Using comparison of structural models and a series of mutations, forefinger loop and an Arg and a Tyr residue of archaeal Pus10 have been determined as critical determinants for its tRNA Ψ54 activity, while a Leu residue and the catalytic Asp are essential for both Ψ54 and Ψ55 activities. It is proposed that archaeal Pus10 uses two distinct mechanisms for substrate uridine recognition and binding, and the two mechanisms share some common features.
Keywords: Pseudouridine synthase, RNA modification, RluA, TruB, Methanocaldococcus, Pyrococcus
Abstract
Pseudouridines (Ψ) are found in structurally and functionally important regions of RNAs. Six families of Ψ synthases, TruA, TruB, TruD, RsuA, RluA, and Pus10 have been identified. Pus10 is present in Archaea and Eukarya. While most archaeal Pus10 produce both tRNA Ψ54 and Ψ55, some produce only Ψ55. Interestingly, human PUS10 has been implicated in apoptosis and Crohn’s and Celiac diseases. Homology models of archaeal Pus10 proteins based on the crystal structure of human PUS10 reveal that there are subtle structural differences in all of these Pus10 proteins. These observations suggest that structural changes in homologous proteins may lead to loss, gain, or change of their functions, warranting the need to study the structure-function relationship of these proteins. Using comparison of structural models and a series of mutations, we identified forefinger loop (reminiscent of that of RluA) and an Arg and a Tyr residue of archaeal Pus10 as critical determinants for its Ψ54, but not for its Ψ55 activity. We also found that a Leu residue, in addition to the catalytic Asp, is essential for both activities. Since forefinger loop is needed for both rRNA and tRNA Ψ synthase activities of RluA, but only for tRNA Ψ54 activity of Pus10, archaeal Pus10 proteins must use a different mechanism of recognition for Ψ55 activity. We propose that archaeal Pus10 uses two distinct mechanisms for substrate uridine recognition and binding. However, since we did not observe any mutation that affected only Ψ55 activity, both mechanisms for archaeal Pus10 activities must share some common features.
INTRODUCTION
Pseudouridine (Ψ), produced by post-transcriptional C5-ribosyl isomerization of uridine (U), is one of the most common modifications in RNA (Charette and Gray 2000; Cantara et al. 2011). Isomerization of U to Ψ in RNA imparts several new structural and physicochemical properties to the nucleotide, including its ability to form an additional hydrogen bond and increase base stacking (Davis 1995; Charette and Gray 2000; Hamma and Ferre-D’Amare 2006). These properties of Ψ can provide increased stability to the RNA. Ψ residues are commonly observed at functionally important regions of RNAs, and possible biological roles of certain Ψ residues in structure stabilization and function of RNAs have been inferred (Charette and Gray 2000; Ofengand et al. 2001; Decatur and Fournier 2002; Lecointe et al. 2002; Namy et al. 2005; Baudin-Baillieu et al. 2009; Karijolich and Yu 2010). Furthermore, a recent report of inducible production of Ψ during stress conditions suggests a regulatory role of this modification (Wu et al. 2011).
Characterization of Escherichia coli Ψ synthases led to classification of these enzymes into five families, namely, TruA, TruB, TruD, RsuA, and RluA (Kaya and Ofengand 2003). Pus10 was later identified as the sixth family present only in Archaea and Eukarya (Watanabe and Gray 2000; McCleverty et al. 2007; Mueller and Ferre-D’Amare 2009). Archaeal Pus10 has been shown to catalyze conversion of the universal U55 to Ψ55 in tRNAs (Roovers et al. 2006; Gurha and Gupta 2008). Additionally, we have previously shown that archaeal Pus10 can also produce Ψ54 in tRNAs (Gurha and Gupta 2008). Interestingly, human PUS10 has been implicated in TRAIL-induced apoptosis (Aza-Blanc et al. 2003; McCleverty et al. 2007) as well as in Crohn’s and Celiac diseases (Festen et al. 2011). Human PUS10 also contains conserved residues present in active sites of all Ψ synthases (McCleverty et al. 2007). This, along with the coincident presence of Ψ54 in a few mammalian tRNAs suggests that human PUS10 may also have Ψ54 synthase activity. These observations suggest that structural changes in homologous Pus10 proteins may lead to loss, gain, or change of their functions, warranting the need to study the structure–function relationship of these proteins.
Unlike Pus10-mediated Ψ55 production in Archaea, Ψ55 is produced in eukaryal and bacterial tRNAs by orthologs of TruB, which do not produce Ψ54 (Nurse et al. 1995; Becker et al. 1997). In spite of a similar function, archaeal Ψ55 synthase, Pus10 is not a TruB homolog; the only known TruB homolog in the archaeal systems is Cbf5. Both archaeal and eukaryal Cbf5 produce Ψ through a ribonucleoprotein complex, where the RNA component of the complex functions as a guide for selection of the target U (Kiss 2002; Decatur and Fournier 2003; Henras et al. 2004; Dennis and Omer 2005; Meier 2005; Yu et al. 2005; Hamma and Ferre-D’Amare 2010; Watkins and Bohnsack 2012; Ge and Yu 2013). Archaeal Cbf5 has also been shown to catalyze Ψ55 formation in tRNAs (and rRNA fragments) in a guide RNA-independent manner in in vitro reactions (Roovers et al. 2006; Gurha et al. 2007; Muller et al. 2007, 2008; Kamalampeta and Kothe 2012). However, tRNAs of a Cbf5 gene-deleted strain of Haloferax volcanii still contain Ψ55 (Blaby et al. 2011), indicating that Cbf5 is most likely not the physiological Ψ55 synthase in Archaea.
Another conserved modification in tRNA is 5-methyluridine (m5U, also known as ribothymidine [rT]) at position 54 (Juhling et al. 2009). trmA (Bjork and Isaksson 1970) and TRM2 (Hopper et al. 1982) produce Ado-Met-dependent tRNA m5U54 methyltransferase in most Bacteria and Eukarya, respectively (Greenberg and Dudock 1980; Ny and Bjork 1980; Nordlund et al. 2000). In some Bacteria, m5U54 is produced by the trmFO product (instead of TrmA), which uses N5,N10-methylene tetrahydrofolate as 1-carbon donor and FADH2 as a reducing agent (Delk et al. 1980; Urbonavicius et al. 2005). In Archaea, only members of the Thermococcales order, including Pyrococcus, harbor a thiolated m5U54 in their tRNAs (Edmonds et al. 1991; Kowalak et al. 1994; Chatterjee et al. 2012). However, the enzyme responsible for this m5U54 modification is an ortholog of a bacterial RlmD (formerly RumA) type methyltransferase, which is responsible for Ado-Met-dependent m5U1939 formation in E. coli 23S rRNA (Urbonavicius et al. 2008). Most archaeal and some eukaryal tRNAs harbor Ψ or 1-methyl-Ψ (m1Ψ) at position 54 instead of m5U (Gupta 1984, 1986; Juhling et al. 2009; Chatterjee et al. 2012).
Previously, we showed that in vitro Pus10 proteins of both Methanocaldococcus jannaschii and Pyrococcus furiosus (MjPus10 and PfuPus10, respectively) can produce tRNA Ψ54 in addition to Ψ55 (Gurha and Gupta 2008). This in vitro Ψ54 activity of MjPus10 is very robust, while that of PfuPus10 is weak and occurs only under certain experimental conditions. However, contributions of structural differences of archaeal Pus10 proteins responsible for these differential Ψ synthase activities have not been elucidated.
Here we determined the major structural features of MjPus10 responsible for its Ψ54 activity and structural differences between MjPus10 and PfuPus10 that are responsible for the reduction and/or loss of Ψ54 activity of PfuPus10. We could not delete the pus10 gene in H. volcanii (Blaby et al. 2011). Hence, we used an E. coli-based heterologous expression system to show that while MjPus10 could produce both tRNA Ψ55 and Ψ54 in an E. coli strain lacking corresponding TruB (Ψ55 synthase) and TrmA (m5U54 methyltransferase) activities, PfuPus10 could form only Ψ55 under identical conditions. Interestingly, MjPus10 could produce tRNA Ψ54 instead of m5U54 in E. coli strains, even in the presence of native TrmA. We created several amino acid substitution and deletion mutants of MjPus10 and assayed them for their tRNA Ψ55 and Ψ54 activities both in vivo and in vitro. MjPus10 proteins that had Ala substitution of the conserved catalytic core and thumb loop residues, and deletions in the forefinger loop region, showed differences in their Ψ55 and Ψ54 activities. Of particular interest was the forefinger loop, also present in RluA, but absent in TruB Ψ synthase families (Hamma and Ferre-D’Amare 2006; McCleverty et al. 2007; Mueller and Ferre-D’Amare 2009). We show here that proper size and configuration of the Pus10 forefinger loop is essential for its Ψ54, but not Ψ55 activity. Hence, we hypothesize that MjPus10 uses a novel combination of an RluA-like mechanism along with a second different mechanism to recognize and modify its substrate uridines at tRNA positions 54 and 55, respectively, and both mechanisms share some common features.
RESULTS
Modifications of conserved catalytic core residues of MjPus10 differentially affect tRNA Ψ54 and Ψ55 synthase activities
A homology model of the MjPus10 protein was prepared based on the C-terminal domain (PDB accession 2V9K: Gly286–Asp528) of the human PUS10 crystal structure (McCleverty et al. 2007). Structural alignment of human PUS10 and MjPus10 showed nearly superimposable catalytic domains (Fig. 1A). To identify functionally important residues, we looked at the amino acids that are conserved within the active sites of all Ψ synthases (McCleverty et al. 2007; Mueller and Ferre-D’Amare 2009). These are: (1) a catalytic Asp, (2) an Arg/Lys that forms a salt bridge with the catalytic Asp, (3) a Tyr/Phe that packs close to the ribose of the target U, (4) a hydrophobic residue, and (5) a Leu. The last two residues are involved in nucleotide stacking. Corresponding MjPus10 residues are Asp275, Lys413, Tyr339, Ile412, and Leu440, respectively (Fig. 1A). Ψ synthases of four families (TruA, RsuA, RluA, and Pus10) also contain an Arg (Arg273) two residues N-terminal to the catalytic Asp (Mueller and Ferre-D’Amare 2009). Furthermore, all Pus10 proteins also have a conserved Asp (Asp277) two residues C-terminal to the catalytic Asp. A second Asp at a similar location is also found in certain members of the RsuA (RsuA, RluE, and RluF) and RluA (RluC and RluD) families (Ofengand et al. 2001). These seven residues are conserved in both MjPus10 and PfuPus10 (Fig. 1A). To explore the importance of these residues toward tRNA Ψ54 and Ψ55 formation, we performed Ala substitutions of each residue in MjPus10 and analyzed pseudouridylation activities of the mutant proteins using H. volcanii tRNATrp (see the sequence in Supplemental Fig. 1A) as substrate. Ala substitutions were chosen to remove the potential key chemical properties of side chains (i.e., charge in Asp and Lys; hydrophobicity and bulk of Tyr, Ile, and Leu) while minimizing the introduction of novel chemical properties (Cunningham and Wells 1989; Morrison and Weiss 2001). We determined end levels of Ψ production by different mutant proteins because of the limitations in accurate quantitation of Ψ at early time points under our experimental conditions (see Materials and Methods) and compared end points rather than the rates of reaction. Furthermore, these reactions can be compared with the in vivo Ψ modifications (see later) that are essentially end point in nature.
