Abstract
A long-term experiment on the effect of chitin addition to soil on the suppression of soilborne pathogens was set up and monitored for 8 years in an experimental field, Vredepeel, The Netherlands. Chitinous matter obtained from shrimps was added to soil top layers on two different occasions, and the suppressiveness of soil toward Verticillium dahliae, as well as plant-pathogenic nematodes, was assessed, in addition to analyses of the abundances and community structures of members of the soil microbiota. The data revealed that chitin amendment had raised the suppressiveness of soil, in particular toward Verticillium dahliae, 9 months after the (second) treatment, extending to 2 years following treatment. Moreover, major effects of the added chitin on the soil microbial communities were detected. First, shifts in both the abundances and structures of the chitin-treated soil microbial communities, both of total soil bacteria and fungi, were found. In addition, the abundances and structures of soil actinobacteria and the Oxalobacteraceae were affected by chitin. At the functional gene level, the abundance of specific (family-18 glycoside hydrolase) chitinase genes carried by the soil bacteria also revealed upshifts as a result of the added chitin. The effects of chitin noted for the Oxalobacteraceae were specifically related to significant upshifts in the abundances of the species Duganella violaceinigra and Massilia plicata. These effects of chitin persisted over the time of the experiment.
INTRODUCTION
Interest in the control of plant pathogens using environmentally friendly approaches has increased greatly in the last decade. For soilborne diseases, a major issue is the suppressiveness of soil toward pathogens that may occur in the soil (1). In this context, it has long been suspected that suppressiveness can be enhanced by adding biopolymers such as chitin and derivatives. For instance, soil treatment with chitin and/or chitosan from shrimp waste has been shown to temporarily increase root growth (e.g., of tomato) and decrease the rate of infection of plant roots by nematodes (2–4). Although not definitely proven in all cases, the mechanisms behind this suppressiveness enhancement most often relate to a change in the structure and/or activity of the microbiota in soil, which thus confers suppression of plant pathogens (1, 5). Presumably, chitinolytic microorganisms, which are capable of hydrolyzing the chitinous hyphae of pathogenic fungi, increased their numbers and/or activities in response to the chitin added. Alternatively, secondary responders to the added chitin confer pathogen suppression.
Chitin is a biopolymer that is distributed among many soil organisms (i.e., it is a major constituent of the cell walls of fungi and the exoskeleton of invertebrates). In soil, chitin can be degraded at a substantial rate. It has been cogitated that due to the enormous abundance and diversity of bacteria in most soils and the presumed presence of chitinases in a considerable fraction of these, chitin degradation is mainly a bacterially driven process (6, 7). However, we still ignore how and to what extent different bacteria with different chitin degradation functions are responsible for the chitinolytic process in soil and whether fungi also cannot have a major role in this process. From previous work based on isolated microbial community members, it has been postulated that the addition of chitin to soil stimulates bacterial communities more than fungal ones (8, 9). Among the bacterial isolates obtained in several studies, members of the genus Streptomyces were dominant (10), followed by Stenotrophomonas and Bacillus (1, 11, 12). The presence of these bacterial chitinolytic groups was confirmed by analyses based on the 16S rRNA and chitinase (chiA) genes (11, 13). Experiments performed in plots of natural fields, in which chitin was placed in litter bags, have addressed and revealed the molecular diversity of the chitinases produced by Streptomyces (14). However, the intricacies of the bacterial responses in soil under chitin amendment, in particular with respect to which bacterial groups function at what point in time and what types of successions may take place, is still poorly understood. Moreover, our understanding of the relationship of chitin and chitin degradation status to crop rotation and soil characteristics is also limited (13, 15). As suggested from the above studies, members of the actinobacteria, which are ubiquitous in, and have been widely isolated from, agricultural soils, have been indicated as key degraders of complex organic molecules like chitin in the field (11, 16). However, in experiments performed in microcosms under laboratory conditions, particular members of the Gamma- and Betaproteobacteria were found to become dominant after the addition of chitin (17). Remarkably, studies of successions in bacterial communities during plant development as affected by compost amendment and seed colonization revealed a role for members of the family Oxalobacteraceae (3, 18). Following their activation by added chitin, such bacteria might also interact with fungi in the soil. Thus, recent studies of bacterium-fungus interactions in soil microcosms showed the role of two bacterial types, i.e., Oxalicibacterium and Streptomyces, in the transformation of calcium oxalate produced by fungi (19, 20).
Given the continued interest in the utilization of chitinous products for enhancing the suppressiveness of soil toward soilborne plant pathogens, the objective of this study was to examine the multiyear effect on the soil microbiota of the addition to soil of a particular chitin product prepared from shrimps on soil in the field. We placed a focus on soil bacteria, soil fungi, and specific soil bacterial groups, i.e., actinobacteria and oxalobacteraceae. The investigation of oxalobacteraceal communities was supported by previous findings in our laboratory of oxalobacteraceal dominance based on 454 pyrosequencing of chitinolytic bacteria. The chiA-gene-based analyses showed that particular oxalobacteraceae may be important colonizers of chitin-amended soils and rhizosphere, as well as other underexplored habitats (21). Additionally, recent reports (22, 23) of the genome sequences of the plant-growth-promoting Herbaspirillum sp. GW103 and the antifungal bacterium Janthinobacterium sp. HH01, both members of the family Oxalobacteraceae, support the hypothesis that members of this group can be involved in soil pathogen suppressiveness. We thus assessed both the suppressiveness and the abundance and diversity of the soil microbial communities over eight (suppressiveness) and three (microbial communities) years. Changes in the structures and diversities of the selected microbial communities were analyzed based on the bacterial 16S rRNA and chiA genes, as well as the 18S rRNA gene, reporting on total fungi.
MATERIALS AND METHODS
Experimental setup and sampling.
The site chosen for the field experiment and subsequent sampling was an agricultural field located at the experimental farm De Vredepeel in the southeast of the Netherlands (51°32′27.10″N and 5°51′14.86″E). The experimental field was in cultivation since 1955 and served for monitoring different agricultural practices. The Vredepeel field contains an agricultural sandy soil used for long-term experiments (http://www.wageningenur.nl/en/location/PPO-Vredepeel-1.htm). The chitin amendment experiment implied the setup of replicate (often triplicate) small (5- by 5-m) soil plots amended with chitin (obtained from shrimp waste) next to unamended control plots. The chitin amendment was initiated in March 2007 in the field monitored since 2005, and the treatment was followed by a second treatment in October 2009. Specifically, the soil was supplemented with 1.8% of shrimp waste chitin (20 tons/ha) calculated over the topsoil (20 cm). Prior to the amendment, the shrimp waste was disinfected overnight by treatment with NaOH and HCl according to a previously described protocol (24, 25). Crops were rotated over the years, as follows: wheat prior to chitin amendment (2006) and potato after the chitin treatments (2007 and 2010). The intercrops were lily (2008), wheat (2009), carrot (2011), and maize (2012).
Soil samples for soil chemistry and suppressiveness testing were taken regularly over the time of the experiment. Those for molecular analyses of the soil microbiota were collected five times over 3 years. The latter sampling times were December 2009 (here referred to as Dec-09), June 2010 (here referred to as June-10), November 2010 (here referred to as Nov-10), March 2011 (here referred to as March-11) and April 2012 (here referred to as April-12). Specifically, in all cases, triplicate 4-kg soil samples were obtained as composites of 8 to 10 individual samples taken from the 10-cm topsoil, both from the amended and nonamended plots. For all sampled, before the analysis, the soil was homogenized by passing it through a 2-mm mesh sieve.
Soil chemical analyses and suppressiveness testing.
Subsamples of 1 kg soil, taken from the composited primary samples, were used to characterize several chemical parameters. Thus, fractions of clay (<2 mm), silt (2 to 50 mm), and sand (50 to 200 mm) were determined. Further, soil samples were dried, ground, and analyzed. Parameters measured included pH, organic matter (OM) (as a percentage), nitrate (NO3− mg/kg soil), and ammonium (NH3 mg/kg soil). The pH was determined as pH-KCl. The OM percentage was calculated as 50% of the measured C content in the soil. C and N contents were determined using a CHN1110 element analyzer (Carlo Erba Instruments, Milan, Italy).
