Abstract
Vibrio cholerae senses its environment, including the surrounding bacterial community, using both the second messenger cyclic di-GMP (c-di-GMP) and quorum sensing (QS) to regulate biofilm formation and other bacterial behaviors. Cyclic di-GMP is synthesized by diguanylate cyclase (DGC) enzymes and degraded by phosphodiesterase (PDE) enzymes. V. cholerae encodes a complex network of 61 enzymes predicted to mediate changes to the levels of c-di-GMP in response to extracellular signals, and the transcription of many of these enzymes is influenced by QS. Because of the complexity of the c-di-GMP signaling system in V. cholerae, it is difficult to determine if modulation of intracellular c-di-GMP in response to different stimuli is driven primarily by changes in c-di-GMP synthesis or hydrolysis. Here, we describe a novel method, named the ex vivo lysate c-di-GMP assay (TELCA), that systematically measures total DGC and PDE cellular activity. We show that V. cholerae grown in different environments exhibits significantly different intracellular levels of c-di-GMP, and we used TELCA to determine that these differences correspond to changes in both c-di-GMP synthesis and hydrolysis. Furthermore, we show that the increased concentration of c-di-GMP at low cell density is primarily due to increased DGC activity due to the DGC CdgA. Our findings highlight the idea that modulation of both total DGC and PDE activity alters the intracellular concentration of c-di-GMP, and we present a new method that is widely applicable to the systematic analysis of complex c-di-GMP signaling networks.
INTRODUCTION
Vibrio cholerae, a Gram-negative marine bacterium, is the causative agent of the human diarrheal disease cholera. In aquatic environments, V. cholerae favors a sessile lifestyle by preferentially forming biofilms on chitinous surfaces (1–3). Upon ingestion by a human host, V. cholerae expresses virulence and colonization factors that lead to severe diarrhea, subsequently reseeding the bacteria back into the environment. The transition between aquatic environments and the human host is mediated both by cyclic di-GMP (c-di-GMP) and quorum sensing (QS), which together regulate biofilm formation, motility, and virulence gene expression (4–6).
c-di-GMP is a bacterial second messenger that regulates many bacterial behaviors, including biofilm formation, motility, virulence, and life cycle progression (7–12). Diguanylate cyclase enzymes (DGCs) synthesize c-di-GMP from two GTP molecules and contain a conserved GGDEF domain (13, 14). Cyclic di-GMP specific phosphodiesterase enzymes (PDEs) hydrolyze c-di-GMP and contain either a conserved EAL or HD-GYP domain (15, 16). It has been suggested that extracellular factors regulate DGCs and PDEs, altering the intracellular concentration of c-di-GMP to adapt to changing environments. Indeed, some environmental signals, such as light, oxygen, amino acids, quorum-sensing autoinducers, and norspermidine, have been shown to alter intracellular c-di-GMP in various bacteria (16–24). However, the vast majority of the regulatory inputs that control DGCs and PDEs remain undetermined.
A characteristic feature of c-di-GMP signaling networks is the relatively large number of DGCs and PDEs that theoretically contribute to changes in the intracellular concentration of c-di-GMP. For example, V. cholerae encodes 61 distinct proteins predicted to modulate c-di-GMP (25). Because of this complexity, c-di-GMP signaling systems are typically redundant, and it is challenging to understand on a systems level how the concentration of c-di-GMP is controlled. Changes in c-di-GMP could be mediated by modulation of total DGC activity, PDE activity, or both.
QS, the process of bacterial cell-cell communication mediated by autoinducers (AI) that are constitutively secreted and accumulate with cell density, is a second fundamental chemical signaling system in bacteria. Sensing of AIs by bacteria provides information on the local population density and composition to appropriately regulate bacterial behaviors such as competence, biofilm formation, and virulence (26–31). In V. cholerae, QS is mediated by two AI molecules (CAI-1 and AI-2) that are produced in tandem (30). In the low-cell-density state, AI concentrations are low and their cognate receptors function as kinases, ultimately leading to phosphorylation of the response regulator LuxO. Phosphorylated LuxO upregulates the transcription of four small regulatory RNAs (sRNAs), which subsequently suppress translation of the master QS regulator HapR (32, 33). When the concentration of AIs increase, they bind the AI receptors, switching them to phosphatases and leading to dephosphorylation of LuxO, deactivation of the small RNAs (sRNAs), and induction of HapR expression (34). In V. cholerae, biofilm formation is repressed in the high-cell-density QS state via direct repression of extracellular polysaccharide synthesis genes by HapR (29, 35).
Both the QS and c-di-GMP signaling systems are inextricably linked in V. cholerae (6). At low cell density, the intracellular c-di-GMP concentration of V. cholerae is relatively high, promoting biofilm formation. Upon transition to the high-cell-density state, the intracellular c-di-GMP concentration is suppressed, decreasing biofilm formation (29, 35). In V. cholerae the transcription of 18 of the 61 predicted c-di-GMP turnover enzymes are differentially regulated at low and high cell density (29, 35, 36). However, because of the complexity of this signaling system it remains unclear if the QS-mediated changes in c-di-GMP levels in V. cholerae are being driven by changes in c-di-GMP synthesis, hydrolysis, or both.
To further our exploration of the environmental modulation of c-di-GMP and the regulatory connections between QS, c-di-GMP, and biofilm formation in V. cholerae, we have developed a novel approach, named the ex vivo lysate c-di-GMP assay (TELCA), which enables the quantification of c-di-GMP synthesis and hydrolysis of a whole-cell lysate. We have shown that growing V. cholerae in divergent environments can lead to a 20-fold difference in the intracellular concentration of c-di-GMP, and these distinct c-di-GMP concentrations can be attributed to changes in both c-di-GMP synthesis and hydrolysis. We also used TELCA to show that alteration of both c-di-GMP synthesis and hydrolysis between the low- and high-cell density QS states contributes to the QS-mediated change of intracellular c-di-GMP. We further show that deletion of the DGC CdgA, which is transcriptionally regulated by HapR and has been predicted to be a key DGC in the induction of biofilm formation (35–38), significantly reduces the elevated DGC activity associated with low cell density, highlighting the sensitivity of TELCA. TELCA provides a new method for studying complex c-di-GMP signaling networks from a systems perspective.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The V. cholerae El Tor biotype strain C6706str2 was used for all experiments (39). The construction of the expression plasmids for VCA0956 and VC1086 is described elsewhere (35, 40). Unless otherwise specified, cultures were grown in Luria-Bertani medium (LB; 1.0% tryptone, 0.5% yeast extract, 1.0% NaCl; Accumedia) at 35°C with shaking at 220 rpm. The compositions of AKI, AB, and M9 media are described elsewhere (41–43). AB and M9 were supplemented with glucose as a carbon source (0.2% and 0.4%, respectively). For experiments involving the expression of DGCs or PDEs from plasmids, media were supplemented with kanamycin (Sigma) at 100 μg/ml and the inducer isopropyl-β-d1-thiogalactopyranoside (IPTG) at 0.1 mM. To estimate the start of stationary phase for Fig. S1 in the supplemental material, we determined the maximum doubling time of V. cholerae grown in each medium. We then determined the specific doubling times between measured time points and compared them to the minimum doubling time. The start of stationary phase was defined as the time when the doubling time exceeded a 50% increase of the minimum doubling time.
