1813K, respectively (Fig. 6C). Both residues are at equivalent positions within their respective domains and have previously been shown to be critical for dsRNA processing by Dicer (Zhang et al. 2004). Importantly, crystallographic study of the bacterial RNase III bearing equivalent E to K substitutions indicated that these mutations prevent Mg2+ and dsRNA binding to the catalytic core without compromising overall structural integrity of the catalytic domain. Instead, the mutated bacterial RNase III adopts an alternative “open” structure with only its dsRBD capable of interacting with dsRNA (Blaszczyk et al. 2004). Localization of the Dicer protein fragment encompassing RNase III and dsRBD domains, and bearing either single point mutations or simultaneous mutations in both RNase III domains, was tested in transfected HeLa cells. A single point mutation in RNase IIIa (E1564K; RNase IIIa mut) was sufficient to allow nuclear accumulation of this C-terminal fragment; mutation in RNase IIIb (E1813K; RNase IIIb mut) also resulted in a weak accumulation in the nucleus. The double RNase III mutation (E1564K/E1813K; RNase IIIa & b mut) had a similar localization to the single mutation in RNase IIIa (Fig. 6A). Surprisingly, in the context of the full-length protein, the E to K mutations in RNase III domains did not lead to appreciable protein relocalization to the nucleus (Fig. 6B).
The helicase domain influences Dicer localization
We next investigated if domains other than RNase IIIa and b that are known to play a role in RNA binding contribute to masking of the dsRBD-NLS. Therefore, additional mutations known to affect RNA binding were introduced in the PAZ (F960A/Y971A/Y972A) (Lingel et al. 2003; Song et al. 2003; Yan et al. 2003) and dsRBD domains (R1895A/I1899A; see above), yielding an “RNA-binding-deficient” full-length Dicer. However, as shown in Figure 7, A and B, introduction of these point mutations did not alter the cytoplasmic localization of the protein. Since the helicase domain of Dicer also contributes to substrate RNA binding (Ma et al. 2008; Soifer et al. 2008; Lau et al. 2012), we also tested the effect of its deletion, in both the context of a wild-type and RNA-binding-deficient mutant Dicer, on localization of the protein. We found that neither of the helicase-domain-deleted proteins accumulated in the nucleus (Fig. 7A,B).
FIGURE 7.
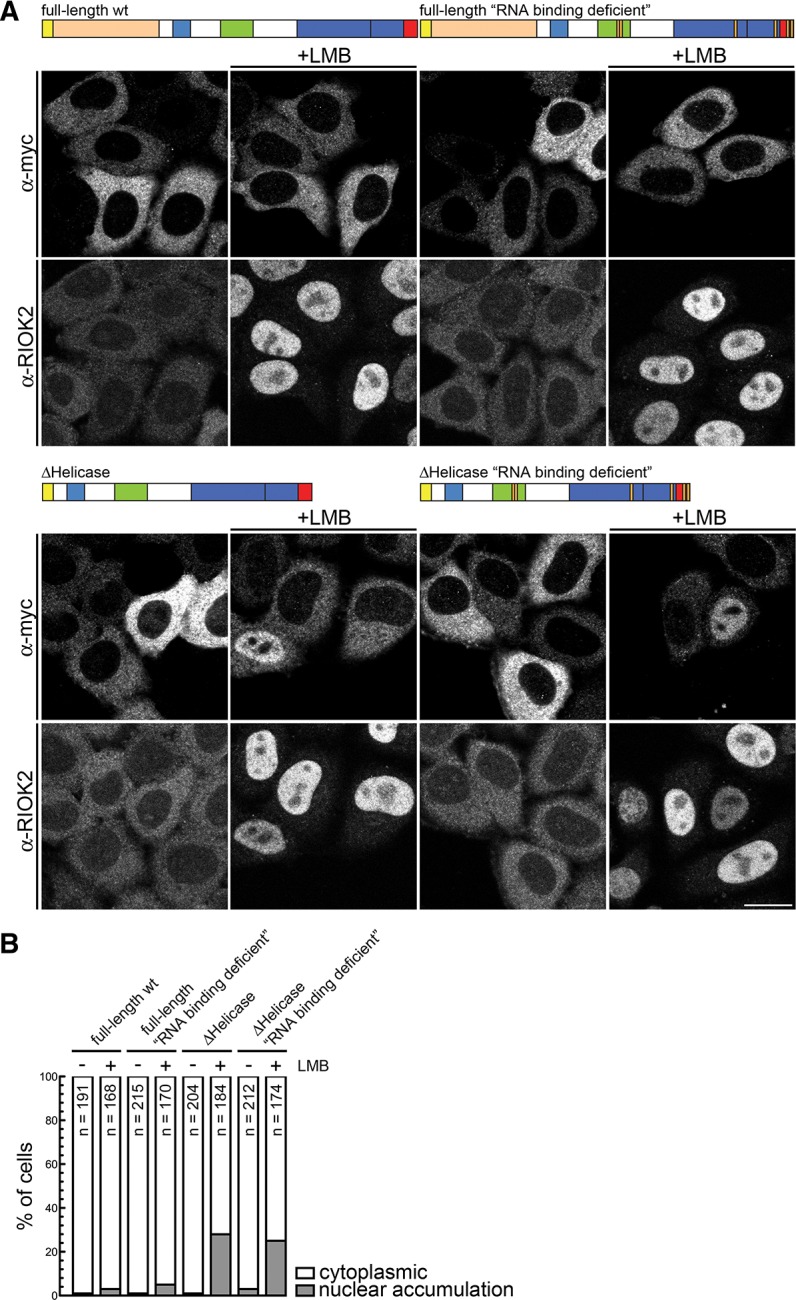
The helicase domain plays a central role in the cytoplasmic retention of Dicer. (A) HeLa cells were transfected with the indicated constructs. Twenty-four hours post transfection, cells were treated with 20 nM LMB or solvent (ethanol) for 10 h and fixed, followed by IF using an α-myc antibody. IF against RIOK2 served as a positive control for the LMB treatment. Bar, 20 μm. (B) Quantification of the experiment shown in A. Cells displaying either cytoplasmic localization or nuclear accumulation of the constructs were counted. (N) Number of counted cells.
