Abstract
The conserved HIV-1 site of coreceptor binding is protected from antibody-directed neutralization by conformational and steric restrictions. While inaccessible to most human antibodies, the coreceptor site has been shown to be accessed by antibody fragments. In this study, we used X-ray crystallography, surface plasmon resonance, and pseudovirus neutralization to characterize the gp120-envelope glycoprotein recognition and HIV-1 neutralization of a heavy chain-only llama antibody, named JM4. We describe full-length IgG2b and IgG3 versions of JM4 that target the coreceptor-binding site and potently neutralize over 95% of circulating HIV-1 isolates. Contrary to established trends that show improved access to the coreceptor-binding region by smaller antibody fragments, the single-domain (VHH) version of JM4 neutralized less well than the full-length IgG2b version of JM4. The crystal structure at 2.1-Å resolution of VHH JM4 bound to HIV-1 YU2 gp120 stabilized in the CD4-bound state by the CD4-mimetic miniprotein, M48U1, revealed a JM4 epitope that combined regions of coreceptor recognition (including the gp120 bridging sheet, V3 loop, and β19 strand) with gp120 structural elements involved in recognition of CD4 such as the CD4-binding loop. The structure of JM4 with gp120 thus defines a novel CD4-induced site of vulnerability involving elements of both coreceptor- and CD4-binding sites. The potently neutralizing JM4 IgG2b antibody that targets this newly defined site of vulnerability adds to the expanding repertoire of broadly neutralizing antibodies that effectively neutralize HIV-1 and thereby potentially provides a new template for vaccine development and target for HIV-1 therapy.
INTRODUCTION
Entry of human immunodeficiency virus type 1 (HIV-1) into host cells requires its gp120 envelope glycoprotein to engage the cell surface CD4 receptor and a coreceptor, either CCR5 or CXCR4 (reviewed in reference 1). Although gp120 shows high variability, functional constraints require conservation of receptor-binding sites, making them targets for neutralizing antibodies. The initial site of CD4 engagement is exposed on the viral spike (2), thereby creating a region of vulnerability for naturally elicited antibodies (3), making the CD4-binding site an attractive target for vaccine design. The coreceptor-binding site, on the other hand, is occluded in the native viral spike by a number of viral defense mechanisms, including conformational masking (4) and steric constraints (5), and it presents itself on the viral surface only following CD4 engagement of the virus (6). Antibodies that bind this region, commonly known as CD4-induced (CD4i) antibodies, show high-affinity binding to monomeric gp120 and neutralize tier 1 HIV-1 isolates but are unable to neutralize tier 2 isolates (4, 5, 7) unless administered in combination with CD4 (6, 8). Therefore, despite a high degree of conservation (6, 9), the coreceptor-binding region of gp120 has not been considered a viable vaccine target.
In a recent study aimed at eliciting neutralizing antibodies by immunizing llamas with HIV-1 envelope immunogens, a single-domain antibody, JM4, was isolated and was found to neutralize HIV-1 isolates from four different clades and bind to gp120 in a CD4-dependent manner (10). JM4 binding to monomeric gp120 was enhanced by CD4, and it competed with CD4i antibodies 17b and X5 for binding to HIV-1 Env, suggesting that JM4 targets the CD4i site of coreceptor engagement on the HIV-1 envelope gp120 glycoprotein. JM4 binding to YU2 gp120 minimal core (coremin) (Fig. 1A) (11) was enhanced more than 10-fold in the presence of soluble CD4 (sCD4), a behavior typical of antibodies targeting the CD4i region of coreceptor binding on gp120 (see Fig. S1 in the supplemental material) (7, 12). JM4 also competed with CD4-binding site antibody b12 for binding to gp120 (10). Surprisingly, JM4 binding to gp120 was not affected by the I420R mutation (10) at the gp120 coreceptor-binding site that knocks out binding to CCR5 and known coreceptor binding site-targeting CD4i antibodies (9, 13). JM4 binding is also not affected by the D368R mutation that knocks out gp120 binding to CD4 and most known CD4-binding site antibodies (10). These data suggested a novel epitope for JM4 that has elements of the CD4i- and CD4-binding site but also differs from each. In this study, we have structurally defined the JM4 epitope on gp120 and have characterized its HIV-1 neutralization properties. Since antibody size has been shown to be a major determinant of virus neutralization at the HIV-1 coreceptor-binding site (5), with smaller fragments achieving greater potency, we have engineered full-length IgG2b and IgG3 versions of JM4 and have measured their ability to neutralize HIV-1.
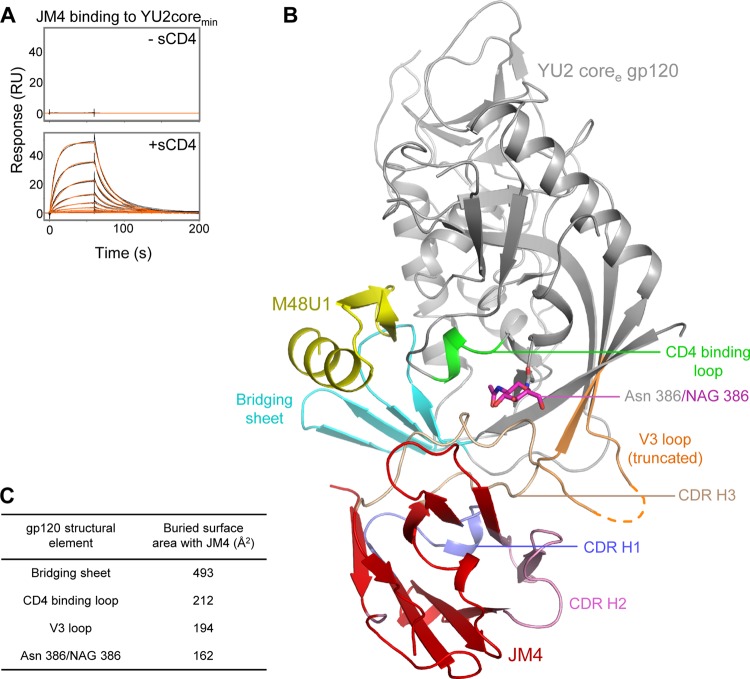
Fig 1.
Crystal structure of llama single-domain CD4i antibody JM4 in complex with HIV-1 YU2 gp120 and CD4-mimetic miniprotein M48U1. (A) JM4 binding to gp120 was measured by surface plasmon resonance, in the absence (top) and presence (bottom) of soluble, two-domain CD4. YU2 coremin gp120 was used for the SPR assays. The coremin (minimal core) gp120 construct has a GAG linker replacing V3 residues 297 to 330. gp120 coremin requires CD4 engagement to form the CD4-induced (CD4i) site for binding to coreceptor-binding site-targeting CD4i antibodies. (B) Single-domain antibody JM4 (red) uses its CDR H1 (slate blue), CDR H2 (pale pink), and CDR H3 (wheat) loops to interact with gp120. The V3 region (orange), bridging sheet (cyan), CD4 binding loop (green), and glycan at position 386 (magenta) of HIV-1 gp120 (gray) interact with JM4. The CD4-mimetic miniprotein M48U1 is colored yellow. The coree (extended core) version of YU2 gp120, which has a GGSGSG linker replacing V3 residues 302 to 323, was used for crystallization. This linker was not visible in the electron density map and is indicated by an orange dashed line. (C) Surface area buried at different gp120 structural elements in the gp120-JM4 complex.
