Abstract
Rapid advances in sequencing technology have changed the experimental landscape of microbial ecology. In the last 10 years, the field has moved from sequencing hundreds of 16S rRNA gene fragments per study using clone libraries to the sequencing of millions of fragments per study using next-generation sequencing technologies from 454 and Illumina. As these technologies advance, it is critical to assess the strengths, weaknesses, and overall suitability of these platforms for the interrogation of microbial communities. Here, we present an improved method for sequencing variable regions within the 16S rRNA gene using Illumina's MiSeq platform, which is currently capable of producing paired 250-nucleotide reads. We evaluated three overlapping regions of the 16S rRNA gene that vary in length (i.e., V34, V4, and V45) by resequencing a mock community and natural samples from human feces, mouse feces, and soil. By titrating the concentration of 16S rRNA gene amplicons applied to the flow cell and using a quality score-based approach to correct discrepancies between reads used to construct contigs, we were able to reduce error rates by as much as two orders of magnitude. Finally, we reprocessed samples from a previous study to demonstrate that large numbers of samples could be multiplexed and sequenced in parallel with shotgun metagenomes. These analyses demonstrate that our approach can provide data that are at least as good as that generated by the 454 platform while providing considerably higher sequencing coverage for a fraction of the cost.
INTRODUCTION
The last 10 years of microbial ecology research have involved a profound shift in focus from observational phylogenetic analyses of as-yet uncultured novel taxa (1) to experimental characterization of the taxonomic shifts in communities through the use of complex experimental designs (2). This shift in focus has been driven by the advent of relatively inexpensive next-generation sequencing approaches and the development of robust bioinformatic tools. The most commonly used sequencing platform has been from 454 (3); however, there is growing interest in using the IonTorrent (4), PacBio (5), and Illumina (6) platforms. This transition between platforms has not been painless, as there has been a steady stream of concerns raised regarding the quality and meaning of sequence data generated by the 454 sequencers (7, 8). Numerous groups have worked to develop bioinformatic solutions that make the 454 platform a robust approach to characterizing microbial communities (9, 10). As other sequencing platforms mature and perhaps replace 454 as the platform of choice for 16S rRNA gene sequencing, it is critical to develop similar solutions so that the field does not sacrifice high data quality for increased sequencing throughput.
A number of considerations must be made when selecting a platform for sequencing the 16S rRNA gene. We contend that sequence quality is the most important consideration, as studies that are built upon data that are unreliable are themselves unreliable (10). A second important consideration is the number of reads that one can obtain per run and per dollar. This is significant, because investigators can titrate the number of samples and reads per sample using multiplexing strategies to fit the number of overall reads. A third consideration for 16S rRNA studies is the length of the sequences, as longer sequences are easier to assign to a taxonomic group using a classifier (11). Finally, the customer and technical support of the companies that manufacture the reagents and instrumentation and the availability of their platform and reagents are significant factors. The goal of this study was to assess the quality of MiSeq-generated data and to determine its advantages and disadvantages compared to 454. Further studies are necessary to make similar comparisons to other platforms.
Illumina-based strategies are able to generate the largest amount of sequence data per dollar, using a chip-based bridge amplification procedure followed by sequencing by synthesis using reversible terminator dye nucleotides (12). Depending on the platform and reagents, one can currently obtain up to 300 and 500 cycles (i.e., nucleotides [nt]) of sequence data on HiSeq and MiSeq platforms, respectively. These cycles are commonly split into two reads, providing paired reads of the same DNA fragment. The platforms also vary in their sequencing throughput, with HiSeq 2500 being capable of generating 600 Gbp using paired 100-nt reads (i.e., 3 billion pairs of reads) or 180 Gbp using paired 150-nt reads (i.e., 1.2 billion pairs of reads), and MiSeq was capable of generating 8.5 Gbp using paired 250-nt reads (i.e., 17 million pairs of reads). Because the HiSeq platform requires one to fill 16 sequencing lanes with the same reagents, logistically it is more difficult for an individual to fill a complete run with a 300-cycle kit when the 200-cycle kit is more commonly used within the genomics field. Reagents for the HiSeq (300 cycle) are approximately $1,500 per lane, and for the MiSeq they are approximately $1,000 per lane. The HiSeq 2500 and MiSeq instruments currently retail for $740,000 and $125,000, respectively. The HiSeq platform has become the standard approach for shotgun metagenomic sequencing because of its increased read depth; however, the MiSeq has greater potential for use with 16S rRNA gene sequence studies, because it generates longer sequence reads, and its performance and cost are tractable to the needs of individual investigators (13).
Until recently, the most significant problem with the Illumina platforms has been the ability to sequence samples with low genetic diversity, such as that commonly found with 16S rRNA gene amplicons. To artificially increase the genetic diversity, it has been common to mix in a control library of genomic DNA from the phage PhiX, such that 50% of the DNA was from PhiX. During the course of this study, Illumina upgraded their image analysis software to overcome this challenge, such that only 5 to 10% PhiX is needed to sufficiently increase the genetic diversity. Another factor that can affect data quality is the amount of DNA loaded onto the flow cell, as this affects the cluster density and the ability of the image analysis software to discriminate between clusters. In this study, we evaluate the effect of the new software on sequencing error rates with various cluster densities.
Previous studies involving 16S rRNA gene sequencing on the Illumina platforms have utilized two approaches. The first approach involves two PCR steps with different primer pairs (for an example, see reference 6). In the first PCR step, the two primers used contain an Illumina sequencing primer, an index (i.e., barcode) sequence, and the gene-specific primer. In the second PCR step, two primers are used that contain the Illumina adapter and sequencing primer sequence. Paired-end sequencing is performed using the built-in Illumina sequencing primers. This approach is limited, because it requires two rounds of PCR, increasing the risks of artifacts commonly observed with large numbers of PCR rounds, and because one must devote 20 to 25 nt to sequencing the index and gene-specific primer. The second approach emulates Illumina's TruSeq genomic library construction protocol, in which a single PCR is used (14). In this approach, the primers contain the Illumina adapter sequence, an index sequence (only for the reverse primer), a 10-nt pad to prevent hairpin formation, a 2-nt linker that is noncomplementary to the 16S rRNA gene, and a gene-specific primer. Sequencing proceeds by (i) using the combined pad-linker-primer as the sequencing primer at the 5′ end to obtain a long read, (ii) using the reverse complement of the combined pad-linker-primer as the sequencing primer at the 3′ end to sequence the index region, and (iii) using the combined pad-linker-primer as the sequencing primer at the 3′ end to obtain a long read. With the 500-cycle reagents, this results in an index sequence and two 250-nt reads. A collection of 2,168 reverse primers with different indices has been published for the V4 region of the 16S rRNA gene (14). Considering these methods were developed using previous Illumina platforms that could only generate 200 or 300 nt and current technology can obtain 500 nt, with anticipated further expansions in sequencing lengths, we sought to develop a dual-index, paired-read approach that could easily be adapted to other regions of the 16S rRNA gene or other genes. The advantage of such an approach is that with dual indices, one could replace the 2,168 previously proposed V4 primers with a total of 94 primers.