The results presented above suggested that domains N-terminal to the RNase IIIa domain inhibit nuclear localization of Dicer independent of the RNA-binding potential of the enzyme. However, these observations do not preclude the scenario that Dicer shuttles between the cytoplasm and the nucleus (scenario III, as discussed above), and its cytoplasmic localization simply reflects the steady-state status of the protein. To address this possibility, we first tested whether blocking CRM1-dependent nuclear export with the drug leptomycin B would result in the nuclear accumulation of full-length Dicer or some of its mutants. No obvious nuclear accumulation of full-length Dicer, either the wild-type or RNA-binding-deficient mutant, was observed upon LMB treatment, whereas the shuttling protein RIO Kinase 2 (RIOK2) that served as a positive control efficiently accumulated in the nucleus (Fig. 7A,B). Interestingly, mutants bearing deletion of the helicase domain, in either wt or RNA-binding-deficient context, showed partial nuclear accumulation of the protein (Fig. 7A,B), indicating that the helicase domain plays an important role in the cytoplasmic accumulation of Dicer. Additionally, this observation suggested the presence of a CRM-1-dependent nuclear export signal (NES) in Dicer protein.
Dicer has the potential to shuttle between the nucleus and the cytoplasm
Our observation that deletion of the helicase domain results in LMB-dependent accumulation of Dicer in the nucleus raised a possibility that Dicer shuttles between the cytoplasm and nucleus. To address this possibility, we made use of the demonstration that Dicer-SV40 and Dicer-dsRBD show nuclear localization. We, therefore, assayed for shuttling of the full-length protein by performing interspecies heterokaryon assays. A GFP version of Dicer-SV40 was cotransfected with either a positive (hnRNPA1) (Fritz et al. 2009) or negative (hnRNPC) (Nakielny and Dreyfuss 1996) protein shuttling control into HeLa cells. After overnight incubation to allow for protein expression, the transfected cells were incubated with cycloheximide to inhibit de novo protein synthesis, then fused with mouse NIH 3T3 cells and incubated for a further 3 h in the presence of cycloheximide. As shown in Figure 8, the Dicer-SV40 protein was able to shuttle between the nucleus and the cytoplasm as indicated by its nearly equal distribution between the HeLa and the NIH 3T3 cell nuclei. A similar relocalization indicative of shuttling was observed for hnRNPA1, while hnRNPC, a nonshuttling protein, was only detectable in HeLa nuclei. The observed ability of Dicer-SV40 to shuttle confirms the presence of a functional NES in Dicer protein.
FIGURE 8.
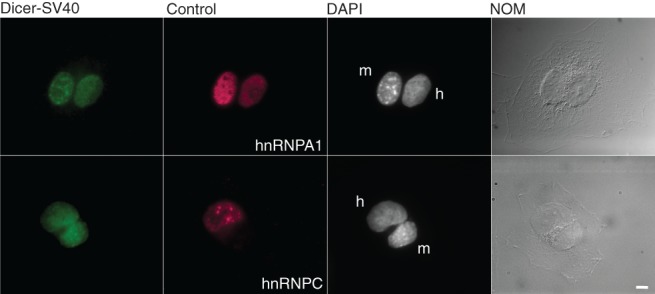
Dicer has the potential to shuttle. Heterokaryon analysis of Dicer-SV40. HeLa cells were cotransfected with plasmids expressing Dicer-SV40 and either hnRNPA1 (positive shuttling control; top panel) or hnRNPC (negative shuttling control; bottom panel). The transfected cells were fused with nontransfected mouse NIH 3T3 cells. Dicer-SV40 was detected in both HeLa and NIH 3T3 nuclei (top and bottom panels) as was hnRNPA1 (top panel). hnRNPC was only detected in the HeLa nuclei (bottom panel). Nuclei were distinguished by DAPI staining and are indicated. (h) Human HeLa cells, (m) mouse NIH 3T3 cells. Bar, 5 µm.
DISCUSSION
In this study, we identified the C-terminal dsRBD of human Dicer as a nonclassical NLS that can efficiently drive the nuclear localization of a reporter construct. Dicer’s dsRBD functions as a NLS independent of its RNA binding potential and interacts with importins β, 7, and 8, which mediate its nuclear import. A physiological function of the identified Dicer NLS remains puzzling given the fact that the endogenous as well as ectopically expressed full-length protein localizes to the cytoplasm (Billy et al. 2001; Provost et al. 2002; this work). However, we found that upon deletion of the N-terminal helicase domain combined with inhibition of nuclear export, the truncated Dicer protein does accumulate in the nucleus. Together with our findings that Dicer fused to the ectopic NLS can shuttle between the nucleus and cytoplasm in heterokaryon assays, these data argue that endogenous Dicer might be a shuttling protein but localizes to the cytoplasm under steady-state conditions. We propose that shuttling of Dicer may be regulated by the helicase domain and in part also by the enzyme’s RNA binding status.
The function of Dicer’s dsRBD
dsRBDs are conserved RNA binding domains found in organisms ranging from yeast to man and feature prominently in proteins involved in the RNAi pathway. Structural data demonstrated that dsRBDs have a canonical αβββα topography and interact with the sugar-phosphate backbone of dsRNA without any obvious sequence specificity (Doyle and Jantsch 2002). Previous analyses of Dicer proteins focused mainly on their biochemical and structural properties, with most attention paid to domains other than the dsRBD, most notably the PAZ and RNase III domains. However, some studies also provide data for the role of the dsRBD within human Dicer. For example, it was shown in biochemical cleavage assays that deletion of the dsRBD lowers enzymatic activity. Given that dsRBDs do not recognize any primary sequence, it was proposed that the domain has a nonspecific role in helping to stabilize the interaction of Dicer with its substrate RNA (Zhang et al. 2004). In a more recent study, it was confirmed that deletion of the dsRBD from human Dicer significantly reduced the ability of the enzyme to cleave substrate RNAs (Ma et al. 2008). However, this study also showed that deletion of the dsRBD did not significantly affect binding of the truncated protein to RNA (Ma et al. 2008). Our data and those of Wostenberg et al. (2012) demonstrate that human Dicer dsRBD binds dsRNA with low micromolar or sub-micromolar affinity for RNA, hence less efficiently when compared to a dsRBD that has robust RNA-binding potential, e.g., dsRBD2 of XlrpA. Therefore, although Dicer’s dsRBD likely contributes to binding of substrate RNA, its exact role in dsRNA processing still remains to be determined.