Soil suppressiveness was tested in triplicate samples per treatment by measuring the efficiency of antagonistic activity against the fungal plant pathogen Verticillium dahliae and against root lesion nematodes of the genus Pratylenchus. These assays have been previously described (26). Briefly, the effect on V. dahliae was estimated based on the average number of microsclerotia per 10 g dry soil. The effect against nematodes was reported as the difference in the average populations of pratylenchidae per 100 ml soil.
Soil DNA extraction.
DNA was isolated from 250 mg of soil, taken from the composited primary samples, using the PowerSoil DNA isolation kit according to the manufacturer's specifications (MoBio Laboratories, Carlsbad, CA, USA). The cells were disrupted by bead beating (bead beater; BioSpec Products) three times for 60 s. Following the extraction, the quality and quantity of DNA extraction were assessed by agarose gel electrophoresis and spectrophotometry (Nanodrop; ThermoFisher Scientific, St. Leon-Rot, Germany) measurements. All subsequent analyses were performed on triplicates.
qPCR.
Absolute quantification by real-time PCR (qPCR) was carried out on the ABI Prism 7300 instrument (Applied Biosystems, Life Technologies Europe BV, Bleiswijk, The Netherlands) using Maxima SYBR green mix (Fermentas, ThermoFisher Scientific, St. Leon-Rot, Germany).
Three technical replicates of DNA were used from each of the three biological replicates.
The genes targeted were general bacterial, actinobacterial, and Oxalobacteraceae 16S rRNA genes, bacterial family 18 chitinase, chiA, and the fungal internal transcribed spacer (ITS). Primers and amplification conditions, previously described (18, 27–30), are detailed in Table 1. Standard curves were obtained using serial dilution of plasmid DNA containing the cloned specific gene (Table 1). Tenfold dilutions of the standard concentrations were estimated to 101 to 108 gene targets per reaction. qPCR analysis was carried out according to the known and well-accepted recommendations (31) as to DNA extraction, technical replicates of biological replicates reagents, instruments, and target gene.
Table 1.
Primers, conditions, and bacterial strains used to generate the standard for qPCR and genes amplified and conditions used for PCR-DGGE
| Gene | qPCR |
PCR-DGGE |
|||||
|---|---|---|---|---|---|---|---|
| Primer | Conditions | Control strain used to generate standard | Reference(s) | PCR annealing temp (°C) | % UF denaturant | Reference(s) | |
| Bacterial 16S rRNA gene | Eub338/Eub518 | 60°C; 40 cycles | Burkholderia terrae BS001 | 27 | Touchdown, 60 to 55 | 40–65 | 32 |
| Oxalobacteraceae, 16S rRNA gene | Ox225Fw/Ox656Rev | 65°C; 40 cycles | Janthinobacterium lividum | 18, 28 | 65 | 20–60 | 18 |
| Actinobacteria, 16S rRNA gene | Actino235/Eub518 | 62°C; 40 cycles | Streptomyces griseus | 27 | 63 | 40–60 | 33 |
| Bacterial chitinase, chiA | GA1F/GA1R | 57°C; 40 cycles | Streptomyces griseus | 29, 30 | 57 | 40–50 | 21 |
| Fungal ITS | 5.8S/ITS1f | 53°C; 40 cycles | Rhizoctonia solani | 27 | 57 | 20–55 | 34, 35, 61 |
PCR-DGGE analysis.
The genetic regions selected for quantitative amplification were also the subject of PCR-denaturing gradient gel electrophoresis (PCR-DGGE) analysis. PCR amplifications were set up for all samples in parallel using the biological triplicate soil DNA (1 to 5 ng/μl).
Bacterial 16S rRNA genes were amplified using the primers F968-GC and R4101.1b (32). Specific 16S rRNA genes of the Oxalobacteraceae were amplified using a PCR system based on the primers OX225F and OX1249R, as previously described (18). Actinobacterial 16S rRNA gene amplicons were obtained using the primers F243 and R513GC (33). The fungal ITS region was first amplified using the primers EF4 and ITS3, followed by a second round of amplification with the primers ITS1f-GC and ITS2 (34, 35). Similarly, the bacterial chiA gene was amplified first using the primers GA1F/GA1R, followed by reamplification with the forward primer equipped with a GC clamp (29). All PCR conditions are described in Table 1. All amplifications were carried out on a Mastercycler-nexus thermocycler (Eppendorf, Nijmegen, The Netherlands).
Approximately 200 ng of PCR product was loaded on a 6% (wt/vol) polyacrylamide gel in the Ingeny Phor-U system (Ingeny International, Goes, The Netherlands). The gels were prepared using an optimized denaturant gradient for each type of gene (Table 1) (18, 21, 32–35). DGGE profiles were obtained using 16 h of electrophoresis at an optimized voltage in 0.5× Tris-acetate-EDTA (TAE) buffer at 60°C. The gels were stained for 60 min in 0.5 μg/liter SYBR gold (Invitrogen, Breda, The Netherlands) and visualized on a UV transilluminator.
Cloning and sequencing of 16S rRNA gene amplicons for the Oxalobacteraceae from selected samples.
Three clone libraries consisting of Oxalobacteraceae 16S rRNA gene fragments were generated using the specific primer set to compare the communities in soils from December 2009, June 2010, and November 2010. The three sampling pointe were selected based on the observed changes in relative abundances of this community. The libraries included clones from the three replicates of unamended and chitin-amended soil. Briefly, after gel purification with Wizard SV gel and the PCR CleanUp system (Promega, Madison, Wi, USA), the amplicons were ligated into pGEM-T Easy vector (pGEM 242-T vector system II; Promega, Madison, WI, USA). Further, the plasmids were introduced into Escherichia coli JM109 cells by transformation according to the manufacturer's recommendations. In total, 288 (48 for each time point, chitin amended, and control) white colonies were picked, and the insert was sequenced using the primer system T7-Sp6 (LGC Genomics, Berlin, Germany).
Analysis of diversity of the Oxalobacteraceae.
A total of 288 sequences were obtained. Sequences were grouped based on their origin (sampling point and treatment) and considered per sample/replicates. Considering their closeness as to relative abundances, we then also considered these as pooled groups over the three replicates per treatment. The sequence qualities were checked manually, and possible chimera formation was assessed using the software program Bellerophon v.3 (36) (http://greengenes.lbl.gov). Eight sequences were removed after the quality checks, and no chimeras were found. The sequences were assigned to operational taxonomic units (OTUs), based on an 80% similarity cutoff, using the program Mothur (37). Sequence alignments were carried out using the Kimura two-parameter algorithm with bootstrap tests of inferred phylogeny with 1,000 replications. The 97% similarity criterion was used to assign an OTU at the “species” level, and 95% was used at the “genus” level. The closest phylogenetic relatedness was determined by BLAST-N versus the nonredundant NCBI database.
Analysis of DGGE fingerprints.
DGGE profiles generated with different primer sets (see above) of all three replicates were analyzed using the GelCompar software program (Applied Maths, Sint-Martens, Latem, Belgium). The diversity of the thus-visualized dominant microbial communities was thus assessed based on the PCR-DGGE profiles of all replicates tested. The patterns were clustered using the unweighted pair group method with arithmetic averages (UPGMA). Similarity matrices were generated using Jaccard correlation coefficient. The Shannon index of bacterial diversity was calculated as H = −ΣPi(log Pi) based on the relative band intensities (Pi) as previously formulated (38). Bacterial (total, Actinobacteria, and Oxalobacteraceae) and fungal abundances were analyzed by one-way analysis of variance (ANOVA) (SPSS Statistic 20; IBM Corp., Armok, NY, USA) to determine the significance of the differences between the sampling points. In addition, the data derived from Jaccard correlation (band-based analysis) were used in principal coordinate analysis (PCoA) performed in the software program Canoco (Canoco 4.55; Plant Research Institute, Wageningen, The Netherlands).
Nucleotide sequence accession numbers.
All sequences generated in this study were deposited in GenBank (http://www.ncbi.nlm.nih.gov/GenBank/index.html) under the numbers KF188803 to KF189070 .
RESULTS
Soil characteristics and suppressiveness toward soilborne plant pathogens.