The construction of the ΔhapR and ΔluxO strains has been described previously (30, 31). To generate the ΔcdgA strain, natural transformation and homologous recombination were used. Briefly, Phusion DNA polymerase (NEB) was used to amplify approximately 500 bp upstream and downstream of cdgA (VCA0074) using the primers 5′-TCGATGTGTCTGATCTGCGTATCCGCGTTGGTAT-3′ and 5′-GAAGCAGCTCCAGCCTACACGGGGCAAAGTTTACCAT-3′ (upstream) and the primers 5′-TAAGGAGGATATTCATATGGGGCTTCTTATGAATCAAAAT-3′ and 5′-GGTTGAGAAGTAGAACACAATCGATATCCACACG-3′ (downstream). Fusion PCR was then used to join these products upstream and downstream of a chloramphenicol resistance cassette (cat) bordered by FLP recombination target (FRT) sites from the plasmid pKD3 (44). This DNA was introduced into V. cholerae by inducing natural transformation using a modification of the protocol of Marvig and Blokesch (45). V. cholerae was grown on chitin flakes in modified M9 medium (43). This medium (M9-T) contained the standard concentration of M9 salts supplemented with 5 mM CaCl2, 3.6 mM MgSO4, and 4.5 mM N-acetyl-d-glucosamine as a carbon source and was balanced to a pH of 6.0. Concentrated DNA was introduced into the culture, and homologous recombination events were selected by growing the culture on LB supplemented with 1 μg/ml chloramphenicol. The cat gene was excised by ectopically expressing the FLP recombinase from the vector pTL17 (46).
Synthesizing [13C]c-di-GMP.
[13C]c-di-GMP was synthesized from [13C]GTP (Sigma) using the purified DGC WspR-R242A as previously described (47). The reaction mixture consisted of 56 μM WspR-R424A, 20 μM [13C]GTP, 24 mM Tris-Cl, 5 mM MgCl2, 45 mM NaCl, and EnzCheck phosphate assay kit reaction buffer mix (Invitrogen) at a total volume of 100 μl, hypothetically yielding 10 μM [13C]c-di-GMP (two GTP molecules are consumed to form one molecule of c-di-GMP). The reaction was run overnight and was followed by treatment for 30 min at 37°C with Antarctic phosphatase (NEB) in its corresponding reaction buffer to remove any excess [13C]GTP. The reaction was heated at 100°C for 10 min to inactivate the Antarctic phosphatase and precipitate protein. After cooling to room temperature, the reaction was centrifuged at maximum speed for 10 min to remove the precipitated protein, and the supernatants were removed and stored at −20°C until use. [13C]c-di-GMP synthesis was confirmed using liquid chromatography with mass spectrometry as described below.
Generation of lysates.
Cell lysates for TELCA were generated by inoculating overnight cultures of V. cholerae 1:1,000 into 30 ml of medium with the appropriate antibiotics or inducer. Cultures grown to the desired optical density at 600 nm (OD600) as indicated in Results were centrifuged at 7,800 rpm to pellet the cells. The pellets were then resuspended in 15 ml TELCA reaction buffer, which consisted of 25 mM Tris, 5 mM MgCl2, and 5 mM NaCl balanced to a pH of 7.5. The cultures were then processed three times using a M110-P processor (Microfluidics) at 20,000 lb/in2, producing a clear lysate. A Bradford protein assay (Bio-Rad) was performed according to the manufacturer's instructions to quantify the total protein content, and each lysate was diluted to 0.1 mg/ml in TELCA reaction buffer. To measure c-di-GMP synthesis, 100 mM [13C]GTP stock was added to a final concentration of 1 mM (10 μl [13C]GTP stock in 990 μl lysate diluent for time course experiments, 1 μl [13C]GTP stock in 99 μl lysate diluent for endpoint experiments). To measure c-di-GMP hydrolysis, 10 μM [13C]c-di-GMP stock was added to a final concentration of 100 nM (10 μl [13C]c-di-GMP stock in 990 μl lysate diluent for time course experiments, 1 μl [13C]c-di-GMP stock in 99 μl lysate diluent for endpoint experiments). The concentrations are within the physiological ranges of the natural concentrations of GTP and c-di-GMP found in a bacterial cell (40, 48). The reaction mixtures were then mixed and incubated at 37°C until the experimental end point, and then the reaction was stopped by mixing 100 μl reaction mixture with 100 μl of phenol-chloroform-isoamyl alcohol (25:24:1). The mixture was centrifuged at maximum speed for 5 min, and then the aqueous phase was removed and stored at −80°C until analysis.
Detection and quantification of c-di-GMP.
For the quantification of intracellular c-di-GMP, overnight cultures grown in the corresponding medium were used to inoculate 2-ml cultures (1:1,000). Cultures were grown to the appropriate OD600 (for AKI, 0.5 to 1.0; for LB, 0.5 to 0.8; for AB, 0.3 to 0.7; for M9, 0.2 to 0.4), and a 1.5-ml aliquot of culture was centrifuged at maximum speed for 30 s. The supernatant was removed, and the pellet was resuspended in 100 μl of cold extraction buffer (40% acetonitrile–40% methanol–0.1 N formic acid). The slurry was incubated at −20°C for 20 min, and then the insoluble fraction was pelleted by centrifugation for 10 min at maximum speed. The supernatant was collected and stored at −80°C. Prior to analysis, the extraction buffer was evaporated using a vacuum manifold, and the pellet was resuspended in 100 μl water. [12C]c-di-GMP and [13C]c-di-GMP were quantified using an Acquity Ultra Performance liquid chromatography system (Waters) coupled with a Quattro Premier XE mass spectrometer (Waters) as previously described (40). Cyclic di-GMP was detected by monitoring the transition of the m/z of the precursor ion to that of the fragmented ion using multiple reaction monitoring ([13C]c-di-GMP, 709.16 > 354.31; [12C]c-di-GMP, 689.16 > 344.31). The concentration of c-di-GMP was determined by generating an 8-point standard curve (1:2 dilutions) of chemically synthesized c-di-GMP (Biolog) ranging from 1.9 nM to 250 nM. The intracellular c-di-GMP concentration was calculated by dividing the c-di-GMP concentration of the sample by the total intracellular volume of bacteria extracted. This was estimated by multiplying the average cell volume of the bacterial cell, determined by measuring individual cell dimensions using differential image contrast microscopy, by the total number of bacterial cells harvested. The c-di-GMP concentration from TELCA reactions was determined using identical liquid chromatography-tandem mass spectrometry (LC-MS/MS) settings.
RESULTS
TELCA quantification of whole-cell DGC activity.
We sought to develop a systems approach to examine if changes in the levels of c-di-GMP in V. cholerae could be attributed to the modulation of net DGC and/or PDE activity. The rationale for developing this method is that current approaches to directly measure c-di-GMP primarily utilize liquid chromatography coupled with tandem mass spectrometry (LC-MS/MS) of cell extracts (49–51) or an in vivo fluorescence resonance energy transfer (FRET) biosensor to quantify c-di-GMP at the single-cell level (52). While these methods allow accurate quantification of the average intracellular c-di-GMP in the cell, they cannot distinguish if differences in the intracellular c-di-GMP are being driven by changes in c-di-GMP synthesis or hydrolysis. Consider a case where the levels of c-di-GMP are elevated by growth in a different environment. This increase in c-di-GMP can be due to increased DGC activity, decreased PDE activity, or modulation of both. One cannot predict a priori solely based on the concentration of c-di-GMP how this change is mediated. Our method, named TELCA, quantifies total DGC or PDE activity of cell lysates using 13C-containing substrates to allow the differentiation of newly synthesized or degraded c-di-GMP from natural occurring c-di-GMP by mass spectrometry. The substrate for the DGC assay measures synthesis of [13C]c-di-GMP from exogenous [13C]GTP, whereas the PDE assay monitors the degradation of exogenous [13C]c-di-GMP.
To assess how effectively TELCA measures cellular DGC activity, extracts were generated from cultures of wild-type (WT) V. cholerae ectopically expressing an active DGC (VCA0956), an active PDE (VC1086), or the vector control. Note that WT V. cholerae has a relatively low intracellular concentration of c-di-GMP at the growth state used for these experiments (mid-exponential-phase growth, with an OD600 of 0.5 to 0.8) due to QS-mediated reduction in c-di-GMP levels at high cell density. We hypothesized that lysates produced from cells expressing VCA0956 would more rapidly synthesize [13C]c-di-GMP from [13C]GTP than lysates with the empty vector control or overproducing the PDE VC1086.
