Abstract
The bacterial pathogen Vibrio vulnificus is found naturally in brackish coastal waters but can be greatly concentrated by filter-feeding organisms such as shellfish. Numerous experiments in which exogenous V. vulnificus cells are added to oysters in an attempt to measure uptake and depuration have been performed. In nearly all cases, results have shown that laboratory-grown bacteria are rapidly taken up by the oysters but ultimately eliminated, while naturally present Vibrio populations in oysters are resistant to depuration. In this study, oysters harvested during winter months, with low culturable Vibrio concentrations, were incubated in aquaria supplemented with strains of V. vulnificus that were either genotypically or phenotypically distinct from the background bacteria. These exogenous cells were eliminated from the oysters, as previously seen, but other vibrios already inhabiting the oysters responded to the V. vulnificus inoculum by rapidly increasing in number and maintaining a large stable population. The presence of such an oyster-adapted Vibrio population would be expected to prevent colonization by exogenous V. vulnificus cells, thus explaining the rapid depuration of these added bacteria.
INTRODUCTION
The Gram-negative bacterium Vibrio vulnificus is an opportunistic pathogen capable of causing gastroenteritis, wound infections, and fatal septicemia in humans (1, 2, 3). Routinely found in waters of estuarine environments as part of the normal microflora, as well as in oysters and other shellfish inhabiting those estuaries, this organism is remarkable as it has the potential for infection through preexisting wounds (e.g., from seawater through a lesion) or through ingestion (primarily oyster meats) (3). V. vulnificus is present in the majority of oysters meant for human consumption, with 67% of raw and 25% of cooked oysters collected from one study of Louisiana restaurants found to be harboring this pathogen (4). Infection caused by V. vulnificus is the leading cause of seafood-borne deaths in the United States, with most resulting from the consumption of raw or undercooked oysters (3). Infections caused by ingesting oysters containing V. vulnificus commonly result in primary septicemia, almost always require hospitalization, and have a fatality rate of greater than 50%, distinguishing V. vulnificus as having the highest case fatality rate of any food-borne pathogen (3, 5, 6, 7).
Many aquatic bacteria and most vibrios (including V. vulnificus) are affected by seasonal environmental shifts, with warmer temperatures correlating with increased isolation frequency and higher concentrations in the water column (8). Conversely, sensitivity to low temperature causes a decrease in the culturable populations of V. vulnificus (9, 10, 11, 12, 13). While some of these decreases can be attributed to decreased survival at the colder temperatures, at least part of our reduced ability to isolate aquatic bacteria in the winter may be due to a phenomenon known as the viable but nonculturable (VBNC) state (14, 15, 16, 17, 18, 19). Many Vibrio spp., including V. vulnificus, enter this state as a response to low temperatures (18, 20, 21, 22, 23). In this state, cells are viable (as confirmed via detection of RNA transcription, intact membranes, and other methods) but cannot be cultured on the routine media normally employed for their isolation (24). Thus, the cells enter a type of dormancy, a result of some form of environmental stress (e.g., cold temperature), and have been shown to acquire resistance to other unrelated stresses, a phenomenon referred to as cross-protection (14, 24, 25). When the initial stress is alleviated, the bacteria can emerge from the VBNC state in a process known as resuscitation (23, 24, 26). In V. vulnificus, the VBNC state can be induced in vitro by a temperature downshift and the bacteria can be resuscitated by a simple temperature upshift (23, 26, 27).
The purpose of the present study was to provide an explanation for the observed transience of exogenous bacteria in oyster uptake experiments. This report suggests that Vibrio cells, either present in the VBNC state or culturable but below the limit of detection, react to exogenous bacteria, potentially preventing their long-term colonization of oysters.
MATERIALS AND METHODS
Bacterial strains and isolation media.
V. vulnificus strain CVD713 possesses a TnphoA transposon that confers kanamycin resistance and alkaline phosphatase activity (28, 29, 30). This strain forms blue colonies when grown on Tn agar, consisting of Luria agar with the addition of 0.2 g liter−1 kanamycin, 2 g liter−1 glucose, and 0.04 g liter−1 5-bromo-4-chloro-3-indolyl phosphate (BCIP). Tn agar selects for the TnphoA-possessing strain via its kanamycin resistance and is differential by means of the presence of BCIP (30). Liquid cultures of this strain were grown in the same medium without the addition of BCIP and agar. V. vulnificus strain VVL1 is a naturally occurring E-genotype strain that is bioluminescent on standard media (31). Studies have shown that this organism does not differ phenotypically from other strains of the species except for its luminescence (31). This strain was grown in Bacto heart infusion (HI) broth (BD) for liquid cultures or with the addition of 1.5% agar for solid medium. Escherichia coli strain K-12 was grown in LB broth or on MacConkey agar (Sigma-Aldrich, St. Louis, MO). All strains were grown at 30°C, with vigorous shaking for liquid cultures. CPC+ agar was employed to isolate V. vulnificus and other vibrios from oyster meat. This medium (32) is an adaptation of CPC agar (33) and is selective for vibrios and differential for V. vulnificus due to the presence of the antibiotics colistin and polymyxin B, which inhibit most bacterial growth, and the fermentation of cellobiose to differentiate colonies of this species from other bacteria and vibrios (32, 33). V. vulnificus colonies growing on CPC+ produce yellow, dark-centered colonies with a yellow zone surrounding them (32, 33). Estuarine agar (34) was employed to measure total culturable heterotrophic bacteria.