The development of bioinformatic solutions for curating sequences generated on the Illumina platforms has been limited. Several studies have insisted that extensive sequence curation and contig formation is unnecessary (14, 15); however, these were largely focused on analyzing the beta-diversity between communities and taxonomic classification to the genus level. These approaches are limited because of the limitations of existing databases and the various levels of diversity across taxonomic lineages. Caporaso and colleagues (14) have utilized a mapping procedure where reads are mapped to a reference database of V4 reads that are not more than 97% similar to each other; if a read is not more than 97% similar to a sequence in the database, it is culled. Although this is clearly a fast approach, a significant number of good reads may be rejected, and it requires the creation of very specialized databases for each region being sequenced. Such a strategy can be impossible should researchers attempt to adapt the sequencing strategy to poorly characterized genes. Others have attempted to use the Phred/Phrap quality scores associated with each base to trim sequence reads in combination with removing rare taxa (16). Unfortunately, no error rates are provided following their sequence-trimming procedure, and removal of rare taxa could be problematic if one is interested in tracking rare populations. Finally, the only published attempt to develop a method of curating paired sequence reads has suggested allowing various numbers of mismatches between the overlapping sequence reads; however, again, final error rates were not provided (6, 17). In the current study, we resequence a mock treatment community where we know the true 16S rRNA gene sequence to assess the effect of various trimming and sequence assembly methods on the overall error rates.
Here, we address several technical and bioinformatic challenges related to employing the MiSeq platform for sequencing of the 16S rRNA gene. First, the recent release of the Illumina MiSeq v. 2.0 platform provides 500 cycles that are typically applied by obtaining paired 250-nt sequences per fragment. This allowed us to determine whether the additional sequence length would allow one to sequence longer regions of the 16S rRNA gene fragment either as a single read or as paired reads. Furthermore, because the sequencing platform is constantly evolving to provide more and longer sequence reads, we developed a sequencing strategy that could easily be adapted when longer reads are possible and reduce the investment in buying large numbers of indexed primers. Second, we evaluated the prospects of sequencing metagenomic shotgun libraries in parallel to 16S rRNA gene amplicons for situations where deep sequencing coverage is not necessary. Third, we developed a sequence curation pipeline that results in a minimal number of sequence reads while producing sequences with error rates comparable to those we have previously observed with 454 data (10). Finally, we reanalyzed a large set of samples that we previously analyzed using the 454 platform using our MiSeq-based approach and observed similar results (18).
MATERIALS AND METHODS
Overall strategy and primer design.
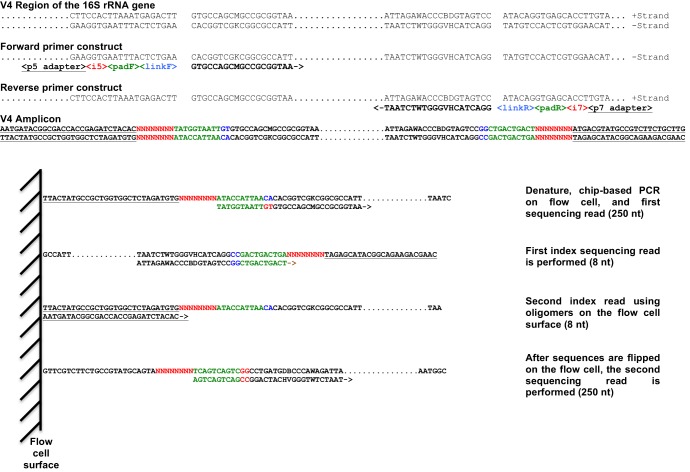
Our dual-index paired-end sequencing approach is analogous to the single-index approach described elsewhere (13, 14). As shown in Fig. 1, each primer consists of the appropriate Illumina adapter, an 8-nt index sequence, a 10-nt pad sequence, a 2-nt linker, and the gene-specific primer. The index sequences were selected to be at least 2 nt different from all other indices in use, and when combined, they provide an equal intensity in the two light channels used by the sequencer (i.e., green channel [G/T] and red channel [A/C]). The index sequences were also at least 2 nt different from the indices that accompany the Nextera library construction kit. The 2-nt linker sequence was selected to not be complementary to the homologous positions in a large collection of 16S rRNA gene sequences (19). Finally, the pad sequence was selected so that the combined pad, linker, and gene-specific primer sequences had an estimated melting temperature between 60 and 65°C. For the development of our method, we used gene-specific primers to amplify the V34, V4, and V45 regions from the bacterial 16S rRNA gene that have been described elsewhere (14, 20). The complete primers were each 63 to 68 bp long.
Fig 1.
Design of dual-index sequencing strategy and schematic describing the four sequencing reads. The primers specific to the 16S rRNA gene are shown in boldface black text, linkers are in blue, pads are in green, the index region is in red, and the adapters are underlined. This schematic is demonstrated using the V4-specific primer sequences and linkers. The PCR and sequencing primers for each of the three regions are provided in the supplemental material.
Two sequence reads, two index reads, and three sequence primers were necessary to sequence each DNA fragment. The first sequence read (250 nt) was obtained using the Read 1 primer, which was the same as the sequence of the combined pad, linker, and gene-specific primer at the 5′ end of the region. The first index, located at the 3′ end of the fragment, was sequenced using the Index primer. The Index primer was the reverse complement of the combined pad linker and gene-specific primer sequence at the 3′ end of the region. After this index read, the platform flips the fragment. The second index read then was performed to obtain the index sequence at the 5′ end of the fragment using the adapter lawn on the surface of the sequencing flow cells. Finally, the second sequence read (250 nt) was obtained using the Read 2 primer, which was the same as the sequence of the combined pad linker, and gene-specific primer sequence at the 3′ end of the region. The overall process of cluster generation, sequencing, image processing, demultiplexing, and quality score calculation was performed on the MiSeq in approximately 40 h.