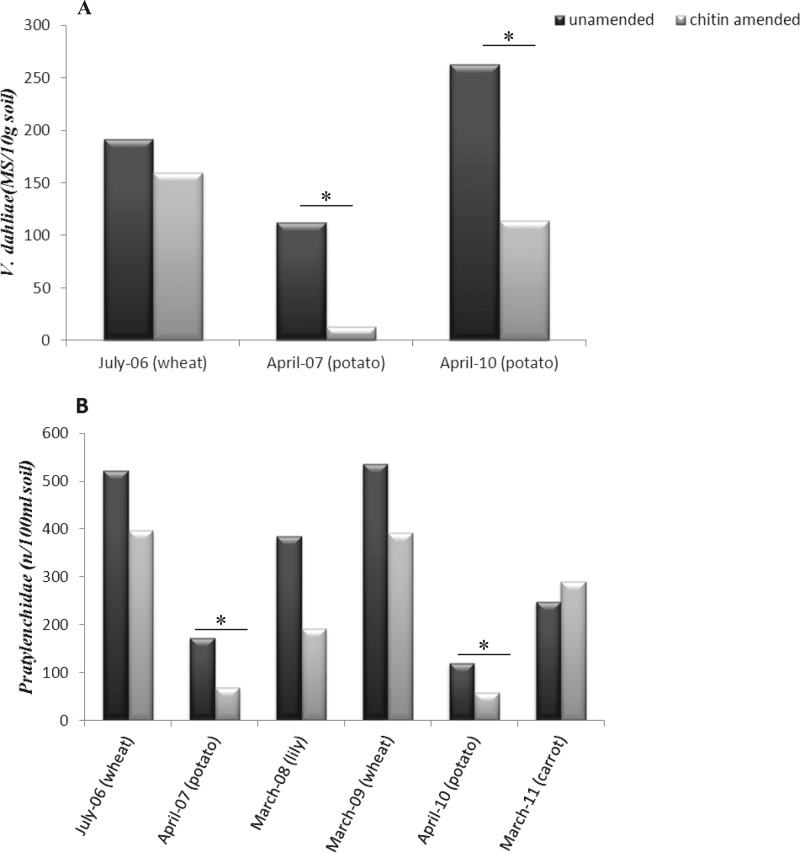
The unamended soil was characterized as a sandy soil with pH 5.7 ± 0.2 and with 3.3% ± 0.1% OM. Chitin addition clearly raised the OM level, to about 3.5% ± 0.1%. Furthermore, it raised the suppressiveness of the soil toward the soilborne pathogens Verticillium dahliae and Pratylenchus in connection to wheat, potato, and lily grown on the field. First, the average number of V. dahliae microsclerotia per 10 g dry soil decreased in all cases in the chitin-amended soil compared to that in the unamended soil. The decrease was significant (P < 0.05) in two of three cases (April-07 and April-10; potato) (Fig. 1A). A similar suppressive effect of the chitin amendment was observed against Pratylenchus nematodes, with average population densities of pratylenchidae decreasing in the chitin-amended soil compared to those in unamended soil (Fig. 1B). Although there was a trend in five out of six samples, the difference was significant in only one (P < 0.05). These observations suggested that the chitin incorporated into the top soil layers is a main driver of suppressiveness against two important potato pathogens, V. dahliae and potato-pathogenic nematodes, irrespective of season.
Fig 1.
Effects of chitin amendment on the soilborne pathogen Verticillium dahliae (A) or Pratylenchidae (nematodes) (B) over sampling points and crops. Averages of triplicate measurements with standard error bars are given; significant values (P < 0.05) are indicated with “∗” (see also reference 26). MS, number of microsclerotia; n, number of individuals.
Dynamics of the abundance of bacterial communities as assessed by 16S rRNA gene-based qPCR.
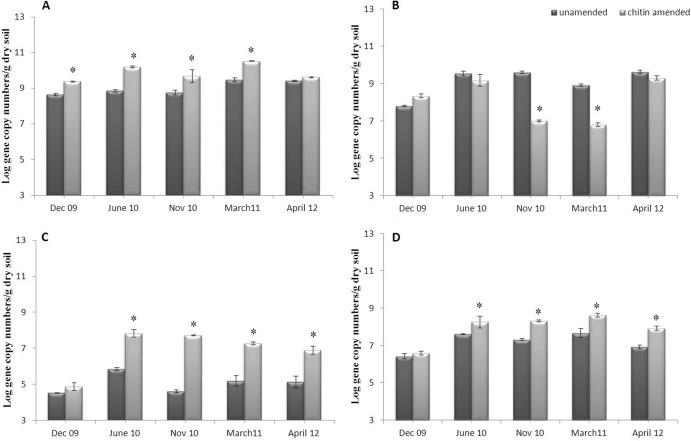
The bacterial abundances in both the chitin-amended and unamended soils, assessed along the five sampling times (Dec-09 through April-12), were consistently in the range of 108 to 1010 gene copies/g dry soil. In the unamended soil (Fig. 2A), these abundances were statistically similar, at about 109/g dry soil, among the samplings (P > 0.05). In the chitin-amended soil, we observed a rapid increase of the 16S rRNA gene copy abundances, i.e., from about 1 × 109 to 1 × 1010 (gene copies/g dry soil), from Dec-09 to June-10 (P = 1.5 × 10−4) (Fig. 2A). After this, the values remained raised, with insignificant variations from the June-10 values, until April 2012. The maximum abundance, i.e., 1.4 × 1010 gene copies/g dry soil, was observed in the March-11 samples. Overall, the abundances of the bacterial communities were significantly higher (P < 0.05) in the chitin-amended soil samples than in the unamended samples, except the April-12 samples. Finally, the bacterial abundance found in the chitin-amended April-12 soil sample was similar to that in the unamended soil (4.5 ± 0.7 × 108 gene copies/g), and this value was not significantly different from that in the Dec-09 unamended soil sample.
Fig 2.
Relative abundance of total bacteria (A), total fungi (B), Oxalobacteraceae (C), or Actinobacteria (D) in unamended and chitin-amended soil. Error bars represents standard errors of the means (geometric) for three replicates, and sampling points indicated with “∗” were significantly different (comparison of chitin-amended with unamended soils, P < 0.05).
Structures of bacterial communities as assessed by 16S rRNA gene based PCR-DGGE.
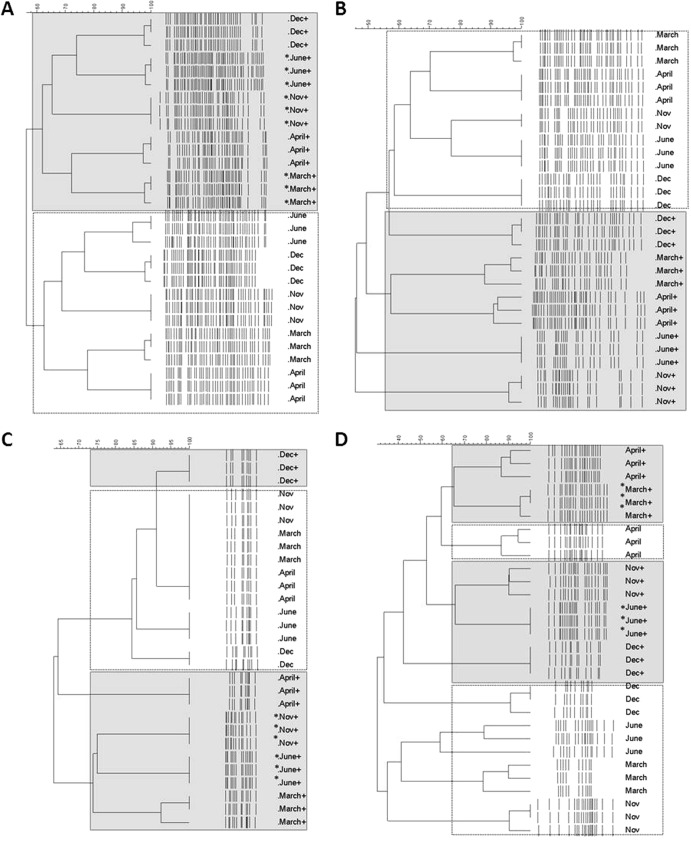
To compare the structures of the bacterial communities in the chitin-amended and unamended soils throughout time, we performed PCR-DGGE analysis based on the 16S rRNA gene. In both treatments, the profiles generated from all three replicate samples for each time point were internally similar, confirming the close similarity of the community structures across replicates. Importantly, clear changes in the community structures as a result of the added chitin were noted throughout. Thus, cluster analysis showed a clear separation of the bacterial community structures in all chitin-treated soil samples from the ones in the unamended soils, since two large clusters that came together at 58% similarity were found (Fig. 3A). The average band numbers in the first cluster, encompassing the profiles of all unamended soil samples, were 45 ± 3. In this cluster, the profiles were divided over two subclusters at 66% similarity, one consisting of the Dec-09, June-10, and Nov-10 samples (72% similarity) and the other of the March-11 and April-12 samples (74% similarity). The profiles of the second (chitin-amended) soil samples, grouping together in the second major cluster at 63% similarity, were divided among three major subclusters. Considering these, the Dec-09 and June-10 profiles (74% similarity) were separate from the Nov-10 ones (64% similarity) and from the March-11 and April-12 ones (72% similarity). On average, the number of bands was 42 ± 5, with raised numbers in June-10 (see Fig. S1A and S2A in the supplemental material). In the latter case, extra bands were observed in the high-GC regions of the profiles compared to findings for the rest of the chitin treatment samples.