The cells from all three cultures were harvested and lysed using a Microfluidics M110-P processor, which exposes the cells to high shear and impact forces, resulting in a uniform lysate. This process causes minimum disruption of protein conformation and thus is well suited to generate lysates for this assay. [13C]GTP was added to 0.1 mg/ml total protein from each extract to a final concentration of 1.0 mM, and aliquots of the reaction were removed at the time points specified in Fig. 2 followed by quenching with a phenol-chloroform extraction. The concentration of newly synthesized [13C]c-di-GMP was quantified using LC-MS/MS. To differentiate [13C]c-di-GMP from naturally occurring [12C]c-di-GMP, we measured a mass-to-charge ratios (m/z) of 709.16 > 354.31 to detect [13C]c-di-GMP and 689.16 > 344.31 for [12C]c-di-GMP, as previously described (Fig. 1A) (40). The TELCA reaction generated a signal at the same retention time of chemically synthesized [12C]c-di-GMP standards at a m/z of 709.16 > 354.31 with no signal at 689.16 > 344.31, indicating that our detection method is sufficient to discriminate between these two carbon isotope forms of c-di-GMP (Fig. 1B).
Fig 2.
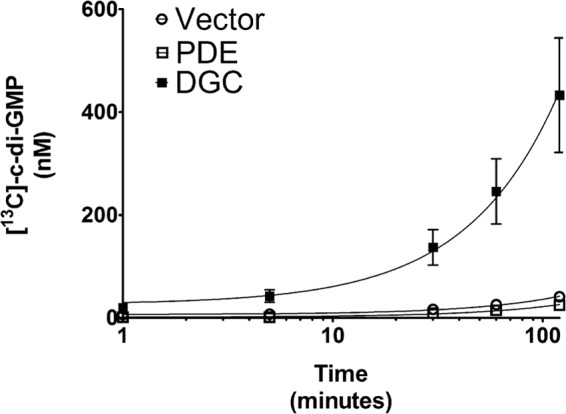
TELCA was applied to lysates of V. cholerae strains ectopically expressing an active DGC (VCA0956; filled squares), an active PDE (VC1086; empty squares), and an empty vector control (empty circles). [13C]GTP was added to each lysate, and [13C]c-di-GMP synthesis was tracked over time using LC-MS/MS. A nonlinear regression using a one-phase exponential decay model was applied to each lysate. Each point represents the mean of three replicates; error bars indicate standard errors.
Fig 1.
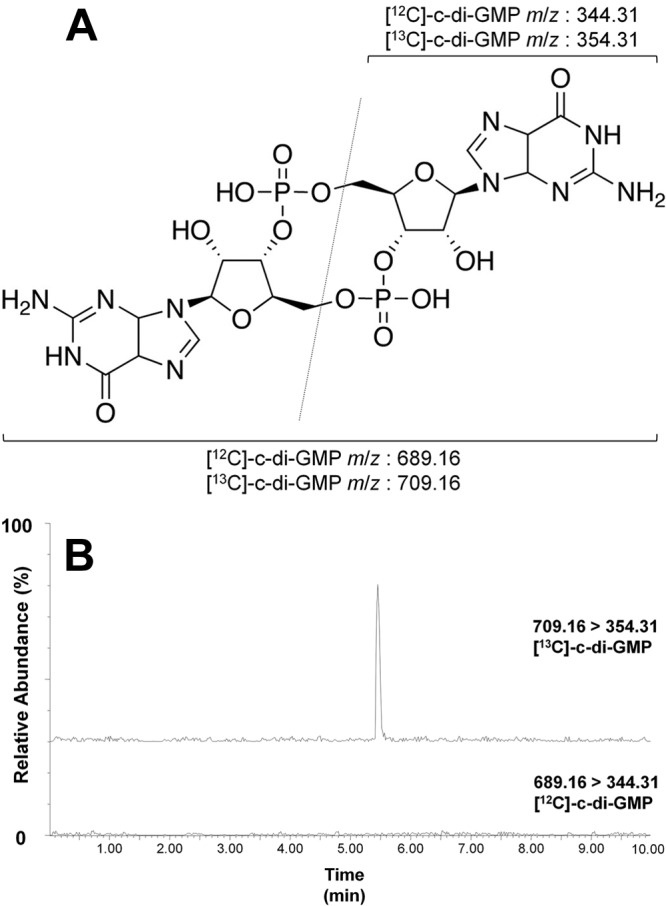
(A) Molecular structure of c-di-GMP with the m/z used for detection and quantification. The top m/z describes the half of the molecule observed after fragmentation (as indicated by the line), while the bottom m/z represent the entire molecule. (B) Spectrum of [13C]c-di-GMP. The top contains a peak at 5.4 min with an m/z of 709.16 > 354.31, indicating the presence of [13C]c-di-GMP. The bottom contains no peak at 5.4 min with an m/z of 689.16 > 354.31, indicating that no [12C]c-di-GMP is present.
A nonlinear regression with a one-phase decay model (GraphPad Prism) was used to analyze the increase in [13C]c-di-GMP concentrations over time. We constrained the maximum yield of the model to 0.5 mM [13C]c-di-GMP, as this is the maximum theoretical yield of the reaction. The lysate from V. cholerae expressing the DGC produced significantly more (two-tailed Student's t test, P < 0.05) [13C]c-di-GMP over time, yielding 10.2-fold and 17.1-fold more c-di-GMP than the vector control and PDE lysates, respectively, after 120 min (Fig. 2). Furthermore, the rate constant for the DGC lysate, which represents the increase of c-di-GMP synthesis per unit time expressed as inverse minutes (6.9 × 10−6 ± 1.1 × 10−6 min−1, R2 = 0.76) was significantly different (sum-of-squares F test, P < 0.05) than those of the vector control lysate (5.9 × 10−7 ± 0.5 × 10−7 min−1, R2 = 0.90) and the PDE lysate (4.1 × 10−7 ± 0.4 × 10−7 min−1, R2 = 0.87). This result indicates that TELCA is able to differentiate the cellular DGC activity of lysates containing a higher concentration of active DGCs. Furthermore, we found that lysates generated from V. cholerae expressing the PDE produced 1.7-fold less [13C]c-di-GMP than the vector control lysate after 120 min and exhibited a lower rate constant, indicating that additional PDEs in the lysate can reduce the net c-di-GMP synthesis by hydrolyzing [13C]c-di-GMP postsynthesis.
Of note, [13C]c-di-GMP accumulated in all three lysates, including the PDE overexpression strain, suggesting that given excess GTP, DGC activity supersedes PDE activity under these conditions over the time course examined. A subset of DGCs exhibit feedback inhibition by an allosteric RXXD inhibition site in close proximity to the DGC active site (53). VCA0956 does not have a predicted inhibition site; however, it is possible that the c-di-GMP synthesis of another DGC subject to feedback inhibition would have an altered synthesis profile. Additionally, it should be noted that we were not able to detect any hybrid c-di-GMP composed of one [12C]GTP and one [13C]c-di-GMP (detection m/z of 699.16 > 344.31), indicating that the native [12C]GTP is depleted in the lysates. Furthermore, there was no [12C]c-di-GMP detected in the vector control lysate or the PDE lysate at any time point. However, in the DGC lysate, we did detect native [12C]c-di-GMP in low abundance, suggesting that if [12C]GTP is to be used in lieu of [13C]GTP, additional measures would be required to compensate for the existence of native c-di-GMP in the lysates.
TELCA quantification of whole-cell PDE activity.