MATERIALS AND METHODS
Protein expression, purification, and complex preparation.
JM4 was purified as previously described (10). HIV-1 clade B YU2 gp120 coree was expressed, purified, and deglycosylated as previously described (14). M48U1 was synthesized by solid-phase methods as described previously (15). The gp120-M48U1-JM4 complex was formed in two sequential steps, first by mixing deglycosylated gp120 with M48U1 (1:2 molar ratio) and then by mixing the gp120-M48U1 complex with JM4 (1:2 molar ratio), purifying the complex at each step by size exclusion chromatography (Hiload 26/60 Superdex S200 prep grade; GE Healthcare) with buffer containing 0.35 M NaCl, 2.5 mM Tris (pH 7.0), and 0.02% NaN3. Fractions with gp120-M48U1-JM4 complexes were concentrated to ∼10 mg/ml and used for crystallization experiments.
Crystallization and X-ray data collection.
Initial screening for crystals was performed in robotic vapor diffusion sitting drop trials with the commercially available screens Hampton Crystal Screen (Hampton Research), Precipitant Wizard Screen (Emerald BioSystems), and Synergy Screen (Emerald BioSystems). Initial crystals obtained were reproduced and optimized using the hanging-drop vapor diffusion method with drops consisting of 0.5 μl protein and 0.5 μl precipitant solution equilibrated against a 500-μl reservoir volume at room temperature. Initial crystals obtained in polyethylene glycol (PEG) 3000 (28%, wt/vol), 0.2 M lithium sulfate monohydrate, and 0.1 M imidazole (pH 8) diffracted to 4.0-Å resolution. Additive screening was performed to improve diffraction quality. 6-Aminohexanoic acid (30%, wt/vol) added to the condition mentioned above resulted in crystals that appeared after 40 days and diffracted to 2.1 Å. Diffraction data were collected under cryogenic conditions using 30% ethylene glycol as a cryoprotectant. X-ray diffraction data were collected at ID-22 beamline (SER-CAT) at the Advanced Photon Source, Argonne National Laboratory, with 1.0000-Å radiation, processed, and reduced with HKL2000 (16).
Structure determination and refinement.
The crystal structure of the gp120-M48U1-JM4 complex was solved by molecular replacement using Phaser in the CCP4 program suite (49). The structure of YU2 gp120 bound to CD4-mimetic miniprotein M48U1 (Protein Data Bank [PDB] code: 4JZZ) was used as a search model. Refinement was carried out with PHENIX (17). Starting with torsion angle-simulated annealing with slow cooling, iterative manual model building was carried out on COOT (18) with maps generated from combinations of standard positional, individual B-factor, translation/libration/screw (TLS) refinement algorithms, and noncrystallographic symmetry (NCS) restraints. Ordered solvents were added during each macrocycle. Throughout the refinement processes, a cross validation (Rfree) test set consisting of 5% of the data was used, and hydrogens were included as a riding model. Structure validations were performed periodically during the model building and refinement process with MolProbity (19) and pdb-care (20). X-ray crystallographic data and refinement statistics are summarized in Table S1 in the supplemental material.
Protein structure analysis and graphical representations.
All superpositions were performed using lsqkab in CCP4. PISA (21) and NCONT were used to perform protein-miniprotein interface analysis. All graphical representations with protein crystal structures were made with PyMOL (www.pymol.org). Per-residue buried surface area on gp120 for the different antibody complexes was computed as the difference between the solvent-accessible area for each gp120 residue in the complex structure versus the solvent-accessible area for the same residue in the unbound gp120 structure (obtained by removing the antibody from the input complex structure). Naccess (22) was used for computing solvent-accessible areas. The Spearman correlation coefficient was used for computing the correlations between per-residue buried surface areas for the different antibodies; only gp120 residues found in at least one epitope (i.e., gp120 residues with nonzero buried surface area for at least one antibody) were included in the correlation computation.
Surface plasmon resonance (SPR).
Experiments were carried out on a Biacore 3000 or Biacore T200 instrument (GE Healthcare). Antibody was covalently coupled to a CM5 chip at 200 resonance units (RU), and a blank surface with no antigen was created under identical coupling conditions for use as a reference. gp120, alone or in the presence of 3 molar excess amounts of soluble, two-domain CD4, was injected over the immobilized antibodies and reference cell at 50 μl/min, using 10 mM HEPES (pH 7.4), 150 mM NaCl, 3 mM EDTA, and 0.05% polysorbate 20 (HBS-EP+) as a running buffer. The data were processed with SCRUBBER-2 and double referenced by subtraction of the blank surface and a blank injection (no analyte). Binding curves were globally fit to a 1:1 binding model.
Assessment of HIV-1 neutralization.
Neutralization was measured using single-round-of-infection HIV-1 Env pseudoviruses and TZM-bl target cells, as described previously (23, 24). Neutralization curves were fit by nonlinear regression using a 5-parameter hill slope equation as described. The 50% inhibitory concentrations (IC50) were reported as the concentrations required to inhibit infection by 50%.
Protein structure accession number.
Atomic coordinates and structure factors have been deposited in the Protein Data Bank under accession number 4LAJ.
RESULTS
Crystal structure of gp120 bound to JM4.
Mutagenesis, competition enzyme-linked immunosorbent assay (ELISA) data (10), and the gp120 binding profile (Fig. 1A) have suggested that JM4 binds to a novel epitope that includes the coreceptor-binding region of gp120. Interpretation of such data, however, can be confounded by conformational effects, with mutations outside the epitope affecting antibody binding. Also, in a competition experiment, binding of gp120 to the antibody of interest might be prevented by an antibody that binds to a different epitope if the two bind different and mutually incompatible gp120 conformations. For instance, based on mutagenesis and competition data, antibody VRC06 was initially thought to recognize a dual epitope comprising the CD4- and coreceptor-binding sites (25). VRC06 was later shown by structural analysis and neutralization fingerprinting to belong to the VRC01 class of CD4-binding site antibodies (26). To gain atomic-level structural information on the gp120 epitope of JM4, we crystallized JM4 in complex with a variable loop-truncated version of YU2 gp120 (YU2coree) (14). YU2coree-M48U1-JM4 crystallized in the P21 space group (see Table S1 in the supplemental material), with four complexes in the asymmetric unit, and diffracted to 2.1 Å. The structure showed JM4 bound to an epitope spanning the gp120 bridging sheet, V3 loop, and the CD4 binding loop (Fig. 1B and C and 2; see also Tables S2 to S4). The JM4 complementarity-determining regions (CDRs)—CDR H1, CDR H2, and CDR H3—interacted with gp120, with CDR H3 accounting for about 60% of the total antibody interaction surface. No interactions were observed between JM4 and the 3-kDa CD4-mimetic miniprotein M48U1, which was used as a CD4 surrogate in the crystallization experiments.
Fig 2.