Fig 3.
Clustering of DGGE profiles based on UPGMA and Jaccard correlation coefficient. Shown are total bacteria (A), total fungi (B), Oxalobacteraceae (C), or Actinobacteria communities (D) in unamended and amended soils. Sampling time points of unamended soils, Dec-09, June-10, Nov-10, March-11, and April-12, are referred to as Dec, June, Nov, March, and April. Sampling time points of chitin-amended soils, Dec-09, June-10, Nov-10, March-11, and April-12, are referred as Dec+, June+, Nov+, March+, and April+. Sampling points indicated with “∗” were significantly different (P < 0.05).
Dynamics of abundances of fungal communities as assessed by internally transcribed spacer (ITS) targeted qPCR.
As expected, the fungal abundances fluctuated (yet did not significantly change) in the unamended soils, revealing values of 107 to 109 gene copies/g dry soil, over experimental time (Fig. 2B). In the chitin-amended soils, the abundances increased slightly, from 108 (Dec-09) to 109 (June-10) gene copies/g dry soil, decreasing significantly (P < 0.05) thereafter, to about 107 and 106 (Nov-10 and March-11, respectively). Significant changes in fungal densities (P < 0.05) were also observed from March-11 to April-12, when the values measured increased from 106 to 109 gene copies/g soil (Fig. 2B).
On average, the abundance of fungi in the chitin-amended soil was 10- to 100-fold lower than that of bacteria. This effect was maintained up to 1.5 years after the amendment.
Structures of fungal communities as assessed by fungus-specific PCR-DGGE.
Analysis of the fungal PCR-DGGE profiles based on the fungal 18S rRNA-ITS gene region showed the occurrence of rather diverse communities in all samples. Small variation among replicates (90% similarity) was observed. At 40% similarity, the fungal community profiles were divided among two major groups. One group encompassed the chitin-amended soil samples of June-10 and Nov-10 (42% similarity), and the second one consisted of all unamended soil samples plus the chitin-amended ones of Dec-10, March-11, and April-12 (44% similarity) (Fig. 3B). Although there was no significant variation in species richness estimated based on band similarity matrices, visual inspection of DGGE profiles showed a decreasing number of bands (from 32 in Dec-09 to 22 in June-10) in the amended soil (see Fig. S1B and S2B in the supplemental material). This suggested a reduction in diversity, which was maintained to up to 2.5 years after the amendment.
Abundances of Actinobacteria and Oxalobacteraceae.
Given the presumed importance of members of the Actinobacteria and Oxalobacteraceae in chitin degradation in soil, the abundances of the relevant specific 16S rRNA genes were measured by qPCR. In the unamended soil, the abundances of both communities fluctuated but showed insignificant changes over the time points, from 2009 to 2012 (Fig. 2C and D). The actinobacterial abundances tended to vary slightly, but not significantly (P > 0.05), from about 106 (Dec-09 and April-12) to about 107 (June-10, Nov-10, and March-11) gene copies/g dry soil. The abundances of Oxalobacteraceae in the unamended soil ranged between about 104 (Dec-09 and Nov-10) and 105 (June-10, March-11, and April-12) gene copies/g dry soil.
In the chitin-amended soil, the abundance of Actinobacteria varied from 106 (Dec-09) to 108 (June-10, Nov-10, and March-11) gene copies/g dry soil (Fig. 2D). In this treatment, the soil Oxalobacteraceae numbers ranged between 104 (Dec-09) and 107 (June-10, Nov-10, and March-11) gene copies/g (Fig. 2C). The abundances of both the Actinobacteria and Oxalobacteraceae were thus positively correlated with the amendment of soil with chitin. Immediately after the second treatment, i.e., in December 2009, we observed a minor increase in the abundances, whereas 8 months after the treatment, in June 2010, both Actinobacteria and Oxalobacteraceae showed significant increases in their abundance in the chitin-amended soil compared to that in the unamended soil (P < 0.05), amounting to up to 2 log units.
Community structures of Actinobacteria and Oxalobacteraceae.
The PCR-DGGE profiles of the Oxalobacteraceae communities revealed simple community structures, with band numbers ranging from 9 (chitin-amended soil, Dec-09) to 15 (amended soil, June-10). Again, all replicates clustered closely together, confirming their close similarity. Cluster analysis of these PCR-DGGE profiles indicated a clear separation of all chitin-treated soil communities from the unamended soil ones (36% similarity), the exception being the Dec-09 unamended soil sample (Fig. 3C). The effect of chitin amendment was most apparent in the June-10, Nov-10, and March-11 samples.
The actinobacterial PCR-DGGE profiles were less uniform. In general, the profiles were rather simple, with numbers of bands varying from 10 (Dec-9, unamended) to 21 (chitin-treated soil, Jun-10 and March-11). Cluster analysis of these profiles suggested that the effect of chitin on the communities of Actinobacteria was gradual. The Dec-09 chitin-amended soil sample separated at 31% similarity from the unamended Dec-09 sample (Fig. 3D). Thereafter, clear shifts in the actinobacterial community structures due to the chitin treatment were observed in the June-10, Nov-10, and March-11 samples. Principal coordinate analysis based on similarities across sample times indicated a clear effect of chitin addition on these communities and persistence of this effect over time (see Fig. S1C, S1D, S2C, and S2D).
Analysis of community composition of the Oxalobacteraceae.
Since the PCR-DGGE analysis had revealed clear effects of chitin on the oxalobacteraceal communities over the first year after the amendment, the chitin-amended and unamended samples from Dec-09, June-10, and Nov-10 were selected for the construction and analysis of oxalobacteraceal 16S rRNA gene clone libraries.
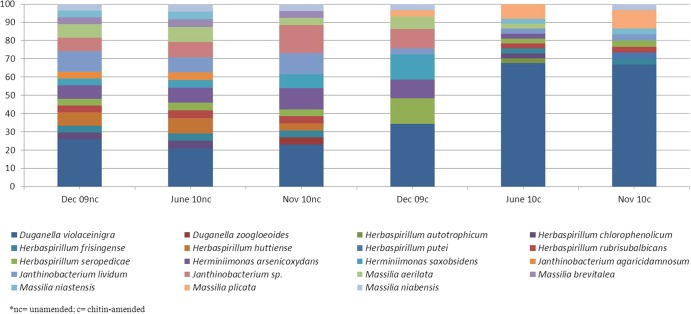
After quality and chimera checks, a total of 280 sequences was obtained (on average, 47 sequences split over triplicate samples/treatment). All sequences were affiliated with Oxalobacteraceae 16S rRNA gene sequences found in the NCBI database. Furthermore, the sequences were, for the greatest part, >97% similar to those of described species. Overall, they covered the following five genera: Duganella, Herbaspirillum, Herminiiomonas, Janthinobacterium, and Massilia. Within these genera, 19 species could be distinguished. A comparison of (relative abundances of) the different oxalobacteraceae in chitin-treated versus untreated samples using nonparametric Chao I analysis (analyzing richness and evenness) showed a very clear effect of chitin amendment (Tables 2 and 3). Using either the genus or species level, we analyzed the relative abundances of the different sequence types in each replicate per treatment and time. First, sequences affiliated with Duganella violaceinigra consistently dominated the Oxalobacteraceae communities in both the chitin-treated and untreated soils, with very little variation across the replicates per treatment. In the unamended soils, the relative D. violaceinigra abundances fluctuated around 29%, with differences remaining insignificant (P > 0.05). However, significant (P < 0.05) increases in these relative abundances were found in the presence of chitin, from about 29% (Dec-09) to up to 64% (June-10 and Nov-10). A second effect of chitin was noted for sequences affiliated with the species Massilia plicata, which showed a consistent increase in relative abundance from below detection in the unamended soil throughout (less than about 2.2% of the total) to values ranging from 6 to 12% (average, 11%) in the chitin-treated samples. Finally, there were no significant increases in the relative abundances of sequences affiliated with those of Herbaspirillum seropedicae, Herminiiomonas saxobsidens, and Herminiiomonas arsenicoxydans. Also, sequences related to the genera Janthinobacterium and Massilia (other than M. plicata) were found throughout, again without significant trends related to treatment or time.