Oyster collection and maintenance.
Oysters (Crassostrea virginica) from a North Carolina Department of Natural Resources-approved shellfishing area were collected by hand from the intertidal zone during cooler months (November to March; temperatures of 6.0 to 11.2°C). Oysters were rinsed and placed into 75-liter holding aquaria to acclimate to laboratory conditions, with a minimal holding time of 1 week and an average holding time of 1.5 months. The tanks contained artificial seawater (ASW) (Instant Ocean; Aquarium Systems, Mentor, OH) adjusted to 20‰ salinity with deionized water and kept at 23°C with constant aeration. Aquaria were filtered with Skilter filters (Danner Mfg. Inc., Islandia, NY), and oysters were removed from tanks and the aquaria cleaned and refilled with fresh ASW every 12 days. Oysters were fed an algal mixture of Skeletonema, Rhodomonas, and Isochrysis grown at room temperature in vented, 1-liter flasks containing F/2 medium and provided with constant fluorescent light (35, 36).
Oyster infection and depuration.
In each experiment, oysters were removed from maintenance aquaria and placed into unfiltered, aerated experimental aquaria containing 5 liters of ASW at room temperature. Twenty-five oysters were placed into each tank, with five oysters sampled, individually, at each time point. Before addition of exogenous bacteria, five oysters were aseptically removed from the tanks using ethanol- and flame-sterilized tongs and sampled to establish a background population count of vibrios and of the marked strains at a time point which is reflected in Fig. 1 to 5 as time zero (see below). Bacterial cells were added at a final concentration of approximately 7.5 × 104 CFU/ml to experimental aquaria containing the oysters. Control oysters received additions of cell-free media (either HI media or phosphate-buffered saline [PBS]) at the same volume as oysters amended with additional bacteria. Cells grown in the presence of antibiotics were washed 3 times with PBS prior to introduction to the aquaria. Oysters were exposed to the bacterially amended water for 24 h. After the initial 24-h exposure, and at days 3 and 6 after bacterial cell addition, the oysters were removed from the tanks and the aquaria were cleaned, sanitized, and refilled with fresh ASW to ensure depurated bacteria were not being reintroduced to oysters. Five oysters from each tank were reserved for analysis, and the remaining oysters were then depurated by replacing into the clean tanks, under the conditions described above. Three repeated experiments using the added bacterium CVD713 were conducted for each tissue type, as well as for whole-oyster sampling; two repeat experiments each were conducted with E. coli and with V. vulnificus strain VVL1.
Fig 1.
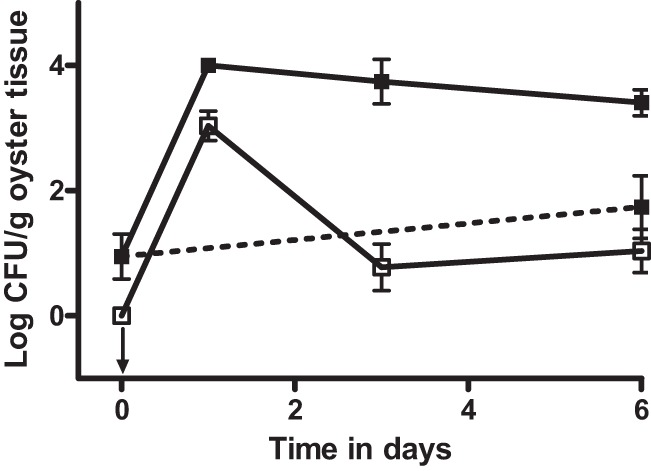
CFU of vibrios isolated on CPC+ (filled squares) and V. vulnificus strain CVD713 isolated on Tn agar (open squares) from oysters inoculated with exogenous V. vulnificus strain CVD713 (solid lines) or oysters treated with cell-free media (dashed line). The arrow pointing down indicates the point that was below the limit of detection. Error bars represent standard errors of the means.
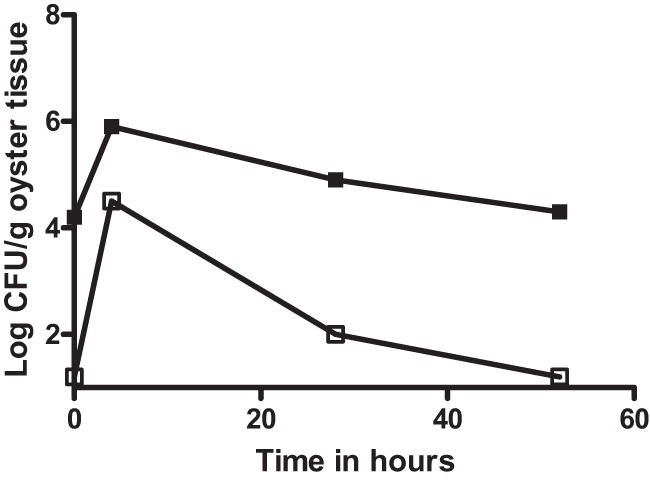
Fig 5.
CFU of vibrios isolated on CPC+ (filled squares) and E. coli isolated on MacConkey agar (open circles) from oysters inoculated with E. coli K-12 (solid lines) or oysters treated with cell-free media (dashed line). Error bars represent standard errors of the means.
Oyster shucking and homogenization.
