Continued studies of group II introns have revealed the existence of new lineages. This paper describes the catalytic properties of two of these lineages, group IIE and group IIF.
Keywords: splicing, RNA structure, transposon, noncoding RNA, RNA catalysis
Abstract
Group II introns are self-splicing, retrotransposable ribozymes that contribute to gene expression and evolution in most organisms. The ongoing identification of new group II introns and recent bioinformatic analyses have suggested that there are novel lineages, which include the group IIE and IIF introns. Because the function and biochemical activity of group IIE and IIF introns have never been experimentally tested and because these introns appear to have features that distinguish them from other introns, we set out to determine if they were indeed self-splicing, catalytically active RNA molecules. To this end, we transcribed and studied a set of diverse group IIE and IIF introns, quantitatively characterizing their in vitro self-splicing reactivity, ionic requirements, and reaction products. In addition, we used mutational analysis to determine the relative role of the EBS-IBS 1 and 2 recognition elements during splicing by these introns. We show that group IIE and IIF introns are indeed distinct active intron families, with different reactivities and structures. We show that the group IIE introns self-splice exclusively through the hydrolytic pathway, while group IIF introns can also catalyze transesterifications. Intriguingly, we observe one group IIF intron that forms circular intron. Finally, despite an apparent EBS2-IBS2 duplex in the sequences of these introns, we find that this interaction plays no role during self-splicing in vitro. It is now clear that the group IIE and IIF introns are functional ribozymes, with distinctive properties that may be useful for biotechnological applications, and which may contribute to the biology of host organisms.
INTRODUCTION
Group II introns are among the largest ribozymes found in nature, and they are present in all kingdoms of life, from bacteria, protozoa, and fungi to plants and simple animals (Pyle et al. 2007; Vallès et al. 2008). Group II introns play a crucial role in genomic regulation (Pyle 2010). They can self-splice by excising themselves from precursor mRNA transcripts and ligating flanking exons. Furthermore, they can function as mobile retroelements by invading genomic DNA at distant sites (Lambowitz and Zimmerly 2011). As they are the likely evolutionary predecessors of spliceosomal introns and retrotransposons in eukaryotes, structural and functional studies of group II introns have contributed considerably toward our understanding of splicing mechanisms and RNA tertiary structure (Cech 1986; Jacquier 1996; Keating et al. 2010). However, recent discoveries identifying new lineages of group II introns suggest that we have only begun to describe the full functional diversity of these ribozymes (Toro et al. 2002; Simon et al. 2008; Toro and Martínez-Abarca 2013).
Group II introns vary widely in primary structure, but they share a common secondary structure consisting of six domains (DI–DVI) that radiate from a central wheel (Michel et al. 1989; Pyle and Lambowitz 2006). DI, the largest domain, forms a scaffold that is required for assembly of the entire intron by engaging in tertiary interactions with other domains (Toor et al. 2010). The series of elaborate tertiary interactions necessary for folding and catalysis have been described (for review, see Pyle 2010; Lambowitz and Zimmerly 2011). Notably, DI recognizes both 5′- and 3′-exon via base-pairing interactions between three motifs (EBS1-3, for exon-binding sites) and complementary sites in the flanking exons (IBS1-3, for intron-binding sites). These interactions are critical for splice site selection and DNA target site recognition during retrohoming (Jacquier and Rosbash 1986; Jacquier and Michel 1987; Yang et al. 1996; Costa et al. 2000; Aizawa et al. 2003; Jiménez-Zurdo et al. 2003).
The central catalytic core of the ribozyme is formed by DV, an RNA hairpin that consists of two helices separated by a bulge and that terminates in a GNRA tetraloop (Costa et al. 1998). Tertiary contacts between the bulge, three nucleotides of the first DV helix, and the J2/3 junction between DII and DIII form a pocket around the 5′-splice site that positions it for nucleophilic attack during the first step of splicing (Toor et al. 2008a,b). In general, during the first step of splicing, attack by the branch-point adenosine creates a 2′-5′ phosphodiester linkage that shapes the intron into a lariat (Peebles et al. 1986). This branching pathway is analogous to the reaction catalyzed by the spliceosome. When a water molecule instead serves as the nucleophile, no lariat forms, and the intron is released as a linear molecule (Peebles et al. 1987; van der Veen et al. 1987; Daniels et al. 1996). This is termed the hydrolytic pathway and occurs in the majority of group II introns in vitro and in vivo (Daniels et al. 1996; Podar et al. 1998; Vogel and Börner 2002). In addition to water and the 2′-OH group of the branch-site, a free 2′-OH on the terminal nucleotide of the intron can also behave as a nucleophile, resulting in intron circles (Murray et al. 2001). This pathway requires the prior release of the 3′-exon presumably by a trans splicing event, and for many introns, circularization products are prominent in vivo and in vitro (Murray et al. 2001; Li-Pook-Than and Bonen 2006; Molina-Sánchez et al. 2006).
During the first step of splicing (by either transesterification or hydrolysis), the 5′-exon is freed from the intron. In the subsequent second step, conformational rearrangement of the J2/3 junction is thought to reposition DVI and make room for the 3′-OH of the 5′-exon to attack the 3′-splice junction (Marcia and Pyle 2012). In vitro, DI and DV are alone sufficient for catalysis of individual hydrolysis events, whereas splicing requires DII, DIII, and DVI. In vivo, an intron-encoded protein (IEP) from DIV is essential for proper folding and catalysis of the intron (Lambowitz and Zimmerly 2004; Solem et al. 2009).
Based on RNA secondary structure motifs of the six functional domains, group II introns are organized into three structural classes: IIA, IIB, and IIC (Granlund et al. 2001; Toor et al. 2001). An alternative classification of group II introns into nine lineages (A, B, C, D, E, F, CL1, CL2, ML) is based on phylogenetic analysis of the open reading frame (ORF) in DIV (Zimmerly et al. 2001; Toro et al. 2002; Simon et al. 2008). The ORF lineages coincide with distinct RNA secondary structures, suggesting that the IEP and the ribozyme have coevolved (Toor et al. 2001). Because of the relatively well-conserved primary structure of the ORF, BLAST searches of the ORF combined with computational folding of flanking sequences are used to identify putative group II introns. This method has become standard for predicting group II-like structures but tells us little about the biochemical capabilities of the predicted introns or the conditions in which they are most active. Moreover, phylogenetic searches may not account for introns that have lost catalytic activity due to degradation in their host genomes. It has been hypothesized that bacterial group II introns, which are generally found in nonessential genomic regions, are probably subject to more negative selective pressures than organellar group II introns, many of which have coevolved with their hosts (Dai and Zimmerly 2002; Chillón et al. 2011; Leclercq and Cordaux 2012). Thus, experimental validation remains a crucial step in assigning function to group II-like intron structures.
The group II intron lineages IIE and IIF have been computationally defined (Toro et al. 2002; Simon et al. 2008). However, to date, none of these bacterial intron lineages have been functionally characterized in vitro or in vivo. A number of features apparent from the predicted secondary structures suggest these introns may exhibit novel behaviors. For example, the essential α′ motif, which is generally located in a bulge loop of DId3, is instead located at the terminus of a stem–loop in DId3 in group IIE and IIF introns. Other idiosyncrasies include an additional bulge loop in DId3 and a 5-nt loop instead of the typical 4-nt ζ′ loop in DV (see Fig. 1 for selected candidates) (Toro et al. 2002; Simon et al. 2008). Apart from these unique features, group IIE and IIF introns are similar to the IIB structural class. But unlike organellar group IIB introns in eukaryotes that have been previously studied, the group IIE and IIF introns are markedly smaller (450–700 nt without the IEP compared to 1200 nt for the ai5γ group IIB intron). Only group IIC introns are smaller (412 nt for the Oceanobacillus iheyensis intron), but group IIC introns lack a portion of the EBS1 motif and the entire EBS2 motif and therefore differ significantly from IIB introns with respect to splice site definition and first step catalysis (Granlund et al. 2001; Toor et al. 2001, 2010). These attributes suggest that group IIE and IIF introns may shed new light on our understanding of RNA molecular recognition and catalysis if they were shown to be active.
FIGURE 1.