Table 2.
Analysis of 16S rRNA gene-based Oxalobacteraceae clone libraries: no. of sequences, levels of similarity, and diversity indices
| Sample | No. of sequences | No. of sequence types | Diversity index value |
||
|---|---|---|---|---|---|
| Chao I | ACEa | Shannon | |||
| Dec-09 | 48 | 14 | 113 | 117 | 2.41 ± 0.02 |
| June-10 | 48 | 14 | 118 | 158 | 3.65 ± 0.04 |
| Nov-10 | 48 | 14 | 115 | 153 | 2.84 ± 0.04 |
| Dec-09+ | 48 | 9 | 87 | 218 | 2.27 ± 0.03 |
| June-10+ | 48 | 10 | 107 | 220 | 2.47 ± 0.01 |
| Nov-10+ | 48 | 14 | 77 | 215 | 2.31 ± 0.01 |
ACE, abundance-based coverage estimation.
Table 3.
Analysis of 16S rRNA gene-based Oxalobacteraceae clone libraries: comparison of chitin-amended soil samples with unamended samples
| Sample | Significance of comparison with samplea |
|||||
|---|---|---|---|---|---|---|
| Dec-09 | June-10 | Nov-10 | Dec-09+ | June-10+ | Nov-10+ | |
| Dec-09 | — | |||||
| June-10 | NS | — | ||||
| Nov-10 | NS | NS | — | |||
| Dec-09+ | NS | NS | NS | — | ||
| June-10+ | ∗ | ∗ | ∗ | ∗ | — | |
| Nov-10+ | ∗ | ∗ | ∗ | ∗ | NS | — |
ANOVA: NS, difference not significant; ∗, difference significant (P < 0.05). “+” indicates a chitin-amended sample.
Overall, the sequence analyses thus enabled visualization of clear positive responses of specific members of the Oxalobacteraceae to the chitin amendment, in particular revealing a progressively increasing effect over time on Duganella violaceinigra and Massilia plicata (Fig. 4).
Fig 4.
Comparison of 16S rRNA gene sequences of the oxalobacteraceal community. The stacked column graph shows relative distribution of different bacterial species based on BLASTN analysis. Average relative abundance from three replicates as the ratio between sequence type abundance and total number of sequences in the group is shown.
Abundances and structures of bacterial chitinolytic communities as assessed on the basis of the chiA gene.
The chiA gene, here used as a proxy for chitinolytic bacteria, has been reported to be the most frequently occurring bacterial gene involved in chitin degradation in soil (29). The abundance of the chiA gene, measured by qPCR, revealed very little variation over time in the unamended soil. In this soil, it consistently fluctuated around 106 gene copies/g soil, irrespective of time. Chitin amendment of the soil clearly exerted a positive effect on this abundance, and the effect was significant (P < 0.05). Specifically, the chiA gene copy number/g dry soil increased, as a corollary of the chitin treatment, from 2 × 106 in June 2010 to 3.5 × 108 in March 2011, and the numbers remained on the order of 108 up to April 2012 (Fig. 5).
Fig 5.
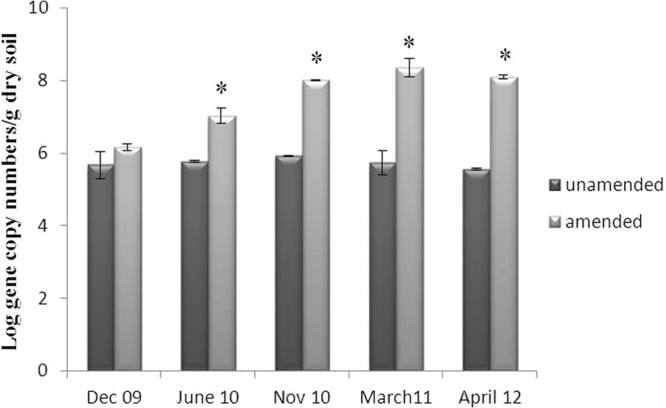
Relative abundances of bacterial chitinolytic communities as assessed on the basis of the chiA gene. Sampling points indicated with “∗” were significantly different between chitin-amended and unamended soils (P < 0.05).
The chiA-based PCR-DGGE patterns showed relatively low numbers of bands, varying on average between 8 ± 2 in the unamended soil to 11 ± 2 in the chitin-amended soil. Within the chitin treatment, the highest numbers of bands were observed in the June-10 and Nov-10 samples, suggesting an activation of diverse chitinolytic community members as a result of the addition of chitin. The profiles of the communities in the unamended soil samples clustered away from those of the chitin-amended ones, at 64% similarity. In both the unamended and amended soils, the profiles formed subgroups defined by the Dec-09/June-10 and March-11/April-12 samples, with an intermediate profile of Nov-10. Overall, chitin treatment was the dominant factor, and time appeared to constitute a secondary driver of the chitinolytic bacterial communities (see Fig. S2E and S3 in the supplemental material). This was consistent with the data obtained with the unamended soil, where the similarity of the samples (80%, except Nov-10) indicated the presence of a stable core group of chitinolytic bacteria.
DISCUSSION
There is still unexplored potential to raise the suppressiveness of soils toward plant pathogens by adding biopolymers such as chitin (4, 39, 40). Previous work already indicated that amendments of soil with chitin modify the soil's chemistry and structure and, importantly, the structures of the microbial communities that occur in association with plants (2, 3). Moreover, the chitin added to soil is subjected to progressive degradation as a result of the activity of a chitinolytic microbial community in the soil. Given the prevalence of chitin-degradative genes across many bacteria, such degradation may be mainly bacterially driven (8, 29, 40). In addition, particular chitinolytic bacteria that are activated might act as suppressive agents of plant pathogens that contain chitinous structures (fungi and nematodes) following their activation. These might even be developed into biological control agents against such fungi, e.g., V. dahliae, or nematodes (11, 40–43, 62). However, in spite of several previous reports on bacterial chitinases in soils (11, 14, 44, 45), the responses of the plethora of soil microorganisms to chitin addition are still largely not understood. This is particularly true for agricultural fields (26, 46), in which we do not quite understand how the effect of chitin may be related to crop rotation, chemical characteristics of the soil, and time.
In this study, we therefore addressed the possible emergence of pathogen (Verticillium dahliae and nematodes of the genus Pratylenchus) suppressiveness in field soil in relation to chitin amendment and the concomitant changes in the microbial communities of the soil. Next to assessing the abundance and diversity of total bacteria and fungi, we placed particular emphasis on soil actinobacteria and oxalobacteraceae in the light of their presumed role in successions related to chitinolysis in soil and their potential to serve as biocontrol agents (3, 17, 47, 48; M. S. Cretoiu, unpublished). We deliberately chose to work with field soil to cover all the variables that affect suppression and microbial communities in a “real-world” situation. Our microbial community measurements were taken upon the observation of enhanced suppression as a result of added chitin in an attempt to link this suppression to changed community structure.
Suppressiveness of soil toward V. dahliae and plant root lesion nematodes of the genus Pratylenchus.
Verticillium wilt, caused mainly by V. dahliae, is among the most important soilborne plant diseases. Traditionally, control of the disease has been based on reducing the population of microsclerotia in the soil, through the use of (volatile) chemicals (fumigation). Recently, the use of organic amendments like green manure and chitin was shown to reduce the population size of V. dahliae in the greenhouse and the field (39, 40, 63). In the current study, a clear and consistent effect of chitin addition on V. dahliae microsclerotia was observed shortly after amendment, and this effect remained present over 1.5 years. Moreover, a positive correlation was observed with the intercrops, meaning that crop rotation in the chitin-amended soil enhanced the suppressiveness against V. dahliae. Effects on plant infection are further studied in the work of G. Korthals (unpublished).
Next to focusing on V. dahliae, we also measured the effects of chitin on the levels of (plant-pathogenic) pratylenchidae. Although the effect was not as large as that measured for V. dahliae, chitin addition did reduce the density of these nematodes for several years. Thus, these observations confirmed the multiyear suppressiveness induced by chitin amendment. This also indicated that in the Vredepeel soil, which constitutes an important Western European agricultural soil, shrimp-derived chitin amendment may offer a robust agricultural system that enables the warding off of key plant (potato) pathogens.