We performed a similar experiment to determine if TELCA is effective at differentiating the PDE activity of different bacterial strains; however, instead of using [13C]GTP as the initial substrate, [13C]c-di-GMP was added. To generate [13C]c-di-GMP, a c-di-GMP synthesis reaction was performed using [13C]GTP and the DGC WspR, which contained a mutation in the RXXD allosteric inhibition site (R242A) (47). The remaining labeled GTP was removed by treating the reaction with Antarctic phosphatase, although quantification of enzymatically synthesized [13C]c-di-GMP using LC-MS/MS indicated that the reaction went to completion, generating the expected hypothetical yield. We observed that the PDE lysate more rapidly degraded [13C]c-di-GMP than both the vector control lysate and the DGC lysate (Fig. 3), as the PDE lysate retained an average of 20.5% [13C]c-di-GMP after 120 min, whereas the vector control lysate and the DGC lysate retained averages of 69.6% and 73.9% of the [13C]c-di-GMP, respectively. The differences between the PDE lysate and both the vector control and DGC lysates were significantly different (two-tailed Student's t test, P < 0.05). Likewise, a nonlinear regression with a one-phase decay model with a plateau constraint of zero was applied to the data, and we found that the rate constant of the PDE lysate (1.9 × 10−2 ± 0.6 × 10−2 min−1, R2 = 0.66) was significantly higher (sum-of-squares F test, P < 0.05) than those of both the vector control lysate (2.5 × 10−3 ± 1.0 × 10−3 min−1, R2 = 0.35) and the DGC lysate (2.0 × 10−3 ± 0.9 × 10−3 min−1, R2 = 0.27). As expected, there were no notable differences between the DGC-expressing lysate and the vector control lysate, suggesting that a difference in DGC activity does not impact the PDE activity. From these data, we conclude that TELCA is sufficient to differentiate both the c-di-GMP synthesis and hydrolysis potential of whole-cell lysates.
Fig 3.
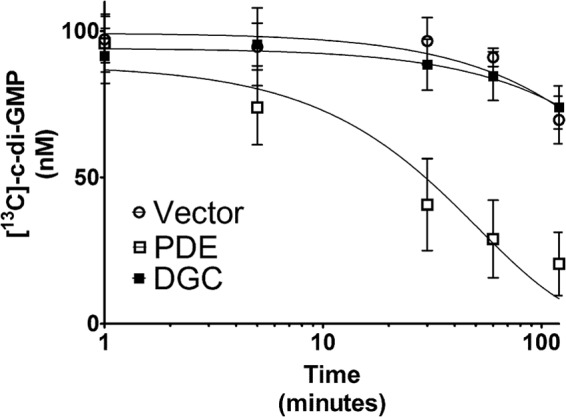
TELCA was applied to lysates of V. cholerae strains ectopically expressing an active DGC (VCA0956; filled squares), an active PDE (VC1086; empty squares), and an empty vector control (empty circles). [13C]c-di-GMP was added to each lysate, and the hydrolysis was tracked over time using LC-MS/MS. A nonlinear regression using a one-phase exponential decay model was applied to each lysate. Each point represents the mean of three replicates; error bars indicate standard errors.
The local environment alters the intracellular concentration of c-di-GMP in V. cholerae.
How the extracellular environment impacts the synthesis and degradation of intracellular c-di-GMP is poorly understood. It has been proposed that extracellular stimuli, such as oxygen, light, amino acids, and QS autoinducers, regulate intracellular c-di-GMP by altering the activity of DGCs and PDEs (16–24). Also, many DGCs and PDEs have domains predicted to be involved in sensing extracellular ligands (54). We hypothesized that V. cholerae growing in different environments would exhibit distinct concentrations of c-di-GMP due to alterations in the environmental inputs controlling DGC and PDE activity. To test this hypothesis, we quantified the intracellular c-di-GMP concentration of V. cholerae in four different growth media using LC-MS/MS. For the purposes of this study, we consider AKI (41) and LB complex, carbon-rich media, while AB (42) and M9 (43) are considered defined, carbon-poor media. We sampled each medium during the latter half of steady-state growth, which we define as 1 to 2 doublings before entry into stationary phase. This time point was determined by generating growth curves for V. cholerae in each different media (see Fig. S1 in the supplemental material).
We found that V. cholerae demonstrated differences of up to 20-fold in the intracellular concentration of c-di-GMP when grown in these different media (Fig. 4). Consistent with prior studies, we measured the concentration of c-di-GMP in LB to be 0.9 ± 0.4 μM (40). The complex, carbon-rich medium AKI also produced relatively low levels of intracellular c-di-GMP that were statistically indistinguishable from those obtained with LB, being measured as 1.1 ± 0.3 μM. In contrast, the carbon-poor media AB and M9 exhibited significantly higher levels of intracellular c-di-GMP at 5.2 ± 2.6 μM and 20.1 ± 5.5 μM, respectively, and M9 had significantly higher intracellular c-di-GMP than AB. Thus, these results support the idea that c-di-GMP signaling systems function primarily as environmental sensors and modulate c-di-GMP accordingly. Moreover, the high concentration of c-di-GMP observed in M9 medium suggests that the range of physiologically relevant concentrations of c-di-GMP experienced by V. cholerae is greater than currently appreciated.
Fig 4.
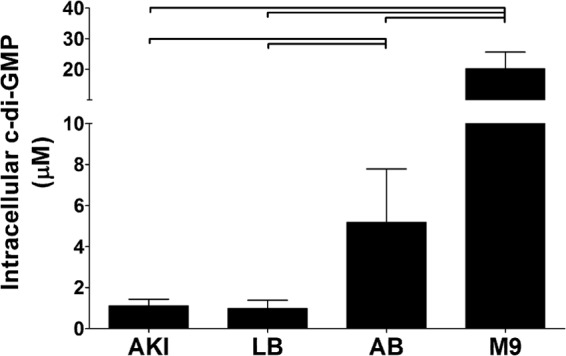
Intracellular c-di-GMP was quantified using LC-MS/MS from V. cholerae grown in four different media to late-exponential-phase growth. Error bars represent standard deviations, and brackets indicate statistical significance, which was determined by using a two-tailed Student t test (P < 0.05).
Growth environments modulate both c-di-GMP synthesis and hydrolysis.
While it is clear that the different growth environments alter intracellular c-di-GMP concentrations of V. cholerae, it is unclear if these changes are driven by modulation of DGC and/or PDE activity. We addressed this question using TELCA on cultures that were grown in the different growth media listed in Fig. 4. We hypothesized that [13C]c-di-GMP synthesis would be greater when cells were grown in both AB and M9 media, as the intracellular c-di-GMP levels of cells grown in these media are elevated. [13C]GTP was added to lysates generated from V. cholerae grown under each environmental condition, and [13C]c-di-GMP synthesis was quantified after 60 min using LC-MS/MS (Fig. 5A). Analogous to the intracellular c-di-GMP measurements, V. cholerae grown in AKI and LB exhibited similar c-di-GMP synthesis. Consistent with our hypothesis, growth of V. cholerae in AB medium exhibited significantly elevated DGC activity, corresponding to its elevated intracellular c-di-GMP concentration. Interestingly, however, cells grown in M9 medium showed a smaller amount of c-di-GMP synthesis than those grown in AB, even though M9 produced the highest intracellular concentration of c-di-GMP that we observed.
Fig 5.