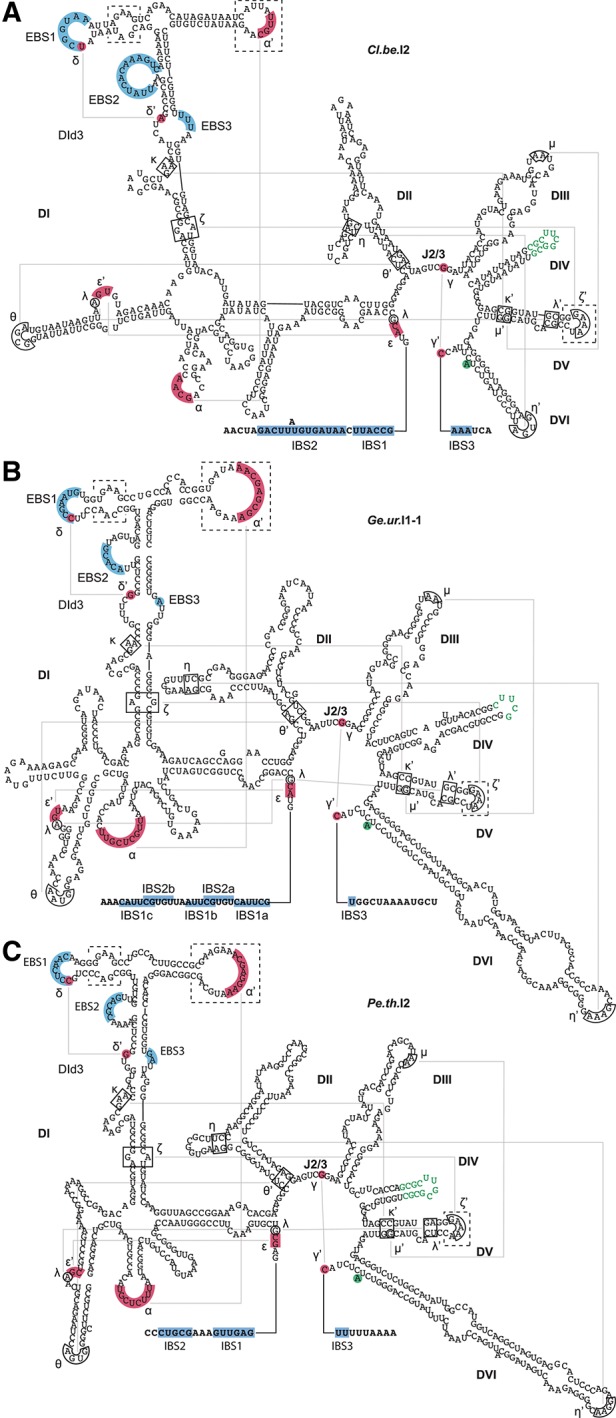
Annotated secondary structures of selected group IIE and IIF introns. Secondary structures of (A) the group IIE intron Cl.be.I2 and the group IIF introns (B) Ge.ur.I1-1 and (C) Pe.th.I2. The folded secondary structures were obtained from the database of mobile group II introns (Dai et al. 2003; Simon et al. 2008). Annotations are based on general conserved structural features of group IIE and IIF introns (Toro et al. 2002; Simon et al. 2008). Conserved sequence elements involved in long-range Watson-Crick and non-Watson-Crick interactions are highlighted in red and boxed, respectively. The unpaired branch-point adenosine is highlighted in green. Residues replacing the extended loop in DIV are indicated in green font. Intron-surrounding exons are indicated by bold lettering, and complementary EBS and IBS regions are indicated by blue shading. Dashed line boxes highlight distinct features of group IIE and IIF introns.
Here, we report the biochemical characterization of group IIE and IIF introns, and we demonstrate their ability to self-splice in vitro. In this study, we elucidated the identity of reaction products and determined reaction rate constants for selected introns under a variety of salt concentrations and temperatures. To gain information about splice site selection, fidelity, and splicing pathway preferences, we subjected intron splicing products to RT-PCR analysis. Finally, to better understand the mechanistic basis for their splicing fidelity, we designed mutant constructs of two selected introns, and we evaluated the importance of EBS–IBS interactions on kinetics and splice site specificity. Taken together, the results show that group IIE and IIF introns are functionally distinct from one another and from other introns characterized to date.
RESULTS
Selection and construct design of group IIE and IIF introns
Sixteen group IIE and six group IIF intron candidates for study were identified by a comprehensive search of the Database for Group II Introns (Dai et al. 2003; Simon et al. 2008). We were primarily interested in candidate group IIE and IIF introns with EBS1 and EBS2 sites located in DId and complementary IBS1 and IBS2 sites located immediately upstream of the predicted 5′-splice site in the 5′-exon. Candidate introns were chosen if they contained individual EBS-IBS pairings that were at least 5-bp long. Our pool of candidates was further reduced by limited access to samples of organisms that encode group IIE and IIF introns. Based on the above criteria, we designed constructs for six group IIE introns from Chlorobium phaeobacteroides (Ch.ph.I2-1), Clostridium beijerincki (Cl.be.I2 and Cl.be.I3), Photorhabdus luminescens (P.l.I1), Prosthecochloris aestuarii (Pr.ae.I1), Syntrophomonas wolfei strain Goettingen (Sy.wo.I1-1), and three group IIF introns from Dechloromonas aromatica (D.a.I1-1), Geobacter uraniumreducens (Ge.ur.I1-1), and Pelotomaculum thermopropionicum strain SI (Pe.th.I2). The large ORF-bearing terminal loop of intron DIV was deleted and replaced with a tetraloop (see Fig. 1, green). To provide an adequate handle for intron binding and to accommodate cryptic intron binding sites that might lie further upstream of the putative 5′-exon-intron junction, we included at least 100 nt of the 5′-exon and 30 nt of the 3′-exon. The splicing precursor constructs were cloned under control of a T7 promoter and transcribed as described previously (Fedorova 2012).
Group IIE and IIF introns are catalytically active in vitro
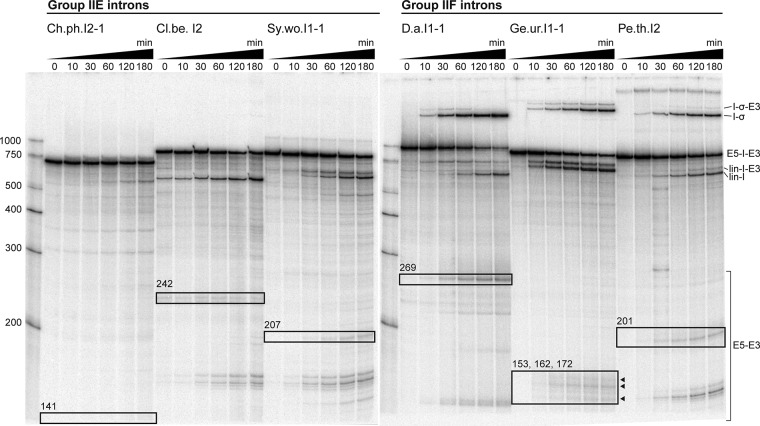
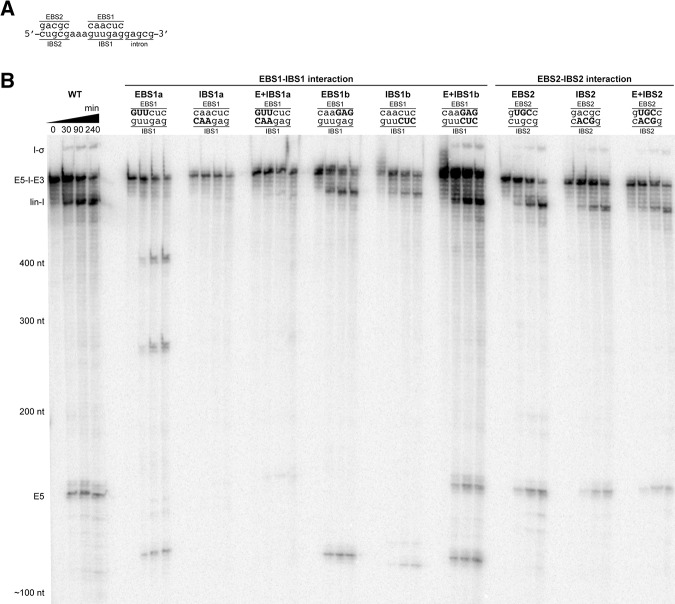
In order to determine if the selected introns are active in vitro, we subjected internally radiolabeled transcripts to high-salt self-splicing conditions [500 mM (NH4)2SO4 or KCl and 100 mM MgCl2 at pH 7.5 and 42°C]. These conditions promote the reactivity of many group II introns in vitro (Jarrell et al. 1988). In this assay, three group IIE introns (Ch.ph.I2-1, Cl.be.I2, and Sy.wo.I1-1) and three group IIF introns (D.a.I1-1, Ge.ur.I1-1, and Pe.th.I2) were observed to undergo both steps of self-splicing, as determined by the disappearance of precursor and appearance of products at predicted electrophoretic mobilities (Fig. 2). Additional reaction products corresponded in size to linear intron-3′-exon, linear intron, and free exons. Importantly, all three group IIF introns tested formed a band of slower electrophoretic mobility than precursor RNA (Fig. 2), suggesting a species corresponding to intron lariat or intron circle (Peebles et al. 1986; Murray et al. 2001). In contrast, the three active group IIE introns did not form any slower mobility products under the conditions employed in this study. Rather, these introns produced exclusively linear intron, which is characteristic of the hydrolytic pathway (Jarrell et al. 1988; Daniels et al. 1996). The remaining three group IIE introns Cl.be.I3, P.l.I1, and Pr.ae.I1 did not splice under any of the conditions tested.
FIGURE 2.