Oysters, once removed from tanks, were rinsed with 70% ethanol and patted dry with paper towels. The oysters were opened with a flame-sterilized oyster knife, and the meat was rinsed with sterile ASW (20‰ salinity) to remove sediment. After shucking, the oyster meat was homogenized in 20‰ ASW at a 1:1 (wt/vol) ratio (minimum 5 ml ASW) using sterile blender cups (Waring, Torrington, CT) and employing 3 bursts of 15 s each, with a 5-s pause between the bursts.
After homogenization, samples were serially diluted in sterile PBS and spread onto CPC+ agar, estuarine agar, and the medium used to specifically select for each genetically marked V. vulnificus or E. coli strain (see above). Total CFU per gram of wet oyster tissue were calculated, with a limit of detection of 10 CFU/g.
Oyster dissection for individual tissues.
Oysters were aseptically opened and washed as described above. Using instruments rinsed with 70% ethanol and flame sterilized, small (ca. 0.3-g) pieces of oyster gill, mantle tissue, and digestive gland were removed and placed into sterile tubes. One milliliter of sterile ASW was added to each tube before homogenization of the tissue using an ethanol- and flame-sterilized Tissue Tearor (Biospec Products, Inc., Bartlesville, OK) until the tissue was liquefied. Samples were serially diluted and spread onto agar plates as described above.
Verification of V. vulnificus.
Cells from presumptive V. vulnificus colonies from oysters sampled at day 6 were picked, moved to HI agar plates, and allowed to grow overnight at 30°C. Using the methods described by Rosche et al. (37), each strain was then subjected to a PCR targeting the vvhA gene (Vibrio vulnificus hemolysin) to confirm the isolate as V. vulnificus. Reactions were performed using GoTaq polymerase (Promega, San Luis Obispo, CA) in a Techne Genius thermal cycler using the parameters suggested by Warner and Oliver (38). PCR products were visualized by gel electrophoresis on 1% agarose gels stained with ethidium bromide.
Sequence-based microbial identification.
Two colonies on CPC+ plates that were isolated from oysters but were not confirmed to be V. vulnificus were subjected to further genetic identification by sequencing of the first 500 bp of the 16S rRNA gene (Accugenix, Newark, DE).
Statistics.
Statistical analyses were performed using a two-way analysis of variance (ANOVA) with Bonferroni posttests using SigmaStat (SPSS Inc.) analysis software. An alpha level of 0.05 was used to determine significant differences in means. Values below the limit of detection were treated as containing 1 CFU/g.
Nucleotide sequence accession numbers.
The 16S rRNA gene sequences determined in this work were deposited into GenBank with accession numbers KF241860 and KF241861.
RESULTS AND DISCUSSION
The uptake and depuration of V. vulnificus in the Eastern oyster, Crassostrea virginica, were examined, in vitro, by collecting oysters from estuarine waters during cooler months followed by treatment with laboratory-grown strains of bacteria. Winter oysters are known to have reduced, or even nondetectable, levels of V. vulnificus and other Vibrio spp. (9, 10, 11, 12, 13). After acclimation to laboratory conditions at room temperature, oysters were placed into aquaria containing laboratory-grown strains of V. vulnificus and bacterial uptake was measured in the oyster meats. Such an experiment has been performed numerous times by several laboratories, including ours (29, 39, 40, 41, 42, 43, 44). For this study, “marked” strains of V. vulnificus were used that could be distinguished from background bacterial populations. The first experiment used strain CVD713, and at the initiation of the study (time zero in Fig. 1), there were no colonies recovered on Tn medium, as expected. After incubation in the seeded water but before they were placed into clean water to allow depuration, oysters were again sampled for the marked strain (Fig. 1; t = 1 day), and ca. 1,100 CFU/gram of oyster tissue was recovered, a significant increase from the untreated oyster value (P < 0.001). Further sampling occurred again at 3 and 6 days after the oysters were allowed to depurate in clean water, during which time the number of CFU/g of the marked strain recovered dropped significantly (P < 0.001). The observation that oysters concentrate the bacteria from the surrounding waters and then quickly clear the added cells upon the removal of the inoculum is consistent with previous reports that utilized protocols which included a method for distinction of bacterial inoculum from naturally occurring bacterial populations (40, 41, 42, 43, 44).
While this study was being performed, oyster homogenates were simultaneously plated onto CPC+ medium to observe any changes in total V. vulnificus populations that occurred during the uptake and depuration phases; these results are also shown in Fig. 1. Prior to incubation in inoculated water, less than 100 CFU V. vulnificus bacteria/gram of oyster tissue were counted. After uptake of the marked V. vulnificus strain, the oysters contained an average of 104 CFU V. vulnificus bacteria/gram when plated onto CPC+ agar, a significant (P < 0.001) increase from the t = 0 values. This increase initially appeared to be a result of the uptake of the marked strain of V. vulnificus, but it was evident upon closer inspection that the number of cells detected on CPC+ at day 1 significantly (P = 0.002) exceeded the number of marked strain counts by nearly an order of magnitude. Furthermore, while the marked strain was depurated by day 3 to a concentration not significantly different from that of untreated oysters (P = 0.634), the number of cells observed on the CPC+ plates, while declining slightly, was still significantly greater (P < 0.001) at day 6 than the number recorded prior to uptake of the laboratory-grown cells. Oysters treated with cell-free media had Vibrio concentrations at day 6 that were no different (P = 0.222) from those of the initial untreated oyster populations (Fig. 1). In a report by Groubert and Oliver (41), those authors observed a similar phenomenon, with the uptake of an exogenous V. vulnificus strain causing a greater increase in total vibrios than could be accounted for by the added strain alone, but did not comment on the findings. A novel figure constructed from the data presented in that publication is shown in Fig. 2.