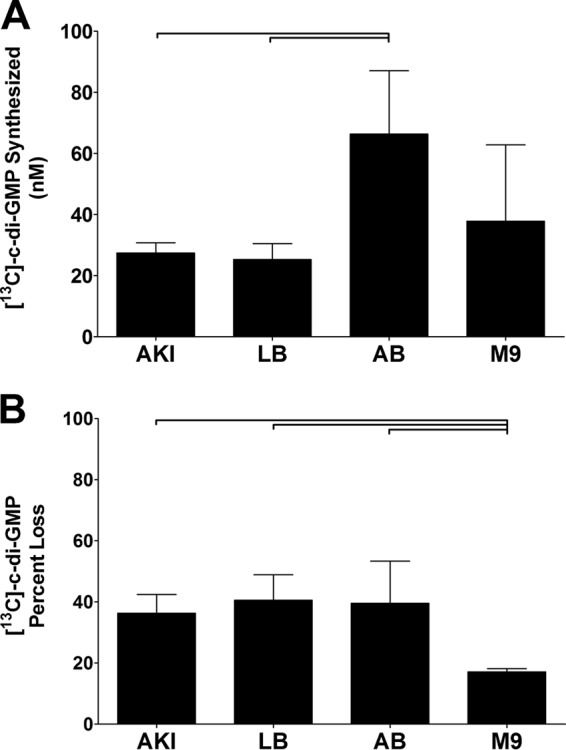
(A) TELCA was applied to lysates of WT V. cholerae grown in different media. [13C]GTP was added to each lysate, and the amount of [13C]c-di-GMP synthesis was quantified using LC-MS/MS. (B) TELCA was applied to lysates of WT and QS mutants of V. cholerae. [13C]c-di-GMP was added to each lysate, and the amount of [13C]c-di-GMP hydrolysis was quantified using LC-MS/MS. The percent loss was calculated by comparing each lysate to the product of a no-protein control reaction. All cultures were grown in triplicate, and the error bars indicate the standard deviations from the mean. Brackets indicate statistical significance determined by using a one-tailed Student t test (P < 0.05).
Based on our analysis of DGC activity, we hypothesized that cells grown in M9 must exhibit lower PDE activity than those grown in the other media to account for the high concentration of c-di-GMP measured for V. cholerae growing in this medium. To test this hypothesis, TELCA was used to quantify [13C]c-di-GMP hydrolysis of cell lysates (Fig. 5B). Consistent with the intracellular c-di-GMP measurements, cells grown in AKI and LB had similar PDE activities; cells grown in AB medium also had c-di-GMP hydrolysis similar to that of cells grown in AKI and AB. However, in support of our hypothesis, cells grown in M9 medium had significantly lower PDE activity than cells grown in the other media. TELCA analysis of V. cholerae grown in these four different media suggests that suppressed PDE activity increases intracellular c-di-GMP in M9 medium, while elevated DGC activity increases intracellular c-di-GMP in AB medium.
QS controls both total DGC and PDE activity in V. cholerae.
As mentioned above, QS modulates the intracellular concentration of c-di-GMP in V. cholerae to regulate biofilm formation, but the specific DGCs and PDEs involved in this regulation are not fully understood. This question is challenging to address, as QS controls the transcription of 18 proteins that potentially alter c-di-GMP levels (29, 35). Moreover, DGCs and PDEs could have differential enzymatic activities at low versus high cell density. To address this question, we used TELCA to determine if QS modulates the levels of c-di-GMP in V. cholerae by altering DGC and/or PDE activity. To analyze DGC and PDE activity of V. cholerae in different QS states, the WT, ΔhapR, and ΔluxO strains were analyzed. The ΔhapR strain is a QS mutant that is locked in low cell density, as HapR is the master high-cell-density transcriptional regulator, whereas the ΔluxO strain is a QS mutant locked in high cell density, as the Qrr sRNAs are never expressed. Importantly, cultures were grown and harvested at an OD600 of 0.2 to 0.3, corresponding to the low-cell-density state. Therefore, we expected the WT strain to be at low cell density and mimic the ΔhapR mutant. Indeed, TELCA analysis indicated that the WT and the ΔhapR lysates exhibited significantly elevated c-di-GMP synthesis compared to the ΔluxO mutant (Fig. 6A), consistent with these strains having a higher concentration of c-di-GMP (29, 35). Analysis of the DGC activity of WT cells grown to high cell density (compare Fig. 5A, LB, to Fig. 6A, WT) indicates that the DGC activity decreases as cells reach a quorum. In fact, the DGC activity of the ΔluxO mutant at low cell density (Fig. 6A) is quite similar to that of WT V. cholerae grown to high cell density (Fig. 5A, LB).
Fig 6.
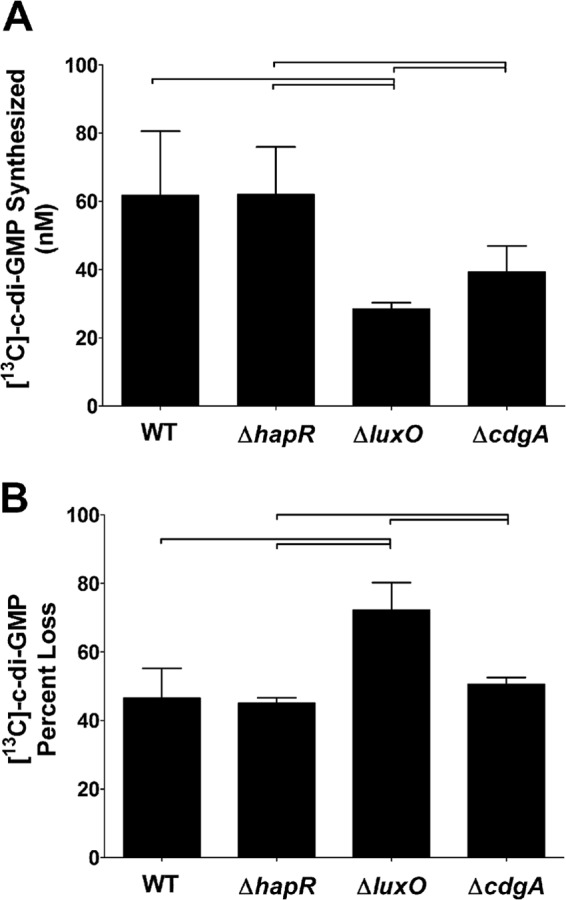
(A) TELCA was applied to lysates of WT V. cholerae and three different mutants. [13C]GTP was added to each lysate, and the amount of [13C]c-di-GMP synthesis was quantified using LC-MS/MS. (B) TELCA was applied to lysates of WT V. cholerae and three different mutants. [13C]c-di-GMP was added to each lysate, and the amount of [13C]c-di-GMP hydrolysis was quantified using LC-MS/MS. The percent loss was calculated by comparing each lysate to the product of a no-protein control reaction. All cultures were grown in triplicate, and the error bars indicate the standard deviations from the mean. Brackets indicate statistical significance determined by using a one-tailed Student t test (P < 0.05).
We performed an analogous experiment to quantify the impact of QS on net PDE activity. The relative PDE activity was determined for each of the lysates from the WT, ΔhapR, and ΔluxO strains by measuring the percent loss of [13C]c-di-GMP. We found that the percent loss of [13C]c-di-GMP was significantly higher in the ΔluxO strain than the ΔhapR mutant and the WT strain, indicating that the locked-high-cell-density mutant had a higher PDE activity (Fig. 6B). The PDE activity of WT cells harvested at low cell density was indistinguishable from that of the ΔhapR mutant. This result suggests that PDE activity is increased as cells transition from low to high cell density. However, a comparison of the WT strain grown to high cell density (Fig. 5B, LB) and the WT strain grown at low cell density (Fig. 6B) reveals similar PDE activities. Indeed, the PDE activity of WT grown to high cell density (Fig. 5B, LB) was lower than that of the ΔluxO mutant (Fig. 6B). This difference in PDE activity between the WT strain and ΔluxO mutant was evident even when the WT strain was grown to stationary phase (data not shown). Thus, in regard to PDE activity, the ΔluxO strain has a more dramatic phenotype than WT V. cholerae growing in LB.
CdgA contributes to the increased DGC activity at low cell density.