FIGURE 1.
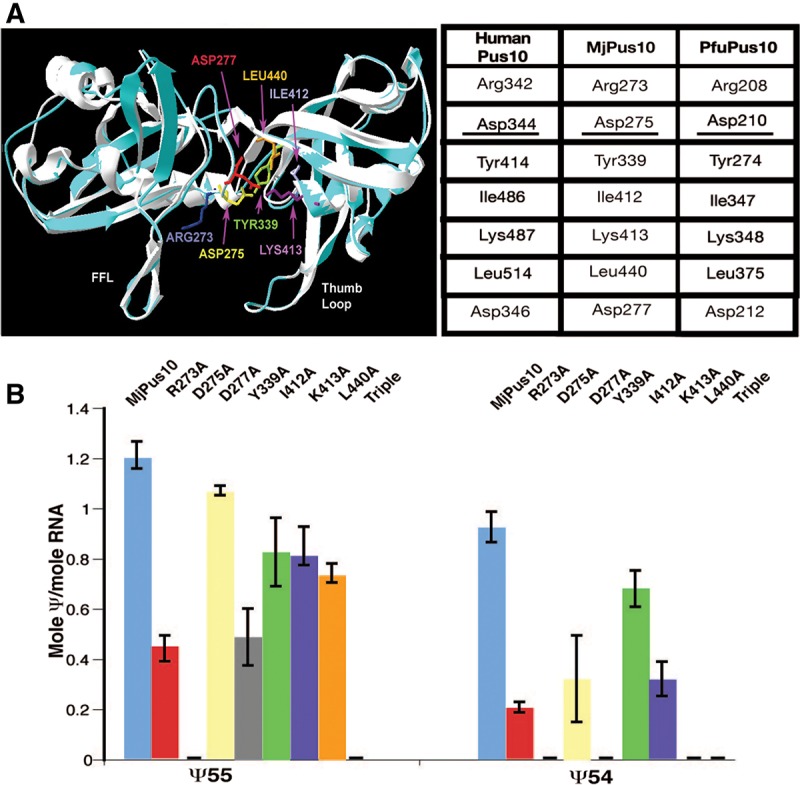
Differences in tRNA Ψ54 and Ψ55 formation by mutants of MjPus10 catalytic core residues. (A) Homology model of a part of the catalytic domain of MjPus10 (blue) superimposed on the C-terminal domain of the human PUS10 crystal structure (white) (McCleverty et al. 2007). Certain residues that are conserved in the catalytic core of most Ψ synthases are indicated. These residues of MjPus10 are shown in different colors and are labeled. (FFL) Forefinger loop. Homologous residues in human PUS10, MjPus10, and PfuPus10 proteins are shown in the table. The catalytic Asp is underlined. (B) Wild-type MjPus10 or its mutants, as indicated, were used to modify in vitro-transcribed [α-32P]UTP or [α-32P]CTP-labeled H. volcanii tRNATrp to determine Ψ54 or Ψ55 production, respectively. Mole Ψ/mole RNA was determined from TLC analysis of RNase T2 digests of the products. “Triple” mutant is a combination of I412A, K413A, and L440A. Error bars represent standard errors of at least two independent experiments.
As expected, mutating the catalytic Asp to Ala (D275A) resulted in the loss of all Ψ activity (Fig. 1B). Most other mutants retained 40%–70% of the wild-type Ψ55 activity (Fig. 1B). In contrast, Ψ54 activity was more severely affected in all cases. Mutant I412A retained ∼70% activity; mutants R273A, D277A, and K413A retained ∼20%–35% activity, whereas mutants Y339A and L440A showed no detectable Ψ54 activity. Next, to determine the synergistic effect of these mutations, we created a triple mutant combining the three mutations (I412A, K413A, and L440A), each of which individually showed ∼70% Ψ55 activity. This mutant lost all Ψ55 and Ψ54 synthase activities (triple in Fig. 1B).
Forefinger and thumb loops are critical structural determinants for tRNA Ψ54 activity of archaeal Pus10 proteins
Next, we wanted to determine the cause of differences in Ψ54 and Ψ55 activities of MjPus10 and PfuPus10. Like human PUS10, homology models of MjPus10 and PfuPus10 sequences have predicted thumb and forefinger loops (Fig. 2A), which in human PUS10 are thought to hold the RNA by “pinching” the major and minor grooves, respectively (McCleverty et al. 2007; Mueller and Ferre-D’Amare 2009). These loops are also observed in members of RluA and TruA families (Hamma and Ferre-D’Amare 2006; Mueller and Ferre-D’Amare 2009). The forefinger loop of the PfuPus10 homology model, however, is predicted to be smaller and lacks sufficient amino acids to form the short antiparallel β sheet secondary structures observed in the human PUS10 and the MjPus10 model (although these structures do not appear in the McCleverty et al. 2007 publication, they do appear in the published structure 2V9K) (Fig. 2A). PfuPus10 forefinger loop region has one large or two smaller deletions when compared with MjPus10 by either sequence or structural alignment (Fig. 2B,C). These are referred to here as ffl1 (Cys227–Arg231) found within the forefinger loop and ffl2 (Cys238–Val249) in the adjacent region. To determine the roles of these segments of the forefinger loop, we created three deletions in MjPus10 to make it more like that of PfuPus10; all of which retained >60% Ψ55 activity (Fig. 2D). However, deletion of ffl1 drastically reduced Ψ54 activity (to ∼5%–6%), while that of ffl2 and the double deletion of ffl1 and ffl2 showed no detectable Ψ54 activity.
FIGURE 2.
Forefinger loop of MjPus10 is a positive determinant of tRNA Ψ54 activity. (A) Homology models for MjPus10 (blue) and PfuPus10 (orange) based on the crystal structure of the C-terminal domain of human PUS10 (white) and overlap of the forefinger loops from the three structures. Both the structure and the size of PfuPus10 forefinger loop (orange loop) are different from those of the other two. (B) Clustal X2 sequence alignment of a region containing the forefinger loop and catalytic Asp residue of Pus10 protein sequences from four animal species (Drosophila melanogaster, Danio rerio, Homo sapiens, Mus musculus), two Pyrococcus species (abyssi and furiosus), Methanocaldococcus jannaschii, and two related Euryarchaeota (Halobacterium sp, Haloarcula marismortui). Position of the forefinger loop is shown relative to the catalytic Asp. Regions corresponding to the ffl1 and ffl2 deletion created in MjPus10 are marked. (C) Structural alignment of regions comparable to those in B of human PUS10, MjPus10, and PfuPus10 proteins. ffl1 and ffl2 indicate residues in the forefinger loop and adjacent region, respectively, that are present in MjPus10, but not in PfuPus10. ffl2 is also not present in human PUS10. “h” and “s” indicate helix and sheet, respectively. Asterisks indicate sequence identity and dots indicate sequence similarity. (D) Ψ54 and Ψ55 production by deletion mutants was determined as in Figure 1B. Δffl1 and Δffl2 are single deletions of ffl1 and ffl2, respectively, and Δffl(1 + 2) is double deletion for both ffl1 and ffl2.
To find additional determinants for the differences in Ψ54 and Ψ55 activities of MjPus10 and PfuPus10, a multiple structural alignment of the forefinger loop to the thumb loop region of Pus10 of P. furiosus, which does not have Ψ54 in its tRNAs, was done with Pus10 proteins from selected Archaea (Thermoplasma acidophilum, Methanobacterium thermoautotrophicum, M. jannaschii, H. volcanii) that are known to have Ψ54 or m1Ψ54 in their tRNAs (Chatterjee et al. 2012) using human PUS10 as a model (Supplemental Fig. 2). The major difference revealed by this alignment was that PfuPus10 contained Asn311 and Ser312 in the thumb loop at the positions where His376 and Arg377 (or similar amino acids) were present in other archaeal and human PUS10. His376 and Arg377 in the thumb loop of MjPus10 create a “hook-like” structure (Fig. 3A), which results in a very narrow gap between the thumb and forefinger loops. However, PfuPus10 lacks this “hook,” which, in addition to the much smaller forefinger loop, results in a wide gap in between these two loops (Fig. 3B). An in silico analysis comparing the molecular surfaces of MjPus10 with its double mutant H376N/R377S, a PfuPus10-like substitution, shows an increased space between the loops (Fig. 3A,C). Substitution of these His and Arg residues by Ala also showed virtually identical structure as Pfu-like substitutions with an increased space between the loops (Fig. 3D). Hence, we replaced these two thumb loop residues of MjPus10 by Ala (H376A/R377A). This double mutant protein showed ∼70% of Ψ55 and 40% of Ψ54 activity (Fig. 3E), suggesting that in vitro (not in vivo, see Table 1) these residues are more critical for Ψ54 activity than Ψ55 activity.
FIGURE 3.
Certain thumb loop and N-terminal Cys residues are important for in vitro Ψ54 activity of MjPus10. Molecular surface of the close up of the substrate-binding regions of (A) MjPus10, (B) PfuPus10, (C) H376N/R377S mutant of MjPus10, and (D) H376A/R377A mutant of MjPus10. Substituting H376 and R377 amino acids in the thumb loop of MjPus10 with N376 and S377 of PfuPus10 residues (or with Ala) results in a large gap between forefinger and thumb loops, as it is in native PfuPus10. (E) Ψ54 and Ψ55 production by H376A/R377A and C106A/C109A mutants was determined as in Figure 1B. Note that the amount of Ψ54 production by C106A/C109A mutant was exactly the same in duplicate experiments, and hence, there is no error bar.