Community DNA.
In the initial studies to develop our sequencing approach, we utilized genomic DNA isolated from four communities that were then sequenced as three technical replicates. The first was termed a “mock community” composed of genomic DNA from 21 bacterial isolates. This mock community is similar to the one that we used previously to assess error rates in 454-generated sequence data: Acinetobacter baumannii ATCC 17978, Actinomyces odontolyticus ATCC 17982, Bacillus cereus ATCC 10987, Bacteroides vulgatus ATCC 8482, Clostridium beijerinckii ATCC 51743, Deinococcus radiodurans ATCC 13939, Enterococcus faecalis ATCC 47077, Escherichia coli ATCC 70096, Helicobacter pylori ATCC 700392, Lactobacillus gasseri ATCC 33323, Listeria monocytogenes ATCC BAA-679, Neisseria meningitidis ATCC BAA-335, Porphyromonas gingivalis ATCC 33277, Propionibacterium acnes DSM 16379, Pseudomonas aeruginosa ATCC 47085, Rhodobacter sphaeroides ATCC 17023, Staphylococcus aureus ATCC BAA-1718, Staphylococcus epidermidis ATCC 12228, Streptococcus agalactiae ATCC BAA-611, Streptococcus mutans ATCC 700610, and Streptococcus pneumoniae ATCC BAA-334. The genomic DNAs were pooled to have an equimolar concentration of 16S rRNA gene copies per genome with a final concentration of 5 ng/μl. Mock community DNA is available through BEI Resources (HM-278D v3.1). Genomic DNAs from the three other communities were obtained using the MO BIO PowerSoil DNA extraction kit with material from mouse and human feces and soil from a residential area. To demonstrate the ability to scale up our method and recapitulate previous results, we reused the DNA from a previous study in which DNA was isolated from mouse feces using the Roche MagnaPure DNA extraction kit. All fecal samples were obtained using protocols that were reviewed and approved by the University Committee on Use and Care of Animals and the Institutional Review Board at the University of Michigan.
Amplicon library construction and sequencing.
16S rRNA gene libraries were constructed using the primers described above to amplify the V34, V4, and V45 regions. Amplicons were generated using a high-fidelity polymerase (AccuPrime; Invitrogen) and then were purified using a magnetic bead capture kit (Ampure; Agencourt) and quantified using a fluorometric kit (QuantIT PicoGreen; Invitrogen). The purified amplicons were then pooled in equimolar concentrations using a SequalPrep plate normalization kit (Invitrogen), and the final concentration of the library was determined using a SYBR green quantitative PCR (qPCR) assay with primers specific to the Illumina adapters (Kappa). Libraries were mixed with Illumina-generated PhiX control libraries and our own genomic libraries and denatured using fresh NaOH. A detailed protocol with primer and index sequences is provided in the supplemental material. We performed 11 sequencing runs for this study. Tables S1 and S2 in the supplemental material provide results from seven sequencing runs performed using Real Time Analysis software (RTA), v. 1.16.18 and 1.17.22, MiSeq Control software (MCS), v. 2.0.5 and 2.1.13, various amounts of a PhiX genomic library control, and various cluster densities. Four sequencing runs were performed with RTA v. 1.17.28, MCS v. 2.2.0, a target of 5% PhiX, and various cluster densities (Table 1).
Table 1.
Summary of operating conditions for various MiSeq sequencing runs and machine-reported quality metricsa
| Run | [DNA] (pM) | Cluster density (103/mm2) | % PhiX | % of bases ≥Q30b | Reported machine error (%) | No. of 16S rRNA pairs (×106) |
|---|---|---|---|---|---|---|
| 130401 | 10.0 | 1,313 | 3.8 | 63.7 | 1.31 | 12.6 |
| 130403 | 5.0 | 1,094 | 3.2 | 70.5 | 0.89 | 12.4 |
| 130417 | 3.0 | 839 | 6.2 | 74.6 | 0.92 | 10.5 |
| 130422 | 1.8 | 690 | 8.0 | 80.1 | 0.65 | 9.0 |
The four runs were performed using RTA v. 1.17.28 and MCS v. 2.2.0. Data for runs performed using previous versions of the software are provided in Tables S1 and S2 in the supplemental material.
Percentage of bases with a quality score of at least 30.
Shotgun library construction.
DNA (2.5 ng) from the mock and human fecal communities used in our amplicon experiments, plus genomic DNA from two Clostridium clostridioforme strains (D4 and CIP110249), were used to generate four shotgun libraries using a customized Nextera XT genomic library construction protocol (Illumina). Our customization involved increasing the amount of input DNA and the amount of transposon-based tagmentation. This modified protocol allowed us to reduce the number of PCR cycles to 10 instead of the recommended 12 while still obtaining a sufficient yield for sequencing. DNA concentration and the length of the fragments were assessed using an Agilent BioAnalyzer. The metagenomes were pooled in equal molar quantities, and the two genomes were pooled at one-half the concentration of the metagenomes. Metagenomic and genomic libraries were quality trimmed using Sickle (https://github.com/najoshi/sickle), assembled using ABySS (21), and filtered to remove contigs smaller than 500 bp.
Bioinformatic analysis and data availability.