In this study, we add a new facet to the functional repertoire of Dicer’s dsRBD. We provide evidence that the dsRBD of human Dicer functions as a NLS as (1) it can drive the nuclear localization of a reporter protein, (2) when duplicated at the end of the full-length protein, it causes it to accumulate in the nucleus, (3) it directly interacts with nuclear import receptors (Impβ, Imp7, and Imp8), and (4) it contains a basic patch of exposed amino acids that are required for NLS activity.
Impβ, Imp7, and Imp8 mediate dsRBD nuclear translocation
We found that Impβ, Imp7, and Imp8, all members of the karyopherin family of importins and exportins, are all involved in mediating import of Dicer’s dsRBD to the nucleus. Only depletion of all three importins strongly affected the localization of the dsRBD reporter protein. Generally, each importin recognizes a distinct set of proteins, RNAs, or complexes, thus allowing multiple transport pathways across the NPC (Xu et al. 2010). However, it is also known that importins can work redundantly with each other, e.g., in nuclear import of ribosomal proteins or core histones (Jakel and Gorlich 1998; Muhlhausser et al. 2001). Our data indicate that Dicer’s dsRBD can enter the nucleus via multiple pathways. It is interesting to note that Imp8 was recently shown to mediate nuclear import of the RISC component Ago2 in human cells (Weinmann et al. 2009). One of the other members of the karyopherin family we tested, transportin 1, did not interact with Dicer dsRBD even though it had been demonstrated to be the import receptor for human ADAR1, whose third dsRBD acts as a NLS (Fritz et al. 2009). Consequently, dsRBD NLSs do not share common import pathways but instead can utilize different ones to enter the nucleus.
With the availability of a crystal structure for the mammalian Dicer dsRBD (Du et al. 2008), it was possible to investigate the structural basis of its function as a NLS, given that primary sequence analysis and reporter assays did not identify an obvious classical NLS. Examination of the surface of this structure revealed a patch of exposed basic amino acids whose mutation abolished NLS activity of the dsRBD. Importantly, these mutations did not affect RNA binding, indicating that the overall folding of the dsRBD remained intact in our assays, letting us conclude that the dsRBD performs two separable functions: RNA-binding and NLS activity. As with other NLS classes, such as the classical NLS and BIB domain (Jakel and Gorlich 1998), Dicer’s dsRBD shares the common characteristic of exposed basic amino acids. As shown by us, these residues are critical for its interaction with Impβ and 7. Although the residues in the Dicer dsRBD NLS are separated from each other in the primary sequence, they are brought into proximity in the three-dimensional structure of the domain.
The helicase domain influences the ability of human Dicer to shuttle between the cytoplasm and nucleus
Our observation that the dsRBD of human Dicer, like that of S. pombe Dcr1 (Emmerth et al. 2010), functions as a NLS led us to question why either endogenous or overexpressed human Dicer appears exclusively cytoplasmic. Through deletion analysis, we identified a minimal fragment composed of RNase III and dsRBD domains that is localized to the cytoplasm but relocalized to the nucleus when the RNA-binding potential of the RNase III domains was compromised. These data suggest that associated RNA was masking the dsRBD NLS. A similar mechanism was proposed for human ADAR1, where RNA bound to the first dsRBD was shown to inhibit the NLS activity of the third dsRBD (Strehblow et al. 2002). However, in the context of full-length Dicer, neither the RNase III mutations nor additional mutations in other domains, including deletion of the helicase domain that affects RNA binding, were sufficient to drive the protein to the nucleus. Interestingly, upon deletion of the helicase domain, Dicer was able to enter the nucleus when CRM-1-dependent nuclear export was blocked with LMB. Of note, deletion or mutation of the helicase domain was demonstrated previously to activate the enzyme endonucleolytic activity by up to 65-fold in vitro (Ma et al. 2008). Our data suggest that, in addition to affecting the enzymatic function of Dicer, the helicase domain may influence the ability of the protein to shuttle between the cytoplasm and the nucleus, possibly by modulating activity of its transport signals.
Dicer accumulation in the cytoplasm could have also been explained by the protein being anchored in this compartment by an unknown factor. However, given that simply duplicating the dsRBD at the end of the protein, or adding an SV40 NLS, allowed nuclear accumulation of Dicer argues against this possibility. Moreover, the observation that duplicating the dsRBD leads to nuclear import proves a role of the dsRBD as a NLS in the context of the full-length protein and supports a model where, under steady-state conditions, the dsRBD is masked and its NLS rendered nonfunctional. The results from the heterokaryon assays clearly demonstrated that a Dicer-SV40 NLS fusion actively shuttles between the nucleus and the cytoplasm. We, therefore, favor a scenario that, under certain circumstances, Dicer enters the nucleus but is rapidly exported back to the cytoplasm and that is what accounts for its steady-state cytoplasmic localization. The observation that Dicer-SV40 shuttles between the nucleus and the cytoplasm provides an indication that human Dicer contains a NES. The observed sensitivity to LMB of Dicer lacking its helicase domain further supports the presence of a NES, and specifically, one that is recognized by the CRM1-dependent export pathway. In the future, it will be interesting to identify sequences in Dicer which carry out NES function. This might contribute to understanding why full-length Dicer protein does not accumulate in the nucleus upon LMB treatment.