TABLE 1.
In vitro and in vivo activities of MjPus10 and its mutants

A zinc atom coordinated by four Cys residues in the N-terminal domain has been observed in the crystal structure of human PUS10 (McCleverty et al. 2007). MjPus10 protein also has four similarly located Cys residues (positions 31, 34, 106, and 109). A mutant protein having two of these four Cys replaced by Ala (C106A/C109A) showed reduced Ψ54 formation in vitro (Fig. 3E). However, this mutant showed both Ψ54 and Ψ55 activity under in vivo conditions (Table 1).
Nucleotides outside of the TΨC arm of tRNA are important for RNA–protein interaction
Previously, we have shown that the minimal substrate for the wild-type MjPus10 Ψ54 and Ψ55 activities is a 17-mer RNA comprising the TΨC stem–loop (TSL) of tRNAs (Gurha and Gupta 2008). To determine whether this is also the case for our mutant proteins, we used this TSL substrate to assay the activity of each mutant protein. The results were often different from that of a full-size tRNA substrate (cf. 17-mer TSL to tRNA in Table 1). Only the thumb loop mutant H376A/R377A and the zinc-binding motif mutant C106A/C109A retained both Ψ54 and Ψ55 activities and only D277A, I412A, and ffl1 deletion produced Ψ55 in the TSL substrate. Loss of mutant protein activity toward TSL is not due to denaturation of TSL at a reaction temperature of 68°C; wild-type protein does show activity on TSL at this temperature (Table 1). This implies that tRNA nucleotides outside of the TSL region are important for overall Pus10 activity, probably by stabilizing the protein–RNA interaction.
MjPus10 can produce tRNA Ψ54 and Ψ55 independent of each other
Our previous work showed that formation of Ψ54 by MjPus10 does not require the presence of U at position 55 in the tRNA (Gurha and Gupta 2008). This U could be changed to C, A, or G. To determine whether the prior presence of Ψ55 has an effect on the Ψ54 formation by MjPus10, tRNA was treated with E. coli TruB to convert U55 to Ψ55, followed by MjPus10 treatment. There was not much difference in Ψ54 formation in both U55 and Ψ55 containing tRNA (Supplemental Fig. 3A). Furthermore, the amount of total modification by MjPus10 was nearly the same for PAB0719 (Pyrococcus abyssi m5U54 methyltransferase) (Urbonavicius et al. 2008), treated and untreated tRNA (Supplemental Fig. 3B). This indicated that the presence of m5U54 does not affect Ψ55 activity of MjPus10. All of these results suggest that Ψ54 and Ψ55 formations by Pus10 are independent of each other.
PfuPus10 can produce tRNA Ψ55 but not Ψ54 in vivo in E. coli, while MjPus10 can produce both
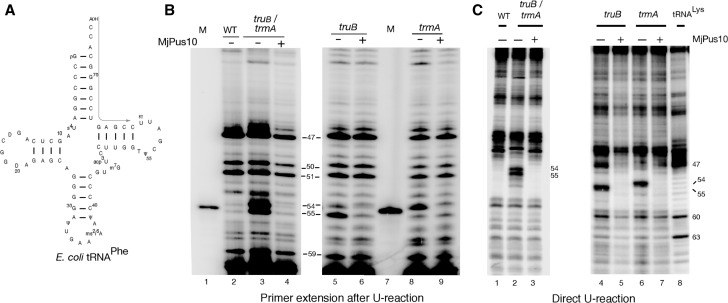
Our in vitro data demonstrated the role of various amino acid residues and regions of Pus10 in its Ψ synthase activities. However, Pus10-mediated in vivo synthesis of Ψ54 and Ψ55 provides a better understanding of the roles of these residues and regions during dynamic interactions of RNA synthesis and modification by Pus10. Therefore, we determined in vivo activities of Pus10 proteins in parallel with their in vitro activities. We cloned MjPus10 and all of its mutants, as well as PfuPus10 in pTrc99a vector, for direct expression of proteins in different E. coli strains. We checked heterologous expression of MjPus10 and PfuPus10 in E. coli strains mutant for trmA (m5U54 methyltransferase) and deleted for truB (Ψ55 synthase) genes separately as well as together. These trmA and truB strains have unmodified U at tRNA positions 54 and 55, respectively, and the double mutant has unmodified U at both positions. Initially U-specific reaction of total tRNA isolated from MjPus10-expressing E. coli cells was followed by primer extension using a primer specific for E. coli tRNAPhe (Fig. 4A). Expression of MjPus10 resulted in the disappearance of unmodified U bands at positions 54 and 55 in all strains (Fig. 4B, lanes 4,6,9), suggesting that MjPus10 can modify both U54 and U55 of tRNAPhe inside E. coli cells. As mentioned in the Materials and Methods section, this procedure can only determine the presence or absence of U, but is not suitable to determine the quantity of leftover unmodified U. We also used direct (i.e., without primer extension) U-specific reactions of 3′-end labeled total tRNA to determine whether this U54 and U55 modification activity of MjPus10 extends to the majority of tRNAs inside the E. coli cells (Fig. 4C). Unlike primer extension of U-specific cleavage of RNA, this method shows direct cleavage of RNA. Since the locations of residues 54 and 55 in all tRNAs are the same from their 3′ ends, we could monitor the modifications at these positions. The results for total tRNA were similar to the ones with tRNAPhe, i.e., MjPus10 modifies both U54 and U55 in (nearly) all tRNAs (unmodified U bands were not observed when MjPus10 was present) (Fig. 4C, cf. lanes 2 and 3, 4 and 5, and 6 and 7).
FIGURE 4.
MjPus10 can modify U54 and U55 of tRNAs in E. coli. (A) Sequence of E. coli tRNAPhe used for primer extension studies. Arrow indicates position of the annealing primer. (B) U-specific reactions of total tRNA isolated from different E. coli strains, followed by primer extension using 5′-labeled tRNAPhe-specific primer. WT (DH5α strain used as wild type), trmA mutant, and truB null strains are indicated above the lanes. Presence (+) or absence (−) of MjPus10 containing plasmid in the cell is also indicated. Numbers on the side mark the positions of modified or unmodified U in tRNAPhe. A dark band at any of these positions suggests the presence of an unmodified U at that position. Dark band at position 47 is due to primer extension stop caused by a hypermodified U (see A). (M) A 22-base labeled primer used as size marker. (C) Total tRNA isolated from different E. coli strains was 3′-end labeled with [5′-32P]pCp and followed by a U-specific reaction. U-specific reaction of 3′-end labeled E. coli tRNALys served as a marker. In addition to positions 54 and 55, certain other positions containing unmodified U in tRNALys are also indicated.
Since the absence of a dark band in the U-specific reaction does not determine the modification to be Ψ or m5U, we utilized a more specific CMCT-based technique to ensure that this MjPus10-mediated U modification is indeed a Ψ. In these reactions, Ψ-specific band at position 55 was observed in all cases except in Pus10-lacking controls when TruB activity was absent, i.e., in truB-deleted and truB/trmA double mutant strains (left panels in Fig. 5,A,C, respectively). The presence of Ψ55-specific bands in these two TruB-lacking strains, when they contained either MjPus10 or PfuPus10 (middle and right panels in Fig. 5, A and C) suggests that both of these proteins can produce tRNA Ψ55 in E. coli. As expected, no Ψ-specific band was observed at position 54 in Pus10-lacking controls (left panels in Fig. 5,A–D) in the CMCT reactions, because these E. coli strains contain either m5U54 in the presence of TrmA or U54 in its absence, none of which would produce band in CMCT reactions. However, Ψ54 bands were observed in TrmA-lacking strains when MjPus10 was present (middle panels in Fig. 5A,B), suggesting that MjPus10 can convert U54 to Ψ54 in these strains. No such Ψ54 activity was observed for PfuPus10 in any strain (right panels in Fig. 5A–D). Again, as mentioned in the Materials and Methods section, this procedure is used to determine Ψ production, but is not suitable for quantitation of Ψ. Overall, these results indicate that MjPus10 can produce both Ψ55 and Ψ54 in vivo in E. coli tRNAs, whereas PfuPus10 can produce only Ψ55. Furthermore, these results also show that both MjPus10 and PfuPus10 proteins are active inside E. coli cells at 37°C, even though M. jannaschii and P. furiosus are routinely grown at 85°C and 90°C, respectively.
FIGURE 5.
MjPus10 can produce both tRNA Ψ54 and Ψ55 in E. coli, even in the presence of TrmA, but PfuPus10 can produce only Ψ55. CMCT analyses of tRNA isolated from E. coli strains mutant for both TruB and TrmA (A), TrmA (B), TruB (C), and wild-type DH5α (D) expressing MjPus10 or PfuPus10. Total tRNA was treated with (+) or without (−) CMCT as indicated (in minutes), followed by alkali (OH−) treatment (+) or no treatment (−). Primer extension was done with primer specific for E. coli tRNAPhe (see Fig. 4A). Dark bands at positions 54 and 55 in 10- and 20-min reactions indicate the presence of Ψ. (E) TLC analysis of [32P]-labeled total tRNA isolated from wild-type (DH5α) or truB strains expressing MjPus10 and digested with nuclease P1. These two strains contain wild-type trmA gene. pA, pG, pC, pU, pΨ, and pm5U indicate 5′-phosphorylated A, G, C, U, Ψ, and m5U (rT), respectively. (Right) The pm5U spots are absent and pΨ spots are darker than left panels.
MjPus10 tRNA Ψ54 activity strongly outcompetes native bacterial TrmA activity in E. coli
Surprisingly, MjPus10 produced Ψ54 in E. coli tRNAPhe instead of m5U54, even when wild-type TrmA was present in the cell (middle panels in Fig. 5C,D). This Ψ54 was produced even in the uninduced DH5α strain, where very low basal levels of MjPus10 were present (data not shown). MjPus10 produces Ψ54 in all tRNAs, as observed by the absence of m5U54 in total tRNA isolated from 32P-labeled E. coli cells containing both MjPus10 and TrmA (Fig. 5E). Here, TLC analyses of nuclease P1 digests of total tRNA isolated from DH5α and truB (both strains contain native trmA) mutant strains show a radioactive spot for m5U in each case (left), which is not observed when these strains express MjPus10 (right). This suggests that m5U54 is not produced in E. coli tRNAs when MjPus10 is present along with TrmA. Furthermore, a concomitant increase in Ψ (derived from Ψ54) is also observed in these cases (Fig. 5E, right). Overall, these results suggest that MjPus10 produces Ψ54 not only in tRNAPhe, but also in nearly all tRNAs, and also blocks m5U54 activity of TrmA when both proteins are present together in E. coli. Similar competition between MjPus10 and TruB, if at all, for Ψ55 production, cannot be determined because they produce indistinguishable Ψ55 products.