All development was carried out using custom Perl and C++ software. The functions required to implement the overall analysis pipeline are available within the mothur software package (v. 1.30) and are illustrated on the mothur website (http://www.mothur.org/wiki/MiSeq_SOP) (22). Contigs between read pairs were assembled as described in this study, and any contigs with an ambiguous base (i.e., N) were culled, as were those sequences where there was no meaningful overlap between sequences. Sequences then were aligned to a reference alignment, and those sequences that did not align to the correct region were culled (23–25). The ends of the sequences were trimmed so that the sequences all started and ended at the same alignment coordinates (25). After identifying the unique sequences and their frequency in each sample, we utilized a preclustering algorithm to further denoise sequences within each sample (10). The resulting sequences were screened for chimeras using UCHIME (26). We then used a naive Bayesian classifier to classify each sequence against the Ribosomal Database Project (RDP) 16S rRNA gene training set (version 9) that was customized to include rRNA gene sequences from mitochondria and Eukaryota. We required an 80% pseudobootstrap confidence score (11). Those sequences that either did not classify to the level of kingdom or that classified as Archaea, Eukaryota, chloroplasts, or mitochondria were culled. Finally, sequences were split into groups corresponding to their taxonomy at the level of order and then assigned to operational taxonomic units (OTUs) at a 3% dissimilarity level; previous experience indicates that the OTU assignments by this approach are equivalent to not splitting the sequences by taxonomic order and has the advantage of parallelization and reduced memory usage (27). Calculations of sequencing error rates and identification of chimeras based on the mock community data were performed as previously described (10). The sequence data used in this study are available at http://www.mothur.org/MiSeqDevelopmentData.html.
RESULTS AND DISCUSSION
Customization of Illumina's MiSeq platform.
The primary objective in customizing the MiSeq platform was to create a dual-index sequencing approach that would allow us to generate a large number of high-quality sequences while minimizing the cost of long, customized primers. To test this platform, we generated an amplicon library using template DNAs isolated from a mock community of genomic DNA with known 16S rRNA gene sequences and natural samples from human feces, mouse feces, and soil. For each sample, we amplified the V4 (length, ca. 250 bp), V34 (ca. 430 bp), and V45 regions (ca. 375 bp). The primers described in Fig. 1 were designed to include the appropriate P5 and P7 Illumina adapter sequences, an 8-nt index sequence, a 10-nt pad sequence, a 2-nt linker sequence, and the gene-specific primer. The 2-nt linker sequence was selected to share a minimum amount of homology with sequences in a reference database (19). The pad sequence was selected so that the combined pad, linker, and gene-specific primer would have a melting temperature over 60°C. When the pad sequence was not included, the sequencing runs failed because of the high annealing temperatures used within the sequencer. We selected 8-nt index sequences that differed by at least 2 nt, and we ensured that each position contained at least one A or C and at least one G or T. This was necessary because the sequencer utilizes two light channels that must be excited in each cycle of sequencing. When we did not utilize both light channels across each base of the index sequences, the sequencing runs failed. For the preliminary analysis we utilized 2 index sequences in the 5′ primer and 6 index sequences at the 3′ end of the sequence. This allowed us to sequence each of the four DNA samples in triplicate to evaluate the effects of different cluster densities on error rates and develop a sequence curation pipeline.
Error profile.
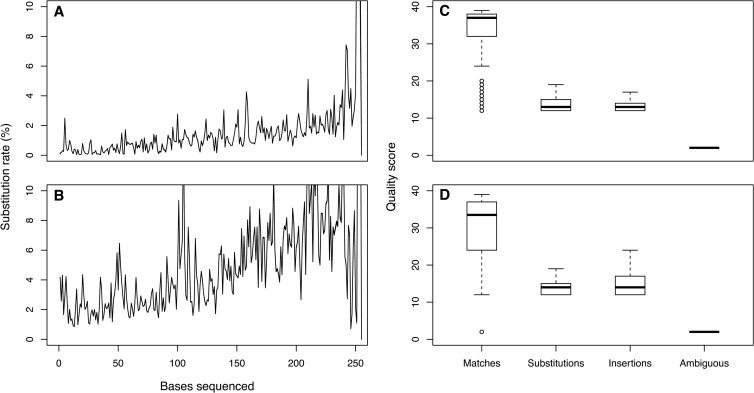
We next sought to develop a strategy to reduce the observed error rates for the two sequencing reads. For all regions of the 16S rRNA gene, the error rates increased over the length of the reads, with the second read having a higher error rate than the first (Fig. 2); substitutions were the primary source of error (99.4%), followed by ambiguous base calls (0.2%), deletions (0.2%), and insertions (0.2%). We were unable to detect a reproducible substitution bias between bases, and all bases were equally likely to be inserted or deleted. To explore whether the quality scores could be used as a reliable surrogate for sequence quality, we categorized each quality score by whether its base call was the expected base (i.e., a match), a substitution, an insertion, or an ambiguous base (Fig. 2). As expected, errors were associated with low-quality scores and rarely had a quality score above 21. Finally, when we performed a regression of the sequencer-generated Phred quality scores (Q) against the observed error rate (P), we observed the expected log-linear relationship (i.e., Q = −10 log10 P) with slopes of −11.7 and −14.3 for the first and second read, respectively. The single-read data indicated that without significantly trimming the length of the reads, it was not possible to obtain the quality of data possible with 454 sequencing; however, the error rates could be improved using the quality scores and building consensus sequences from the paired reads.
Fig 2.
Profile of sequencing errors in the first and second read (A and C) and the quality scores associated with different types of errors in the first and second read (B and D) using data from run 130403.
Denoising via consensus.