Splicing of group IIE and IIF introns. Splicing products of three group IIE and three group IIF introns. Reaction time points and nucleotide size markers are indicated on top and to the left of the gel, respectively. Precursors (E5-I-E3) and linear introns (lin-I) are labeled according to their predicted sizes. Ligated exon species (E5-E3) were verified by sequencing. Ligated exon bands are boxed, and product sizes are indicated. Intron lariat/intron circle species (I-σ) were verified by sequencing for Ge.ur.I1-1 and Pe.th.I2. Presumable splicing intermediates, where the 3′-exon is still attached to the linear intron (lin-I-E3) and intron lariat (I-σ-E3), are also marked. The group IIE introns were reacted in 500 mM KCl. Although Ch.ph.I2-1 and Cl.be.I2 also spliced in (NH4)2SO4, they reacted much more slowly and did not form slower mobility products that correspond to intron lariat or intron circle (data not shown). Sy.wo.I1-1 was inactive in (NH4)2SO4. We show activity of the group IIF introns in (NH4)2SO4. All Group IIF introns were also active in KCl, but formation of slower mobility products was reduced (data not shown). The splicing rate constant for the D.a. intron was 0.012 min−1 under conditions shown. See Figure 5 for kinetic analysis of other introns. For detailed splicing conditions see Materials and Methods.
Investigation of splice site fidelity of group IIE and IIF introns
With one exception, Ge.ur.I1-1, all the introns found to splice in this study produced a single distinct band migrating near the expected molecular weight of ligated exons (Fig. 2). To confirm that the second step of splicing occurred, bands of appropriate mobility were excised, and RT-PCR was performed on the sequence spanning the ligation point (Fig. 3A). The RT-PCR products were subsequently cloned and sequenced. We selected the group IIE intron Cl.be.I2 and the group IIF introns Ge.ur.I1-1 and Pe.th.I2 for this analysis. For the Cl.be.I2 and Pe.th.I2 introns, sequencing of the resultant clones confirmed that exon ligation occurred at the predicted splice junctions and resulted in a single product (Fig. 3B,C).
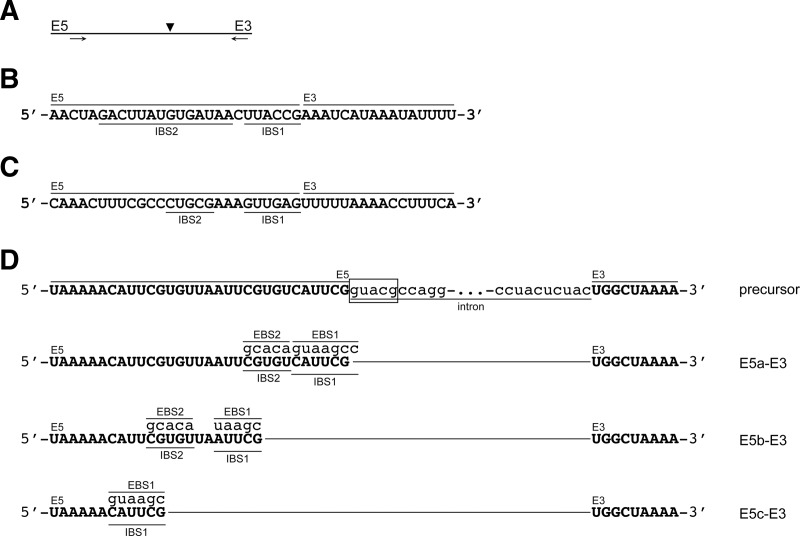
FIGURE 3.
Sequences and identification of ligated exon species. (A) Schematic of the RT-PCR across the splice site of ligated exons. Arrows indicate the primers utilized and the arrowhead indicates the splice site. Sequences of RT-PCR products for the exon–exon junctions of spliced exons for (B) Cl.be.I2, (C) Pe.th.I2, and (D) Ge.ur.I1-1 are shown. The 5′-exons (E5), 3′-exons (E3), IBS1, and IBS2 regions are indicated. The three different exon–exon junctions (E5a-c-E3) and EBS-IBS interactions of Ge.ur.I1-1 are shown in relation to the excised introns in D. The precursor of Ge.ur.I1-1 is depicted for reference, and the predicted splice site is boxed.
In contrast, splicing of Ge.ur.I1-1 results in three bands that migrate approximately at the expected position of ligated exons. We isolated one gel section containing all three bands and subjected the resulting RNA mixture to a single RT-PCR. Sequencing of the cloned RT-PCR products revealed one major and two minor ligation products composed of an intact 3′-exon joined to varying 5′-exon species that differ by several nucleotides in length (Fig. 3D). One of the minor products, E5a-E3, corresponds to expected excision of the 5′-exon at the predicted 5′-splice site based on similarity to the group IIB consensus 5′-GUGYG splice site, thereby representing correctly spliced exons.
The major product of Ge.ur.I1-1 splicing, E5b-E3, corresponds to excision of the 5′-exon at a site that is located nine nucleotides upstream of the predicted 5′-splice site (within the 5′-exon). Careful examination of the 5′-exon at this position revealed the presence of an alternative intron binding site. For example, recognition of 5′-AUUCG and 5′-CGUGU by EBS1 and EBS2 results in the cleavage of the precursor at alternative 5′-splice site 5′-UGUCA (Fig. 3D). A second minor product, E5c-E3, is formed from an EBS1 interaction with the 5′-CAUUCG sequence located 19 nucleotides upstream of the expected splice site, resulting in excision at 5′-UGUUA (Fig. 3D). All three ligated exon species share the same 3′-splice site, 5′-AC, which is consistent with the group IIB consensus 3′-splice site 5′-AY. Taken together, these results show that Group IIE and IIF introns undergo the second step of splicing with proper fidelity, forming ligated exon products.
Pe.th.I2 forms an intron lariat, and Ge.ur.I1-1 1 forms a circular intron
It was important to establish whether the slow-mobility products of group IIF splicing correspond to excised lariat or circular introns (Murray et al. 2001). To address this question, reaction products from two representative cases (Ge.ur.I1-1 and Pe.th.I2) were isolated and analyzed in detail. In order to distinguish between lariat and circular species, RT-PCR was used to amplify the sequences that flank the intron branch-site in both lariat and circular molecules. Specifically, we designed a reverse primer to bind within intron DI and a forward primer to bind toward the 3′-end of the intron, amplifying a ∼300-nt region that spans any junction between these domains. (Fig. 4A). Resulting RT-PCR products were then incorporated within a plasmid and subsequently sequenced.
FIGURE 4.
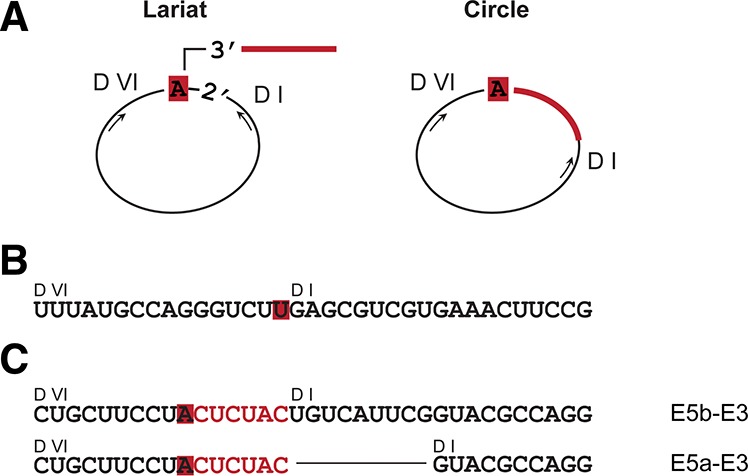
Characterization of slow-mobility reaction products. (A) Schematic of the RT-PCR across the branch point of an intron lariat and the circularization point of a circular intron. RNA sequences of the corresponding RT-PCR products for (B) Pe.th.I2 and (C) Ge.ur.I1-1 are illustrated. The unpaired branch-point adenosine in DVI and the 3′-tail are highlighted in red, and DI and DVI are indicated. For Ge.ur.I1-1, the corresponding alternative ligated exons species are indicated to the right.
Sequencing of the cloned Pe.th.I2 RT-PCR product indicates a direct connection between the branch-site and the first nucleotide of D1 and a lack of the six nucleotides between the branch-site and the 3′-splice site (Fig. 4B). In addition, a thymidine is located in place of the branch-point adenosine (Fig. 4B), a polymerase error that commonly results from the reduced fidelity of reverse transcriptase over 2′-3′-5′ phosphodiester linkages (Vogel et al. 1997). Taken together, these results indicate that Pe.th.I2 reacts through branching and forms proper lariats.
In contrast, sequences of the cloned Ge.ur.I1-1 RT-PCR product show that the six nucleotides that connect the branch point to the 3′-splice site have been included, indicating that a 2′-5′ phosphodiester bond has formed between the first and last nucleotides of the intron to produce true circles (Fig. 4C). Because Ge.ur.I1-1 uses multiple 5′-splice sites within the 5′-exon, the sequences of the RT-PCR products differ in inclusion of various upstream sequences, as expected from mapping of the ligated exon products. Interestingly, the sequences obtained from RT-PCR of cyclic intron products correspond to the alternatively spliced 5′-exons E5a and, more frequently, to E5b (Fig. 3D). We did not observe circular intron sequences that correspond to the 5′-exon E5c.