The above-described experiments suggest that increased DGC activity at low cell density contributes to the higher concentration of c-di-GMP. CdgA (VCA0074) is a well-studied DGC that is repressed in the high-cell-density state and is important for biofilm formation (35, 37, 55–58). Based on a large body of evidence that indicates that CdgA plays an important role in bridging c-di-GMP signaling and QS (see Discussion), we hypothesized that CdgA significantly contributed to the increased DGC activity we observed at low cell density. To test this hypothesis and determine if TELCA can detect the contribution of a single DGC to total cellular activity, lysates from a ΔcdgA mutant grown to low cell density were analyzed. We observed a 36.4% decrease in DGC activity in the ΔcdgA strain compared to that in the WT strain, although the differences did not quite reach statistical significance (P = 0.06) (Fig. 6A). As expected, there was no significant difference in the net PDE activity of the ΔcdgA strain and the WT strain (Fig. 6B). These results confirm previous genetic studies that implicate CdgA as a major contributor to the increased concentration of c-di-GMP observed in the low-cell-density state in V. cholerae and further illustrate the applicability of TELCA to analyze complex c-di-GMP signaling pathways.
DISCUSSION
The bacterial second messenger c-di-GMP is a central regulator of biofilm formation and motility in bacteria. Indeed, bioinformatic studies indicate that DGCs and PDEs are encoded by more than 80% of all sequenced bacteria (59). Cyclic di-GMP controls many important phenotypes in bacteria, including biofilm formation, motility, production of virulence factors, and life cycle progression, and this list continues to grow (7–12).
Net changes in DGC or PDE activity leading to different intracellular c-di-GMP concentrations can be difficult to parse, as bacteria often encode multiple DGCs and PDEs. Furthermore, it is often challenging to resolve differences in c-di-GMP signaling pathways in single DGC or PDE mutants due to this redundancy and the inherently low concentration of c-di-GMP in many bacterial species (for example, see reference 60). To circumvent these challenges, we developed TELCA to quantify total cellular c-di-GMP synthesis and hydrolysis activities. The 13C-labeled substrates that TELCA utilizes are stable and safe to use and can be distinguished from naturally occurring [12C]c-di-GMP using mass spectrometry.
To test TELCA, we examined strains of V. cholerae ectopically expressing either a GGDEF or an EAL enzyme and observed enhanced DGC and PDE activity, respectively. These experiments validated the approach and demonstrated that we can differentiate lysates with increased synthesis and hydrolysis activities. As an in vitro assay, disruption of the cell might alter the activity of localized or membrane bound proteins in the lysate. However, it should be noted that CdgA is a predicted integral inner membrane protein, yet we were able to quantify a loss in cellular DGC activity of a ΔcdgA mutant using TELCA, showing that CdgA remained active during the assay.
Regulatory inputs of the c-di-GMP signaling network remain undercharacterized. The consensus is that c-di-GMP turnover enzymes bind and respond to environmental ligands or host-derived metabolites or are directly modified by posttranslational regulatory mechanisms to regulate c-di-GMP synthesis or hydrolysis, thus enabling the bacterium to fine-tune intracellular c-di-GMP to its environment (61). Indeed, we showed that when V. cholerae is cultured in different media, the intracellular c-di-GMP level varies significantly (Fig. 4). Surprisingly, the changes in the c-di-GMP levels were dramatic, with cells grown in M9 exhibiting 20 times more c-di-GMP than cells grown in LB. This result suggests that V. cholerae, and potentially other bacteria, experiences a much wider range of c-di-GMP concentrations than previously appreciated. Moreover, cells grown in defined, carbon-poor media demonstrated lower growth rates and higher levels of c-di-GMP than those grown in complex, carbon-rich media. We are investigating if c-di-GMP and bacterial growth rate are linked or, alternatively, if induction of c-di-GMP is a stress response to starvation conditions.
The increased c-di-GMP level observed in AB and M9 could be generated by three distinct processes: (i) increased DGC activity, (ii) decreased PDE activity, or (iii) modulation of both DGC and PDE activity. TELCA analysis revealed that c-di-GMP in AB medium was driven by increased DGC activity, while the c-di-GMP increase in M9 was driven by reduced PDE activity. This result highlights the fact that bacteria can and do control intracellular c-di-GMP by regulating both c-di-GMP synthesis and hydrolysis. Moreover, one cannot determine the relative DGC and PDE activities based solely on measurements of the intracellular concentration of c-di-GMP.
We applied TELCA to further understand how QS controls intracellular c-di-GMP in V. cholerae. By analyzing mutants that lock the bacteria in the low- or high-cell-density state, we determined that DGC activity is increased at low cell density and decreased as the cells reach a quorum. Alternatively, we observed increased PDE activity of the ΔluxO high-cell-density mutant, compared to a locked low-cell-density mutant and the WT strain. However, we did not observe changes in PDE activity of the WT strain at different cell densities; thus, the ΔluxO mutant exhibits a more pronounced modulation of PDE activity than the WT strain. A discrepancy in phenotypic expression of the WT and ΔluxO mutant is not unprecedented. For example, the ΔluxO mutant is deficient for colonization of the infant mouse, whereas the WT is proficient (31). Therefore, these strains should not be considered equivalent. The analysis of the ΔluxO mutant does indicate that QS in V. cholerae has the potential to modulate cellular PDE activity. We speculate that under other conditions not examined here, in which the Qrr sRNAs are more fully repressed at a high cell density, the WT strain would exhibit density-dependent PDE activity. Our results clearly show that in LB, V. cholerae QS control of c-di-GMP occurs through modulation of c-di-GMP synthesis but not c-di-GMP degradation.
One protein that has been implicated in QS-mediated control of c-di-GMP is the DGC CdgA. It has been previously shown that CdgA is capable of synthesizing c-di-GMP in V. cholerae (40). The regulation of cdgA is controlled by QS, as cdgA is repressed at high cell density via direct binding of HapR to the promoter region (35–37). Furthermore, CdgA actively regulates biofilm formation, as a ΔcdgA mutant strain demonstrates decreased colony rugosity (38), and ectopic expression of cdgA leads to increased biofilm formation (40). Also of note, a ΔhapR ΔcdgA double mutant strain has reduced biofilm formation compared to the ΔhapR mutant, indicating that CdgA is involved in the HapR mediated increase in biofilm formation (62). Taken together, this evidence suggested that repression of cdgA by HapR is one mechanism by which QS controls the intracellular concentration of c-di-GMP and biofilm formation. To confirm these previous genetic studies and quantify the contribution of CdgA to total DGC activity, we created a deletion mutant of cdgA and measured the relative c-di-GMP synthesis and hydrolysis potential using TELCA. Our results are consistent with previous findings, as the ΔcdgA strain had reduced c-di-GMP synthesis compared to the ΔhapR and WT strains while having c-di-GMP synthesis similar to that of the ΔluxO mutant (Fig. 4). Importantly, this finding demonstrates that TELCA is able to distinguish the input of one DGC in total cellular DGC activity.
TELCA is a straightforward technique that can be utilized to quantify total DGC or PDE activity for any bacterium. TELCA can also be useful for a number of other purposes. For example, we have been unable to detect c-di-GMP in multiple bacterial species using LC-MS/MS, presumably due to the low concentration of this signal molecule (our unpublished results). In these cases, TELCA can function as an additional approach to determine the impact of DGCs and PDEs on c-di-GMP signaling pathways in these bacteria. TELCA will also be useful for identifying external factors or signals that impact total cellular DGC or PDE activity in lysates. TELCA allows a systems-level analysis of total c-di-GMP synthesis or degradation activity and can serve as a complementary approach to the current methods of c-di-GMP detection to further unravel complex c-di-GMP signaling networks.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the NIH grant U19AI090872 and membership and support from the Region V ‘Great Lakes' RCE (NIH award U54AI057153). We also acknowledge the Rudolph Hugh Fellowship and the Russell B. DuVall Scholarship to B.J.K.