In S. pombe, Dicer is predominately localized to the nucleus, where it colocalizes with NPCs (Emmerth et al. 2010). Interestingly, human Dicer has also been shown to associate with a component of the NPC, NUP153, suggesting its role in Dicer’s nuclear localization (Ando et al. 2011). As for human Dicer, the dsRBD of S. pombe Dicer acts as a NLS and contributes to shuttling. However, in S. pombe Dicer, the C-terminal 33 aa (C33) adjacent to the dsRBD also contribute to nuclear localization. It has been suggested that the dsRBD-C33 interface (Barraud et al. 2011) mediates nuclear retention of the enzyme through an interaction with an unidentified nuclear protein. Notably, the C33-like extension is absent in human Dicer. The lack of such a retention signal in human Dicer supports the assumption that, if endogenous human Dicer is a shuttling protein, it is not retained in the nucleus.
The function of nuclear Dicer
As our results suggest that human Dicer is a shuttling protein, the question arises as to its nuclear function. Many observations in mammalian cells might be attributed to a function of Dicer in the nucleus, but direct evidence to support this remains limited (see Introduction). In nonvertebrate organisms such as A. thaliana, S. pombe, D. melanogaster, and C. elegans, Dicers, in addition to their cytoplasmic roles, have been shown to both localize and function in the nucleus. Here, they are involved in epigenetic regulation, including RNA-directed DNA methylation (RdDM) (Cernilogar et al. 2011; Doyle et al. 2012). It is likely that, also in vertebrate cells, Dicer functions in the nucleus in epigenetic regulation. This is supported by recent findings implicating Dicer in silencing of genes expressing convergent transcripts and in processing of dsRNA in the nucleus of mammalian cells (Gullerova and Proudfoot 2012). Alternatively, Dicer might participate in either nuclear processing or nuclear-cytoplasmic transport of selected miRNA precursors or other RNAs bearing extended double-stranded RNA regions. Recently, mammalian Dicer was shown to localize to ribosomal DNA repeats (Sinkkonen et al. 2010), possibly maintaining their integrity, as is the case of Dicers and RNAi machinery in S. pombe and D. melanogaster (Cam et al. 2005; Peng and Karpen 2007). Regardless of its potential function in the nucleus, our data suggest that human Dicer has the ability to enter and exit the nucleus and exhibits characteristics of a shuttling protein.
MATERIALS AND METHODS
Molecular cloning and mutagenesis
A previously described C-terminal myc-tagged pyruvate kinase construct was used as the basis to generate PK fusions (Eckmann et al. 2001). We replaced the original polylinker with adaptor oligonucleotides containing 5′ NotI and 3′ HindIII sites and cloned regions to be tested for NLS activity via these sites. As a positive control, we inserted complementary oligonucleotides coding for an SV40 NLS (PKKKRKV) via these sites. To generate C-terminal PK fusions, a unique XhoI restriction site immediately downstream from the PK ORF and before the start of the myc tags was used to insert sequences of interest, as well as an SV40 NLS. Using the PSORT II server (http://psort.nibb.ac.jp/form2.html), three NLSs were predicted starting at (1) amino acid 740, having the sequence PGSTKRR, (2) amino acid 788, having the sequence RRRK, and (3) amino acid 1349, having the sequence KKVSNCNLYRLGKKKGL. N-terminal tagged versions of Dicer in pCI-neo were generated by replacing the polylinker of pCI-neo with complementary oligonucleotides containing an AUG with Kozak’s consensus, followed by unique restriction sites (5′ XhoI, NheI, SalI, and NotI 3′) absent in Dicer cDNA. Tags were cloned immediately downstream from the AUG via NheI/SalI (GFP) or XhoI/SalI (4xmyc) sites. A previously described full-length Dicer cDNA (Billy et al. 2001) was introduced via SalI and NotI, thus yielding myc-Dicer and GFP-Dicer. These constructs maintain the native stop codon of Dicer immediately before the NotI site. We generated a second version of these constructs that had the stop codon removed, thus allowing C-terminal insertions at the end of Dicer (myc-Dicer-Δstop and GFP-Dicer-Δstop). Tagged deletion constructs were made by amplifying regions of myc-Dicer using 5′ SalI and 3′ NotI primers, and switching the product with that of myc-Dicer. The ΔHelicase constructs were generated as previously described (Lau et al. 2012). Myc-Dicer-SV40 and GFP-Dicer-SV40 were generated by the introduction of complementary oligonucleotides coding for an SV40 NLS (with stop codon) via NotI overhangs into myc-Dicer-Δstop and GFP-Dicer-Δstop digested with NotI. To generate the dsRBD duplication construct, the dsRBD from myc-Dicer was amplified. The 5′ primer contained a NotI site and started immediately at the end of the RNase IIIb domain and the 3′ primer amplified back into Dicer over the NotI site and stop codon. This fragment was cloned via NotI into myc-Dicer-Δstop, thus generating Dicer-dsRBD. GST-fusion constructs were generated by cloning PCR-generated inserts into the previously described pGEX-2 vector (Krovat and Jantsch 1996). The plasmid expressing GST-tagged dsRBD2 of XlrbpA was described previously (Krovat and Jantsch 1996). All mutagenesis was carried out according to Zheng et al. (2004). The integrity of all constructs was verified by sequencing.
Transfections and immunofluorescence
For pyruvate kinase reporter assays, HeLa cells grown on coverslips were transiently transfected using Nanofectin (PAA) according to the manufacturer’s instructions. After 24 h, cells were fixed with 2% paraformaldehyde diluted in 1×PBS and 0.05% Triton X-100 (PBS-T) for 10 min and then permeabilized with ice-cold methanol. Cells were blocked with 10% horse serum diluted in PBS-T and stained with antibodies diluted with 2.5% horse serum in PBS-T. Coverslips were mounted with DAPI/Prolong Gold (Molecular Probes). All images were acquired on an Axiovert 200 (Zeiss) using MetaMorph. For the analysis of dsRBD basic patch, transient transfections were performed using FuGENE reagent (Roche). After 24 h, cells were fixed in 4% PFA/PBS for 12 min and permeabilized in 0.5% Triton X-100 for 15 min. Blocking was performed in 10% goat serum in 2% BSA/PBS for 30 min, and primary antibodies diluted in blocking solution were applied for 1 h at room temperature. Next, cells were washed three times with 2% BSA/PBS and incubated with secondary antibody diluted in blocking solution for 45 min. Cells were washed three times with BSA/PBS, followed by one washing step in H2O, and cover slips were mounted in VectaShield (Vector Laboratories). In Figure 7, transient transfections were performed by using X-tremeGENE 9 DNA Transfection Reagent according to the manufacturer’s protocol.