Effects of chitin on soil microbial communities.
The population densities of bacteria and fungi estimated by qPCR were found to be within the range reported for other soil systems (49, 50). While the analysis of the unamended soil samples indicated that the community sizes did not change significantly over the seasons, the addition of chitin raised both microbial densities. Strikingly, the average bacterial densities under the chitin amendment were orders of magnitude higher than the fungal ones, and this effect persisted over time (up to 2 years). Critical changes in the abundances were observed in the time interval June to November 2010. In the June-10 samples, the bacterial abundances in the chitin-amended soil were 10-fold higher than those in the unamended soil, while the fungal abundances decreased 10 times. These observations are in accordance with reports from microcosm experiments in which soil was incubated for up to 60 days with chitin (15, 17). Moreover, the soil bacterial community structures were also strongly affected by the chitin amendments, as evidenced by PCR-DGGE, in which the amended soil profiles clustered away from the unamended ones. This suggests that chitin added to soil induces quite persistent changes in the local bacterial communities, probably by creating local nutritive and other conditions that select particular microbial types.
The clear effect of chitin was also observed at the level of the chiA gene, here used as a proxy for the bacterial chitinolytic communities. The abundance of the chiA gene increased over time, indicating the positive selection of the respective chiA hosts. It is possible that chitin derivatives of lowered complexity become available after first rounds of degradation and that, hence, secondary responders to such compounds get stimulated. Other reports of chitinolysis taking place in the field have shown that such processes may accelerate after successive additions of chitin (E. M. H. Wellington, unpublished data). In our experiment, the relatively stable level of chiA-carrying cells in the chitin-treated soils at later sampling points may indicate the then-stable presence of a chitinolytic community, which is useful considering the longevity of the suppressiveness of the soil.
Effects of added chitin on soil actinobacteria and oxalobacteraceae.
Members of the actinobacteria have been indicated as major chitin degraders in soil and soil-like environments (14). Studies of the rhizosphere have shown that the Streptomyces group is often more abundant in rhizophere than in nonrhizosphere soils (51, 52). There are leads pointing at an important role of such actinomycetal chitinolysis in soil treated with chitin (6, 8), while other reports indicate their secondary role (46). Moreover, molecular and genetic studies have revealed the presence of chitinolytic genes among important members of the Oxalobacteraceae (23, 51–53, 60). Thus, the genus Streptomyces, along with Oxalicibacterium (in the presence of fungi), might play key roles in disease-suppressive soils as being part of the amendment-reactive microbiota (5, 19, 20, 54). A further lead, obtained from chiA-based Roche 454 deep pyrosequencing analysis of the June-10 chitin-amended field and soil from a microcosm experiment (17, 21), recently indicated an increase in the abundance of Oxalobacteraceae-like sequences. In the present study, the relative abundances of both groups, Actinobacteria and Oxalobacteraceae, were positively correlated with chitin amendment over experimental time. The community size increases of both groups, observed from Dec-09 until June-10, suggested that some of their members may be among the key responders to chitin addition. In addition, at three sampling times (June 2010, November 2010, and March 2011), the fungal abundances were negatively correlated with those of Actinobacteria and Oxalobacteraceae. These decreases in fungal abundances may have been a consequence of increased bacterial chitinolytic activity, in particular due to bacterial β-N-acetylglucosaminidases and chitobiosidases.
At the level of the Actinobacteria, the changes in the community structures were fast, since significant community shifts were found shortly after treatment. It is likely that some Actinobacteria, in particular members of the Streptomyces group, colonized the chitin fibers (conglomerates) in soil (8, 14). The actinobacterial species richness increases over time, from autumn-winter to spring-summer, might reflect germination and outgrowth processes that took place related to temperature rises. The Oxalobacteraceae community showed a structure of low diversity based on the analysis of bands. A core community becoming abundant was observed in the chitin-amended soils 9 months after the treatment, and this core community persisted for more than 2 years. Thus, the concomitant occurrence of clear shifts in both the oxalobacteraceal and actinobacterial communities and enhanced pathogen suppressiveness as a result of added chitin pinpointed chitin as a main factor inducing microbially based suppressiveness.
Temporal variation of Oxalobacteraceae.
Members of the Oxalobacteraceae have recently been described as being involved in seed and root colonization, potentially rising to high abundance in the rhizospheres of particular plants (18, 55). We assessed the Oxalobacteraceae communities at the levels of genus and species. Among the five different genera detected, Duganella, Janthinobacterium, and Massilia varied under chitin amendment and over time. In particular, Duganella violaceinigra and Massilia plicata showed clear positive responses to the added chitin. Massilia, Duganella, and Janthinobacterium constitute genera that encompass species that produce and secrete chitinases. Furthermore, Massilia contains seed- and root-colonizing organisms that can proliferate rapidly when attached to plant surfaces (3, 18, 55–57). Remarkably, a slight decrease in the abundance of Janthinobacterium, albeit insignificant, correlated with the increase of the above Massilia and Duganella species. Soil temperature and the availability of chitin oligomers may have been the main factors affecting these relative abundances. Considering Janthinobacterium, its type species, J. lividum, was first isolated from a cold soil (58). The genus is known to encompass key producers of chitinases (57). The recently published genome sequence of Janthinobacterium sp. HH01 revealed the presence of four putative chitinase genes (23). These features are to be considered when the Janthinobacterium relative abundance in soil in connection to soil chitin amendment is evaluated.
Previous studies of plant root-associated Oxalobacteraceae (3, 56) suggested the existence of two groups of Oxalobacteraceae, i.e., the so-called “short-term active” ones, with as key types Herminiimonas saxobsidens and Herbaspirillum seropedicae, and the “long-term active” ones, i.e., Duganella violaceinigra and Massilia plicata. The former group can be compared to classical copiotrophic (r strategist) and the latter to oligotrophic (K strategist) organisms. The significant stimulus of both of the latter long-term active responders in our soil months after chitin addition is consistent with their presumed slow metabolisms. Moreover, Roche 454 pyrosequencing of total bacterial 16S rRNA gene amplicons also revealed an increase in relative abundances of Duganella and Massilia types (from below 1% to 20%) in the chitin-amended soil compared to those in the unamended soil (June-10; M. S. Cretoiu, A. M. Kielak, A. Schlueter, and J. D. van Elsas, unpublished data). On the other hand, the conspicuous absence of Collimonas-like sequences in our here-reported data set was consistent with these deep sequencing data, which also showed the level of members of this bacterial genus to be below 1% (which is undetectable by our clone library approach). Collimonas encompasses soil bacteria that have the capacity of growing at the expense of intact, living fungal hyphae (59, 60). Thus, we surmised that the chitin treatment (in which tillage was used) did not substantially favor fungal hosts that might have supported Collimonas to build up high cell densities.
Relationship between chitin amendment, microbial community shifts, and soil suppressiveness.
Across the 2.5 years of monitoring, a clear effect of soil chitin amendment on both suppressiveness toward two key plant pathogens and selected microbial groups was observed in the experimental field. Thus, parameters such as the abundances of total bacteria, of total fungi, and of actinobacteria and oxalobacteria, next to the respective community structures, were clearly correlated with the suppressiveness of soil toward V. dahliae and particular Pratylenchus types. However, without a deeper understanding of the mechanisms behind pathogen suppression, it is difficult to discern the direct links between the two types of observations. It is likely that the responses found in the chitin-amended soils related to effect of chitin on the growth and survival of the affected communities, in particular the bacterial ones. Whether the rise of such bacterial empires relates, in a direct and mechanistic sense, to the suppression is a question for future research. In such work, a focus on particular members of the Actinobacteria next to members of the underexplored Oxalobacteraceae, in particular Duganella violaceinigra and Massilia plicata, is warranted.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the METAEXPLORE project, awarded to J. D. van Elsas. G. W. Korthals and J. H. M. Visser were supported by the Ministry of EZ.
Footnotes
Published ahead of print 28 June 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01361-13.