These results demonstrate that, as a class, group IIF introns use three pathways for cleavage at the 5′-splice site: They can react via hydrolysis, as observed in all cases, or they can undergo transesterification whereby the nucleophile is either the branch-point adenosine (as with Pe.th.I2), or the terminal intron nucleotide (in the case of Ge.ur.I1-1).
Kinetic analysis of group IIE and IIF introns
To evaluate the relative reactivity of each intron, we analyzed the splicing reaction kinetics under a diverse set of experimental conditions. However, it was first essential to identify optimal conditions for in vitro splicing of each intron. To this end, we varied concentrations of monovalent salt [KCl or (NH4)2SO4] from 0 to 500 mM, concentrations of MgCl2 from 8 to 100 mM, and temperatures from 37°C to 55°C, while monitoring the appearance of splicing products over time. Rate constants were calculated by fitting the relative fractions of precursor RNA remaining at each time point to a single exponential decay equation (see Materials and Methods).
We first conducted an overview of ionic conditions for group IIE and IIF splicing. Under conditions of high MgCl2 concentration (80–100 mM), introns from both classes spliced in either type of monovalent salt employed in this study (Fig. 2 and legend) except for Sy.wo.I1-1, which reacts only in the presence of KCl. Generally, the introns spliced most rapidly at high concentrations of KCl (500 mM) and MgCl2 (80–100 mM). An exception is D.a.I1-1, which spliced primarily through transesterification and which spliced most rapidly at high concentrations of (NH4)2SO4 (Fig. 2). When we tested D.a.I1-1, Ge.ur.I1-1, Ch.ph.I2-1, and Cl.be.I2 in the presence of low concentrations of MgCl2 (8–10 mM) and 40 mM MOPS, pH 7.5 without additional monovalent ion, we observed splicing by hydrolysis for all four introns, albeit with generally much slower reactivity (data not shown). An important exception is Ge.ur.I1-1, which was able to splice about twice as fast under these low salt conditions (0.022 ± 0.0036 min−1) compared to high salt conditions (0.011 ± 0.0015 min−1). However, a substantial decrease in amplitude from 0.88 ± 0.043 (100 mM MgCl2, 500 mM KCl) to 0.58 ± 0.025 (10 MgCl2) suggests that, in the absence of monovalent ions, a fraction of Ge.ur.I1-1 introns is misfolded or otherwise kinetically trapped.
An overview of temperature dependence for the Cl.be.I2 and Pe.th.I2 introns suggests differences in relative stability and reactivity. For example, we observed that the Cl.be.I2 intron remained stable at 50°C and spliced 10-fold more rapidly than at 42°C under the same salt conditions (data not shown). When the temperature was raised to 55°C, a further increase in Cl.be.I2 splicing rate was observed, but it was accompanied by RNA degradation. The observed temperature optimum for the Pe.th.I2 intron was 42°C, and the reaction rate did not increase at higher temperatures.
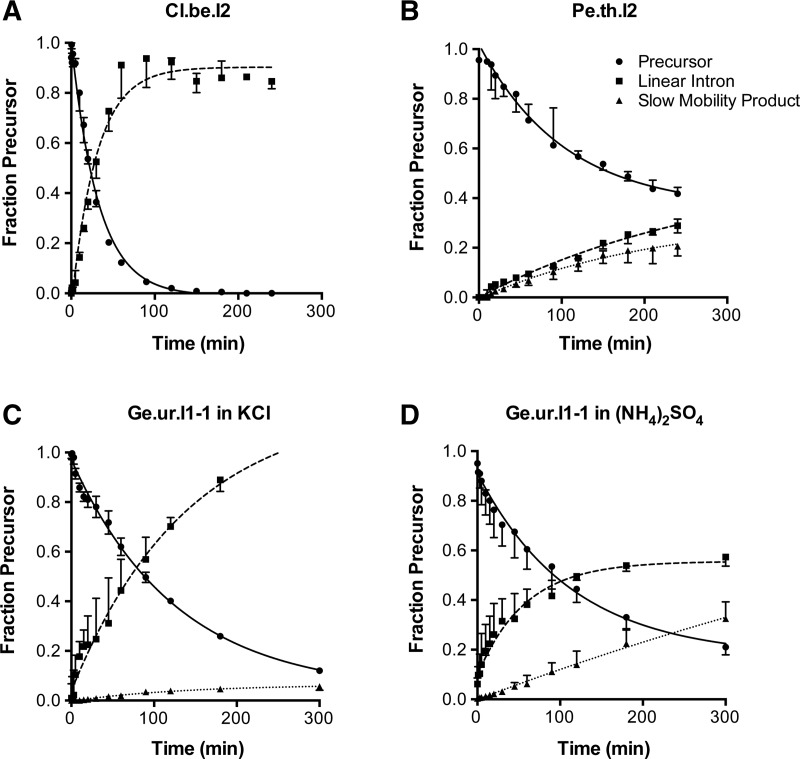
For selected introns (Cl.b.e.I2, Ge.ur.I1-1 and Pe.th.I2), we collected time courses at intervals ranging from 0.5 to 300 min in order to calculate rate constants for the splicing reaction. Maximal rate constants were calculated for reactions under optimized conditions (Fig. 5). The fraction of precursor at each time point was fit to a single exponential decay equation in order to determine the total rate constant for reaction via all splicing pathways (ktotal). For example, Cl.be.I2 reacted to completion by 2 h via the hydrolytic pathway, with a rate constant of 0.032 min−1 (Fig. 5A). The Pe.th.I2 intron reacted about 1.5 times faster in the presence of KCl as a monovalent (0.012 min−1) (time course not shown) than in (NH4)2SO4 (0.0083 min−1) (Fig. 5B), but branching was reduced in KCl. Notably, the formation of circles was significantly increased when Ge.ur.I1-1 was reacted in (NH4)2SO4 compared to reactions in KCl without a significant change in ktotal (0.010 min−1 and 0.011 min−1, respectively) (Fig. 5C,D). Overall, we observe that the group IIE and IIF introns studied here react with an efficiency similar to that of previously studied introns (Matsuura et al. 2001; Adamidi et al. 2003; Zingler et al. 2010).
FIGURE 5.
Kinetics of representative group IIE and IIF introns. Time courses for self-splicing reactions were performed under optimized conditions for each intron (see Materials and Methods for conditions). Experimental data for the disappearance of precursor RNA over time were fit to a single exponential equation with endpoint correction. We obtained rate constants of (A) 0.032 ± 0.0017 min−1 for Cl.be.I2, (B) 0.0083 ± 0.00077 min−1 for Pe.th.I2, (C) 0.011 ± 0.0015 for Ge.ur.I1-1 in KCl, and (D) 0.01 ± 0.00075 for Ge.ur.I1-1 in (NH4)2SO4. Rate constants derived from three independent experiments were averaged. Data points and curve fits are derived from a single representative trial. Error bars represent the standard error of the mean over three trials. Outliers have been omitted in the calculation of the mean and errors.
Relative role of the EBS-IBS interactions in group IIE and IIF introns reactivity and fidelity
Long-range interactions between recognition elements in the 5′-exon (the IBS sequences) and within intron DI (the EBS sequences) play a crucial role in properly positioning the 5′-exon during the two steps of splicing (Jacquier and Michel 1987; Xiang et al. 1998; Su et al. 2001). For each intron characterized in this study, we identified potential base-pairing interactions between sequences in two distinct loops of DI and the 5′-exon, suggesting the existence of putative EBS1 and EBS2 structures. To determine whether these EBS-IBS interactions contribute to in vitro self-splicing, we designed sets of mutants in which three consecutive base pairs at either the proximal or distal terminus of the EBS1-IBS1 duplex (relative to the 5′-splice site) were disrupted by mutations in EBS1 and in IBS1 of Cl.be.I2 and Pe.th.I2, which were chosen as representative IIE and IIF introns for this part of the study. Compensatory mutants that restore disrupted base-pairings were also created (Figs. 6, 7). Mutational analysis was not performed on Ge.ur.I1-1 owing to the presence of multiple EBS-IBS interactions (see above).
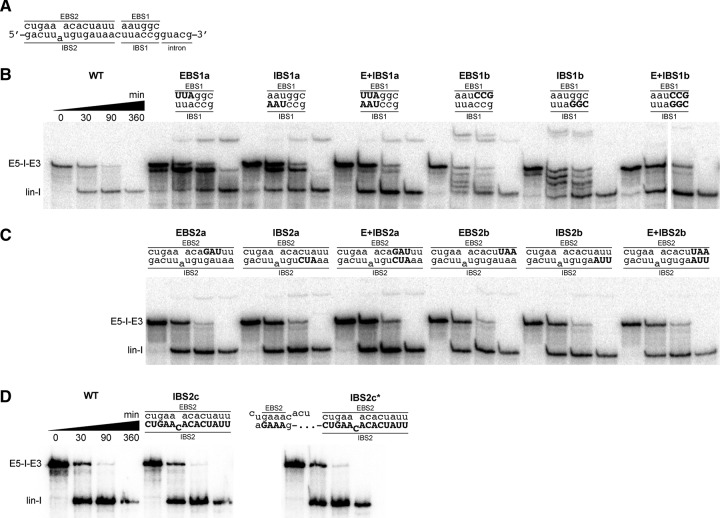
FIGURE 6.