RNAi
RNAi oligonucleotides were transfected with INTERFERin (Polyplus) and incubated for 72 h at 3.3 nM. The following siRNA oligonucleotides, all from Qiagen, were used (sense sequences are shown):
si-Importin β (5′-TCGGTTATATTTGCCAAGATA),
si-Importin 7 (5′-CACCTACTACTCAATACCTTA),
si-Importin 8 (5′-CAGGTCTGTGCTACTAGACAA),
AllStars (negative control).
Confocal microscopy
Images from fixed cells were taken at room temperature with a confocal scanning system (SP1; Leica) using a HCX Pl APO Ibd.Bl. 63×, NA 1.32 oil immersion lens.
Antibodies
Antibodies against Impβ, Imp7, and Imp5 were kind gifts of D. Görlich (MPI Göttingen, Germany). Imp8 antibody was a gift of G. Meister (University Regensburg, Germany). RIOK2 antibody was previously described (Zemp et al. 2009). Anti-β-actin was purchased from Sigma-Aldrich and anti-myc (9E10) from Santa Cruz Biology. Secondary antibodies for immunofluorescence were purchased from Invitrogen and for Western blotting from Sigma-Aldrich.
Cell lines and reagents
The tet-inducible HeLa K dsRBD-PK-myc cell line was established as previously described (Zemp et al. 2009). DsRBD-PK-myc (cloned into the BamHI and NotI sites of pcDNA/FRT/TO; Invitrogen) was integrated into the FRT site by cotransfection with pOG44 (Invitrogen). Expression of the reporter protein was induced for 12 h with tet (125 ng/mL). Cells were treated with 20 nM LMB (LC Laboratories) or solvent (ethanol) for 10 h.
Native gel shifts
GST-fusion proteins were expressed in E. coli BL21-CodonPlus(DE3)-RIPL cells (Stratagene), purified using GSH-sepharose (Amersham) according to the manufacturer’s instructions, and dialyzed against buffer containing 20 mM Tris-HCl pH 7.5, 100 mM NaCl, 1 mM MgCl2, 50% glycerol. The purity of proteins was analyzed by 15% PAGE. Protein concentration was determined by the Bradford method with BSA as a standard. The internally 32P-labeled 30-bp, 50-bp, 70-bp, and 130-bp dsRNAs were prepared as described before (Zhang et al. 2002). RNAs were synthesized by the T7 polymerase in vitro transcription, using the Ambion T7 MaxiScript transcription kit and [α-32P]UTP. After transcription, samples were purified by denaturing 8% PAGE. Complementary RNA strands were annealed at 95°C for 3 min in 20 mM NaCl, transferred to 75°C, and then slowly cooled down to 20°C. The 5′-end-labeling of siRNA, using T4 polynucleotide kinase (New England Biolabs) and either [γ-32P]ATP or cold ATP, was carried out according to the manufacturer’s protocol.
Ten-μL reactions containing 300 pM labeled dsRNA and varying protein concentrations were incubated for 30 min at 37°C in buffer containing a final concentration of 30 mM Tris-HCl, pH 8.0, 50 mM NaCl, 1 mM MgCl2, and 25% glycerol. Complexes were analyzed on 5% native polyacrylamide gels with acrylamide/bis-acrylamide ratio of 19:1. Gels were electrophoresed in 1×TBE buffer at 4°C, dried and quantified by Phosphorimager. The fraction bound was calculated using Molecular Dynamics Image Quant 5. Radioactivity corresponding to free (nonbound) dsRNA and total dsRNA (total radioactivity in the entire lane) was quantified. Apparent dissociation constants (Kd) were calculated by fitting the experimental data by nonlinear least-squares regression to the single-site binding isotherm: % free RNA = Kd[app]/(Kd[app] + [protein]). From this equation, the apparent Kd corresponds to the protein concentration at which half of the RNA is bound. Fitting of the data was done using Prism 5 (GraphPad).
Sequences of RNAs used:
19 + 2(a):
upper strand: 5′-GCAGCACGACUUCUUCAAGUU-3′
lower strand: 5′-CUUGAAGAAGUCGUGCUGCUU-3′
19 + 2(b):
upper strand: 5′-GUCACAUUGCCCAAGUCUCUU-3′
lower strand: 5′-GAGACUUGGGCAAUGUGACUU-3′
21blunt:
upper strand: 5′-AAGUCACAUUGCCCAAGUCUC-3′
lower strand: 5′-GAGACUUGGGCAAUGUGACUU-3′
Pull-down experiments
GST-dsRBD and GST-dsRBD NLSmut used in pull-down experiments were expressed in E. coli BLR (pRep4) cells at 25°C by induction with 2 mM IPTG for 4 h. Cells were lysed by sonication in 50 mM Tris-HCl, pH 7.5, 200 mM NaCl, 2 mM MgCl2, 5% glycerol, 1 mM 2-mercaptoethanol, 0.1 mM PMSF. The lysate was cleared by ultracentrifugation and supplemented with 250 mM sucrose. The expression and purification of Impβ (Kutay et al. 1997), RanQ69L (Izaurralde et al. 1997), and Imp7 (Jakel and Gorlich 1998) have been described.
For pull-down experiments from HeLa cell low-salt extracts (LSE, prepared as described [Kutay et al. 1998]), GST-dsRBD was immobilized on GSH-sepharose beads for 2–4 h at 4°C. Beads were washed three times in binding buffer (50 mM Tris-HCl, pH 7.5, 200 mM NaCl, 2 mM MgCl2) and incubated with 400 μL LSE at 4°C for 3 h. Beads were washed three times in binding buffer, and elution was performed in SDS sample buffer to which DTT was added after the elution step. For binding experiments using purified import factors, GST-dsRBD constructs were immobilized as described above. To test the interaction between GST-dsRBD and Impβ or Imp7 (Fig. 4A), each sample contained 100 μL of E. coli lysate (in 50 mM Tris-HCl, pH 7.5, 100 mM KCl, 2 mM MgCl2) supplemented with 0.5 μM purified recombinant importin, and, if indicated, 2.5 μM RanQ69L(GTP). To test the influence of the NLSmut (Fig. 4C), 180 μL of E. coli lysate was used per sample and supplemented with 1 μM of the indicated importin. In both cases, binding was performed for 2 h at 4°C with gentle mixing, and elution was completed as described above.