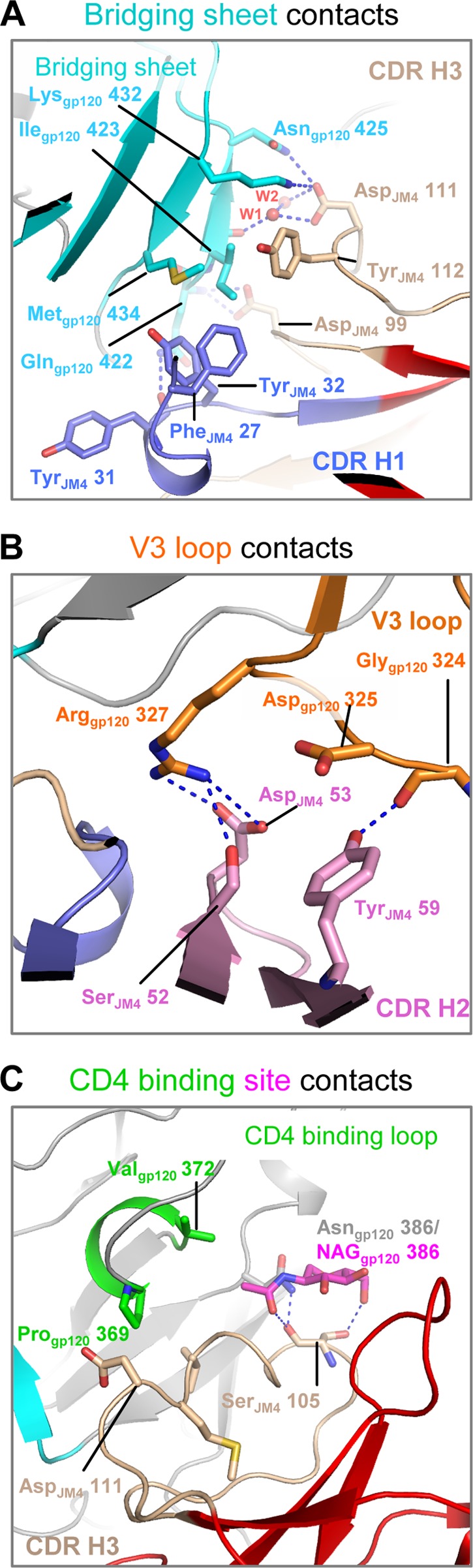
JM4 epitope on gp120 includes elements of the coreceptor-binding site and critical elements of the CD4-binding region. Shown are details of JM4 interaction with different gp120-binding regions. The JM4 CDR H1 (slate) and CDR H3 (wheat) loops make hydrogen bonds and hydrophobic interactions with the gp120 bridging sheet (cyan) (A), the JM4 CDR H2 loop (pink) interacts with the gp120 V3 loop (orange) (B), and the JM4 CDR H3 (wheat) loop interacts with the gp120 CD4 binding loop (green) and NAG 386 (magenta) (C). Blue dashed lines indicate hydrogen bonds.
JM4 interacts with the gp120 coreceptor-binding region.
The gp120 coreceptor-binding region extends from the bridging sheet to the V3 loop (9, 13, 27). JM4 interacts with the gp120 bridging sheet with its CDR H3 and CDR H1 loops (Fig. 2A). JM4 CDR H3 interacts via a salt bridge (between Asp111JM4 and Lys432gp120), hydrogen bonds (between the side chains of Asp111JM4 and Asn425gp120 and between the Asp99JM4 side chain and Gln422gp120 main chain nitrogen), water-mediated hydrogen bonds (between the Asp111JM4 side chain and Ile423gp120 main chain carbonyl), and hydrophobic interactions (between the side chains of Tyr112JM4 and Ile423gp120) (for clarity, residue numbers are subscript labeled with the macromolecule of which they are a part). Ile423gp120 and Met434gp120 also make hydrophobic contacts with Phe27JM4 and Tyr32JM4 from the CDR H1 loop. The side chain of Gln422gp120 hydrogen bonds with the main chain carbonyl of Tyr31JM4. These extensive bridging sheet interactions are reminiscent of the interactions of CD4i antibodies with gp120 (27–31) and agree with biochemical data that demonstrate the requirement of an assembled CD4-induced epitope for JM4 binding (Fig. 1A) (10).
JM4 interactions with the gp120 V3 loop are mediated by its CDR H2 loop (Fig. 2B). Arg327gp120 makes a salt bridge with Asp53JM4 and a hydrogen bond with the side chain hydroxyl group of Ser52JM4. In addition, Tyr59JM4 makes a hydrogen bond with Gly324gp120 and hydrophobic interaction with the alkyl region of the Asp325gp120 side chain. In summary, JM4 interacts extensively with the coreceptor-binding region of gp120, making contacts with residues that are known to be critical for coreceptor binding (9, 28).
JM4 interacts with the gp120 CD4-binding region.
The CD4 binding loop is a central feature of the CD4-binding site and contains elements critical for binding to CD4 and CD4 binding-site antibodies (2, 30, 32, 33). JM4 CDR H3 makes hydrophobic interactions with residues Pro369gp120, Val372gp120, and Thr373gp120 in the gp120 CD4 binding loop (Fig. 2C) and hydrophobic and hydrogen bonding contacts with the glycan at Asn386gp120. This interaction is reminiscent of the CD4-binding site antibodies b12 and b13, which also interact with the glycan at Asn386gp120 (2, 32).
Therefore, JM4 interacts with critical structural elements in the CD4-binding region of gp120, although its binding remains compatible with CD4 binding to gp120 as shown by SPR assays, which demonstrate binding of JM4 to gp120 in the presence of CD4.
JM4 targets a hybrid epitope on gp120 that combines elements from both the CD4-binding region and the coreceptor binding surface.