Heterologous in vivo activities of most MjPus10 mutants are similar to their in vitro activities
To study the effects of mutations of Pus10 under in vivo conditions, mutant versions of MjPus10 used for in vitro studies were also produced and expressed in our heterologous in vivo system. Initially, the mutants were expressed in E. coli strains lacking both TruB and TrmA activities, which normally contain unmodified U54 and U55 in their tRNAs. Results are shown in Figure 6 and summarized in Table 1. Unlike in vitro reactions, in vivo activities of mutants can only be scored as presence or absence of Ψ, since these cannot be quantified by our techniques (see Materials and Methods). Ψ54 and Ψ55 activities of all mutants, except R273A and L440A, were similar to their in vitro activities with the full-size tRNA. Even the mutants that had low Ψ54 activity in vitro showed complete Ψ54 conversion of native substrate in vivo. A partial in vivo production of Ψ with a substantial amount of leftover unmodified U shows dark bands in both CMCT and U-specific reactions (M Majumder and R Gupta, unpubl.), which was not observed with any mutant in the present study. Either the mutant produced a Ψ or showed an unmodified U at a particular position. As expected, the catalytic Asp mutant D275A could not rescue either Ψ formation. The mutant Y339A, which only had Ψ55 activity in vitro, showed the same result in vivo as well. The R273A mutant did not show any Ψ54 activity in vivo, although it had a low amount of this activity in vitro. The L440A mutant had robust Ψ55 activity, but no Ψ54 activity in vitro. However, in vivo it showed neither Ψ55 nor Ψ54 activity. This absence of Ψ activity was not due to the absence of mutant protein in the cell, as expression of L440A and D275A (the two proteins that did not show any Ψ activity), was comparable to the expression of wild-type MjPus10 under similar conditions as estimated by SDS-PAGE of induced cell lysates (data not shown). Overall, these data suggest that except for R273A and L440A, the functions of the mutant proteins are similar under in vitro and in vivo conditions.
FIGURE 6.
In vivo activities of MjPus10 mutants in the heterologous E. coli system. Primer extension following CMCT (top) and U-specific (bottom) reactions were done (as in Figs. 5 and 4, respectively) on total tRNA from E. coli strains lacking both TruB and TrmA activities (normally containing unmodified U54 and U55 in their tRNAs), expressing different mutants of MjPus10. Lanes 1 and 2 in U-specific reactions are primer extensions without and with U reactions, respectively. Unmodified U51 and U50 of E. coli tRNAPhe serve as internal controls. Asterisks are against the bands at positions 54 and 55. pTrc99a (vector only), D275A, and L440A do not show any Pus10 activity (U54 and U55). MjPus10 (wild-type), D277A, I412A, K413A, Δffl1, H376A/R377A, and C106A/C109A show both activities (Ψ54 and Ψ55). R273A, Y339A, Δffl2, and Δffl(1 + 2) show only Ψ55 activity (U54 and Ψ55).
As shown above, tRNAs of wild-type E. coli strain (DH5α) contain Ψ54 instead of m5U54 when wild-type MjPus10 is present in the cell. To test the behavior of mutant MjPus10 proteins in the presence of native TrmA, all mutant proteins that were previously expressed in TruB and TrmA lacking E. coli strains were also expressed in a DH5α strain of E. coli. Behaviors of all mutants for their Ψ54 activities were identical in the presence and absence of TrmA (cf. in vivo Ψ54 activity in DH5α and truB/trmA strains shown in Supplemental Figs. 4 and 6, respectively, as well as summary in Table 1), suggesting that all mutant proteins that had any Ψ54 activity in TrmA-deleted strains retained their ability to block native TrmA activity in its presence. Furthermore, the presence in E. coli of those mutant proteins that did not show in vivo Ψ54 activity (R273A, D275A, Y339A, L440A, and ffl2 deletions, listed in Table 1) did not affect native TrmA activity in DH5α cells. The tRNA in these cases contained m5U54, as observed by the absence of an unmodified U54 in U-specific reactions and the absence of Ψ54 in CMCT reactions (Supplemental Fig. 4). These data indicate that unmodified U54 is not available to native TrmA when functional MjPus10 is present in the cell.
DISCUSSION
MjPus10 is a multi-site Ψ synthase, but PfuPus10 is specific only for a single site in archaeal tRNAs in vivo
There is a strong indication that most Archaea that harbor the pus10 gene contain Ψ or m1Ψ at tRNA position 54 (Urbonavicius et al. 2008; Chatterjee et al. 2012). The only exceptions are members of order Thermococcales, like that of Pyrococcus, which contain a modified m5U54 in their tRNAs (Chatterjee et al. 2012). This coincidence of weak in vitro Ψ54 activity of PfuPus10 (Gurha and Gupta 2008) and the simultaneous presence of m5U54 in Pyrococcus tRNAs (Urbonavicius et al. 2008) suggests that PfuPus10 functions as a single-site (for Ψ55) Ψ synthase under native conditions. This is in contrast to other archaeal Pus10 proteins such as MjPus10, which produces both Ψ54 and Ψ55 in vitro as shown earlier (Gurha and Gupta 2008) and in vivo, as shown here by heterologous expression of these proteins in E. coli. During evolution, probably members of the Thermococcales order lost Ψ54 activity of Pus10 when their ancestor acquired an ortholog of bacterial RlmD methyltransferase by horizontal gene transfer (Urbonavicius et al. 2008). This rRNA methyltransferase evolved to produce tRNA m5U54, and m5U further thiolated to s2m5U as an adaptation to hyperthermophilic conditions of growth of these organisms (Kowalak et al. 1994; Grosjean and Oshima 2007).
MjPus10 can produce Ψ54 in E. coli even in presence of native trmA, outcompeting its product tRNA-m5U54 methyltransferase (Fig. 5). These effects are not due to excess Pus10 proteins, because even the basal levels of MjPus10 under uninduced conditions can block activity of this enzyme in DH5α cells (data not shown). Furthermore, all mutants of MjPus10 that are functional for Ψ54 also out compete this methyltransferase (tRNAs have Ψ54), but mutants that have no Ψ54 activity do not (tRNAs have m5U54). Probably, MjPus10-mediated Ψ54 production occurs before TrmA can convert U54 to m5U54 during tRNA biogenesis. Once a Ψ is produced in RNA, it cannot be converted to m5U by methylation. We have shown that in vitro Ψ54 and Ψ55 activities of MjPus10 are independent. However, since Pus10 proteins can produce Ψ55 in E. coli (see Fig. 5), it is also possible that during tRNA biogenesis MjPus10 binds a tRNA substrate and converts both U54 and U55 to Ψ (which may occur sequentially) before releasing it, thus not allowing m5U54 production. However, in such a case one has to assume that MjPus10 can also outcompete native Ψ55 synthase (truB product). Our study cannot determine whether MjPus10 or TruB produces Ψ55 in E. coli, when both coexist simultaneously, because in either case the end product will be Ψ55.
tRNA Ψ54 activity of MjPus10 is more sensitive to structural perturbations of the protein than its Ψ55 activity
We compared the in vitro activities of our wild-type and mutant Pus10 proteins with their in vivo activities. Since all of our mutant (except catalytic D275A) proteins retained substantial Ψ55 activity (Table 1), it is safe to assume that the mutant proteins retain similar structures and do not have significant misfolding. In vitro activities of mutant proteins were determined under single turnover conditions. We have previously shown that most of the Ψ54 and Ψ55 were produced by MjPus10 within the first 2 min (reached maximum in 5 min) of reaction under similar conditions (Gurha and Gupta 2008). In the present work, end levels (after 1 h) of reactions were used to determine the activities of mutant proteins. Although these reaction conditions mask kinetic effects of mutations, these are theoretically comparable to activities under in vivo conditions. In principle, in vivo Ψ modifications can be defined as end points of the reactions occurring in the cytoplasm within generation time of E. coli at 37°C.
Mutant MjPus10, where catalytic Asp275 was changed to Ala, lost all of its Ψ synthase activity. All other mutants retained at least about half of the wild-type Ψ55 activity under in vitro conditions (Table 1). This was observed even when some mutants lost Ψ54 activity. In vitro effects of all mutations on Ψ54 activity were more drastic than on their Ψ55 activities. Overall, these observations suggest that Ψ54 activity of MjPus10 is more sensitive to structural perturbations of the protein than its Ψ55 activity. Mutants I412A, K413A, and L440A, which individually retained ∼70% of the wild-type activity, lost all Ψ synthase activity when combined together (Fig. 1B). This suggests that each conserved residue plays some quantitative role in enzyme activity that becomes significant when combined together, but none of these by itself is essential for Ψ55 production.
In vitro Ψ activity of all mutants (except for R273A and L440A) were similar to their Ψ activity in vivo in E. coli. Ψ conversion in vivo was complete even if it was limited in amount in vitro (Table 1). Mutants that lost Ψ54 activity in vitro also lost this activity in vivo (see Y339A, L440A, and deletions of forefinger loop segment ffl2 in Table 1). Overall, these results suggest that several conserved catalytic core residues and configuration and size of forefinger loop of MjPus10 are major determinants for its Ψ54 activity, but not for Ψ55 activity. Furthermore, the lack of any one mutant that affected U55 modification without affecting U54 suggests that U54 modification relies on shared components of U55 modification in addition to other structural features.
Apparent improvement in in vivo Ψ54 activity of mutants (when compared with their in vitro activities) might be due to more stable interactions of mutant proteins with tRNA under in vivo environment than under our in vitro reaction conditions using purified RNA and protein components. This improvement in in vivo activity over in vitro activity was also noted for Ψ55 formation. These observations suggest that individual mutations of certain residues that are conserved among homologous proteins do not affect overall activity of the protein inside the cell, even when the same mutation reduces activity in vitro. This property may be useful for evolutionary divergence of homologous proteins.