Mutational analysis of EBS-IBS interactions in Cl.be.I2. (A) Wild type EBS1-IBS1 and EBS2-IBS2 regions are shown. Splicing of Cl.be.I2 variants with sets of three consecutive point mutations of (B) the EBS1-IBS1 and (C) the EBS2-IBS2 interactions. (D) Splicing of variants with disruption of the entire IBS (IBS2c) and the entire IBS in addition to an alternative IBS2 region 95 nt upstream of the splice site (IBS2c*). Note that the experiments in D were done on a different day, whereas experiments in B and C were done on the same day. Mutated residues are highlighted in capitalized bold characters. Time points of the splicing reaction indicated for wild type (WT) also apply for mutant splicing reactions in B, C, and D. Splicing products are labeled as in Figure 2.
FIGURE 7.
Mutational analysis of EBS-IBS interactions in Pe.th.I2. (A) Wild type EBS1-IBS1 and EBS2-IBS2 regions are shown. (B) Splicing of Pe.th.I2 variants with sets of three consecutive point mutations of the EBS1-IBS1 and EBS2-IBS2 interactions. Mutated residues are highlighted in capitalized bold characters. Time points of the splicing reaction indicated for WT also apply for mutant splicing reactions. Precursors (E5-I-E3), linear introns (lin-I), and 5′-exon (E5) are labeled according to their predicted sizes.
Under the optimized reaction conditions, wild type (WT) Cl.be.I2 spliced to completion within two hours via the hydrolytic pathway (Fig. 6A,B). Under the same conditions, mutations at the distal end of EBS1 (EBS1a) diminished overall reactivity and resulted in the formation of a slowly migrating splicing product (Fig. 6B). This product also formed when IBS1 was mutated at the corresponding positions (IBS1a). Interestingly, compensatory mutations that restore disrupted base-pairing (E + IBS1a) did not reverse this effect, and the slower mobility product still formed with roughly the same efficiency (Fig. 6B). In addition to the formation of a slower mobility product, the EBS1a and IBS1a mutants formed an additional linear product that migrates just below the precursor band. These results suggest that disruption of the three EBS/IBS base pairs farthest from the 5′-splice site disrupt both the efficiency and fidelity of initial 5′-splice site selection. Reduction in fidelity was particularly severe with mutant combinations EBS1b and IBS1b, which disrupt the three base pairs closest to the 5′-splice site, resulting in formation of multiple additional products (Fig. 6B). In this case, unlike the case of E + IBS1a, the compensatory mutations, E + IBS1b, fully restored WT splicing behavior.
Strikingly, we did not observe negative effects on splicing when we disrupted three consecutive base pairs within the EBS2-IBS2 interaction of Cl.be.I2 (Fig. 6C). To probe the EBS2-IBS2 interaction further, we mutated the entire IBS2 site. Neither mutation of the entire IBS2 site (IBS2c) nor additional mutation of a potential alternative IBS2 95 nucleotides upstream of the 5′-splice site (IBS2c*) led to the formation of additional splicing products or diminution of reactivity compared to WT Cl.be.I2 (Fig. 6D). This suggests that the EBS2-IBS2 interaction is dispensable for splice site recognition and catalysis.
A similar mutagenesis analysis of Pe.th.I2 was performed in which three consecutive base pairs at either the proximal or distal terminus (relative to the 5′-splice site) of the EBS1-IBS1 duplex were disrupted by respective mutations either in EBS1 or in IBS1 (Fig. 7A,B). Reactions were carried out in the presence of KCl to enhance overall reactivity, despite the fact that these conditions reduce the branching pathway. Under these conditions, WT Pe.th.I2 appears to form linear intron, lariat, and free 5′-exons based on product sizes. The second step of splicing appears to proceed too slowly for the accumulation of ligated exons (Fig. 7B). Mutations at the distal end of EBS1 (EBS1a) diminished reactivity and resulted in three products migrating at ∼400, 250, and 120 nt (Fig. 7B). Mutation of IBS1 at the corresponding positions (IBS1a) completely abolished catalytic activity. These findings suggest that mutation of EBS1-IBS1 base-pairings that are relatively distant from the 5′-splice site impede splice site selection, perhaps because they weaken proper anchoring of the 5′-splice site in a unique spot.
Variants of Pe.th.I2 in which we mutated three base pairs of the EBS1-IBS1 closest to the splice site (EBS1b and IBS1b) were unable to splice by branching. These mutants formed one product that migrated just above the linear intron of the WT Pe.th.I2 reaction and a second product migrating at ∼120 nucleotides in size (Fig. 7B). The compensatory mutant E + IBS1b restored the splicing products observed in the WT Pe.th.I2 reaction. These findings suggest that disrupting the base-pairings close to the splice site of the EBS1-IBS1 duplex affects fidelity of 5′-splice site selection. Mutants of Pe.th.I2 in which three consecutive base-pairings in the center of EBS2-IBS2 interaction were disrupted showed no effect on splicing (Fig. 7B).
Taken together, we conclude that the EBS1-IBS1 interaction of Cl.be.I2 and Pe.th.I2 is crucial for splice site fidelity as well as for the catalytic reactivity of the introns. In contrast, the EBS2-IBS2 interaction does not contribute to the mechanism of self-splicing by either intron.
DISCUSSION
We have, for the first time, characterized the biochemical activity of novel group IIE and IIF intron lineages that had been identified computationally in previous studies (Toro et al. 2002; Simon et al. 2008). We show now that both types of introns comprise fully functional ribozymes that self-splice independently of protein factors in vitro. The group IIE introns that we analyzed splice predominantly by hydrolysis, while the group IIF introns can also react through transesterification, producing lariats or circular introns. We found that Cl.be.I2 (IIE) and Pe.th.I2 (IIF) produce accurate splicing products based on canonical splice site recognition, whereas Ge.ur.I1-1 (IIF) can also use alternative 5′-splice sites. Intriguingly, we find that the apparent EBS2-IBS2 interaction in Cl.be.I2 (IIE) and Pe.th.I2 (IIF) plays no role in the fidelity or efficiency of self-splicing, whereas the EBS1-IBS1 interaction is crucial for splice site recognition and catalysis.
Group IIE and IIF introns are highly reactive ribozymes
The predicted group IIE and IIF introns tested here are reactive over a wide range of salt concentrations and temperatures with reaction rates comparable to previously studied group IIB introns (Matsuura et al. 2001; Adamidi et al. 2003; Zingler et al. 2010). We found group IIE and IIF introns that retained activity under conditions of very low salt concentrations (10 mM MgCl2 without additional monovalent salt). It is remarkable that these introns are able to form a stable and catalytically active tertiary structure under these types of conditions. Even in the presence of protein, salt requirements usually remain relatively high for most introns (Noah and Lambowitz 2003). The group II intron from brown algae Pylaiella littoralis is similarly reactive under low magnesium concentrations; however, it maintains a requirement for high concentrations of monovalent salts (Costa et al. 1997). Indeed, the ionic requirements of the Ge.ur.I1-1 intron are most similar to those of certain group IIC introns, such as the Oceanobacillus iheyensis intron, which splices efficiently under physiological conditions (Toor et al. 2008a).
Interestingly, we found that the Cl.be.I2 intron is most reactive at elevated temperatures, although the host organism has an optimal temperature for growth at 37°C. In vivo, the IEP or other host-encoded factors may facilitate reactivity of Cl.be.I2 at physiological temperatures. Alternatively, the intron could be “silent” and only be activated upon dramatic changes in temperature. Activation of a group II intron at elevated temperatures has been observed for an intron in Azotobacter vinelandii. The intron is located in a heat-shock protein and possibly regulates a pathway of heat shock response (Adamidi et al. 2003).
Group IIE and IIF introns utilize different splicing pathways
Group IIE and IIF introns tested in our study display clear differences in the mechanism for the first step of splicing, with the former preferring hydrolysis while the latter can also catalyze transesterification. The group IIF introns perform transesterification reactions and hydrolysis competitively as previously observed for group II introns (Daniels et al. 1996). We detected intron lariat for Pe.th.I2 and intron circles for Ge.ur.I1-1. For many years, circular introns were presumed to be byproducts or reaction artifacts of a minority of introns. However, recent studies demonstrate that circles are a direct result of the splicing reaction in vivo (Vogel and Börner 2002; Li-Pook-Than and Bonen 2006; Molina-Sánchez et al. 2006) and in vitro (Murray et al. 2001; Stabell et al. 2007). Although we identified only true circles for Ge.ur.I1-1, this group IIF intron may also form intron lariat in vitro. There are cases in which reverse transcriptase may be strongly stopped by the 2′-3′-5′ phosphodiester linkage of the branched adenosine (Murray et al. 2001). If this were true for Ge.ur.I1-1, an intron lariat would fail to amplify in our RT-PCR assay.