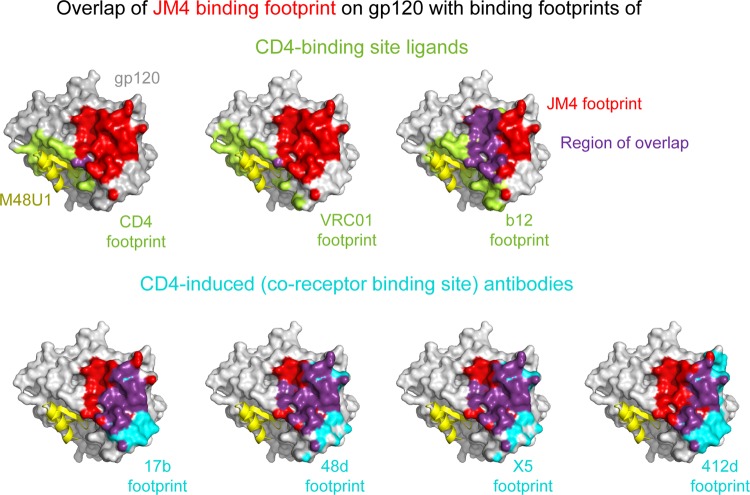
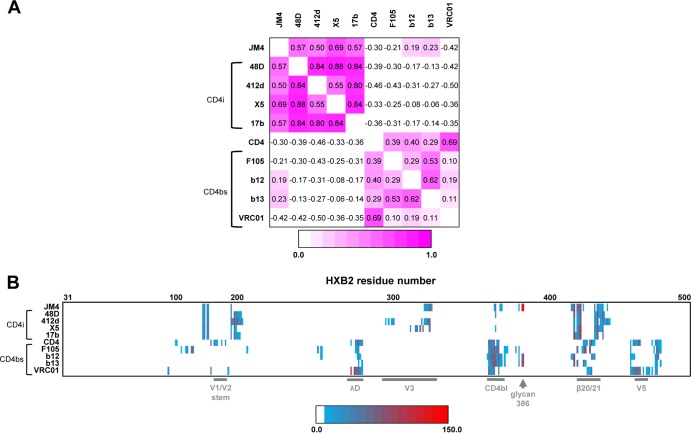
To place the JM4 epitope on gp120 in the context of epitopes of known binders, we analyzed the extent of overlap of the JM4 epitope with molecules that target the CD4-binding site and those that target the coreceptor-binding regions, using multiple complementary approaches. In the first approach, the overlap of the JM4 epitope on gp120 with the binding surfaces of CD4-binding site molecules CD4, VRC01, and b12 (2, 30, 33) and CD4i antibodies 17b, 48d, X5, and 412d (27, 28, 30, 31) were plotted on the JM4-bound gp120 surface (Fig. 3). The binding surfaces of each ligand were defined as the region on gp120 within 5 Å of the binding partner and were plotted on the surface of JM4/M48U1-bound gp120. Among the CD4-binding site ligands analyzed, the b12 epitope overlapped most extensively with that of JM4. This is consistent with the observation that b12 competes with JM4 for gp120 binding (10). The JM4 epitope shows very little overlap the VRC01 epitope or the CD4 binding surface, although the binding surfaces are in close proximity. This is consistent with the observation that JM4 binding does not compete with CD4 binding. In a second, more quantitative approach, per-residue buried surface area was calculated for each gp120 residue involved in an interaction (Fig. 4). Correlations between per-residue buried surface areas were calculated for each pair of antibodies. The correlations sorted into two groups—the CD4-binding site group and the CD4i group—showed strong intragroup and weak intergroup correlation. JM4 showed strong correlation with the CD4i group and also showed a small correlation with two CD4-binding site antibodies, b12 and b13 (2, 32).
Fig 3.
Comparison of JM4 epitope with that of other molecules that target the CD4 and coreceptor binding surfaces on gp120. Shown is the overlap of the JM4 footprint with binding surfaces of CD4-binding site molecules (top) and CD4i antibodies (bottom). The footprints of the antibodies are projected onto the JM4-bound gp120 structure, shown as a gray surface with the CD4-binding site antibody footprints colored lime green and the CD4i antibody footprints colored cyan. The region of overlap of JM4 footprint with that of each antibody is colored deep purple. M48U1 is shown in yellow.
Fig 4.
Comparison of per-residue buried gp120 surface area upon binding of JM4 and a set of CD4i and CD4-binding site (CD4bs) antibodies. (A) Correlation of per-residue buried gp120 surface area for each antibody pair. The Spearman correlation coefficients are shown, with coefficients between 0 (white) and 1 (pink) shown as a heat map. (B) Heat map of the per-residue buried gp120 surface area for each antibody (y axis). Each residue (x axis) is colored according to magnitude of buried gp120 surface area, ranging from blue (low) to red (high), while residues not part of the epitope for a given antibody are colored in white. gp120 regions of interest are highlighted in gray.
Figure 4B presents a graphical representation of the binding footprint of each molecule on gp120 sorted by gp120 region, and it shows that JM4 shares binding features of both CD4i antibodies, such as contacts at the V1/V2 stem, the β20-21 strand, and the V3 loop, as well as the binding features of CD4-binding site molecules such as contacts at the CD4-binding loop and NAG 386. Taken together, these results reveal a CD4i epitope extending toward and making additional contacts with the gp120 CD4 binding loop.
HIV-1 neutralization.
To quantify overall neutralization breadth and potency of JM4, we assessed HIV-1 neutralization by JM4 on a panel of 30 HIV-1 isolates from different clades (Fig. 5; see also Table S5 in the supplemental material). These included 12 viruses that are resistant to the broadly neutralizing CD4-binding site antibody VRC01. JM4 showed neutralization breadth and geometric mean IC50 of 60% and 1.12 μg/ml, respectively (Fig. 5B). JM4 neutralized HIV-1 with greater efficacy than known human CD4i IgGs as well as their antigen binding (Fab) fragments (see Table S6 in the supplemental material).
Fig 5.
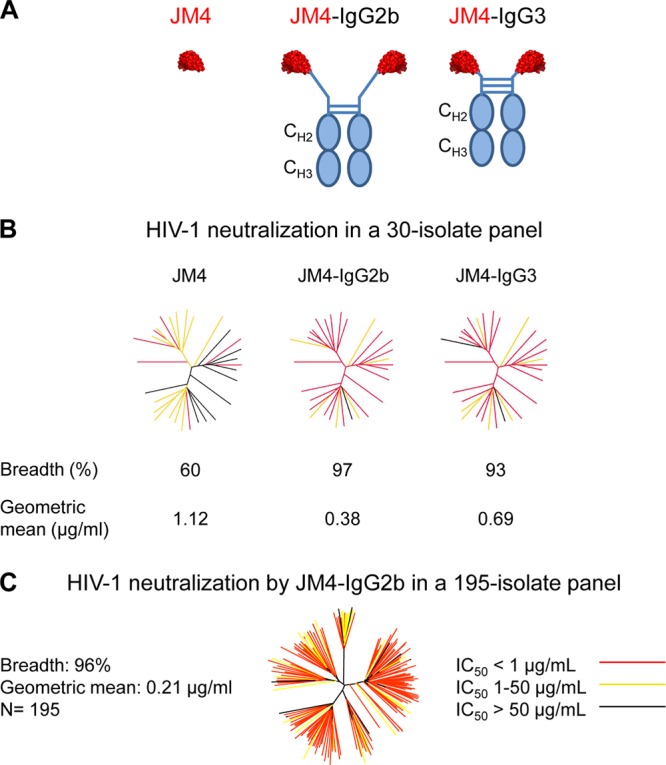
HIV-1 neutralization by single-domain antibody JM4 and its full-length variants JM4 IgG2b and JM4 IgG3. (A) Schematic representations of JM4, JM4 IgG2b, and JM4 IgG3. (B) HIV-1 neutralization was first assessed in a 30-isolate panel of viruses. Results are displayed as color-coded dendrograms. Strikingly, the full-length antibody variants were found to be more potent and broad than single-domain JM4. (C) The best neutralizer, JM4 IgG2b, was further assessed in a 195-member representative panel of circulating HIV-1 tier 1 and tier 2 isolates. JM4 IgG2b showed near-pan neutralization with high overall potency.
It is known that size plays a critical role in determining neutralization efficacy at the gp120 coreceptor-binding region (5), with smaller antibody fragments showing greater neutralization than full-length IgGs. In a study done with human single-domain CD4i antibody m36, it was observed that IgG variants of this antibody showed reduction in overall neutralization efficacy, although some strain dependence was observed, with 3 isolates neutralized similarly by both the single-domain and the IgG versions of m36 (34). To assess the effect of size in targeting this newly defined hybrid epitope, we grafted JM4 on an IgG2b or IgG3 scaffold that has a 29- and 12-amino-acid-long hinge region, respectively (Fig. 5A) (35). Interestingly, and contrary to what we had expected, both IgG2b and IgG3 showed dramatic improvement in breadth and potency, with IgG2b neutralizing all but one virus in the 30-isolate panel, with an overall geometric mean IC50 of 0.38 μg/ml (Fig. 5B). To obtain a comprehensive estimate of its neutralization efficacy, we tested JM4 IgG2b on an expanded panel of 195 HIV-1 isolates and found that it was near-pan reactive, neutralizing 96% of the panel with a geometric IC50 mean of 0.21 μg/ml (Fig. 5C).