REFERENCES
- 1.Weller DM, Raaijmakers JM, McSpadden Gardener BB, Thomashow LS. 2002. Microbial populations responsible for specific soil suppressiveness to plant pathogens. Annu. Rev. Phytopathol. 40:309–348 [DOI] [PubMed] [Google Scholar]
- 2.Sarathchandra SU, Watson RN, Cox NR, di Menna ME, Brown JA, Burch G, Neville FJ. 1996. Effects of chitin amendment of soil on microorganisms, nematodes, and growth of white clover (Trifolium repens L.) and perennial ryegrass (Lolium perenne L.). Biol. Fertil. Soils 22:221–226 [Google Scholar]
- 3.Green SJ, Inbar E, Michel FC, Jr, Hadar Y, Minz D. 2006. Succession of bacterial communities during early plant development: transition from seed to root and effect of compost amendment. Appl. Environ. Microbiol. 72:3975–3983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Radwan MA, Farrag SAA, Abu-Elamayem MM, Ahmed NS. 2012. Extraction, characterization, and nematicidal activity of chitin and chitosan derived from shrimp shell waste. Biol. Fertil. Soils 48:463–468 [Google Scholar]
- 5.Mendes R, Kruijt M, de Bruijn I, Dekkers E, van der Voort M, Schneider JHM, Piceno YM, DeSantis TZ, Andersen GL, Bakker PAHM, Raaijmakers JM. 2011. Deciphering the rhizosphere microbiome for disease-suppressive bacteria. Science 332:1097–1100 [DOI] [PubMed] [Google Scholar]
- 6.Gooday GW. 1990. The ecology of chitin degradation. Adv. Microb. Ecol. 11:387–430 [Google Scholar]
- 7.Pillai CKS, Paul W, Sharma CP. 2009. Chitin and chitosan polymers: chemistry, solubility and fiber formation. Prog. Polymer Sci. 34:641–678 [Google Scholar]
- 8.Manucharova NA, Vlasenko AN, Stepanov AL. 2007. Temperature as an autoecological factor of chitinolytic microbial complex formation in soils. Biol. Bull. 34:163–169 [PubMed] [Google Scholar]
- 9.Gooday GW. 1990. Physiology of microbial degradation of chitin and chitosan. Biodegradation 1:177–190 [Google Scholar]
- 10.Manucharova NA, Yaroslavtsev AM, Senchenko DV, Stepanov AL, Zvyagintsev DG. 2006. Microbial transformation of chitin in soil under anaerobic conditions. Biol. Bull. 33:191–194 [PubMed] [Google Scholar]
- 11.Hjort K, Bergstrom M, Adesina MF, Jansson JK, Smalla K, Sjoling S. 2010. Chitinase genes revealed and compared in bacterial isolates, DNA extracts and a metagenomic library from a phytopathogen-suppressive soil. FEMS Microbiol. Ecol. 71:197–207 [DOI] [PubMed] [Google Scholar]
- 12.Whips JM. 2001. Microbial interactions and biocontrol in the rhizosphere. J. Exp. Bot. 52:487–511 [DOI] [PubMed] [Google Scholar]
- 13.Krsek M, Wellington EMH. 2001. Assessment of chitin decomposer diversity within an upland grassland. Antonie Van Leeuwenhoek 79:261–267 [DOI] [PubMed] [Google Scholar]
- 14.Metcalfe AC, Krsek M, Gooday GW, Prosser JI, Wellington EMH. 2002. Molecular analysis of a bacterial chitinolytic community in an upland pasture. Appl. Environ. Microbiol. 68:5042–5050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hallmann J, Rodriguez-Kabana R, Kloepper JW. 1999. Chitin-mediated changes in bacterial communities of the soil, rhizosphere and whitin roots of cotton in relation to nematode control. Soil. Biol. Biochem. 31:551–560 [Google Scholar]
- 16.Kawase T, Yokokawa S, Saito A, Fuji T, Nikaidou N, Miyashita K, Watanabe T. 2006. Comparison of enzymatic and antifungal properties between family 18 and 19 chitinases from S. coelicolor A3(2). Biosci. Biotechnol. Biochem. 70:988–998 [DOI] [PubMed] [Google Scholar]
- 17.Kielak AM, Cretoiu MS, Semenov A, Sorensen S, van Elsas JD. 2013. Bacterial chitinolytic communities respond to chitin and pH alteration in soil. Appl. Environ. Microbiol. 79:263–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Green SJ, Michel FC, Jr, Hadar Y, Minz D. 2007. Contrasting patterns of seed and root colonization by bacteria from the genus Chryseobacterium and from the family Oxalobacteraceae. ISME J. 1:291–299 [DOI] [PubMed] [Google Scholar]
- 19.Martin G, Guggiari M, Bravo D, Zopfi J, Cailleau G, Aragno M, Job D, Verrecchia E, Junier P. 2012. Fungi, bacteria and soil pH: the oxalate-carbonate pathway as a model for metabolic interaction. Environ. Microbiol. 14:2960–2970 [DOI] [PubMed] [Google Scholar]
- 20.Junier P, Caileau G, Martin G, Guggiari M, Bravo D, Clerc M, Aragno M, Job D, Verrechia E. 2012. The oxalate-carbonate pathway: at the interface between biology and geology. Geophys. Res. Abstr. 14:EGU2012–5673 [Google Scholar]
- 21.Cretoiu MS, Kielak AM, Abu Al-Soud W, Sorensen SJ, van Elsas JD. 2012. Mining of unexplored habitats for novel chitinases—chiA as a helper gene proxy in metagenomics. Appl. Microbiol. Biotechnol. 94:1347–1358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee GW, Lee KJ, Chae JC. 2012. Genome sequence of Herbaspirillum sp. strain GW103, a plant growth-promoting bacterium. J. Bacteriol. 194:4150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hornung C, Poehlein A, Haack FS, Schmidt M, Dierking K, Pohlen A, Schulenburg H, Blokesch M, Plener L, Jung K, Bonge A, Khron-Molt I, Utpatel C, Timmermann G, Spieck E, Pommerening-Roser A, Bode E, Bode HB, Daniel R, Schmeisser Streit WR. 2013. The Janthinobacterium sp. HH01 genome encodes a homologue of the V. cholerae CqsA and L. pneumophila LqsA autoinducer synthases. PLoS One 8:e55045. 10.1371/journal.pone.0055045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu Y, Gallert C, Winter J. 2008. Chitin purification from shrimp wastes by microbial deproteination and decalcification. Appl. Microbiol. Biotechnol. 79:687–697 [DOI] [PubMed] [Google Scholar]
- 25.Kjartansson GT, Zivanovic S, Kristberg K, Weiss J. 2006. Sonication-assisted extraction of chitin from North Atlantic shrimps (Pandalus borealis). J. Agric. Food Chem. 54:5894–5902 [DOI] [PubMed] [Google Scholar]
- 26.Korthals GW, de Boer M, Visser JHM, Molendijk LPG. 2010. Bodemgezondheid binnen bedrijfssytemen. Mededelingenglad Koninklijke Nederlandse Plantenziektekundige Vereninging 41:281–284 [Google Scholar]
- 27.Fierer N, Jackson JA, Vilgalys R, Jackson RB. 2005. Assessment of soil microbial community structure by use taxon-specific quantitative PCR assay. Appl. Environ. Microbiol. 71:4117–4120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dohrmann AB, Tebbe CC. 2005. Effect of elevated tropospheric ozone on the structure of bacterial communities inhabiting the rhizosphere of herbaceous plants native to Germany. Appl. Environ. Microbiol. 71:7750–7758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Williamson N, Brian P, Wellington EMH. 2000. Molecular detection of bacterial and streptomycete chitinase in the environment. Antonie Van Leeuwenhoek 78:315–321 [DOI] [PubMed] [Google Scholar]
- 30.Yergeau E, Kang S, He Z, Zhou J, Kowalchuk GA. 2007. Functional microarray analysis of nitrogen and carbon cycling genes across an Antarctic latitudinal transect. ISME J. 1:163–179 [DOI] [PubMed] [Google Scholar]
- 31.Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL, Vandesompele J, Wittwer CT. 2009. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 55:611–622 [DOI] [PubMed] [Google Scholar]
- 32.Brons J, van Elsas JD. 2008. Analysis of bacterial communities in soil by use of denaturing gradient gel electrophoresis and clone libraries, as influenced by different reverse primers. Appl. Environ. Microbiol. 74:2717–2727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heuer H, Krsek M, Baker P, Smalla K, Wellington EHM. 1997. Analysis of actinomycete communities by specific amplification of genes encoding 16S rRNA and gel-electrophoretic separation in denaturing gradients. Appl. Environ. Microbiol. 63:3233–3241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.White TJ, Bruns T, Lee S, Taylor JW. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics, p 315–322 In Innis MA, Gelfand DH, Sninsky JJ, White TJ. (ed), PCR protocols: a guide to methods and applications. Academic Press, New York, NY [Google Scholar]