We have attempted to rationalize why group IIE and IIF introns prefer different splicing pathways by comparing the structural features of the two group II intron lineages. Despite structural differences between consensus secondary structures of group IIE and IIF introns (Simon et al. 2008), there is no single feature that clearly explains the mechanistic differences we observe. However, the IIF introns tend to have long, highly structured D6 stems, which may stabilize the branch-site and promote transesterification reactions by these introns. The long D6 stems of IIF introns may also engage in additional tertiary interactions that stabilize the intron core and shield the active site from water, which is the predominant nucleophile in reactions catalyzed by the group IIE introns studied here.
Investigation of more group IIE and IIF introns will be necessary to verify our observed trends in splicing preference. For example, some group IIF introns may splice predominantly through hydrolysis, whereas other group IIE introns may perform catalysis through transesterification, particularly in the presence of their encoded maturase proteins. It is possible that group IIE introns are highly dependent on protein factors to facilitate branching or circularization, and these may be provided in vivo. The IEP lineages E and F do not appear to be closely related and may have distinct roles in splicing (Simon and Zimmerly 2008). For instance, IEPs of lineage F could be limited to assistance in folding of the group IIF intron at physiological conditions, whereas IEPs of lineage E could play an additional role in facilitating transesterification reactivity of the intron.
Splice site recognition of group IIE and IIF introns
The group IIE intron Cl.be.I2 and the group IIF intron Pe.th.I2 utilize canonical splice sites in vitro. For these introns, the EBS1-IBS1 interaction is crucial to splice site recognition and catalysis, as observed for other group II introns (Jacquier and Michel 1987). Interestingly, we observed that compensatory mutants in both Cl.be.I2 and Pe.th.I2, that restore disrupted base-pairing at the distal end of the EBS1-IBS1 duplex (E + IBS1a), fail to completely reverse the effect of base pair disruption. This suggests that correct base-pairing in the EBS1-IBS1 duplex is not sufficient for positioning the 5′-splice site correctly. Instead, correct splice site selection may involve additional structural components that recognize a specific sequence of the EBS1-IBS1 duplex.
Strikingly, we found that the well-conserved and clearly defined EBS2-IBS2 interaction in Cl.be.I2 and Pe.th.I2 is completely dispensable for catalysis and splice site fidelity. Possibly, the EBS2-IBS2 interaction in group IIE and IIF introns is instead important for retrohoming and would allow highly site-specific reintegration into the genome as demonstrated for the Lactococcus lactis Ll.LtrB group IIA intron or the Sinorhizobium meliloti RmInt1 group IIB intron (Mohr et al. 2000; Barrientos-Durán et al. 2011). In contrast, group IIC introns, which naturally lack the EBS2-IBS2 interaction, are highly mobile, and they retrotranspose without high sequence specificity (Dai and Zimmerly 2002; Robart et al. 2007). Our findings indicate that the appearance of an EBS2-IBS2 interaction within the secondary structure of a group II intron does not mean that this interaction forms, or that it contributes to splicing; but the persistence of this covarying element in sequences of group IIE and IIF introns implies that the EBS2-IBS2 interaction is likely to be important for some aspect of function, such as retrotransposition. This implies that different stages of the group II intron “lifecycle” may have differential dependence on specific intron substructures.
Analysis of splicing products revealed that the group IIF intron Ge.ur.I1-1 uses two alternative 5′-splice sites (E5b-E3 and E5c-E3) in addition to the canonical splice site (E5a-E3) (Fig. 3D). The three splice sites are directly preceded by a variant of the IBS1 motif, which would allow sampling of the splice sites by different EBS1-IBS1 interactions. But only E5a-E3 and E5b-E3 are associated with an IBS2 motif that is directly upstream of the respective IBS1. Strikingly, despite the missing IBS2 motif and the distance to the intron domains, the E5c-E3 splice site seems to be favored over the conserved splice site E5a-E3 (2, 25, and 14 clones for E5a-E3, E5b-E3, and E5c-E3, respectively). This independently corroborates the observation that the IBS2 motif in the IIE and IIF introns is dispensable. Though band intensities correlate with our RT-PCR analysis, we note that it is difficult to precisely quantitate the splicing products by counting clones due to possible variation in the efficiency of reverse transcription, PCR, and cloning for the three spliced exon products. Interestingly, only the spliced exon species E5a-E3 and E5b-E3 coincide with the circular intron species we identified (2 and 16 sequences for E5a-E3 and E5b-E3, respectively) (Fig. 4B). Possibly, recognition of the E5c-E3 splice site triggers exclusively hydrolysis, whereas the canonical splice site and the alternative splice site immediately adjacent to it allow circularization.
Alternative splice site recognition has been observed in introns of other classes (Tourasse et al. 2005; Costa et al. 2006a,b; Toor et al. 2006). For the group IIC intron B.h.I1, alternative 5′-splice site selection was observed in vitro but not in vivo (Toor et al. 2006). Whether these alternative splicing events actually occur under physiological conditions in vivo, and what role they may play in Geobacter uraniumreducens biology, remains to be investigated.
CONCLUSION
We have shown that members of the group IIE and IIF intron families are catalytically active, self-splicing ribozymes in vitro. Although they have some distinct characteristics, these introns display many properties that are typical of other group II introns tested previously. For example, in the absence of the maturase, and without stabilizing host proteins such as Mss116 (Karunatilaka et al. 2010; Zingler et al. 2010; Fedorova and Pyle 2012), most of the group IIE and IIF introns are optimally reactive under high salt conditions. For the most part, the group IIE and IIF introns display appropriate splice site selection fidelity, with exceptions occurring only in cases where alternative EBS interactions are possible within the 5′-exon. The group IIE and IIF introns have properties that suggest that they are distinct from one another, as indicated by phylogenetic analyses and inspection of their secondary structures. The group IIE introns use the hydrolytic pathway exclusively during the first step of splicing in vitro, generating only linear intron products. In contrast, the group IIF introns behave more like eukaryotic group IIB introns in that they can self-splice via either the branching or hydrolysis pathways. In addition, one group IIF intron forms circular products, indicating that the IIF family catalyzes multiple transesterification reactions. Finally, we observe a parallel with group IIC introns in that the group IIE and IIF introns only utilize the EBS1-IBS1 interaction to define the 5′-splice site. Although an EBS2-IBS2 interaction is apparent from the secondary structures of group IIE and IIF introns, this motif plays no apparent role during in vitro splicing, perhaps serving to mediate the specificity of homing during subsequent retrotransposition reactions in vivo. Group IIE and IIF introns can now be considered functional ribozymes with distinct properties, having potential utility in structural biology, biotechnology, in medicine, and in studies of molecular evolution.
MATERIALS AND METHODS
Generation of intron constructs
We obtained information about secondary structures and sequences of group IIE and IIF introns from the website for mobile group II introns (http://www.fp.ucalgary.ca/group2introns/) (Dai et al. 2003; Simon et al. 2008). We used genomic DNA of Chlorobium phaeobacteroides (DSM 266), Dechloromonas aromatica (CP000089.1), Geobacter uraniumreducens Rf4 (NZ_AAON01000001.1), Pelotomaculum thermopropionicum (DSM 13744 from DSMZ), and Prosthecochloris aestuarii (DSM 271), and cells of Clostridium beijerinckii (NCIMB 8052 from NCIMB), Photorhabdus luminescens (NCIMB 14339 from NCIMB), and Syntrophomonas wolfei str. Goettingen (DSM 2245B from DSMZ) as templates for amplification of the group II introns. The region upstream of the distal loop of DIV (product A) was amplified using oligonucleotides, A-for and A-rev; and the downstream region (product B) was amplified using oligonucleotides B-for and B-rev from 1 µL liquid culture or 0.1 µg genomic DNA (Supplemental Table 1). Oligonucleotides A-rev and B-for contained overlapping regions to join product A and B by overlap extension PCR using primer A-for and B-rev. The resulting constructs including the intron and flanking exon regions were cloned downstream from a T7 promoter in pBluescript II SK (+) using HindIII and BamHI restriction enzymes. Construct sequences were verified by sequencing.
Mutant Cl.be.I2 and Pe.th.I2 constructs were generated by QuikChange site-directed mutagenesis (Stratagene) of the WT constructs (Supplemental Table 1). The Cl.be.I2 mutant IBS2c was generated by two sequential mutageneses using primers IBS2c-1-for and IBS2c-1-rev to mutate the first half of IBS2. In a second step, the resulting construct was mutated using primers IBS2c-2-for and IBS2c-2-rev to alter the second half of IBS2. The Cl.be.I2 IBS2c* mutant was generated from the IBS2c mutant. Sequences of mutant constructs were verified by sequencing.