The crystal structure of JM4 bound to gp120 shows that JM4 interacts with the glycan at position N386 (Fig. 3C). Since approximately 17% of the HIV-1 isolates tested in this study lacked a glycan at this position (see Table S7 in the supplemental material), we asked whether the presence or absence of a glycan at position 386 affected the neutralization sensitivity of the virus. We found no significant difference in the neutralization sensitivities between viruses that have a glycan at position 386 and those that do not (see Fig. S2 in the supplemental material), suggesting redundancy in the JM4 interactions with HIV-1 gp120, with loss of the interaction at glycan 386 still allowing antibody binding to HIV-1 envelope, leading to virus neutralization.
Taken together, these results show that the JM4 epitope on the native HIV-1 spike is accessible not only to a single-domain antibody but also to a full-length antibody molecule that is about 7 times its size. Moreover, the full-length, bivalent antibody neutralizes with increased breadth and potency. This is strikingly different from published trends on other known CD4i antibodies, where smaller antibody fragments show better neutralization than their full-length IgG counterparts (5, 34).
DISCUSSION
Despite being highly conserved, conformational and steric restrictions on the CD4i site of coreceptor engagement on the HIV-1 spike have prevented its utilization as a vaccine target (5). CD4i antibodies, although reactive to monomeric gp120 and effective against laboratory-adapted isolates, have generally been ineffective in neutralizing primary tier 2 HIV-1 isolates (4, 36). In this study, we have characterized the structural and neutralization properties of a single-domain CD4i antibody elicited by immunizing llamas. While immunization protocols in humans and most animal models have generally yielded only partial success toward eliciting antibodies that neutralize HIV-1 broadly, the ability of llamas to produce the smaller-size VHH-only antibodies perhaps lowers the bar for elicitation against neutralization targets that are protected by various defense mechanisms and allows elicitation of antibodies against regions where the requirement for precise targeting might pose an insurmountable barrier for a full-length human antibody. Immunizing llamas has consequently yielded a number of antibodies that neutralize HIV-1 effectively (10, 37), and these have helped in answering critical questions related to the epitope landscape on the HIV-1 envelope. Analysis of broadly neutralizing antibodies isolated from HIV-1-infected patients suggests that a significant degree of affinity maturation is necessary to produce effective neutralization (33, 38, 39). Similarly, CD4-binding site broadly neutralizing antibody J3, isolated from llamas immunized with HIV-1 Env, has been shown to have a shortened CDR2 as well as 25 separate sites where residues deviate from the germ line sequence (37). Antibody A12, which is not as potent and broad as J3, has residue changes at only 10 sites, suggesting a potential requirement for affinity maturation and recombination for improved breadth and potency of HIV-1-targeting antibodies elicited in llamas (37). In comparison, JM4 shows 15 amino acid changes from the germ line (see Fig. S3 in the supplemental material). The CDR2 length of JM4 is similar to that of A12 and two residues longer than that of J3, and its CDR3, at 15 residues, is shorter than the CDR3 of both J3 (16 residues) and A12 (17 residues).
Whether JM4-like antibodies that target the CD4i site of coreceptor engagement on gp120 and neutralize HIV-1 can be elicited in humans remains to be determined. Compared to a human antibody, absence of light chain and substitution of the bulky CH1 region by a long and flexible linker in JM4 IgG2b might play a role is allowing better accessibility to the newly defined epitope. Recent studies, however, report the isolation of neutralizing antibodies that target an epitope spanning the CD4i site of coreceptor binding from HIV-1-infected patients (40). While structural data on the epitope of these antibodies are not yet available, mutagenesis results suggest that JM4 shares gp120 binding properties with these naturally elicited antibodies. Additionally, immunization studies with animal models have shown that given the right immunogen, CD4i antibodies can be elicited with relative ease (7). Furthermore, the JM4 footprint on gp120 measures about 836 Å2, similar to the footprints of human CD4i antibodies 17b, 48d, X5, and 412d, which measure 853 Å2, 771 Å2, 846 Å2, and 1,072 Å2, respectively (27, 28, 30, 31). Taken together, these results suggest that JM4-like CD4i antibodies that potently neutralize HIV-1 have potential to be elicited in a vaccine regimen, and the structural studies described here may allow the design of immunogens targeting this newly defined epitope on gp120.
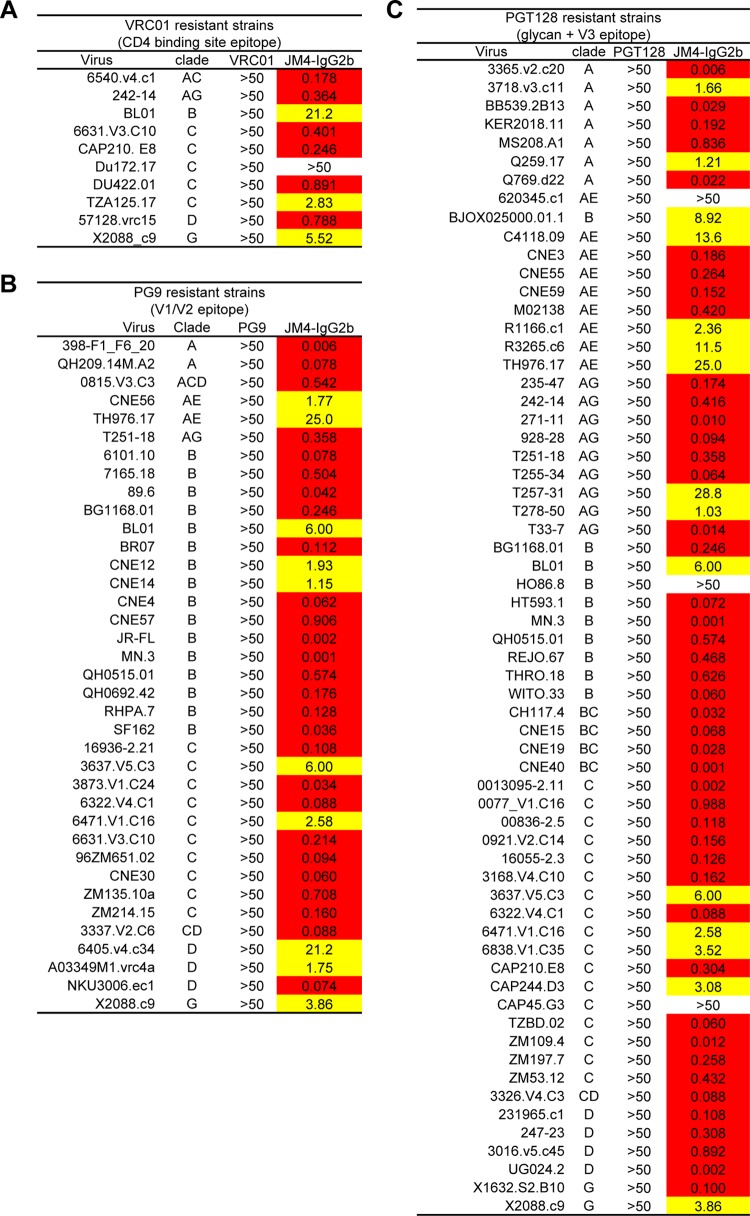
Recent studies have highlighted the utility of multiple antibodies targeting different epitopes in overall neutralization potency and breadth both in passive transfer approaches (41), where antibody monotherapy is often ineffective due to emergence of resistant isolates (42, 43), and in natural elicitation, where sera with multiple neutralization specificities contributing to overall efficacy have been identified (44). While in recent years, a number of broadly neutralizing antibodies have been identified that target different epitopes on the HIV-1 spike (38, 39, 45–47), antibodies targeting the CD4i coreceptor-binding site have not been part of this repertoire. We found that JM4 IgG2b was able to potently neutralize HIV-1 isolates that were resistant to other broadly neutralizing antibodies (Fig. 6). JM4 and its full-length derivative, JM4 IgG2b, have potential applications in HIV-1 therapy by providing the means to target another site of vulnerability on the HIV-1 spike.