Fig 2.
Total CFU of vibrios isolated from thiosulfate-citrate-bile salts-sucrose (TCBS) medium (filled squares) and V. vulnificus strain CVD713 isolated on Tn agar from oysters inoculated with exogenous V. vulnificus (open squares), created from data presented by Groubert and Oliver (41).
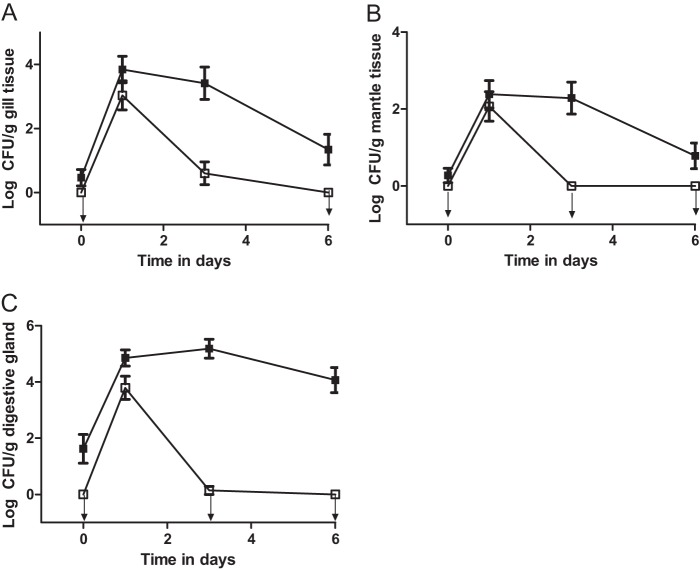
The uptake/depuration experiments were repeated, examining individual oyster tissues (gill, mantle, and digestive gland) rather than the whole-oyster meat. The results for the individual oyster tissues were similar to those seen with the whole-oyster homogenates and are represented in Fig. 3. Total culturable bacteria were also examined, but the starting concentration of these bacteria was very high, at as much as 105 CFU/g, and no significant difference between the concentrations of total natural bacteria before and after the addition of exogenous bacteria was detected (data not shown).
Fig 3.
CFU of vibrios isolated on CPC+ (filled squares) and V. vulnificus strain CVD713 isolated on Tn agar (open squares) from oyster gill tissue (A), mantle tissue (B), or digestive gland tissue (C) following inoculation with exogenous V. vulnificus strain CVD713. Arrows pointing down indicate the points that were below the limit of detection. Error bars represent standard errors of the means.
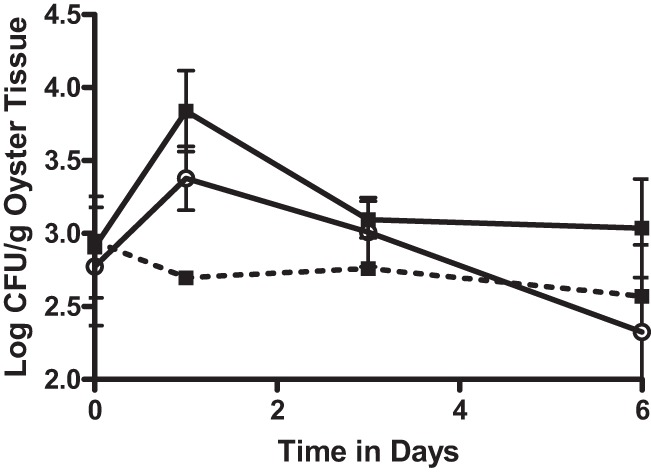
The transposon present in strain CVD713 has been shown to be stable for at least 10 days in artificial seawater (29). However, while the experiments in this study lasted only 6 days, there are no published data showing the stability of this transposon in cells introduced into oysters. To ensure that there had been a true increase of number of total vibrios after the depuration of the marked V. vulnificus strain and not simply an increase in the number of cells of the marked strain that had lost the transposon, the study was repeated with an additional V. vulnificus strain. V. vulnificus VVL1, a naturally occurring bioluminescent strain (31) which can be differentiated from the background bacterial population by viewing and counting the cells in the dark, was used. Detection of this strain thus relies neither on genetic manipulation nor on antibiotic resistance. Similar to the results observed with CVD713, this strain was significantly (P = 0.001) taken up and concentrated by the oysters and was completely depurated (P = 0.003) once the oysters were placed into clean water (Fig. 4). Despite this, there was continued observation of colonies on CPC+ at 3 days at levels that were significantly (P = 0.006) greater than those present in oysters before the marked strain was introduced. These cells remained present in the oysters even though the marked strain had been completely depurated at day 3 and day 6 (P < 0.001 and P = 0.018, respectively). This strain provided the best evidence that the phenomenon observed was not due to the loss of antibiotic resistance genes on the transposon in CVD713, as the luminescence of this bacterium occurs naturally and is not the consequence of the presence of a mobile genetic element.
Fig 4.