Transcription and splicing
Constructs were transcribed from 4 µg BamHI linearized plasmids. Transcription reactions were performed in 50-µL reactions in the presence of 40 mM Tris-HCl pH 7.5, 15 mM MgCl2, 2 mM spermidine, 15 mM dithiothreitol (DTT), 1 mM each ATP, GTP, CTP, 65 µM UTP, 0.2 µM (0.6 µCi) [α-32P]UTP, 2 µL RNase inhibitor, and 3 µL T7 RNA polymerase for 1.5 h at 37°C, resulting in internally 32P-labeled precursor RNA. Reactions were spun over an Illustra MicroSpin G-25 or G-50 column (GE Healthcare) to separate reaction products from unincorporated nucleotides. Products were resolved on a 5% denaturing 7 M urea polyacrylamide gel. The major reaction product of each construct was cut out from the gel, the gel pieces were crushed, 500 µL elution buffer (10 mM MOPS pH 6.0, 300 mM NaCl, 1 mM EDTA) were added, and samples were frozen and thawed before eluting for 3 h at 4°C while shaking. The solution was filtered to remove remaining gel pieces. RNA was precipitated at −80°C for at least 1 h upon adding 1 ml cold ethanol and 30 µg glycogen. RNA pellets were dried, washed with 70% ethanol, and dissolved in 10–50 µL storage buffer (10 mM MOPS pH 6.0, 1 mM EDTA). In some cases, instead of gel purification, precursor RNA was purified using RNeasy columns (Qiagen), following manufacturer’s protocols and eluted with 10 mM MOPS, pH 6.0, 1 mM EDTA.
Splicing reactions were performed in 40 mM MOPS pH 7.5. Precursor RNA was preincubated for 1 min at 95°C and cooled for 2 min at room temperature. Monovalent salt and MgCl2 were added to start the reaction. In time-course experiments, 2 µL of the splicing reaction were added to 5 µL quench buffer (0.18× TBE, 3.6% sucrose [w/v], 72% formamide [v/v], 50 mM EDTA, 0.04% bromophenol blue [w/v], 0.04% xylene cyanol [w/v]) to stop the reaction at different time points. Splicing products were separated on a 5% denaturing 7 M urea polyacrylamide gel. Gels were then dried, exposed to a phosphor screen (GE Healthcare), and scanned with a Storm 860 phosphor imager (GE Healthcare). For isolation and characterization of splicing products, 25 µL transcript RNA was reacted for 4 h in a total volume of 50 µL. The reactions were stopped by adding 100 µL quench buffer.
Introns shown in Figure 2 were reacted in the presence of 500 mM monovalent salt and 100 mM MgCl2 at 42°C. For isolation and characterization of splicing products, Ge.ur.I1-1 was spliced in the presence of 500 mM (NH4)2SO4 and 100 mM MgCl2 at 42°C, Pe.th.I2 was spliced in 500 mM (NH4)2SO4 and 80 mM MgCl2 at 42°C, and Cl.be.I2 in 500 mM KCl and 80 mM MgCl2 at 50°C. Reaction rate constants were determined for time-course experiments in the presence of 500 mM KCl and 80 mM MgCl2 at 50°C (Cl.be.I2), 500 mM (NH4)2SO4 and 100 mM MgCl2 at 42°C (Pe.th.I2) and 500 mM monovalent salt and 100 mM MgCl2 at 42°C (Ge.ur.I1-1). Splicing of Cl.be.I2 mutants and Pe.th.I2 mutants was performed in the presence of 500 mM KCl and 80 mM MgCl2. The Cl.be.I2 mutants were reacted at 50°C and Pe.th.I2 mutants at 42°C.
Kinetics
The precursor and product bands were quantified using the program ImageQuant TL 7.0. Band signal intensities were adjusted for the uridine-content of each species, and the fraction of precursor, lariat, and linear intron calculated relative to starting material. Fractions of intensities were plotted over time. Using Graphpad Prism, the data were fit to a single exponential decay equation with endpoint correction of the form (y0 – yfinal) · e[–kobs (t)] + yfinal, where y0 is the starting point, yfinal is the endpoint, and kobs is the reaction rate. Values of kobs are reported as averages of three independent time courses with standard errors.
Characterization of splicing products
Constructs were transcribed in a standard labeling reaction and subsequently spliced as described above. Splicing products were resolved on a 5% denaturing 7 M urea polyacrylamide gel. The reaction product that corresponded in size to ligated exons and the major product of slower electrophoretic mobility than precursor RNA were cut out from the gel, eluted, filtered, precipitated, and dissolved as described for transcription products.
Reverse transcriptions were set up in a total volume of 20 µL. Reactions containing 5 µL RNA, 0.5 µM reverse primer, and 500 µM dNTP each were preincubated for 5 min at 65°C and cooled on ice. Then, 4 µL 5× SuperScript II RT reaction buffer (Invitrogen), 10 mM DTT, and 2 µL Protector RNase inhibitor (Roche) were added, and reactions were incubated for 2 min at 42°C, followed by addition of 2 µL SuperScript II RT (Invitrogen) and incubation for 35 min at 42°C and 15 min at 70°C. RT reaction products were amplified by PCR using 1 µL template and 0.5 µM each of forward and reverse primers. RT-PCR reactions of ligated exons were performed with reverse primers B-rev and forward primers A-for. Primers DI-rev and RT-PCR-for were used to amplify slower mobility splicing products (Supplemental Table 1). PCR products were extracted from an agarose gel and blunt-end ligated into pJET-1.2 (Fermentas). After transformation, plasmid DNA of several clones was purified for each splicing product and analyzed by sequencing.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
ACKNOWLEDGMENTS
We thank Donald Bryant at Penn State University, BMB, University Park, Pennsylvania, for genomic DNA of Ch.ph. and Pr.ae.; John Coates at University of California, PMB, Berkeley, California, for genomic DNA of D.a.; and Evgenya Shelobolina, University of Wisconsin-Madison, Geology and Geophysics, Madison, Wisconsin, for genomic DNA of Ge.ur. We thank Olga Fedorova for critically reading the manuscript, for helpful discussion, and for technical advice. N.P. was funded by NIH MSTP TG-T32GM007205; K.I.Z. was funded by the Yale Science Scholars Award; and J.L. was funded by the Yale College Dean’s Research Fellowship in the Sciences. This work was supported by grant GM50313 from the National Institutes of Health. A.M.P. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Freely available online through the RNA Open Access option.
REFERENCES
- Adamidi C, Fedorova O, Pyle AM 2003. A group II intron inserted into a bacterial heat-shock operon shows autocatalytic activity and unusual thermostability. Biochemistry 42: 3409–3418 [DOI] [PubMed] [Google Scholar]
- Aizawa Y, Xiang Q, Lambowitz AM, Pyle AM 2003. The pathway for DNA recognition and RNA integration by a group II intron retrotransposon. Mol Cell 11: 795–805 [DOI] [PubMed] [Google Scholar]
- Barrientos-Durán A, Chillón I, Martínez-Abarca F, Toro N 2011. Exon sequence requirements for excision in vivo of the bacterial group II intron RmInt1. BMC Mol Biol 12: 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cech TR 1986. The generality of self-splicing RNA: Relationship to nuclear mRNA splicing. Cell 44: 207–210 [DOI] [PubMed] [Google Scholar]
- Chillón I, Martínez-Abarca F, Toro N 2011. Splicing of the Sinorhizobium meliloti RmInt1 group II intron provides evidence of retroelement behavior. Nucleic Acids Res 39: 1095–1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa M, Fontaine JM, Loiseaux-de Goër S, Michel F 1997. A group II self-splicing intron from the brown alga Pylaiella littoralis is active at unusually low magnesium concentrations and forms populations of molecules with a uniform conformation. J Mol Biol 274: 353–364 [DOI] [PubMed] [Google Scholar]
- Costa M, Christian EL, Michel F 1998. Differential chemical probing of a group II self-splicing intron identifies bases involved in tertiary interactions and supports an alternative secondary structure model of domain V. RNA 4: 1055–1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa M, Michel F, Westhof E 2000. A three-dimensional perspective on exon binding by a group II self-splicing intron. EMBO J 19: 5007–5018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa M, Michel F, Molina-Sánchez MD, Martinez-Abarca F, Toro N 2006a. An alternative intron–exon pairing scheme implied by unexpected in vitro activities of group II intron RmInt1 from Sinorhizobium meliloti. Biochimie 88: 711–717 [DOI] [PubMed] [Google Scholar]
- Costa M, Michel F, Toro N 2006b. Potential for alternative intron–exon pairings in group II intron RmInt1 from Sinorhizobium meliloti and its relatives. RNA 12: 338–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai L, Zimmerly S 2002. Compilation and analysis of group II intron insertions in bacterial genomes: Evidence for retroelement behavior. Nucleic Acids Res 30: 1091–1102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai L, Toor N, Olson R, Keeping A, Zimmerly S 2003. Database for mobile group II introns. Nucleic Acids Res 31: 424–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels DL, Michels WJ Jr, Pyle AM 1996. Two competing pathways for self-splicing by group II introns: A quantitative analysis of in vitro reaction rates and products. J Mol Biol 256: 31–49 [DOI] [PubMed] [Google Scholar]
- Fedorova O 2012. Kinetic characterization of group II intron folding and splicing. Methods Mol Biol 848: 91–111 [DOI] [PubMed] [Google Scholar]