Fig 6.
Heat map showing neutralization activity of JM4-IgG2b on panels of viruses resistant to known broadly HIV-1-neutralizing antibodies. (A) Viruses resistant to VRC01. (B) Viruses resistant to PG9. (C) Viruses resistant to PGT128. IC50 are color-coded: red, IC50 < 1.0 μg/ml; yellow, IC50 between 1.0 and 50 μg/ml; white, IC50 > 50 μg/ml.
Size has been a limiting factor toward antibodies targeting the coreceptor-binding site. While inaccessible to full-length antibodies, the CD4i site of coreceptor binding is accessible to antibody fragments and small molecules (5, 48). This study demonstrates a striking enhancement of potency and breadth of JM4 IgG2b versus single-domain JM4, and thus challenges this established paradigm that inversely correlates overall size of the molecule with its ability to neutralize by targeting this site.
In summary, we have structurally characterized a CD4-induced epitope with elements of both the coreceptor-binding site and the CD4-binding site. We have achieved near-pan neutralization by an engineered full-length derivative of a llama single-headed coreceptor-binding site-targeting antibody. These findings provide insight into how ligands that recognize a CD4-induced site of vulnerability might effectively neutralize HIV-1 and have implications for vaccine development and passive HIV therapy.
Supplementary Material
ACKNOWLEDGMENTS
We thank members of the Structural Biology and Structural Bioinformatics Core Sections, Vaccine Research Center, for discussions and comments on the manuscript and J. Stuckey for assistance with figures. sCD4 was obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH. We thank J. Baalwa, D. Ellenberger, D. Gabuzda, F. Gao, B. Hahn, K. Hong, J. Kim, F. McCutchan, D. Montefiori, L. Morris, J. Overbaugh, E. Sanders-Buell, G. Shaw, R. Swanstrom, M. Thomson, S. Tovanabutra, C. Williamson, and L. Zhang for contributing the HIV-1 envelope plasmids used in our neutralization panels.
Support for this work was provided by the Intramural AIDS Targeted Anti-retroviral Program (IATAP), NIH, the Intramural Research Program of the Vaccine Research Center, National Institute of Allergy and Infectious Diseases, NIH, the Framework Program (FP7/2010-2014) under grant agreement no. 242135 (CHAARM), and Agence Nationale de Recherche sur le SIDA (ANRS). J.M. was supported by a fellowship from ANRS. D.B. and P.C. are members of the IBiSA nanobody platform of CRCM, supported by INSERM transfert, OSEO, and FP7 program NAMDIATREAM. Use of sector 22 (Southeast Region Collaborative Access team) at the Advanced Photon Source was supported by the U.S. Department of Energy, Basic Energy Sciences, Office of Science, under contract number W-31-109-Eng-38.
Footnotes
Published ahead of print 10 July 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.01332-13.
REFERENCES
- 1. Wyatt R, Sodroski J. 1998. The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens. Science 280:1884–1888 [DOI] [PubMed] [Google Scholar]
- 2. Zhou T, Xu L, Dey B, Hessell AJ, Van Ryk D, Xiang SH, Yang X, Zhang MY, Zwick MB, Arthos J, Burton DR, Dimitrov DS, Sodroski J, Wyatt R, Nabel GJ, Kwong PD. 2007. Structural definition of a conserved neutralization epitope on HIV-1 gp120. Nature 445:732–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lynch RM, Tran L, Louder MK, Schmidt SD, Cohen M, Dersimonian R, Euler Z, Gray ES, Abdool Karim S, Kirchherr J, Montefiori DC, Sibeko S, Soderberg K, Tomaras G, Yang ZY, Nabel GJ, Schuitemaker H, Morris L, Haynes BF, Mascola JR. 2012. The development of CD4 binding site antibodies during HIV-1 infection. J. Virol. 86:7588–7595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kwong PD, Doyle ML, Casper DJ, Cicala C, Leavitt SA, Majeed S, Steenbeke TD, Venturi M, Chaiken I, Fung M, Katinger H, Parren PW, Robinson J, Van Ryk D, Wang L, Burton DR, Freire E, Wyatt R, Sodroski J, Hendrickson WA, Arthos J. 2002. HIV-1 evades antibody-mediated neutralization through conformational masking of receptor-binding sites. Nature 420:678–682 [DOI] [PubMed] [Google Scholar]
- 5. Labrijn AF, Poignard P, Raja A, Zwick MB, Delgado K, Franti M, Binley J, Vivona V, Grundner C, Huang CC, Venturi M, Petropoulos CJ, Wrin T, Dimitrov DS, Robinson J, Kwong PD, Wyatt RT, Sodroski J, Burton DR. 2003. Access of antibody molecules to the conserved coreceptor binding site on glycoprotein gp120 is sterically restricted on primary human immunodeficiency virus type 1. J. Virol. 77:10557–10565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Decker JM, Bibollet-Ruche F, Wei X, Wang S, Levy DN, Wang W, Delaporte E, Peeters M, Derdeyn CA, Allen S, Hunter E, Saag MS, Hoxie JA, Hahn BH, Kwong PD, Robinson JE, Shaw GM. 2005. Antigenic conservation and immunogenicity of the HIV coreceptor binding site. J. Exp. Med. 201:1407–1419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dey AK, Burke B, Sun Y, Sirokman K, Nandi A, Hartog K, Lian Y, Geonnotti AR, Montefiori D, Franti M, Martin G, Carfi A, Kessler P, Martin L, Srivastava IK, Barnett SW. 2012. Elicitation of neutralizing antibodies directed against CD4-induced epitope(s) using a CD4 mimetic cross-linked to a HIV-1 envelope glycoprotein. PLoS One 7:e30233. 10.1371/journal.pone.0030233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dey B, Del Castillo CS, Berger EA. 2003. Neutralization of human immunodeficiency virus type 1 by sCD4-17b, a single-chain chimeric protein, based on sequential interaction of gp120 with CD4 and coreceptor. J. Virol. 77:2859–2865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rizzuto CD, Wyatt R, Hernandez-Ramos N, Sun Y, Kwong PD, Hendrickson WA, Sodroski J. 1998. A conserved HIV gp120 glycoprotein structure involved in chemokine receptor binding. Science 280:1949–1953 [DOI] [PubMed] [Google Scholar]
- 10. Matz J, Kessler P, Bouchet J, Combes O, Ramos OH, Barin F, Baty D, Martin L, Benichou S, Chames P. 2013. Straightforward selection of broadly neutralizing single-domain antibodies targeting the conserved CD4 and coreceptor binding sites of HIV-1 gp120. J. Virol. 87:1137–1149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kwong PD, Wyatt R, Majeed S, Robinson J, Sweet RW, Sodroski J, Hendrickson WA. 2000. Structures of HIV-1 gp120 envelope glycoproteins from laboratory-adapted and primary isolates. Structure 8:1329–1339 [DOI] [PubMed] [Google Scholar]