- 35.Gardes M, Bruns TD. 1993. ITS primers with enhanced specificity for basidiomycetes—application to the identification of mycorrhizae and rusts. Mol. Ecol. 2:113–118 [DOI] [PubMed] [Google Scholar]
- 36.Huber T, Faulkner G, Hugenholtz P. 2004. Bellerophon; a program to detect chimeric sequences in multiple sequence alignments. Bioinformatics 20:2317–2319 [DOI] [PubMed] [Google Scholar]
- 37.Schloss PD, Westcott S, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, van Horn DJ, Weber CF. 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75:7537–7541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Eichner CA, Erb RW, Timmis KN, Wagner-Dobler I. 1999. Thermal gradient gel electrophoresis analysis of bioprotection from pollutant shocks in the activated sludge microbial community. Appl. Environ. Microbiol. 65:102–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Giotis C, Theodoropoulou A, Cooper J, Hodgson R, Shotton P, Shiel R, Eyre M, Wilcockson S, Markellou E, Liopa-Tsakalidis A, Volakakis N, Leifert C. 2012. Effect of variety choice, resistant rootstocks and chitin soil amendments on soil-borne diseases in soil-based, protected tomato production systems. Eur. J. Plant Pathol. 134:605–617 [Google Scholar]
- 40.Ladner DC, Tchounwou PB, Lawrence GW. 2008. Evaluation of the effect of Ecologic on root knot nematode, Meloidogyne incognita, and tomato plant, Lycopersicon esculenum. Int. J. Environ. Res. Public Health 5:104–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Downing K, Thomson JA. 2000. Introduction of the Serratia marcescens chiA gene into an endophytic Pseudomonas fluorescens for the biocontrol of phytopathogenic fungi. Can. J. Microbiol. 46:363–369 [DOI] [PubMed] [Google Scholar]
- 42.Kobayashi DY, Reedy RM, Bick J, Oudemans PV. 2002. Characterization of a chitinase gene from Stenotrophomonas maltophilia strain 34S1 and its involvement in biological control. Appl. Environ. Microbiol. 68:1047–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kotan R, Dikbas N, Bostan H. 2009. Biological control of post-harvest disease caused by Aspergillus flavus on stored lemon fruits. Afr. J. Biotechnol. 8:209–214 [Google Scholar]
- 44.Terahara T, Ikeda S, Noritake C, Minamisawa K, Ando K, Tsuneda S, Harayama S. 2009. Molecular diversity of bacterial chitinases in arable soil and the effects of environmental factors of the chitinolytic bacterial community. Soil Biol. Biochem. 41:473–480 [Google Scholar]
- 45.Sato K, Azama Y, Nogawa M, Taguchi G, Shimosaka M. 2010. Analysis of a change in bacterial community in different environments with addition of chitin or chitosan. J. Biosci. Bioeng. 109:472–478 [DOI] [PubMed] [Google Scholar]
- 46.Timmer RD, Korthals GW, Molendijk LPG. 2003. Groebenmesters. Van teelttechniek tot ziekten en plagen. Brochure PPO 316:59 [Google Scholar]
- 47.Leveau JHJ, Uroz S, de Boer W. 2010. The bacterial genus Collimonas: mycophagy, weathering and other adaptive solutions to life in oligotrophic soil environments. Environ. Microbiol. 12:281–292 [DOI] [PubMed] [Google Scholar]
- 48.De Boer W, Gerards S, Klein Gunnewiek PJA, Modderman R. 1999. Response of the chitinolytic microbial community to chitin amendments of dune soils. Biol. Fertil. Soils 29:170–177 [Google Scholar]
- 49.Rousk J, Bååth E, Brookes PC, Lauber CL, Lozupone C, Caporoso JG, Knight R, Fierer N. 2010. Soil bacterial and fungal communities across a pH gradient in an arable soil. ISME J. 4:1340–1351 [DOI] [PubMed] [Google Scholar]
- 50.Bailey VL, Smith JL, Bolton H. 2002. Fungal-to-bacterial ratios in soils investigated for enhanced C sequestration. Soil Biol. Biochem. 34:997–1007 [Google Scholar]
- 51.Knutson DM, Stiebrs Hutchins A, Cromack K., Jr 1980. The association of calcium oxalate-utilizing Streptomyces with conifer ectomycorrhizae. Antonie Van Leeuwenhoek 46:611–619 [Google Scholar]
- 52.Lopez-Bucio J, Nieto-Jacobo MF, Ramirez-Rodriguez V, Herrera Estrella L. 2000. Organic acid metabolism in plants: from adaptive physiology to transgenic varieties for cultivation in extreme soils. Plant Sci. 160:1–13 [DOI] [PubMed] [Google Scholar]
- 53.Johnsen MG, Hansen OC, Stougaard P. 2010. Isolation, characterization and heterologous expression of a novel chitosanase from Janthinobacterium sp. strain 4239. Microb. Cell Fact. 9:5. 10.1186/1475-2859-9-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Khammar N, Martin G, Ferro K, Job D, Aragno M, Verrecchia E. 2009. Use of the frc gene as a molecular marker to characterize oxalate-oxidizing bacterial abundance and diversity structure in soil. J. Microbiol. Methods 76:120–127 [DOI] [PubMed] [Google Scholar]
- 55.Ofek M, Hadar Y, Minz D. 2009. Comparison of effects of compost amendment and of single-strain inoculation on root bacterial communities of young cucumber seedlings. Appl. Environ. Microbiol. 75:6441–6450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ofek M, Hadar Y, Minz D. 2012. Ecology of root colonizing Massilia. PLoS One 7:e40117. 10.1371/journal.pone.0040117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gleave AP, Taylor RK, Morris BA, Greenwood DR. 1995. Cloning and sequencing of a gene encoding the 69-kDa extracellular chitinase of Janthinobacterium lividum. FEMS Microbiol. Lett. 131:279–288 [DOI] [PubMed] [Google Scholar]
- 58.Shivaji S, Ray MK, Seshu Kumar G, Reddy GSN, Saisree L, Wynn-Williams DD. 1991. Identification of Janthinobacterium lividum from the soils of the islands of Scotia Ridge and from Antarctic peninsula. Polar Biol. 11:267–271 [Google Scholar]
- 59.de Boer W, Leveau JH, Kowalchuk GA, Klein Gunnewiek PJ, Abeln EC, Figge MJ, Sjollema K, Janse JD, van Veen JA. 2004. Collimonas fungivorans gen. nov., sp. nov., a chitinolytic soil bacterium with the ability to grow on living fungal hyphae. Int. J. Syst. Evol. Microbiol. 54:857–864 [DOI] [PubMed] [Google Scholar]
- 60.Höppener-Ogawa S, Leveau JH, Hundscheid MP, van Veen JA, de Boer W. 2009. Impact of Collimonas bacteria on community composition of soil fungi. Environ. Microbiol. 11:1444–1452 [DOI] [PubMed] [Google Scholar]
- 61.Smit E, Leeflang P, Glandorf B, van Elsas JD, Wernars K. 1999. Analysis of fungal diversity in the wheat rhizosphere by sequencing of cloned PCR-amplified genes encoding 18S rRNA and temperature gradient gel electrophoresis. Appl. Environ. Microbiol. 65:2614–2621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gomes RC, Semedo LTAS, Soares RMA, Linhares LF, Ulhoa CJ, Alviano CS, Coelho RRR. 2001. Purification of thermostable endochitinase from Streptomyces RC1071 isolated from a cerrado soil and its antagonism against phytopathogenic fungi. J. Appl. Microbiol. 90:653–661 [DOI] [PubMed] [Google Scholar]
- 63.Larkin RP, Honeycutt CW, Olanya OM. 2011. Management of Verticillium wilt of potato with disease-suppressive green manures and as affected by previous cropping history. Plant Dis. 95:568–576 [DOI] [PubMed] [Google Scholar]
- 64.Fritsche K, de Boer W, Gerards S, van den Berg M, van Veen JA, Leveau JH. 2008. Identification and characterization of genes underlying chitinolysis in Collimonas fungivorans Ter331. FEMS Microbiol. Ecol. 66:123–135 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.