- Fedorova O, Pyle AM 2012. The brace for a growing scaffold: Mss116 protein promotes RNA folding by stabilizing an early assembly intermediate. J Mol Biol 422: 347–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granlund M, Michel F, Norgren M 2001. Mutually exclusive distribution of IS1548 and GBSi1, an active group II intron identified in human isolates of group B streptococci. J Bacteriol 183: 2560–2569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquier A 1996. Group II introns: Elaborate ribozymes. Biochimie 78: 474–487 [DOI] [PubMed] [Google Scholar]
- Jacquier A, Michel F 1987. Multiple exon-binding sites in class II self-splicing introns. Cell 50: 17–29 [DOI] [PubMed] [Google Scholar]
- Jacquier A, Rosbash M 1986. Efficient trans-splicing of a yeast mitochondrial RNA group II intron implicates a strong 5′ exon–intron interaction. Science 234: 1099–1104 [DOI] [PubMed] [Google Scholar]
- Jarrell KA, Peebles CL, Dietrich RC, Romiti SL, Perlman PS 1988. Group II intron self-splicing. Alternative reaction conditions yield novel products. J Biol Chem 263: 3432–3439 [PubMed] [Google Scholar]
- Jiménez-Zurdo JI, García-Rodríguez FM, Barrientos-Durán A, Toro N 2003. DNA target site requirements for homing in vivo of a bacterial group II intron encoding a protein lacking the DNA endonuclease domain. J Mol Biol 326: 413–423 [DOI] [PubMed] [Google Scholar]
- Karunatilaka KS, Solem A, Pyle AM, Rueda D 2010. Single-molecule analysis of Mss116-mediated group II intron folding. Nature 467: 935–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keating KS, Toor N, Perlman PS, Pyle AM 2010. A structural analysis of the group II intron active site and implications for the spliceosome. RNA 16: 1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambowitz AM, Zimmerly S 2004. Mobile group II introns. Annu Rev Genet 38: 1–35 [DOI] [PubMed] [Google Scholar]
- Lambowitz AM, Zimmerly S 2011. Group II introns: Mobile ribozymes that invade DNA. Cold Spring Harb Perspect Biol 3: a003616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leclercq S, Cordaux R 2012. Selection-driven extinction dynamics for group II introns in Enterobacteriales. PLoS One 7: e52268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li-Pook-Than J, Bonen L 2006. Multiple physical forms of excised group II intron RNAs in wheat mitochondria. Nucleic Acids Res 34: 2782–2790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcia M, Pyle AM 2012. Visualizing group II intron catalysis through the stages of splicing. Cell 151: 497–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuura M, Noah JW, Lambowitz AM 2001. Mechanism of maturase-promoted group II intron splicing. EMBO J 20: 7259–7270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel F, Umesono K, Ozeki H 1989. Comparative and functional anatomy of group II catalytic introns—a review. Gene 82: 5–30 [DOI] [PubMed] [Google Scholar]
- Mohr G, Smith D, Belfort M, Lambowitz AM 2000. Rules for DNA target-site recognition by a lactococcal group II intron enable retargeting of the intron to specific DNA sequences. Genes Dev 14: 559–573 [PMC free article] [PubMed] [Google Scholar]
- Molina-Sánchez MD, Martinez-Abarca F, Toro N 2006. Excision of the Sinorhizobium meliloti group II intron RmInt1 as circles in vivo. J Biol Chem 281: 28737–28744 [DOI] [PubMed] [Google Scholar]
- Murray HL, Mikheeva S, Coljee VW, Turczyk BM, Donahue WF, Bar-Shalom A, Jarrell KA 2001. Excision of group II introns as circles. Mol Cell 8: 201–211 [DOI] [PubMed] [Google Scholar]
- Noah JW, Lambowitz AM 2003. Effects of maturase binding and Mg2+ concentration on group II intron RNA folding investigated by UV cross-linking. Biochemistry 42: 12466–12480 [DOI] [PubMed] [Google Scholar]
- Peebles CL, Perlman PS, Mecklenburg KL, Petrillo ML, Tabor JH, Jarrell KA, Cheng HL 1986. A self-splicing RNA excises an intron lariat. Cell 44: 213–223 [DOI] [PubMed] [Google Scholar]
- Peebles CL, Benatan EJ, Jarrell KA, Perlman PS 1987. Group II intron self-splicing: Development of alternative reaction conditions and identification of a predicted intermediate. Cold Spring Harb Symp Quant Biol 52: 223–232 [DOI] [PubMed] [Google Scholar]
- Podar M, Chu VT, Pyle AM, Perlman PS 1998. Group II intron splicing in vivo by first-step hydrolysis. Nature 391: 915–918 [DOI] [PubMed] [Google Scholar]
- Pyle AM 2010. The tertiary structure of group II introns: Implications for biological function and evolution. Crit Rev Biochem Mol Biol 45: 215–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyle AM, Lambowitz AM 2006. Group II introns: Ribozymes that splice RNA and invade DNA. In The RNA world, 3rd ed (ed. Gesteland RF, et al. ), pp. 469–505 Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Pyle AM, Fedorova O, Waldsich C 2007. Folding of group II introns: A model system for large, multidomain RNAs? Trends Biochem Sci 32: 138–145 [DOI] [PubMed] [Google Scholar]
- Robart AR, Seo W, Zimmerly S 2007. Insertion of group II intron retroelements after intrinsic transcriptional terminators. Proc Natl Acad Sci 104: 6620–6625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon DM, Zimmerly S 2008. A diversity of uncharacterized reverse transcriptases in bacteria. Nucleic Acids Res 36: 7219–7229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon DM, Clarke NA, McNeil BA, Johnson I, Pantuso D, Dai L, Chai D, Zimmerly S 2008. Group II introns in eubacteria and archaea: ORF-less introns and new varieties. RNA 14: 1704–1713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solem A, Zingler N, Pyle AM, Li-Pook-Than J 2009. Group II introns and their protein collaborators. In Non-protein coding RNAs (ed. Walter NG, et al. ), Vol. 13, pp. 167–182 Springer-Verlag, New York [Google Scholar]
- Stabell FB, Tourasse NJ, Ravnum S, Kolstø AB 2007. Group II intron in Bacillus cereus has an unusual 3′ extension and splices 56 nucleotides downstream of the predicted site. Nucleic Acids Res 35: 1612–1623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su LJ, Qin PZ, Michels WJ, Pyle AM 2001. Guiding ribozyme cleavage through motif recognition: The mechanism of cleavage site selection by a group II intron ribozyme. J Mol Biol 306: 655–668 [DOI] [PubMed] [Google Scholar]
- Toor N, Hausner G, Zimmerly S 2001. Coevolution of group II intron RNA structures with their intron-encoded reverse transcriptases. RNA 7: 1142–1152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toor N, Robart AR, Christianson J, Zimmerly S 2006. Self-splicing of a group IIC intron: 5′ exon recognition and alternative 5′ splicing events implicate the stem–loop motif of a transcriptional terminator. Nucleic Acids Res 34: 6461–6471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toor N, Keating KS, Taylor SD, Pyle AM 2008a. Crystal structure of a self-spliced group II intron. Science 320: 77–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toor N, Rajashankar K, Keating KS, Pyle AM 2008b. Structural basis for exon recognition by a group II intron. Nat Struct Mol Biol 15: 1221–1222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toor N, Keating KS, Fedorova O, Rajashankar K, Wang J, Pyle AM 2010. Tertiary architecture of the Oceanobacillus iheyensis group II intron. RNA 16: 57–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toro N, Martínez-Abarca F 2013. Comprehensive phylogenetic analysis of bacterial group II intron-encoded ORFs lacking the DNA endonuclease domain reveals new varieties. PLoS One 8: e55102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toro N, Molina-Sánchez MD, Fernández-López M 2002. Identification and characterization of bacterial class E group II introns. Gene 299: 245–250 [DOI] [PubMed] [Google Scholar]
- Tourasse NJ, Stabell FB, Reiter L, Kolstø AB 2005. Unusual group II introns in bacteria of the Bacillus cereus group. J Bacteriol 187: 5437–5451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallès Y, Halanych KM, Boore JL 2008. Group II introns break new boundaries: Presence in a bilaterian’s genome. PLoS One 3: e1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Veen R, Kwakman JH, Grivell LA 1987. Mutations at the lariat acceptor site allow self-splicing of a group II intron without lariat formation. EMBO J 6: 3827–3831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel J, Börner T 2002. Lariat formation and a hydrolytic pathway in plant chloroplast group II intron splicing. EMBO J 21: 3794–3803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel J, Hess WR, Börner T 1997. Precise branch point mapping and quantification of splicing intermediates. Nucleic Acids Res 25: 2030–2031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Q, Qin PZ, Michels WJ, Freeland K, Pyle AM 1998. Sequence specificity of a group II intron ribozyme: Multiple mechanisms for promoting unusually high discrimination against mismatched targets. Biochemistry 37: 3839–3849 [DOI] [PubMed] [Google Scholar]
- Yang J, Zimmerly S, Perlman PS, Lambowitz AM 1996. Efficient integration of an intron RNA into double-stranded DNA by reverse splicing. Nature 381: 332–335 [DOI] [PubMed] [Google Scholar]
- Zimmerly S, Hausner G, Wu X 2001. Phylogenetic relationships among group II intron ORFs. Nucleic Acids Res 29: 1238–1250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zingler N, Solem A, Pyle AM 2010. Dual roles for the Mss116 cofactor during splicing of the ai5γ group II intron. Nucleic Acids Res 38: 6602–6609 [DOI] [PMC free article] [PubMed] [Google Scholar]