L440A mutant (in addition to catalytic D275A mutant) did not show any Ψ synthase activity in vivo, although it had substantial Ψ55 activity in vitro. R273A did not show Ψ54 activity in vivo, although it had both activities in vitro. One can speculate that these amino acids are necessary under natural reaction conditions inside the cell. On the other hand, in vitro, if these amino acids are absent, certain other amino acids of the protein may provide similar functionality, especially when purified reaction components and higher enzyme concentrations (under higher salt concentrations than in the E. coli cytoplasm) are used in the reaction. The essential nature of Leu440 under in vivo conditions (and also for Ψ54 activity in vitro) was a surprising result. Although this residue is conserved in most Ψ synthases, no specific role for this residue has been reported. It is considered to provide general hydrophobicity to the active site and supposedly involved in nucleotide stacking (McCleverty et al. 2007; Mueller and Ferre-D’Amare 2009).
Archaeal Pus10 protein–tRNA interaction: Plausible mechanism
Based on our present study and the information available about various Ψ synthases, we propose that archaeal Pus10 proteins use a combination of two mechanisms for recognition of their substrate uridines and binding to tRNAs. These mechanisms are similar to those of RluA and TruB in certain aspects. There are several reasons for our proposal. The catalytic domain of human PUS10 can superpose that of TruB and RluA (McCleverty et al. 2007). Bacterial TruB produces Ψ55 in almost all tRNAs, while bacterial RluA produces Ψ746 (E. coli position) in 23S rRNA and Ψ32 in certain specific tRNAs. On the other hand, most archaeal Pus10 produce Ψ54 and Ψ55 in nearly all tRNAs of the cell (Gupta 1984, 1986; Gurha and Gupta 2008; Chatterjee et al. 2012). Many MjPus10 mutants, including deletions in the forefinger loop region, seem to affect Ψ54 activity specifically, but not Ψ55 activity (Table 1). All three proteins (TruB, RluA, and Pus10) convert U to Ψ in a 7-base loop of a stem–loop structure. Consensus sequence of this loop for the three proteins is 5′-UUNNANN-3′, where RluA modifies the first U, TruB modifies the second U, and Pus10 modifies both U’s. Conserved A of this sequence is involved in alternate reverse-Hoogstein pair with the first or second U for the activity of TruB or RluA, respectively. Crystal structures of TruB and RluA with and without their substrate RNAs (Hoang and Ferre-D’Amare 2001; Pan et al. 2003; Chaudhuri et al. 2004; Phannachet and Huang 2004; Hoang et al. 2005, 2006) have helped in determining the mechanisms of substrate recognition and binding by these two Ψ synthases. Both TruB and RluA flip out three bases of a 7-base loop. Furthermore, docking of tRNA into human PUS10 using the TruB-TSL cocrystal as a model has shown that the TSL can fit into the basic cleft of the catalytic domain of the protein, and flipped out U55 of the TSL protrudes close to the “catalytic” Asp in the active site (McCleverty et al. 2007). For these reasons, we hypothesize that archaeal Pus10 recognizes the TΨC loop of tRNA in a manner that shows some resemblance to that of TruB, and converts U55 to Ψ. This is followed by the induction of changes in the TΨC loop somewhat similar to that of RluA, and then conversion of U54 to Ψ. These changes in the TΨC loop are shown in the cartoon form in Figure 7.
FIGURE 7.

Proposed model for changes in TΨC-loop of tRNA during its interaction with archaeal Pus10. (First panel) TΨC-loop contains a reverse-Hoogstein U54•A58 (double lines) pair. (Second panel) Pus10 recognizes the loop by shape complementarity and flips out U55, C56, and R57. (Third panel) After conversion of U55, newly formed Ψ55 and R57 revert to original position, a new reverse-Hoogstein Ψ55•A58 (double lines) pair is formed and U54, C56 (pre-existing), and N59 flip out. (Fourth panel) Newly formed Ψ54 and other residues revert to their original positions and reverse-Hoogstein Ψ54•A58 (double lines) pair is reformed. Note: C56 flips out in both interactions.
The consensus sequence of the TΨC-loop of the tRNA is UUCRANY, where TruB converts the second U (bold italic, at position 55 of tRNA) to Ψ. TruB recognizes the prefolded structure of this loop by shape complementarity (schematically shown in the first panel in Fig. 7) (Hamma and Ferre-D’Amare 2006; Mueller and Ferre-D’Amare 2009). In addition, the protein makes sequence-specific contact with invariant C56 of the loop. The thumb loop of TruB undergoes a large conformation change upon binding the major groove of the substrate RNA. In TruB-bound tRNA, the target U55, as well as C56 and R57, are flipped out of the helical stack (as in second panel of Fig. 7). U54 and A58 (underlined in above sequence) form a conserved reverse-Hoogstein base pair. Based on the above-mentioned docking model of tRNA into human PUS10 (McCleverty et al. 2007), archaeal Pus10, in principle, should also be able to recognize the prefolded structure of the TΨC loop, contact its C56, and use its thumb loop to bind to a major groove of the substrate, as is the case for TruB. However, the mechanisms of binding to RNA and changes in RNA structure have to differ between TruB and Pus10 due to the following reasons. TruB family members contain a conserved His, five residues N-terminal to the catalytic Asp (active-site consensus sequence of TruB is HXGXLD, catalytic Asp underlined). This His residue stacks underneath the U54•A58 reverse-Hoogstein base pair. Pus10 does not contain this His. A Tyr residue in TruB that corresponds to Tyr339 of MjPus10, when mutated to Ala, Leu, or Phe, lost all of its Ψ55 synthase activity (Phannachet et al. 2005). It has been proposed that this Tyr in TruB maintains the hydrophobic nature of the active site and its OH group is involved in catalysis (Phannachet et al. 2005). Mutations of the corresponding Tyr in human PUS1 (member of TruA family) also show significant reduction of activity (Sibert et al. 2008). However, the Y339A mutant of MjPus10 produces Ψ55 both in vitro and in vivo. Therefore, the absence of His in MjPus10 that corresponds to conserved His of TruB, and lack of importance of Tyr339 for the Ψ55 activity of MjPus10, suggests that the mechanisms used by TruB and Pus10 for their Ψ55 activities are significantly different, even if these may initially recognize the TΨC loop in a similar manner. Probably, as yet unidentified amino acids of MjPus10 take over the roles of conserved His and Tyr of TruB for its Ψ55 activity.
The rRNA and tRNA target loops for RluA-mediated modifications share local sequence and structural similarity. Consensus sequence for these loops is UUNNAAA, where the first U (bold italic) is modified to Ψ. RluA binding induces some changes in the secondary structure of the loop (Hamma and Ferre-D’Amare 2006; Mueller and Ferre-D’Amare 2009) that include flipping out of the first, third, and sixth nucleotide from the loop (as in the third panel of Fig. 7), formation of a new stack by the second, fourth, and fifth nucleotides, and formation of a new reverse-Hoogstein U•A (underlined in the sequence) base pair. RluA recognizes this induced structure by shape complementarity. An Arg, two residues N-terminal to catalytic Asp of RluA intercalates between the newly formed reverse-Hoogstein pair and the Watson-Crick pair closing the target 7-base loop. This Arg is conserved in members of RluA, RsuA, and TruA (as well as in Pus10, above-mentioned Arg273 in MjPus10) families (Hamma and Ferre-D’Amare 2006). (Active site consensus sequence for RluA and Pus10 are HRLD and GREDXD, respectively. Catalytic Asp is underlined). This Arg in all of these enzymes has been suggested to take the place of target uridine when it is flipped out for catalysis, a role analogous to the conserved His of TruB (Hamma and Ferre-D’Amare 2006). RluA binds the major groove of RNA via its thumb loop, analogous to TruB. However, RluA also has a forefinger loop, which binds the minor groove, and together with the thumb loop, “pinches” the loop of the target RNA and places it in proper orientation with respect to the catalytic core. We hypothesize that forefinger and thumb loops of MjPus10 can also similarly “pinch” the tRNA and induce changes in the loop of TSL analogous to those induced by RluA. Deletions in the forefinger loop region (ffl2 deletions) abolish Ψ54 activity of MjPus10 (Figs. 1B, 2D), probably due to the inability of these mutant proteins to “pinch” and present the RNA substrate in proper orientation relative to the catalytic core. Furthermore, we hypothesize that MjPus10 Arg273 may then intercalate between the newly formed U55•A58 reverse-Hoogstein pair and the conserved G53–C61 Watson-Crick pair of tRNA, which closes the 7-base loop (in the gap formed by flipping out of U54) (Fig. 7, see third panel). This role of Arg273 would be analogous to the corresponding Arg of RluA (and as suggested for RsuA and TruA families). R273A mutant is unique; it shows very low Ψ54 activity in vitro but none in vivo, suggesting that it is an essential amino acid for Ψ54 activity of MjPus10. It seems that forefinger loop, and Arg273, Tyr339, and Leu440 are the most important determinants of MjPus10 Ψ54 activity (or protein-induced conformational change), because all other mutants show in vivo Ψ54 activity even when they have only trace amounts of in vitro activity (Fig. 2D; Table 1). Tyr339 of MjPus10 may play a role in its Ψ54 activity, but not, as mentioned above, in its Ψ55 activity. This role in Ψ54 activity may be analogous to the role of conserved Tyr in Ψ55 activity of TruB, i.e., maintaining hydrophobicity of the active site and involvement of the OH group in catalysis. In such a scenario, one would have to assume that active sites for the two activities of MjPus10 and the amino acids involved in the two reactions (except perhaps the catalytic Asp 275) are not all identical.
As mentioned above, TruB contains a His five residues N-terminal to the catalytic Asp, and RluA contains an Arg two residues N-terminal to the catalytic Asp. These two amino acid residues occupy the positions vacated by flipping out of their corresponding uridines (Mueller and Ferre-D’Amare 2009). Similar to RluA, MjPus10 (and PfuPus10) also contain an Arg two residues N-terminal to the catalytic Asp. This Arg is present in Pus10 of all sequenced archaeal species that contain conserved core catalytic sequence GREDXD (catalytic Asp underlined) (see Supplemental Fig. 5). Furthermore, a His that is six residues N-terminal to the catalytic Asp is also present in these Archaea, except for the members of order Thermococcales (Pyrococcus and Thermococcus), where it is substituted by a Lys. No other amino acid in the region near the catalytic Asp showed a similar pattern of conservation between Pus10 proteins expected to have dual (Ψ55 and Ψ54) activities and those expected to have single (Ψ55) activity. Therefore, we speculate that these Arg and His residues near the catalytic Asp of archaeal Pus10 may perform flipping out of the uridines, as are the cases for these residues in RluA and TruB.