- 12. Huang CC, Stricher F, Martin L, Decker JM, Majeed S, Barthe P, Hendrickson WA, Robinson J, Roumestand C, Sodroski J, Wyatt R, Shaw GM, Vita C, Kwong PD. 2005. Scorpion-toxin mimics of CD4 in complex with human immunodeficiency virus gp120 crystal structures, molecular mimicry, and neutralization breadth. Structure 13:755–768 [DOI] [PubMed] [Google Scholar]
- 13. Xiang SH, Wang L, Abreu M, Huang CC, Kwong PD, Rosenberg E, Robinson JE, Sodroski J. 2003. Epitope mapping and characterization of a novel CD4-induced human monoclonal antibody capable of neutralizing primary HIV-1 strains. Virology 315:124–134 [DOI] [PubMed] [Google Scholar]
- 14. Kwon YD, Finzi A, Wu X, Dogo-Isonagie C, Lee LK, Moore LR, Schmidt SD, Stuckey J, Yang Y, Zhou T, Zhu J, Vicic DA, Debnath AK, Shapiro L, Bewley CA, Mascola JR, Sodroski JG, Kwong PD. 2012. Unliganded HIV-1 gp120 core structures assume the CD4-bound conformation with regulation by quaternary interactions and variable loops. Proc. Natl. Acad. Sci. U. S. A. 109:5663–5668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Van Herrewege Y, Morellato L, Descours A, Aerts L, Michiels J, Heyndrickx L, Martin L, Vanham G. 2008. CD4 mimetic miniproteins: potent anti-HIV compounds with promising activity as microbicides. J. Antimicrob. Chemother. 61:818–826 [DOI] [PubMed] [Google Scholar]
- 16. Otwinowski Z, Minor W. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276:307–326 [DOI] [PubMed] [Google Scholar]
- 17. Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. 2010. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66:213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Emsley P, Lohkamp B, Scott WG, Cowtan K. 2010. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66:486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chen VB, Arendall WB, III, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. 2010. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66:12–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lütteke T, von der Lieth CW. 2004. pdb-care (PDB carbohydrate residue check): a program to support annotation of complex carbohydrate structures in PDB files. BMC Bioinformatics 5:69. 10.1186/1471-2105-5-69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Krissinel E, Henrick K. 2007. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372:774–797 [DOI] [PubMed] [Google Scholar]
- 22. Hubbard SJ, Thornton JM. 1993. ‘NACCESS,' computer program. Department of Biochemistry and Molecular Biology, University College London, London, United Kingdom [Google Scholar]
- 23. Li M, Gao F, Mascola JR, Stamatatos L, Polonis VR, Koutsoukos M, Voss G, Goepfert P, Gilbert P, Greene KM, Bilska M, Kothe DL, Salazar-Gonzalez JF, Wei X, Decker JM, Hahn BH, Montefiori DC. 2005. Human immunodeficiency virus type 1 env clones from acute and early subtype B infections for standardized assessments of vaccine-elicited neutralizing antibodies. J. Virol. 79:10108–10125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Seaman MS, Janes H, Hawkins N, Grandpre LE, Devoy C, Giri A, Coffey RT, Harris L, Wood B, Daniels MG, Bhattacharya T, Lapedes A, Polonis VR, McCutchan FE, Gilbert PB, Self SG, Korber BT, Montefiori DC, Mascola JR. 2010. Tiered categorization of a diverse panel of HIV-1 Env pseudoviruses for assessment of neutralizing antibodies. J. Virol. 84:1439–1452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Li Y, O'Dell S, Wilson R, Wu X, Schmidt SD, Hogerkorp CM, Louder MK, Longo NS, Poulsen C, Guenaga J, Chakrabarti BK, Doria-Rose N, Roederer M, Connors M, Mascola JR, Wyatt RT. 2012. HIV-1 neutralizing antibodies display dual recognition of the primary and coreceptor binding sites and preferential binding to fully cleaved envelope glycoproteins. J. Virol. 86:11231–11241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Georgiev IS, Doria-Rose NA, Zhou T, Kwon YD, Staupe RP, Moquin S, Chuang GY, Louder MK, Schmidt SD, Altae-Tran HR, Bailer RT, McKee K, Nason M, O'Dell S, Ofek G, Pancera M, Srivatsan S, Shapiro L, Connors M, Migueles SA, Morris L, Nishimura Y, Martin MA, Mascola JR, Kwong PD. 2013. Delineating antibody recognition in polyclonal sera from patterns of HIV-1 isolate neutralization. Science 340:751–756 [DOI] [PubMed] [Google Scholar]
- 27. Huang CC, Tang M, Zhang MY, Majeed S, Montabana E, Stanfield RL, Dimitrov DS, Korber B, Sodroski J, Wilson IA, Wyatt R, Kwong PD. 2005. Structure of a V3-containing HIV-1 gp120 core. Science 310:1025–1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Huang CC, Lam SN, Acharya P, Tang M, Xiang SH, Hussan SS, Stanfield RL, Robinson J, Sodroski J, Wilson IA, Wyatt R, Bewley CA, Kwong PD. 2007. Structures of the CCR5 N terminus and of a tyrosine-sulfated antibody with HIV-1 gp120 and CD4. Science 317:1930–1934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Huang CC, Venturi M, Majeed S, Moore MJ, Phogat S, Zhang MY, Dimitrov DS, Hendrickson WA, Robinson J, Sodroski J, Wyatt R, Choe H, Farzan M, Kwong PD. 2004. Structural basis of tyrosine sulfation and VH-gene usage in antibodies that recognize the HIV type 1 coreceptor-binding site on gp120. Proc. Natl. Acad. Sci. U. S. A. 101:2706–2711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kwong PD, Wyatt R, Robinson J, Sweet RW, Sodroski J, Hendrickson WA. 1998. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 393:648–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pancera M, Majeed S, Ban YE, Chen L, Huang CC, Kong L, Kwon YD, Stuckey J, Zhou T, Robinson JE, Schief WR, Sodroski J, Wyatt R, Kwong PD. 2010. Structure of HIV-1 gp120 with gp41-interactive region reveals layered envelope architecture and basis of conformational mobility. Proc. Natl. Acad. Sci. U. S. A. 107:1166–1171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chen L, Kwon YD, Zhou T, Wu X, O'Dell S, Cavacini L, Hessell AJ, Pancera M, Tang M, Xu L, Yang ZY, Zhang MY, Arthos J, Burton DR, Dimitrov DS, Nabel GJ, Posner MR, Sodroski J, Wyatt R, Mascola JR, Kwong PD. 2009. Structural basis of immune evasion at the site of CD4 attachment on HIV-1 gp120. Science 326:1123–1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhou T, Georgiev I, Wu X, Yang ZY, Dai K, Finzi A, Kwon YD, Scheid JF, Shi W, Xu L, Yang Y, Zhu J, Nussenzweig MC, Sodroski J, Shapiro L, Nabel GJ, Mascola JR, Kwong PD. 2010. Structural basis for broad and potent neutralization of HIV-1 by antibody VRC01. Science 329:811–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chen W, Zhu Z, Feng Y, Dimitrov DS. 2008. Human domain antibodies to conserved sterically restricted regions on gp120 as exceptionally potent cross-reactive HIV-1 neutralizers. Proc. Natl. Acad. Sci. U. S. A. 105:17121–17126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Vu KB, Ghahroudi MA, Wyns L, Muyldermans S. 1997. Comparison of llama VH sequences from conventional and heavy chain antibodies. Mol. Immunol. 34:1121–1131 [DOI] [PubMed] [Google Scholar]
- 36. Dey B, Pancera M, Svehla K, Shu Y, Xiang SH, Vainshtein J, Li Y, Sodroski J, Kwong PD, Mascola JR, Wyatt R. 2007. Characterization of human immunodeficiency virus type 1 monomeric and trimeric gp120 glycoproteins stabilized in the CD4-bound state: antigenicity, biophysics, and immunogenicity. J. Virol. 81:5579–5593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. McCoy LE, Quigley AF, Strokappe NM, Bulmer-Thomas B, Seaman MS, Mortier D, Rutten L, Chander N, Edwards CJ, Ketteler R, Davis D, Verrips T, Weiss RA. 2012. Potent and broad neutralization of HIV-1 by a llama antibody elicited by immunization. J. Exp. Med. 209:1091–1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Scheid JF, Mouquet H, Ueberheide B, Diskin R, Klein F, Oliveira TY, Pietzsch J, Fenyo D, Abadir A, Velinzon K, Hurley A, Myung S, Boulad F, Poignard P, Burton DR, Pereyra F, Ho DD, Walker BD, Seaman MS, Bjorkman PJ, Chait BT, Nussenzweig MC. 2011. Sequence and structural convergence of broad and potent HIV antibodies that mimic CD4 binding. Science 333:1633–1637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Walker LM, Phogat SK, Chan-Hui PY, Wagner D, Phung P, Goss JL, Wrin T, Simek MD, Fling S, Mitcham JL, Lehrman JK, Priddy FH, Olsen OA, Frey SM, Hammond PW, Kaminsky S, Zamb T, Moyle M, Koff WC, Poignard P, Burton DR. 2009. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science 326:285–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Klein F, Gaebler C, Mouquet H, Sather DN, Lehmann C, Scheid JF, Kraft Z, Liu Y, Pietzsch J, Hurley A, Poignard P, Feizi T, Morris L, Walker BD, Fatkenheuer G, Seaman MS, Stamatatos L, Nussenzweig MC. 2012. Broad neutralization by a combination of antibodies recognizing the CD4 binding site and a new conformational epitope on the HIV-1 envelope protein. J. Exp. Med. 209:1469–1479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Klein F, Halper-Stromberg A, Horwitz JA, Gruell H, Scheid JF, Bournazos S, Mouquet H, Spatz LA, Diskin R, Abadir A, Zang T, Dorner M, Billerbeck E, Labitt RN, Gaebler C, Marcovecchio PM, Incesu RB, Eisenreich TR, Bieniasz PD, Seaman MS, Bjorkman PJ, Ravetch JV, Ploss A, Nussenzweig MC. 2012. HIV therapy by a combination of broadly neutralizing antibodies in humanized mice. Nature 492:118–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Poignard P, Sabbe R, Picchio GR, Wang M, Gulizia RJ, Katinger H, Parren PW, Mosier DE, Burton DR. 1999. Neutralizing antibodies have limited effects on the control of established HIV-1 infection in vivo. Immunity 10:431–438 [DOI] [PubMed] [Google Scholar]
- 43. Wei X, Decker JM, Wang S, Hui H, Kappes JC, Wu X, Salazar-Gonzalez JF, Salazar MG, Kilby JM, Saag MS, Komarova NL, Nowak MA, Hahn BH, Kwong PD, Shaw GM. 2003. Antibody neutralization and escape by HIV-1. Nature 422:307–312 [DOI] [PubMed] [Google Scholar]
- 44. Bonsignori M, Montefiori DC, Wu X, Chen X, Hwang KK, Tsao CY, Kozink DM, Parks RJ, Tomaras GD, Crump JA, Kapiga SH, Sam NE, Kwong PD, Kepler TB, Liao HX, Mascola JR, Haynes BF. 2012. Two distinct broadly neutralizing antibody specificities of different clonal lineages in a single HIV-1-infected donor: implications for vaccine design. J. Virol. 86:4688–4692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Huang J, Ofek G, Laub L, Louder MK, Doria-Rose NA, Longo NS, Imamichi H, Bailer RT, Chakrabarti B, Sharma SK, Alam SM, Wang T, Yang Y, Zhang B, Migueles SA, Wyatt R, Haynes BF, Kwong PD, Mascola JR, Connors M. 2012. Broad and potent neutralization of HIV-1 by a gp41-specific human antibody. Nature 491:406–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Pejchal R, Doores KJ, Walker LM, Khayat R, Huang PS, Wang SK, Stanfield RL, Julien JP, Ramos A, Crispin M, Depetris R, Katpally U, Marozsan A, Cupo A, Maloveste S, Liu Y, McBride R, Ito Y, Sanders RW, Ogohara C, Paulson JC, Feizi T, Scanlan CN, Wong CH, Moore JP, Olson WC, Ward AB, Poignard P, Schief WR, Burton DR, Wilson IA. 2011. A potent and broad neutralizing antibody recognizes and penetrates the HIV glycan shield. Science 334:1097–1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wu X, Yang ZY, Li Y, Hogerkorp CM, Schief WR, Seaman MS, Zhou T, Schmidt SD, Wu L, Xu L, Longo NS, McKee K, O'Dell S, Louder MK, Wycuff DL, Feng Y, Nason M, Doria-Rose N, Connors M, Kwong PD, Roederer M, Wyatt RT, Nabel GJ, Mascola JR. 2010. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science 329:856–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Acharya P, Dogo-Isonagie C, LaLonde JM, Lam SN, Leslie GJ, Louder MK, Frye LL, Debnath AK, Greenwood JR, Luongo TS, Martin L, Watts KS, Hoxie JA, Mascola JR, Bewley CA, Kwong PD. 2011. Structure-based identification and neutralization mechanism of tyrosine sulfate mimetics that inhibit HIV-1 entry. ACS Chem. Biol. 6:1069–1077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AGW, McCoy A, McNicholas SJ, Murshudov GN, Pannu NS, Potterton EA, Powell HR, Read RJ, Vagin A, Wilson KS. 2011. Overview of the CCP4 suite and current developments. Acta Cryst. D Biol. Crystallogr. 67(Pt. 4):235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.