We are grateful for assistance from the MSU mass spectrometry facility in quantifying intracellular c-di-GMP.
Footnotes
Published ahead of print 21 June 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01596-13.
REFERENCES
- 1.Huq A, Small EB, West PA, Huq MI, Rahman R, Colwell RR. 1983. Ecological relationships between Vibrio cholerae and planktonic crustacean copepods. Appl. Environ. Microbiol. 45:275–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tamplin ML, Gauzens AL, Huq A, Sack DA, Colwell RR. 1990. Attachment of Vibrio cholerae serogroup-O1 to zooplankton and phytoplankton of Bangladesh waters. Appl. Environ. Microbiol. 56:1977–1980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Halpern M, Broza YB, Mittler S, Arakawa E, Broza M. 2004. Chironomid egg masses as a natural reservoir of Vibrio cholerae non-O1 and non-O139 in freshwater habitats. Microb. Ecol. 47:341–349 [DOI] [PubMed] [Google Scholar]
- 4.Ng WL, Bassler BL. 2009. Bacterial quorum-sensing network architectures. Annu. Rev. Genet. 43:197–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cotter PA, Stibitz S. 2007. c-di-GMP-mediated regulation of virulence and biofilm formation. Curr. Opin. Microbiol. 10:17–23 [DOI] [PubMed] [Google Scholar]
- 6.Srivastava D, Waters CM. 2012. A tangled web: regulatory connections between quorum sensing and cyclic di-GMP. J. Bacteriol. 194:4485–4493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tischler AD, Camilli A. 2004. Cyclic diguanylate (c-di-GMP) regulates Vibrio cholerae biofilm formation. Mol. Microbiol. 53:857–869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Simm R, Morr M, Kader A, Nimtz M, Romling U. 2004. GGDEF and EAL domains inversely regulate cyclic di-GMP levels and transition from sessility to motility. Mol. Microbiol. 53:1123–1134 [DOI] [PubMed] [Google Scholar]
- 9.Boehm A, Kaiser M, Li H, Spangler C, Kasper CA, Ackermann M, Kaever V, Sourjik V, Roth V, Jenal U. 2010. Second messenger-mediated adjustment of bacterial swimming velocity. Cell 141:107–116 [DOI] [PubMed] [Google Scholar]
- 10.Kulasakara H, Lee V, Brencic A, Liberati N, Urbach J, Miyata S, Lee DG, Neely AN, Hyodo M, Hayakawa Y, Ausubel FM, Lory S. 2006. Analysis of Pseudomonas aeruginosa diguanylate cyclases and phosphodiesterases reveals a role for bis-(3′-5′)-cyclic-GMP in virulence. Proc. Natl. Acad. Sci. U. S. A. 103:2839–2844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hecht GB, Newton A. 1995. Identification of a novel response regulator required for the swarmer-to-stalked-cell transition in Caulobacter crescentus. J. Bacteriol. 177:6223–6229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aldridge P, Paul R, Goymer P, Rainey P, Jenal U. 2003. Role of the GGDEF regulator PleD in polar development of Caulobacter crescentus. Mol. Microbiol. 47:1695–1708 [DOI] [PubMed] [Google Scholar]
- 13.Ausmees N, Mayer R, Weinhouse H, Volman G, Amikam D, Benziman M, Lindberg M. 2001. Genetic data indicate that proteins containing the GGDEF domain possess diguanylate cyclase activity. FEMS Microbiol. Lett. 204:163–167 [DOI] [PubMed] [Google Scholar]
- 14.Ryjenkov DA, Tarutina M, Moskvin OV, Gomelsky M. 2005. Cyclic diguanylate is a ubiquitous signaling molecule in bacteria: Insights into biochemistry of the GGDEF protein domain. J. Bacteriol. 187:1792–1798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schmidt AJ, Ryjenkov DA, Gomelsky M. 2005. The ubiquitous protein domain EAL is a cyclic diguanylate-specific phosphodiesterase: enzymatically active and inactive EAL domains. J. Bacteriol. 187:4774–4781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ryan RP, Fouhy Y, Lucey JF, Crossman LC, Spiro S, He YW, Zhang LH, Heeb S, Camara M, Williams P, Dow JM. 2006. Cell-cell signaling in Xanthomonas campestris involves an HD-GYP domain protein that functions in cyclic di-GMP turnover. Proc. Natl. Acad. Sci. U. S. A. 103:6712–6717 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 17.Bernier SP, Ha DG, Khan W, Merritt JH, O'Toole GA. 2011. Modulation of Pseudomonas aeruginosa surface-associated group behaviors by individual amino acids through c-di-GMP signaling. Res. Microbiol. 162:680–688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carlson HK, Vance RE, Marletta MA. 2010. H-NOX regulation of c-di-GMP metabolism and biofilm formation in Legionella pneumophila. Mol. Microbiol. 77:930–942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kanazawa T, Ren S, Maekawa M, Hasegawa K, Arisaka F, Hyodo M, Hayakawa Y, Ohta H, Masuda S. 2010. Biochemical and physiological characterization of a BLUF protein-EAL protein complex involved in blue light-dependent degradation of cyclic diguanylate in the purple bacterium Rhodopseudomonas palustris. Biochemistry 49:10647–10655 [DOI] [PubMed] [Google Scholar]
- 20.Ryan RP, McCarthy Y, Andrade M, Farah CS, Armitage JP, Dow JM. 2010. Cell-cell signal-dependent dynamic interactions between HD-GYP and GGDEF domain proteins mediate virulence in Xanthomonas campestris. Proc. Natl. Acad. Sci. U. S. A. 107:5989–5994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Trimble MJ, McCarter LL. 2011. Bis-(3′-5′)-cyclic dimeric GMP-linked quorum sensing controls swarming in Vibrio parahaemolyticus. Proc. Natl. Acad. Sci. U. S. A. 108:18079–18084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tuckerman JR, Gonzalez G, Sousa EHS, Wan XH, Saito JA, Alam M, Gilles-Gonzalez MA. 2009. An oxygen-sensing diguanylate cyclase and phosphodiesterase couple for c-di-GMP control. Biochemistry 48:9764–9774 [DOI] [PubMed] [Google Scholar]
- 23.Wan XH, Tuckerman JR, Saito JA, Freitas TAK, Newhouse JS, Denery JR, Galperin MY, Gonzalez G, Gilles-Gonzalez MA, Alam M. 2009. Globins synthesize the second messenger bis-(3′-5′)-cyclic diguanosine monophosphate in bacteria. J. Mol. Biol. 388:262–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Karatan E, Duncan TR, Watnick PI. 2005. NspS, a predicted polyamine sensor, mediates activation of Vibrio cholerae biofilm formation by norspermidine. J. Bacteriol. 187:7434–7443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Galperin MY, Nikolskaya AN, Koonin EV. 2001. Novel domains of the prokaryotic two-component signal transduction systems. FEMS Microbiol. Lett. 203:11–21 [DOI] [PubMed] [Google Scholar]
- 26.Meibom KL, Blokesch M, Dolganov NA, Wu CY, Schoolnik GK. 2005. Chitin induces natural competence in Vibrio cholerae. Science 310:1824–1827 [DOI] [PubMed] [Google Scholar]
- 27.Atkinson S, Throup JP, Stewart G, Williams P. 1999. A hierarchical quorum-sensing system in Yersinia pseudotuberculosis is involved in the regulation of motility and clumping. Mol. Microbiol. 33:1267–1277 [DOI] [PubMed] [Google Scholar]
- 28.Hammer BK, Bassler BL. 2003. Quorum sensing controls biofilm formation in Vibrio cholerae. Mol. Microbiol. 50:101–114 [DOI] [PubMed] [Google Scholar]
- 29.Hammer BK, Bassler BL. 2009. Distinct sensory pathways in Vibrio cholerae El Tor and classical biotypes modulate cyclic dimeric GMP levels to control biofilm formation. J. Bacteriol. 191:169–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miller MB, Skorupski K, Lenz DH, Taylor RK, Bassler BL. 2002. Parallel quorum sensing systems converge to regulate virulence in Vibrio cholerae. Cell 110:303–314 [DOI] [PubMed] [Google Scholar]