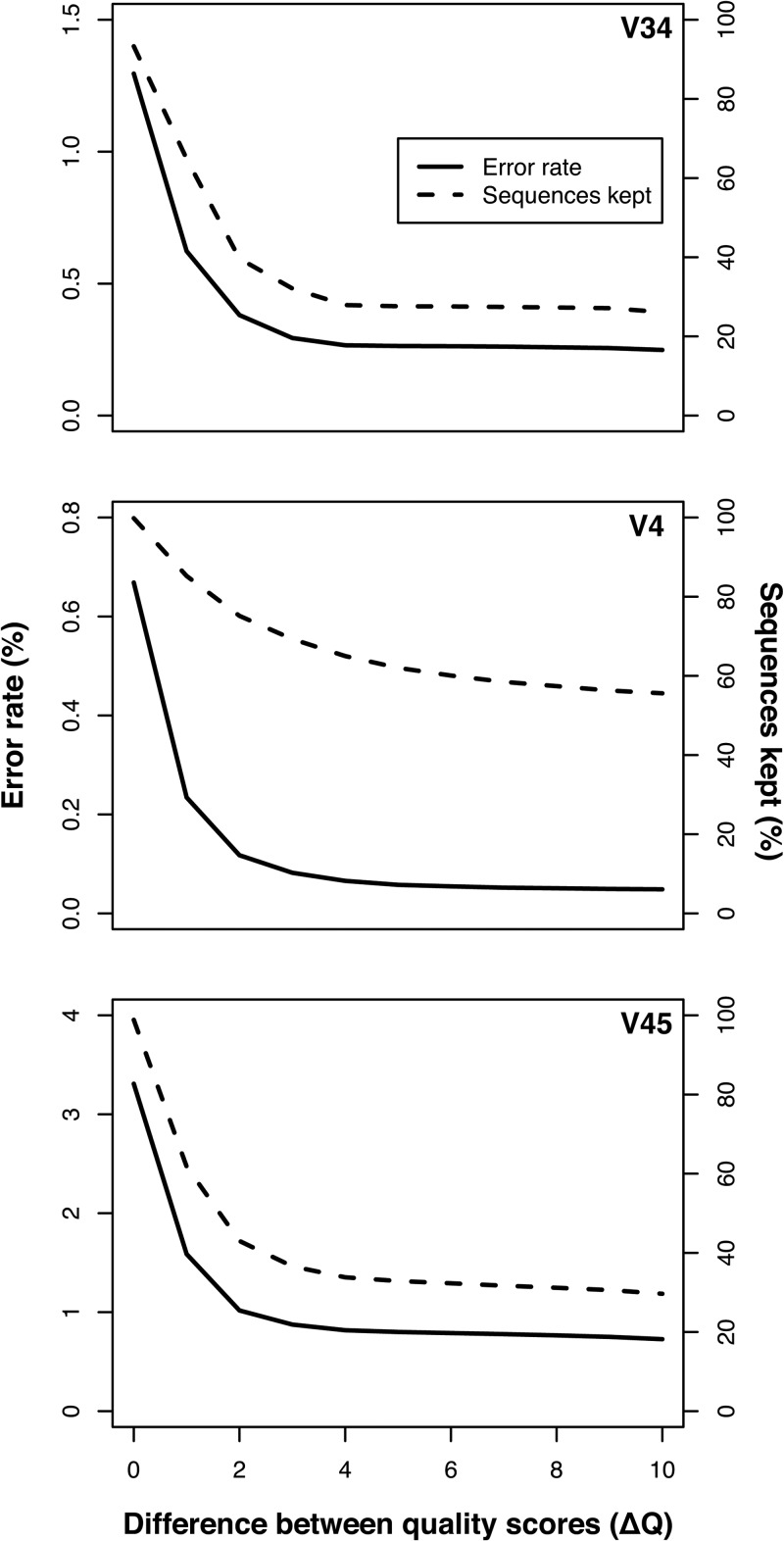
Building upon the observation that the quality scores provided meaningful information, we developed a denoising strategy that utilized that quality score information. Furthermore, by sequencing the three regions we were able to explore the value of building consensus sequences with various lengths of overlap. The entire V4 region was covered by two reads, and the V45 and V34 regions had 125 and 70 overlapping nucleotides, respectively. To assess the error rates of these three regions, we removed any consensus sequences that contained an ambiguous base call or that were considerably shorter than predicted. In the rare cases where the alignment of the two reads suggested a putative insertion or deletion, we required the quality score to be greater than 25; if it was less than this threshold, then the base was erased. To resolve differences in base calls in the overlapping region, we defined a parameter, ΔQ, which represented the difference in the quality scores of the two reads at that position in the sequence. Based on the empirical definition of the quality scores, we expected the fold reduction in the error rate of the base in question (ΔP) to be proportional to 10(ΔQ/10). We varied the minimum ΔQ between 0 and 10, and if the observed ΔQ value fell below the specified minimum, the read was culled from the data set. When this approach was applied to the V4 region (Fig. 3), we observed a significant reduction in the error rate as we increased ΔQ. The error rate did not change by more than 0.01% for values of ΔQ greater than 6, for a theoretical 4-fold reduction in the error rate. For the V4 data set, the basic error rate (i.e., ΔQ = 0) was proportional to the cluster density (range, 0.25 to 1.08%); however, when the value of ΔQ was set at 6, the error rate dropped to 0.05 to 0.06% (Table 2). For the V34 and V45 data sets, the initial error rate again varied with cluster density. Applying the same ΔQ to data from the V34 and V45 data sets reduced the error rates to 0.29 and 0.58%, respectively, when the lowest cluster density was used (Table 2). It was surprising that the shorter V45 region actually had a larger error rate than the V34 region. One hypothesis is that this was due to the number of sites within the V34 (43 forward and 51 reverse) and V45 (81 forward and 29 reverse) regions that lacked heterogeneity between the two imaging channels. Taken together, these data demonstrate that for the V4 data, the fraction of sequences retained and length of sequences (i.e., ca. 250 nt) was comparable to our previous results using the PyroNoise algorithm on 454 flowgrams trimmed to 450 flows (i.e., ca. 260 nt) (10).
Fig 3.
Relationship between the error rate and the fraction of sequences kept as a function of the ΔQ value for the V34, V4, and V45 regions using data from run 130403.
Table 2.
Summary of the error rates and number of observed OTUs for the sequencing runs described in Table 1
| Region and run | Error rate (%) for: |
% reads remaining from basic (ΔQ = 0) | Average no. of OTUsa |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Basic | ΔQ = 6 | Precluster | Mockb | Mockc | Soil | Mouse | Human | ||
| V34 | |||||||||
| 130401 | 2.14 | 0.37 | 0.21 | 10.3 | 26.9 | 49.6 | 1,110.6 | 175.1 | 187.5 |
| 130403 | 1.30 | 0.26 | 0.12 | 27.6 | 31.1 | 47.8 | 1,095.8 | 158.2 | 164.1 |
| 130417 | 1.12 | 0.24 | 0.10 | 27.9 | 35.1 | 52.2 | 1,038.6 | ND | 142.6 |
| 130422 | 0.91 | 0.29 | 0.17 | 47.5 | 41.4 | 54.3 | 1,053.0 | 162.5 | 145.1 |
| V4 | |||||||||
| 130401 | 1.08 | 0.06 | 0.01 | 44.2 | 23.5 | 43.4 | 1,248.2 | 136.1 | 115.4 |
| 130403 | 0.67 | 0.05 | 0.01 | 60.1 | 23.5 | 40.9 | 1,261.8 | 133.9 | 117.8 |
| 130417 | 0.40 | 0.05 | 0.01 | 69.3 | 22.8 | 37.5 | 1,257.3 | 135.5 | 117.2 |
| 130422 | 0.28 | 0.05 | 0.01 | 78.4 | 23.2 | 37.2 | 1,256.2 | 132.8 | 117.3 |
| V45 | |||||||||
| 130401 | 4.60 | 0.87 | 0.64 | 13.5 | 191.9 | 271.7 | 1,462.8 | 198.0 | 312.9 |
| 130403 | 3.31 | 0.79 | 0.56 | 32.3 | 180.4 | 246.1 | 1,519.7 | 213.3 | 324.0 |
| 130417 | 2.38 | 0.66 | 0.43 | 36.5 | 110.8 | 158.3 | ND | 180.4 | 242.1 |
| 130422 | 1.67 | 0.58 | 0.36 | 56.4 | 98.0 | 131.6 | 1,403.3 | 186.3 | 227.2 |
The average number of OTUs is based on rarefaction of each sample to 5,000 sequences per sample; cells labeled ND reflect samples that did not have at least one replicate with more than 5,000 sequences.