Heterokaryon fusion assays
HeLa cells were cotransfected with plasmids expressing GFP-Dicer-SV40 and either myc-tagged hnRNPA1 or hnRNPC. Cells were incubated for 24 h then trypsinized and coplated on coverslips with an equal number of mouse NIH 3T3 cells. Cells were treated with 75 μg of cycloheximide per ml medium to inhibit translation for 2 h before inducing fusion with polyethylene glycol 6000 (1 g/mL, in serum-free medium) for 2 min at 37°C. Cells were washed three times with prewarmed 1×PBS, returned to standard medium, and incubated in the presence of cycloheximide for 3 h before being fixed and stained. Coverslips were mounted with DAPI/Prolong Gold (Molecular Probes). HeLa and NIH 3T3 nuclei were distinguished by DAPI staining. All images were acquired on Axioimager Z1 (Zeiss) using MetaMorph.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
ACKNOWLEDGMENTS
M.D. was initially supported by the Austrian Science Fund’s (FWF) Erwin Schrödinger Stipendium (J2436) and a long-term fellowship from the Human Frontiers Science Organization (LT00199/2005-L). The authors thank Drs. Gideon Dreyfuss and Narry Kim for the plasmids encoding hnRNPC and Drosha, respectively. We also thank Dr. Michael Jantsch for the XlhnRNPA1 and pGEX-XlrbpA dsRBD2 plasmids and for communicating results prior to publication. Antibodies against Impβ, Imp5, and Imp7 were kind gifts from Dr. Dirk Görlich (MPI Göttingen, Germany). Imp8 antibody was kindly provided by Dr. Gunter Meister (University Regensburg, Germany). This work was supported by the EC FP6 Program “Sirocco” to W.F. as well as by the Swiss National Science Foundation and the ETH Zurich (ETHIIRA) to U.K. The FMI is part of the Novartis Research Foundation.
REFERENCES
- Ando Y, Tomaru Y, Morinaga A, Burroughs AM, Kawaji H, Kubosaki A, Kimura R, Tagata M, Ino Y, Hirano H, et al. 2011. Nuclear pore complex protein mediated nuclear localization of dicer protein in human cells. PLoS One 6: e23385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barraud P, Emmerth S, Shimada Y, Hotz HR, Allain FH, Buhler M 2011. An extended dsRBD with a novel zinc-binding motif mediates nuclear retention of fission yeast Dicer. EMBO J 30: 4223–4235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benetti R, Gonzalo S, Jaco I, Munoz P, Gonzalez S, Schoeftner S, Murchison E, Andl T, Chen T, Klatt P, et al. 2008. A mammalian microRNA cluster controls DNA methylation and telomere recombination via Rbl2-dependent regulation of DNA methyltransferases. Nat Struct Mol Biol 15: 268–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billy E, Brondani V, Zhang H, Muller U, Filipowicz W 2001. Specific interference with gene expression induced by long, double-stranded RNA in mouse embryonal teratocarcinoma cell lines. Proc Natl Acad Sci 98: 14428–14433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaszczyk J, Gan J, Tropea JE, Court DL, Waugh DS, Ji X 2004. Noncatalytic assembly of ribonuclease III with double-stranded RNA. Structure 12: 457–466 [DOI] [PubMed] [Google Scholar]
- Cam HP, Sugiyama T, Chen ES, Chen X, FitzGerald PC, Grewal SI 2005. Comprehensive analysis of heterochromatin- and RNAi-mediated epigenetic control of the fission yeast genome. Nat Genet 37: 809–819 [DOI] [PubMed] [Google Scholar]
- Cernilogar FM, Onorati MC, Kothe GO, Burroughs AM, Parsi KM, Breiling A, Lo Sardo F, Saxena A, Miyoshi K, Siomi H, et al. 2011. Chromatin-associated RNA interference components contribute to transcriptional regulation in Drosophila. Nature 480: 391–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravarthy S, Sternberg SH, Kellenberger CA, Doudna JA 2010. Substrate-specific kinetics of Dicer-catalyzed RNA processing. J Mol Biol 404: 392–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colmenares SU, Buker SM, Buhler M, Dlakic M, Moazed D 2007. Coupling of double-stranded RNA synthesis and siRNA generation in fission yeast RNAi. Mol Cell 27: 449–461 [DOI] [PubMed] [Google Scholar]
- Doyle M, Jantsch MF 2002. New and old roles of the double-stranded RNA-binding domain. J Struct Biol 140: 147–153 [DOI] [PubMed] [Google Scholar]
- Doyle M, Jaskiewicz L, Filipowicz W 2012. Dicer proteins and their role in gene silencing pathways. In The enzymes (ed. Guo F), Vol. 32, pp. 1–35 Elsevier, Philadelphia, PA [Google Scholar]
- Du Z, Lee JK, Tjhen R, Stroud RM, James TL 2008. Structural and biochemical insights into the dicing mechanism of mouse Dicer: A conserved lysine is critical for dsRNA cleavage. Proc Natl Acad Sci 105: 2391–2396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckmann CR, Neunteufl A, Pfaffstetter L, Jantsch MF 2001. The human but not the Xenopus RNA-editing enzyme ADAR1 has an atypical nuclear localization signal and displays the characteristics of a shuttling protein. Mol Biol Cell 12: 1911–1924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emmerth S, Schober H, Gaidatzis D, Roloff T, Jacobeit K, Buhler M 2010. Nuclear retention of fission yeast dicer is a prerequisite for RNAi-mediated heterochromatin assembly. Dev Cell 18: 102–113 [DOI] [PubMed] [Google Scholar]
- Ender C, Krek A, Friedlander MR, Beitzinger M, Weinmann L, Chen W, Pfeffer S, Rajewsky N, Meister G 2008. A human snoRNA with microRNA-like functions. Mol Cell 32: 519–528 [DOI] [PubMed] [Google Scholar]
- Fabian MR, Sonenberg N, Filipowicz W 2010. Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem 79: 351–379 [DOI] [PubMed] [Google Scholar]