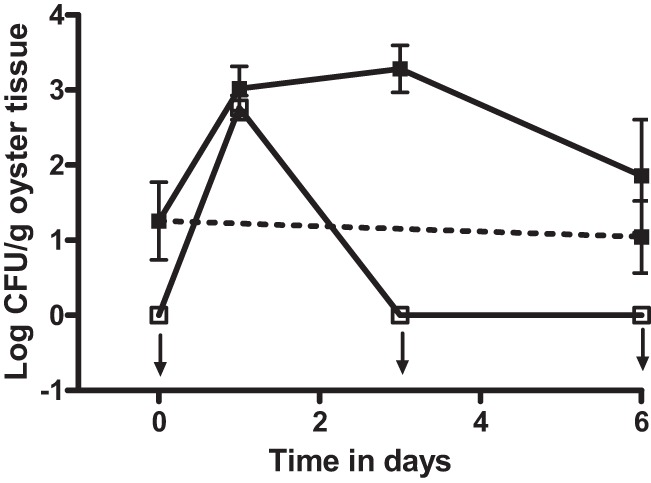
CFU of vibrios isolated on CPC+ (filled squares) and V. vulnificus strain VVL1 isolated on estuarine agar (open squares) from oysters inoculated with exogenous V. vulnificus strain VVL1 (solid lines) or oysters treated with cell-free media (dashed line). Arrows pointing down indicate the point was below the limit of detection. Error bars represent standard errors of the means.
Several of the presumptive colonies recovered from CPC+ in these studies were isolated and subjected to PCR analysis to confirm that they were V. vulnificus. Only 4 of 144 (2.8%) colonies tested were found to be V. vulnificus. This was surprising, given the selective nature of CPC+, but provided strong evidence that the cells appearing on CPC+ were not the marked V. vulnificus cells added to the oyster aquaria. Subsequent sequencing of the 16S rRNA gene of two of these unknown bacteria indicated again that neither was V. vulnificus (>7% sequence mismatches) but were likely V. coralliilyticus and V. mediterranei. It has been demonstrated that when V. vulnificus populations are low, as in higher-salinity environments such as the Mediterranean Sea, competing vibrios can be commonly isolated on CPC+ (45, 46, 47). North Carolina oysters were recently shown to be colonized by vibrios with greater salt tolerance than V. vulnificus following an extreme drought (47). A study employing mCPC agar observed bacterial isolates from oysters and water at the Gulf of Mexico that appeared to be V. vulnificus but were confirmed to be V. sinaloensis instead (48). Furthermore, an extensive study of laboratory-grown strains showed that, while CPC+ does eliminate most Vibrio bacteria, V. alginolyticus and V. harveyi may also appear similar to V. vulnificus in some instances (49). Thus, after multiple methods of confirmation, it was evident that the addition of exogenous V. vulnificus cells to oysters was associated with an increase in a bacterial population of a different Vibrio species.
Further evidence that the sudden increase in the number of detectable bacteria observed upon addition of V. vulnificus was due to the rapid response of an endogenous population, and not to the presence of the exogenously added bacteria, came when the study was repeated using E. coli as the added bacterium. The E. coli K-12 strain used does not grow on CPC+ but can be detected using MacConkey agar. When oysters were incubated with E. coli, results similar to those seen when the marked V. vulnificus strains were added were observed, with colonies recovered from CPC+ increasing from 2.9 log initially to 3.8 log after 24 h, even though no vibrios were added to the oysters (Fig. 5). This increase was significant (P = 0.021) compared to the results seen with control oysters that were not inoculated with E. coli (Fig. 5).
In all experiments, control oysters that received identical experimental treatments but were exposed to cell-free media did not show any changes on CPC+ but maintained a constant concentration throughout the 6-day study period (Fig. 1, 4, and 5; P = 0.222, P = 0.653, and P = 1.000, respectively).
As numerous studies have shown, while adult oysters are able to concentrate bacteria from the surrounding water, this results in a transient population that is rapidly depurated when the oysters are removed to waters with fewer bacteria (40, 41, 42, 43, 44, 47, 50). This is in contrast to the established gut microflora, which likely develops during the larval stage of oyster development (M. P. Doyle and J. D. Oliver, unpublished data) and which does not readily depurate (39, 41, 43). It thus appears that an established population of bacteria in oysters prevents exogenous bacteria from permanently colonizing oyster tissue, possibly through competition for adhesion sites on gut tissues or other surfaces, an effect that provides the basis for the probiotic prophylaxis against exogenous bacteria (51, 52, 53, 54). This does not explain, however, how oysters that initially appeared to contain very low numbers of vibrios suddenly contained several logs more of such cells after exposure to a different bacterial genus or species. Such rapid development of this population could be due an extremely low number of detectable cells that rapidly multiply in response to exogenous bacteria. Alternatively, this could also be a result of a natural microflora population present in a viable but nonculturable state during the winter months (14, 24, 26, 55). It is possible that the established microbiota enters this state as a mechanism to survive the reduced temperatures. Indeed, numerous studies have documented the lack of culturable vibrios in oysters during cold-water months, and their “reappearance” when the waters become warmer (9, 10, 11, 12, 13, 27, 55, 56, 57, 58, 59). If the cells are in the VBNC state, then we suggest that, in response to the addition of a substantial population of exogenous, actively metabolizing bacteria, these VBNC cells resuscitate (23, 26, 60, 61). Perhaps the presence of a large population of exogenous bacteria is a signal to the VBNC cells that the environment is now better able to support their existence without the protective effects of the nonculturable state, a communication possibly achieved though cell-to-cell signaling molecules such as autoinducers or resuscitation-promoting factors (62, 63). Such a scenario could involve the “scout cells” described by Epstein (64). This was further suggested in the present study, in which oysters were collected from cold waters, transported to the laboratory, and kept in aquaria at warm (27°C) temperatures for over a month. These oysters, and their bacterial populations, were no longer at inhibitory temperatures and yet when sampled were still low in culturable Vibrio populations. These oysters, upon the addition of V. vulnificus, developed the large and stable population of non-V. vulnificus vibrios. These bacteria must therefore have been present in the oysters from the outset but resuscitated in response not solely to a temperature increase but also to an unknown factor associated with the exogenously added, culturable bacteria. The findings presented here thus offer further understanding of how and why cells that are present in oysters but in a dormant state become readily detectable when water temperatures, and bacterial populations in those waters, increase in spring and summer months.