Archaeal tRNAs contain 2′-O-modified C56 unlike an unmodified C present in all bacterial and eukaryal tRNAs (Juhling et al. 2009). This modification may affect flipping out of this residue, which is common in both TruB and RluA mechanisms. However, it is not known whether in vivo C56 is modified before or after Pus10-mediated modifications. In vitro activities of archaeal Pus10 have been tested only with unmodified C56 (Roovers et al. 2006; Gurha and Gupta 2008), and E. coli used for in vivo studies here also contains unmodified C56 in its tRNAs. Furthermore, absolute requirement of A58 in forming a reverse-Hoogstein pair with U54 and U55 need to be relaxed, at least in one case. Primary sequence of the T-loop of H. volcanii elongator methionine tRNA is UUCGGAG and both U’s are normally converted to Ψ (Gupta 1984). In this case, there is G58 instead of A58. Whether A59 (instead of A58) can form the reverse-Hoogstein pairs, if necessary for Pus10 activity, remains an open question.
Although we have shown that Ψ54 and Ψ55 activities of MjPus10 are independent of each other, we believe that inside the cell these occur sequentially. The reason for this is that we did not observe any incidence where a mutant MjPus10 lost its Ψ55 activity while still retaining Ψ54 activity (or any case where Ψ55 activity was more severely affected than Ψ54 activity). We think that these Pus10-mediated reactions occur very early during tRNA biogenesis, because in E. coli, even in the presence of native TrmA, MjPus10 produces Ψ54 and does not leave any (substantial) unmodified U54 to be converted to m5U54. We believe that Pyrococcus Pus10 lost its Ψ54 activity during evolution. Members of the order Thermococcales (which includes Pyrococcus) use aTrmU54, an ortholog of bacterial RlmD, an rRNA methyltransferase (responsible for m5U1939 in E. coli 23S rRNA) to produce m5U54 in the tRNA (Urbonavicius et al. 2008). Once members of the order Thermococcales acquired a bacterial RlmD gene by horizontal gene transfer (Urbonavicius et al. 2008) and changed its specificity to produce tRNA m5U54, there was no need for them to retain Ψ54 activities of their Pus10 proteins. Structural changes, especially in the forefinger region of Pus10, may have aided in the loss of natural Ψ54 activity of PfuPus10. (Consistent with this hypothesis, we observed loss of Ψ54 activity of ffl2-deleted MjPus10 protein.) However, these changes did not affect Ψ55 activity of the PfuPus10 protein, because forefinger loop has a limited role, if any, in Ψ55 formation. Indeed TruB, the bacterial Ψ55 synthase, does not contain forefinger loop, and in our work changes in the forefinger loop of MjPus10 did not affect its Ψ55 activity. In conclusion, we propose that a unique combination of two mechanisms dictates Ψ55 and Ψ54 formation by archaeal Pus10. Future studies with cocrystal structures of PfuPus10 and MjPus10 with tRNA (or TSL) are needed to validate these observations.
MATERIALS AND METHODS
Standard molecular biological protocols were used where not specifically mentioned.
Strains, plasmids, media, and oligonucleotides
E. coli DH5α was used as wild-type strain. E. coli mutant strains for truB, trmA, and double mutant truB/trmA were GOB113 (Hfr P4X sdr_ truB2422::mini-Tn10Cm), GRB1882 (Hfr P4X trmA5), and GRB1887 (Hfr P4X truB2422::mini-Tn10Cm trmA5), respectively (Urbonavicius et al. 2002). pTrc99a (Pharmacia/GE Healthcare) was used as a cloning vector for expression of Pus10 genes in E. coli. The pTrc series of vectors contain a hybrid trp and lac (trc) promoter and are IPTG inducible (Amann et al. 1988). Leaky expression can be minimized by addition of 2% glucose in the medium. E. coli cells were routinely grown in LB medium at 37°C supplemented with ampicillin (100 μg/mL) and chloramphenicol (34 μg/mL) as required. DNA oligonucleotides used here are listed in Supplemental Table 1 and their specific use is mentioned in the table.
Preparation of recombinant Pus10 proteins
Recombinant Pus10 proteins were prepared as described previously (Gurha et al. 2007; Gurha and Gupta 2008). Ala-substituted and partly deleted versions of recombinant proteins were prepared after site-directed mutagenesis of the corresponding genes. The DNA sequence of each mutant was checked and confirmed. Proteins were quantitated either by Coomassie blue staining of the gels after SDS-PAGE or by using BCA protein assay kit (Pierce). Bovine serum albumin was used as standard in both cases.
Pseudouridylation assays and quantitation of Ψ
Preparation of radiolabeled transcripts, pseudouridylation assays, thin-layer chromatography (TLC) analyses, and quantitation of Ψ were done as described before (Gurha et al. 2007; Gurha and Gupta 2008). Pseudouridylation assays were done by incubating equimolar (1:1) amounts of in vitro-transcribed radiolabeled H. volcanii tRNATrp (see sequence in Supplemental Fig. 1A) with wild-type MjPus10 or its mutants. Briefly, a typical 20-μL reaction consisted of 240 nM RNA and 240 nM of protein in a buffer containing 20 mM Tris-Cl (pH 7.5), 300 mM NaCl, 0.75 mM DTT, 1.5 mM MgCl2, 0.1 mM EDTA, and 10% glycerol at 68°C for 1 h. The modified RNA was purified by phenol/chloroform extraction and ethanol precipitation and digested with RNase T2 or nuclease P1. Digests were resolved by TLC (see details of TLC analyses in Supplemental Fig. 1). Radioactivity in the TLC spots was quantitated by phosphorimaging. The amount of Ψ produced in RNase T2 digests was determined as (radioactivity in Ψp spot × number of U’s in the RNA preceding the labeled nucleotide used to produce transcript)/(sum of the radioactivity in Up and Ψp spots). The amount of Ψ produced in nuclease P1 digests was determined (radioactivity in Ψp spot × number of U’s in the RNA)/(sum of the radioactivity in pU and pΨ spots). Production of tRNA Ψ54 and Ψ55 was determined by using RNase T2 digestion of substrates labeled with [α-32P]UTP and [α-32P]CTP, respectively, whereas nuclease P1 digests of [α-32P]UTP-labeled substrates were used to determine total Ψ (sum of Ψ54 and Ψ55) production. Reaction products of same [α-32P]UTP-labeled transcripts were used to determine total Ψ and Ψ54 production. Each assay was repeated at least twice using independently transcribed RNAs. To determine whether there was any nonspecific production of Ψ by a protein, its total Ψ production was compared with the sum of independently calculated amounts of Ψ54 and Ψ55 produced by this protein. To determine independence of Ψ54 and Ψ55 activities of MjPus10, [α-32P]UTP-labeled transcripts were treated either with PAB0179 protein of P. abyssi (Urbonavicius et al. 2008) or with E. coli TruB before treatment with MjPus10. m5U production in PAB0179-treated transcripts was also determined by nuclease P1 digestion as for the Ψ production.
TLC analyses of different digests (nuclease P1 and RNase T2) of a 32P-labeled reaction product of a single reaction is a sensitive technique that can provide information both about total Ψ production at multiple sites (after nuclease P1) as well as Ψ at a single specific position (nearest-neighbor transfer after RNase T2). However, these analyses, especially the two-dimensional ones, have limited dynamic range for quantitative purposes. Since we manually outline a specific spot to be quantitated, parameters like size, shape, intensity, and presence of another nearby spot, and sometimes even a dark background (probably due to some impurity in the solvents), affect quantitation of a particular spot (see spots for U and Ψ nucleotides in Fig. 5E and Supplemental Fig. 1C,D). We have also observed differences in the separation of nucleotides between different TLC plates, especially among different batches of the plates. Furthermore, amounts of modification can be significantly underestimated, when high specific activity transcripts are used in the reaction, especially for low-activity proteins. Radiodecay causes breakdown of transcripts within a short time. These breakdown products do not serve as substrates for the reaction, but they do contribute to radioactivity of unmodified spots on the TLC. Due to these technical challenges, our quantitation of Ψ production, especially when Ψ production is low, can only be considered as an approximate measure. These reasons limited our ability to accurately determine the rate of reactions of mutants using time course studies. So we restricted our analyses to comparisons of the end points of the reactions.
Cloning and expression of Pus10 proteins for in vivo analyses
The genes encoding MjPus10 and PfuPus10 were PCR amplified from M. jannaschii and P. furiosus genomic DNA, respectively, using primers containing EcoRI and KpnI sites at the 5′ and 3′ ends, respectively, and cloned in pTrc99a. Resulting Pus10 genes contained two additional codons, for Glu and Phe, between the first and second amino acids of the native gene. Ala substituted and partly deleted versions of these genes were created by site-directed mutagenesis as done for in vitro studies. MjPus10 or PfuPus10 and their mutants were expressed in different E. coli strains by growing the culture with appropriate antibiotics to A600 of 0.5, followed by induction with 0.3–1.0 mM IPTG for 4 h at 37°C, or overnight at 16°C. The levels of expression of all mutant proteins in E. coli were checked by SDS-PAGE and compared with the expression of wild-type proteins.
Determination of Ψ and U at positions 54 and 55 of tRNAPhe and total tRNA of E. coli
Total tRNA was isolated from E. coli by phenol extraction of intact cell suspension following published procedures (Ellis et al. 1986). Then the presence or absence of Ψ at positions 54 and 55 of tRNAPhe was determined by CMCT modification of total tRNA, followed by primer extension using [5′-32P]-labeled primer as described before (Motorin et al. 2007; Blaby et al. 2011). This primer pairs with residues at position 76–61 of E. coli tRNAPhe (see Fig. 4A). Essentially, in this method RNA is treated with 1-cyclohexyl-3-(2-morpholinoethyl) carbodiimide metho-p-toluenesufonate, ethanol precipitated, and then treated with alkali before primer extension. Ψ-CMCT adducts are more stable to alkali hydrolysis than other CMCT adducts (Motorin et al. 2007). This method is used to determine Ψ production, but is not suitable for quantitation of Ψ. Partial reaction conditions used in these methods produce ladder-like bands in thin denaturing gels, and the presence or absence of Ψ at a particular position is indicated by the presence or absence of a dark band at that position. Furthermore, reverse transcriptase reactions, in general, are sensitive to secondary structure and modified bases present in the tRNAs and would not proceed beyond the first Ψ if every Ψ in the tRNA had a CMCT adduct.