Number of OTUs in the mock community when all chimeras were removed; in the absence of chimeras and sequencing errors, there should be 20 OTUs for all three regions.
Number of OTUs in the mock community when chimeras were removed using UCHIME.
Preclustering sequences.
We previously showed that a preclustering step could further reduce the sequencing error and number of unique sequences (10). Briefly, sequences are sorted in decreasing abundance and then are sequentially compared to each of the rarer sequences. If a rare sequence is less than a specified number of bases different from the more abundant sequence, the rare sequence is removed from the data set and its abundance is added to the more abundant sequence. We found that allowing a 1-nt difference per 100 nt of sequence was the most appropriate threshold. For the V4 data, the error rate decreased to 0.01% for each of the four cluster densities, and it was reduced to between 0.10 and 0.21% for the V34 data set and to between 0.36 and 0.64% for the V45 data set (Table 2). Again, the error rates for the V4 data were at least as good as what we have observed using these approaches with 454 data (10).
OTU assignment.
Our analysis focused on optimizing a sequence curation pipeline to minimize error rates. Another popular metric of sequence quality is the number of spurious OTUs that were generated compared to the number of OTUs that would have been generated using perfect sequences (7, 8). Application of this approach is generally flawed, because the number of spurious OTUs is a product of the complexity of the mock community and the number of sequence reads being analyzed. Regardless, we performed two OTU-based analyses, expecting 20 OTUs from each region in the absence of any sequencing errors or chimeras. In the first analysis, we rarefied the data to 5,000 sequences per sample and assumed perfect removal of all chimeras. Using the V4 data set, we observed between 22.8 and 23.5 OTUs (i.e., 2.8 to 3.5 spurious OTUs). In the second analysis, we used UCHIME to identify chimeric sequences and rarefied the data to 5,000 sequences per sample. We observed between 37.2 and 43.4 OTUs (i.e., 17.2 to 23.4 spurious OTUs) (Table 2). When we replicated this analysis for the V34 and V45 data sets, we observed significantly more OTUs (Table 2). In general, the number of spurious OTUs was correlated with the error rate of the data set.
We next applied our sequence curation procedures to DNA isolated from soil and feces collected from a mouse and a human. In general, the relationships we saw with the mock community data held for these natural communities. Interestingly, the three regions did not provide consistent relationships between the samples. Comparing the mouse and human samples suggested that the mouse had more OTUs than the human within the V4 region, the human had more OTUs than the mouse in the V45 region, and they had similar numbers of OTUs in the V34 region. These differences could be due to previously described variation in rates of evolution between the regions or differences in error rates (23). In spite of these differences, it was clear that the numbers of OTUs per community generally were consistent. These data indicate that the method is robust across numerous environments and that caution is necessary before comparing data collected from different regions.
Scaling up.
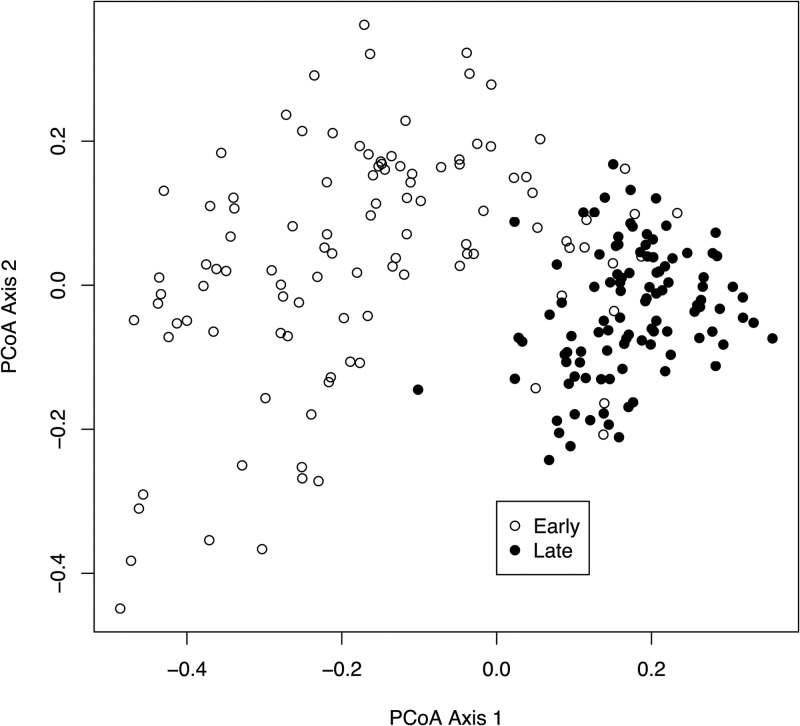
The advantage of the dual-index approach is that a large number of samples can be sequenced using a number of primers equal to only twice the square root of the number of samples. To fully evaluate this approach, we resequenced the V4 region of 360 samples that were previously described by sequencing the distal end of the V35 region on the 454 GS-FLX Titanium platform (18). In that study, we observed a clear separation between murine fecal samples obtained from 8 C57BL/6 mice at 0 to 9 (early) and 141 to 150 (late) days after weaning, and there was significantly less variation between the late samples than the early samples. In addition to the mouse fecal samples, we allocated 2 pairs of indices to resequence our mock community. We generated 4.3 million pairs of sequence reads from the 16S rRNA gene with an average coverage of 9,913 pairs of reads per sample (95% of the samples had more than 2,454 pairs of sequences) using a new collection of 8-nt indices (see the supplemental material). Although individual samples were expected to have various amplification efficiencies, analysis of the number of reads per index did not suggest a systematic positive or negative amplification bias that could be attributed to the indices. The combined error rate for the two mock communities was 0.07% before preclustering and 0.01% after (n = 14,094 sequences). When we used UCHIME to remove chimeras and rarefied to 5,000 sequences, there was an average of 30.4 OTUs (i.e., 10.4 spurious OTUs). Similar to our previous results, ordination of the mouse fecal samples again showed the separation between the early and late periods and increased stabilization with age (Fig. 4) (Mantel test coefficient, 0.81; P < 0.001). These results clearly indicate that our approach can be scaled to multiplex large numbers of samples.