Future work will include next-generation sequencing techniques to observe the oyster microflora over different time points. This will permit monitoring of Vibrio populations over time in response to exogenous bacteria.
ACKNOWLEDGMENTS
We thank Tiffany Williams for oyster collection and tank maintenance and Amy Ringwood for remote laboratory facilities.
Funding for this project was provided by a grant from the USDA (number 2007-35201-18381).
Footnotes
Published ahead of print 21 June 2013
REFERENCES
- 1.Johnston JM, Becker SF, McFarland LM. 1986. Gastroenteritis in patients with stool isolates of Vibrio vulnificus. Am. J. Med. 80:336–338 [DOI] [PubMed] [Google Scholar]
- 2.Oliver JD. 1989. Vibrio vulnificus, p 569–599 In Doyle MP. (ed), Foodborne bacterial pathogens. Marcel Dekker, Inc, New York, NY [Google Scholar]
- 3.Oliver JD. 2006. Vibrio vulnificus, p 349–366 In Thompson FL, Austin B, Swings J. (ed), The biology of vibrios. American Society of Microbiology, Washington, DC [Google Scholar]
- 4.Lowry PW, McFarland LM, Peltier BH, Roberts NC, Bradford HB, Herndon JL, Stroup DF, Mathison JB, Blake PA, Gunn RA. 1989. Vibrio gastroenteritis in Louisiana: a prospective study among attendees of a scientific congress in New Orleans. J. Infect. Dis. 160:978–984 [DOI] [PubMed] [Google Scholar]
- 5.Centers for Disease Control and Prevention 2007. Summary of human Vibrio cases reported to CDC. The Cholera and Other Vibrio Illness Surveillance System. Centers for Disease Control and Prevention, Atlanta, GA [Google Scholar]
- 6.Oliver JD. 2005. Vibrio vulnificus, p 253–276 In Belkin S, Colwell RR. (ed), Oceans and health: pathogens in the marine environment. Springer, New York, NY [Google Scholar]
- 7.Mead PS, Slusker L, Dietz V, McCaig LF, Bresee JS, Shapiro C, Griffin PM, Tauxe RBV. 1999. Food-related illness and death in the United States. Emerg. Infect. Dis. 5:607–625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Oliver JD, Kaper JB. 2001. Vibrio species, p 263–300 In Doyle MP, Beuchat LR, Montville TJ. (ed), Food microbiology: fundamentals and frontiers. ASM Press, Washington, DC [Google Scholar]
- 9.Pfeffer CS, Hite MF, Oliver JD. 2003. Ecology of Vibrio vulnificus in estuarine waters of eastern North Carolina. Appl. Environ. Microbiol. 69:3526–3531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kelly MT. 1982. Effect of temperature and salinity on Vibrio (Beneckea) vulnificus occurrence in a Gulf Coast environment. Appl. Environ. Microbiol. 44:820–824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kaspar CW, Tamplin ML. 1993. Effects of temperature and salinity on the survival of Vibrio vulnificus in seawater and shellfish. Appl. Environ. Microbiol. 59:2425–2429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Randa MA, Polz MF, Lim E. 2004. Effects of temperature and salinity on Vibrio vulnificus population dynamics as assessed by quantitative PCR. Appl. Environ. Microbiol. 70:5469–5476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Motes ML, DePaola A, Cook DW, Veazey JE, Hunsucker JC, Garthright WE, Blodgett RJ, Chirtel SJ. 1998. Influence of water temperature and salinity on Vibrio vulnificus in northern gulf and Atlantic coast oysters (Crassostrea virginica). Appl. Environ. Microbiol. 64:1459–1465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oliver JD. 1993. Formation of viable nut nonculturable cells, p 239–272 In Kjelleberg S. (ed), Starvation in bacteria. Plenum Press, New York, NY [Google Scholar]
- 15.Habibur Rahman M, Suzuki S, Kawai K. 2001. Formation of viable but non-culturable state (VBNC) of Aeromonas hydrophila and its virulence in goldfish, Carassius auratus. Microbiol. Res. 156:103–106 [DOI] [PubMed] [Google Scholar]
- 16.Ilari JL. 2002. Flow properties of industrial dairy powders. Lait 82:383–399 [Google Scholar]
- 17.Ohtomo R, Saito M. 2001. Increase in the culturable cell number of Escherichia coli during recovery from saline stress: possible implication for resuscitation from the VBNC State. Microb. Ecol. 42:208–214 [DOI] [PubMed] [Google Scholar]
- 18.Roszak D, Colwell R. 1987. Survival strategies of bacteria in the natural environment. Microbiol. Rev. 51:365–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Colwell RR, Grimes DJ. (ed). 2000. Nonculturable microorganisms in the environment. ASM Press, Washington, DC [Google Scholar]
- 20.Banakar V, Constantin de Magny G, Jacobs J, Murtugudde R, Huq A, Wood RJ, Colwell RR. 2011. Temporal and spatial variability in the distribution of Vibrio vulnificus in the Chesapeake Bay: a hindcast study. EcoHealth 8:456–467 [DOI] [PubMed] [Google Scholar]