The presence or absence of unmodified U at positions 54 and 55 of tRNAPhe was determined by U-specific chemical RNA-sequencing reactions as described previously (Peattie 1979; Gupta 1984; Blaby et al. 2011) on total tRNA followed by primer extension as done after CMCT modification. Uridines are hydrazinolysed in U-specific reactions using aqueous hydrazine, which is followed by strand scission at the position of the damaged base using acidified aniline (Peattie 1979; Motorin et al. 2007). The products were resolved by 10% or 12% denaturing PAGE. Again, the conditions used are for partial hydrazinolysis. The ideal conditions would be to produce one cleavage per molecule. Therefore, this method is also not suitable to determine the quantity of leftover unmodified U.
The presence or absence of unmodified U at positions 54 and 55 in total cellular tRNAs was determined by isolation of tRNA from different strains, as mentioned above. The total tRNA so obtained was 3′ end labeled with [5′-32P]pCp, followed by U-specific reaction and resolution by 15% denaturing PAGE. Similarly treated 3′ end-labeled E. coli tRNALys (Sigma) served as marker.
In vivo labeling and thin layer chromatography
A single colony of appropriate E. coli strain was inoculated into 5-mL phosphate-depleted LB medium (Gupta 1984). After overnight passage the culture was reinoculated in 5-mL phosphate-depleted LB medium, grown to A600 of 0.5, induced by 1 mM IPTG, followed by the addition of 0.5 mCi of [32P]-phosphoric acid. The culture was grown for another 1–2 h, and uniformly labeled total tRNA was isolated from these cells as described above. Purified tRNA was digested with nuclease P1, and digests were resolved by two-dimensional TLC on cellulose plates (Cel 300, Macherey-Nagel) using isobutyric acid/0.5 N NH4OH (5:3, v/v) for the first dimension and 0.1 M sodium phosphate, pH 6.8/ammonium sulfate/n-propanol (100:60:2, v/w/v) for the second (Gupta 1984). Radioactivity in TLC spots was visualized by phosphorimaging.
Homology modeling
Homology modeling was used to create potential 3D structures for M. jannaschii and P. furious Pus10 based on the existing crystal structure for human PUS10 (2V9K) as a starting point and use of energy minimization of carbon–carbon bonds to calculate the predicted structure. The sequence identity of MjPus10 and PfuPus10 are 25.6% and 27%, respectively, with respect to human PUS10. A pairwise alignment of human PUS10 and MjPus10 resulted in an alignment size of 206 aa with 35% identity and 55.8% similarity. PfuPus10 alignment size was 241 aa with 34% identity and 54.8% similarity. This was sufficient alignment length and identity for 3D-JIGSAW (version 2.0; http://bmm.cancerresearchuk.org/~3djigsaw/) to model the aligned portions; M. jannaschii (Asn205–Leu451) and P. furiosus (Asn158–Asn388) were successfully fitted to the catalytic domain of Human PUS10 2V9K (Gly286–Asp528) (Contreras-Moreira and Bates 2002). The catalytic domain models contained all relevant features (forefinger loop, catalytic region with its Asp, and the thumb loop). Structures were visualized, structurally aligned, and manipulated in Swiss-PDBViewer version 3.7 (Bordoli et al. 2009; Guex et al. 2009). Multiple sequence alignment was performed on full-length protein sequences obtained from NCBI of Drosophila melanogaster, Danio rerio, Homo sapiens, Mus musculus, Pyrococcus abyssi, Pyrococcus furiosus, Methanocaldococcus jannaschii, Halobacterium sp, and Haloarcula marismortui using ClustalX2 (www.clustal.org; Larkin et al. 2007), with the region corresponding to the forefinger loop and catalytic site shown in Figure 2. Multiple structural alignment analysis of T. acidophilum, T. thermoautotrophicum, M. jannaschii, H. volcanii, P. furiosus, and human PUS10 was performed using the MuliProt server to identify key conserved amino acid’s within the Archaea (Supplemental Fig. 2; Shatsky et al. 2004).
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
ACKNOWLEDGMENTS
We thank Glenn Bjork (University of Umea) for providing E. coli mutant strains GOB113, GRB1882, and GRB1887 and pTrc99A vector, Henri Grosjean (Université Paris) for PAB0719 clone, and Geena Skariah for recombinant TruB protein. This work was supported by NIH grant GM55045 to R.G.
REFERENCES
- Amann E, Ochs B, Abel KJ 1988. Tightly regulated tac promoter vectors useful for the expression of unfused and fused proteins in Escherichia coli. Gene 69: 301–315 [DOI] [PubMed] [Google Scholar]
- Aza-Blanc P, Cooper CL, Wagner K, Batalov S, Deveraux QL, Cooke MP 2003. Identification of modulators of TRAIL-induced apoptosis via RNAi-based phenotypic screening. Mol Cell 12: 627–637 [DOI] [PubMed] [Google Scholar]
- Baudin-Baillieu A, Fabret C, Liang XH, Piekna-Przybylska D, Fournier MJ, Rousset JP 2009. Nucleotide modifications in three functionally important regions of the Saccharomyces cerevisiae ribosome affect translation accuracy. Nucleic Acids Res 37: 7665–7677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker HF, Motorin Y, Planta RJ, Grosjean H 1997. The yeast gene YNL292w encodes a pseudouridine synthase (Pus4) catalyzing the formation of Ψ55 in both mitochondrial and cytoplasmic tRNAs. Nucleic Acids Res 25: 4493–4499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjork GR, Isaksson LA 1970. Isolation of mutants of Escherichia coli lacking 5-methyluracil in transfer ribonucleic acid or 1-methylguanine in ribosomal RNA. J Mol Biol 51: 83–100 [DOI] [PubMed] [Google Scholar]
- Blaby IK, Majumder M, Chatterjee K, Jana S, Grosjean H, de Crecy-Lagard V, Gupta R 2011. Pseudouridine formation in archaeal RNAs: The case of Haloferax volcanii. RNA 17: 1367–1380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordoli L, Kiefer F, Arnold K, Benkert P, Battey J, Schwede T 2009. Protein structure homology modeling using SWISS-MODEL workspace. Nat Protoc 4: 1–13 [DOI] [PubMed] [Google Scholar]
- Cantara WA, Crain PF, Rozenski J, McCloskey JA, Harris KA, Zhang X, Vendeix FA, Fabris D, Agris PF 2011. The RNA Modification Database, RNAMDB: 2011 update. Nucleic Acids Res 39: D195–D201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charette M, Gray MW 2000. Pseudouridine in RNA: What, where, how, and why. IUBMB Life 49: 341–351 [DOI] [PubMed] [Google Scholar]
- Chatterjee K, Blaby IK, Thiaville PC, Majumder M, Grosjean H, Yuan YA, Gupta R, de Crecy-Lagard V 2012. The archaeal COG1901/DUF358 SPOUT-methyltransferase members, together with pseudouridine synthase Pus10, catalyze the formation of 1-methylpseudouridine at position 54 of tRNA. RNA 18: 421–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri BN, Chan S, Perry LJ, Yeates TO 2004. Crystal structure of the apo forms of Ψ55 tRNA pseudouridine synthase from Mycobacterium tuberculosis: A hinge at the base of the catalytic cleft. J Biol Chem 279: 24585–24591 [DOI] [PubMed] [Google Scholar]
- Contreras-Moreira B, Bates PA 2002. Domain fishing: A first step in protein comparative modelling. Bioinformatics 18: 1141–1142 [DOI] [PubMed] [Google Scholar]
- Cunningham BC, Wells JA 1989. High-resolution epitope mapping of hGH-receptor interactions by alanine-scanning mutagenesis. Science 244: 1081–1085 [DOI] [PubMed] [Google Scholar]
- Davis DR 1995. Stabilization of RNA stacking by pseudouridine. Nucleic Acids Res 23: 5020–5026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decatur WA, Fournier MJ 2002. rRNA modifications and ribosome function. Trends Biochem Sci 27: 344–351 [DOI] [PubMed] [Google Scholar]
- Decatur WA, Fournier MJ 2003. RNA-guided nucleotide modification of ribosomal and other RNAs. J Biol Chem 278: 695–698 [DOI] [PubMed] [Google Scholar]
- Delk AS, Nagle DP Jr, Rabinowitz JC 1980. Methylenetetrahydrofolate-dependent biosynthesis of ribothymidine in transfer RNA of Streptococcus faecalis. Evidence for reduction of the 1-carbon unit by FADH2. J Biol Chem 255: 4387–4390 [PubMed] [Google Scholar]
- Dennis PP, Omer A 2005. Small non-coding RNAs in Archaea. Curr Opin Microbiol 8: 685–694 [DOI] [PubMed] [Google Scholar]
- Edmonds CG, Crain PF, Gupta R, Hashizume T, Hocart CH, Kowalak JA, Pomerantz SC, Stetter KO, McCloskey JA 1991. Posttranscriptional modification of tRNA in thermophilic archaea (Archaebacteria). J Bacteriol 173: 3138–3148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis SR, Morales MJ, Li JM, Hopper AK, Martin NC 1986. Isolation and characterization of the TRM1 locus, a gene essential for the N2,N2-dimethylguanosine modification of both mitochondrial and cytoplasmic tRNA in Saccharomyces cerevisiae. J Biol Chem 261: 9703–9709 [PubMed] [Google Scholar]
- Festen EA, Goyette P, Green T, Boucher G, Beauchamp C, Trynka G, Dubois PC, Lagace C, Stokkers PC, Hommes DW, et al. 2011. A meta-analysis of genome-wide association scans identifies IL18RAP, PTPN2, TAGAP, and PUS10 as shared risk loci for Crohn’s disease and celiac disease. PLoS Genet 7: e1001283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge J, Yu YT 2013. RNA pseudouridylation: New insights into an old modification. Trends Biochem Sci 38: 210–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg R, Dudock B 1980. Isolation and chracterization of m5U-methyltransferase from Escherichia coli. J Biol Chem 255: 8296–8302 [PubMed] [Google Scholar]
- Grosjean H, Oshima T 2007. How nucleic acids cope with high temperature. In Physiology and biochemistry of extremophiles (ed. Gerday C, Glansdorff N), pp. 39–56 ASM Press, Washington, DC [Google Scholar]
- Guex N, Peitsch MC, Schwede T 2009. Automated comparative protein structure modeling with SWISS-MODEL and Swiss-PdbViewer: A historical perspective. Electrophoresis 30: S162–S173 [DOI] [PubMed] [Google Scholar]