Fig 4.
Principal coordinate ordination of ϴYC values (28) relating the community structures of the fecal microbiota from 12 mice collected on days 0 through 9 (Early) and days 141 through 150 (Late) after weaning.
Titrating the number of 16S rRNA sequence reads.
With 384 samples there is the potential to obtain an average of more than 20,000 sequences per sample. For some studies, this may be an excessive amount of sequence coverage. If the investigator does not have access to additional samples, one option would be to replace this coverage with metagenomic shotgun libraries. As a proof of concept, we pooled Nextera-based shotgun libraries constructed from the genomic DNAs of the mock community, human feces, and two cultured isolates from mouse feces. We then repeated the scaled-up analysis using a target of 5% PhiX, 50% V4 amplicons, and 45% metagenomes with a cluster density of 718,000 clusters/mm2. The increased fraction of sequencing allocated to the V4 amplicons allowed us to have an average library coverage of 13,980 pairs of reads per sample (95% of the samples had more than 3,437 pairs of sequences). The mock community amplicons again gave a final error rate of 0.01%, and the V4 analysis was unchanged. The assemblies of the four shotgun samples demonstrated that excess 16S rRNA gene sequencing coverage could be replaced by sequencing several bacterial genomes or one or two metagenomes (Table 3). Although the HiSeq platform is considerably more efficient for sequencing metagenomes, this application demonstrates that genome and metagenome sequencing on the MiSeq platform can be used to complement 16S rRNA gene sequencing.
Table 3.
Summary of assemblies using shotgun sequence data generated in parallel to 16S rRNA gene sequences
| Library | No. of reads (×106) | No. of bases (×106 bp) | No. of contigs (≥500 bp each) | N50a (×103 bp) | Reads that mapped to contigs (%) |
|---|---|---|---|---|---|
| Clostridium clostridioforme D4 | 1.44 | 360 | 317 | 37.88 | 97 |
| C. clostridioforme CIP110249 | 1.54 | 380 | 323 | 43.80 | 96 |
| Mock community | 2.40 | 600 | 31,946 | 1.46 | 66 |
| Human feces | 5.79 | 1,450 | 27,321 | 1.00 | 49 |
The contig length where all contigs of that length or longer contain more than 50% of the bases found across all contigs.
Conclusions.
The results of our analysis allowed us to evaluate Illumina's MiSeq (v. 2.0) platform as an alternative to the 454 platform for sequencing the 16S rRNA gene. First, we showed that MiSeq-generated 16S rRNA gene sequence data can be curated to be at least as good as the data we are able to obtain using the 454 platform (10). Second, the 10-fold increase in read depth provided by Illumina's MiSeq platform over 454's GS-FLX Titanium platform would allow one to either obtain 10-fold more sequences per sample or to sequence 10-fold more samples. That the MiSeq reagents and instrumentation are considerably cheaper than the 454's would allow even more depth per dollar. Finally, our experiences with 454 and Illumina have shown that both have difficulties maintaining reagent quality; however, the simplicity of MiSeq library construction compared to the emulsion PCRs required by 454 make MiSeq a clear favorite. Thus, the MiSeq platform satisfies the desire for economically generating a large number of high-quality sequence reads that can easily be distributed across a large number of samples.
Determining the amount of DNA to load onto the flow cell to achieve a desired cluster density, as well as the PhiX concentration, is an empirical process that appears to be dependent on the sequencer software and fragment length. This underscores the importance of resequencing mock communities to identify the proper parameters that will minimize the error rate and maximize the number of reads generated. We observed that the fraction of sequences remaining after applying the ΔQ value was inversely proportional to the cluster density, and that the error rate observed after preclustering was independent of the cluster density for the V4 data. To assess the trade-off between number of usable reads and cluster density, we multiplied the number of 16S rRNA gene sequences (Table 1) by the percentage of reads that passed the threshold (Table 2). This demonstrated that the actual number of sequences obtained when the cluster density was between 690 and 1,094 K clusters/mm2 yielded 7.0 to 7.5 million contigs. If this sequencing depth were achieved when sequencing 384 samples, one would expect an average of 18,000 to 20,000 reads per sample. Subsequent experience sequencing samples from other projects suggests that the low complexity of the mock community artificially reduces the number of reads that pass the criteria laid out in the present analysis. Thus, this yield represents a low end to what would be expected for sequencing runs with only samples from natural communities.
Previous demonstrations of the Illumina-based platforms have focused primarily on quantifying the beta-diversity between communities using database-dependent methods (13–15). Although beta-diversity is an important metric for comparing communities, its use is limited to comparisons where there are clear differences between communities, and it does little to inform one of the details of the differences between the communities. Considering the deep coverage of individual samples, database-dependent methods are limited, because they will not have sufficient representation of many rare and novel populations that the extended coverage will likely detect. The OTU-based approach described here has been facilitated by reducing the sequencing error rate from 1.08 to 0.01%, resulting in a reduction in the number of unique sequences that need to be processed. As sequence lengths continue to increase, it will become possible to reliably sequence longer regions of the 16S rRNA gene fragment; however, based on this analysis, it is critical that the fragments fully overlap. Although clearly an idealized community, the sequencing and analysis of mock community DNA in parallel with the biological samples of interest has allowed us to optimize our curation steps and choice of variable region within the 16S rRNA gene by minimizing the overall error rates. We encourage others to include mock community samples as a standard control in all sequencing analyses.