- 21.Linder K, Oliver JD. 1989. Membrane fatty acid and virulence changes in the viable but nonculturable state of Vibrio vulnificus. Appl. Environ. Microbiol. 55:2837–2842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brauns LA, Hudson MC, Oliver JD. 1991. Use of the polymerase chain reaction in detection of culturable and nonculturable Vibrio vulnificus cells. Appl. Environ. Microbiol. 57:2651–2655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nilsson L, Oliver JD, Kjelleberg S. 1991. Resuscitation of Vibrio vulnificus from the viable but nonculturable state. J. Bacteriol. 173:5054–5059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oliver JD. 1995. The viable but non-culturable state in the human pathogen Vibrio vulnificus. FEMS Microbiol. Lett. 133:203–208 [DOI] [PubMed] [Google Scholar]
- 25.Nowakowska J, Oliver JD. 2013. Resistance to environmental stresses by Vibrio vulnificus in the viable but noncultureble state. FEMS Microbiol. Ecol. 84:213–222 [DOI] [PubMed] [Google Scholar]
- 26.Oliver JD, Hite F, McDougald D, Andon N, Simpson L. 1995. Entry into, and resuscitation from, the viable but nonculturable state by Vibrio vulnificus in an estuarine environment. Appl. Environ. Microbiol. 61:2624–2630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wolf P, Oliver JD. 1992. Temperature effects on the viable but nonculturable state of Vibrio vulnificus. FEMS Microbiol. Lett. 100:205–210 [DOI] [PubMed] [Google Scholar]
- 28.Morris JG, Wright AC, Roberts DM, Wood PK, Simpson LM, Oliver JD. 1987. Identification of environmental Vibrio vulnificus isolates with a DNA probe for the cytotoxin-hemolysin gene. Appl. Environ. Microbiol. 53:193–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Murphy SK, Oliver JD. 1992. Effects of temperature abuse on survival of Vibrio vulnificus in oysters. Appl. Environ. Microbiol. 58:2771–2775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wright AC, Simpson LM, Oliver JD, Morris JG. 1990. Phenotypic evaluation of acapsular transposon mutants of Vibrio vulnificus. Infect. Immun. 58:1769–1773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oliver JD, Roberts DM, White VK, Dry MA, Simpson LM. 1986. Bioluminescence in a strain of the human pathogenic bacterium Vibrio vulnificus. Appl. Environ. Microbiol. 52:1209–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Warner EB, Oliver JD. 2007. Refined medium for direct isolation of Vibrio vulnificus from oyster tissue and seawater. Appl. Environ. Microbiol. 73:3098–3100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Massad G, Oliver JD. 1987. New selective and differential medium for Vibrio cholerae and Vibrio vulnificus. Appl. Environ. Microbiol. 53:2262–2264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weiner RM, Hussong D, Colwell RR. 1980. An estuarine agar medium for enumeration of aerobic heterotrophic bacteria associated with water, sediment, and shellfish. Can. J. Microbiol. 26:1366–1369 [DOI] [PubMed] [Google Scholar]
- 35.American Public Health Association 1976. Compendium of methods for microbiological examination of food. American Public Health Association, Washington, DC [Google Scholar]
- 36.James DE. 1978. Culturing algae. Carolina Biological Supply Company, Burlington, NC [Google Scholar]
- 37.Rosche TM, Yano Y, Oliver JD. 2005. A rapid and simple PCR analysis indicates there are two subgroups of Vibrio vulnificus which correlate with clinical and environmental isolation. Microbiol. Immunol. 49:381–389 [DOI] [PubMed] [Google Scholar]
- 38.Warner EB, Oliver JD. 2008. Multiplex PCR assay for detection and simultaneous differentiation of genotypes of Vibrio vulnificus biotype 1. Foodborne Pathog. Dis. 5:691–693 [DOI] [PubMed] [Google Scholar]
- 39.Chae MJ, Cheney D, Su YC. 2009. Temperature effects on the depuration of Vibrio parahaemolyticus and Vibrio vulnificus from the American Oyster (Crassostrea virginica). J. Food Sci. 74:M62–M66 [DOI] [PubMed] [Google Scholar]
- 40.Froelich B, Ringwood A, Sokolova I, Oliver J. 2010. Uptake and depuration of the C- and E-genotypes of Vibrio vulnificus by the Eastern Oyster (Crassostrea virginica). Environ. Microbiol. Rep. 2:112–115 [DOI] [PubMed] [Google Scholar]
- 41.Groubert TN, Oliver JD. 1994. Interaction of Vibrio vulnificus and the Eastern oyster, Crassostrea virginica. J. Food Prot. 57:224–228 [DOI] [PubMed] [Google Scholar]
- 42.Kelly MT, Dinuzzo A. 1985. Uptake and clearance of Vibrio vulnificus from Gulf coast oysters (Crassostrea virginica). Appl. Environ. Microbiol. 50:1548–1549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Paranjpye RN, Johnson AB, Baxter AE, Strom MS. 2007. Role of type IV pilins of Vibrio vulnificus in persistence in oysters, Crassostrea virginica. Appl. Environ. Microbiol. 73:5041–5044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Srivastava M, Tucker MS, Gulig PA, Wright AC. 2009. Phase variation, capsular polysaccharide, pilus and flagella contribute to uptake of Vibrio vulnificus by the Eastern oyster (Crassostrea virginica). Environ. Microbiol. 11:1934–1944 [DOI] [PubMed] [Google Scholar]