- Gupta R 1984. Halobacterium volcanii tRNAs. Identification of 41 tRNAs covering all amino acids, and the sequences of 33 class I tRNAs. J Biol Chem 259: 9461–9471 [PubMed] [Google Scholar]
- Gupta R 1986. Transfer RNAs of Halobacterium volcanii: Sequences of five leucine and three serine tRNAs. Syst Appl Microbiol 7: 102–105 [Google Scholar]
- Gurha P, Gupta R 2008. Archaeal Pus10 proteins can produce both pseudouridine 54 and 55 in tRNA. RNA 14: 2521–2527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurha P, Joardar A, Chaurasia P, Gupta R 2007. Differential roles of archaeal box H/ACA proteins in guide RNA-dependent and independent pseudouridine formation. RNA Biol 4: 101–109 [DOI] [PubMed] [Google Scholar]
- Hamma T, Ferre-D’Amare AR 2006. Pseudouridine synthases. Chem Biol 13: 1125–1135 [DOI] [PubMed] [Google Scholar]
- Hamma T, Ferre-D’Amare AR 2010. The box H/ACA ribonucleoprotein complex: Interplay of RNA and protein structures in post-transcriptional RNA modification. J Biol Chem 285: 805–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henras AK, Dez C, Henry Y 2004. RNA structure and function in C/D and H/ACA s(no)RNPs. Curr Opin Struct Biol 14: 335–343 [DOI] [PubMed] [Google Scholar]
- Hoang C, Ferre-D’Amare AR 2001. Cocrystal structure of a tRNA Ψ55 pseudouridine synthase: Nucleotide flipping by an RNA-modifying enzyme. Cell 107: 929–939 [DOI] [PubMed] [Google Scholar]
- Hoang C, Hamilton CS, Mueller EG, Ferre-D’Amare AR 2005. Precursor complex structure of pseudouridine synthase TruB suggests coupling of active site perturbations to an RNA-sequestering peripheral protein domain. Protein Sci 14: 2201–2206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoang C, Chen J, Vizthum CA, Kandel JM, Hamilton CS, Mueller EG, Ferre-D’Amare AR 2006. Crystal structure of pseudouridine synthase RluA: Indirect sequence readout through protein-induced RNA structure. Mol Cell 24: 535–545 [DOI] [PubMed] [Google Scholar]
- Hopper AK, Furukawa AH, Pham HD, Martin NC 1982. Defects in modification of cytoplasmic and mitochondrial transfer RNAs are caused by single nuclear mutations. Cell 28: 543–550 [DOI] [PubMed] [Google Scholar]
- Juhling F, Morl M, Hartmann RK, Sprinzl M, Stadler PF, Putz J 2009. tRNAdb 2009: Compilation of tRNA sequences and tRNA genes. Nucleic Acids Res 37: D159–D162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamalampeta R, Kothe U 2012. Archaeal proteins Nop10 and Gar1 increase the catalytic activity of Cbf5 in pseudouridylating tRNA. Sci Rep 2: 663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karijolich J, Yu YT 2010. Spliceosomal snRNA modifications and their function. RNA Biol 7: 192–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaya Y, Ofengand J 2003. A novel unanticipated type of pseudouridine synthase with homologs in bacteria, archaea, and eukarya. RNA 9: 711–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss T 2002. Small nucleolar RNAs: An abundant group of noncoding RNAs with diverse cellular functions. Cell 109: 145–148 [DOI] [PubMed] [Google Scholar]
- Kowalak JA, Dalluge JJ, McCloskey JA, Stetter KO 1994. The role of posttranscriptional modification in stabilization of transfer RNA from hyperthermophiles. Biochemistry 33: 7869–7876 [DOI] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947–2948 [DOI] [PubMed] [Google Scholar]
- Lecointe F, Namy O, Hatin I, Simos G, Rousset JP, Grosjean H 2002. Lack of pseudouridine 38/39 in the anticodon arm of yeast cytoplasmic tRNA decreases in vivo recoding efficiency. J Biol Chem 277: 30445–30453 [DOI] [PubMed] [Google Scholar]
- McCleverty CJ, Hornsby M, Spraggon G, Kreusch A 2007. Crystal structure of human Pus10, a novel pseudouridine synthase. J Mol Biol 373: 1243–1254 [DOI] [PubMed] [Google Scholar]
- Meier UT 2005. The many facets of H/ACA ribonucleoproteins. Chromosoma 114: 1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison KL, Weiss GA 2001. Combinatorial alanine-scanning. Curr Opin Chem Biol 5: 302–307 [DOI] [PubMed] [Google Scholar]
- Motorin Y, Muller S, Behm-Ansmant I, Branlant C 2007. Identification of modified residues in RNAs by reverse transcription-based methods. Methods Enzymol 425: 21–53 [DOI] [PubMed] [Google Scholar]
- Mueller EG, Ferre-D’Amare AR 2009. Pseudouridine formation, the most common transglycosylation in RNA. In DNA and RNA modification enzymes: structure, mechanism, function and evolution (ed. Grosjean H), pp. 363–376 Landes Bioscience, Austin, TX [Google Scholar]
- Muller S, Fourmann JB, Loegler C, Charpentier B, Branlant C 2007. Identification of determinants in the protein partners aCBF5 and aNOP10 necessary for the tRNA:Ψ55-synthase and RNA-guided RNA:Ψ-synthase activities. Nucleic Acids Res 35: 5610–5624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller S, Leclerc F, Behm-Ansmant I, Fourmann JB, Charpentier B, Branlant C 2008. Combined in silico and experimental identification of the Pyrococcus abyssi H/ACA sRNAs and their target sites in ribosomal RNAs. Nucleic Acids Res 36: 2459–2475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namy O, Lecointe F, Grosjean H, Rousset J-P 2005. Translational recoding and RNA modifications. In Fine-tuning of RNA function by modification and editing, topics in current genetics, Vol. 12 (ed. Grosjean H), pp. 309–340 Springer-Verlag, Berlin-Heidelberg [Google Scholar]
- Nordlund ME, Johansson JO, von Pawel-Rammingen U, Bystrom AS 2000. Identification of the TRM2 gene encoding the tRNA(m5U54)methyltransferase of Saccharomyces cerevisiae. RNA 6: 844–860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurse K, Wrzesinski J, Bakin A, Lane BG, Ofengand J 1995. Purification, cloning, and properties of the tRNA Ψ55 synthase from Escherichia coli. RNA 1: 102–112 [PMC free article] [PubMed] [Google Scholar]
- Ny T, Bjork GR 1980. Cloning and restriction mapping of the trmA gene coding for transfer ribonucleic acid (5-methyluridine)-methyltransferase in Escherichia coli K-12. J Bacteriol 142: 371–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ofengand J, Malhotra A, Remme J, Gutgsell NS, Del Campo M, Jean-Charles S, Peil L, Kaya Y 2001. Pseudouridines and pseudouridine synthases of the ribosome. Cold Spring Harb Symp Quant Biol 66: 147–159 [DOI] [PubMed] [Google Scholar]
- Pan H, Agarwalla S, Moustakas DT, Finer-Moore J, Stroud RM 2003. Structure of tRNA pseudouridine synthase TruB and its RNA complex: RNA recognition through a combination of rigid docking and induced fit. Proc Natl Acad Sci 100: 12648–12653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peattie DA 1979. Direct chemical method for sequencing RNA. Proc Natl Acad Sci 76: 1760–1764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phannachet K, Huang RH 2004. Conformational change of pseudouridine 55 synthase upon its association with RNA substrate. Nucleic Acids Res 32: 1422–1429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phannachet K, Elias Y, Huang RH 2005. Dissecting the roles of a strictly conserved tyrosine in substrate recognition and catalysis by pseudouridine 55 synthase. Biochemistry 44: 15488–15494 [DOI] [PubMed] [Google Scholar]
- Roovers M, Hale C, Tricot C, Terns MP, Terns RM, Grosjean H, Droogmans L 2006. Formation of the conserved pseudouridine at position 55 in archaeal tRNA. Nucleic Acids Res 34: 4293–4301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shatsky M, Nussinov R, Wolfson HJ 2004. A method for simultaneous alignment of multiple protein structures. Proteins 56: 143–156 [DOI] [PubMed] [Google Scholar]
- Sibert BS, Fischel-Ghodsian N, Patton JR 2008. Partial activity is seen with many substitutions of highly conserved active site residues in human Pseudouridine synthase 1. RNA 14: 1895–1906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbonavicius J, Durand JM, Bjork GR 2002. Three modifications in the D and T arms of tRNA influence translation in Escherichia coli and expression of virulence genes in Shigella flexneri. J Bacteriol 184: 5348–5357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbonavicius J, Skouloubris S, Myllykallio H, Grosjean H 2005. Identification of a novel gene encoding a flavin-dependent tRNA:m5U methyltransferase in bacteria—evolutionary implications. Nucleic Acids Res 33: 3955–3964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbonavicius J, Auxilien S, Walbott H, Trachana K, Golinelli-Pimpaneau B, Brochier-Armanet C, Grosjean H 2008. Acquisition of a bacterial RumA-type tRNA(uracil-54, C5)-methyltransferase by Archaea through an ancient horizontal gene transfer. Mol Microbiol 67: 323–335 [DOI] [PubMed] [Google Scholar]
- Watanabe Y, Gray MW 2000. Evolutionary appearance of genes encoding proteins associated with box H/ACA snoRNAs: Cbf5p in Euglena gracilis, an early diverging eukaryote, and candidate Gar1p and Nop10p homologs in archaebacteria. Nucleic Acids Res 28: 2342–2352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins NJ, Bohnsack MT 2012. The box C/D and H/ACA snoRNPs: Key players in the modification, processing and the dynamic folding of ribosomal RNA. Wiley Interdiscip Rev RNA 3: 397–414 [DOI] [PubMed] [Google Scholar]
- Wu G, Xiao M, Yang C, Yu YT 2011. U2 snRNA is inducibly pseudouridylated at novel sites by Pus7p and snR81 RNP. EMBO J 30: 79–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu YT, Terns RM, Terns MP 2005. Mechanisms and functions of RNA-guided RNA modification. In Fine-tuning of RNA functions by modification and editing (ed. Grosjean H), pp. 223–262 Springer-Verlag, New York [Google Scholar]