- 31.Zhu J, Miller MB, Vance RE, Dziejman M, Bassler BL, Mekalanos JJ. 2002. Quorum-sensing regulators control virulence gene expression in Vibrio cholerae. Proc. Natl. Acad. Sci. U. S. A. 99:3129–3134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lenz DH, Mok KC, Lilley BN, Kulkarni RV, Wingreen NS, Bassler BL. 2004. The small RNA chaperone Hfq and multiple small RNAs control quorum sensing in Vibrio harveyi and Vibrio cholerae. Cell 118:69–82 [DOI] [PubMed] [Google Scholar]
- 33.Hammer BK, Bassler BL. 2007. Regulatory small RNAs circumvent the conventional quorum sensing pathway in pandemic Vibrio cholerae. Proc. Natl. Acad. Sci. U. S. A. 104:11145–11149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Freeman JA, Bassler BL. 1999. A genetic analysis of the function of LuxO, a two-component response regulator involved in quorum sensing in Vibrio harveyi. Mol. Microbiol. 31:665–677 [DOI] [PubMed] [Google Scholar]
- 35.Waters CM, Lu WY, Rabinowitz JD, Bassler BL. 2008. Quorum sensing controls biofilm formation in Vibrio cholerae through modulation of cyclic Di-GMP levels and repression of vpsT. J. Bacteriol. 190:2527–2536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yildiz FH, Liu XS, Heydorn A, Schoolnik GK. 2004. Molecular analysis of rugosity in a Vibrio cholerae O1 El Tor phase variant. Mol. Microbiol. 53:497–515 [DOI] [PubMed] [Google Scholar]
- 37.Zhu J, Mekalanos JJ. 2003. Quorum sensing-dependent biofilms enhance colonization in Vibrio cholerae. Dev. Cell 5:647–656 [DOI] [PubMed] [Google Scholar]
- 38.Lim B, Beyhan S, Meir J, Yildiz FH. 2006. Cyclic-diGMP signal transduction systems in Vibrio cholerae: modulation of rugosity and biofilm formation. Mol. Microbiol. 60:331–348 [DOI] [PubMed] [Google Scholar]
- 39.Thelin KH, Taylor RK. 1996. Toxin-coregulated pilus, but not mannose-sensitive hemagglutinin, is required for colonization by Vibrio cholerae O1 El Tor biotype and O139 strains. Infect. Immun. 64:2853–2856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Massie JP, Reynolds EL, Koestler BJ, Cong J-P, Agostoni M, Waters CM. 2012. Quantification of high-specificity cyclic diguanylate signaling. Proc. Natl. Acad. Sci. U. S. A. 109:12746–12751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Iwanaga M, Yamamoto K. 1985. New medium for the production of cholera toxin by Vibrio cholerae O1 biotype El Tor. J. Clin. Microbiol. 22:405–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Clark DJ, Maaløe O. 1967. DNA replication and the division cycle in Escherichia coli. J. Mol. Biol. 23:99–112 [Google Scholar]
- 43.Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual, 3rd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 44.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Marvig RL, Blokesch M. 2010. Natural transformation of Vibrio cholerae as a tool—optimizing the procedure. BMC Microbiol. 10:155. 10.1186/1471-2180-10-155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Long T, Tu KC, Wang YF, Mehta P, Ong NP, Bassler BL, Wingreen NS. 2009. Quantifying the integration of quorum-sensing signals with single-cell resolution. PLoS Biol. 7:640–649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sambanthamoorthy K, Sloup RE, Parashar V, Smith JM, Kim EE, Semmelhack MF, Neiditch MB, Waters CM. 2012. Identification of small molecules that antagonize diguanylate cyclase enzymes to inhibit biofilm formation. Antimicrob. Agents Chemother. 56:5202–5211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bennett BD, Kimball EH, Gao M, Osterhout R, Van Dien SJ, Rabinowitz JD. 2009. Absolute metabolite concentrations and implied enzyme active site occupancy in Escherichia coli. Nat. Chem. Biol. 5:593–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bobrov AG, Kirillina O, Ryjenkov DA, Waters CM, Price PA, Fetherston JD, Mack D, Goldman WE, Gomelsky M, Perry RD. 2011. Systematic analysis of cyclic di-GMP signalling enzymes and their role in biofilm formation and virulence in Yersinia pestis. Mol. Microbiol. 79:533–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bellows LE, Koestler BJ, Karaba SM, Waters CM, Lathem WW. 2012. Hfq-dependent, co-ordinate control of cyclic diguanylate synthesis and catabolism in the plague pathogen Yersinia pestis. Mol. Microbiol. 86:661–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Newell PD, Yoshioka S, Hvorecny KL, Monds RD, O'Toole GA. 2011. Systematic analysis of diguanylate cyclases that promote biofilm formation by Pseudomonas fluorescens Pf0-1. J. Bacteriol. 193:4685–4698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Christen M, Kulasekara HD, Christen B, Kulasekara BR, Hoffman LR, Miller SI. 2010. Asymmetrical distribution of the second messenger c-di-GMP upon bacterial cell division. Science 328:1295–1297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Christen B, Christen M, Paul R, Schmid F, Folcher M, Jenoe P, Meuwly M, Jenal U. 2006. Allosteric control of cyclic di-GMP signaling. J. Biol. Chem. 281:32015–32024 [DOI] [PubMed] [Google Scholar]
- 54.Romling U, Gomelsky M, Galperin MY. 2005. C-di-GMP: the dawning of a novel bacterial signaling system. Mol. Microbiol. 57:629–639 [DOI] [PubMed] [Google Scholar]
- 55.Beyhan S, Odell LS, Yildiz FH. 2008. Identification and characterization of cyclic diguanylate signaling systems controlling rugosity in Vibrio cholerae. J. Bacteriol. 190:7392–7405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fong JCN, Yildiz FH. 2008. Interplay between cyclic AMP-cyclic AMP receptor protein and cyclic di-GMP signaling in Vibrio cholerae biofilm formation. J. Bacteriol. 190:6646–6659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mueller RS, Beyhan S, Saini SG, Yildiz FH, Bartlett DH. 2009. Indole acts as an extracellular cue regulating gene expression in Vibrio cholerae. J. Bacteriol. 191:3504–3516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shikuma NJ, Fong JCN, Yildiz FH. 2012. Cellular levels and binding of c-di-GMP control subcellular localization and activity of the Vibrio cholerae transcriptional regulator VpsT. PLoS Pathog. 8:e1002719. 10.1371/journal.ppat.1002719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Seshasayee ASN, Fraser GM, Luscombe NM. 2010. Comparative genomics of cyclic-di-GMP signaling in bacteria: post-translational regulation and catalytic activity. Nucleic Acids Res. 38:5970–5981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Edmunds AC, Castiblanco LF, Sundin GW, Waters CM. 2013. Cyclic di-GMP modulates the disease progression of Erwinia amylovora. J. Bacteriol. 195:2155–2165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ryan RP, Fouhy Y, Lucey JF, Dow JM. 2006. Cyclic Di-GMP signaling in bacteria: recent advances and new puzzles. J. Bacteriol. 188:8327–8334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Beyhan S, Bilecen K, Salama SR, Casper-Lindley C, Yildiz FH. 2007. Regulation of rugosity and biofilm formation in Vibrio cholerae: comparison of VpsT and VpsR regulons and epistasis analysis of vpsT, vpsR, and hapR. J. Bacteriol. 189:388–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