- 45.Macián M, Arias C, Aznar R, Garay E, Pujalte M. 2000. Identification of Vibrio spp. (other than V. vulnificus) recovered on CPC agar from marine natural samples. Int. Microbiol. 3:51–53 [PubMed] [Google Scholar]
- 46.Williams TC, Froelich B, Oliver JD. 2010. A new culture-based method for the improved identification of Vibrio vulnificus from environmental samples, reducing the need for molecular confirmation. J. Microbiol. Methods 93(3):277–283. 10.1016/j.mimet.2013.03.023 [DOI] [PubMed] [Google Scholar]
- 47.Froelich BA, Williams T, Noble R, Oliver JD. 2012. Apparent loss of Vibrio vulnificus in North Carolina oysters coincides with drought-induced increase in salinity. Appl. Environ. Microbiol. 78:3885–3889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Staley C, Chase E, Harwood VJ. 2013. Detection and differentiation of Vibrio vulnificus and V. sinaloensis in water and oysters of a Gulf of Mexico estuary. Environ. Microbiol. 15:623–633 [DOI] [PubMed] [Google Scholar]
- 49.Williams TC, Froelich B, Oliver JD. 2013. A new culture-based method for the improved identification of Vibrio vulnificus from environmental samples, reducing the need for molecular confirmation. J. Microbiol. Methods 93:277–283 [DOI] [PubMed] [Google Scholar]
- 50.Motes ML, DePaola A. 1996. Offshore suspension relaying to reduce levels of Vibrio vulnificus in oysters (Crassostrea virginica). Appl. Environ. Microbiol. 62:3875–3877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Garcia T, Kjelleberg OKS, Nelson DR. 1997. Growth of Vibrio anguillarum in salmon intestinal mucus. Appl. Environ. Microbiol. 63:1034–1039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Krovacek K, Faris A, Ahne W, Mansson I. 1987. Adhesion of Aeromonas hydrophila and Vibrio anguillarum to fish cells and to mucus coated glass slides. FEMS Microbiol. Lett. 42:85–89 [Google Scholar]
- 53.Olsson JC, Westerdahl A, Conway P, Kjelleberg S. 1992. Intestinal colonization potential of turbot (Scophthalmus maximus)- and dab (Limanda limanda)-associated bacteria with inhibitory effects against Vibrio anguillarum. Appl. Environ. Microbiol. 58:551–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Verschuere L, Rombaut G, Sorgeloos P, Verstraete W. 2000. Probiotic bacteria as biological control agents in aquaculture. Microbiol. Mol. Biol. Rev. 64:655–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nguyen K, LaRock P, La Peyre J. 2001. Vibrio vulnificus infection of oyster cells: a possible survival mechanism, p 475 Abstr. Aquaculture 2001. [Google Scholar]
- 56.Warner EB, Oliver JD. 2008. Population structures of two genotypes of Vibrio vulnificus in oysters (Crassostrea virginica) and seawater. Appl. Environ. Microbiol. 74:80–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.O'Neill KR, Jones SH, Grimes DJ. 1992. Seasonal incidence of Vibrio vulnificus in the Great Bay estuary of New Hampshire and Maine. Appl. Environ. Microbiol. 58:3257–3262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lin M, Schwarz JR. 2003. Seasonal shifts in population structure of Vibrio vulnificus in an estuarine environment as revealed by partial 16S ribosomal DNA sequencing. FEMS Microbiol. Ecol. 45:23–27 [DOI] [PubMed] [Google Scholar]
- 59.Wright A, Hill R, Johnson J, Roghman M, Colwell R, Morris J., Jr 1996. Distribution of Vibrio vulnificus in the Chesapeake Bay. Appl. Environ. Microbiol. 62:717–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Whitesides M, Oliver J. 1997. Resuscitation of Vibrio vulnificus from the viable but nonculturable state. Appl. Environ. Microbiol. 63:1002–1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Birbari W, Rodrick GE, Oliver JD. 2000. Uptake and resuscitation of viable but non-culturable Vibrio vulnificus by Mercenaria campechiensis. Abstr. 16th Annu. Trop. Subtrop. Fish. Technol. Conf. Am. Seafood Sci. Technol. Soc. Am [Google Scholar]
- 62.Nikitushkin V, Demina G, Shleeva M, Kaprelyants A. 2013. Peptidoglycan fragments stimulate resuscitation of “non-culturable” mycobacteria. Antonie Van Leeuwenhoek 103:37–46 [DOI] [PubMed] [Google Scholar]
- 63.Pereira CS, McAuley JR, Taga ME, Xavier KB, Miller ST. 2008. Sinorhizobium meliloti, a bacterium lacking the autoinducer-2 (AI-2) synthase, responds to AI-2 supplied by other bacteria. Mol. Microbiol. 70:1223–1235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Epstein SS. 2009. Microbial awakenings. Nature 457:1083. 10.1038/4571083a [DOI] [PubMed] [Google Scholar]