- Fried H, Kutay U 2003. Nucleocytoplasmic transport: Taking an inventory. Cell Mol Life Sci 60: 1659–1688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritz J, Strehblow A, Taschner A, Schopoff S, Pasierbek P, Jantsch MF 2009. RNA-regulated interaction of transportin-1 and exportin-5 with the double-stranded RNA-binding domain regulates nucleocytoplasmic shuttling of ADAR1. Mol Cell Biol 29: 1487–1497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukagawa T, Nogami M, Yoshikawa M, Ikeno M, Okazaki T, Takami Y, Nakayama T, Oshimura M 2004. Dicer is essential for formation of the heterochromatin structure in vertebrate cells. Nat Cell Biol 6: 784–791 [DOI] [PubMed] [Google Scholar]
- Gullerova M, Proudfoot NJ 2012. Convergent transcription induces transcriptional gene silencing in fission yeast and mammalian cells. Nat Struct Mol Biol 19: 1193–1201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haase AD, Jaskiewicz L, Zhang H, Laine S, Sack R, Gatignol A, Filipowicz W 2005. TRBP, a regulator of cellular PKR and HIV-1 virus expression, interacts with Dicer and functions in RNA silencing. EMBO Rep 6: 961–967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haussecker D, Proudfoot NJ 2005. Dicer-dependent turnover of intergenic transcripts from the human β-globin gene cluster. Mol Cell Biol 25: 9724–9733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huntzinger E, Izaurralde E 2011. Gene silencing by microRNAs: Contributions of translational repression and mRNA decay. Nat Rev Genet 12: 99–110 [DOI] [PubMed] [Google Scholar]
- Izaurralde E, Kutay U, von Kobbe C, Mattaj IW, Gorlich D 1997. The asymmetric distribution of the constituents of the Ran system is essential for transport into and out of the nucleus. EMBO J 16: 6535–6547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakel S, Gorlich D 1998. Importin β, transportin, RanBP5 and RanBP7 mediate nuclear import of ribosomal proteins in mammalian cells. EMBO J 17: 4491–4502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakel S, Albig W, Kutay U, Bischoff FR, Schwamborn K, Doenecke D, Gorlich D 1999. The importin β/importin 7 heterodimer is a functional nuclear import receptor for histone H1. EMBO J 18: 2411–2423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, Doudna JA 2009. A three-dimensional view of the molecular machinery of RNA interference. Nature 457: 405–412 [DOI] [PubMed] [Google Scholar]
- Kanellopoulou C, Muljo SA, Kung AL, Ganesan S, Drapkin R, Jenuwein T, Livingston DM, Rajewsky K 2005. Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev 19: 489–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanellopoulou C, Muljo SA, Dimitrov SD, Chen X, Colin C, Plath K, Livingston DM 2009. X chromosome inactivation in the absence of Dicer. Proc Natl Acad Sci 106: 1122–1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim VN, Han J, Siomi MC 2009. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol 10: 126–139 [DOI] [PubMed] [Google Scholar]
- Kok KH, Ng MH, Ching YP, Jin DY 2007. Human TRBP and PACT directly interact with each other and associate with dicer to facilitate the production of small interfering RNA. J Biol Chem 282: 17649–17657 [DOI] [PubMed] [Google Scholar]
- Krovat BC, Jantsch MF 1996. Comparative mutational analysis of the double-stranded RNA binding domains of Xenopus laevis RNA-binding protein A. J Biol Chem 271: 28112–28119 [DOI] [PubMed] [Google Scholar]
- Kutay U, Bischoff FR, Kostka S, Kraft R, Gorlich D 1997. Export of importin α from the nucleus is mediated by a specific nuclear transport factor. Cell 90: 1061–1071 [DOI] [PubMed] [Google Scholar]
- Kutay U, Lipowsky G, Izaurralde E, Bischoff FR, Schwarzmaier P, Hartmann E, Gorlich D 1998. Identification of a tRNA-specific nuclear export receptor. Mol Cell 1: 359–369 [DOI] [PubMed] [Google Scholar]
- Lau PW, Potter CS, Carragher B, MacRae IJ 2009. Structure of the human Dicer-TRBP complex by electron microscopy. Structure 17: 1326–1332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau PW, Guiley KZ, De N, Potter CS, Carragher B, MacRae IJ 2012. The molecular architecture of human Dicer. Nat Struct Mol Biol 19: 436–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lejeune E, Bayne EH, Allshire RC 2010. On the connection between RNAi and heterochromatin at centromeres. Cold Spring Harb Symp Quant Biol 75: 275–283 [DOI] [PubMed] [Google Scholar]
- Lingel A, Simon B, Izaurralde E, Sattler M 2003. Structure and nucleic-acid binding of the Drosophila Argonaute 2 PAZ domain. Nature 426: 465–469 [DOI] [PubMed] [Google Scholar]
- Ma E, MacRae IJ, Kirsch JF, Doudna JA 2008. Autoinhibition of human dicer by its internal helicase domain. J Mol Biol 380: 237–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma E, Zhou K, Kidwell MA, Doudna JA 2012. Coordinated activities of human dicer domains in regulatory RNA processing. J Mol Biol 422: 466–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacRae IJ, Zhou K, Li F, Repic A, Brooks AN, Cande WZ, Adams PD, Doudna JA 2006. Structural basis for double-stranded RNA processing by Dicer. Science 311: 195–198 [DOI] [PubMed] [Google Scholar]
- MacRae IJ, Zhou K, Doudna JA 2007. Structural determinants of RNA recognition and cleavage by Dicer. Nat Struct Mol Biol 14: 934–940 [DOI] [PubMed] [Google Scholar]