Supplementary Material
ACKNOWLEDGMENTS
We appreciate the generous guidance of numerous individuals who work within Illumina's technical support department. The genomic DNA from microbial mock community A (even, low concentration, v3.1; HM-278D) was obtained through the NIH Biodefense and Emerging Infections Research Resources Repository, NIAID, NIH, as part of the Human Microbiome Project.
This study was supported by grants from the NIH (R01HG005975, 5R01GM099514, and P30DK034933 to P.D.S. and U54HG004973 to S.K.H.).
Footnotes
Published ahead of print 21 June 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01043-13.
REFERENCES
- 1.Hugenholtz P, Pitulle C, Hershberger KL, Pace NR. 1998. Novel division level bacterial diversity in a Yellowstone hot spring. J. Bacteriol. 180:366–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.The Human Microbiome Consortium 2012. Structure, function and diversity of the healthy human microbiome. Nature 486:207–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sogin ML, Morrison HG, Huber JA, Welch DM, Huse SM, Neal PR, Arrieta JM, Herndl GJ. 2006. Microbial diversity in the deep sea and the underexplored “rare biosphere.”. Proc. Natl. Acad. Sci. U. S. A. 103:12115–12120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Junemann S, Prior K, Szczepanowski R, Harks I, Ehmke B, Goesmann A, Stoye J, Harmsen D. 2012. Bacterial community shift in treated periodontitis patients revealed by Ion Torrent 16S rRNA gene amplicon sequencing. PLoS One 7:e41606. 10.1371/journal.pone.0041606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fichot EB, Norman RS. 2013. Microbial phylogenetic profiling with the Pacific Biosciences sequencing platform. Microbiome 1:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gloor GB, Hummelen R, Macklaim JM, Dickson RJ, Fernandes AD, MacPhee R, Reid G. 2010. Microbiome profiling by Illumina sequencing of combinatorial sequence-tagged PCR products. PLoS One 5:e15406. 10.1371/journal.pone.0015406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huse SM, Welch DM, Morrison HG, Sogin ML. 2010. Ironing out the wrinkles in the rare biosphere through improved OTU clustering. Environ. Microbiol. 12:1889–1898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kunin V, Engelbrektson A, Ochman H, Hugenholtz P. 2010. Wrinkles in the rare biosphere: pyrosequencing errors can lead to artificial inflation of diversity estimates. Environ. Microbiol. 12:118–123 [DOI] [PubMed] [Google Scholar]
- 9.Quince C, Lanzen A, Davenport RJ, Turnbaugh PJ. 2011. Removing noise from pyrosequenced amplicons. BMC Bioinformatics 12:38. 10.1186/1471-2105-12-38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schloss PD, Gevers D, Westcott SL. 2011. Reducing the effects of PCR amplification and sequencing artifacts on 16S rRNA-based studies. PLoS One 6:e27310. 10.1371/journal.pone.0027310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Q, Garrity GM, Tiedje JM, Cole JR. 2007. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73:5261–5267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bentley DR, Balasubramanian S, Swerdlow HP, Smith GP, Milton J, Brown CG, Hall KP, Evers DJ, Barnes CL, Bignell HR, Boutell JM, Bryant J, Carter RJ, Keira Cheetham R, Cox AJ, Ellis DJ, Flatbush MR, Gormley NA, Humphray SJ, Irving LJ, Karbelashvili MS, Kirk SM, Li H, Liu X, Maisinger KS, Murray LJ, Obradovic B, Ost T, Parkinson ML, Pratt MR, Rasolonjatovo IM, Reed MT, Rigatti R, Rodighiero C, Ross MT, Sabot A, Sankar SV, Scally A, Schroth GP, Smith ME, Smith VP, Spiridou A, Torrance PE, Tzonev SS, et al. 2008. Accurate whole human genome sequencing using reversible terminator chemistry. Nature 456:53–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, Owens SM, Betley J, Fraser L, Bauer M, Gormley N, Gilbert JA, Smith G, Knight R. 2012. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 6:1621–1624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ, Fierer N, Knight R. 2011. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. U. S. A. 108(Suppl. 1):4516–4522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Werner JJ, Zhou D, Caporaso JG, Knight R, Angenent LT. 2012. Comparison of Illumina paired-end and single-direction sequencing for microbial 16S rRNA gene amplicon surveys. ISME J. 6:1273–1276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bokulich NA, Subramanian S, Faith JJ, Gevers D, Gordon JI, Knight R, Mills DA, Caporaso JG. 2013. Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat. Methods 10:57–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Masella AP, Bartram AK, Truszkowski JM, Brown DG, Neufeld JD. 2012. PANDAseq: paired-end assembler for Illumina sequences. BMC Bioinformatics 13:31. 10.1186/1471-2105-13-31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schloss PD, Schubert AM, Zackular JP, Iverson KD, Young VB, Petrosino JF. 2012. Stabilization of the murine gut microbiome following weaning. Gut Microbes 3:383–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J, Glockner FO. 2007. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 35:7188–7196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haas BJ, Gevers D, Earl AM, Feldgarden M, Ward DV, Giannoukos G, Ciulla D, Tabbaa D, Highlander SK, Sodergren E, Methé B, DeSantis TZ, Human Microbiome Consortium. Petrosino JF, Knight R, Birren BW. 2011. Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res. 21:494–504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Simpson JT, Wong K, Jackman SD, Schein JE, Jones SJ, Birol I. 2009. ABySS: a parallel assembler for short read sequence data. Genome Res. 19:1117–1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF. 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75:7537–7541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schloss PD. 2010. The effects of alignment quality, distance calculation method, sequence filtering, and region on the analysis of 16S rRNA gene-based studies. PLoS Comput. Biol. 6:e1000844. 10.1371/journal.pcbi.1000844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schloss PD. 2009. A high-throughput DNA sequence aligner for microbial ecology studies. PLoS One 4:e8230. 10.1371/journal.pone.0008230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schloss PD. 2013. Secondary structure improves OTU assignments of 16S rRNA gene sequences. ISME J. 7:457–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. 2011. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27:2194–2200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schloss PD, Westcott SL. 2011. Assessing and improving methods used in operational taxonomic unit-based approaches for 16S rRNA gene sequence analysis. Appl. Environ. Microbiol. 77:3219–3226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yue JC, Clayton MK. 2005. A similarity measure based on species proportions. Commun. Stat. Theory Methods 34:2123–2131 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.