- Muhlhausser P, Muller EC, Otto A, Kutay U 2001. Multiple pathways contribute to nuclear import of core histones. EMBO Rep 2: 690–696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murchison EP, Stein P, Xuan Z, Pan H, Zhang MQ, Schultz RM, Hannon GJ 2007. Critical roles for Dicer in the female germline. Genes Dev 21: 682–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakielny S, Dreyfuss G 1996. The hnRNP C proteins contain a nuclear retention sequence that can override nuclear export signals. J Cell Biol 134: 1365–1373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesterova TB, Popova BC, Cobb BS, Norton S, Senner CE, Tang YA, Spruce T, Rodriguez TA, Sado T, Merkenschlager M, et al. 2008. Dicer regulates Xist promoter methylation in ES cells indirectly through transcriptional control of Dnmt3a. Epigenetics Chromatin 1: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa Y, Sun BK, Lee JT 2008. Intersection of the RNA interference and X-inactivation pathways. Science 320: 1336–1341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohrt T, Muetze J, Svoboda P, Schwille P 2012. Intracellular localization and routing of miRNA and RNAi pathway components. Curr Top Med Chem 12: 79–88 [DOI] [PubMed] [Google Scholar]
- Park JE, Heo I, Tian Y, Simanshu DK, Chang H, Jee D, Patel DJ, Kim VN 2011. Dicer recognizes the 5′ end of RNA for efficient and accurate processing. Nature 475: 201–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng JC, Karpen GH 2007. H3K9 methylation and RNA interference regulate nucleolar organization and repeated DNA stability. Nat Cell Biol 9: 25–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Provost P, Dishart D, Doucet J, Frendewey D, Samuelsson B, Radmark O 2002. Ribonuclease activity and RNA binding of recombinant human Dicer. EMBO J 21: 5864–5874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryter JM, Schultz SC 1998. Molecular basis of double-stranded RNA-protein interactions: Structure of a dsRNA-binding domain complexed with dsRNA. EMBO J 17: 7505–7513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinkkonen L, Hugenschmidt T, Filipowicz W, Svoboda P 2010. Dicer is associated with ribosomal DNA chromatin in mammalian cells. PLoS One 5: e12175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soifer HS, Sano M, Sakurai K, Chomchan P, Saetrom P, Sherman MA, Collingwood MA, Behlke MA, Rossi JJ 2008. A role for the Dicer helicase domain in the processing of thermodynamically unstable hairpin RNAs. Nucleic Acids Res 36: 6511–6522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song JJ, Liu J, Tolia NH, Schneiderman J, Smith SK, Martienssen RA, Hannon GJ, Joshua-Tor L 2003. The crystal structure of the Argonaute2 PAZ domain reveals an RNA binding motif in RNAi effector complexes. Nat Struct Biol 10: 1026–1032 [DOI] [PubMed] [Google Scholar]
- St. Johnston D, Brown NH, Gall JG, Jantsch M 1992. A conserved double-stranded RNA-binding domain. Proc Natl Acad Sci 89: 10979–10983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strehblow A, Hallegger M, Jantsch MF 2002. Nucleocytoplasmic distribution of human RNA-editing enzyme ADAR1 is modulated by double-stranded RNA-binding domains, a leucine-rich export signal, and a putative dimerization domain. Mol Biol Cell 13: 3822–3835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taft RJ, Glazov EA, Lassmann T, Hayashizaki Y, Carninci P, Mattick JS 2009. Small RNAs derived from snoRNAs. RNA 15: 1233–1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsutsumi A, Kawamata T, Izumi N, Seitz H, Tomari Y 2011. Recognition of the pre-miRNA structure by Drosophila Dicer-1. Nat Struct Mol Biol 18: 1153–1158 [DOI] [PubMed] [Google Scholar]
- Wang HW, Noland C, Siridechadilok B, Taylor DW, Ma E, Felderer K, Doudna JA, Nogales E 2009. Structural insights into RNA processing by the human RISC-loading complex. Nat Struct Mol Biol 16: 1148–1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg DE, Nakanishi K, Patel DJ, Bartel DP 2011. The inside-out mechanism of Dicers from budding yeasts. Cell 146: 262–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinmann L, Hock J, Ivacevic T, Ohrt T, Mutze J, Schwille P, Kremmer E, Benes V, Urlaub H, Meister G 2009. Importin 8 is a gene silencing factor that targets argonaute proteins to distinct mRNAs. Cell 136: 496–507 [DOI] [PubMed] [Google Scholar]
- Wostenberg C, Lary JW, Sahu D, Acevedo R, Quarles KA, Cole JL, Showalter SA 2012. The role of human Dicer-dsRBD in processing small regulatory RNAs. PLoS One 7: e51829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D, Farmer A, Chook YM 2010. Recognition of nuclear targeting signals by Karyopherin-β proteins. Curr Opin Struct Biol 20: 782–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita S, Nagata T, Kawazoe M, Takemoto C, Kigawa T, Guntert P, Kobayashi N, Terada T, Shirouzu M, Wakiyama M, et al. 2011. Structures of the first and second double-stranded RNA-binding domains of human TAR RNA-binding protein. Protein Sci 20: 118–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan KS, Yan S, Farooq A, Han A, Zeng L, Zhou MM 2003. Structure and conserved RNA binding of the PAZ domain. Nature 426: 468–474 [DOI] [PubMed] [Google Scholar]
- Zemp I, Wild T, O’Donohue MF, Wandrey F, Widmann B, Gleizes PE, Kutay U 2009. Distinct cytoplasmic maturation steps of 40S ribosomal subunit precursors require hRio2. J Cell Biol 185: 1167–1180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Kolb FA, Brondani V, Billy E, Filipowicz W 2002. Human Dicer preferentially cleaves dsRNAs at their termini without a requirement for ATP. EMBO J 21: 5875–5885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Kolb FA, Jaskiewicz L, Westhof E, Filipowicz W 2004. Single processing center models for human Dicer and bacterial RNase III. Cell 118: 57–68 [DOI] [PubMed] [Google Scholar]
- Zheng L, Baumann U, Reymond JL 2004. An efficient one-step site-directed and site-saturation mutagenesis protocol. Nucleic Acids Res 32: e115. [DOI] [PMC free article] [PubMed] [Google Scholar]